Nicotine has been studied as a model for toxic N-heterocyclic aromatic compounds. Microorganisms can catabolize nicotine via various pathways and conserve energy from its oxidation. Although several oxidoreductases have been characterized to participate in nicotine degradation, the electron transfer involved in these processes is poorly understood. In this study, we found that 6-hydroxypseudooxynicotine dehydrogenase, a key enzyme in the hybrid pyridine and pyrrolidine pathway for nicotine degradation in Agrobacterium tumefaciens S33, utilizes EtfAB as a physiological electron acceptor. Catalyzed by the membrane-associated electron transfer flavoprotein:ubiquinone oxidoreductase, the electrons are transferred from the reduced EtfAB to coenzyme Q, which then could enter into the classic ETC. Thus, the route for electron transport from the substrate to O2 could be constructed, by which ATP can be further sythesized via chemiosmosis to support the baterial growth. These findings provide new knowledge regarding the catabolism of N-heterocyclic aromatic compounds in microorganisms.
KEYWORDS: 6-hydroxypseudooxynicotine dehydrogenase, Agrobacterium tumefaciens, nicotine degradation, electron acceptor, electron transfer flavoprotein, electron transfer flavoprotein:ubiquinone oxidoreductase
ABSTRACT
Agrobacterium tumefaciens S33 degrades nicotine via a novel hybrid of the pyridine and the pyrrolidine pathways. The hybrid pathway consists of at least six steps involved in oxidoreductive reactions before the N-heterocycle can be broken down. Collectively, the six steps allow electron transfer from nicotine and its intermediates to the final acceptor O2 via the electron transport chain (ETC). 6-Hydroxypseudooxynicotine oxidase, renamed 6-hydroxypseudooxynicotine dehydrogenase in this study, has been characterized as catalyzing the fourth step using the artificial electron acceptor 2,6-dichlorophenolindophenol. Here, we used biochemical, genetic, and liquid chromatography-mass spectrometry (LC-MS) analyses to determine that 6-hydroxypseudooxynicotine dehydrogenase utilizes the electron transfer flavoprotein (EtfAB) as the physiological electron acceptor to catalyze the dehydrogenation of pseudooxynicotine, an analogue of the true substrate 6-hydroxypseudooxynicotine, in vivo, into 3-succinoyl-semialdehyde-pyridine. NAD(P)+, O2, and ferredoxin could not function as electron acceptors. The oxygen atom in the aldehyde group of the product 3-succinoyl-semialdehyde-pyridine was verified to be derived from H2O. Disruption of the etfAB genes in the nicotine-degrading gene cluster decreased the growth rate of A. tumefaciens S33 on nicotine but not on 6-hydroxy-3-succinoylpyridine, an intermediate downstream of the hybrid pathway, indicating the requirement of EtfAB for efficient nicotine degradation. The electrons were found to be further transferred from the reduced EtfAB to coenzyme Q by the catalysis of electron transfer flavoprotein:ubiquinone oxidoreductase. These results aid in an in-depth understanding of the electron transfer process and energy metabolism involved in the nicotine oxidation and provide novel insights into nicotine catabolism in bacteria.
IMPORTANCE Nicotine has been studied as a model for toxic N-heterocyclic aromatic compounds. Microorganisms can catabolize nicotine via various pathways and conserve energy from its oxidation. Although several oxidoreductases have been characterized to participate in nicotine degradation, the electron transfer involved in these processes is poorly understood. In this study, we found that 6-hydroxypseudooxynicotine dehydrogenase, a key enzyme in the hybrid pyridine and pyrrolidine pathway for nicotine degradation in Agrobacterium tumefaciens S33, utilizes EtfAB as a physiological electron acceptor. Catalyzed by the membrane-associated electron transfer flavoprotein:ubiquinone oxidoreductase, the electrons are transferred from the reduced EtfAB to coenzyme Q, which then could enter into the classic ETC. Thus, the route for electron transport from the substrate to O2 could be constructed, by which ATP can be further sythesized via chemiosmosis to support the baterial growth. These findings provide new knowledge regarding the catabolism of N-heterocyclic aromatic compounds in microorganisms.
INTRODUCTION
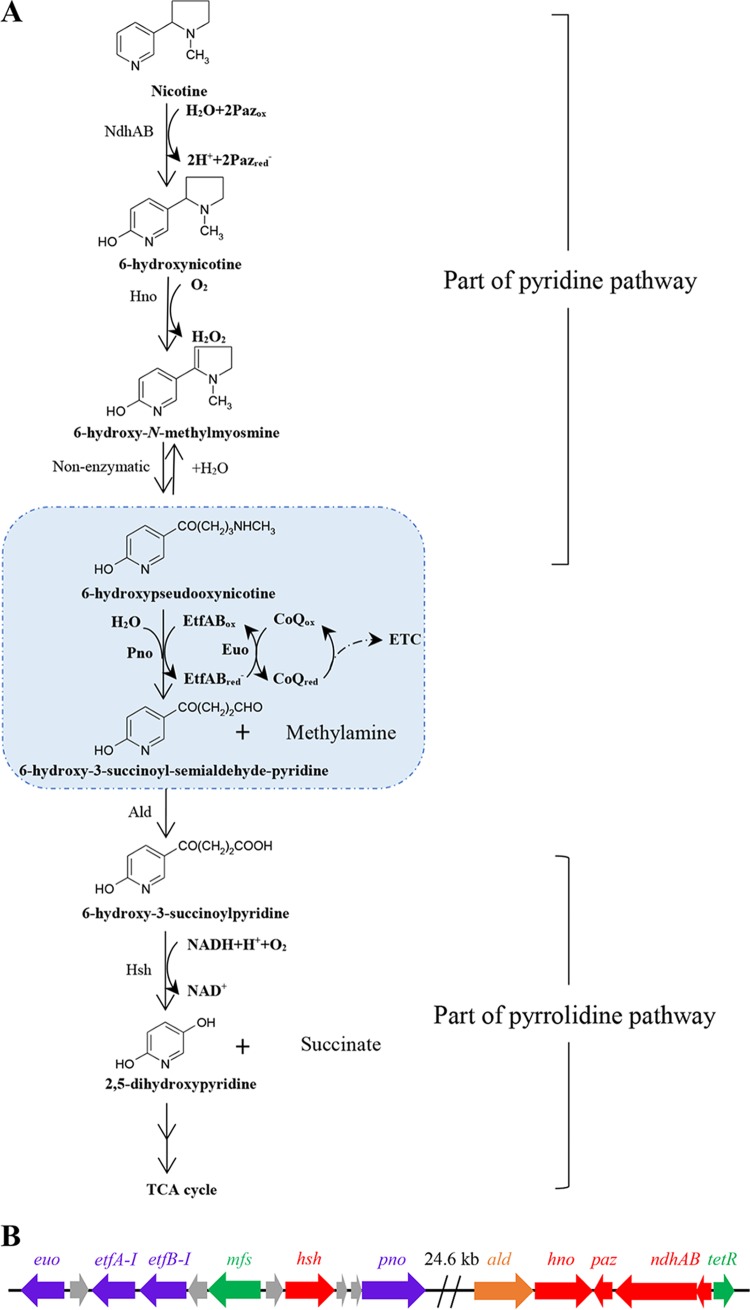
Nicotine is the main toxic N-heterocyclic aromatic compound in tobacco and its waste products (1–3). The tobacco manufacturing process produces significant amounts of wastes that contain nicotine, which can pollute the environment and affect the human health where there is improper disposal. Nicotine, a constituent of these manufacturing wastes, has been designated by the U.S. Environmental Protection Agency (EPA) as a Toxics Release Inventory (TRI) chemical. Currently, the larvaceous ecotoxicological risk caused by tobacco wastes has been demonstrated by the detection of nicotine in industrial effluents, municipal and treated wastewater, surface water, groundwater, and even bottled mineral water (4–7). The biodegradation of nicotine by microorganisms has attracted significant attention as an effective and economical method for the disposal of toxic chemicals without causing harm to the environment. Two biochemical pathways, namely, the pyridine pathway found in the Gram-positive bacteria Arthrobacter spp. and the pyrrolidine pathway identified in the Gram-negative bacteria Pseudomonas spp., are well known to be involved in the bacterial degradation of nicotine (7–11). Recently, a novel nicotine oxidative degradation pathway (Fig. 1A), a hybrid form of the pyridine and pyrrolidine pathways, was discovered in Agrobacterium tumefaciens S33 (12, 13). In the hybrid pathway, nicotine is first degraded into 6-hydroxypseudooxynicotine through 6-hydroxynicotine and 6-hydroxy-N-methylmyosmine via the pyridine pathway, and 6-hydroxypseudooxynicotine is then transformed into 6-hydroxy-3-succinoylpyridine and 2,5-dihydroxypyridine along the pyrrolidine pathway. The same pathway was also discovered later in Shinella sp. strain HZN7 (14) and Ochrobactrum sp. strain SJY1 (15). In order to elucidate the biochemical and molecular details of this hybrid pathway, much work has been carried out on the three strains in recent years, including genomic and transcriptomic analyses, as well as purification and characterization of key enzymes (16–25). However, the mechanistic details of the electron transport involved in the process of oxidative degradation of nicotine are not clearly understood.
FIG 1.
