Abstract
The Million Veteran Program (MVP) was established in 2011 as a national research initiative to determine how genetic variation influences the health of U.S. military veterans. We genotyped 312,571 MVP participants using a custom biobank array and linked the genetic data to laboratory and clinical phenotypes extracted from electronic health records covering a median of 10.0 years of follow-up. Among 297,626 veterans with at least 1 blood lipid measurement including 57,332 blacks and 24,743 Hispanics, we tested up to ~32 million variants for association with lipid levels and identified 118 novel genome-wide significant loci after meta-analysis with data from the Global Lipids Genetics Consortium (total N > 600,000). Through a focus on mutations predicted to result in a loss of gene function and a phenome-wide association study, we propose novel indications for pharmaceutical inhibitors targeting PCSK9 (abdominal aortic aneurysm), ANGPTL4 (type 2 diabetes), and PDE3B (triglycerides and coronary disease).
Keywords: Lipids, population genetics, genome-wide association studies, coronary artery disease
Introduction
Large-scale biobanks offer the potential to link genes to health traits documented in electronic health records (EHR) with unprecedented power1. In turn, these discoveries are expected to improve our understanding of the etiology of common and complex diseases as well as our ability to treat and prevent these conditions. To this end, the Million Veteran Program (MVP) was established in 2011 by the Veteran Affairs (VA) Office of Research and Development as a nationwide research program within the VA healthcare system2. The overarching goal of MVP is to reveal new biologic insights and clinical associations broadly relevant to human health and to enhance the care of veterans (former U.S. military personnel) through precision medicine.
Blood concentrations of low-density lipoprotein cholesterol (LDL-C), triglycerides, total cholesterol, and high-density lipoprotein cholesterol (HDL-C) are heritable risk factors for atherosclerotic cardiovascular disease3, a highly prevalent condition among U.S. veterans. Genome-wide association studies (GWAS) to date have identified at least 268 loci that influence these levels4-12, many of which are under investigation as potential therapeutic targets13,14. However, off-target effects have dampened enthusiasm for some of these molecules15,16. Understanding the full spectrum of clinical consequences of a genetic variant through phenome-wide association scanning (“PheWAS”17) may shed light on potential unintended effects as well as novel therapeutic indications for some of these molecules.
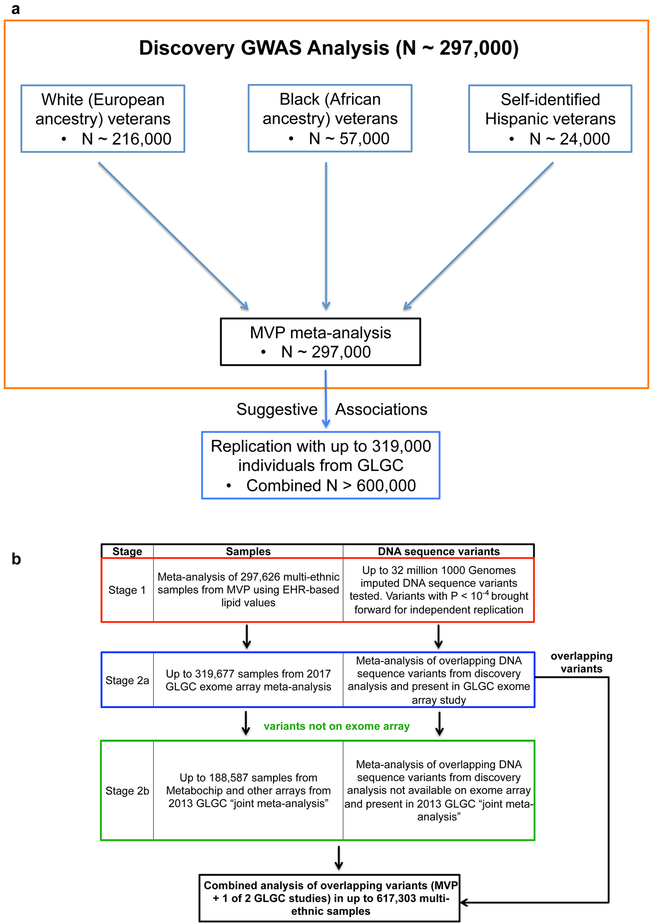
We first performed a GWAS including a discovery phase in MVP and a replication phase in the Global Lipids Genetics Consortium (GLGC) (Fig. 1). In the discovery phase (Stage 1), we performed association testing among 297,626 white (European ancestry), black (African ancestry), and Hispanic MVP participants with blood lipids stratified by ethnicity followed by a meta-analysis of results across all three groups. Replication of MVP findings was conducted in Stages 2a or 2b with data from either one of two independent studies from the GLGC. Next, we leveraged the results of our discovery and meta-analysis to i. estimate the variance explained by known and newly discovered lipid loci, ii. assess the potential of the use of multiple lipid measurements in discovery within MVP, iii. perform a transcriptome-wide association study (TWAS), a competitive gene-set pathway analysis, and a tissue-expression analysis. We then focused on novel, genome-wide lipid-associated, low-frequency missense variants unique to our non-European populations as well as predicted loss of gene function (pLoF) mutations across all ethnic groups, as these associations have revealed target pathways for pharmacologic inactivation and modulation of cardiovascular risk14,18,19. Lastly, we performed a PheWAS for a set of DNA sequence variants within genes that have already emerged as therapeutic targets for lipid modulation, leveraging the full catalog of ICD-9 diagnosis codes in the VA EHR to better understand the potential consequences of pharmacologic modulation of these genes or their products. We followed up significant findings from our PheWAS with multivariate Mendelian randomization analyses.
Figure 1. GWAS Study Design.
a) DNA sequence variants across 3 separate ancestry groups in the Million Veteran Program were meta-analyzed using an inverse-variance weighted fixed effects method in the discovery phase (Stage 1). Variants with suggestive association were then brought forward for independent replication.
b) DNA sequence variants with suggestive association (two-sided linear regression P < 10−4) in discovery (Stage 1) were brought forward for independent replication and tested using summary statistics from the 2017 exome-array focused GLGC meta-analysis (Stage 2a). Only variants with suggestive association in Stage 1 that were not present in the GLGC 2017 exome-array study (Stage 2a) were alternatively replicated in the 2013 GLGC “joint meta-analysis” (Stage 2b).
Abbreviations: MVP, Million Veteran Program; GWAS, genome-wide association study; EHR, electronic health record; GLGC, Global Lipids Genetics Consortium
Results
Demographics of Genotyped MVP Participants
A total of 353,323 veterans had genetic data available in MVP, with clinical phenotypes recorded in the VA EHR over 3,088,030 patient-years prior to enrollment (median of 10.0 years per participant) and 61,747,974 distinct clinical encounters (median of 99 per participant). We categorized veterans into three mutually exclusive ancestral groups for association analysis: 1) non-Hispanic whites, 2) non-Hispanic blacks, and 3) Hispanics. Admixture plots depicting the genetic background of the black and Hispanic groups are shown in Supplementary Figures 1 and 2. Demographics and participant counts for a number of cardiometabolic traits for the 312,571 white, black, and Hispanic MVP participants that passed our quality control are depicted in Table 1.
Table 1.
Demographic and clinical characteristics of black, white, and Hispanic individuals passing quality control in the Million Veteran Program
| Basic Demographics | Genotyped Veterans |
|---|---|
| N | 312,571 |
| Age at Enrollment ± SD, years | 62.4 ± 13.5 |
| Male, n (%) | 287,441 (92.0%) |
| Body Mass Index ± SD, kg/m2 | 30.3 ± 6.0 |
| Current Smoker, n (%) | 59,385 (19.0%) |
| Former Smoker, n (%) | 159,459 (51.0%) |
| N with ≥ 1 Measurement of Plasma Lipids, (%) | 297,626 (95.2%) |
| Number of Lipid Measurements, (Median Per Lipid Fraction) | 15,456,328 (12) |
| Race/Ethnicity | |
| Black, n (%) | 59,007 (18.9%) |
| White, n (%) | 227,817 (72.8%) |
| Hispanic, n (%) | 25,747 (8.1%) |
| Cardiometabolic Disease at Enrollment* | |
| Coronary Artery Disease, n (%) | 67,912 (21.7%) |
| Type 2 Diabetes, n (%) | 92,079 (29.5%) |
| Peripheral Artery Disease, n (%) | 21,418 (6.9%) |
| Abdominal Aortic Aneurysm, n (%) | 5,618 (1.8%) |
| Deep Venous Thrombosis or Pulmonary Embolism, n (%) | 7,009 (2.2%) |
Diseases are defined by International Classification of Disease, Ninth Edition (ICD-9) diagnosis codes.
