Abstract
TEN1 protein is a key component of CST complex, implicated in maintaining the telomere homeostasis, and provides stability to the eukaryotic genome. Mutations in TEN1 gene have higher chances of deleterious impact; thus, interpreting the number of mutations and their consequential impact on the structure, stability, and function is essentially important. Here, we have investigated the structural and functional consequences of nsSNPs in the TEN1 gene. A wide array of sequence- and structure-based computational prediction tools were employed to identify the effects of 78 nsSNPs on the structure and function of TEN1 protein and to identify the deleterious nsSNPs. These deleterious or destabilizing nsSNPs are scattered throughout the structure of TEN1. However, major mutations were observed in the α1-helix (12–16 residues) and β5-strand (88–96 residues). We further observed that mutations at the C-terminal region were having higher tendency to form aggregate. In-depth structural analysis of these mutations reveals that the pathogenicity of these mutations are driven mainly through larger structural changes because of alterations in non-covalent interactions. This work provides a blueprint to pinpoint the possible consequences of pathogenic mutations in the CST complex subunit TEN1.
Keywords: OB fold protein, Pathogenic mutations, Structural genomics, SNP, Sequence analysis
Introduction
Telomeres consist of non-coding ends of eukaryotic linear chromosomes and play a vital role in the replication, regulation, and protection of genome [1,2]. Ends of eukaryotic chromosomes can be identified by recombination and repair system of the cells as DNA strand breaks that often proceed to end-to-end fusion and instability of genome [3,4]. Shelterin complex is composed of six subunits (TRF1, TRF2, RAP1, TIN2, TPP1, and POT1) which are located primarily to single- and double-stranded telomeric DNA [5]. In addition to repressing DDR and chromosome fusion, shelterin complex also caps the telomeric ends by facilitating the formation of T-loop. It is also acting as a processivity factor via recruiting telomerase to chromosomes end [6,7].
The CST complex is composed of three subunits, conserved telomere maintenance component 1 (CTC1), suppressor of CDC thirteen homolog (STN1), and telomere length regulation protein TEN1 homolog (TEN1) [8], specifically localizes to the ssDNA) of telomere and is involved in telomere capping and regulation of telomere length [9–11]. However, increasing evidence has demonstrated that the STN1-TEN1 complex possesses some extra telomeric functions. It is involved in resolving replication fork stalling during replication stress [12,13]. CST complex is also involved in the removal of G-quadruplexes (G4: G-rich repeats) [14]. The G-rich region of the telomere is very prone to form G4 throughout telomeric DNA and poses severe challenges for telomere replication machinery [15]. In addition, CST complex binds to the 3′ ends of telomeres and regulates polymerase α-mediated syntheses of C-strand [16]. Some important reports on the structure and function of the CST complex can be found elsewhere [17–20].
In addition to polymerase α-mediated syntheses of C-strand, a subunit of CST (CTC1-STN1) regulates telomerase-mediated extension of G-rich overhang which is critical for the cell proliferation. Deficiency of CTC1-STN1 complex leads to overextension of G-rich overhangs which initiate DDR [21,22]. In this process, the role of TEN1 is indispensable as it is essential to provide stability to CTC1-STN1 complex. Disruption of TEN1 results in progressive shortening of telomere more like caused by telomerase deficiency. As telomere maintenance is paramount to genome stability, mutations in the genes encoding essential components of CST are associated with varieties of genetic abnormalities including cancer [23], coat plus [24–26], and dyskeratosis congenita [27,28].
Prediction of nsSNPs affecting protein structure and function in detail may be investigated by the aid of cutting-edge computational methods. In many cases, nsSNPs have little or no effect on protein structure and functions, but often a single mutation is highly lethal [29]. Experimental studies suggested that about one-third of nsSNPs are deleterious to human health [30]. Thus, identification of such deleterious nsSNPs is of serious concern in terms of diagnosis and therapeutic perspective. In vitro mutational studies by Bryan et al., suggested that mutation in some important residue in TEN1 gene directly affect the interaction with STN1 to many fold. During structure analysis, we have observed that mutant R27Q caused a marked reduction in the polar interactions between TEN1-STN1complex (Supplementary Figure S1). Disruption in TEN1-STN1 interaction leads to the development telomere malfunctions and thus telomeropathies [31]. A little report is available on the mutational analysis of nsSNPs in TEN1 gene. Taking this opportunity into consideration and the fact that TEN1 plays crucial role in the telomere maintenance; we have predicted the structural and functional effects of about 78 nsSNPs in the coding region of TEN1 gene. The present study will offer in-depth understanding of the role of nsSNPs on the structure and function of TEN1 protein.
Materials and methods
Data collection
Distribution of nsSNPs in human TEN1 gene was retrieved from dbSNP [32], Ensembl [33], and HGMD [34] databases. Data enrichment was carried out by removing the variant duplicates of different databases. The human TEN1 amino acid sequence was obtained in FASTA format from UniProt database (UniProt ID: Q86WV5) (http://www.uniprot.org/). A 3D structure of TEN1 (PDB ID: 4JOI) was downloaded from the Protein Data Bank (PDB) [35]. Functional annotations of all SNPs were extracted from the dbSNP database; for example, whether the SNPs present in an intron or exon, in the 3′ or 5′-UTR, or downstream or upstream of the TEN1 gene.
Sequence-based prediction of deleterious nsSNPs
Sorting Intolerant from Tolerant (SIFT) (http://sift.jcvi.org/) algorithm was used to predict the amino acid substitution as tolerable and intolerable depending upon the physical and sequence-homology features. Substitutions with normalized probabilities of ≥0.05 and ≤0.05 were predicted as tolerated and deleterious, respectively [36,37]. There were about 78 nsSNPs identified from Ensembl and dbSNP databases. Prediction of tolerated and deleterious effect of these nsSNPs in human TEN1 gene was predicted using SIFT. Protein variation effect analyzer (PROVEAN) (http://provean.jcvi.org/) tool was used to predict the consequences of amino acid substitution on protein function [38]. It predicts nsSNPs as ‘deleterious’ if the score is less than the threshold value (cutoff is −2.5), and ‘neutral’ if the predicted score is more than the cut-off value. All the nsSNPs in human TEN1 gene were calculated and analyzed using this cut-off value.
PolyPhen-2 (polymorphism phenotyping-2) (http://genetics.bwh.harvard.edu/pph2/) was used to calculate functional predictions of coding variants. It uses a particular empirical rule comprises of both comparative and physical considerations to predict the probable functional impacts of mutation on the structure–function relationship. FASTA format of protein sequence was used as input to calculate the effects of a particular substitution [39]. It calculates a position-specific independent count (PSIC) score for each substitution and then estimates the score deviations. A mutation is considered as possibly destructive mutation if the PSIC score is ≥0.9.
Structure-based prediction of destabilizing nsSNPs
STRUM (https://zhanglab.ccmb.med.umich.edu/STRUM/) tool was used to predict the stability differences between WT and mutant proteins. Initially, from protein sequences, a 3D model was generated by I-TASSER simulation and used to train STRUM model through gradient boosting regression. STRUM predicts the possible effects of nsSNPs on the structure and function of a protein using conservation score from an alignment of the multiple-threading template. The query sequence used as input in FASTA format and calculated the impact of a particular substitution in a given sequence [40]. SDM2 (site direct mutator 2) (http://structure.bioc.cam.ac.uk/sdm2) is a knowledge-based tool used to estimate the impact of mutations on the stability of protein [41]. It uses constrained environment-specific substitution tables (ESSTs) to calculate the differences in the protein stability upon mutation [41,42]. SDM2 uses PDB as an input file, and point variants to estimate the stability difference score between the WT and mutants.
