Abstract
Molecular pathological epidemiology (MPE) is an integrative transdisciplinary field that addresses heterogeneous effects of exogenous and endogenous factors (collectively termed “exposures”), including microorganisms, on disease occurrence and consequence utilising molecular pathological signatures of the disease. In parallel with the paradigm of precision medicine, findings from MPE research can provide aetiological insights into tailored strategies of disease prevention and treatment. Due to the availability of molecular pathological tests on tumours, the MPE approach has been utilised predominantly in research on cancers including breast, lung, prostate, and colorectal carcinomas. Mounting evidence indicates that the microbiome (inclusive of viruses, bacteria, fungi, and parasites) plays an important role in a variety of human diseases including neoplasms. An alteration of the microbiome may be not only a cause of neoplasia but also an informative biomarker that indicates or mediates the association of an epidemiological exposure with health conditions and outcomes. To adequately educate and train investigators in this emerging area, we herein propose the integration of microbiology into the MPE model (termed “microbiology-MPE”), which can improve our understanding of the complex interactions of environment, tumour cells, the immune system, and microbes in the tumour microenvironment during the carcinogenic process. Using this approach, we can examine how lifestyle factors, dietary patterns, medications, environmental exposures, and germline genetics influence cancer development and progression through impacting the microbial communities in the human body. Further integration of other disciplines (e.g. pharmacology, immunology, nutrition) into microbiology-MPE would expand this developing research frontier. With the advent of high-throughput next-generation sequencing technologies, researchers now have increasing access to large-scale metagenomics as well as other omics data (e.g. genomics, epigenomics, proteomics, and metabolomics) in population-based research. The integrative field of microbiology-MPE will open new opportunities for personalised medicine and public health.
Keywords: biobank, bioinformatics, causal inference, cohort study, immunity, inflammation, microbiota, population health science, statistics, translational research
The role of the microbiome in carcinogenesis
Microbiology characterises small organisms such as viruses, bacteria, fungi, archaea, and protozoa, and links them to the pathogenesis of human diseases. The human microbiome represents an interactive ecosystem consisting of numerous microorganisms, which continuously interacts with the environment and host, especially the immune system [1–4]. Accumulating evidence points to the role of endogenous and exogenous microorganisms in the pathogenesis of various neoplasms [5–8] as well as non-neoplastic diseases [4,9–13]. Microorganisms with established or possible carcinogenic effects include Helicobacter pylori and Epstein-Barr virus for gastric carcinoma [14]; hepatitis B and C viruses (HBV and HCV) for hepatocellular carcinoma (HCC) [15,16]; human herpesvirus-8 for Kaposi’s sarcoma [17]; human immunodeficiency virus for Kaposi’s sarcoma, aggressive B-cell non-Hodgkin lymphoma, and cervical carcinoma [17]; HPV for uterine cervical, anal, and oropharyngeal carcinomas [18,19]; human T-lymphotropic virus type 1 for adult T-cell leukaemia / lymphoma [20]; and Fusobacterium nucleatum (F. nucleatum) for colorectal carcinoma [21–23]. Furthermore, tumours arising among carriers of these pathogenic microorganisms may have distinct molecular pathological features compared to tumours arising among non-carriers [14,16,19,24–26]. Accumulating evidence indicates that alterations of the microbial ecosystem also play a major role in the pathogenesis of various neoplasms. Studies have associated altered microbial communities not only with tumours arising in the affected organs, but also with tumours arising in distant organs (e.g. the colorectal microbiome and non-colorectal gastrointestinal cancers [5,27–31]). In the tumour microenvironment, there is a dynamic interactive network that includes neoplastic cells, microorganisms, and immune cells, all of which are affected by the genetic architecture and epidemiological factors including aging, diet, nutrition, smoking, alcohol, adiposity, diabetes mellitus, physical exercise, and medication (Figure 1) [1–3,7,32–36]. Therefore, an integrative approach is required to elucidate the role of microbial communities in the pathogenic process of human cancers.
Figure 1.
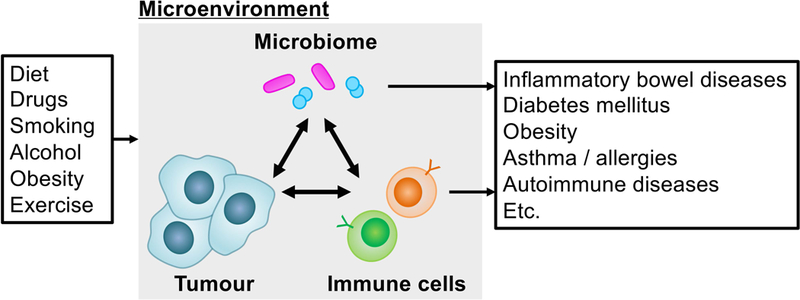
Interactions of the microbiome, host immune system, and tumour in the microenvironment. The imbalance of microbial communities may cause neoplasms as well as non-neoplastic diseases.
Molecular pathological epidemiology (MPE) in the era of precision medicine
Epidemiology provides conceptual and analytical frameworks for investigations of an association of an endogenous or exogenous factor (termed “exposure”) with incidence of a disease or its consequence. Epidemiology aims to identify determinants of diseases and ultimately contribute to human health. Conventional epidemiological research has utilised organ- and/or function-based disease classifications [e.g. the International Statistical Classification of Diseases and Related Health Problems (ICD)] and addressed the relationship of an exposure with a specific disease entity or a collective group of similar disease entities. Owing to the availability of molecular diagnostic tests [37], molecular pathology has become a major subfield of pathology. Molecular pathological epidemiology (MPE) has been proposed as an integrative research field, which utilises molecular pathological signatures for disease sub-classification and addresses inter-individual differences in disease phenotypes in relation to specific epidemiological factors in human populations [35,38,39]. MPE studies examine occurrence of disease subtype or its consequence as an outcome, and assess a difference between subtypes of a single disease entity (Figure 2) [40]. “The unique disease (tumour) principle [41–43]” has paved the way for the MPE paradigm by highlighting the uniqueness of disease pathogenesis within each individual. Of note, MPE analyses can provide evidence for links between aetiological factors and pathogenic signatures, which can augment causal inference [39,44]. MPE analyses compute risk estimates for specific disease subtypes, thereby contributing to the advancement of personalised management strategies [45]. This echoes the aim of precision medicine for developing personalised prevention and treatment strategies to optimise the risk-benefit balance [46–50]. Due to the wide availability of established molecular pathology diagnostics and tumour tissue specimens for research, the MPE approach has been most commonly utilised in epidemiological studies of breast, colorectal, lung, and prostate cancers; however, this MPE model can be readily applied to research on any neoplastic and non-neoplastic diseases that represent substantial interpersonal heterogeneity [51,52]. The emerging MPE framework has been recognised and discussed in international conferences [53–56] and publications [6,7,57–98].
Figure 2.
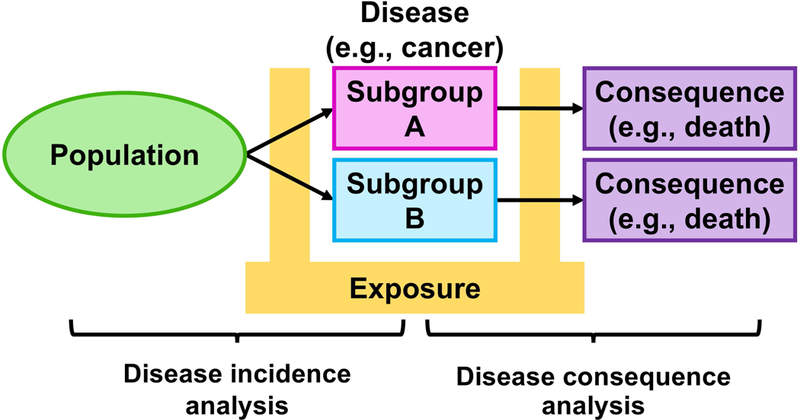
Disease incidence and consequence analyses in molecular pathological epidemiology (MPE) research. Considering different characteristics of disease subtypes classified by molecular pathological signatures, incidence and consequence analyses assess the heterogeneity in associations of an exposure of interest with disease incidence and consequence, respectively. For a simple illustration, a disease with two subtypes is exemplified, but more than two subtypes can be modelled in MPE studies. Arrows indicate the time course of disease subtypes.
Integration of microbiology and MPE
Emerging evidence attests to not only the carcinogenic effects of the altered microbiome but also the distinctive phenotypes of tumours arising in the presence of specific microorganisms [14,16,19,24–26]. Microorganisms, the immune system, and tumour cells interact each other in complex manners. Hence, to better understand cancer aetiologies and their consequences in populations, analyses of the microbiome in various body sites including pathologically-altered tissue (such as tumour) should be adequately integrated into MPE (referred to as “microbiology-MPE” [40]). Microbiology-MPE provides a promising approach to explore the interpersonal heterogeneity of the carcinogenic process in relation to the altered microbial composition and to generate evidence for the role of microorganisms in specific processes of tumour initiation and progression [40]. While mechanistic studies have been a major part of microbiology research on carcinogenesis, any in vitro or in vivo model can never exactly replicate the complex molecular and cellular network in the tumour microenvironment in the human [99]. Therefore, insights from microbiology-MPE research would serve as particularly valuable evidence for the microbial aetiologies and pathogenesis of human neoplasms.
Study design
The epidemiological term “exposure” indicates a factor that may (or may not) cause, prevent, or influence an outcome such as incidence or consequence of a disease of interest. In microbiology-MPE research, the microbial profile is incorporated as an exposure or outcome variable. Namely, the microbial profile in non-neoplastic tissue or biospecimen obtained before cancer diagnosis can be examined as an exposure in relation to disease incidence or consequence as an outcome. The microbial profile in non-neoplastic or pre-malignant biospecimens can also serve as an intermediary outcome variable, particularly under a hypothesis that a certain exposure can cause the disease through altering the microbiome. Notably, the microbial profile in neoplastic tissue can serve as an outcome variable used to subclassify a tumour of interest in incidence analyses, or an exposure variable in consequence analyses. Nonetheless, we should be aware that the detection of a certain microorganism may suggest its causal relationship with the tumour or may merely be a consequence of tumour development.
