Abstract
Significance: In addition to their classical role in cellular ATP production, mitochondria are of key relevance in various (patho)physiological mechanisms including second messenger signaling, neuro-transduction, immune responses and death induction.
Recent Advances: Within cells, mitochondria are motile and display temporal changes in internal and external structure (“mitochondrial dynamics”). During the last decade, substantial empirical and in silico evidence was presented demonstrating that mitochondrial dynamics impacts on mitochondrial function and vice versa.
Critical Issues: However, a comprehensive and quantitative understanding of the bidirectional links between mitochondrial external shape, internal structure and function (“morphofunction”) is still lacking. The latter particularly hampers our understanding of the functional properties and behavior of individual mitochondrial within single living cells.
Future Directions: In this review we discuss the concept of mitochondrial morphofunction in mammalian cells, primarily using experimental evidence obtained within the last decade. The topic is introduced by briefly presenting the central role of mitochondria in cell physiology and the importance of the mitochondrial electron transport chain (ETC) therein. Next, we summarize in detail how mitochondrial (ultra)structure is controlled and discuss empirical evidence regarding the equivalence of mitochondrial (ultra)structure and function. Finally, we provide a brief summary of how mitochondrial morphofunction can be quantified at the level of single cells and mitochondria, how mitochondrial ultrastructure/volume impacts on mitochondrial bioreactions and intramitochondrial protein diffusion, and how mitochondrial morphofunction can be targeted by small molecules.
Keywords: mitochondrial dynamics, ultrastructure, quantitative live-cell microscopy
Table Of Contents
I. Introduction
Virtually every mammalian cell contains mitochondria, which are classically recognized as key generators of cellular energy in the form of adenosine triphosphate (ATP) (99). In addition, mitochondria constitute an integral part of the mechanisms that control cell functioning and survival. This is exemplified by their established physiological role in cell differentiation (289), immune cell function (40, 252, 261), cell death regulation (20, 123), calcium homeostasis (77, 109, 110, 434), and neurogenesis (175). In addition, mitochondrial dysfunction is associated with a multitude of pathophysiological conditions, including metabolic disorders (196), cancer (6, 53), diabetes (336, 436), and neurodegeneration (188). Proper mitochondrial function in mammals requires ∼1200 genes [MitoCarta 2.0 (43) and MitoMiner 4.0 (369)].
Only a small fraction of these mitochondrial proteins is encoded by the mitochondrial DNA (mtDNA), of which multiple copies are present in each individual mitochondrion. The mtDNA consists of a light (L) and heavy (H) strand, has a size of 16,569 base pairs, and contains 37 genes (9 on the L-strand and 28 on the H-strand). These genes encode 13 proteins [all of which are subunits of the mitochondrial oxidative phosphorylation (OXPHOS) system; see section I.A], 22 transfer RNAs, and 2 subunits of the mitochondrial ribosome (135). The nuclear DNA (nDNA) encodes the large majority of mitochondrial proteins, which are synthesized in the cytosol and imported into the mitochondrion. This import is carried out by a dedicated machinery of protein translocases in the mitochondrial outer membrane (TOM) and inner membrane (TIM), as reviewed in detail elsewere (172, 422).
A. Mitochondrial adenosine triphosphate production
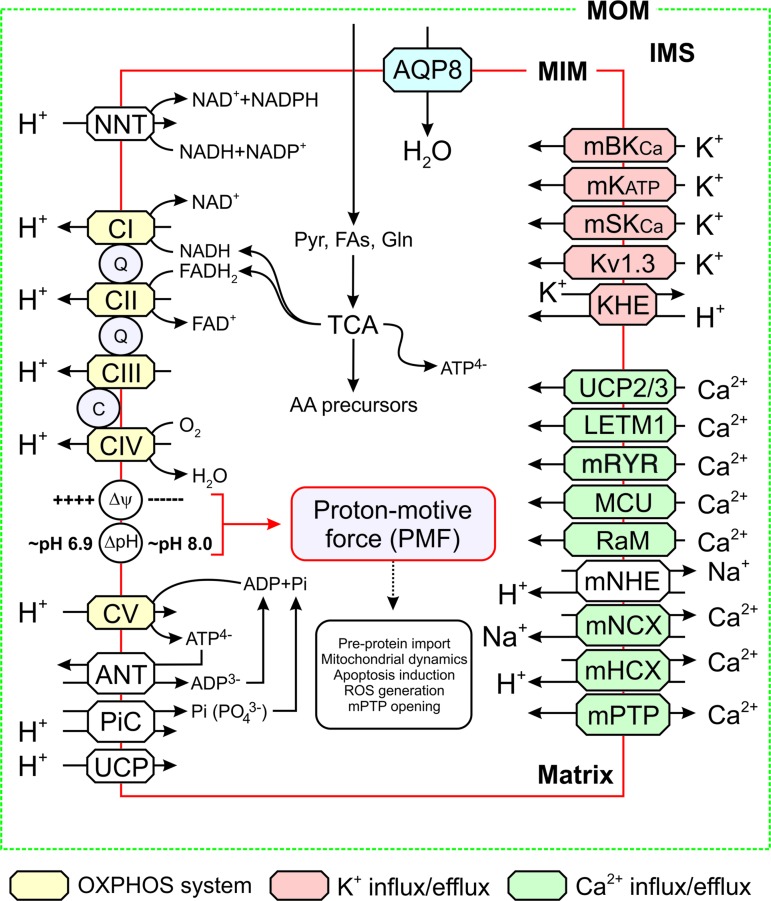
Mitochondrial adenosine triphosphate (ATP) is primarily generated by the action of the mitochondrial OXPHOS system (Fig. 1). This system is embedded in the mitochondrial inner membrane (MIM), consists of five multi-subunit complexes (CI–CV), and is fueled by NADH and FADH2 provided by the glycolysis pathway in the cytosol and the tricarboxylic acid (TCA) cycle in the mitochondrial matrix (234, 397). Functionally, the OXPHOS system consists of the electron transport chain (ETC; comprising CI-CIV) and the ATP-generating FoF1-ATP synthase (“CV”). ETC assembly from its nDNA- and mtDNA-encoded subunits requires assistance of at least 33 nDNA-encoded assembly factors (188). At a higher structural level, the ETC can be organized in respiratory supercomplexes (98, 260).
FIG. 1.
Mitochondrial energy metabolism and solute exchange. Schematic visualization of the ATP-generating mitochondrial oxidative phosphorylation system (yellow) and TCA cycle. Solute exchangers are indicated in different colors. See main text for details. This figure was compiled by integrating information from (160, 188, 185, 227, 315, 335). Δψ, trans-MIM electrical gradient; ΔpH, trans-MIM pH gradient; AA, amino acids; ADP, adenosine diphosphate; ANT, adenine nucleotide translocator; AQP8, aquaporin-8; ATP, adenosine triphosphate; C, cytochrome-c; CI-CV, complex I–V; FAs, fatty acids; Gln, glutamine; IMS, intermembrane space; KHE, K+/H+ exchanger; Kv1.3, mitochondrial voltage-activated K+ channel of the Kv1.3 type; LETM1, leucine zipper-EF-hand-containing transmembrane protein 1; mBKCa, mitochondrial Ca2+-regulated K+ large conductance channel; MCU, mitochondrial calcium uniporter; mHCX, mitochondrial H+/Ca2+ exchanger; MIM, mitochondrial inner membrane; mKATP, mitochondrial ATP-activated K+ channel; mNCX, mitochondrial Na+/Ca2+ exchanger; mNHE, mitochondrial Na+/H+ exchanger; MOM, mitochondrial outer membrane; mPTP, mitochondrial permeability transition pore; mRYR, mitochondrial ryanodine receptor; mSKCa, mitochondrial small conductance Ca2+-activated K+ channel; NNT, nicotinamide nucleotide transhydrogenase; OXPHOS, oxidative phosphorylation; PiC, Pi/H+ symporter; PMF, proton-motive force; Pyr, pyruvate; Q, Coenzyme Q10; RaM, rapid mode of Ca2+ uptake; ROS, reactive oxygen species; TCA, tricarboxylic acid; UCP, uncoupling protein. Color images are available online.
Within the ETC, CI and CII abstract electrons from NADH and FADH2, respectively, and donate them to Coenzyme Q10 (“Q”). The latter molecule transports electrons to CIII, from where they are conveyed to CIV by cytochrome-c (“c”). At CIV, the electrons are donated to molecular oxygen to form water. As an alternative to CI, CII, and CIII, various other MIM-associated enzymes can donate electrons to Q (241, 284). For instance, by metabolizing: (i) acetyl coenzyme A (acyl-CoA) (by electron transfer flavoprotein-ubiquinone oxidoreductase or ETFQ), (ii) glycerol-3-phosphate (s,n-Glycerol-3-phosphate dehydrogenase or G3PDH), (iii) proline (proline dehydrogenase or PRODH), (iv) dihydroorotate (dihydroorotate dehydrogenase or DHODH), and (v) hydrogen sulfide (succinate:quinone reductase or SQR). Moreover, electrons can be donated to cytochrome-c by Mo-pterin and B-type heme. In this sense, Q and cytochrome-c can be regarded as “junctions,” on which different electron-donating systems converge to feed electrons into the ETC (213). It appears that the “alternative” electron donors do not simultaneously supply electrons to the ETC. Moreover, these enzymes display tissue and species-specific expression (241). During electron transport, energy is gradually released and used (at CI, CIII, and CIV) to expel protons (H+) from the mitochondrial matrix across the MIM. As a consequence, an inward-directed trans-MIM proton-motive force (PMF) is generated, consisting of an electrical (Δψ) and chemical (ΔpH) component (448). The PMF is utilized by CV to catalyze the formation of ATP from adenosine diphosphate (ADP) and inorganic phosphate (Pi) by allowing the controlled re-entry of protons into the matrix (267, 410). This ATP generation requires Pi import in the form of PO43− by the Pi/H+ symporter (PiC) and the electrogenic exchange of ADP3− (import) against ATP4− (export) by the adenine nucleotide translocator (ANT; Fig. 1). This combined (forward) action of CV and ANT will depolarize Δψ, which is counterbalanced by ETC action. Under pathological conditions, CV can also hydrolyze ATP and expel protons from the mitochondrial matrix to sustain Δψ (285). This mechanism requires transport of ATP generated in the cytosol, for instance by the glycolysis pathway, into the mitochondrial matrix by ANT reverse mode action.
It is well established that the ETC plays a key role in the production of mitochondrial reactive oxygen species (ROS), particularly under pathological conditions. Information about how ETC-mediated ROS production relates to: (i) other sources of mitochondrial and cellular ROS, (ii) the spatial aspects of ROS action, (iii) oxidative stress induction, and (iv) ROS signaling is discussed in detail elsewhere (21, 89, 190, 241, 365, 425). Regarding the link between the ETC and redox metabolism, the mitochondrial nicotinamide nucleotide transhydrogenase (NNT) directly couples the trans-MIM influx of H+ to the transfer of electrons from NADH to NADP (Fig. 1). This coupling keeps the mitochondrial NADP/NADPH pool in a reduced state, which protects mitochondria against oxidative damage (273). The NNT can also operate in reverse mode, thereby oxidizing the NADP/NADPH pool and disrupting antioxidant defense (286). Both NAD+/NADH and NADP+/NADPH play important (regulatory) roles in mitochondrial/cellular metabolism and redox homeostasis. These roles, as well as their mechanistic connection and signaling function in health and disease are discussed in detail elsewhere (140, 145, 161, 435).
B. Cellular ATP production displays metabolic flexibility
In addition to mitochondrial OXPHOS, the glycolysis pathway also generates ATP by converting glucose (taken up by the cell via glucose transporters) into pyruvate. The latter is either converted into lactate (which can be released into the extracellular medium) or enters the mitochondrial matrix to form acyl-CoA as a TCA cycle substrate yielding additional ATP (Fig. 1). In addition, also fatty acids (FAs) and glutamine (Gln) can serve as TCA substrates (397). Cells display a substantial degree of metabolic flexibility, meaning that the balance between glycolysis- and OXPHOS-derived ATP generation is variable (394). This is exemplified by studies in C2C12 mouse myoblasts demonstrating that acute OXPHOS inhibition rapidly increases steady-state glucose uptake/consumption and that this increase fully compensates for the reduction in mitochondrial ATP production (219, 220). Alternatively, ATP can be generated by substrate-level phosphorylation within the mitochondrial matrix. In this process, a phosphorylated biomolecule directly transfers a PO32− (phosphoryl) group to ADP or guanosine diphosphate (GDP) to form ATP or guanosine triphosphate (GTP) (61).
C. ETC function is not only essential for mitochondrial ATP production
In addition to ATP generation, Δψ and/or ΔpH are crucial for the activity of transporters that exchange metabolites and ions between the cytosol, intermembrane space (IMS), and mitochondrial matrix compartment (Fig. 1) (302, 379). This exchange is of key importance, for instance to maintain Ca2+ homeostasis and viability in the heart (235). Further, Δψ plays a dual role in the trans-MIM import of mitochondrial preproteins via the TOM/TIM system (422). Given the fact that Δψ is negative on the matrix-facing side of the MIM, it exerts an electrophoretic effect on the N-terminal mitochondrial targeting sequence (or “presequence”), which often carries a positive charge (247, 392). In addition, Δψ directly activates a key TIM protein (TIM23), which translocates cleavable preproteins into the IMS or mitochondrial matrix (253).
A less negative (depolarized) Δψ has been associated with reduced mitochondrial ATP production and also connects to various other (patho) physiological phenomena by: (i) increasing cellular ROS levels (190, 192, 193), (ii) preventing fusion of the MIM but not of the mitochondrial outer membrane (MOM; see section III) (243), (iii) increasing the opening probability of the mitochondrial permeability transition pore (mPTP), ultimately leading to apoptosis induction (26), and (iv) stimulating mitochondrial degradation by organelle-specific autophagy (“mitophagy”) (137, 382, 393).
D. Mitochondria physically and functionally interact with other cell constituents
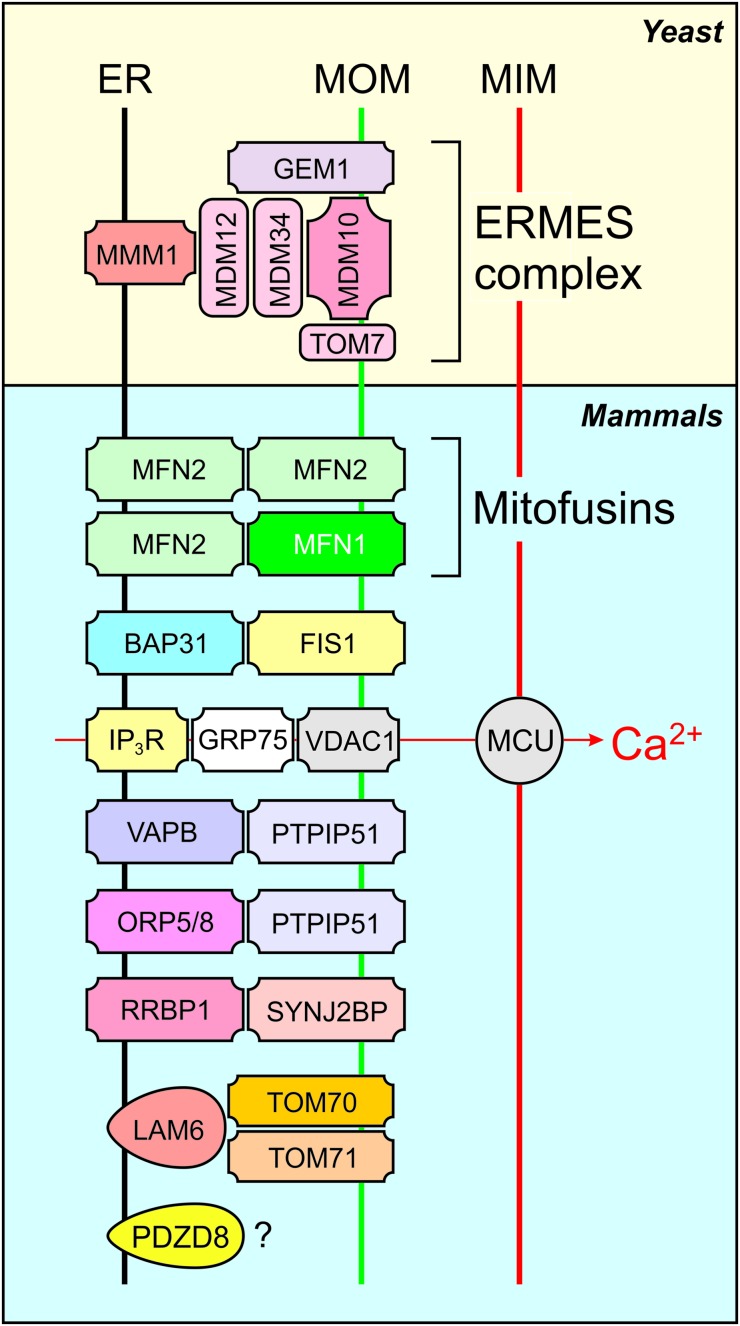
Further emphasizing their central role in cell physiology, mitochondria physically and functionally interact with various other cell components, including the cytoskeleton, plasma membrane (PM), endoplasmic reticulum (ER), Golgi apparatus, lipid droplets (LDs), peroxisomes, and lysosomes (316, 395, 428). With respect to peroxisomes, mitochondria are essential for their de novo biosynthesis from mitochondrial and ER-derived pre-peroxisomes (376). Mitochondria-ER interactions are currently the best studied and are mediated by mitochondria-associated membranes. The latter are at the center of intracellular signaling and other pathways, including cholesterol and phospholipid synthesis, ROS production, and mitochondrial calcium uptake/release. In this respect, various tethering proteins involved in mito-ER coupling have been identified in yeast and mammalian systems (Fig. 2) (118, 141, 224). Further detailed information about these protein–protein interactions, as well as their functional relevance, is presented elsewhere (50, 72, 118, 121, 141, 190, 224).
FIG. 2.
Selected proteins involved in mitochondria-ER tethering. Tethering protein complexes include the ER-ERMES, MFNs, and the VDAC-containing complex involved in mitochondrial calcium uptake. BAP31, B cell receptor associated protein 31; ER, endoplasmic reticulum; FIS1, fission protein 1; GEM1, GTPase EF-hand protein of mitochondria 1; GRP75, glucose-regulated protein of 75 kD; IP3R, inositol 1,4,5-trisphosphate receptor; LAM6, lipid transfer protein anchored at membrane contact site 6; MFNs, mitofusins; MMM1, maintenance of mitochondrial morphology 1; MDM10, mitochondrial distribution and morphology 10; MDM12, mitochondrial distribution and morphology 12; MDM34, mitochondrial distribution and morphology 34; PDZD8, PDZ domain-containing 8; ORP5/8, oxysterol-binding protein-related proteins 5 and 8; PTPIP51, protein tyrosine phosphatase-interacting protein 51; RRBP1, ribosome-binding protein 1; SYNJ2BP, synaptojanin 2 binding protein; TOM7, translocase of the outer mitochondrial membrane 7; TOM70, translocase of the outer mitochondrial membrane 70; TOM71, translocase of the outer mitochondrial membrane 71; VAPB, VAMP (vesicle-associated membrane protein) associated protein B and C; VDAC, voltage-dependent anion channel. Color images are available online.
II. Mitochondrial (Ultra)Structural Dynamics
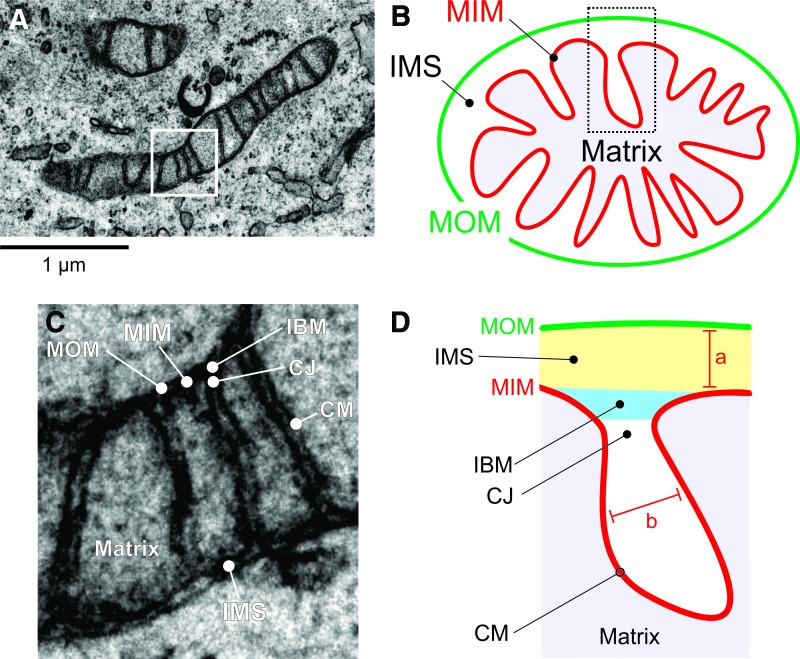
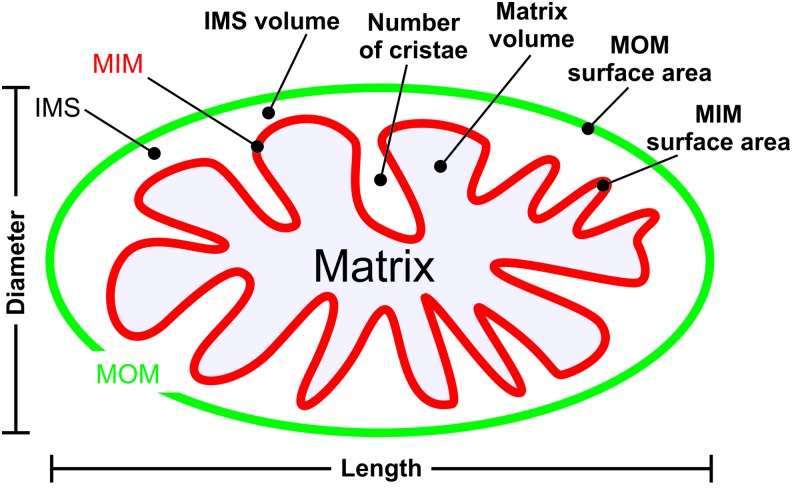
Mitochondria consist of a double membrane system, in which the MOM surrounds the MIM (Fig. 3A, B). The latter constitutes the boundary of the mitochondrial matrix compartment and contains many folds (cristae) that protrude into this compartment, thereby enlarging the MIM surface area. The MIM and MOM are separated by the mitochondrial IMS and are partially connected via contact sites that are involved in cristae organization (309). Important structural features of the mitochondrial matrix and cristae system include the inner boundary membrane (IBM), cristae junction (CJ), and cristae membrane (CM; Fig. 3C, D). Parameters describing mitochondrial (ultra)structure include the distance between the MIM and MOM (marked “a”), the intracristae space (marked “b”), and the distance between adjacent cristae (“inter-cristae space”) (403).
FIG. 3.
Mitochondrial ultrastructure. (A) EM image depicting a mitochondrion (HeLa cell). The white box indicates the magnified view in (C). (B) Schematic representation of mitochondrial ultrastructure showing the mitochondrial matrix enveloped by the MIM and MOM. Between the MIM and MOM lies the IMS. (C) Magnification of the box in (A) to illustrate mitochondrial ultrastructure depicting the IBM, CJ, and CM. (D) Schematic representation of mitochondrial ultrastructure. Structural parameters include the distance between the MIM and MOM (marked “a”) and the intracristae space (marked “b”). (D) was adapted from (404). CJ, cristae junction; CM, cristae membrane; EM, electron microscopy; IBM, inner boundary membrane. Color images are available online.
Maintaining CM integrity is of key importance for proper OXPHOS functioning (68, 149). The latter is illustrated by analyis of the yeast Saccharomyces cerevisiae using superresolution microscopy (see section IV.D) and cryo-immunogold electron microscopy (EM). This revealed that mtDNA-encoded subunits of CIII and CIV are inserted at different sites of the MIM when compared with CV subunits (374). In addition, it was found that early (but not late) steps in CIII and CIV assembly occur at the IBM, whereas CV assembly is carried out in the CM. This suggests that alterations in cristae structure directly but differentially affect the assembly of OXPHOS complexes. Net mitochondrial external/internal morphology and positioning greatly differs between cell types (39, 69, 201, 389) and dynamically changes over time (Fig. 4; “mitochondrial dynamics”). These changes require mitochondrial movement via mechanical interactions with the cytoskeleton, the functional impact and molecular mechanisms of which are discussed elsewhere (29, 223, 264, 362). In addition, mitochondrial external and internal morphology is affected by: (i) fission/fusion events (51, 297, 392, 420), (ii) chemiosmosis-related swelling/shrinking (160), (iii) the composition and physicochemical properties of the MOM and MIM (131), and (iv) the nature of the extracellular matrix (19).
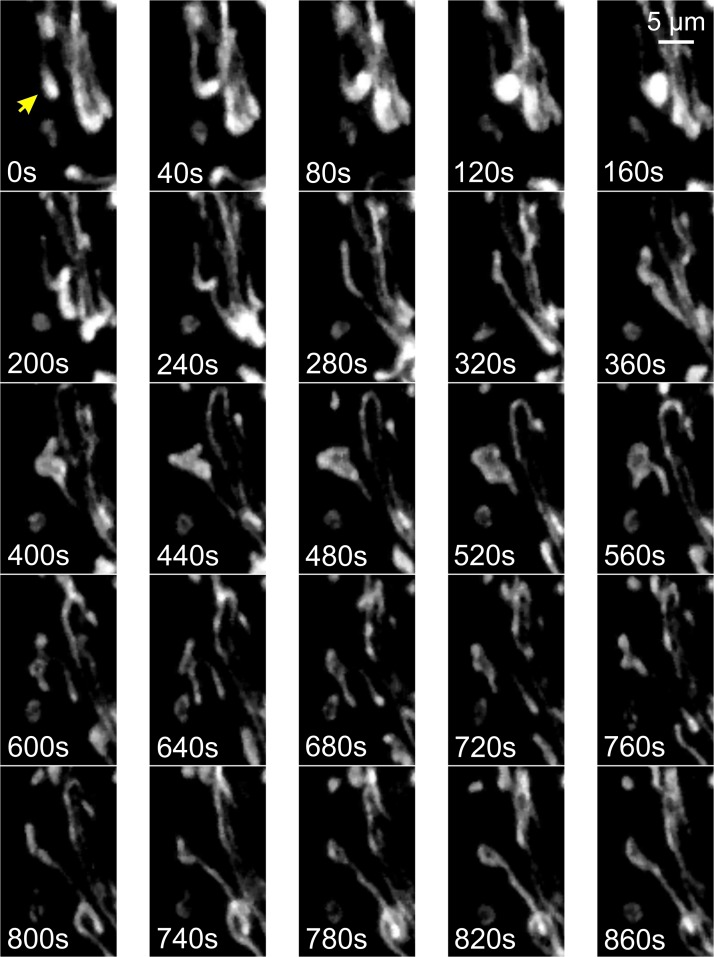
FIG. 4.
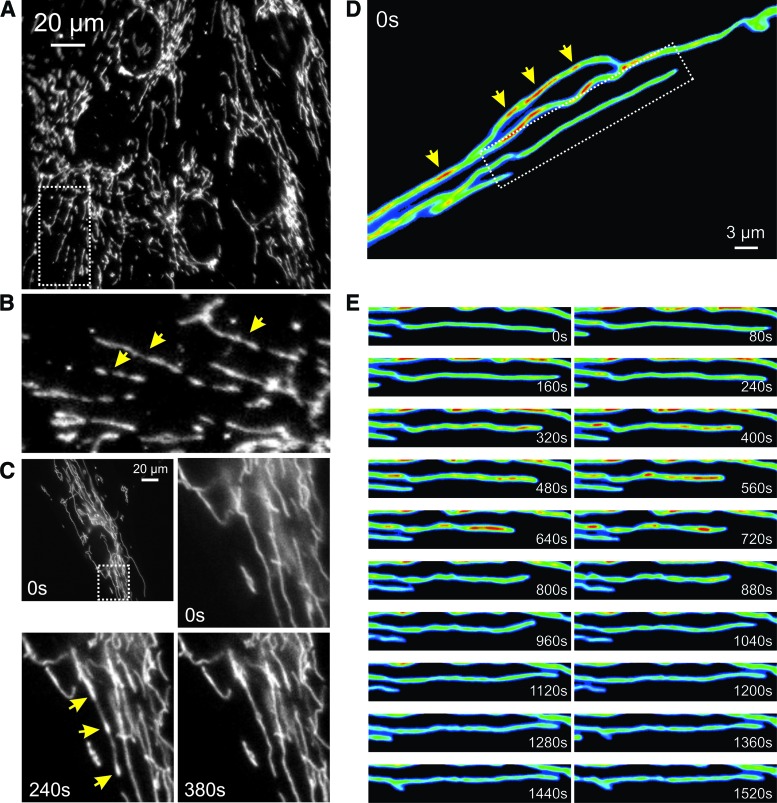
Live-cell dynamics of mitochondrial external structure. Primary mouse myoblasts from skeletal muscle were stained with rhodamine 123 and visualized by using fluorescence microscopy at an interval of 40 s. Individual mitochondria (arrow) were motile and displayed volume and structural changes. Color images are available online.