PoPMuSiC (http://babylone.ulb.ac.be/popmusic/) tool was used to predict changes in thermodynamic stability upon mutation. PoPMuSiC employing a linear combination of statistical potentials whose coefficients depend on the solvent accessibility of the substituted residues. It uses PDB as an input file. DUET server was used to predict the impact of mutations on the stability of TEN1 protein using PDB code. DUET calculates a combined or consensus predictions of SDM and mCSM (mutation Cutoff Scanning Matrix) using support vector machines (SVMs) in a non-linear regression fashion. The output it provides is in the form of change in Gibbs free energy (∆∆G), where negative sign indicates destabilizing mutation [43]. MCSM was implicated to predict the impact of mutations on the stability of proteins using graph-based structural signatures. It predicts protein–protein and protein–nucleic acid interaction [44].
Identification of diseased phenotype
MutPred (http://mutpred.mutdb.org/) tool was used to predict the association of nsSNPs with disease phenotype [45]. It employs several attributes associated with structure, function, and evolution using PSI-BLAST [46], SIFT [36], and Pfam profiles [47] together with structure disorder prediction tools such as TMHMM [48], DisProt [49], and MARCOIL [50]. Score with g-value more than 0.75 and a p-value less than 0.05 is considered as a confident hypothesis. PhD-SNP, (http://snps.biofold.org/phd-snp/phd-snp.html) is online SVM based prediction tool, was used to predict the pathological effects of a given mutation [51].
Aggregation propensity analysis
SODA (protein solubility from disorder and aggregation propensity) was used to predict the change in protein solubility upon mutation by comparing the sequence profile of WT and mutants. The aggregation or intrinsic disorder score obtained from PASTA [52], and ESpritz [53], and a combined result obtained from Kyte-Doolittle [54] and FELLS [55]. SODA also predicts types of variation, including insertion and deletion in a given sequence [56].
Sequence conservation analysis
The importance of a particular amino acid in the structure and functions of protein can be generally retrieved from its conservation score using multiple sequence alignment. The blueprint of amino acid conservation was identified by ConSurf tool, which measures the degree of conservation of each amino acid at a particular position along with the evolutionary profile of amino acid sequence [57]. Conservation score ranged from 1 to 9, where 1 depicts rapidly evolving (variable), 5 indicates region which is evolving moderately, and 9 shows slowly evolving (evolutionarily conserved) position. Exposed residues with high conservation score are being considered as functional whereas buried residues with high conservation score are believed as structural residues.
Analysis of solvent accessibility
Relative side-chain solvent accessibility (RSA), residue depth and residue-occluded packing density (OSP) of WT, and mutant TEN1 protein have been performed using SDM2 server [41]. It uses ESSTs table to calculate the differences in their RSA, residue depth, and OSP of WT and mutant proteins. RSA has been calculated based on Lee and Richards method [58]. Three classes of relative RSA were defined based on the method of Lee and Richards, whereby a probe of given radius is rolled around the surface of the molecule [58].
Results and discussion
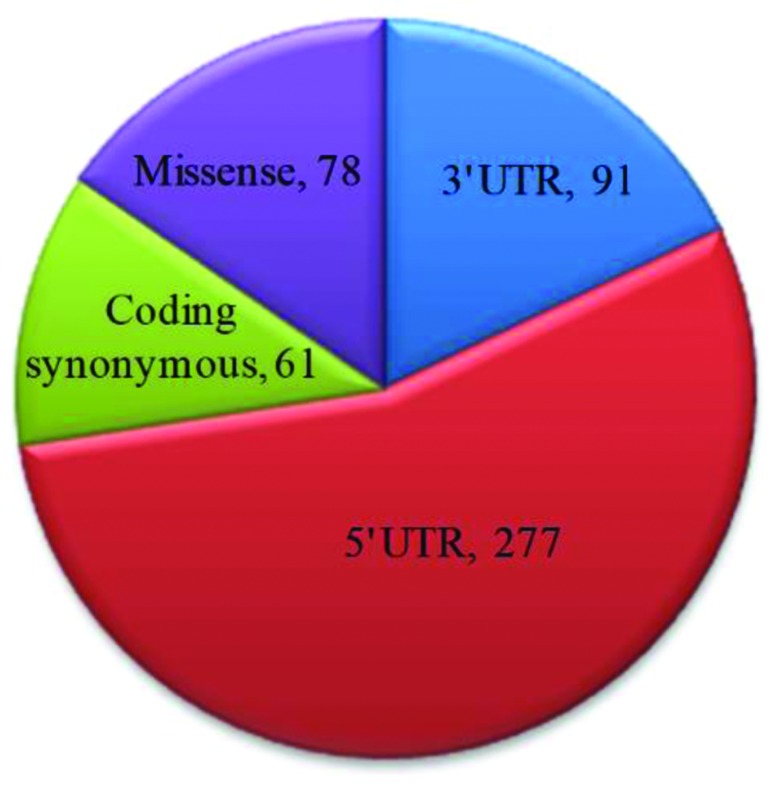
All reported SNPs of TEN1 gene was extracted from Ensembl (http://www.ensembl.org/) and dbSNP databases (http://www.ncbi.nlm.nih.gov/snp). A total of about 5712 SNPs were mapped and classified into nine different functional classes. Four major classes of SNPs in TEN1 gene are shown in Figure 1. About 5250 SNPs were mapped in the intronic region and approximately 78 were found in the coding non-synonymous/missense region. The 5′- and 3′-UTR regions have 277 and 91 SNPs, respectively. In addition, 61 SNPs in coding synonymous, five SNPs in frameshift, and three SNPs in each 3′ and 5′ splice site regions are also observed. The present study focuses only on missense mutations mapped in the coding region. A total of 78 nsSNPs were taken for further analysis.
Figure 1. Representation of number of SNPs in TEN1 gene using dbSNP database.
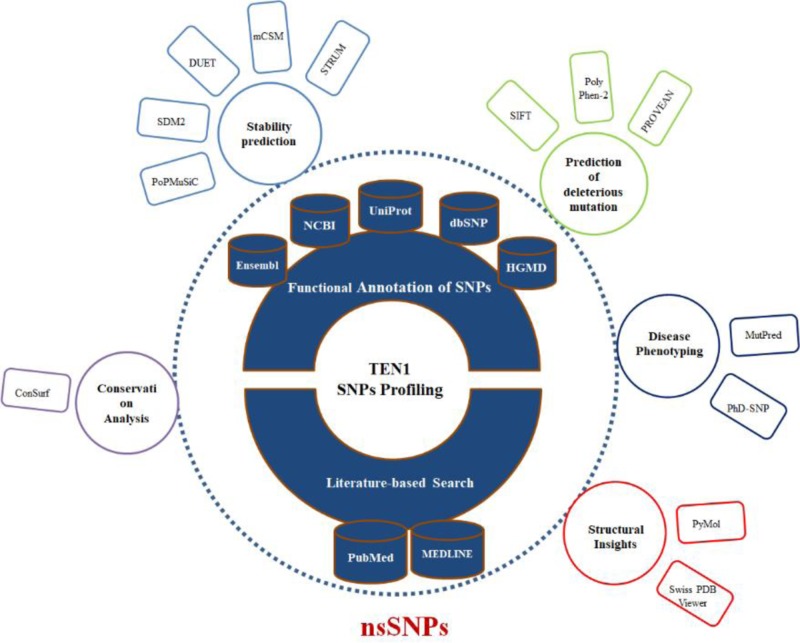
To identify the structural and functional impact on missense mutations in TEN1 gene, we have employed a multi-tier approach. To collect high confidence nsSNPs in the TEN1 gene, all mapped TEN1 nsSNPs were first subjected to sequence-based prediction using PolyPhen-2, PROVEAN and SIFT, followed by structure-based stability predictions using PoPMuSiC, SDM2, DUET, mCSM, and STRUM web-servers. Further, distributions of high confidence nsSNPs were analyzed on the basis of their structure descriptors and phenotypic association. In consistence, we discuss pathogenic mutations in relation to their sequence conservation, functional importance, and aggregation propensities. Finally, we expand our analysis and extensively analyzed the structural and functional impact of pathogenic mutations on the local environment of the TEN1 protein. An overview of computational methods used in the present study is depicted in Figure 2.