(A and B) The hybrid pathway of nicotine oxidative degradation by A. tumefaciens S33 (A) and the arrangement of pno, etfAB-I, and euo genes in the genome of A. tumefaciens S33 (B). NdhAB, nicotine dehydrogenase; Paz, pseudoazurin; Hno, 6-hydroxynicotine oxidase; Pno, 6-hydroxypseudooxynicotine oxidase (renamed 6-hydroxypseudooxynicotine dehydrogenase in this study); EtfAB-I, electron transfer flavoprotein; Euo, electron transfer flavoprotein:ubiquinone oxidoreductase; ETC, electron transport chain; Ald, predicted aldehyde dehydrogenase; Hsh, 6-hydroxy-3-succinoylpyridine hydroxylase; mfs, predicted major facilitator superfamily transporter gene; TCA, tricarboxylic acid; tetR, predicted TetR family transcriptional regulator gene. Purple arrows, genes encoding key enzymes in nicotine degradation characterized in this study; red arrows, genes encoding enzymes characterized previously; orange arrow, predicted gene encoding key enzyme for nicotine degradation; green arrows, predicted transporter and regulatory genes; and gray arrows, genes with unknown function.
A. tumefaciens S33 grows on nicotine as the sole source of carbon and nitrogen by conserving energy through the oxidation of nicotine. The hybrid nicotine degradation pathway contains at least six steps that are involved in oxidoreductive reactions before the N-heterocycle is broken down, wherein electrons can be delivered from nicotine and its intermediates to the final acceptor O2 via the classic electron transport chain (ETC). Thus, electron transfer during nicotine degradation becomes a key challenge in understanding the bacterial energy metabolism. Electron transfer processes catalyzed by several key enzymes involved in nicotine oxidative degradation in A. tumefaciens S33 have been studied. Nicotine dehydrogenase (NdhAB; 82.4 kDa for NdhA and 17.1 kDa for NdhB) is a molybdenum-containing and flavin-lacking hydroxylase. It catalyzes the initial step of nicotine hydroxylation to 6-hydroxynicotine with pseudoazurin (Paz), a small blue copper-binding protein, as its natural electron acceptor in the periplasmic space, which might further transfer electrons to O2 directly or via cytochrome c (17). 6-Hydroxynicotine oxidase (Hno), a flavin adenine dinucleotide (FAD)-containing oxidoreductase, oxidizes 6-hydroxynicotine to 6-hydroxy-N-methylmyosmine with O2 as its electron acceptor, forming H2O2 (11, 21). 6-Hydroxy-3-succinoylpyridine hydroxylase (Hsh; 45 kDa) is an FAD-containing homodimeric monooxygenase, which catalyzes the 6-hydroxy-3-succinoylpyridine (HSP) to 2,5-dihydroxypyridine and succinic acid by oxidative decarboxylation in the presence of O2 and NADH (18). 6-Hydroxypseudooxynicotine oxidase (Pno; 73.3 kDa), a novel iron-sulfur flavoprotein harboring a flavin mononucleotide (FMN) and a [4Fe4S] cluster, catalyzes the oxidative deamination of 6-hydroxypseudooxynicotine to 6-hydroxy-3-succinoyl-semialdehyde-pyridine (16). The enzymatic activity of Pno, however, has only been determined with the artificial electron acceptor 2,6-dichlorophenolindophenol (DCPIP), mainly since the common acceptor O2 was nonfunctional. Thus, the physiological electron acceptor of Pno activity awaits identification, which will be crucial in understanding the mechanism of electron transfer in the reaction.
Our previous study showed that the core nicotine-degrading genes, including ndhAB, paz, hno, pno, and hsh, form a large cluster located on a genomic island in the circular chromosome of A. tumefaciens S33 (19) (Fig. 1B). Three open reading frames (ORFs) in the upstream region of hsh were annotated to encode the electron transfer flavoprotein (EtfAB-I) and electron transfer flavoprotein:ubiquinone oxidoreductase (Euo). Interestingly, the transcription levels of etfAB-I and euo were differentially upregulated along with the core nicotine-degrading genes when strain S33 was cultured on nicotine compared to glucose plus ammonium sulfate (19). It is well established that EtfAB can accept electrons from the dehydrogenation reaction catalyzed by various dehydrogenases, and Euo is a membrane-associated protein (26) that catalyzes the electron transfer from reduced EtfAB to coenzyme Q (CoQ) (27, 28). Thus, the pattern of genetic organization along with transcriptomic analysis suggests that etfAB-I and euo may encode proteins that partake in the electron transport process in nicotine degradation. Among the enzymes that are encoded by core nicotine-degrading genes, Pno is the only enzyme whose physiological electron acceptor is still unknown (note that ald, an additional gene, which was predicted to encode a NADP-dependent aldehyde dehydrogenase catalyzing the fifth step of oxidation of 6-hydroxy-3-succinoylsemialdehyde-pyridine into 6-hydroxy-3-succinoylpyridine, is being verified experimentally). Pno from A. tumefaciens S33 exhibits 48%, 40%, and 39% sequence identity to histamine dehydrogenase from Pimelobacter simplex, the trimethylamine dehydrogenase from Methylophilus methylotrophus W3A1, and the dimethylamine dehydrogenase from Hyphomicrobium sp. strain X, respectively (16). It is reported that both trimethylamine dehydrogenase and dimethylamine dehydrogenase use EtfAB as the physiological electron acceptor (29, 30). Therefore, we predict that Pno from strain S33 most likely catalyzes the electron transfer from 6-hydroxypseudooxynicotine to EtfAB, and the reduced EtfAB can be reoxidized by Euo to transfer electrons to coenzyme Q (CoQ). In addition, there is a second set of etfAB genes (etfAB-II) in the genome of strain S33 which is outside the nicotine-degrading gene cluster and should also be considered.
In this study, we investigated the details of the catalytic step involving Pno and experimentally verified our hypothesis. We observed that Pno is capable of using pseudooxynicotine, an analogue of the true substrate 6-hydroxypseudooxynicotine in vivo, as an alternative substrate. Based on the biochemical analysis and gene disruption, EtfAB and Euo were found to play a critical role in the oxidative deamination of pseudooxynicotine to 3-succinoyl-semialdehyde-pyridine. EtfAB functioned as the physiological electron acceptor of Pno, and the electrons were further transferred to CoQ by the catalysis of Euo, which then could enter the ETC. These findings provide novel evidence for electron transfer in the hybrid pathway of nicotine oxidative degradation in A. tumefaciens S33.
RESULTS
Pseudooxynicotine may replace 6-hydroxypseudooxynicotine as the substrate of Pno.
Since 6-hydroxypseudooxynicotine is not commercially available, we tested whether the analogue of 6-hydroxypseudooxynicotine, pseudooxynicotine, is able to function as the substrate of Pno. As shown in Fig. S1A in the supplemental material, DCPIP could be reduced in the presence of pseudooxynicotine and the electron transfer mediator phenazine methosulfate (PMS) at an optimum reaction pH of 8.5. Under the conditions of pH 8.5 in 50 mM Tris-HCl buffer at 30°C, Pno presented a Vmax of 52.8 U/mg and an apparent Km value of 0.42 mM for pseudooxynicotine (Fig. S1B), which were similar to the previously reported enzyme activity (32.3 U/mg) and the apparent Km value (0.37 mM) of 6-hydroxypseudooxynicotine (16). None of the common electron acceptors, such as O2, NAD(P)+, or ferredoxin, allowed pseudooxynicotine oxidation activity. The commonly used artificial electron acceptors, such as ferricyanide, cytochrome c, DCPIP (without PMS), methylene blue, 2,3,5-triphenyltetrazolium chloride (TTC), and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT), showed minor activity (less than 0.9 U/mg). Additionally, the purified Pno could be directly reduced by pseudooxynicotine (Fig. S2), as analyzed by a UV-visible spectrophotometer. The absorption spectra showed that characteristic absorption peaks (maximum absorption at 375 nm and 450 nm) for its cofactors of a FMN and a [4Fe4S] cluster (16) disappeared after treatment with pseudooxynicotine. The final products of reaction with DCPIP plus PMS as the electron acceptor were identified using liquid chromatography-mass spectrometry (LC-MS) (Fig. S3). LC-MS profiles showed that Pno was able to catalyze the conversion of pseudooxynicotine to 3-succinoyl-semialdehyde-pyridine. These results indicate that pseudooxynicotine could be catalyzed by Pno in the same way as 6-hydroxypseudooxynicotine (16). Thus, we used pseudooxynicotine in lieu of 6-hydroxypseudooxynicotine in order to simplify the enzyme assay.
Heterologous expression of EtfAB and the activity of Pno with EtfAB as the electron acceptor.