As the integrated result of the mechanisms described earlier, mitochondrial external morphology ranges from elongated tubular structures (Fig. 5A–C) to donut-shaped organelles (Fig. 5D, arrows) and often displays heterogeneity within the same cell (Fig. 5C–E). It was proposed that this morphological plasticity allows: (i) mixing of mitochondrial content, (ii) redistribution of damaged proteins and lipids between mitochondria in stress reduction, (iii) local functioning of (subsets of) mitochondria within the cell, and (iv) mitophagy (179). Importantly, mitochondria cannot be generated de novo (363). This means that mitochondrial fission is crucial to allow their inheritance by both daughter cells during cell division.
FIG. 5.
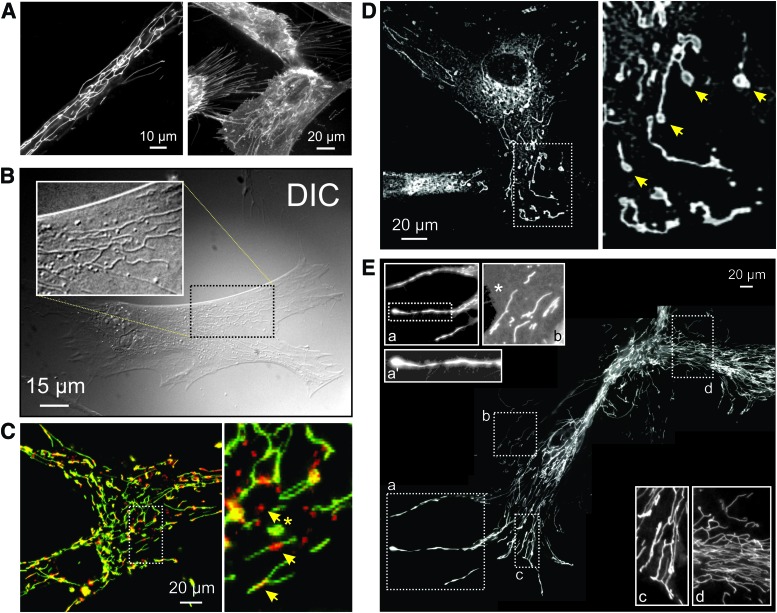
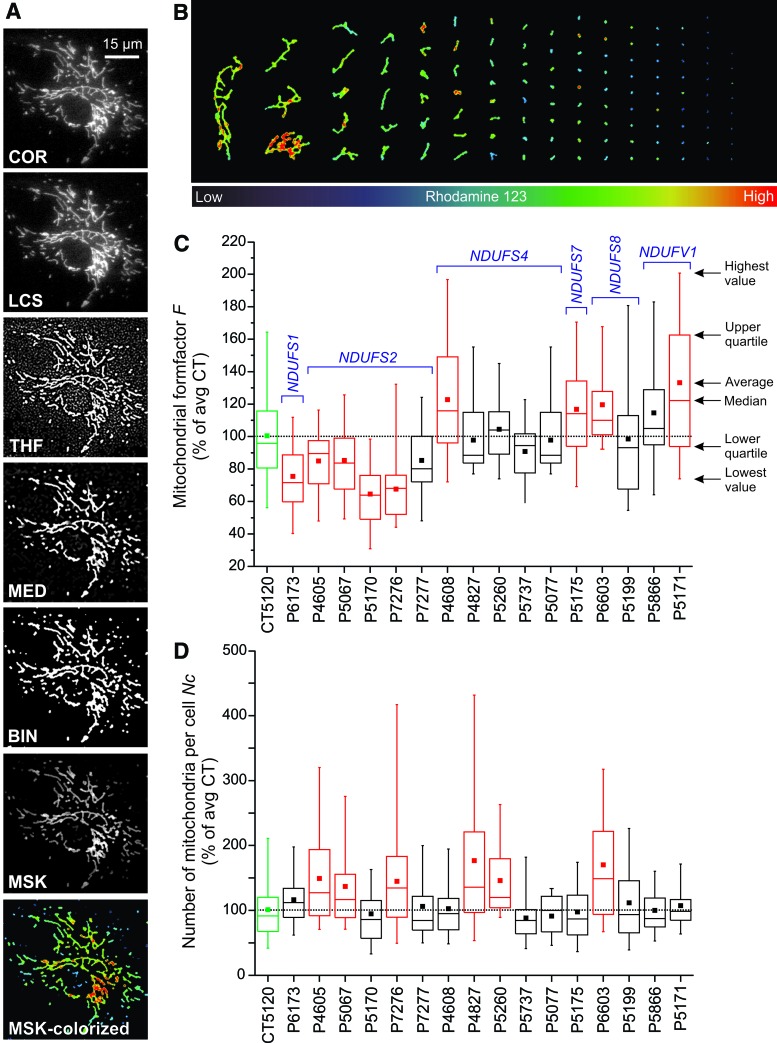
Mitochondrial morphology visualized by using live-cell fluorescence microscopy. (A) Left panel: visualization of mitochondrial structure in a primary fibroblast of a patient carrying a compound heterozygous mutation (R59X/T423M) in the gene encoding the NDUFV1 subunit of CI (left panel). Cells were stained with the mitochondria-accumulating cation rhodamine 123. Right panel: PHSFs (control CT5120) co-stained with rhodamine 123 and the membrane-staining reporter molecule FM1-43. (B) Visualization of mitochondrial structure in a PHSF (control CT5120) using DIC imaging (also known as Nomarski microscopy). Cells were cultured in the presence of the CI inhibitor rotenone (100 nM, 72 h). The white box shows a zoom-in of the indicated region. (C) Confocal image of a control PHSF stained with JC-1, which accumulates in the mitochondrial matrix according to the Δψ across the MIM. The right part of the image shows a zoom-in of the indicated region. Arrows mark regions of JC-1 aggregation (top arrow; red), suggesting a more negative Δψ, and inhomogeneous JC-1 signals across mitochondrial filaments (middle and lower arrow). (D) Microscopy image of a PHSF (control CT5120) stained with MitoTracker Green FM. This image was obtained by using video-rate microscopy and deconvolved off-line. Arrows mark “donut-shaped” mitochondria. (E) Collage of microscopy images visualizing a large mitochondrial network in rhodamine 123-stained PHSFs of a patient carrying a mutation (R228Q) in the gene encoding the NDUFS2 subunit of CI. These cells were imaged following immortalization and chromosome transfer. Magnifications (a, a', b, c, d) show mitochondrial morphologies throughout the cell. The magnified images were contrast-optimized for visualization purposes (an asterisk shows the cytosol). DIC, differential interference contrast; PHSFs, primary human skin fibroblasts. Color images are available online.
In vivo mouse studies revealed that mtDNA-devoid tumor cells acquired mtDNA from host cells, suggesting transfer of whole mitochondria and/or their content between cells (381). In agreement with this idea, transcellular exchange of individual mitochondria via nanotubular structures (“nanotunneling”) has been demonstrated under certain conditions (255, 338, 404). On the other hand, it appears that mtDNA can be transferred between cells via exchange of extracellular vesicles (343). Also, the MIM displays heterogeneity and dynamic behavior (Fig. 6) and, although a rare event, cristae can disappear or form within a matter of seconds (87). Fission and fusion of the MOM and MIM are executed by a core machinery of nine currently identified proteins (Fig. 7A) (51, 308, 337). Directly below, we discuss these fission and fusion mechanisms in more detail.
FIG. 6.
Mitochondrial heterogeneity and live-cell dynamics of mitochondrial internal structure. (A) Visualization of mtHSP70 antibody staining localized to mitochondria in PHSFs (control CT5120) by using fluorescence microscopy. (B) Magnification of a region of interest in (A) (dotted lines). The image was rotated 90 degrees clock-wise for visualization purposes. Arrows indicate inhomogeneous mitochondrial mtHSP70 staining. (C) First fluorescence microscopy image from a time-lapse recording of PHSFs (control CT5120) expressing a mitochondria-targeted fluorescent protein (COX8-EYFP). Arrows indicate inhomogeneous mitochondrial EYFP fluorescence signals. (D) Fluorescence microscopy images depicting a time-lapse recording from part of a primary fibroblast of a patient carrying a compound heterozygous mutation (R59X/T423M) in the gene encoding the NDUFV1 subunit of CI. This cell expressed COX8-EYFP and was visualized by using video-rate confocal microscopy (1 s interval; 30 images/s were averaged). Arrows indicate inhomogeneous mitochondrial EYFP fluorescence signals. (E) Magnification of a region of interest in (D) (dotted lines). The image was put in a horizontal position for visualization purposes. Dynamic changes in the mitochondrial EYFP fluorescence signal were observed. Color images are available online.
FIG. 7.
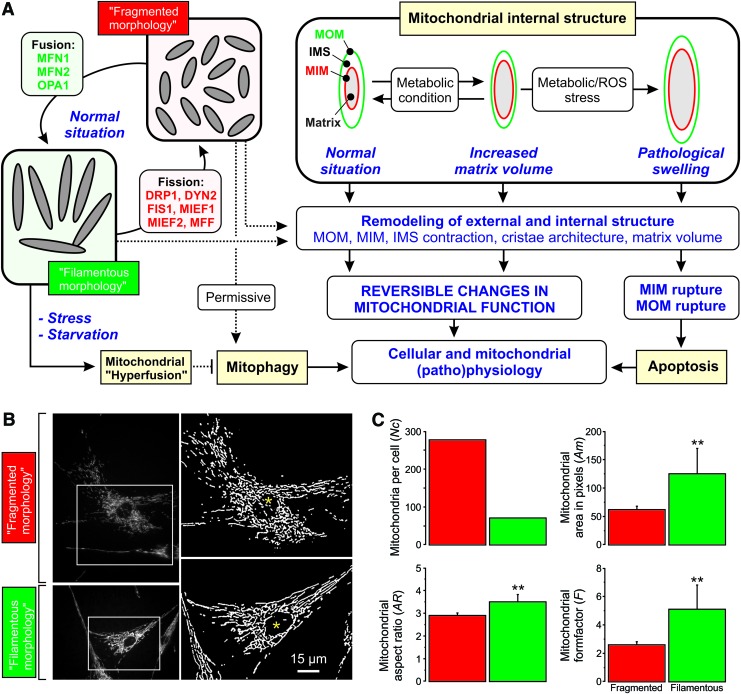
Key mechanisms that govern dynamic changes in mitochondrial internal/external structure and volume. (A) Mitochondrial fission and fusion is carried out by a set of nine proteins (MFN1, MFN2, OPA1, DRP1, DYN2, FIS1, MIEF1, MIEF2, and MFF). A fragmented mitochondrial morphology allows mitochondria to undergo mitophagy whereas the latter is prevented by mitochondrial elongation and hyperfusion (observed during stress and starvation conditions). In addition, mitochondrial matrix volume can increase as a function of metabolic demand and/or as a consequence of stress. Ultimately, the latter will lead to mitochondrial (ultra)structural remodeling and apoptosis induction. During (patho)physiological conditions (reversible) remodeling of mitochondrial internal and external structure is linked to (reversible) changes in mitochondrial function (see section IV for further details). (B) Typical examples of PHSFs (control CT5120) stained with TMRM and visualized by using fluorescence microscopy (left panels; contrast-optimized). Image processing was applied to obtain “binary” images depicting white mitochondrial objects on a black background (right panels, see Fig. 13A). The cells displayed a “fragmented” mitochondrial morphology (upper panels; cells treated with 12.5 μM of the glutathione-neosynthesis inhibitor L-Buthionine-sulfoximine during 24 h) and a “filamentous” mitochondrial morphology (lower panels; vehicle-treated cells). (C) Quantitative analysis of the mitochondrial morphology for the cells (marked with asterisks) in (B). Relative to the filamentous mitochondrial phenotype (green), the fragmented (L-Buthionine-sulfoximine-induced) phenotype 9 (red) was characterized by an increased number of mitochondria per cell (Nc) and reduced mitochondrial area (Am; a measure of mitochondrial size/volume; **p = 0.004), aspect ratio (AR; a measure of mitochondrial length; **p = 0.001), and formfactor (F; a combined measure of mitochondrial length and degree of branching; **p = 0.003). DRP1, dynamin-related protein 1; DYN2, dynamin 2; MFF, mitochondrial fission factor; MFN1/MFN2, mitofusin 1 and 2; MIEF1/MID51, mitochondrial elongation factor 1; MIEF2/MID49, mitochondrial elongation factor 2; OPA1, optic atrophy protein 1; TMRM, tetramethylrhodamine. Color images are available online.
A. Fission of the mitochondrial outer membrane
MOM fission is carried out by dynamin-related protein 1 (DRP1; Fig. 8), a cytosolic GTPase (314, 367). This protein also mediates peroxisome fission (see section II.F). The large majority of DRP1 protein appears to reside in the cytosol, from which it is recruited to the MOM by four currently unknown adaptor proteins (Fig. 8): (i) fission protein 1 (FIS1) (230, 300, 437), (ii) mitochondrial fission factor (MFF) (230, 299, 300), and (iii) mitochondrial elongation factor 1 (MIEF1/MID51) and mitochondrial elongation factor 2 (MIEF2/MID49) (229, 230, 300, 301, 331, 439, 445).
FIG. 8.
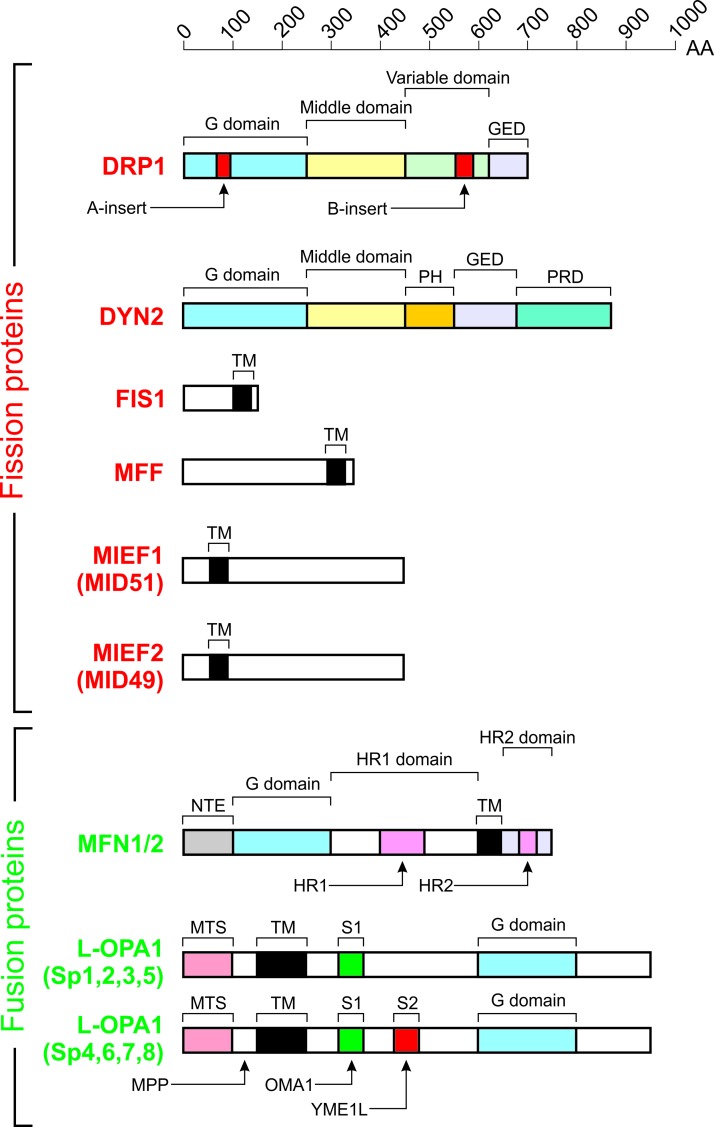
Core mitochondrial fission and fusion proteins. Domain structure of the nine proteins controlling mitochondrial fission (red) and fusion (green). Each protein is drawn to scale according to its length in AA. Further details are provided in the main text. This figure was compiled by integrating information from (206, 208, 238, 425, 449). G-domain, GTPase domain; GED, GTPase effector domain; MPP, matrix-soluble protein peptidase; HR1, heptad repeat domain 1; HR2, heptad repeat domain 2; MTS, mitochondrial targeting sequence; PH, pleckstrin-homology domain; PRD, proline-rich domain; S1, protease cleavage site 1 for OMA1; S2, protease cleavage site 2 for YME1L; Sp, splice variant; TM, transmembrane domain. Color images are available online.
In principle, mitochondrial fission is a stochastic process (200) during which DRP1 is recruited to the MOM. Once recruited, DRP1 monomers oligomerize into a contractile ring-like structure. On GTP hydrolysis, this structure decreases its diameter, thereby inducing mitochondrial fission (41, 112, 250). Quantitative analysis of DRP1 protein distribution in HeLa cells expressing GFP-tagged DRP1 suggests that cytosolic GFP-DRP1 predominantly exists in a tetrameric form and constitutes ∼50% of the total GFP-DRP1 pool (258). The latter study estimated that a functional DRP1 fission complex contains ∼100 DRP1 molecules. A population of DRP1 oligomers was detected on ER membranes, being distinct from mitochondrial or peroxisome-associated oligomers (162). On MFF knockdown, DRP1 oligomer levels at the ER, mitochondria, and peroxisomes were reduced, suggesting that DRP1 can use these three organelle membranes as oligomerization platforms.
ER tubules are involved in marking the site of mitochondrial division (111) and mtDNA replication (216). Mitochondrial ER-mediated constriction further involves inverted formin 2, an ER-associated actin modulator that induced actin polymerization at the ER-mitochondrial contact site (197). Additional actin-binding proteins (e.g. SPIRE1C; 211, 245) and lysosome-mitochondrial contacts also play a role in marking DRP1- and ER-positive sites of mitochondrial fission (428). Mechanistically, these contacts were promoted by active (GTP-bound) RAB7, a lysosomal GTPase that is a member of the RAS oncogene family. Interestingly, disconnection of lysosome-mitochondria contacts involved FIS1-mediated recruitment of TBC1D15 (TBC1 Domain Family Member 15). The latter is a GAP (GTPase-accelerating protein) of RAB7, which deactivates RAB7 by promoting its GTP hydrolyzing activity (428). In yeast, FIS1 was identified as a factor involved in the mitochondrial recruitment of DRP1 (DNM1) (202). However, it appears that FIS1 is not essential for mitochondrial fission in mammals (296, 299) but binds to MFF on the MOM, which, subsequently, attaches to a complex containing FIS1 and ER proteins at the mitochondria-ER interface (360). This study also provided evidence that FIS1 is not strictly required for mitochondrial fission but is involved in mitophagy. In this sense, inactivation of FIS1 gives rise to formation of LGG-1/LC3 aggregates (360).
Taken together, the current mechanistic insights suggest that MFF is the main DRP1 adaptor protein that, together with MIEF1/MID51 and MIEF2/MID49, controls DRP1 ring formation and constriction (169, 200). It was suggested that MIEF1/MID51 and MIEF2/MID49 also regulate the association of DRP1 with MFF in a trimeric DRP1-MIEF/MID-MFF complex (439). In addition, evidence was provided that MFF can act as a biomechanical membrane-bound force sensor (139). This would allow recruitment of the fission machinery to mechanically strained sites of the MOM. Recently, a new member of the mitochondrial division machinery, dynamin 2 (DYN2; Fig. 8), was discovered (210). DYN2 is ubiquitously expressed and acts together with DRP1 in sequential constriction, ultimately leading to a final DYN2-mediated step in mitochondrial fission (210).
B. Fusion of the MOM
Mitochondrial fusion requires sequential merging of the MOM and MIM of the two precursor mitochondria (243, 372). MOM fusion is mediated by the GTPases mitofusin 1 and 2 (MFN1/MFN2) (46, 212, 334, 344, 431). This process is well studied (100, 211) and is mediated by homotypic and heterotypic interactions between MFN1 and MFN2 on opposing mitochondria (56). These interactions are believed to involve (indirect) formation of disulfide bridges (249, 364). Both mitofusins (Fig. 8) contain a GTPase (G) domain, a coiled-coil domain (heptad repeat domain 1 or HR1), an MOM-spanning transmembrane domain (TM), and a second IMS-protruding coiled-coil domain (heptad repeat domain 2) (116). It was recently demonstrated that mitofusins are single MOM-spanning proteins with their N-terminus facing outward into the cytosol and their C-terminus protruding into the IMS (249). Analysis of the MFN1 crystal structure suggests that it forms homodimers during mitochondrial fusion by conformational changes that promote GTPase-domain (G-domain) dimerization (46, 321). This is in contrast with previous evidence suggesting that the C-terminal antiparallel coiled-coil domain of MFN1 plays a key role in tethering (105, 199). It was further suggested that MFN1 might use a two-step tethering mechanism consisting of a nucleotide-regulated dimerization of the G domains and subsequent interaction between the coiled-coil domains (46). Using in situ and in vitro fusion assays, it was demonstrated that interaction with the surface of the lipid bilayer induces folding of a conserved amphipathic helix in HR1 (75). This suggests a mechanism in which HR1 destabilizes the MOM lipid bilayer, especially in membrane regions displaying defects in lipid packing, to facilitate MOM fusion.
C. Mitochondrial inner membrane fusion
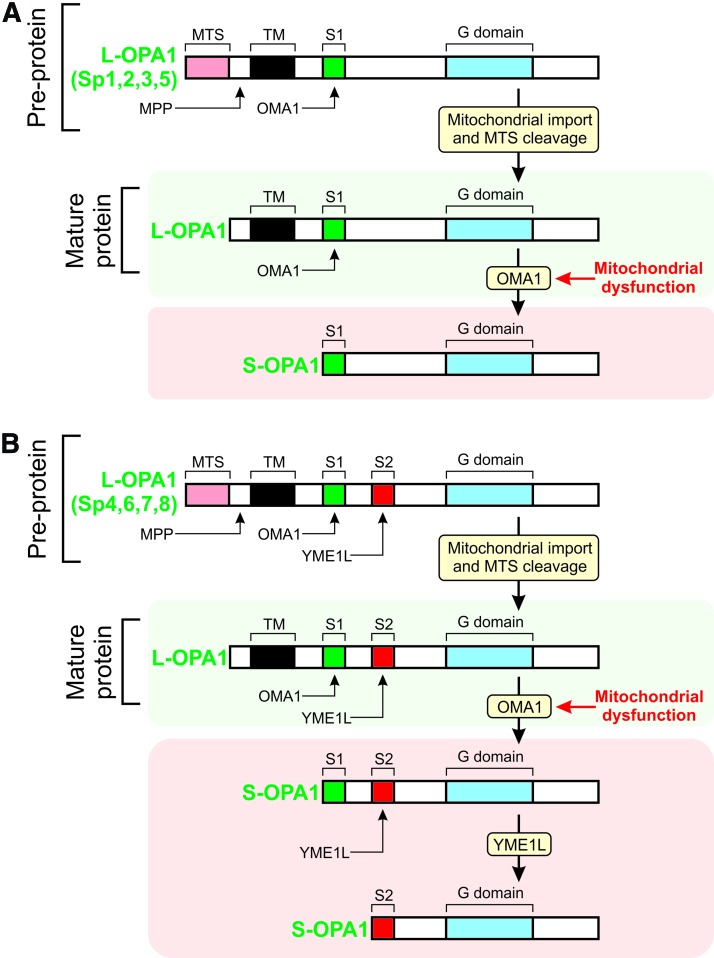
MIM fusion requires the action of the GTPase optic atrophy protein 1 (OPA1) (108, 128, 293, 371). GTP is supplied to OPA1 by nucleosidediphosphate kinases, which produce GTP through ATP-driven conversion of GDP (32). The OPA1 protein exists in eight different isoforms (splice variants, Sp1–Sp8) due to differential splicing (Fig. 8) (51). Each OPA1 isoform contains various domains including an N-terminal mitochondrial targeting sequence (MTS), a TM, and a protease cleavage site (S1). Four of the eight OPA1 isoforms also contain an additional (S2) protease cleavage site (51). After mitochondrial import of the OPA1 precursor, its MTS is cleaved off by the matrix-soluble protein peptidase, yielding an MIM-anchored “Long” OPA1 form (“L-OPA1”; Fig. 9). Cleavage of the OPA1 precursor at S1 or S2 yields “Short” OPA1 forms (“S-OPA1”). Proteolytic processing of OPA1 at S1 and S2 constitutes a major regulatory mechanism (371) that is carried out by a complex proteolysis network (51). Two mitochondrial proteases, OMA1 (“overlapping activity with m-AAA protease”) and YME1L (“Human yme1-like protein”), are responsible for L-OPA1 cleavage at S1 and S2, respectively (Fig. 9) (8, 51, 265, 323). Because both L-OPA1 and S-OPA1 forms need to be present in a proper ratio for MIM fusion, it appears that some OPA1 processing is always required.
FIG. 9.
Proteolytic processing of OPA1 isoforms. As a result of differential splicing, the OPA1 protein exists in eight different isoforms (Sp1-Sp8). (A) Four OPA1 splice variants (Sp1, Sp2, Sp3, and Sp5) contain a single protease cleavage site (S1). OPA1 is imported into the mitochondrion as a pre-protein, after which its targeting sequence (MTS) is cleaved off by the MPP. This yields a “long” OPA1 form (L-OPA1), which is MIM-anchored and processed by OMA1 on mitochondrial dysfunction. This yields a “short” OPA1 form (S-OPA1), which is not MIM-anchored. (B) Similar to panel A, but now for the four other OPA1 splice variants (Sp4, Sp6, Sp7, and Sp8). In addition to S1, these contain a second protease cleavage site (S2), which is processed by YME1L on mitochondrial dysfunction. Further details are provided in the main text. This figure was compiled by integrating information from (151, 238, 294, 371). Color images are available online.
D. Regulation and maintenance of cristae structure
1. Role of OPA1
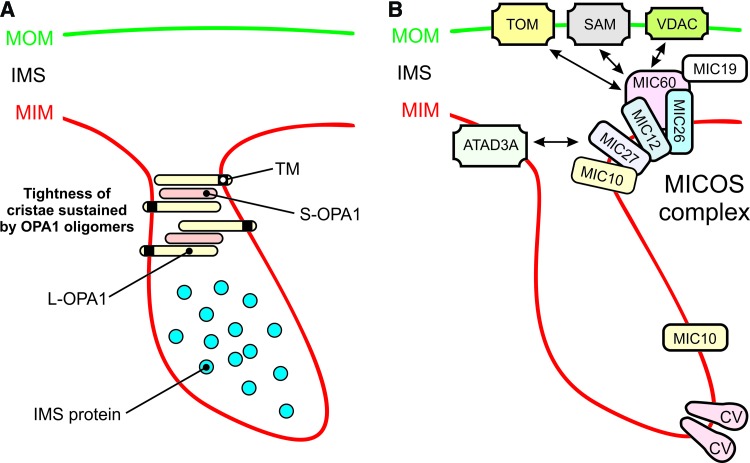
Experimental studies in HeLa cells revealed that OPA1 knockdown induced mitochondrial fragmentation, Δψ dissipation, and cristae disorganization, which were associated with cytochrome-c release and caspase-dependent apoptotic cell (AC) death (294). These observations are compatible with the observation that OPA1, independently of its role in MIM fusion and without interfering with activation of BAX and BAK, prevents mitochondrial cytochrome-c release, thereby protecting against apoptosis induction (108). Mechanistically, this is achieved by oligomerization of MIM-anchored L-OPA1 and soluble S-OPA1, which maintains tightness of the CJs during apoptosis (Fig. 10A).
FIG. 10.
Role of OPA1, the MICOS complex and CV in cristae shaping. (A) Tightening of cristae by OPA1 oligomers. See main text for details. (B) Structural shaping of the MIM by the MICOS complex and CV (arrow indicates potential interactions). Additional MICOS complex subunits (MICs) and further details are presented in the main text and Supplementary Table. This figure was compiled by integrating information from (238, 308, 327, 427). ATAD3A, MIM ATPase; CV, OXPHOS (oxidative phosphorylation) complex V; L-OPA1, long form of OPA1; MIC10, MICOS complex subunit 10; MIC12, MICOS complex subunit 12; MIC19, MICOS complex subunit 19 (or CHCHD3); MIC26, MICOS complex subunit 26; MIC27, MICOS complex subunit 27; MIC60, MICOS complex subunit 60 (or Mitofilin, IMMT); MICOS, mitochondrial contact site and cristae organization; S-OPA1, short form of OPA1; SAM, protein sorting and assembly machinery; TOM, protein translocase of the MOM. Color images are available online.
The Presenilin-associated rhomboid-like (PARL) protease is involved in this OPA1-dependent protection, since Parl−/− mice displayed reduced levels of S-OPA1 and PARL−/− mitochondria exhibited accelerated cristae remodeling and cytochrome-c release during apoptosis (65). Knockdown of OPA1 in HeLa cells specifically reduced CM dynamics, whereas the IBM remained flexible (124). Recently, an elegant mechanistic framework was presented that integrates these earlier observations with the existence of OPA1 isoforms and OMA1/YME1L-mediated OPA1 processing (79–81). In this model, all eight OPA1 isoforms (Fig. 9) are proposed to be involved in organization of OXPHOS supercomplexes, cristae shaping, and mtDNA maintenance. Independent of mitochondrial morphology, L-OPA1 and S-OPA1 isoforms support mitochondrial fusion and preserve mitochondrial bioenergetics, respectively. In addition to OMA1, YME1L, and PARL, various other biomolecules interact with or affect OPA1 function to regulate or maintain MIM structure. These include MFN1, MFN2, BAX/BAK, Paraplegin (SPG7), prohibitin 2 (PHB2), and mtDNA polymerase gamma. Detailed information on these interactions and their functional impact is provided in the Supplementary Table and elsewhere (67, 74, 447).