Designs of microbiology-MPE studies are illustrated in Figures 3 and 4. In disease incidence analyses, the case-control study design has often been adopted because of the limited availability of prospective cohort studies with microbial data. Although there are advantages in the case-control study design including no requirement for follow-up of participants, lower costs compared to prospective cohort studies, and the ability to investigate rare tumours, special considerations are needed for potential biases such as recall and selection biases. In particular, selection bias may substantially affect the generalisability of study findings. In case-control studies examining tumour subtypes classified by the microbial profile, we compare proportions or levels of an exposure (which can be a microbial exposure) between patients with each subtype and controls, and assess a differential association between the exposure and subtypes (Figure 3A). In case-control studies examining the microbial profile as an exposure, we examine differences in the microbial communities between molecularly- or pathologically-classified subtypes and controls, and assess a differential association between the microbial profile and subtypes (Figure 3B). In contrast, the strengths of prospective cohort studies are that the source population that has given rise to tumours is well-defined, which diminishes issues due to selection bias. Recall bias can also be reduced by prospective data collection. When examining tumour subtypes classified by the microbial profile in a prospective cohort design, we can examine the association of an exposure to a specific factor (which can be microbes) and tumour subtypes (Figure 3C). If the microbial data before cancer diagnosis are available, we can link a microbe with the incidence of tumour subtypes classified by molecular pathological signatures of the tumour (Figure 3D).
Figure 3.
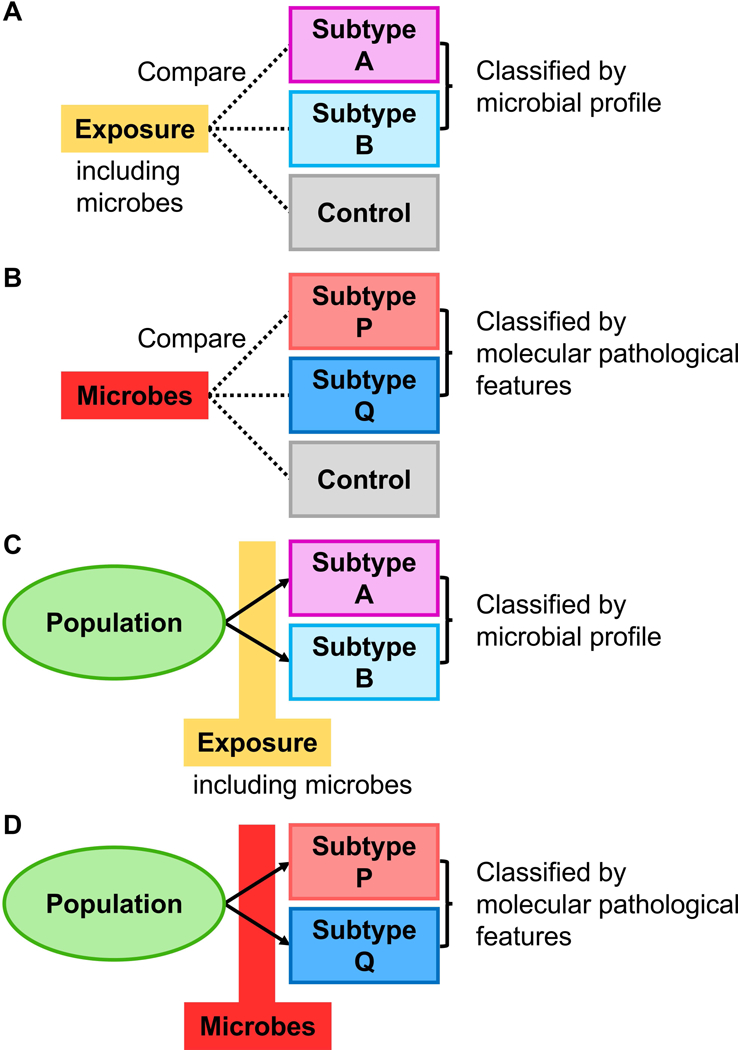
Disease incidence analyses in microbiology-molecular pathological epidemiology (microbiology-MPE) research. Arrows indicate the time course of disease subtypes. A. Case-control study examining a between-group difference in an exposure (which can be microbes) according to the microbial profile of the tumour (as an outcome variable). B. Case-control study examining a between-group difference in the microbial profile (as an exposure variable) according to molecular pathological signatures of the tumour. C. Prospective study examining the association of an exposure (which can be microbes) with incidence of tumours characterised by the microbial profile (as an outcome variable). D. Prospective study examining the association of the microbial profile (as an exposure variable) with incidence of tumours characterised by molecular pathological signatures of the tumour.
Figure 4.
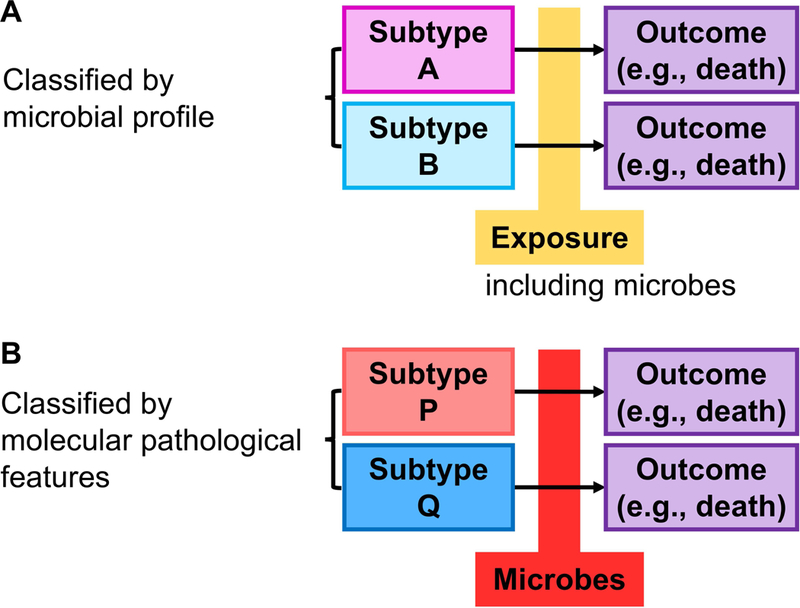
Disease consequence analyses in microbiology-molecular pathological epidemiology (microbiology-MPE) research. Arrows indicate the time course of disease subtypes. A. Study examining the association of an exposure (which can be microbes) with disease consequence according to the microbial profile. B. Study examining the association of the microbial profile with disease consequence according to molecular pathological signatures of the tumour.
In disease consequence analyses, the microbial data can be analysed as an exposure and/or be used to sub-classify a tumour if a microbe of interest is a constituent of the tumour. Effects of epidemiological factors including microbial exposures on tumour progression may differ by the presence of other microbes that are associated with altered anti-tumour immunity and/or specific tumour molecular pathological features (Figure 4A). Microbes may exert promoting or suppressing effects on tumour growth through modulating specific signalling pathways; therefore, the association of specific microbes with tumour behaviour after clinical diagnosis may differ by molecular pathological signatures of the tumour (Figure 4B). As an experimental design, interventional trials using microbial manipulation strategies such as antibiotics, probiotics, prebiotics, and synbiotics targeting a specific tumour subtype can be conceived [100] though there exist concerns including emergence of antibiotic resistance.
In microbiology-MPE studies, specimen types for acquisition of microbial data should be discussed. In addition to tissue specimens, we can utilise various biospecimens including stool. There is an advantage of microbial analyses utilising tissue specimens. Compared to any microbe in stool, a microbe detected in tumour tissue is more likely to have a causal association with tumour development. Fresh frozen tissue specimens are amenable to various laboratory assays and metagenomic analyses for high resolution mapping of tissue microorganisms; however, their collection is not part of routine clinical practice. In contrast, formalin-fixed paraffin-embedded (FFPE) tissue blocks are often utilised, especially in population-based studies. Limitations of FFPE tissue specimens include contamination with non-pathogenic microorganisms and alteration of microbial composition due to fixation, processing, and storage of the tissue.
Study examples and proposals
In this section, we discuss microbiology-MPE studies that have examined intratumoural microorganisms in relation to various exposures, and successfully provided novel insights into cancer aetiologies in the human. There have been many epidemiological studies that examined microorganisms as exposures; most of those studies dealt with a disease entity of interest (e.g. organ-specific cancer) as a single outcome variable. Here, we focus on studies that investigated biological heterogeneity of tumour subtypes.
Human papillomavirus (HPV) is a sexually-transmitted DNA virus and has been involved in carcinogenesis of uterine cervical and anal cancers, and head and neck squamous cell carcinomas (particularly high-risk oncogenic types 16 and 18) [18,19]. In addition to distinctive characteristics of HPV-related carcinomas [19,101,102], evidence suggests differential responses to cancer treatment by HPV viral load [102–106]. HPV-positive subtypes may have a reduced ability to repair DNA damage, potentially representing better response to chemoradiotherapy which induces high-degree DNA damage and promotes apoptosis in neoplastic cells [102,106–108]. Intriguingly, studies have reported that epidemiological factors (e.g. caffeine) may exert radio-sensitizing effects in cancer cells through inhibition of DNA repair mechanisms (e.g. ATM, ATR) [109,110]; therefore, high HPV load may enhance the effects of those epidemiological exposures on clinical outcomes of cancer patients. In addition, viral pathogens including HPV contribute to high levels of epitopes in the tumour microenvironment [111,112], which potentially lead to an immunogenic tumour microenvironment and better response to immune checkpoint blockade. Accordingly, immune checkpoint blockade may synergise with HPV-16 vaccination to enhance their anti-tumour effects [113]. In HPV-negative head and neck squamous cell carcinomas, lifestyle factors such as smoking and heavy alcohol consumption play a major role in carcinogenesis [114,115], whereas HPV carriers are more common in current smokers than in non-current smokers [116]. In addition, genetic variants at the HLA region may be differentially associated with incidence of oropharyngeal carcinomas by HPV positivity [117]. Therefore, an integrative analysis of host and epidemiological factors incorporating tumour subtyping based on HPV infection status is mandatory when examining risk factors for head and neck carcinomas [115,118,119]. In aggregate, there are expected to be open opportunities for microbiology-MPE research in HPV-related malignancies.