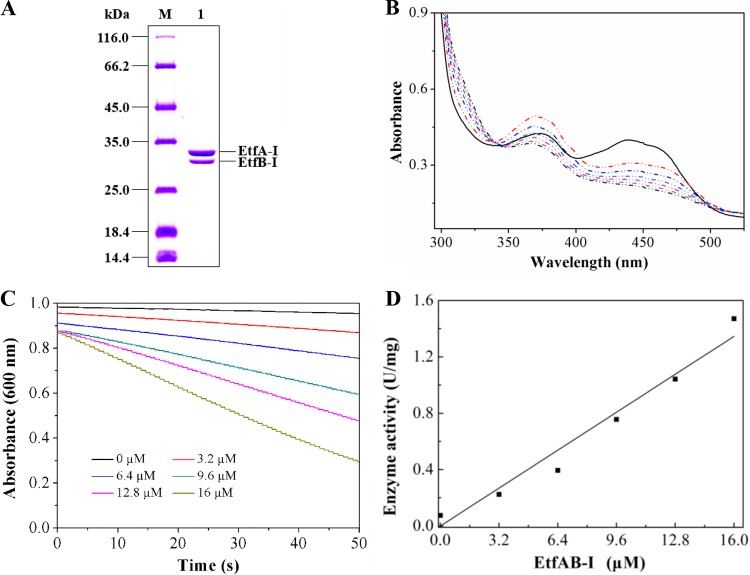
etfA-I and etfB-I were cloned together, and their encoded products were expressed in Escherichia coli BL21(DE3). The recombinant target protein was purified by a HisTrap column and analyzed by SDS-PAGE. The purified protein presented a light-yellow color after concentration. Two distinct bands with molecular masses of 31 kDa for EtfA-I and 27 kDa for EtfB-I were observed on SDS-PAGE (Fig. 2A), which were in agreement with the calculated masses and within reported molecular masses ranging from 31 to 42 kDa for EtfA and 25 to 38 kDa for EtfB from other organisms (31). Gel filtration showed that the purified protein sample was a mixture of heterodimer (EtfAB-I, 58.3 kDa, as determined in this study) and tetramer [(EtfAB-I)2, 116.6 kDa, as determined in this study] (Fig. S4A and C). The UV-visible absorption spectrum showed two typical absorption peaks (maximum absorption at 375 nm and 442 nm) (Fig. S5A) identical to those of EtfAB from other organisms (32). This was in agreement with the cofactor measurement that 1 mol purified EtfAB-I contained 0.64 mol FAD and 0.80 mol AMP, suggesting that EtfAB-I from strain S33 harbors one FAD and one AMP.
FIG 2.
Heterologous expression and purification of EtfAB-I and the pseudooxynicotine oxidation catalyzed by Pno with purified EtfAB-I as the electron acceptor. (A) SDS-PAGE analysis of purified His-tagged EtfAB-I. M, protein marker; 1, purified protein. (B) UV-visible absorption spectra of purified EtfAB-I (solid line) and spectrophotometric changes from 300 nm to 550 nm during the reduction of EtfAB-I (dash-dot-dot line) with pseudooxynicotine by Pno. Pseudooxynicotine has no absorption in the wavelength window. The reaction mixture contained 89 μM EtfAB-I, 1 mM pseudooxynicotine, and 20 mM sodium phosphate buffer (pH 7.4). The reaction was initiated by adding 0.03 μM Pno. The red dash-dot-dot line is the first record after the addition of Pno. The time between the first two records (between black line and red dash-dot-dot line) was 45 s. The rest of the records were done every 45 s, where the absorption continuously decreased. (C) The effect of different concentrations of EtfAB-I (0 to 16 μM) on the reduction of DCPIP by Pno (0.28 μM) in the presence of 1 mM pseudooxynicotine monitored at 600 nm. (D) The relationship between the concentration of EtfAB-I and DCPIP reduction activity (in units per milligram) catalyzed by Pno. Data were used from panel C.
Then, EtfAB-I was tested as the electron acceptor instead of DCPIP and PMS to determine the activity of Pno. As shown in Fig. 2B, the absorption spectrum of purified EtfAB-I changed rapidly upon the addition of Pno; the peak at 442 nm decreased significantly, whereas the peak at 370 nm showed an initial increase followed by a decrease, which was similar to the pattern of EtfAB reduction by trimethylamine dehydrogenases (32, 33). This indicated that EtfAB-I was first reduced to form the red anionic semiquinone with pseudooxynicotine by Pno, and the subsequent decrease of absorption at 370 nm was caused by the formation of fully reduced hydroquinone (33). In order to quantify the activity of EtfAB-I, we added DCPIP to investigate the effects of EtfAB-I on the reduction of DCPIP with pseudooxynicotine by Pno (where PMS was omitted), as described previously (34). Increased DCPIP-reducing activity was observed when purified EtfAB-I was added (Fig. 2C), and activity increased proportional to increasing concentrations of EtfAB-I (Fig. 2D), indicating that EtfAB-I can efficiently mediate the electron transfer to DCPIP.
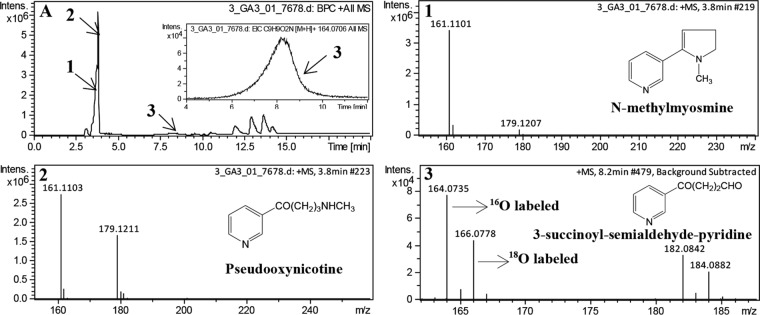
Further, we determined the reaction products with EtfAB-I as the sole electron acceptor in the reaction mixture (where both DCPIP and PMS were omitted) using LC-MS. 18O-labeled water was added as one of the components in the reaction mixture to determine the source of oxygen atom in the expected product 3-succinoyl-semialdehyde-pyridine. Both 16O-labeled and 18O-labeled 3-succinoyl-semialdehyde-pyridine were detected in the LC-MS analysis (Fig. 3). This confirmed that EtfAB-I indeed functions as the electron acceptor of Pno under physiological conditions. It also demonstrated that the oxygen atom in the reaction product 3-succinoyl-semialdehyde-pyridine was not derived from O2 but from the water molecules, which was similar to the reactions catalyzed by histamine dehydrogenase and trimethylamine dehydrogenase (35, 36).
FIG 3.
LC-MS profiles of the reaction catalyzed by Pno using purified EtfAB-I as the sole electron acceptor in the presence of both H216O and H218O. The reaction mixture contained 20 mM phosphate buffer (pH 7.4), 1 mM pseudooxynicotine, 89 μM purified EtfAB-I, 0.03 μM purified Pno, and 40% (vol/vol) H218O. (A) LC profile of the reaction, where an enlargement of peak 3 is shown in the inset. (1) Mass spectrum of N-methylmyosmine (m/z 161.1101), which was the product of spontaneous dehydration reaction from the substrate pseudooxynicotine. There is a hydrolysis equilibrium between N-methylmyosmine and pseudooxynicotine. (2) Mass spectra of the excess substrate pseudooxynicotine (m/z 179.1211) and its dehydrated product N-methylmyosmine (m/z 161.1103) with retention time at 3.8 min. (3) Mass spectra of the product 3-succinoyl-semialdehyde-pyridine (m/z 164.0735 for 16O-labeled, m/z 166.0778 for 18O-labeled) and its adduct of water (m/z 182.0842 for 16O-labeled, m/z 184.0882 for 18O-labeled). The ratio of the two peak areas for 16O- and 18O-labeled products (∼1.5) is identical to the ratio of H216O and H218O in the reaction mixture.
To explore the role of the etfAB-II genes in the S33 genome, heterologous expression and purification of EtfAB-II were performed. SDS-PAGE analysis showed two distinct bands with molecular masses of 31 kDa for EtfA-II and 26 kDa for EtfB-II (Fig. S6A). The purified EtfAB-II was a heterodimer with a molecular mass of 57 kDa, as determined by gel filtration (Fig. S4A and C). It contained 0.56 mol FAD and 0.81 mol AMP. Meanwhile, EtfAB-II presented a UV-visible absorption spectrum very similar to that of EtfAB-I and could also be reduced with pseudooxynicotine by Pno, indicating that it can function as the electron acceptor of a Pno like EtfAB-I (Fig. S6B to D).
Euo catalyzes electron transfer from EtfAB-I to CoQ.
euo was cloned into plasmid pET28b(+), and the encoded protein was expressed in E. coli C41(DE3). The recombinant Euo was purified by a HisTrap column, where 0.2% (vol/vol) Triton X-100 was added to the binding buffer and elution buffer to prevent Euo from precipitating. The purified recombinant protein presented a brown color and three characteristic absorption peaks at 379 nm, 412 nm, and 460 nm (shoulder peak) in the UV-visible absorption spectrum (Fig. S5B), which was identical to the spectrum of Euo from beef heart mitochondria (37). The determination of cofactors showed that 1 mol purified Euo contained 3.27 mol Fe and 0.80 mol FAD, which was in agreement with the finding that Euo harbors one [4Fe4S] and one FAD as reported for Euo from other organisms (37). SDS-PAGE analysis of purified Euo presented a distinct band corresponding to a molecular mass of 60 kDa (Fig. 4A), which was in agreement with the calculated theoretical mass. Gel filtration showed that the purified Euo was a trimer with a determined molecular mass of 180 kDa (Fig. S4B and C).
FIG 4.
