Abstract
Mitochondrial dysfunction has consequences not only for cellular energy output but also for cellular signaling pathways. Mitochondrial dysfunction, often based on inherited gene variants, plays a role in devastating human conditions such as mitochondrial neuropathies, myopathies, cardiovascular disorders, and Parkinson and Alzheimer diseases. Of the proteins essential for mitochondrial function, more than 98% are encoded in the cell nucleus, translated in the cytoplasm, sorted based on the presence of encoded mitochondrial targeting sequences (MTSs), and imported to specific mitochondrial sub-compartments based on the integrated activity of a series of mitochondrial translocases, proteinases, and chaperones. This import process is typically dynamic; as cellular homeostasis is coordinated through communication between the mitochondria and the nucleus, many of the adaptive responses to stress depend on modulation of mitochondrial import. We here describe an emerging class of disease-linked gene variants that are found to impact the mitochondrial import machinery itself or to affect the proteins during their import into mitochondria. As a whole, this class of rare defects highlights the importance of correct trafficking of mitochondrial proteins in the cell and the potential implications of failed targeting on metabolism and energy production. The existence of this variant class could have importance beyond rare neuromuscular disorders, given an increasing body of evidence suggesting that aberrant mitochondrial function may impact cancer risk and therapeutic response.
Main Text
Although best known for their role as the cellular powerhouse, mitochondria also have key functions as signaling hubs. Under conditions of altered substrate availability or changing environmental demands, reprogrammed signaling in mitochondria plays a crucial role in maintaining metabolic flexibility.1, 2 Dynamic regulation of mitochondrial activity is essential under normal physiological conditions and is frequently altered in human disease.3 Mitochondrial failure leads to common devastating conditions such as mitochondrial neuropathies, myopathies, cardiovascular disorders, and Parkinson and Alzheimer diseases, all reflecting the high levels of energy production and consumption required in neuronal tissues, muscles, and the heart. Increasingly, mitochondrial defects have been linked to additional pathological states, including diabetes and cancer, for example when tumor cells adapt to a metastatic or a drug-resistant phenotype by reprogramming metabolism.4 Rather than solely reflecting the poor performance of dysfunctional mitochondria, these diseases may also involve pathological disease-associated mitochondrial signaling.5
In humans, the mitochondrial genome encodes 13 proteins involved in oxidative phosphorylation (OXPHOS), 22 mitochondrial (mt)-tRNAs that participate in translation, the 12S and 16S mitochondrial rRNAs, and mitochondrial-derived peptides (MDPs) such as humanin, MOTS-c, and SHLPs produced from short open reading frames within the 16S and 12S rDNA regions. In contrast, the nuclear genome encodes more than 98% of the mitochondrial proteome (∼1,500 nuclear-encoded proteins). Because of the need to unfold and refold proteins as they cross mitochondrial membrane barriers and to promote assembly of oligomeric complexes with mitochondrially encoded components, the import process is typically supported by chaperones and subject to tight regulation.
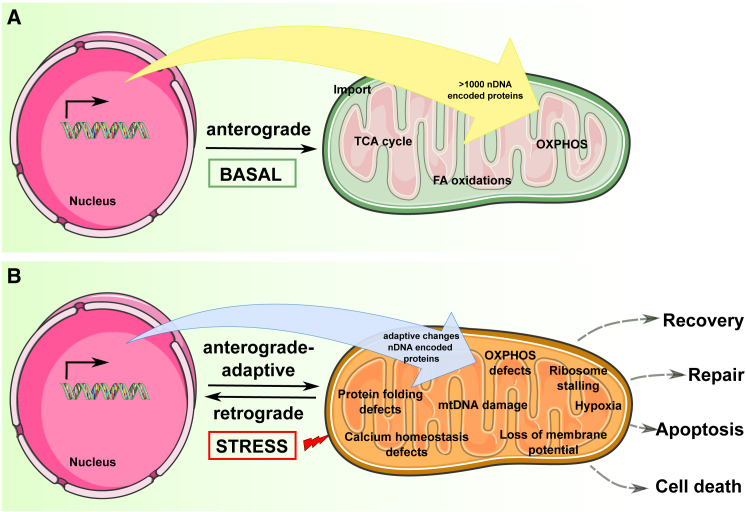
Mitonuclear signaling is bidirectional. In anterograde signaling, nuclear-encoded transcription factors and other proteins provide information regulating mitochondrial biogenesis and other functions. Retrograde signaling is often associated with mitochondrial distress, based on stimuli such as accumulation of reactive oxygen species (ROS), loss of membrane potential, or defects in mtDNA. Distressed mitochondria send signals to the nucleus that reconfigure the nuclear transcriptome6 and trigger cytosolic changes in translation, proteasomal assembly, and folding7 that together support cell adaptation or trigger cell death via reactive anterograde signaling (Figure 1). Therefore, besides being crucial for mitochondrial biogenesis, regulation of mitochondrial import is involved in the adaptive response to mitochondrial stress. For example, the UPRmt (unfolded protein response associated with mitochondria) triggers expression of genes coding for proteases and chaperones that need to be imported when mitochondrial protein folding is defective8 or when mitochondrial translation is otherwise affected.9 The import machinery may be exploited to dispose of cytosolic protein aggregates resulting from misfolding.10 Import controls also govern the targeting of severely dysfunctional mitochondria to the autophagic system, for destruction via a PINK1-Parkin relay system. In this case, upon lethal loss of mitochondrial membrane potential or toxic ROS accumulation, PINK1 is redirected upon import from the inner to the outer mitochondrial membrane, where it recruits the E3 ubiquitin ligase Parkin, targeting the mitochondria for autophagosome engulfment.11 Finally, separate from these complex signaling processes but also involving transport during stress response, is the process of inter-organellar coordination mediated by the direct redistribution of some nuclear or mitochondrial proteins between the two compartments.12 Nuclear proteins with partial mitochondrial distribution are mainly estrogen, glucocorticoid, and thyroid hormone receptors or transcription factors (e.g., NF-κB, CREB, p53, STAT3). These proteins translocate to mitochondria upon a stress stimulus or reside in mitochondria under steady-state conditions and are activated upon stress. Nuclear-encoded proteins that predominantly reside in the mitochondria under normal growth conditions but translocate to the nucleus in response to stress are mainly transcription factors with roles both in mitochondrial and nuclear DNAs (e.g., TFAM, ATFs), biosynthetic enzymes (e.g., CLK-1), or pro-apoptotic factors (e.g., AIF).12 Although beyond the scope of this review, insights into the mechanisms of mitochondrial transport of nuclear receptors and transcription factors that differ from the canonical import linked to mitochondrial biosynthesis are detailed in Sepuri et al.13
Figure 1.
Anterograde and Retrograde Signaling in Mitonuclear Communication
(A) Under basal conditions.
(B) Under stress.
Over the past several decades, a growing number of genetic disorders have been recognized as linked to some form of mitochondrial dysfunction. The first DNA variation in a nuclear gene responsible for a mitochondrial respiratory chain deficiency in humans was discovered in 1995 in two siblings from a consanguineous family diagnosed with Leigh syndrome (MIM: 256000), characterized by severe psychomotor regression usually in the first year of life and death within 2–3 years. These siblings were homozygous for the GenBank: NM_004168.3; c.1660C>T (p.Arg554Trp) variant (rs9809219) in SDHA (MIM: 600857), a subunit of the succinate dehydrogenase (SDH). The deleterious effect of this homozygous variant was confirmed by comparing the SDHA activity in an SDHA (sdh1)-deficient yeast strain transformed with wild-type or c.1660C>T SDHA cDNA.14
Causal defects linked to diseases of mitochondrial dysfunction have now been identified in ∼300 of the ∼1,200 nuclear genes that encode mitochondrial proteins, with a surge in discovery in recent years based on successful application of whole-exome or whole-genome sequencing.15, 16 In 2015, it was proposed that the genetic variants that cause mitochondrial disorders could be classified in seven categories according to their impact on specific cell processes: (1) oxidative phosphorylation, (2) mitochondrial DNA maintenance and gene expression, (3) mitochondrial quality control, (4) iron-sulfur cluster homeostasis, (5) mitochondrial dynamics of fission and fusion, (6) phospholipid metabolism, and (7) mitochondrial import.17 As more is learned about the intricacy of the various processes required for mitochondrial biogenesis and function, these categories are likely to evolve. For example, a recent review on membrane dynamics considered defects of membrane fusion as a subgroup of mtDNA maintenance defects.18 As discussed below, additional functional classes of genes may also significantly modify mitochondrial function.
Among the categories of defects associated with mitochondrial disorder, the variants in genes involved in mitochondrial import are of particular interest because, as all nuclear-encoded proteins involved in these various mitochondrial functions have to be imported themselves, defects in mitochondrial import can potentially induce all classes of dysfunction, depending on which proteins experience faulty import. For example, while it could be argued that the disease-associated p.Val10Gly-iron-sulfur cluster assembly 1 (ISCA1) (MIM: 611006) acts by impairing the iron-sulfur clusters of metabolic and respiratory enzymes, the mechanistic basis for the defect is impaired import and protein stability.19 As another example, human syndromes arising from variants in genes encoding mt-tRNA-modifying proteins (e.g., MTO1 [MIM: 614667], GTPBP3 [MIM: 608536], MTFMT [MIM: 611766], and TRIT1 [MIM: 617840]) were described as mt-translation disorders according to another simplified classification of mitochondrial diseases.20 However, cellular and mouse models of Mto1 deficiency both manifested an abnormal mitochondrial morphology, associated with aberrant trafficking of normally imported proteins. This import defect leads mitochondrial proteins to aggregate and misfold in the cytoplasm, inducing a cytotoxic unfolding protein response.21 Rather than separating into classes, it may therefore be beneficial to look at the consequences of genetic defects in mitochondrial import more broadly.
Given the increasing number of diseases linked to defects in mitochondrial import, in this review, we focus on the mitochondrial import pathway and its signaling, particularly as this information impacts inherited disease-associated gene variants. After providing a basic overview of the current understanding of this pathway, we provide illustrative examples of disease-causing variants that highlight import defects that may underlie some general mechanisms at play in various inherited diseases. We also discuss the ways the findings may impact understanding of diseases such as cancer, in which altered mitochondrial bio-energetic processes are typical, and where changes in mitochondrial import are being linked to tumorigenesis. We conclude with some challenges faced by the variant curator trying to decipher the functional impact of a DNA variant coding for a protein that localizes to mitochondria, including examples of DNA variants that would not typically be classified as disease causing (as they are not rare in the general population) but are nevertheless associated with disease based on roles potentiating rare variants. An extended list of citations related to the work discussed herein is provided in the Supplemental Data.