Figure 2. Overview of computational approaches used to identify the deleterious or pathogenic mutations in the TEN1 protein at structural and functional level.
Identification of deleterious nsSNPs
To pinpoint the structural and functional consequences of nsSNPs in TEN1 gene, we have performed an extensive structural analysis. The reason for using multiple tools is to improve the confidence level of prediction. Accumulation of deleterious nsSNPs using a single approach may not always be satisfactory as some mutations that have scored very close to cut-off value are prone to false prediction. Therefore, using multiple tools in both sequence- and structure-based predictions may provide an accurate result. The nsSNPs predicted to be deleterious in at least two methods from sequence-based prediction methods and three tools depict destabilizing effects from structure-based prediction were collected and termed as ‘high confidence nsSNPs’.
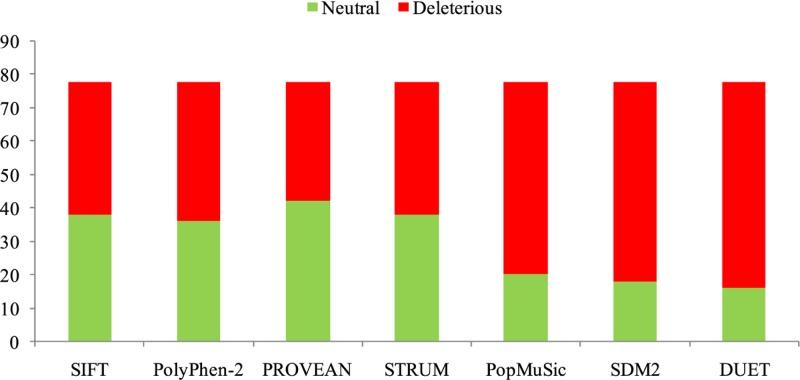
Sequence-based prediction of all nsSNPs in TEN1 gene was calculated by SIFT, PROVEAN, and PolyPhen-2. A total of 78 nsSNPs of human TEN1 gene were considered for analysis. Sequence-and structure-based predictions are listed in Supplementary Table S1 and 2. SIFT, PolyPhen-2, and PROVEAN predicted that out of 78 nsSNPs, 40 (51%), 42 (53%), 36 (46%) nsSNPs, respectively, were deleterious (Figure 3). Similarly, STRUM, mCSM, DUET, SDM2, and PoPMuSiC predicted that 40 (51%), 70 (89%), 62 (79%), 60 (76%), and 58 (74%) nsSNPs, respectively, as protein destabilizing (Figure 3). We have further focussed only on those mutations which are predicted to be deleterious and identified 34 mutations showing a destabilizing behavior.
Figure 3. Distribution of predicted deleterious (red) and neutral (green) nsSNPs in TEN1 gene.
Sequence conservation analysis
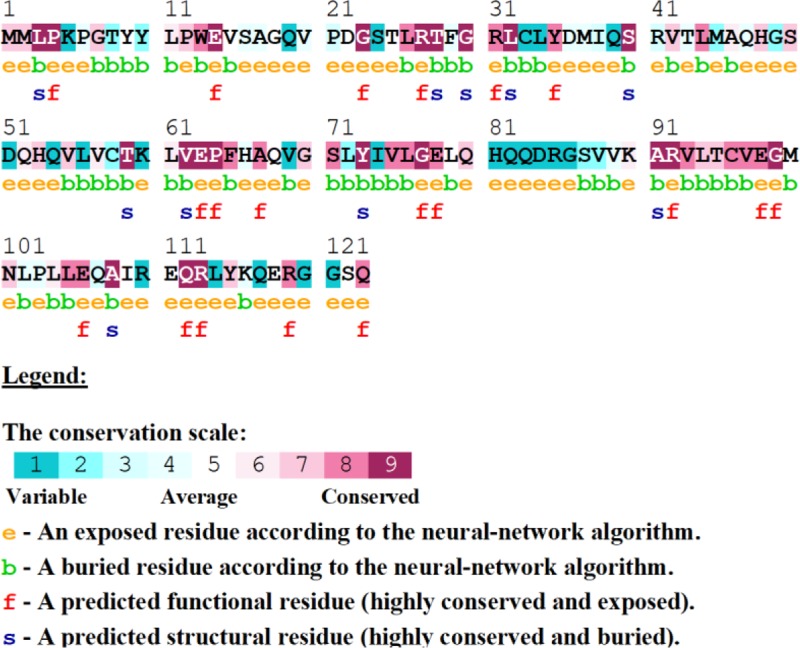
A relative analysis of amino acid residue conservation based on protein sequence provides an understanding of the significance of particular amino acid residue and reveals its localized evolution. ConSurf results indicate that the amino acid residues stretch ranges, 26–32, 62–65, 75–78, and 91–99, were highly conserved (Figure 4). The stretches of amino acids residues range, 32–61 and 100–121, are not conserved. Further, structure-based conservation analysis suggested that amino acid residue belongs to β1 (25–36 residues) and L1-2 (37–40 residues) (loop connecting β1 and β2), β4 (72–80 residues) and β5 (88–96 residues) are more conserved than β2 (41–48 residues) and β3 (51–58 residues) of TEN1 protein. Amongst these structural components, β5 (88-96 residues) is highly conserved while L4-5 (81–87 residues) (loop connecting β4 and β5) is the least conserved. Figure 5.
Figure 4. Conservation analysis of the TEN1 protein using ConSurf.
ConSurf analysis also entails structural importance of a particular residue along with conservation score.
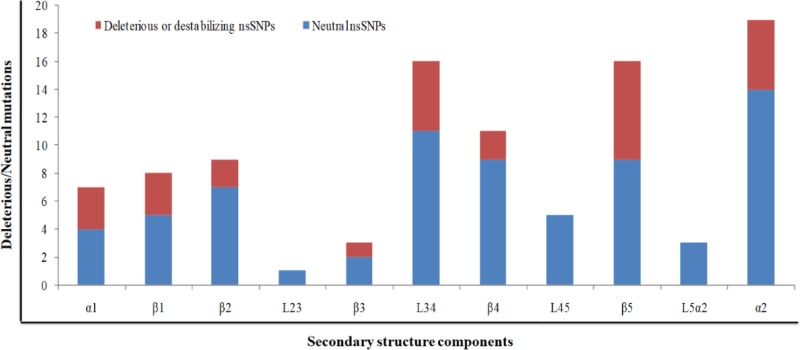
Figure 5. Distribution of deleterious/destabilizing and neutral nsSNPs in different structural components in TEN1 protein.