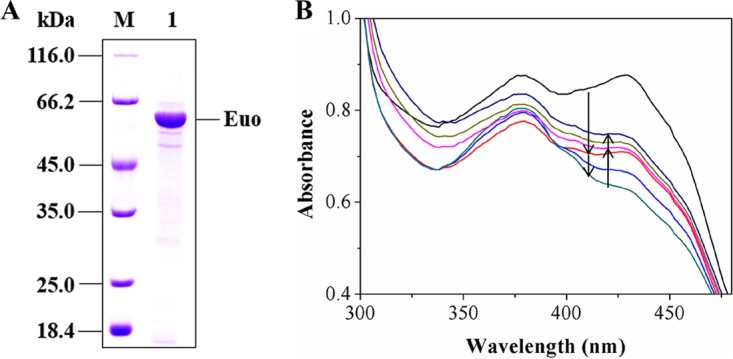
(A) SDS-PAGE analysis of the purified recombinant Euo. M, protein marker; 1, purified protein. (B) Changes in UV-visible absorption spectra during the reduction of CoQ1 with reduced EtfAB-I by Euo coupling with the reduction of EtfAB-I with pseudooxynicotine by Pno. Initially, the reaction mixture contained 50 mM Tris-HCl (pH 8.5), 1 mM pseudooxynicotine, 0.05 μM purified Pno, and 30 μM purified Euo (black line). The mixture was then treated by adding 8 μM purified EtfAB-I (red line). The reduction of Euo proceeded with time (light-blue and dark-green lines). Upon stabilization of the spectrum, 32 μM CoQ1 was added and Euo was reoxidized (pink line). The reoxidization proceeded with time (light-green and dark-blue lines). The downward arrows indicate the reduction of Euo after the addition of EtfAB-I, whereas the upward arrows indicate the reoxidation of Euo after the addition of CoQ1. Neither oxidized nor reduced CoQ1 has absorption in the wavelength window. The time between the first two records (between the black line and dark-blue line) was 45 s. The rest of the records were done every 45 s.
Euo is known to oxidize the reduced EtfAB by using CoQ as the electron acceptor and deliverer of electrons to ETC (27). Thus, we determined the reducing activity of EtfAB-I on CoQ1 by coupling Euo with the reduction of EtfAB-I with pseudooxynicotine by Pno. As monitored by UV-visible photometry (Fig. 4B), the absorption decreased rapidly when purified EtfAB-I was added to the reaction mixture containing 50 mM Tris-HCl, purified Pno, pseudooxynicotine, and purified Euo, indicating that purified Euo was reduced by EtfAB-I and that EtfAB-I itself was initially reduced with pseudooxynicotine by Pno. After the completion of the reaction (indicated by a stable absorption spectrum), CoQ1 was added to the reaction mixture, and the absorption was observed to increase gradually (Fig. 4B), indicating that the reduced Euo was reoxidized with CoQ1. We analyzed the final reaction mixture using LC-MS. Both of the expected products, namely, 3-succinoyl-semialdehyde-pyridine and reduced CoQ1, were detected (Fig. S7A). In contrast, reduced CoQ1 was not detected in the control group without the addition of purified Euo (Fig. S7B). These results clearly indicated that Euo played an essential role in the reoxidization of EtfAB-I and electron transfer to the ETC.
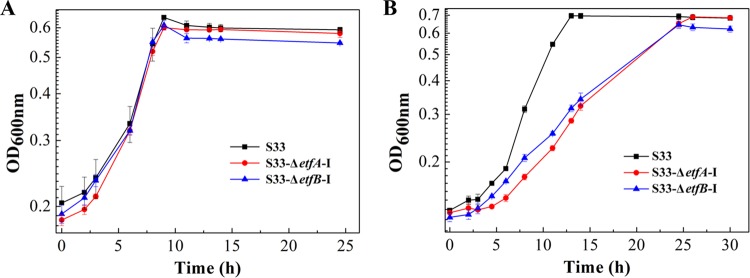
Growth of strain S33 on nicotine was negatively affected by disruption of the etfAB-I genes.
etfA-I and etfB-I were disrupted to confirm their role in nicotine degradation by strain S33. Wild-type A. tumefaciens S33 and the S33-ΔetfA-I and S33-ΔetfB-I mutant strains were inoculated in HSP and nicotine media, respectively, to test their growth. As shown in Fig. 5, both the S33-ΔetfA-I and S33-ΔetfB-I mutants grew almost as well as the wild-type in HSP medium; however, their growth on nicotine as the substrate was negatively affected. This indicated that etfA-I and etfB-I were involved in oxidative degradation of nicotine by A. tumefaciens S33. According to our previous study, the etfAB-II genes (locus tags AWN88_RS08490 and AWN88_RS08495), which are located outside the nicotine-degrading gene cluster in strain S33, are transcribed at a similar moderate level when strain S33 was grown on both nicotine and glucose-ammonium media (19). This might explain the utilization of nicotine by the S33-ΔetfA-I and S33-ΔetfB-I mutants.
FIG 5.
Growth of wild-type A. tumefaciens S33 (black line), S33-ΔetfA-I mutant strain (red line), and S333-ΔetfB-I mutant strain (blue line) in HSP medium (A) and nicotine medium (B). HSP (0.5 g/liter) and nicotine (1 g/liter) were used as the sole sources of carbon and nitrogen in the two media, respectively. Data in figures are the means of the results from triple repeated experiments, and the error bars indicate the standard deviations of the data.
To verify this hypothesis, quantitative real-time reverse transcription-PCR (qRT-PCR) analysis was performed. As shown in Fig. 6, etfA-II showed a moderate expression in both the wild-type strain and the S33-ΔetfA-I and S33-ΔetfB-I mutants no matter whether grown in LB medium or nicotine medium. Contrarily, in LB medium, etfA-I had almost no expression, while in nicotine medium, it had a high expression in the wild-type strain and S33-ΔetfB-I mutant strain. This suggested that the moderate expression of etfA-II can make the S33-ΔetfA-I mutant survive in nicotine medium, and etfAB-I plays a major role in the growth of A. tumefaciens S33 utilizing nicotine as the substrate.
FIG 6.
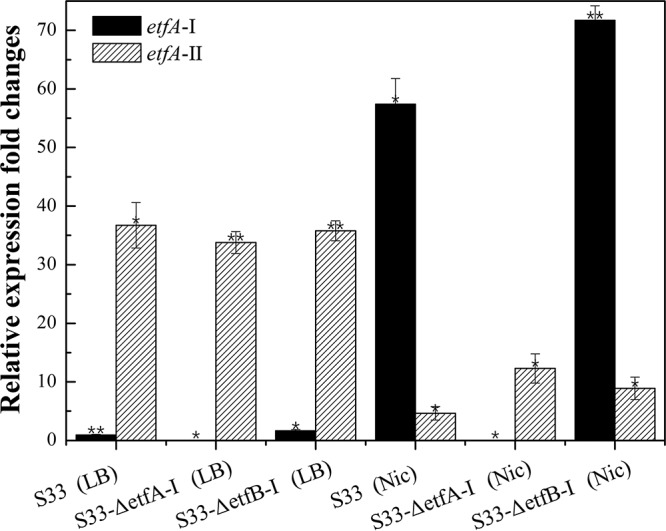
Expression levels of etfA-I and etfA-II in wild-type strain A. tumefaciens S33, S33-ΔetfA-I, and S333-ΔetfB-I cultured in LB medium (LB) and nicotine medium (Nic) determined by qRT-PCR analysis. The 16S rRNA gene was used as an internal reference, and the CT values of etfA-I in the wild-type strain cultured in LB medium were used as a baseline. Statistical analysis was performed on 2−ΔΔCT values with normalization to the 16S rRNA gene using paired Student’s t test by SPSS. **, P < 0.001; *, P < 0.05.
DISCUSSION
Previously, we copurified Pno with NdhAB from crude cell extracts of strain S33 and identified the localization of Pno in the cytoplasm and NdhAB in the periplasmic space (16, 17). Using DCPIP as the artificial electron acceptor, we showed that Pno catalyzes the oxidative deamination of 6-hydroxypseudooxynicotine to 6-hydroxy-3-succinoyl-semialdehyde-pyridine and previously believed it to function as an oxidase. However, in the current study, we discovered that the enzyme functions as a dehydrogenase. This conclusion is based on the finding that Pno utilizes EftAB as the physiological electron acceptor. It could clearly be shown that (i) the etfAB-I genes are clustered with the core nicotine-degrading genes in the genome and their transcriptional level is differentially upregulated when strain S33 is cultured on nicotine (19); (ii) both EtfAB-I and EtfAB-II could be reduced with pseudooxynicotine by Pno, while O2, NAD(P)+, or ferredoxin cannot serve as the electron acceptor for Pno; (iii) the oxygen atom in the aldehyde group of the product 3-succinoyl-semialdehyde-pyridine is derived from H2O; and (iv) disruption of etfAB-I genes decelerates the growth of strain S33 on nicotine. Based on these factual evidences, we have to rename the enzyme 6-hydroxypseudooxynicotine dehydrogenase. According to its enzymatic reaction, we classify it as a new member of EC 1.5.8., which is a group of enzymes acting on the CH-NH group of donors with a flavin as acceptor. In the Brenda enzyme database (https://www.brenda-enzymes.org/), there have been reported so far four members in this group, dimethylamine dehydrogenase (EC 1.5.8.1), trimethylamine dehydrogenase (EC 1.5.8.2), sarcosine dehydrogenase (EC 1.5.8.3), and dimethylglycine dehydrogenase (EC 1.5.8.4). All use EtfAB as an electron acceptor, which links their activities to the ETC.
