Abstract
Mutations in oncogenes and tumor suppressor genes engender unique metabolic phenotypes crucial to the survival of tumor cells. Epidermal growth factor receptor (EGFR) signaling has been linked to the rewiring of tumor metabolism in non-small cell lung cancer (NSCLC). We have integrated the use of a functional genomics screen and metabolomics to identify metabolic vulnerabilities induced by EGFR inhibition. These studies reveal that following EGFR inhibition, EGFR-driven NSCLC cells become dependent on the urea cycle and in particular, the urea cycle enzyme CPS1. Combining knockdown of CPS1 with EGFR inhibition further reduces cell proliferation and impedes cell cycle progression. Profiling of the metabolome demonstrates that suppression of CPS1 potentiates the effects of EGFR inhibition on central carbon metabolism, pyrimidine biosynthesis, and arginine metabolism, coinciding with reduced glycolysis and mitochondrial respiration. We show that EGFR inhibition and CPS1 knockdown lead to a decrease in arginine levels and pyrimidine derivatives, and the addition of exogenous pyrimidines partially rescues the impairment in cell growth. Finally, we show that high expression of CPS1 in lung adenocarcinomas correlated with worse patient prognosis in publically available databases. These data collectively reveal that NSCLC cells have a greater dependency on the urea cycle to sustain central carbon metabolism, pyrimidine biosynthesis, and arginine metabolism to meet cellular energetics upon inhibition of EGFR.
Keywords: Urea cycle, CPS1, erlotinib, EGFR, NSCLC
Introduction
Lung cancer remains the leading cause of cancer-related deaths worldwide. In the United States, over 230,000 new cases are expected to be diagnosed in 20181. Lung cancer is often diagnosed at late stages contributing to a dismal 5-year relative survival rate of 18%. Approximately 84% of lung cancers are NSCLC. The most common histological type of NSCLC is adenocarcinoma which has been associated with overexpression and activating mutations in EGFR2,3. The identification of molecular drivers and the introduction of targeted therapies including the use of EGFR tyrosine kinase inhibitors (TKIs), such as erlotinib, have significantly improved the overall survival rate and response rates compared to standard chemotherapy for patients with EGFR mutant lung cancer. While advanced NSCLC patients with EGFR mutant tumors initially respond to TKIs, after 10–14 months almost all patients start to develop resistance to the drug and eventually relapse4,5. Multiple mechanisms of resistance to EGFR TKIs have been identified including secondary mutation in EGFR (T790M)6, activation of compensatory signaling (cMET, AXL, FGFR)7–9 and transition to a mesenchymal phenotype10. Moreover, mechanisms of intrinsic resistance including the crosstalk between EGFR and Wnt11, expression of receptor tyrosine kinase ligands12, and additional mechanisms described to hinder the effectiveness of EGFR inhibitors13,14. Identifying other potential mechanisms of adaptation or intrinsic resistance following EGFR inhibition may reveal strategies to further reduce tumor burden, limiting the fraction of NSCLC cells that may persists in the presence of EGFR inhibitors.
Various studies have shown that activation and/or mutations in oncogenes can influence the metabolic reprogramming of tumor cells15,16. EGFR enhances glycolysis through PI3K/AKT activation and the promotion of glycolytic gene expression mediated by c-Myc17,18. In addition to glycolysis, EGFR signaling has also been reported to be specifically involved in regulating the pentose phosphate pathway, glutaminolysis and pyrimidine biosynthesis in EGFR mutant lung cancer cells19. While EGFR signaling has been associated with the rewiring of tumor metabolism, the metabolic dependencies that arise upon EGFR inhibition are largely unknown.
The urea cycle is an essential pathway involved in the conversion of toxic ammonia generated from amino acid breakdown and glutaminolysis activity20,21, into the less toxic urea in mammals. Carbamoyl phosphate synthetase 1 (CPS1) is a mitochondrial rate-limiting enzyme in the urea cycle which converts bicarbonate and ammonia into carbamoyl phosphate, in turn depleting the amount of ammonia in the cell. Carbamoyl phosphate plays a crucial role in arginine metabolism and pyrimidine biosynthesis, serving as a precursor for both processes22. CPS1 has been shown to play a role in metabolism and cell growth of LKB1-inactivated lung adenocarcinomas and CPS1 expression in lung adenocarcinoma tumors has been associated with worse overall survival23. Mechanistically, CPS1 has been shown to sustain pyrimidine levels and DNA synthesis in KRAS/LKB1 lung cancer cells24. Moreover, overexpression of CPS1 in colorectal cancer patients correlated with shorter disease specific survival, shorter metastatic free survival and poor therapeutic responses25. In contrast to CPS1, another urea cycle enzyme, argininosuccinate synthase (ASS1) has been reported to be repressed in several types of cancers including osteosarcomas, melanoma, and hepatocellular carcinomas26. Additionally, decreased ASS1 activity promoted cancer cell growth by increasing pyrimidine biosynthesis27. To identify metabolic phenotypes underlying the inability of EGFR inhibitors to completely eliminate NSCLC cells, we performed a metabolic shRNA screen to identify metabolic genes whose inhibition could further sensitize EGFR mutant NSCLC cells to EGFR inhibitors. In this study, we identified the urea cycle as one of the most critical metabolic pathways in the context of EGFR inhibition in EGFR-driven NSCLC and potentially a novel metabolic vulnerability in NSCLC.
Materials and Methods
Cell culture and generation of knockdown cell lines
NSCLC cells lines H322C and H1650 and 293FT were acquired from the University of Colorado Tissue Culture Shared Resource. PC9 were provided in 2006 and HCC4006 cells were provided in 2013 by Drs. John Minna and Adi Gazdar (University of Texas Southwestern Medical School, Dallas, USA). PC9 T790M and H1975 were provided by Dr. Lynn Heasley (University of Colorado, Denver, USA) in 2013. H3122 was provided by Dr. Robert Doebele (University of Colorado, Denver, USA) in 2017. All cell lines were authenticated within six months prior to experimental use by short tandem repeat (STR) analysis by the Barbara Davis Molecular Biology Service Center. All NSCLC cell lines were cultured in RPMI supplemented with 10% FBS (Sigma, St, Louis, USA) and 1% antibiotic-antimycotic (anti-anti) from Gibco at 37°C in 5% CO2 incubator. 293FT cells were cultured in DMEM supplemented with 10% FBS and 1% anti-anti. Lentiviruses were generated using pLKO.1 or pLKO.2 vectors (Sigma-Aldrich, obtained through the University of Colorado Functional and Genomics Shared Resource, USA, Supplementary Table S2) and were used to transduce cells as previously described11. Cells were selected in 1 µg/mL puromycin. All cells were routinely monitored for mycoplasma contamination using e-Myco plus Mycoplasma PCR detection kit (iNtRON biotechnology) according to the manufacturer’s directions.
Metabolic shRNA functional screening
2 × 106 cells from each cell line were transduced with the metabolic shRNA library (obtained from the University of Colorado Functional and Genomics Core facility as requested). The metabolic shRNA library comprised of 535 shRNAs (~5 shRNA/gene), targeting 104 genes expressing metabolic genes. Forty-eight hours after transduction, cells were selected in 1 µg/mL puromycin for 7 days-10 days until enough cells were obtained (10–15 million) for plating. Cells (2×106) were treated in replicates of 5 with vehicle (DMSO) or increasing doses of erlotinib (10nM, 30nM, 60 nM, 90nM, 270 nM) for 72h followed by culture for 72h without drugs. Genomic DNA was isolated using the Qiagen DNeasy blood and tissue kit as instructed. The shRNA was amplified using customized ultramer oligos with addition of Illumina-specific adapter sequences as previously described47. The samples were sequenced on an Illumina HiSeq Analyzer and shRNA sequences were identified and normalized counts subjected to linear regression analysis.
Analysis of shRNA sequencing data
shRNA in all sequencing samples were counted by querying for a direct sequence match to the query library shRNA. This approach allows avoiding errors and biases introduced by other matching algorithms, as the sequencing error rates are low and should be randomly distributed among shRNAs so that all shRNAs have similar enough chances of being unaccounted due to 1 or more mismatches. All samples were normalized to account for the differences in total read counts using the equation (A/T)*C, where A is the average total sample read count across samples, T is the total read count in the analyzed sample, and C is the read count for particular shRNA in the analyzed sample.