Hepatocellular carcinoma (HCC) is the most common primary liver cancer and a collection of pathogenically heterogeneous carcinomas [15,16,120]. HBV and HCV are DNA and RNA viruses, respectively, which have been established as pathogens involved in chronic inflammation, cirrhotic changes, and carcinogenesis in the liver [15,16]. Heavy alcohol drinking is a major risk factor for incidence of non-viral HCC [15,121]. Recently, steatohepatitis among individuals with non-alcoholic fatty liver disease has gained attention as an alternative pathogenic mechanism of HCC [122–124]. It is evident that the incidence of HCC derived from non-alcoholic steatohepatitis (NASH) increases by the presence of metabolic risk factors including type 2 diabetes, obesity and, collectively, the metabolic syndrome. Emerging data indicate that dysbiosis of the gut microbiome may be associated with NASH and HCC [27–30,125,126]. Therefore, intervention strategies modulating lifestyle and the microbiota for prevention of HCC can be indicated for individuals with NASH [121,127–129]. Interestingly, among HBV carriers, the association of metabolic risk factors or insulin resistance with HCC incidence may be stronger in individuals with lower HBV load than in those with higher HBV load [130]. On the other hand, studies suggest that the association of epidemiological factors (e.g. smoking) with patient survival may differ by viral subtypes of HCC [131]. Taken together, the paradigm of microbiology-MPE would enrich cohort studies investigating incidence and mortality of HCC [71].
The colorectum is an organ that hosts the most abundant and diverse microorganisms in the human body, and dysregulation of the gut microbial ecosystem may result in colorectal carcinogenesis through impairing intestinal immune status, provoking a chronic proinflammatory reaction and affecting host metabolism [132–139]. Influences of the gut microbiome on carcinogenesis likely underlie the continuum of changes in colorectal tumour characteristics [such as microsatellite instability (MSI) status, CpG island methylator phenotype (CIMP), BRAF and PIK3CA mutations, and abundant intratumour F. nucleatum] along the detailed sublocations from caecum or ascending colon to rectum [140–143]. These findings led to the “colorectal continuum” model, implicating the pathogenic influences of the gut microbiome on colorectal tumours [144] (Figure 5). Compelling evidence indicates considerable heterogeneity between colorectal cancer subtypes due to underlying genetic, epigenetic, and microbial statuses in each tumour [39,145–148]. Accordingly, colon and rectal carcinomas have served as a practical disease model for microbiology-MPE research. F. nucleatum is a microbial pathogen that has been implicated in tumourigenesis and progression of colorectal cancer [21–23,26,149–151]. In two studies using a sample size of at least 200 cases of colorectal cancer (with available FFPE tissue specimens), intratumour F. nucleatum was detected in 9–13% patients [26,152]. Mechanistic studies indicate that F. nucleatum may have carcinogenic properties through up-regulating signalling pathways such as the CTNNB1 (beta-catenin)-WNT pathway [153] and also confer a metastatic potential to colorectal cancer [154]. In U.S. population-based studies, researchers explored the heterogeneity in associations of dietary patterns with colorectal cancer sub-classified by the presence of F. nucleatum in tumour tissue. A so-called prudent dietary pattern (rich in vegetables, whole grains, fish, fruits, and poultry) has been associated with a lower risk of F. nucleatum-positive colorectal cancer, but not with a risk of F. nucleatum-negative cancer [155]. In contrast, an inflammatory dietary pattern (rich in red and processed meat, refined grains, and sugar) has been associated with a higher risk of F. nucleatum-positive colorectal cancer, but not with a risk of F. nucleatum-negative cancer, and this differential association is pronounced for proximal colon cancer [156]. These studies provide population-based evidence for the potential role of the microbiome in mediating the relationship between diet and colorectal carcinogenesis. Studies support the roles of viruses and other bacteria, such as enterotoxigenic Bacteroides fragilis, pks-positive Escherichia coli, Enterococcus faecalis, Lactobacillus, and Bifidobacterium in colorectal cancer [5,157]. Therefore, investigation of these bacteria and a microbial community as a whole in relation to incidence and progression of colorectal cancer is also warranted.
Figure 5.
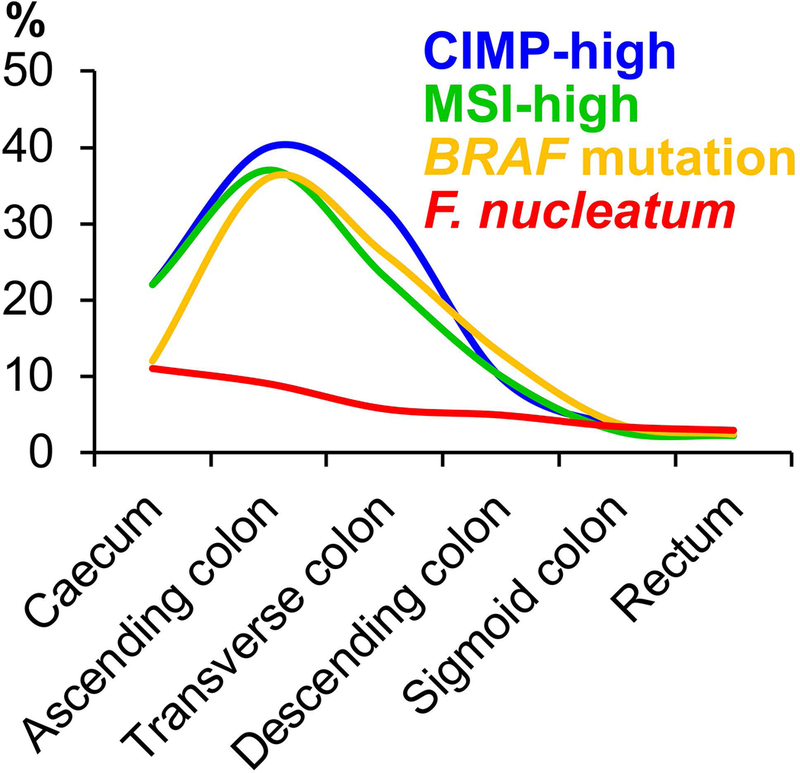
The colorectal continuum theory that highlights the interaction between the gut microbiome and immune cells in the colorectal microenvironment. Certain characteristics of colorectal cancer [e.g. abundant intratumour Fusobacterium nucleatum (F. nucleatum)] represent a gradual transition along the colorectal axis without a clear cut-off at the splenic flexure [140,143]. CIMP, CpG island methylator phenotype; MSI, microsatellite instability.
The immune system functions as the body’s defence mechanism against non-self, including microorganisms, foreign objects, and tumours with immunogenic peptides (“neoantigens”) [111,158–160]. Therefore, immunology has close ties to both microbiology and oncology [161–164]. Anti-tumour immune response is modified by not only immune cells but also tumour and microbial factors [3,33–36,165–168]. Immunology-MPE has been derived from an integration of immunology into MPE research to examine inter-individual heterogeneity in disease process by host immune status [3,169]. The immunology-MPE approach has been successfully utilised to investigate immune-enhancing or -suppressing effects of epidemiological factors in relation to the incidence of colorectal cancer and patient survival [170–180]. The data supporting dietary components and medications as immunomodulators can inform immunoprevention strategies against cancers [181–184]. In addition to the carcinogenic effects of F. nucleatum, evidence suggests molecular and pathological characteristics of F. nucleatum-positive colorectal cancer, including high levels of MSI and CIMP [26,152,185]. In MSI-high colorectal cancer, abundant neoantigens due to numerous frameshift mutations result in vigorous immune response to the tumour, potentially contributing to the enhanced efficacy of the immune checkpoint inhibitors [159,186–190]. On the other hand, immunosuppressive effects of F. nucleatum have been implicated as an alternative mechanism through which this microbe can exert carcinogenic effects [26,191–194]. Evidence suggests that the virulence factor FAP2 of F. nucleatum binds to the host factor Gal-GalNAc and thereby interacts with the inhibitory receptor TIGIT on T lymphocytes [193,195,196]. In our previous study, the enrichment of F. nucleatum in colorectal carcinoma tissue was negatively associated with the density of CD3+ pan-T cells infiltrating the tumour [26]. To address these seemingly conflicting findings (Figure 6A), a microbiology-MPE study was conducted based on large U.S. prospective cohort studies [197]. The presence of F. nucleatum was inversely associated with levels of tumour-infiltrating lymphocytes (TIL) in MSI-high colorectal tumours, but positively associated with TIL levels in non-MSI-high tumours. Based on these findings, it is speculated that the immunosuppressive effects of F. nucleatum may dominate in MSI-high colorectal cancer, whereas its proinflammatory effects may dominate in non-MSI-high cancer (Figure 6B) [192,194,198]. The data support interactive effects of tumour microbiota and molecular features on anti-tumour immunity and the potential role of F. nucleatum as a new immunotherapeutic target.
Figure 6.