Distribution of deleterious or destabilizing nsSNPs
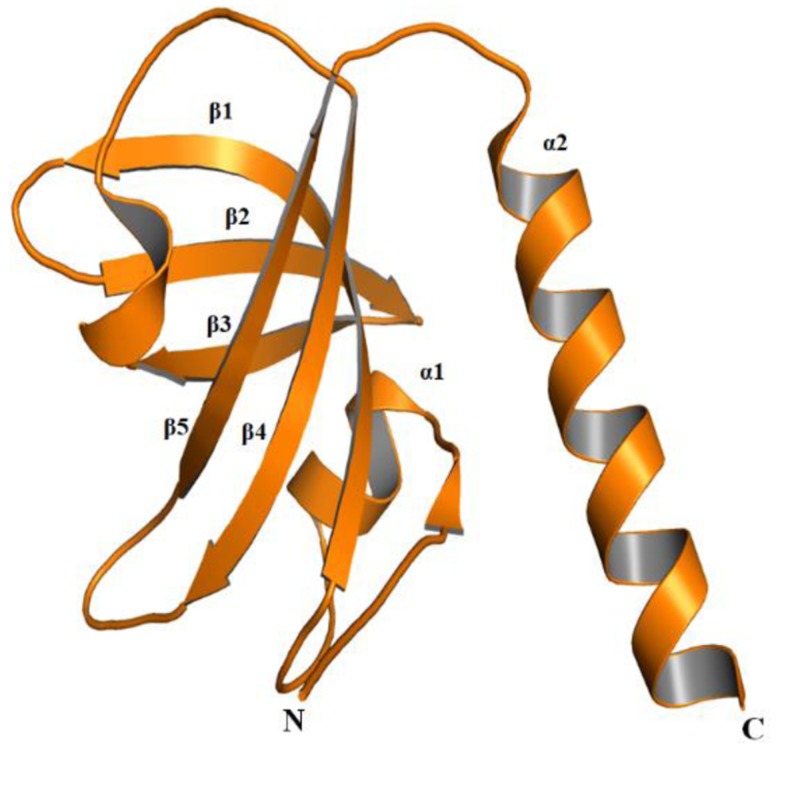
TEN1 comprises of 123 amino acid residues and have one OB (oligonucleotide or oligosaccharides)-fold domain (Figure 6) [31]. The OB-folds domain was originally identified from a group of yeast and bacteria [59]. The OB-fold domain can bind and establish protein-DNA, protein-RNA, and protein–protein interactions [60,61]. Amongst these functions, the interaction of OB-folds with ssDNA is extensively studied and characterized [13,62]. Structurally, the OB-folds are β-barrel consisting of five antiparallel β-strands capped by one α-helix at one end has a binding cleft on the other end. The variability in length amongst OB-folds domain is mainly due to the differences in the lengths of variable loops connecting the conserved secondary structure elements [62].
Figure 6. Cartoon representation of TEN1 protein (PDB ID: 4JOI).
Identification of relative percentage of high confidence nsSNPs in the OB-fold of TEN1 protein provides information about the relationship of a particular secondary structure component to be neutral or pathogenic. The secondary structure components; α1, β1, β2, β3, L3-4 (loop connecting the β3 and β4), β4, β5, α2, respectively have 75, 60, 28, 50, 45, 22, 77, and 35% deleterious or destabilizing mutations (Figure 6). Mutations in the α1 and β5 are having more than 75% chance to be deleterious, while β1, β3, and L3-4 have about 50% chance. In addition, mutations in L1-2, L2-3, L4-5, and L5-α2 (loop connecting β5 and α2) suggested that nsSNPs occurring in these region have negligible chance to be deleterious. From these results, we can suggest that mutations in the α1, β1, and β5 are possibly more lethal than in other parts of TEN1. These observations were further complemented by sequence conservation analysis, which suggested that residues belonging to α1, β1, and β5 of TEN1 are highly conserved.
Evaluation of disease phenotype
High confidence nsSNPs (deleterious and destabilizing) were analyzed for their phenotypic association using MutPred and PhD-SNP methods (Table 1). These methods predict a particular mutation as benign or pathogenic based on prediction score. MutPred and PhD-SNP methods depict 14 (58%) and 10 (29%), respectively mutations are associated with the disease phenotype. Of the 34 high confidence nsSNPs, we have identified only eight (24%) mutations (W13G, L26P, C58Y, G70A, G77R, R92H, R92C, and C96Y) as pathogenic from both prediction methods. We can conclude that eight (10%) of the total mutation, 78 (100%), found in TEN1 gene are pathogenic in nature.
Table 1. Prediction of disease phenotype analysis of high confidence nsSNPs in TEN1 gene using PhD-SNP and MutPred prediction tools.
| S. No. | Variant ID | Variants | PhD-SNP | MutPred2 | |
|---|---|---|---|---|---|
| Remark | Score | Remark | |||
| 1. | rs1322628164 | M2V | Neutral | 0.329 | Benign |
| 2. | rs892524367 | P4L | Neutral | 0.543 | Pathogenic |
| 3. | rs1212831970 | Y9C | Disease | 0.326 | Benign |
| 4. | rs1333358260 | W13G | Disease | 0.684 | Pathogenic |
| 5. | rs1224481693 | E14D | Neutral | 0.528 | Pathogenic |
| 6. | rs1175908725 | V15 F | Neutral | 0.584 | Pathogenic |
| 7. | rs1328038606 | G18V | Disease | 0.325 | Benign |
| 8. | rs964588646 | G23E | Neutral | 0.744 | Pathogenic |
| 9. | rs376979590 | T25M | Neutral | 0.171 | Benign |
| 10. | rs1262136645 | L26P | Disease | 0.855 | Pathogenic |
| 11. | rs1223059981 | D36N | Neutral | 0.301 | Benign |
| 12. | rs1250997925 | R41S | Neutral | 0.221 | Benign |
| 13. | rs1178755431 | L44V | Neutral | 0.286 | Benign |
| 14. | rs1412009927 | C58Y | Disease | 0.581 | Pathogenic |
| 15. | rs977512123 | L61M | Neutral | 0.168 | Benign |
| 16. | rs1032051988 | L61W | Neutral | 0.575 | Pathogenic |
| 17. | rs889310547 | P64T | Neutral | 0.510 | Pathogenic |
| 18. | rs951187486 | G70A | Disease | 0.482 | Pathogenic |
| 19. | rs1180274799 | G70S | Neutral | 0.545 | Pathogenic |
| 20. | rs1445270614 | Y73C | Neutral | 0.901 | Pathogenic |
| 21. | rs1358892195 | G77R | Disease | 0.880 | Pathogenic |
| 22. | rs562062613 | V88G | Neutral | 0.488 | Benign |
| 23. | rs1401886733 | A91V | Neutral | 0.831 | Pathogenic |
| 24. | rs1016457057 | R92H | Disease | 0.831 | Pathogenic |
| 25. | rs759839415 | R92C | Disease | 0.909 | Pathogenic |
| 26. | rs905216603 | V93M | Neutral | 0.543 | Pathogenic |
| 27. | rs1286634889 | C96Y | Disease | 0.922 | Pathogenic |
| 28. | rs1286634889 | C96F | Neutral | 0.906 | Pathogenic |
| 29. | rs368827427 | V97M | Neutral | 0.707 | Pathogenic |
| 30. | rs1216398771 | E106D | Neutral | 0.226 | Benign |
| 31. | rs1230794805 | R110W | Neutral | 0.130 | Benign |
| 32. | rs1158635929 | E111G | Neutral | 0.280 | Benign |
| 33. | rs772974788 | R119G | Neutral | 0.382 | Benign |
| 34. | rs772974788 | R119W | Neutral | 0.277 | Benign |
Analysis of conformational changes in protein structure
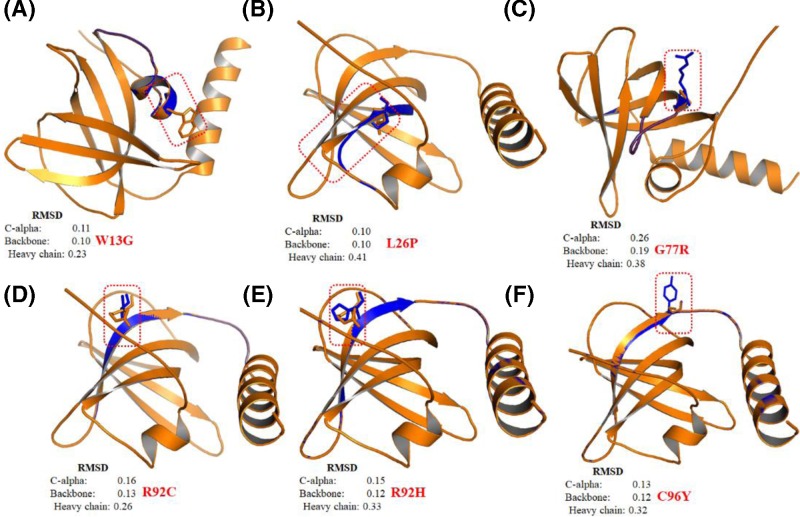
Root mean square deviation (RMSD) is a commonly used quantitative measure of the similarity between two superimposed atomic coordinates, considered as a relative measure of structural and conformational changes in a given protein structure [63]. We have performed a comparative analysis of modeled tertiary structure of mutant proteins with the WT to deduce possible structural and functional consequences imposed by pathogenic nsSNPs in TEN1 protein. We have superimposed the six pathogenic mutants (W13G, L26P, G77R, R92H, R92C, and C96Y) of TEN1 protein onto the structure of WT protein using PyMol (Figure 7A–F). Mutation G77R in the β4-strand of TEN1 protein showed a remarkable conformational change with the highest RMSD values in comparison with other mutations (Figure 7C). R92H and R92C mutations are involving the substitution of arginine by a small histidine and cysteine, thus expecting to affect the conformation of TEN1 protein which is evident from changes in RMSD values of backbone atoms (Figure 7D,E). Other three pathogenic mutations (W13G, L26P, and C96Y) are also showing a considerable structural change in the local structure as compared to WT.
