Abstract
Over the last several decades, an impressive array of advanced microscopic and analytical tools, such as single particle tracking and nanoscopic fluorescence correlation spectroscopy, has been applied to characterize the lateral organization and mobility of components in the plasma membrane. Such analysis can tell researchers about the local dynamic composition and structure of membranes and is important for predicting the outcome of membrane-based reactions. However, owing to the unresolved complexity of the membrane and the structures peripheral to it, identification of the detailed molecular origin of the interactions that regulate the organization and mobility of the membrane has not proceeded quickly. This review presents an overview of how cell surface structure may give rise to the types of lateral mobility that are observed and some potentially fruitful future directions to elucidate the architecture of these structures in more molecular detail.
Keywords: plasma membrane, lateral mobility, membrane proteins, rafts, membrane dynamics
Introduction
Biological membranes, in general, are fluids meaning that the component proteins and lipids can translate laterally in the plane of the membrane. Measurements of the lateral mobility of membrane proteins and lipids, which includes both random and directed movements, provide information that is useful in two ways (Zhang et al., 1993). First, functionally speaking, the lateral mobility of membrane proteins and lipids is often required, and in some cases, is rate-limiting, in membrane-based reactions. For example, some ligands bound to receptors must laterally diffuse to coated pits prior to receptor mediated endocytosis. Second, lateral mobility is related to, and therefore provides a probe for, the local dynamic composition and structure of the membrane. Indeed, five decades of research have demonstrated that membrane associated cytoskeleton and pericellular matrix immediate external to the cell restrain lateral mobility in the plasma membrane as recognized early on (Nicolson, 1976). Exploration of these factors has been enhanced by numerous new imaging and analytic techniques, including microscopic techniques such as single particle tracking and nanoscopic fluorescence correlation spectroscopy (FCS), and sophisticated data analysis tools.
Despite progress, it remains difficult to identify the molecular origin of many of those interactions that dictate the dynamic lateral organization and mobility of membrane proteins. This lack of molecular detail contrasts with other areas of cell biology where impressive progress has been made in identifying molecules involved in many phenomena. To a large extent, this impasse is due to the inherent and unresolved complexity of the composition and dynamic structure of the membrane, including the impact of membrane trafficking (Gheber, 2018; Goiko et al., 2018), and the subjacent actomyosin cortex and pericellular matrix (Figure 1). Even when some generic structural specificity is conferred on models for membrane organization on the nanoscale and the lateral mobility of membrane components, the molecular nature of the restraining structures cannot be specified in much detail. In this review, we present current views of the organization of plasma membranes, indicating interpretations in need of modification to accommodate the full range of observations, and the challenge of elucidating the molecular structures responsible for this dynamic organization together with some avenues that are likely to yield progress.
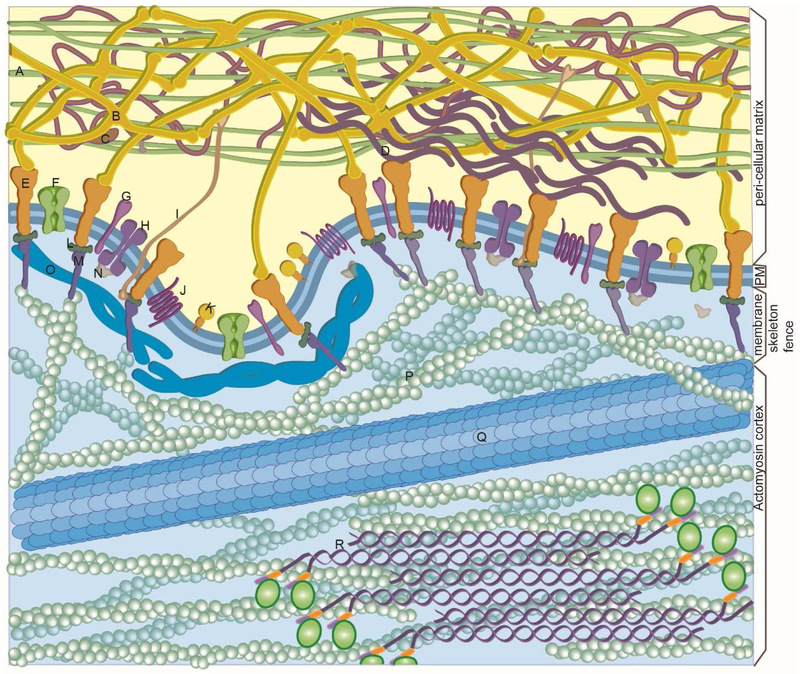
Figure 1. The Plasma Membrane.
The cell surface defined as the plasma membrane and associated peripheral structures: schematic cross-sectional view of the plasma membrane, the subjacent membrane skeleton fence and the associated actin cortex, and the pericellular matrix. Components labeled by letters include: A, collagen. B, cross-shaped laminin. C, proteoglycan. D, fibronectin. E, integrin. F. ion channel. G, single pass transmembrane protein. H, transmembrane protein dimers. I, integral membrane proteoglycan J, GCPR. K, GPI-AP. L, M and N, adaptor proteins. O, spectrin. P, F actin. Q, MT. R, non-muscle myosin. This figure has been inspired and adapted, in part, from the drawings of David Goodsell (The Machinery of Life, second edition, Copernicus Books, NY).
Plasma membrane lateral organization and dynamics
Several excellent and comprehensive reviews of membrane structure have recently been published (Kusumi et al., 2012; Nicolson, 2014; Sezgin et al., 2017). Membrane components exist in a variety of motional states ranging from immobilized and, in some cases, essentially stationary, to fully laterally and rotationally mobile (with axis of rotation normal to the membrane plane) as was emphasized in the original Singer-Nicolson Fluid Mosaic model in which integral membrane proteins were envisioned to be embedded in a lipid bilayer sea (Singer and Nicolson, 1972). These motional states are more fully described below in the section “Types of Lateral Mobility” and in Figure 2. The organization and mobility of membrane components is dictated by both intramembrane lipid-protein interactions and associations with structures peripheral to the membrane: the actomyosin cortex underlying the plasma membrane and the pericellular matrix abutting the external face of the membrane (Figure 1).
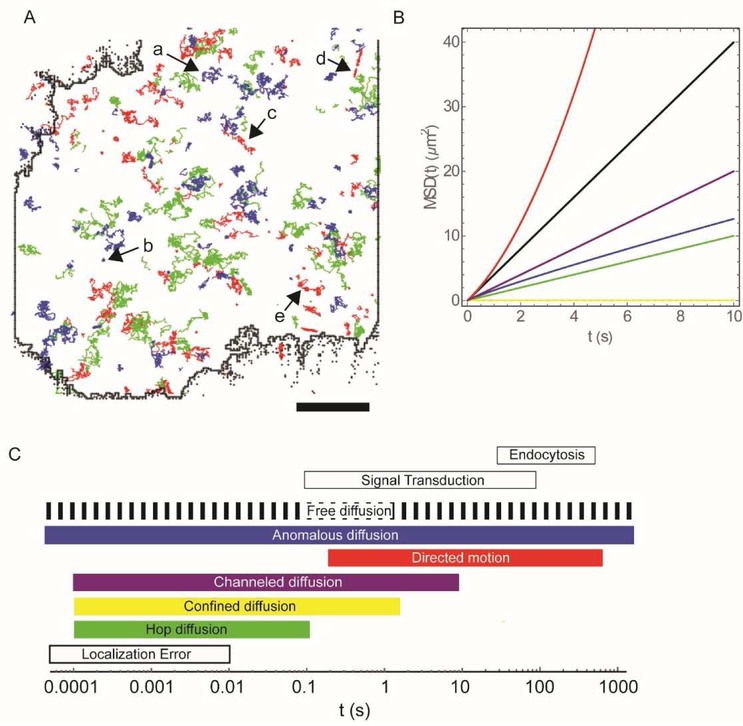
Figure 2. Modes of lateral mobility in the plasma membrane of live cells.
(A) Single particle tracking measurements in live cells indicate that the lateral mobility in the plasma membrane is heterogeneous. This is exemplified by the experimental trajectories in the plasma membrane of a live NIH 3T3 fibroblast of the sphingolipid GM1, (red); CD59 (green), and the transmembrane protein epidermal growth factor receptor (EGFR) (blue). These molecules were simultaneously labelled, with distinct, differently emitting Qdot conjugates and imaged as described previously (Clausen et al., 2014). This data set, from a portion of the cell whose edges are indicated in a black solid line, includes apparent examples of (a) free diffusion, (b) confined diffusion, (c) channeled diffusion, (d) directed motion, and (e) diffusion interrupted by periods of transient anchorage (STALL) (Bar = 5 μm). (B) Different modes of lateral mobility can be quantitatively differentiated in traditional MSD versus time plots and subsequent analysis by curve fitting to a range of diffusion models. Shown are the cases for Brownian (free) diffusion (black); directed flow (MSD is proportional to t2; red); anomalous sub diffusion (blue); effectively 1-dimensional channeled diffusion (purple); transiently confined diffusion (green), and the extreme case of totally confined diffusion (yellow). (C) Time ranges over which different modes of lateral mobility can be observed in relation to physiological processes occurring at the plasma membrane. For flow (directed motion), the lower limit of time range is estimated for a particle with a velocity of ~2.5 μm/s (Liu et al, 2017) to move a detectable 500nm; the upper limit is for a particle undergoing retrograde flow at 1μm/min for a distance of 10μm (as in capping of antibody cross-linked antigens). The rectangular boxes at the top indicate order of magnitude time ranges for signal transduction events across the plasma membrane and for endo-and exocytotic events at membrane spanning the range from receptor mediated endocytosis to phagocytosis. For most of these phenomena, lateral diffusion (e.g. soluble membrane ligand finding membrane receptor or membrane receptor moving to site of endocytosis) is an obligatory, but not necessarily a rate limiting, step.
Plasma membrane lateral organization
In general, because the interaction affinities of various components are not identical, the membrane is not a homogeneous random mixture of lipids and proteins; rather, it is laterally heterogeneous in that the membrane is characterized by compartmentalization into domains in which the local composition, lateral organization and/or dynamics differ in some way from the average membrane properties. Domain dimensions can range from ~ 100 nm or less (e.g. clathrin coated pits and lipid envelope virus budding sites) to several microns (e.g. focal adhesions, the immune synapse, junctional complexes and apical and basolateral domains in epithelial cells). Domain constituents enter or leave on a range of time scales depending on the function of the domain; for example, the aligned connexon pores permitting passage of small molecules from one cell to another in gap junctions must be stable whereas the receptor and ligand complex must enter coated pits on a much shorter time scale for efficient receptor mediated endocytosis.
For over two decades, the study of nanoscopic lipid lateral heterogeneity has been motivated by the concept of lipid rafts: lipid-driven membrane regions whose lipid component, enriched in saturated lipids, cholesterol and sphingomyelin, is in the liquid-ordered state (Lingwood and Simons, 2010). It now appears that lipid rafts in the plasma membranes are predominantly very small (10–200nm) (Pike, 2006), transient, or both (Eggeling et al., 2009) but that these can increase in size and stability during membrane trafficking, signal transduction, or the addition of clustering agents that bring small domains together (Raghunathan and Kenworthy, 2018; Sezgin et al., 2017). Thus, these tiny clusters in resting cells probably arise from short-range transient ordering imposed upon lipids by a high concentration of transmembrane proteins (Jacobson et al., 2007) as opposed to discrete lipid phase separations. In general, transmembrane proteins must be solvated in the bilayer and the molecular details of such lipid solvation--ranging from specifically bound lipids to much less specific annular lipid--have been reviewed recently (Lee, 2011).
