Abstract
Pulmonary arterial hypertension (PAH) is characterized by increased pulmonary artery pressure and vascular resistance, typically leading to right heart failure and death. Current therapies improve quality of life of the patients but have a modest effect on long-term survival. A detailed transcriptomics and systems biology view of the PAH lung is expected to provide new testable hypotheses for exploring novel treatments. We completed transcriptomics analysis of PAH and control lung tissue to develop disease-specific and clinical data/tissue pathology gene expression classifiers from expression datasets. Gene expression data were integrated into pathway analyses. Gene expression microarray data were collected from 58 PAH and 25 control lung tissues. The strength of the dataset and its derived disease classifier was validated using multiple approaches. Pathways and upstream regulators analyses was completed with standard and novel graphical approaches. The PAH lung dataset identified expression patterns specific to PAH subtypes, clinical parameters, and lung pathology variables. Pathway analyses indicate the important global role of TNF and transforming growth factor signaling pathways. In addition, novel upstream regulators and insight into the cellular and innate immune responses driving PAH were identified. Finally, WNT-signaling pathways may be a major determinant underlying the observed sex differences in PAH. This study provides a transcriptional framework for the PAH-diseased lung, supported by previously reported findings, and will be a valuable resource to the PAH research community. Our investigation revealed novel potential targets and pathways amenable to further study in a variety of experimental systems.
Keywords: pulmonary arterial hypertension, lung transcriptomics, bioinformatics
Clinical Relevance
Our lung transcriptome study data in pulmonary arterial hypertension (PAH) were analyzed in a variety of ways, relating transcriptional changes with pulmonary pathological observations, current targeted therapies, and the potential role of inflammation and novel pathways and regulators in this disease. We found strong evidence for the estrogen receptor pathway, inflammation modulators, innate immunity, and WNT pathway processes in the PAH transcriptome. Our work represents the largest PAH lung transcriptome study to date and provides insights into current therapies and generates new hypotheses for preclinical testing.
Pulmonary arterial hypertension (PAH) is characterized by mean pulmonary artery pressure (mPAP) of 25 mm Hg or greater, though the majority of patients are referred for a formal clinical work-up after finding elevated right ventricular systolic pressure by echocardiogram studies (see Reference 1 for a recent review). PAH is a rare pulmonary disease, with an incidence of 2–10 cases/million in the United States and Europe, and is clinically subdivided into 5 groups. Within group 1 PAH are idiopathic PAH (IPAH), associated PAH (APAH), and heritable PAH (HPAH). APAH can be further subdivided into associated phenotypes, such as connective tissue disease, congenital heart defects, and anorexigen/stimulant drug use. HPAH most frequently involves bone morphogenetic protein (BMP) receptor (BMPR) 2 mutations/deletions, though other transforming growth factor-β (TGFB)/BMP superfamily member mutations have been described (2). Recently, a whole-genome sequencing project of 1,038 patients with PAH/6,385 control subjects was completed, notably adding SRY-box 17 (SOX17) and growth differentiation factor 2 as potential drivers of PAH (3). With current clinical management, diagnostics, and therapeutics, patient 5-year survival has improved to 50–60% (1, 4, 5). Women are two to three times more likely to be diagnosed with PAH, though, interestingly, they exhibit longer survival (6, 7). Group 1 PAH is characterized by an endothelial cell hyperproliferative phenotype in the lung vasculature creating obstructions, vascular lesions (identified as plexiform lesions), and vasoconstriction (8, 9). The physical blocking of normal pulmonary blood flow results in increased pulmonary vascular resistance and right ventricular maladaptation, ultimately leading to right heart failure as the most common cause of death. Current treatment options typically focus on symptomatic relief rather than disease progression intervention (10).
Diseases such as PAH are driven by a complicated network of molecular processes (11–14). Data supporting this notion are accumulating from large-scale “-omics” profiling in numerous disease conditions (15–17). Methods are currently emerging to employ molecular profiling to understand more expansive systems-level pathways in human disease contexts. However, for PAH, advances have been slowed by anatomic inaccessibility of diseased pulmonary tissue and consequent inability to develop relevant computational tools for these transcriptomic datasets. Our work uses established and novel computational analyses of gene expression applied to a large number of PAH lung tissue specimens in comparison to controls. These results provide validation of current therapeutic approaches as well as predictions of both the connections and rewiring of pathways with potential new insights in to pathogenesis and treatment.
Methods
Lung tissues preserved in RNALater (Ambion) were provided by the Pulmonary Hypertension Breakthrough Initiative (PHBI; https://www.ipahresearch.org/services.html). The PHBI is a multicenter network of lung transplant centers, the goal of which is the accrual of clinical data and lung biospecimens from patients with PAH at transplant and control subjects (failed donors [FD]). FD control lung samples were obtained from patients not found to have an appropriate recipient, but still meeting physiologic standards. The tissue procurement protocol was previously described (see Reference 18 and the data supplement), and application for lung tissue, primary cell lines, and genomics datasets can be made at http://phbi.org/index.do. A summary of the patient demographics is shown in Table 1 and a detailed table in the data supplement (Table E1). Total RNA was prepared and analyzed using standard methods (see the data supplement), and the microarray CEL files and associated data are available at NCBI GEO as GSE117261.
Table 1.
Demographics of Pulmonary Hypertension Breakthrough Initiative Genomics Samples
| FD | PAH | P Value | |
|---|---|---|---|
| Samples, n | 25 | 58 | — |
| Age range, yr | 1–64 | 7–79 | NS |
| Sex | |||
| Male | 18 | 15 | 0.0001 |
| Female | 7 | 43 | — |
| Race | |||
| White | 21 | 40 | NS |
| Black | 4 | 6 | — |
| Hispanic | 0 | 7 | — |
| Asian | 0 | 5 | — |
| PAH subtypes | |||
| IPAH | 31 | — | |
| APAH | 18 | — | |
| HPAH | 5 | — | |
| Other | 4 | — |
Definition of abbreviations: APAH = associated pulmonary arterial hypertension; FD = failed donor; HPAH = hereditary pulmonary arterial hypertension; IPAH = idiopathic pulmonary arterial hypertension; NS = not significant; PAH = pulmonary arterial hypertension.
Microarray CEL files were imported into a Partek Genomics Suite 6.6 and analyzed as described in detail in the data supplement. Briefly, the 83 microarrays were imported to visualize batch effects. As the microarrays were collected over a 4-year period in three separate batches, “batch” became the biggest confounding variable (see Figure E1A in the data supplement). In addition, PAH disease has a female bias (3:1 female:male, as seen in our samples), whereas the FDs show a male bias (1:2; overall distribution χ2 < 0.0001). Thus, batches 1–3 were used, after correcting for batch and sex (Partek’s “batch remove” function, Figure E1B). The microarray cohort included 58 subjects with PAH (group 1 clinical subtypes) and 25 FD patients. Differential gene expression analysis (Partek ANOVA model after batch/sex correction) generated 1,140 transcript IDs (using false discovery rate [FDR] q value < 0.001 [19]; Figure E2 and Table E2).
Pathway analysis of the gene list was performed in three ways. First, the online freeware tool, Enrichr (http://amp.pharm.mssm.edu/Enrichr/ [20]) interrogated standard Gene Ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analyses. Ingenuity pathway analysis (IPA; application build 463341M, March 9, 2018) helped to visualize pathway overrepresentation, as well as to identify potential upstream regulators along with predicting changed activity in their pathways. In addition, considering complex molecular mechanisms underlying diseases, such as cancer and PAH, a novel, network-based computational statistical approach, Evaluation of Differential DependencY (EDDY; see additional details in the data supplement), was used. EDDY combines pathway-guided and differential dependency analyses with a probabilistic framework (21, 22) to identify rewired pathways in PAH.
