Abstract
Background
The functional determinants of H3K4me3, their potential dependency on histone H2B monoubiquitination, and their contribution to defining transcriptional regimes are poorly defined in plant systems. Unlike in Saccharomyces cerevisiae, where a single SET1 protein catalyzes H3K4me3 as part of COMPlex of proteins ASsociated with Set1 (COMPASS), in Arabidopsis thaliana, this activity involves multiple histone methyltransferases. Among these, the plant-specific SET DOMAIN GROUP 2 (SDG2) has a prominent role.
Results
We report that SDG2 co-regulates hundreds of genes with SWD2-like b (S2Lb), a plant ortholog of the Swd2 axillary subunit of yeast COMPASS. We show that S2Lb co-purifies with the AtCOMPASS core subunit WDR5, and both S2Lb and SDG2 directly influence H3K4me3 enrichment over highly transcribed genes. S2Lb knockout triggers pleiotropic developmental phenotypes at the vegetative and reproductive stages, including reduced fertility and seed dormancy. However, s2lb seedlings display little transcriptomic defects as compared to the large repertoire of genes targeted by S2Lb, SDG2, or H3K4me3, suggesting that H3K4me3 enrichment is important for optimal gene induction during cellular transitions rather than for determining on/off transcriptional status. Moreover, unlike in budding yeast, most of the S2Lb and H3K4me3 genomic distribution does not rely on a trans-histone crosstalk with histone H2B monoubiquitination.
Conclusions
Collectively, this study unveils that the evolutionarily conserved COMPASS-like complex has been co-opted by the plant-specific SDG2 histone methyltransferase and mediates H3K4me3 deposition through an H2B monoubiquitination-independent pathway in Arabidopsis.
Electronic supplementary material
The online version of this article (10.1186/s13059-019-1705-4) contains supplementary material, which is available to authorized users.
Background
Dynamic changes in chromatin organization and composition rely on many activities, such as the remodeling of nucleosome positioning and the incorporation of histone variants, DNA methylation, and histone post-translational modifications (PTMs) [1–4]. Genome-wide profiling of histone PTMs in the Arabidopsis thaliana plant species has established that transcriptionally active genes are typically marked by acetylated histones H3 and H4, monoubiquitinated histone H2B (H2Bub), and di/trimethylated histone H3 at lysine residues such as Lys-4 and Lys-36 (H3K4me2/3, H3K36me2/3) [5–7]. Combinations of histone modifications contribute to create a multilayered system of chromatin states and transcriptional activity [8]. In plant systems, molecular mechanisms underpinning the functional interdependencies between different histone modifications have recently been reported for Polycomb-mediated gene repression [9] but have not been characterized for active transcription.
One of the best-described cases of functional trans-histone crosstalk is that of H2Bub promotion of histone H3K4me3 deposition on actively transcribed genes in yeast [10] and metazoans [11]. In Saccharomyces cerevisiae, H3K4me3 deposition is catalyzed by the SET1 histone methyltransferase (HMT) embedded in a so-called COMPlex of Proteins Associated with Set1 (COMPASS), which also contains the WD40 repeats-containing proteins Swd1, Swd2, and Swd3 as well as Bre2, Spp1, and Sdc1 subunits (reviewed in [12, 13]). Tethering of Swd2 on H2Bub-modified nucleosomes is proposed to recruit yCOMPASS, polymerase-associated factor 1 complex (PAF1c), and RNA polymerase II (RNPII) to promote histone H3 Lys-4 trimethylation [14–17]. Hence, prior monoubiquitination of H2B on Lys-123 by the Rad6/Bre1 ubiquitin conjugase and E3 ligase is a prerequisite for H3K4me3 deposition by Set1 in budding yeast [10, 18–21].
COMPASS-like H3K4me3 HMT activity is evolutionarily conserved in eukaryotes with for example Trithorax (Trx) in Drosophila and mixed lineage leukemia 1 (MLL1) in humans [12, 22]. H3K4me3 is usually found on a limited number of nucleosomes surrounding the transcription start site (TSS) and is functionally linked to RNPII transcriptional activation and the switch to elongation in many eukaryotes including plants [5, 23–28].
As many as 47 distinct SET-domain proteins are encoded in the Arabidopsis genome, together forming the SET domain protein group (SDG) [29–31]. Among them, ATX1 (ARABIDOPSIS TRITHORAX1) [32] and ATXR7 (ARABIDOPSIS TRITHORAX-RELATED7) [33, 34] appear to target highly specific genomic loci or to be cell type specific whereas the plant-specific SET DOMAIN GROUP 2 (SDG2)/ARABIDOPSIS TRITHORAX RELATED 3 HMT presumably targets a broad repertoire of genes [35, 36]. Notwithstanding, while the influence of H3 Lys-4 trimethylation on transcription activation/elongation in plants has begun to emerge [37–40], the Arabidopsis genomic loci targeted by SDG2 as well as the mechanisms determining its specificity remain undetermined. ATX1 has been shown to have a high affinity for Ser5-phosphorylated RNPII, a property enabling this COMPASS-associated HMT to facilitate RNPII exit from to the promoter proximal pause region to favor transcription elongation [37, 41]. A molecular crosstalk mechanistically linking H3K4me3 and H2Bub deposition has however not been established in plants [42].
In addition to HMT diversification, Arabidopsis also possesses homologs for all known COMPASS subunits such as WDR5a and WDR5b playing the role of the yeast Swd3 core component, potentially forming several COMPASS-like complexes. More generally, all structural components of the yeast COMPASS (yCOMPASS) complex subunits appear to be conserved in Arabidopsis, such as a RbBP5-LIKE (RBL), a Swd3 homolog (WDR5a/b), and a Bre2 homolog (ARABIDOPSIS Ash2 RELATIVE or ASH2R) [40, 43–45]. They contribute to flowering time control by allowing H3K4me3 deposition on the FLOWERING-LOCUS C (FLC) regulatory gene [43, 44] but also to drought stress tolerance [46] and endoplasmic reticulum stress response [47], presumably as a consequence of a general influence on RNPII activity during cellular transitions or in response to environmental signals. Accordingly, knocking out AtCOMPASS-like core subunit genes is lethal [43, 44], indicating a fundamental role in plant development.
Here, we first identify S2Lb as an Arabidopsis homolog of the Swd2 COMPASS-associated subunit, which acts as a key component of the H2Bub-H3K4me3 trans-histone crosstalk in S. cerevisiae [14, 17]. We report that S2Lb is a euchromatic protein that functionally associates with an AtCOMPASS-like complex and with the plant-specific SDG2 HMT to broaden H3K4me3 enrichment over most transcribing genes, especially those abundantly occupied by RNPII. Using HUB1 loss-of-function plants in which H2Bub deposition is abolished [48, 49], we further unveil that COMPASS-like activities mediate H3-Lys4 trimethylation largely independently from histone H2B monoubiquitination in Arabidopsis.
Results
Two evolutionarily conserved SWD2-LIKE genes encode euchromatic proteins in A. thaliana
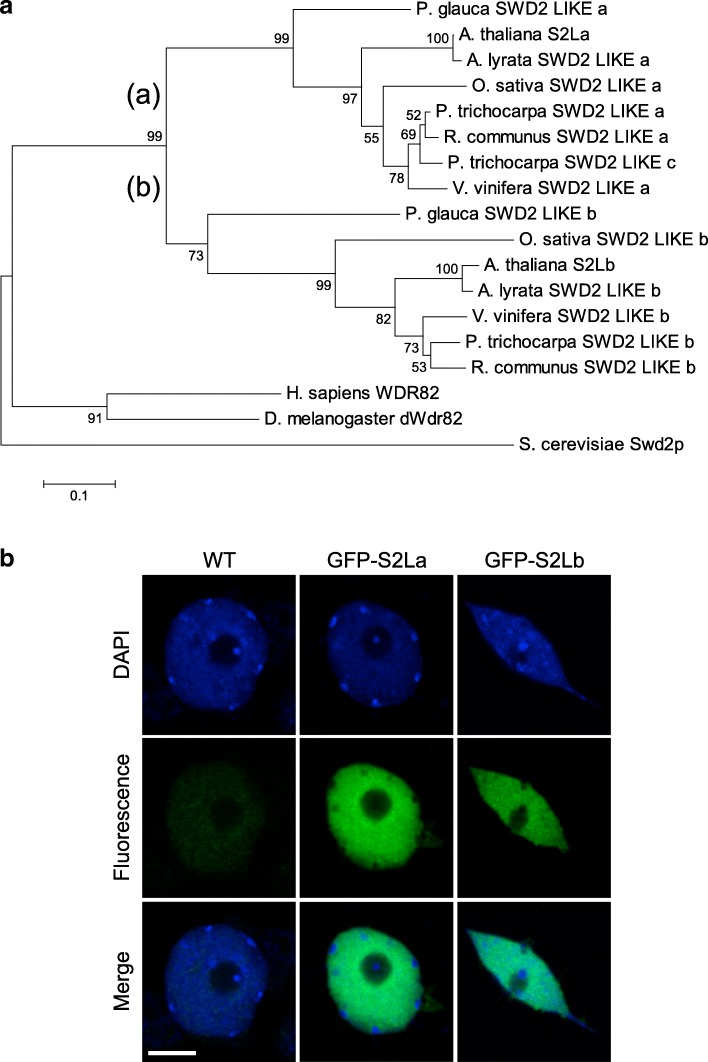
Phylogenetic analysis of the S. cerevisiae Swd2 protein sequence and putative homologs in human, Drosophila, and representative plant species revealed that plant SWD2-like genes form a distinct clade from metazoan orthologs (Fig. 1a; Additional file 1). The presence of two or more SWD2-LIKE genes suggests that a gene duplication event occurred before the separation of gymnosperms and angiosperms. In Arabidopsis, At5g14530 and At5g66240, designated here as SWD2-LIKE-a (S2La) and S2Lb, encode such predicted paralogs with high amino acids sequence similarity with yeast Swd2 (45.3% and 43.4%, respectively). These two genes were first identified for their influence on Arabidopsis flowering time as Anthesis promoting factor 1 (APRF1; [50]) and ubiquitin ligase complex subunit 1 ([51]), respectively. RT-qPCR analysis of whole seedlings and of different adult plant organs showed that both genes are broadly expressed, S2Lb being usually expressed to a much higher level than S2La (Additional file 2: Figure S1). This difference is also apparent in publicly available anatomy-related transcriptomes [52], with S2La mRNA being mildly detected in all analyzed samples except in senescent leaves (Additional file 2: Figure S1). In addition to this differential regulation, structural variations can be identified between S2La and S2Lb, and with their yeast and human orthologs. Six canonical WD40 repeats can be identified in S2La and S2Lb versus seven in Swd2 and in the human homolog Wdr82 (Additional file 2: Figure S2). The WD40 repeat IV was not detected in the central part of S2La, and the S2Lb carboxy-terminal domain carries divergent WD40 repeats. Sub-nuclear immunolocalization of GFP-tagged S2La and S2Lb stably expressed in planta further showed that both proteins localize to euchromatin and are excluded from the nucleolar compartment and from all densely packed heterochromatic foci known as chromocenters (Fig. 1b), in agreement with a potential role linked to RNPII transcription.
Fig. 1.
Two Swd2-like euchromatin proteins in Arabidopsis. a Neighbor-joining tree of full-length Swd2-like proteins from representative species. The plant clade can be divided into two sub-clades containing each a Swd2-Like isoform, (a) and (b). The numbers indicate bootstrap values from 1000 replicates. Scale for sequence divergence is represented by a bar. Gene and protein IDs are given in Additional file 1: Table S1. b S2La and S2Lb are euchromatic proteins. Immunodetection of GFP-tagged S2La and S2Lb proteins in interphase cotyledon nuclei (green). DNA is counterstained with DAPI (blue). Scale bar, 2 μm
S2Lb but not S2La loss of function causes pleiotropic developmental defects
To explore the function of S2La or S2Lb in planta, T-DNA insertion lines interrupting each gene were obtained from public collections, which we named s2la-1 [previously described as aprf1-9 in [50]] and s2lb-1, respectively (Additional file 2: Figure S3). Being in a Nossen background, the s2lb-1 allele was introgressed in a Col-0 background through five successive backcrosses to generate the s2lb-2 line.
As described earlier [50], s2la-1 plants exhibited no apparent developmental abnormalities under laboratory growth conditions at the vegetative stage. In contrast, S2Lb loss-of-function in both Nossen and Col-0 backgrounds triggered significant growth defects resulting in small leaf size, small rosette diameter, shorter roots, and reduced number of lateral roots (Fig. 2a–e). Such defects were not observed in heterozygous plants for the s2lb-2 allele. Homozygous plants could efficiently be rescued by stably expressing GFP-tagged or native S2Lb proteins under control of the S2Lb endogenous promoter (Additional file 2: Figure S4), thus confirming the specific and recessive property of the mutation effect on plant growth.
Fig. 2.