Figure 7. Structural superimposition of Wild-type (Tan color) and mutant (Blue color) TEN1 proteins using PyMol.
(A) W13G, (B) L26P, (C) G77R, (D) R92C, (E) R92H, and (F) C96Y.
Aggregation propensities analysis
Protein solubility is one of the critical attribute primarily related to its function [64,65]. Insoluble parts in proteins often tend to form an aggregate which leads to development of many diseases including, amyloidoses [66], Alzheimer’s [67], and Parkinson diseases [68]. Aggregation propensity analysis was performed in the context of identification of a disease or pathogenic SNPs. SODA classifies SNPs based on changes in α-helix and β-strand propensities; aggregation and disorder score, etc. Out of eight pathogenic mutations obtained from MutPred and PhD-SNP tools, six (75%) were found to have an increased tendency to form an aggregate (Table 2). These aggregate forming potential of amino acid residues are primarily located at the C-terminal of TEN1 protein. Replacement of Arg92 by cysteine or histidine is considerably more prone to form an aggregate in comparison with other pathogenic mutations.
Table 2. Predicted aggregation scores of wild-type and mutant TEN1 proteins using SODA server.
| Variants | Helix | Strand | Aggregation | Disorder | SODA | Remark |
|---|---|---|---|---|---|---|
| 4JOI* | 0.293 | 0.316 | −4.44 | 0.089 | ||
| W13G | −0.211 | −0.75 | 4.7 | 0.748 | 4.072 | More soluble |
| L26P | −1.086 | −0.695 | 8.87 | 0.339 | 8.18 | More soluble |
| C58Y | −0.139 | 0.259 | −10.084 | −0.059 | −8.416 | Less soluble |
| G70A | 5.76 | −4.364 | −12.264 | 0.021 | −10.323 | Less soluble |
| G77R | 7.374 | −5.892 | −6.704 | 0.166 | −1.741 | Less soluble |
| R92H | 0.768 | −0.72 | −15.677 | −0.041 | −16.19 | Less soluble |
| R92C | 1.575 | −1.358 | −42.972 | 0.114 | −45.157 | Less soluble |
| C96Y | 1.861 | −1.429 | −8.643 | 0.067 | −6.102 | Less soluble |
4JOI* = PDB ID of wild-type
Structural and functional consequence of mutations
The OB-fold of TEN1 comprises of five antiparallel β-strands folded into a complex β-barrel flanked by two α-helices. N-terminal residues forming a long coil and plays a crucial role in STN1-TEN1 complex formation. Following N-terminal coil, there is a short α-helix (α1) located at an interface of two β-sheets known to provide stability to the structure. However, the C-terminal α-helix (α2) is situated at the opposite end of the β-barrel and spans the whole length of the structure. The N-terminal of STN1 forms a stable heterodimer complex with TEN1. Complex formation between these two proteins is mediated by extensive interactions between the α2- and α3-helices of TEN1 and STN1, respectively (Supplementary Figure S2A). In addition to α-helices, β-barrels of TEN1 and STN1 also form extensive contacts (Supplementary Figure S2B) [31].
Some important amino acid residues, including Val159, Trp160, Ile164, Met167, and Leu168, of α3-helix and some region of flanking coils of STN1 form extensive hydrophobic contacts with the amino acid residues, Met100, Leu104, Leu105, and Ile109, of α2 of TEN1 (Supplementary Figure S2C). Additional interactions between the STN1 and TEN1 are mainly mediated by the conserved Tyr115 of TEN1 α2. Tyr115 is found at the interface of the STN1 and TEN1 and known to form extensive hydrophobic contacts with the side chains of Tyr49, Pro171, and Tyr174 of STN1. Similarly, interactions between the STN1 and TEN1 involve the surface of the β-barrels and the N-terminal tail of TEN1, that runs along the interface of the two domains and form extensive contacts with both of these two proteins (Supplementary Figure S2D). In particular, Arg27 of β1-strand and Arg119 of α2 of TEN1 make an important salt bridge with Asp78 of β2-strand and Asp33 of α2 of STN1, respectively. Further, Met167 of STN1 spans toward α2 and β-barrel interface of TEN1 and form extensive interactions with Leu105, Ala108, and Ile109 of α2 and Tyr9 of the N-terminal coil. It is fascinating that the STN1-TEN1 complex positions the ligand-binding pockets of each subunit on the same side of the heterodimer, forming an extensive ligand-binding pocket [31].
Mutations in protein are often coupled with destabilization or some time associated with disease pathogenesis. Previous studies on mutational analysis demonstrated that the effects of mutations on the stability of protein are primarily owing to changes in hydrophobic contacts [69–71]. However, subsequent studies in a number of cases revealed that substitutions of a large amino acid with smaller ones are usually accompanied by the formation of cavity and effect residue depth and solvent accessibility [72–75]. To find out the impact of a particular mutation on the local and global environment of TEN1 protein structure, we have calculated van der Waals, hydrogen bonding, electrostatic and hydrophobic interactions in WT, and mutant TEN1 using Arpeggio web server (Table 3) [76]. We have estimated the change in the RSA, OSP, and residue depth of wild-type and mutant TEN1 proteins (Figure S3).
Table 3. Predictions of non-covalent interactions in wild-type and mutant TEN1 proteins using Arpeggio web server.
| Variants | van der Waals interactions | Hydrogen bonds | Ionic interactions | Aromatic contacts | Hydrophobic contacts |
|---|---|---|---|---|---|
| 4JOI* | 64 | 101 | 18 | 27 | 235 |
| P4L | 64 | 102 | 18 | 27 | 235 |
| W13G | 67 | 102 | 18 | 7 | 218 |
| E14D | 64 | 101 | 13 | 27 | 236 |
| V15F | 63 | 102 | 18 | 27 | 263 |
| G23E | 63 | 102 | 18 | 27 | 235 |
| L26P | 63 | 101 | 18 | 27 | 231 |
| C58Y | 64 | 101 | 18 | 27 | 242 |
| L61W | 64 | 103 | 18 | 27 | 240 |
| P64T | 64 | 102 | 18 | 27 | 235 |
| G70A | 64 | 102 | 18 | 27 | 235 |
| G70S | 64 | 102 | 18 | 27 | 235 |
| Y73C | 65 | 101 | 18 | 27 | 222 |
| G77R | 65 | 102 | 18 | 27 | 239 |
| A91V | 65 | 102 | 18 | 27 | 246 |
| R92H | 65 | 100 | 14 | 27 | 235 |
| R92C | 64 | 100 | 12 | 27 | 235 |
| V93M | 64 | 102 | 18 | 27 | 234 |
| C96Y | 65 | 101 | 18 | 35 | 237 |
| C96F | 65 | 101 | 18 | 28 | 243 |
| V97M | 64 | 102 | 18 | 27 | 240 |
4JOI* = PDB ID of wild-type TEN1 protein.