It has been reported that a large number of dehydrogenases use Etfs as their physiological electron acceptor. Etf is composed of two subunits, and at least one of these subunits harbors a molecule of FAD. Based on sequence identity and functional differences, Etf homologs can be divided into five groups (38). Group I Etfs bind one FAD and one AMP and are found in Proteobacteria such as Paracoccus denitrificans and human mitochondria, and they are capable of passing electrons from at least nine mitochondrial matrix flavoprotein dehydrogenases (39–41) and bacterial dehydrogenases, like glutaryl-coenzyme A (glutaryl-CoA) dehydrogenase from P. denitrificans (42), to membrane-associated Euo. Bradyrhizobium japonicum, a nitrogen-fixing bacterium, has two sets of Etf-like genes, as follows: the first set of genes encoding EtfSL belonging to group I are expressed under aerobic conditions, while the second set of genes encoding FixAB belonging to group II are expressed under anaerobic conditions and are involved in nitrogen fixation (43). Group II Etfs function through a novel mechanism of electron bifurcation like Etfs from Clostridium kluyveri (44), Clostridium ljungdahlii, and Clostridioides difficile (38), and coordinate two FAD. Group III Etfs exist in a variety of bacterial and archaeal phyla. EtfAB from M. methylotrophus W3A1 belongs to this group and functions exclusively in the presence of trimethylamine dehydrogenase. Group IV and group V Etfs are confined to the bacterial phyla Bacteroidetes and Firmicutes, respectively. We aligned the two sets of EtfAB of strain S33 with Etfs from Pseudomonas stutzeri A1501 (34), M. methylotrophus W3A1, B. japonicum, P. denitrificans, humans, Sulfolobus solfataricus (45), C. kluyveri (44), E. coli, and Bacillus subtilis (46) (Fig. S8). It has been shown that EtfAB-I and EtfAB-II from A. tumefaciens S33 contain the FAD-binding consensus sequence PX[L,I,V]Y[L,I,V]AXGISGX[L,I,V]QHX2G in EtfA, the conserved signature sequence of EtfB, namely, VXRX2[E,D]X3[E,Q]X[L,I,V]X3LP[C,A][L,I,V]2, and the conserved AMP-binding sites (corresponding to V120 through D143 of EtfB from humans) which exist in group I Etfs only (40, 47, 48). In addition, EtfBs from A. tumefaciens S33 also contain the highly conserved partner region DLRLNEPRYA[S/T]LPNIMKAKKK of group I EtfBs. A further phylogenetic analysis (Fig. S9) showed that both EtfABs from strain S33 belong to group I. In spite of a high sequence identity (84% and 88% for EtfA-I and EtfA-II, and EtfB-I and EtfB-II, respectively), transcriptional analysis of A. tumefaciens S33 showed that only the EtfAB-I encoding genes had significantly higher expression levels in nicotine medium than in glucose-ammonium medium (19) and LB medium (Fig. 6). EtfAB-I-encoding genes are localized in close proximity to the pno gene in the nicotine-degrading gene cluster (Fig. 1B). The genes of EtfAB-II are outside the nicotine-degrading gene cluster and are expressed at a moderate level (19). This suggests that EtfAB-I plays a pivotal function as the electron acceptor of Pno in vivo. Upon disruption of EtfAB-I-encoding genes, EtfAB-II was likely able to aid in strain survival on nicotine medium. However, the low growth rate of the mutants on nicotine suggested that EtfAB-II complementation was not very effective, which could be explained by its moderate-level expression (Fig. 6).
Similar to other group I EtfABs from humans, P. denitrificans, and P. stutzeri, electrons from the reduced EtfAB in strain S33 are also transferred to CoQ by Euo-mediated catalysis (37, 49). This was verified by coupling the reduction of EtfAB-I with pseudooxynicotine by Pno to the reduction of CoQ with reduced EtfAB-I by Euo (Fig. 4B and S7). The location of euo together with etfAB-I and other nicotine-degrading genes and their coincident transcription pattern in nicotine medium also support these results (19). A genome search showed that strain S33 also harbors a second Euo-encoding gene (locus tag AWN88_24580, with 88% identity in protein sequence to the Euo characterized in this study), which is also outside the nicotine-degrading gene cluster. Protein sequence alignment (Fig. S10) showed that both Euo proteins from strain S33 are highly similar to those from humans, pigs, Drosophila melanogaster, and Rhodobacter sphaeroides. The six sequences have 92.4% consensus positions and 35.5% identity positions. Structurally, the Euo polypeptide does not traverse the entire membrane, but two highly hydrophobic peptide segments (β-hairpin [F114 to L131, in the case of Euo from pigs, here] and α-helix [G427 to W451]) form a “hydrophobic plateau” at the surface of the molecule, which may bind to the membrane. In addition, the FAD-binding domain (D37 to L57), the ubiquinone-binding domain (F114 to L131, G427 to W451, and T270 to H278), and the [4Fe4S]-binding domain (Q525 to I568) are also highly conserved (26, 49). However, the second euo gene in strain S33 has no significant expression level change in nicotine medium compared to glucose-ammonium medium (19), suggesting that it does not play a major role in nicotine catabolism. Thus, under the catalysis of Euo encoded by the gene in the nicotine-degrading gene cluster, the electrons are transferred from the reduced EtfAB to CoQ, which then could enter into the classic ETC. The route for electron transport from pseudooxynicotine to O2 could be constructed, by which ATP can be sythesized via chemiosmosis to support baterial growth.
Like Pno in this study, the pseudooxynicotine amine oxidase (Pao or Pnao) from Pseudomonas sp. strain HZN6 (10) and Pseudomonas putida S16 (50) also catalyzes the oxidative deamination of pseudooxynicotine with the formation of 3-succinoylsemialdehyde-pyridine, and the oxygen atom in the aldehyde group of the product 3-succinoyl-semialdehyde-pyridine is derived from H2O. However, there is almost no identity (less than 12%) in protein sequence between Pno and Pao/Pnao. Differences include that (i) Pno contains an FMN, a [4Fe4S], and an ADP (as indicated by the determination that 1 mol Pno contains 0.73 mol ADP), while Pao/Pnao binds only an FAD; and (ii) Pao/Pnao uses molecular oxygen rather than EtfAB as an electron acceptor, forming H2O2. It is known that Pao/Pnao is one of the crucial enzymes in the pyrrolidine pathway for nicotine degradation by Pseudomonas sp. strain HZN6 and P. putida S16, while strain S33 harboring Pno degrades nicotine via a novel hydrid of the pyrrolidine and the pyridine pathways, indicating the independent and diverse evolution of the biochemical pathways. Indeed, Shinella sp. strain HZN7 and Ochrobactrum sp. strain SJY1, also catabolizing nicotine via the hybrid pathway, harbor pno-etfAB-euo genes almost identical to those of strain S33, which do not exist in bacteria degrading nicotine by the pyrrolidine or the pyridine pathways (19).
Although all experiments in this study were performed using pseudooxynicotine, based on the very similar kinetic parameters and reaction products, these results may be assumed to be identical to potentially those obtained using the true substrate of Pno, namely, 6-hydroxypseudooxynicotine (commercially unvailable) (Fig. S1) (16). Thus, the current study provides novel insights in the nicotine oxidation process toward an in-depth understanding of the electron transfer reaction and energy metabolism involved in the catabolism of N-heterocyclic aromatic compounds in bacteria.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and materials.
A. tumefaciens S33 was deposited at the China Center for Type Culture Collection (CCTCC) under accession number CCTCC AB 2016054 (originally CCTCC M 206131) (13). It was incubated aerobically in nicotine or HSP medium at 30°C, as described previously (18), where nicotine and HSP were used as the sole source of carbon and nitrogen. E. coli BL21(DE3) [F− ompT hsdS(rB− mB−) gal dcm lacY1(DE3); Novagen], E. coli C43(DE3) [F− ompT hsdS(rB− mB−) gal dcm(DE3), harboring pCodonPlus (chloramphenicol resistant {Cmr}) and pRKISC (tetracycline resistant {Tcr}); gifted by Yasuhiro Takahashi from Osaka University], and E. coli S17-1 (recA pro hsdR17 RP4-2-Tcr::Mu-Kmr::Tn7 integrated into the chromosome) (51) were cultured in LB medium at 37°C. DCPIP, PMS, MTT, TTC, FAD, ADP, AMP, ferene [3-(2-pyridyl)-5,6-bis(5-sulfo-2-furyl)-1,2,4-triazinedisodium trihydrate], and coenzyme Q1 were purchased from Sigma-Aldrich (St. Louis, MO). H218O (97% purity) was obtained from Aladdin (Shanghai, China). Pseudooxynicotine was purchased from Toronto Research Chemicals, Inc. (Toronto, Ontario, Canada). HSP was prepared from (S)-nicotine by using whole cells of Pseudomonas putida S16 (11). Tetracycline (10 mg/liter), chloramphenicol (25 mg/liter), ampicillin (100 mg/liter), gentamicin sulfate (50 mg/liter), and kanamycin (50 mg/liter or 25 mg/liter) were used as required.
Activity of Pno with pseudooxynicotine as the substrate and identification of the reaction products.
The methods for preparing the recombinant Pno and measuring Pno activity were as previously described (16). The reaction was measured at 30°C by monitoring the reduction of DCPIP at 600 nm (ε = 21 mM−1·cm−1). In this study, we substituted 6-hydroxypseudooxynicotine with pseudooxynicotine as the substrate. The assay mixture contained 50 mM Tris-HCl buffer (pH 8.5), 1 mM pseudooxynicotine, 0.5 mM PMS, and 0.05 mM DCPIP. The optimum reaction pH was measured using the buffers prepared with 50 mM sodium phosphate or Tris-HCl with pH in the range of 7.5 to 9.5. One unit (U) was defined as the reduction of 1 μmol DCPIP per min. The Km value for the pseudooxynicotine in the reaction system was determined at the optimum pH. To identify the products of the reaction, the final mixture was analyzed using LC-MS (see “Analytical methods,” below, for details).
Heterologous expression and purification of EtfAB and Euo.
