Significance
GGGGCC (G4C2) repeat expansion in the C9ORF72 gene is the most common genetic cause of both amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Several dysregulated downstream molecular pathways have been identified; however, it is not known which pathway is primarily responsible for neurodegeneration, and effective therapeutic approaches remain elusive. In cellular and animal models of C9ORF72-ALS/FTD, we found that partial inhibition of an overactivated DNA repair pathway, and of Ku80 in particular, suppresses a cell death pathway, suggesting a therapeutic approach in C9ORF72-ALS/FTD.
Keywords: ALS/FTD, C9ORF72, Drosophila, iPSC, DNA damage
Abstract
GGGGCC (G4C2) repeat expansion in C9ORF72 is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). One class of major pathogenic molecules in C9ORF72-ALS/FTD is dipeptide repeat proteins such as poly(GR), whose toxicity has been well documented in cellular and animal models. However, it is not known how poly(GR) toxicity can be alleviated, especially in patient neurons. Using Drosophila as a model system in an unbiased genetic screen, we identified a number of genetic modifiers of poly(GR) toxicity. Surprisingly, partial loss of function of Ku80, an essential DNA repair protein, suppressed poly(GR)-induced retinal degeneration in flies. Ku80 expression was greatly elevated in flies expressing poly(GR) and in C9ORF72 iPSC-derived patient neurons. As a result, the levels of phosphorylated ATM and P53 as well as other downstream proapoptotic proteins such as PUMA, Bax, and cleaved caspase-3 were all significantly increased in C9ORF72 patient neurons. The increase in the levels of Ku80 and some downstream signaling proteins was prevented by CRISPR-Cas9-mediated deletion of expanded G4C2 repeats. More importantly, partial loss of function of Ku80 in these neurons through CRISPR/Cas9-mediated ablation or small RNAs-mediated knockdown suppressed the apoptotic pathway. Thus, partial inhibition of the overactivated Ku80-dependent DNA repair pathway is a promising therapeutic approach in C9ORF72-ALS/FTD.
Frontotemporal dementia (FTD), the second most common form of presenile dementia, is caused by focal degeneration of the prefrontal and/or temporal cortex, resulting in changes in personality, social behaviors, and other executive functions (1). The notion that FTD and the motor neuron disease amyotrophic lateral sclerosis (ALS) are part of a spectrum disorder is supported in large part by the fact that mutations in a number of genes cause both clinical conditions (2). In particular, a GGGGCC (G4C2) repeat expansion in C9ORF72 is the most common genetic cause of both ALS and FTD (3, 4). Despite progress in understanding the pathogenic mechanisms of C9ORF72-ALS/FTD, effective therapies remain elusive.
One class of major pathogenic molecules associated to the C9ORF72 repeat expansion is dipeptide repeat (DPR) proteins synthesized from both sense and antisense repeat RNAs via repeat-associated non-AUG (RAN) translation or other mechanisms (5–7). Of all DPR proteins, only poly(glycine arginine) [poly(GR)] accumulation correlates well with neurodegeneration in C9ORF72-ALS/FTD patients (8, 9). Moreover, ectopic expression of poly(GR) in yeast, cultured mammalian cells, fruit flies, or mice induces cell death (10–16). Although multiple dysregulated molecular pathways have been identified in C9ORF72-ALS/FTD (17), it is not known which pathway is primarily responsible for disease initiation and thus can be targeted more effectively for therapeutic benefits.
Increased DNA damage is associated with many neurodegenerative diseases (18). Long-lived postmitotic neurons are especially susceptible to various forms of DNA damage, and the nonhomologous end joining (NHEJ) pathway has an important role in repair of double-strand DNA breaks. The Ku80/Ku70 heterodimer binds to broken DNA ends and initiates repair through the NHEJ pathway (19). In the DNA damage response pathway, ATM phosphorylation is an essential signaling event that results in the phosphorylation of other important proteins, such as P53 and the histone H2AX (20). In C9ORF72-ALS/FTD, there is an age-dependent increase in DNA damage in motor neurons derived from C9ORF72-induced pluripotent stem cells (iPSCs) (21). Moreover, elevated DNA damage is found in postmortem brain and spinal cord tissues of C9ORF72 patients (22, 23). However, it is unknown to what extent DNA damage contributes to neurodegeneration in C9ORF72-ALS/FTD.
In this study, we took advantage of the genetic power of Drosophila and the disease relevance of C9ORF72 patient-specific iPSC-derived neurons to identify and investigate modifier genes of poly(GR) toxicity. We reveal an unexpected detrimental role of overactivated DNA repair machinery, and of Ku80 in particular, in C9ORF72-ALS/FTD. These findings suggest that partial inhibition of Ku80 is a promising therapeutic approach in these disorders.
Results
Identification of Genetic Modifiers of Poly(GR) Toxicity in Drosophila.