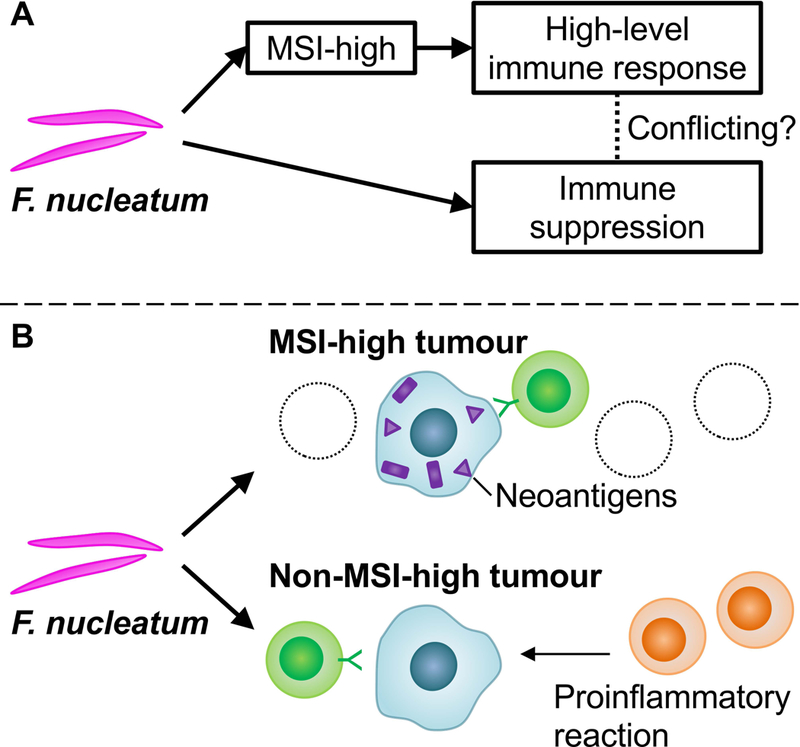
The association between Fusobacterium nucleatum (F. nucleatum) and immune response to colorectal carcinoma depends on tumour microsatellite instability (MSI) status. A. The seemingly paradoxical relationship of F. nucleatum, tumour MSI status, and T lymphocyte infiltrates. B. The relationship of F. nucleatum with lymphocytic reaction according to tumour MSI status. F. nucleatum may exert immunosuppressive effects in MSI-high colorectal cancer, whereas F. nucleatum is associated with high-level lymphocytic reaction in non-MSI-high cancer in which immune reaction is generally inactive and the proinflammatory properties of F. nucleatum are thought to be dominant. MSI, microsatellite instability.
Of note, survival benefits from chemotherapy, radiotherapy, and immunotherapy may be modulated by the gut microbiome [8,93,199,200]. Using clinical faecal specimens of patients receiving anti-PDCD1 (PD-1) immunotherapy, studies have identified several candidate bacteria that may play a particular role in modulating benefits from the therapy [201–204]. The enrichment of specific bacteria in the gut (e.g. Faecalibacterium genus, Bifidobacterium genus) has been correlated with response to anti-PDCD1 blockade therapy. Compared to mice receiving faecal microbiota transplantation from non-responders, mice receiving transplantation from responders were associated with higher levels of tumour CD8+ cytotoxic T cells [201–203]. Therefore, immunomodulatory factors may affect the survival of cancer patients differentially by the microbial repertoire in the host.
Challenges in microbiology-MPE research
There exist challenges in the field of microbiology-MPE, some of which are attributed to epidemiological and MPE research in general as previously discussed [39]. To address the general issue of small sample sizes due to limited availability of specimens, MPE researchers should make efforts to maximise the number of cases available for research. The recent trend in data sharing and world-wide collaborative consortia may help collect large-scale data from different settings to increase statistical power and generalizability of study findings [205–208]. In order to facilitate collaborative projects on microbiology-MPE, we need to standardise procedures for collecting, processing, and storing biospecimens, because microbial compositions change due to those preanalytical factors. It should also be noted that certain preanalytical variables are uncontrollable especially when archival tissue specimens are used. For enhancement of robust data analyses, statistical methods specific for MPE analyses have been developed to assess disease heterogeneity in various study designs [80,209–216] and to address a bias due to missing data [217,218]. In addition, statistical methods such as inverse probability weighting and multiple imputation can be used to mitigate selection bias due to the availability of tissue specimens [218–222]. We need to establish certain general scientific standards across many fields for overall scientific rigour and reproducibility, while there is also a great need for standardisation of methodologies in specific fields.
There are also challenges specific to microbiological research. When a number of microorganisms or microbial communities as a whole are examined, microbiological studies may be prone to false positive findings if multiple hypothesis testing is not taken into account. In microbiology-MPE research, FFPE tissue specimens are often utilised due to relatively easy accessibility; however, the microbial compositions in those specimens might be different from those in vivo or in fresh tissue specimens. Finally, the necessity for transdisciplinary education to cover all component fields of microbiology-MPE is even more difficult to address [223]. Due to the nature of microbiology-MPE, it is of considerable importance to set up transdisciplinary research teams. Nonetheless, researchers in microbiology-MPE should at least obtain fundamental knowledge of the principles of the component fields. It should be noted that the new conceptual and methodological developments in MPE have only been accelerated after 2010. Integrative expertise and viewpoints have been essential in these developments. Essentially, collaboration of an expert in pathology and another expert in epidemiology (which in fact has been ongoing since the 1990s) cannot match a single expert who has obtained adequate knowledge in MPE resulting from appropriate training. A reformation of school curricula may be considered. For example, lectures on pathology and training at laboratories of clinical pathology and microbiology would be useful for students at schools of public health. We may implement training programmes of molecular pathology, epidemiology, microbiology, and immunology for physicians and researchers (e.g. lectures and/or hands-on training of epidemiology and biostatistics in departments of pathology). In addition, the International MPE Meeting series has successfully provided diverse types of researchers with the latest information on the methodology and findings of interdisciplinary MPE research [224,225], and can continue to be an educational resource. To address all of these challenges, we should develop educational programmes that integrate training for microbiology, molecular pathology, and epidemiology.
Future perspectives and conclusions
The integrative approach of microbiology-MPE can be a powerful tool that potentially expands our knowledge of the aetiologies and pathogenesis of carcinomas evolving through dysregulated microbiota [40,226]. While our discussion primarily focuses on neoplastic diseases, this methodology can be readily extrapolated to non-neoplastic diseases that have substantial inter-personal heterogeneity in the disease process and a potential link to the microbiome. It should be noted that the purpose of using the name of a particular scientific discipline is to clarify the need for adequate education, training, knowledge, and expertise in order to properly conduct high-quality research in the discipline. That is, to conduct microbiology-MPE research, one should be well trained in all of the component fields, i.e. microbiology, molecular pathology, and epidemiology.
Due to the nature of MPE as a method-based discipline (but not a disease- or organ-specific discipline), it would be possible to further integrate other important disciplines, including immunology, pharmacology, and nutritional science, and optimise the potential of microbiology-MPE. Preclinical studies suggest the modulatory effects of common medications (e.g. aspirin, statins, and metformin) [227–231] and dietary and nutritional factors (e.g. vitamins, omega-3 polyunsaturated fatty acids, red and processed meat, coffee, alcohol, fibre, and sugar) [231–238] on the microbiota and host immune response. Owing to the advance and cost reduction in high-throughput sequencing technologies and analytical platforms for large-scale data, multi-omics data are increasingly available for epidemiology studies [239]. Sequencing-based analyses including RNA transcriptomic sequencing, metagenomic analyses [240–243] and single cell sequencing [244–246] will allow us to examine the complex relationship of the microbiota, immune cells, and tumour cells in a more comprehensive fashion.
In conclusion, microbiology-MPE can provide a novel methodological framework to gain insights into the tumour-immune-microbiome interaction from human tissue and population data, thereby informing targeted microbiome-modulating strategies for cancer prevention and treatment. Given the increasing availability of omics data on host and tumour with microbial and immune profiles, this new approach should further promote the global trend of precision medicine. Nonetheless, analytical frameworks and educational programmes should be urgently refined specifically for MPE and microbiology-MPE research.
Why do we need names for scientific fields?
The primary purpose of this article is to discuss an integration of microbiology, molecular pathology, and epidemiology. Why do these fields need names? The existence of the name of a scientific field implies that a specific set of education, training, knowledge, and expertise is needed to conduct research in that field. For example, if one investigates the epidemiology of human papillomavirus (HPV), one needs to gain adequate knowledge and expertise in both microbiology and epidemiology through appropriate education and training. However, not all researchers who study the epidemiology of HPV may have proper expertise in both microbiology and epidemiology. There are substantial pitfalls in non-experts conducting research studies. Hence, the integration of microbiology, molecular pathology, and epidemiology necessitates knowledge and expertise in all of these fields. The broader goal of this article is to illustrate an increasing need for transdisciplinary education and training systems for future science.
Acknowledgements
This work was supported by U.S. National Institutes of Health (NIH) grants (K99 CA215314 and R00 CA215314 to MS, R35 CA197735 to SO); by the American Cancer Society (MRSG-17–220-01-NEC to MS); by Nodal Award (2016–02) from the Dana-Farber Harvard Cancer Center (to SO); and by The Friends of the Dana-Farber Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funding source had no role in decision to submit the manuscript to publication or preparation, review, and approval of the manuscript.
Funding: This work was supported by U.S. National Institutes of Health (NIH) grants (K99 CA215314 and R00 CA215314 to MS, R35 CA197735 to SO); by the American Cancer Society (MRSG-17–220-01-NEC to MS); by Nodal Award (2016–02) from the Dana-Farber Harvard Cancer Center (to SO); and by The Friends of the Dana-Farber Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funding source had no role in decision to submit the manuscript to publication or preparation, review, and approval of the manuscript.
Abbreviations:
- CIMP
CpG island methylator phenotype
- FFPE
formalin-fixed paraffin-embedded
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HPV
human papillomavirus
- MPE
molecular pathological epidemiology
- MSI
microsatellite instability
- NASH
non-alcoholic steatohepatitis
- TIL
tumour-infiltrating lymphocytes
Footnotes
Conflicts of interest: The authors declare that they have no conflicts of interest.
Use of standardised official symbols: We use HUGO (Human Genome Organisation)-approved official symbols (or root symbols) for genes, gene families, and gene products, including ATM, ATR, BRAF, CD3, CD8, CTNNB1, HLA, PDCD1, PIK3CA, TIGIT, and WNT; all of which are described at www.genenames.org. Gene names are italicised, and gene product names are non-italicised.