Nanoclusters of membrane proteins interacting with lipids have emerged as a dominant theme in the lateral organization of membranes on the sub 100nm length scale; plausible ideas exist concerning the functional significance of such clusters (Garcia-Parajo et al., 2014), particularly with respect to the immune response (Dustin and Choudhuri, 2016) and Ras signaling (Zhou and Hancock, 2015). For example, transmembrane C-type lectin receptors are found in nanoclusters and provide immune recognition of a variety of pathogens on dendritic cells by binding specific polysaccharide structures on a wide range of pathogenic agents. One such receptor, dendritic cell-SIGN or DC-SIGN (specific intercellular adhesion molecule-3-grabbing non-integrin, also termed CD209), is found near the leading edge of dendritic cells patrolling peripheral tissues where they first encounter pathogens (Neumann et al., 2008). The extracellular repeat domains of DC-SIGN are thought to promote its tetramerization (Feinberg et al., 2005). In the plane of the plasma membrane, a hierarchical organization of DC-SIGN exists (Garcia-Parajo et al., 2014) ranging from nanoclusters containing 1–3 tetramers (Itano et al., 2012) to mesoscale microdomains over a micron in dimension (Neumann et al., 2008). Clustering tetramers into nanodomains may enhance the capture of small viruses (Manzo et al., 2012) and further clustering of dendritic cell-SIGN nanodomains into microdomains appears to be required for efficient pathogen binding and internalization (Cambi et al., 2004; Itano et al., 2014), presumably because it increases the avidity to multivalent ligands that are presented by pathogens.
Glycosyl phosphatidylinositol-anchored proteins (GPI-APs) are a broad group of lipid-linked proteins, including the folate receptor and Thy-1, that are tethered to the outer-leaflet of the plasma membrane by the phospholipid tail; this class of membrane proteins is often found in nanoclusters (Varma and Mayor, 1998; van Zanten et al., 2009). An important question is what gives rise to these nanoclusters in addition to membrane lipid-protein interactions. Gowrishankar et al. (Gowrishankar et al., 2012) proposed that F-actin asters just beneath the plasma membrane drive lipid-anchored proteins into nanoclusters, perhaps via myosin-based contraction. Asters are composed of short (~1 μm), dynamic F-actin filaments arrayed in a spoke-like fashion about a nucleating center containing the Arp2/3 complex (Fritzsche et al., 2017). Raghupathy et al. (Raghupathy et al., 2015) showed that longer chain outer leaflet GPI-APs are coupled to complementary inner leaflet nanodomains consisting of lipids such as phosphatidylserine via chain interdigitation. However, the nature of the in vivo coupling between inner leaflet and actin, including possible adaptor proteins, remains to be elucidated (Sezgin et al., 2017).
Structures peripheral to the plasma membrane
Membrane components must interact with the subjacent actomyosin cortex and the pericellular matrix; thus, these peripheral structures will play a role in the lateral organization and dynamics (Figure 1). Indeed, the coupling of peripheral structures to the plasma membrane strongly suggests a continuum of connectivity between the extracellular and intracellular environments (Nicolson, 2014). At the inner leaflet of the plasma membrane, part of this connectivity is mediated by the plethora of regulatory proteins competing for sites on the cytoplasmic domains of transmembrane proteins and phosphosinostide lipid head groups (Sheetz, 2001). A consequence of this connectivity is that the active cortex plays a role in laterally organizing components of the plasma membrane (Krapf, 2018; Rao and Mayor, 2014).
Detailed knowledge of the cortical layer immediately beneath the plasma membrane originated with studies of the red cell cytoskeleton, a well-defined meshwork of spectrin and actin that underlies the erythrocyte (red blood cell) plasma membrane (Branton et al., 1981; Fowler, 2013). An analogous structure, termed the “membrane skeleton fence”, has also been observed by electron microscopy in nucleated mammalian cells (Kusumi et al., 2012). A different but related structure underlies the axonal plasma membrane in which rings of F-actin, perpendicular to the long axis of the axon, are periodically spaced alone the axon as observed using 3D STORM (Stochastic Optical Reconstruction Microscopy) super resolution microscopy (Xu et al., 2013). Spectrin tetramers oriented parallel to the long axis appear to connect these rings. Finally, septin filaments assemble at the inner leaflet of the membrane possibly to sense curvature and establish diffusion barriers (Mostowy and Cossart, 2012). Families of adaptor proteins serve to connect the membrane associated cytoskeleton to the plasma membrane. Chief among these are the ankryin family (Bennett and Lorenzo, 2016), the ezrin-radixin-moesin (ERM) family (Fehon et al., 2010), and the catenin families (Ostrowski et al., 2016).
By contrast, progress in understanding the structure and properties of the more amorphous pericellular matrix or glycocaylx has been made but not on the molecular scale (e.g (Wilusz et al., 2014)). The pericellular matrix consists of a collection of macromolecules whose proportion depends on the cell and tissue context; these include collagens, long chain polysaccharides such as hyaluronic acid as well as proteoglycans, including heparin sulfate proteoglycan and glycoproteins such as fibronectin and heavily glycosylated mucins. Such components are often linked to receptors in the plasma membrane. The pericellular matrix may affect the organization and mobility of membrane components. For example, the complete loss of observable DC-SIGN microdomains on the cell surface was observed following removal of the carbohydrate recognition domain in its ectodomain, suggesting that this domain might interact directly with components of the pericellular matrix while removal of the cytoplasmic domain had little effect on microdomain formation (Liu et al., 2012). Binding of largely immobile, pericellular hyaluronic acid bound to its receptor, CD44, was also found to reduce the mobility of Fc receptors in macrophages (Freeman et al., 2018).
These examples illustrate the importance of structures peripheral to the plasma membrane in regulating its dynamic organization.
Types of lateral mobility
Lateral heterogeneity in membrane organization presumably gives rise to different modes of lateral mobility (Figure 2). The visual appearance of a given trajectory is dependent on how frequently and for how long the particle path is sampled (Figure 3). In addition, the appearance of the trajectory is also dependent on how precisely the particle position can be measured; the impact of finite spatial precision in ‘blurring’ the trajectory is discussed in Supplemental Information.
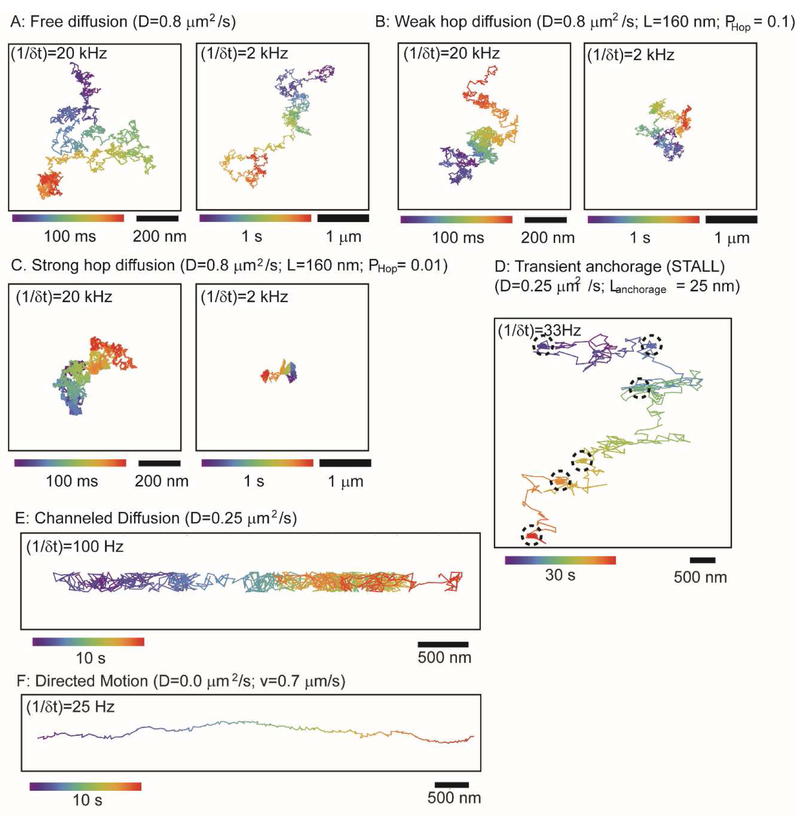
Figure 3. Simulated trajectories for various modes of lateral mobility.
(A) ‘free’ diffusion, (B) weak hop diffusion, (C) strong hop diffusion, (D) diffusion interrupted by periods of transient anchorage (STALL; dashed circles), (E) channeled diffusion, and (F) directed motion. All trajectories were simulated in Mathematica for the specified sampling frequencies without positional uncertainty error. Parameters (as noted in the Figure) are chosen to resemble various examples from literature for cases of hop diffusion (Kusumi et al., 2012; Lagerholm et al., 2017), transient anchorage/STALL (with anchorage times ~2s) (Chen et al., 2009; Suzuki et al., 2007a), channeled diffusion (Jaqaman et al., 2011), and directed motion (Liu et al., 2017). Scale bars as indicated; time evolution of trajectory indicated by color bars.
The most elementary mode of mobility, and which we would expect in, for example, a perfectly homogeneous membrane, is simple Brownian translational diffusion in the plane of the membrane, colorfully described as the random walk of a drunken sailor around a lamp post (Figure 3 A). A measure of the distance moved by the diffusing particle in its random walk during a given time interval is the square root of the mean squared displacement (MSD) over that interval. (The squared displacement is calculated because over a long enough time, the net displacement will be zero.) The relationship between the MSD and the time interval over which the particle is observed in the case of a random walk in a two dimensional plane is:
(1) where D is the diffusion coefficient, n is the number of time lags, tlag, between specific positions of a diffusant that were observed at a sampling frequency of (1/tlag). The product n ttag is the time over which the diffusion particle is observed. In the case of pure Brownian motion, D is thus independent of the how frequently or for how long the trajectory is sampled. In such cases, results between different studies can be compared even though the data may have been obtained at vastly different sampling frequencies.
The observation of time-independent diffusion of molecules in the plasma membrane is, however, the exception rather than the rule because in practice many impediments to Brownian diffusion exist. These impediments result in more complex mean squared displacement versus time curves (Figure 2 and Supplementary Table 1). Important for these more complex cases is that the resulting diffusion coefficient is time-dependent such that quantitative comparisons between results, that have been obtained by different fitting models (Supplementary Table 1), is only possible at specific time points.
Examples of more complex lateral diffusion include the extreme case of completely confined or corralled diffusion such that diffusant is laterally mobile but confined to a domain where escape is not possible. A more typical observed result is a form of anomalous diffusion that will occur if the diffusant encounters, in its trajectory, a wide distribution of trapping or binding times. For example, the diffusant may encounter obstructions (Saxton and Jacobson, 1997) that are caused by high concentrations of membrane proteins; some of these may be anchored or very slowly diffusing and serve to impede the diffusant by either sterically trapping it or transiently binding to it.
Cytoskeletal barriers arranged as a network of juxtaposed corrals just under the membrane may also obstruct ‘free’ diffusion by virtue of the cytoplasmic domains of transmembrane proteins interacting with the filaments composing the network; a case in point is hop diffusion discussed below where the diffusant is transiently trapped within a corral with a probability of escape (PHop) that that distinguishes weak (Figure 3B) and strong hop diffusion (Figure 3C) (Lagerholm et al., 2017). Note that weak hop diffusion (PHop =0.1; Figure 3B) can be difficult to discern from free diffusion (Figure 3A) from the appearance of the trajectory alone at fast sampling frequencies and short time duration (compare left panels of Figure 3A and 3B) but that the difference is much more apparent at lower sampling frequencies and longer time durations (compare right panel of Figure 3A to right panel of Figure 3B); by contrast, strong hop diffusion (PHop =0.01; Figure 3C) can be readily discerned from free diffusion in both cases that are shown. Cytoskeletal factors can also transiently anchor the diffusant (Figure 3D). Diffusion can also be obstructed such that it is effectively confined to one dimension, termed channeled diffusion (Figure 3E). In some cases, the diffusing particle goes farther than expected when compared to free diffusion; this can be due to an underlying flow augmenting diffusive transport or completely directed movement (Figure 3F), say, powered by a cytoplasmic motor associated with an underlying filament.