Additional methods information and details are available in the data supplement.
Results
Characterization of the Microarray Dataset
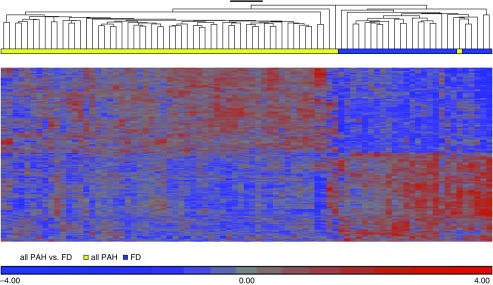
Patient demographics (Table 1) did not show a significant difference in range of ages or ethnicity among the PHBI cohort. The FD control subjects were provided with minimum clinical data (age, sex, and race/ethnicity) and no previous medical history information. As seen with end-stage PAH disease, the vast majority of patients were receiving triple therapy. A total of 76% of patients were on triple therapy (phosphodiesterase [PDE] 5A inhibitor, ERA [endothelin receptor antagonist], and prostanoid) with another 17% on a double therapy that included a prostanoid. As outlined in the data supplement, the microarray dataset had large batch and sex effects, which were corrected by Partek’s “batch-remove” feature. After batch/sex correction, the complete microarray dataset (n = 33,297 transcript IDs) was analyzed by ANOVA modeling (Partek) for differentially expressed (DE) genes between PAH and FD, followed by q value FDR correction. There were 1,140 transcript IDs meeting an FDR q value less than 0.001 cut-off (termed PAH classifier, Table E2), and is displayed as a supervised expression heatmap and volcano plot (Figure 1 and Figure E2). We tested the PAH classifier on an independent control versus PAH lung expression dataset (23) and by using a literature-derived PAH gene network (see Reference 14 and Figures E3 and E4). In addition, we also validated specific gene expression differences predicted from the microarray analysis by qRT-PCR (Figure E5).
Figure 1.
Supervised hierarchical clustering of PHBI (Pulmonary Hypertension Breakthrough Initiative) classifier discovery genelist. The PHBI classifier is visualized by supervised hierarchical clustering of the PHBI dataset showing 99% correct identification of the pulmonary arterial hypertension (PAH) and failed donor (FD) samples into separate branches of the dendrogram. One PAH (idiopathic PAH subgroup) sample was misclassified as FD. Figures E1 and E2 show the batch and sex correction process, as well as independent validation of the PHBI classifier on independent lung PAH transcriptome datasets and literature-derived PAH-specific gene pathways.
Current PAH treatments generally target vasoconstriction by three different modalities: nitric oxide→soluble guanylate cyclase→cGMP levels, and the endothelin (EDN) and prostacyclin pathways (1, 8, 24). For the small subset of patients with PAH that exhibit vasoreactivity (13, 25), voltage-gated calcium channel blockers can be effective. Evidence within the PAH lung transcriptome support the importance of these three therapeutic options. PDE5A and other PDE family members were found to be significantly upregulated (Table 2 and Figure E5). An alternative treatment strategy targets soluble guanylate cyclase with stimulators (e.g., riociguat), with a subunit of soluble guanylate cyclase (guanylate cyclase 1 soluble subunit Beta 1 [GUCY1B1]) being downregulated (Table 2). EDN pathway antagonists (e.g., bosentan) are another established treatment for PAH. EDN1 and EDN receptor A were both upregulated (Table 2). Finally, the L-type voltage-gated calcium channel, CACNA1C, the target for dihydropyridine-class drugs, amlodipine and nifedipine, used in patients with PAH that exhibit vasoreactivity, was significantly upregulated (Table 2). Additional listing of these gene families, along with BMPs/BMPR1A, S100 calcium binding proteins (S100 s), and Toll-like receptors (TLRs) are shown in Table E3.
Table 2.
Differentially Regulated Drug Targets in PAH
| Fold Change (PAH/FD) | Adjusted P Value | |
|---|---|---|
| Endothelin pathway | ||
| Endothelin 1 (EDN1) | 2.60 | 6.74E−05 |
| Endothelin receptor type A (EDNRA) | 1.66 | 2.64E−04 |
| Phosphodiesterase family | ||
| Phosphodiesterase 5A (PDE5A) | 1.48 | 1.30E−03 |
| Guanylate cyclase 1 β subunit 1 (GUCY1B1) | 0.84 | 3.70E−02 |
| Prostanoid pathway proteins | ||
| Prostaglandin I2 (prostacyclin) receptor (PTGIR) | 1.21 | 9.34E−02 |
| Prostaglandin I2 (prostacyclin) synthase (PTGIS) | 1.26 | 8.23E−02 |
| Voltage-gated calcium channels (vasoreactive-PAH) | ||
| Calcium voltage-gated channel subunit A1C (CACNA1C) | 1.22 | 3.60E−02 |
Geneset Enrichment Analysis of the PAH Classifier
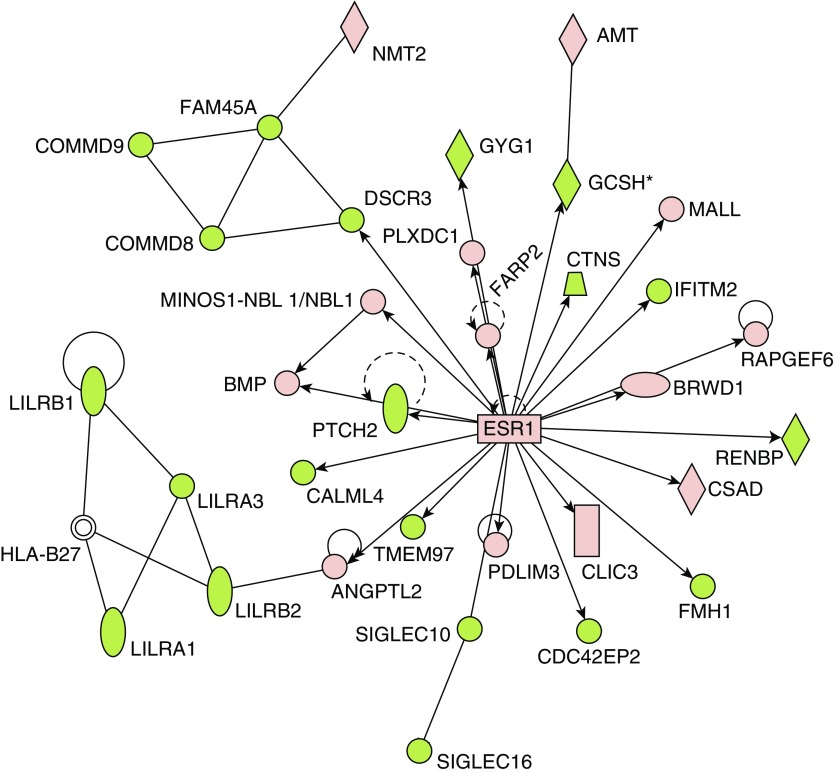
The PAH classifier was imported into IPA for pathway analysis (Table 3 and Table E4). The top four canonical pathways described by the PAH classifier included G protein–coupled receptors (Fisher exact P = 1 × 10−6) and three immunological responses, IL-8 signaling (P = 4.6 × 10−5) and two innate pathways (phagosome formation and role of pattern recognition receptors; P = 8.5 × 10−7 and 6.8 × 10−6). Using its extensive database of published data as well as natural language processing and curated text mining of the published literature, IPA can place an identified gene list into the context of potential upstream regulators. The predicted upstream regulators may themselves be DE, though this is not a criterion for inclusion. Estrogen receptor 1 (ESR1) was one specific example of a predicted upstream regulator that itself was found to be upregulated in PAH (Figure 2) and is thought to play a major role in PAH, vis a vis the female bias in PAH incidence as well as the better survival of females (26, 27). The female hormone, β-estradiol, was identified as a potential upstream regulator suggesting a potentially pleiotropic role.