Statistical significance of the effect of an experimental condition was tested by linear regression of shRNA counts to the condition intensity (drug dose in a multi-dose drug application setting). Linear regression was chosen in order to disregard dose-specific non-linear effects of the drug on particular genes, addressing whether the presence of a particular drug creates directional selection on a particular gene and what is the directionality of selection (a gene’s net enrichment or depletion in a cell culture as a result of drug application). Combined with linear regression, multi-dose testing results in better statistical resolution compared to single-dose treatment vs control tests per the same total number of samples. We used the standard implementation of linear regression in Matlab (MathWorks Inc, Massachusetts).
Pharmacological agents
Osimertinib (AZD9291) was provided by Dr. Lynn Heasley. Erlotinib was purchased from Tocris pharmaceuticals. Crizotinib was provided by Dr. Robert Doebele. Each of these compounds was resuspended in DMSO at 10 mM, and diluted in culture media for experimental studies. Cisplatin and doxorubicin were obtained from Sigma-Aldrich.
Metabolomics Analysis
Samples were prepared for UHPLC-MS metabolomics in the following manner. Cells pellets were extracted in ice-cold lysis/extraction buffer (methanol:acetonitrile:water 5:3:2) at 2–3×106 cells/mL lysis buffer. Samples were agitated (30 min, 4°C) followed by centrifuged (12,000 g, 10 min, 4 °C). Protein pellets were discarded, and supernatants were stored at −80°C prior to metabolomic analyses. Cell extracts were injected (10 or 15 µL) into a Thermo Vanquish UHPLC system (San Jose, CA, USA) coupled to a Thermo Q Exactive mass spectrometer (Bremen, Germany). Metabolites were separated on a Kinetex C18 column (150 × 2.1 mm, 1.7 µm – Phenomenex, Torrance, CA, USA ) at 25°C using a three minute isocratic method at 250 µl/min and 95% A (A: water/0.1% formic acid; B: acetonitrile/0.1% formic acid) for positive ion mode. The negative ion mode used a three-minute isocratic method at 250 µl/min and 100% A (A: 95/5 water/acetonitrile 1mM NH4OAc). Technical mixes were generated by pooling aliquots of cell extracts, and were run every 16–20 analytical runs, to control for technical variability, as judged by coefficients of variation (CV). CV were determined by calculating the ratios of standard deviations divided by mean measurements for compounds of interest across all technical mix runs. Data files were converted to .mzXML format followed by analysis in Maven (Princeton, NJ, USA) and metabolites were identified and validated as previously described48,49. Integrated peak areas were exported into Excel (Microsoft) and elaborated for statistical analysis (t test, ANOVA), hierarchical clustering analysis (HCA) and pathway analysis through MetaboAnalyst (Statistics Software) and GENE E (Broad Institute).
Public Database analysis
Oncomine™ (Compendia Bioscience, Ann Arbor, MI) was used for analysis and visualization using the Garber lung and Staunton cancer cell line databases. The Bild tumor lung microarray data was generated from the R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl). Kaplan-Meier Plotter was used to compare CPS1 expression between different patient subgroups resulting in survival curves, hazard ratios, and p values (obtained via the log-rank test). CPS1 probe set 217564_s_at was used for analysis.
Statistical analysis
Standard deviation is shown for all error and is based on biological replicates. Unless otherwise indicated, one-way ANOVA with Tukey post-test was used to compute p-values using GraphPad Prism. For Seahorse assay analysis, LOESS regression was used to fit data, the effect of glycolysis and oxygen consumption over time was assessed by the area under the curve (AUC). A two sample mean difference Welch’s t-test for unknown and unequal variances was used to evaluate the differences between two AUCs.
See Supplemental Methods for additional procedures.
Results
Metabolic shRNA screen identifies inhibition of urea cycle enzymes as synthetic lethal with EGFR inhibition.
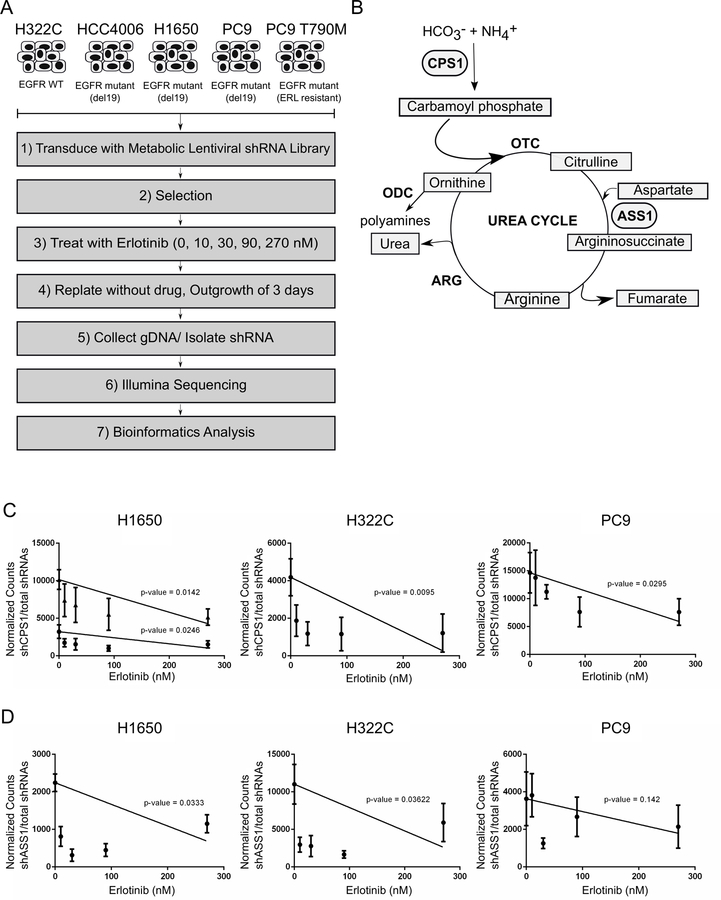
To uncover potential metabolic vulnerabilities of NSCLC in the context of EGFR inhibition, we designed a synthetic lethal screen containing shRNAs targeting rate-limiting metabolic enzymes encompassing all metabolic pathways in addition to key metabolic enzymes previously identified as being implicated in cancer (Figure 1A). We utilized EGFR mutant cell lines (PC9, HCC4006, H1650), an EGFR wild type cell line responsive to EGFR inhibition at higher doses (H322C), and an EGFR mutant cell line that is resistant to erlotinib treatment (PC9 T790M) to generate NSCLC cell lines expressing the library of lentiviral-encoded shRNAs targeting metabolic enzymes. The cells were treated with increasing concentrations of erlotinib for 72 hours, followed by 72 hours cultured without the presence of drug to allow cells to rebound. We then isolated genomic DNA, shRNA sequences were amplified, identified and analyzed using linear regression analysis as described in Materials and Methods.
Figure 1: Metabolic screen uncovers CPS1 as a synthetic lethal hit with EGFR inhibition in NSCLC cells.
A. Experimental schematic of metabolic shRNA screen and data analyses. B. Diagram of urea cycle with synthetic lethal hits highlighted. NSCLC cells transduced with shRNA library were treated in replicates of 5 with vehicle (DMSO) or increasing concentrations of erlotinib (10 nM, 30 nM, 90 nM, 270 nM). Normalized shRNA counts represented across multiple doses of erlotinib are shown. C. Linear regression analysis of shRNA(s) targeting CPS1 in H1650, H322C, and PC9 NSCLC cell lines with p-values indicated. D. Linear regression analysis of shRNA(s) targeting ASS1 in H1650, H322C, and PC9 NSCLC cell lines with p-values indicated.
Genes were deemed synthetic lethal if targeting shRNAs exhibited significance based on linear regression analysis (p≤0.05) and fold change > 2 when comparing vehicle treated to 90 nM erlotinib, which is the minimal concentration to maximally inhibit kinase activity (55, 56, 34, and 32 shRNAs were identified for HCC4006, H1650, and H322C and PC9 cells, respectively; Supplementary Table S1). For PC9 T790M cells, which are resistant to erlotinib, we identified 71 shRNAs from linear regression analysis. These shRNAs were cross-referenced with synthetic lethal targets from PC9 (erlotinib sensitive cell line) in order to eliminate shRNAs that were independent of erlotinib sensitivity. Of the genes identified as synthetic lethal hits, several have been previously reported to play a role in EGFR-dependent NSCLC providing confidence in the screen results (Supplementary Figure S1A-B). For instance, glutaminase (GLS1), which is rate-limiting for glutaminolysis, has also been shown to be protective in EGFR-dependent NSCLC cells following treatment with EGFR inhibitors28. Moreover, inhibiting the rate limiting enzyme for cholesterol synthesis, MSMO1, was previously found to synergistically eliminate NSCLC cells with EGFR inhibition29. Based on the results from the screen, we identified genes involved in the urea cycle, CPS1 and ASS1, as synthetic lethal in a number of EGFR mutant cell lines (Figure 1B). Two individual shRNAs targeting CPS1 were underrepresented in our treatment conditions compared to vehicle treated cells in the EGFR mutant cell line H1650. We also identified CPS1 as synthetic lethal in the EGFR mutant cell line PC9 and in H322C cells, which overexpress WT EGFR but are sensitive to high dose erlotinib (Figure 1C). ASS1 was deemed synthetic lethal in both the mutant EGFR H1650 and the WT EGFR H322C cell lines (Figure 1D).