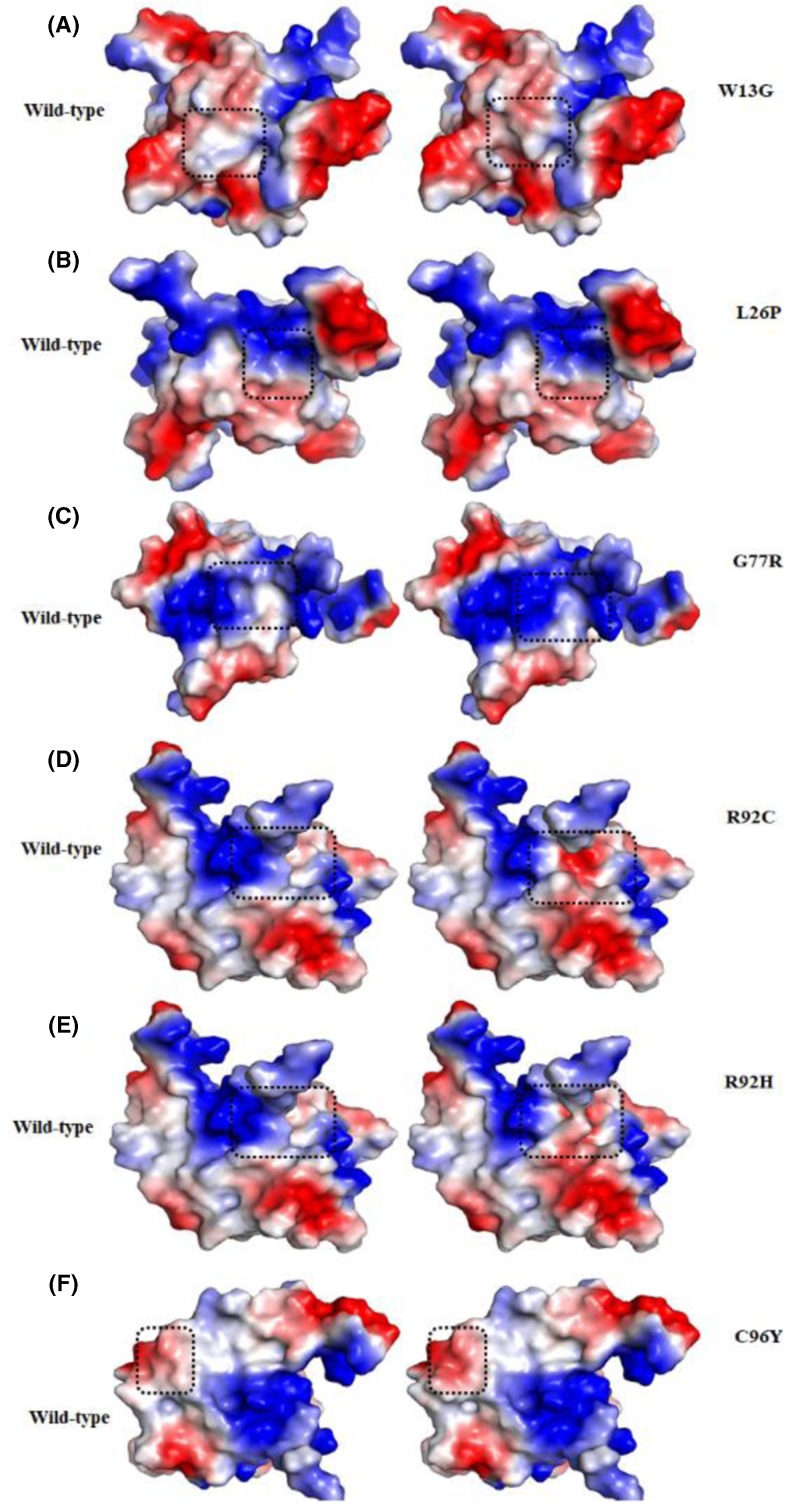
Trp13 is a highly conserved and a buried residue of the N-terminal flanking coil and plays important role in STN1-TEN1 complex formation. Substitutions of larger bulky and highly hydrophobic Trp13 by small, less hydrophobic glycine does not change van der Waals and hydrogen bond interactions significantly, while a large decrease in stacking and hydrophobic interactions are observed (Table 3). Differences in the size and polarity of Trp and glycine affecting the RSA, OSP, and residue depth of protein. Increased RSA value in the Trp13Gly substitution suggested that the substituted residue at Trp13 becomes more accessible to solvent, which is further supported by a decrease in packing density (Supplementary Figure S3). The Surface potential analysis shows a decrease in hydrophobicity in Trp13Gly substitution (Figure 8A). The results suggested that the substitution of Trp13 with the glycine seems indispensable for the stability of TEN1 structure.
Figure 8. Surface potential representations of WT (left panel) and mutant (right panel) TEN1 proteins.
(A) W13G, (B) L26P, (C) G77R, (D) R92C, (E) R92H and (F) C96Y. The color ramp for the electrostatic surface potential ranges from blue (most positive) to red (most negative). Surface potential of WT and mutant residues are highlighted by dashed square.
Similarly, Leu26 is a highly conserved and buried residue found at the β1-strand of TEN1. Substitutions of hydrophobic Leu26 by a less hydrophobic proline effects only van der Waals and hydrophobic interaction at a little extent (Table 1). However, no significant change was observed for RSA, OSP, surface potential, and residue depth by Leu26Pro mutation (Figure 8B). We may conclude that the incorporation of imino group as a side chain of proline may interfere with the folding pathway of TEN1 without effecting non-covalent interactions.
Gly77 is located in the β4-strand of TEN1 and plays an important role in maintaining the structure and stability (Figure 7D). Substitution of small, hydrophobic, highly conserved, exposed, and functional Gly77 by a large and least hydrophobic, positively charged arginine shows an increase in the van der Waals, hydrogen bonding, and hydrophobic interactions. In consistence, Gly77Arg mutation shows an increase in RSA, a subtle decrease in OSP and residue depth. Gly77Arg mutation increases positively charge environment in the vicinity of Gly77 (Figure 8C). Lethality of Gly77Arg mutation is associated with the changes in RSA of surrounding residues which are critical to maintaining the TEN1 stability.
Arg92 is belonging to the β4-strand of TEN1 and is important for the stability. Substitution of large, highly hydrophilic, conserved, exposed, and functional Arg92 by a small less hydrophilic, positively charged (histidine), and uncharged (cysteine) shows a disruption of one hydrogen bond and a large decrease in the ionic interactions. While, no significant change was observed in the van der Waals, stacking and hydrophobic interactions. Similarly, Arg92His and Arg92Cys mutations show an increase in RSA, and a slight decrease in the OSP and residue depth. The increase in RSA suggesting that the substitution of Arg92 may increase the solvent accessibility of newly incorporated residues. A marked change in surface potential has been observed in Arg92His and Arg92Cys mutations (Figure 8D,E). These results indicate that the lethal effect of Arg92His and Arg92Cys mutations is primarily associated with the changes in hydrogen bonding, ionic interactions, and RSA and thus protein stability.