Table 3.
Top Ranked Pathways and Upstream Regulators from Ingenuity Pathways Analysis
| P Value | z-Score | ||
|---|---|---|---|
| Ingenuity canonical pathways | |||
| Phagosome formation | 7.41E−07 | — | |
| G protein–coupled receptor signaling | 1.17E−06 | — | |
| IL-8 signaling | 4.57E−06 | −1.04 | |
| Role of pattern recognition receptors in recognition of bacteria and viruses | 6.76E−06 | −0.53 | |
| Complement system | 2.04E−05 | 1.34 | |
| Osteoarthritis pathway | 4.68E−05 | −0.23 | |
| cAMP-mediated signaling | 4.90E−05 | −0.63 | |
| tRNA splicing | 5.01E−05 | — | |
| Hepatic fibrosis/hepatic stellate cell activation | 6.03E−05 | — | |
| Upstream regulator | P value of overlap | Activation z-score | Predicted activation state |
| TNF | 1.24E−14 | −0.97 | — |
| CSF3 | 3.85E−12 | −2.42 | Inhibited |
| β-estradiol | 1.29E−11 | −0.85 | — |
| LPS | 4.00E−11 | −1.41 | — |
| Ig | 1.47E−10 | 3.64 | Activated |
| IL-13 | 2.19E−10 | 1.37 | — |
| TGFB1 | 7.02E−10 | 0.93 | — |
| TCL1A | 1.35E−09 | −2.00 | Inhibited |
| Tretinoin | 2.71E−09 | −1.72 | — |
| Dexamethasone | 1.21E−08 | −1.39 | — |
| TGM2 | 1.98E−08 | −4.03 | Inhibited |
| ESR1 | 2.70E−08 | 1.51 | — |
| GNA12 | 3.06E−08 | 1.14 | — |
| IL-10RA | 3.82E−08 | 2.47 | Activated |
| OSM | 7.60E−08 | 1.62 | — |
| IL-10 | 8.48E−08 | −2.39 | Inhibited |
Definition of abbreviations: CSF = colony-stimulating factor; ESR = estrogen receptor; GNA = G protein subunit α; IL-10RA = IL-10 receptor A; OSM = oncostatin M; TCL = T cell leukemia/lymphoma; TGF = transforming growth factor; TGM = transglutaminase.
Figure 2.
Estrogen receptor (ESR) 1 is a predicted upstream regulator in the PAH transcriptome. ESR1 was identified from the PHBI classifier using Ingenuity Pathways Analysis (IPA) upstream regulator algorithm (enrichment P = 2.70E−08). Upregulated (red) or downregulated (green) genes from the PHBI classifier are indicated in the network surrounding ESR1. ESR1 itself was slightly upregulated in PAH compared with FD (×1.12; q value = 6.16E−05).
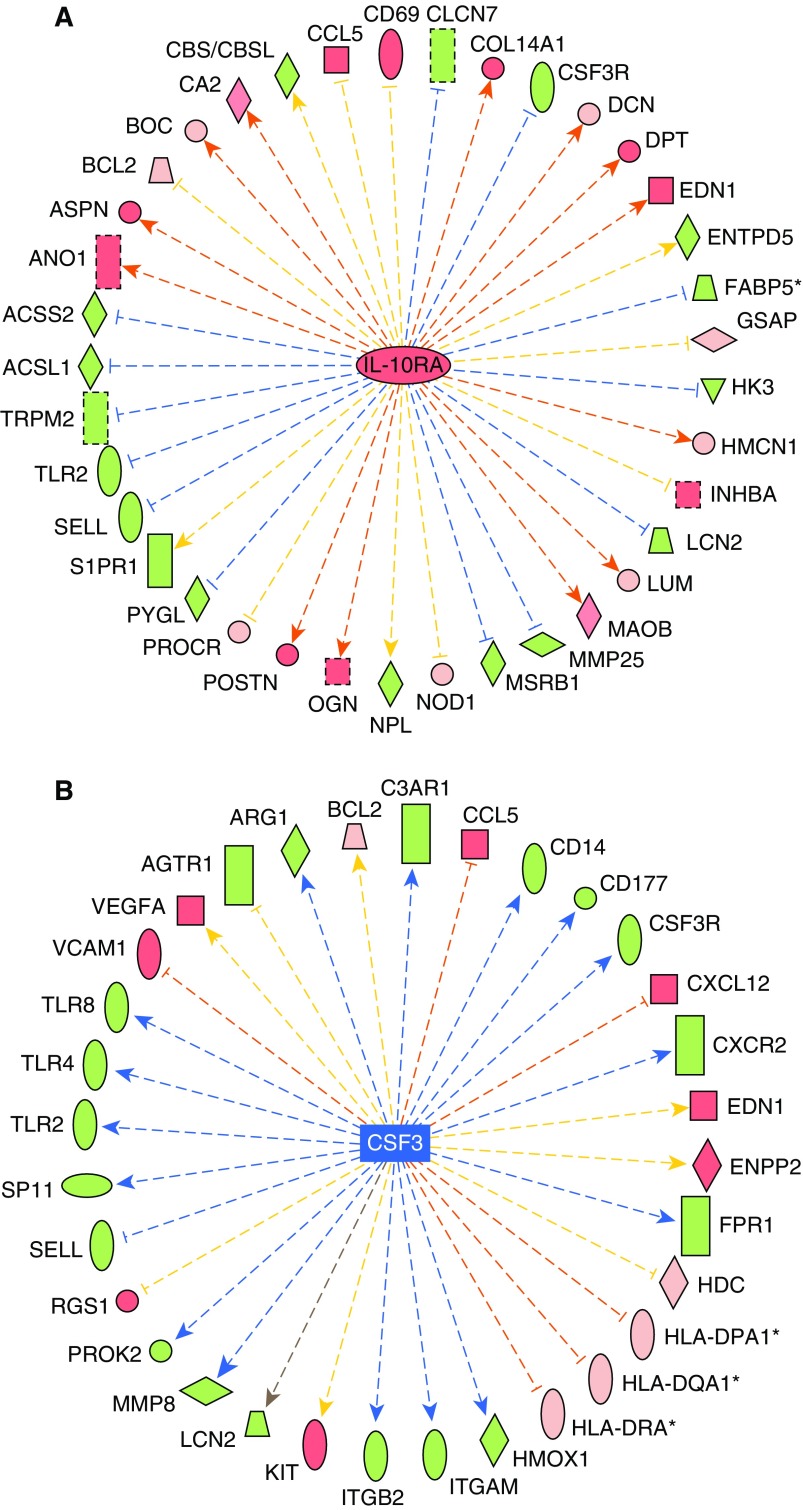
Most consistently, the PAH classifier upstream regulators were found to correspond to immunological roles, including known genes (see Reference 28; TNF, high-resolution Figure E6), colony-stimulating factor (CSF) 3, and IL-10 receptor A (IL-10RA; Figures 3A and 3B), as well as small molecules (β-estradiol, LPS, and dexamethasone). Statistical analysis identified TNF having the lowest P value (1.2 × 10−14), whereas CSF3, IL-10RA, and IL-10 displayed small P values (3.8 × 10−12, 3.8 × 10−8, and 8.5 × 10−8, respectively), and were scored as inhibited (CSF3 z-score = −2.4; IL-10 z-score = −2.4) or activated (IL-10RA z-score = +2.5) in IPA. Upstream pathway status is inferred in IPA by considering the observed effects in their downstream genes derived from the PAH classifier. Other top-ranked upstream regulators identified include additional cytokines (IL-13, IL-10, IL-5, IL-15, and IFN-γ), two sterol regulatory element binding transcription factors (SREBF1 and -2), and TGFB1.