2. Role of mitochondrial contact site and cristae organization
In addition to OPA1, another key player in mitochondrial ultrastructural maintenance is the mitochondrial contact site and cristae organization (MICOS) system (Fig. 10B). Currently, this system is best understood in yeast (327, 398, 427). The MICOS complex impacts metabolite handling, cellular signaling, mitochondrial membrane architecture, and mitochondrial protein import. This coupling is mediated by MICOS interactions with: (i) the MOM protein Porin/VDAC (voltage-dependent anion channel; interaction with the MICOS subunit MIC60), (ii) the protein sorting and assembly machinery (SAM; MIC60 interaction), (iii) the protein translocase of the MOM (TOM; MIC60 interaction), (iv) the presequence translocase of the MIM (TIM23), (v) mtDNA, (vi) the ETC, and (vii) CV (352). Recent evidence suggests that MICOS also interacts with the MIM ATPase ATAD3A, and that the latter protein assists in stabilizing mitochondrial cristae, potentially via interactions with the MOM (307). Subunit MIC10 plays a central role in the yeast MICOS system (31), and MIC10 oligomerization induces MOM bending at CJs (17). Electron tomography analysis revealed that MIC19 and MIC60 were enriched inside cristae and localized at CJs (346). Moreover, the latter study provided evidence that MIC19 interacted with subunit IV of CIV. Various interactions between MICOS subunits and mitochondrial fission/fusion proteins have been reported (Supplementary Table). For instance, the core MICOS protein MIC60 binds to OPA1 and together they control the number and stability of CJs (119).
3. Role of CV
Another important determinant of mitochondrial ultrastructure is CV of the OXPHOS system, the dimerization of which induces MIM curvature (Fig. 10B) (133). Dissociation of CV dimers into monomers was paralleled by inversion of MOM curvature at the CV site, MOM vesiculation, and cristae structural aberrations during aging in the model organism Podospora anserina (76). Evidence was provided that CV dimerization is mediated by interactions between its Fo and F1 domains and promotes cristae formation (262). Alternatively, it was proposed that CV dimerization is mediated by connecting its two F1 moieties by a protein bridge formed by ATPase inhibitory factor 1 (IF1) (262). Compatible with this connecting function, IF1 overexpression and knockdown increased and decreased mitochondrial volume and mitochondrial cristae density, respectively (44, 45). IF1 limits mitochondrial ATP consumption when mitochondrial respiration is impaired by inhibiting CV reverse-mode action (44). Moreover, IF1 inhibits OMA1-mediated processing of OPA1, thereby impeding cristae remodeling during apoptosis (101). Evidence in yeast suggests that the extent of CV dimerization and/or clustering depends on the cellular demand for OXPHOS-generated ATP (164).
4. Role of MitoNEET
Interestingly, adjacent mitochondria display a coordinated cristae architecture at inter-mitochondrial junctions (IMJs), where cristae form parallel arrays that are perpendicular to the IMJ, suggesting a role in electrochemical coupling (311). This study also provided evidence that MFNs are not required for IMJ formation and that IMJs are dynamic structures, the amount and function of which are dynamically regulated by the bioenergetic state of the mitochondrion. It was recently demonstrated that knockdown of MitoNEET (CDGSH iron-sulfur domain-containing protein 1 or CISD1), a dimeric outer membrane protein, reduced IMJ frequency (402). This suggests that MitoNEET plays a role in the regulation of IMJ formation.
E. Role of “non-core” proteins in mitochondrial fission, fusion, and ultrastructure
In addition to the “well-studied” mechanisms described earlier, there is a large body of empirical evidence in higher eukaryotes reporting additional proteins and pathways that affect mitochondrial (ultra)structure. These effects are mediated not only by these proteins interacting with core fission/fusion proteins (Supplementary Table) but also by other mechanisms. For instance, the mitochondrial protein TMEM11 (transmembrane protein 11) regulated mitochondrial morphology by a mechanism that was independent of DRP1/MFN (332). TMEM11 is the human orthologue of Drosophila PM1 (pantagruelian mitochondrion 1), which controls cristae biogenesis and mitochondrial diameter in flies (236). Also, TMEM135 (transmembrane protein 135) localized to mitochondria and its overexpression and/or mutation disturbed mitochondrial dynamics (206).
Overexpression and knockdown of optic atrophy 3 (OPA3), an integral MOM protein of which the gene is mutated in hereditary optic neuropathies, induced mitochondrial fragmentation and elongation, respectively (340). OPA3-overexpressing cells did not display spontaneous AC death but were sensitized to apoptosis induction by staurosporine- and tumor necrosis factor-related apoptosis-inducing ligand. This suggests that OPA1 and OPA3 exert different functions in apoptosis. OPA3 gene mutations were associated with mitochondrial fragmentation in patient fibroblasts (125). Mitochondrial fragmentation and spontaneous apoptosis were also induced after overexpression of a pathological OPA3 mutant (G93S), suggesting that OPA3 plays a role in mitochondrial fission and links this process to optic atrophy (340).
Another protein involved in mitochondrial morphology is mitochondrial fission process protein 1 (MTFP1). Knockdown of MTFP1 inhibited DRP1-mediated mitochondrial fission, leading to mitochondrial hyperfusion (386, 387, 407). Translation of MTFP1, and thereby the mitochondrial recruitment of DRP1, is regulated by the nutrient-sensing complex mechanistic/mammalian target of rapamycin complex 1 (270). This mechanism provides a link between environmental/intracellular stimuli and mitochondrial morphofunction. With respect to calcium homeostasis, mitochondria can undergo “mitochondrial shape transition” (MiST), which is distinct from mitochondrial fission/swelling and mediated by the mitochondrial Rho GTPase 1 (MIRO1) (280). MiST was induced by increased calcium levels in the cytosol, but not by mitochondrial calcium uptake by the mitochondrial calcium uniporter (MCU). It was further demonstrated that autophagy/mitophagy induction requires MIRO1-dependent MiST.
F. Secondary functions of mitochondrial fission and fusion proteins
It is often overlooked that mitochondrial fission/fusion proteins also exert important “secondary” functions in cell physiology (412). For instance, the Mfn2 gene is also known as Hsg (hyperplasia suppressor gene) and is involved in the control of cell proliferation and apoptosis. The MFN2 protein exerts its antiproliferative effect by inhibiting the RAS-RAF-ERK signaling cascade by N-terminally interacting with RAF-1 and C-terminally with RAF (55). In brown adipose tissue (BAT), MFN2 was involved in the docking of mitochondria to LDs, allowing efficient FA transfer to mitochondria for β-oxidation (37). In mice, MFN2 in BAT was required for cold-induced thermogenesis and promoted insulin resistance in obese animals (240). On dysfunctional mitochondria, MFN2 can also act as a Parkin-receptor, an E3 ubiquitin ligase implicated in Parkinson's disease (57). Mechanistically, it was proposed that mitochondrial Δψ depolarization leads to stabilization of PTEN-induced putative kinase 1 (PINK1) at the MOM. After this stabilization, PINK1 phosphorylates MFN2, thereby increasing the binding of Parkin to stimulate mitophagy. In this way, absence of MFN2 interrupted the PINK1-Parkin mitochondrial quality control mechanism, leading to accumulation of dysfunctional mitochondria (57). In addition to the pathway just described, there is considerable additional crosstalk between mitochondrial fusion and Parkin-mediated mitophagy (92, 382). Evidence in adipocytes suggests that OPA1 also can reside outside mitochondria on LDs to act as an A-kinase anchoring protein (313). It was proposed that OPA1 is involved in lipolysis of neutral lipids stored in the LDs. However, this mechanism is still controversial (23).
Other “secondary” functions of mitochondrial fission/fusion proteins include (Table 1): (i) execution of peroxisome fission (DRP1, FIS1, MFF) (181, 353, 354), (ii) MOM protein distribution (MFNs) (418), (iii) ER morphology regulation and ER-mitochondria interactions (MFN2) (77, 102), (iv) the unfolded protein response (MFN2) (272), (v) apoptosis (DRP1, FIS1, OPA1) (108, 378, 416), (vi) metabolic regulation (MFN1, MFN2) (86, 312), (vii) cell cycle and division (hFIS1, OPA1, MFN1) (208), (viii) mPTP opening (hFIS1, DRP1) (184), (ix) endocytosis (DRP1) (217), (x) calcium homeostasis (MFN2, DRP1) (54, 77, 102, 378), (xi) antiviral signaling (MFN2) (433), (xii) melanosome biogenesis (MFN2) (73), and (xiii) cell senescence (hFIS1, OPA1) (207). This multi-functionality is possibly responsible for apparent discrepancies in experimental results between various studies. For example, stimulation of mitochondrial fusion in Drosophila dramatically reduced ROS levels (276), whereas stimulation of mitochondrial fission suppressed oxidative damage in murine postmitotic neurons (168). This means that similar alterations in mitochondrial morphology (e.g., fragmentation or filamentation; Fig. 7B, C) can serve a different purpose depending on the cell type, upstream signaling pathway, metabolic state, and physiological context. Obviously, the “secondary” functions of mitochondrial fission and fusion proteins need to be taken into account for proper interpretation of experimental results.
Table 1.
Other Selected Functions of Core Mitochondrial Fusion and Fission Proteins
| Secondary function | Refs |
|---|---|
| DRP1 | |
| Peroxisome fission. | (354) |
| Binds to the ER and regulates ER morphology. | (314, 438) |
| Stimulates tBID-induced BAX oligomerization and cytochrome-c release by promoting tethering and hemifusion of membranes in vitro. | (269) |
| Localizes to the Golgi complex in some cell lines, potentially acting as a component of the apical sorting machinery at the trans-Golgi network. | (33) |
| During synaptic stimulation, BCL-XL translocates to clathrin-coated pits in a calmodulin-dependent manner and forms a complex with DRP1, MFF, and clathrin. | (217) |
| The DRP1-ABCD isoform is located at lysosomes and enriched at the inter-organelle interface between mitochondria and lysosomes/late endosomes. This enrichment depends on lysosomal pH. | (153) |
| FIS1 | |
| Peroxisomal adaptor for DRP1 | (183) |
| Interacts with BAP31 at the ER membrane to create a platform for activation of procaspase-8 during apoptosis. | (154) |
| MFF | |
| Peroxisomal adaptor for DRP1 | (114) |
| During synaptic stimulation, BCL-XL translocates to clathrin-coated pits in a calmodulin-dependent manner and forms a complex with DRP1, MFF, and clathrin. | (217) |
| MFN1 | |
| ER-MOM tethering. MFN2 on the ER engages heterotypic complexes with MFN1 on the surface of mitochondria. | (77) |
| MFN2 | |
| ER-MOM tethering. MFN2 on the ER engages in homotypic and heterotypic complexes with MFN1 or MFN2 on the surface of mitochondria. MFN2 knockdown increased Ca2+ transfer from the ER to mitochondria. | (14, 77, 203) |
| Suppresses vascular smooth muscle cell proliferation independent of its role in mitochondrial fusion. This growth suppression is inhibited by PKA-mediated phosphorylation of the Ser442 residue of MFN2. | (446) |
| Enhances mitochondrial metabolism. | (312) |
| MFN2 but not MFN1 is an ER stress-inducible protein that is required for the proper temporal sequence of the ER stress response. | (282) |
| MFN2 physically interacts with and is an upstream modulator of PERK, a protein involved in the response to ER stress stimuli by triggering the UPR. | (272) |
| In the heart, MFN2 serves as an adaptor protein to mediate fusion of autophagosomes and lysosomes. | (444) |
| Involved in trafficking of STIM1 to the ER-PM junction and subsequent activation of CRAC channel activity after Δψ depolarization. | (366) |
| Interacts with MIRO and MILTON to allow transport of axonal mitochondria. | (263) |
| MFN2 overexpression induces apoptosis in cardiac myocytes. | (361) |
| MFN2 exerts antiproliferative effects. | (55) |
| In BAT, MFN2 mediates the docking of mitochondria to LDs, allowing an efficient fatty acid transfer to mitochondria for β-oxidation. | (37) |
| MFN2 acts as a Parkin receptor on dysfunctional mitochondria. | (57) |
| MFN2 negatively regulates NFAT in the maintenance of HSCs with extensive lymphoid potential. In these cells, MFN2 increases ER-mitochondria tethering and therefore the cytosolic Ca2+-buffering capacity to inhibit NFAT activity. | (232) |
| MFN2 is important in maintaining coenzyme Q levels and might play a role in mevalonate synthesis. | (271) |
| OPA1 | |
| Organizes a supramolecular complex containing both PKA and Perilipin (PLIN1). OPA1 targeting of PKA to LDs is necessary for hormonal control of perilipin phosphorylation and lipolysis. | (313) |
BAP31, B cell receptor associated protein 31; BAT, brown adipose tissue; CRAC, calcium release-activated calcium channel; DRP1, dynamin-related protein 1; ER, endoplasmic reticulum; FIS1, fission protein 1; HSC, hematopoietic stem cell; LD, lipid droplet; MFF, mitochondrial fission factor; MFN1/MFN2, mitofusin 1 and 2; MIRO, mitochondrial RHO GTPase; MOM, mitochondrial outer membrane; NFAT, nuclear factor of activated T cells; OPA1, optic atrophy protein 1; PERK, protein kinase RNA (PKR)-like ER kinase; PKA, protein kinase A; PM, plasma membrane; STIM1, stromal interaction molecule 1; UPR, unfolded protein response.
III. Regulation of Mitochondrial (Ultra)Structure
The regulation of mitochondrial morphology is a subject of intense study. This control is primarily exerted by modifying the function of mitochondrial fission and fusion proteins at various levels. This includes microRNAs (Supplementary Table), which act as negative regulators of gene expression by inhibiting mRNA translation or promoting mRNA degradation (205). Here, we focus on the role of post-translational modifications (PTMs) and mitochondrial lipids.
A. Regulation by post-translationlal modifications of fission and fusion proteins
Mitochondrial fission and fusion proteins contain various (predicted) PTM sites, allowing their phosphorylation, S-nitrosilation, sumoylation, ubiquitination, tyrosine sulfation, acetylation, and S-palmitoylation (5, 70, 279, 425). Depending on the nature and location of the PTM, either mitochondrial fission or fusion is stimulated (51, 103, 289). These mechanisms are explained in more detail below (sections III.A.1–12).
1. Stimulating fission by DRP1 phosphorylation at S616 (human isoform 1)
S616 corresponds to S579 in rats and mice. In mitotic cells and neurons, DRP1 phosphorylation at S616 is stimulated by cyclin-dependent kinases (CDKs) (63, 375, 380) and protein kinase Cδ (319). This phosphorylation stimulates DRP1 activity and mitochondrial fission. To the best of our knowledge, no phosphatases are currently described for this phosphorylation site. Mammalian DRP1 genes include three alternative exons and evidence was provided that alternative splicing generates a cytoskeletal DRP1 “reserve pool,” which translocates to the MOM on CDK-induced phosphorylation (375). This means that alternative splicing and phosphorylation might function as an isoform-specific mechanism that (co)controls DRP1 subcellular localization and mitochondrial fission. Remarkably, S616 phosphorylation by CDK5 inhibited DRP1, thereby contributing to mitochondrial elongation during neuronal maturation under physiological conditions (63). It was proposed that CDK5 reduces the oligomerization activity of DRP1.
2. Stimulating fission by DRP1 phosphorylation at S585
Analysis of a mouse model revealed that neuronal injury induced by N-methyl-D-aspartate was associated with DRP1 phosphorylation by CDK5 at S585 and mitochondrial fission (156).
3. Stimulating fission by DRP1 SUMOylation
SUMO (small ubiquitin-like modifier) proteins are attached to lysine residues of other proteins by the combined action of an E1 complex, a single E2, ubiquitin carrier protein 9, and several E3 enzymes (424). SUMOylation of DRP1 potentially stimulates its oligomerization at the MOM, leading to increased mitochondrial fission (416).
4. Stimulating fission by DRP1 glycosylation
In mouse cardiomyocytes, pharmacological inhibition of N-acetyl-glucosaminidase (OGA), which removes O-GlcNAc residues from Thr and Ser residues, resulted in increased O-linked-N-acetyl-glucosamine glycosylation of DRP1 at T585 and T586 (and possibly also S616 and S637) (115). Increased DRP1 glycosylation enhances GTP binding and MOM recruitment of this protein, paralleled by mitochondrial fragmentation and Δψ depolarization. Evidence was provided that increased O-GlcNAcylation decreases the phosphorylation of DRP1 at S637, compatible with stimulation of mitochondrial fission.
5. Stimulating fission by MFF phosphorylation at S155 and S172
MFF phosphorylation at S155 and S172 is mediated by AMP-activated protein kinase A (AMPK). This phosporylation stimulates mitochondrial fission by promoting DRP1 recruitment to the MOM (388).
6. Stimulating fission by MFN1 phosphorylation
ERK2 or MAPK1-mediated MFN1 phosphorylation inhibits MFN1 oligomerization, thereby stimulating mitochondrial fission (318).
7. Stimulating fission by MFN1 acetylation
MFN1 acetylation triggers its degradation via ubiquitination mediated by membrane-associated ring finger (C3HC4) 5 (MARCH5 or MITOL), thereby stimulating mitochondrial fission (304).
8. Stimulating fission by MFN2 phosphorylation at S27
During stress conditions (e.g., treatment with doxorubicin, cis-platinum, or tunicamycin), MFN2 is phosphorylated at S27 by c-JUN N-terminal kinase (JNK), which leads to recruitment of the ubiquitin ligase (E3) HUWE1/MULR/ARF-BP1/HECTH9/E3Histone/LASU1 to MFN2 (204). As a consequence, MFN2 is ubiquitinated and subsequently degraded, leading to mitochondrial fragmentation and enhanced AC death.
9. Stimulating fusion by DRP1 ubiquitination
Poly-ubiquitination of DRP1 leads to its degradation and is promoted by MARCH5 (278), the cytosolic E3 ubiquitin ligase Parkin (413), and anaphase-promoting complex/cyclosome and its coactivator CDH1 (APC/CCDH1) E3 ubiquitin ligase complex (144). MARCH5 is an integral MOM protein that interacts with ubiquitinated DRP1 and its overexpression stimulates mitochondrial filamentation (278). Parkin interacts with and ubiquitinates DRP1 to stimulate its proteasome-dependent degradation (413). During cell division, changes in mitochondrial morphology involve APC/CCDH1-driven DRP1 ubiquitination and its subsequent proteasomal degradation. Inhibition of this process prevents normal G1 phase regrowth of mitochondrial networks after cell division (144).
10. Stimulating fusion by DRP1 phosphorylation at S637 (human isoform 1)
S637 corresponds to S656 (in rat) and S600 (in mouse). This phosphorylation is stimulated by: (i) the proto-oncogene PIM1 (88), (ii) protein kinase A/A kinase anchor protein 1 (PKA/AKAP1) (52, 71), (iii) the rho-associated coiled coil-containing protein kinase 1 (ROCK1) (415), and (iv) calcium/calmodulin-dependent kinase 1α (CAMK1α) (136). The functional role of S637 phosphorylation is somewhat enigmatic. Under conditions of simulated ischemia, PIM1-induced S637 phosphorylation preserved mitochondrial filamentation by preventing DRP1 translocation to the MOM (88). Compatible with this result, PKA-induced phosphorylation of S637 decreased DRP1 GTPase activity (52, 71). In contrast, ROCK1-induced S637 phosphorylation stimulated DRP1 translocation to the MOM in a hyperglycemic mouse model (415). Similarly, Ca2+ influx through voltage-operated calcium channels blocked mitochondrial movement and stimulated CAMK1α-mediated S637 phosphorylation, associated with increased DRP1 recruitment to the MOM and mitochondrial fission in neurons (136). It was proposed that the functional consequences of S637 phosphorylation might be highly cell type and/or stimulus dependent (415).
Dephosphorylation of S637 involves: (i) phosphoglycerate mutase family member 5 (171), (ii) protein phosphatase 2A/Bβ2 (PP2A/Bβ2) (82), and (iii) calcineurin (CaN) (71). For these phosphatases, S637 dephosphorylation was associated with a stimulation of mitochondrial fission. Translocation of PPA2/Bβ2 to the MOM is regulated by its N-terminal phosphorylation (257). The Bβ2 regulatory subunit targets the scaffolding (A) and catalytic (C) subunits of PP2A to the MOM via transient association with receptor components of the TOM complex (426). In addition to PKA, AKAP1 also recruits other signaling enzymes to the MOM, including protein phosphatase 1 and CaN (426). Inducers of Δψ depolarization trigger Ca2+/CaN-dependent S637 dephosphorylation, which stimulates DRP1 translocation to the MOM and mitochondrial fission (49). Interestingly, it was recently demonstrated in mouse cells and brain lysates that the levels of S637-phosphorylated DRP1 displayed a 24-h rhythm. This was paralleled by rhythmic stimulation of mitochondrial fusion and circadian ATP production (349). This suggests that oxidative metabolism is regulated by the circadian rhythm in non-dividing cells mediated by cyclic DRP1 phosphorylation and inactivation.
11. Redox-dependent PTMs
Especially during pathological conditions, mitochondria are considered significant intracellular sources of ROS, which can not only potentially damage biomolecules but also play crucial signaling roles (188, 241, 357). Also, mitochondrial fission and fusion proteins constitute targets of various redox pathways, thereby allowing regulation of mitochondrial morphology (425): (i) nitration of Paraplegin and PKA/AKAP1 (potentially affecting DRP1/MFN2 activity and OPA1 processing); (ii) S-nitrosylation of DRP1 at C644 (potentially increasing its activity), OPA1 (potentially affecting its activity), Caspase 3 (inhibiting OPA1 processing), CDK5 (inhibiting DRP1 activity), Parkin (stimulating DRP1 and MFN2 activity), and JNK (stimulating MFN2 activity); (iii) disulfide bond alterations in amyloid-β (stimulating DRP1 activity), PKA/AKAP1 (inhibiting DRP1 activity), TIMM8A (potentially affecting DRP1 activity), and ROMO1 (stimulating OPA1 cleavage); and (iv) ROS-induced Δψ depolarization stimulating hydroxyl radical production, lipid peroxidation, protein carbonylation, and OPA1 cleavage.
Disulfide bond-mediated crosslinking between MFNs in a glutathione and GTP-dependent manner might contribute to mitochondrial hyperfusion that protects against oxidative stress (364). This protection might be exerted by sharing ROS-induced damage and/or mitochondrial antioxidants between individual mitochondria by stimulating fusion between “damaged” and “healthy” organelles (281). Recently, it was demonstrated that protein disulfide isomerase A1 (PDIA1) can function as a DRP1 thiol reductase (176). PDIA1 depletion stimulates DRP1 sulfenylation, thereby stimulating mitochondrial fragmentation.
12. Other PTMs
In case of MFN2, evidence was provided that its Parkin-mediated ubiquitination depends on phosphorylation of MFN2 residues T111/S442 by the mitochondrial kinase PINK1 (57). The same study demonstrated that MFN2 ablation prevents Parkin translocation to the MOM and suppresses mitophagy in mouse cardiac myocytes during Δψ depolarization. S442 can also be phosphorylated by PKA but this appears to play a role in MFN2-mediated suppression of vascular smooth muscle cell growth independent of mitochondrial morphology (446). MARCH5 binds and ubiquitinates mitochondrial MFN2 but not ER-associated MFN2, thereby regulating ER tethering to mitochondria by activating MFN2 via K192 ubiquitination (377). Polyubiquitinated MFN2 is then activated, leading to an increase in GTP-binding ability and is associated with the ER-localized MFN2, resulting in the oligomerization of MFN2 and attachment of mitochondria to the ER (277).
Evidence in yeast demonstrated the existence of two independent pathways for activation and degradation of MFNs to control MOM fusion (9, 423). In this mechanism, MOM fusion is activated by addition of stabilizing ubiquitin chains to MFN by an E3 ubiquitin ligase. Removal of these chains by a deubiquitylase (UBP12) then impairs MOM fusion and promotes mitochondrial fragmentation. In contrast, another E3 ligase attaches destabilizing ubiquitin chains to MFN, which are removed by a different deubiquitylase (UBP2) to support outer membrane fusion.
B. Role of mitochondrial lipids in mitochondrial (ultra)structure
Mitochondria display a distinct lipid composition when compared with other cell constituents (ER, PM, Golgi, and late endosomes) (399). More specifically, the MOM and MIM display a high content of non-bilayer phospholipids such as cardiolipin (CL) and phosphatidylethanolamine (PE) (384). Lipidomics analysis of mouse liver mitochondria revealed the following lipid composition: 25% PC (phosphatidylcholine), 18% CL, 14% PE, 10% phosphatidylglycerol, 10% phosphatidylinositol, 6% phosphatidylserine, 5% lysophosphatidylethanolamine, 5% sphingomyelin, 3% CER (ceramide), 3% lysocardiolipin, and 2% PA (phosphatidic acid) (13). In yeast, the MIM displayed a higher CL content than the MOM, whereas CL was virtually absent from the ER membrane (384). Mitochondrial membrane lipids are delivered by the ER or synthesized within mitochondria from ER-derived precursor lipids (347, 384). Inside the mitochondrion, phospholipids can be transported between the MIM and MOM and also be exchanged with the ER membrane. For instance, the lipid transfer protein STARD7 shuttles PC from the MOM to the MIM (342).
Various lipid-mediated signaling mechanisms are involved in the regulation of mitochondrial function. Examples include (254, 287): (i) mitophagy, where CL translocates from the MIM to the MOM to bind LC3 and LC3-B-II in collaboration with CER to induce autophagosomal mitochondrial recruitment; (ii) ETC assembly, where the chaperone PHB2 involved in CIV assembly interacts with sphingosine-1-phosphate (S1P); and (iii) apoptosis, where CER accumulates and acts together with Bax to induce pore formation, allowing cytochrome-c exit and apoptosis activation.
With respect to mitochondrial function, CL is the best studied mitochondrial lipid. CL has a dimeric structure, consists of four acyl chains and two phosphate groups, and plays a role in mitochondrial cristae organization. The latter is probably linked to the observation that the curvature of the MIM induces an asymmetric distribution of CL and PE (these segregate into the negatively curved monolayer leaflet facing the crista lumen) versus PC (this segregates into the positively curved monolayer leaflet facing the mitochondrial matrix (149). On CL incorporation, membrane bilayer deformability increases and CL becomes enriched in regions with a highly negative curvature (38).
Interestingly, specific proteins have been described that sense membrane curvature (membrane curvature sensing or MCS proteins) via their amphipathic helices (AHs) and Bin/Amphiphysin/Rcs BAR domains (239). Mechanistically, it was proposed that AHs detect and fill up lipid packing defects by inserting their hydrophobic part whereas BAR domains sense membrane curvature via electrostatic interactions (239). It is tempting to speculate that (CL-induced) changes in MIM curvature affect the stability and/or activity of integral membrane proteins (IMPs). Such an IMP-stabilizing effect might also be linked to the established role of CL in mitochondrial protein transport, MCU-mediated mitochondrial calcium uptake, and ETC functioning (35, 93). Indeed, CL-induced IMP stabilization has been described for individual ETC complexes in bovine and yeast as well as ETC supercomplexes and uncoupling protein 1 (274). Studying isolated CI revealed that its catalytic activity strongly depends on the phospholipid content of the preparation (359). CL was bound to CI, possibly playing a stabilizing role, and CI catalytic activity was influenced by binding of PE and PC to the complex (359).
Mitochondrial lipids have also been implicated in the regulation of mitochondrial (ultra)structure (131). CL and PA coordinate mitochondrial dynamics by interacting with DRP1, MFN, and OPA1 (16, 170). Using cryo-EM analysis of DRP1 on nanotubes with a distinct lipid composition, evidence was provided that DRP1-CL interactions activate DRP1 oligomers (106). These authors further suggested that MIM-to-MOM translocation of CL enhances mitochondrial fission. PA inhibits DRP1 activity (2, 174, 405). A mitochondrial phospholipase D (mitoPLD) was detected on the MOM to promote transmitochondrial membrane adherence by a mechanism that was MFN-dependent and involved CL hydrolysis to form PA (64). It was suggested that PA could play the following roles during mitochondrial fusion: (i) to stimulate opposing membrane fusion by generating negative membrane curvature, (ii) to recruit additional biomolecules involved in fusion, and (iii) to be converted in another fusogenic lipid. A later study suggested that a PA-preferring phospholipase A1 (PA-PLA1) plays a regulatory role in mitochondrial morphology regulation since its overexpression and knockdown induced mitochondrial fragmentation and elongation, respectively (14). In this mechanism, increased PA-PLA1 activity (which leads to lower PA levels to inhibit mitochondrial fusion) counterbalances mitoPLD activity (which increases PA levels to stimulate mitochondrial fusion).