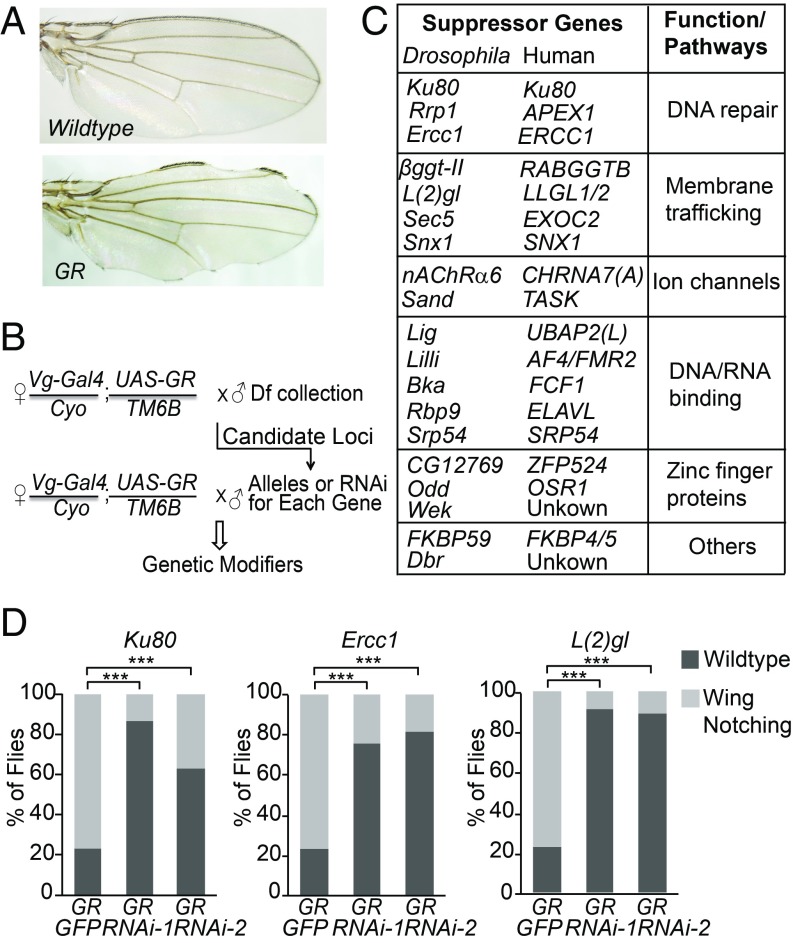
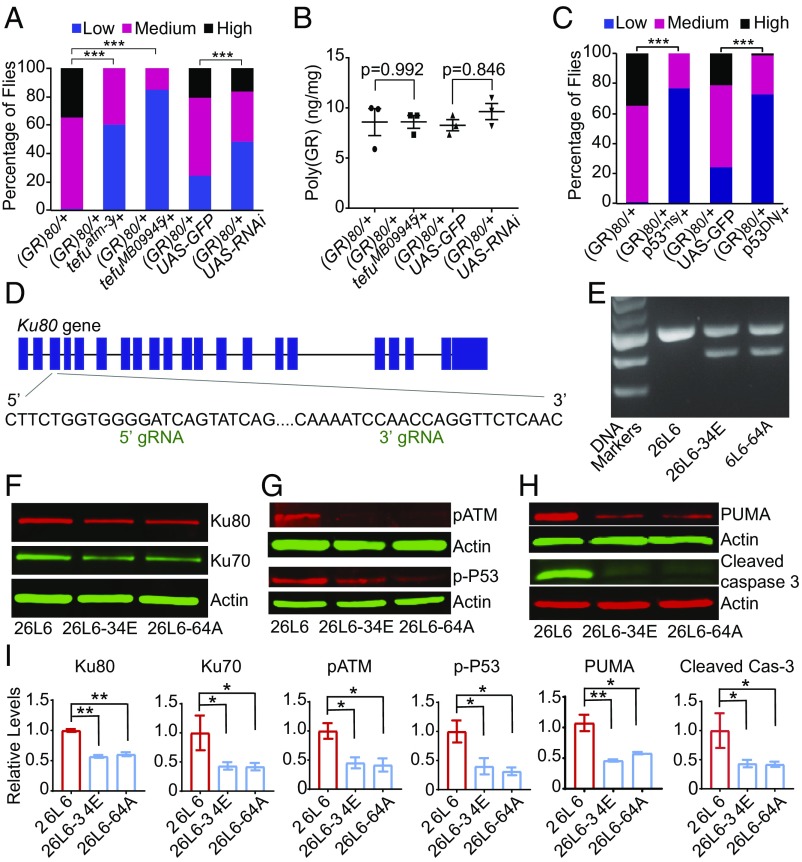
To identify genes whose partial loss of function may modify disease phenotypes in C9ORF72-ALS/FTD, we first took advantage of the powerful genetic approaches available in Drosophila, a model organism that has been widely used to understand various neurodegenerative diseases (24). We previously reported that Vg-Gal4-driven expression of (GR)80 in the wing caused a modest and quantifiable loss of nonneuronal cells (14). Using this phenotype as an assay (Fig. 1A), we performed an unbiased genetic screen and identified 20 suppressor and 15 enhancer deficiencies on the second chromosome (Fig. 1B and SI Appendix, Table S1). Using the genetic alleles and available RNAi lines targeting genes in some deficiency regions (SI Appendix, Table S2), we found 19 genes whose partial loss of function suppressed poly(GR) toxicity (Fig. 1 C and D and SI Appendix, Fig. S1). Interestingly, some of these genes function in the same pathway, such as DNA repair, membrane trafficking, and DNA/RNA binding (Fig. 1C), including Lig, the Drosophila homolog of UBAP2(L), a recently identified novel stress granule protein (25). Since DNA damage is increased in aged neurons derived from C9ORF72 iPSCs and in brain and spinal cord tissues of C9ORF72-ALS/FTD patients (21–23), it was surprising that partial loss of function in the key DNA repair gene Ku80 also suppressed poly(GR) toxicity. Ku80 directly binds to the ends of DNA double-strand breaks (19) and interacts with the RNA-binding protein RBM14 in the NHEJ DNA repair pathway (26) that has been implicated in a number of neurodegenerative diseases (18). This piece of genetic evidence raises the intriguing possibility that overactivation of this DNA repair pathway is detrimental in C9ORF72-ALS/FTD.
Fig. 1.
Identification of genetic suppressors of poly(GR) toxicity in Drosophila nonneuronal cells. (A) Representative images of adult fly wings of UAS-(GR)80/+ and Vg-Gal4/+; UAS-(GR)80/+ flies. (B) Schematic of primary and secondary genetic modifier screens. Df, deficiency lines. (C) Summary table of newly identified genes that suppress poly(GR) toxicity in Drosophila. (D) Quantification of the effects of some suppressor genes on poly(GR)-induced loss of nonneuronal cells. ***P < 0.001 by χ2 analysis. Flies (80–130) were examined for each genotype.
Elevated Ku80 Expression Contributes to Poly(GR)-Induced Neurodegeneration in Drosophila.