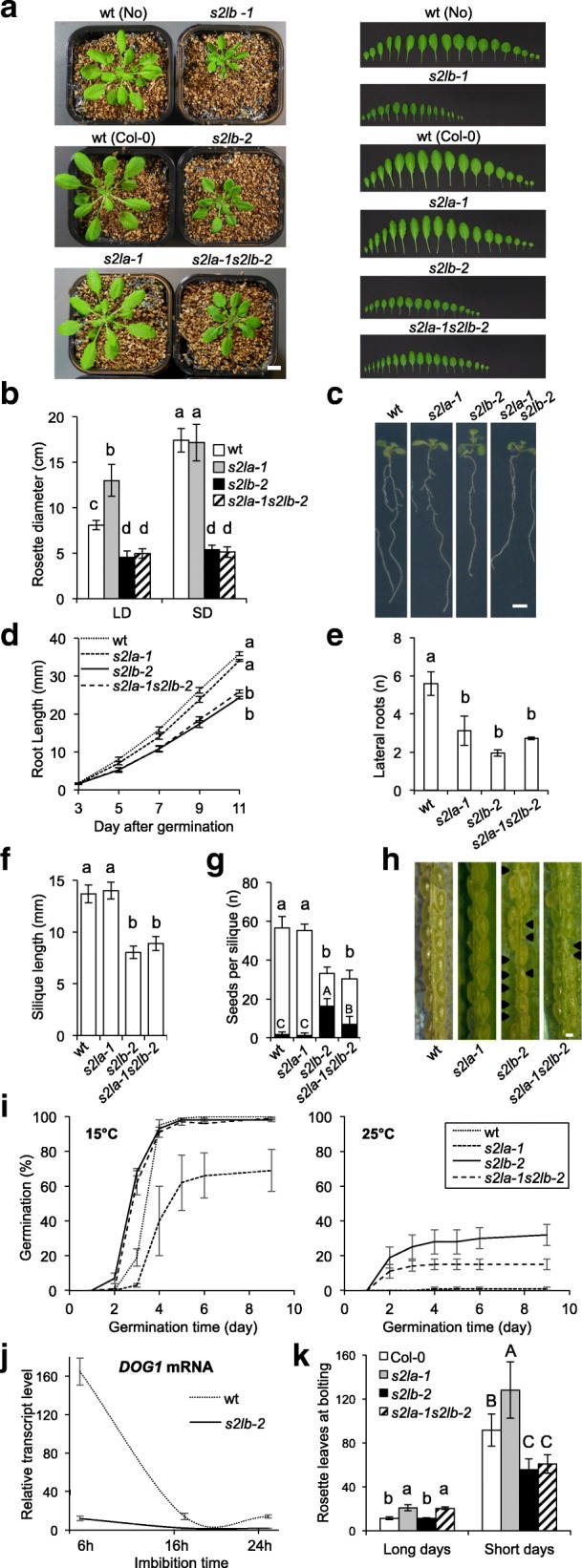
Growth defects of s2l mutant plants at the reproductive and vegetative stages. a Representative phenotypes of 4-week-old adult plants (left panel) and of dissected leaves (right panel) from plants grown under short-day conditions. Scale bar, 1 cm. b Rosette diameter at bolting of plants grown under-short (SD) or long-day (LD) photoperiods. Error bars indicate SD (n > 12). c Representative root phenotypes of 11-day-old seedlings. Scale bar, 0.5 cm. d Primary root length from 3 to 11 dpi. Error bars indicate SD from two biological replicates. e Seedling lateral root number at 11 dpi. Error bars indicate SD from two biological replicates. f Mean silique length and g number of seeds (or ovules) per silique in selfed wild-type and s2la/b mutant plants prior to seed desiccation. Black bars represent the mean number of undeveloped seeds per silique. Error bars indicate SD (n > 12 siliques each, from three individuals). h Enlarged view on a representative silique for each genotype. Arrows indicate undeveloped seeds/ovules. i Dormancy defects induced by S2Lb loss-of-function. The graph shows the percentage of freshly harvested germinating seeds from wild type, s2la, s2lb, and double s2las2lb mutant plants at 15 °C (vigor test, left graph) and 25 °C (dormancy test, right graph) in darkness. Error bars indicate SD between three biological replicates using two independent seed propagation sets (n = 50 seeds each). j RT-qPCR analysis of DOG1 transcripts level in seeds of wild-type and s2lb mutant plants upon imbibition. Total RNAs were extracted at the indicated time points. DOG1 expression level relative to housekeeping genes is given as the mean of three independent biological replicates. k Flowering time phenotype analyzed as the number of leaves at bolting under SD or LD conditions. Error bars indicate SD (n > 12). The letters indicate significant differences among sample means in ANOVA analysis followed by Tukey’s range test (p < 0.05)
At the reproductive stage, s2lb-2 mutant plants display fertile flowers, but the resulting siliques are short and contain a low number of ovules leading to ~ 50% arrested seed development (Fig. 2f–h). Interestingly, however, freshly harvested s2lb-2 viable seeds reproducibly had a high germination capacity at 25 °C, but both genotypes displayed high seed vigor at 15 °C, a temperature at which dormancy is not induced [53, 54]. These observations indicate that establishment or completion of the dormancy process is deficient in s2lb-2 seeds (Fig. 2i).
Considering the influence of HUB1 on the DELAY OF GERMINATION1 (DOG1) gene [48, 55], possibly via a related chromatin mechanism, we further investigated the expression of this master regulator of dormancy. As expected [56], DOG1 was strongly expressed in imbibed seeds and subsequently downregulated during germination in wild-type plants. In contrast, DOG1 transcripts were barely detected in s2lb-2 seeds (Fig. 2j). Hence, incapacity in inducing DOG1 upon imbibition might on its own be responsible for the dormancy phenotype.
To test for potential redundancy of S2La and S2Lb function, s2la-1 s2lb-2 double mutant plants were generated, showing largely similar vegetative phenotypic defects as s2lb-2 single mutants (Fig. 2a–e). The double mutants nonetheless generated slightly longer siliques than s2lb-2 mutant plants (Fig. 2f), producing more viable seeds (Fig. 2h) with an intermediate dormancy phenotype between wild-type and s2lb-2 seeds (Fig. 2i). Strikingly, s2la-1 and s2lb-2 mutations also have distinct effects on flowering time control under a long-day photoperiod [as shown previously with s2la-1 (aprf1-9 in [50]) and S2Lb RNAi lines [51]]. Under a short-day photoperiod, the s2la-1 plants exhibited a late flowering phenotype while s2lb-2 and double mutant plants were early flowering (Fig. 2k). Collectively, these analyses indicate that (i) S2Lb is more expressed than S2La, (ii) has a more pleiotropic role than S2La in vegetative and reproductive phases of plant development, and (iii) some of the phenotypes observed in s2lb mutant can be partially rescued by S2La loss-of-function.
S2Lb is a major determinant of the H3K4me3 landscape
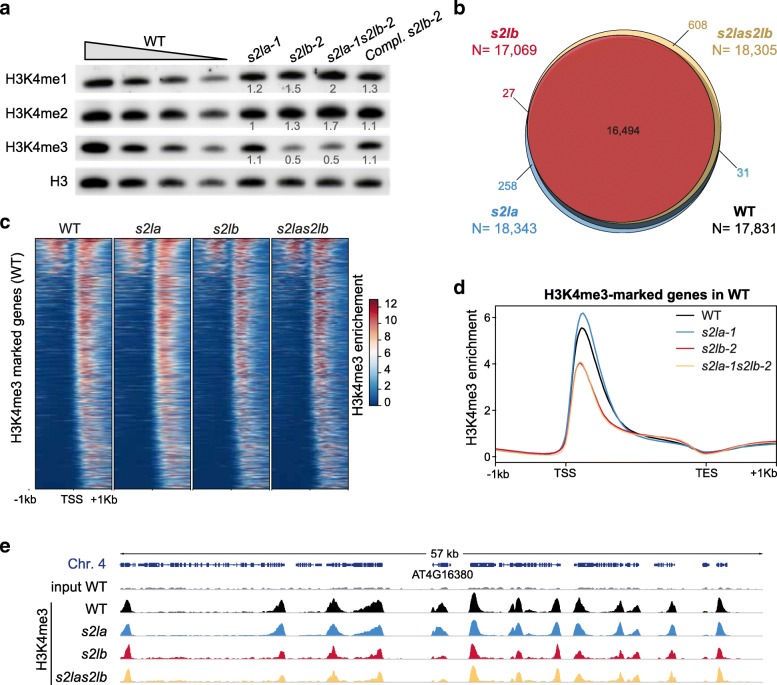
To explore the potential influence of S2L proteins in COMPASS-like activities, we first determined H3K4me1/2/3 global levels in s2la and s2lb lines by immunoblot analysis of chromatin extracts. No significant alterations could be detected in the s2la-1 line. By contrast, in both Col-0 and Nossen backgrounds, S2Lb loss-of-function triggered a ~ 2-fold decrease of H3K4me3 relative to total histone H3 levels, but not of H3K4me1 and H3K4me2 enrichment (Fig. 3a, Additional file 2: Figure S5). This defect was similarly observed in s2la-1 s2lb-2 and was rescued upon complementation of the s2lb-2 line by a S2Lb-GFP transgene, indicating that S2Lb but not S2La has a prominent influence on global H3-Lys4 trimethylation. Hence, we conclude that, similarly to the WDR5a AtCOMPASS core subunit [43, 44] and the SDG2 HMT [35, 36], S2Lb represents a major contributor to H3K4me3 deposition in Arabidopsis.
Fig. 3.
S2Lb is a major determinant of the H3K4me3 landscape. a Immunoblot analysis of chromatin extracts from rosette leaves of wild-type, s2la-1, s2lb-2, s2la-1 s2lb-2, and complemented s2lb-2/S2Lb::S2Lb-GFP plant lines. Histone H3 methylation was detected using the indicated antibodies. Decreasing quantities of chromatin extracts from wild-type plants are shown for comparison, and detection of total histone H3 is used as a loading control. Quantification of H3-Lys4 methylation signals in S2L loss-of-function plants is given relative to their corresponding histone H3 levels. For each histone mark, levels in wild-type plants are arbitrarily set to 1. b ChIP-seq identification of the genes marked by H3K4me3 in WT, s2la-1, s2lb-2, and s2la-1 s2lb-2 seedlings in two independent biological replicates. The corresponding gene IDs are listed in Additional file 3: Table S2. c H3K4me3 enrichment over the genes marked in WT plants (N = 17,831). Genes are equally ranked from top to bottom in each plant line according to the H3K4me3 median enrichment in the WT. d H3K4me3 median enrichment over the genes marked in WT plants (N = 17,831). e Genome browser view of H3K4me3 enrichment over a representative region of chromosome 4. At4g16380 is shown as an example of a gene called by MACS2 as marked by H3K4me3 in WT and s2la-1 but not in s2lb-2 and s2la-1 s2lb-2 mutant lines. Each track represents the average data of two independent biological replicates. All tracks are equally scaled
To determine more precisely the genomic loci impacted by S2L proteins, we first conducted a ChIP-seq analysis of the H3K4me3 landscape in 6-day-old s2la-1, s2lb-2, and s2la-1 s2lb-2 mutant seedlings. At this early stage, mutant and wild-type phenotypes were not visibly distinguishable. In accordance with previous H3K4me3 profiling [5, 28], H3K4me3 was mostly enriched downstream of the transcription start sites (TSS) of ~ 18,000 genes in wild-type plants (Fig. 3b–d; Additional file 2: Figure S6). H3K4me3 enrichment in s2la-1 was similar to the wild-type plants in terms of the number of marked genes and the overall profile. H3K4me3 peaks could also be detected over the same repertoire of genes in s2lb-2 and s2la-1 s2lb-2 mutants (WT-marked genes, Fig. 3b), although H3K4me3 peaks are lower and/or narrower in the absence of S2Lb function (Fig. 3d, e). Overall, these analyses showed that S2Lb, but not S2La, is required for increasing or possibly broadening H3K4me3 enrichment over most genes.