Abbreviations: SD, Standard Deviation
A subset of 297,626 participants passing quality control had at least 1 laboratory measurement of blood lipids in their EHR. These individuals collectively had a total of 15,456,328 lab entries for blood lipids, or a median of 12 measures per lipid fraction per participant. To minimize potential confounding from the use of lipid-altering agents with variable adherence, we selected a participant’s maximum LDL-C, triglycerides, and total cholesterol as well as his or her minimum HDL-C for genetic association analysis20. Table 2 summarizes characteristics at enrollment and the distribution lipid levels for MVP participants included in our analysis. As expected, participants were largely male but 28% were of non-European ancestry. While approximately 45% had evidence of a statin prescription at the time of enrollment, only 8 to 9% participants had such evidence at the time of their maximum LDL-C or total cholesterol measurement used for our GWAS analysis.
Table 2.
Demographic and clinical characteristics for 297,626 veterans in the Million Veteran Program lipids analysis
| White | Black | Hispanic | |
|---|---|---|---|
| Veterans, N (%) | 215,551 (72.4%) | 57,332 (19.3%) | 24,743 (8.3%) |
| Age at Enrollment ± SD, years | 64.2 ± 13 | 57.7 ± 11.8 | 56.3 ± 15.0 |
| Male, n (%) | 200,900 (93.2%) | 50,059 (87.3%) | 22,601 (91.3%) |
| Body Mass Index ± SD, kg/m2 | 30.1 ± 5.9 | 30.4 ± 6.3 | 30.7 ± 5.8 |
| Statin Therapy Prescription at Enrollment, n (%) | 100,024 (46.4%) | 23,302 (40.6%) | 9,646 (39.0%) |
| Statin Therapy Prescription at time of Max LDL-C Blood Draw, n (%) | 18,818 (8.7%) | 5,024 (8.8%) | 2,262 (9.1%) |
| Statin Therapy Prescription at time of Max TC Blood Draw, n (%) | 18,433 (8.6%) | 5,027 (8.8%) | 2,162 (8.7%) |
| Mean Min HDL-C ± SD, mg/dL | 36.2 ± 11.4 | 38.9 ± 12.8 | 36.4 ± 11.0 |
| Mean Max LDL-C ± SD, mg/dL | 139 ± 38.4 | 142.2 ± 40.7 | 141.3 ± 38.1 |
| Median Max TG ± IQR, mg/dL | 211 ± 174 | 179 ± 149 | 221 ± 184 |
| Mean Max TC ± SD, mg/dL | 218.6 ± 46.7 | 220.8 ± 47.2 | 221.9 ± 48.0 |
| Variants Included in Analysis | 19,342,852 | 31,448,849 | 30,455,745 |
Abbreviations: Min, Minimum; Max, Maximum; SD, Standard Deviation; HDL-C, High-Density Lipoprotein Cholesterol; LDL-C, Low-Density Lipoprotein Cholesterol; TG, Triglycerides; TC, Total Cholesterol
Lipid Genetic Association and Conditional Analyses
We successfully imputed [INFO > 0.3, minor allele frequency (MAF) > 0.0003] 19.3, 31.4, and 30.4 million variants in white, black, and Hispanic veterans, respectively, using the 1000 Genomes Project21 reference panel (Table 2). Black and Hispanic participants had substantially more variants available for analysis, reflecting the known greater genetic diversity within these populations21,22. We also identified 6,657 pLoF variants in 4,294 genes across the three ethnicities (Supplementary Fig. 3).
We compared the Z scores and effect estimates from the published literature with those observed in MVP for 444 previously reported11 exome-wide significant variants for lipids. We found a strong correlation of genetic associations across all four traits, validating the lipid data secured through the EHR (Supplementary Fig. 4, 5).
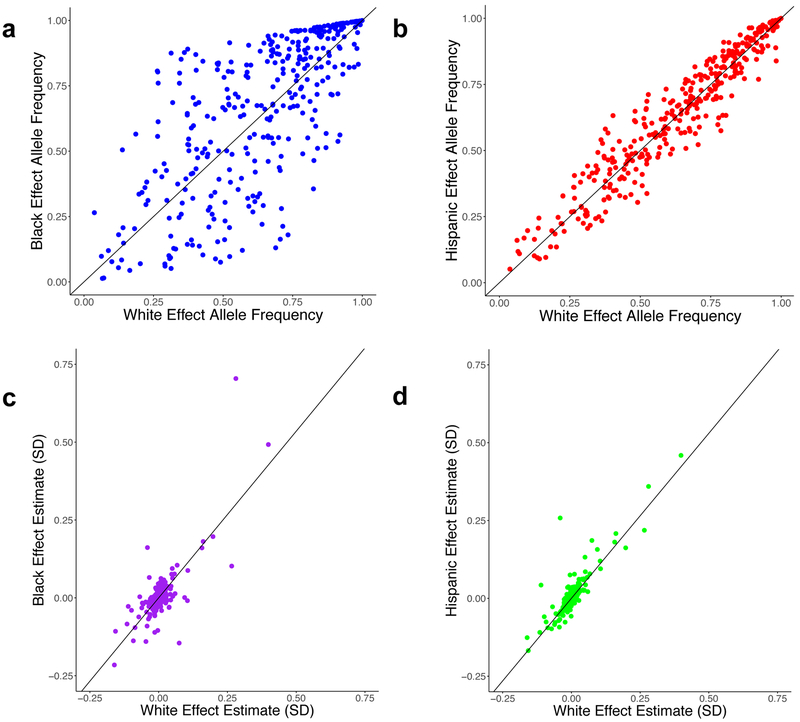
We performed association testing separately among individuals of each of three ancestries (whites, blacks, and Hispanics) in our initial discovery analysis and then meta-analyzed results across ancestry groups using an inverse variance-weighted fixed effects method (Fig. 1a, Supplementary Fig. 6). Following trans-ethnic meta-analysis in the discovery phase of our study (Stage 1), a total of 46,526 variants at 188 of the 268 known loci for lipids met the genome-wide significance threshold (P < 5×10−8) (Supplementary Tables 1-4). We performed pairwise comparisons of the allele frequencies and effect estimates between whites and blacks as well as between whites and Hispanics for 354 of the 444 previously established independent variants for lipids which were well imputed in all three ancestral groups in MVP (Fig. 2)11. We observed a much stronger correlation between white and Hispanic effect allele frequencies (Pearson correlation coefficient R = 0.96) than between whites and blacks (R = 0.72), likely reflecting the greater European admixture in the MVP Hispanic participants. The effect estimates among the three ethnicities varied by lipid trait (Fig. 2, Supplementary Fig. 7).
Figure 2. Comparison of 354 Independent Lipid Associated Variants Across Ethnicities.
Allele frequencies observed in white individuals (n=215,196; x-axes) compared to black (a, n=57,280; R = 0.72,) or Hispanic (b, n=24,742; R = 0.96) individuals for lipid-associated variants are shown. Effect estimates for LDL-C association in white individuals (n = 215,196; x-axes) compared to black (c, n = 57,280; β = 1.07) or Hispanic (d, n = 24,742; β = 1.06) individuals are also depicted.