Regulation of lateral mobility in the plasma membrane
The anchored picket fence model and hop diffusion
The anchored picket fence model has been the most prominent scheme to explain the regulation of plasma membrane lateral mobility for two decades. The attributes of this model, introduced by Kusumi and his co-workers, have been documented in a series of seminal and elegant papers and authoritative reviews on the topic that offer interesting speculations (e.g. (Kusumi et al., 2012)). This model had forerunners, one being the relationship of the red cell spectrin-actin cytoskeleton to its plasma membrane (Branton et al., 1981; Sheetz et al., 1980). And, it was in this era that the notion of membrane posts was first proposed though in a somewhat different context. Picket-like posts, termed “protein-X”, were first proposed by Bourguignon and Singer (Bourguignon and Singer, 1977) to trap or otherwise interact with patches of cross-linked membrane proteins serving to couple the patches to a retrograde flowing actin cytoskeleton.
The anchored picket fence model by Kusumi et al. (Kusumi et al., 2012) envisioned a relatively static network of adjacent, connected corrals bounded by cortical filaments immediately underneath the membrane that undergo thermally driven fluctuations. This view was bolstered by electron microscopy studies showing that apparent corrals bounded by cytoskeletal filaments, presumably F-actin, had a similar size distribution as those determined by single particle tracking studies (Fujiwara et al., 2016; Kusumi et al., 2012). The fundamental “membrane skeleton fence” was postulated to cover the entire cell surface with a network of corrals and, within the corrals, diffusion (as measured at sub millisecond time scales) was controversially interpreted as very rapid, approaching or even exceeding values for membrane proteins and lipids in pure bilayers, i.e. 5–10 μm2/s. By contrast, diffusion over larger spatiotemporal scales (from about a few tens to over a hundred nanometers and a few to tens of milliseconds) was ~20-fold slower because the diffusant had to surmount the membrane skeleton fence barriers.
To account for the fact that lipids also displayed hop diffusion (Fujiwara et al., 2002), the model further postulated that subpopulation of largely unidentified transmembrane proteins (see below) are attached to the membrane skeleton fence and formed “pickets”, i.e. the anchored picket fence. Hop diffusion of lipids is caused by the picket proteins in three possible ways. First, the pickets create steric hindrance to the diffusing lipid. Second, the diffusing lipid may transiently bind to the pickets. Third, because the immobilized picket proteins may bind lipid, the inter-picket space was assumed to be of greater viscosity than other parts of the membrane further contributing to the barrier action of the picket fence. Importantly, the anchored picket fence model does not rule out the existence of other plasma membrane organizing principles such as lipid rafts (Lingwood and Simons, 2010) but the actin-based membrane skeleton fence and transmembrane protein pickets anchored to the fence provides an effective barrier against large-scale coalescence of lipid rafts (Kusumi et al., 2012). Indeed, large scale phase separation of proteins and lipids can occur in giant plasma membrane vesicles that are devoid of cortical actin (Baumgart et al., 2007).
Much of the definitive work by Kusumi and colleagues that produced the anchored picket fence model was conducted using 40nm gold particle tracking. The “hopping” transitions between adjacent corrals could only be detected in gold single particle tracking data acquired at sufficiently fast sampling frequencies. Single molecule tracking which measures diffusion coefficients on a longer spatial scale (~ tens of milliseconds to seconds) is then used to back calculate the hopping probability between corrals (see, for example, (Fujiwara et al., 2016)); it is the hopping between corrals that is the rate limiting step which determines the magnitude of the long-range (over 100s of nm to μm) lateral diffusion coefficient. Thus, the proposed upwards of 20-fold difference between the intra-corral diffusion coefficient and the inter-corral diffusion coefficient suggests very tight confinement resembling the strong hop diffusion case in Figure 3C with a hopping probability PHop=0.01, i.e. that only 1 out of 100 collisions with the corral boundary results in an escape from a confining corral, severely limiting long-range diffusion.
There have been relatively few efforts to independently corroborate Kusumi and colleagues’ findings. Wieser et al measured (Wieser et al., 2007) the plasma membrane diffusion of a GPI-AP, CD59, by single molecule tracking, using Fab fragments to label the protein. While these investigators did not directly observe hop diffusion, they concluded that this could be due to two possible scenarios: either strong confinement existed but with a confinement size <60nm or weak confinement and much larger confinement zones occurred. A more recent effort, using the complementary technique, fluorescence correlation spectroscopy using the super-resolution modality, stimulated emission-depletion microscopy (STED-FCS), with variable beam radii (Andrade et al., 2015), modeled the results by constructing a simulation of compartmentalized diffusion with different compartment sizes and barrier hopping probabilities. Importantly, this study employed only fluorescent lipid analogs so no larger particle label was involved. The results were consistent with compartmentalized hop diffusion but with much weaker confinement (i.e. the probability of lipid crossing a cytoskeletal barrier was higher than previously indicated by Kusumi et al. and resembled the weak hop diffusion case in Figure 3B). Part of these results have also been cross-validated by single particle tracking with Quantum dot (Qdot) tracking (Lagerholm et al., 2017). This study indicated that faster, intracorral diffusion, occurred over distances of about 100nm and times less than ~ 5ms time whereas above this spatiotemporal regime slower, intercorral diffusion, was measured as a consequence of the rate limiting step of crossing barriers between corrals. In a second example, diffusion of ectopically expressed GPI-APs in the axon initial segment is largely confined to the 190nm spectrin-rich spacing between actin rings as discussed above; this confinement reduces the macroscopic diffusion coefficient by up to an order of magnitude compared that between the rings (Albrecht et al., 2016). Thus, studies on lipid and lipid-anchored protein diffusion at least partially corroborate the anchored picket fence model but plausibly suggest that, in some cases, experiments using 40nm gold particles at very fast sampling frequencies significantly exaggerates the degree of confinement, presumably because the size of the gold particle-antibody complex sterically hinders passage through the picket fence and possibly by under-estimating the influence of experimental errors in the data analysis.
Recent studies on transmembrane proteins have provided further corroboration for the anchored picket fence model. Sadegh et al., (2017), using Qdot tracking, albeit at much reduced sampling frequency of 50Hz, and molecular localization microscopy, presented a convincing demonstration of how lateral diffusion of a plasma membrane protein, the potassium channel, expressed in HEK cells, is obstructed by interactions with the subjacent and compartmentalized actin cytoskeleton. Confined compartment trajectories, in which the diffusant sometimes hops to an adjacent compartment, are suggestively delimited by cortical actin filaments visualized by photoactivation localization microscopy (PALM). Hop diffusion was also observed for Qdot labelled G protein coupled receptors (GPCR) in primary ciliary membranes with large intracompartment D values (~3 um/sec2) but larger residence times (~700ms) than Kusumi et al (Kusumi et al., 2012) found for the basic corrals; surprisingly, actin disruption did not affect corral size but rather increased hop rate (Lee et al., 2018). These studies offer both intuitive and quantitative support for membrane skeleton fence regulation of the lateral diffusion of transmembrane proteins, albeit with some unexpected results that await further explanation.
Transient anchorage
The lateral diffusion of complexes in the plasma membrane may be transiently anchored (i.e.transiently immobilized) (Figure 3 D), likely as a prelude to a signal transduction event. Suzuki et al (Suzuki et al., 2007b) showed that a low degree of clustering (<10 molecules) of outer leaflet GPI-AP CD59 mediated by 40nm gold particles, induces transient association of inner leaflet Lyn kinase with the cluster. Giα2 also briefly associates with the cluster and when it does, it activates Lyn and shortly thereafter (within fractions of a second) a STALL (stimulation-induced temporary arrest of lateral diffusion) event occurs whereby the cluster is transiently immobilized. This event presumably requires linkage to the underlying cytoskeleton by yet undefined mechanisms. A STALL event leads to calcium signaling via PLCγ recruitment to the cluster (Suzuki et al., 2007a).
Chen et al (Chen et al., 2009) partially elucidated how transient anchorage occurs in molecular terms. Clusters of cross-linked Thy-1, a GPI-AP, formed on the addition of 40 nm streptavidin coated gold particles or Qdots to cells incubated with biotinconjugated anti Thy-1 antibody followed by addition of an anti Ig antibody. The lateral diffusion of such complexes was interrupted by episodes of transient anchorage of similar durations as STALLs; these events were mediated by a Src-regulatable transmembrane adaptor, csk binding protein (CBP), that was linked to ezrin binding protein 50 and presumably to the cytoskeleton via ERM proteins. Similar to STALL, Thy-1 transient anchorage was dependent on cholesterol.
Channeled diffusion
An example of channeled diffusion, in which the diffusing particle undergoes effectively one dimensional diffusion confined by parallel barriers, was discovered by McConnell and co-workers who found the diffusion of lectin receptors was highly anisotropic with the fast direction being parallel to actin stress fibers (Smith et al., 1979). Much more recently, Jaqaman et al. also found that the diffusion of a fraction of the population of CD36, a scavenger receptor, was likewise channeled possibly by microtubules proximate to the inner leaflet of the plasma membrane (Jaqaman et al., 2011) (Figure 3E).
Active, directed movement
Directed lateral movement of cross-linked membrane proteins has been observed in the plasma membrane for nearly half a century. In the case of relatively slow actin associated directed movements, such as the phenomenon of capping, cross-linked membrane protein patches (and even lipids (Schroit and Pagano, 1981) undergo active, retrograde directed movement at speeds of ~1 um/min in the plane of membrane towards the posterior portion of lymphocytes (Taylor et al., 1971) or the boundary between the lamella and perinuclear zone, termed the null border, of mesenchymal cells (Holifield et al., 1990).
Much more rapid directed movement in the plane of the membrane has been observed that appears to be associated with microtubules just under the cytoplasmic leaflet of the plasma membrane; this movement is distinct from rapid intracellular vesicular movement involving microtubules. Intraflagellar transport (Kozminski et al., 1993), where transport of TRPV (transient receptor potential channels of the vanilloid subtype) channels in the membranes of cilia occurs with velocities exceeding 1000 nm/s (Qin et al., 2005), is the most prominent example and is dependent on ciliary microtubules.
Other examples of this intriguing, very rapid directed movement exist. After T cells recognize foreign peptides displayed by major histocompatibility complexes on antigen presenting cells, T-cell receptors move in a highly directed fashion and with initially very high velocities towards the center of the immune synapse; this directed movement is along microtubule tracks and driven by the microtubule motor, dynein (Hashimoto-Tane et al., 2011). In addition, surface DC-SIGN nanoclusters were observed to undergo directed retrograde transport at high speeds of ~ 1000 nm/s (Neumann et al., 2008). A small fraction of dendritic cell- SIGN clusters underwent microtubule-dependent directed transport in the plane of the plasma membrane at speeds well over 1000 nm/s in both dendritic cells and in murine fibroblasts expressing human DC-SIGN (Liu et al., 2017) (Figure 3F). Rapid, directed rearward movement of DC-SIGN could bring bound pathogens on the leading edge and projections of dendritic cells to the perinuclear region for internalization and processing in a manner that is much more efficient than lateral diffusion.
Fundamental questions about the dynamic organization of the plasma membrane
While the anchored picket fence model may represent the most general way to view control of lateral mobility in the plasma membrane, at least at small spatiotemporal scales, there are several key issues that must be resolved to produce a more complete understanding of the plasma membrane in terms of its dynamic structure. These include whether the lateral density of proteins in the membrane is sufficient to provide the number of pickets required to form the picket fence and the identity of pickets. And, how a universal anchored picket fence, with small inter-picket spacings, can accommodate the lateral diffusion of nanoclusters and the directed transport of membrane components.