S2Lb associates with highly transcribed genes and is required for optimal gene inducibility
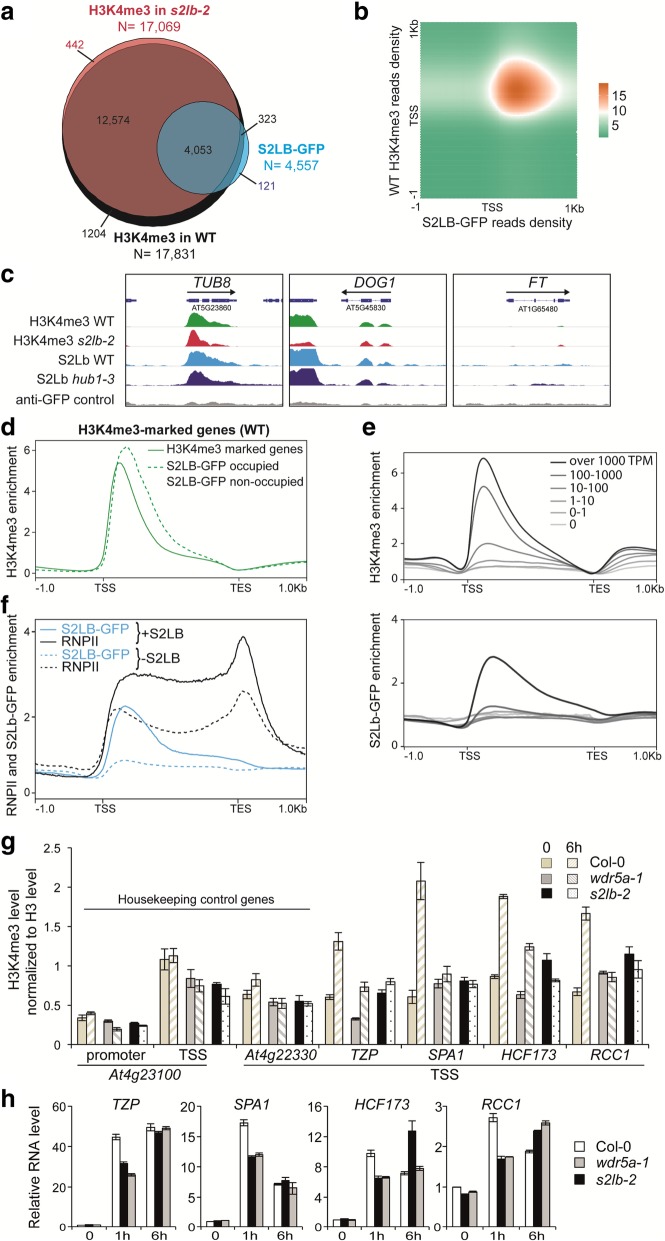
To define which genes are directly targeted by S2Lb, we conducted an anti-GFP ChIP-seq analysis of a s2lb-2/S2Lb::S2Lb-GFP complemented line (4a). An EGS/formaldehyde double crosslinking allowed us to obtain robust signals with discrete peaks (see Additional file 2: Figure S6 and Methods) and no significant background in wild-type plants used as a negative control (Additional file 2: Figure S7). Remarkably, among the 4557 S2Lb-GFP peaks, 97% matched H3K4me3-marked genes, altogether targeting one quarter of them (Fig. 4a; Additional files 3 and 4). More precisely, profiling of reads density showed a clear tendency for co-occurrence of S2Lb-GFP and H3K4me3 in the region just downstream of TSS (Fig. 4a–c). For example, S2Lb-GFP profile perfectly matched H3K4me3 domains over housekeeping genes like TUBULIN8 but was not detected over non-expressed genes like FT (Fig. 4c). Of note, similar S2Lb-GFP and H3K4me3 peaks of low intensity were found at different locations along the dormancy gene DOG1, which likely result from sense and antisense transcription start sites [57]. Also in agreement with a potential direct link between S2Lb and H3K4 trimethylation, S2Lb-GFP tends to occupy genes that are highly enriched in H3K4me3 (Fig. 4d). Moreover, H3K4me3 levels over S2Lb-targeted genes were particularly decreased in the s2lb-2 mutant line (Additional file 2: Figure S7). Finally, we also noted that S2Lb-occupied genes typically displayed a 3′-shift of their H3K4me3 peak as compared to other H3K4me3-marked genes non-targeted by S2Lb (Fig. 4d; Additional file 2: Figure S7). These observations reveal a direct link between S2Lb and H3K4me3 enrichment over a large gene set.
Fig. 4.
S2Lb-GFP is enriched over highly transcribed genes. a About 96% of the S2Lb-GFP target genes are also occupied by H3K4me3. The Venn diagram displays the overlap between genes marked by H3K4me3 in wild-type and s2lb-2 seedlings and those occupied by S2Lb-GFP in wild-type plants. Gene IDs are listed in Additional files 3 and 4. b Density matrix showing the co-occurrence frequency of H3K4me3 and S2Lb-GFP along S2Lb-occupied gene annotations (N = 4557). c Genome browser view of H3K4me3 and S2Lb-GFP over the TUBULIN8, DOG1, and FT genes. H3K4me3 and GFP signals in different genetic backgrounds are equally scaled. d Wild-type H3K4me3 median enrichment over the genes marked by H3K4me3 (dark green line; N = 17,831) and those occupied by S2Lb-GFP (dashed green line; N = 4557) or not (pale green line; N = 13,455). e Median enrichment of H3K4me3 (upper metagene plot; N = 17,831) or S2Lb-GFP (lower metagene plot; N = 4557) on TAIR10 genes split into six classes ranked by expression level in wild-type seedlings. f Median enrichment of S2Lb and RNA Pol II over the 4557 genes targeted by S2Lb (+S2Lb) versus the genes targeted by RNPII but not by S2Lb (-S2Lb; N = 11,735). RNPII-occupied gene IDs are given in Additional file 6: Table S5. g Both S2Lb and AtCOMPASS-like activities are required for H3K4me3 enrichment and induction dynamics of light-responsive genes during de-etiolation. The histogram shows H3K4me3 enrichment over several light-responsive genes in 3-day-old etiolated seedlings upon transfer from dark to light for 1 or 6 h. ChIP-qPCR analyses were performed with an anti-H3K4me3 antibody and with anti-histone H3 to normalize levels to nucleosome occupancy. H3K4me3/H3 levels are given as percentages of IP/input input relative to the mean signals over two housekeeping genes (At4g23100 and At4g22330) with no change in expression and in H3K4me3 level during the transition [58]. Error bars correspond to standard deviations from two replicates. h RT-qPCR analysis of light-responsive gene expression in samples in f. RNA levels are given relative to the wild-type dark sample (arbitrarily set to 1) normalized to At5g13440 and At2g36060 transcript levels. Error bars correspond to standard deviations from two replicates
To further assess a potential link between S2Lb function and transcription, we compared wild-type and s2lb-2 expression patterns by RNA-seq analysis. Genes misregulated in s2lb-2 seedlings are prevalently involved in plant adaptive responses to biotic and abiotic environmental cues (Additional file 2: Figure S8 and Additional file 5: Table S4). In good agreement with the hypothesized influence of S2Lb on RNPII progression, s2lb-2 misregulated genes display a biased proportion between down- and upregulated genes (60% vs 40%, respectively; Additional file 2: Figure S8). Secondly, we assessed the relationship between S2Lb chromatin association and gene expression by quantifying its occupancy over classes of genes having different transcript levels. This showed that H3K4me3 levels correlate positively with transcript abundance while, in contrast, S2Lb-GFP targeted genes correspond to the most highly expressed genes (Fig. 4e).
These observations may underscore a strict correlation between H3K4me3 marking and transcription in the bulk of different cell populations of seedlings, while in contrast S2Lb might only be detected on the most frequently transcribed genes in those various cell types. To test this hypothesis, we compared the occupancy of S2Lb-GFP and RNPII along the genome (Additional file 2: Figure S9). RNPII ChIP-seq profiling identified about 16,000 genes that, as expected, were usually marked by H3K4me3 (88% of them) and overlapped almost entirely with S2Lb-GFP occupied genes (94% of them; Additional file 2: Figure S9). As reported earlier in various species [8], RNPII was typically enriched along the transcribed domains, with a peak at the transcription elongation stop (TES; Fig. 4e; Additional file 2: Figure S9). Of note, RNPII enrichment was much higher on S2Lb-GFP occupied genes than on other genes (Fig. 4f) as observed with H3K4me3 (Fig. 4d). We concluded from these observations that S2Lb prevalently occupies genes when they are highly transcribed.
Considering these findings, the number of misregulated genes in s2lb-2 (N = 674) appears small as compared to the number of genes occupied by S2Lb-GFP (N = 4557). This contrast may suggest that H3K4me3 deposition is not sufficiently decreased upon S2Lb knockout to impair their transcription or that H3K4me3 enrichment is dispensable for efficient gene expression. Another possibility is that biological deficiencies linked to H3-Lys4 trimethylation function on transcription elongation or mRNA processing would be more easily detectable during a cellular transition when genes are upregulated rather than long after reaching steady-state expression levels. To assess whether S2Lb, and more generally AtCOMPASS activity, impacts gene induction dynamics, we monitored the expression of representative light-responsive genes during de-etiolation. This morphogenic transition involves a global increase of transcription when dark-grown, etiolated seedlings are exposed to light for the first time [59]. We tested candidate genes (TZP, SPA1, HCF173, and RCC1) that were previously identified as being subject to histone H2Bub dynamics for optimal inducibility by light [60]. ChIP-qPCR and RT-qPCR showed that these genes are subject to H3K4me3 enrichment within the first 6 h of the dark-to-light transition (Fig. 4g). As expected, H3K4me3 levels displayed slower induction dynamics in s2lb-2 seedlings and in the wdr5a-1 RNAi line [43] than in wild-type plants. This deficiency was also true for dynamic changes in transcript levels (Fig. 4h). We conclude from these analyses that, as previously proposed for H2Bub dynamics [60], H3-Lys4 trimethylation and/or AtCOMPASS-like activity is required for optimal gene upregulation in Arabidopsis but marginally impair mRNA steady-state levels.
S2Lb co-regulates a large set of genes with AtCOMPASS-like complexes and with SDG2
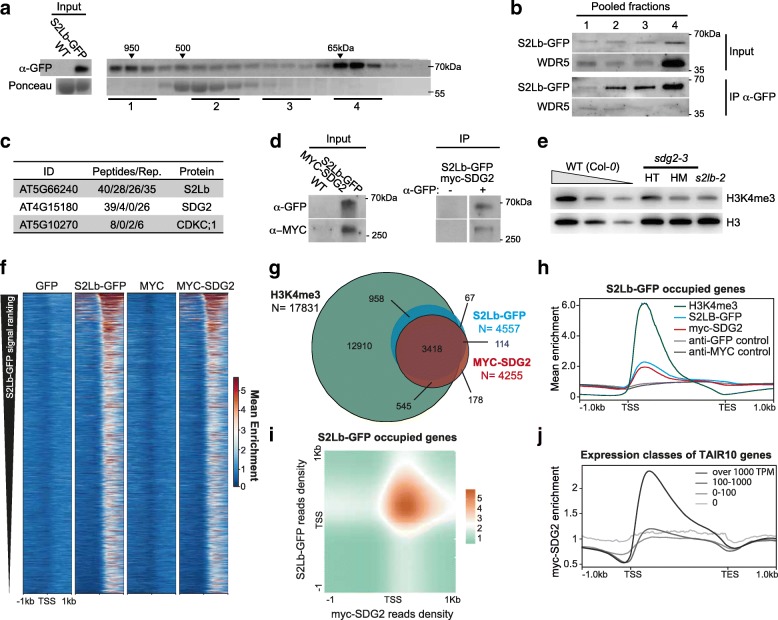
To test whether S2Lb acts as a COMPASS-like associated factor, we first assessed whether it was part of a high-molecular weight (HMW) complex by size-exclusion chromatography of soluble protein extracts from S2Lb::S2Lb-GFP plants. S2Lb-GFP eluted in two main peaks, the first one likely corresponding to its ~ 65 kDa monomeric form and the second one to a high-molecular weight complex of ~ 900 kDa or more (Fig. 5a). To test whether these HMW fractions correspond to COMPASS-like complexes, S2Lb-GFP was immunoprecipitated from different fraction pools and tested for the presence of WDR5. This core subunit of AtCOMPASS-like complexes is also essential for a large fraction of H3K4me3 deposition in Arabidopsis [43–45]. S2Lb-GFP and WDR5 were both initially more abundant in monomeric fractions (input, pool 4), but WDR5 mainly immunoprecipitated with S2Lb-GFP from HMW fractions (pools 1 and 2; Fig. 5b). Hence, we conclude that S2Lb and WDR5 can associate within one or more HMW complexes in planta. Given the influence of S2Lb and WDR5a in H3K4me3, these HMW complexes likely correspond to AtCOMPASS-like activities.
Fig. 5.
S2Lb and SDG2 associate over a large gene repertoire. a Size-exclusion chromatography of S2Lb-GFP complex size in S2Lb::S2Lb-GFP plant extracts. FPLC fractions were analyzed by anti-GFP immunoblot. Arrows indicate the elution peaks of molecular-weight standards in the same conditions. b Endogenous WDR5 co-immunoprecipitates with S2Lb-GFP in HMW fractions. Pooled fractions from samples in a were used for anti-GFP immunoprecipitation and analyzed using anti-GFP and anti-WDR5 antibodies. c IP-MS identification of S2Lb-GFP interacting proteins. The table gives the number of peptides from the indicated proteins detected using plant extracts from S2Lb-GFP plants but not from wild-type Col-0 plants in four biological replicates. d Co-immunoprecipitation assay on SDG2::MYC-SDG2;S2Lb::S2Lb-GFP plants with anti-GFP coated (+) or uncoated beads (−). Immunoprecipitated S2Lb-GFP and MYC-SDG2 proteins were detected by anti-GFP and anti-MYC immunoblots, respectively. e Chromatin extracts from S2Lb and SDG2 loss-of-function plants display similar defects in H3K4me3 global level. Immunoblot analysis of WT, s2lb-2, and heterozygote (HT) or homozygote (HM) sdg2-1 seedlings was performed using antibodies recognizing total histone H3 and H3K4me3. H3 is used as a loading control. f The heatmap shows S2Lb-GFP and MYC-SDG2 enrichment over S2Lb-GFP occupied genes (N = 4557). Genes are equally ranked from top to bottom in each line according to S2LB-GFP median enrichment. g Overlap between genes marked by H3K4me3 and genes occupied by MYC-SDG2 or S2Lb-GFP. The overlap between S2Lb-GFP and MYC-SDG2: × 5.7 p value < 0e+00; overlap between S2Lb-FP and H3K4me3: × 1.7 p value < 0e+00; overlap between MYC-SDG2 ad H3K4me3: × 1.6 p value < 0e+00. The corresponding gene IDs are listed in Additional files 4 and 7. h Median enrichment of S2Lb-GFP, MYC-SDG2, and H3K4me3 over S2Lb-GFP occupied genes (N = 4557). i Density matrix showing the co-occurrence of S2LB-GFP and MYC-SDG2 along S2Lb target genes (N = 4557). j Median enrichment of MYC-SDG2 on TAIR10 genes split into four classes ranked by expression level in wild-type seedlings
To gain better insights into S2Lb complex activity, we conducted mass spectrometry analysis of proteins co-immunoprecipitating with GFP-S2Lb from S2Lb::S2Lb-GFP seedlings using wild-type plants as negative control. GFP-SL2b was efficiently retrieved in each of four biological replicates. Under the mild detergent conditions used, this analysis did not allow the recovery of WDR5 or other known COMPASS subunits; however, the most abundantly detected protein that was robustly co-immunoprecipitated was SDG2 (Fig. 5c). Of note, the CDKC;1 protein was also significantly detected in three out of four biological replicates, although with low peptide numbers. This homolog of human CDK9 belongs to the CDK9/CycT complex of P-TEFb and mediates RNPII CTD Ser-2 phosphorylation in Arabidopsis [61].