Abbreviations: SD, Standard Deviations; LDL-C, Low-Density Lipoprotein Cholesterol; R = Pearson correlation coefficient
We sought replication for variants within MVP with suggestive associations (P < 1×10−4) in either Stages 2a or 2b (Fig. 1b). We first attempted replication of these variants using summary statistics from the 2017 GLGC exome array meta-analysis (Stage 2a)11. If association statistics for promising DNA sequence variants from Stage 1 were not available for replication in the 2017 exome array-focused study, we sought replication of these variants in publicly available summary statistics from the 2013 GLGC “joint meta-analysis” (Stage 2b). We did not attempt replication of any variant in both studies given the substantial overlap of participants in these two studies. A total of 170,925 variants demonstrated suggestive association (P<10−4) in the MVP discovery analysis. Among these variants, 39,663 were also available for in silico replication in either Stage 2a (GLGC 2017) or Stage 2b (GLGC 2013). We defined significant novel associations as those that were at least nominally significant in replication (P<0.05) with consistent direction of effect and had an overall P < 5×10−8 (genome-wide significance) in the discovery and replication cohorts combined. Following replication, 118 novel loci (from 142 lead variants) exceeded genome-wide significance (P < 5×10−8, Supplementary Tables 5-8). MAF of lead variants ranged from 0.08% to 49.9%, with effect sizes ranging from 0.01 to 0.243 standard deviations. For example, carriers of a rare missense mutation in the gene encoding Sorting Nexin-8 [SNX8 p.Ile414Thr, (rs144787122, NC_000007.13:g.2296552A>G) MAF = 0.35% in MVP] demonstrated a 0.10 standard deviation (3.8 mg/dL) higher plasma LDL-C after testing in 587,481 individuals.
More than one variant may independently affect plasma lipid levels at any given genetic locus. We performed a conditional analysis using combined summary statistics from MVP and publicly available data from GLGC for each lipid trait (Supplementary Fig. 8) and identified a total of 826 independently associated lipid variants across 118 novel and 268 previously identified loci (Supplementary Table 9).
Variance Explained Using Multiple Lipid Measurements
The previously mapped 444 lipid variants explain about 7.5-10.5% of the phenotypic variance in lipid levels in the MVP population. The 118 novel loci in our study explain an additional 0.38-0.74% in phenotypic variance, and the 826 independent variants identified in our conditional analysis increase the overall phenotypic variance explained to 8.8-12.3% (Supplementary Table 10).
We subsequently explored the impact of multiple lipid measurements in an analysis restricted to 171,314 European MVP participants with ≥ 5 lipid measurements in their EHR. We constructed a weighted genetic risk score (GRS) of 223 variants across 268 of the previously mapped loci with effect estimates available in the 2017 GLGC exome array analysis summary statistics (Supplementary Table 11)11. Generally across the four lipid traits, the GRS explained a larger proportion of the phenotypic variance with an increasing number of lipid measurements included in the analysis (Supplementary Table 12). In addition, when the maximal/minimal lipid values were used as in our discovery GWAS, the GRS explained more total variance than when using up to 5 lipid measurements for the LDL-C, triglycerides, and total cholesterol phenotypes.
Transcriptome-wide Association Study
We next performed a TWAS23 using: 1) pre-computed weights from expression array data measured in peripheral blood from 1,245 unrelated control individuals from the Netherlands Twin Registry (NTR)24, RNA-seq data measured in adipose tissue from 563 control individuals from the Metabolic Syndrome in Men study (METSIM)23, and RNA-seq data from post-mortem liver (97 individuals) and tibial artery (285 individuals) tissue from the Genotype-Tissue Expression project25 (GTEx V6), and 2) combined MVP and GLGC summary statistics for each of the four lipid traits (Supplementary Fig. 8). Briefly, this approach integrates information from expression reference panels (variant–expression correlation), GWAS summary statistics (variant–trait correlation), and linkage disequilibrium (LD) reference panels (variant–variant correlation) to assess the association between the cis-genetic component of expression and phenotype23. The results yield candidate causal genes from the GWAS results under the assumption that the causal mechanism of the tested genes involves changes in cis-expression.
Our TWAS identified a total of 655 genome-wide significant (P < 5×10−8) gene-lipid associations (summed across expression reference panels) in 333 distinct genes, including 194 that were significant in more than one tissue or lipid trait (Supplementary Tables 13-16, Supplementary Fig. 9-10). The 333 distinct genes fell within 122 genomic loci, 117 of which were within a lipid GWAS region (± 1mB around a mapped sentinel GWAS variant) identified in either a prior analysis or in the current study. However, 5 genes identified with TWAS fell outside of previously mapped GWAS regions, representing potentially novel genomic loci for lipids (Supplementary Table 17). Previous work has suggested that future lipid GWAS with larger sample sizes will likely confirm the novel lipid loci identified by our TWAS26. Results from additional competitive gene-set pathway and tissue expression analyses are available in the supplementary note.
Non-European Low-Frequency Missense Variant Associations
We next focused on ancestry specific low-frequency (MAF < 5%) missense variants, as these variants have been suggested to have a higher likelihood of causality27,28. We identified several novel low-frequency missense variants associated with one or more lipid levels at genome-wide significance that were specific to blacks or Hispanics. We found a total of 5 variants associated with LDL-C and/or total cholesterol among blacks (Supplementary Table 18) and2 associated with HDL-C and/or total cholesterol among Hispanics (Supplementary Table 19) in PCSK9, LDLR, APOB, and ABCA1. All 10 associations were directionally consistent in the 2017 GLGC exome chip meta-analysis with 9 reaching nominal significance (p < 0.05) among 17,009 blacks and 5,084 Hispanics included in the GLGC study. In addition, the 7 variants we identified were either monomorphic or had a MAF of < 0.0005 in the ~215,000 white veterans in MVP. Of note, we observed the low-frequency 443Thr allele in PCSK9 within Hispanics to be 8 fold more common in blacks (MAF = 0.011 in Hispanics versus 0.092 in blacks). We also found this variant to be associated with total cholesterol in blacks at genome-wide significance.
Predicted Loss of Gene Function Lipid Associations
We focused next on the subset of genotyped or imputed pLoF variants [variants annotated as: premature stop (nonsense), canonical splice-sites (splice-donor or splice-acceptor) or insertion/deletion variants that shifted frame (frameshift) by the Variant Effect Predictor software29]. A total of 15 distinct pLoF variants demonstrated genome-wide significant lipid associations across individuals of all three ethnic groups (Supplementary Table 20). We replicated known pLoF associations at PCSK919, APOC318, ANGPTL88, LPL30, CD3631, and HBB32, and we observed genome-wide significant associations of comparable magnitude of effect in each of the three ethnic groups for 2 pLoF variants: APOC3 c.55+1G>A and LPL p.Ser747Ter.