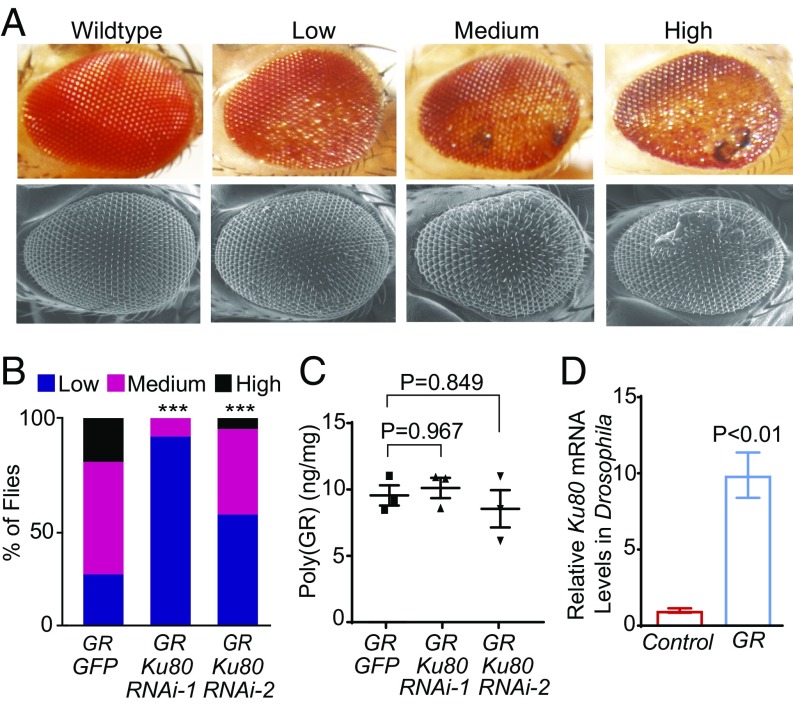
Although other nonneuronal cells such as yeast have also been used to investigate the toxicity and genetic modifiers of human disease proteins such as poly(GR) (10), it is important to confirm that partial loss of Ku80 also suppresses poly(GR) toxicity in neuronal cells. To this end, we first used the Drosophila photoreceptor neurons as a model system. (GR)80 expression driven by GMR-Gal4 caused a severe degenerative eye phenotype and a semilethal adult phenotype (14), making this model unsuitable for genetic screens. We therefore established a model in which temperature-sensitive Gal80 (Gal80ts) was included to reduce GMR-Gal4-driven poly(GR) expression. These flies were viable when grown at 23 °C and had a moderate, quantifiable degenerative eye phenotype (Fig. 2A). Ku80 expression was reduced by about 50% in the fly eye by expressing two different Ku80-specific RNAi (SI Appendix, Fig. S2A). Coexpression of Ku80 RNAi in the fly eye suppressed neurodegeneration caused by poly(GR) (Fig. 2B). To exclude the possibility that this genetic suppression was due to a reduced poly(GR) level, we measured poly(GR) levels in fly head lysates with an immunoassay (SI Appendix, Fig. S2B). The expression level of poly(GR) in the fly eye seemed to be overexpressed since its expression level was about 30-fold higher than in 2-mo-old C9ORF72 iPSC-derived motor neuron cultures (Fig. 2C and SI Appendix, Fig. S2C). We found that partial loss of Ku80 did not affect the level of poly(GR) (Fig. 2C). Thus, Ku80 functions downstream of poly(GR) toxicity and partial loss of Ku80 blocks poly(GR)-induced neuronal cell death. In exploring the potential mechanism, we found that poly(GR) expression in the eye greatly increased Ku80 expression (Fig. 2D). The Ku80 protein level was also significantly elevated in the Drosophila eye expressing 58 copies of G4C2 repeats (SI Appendix, Fig. S3), whose toxicity is likely in part due to poly(GR) synthesized from the repeat RNA (15). These results suggest that the DNA repair pathway, at least in the case of Ku80, is overactivated and that partial inhibition of Ku80 activity suppresses poly(GR)-induced neurodegeneration in the fly model.
Fig. 2.
Elevated Ku80 expression contributes to poly(GR) toxicity in Drosophila neuronal cells. (A) Representative images of Drosophila eye with various degrees of poly(GR) toxicity, as revealed by dissecting microscope (Top row) and scanning electron microscope (Bottom row). (B) Partial loss of Ku80 activity suppresses poly(GR)-induced retinal degeneration. ***P < 0.001 by χ2 analysis. (C) ELISA analysis of poly(GR) protein levels in the eyes of 1-wk-old flies. Values are mean ± SEM of three independent experiments, analyzed by one-way ANOVA and Tukey’s test. (D) Ku80 mRNA level is elevated by poly(GR) expression. Values are mean ± SD from three independent experiments, analyzed by Student’s t test.
Ku80 Expression Is Greatly Elevated in Aged C9ORF72 iPSC-Derived Motor Neurons.
