Abstract
RNA localization is a fundamental mechanism for controlling cell structure and function. Early development in fish and amphibians requires the localization of specific mRNAs to establish the initial differences in cell fates prior to the onset of zygotic genome activation. RNA localization in these oocytes (e.g., Xenopus and zebrafish) requires that animal-vegetal polarity be established early in oogenesis, mediated by formation of the Balbiani body/mitochondrial cloud. This structure serves as a platform for assembly and transport of germline determinants to the future vegetal pole and also sets up the machinery for the localization of non-germline transcripts later in oogenesis. Understanding these polarization and localization mechanisms is critical for understanding the basis for early embryonic development in these organisms and also for understanding the role of RNA compartmentalization in animal gametogenesis. Here we outline recent advances in elucidating the molecular basis for the establishment of oocyte polarity at the level of Balbiani body assembly as well as the formation of RNP assemblies for early and late pathway mRNA localization in the oocyte.
1. Introduction
The eggs of many animals are highly polarized, often leading to different cell fates arising from different regions of the egg. The amphibian egg is a long-studied example of a polarized egg: the pigmented animal pole region, from which the polar body is extruded, forms the nervous system and epidermis, whereas the gut derivatives are derived from the vegetal pole. Additionally, in anuran amphibians such as Xenopus, the future germline arises from vegetal cells through the inheritance of germ plasm, a cytologically distinct collection of mitochondria and other organelles, along with dense germ granules. Classic amphibian embryological experiments have demonstrated the general importance of regional vegetal factors in critical aspects of development, including dorsal axis formation, germ layer establishment, and primordial germ cell (PGC) fate. Relative displacement of the vegetal cortex was implicated in dorsoventral axis patterning (Ancel and Vintembenger 1948), and vegetal cells induce mesoderm fate in more equatorial and animal cells (Nieuwkoop 1969; Sudarwati and Nieuwkoop 1971). Additionally, ultraviolet irradiation of the vegetal pole (but not the animal pole) leads to loss of PGCs and sterility that is rescuable by transplantation of vegetal cytoplasm (Bounoure 1937; Bounoure et al. 1954; Smith 1966; Ikenishi et al. 1974; Züst and Dixon 1975; Wakahara 1977).
Since these seminal experiments, a number of equally important studies have established primary roles for individual maternal messenger RNAs (mRNAs) localized during oogenesis in mediating germ layer and PGC fates in Xenopus development. Since the cloning of gdf1 (alias vg1), the first localized mRNA to be identified in any organism (Rebagliati et al. 1985; Weeks and Melton 1987), improvements in technology and the sequencing of Xenopus genomes (Hellsten et al. 2010; Session et al. 2016) have allowed the large-scale characterization and annotation of vegetally localized mRNAs. Subtractive hybridization, different microarray strategies, and RNA sequencing have all been used and have generated quite consistent and progressively more complete results (reviewed in Houston 2013). Estimates of the total number of vegetally localized RNAs, around 400 (out of 10,000 maternally expressed mRNAs), are satisfyingly close to the original estimates of vegetal RNA localization obtained from kinetic hybridization experiments [~4% of total poly(A) RNA (Carpenter and Klein 1982; Cuykendall and Houston 2010)]. Additionally, specific pathways mediating the localization of transcripts to the vegetal pole have been characterized, involving regulation of the cytoskeleton and specific RNA-binding proteins. In some cases, homologous proteins mediate RNA localization in somatic cells, suggesting the existence of a conserved core mechanism for RNA transport in cells (Kloc et al. 2002b; King et al. 2005; Houston 2013).
In Xenopus and zebrafish, animal-vegetal polarity is established early in oogenesis, in conjunction with the formation of the conspicuous Balbiani body/mitochondrial cloud. This structure serves as a hub for localization for the so-called early pathway/METRO RNAs and is the main pathway for localization to the germ plasm (Forristall et al. 1995; Kloc and Etkin 1995). During the vitellogenic stages, this mitochondrial cloud breaks down, and the fragments localize to the future vegetal pole cortex, establishing a definitive structural polarity to the oocyte. This fragmenting cloud sets up the path for “late pathway” mRNAs to localize to the cortex (reviewed in Kloc et al. 2002b). These mRNAs generally lack strict commonality of function but rather seem to act broadly in germ layer induction and patterning in the vegetal hemisphere. A subset of mRNAs can localize to the germ plasm but also more widely form a little understood “intermediate pathway” (reviewed in Houston 2013). This review will highlight recent advances in understanding the establishment of animal-vegetal polarity and formation of the Balbiani body in both Xenopus and zebrafish as well as the formation of ribonucleoprotein (RNP) granules involved in RNA localization and their roles in the various localization pathways.
2. The Origin of Animal-Vegetal Polarity During Oogenesis
2.1. Oocyte Formation and Polarization
Oogenesis in vertebrates begins when PGCs reach the genital ridge and form germline cysts, with oogonia remaining connected by ring canals after incomplete cell divisions (Pepling et al. 1999). These ring canals are intercellular bridges thought to be important in distributing nutrients and maintaining synchronous development signals in the premeiotic oogonia. In Xenopus and also zebrafish, oogenesis is a continuous process in the adult, with cysts arising from germline stem cells at different time points and resulting in a collection of asynchronous stages in the mature female ovary (Pelegri 2003; Rasar and Hammes 2006). Cysts form through a series of four mitotic divisions in the nest providing 16 cystocytes all of which will eventually form oocytes (Kloc et al. 2004a). When the ring canals are broken down, the cystocytes are disconnected and enter pre-stage I of oogenesis where prophase of meiosis I begins (al-Mukhtar and Webb 1971; Coggins 1973).
Subsequently, the Balbiani body develops from one of many “premitochondrial clouds” (Heasman et al. 1984; Kloc et al. 1996) adjacent to the germinal vesicle (GV; i.e., the nucleus in oocytes) and becomes positioned toward the future vegetal pole. This marks the first incidence of asymmetry and the orientation of AV polarity in a cell that must expand at least 100 times in size (Dumont 1972; Selman et al. 1993). As the oocyte concludes stage I and transitions into stage II, the oocyte is arrested in diplotene of meiosis I. During the subsequent progression through oogenesis (Dumont 1972; Selman 1993), there is a massive growth phase to accommodate the accumulation of yolk, synthesis of proteins and RNAs, as well as a drastic rearrangement of cytoskeleton.
The formation and function of the Balbiani body,1 or mitochondrial cloud, in early oocytes have remained largely mysterious since its discovery in myriapods and arachnids (reviewed in Wilson 1928; Kloc et al. 2004b). The Balbiani body appears in early oocytes of many animals, including the mouse, which was thought to be a notable exception until recently (Guraya 1979; Kloc et al. 2004b, 2008; Pepling et al. 2007). In organisms with maternal inheritance of germ plasm, the Balbiani body serves as the source for this material (Heasman et al. 1984), whereas the structure uniformly disperses in mammals (Pepling et al. 2007; Kloc et al. 2008) and presumably other organisms as well. The ultrastructure of the Balbiani body is similar to other intracellular “bodies” including the chromatoid body in spermatids, the germ plasm in oocytes, and other RNA-processing structures (e.g., P bodies, stress granules). It is likely that the assembly of such a structure in germ cells is essential for gametogenesis. And, this structure may have been adapted to localize and store mRNAs and contribute to germ plasm in diverse animal lineages where it has evolved (reviewed in Extavour and Akam 2003). Additionally, the Balbiani body may act to select and concentrate the more active (i.e., “healthy”) mitochondria (Kogo et al. 2011; Boke et al. 2016), a feature that would also be useful for future PGCs inheriting germ plasm, since early embryos typically do not generate new mitochondria [Xenopus: (El Meziane et al. 1989); mouse: (Shoubridge and Wai 2007)].