The Basic Machinery for Mitochondrial Import
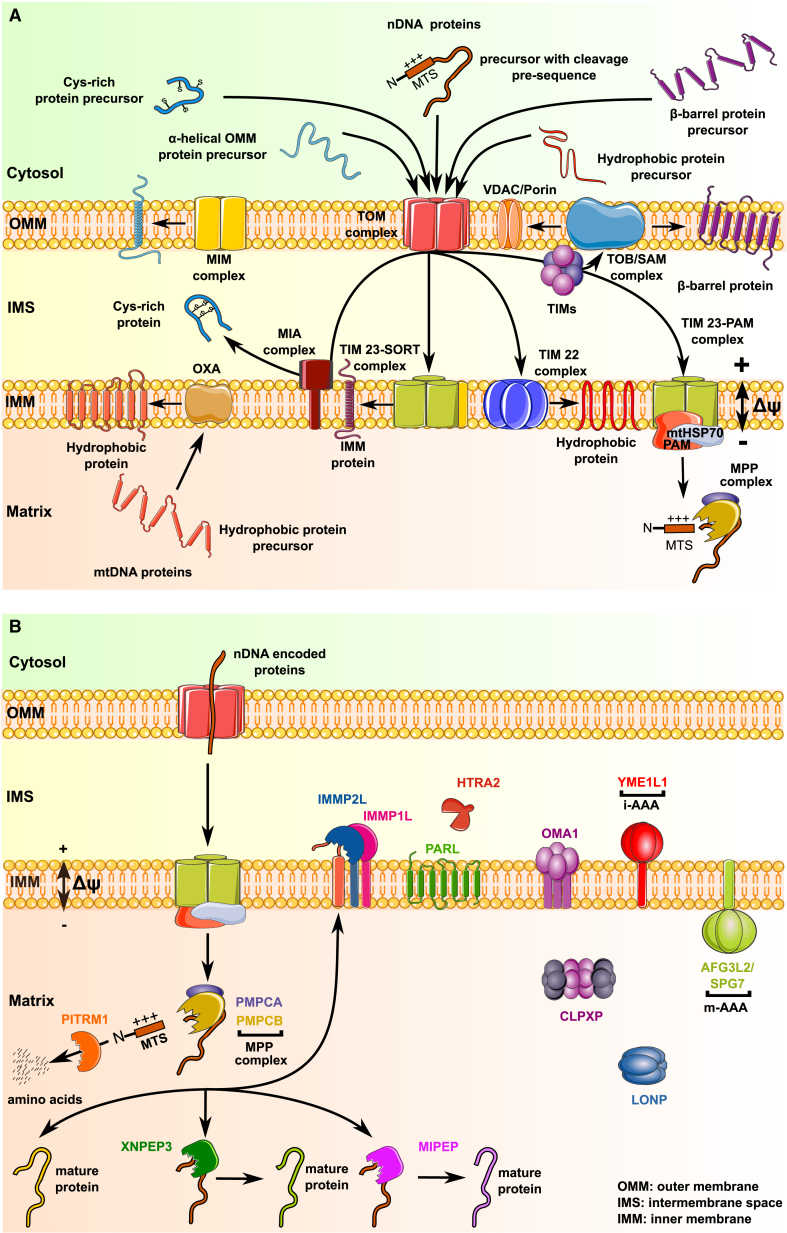
The mitochondrial protein import pathway has recently been thoroughly reviewed.1 Figure 2 illustrates the key components of the mitochondrial import machinery, which we briefly present here to provide context for discussion of DNA-damaging variants. Mitochondrial proteins that are synthesized in the cytosol are initially recognized by the translocase of the outer membrane (TOM) complex, which mediates the initial entry through the outer mitochondrial membrane (OMM) into the intermembrane space (IMS). Its components include the pore forming β-barrel TOM40, which is membrane anchored with cytosol-exposed C-terminals that serve as receptors for the incoming complex components TOM20 and TOM70. The TOM complex also contains the architectural component TOM22, which provides further binding sites for many precursor proteins on both sides of the OMM. Subsequent sorting and transport of these proteins to their final destination (which may be the OMM, the IMS, the inner mitochondrial membrane [IMM], or the mitochondrial matrix) require the activity of additional complexes. Essential elements for appropriate localization of mitochondrial proteins include (1) a set of complexes mediating import of proteins to different regions of the mitochondria, (2) mitochondria-targeting sequences on the imported proteins, and (3) proteases that modify, and chaperones that sustain, proteins concomitant with their mitochondrial import.
Figure 2.
Overview of Proteins Involved in Mitochondrial Import, Protein Sorting, and Processing
(A) Individual proteins and protein complexes involved in import and sorting.
(B) Steps in processing of imported proteins, indicating localization of proteases in mitochondrial sub-compartments. See main text for detailed description of represented pathways.
Complexes Mediating Import of Proteins to Mitochondria
Insertion of Nuclear-Encoded Proteins into the OMM. After translocation through the TOM complex, proteins with β-barrel topology interact with the small translocases of inner membrane (TIM) chaperones (TIM8/13 and TIM9/10), which deliver clients to the topogenesis of mitochondrial outer membrane β-barrel proteins/sorting and assembly machinery (TOB/SAM) complex, which helps insert them in the OMM. Proteins transported by the TOB/SAM complex include the voltage-dependent anion channel (VDAC/Porin), the major channel for import of small hydrophilic molecules, and components of the TOM complex itself, such as TOM40 (Figure 2A). Of note, not all OMM proteins require the TOM and TOB/SAM complexes. For example, some α-helical OMM proteins, such as the outer membrane protein of 45 kDa (OM45), are directly inserted into the OMM by the mitochondrial import (MIM) complex, a 200 kDa oligomeric assembly of two small single-spanning outer membrane proteins, MIM1 and MIM2.
Insertion of Nuclear-Encoded Proteins into the IMM and IMS. The TIM22 and TIM23 complexes localize to the IMM, where their transport activity is driven by mitochondrial membrane potential. Besides their activity in transporting β-barrel proteins to the TOB/SAM complex, the small TIM chaperones also deliver hydrophobic proteins such as those of the mitochondrial carrier family (e.g., the SCL25 family) to TIM22 for insertion into the IMM. The TIM23 complex consists of the three essential components TIMM23, TIMM17A, and TIMM50. TIMM23 and TIMM17A are each tightly anchored in the membrane by four transmembrane helices, while TIMM50 is integrated in the membrane by a single helix, with an extended domain exposed in the IMS. Through dynamic associations with different partners, the TIM23 complex forms two non-identical complexes responsible for directing proteins to distinct compartments. The TIM23-SORT complex is used for protein insertion into the IMM. A subset of the nuclear-encoded mitochondrial proteins bearing a cleavage pre-sequence initially enter the matrix through TIM23-PAM (presequence translocase associated motor); some of these are subsequently redirected to the IMM through the oxidase assembly (OXA) translocase.
Small nuclear-encoded proteins with multiple cysteine residues enter the IMS after transport through TOM and are sorted by the mitochondrial intermembrane space assembly (MIA) complex for oxidative protein folding. The MIA complex consists of the oxidoreductase CHCHD4 (MIA40) and the flavin adenine dinucleotide (FAD)-dependent sulfhydryl oxidase ALR (augmenter of liver regeneration) encoded by GFER (MIM: 600924). Both factors form a disulfide relay system in which CHCHD4 functions as a receptor that transiently interacts with incoming polypeptides via disulfide bonds.
Delivery of Nuclear-Encoded Proteins to the Mitochondrial Matrix. Upon association with mtHSP70 and additional co-chaperones, the TIM23-PAM complex guides precursors with a cleavable N-terminal mitochondrial targeting signal (MTS) to the mitochondrial matrix, where mitochondrial processing proteases and peptidases remove and degrade the MTS, as discussed below.
Insertion of Mitochondria-Encoded Proteins into the IMM. The membrane integral core subunits of the oxidative phosphorylation system are encoded by mtDNA. These typically hydrophobic proteins are synthesized in the mitochondrial matrix by mitochondrial ribosomes and exported into the IMM by the cytochrome oxidase assembly (OXA) translocase.
Mitochondrial Targeting Sequence (MTS)
The TOM40 and TIM23 complexes transport precursor proteins with positively charged N-terminal stretches which are present in precursor forms of soluble proteins targeted to the matrix, in the proteins targeted to the IMS, and in transmembrane-domain-bearing proteins targeted to the IMM. In this last case, the targeting signal is sometimes referred to as bipartite, as it consists of the MTS followed by a hydrophobic signal. Because this N-terminal signal gets cleaved from the proteins that reach the matrix, the generic term of pre-sequence has been widely used to describe it. However, the more precise designation as a mitochondrial targeting sequence (MTS) has been recommended as a possible source of higher reliability in data-mining analyses.22 The initial step of import is also driven by non-N-terminal signals distributed throughout imported proteins. These have been referred as internal MTS-like signals, as they share some sequence properties with the MTS. They mediate binding to TOM70 in a manner that prevents premature folding and aggregation, enhancing efficient import.23
Initial low throughput sequence analyses of the N termini of mitochondrial precursor proteins based on Edman sequencing revealed four main features of the MTS: (1) a length of 20 to 40 amino acids, (2) an overall positive charge, (3) the ability to form an amphiphilic helix, and (4) in many cases, the presence of an arginine in position −2, −3, or −10 from the cleavage site, surrounded by specific flanking sequences.24 In the genomic/proteomic era, this work has been greatly enhanced by global determination of the (mitochondrial) N-terminome.25
An extensive survey of mitochondrial precursors has shown that the specific flanking sequences around arginine motifs, initially described as characteristic of the MTS, can be found in degenerate forms, with 15%–25% of experimentally defined cleavage sites not matching previously described motifs.25 Approximately 30% of pre-sequences are now recognized as longer than 40 residues, with ability to form more than one helix.11, 26 The great diversity of import sequences has made development of computational methods for prediction of mitochondrial localization challenging and sometimes controversial, as we discuss later in this review.11, 27
Transport-Associated Protein Processing
Import of nuclear proteins into the mitochondria is supported by their interaction with various chaperones and often accompanied by transport-associated folding and processing. Components of the proteolytic system include the ubiquitin-proteasome system and a group of sub-compartment-specific proteases, which may have additional chaperone activities (Figure 2B). Of the nearly 600 proteases of the human degradome, about 30 localize to the mitochondrial matrix or are anchored in the IMM or OMM.28 They fall into three classes: (1) processing peptidases for the maturation of the mitochondrial proteome, (2) quality control proteases for degradation of damaged or misfolded proteins and regulation of mitochondrial functions, and (3) oligopeptidases that further degrade residual peptides produced by action of the other two classes.
MTS-containing precursor proteins that enter the matrix are subject to maturation proteolytic steps that support appropriate protein stability. The most common step involves the cleavage of the MTS by the matrix-localized mitochondrial processing peptidase (MPP) complex (comprised of mitochondrial-processing peptidase subunits α and β, encoded by PMPCA [MIM: 613036] and PMPCB [MIM: 603131]). Additional processing by the mitochondrial intermediate peptidase (MIPEP/MIP/Oct1) which cleaves an octapeptide, or X-Prolyl aminopeptidase 3 (XPNPEP3/Icp55), which removes a single N-terminal amino acid, replace destabilizing N-terminal amino acids that would lead to degradation by amino acids associated with longer half-life.29 Alternatively, after cleavage by MPP, the IMM protease subunits 1 and 2 (encoded by IMMP1L [MIM: 612323] and IMMP2L [MIM: 605977]) remove hydrophobic sorting signals for release to the IMS.
A thorough review of the mitochondrial quality control (QC) proteases and their crucial role as regulators of the integrity of the mitochondrial proteome and morphology and key players in apoptotic signaling and stress response has recently been provided.30 For the present discussion, we note that the main players in mitochondrial QC (the i-AAA protease YME1L1, the m-AAA protease AFG3L2/SPG7, LONP, CLPXP [hetero-oligomer of CLPX and CLPP], HTRA2, OMA1, and PARL) are themselves clients of the import machinery. Therefore, any defect affecting import of these proteins could have more global damaging consequences.
Finally, the principal oligopeptidase in the matrix is pitrilysin metallopeptidase 1 (PITRM1). Known substrates include the cleaved MTS produced during the processing of precursor proteins and several forms of amyloid-beta peptide (Aβ) associated with Alzheimer disease.
Rare Disorders Caused by Aberrant Mitochondrial Import
A growing number of cases of defects in proteins involved in mitochondrial import have been associated with rare diseases. Some examples are presented here, organized by mitochondrial import process. An extended list of human genes with variants associated with defective import is presented as Table S1.
Pathological Genetic Variation Affecting the TIM and OXA Complexes
Rare variants in genes related to the TIM (translocases of inner membrane) and OXA complexes (see Figure 2A) may manifest as clinically severe childhood syndromes involving encephalopathy, 3-methylglutaconic aciduria (3-MGA-uria), and cardiomyopathy due to diverse activities of these complexes in oxidative phosphorylation, as well as mitochondrial functions involving and mediated by cardiolipin.
Several recent publications describe defects in TIMM50, TIMM22, and OXA1L. For example, homozygous missense variations in TIMM50 (MIM: 607381) (GenBank: NM_001001563.3; c.649C>T [p.Arg217Trp] [rs1300848445] and c.755C>T [p.Thr252Met] [rs1244226820]) were reported in individuals who exhibited epileptic encephalopathy with 3-MGA-uria.31 In another report, compound heterozygous variants in TIMM50 (c.335C>A [p.Ser112∗] [rs35135520] and c.569G>C [p.Gly190Ala] [rs776019250]) were identified as disease associated in an infant with rapidly progressive severe encephalopathy. Here, the patient’s fibroblasts presented low levels of TIMM50 and other components of the TIM23 complex, lower mitochondrial membrane potential, and impaired TIM23-dependent protein import impacting mitochondrial oxidative phosphorylation.32 A case of compound heterozygous variants in TIMM22 (MIM: 607251) (GenBank: NM_013337.3; c.75C>A [p.Tyr25∗] [rs754537066] and c.97G>C [p.Val33Leu] [rs149879547]) was also recently identified in an individual exhibiting a neuromuscular phenotype associated with elevated lactate levels. Biochemical analyses of fibroblasts from this individual revealed reduced TIM22 protein levels, deficiency of TIM22 complex formation, a compromised oxidative capacity, and altered mitochondrial morphology.33 Affecting a different part of the import machinery, biallelic disease-associated variants in OXA1L (MIM: 601066) (GenBank: NM_005015.3; c.500_507dup [p.Ser170Glnfs∗18] and c.620G>T [p.Cys207Phe] [rs772751581]) were identified in an individual presenting with severe encephalopathy, hypotonia, and developmental delay in early childhood. These symptoms were associated with decreased production of OXA1L and subunits of mitochondrial oxidative phosphorylation complexes IV and V, and deficient activity of the respiratory chain.34
It is possible that additional proteins that modulate TIM and OXA complex function will be identified. For example, in 2017, the mitochondrial acylglycerol kinase AGK was described as a component of the TIM22 complex, required for mitochondrial import of IMM transmembrane proteins, including members of the SLC25A family.35, 36 AGK (MIM: 610345) defects are associated with Sengers syndrome (MIM: 212350) which is characterized by congenital cataracts, hypertrophic cardiomyopathy, mitochondrial myopathy, and lactic acidosis. Sengers syndrome has also been recognized as one of many mitochondrial DNA depletion syndromes (MTDPS10).37 Recognizing that AGK is a component of TIM22 provides the molecular basis for the decrease in SLC25A4 (also called ANT1; adenine nucleotide translocator 1) protein levels observed in the muscle tissues of two unrelated individuals with Sengers syndrome.38 Hence, AGK is positioned to influence mtDNA abundance, at least in part by controlling SLC25A4 abundance, with AGK deficiency potentially contributing to mitochondrial depletion.