The etfA-I gene (locus tag AWN88_RS01180) and etfB-I gene (locus tag AWN88_RS01185) from A. tumefaciens S33 were amplified together with the genomic DNA of A. tumefaciens S33 as the template. The following primers were used: 5′-CTAGCTAGCATGCAAATAAACTCCGGCATT-3′ (forward, NheI recognition site is underlined) and 5′-CCGCTCGAGAAGCGCTTTCTGCAGTTCC-3′ (reverse, XhoI recognition site is underlined). The etfAB-I gene was cloned into plasmid pET28b(+) at the NheI and XhoI sites, and the construct was transformed into E. coli BL21(DE3) cells. The cells were cultured to an optical density at 600 nm (OD600) of 0.8 to 1.0 at 37°C in LB medium containing kanamycin (50 mg/liter) and riboflavin (50 mg/liter). Subsequently, 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture, and cells were grown overnight at 30°C. Cells were collected by centrifugation and resuspended with buffer (pH 7.4, 20 mM sodium phosphate, 0.28 M NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 10% glycerol) (52). Cells were broken by low-temperature ultrahigh-pressure continuous flow cell disruption using JN-02C (JNBIO, Guangzhou, China). The disrupted cells were centrifuged at 30,000 × g for 30 min at 4°C. The supernatant was loaded on a 5-ml HisTrap column (GE Healthcare, Little Chalfont, Buckinghamshire, UK) preequilibrated with binding buffer (20 mM sodium phosphate and 0.28 M NaCl [pH 7.4]). The target protein was eluted with 30% elution buffer (20 mM sodium phosphate, 0.28 M NaCl, and 0.5 M imidazole [pH 7.4]) and stored anaerobically at 4°C after concentration. The etfA-II gene (locus tag AWN88_RS08490) and etfB-II gene (locus tag AWN88_RS08495) were treated in the same way as the etfA-I gene and etfB-I gene. The following primers were used: 5′-CCGGAATTCGATGAAAATCCTTGTCCCCGTTAAAC-3′ (forward, EcoRI recognition site is underlined) and 5′-CCGCTCGAGGAGGGCCTTTTGCAGTTCC-3′ (reverse, XhoI recognition site is underlined).
The euo gene (locus tag AWN88_RS01170) from A. tumefaciens S33 was amplified with the following primers: 5′-CTGACCATATGAGCGAAGAAATGGAACTCCC-3′ (forward, NdeI recognition site is underlined) and 5′-CAAAGTAAGCTTTCACATGTTCGGATAGACCGGC-3′ (reverse, HindIII recognition site is underlined). The euo gene was cloned into plasmid pET28b(+) at the NdeI and HindIII sites and transformed into E. coli C43(DE3) cells. The cells were grown to an OD600 of 0.6 to 1.0 at 37°C in LB medium containing kanamycin (25 mg/liter), tetracycline (10 mg/liter), chloromycetin (25 mg/liter), riboflavin (2 µM), Fe3+(40 µM), and 8-hydroxyquinoline (40 µM) (53). Cells were grown overnight at 30°C after the addition of 0.5 mM IPTG. Cell collection, disruption, and subsequent centrifugation were performed as described above, with the exception of the addition of 0.2% (vol/vol) Triton X-100 to the binding buffer. Recombinant Euo was also purified as described above, with the exception of the addition of 0.2% (vol/vol) Triton X-100 to the binding and elution buffer. The target protein was eluted with 25 to 50% elution buffer and stored at 4°C after concentration.
Assay of Pno activity with EtfAB as the electron acceptor.
To determine the effect of EtfAB on Pno activity, different concentrations of purified EtfAB-I (0 to 16 μM) or EtfAB-II (0 to 7.5 μM) were added to the enzyme reaction system mentioned above in lieu of PMS. The DCPIP was still added, but its concentration was set to 0.1 mM. The reaction was monitored spectrophotometrically.
In order to determine the EtfAB reduction as the sole electron acceptor of Pno, the reaction was performed by adding purified EtfAB-I at a final concentration of 89 μM to a mixture containing 0.03 μM purified Pno and 1 mM pseudooxynicotine in 20 mM phosphate buffer (pH 7.4); DCPIP and PMS were not added to the reaction mixture. The reaction was monitored spectrophotometrically. The same experiment was carried out with EtfAB-II at a final concentration of 100 μM. The reaction system also included 0.05 μM purified Pno and 1 mM pseudooxynicotine in 20 mM phosphate buffer (pH 7.4).
Determination of the function of Euo in converting pseudooxynicotine to 3-succinoyl-semialdehyde-pyridine.
The reaction was performed by coupling with the transformation of pseudooxynicotine by Pno. The reaction mixture contained 50 mM Tris-HCl (pH 8.5), 1 mM pseudooxynicotine, 8 μM purified EtfAB-I, 0.05 μM purified Pno, and 32 μM CoQ1. The assay was started by adding 30 μM Euo at 30°C and monitored using a UV-visible spectrophotometer. The final reaction mixture was treated to further identify the products using LC-MS.
Analytical methods.
Protein concentration was determined using the Bradford assay, with bovine serum albumin as the standard (54). SDS-PAGE was performed using a 12.5% gel with the Bio-Rad Mini-Protean III cell. All enzymatic assays were carried out in quartz cuvettes (1-cm light path) filled with 500 μl of reaction mixture using a UV-visible Ultrospec 2100 Pro spectrophotometer (GE Healthcare, USA).
The reaction products were analyzed and identified using LC-MS. Briefly, the reaction mixtures were mixed with an equal volume of absolute ethyl alcohol (AR). After incubating for 3 h at room temperature, the mixture was centrifuged at 50,000 × g and 4°C for 30 min. The supernatant was used for injection. High-performance liquid chromatography–tandem mass spectrometry (HPLC-MS/MS) was performed on a rapid-separation liquid chromatography system (UltiMate3000 ultra-HPLC [UHPLC]; Dionex) coupled with an electrospray ionization-quadrupole time of flight (ESI-Q-TOF) mass spectrometer (Impact HD; Bruker Daltonics). Chromatographic separations were performed on a Zorbax Eclipse XDB-C18 column (250 mm by 4.6 mm, particle size 5 μm; Agilent) at 30°C, with a mobile phase system containing 8 mM formic acid in Milli-Q filtered water (A) and acetonitrile (B). For determining the products of oxidation of pseudooxynicotine by Pno with DCPIP as the electron acceptor, the following gradient program was applied at a flow rate of 0.5 ml/min: 0 to 20 min for 95% to 50% A plus 5% to 50% B, and 20 to 30 min for 50% to 95% A plus 50% to 5% B, and the injection volume was 5 μl. For determining the products of oxidation of pseudooxynicotine by Pno with EtfAB-I as the electron acceptor, the following gradient program was applied at a flow rate of 0.75 ml/min: 0 to 20 min for 95% to 50% A plus 5% to 50% B, and 20 to 45 min for 50% to 0% A plus 50% to 100% B, and the injection volume was 10 μl. MS analysis was conducted using the OTOF Control software (Bruker Daltonics) under the following conditions: ESI-positive mode for scanning, enhanced quadratic mode for calibration, nebulizer nitrogen gas with 105 Pa, dry gas nitrogen with 8 liters/min, probe temperature of 180°C, and full scan mass range from m/z 80 to 1,300. The data were analyzed using data analysis software (Bruker Daltonics).
The cofactors of the enzymes were measured by HPLC. For determining ADP and AMP, a mobile phase containing 15 mM KH2PO4 and methanol (90:10 [vol/vol]) was pumped through a Zorbax Eclipse XDB-C18 column (250 mm by 4.6 mm; particle size, 5 μm; Agilent) at a flow rate of 0.5 ml·min−1 using a submergible photon detector (SPD). The detection wavelength was set at 254 nm (55). For FAD and FMN, HPLC was performed on the same column with a mobile-phase system containing 5 mM ammonium acetate in Milli-Q filtered water and methanol (75:25 [vol/vol]) at a flow rate of 1 ml·min−1 using a fluorescence detector. The excitation wavelength was 450 nm, while the emission wavelength was 520 nm (56).
The iron content of the purified Euo was measured colorimetrically with ferene (57).
The relative molecular masses of the purified enzymes were measured by gel filtration on a GE Superdex G200 column (10 mm by 300 mm) calibrated with dextran blue 2000 (2,000 kDa; Sigma), ferritin from equine spleen (440 kDa; Sigma), γ-globulins from bovine blood (150 kDa, Solarbio), bovine serum albumin (66 kDa; Solarbio), albumin from chicken egg white (44.3 kDa; Sigma), myoglobin (16.7 kDa; Sigma), and vitamin B12 (1,360 Da; Sinopharm Chemical Reagent Co., Ltd., China). The system was equilibrated with 50 mM Tris-HCl buffer containing 150 mM NaCl (pH 7.4) and run at a flow rate of 0.5 ml·min−1.
Disruption of etfA and etfB genes.