Important, broader issues exist that we will not discuss in detail. First, whether the anchored picket fence model applies similarly to cells of mesenchymal, epithelial and myeloid origin where the membrane skeleton may be more static or dynamic. Second, how are meso-microscale specializations constructed in the presence of the anchored picket fence? For example, the cell surface must be locally modified to permit phagocytosis of larger particles (Ostrowski et al., 2016); by contrast, micron-sized spectrin-ankryin-G domains patrolling the lateral membrane of epithelial cells resist endocytosis (Jenkins et al., 2015). As another example, micron-sized preferential localizations of DC-SIGN have been discovered by molecular cartography (Garcia-Parajo et al., 2014). Third, the discovery of local cortical actin asters coupled to the plasma membrane thereby forming an “active membrane composite” (Gowrishankar et al., 2012; Rao and Mayor, 2014) prompts the question of whether such composites may be related to an interconnected membrane associated cytoskeleton that can, via local stimuli, propagate changes in lateral mobility over the entire cell surface; this latter phenomenon has been termed “anchorage modulation” (Henis and Elson, 1981; Rutishauser et al., 1974). And lastly, how different are plasma membrane organization and dynamics in differentiated cells in tissue (Gall and Edelman, 1981) from commonly used secondary cell culture (Bennett and Lorenzo, 2016).
The high concentration of membrane proteins: implications for plasma membrane organization
The original Singer-Nicolson Fluid Mosaic model (Singer and Nicolson, 1972) and numerous subsequent schematic illustrations depict a concentration of membrane proteins that is far too low (Engleman, 2005; Yeagle, 1989). When protein concentration estimates taken from literature (Jacobson et al., 2007) are considered, in fact, the lipid bilayer is chock full of membrane proteins (Dupuy and Engelman, 2008; Takamori et al., 2006) such that little “free” lipid outside of the solvating lipid shell (Lee, 2011) exists. Roughly 30,000 proteins/μm2 are in the plasma membrane of a typical cultured mammalian cell (Jacobson et al., 2007). Thus, the membrane should be envisioned as a “protein-lipid composite material” with little “free lipid” as opposed to a lipid bilayer with proteins embedded at a relatively low lateral concentration, as is frequently depicted. One consequence of this high density of membrane proteins, particularly those that are attached to the membrane associated cytoskeleton, is that membrane lipids largely resist flow leading to the proposition that the plasma membrane, while fluid on the nanoscale, is more gel-like on the mesoscale; as a result, local tension changes are not propagated globally via the plasma membrane but rather are transmitted by cytoplasmic mediators such as calcium (Shi et al., 2018).
While biophysical studies with pure and mixed bilayers are obviously important to investigate the properties of liquid crystalline systems and to provide mechanistic insights into reactions occurring at the membrane (McLean et al., 2018; Nair et al., 2011), they cannot fully capture the general reality of biomembrane organization. Clearly, if we are to use the model systems to gain insight into biomembranes, these systems should reflect the presumed high lateral concentration of proteins, as giant plasma membrane vesicles probably do (Sezgin et al., 2017). As a consequence, domains of selected components must exist within these protein-lipid composites rather than as isolated entities in a lipid bilayer sea.
What molecular details are lacking in the anchored picket fence model?
Number of pickets required to form the picket fence:
A back of the envelope calculation suggests that roughly 3×106 pickets would be needed for a “square” fibroblast 40μm on a side covered by 100nm square compartments underlying the plasma membrane. This translates to ~ 20 picket proteins per compartment because of common sides, assuming each picket is 2nm in diameter and is 8nm from the next picket (Kusumi et al., 2012). Thus, even though a putative picket, CD44 (see below) is expressed at about 106 copies per cell (Freeman et al., 2018), it is likely that different types of membrane proteins must also be able to play the role of pickets. Using the estimate above (Jacobson et al., 2007), there are ~ 300 proteins in square compartments 100nm on a side meaning that pickets comprise only a few percent of the total membrane proteins in 100nm square compartments. This suggests that the very high intracompartment lipid diffusion coefficients reported by Kusumi et al. (Kusumi et al., 2012) are probably overestimated; this is possibly the result of the difficulty in obtaining accurate diffusion coefficients at faster sampling rates due to increased influence of measurement noise (Supplementary Information). Indeed, more recent diffusion measurements that yield lower numbers probably reflect diffusion through the smallest, protein-rich compartments defined by the anchored picket fence (Andrade et al., 2015; Lagerholm et al., 2017; Wieser et al., 2007).
Identity of pickets and adaptors:
Recently, Freeman et al (Freeman et al., 2018) found that anchored CD-44 pickets, in macrophages, interact with diffusing proteins sterically and/or via chemical interactions to hinder diffusion and sometimes corral the diffusant. While the size of the corrals is similar to those in the anchored picket fence model (Kusumi et al., 2012), they seemingly occur “randomly” presumably due to their transient nature dictated by cytoskeletal dynamics as noted by the authors. This begs the question of what determines where CD44 mediated corrals appear.
The overall dynamics that permit lateral diffusion of both non-picket and picket proteins are complex, involving thermally derived fluctuations of the membrane-fence separation as well as association-dissociation kinetics of the ezrin mediated fence-CD44 linkage. Thus, CD44 exists in three states with respect to lateral diffusion: it can be an immobile membrane picket but, when released from the fence, it may be transiently confined to corrals consisting of other CD44 pickets and/or other candidate picket proteins. Between states of transient confinement, it may also undergo long-range “freely mobile Brownian diffusion” with D ~ 0.1 μm2/s. Proponents of the anchored picket fence model would argue that the “freely mobile” CD44 diffusion is not really free but actually consists of hop diffusion (Kusumi et al., 2012) obscured by the lower time resolution of the authors’ measurements [(30ms (Qdot single particle tracking) & 100 ms (single molecule tracking) vs 25 μs (for gold single particle tracking)]. As mentioned above, it is important to inquire whether the anchored picket fence model applies not only to mesenchymal cells but also to cells of myeloid and epithelial origin. In this regard, a key test will be to see if, at high time resolution, CD44 hop diffusion, prevails in those parts of the trajectories that exhibit, at lower time resolution, “freely mobile” diffusion in macrophages which are of myeloid origin.
Could cell surface topography compartmentalize diffusion?
Other effects could replace or augment the anchored picket fence in producing compartmentalizing diffusion. For example, blebs, largely devoid of any membrane associated cytoskeleton, are initiated by the local loss of linkages between the actomyosin cortex and the plasma membrane (Charras, 2008). In addition, especially in cells that are not attached, excess surface area is stored in complex bleb-like morphologies as seen in mitotic cells and cells that have been detached from the substrate (Kapustina et al., 2016). Such morphologies could in effect produce compartmentalized diffusion: diffusion would be more rapid in a bleb or bleb-like protrusion than near the base of these structures where they connect to the cortex proper in regions of higher curvature (Figure 4A, B). It is also possible that the membrane skeleton fence could imprint the plasma membrane with subtle curvature induced where the fence is closest to the inner leaflet to provide weak confinement that would complement the effects of pickets (Adler et al., 2010).
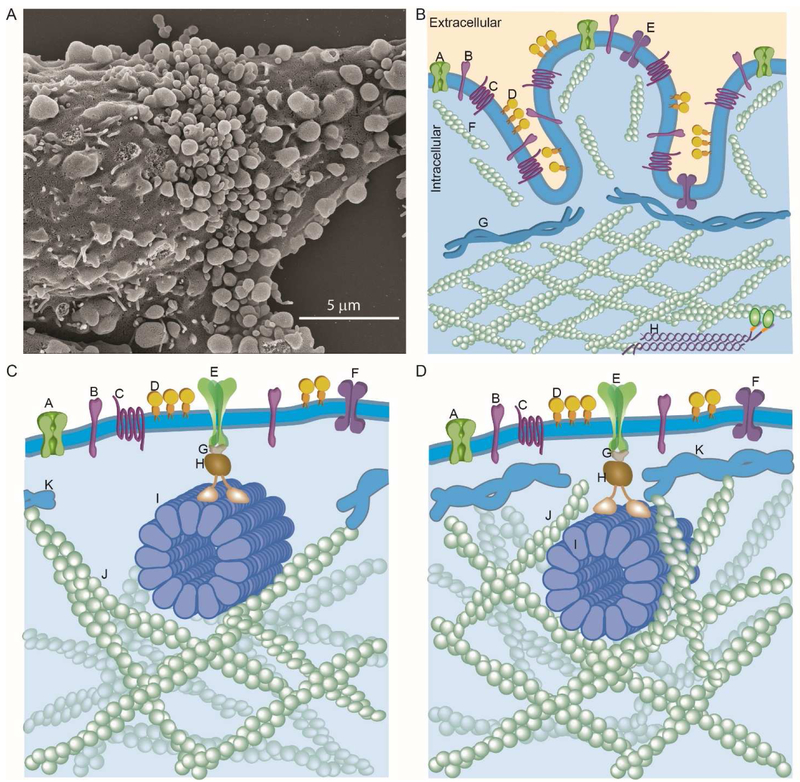
Figure 4. The cortical cytoskeleton and its effects on mobility.
A. Bleb-like protrusions on the surface of CHO cells in the process of spreading on a substrate as imaged by scanning electron microscopy. B. How blebs and/or bleb-like protrusions could compartmentalize lateral diffusion (see text for discussion). The labels in panel B are: A, ion channel; B, single pass transmembrane protein; C, GCPR; D, GPI-AP; E, transmembrane protein dimer; F, F-actin; G, spectrin; H, non-muscle myosin. C,D. How rapid, MT-based directed transport might occur in the presence of the membrane skeleton fence (see text for discussion). C. Cortex deforms to allow MT and its associated motor(s) direct access to PM protein cluster. The labels in panel C are: A-D are the same as in panel B; E, oligomeric transmembrane protein, such as DC-SIGN, coupled to a MT motor; F, transmembrane protein dimer; G, adaptor protein, H, motor protein; I, MT; J, F-actin; K, spectrin. D. MT and associated motor (s) proximate to the inner leaflet is able to slice through the membrane skeleton fence due to motor force and thermally activated breaks in the fence. The labels in panel D are: A-K are the same as in panel C. Panels B,C and D have been inspired and adapted, in part, from the drawings of David Goodsell (The Machinery of Life, second edition, Copernicus Books, NY)
To what extent do nanoclusters hop diffuse?
Given the apparent prevalence of nanoclusters of membrane proteins (Garcia-Parajo et al., 2014), how do these clusters laterally diffuse to subserve biological functions in the context of the anchored picket fence model? Indeed, a main tenet of this model is that clustering leads to oligomerization-induced trapping whereby oligomers are too big to squeeze through the interpacket spaces leading to trapping within the compartments defined by the anchored picket fence. This effective immobilization is hypothesized to provide a way to localize membrane components for downstream signal transduction events (Kusumi et al., 2012). And, a recent single molecule tracking study showing compartmentalization for GPCRs and Gα by both actin, clathrin coated pits and microtubules furnished independent support for this notion (Sungkaworn et al., 2017).
But some nanoclusters can laterally diffuse. For example, DC-SIGN nanoclusters laterally diffuse with a median diffusion coefficient of ~0.065 μm2/s (Manzo et al., 2012) yet these nanoclusters are composed of, on average, about two tetramers (Itano et al., 2012) having a minimal diameter of ~20nm. The formula for lattice diffusion in two dimensions, D=1/4 νλ2, where ν is the hop frequency from lattice site to lattice site and λ is the lattice constant, can be used to estimate hop frequencies between adjacent compartments given D~ 0.065 μm2/s; this yields a residence time (ν−1) in a 100nm corral provided by the anchored picket fence of ~40 ms. This dwell time in a corral is similar to the longer times found for membrane proteins in the presence of a complete picket fence (Kusumi et al., 2012). Thus, for long range diffusion to occur, selected pickets must be dynamically or permanently removed to permit passage of a DC-SIGN nanocluster from one compartment to an adjacent one at a frequency of ~25 times per second.