Figure 3.
IL-10 receptor A (IL-10RA) and colony-stimulating factor (CSF) 3 are predicted upstream regulators driving pathway activation or inhibition. IL-10RA was predicted as an upstream regulator driving an activated pathway (A; enrichment P = 3.82E−08; activation z-score = +2.47), whereas CSF3 was predicted as an upstream regulator of an inhibited pathway (B; enrichment P = 3.85E−12; inhibition z-score = −2.42) by IPA. Genes are colored as in Figure 2, whereas the arrows denote predicted upregulation (orange) or downregulation (blue) by the central upstream regulator. z-Scores are generated by the Upstream Regulator algorithm by calculating the concordance of red genes (orange arrows) and green genes (blue arrows) in comparison to incorrect directionality of the relationships (yellow). The gray arrow indicates no known interaction between genes.
Correlation of Gene Expression to Clinical and Pathological Information
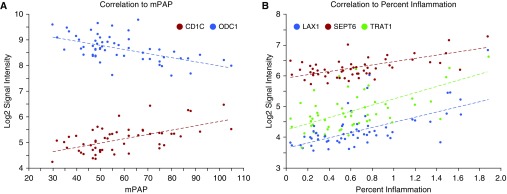
One unique resource from the PHBI program is the combination of lung transcriptome data with a detailed pathophysiological evaluation of lung tissue (18) from the same individuals. A total of 53 patients with PAH had both data types, and the basic clinical and pathological information was imported into Partek. Gene expression data was then correlated pairwise to each individual's clinical and pathological data (using a cut-off r > 0.50; Table E5). Several examples of clinical and pathologic variables are shown in Figure 4. CD1C and ornithine decarboxylase (ODC) 1 expression have significant correlations to mPAP (Figure 4A; CD1C correlation r = +0.58, P = 2.6 × 10−6, ODC1 r = −0.56, P = 6.5 × 10−6), whereas CD27, septin (SEPT) 6, and T cell receptor–associated adapter (TRAT) 1 were correlated to percent inflammation scoring (Figure 4B; CD27 correlation r = +0.73, P = 7.3 × 10−11, SEPT6 r = +0.63, P = 1.1 × 10−7, TRAT1 r = +0.60, P = 8.0 × 10−7). The correlation analysis to percent inflammation retrieved 164 Transcript IDs, which were imported into Enrichr for GO Biological Processes (GO-BP) categorization. Not surprisingly, this gene list highlights immune response GO-BP categories, but, interestingly, the top 10 categories (Table E6; adjusted P < 2.0 × 10−6) focused on T cell–related processes, supported by recent PAH and control lung immunophenotyping (29–31).
Figure 4.
Gene expression correlation to clinical and histological parameters. Mean pulmonary arterial pressure (mPAP; A) and the histological variable percent inflammation (B) were correlated to gene expression data on a per–patients with PAH basis from an extensively characterized subset of the Pulmonary Hypertension Breakthrough Initiative biobank. Representative gene expression patterns are shown: CD1C and ODC1 correlations to mPAP (CD1C r = +0.58, P = 2.56E−06; ODC1 r = −0.56, P = 6.51E−06) and SEPT6, TRAT1, and LAX1 correlations to percent inflammation (SEPT6 r = +0.63, P = 1.12E−07; TRAT1 r = +0.60, P = 8.00E−07; LAX1 r = +0.68, P = 6.21E−09). These genes include those that were significant differentially expressed genes between PAH and failed donors (CD1C q value = 0.00016; SEPT6 q value = 0.00059; TRAT1 q value = 0.0048; LAX1 q value = 0.03). A more complete listing can be found in Table E5.
The PHBI program offered sufficient numbers of samples from PAH clinical subgroups, APAH (n = 18) and IPAH (n = 31), to complete clinical subgroup DE and pathway analysis (Table 4). The HPAH subgroup had a small number of samples (n = 6) generating a short DE gene list insufficient for pathway analysis. PAH subgroup analysis showed a stronger reliance on BMP3, hepatocyte growth factor (HGF), and TGFB3 in APAH, C-X-C chemokine ligand (CXCL) 9 and early growth response (EGR) 1 in IPAH, and PDE8B, carbonic anhydrase (CA) 1, enhancer of polycomb 1 (EPC1), and Pim-2 proto-oncogene kinase (PIM) 2 in HPAH. Table 4 also includes PDE4B as APAH specific in contrast to PDE8B as IPAH specific, though its fold-change (FC) was only increased 1.26×. A stringent q value cut-off (q value < 0.001) was used for the APAH and IPAH comparisons in Enrichr for GO-PB gene set enrichment analysis, consistent with the PAH classifier cut-off (Table E7). The APAH-to-FD comparison found 82 APAH-specific DE genes, the IPAH-to-FD comparison found 308 IPAH-specific DE genes, and there were 119 DE genes in common. Table 5 highlights the top five GO-BP categories (Enrichr combined score > 20) overrepresented by each of these gene lists. IPAH-specific DE genes are strongly overrepresented in neutrophil and dendritic immune cell types. APAH-specific DE genes are strongly overrepresented in phospholipase C activation and extracellular matrix organization. The DE genes in common to both IPAH and APAH suggested a role for innate immunity, as a wide array of TLRs were represented.
Table 4.
PAH Subtype–Specific Differentially Regulated Genes
| PAH Subtype | Differentially Expressed Ratio |
|---|---|
| APAH specific (n = 49*) | APAH/FD ratio |
| BMP3 | 1.58 |
| HGF (hepatocyte growth factor) | 1.84 |
| TGFB3 | 1.95 |
| PDE4B | 1.26 |
| IPAH specific (n = 67*) | IPAH/FD ratio |
| CXCL9 | 1.98 |
| EGR1 (early growth response 1) | 1.95 |
| PDE8B | 1.53 |
| HPAH specific (n = 40*) | HPAH/FD ratio |
| CA1 | 2.22 |
| EPC1 (enhancer of polycomb homolog 1) | 3.05 |
| PIM2 (PIM2 proto-oncogene STK) | 2.83 |
Definition of abbreviations: BMP = bone morphogenetic protein; CA = carbonic anhydrase; PDE = phosphodiesterase; PIM = Pim-2 proto-oncogene; STK = serine/threonine kinase.
q value < 0.05 and ratio > 1.5×.
Table 5.