The potential association of SDG2 with S2Lb was confirmed by co-immunoprecipitation of S2Lb-GFP and MYC-SDG2 tagged proteins stably expressed in Arabidopsis under the control of their endogenous promoters (Fig. 5d). Furthermore, comparison of global H3K4me3 levels in s2lb-2 and sdg2-3 mutant plants by immunoblot showed comparable defects (Fig. 5e). Further comparison of the s2lb-2 transcriptomic profile with all available Genevestigator datasets [52] using the Signature tool identified the sdg2-3 profile [36] as being the most similar among all available transcriptome datasets (Additional file 2: Figure S10). Even though distinct transcriptomic methodologies were used, direct comparison of misregulated genes in both transcriptome analyses showed that a majority of the s2lb-2 misregulated genes display a similar trend in sdg2-3 seedlings (55%; Additional file 2: Figure S10).
We further examined whether S2Lb shares any functional properties with other known H3-Lys4 HMTs such as ATXR7 [33, 34] and ATX1 [32], but we identified no significant similarity with atxr7-1 and atx1 transcriptomic data (about 9% maximum overlap; Additional file 7: Table S6). We conclude from these analyses that S2Lb and SDG2 directly or indirectly associate to regulate a common set of genes, possibly acting together at the chromatin level.
Finally, to ascertain whether S2Lb and SDG2 co-regulate genes in situ, we determined the SDG2 chromatin profile by anti-MYC ChIP-seq analysis of SDG2::MYC-SDG2 seedlings (Fig. 5f; Additional file 9: Table S8). This unveiled that SDG2 associates with a similar number of chromatin loci than SL2b (4255 vs 4557), 80% of them occurring on the same genes (Fig. 5g). Metagene plot analyses showed very similar enrichment profiles over gene bodies (Fig. 5h), mainly co-occurring over 3′ domains of TSS (Fig. 5i). As described above for S2Lb-GFP, almost all genes occupied with MYC-SDG2 were marked by H3K4me3 (Fig. 5g) and tend to be highly expressed (Fig. 5j). Searching for potential DNA sequence contexts underlying S2Lb and/or SDG2 association with chromatin, we were not able to identify a specific set of transcription factor binding motifs, apart from diverse forms of GA stretches (Additional file 8: Table S7). Collectively, these findings show that S2Lb associates with one or more AtCOMPASS-like complexes and co-regulate multiple genes in association with the SDG2 histone methyltransferase.
S2Lb chromatin association and H3K4me3 deposition do not depend on prior histone H2B monoubiquitination
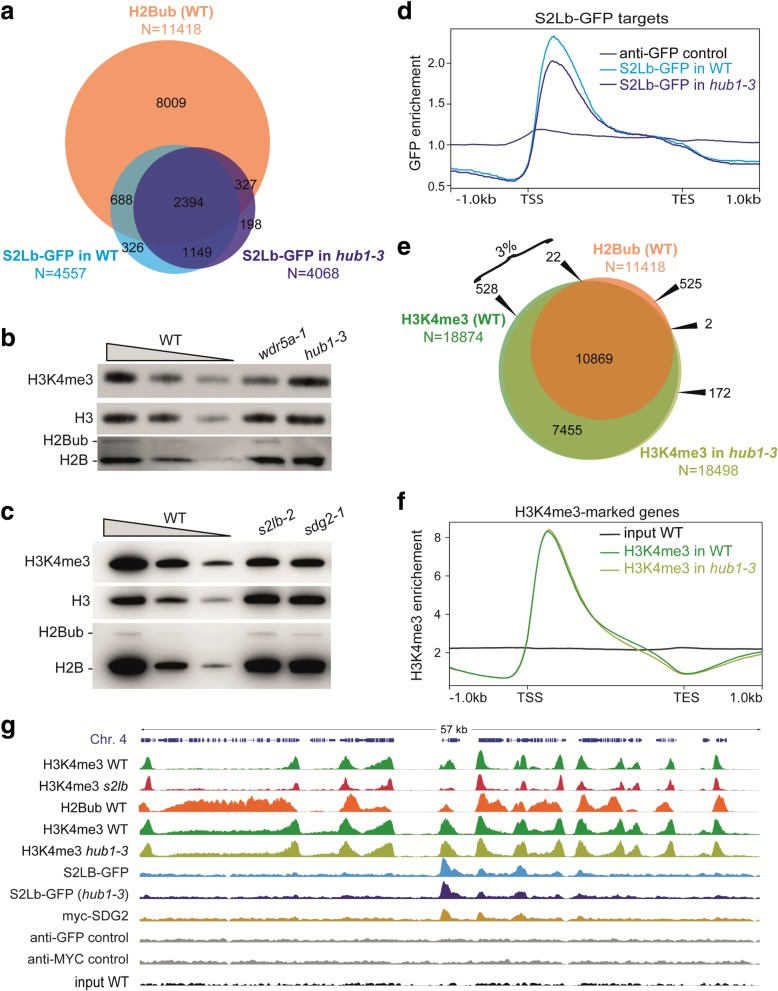
The yeast Swd2 homolog of S2Lb is thought to drive yCOMPASS-mediated H3K4me3 deposition on H2Bub-modified nucleosomes [14–16]. Conservation of COMPASS-like activities in Arabidopsis may suggest that S2Lb is also required for H3K4me3 deposition or maintenance through a similar mechanism of action. Accordingly, genome profiling showed a clear tendency of S2Lb-GFP to occupy H2Bub-marked genes (Fig. 6a), suggesting that some H2Bub-enriched domains allow recruitment of COMPASS activity in Arabidopsis as well. Nevertheless, differently from yeast, the bulk of H3K4me3 is maintained in mutant plants lacking or over-accumulating H2Bub (Fig. 6b and [63–67]). Vice versa, H2Bub levels are not visibly affected in s2lb-2 and sdg2-3 plants defective in H3K4me3 deposition (Fig. 6c). This suggests an absence, or at most only limited, AtCOMPASS-mediated H2Bub/H3K4me3 crosstalk in Arabidopsis, and further interrogates (1) whether H3K4me3 patterns along the genome rely, even partially, on histone H2B monoubiquitination and (2) how S2Lb, AtCOMPASS, and SDG2 are recruited to chromatin loci.
Fig. 6.
S2Lb chromatin association and H3K4me3 deposition are usually independent from histone H2B monoubiquitination. a Overlap between genes marked by H2Bub (from [62]) and by S2LB-GFP in wild-type (WT) or in hub1-3 seedlings. For proper comparison of S2Lb-GFP enrichment, WT/S2Lb::S2Lb-GFP and hub1-3/S2Lb::S2Lb-GFP seedlings were grown and processed for ChIP-seq in parallel. The corresponding gene IDs are listed in Additional file 4: Table S3. b Wild-type H3K4me3 levels in chromatin extracts of HUB loss-of-function plants. Decreasing quantities of chromatin extracts from wild-type plants are shown for comparison, and detection of total histone H3 and H2B forms is used as loading controls. c Wild-type H2Bub levels in S2Lb and SDG2 loss-of-function plants. Immunoblots were performed as in b. d S2Lb-GFP median enrichment in WT/S2Lb::S2Lb-GFP and hub1-3/S2Lb::S2Lb-GFP seedlings on the S2Lb-GFP targeted genes (N = 4557). e Overlap between H2Bub-marked genes (from [62]) and H3K4me3-marked genes in wild-type (WT) or in hub1-3 seedlings (this study). For proper comparison of H3K4me3 marking, wild-type and hub1-3 seedlings were grown and processed for ChIP-seq in parallel, independently from other ChIP-seq in Figs. 3, 4, and 5. The corresponding gene IDs are listed in Additional file 9. f H3K4me3 median enrichment in WT and hub1-3 seedlings on the genes marked by H3K4me3 in WT (N = 18,874). g Genome browser view of H3K4me3, S2Lb-GFP, and MYC-SDG2 over the same region of chromosome 4 shown in Fig. 3. H3K4me3 and GFP or MYC signals in different genetic backgrounds are equally scaled
To address these questions, we took advantage of hub1-3 mutant plants in which the deposition of histone H2B monoubiquitination is abolished to test whether S2Lb-GFP recruitment and H3K4me3 enrichment rely on H2Bub. H2Bub levels are undetectable in homozygous hub1-3 seedlings ([48, 49, 60] and Fig. 6b). Upon introgression of S2Lb::S2Lb-GFP in the hub1-3 background, ChIP-seq analysis of S2Lb-GFP showed that about one third of the SL2b-targeted genes were different in the hub1-3 plants, with a tendency to be marked in WT but not in hub1-3 background (1014 vs 525 genes; Fig. 6a). SL2b-GFP enrichment over gene bodies was slightly decreased in hub1-3 plants, still with a similar profile (Fig. 6d). This analysis showed that S2Lb-GFP can be recruited over many H2Bub-marked genes largely independently of this histone, whereas a minority of genes might be subjected to a COMPASS-based histone crosstalk. To test this second hypothesis, we compared the set of genes that lost both S2Lb-GFP occupancy and H3K4me3 marking in hub1-3 mutant plants. Only nine genes corresponding to this criterion could be identified (Additional file 2: Figure S11). We concluded that S2Lb-GFP and H3K4me3 may aberrantly target other genes in hub1-3 plants, possibly as a consequence of mild transcriptomic and phenotypic variations induced by HUB1 loss-of-function [48, 49, 60].
Direct comparison of H3K4me3 profiles in wild-type and hub1-3 seedlings further showed that only ~ 3% of the genes lose H3K4me3 enrichment in the absence of H2Bub (Fig. 6e; Additional file 10: Table S9). Moreover, contrasting with PAF1c mutant plants [68], global H3K4me3 enrichment and positioning along the 5′ domains of gene bodies were not detectably affected by loss of HUB activity (Fig. 6f; Additional file 2: Figure S12), again supporting that the vast majority of genes are subject to H3-Lys4 trimethylation independently from H2Bub deposition.
The genome-wide profiles obtained in this study confirmed the spatial correlation between S2Lb-GFP and MYC-SDG2 peaks over about one third of the H3K4me3-marked genes (Fig. 6g). H3K4me3 enrichment was robustly diminished in the s2lb-2 line while, in contrast, both H3K4me3 and S2Lb-GFP occupancy were typically unaffected in the hub1-3 line (Additional file 2: Figure S6). Collectively, we conclude that AtCOMPASS and SDG2 mainly drive H3-Lys4 trimethylation through H2Bub-independent pathways in Arabidopsis.
Discussion
S2Lb and COMPASS-like proteins as partners of the plant-specific histone methyltransferase SDG2
In this study, we first report that S2Lb, a homolog of the yCOMPASS-associated subunit, is a major actor of H3-Lys4 trimethylation with SDG2 despite the absence of clear H2Bub-H3K4me3 histone crosstalk in Arabidopsis. A series of complementary evidence points towards a functional partnership between S2Lb and SDG2 with one or more COMPASS-like complexes in Arabidopsis. Firstly, GFP-tagged S2Lb resides only on H3K4me3-enriched genes, mostly those also displaying the H3K4me3 HMT protein SDG2 [35, 36]. ChIP-seq analyses indeed showed a tight co-occurrence of H3K4me3, S2Lb-GFP, and MYC-SDG2 just downstream from the TSS of more than four thousands of genes, in particular those displaying high RNPII occupancy and elevated transcript levels. Our ChIP analyses do not allow us to ascertain whether S2Lb, SDG2, and RNPII physically associate onto the same chromatin fragments, but in support of this possibility, SDG2 was the most abundantly co-purifying protein with S2Lb. Secondly, S2Lb and SDG2 are both important for establishing or maintaining H3K4 trimethylation levels since their loss-of-function leads to a similar 50–70% decrease of the bulk of H3K4me3 [35, 36]. Thirdly, S2Lb and SDG2 loss-of-function plants share several related phenotypic defects throughout the life cycle including dwarfism, short roots, loss of apical dominance, and impaired fertility [35, 36, 69, 70]. Accordingly, transcriptomic analyses revealed a striking overlap between genes misregulated in s2lb-2 and in sdg2-3, indicating that S2Lb and SDG2 regulate expression of a common gene set. Still, not all H3K4me3-marked genes are enriched in S2Lb or SDG2, as a majority of H3K4me3 peaks do not overlap with S2Lb or SDG2 peaks. S2Lb and SDG2 might only be detected over the genes most frequently transcribed among the various seedling cell types, or formerly transcribed genes might retain H3K4me3 marking but not S2Lb proteins.