Cys96 is situated in the β5-strand of TEN1. Substitution of small, less hydrophobic, highly conserved and buried Cys96 by a large and more hydrophobic tyrosine show an increase in the stacking and hydrophobic interactions, while no change was observed in other interactions. Cys96Tyr mutation shows an increase in the RSA and decrease in OSP. No significant change in surface potential has been found except an increase in the hydrophobicity (Figure 8F). Our findings suggest that Cys96Tyr mutation may increase the important hydrophobic and stacking interactions which are being considered as a driving force for protein stability. These increase in stability possibly overcome due to disruptions some important interaction Cys96.
Conclusion
SNPs are considered as one of the most recurring genetic variants associated with a number of diseases. In the present study, we have examined the consequences of nsSNPs in TEN1 gene using advanced integrated bioinformatics approach. We have identified a large number of deleterious and destabilizing nsSNPs, which are scattered in different secondary structural components of TEN1 with a high chance of occurring in α1-helix and β5-strands. Aggregation propensity analysis of pathogenic mutation shows that 75% of pathogenic mutations in TEN1 have a tendency to form aggregate and located at C-terminal of TEN1. In-depth structural analysis of these mutations reveals that the pathogenicity of these mutations may be driven through a large structural changes caused by loss/gain of non-covalent intramolecular interactions. The present study provides a mechanistic insight into the understanding of pathogenic mutations in TEN1 gene and their possible consequences.
Supporting information
Supplementary Figure S1.
Supplementary Figure S2.
Supplementary Figure S3.
Supplemental Table S1. Sequence-based predictions of deleterious nsSNPs in TEN1 gene using SIFT, PolyPhen-2 and PROVEAN.
Supplemental Table S2. Structure-based predictions of destabilizing nsSNPs in TEN1 gene using STRUM, mCSM, SDM2, DUET and PopMuSic.
Acknowledgments
Authors are highly thankful to Department of Science and Technology, and Council of Scientific and Industrial Research, India, for financial support. M.F.A., M.T.R., and A.H. are thankful to the generous support from the Deanship of Scientific Research at King Saud University, Riyadh, Kingdom of Saudi Arabia (grant number RGP-150). F.A. is grateful to Indian National Science Academy for INSA senior scientist fellowship.
Abbreviations
- CC1
conserved telomere maintenance component 1
- ESST
environment-specific substitution table
- OSP
occluded packing density
- PDB
protein data bank
- PolyPhen-2
polymorphism phenotyping-2
- PROVEAN
protein variation effect analyzer
- PSIC
position-specific independent count
- RMSD
root mean square deviation
- RSA
relative side-chain solvent accessibility
- SDM2
site direct mutator 2
- SIFT
sorting intolerant from tolerant
- SODA
protein solubility from disorder and aggregation
- SVM
support vector machine
Funding
M.A. is thankful to Council of Scientific and Industrial Research, India for the award of senior research fellowship [grant number 09/466(0197)2K18 EMR-7].
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
M.A., V.K., T.M. and M.I.H. prepared the manuscript, and conceived and designed the study. R.D., M.T.R., M.F.A., and A.H. contributed to data analysis/interpretation. F.A. and M.A. contributed to the literature research. V.K., T.M., and M.I.H. contributed to the experimental studies, and data collection. M.I.H. was the guarantor of integrity of the entire study and responsible for the manuscript preparation, editing, and review. All authors approved the final version of the manuscript for submission.
References
- 1.Blackburn E.H. and Gall J.G. (1978) A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 120, 33–53 10.1016/0022-2836(78)90294-2 [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. (2009) How telomeres solve the end-protection problem. Science 326, 948–952 10.1126/science.1170633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longhese M.P. (2008) DNA damage response at functional and dysfunctional telomeres. Genes Dev. 22, 125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maser R.S. and DePinho R.A. (2004) Telomeres and the DNA damage response: why the fox is guarding the henhouse. DNA Repair 3, 979–988 10.1016/j.dnarep.2004.05.009 [DOI] [PubMed] [Google Scholar]
- 5.De Lange T. (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 6.Wang F. and Lei M. (2011) Human Telomere POT1-TPP1 Complex and Its Role in Telomerase Activity Regulation, In Telomeres and Telomerase, pp. 173–187, Springer; [DOI] [PubMed] [Google Scholar]
- 7.Wang F. et al. (2007) The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature 445, 506 10.1038/nature05454 [DOI] [PubMed] [Google Scholar]
- 8.Stewart J.A. et al. (2018) Emerging roles of CST in maintaining genome stability and human disease. Front. Biosci. (Landmark Ed.) 23, 1564–1586 10.2741/4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wellinger R.J. (2009) The CST complex and telomere maintenance: the exception becomes the rule. Mol. Cell 36, 168–169 10.1016/j.molcel.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 10.Nakaoka H. et al. (2012) Xenopus laevis Ctc1-Stn1-Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J. Biol. Chem. 287, 619–627 10.1074/jbc.M111.263723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L.-Y., Redon S. and Lingner J. (2012) The human CST complex is a terminator of telomerase activity. Nature 488, 540 10.1038/nature11269 [DOI] [PubMed] [Google Scholar]
- 12.Ganduri S. and Lue N.F. (2017) STN1–POLA2 interaction provides a basis for primase-pol α stimulation by human STN1. Nucleic Acids Res. 45, 9455–9466 10.1093/nar/gkx621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharjee A. et al. (2016) STN1 OB fold mutation alters DNA binding and affects selective aspects of CST function. PLoS Genet. 12, e1006342 10.1371/journal.pgen.1006342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharjee A. et al. (2017) Dynamic DNA binding, junction recognition and G4 melting activity underlie the telomeric and genome-wide roles of human CST. Nucleic Acids Res. 45, 12311–12324 10.1093/nar/gkx878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson E. et al. (1987) Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine· guanine base pairs. Cell 51, 899–908 10.1016/0092-8674(87)90577-0 [DOI] [PubMed] [Google Scholar]
- 16.Lue N.F. (2018) Evolving linear chromosomes and telomeres: a C-strand-centric view. Trends Biochem. Sci. 43, 314–326 10.1016/j.tibs.2018.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chastain M. et al. (2016) Human CST facilitates genome-wide RAD51 recruitment to GC-rich repetitive sequences in response to replication stress. Cell Rep. 16, 1300–1314 10.1016/j.celrep.2016.06.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart J.A. et al. (2012) Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 31, 3537–3549 10.1038/emboj.2012.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C., Dai X. and Chai W. (2012) Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 22, 1681 10.1038/cr.2012.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyake Y. et al. (2009) RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36, 193–206 10.1016/j.molcel.2009.08.009 [DOI] [PubMed] [Google Scholar]
- 21.Feng X. et al. (2017) CTC1-mediated C-strand fill-in is an essential step in telomere length maintenance. Nucleic Acids Res. 45, 4281–4293 10.1093/nar/gkx125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan X. and Price C.M. (1997) Coordinate regulation of G-and C strand length during new telomere synthesis. Mol. Biol. Cell 8, 2145–2155 10.1091/mbc.8.11.2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Y. et al. (2014) CTC1 increases the radioresistance of human melanoma cells by inhibiting telomere shortening and apoptosis. Int. J. Mol. Med. 33, 1484–1490 10.3892/ijmm.2014.1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bisserbe A. et al. (2015) Cerebro-retinal microangiopathy with calcifications and cysts due to recessive mutations in the CTC1 gene. Rev. Neurol. (Paris) 171, 445–449 10.1016/j.neurol.2015.01.566 [DOI] [PubMed] [Google Scholar]