PAH Subtype Gene Ontology–Biological Process Categories
| Adjusted P Value | z-Score | Combined Score | |
|---|---|---|---|
| Term (APAH and IPAH overlap genes) | |||
| Toll-like receptor 4 signaling pathway (GO:0034142) | 3.50E−05 | −2.553 | 44.469 |
| Toll-like receptor 15 signaling pathway (GO:0035681) | 3.50E−05 | −2.496 | 38.157 |
| TIRAP-dependent toll-like receptor signaling pathway (GO:0035664) | 3.50E−05 | −2.492 | 38.090 |
| Toll-like receptor 6 signaling pathway (GO:0034150) | 3.50E−05 | −2.491 | 38.080 |
| Toll-like receptor 11 signaling pathway (GO:0034170) | 3.50E−05 | −2.490 | 38.066 |
| Term (IPAH-specific genes) | |||
| Neutrophil degranulation (GO:0043312) | 2.50E−06 | −5.241 | 111.566 |
| Positive regulation of chronic inflammatory response (GO:0002678) | 0.059 | −2.566 | 26.995 |
| Plasmacytoid dendritic cell chemotaxis (GO:0002410) | 0.066 | −3.290 | 30.770 |
| Myeloid dendritic cell chemotaxis (GO:0002408) | 0.066 | −3.256 | 30.450 |
| Dendritic cell chemotaxis (GO:0002407) | 0.066 | −3.244 | 30.339 |
| Term (APAH-specific genes) | |||
| Phospholipase C–activating dopamine receptor signaling pathway (GO:0060158) | 0.012 | −2.530 | 28.422 |
| Phospholipase C–activating G protein–coupled acetylcholine receptor signaling pathway (GO:0007207) | 0.012 | −2.408 | 28.055 |
| Phospholipase C–activating serotonin receptor signaling pathway (GO:0007208) | 0.022 | −2.377 | 22.887 |
| Phospholipase C–activating G protein–coupled receptor signaling pathway (GO:0007200) | 0.022 | −2.375 | 22.865 |
| Phospholipase C–activating angiotensin-activated signaling pathway (GO:0086097) | 0.022 | −2.413 | 22.538 |
Definition of abbreviations: GO = Gene Ontology; TIRAP = TIR domain containing adapter protein.
Discovery of Pathway Rewiring in PAH Using EDDY
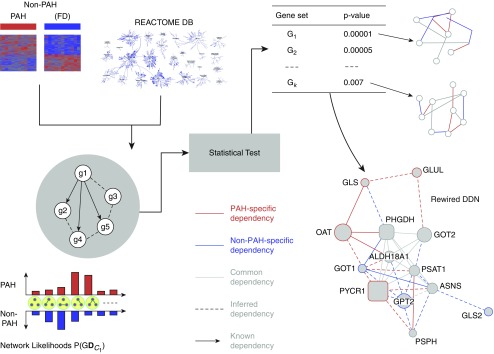
The gene expression data preprocessed, as described in the Methods section, were analyzed using the EDDY algorithm (see Reference 21, Figure 5, and Table 6) to identify known biological pathways enriched with differential dependencies between PAH and FD control lungs. Known biological pathways (472 in total) represented in the REACTOME (32) database were interrogated by EDDY in the context of the PAH and donor expression array datasets. Across the 16 REACTOME pathways of statistical significance, the level of rewiring was substantial, ranging from 46% to 100% of the entire pathways displaying some degree of alterations of gene interdependency (Figure 6A). Furthermore, in four of the identified differential dependency networks, greater than 40% of dependencies were previously unknown, but carried statistically derived linkages never before cataloged in prior studies.
Figure 5.
Knowledge-assisted Evaluation of Differential DependencY (EDDY) workflow. By analyzing transcriptomic gene expression levels, EDDY interrogates gene sets (in this case, pathways defined by the REACTOME database) for dependencies evident between any given two pathway genes and present in either or both conditions tested (PAH vs. FD control subjects). To do so, EDDY compares network likelihood distribution over multiple networks constructed for each condition via resampling, and the statistical significance of the divergence is estimated using a permutation test. Notably, using prior knowledge of REACTOME pathways, previously defined gene dependencies that result from EDDY analysis can be distinguished from statistically inferred dependencies that have never been reported previously. Differential dependency networks can then be constructed for gene sets that display statistically significant rewiring between patients with PAH versus FD control subjects. DB = database.
Table 6.
EDDY Analysis of the PAH Transcriptome
| Pathway | DB | No. Genes | P Value | JS | New Dependency | Rewiring | Essentiality Mediators | Specificity Mediators |
|---|---|---|---|---|---|---|---|---|
| Formation of ATP by chemiosmotic coupling | R | 16 | 0.0053 | 0.9675 | 0 | 0.62 | ATP5H ATP5A1 ATP5C1 ATP5B | — |
| Mitochondrial tRNA aminoacylation | R | 21 | 0.0084 | 0.9882 | 0.47 | 0.8 | VARS2 CARS2 HARS2 | — |
| TGF-β receptor signaling in EMT | R | 16 | 0.0104 | 0.9441 | 0 | 0.79 | ARHGEF18 CGN | — |
| Translocation of ZAP70 to immunological synapse | R | 14 | 0.0164 | 0.922 | 0 | 0.46 | HLA-DQA1 ZAP70 | CD3E CD4 |
| RNA Pol I transcription initiation | R | 25 | 0.0231 | 0.9705 | 0.08 | 0.72 | GTF2H3 CDK7 GTF2H4 | RRN3 |
| CDC6 association with the ORC origin complex | R | 11 | 0.0268 | 0.9471 | 0.14 | 0.75 | ORC1 | — |
| DCC-mediated attractive signaling | R | 13 | 0.0296 | 0.9604 | 0.04 | 0.67 | DOCK1 WASL NTN1 | — |
| Amino acid synthesis and interconversion transamination | R | 17 | 0.0337 | 0.9196 | 0.65 | 0.68 | PYCR1 PHGDH | — |
| Activation of IRF3/IRF7 mediated by TBK1 IKKϵ | R | 14 | 0.0341 | 0.9647 | 0.05 | 0.89 | TLR3 TICAM1 RPS27A | — |
| tRNA aminoacylation | R | 42 | 0.0342 | 0.9669 | 0.42 | 0.57 | DARS MARS VARS2 YARS YARS2 EPRS | CARS2 MARS2 TARS2 VARS2 SARS2 NARS2 FARS2 PARS2 IARS2 LARS2 WARS2 AARS2 QARS |
| Resolution of AP sites via the single nucleotide replacement | R | 12 | 0.0348 | 0.9819 | 0.43 | 1 | MPG LIG3 | — |
| mRNA decay by 3 to 5 exornase | R | 11 | 0.0382 | 0.938 | 0.06 | 0.71 | EXOSC6 | — |
| Regulation of hypoxia inducible factor HIF by oxygen | R | 25 | 0.0383 | 0.9864 | 0.14 | 0.78 | RBX1 CREBBP HIF3A | — |
| Phosphorylation of CD3 and TCRζ chains | R | 16 | 0.0421 | 0.8915 | 0 | 0.57 | LCK CD4 | CD3E |
| Activated TAK1 mediates p38 MAPK activation | R | 18 | 0.0482 | 0.9482 | 0.16 | 0.75 | RIPK2 MAP3K7 IRAK2 | — |
| mTORC1-mediated signaling | R | 11 | 0.0489 | 0.9736 | 0.12 | 0.81 | EIF4E |
Definition of abbreviations: AP = apurinic/apyrimidinic; DB = database; DCC = deleted in colorectal cancer; EDDY = Evaluation of Differential DependencY; EMT = epithelial-to-mesenchymal transition; HIF = hypoxia-inducible factor; IRF = IFN-regulatory factor; JS = Jensen-Shannon divergence estimate; MAPK = mitogen-activated protein kinase; mTORC = mTOR complex; ORC = origin complex; Pol = polymerase; TAK = mitogen-activated protein kinase kinase kinase 7 (MAP3K7/TAK1); TBK = TANK binding kinase; TCRζ = T-cell receptor; ZAP = Zeta chain of T cell receptor associated protein kinase 70.
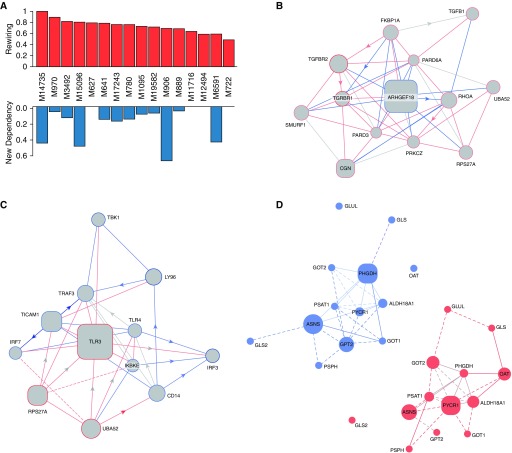
Figure 6.