The etfA-I and etfB-I genes in the genome of A. tumefaciens S33 were disrupted by using the suicide plasmid pJQ200SK by a homologous recombination method described previously (13). The upstream products amplified by primers A and B and downstream products amplified by primers C and D were recombined to obtain truncated ΔetfA-I and ΔetfB-I fragments via a third PCR step using primers B and C. The primer sequences for etfA-I gene disruption are as follows: 5′-CGCGGATCCATGGCCATTCTTCTTCTG-3′ (primer A, BamH I recognition site is underlined), 5′-GGAACTTCTCCGACAGGGCGGCAACG-3′ (primer B), 5′-CGTTGCCGCCCTGTCGGAGAAGTTCC-3′ (primer C), and 5′-CCGCTCGAGTCAAAGCGCTTTCTGCAGC-3′ (primer D, XhoI recognition site is underlined). The primer sequences for disruption of the etfB-I gene are as follows: 5′-CGCGGATCCATGCAAATAAACTCCGGCATTAAG-3′ (primer A, BamHI recognition site is underlined), 5′-TGCAGACCACCCACGGCGCGATCC-3′ (primer B), 5′-GGATCGCGCCGTGGGTGGTCTGCA-3′ (primer C), and 5′-CCGCTCGAGTTAGAGCACGCCGGCTTC-3′ (primer D, XhoI recognition site is underlined). The PCR products were cloned into the suicide plasmid pJQ200SK to generate deletion vectors. They were then transferred into A. tumefaciens S33 with the help of E. coli S17-1 through biparental mating. Single-crossover mutants were selected on HSP medium and via gentamicin (Gm) resistance. The selected single-crossover mutants were cultured in HSP medium containing sucrose (200 g/liter) to obtain double-crossover mutants S33-ΔetfA-I and S33-ΔetfB-I. To evaluate the effects of gene disruption on cell growth, the S33-ΔetfA-I and S33-ΔetfB-I mutant strains were grown in HSP and nicotine medium at 30°C, respectively. The wild-type strain A. tumefaciens S33, used as a control, was grown under identical conditions.
qRT-PCR analysis.
The expression levels of etfA-I and etfA-II in the wild-type strain and the S33-ΔetfA-I and S33-ΔetfB-I mutant strains grown in LB and nicotine medium were calculated by the relative quantitative PCR (qPCR). A. tumefaciens S33, S33-ΔetfA-I, and S33-ΔetfB-I were cultured in LB and nicotine medium, respectively, at 30°C to logarithmic-growth period. Harvested cells were used for RNA extraction using the EasyPure RNA kit (Transgen Biotech). The cDNA was synthesized by 5× All-In-One MasterMix with the AccuRT genomic DNA removal kit (ABM) and used as the templates for qPCR analysis. qPCR was carried out in a LightCycler 480 with TransStart Top Green qPCR supermix (Transgen Biotech). The primers for etfA-I are as follows: 5′-GAAATCATTGAGGTCGTCTCTGCTG-3′ (forward primer) and 5′-GGTCTCGACCGAAGCGCTG-3′ (reverse primer). The primers for etfA-II are as follows: 5′-GGCCAGGGCACCTTCG-3′ (forward primer) and 5′-GTTCAAACGAAGATCGCTGG-3′ (reverse primer). The primers for 16S rRNA gene are as follows: 5′-ACTCTGGAACTGCCTTTGATA-3′ (forward primer) and 5′-CGTTTACGGCGTGGACTA-3′ (reverse primer). The 16S rRNA gene was used as an internal reference. Three repeats for each sample were used for qPCR, after which the average threshold cycle (CT) was calculated for each sample. By using the CT values of etfA-I in the wild-type strain cultured in LB medium as a baseline, the relative fold changes in gene expression were calculated using the 2−ΔΔCT method. Statistical analysis was performed on 2−ΔΔCT values using paired Student’s t test by Statistical Product and Service Solutions (SPSS Statistics 22.0; IBM).
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by a grant from National Natural Science Foundation of China (grant 31470167).
We thank Ping Xu from Shanghai Jiao Tong University for his valuable support. We are also grateful to the anonymous reviewers for their critical comments and helpful suggestions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00454-19.
REFERENCES
- 1.Brandsch R. 2006. Microbiology and biochemistry of nicotine degradation. Appl Microbiol Biotechnol 69:493–498. doi: 10.1007/s00253-005-0226-0. [DOI] [PubMed] [Google Scholar]
- 2.Saitoh F, Noma M, Kawashima N. 1985. The alkaloid contents of sixty Nicotiana species. Phytochemistry 24:477–480. doi: 10.1016/S0031-9422(00)80751-7. [DOI] [Google Scholar]
- 3.Schwarz G, Lingens F. 1994. Bacterial degradation of N-heterocyclic compounds, p 459–486. In Ratledge C. (ed), Biochemistry of microbial degradation. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 4.Valcárcel Y, González Alonso S, Rodríguez-Gil JL, Gil A, Catalá M. 2011. Detection of pharmaceutically active compounds in the rivers and tap water of the Madrid Region (Spain) and potential ecotoxicological risk. Chemosphere 84:1336–1348. doi: 10.1016/j.chemosphere.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 5.González Alonso S, Valcarcel Y, Montero J, Catalá M. 2012. Nicotine occurrence in bottled mineral water: analysis of 10 brands of water in Spain. Sci Total Environ 416:527–531. doi: 10.1016/j.scitotenv.2011.11.046. [DOI] [PubMed] [Google Scholar]
- 6.Buerge I, Kahle M, Buser H-R, D Müller M, Poiger T. 2008. Nicotine derivatives in wastewater and surface waters: application as chemical markers for domestic wastewater. Environ Sci Technol 42:6354–6360. doi: 10.1021/es800455q. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Ma G, Chen T, Hou Y, Yang S, Zhang KQ, Yang J. 2015. Nicotine-degrading microorganisms and their potential applications. Appl Microbiol Biotechnol 99:3775–3785. doi: 10.1007/s00253-015-6525-1. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Li X, Duan Y, Zhang K, Yang J. 2010. Biotransformation of nicotine by microorganism: the case of Pseudomonas spp. Appl Microbiol Biotechnol 86:11–17. doi: 10.1007/s00253-009-2427-4. [DOI] [PubMed] [Google Scholar]
- 9.Brühmüller M, Möhler H, Decker K. 1972. Covalently bound flavin in d-6-hydroxynicotine oxidase from Arthrobacter oxidans. Eur J Biochem 29:143–151. doi: 10.1111/j.1432-1033.1972.tb01968.x. [DOI] [PubMed] [Google Scholar]
- 10.Qiu J, Ma Y, Wen Y, Chen L, Wu L, Liu W. 2012. Functional identification of two novel genes from Pseudomonas sp. strain HZN6 involved in the catabolism of nicotine. Appl Environ Microbiol 78:2154–2160. doi: 10.1128/AEM.07025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang SN, Xu P, Tang HZ, Meng J, Liu XL, Ma CQ. 2005. “Green” route to 6-hydroxy-3-succinoyl-pyridine from (S)-nicotine of tobacco waste by whole cells of a Pseudomonas sp. Environ Sci Technol 39:6877–6880. doi: 10.1021/es0500759. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Huang H, Xie K, Xu P. 2012. Identification of nicotine biotransformation intermediates by Agrobacterium tumefaciens strain S33 suggests a novel nicotine degradation pathway. Appl Microbiol Biotechnol 95:1567–1578. doi: 10.1007/s00253-012-4007-2. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Liu Z, Xu P. 2009. Biodegradation of nicotine by a newly isolated Agrobacterium sp. strain S33. J Appl Microbiol 107:838–847. doi: 10.1111/j.1365-2672.2009.04259.x. [DOI] [PubMed] [Google Scholar]
- 14.Ma Y, Wei Y, Qiu J, Wen R, Hong J, Liu W. 2014. Isolation, transposon mutagenesis, and characterization of the novel nicotine-degrading strain Shinella sp. HZN7. Appl Microbiol Biotechnol 98:2625–2636. doi: 10.1007/s00253-013-5207-0. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Tang H, Zhu X, Li Y, Xu P. 2015. Molecular mechanism of nicotine degradation by a newly isolated strain, Ochrobactrum sp. strain SJY1. Appl Environ Microbiol 81:272–281. doi: 10.1128/AEM.02265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Xie K, Yu W, Hu L, Huang H, Xie H, Wang S. 2016. Nicotine dehydrogenase complexed with 6-hydroxypseudooxynicotine oxidase involved in the hybrid nicotine-degrading pathway in Agrobacterium tumefaciens S33. Appl Environ Microbiol 82:1745–1755. doi: 10.1128/AEM.03909-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu W, Wang R, Huang H, Xie H, Wang S. 2017. Periplasmic nicotine dehydrogenase NdhAB utilizes pseudoazurin as its physiological electron acceptor in Agrobacterium tumefaciens S33. Appl Environ Microbiol 83:e01050-17. doi: 10.1128/AEM.01050-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Xie K, Huang H, Wang S. 2014. 6-Hydroxy-3-succinoylpyridine hydroxylase catalyzes a central step of nicotine degradation in Agrobacterium tumefaciens S33. PLoS One 9:e103324. doi: 10.1371/journal.pone.0103324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H, Yu W, Wang R, Li H, Xie H, Wang S. 2017. Genomic and transcriptomic analyses of Agrobacterium tumefaciens S33 reveal the molecular mechanism of a novel hybrid nicotine-degrading pathway. Sci Rep 7:4813. doi: 10.1038/s41598-017-05320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu W, Wang R, Li H, Liang J, Wang Y, Huang H, Xie H, Wang S. 2017. Green route to synthesis of valuable chemical 6-hydroxynicotine from nicotine in tobacco wastes using genetically engineered Agrobacterium tumefaciens S33. Biotechnol Biofuels 10:288. doi: 10.1186/s13068-017-0976-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu J, Wei Y, Ma Y, Wen R, Wen Y, Liu W. 2014. A novel (S)-6-hydroxynicotine oxidase gene from Shinella sp. strain HZN7. Appl Environ Microbiol 80:5552–5560. doi: 10.1128/AEM.01312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu J, Liu M, Wen R, Zhang D, Hong J. 2015. Regulators essential for nicotine degradation in Shinella sp. HZN7. Process Biochem 50:1947–1950. doi: 10.1016/j.procbio.2015.07.013. [DOI] [Google Scholar]