Evidence in a fly model and mammalian cells revealed that loss of mitoguardin (MIGA) induces mitochondrial fragmentation via its interaction with mitoPLD (443). This study provided evidence that MIGA functions downstream of MFN by stabilizing mitoPLD and stimulating MFN dimerization. Also, PE appears to play a role in this process since a moderate reduction (<30%) in mitochondrial PE levels induced mitochondrial fragmentation, impairment of mitochondrial function and reduced cell growth (383). Similarly, a role for lysophosphatidic acid (LPA) was proposed in mitochondrial fusion (292). This study revealed that mitochondrial GPAT (glycerol-3-phosphate acyltransferase; mitoGPAT) catalyzes the initial and rate-limiting step in LPA synthesis. Analyses in Caenorhabditis elegans and mammalian cells demonstrated that mitoGPAT depletion induced mitochondrial fragmentation, which was rescued by LPA. This suggests that mitoGPAT-mediated LPA-formation plays a role in mitochondrial fusion (292).
IV. The Concept of Mitochondrial Morphofunction
Changes in mitochondrial (ultra)structure and function are bidirectionally linked in many ways, and there are numerous studies in which (ultra)structural modulation induces functional changes and vice versa. The strong coupling between mitochondrial (internal) structure and function suggests that they are equivalent, thereby giving rise to the concept of mitochondrial “morphofunction.” This term was first proposed by Benard and Rossignol (24) when they defined “mitochondrial morphofunctional analysis” as: “the simultaneous quantification of mitochondrial morphological and functional readouts.” Later, we illustrate the importance of the mitochondrial morphofunctional concept by presenting results obtained in knockout (KO) animals and patients with mutations in mitochondrial fission/fusion proteins. Next, we discuss various other experimental studies that highlight the bidirectional link between mitochondrial (ultra)structure and function. Finally, we briefly summarize how mitochondrial morphofunction can be quantified in living cells, how mitochondrial (ultra)structure links to mitochondrial bioreactions, and how mitochondrial morphology can be targeted by small molecules.
A. Studies in knockout animals
Animal models have delivered valuable insights into the physiological and functional role of fission and fusion proteins. For instance, Drp1 gene KO mice display developmental abnormalities, particularly in the forebrain, and die after embryonic day 12.5 (152). The same study revealed that neural cell-specific Drp1-null animals die shortly after birth due to brain hypoplasia and apoptosis. Compatible with the dual role of DRP1 in mitochondrial and peroxisomal fission (see section III and Table 1), Drp1-null cells contain extensive mitochondrial networks and elongated peroxisomes, although a reduction in cellular ATP level was not detected (408). In postmitotic Purkinje cells of mouse cerebellum, Drp1 deletion induced mitochondrial elongation followed by formation of large spherical mitochondria due to oxidative stress (168). These mitochondrial spheres lost respiratory function, and accumulated ubiquitin and markers of mitophagy, leading to neurodegeneration. It was concluded that fission of mitochondria prevents oxidative damage, thereby contributing to neuronal survival. This is compatible with the idea presented earlier that fusion of individual mitochondria allows sharing of ROS-induced damage and/or antioxidants within the mitochondrial population (281).
MFN1 and MFN2 are of key importance for proper embryonic development, and KO mice die before mid-gestation (56). Loss of Mfn2 resulted in progressive, retrograde degeneration of dopaminergic neurons in the nigrostriatal circuit (310). The current experimental evidence in various pathological models suggests that increased levels of L-OPA1 are protective (238). Such a protection was induced by Oma1 KO (198, 429), increased OMA1 degradation (432), or OPA1 overexpression (400). Compatible with the latter, increased L-OPA1 processing was not detected during OPA1 overexpression associated with (mild) improvement of mitochondrial disease phenotypes in (CI-deficient) Ndufs4 KO mice and (CIV-deficient) Cox15 muscle-specific KO mice (66). In adult cardiomyocyte-specific Yme1l KO mice, L-OPA1-mediated mitochondrial fusion was required to preserve cardiac function whereas stress-induced OMA1-mediated processing of L-OPA1 and accompanying mitochondrial fragmentation induced dilated cardiomyopathy and heart failure (406). In this sense, Oma1 gene deletion and normalization of mitochondrial structure protected against cell death and heart failure. Interestingly, mitochondrial fragmentation triggered a switch in metabolism from FA to glucose utilization and reversing this switch preserved normal heart function even though mitochondrial fragmentation was not normalized (406). This demonstrates that in the absence of YME1L, heart function can be restored by preventing OMA1-mediated L-OPA1 processing and restoring mitochondrial morphology, or by normalizing metabolism that bypasses the deleterious effects of altered mitochondrial morphology on heart metabolic function (406).
B. Patients carrying mutations in mitochondrial fission and fusion proteins
Mutations in mitochondrial fission/fusion proteins induce human disease (11). This includes congenital microcephaly, lactic acidosis and sudden death (DRP1) (417), Leigh-like encephalopathy, optic atrophy and peripheral neuropathy (MFF) (182), Charcot-Marie-Tooth neuropathy type 2A (MFN2) (450), and optic atrophy (OPA1) (4). In addition, various alterations in mitochondrial fission and fusion proteins were demonstrated in experimental models of neurodegeneration (103, 180): (i) The levels/activity of DRP1 and mitochondrial fission were increased in a rat model of amyotrophic lateral sclerosis (various gene mutations), a mouse model of Alzheimer's disease (AD; various gene mutations), and human/mouse/rat models of Huntington's disease (HD; Huntingtin mutation); (ii) the levels/activity of DRP1 and mitochondrial fission were decreased in human and mouse models of autosomal-recessive spastic ataxia of Charlevoix-Sanguenay disease (SACS/Sacs gene mutation); (iii) the levels/activity of ganglioside-induced differentiation-associated protein 1 (GDAP1) and mitochondrial fission were reduced in human models of autosomal dominant Charcot–Marie–Tooth disease, axonal (CMT2K; GDAP1 gene mutation), autosomal recessive Charcot–Marie–Tooth disease, axonal, type 2K (ARCMT2K; GDAP1 gene mutation), and autosomal recessive Charcot–Marie–Tooth disease, type 4A, demyelinating (CMT4A; GDAP1 gene mutation); (iv) the levels/activity of MFN2 and mitochondrial fusion were reduced in human and rat models of autosomal dominant Charcot–Marie–Tooth disease, axonal, type 2A2 (CMT2A2; MFN2/Mfn2 gene mutation); and (v) the levels/activity of OPA1 and mitochondrial fusion were reduced in human and rat models of autosomal dominant optic atrophy (OPA1/Opa1 gene mutation).
With respect to CMT2A2, the consequences of its mutations on MFN2 activity and neuronal function were recently investigated in a Drosophila fly model (96). Analysis of four pathological mutations (R94Q, R364W, T105M, and L76P) revealed that they all increased mtDNA mutations, decreased oxidative metabolism, and induced mitochondrial depletion at neuromuscular junctions. R94Q and T105M mutations induced aggregation of nonfused mitochondria and their loss of function. Remarkably, R364W and L76P mutations stimulated mitochondrial fusion. It was proposed that both excessive mitochondrial aggregation and fusion can underlie CMT2A pathophysiology (96). Changes in mitochondrial (ultra)structure and mitochondrial function have also been described in human aging (356) as well as in a plethora of other human pathologies, including: (i) Parkinson's disease (339), (ii) AD (126), (iii) HD (130), (iv) ischemia/reperfusion injury (215), (v) mitochondrial metabolic disorders (196), (vi) cardiac disease (92), (vii) kidney disease (27), (viii) cell starvation (325), (ix) diabetes (336), (x) cancer (390), (xi) sepsis (122), and (xii) the effects of environmental toxins (163).
C. Bidirectional links between mitochondrial (ultra)structure and function
Next, we further elaborate on the concept of mitochondrial morphofunction by presenting experimental evidence demonstrating that: (i) mitochondrial (ultra)structure affects mitochondrial function, (ii) mitochondrial function affects mitochondrial (ultra)structure, (iii) mitochondrial internal structure affects mitochondrial external structure, (iv) mitochondrial external structure affects mitochondrial internal structure, (v) mitochondrial morphofunction affects cell function, and (vi) cell function affects mitochondrial morphofunction.
1. Mitochondrial (ultra)structure affects mitochondrial function
It is firmly established that MIM dynamics and topology impact on mitochondrial bioenergetics (244, 306–308). For instance, changes in mitochondrial morphology and organization can enhance energy supply from OXPHOS in diabetic cardiomyopathy (159). Analysis of heteroplasmic human rhabdomyosarcoma cells harboring the pathological A3243G mtDNA mutation, associated with the neuromuscular syndrome MELAS (mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes), revealed that knockdown of DRP1 or FIS1 leads to increased levels of mutated mtDNA (242). This increase was not observed on knockdown of OPA1, suggesting that OPA1-dependent MIM fission and cristae organization are not involved in segregation of mutant and wild-type mtDNA (242). Supporting these results, MFN-mediated mitochondrial fusion protected against neurodegeneration in the cerebellum (58) and was required for mtDNA stability and tolerance of mtDNA mutations in skeletal muscle (59). MFN2-induced mitochondrial elongation protects against ROS-induced mPTP opening and represses staurosporin-induced BAX and cytochrome-c release (281).
During starvation-induced autophagy, the MOM participates in autophagosome formation by supplying membranes (134) and DRP1 was phosphorylated in a cAMP/PKA-dependent manner (120). This prevented DRP1 translocation to the MOM and induced formation of elongated mitochondria, which maintained their ATP production, contained more cristae, displayed increased CV levels/activity, and were not undergoing mitophagy. As a consequence, these elongated mitochondria were protected from autophagosomal degradation (326). Similarly, DRP1 knockdown and mitochondrial elongation protected melanoma cells from mitophagy-mediated death induced by CI inhibition (20).
Using a cell starvation model, evidence was provided that the autophagic process provides FAs for LDs, which transfer into mitochondria only if the latter have a tubulated morphology (325). This mechanism, which also required LD lipolysis, allows cells to survive during starvation by shifting their metabolism toward FA oxidation (409). Later studies argued that LDs are not required for FA delivery to mitochondria but instead function to prevent acylcarnitine accumulation and lipotoxic dysregulation of mitochondria (283). Further, a DRP1-independent mitophagy pathway was described (430). In this respect, inhibition and stimulation of the autophagy pathway were associated with increased and decreased DRP1 levels, respectively, suggesting that DRP1 is targeted by the autophagy process for lysosomal degradation (317).
Interestingly, both mitochondrial hyperfusion and hyperfragmentation protected cells against BAX-mediated apoptosis induction, suggesting that BAX requires a distinct mitochondrial shape to induce mitochondrial outer membrane permeabilization (330). The latter study proposed three factors that regulate BAX-mediated apoptosis: (i) a combination of stress-specific BH3-only proteins that induce apoptosis, (ii) an MOM composition that is regulated by lipid metabolic pathways, and (iii) a distinct net mitochondrial morphology resulting from the combined action of mitochondrial dynamics proteins that allows proper BAX integration and pore formation. In this sense, the large variety in mitochondrial morphology observed using different cell types and (patho)physiological conditions suggests that this structural diversity contributes to the differences in apoptosis sensitivity in these systems (330). The mitochondrial content of cells depends on cell size (177) and impacts mitochondrial function (259). In this context, single-cell analysis revealed that mitochondrial content affects the level of apoptotic proteins and thus cell sensitivity to apoptotic stimuli, meaning that cells with more mitochondria were more prone to cell death induction (246).
2. Mitochondrial function affects mitochondrial (ultra)structure
Mitochondrial Δψ regulates the configuration of the mitochondrial matrix and cytochrome-c release during apoptosis (123). Classical studies revealed that alterations in mitochondrial functional state, as typically observed in a wide range of (patho)physiological conditions and human diseases, are not only associated with altered mitochondrial dynamics but also affect the ultrastructure of the MIM. In response to low ADP concentrations, the structure of the MIM changed from a condition in which the mitochondrial matrix was contracted and dense (the “condensed state”), to a less dense and more expanded state (“orthodox”), associated with a more compact cristae compartment (132).
Inactivation of OPA1 by proteolytic cleavage mediated by OMA1 and YME1L is induced during cellular stress, leading to stimulation of mitochondrial fission (238). OPA1 cleavage also depends on mitochondrial ATP levels and bivalent metals (18). Both OMA1 and YME1L are activated by Δψ depolarization, thereby inducing OPA1 cleavage, mitochondrial fragmentation, and removal of damaged mitochondria by mitophagy (138, 238, 441). Inhibitory OMA1-mediated processing of OPA1 is also stimulated by knockdown of the m-AAA proteases AFG3L1 and AFG3L2 (95). Moreover, in the absence of the MIM PHB2, L-OPA1 is lost and mitochondrial fusion is defective (256). An MIM-specific effect on OPA1 cleavage was demonstrated in cells from patients with mtDNA mutations. This effect was mediated by YME1L, which cleaved OPA1 more efficiently under conditions at which OXPHOS activity was high (265). When Δψ drops below a certain threshold, DRP1-induced mitochondrial fission and OMA1-mediated cleavage of OPA1 are activated (165). OPA1 processing mediated by YME1L was modulated by Δψ, and OPA1 isoforms interact with MFN1 and MFN2 in a Δψ-independent manner (129). In combination with cellular ATP depletion, Δψ depolarization is paralleled by OMA1 activation and YME1L degradation (324). This differential degradation of OMA1 and YME1L affects the proteolytic processing of OPA1, which might allow cells to adjust mitochondrial morphology and function to different types of stress (324). Evidence was provided that Δψ-dependent processing of OPA1 impacts mitochondrial function (152, 294). In mouse muscle, age-associated loss of OPA1 can activate a signaling cascade starting from the ER that systemically affects metabolism and aging (385).
3. Mitochondrial internal structure affects mitochondrial external structure
A recent study in neuronal cells demonstrated that spontaneous and repetitive constrictions of the mitochondrial inner compartment (CoMIC) were closely associated with subsequent division of the mitochondrion (62). CoMIC was stimulated by increased matrix Ca2+ levels, mitoKCa-mediated ultrastructural changes, and Δψ depolarization and OMA1-catalyzed accumulation of S-OPA1, suggesting that these intra-mitochondrial constrictions prime mitochondrial division.
4. Mitochondrial external structure affects mitochondrial internal structure
Interestingly, cristae remodeling during apoptosis requires mitochondrial fission mediated by DRP1, MIEF1/MID51, and MIEF2/MID49 (298). This study revealed that during intrinsic apoptosis Mief1/MiD51-Mief2/Mid49-KO, and Drp1-KO cells did not exhibit cristae remodeling and cytochrome-c release, whereas this was not the case in Mff-KO cells.
5. Mitochondrial morphofunction affects cell function
Fis1 overexpression in HeLa cells induced fragmentation and perinuclear clustering of mitochondria that maintained a normal Δψ and normally sequestered Ca2+ released from the ER (110). In contrast, Ca2+ entering across the PM was taken up more slowly by these mitochondria, compatible with a role of subplasmalemmal mitochondria in modulating the activity of PM Ca2+ATPases (109). Supported by other studies (77, 434), this demonstrates that mitochondrial morphological aberrations affect (local) cellular Ca2+ homeostasis. Continued uptake of multiple ACs by phagocytes (a process known as efferocytosis) requires DRP1-dependent mitochondrial fragmentation in the phagocytes (414). The latter study demonstrated that mitochondrial fragmentation allows the release of Ca2+ from the ER into the cytosol to allow phagocytosis by stimulating Ca2+-mediated vesicle trafficking.
MFN2 KO in POMC (pro-opiomelanocortin) neurons (involved in the regulation of energy and glucose metabolism) induced defective POMC processing, loss of mitochondria-ER contacts, ER stress-induced leptin resistance, hyperphagia, reduced energy expenditure, and obesity (350). In agreement with these findings, DRP1-induced mitochondrial fission reduces the sensitivity of POMC neurons for leptin and glucose (345).
Mitochondrial structure affects cell cycle phase and vice versa (143). For instance, CDK5 inhibits DRP1 by phosphorylation (63) and mitochondria form a hyperfused network at the G1-to-S transition associated with buildup of cyclin E (268). The latter study also revealed that Δψ depolarization at early G1 prevents cell cycle entry into S phase. Perhaps related to this phenomenon, Δψ loss in HeLa cells was linked to homogenization and decompaction of chromatin structure, suggesting that the latter is linked to mitochondrial function (7).
6. Cell function affects mitochondrial morphofunction
Mitochondrial fusion appears to be ATP dependent (251), suggesting that mitochondria-generated or glycolytic ATP might (locally) affect mitochondrial fusion dynamics. In this context, high glucose induced mitochondrial fragmentation paralleled by increased ROS levels (391, 440) and inhibition of glycolysis impacted MIM fusion more than it did MOM fusion, whereas Δψ dissipation prevented MIM fusion but not MOM fusion (342).
In endothelial cells, mitochondrial morphology in situ was altered by Ca2+ overload (36). Using the same cell type, it was observed that KO of ROS-sensitive TRPM2 (transient receptor potential cation channel M2) channels prevented glucose-induced mitochondrial fission (1). The latter study proposed a mechanism in which: (i) TRPM2-mediated Ca2+ entry induces permeabilization of lysosomal membranes and lysosomal Zn2+ release, and (ii) the released Zn2+ stimulates mitochondrial DRP1 recruitment to induce fission (1).
AMPK is a key regulatory protein in cellular energy homeostasis (221). Mitochondrial morphofunction is linked to cellular bioenergetics via AMPK, which phosphorylates MFF to enhance DRP1 recruitment and induce mitochondrial fission (388). Interestingly, unphosphorylated MFF mutants inhibit mitophagy, connecting AMPK to mitochondrial fission and mitophagy regulation (388, 442).
Mitochondrial morphology changes were also induced via altered fusion dynamics during hypoxia-reoxygenation stress (222). These changes involved formation of donut-like (toroidal) mitochondrial structures during hypoxia in glucose-free media and reoxygenation-induced formation of these structures in glucose-containing medium. Formation of these donut-like morphologies involved partial mitochondrial detachment from the cytoskeleton and mitochondrial swelling, involving opening of the mPTP or mitochondrial K+ channels, allowing “autofusion” (222). Computational analysis suggested that an increase in osmotic potential during stress conditions induces donut formation and that this process is reversible (228).
D. Quantification of mitochondrial morphofunction in living cells
Although substantial progress has been made (24, 142, 266, 355, 407), we are still far from understanding how the morphofunctional heterogeneity of individual mitochondria within a single cell (Figs. 5, 7B, and 11) is maintained and how this heterogeneity connects to mitochondrial and cellular physiology. This lack of insight not only limits our understanding of mitochondrial morphofunction at the mechanistic level but also hampers the development and (large-scale) screening of morphofunction-targeting drugs. Given the tight connection between mitochondrial and cellular function, physiological analysis of mitochondrial morphofunction is ideally carried out within the context of a living cell (147, 148, 188, 196). If required, this can also entail analysis of mitochondrial fusion dynamics and/or motility using photoactivatable proteins (173, 231, 248). Micropatterned coverslips have been used to “standardize” cell size and shape and allow analysis of mitochondrial structure under controlled conditions (60). However, alterations in cell shape and cell-substrate adherence might affect mitochondrial mechanical interactions with the cytoskeleton and therefore mitochondrial (ultra)structure (19).
FIG. 11.
Mitochondrial morphofunctional heterogeneity. (A) Summarizing scheme for simultaneous quantification of mitochondrial free ATP levels, membrane potential (Δψ), and mitochondrial morphology by single-cell fluorescence microscopy. Bold symbols (i.e., B, C, D, E, F, G, N) refer to the corresponding panels to the right. PHSFs (control CT5120) were transfected with a mitochondria-targeted variant of “ATeam” (Cox8-ATeam 1.03 or “MitoATeam”), an ATP-sensing CFP/YFP-based FRET (fluorescence resonance energy transfer) reporter molecule (150, 218). The YFP/CFP emission ratio of ATeam can be used as a measure of free ATP concentration. Subsequently, MitoATeam-expressing cells were stained with TMRM (that accumulates in mitochondria in a Δψ-dependent fashion). Next, the acquired images were processed and quantified to assess ATeam and TMRM signals for individual mitochondria and to allow multiparameter analysis. (B) Background-corrected (COR) image depicting the ATeam YFP signal. (C) Binarized (black-and-white; BIN) version of (B) obtained after image processing (explained in Fig. 13). (D) Version of the YFP-BIN image in which non-mitochondrial objects with an area <10 pixels (noise pixels) were removed by thresholding. (E) Masked (MSK) version of (B) obtained by applying a Boolean AND operation to the YFP-COR (B) and YFP-BIN image (C). (F) Same as (E), but now for the CFP-COR (not shown) and YFP-BIN image (C). (G) Same as (E), but now for the TMRM-COR (not shown) and YFP-BIN image (C). (H) ATeam ratio image obtained by dividing the YFP-MSK image (E) by the CFP-MSK image (F). To allow visualization, the ATeam ratio value for each pixel was multiplied by the number 20. (I) Colorized version of (F). Pixels were colorized according to their ratio value (blue = low; red = high). (J) Same as (I), but now for (G). (K) Magnification of the region in (H) depicting individual mitochondria. (L) Same as (K), but now showing a magnification of (I). (M) Same as (K), but now showing a magnification of (J). (N) 3D scatter plot of the ATeam ratio signal versus mitochondrial TMRM signal versus mitochondrial size (area) of the mitochondrial objects in (M). Each dot represents an individual mitochondrial object. Color images are available online.
Quantification of mitochondrial internal and external structural parameters (Fig. 12) is not trivial, given the small dimensions of this organelle (diameter typically <1 μm). For this reason, mitochondrial ultrastructure has primarily been visualized by using EM-based approaches, although only in fixed cell specimens (107, 113, 233). In addition, various superresolution microscopy techniques (104, 396) have been applied to mitochondrial (ultra)structural analysis (157). These include: structured illumination microscopy (124, 358), 4Pi-confocal microscopy (94), STimulated Emission Depletion microscopy (158), Gated STimulated Emission Depletion microscopy (28), stochastic optical reconstruction microscopy (STORM), 3D-STORM, and direct STORM (146, 178) and the “Zernike Optimized Localisation approach in 3D” (12). Unfortunately, the superresolution approaches that are currently available, have not been optimized for live-cell imaging and lack a sufficiently high acquisition rate (i.e., with low subsecond time resolution) and axial imaging depth to allow real-time analysis of mitochondrial ultrastructural dynamics in living cells (104).
FIG. 12.
Mitochondrial ultrastructural parameters related to mitochondrial function. Various quantitative parameters of mitochondrial ultrastructure affecting mitochondrial bioreactions. This figure was created by using information from (403). Color images are available online.
Currently, live-cell visualization of mitochondrial morphology and function is primarily carried out by combining fluorescent cations and/or mitochondria-targeted fluorescent proteins (FPs) with confocal laser scanning microscopy (CLSM) or epifluorescence microscopy. During the past decade, we and others had developed integrated experimental and computational strategies for semiautomatic quantification of mitochondrial morphology (Fig. 13) (78, 192, 194, 389). Our approach is optimized for analysis of relatively large flat cells such as primary human skin fibroblasts (PHSFs) but, after optimization, is also applicable to other cell types (147). As an alternative, (fixed) non-flat cell types and/or tissues can be analyzed by using 3D-CLSM (22, 288). In case of PHSFs, high-content microscopy images are acquired by using cells co-stained with: (i) tetramethylrhodamine (TMRM), which accumulates in mitochondria in a Δψ-dependent fashion; (ii) Calcein-AM, which accumulates in the cytosol; and (iii) Hoechst 33258, to stain nucleic acids (dsDNA). In a collaborative effort, we also developed high-content protocols for cells co-stained with TMRM and 5-(and-6)-chloromethyl-2’,7’-dichlorodihydrofluorescein (CM-H2DCFDA), a reporter of cellular oxidant levels (365). The acquired images are then processed (Fig. 14A) to allow extraction of various descriptors of mitochondrial morphofunction (e.g., number of mitochondria per cell, mitochondrial size, TMRM staining intensity; see Table 2). These descriptors then can be used for quality control and further evaluated by principal component analysis, univariate/bivariate/multivariate methods, and machine-learning strategies (Fig. 14) (3, 22, 30, 214, 329). For example, quantitative analysis and machine learning were applied to determine whether certain mitochondrial morphologies pre-disposed individual mitochondria to undergo subsequent fission or fusion (421). This revealed that mitochondrial perimeter and solidity (i.e., the compactness of its shape) positively and negatively correlated with the occurrence of a mitochondrial fission or fusion event, respectively. It was concluded that the mechanical properties of the MOM might influence mitochondrial fission and fusion probability (421).
FIG. 13.
Analysis of mitochondrial morphology in fibroblasts with isolated CI deficiency by microscopy image processing and quantification. PHSFs were stained with the mitochondria-accumulating cation rhodamine 123 and visualized by using video-rate confocal microscopy (1 s image acquisition time; 30 images/s were averaged). (A) Illustration of the image processing strategy using control CT5120 cells. The acquired images were subsequently background corrected (COR), subjected to an LCS operation, processed by a THF, processed by a median (MED) filter, and thresholded to obtain a binarized (BIN) black-and-white image. The COR image was masked by the BIN image, yielding a masked (MSK) image that was colorized (MSK-colorized; blue = low and red = high fluorescence intensity). This processing strategy was described in detail elsewhere (186, 192, 194). (B) Image depicting the mitochondrial objects in the MSK-colorized image sorted according to their size (area) from left to right. This “mitogram” reveals considerable heterogeneity in rhodamine 123 fluorescence signal between and within individual mitochondrial objects. (C) Application of the strategy in (B) on primary skin fibroblasts of control subjects (CT5120; green) and patients (P) with isolated CI deficiency. The latter carried mutations in various CI subunit-encoding nDNA genes (NDUFS1, NDUFS2, NDUFS4, NDUFS7, NDUFS8, and NDUFV1). The boxplot reveals that the mitochondrial formfactor (F; a combined measure of mitochondrial length and degree of branching) is significantly altered (colorized red) or similar (black) to CT5120 cells (green). (D) Similar to (C), but now depicting the number of mitochondria per cell (Nc). The interpretation and implications of this data is presented elsewhere (187, 193, 195). LCS, linear contrast stretch; THF, top-hat filter. Color images are available online.
FIG. 14.
High-content analysis (“cellomics”) of mitochondrial morphofunction. PHSFs (CT5120) were stained with the mitochondria-accumulating cation TMRM and visualized by using fluorescence microscopy. The acquired images were processed as described in Figure 13A. (A) Typical TMRM fluorescence image used to illustrate the high-content analysis. (B) Heat map depicting the numerical value of 42 morphofunctional descriptors (horizontal axis; Table 2) for each of the 217 mitochondrial objects in (A) (vertical axis). These data were sorted from large (top) to small (bottom) values of mitochondrial size (descriptor “Area”; marked in red) and Log10 scaled. Individual horizontal rows represent the morphofunctional “fingerprint” for each mitochondrial object (30), which can be averaged at the level of individual cells and cell populations to obtain information at different complexity levels. (C) Degree of linear correlation (calculated by using Pearson's R) between the individual descriptors in (B). Negative and positive correlations are depicted in blue and red, respectively. This correlation plot also can be considered a cellular fingerprint of mitochondrial morphofunction in the analyzed cell. The data presented in this figure can be explored and classified by using various computational techniques (147, 411), allowing time analysis, comparison of different cells and cell types, culture conditions, and modulation strategies. Color images are available online.
Table 2.