Transcriptomic pathway rewiring in PAH. (A) Rewiring of pathways. Across the 16 REACTOME pathways of statistical significance, the level of rewiring was substantial, ranging from 46% to 100% of the entire pathways, displaying some degree of alterations of gene interdependency (top row). Notably, four of the identified differential dependency networks (DDNs) displayed rewiring only of known interactions previously cataloged by prior studies. Conversely, in four of the DDNs, over 40% of the dependencies involved previously unknown interactions that carried statistically derived linkages never before cataloged in prior studies (bottom row). (B–D) In each DDN, the gene interdependencies are color coded based on their presence in PAH lung (edges in red), in failed donor lung (edges in blue), or both (edges in gray). Known functional interactions are denoted by solid lines, whereas statistical dependencies, as revealed by EDDY analysis, are displayed by dashed lines. Size of nodes reflects the degree of betweenness centrality. Square nodes represent essentiality or specificity mediators of the DDN; circular nodes represent other genes in the pathway. (B) DDN for the pathway “transforming growth factor-β receptor signaling in epithelial-to-mesenchymal transition.” (C) DDN for the pathway “Activation of IFN regulatory factor (IRF)-3 and IRF7 mediated by TANK binding kinase 1 (TBK1) and inhibitor of nuclear factor kappa B kinase subunit epsilon (IKKE).” (D) Control-specific (left) and PAH-specific (right) gene dependency networks for the pathway “Amino Acid Synthesis and Interconversion Transamination.”
For some critical pathways known in PAH, such as TGF signaling, the breadth of rewiring relied solely on alterations of known gene interactions, which have never been specifically defined in PAH. Figure 6B provides such an example where the changing landscape of PAH-dependent gene interdependencies are defined for SMAD-specific E3 ubiquitin ligase (SMURF) 1, a gene that only recently has been linked to PAH (33). EDDY identified TLR3 and TLR4 as central to the REACTOME pathway describing IFN regulatory factor (IRF)-3 and -7 activation, potential targets for therapeutic development (Figure 6C). Furthermore, rewiring was specifically observed in other pathways more recently described to be important in PAH pathogenesis, such as glutamine metabolism and its pathogenic reliance on the enzyme glutaminase (GLS) 1 (see Reference 34, Figure 6D). As illustrated specifically in the gene dependency network of FD tissue, our findings identified an as-of-yet undescribed gene connection between GLS and an essentiality mediator gene, phosphoglycerate dehydrogenase (PHGDH). In the gene dependency network of PAH tissue; however, this link disappeared and was replaced by a known connection between GLS and ornithine aminotransferase (OAT) and a novel connection to glutamate-ammonia ligase (GLUL). These pathways results are publicly available at the online (http://www.sychanlab.pitt.edu/sysbio/eddy/phbi/).
Characterization of Sex-Specific Gene Expression in PAH
Because of the extreme sex imbalance of the PHBI patient collection, we corrected for sex along with batch in the initial analysis, making determination of PAH sex-based gene expression difficult. To circumvent this difficulty, a supervised analysis using sex-based DE in human lung tissue (n = 213 gene [35]) was completed. Interestingly, there were 71 DE genes that were different between patients with PAH and FD patients (P < 0.05; Table E7). As expected for our corrected dataset, none of these genes had residual significance when analyzed by sex. When this gene list was imported into Enrichr for GO-BP enrichment, the top-ranked category was “response to hydrogen peroxide,” whereas there was a notable pattern of negative regulation of WNT-related processes (Table 7 and Table E8).
Table 7.
Sex-biased Genes in PAH Gene Ontology–Biological Process Categories
| Term | Adjusted P Value | z-Score | Combined Score |
|---|---|---|---|
| Response to hydrogen peroxide (GO:0042542) | 0.002 | −2.671 | 36.669 |
| Positive regulation of heart induction by negative regulation of canonical Wnt signaling pathway (GO:0090082) | 0.024 | −3.212 | 29.290 |
| Negative regulation of canonical Wnt signaling pathway involved in heart development (GO:1905067) | 0.024 | −3.187 | 29.167 |
| Negative regulation of canonical Wnt signaling pathway involved in neural crest cell differentiation (GO:0072336) | 0.024 | −3.185 | 29.144 |
| Negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation (GO:0060829) | 0.024 | −3.196 | 29.139 |
| Positive regulation of spinal cord association neuron differentiation by negative regulation of canonical Wnt signaling pathway (GO:1902844) | 0.024 | −3.272 | 28.999 |
| Negative regulation of canonical Wnt signaling pathway involved in osteoblast differentiation (GO:1905240) | 0.024 | −3.407 | 28.882 |
| Negative regulation of Wnt signaling pathway by Wnt receptor internalization (GO:0038012) | 0.024 | −2.606 | 22.318 |
| Negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification (GO:2000054) | 0.024 | −2.632 | 22.309 |
| Negative regulation of Wnt-mediated midbrain dopaminergic neuron differentiation (GO:1905425) | 0.024 | −2.601 | 22.273 |
| Negative regulation of Wnt signaling pathway involved in digestive tract morphogenesis (GO:2000057) | 0.024 | −2.598 | 22.252 |
| Negative regulation of Wnt signaling pathway involved in heart development (GO:0003308) | 0.024 | −2.559 | 21.688 |
Discussion
Until now, the comprehensive gene expression landscape across a large number of human group 1 PAH lung tissues had not been reported, limited greatly by the small number of tissues accessible for this type of analysis. Hoffmann and colleagues (36) provided an overview of microarray studies in pulmonary hypertension using a variety of source tissues, where other previously reported lung-based expression studies included patients with pulmonary fibrosis. By a number of quality control measures and independent validation, we have shown that the multicenter PHBI gene expression dataset is robust at discriminating the transcriptomic landscape of lung PAH tissue compared with FD tissue. Specific genes and pathways previously implicated in the PAH disease process were found altered in the transcriptome analysis, with the effects in the direction expected from extensive literature findings and approved therapeutics (1, 17). Of course, mRNA levels are not perfectly reflective of protein levels. The current best estimate for a variety of human tissues shows a correlation of r = +0.40 for mRNA and protein levels (37). This correlation can be improved to r = +0.88 by normalizing across the 12 different tissues they analyzed. We compared the PHBI transcriptomic dataset to a published PAH versus control lung homogenate proteomic study (38). In line with the expectation from other tissues, our data gave 38% agreement in direction of change in expression with the proteomic data.
The potential power of pathway analysis is looking at groups of biologically connected genes without necessarily relying on single gene measurements. Numerous applications of validated computations algorithms applied to this dataset revealed both expected and surprising gene and pathway connections to PAH, including immune, sex, metabolic, and BMP signaling. Thus, we envision these data and analyses as a unique and valuable resource for traditional molecular discovery and for reference and analytic overlay as further -omics analyses emerge in this historically neglected condition (39).
Due to the size of the PHBI dataset, we were also able to demonstrate, for the first time, APAH- and IPAH-specific gene expression profiles. This analysis pointed toward the importance of phospholipase C activation of a variety of signaling pathways in APAH versus a neutrophil and dendritic immune cellular response in IPAH. Further subdivision with clinical subtypes was not possible, due to the limited number of treatment options and similar comorbidities that existed within this end-stage patient population. In addition, for the first time, we showed lung gene expression correlation to clinical parameters (mPAP) and semiquantitative pathology analysis (percent inflammation) of the PAH lungs from the same patients (18). Beyond the scope of this paper, the data could potentially be used to develop a blood-based PAH biomarker to more easily follow a patient’s therapeutic response and/or disease progression.