How does directed, active lateral transport in the plasma membrane occur in the presence of the underlying membrane skeleton fence?
Actin or microtubule based directed movements raise the issue of how such movement occurs in the presence of a universal, subjacent membrane cytoskeleton fence. Actomyosin- based capping of cross-linked (patched) antigens proceeds with a relatively slow velocity (~1 μm/min), a time scale that permits cytoskeletal remodeling. In this case, two extreme possibilities exist. First, the entire fence could move being coupled to a retrograde moving cortical cytoskeleton in which patched antigens are entrapped akin to “oligomerization induced trapping”. Alternatively, a variety of patched antigens could be indirectly coupled to a retrograde cortical cytoskeletal flow through lateral interaction with a subset of integral membrane proteins that possess a specific, stable anchorage to moving cytoskeletal elements that function like the tines of a rake (Bourguignon and Singer, 1977; Holifield et al., 1990). Patches could be moved if such “tine” membrane proteins were entrapped within the patch of immunoaggregated protein during its formation or simply caught between the tines of the rake; however, the tines would need to slice through the membrane skeleton fence, a task made possible by the thermally labile fence structure ((Kusumi et al., 2012) and see below).
In the case of the more rapid microtubule-base transport, presumably microtubules must be close enough to the plasma membrane so that putative microtubule motors can interact with the proteins to be transported. How might this occur? One possibility is that membrane skeleton fence coupled to the underlying cortex is locally dissociated from the plasma membrane creating narrow channels that contain the microtubules and the motor driving DC-SIGN cluster directed movement (Figure 4C). In this regard, effective one-dimensional diffusion of CD36 is mediated by channels in the cortex that are associated with membrane proximate microtubules (Jaqaman et al., 2011). Another possibility (Figure 4D) is based on the fact that network connections within the membrane skeleton fence are opening and closing owing to thermal energy so that the microtubule motor could drive the cluster through the anchored picket fence (Kusumi et al., 2012; Liu et al., 2017). This is plausible because microtubule motors can generate typically 5–7pN of force (Nicholas et al., 2015; Valentine et al., 2006) but it only requires forces as weak as 0.25 pN to drag the transferrin receptor through the labile membrane skeleton fence (Sako and Kusumi, 1995).
Future directions
Composition and structure of stable nanodomains.
With respect to determining molecular composition of plasma membranes and their associated peripheral structures, there seem to be few alternatives to well-designed, validated proteomics efforts or screens especially in the case of stable membrane protein clusters or isolated, authentic membrane domains. And new reagents including amphipathic copolymers promise more faithful extraction of membrane protein- lipid complexes than detergents (Jamshad et al., 2011). The affinity purification-mass spectrometry/proteomic approach has, for example, identified and corroborated binding partners of B-cell receptor clusters, including dynein, dynactin and tubulin subunits (Schnyder et al., 2011). Complementing this approach, there are several enzyme catalyzed proximity-based labeling approaches to detect nearest neighbors in vivo (Rees et al., 2015). Such methods promiscuously label neighboring proteins making them suitable for detecting weak, transient, or even indirect protein-protein interactions within the surface nanodomains; however, this approach can result in many species being labelled resulting in the need for a biased pare down of possibly relevant targets for further analysis. Thus, the use of complementary methods to confirm the importance of certain binding partners is desirable (Rees et al., 2015).
In certain cases, membrane domains can be assembled or isolated and peripheral binding partners screened or identified. For example, expression of caveolae I in the cell free system results in the assembly of caveolae like structures in which caveolin 1 is correctly inserted in membranes (Jung et al., 2018). Putative caveolin 1 binding partners could then be tested for actual caveolin 1 binding in a native environment. Membrane blebs can be generated and harvested to ask what peripheral, cortical components are bound to the bleb membrane by proteomic analysis (Biro et al., 2013) followed by RNA interference screens to test for functionality of candidate proteins (Bovellan et al., 2014; Chugh et al., 2017)
On the horizon are non-microscopic methods, involving DNA origami technology, for detecting components associated with the protein of interest and their spatial arrangements on the 100 nm scale. These methods produce scaffolds of known architecture to which other nucleic acid molecules and antibodies can be conjugated at known positions (Shaw et al., 2014). The new technology shifts the analysis from the membrane proteins directly as in microscopy and proteomics to the analysis of the released origami structure and the tags affixed to it that encode positional and component information employing the sequencing methods now available. Thus, the DNA origami structures serve as nanoscale templates of defined size and provide a novel way to approach the stoichiometry, relative size and organization of clustered membrane proteins on the 100nm length scale but in a high throughput fashion, capable of resolving different organizational states of the nanoclusters as population averages.
Elucidation of the structures peripheral to the membrane
In general, the overall structure of the cortex underlying the plasma membrane is far from being completely understood (Chugh and Paluch, 2018; Svitkina, 2018) although progress has been made on the membrane associated cytoskeleton (Kusumi et al., 2012; Xu et al., 2013). Ultrastructural studies of the actin network near the plasma membrane using electron microscopy began almost a half century ago. But now we can expect recent developments in cryoelectron microscopy (Mahamid et al., 2016) will afford the opportunity to understand the cortical structures underlying the plasma membrane (and the pericellular matrix) in much greater detail. First, vitrification by rapid freezing so that water does not crystallize will better preserve the living state largely removing fixation artifacts. Second, focused ion beam milling permits sections previously too thick for imaging to be thinned so that they can be examined (Mahamid et al., 2016) and new contrast enhancement techniques for lower spatial frequencies improves the interpretation of the images (Fukuda et al., 2015). Lastly, while electron microscopy has provided important insights into actin-filament organization, standard TEM images are two-dimensional projections where features are superimposed upon one another in the direction of the electron beam; this limits resolution and makes it difficult to study the three-dimensional organization of complex filament networks. By contrast, tomographic techniques acquire projections of the object as viewed from different directions and then merge them computationally into a 3D reconstruction. Such tomograms provide an unprecedented look into the 3D structure of the cortex underlying the plasma membrane (Fujiwara et al., 2016). Combining immunogold and aptamer labeling with such tomographic images will permit the location of specific components, including those identified in proteomic searches and shRNA screens.
Closing remarks
At this juncture, it is clear that there are experimental and computational roads (Ingólfsson et al., 2016; Sezgin et al., 2017) to investigating the structure and composition of the membrane and structures peripheral to it that remain to be fully exploited and thereby complement the outstanding progress in the biophysical analysis of restraints to lateral mobility. Such knowledge will be an important precursor to the clearly crucial topic of how the non-equilibrium (Rao and Mayor, 2014) membrane dynamic structure changes directly via mechanical and electrical ques and indirectly through metabolism affecting lipid and protein composition.
Supplementary Material
Supplemental Figure 1. Effect of spatial measurement error on the appearance of single particle trajectories. Related to Figure 3. Experimental single particle tracking data contains a combination of spatial localization and camera blur error that has to be accounted for in the analysis. The spatial localization error occurs because the position of a diffusant at a specific time point can only be determined experimentally to within some finite precision, ±δ (Martin et al., 2002). This effect is illustrated on simulated trajectories of various experimentally observed motion modes at relevant observation durations and sampling frequencies with (below diagonal) and without (above diagonal) the imposition of typical measurement uncertainty in the particle position. The trajectories without imposition of spatial measurement imprecision are from Figure 3. (A) ‘free’ diffusion, (B) weak hop diffusion, (C) strong hop diffusion, (D) diffusion interrupted by periods of transient anchorage (STALL; dashed circles), (E) channeled diffusion, and (F) directed. All trajectories were simulated in Mathematica for the specified sampling frequencies with and without a 20 nm positional uncertainty error as indicated. Scale bars as indicated; time evolution of trajectory indicated by color bars. The simulated noise has an overall blurring effect upon the trajectories that is in particular very noticeable at the faster sampling frequency of 20 kHz for the cases of free diffusion (Supplemental Figure 1A) and weak hop diffusion (Supplemental Figure 1B). By contrast, the effect is hardly noticeable at sampling frequencies of 2 kHz or lower (see examples in Supplemental Figures 1 A-F). This is because the magnitude of the simulated measurement error at the simulated parameters and a sampling frequency of 20 kHz is of the same scale as the characteristic unhindered mean diffusion length ((4 D tlag)1/2≈13 nm) whereas it is much less for all other cases shown. In particular, note that the trajectory example for free diffusion with simulated measurement errors (Supplemental Figure 1A) very strongly resembles the appearance of the example for weak hop diffusion (Phop=0.1) without measurement errors (Supplemental Figure 1B) and also that the same is true for the case of weak hop diffusion (Phop=0.1) with measurement errors (Supplemental Figure 1B) and strong hop diffusion (Phop=0.01) without measurement errors (Supplemental Figure 1A). Thus it is very difficult to discern these cases by visual inspection of the trajectories alone.
Supplemental Figure 2. Effect of spatial measurement error on the appearance of MSD versus time and MSD/(4t) versus time plots. Related to Figure 2. A consequence of spatial measurement error is that even the simplest case of Brownian diffusion results in a complex non-linear apparent time-dependent diffusion coefficient, in particular when fast sampling frequencies are employed. The effect of spatial measurement error on traditional MSD vs time (n tlag) plots are shown in Supplemental Figure 2A where solid lines represent the ideal situation with no localization noise (δx,y=0 nm) and the dotted lines show the effect of added localization noise (δx,y=20 nm) on the same diffusion cases as in Figure 2 (i.e. directed motion (red); Brownian diffusion (black), anomalous sub-diffusion (blue), channeled diffusion (purple), transiently confined diffusion (green), and confined diffusion (yellow)). These measurement errors, if either left unaccounted for or if underestimated, will artificially increase the experimentally determined D because the jiggle in position of the diffusant is only due the uncertainty in determining position, not true movement (Lagerholm et al., 2017; Martin et al., 2002; Savin and Doyle, 2005). The effect of camera blur, on the other hand, depreciates the experimental D at short times because the diffusant is mobile during the camera integration time and blur artificially is counted as a contribution to the MSD (Savin and Doyle, 2005). The equivalent relation for the experimentally determined MSD [n tlag] after accounting for these errors in the case of free diffusion is , or alternatively in generic terms (Berglund, 2010; Lagerholm et al., 2017) where R is a motion blur constant with values of 0 ≤ R ≤ 1/4 and, specifically, R=1/6 for full frame averaging (Savin and Doyle, 2005), d is the dimensionality of the diffusion process, D is the diffusion coefficient, n is the number of time lags, tlag, between specific positions of a diffusant that were observed at a sampling frequency of (1/tlag), and δx,y is the localization noise along the x- and y-axis. By using the latter equation, it is subsequently possible to accurately extract a variety of parameters describing a diffusion process by curve fitting to a range of diffusion models (Supplementary Table 1). This approach is, however, hampered because it is only possible to compare the different models in terms of the diffusion coefficient after curve fitting. Furthermore, the effect of the noise on the magnitude of the MSD plots is deceptively small such that it is very difficult to correctly assess the extent of the impact that the noise can have on the interpretation of the fitted results for the diffusion coefficient. A preferred method for direct differentiation of the different types of lateral mobility is to instead plot MSD/(2 d n tlag) versus time as this effectively directly shows the time evolution of the raw data for the diffusion process in terms of an apparent diffusion coefficient This is shown in Supplemental Figure 2B for same diffusion cases as in Supplemental Figure 2A where solid lines represent the ideal situation with no localization noise (δx,y=0 nm)and the dotted lines show the effect of added localization noise (δx,y=20 nm). In this plot format it is possible to directly assess the impact that localization noise has for data acquired at fast sampling frequencies (≈>100 Hz). Furthermore, by employing this approach, it is possible to directly observe the time evolution of the diffusion coefficient for a particular diffusion process; in particular, the relevant time scales required to observe the different types of lateral motion shown in Figure 2 can be efficiently distinguished. It is evident from the latter equation that the non-linearity that is introduced by positional uncertainty, but not camera blur, becomes more prominent at higher sampling frequencies, (1/tlag), especially where analysis is performed exclusively on only the initial few data points, n. For example, consider diffusion in a xy-plane (d=2) with symmetric spatial localization error along each axis (δx = δy =δx,y and δr2 =2 δx,y 2), full frame averaging (R=1/6), D=1 μm2/s and δx,y=10 nm. The characteristic sampling frequency where the noise contribution to Dapp[n tlag] for the first data point (n=1) exceeds the real diffusion contribution, D, by greater than 10% is (1/ tlag) ≈ 0.7 kHz or tlag ≈ 1.5 ms; the sampling frequency where the noise contribution to D for the first data point is greater than 100% is (1/ tlag) ≈ 6.7 kHz or tlag ≈ 0.15 ms. This calculation illustrates the particular challenge of accurate analysis of high-speed single particle tracking data; a problem that is further complicated because the uncertainty in spatial position, δx,y, is inversely proportional to the square root of the number of collected photons which decreases as the sampling frequency increases (Thompson et al., 2002).