Suppression of CPS1 sensitizes NSCLC cells to EGFR inhibition.
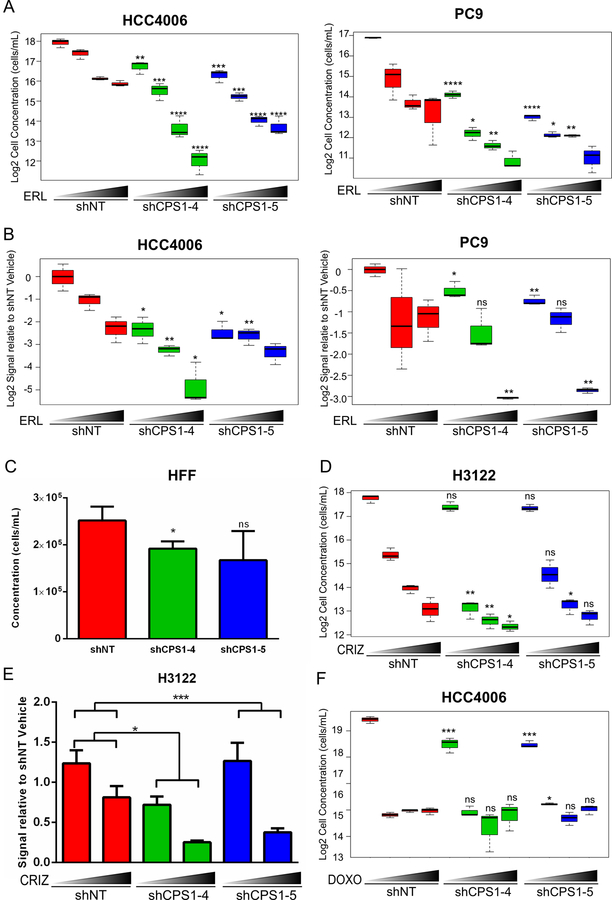
We chose to focus on CPS1 since it is the rate-limiting enzyme for the urea cycle. To validate CPS1 as synthetic lethal with erlotinib, HCC4006 and PC9 cells were transduced with individual shRNA constructs targeting CPS1 or a non-targeting shRNA control (see Supplementary Table S2 for sequences). CPS1 knockdown was confirmed both at the mRNA and protein levels (Figure S2A). While knockdown of CPS1 significantly decreased cell proliferation, the addition of erlotinib greatly exacerbated this effect (Figure 2A). Similar results were observed with a third generation EGFR inhibitor, osimertinib (effective in NSCLC with EGFR sensitizing mutations and T790M gatekeeper mutations) treatment of HCC4006, PC9 and H1975 EGFR T790M cell lines (Supplementary Figure S2B). We further evaluated the effect of CPS1 knockdown on the ability of cells to form colonies by performing colony forming assays. Knockdown of CPS1 had a significant effect on the ability of EGFR mutant NSCLC cells to form colonies, and treatment with erlotinib further impaired colony growth (Figure 2B, Supplementary Figure S2C). To determine the effect of CPS1 knockdown in normal cells, expression of CPS1 was knocked-down in primary human foreskin fibroblasts (HFF) (Supplementary Figure S2D). Knockdown of CPS1 had a modest effect on cell proliferation (Figure 2C), noting that CPS1 homozygous knockout mice die within one day of birth30. We found that EGFR mutant NSCLC cells display varying levels of CPS1 (Supplementary Figure S2E). We also determined whether knockdown of CPS1 would decrease cell growth across multiple NSCLC cell lines. Interestingly, multiple EGFR mutant also displayed sensitivity to CPS1 knockdown suggesting a potential role for CPS1 in EGFR mutant cells (Supplementary Figure S2F).
Figure 2: Inhibition of EGFR and CPS1 knockdown leads to combinatorial inhibition of NSCLC cells.
A. HCC4006 and PC9 cells expressing either non-targeting control shRNA (shNT) or shRNAs targeting CPS1 (CPS1–4, CPS1–5) were treated in triplicate with vehicle (DMSO) or increasing concentrations (30, 60, 90 nM) of erlotinib (ERL) for 3 days, followed by replating without the presence of drug for 3 additional days. The number of viable cells was determined by flow cytometry using PI-exclusion. Statistical comparison of shNT to shCPS1 with and without each erlotinib dose is shown (similar comparisons are made for other drug treatments below). B. HCC4006 and PC9 cells expressing shNT or shCPS1 were treated in triplicate with DMSO or erlotinib (50 nM and 100 nM) for 3 days, and then replated without drug for colony forming assays. C. HFF cells expressing shNT or shCPS1 were plated for 3 days. Viable cells were counted by flow cytometry. D. H3122 cells transduced with shNT or shCPS1 were treated in triplicate with vehicle (DMS0) or crizotinib (100, 500, 1000 nM) for 3 days, followed by replating without the presence of drug for 3 more days. Viable cells were determined by flow cytometry. E. H3122 cells expressing shNT or shCPS1 were treated with vehicle or 500 nM crizotinib for 3 days in triplicate and then replated without drug for colony forming assays. F. HCC4006 expressing shNT or shCPS1 were treated with vehicle (DMSO) or increasing concentrations of doxorubicin (250 nM, 500 nM, 1000 nM) for 3 days, followed by replating without drug for 3 more days. Viable cells were counted by flow cytometry. (ANOVA; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001)
We next asked whether the potentiating effect in decreasing cell growth with CPS1 knockdown was specific to EGFR inhibition or could be observed in the context of other TKIs. We transduced EML4-ALK driven H3122 cells with shRNA constructs targeting CPS1. Interestingly, knockdown of CPS1 alone had a modest effect on cell proliferation (Figure 2D) and clonogenic outgrowth (Figure 2E, Supplementary Figure S2G) but the addition of an ALK kinase inhibitor, crizotinib, further suppressed the ability of these cells to expand and form colonies. These data indicate that knockdown of CPS1 may be beneficial for the elimination of other tyrosine kinase driven lung cancers. In order to test whether the effect of CPS1 inhibition could be advantageous in the setting of chemotherapy, CPS1 knockdown cells were treated with increasing concentrations of both doxorubicin and cisplatin independently. As expected, treatment of either drug impaired cell expansion, however, knockdown of CPS1 did not further enhance this effect (Figure 1F, Supplementary Figure S2H), indicating that the combinatorial effect of CPS1 suppression in mediating the elimination of NSCLC cells is specific to tyrosine kinase inhibition.
CPS1 knockdown in combination with EGFR inhibition affects cell cycle progression and metabolic energetics.