The various approaches to pathway analysis highlight different aspects of the PAH lung transcriptome. Standard pathway enrichment approaches point to an under-appreciated role of the immune response, specifically the innate, TLR-dependent pathway (Enrichr and IPA). The DE genes downstream of the ESR1 suggest a bigger role in estrogen signaling (IPA), which is well supported in the rodent models of PAH (40). This may help explain some of the sex-bias paradox of greater female incidence in PAH and/or better long-term survival of female patients. Upstream activator predictions point to a global role for TNF, as well as predictions of increased activation state of IL-10RA and decreased activation of CSF3 and IL-10 (IPA).
Finally, the application of EDDY, a novel pathway analysis tool, to analyze topological characteristics of gene differential dependency networks previously has been shown to be capable of defining biological network dependencies not evident with similar analytic tools. It has been successfully applied to glioblastoma (21) and adrenocortical carcinoma (41), but never cardiopulmonary vascular diseases, such as PAH. Here, EDDY analysis not only revealed a number of notable molecular insights, but, in so doing, further supported the use of advanced statistical approaches of big data to uncover hidden pathogenic mechanisms in PAH, even in the absence of larger datasets available in cancer and other, more prevalent conditions. Of the 16 REACTOME pathways with statistically significant rewiring, some have previously been defined as critical to PAH pathogenesis, thus offering a reassuring level of credibility for EDDY-based statistical analyses. Among these included TGF-β signaling pathways, particularly as they relate to BMPR2 signaling (42), hypoxic signaling (24), mechanistic target of rapamycin kinase (mTOR) transcriptional activity (43), and amino acid synthesis mediated by glutamine metabolism (34, 44), to name a few. However, beyond this already-known biology, EDDY-based analysis delineated a specific roadmap of rewiring events that have not been fully cataloged for PAH. As -omics data continue to expand in PAH beyond genomics and traditional transcriptomics, we expect that algorithms such as EDDY may prove to be crucial in charting pathogenic relationships among multiple molecular landscapes in attempts to “reverse engineer” the molecular steps that sit at the origin of this enigmatic disease.
One caveat of our study that is not readily addressed is the use of end-stage PAH disease lung and whether this confounds interpretation of the disease’s pathophysiological development. In addition, perhaps the transcriptomic changes are a result of long-term patient use of many different therapeutic interventions used to maintain their quality of life up to time of transplantation. This is unlikely to bias the overall analysis, as, from within the PAH classifier, one can find many specific genes reported in literature results from patient lung tissue, PAH primary cells, choice of patient therapeutics, and from the mouse and rat animal model studies.
In conclusion, this study has improved the understanding of the PAH lung transcriptome, in relation to control lung tissue, highlighting new potential upstream regulators and significant pathway rewiring in the disease state. We anticipate this dataset to be useful in increasing the understanding of the PAH lung transcriptome and its associated changes in transcriptional regulation and downstream signaling pathways. Our dataset provides several transcription-based validations of currently approved therapeutic options, as well as suggesting a variety of potential targets to be investigated. From a basic science perspective, we envision that this hypothesis-generating resource will broaden the approaches used in both cell culture and animal model systems for PAH.
Supplementary Material
Footnotes
Supported by Chancellor’s Research Initiative funding for the Center for Computational Systems Biology at the Prairie View A&M University (S.K.) and a grant from the NVIDIA Foundation Compute the Cure initiative and the Silicon Valley Community Foundation (G.S. and S.K.), National Institutes of Health grants R01 HL124021, HL 122596, HL 138437, UH2 TR002073, and American Heart Association Established Investigator Award 18EIA33900027 (S.Y.C.); data and tissue samples were provided by the Pulmonary Hypertension Breakthrough Initiative; funding for the Pulmonary Hypertension Breakthrough Initiative is provided under a National Heart, Lung, and Blood Institute R24 grant R24HL123767 (M.W.G. [Principal Investigator]), and by the Cardiovascular Medical Research and Education Fund.
Author Contributions: R.S.S., B.B.G., S.K., S.Y.C., and M.W.G. conceived the project and designed the experiments; R.S.S., Q.M.B., G.S., A.H., A.R.C., B.B.G., S.K., E.A.M., and R.M.T. conducted experiments or helped with analysis; R.S.S., S.K., S.Y.C., and M.W.G. wrote the manuscript; all authors read and approved the manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0368OC on December 18, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Thenappan T, Ormiston ML, Ryan JJ, Archer SL. Pulmonary arterial hypertension: pathogenesis and clinical management. BMJ. 2018;360:j5492. doi: 10.1136/bmj.j5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machado RD, Southgate L, Eichstaedt CA, Aldred MA, Austin ED, Best DH, et al. Pulmonary arterial hypertension: a current perspective on established and emerging molecular genetic defects. Hum Mutat. 2015;36:1113–1127. doi: 10.1002/humu.22904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gräf S, Haimel M, Bleda M, Hadinnapola C, Southgate L, Li W, et al. Identification of rare sequence variation underlying heritable pulmonary arterial hypertension. Nat Commun. 2018;9:1416. doi: 10.1038/s41467-018-03672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaine S, McLaughlin V. Pulmonary arterial hypertension: tailoring treatment to risk in the current era. Eur Respir Rev. 2017;26:pii:170095. doi: 10.1183/16000617.0095-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghataorhe P, Rhodes CJ, Harbaum L, Attard M, Wharton J, Wilkins MR. Pulmonary arterial hypertension—progress in understanding the disease and prioritizing strategies for drug development. J Intern Med. 2017;282:129–141. doi: 10.1111/joim.12623. [DOI] [PubMed] [Google Scholar]
- 6.Batton KA, Austin CO, Bruno KA, Burger CD, Shapiro BP, Fairweather D. Sex differences in pulmonary arterial hypertension: role of infection and autoimmunity in the pathogenesis of disease. Biol Sex Differ. 2018;9:15. doi: 10.1186/s13293-018-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs W, van de Veerdonk MC, Trip P, de Man F, Heymans MW, Marcus JT, et al. The right ventricle explains sex differences in survival in idiopathic pulmonary arterial hypertension. Chest. 2014;145:1230–1236. doi: 10.1378/chest.13-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemnes AR, Humbert M. Pathobiology of pulmonary arterial hypertension: understanding the roads less travelled. Eur Respir Rev. 2017;26:pii:170093. doi: 10.1183/16000617.0093-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuder RM, Archer SL, Dorfmüller P, Erzurum SC, Guignabert C, Michelakis E, et al. Relevant issues in the pathology and pathobiology of pulmonary hypertension. J Am Coll Cardiol. 2013;62(Suppl)(25):D4–D12. doi: 10.1016/j.jacc.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burks M, Stickel S, Galiè N. Pulmonary arterial hypertension: combination therapy in practice. Am J Cardiovasc Drugs. 2018;18:249–257. doi: 10.1007/s40256-018-0272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertero T, Lu Y, Annis S, Hale A, Bhat B, Saggar R, et al. Systems-level regulation of microRNA networks by miR-130/301 promotes pulmonary hypertension. J Clin Invest. 