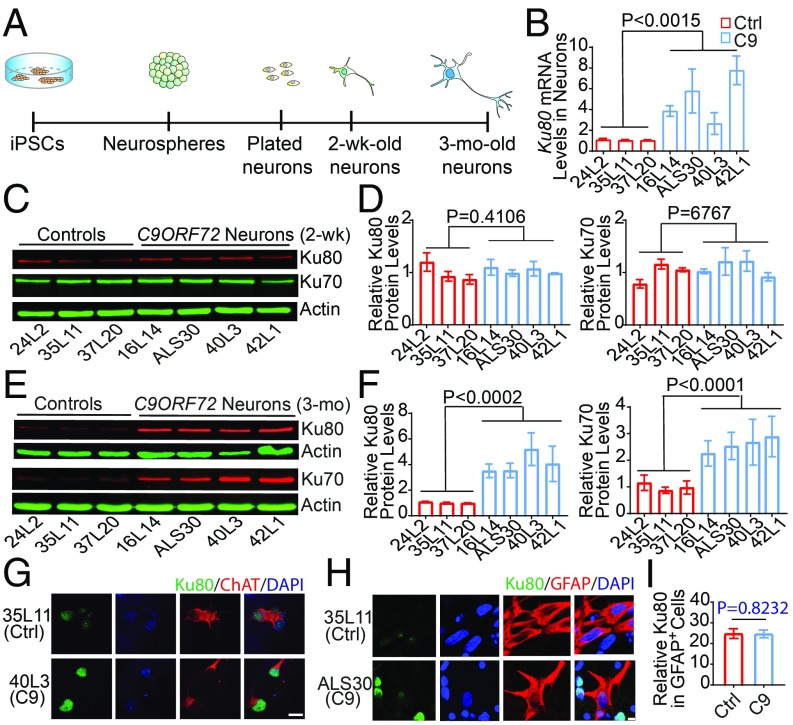
We differentiated iPSC lines derived from three control subjects and four C9ORF72 patients into motor neuron cultures as described (21) (Fig. 3A). The age of neurons was counted after plating neurospheres for terminal differentiation. The numbers of G4C2 repeats in lines 16L14, 40L3, 42L1, and ALS30 are ∼590, ∼1000, ∼1100, and ∼70, respectively (21, 27). The Ku80 mRNA level was much higher in 3-mo-old C9ORF72 motor neuron cultures than in control cultures (Fig. 3B), suggesting that Ku80 expression is up-regulated at the transcriptional level. Because we used iPSC lines from multiple control subjects and C9ORF72 patients, the difference in Ku80 mRNA levels is unlikely due to variability in iPSC differentiation. Nonetheless, to further confirm this finding, we did Western blot analysis of motor neurons derived from three independent differentiations. We found that the level of Ku80 protein was indeed consistently higher in 3-mo-old but not in 2-wk-old C9ORF72 motor neuron cultures than in control cultures (Fig. 3 C–F). This finding is consistent with the timeline of increased DNA damage in C9ORF72 motor neurons as we reported before (21). Interestingly, Ku70 also accumulated in 3-mo-old C9ORF72 motor neuron cultures (Fig. 3 C–F), likely because it forms a dimeric complex with Ku80 that binds to the ends of DNA double-strand breaks and initiates the NHEJ pathway of DNA repair (19). To demonstrate that Ku80 is specifically elevated in motor neurons, we did an immunostaining experiment, which showed increased Ku80 expression in ChAT-positive motor neurons (Fig. 3G), but not in GFAP-positive astrocytes (Fig. 3 H and I).
Fig. 3.
Ku80 levels are greatly elevated in C9ORF72 iPSC-derived motor neurons. (A) Schematic of the motor neuron differentiation. (B) Ku80 mRNA levels in control and C9ORF72 motor neurons. The average of Ku80 mRNA levels in all control neuron cultures was defined as 1. Values for each iPSC line are mean ± SEM from two independent differentiations. (C–F) Western blot analysis and quantification of Ku80 and Ku70 protein levels in 2-wk-old (C and D) and 3-mo-old (E and F) motor neurons. The average of Ku80 protein levels in all control neuron cultures was defined as 1. Values for each column are mean ± SEM from three independent differentiations. Two-tailed Student’s t test was used in B, D, and F to compare three control subjects and four C9ORF72 patients. (G) Increased level of Ku80 in C9ORF72 iPSC-derived ChAT-positive motor neurons. (Scale bar, 20 µm.) (H and I) Ku80 immunostaining (H) and quantification (40 cells per line) (I) in control and C9ORF72 astrocytes positive for glial fibrillary acidic protein (GFAP). Values are mean ± SEM analyzed by two-tailed Student’s t test. (Scale bar, 10 µm.)
Activation of a Proapoptotic Pathway in C9ORF72 iPSC-Derived Motor Neurons.
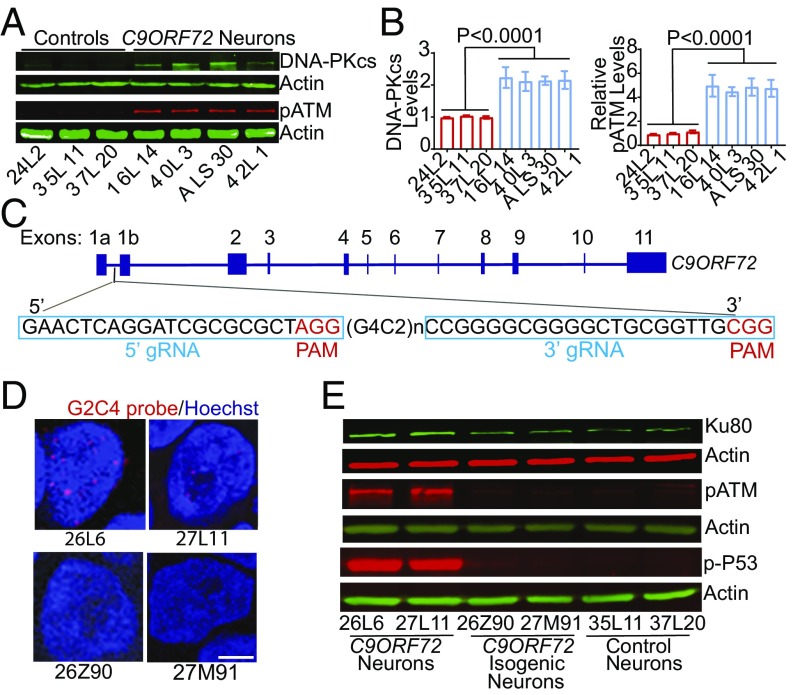
The Ku80/Ku70 dimer recruits other DNA repair proteins such as DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and through phosphorylation activates another important kinase, ataxia telangiectasia mutated (ATM) (19). Indeed, the levels of DNA-PKcs and phosphorylated ATM (pATM) in 3-mo-old C9ORF72 motor neuron cultures were substantially higher than of control subjects (Fig. 4A). The results from two (in the case of DNA-PKcs) or three (in the case of pATM) independent iPSC differentiations were quantified, and the averages of four C9ORF72 patients and three control subjects were compared for statistical analysis (Fig. 4B). This increase in pATM level is likely a direct consequence of elevated DNA damage, since treatment with doxorubicin, which induces DNA damage, increased the levels of pATM in both control and C9ORF72 motor neuron cultures (SI Appendix, Fig. S4).
Fig. 4.
Activation of the DNA damage pathway in C9ORF72 iPSC-derived neurons. (A) Western blot analysis of DNA-PKcs and pATM in control and C9ORF72 iPSC-derived motor neuron cultures. (B) DNA-PKcs protein levels from two independent differentiations and pATM protein levels from three independent differentiations. Values for each iPSC line are mean ± SEM. Two-tailed Student’s t test was used to compare three control subjects and four C9ORF72 patients. (C) Schematic representation of the CRISPR-Cas9 strategy to delete the expanded G4C2 repeats in C9ORF72 iPSC lines. (D) Nuclear RNA foci are present in 26L6 and 27L11, but not in 26Z90 and 27M91 iPSCs. (Scale bar, 5 μm.) (E) Western blot analysis of Ku80, pATM, and p-P53 in iPSC-derived motor neuron cultures.
