Abstract
Bacteriophage (“bacteria eaters”) or phage is the collective term for viruses that infect bacteria. While most phages are pathogens that kill their bacterial hosts, the filamentous phages of the sub‐class Inoviridae live in cooperative relationships with their bacterial hosts, akin to the principal behaviours found in the modern‐day sharing economy: peer‐to‐peer support, to offset any burden. Filamentous phages impose very little burden on bacteria and offset this by providing service to help build better biofilms, or provision of toxins and other factors that increase virulence, or modified behaviours that provide novel motile activity to their bacterial hosts. Past, present and future biotechnology applications have been built on this phage–host cooperativity, including DNA sequencing technology, tools for genetic engineering and molecular analysis of gene expression and protein production, and phage‐display technologies for screening protein–ligand and protein–protein interactions. With the explosion of genome and metagenome sequencing surveys around the world, we are coming to realize that our knowledge of filamentous phage diversity remains at a tip‐of‐the‐iceberg stage, promising that new biology and biotechnology are soon to come.
Keywords: filamentous phage, phage, procoat protein, secretin, Zot
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Structural Biology; Synthetic Biology & Biotechnology
Glossary
- CRISPR
clustered regularly interspaced short palindromic repeats
- Ff
collective term for near‐identical phages M13, fd and f1
- MRSA
methicillin‐resistant Staphylococcus aureus
- RF
replicative form
- T2SS
bacterial type 2 secretion system
- T3SS
bacterial type 3 secretion system
- T4P
bacterial type 4 pili
Introduction
Overview of phage diversity and their application in biotechnology
Phages come in diverse morphological forms and show huge diversity in their genome size, structure and sequences. They are the predominant biological entity on Earth, with the exponential acquisition in genome and metagenome sequence data now making estimates of their diversity and impact undeniable 1, 2, 3, 4. In addition to their impact on shaping bacterial communities including our own microbiomes, the knowledge acquired from phages of various families (Fig 1A) has provided immeasurable benefit to biotechnology.
Figure 1. Filamentous phages: classification and applications in biotechnology.
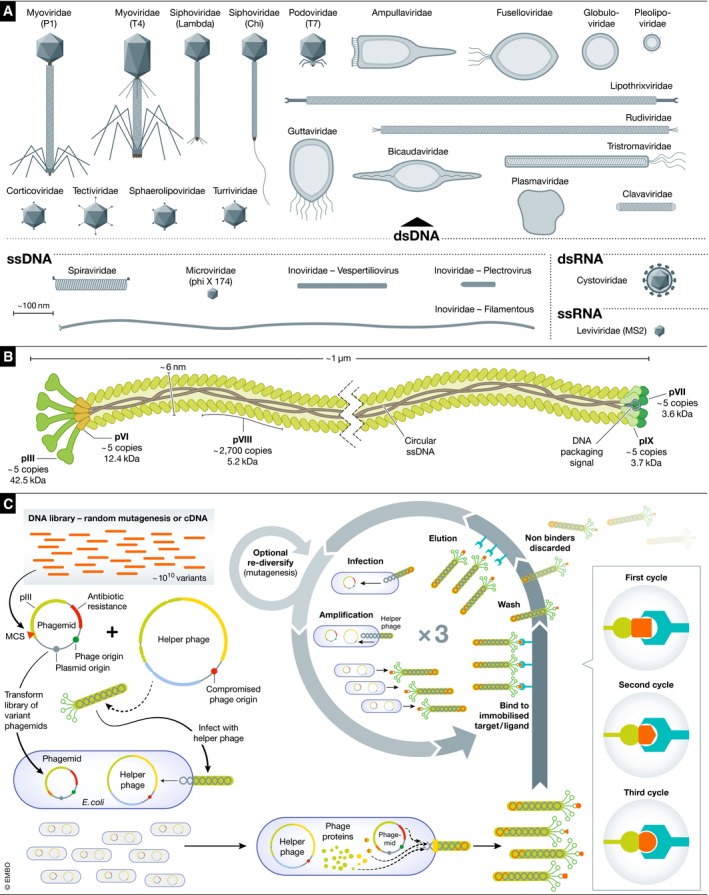
(A) Bacterial and Archaeal virus sub‐families are represented and grouped based on their Baltimore classification. Relative sizes and symmetries are approximate. (B) Schematic representation of the Escherichia coli Ff phage showing the overall architecture and copy number of the structural proteins. (C) In phage‐display screening, a natural or synthetic DNA library is cloned between the signal peptide and mature pIII‐encoding gene on a phagemid vector containing an Ff origin of replication, a plasmid origin of replication, and a selectable marker. The phagemid pool is transformed into E. coli infected with a helper phage containing a compromised Ff origin. The helper phage produces all the machinery required for phage replication and assembly, and the phagemid produces modified pIII capsids. Phages are assembled and secreted with a subpopulation of the pIII capsid proteins containing the insert. The phage library them undergoes multiple rounds of “panning”: (i) phages are applied to a matrix with immobilized ligand or target, those phages displaying peptides which bind to or are recognized by the target/ligand are bound while the non‐binding phages are washed away, and then, (ii) bound phages are eluted and used to infect E. coli cells, which are then pooled and infected with a helper phage to amplify the library and produce phages for a subsequent round of panning. Multiple cycles of panning can produce peptides with increased affinity.