2.2. Molecular Regulation of Balbiani Body Assembly
Maternal effect genetic screens in zebrafish have provided critical insight into Balbiani body assembly (Dosch et al. 2004; Wagner et al. 2004). Mutants in the buckyball (buc) locus showed a loss of AV polarity during oogenesis, failure of Balbiani body formation, and concomitant loss of germ plasm RNA localization and animal pole cytoplasmic streaming and blastodisc formation (Dosch et al. 2004; Marlow and Mullins 2008; Bontems et al. 2009). Buc encodes a rapidly evolving novel vertebrate protein with homology to Velo1, the product of a localized mRNA in Xenopus [(Claussen and Pieler 2004; Bontems et al. 2009); Velo1 hereafter]. In fish embryos, overexpressed Velo1 can induce Balbiani body-like structures competent to recruit localized mRNAs (Bontems et al. 2009), suggesting that Velo1 may function analogously to Drosophila oskar in assembling the germ plasm, although there is no sequence or structural homology. Recent data suggest that Velo1 contributes to Balbiani body assembly by forming amyloid-like fibrils (Boke et al. 2016; see below).
Zebrafish magellan (mgn) mutants also exhibit Balbiani body defects (Dosch et al. 2004; Gupta et al. 2010), although in this case the Balbiani body is enlarged and mispositioned. Germ plasm RNAs can localize but are not distributed vegetally. The mgn locus maps to microtubule-actin cross-linking factor 1 (macf1; (Gupta et al. 2010), which encodes a large and conserved spectraplakin-related protein, distantly related to the Shortstop protein in Drosophila, a component of the organellar fusome involved in germline cyst formation (Bottenberg et al. 2009). Macf1 has several functional motifs and can interact with F-actin, microtubules, and intermediate filaments to regulate cytoskeletal organization and organelle trafficking (Suozzi et al. 2012). Recent data suggest that Macf1 is required for basal body positioning and maintenance of polarity in retinal photoreceptors, suggesting similar mechanisms may mediate oocyte polarity and apicobasal polarity. Fusome material (likely containing Macf1) is clustered with the centrosome and premitochondrial cloud aggregates in Xenopus germline cysts (Kloc et al. 2004a), providing a site of possible functional interaction that might explain the apparent opposing effects for Macf1 and Velo1. Also, the Balbiani body becomes surrounded by a “net” of cytokeratin (Heasman et al. 1984; Gard et al. 1997), which could interact with Macf1 during its localization or disassembly.
Evidence suggests that Balbiani body assembly involves a feedback localization loop with velo1 mRNA and protein and self-organizing properties of Velo1 protein itself. In zebrafish, Velo1 protein is localized asymmetrically and perinuclearly in premitochondrial cloud-stage oocytes [zygotene; prior to velo1 mRNA localization; (Heim et al. 2014)]. Also, Velo1 protein binds to putative translation-promoting RNA-binding proteins Dazl and Rbpms2 (alias Hermes), which themselves bind velo1 mRNA (as well as other mRNAs, including possibly their own) (Heim et al. 2014)—thus, a local stochastic activation of Velo1 translation would be sufficient to recruit additional Velo1 and amplify its own expression and localization. However, velo1 RNA is not localized to the mitochondrial cloud in Xenopus, so it is possible that recruitment of the protein is more critical.
Interestingly, Elkouby et al. (2016) show that the Balbiani body in zebrafish arises subsequent to the formation of the “chromosome bouquet,” a polarized arrangement of telomeres at the nuclear envelope, during zygotene of meiosis I (Fig. 1). The bouquet is formed during the early stages of prophase and is thought to function in homologous chromosome pairing (Zickler and Kleckner 1998; Scherthan 2001; Harper et al. 2004). Both the bouquet and Balbiani body are conserved in animal meiosis but had not previously been linked together. Co-localization studies showed that Balbiani body/germ plasm components localize to the bouquet at the nuclear envelope during zygotene, along with mitochondria, the centrosome, and nuclear pore proteins (Elkouby et al. 2016). These germ plasm components include dazl mRNA (see above), Velo1 and Ddx4 (alias Vasa) proteins, as well as Piwi pathway proteins (Piwil1/2, alias Ziwi/Zili). Analysis of buc mutant zebrafish showed that bouquet formation and initial polarization are normal in buc mutants and that AV polarity is eventually lost in the absence of Balbiani body assembly. Importantly, inhibition of microtubule polymerization (nocodazole) blocked the clustering of telomeres, centrosomes and Balbiani components, suggesting that bouquet microtubules are the critical connection between nuclear asymmetry and the assembly of the Balbiani body and establishment of the future vegetal pole (Elkouby et al. 2016). The role of microtubules in this process remains unclear, as does the ultimate source of asymmetry, although Elkouby et al. (2016) hint that the final cell division plane in the germline cyst may be involved.
Fig. 1.
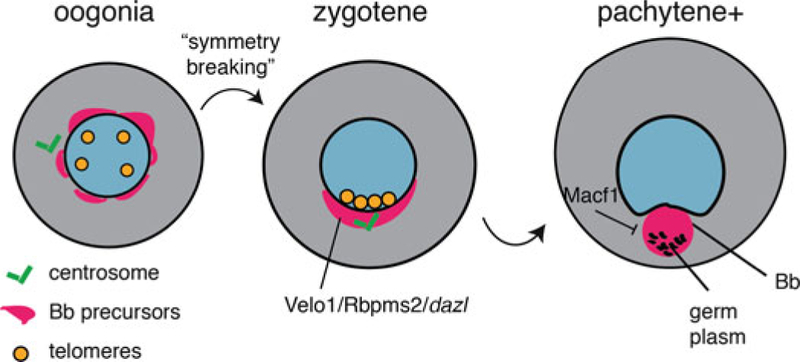
Generalized stages of Balbiani body assembly (Bb). Stages of oogenesis are indicated above each panel. Early oogonia are not polarized and undergo a symmetry-breaking event upon meiotic entry, leading to chromosomal bouquet formation, clustering of telomeres, and alignment with the centrosome. Subsequently, in zygotene of meiosis I, Bb precursors are polarized, including Velo1 protein and RNA, Rbpms2, and dazl RNA. Velo1 self-organizes into an amyloid scaffold for the Bb, and localized RNAs are recruited and assembled into germ plasm/germ granules in a subregion of the Bb. Macf1 plays an inhibitory role in Bb assembly
2.3. Structural Role of Velo1 in the Balbiani Body and Germ Plasm
Recent data suggest that Velo1 likely plays a key structural role in the formation and possibly the dispersal of the Balbiani body and germ plasm. Full-length Velo1 protein is highly enriched in the Balbiani body (Nijjar and Woodland 2013a; Heim et al. 2014; Boke et al. 2016). In Xenopus there is also a shorter variant lacking a region in the C-terminal intrinsically disordered region (IDR) that does not localize to the Balbiani body but accumulates in later-stage germ plasm islands (Nijjar and Woodland 2013a). Depletion of this short velo1 isoform RNA with antisense oligos (but not the full-length RNA) resulted in dispersal of germ plasm islands in cultured stage VI oocytes (Nijjar and Woodland 2013a). Additionally, Balbiani bodies bind Thioflavin T, a fluorescent probe specific for amyloid fibrils (Boke et al. 2016), suggesting a role for these structures in the Balbiani body. The N-terminus of Velo1 contains a prion-like domain (PLD), a sequence involved in promoting conformational changes to form amyloid fibers (Alberti et al. 2009). Tagged Velo1 can localize to the Balbiani body in a manner dependent on its specific PLD (Nijjar and Woodland 2013a; Boke et al. 2016). Also, Velo1 protein can form amyloid fibrils in vitro and can also directly mediate clustering of mitochondria and RNA. The latter activity was non-sequence-specific however (Boke et al. 2016).