References
- 1.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capurso G, Lahner E. The interaction between smoking, alcohol and the gut microbiome. Best Pract Res Clin Gastroenterol 2017; 31: 579–588. [DOI] [PubMed] [Google Scholar]
- 3.Ogino S, Nowak JA, Hamada T, et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut 2018; 67: 1168–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Q, Chen WD, Wang YD. Gut Microbiota: An Integral Moderator in Health and Disease. Front Microbiol 2018; 9: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Domingue JC, Sears CL. Microbiota dysbiosis in select human cancers: Evidence of association and causality. Semin Immunol 2017; 32: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajpoot M, Sharma AK, Sharma A, et al. Understanding the microbiome: Emerging biomarkers for exploiting the microbiota for personalized medicine against cancer. Semin Cancer Biol 2018; 52: 1–8. [DOI] [PubMed] [Google Scholar]
- 7.Morgillo F, Dallio M, Della Corte CM, et al. Carcinogenesis as a Result of Multiple Inflammatory and Oxidative Hits: a Comprehensive Review from Tumor Microenvironment to Gut Microbiota. Neoplasia 2018; 20: 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopalakrishnan V, Helmink BA, Spencer CN, et al. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018; 33: 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Healey GR, Murphy R, Brough L, et al. Interindividual variability in gut microbiota and host response to dietary interventions. Nutr Rev 2017; 75: 1059–1080. [DOI] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ. Microbes and Diet-Induced Obesity: Fast, Cheap, and Out of Control. Cell Host Microbe 2017; 21: 278–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Routy B, Gopalakrishnan V, Daillere R, et al. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol 2018; 15: 382–396. [DOI] [PubMed] [Google Scholar]
- 12.Zhernakova DV, Le TH, Kurilshikov A, et al. Individual variations in cardiovascular-disease-related protein levels are driven by genetics and gut microbiome. Nat Genet 2018; 50: 1524–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vatanen T, Franzosa EA, Schwager R, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018; 562: 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet 2016; 388: 2654–2664. [DOI] [PubMed] [Google Scholar]
- 15.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018; 391: 1301–1314. [DOI] [PubMed] [Google Scholar]
- 16.Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer 2013; 13: 123–135. [DOI] [PubMed] [Google Scholar]
- 17.Yarchoan R, Uldrick TS. HIV-Associated Cancers and Related Diseases. N Engl J Med 2018; 378: 1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serrano B, Brotons M, Bosch FX, et al. Epidemiology and burden of HPV-related disease. Best Pract Res Clin Obstet Gynaecol 2018; 47: 14–26. [DOI] [PubMed] [Google Scholar]
- 19.Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 2018; 18: 269–282. [DOI] [PubMed] [Google Scholar]
- 20.Ishitsuka K, Tamura K. Human T-cell leukaemia virus type I and adult T-cell leukaemia-lymphoma. Lancet Oncol 2014; 15: e517–526. [DOI] [PubMed] [Google Scholar]
- 21.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012; 22: 292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 2012; 22: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gholizadeh P, Eslami H, Kafil HS. Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomed Pharmacother 2017; 89: 918–925. [DOI] [PubMed] [Google Scholar]
- 24.Morris LG, Chandramohan R, West L, et al. The Molecular Landscape of Recurrent and Metastatic Head and Neck Cancers: Insights From a Precision Oncology Sequencing Platform. JAMA Oncol 2017; 3: 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols AC, Palma DA, Chow W, et al. High frequency of activating PIK3CA mutations in human papillomavirus-positive oropharyngeal cancer. JAMA Otolaryngol Head Neck Surg 2013; 139: 617–622. [DOI] [PubMed] [Google Scholar]
- 26.Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol 2015; 1: 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren Z, Li A, Jiang J, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cani PD, Jordan BF. Gut microbiota-mediated inflammation in obesity: a link with gastrointestinal cancer. Nat Rev Gastroenterol Hepatol 2018; 15: 671–682. [DOI] [PubMed] [Google Scholar]
- 29.Schramm C Bile Acids, the Microbiome, Immunity, and Liver Tumors. N Engl J Med 2018; 379: 888–890. [DOI] [PubMed] [Google Scholar]
- 30.Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol 2018; 15: 111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akshintala VS, Talukdar R, Singh VK, et al. The Gut Microbiome in Pancreatic Disease. Clin Gastroenterol Hepatol 2019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Keefe SJ, Li JV, Lahti L, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun 2015; 6: 6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo E, Taddei A, Ringressi MN, et al. The interplay between the microbiome and the adaptive immune response in cancer development. Therap Adv Gastroenterol 2016; 9: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer 2018; 18: 139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogino S, Nowak JA, Hamada T, et al. Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu Rev Pathol 2019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sethi V, Kurtom S, Tarique M, et al. Gut Microbiota Promotes Tumor Growth in Mice by Modulating Immune Response. Gastroenterology 2018; 155: 33–37 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurnit KC, Dumbrava EEI, Litzenburger B, et al. Precision Oncology Decision Support: Current Approaches and Strategies for the Future. Clin Cancer Res 2018; 24: 2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: the evolving field of molecular pathological epidemiology. J Natl Cancer Inst 2010; 102: 365–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogino S, Chan AT, Fuchs CS, et al. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut 2011; 60: 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamada T, Keum N, Nishihara R, et al. Molecular pathological epidemiology: new developing frontiers of big data science to study etiologies and pathogenesis. J Gastroenterol 2017; 52: 265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn 2012; 12: 621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogino S, Giovannucci E. Commentary: Lifestyle factors and colorectal cancer microsatellite instability--molecular pathological epidemiology science, based on unique tumour principle. Int J Epidemiol 2012; 41: 1072–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogino S, Lochhead P, Chan AT, et al. Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease. Mod Pathol 2013; 26: 465–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishihara R, VanderWeele TJ, Shibuya K, et al. Molecular pathological epidemiology gives clues to paradoxical findings. Eur J Epidemiol 2015; 30: 1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haendel MA, Chute CG, Robinson PN. Classification, Ontology, and Precision Medicine. N Engl J Med 2018; 379: 1452–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kensler TW, Spira A, Garber JE, et al. Transforming Cancer Prevention through Precision Medicine and Immune-oncology. Cancer Prev Res (Phila) 2016; 9: 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spira A, Yurgelun MB, Alexandrov L, et al. Precancer Atlas to Drive Precision Prevention Trials. Cancer Res 2017; 77: 1510–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dienstmann R, Vermeulen L, Guinney J, et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017; 17: 268. [DOI] [PubMed] [Google Scholar]
- 49.Michel P, Boige V, Andre T, et al. Aspirin versus placebo in stage III or high-risk stage II colon cancer with PIK3CA mutation: A French randomised double-blind phase III trial (PRODIGE 50-ASPIK). Dig Liver Dis 2018; 50: 305–307. [DOI] [PubMed] [Google Scholar]
- 50.Hamada T, Giannakis M, Ogino S. Aspirin in the era of immunotherapy. Oncotarget 2017; 8: 73370–73371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Field AE, Camargo CA Jr., Ogino S. The merits of subtyping obesity: one size does not fit all. JAMA 2013; 310: 2147–2148. [DOI] [PubMed] [Google Scholar]
- 52.Ogino S, Nishihara R, VanderWeele TJ, et al. Review Article: The Role of Molecular Pathological Epidemiology in the Study of Neoplastic and Non-neoplastic Diseases in the Era of Precision Medicine. Epidemiology 2016; 27: 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogino S Molecular pathological epidemiology (MPE): Overview of its paradigm and wide applicability even without tumor tissue [abstract]. Proceedings of the Twelfth Annual AACR International Conference on Frontiers in Cancer Prevention Research National Harbor, MD, USA Cancer Prev Res (Phila) 2013; 27–30 Oct 2013; 6: CN06–1. [Google Scholar]
- 54.Kuller LH, Bracken MB, Ogino S, et al. The role of epidemiology in the era of molecular epidemiology and genomics: Summary of the 2013 AJE-sponsored Society of Epidemiologic Research Symposium. Am J Epidemiol 2013; 178: 1350–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Epplein M, Bostick RM, Mu L, et al. Challenges and opportunities in international molecular cancer prevention research: An ASPO Molecular Epidemiology and the Environment and International Cancer Prevention Interest Groups Report. Cancer Epidemiol Biomarkers Prev 2014; 23: 2613–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogino S Molecular pathological epidemiology of risk factors and CRC microbial and immune characteristics [abstract]. Proceedings of the AACR Special Conference on Colorectal Cancer: From Initiation to Outcomes. Tampa, FL, USA Cancer Res 2017; 17–20 Sep 2016; 77: IA28. [Google Scholar]
- 57.Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Pathol Res Int 2011; 2011: 902674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes LA, Khalid-de Bakker CA, Smits KM, et al. The CpG island methylator phenotype in colorectal cancer: Progress and problems. Biochim Biophys Acta 2012; 1825: 77–85. [DOI] [PubMed] [Google Scholar]
- 59.Chia WK, Ali R, Toh HC. Aspirin as adjuvant therapy for colorectal cancer-reinterpreting paradigms. Nat Rev Clin Oncol 2012; 9: 561–570. [DOI] [PubMed] [Google Scholar]
- 60.Bae JM, Kim JH, Cho NY, et al. Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. Br J Cancer 2013; 109: 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buchanan DD, Win AK, Walsh MD, et al. Family History of Colorectal Cancer in BRAF p.V600E mutated Colorectal Cancer Cases. Cancer Epidemiol Biomarkers Prev 2013; 22: 917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin JH, Giovannucci E. Environmental exposure and tumor heterogeneity in colorectal cancer risk and outcomes. Curr Colorectal Cancer Rep 2014; 10: 94–104. [Google Scholar]
- 63.Campbell PT, Deka A, Briggs P, et al. Establishment of the Cancer Prevention Study II Nutrition Cohort Colorectal Tissue Repository. Cancer Epidemiol Biomarkers Prev 2014; 23: 2694–2702. [DOI] [PubMed] [Google Scholar]