IP-MS attempts to recover WDR5 or other known COMPASS subunits by IP/MS of GFP-S2Lb were unsuccessful, possibly because of the mild detergent conditions used in these assays as a consequence of the large MYC-SDG2 protein (~ 300 kDa migrating form) being largely insoluble in plant extracts. Notwithstanding, S2Lb and WDR5 successfully pulled down from one or more HMW complexes from soluble plant extracts. They presumably interact indirectly as shown for yeast Swd2 and Swd3 [71]. Their association was somehow expected given the high conservation of COMPASS subunits from yeast to plants and mammals [39, 72, 73], but less so for a plant-specific protein like SDG2.
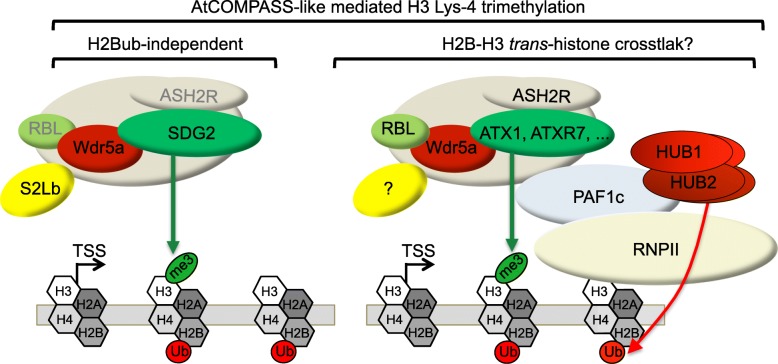
We have been able to identify a high overlap between the misregulated gene repertoire in s2lb seedlings and SDG2 loss-of-function seedlings but not for other H3K4me3 HMTs such as ATX1 and ATXR7. S2Lb therefore seems to have a certain level of specificity for SDG2, which may relate to the wide expression pattern of these two genes throughout plant development. Hence, the evolutionarily conserved COMPASS-like complexes not only act with Trithorax-like proteins such as ATX1, SDG14, and SDG16 in Arabidopsis as in other species [43, 44] but also appears to have been co-opted by the plant-specific SDG2 HMT (Fig. 7).
Fig. 7.
Working model for S2Lb activity with AtCOMPASS-like and SDG2 in histone H3-Lys4 trimethylation. In a major pathway, S2Lb and SDG2 target highly transcribed genes for H3-Lys4 trimethylation as part of a COMPASS-like complex acting independently of H2B monoubiquitination. S2Lb might as well be part of other COMPASS-like complexes with other histone methyltransferases such as ATX1 and ATXR7. Arabidopsis WDR5a, RbBP5-like (RBL), and Ash2-related (ASHR) are core components of AtCOMPASS-like complexes [37, 43–45] but the nature of their physical association with SDG2 remains to be determined. SDG2 might additionally act independently from S2Lb in different genic or cellular contexts, possibly with S2La. Whether histone H2B monoubiquitination is required for other types of At-COMPASS complexes activity and acts co-transcriptionally with PAF1c as in other species also remains to be resolved
The observation that both SDG2- and WDR5-null mutations are sterile while mutant plants combining S2la and S2Lb knockout are partially fertile suggests that S2L proteins are less essential than SDG2 and AtCOMPASS complexes. SDG2 may either catalyze H3K4 trimethylation alone or with COMPASS-like independently from S2Lb, potentially having a residual activity at specific chromatin loci or in specific cell types such as in male and/or female gametophytes. Whether S2Lb physically interacts with SDG2 remains to be resolved, as does the question of whether SDG2 associates with WDR5a within an AtCOMPASS super-complex with other histone modifying and remodeling activities, as recently identified for the AtCOMPASS-FRIGIDA complex [74].
H2Bub-independent AtCOMPASS-like activity
Several independent studies have revealed that the bulk of H3K4me3 is retained in mutant plants lacking or over-accumulating H2Bub and in Paf1c mutants [68], as reproducibly shown by immunoblot analyses here and by several other studies ([63–67] and Fig. 6b). Targeted ChIP-qPCR has also been conducted over a handful of genes in hub mutant plants, such as the flowering regulatory genes FLC [63], SOC1, FT, and MAF4 [67] and the clock component genes CCA1, TOC1, and ELF4 [75], showing in all cases that H3K4me3 level was lower than in wild-type plants. Genetic approaches combining mutations impairing H2B monoubiquitination and histone methylation identified both additive and synergistic effects on Arabidopsis phenotypic quantitative traits, suggesting the existence of interplays among different histone modifications [67]. Still, lack of mechanistic assessments and of genome-wide resolution have not allowed an unambiguous evaluation of whether an H2Bub-H3K4me3 trans-histone crosstalk is at play in plants. Here, we first observed using two independent seed batches and upon certifying that homozygous seeds were used that H3K4me3 profiles were quasi indistinguishable between wild-type plants and hub1-3 mutant plants lacking detectable H2Bub. Using other harvesting daytime, growth conditions or developmental stages might possibly be more accurate to compare our results with former studies. Nevertheless, considering that Swd2 allows tethering COMPASS on H2Bub-modified nucleosomes in other species [14–16], our second approach consisted in assessing whether S2Lb is recruited onto the epigenome in the absence of H2B monoubiquitination as a proxy to test for a potential AtCOMPASS-mediated H3K4me3-H2Bub crosstalk. Although enrichment levels were slightly weaker, the vast majority of S2Lb target genes were occupied by S2Lb-GFP in both wild-type and hub1-3 seedlings. Hence, our two complementary approaches point towards a role for AtCOMPASS/SDG2 in driving H3-Lys4 trimethylation that is largely independent of histone H2B monoubiquitination in Arabidopsis.
A 3′-shift of the H3K4me3 peak was observed on S2Lb-targeted genes. A similar shift of H3K4me3 has been reported in Arabidopsis PAF1c mutant seedlings and proposed to result from an irregular transition from the Ser-5 to Ser-2 phosphorylated form of RNPII [68]. Considering that most genes were still marked by H3K4me3 in S2Lb mutant plants, albeit to a lower extent, we propose that S2Lb is required for the maintenance or broadening of the H3K4me3 landscape during RNPII transition into productive elongation while it might not be involved in its nucleation. If true, this hypothesis would provide a rationale for conserving SWD2-like activities in plants despite not contributing to a recognizable trans-histone crosstalk function.
These findings add to our former report that H3K4me3 is efficiently established over light-responsive genes in hub1-3 seedlings upon their induction [60]. In the absence of H2Bub-H3K4me3 trans-histone crosstalk, AtCOMPASS complexes might rather be recruited onto chromatin loci in a sequence-specific manner and in response to specific signals by means of transcription factors. Accordingly, a targeting mechanism by transcription factors such as bZIP28 and bZIP60 has recently been unveiled for the regulation of endoplasmic reticulum stress-responsive genes [47].
Complex relationships between histone H3 Lys-K4 trimethylation and histone H2B monoubiquitination with transcription regulation in Arabidopsis
RNA-seq analysis showed that only a small subset of H3K4me3-marked and of S2Lb-targeted genes was misregulated in young S2Lb knockout seedlings. This is line with the apparent wild-type phenotype of s2lb seedlings at this early developmental stage but also appears counter intuitive with the proposed role of S2Lb in AtCOMPASS activity and with the instructive role of H3 Lys-4 trimethylation on RNPII processivity. Still, as for S2Lb, both HUB and PAF1c loss-of-function trigger weak transcriptomic defects in Arabidopsis [49, 63, 68]. Depletion of H3K4me3 has only marginal effects on gene expression in other species as well [12]. Hence, H3K4me3 may contribute to the reinforcement of the active state of transcription [68] and to fine-tuning genome expression during plant development and adaptive responses [67]. In line with this proposed function, we observed that AtCOMPASS-like-deficient plants are impaired in the accurate inducibility of light-regulated genes. Investigating more precisely the effect of S2Lb or other COMPASS subunits on transcription efficiency would probably require a quantification of nascent transcripts production in a dynamic system such as de-etiolation or another cellular adaptive response.
The CDKC;1 protein was detected as co-purifying with S2Lb in our IP-MS analyses, although not systematically and with low peptide numbers. This association might be functionally meaningful, as CDKC;1 mediates RNPII CTD Ser-2 phosphorylation in Arabidopsis [61, 76–78] and acts as an activator of transcription in plants [79]. CDKC;2, another cyclin-dependent kinase involved in RNPII regulation, has also been found recently to co-purify with HMTs and chromatin remodeling factors using similar approaches [80]. Such interactions potentially link S2Lb to the regulation of RNPII CTD phosphorylation and therefore to the transition towards transcription elongation.
A diversity of COMPASS-like complexes in A. thaliana
A. thaliana encodes two paralogs of the S. cerevisiae SWD2 gene with similar expression patterns in most organs. S2La is expressed to a much lower level than S2Lb, possibly targeting only a few genes or acting in a few cell types. This is also the case for ATX1 [35, 81], which presumably targets a few specialized genes on which it helps recruiting a COMPASS-like complex and promotes assembly of the RNPII pre-initiation complex [37]. Both S2L genes encode euchromatic proteins that differ in their structure. Despite our analysis used plants originating from different genetic backgrounds, S2La disruption detectably aggravated neither s2lb-2 morphological phenotypes nor its H3K4me3 defects. Hence, at this stage, we cannot exclude that lowly expressed S2La might also work in a H3K4me3 deposition pathway, possibly contributing to a minor trans-histone crosstalk with H2Bub, or rather act in other histone modifications.
In contrast to S2La, S2Lb is strongly and widely expressed in the Col-0 accession, and independent attempts to isolate null T-DNA mutations in this genetic background have been unsuccessful [51]. This suggests that S2Lb function is more essential in Col-0 than in Nossen; an hypothesis also supported by our observation that successive introgressions of s2lb-1 in Col-0 to generate s2lb-2 showed aggravated phenotypes as compared to the original effect in Nossen background.
S2La and S2Lb polymorphic WD40 repeat domains may underpin different protein association capacities, for example, influencing their association with different transcription factors targeting SDG2 or other HMTs to distinct loci, or with other protein complexes. Noteworthy, yeast SWD2 is also an integral subunit of the cleavage and polyadenylation factor (CPF) complex involved in 3’end mRNA processing [82, 83]. In agreement with its role in H3K4me3 deposition, predominant phenotypes induced by S2Lb loss-of-function are shared with COMPASS [40, 43–45] and SDG2 phenotypes, both impaired in H3K4 trimethylation: dwarfism, impaired fertility, and early flowering [35, 36]. In contrast, s2la-1 plants are late flowering like CPF subunit mutants [84]. Hence, two SWD2 paralogs might be specialized in Arabidopsis, a situation previously identified in Schizosaccharomyces pombe [85]. Given the ancient origin of the duplication event of S2La and S2Lb genes in the plant lineage, the examination of their functional diversification represents an interesting aspect to decipher in future studies.
Conclusion
By contrast with S. cerevisiae in which a single SET1 protein catalyzes histone H3 Lys-4 trimethylation as part of COMPASS acting upon histone H2B monoubiquitination, in Arabidopsis H3K4me3 deposition is mediated by multiple ubiquitous or cell-specific histone methyltransferases (HMT). Here, we show that a major pathway for H3 Lys-4 trimethylation involves the plant-specific HMT SDG2 acting in the context of an evolutionarily conserved COMPASS-like activity in Arabidopsis. In addition, we report that a Swd2-like (S2Lb) COMPASS axillary subunit is recruited onto most transcribed genes along with SDG2 and allows increasing H3K4me3 occupancy in wild-type plants but also in plants lacking H2Bub. Collectively, this study sheds light on the evolution of SWD2-like proteins and COMPASS-like activity, which might underpin an atypical and H2Bub-independent pathway driving most H3K4me3 deposition in plants.
Methods
Plant material and growth conditions
All Arabidopsis lines used in this study are in the Col-0 background except s2lb-1 and its parental line Ds1-388-5 that are in a Nossen background. The s2la-1 T-DNA insertion line (WiscDsLox489-492 K11) described in [50] was obtained from NASC [86]. The s2lb-2 line (RATM54-3645-1) was obtained from the RIKEN Institute [87] and subjected to five successive backcrossing with Col-0 wild-type plants as female counterparts to generate s2lb-2 plants. The wdr5a-1 RNAi line and the sdg2/SDG2::myc-SDG2 line have previously been described [36, 43]. Plants were grown under 100 μmol m2 s−1 light in soil or in vitro under long-day (16 h day 23 °C/8 h night 19 °C) conditions (except for the indicated flowering time experiments). For in vitro growth, seeds were surface sterilized and plated on MS medium containing 0.9% agar and stratified for 3 days at 4 °C before transfer to growth chambers. Root length was determined on seedlings grown in vitro on vertical MS plates supplemented with 1% sucrose. Position of root tips was marked every 2 days from day 3 to day 11 post-germination. Plates were scanned at day 11, and root length was measured using ImageJ [88]. De-etiolation experiments were conducted as in [60].