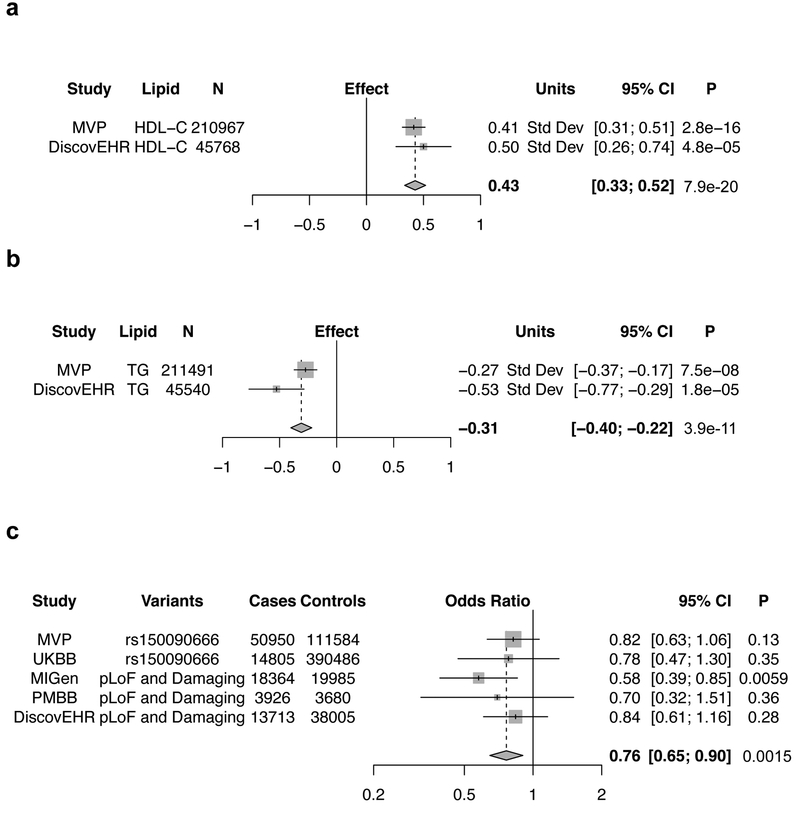
We identified one novel pLoF association. Among white MVP participants, carriers of a rare stop-gain mutation in PDE3B (p.Arg783Ter; carrier frequency of 1 in 625), exhibited a 4.72 mg/dL (0.41 standard deviations) higher blood HDL-C (P < 2.8 × 10−16) and 43.3 mg/dL (−0.27 standard deviations) lower blood triglycerides (P = 7.5×10−8). We found this signal to be independent of a previously reported genome-wide significant association in the region involving a common polymorphism, rs103737811 (p.Arg783Ter conditional analysis P = 6.3 × 10−16 for HDL-C, and P = 8.91 × 10−8 for triglycerides). We also identified one individual who was homozygous for p.Arg783Ter. This PDE3B “human knockout” was in his sixth decade of life and had HDL-C and triglycerides levels of 73 and 56 mg/dL, respectively. He was not on lipid-lowering medication and was free of coronary artery disease (CAD). We replicated the triglyceride and HDL-C associations for this pLoF variant in an independent sample of ~45,000 participants of the DiscovEHR study (Fig. 3a,b).
Figure 3. PDE3B Loss of Gene Function, Lipids, and Coronary Disease.
Linear regression results for the association of the predicted loss of function mutation p.Arg783Ter in PDE3B with HDL-C (a) and triglycerides (b) for white veterans in MVP with independent replication in the DiscovEHR study. Two-sided P values are displayed.
c) Meta-analysis of the association of damaging PDE3B mutations and coronary artery disease across five studies, including three (MIGen, PMBB, DiscovEHR) with exome sequencing. Logistic regression results were pooled in an inverse-variance weighted fixed effects meta-analysis. Minimal evidence of heterogeneity across cohorts was observed (I2 = 0%). Two-sided P values are displayed.
Abbreviations: MVP, Million Veteran Program; HDL-C, High-Density Lipoprotein Cholesterol; TG, Triglycerides; UKBB, UK Biobank; MIGen, Myocardial Infarction Genetics Consortium; PMBB, Penn Medicine Biobank
Loss of PDE3B function and risk of Coronary Artery Disease
Hypothesizing that mutations damaging or causing a loss of function in PDE3B could protect against the development of CAD based on their association with lifelong lower levels of triglycerides in blood, we conducted a case-control study of CAD involving 5 cohorts: MVP, UK Biobank, Myocardial Infarction Genetics Consortium (MIGen), Penn Medicine Biobank (PMBB), and DiscovEHR. For 3 studies that underwent exome sequencing (MIGen, PMBB, DiscovEHR), we combined pLoF variants with missense variants predicted to be damaging or possibly damaging by each of 5 computer prediction algorithms (LRT score, MutationTaster, PolyPhen-2, HumDiv, PolyPhen-2 HumVar, and SIFT) as performed previously30,33. Because damaging mutations are individually rare, we aggregated them in subsequent association analysis with CAD (Supplementary Table 21). Among 103,580 individuals with CAD and 566,813 controls available for meta-analysis in these 5 cohorts, carriers of damaging PDE3B mutations were found to have a 24% decreased risk of CAD (OR = 0.76, 95% CI = 0.65-0.90, P = 0.0015, Fig. 3c). Data from an additional analysis examining the association of all novel lipid loci identified in our study with CAD is available in the supplementary note.
PheWAS of Variants in Genes Targeted by Lipid Therapies
We leveraged a median of 65 unique ICD-9 diagnosis codes per participant prior to enrollment in MVP to explore the spectrum of phenotypic consequences of genetic variation within genes targeted by lipid-lowering medicines. We selected five lipid genes currently being targeted by pharmaceutical agents and identified functional variants in these genes: two nonsense variants (LPL p.Ser474Ter, ANGPTL8 p.Gln121Ter) and three missense variants (ANGPTL4 p.Glu40Lys, APOA5 p.Ser19Trp, PCSK9 p.Arg46Leu). We considered phenotypes to be significantly associated with a variant if they met a Bonferroni corrected P < 4.98 × 10−5 [0.05/1004 traits], a conservative threshold given the correlation structure present among PheWAS phenotypes34.
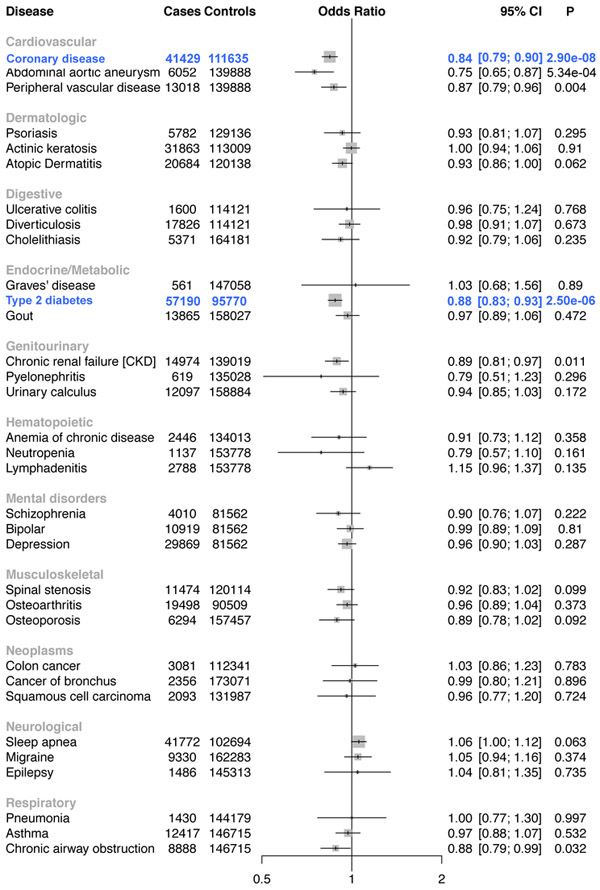
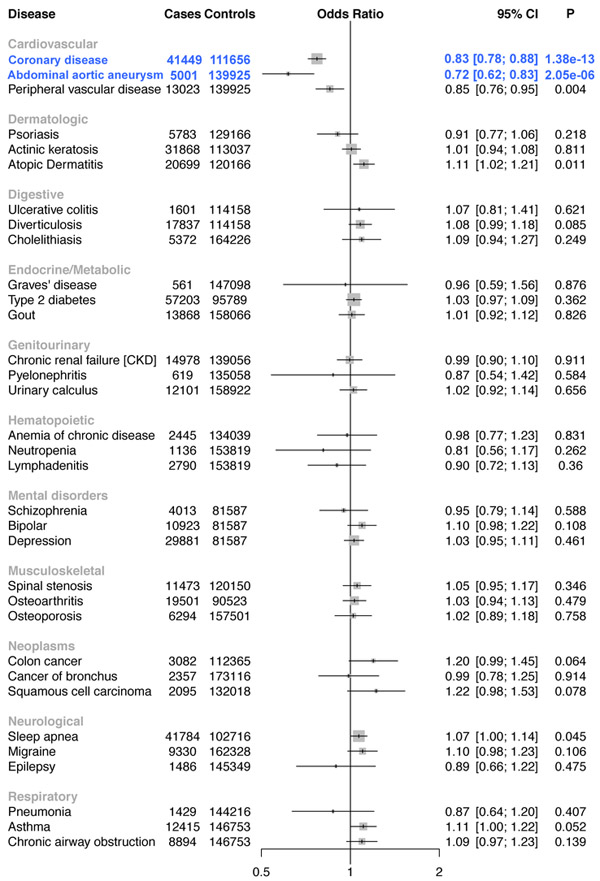
A total of 176,913 white veterans were available for analysis after quality control. Among these individuals, we identified 33 statistically significant phenotypic associations across the 5 variants, all of which are correlated with lipids (Supplementary Table 22). We replicated known associations with CAD for LPL30, ANGPTL414, and PCSK919. Notably, carriers of triglyceride-lowering/HDL-C-raising mutations in ANGPTL4 (p.Glu40Lys, 7,013 carriers) were also found to have a reduced risk of type 2 diabetes (Fig. 4). We replicated the type 2 diabetes association for the ANGPTL4 p.E40K variant in an independent sample of ~452,000 participants in the recently published trans-ethnic diabetes GWAS35[(OR =0.89, 95% CI = 0.86-0.93, P =9.24×10−10, Supplementary Fig. 11). In addition, carriers of LDL-C-lowering mutations in PCSK9 (p.Arg46Leu, 5,537 carriers) also demonstrated a reduced risk of AAA (Fig. 5).