- 64.Li W, Qiu T, Ling Y, et al. Molecular pathological epidemiology of colorectal cancer in Chinese patients with KRAS and BRAF mutations. Oncotarget 2015; 6: 39607–39613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ng JM, Yu J. Promoter hypermethylation of tumour suppressor genes as potential biomarkers in colorectal cancer. Int J Mol Sci 2015; 16: 2472–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campbell PT, Newton CC, Newcomb PA, et al. Association between body mass index and mortality for colorectal cancer survivors: overall and by tumor molecular phenotype. Cancer Epidemiol Biomarkers Prev 2015; 24: 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuipers EJ, Grady WM, Lieberman D, et al. Colorectal cancer. Nat Rev Dis Primers 2015; 1: 15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez-Useros J, Garcia-Foncillas J. Obesity and colorectal cancer: molecular features of adipose tissue. J Transl Med 2016; 14: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuroiwa-Trzmielina J, Wang F, Rapkins RW, et al. SNP rs16906252C>T is an expression and methylation quantitative trait locus associated with an increased risk of developing MGMT-methylated colorectal cancer. Clin Cancer Res 2016; 22: 6266–6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alnabulsi A, Murray GI. Integrative analysis of the colorectal cancer proteome: potential clinical impact. Expert Rev Proteomics 2016; 13: 917–927. [DOI] [PubMed] [Google Scholar]
- 71.Gao C Molecular pathological epidemiology in diabetes mellitus and risk of hepatocellular carcinoma. World J Hepatol 2016; 8: 1119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li SK, Martin A. Mismatch Repair and Colon Cancer: Mechanisms and Therapies Explored. Trends Mol Med 2016; 22: 274–289. [DOI] [PubMed] [Google Scholar]
- 73.Lee DH, Keum N, Giovannucci EL. Colorectal Cancer Epidemiology in the Nurses’ Health Study. Am J Public Health 2016; 106: 1599–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li YY, Ge QX, Cao J, et al. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol 2016; 22: 3227–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rescigno T, Micolucci L, Tecce MF, et al. Bioactive nutrients and nutrigenomics in age-related diseases. Molecules 2017; 22: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hughes LA, Simons CC, van den Brandt PA, et al. Lifestyle, diet, and colorectal cancer risk according to (epi)genetic instability: current evidence and future direction of molecular pathological epidemiology. Curr Colorectal Cancer Rep 2017; 13: 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang M, Dai J, Gu D, et al. Aspirin in pancreatic cancer: chemopreventive effects and therapeutic potentials. BBA Rev Cancer 2017; 1866: 163–176. [DOI] [PubMed] [Google Scholar]
- 78.Serafino A, Sferrazza G, Colini Baldeschi A, et al. Developing drugs that target the Wnt pathway: recent approaches in cancer and neurodegenerative diseases. Expert Opin Drug Discov 2017; 12: 169–186. [DOI] [PubMed] [Google Scholar]
- 79.Slattery ML, Lee FY, Pellatt AJ, et al. Infrequently expressed miRNAs in colorectal cancer tissue and tumor molecular phenotype. Mod Pathol 2017; 30: 1152–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Richiardi L, Barone-Adesi F, Pearce N. Cancer subtypes in aetiological research. Eur J Epidemiol 2017; 32: 353–361. [DOI] [PubMed] [Google Scholar]
- 81.Patil H, Saxena SG, Barrow CJ, et al. Chasing the personalized medicine dream through biomarker validation in colorectal cancer. Drug Discov Today 2017; 22: 111–119. [DOI] [PubMed] [Google Scholar]
- 82.Carr PR, Alwers E, Bienert S, et al. Lifestyle factors and risk of sporadic colorectal cancer by microsatellite instability status: a systematic review and meta-analysis. Ann Oncol 2018; 29: 825–834. [DOI] [PubMed] [Google Scholar]
- 83.Benelli R, Vene R, Ferrari N. Prostaglandin-endoperoxide synthase 2 (cyclooxygenase-2), a complex target for colorectal cancer prevention and therapy. Transl Res 2018; 196: 42–61. [DOI] [PubMed] [Google Scholar]
- 84.Zhang S, Cai S, Ma Y. Association between Fusobacterium nucleatum and colorectal cancer: Progress and future directions. J Cancer 2018; 9: 1652–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waluga M, Zorniak M, Fichna J, et al. Pharmacological and dietary factors in prevention of colorectal cancer. J Physiol Pharmacol 2018; 69. [DOI] [PubMed] [Google Scholar]
- 86.Zakhari S, Hoek JB. Epidemiology of moderate alcohol consumption and breast cancer: association or causation? Cancers 2018; 10: 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murphy N, Jenab M, Gunter MJ. Adiposity and gastrointestinal cancers: epidemiology, mechanisms and future directions. Nat Rev Gastroenterol Hepatol 2018; 15: 659–670. [DOI] [PubMed] [Google Scholar]
- 88.Myte R, Gylling B, Haggstrom J, et al. One-carbon metabolism biomarkers and genetic variants in relation to colorectal cancer risk by KRAS and BRAF mutation status. PLoS One 2018; 13: e0196233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alexander JL, Scott AJ, Pouncey AL, et al. Colorectal carcinogenesis: an archetype of gut microbiota-host interaction. ecancer 2018; 12: 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Valeria M, Daniela DA, Francesca M, et al. Epigenetics and Infectious Disease: State-of-the-Art and Perspectives in New Generation Therapies. OBM Genetics 2018; 2. [Google Scholar]
- 91.Molloy MP, Engel A. Precision medicine beyond medical oncology: using molecular analysis to guide treatments of colorectal neoplasia. Expert Review of Gastroenterology & Hepatology 2018; 12: 1179–1181. [DOI] [PubMed] [Google Scholar]
- 92.Brenner H, Chen C. The colorectal cancer epidemic: challenges and opportunities for primary, secondary and tertiary prevention. Br J Cancer 2018; 119: 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ding C, Tang W, Fan X, et al. Intestinal microbiota: a novel perspective in colorectal cancer biotherapeutics. Onco Targets Ther 2018; 11: 4797–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Galipeau PC, Oman KM, Paulson TG, et al. NSAID use and somatic exomic mutations in Barrett’s esophagus. Genome Med 2018; 10: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Coleman HG, Xie SH, Lagergren J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology 2018; 154: 390–405. [DOI] [PubMed] [Google Scholar]
- 96.Spallanzani A, Gelsomino F, Caputo F, et al. Immunotherapy in the treatment of colorectal cancer: a new kid on the block. J Cancer Metastasis Treat 2018; 4: 28. [Google Scholar]
- 97.Fathi N, Ahmadian E, Shahi S, et al. Role of vitamin D and vitamin D receptor (VDR) in oral cancer. Biomed Pharmacother 2019; 109: 391–401. [DOI] [PubMed] [Google Scholar]
- 98.Amitay EL, Carr PR, Jansen L, et al. Association of Aspirin and Nonsteroidal Anti-Inflammatory Drugs With Colorectal Cancer Risk by Molecular Subtypes. J Natl Cancer Inst 2019; 111: djy170. [DOI] [PubMed] [Google Scholar]
- 99.Tytgat HLP, Nobrega FL, van der Oost J, et al. Bowel Biofilms: Tipping Points between a Healthy and Compromised Gut? Trends Microbiol 2019; 27: 17–25. [DOI] [PubMed] [Google Scholar]
- 100.Hendler R, Zhang Y. Probiotics in the Treatment of Colorectal Cancer. Medicines (Basel) 2018; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Franzen A, Vogt TJ, Muller T, et al. PD-L1 (CD274) and PD-L2 (PDCD1LG2) promoter methylation is associated with HPV infection and transcriptional repression in head and neck squamous cell carcinomas. Oncotarget 2018; 9: 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu C, Mann D, Sinha UK, et al. The molecular mechanisms of increased radiosensitivity of HPV-positive oropharyngeal squamous cell carcinoma (OPSCC): an extensive review. J Otolaryngol Head Neck Surg 2018; 47: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Psyrri A, Rampias T, Vermorken JB. The current and future impact of human papillomavirus on treatment of squamous cell carcinoma of the head and neck. Ann Oncol 2014; 25: 2101–2115. [DOI] [PubMed] [Google Scholar]
- 104.Jacobi C, Ayx I, Fritsche K, et al. Potential impact of human papilloma virus on survival of basaloid squamous carcinoma of the head and neck. Oncotarget 2015; 6: 3462–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dillon MT, Harrington KJ. Human Papillomavirus-Negative Pharyngeal Cancer. J Clin Oncol 2015; 33: 3251–3261. [DOI] [PubMed] [Google Scholar]
- 106.Fung N, Faraji F, Kang H, et al. The role of human papillomavirus on the prognosis and treatment of oropharyngeal carcinoma. Cancer Metastasis Rev 2017; 36: 449–461. [DOI] [PubMed] [Google Scholar]
- 107.Wieringa HW, van der Zee AG, de Vries EG, et al. Breaking the DNA damage response to improve cervical cancer treatment. Cancer Treat Rev 2016; 42: 30–40. [DOI] [PubMed] [Google Scholar]
- 108.Low GM, Thylur DS, V NY, et al. The effect of human papillomavirus on DNA repair in head and neck squamous cell carcinoma. Oral Oncol 2016; 61: 27–30. [DOI] [PubMed] [Google Scholar]
- 109.Sarkaria JN, Busby EC, Tibbetts RS, et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res 1999; 59: 4375–4382. [PubMed] [Google Scholar]
- 110.Tsabar M, Eapen VV, Mason JM, et al. Caffeine impairs resection during DNA break repair by reducing the levels of nucleases Sae2 and Dna2. Nucleic Acids Res 2015; 43: 6889–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015; 348: 69–74. [DOI] [PubMed] [Google Scholar]
- 112.Outh-Gauer S, Alt M, Le Tourneau C, et al. Immunotherapy in head and neck cancers: A new challenge for immunologists, pathologists and clinicians. Cancer Treat Rev 2018; 65: 54–64. [DOI] [PubMed] [Google Scholar]
- 113.Massarelli E, William W, Johnson F, et al. Combining Immune Checkpoint Blockade and Tumor-Specific Vaccine for Patients With Incurable Human Papillomavirus 16-Related Cancer: A Phase 2 Clinical Trial. JAMA Oncol 2019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rettig EM, D’Souza G. Epidemiology of head and neck cancer. Surg Oncol Clin N Am 2015; 24: 379–396. [DOI] [PubMed] [Google Scholar]
- 115.Wang C, Dickie J, Sutavani RV, et al. Targeting Head and Neck Cancer by Vaccination. Front Immunol 2018; 9: 830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fakhry C, Gillison ML, D’Souza G. Tobacco use and oral HPV-16 infection. JAMA 2014; 312: 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lesseur C, Diergaarde B, Olshan AF, et al. Genome-wide association analyses identify new susceptibility loci for oral cavity and pharyngeal cancer. Nat Genet 2016; 48: 1544–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schwartz SM, Daling JR, Doody DR, et al. Oral cancer risk in relation to sexual history and evidence of human papillomavirus infection. J Natl Cancer Inst 1998; 90: 1626–1636. [DOI] [PubMed] [Google Scholar]