Balbiani body Velo1 is thought to form a stable matrix, as shown through low mobility in FRAP experiments. Interestingly, however, this behavior may change during Balbiani fragmentation during oogenesis. The Velo1 PLD was also necessary and sufficient for localization to germ plasm islands when injected into stage VI oocytes, whereas the amyloid-forming IDR was dispensable (Nijjar and Woodland 2013a). Furthermore, Velo1 may be phosphorylated in eggs and is likely not present as amyloid (Boke et al. 2016). Correspondingly, Velo1 and other proteins are mobile in stage VI germ plasm, which would also be inconsistent with an inert amyloid structure (Nijjar and Woodland 2013a). The significance of these differences in Velo1 behavior are not clear, but it has been known for some time, based on ultrastructural and mRNA localization studies, that germ plasm assembles in a specific vegetal subregion of the Balbiani body (Kloc and Etkin 1995; Kloc et al. 2002b). Thus, the formation of bona fide germ plasm may involve different structural forms of Velo1, likely in addition to specific protein-protein interactions and the formation of germ plasm RNP granules [i.e., see (Nijjar and Woodland 2013a; Aguero et al. 2016)]. These mechanisms will be briefly reviewed in the following section. It is interesting to note however that these RNPs are thought to form via structural transition states involving disordered protein domains, although involving hydrogel states as opposed to amyloid.
Taken together, these studies suggest that the Balbiani body is a semi-inert mass of amyloid and other proteins, mitochondria, and RNAs that is localized by nuclear asymmetry in early meiosis and formed through localized self-organization. The Balbiani body may contribute to general early oogenesis and also serve as the site for RNA localization and germ plasm assembly in animals that possess it.
3. RNA Localization Mechanisms
3.1. RNA Localization Pathways in Xenopus
Two main general mechanisms for RNA localization in Xenopus oocytes have been identified (Forristall et al. 1995; Kloc and Etkin 1995): an early pathway in stage I oocytes which localizes RNAs to the germ plasm for germline specification and axis formation and a late pathway occurring around stage III that localizes RNAs via microtubules to the cortex throughout the vegetal hemisphere for somatic fate (reviewed in Houston 2013; summarized in Fig. 2). A number of RNA sequence motifs within larger localization elements (LE) have been identified in addition to required RNA-binding proteins; however, the same cis and trans-acting components appear to be used by both localization pathways. These elements generally include superclusters of UUCAC (VM1) and UUUCU (E2) motifs, which are bound by Igf2bp3 and Ptbp1 proteins, respectively (reviewed in Houston 2013). However, these motifs may only be a requirement for a distinct population of RNAs, including the well-studied gdf1 and vegt. Additional localized RNAs including dnd1, velo1, and plin2 do not possess clustered UUCAC and UUUCU motifs, yet they continue to localize to the vegetal pole (Chan et al. 1999; Claussen et al. 2004; Horvay et al. 2006), indicating that much remains unknown about the ultimate determinants of how RNAs are recognized for transport.
Fig. 2.
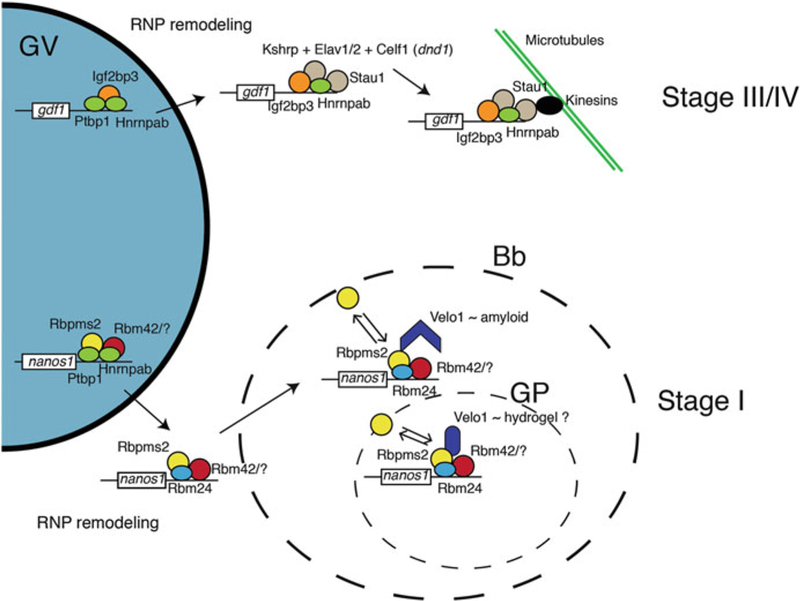
Schematic of RNA localization pathways in the Xenopus oocyte. For the late localization pathway (stage III/IV), Ptbp1 and Hnrnpab associate with target RNAs in the germinal vesicle (GV, e.g., gdf1) along with Igf2bp3. Upon nuclear export, RNPs are remodeled to remove Ptbp1and allow binding of Igf2bp3 to RNA. Other factors also associate with the complex, including Kshrp and Elav proteins and/or Celf1 in the case of dnd1 mRNA localization. Also, Stau1 binds late localizing RNAs in the cytoplasm and couples them to the microtubule cytoskeleton. For the early pathway (stage I), RNPs are likely nucleated in the GV by Ptbp1 and Hnrnpab, in common with the late pathway, along with Rbpms2/Hermes and an undefined RNA-binding protein partner (e.g., Rbm42 and others). In the cytoplasm, RNPs are remodeled to remove Ptbp1and Hnrnpab and incorporate other RNA-binding proteins (e.g., Rbm24). These RNPs then associate with the Bb and then the germ plasm (GP) by diffusion-entrapment, likely mediated by Rbpms2 interaction with different conformational aggregate forms of the Velo1 protein. Double arrows indicate that Rbpms2 is likely in equilibrium in these complexes
One of the main roles of mRNA localization is to spatially and temporally regulate translation of transcripts to restrict protein expression. Although long associated with the germline, it is becoming increasingly apparent that the compartmentalization of various aspects of RNA metabolism into large-scale macroscopic bodies or assemblages is critical for normal cell structure and function (reviewed in Voronina et al. 2011; Toretsky and Wright 2014; Courchaine et al. 2016). Additionally, these bodies lack surrounding membranes and are thought to form “droplets” by what is termed liquid-liquid phase separation (Courchaine et al. 2016). Such bodies include in the nucleus, the nucleoli, Cajal bodies, and nuclear speckles, and in the cytoplasm, the P bodies, stress granules, and germ granules (Voronina et al. 2011). Centrosomes likely also exhibit this behavior (Zwicker et al. 2014). Liquid phase transition behavior was first seen for nematode P granules, homologues of the germ granules (Brangwynne et al. 2009). This transition has since been shown to depend on low complexity/intrinsically disordered protein regions (LC/IDRs) present in many RNA-binding proteins, with droplet formation occurring at high local concentrations of proteins and RNAs (Kato et al. 2012; Han et al. 2012). Additionally, because these structures are dynamic, their regulated phase transitions would be a compelling mechanism for the packaging and unpacking of cell fate determinants. Table 1 summarizes the RNA-binding proteins discussed in this review and identifies their propensity to oligomerize.
Table 1.