Descriptors of Mitochondrial Morphofunction for Tetramethylrhodamine
| No.a | Descriptor | Symbol | Definition/meaning of descriptorb |
|---|---|---|---|
| 1 | Aspect ratio | AR | Ratio between major axis and minor axis of an ellipse equivalent to object: measure of mitochondrial length. |
| 2 | Area/Box | Ab | Ratio between the area of an object and the area of its bounding box. |
| 3 | Box XonY | Bxy | Ratio between the width and the height of an object's bounding box. |
| 4 | Density mean | Dm | Average intensity of object (grayvalue): average mitochondrial intensity of TMRM signal. |
| 5 | Axis minor | Axm | Length of minor axis of ellipse with same moments of order 1 and 2 as object (pixels). |
| 6 | Diameter maximum | Dimax | Length of the longest line joining two points of the object's outline and passing through the object's centroid (pixels). |
| 7 | Diameter minimum | Dimin | Length of the shortest line joining two points of the object's outline and passing through the object's centroid (pixels). |
| 8 | Diameter mean | Dim | Average length of diameters measured at 2-degree intervals and passing through the object's centroid. |
| 9 | Radius maximum | Rmax | Minimum distance between the object's centroid and outline (pixels). |
| 10 | Radius minimum | Rmin | Maximum distance between the object's centroid and outline (pixels). |
| 11 | Perimeter ellipse | P1 | The perimeter of the ellipse surrounding the outline of each object (pixels). |
| 12 | Radius ratio | Rr | Ratio between Rmax and Rmin. |
| 13 | Roundness | F | Perimeter2/(4*π*Area; or Formfactor or F): measure of mitochondrial length and degree of branching. |
| 14 | Length | Le | Feret diameter (caliper length) along the major axis of the object (pixels). |
| 15 | Width | Wi | Feret diameter (caliper length) along the minor axis of the object (pixels). |
| 16 | Perimeter2 | P2 | Chain code length of the outline (pixels). |
| 17 | IOD | IOD | Integrated optical density of all objects (grayvalue): sum of all mitochondrial TMRM intensity values. |
| 18 | Perimeter convex | Pc | Perimeter of the convex outline of the object (pixels). |
| 19 | Perimeter | P3 | Length of the object's outline (pixels). |
| 20 | Perimeter ratio | Pr | Ratio of convex perimeter to perimeter. |
| 21 | Area polygon | Ap | Area included in the polygon defining the object's outline (same polygon as used for perimeter). |
| 22 | Count | Cou | Size-weighted object “count” (number of objects). |
| 23 | Box width | Bw | Width of the object's bounding box (pixels). |
| 24 | Box height | Bh | Height of the object's bounding box (pixels). |
| 25 | Feret minimum | Fmin | Smallest feret (caliper) length (pixels). |
| 26 | Feret maximum | Fmax | Longest feret (caliper) length (pixels). |
| 27 | Feret mean | Fm | Average feret (caliper) length (pixels). |
| 28 | Density minimum | Dmin | Minimum density inside object (grayvalue). |
| 29 | Density maximum | Dmax | Maximum density inside object (grayvalue). |
| 30 | Density standard deviation | Dstdv | Standard deviation of intensity or density inside object (grayvalue). |
| 31 | Margination | Ma | Distribution of intensity between the center of an object and the edge of the object. |
| 32 | Area | Area | Area of the object (pixels). |
| 33 | Center X | CX | X coordinate of the object's centroid (pixel position). |
| 34 | Center Y | CY | Y coordinate of the object's centroid (pixel position). |
| 35 | Angle | Angle | Angle between the major axis of the object and the vertical. |
| 36 | Axis (major) | Axmaj | Length of the major axis of the ellipse with the same moments of order 1 and 2 as the object. |
| 37 | Fractal dimension | FD | Fractal dimension of the object's outline. |
| 38 | Center X (mass) | CmassX | Intensity-weighed centroid X-position (pixel position). |
| 39 | Center Y (mass) | CmassY | Intensity-weighed centroid Y-position (pixel position). |
| 40 | Heterogeneity | Het | Fraction of pixels that deviate more than a certain range (here 10%) from the average intensity. |
| 41 | Density Sum | Dsum | Sum of intensity inside the object (grayvalue). |
| 42 | Perimeter3 | P4 | Corrected chain code perimeter (pixels). |
E. A brief primer on the role of mitochondrial (ultra)structure in mitochondrial bioreactions
Understanding the dynamics and design principles of biochemical reactions within an inhomogeneous cell compartment such as the mitochondrion still represents a major challenge (127, 373). This is due to the fact that mitochondrial bioreactions are spatially compartmentalized, meaning that they generally involve the conversion of (im)mobile substrates by (im)mobile enzymes into (im)mobile products. As a consequence, these reaction systems require biomolecule transport and/or diffusion within or between mitochondria and, therefore, constitute “reaction-diffusion” systems (226, 419).
It is higly likely that changes in mitochondrial (ultra)structure affect mitochondrial function by modulating mitochondrial bioreactions, for instance by: (i) changes in MOM surface area and matrix volume, and (ii) changes in the extent of MIM folding, which affects cristae number/morphology/dimensions/topology. In addition, these bioreactions will be affected by alterations in matrix solvent properties, including ionic strength, viscosity, and macromolecular crowding. In this manner, changes in mitochondrial (ultra)structure and matrix physicochemical parameters could affect many diffusion-influenced mitochondrial reactions, including: (i) metabolic reactions (ATP, metabolites), (ii) protein folding, (iii) protein–protein interactions (signaling and macromolecular assembly), and (iv) mtDNA replication/translation. In addition, physicochemical differences between individual mitochondria could be underlying Δψ and functional inhomogeneities between and within single mitochondria.
Although the general properties of reaction-diffusion systems are relatively well understood, detailed quantitative information on solute diffusion and biomolecular interactions in the mitochondrial matrix is currently lacking (25, 91, 97, 155, 295, 351). The analysis of protein diffusion in the mitochondrial matrix generally involves expression of an FP with favorable spectral and biophysical properties. FP mobility then can be determined by using fluorescence recovery after photobleaching (FRAP), fluorescence loss in photobleaching (FLIP; Fig. 15), and/or fluorescence correlation spectroscopy (FCS) analysis (10, 85, 187, 189). Given the complex nature of FP diffusion within the matrix (i.e., it is affected by the molecular weight (MW) of the FP, cristae diffusion barriers, mitochondrial length and diameter, the size of the bleach region, and solvent viscosity), one will always measure an “apparent” FP fluorescence recovery time (FRAP/FLIP) or diffusion constant (FCS). This necessitates the use of a diffusion model in which various parameters are experimentally constrained (i.e., mitochondrial dimensions, number of cristae, FP concentration, and FRAP region size) (84). Applying this strategy in HEK293 cells, we demonstrated that, as expected (303), FP diffusion in the mitochondrial matrix is MW-dependent. Unexpectedly (305, 401), our experiments also revealed that cristae severely hinder FP diffusion. This suggests that differences in mitochondrial ultrastructure, as observed between cell types and between healthy and pathological conditions (403, 447) affect diffusion-limited reactions in the mitochondrial matrix. Moreover, it is highly probable that changes in mitochondrial metabolic state accompanied by orthodox-to-condensed transitions (132, 335) will also affect intra-matrix solute diffusion. This implies that alterations in mitochondrial nanostructure, as observed during numerous (patho)physiological conditions, can affect the properties of intramatrix reaction-diffusion systems and therefore mitochondrial and cellular function (84).
FIG. 15.
FLIP analysis of single mitochondrion in HEK293 cells. HEK293 cell lines were created that stably expressed mitochondria-targeted GFP (COX8-AcGFP1) or a GFP-tagged version of the NDUFS3 subunit of CI (NDUFS3-AcGFP1). Analysis of single mitochondria by FRAP revealed that AcGFP1 was fully mobile, whereas NDUFS3-AcGFP1 was partially immobilized in the mitochondrial matrix (85). Here, we used FLIP analysis to determine the submitochondrial localization of immobile NDUFS3-AcGFP1. (A) In COX8-AcGFP1 cells, repetitive bleaching of mitochondrial AcGFP1 using a 1.4 μm square bleach region induced complete loss of fluorescence in the bleach and FLIP region within 12 s. Visual inspection revealed that AcGFP1 fluorescence was completely lost from the mitochondrial filament (inset). (B) In contrast, for NDUFS3-AcGFP1 cells, repetitive AcGFP1 bleaching induced complete fluorescence loss in the bleach region but not in the FLIP region (arrowhead). In this case, visual inspection confirmed that NDUFS3-AcGFP1 fluorescence was absent in the bleach region but not outside this region. (C) Explanation of the obtained results with NDUFS3-AcGFP1 cells. Repetitive photobleaching induces loss of fluorescence for the mobile fraction of NDUFS3-AcGFP1 but not for the immobile fraction of NDUFS3-AcGFP1, which appears to be localized (arrowheads). (D) Magnification of a typical FLIP experiment illustrating the localization of the immobile NDUFS3-AcGFP1 fluorescence fraction (arrowhead). This might represent local compartments of the mitochondrial matrix that restrict NDUFS3-AcGFP1 diffusion and/or NDUFS3-AcGFP1 that is membrane bound in CI (sub)complexes or ETC supercomplexes. (C) and (D) were taken from (83). ETC, electron transport chain; FLIP, fluorescence loss in photobleaching. Color images are available online.
In contrast to FPs in the matrix, early evidence suggested that MIM-embedded ETC complexes exhibit restricted diffusion (117). Indeed, after artificial cell fusion, FP-labeled ETC complexes and CV displayed a patterned arrangement, possibly as a result of their incorporation in supercomplexes within cristae (275). This suggests that after mitochondrial fusion complete equilibration of ETC complexes, being MIM-embedded, is slow relative to FP diffusion in the mitochondrial matrix.
The volume of the reaction compartment is a key parameter in chemical reaction dynamics. Changes in mitochondrial volume (Fig. 7A) were induced by alterations in OXPHOS function, ischemia/reperfusion, anoxia, and apoptotic signaling. Evidence was provided that these volume changes (co)control mitochondrial function (48, 225, 291). Mitochondrial volume depends on the osmotic balance between the cytosol and the mitochondrion (167) and is mainly regulated by potassium (K+) ions (Fig. 1), which enter and leave the mitochondrial matrix in a Δψ-dependent manner (341). The latter study revealed that Δψ depolarization induced mitochondrial swelling and impaired mitochondrial motility in neuronal cells. Together with K+, also Ca2+ ions are important in triggering mitochondrial swelling, especially when the matrix influx of these ions is increased and/or their matrix efflux is decreased (160). Under these conditions, the osmotic pressure and water accumulation in the mitochondrial matrix is increased, leading to mitochondrial swelling. Importantly, the rate at which water enters the mitochondrial matrix is dependent not only on the osmotic gradient with the cytosol but also on the water permeability of the MIM (167). In addition, it was suggested that metabolic water produced by the OXPHOS system is involved in the regulation of mitochondrial volume (48). A water channel (aquaporin-8/AQP8; Fig. 1) was detected in the MIM of rat liver mitochondria and proposed to mediate rapid matrix expansion, thereby regulating the activity of the ETC and CV (42). Whether this AQP8-mediated influx is a general mechanism across tissues and whether AQP8 also participates in the trans-MIM transport of other molecules still remains to be determined (160, 209).
Physiologically relevant changes in matrix volume (Fig. 7A) are moderate, reversible, and linked to regulation of the ETC and mitochondrial ATP production (160). It appears that mitochondrial osmotic pressure regulates mPTP opening (15) and that massive swelling of the mitochondrial matrix associated with long-term (Ca2+-induced) mPTP opening leads to cell death by compromising mitochondrial function (291). Mechanistically, evidence in rat liver mitochondria suggests that this mPTP regulation involves volume-induced changes in matrix concentration of mPTP regulatory factors (290). In this context, the latter study revealed that mitochondrial shrinkage stimulated mPTP inhibition by Mg2+ and ubiquinone 0 (Ub0) and decreased the effect of the mPTP-inhibitor Cyclosporin A. It is currently unknown how the transition from physiological to pathological volume changes (Fig. 7A) is exactly regulated, although a role for ROS and metabolic stress was proposed (160).
F. Small-molecule targeting of mitochondrial morphology
The concept of mitochondrial morphofunction strongly suggests that mitochondrial function can be modulated by rational manipulation of mitochondrial (ultra)structure and vice versa. This is of great relevance for the large number of pathological conditions in humans where the mitochondrial structure-function relationship is disturbed (11, 328).
Various small molecules have been developed that inhibit DRP1 activity. These include Dynasore (237), Mdivi1 (47), and the P110-TAT peptide (130, 166, 320). All three of these molecules inhibited mitochondrial fission in various studies, suggesting that they might be useful to reduce excessive mitochondrial fission and oxidative stress in neurons affected in AD, Parkinson's disease, and HD (328). However, in case of Mdivi1, off-target effects have been described, including inhibition of K+ channels and membrane potential in HL-1 murine atrial cardiomyocytes (370) as well as CI inhibition and modulation of mitochondrial ROS production (34, 322). The pros and cons of the use of Mdivi1 as a specific DRP1 inhibitor were recently discussed (368). It was concluded that although the current evidence supports the previously described Mdivi1 bioactivity, use of this molecule requires inclusion of stringent positive controls that directly demonstrate its effect on mitochondrial fission. Importantly, DRP1 also mediates peroxisomal fission (353, 354) and exerts various other functions (Table 1). This means that mitochondrial fission cannot be specifically modulated by targeting of DRP1.
In addition to DRP1-targeting molecules, a cell-permeant fusogenic peptide (TAM-MP1Gly) was developed that destabilized the fusion-constrained configuration of MFN2, thereby promoting the fusion-permissive conformation (105). Interestingly, a small-molecule mimetic of the MFN2 peptide-peptide interface (“Chimera B-A/long”) allosterically activated MFN2, thereby promoting mitochondrial fusion (333). This small molecule mitigated mitochondrial clumping, Δψ depolarization, fragmentation, and dysmotility in CMT2A mutant neurons. The strategies described earlier focus on inhibition and stimulation of mitochondrial fission and fusion, respectively, to improve mitochondrial function. However, altering mitochondrial (ultra)structure by exogenous means also affects the cell's susceptibility to death-inducing agents. The latter is likely to be cell type- and condition-dependent. For example, mitochondrial fragmentation induced by DRP1 overexpression protected against the efficacy of CER-induced, Ca2+-dependent, apoptosis (378). In contrast, DRP1 knockdown induced mitochondrial filamentation and protected against necroptosis and apoptosis induced by CI inhibition in melanoma cell models (20).
In our own studies, we observed that Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid), a water-soluble vitamin E-derived antioxidant, increased the levels of fully assembled and active CI in primary skin fibroblasts of children with isolated CI deficiency (191). In these patient cells, Trolox displayed pleiotropic effects since it: (i) reduced ROS levels, (ii) normalized aberrant cytosolic Ca2+/ATP handling, and (iii) restored Δψ depolarization (90, 191). Mechanistic studies in healthy primary skin fibroblasts revealed that Trolox: (i) reduced ROS-induced formation of chloromethyl 2′,7′-dichlorofluorescein, (ii) did not affect oxidation of the ROS sensor HEt (hydroethidium), (iii) did not affect mitochondrial NAD(P)H levels or Δψ, (iv) reduced lipid peroxidation, (v) increased the expression and activity of all OXPHOS complexes, (vi) stimulated mitochondrial routine and maximal oxygen consumption, and (vii) prevented hydrogen peroxide-induced aberrations in cytosolic Ca2+ handling (89). Within the context of this review, it is important that the latter study demonstrated that Trolox stimulated the protein levels of MFN2 but not of DRP1 and FIS1 in mitochondria-enriched fractions from PHSFs and Chinese Hamster Ovary cells. Moreover, Trolox-induced stimulaton of mitochondrial filamentation was glutathione-dependent and absent in Mfn1−/−, Mfn2−/−, and Mfn1−/−Mfn2−/− cells. Given the fact that the Trolox effects were observed in healthy fibroblasts, we proposed that Trolox reduces the levels of endogenous CM-H2DCF oxidizing signaling ROS, which increases the levels of mitochondria-attached mitofusins and therefore induces mitochondrial filamentation (89). Functionally, this filamentation is associated with increased mitochondrial function as exemplified by increased activity of OXPHOS complexes and higher mitochondrial oxygen consumption. Given these pleiotropic effects, it remains to be determined whether Trolox and other antioxidants can exert positive effects by targeting aberrant mitochondrial (ultra)structure in human disease (21, 185, 348).
V. Conclusions
In this review, we highlighted the basic mechanisms and experimental evidence, demonstrating a tight and multidirectional connection between mitochondrial internal structure, external structure, and function. This supports the novel concept of “mitochondrial morphofunction.” Currently, a comprehensive understanding of mitochondrial morphofunction is still lacking, and obtaining this information requires quantitative information at the level of individual mitochondria within single living cells. In addition, more detailed insights are required into how the volume and complex nanoarchitecture of the mitochondrion impacts its biochemistry. It is expected that a detailed understanding of mitochondrial morphofunction will significantly contribute to deciphering: (i) why individual mitochondria display heterogeneous (ultra)structural and functional properties between and within cells, (ii) how mitochondrial (ultra)structure affects mitochondrial and cellular bioreactions and therefore mitochondrial and cellular functioning, (iii) how exogenous manipulation of mitochondrial (ultra)structure can be used to “control” mitochondrial function, and (iv) whether such manipulation is therapeutically relevant in human diseases with disturbed mitochondrial morphofunction.
Supplementary Material
Acknowledgments
The authors apologize to those authors whose articles they were unable to cite because of space limitations. This research was supported by an RIMLS junior researcher project granted to W.J.H.K. The authors are grateful to Dr. J. Esseling (Department of Biochemistry) for Differential Interference Contrast microscopy (supported by the Dutch Ministry of Economic Affairs; IOP Grant No. IGE05003), Ing. H.G. Swarts (Department of Biochemistry) for generating baculoviruses, Dr. J.A. Fransen and Ing. M.M. Kea-te Lindert (Department of Cell Biology) for electron microscopy analysis, and Dr. C.E.J. Dieteren (Department of Biochemistry) for creating inducible HEK293 cell lines and performing photobleaching experiments (supported by a joint grant of the “Nijmegen Center for Mitochondrial Disorders” to W.J.H.K. and Dr. L.G.J. Nijtmans from the Department of Pediatrics).
Abbreviations Used
- AC
apoptotic cell
- acyl-CoA
acetyl coenzyme A
- AD
Alzheimer's disease
- ADP
adenosine diphosphate
- AHs
amphipathic helices
- AMPK
AMP-activated protein kinase A
- ANT
adenine nucleotide translocator
- AQP8
aquaporin-8
- ATP
adenosine triphosphate
- BAT
brown adipose tissue
- CAMK1α
calcium/calmodulin-dependent kinase 1α
- CaN
calcineurin
- CDKs
cyclin-dependent kinases
- CJ
cristae junction
- CL
cardiolipin
- CLSM
confocal laser scanning microscopy
- CM
cristae membrane
- CoMIC
constrictions of the mitochondrial inner compartment
- DRP1
dynamin-related protein 1
- DYN2
dynamin 2
- EM
electron microscopy
- ER
endoplasmic reticulum
- ETC
electron transport chain
- FAs
fatty acids
- FCS
fluorescence correlation spectroscopy
- FIS1
fission protein 1
- FLIP
fluorescence loss in photobleaching
- FPs
fluorescent proteins
- FRAP
fluorescence recovery after photobleaching
- GDAP1
ganglioside-induced differentiation-associated protein 1
- G-domain
GTPase domain
- GDP
guanosine diphosphate
- Gln
glutamine
- GPAT
glycerol-3-phosphate acyltransferase
- GTP
guanosine triphosphate
- HD
Huntington's disease
- HR1
heptad repeat domain 1
- IBM
inner boundary membrane
- IF1
inhibitory factor 1
- IMJs
inter-mitochondrial junctions
- IMPs
integral membrane proteins
- IMS
intermembrane space
- JNK
c-JUN N-terminal kinase
- KO
knockout
- LD
lipid droplet
- LPA
lysophosphatidic acid
- MCU
mitochondrial calcium uniporter
- MFF
mitochondrial fission factor
- MFN1/MFN2
mitofusin 1 and 2
- MFNs
mitofusins
- MIC10
MICOS complex subunit 10
- MICOS
mitochondrial contact site and cristae organization
- MIEF1/MID51
mitochondrial elongation factor 1
- MIEF2/MID49
mitochondrial elongation factor 2
- MIGA
mitoguardin
- MIM
mitochondrial inner membrane
- MIRO1
mitochondrial Rho GTPase 1
- MiST
“mitochondrial shape transition”
- mitoPLD
mitochondrial phospholipase D
- MOM
mitochondrial outer membrane
- MPP
matrix-soluble protein peptidase
- mPTP
mitochondrial permeability transition pore
- mtDNA
mitochondrial DNA
- MTFP1
mitochondrial fission process protein 1
- MTS
mitochondrial targeting sequence
- MW
molecular weight
- nDNA
nuclear DNA
- NNT
nicotinamide nucleotide transhydrogenase
- OPA1
optic atrophy protein 1
- OPA3
optic atrophy 3
- OXPHOS
oxidative phosphorylation
- PA
phosphatidic acid
- PA-PLA1
PA-preferring phospholipase A1
- PARL
Presenilin-associated rhomboid-like
- PC
phosphatidylcholine
- PDIA1
protein disulfide isomerase A1
- PE
phosphatidylethanolamine
- PHB2
prohibitin 2
- PHSFs
primary human skin fibroblasts
- PiC
Pi/H+ symporter
- PINK1
PTEN-induced putative kinase 1
- PKA/AKAP1
protein kinase A/A kinase anchor protein 1
- PM
plasma membrane
- PMF
proton-motive force
- POMC
pro-opiomelanocortin
- PP2A/Bβ2
protein phosphatase 2A/Bβ2
- PTMs
post-translational modifications
- Q
Coenzyme Q10
- ROCK1
rho-associated coiled coil-containing protein kinase 1
- ROS
reactive oxygen species
- SAM
protein sorting and assembly machinery
- Sp
splice variant
- STORM
stochastic optical reconstruction microscopy
- TCA
tricarboxylic acid
- TIM
protein translocase of the MIM
- TM
transmembrane domain
- TMEM11
transmembrane protein 11
- TMRM
tetramethylrhodamine
- TOM
protein translocase of the MOM
- TRPM2
transient receptor potential cation channel M2
- VDAC
voltage-dependent anion channel
Author Disclosure Statement
W.J.H.K. and P.H.G.M.W. are scientific advisors of Khondrion B.V. (Nijmegen, The Netherlands). W.J.H.K. is scientific advisor of Mitoconix Bio., Ltd. (Ness Ziona, Israel) and of Fortify Therapeutics (Palo Alto, California). These SMEs had no involvement in the data collection, analysis and interpretation, writing of the article, and in the decision to submit the article for publication.