3-MGA-Uria Is Associated with Multiple Factors Affecting Mitochondrial Import
Interactions between proteins and lipids regulate mitochondrial import. Elevated urinary excretion of 3-MGA (or 3-MGA-uria) is a hallmark of a genetically and phenotypically heterogeneous group of inherited metabolic syndromes. A long-standing OMIM Phenotypic Series (MIM: PS250950) classification scheme for this disorder describes nine different types of 3-MGA-uria (Table S2A). More recently, a new classification based on patho-mechanisms and genetics was proposed, with “primary” 3-MGA-uria associated with variants in AUH (required for leucine catabolism), multiple “secondary” subtypes of 3-MGA-uria associated with defective phospholipid remodeling (for defects in TAZ and SERAC1) or mitochondrial membrane associated disorder (for defects in OPA3, DNAJC19, and TMEM70), and the “not otherwise specified (NOS)” 3-MGA-uria for which no patho-mechanism has been identified.39 Evaluation of the set of genes associated with this syndrome exemplifies the complexity of signaling that can impact mitochondrial import.
While the metabolic origin of 3-MGA in the urine, independent of a leucine catabolism defect, has long been unknown, it was recently proposed that the 3-MGA pathway functions as an overflow valve generating organic waste when acetyl-CoA exceeds utilization capacity.40 Diversion of the intra-mitochondrial acetyl-CoA to organic waste occurs under conditions of reduced electron transport chain (ETC) activity. Reduced ETC is associated with the accumulation of NADH in the matrix space, which leads to inhibition of key enzymes of the TCA cycle, driving a compensatory increase in anerobic glycolysis. As the ETC machinery is embedded in the IMM, its activity is highly susceptible to any perturbation of its lipid composition, with negative consequences for the import of mitochondrial matrix-targeted proteins, as import requires the electrochemical membrane potential generated by a functional ETC across the IMM. We hypothesize that these close relationships between control of lipid composition, and functionality of the import machinery indicate a common mechanism linking many of the genetic defects that result in 3-MGA-uria.
An example of IMM lipid perturbation is seen in individuals with Barth syndrome, associated with TAZ variants, who present with a characteristic lipid profile marked by decreased cardiopin (CL) levels, mainly of (poly)unsaturated species, and elevated levels of the remodeling intermediate monolysocardiolipin. How these anomalies in CL and other lipids in mitochondrial membranes impact ETC and lead to 3-MGA-uria is elaborated in Ikon and Ryan.41
Interestingly, we note many of the genes associated with secondary 3-MGA-uria (Table S2A) are linked to CL. CL comprises ∼20% of the IMM, has a critical role in IMM dynamics, and associates with many IMM proteins, including those encoded by TAZ, DNAJC19, and SERAC1, as well as the prohibitins (PHB1 and 2), which function as protein and lipid scaffolds to ensure the integrity and functionality of the IMM. TAZ (MIM: 300394) encodes tafazzin, a phospholipid transacylase involved in CL remodeling, a process of sequential deacylation-reacylation/transacylation reactions that establish the final composition of the acyl chains of CL. DNAJC19 (MIM: 608977) encodes a mitochondrial co-chaperone that associates with prohibitin to regulate CL remodeling by taffazin. Phospholipid exchange also depends on activity of the SERAC1 (MIM: 614725)-encoded phosphatidylglycerol remodeling protein found at the interface of mitochondria and endoplasmic reticula. In addition, analyses performed in vivo, in isolated mitochondria, and in model membrane systems demonstrate that the soluble receptor domain of TIMM50 interacts with membranes and with specific sites on the TIM23 channel in a manner that is directly modulated by CL.
Other genes associated with mitochondrial import have been linked to control of 3-MGA-uria and ETC. These include AGK, already described above as encoding a component of the TIM22 complex, and genes encoding components of complex V (ATP synthase), the complex V associated factor TMEM70 (MIM: 612418) and QIL1 encoded by MICOS13 (MIM: 616658) (Table S2B). AGK exerts an essential function in defining membrane lipid composition, including CL content. CL is a component of the cristae in which complex V is embedded. TMEM70 and QIL1 are necessary for cristae formation. Globally, based on these various functional interactions, 3-MGA-uria emerges as linked to defects in the ultrastructure of the inner membrane in a manner that disrupts mitochondrial import.
Pathological Genetic Variation in the MIA Pathway
Amyotrophic lateral sclerosis (ALS) (MIM: PS105400) is a neurodegenerative disease affecting motor neurons in the brain and spinal cord, leading to muscle weakness. Familial ALS constitutes ∼10% of case subjects. Variants in at least 15 different genes or loci have been identified as disease causing, with the most common in SOD1 (MIM: 147450) and C9orf72 (MIM: 614260). Variants in CHCHD10 (MIM: 615903) also have been reported in families with autosomal-dominant ALS.42 CHCHD10 encodes a soluble IMS-localized member of the coiled-coil-helix-coiled-coil-helix domain-containing protein family which participates in a system that stabilizes membrane curvature and forms contact sites between the IMM and the OMM. The disease-associated CHCHD10 variant c.323A>C (p.Gln108Pro) (GenBank: NM_213720.2; rs889489701) blocks CHCHD10 import to the IMS, thereby promoting its destruction.42 The dramatic reduction of CHCHD10 import, also observed upon CHCHD4 knockdown, supports the idea that import of MIA substrates into the IMS competes with their ubiquitin-dependent degradation in the cytosol.43, 44
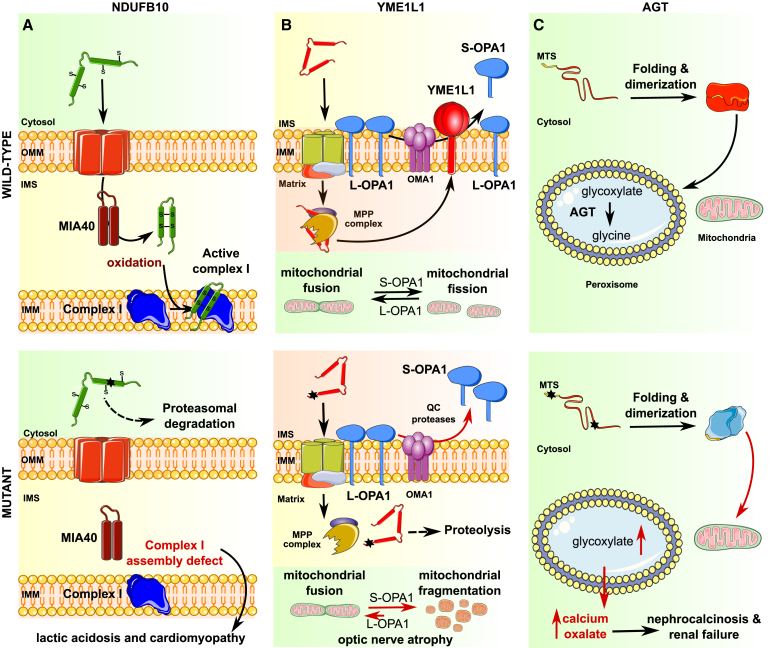
In two additional examples, rare myopathies are caused by autosomal-recessive inheritance of components of the MIA pathway. In one example, an autosomal-recessive myopathy (MIM: 600924) is associated with an inherited variant in the core component of the MIA pathway ALR (GFER/ERV1).45 Three children born to healthy consanguineous parents presented with progressive myopathy and partial combined respiratory-chain deficiency, congenital cataract, sensorineural hearing loss, and developmental delay. The homozygous c.581G>A (p.Arg194His) variant (GenBank: NM_005262.2; rs121908192) was found to affect ALR protein stability and FAD binding, causing reduced levels of several substrates of the disulfide relay.46 In another example, compound heterozygosity of the NDUFB10 (MIM: 603843) variants GenBank: NM_004548.3; c.207dup (p.Glu70∗) and c.319T>A, (p.Cys107Ser) has been identified as a cause of fatal infantile lactic acidosis and cardiomyopathy.47 NDUFB10 encodes an accessory subunit of respiratory chain complex I that is a substrate of the MIA pathway. At autopsy, diminished NDUFB10 protein levels were observed, likely the result of the altered protein becoming a poor CHCHD4 substrate. Further, an in vitro assay indicated that the cysteine to serine substitution in position 107 resulted in impaired oxidation by CHCHD4. As with many mitochondrial diseases,48 this variant had tissue-specific manifestation; while NDUFB10 and complex I activity were almost completely absent in muscle and heart, protein level and activity were almost normal in fibroblasts, implying that despite the variation, the protein could be imported and folded under certain environmental conditions (Figure 3A). For further reference, an extended discussion of disease-associated variants affecting the MIA machinery or its substrates is provided in Ikon and Ryan.49
Figure 3.
Schematic Representation of Pathogenic Signaling Associated with Disease-Associated Variants
(A) Effect of MIA pathway defects on maturation of NDUFB10 associated with lactic acidosis and cardiomyopathy.
(B) Deficient processing of YME1L1 by MPP upon import to the mitochondrial matrix causes defects in OPA1 cleavage, resulting in mitochondrial fragmentation and atrophy of optic nerves.
(C) Combination of genetic variants weakening AGT peroxisomal targeting and protein folding together lead to inappropriate localization to the mitochondria, causing deficient glycoxylate metabolism, nephrocalcinosis, and renal failure. For each example, top panel indicates wild-type function, and bottom panel illustrates defect associated with mitochondrial import variant. For full description of the variants, see text.
Pathological Genetic Variation Affecting the MTS
Genetic variants in the mitochondrial targeting sequences of proteins that are translated in the cell nucleus and then targeted to the mitochondria have been shown to manifest clinically as deficiencies in Krebs cycle components (pyruvate dehydrogenase and malonyl-CoA decarboxylase), and in one report, as leukodystrophy.
Pyruvate dehydrogenase complex (PDC) deficiency is one of the most common neurodegenerative disorders associated with abnormal mitochondrial metabolism. A genetically heterogeneous syndrome, PDC deficiency is characterized by progressive neurological and neuromuscular degeneration, lactic acidosis, and often death in childhood. Among the six different forms of PDC deficiency (MIM: PS312170), recessive disease is associated with defects in autosomally encoded subunits of PDC, while the most common form arises from defects in PDHA1 (MIM: 300502), located on the X chromosome.50 A PDHA1 variant, p.Arg10Pro (GenBank: NM_000284.3; c.29G>C [rs137853257]), was identified in a 10-month-old boy and his brother, both with pyruvate dehydrogenase deficiency, as well as in their more mildly affected mother. This was the first description of an amino acid substitution in a mitochondrial targeting sequence resulting in a human genetic disease.51 Defective import of proteins into the mitochondria was thought responsible for the reduced pyruvate dehydrogenase levels, as in vitro experiments indicated that the mitochondrial matrix protein ornithine transcarbamylase (OTC) was translocated at a reduced rate to the matrix when attached to the mutant p.Arg10Pro PDHA1-MTS.