RNA localization proteins
| Protein | Family | RNA-binding domains | Oligomerization | References |
|---|---|---|---|---|
| Celf1 | ELAV/Hu like | 3 RRMs | Yes | Bauermeister et al. (2015), Arthur et al. (2009) |
| Dazl | RRM containing | 1 RRM | Yes | Heim et al. (2014), Fu et al. (2015) |
| Ddx4 (Vasa) | DEAD box RNA helicase | DEAD domain | Yes | Nott et al. (2015) |
| Dnd1 | RRM containing | 2 RRMs | No | Kedde et al. (2007), Koebernick et al. (2010) |
| Elav1/2 | ELAV/Hu like | 3 RRMs | Yes | Cosson et al. (2006), Devaux et al. (2006) |
| Hnrnpab | hnRNP/RRM | 2 RRMs | Yes | Xiang et al. (2015), Aguero et al. (2016) |
| Igf2bp3 (Vera) | IMP/VICKZ | KH þ RRM | No | Deshler et al. (1997), Havin et al. (1998) |
| Ptbp1 | hnRNP/RRM | 4 RRMs | No | Monie et al. (2005) |
| Rbm24b | RRM containing | 1 RRM | Yes | Nijjar and Woodland (2013a) |
| Rbm42b | RRM containing | 1 RRM | Yes | Nijjar and Woodland (2013a) |
| Rbpms2 (Hermes) | RRM containing | 1 RRM | Yes | Heim et al. (2014), Sagnol et al. (2014) |
| Staufen1 | Double-stranded RNA binding | DSRM | Yes | Martel et al. (2010) |
| Velo1 (Buc) | YppG like | Predicted RNA binding C-terminal (K/R rich) | Yes | Boke et al. (2016) |
Characteristics of RNA-binding proteins involved in RNA localization and discussed in this review. The general mode of RNA binding is noted, as is the ability for self-interaction (oligomerization). All of these proteins contain IDRs either at the N- and C-termini and/or interspersed between the RNA-binding domains (our unpublished observations)
3.2. Germ Plasm RNA Localization
RNA localization to the germ plasm and germ granules is thought to occur by a diffusion/entrapment-mediated mechanism (Zhou and King 1996; Kloc et al. 1996; Chang et al. 2004), but the details of this mechanism is unclear. Cytoskeletal reorganization is thought to be dispensable for direct RNA localization to the germ plasm (Chang et al. 2004), but there may be a requirement indirectly through organelle trafficking (Heinrich and Deshler 2009). Structure-function analysis of the nanos1 30UTR identified distinct motifs required for localization to the mitochondrial cloud/Balbiani body and to germ granules (Zhou and King 1996; Kloc et al. 2000, 2002a; Chang et al. 2004). However, these motifs are bound by similar proteins involved in late pathway RNAs [i.e., Igf2bp3 and Ptbp1 (Chang et al. 2004)], suggesting that other proteins might provide the germ plasm specificity.
A series of recent studies have implicated the RNA-binding protein Rbpms2 (alias Hermes) in germ plasm RNA localization and germ granule formation. Rbpms2 is an RRM-containing RNA-binding protein initially identified in heart development but also as a vegetally and Balbiani body localized mRNA in Xenopus (Gerber et al. 2002; Zearfoss et al. 2004) and zebrafish (Kosaka et al. 2007; Marlow and Mullins 2008). In addition to having likely roles in controlling the cell cycle, Rbpms2 binds to nanos1 mRNA (likely with other RNA-binding protein partners; see below) and localizes to germ granules in forming and maturing Balbiani bodies, as shown by immunoelectron microscopy (Song et al. 2007). Rbpms2 and nanos1 are co-localized in stage VI oocyte germ plasm islands, along with another germ plasm RNA, pgat (alias xpat) (Nijjar and Woodland 2013b). These structures likely represent germ granules, since this localization has been demonstrated by electron microscopy, along with ddx25 (Kloc et al. 2002a). Interestingly, Rbpms2 showed rapid recovery in germ plasm in FRAP experiments, whereas nanos1 did not, suggesting that Rbpms2 may shuttle nanos1 into more stable germ granules (Nijjar and Woodland 2013b). Relatedly, Rbpms2 can bind to velo1 RNA and protein, and this interaction may facilitate recruitment to the Balbiani body to initiate germ plasm assembly (Heim et al. 2014).
Rbpms2 also binds other RNA-binding proteins, Rbm24b and Rbm42b, as well as the Velo1 isoforms. Bimolecular fluorescence complementation analysis suggests that these interact cytoplasmically within the germ plasm and that Rbpms2 and Rbm42b interact in the germinal vesicle (Nijjar and Woodland 2013b). Additionally, Rbpms2 likely initiates nanos1 RNP granule formation in the germinal vesicle. Rbpms2 requires other oocyte factors for RNA binding and self-multimerizes upon association with RNA (Gerber et al. 2002). One of these requisite oocyte proteins could be Rbm42b (see above). Recent data show that Rbpms2 can bind nanos1 in the germinal vesicle and can also interact with Ptbp1 (alias hnRNPI), likely also in the GV (Aguero et al. 2016). Ptbp1 has been implicated in nuclear RNP formation and remodeling in the context of gdf1 localization (Lewis et al. 2008). It is tempting to speculate that differential remodeling might occur depending on the ultimate localization pathway, with Rbpms2 serving as a key Balbiani body determinant in the vertebrate oocyte. Also, many of these proteins are predicted to undergo liquid–liquid phase separation (Courchaine et al. 2016), although biochemical studies are lacking in the Xenopus system.
Recent studies in cultured cells have shown that the DEAD-box helicase 4 (Ddx4), a conserved component of germ plasm-related granules, undergoes similar liquid phase separation events (Nott et al. 2015). The N-terminus of Ddx4 is an intrinsically disordered region (IDR) and mediates the phase transition. Interestingly, these Ddx4 droplets are less stable than and distinct from amyloid and are dependent on electrostatic interactions (as opposed to hydrogen bonding between amyloid beta-sheets). Arginine methylation and other perturbations could destabilize the droplets, suggesting a mechanism for dynamic regulation. Additionally, the droplets excluded double-stranded DNA and only incorporated RNAs, consistent with the formation of RNP-rich germ granules (Nott et al. 2015).
Overall, these observations suggest that germ plasm RNAs are nucleated into granules in the germinal vesicle, possibly by association of the Rbpms2 protein. These proto-germ plasm granules would then diffuse until anchored to the Balbiani body and ultimately incorporated into the germ plasm. It is unclear however how these would be distinguished from other localized mRNA RNP granules or what would mediate their organization within the proposed amyloid Balbiani body. Differential use of Velo1 isoforms or Velo1 phosphorylation may be involved (Nijjar and Woodland 2013a; Boke et al. 2016).
3.3. Late Pathway RNAs and RNP Formation
Late pathway RNA localization differs from the early pathway mainly in that localization occurs after translocation of the Balbiani body vegetally and requires microtubule transport. Indeed, these RNAs likely follow in the wake of the localizing Balbiani body, possibly using transport apparatus assembled for that purpose. Numerous studies have identified a core set of proteins that bind vegetally localized mRNAs, although in many cases, these proteins do not distinguish between different pathways (reviewed in Houston 2013). Recent work by Snedden et al. (2013) has demonstrated that these proteins, which include Igf2bp3, Ptbp1, Hnrnpab, Prrp, Khsrp, and possibly Elav family proteins, bind RNAs that are targeted to both the animal and vegetal hemispheres. Intriguingly, phax (phosphorylated adaptor for RNA export), an animally localized RNA, was found to contain vegetal localization motifs (Snedden et al. 2013), suggesting that these are not the main determinant for vegetal localization. However, the presence of Staufen1 (Stau1), a ds-RNA-binding protein and a kinesin motor protein linker (Yoon and Mowry 2004; Allison et al. 2004), was implicated in late pathway localization, as cross-linking experiments demonstrated the absence of Stau1 from complexes bound to early pathway RNAs and animal hemisphere transcripts (Snedden et al. 2013). Stau1 thus might provide the link to kinesin-based transport and is thought to associate with gdf1 RNPs upon nuclear export (Kress et al. 2004). The specificity of Stau1 association with late localizing transcripts is not well understood, and coupling with nuclear export may not be required, since cytoplasmically injected RNAs are still able to achieve proper localization.