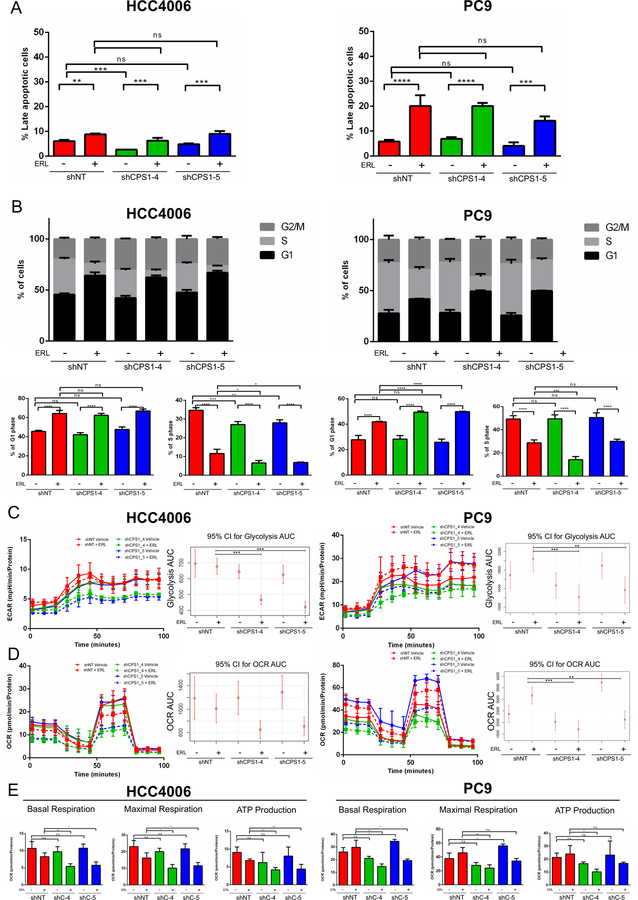
While knockdown of CPS1 in combination with erlotinib did not affect the presence of apoptotic cells by annexin-V staining (Figure 3A), it significantly impacted cell cycle profiles of both PC9 and HCC4006 EGFR mutant cell lines. Knockdown of CPS1 by itself did not significantly affect cell cycle progression, while treatment with erlotinib led to an increase in G1 phase and a decrease in S phases as has been shown previously31. Importantly, knockdown of CPS1 with EGFR inhibition led to a further increase of cells in G1 phase in PC9 and a further decrease in S phase in HCC4006 and to a lesser extent PC9 cells (Figure 3B). We also performed EdU analysis and observed a significant decrease in the percentage of EdU-positive cells with CPS1 knockdown in combination with erlotinib (Supplementary Figure S3A). While CPS1 knockdown enhanced erlotinib mediated effects on cell cycle progression, it did not affect phosphorylation of EGFR and its downstream targets (Supplementary Figure S3B) suggesting that CPS1 knockdown is acting through a mechanism independent of the EGFR signaling axis. Interestingly, treatment with an EGFR inhibitor led to an increase in CPS1 expression possibly suggesting that upon EGFR inhibition, CPS1 is upregulated as an adaptive mechanism to permit cell survival (Supplementary Figure S3C).
Figure 3: Combined EGFR inhibition and CPS1 knockdown leads to a decrease in central carbon metabolism and to G1 arrest.
A. HCC4006 and PC9 expressing shNT or shCPS1 cells were treated with vehicle (DMSO) or 100 nM erlotinib for 3 days in triplicate and apoptosis was measured by Annexin V staining by flow cytometry. B. HCC4006 and PC9 expressing shNT or shCPS1 cells were treated with vehicle (DMSO) or 100 nM erlotinib for 24h in triplicate and stained with PI for cell cycle analysis. Full cell cycle panels are shown with smaller panels depicting G and S phases below. HCC4006 and PC9 expressing shNT or shCPS1 cells were plated on microplates and treated with vehicle (DMSO) or 100 nM erlotinib for 24 hours. C. Extracellular acidification rate or glycolysis rate was measured at indicated time points and 95% confidence intervals were calculated for the area under the curve for each condition. D. Oligomycin-A (OA), FCCP, rotenone (Rot)/ antimycin A (Anti) were sequentially added and the oxygen consumption rate measured with Agilent Seahorse instrument. The 95% confidence intervals were calculated for the area under the curve for each condition across the course of the experiment. E. Basal respiration, maximal respiration and ATP production were determined for each condition. (ANOVA; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
We next asked whether elevated ammonia levels could contribute to growth impairment in CPS1 knockdown cells since CPS1 catalyzes the conversion of ammonia to carbamoyl phosphate and knockdown of CPS1 would presumably lead to a buildup of ammonia levels. However, CPS1 knockdown and the addition of erlotinib did not further increase ammonia levels, indicating that ammonia buildup does not account for the reduction in cell proliferation (Supplementary Figure S3D). Interestingly, it has been reported that ammonia can facilitate the proliferation of breast cancer cells by acting as a nitrogen source to be recycled into central amino acid metabolism32. Unlike breast cancer cells, NSCLC cells do not proliferate faster when cultured in the presence of extra ammonia (data not shown) and ammonia levels are indeed toxic at 1mM. These results indicate a minimal role for ammonia in regulating cell growth through recycling of ammonia in EGFR mutant NSCLC cells.
Given that we observed a substantial impairment in cell cycle progression in NSCLC cells with CPS1 knockdown under treatment with erlotinib, we asked whether CPS1 knockdown in combination with erlotinib may affect cellular energetics and dampen metabolic activity. We used a Seahorse assay to measure the extracellular acidification rate (ECAR) to assess glycolytic activity and oxygen consumption rate (OCR) in HCC4006 and PC9 cells with and without CPS1 knockdown and erlotinib treatment. Knockdown of CPS1 alone or treatment with erlotinib alone had minimal effects on ECAR, but the addition erlotinib with CPS1 knockdown significantly reduced ECAR (Figure 3C). Multiple parameters from the assay were used to evaluate mitochondrial function by introducing oligomycin-A (ATP synthase inhibitor), FCCP (protonophoric uncoupler), antimycin A and rotenone (electron transport chain inhibitors) to the NSCLC cells while measuring OCR. Total OCR was not significantly changed with either CPS1 knockdown or EGFR inhibition alone, but was reproducibly and substantially suppressed by combined inhibition of EGFR and CPS1, suggesting compensatory roles in the TCA cycle and the mitochondrial respiratory pathway (Figure 3D). Moreover, all ETC inhibitors affected OCR as expected, and the combination of CPS1 knockdown with erlotinib led to a further reduction in OCR upon treatment of each ETC inhibitor [95% confidence intervals (CI) were calculated for the area under the curve (AUC) for each treatment interval for HCC4006 (Supplementary Figure S3E) and PC9 cells (Supplementary Figure S3F)]. Basal and maximal respiration is further reduced by CPS1 knockdown in both cell lines upon erlotinib treatment. Additionally, ATP production reflected OCR results, with a substantial decrease in ATP levels only observed consistently in CPS1 knockdown cells treated with erlotinib (Figure 3E). ATP levels were also measured using a colorimetric assay which further confirmed the ATP measurements from the Seahorse assay (Supplementary Figure S3G). Given that we observed significant changes in ATP levels in CPS1 knockdown cells upon EGFR inhibition, we asked whether redox state was affected by measuring mitochondrial reactive oxygen species (ROS) levels. Mitochondrial ROS were not affected with EGFR inhibition or in CPS1 knockdown cells (data not shown) indicating that the synergistic elimination of CPS1 knockdown cells treated with erlotinib is not due to an induction of mitochondrial ROS. These data demonstrate that CPS1 inhibition further sensitizes NSCLC cells to EGFR inhibition by impeding cell cycle progression and impacting cellular energetics.
The effects of inhibition of EGFR on central carbon metabolism are further enhanced with loss of CPS1.
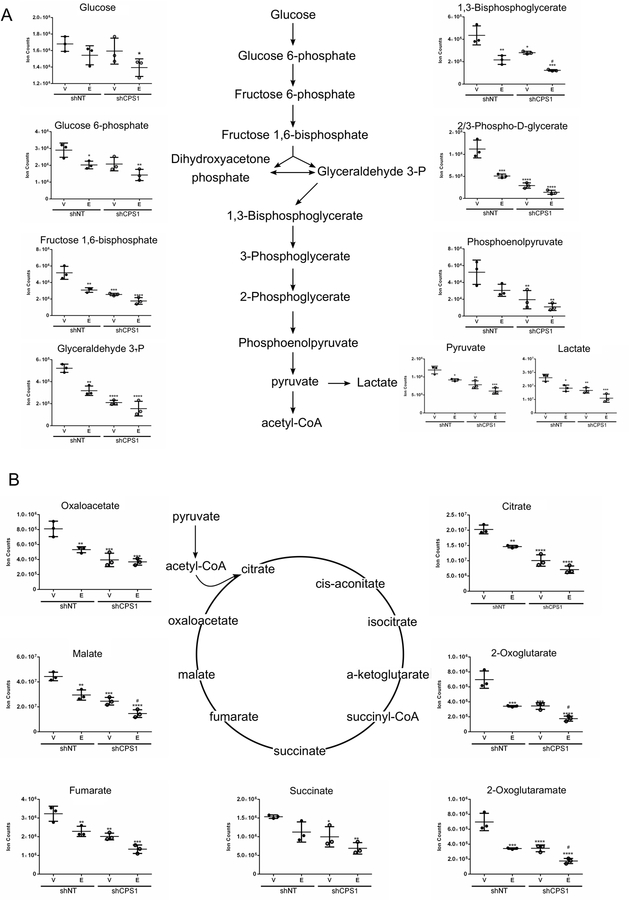
Given that we observed a reduction in glycolysis, oxygen consumption and ATP levels when both EGFR and CPS1 were inhibited, we wanted to assess whether knockdown of CPS1 could cause global metabolic changes, with or without EGFR inhibition. We hypothesized that CPS1 knockdown in combination with erlotinib could further influence metabolic pathways, which may account for the decrease in cell proliferation of NSCLC cells. To address this, HCC4006 cell lines expressing CPS1 shRNAs or a control shRNA were treated with erlotinib for 22 hours. Knockdown of CPS1 was confirmed at the mRNA level by real time PCR (Supplementary Figure S4A). We chose 22 hours of treatment; a time point when neither apoptosis nor total cellular protein level reductions had been induced by treatment of erlotinib. Cell extracts were analyzed using ultra-high-performance LC-tandem mass spectrometry (UPLC-MS/MS). Unsupervised principal component analysis (PCA) showed clustering among control shRNA cells and CPS1 knockdown cells, as well as non-treated and erlotinib-treated samples (Supplementary Figure S4B). Treatment with erlotinib led to reductions in numerous metabolites including those associated with glycolysis, TCA cycle, and nucleotide metabolism (Supplementary Figure S4C). As expected, knockdown of CPS1 alone led to a decrease in the level of downstream metabolites in the urea cycle (arginine) and related pathways (fumarate). Interestingly, metabolites involved in early steps in the urea cycle modestly accumulated following CPS1 knockdown (citrulline, aspartate, and argininosuccinate); with the caveat that these results are based on steady state measurements, these results are suggestive of a blockade in the urea cycle (Supplementary Figure S4D).