Figure 4. ANGPTL4 40Lys Carrier Disease Associations.
Forest plot for a representative 33 of the 1004 disorders tested in the ANGPTL4 p.Glu40Lys PheWAS. Statistically significant logistic regression associations are shown in blue. Two-sided P values are displayed.
Figure 5. PCSK9 46Leu Carrier Disease Associations.
Forest plot for a representative 33 of the 1004 disorders tested in the PCSK9 p.Arg46Leu PheWAS. Statistically significant logistic regression associations are shown in blue. Two-sided P values are displayed.
Lipids and AAA Mendelian Randomization Analysis
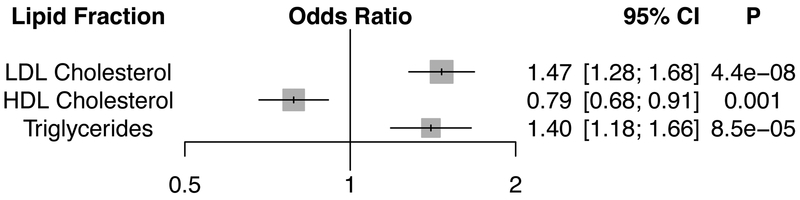
To further explore the causal relationship of lipids on AAA development, we performed a multivariate Mendelian randomization analysis using a weighted GRS of 223 lipid associated variants and summary data from a GWAS of 5,002 AAA cases and 139,968 controls in MVP. Consistent with our PheWAS results, a 1-standard deviation genetically elevated LDL-C was associated with an increased risk of AAA (OR = 1.47, 95% CI =1.28-1.68, P = 4.4×10−8). Furthermore, a 1-standard deviation genetically elevated HDL-C was associated with a decreased risk of AAA (OR = 0.79, 95% CI = 0.68-0.91, P = 0.001); and a 1-standard deviation genetically elevated triglycerides was associated with an increased risk of AAA (OR = 1.40, 95% CI = 1.18–1.66, P = 8.5×10−5, Fig. 6). An MR-Egger analysis36 indicated no pleiotropic bias of our lipid genetic instruments [MR-Egger intercept P > 0.05 for all 3 lipid fractions (Supplementary Table 23)].
Figure 6. Lipid Associations with Abdominal Aortic Aneurysm.
Logistic regression association results of the 223 variant lipid genetic risk score with abdominal aortic aneurysm in a multivariable Mendelian randomization analysis. Odds ratios are displayed per 1-standard deviation genetically increased lipid fraction. Two-sided P values are displayed.
Abbreviations: HDL, High-Density Lipoprotein; LDL, Low-Density Lipoprotein
Discussion
We leveraged clinical and genetic data from the Million Veteran Program to investigate the inherited basis of blood lipids in nearly 300,000 U.S. veterans. Our investigation resulted in several key findings. First, we robustly confirmed 188 previously identified loci while concurrently uncovering an additional 118 novel genome-wide significant loci. Next, we identified a total of 826 independent lipid associated variants increasing the phenotypic variance explained by nearly 2%. We performed a TWAS in four tissues identifying 5 additional novel lipid loci at a genome-wide level of significance, and performed a pathway analysis highlighting lipid transport mechanisms in our GWAS results. We identified ancestry-specific effects of rare coding variation on lipids among white, black, and Hispanic participants, and observed 15 pLoF mutations associated with lipids at a genome-wide level of significance, including a protein-truncating variant in PDE3B that lowers triglycerides, raises HDL-C, and protects against CAD. Finally, we examined the full spectrum of phenotypic consequences for mutations in lipid genes emerging as therapeutic targets, identifying protective effects of functional mutations in PCSK9 for abdominal aortic aneurysm and in ANGPTL4 for type 2 diabetes.
We glean four main insights through our findings. First, we confirm the enormous potential of a large-scale multi-ethnic biobank built within an integrated health care system in the discovery of the genetic basis of human traits. Specifically, we leveraged the VA’s mature nationwide EHR to efficiently extract existing repeated laboratory measures of lipids collected during the course of clinical care in nearly 300,000 veterans over a median of 10 years for GWAS analysis. Our results highlight the expected increase in variance explained by known loci when repeated lipid measurements are considered but also demonstrate the efficiency of examining the single most extreme lipid value least likely influenced by the use of lipid altering medications. Subsequent meta-analysis (combined N>600,000) with existing datasets increased the number of known independent genetic lipid loci to nearly 400 including several lipid pathways with links to human disease. For example, common variants near genes such as COL4A2 and ITGA1 identified for LDL-C/total cholesterol suggest links to extracellular matrix and cell adhesion biology, two pathways recently implicated by GWAS of CAD37,38. We also demonstrated that carriers of a rare missense mutation in the gene encoding Perilipin-1 (PLIN1 p.Leu90Pro) possess a markedly higher plasma HDL-C (0.243 standard deviations). In humans, Perilipin-1 is required for lipid droplet formation, triglyceride storage, as well as free fatty acid metabolism, and frameshift pLoF mutations in the PLIN1 gene have been reported to result in severe lipodystrophy39. A variant downstream of BDNF (encoding Brain-Derived Neurotrophic Factor) was found to be associated with HDL-C and triglycerides levels, supporting recent evidence linking this gene with metabolic syndrome and diabetes40. These findings not only improve our understanding of the genetic basis of dyslipidemia, but also provide insights into targets for the development of novel therapeutic agents.
Our second insight embraces the benefit of studying individuals with a diverse ethnic background. Such a design can provide valuable incremental information on the nature of previously identified human genetic associations. In MVP, we examined nearly 60,000 black and 25,000 Hispanic veterans for analysis, representing one of the largest - if not the largest - single-cohort GWAS to date for these ethnic groups for any trait. Among these individuals, we compared the effect estimates and allele frequencies of lipid-associated variants across ancestral groups and identified 7 novel low-frequency coding variants associated with lipids only in non-European populations. Conversely, we also confirmed a shared genetic architecture across all three racial groups for pLoF variation at the LPL and APOC3 loci. Previous work identifying low-frequency missense and pLoF variation in lipid genes have led to the development of the next generation of pharmaceutical agents for cardiovascular disease14,15,41,42. Expansion of these efforts to larger sample sizes and additional ancestries may help explain differences in blood lipid levels and risk of atherosclerosis among select populations.
Our third insight centers around our findings for the deleterious exonic variants within PDE3B. These findings lend human genetic support to PDE3B inhibition as a therapeutic strategy for atherosclerosis. Cilostazol, an inhibitor of both the 3A and 3B isoforms of the phosphodiesterase enzyme, is known to have anti-platelet43, vasodilatory44, and inotropic45 effects via inhibition of PDE3A, and also has well-documented, substantial effects on triglycerides and HDL cholesterol levels46 — likely through antagonism of PDE3B. We demonstrate that a PDE3B pLoF variant recapitulates the known lipid effects of cilostazol, and extend these findings to show that damaging PDE3B mutations are also associated with reduced risk of CAD. Randomized control trials to date have demonstrated cilostazol’s efficacy in intermittent claudication46 and prevention of restenosis following percutaneous coronary intervention47. The drug is also currently used off-label for the prevention of stroke recurrence through a presumed anti-platelet effect48. We note that mice genetically deficient in Pde3b display reduced atherosclerosis49 as well as decreased infarct size and improved cardiac function following experimental coronary artery ligation50. In light of our findings, use of cilostazol, or one of its derivatives, for the primary or secondary prevention of CAD deserves further consideration.