Biallelic variants in the MTS of MLCYD (MIM: 606761), located in residues 1 to 39, cause an autosomal-recessive form of malonyl-CoA decarboxylase deficiency (MIM: 248360). The resultant disease is characterized by malonic aciduria, developmental delay, seizure disorder, hypoglycemia, and cardiomyopathy. MLYCD catalyzes the decarboxylation of malonyl-CoA to acetyl-CoA, an important step in the breakdown of fatty acids. Immunocytochemistry performed on the fibroblasts of a MLYCD-deficient individual homozygous for the variant GenBank: NM_012213.2; c.8G>A (p.Gly3Asp) (rs121908081) revealed dense aggregates of MLYCD at the plasma membrane, indicative of mistargeting.52
Leukodystrophies are a group of rare, progressive, metabolic, genetic diseases that impact the white matter of the central nervous system. A case of severe early-onset leukodystrophy with multiple defects of respiratory complexes and a severe impairment of lipoic acid synthesis has been linked to a homozygous DNA variant, GenBank: NM_030940.3; c.29T>G (p.Val10Gly), in the MTS in the ISCA1 protein, a component of the mitochondrial Fe-S biogenesis machinery. This homozygous variant is predicted to reduce mitochondrial targeting, resulting in drastically reduced ISCA1 protein levels and the severe neurologic phenotype observed.19
The recognition that some mitochondrial proteins have internal MTS signals23 may help explain the function of some previously defined variants. For example, Kobayashi et al. described the variant GenBank: NM_000274.3; c.268C>G (p.Gln90Glu) (rs121965060) in ornithine aminotransferase (OAT) (MIM: 613349) in a person with autosomal-recessive gyrate atrophy (MIM: 258870). Although far removed from the OAT N terminus, this variant was shown to obstruct the mitochondrial entry of the precursor, supporting that it was required in addition to the N-terminal signal sequence for mitochondrial entry.53
Some MTS variants are associated with therapeutic response, rather than disease onset. SOD2 (MIM: 147460) encodes the antioxidant enzyme that affords the major defense against ROS. The SOD2 Val16Ala variant is reported in ClinVar (ID: 14751) as significant for drug response, as it affects the survival of individuals with breast cancer receiving cyclophosphamide-containing chemotherapy.54 Amino acid position 16 in the MTS of SOD2 was the first reported dimorphic site in an MTS (GenBank: NM_000636.3; c.47T>C [p.Val16Ala] [rs4880]) and has been much studied.55 According to the gnomAD database, this variant is present in close to half of the individuals in all populations except East Asians, where it is observed at a frequency of 0.12. Notably, position 16 is located in a region of low evolutionary conservation. Although high prevalence and low conservation often causes dismissal of possible pathogenicity, there are a growing number of examples in which a disease-causing variant corresponds to the wild-type allele in another species,56 emphasizing the need for caution in interpretation.
Pathological Genetic Variation Affecting Mitochondrial Proteases
Variants leading to changes in mitochondrial proteases have been identified in families with inherited forms of infantile neurodegeneration, Perrault syndrome (characterized by sensorineural hearing loss, additional neurological defects, and other symptoms), ocular ataxia, and cerebellar ataxia, reflecting the importance mitochondrial protease processing in neuronal tissues (see Table 1).
Table 1.
Reported Disease-Causing Variants Associated with Defects in Mitochondrial Proteases
| Gene, Function (RefSeq#)a | Variant Effect on DNA and Protein Sequence (Linked SNPs) | Clinical Phenotype (OMIM#) | Evidence for Pathogenicity | Ref. |
|---|---|---|---|---|
| PMPCA, presequence cleavage (NM_015160.2) | 1. c.[1129G>A];[1129G>A],b p.[Ala377Thr];[Ala377Thr]; (rs753611141) 2. c.[287C>T];[1543G>A], p.[Ser96Leu];[Gly515Arg]; (rs869025292); (rs869025293) |
cerebellar ataxia (MIM: 213200) | Allelic frequency: 0%c p.Ala377Thr associated with deficient MPP activity in patient-derived cell lines; predicted to cause a catalytic activity or substrate binding defect p.Ser96Leu and p.Gly515Arg in structurally conserved domains |
Jobling et al.59 |
| PMPCB, presequence cleavage (NM_004279.2) | 1. c.[523C>T];[601G>C], p.[Arg175Cys];[Ala201Pro]; (rs145596167);(rs146343535) 2. c.[524G>A];[530T>G], p.[Arg175His];[Val177Gly]; (rs200188353); (rs1436866272) 3. c.[1265T>C];[1265T>C], p.[Ile422Thr];[Ile422Thr]; (rs1461200360) |
infantile neurodegeneration (MIM: 29576218) | p.Arg175Cys, p. Ala201Pro, and p.Ile422Thr decrease protein level and activity in fibroblast and/or hiPSCs/NESs patient cell lines, and severe growth and/or MPP processing defects in yeast complementation. p.Arg175His, p.Ala201Pro, p.Ile422Thr, and p.Val177Gly predicted to cause protein stability, active site, and/or dimerization defects |
Vögtle et al.57 |
| MIPEP,presequence trimming (NM_005932.3) | 1. c.[212T>A];[1745T>G], p.[Leu71Gln]; [Leu582Arg]; (rs10575118740); (rs1057518739) 2. c.[916C>T];[1804G>T], p.[Leu306Phe];[Glu602∗]; (rs143912947); (rs114638163) 3. c.[1027A>G];[1027A>G], p.[Lys343Glu];[Lys343Glu]; (rs1057518741) 4. c.[1534C>G]; + ch13: 1.4 Mb CNV_del, p.[His512Asp] |
left ventricular non-compaction (MIM: 602241) | Allelic frequency: 0%d for all variants except p.Leu306Phe = 8.2 × 10−6, p. pHis512Asp = 3.2 × 10−5 p.Leu71Gln and p.Lys343Glu and orthologous yeast mutants have decreased enzymatic activity p.Leu71Gln, p.Leu582Arg, p.Leu306Phe, p.Lys343Glu, and p.His512Asp predicted pathogenic |
Eldomery et al.105 |
| XPNPEP3, presequence trimming (NM_022098.3) | 1. c.[1357G>T]; [1357G>T], p.[Gly453Cys];[Gly453Cys]; (rs267607179)e 2. c.[931_934del];[931_934del], p.[Asn311Leufs∗5)];[(Asn311Leufs∗5]f |
nephronophthisis-like nephropathy-1 (MIM: 613159) | Allelic frequency: 0%g p.Gly453Cys has been showed to cause a splicing defect on patient’s mRNA | O’Toole et al.106 |
| PITRM1, post cleavage MTS degradation (NM_014889.3) | c.[548G>A];[548G>A], p.[Arg183Gln];[Arg183Gln]; (rs1249144069) | mental retardation, spinocerebellar ataxia, cognitive decline and psychosis | Allelic frequency: 0%h Protein instability, impaired cell proliferation, and mitochondrial function have been detected on patients’ fibroblasts and in vitro functional assay |
Brunetti et al.107 |
| HTRA2, intermembrane space protease (NM_013247.4) | 1. c.[1211G>A];[1211G>A], p.[Arg404Gln];[Arg404Gln]; (rs767006508) 2. c.[1316_1320del];[1316_1320del], p.[Ala439Aspfs∗15];[Ala439Aspfs∗15]; (rs1057519080) |
3-methylglutaronic aciduria type VIII, MGCA8 (MIM: 617248) | Allelic frequency: 0%3 Both variants cause complete absence of the protein; fibroblasts with p.Arg404Gln have impaired cell growth |
Mandel et al.108 |
| LONP1, matrix protease/chaperone (NM_004793.3) | 1. c.[2245C>T];[2300G>A], p.[Pro749Ser];[Gly767Glu]f 2. c.[1427A>C];[2245C>T], p.[Glu476Ala];[Pro749Ser] 3. c.[2009C>T];[2780_2782del], p.[Ala670Val];[Ile927del]f 4. c.[2014C>T];[2014C>T], p.[(Arg672Cys)];[(Arg672Cys)]f 5. c.[2009C>T];[2009C>T], p.[(Ala670Val)];[(Ala670Val)]f 6. c.[1392G>A]; [2036G>A], p.[(Trp464∗)f];[(Arg679His)]; (rs549574673) |
cerebral, ocular, dental, auricular, skeletal anomalies (CODAS) syndrome (MIM:600373) | Allelic frequency: 0%c All variants except p.Arg679His predicted as pathogenic and to cause structural defects |
Dikoglu et al.109 |
| CLPP, matrix protease/chaperone (NM_006012.2) | 1. c.[433A>C];[433A>C], p.[Thr145Pro];[Thr145Pro]; (rs398123033) 2. c.[440G>C];[440G>C], p.[Cys147Ser];[Cys147Ser]; (rs398123034) 3. c.[270+4A>G];[270+4A>G]; (rs398123035) |
Perrault syndrome (MIM: 23541340) | Allelic frequency: 0%i p.Thr145Pro and p.Cys147Ser predicted as pathogenic and to cause folding defect c.270+4A>G caused splicing defect in vitro |
Jenkinson et al.110 |
| YME1L1, i-AAA-protease (NM_014263.3) | c.[445C>T];[445C>T], p.[Arg149Trp];[Arg149Trp]; (rs1057519312) | optic atrophy-11 (MIM: 617302) | Allelic frequency: 0%j Variant causes protein degradation, abnormal substrate processing, and other functional defects |
Hartmann et al.26 |
Abbreviations: hiPSC, human induced pluripotent stem cells; MPP, mitochondrial processing peptidase; NES, neuroepithelial stem cells.
All the identified DNA variants cosegregate with disease phenotype.
Founder mutation in a Lebanese Christian Maronite population.
Allelic frequency is based on control group from ExAC.
Allelic frequency is based on control groups from ExAC and Baylor-CMG exome.
Founder mutation in a Northern Finnish population.
dbSNP ID not applicable.
Allelic frequency is based on in-house control database, ethnicity or geographic-matched controls.
Allelic frequency is based on 1000 Genomes.
Allelic frequency is based on NHLBI Exome Sequencing Project (ESP) Exome Variant Server.
Allelic frequency is based on ExAC, 1000 Genomes, NHLBI Exome Sequencing Project (ESP) Exome Variant Server, dbSNP.
In one such example, biallelic variants in the gene encoding the protease PMPCB have been identified as a cause of early childhood neurodegeneration. Their presence caused defective proteolytic activity manifested by an accumulation of a processing intermediate of frataxin, a defined PMPCB substrate and iron chaperone involved in iron-sulfur cluster biogenesis and heme biosynthesis. This incomplete processing of frataxin, observed in vivo in mitochondria isolated from fibroblasts from the affected individuals, led to a dysregulation of iron-sulfur cluster biogenesis that caused the neurological phenotype57.
In another example, optic atrophy has been associated with biallelic variants in YME1L1 (MIM: 607472) (GenBank: NM_014263.3; c.445C>T [p.Arg149Trp] [rs1057519312]). YME1L1 is an IMM-associated quality control protease; pathogenic variation impairs YME1L1 cleavage by MPP, causing YME1L1 to undergo rapid proteolysis and reducing its steady-state level. Consequently, skin fibroblasts derived from affected individuals poorly process two substrates: PRELID1, a lipid transfer protein for phosphatidic acid in the IMS, and OPA1, a protein important for balancing fusion and fission in mitochondrial dynamics. After import to the mitochondria, the precursor OPA1 protein undergoes complex proteolytic processing by the mitochondrial QC proteases (giving rise to IMM-anchored L-OPA1 involved in fusion, which may be further cleaved to generate S-OPA1 involved in mitochondrial fission), making it vulnerable to processing defects (Figure 3B). The observed proliferation defects and mitochondrial network fragmentation in cells from affected family members are consistent with an abnormal processing of OPA1.26 Genetic variations in OPA1 are also a known cause of optic atrophy.58
As a cautionary note, the complete understanding of the molecular functions of the proteases discussed here depends on the identification of their substrates/clients. However, these may be hard to discern, as the rules for substrate selection by the proteases are not well established. Although specific degron sequences have been identified for YMEL1 and AFG3L2 in the unstructured N termini of some substrates, the full spectrum of substrates remains poorly defined. Known substrates are used for functional variant validation although they may not be relevant for the cellular context or physiological conditions in which a new variant is identified (e.g., frataxin [FXN] or malate dehydrogenase [MDH2] are typically used to access the impact of variations in PMPCA59, 60 or PMPCB,57 and OPA1 for variations in YME1L126). Several known substrates are routinely tested as only some may be affected.57, 60 In the case of ClpP, studies have demonstrated that the preference toward certain amino acids adjacent to the cleavage sites observed when using a tailored fluorogenic substrate library was not retained during proteolysis of endogenous substrates.