EGFR inhibition decreases aerobic glycolysis via suppression of the PI3K pathway33,34. Notably, combined treatment with erlotinib and CPS1 knockdown induced dramatic changes in the metabolome. For instance, treatment of erlotinib or CPS1 knockdown alone led to a reduction in glycolytic metabolites, which was exacerbated with the combination of erlotinib and CPS1 knockdown, suggesting a disruption in the maintenance of glucose homeostasis (Figure 4A). Additionally, TCA cycle intermediates were decreased with erlotinib treatment or CPS1 knockdown alone, while the combination of EGFR and CPS1 inhibition further decreased TCA cycle metabolites (Figure 4B). Taken together, these results demonstrate that combined inhibition of EGFR and CPS1 significantly impairs central carbon metabolism which contributes to the combinatorial effect of CPS1 knockdown and EGFR inhibition in eliminating EGFR mutant NSCLC cells.
Figure 4. Metabolites involved in carbon metabolism are decreased by CPS1 knockdown with further exacerbation upon EGFR inhibition.
HCC4006 cells were treated with vehicle (DMSO) or 100 nM erlotinib for 22 hours in replicates of three. Cell extracts were processed for metabolic profiling using UPLC-MS/MS. A. Levels of glycolysis metabolites B. Levels of TCA cycle intermediates. ANOVA; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 for comparisons of all samples to vehicle-treated control shNT cells, and pound signs indicate statistical significance (#P ≤ 0.05, ##P ≤ 0.01, ###P ≤ 0.001) for comparisons of CPS1 knockdown cells to control shNT cells under the same treatment conditions.
Knockdown of CPS1 in combination with EGFR inhibition affects pyrimidine biosynthesis.
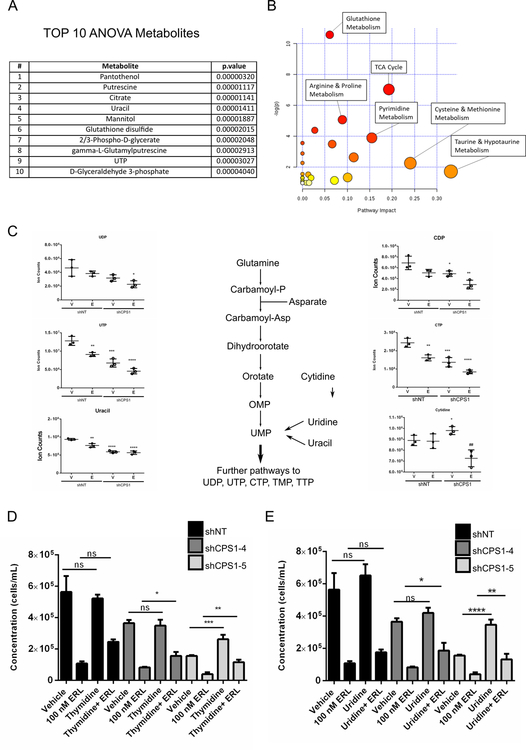
Carbamoyl phosphate serves as a precursor for the de novo synthesis of pyrimidines, thus linking the urea cycle to the pyrimidine anabolic pathway. One of the most severely affected metabolic pathways identified from our metabolomics analysis was the pyrimidine pool, with UTP and uracil represented in the top significantly altered metabolites (Figure 5A). Additionally, when the top 25 significant metabolites were utilized to perform pathway analysis, the pyrimidine metabolism pathway came up as one of the most prominent pathways altered in CPS1 knockdown cells treated with erlotinib (Figure 5B). Upon further analysis, metabolites involved in pyrimidine biosynthesis including UTP, uracil CDP and CTP were substantially decreased in CPS1 knockdown cells and were further reduced with erlotinib treatment (Figure 5C). CPS1 has been reported to play a role in pyrimidine biosynthesis in LKB1 mutant KRAS lung cancers by replenishing cytosolic levels of carbamoyl phosphate24. Treatment with erlotinib also reduced pyrimidine metabolites as expected since EGFR inhibition has been shown to inhibit the rate limiting enzyme of pyrimidine biosynthesis, CAD19. Strikingly, the combined effect of CPS1 knockdown and erlotinib substantially depleted levels of pyrimidine metabolites compared to knockdown or treatment alone, indicating that dampening of pyrimidine biosynthesis may account for the underlying mechanism leading to the combinatorial effect of CPS1 and EGFR inhibition on cell proliferation. Levels of purines increased in vehicle-treated CPS1 knockdown cells indicative of a shift in the ratio between pyrimidines and purines, although interestingly erlotinib treatment reversed this increase (Supplementary Figure S5A).
Figure 5: Pyrimidine biosynthesis metabolites are reduced by combined CPS1 knockdown cells with erlotinib treatment and exogenous pyrimidines can partially rescue cell proliferation.
HCC4006 cells were treated with erlotinib for vehicle (DMSO) or 100 nM erlotinib for 22 and were processed using UPLC-MS/MS for metabolic profiling as in figure 4 A. Table showing the TOP ten metabolites identified using ANOVA. B. Pathway analysis performed showing the most affected metabolic pathways based on the top 25 significantly changed metabolites. C. Pyrimidine metabolite levels in HCC4006 cell treated with 100 nM erlotinib for 22 hours. D. HCC4006 shNT control or shCPS1 cells treated in triplicate with vehicle (DMSO), 100 nM erlotinib, 1 mM thymidine or thymidine and erlotinib for 6 days. Cell viability was determined by flow cytometry using PI. E. As in D, but with vehicle (DMSO), 100 nM erlotinib, 1 mM uridine or uridine and erlotinib for 6 days. Cell viability was determined by flow cytometry using PI. (ANOVA; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
We next asked whether adding back pyrimidine nucleosides could rescue the effect of CPS1 inhibition with or without erlotinib treatment. Exogenous thymidine (Figure 5D) and uridine (Figure 5E) partially rescued growth impairment mediated by CPS1 knockdown combined with erlotinib treatment in HCC4006 cells. Similar rescue was observed for combined CPS1 knockdown and osimertinib treatment (Supplementary Figure S5B). Adding back pyrimidine nucleosides was also sufficient to partially rescue colony formation in HCC4006 and PC9 cells with CPS1 knockdown treated with erlotinib (Supplementary Figure S5C-D). These data suggest that erlotinib treatment alone leads to a decrease in pyrimidine biosynthesis and when combined with CPS1 knockdown leads to a further decrease of pyrimidines, contributing to a decrease in NSCLC cell proliferation.
Combinatorial inhibition of CPS1 and EGFR impairs arginine metabolism.