Supplemental Table 1. Mathematical expressions for a selection of diffusion models. Related to Figures 2 and 3.
Footnotes:
a The following parameters are defined for all diffusion models as follows: MSD is the mean squared displacement, d is the dimensionality of the diffusion process, i.e. d=2 for diffusion in a 2-dimensional plane, D is the diffusion coefficient, n is the number of experimental data points, tlag is the lag time between consecutive data points, T is a specific time point n tlag, v is the directed flow velocity, and α is an anomaly constant where α < 1 for sub-diffusion and α>1 for super-diffusion. The remaining parameters are defined below.
b The typical main aim in investigations of lateral dynamics is to determine the time- dependence of the diffusion process and the magnitude of the diffusion coefficient, D. A standard approach for this is to perform a curve fitting of the time-dependence of the MSD (n tlag) to a suitable diffusion model. It has in this case further become customary to also include plots of the time dependence of the raw MSD (n tlag) data in order to directly be able to compare lateral dynamics data from different experimental conditions. However, this approach suffers from that it is not amendable for a direct, model-independent, assessment of the time-dependence of the diffusion process in terms of D, and furthermore that it is very difficult to directly assess the impact of experimental noise for data acquired at fast sampling frequencies (Supplemental Figure 1).
c An alternative method that is better suited for direct comparison of lateral dynamics data in terms of the time-dependence of D is to instead define a time dependent diffusion coefficient, D(n tlag), as given in the table. This approach has the advantage that it is then possible to directly evaluate the time-dependence of the lateral dynamics data in terms of D in a model independent way, and furthermore to directly assess the impact of experimental noise in the measurements (Supplemental Figure 2).
d Constant diffusion processes are the only type of diffusion processes for which the magnitude of D can be directly compared between studies. This is because this is the only case where the magnitude of D is independent of the sampling time n tlag.
e Continuous time-dependent diffusion processes have a time-dependent diffusion coefficient, D(n tlag), that varies continuously with the sampling time n tlag. Direct comparisons of the magnitude of the time-dependent diffusion coefficient, D (n tlag), from different studies in this case is thus only possible at specific times, T=n tlag.
f Transient time-dependent diffusion processes are defined by having a diffusion coefficient, D(n tlag), that approaches constant, but separate, magnitudes in the limits of time n tlag → 0 and n tlag → ∞, with some intermediate temporal transition zone. It is in this case possible to compare the magnitude of D but only then at the limiting values or at specific times, T=n tlag.
g This conventional definition of the anomalous diffusion has the disadvantage that D has units that is also dependent on the magnitude of the anomaly constant α. It is thus not possible to compare the magnitude of D unless the magnitude of the anomaly constant α is also considered.
h This revised time normalized anomalous diffusion model has the advantage that the units of D are always in the traditional format of (distance)2/time for all values of the anomaly constant α and the time constant T. This expression is furthermore directly amendable for calculating the magnitude of D independent of the anomaly constant α for any specific value of the time T=n tlag, and is thus also suitable for direct comparison of the magnitude of D at specific times.
I This expression is for completely confined diffusion in a confinement area that is given by L2=12 Dμ T where L is the linear confinement dimension, Dμ is the unhindered diffusion coefficient within the confinement area and T is a time constant at which the magnitude of Dμ has been reduced by a factor of 1/e.
j This expression is for transiently confined diffusion, or so called hop-diffusion, where the time-dependent diffusion coefficient, D(n tlag), consists of a long-range, intracompartmental, time-independent, diffusion coefficient component, DM, and a short-range, intercompartmental, time-dependent diffusion coefficient component, Dμ. The confinement area is given, as above, by L2=12 Dμ T where T is a time constant at which the magnitude of Dμ has been reduced by a factor of 1/e.
k This expression is for channeled diffusion, where a molecule is confined along one spatial dimension, but diffuses freely along the other spatial dimensions. The channel width is given by L=(12 DT)1/2 where T is a time constant at which the magnitude of D, in a direction that is perpendicular to the channel has been reduced by a factor of 1/e
Acknowlegements
This work was supported by NIGMS 41402. Because this review covers a span of nearly five decades, we have often had to refer to reviews as opposed to the original papers. For this we apologize to the primary authors, many of whom have provided invaluable advice over nearly five decades (KJ). We thank Joe Costello for helpful comments and Maryna Kapustina for provision of Figure 4A and Mathias Porsmose Clausen for provision of Figure 2A. KJ also thanks NIGMS for many years of support and, in particular, Dr. Jean Chin, formerly of NIGMS, for her patient counsel over the years in navigating the grant landscape. B.C.L. acknowledge support from the Medical Research Council (MC_UU_12009 and MC_UU_12010). P.L. acknowledges support from the Bioimaging Core Facility of WNLO-HUST.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no conflicts of interests.
Literature Cited
- Adler J, Shevchuk AI, Novak P, Korchev YE, and Parmryd I (2010). Plasma membrane topography and interpretation of single-particle tracks. Nat. Methods 7, 170–171. [DOI] [PubMed] [Google Scholar]
- Albrecht D, Winterflood CM, Sadeghi M, Tschager T, Noé F, and Ewers H (2016). Nanoscopic compartmentalization of membrane protein motion at the axon initial segment. J. Cell Biol 215, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade DM, Clausen MP, Keller J, Mueller V, Wu C, Bear JE, Hell SW, Lagerholm BC, and Eggeling C (2015). Cortical actin networks induce spatio-temporal confinement of phospholipids in the plasma membrane--a minimally invasive investigation by STED-FCS. Sci. Rep 5, 11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, and Webb WW (2007). Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci U A 104, 3165–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V, and Lorenzo DN (2016). Chapter Five - An Adaptable Spectrin/Ankyrin-Based Mechanism for Long-Range Organization of Plasma Membranes in Vertebrate Tissues In Current Topics in Membranes, Bennett V, ed. (Academic Press; ), pp. 143–184. [DOI] [PubMed] [Google Scholar]
- Biro M, Romeo Y, Kroschwald S, Bovellan M, Boden A, Tcherkezian J, Roux PP, Charras G, and Paluch EK (2013). Cell cortex composition and homeostasis resolved by integrating proteomics and quantitative imaging. Cytoskelet. Hoboken NJ 70, 741–754. [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, and Singer SJ (1977). Transmembrane interactions and the mechanism of capping of surface receptors by their specific ligands. Proc. Natl. Acad. Sci. U. S. A 74, 5031–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, Vaghela M, Fritzsche M, Moulding D, Thorogate R, et al. (2014). Cellular Control of Cortical Actin Nucleation. Curr. Biol 24, 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branton D, Cohen CM, and Tyler J (1981). Interaction of cytoskeletal proteins on the human erythrocyte membrane. Cell 24, 24–32. [DOI] [PubMed] [Google Scholar]
- Cambi A, Lange F. de, Maarzeveen N. van, Nijhuis M, Joosten B, Dijk E. van, Bakker B. de, Fransen J, Bovee-Geurts P, Leeuwen F. van, et al. (2004). Microdomains of the C-typle lectin DC-SIGN are portals for virus entry into dendritic cells. J Cell Biol 164, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras GT (2008). A short history of blebbing. J. Microsc 231, 466–478. [DOI] [PubMed] [Google Scholar]
- Chen Y, Veracini L, Benistant C, and Jacobson K (2009). The transmembrane protein CBP plays a role in transiently anchoring small clusters of Thy-1, a GPI-anchored protein, to the cytoskeleton. J Cell Sci 122, 3966–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P, and Paluch EK (2018). The actin cortex at a glance. J Cell Sci 131, jcs186254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P, Clark AG, Smith MB, Cassani DAD, Dierkes K, Ragab A, Roux PP, Charras G, Salbreux G, and Paluch EK (2017). Actin cortex architecture regulates cell surface tension. Nat. Cell Biol 19, 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AD, and Engelman DM (2008). Protein area occupancy at the center of the red blood cell membrane. Proc. Natl. Acad. Sci 105, 2848–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML, and Choudhuri K (2016). Signaling and Polarized Communication Across the T Cell Immunological Synapse. Annu. Rev. Cell Dev. Biol 32, 303–325. [DOI] [PubMed] [Google Scholar]
- Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schonle A, et al. (2009). Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1162. [DOI] [PubMed] [Google Scholar]
- Engleman DM (2005). Membranes are more mosaic than fluid. Nature 438, 578–580. [DOI] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, and Bretscher A (2010). Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol 11, 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg H, Guo Y, Mitchell DA, Drickamer K, and Weis WI (2005). Extended neck regions stabilize tetramers of the receptors DC-SIGN and DC-SIGNR. J Biol Chem 280, 1327–1335. [DOI] [PubMed] [Google Scholar]
- Fowler VM (2013). Chapter Two - The Human Erythrocyte Plasma Membrane: A Rosetta Stone for Decoding Membrane–Cytoskeleton Structure In Current Topics in Membranes, Bennett V, ed. (Academic Press; ), pp. 39–88. [DOI] [PubMed] [Google Scholar]
- Freeman SA, Vega A, Riedl M, Collins RF, Ostrowski PP, Woods EC, Bertozzi CR, Tammi MI, Lidke DS, Johnson P, et al. (2018). Transmembrane Pickets Connect Cyto- and Pericellular Skeletons Forming Barriers to Receptor Engagement. Cell 172, 305–317.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche M, Li D, Colin-York H, Chang VT, Moeendarbary E, Felce JH, Sezgin E, Charras G, Betzig E, and Eggeling C (2017). Self-organizing actin patterns shape membrane architecture but not cell mechanics. Nat. Commun 8, 14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, and Kusumi A (2002). Phospholipids undergo hop diffusion in compartmentalized cell membrane. J Cell Biol 157, 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara TK, Iwasawa K, Kalay Z, Tsunoyama TA, Watanabe Y, Umemura YM, Murakoshi H, Suzuki KGN, Nemoto YL, Morone N, et al. (2016). Confined diffusion of transmembrane proteins and lipids induced by the same actin meshwork lining the plasma membrane. Mol. Biol. Cell 27, 1101–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y, Laugks U, Lučić V, Baumeister W, and Danev R (2015). Electron cryotomography of vitrified cells with a Volta phase plate. J. Struct. Biol 190, 143–154. [DOI] [PubMed] [Google Scholar]
- Gall WE, and Edelman GM (1981). Lateral diffusion of surface molecules in animal cells and tissues. Science 213, 903–905. [DOI] [PubMed] [Google Scholar]
- Garcia-Parajo MF, Cambi A, Torreno-Pina JA, Thompson N, and Jacobson K (2014). Nanoclustering as a dominant feature of plasma membrane organization. J. Cell Sci 127, 4995–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheber LA (2018). The Life of a Membrane Protein. Biophys. J 114, 2762–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goiko M, de Bruyn JR, and Heit B (2018). Membrane Diffusion Occurs by Continuous-Time Random Walk Sustained by Vesicular Trafficking. Biophys. J 114, 2887–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar K, Ghosh S, Saha S, C R, Mayor S, and Rao M (2012). Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell 149, 1353–1367. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Tane A, Yokosuka T, Sakata-Sogawa K, Sakuma M, Ishihara C, Tokunaga M, and Saito T (2011). Dynein-Driven Transport of T Cell Receptor Microclusters Regulates Immune Synapse Formation and T Cell Activation. Immunity 34, 919–931. [DOI] [PubMed] [Google Scholar]