Rare Pathologic Genetic Variation Leading to Protein Mistargeting to the Mitochondria
MTSs bear similarity to other targeting signals, including peroxisomal targeting signal type 2 (PTS2). PTS2 mediates the import of a fraction of soluble proteins to the peroxisome. Osumi et al. first showed, by site-directed mutagenesis, that the replacement of His17 in the N-terminal signal peptide of rat peroxisomal 3-ketoacyl-CoA thiolase encoded by Acaa1a by Arg, Lys, Leu, or Val results in mitochondrial localization of this protein.61
AGXT (MIM: 604285) and EHHADH (MIM: 607037) are two examples of genes encoding proteins normally destined to the peroxisomes for which disease-causing mislocalization in the mitochondrial matrix has been reported. In rare individuals, mislocalization of the protein products of AGXT and EHHADH to the mitochondrial matrix rather than the peroxisome can cause severe renal disease. Primary hyperoxaluria type I (PH1) (MIM: 259900) is an autosomal-recessive disorder of the glyoxylate metabolism leading to overproduction of oxalate, which manifests as renal stones and/or nephrocalcinosis. PH1 is caused by mutations in AGTX, which encodes the hepatic, peroxisomal alanine-glyoxylate aminotransferase (AGT). Mitochondrial targeting of AGTX occurs in persons homozygous for the variant c.32C>T (p.Pro11Leu) (GenBank: NM_000030.2, rs34116584) but, as described below, this variant is ineffective by itself. Mistargeting of AGT leads to poor efficiency of glyoxylate detoxification and accumulation of calcium oxalate in various bodily tissues, especially the kidney, leading to renal failure62 (Figure 3C), although at this time conflicting interpretations of the pathogenicity of this variant are found in ClinVar (ID: 5641).The relatively common p.Pro11Leu variant is reported in the gnomAD database at an allelic frequency of ∼0.15 in the general population (ranging from ∼0.20 in Europeans to ∼0.0004 in East Asians), and hence is clearly not sufficient as a monogenic source of dysfunction. A possible molecular explanation for the relationship of this specific variant to control of mitochondrial import and disease lies in the fact that the leucine residue introduces a weak, non-cleavable putative mitochondrial targeting sequence at the N terminus of the molecule (MASHKLLVTPLKALLKPLSC). Because the dimerization of AGT that rapidly follows its folding results in a tight folded structure with a buried N terminus, incompatible with mitochondrial import, the variant is functionally ineffective by itself. However, present in cis with rare variants that disrupt the folding/dimerization process and expose the N terminus (e.g., c.508G>A [p.Gly170Arg], rs121908529; c.731T>C [p.Ile244Thr], rs121908525; c.454T>A [p.Phe152Ile], rs121908524, and c.121G>A [p.Gly41Arg], rs121908523), this variant allows preferential mitochondrial import.63 Mild translation inhibition with emetine was recently found to correct the mislocalization of the mutant protein Pro11Leu;Gly170Arg, likely because slowing translation improved folding.64
EHHADH encodes one of the four enzymes of the peroxisomal β-oxidation pathway and has also been linked to disease due to mitochondrial mistargeting. EHHADH is a bifunctional protein; its N-terminal region contains enoyl-CoA hydratase activity while its C-terminal region contains 3-hydroxyacyl-CoA dehydrogenase activity. The EHHADH variant GenBank: NM_001966.3; c.7G>A (p.Glu3Lys) (rs398124646) has been associated with a familial case of autosomal-dominant renal Fanconi syndrome (MIM: 615605),65 manifesting as rickets, impaired growth, glucosuria, generalized aminoaciduria, phosphaturia, metabolic acidosis, and low-molecular-weight proteinuria. This variant generates a de novo MTS which is cleaved upon import to the mitochondria. The neoprotein is then incorporated into mitochondrial trifunctional protein (MTP), an IMM-bound hetero-octamer encoded by HADHA (MIM: 600890) and HADHB (MIM: 143450), which catalyze the final steps of mitochondrial long-chain fatty acid β-oxidation. The improper incorporation of the EHHADH protein resulting from the DNA variation impairs the normal catalytic process for fatty acid β-oxidation and also impairs mitochondrial respiration due to the coupling of these processes by MTP.66
Tail-anchored (TA) proteins are also aberrantly targeted to the mitochondria. This unique class of functionally diverse membrane proteins is defined by their single C-terminal transmembrane domain and their ability to insert post-translationally into specific organelles (including endoplasmic reticulum [ER], mitochondria, and peroxisomes) with N termini exposed to cytoplasm and C termini within the organelle. Mislocalized tail-anchored (TA) proteins of the OMM are cleared by the quality control pathway involving the conserved eukaryotic protein Msp1 (ATAD1/Thorase in humans),67 a signal-anchored membrane protein comprising an N-terminal transmembrane domain that tethers its soluble ATPase domain to the cytosolic face of the OMM and peroxisome. ATAD1 is also known as a mediator of recycling of the glutamate-gated channels α-amino-3-hydroxy-5-methylisoxazole-4-proprionate (AMPA) receptors, thereby regulating synaptic plasticity and learning and memory.68 Several recent studies have reported lethal encephalopathy cases with defects in AMPAR recycling linked to ATAD1 (MIM: 614452) variants.69, 70 It is interesting to note that Msp1 also is involved in the mitoCPR pathway in yeast. This pathway resolves mitochondrial import defects occurring at the intermembrane level;71 Msp1 mediates clearance and proteasomal degradation of unimported precursors from the mitochondrial surface. The recognition that Msp1/MitoCPR can reduce mitochondrial mistargeting in yeast suggests that further studies of its human homolog ATAD1 could elucidate similar or expanded functionality in mammalian cells.72
A final example of protein mistargeting to the mitochondria involves the variant GenBank: NM_007262.4; c.497T>C (p.Leu166Pro) variant (rs28938172) in DJ-1/PARK7 (MIM: 602533). This variant has been linked to early-onset familial Parkinson disease and appears to cause mistargeting of the normally cytoplasmic localized protein to the mitochondria. In the mitochondria, DJ-1/PARK7 impairs cell growth by differentially regulating mitochondrial Bax/Bcl-XL functions. The variation also reduces stability of DJ-1 by allowing it to directly bind to the mitochondrial serine protease HTRA2.73
Deregulated Mitochondrial Import in Cancer
As discussed in the preceding sections, most of the genetic diseases associated with defects of mitochondrial import involve neurologic and neuromuscular degeneration. As with neurodegenerative diseases, the physiologic changes that accompany and support cancer involve altered abundance of reactive oxygen species (ROS), altered control of protein production and degradation, and altered mitochondrial activity, albeit with some changes being opposite to those seen in neurodegenerative disease. For example, most cancer cells preferentially use glycolysis as an energy source (the Warburg effect), while neuronal cells bearing genetic variants associated with neurodegeneration upregulate proteins required for oxidative phosphorylation, in what has sometimes been termed an inverse Warburg effect.74 These relationships have raised the interesting possibility that inherited damaging genetic variants affecting import of proteins required for normal mitochondrial activity may in some cases reduce the risk of some forms of cancer through alteration of necessary energy resources in the cancer cell.75 Conversely, the enhanced production or activity of proteins promoting mitochondrial import, and thereby supporting mitochondrial biogenesis or function, may characterize some tumors with particularly high energy needs.
While the relative rarity of known disease-associated variants affecting the import machinery or import signals makes it difficult to assess the impact of such variants on cancer incidence or other clinical correlates of cancer, studies of tumors have revealed intriguing patterns of changes affecting subsets of mitoproteases,28 translocases,76 and mitochondrial chaperones.77 Although no global analyses have been reported, multiple components of the IMM and OMM import complexes have been shown to be present at elevated protein levels in some tumor types; for example, aberrant overproduction of MIA40 has been seen in multiple forms of cancer and has been associated with poor prognosis. Similarly, the viral oncogenic VacA protein of Helicobacter pylori has also been shown to rapidly induce production of mitochondrial translocases, concomitant with the progression of gastric lesions to a highly malignant state, suggesting a direct pathogenic role of upregulation of mitochondrial components. In some cases, proteins typically thought to serve as elements of oncogenic signaling cascades are emerging as regulators of mitochondrial import. For example, phosphorylation of the chaperone HSP70 by AKT1 was shown to regulate the mitochondrial import of SOD2, thereby affecting cellular redox balance.78 We note that this finding emphasizes the difficulty in exactly defining the import machinery as it uses many accessory components in addition to the core components.
The studies mentioned above demonstrating elevated levels of proteins associated with mitochondrial import in cancer align with a growing number of studies that emphasize that mitochondrial import of proteins normally thought of as cytoplasmic or nuclear may contribute to tumor growth (e.g., MDM279). Moreover, a substantial literature addresses the roles of mitochondrial proteases such as HTRA2 and LONP in cancer (reviewed in Quiros et al.28). For example, heterozygous LONP deletion in mouse models for colorectal and skin cancer has been shown to attenuate tumor formation; elevated LONP in human specimens is associated with poor prognosis for these cancers;80 and H. pylori infection was also shown to strongly induce LONP protein production in gastric cancer. In each of these examples, increased LONP activity was also associated with increased glycolysis.
Certainly, the pattern of mitochondrial protein production observed in cancer is relevant to expected requirements for metabolic reprogramming and mitochondrial biogenesis. However, we note, for the examples cited (and numerous other emerging examples), the degree to which these phenotypes are dependent on amplification of normal function of abnormally increased levels of proteins involved in mitochondrial import and related quality control, versus neomorphic activities remains to be established. From the point of view of the geneticist, these examples suggest that it may be of interest to determine whether known polymorphisms or rare DNA variants are associated with elevated levels of proteins of the import machinery, as these may emerge as relevant to elevated cancer risk.
Finally, it is possible that underlying or induced variance in mitochondrial import capacity may influence efficacy of cancer therapy.81 Based on the recognition that tumors differ significantly from untransformed cells in utilization of oxidative phosphorylation, some therapeutic approaches may act by selectively inhibiting energy production in cancer cells (for example, through reduced mitochondrial import, discussed in Zhu et al.81). The natural product stendomycin, long used as an antifungal, was recently identified as an inhibitor of the TIM23-dependent import machinery82 and may be useful in this regard. Conversely, some existing therapies may in part function via interaction with the mitochondrial import machinery. For example, the multikinase inhibitor sorafenib was found to unexpectedly cause inhibition of complex II/III of the mitochondrial electron transport chain. This activity was due to sorafenib stabilization of PINK1 on the mitochondrial outer membrane, and recruitment of Parkin,83 associated with changes in import. In another example, reduced levels of proteins required for mitochondrial import (TOM40, TIM23, and ALR) were selectively cytotoxic in acute myeloid leukemia (AML) and in stem cells, in contrast to non-transformed hematopoietic cells, reflecting the greater OXPHOS requirements of AML.84 Overall, these factors reinforce the clinical value of gaining insights into mitochondrial function through consideration of the features of monogenic diseases involving mitochondrial import.
Diagnosis and Potential Implications of Altered Mitochondrial Import and Targeting
In the diagnosis of rare disorders such as many of those described here, traditional approaches to diagnosis through careful phenotypic characterization and tissue biopsy are being replaced by a “genotype first” approach to diagnosis by way of reverse phenotyping. With growing use and availability of NGS-based exome and whole-genome testing and array technologies, it is likely that an increasing number of variants within mitochondrial genes and gene products associated with mitochondrial function, transport, and targeting will be identified across a wide range of human diseases including common diseases like cardiovascular disease and cancer. While at present many of the examples we discuss are supported only by limited numbers of case studies, we anticipate the number of observed cases will rapidly increase. The examples cited above make it clear that the process of mitochondrial targeting is vulnerable at numerous steps and that targeting defects may explain at least some common features of otherwise distinct genetic disorders.
The scarcity of established pathogenic variants in genes relevant for mitochondrial import has been proposed to reflect their essentiality and intolerance to variation.85, 86 Indeed, only ∼5% of known mitochondrial disorders are autosomal dominant (AD) (see Table S1).87 Moreover, genetic and other high-throughput screens in yeast47, 88, 89 and targeted studies performed in mice and in other model organisms (e.g., Sokol et al.76) have since demonstrated the essentiality of a number of the genes of the import machinery. Further, although lethality is typically associated with homozygous loss, compatible with a recessive phenotype, some of these genes are at least partially haploinsufficient. For example, while homozygous mutant mice in Lonp1 or Tmem70 are not viable (Table S1), Lonp1+/− and Tmem70+/− heterozygous mice display altered susceptibility to some forms of cancer and mild deterioration of heart function, respectively. Ongoing efforts to identify genes essential in humans by observing intolerance to loss-of-function variants across large numbers of sequenced genomes90 will provide clarity as to whether genes involved in mitochondrial import are unusually sensitive to function-disrupting gene variants.