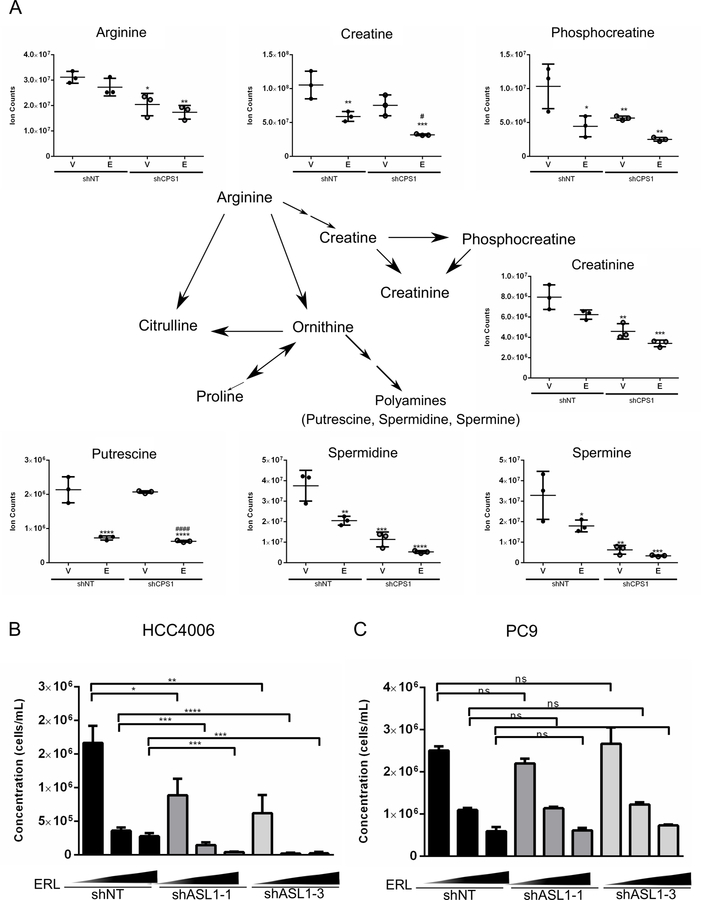
In addition to pyrimidine metabolism, we also identified metabolites involved in arginine metabolism, which utilizes carbamoyl phosphate as a precursor from the urea cycle, to be substantially affected by CPS1 knockdown. From the pathway analysis of the 25 most significantly altered metabolites in HCC4006 CPS1 knockdown cells treated with erlotinib, arginine and proline metabolism was one of the most affected pathways (Figure 5B). As shown in Figure 6A, knockdown of CPS1 significantly decreased levels of intracellular arginine, as well as downstream metabolites, phosphocreatine and creatinine. Furthermore, polyamines such as putrescine, spermidine and spermine, which are important for cell survival and proliferation, were also found to be reduced in CPS1 knockdown cells. Combined erlotinib treatment with CPS1 knockdown led to a further reduction of metabolites involved in arginine metabolism. We then tested whether suppression of argininosuccinate lyase (ASL), involved in the generation of arginine, would further sensitize NSCLC cells to EGFR inhibition. ASL is responsible for catalyzing the conversion of argininosuccinate to produce fumarate and arginine in the urea cycle. Knockdown of ASL (Supplementary Figure S6A) synergistically reduced the expansion of HCC4006 cells when combined with EGFR inhibition (Figure 6B). However, knockdown of ASL in PC9 cells in combination with erlotinib had no additive effect on cell proliferation (Figure 6C). Interestingly, knockdown of ASL alone had no effect on the survival of PC9 cells, suggesting that these cells may not rely on de novo arginine synthesis but are instead dependent on the uptake of extracellular arginine. We also determined the effect of suppressing other urea cycle enzymes in combination with EGFR inhibition. ASS1 was also identified as a synthetic lethal hit from our metabolic screen. The knockdown of ASS1 alone led to a decrease in the number of viable cells and the addition of erlotinib further inhibited the expansion of NSCLC cells (Supplementary Figure S6B). Similarly, knockdown of urea cycle enzymes ornithine transcarbamylase (OTC) and arginase (ARG2) also sensitized NSCLC cells to EGFR inhibition, suggesting that multiple branches in the urea cycle are crucial for survival of NSCLC cells following treatment with erlotinib (Supplementary Figures S6C, S6D). Taken together, these results suggest that impairment of arginine production may play an important (albeit partial) role in the detrimental effects of combinatorial inhibition of CPS1 and EGFR in NSCLC cells.
Figure 6: Arginine metabolites are decreased with combined CPS1 knockdown and EGFR inhibition.
A. Arginine metabolite levels in HCC4006 cells treated with 100 nM erlotinib for 22 hours B. HCC4006 expressing shNT control or shASL1 treated with vehicle (DMSO) or increasing dose of erlotinib (50 nM and 100 nM) in triplicate for 3 days, followed by 3 days without drug. Cell viability was determined by flow cytometry. C. PC9 expressing shNT control or shASL1 were treated with vehicle (DMSO) or increasing dose of erlotinib (50 nM and 100 nM) in triplicate for 3 days, followed by 3 days without drug. Cell viability was determined by flow cytometry. (ANOVA; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
Association of CPS1 expression with lung adenocarcinoma (LADC) patient survival.
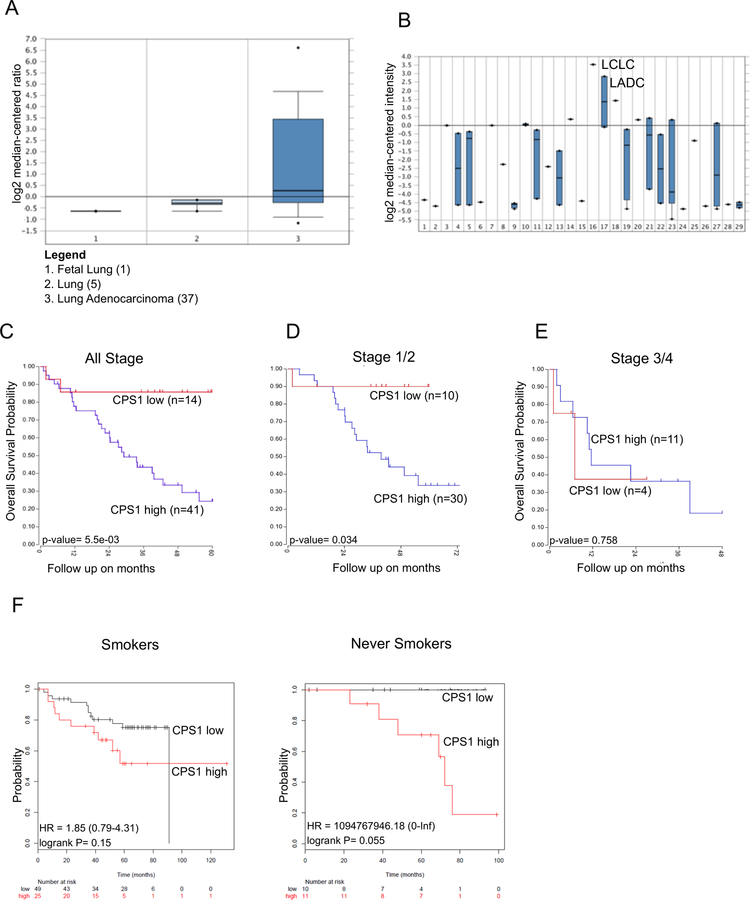
It has been shown that in human LADC, CPS1 mRNA expression was highest in tumors with LKB1 loss24. Similarly, LADC with LKB1 loss exhibited higher CPS1 protein expression by analyzing a tissue microarray23. Furthermore, high expression of CPS1 was found to be associated with worse overall survival. We mined several patient datasets using Oncomine and the R2 Genomics Platform to further evaluate the relevance of CPS1 in LADC. We first asked whether expression of CPS1 was higher in LADC compared to normal tissues. As shown in Figure 7A, expression of CPS1 mRNA was significantly higher in LADC relative to both fetal and normal lung tissue consistent with a proteomic analysis of normal lung versus NSCLC adenocarcinoma35. Additionally, while mining patient datasets across a spectrum of cancers, we identified that CPS1 expression was high in large cell lung carcinomas and lung adenocarcinomas compared to other cancer types (Figure 7B). Moreover, when we determined the 5-year overall survival of lung patients with adenocarcinomas, high expression of CPS1 greatly diminished overall survival compared to patients with low CPS1 expression (Figure 7C). Interestingly, expression of CPS1 correlated with worse survival during early stage LADC (Figure 7D) but not in later stages of lung cancer (Figure 7E), potentially indicating a more prominent role of CPS1 during early stages of lung cancer evolution. Lastly, we used KM Plotter36 to evaluate the expression of CPS1 in never-smokers, as EGFR mutations as well as other tyrosine kinase drivers are more commonly found in never-smokers37. As shown in Figure 7F, we found that never-smoker patients with high expression of CPS1 did exhibit a strong trend indicating substantially worse overall survival compared to patients with low CPS1 expression, which was apparent despite the low numbers of patients analyzed (11 and 10 for the high and low CPS1 groups, respectively). These analyses indicate that CPS1 may be a strong negative prognostic indicator in NSCLC patients, which may be particularly relevant for never-smokers and for those with LADC bearing mutations activating receptor tyrosine kinases.