To further confirm that expanded G4C2 repeats are the root cause of the increased expression of Ku80, we used CRISPR-Cas9 technology to delete expanded G4C2 repeats from iPSC lines derived from two C9ORF72 patients, 26L6 and 27L11 (28). In the two resulting isogenic iPSC lines, 26Z90 and 27M91, respectively (Fig. 4C), the deletion did not affect the expression of several stem cell markers or their karyotypes (SI Appendix, Fig. S5). As expected, nuclear RNA foci in C9ORF72 isogenic iPSC lines were no longer present (Fig. 4D). Moreover, the Ku80 level was similar in isogenic C9ORF72 motor neuron cultures and controls (Fig. 4E). The deletion also prevented increases in the levels of some signaling molecules downstream of Ku80 such as pATM and phosphorylated P53 (p-P53) in 3-mo-old C9ORF72 motor neuron cultures (Fig. 4E).
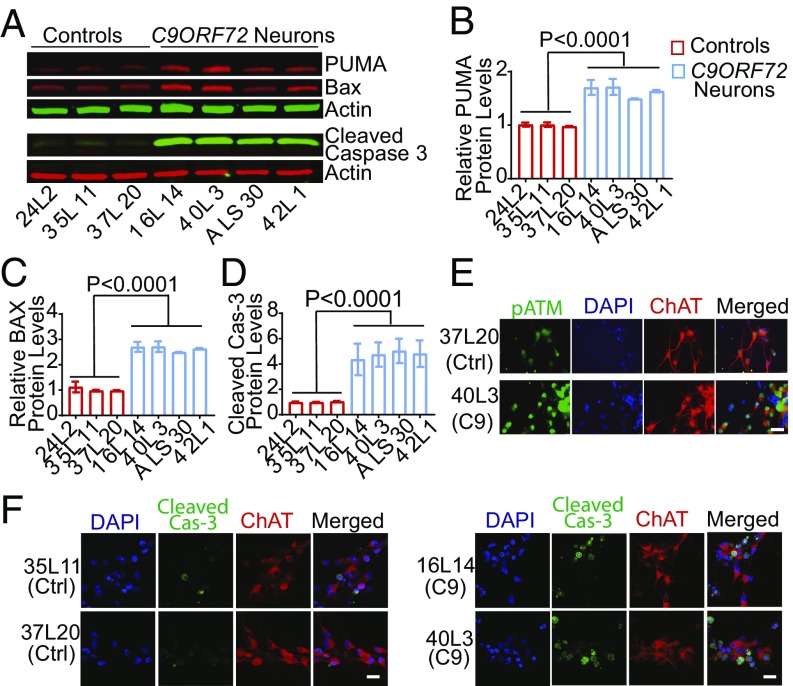
Persistent activation of ATM leads to P53 phosphorylation, which up-regulates proapoptotic proteins in the Bcl-2 gene family, such as PUMA and BAX (20). Accordingly, in C9ORF72 iPSC-derived motor neuron cultures, PUMA and BAX mRNA levels were higher than in control neurons (SI Appendix, Fig. S6 A and B). Similarly, PUMA and BAX levels were also elevated, as shown by Western blot analysis of C9ORF72 motor neuron cultures derived from three independent iPSC differentiations (Fig. 5 A–D). The increase of cleaved caspase-3 in C9ORF72 motor neuron cultures (Fig. 5A) indicates an activated proapoptotic pathway. Consistent with C9ORF72 motor neuron-specific increase in Ku80 (Fig. 3G), expression of pATM (Fig. 5E) and cleaved caspase-3 (Fig. 5F and SI Appendix, Fig. S6C) were increased in ChAT-positive C9ORF72 motor neurons as well.
Fig. 5.
Activation of the proapoptotic pathway in C9ORF72 iPSC-derived motor neurons. (A) Western blot analysis of apoptotic markers PUMA, BAX, and cleaved caspase 3 in iPSC-derived motor neuron cultures. (B–D) Levels of PUMA (B), Bax (C), and cleaved caspase-3 (D) protein in iPSC-derived motor neuron cultures from three independent differentiations. Values for each iPSC line are mean ± SEM. Two-tailed Student’s t test was used to compare three control subjects and four C9ORF72 patients. (E and F) Immunostaining analysis of increased pATM (E) and cleaved caspase 3 (F) levels in C9ORF72 iPSC-derived ChAT-positive motor neurons. (Scale bar, 20 μm.)
Genetic Inhibition of ATM and P53 Activities Rescues Poly(GR)-Induced Neurodegeneration in Drosophila.
To further show that DNA repair pathways contribute to neurodegeneration in C9ORF72-ALS/FTD, we first used a genetic null allele and a UAS-RNAi line to reduce the activity of the Drosophila homolog of ATM, telomere fusion (Tefu). These genetic manipulations suppressed poly(GR) toxicity in the eye (Fig. 6A), which was not due to reduced poly(GR) expression, as shown by MSD immunoassay (Fig. 6B). This result supports the notion that activation of DNA repair pathways has a key role in poly(GR) toxicity in vivo, even though the DNA-PK component of the Ku80/Ku70 complex is absent in Drosophila. Thus, Tefu activation is unlikely to act directly downstream of Ku80 signaling (29). ATM inhibition also rescues neurodegeneration in cellular and mouse models of Huntington disease (30). Because the level of p-P53 is elevated in aged C9ORF72 iPSC-derived motor neurons (21), we also tested the role of P53 in poly(GR) toxicity in Drosophila. Partial reduction of P53 activity suppressed retinal degeneration in poly(GR) flies (Fig. 6C), consistent with our earlier finding that loss of P53 activity also suppressed poly(GR) toxicity in Drosophila nonneuronal cells (21).
Fig. 6.