References
- 1. Abuarab N, Munsey TS, Jiang LH, Li J, and Sivaprasadarao A. High glucose-induced ROS activates TRPM2 to trigger lysosomal membrane permeabilization and Zn2+-mediated mitochondrial fission. Sci Signal 10: eaal4161, 2017 [DOI] [PubMed] [Google Scholar]
- 2. Adachi Y, Itoh K, Yamada T, Cerveny KL, Suzuki TL, Macdonald P, Frohman MA, Ramachandran R, Iijima M, and Sesaki H. Coincident phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol Cell 63: 1034–1043, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahmad T, Aggarwal K, Pattnaik B, Mukherjee S, Sethi T, Tiwari BK, Kumar M, Micheal A, Mabalirajan U, Ghosh B, Sinha Roy S, and Agrawal A. Computational classification of mitochondrial shapes reflects stress and redox state. Cell Death Dis 4: e461, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, Bhattacharya SS, and Wissinger B. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet 26: 211–215, 2000 [DOI] [PubMed] [Google Scholar]
- 5. Ali S. and McStay GP. Regulation of mitochondrial dynamics by proteolytic processing and protein turnover. Antioxidants (Basel) 7: E105, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alirol E. and Martinou JC. Mitochondria and cancer: is there a morphological connection? Oncogene 25: 4706–4716, 2006 [DOI] [PubMed] [Google Scholar]
- 7. Almassalha LM, Bauer GM, Chandler JE, Gladstein S, Cherkezyan L, Stypula-Cyrus Y, Weinberg S, Zhang D, Thusgaard Ruhoff P, Roy HK, Subramanian H, Chandel NS, Szleifer I, and Backman V. Label-free imaging of the native, living cellular nanoarchitecture using partial-wave spectroscopic microscopy. Proc Natl Acad Sci U S A 113: E6372–E6381, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E, and Langer T. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol 204: 919–929, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anton F, Dittmar G, Langer T, and Escobar-Henriques M. Two deubiquitylases act on mitofusin and regulate mitochondrial fusion along independent pathways. Mol Cell 49: 487–498, 2013 [DOI] [PubMed] [Google Scholar]
- 10. Appelhans T. and Busch KB. Dynamic imaging of mitochondrial membrane proteins in specific sub-organelle membrane locations. Biophys Rev 9: 345–352, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Archer SL. Mitochondrial dynamics - mitochondrial fission and fusion in human diseases. N Engl J Med 369: 2236–2251, 2013 [DOI] [PubMed] [Google Scholar]
- 12. Aristov A, Lelandais B, Rensen E, and Zimmer C. ZOLA-3D allows flexible 3D localization microscopy over an adjustable axial range. Nat Commun 9: 2409, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aviram R, Manella G, Kopelman N, Neufeld-Cohen A, Zwighaft Z, Elimelech M, Adamovich Y, Golik M, Wang C, Han X, and Asher G. Lipidomics analyses reveal temporal and spatial lipid organization and uncover daily oscillations in intracellular organelles. Mol Cell 62: 636–648, 2016 [DOI] [PubMed] [Google Scholar]
- 14. Baba T, Kashiwagi Y, Arimitsu N, Kogure T, Edo A, Maruyama T, Nakao K, Nakanishi H, Kinoshita M, Frohman MA, Yamamoto A, and Tani K. Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. J Biol Chem 289: 11497–11511, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baev AY, Elustondo PA, Negoda A, and Pavlov EV. Osmotic regulation of the mitochondrial permeability transition pore investigated by light scattering, fluorescence and electron microscopy techniques. Anal Biochem 552: 38–44, 2018 [DOI] [PubMed] [Google Scholar]
- 16. Ban T, Ishihara T, Kohno H, Saita S, Ichimura A, Maenaka K, Oka T, Mihara K, and Ishihara N. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat Cell Biol 19: 856–863, 2017 [DOI] [PubMed] [Google Scholar]
- 17. Barbot M, Jans DC, Schulz C, Denkert N, Kroppen B, Hoppert M, Jakobs S, and Meinecke M. MIC10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab 21: 756–763, 2015 [DOI] [PubMed] [Google Scholar]
- 18. Baricault L, Ségui B, Guégand L, Olichon A, Valette A, Larminat F, and Lenaers G. OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp Cell Res 313: 3800–3808, 2007 [DOI] [PubMed] [Google Scholar]
- 19. Bartolák-Suki E, Imsirovic J, Nishibori Y, Krishnan R, and Suki B. Regulation of mitochondrial structure and dynamics by the cytoskeleton and mechanical factors. Int J Mol Sci 18: pii: , 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Basit F, van Oppen LM, Schöckel L, Bossenbroek HM, van Emst-de Vries SE, Hermeling JC, Grefte S, Kopitz C, Heroult M, Hgm Willems P, and Koopman WJ. Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells. Cell Death Dis 8: e2716, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bast A. and Haenen GR. Ten misconceptions about antioxidants. Trends Pharm Sci 34: 430–436, 2011 [DOI] [PubMed] [Google Scholar]
- 22. Baumuratov AS, Antony PM, Ostaszewski M, He F, Salamanca L, Antunes L, Weber J, Longhino L, Derkinderen P, Koopman WJ, and Diederich NJ. Enteric neurons from Parkinson's disease patients display ex vivo aberrations in mitochondrial structure. Sci Rep 6: 33117, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Belenguer P. and Pellegrini L. The dynamin GTPase OPA1: more than mitochondria? Biochim Biophys Acta 1833: 176–183, 2013 [DOI] [PubMed] [Google Scholar]
- 24. Benard G. and Rossignol R. Ultrastructure of the mitochondrion and its bearing on function and bioenergetics. Antioxid Redox Signal 10: 1313–1342, 2008 [DOI] [PubMed] [Google Scholar]
- 25. Bénichou O, Chevalier C, Klafter J, Meyer B, and Voituriez R. Geometry-controlled kinetics. Nat Chem 2: 472–477, 2010 [DOI] [PubMed] [Google Scholar]
- 26. Bernardi P, Rasola A, Forte M, and Lippe G. The mitochondrial permeability transition pore: channel formation by F-ATP synthase, integration in signal transduction, and role in pathophysiology. Physiol Rev 95: 1111–1155, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhargava P. and Schnellmann RG. Mitochondrial energetics in the kidney. Nat Rev Nephrol 13: 629–646, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhuvanendran S, Salka K, Rainey K, Sreetama SC, Williams E, Leeker M, Prasad V, Boyd J, Patterson GH, Jaiswal JK, and Colberg-Poley AM. Superresolution imaging of human cytomegalovirus vMIA localization in sub-mitochondrial compartments. Viruses 6: 1612–1636, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Birsa N, Norkett R, Higgs N, Lopez-Domenech G, and Kittler JT. Mitochondrial trafficking in neurons and the role of the Miro family of GTPase proteins. Biochem Soc Trans 41: 1525–1531, 2013 [DOI] [PubMed] [Google Scholar]
- 30. Blanchet L, Smeitink JA, van Emst-de Vries SE, Vogels C, Pellegrini M, Jonckheere AI, Rodenburg RJ, Buydens LM, Beyrath J, Willems PH, and Koopman WJ. Quantifying small molecule phenotypic effects using mitochondrial morpho-functional fingerprinting and machine learning. Sci Rep 5: 8035, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bohnert M, Zerbes RM, Davies KM, Mühleip AW, Rampelt H, Horvath SE, Boenke T, Kram A, Perschil I, Veenhuis M, Kühlbrandt W, van der Klei IJ, Pfanner N, and van der Laan M. Central role of MIC10 in the mitochondrial contact site and cristae organizing system. Cell Metab 21: 747–755, 2015 [DOI] [PubMed] [Google Scholar]
- 32. Boissan M, Montagnac G, Shen Q, Griparic L, Guitton J, Romao M, Sauvonnet N, Lagache T, Lascu I, Raposo G, Desbourdes C, Schlattner U, Lacombe ML, Polo S, van der Bliek AM, Roux A, and Chavrier P. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science 344: 1510–1515, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bonekamp NA, Vormund K, Jacob R, and Schrader M. Dynamin-like protein 1 at the Golgi complex: a novel component of the sorting/targeting machinery en route to the plasma membrane. Exp Cell Res 316: 3454–3467, 2010 [DOI] [PubMed] [Google Scholar]
- 34. Bordt EA, Clerc P, Roelofs BA, Saladino AJ, Tretter L, Adam-Vizi V, Cherok E, Khalil A, Yadava N, Ge SX, Francis TC, Kennedy NW, Picton LK, Kumar T, Uppuluri S, Miller AM, Itoh K, Karbowski M, Sesaki H, Hill RB, and Polster BM. The putative Drp1 inhibitor mdivi-1 is a reversible mitochondrial complex I inhibitor that modulates reactive oxygen species. Dev Cell 40: 583.e6–594.e6, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Böttinger L, Ellenrieder L, and Becker T. How lipids modulate mitochondrial protein import. J Bioenerg Biomembr 48: 125–135, 2016 [DOI] [PubMed] [Google Scholar]
- 36. Boustany NN, Drezek R, and Thakor NV. Calcium-induced alterations in mitochondrial morphology quantified in situ with optical scatter imaging. Biophys J 83: 1691–1700, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boutant M, Kulkarni SS, Joffraud M, Ratajczak J, Valera-Alberni M, Combe R, Zorzano A, and Cantó C. Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J 36: 1543–1558, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boyd KJ, Alder NN, and May ER. Buckling under pressure: curvature-based lipid segregation and stability modulation in cardiolipin-containing bilayers. Langmuir 33: 6937–6946, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bruton J, Jeffries GD, and Westerblad H. Usage of a localised microflow device to show that mitochondrial networks are not extensive in skeletal muscle fibres. PLoS One 9: e108601, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buck MD, O'Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJ, Chen Q, Huang SC, O'Neill CM, Edelson BT, Pearce EJ, Sesaki H, Huber TB, Rambold AS, and Pearce EL. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166: 63–76, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bui HT. and Shaw JM. Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr Biol 23: R891–R899, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Calamita G, Ferri D, Gena P, Liquori GE, Cavalier A, Thomas D, and Svelto M. The inner mitochondrial membrane has aquaporin-8 water channels and is highly permeable to water. J Biol Chem 280: 17149–17153, 2005 [DOI] [PubMed] [Google Scholar]
- 43. Calvo SE, Clauser KR, and Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44: D1251–D1257, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Campanella M, Casswell E, Chong S, Farah Z, Wieckowski MR, Abramov AY, Tinker A, and Duchen MR. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab 8: 13–25, 2008 [DOI] [PubMed] [Google Scholar]
- 45. Campanella M, Seraphim A, Abeti R, Casswell E, Echave P, and Duchen MR. IF1, the endogenous regulator of the F1Fo-ATPsynthase, defines mitochondrial volume fraction in HeLa cells by regulating autophagy. Biochim Biophys Acta 1787: 393–401, 2009 [DOI] [PubMed] [Google Scholar]
- 46. Cao YL, Meng S, Chen Y, Feng JX, Gu DD, Yu B, Li YJ, Yang JY, Liao S, Chan DC, and Gao S. MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature 542: 372–376, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, and Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell 14: 193–204, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Casteilla L, Devin A, Carriere A, Salin B, Schaeffer J, and Rigoulet M. Control of mitochondrial volume by mitochondrial metabolic water. Mitochondrion 11: 862–866, 2011 [DOI] [PubMed] [Google Scholar]
- 49. Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, and Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A 105: 15803–15808, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cerqua C, Anesti V, Pyakurel A, Liu D, Naon D, Wiche G, Baffa R, Dimmer KS, and Scorrano L. Trichoplein/mitostatin regulates endoplasmic reticulum-mitochondria juxtaposition. EMBO Rep 11: 854–860, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet 46: 265–287, 2012 [DOI] [PubMed] [Google Scholar]
- 52. Chang CR. and Blackstone C. Drp1 phosphorylation and mitochondrial regulation. EMBO Rep 8: 1088–1089, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen H. and Chan DC. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stem cells. Cell Metab 26: 39–48, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen Y, Csordás G, Jowdy C, Schneider TG, Csordás N, Wang W, Liu Y, Kohlhaas M, Meiser M, Bergem S, Nerbonne JM, Dorn GW, and Maack C., 2nd Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetics responses via interorganelle Ca2+ cross talk. Circ Res 111: 863–875, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen KH, Dasgupta A, Ding J, Indig FE, Ghosh P, and Longo DL. Role of mitofusin 2 (Mfn2) in controlling cellular proliferation. FASEB J 28: 382–394, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, and Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 160: 189–200, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen Y. and Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 340: 471–475, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen H, McCaffery JM, and Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell 130: 548–562, 2007 [DOI] [PubMed] [Google Scholar]
- 59. Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, and Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141: 280–289, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chevrollier A, Cassereau J, Ferré M, Alban J, Desquiret-Dumas V, Gueguen N, Amati-Bonneau P, Procaccio V, Bonneau D, and Reynier P. Standardized mitochondrial analysis gives new insights into mitochondrial dynamics and OPA1 function. Int J Biochem Cell Biol 44: 980–988, 2012 [DOI] [PubMed] [Google Scholar]
- 61. Chinopoulos C, Gerencser AA, Mandi M, Mathe K, Töröcsik B, Doczi J, Turiak L, Kiss G, Konràd C, Vajda S, Vereczki V, Oh RJ, and Adam-Vizi V. Forward operation of adenine nucleotide translocase during F0F1-ATPase reversal: critical role of matrix substrate-level phosphorylation. FASEB J 24: 2405–2416, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cho B, Cho HM, Jo Y, Kim HD, Song M, Moon C, Kim H, Kim K, Sesaki H, Rhyu IJ, Kim H, and Sun W. Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat Commun 8: 15754, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cho B, Cho HM, Kim HJ, Jeong J, Park SK, Hwang EM, Park JY, Kim WR, Kim H, and Sun W. CDK5-dependent inhibitory phosphorylation of Drp1 during neuronal maturation. Exp Mol Med 46: e105, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, and Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol 8: 1255–1262, 2006 [DOI] [PubMed] [Google Scholar]
- 65. Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D'Adamio L, Derks C, Dejaegere T, Pellegrini L, D'Hooge R, Scorrano L, and De Strooper B. Mitochondrial rhomboid PARL regulates cytochrome-c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126: 163–175, 2006 [DOI] [PubMed] [Google Scholar]
- 66. Civiletto G, Varanita T, Cerutti R, Gorletta T, Barbaro S, Marchet S, Lamperti C, Viscomi C, Scorrano L, and Zeviani M. Opa1 overexpression ameliorates the phenotype of two mitochondrial disease mouse models. Cell Metab 21: 845–854, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cogliati S, Enriquez JA, and Scorrano L. Mitochondrial cristae: where beauty meets functionality. Trends Biochem Sci 41: 261–273, 2016 [DOI] [PubMed] [Google Scholar]
- 68. Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC, Perales-Clemente E, Salviati L, Fernandez-Silva P, Enriquez JA, and Scorrano L. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155: 160–171, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Collins TJ. and Bootman MD. Mitochondria are morphologically heterogeneous within cells. J Exp Biol 206: 1993–2000, 2003 [DOI] [PubMed] [Google Scholar]
- 70. Covill-Cooke C, Howden JH, Birsa N, and Kittler JT. Ubiquitination at the mitochondria in neuronal health and disease. Neurochem Int 117: 55–64, 2018 [DOI] [PubMed] [Google Scholar]
- 71. Cribbs JT. and Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep 8: 939–944, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Csordás G, Weaver D, and Hajnóczky G. Endoplasmic reticular-mitochondrial contactology: structure and signaling functions. Trends Cell Biol 28: 523–540, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Daniele T, Hurbain I, Vago R, Casari G, Raposo G, Tacchetti C, and Schiaffino MV. Mitochondria and melanosomes establish physical contacts modulated by Mfn2 and involved in organelle biogenesis. Curr Biol 24: 393–403, 2014 [DOI] [PubMed] [Google Scholar]
- 74. Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, and Taylor SS. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem 286: 2918–2932, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Daste F, Sauvanet C, Bavdek A, Baye J, Pierre F, Le Borgne R, David C, Rojo M, Fuchs P, and Tareste D. The heptad repeat domain 1 of Mitofusin has membrane destabilization function in mitochondrial fusion. EMBO Rep 19: e43637, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Daum B, Walter A, Horst A, Osiewacz HD, and Kühlbrandt W. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc Natl Acad Sci U S A 110: 15301–15306, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. de Brito OM. and Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456: 605–610, 2008 [DOI] [PubMed] [Google Scholar]
- 78. De Vos KJ. and Sheetz MP. Visualization and quantification of mitochondrial dynamics in living animal cells. Methods Cell Biol 80: 627–682, 2007 [DOI] [PubMed] [Google Scholar]
- 79. Del Dotto V, Fogazza M, Carelli V, Rugolo M, and Zanna C. Eight human OPA1 isoforms, long and short: what are they for? Biochim Biophys Acta 1859: 263–269, 2018 [DOI] [PubMed] [Google Scholar]
- 80. Del Dotto V, Fogazza M, Lenaers G, Rugolo M, Carelli V, and Zanna C. OPA1: how much do we know to approach therapy? Pharmacol Res 131: 199–210, 2018 [DOI] [PubMed] [Google Scholar]
- 81. Del Dotto V, Mishra P, Vidoni S, Fogazza M, Maresca A, Caporali L, McCaffery JM, Cappelletti M, Baruffini E, Lenaers G, Chan D, Rugolo M, Carelli V, and Zanna C. OPA1 isoforms in the hierarchical organization of mitochondrial functions. Cell Rep 19: 2557–2571, 2017 [DOI] [PubMed] [Google Scholar]
- 82. Dickey AS. and Strack S. PKA/AKAP1 and PP2A/Bβ2 regulate neuronal morphogenesis via Drp1 phosphorylation and mitochondrial bioenergetics. J Neurosci 31: 15716–15726, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dieteren CEJ. Mitochondrial complex I assembly: an exploration in the living cell [PhD thesis]. Nijmegen (The Netherlands): Radboud University; 2010 [Google Scholar]
- 84. Dieteren CE, Gielen SC, Nijtmans LG, Smeitink JA, Swarts HG, Brock R, Willems PH, and Koopman WJ. Solute diffusion is hindered in the mitochondrial matrix. Proc Natl Acad Sci U S A 108: 8657–8662, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dieteren CE, Willems PH, Vogel RO, Swarts HG, Fransen J, Roepman R, Crienen G, Smeitink JA, Nijtmans LG, and Koopman WJ. Subunits of mitochondrial complex I exist as part of matrix- and membrane-associated subcomplexes in living cells. J Biol Chem 283: 34753–34761, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dietrich MO, Liu ZW, and Horvath TL. Mitochondrial dynamics controlled by mitofusins regulate Agrp neuronal activity and diet-induced obesity. Cell 155: 188–199, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dikov D. and Bereiter-Hahn J. Inner membrane dynamics in mitochondria. J Struct Biol 183: 455–466, 2013 [DOI] [PubMed] [Google Scholar]
- 88. Din S, Mason M, Völkers M, Johnson B, Cottage CT, Wang Z, Joyo AY, Quijada P, Erhardt P, Magnuson NS, Konstandin MH, and Sussman MA. Pim-1 preserves mitochondrial morphology by inhibiting dynamin-related protein 1 translocation. Proc Natl Acad Sci U S A 110: 5969–5974, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Distelmaier F, Valsecchi F, Forkink M, van Emst-de Vries S, Swarts HG, Rodenburg RJ, Verwiel ET, Smeitink JA, Willems PH, and Koopman WJ. Trolox-sensitive reactive oxygen species regulate mitochondrial morphology, oxidative phosphorylation and cytosolic calcium handling in healthy cells. Antioxid Redox Signal 17: 1657–1669, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Distelmaier F, Visch HJ, Smeitink JA, Mayatepek E, Koopman WJ, and Willems PH. The antioxidant Trolox restores mitochondrial membrane potential and Ca2+-stimulated ATP production in human complex I deficiency. J Mol Med (Berl) 87: 515–522, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dix JA. and Verkman AS. Crowding effects on diffusion in solutions and cells. Annu Rev Biophys 37: 247–263, 2008 [DOI] [PubMed] [Google Scholar]
- 92. Dorn GW., 2nd Mitochondrial dynamism and heart disease: changing shape and shaping change. EMBO Mol Med 7: 865–877, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dudek J. Role of Cardiolipin in mitochondrial signaling pathways. Front Cell Dev Biol 5: 90, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Egner A, Jakobs S, and Hell SW. Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. Proc Natl Acad Sci U S A 99: 3370–3375, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, and Langer T. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol 187: 1023–1036, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. El Fissi N, Rojo M, Aouane A, Karatas E, Poliacikova G, David C, Royet J, and Rival T. Mitofusin gain and loss of function drive pathogenesis in Drosophila models of CMT2A neuropathy. EMBO Rep 19: e45241, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci 26: 597–604, 2001 [DOI] [PubMed] [Google Scholar]
- 98. Enríquez JA. Supramolecular organization of respiratory complexes. Annu Rev Physiol 78: 533–561, 2016 [DOI] [PubMed] [Google Scholar]
- 99. Ernster L. and Schatz G. Mitochondria: a historical review. J Cell Biol 91: 227s–255s, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Escobar-Henriques M. and Anton F. Mechanistic perspective of mitochondrial fusion: tubulation vs. fragmentation. Biochim Biophys Acta 1833: 162–175, 2013 [DOI] [PubMed] [Google Scholar]
- 101. Faccenda D, Nakamura J, Gorini G, Dhoot GK, Piacentini M, Yoshida M, and Campanella M. Control of mitochondrial remodeling by the ATPase inhibitory factor 1 unveils a pro-survival relay via OPA1. Cell Rep 18: 1869–1883, 2017 [DOI] [PubMed] [Google Scholar]
- 102. Filadi R, Greotti E, Turacchio G, Luini A, Pozzan T, and Pizzo P. Mitofusin 2 ablation increases endoplasmic reticulum-mitochondria coupling. Proc Natl Acad Sci U S A 112: E2174–E2181, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Flippo KH. and Strack S. Mitochondrial dynamics in neuronal injury, development and plasticity. J Cell Sci 130: 671–681, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Fornasiero EF. and Opazo F. Super-resolution imaging for cell biologists: concepts, applications, current challenges and developments. Bioessays 37: 436–451, 2015 [DOI] [PubMed] [Google Scholar]
- 105. Franco A, Kitsis RN, Fleischer JA, Gavathiotis E, Kornfeld OS, Gong G, Biris N, Benz A, Qvit N, Donnelly SK, Chen Y, Mennerick S, Hodgson L, Mochly-Rosen D, and Dorn GW., II Correcting mitochondrial fusion by manipulating mitofusin conformations. Nature 540: 74–79, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Francy CA, Clinton RW, Fröhlich C, Murphy C, and Mears JA. Cryo-EM studies of Drp1 reveal cardiolipin interactions that activate the helical oligomer. Sci Rep 7: 10744, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Frey TG, Perkins GA, and Ellisman MH. Electron tomography of membrane-bound cellular organelles. Annu Rev Biophys Biomol Struct 35: 199–224, 2006 [DOI] [PubMed] [Google Scholar]
- 108. Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, and Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126: 177–189, 2006 [DOI] [PubMed] [Google Scholar]
- 109. Frieden M, Arnaudeau S, Castelbou C, and Demaurex N. Subplasmalemmal mitochondria modulate the activity of plasma membrane Ca2+-ATPases. J Biol Chem 280: 43198–43208, 2005 [DOI] [PubMed] [Google Scholar]
- 110. Frieden M, James D, Castelbou C, Danckaert A, Martinou JC, and Demaurex N. Ca2+ homeostasis during mitochondrial fragmentation and perinuclear clustering induced by hFis1. J Biol Chem 279: 22704–22714, 2004 [DOI] [PubMed] [Google Scholar]
- 111. Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, and Voeltz GK. ER tubules mark sites of mitochondrial division. Science 334: 358–362, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Fröhlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, and Daumke O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J 32: 1280–1292, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Fujioka H, Tandler B, Consolo MC, and Karnik P. Division of mitochondria in cultured human fibroblasts. Microsc Res Tech 76: 1213–1216, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gandre-Babbe S. and van der Bliek AM. The novel tail-anchored membrane protein MFF controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 19: 2402–2412, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Gawlowski T, Suarez J, Scott B, Torres-Gonzalez M, Wang H, Schwappacher R, Han X, Yates JR., 3rd Hoshijima M, and Dillmann W. Modulation of dynamin-related protein 1 (DRP1) function by increased O-linked-β-N-acetylglucosamine modification (O-GlcNAc) in cardiac myocytes. J Biol Chem 287: 30024–30034, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Giacomello M. and Scorrano L. The INs and OUTs of mitofusins. J Cell Biol 217: 439–440, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Gilkerson RW, Selker JM, and Capaldi RA. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett 546: 355–358, 2003 [DOI] [PubMed] [Google Scholar]
- 118. Giorgi C, Missiroli S, Patergnani S, Duszynski J, Wieckowski MR, and Pinton P. Mitochondria-associated membranes: composition, molecular mechanisms, and physiopathological implications. Antioxid Redox Signal 22: 995–1019, 2015 [DOI] [PubMed] [Google Scholar]
- 119. Glytsou C, Calvo E, Cogliati S, Mehrotra A, Anastasia I, Rigoni G, Raimondi A, Shintani N, Loureiro M, Vazquez J, Pellegrini L, Enriquez JA, Scorrano L, and Soriano ME. Optic atrophy 1 is epistatic to the core MICOS component MIC60 in mitochondrial cristae shape control. Cell Rep 17: 3024–3034, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Gomes LC, Di Benedetto G, and Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 13: 589–598, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gómez-Suaga P, Bravo-San Pedro JM, González-Polo RA, Fuentes JM, and Niso-Santano M. ER-mitochondria signaling in Parkinson's disease. Cell Death Dis 9: 337, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gonzalez AS, Elguero ME, Finocchietto P, Holod S, Romorini L, Miriuka SG, Peralta JG, Poderoso JJ, and Carreras MC. Abnormal mitochondrial fusion-fission balance contributes to the progression of experimental sepsis. Free Radic Res 48: 769–783, 2014 [DOI] [PubMed] [Google Scholar]
- 123. Gottlieb E, Armour SM, Harris MH, and Thompson CB. Mitochondrial membrane potential regulates matrix configuration and cytochrome-c release during apoptosis. Cell Death Differ 10: 709–717, 2003 [DOI] [PubMed] [Google Scholar]
- 124. Gottschalk B, Klec C, Waldeck-Weiermair M, Malli R, and Graier WF. Intracellular Ca2+ release decelerates mitochondrial cristae dynamics within the junctions to the endoplasmic reticulum. Pflugers Arch 470: 1193–1203, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Grau T, Burbulla LF, Engl G, Delettre C, Delprat B, Oexle K, Leo-Kottler B, Roscioli T, Krüger R, Rapaport D, Wissinger B, Schimpf-Linzenbold S. A novel heterozygous OPA3 mutation located in the mitochondrial target sequence results in altered steady-state levels and fragmented mitochondrial network. J Med Genet 50: 848–858, 2013 [DOI] [PubMed] [Google Scholar]
- 126. Gray NE. and Quinn JF. Alterations in mitochondrial number and function in Alzheimer's disease fibroblasts. Metab Brain Dis 30: 1275–1278, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Grima R. and Schnell S. How reaction kinetics with time-dependent rate coefficients differs from generalized mass action. Chemphyschem 7: 1422–1424, 2006 [DOI] [PubMed] [Google Scholar]
- 128. Griparic L, Kanazawa T, and van der Bliek AM. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol 178: 757–764, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Guillery O, Malka F, Landes T, Guillou E, Blackstone C, Lombès A, Belenguer P, Arnoult D, and Rojo M. Metalloprotease-mediated OPA1 processing is modulated by the mitochondrial membrane potential. Biol Cell 100: 315–325, 2008 [DOI] [PubMed] [Google Scholar]
- 130. Guo X, Disatnik MH, Monbureau M, Shamloo M, Mochly-Rosen D, and Qi X. Inhibition of mitochondrial fragmentation diminishes Huntington's disease-associated neurodegeneration. J Clin Invest 123: 5371–5388, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ha EE. and Frohman MA. Regulation of mitochondrial morphology by lipids. Biofactors 40: 419–424, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hackenbrock CR. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. I. Reversible ultrastructural changes with change in metabolic steady state in isolated liver mitochondria. J Cell Biol 30: 269–297, 1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Hahn A, Parey K, Bublitz M, Mills DJ, Zickermann V, Vonck J, Kühlbrandt W, and Meier T. Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol Cell 63: 445–456, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, and Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141: 656–667, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Hällberg BM. and Larsson NG. Making proteins in the powerhouse. Cell Metab 20: 226–240, 2014 [DOI] [PubMed] [Google Scholar]
- 136. Han XJ, Lu YF, Li SA, Kaitsuka T, Sato Y, Tomizawa K, Nairn AC, Takei K, Matsui H, and Matsushita M. CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol 182: 573–585, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Harper JW, Ordureau A, and Heo JM. Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol 19: 93–108, 2018 [DOI] [PubMed] [Google Scholar]
- 138. Head B, Griparic L, Amiri M, Gandre-Babbe S, and van der Bliek AM. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol 187: 959–966, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Helle SCJ, Feng Q, Aebersold MJ, Hirt L, Grüter RR, Vahid A, Sirianni A, Mostowy S, Snedeker JG, Šarić A, Idema T, Zambelli T, and Kornmann B. Mechanical force induces mitochondrial fission. Elife 6: e30292, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Hershberger KA, Martin AS, and Hirschey MD. Role of NAD+ and mitochondrial sirtuins in cardiac and renal diseases. Nat Rev Nephrol 13: 213–225, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Hirabayashi Y, Kwon SK, Paek H, Pernice WM, Paul MA, Lee J, Erfani P, Raczkowski A, Petrey DS, Pon LA, and Polleux F. ER-mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science 358: 623–630, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Hoitzing H, Johnston IG, and Jones NS. What is the function of mitochondrial networks? A theoretical assessment of hypotheses and proposal for future research. Bioessays 37: 687–700, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Horbay R. and Bilyy R. Mitochondrial dynamics during cell cycling. Apoptosis 21: 1327–1335, 2016 [DOI] [PubMed] [Google Scholar]
- 144. Horn SR, Thomenius MJ, Johnson ES, Freel CD, Wu JQ, Coloff JL, Yang CS, Tang W, An J, Ilkayeva OR, Rathmell JC, Newgard CB, and Kornbluth S. Regulation of mitochondrial morphology by APC/CCdh1-mediated control of Drp1 stability. Mol Biol Cell 22: 1207–1216, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Houtkooper RH, Cantó C, Wanders RJ, and Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 31: 194–223, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Huang B, Jones SA, Brandenburg B, and Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods 5: 1047–1052, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Iannetti EF, Smeitink JA, Beyrath J, Willems PH, and Koopman WJ. Multiplexed high-content analysis of mitochondrial morphofunction using live-cell microscopy. Nat Protoc 11: 1693–1710, 2016 [DOI] [PubMed] [Google Scholar]
- 148. Iannetti EF, Willems PH, Pellegrini M, Beyrath J, Smeitink JA, Blanchet L, and Koopman WJ. Toward high-content screening of mitochondrial morphology and membrane potential in living cells. Int J Biochem Cell Biol 63: 66–70, 2015 [DOI] [PubMed] [Google Scholar]
- 149. Ikon N, and Ryan RO. Cardiolipin and mitochondrial cristae organization. Biochim Biophys Acta, 1859: 1156–1163 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, and Noji H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci U S A 106: 15651–15656, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ishihara N, Fujita Y, Oka T, and Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J 25: 2966–2977, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, and Mihara K. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol 11: 958–966, 2009 [DOI] [PubMed] [Google Scholar]