Our final insight highlights the potential benefit of phenome-wide association scanning across a large-scale EHR-based biobank to predict both potentially adverse as well as beneficial consequences of artificially inhibiting gene function. Here, we provide evidence that pharmacologic PCSK9 inhibition may reduce abdominal aortic aneurysm risk in addition to its known effects on atherosclerotic cardiovascular disease13. This finding is further supported by: our Mendelian randomization results; a recently published analysis using an independent AAA dataset51; and a recent report demonstrating that a PCSK9 gain-of-function mutation augments AAA development in a mouse model52. However, we also recognize the possibility that these results may be a consequence of pleiotropic effects induced by a high phenotypic correlation between AAA and the presence of advanced atherosclerotic disease. Thus, additional studies are necessary before definitive conclusions can be made on causality. Similarly, we expand on the potential indications for ANGPTL4 inhibition to include type 2 diabetes. Future PheWAS efforts may reveal associations that facilitate prioritization of drugs currently in development, repurposing of therapies already in clinical use, or prediction of adverse or off-target effects prior to investigation through expensive and time-consuming clinical trials.
Several limitations deserve mention. First, our MVP lipid phenotype definitions are based entirely on EHR data with a high prevalence of use of lipid-lowering therapy at enrollment. We used maximum or minimum values to capture untreated lipid levels, but the possibility of misclassification of lipid levels remains for participants entering the VA healthcare system on therapy. Such misclassification, however, would be expected to generally reduce our power to detect genetic associations. Second, participants in MVP are overwhelmingly male. Although almost 25,000 women were included in our discovery analysis, we did not attempt to detect genetic associations specific to females or heterogeneity of effects between sexes due to suspected limited power. Third, our TWAS identifies candidate causal genes under the assumption that the causal mechanism of the tested genes involves changes in cis-expression. However, we are unable to discriminate between instances of pleiotropy (when a given variant may alter gene expression and affect lipid levels independently) with TWAS alone and further functional analysis may be necessary. Fourth, our analysis demonstrating a lack of association between HDL-C raising alleles and CAD risk may be underpowered given the small number of alleles examined, though this finding has been demonstrated consistently in previous studies53,54. Lastly, power to detect associations with less common diseases in our PheWAS may also be limited despite the overall number of participants included in the analysis.
In conclusion, we identified >100 new genetic signals for blood lipid levels utilizing a biobank that exploits existing EHRs of U.S. veterans. We demonstrate the potential of this approach in the discovery of novel genetic associations and the development of novel therapeutic agents.
Online Methods
The design of the Million Veteran Program (MVP) has been previously described2. Briefly, individuals aged 19 to 104 years have been recruited from more than 50 VA Medical Centers nationwide since 2011. Each veteran’s EHR data are being integrated into the MVP biorepository, including inpatient International Classification of Diseases (ICD-9) diagnosis codes, Current Procedural Terminology (CPT) procedure codes, clinical laboratory measurements, and reports of diagnostic imaging modalities. The MVP received ethical and study protocol approval from the VA Central Institutional Review Board (IRB) in accordance with the principles outlined in the Declaration of Helsinki. Informed consent was obtained from all participants of the MVP study.
Genetic Data
DNA extracted from whole blood was genotyped using a customized Affymetrix Axiom biobank array, the MVP 1.0 Genotyping Array. With 723,305 total DNA sequence variants, the array is enriched for both common and rare variants of clinical significance in different ethnic backgrounds. Veterans of three mutually exclusive ethnic groups were identified for analysis: 1) non-Hispanic whites (European ancestry), 2) non-Hispanic blacks (African ancestry), and 3) Hispanics. Further details of methods used to assign ancestry and perform sample quality control are described in the supplementary note.
Variant Quality Control
Prior to imputation, variants that were poorly called (genotype missingness > 5%) or that deviated from their expected allele frequency based on reference data from the 1000 Genomes Project21 were excluded. After pre-phasing using EAGLE55 v2, genotypes from the 1000 Genomes Project21 phase 3, version 5 reference panel were imputed into Million Veteran Program (MVP) participants via Minimac3 software56. Ethnicity-specific principal component analysis was performed using the EIGENSOFT software57.
Following imputation, variant level quality control was performed using the EasyQC R package58 (see URLs), and exclusion metrics included: ancestry specific Hardy-Weinberg equilibrium59 P <1×10−20, posterior call probability < 0.9, imputation quality/INFO <0.3, minor allele frequency (MAF) < 0.0003, call rate < 97.5% for common variants (MAF > 1%), and call rate < 99% for rare variants (MAF < 1%). Variants were also excluded if they deviated > 10% from their expected allele frequency based on reference data from the 1000 Genomes Project21.
EHR-Based Lipid Phenotypes
EHR clinical laboratory data were available for MVP participants from as early as 2003. We extracted the maximum LDL-C/triglycerides/total cholesterol, and minimum HDL-C for each participant for analysis. These extreme values were selected to approximate plasma lipid concentrations in the absence of lipid lowering therapy as described previously20. For each phenotype (LDL-C, natural log transformed triglycerides, HDL-C, and total cholesterol), residuals were obtained after regressing on age, age2, sex, and 10 principal components of ancestry. Residuals were subsequently inverse normal transformed for association analysis. Statin therapy prescription at enrollment was defined as the presence of a statin prescription in the EHR within 90 days before or after enrollment in MVP. Statin therapy prescription at the maximum lipid measurement was defined as the presence of a statin prescription in the EHR within 90 days prior to the maximum lipid laboratory measurement used in our GWAS analysis. Further details of lipid phenotype quality control are described in the supplementary note.
MVP Association Analysis
Genotyped and imputed DNA sequence variants with a MAF > 0.0003 were tested for association with the inverse normal transformed residuals of lipid values through linear regression assuming an additive genetic model. In our initial discovery analysis (Stage 1), we performed association testing separately among individuals of each of three genetic ancestries (whites, blacks, and Hispanics) and then meta-analyzed results across ethnic groups using an inverse variance-weighted fixed effects method. For variants with suggestive associations (association P < 10−4), we sought replication of our findings in one of two independent studies: the 2017 GLGC exome array meta-analysis11 (Stage 2a) or the 2013 GLGC “joint meta-analysis5“ (Stage 2b). Replication was first attempted using summary statistics from the 2017 GLGC exome array study (Stage 2a). A total of 242,289 variants in up to 319,677 individuals were analyzed after quality control and were available for replication. If a DNA sequence variant was not available for replication in the above exome array-focused study, we sought replication from publicly available summary statistics from the 2013 GLGC “joint meta-analysis” (Stage 2b). An additional 2,044,165 variants in up to 188,587 individuals were available for replication in this study. In total, 2,286,454 DNA sequence variants in up to 319,677 individuals were available for independent replication in either Stage 2a or Stage 2b. We emphasize that if a variant was available for replication in both studies, replication was performed only using summary statistics from the 2017 GLGC exome array study given its larger sample size. We defined significant novel associations as those that were at least nominally significant in replication (P<0.05) and had an overall P < 5 ×10−8 (genome-wide significance) in the discovery and replication cohorts combined. Novel loci were defined as being greater than 1 mB away from a known lipid genome-wide associated lead variant. Additionally, linkage disequilibrium information from the 1000 Genomes Project21 was used to determine independent variants where a locus extended beyond 1 mB. All association P values were two-sided. Further details of the association analysis are described in the supplementary note.
Conditional Analysis
We used the COJO-GCTA software (see URLs) to perform an approximate, stepwise conditional analysis to identify independent variants within lipid-associated loci given that individual level data for the prior GLGC lipid analyses are not publicly available. We used summary statistics of ~1.9 million overlapping variants that we meta-analyzed across either one of the two GLGC datasets (predominantly European) and the European MVP dataset to conduct this analysis (Supplementary Figure 8) combined with an LD-matrix obtained from 10,000 unrelated European individuals randomly sampled from the UK Biobank interim release.