Genetic inhibition of the overactivated Ku80-dependent DNA repair pathway rescues neurodegeneration in C9ORF72-ALS/FTD. (A) Suppression of poly(GR) toxicity in the eye by partial loss of function of Tefu, the Drosophila homolog of ATM. ***P < 0.001 by χ2 analysis. Flies (80–130) were analyzed for each genotype. (B) ELISA analysis of poly(GR) protein level. Values are mean ± SEM of three independent experiments analyzed by one-way ANOVA and Tukey’s test. (C) Partial loss of p53 activity rescues poly(GR) toxicity in the fly eye. ***P < 0.001 by χ2 analysis. Flies (80–130) were analyzed for each genotype. (D) Schematic representation of CRISPR-Cas9 deletion of one copy of Ku80. (E) PCR analysis of Ku80 deletion. (F–H) Western blot analysis of the relative levels of several proteins in motor neurons derived from the parental C9ORF72 iPSC line 26L6 and its isogenic daughter lines 26L6-34E and 26L6-64A. (I) Quantification of Western blot analysis. Values are mean ± SEM of two independent differentiation experiments. *P < 0.05, **P < 0.01 by one-way ANOVA and Tukey’s test.
Partial Loss of Function of Ku80 Through CRISPR-Cas9 or Small RNA-Mediated Knockdown Suppresses Cell Death Pathway in C9ORF72 Patient Neurons.
Based on our unexpected findings in the fly model (Fig. 2), we hypothesize that partial loss of Ku80 activity in C9ORF72 iPSC-derived motor neurons also suppresses the apoptotic pathway. To test this hypothesis, we first used CRISPR-Cas9 technology to delete one copy of Ku80 in a C9ORF72 iPSC line (Fig. 6D); the deletion was confirmed by PCR analysis (Fig. 6E). The resulting isogenic iPSC lines still expressed the same levels of stem cell markers as the parental line (SI Appendix, Fig. S7 A and B) and had normal karyotypes (SI Appendix, Fig. S7C). As expected, Ku80 mRNA and Ku80 protein levels were reduced by ∼50% in two Ku80 heterozygous C9ORF72 iPSC lines (SI Appendix, Fig. S7 D and E). Moreover, CRISPR-Cas9 deletion of one copy of Ku80 did not affect the poly(GR) level in 2-mo-old neurons (SI Appendix, Fig. S7F). Ku80 protein levels were also reduced in human motor neurons derived from these two iPSC lines—both differentiated independently twice—as were the levels of Ku70, which forms a heterodimer with Ku80 (Fig. 6 F and I). However, the extent of DNA double-strand breaks measured by comet assay was not increased (SI Appendix, Fig. S8), suggesting that the remaining Ku80/Ku70 complex is sufficient to repair DNA damage. More importantly, partial loss of Ku80 significantly decreased the levels of pATM, p-P53, PUMA, and cleaved caspase 3 (Fig. 6 G–I)—evidence that excess Ku80 is causal to neuronal cell death and that partial loss of Ku80 suppresses the proapoptotic pathway regulated by ATM and P53 in C9ORF72 iPSC-derived neurons.
To further assess this possibility and to explore potential therapeutic approaches, we also used lentiviral short hairpin RNA (shRNA) (SI Appendix, Fig. S9A) and small self-deliverable interference RNA (sdRNA) (SI Appendix, Fig. S9G) to knock down Ku80 in C9ORF72 neurons from two independent differentiations. After a single administration, sdRNAs with lipophilic conjugates showed sustained, multimonth efficiency in vivo and are thus a promising therapeutic approach (31). Because Ku80 levels were about fourfold higher in C9ORF72 neuron cultures than in controls (Fig. 3F), its levels in C9ORF72 neurons after about 50% knockdown were still higher than that in control neurons. The small RNA-mediated knockdown of Ku80 also significantly inhibited the proapoptotic pathway in C9ORF72 neurons, as shown by reduced levels of pATM (SI Appendix, Fig. S9 B and E), PUMA (SI Appendix, Fig. S9 C, F, H, and K), and cleaved caspase 3 (SI Appendix, Fig. S9 I and L). The levels of these proteins in C9ORF72 neurons after Ku80 knockdown were estimated to be similar or higher than that in control neurons based on their relative levels (SI Appendix, Fig. S9 vs. Figs. 4 and 5). These results suggest that using small RNAs to target Ku80 is a potential therapeutic approach for C9ORF72-ALS/FTD.
Discussion
In this study, we performed an unbiased genetic screen and identified suppressor genes whose partial loss of function decreased poly(GR) toxicity in Drosophila. Increased DNA damage is a primary driver of neuronal cell death in many neurodegenerative diseases (18). Thus, it is thought that the DNA repair pathway is beneficial to neuronal survival. Paradoxically, our findings show that the Ku80 level is greatly elevated in flies expressing poly(GR) and that partial knockdown of Ku80 or P53 suppresses retinal degeneration, suggesting that the overactivated Ku80-dependent DNA repair pathway contributes to neurodegeneration in this fly model of C9ORF72-ALS/FTD. More importantly, partial reduction in Ku80 expression in C9ORF72 iPSC-derived patient neurons through multiple approaches, including small RNA-mediated knockdown and CRISPR-Cas9 deletion of one copy of Ku80, prevented the activation of a proapoptotic pathway. Thus, genes in the DNA repair and damage response pathways are potential therapeutic targets to reduce poly(GR) toxicity in C9ORF72-ALS/FTD.
Our genetic screen in Drosophila also identified other genes that suppress poly(GR) toxicity and whose protein products participate in DNA/RNA binding, membrane trafficking, signal transduction, and other molecular processes. For instance, partial loss of function of Lig, the Drosophila homolog of UBAP2(L), a stress granule protein (25), suppressed poly(GR) toxicity, consistent with the notion that dysregulated stress granule dynamics is another major pathogenic mechanism in C9ORF72-ALS/FTD (32, 33). These findings confirm the validity of our genetic screen in Drosophila and suggest that further investigation of other genetic suppressor genes identified here will provide additional insights into C9ORF72-ALS/FTD pathogenesis.