A number of variant databases are available to assist clinical researchers address disorders associated with mitochondrial import. Some have general scope (e.g., the Mitochondrial Disease Sequence Data Resource consortium [MSeqDR]), while others focus on a specific gene (e.g., POLG). The availability of public databases including NCBI LitVar (which automatically extracts variant data from PubMed articles), NCBI ClinVar (which reports of the relationships among human variations and phenotypes), and gnomAD (providing allele frequencies in the context of variance among ethnic groups, based on exome data from >120,000 individuals and whole-genome data from >15,000 individuals), can help in identifying disease-associated variants. This analysis suffers the general pitfalls involving any variant curation such as the unfortunate bias in databases toward populations of European descent and the limitations of self-reported race, ethnicity, and genetic ancestry91 that may lead to worse clinical care and outcome for under-represented populations but also variant pathogenicity misclassifications.92
For a significant number of variants, these methods can be complemented by computational approaches to predict the consequences of an amino-acid replacement. For the cases of mitochondrial targeting defects, algorithms that predict the effect of amino acid substitutions can be useful in cases where a variant affects a folded domain of a protein with a high degree of evolutionary conservation and well-defined structure-function relationship. However, for variants affecting the MTS, these programs can have limited utility, and there is clear need for experimental validation of predictions. Similar to other targeting signals that are involved in promiscuous recognition by a common machinery (e.g., in the secretory pathway),93 MTS may be conserved between species in terms of general structural properties (such as the ability to form an α-helix) but not in primary amino acid sequences.25 This low conservation has practical implications for assessment of gene variants potentially associated with genetic disease, as low conservation may result in poor scores by some of the most frequently used pathogenicity predictor programs (e.g., SIFT and PolyPhen), which are based on the principle that evolutionary constrained regions are of critical importance for protein function. This may explain why only 0.9% of disease variations reported in UniProt fall within annotated sequences or motifs targeting signals for nuclear import or export, mitochondrial localization, or the secretory pathway.94 A number of groups have proposed specific structural features and lists of MTS sequences, but for many proteins, these sequences remain cryptic. Hence, empirical investigations that yield a mechanistic understanding of how specific variants relate to biology is highly desirable, particularly for suspected targeting variants (see discussion in Gonzalez-Sanchez et al.95). As pointed out in a recent review of mechanisms by which coding sequence variations disrupt a protein function, particularly common defects include folding disruption, subcellular localization alteration, and rewiring of an interaction network.94 As illustrated above, all three classes of defect pertain to variants of proteins involved in mitochondrial import.
An issue unique to mitochondrial dysfunction, and potentially important in assessment of inherited variants, is the involvement of both the nuclear and the mitochondrial genomes in the mitochondrial proteome. The human population is subdivided into a small number of mitochondrial haplogroups created by variations in the mtDNA that have accumulated throughout human history. Cases of incompatibility between mitochondrial and nuclear genomes have been reported in various organisms (e.g., in Drosophila). In humans, divergent patterns of mitochondrial and nuclear ancestry are associated with the risk for preterm birth and it has been suggested that deleterious interactions between the mitochondrial and nuclear genomes could pose a problem for mitochondrial replacement therapy (MRT) (reviewed in Eyre-Walker96). Examples of nuclear modifier genes that would explain the variable penetrance of disorders caused by mitochondrial variants have been presented.97, 98 Conversely, it was recently shown that the severity of the cardiomyopathy caused by variations in ANT1 can be modulated by the presence of otherwise mild mtDNA variants.99 Studies on the potential effects of nuclear polymorphism-mitochondrial haplotype interactions are emerging.100
Finally, it may be possible to develop therapies for mitochondrial diseases or diseases that are in part driven by aberrant mitochondrial function or energy production based on targeting the mitochondrial import and transport processes. A number of clinical trials are ongoing or planned for treatment of mitochondrial diseases (discussed in El-Hattab et al.101 and Hirano et al.102); for example, use of elamipretide (MTP 131) for cardiolipin protection from oxidation and stabilization is proposed in cases of mitochondrial myopathy. Pyridoxine is being commonly used for treatment of PH1; in vitro, pyridoxine has been shown to increase AGT levels and enhance peroxisomal localization of some common disease-associated variants;103 conversely, other compounds, such as dequalinium chloride, block AGT import into mitochondria.104 Extensions and variations of such approaches to target mitochondrial function offer future promise for treatment of the rare disorders described here and more common conditions that may, in part, be susceptible to therapies targeting mitochondrial protein trafficking.
Acknowledgments
The authors were supported by NIH R01 DK108195 (to E.A.G.) and by NIH Core Grant CA006927 (to Fox Chase Cancer Center). The authors would like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at https://gnomad.broadinstitute.org/about.
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.019.
Web Resources
1000 Genomes, http://www.internationalgenome.org/1000-genomes-browsers
Baylor-Hopkins Center for Mendelian Genomics, https://mendeliangenomics.org/
dbSNP (build 138), https://www.ncbi.nlm.nih.gov/projects/SNP/
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, https://gnomad.broadinstitute.org/
Human DNA Polymerase Gamma Mutation Database, https://tools.niehs.nih.gov/polg/
International Mouse Phenotyping Consortium, http://www.mousephenotype.org/data/genes/
LitVar, https://www.ncbi.nlm.nih.gov/CBBresearch/Lu/Demo/LitVar/
Mouse Genome Informatics, http://www.informatics.jax.org/
MSeqDR, https://mseqdr.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Pfanner N., Warscheid B., Wiedemann N. Mitochondrial proteins: from biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019 doi: 10.1038/s41580-018-0092-0. Published online January 9, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith R.L., Soeters M.R., Wüst R.C.I., Houtkooper R.H. Metabolic flexibility as an adaptation to energy resources and requirements in health and disease. Endocr. Rev. 2018;39:489–517. doi: 10.1210/er.2017-00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porporato P.E., Filigheddu N., Pedro J.M.B., Kroemer G., Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018;28:265–280. doi: 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerra F., Guaragnella N., Arbini A.A., Bucci C., Giannattasio S., Moro L. Mitochondrial dysfunction: A novel potential driver of epithelial-to-mesenchymal transition in cancer. Front. Oncol. 2017;7:295. doi: 10.3389/fonc.2017.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raimundo N. Mitochondrial pathology: stress signals from the energy factory. Trends Mol. Med. 2014;20:282–292. doi: 10.1016/j.molmed.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Bohovych I., Khalimonchuk O. Sending out an SOS: Mitochondria as a signaling hub. Front. Cell Dev. Biol. 2016;4:109. doi: 10.3389/fcell.2016.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samluk L., Chroscicki P., Chacinska A. Mitochondrial protein import stress and signaling. Curr. Opin. Physiol. 2018;3:41–48. [Google Scholar]
- 8.Jovaisaite V., Mouchiroud L., Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J. Exp. Biol. 2014;217:137–143. doi: 10.1242/jeb.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callegari S., Dennerlein S. Sensing the stress: A role for the UPRmt and UPRam in the quality control of mitochondria. Front. Cell Dev. Biol. 2018;6:31. doi: 10.3389/fcell.2018.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruan L., Zhou C., Jin E., Kucharavy A., Zhang Y., Wen Z., Florens L., Li R. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature. 2017;543:443–446. doi: 10.1038/nature21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekine S., Youle R.J. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol. 2018;16:2. doi: 10.1186/s12915-017-0470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lionaki E., Gkikas I., Tavernarakis N. Differential protein distribution between the nucleus and mitochondria: Implications in aging. Front. Genet. 2016;7:162. doi: 10.3389/fgene.2016.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sepuri N.B.V., Tammineni P., Mohammed F., Paripati A. Nuclear transcription factors in the mitochondria: A new paradigm in fine-tuning mitochondrial metabolism. Handb. Exp. Pharmacol. 2017;240:3–20. doi: 10.1007/164_2016_3. [DOI] [PubMed] [Google Scholar]
- 14.Bourgeron T., Rustin P., Chretien D., Birch-Machin M., Bourgeois M., Viegas-Péquignot E., Munnich A., Rötig A. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet. 1995;11:144–149. doi: 10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Marmiesse A., Gouveia S., Couce M.L. NGS technologies as a turning point in rare disease research, diagnosis and treatment. Curr. Med. Chem. 2018;25:404–432. doi: 10.2174/0929867324666170718101946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenton S.L., Prokisch H. Advancing genomic approaches to the molecular diagnosis of mitochondrial disease. Essays Biochem. 2018;62:399–408. doi: 10.1042/EBC20170110. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y.W., Claypool S.M. Disorders of phospholipid metabolism: an emerging class of mitochondrial disease due to defects in nuclear genes. Front. Genet. 2015;6:3. doi: 10.3389/fgene.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Hattab A.W., Suleiman J., Almannai M., Scaglia F. Mitochondrial dynamics: Biological roles, molecular machinery, and related diseases. Mol. Genet. Metab. 2018;125:315–321. doi: 10.1016/j.ymgme.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Torraco A., Stehling O., Stumpfig C., Rosser R., De Rasmo D., Fiermonte G., Verrigni D., Rizza T., Vozza A., Di Nottia M. ISCA1 mutation in a patient with infantile-onset leukodystrophy causes defects in mitochondrial [4Fe-4S] proteins. Hum. Mol. Genet. 2018;27:2739–2754. doi: 10.1093/hmg/ddy183. [DOI] [PubMed] [Google Scholar]
- 20.Lightowlers R.N., Taylor R.W., Turnbull D.M. Mutations causing mitochondrial disease: What is new and what challenges remain? Science. 2015;349:1494–1499. doi: 10.1126/science.aac7516. [DOI] [PubMed] [Google Scholar]
- 21.Fakruddin M., Wei F.Y., Suzuki T., Asano K., Kaieda T., Omori A., Izumi R., Fujimura A., Kaitsuka T., Miyata K. Defective mitochondrial tRNA taurine modification activates global proteostress and leads to mitochondrial disease. Cell Rep. 2018;22:482–496. doi: 10.1016/j.celrep.2017.12.051. [DOI] [PubMed] [Google Scholar]
- 22.Miernyk J.A. Getting into mitochondria. Biochem. J. 2016;473:3755–3758. doi: 10.1042/BCJ20160667C. [DOI] [PubMed] [Google Scholar]