Recent work studying the localization of dnd1, whose 30UTR localization element lacks obvious similarity to that of gdf1, identified novel candidate localization proteins of the Elav family, including Elavl1/2 (ELAV like neuron-specific RNA-binding proteins 1 and 2; (Arthur et al. 2009) and Celf1 [CUGBP, Elav-like family member 1; (Bauermeister et al. 2015)]. These proteins are conserved RRM-type RNA-binding proteins involved in RNA processing at various levels, including mRNA stability, splicing, polyadenylation, and translational control (reviewed in (Hinman and Lou 2008; Dasgupta and Ladd 2012). Celf1 also has roles in regulating mRNA deadenylation in Xenopus embryos through U(A/G)-rich EDEN (embryo deadenylation) elements (Paillard et al. 1998). These proteins bind to a central region of the dnd1 30UTR enriched in uracil and guanine, although no specific sequences were identified, and both Elavl1/2 and Celf1 could co-immunoprecipitate with each other and with core RNP components Stau1, Igf2bp3, and Hnrnpab, likely in an RNA-dependent manner (Arthur et al. 2009; Bauermeister et al. 2015).
RNA binding of Celf1 was suggested to be in low affinity (Bauermeister et al. 2015) and Celf1 bound a subset, but not all vegetally-localizing RNAs, including vegt, trim36, and spire1. All of these RNAs appear to be able to localize through the late pathway (Zhang and King 1996; Le Goff et al. 2006; Cuykendall and Houston 2009), although dnd1, trim36, and spire1 likely belong to the “intermediate” class of localized RNAs since they also preferentially accumulate in the germ plasm (Horvay et al. 2006; Le Goff et al. 2006; Cuykendall and Houston 2009). Interestingly, vegt mRNA can also accumulate somewhat in the germ plasm (Sudou et al. 2016), suggesting that many localizing RNAs may have this property, possibly through nonspecific RNP droplet association. Curiously, vegt undergoes a “relocalization” during the cleavage stages and becomes enriched dorsoanimally and accumulates around vegetal nuclei (Sudou et al. 2016), although the specificity and significance of this behavior is unclear.
Elavl2 and Celf1 are able to oligomerize (Devaux et al. 2006; Cosson et al. 2006), suggesting these proteins may be involved in forming RNP granules through liquid–liquid phase transitions. These proteins are also required for localization of the RNAs they bind, but not unbound ones (Arthur et al. 2009; Bauermeister et al. 2015), and their mutual interactions suggest that these proteins may form heterogeneous RNP granules for the localization of a subset of vegetal transcripts. Oligomerization has been shown to be required for Celf1-mediated deadenylation (Cosson et al. 2006) and may be true for RNA localization. Celf1 along with Igf2bp3 may also have a role in anchoring at the vegetal cortex (Git et al. 2009; Bauermeister et al. 2015). Elavl1/2 along with Dnd1 itself are thought to protect dnd1 and other RNAs from miRNA-mediated degradation in germ plasmcontaining cells (Kedde et al. 2007; Koebernick et al. 2010). In general, RNA localization is correlated with translation repression and stabilization, but the interplay of the various RNA-binding proteins involved in these processes and their regulation spatiotemporally are not well understood mechanistically.
4. Conclusions and Future Directions
Localized determinants in the egg play critical roles in development. Recent years have seen a great deal of progress in not only identifying many of these determinants but also in discovering how cell polarity is established in the oocyte and key determinants are localized in the first place.
The Balbiani body has garnered recent attention in this regard. This structure is conspicuous in the oocytes of animals and its role in servingas the source of germ plasm (Heasman et al. 1984), and a hub for localized RNAs (Forristall et al. 1995; Kloc and Etkin 1995) had been well studied. However, not all animals use germ plasm for specification of the germline yet still develop Balbiani bodies, including myriapods (where the Balbiani body was first observed) (Extavour and Akam 2003) and mouse (Pepling et al. 2007). It has been speculated that the Balbiani body sequesters high-quality mitochondria and suppresses respiration, thus limiting potential oxidative damage during oocyte dormancy and passage to the next generation (Kogo et al. 2011; Boke et al. 2016). However, there is a similar structure formed in sperm, the chromatoid body, which does not pass on mitochondria. Thus, it is likely the Balbiani body minimally serves a more general function in gametogenesis, regulating cytoplasmic RNA processing and storage and transposon silencing. And, this body could then serve as a scaffold allowing for the evolution of novel functions, including the germ plasm in multiple lineages where this occurs.
The demonstration that the Balbiani body forms in association with the chromosomal bouquet in meiosis shows that nuclear asymmetry can drive oocyte polarity (Elkouby et al. 2016). However, it remains unclear to what extent bouquet formation occurs stochastically along the nuclear envelope or is itself directed by existing polarity determinants in the oogonia, such as the centrosome or mitochondrial aggregates. If this were the case, then vegetal polarity would be carried over from the polarity in the progenitor cells. In either case, the mechanisms linking the chromosomal bouquet to localizing Velo1 and nascent Balbiani body components need to be elucidated; these could include the SUN-KASH domain proteins involved in homolog pairing in meiosis (Starr and Fridolfsson 2010). Additionally, the mechanisms controlling dazl mRNA localization to the bouquet would be interesting to determine, since Dazl plays numerous roles during germline development (Fu et al. 2015). Rbpms2 may also have a critical role in this early localization, given its role in later RNA localization events.
An association of the Balbiani body and the chromosomal bouquet was also seen in a primitive insect Thermobia (Tworzydlo et al. 2016; Bilinski et al. 2017), suggesting its conservation in animals. The significance of coupling bouquet formation with Balbiani assembly is not known, but linking a meiotic event with Balbiani assembly may ensure that its formation is limited to germ cells. Interestingly, Balbiani body assembly occurs within a “nuclear cleft” (Elkouby et al. 2016) which is likely similar to the classically characterized “nuclear bay” concavity that typically surrounds the centrosome (Wilson 1928).2 Recently these nuclear bays have been shown to accumulate proteins destined for degradation in the pericentriolar material (Fuentealba et al. 2008), potentially implicating these structures in protein turnover during oocyte polarity and Balbiani body formation. Additionally, the ability of Velo1 to assemble into amyloid aggregates and co-aggregate mitochondria and RNA (Boke et al. 2016) suggests that the process is largely selforganizing and does not require specific directed transport and assembly.
The Balbiani body serves as the site for germ plasm assembly, which also likely occurs through self-organization. RNAs have been shown to localize to germ plasm through diffusion-entrapment, and recent data suggest a key role for Rbpms2 as a shuttling protein. Although different pathways for RNA localization have been known for some time, involving specific cis-RNA sequence elements, identifying specific trans-acting proteins has been more problematic. Definitive germ plasm assembly may involve phase transitions of Velo1 from the amyloid structure used in Balbiani body assembly to hydrogel or other forms. Velo1 can associate with RNAs but nonspecifically and may thus provide an initial low-affinity step in RNA localization, after which more specific interactions mediate the localization into germ plasm/germ granules. This latter process is not well understood; only one RNA element and no trans-acting factors have been identified.
Rbpms2 may be a key determinant for the early pathway since it can interact with nanos1 RNA and Velo1 protein, although how it recognizes specific RNAs is unclear. Similarly, StauI appears specific for late pathway RNAs, and the specificity of its association is similarly not understood. Since StauI is a dsRNA-binding protein, it is tempting to speculate that RNP assembly and remodeling create a double-stranded region recognized by StauI and coupled to the transport machinery.