- 119.Kjellberg L, Hallmans G, Ahren AM, et al. Smoking, diet, pregnancy and oral contraceptive use as risk factors for cervical intra-epithelial neoplasia in relation to human papillomavirus infection. Br J Cancer 2000; 82: 1332–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sia D, Jiao Y, Martinez-Quetglas I, et al. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology 2017; 153: 812–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fujiwara N, Friedman SL, Goossens N, et al. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol 2018; 68: 526–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Degasperi E, Colombo M. Distinctive features of hepatocellular carcinoma in non-alcoholic fatty liver disease. Lancet Gastroenterol Hepatol 2016; 1: 156–164. [DOI] [PubMed] [Google Scholar]
- 123.Younossi ZM, Koenig AB, Abdelatif D, et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016; 64: 73–84. [DOI] [PubMed] [Google Scholar]
- 124.Younossi Z, Tacke F, Arrese M, et al. Global Perspectives on Non-alcoholic Fatty Liver Disease and Non-alcoholic Steatohepatitis. Hepatology 2019. in press. [DOI] [PubMed] [Google Scholar]
- 125.Schwenger KJP, Bolzon CM, Li C, et al. Non-alcoholic fatty liver disease and obesity: the role of the gut bacteria. Eur J Nutr 2019. in press. [DOI] [PubMed] [Google Scholar]
- 126.Hoyles L, Fernandez-Real JM, Federici M, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med 2018; 24: 1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brandi G, De Lorenzo S, Candela M, et al. Microbiota, NASH, HCC and the potential role of probiotics. Carcinogenesis 2017; 38: 231–240. [DOI] [PubMed] [Google Scholar]
- 128.Delaune V, Orci LA, Lacotte S, et al. Fecal microbiota transplantation: a promising strategy in preventing the progression of non-alcoholic steatohepatitis and improving the anti-cancer immune response. Expert Opin Biol Ther 2018; 18: 1061–1071. [DOI] [PubMed] [Google Scholar]
- 129.Bomhof MR, Parnell JA, Ramay HR, et al. Histological improvement of non-alcoholic steatohepatitis with a prebiotic: a pilot clinical trial. Eur J Nutr 2019. in press. [DOI] [PubMed] [Google Scholar]
- 130.Yu MW, Lin CL, Liu CJ, et al. Influence of Metabolic Risk Factors on Risk of Hepatocellular Carcinoma and Liver-Related Death in Men With Chronic Hepatitis B: A Large Cohort Study. Gastroenterology 2017; 153: 1006–1017 e1005. [DOI] [PubMed] [Google Scholar]
- 131.Kai K, Komukai S, Koga H, et al. Correlation between smoking habit and surgical outcomes on viral-associated hepatocellular carcinomas. World J Gastroenterol 2018; 24: 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brennan CA, Garrett WS. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu Rev Microbiol 2016; 70: 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lasry A, Zinger A, Ben-Neriah Y. Inflammatory networks underlying colorectal cancer. Nat Immunol 2016; 17: 230–240. [DOI] [PubMed] [Google Scholar]
- 134.O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol 2016; 13: 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen J, Pitmon E, Wang K. Microbiome, inflammation and colorectal cancer. Semin Immunol 2017; 32: 43–53. [DOI] [PubMed] [Google Scholar]
- 136.Mima K, Ogino S, Nakagawa S, et al. The role of intestinal bacteria in the development and progression of gastrointestinal tract neoplasms. Surg Oncol 2017; 26: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Alexander JL, Scott AJ, Pouncey AL, et al. Colorectal carcinogenesis: an archetype of gut microbiota–host interaction. ecancermedicalscience 2018; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Niederreiter L, Adolph TE, Tilg H. Food, microbiome and colorectal cancer. Dig Liver Dis 2018; 50: 647–652. [DOI] [PubMed] [Google Scholar]
- 139.Tilg H, Adolph TE, Gerner RR, et al. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018; 33: 954–964. [DOI] [PubMed] [Google Scholar]
- 140.Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut 2012; 61: 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rosty C, Young JP, Walsh MD, et al. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol 2013; 26: 825–834. [DOI] [PubMed] [Google Scholar]
- 142.Phipps AI, Buchanan DD, Makar KW, et al. BRAF mutation status and survival after colorectal cancer diagnosis according to patient and tumor characteristics. Cancer Epidemiol Biomarkers Prev 2012; 21: 1792–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mima K, Cao Y, Chan AT, et al. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol 2016; 7: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yamauchi M, Lochhead P, Morikawa T, et al. Colorectal cancer: a tale of two sides or a continuum? Gut 2012; 61: 794–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol 2017; 14: 235–246. [DOI] [PubMed] [Google Scholar]
- 146.Dienstmann R, Vermeulen L, Guinney J, et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017; 17: 79–92. [DOI] [PubMed] [Google Scholar]
- 147.Inamura K Colorectal Cancers: An Update on Their Molecular Pathology. Cancers (Basel) 2018; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Riley JM, Cross AW, Paulos CM, et al. The clinical implications of immunogenomics in colorectal cancer: A path for precision medicine. Cancer 2018; 124: 1650–1659. [DOI] [PubMed] [Google Scholar]
- 149.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016; 65: 1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017; 170: 548–563 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yamaoka Y, Suehiro Y, Hashimoto S, et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J Gastroenterol 2018; 53: 517–524. [DOI] [PubMed] [Google Scholar]
- 152.Nosho K, Sukawa Y, Adachi Y, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol 2016; 22: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 2013; 14: 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017; 358: 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mehta RS, Nishihara R, Cao Y, et al. Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol 2017; 3: 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu L, Tabung FK, Zhang X, et al. Diets That Promote Colon Inflammation Associate With Risk of Colorectal Carcinomas That Contain Fusobacterium nucleatum. Clin Gastroenterol Hepatol 2018; 16: 1622–1631 e1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kosumi K, Hamada T, Koh H, et al. The Amount of Bifidobacterium Genus in Colorectal Carcinoma Tissue in Relation to Tumor Characteristics and Clinical Outcome. Am J Pathol 2018; 188: 2839–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Germano G, Lamba S, Rospo G, et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 2017; 552: 116–120. [DOI] [PubMed] [Google Scholar]
- 159.Giannakis M, Mu XJ, Shukla SA, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep 2016; 15: 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Efremova M, Finotello F, Rieder D, et al. Neoantigens Generated by Individual Mutations and Their Role in Cancer Immunity and Immunotherapy. Front Immunol 2017; 8: 1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zitvogel L, Pietrocola F, Kroemer G. Nutrition, inflammation and cancer. Nat Immunol 2017; 18: 843–850. [DOI] [PubMed] [Google Scholar]
- 162.Fridman WH, Zitvogel L, Sautes-Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017; 14: 717–734. [DOI] [PubMed] [Google Scholar]
- 163.Taube JM, Galon J, Sholl LM, et al. Implications of the tumor immune microenvironment for staging and therapeutics. Mod Pathol 2018; 31: 214–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Grizzi F, Basso G, Borroni EM, et al. Evolving notions on immune response in colorectal cancer and their implications for biomarker development. Inflamm Res 2018; 67: 375–389. [DOI] [PubMed] [Google Scholar]
- 165.Masugi Y, Nishihara R, Hamada T, et al. Tumor PDCD1LG2 (PD-L2) Expression and the Lymphocytic Reaction to Colorectal Cancer. Cancer Immunol Res 2017; 5: 1046–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Masugi Y, Nishihara R, Yang J, et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut 2017; 66: 1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cremonesi E, Governa V, Garzon JFG, et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 2018; 67: 1984–1994. [DOI] [PubMed] [Google Scholar]
- 168.Grasso CS, Giannakis M, Wells DK, et al. Genetic Mechanisms of Immune Evasion in Colorectal Cancer. Cancer Discov 2018; 8: 730–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ogino S, Galon J, Fuchs CS, et al. Cancer immunology--analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol 2011; 8: 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ogino S, Giannakis M. Immunoscore for (colorectal) cancer precision medicine. Lancet 2018; 391: 2084–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Khalili H, Gong J, Brenner H, et al. Identification of a common variant with potential pleiotropic effect on risk of inflammatory bowel disease and colorectal cancer. Carcinogenesis 2015; 36: 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cao Y, Nishihara R, Qian ZR, et al. Regular Aspirin Use Associates With Lower Risk of Colorectal Cancers With Low Numbers of Tumor-Infiltrating Lymphocytes. Gastroenterology 2016; 151: 879–892 e874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hanyuda A, Ogino S, Qian ZR, et al. Body mass index and risk of colorectal cancer according to tumor lymphocytic infiltrate. Int J Cancer 2016; 139: 854–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Song M, Nishihara R, Cao Y, et al. Marine omega-3 Polyunsaturated Fatty Acid Intake and Risk of Colorectal Cancer Characterized by Tumor-Infiltrating T Cells. JAMA Oncol 2016; 2: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Song M, Nishihara R, Wang M, et al. Plasma 25-hydroxyvitamin D and colorectal cancer risk according to tumour immunity status. Gut 2016; 65: 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Liu L, Nishihara R, Qian ZR, et al. Association Between Inflammatory Diet Pattern and Risk of Colorectal Carcinoma Subtypes Classified by Immune Responses to Tumor. Gastroenterology 2017; 153: 1517–1530.e1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Berntsson J, Svensson MC, Leandersson K, et al. The clinical impact of tumour-infiltrating lymphocytes in colorectal cancer differs by anatomical subsite: A cohort study. Int J Cancer 2017; 141: 1654–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hamada T, Liu L, Nowak JA, et al. Vitamin D status after colorectal cancer diagnosis and patient survival according to immune response to tumour. Eur J Cancer 2018; 103: 98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Koh H, Hamada T, Song M, et al. Physical Activity and Colorectal Cancer Prognosis According to Tumor-infiltrating T Cells. JNCI Cancer Spectr 2019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hamada T, Nowak JA, Masugi Y, et al. Smoking and Risk of Colorectal Cancer Sub-Classified by Tumor-Infiltrating T Cells. J Natl Cancer Inst 2019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Janakiram NB, Mohammed A, Madka V, et al. Prevention and treatment of cancers by immune modulating nutrients. Mol Nutr Food Res 2016; 60: 1275–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pandolfi F, Franza L, Mandolini C, et al. Immune Modulation by Vitamin D: Special Emphasis on Its Role in Prevention and Treatment of Cancer. Clin Ther 2017; 39: 884–893. [DOI] [PubMed] [Google Scholar]