- 23.Backes S., Hess S., Boos F., Woellhaf M.W., Gödel S., Jung M., Mühlhaus T., Herrmann J.M. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J. Cell Biol. 2018;217:1369–1382. doi: 10.1083/jcb.201708044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrick J.P., Hodges P.E., Rosenberg L.E. Survey of amino-terminal proteolytic cleavage sites in mitochondrial precursor proteins: leader peptides cleaved by two matrix proteases share a three-amino acid motif. Proc. Natl. Acad. Sci. USA. 1989;86:4056–4060. doi: 10.1073/pnas.86.11.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvo S.E., Julien O., Clauser K.R., Shen H., Kamer K.J., Wells J.A., Mootha V.K. Comparative analysis of mitochondrial N-termini from mouse, human, and yeast. Mol. Cell. Proteomics. 2017;16:512–523. doi: 10.1074/mcp.M116.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartmann B., Wai T., Hu H., MacVicar T., Musante L., Fischer-Zirnsak B., Stenzel W., Gräf R., van den Heuvel L., Ropers H.H. Homozygous YME1L1 mutation causes mitochondriopathy with optic atrophy and mitochondrial network fragmentation. eLife. 2016;5:5. doi: 10.7554/eLife.16078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun S., Habermann B.H. A guide to computational methods for predicting mitochondrial localization. Methods Mol. Biol. 2017;1567:1–14. doi: 10.1007/978-1-4939-6824-4_1. [DOI] [PubMed] [Google Scholar]
- 28.Quirós P.M., Langer T., López-Otín C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- 29.Dougan D.A., Varshavsky A. Understanding the Pro/N-end rule pathway. Nat. Chem. Biol. 2018;14:415–416. doi: 10.1038/s41589-018-0045-0. [DOI] [PubMed] [Google Scholar]
- 30.Lebeau J., Rainbolt T.K., Wiseman R.L. Coordinating mitochondrial biology through the stress-responsive regulation of mitochondrial proteases. Int. Rev. Cell Mol. Biol. 2018;340:79–128. doi: 10.1016/bs.ircmb.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shahrour M.A., Staretz-Chacham O., Dayan D., Stephen J., Weech A., Damseh N., Pri Chen H., Edvardson S., Mazaheri S., Saada A., NISC Intramural Sequencing Mitochondrial epileptic encephalopathy, 3-methylglutaconic aciduria and variable complex V deficiency associated with TIMM50 mutations. Clin. Genet. 2017;91:690–696. doi: 10.1111/cge.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reyes A., Melchionda L., Burlina A., Robinson A.J., Ghezzi D., Zeviani M. Mutations in TIMM50 compromise cell survival in OxPhos-dependent metabolic conditions. EMBO Mol. Med. 2018;10:e8698. doi: 10.15252/emmm.201708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pacheu-Grau D., Callegari S., Emperador S., Thompson K., Aich A., Topol S.E., Spencer E.G., McFarland R., Ruiz-Pesini E., Torkamani A. Mutations of the mitochondrial carrier translocase channel subunit TIM22 cause early-onset mitochondrial myopathy. Hum. Mol. Genet. 2018;27:4135–4144. doi: 10.1093/hmg/ddy305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson K., Mai N., Oláhová M., Scialó F., Formosa L.E., Stroud D.A., Garrett M., Lax N.Z., Robertson F.M., Jou C. OXA1L mutations cause mitochondrial encephalopathy and a combined oxidative phosphorylation defect. EMBO Mol. Med. 2018;10:e9060. doi: 10.15252/emmm.201809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang Y., Stroud D.A., Baker M.J., De Souza D.P., Frazier A.E., Liem M., Tull D., Mathivanan S., McConville M.J., Thorburn D.R. Sengers syndrome-associated mitochondrial acylglycerol kinase is a subunit of the human TIM22 protein import complex. Mol. Cell. 2017;67:457–470.e5. doi: 10.1016/j.molcel.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Vukotic M., Nolte H., König T., Saita S., Ananjew M., Krüger M., Tatsuta T., Langer T. Acylglycerol kinase mutated in Sengers syndrome is a subunit of the TIM22 protein translocase in mitochondria. Mol. Cell. 2017;67:471–483.e7. doi: 10.1016/j.molcel.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 37.Calvo S.E., Compton A.G., Hershman S.G., Lim S.C., Lieber D.S., Tucker E.J., Laskowski A., Garone C., Liu S., Jaffe D.B. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci. Transl. Med. 2012;4:118ra10. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jordens E.Z., Palmieri L., Huizing M., van den Heuvel L.P., Sengers R.C., Dörner A., Ruitenbeek W., Trijbels F.J., Valsson J., Sigfusson G. Adenine nucleotide translocator 1 deficiency associated with Sengers syndrome. Ann. Neurol. 2002;52:95–99. doi: 10.1002/ana.10214. [DOI] [PubMed] [Google Scholar]
- 39.Wortmann S.B., Duran M., Anikster Y., Barth P.G., Sperl W., Zschocke J., Morava E., Wevers R.A. Inborn errors of metabolism with 3-methylglutaconic aciduria as discriminative feature: proper classification and nomenclature. J. Inherit. Metab. Dis. 2013;36:923–928. doi: 10.1007/s10545-012-9580-0. [DOI] [PubMed] [Google Scholar]
- 40.Ikon N., Ryan R.O. On the origin of 3-methylglutaconic acid in disorders of mitochondrial energy metabolism. J. Inherit. Metab. Dis. 2016;39:749–756. doi: 10.1007/s10545-016-9933-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikon N., Ryan R.O. Barth syndrome: Connecting cardiolipin to cardiomyopathy. Lipids. 2017;52:99–108. doi: 10.1007/s11745-016-4229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehmer C., Schludi M.H., Ransom L., Greiling J., Junghänel M., Exner N., Riemenschneider H., van der Zee J., Van Broeckhoven C., Weydt P. A novel CHCHD10 mutation implicates a Mia40-dependent mitochondrial import deficit in ALS. EMBO Mol. Med. 2018;10:10. doi: 10.15252/emmm.201708558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kowalski L., Bragoszewski P., Khmelinskii A., Glow E., Knop M., Chacinska A. Determinants of the cytosolic turnover of mitochondrial intermembrane space proteins. BMC Biol. 2018;16:66. doi: 10.1186/s12915-018-0536-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zöller E., Todd Alexander R., Herrmann J.M. Proteasomal degradation competes with Mia40-mediated import into mitochondria. BMC Biol. 2018;16:63. doi: 10.1186/s12915-018-0537-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Fonzo A., Ronchi D., Lodi T., Fassone E., Tigano M., Lamperti C., Corti S., Bordoni A., Fortunato F., Nizzardo M. The mitochondrial disulfide relay system protein GFER is mutated in autosomal-recessive myopathy with cataract and combined respiratory-chain deficiency. Am. J. Hum. Genet. 2009;84:594–604. doi: 10.1016/j.ajhg.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ceh-Pavia E., Ang S.K., Spiller M.P., Lu H. The disease-associated mutation of the mitochondrial thiol oxidase Erv1 impairs cofactor binding during its catalytic reaction. Biochem. J. 2014;464:449–459. doi: 10.1042/BJ20140679. [DOI] [PubMed] [Google Scholar]
- 47.Friederich M.W., Erdogan A.J., Coughlin C.R., 2nd, Elos M.T., Jiang H., O’Rourke C.P., Lovell M.A., Wartchow E., Gowan K., Chatfield K.C. Mutations in the accessory subunit NDUFB10 result in isolated complex I deficiency and illustrate the critical role of intermembrane space import for complex I holoenzyme assembly. Hum. Mol. Genet. 2017;26:702–716. doi: 10.1093/hmg/ddw431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pacheu-Grau D., Rucktäschel R., Deckers M. Mitochondrial dysfunction and its role in tissue-specific cellular stress. Cell Stress. 2018;2:184–199. doi: 10.15698/cst2018.07.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Habich M., Salscheider S.L., Riemer J. Cysteine residues in mitochondrial intermembrane space proteins: more than just import. Br. J. Pharmacol. 2019;176:514–531. doi: 10.1111/bph.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel K.P., O’Brien T.W., Subramony S.H., Shuster J., Stacpoole P.W. The spectrum of pyruvate dehydrogenase complex deficiency: clinical, biochemical and genetic features in 371 patients. Mol. Genet. Metab. 2012;105:34–43. doi: 10.1016/j.ymgme.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takakubo F., Cartwright P., Hoogenraad N., Thorburn D.R., Collins F., Lithgow T., Dahl H.H. An amino acid substitution in the pyruvate dehydrogenase E1 alpha gene, affecting mitochondrial import of the precursor protein. Am. J. Hum. Genet. 1995;57:772–780. [PMC free article] [PubMed] [Google Scholar]
- 52.Wightman P.J., Santer R., Ribes A., Dougherty F., McGill N., Thorburn D.R., FitzPatrick D.R. MLYCD mutation analysis: evidence for protein mistargeting as a cause of MLYCD deficiency. Hum. Mutat. 2003;22:288–300. doi: 10.1002/humu.10264. [DOI] [PubMed] [Google Scholar]
- 53.Kobayashi T., Ogawa H., Kasahara M., Shiozawa Z., Matsuzawa T. A single amino acid substitution within the mature sequence of ornithine aminotransferase obstructs mitochondrial entry of the precursor. Am. J. Hum. Genet. 1995;57:284–291. [PMC free article] [PubMed] [Google Scholar]
- 54.Glynn S.A., Boersma B.J., Howe T.M., Edvardsen H., Geisler S.B., Goodman J.E., Ridnour L.A., Lønning P.E., Børresen-Dale A.L., Naume B. A mitochondrial target sequence polymorphism in manganese superoxide dismutase predicts inferior survival in breast cancer patients treated with cyclophosphamide. Clin. Cancer Res. 2009;15:4165–4173. doi: 10.1158/1078-0432.CCR-09-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bresciani G., Cruz I.B., de Paz J.A., Cuevas M.J., González-Gallego J. The MnSOD Ala16Val SNP: relevance to human diseases and interaction with environmental factors. Free Radic. Res. 2013;47:781–792. doi: 10.3109/10715762.2013.836275. [DOI] [PubMed] [Google Scholar]
- 56.Azevedo L., Mort M., Costa A.C., Silva R.M., Quelhas D., Amorim A., Cooper D.N. Improving the in silico assessment of pathogenicity for compensated variants. Eur. J. Hum. Genet. 2016;25:2–7. doi: 10.1038/ejhg.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vögtle F.N., Brändl B., Larson A., Pendziwiat M., Friederich M.W., White S.M., Basinger A., Kücükköse C., Muhle H., Jähn J.A. Mutations in PMPCB encoding the catalytic subunit of the mitochondrial presequence protease cause neurodegeneration in early childhood. Am. J. Hum. Genet. 2018;102:557–573. doi: 10.1016/j.ajhg.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burté F., Carelli V., Chinnery P.F., Yu-Wai-Man P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 2015;11:11–24. doi: 10.1038/nrneurol.2014.228. [DOI] [PubMed] [Google Scholar]
- 59.Jobling R.K., Assoum M., Gakh O., Blaser S., Raiman J.A., Mignot C., Roze E., Dürr A., Brice A., Lévy N. PMPCA mutations cause abnormal mitochondrial protein processing in patients with non-progressive cerebellar ataxia. Brain. 2015;138:1505–1517. doi: 10.1093/brain/awv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choquet K., Zurita-Rendón O., La Piana R., Yang S., Dicaire M.J., Boycott K.M., Majewski J., Shoubridge E.A., Brais B., Tétreault M., Care4Rare Consortium Autosomal recessive cerebellar ataxia caused by a homozygous mutation in PMPCA. Brain. 2016;139:e19. doi: 10.1093/brain/awv362. [DOI] [PubMed] [Google Scholar]
- 61.Osumi T., Tsukamoto T., Hata S. Signal peptide for peroxisomal targeting: replacement of an essential histidine residue by certain amino acids converts the amino-terminal presequence of peroxisomal 3-ketoacyl-CoA thiolase to a mitochondrial signal peptide. Biochem. Biophys. Res. Commun. 1992;186:811–818. doi: 10.1016/0006-291x(92)90818-6. [DOI] [PubMed] [Google Scholar]
- 62.Montioli R., Fargue S., Lewin J., Zamparelli C., Danpure C.J., Borri Voltattorni C., Cellini B. The N-terminal extension is essential for the formation of the active dimeric structure of liver peroxisomal alanine:glyoxylate aminotransferase. Int. J. Biochem. Cell Biol. 2012;44:536–546. doi: 10.1016/j.biocel.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 63.Fargue S., Lewin J., Rumsby G., Danpure C.J. Four of the most common mutations in primary hyperoxaluria type 1 unmask the cryptic mitochondrial targeting sequence of alanine:glyoxylate aminotransferase encoded by the polymorphic minor allele. J. Biol. Chem. 2013;288:2475–2484. doi: 10.1074/jbc.M112.432617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Belostotsky R., Lyakhovetsky R., Sherman M.Y., Shkedy F., Tzvi-Behr S., Bar R., Hoppe B., Reusch B., Beck B.B., Frishberg Y. Translation inhibition corrects aberrant localization of mutant alanine-glyoxylate aminotransferase: possible therapeutic approach for hyperoxaluria. J. Mol. Med. (Berl.) 2018;96:621–630. doi: 10.1007/s00109-018-1651-8. [DOI] [PubMed] [Google Scholar]
- 65.Klootwijk E.D., Reichold M., Helip-Wooley A., Tolaymat A., Broeker C., Robinette S.L., Reinders J., Peindl D., Renner K., Eberhart K. Mistargeting of peroxisomal EHHADH and inherited renal Fanconi’s syndrome. N. Engl. J. Med. 2014;370:129–138. doi: 10.1056/NEJMoa1307581. [DOI] [PubMed] [Google Scholar]