- 153. Itoh K, Adachi Y, Yamada T, Suzuki TL, Otomo T, McBride H, Yoshimori T, Iijima M, and Sesaki H. A brain-enriched Drp1 isoform associates with lysomes, late endosomes and the plasma membrane. J Biol Chem 293: 11809–11822, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Iwasawa R, Mahul-Mellier AL, Datler C, Pazarentzos E, and Grimm S. Fis1 and BAP31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J 30: 556–568, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Jacob M, Schindler T, Balbach J, and Schmid FX. Diffusion control in an elementary protein folding reaction. Proc Natl Acad Sci U S A 94: 5622–5627, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Jahani-Asl A, Huang E, Irrcher I, Rashidian J, Ishihara N, Lagace DC, Slack RS, and Park DS. CDK5 phosphorylates DRP1 and drives mitochondrial defects in NMDA-induced neuronal death. Hum Mol Genet 24: 4573–4583, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Jakobs S. and Wurm CA. Super-resolution microscopy of mitochondria. Curr Opin Chem Biol 20: 9–15, 2014 [DOI] [PubMed] [Google Scholar]
- 158. Jans DC, Wurm CA, Riedel D, Wenzel D, Stagge F, Deckers M, Rehling P, and Jakobs S. STED super-resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc Natl Acad Sci U S A 110: 8936–8941, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Jarosz J, Ghosh S, Delbridge LM, Petzer A, Hickey AJ, Crampin EJ, Hanssen E, and Rajagopal V. Changes in mitochondrial morphology and organization can enhance energy supply from mitochondrial oxidative phosphorylation in diabetic cardiomyopathy. Am J Physiol Cell Physiol 312: C190–C197, 2017 [DOI] [PubMed] [Google Scholar]
- 160. Javadov S, Chapa-Dubocq X, and Makarov V. Different approaches to modeling analysis of mitochondrial swelling. Mitochondrion 38: 58–70, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Jeon SM, Chandel NS, and Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature 485: 661–665, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Ji WK, Chakrabarti R, Fan X, Schoenfeld L, Strack S, and Higgs HN. Receptor-mediated Drp1 oligomerization on endoplasmic reticulum. J Cell Biol 216: 4123–4139, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Jia G, Aroor AR, Martinez-Lemus LA, and Sowers JR. Mitochondrial functional impairment in response to environmental toxins in the cardiorenal metabolic syndrome. Arch Toxicol 89: 147–153, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Jimenez L, Laporte D, Duvezin-Caubet S, Courtout F, and Sagot I. Mitochondrial ATP synthases cluster as discrete domains that reorganize with the cellular demand for oxidative phosphorylation. J Cell Sci 127: 719–726, 2014 [DOI] [PubMed] [Google Scholar]
- 165. Jones E, Gaytan N, Garcia I, Herrera A, Ramos M, Agarwala D, Rana M, Innis-Whitehouse W, Schuenzel E, and Gilkerson R. A threshold of transmembrane potential is required for mitochondrial dynamic balance mediated by DRP1 and OMA1. Cell Mol Life Sci 74: 1347–1363, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Joshi AU, Saw NL, Vogel H, Cunnigham AD, Shamloo M, and Mochly-Rosen D. Inhibition of Drp1/Fis1 interaction slows progression of amyotrophic lateral sclerosis. EMBO Mol Med 10: e8166, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Kaasik A, Safiulina D, Zharkovsky A, and Veksler V. Regulation of mitochondrial matrix volume. Am J Physiol Cell Physiol 292: C157–C163, 2007 [DOI] [PubMed] [Google Scholar]
- 168. Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, and Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. J Cell Biol 197: 535–551, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Kalia R, Wang RY, Yusuf A, Thomas PV, Agard DA, Shaw JM, and Frost A. Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature 558: 401–405, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Kameoka S, Adachi Y, Okamoto K, Iijima M, and Sesaki H. Phosphatidic acid and cardiolipin coordinate mitochondrial dynamics. Trends Cell Biol 28: 67–76, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Kanamaru Y, Sekine S, Ichijo H, and Takeda K. The phosphorylation-dependent regulation of mitochondrial proteins in stress responses. J Signal Transduct 2012: 931215, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Kang Y, Fielden LF, and Stojanovski D. Mitochondrial protein transport in health and disease. Semin Cell Dev Biol 76: 142–153, 2018 [DOI] [PubMed] [Google Scholar]
- 173. Karbowski M, Cleland MM, and Roelofs BA. Photoactivatable green fluorescent protein-based visualization and quantification of mitochondrial fusion and mitochondrial network complexity in living cells. Methods Enzymol 547: 57–73, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Kashatus DF. Restraining the divider: a Drp1-phospholipid interaction inhibits Drp1 activity and shifts the balance from mitochondrial fission to fusion. Mol Cell 63: 913–915, 2016 [DOI] [PubMed] [Google Scholar]
- 175. Khacho M. and Slack RS. Mitochondrial dynamics in the regulation of neurogenesis: from development to the adult brain. Dev Dyn 247: 47–53, 2018 [DOI] [PubMed] [Google Scholar]
- 176. Kim YM, Youn SW, Sudhahar V, Das A, Chandhri R, Cuervo Grajal H, Kweon J, Leanhart S, He L, Toth PT, Kitajewski J, Rehman J, Yoon Y, Cho J, Fukai T, and Ushio-Fukai M. Redox regulation of mitochondrial fission protein Drp1 by protein disulfide isomerase limits endothelial senescence. Cell Rep 23: 3565–3578, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Kitami T, Logan DJ, Negri J, Hasaka T, Tolliday NJ, Carpenter AE, Spiegelman BM, and Mootha VK. A chemical screen probing the relationship between mitochondrial content and cell size. PLoS One 7: e33755, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Klotzsch E, Smorodchenko A, Löfler L, Moldzio R, Parkinson E, Schütz GJ, and Pohl EE. Superresolution microscopy reveals spatial separation of UCP4 and F0F1-ATP synthase in neuronal mitochondria. Proc Natl Acad Sci U S A 112: 130–135, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Kluge MA, Fetterman JL, and Vita JA. Mitochondria and endothelial function. Circ Res 112: 1171–1188, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Knott AB, Perkins G, Schwarzenbacher R, and Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci 9: 505–518, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Koch J. and Brocard C. PEX11 proteins attract MFF and human Fis1 to coordinate peroxisomal fission. J Cell Sci 125: 3813–3826, 2012 [DOI] [PubMed] [Google Scholar]
- 182. Koch J, Feichtinger RG, Freisinger P, Pies M, Schrödl F, Iuso A, Sperl W, Mayr JA, Prokisch H, and Haack TB. Disturbed mitochondrial and peroxisomal dynamics due to loss of MFF causes Leigh-like encephalopathy, optic atrophy and peripheral neuropathy. J Med Genet 53: 270–278, 2016 [DOI] [PubMed] [Google Scholar]
- 183. Koch A, Yoon Y, Bonekamp NA, McNiven MA, and Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 16: 5077–5086, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Kong D, Xu L, Yu Y, Zhu W, Andrews DW, Yoon Y, and Kuo TH. Regulation of Ca2+-induced permeability transition by Bcl-2 is antagonized by Drpl and hFis1. Mol Cell Biochem 272: 187–199, 2005 [DOI] [PubMed] [Google Scholar]
- 185. Koopman WJ, Beyrath J, Fung CW, Koene S, Rodenburg RJ, Willems PH, and Smeitink JA. Mitochondrial disorders in children: toward development of small-molecule treatment strategies. EMBO Mol Med 8: 311–327, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Koopman WJ, Distelmaier F, Esseling JJ, Smeitink JA, and Willems PH. Computer-assisted live cell analysis of mitochondrial membrane potential, morphology and calcium handling. Methods 46: 304–311, 2008 [DOI] [PubMed] [Google Scholar]
- 187. Koopman WJ, Distelmaier F, Hink MA, Verkaart S, Wijers M, Fransen J, Smeitink JA, and Willems PH. Inherited complex I deficiency is associated with faster protein diffusion in the matrix of moving mitochondria. Am J Physiol Cell Physiol 294: C1124–C1132, 2008 [DOI] [PubMed] [Google Scholar]
- 188. Koopman WJ, Distelmaier F, Smeitink JA, and Willems PH. OXPHOS mutations and neurodegeneration. EMBO J 32: 9–29, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Koopman WJ, Hink MA, Verkaart S, Visch HJ, Smeitink JA, and Willems PH. Partial complex I inhibition decreases mitochondrial motility and increases matrix protein diffusion as revealed by fluorescence correlation spectroscopy. Biochim Biophys Acta 1767: 940–947, 2007 [DOI] [PubMed] [Google Scholar]
- 190. Koopman WJ, Nijtmans LG, Dieteren CE, Roestenberg P, Valsecchi F, Smeitink JA, and Willems PH. Mammalian mitochondrial complex I: biogenesis, regulation, and reactive oxygen species generation. Antioxid Redox Signal 12: 1431–1470, 2010 [DOI] [PubMed] [Google Scholar]
- 191. Koopman WJ, Verkaart S, van Emst-de Vries SE, Grefte S, Smeitink JA, Nijtmans LG, and Willems PH. Mitigation of NADH: ubiquinone oxidoreductase deficiency by chronic Trolox treatment. Biochim Biophys Acta 1777: 853–859, 2008 [DOI] [PubMed] [Google Scholar]
- 192. Koopman WJ, Verkaart S, Visch HJ, van der Westhuizen FH, Murphy MP, van den Heuvel LW, Smeitink JA, and Willems PH. Inhibition of complex I of the electron transport chain causes O2−·-mediated mitochondrial outgrowth. Am J Physiol Cell Physiol 288: C1440–C1450, 2005 [DOI] [PubMed] [Google Scholar]
- 193. Koopman WJ, Verkaart S, Visch HJ, van Emst-de Vries S, Nijtmans LG, Smeitink JA, and Willems PH. Human NADH: ubiquinone oxidoreductase deficiency: radical changes in mitochondrial morphology? Am J Physiol Cell Physiol 293: C22–C29, 2007 [DOI] [PubMed] [Google Scholar]
- 194. Koopman WJ, Visch HJ, Smeitink JA, and Willems PH. Simultaneous quantitative measurement and automated analysis of mitochondrial morphology, mass, potential, and motility in living human skin fibroblasts. Cytometry A 69: 1–12, 2006 [DOI] [PubMed] [Google Scholar]
- 195. Koopman WJ, Visch HJ, Verkaart S, van den Heuvel LW, Smeitink JA, and Willems PH. Mitochondrial network complexity and pathological decrease in complex I activity are tightly correlated in isolated human complex I deficiency. Am J Physiol Cell Physiol 289: C881–C890, 2005 [DOI] [PubMed] [Google Scholar]
- 196. Koopman WJ, Willems PH, and Smeitink JA. Monogenic mitochondrial disorders. N Engl J Med 366: 1132–1141, 2012 [DOI] [PubMed] [Google Scholar]
- 197. Korobova F, Ramabhadran V, and Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339: 464–467, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Korwitz A, Merkwirth C, Richter-Dennerlein R, Tröder SE, Sprenger HG, Quirós PM, López-Otín C, Rugarli EI, and Langer T. Loss of OMA1 delays neurodegeneration by preventing stress-induced OPA1 processing in mitochondria. J Cell Biol 212: 157–166, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, and Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science 305: 858–862, 2004 [DOI] [PubMed] [Google Scholar]
- 200. Kraus F. and Ryan MT. The constriction and scission machineries involved in mitochondrial fission. J Cell Sci 130: 2953–2960, 2017 [DOI] [PubMed] [Google Scholar]
- 201. Kuznetsov AV, Hermann M, Saks V, Hengster P, and Margreiter R. The cell-type specificity of mitochondrial dynamics. Int J Biochem Cell Biol 41: 1928–1939, 2009 [DOI] [PubMed] [Google Scholar]
- 202. Lackner LL, Horner JS, and Nunnari J. Mechanistic analysis of a dynamin effector. Science 325: 874–877, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Leal NS, Schreiner B, Pinho CM, Filadi R, Wiehager B, Karlström H, Pizzo P, and Ankarcrona M. Mitofusin-2 knockdown increases ER-mitochondria contact and decreases amyloid β-peptide production. J Cell Mol Med 20: 1686–1695, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, and Weissman AM. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol Cell 47: 547–557, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, and Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419, 2003 [DOI] [PubMed] [Google Scholar]
- 206. Lee WH, Higuchi H, Ikeda S, Macke EL, Takimoto T, Pattnaik BR, Liu C, Chu LF, Siepka SM, Krentz KJ, Rubinstein CD, Kalejta RF, Thomson JA, Mullins RF, Takahashi JS, Pinto LH, and Ikeda A. Mouse TMEM135 mutation reveals a mechanism involving mitochondrial dynamics that leads to age-dependent retinal pathologies. Elife 5: e19264, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Lee S, Jeong SY, Lim WC, Kim S, Park YY, Sun X, Youle RJ, and Cho H. Mitochondrial fission and fusion mediators, hFis1 and OPA1, modulate cellular senescence. J Biol Chem 282: 22977–22983, 2007 [DOI] [PubMed] [Google Scholar]
- 208. Lee S, Park YY, Kim SH, Nguyen OT, Yoo YS, Chan GK, Sun X, and Cho H. Human mitochondrial Fis1 links to cell cycle regulators at G2/M transition. Cell Mol Life Sci 71: 711–725, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Lee WK. and Thévenod F. A role for mitochondrial aquaporins in cellular life-and-death decisions? Am J Physiol Cell Physiol 291: C195–C202, 2006 [DOI] [PubMed] [Google Scholar]
- 210. Lee JE, Westrate LM, Wu H, Page C, and Voeltz GK. Multiple dynamin family members collaborate to drive mitochondrial division. Nature 540: 139–143, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Lee H. and Yoon Y. Mitochondrial fission and fusion. Biochem Soc Trans 44: 1725–1735, 2016 [DOI] [PubMed] [Google Scholar]
- 212. Legros F, Lombès A, Frachon P, and Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell 13: 4343–4354, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Lemieux H, Blier PU, and Gnaiger E. Remodeling pathway control of mitochondrial respiratory capacity by temperature in mouse heart: electron flow through the Q-junction in permeabilized fibers. Sci Rep 7: 2840, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Leonard AP, Cameron RB, Speiser JL, Wolf BJ, Peterson YK, Schnellmann RG, Beeson CC, and Rohrer B. Quantitative analysis of mitochondrial morphology and membrane potential in living cells using high-content imaging, machine learning, and morphological binning. Biochim Biophys Acta 1853: 348–360, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Lesnefsky EJ, Chen Q, Tandler B, and Hoppel CL. Mitochondrial dysfunction and myocardial Ischemia-Reperfusion: implications for novel therapies. Annu Rev Pharmacol Toxicol 57: 535–565, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Lewis SC, Uchiyama LF, and Nunnari J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353: aaf5549, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Li H, Alavian KN, Lazrove E, Mehta N, Jones A, Zhang P, Licznerski P, Graham M, Uo T, Guo J, Rahner C, Duman RS, Morrison RS, and Jonas EA. A Bcl-xL-Drp1 complex regulates synaptic vesicle membrane dynamics during endocytosis. Nat Cell Biol 15: 773–785, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Liemburg-Apers DC, Imamura H, Forkink M, Nooteboom M, Swarts HG, Brock R, Smeitink JA, Willems PH, and Koopman WJ. Quantitative glucose and ATP sensing in mammalian cells. Pharm Res 28: 2745–2757, 2011 [DOI] [PubMed] [Google Scholar]
- 219. Liemburg-Apers DC, Schirris TJ, Russel FG, Willems PH, and Koopman WJ. Mitoenergetic dysfunction triggers a rapid compensatory increase in steady-state glucose flux. Biophys J 109: 1372–1386, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Liemburg-Apers DC, Wagenaars JA, Smeitink JA, Willems PH, and Koopman WJ. Acute stimulation of glucose influx upon mitoenergetic dysfunction requires LKB1, AMPK, Sirt2 and mTOR-RAPTOR. J Cell Sci 129: 4411–4423, 2016 [DOI] [PubMed] [Google Scholar]
- 221. Lin SC. and Hardie DG. AMPK: sensing glucose as well as cellular energy status. Cell Metab 27: 299–313, 2018 [DOI] [PubMed] [Google Scholar]
- 222. Liu X. and Hajnóczky G. Altered fusion dynamics underlie unique morphological changes in mitochondria during hypoxia-reoxygenation stress. Cell Death Differ 18: 1561–1572, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Liu X, Weaver D, Shirihai O, and Hajnóczky G. Mitochondrial ‘kiss-and-run’: interplay between mitochondrial motility and fusion-fission dynamics. EMBO J 28: 3074–3089, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Liu Y. and Zhu X. Endoplasmic reticulum-mitochondria tethering in neurodegenerative diseases. Transl Neurodegener 6: 21, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Lizana L, Bauer B, and Orwar O. Controlling the rates of biochemical reactions and signaling networks by shape and volume changes. Proc Natl Acad Sci U S A 105: 4099–4104, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Lizana L, Konkoli Z, Bauer B, Jesorka A, and Orwar O. Controlling chemistry by geometry in nanoscale systems. Annu Rev Phys Chem 60: 449–468, 2009 [DOI] [PubMed] [Google Scholar]
- 227. Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, and Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci U S A 95: 6803–6808, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Long Q, Zhao D, Fan W, Yang L, Zhou Y, Qi J, Wang X, and Liu X. Modeling of Mitochondrial Donut Formation. Biophys J 109: 892–899, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Losón OC, Meng S, Ngo H, Liu R, Kaiser JT, and Chan DC. Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1. Protein Sci 24: 386–394, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Losón OC, Song Z, Chen H, and Chan DC. Fis1, MFF, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell 24: 659–667, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Lovy A, Molina AJ, Cerqueira FM, Trudeau K, and Shirihai OS. A faster, high resolution, mtPA-GFP-based mitochondrial fusion assay acquiring kinetic data of multiple cells in parallel using confocal microscopy. J Vis Exp 65: e3991, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Luchsinger LL, de Almeida MJ, Corrigan DJ, Mumau M, and Snoeck HW. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature 529: 528–531, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Lučič V, Rigort A, and Baumeister W. Cryo-electron tomography: the challenge of doing structural biology in situ. J Cell Biol 202: 407–419, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Lunt SY. and Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 27: 441–464, 2011 [DOI] [PubMed] [Google Scholar]
- 235. Luongo TS, Lambert JP, Gross P, Nwokedi M, Lombardi AA, Shanmughapriya S, Carpenter AC, Kolmetzky D, Gao E, van Berlo JH, Tsai EJ, Molkentin JD, Chen X, Madesh M, Houser SR, and Elrod JW. The mitochondrial Na+/Ca2+ exchanger is essential for Ca2+ homeostasis and viability. Nature 545: 93–97, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Macchi M, El Fissi N, Tufi R, Bentobji M, Liévens JC, Martins LM, Royet J, and Rival T. The Drosophila inner-membrane protein PMI controls crista biogenesis and mitochondrial diameter. J Cell Sci 126: 814–824, 2013 [DOI] [PubMed] [Google Scholar]
- 237. Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, and Kirchhausen T. Dynasore, a cell permeable inhibitor of dynamin. Dev Cell 10: 839–850, 2006 [DOI] [PubMed] [Google Scholar]
- 238. MacVicar T. and Langer T. OPA1 processing in cell death and disease—the long and short of it. J Cell Sci 129: 2297–2306, 2016 [DOI] [PubMed] [Google Scholar]
- 239. Madsen KL, Bhatia VK, Gether U, and Stamou D. BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett 584: 1848–1855, 2010 [DOI] [PubMed] [Google Scholar]
- 240. Mahdaviani K, Benador IY, Su S, Gharakhanian RA, Stiles L, Trudeau KM, Cardamone M, Enríquez-Zarralanga V, Ritou E, Aprahamian T, Oliveira MF, Corkey BE, Perissi V, Liesa M, and Shirihai OS. Mfn2 deletion in brown adipose tissue protects from insulin resistance and impairs thermogenesis. EMBO Rep 18: 1123–1138, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Mailloux RJ. Teaching the fundamentals of electron transfer reactions in mitochondria and the production and detection of reactive oxygen species. Redox Biol 4: 381–398, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Malena A, Loro E, Di Re M, Holt IJ, and Vergani L. Inhibition of mitochondrial fission favours mutant over wild-type mitochondrial DNA. Hum Mol Genet 18: 3407–3416, 2009 [DOI] [PubMed] [Google Scholar]
- 243. Malka F, Guillery O, Cifuentes-Diaz C, Guillou E, Belenguer P, Lombès A, and Rojo M. Separate fusion of outer and inner mitochondrial membranes. EMBO Rep 6: 853–859, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Mannella CA, Lederer WJ, and Jafri MS. The connection between inner membrane topology and mitochondrial function. J Mol Cell Cardiol 62: 51–57, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, Spudich J, and Lippincott-Schwartz J. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife 4: e08828, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Márquez-Jurado S, Díaz-Colunga J, das Neves RP, Martinez-Lorente A, Almazán F, Guantes R, and Iborra FJ. Mitochondrial levels determine variability in cell death by modulating apoptotic gene expression. Nat Commun 9: 389, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Martin J, Mahlke K, and Pfanner N. Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J Biol Chem 266: 18051–18057, 1991 [PubMed] [Google Scholar]
- 248. Matsuda T. and Nagai T. Quantitative measurement of intracellular protein dynamics using photobleaching or photoactivation of fluorescent proteins. Microscopy (Tokyo) 63: 403–408, 2014 [DOI] [PubMed] [Google Scholar]
- 249. Mattie S, Riemer J, Wideman JG, and McBride HM. A new mitofusin topology places the redox-regulated C terminus in the mitochondrial intermembrane space. J Cell Biol 217: 507–515, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, and Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol 18: 20–26, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Meeusen S, McCaffery JM, and Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science 305: 1747–1752, 2004 [DOI] [PubMed] [Google Scholar]
- 252. Mehta MM, Weinberg SE, and Chandel NS. Mitochondrial control of immunity: beyond ATP. Nat Rev Immunol 17: 608–620, 2017 [DOI] [PubMed] [Google Scholar]
- 253. Meinecke M, Wagner R, Kovermann P, Guiard B, Mick DU, Hutu DP, Voos W, Truscott KN, Chacinska A, Pfanner N, and Rehling P. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312: 1523–1526, 2006 [DOI] [PubMed] [Google Scholar]
- 254. Mejia EM. and Hatch GM. Mitochondrial phospholipids: role in mitochondrial function. J Bioenerg Biomembr 48: 99–112, 2016 [DOI] [PubMed] [Google Scholar]
- 255. Melcher M, Danhauser K, Seibt A, Degistirici Ö, Baertling F, Kondadi AK, Reichert AS, Koopman WJH, Willems PHGM, Rodenburg RJ, Mayatepek E, Meisel R, and Distelmaier F. Modulation of oxidative phosphorylation and redox homeostasis in mitochondrial NDUFS4 deficiency via mesenchymal stem cells. Stem Cell Res Ther 8: 150, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Löwer B, Wunderlich FT, von Kleist-Retzow JC, Waisman A, Westermann B, and Langer T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev 22: 476–488, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Merrill RA, Slupe AM, and Strack S. N-terminal phosphorylation of protein phosphatase 2A/Bβ2 regulates translocation to mitochondria, dynamin-related protein 1 dephosphorylation, and neuronal survival. FEBS J 280: 662–673, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Michalska BM, Kwapiszewska K, Szczepanowska J, Kalwarczyk T, Patalas-Krawczyk P, Szczepański K, Hołyst R, Duszyński J, and Szymański J. Insight into the fission mechanism by quantitative characterization of Drp1 protein distribution in the living cell. Sci Rep 8: 8122, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Miettinen TP. and Björklund M. Mitochondrial function and cell size: an allometric relationship. Trends Cell Biol 27: 393–402, 2017 [DOI] [PubMed] [Google Scholar]
- 260. Milenkovic D, Blaza JN, Larsson NG, and Hirst J. The enigma of the respiratory chain supercomplex. Cell Metab 25: 765–776, 2017 [DOI] [PubMed] [Google Scholar]
- 261. Mills EL, Kelly B, and O'Neill LAJ. Mitochondria are the powerhouses of immunity. Nat Immunol 18: 488–498, 2017 [DOI] [PubMed] [Google Scholar]
- 262. Minauro-Sanmiguel F, Wilkens S, and García JJ. Structure of dimeric mitochondrial ATP synthase: novel Fo bridging features and the structural basis of mitochondrial cristae biogenesis. Proc Natl Acad Sci U S A 102: 12356–12358, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Misko A, Jiang S, Wegorzewska I, Milbrandt J, and Baloh RH. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J Neurosci 30: 4232–4240, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Misgeld T, Kerschensteiner M, Bareyre FM, Burgess RW, and Lichtman JW. Imaging axonal transport of mitochondria in vivo. Nat Methods 4: 559–561, 2007 [DOI] [PubMed] [Google Scholar]
- 265. Mishra P, Carelli V, Manfredi G, and Chan DC. Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell Metab 19: 630–641, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Mishra P. and Chan DC. Metabolic regulation of mitochondrial dynamics. J Cell Biol 212: 379–387, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemiosmotic type of mechanism. Nature 191: 144–148, 1961 [DOI] [PubMed] [Google Scholar]
- 268. Mitra K, Wunder C, Roysam B, Lin G, and Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A 106: 11960–11965, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basañez G, Meda P, and Martinou JC. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell 142: 889–901, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Morita M, Prudent J, Basu K, Goyon V, Katsumura S, Hulea L, Pearl D, Siddiqui N, Strack S, McGuirk S, St-Pierre J, Larsson O, Topisirovic I, Vali H, McBride HM, Bergeron JJ, and Sonenberg N. mTOR controls mitochondrial dynamics and cell survival via MTFP1. Mol Cell 67: 922.e5–935.e5, 2017 [DOI] [PubMed] [Google Scholar]
- 271. Mourier A, Motori E, Brandt T, Lagouge M, Atanassov I, Galinier A, Rappl G, Brodesser S, Hultenby K, Dieterich C, and Larsson NG. Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. J Cell Biol 208: 429–442, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Muñoz JP, Ivanova S, Sánchez-Wandelmer J, Martínez-Cristóbal P, Noguera E, Sancho A, Díaz-Ramos A, Hernández-Alvarez MI, Sebastián D, Mauvezin C, Palacín M, and Zorzano A. Mfn2 modulates the UPR and mitochondrial function via repression of PERK. EMBO J 32: 2348–2361, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Murphy MP. Redox modulation by reversal of the mitochondrial nicotinamide nucleotide transhydrogenase. Cell Metab 22: 363–365, 2015 [DOI] [PubMed] [Google Scholar]
- 274. Musatov A. and Sedlák E. Role of cardiolipin in stability of integral membrane proteins. Biochimie 142: 102–111, 2017 [DOI] [PubMed] [Google Scholar]
- 275. Muster B, Kohl W, Wittig I, Strecker V, Joos F, Haase W, Bereiter-Hahn J, and Busch K. Respiratory chain complexes in dynamic mitochondria display a patchy distribution in life cells. PLoS One 5: e11910, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Nagaraj R, Gururaja-Rao S, Jones KT, Slattery M, Negre N, Braas D, Christofk H, White KP, Mann R, and Banerjee U. Control of mitochondrial structure and function by the Yorkie/YAP oncogenic pathway. Genes Dev 26: 2027–2037, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Nagashima S, Tokuyama T, Yonashiro R, Inatome R, and Yanagi S. Roles of mitochondrial ubiquitin ligase MITOL/MARCH5 in mitochondrial dynamics and diseases. J Biochem 155: 273–279, 2014 [DOI] [PubMed] [Google Scholar]
- 278. Nakamura N, Kimura Y, Tokuda M, Honda S, and Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep 7: 1019–1022, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Napoli E, Song G, Liu S, Espejo A, Perez CJ, Benavides F, and Giulivi C. Zdhhc13-dependent Drp1 S-palmitoylation impacts brain bioenergetics, anxiety, coordination and motor skills. Sci Rep 7: 12796, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Nemani N, Carvalho E, Tomar D, Dong Z, Ketschek A, Breves SL, Jaña F, Worth AM, Heffler J, Palaniappan P, Tripathi A, Subbiah R, Riitano MF, Seelam A, Manfred T, Itoh K, Meng S, Sesaki H, Craigen WJ, Rajan S, Shanmughapriya S, Caplan J, Prosser BL, Gill DL, Stathopulos PB, Gallo G, Chan DC, Mishra P, and Madesh M. MIRO-1 Determines mitochondrial shape transition upon GPCR activation and Ca2+ stress. Cell Rep 23: 1005–1019, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Neuspiel M, Zunino R, Gangaraju S, Rippstein P, and McBride H. Activated mitofusin 2 signals mitochondrial fusion, interferes with Bax activation, and reduces susceptibility to radical induced depolarization. J Biol Chem 280: 25060–25070, 2005 [DOI] [PubMed] [Google Scholar]
- 282. Ngoh GA, Papanicolaou KN, and Walsh K. Loss of mitofusin 2 promotes endoplasmic reticulum stress. J Biol Chem 287: 20321–20332, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Nguyen TB, Louie SM, Daniele JR, Tran Q, Dillin A, Zoncu R, Nomura DK, and Olzmann JA. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev Cell 42: 9.e5–21.e5, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Nicholls DG. and Ferguson SJ. Bioenergetics 4. Amsterdam: Academic Press, Elsevier, 2013 [Google Scholar]
- 285. Nicholls DG. and Lindberg O. Inhibited respiration and ATPase activity of rat liver mitochondria under conditions of matrix condensation. FEBS Lett 25: 61–64, 1972 [DOI] [PubMed] [Google Scholar]
- 286. Nickel AG, von Hardenberg A, Hohl M, Löffler JR, Kohlhaas M, Becker J, Reil JC, Kazakov A, Bonnekoh J, Stadelmaier M, Puhl SL, Wagner M, Bogeski I, Cortassa S, Kappl R, Pasieka B, Lafontaine M, Lancaster CR, Blacker TS, Hall AR, Duchen MR, Kästner L, Lipp P, Zeller T, Müller C, Knopp A, Laufs U, Böhm M, Hoth M, and Maack C. Reversal of mitochondrial transhydrogenase causes oxidative stress in heart failure. Cell Metab 22: 472–484, 2015 [DOI] [PubMed] [Google Scholar]
- 287. Nielson JR. and Rutter JP. Lipid-mediated signals that regulate mitochondrial biology. J Biol Chem 293: 7517–7521, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Nikolaisen J, Nilsson LI, Pettersen IK, Willems PH, Lorens JB, Koopman WJ, and Tronstad KJ. Automated quantification and integrative analysis of 2D and 3D mitochondrial shape and network properties. PLoS One 9: e101365, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Noguchi M. and Kasahara A. Mitochondrial dynamics coordinate cell differentiation. Biochem Biophys Res Commun 500: 59–64, 2018 [DOI] [PubMed] [Google Scholar]
- 290. Nogueira V, Devin A, Walter L, Rigoulet M, Leverve X, and Fontaine E. Effects of decreasing mitochondrial volume on the regulation of the permeability transition pore. J Bioenerg Biomembr 37: 25–33, 2005 [DOI] [PubMed] [Google Scholar]
- 291. Nowikovsky K, Schweyen RJ, and Bernardi P. Pathophysiology of mitochondrial volume homeostasis: potassium transport and permeability transition. Biochim Biophys Acta 1787: 345–350, 2009 [DOI] [PubMed] [Google Scholar]
- 292. Ohba Y, Sakuragi T, Kage-Nakadai E, Tomioka NH, Kono N, Imae R, Inoue A, Aoki J, Ishihara N, Inoue T, Mitani S, and Arai H. Mitochondria-type GPAT is required for mitochondrial fusion. EMBO J 32: 1265–1279, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, and Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem 278: 7743–7746, 2003 [DOI] [PubMed] [Google Scholar]
- 294. Olichon A, Elachouri G, Baricault L, Delettre C, Belenguer P, and Lenaers G. OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ 14: 682–692, 2007 [DOI] [PubMed] [Google Scholar]
- 295. Olveczky BP. and Verkman AS. Monte Carlo analysis of obstructed diffusion in three dimensions: application to molecular diffusion in organelles. Biophys J 74: 2722–2730, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Osellame LD, Singh AP, Stroud DA, Palmer CS, Stojanovski D, Ramachandran R, and Ryan MT. Cooperative and independent roles of the Drp1 adaptors MFF, MiD49 and MiD51 in mitochondrial fission. J Cell Sci 129: 2170–2181, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Otera H, Ishihara N, and Mihara K. New insights into the function and regulation of mitochondrial fission. Biochim Biophys Acta 1833: 1256–1268, 2013 [DOI] [PubMed] [Google Scholar]
- 298. Otera H, Miyata N, Kuge O, and Mihara K. Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J Cell Biol 212: 531–544, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, and Mihara K. MFF is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 191: 1141–1158, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Palmer CS, Elgass KD, Parton RG, Osellame LD, Stojanovski D, and Ryan MT. Adaptor proteins MiD49 and MiD51 can act independently of MFF and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J Biol Chem 288: 27584–27593, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, and Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep 12: 565–573, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Palmieri F. Diseases caused by defects of mitochondrial carriers: a review. Biochim Biophys Acta 1777: 564–578, 2008 [DOI] [PubMed] [Google Scholar]