- Henis YI, and Elson EL (1981). Inhibition of the mobility of mouse lymphocyte surface immunoglobulins by locally bound concanavalin A. Proc. Natl. Acad. Sci 78, 1072–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holifield BF, Ishihara A, and Jacobson K (1990). Comparative behavior of membrane protein-antibody complexes on motile fibroblasts: implications for a mechanism of capping. J Cell Biol 111, 2499–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson HI, Arnarez C, Periole X, and Marrink SJ (2016). Computational ‘microscopy’ of cellular membranes. J Cell Sci 129, 257–268. [DOI] [PubMed] [Google Scholar]
- Itano MS, Steinhauer C, Schmied JJ, Forthmann C, Liu P, Neumann AK, Thompson NL, Tinnefeld P, and Jacobson K (2012). Super-resolution imaging of C-type lectin and influenza hemagglutinin nanodomains on plasma membranes using blink microscopy. Biophys. J 102, 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itano MS, Graus MS, Pehlke C, Wester MJ, Liu P, Lidke KA, Thompson NL, Jacobson K, and Neumann AK (2014). Super-resolution imaging of C-type lectin spatial rearrangement within the dendritic cell plasma membrane at fungal microbe contact sites. Membr. Physiol. Membr. Biophys 2, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K, Mouritsen OG, and Anderson RG (2007). Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol 9, 7–14. [DOI] [PubMed] [Google Scholar]
- Jamshad M, Lin Y-P, Knowles TJ, Parslow RA, Harris C, Wheatley M, Poyner DR, Bill RM, Thomas ORT, Overduin M, et al. (2011). Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem. Soc. Trans 39, 813–818. [DOI] [PubMed] [Google Scholar]
- Jaqaman K, Kuwata H, Touret N, Collins R, Trimble WS, Danuser G, and Grinstein S (2011). Cytoskeletal control of CD36 diffusion promotes its receptor and signaling function. Cell 146, 593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins PM, He M, and Bennett V (2015). Dynamic spectrin/ankyrin-G microdomains promote lateral membrane assembly by opposing endocytosis. Sci. Adv 1, e1500301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W, Sierecki E, Bastiani M, O’Carroll A, Alexandrov K, Rae J, Johnston W, Hunter DJB, Ferguson C, Gambin Y, et al. (2018). Cell-free formation and interactome analysis of caveolae. J. Cell Biol 217, 2141–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapustina M, Tsygankov D, Zhao J, Wessler T, Yang X, Chen A, Roach N, Elston TC, Wang Q, Jacobson K, et al. (2016). Modeling the Excess Cell Surface Stored in a Complex Morphology of Bleb-Like Protrusions. PLOS Comput. Biol 12, e1004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, and Rosenbaum JL (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. U. S. A 90, 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapf D (2018). Compartmentalization of the plasma membrane. Curr. Opin. Cell Biol. 53, 15–21. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, and Suzuki KGN (2012). Dynamic Organizing Principles of the Plasma Membrane that Regulate Signal Transduction: Commemorating the Fortieth Anniversary of Singer and Nicolson’s Fluid-Mosaic Model. Annu. Rev. Cell Dev. Biol 28, 215–250. [DOI] [PubMed] [Google Scholar]
- Lagerholm BC, Andrade DM, Clausen MP, and Eggeling C (2017). Convergence of lateral dynamic measurements in the plasma membrane of live cells from single particle tracking and STED-FCS. J. Phys. Appl. Phys 50, 063001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AG (2011). Biological membranes: the importance of molecular detail. Trends Biochem. Sci 36, 493–500. [DOI] [PubMed] [Google Scholar]
- Lee S, Tan HY, Geneva II, Kruglov A, and Calvert PD (2018). Actin filaments partition primary cilia membranes into distinct fluid corrals. J. Cell Biol 217, 2831–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, and Simons K (2010). Lipid rafts as a membrane-organizing principle. Science 327, 46–50. [DOI] [PubMed] [Google Scholar]
- Liu P, Wang X, Itano MS, Neumann AK, Jacobson K, and Thompson NL (2012). The formation and stability of DC-SIGN microdomains require its extracellular moiety. Traffic Cph. Den 13, 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Weinreb V, Ridilla M, Betts L, Patel P, Silva A.M. de, Thompson NL, and Jacobson K (2017). Rapid, directed transport of DC-SIGN clusters in the plasma membrane. Sci. Adv 3, eaao1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, and Baumeister W (2016). Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972. [DOI] [PubMed] [Google Scholar]
- Manzo C, Torreno-Pina JA, Joosten B, Reinieren-Beeren I, Gualda EJ, Loza-Alvarez P, Figdor CG, Garcia-Parajo MF, and Cambi A (2012). The neck region of the C-type lectin DC-SIGN regulates its surface spatiotemporal organization and virus-binding capacity on antigen-presenting cells. J. Biol. Chem 287, 38946–38955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean MA, Gregory MC, and Sligar SG (2018). Nanodiscs: A Controlled Bilayer Surface for the Study of Membrane Proteins. Annu. Rev. Biophys [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, and Cossart P (2012). Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol 13, 183–194. [DOI] [PubMed] [Google Scholar]
- Nair PM, Salaita K, Petit RS, and Groves JT (2011). Using patterned supported lipid membranes to investigate the role of receptor organization in intercellular signaling. Nat. Protoc 6, 523–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann AK, Thompson NL, and Jacobson K (2008). Distribution and lateral mobility of DC-SIGN on immature dendritic cells--implications for pathogen uptake. J. Cell Sci 121, 634–643. [DOI] [PubMed] [Google Scholar]
- Nicholas MP, Höök P, Brenner S, Wynne CL, Vallee RB, and Gennerich A (2015). Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat. Commun 6, 6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson GL (1976). Trans-membrane control of the receptors on normal and tumor cells II. Surface changes associated with transformation and malignancy. Biochim. Biophys. Acta BBARev. Cancer 458, 1–72. [DOI] [PubMed] [Google Scholar]
- Nicolson GL (2014). The Fluid—Mosaic Model of Membrane Structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40years. Biochim. Biophys. Acta BBA - Biomembr. 1838, 1451–1466. [DOI] [PubMed] [Google Scholar]
- Ostrowski PP, Grinstein S, and Freeman SA (2016). Diffusion Barriers, Mechanical Forces, and the Biophysics of Phagocytosis. Dev. Cell 38, 135–146. [DOI] [PubMed] [Google Scholar]
- Pike LJ (2006). Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res 47, 1597–1598. [DOI] [PubMed] [Google Scholar]
- Qin HM, Burnette DT, Bae YK, Forscher P, Barr MM, and Rosenbaum JL (2005). Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr. Biol 15, 1695–1699. [DOI] [PubMed] [Google Scholar]
- Raghunathan K, and Kenworthy AK (2018). Dynamic pattern generation in cell membranes: Current insights into membrane organization. Biochim. Biophys. Acta BBA - Biomembr. 1860, 2018–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghupathy R, Anilkumar AA, Polley A, Singh PP, Yadav M, Johnson C, Suryawanshi S, Saikam V, Sawant SD, Panda A, et al. (2015). Transbilayer Lipid Interactions Mediate Nanoclustering of Lipid-Anchored Proteins. Cell 161, 581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, and Mayor S (2014). Active organization of membrane constituents in living cells. Curr. Opin. Cell Biol 29, 126–132. [DOI] [PubMed] [Google Scholar]
- Rees JS, Li X-W, Perrett S, Lilley KS, and Jackson AP (2015). Protein Neighbors and Proximity Proteomics. Mol. Cell. Proteomics MCP 14, 2848–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser U, Yahara I, and Edelman GM (1974). Morphology, Motility, and Surface Behavior of Lymphocytes Bound to Nylon Fibers. Proc. Natl. Acad. Sci 71, 1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadegh S, Higgins JL, Mannion PC, Tamkun MM, and Krapf D (2017). Plasma Membrane is Compartmentalized by a Self-Similar Cortical Actin Meshwork. Phys. Rev. X 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, and Kusumi A (1995). Barriers for lateral diffusion of transferrin receptor in the plasma membrane as characterized by receptor dragging by laser tweezers: fence versus tether. J. Cell Biol 129, 1559–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ, and Jacobson K (1997). Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct 26, 373–399. [DOI] [PubMed] [Google Scholar]
- Schnyder T, Castello A, Feest C, Harwood NE, Oellerich T, Urlaub H, Engelke M, Wienands J, Bruckbauer A, and Batista FD (2011). B Cell Receptor-Mediated Antigen Gathering Requires Ubiquitin Ligase Cbl and Adaptors Grb2 and Dok-3 to Recruit Dynein to the Signaling Microcluster. Immunity 34, 905–918. [DOI] [PubMed] [Google Scholar]
- Schroit AJ, and Pagano RE (1981). Capping of a phospholipid analog in the plasma membrane of lymphocytes. Cell 23, 105–112. [DOI] [PubMed] [Google Scholar]
- Sezgin E, Levental I, Mayor S, and Eggeling C (2017). The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol 18, 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A, Lundin V, Petrova E, Fördős F, Benson E, Al-Amin A, Herland A, Blokzijl A, Högberg B, and Teixeira AI (2014). Spatial control of membrane receptor function using ligand nanocalipers. Nat. Methods 11, 841–846. [DOI] [PubMed] [Google Scholar]
- Sheetz MP (2001). Cell control by membrane-cytoskeleton adhesion. Nat Rev Mol Cell Biol 2, 392–396. [DOI] [PubMed] [Google Scholar]
- Sheetz MP, Schindler M, and Koppel DE (1980). Lateral mobility of integral membrane proteins is increased in spherocytic erythrocytes. Nature 285, 510–511. [DOI] [PubMed] [Google Scholar]
- Shi Z, Graber ZT, Baumgart T, Stone HA, and Cohen AE (2018). Cell Membranes Resist Flow. Cell 175, 1769–1779.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SJ, and Nicolson GL (1972). The fluid mosaic model of the structure of cell membranes. Science 175, 720–731. [DOI] [PubMed] [Google Scholar]
- Smith BA, Clark WR, and McConnell HM (1979). Anisotropic molecular motion on cell surfaces. Proc. Natl. Acad. Sci. U. S. A 76, 5641–5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungkaworn T, Jobin M-L, Burnecki K, Weron A, Lohse MJ, and Calebiro D (2017). Single-molecule imaging reveals receptor–G protein interactions at cell surface hot spots. Nature 550, 543–547. [DOI] [PubMed] [Google Scholar]
- Suzuki KG, Fujiwara TK, Edidin M, and Kusumi A (2007a). Dynamic recruitment of phospholipase C gamma at transiently immobilized GPI-anchored receptor clusters induces IP3-Ca2+ signaling: single-molecule tracking study 2. J Cell Biol 177, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki KGN, Fujiwara TK, Sanematsu F, Iino R, Edidin M, and Kusumi A (2007b). GPI-anchored receptor clusters transiently recruit Lyn and G alpha for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol 177, 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM (2018). Ultrastructure of the actin cytoskeleton. Curr. Opin. Cell Biol 54, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, et al. (2006). Molecular anatomy of a trafficking organelle. Cell 127, 831–846. [DOI] [PubMed] [Google Scholar]
- Taylor RB, Duffus WPH, Raff MC, and de PETRIS S (1971). Redistribution and Pinocytosis of Lymphocyte Surface Immunoglobulin Molecules Induced by Anti-Immunoglobulin Antibody. Nature 233, 225–229. [DOI] [PubMed] [Google Scholar]
- Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, and Block SM (2006). Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol 8, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma R, and Mayor S (1998). GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394, 798–801. [DOI] [PubMed] [Google Scholar]
- Wieser S, Moertelmaier M, Fuertbauer E, Stockinger H, and Schütz GJ (2007). (Un)confined diffusion of CD59 in the plasma membrane determined by high-resolution single molecule microscopy. Biophys. J 92, 3719–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz RE, Sanchez-Adams J, and Guilak F (2014). The structure and function of the pericellular matrix of articular cartilage. Matrix Biol. 39, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhong G, and Zhuang X (2013). Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeagle PL (1989). Lipid regulation of cell membrane structure and function. FASEB J. 3, 1833–1842. [PubMed] [Google Scholar]
- van Zanten TS, Cambi A, Koopman M, Joosten B, Figdor CG, and Garcia-Parajo MF (2009). Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc. Natl. Acad. Sci. U. S. A 106, 18557–18562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lee GM, and Jacobson K (1993). Protein lateral mobility as a reflection of membrane microstructure. Bioessays 15, 579–588. [DOI] [PubMed] [Google Scholar]