- 25.Gu P. and Chang S. (2013) Functional characterization of human CTC 1 mutations reveals novel mechanisms responsible for the pathogenesis of the telomere disease C oats plus. Aging Cell 12, 1100–1109 10.1111/acel.12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y. and Chai W. (2018) Pathogenic CTC1 mutations cause global genome instabilities under replication stress. Nucleic Acids Res. 46, 3981–3992 10.1093/nar/gky114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller R.B. et al. (2012) CTC1 Mutations in a patient with dyskeratosis congenita. Pediatr. Blood Cancer 59, 311–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wartiovaara-Kautto U. et al. (2016) A novel homozygous CTC1 germline mutation associated with bone marrow failure. Blood 128, 1508 [Google Scholar]
- 29.Yates C.M. and Sternberg M.J. (2013) The effects of non-synonymous single nucleotide polymorphisms (nsSNPs) on protein–protein interactions. J. Mol. Biol. 425, 3949–3963 10.1016/j.jmb.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 30.Tokuriki N. and Tawfik D.S. (2009) Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 19, 596–604 10.1016/j.sbi.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 31.Bryan C. et al. (2013) Structure of the human telomeric Stn1-Ten1 capping complex. PLoS ONE 8, e66756 10.1371/journal.pone.0066756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherry S.T. et al. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubbard T. et al. (2002) The Ensembl Genome Database Project. Nucleic Acids Res. 30, 38–41 10.1093/nar/30.1.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenson P.D. et al. (2009) The human gene mutation database: 2008 update. Genome Med. 1, 13 10.1186/gm13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berman H.M. et al. (2000) The protein data bank. Nucleic Acids Res. 28, 235–242 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng P.C. and Henikoff S. (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar P., Henikoff S. and Ng P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 38.Choi Y. and Chan A. (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramensky V., Bork P. and Sunyaev S. (2002) Human non‐synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900 10.1093/nar/gkf493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quan L., Lv Q. and Zhang Y. (2016) STRUM: structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics 32, 2936–2946 10.1093/bioinformatics/btw361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandurangan A.P. et al. (2017) SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res. 45, W229–W235 10.1093/nar/gkx439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overington J. et al. (1992) Environment‐specific amino acid substitution tables: tertiary templates and prediction of protein folds. Protein Sci. 1, 216–226 10.1002/pro.5560010203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pires D.E., Ascher D.B. and Blundell T.L. (2014) DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 42, W314–W319 10.1093/nar/gku411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pires D.E., Ascher D.B. and Blundell T.L. (2013) mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 30, 335–342 10.1093/bioinformatics/btt691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B. et al. (2009) Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25, 2744–2750 10.1093/bioinformatics/btp528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul S.F. et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finn R.D. et al. (2015) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krogh A. et al. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 49.Sickmeier M. et al. (2006) DisProt: the database of disordered proteins. Nucleic Acids Res. 35, D786–D793 10.1093/nar/gkl893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delorenzi M. and Speed T. (2002) An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625 10.1093/bioinformatics/18.4.617 [DOI] [PubMed] [Google Scholar]
- 51.Capriotti E., Calabrese R. and Casadio R. (2006) Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 22, 2729–2734 10.1093/bioinformatics/btl423 [DOI] [PubMed] [Google Scholar]
- 52.Walsh I. et al. (2014) PASTA 2.0: an improved server for protein aggregation prediction. Nucleic Acids Res. 42, W301–W307 10.1093/nar/gku399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walsh I. et al. (2011) ESpritz: accurate and fast prediction of protein disorder. Bioinformatics 28, 503–509 10.1093/bioinformatics/btr682 [DOI] [PubMed] [Google Scholar]
- 54.Kyte J. and Doolittle R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- 55.Piovesan D. et al. (2017) FELLS: fast estimator of latent local structure. Bioinformatics 33, 1889–1891 10.1093/bioinformatics/btx085 [DOI] [PubMed] [Google Scholar]
- 56.Paladin L., Piovesan D. and Tosatto S.C. (2017) SODA: prediction of protein solubility from disorder and aggregation propensity. Nucleic Acids Res. 45, W236–W240 10.1093/nar/gkx412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashkenazy H. et al. (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 10.1093/nar/gkw408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee B. and Richards F.M. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55, 379–IN4 10.1016/0022-2836(71)90324-X [DOI] [PubMed] [Google Scholar]
- 59.Murzin A.G. (1993) OB (oligonucleotide/oligosaccharide binding)‐fold: common structural and functional solution for non‐homologous sequences. EMBO J. 12, 861–867 10.1002/j.1460-2075.1993.tb05726.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agrawal N. et al. (2003) RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 67, 657–685 10.1128/MMBR.67.4.657-685.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arcus V. (2002) OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 12, 794–801 10.1016/S0959-440X(02)00392-5 [DOI] [PubMed] [Google Scholar]
- 62.Theobald D.L., Mitton-Fry R.M. and Wuttke D.S. (2003) Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 32, 115–133 10.1146/annurev.biophys.32.110601.142506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maiorov V.N. and Crippen G.M. (1994) Significance of root-mean-square deviation in comparing three-dimensional structures of globular proteins. 235, 625–634 [DOI] [PubMed] [Google Scholar]
- 64.Balch W.E. et al. (2008) Adapting proteostasis for disease intervention. Science 319, 916–919 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- 65.Ciryam P. et al. (2013) Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 5, 781–790 10.1016/j.celrep.2013.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knowles T.P., Vendruscolo M. and Dobson C.M. (2014) The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384 10.1038/nrm3810 [DOI] [PubMed] [Google Scholar]
- 67.Thal D.R. et al. (2015) Neuropathology and biochemistry of Aβ and its aggregates in Alzheimer’s disease. Acta Neuropathol. 129, 167–182 10.1007/s00401-014-1375-y [DOI] [PubMed] [Google Scholar]
- 68.Tan J.M., Wong E.S. and Lim K.-L. (2009) Protein misfolding and aggregation in Parkinson's disease. Antioxid. Redox Signal. 11, 2119–2134 [DOI] [PubMed] [Google Scholar]
- 69.Pace C.N. (1992) Contribution of the hydrophobic effect to globular protein stability. J. Mol. Biol. 226, 29–35 10.1016/0022-2836(92)90121-Y [DOI] [PubMed] [Google Scholar]
- 70.Sturtevant J.M. (1994) The thermodynamic effects of protein mutations. Curr. Opin. Struct. Biol. 4, 69–78 10.1016/S0959-440X(94)90062-0 [DOI] [Google Scholar]
- 71.Matthews B.W. (1995) Studies on Protein Stability with T4 Lysozyme, in Advances in Protein Chemistry, pp. 249–278, Elsevier; [DOI] [PubMed] [Google Scholar]
- 72.Varadarajan R., Richards F.M. and Connelly P.R. (1990) Proteins: the hard sphere, structure, and energetics. Curr. Sci. 59, 819–824 [Google Scholar]
- 73.Eriksson A.E. et al. (1992) Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science 255, 178–183 10.1126/science.1553543 [DOI] [PubMed] [Google Scholar]
- 74.Richards F.M. and Lim W.A. (1993) An analysis of packing in the protein folding problem. Q. Rev. Biophys. 26, 423–498 10.1017/S0033583500002845 [DOI] [PubMed] [Google Scholar]
- 75.Jackson S.E. et al. (1993) Effect of cavity-creating mutations in the hydrophobic core of chymotrypsin inhibitor 2. Biochemistry 32, 11259–11269 10.1021/bi00093a001 [DOI] [PubMed] [Google Scholar]
- 76.Jubb H.C. et al. (2017) Arpeggio: a web server for calculating and visualising interatomic interactions in protein structures. J. Mol. Biol. 429, 365–371 10.1016/j.jmb.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]