It is critically important that findings in Drosophila overexpression models of neurodegeneration be validated in more disease-relevant models such as patient-specific iPSC-derived neurons, where disease genes are expressed in their native genetic contexts (34). Therefore, we also examined the DNA repair pathway in C9ORF72 iPSC-derived neurons and found greatly increased expression of Ku80. This phenotype is a molecular consequence of expanded G4C2 repeats because it was prevented by CRISPR-Cas9-mediated deletion of the repeats in C9ORF72 iPSC-derived neurons (Fig. 4). Since poly(GR) alone induced Ku80 expression in flies (Fig. 2D) and can induce DNA damage in human control neurons (21), it is highly likely that endogenously expressed poly(GR) contributes to the increase in Ku80 expression and DNA damage in C9ORF72 iPSC-derived neurons. A central role for poly(GR) in C9ORF72-ALS/FTD pathogenesis is also supported by the positive correlation between poly(GR) pathology and neurodegeneration in multiple C9ORF72-ALS/FTD patients (8, 9).
More importantly, partial reduction of Ku80 activity in C9ORF72 iPSC-derived neurons through CRISPR/Cas9-mediated genetic ablation or sdRNA inhibition suppressed neuronal cell death, as indicated by reduced expression of several proapoptotic proteins (Fig. 6), and did so without increasing DNA double-strand breaks in C9ORF72 neurons (SI Appendix, Fig. S6). These findings indicate that overactivation of Ku80 is detrimental. DNA damage repair as well as many other molecular pathways including nucleocytoplasmic transport, nucleolar stress, and autophagy are dysregulated in C9ORF72-ALS/FTD; however, it remains to be determined which pathway is primarily responsible for neurodegeneration (17). These pathways might interact to promote disease progression. For instance, poly(GR) increases oxidative stress, which contributes to increased DNA damage in C9ORF72 iPSC-derived motor neurons (21). Oxidative damage may also compromise the function of long-lived nuclear pore proteins in postmitotic cells in an age-dependent manner (35). Importantly, nuclear pore complexes (NPCs) have a key role in maintaining genomic integrity (36). Despite this complexity, our results suggest that an overactivated Ku80-dependent DNA repair pathway is a key contributing factor in C9ORF72-ALS/FTD. Thus, proteins in this pathway, and Ku80 in particular, are promising molecular targets for therapeutic intervention.
Materials and Methods
Drosophila Strains and Genetics.
Flies were raised at 25 °C or 23 °C. GMR-Gal4, Tub-Gal80ts, deficiency stocks, mutant and RNAi lines were from the Bloomington Drosophila Stock Center or Vienna Drosophila RNAi Center. The Vg-Gal4/CyO; UAS-(GR)80/TM6,Tb fly, GMR-Gal4, Tub-Gal80ts,UAS-(GR)80/CyO and GMR-Gal4, Tub-GAL80ts/CyO; UAS-(GR)80-Control/TM6B lines were generated in our laboratory. The UAS-(G4C2)58 line and the GMR-Gal4/CyO; UAS-(G4C2)58/TM6B,Tb line were reported previously (15).
Genetic Modifier Screen.
A genetic modifier screen was carried out by crossing Vg-Gal4/CyO; UAS-(GR)80/TM6B flies to each of 169 chromosome deficiency lines. Adult (3–5 d old) F1 Vg-Gal4/Df; UAS-(GR)80/+ flies were examined with a dissecting microscope as described (14). The modifier genes were further recovered by using genetic mutant alleles and/or RNAi lines of candidate genes within some deficiency regions.
Drosophila Eye Phenotype Analysis.
GMR-Gal4, UAS-(GR)80, Gal80ts/CyO flies were crossed to different lines at 23 °C. Five-day-old adult F1 flies were examined with a dissecting microscope or a scanning electron microscope (FEI Quanta 200 FEG MKII, Field Electron and Ion Company). Adult flies were separated into four groups based on the severity of the rough eye phenotype: wild type, low, medium, and high. The percentage of flies in each category was calculated. The results were analyzed by χ2 test.
Deletion of the C9ORF72 Expanded Repeats by CRISPR-Cas9.
The CRISPR-Cas9 system was used to delete expanded G4C2 repeats in two iPSC lines generated from two C9ORF72 patients. Four guide RNAs (gRNAs) were used for CRISPR-Cas9 deletion of the repeats. For the upstream target, oligos are 5′-CACCGAACTCAGGAGTCGCGCGCT-3′ and 5′-AAACAGCGCGCGACTCCTGAGTTC-3′ and the downstream target oligos are 5′-CACCGCGGGGCGGGGCTGCGGTTG-3′ and 5′-AAACCAACCGCAGCCCCGCCCCGC-3′. The oligos were individually cloned into the BbsI-digested plasmid px330 plasmid #42230 (Addgene) that contains both SpCas9 nuclease and the gRNA. To transfect the plasmid, 1 × 106 iPSCs were dissociated into single cells with Accutase (Life Technologies) and nucleofected using the Amaxa Human Stem Cell Nucleofector Kit 2 (Lonza) with program A-23. After electroporation, cells were plated with ROCK inhibitor at clonal density to derive individual lines.
Generation of Ku80 Heterozygous Lines by CRISPR/Cas9.
The CRISPR-Cas9 system was used to delete one copy of Ku80 in a C9ORF72 iPSC line (26L6). The Invitrogen Neon System was used to transfect the plasmid (PX459, Addgene) containing one of two gRNAs (SI Appendix, Table S3). One microgram of the two gRNA-Cas9 plasmids were electroporated into 0.5 × 106 iPSCs with 1 at 1050 V (30 ms, two pulses). Then we plated single cells by serial dilution on 96-well plates to derive individual lines. Quality control assays for newly generated isogenic lines with CRISPR/Cas9-mediated deletion of expanded G4C2 repeats or Ku80 are summarized in SI Appendix, Table S4).
Analysis of Poly(GR) by ELISA.
A poly(GR) sandwich immunoassay was established by using newly generated, affinity-purified polyclonal rabbit antibody against (GR)8. For the assay, iPSC-derived neurons or ∼25–30 frozen heads of 1-wk-old flies were homogenized in Tris-lysis buffer and sonicated followed by centrifugation at 16,000× g for 15 min at 4 °C. The supernatant was collected in a fresh tube. The protein concentration of the lysates was determined with the BCA protein assay (Pierce). Lysates were diluted to the same concentration (50 µg/well) with Tris-buffered saline (TBS) and tested in duplicate wells. Immunoassay from Meso Scale Discovery was used to measure Poly(GR) levels.