Dormancy assay
Dormancy was measured on seeds issued from 3 independent productions after plant growth at 20–22 °C under a long-day photoperiod. At full maturity, seeds were harvested and germination was assessed at 15 °C and 25 °C in darkness in 3 biological replicates of 50 seeds for each genotype. Experiments were conducted in 9 cm Petri dishes on a layer of cotton wool covered by a filter paper sheet soaked with water. A seed was considered as germinated when the radicle has protruded through the testa. Germination was scored daily for 10 days, and the results presented correspond to the mean of the germination.
Plasmid construction
The p35S::GFP-S2La construct was generated by inserting the entire coding sequence of S2La (including stop codon) amplified from wild-type Col-0 cDNA downstream of the GFP coding sequence in the pB7WGF2 plasmid (Ghent plasmids collection, https://gateway.psb.ugent.be) via Gateway technology (Invitrogen). The same was done for the p35S::GFP-S2Lb construct, except that the entire coding sequence of S2Lb was obtained from the U16729 pENTR-D-TOPO plasmid (ABRC). The pS2Lb::S2Lb-GFP and pS2Lb::S2Lb constructs were generated by inserting a PCR-amplified 3.1 kb S2Lb genomic fragment (entire genomic coding sequence and 1 kb of promoter region) in frame with a downstream GFP reporter gene in the pB7FWG,0 plasmid or in the pB7WG plasmid, respectively (Ghent plasmids collection, https://gateway.psb.ugent.be) via Gateway technology (Invitrogen). As the S2Lb fragment was cloned without STOP codon, a TAG codon was then introduced in the pS2Lb::S2Lb construct by changing one nucleotide using a site-specific mutagenesis kit (QuikChange XL Site-directed mutagenesis kit, Agilent).
In situ immunolocalization
Five-day-old wild-type p35S::GFP-S2La, p35S::GFP-S2Lb, and pS2Lb::S2Lb-GFP seedlings were vacuum infiltrated in 4% formaldehyde, 10 mM Tris-HCL pH 7.5, 10 mM EDTA, and 100 mM NaCl for 30 min and washed with Tris buffer. Cotyledons were chopped in ice-cold LB01 buffer (15 mM Tris-HCl at pH 7.5, 2 mM EDTA, 0.5 mM spermine, 80 mM KCl, 20 mM NaCl, 0.1% Triton X-100), and the nuclei were isolated using a Douncer (Wheaton), filtered through a 50-μm nylon mesh, centrifuged at 500g for 5 min at 4 °C, spread, and air dried on APTES/glutaraldehyde-treated slides. Slides were post-fixed in methanol-acetone 1:1 solution for 10 min and blocked in PEMSB for 2 h at room temperature. The slides were incubated overnight at 4 °C with a primary antibody specific to GFP (1/200, Life Technologies, A-11122) then for 2 h with goat-anti-rabbit-AlexaFluor488 secondary antibody (Life Technologies, A-11008). The slides were washed and mounted in Vectashield with 2 μg/μl DAPI. Images were taken using a confocal laser scanning microscope (SP5, Leica).
Immunoblot analyses
Soluble protein samples were obtained using the indicated methods, and chromatin extracts were obtained as previously described [60]. Unless stated, 10 μg of protein samples were loaded on 14% LiDS Tris-Tricine gels and blotted onto PVDF membranes before immunodetection and analysis using a LAS4000 luminescence imager (Fuji). The following antibodies were used: anti-H3 (Millipore #07-690), anti-H3K4me1 (Active Motifs #39297), anti-H3K4me2 (Millipore #07-030), anti-H3K4me3 (Millipore #05-745), anti-GFP (Clontech #632381), anti-MYC antibodies (Millipore #05-724), or custom-designed anti-rice histone H2B [60]. Anti-WDR5 serum was obtained by immunization of a rabbit with a 50-amino-acid synthetic peptide corresponding to amino acids 42–91 of the Arabidopsis WDR5a protein and affinity purification by the SDIX company (USA). All uncropped blots are given in Additional file 12.
Gel filtration
Size exclusion chromatography was performed as previously described [89]. Elution fractions were either analyzed by immunodetection of S2Lb-GFP on 40 μl samples or pooled as indicated before immunoprecipitation using a Crosslink IP kit (Thermo Scientific) and an anti-GFP antibody (ThermoFisher #A-11122).
In vivo pull-down assays
In vivo pull-down assays were performed on 1 mg of protein extracts from 10-day-old seedlings. Proteins from pSDG2::myc-SDG2/pS2Lb::S2Lb-GFP homozygous plants obtained by crossing the two respective lines were extracted using a modified RIPA buffer (Tris pH 7.6 25 mM, NaCl 150 mM, NP40 1%, sodium deoxycholate 1%, SDS 0.1%, and protease inhibitors). After clearing the samples with uncoupled beads (ChIP Adembeads, Ademtech), S2Lb-GFP proteins were immunoprecipitated for 2 h using a GFP-Trap system (Chromotek) coupled to magnetic beads. A mock was done using uncoupled beads. Beads were washed with RIPA buffer without detergents before elution with 2× Laemmli buffer. Eluates were analyzed on 8% SDS-PAGE gels and blotted onto PVDF membranes before immunodetection.
SDG2 and S2Lb affinity purification and mass spectrometry
For each biological replicate, protein samples were immuno-isolated from 2 g of either wild-type, pSDG2::myc-SDG2, or pS2Lb::S2Lb-GFP 10-day-old seedlings as described above using either GFP-or MYC-trap slurries (Chromotek # gtma-20 and #yta-20, respectively) in modified RIPA buffer to allow for SDG2 affinity purification. For mass spectrometry, SDS/PAGE was used without separation as a clean-up step, and only one gel slice was excised. Gel slices were washed, and proteins were reduced with 10 mM DTT before alkylation with 55 mM iodoacetamide. After washing and shrinking the gel pieces with 100% (v/v) MeCN, in-gel digestion was performed using trypsin/LysC (Promega) overnight in 25 mM NH4HCO3 at 30 °C. Peptides were analyzed by LC-MS/MS using an RSLCnano system (Ultimate 3000, Thermo Scientific) coupled to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific). Peptides were loaded onto a C18-reversed phase column (75-μm inner diameter × 2 cm; nanoViper Acclaim PepMap™ 100, Thermo Scientific), separated, and MS data acquired using Xcalibur software. Peptide separation was performed over a linear gradient of 100 min from 5 to 30% (v/v) acetonitrile (75-μm inner diameter × 50 cm; nanoViper C18, 2 μm, 100 Å, Acclaim PepMap™ RSLC, Thermo Scientific). Full scan MS was acquired in the Orbitrap analyzer with a resolution set to 120,000, and ions from each full scan were HCD fragmented and analyzed in the linear ion trap. For identification, the data were searched against the Arabidopsis thaliana TAIR10 database (2016) using Mascot 2.5.1 (Matrix Science). Enzyme specificity was set to trypsin and a maximum of two missed cleavage sites were allowed. Oxidized methionine, N-terminal acetylation, and carbamidomethyl cysteine were set as variable modifications. Maximum allowed mass deviation was set to 10 ppm for monoisotopic precursor ions and 0.4 Da for MS/MS peaks. The resulting files were further processed using myProMS v3.0 [90]. FDR calculation used Percolator and was set to 1% at the peptide level for the whole study. Unless indicated otherwise, a protein was considered present if at least three peptides in all three biological replicates were detected for qualitative analysis of immuno-isolated samples.
Protein sequence analyses
Full-length protein sequences were aligned with ClustalW using default parameters. The alignment was used to construct a neighbor-joining tree using MEGA4. Bootstrap values were obtained after 1000 permutation replicates. WD40 repeats were determined using the WDSP predicting software [91, 92].
RT-PCR analyses
For seedlings, total RNA was extracted using NucleoSpin RNA Plant (Macherey-Nagel). For seeds, 70 mg aliquots of seeds were ground in liquid nitrogen, and total RNA was extracted using a modified CTAB method [93]. Reverse transcription and subsequent quantitative PCR were performed on 1 μg of DNaseI-treated (Invitrogen, Amplification Grade DNaseI) RNAs using random hexamers and a cDNA reverse transcription kit (Applied Biosystems). Quantitative PCR was performed using LightCycler 480 SYBR green I Master mix and a LightCycler 480 (Roche). To confirm the absence of contamination of the samples by genomic DNA, PCR was also performed using primers flanking one intron of ACTIN2 and the size of the amplicons was checked on agarose gels. Data were normalized relative to genes with invariable expression as indicated in the figure legends. Primers sequences are given in Additional file 11: Table S10.
RNA sequencing and bioinformatics
Wild-type Col-0 and s2lb-2 seedlings were grown in vitro under long-day conditions and harvested 6 days after germination at 8 ZT. Two independent biological replicates for each genotype were produced using different seed batches. Total RNA was extracted using NucleoSpin RNA Plant (Macherey-Nagel). Messenger (polyA+) RNAs were purified from 1 μg of total RNA using oligo(dT). Libraries were prepared using the strand-specific RNA-Seq library preparation TruSeq Stranded mRNA kit (Illumina). Libraries were multiplexed by 4 on 1 flowcell lane. A 50-bp single-read end sequencing was performed on a HiSeq 1500 device (Illumina). A minimum of 37 million passing Illumina quality filter reads were obtained for each of the 4 samples. TruSeq adapters were removed with trimmomatic v0.36 [94] using the parameters “ILLUMINACLIP:TruSeq3-SE.fa:2:30:10 LEADING:5 TRAILING:5 MINLEN:20.” Reads were mapped on TAIR10 genome assembly of A. thaliana genome providing the gene annotation obtained from Araport11 [95] using STAR [96]. The command used is “STAR --genomeDir STAR_2.5.4b.TAIR10 --quantMode GeneCounts --outSAMstrandField intronMotif --sjdbOverhang 100 --sjdbGTFfile Araport11_GFF3_genes_transposons.201606.gtf --outSAMtype BAM SortedByCoordinate --outFilterIntronMotifs RemoveNoncanonical.” Differentially expressed genes were identified with DESeq2 (adj. p value < 0.01). Genes were split into 4 groups based on the normalized read counts in wild type (TPM equal to 0, TPM from 0 to 100, TPM from 100 to 1000, TPM above 1000) and used for Fig. 5j.
ChIP-qPCR, ChIP-sequencing, and ChIP bioinformatics
Plants were grown in vitro under long-day conditions, and whole seedlings were harvested 6 days after germination at 8 ZT. Chromatin extraction and immunoprecipitation of histones were performed as previously described [62]. Sequences of primers used for ChIP-qPCR are given in Additional file 11: Table S10. Quantitative analyses were performed as for RT-qPCR experiments using technical triplicate PCR samples. For ChIP-sequencing, a first ChIP series was performed using 5-day-old wild-type Col-0, s2la-1, s2lb-2, and s2la-1 s2lb-2 seedlings; a second series was performed using 5-day-old wild-type Col-0 and hub1-3 [49] plants, with an anti-H3K4me3 (Millipore #07–473) antibody; and a third series was performed using 14-day-old wild-type Col-0 seedlings with anti-RNPII (Abcam ab817) before library preparation and Illumina sequencing. To ascertain that hub1-3 plants used in these analyses displayed homozygous mutant alleles, seed batches from each corresponding stock were genotyped and “epigenotyped,” the HUB1 gene being reproducibly found among the few genes gaining H3K4me3 in hub1-3 plants presumably because of T-DNA based ectopic transcription (Additional file 2: Figure S13). Profiling of S2Lb-GFP and MYC-SDG2 was performed using anti-GFP (Life Technologies #11122) and anti-MYC (Ozyme #71D10) antibodies, respectively, and two crosslink steps. As recently described [58], samples were crosslinked first with 1.5 mM ethylene glycol bis(succinimidyl succinate) for 20 min and then with 1% formaldehyde for 10 min at room temperature. Crosslinking was stopped by adding 1.7 mL of 2 M glycine and incubating for 10 min. Libraries were prepared using 1 to 10 ng of input or IP DNA as described in the corresponding NCBI accession Super-Series GSE124319 datasets. TruSeq adapters were removed from the sequenced short reads with trimmomatic v0.36 [94] using the following different parameters for each ChIP type: (1) for H3K4me3 in s2l mutants: “-phred33 LEADING:5 TRAILING:5.” Dataset-specific parameters were also used: “PE --validatePairs ILLUMINACLIP:TruSeq2-PE.fa:2:30:10 MINLEN:20.” (2) for H3K4me3 in hub1.3 mutant: “SE ILLUMINACLIP:TruSeq3-SE.fa:2:30:10 MINLEN:30,” (3) for S2Lb-GFP and MYC-SDG2: “PE -validatePairs ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 MINLEN:20.” Reads from all ChIP-Seq experiments were aligned to TAIR10 genome assembly with Bowtie2 v.2.3.3 [97] with “--very-sensitive” setting. Peaks were identified with MACS2 v2.1.1 [98] with the command MACS2 callpeak and different parameters for each ChIP-seq type: (1) for H3K4me3 in s2lb mutants: “-f BAMPE --nomodel -q 0.01 -g 120e6 --bw 300,” (2) for H3K4me3 in hub1-3 mutant: “macs2 callpeak -f BAM -q 0.01 --bdg -g 120e6 --bw 300 --verbose 3 --nomodel --extsize 200,” and (3) for S2Lb-GFP and MYC-SDG2: “-f BAMPE --nomodel -q 0.05 --bdg -g 120e6 --bw 300.” For S2Lb-GFP and MYC-SDG2 peak detection, the peaks found in the wild-type negative controls were used to clean up the peak lists of S2Lb and SDG2 profiles. H3K4me3 peaks obtained from two independent biological replicates were merged with bedtools v2.27.1 intersect [99]. Peaks from each experiment were annotated with Araport11 genes bedtools v2.27.1 intersect. Genes were considered marked by H3K4me3, S2Lb-GFP, or MYC-SDG2 if overlapping for at least 150 bp with a relevant peak. To include nucleosomes in close proximity of the TSS, an upstream region of 250 bp was also considered for the overlap. Depth-normalized average values of the read densities were computed over 10 bp non-overlapping genomic bins with Deeptools v3.1.0 bamCoverage [100] and used to draw the metagene plots and heatmaps with Deeptools computeMatrix, plotHeatmap, and plotProfile. The normalized read densities of S2Lb-GFP, MYC-SDG2, and H3K4me3 in wild type were also used to generate co-occurrence plots over the TSS of S2Lb-occupied genes using R 3.4.3 (www.R-project.org) and the package ggplot2 v3.1. 0 (www.github.com). Gene ontology analysis was performed using the GO-TermFinder software [101] via the Princeton GO-TermFinder interface (http://go.princeton.edu/cgi-bin/GOTermFinder). GO categories were filtered with the REVIGO platform [102].