The number of LE studied mechanistically has been increasing but is still limited and has identified divergent LE using a set of overlapping general RNA-binding proteins. Unifying these mechanisms as well as identifying distinct cellular “addresses” in the oocyte for various localized RNAs will be one of the future challenges. RNA localization research suffers several inherent technical limitations, including the promiscuous nature of many RNA-protein interactions and the propensity of RNPs to interact in liquid phase granules. Also, many studies have relied on overexpressed proteins and RNAs, which, while informative in general, may fail to uncover subtle features of localization.
Increasingly sophisticated technological advancements are becoming available and could shed light on RNA localization mechanisms in the oocyte. These methods include high-throughput analysis of RNAs bound to specific proteins using various “Clip-omics” (cross-linking and immunoprecipitation) approaches [i.e., PAR-Clip, HITS-Clip, iClip; e.g., (Ascano et al. 2011)], similar proteomics strategies to identify interacting proteins, and computational prediction of RNA-binding protein binding sites (Li et al. 2013). Additionally, genome editing should allow more robust labeling and tagging of localized RNAs in vivo to gain a more precise picture of endogenous RNA localization mechanisms [e.g., (Nelles et al. 2016)]. Last, the increasing applicability of single-molecule analysis and biochemistry is likely to enable unforeseen detail of molecular interactions in the assembly of localization granules. Xenopus has proven especially suitable for studying RNA localization, owing to the large size and accessibility of oocytes and abundant material for biochemical or physical fractionation. Many of these newer methods are only now being applied to oocyte RNA localization, and given the large number of localized RNAs identified along different pathways in Xenopus, there is considerable opportunity to elucidate many novel basic aspects of RNA localization.
Acknowledgments
This work was supported by the University of Iowa and by NIH grant GM083999 (DWH).
Footnotes
The Balbiani body was named in honor of Édouard Gérard Balbiani, a Haitian-born French naturalist and embryologist, by his student Louis Félix Henneguy (la vesicule de Balbiani/le corps vitellin de Balbiani). Balbiani studied the structure, which was first identified by von Wittich and Carus in 1845 and 1850, respectively, and termed the “yolk/vitellin nucleus”, Dotterkern, or corps vitellin in older literature (Wilson 1928; Kloc et al. 2004b).
Curiously, Wilson, a leading figure in the early years of cell and developmental biology, considered the term centrosome “undesirable”, apparently preferring “central body” instead (Wilson 1928, p. 30).
References
- Aguero T, Zhou Y, Kloc M et al. (2016) Hermes (Rbpms) is a critical component of RNP complexes that sequester germline RNAs during oogenesis. J Dev Biol 4:2 10.3390/jdb4010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O et al. (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137:146–158. 10.1016/j.cell.2009.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison R, Czaplinski K, Git A et al. (2004) Two distinct Staufen isoforms in Xenopus are vegetally localized during oogenesis. RNA 10:1751–1763. 10.1261/rna.7450204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- al-Mukhtar K, Webb A (1971) An ultrastructural study of primordial germ cells, oogonia and early oocytes in Xenopus laevis. J Embryol Exp Morphol 26:195–217 [PubMed] [Google Scholar]
- Ancel P, Vintembenger P (1948) Recherches sur le déterminisme de la symétrie bilatérale dans l’œuf des amphibiens. Bull Biol Fr Belg 31(Suppl):1–182 [Google Scholar]
- Arthur PK, Claussen M, Koch S et al. (2009) Participation of Xenopus Elr-type proteins in vegetal mRNA localization during oogenesis. J Biol Chem 284:19982–19992. 10.1074/jbc.M109.009928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M, Hafner M, Cekan P et al. (2011) Identification of RNA-protein interaction networks using PAR-CLIP. WIREs RNA 3:159–177. 10.1002/wrna.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauermeister D, Claussen M, Pieler T (2015) A novel role for Celf1 in vegetal RNA localization during Xenopus oogenesis. Dev Biol 405:214–224. 10.1016/j.ydbio.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Bilinski SM, Jaglarz MK, Tworzydlo W (2017) The pole (germ) plasm in insect oocytes. In: Kloc M, Kubiak JZ (eds) Oocytes: maternal information and functions, Results and problems in cell differentiation Springer, Cham: [DOI] [PubMed] [Google Scholar]
- Boke E, Ruer M, Wühr M et al. (2016) Amyloid-like self-assembly of a cellular compartment. Cell 166:637–650. 10.1016/j.cell.2016.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontems F, Stein A, Marlow F et al. (2009) Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol 19:414–422. 10.1016/j.cub.2009.01.038 [DOI] [PubMed] [Google Scholar]
- Bottenberg W, Sanchez-Soriano N, Alves-Silva J et al. (2009) Context-specific requirements of functional domains of the Spectraplakin short stop in vivo. Mech Dev 126:489–502. 10.1016/j.mod.2009.04.004 [DOI] [PubMed] [Google Scholar]
- Bounoure L (1937) Le sort de la ligneé germinale chez la Grenouille rousse après l’action des rayons ultra-violets sur le poˆle inférieur de l’oeuf. CR Acad Sci Paris 204:1837 [Google Scholar]
- Bounoure L, Aubry R, Huck ML (1954) Nouvelles recherches experimentales sur les origines de la lignee reproductrice chez la Grenouille rousse. J Embryol Exp Morph 2:245–263 [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS et al. (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324:1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Carpenter CD, Klein WH (1982) A gradient of poly(A) RNA sequences in Xenopus laevis eggs and embryos. Dev Biol 91:43–49 [DOI] [PubMed] [Google Scholar]
- Chan AP, Kloc M, Etkin LD (1999) fatvg encodes a new localized RNA that uses a 25-nucleotide element (FVLE1) to localize to the vegetal cortex of Xenopus oocytes. Development 126: 4943–4953 [DOI] [PubMed] [Google Scholar]
- Chang P, Torres J, Lewis RA et al. (2004) Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol Biol Cell 15:4669–4681. 10.1091/mbc.E04-03-0265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen M, Pieler T (2004) Xvelo1 uses a novel 75-nucleotide signal sequence that drives vegetal localization along the late pathway in Xenopus oocytes. Dev Biol 266:270–284 [DOI] [PubMed] [Google Scholar]
- Claussen M, Horvay K, Pieler T (2004) Evidence for overlapping, but not identical, protein machineries operating in vegetal RNA localization along early and late pathways in Xenopus oocytes. Development 131:4263–4273. 10.1242/dev.01283 [DOI] [PubMed] [Google Scholar]
- Coggins LW (1973) An ultrastructural and radioautographic study of early oogenesis in the toad Xenopus laevis. J Cell Sci 12:71–93 [DOI] [PubMed] [Google Scholar]
- Cosson B, Gautier-Courteille C, Maniey D et al. (2006) Oligomerization of EDEN-BP is required for specific mRNA deadenylation and binding. Biol Cell 98:653–665. 10.1042/BC20060054 [DOI] [PubMed] [Google Scholar]
- Courchaine EM, Lu A, Neugebauer KM (2016) Droplet organelles? EMBO J 35:1603–1612. 10.15252/embj.201593517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuykendall TN, Houston DW (2009) Vegetally localized Xenopus trim36 regulates cortical rotation and dorsal axis formation. Development 136:3057–3065. 10.1242/dev.036855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuykendall TN, Houston DW (2010) Identification of germ plasm-associated transcripts by microarray analysis of Xenopus vegetal cortex RNA. Dev Dyn 239:1838–1848. 10.1002/dvdy.22304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T, Ladd AN (2012) The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. WIREs RNA 3:104–121. 10.1002/wrna.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler JO, Highett MI, Schnapp BJ (1997) Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science 276:1128–1131 [DOI] [PubMed] [Google Scholar]