Variance Explained Using Multiple Lipid Measurements
We estimated the proportion of variance explained by the set of 444 previously mapped independent lipid variants, the 118 novel lipid loci identified in our study, and the 826 independent lipid variants identified from conditional analysis using ridge regression with the glmnet R package. The variance explained was determined after tuning the hyperparameter (lambda) to approximate an optimal value, and then calculating the model R2 after performing linear regression with the inverse normal transformed lipid outcome and each set (444, 118, 826) of independent genome-wide variants as predictors.
We estimated the variance explained for a GRS of 223 previously described GWAS lipid variants weighted by their previously reported effect sizes11 (Supplementary Table 11) as a function of the number of lipid measurements in MVP to assess the potential impact of using multiple lipid measurements in discovery. We performed this analysis using the mean of one, two, three, four, and five lipid measurements for each individual starting with their measurement closest to enrollment and moving backward in time. To account for the use of statin therapy, individuals with evidence of a statin prescription in their EHR at the time of enrollment had their LDL-C/total cholesterol values adjusted by dividing by 0.7/0.8, respectively as previously described5. In addition, we also calculated the variance explained by the single maximal triglycerides, LDL-C/total cholesterol, and minimal HDL-C from the EHR without adjustment for lipid lowering therapy. Our analyses were restricted to a subset of 171,314 European MVP participants with ≥ 5 lipid measurements.
Lipids Transcriptome-wide Association Study
We performed a TWAS using summary statistics after a meta-analysis of ~1.9 million overlapping variants among GLGC (predominantly European) and European MVP datasets (Supplementary Figure 8) and four gene-expression reference panels (NTR whole blood, METSIM adipose tissue, and tibial artery and liver from GTEx) in independent samples as previously described23. In brief, for a given gene, variant-expression weights in the 1-mB cis locus were first computed with the BSLMM60, which: “models effects on expression as a mixture of normal distributions to account for the sparse expression architecture. Given weights w, lipid Z scores Z, and variant-correlation (LD) matrix D; the association between predicted expression and lipids (i.e., the TWAS statistic) was estimated as ZTWAS = w’Z/(w’Dw)1/2 (details in ref. 23).” We computed TWAS statistics by using either the variants genotyped in each expression reference panel or imputed HapMap3 variants. To account for multiple hypotheses we applied a genome-wide significant P value threshold (two-sided P < 5 ×10−8), significantly more stringent than previously used Bonferroni corrections in prior TWAS26. We defined novel TWAS loci as a TWAS gene falling outside of a previously identified lipid GWAS region (± 1mB around a mapped sentinel GWAS variant).
Identification of Independent Low-Frequency Coding Variant Lipid Associations Specific to Blacks and Hispanics
We used the P value and linkage disequilibrium-driven clumping procedure in PLINK version 1.90b (--clump) to identify associations between low-frequency coding variants and lipids specific to blacks and Hispanics. Input included summary lipid association statistics from our MVP 1000 Genomes imputed genome-wide association study of black and Hispanic individuals, and reference linkage disequilibrium panels of 661 African (AFR) and 347 Ad Mixed American (AMR) samples from 1000 Genomes phase 3 whole genome sequencing data. Variants were clumped with stringent r2 (<0.01) and P (< 5 × 10−8) thresholds in a 1mB region surrounding the lead variant at each locus to reveal independent index variants at genome-wide significance. From this list of independent variants, we report novel protein-altering variants specific to blacks and Hispanics at a MAF < 0.05.
Loss of Gene Function Analysis
We used the Variant Effect Predictor29 software to identify pLoF DNA sequence variants defined as: premature stop (nonsense), canonical splice-sites (splice-donor or splice-acceptor) or insertion/deletion variants that shifted frame (frameshift). For the pLoF lipids analysis, we then merged these variants with data from the Exome Aggregation Consortium27 (Version 0.3.1, see URLs), a publicly available catalogue of exome sequence data to confirm consistency in variant annotation. We required that pLoF DNA sequence variants be observed in at least 50 individuals, and set a statistical significance threshold of P < 5 × 10−8 (genome-wide significance).
Loss of PDE3B Gene Function and Coronary Artery Disease
We identified a novel lipid association for a pLoF mutation in the PDE3B gene (rs150090666, p.Arg783Ter). For carriers of damaging mutations in Phosphodiesterase 3B, we examined the mutation’s effects on risk for CAD using logistic regression in five separate cohorts: MVP, UK Biobank, and 3 cohorts with exome sequencing: the Myocardial Infarction Genetics Consortium (MIGen), the Penn Medicine Biobank (PMBB), and DiscovEHR. In studies with exome sequencing, we combined pLoF variants with missense variants predicted to be damaging or possibly damaging by each of 5 computer prediction algorithms (LRT score, MutationTaster, PolyPhen-2, HumDiv, PolyPhen-2 HumVar, and SIFT) as performed previously30,33. Because any individual damaging mutation was rare, variants were aggregated together for subsequent phenotypic analysis. We performed logistic regression on disease status, adjusting for age, sex, and principal components of ancestry as appropriate. Effects of PDE3B damaging mutations were pooled across studies using an inverse-variance weighted fixed effects meta-analysis. Further details of participating cohorts and CAD case definitions are described in the supplementary note. We set a two-sided P < 0.05 threshold for statistical significance.
PheWAS of Variation in Genes Targeted by Lipid Lowering Therapies
For a set of DNA sequence variants within genes targeted by lipid-lowering medicines, we performed a PheWAS leveraging the full catalog of EHR ICD-9 diagnosis codes. We selected five lipid genes currently being targeted by pharmaceutical agents and identified functional variants in these genes: two nonsense variants (LPL p.Ser474Ter, ANGPTL8 p.Gln121Ter) and three missense variants (ANGPTL4 p.Glu40Lys, APOA5 p.Ser19Trp, PCSK9 p.Arg46Leu). Details of PheWAS quality control, case definitions, and association analysis are described in the supplementary note. We considered phenotypes to be significantly associated with a variant if they met a Bonferroni corrected two-sided P < 4.98 × 10−5 [0.05/1004 traits]. For replication of our ANGPTL4 p.E40K type 2 diabetes finding, we combined the PheWAS results with publicly available data from the recently published trans-ethnic type 2 diabetes GWAS35 using an inverse variance-weighted fixed effects method.
Lipids and Abdominal Aortic Aneurysm Mendelian Randomization Analysis
Summary-level data for 223 genome-wide lipids-associated variants were obtained from publicly available data from the Global Lipids Genetics Consortium11. We then utilized results from a GWAS of 5,002 AAA cases and 139,968 controls performed in white MVP participants using the definition proposed by Denny et al17. The effect alleles were matched with all lipid and AAA summary data and 3 different Mendelian randomization analyses were performed: 1) inverse variance–weighted; 2) multivariable; 3) MR-Egger to account for pleiotropic bias. First, we performed inverse-variance–weighted Mendelian randomization using each set of variants for each lipid trait as instrumental variables. This method, however, does not account for possible pleiotropic bias. Therefore, we next performed inverse-variance–weighted multivariable Mendelian randomization. This method adjusts for possible pleiotropic effects across the included lipid traits in our analyses using effect estimates from the variant-AAA outcome and effect estimates from variant-LDL-C, variant-HDL-C, and variant-triglycerides as predictors in 1 multivariable model. We additionally performed MR-Egger as previously described36. This technique can be used to detect bias secondary to unbalanced pleiotropy in Mendelian randomization studies. In contrast to inverse variance–weighted analysis, the regression line is unconstrained, and the intercept represents the average pleiotropic effects across all variants. Bonferroni-corrected two-sided P values (P=0.016; 0.05/3) for 3 tests were used to declare statistical significance.
Reporting Summary
Further information on experimental design is available in the Nature Research Life Sciences Reporting Summary linked to this article.
Data availability.