Additional files
Table S1. Protein IDs used for phylogenetic analyses. (XLSX 10 kb)
Figure S1. S2La and S2Lb mRNA levels in plant organs. Figure S2. Amino acid sequence alignment of full-length S2La and S2Lb with Swd2 (S. cerevisiae) and Wdr82 (H. sapiens) proteins. Figure S3. Expression of S2La and S2Lb genes in s2la-1 and s2lb-1 mutant plants. Figure S4. Complementation of s2lb-2 morphological phenotypes by pS2Lb::S2Lb-GFP. Figure S5. Decreased H3K4me3 level in S2Lb loss-of-function plants. Figure S6. ChIP-seq quality control and statistical analyses. Figure S7. H3K4me3 over S2Lb-GFP targeted genes. Figure S8. RNA-seq analysis of s2lb-2 mutant seedlings. Figure S9. S2Lb-GFP expression and RNA-seq. Figure S10. S2Lb and SDG2 co-regulate a large set of genes and associate within a high molecular weight complex. Figure S11. Detection of genes losing H3K4me3 enrichment in hub1-3 seedlings targeted by S2Lb-GFP. Figure S12. Genome browser snapshots of H3K4me3 profiles over representative genes. Figure S13. PCR genotyping and ChIP epigenotyping of the hub1-3 plants in the seed batches used for ChIP-seq analyses in this study. (PDF 16889 kb)
Table S2. Gene lists summarizing ChIP-seq analysis of H3K4me3 levels in WT, s2la-1, s2lb-2, and s2la-1s2lb-2 seedlings. (XLSX 579 kb)
Table S3. Gene lists summarizing ChIP-seq analysis of S2Lb-GFP occupancy in WT and hub1-3 seedlings. (XLSX 96 kb)
Table S4. RNA-seq identification of genes differentially expressed in WT and s2lb seedlings by DESeq2. (XLSX 2678 kb)
Table S5. Gene lists summarizing ChIP-seq analysis of RNA Pol II occupancy in wild-type seedlings. (XLSX 180 kb)
Table S6. Comparative analysis of mRNA-seq dataset for s2lb-2 mutant with transcriptomic data from sdg2-3, atx1, and atxr7-1 mutants. (XLSX 168 kb)
Table S7. MEME/TOMTOM analysis of S2Lb-GFP and MYC-SDG2 peak sequences. (XLSX 560 kb)
Table S8. Gene lists summarizing ChIP-seq analysis of MYC-SDG2 occupancy in WT seedlings. (XLSX 61 kb)
Table S9. Gene lists summarizing ChIP-seq analysis of H3K4me3 levels in WT and hub1-3 seedlings. (XLSX 346 kb)
Table S10. List of primers used for ChIP-PCR and RT-qPCR analyses. (XLSX 11 kb)
Uncropped blots from Figure.3, 5, 6, and S5. (PDF 9903 kb)
Review history. (DOCX 57 kb)
Acknowledgements
We kindly thank Prof. Yuehui He (Shanghai Center for Plant Stress Biology, China) for providing wdr5a-1 seeds, Prof. Xiaoyu Zhang (University of Georgia, USA) for providing sdg2-3/SDG2::myc-SDG2 line, and Dr. Olivier Mathieu (CNRS, GreD, France) for providing the Ds1-388-5 parental line. The authors are also much indebted to Alexandre Berr and Wen-Hui Shen (CNRS, Strasbourg, France) for the critical reading of the manuscript and Vincent Colot (IBENS, Paris, France) for the helpful discussions. They also thank the European Union COST Actions CA16212 INDEPTH and BM1307 PROTEOSTASIS as well as the Groupe De Recherche EPIPLANT (France).
Review history
The review history is available as Additional file 13.
Funding
This work was supported by grant ANR-11-JSV2-003-01 from the Agence Nationale pour la Recherche (ANR) to FB, by the Investissements d’Avenir program launched by the French Government and implemented by ANR (ANR-10-LABX-54 MEMOLIFE and ANR-10-IDEX-0001-02 PSL Research University) to IBENS, and PhD fellowships from the Université Paris-Sud Doctoral School in Plant Sciences to ASF, CB, and MR. IBENS genomic core facility was supported by the France Génomique national infrastructure, funded as part of the “Investissements d’Avenir” program (ANR-10-INBS-09). The Curie Institute Proteomics facility work was supported by “Région Ile-de-France” and Fondation pour la Recherche Médicale grants to DL.
Availability of data and materials
Uncropped blots are given in Additional file 2: Figure S14. The mass spectrometry proteomics data are available via ProteomeXchange with identifier PXD012292 [103]. All ChIP-seq and RNA-seq gene lists described in this study are directly available in Additional files 3, 4, 5, and 6: Tables S2 to S5. All ChIP-seq and RNA-seq raw data generated in this work are publicly accessible through GEO accession super-series GSE124319 [104]. The RNPII ChIP raw data are publicly available through GEO accession series GSM2028113 [105] and GSM2028107 [106]. All other processed datasets produced in this work will be made available on request.
Authors’ contributions
All authors contributed reagents, materials, or analysis tools. Protein analyses have been performed by ASF, MR, GZ, DS, and AFD, except for MS analyses that were carried out by BL and supervised by DL. ASF performed the DNA cloning, protein sequence analyses, and microscopy and conducted the RNA-seq experiments and all plant phenotyping analyses except dormancy assays that were conducted by EL and supervised by ChBa. RT-qPCR experiments were conducted by ASF and ClB. ChIPs were performed by ASF, ClB, and MR. DL, AM, and SKK prepared the ChIP libraries and performed the ChIP Illumina sequencing. RNA-seq library preparation and Illumina sequencing were performed at the Ecole normale superieure Genomic Platform (Paris, France). LC and ClB performed the bioinformatics analyses. ASF, ClB, MB, ChBo, and FB conceived the experiments. FB, MB, and ChBo rose funding. ASF, ClB, ChBo, and FB wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Ramaswamy A, Ioshikhes I. Dynamics of modeled oligonucleosomes and the role of histone variant proteins in nucleosome organization. Adv Protein Chem Struct Biol. 2013;90:119–149. doi: 10.1016/B978-0-12-410523-2.00004-3. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 4.Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta. 2014;1839:627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sequeira-Mendes J, Araguez I, Peiro R, Mendez-Giraldez R, Zhang X, Jacobsen SE, Bastolla U, Gutierrez C. The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell. 2014;26:2351–2366. doi: 10.1105/tpc.114.124578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vergara Z, Gutierrez C. Emerging roles of chromatin in the maintenance of genome organization and function in plants. Genome Biol. 2017;18:96. doi: 10.1186/s13059-017-1236-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen FX, Smith ER, Shilatifard A. Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2018;19:464–478. doi: 10.1038/s41580-018-0010-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, Romero-Campero FJ, Gomez-Zambrano A, Turck F, Calonje M. H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol. 2017;18:69. doi: 10.1186/s13059-017-1197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 15.Zheng S, Wyrick JJ, Reese JC. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol Cell Biol. 2010;30:3635–3645. doi: 10.1128/MCB.00324-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soares LM, Buratowski S. Yeast Swd2 is essential because of antagonism between Set1 histone methyltransferase complex and APT (associated with Pta1) termination factor. J Biol Chem. 2012;287:15219–15231. doi: 10.1074/jbc.M112.341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thornton JL, Westfield GH, Takahashi YH, Cook M, Gao X, Woodfin AR, Lee JS, Morgan MA, Jackson J, Smith ER, et al. Context dependency of Set1/COMPASS-mediated histone H3 Lys4 trimethylation. Genes Dev. 2014;28:115–120. doi: 10.1101/gad.232215.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorstensen T, Grini PE, Aalen RB. SET domain proteins in plant development. Biochim Biophys Acta. 2011;1809:407–420. doi: 10.1016/j.bbagrm.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 24.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/S1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 25.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 26.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, Kaeppler HF, Kaeppler SM. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003;132:907–925. doi: 10.1104/pp.102.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Ma H. Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol. 2012;195:248–263. doi: 10.1111/j.1469-8137.2012.04143.x. [DOI] [PubMed] [Google Scholar]
- 32.Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol. 2003;13:627–637. doi: 10.1016/S0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- 33.Tamada Y, Yun JY, Woo SC, Amasino RM. ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell. 2009;21:3257–3269. doi: 10.1105/tpc.109.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berr A, Xu L, Gao J, Cognat V, Steinmetz A, Dong A, Shen WH. SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 2009;151:1476–1485. doi: 10.1104/pp.109.143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berr A, McCallum EJ, Menard R, Meyer D, Fuchs J, Dong A, Shen WH. Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell. 2010;22:3232–3248. doi: 10.1105/tpc.110.079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo L, Yu Y, Law JA, Zhang X. SET DOMAIN GROUP2 is the major histone H3 lysine [corrected] 4 trimethyltransferase in Arabidopsis. Proc Natl Acad Sci U S A. 2010;107:18557–18562. doi: 10.1073/pnas.1010478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding Y, Ndamukong I, Xu Z, Lapko H, Fromm M, Avramova Z. ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet. 2012;8:e1003111. doi: 10.1371/journal.pgen.1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fromm M, Avramova Z. ATX1/AtCOMPASS and the H3K4me3 marks: how do they activate Arabidopsis genes? Curr Opin Plant Biol. 2014;21:75–82. doi: 10.1016/j.pbi.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Xiao J, Lee US, Wagner D. Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr Opin Plant Biol. 2016;34:41–53. doi: 10.1016/j.pbi.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Fletcher JC. State of the art: trxG factor regulation of post-embryonic plant development. Front Plant Sci. 2017;8:1925. doi: 10.3389/fpls.2017.01925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding Y, Avramova Z, Fromm M. Two distinct roles of ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1) at promoters and within transcribed regions of ATX1-regulated genes. Plant Cell. 2011;23:350–363. doi: 10.1105/tpc.110.080150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng J, Shen WH. Dynamic regulation and function of histone monoubiquitination in plants. Front Plant Sci. 2014;5:83. doi: 10.3389/fpls.2014.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang D, Gu X, He Y. Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell. 2009;21:1733–1746. doi: 10.1105/tpc.109.067967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang D, Kong NC, Gu X, Li Z, He Y. Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 2011;7:e1001330. doi: 10.1371/journal.pgen.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aquea F, Johnston AJ, Canon P, Grossniklaus U, Arce-Johnson P. TRAUCO, a Trithorax-group gene homologue, is required for early embryogenesis in Arabidopsis thaliana. J Exp Bot. 2010;61:1215–1224. doi: 10.1093/jxb/erp396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu WC, Li YH, Yuan HM, Zhang BL, Zhai S, Lu YT. WD40-REPEAT 5a functions in drought stress tolerance by regulating nitric oxide accumulation in Arabidopsis. Plant Cell Environ. 2017;40:543–52. [DOI] [PubMed]
- 47.Song ZT, Sun L, Lu SJ, Tian Y, Ding Y, Liu JX. Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc Natl Acad Sci U S A. 2015;112:2900–2905. doi: 10.1073/pnas.1419703112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Koornneef M, Soppe WJ. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell. 2007;19:433–444. doi: 10.1105/tpc.106.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, Beemster GT, Neyt P, Anami S, Robles P, et al. The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell. 2007;19:417–432. doi: 10.1105/tpc.106.041319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kapolas G, Beris D, Katsareli E, Livanos P, Zografidis A, Roussis A, Milioni D, Haralampidis K. APRF1 promotes flowering under long days in Arabidopsis thaliana. Plant Sci. 2016;253:141–153. doi: 10.1016/j.plantsci.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 51.Beris D, Kapolas G, Livanos P, Roussis A, Milioni D, Haralampidis K. RNAi-mediated silencing of the Arabidopsis thaliana ULCS1 gene, encoding a WDR protein, results in cell wall modification impairment and plant infertility. Plant Sci. 2016;245:71–83. doi: 10.1016/j.plantsci.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Basbouss-Serhal I, Leymarie J, Bailly C. Fluctuation of Arabidopsis seed dormancy with relative humidity and temperature during dry storage. J Exp Bot. 2016;67:119–130. doi: 10.1093/jxb/erv439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leymarie J, Vitkauskaite G, Hoang HH, Gendreau E, Chazoule V, Meimoun P, Corbineau F, El-Maarouf-Bouteau H, Bailly C. Role of reactive oxygen species in the regulation of Arabidopsis seed dormancy. Plant Cell Physiol. 2012;53:96–106. doi: 10.1093/pcp/pcr129. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, Geyer R, Brambilla V, Nakabayashi K, Soppe WJ. Chromatin dynamics during seed dormancy. Methods Mol Biol. 2011;773:239–257. doi: 10.1007/978-1-61779-231-1_15. [DOI] [PubMed] [Google Scholar]