- 303. Papadopoulos S, Jürgens KD, and Gros G. Protein diffusion in living skeletal muscle fibers: dependence on protein size, fiber type, and contraction. Biophys J 79: 2084–2094, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Park YY, Nguyen OT, Kang H, and Cho H. MARCH5-mediated quality control on acetylated Mfn1 facilitates mitochondrial homeostasis and cell survival. Cell Death Dis 5: e1172, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Partikian A, Olveczky B, Swaminathan R, Li Y, and Verkman AS. Rapid diffusion of green fluorescent protein in the mitochondrial matrix. J Cell Biol 140: 821–829, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Patten DA, Wong J, Khacho M, Soubannier V, Mailloux RJ, Pilon-Larose K, MacLaurin JG, Park DS, McBride HM, Trinkle-Mulcahy L, Harper ME, Germain M, and Slack RS. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J 33: 2676–2691, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Peralta S, Goffart S, Williams SL, Diaz F, Garcia S, Nissanka N, Area-Gomez E, Pohjoismäki J, and Moraes CT. ATAD3 controls mitochondrial cristae structure, influencing mtDNA replication and cholesterol levels in muscle. J Cell Sci 131, , 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Pernas L. and Scorrano L. Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu Rev Physiol 78: 505–531, 2016 [DOI] [PubMed] [Google Scholar]
- 309. Pfanner N, van der Laan M, Amati P, Capaldi RA, Caudy AA, Chacinska A, Darshi M, Deckers M, Hoppins S, Icho T, Jakobs S, Ji J, Kozjak-Pavlovic V, Meisinger C, Odgren PR, Park SK, Rehling P, Reichert AS, Sheikh MS, Taylor SS, Tsuchida N, van der Bliek AM, van der Klei IJ, Weissman JS, Westermann B, Zha J, Neupert W, and Nunnari J. Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J Cell Biol 204: 1083–1086, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Pham AH, Meng S, Chu QN, and Chan DC. Loss of Mfn2 results in progressive, retrograde degeneration of dopaminergic neurons in the nigrostriatal circuit. Hum Mol Genet 21: 4817–4826, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Picard M, McManus MJ, Csordás G, Várnai P, Dorn GW., 2nd Williams D. Hajnóczky G, and Wallace DC. Trans-mitochondrial coordination of cristae at regulated membrane junctions. Nat Commun 6: 6259, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Pich S, Bach D, Briones P, Liesa M, Camps M, Testar X, Palacín M, and Zorzano A. The Charcot–Marie–Tooth type 2A gene product, Mfn2, up-regulates fuel oxidation through expression of OXPHOS system. Hum Mol Genet 14: 1405–1415, 2005 [Google Scholar]
- 313. Pidoux G, Witczak O, Jarnæss E, Myrvold L, Urlaub H, Stokka AJ, Küntziger T, and Taskén K. Optic atrophy 1 is an A-kinase anchoring protein on lipid droplets that mediates adrenergic control of lipolysis. EMBO J 30: 4371–4386, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Pitts KR, Yoon Y, Krueger EW, and McNiven MA. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol Biol Cell 10: 4403–4417, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Porcelli AM, Ghelli A, Zanna C, Pinton P, Rizzuto R, and Rugolo M. pH difference across the outer mitochondrial membrane measured with a green fluorescent protein mutant. Biochem Biophys Res Commun 326: 799–804, 2005 [DOI] [PubMed] [Google Scholar]
- 316. Prinz WA. Bridging the GAP: membrane contact sites in signaling, metabolism, and organelle dynamics. J Cell Biol 205: 759–769, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Purnell PR. and Fox HS. Autophagy-mediated turnover of dynamin-related protein 1. BMC Neurosci 14: 86, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Pyakurel A, Savoia C, Hess D, and Scorrano L. Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol Cell 58: 244–254, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Qi X, Disatnik MH, Shen N, Sobel RA, and Mochly-Rosen D. Aberrant mitochondrial fission in neurons induced by protein kinase Cδ under oxidative stress conditions in vivo. Mol Biol Cell 22: 256–265, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Qi X, Qvit N, Su YC, and Mochly-Rosen D. A novel Drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. J Cell Sci 126: 789–802, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Qi Y, Yan L, Yu C, Guo X, Zhou X, Hu X, Huang X, Rao Z, Lou Z, and Hu J. Structures of human mitofusin 1 provide insight into mitochondrial tethering. J Cell Biol 215: 621–629, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Qian W, Wang J, Roginskaya V, McDermott LA, Edwards RP, Stolz DB, Llambi F, Green DR, and Van Houten B. Novel combination of mitochondrial division inhibitor 1 (mdivi-1) and platinum agents produces synergistic pro-apoptotic effect in drug resistant tumor cells. Oncotarget 5: 4180–4194, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Quirós PM, Ramsay AJ, Sala D, Fernández-Vizarra E, Rodríguez F, Peinado JR, Fernández-García MS, Vega JA, Enríquez JA, Zorzano A, and López-Otín C. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J 31: 2117–2133, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Rainbolt TK, Lebeau J, Puchades C, and Wiseman RL. Reciprocal degradation of YME1L and OMA1 adapts mitochondrial proteolytic activity during stress. Cell Rep 14: 2041–2049, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Rambold AS, Cohen S, and Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell 32: 678–692, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Rambold AS, Kostelecky B, Elia N, and Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A 108: 10190–10195, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Rampelt H, Zerbes RM, van der Laan M, and Pfanner N. Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics. Biochim Biophys Acta 1864: 737–746, 2017 [DOI] [PubMed] [Google Scholar]
- 328. Reddy PH. Inhibitors of mitochondrial fission as a therapeutic strategy for diseases with oxidative stress and mitochondrial dysfunction. J Alzheimers Dis 40: 245–256, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Reis Y, Bernardo-Faura M, Richter D, Wolf T, Brors B, Hamacher-Brady A, Eils R, and Brady NR. Multi-parametric analysis and modeling of relationships between mitochondrial morphology and apoptosis. PLoS One 7: e28694, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Renault TT, Floros KV, Elkholi R, Corrigan KA, Kushnareva Y, Wieder SY, Lindtner C, Serasinghe MN, Asciolla JJ, Buettner C, Newmeyer DD, and Chipuk JE. Mitochondrial shape governs BAX-induced membrane permeabilization and apoptosis. Mol Cell 57: 69–82, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Richter V, Palmer CS, Osellame LD, Singh AP, Elgass K, Stroud DA, Sesaki H, Kvansakul M, and Ryan MT. Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission. J Cell Biol 204: 477–486, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Rival T, Macchi M, Arnauné-Pelloquin L, Poidevin M, Maillet F, Richard F, Fatmi A, Belenguer P, and Royet J. Inner-membrane proteins PMI/TMEM11 regulate mitochondrial morphogenesis independently of the DRP1/MFN fission/fusion pathways. EMBO Rep 12: 223–230, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Rocha AG, Franco A, Krezel AM, Rumsey JM, Alberti JM, Knight WC, Biris N, Zacharioudakis E, Janetka JW, Baloh RH, Kitsis RN, Mochly-Rosen D, Townsend RR, Gavathiotis E, and Dorn GW. 2nd MFN2 Agonist reverse mitochondrial defects in preclinical models of Charcot–Marie–Tooth disease type 2A. Science 360: 336–341, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Rojo M, Legros F, Chateau D, and Lombès A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci 115: 1663–1674, 2002 [DOI] [PubMed] [Google Scholar]
- 335. Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, and Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res 64: 985–993, 2004 [DOI] [PubMed] [Google Scholar]
- 336. Rovira-Llopis S, Bañuls C, Diaz-Morales N, Hernandez-Mijares A, Rocha M, and Victor VM. Mitochondrial dynamics in type 2 diabetes: pathophysiological implications. Redox Biol 11: 637–645, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Roy M, Reddy PH, Iijima M, and Sesaki H. Mitochondrial division and fusion in metabolism. Curr Opin Cell Biol 33: 111–118, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Rustom A, Saffrich R, Markovic I, Walther P, and Gerdes HH. Nanotubular highways for intercellular organelle transport. Science 303: 1007–1010, 2004 [DOI] [PubMed] [Google Scholar]
- 339. Ryan BJ, Hoek S, Fon EA, and Wade-Martins R. Mitochondrial dysfunction and mitophagy in Parkinson's: from familial to sporadic disease. Trends Biochem Sci 40: 200–210, 2015 [DOI] [PubMed] [Google Scholar]
- 340. Ryu SW, Jeong HJ, Choi M, Karbowski M, and Choi C. Optic atrophy 3 as a protein of the mitochondrial outer membrane induces mitochondrial fragmentation. Cell Mol Life Sci 67: 2839–2850, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Safiulina D, Veksler V, Zharkovsky A, and Kaasik A. Loss of mitochondrial membrane potential is associated with increase in mitochondrial volume: physiological role in neurones. J Cell Physiol 206: 347–353, 2006 [DOI] [PubMed] [Google Scholar]
- 342. Saita S, Tatsuta T, Lampe PA, König T, Ohba Y, and Langer T. PARL partitions the lipid transfer protein STARD7 between the cytosol and mitochondria. EMBO J 37: e97909, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, Strillacci A, Stepanova A, Iommarini L, Mastroleo C, Daly L, Galkin A, Thakur BK, Soplop N, Uryu K, Hoshino A, Norton L, Bonafé M, Cricca M, Gasparre G, Lyden D, and Bromberg J. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci U S A 114: E9066–E9075, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Santel A. and Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci 114: 867–874, 2001 [DOI] [PubMed] [Google Scholar]
- 345. Santoro A, Campolo M, Liu C, Sesaki H, Meli R, Liu ZW, Kim JD, and Diano S. DRP1 suppresses leptin and glucose sensing of POMC neurons. Cell Metab 25: 647–660, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Sastri M, Darshi M, Mackey M, Ramachandra R, Ju S, Phan S, Adams S, Stein K, Douglas CR, Kim JJ, Ellisman MH, Taylor SS, and Perkins GA. Sub-mitochondrial localization of the genetic-tagged mitochondrial intermembrane space-bridging components Mic19, Mic60 and Sam50. J Cell Sci 130: 3248–3260, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Schenkel LC. and Bakovic M. Formation and regulation of mitochondrial membranes. Int J Cell Biol 2014: 709828, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Schmidt HH, Stocker R, Vollbracht C, Paulsen G, Riley D, Daiber A, and Cuadrado A. Antioxidants in translational medicine. Antioxid Redox Signal 23: 1130–1143, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349. Schmitt K, Grimm A, Dallmann R, Oettinghaus B, Restelli LM, Witzig M, Ishihara N, Mihara K, Ripperger JA, Albrecht U, Frank S, Brown SA, and Eckert A. Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell Metab 27: 657.e5–666.e5, 2018 [DOI] [PubMed] [Google Scholar]
- 350. Schneeberger M, Dietrich MO, Sebastián D, Imbernón M, Castaño C, Garcia A, Esteban Y, Gonzalez-Franquesa A, Rodríguez IC, Bortolozzi A, Garcia-Roves PM, Gomis R, Nogueiras R, Horvath TL, Zorzano A, and Claret M. Mitofusin 2 in POMC neurons connects ER stress with leptin resistance and energy imbalance. Cell 155: 172–187, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Schnell S. and Turner TE. Reaction kinetics in intracellular environments with macromolecular crowding: simulations and rate laws. Prog Biophys Mol Biol 85: 235–260, 2004 [DOI] [PubMed] [Google Scholar]
- 352. Schorr S. and van der Laan M. Integrative functions of the mitochondrial contact site and cristae organizing system. Semin Cell Dev Biol 76: 191–200, 2018 [DOI] [PubMed] [Google Scholar]
- 353. Schrader M, Bonekamp NA, and Islinger M. Fission and proliferation of peroxisomes. Biochim Biophys Acta 1822: 1343–1357, 2012 [DOI] [PubMed] [Google Scholar]
- 354. Schrader M, Costello JL, Godinho LF, Azadi AS, and Islinger M. Proliferation and fission of peroxisomes—an update. Biochim Biophys Acta 1863: 971–983, 2016 [DOI] [PubMed] [Google Scholar]
- 355. Schrepfer E. and Scorrano L. Mitofusins, from mitochondria to metabolism. Mol Cell 61: 683–694, 2016 [DOI] [PubMed] [Google Scholar]
- 356. Sebastián D, Palacín M, and Zorzano A. Mitochondrial dynamics: coupling mitochondrial fitness with healthy aging. Trends Mol Med 23: 201–215, 2017 [DOI] [PubMed] [Google Scholar]
- 357. Sena LA. and Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell 48: 158–167, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. Shao L, Kner P, Rego EH, and Gustafsson MG. Super-resolution 3D microscopy of live whole cells using structured illumination. Nat Methods 8: 1044–1046, 2011 [DOI] [PubMed] [Google Scholar]
- 359. Sharpley MS, Shannon RJ, Draghi F, and Hirst J. Interactions between phospholipids and NADH: ubiquinone oxidoreductase (complex I) from bovine mitochondria. Biochemistry 45: 241–248, 2006 [DOI] [PubMed] [Google Scholar]
- 360. Shen Q, Yamano K, Head BP, Kawajiri S, Cheung JT, Wang C, Cho JH, Hattori N, Youle RJ, and van der Bliek AM. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol Biol Cell 25: 145–159, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. Shen T, Zheng M, Cao C, Chen C, Tang J, Zhang W, Cheng H, Chen KH, and Xiao RP. Mitofusin-2 is a major determinant of oxidative stress-mediated heart muscle cell apoptosis. J Biol Chem 282: 23354–23361, 2007 [DOI] [PubMed] [Google Scholar]
- 362. Sheng ZH. and Cai Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci 13: 77–93, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Shiota T, Traven A, and Lithgow T. Mitochondrial biogenesis: cell-cycle dependent investment in making mitochondria. Curr Biol 25: R78–R80, 2015 [DOI] [PubMed] [Google Scholar]
- 364. Shutt T, Geoffrion M, Milne R, and McBride HM. The intracellular redox state is a core determinant of mitochondrial fusion. EMBO Rep 13: 909–915, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Sieprath T, Corne TD, Willems PH, Koopman WJ, and De Vos WH. Integrated high-content quantification of intracellular ROS levels and mitochondrial morphofunction. Adv Anat Embryol Cell Biol 219: 149–177, 2016 [DOI] [PubMed] [Google Scholar]
- 366. Singaravelu K, Nelson C, Bakowski D, de Brito OM, Ng SW, Di Capite J, Powell T, Scorrano L, and Parekh AB. Mitofusin 2 regulates STIM1 migration from the Ca2+ store to the plasma membrane in cells with depolarized mitochondria. J Biol Chem 286: 12189–12201, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Smirnova E, Shurland DL, Ryazantsev SN, and van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol 143: 351–358, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Smith G. and Gallo G. To mdivi-1 or not to mdivi-1: Is that the question? Dev Neurobiol 77: 1260–1268, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Smith AC, and Robinson AJ. MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucl Acid Res 44: D1258–D1261, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. So EC, Hsing CH, Liang CH, and Wu SN. The actions of mdivi-1, an inhibitor of mitochondrial fission, on rapidly activating delayed-rectifier K+ current and membrane potential in HL-1 murine atrial cardiomyocytes. Eur J Pharmacol 683: 1–9, 2012 [DOI] [PubMed] [Google Scholar]
- 371. Song Z, Chen H, Fiket M, Alexander C, and Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol 178: 749–755, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Song Z, Ghochani M, McCaffery JM, Frey TG, and Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell 20: 3525–3532, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Spruijt E, Sokolova E, and Huck WT. Complexity of molecular crowding in cell-free enzymatic reaction networks. Nat Nanotechnol 9: 406–407, 2014 [DOI] [PubMed] [Google Scholar]
- 374. Stoldt S, Wenzel D, Kehrein K, Riedel D, Ott M, and Jakobs S. Spatial orchestration of mitochondrial translation and OXPHOS complex assembly. Nat Cell Biol 20: 528–534, 2018 [DOI] [PubMed] [Google Scholar]
- 375. Strack S, Wilson TJ, and Cribbs JT. Cyclin-dependent kinases regulate splice-specific targeting of dynamin-related protein 1 to microtubules. J Cell Biol 201: 1037–1051, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Sugiura A, Mattie S, Prudent J, and McBride HM. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 542: 251–254, 2017 [DOI] [PubMed] [Google Scholar]
- 377. Sugiura A, Nagashima S, Tokuyama T, Amo T, Matsuki Y, Ishido S, Kudo Y, McBride HM, Fukuda T, Matsushita N, Inatome R, and Yanagi S. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol Cell 51: 20–34, 2013 [DOI] [PubMed] [Google Scholar]
- 378. Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, and Rizzuto R. Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol Cell 16: 59–68, 2004 [DOI] [PubMed] [Google Scholar]
- 379. Szabo I. and Zoratti M. Mitochondrial channels: ion fluxes and more. Physiol Rev 94: 519–608, 2014 [DOI] [PubMed] [Google Scholar]
- 380. Taguchi N, Ishihara N, Jofuku A, Oka T, and Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem 282: 11521–11529, 2007 [DOI] [PubMed] [Google Scholar]
- 381. Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, Pesdar EA, Sobol M, Filimonenko A, Stuart S, Vondrusova M, Kluckova K, Sachaphibulkij K, Rohlena J, Hozak P, Truksa J, Eccles D, Haupt LM, Griffiths LR, Neuzil J, and Berridge MV. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab 21: 81–94, 2015 [DOI] [PubMed] [Google Scholar]
- 382. Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, and Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol 191: 1367–1380, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383. Tasseva G, Bai HD, Davidescu M, Haromy A, Michelakis E, and Vance JE. Phosphatidylethanolamine deficiency in Mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J Biol Chem 288: 4158–4173, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384. Tatsuta T. and Langer T. Intramitochondrial phospholipid trafficking. Biochim Biophys Acta 1862: 81–89, 2017 [DOI] [PubMed] [Google Scholar]
- 385. Tezze C, Romanello V, Desbats MA, Fadini GP, Albiero M, Favaro G, Ciciliot S, Soriano ME, Morbidoni V, Cerqua C, Loefler S, Kern H, Franceschi C, Salvioli S, Conte M, Blaauw B, Zampieri S, Salviati L, Scorrano L, and Sandri M. Age-associated loss of OPA1 in muscle impacts muscle mass, metabolic homeostasis, systemic inflammation, and epithelial senescence. Cell Metab 25: 1374–1389.e6, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386. Tondera D, Czauderna F, Paulick K, Schwarzer R, Kaufmann J, and Santel A. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J Cell Sci 118: 3049–3059, 2005 [DOI] [PubMed] [Google Scholar]
- 387. Tondera D, Santel A, Schwarzer R, Dames S, Giese K, Klippel A, and Kaufmann J. Knockdown of MTP18, a novel phosphatidylinositol 3-kinase-dependent protein, affects mitochondrial morphology and induces apoptosis. J Biol Chem 279: 31544–31555, 2004 [DOI] [PubMed] [Google Scholar]
- 388. Toyama EQ, Herzig S, Courchet J, Lewis TL, Jr, Losón OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, and Shaw RJ. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 351: 275–281, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Tronstad KJ, Nooteboom M, Nilsson LI, Nikolaisen J, Sokolewicz M, Grefte S, Pettersen IK, Dyrstad S, Hoel F, Willems PH, and Koopman WJ. Regulation and quantification of cellular mitochondrial morphology and content. Curr Pharm Des 20: 5634–5652, 2014 [DOI] [PubMed] [Google Scholar]
- 390. Trotta AP. and Chipuk JE. Mitochondrial dynamics as regulators of cancer biology. Cell Mol Life Sci 74: 1999–2017, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391. Trudeau K, Molina AJ, and Roy S. High glucose induces mitochondrial morphology and metabolic changes in retinal pericytes. Invest Ophthalmol Vis Sci 52: 8657–8664, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392. Turakhiya U, von der Malsburg K, Gold VAM, Guiard B, Chacinska A, van der Laan M, and Ieva R. Protein import by the mitochondrial presequence translocase in the absence of a membrane potential. J Mol Biol 428: 1041–1052, 2016 [DOI] [PubMed] [Google Scholar]
- 393. Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, and Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433–446, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394. Urra FA, Weiss-López B, and Araya-Maturana R. Determinants of anti-cancer effect of mitochondrial electron transport chain inhibitors: bioenergetic profile and metabolic flexibility of cancer cells. Curr Pharm Des 22: 5998–6008, 2016 [DOI] [PubMed] [Google Scholar]
- 395. Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, and Lippincott-Schwartz J. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546: 162–167, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396. Vandenberg W, Leutenegger M, Lasser T, Hofkens J, and Dedecker P. Diffraction-unlimited imaging: from pretty pictures to hard numbers. Cell Tissue Res 360: 151–178, 2015 [DOI] [PubMed] [Google Scholar]
- 397. Vander Heiden MG, Cantley LC, and Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398. Van der Laan M, Horvath SE, and Pfanner N. Mitochondrial contact site and cristae organizing system. Curr Opin Cell Biol 41: 33–42, 2016 [DOI] [PubMed] [Google Scholar]
- 399. Van Meer G, Voelker DR, and Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9: 112–124, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400. Varanita T, Soriano ME, Romanello V, Zaglia T, Quintana-Cabrera R, Semenzato M, Menabò R, Costa V, Civiletto G, Pesce P, Viscomi C, Zeviani M, Di Lisa F, Mongillo M, Sandri M, and Scorrano L. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab 21: 834–844, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401. Verkman AS. Solute and macromolecule diffusion in cellular aqueous compartments. Trends Biochem Sci 27: 27–33, 2002 [DOI] [PubMed] [Google Scholar]
- 402. Vernay A, Marchetti A, Sabra A, Jauslin TN, Rosselin M, Scherer PE, Demaurex N, Orci L, and Cosson P. MitoNEET-dependent formation of intermitochondrial junctions. Proc Natl Acad Sci U S A 114: 8277–8282, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403. Vincent AE, Ng YS, White K, Davey T, Mannella C, Falkous G, Feeney C, Schaefer AM, McFarland R, Gorman GS, Taylor RW, Turnbull DM, and Picard M. The spectrum of mitochondrial ultrastructural defects in mitochondrial myopathy. Sci Rep 6: 30610, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404. Vincent AE, Turnbull DM, Eisner V, Hajnóczky G, and Picard M. Mitochondrial nanotunnels. Trends Cell Biol 27: 787–799, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405. Vögtle FN, Keller M, Taskin AA, Horvath SE, Guan XL, Prinz C, Opalińska M, Zorzin C, van der Laan M, Wenk MR, Schubert R, Wiedemann N, Holzer M, and Meisinger C. The fusogenic lipid phosphatidic acid promotes the biogenesis of mitochondrial outer membrane protein Ugo1. J Cell Biol 210: 951–960, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406. Wai T, García-Prieto J, Baker MJ, Merkwirth C, Benit P, Rustin P, Rupérez FJ, Barbas C, Ibañez B, and Langer T. Imbalanced OPA1 processing and mitochondrial fragmentation cause heart failure in mice. Science 350: aad0116, 2015 [DOI] [PubMed] [Google Scholar]
- 407. Wai T. and Langer T. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol Metab 27: 105–117, 2016 [DOI] [PubMed] [Google Scholar]
- 408. Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, and Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol 186: 805–816, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409. Walch L, Čopič A, and Jackson CL. Fatty acid metabolism meets organelle dynamics. Dev Cell 32: 657–658, 2015 [DOI] [PubMed] [Google Scholar]
- 410. Walker JE. The ATP synthase: the understood, the uncertain and the unknown. Biochem Soc Trans 41: 1–16, 2013 [DOI] [PubMed] [Google Scholar]
- 411. Walter T, Shattuck DW, Baldock R, Bastin ME, Carpenter AE, Duce S, Ellenberg J, Fraser A, Hamilton N, Pieper S, Ragan MA, Schneider JE, Tomancak P, and Hériché JK. Visualization of image data from cells to organisms. Nat Methods 7: S26–S41, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412. Wang W, Fernandez-Sanz C, and Sheu SS. Regulation of mitochondrial bioenergetics by the non-canonical roles of mitochondrial dynamics proteins in the heart. Biochim Biophys Acta 1864: 1991–2001, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413. Wang H, Song P, Du L, Tian W, Yue W, Liu M, Li D, Wang B, Zhu Y, Cao C, Zhou J, and Chen Q. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem 286: 11649–11658, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414. Wang Y, Subramanian M, Yurdagul A, Jr, Barbosa-Lorenzi VC, Cai B, de Juan-Sanz J, Ryan TA, Nomura M, Maxfield FR, and Tabas I. Mitochondrial fission promotes the continued clearance of apoptotic cells by macrophages. Cell 171: 331.e22–345.e22, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415. Wang W, Wang Y, Long J, Wang J, Haudek SB, Overbeek P, Chang BH, Schumacker PT, and Danesh FR. Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell Metab 15: 186–200, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416. Wasiak S, Zunino R, and McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol 177: 439–450, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417. Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, and Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med 356: 1736–1741, 2007 [DOI] [PubMed] [Google Scholar]
- 418. Weaver D, Eisner V, Liu X, Várnai P, Hunyady L, Gross A, and Hajnóczky G. Distribution and apoptotic function of outer membrane proteins depend on mitochondrial fusion. Mol Cell 54: 870–878, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419. Weiss M. Crowding, diffusion, and biochemical reactions. Int Rev Cell Mol Biol 307: 383–417, 2014 [DOI] [PubMed] [Google Scholar]
- 420. Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11: 872–884, 2010 [DOI] [PubMed] [Google Scholar]
- 421. Westrate LM, Drocco JA, Martin KR, Hlavacek WS, and MacKeigan JP. Mitochondrial morphological features are associated with fission and fusion events. PLoS One 9: e95265, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422. Wiedemann N. and Pfanner N. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem 86: 685–714, 2017 [DOI] [PubMed] [Google Scholar]
- 423. Wiedemann N, Stiller SB, and Pfanner N. Activation and degradation of mitofusins: two pathways regulate mitochondrial fusion by reversible ubiquitylation. Mol Cell 49: 423–425, 2013 [DOI] [PubMed] [Google Scholar]
- 424. Wilkinson KA. and Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428: 133–145, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425. Willems PH, Rossignol R, Dieteren CE, Murphy MP, and Koopman WJ. Redox homeostasis and mitochondrial dynamics. Cell Metab 22: 207–218, 2015 [DOI] [PubMed] [Google Scholar]
- 426. Wilson TJ, Slupe AM, and Strack S. Cell signaling and mitochondrial dynamics: implications for neuronal function and neurodegenerative disease. Neurobiol Dis 51: 13–26, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427. Wollweber F, von der Malsburg K, and van der Laan M. Mitochondrial contact site and cristae organizing system: a central player in membrane shaping and crosstalk. Biochim Biophys Acta 1864: 1481–1489, 2017 [DOI] [PubMed] [Google Scholar]
- 428. Wong YC, Ysselstein D, and Krainc D. Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554: 382–386, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429. Xiao X, Hu Y, Quirós PM, Wei Q, López-Otín C, and Dong Z. OMA1 mediates OPA1 proteolysis and mitochondrial fragmentation in experimental models of ischemic kidney injury. Am J Physiol Renal Physiol 306: F1318–F1326, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430. Yamashita SI, Jin X, Furukawa K, Hamasaki M, Nezu A, Otera H, Saigusa T, Yoshimori T, Sakai Y, Mihara K, and Kanki T. Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J Cell Biol 215: 649–665, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431. Yan L, Qi Y, Huang X, Yu C, Lan L, Guo X, Rao Z, Hu J, and Lou Z. Structural basis for GTP hydrolysis and conformational change of MFN1 in mediating membrane fusion. Nat Struct Mol Biol 25: 233–243, 2018 [DOI] [PubMed] [Google Scholar]
- 432. Yang F, Wu R, Jiang Z, Chen J, Nan J, Su S, Zhang N, Wang C, Zhao J, Ni C, Wang Y, Hu W, Zeng Z, Zhu K, Liu X, Hu X, Zhu W, Yu H, Huang J, and Wang J. Leptin increases mitochondrial OPA1 via GSK3-mediated OMA1 ubiquitination to enhance therapeutic effects of mesenchymal stem cell transplantation. Cell Death Dis 9: 556, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433. Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata S, and Koshiba T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal 2: ra47, 2009 [DOI] [PubMed] [Google Scholar]
- 434. Yi M, Weaver D, and Hajnóczky G. Control of mitochondrial motility and distribution by the calcium signal: a homeostatic circuit. J Cell Biol 167: 661–672, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435. Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal 10: 179–206, 2008 [DOI] [PubMed] [Google Scholar]
- 436. Yoon Y, Galloway CA, Jhun BS, and Yu T. Mitochondrial dynamics in diabetes. Antioxid Redox Signal 14: 439–457, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437. Yoon Y, Krueger EW, Oswald BJ, and McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol 23: 5409–5420, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438. Yoon Y, Pitts KR, Dahan S, and McNiven MA. A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J Cell Biol 140: 779–793, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439. Yu R, Liu T, Jin SB, Ning C, Lendahl U, Nistér M, and Zhao J. MIEF1/2 function as adaptors to recruit Drp1 to mitochondria and regulate the association of Drp1 with MFF. Sci Rep 7: 880, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440. Yu T, Robotham JL, and Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc Natl Acad Sci U S A 103: 2653–2658, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441. Zhang K, Li H, and Song Z. Membrane depolarization activates the mitochondrial protease OMA1 by stimulating self-cleavage. EMBO Rep 15: 576–585, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442. Zhang CS. and Lin SC. AMPK promotes autophagy by facilitating mitochondrial fission. Cell Metab 23: 399–401, 2016 [DOI] [PubMed] [Google Scholar]
- 443. Zhang Y, Liu X, Bai J, Tian X, Zhao X, Liu W, Duan X, Shang W, Fan HY, and Tong C. Mitoguardin regulates mitochondrial fusion through MitoPLD and is required for neuronal homeostasis. Mol Cell 61: 111–124, 2016 [DOI] [PubMed] [Google Scholar]
- 444. Zhao T, Huang X, Han L, Wang X, Cheng H, Zhao Y, Chen Q, Chen J, Cheng H, Xiao R, and Zheng M. Central role of mitofusin 2 in autophagosome-lysosome fusion in cardiomyocytes. J Biol Chem 287: 23615–23625, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445. Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlén P, Tomilin N, Shupliakov O, Lendahl U, and Nistér M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J 30: 2762–2778, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446. Zhou W, Chen KH, Cao W, Zeng J, Liao H, Zhao L, and Guo X. Mutation of the protein kinase A phosphorylation site influences the anti-proliferative activity of mitofusin 2. Atherosclerosis 211: 216–223, 2010 [DOI] [PubMed] [Google Scholar]
- 447. Zick M, Rabl R, and Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta 1793: 5–19, 2009 [DOI] [PubMed] [Google Scholar]
- 448. Zorova LD, Popkov VA, Plotnikov EY, Silachev DN, Pevzner IB, Jankauskas SS, Babenko VA, Zorov SD, Balakireva AV, Juhaszova M, Sollott SJ, and Zorov DB. Mitochondrial membrane potential. Anal Biochem 552: 50–59, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449. Zorzano A, Liesa M, Sebastián D, Segalés J, and Palacín M. Mitochondrial fusion proteins: dual regulators of morphology and metabolism. Semin Cell Dev Biol 21: 566–574, 2010 [DOI] [PubMed] [Google Scholar]
- 450. Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schröder JM, and Vance JM. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet 36: 449–451, 2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.