Ku80 Knockdown with Lentiviral shRNA and sdRNA.
Human C9ORF72 iPSC-derived motor neuron cultures were treated with control or Ku80 sdRNAs. Three-month-old neurons were treated for 3 wk by adding 1.5 μM of sdRNA directly to the medium every 5 d, and RNA proteins were isolated for further analysis. Lentiviral particles expressing Ku80 shRNA from TRC (N0000039838) were generated and transduced into cultures of C9ORF72 iPSC-derived motor neurons at a multiplicity of infection of 10. After 3 wk, proteins were isolated to verify knockdown efficiency.
Motor Neuron Differentiation from iPSC Lines.
We used one iPSC line per donor. Clinical information and quality control assays are summarized in SI Appendix, Table S5). These iPSCs were only a few passages after the initial karyotyping analysis was done. All of the control and C9ORF72 iPSC lines as well as CRISPR-Cas9 modified isogenic lines used in this study were tested for quality control, such as qPCR-based genetic analysis and Western blot analysis of phosphorylated P53. Moreover, short tandem repeat analysis was performed for parental C9ORF72 iPSC lines and newly generated CRISPR-Cas9-modified isogenic iPSC lines. All these analyses did not detect any abnormalities (SI Appendix, Figs. S10 and S11). Motor neurons were differentiated as reported (21) and also as described in the SI Appendix.
Other Methods.
Western blot analysis (including all Western blot raw data in SI Appendix, Fig. S12), immunostaining, comet assay, RNA extraction and quantitative real-time PCR, as well as genomic analyses of iPSC lines are described in the SI Appendix.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center for some fly lines. This work was supported by grants from the NIH [R37NS057553, R01NS101986, and R01NS093097 (to F.-B.G.) and UL1TR000124, R01AG035610, P30NS062691 (to G.C.)], the Muscular Dystrophy Association, Packard Center for ALS Research and Target ALS (to F.-B.G.), the John Douglas French Alzheimer’s Foundation (to G.C.), Alzheimer’s Association [2018-AARFD-592264 (to R.L.-G.)], and ALS Association [16-11P-246 (to D.Y.)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901313116/-/DCSupplemental.
References
- 1.Olney NT, Spina S, Miller BL. Frontotemporal dementia. Neurol Clin. 2017;35:339–374. doi: 10.1016/j.ncl.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen HP, Van Broeckhoven C, van der Zee J. ALS genes in the genomic era and their implications for FTD. Trends Genet. 2018;34:404–423. doi: 10.1016/j.tig.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 3.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renton AE, et al. ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 6.Ash PE, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zu T, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saberi S, et al. Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol. 2018;135:459–474. doi: 10.1007/s00401-017-1793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakae N, et al. Poly-GR dipeptide repeat polymers correlate with neurodegeneration and Clinicopathological subtypes in C9ORF72-related brain disease. Acta Neuropathol Commun. 2018;6:63. doi: 10.1186/s40478-018-0564-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jovičić A, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon I, et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen X, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizielinska S, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang D, et al. FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathol. 2015;130:525–535. doi: 10.1007/s00401-015-1448-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freibaum BD, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang YJ, et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat Med. 2018;24:1136–1142. doi: 10.1038/s41591-018-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao FB, Almeida S, Lopez-Gonzalez R. Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder. EMBO J. 2017;36:2931–2950. doi: 10.15252/embj.201797568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madabhushi R, Pan L, Tsai LH. DNA damage and its links to neurodegeneration. Neuron. 2014;83:266–282. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rulten SL, Grundy GJ. Non-homologous end joining: Common interaction sites and exchange of multiple factors in the DNA repair process. BioEssays. January, 30 2017 doi: 10.1002/bies.201600209. [DOI] [PubMed] [Google Scholar]
- 20.Shiloh Y, Ziv Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 21.Lopez-Gonzalez R, et al. Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron. 2016;92:383–391. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farg MA, Konopka A, Soo KY, Ito D, Atkin JD. The DNA damage response (DDR) is induced by the C9orf72 repeat expansion in amyotrophic lateral sclerosis. Hum Mol Genet. 2017;26:2882–2896. doi: 10.1093/hmg/ddx170. [DOI] [PubMed] [Google Scholar]
- 23.Walker C, et al. C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat Neurosci. 2017;20:1225–1235. doi: 10.1038/nn.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGurk L, Berson A, Bonini NM. Drosophila as an in vivo model for human neurodegenerative disease. Genetics. 2015;201:377–402. doi: 10.1534/genetics.115.179457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markmiller S, et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172:590–604.e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kai M. Roles of RNA-binding proteins in DNA damage response. Int J Mol Sci. 2016;17:310, and erratum (2016) 17:E604. doi: 10.3390/ijms17040604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sareen D, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almeida S, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekelsky J. DNA repair in Drosophila: Mutagens, models, and missing genes. Genetics. 2017;205:471–490. doi: 10.1534/genetics.116.186759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu XH, et al. Targeting ATM ameliorates mutant Huntingtin toxicity in cell and animal models of Huntington’s disease. Sci Transl Med. 2014;6:268ra178. doi: 10.1126/scitranslmed.3010523. [DOI] [PubMed] [Google Scholar]
- 31.Osborn MF, Khvorova A. Improving siRNA delivery in vivo through lipid conjugation. Nucleic Acid Ther. 2018;28:128–136. doi: 10.1089/nat.2018.0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee KH, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–788.e17. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boeynaems S, et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell. 2017;65:1044–1055.e5. doi: 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuva-Aydemir Y, Almeida S, Gao FB. Insights into C9ORF72-related ALS/FTD from Drosophila and iPSC models. Trends Neurosci. 2018;41:457–469. doi: 10.1016/j.tins.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Géli V, Lisby M. Recombinational DNA repair is regulated by compartmentalization of DNA lesions at the nuclear pore complex. BioEssays. 2015;37:1287–1292. doi: 10.1002/bies.201500084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.