Figure 7: CPS1 expression is high in LADC and correlated with worse overall patient survival.
A. Oncomine analysis of CPS1 expression in fetal lung, normal lung, and lung adenocarcinoma. B. Oncomine analysis of CPS1 expression across cancers (Legend in Supplemental Table 5) with LADC highlighted (Group 17). C. High and low CPS1 expression correlated with overall survival probability in LADC patients across all stages. D. High and low CPS1 expression correlated with overall survival probability in LADC patients in stages 1/2. E. High and low CPS1 expression correlated with overall survival probability in LADC patients in stages 3/4. F. Overall survival probability for smoker and never smoker patients with LADC exhibiting high or low CPS1 expression.
Discussion
Our results reveal an important role of the urea cycle for the survival of NSCLC cells upon EGFR inhibition. EGFR inhibitors have been effective in patients harboring EGFR mutations, significantly extending overall survival and improving quality of life compared to conventional chemotherapy. Yet, relapse is almost inevitable. Recently, urea cycle dysregulation has been reported to be a common phenomenon in tumorigenesis, leading to the ability to detect both biochemical and molecular signatures in patient samples38. Interestingly, it has been shown that urea cycle dysregulation correlates with better responses to immune checkpoint therapies due to pyrimidine-rich transversion mutational bias as a result of elevated pyrimidine synthesis. Our studies have shown that the urea cycle may serve as a key metabolic vulnerability in the context of EGFR inhibition.
Using a functional metabolic shRNA screen, we identified the urea cycle enzyme CPS1 as synthetic lethal with erlotinib treatment in EGFR mutant NSCLC. We validated that suppression of CPS1 and EGFR inhibition led to a further decrease in the expansion of NSCLC cells compared to knockdown or treatment alone. We did not observe significant combinatorial efficacy for CPS1 knockdown with conventional chemotherapy agents. Notably, Çeliktaş et al. demonstrated an additive effect of CPS1 knockdown with chemotherapy agents in LKB1 mutant NSCLC cells23, suggesting that the role of CPS1 may be distinct in EGFR or EML4-ALK-driven lung cancer. In normal tissues, CPS1 is expressed abundantly in the liver and gastrointestinal tract39,40; however, CPS1 has been reported to exhibit higher expression in multiple cancer types25,41. More recently, CPS1 expression has been associated with KRAS/LKB1 mutant lung cancer cell growth24. We find that the growth impairment mediated by CPS1 knockdown alone is consistent with studies addressing CPS1 dependency in KRAS/LKB1 mutant lung cancer cells. Our studies further suggest a role for CPS1 in EGFR mutant lung cancers following treatment with an EGFR TKI. The treatment of NSCLC cells with erlotinib inhibits glycolysis42, mirroring poor glucose availability, a phenotype that is also observed during caloric restriction43. During caloric restriction, the NAD+ -dependent deacetylases sirtuins are activated to promote the urea cycle to facilitate mitochondrial adaptation when energy levels are low44. Interestingly, it has been demonstrated that Sirt5 can regulate the urea cycle by deacetylating CPS1, and in turn, upregulating its activity45. Upon erlotinib treatment, a fraction of cancer cells may be able to adapt to erlotinib-mediated glycolysis inhibition by generating tricyclic acid cycle intermediates to support mitochondrial metabolism and increased levels of ATP to permit cell survival by activating CPS1, engendering a greater dependency on the urea cycle. Our data demonstrate that CPS1 inhibition further sensitizes NSCLC cells to EGFR inhibition, decreasing cell proliferation by potentiating EGFR inhibition, promoting G1 arrest and dampening central carbon metabolism.
Interestingly, CPS1 knockdown in combination with erlotinib led to dramatic changes in cellular metabolism. We showed that ECAR and OCR were significantly decreased only when erlotinib treatment was combined with CPS1 knockdown. EGFR inhibition also decreased metabolites associated with glycolysis and the TCA cycle, and these effects were amplified with CPS1 knockdown consistent with the diminished ECAR and OCR observed. That EGFR inhibition alone was unable to reduce ECAR and OCR reveals a critical role for CPS1 and the urea cycle in buffering the metabolic impact of TKI treatment. While glycolytic and TCA cycle intermediates are reduced by either EGFR inhibition or CPS1 knockdown, the functional impacts (at least as measured using Seahorse assays) are only apparent upon combined inhibitions. Our metabolomics data also indicate that EGFR inhibition decreased pyrimidines, as expected as EGFR inhibition has been reported to decrease activation of CAD, the enzyme responsible for de novo pyrimidine biosynthesis19. Similarly, CPS1 knockdown also reduced pyrimidine related metabolites as previously reported23,24, however, the effect was the most pronounced with both EGFR inhibition and CPS1 knockdown, suggesting that these roles are non-epistatic. Exogenous levels of pyrimidine nucleosides partially rescued the growth impairment mediated by CPS1 and EGFR inhibition. These findings suggest a potential for combining EGFR inhibitors with pharmacological agents targeting nucleic acid biosynthesis could be beneficial.
We also observed that CPS1 knockdown reduced levels of metabolites associated with arginine metabolism. While erlotinib treatment alone had no significant effect on arginine levels and a modest effect on metabolites derived from arginine, the addition of CPS1 knockdown further diminished arginine levels and polyamines indicating that the reduction in cell growth may in part be attributed to reductions in arginine and its metabolites. Arginine is a semi-essential amino acid that is an intermediate in the urea cycle. It has multiple roles in metabolism including polyamine, creatine and nitric oxide (NO) biosynthesis46. We further showed that knockdown of ASL, the enzyme that catalyzes the reaction to form arginine and fumarate, was synergistic with EGFR inhibition in HCC4006 cells. Our findings suggest that arginine and arginine-derived metabolites contribute to the maintenance of cell proliferation in the context of EGFR inhibition in EGFR dependent NSCLC cells.
Overall, our studies demonstrate that CPS1 and the urea cycle play an essential role in maintaining central carbon metabolism and energetics in EGFR-dependent NSCLC cells treated with TKI. As further indication of the clinical relevance of these findings, we showed that higher levels of CPS1 correlated with worse overall survival compared to patients with low CPS1 expression. These analyses indicate that CPS1 may be a strong negative prognostic indicator in NSCLC patients, and possibly more beneficial in patients with EGFR mutant tumors treated with a TKI.
Supplementary Material
Implications:
Our results reveal that the urea cycle may be a novel metabolic vulnerability in the context of EGFR inhibition, providing an opportunity to develop rational combination therapies with EGFR inhibitors for the treatment of EGFR-driven NSCLC.
Acknowledgements:
We thank Hannah Scarborough for suggestions and critical review of the text. Additionally, we thank the University of Colorado Cancer Center Functional Genomics, Genomics, Biostatistics/Bioinformatics and Flow Cytometry Shared Resources.
Financial support: Studies were supported by RO1-CA157850 and NCI SPORE P50-CA058187, and the Courtenay C. and Lucy Patten Davis Endowed Chair in Lung Cancer Research to J.D. Cancer Center Shared Resources are supported by NIH grant 2-P30-CA46934.