- 66.Assmann N., Dettmer K., Simbuerger J.M.B., Broeker C., Nuernberger N., Renner K., Courtneidge H., Klootwijk E.D., Duerkop A., Hall A. Renal Fanconi syndrome is caused by a mistargeting-based mitochondriopathy. Cell Rep. 2016;15:1423–1429. doi: 10.1016/j.celrep.2016.04.037. [DOI] [PubMed] [Google Scholar]
- 67.Wohlever M.L., Mateja A., McGilvray P.T., Day K.J., Keenan R.J. Msp1 is a membrane protein dislocase for tail-anchored proteins. Mol. Cell. 2017;67:194–202.e6. doi: 10.1016/j.molcel.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y.C., Umanah G.K., Dephoure N., Andrabi S.A., Gygi S.P., Dawson T.M., Dawson V.L., Rutter J. Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 2014;33:1548–1564. doi: 10.15252/embj.201487943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahrens-Nicklas R.C., Umanah G.K., Sondheimer N., Deardorff M.A., Wilkens A.B., Conlin L.K., Santani A.B., Nesbitt A., Juulsola J., Ma E. Precision therapy for a new disorder of AMPA receptor recycling due to mutations in ATAD1. Neurol Genet. 2017;3:e130. doi: 10.1212/NXG.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Piard J., Umanah G.K.E., Harms F.L., Abalde-Atristain L., Amram D., Chang M., Chen R., Alawi M., Salpietro V., Rees M.I. A homozygous ATAD1 mutation impairs postsynaptic AMPA receptor trafficking and causes a lethal encephalopathy. Brain. 2018;141:651–661. doi: 10.1093/brain/awx377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weidberg H., Amon A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science. 2018;360:360. doi: 10.1126/science.aan4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Desai R., Campanella M. MitoCPR: Meticulous monitoring of mitochondrial proteostasis. Mol. Cell. 2018;71:8–9. doi: 10.1016/j.molcel.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 73.Fu K., Wang Y., Guo D., Wang G., Ren H. Familial Parkinson’s disease-associated L166P mutant DJ-1 is cleaved by mitochondrial serine protease Omi/HtrA2. Neurosci. Bull. 2017;33:685–694. doi: 10.1007/s12264-017-0196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Demetrius L.A., Simon D.K. An inverse-Warburg effect and the origin of Alzheimer’s disease. Biogerontology. 2012;13:583–594. doi: 10.1007/s10522-012-9403-6. [DOI] [PubMed] [Google Scholar]
- 75.Driver J.A. Inverse association between cancer and neurodegenerative disease: review of the epidemiologic and biological evidence. Biogerontology. 2014;15:547–557. doi: 10.1007/s10522-014-9523-2. [DOI] [PubMed] [Google Scholar]
- 76.Sokol A.M., Sztolsztener M.E., Wasilewski M., Heinz E., Chacinska A. Mitochondrial protein translocases for survival and wellbeing. FEBS Lett. 2014;588:2484–2495. doi: 10.1016/j.febslet.2014.05.028. [DOI] [PubMed] [Google Scholar]
- 77.Czarnecka A.M., Campanella C., Zummo G., Cappello F. Mitochondrial chaperones in cancer: from molecular biology to clinical diagnostics. Cancer Biol. Ther. 2006;5:714–720. doi: 10.4161/cbt.5.7.2975. [DOI] [PubMed] [Google Scholar]
- 78.Zemanovic S., Ivanov M.V., Ivanova L.V., Bhatnagar A., Michalkiewicz T., Teng R.J., Kumar S., Rathore R., Pritchard K.A., Jr., Konduri G.G., Afolayan A.J. Dynamic phosphorylation of the C terminus of Hsp70 regulates the mitochondrial import of SOD2 and redox balance. Cell Rep. 2018;25:2605–2616.e7. doi: 10.1016/j.celrep.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arena G., Cissé M.Y., Pyrdziak S., Chatre L., Riscal R., Fuentes M., Arnold J.J., Kastner M., Gayte L., Bertrand-Gaday C. Mitochondrial MDM2 regulates respiratory complex I activity independently of p53. Mol. Cell. 2018;69:594–609.e8. doi: 10.1016/j.molcel.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quirós P.M., Español Y., Acín-Pérez R., Rodríguez F., Bárcena C., Watanabe K., Calvo E., Loureiro M., Fernández-García M.S., Fueyo A. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014;8:542–556. doi: 10.1016/j.celrep.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 81.Zhu Y., Dean A.E., Horikoshi N., Heer C., Spitz D.R., Gius D. Emerging evidence for targeting mitochondrial metabolic dysfunction in cancer therapy. J. Clin. Invest. 2018;128:3682–3691. doi: 10.1172/JCI120844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Filipuzzi I., Steffen J., Germain M., Goepfert L., Conti M.A., Potting C., Cerino R., Pfeifer M., Krastel P., Hoepfner D. Stendomycin selectively inhibits TIM23-dependent mitochondrial protein import. Nat. Chem. Biol. 2017;13:1239–1244. doi: 10.1038/nchembio.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang C., Liu Z., Bunker E., Ramirez A., Lee S., Peng Y., Tan A.C., Eckhardt S.G., Chapnick D.A., Liu X. Sorafenib targets the mitochondrial electron transport chain complexes and ATP synthase to activate the PINK1-Parkin pathway and modulate cellular drug response. J. Biol. Chem. 2017;292:15105–15120. doi: 10.1074/jbc.M117.783175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jeyaraju D.V., Voisin V., Xu C., Hurren R., Gronda M., Wang X., Jitkova Y., Soriano J., MacLean N., Bader G.D. Inhibition of the mitochondrial protein import machinery is selectively cytotoxic to acute myeloid leukemia (AML) cells and stem cells. Blood. 2016;128 1572–1572. [Google Scholar]
- 85.Fenton W.A. Mitochondrial protein transport--a system in search of mutations. Am. J. Hum. Genet. 1995;57:235–238. [PMC free article] [PubMed] [Google Scholar]
- 86.DiMauro S. Mitochondrial diseases. Biochim. Biophys. Acta. 2004;1658:80–88. doi: 10.1016/j.bbabio.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 87.Ajroud-Driss S., Fecto F., Ajroud K., Lalani I., Calvo S.E., Mootha V.K., Deng H.X., Siddique N., Tahmoush A.J., Heiman-Patterson T.D., Siddique T. Mutation in the novel nuclear-encoded mitochondrial protein CHCHD10 in a family with autosomal dominant mitochondrial myopathy. Neurogenetics. 2015;16:1–9. doi: 10.1007/s10048-014-0421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Altmann K., Westermann B. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:5410–5417. doi: 10.1091/mbc.E05-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lasserre J.P., Dautant A., Aiyar R.S., Kucharczyk R., Glatigny A., Tribouillard-Tanvier D., Rytka J., Blondel M., Skoczen N., Reynier P. Yeast as a system for modeling mitochondrial disease mechanisms and discovering therapies. Dis. Model. Mech. 2015;8:509–526. doi: 10.1242/dmm.020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bartha I., di Iulio J., Venter J.C., Telenti A. Human gene essentiality. Nat. Rev. Genet. 2018;19:51–62. doi: 10.1038/nrg.2017.75. [DOI] [PubMed] [Google Scholar]
- 91.Mersha T.B., Abebe T. Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities. Hum. Genomics. 2015;9:1. doi: 10.1186/s40246-014-0023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hindorff L.A., Bonham V.L., Brody L.C., Ginoza M.E.C., Hutter C.M., Manolio T.A., Green E.D. Prioritizing diversity in human genomics research. Nat. Rev. Genet. 2018;19:175–185. doi: 10.1038/nrg.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kohda D. “Multiple partial recognitions in dynamic equilibrium” in the binding sites of proteins form the molecular basis of promiscuous recognition of structurally diverse ligands. Biophys. Rev. 2018;10:421–433. doi: 10.1007/s12551-017-0365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taipale M. Disruption of protein function by pathogenic mutations: common and uncommon mechanisms. Biochem. Cell Biol. 2018;97:46–57. doi: 10.1139/bcb-2018-0007. [DOI] [PubMed] [Google Scholar]
- 95.González-Sánchez J.C., Raimondi F., Russell R.B. Cancer genetics meets biomolecular mechanism-bridging an age-old gulf. FEBS Lett. 2018;592:463–474. doi: 10.1002/1873-3468.12988. [DOI] [PubMed] [Google Scholar]
- 96.Eyre-Walker A. Mitochondrial replacement therapy: Are mito-nuclear interactions likely to be a problem? Genetics. 2017;205:1365–1372. doi: 10.1534/genetics.116.196436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen C., Chen Y., Guan M.X. A peep into mitochondrial disorder: multifaceted from mitochondrial DNA mutations to nuclear gene modulation. Protein Cell. 2015;6:862–870. doi: 10.1007/s13238-015-0175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiang P., Jin X., Peng Y., Wang M., Liu H., Liu X., Zhang Z., Ji Y., Zhang J., Liang M. The exome sequencing identified the mutation in YARS2 encoding the mitochondrial tyrosyl-tRNA synthetase as a nuclear modifier for the phenotypic manifestation of Leber’s hereditary optic neuropathy-associated mitochondrial DNA mutation. Hum. Mol. Genet. 2016;25:584–596. doi: 10.1093/hmg/ddv498. [DOI] [PubMed] [Google Scholar]
- 99.McManus M.J., Picard M., Chen H.W., De Haas H.J., Potluri P., Leipzig J., Towheed A., Angelin A., Sengupta P., Morrow R.M. Mitochondrial DNA variation dictates expressivity and progression of nuclear DNA mutations causing cardiomyopathy. Cell Metab. 2018;29:78–90. doi: 10.1016/j.cmet.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Smieszek S., Jia P., Samuels D.C., Zhao Z., Barnholtz-Sloan J., Kaur H., Letendre S., Ellis R., Franklin D.R., Hulgan T., CHARTER Study Group Nuclear-mitochondrial interactions influence susceptibility to HIV-associated neurocognitive impairment. Mitochondrion. Published July 17, 2018. 2018 doi: 10.1016/j.mito.2018.07.004. S1567-7249(17)30356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.El-Hattab A.W., Zarante A.M., Almannai M., Scaglia F. Therapies for mitochondrial diseases and current clinical trials. Mol. Genet. Metab. 2017;122:1–9. doi: 10.1016/j.ymgme.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hirano M., Emmanuele V., Quinzii C.M. Emerging therapies for mitochondrial diseases. Essays Biochem. 2018;62:467–481. doi: 10.1042/EBC20170114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cellini B., Montioli R., Oppici E., Astegno A., Voltattorni C.B. The chaperone role of the pyridoxal 5′-phosphate and its implications for rare diseases involving B6-dependent enzymes. Clin. Biochem. 2014;47:158–165. doi: 10.1016/j.clinbiochem.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 104.Miyata N., Steffen J., Johnson M.E., Fargue S., Danpure C.J., Koehler C.M. Pharmacologic rescue of an enzyme-trafficking defect in primary hyperoxaluria 1. Proc. Natl. Acad. Sci. USA. 2014;111:14406–14411. doi: 10.1073/pnas.1408401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Eldomery M.K., Akdemir Z.C., Vögtle F.N., Charng W.L., Mulica P., Rosenfeld J.A., Gambin T., Gu S., Burrage L.C., Al Shamsi A. MIPEP recessive variants cause a syndrome of left ventricular non-compaction, hypotonia, and infantile death. Genome Med. 2016;8:106. doi: 10.1186/s13073-016-0360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.O’Toole J.F., Liu Y., Davis E.E., Westlake C.J., Attanasio M., Otto E.A., Seelow D., Nurnberg G., Becker C., Nuutinen M. Individuals with mutations in XPNPEP3, which encodes a mitochondrial protein, develop a nephronophthisis-like nephropathy. J. Clin. Invest. 2010;120:791–802. doi: 10.1172/JCI40076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brunetti D., Torsvik J., Dallabona C., Teixeira P., Sztromwasser P., Fernandez-Vizarra E., Cerutti R., Reyes A., Preziuso C., D’Amati G. Defective PITRM1 mitochondrial peptidase is associated with Aβ amyloidotic neurodegeneration. EMBO Mol. Med. 2016;8:176–190. doi: 10.15252/emmm.201505894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mandel H., Saita S., Edvardson S., Jalas C., Shaag A., Goldsher D., Vlodavsky E., Langer T., Elpeleg O. Deficiency of HTRA2/Omi is associated with infantile neurodegeneration and 3-methylglutaconic aciduria. J. Med. Genet. 2016;53:690–696. doi: 10.1136/jmedgenet-2016-103922. [DOI] [PubMed] [Google Scholar]
- 109.Dikoglu E., Alfaiz A., Gorna M., Bertola D., Chae J.H., Cho T.J., Derbent M., Alanay Y., Guran T., Kim O.H. Mutations in LONP1, a mitochondrial matrix protease, cause CODAS syndrome. Am. J. Med. Genet. A. 2015;167:1501–1509. doi: 10.1002/ajmg.a.37029. [DOI] [PubMed] [Google Scholar]
- 110.Jenkinson E.M., Rehman A.U., Walsh T., Clayton-Smith J., Lee K., Morell R.J., Drummond M.C., Khan S.N., Naeem M.A., Rauf B., University of Washington Center for Mendelian Genomics Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am. J. Hum. Genet. 2013;92:605–613. doi: 10.1016/j.ajhg.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.