The full summary level association data from the trans-ancestry meta-analysis for each lipid trait from this report are available through dbGaP, accession code __.
Supplementary Material
Acknowledgements:
Data on coronary artery disease/myocardial infarction have been contributed by the CARDIoGRAMplusC4D investigators and the Myocardial Infarction Genetics and CARDIoGRAM Exome investigators. Both datasets were obtained online (see URLs). This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by the Department of Veterans Affairs (VA) Cooperative Studies Program (CSP) award #G002. This research was also supported by three additional Department of Veterans Affairs awards (I01-01BX03340, I01-BX002641, and I01-CX001025) and the NIH (T32 HL007734, K01HL125751, R01HL127564). The content of this manuscript does not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
Competing Interests: Dr. Kathiresan reports grant support from Regeneron and Bayer, grant support and personal fees from Aegerion, personal fees from Regeneron Genetics Center, Merck, Celera, Novartis, Bristol-Myers Squibb, Sanofi, AstraZeneca, Alnylam, Eli Lilly, and Leerink Partners, personal fees and other support from Catabasis, and other support from San Therapeutics outside the submitted work. He is also the chair of the scientific advisory board at Genomics plc. Drs. Teslovich, Alex Li, Baras, Dewey, and Carey are employees of Regeneron Pharmaceuticals. Dr. Abecasis has received consulting income from Regeneron Genetics Center, 23andme, and Helix. Dr. Duvall has received research grant support from the following for-profit companies through the University of Utah or the Western Institute for Biomedical Research (VA Salt Lake City’s affiliated non-profit): AbbVie Inc., Anolinx LLC, Astellas Pharma Inc., AstraZeneca Pharmaceuticals LP, Boehringer Ingelheim International GmbH, Celgene Corporation, Eli Lilly and Company, Genentech Inc., Genomic Health, Inc., Gilead Sciences Inc., GlaxoSmithKline PLC, Innocrin Pharmaceuticals Inc., Janssen Pharmaceuticals, Inc., Kantar Health, Myriad Genetic Laboratories, Inc., Novartis International AG, and PAREXEL International Corporation.
URLs
R Statistical Software, www.R-project.org. EasyQC, https://www.uni-regensburg.de/medizin/epidemiologie-praeventivmedizin/genetische-epidemiologie/software/. PheWAS. https://github.com/PheWAS/PheWAS. GCTA, http://cnsgenomics.com/software/gcta/#Overview. FUMA, http://fuma.ctglab.nl/. ExAC Browser, http://exac.broadinstitute.org/. SNPTEST software program, http://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html; CARDIoGRAMplusC4D and MIGen and CARDIoGRAM Exome investigators datasets, www.CARDIOGRAMPLUSC4D.ORG.
References:
- 1.Collins R What makes UK Biobank special? The Lancet 379, 1173–1174 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Gaziano JM et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–23 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Di Angelantonio E et al. Major lipids, apolipoproteins, and risk of vascular disease. Jama 302, 1993–2000 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teslovich TM et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Global Lipids Genetics Consortium et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 45, 1274–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chasman DI et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet 5, e1000730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albrechtsen A et al. Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia 56, 298–310 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peloso GM et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet 94, 223–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asselbergs FW et al. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am J Hum Genet 91, 823–38 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Below JE et al. Meta-analysis of lipid-traits in Hispanics identifies novel loci, population-specific effects, and tissue-specific enrichment of eQTLs. Sci Rep 6, 19429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu DJ et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu X et al. Exome chip meta-analysis identifies novel loci and East Asian-specific coding variants that contribute to lipid levels and coronary artery disease. Nat Genet (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabatine MS et al. Evolocumab and Clinical Outcomes in Patients with Cardiovascular Disease. N Engl J Med (2017). [DOI] [PubMed] [Google Scholar]
- 14.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med 374, 1134–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewey FE et al. Inactivating Variants in ANGPTL4 and Risk of Coronary Artery Disease. N Engl J Med 374, 1123–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barter PJ et al. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med 357, 2109–22 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Denny JC et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 31, 1102–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosby J et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med 371, 22–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen JC, Boerwinkle E, Mosley TH Jr. & Hobbs HH Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 354, 1264–72 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Abul-Husn NS et al. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science 354(2016). [DOI] [PubMed] [Google Scholar]
- 21.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tishkoff SA et al. The genetic structure and history of Africans and African Americans. Science 324, 1035–44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gusev A et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48, 245–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright FA et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet 46, 430–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancuso N et al. Integrating Gene Expression with Summary Association Statistics to Identify Genes Associated with 30 Complex Traits. Am J Hum Genet 100, 473–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marouli E et al. Rare and low-frequency coding variants alter human adult height. Nature (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaren W et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26, 2069–70 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khera AV et al. Association of Rare and Common Variation in the Lipoprotein Lipase Gene With Coronary Artery Disease. JAMA 317, 937–946 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dewey FE et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 354(2016). [DOI] [PubMed] [Google Scholar]
- 32.Sidore C et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet 47, 1272–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell SM et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diogo D et al. Phenome-wide association studies (PheWAS) across large “real-world data” population cohorts support drug target validation. bioRxiv (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahajan A et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet 50, 559–571 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden J, Davey Smith G & Burgess S Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44, 512–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klarin D et al. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson CP et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet 49, 1385–1391 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Gandotra S et al. Perilipin deficiency and autosomal dominant partial lipodystrophy. N Engl J Med 364, 740–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rani J et al. T2DiACoD: A Gene Atlas of Type 2 Diabetes Mellitus Associated Complex Disorders. Sci Rep 7, 6892 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musunuru K et al. Exome Sequencing, ANGPTL3 Mutations, and Familial Combined Hypolipidemia New England Journal of Medicine (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham MJ et al. Cardiovascular and Metabolic Effects of ANGPTL3 Antisense Oligonucleotides. N Engl J Med (2017). [DOI] [PubMed] [Google Scholar]
- 43.Zhang W & Colman RW Thrombin regulates intracellular cyclic AMP concentration in human platelets through phosphorylation/activation of phosphodiesterase 3A. Blood 110, 1475–82 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maass PG et al. PDE3A mutations cause autosomal dominant hypertension with brachydactyly. Nat Genet 47, 647–53 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Vandeput F et al. Selective regulation of cyclic nucleotide phosphodiesterase PDE3A isoforms. Proc Natl Acad Sci U S A 110, 19778–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bedenis R et al. Cilostazol for intermittent claudication. Cochrane Database Syst Rev, Cd003748 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuchikane E et al. Impact of cilostazol on restenosis after percutaneous coronary balloon angioplasty. Circulation 100, 21–6 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Shinohara Y et al. Cilostazol for prevention of secondary stroke (CSPS 2): an aspirin-controlled, double-blind, randomised non-inferiority trial. Lancet Neurol 9, 959–68 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Ahmad F et al. Phosphodiesterase 3B (PDE3B) regulates NLRP3 inflammasome in adipose tissue. Sci Rep 6, 28056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung YW et al. Targeted disruption of PDE3B, but not PDE3A, protects murine heart from ischemia/reperfusion injury. Proc Natl Acad Sci U S A 112, E2253–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrison SC et al. Genetic Association of Lipids and Lipid Drug Targets With Abdominal Aortic Aneurysm: A Meta-analysis. JAMA Cardiol 3, 26–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu H et al. Hypercholesterolemia Induced by a PCSK9 Gain-of-Function Mutation Augments Angiotensin II-Induced Abdominal Aortic Aneurysms in C57BL/6 Mice-Brief Report. Arterioscler Thromb Vasc Biol 36, 1753–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voight BF et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. The Lancet 380, 572–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Do R et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet 45, 1345–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online Methods References
- 55.Loh PR, Palamara PF & Price AL Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet 48, 811–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howie B, Fuchsberger C, Stephens M, Marchini J & Abecasis GR Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44, 955–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–9 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Winkler TW et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 9, 1192–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyde CL et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 48, 1031–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou X, Carbonetto P & Stephens M Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet 9, e1003264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full summary level association data from the trans-ancestry meta-analysis for each lipid trait from this report are available through dbGaP, accession code __.