- Devaux A, Colegrove-Otero LJ, Standart N (2006) Xenopus ElrB, but not ElrA, binds RNA as an oligomer: possible role of the linker. FEBS Lett 580:4947–4952. 10.1016/j.febslet.2006.08.012 [DOI] [PubMed] [Google Scholar]
- Dosch R, Wagner DS, Mintzer KA et al. (2004) Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev Cell 6:771–780. 10.1016/j.devcel.2004.05.002 [DOI] [PubMed] [Google Scholar]
- Dumont JN (1972) Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J Morphol 136:153–179. 10.1002/jmor.1051360203 [DOI] [PubMed] [Google Scholar]
- El Meziane A, Callen JC, Mounolou JC (1989) Mitochondrial gene expression during Xenopus laevis development: a molecular study. EMBO J 10.1371/journal.pgen.1005019.s006 [DOI] [PMC free article] [PubMed]
- Elkouby YM, Jamieson-Lucy A, Mullins MC (2016) Oocyte polarization is coupled to the chromosomal bouquet, a conserved polarized nuclear configuration in meiosis. PLoS Biol 14:e1002335 10.1371/journal.pbio.1002335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour CG, Akam M (2003) Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130:5869–5884. 10.1242/dev.00804 [DOI] [PubMed] [Google Scholar]
- Forristall C, Pondel M, Chen L, King M (1995) Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development 121:201–208 [DOI] [PubMed] [Google Scholar]
- Fu X-F, Cheng S-F, Wang L-Q et al. (2015) DAZ family proteins, key players for germ cell development. Int J Biol Sci 11:1226–1235. 10.7150/ijbs.11536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Geissert D et al. (2008) Asymmetric mitosis: unequal segregation of proteins destined for degradation. Proc Natl Acad Sci 105:7732–7737. 10.1073/pnas.0803027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Cha BJ, King E (1997) The organization and animal-vegetal asymmetry of cytokeratin filaments in stage VI Xenopus oocytes is dependent upon F-actin and microtubules. Dev Biol 184:95–114. 10.1006/dbio.1997.8508 [DOI] [PubMed] [Google Scholar]
- Gerber WV, Vokes SA, Zearfoss NR, Krieg PA (2002) A role for the RNA-binding protein, hermes, in the regulation of heart development. Dev Biol 247:116–126. 10.1006/dbio.2002.0678 [DOI] [PubMed] [Google Scholar]
- Git A, Allison R, Perdiguero E et al. (2009) Vg1RBP phosphorylation by Erk2 MAP kinase correlates with the cortical release of Vg1 mRNA during meiotic maturation of Xenopus oocytes. RNA 15:1121–1133. 10.1261/rna.1195709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta T, Marlow FL, Ferriola D et al. (2010) Microtubule actin crosslinking factor 1 regulates the Balbiani body and animal-vegetal polarity of the zebrafish oocyte. PLoS Genet 6:e1001073 10.1371/journal.pgen.1001073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guraya SS (1979) Recent advances in the morphology, cytochemistry, and function of Balbiani’s vitelline body in animal oocytes. Int Rev Cytol 59:249–321 [DOI] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S et al. (2012) Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149:768–779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Harper L, Golubovskaya I, Cande WZ (2004) A bouquet of chromosomes. J Cell Sci 117:4025–4032. 10.1242/jcs.01363 [DOI] [PubMed] [Google Scholar]
- Havin L, Git A, Elisha Z et al. (1998) RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev 12:1593–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Quarmby J, Wylie CC (1984) The mitochondrial cloud of Xenopus oocytes: the source of germinal granule material. Dev Biol 105:458–469 [DOI] [PubMed] [Google Scholar]
- Heim AE, Hartung O, Rothhämel S et al. (2014) Oocyte polarity requires a Bucky ball-dependent feedback amplification loop. Development 141:842–854. 10.1242/dev.090449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich B, Deshler JO (2009) RNA localization to the Balbiani body in Xenopus oocytes is regulated by the energy state of the cell and is facilitated by kinesin II. RNA 15:524–536. 10.1261/rna.975309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ et al. (2010) The genome of the Western clawed frog Xenopus tropicalis. Science 328:633–636. 10.1126/science.1183670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman MN, Lou H (2008) Diverse molecular functions of Hu proteins. Cell Mol Life Sci 65: 3168–3181. 10.1007/s00018-008-8252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvay K, Claussen M, Katzer M et al. (2006) Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev Biol 291:1–11. 10.1016/j.ydbio.2005.06.013 [DOI] [PubMed] [Google Scholar]
- Houston DW (2013) Regulation of cell polarity and RNA localization in vertebrate oocytes. Int Rev Cell Mol Biol 306:127–185. 10.1016/B978-0-12-407694-5.00004-3 [DOI] [PubMed] [Google Scholar]
- Ikenishi K, Kotani M, Tanabe K (1974) Ultrastructural changes associated with UV irradiation in the “germinal plasm” of Xenopus laevis. Dev Biol 36:155–168 [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S et al. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149:753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B et al. (2007) RNA-binding protein Dnd1 inhibits MicroRNA access to target mRNA. Cell 131:1273–1286. 10.1016/j.cell.2007.11.034 [DOI] [PubMed] [Google Scholar]
- King ML, Messitt TJ, Mowry KL (2005) Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell 97:19–33. 10.1042/BC20040067 [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD (1995) Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development 121:287–297 [DOI] [PubMed] [Google Scholar]
- Kloc M, Larabell C, Etkin LD (1996) Elaboration of the messenger transport organizer pathway for localization of RNA to the vegetal cortex of Xenopus oocytes. Dev Biol 180:119–130. 10.1006/dbio.1996.0289 [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Pui-Yee Chan A, Etkin LD (2000) The targeting of Xcat2 mRNA to the germinal granules depends on a cis-acting germinal granule localization element within the 30UTR. Dev Biol 217:221–229. 10.1006/dbio.1999.9554 [DOI] [PubMed] [Google Scholar]
- Kloc M, Dougherty MT, Bilinski S et al. (2002a) Three-dimensional ultrastructural analysis of RNA distribution within germinal granules of Xenopus. Dev Biol 241:79–93. 10.1006/dbio.2001.0488 [DOI] [PubMed] [Google Scholar]
- Kloc M, Zearfoss NR, Etkin LD (2002b) Mechanisms of subcellular mRNA localization. Cell 108:533–544 [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Dougherty MT et al. (2004a) Formation, architecture and polarity of female germline cyst in Xenopus. Dev Biol 266:43–61 [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Etkin LD (2004b) The Balbiani body and germ cell determinants: 150 years later. Curr Top Dev Biol 59:1–36. 10.1016/S0070-2153(04)59001-4 [DOI] [PubMed] [Google Scholar]
- Kloc M, Jaglarz M, Dougherty M et al. (2008) Mouse early oocytes are transiently polar: three-dimensional and ultrastructural analysis. Exp Cell Res 314:3245–3254. 10.1016/j.yexcr.2008.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebernick K, Loeber J, Arthur PK et al. (2010) Elr-type proteins protect Xenopus Dead end mRNA from miR-18-mediated clearance in the soma. Proc Natl Acad Sci 107:16148–16153. 10.1073/pnas.1004401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo N, Tazaki A, Kashino Y et al. (2011) Germ-line mitochondria exhibit suppressed respiratory activity to support their accurate transmission to the next generation. Dev Biol 349:462–469. 10.1016/j.ydbio.2010.11.021 [DOI] [PubMed] [Google Scholar]
- Kosaka K, Kawakami K, Sakamoto H, Inoue K (2007) Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis. Mech Dev 124:279–289. 10.1016/j.mod.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Kress TL, Yoon YJ, Mowry KL (2004) Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J Cell Biol 165:203–211. 10.1083/jcb.200309145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff C, Laurent V, Le Bon K et al. (2006) pEg6, a spire family member, is a maternal gene encoding a vegetally localized mRNA in Xenopus embryos. Biol Cell 98:697–708. 10.1042/BC20050095 [DOI] [PubMed] [Google Scholar]