- Zhou Y, and Hancock JF (2015). Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim. Biophys. Acta 1853, 841–849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Effect of spatial measurement error on the appearance of single particle trajectories. Related to Figure 3. Experimental single particle tracking data contains a combination of spatial localization and camera blur error that has to be accounted for in the analysis. The spatial localization error occurs because the position of a diffusant at a specific time point can only be determined experimentally to within some finite precision, ±δ (Martin et al., 2002). This effect is illustrated on simulated trajectories of various experimentally observed motion modes at relevant observation durations and sampling frequencies with (below diagonal) and without (above diagonal) the imposition of typical measurement uncertainty in the particle position. The trajectories without imposition of spatial measurement imprecision are from Figure 3. (A) ‘free’ diffusion, (B) weak hop diffusion, (C) strong hop diffusion, (D) diffusion interrupted by periods of transient anchorage (STALL; dashed circles), (E) channeled diffusion, and (F) directed. All trajectories were simulated in Mathematica for the specified sampling frequencies with and without a 20 nm positional uncertainty error as indicated. Scale bars as indicated; time evolution of trajectory indicated by color bars. The simulated noise has an overall blurring effect upon the trajectories that is in particular very noticeable at the faster sampling frequency of 20 kHz for the cases of free diffusion (Supplemental Figure 1A) and weak hop diffusion (Supplemental Figure 1B). By contrast, the effect is hardly noticeable at sampling frequencies of 2 kHz or lower (see examples in Supplemental Figures 1 A-F). This is because the magnitude of the simulated measurement error at the simulated parameters and a sampling frequency of 20 kHz is of the same scale as the characteristic unhindered mean diffusion length ((4 D tlag)1/2≈13 nm) whereas it is much less for all other cases shown. In particular, note that the trajectory example for free diffusion with simulated measurement errors (Supplemental Figure 1A) very strongly resembles the appearance of the example for weak hop diffusion (Phop=0.1) without measurement errors (Supplemental Figure 1B) and also that the same is true for the case of weak hop diffusion (Phop=0.1) with measurement errors (Supplemental Figure 1B) and strong hop diffusion (Phop=0.01) without measurement errors (Supplemental Figure 1A). Thus it is very difficult to discern these cases by visual inspection of the trajectories alone.
Supplemental Figure 2. Effect of spatial measurement error on the appearance of MSD versus time and MSD/(4t) versus time plots. Related to Figure 2. A consequence of spatial measurement error is that even the simplest case of Brownian diffusion results in a complex non-linear apparent time-dependent diffusion coefficient, in particular when fast sampling frequencies are employed. The effect of spatial measurement error on traditional MSD vs time (n tlag) plots are shown in Supplemental Figure 2A where solid lines represent the ideal situation with no localization noise (δx,y=0 nm) and the dotted lines show the effect of added localization noise (δx,y=20 nm) on the same diffusion cases as in Figure 2 (i.e. directed motion (red); Brownian diffusion (black), anomalous sub-diffusion (blue), channeled diffusion (purple), transiently confined diffusion (green), and confined diffusion (yellow)). These measurement errors, if either left unaccounted for or if underestimated, will artificially increase the experimentally determined D because the jiggle in position of the diffusant is only due the uncertainty in determining position, not true movement (Lagerholm et al., 2017; Martin et al., 2002; Savin and Doyle, 2005). The effect of camera blur, on the other hand, depreciates the experimental D at short times because the diffusant is mobile during the camera integration time and blur artificially is counted as a contribution to the MSD (Savin and Doyle, 2005). The equivalent relation for the experimentally determined MSD [n tlag] after accounting for these errors in the case of free diffusion is , or alternatively in generic terms (Berglund, 2010; Lagerholm et al., 2017) where R is a motion blur constant with values of 0 ≤ R ≤ 1/4 and, specifically, R=1/6 for full frame averaging (Savin and Doyle, 2005), d is the dimensionality of the diffusion process, D is the diffusion coefficient, n is the number of time lags, tlag, between specific positions of a diffusant that were observed at a sampling frequency of (1/tlag), and δx,y is the localization noise along the x- and y-axis. By using the latter equation, it is subsequently possible to accurately extract a variety of parameters describing a diffusion process by curve fitting to a range of diffusion models (Supplementary Table 1). This approach is, however, hampered because it is only possible to compare the different models in terms of the diffusion coefficient after curve fitting. Furthermore, the effect of the noise on the magnitude of the MSD plots is deceptively small such that it is very difficult to correctly assess the extent of the impact that the noise can have on the interpretation of the fitted results for the diffusion coefficient. A preferred method for direct differentiation of the different types of lateral mobility is to instead plot MSD/(2 d n tlag) versus time as this effectively directly shows the time evolution of the raw data for the diffusion process in terms of an apparent diffusion coefficient This is shown in Supplemental Figure 2B for same diffusion cases as in Supplemental Figure 2A where solid lines represent the ideal situation with no localization noise (δx,y=0 nm)and the dotted lines show the effect of added localization noise (δx,y=20 nm). In this plot format it is possible to directly assess the impact that localization noise has for data acquired at fast sampling frequencies (≈>100 Hz). Furthermore, by employing this approach, it is possible to directly observe the time evolution of the diffusion coefficient for a particular diffusion process; in particular, the relevant time scales required to observe the different types of lateral motion shown in Figure 2 can be efficiently distinguished. It is evident from the latter equation that the non-linearity that is introduced by positional uncertainty, but not camera blur, becomes more prominent at higher sampling frequencies, (1/tlag), especially where analysis is performed exclusively on only the initial few data points, n. For example, consider diffusion in a xy-plane (d=2) with symmetric spatial localization error along each axis (δx = δy =δx,y and δr2 =2 δx,y 2), full frame averaging (R=1/6), D=1 μm2/s and δx,y=10 nm. The characteristic sampling frequency where the noise contribution to Dapp[n tlag] for the first data point (n=1) exceeds the real diffusion contribution, D, by greater than 10% is (1/ tlag) ≈ 0.7 kHz or tlag ≈ 1.5 ms; the sampling frequency where the noise contribution to D for the first data point is greater than 100% is (1/ tlag) ≈ 6.7 kHz or tlag ≈ 0.15 ms. This calculation illustrates the particular challenge of accurate analysis of high-speed single particle tracking data; a problem that is further complicated because the uncertainty in spatial position, δx,y, is inversely proportional to the square root of the number of collected photons which decreases as the sampling frequency increases (Thompson et al., 2002).
Supplemental Table 1. Mathematical expressions for a selection of diffusion models. Related to Figures 2 and 3.
Footnotes:
a The following parameters are defined for all diffusion models as follows: MSD is the mean squared displacement, d is the dimensionality of the diffusion process, i.e. d=2 for diffusion in a 2-dimensional plane, D is the diffusion coefficient, n is the number of experimental data points, tlag is the lag time between consecutive data points, T is a specific time point n tlag, v is the directed flow velocity, and α is an anomaly constant where α < 1 for sub-diffusion and α>1 for super-diffusion. The remaining parameters are defined below.
b The typical main aim in investigations of lateral dynamics is to determine the time- dependence of the diffusion process and the magnitude of the diffusion coefficient, D. A standard approach for this is to perform a curve fitting of the time-dependence of the MSD (n tlag) to a suitable diffusion model. It has in this case further become customary to also include plots of the time dependence of the raw MSD (n tlag) data in order to directly be able to compare lateral dynamics data from different experimental conditions. However, this approach suffers from that it is not amendable for a direct, model-independent, assessment of the time-dependence of the diffusion process in terms of D, and furthermore that it is very difficult to directly assess the impact of experimental noise for data acquired at fast sampling frequencies (Supplemental Figure 1).
c An alternative method that is better suited for direct comparison of lateral dynamics data in terms of the time-dependence of D is to instead define a time dependent diffusion coefficient, D(n tlag), as given in the table. This approach has the advantage that it is then possible to directly evaluate the time-dependence of the lateral dynamics data in terms of D in a model independent way, and furthermore to directly assess the impact of experimental noise in the measurements (Supplemental Figure 2).
d Constant diffusion processes are the only type of diffusion processes for which the magnitude of D can be directly compared between studies. This is because this is the only case where the magnitude of D is independent of the sampling time n tlag.
e Continuous time-dependent diffusion processes have a time-dependent diffusion coefficient, D(n tlag), that varies continuously with the sampling time n tlag. Direct comparisons of the magnitude of the time-dependent diffusion coefficient, D (n tlag), from different studies in this case is thus only possible at specific times, T=n tlag.
f Transient time-dependent diffusion processes are defined by having a diffusion coefficient, D(n tlag), that approaches constant, but separate, magnitudes in the limits of time n tlag → 0 and n tlag → ∞, with some intermediate temporal transition zone. It is in this case possible to compare the magnitude of D but only then at the limiting values or at specific times, T=n tlag.
g This conventional definition of the anomalous diffusion has the disadvantage that D has units that is also dependent on the magnitude of the anomaly constant α. It is thus not possible to compare the magnitude of D unless the magnitude of the anomaly constant α is also considered.
h This revised time normalized anomalous diffusion model has the advantage that the units of D are always in the traditional format of (distance)2/time for all values of the anomaly constant α and the time constant T. This expression is furthermore directly amendable for calculating the magnitude of D independent of the anomaly constant α for any specific value of the time T=n tlag, and is thus also suitable for direct comparison of the magnitude of D at specific times.
I This expression is for completely confined diffusion in a confinement area that is given by L2=12 Dμ T where L is the linear confinement dimension, Dμ is the unhindered diffusion coefficient within the confinement area and T is a time constant at which the magnitude of Dμ has been reduced by a factor of 1/e.
j This expression is for transiently confined diffusion, or so called hop-diffusion, where the time-dependent diffusion coefficient, D(n tlag), consists of a long-range, intracompartmental, time-independent, diffusion coefficient component, DM, and a short-range, intercompartmental, time-dependent diffusion coefficient component, Dμ. The confinement area is given, as above, by L2=12 Dμ T where T is a time constant at which the magnitude of Dμ has been reduced by a factor of 1/e.
k This expression is for channeled diffusion, where a molecule is confined along one spatial dimension, but diffuses freely along the other spatial dimensions. The channel width is given by L=(12 DT)1/2 where T is a time constant at which the magnitude of D, in a direction that is perpendicular to the channel has been reduced by a factor of 1/e