- 23.Qiu J, Yang Y, Zhang J, Wang H, Ma Y, He J, Lu Z. 2016. The complete genome sequence of the nicotine-degrading bacterium Shinella sp. HZN7. Front Microbiol 7:1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, Tang H, Li Y, Xu P. 2015. Molybdenum-containing nicotine hydroxylase genes in a nicotine degradation pathway that is a variant of the pyridine and pyrrolidine pathways. Appl Environ Microbiol 81:8330–8338. doi: 10.1128/AEM.02253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H, Li Y, Tang H, Xu P. 2014. Genome sequence of a newly isolated nicotine-degrading bacterium, Ochrobactrum sp. SJY1. Genome Announc 2:e00720-14. doi: 10.1128/genomeA.00720-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Frerman FE, Kim J-J. 2006. Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. Proc Natl Acad Sci 103:16212–16217. doi: 10.1073/pnas.0604567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramsay RR, Steenkamp DJ, Husain M. 1987. Reactions of electron-transfer flavoprotein and electron-transfer flavoprotein: ubiquinone oxidoreductase. Biochem J 241:883–892. doi: 10.1042/bj2410883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beckmann JD, Frerman FE. 1985. Reaction of electron-transfer flavoprotein with electron-transfer flavoprotein-ubiquinone oxidoreductase. Biochemistry 24:3922–3925. doi: 10.1021/bi00336a017. [DOI] [PubMed] [Google Scholar]
- 29.Steenkamp DJ, Beinert H. 1982. Mechanistic studies on the dehydrogenases of methylotrophic bacteria. 2. Kinetic studies on the intramolecular electron transfer in trimethylamine and dimethylamine dehydrogenase. Biochem J 1:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White SA, Mathews FS, Rohlfs RJ, Hille R. 1994. Crystallization and preliminary crystallographic investigation of electron-transfer flavoprotein from the bacterium Methylophilus W3A1. J Mol Biol 240:265–266. doi: 10.1006/jmbi.1994.1440. [DOI] [PubMed] [Google Scholar]
- 31.Chen D, Swenson RP. 1994. Cloning, sequence analysis, and expression of the genes encoding the two subunits of the methylotrophic bacterium W3A1 electron transfer flavoprotein. J Biol Chem 269:32120–32130. [PubMed] [Google Scholar]
- 32.Davidson VL, Husain M, Neher JW. 1986. Electron transfer flavoprotein from Methylophilus methylotrophus: properties, comparison with other electron transfer flavoproteins, and regulation of expression by carbon source. J Bacteriol 166:812–817. doi: 10.1128/jb.166.3.812-817.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jang MH, Scrutton NS, Hille R. 2000. Formation of W3A1 electron-transferring flavoprotein (ETF) hydroquinone in the trimethylamine dehydrogenase-ETF protein complex. J Biol Chem 275:12546–12552. doi: 10.1074/jbc.275.17.12546. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Zhang M, Gao C, Zhang Y, Ge Y, Guo S, Guo X, Zhou Z, Liu Q, Zhang Y, Ma C, Tao F, Xu P. 2017. Coupling between d-3-phosphoglycerate dehydrogenase and d-2-hydroxyglutarate dehydrogenase drives bacterial l-serine synthesis. Proc Natl Acad Sci U S A 114:E7574–E7582. doi: 10.1073/pnas.1619034114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujieda N, Satoh A, Tsuse N, Kano K, Ikeda T. 2004. 6-S-Cysteinyl flavin mononucleotide-containing histamine dehydrogenase from Nocardioides simplex: molecular cloning, sequencing, overexpression, and characterization of redox centers of enzyme. Biochemistry 43:10800–10808. doi: 10.1021/bi049061q. [DOI] [PubMed] [Google Scholar]
- 36.Jones M, Talfournier F, Bobrov A, Günter Grossmann J, Vekshin N, Sutcliffe MJ, Scrutton NS. 2002. Electron transfer and conformational change in complexes of trimethylamine dehydrogenase and electron transferring flavoprotein. J Biol Chem 277:8457–8465. doi: 10.1074/jbc.M111105200. [DOI] [PubMed] [Google Scholar]
- 37.Ruzicka FJ, Beinert H. 1977. A new iron-sulfur flavoprotein of the respiratory chain. J Biol Chem 252:8440–8445. [PubMed] [Google Scholar]
- 38.Garcia Costas AM, Poudel S, Miller A-F, Schut GJ, Ledbetter RN, Fixen KR, Seefeldt LC, Adams MWW, Harwood CS, Boyd ES, Peters JW. 2017. Defining electron bifurcation in the electron-transferring flavoprotein family. J Bacteriol 199:e00440-17. doi: 10.1128/JB.00440-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker AR. 2003. Binding of the human “electron transferring flavoprotein” (ETF) to the medium chain acyl-CoA dehydrogenase (MCAD) involves an arginine and histidine residue. J Enzyme Inhib Med Chem 18:453–462. doi: 10.1080/1475636031000138741. [DOI] [PubMed] [Google Scholar]
- 40.Roberts DL, Frerman FE, Kim JJ. 1996. Three-dimensional structure of human electron transfer flavoprotein to 2.1-Å resolution. Proc Natl Acad Sci U S A 93:14355–14360. doi: 10.1073/pnas.93.25.14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crane FL, Mii S, Hauge JG, Green DE, Beinert H. 1956. On the mechanism of dehydrigenation of fatty acyl derivates of coenzyme A. I. The general fatty acyl coenzyme A dehydrogenase. J Biol Chem 218:701–716. [PubMed] [Google Scholar]
- 42.Roberts DL, Salazar D, Fulmer JP, Frerman FE, Kim J. 1999. Crystal structure of Paracoccus denitrificans electron transfer flavoprotein: structural and electrostatic analysis of a conserved flavin binding domain. Biochemistry 38:1977–1989. doi: 10.1021/bi9820917. [DOI] [PubMed] [Google Scholar]
- 43.Weidenhaupt M, Rossi P, Beck C, Fischer HM, Hennecke H. 1996. Bradyrhizobium japonicum possesses two discrete sets of electron transfer flavoprotein genes: fixA, fixB and etfS, etfL. Arch Microbiol 165:169–178. [DOI] [PubMed] [Google Scholar]
- 44.Li F, Hinderberger J, Seedorf H, Zhang J, Buckel W, Thauer RK. 2008. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol 190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quaiser A, Ochsenreiter T, Klenk HP, Kletzin A, Treusch AH, Meurer G, Eck J, Sensen CW, Schleper C. 2002. First insight into the genome of an uncultivated crenarchaeote from soil. Environ Microbiol 4:603–611. doi: 10.1046/j.1462-2920.2002.00345.x. [DOI] [PubMed] [Google Scholar]
- 46.Matsuoka H, Hirooka K, Fujita Y. 2007. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J Biol Chem 282:5180–5194. doi: 10.1074/jbc.M606831200. [DOI] [PubMed] [Google Scholar]
- 47.Toogood HS, Leys D, Scrutton NS. 2007. Dynamics driving function–new insights from electron transferring flavoproteins and partner complexes. FEBS J 274:5481–5504. doi: 10.1111/j.1742-4658.2007.06107.x. [DOI] [PubMed] [Google Scholar]
- 48.Tsai MH, Saier MH. 1995. Phylogenetic characterization of the ubiquitous electron transfer flavoprotein families ETF-α and ETF-β. Res Microbiol 146:397–404. doi: 10.1016/0923-2508(96)80285-3. [DOI] [PubMed] [Google Scholar]
- 49.Watmough NJ, Frerman FE. 2010. The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim Biophys Acta 1797:1910–1916. doi: 10.1016/j.bbabio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 50.Hu H, Wang W, Tang H, Xu P. 2015. Characterization of pseudooxynicotine amine oxidase of Pseudomonas putida S16 that is crucial for nicotine degradation. Sci Rep 5:17770. doi: 10.1038/srep17770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 52.Steenkamp DJ, Gallup M. 1978. The natural flavoprotein electron acceptor of trimethylamine dehydrogenase. J Biol Chem 253:4086–4089. [PubMed] [Google Scholar]
- 53.Usselman RJ, Fielding AJ, Frerman FE, Watmough NJ, Eaton GR, Eaton SS. 2008. Impact of mutations on the midpoint potential of the [4Fe-4S]+1,+2 cluster and on catalytic activity in electron transfer flavoprotein-ubiquinone oxidoreductase (ETF-QO). Biochemistry 47:92–100. doi: 10.1021/bi701859s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 55.Zheng Y, Chen S, Ye W, Lu J. 2012. Establishment of high-performance liquid chromatography for energy metabolic materials assay and its applications. Bull Sci Technol 28:46–50. [Google Scholar]
- 56.Wei J, Guo C, Xu J, Yang J, Wei W, Xu Q. 2006. Determination of riboflavin, FMN and FAD in human plasma and erythrocytes by high performance liquid chromatography. Acta Nutr Sin 28:79–82. [Google Scholar]
- 57.Pierik AJ, Wolbert RBG, Mutsaers PHA, Hagen WR, Veeger C. 1992. Purification and biochemical characterization of a putative [6Fe-6S] prismane-cluster-containing protein from Desulfovibrio vulgaris (Hildenborough). Eur J Biochem 206:697–704. doi: 10.1111/j.1432-1033.1992.tb16976.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.