- Lewis RA, Gagnon JA, Mowry KL (2008) PTB/hnRNP I is required for RNP remodeling during RNA localization in Xenopus oocytes. Mol Cell Biol 28:678–686. 10.1128/MCB.00999-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kazan H, Lipshitz HD, Morris QD (2013) Finding the target sites of RNA-binding proteins. WIREs RNA 5:111–130. 10.1002/wrna.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow FL, Mullins MC (2008) Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Dev Biol 321:40–50. 10.1016/j.ydbio.2008.05.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Dugré-Brisson S, Boulay K et al. (2010) Multimerization of Staufen1 in live cells. RNA 16:585–597. 10.1261/rna.1664210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monie TP, Hernandez H, Robinson CV et al. (2005) The polypyrimidine tract binding protein is a monomer. RNA 11:1803–1808. 10.1261/rna.2214405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR et al. (2016) Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165:488–496. 10.1016/j.cell.2016.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD (1969) The formation of mesoderm in Urodelean amphibians. I. Induction by the endoderm. Wilhelm Roux’ Arch EntwMech Org 162:341–373 [DOI] [PubMed] [Google Scholar]
- Nijjar S, Woodland HR (2013a) Protein interactions in Xenopus germ plasm RNP particles. PLoS One 8:e80077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijjar S, Woodland HR (2013b) Localisation of RNAs into the germ plasm of vitellogenic Xenopus oocytes. PLoS One 8:e61847 10.1371/journal.pone.0061847.t002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P et al. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57:936–947. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard L, Omilli F, Legagneux V et al. (1998) EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J 17:278–287. 10.1093/emboj/17.1.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegri F (2003) Maternal factors in zebrafish development. Dev Dyn 228:535–554. 10.1002/dvdy.10390 [DOI] [PubMed] [Google Scholar]
- Pepling ME, de Cuevas M, Spradling AC (1999) Germline cysts: a conserved phase of germ cell development? Trends Cell Biol 9:257–262 [DOI] [PubMed] [Google Scholar]
- Pepling ME, Wilhelm JE, O’Hara AL et al. (2007) Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proc Natl Acad Sci 104:187–192. 10.1073/pnas.0609923104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasar MA, Hammes SR (2006) The physiology of the Xenopus laevis ovary. Methods Mol Biol 322:17–30. 10.1007/978-1-59745-000-3_2 [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Weeks DL, Harvey RP, Melton DA (1985) Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell 42:769–777 [DOI] [PubMed] [Google Scholar]
- Sagnol S, Yang Y, Bessin Y et al. (2014) Homodimerization of RBPMS2 through a new RRM-interaction motif is necessary to control smooth muscle plasticity. Nucleic Acids Res 42:10173–10184. 10.1093/nar/gku692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H (2001) A bouquet makes ends meet. Nat Rev Mol Cell Biol 10.1038/35085086 [DOI] [PubMed]
- Selman K, Wallace RA, Sarka A, Qi X (1993) Stages of oocyte development in the zebrafish, Brachydanio rerio. J Morphol 218:203–224 [DOI] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T et al. (2016) Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538:336–343. 10.1038/nature19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge EA, Wai T (2007) Mitochondrial DNA and the mammalian oocyte. Curr Top Dev Biol 77:87–111. 10.1016/S0070-2153(06)77004-1 [DOI] [PubMed] [Google Scholar]
- Smith LD (1966) The role of a “germinal plasm” in the formation of primordial germ cells in Rana pipiens. Dev Biol 14:330–347 [DOI] [PubMed] [Google Scholar]
- Snedden DD, Bertke MM, Vernon D, Huber PW (2013) RNA localization in Xenopus oocytes uses a core group of trans-acting factors irrespective of destination. RNA 19:889–895. 10.1261/rna.038232.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H-W, Cauffman K, Chan AP et al. (2007) Hermes RNA-binding protein targets RNAs-encoding proteins involved in meiotic maturation, early cleavage, and germline development. Differentiation 75:519–528. 10.1111/j.1432-0436.2006.00155.x [DOI] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN (2010) Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 26:421–444. 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarwati S, Nieuwkoop PD (1971) Mesoderm formation in the anuran Xenopus laevis (Daudin). Dev Genes Evol 166:189–204. 10.1007/BF00650029 [DOI] [PubMed] [Google Scholar]
- Sudou N, Garcés-Vásconez, López-Latorre MA et al. (2016) Transcription factors Mix1 and VegT, relocalization of vegt mRNA, and conserved endoderm and dorsal specification in frogs. Proc Natl Acad Sci 113:5628–5633. 10.1073/pnas.1605547113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suozzi KC, Wu X, Fuchs E (2012) Spectraplakins: master orchestrators of cytoskeletal dynamics. J Cell Biol 197:465–475. 10.1083/jcb.201112034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toretsky JA, Wright PE (2014) Assemblages: functional units formed by cellular phase separation. J Cell Biol 206:579–588. 10.1083/jcb.201404124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tworzydlo W, Marek M, Kisiel E, Bilinski SM (2016) Meiosis, Balbiani body and early asymmetry of Thermobia oocyte. Protoplasma 10.1007/s00709-016-0978-7 [DOI] [PMC free article] [PubMed]
- Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I (2011) RNA granules in germ cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a002774 [DOI] [PMC free article] [PubMed]
- Wagner DS, Dosch R, Mintzer KA et al. (2004) Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev Cell 6:781–790. 10.1016/j.devcel.2004.04.001 [DOI] [PubMed] [Google Scholar]
- Wakahara M (1977) Partial characterization of “primordial germ cell-forming activity” localized in vegetal pole cytoplasm in anuran eggs. Development 39:221–233 [PubMed] [Google Scholar]
- Weeks DL, Melton DA (1987) A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-beta. Cell 51:861–867 [DOI] [PubMed] [Google Scholar]
- Wilson E (1928) The cell in development and heredity, 3rd edn. Macmillian, New York [Google Scholar]
- Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL (2015) The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell 163:829–839. 10.1016/j.cell.2015.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YJ, Mowry KL (2004) Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development 131:3035–3045. 10.1242/dev.01170 [DOI] [PubMed] [Google Scholar]
- Zearfoss NR, Chan AP, CF W et al. (2004) Hermes is a localized factor regulating cleavage of vegetal blastomeres in Xenopus laevis. Dev Biol 267:60–71. 10.1016/j.ydbio.2003.10.032 [DOI] [PubMed] [Google Scholar]
- Zhang J, King ML (1996) Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development 122:4119–4129 [DOI] [PubMed] [Google Scholar]
- Zhou Y, King ML (1996) Localization of Xcat-2 RNA, a putative germ plasm component, to the mitochondrial cloud in Xenopus stage I oocytes. Development 122:2947–2953 [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N (1998) The leptotene-zygotene transition of meiosis. Annu Rev Genet 32: 619–697. 10.1146/annurev.genet.32.1.619 [DOI] [PubMed] [Google Scholar]
- Züst B, Dixon K (1975) The effect of u.v. irradiation of the vegetal pole of Xenopus laevis eggs on the presumptive primordial germ cells. J Embryol Exp Morph 34:209–220 [PubMed] [Google Scholar]
- Zwicker D, Decker M, Jaensch S et al. (2014) Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc Natl Acad Sci 111:E2636–E2645. 10.1073/pnas.1404855111 [DOI] [PMC free article] [PubMed] [Google Scholar]