- 56.Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yatusevich R, Fedak H, Ciesielski A, Krzyczmonik K, Kulik A, Dobrowolska G, Swiezewski S. Antisense transcription represses Arabidopsis seed dormancy QTL DOG1 to regulate drought tolerance. EMBO Rep. 2017;18:2186–2196. doi: 10.15252/embr.201744862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bourbousse C, Vegesna N, Law JA. SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc Natl Acad Sci U S A. 2018;115:E12453–E12462. doi: 10.1073/pnas.1810582115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perrella G, Kaiserli E. Light behind the curtain: photoregulation of nuclear architecture and chromatin dynamics in plants. New Phytol. 2016;212:908–919. doi: 10.1111/nph.14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bourbousse C, Ahmed I, Roudier F, Zabulon G, Blondet E, Balzergue S, Colot V, Bowler C, Barneche F. Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet. 2012;8:e1002825. doi: 10.1371/journal.pgen.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li F, Cheng C, Cui F, de Oliveira MV, Yu X, Meng X, Intorne AC, Babilonia K, Li M, Li B, et al. Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe. 2014;16:748–758. doi: 10.1016/j.chom.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nassrallah A, Rougee M, Bourbousse C, Drevensek S, Fonseca S, Iniesto E, Ait-Mohamed O, Deton-Cabanillas AF, Zabulon G, Ahmed I, et al. DET1-mediated degradation of a SAGA-like deubiquitination module controls H2Bub homeostasis. Elife. 2018;7:e37892. [DOI] [PMC free article] [PubMed]
- 63.Cao Y, Dai Y, Cui S, Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell. 2008;20:2586–2602. doi: 10.1105/tpc.108.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gu X, Jiang D, Wang Y, Bachmair A, He Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009;57:522–533. doi: 10.1111/j.1365-313X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- 65.Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM. Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol. 2009;149:1196–1204. doi: 10.1104/pp.108.131508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T. HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell. 2009;21:1000–1019. doi: 10.1105/tpc.108.062364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao W, Neyt P, Van Lijsebettens M, Shen WH, Berr A. Interactive and noninteractive roles of histone H2B monoubiquitination and H3K36 methylation in the regulation of active gene transcription and control of plant growth and development. New Phytol. 2018;221:1101–1116. doi: 10.1111/nph.15418. [DOI] [PubMed] [Google Scholar]
- 68.Oh S, Park S, van Nocker S. Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 2008;4:e1000077. doi: 10.1371/journal.pgen.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao X, Feng H, Yu Y, Dong A, Shen WH. SDG2-mediated H3K4 methylation is required for proper Arabidopsis root growth and development. PLoS One. 2013;8:e56537. doi: 10.1371/journal.pone.0056537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yun JY, Tamada Y, Kang YE, Amasino RM. Arabidopsis trithorax-related3/SET domain GROUP2 is required for the winter-annual habit of Arabidopsis thaliana. Plant Cell Physiol. 2012;53:834–846. doi: 10.1093/pcp/pcs021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tuukkanen A, Huang B, Henschel A, Stewart F, Schroeder M. Structural modeling of histone methyltransferase complex Set1C from Saccharomyces cerevisiae using constraint-based docking. Proteomics. 2010;10:4186–4195. doi: 10.1002/pmic.201000283. [DOI] [PubMed] [Google Scholar]
- 72.Crevillen P, Dean C. Regulation of the floral repressor gene FLC: the complexity of transcription in a chromatin context. Curr Opin Plant Biol. 2011;14:38–44. doi: 10.1016/j.pbi.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 73.Berr A, Shafiq S, Shen WH. Histone modifications in transcriptional activation during plant development. Biochim Biophys Acta. 2011;1809:567–576. doi: 10.1016/j.bbagrm.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 74.Li Z, Jiang D, He Y. FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat Plants. 2018;4:836–846. doi: 10.1038/s41477-018-0250-6. [DOI] [PubMed] [Google Scholar]
- 75.Himanen K, Woloszynska M, Boccardi TM, De Groeve S, Nelissen H, Bruno L, Vuylsteke M, Van Lijsebettens M. Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J. 2012;72:249–260. doi: 10.1111/j.1365-313X.2012.05071.x. [DOI] [PubMed] [Google Scholar]
- 76.Barroco RM, De Veylder L, Magyar Z, Engler G, Inze D, Mironov V. Novel complexes of cyclin-dependent kinases and a cyclin-like protein from Arabidopsis thaliana with a function unrelated to cell division. Cell Mol Life Sci. 2003;60:401–412. doi: 10.1007/s000180300033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cui X, Fan B, Scholz J, Chen Z. Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell. 2007;19:1388–1402. doi: 10.1105/tpc.107.051375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang ZW, Wu Z, Raitskin O, Sun Q, Dean C. Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc Natl Acad Sci U S A. 2014;111:7468–7473. doi: 10.1073/pnas.1406635111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fulop K, Pettko-Szandtner A, Magyar Z, Miskolczi P, Kondorosi E, Dudits D, Bako L. The Medicago CDKC;1-CYCLINT;1 kinase complex phosphorylates the carboxy-terminal domain of RNA polymerase II and promotes transcription. Plant J. 2005;42:810–820. doi: 10.1111/j.1365-313X.2005.02421.x. [DOI] [PubMed] [Google Scholar]
- 80.Antosz W, Pfab A, Ehrnsberger HF, Holzinger P, Kollen K, Mortensen SA, Bruckmann A, Schubert T, Langst G, Griesenbeck J, et al. The composition of the Arabidopsis RNA polymerase II transcript elongation complex reveals the interplay between elongation and mRNA processing factors. Plant Cell. 2017;29:854–870. doi: 10.1105/tpc.16.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alvarez-Venegas R, Avramova Z. Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 2005;33:5199–5207. doi: 10.1093/nar/gki830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dichtl B, Aasland R, Keller W. Functions for S. cerevisiae Swd2p in 3′ end formation of specific mRNAs and snoRNAs and global histone 3 lysine 4 methylation. RNA. 2004;10:965–977. doi: 10.1261/rna.7090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cheng H, He X, Moore C. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol Cell Biol. 2004;24:2932–2943. doi: 10.1128/MCB.24.7.2932-2943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 85.Roguev A, Shevchenko A, Schaft D, Thomas H, Stewart AF, Shevchenko A. A comparative analysis of an orthologous proteomic environment in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Mol Cell Proteomics. 2004;3:125–132. doi: 10.1074/mcp.M300081-MCP200. [DOI] [PubMed] [Google Scholar]
- 86.Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ. The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res. 2007;120:157–165. doi: 10.1007/s10265-006-0048-x. [DOI] [PubMed] [Google Scholar]
- 87.Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 2004;37:897–905. doi: 10.1111/j.1365.313X.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- 88.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Castells E, Molinier J, Benvenuto G, Bourbousse C, Zabulon G, Zalc A, Cazzaniga S, Genschik P, Barneche F, Bowler C. The conserved factor DE-ETIOLATED 1 cooperates with CUL4-DDB1DDB2 to maintain genome integrity upon UV stress. EMBO J. 2011;30:1162–1172. doi: 10.1038/emboj.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Poullet P, Carpentier S, Barillot E. myProMS, a web server for management and validation of mass spectrometry-based proteomic data. Proteomics. 2007;7:2553–2556. doi: 10.1002/pmic.200600784. [DOI] [PubMed] [Google Scholar]
- 91.Wang Y, Hu XJ, Zou XD, Wu XH, Ye ZQ, Wu YD. WDSPdb: a database for WD40-repeat proteins. Nucleic Acids Res. 2015;43:D339–D344. doi: 10.1093/nar/gku1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y, Jiang F, Zhuo Z, Wu XH, Wu YD. A method for WD40 repeat detection and secondary structure prediction. PLoS One. 2013;8:e65705. doi: 10.1371/journal.pone.0065705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- 94.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89:789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- 96.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Loew D. and Arras G. (2019) SDG2 and S2Lb affinity purification and mass spectrometry. Datasets. https://www.ebi.ac.uk/pride/archive/projects/PXD012292. Accessed 26 Apr 2019.
- 104.Concia L. (2019) ChIP-seq and RNA-seq in HUB1 and SWD2 Arabidopsis mutant seedlings. Datasets: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124319. Accessed 2 May 2019.
- 105.Veluchamy A. (2016) RNAPol-II. Gene expression omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM2028113. Accessed 2 May 2019.
- 106.Veluchamy A. (2016) RNAPol-II. Gene Expression omnibus https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM2028107. Accessed 2 May 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Protein IDs used for phylogenetic analyses. (XLSX 10 kb)
Figure S1. S2La and S2Lb mRNA levels in plant organs. Figure S2. Amino acid sequence alignment of full-length S2La and S2Lb with Swd2 (S. cerevisiae) and Wdr82 (H. sapiens) proteins. Figure S3. Expression of S2La and S2Lb genes in s2la-1 and s2lb-1 mutant plants. Figure S4. Complementation of s2lb-2 morphological phenotypes by pS2Lb::S2Lb-GFP. Figure S5. Decreased H3K4me3 level in S2Lb loss-of-function plants. Figure S6. ChIP-seq quality control and statistical analyses. Figure S7. H3K4me3 over S2Lb-GFP targeted genes. Figure S8. RNA-seq analysis of s2lb-2 mutant seedlings. Figure S9. S2Lb-GFP expression and RNA-seq. Figure S10. S2Lb and SDG2 co-regulate a large set of genes and associate within a high molecular weight complex. Figure S11. Detection of genes losing H3K4me3 enrichment in hub1-3 seedlings targeted by S2Lb-GFP. Figure S12. Genome browser snapshots of H3K4me3 profiles over representative genes. Figure S13. PCR genotyping and ChIP epigenotyping of the hub1-3 plants in the seed batches used for ChIP-seq analyses in this study. (PDF 16889 kb)
Table S2. Gene lists summarizing ChIP-seq analysis of H3K4me3 levels in WT, s2la-1, s2lb-2, and s2la-1s2lb-2 seedlings. (XLSX 579 kb)
Table S3. Gene lists summarizing ChIP-seq analysis of S2Lb-GFP occupancy in WT and hub1-3 seedlings. (XLSX 96 kb)
Table S4. RNA-seq identification of genes differentially expressed in WT and s2lb seedlings by DESeq2. (XLSX 2678 kb)
Table S5. Gene lists summarizing ChIP-seq analysis of RNA Pol II occupancy in wild-type seedlings. (XLSX 180 kb)
Table S6. Comparative analysis of mRNA-seq dataset for s2lb-2 mutant with transcriptomic data from sdg2-3, atx1, and atxr7-1 mutants. (XLSX 168 kb)
Table S7. MEME/TOMTOM analysis of S2Lb-GFP and MYC-SDG2 peak sequences. (XLSX 560 kb)
Table S8. Gene lists summarizing ChIP-seq analysis of MYC-SDG2 occupancy in WT seedlings. (XLSX 61 kb)
Table S9. Gene lists summarizing ChIP-seq analysis of H3K4me3 levels in WT and hub1-3 seedlings. (XLSX 346 kb)
Table S10. List of primers used for ChIP-PCR and RT-qPCR analyses. (XLSX 11 kb)
Uncropped blots from Figure.3, 5, 6, and S5. (PDF 9903 kb)
Review history. (DOCX 57 kb)
Data Availability Statement
Uncropped blots are given in Additional file 2: Figure S14. The mass spectrometry proteomics data are available via ProteomeXchange with identifier PXD012292 [103]. All ChIP-seq and RNA-seq gene lists described in this study are directly available in Additional files 3, 4, 5, and 6: Tables S2 to S5. All ChIP-seq and RNA-seq raw data generated in this work are publicly accessible through GEO accession super-series GSE124319 [104]. The RNPII ChIP raw data are publicly available through GEO accession series GSM2028113 [105] and GSM2028107 [106]. All other processed datasets produced in this work will be made available on request.