Footnotes
Conflicts of interest: The authors declare that they have no conflict of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S et al. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science (80- ) 2004; 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non–Small-Cell Lung Cancer to Gefitinib. N Engl J Med 2004; 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- 4.Mok TS, Wu Y-L, Thongprasert S, Yang C-H, Chu D-T, Saijo N et al. Gefitinib or Carboplatin–Paclitaxel in Pulmonary Adenocarcinoma. N Engl J Med 2009; 361: 947–957. [DOI] [PubMed] [Google Scholar]
- 5.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011; 12: 735–742. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M et al. EGFR Mutation and Resistance of Non–Small-Cell Lung Cancer to Gefitinib. N Engl J Med 2005; 352: 786–792. [DOI] [PubMed] [Google Scholar]
- 7.Turke AB, Zejnullahu K, Wu Y-L, Song Y, Dias-Santagata D, Lifshits E et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010; 17: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghiso E, Migliore C, Ciciriello V, Morando E, Petrelli A, Corso S et al. YAP-Dependent AXL Overexpression Mediates Resistance to EGFR Inhibitors in NSCLC. Neoplasia 2017; 19: 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ware KE, Marshall ME, Heasley LR, Marek L, Hinz TK, Hercule P et al. Rapidly Acquired Resistance to EGFR Tyrosine Kinase Inhibitors in NSCLC Cell Lines through De-Repression of FGFR2 and FGFR3 Expression. PLoS One 2010; 5: e14117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suda K, Tomizawa K, Fujii M, Murakami H, Osada H, Maehara Y et al. Epithelial to Mesenchymal Transition in an Epidermal Growth Factor Receptor-Mutant Lung Cancer Cell Line with Acquired Resistance to Erlotinib. J Thorac Oncol 2011; 6: 1152–1161. [DOI] [PubMed] [Google Scholar]
- 11.Casás-Selves M, Kim J, Zhang Z, Helfrich BA, Gao D, Porter CC et al. Tankyrase and the Canonical Wnt Pathway Protect Lung Cancer Cells from EGFR Inhibition. Cancer Res 2012; 72: 4154–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 2012; 487: 505–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarborough HA, Bunn PA, DeGregori J. Personalized one-two punches for lung cancer. Cancer Res 2015; 25: 269–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garraway LA, Jänne PA. Circumventing Cancer Drug resistance in the era of Personalized Medicine. Cancer Discov 2012; 2: 214–226. [DOI] [PubMed] [Google Scholar]
- 15.Levine AJ, Puzio-Kuter AM. The Control of the Metabolic Switch in Cancers by Oncogenes and Tumor Suppressor. Science 2010; 330:1340–4. [DOI] [PubMed] [Google Scholar]
- 16.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009; 324: 1029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR et al. Akt Stimulates Aerobic Glycolysis in Cancer Cells. Cancer Res 2004; 64: 3892–3899. [DOI] [PubMed] [Google Scholar]
- 18.Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B et al. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab 2013; 17: 1000–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makinoshima H, Takita M, Matsumoto S, Yagishita A, Owada S, Esumi H et al. Epidermal Growth Factor Receptor (EGFR) Signaling Regulates Global Metabolic Pathways in EGFR-mutated Lung Adenocarcinoma. J Biol Chem 2014; 289: 20813–20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deberardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010; 29: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimski DS. Ammonia Metabolism and the Urea Cycle: Function and Clinical Implications. J Vet Intern Med 1994; 8:73–8. [DOI] [PubMed] [Google Scholar]
- 22.Holden HM, Thodent JB, Raushel FM. Carbamoyl phosphate synthetase: a tunnel runs through it. Current Opinion in Structural Biology 1998, 8:679–685 [DOI] [PubMed] [Google Scholar]
- 23.Çeliktaş M, Tanaka I, Chandra Tripathi S, Fahrmann JF, Aguilar-Bonavides C, Villalobos P et al. Role of CPS1 in Cell Growth, Metabolism, and Prognosis in LKB1-Inactivated Lung Adenocarcinoma. J Natl Cancer Inst 2017; 109: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Hu Z, Cai L, Li K, Choi E, Faubert B et al. CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nat Publ Gr 2017; 546: 168–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y-Y, Li C-F, Lin C-Y, Lee S-W, Sheu M-J, Lin L-C et al. Overexpression of CPS1 is an independent negative prognosticator in rectal cancers receiving concurrent chemoradiotherapy. Tumor Biol 2014; 35: 11097–11105. [DOI] [PubMed] [Google Scholar]
- 26.Delage B, Fennell DA, Nicholson L, Mcneish I, Lemoine NR, Crook T et al. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. UICC Int J Cancer IJC 2010; 126: 2762–2772. [DOI] [PubMed] [Google Scholar]
- 27.Rabinovich S, Adler L, Yizhak K, Sarver A, Silberman A, Agron S et al. Diversion of aspartate in ASS1-deficient tumors fosters de novo pyrimidine synthesis. Nature 2015; 527: 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Momcilovic M, Bailey ST, Lee JT, Fishbein MC, Magyar C, Braas D et al. Targeted Inhibition of EGFR and Glutaminase Induces Metabolic Crisis in EGFR Mutant Lung Cancer. Cell Rep 2017; 18: 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sukhanova A, Gorin A, Serebriiskii IG, Gabitova L, Zheng H, Restifo D et al. Targeting C4-Demethylating Genes in the Cholesterol Pathway Sensitizes Cancer Cells to EGF Receptor Inhibitors via Increased EGF Receptor Degradation. Cancer Discov 2013; 3: 96–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schofield JP, Schofield JP, Cox TM, Caskey CT, Wakamiya M. Mice deficient in the urea-cycle enzyme, carbamoyl phosphate synthetase i, die during the early neonatal period from hyperammonemia. Hepatology 1999; 29: 181–185. [DOI] [PubMed] [Google Scholar]
- 31.Scarborough HA, Helfrich BA, Casás-Selves M, Schuller AG, Grosskurth SE, Kim J et al. AZ1366: An Inhibitor of Tankyrase and the Canonical Wnt Pathway that Limits the Persistence of Non-Small Cell Lung Cancer Cells Following EGFR Inhibition. Clin Cancer Res 2017; 23: 1531–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spinelli JB, Spinelli JB, Yoon H, Ringel AE, Jeanfavre S, Clish CB et al. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass 2017; 9305: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson JE, Thompson CB. Putting the rap on Akt. J Clin Oncol 2004; 22: 4217–26. [DOI] [PubMed] [Google Scholar]
- 34.Saruwatari K, Umemura S, Obata Y, Ishii G, Matsumoto S, Sugiyama E et al. Signaling through the Phosphatidylinositol 3-Kinase (PI3K)/ Mammalian Target of Rapamycin (mTOR) Axis Is Responsible for Aerobic Glycolysis mediated by Glucose Transporter in Epidermal Growth Factor Receptor (EGFR)-mutated Lung Adenocarcinoma. J Biol Chem 2015; 290: 17495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kikuchi T, Hassanein M, Amann JM, Liu Q, Slebos RJC, Rahman SMJ et al. In-depth proteomic analysis of nonsmall cell lung cancer to discover molecular targets and candidate biomarkers. Mol Cell Proteomics 2012; 11: 916–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online Survival Analysis Software to Assess the Prognostic Value of Biomarkers Using Transcriptomic Data in Non-Small-Cell Lung Cancer. PLoS One 2013; 8: e82241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couraud S, Zalcman G, Milleron B, Morin F, Souquet P-J. Lung cancer in never smokers – A review. Eur J Cancer 2012; 48: 1299–1311. [DOI] [PubMed] [Google Scholar]
- 38.Lee JS, Adler L, Karathia H, Hannenhalli S, Ruppin E, Erez A. Urea Cycle Dysregulation Generates Clinically Relevant Genomic and Biochemical Signatures. Cell 2018; 174: 1559–1570.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butler SL, Dong H, Cardona D, Jia M, Zheng R, Zhu H et al. The antigen for Hep Par 1 antibody is the urea cycle enzyme carbamoyl phosphate synthetase 1. Lab Investig 2008; 88: 78–88. [DOI] [PubMed] [Google Scholar]
- 40.Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 2014; 13: 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiaoguang Z, Meirong L, Jingjing Z, Ruishen Z, Qing Z, Xiaofeng T. Long Noncoding RNA CPS1-IT1 Suppresses Cell Proliferation and Metastasis in Human Lung Cancer. Oncol Res 2017; 25: 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poliaková M, Aebersold DM, Zimmer Y, Medová M. The relevance of tyrosine kinase inhibitors for global metabolic pathways in cancer. Mol Cancer 2018; 17:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingram DK, Roth GS. Glycolytic inhibition as a strategy for developing calorie restriction mimetics. Exp Gerontol 2011; 46: 148–154. [DOI] [PubMed] [Google Scholar]
- 44.Hallows WC, Yu W, Smith BC, Devries MK, Devires MK, Ellinger JJ et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell 2011; 41: 139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates Carbamoyl Phosphate Synthetase 1 and Regulates the Urea Cycle. Cell 2009;137: 560–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knowledge O, Morris SM. Arginine Metabolism : Boundaries of our knowledge J Nutr 2007; 137: 1602S–1609S. [DOI] [PubMed] [Google Scholar]
- 47.Zhang G, Scarborough H, Kim J, Rozhok AI, Chen YA, Zhang X et al. Coupling an EML4-ALK-centric interactome with RNA interference identifies sensitizers to ALK inhibitors. Sci Signal 2016; 9:rs12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nemkov T, Hansen KC, D’Alessandro A. A three-minute method for high-throughput quantitative metabolomics and quantitative tracing experiments of central carbon and nitrogen pathways. Rapid Commun Mass Spectrom 2017; 31: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nemkov T, D’Alessandro A, Hansen KC. Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole orbitrap mass spectrometry. Amino Acids 2015; 47: 2345–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.