- 183.Nosrati N, Bakovic M, Paliyath G. Molecular Mechanisms and Pathways as Targets for Cancer Prevention and Progression with Dietary Compounds. Int J Mol Sci 2017; 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fletcher R, Wang YJ, Schoen RE, et al. Colorectal cancer prevention: Immune modulation taking the stage. Biochim Biophys Acta Rev Cancer 2018; 1869: 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 2014; 33: 1381–1390. [DOI] [PubMed] [Google Scholar]
- 186.Ogino S, Nosho K, Irahara N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res 2009; 15: 6412–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nosho K, Baba Y, Tanaka N, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol 2010; 222: 350–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017; 357: 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol 2017; 18: 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kaplan CW, Ma X, Paranjpe A, et al. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun 2010; 78: 4773–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013; 14: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015; 42: 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bashir A, Miskeen AY, Hazari YM, et al. Fusobacterium nucleatum, inflammation, and immunity: the fire within human gut. Tumour Biol 2016; 37: 2805–2810. [DOI] [PubMed] [Google Scholar]
- 195.Abed J, Emgard JE, Zamir G, et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016; 20: 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Manieri NA, Chiang EY, Grogan JL. TIGIT: A Key Inhibitor of the Cancer Immunity Cycle. Trends Immunol 2017; 38: 20–28. [DOI] [PubMed] [Google Scholar]
- 197.Hamada T, Zhang X, Mima K, et al. Fusobacterium nucleatum in Colorectal Cancer Relates to Immune Response Differentially by Tumor Microsatellite Instability Status. Cancer Immunol Res 2018; 6: 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ye X, Wang R, Bhattacharya R, et al. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev Res (Phila) 2017; 10: 398–409. [DOI] [PubMed] [Google Scholar]
- 199.Alexander JL, Wilson ID, Teare J, et al. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol 2017; 14: 356–365. [DOI] [PubMed] [Google Scholar]
- 200.Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer 2017; 17: 271–285. [DOI] [PubMed] [Google Scholar]
- 201.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018; 359: 91–97. [DOI] [PubMed] [Google Scholar]
- 202.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018; 359: 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018; 359: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zitvogel L, Ma Y, Raoult D, et al. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018; 359: 1366–1370. [DOI] [PubMed] [Google Scholar]
- 205.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature 2007; 449: 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Peterson J, Garges S, Giovanni M, et al. The NIH Human Microbiome Project. Genome Res 2009; 19: 2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ogino S, Lochhead P, Giovannucci E, et al. Discovery of colorectal cancer PIK3CA mutation as potential predictive biomarker: power and promise of molecular pathological epidemiology. Oncogene 2014; 33: 2949–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wang J, Kurilshikov A, Radjabzadeh D, et al. Meta-analysis of human genome-microbiome association studies: the MiBioGen consortium initiative. Microbiome 2018; 6: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chatterjee N, Sinha S, Diver WR, et al. Analysis of cohort studies with multivariate and partially observed disease classification data. Biometrika 2010; 97: 683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rosner B, Glynn RJ, Tamimi RM, et al. Breast cancer risk prediction with heterogeneous risk profiles according to breast cancer tumor markers. Am J Epidemiol 2013; 178: 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Begg CB, Zabor EC, Bernstein JL, et al. A conceptual and methodological framework for investigating etiologic heterogeneity. Stat Med 2013; 32: 5039–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Begg CB, Seshan VE, Zabor EC, et al. Genomic investigation of etiologic heterogeneity: methodologic challenges. BMC Med Res Methodol 2014; 14: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Begg CB, Orlow I, Zabor EC, et al. Identifying Etiologically Distinct Sub-Types of Cancer: A Demonstration Project Involving Breast Cancer. Cancer Med 2015; 4: 1432–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang M, Kuchiba A, Ogino S. A Meta-Regression Method for Studying Etiological Heterogeneity Across Disease Subtypes Classified by Multiple Biomarkers. Am J Epidemiol 2015; 182: 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wang M, Spiegelman D, Kuchiba A, et al. Statistical methods for studying disease subtype heterogeneity. Stat Med 2016; 35: 782–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zabor EC, Begg CB. A comparison of statistical methods for the study of etiologic heterogeneity. Stat Med 2017; 36: 4050–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Nevo D, Nishihara R, Ogino S, et al. The competing risks Cox model with auxiliary case covariates under weaker missing-at-random cause of failure. Lifetime Data Anal 2018; 24: 425–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Liu L, Nevo D, Nishihara R, et al. Utility of inverse probability weighting in molecular pathological epidemiology. Eur J Epidemiol 2018; 33: 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Seaman SR, White IR. Review of inverse probability weighting for dealing with missing data. Stat Methods Med Res 2013; 22: 278–295. [DOI] [PubMed] [Google Scholar]
- 220.Cummings P Missing data and multiple imputation. JAMA Pediatr 2013; 167: 656–661. [DOI] [PubMed] [Google Scholar]
- 221.Phipps AI, Limburg PJ, Baron JA, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 2015; 148: 77–87 e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hamada T, Cao Y, Qian ZR, et al. Aspirin Use and Colorectal Cancer Survival According to Tumor CD274 (Programmed Cell Death 1 Ligand 1) Expression Status. J Clin Oncol 2017; 35: 1836–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ogino S, King EE, Beck AH, et al. Interdisciplinary education to integrate pathology and epidemiology: towards molecular and population-level health science. Am J Epidemiol 2012; 176: 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ogino S, Campbell PT, Nishihara R, et al. Proceedings of the second international molecular pathological epidemiology (MPE) meeting. Cancer Causes Control 2015; 26: 959–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Campbell PT, Rebbeck TR, Nishihara R, et al. Proceedings of the third international molecular pathological epidemiology (MPE) meeting. Cancer Causes Control 2017; 28: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kosumi K, Mima K, Baba H, et al. Dysbiosis of the gut microbiota and colorectal cancer: the key target of molecular pathological epidemiology. J Lab Precis Med 2018; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gronich N, Rennert G. Beyond aspirin-cancer prevention with statins, metformin and bisphosphonates. Nat Rev Clin Oncol 2013; 10: 625–642. [DOI] [PubMed] [Google Scholar]
- 228.Ogino S, Jhun I, Mata DA, et al. Integration of pharmacology, molecular pathology, and population data science to support precision gastrointestinal oncology. NPJ Precis Oncol 2017; 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Maier L, Typas A. Systematically investigating the impact of medication on the gut microbiome. Curr Opin Microbiol 2017; 39: 128–135. [DOI] [PubMed] [Google Scholar]
- 230.Jackson MA, Verdi S, Maxan ME, et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun 2018; 9: 2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Dong TS, Gupta A. Influence of Early Life, Diet, and the Environment on the Microbiome. Clin Gastroenterol Hepatol 2019. in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Song M, Chan AT. The Potential Role of Exercise and Nutrition in Harnessing the Immune System to Improve Colorectal Cancer Survival. Gastroenterology 2018; 155: 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zinocker MK, Lindseth IA. The Western Diet-Microbiome-Host Interaction and Its Role in Metabolic Disease. Nutrients 2018; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hadrich D Microbiome Research Is Becoming the Key to Better Understanding Health and Nutrition. Front Genet 2018; 9: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Song M, Chan AT. Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clin Gastroenterol Hepatol 2019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Chen L, He Z, Iuga AC, et al. Diet Modifies Colonic Microbiota and CD4(+) T-Cell Repertoire to Induce Flares of Colitis in Mice With Myeloid-Cell Expression of Interleukin 23. Gastroenterology 2018; 155: 1177–1191 e1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Levine A, Sigall Boneh R, Wine E. Evolving role of diet in the pathogenesis and treatment of inflammatory bowel diseases. Gut 2018; 67: 1726–1738. [DOI] [PubMed] [Google Scholar]
- 238.Song M, Garrett WS, Chan AT. Nutrients, foods, and colorectal cancer prevention. Gastroenterology 2015; 148: 1244–1260 e1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Petrosino JF. The microbiome in precision medicine: the way forward. Genome Med 2018; 10: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Osman MA, Neoh HM, Ab Mutalib NS, et al. 16S rRNA Gene Sequencing for Deciphering the Colorectal Cancer Gut Microbiome: Current Protocols and Workflows. Front Microbiol 2018; 9: 767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Fuks G, Elgart M, Amir A, et al. Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome 2018; 6: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Bullman S, Meyerson M, Kostic AD. Emerging Concepts and Technologies for the Discovery of Microorganisms Involved in Human Disease. Annu Rev Pathol 2017; 12: 217–244. [DOI] [PubMed] [Google Scholar]
- 244.Prakadan SM, Shalek AK, Weitz DA. Scaling by shrinking: empowering single-cell ‘omics’ with microfluidic devices. Nat Rev Genet 2017; 18: 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Finotello F, Eduati F. Multi-Omics Profiling of the Tumor Microenvironment: Paving the Way to Precision Immuno-Oncology. Front Oncol 2018; 8: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Nguyen A, Khoo WH, Moran I, et al. Single Cell RNA Sequencing of Rare Immune Cell Populations. Front Immunol 2018; 9: 1553. [DOI] [PMC free article] [PubMed] [Google Scholar]