2014;124:3514–3528. doi: 10.1172/JCI74773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu M, Shao NY, Sa S, Li D, Termglinchan V, Ameen M, et al. Patient-specific iPSC-derived endothelial cells uncover pathways that protect against pulmonary hypertension in BMPR2 mutation carriers. Cell Stem Cell. 2017;20:490–504, e5. doi: 10.1016/j.stem.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemnes AR, Zhao M, West J, Newman JH, Rich S, Archer SL, et al. Critical genomic networks and vasoreactive variants in idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med. 2016;194:464–475. doi: 10.1164/rccm.201508-1678OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao M, Austin ED, Hemnes AR, Loyd JE, Zhao Z. An evidence-based knowledgebase of pulmonary arterial hypertension to identify genes and pathways relevant to pathogenesis. Mol Biosyst. 2014;10:732–740. doi: 10.1039/c3mb70496c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kan M, Shumyatcher M, Himes BE. Using omics approaches to understand pulmonary diseases. Respir Res. 2017;18:149. doi: 10.1186/s12931-017-0631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lythgoe MP, Rhodes CJ, Ghataorhe P, Attard M, Wharton J, Wilkins MR. Why drugs fail in clinical trials in pulmonary arterial hypertension, and strategies to succeed in the future. Pharmacol Ther. 2016;164:195–203. doi: 10.1016/j.pharmthera.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Savoia C, Volpe M, Grassi G, Borghi C, Agabiti Rosei E, Touyz RM. Personalized medicine—a modern approach for the diagnosis and management of hypertension. Clin Sci (Lond) 2017;131:2671–2685. doi: 10.1042/CS20160407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stacher E, Graham BB, Hunt JM, Gandjeva A, Groshong SD, McLaughlin VV, et al. Modern age pathology of pulmonary arterial hypertension. Am J Respir Crit Care Med. 2012;186:261–272. doi: 10.1164/rccm.201201-0164OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung S, Kim S. EDDY: a novel statistical gene set test method to detect differential genetic dependencies. Nucleic Acids Res. 2014;42:e60. doi: 10.1093/nar/gku099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speyer G, Kiefer J, Dhruv H, Berens M, Kim S. Knowledge-assisted approach to identify pathways with differential dependencies. Pac Symp Biocomput. 2016;21:33–44. [PMC free article] [PubMed] [Google Scholar]
- 23.Rajkumar R, Konishi K, Richards TJ, Ishizawar DC, Wiechert AC, Kaminski N, et al. Genomewide RNA expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. Am J Physiol Heart Circ Physiol. 2010;298:H1235–H1248. doi: 10.1152/ajpheart.00254.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kylhammar D, Rådegran G. The principal pathways involved in the in vivo modulation of hypoxic pulmonary vasoconstriction, pulmonary arterial remodelling and pulmonary hypertension. Acta Physiol (Oxf) 2017;219:728–756. doi: 10.1111/apha.12749. [DOI] [PubMed] [Google Scholar]
- 25.Sitbon O, Humbert M, Jaïs X, Ioos V, Hamid AM, Provencher S, et al. Long-term response to calcium channel blockers in idiopathic pulmonary arterial hypertension. Circulation. 2005;111:3105–3111. doi: 10.1161/CIRCULATIONAHA.104.488486. [DOI] [PubMed] [Google Scholar]
- 26.Frump AL, Selej M, Wood JA, Albrecht M, Yakubov B, Petrache I, et al. Hypoxia upregulates estrogen receptor β in pulmonary artery endothelial cells in a HIF-1α–dependent manner. Am J Respir Cell Mol Biol. 2018;59:114–126. doi: 10.1165/rcmb.2017-0167OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahm T, Frump AL, Albrecht ME, Fisher AJ, Cook TG, Jones TJ, et al. 17β-estradiol mediates superior adaptation of right ventricular function to acute strenuous exercise in female rats with severe pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2016;311:L375–L388. doi: 10.1152/ajplung.00132.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurst LA, Dunmore BJ, Long L, Crosby A, Al-Lamki R, Deighton J, et al. TNFα drives pulmonary arterial hypertension by suppressing the BMP type-II receptor and altering NOTCH signalling. Nat Commun. 2017;8:14079. doi: 10.1038/ncomms14079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen R, Yan J, Liu P, Wang Z, Wang C, Zhong W, et al. The role of nuclear factor of activated T cells in pulmonary arterial hypertension. Cell Cycle. 2017;16:508–514. doi: 10.1080/15384101.2017.1281485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh LM, Jandl K, Grünig G, Foris V, Bashir M, Ghanim B, et al. The inflammatory cell landscape in the lungs of patients with idiopathic pulmonary arterial hypertension. Eur Respir J. 2018;51:pii:1701214. doi: 10.1183/13993003.01214-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamosiuniene R, Manouvakhova O, Mesange P, Saito T, Qian J, Sanyal M, et al. Dominant role for regulatory T cells in protecting females against pulmonary hypertension. Circ Res. 2018;122:1689–1702. doi: 10.1161/CIRCRESAHA.117.312058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothman AM, Arnold ND, Pickworth JA, Iremonger J, Ciuclan L, Allen RM, et al. MicroRNA-140-5p and SMURF1 regulate pulmonary arterial hypertension. J Clin Invest. 2016;126:2495–2508. doi: 10.1172/JCI83361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertero T, Oldham WM, Cottrill KA, Pisano S, Vanderpool RR, Yu Q, et al. Vascular stiffness mechanoactivates YAP/TAZ-dependent glutaminolysis to drive pulmonary hypertension. J Clin Invest. 2016;126:3313–3335. doi: 10.1172/JCI86387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dugo M, Cotroneo CE, Lavoie-Charland E, Incarbone M, Santambrogio L, Rosso L, et al. Human lung tissue transcriptome: influence of sex and age. PLoS One. 2016;11:e0167460. doi: 10.1371/journal.pone.0167460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann J, Wilhelm J, Olschewski A, Kwapiszewska G. Microarray analysis in pulmonary hypertension. Eur Respir J. 2016;48:229–241. doi: 10.1183/13993003.02030-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, et al. Mass-spectrometry–based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 38.Abdul-Salam VB, Wharton J, Cupitt J, Berryman M, Edwards RJ, Wilkins MR. Proteomic analysis of lung tissues from patients with pulmonary arterial hypertension. Circulation. 2010;122:2058–2067. doi: 10.1161/CIRCULATIONAHA.110.972745. [DOI] [PubMed] [Google Scholar]
- 39.Hemnes AR, Beck GJ, Newman JH, Abidov A, Aldred MA, Barnard J, et al. PVDOMICS Study Group. PVDOMICS: a multi-center study to improve understanding of pulmonary vascular disease through phenomics. Circ Res. 2017;121:1136–1139. doi: 10.1161/CIRCRESAHA.117.311737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frump AL, Albrecht ME, McClintick JN, Lahm T. Estrogen receptor–dependent attenuation of hypoxia-induced changes in the lung genome of pulmonary hypertension rats. Pulm Circ. 2017;7:232–243. doi: 10.1177/2045893217702055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng S, Cherniack AD, Dewal N, Moffitt RA, Danilova L, Murray BA, et al. Cancer Genome Atlas Research Network. Comprehensive pan-genomic characterization of adrenocortical carcinoma. Cancer Cell. 2016;29:723–736. doi: 10.1016/j.ccell.2016.04.002. [Published erratum appears in Cancer Cell 30:363.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Austin ED, Loyd JE. The genetics of pulmonary arterial hypertension. Circ Res. 2014;115:189–202. doi: 10.1161/CIRCRESAHA.115.303404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goncharova EA. mTOR and vascular remodeling in lung diseases: current challenges and therapeutic prospects. FASEB J. 2013;27:1796–1807. doi: 10.1096/fj.12-222224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dumas SJ, Bru-Mercier G, Courboulin A, Quatredeniers M, Rücker-Martin C, Antigny F, et al. NMDA-type glutamate receptor activation promotes vascular remodeling and pulmonary arterial hypertension. Circulation. 2018;137:2371–2389. doi: 10.1161/CIRCULATIONAHA.117.029930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.