Much within the toolbox of modern‐day molecular biology derived from researchers working towards an understanding of bacteriophage lambda, a member of the Siphoviridae family of non‐contractile tailed phages (reviewed in 5). Phage T7 is a member of the Podoviridae family (Fig 1A), and the “Sequenase” reagent that revolutionized DNA sequencing is a slightly modified form of phage T7 DNA polymerase 6, while the phage T7 RNA polymerase system has proved to be a workhorse for heterologous protein expression as well as enabling some of the first demonstrations and applications of in vitro transcription systems 7.
The family Microviridae includes the phage phiX174 (Fig 1A), famed as the first genome sequenced by Fred Sanger 8; as the model system Arthur Kornberg used to prove that DNA synthesized in vitro using purified enzymes generated a biologically active entity 9; and as the paradigm in which Craig Venter showed that a genome created from synthetic oligonucleotides was both necessary and sufficient to produce biologically active entities 10.
The family Leviviridae includes bacteriophage MS2 (Fig 1A), a minimalist virus that encodes only four proteins and which infects Escherichia coli. The MS2 coat protein in conjunction with the hairpin sequence derived from the MS2 operator sequence forms the basis of a revolutionary system for live‐cell imaging of specific RNAs in eukaryotic cells 11.
Four Siphoviridae prophages, φNM1 to φNM4, detected in the genome of the Gram‐positive bacterium Staphylococcus aureus 12, led to studies in phage‐based payload delivery: a small set of genes from the φNM1 phage were then used to create a packaging capsid for the delivery of a CRISPR/Cas9 system directed against S. aureus, and this phage therapy has been applied to kill MRSA, an antibiotic‐resistant form of S. aureus 13.
The family Inoviridae includes phage sub‐families with diverse structures and lifestyles (Fig 1A). One of these subgroups, the filamentous phages, includes species that can be longer than the bacteria that they infect, ranging in length from 800 nm to 4 μm (Fig 1B). Most of our understanding of the biology of filamentous phages comes from a group of closely related Escherichia coli phages called Ff filamentous phages. These phages, historically called M13, fd and f1, have 98% DNA sequence identity, and their replication mechanisms are identical. They were independently discovered in sewage samples in the early 1960s 14, 15, 16, 17, and they will here be collectively termed E. coli Ff phages. One characteristic feature of Ff phages (and most filamentous phages) is that they do not lyse the host. Indeed, when isolated, they do not form typical lytic plaques on bacterial lawns, but rather opaque zones of reduced growth. Much of what we know about the biology of filamentous phages comes from experiments on these Ff phages, but the Ff phages are by no means representative of the vast diversity observed in the filamentous phage family.
Ff phages gave rise to one of the early cloning vectors for DNA sequencing (M13 sequencing: reviewed by 18), and Ff served as the basis for phage‐display protein–protein interaction screening and the maturation of protein, peptide, antigen or antibody libraries. Several features of Ff facilitated its use as the basis for phage‐display technology: (i) assembly of these virions occurs without lysis of the E. coli host, allowing secretion of phage particles throughout the culture to the highest titres of any phage (up to 1013 virions per ml of culture); (ii) the capsid proteins are amenable to genetic fusion to other proteins, and thus the display of a foreign protein on the surface of the virion; and (iii) the Ff genome is small and easily modifiable, and most importantly, the modified genome is packaged into virions displaying corresponding modified capsid protein. This physical link between protein and corresponding DNA allows isolation of a desired protein/peptide/antigen/antibody from a library along with the gene encoding that specific variant 19, with sequential screening improving the affinity of the interactions (Fig 1C). In 2018, the Nobel Prize in Chemistry was awarded for advances in directed evolution, incorporating the work by Sir Gregory Winter and George Smith on phage‐display in the directed evolution of new proteins, particularly directed at the production of antibody‐based therapeutics 20, 21, 22.
Escherichia coli Ff: the archetypal filamentous phage
Structurally, the two best‐studied filamentous phage virions are E. coli Ff and the Pseudomonas Pf1 phages. Both are ~6 nm in diameter, with the Ff phage virion approximately 1 μm long and the Pf1 virion approximately 2 μm long. For point of reference, E. coli cells are 2–3 μm long. The Ff phage filament has a helical structure with a fivefold rotational axis with a twofold screw axis (C5S2) termed Class I, and Pf1 phages have a more complex structure (C1S5.4) termed Class II (see 23 and 24 for detailed reviews). It had been thought that filamentous phages with larger genomes have longer virions. However, as more and diverse virions are studied, this generalization seems to be overly simplistic. Ff and Pf1 have similar genome sizes (6,408 and 7,349 bp, respectively), but the Pf1 filament is more than double the length of Ff. The lumen of the Ff filament has more positive charges per capsid subunit, and modifications to charged residues in the lumen do indeed seem to alter the DNA packing and therefore the virion length 23, 24, 25, 26, 27, 28. This suggests that both size and physicochemical properties matter when it comes to packaging phage genomes. The Ff virion is made up of numerous copies of five different proteins (Fig 1B). The major capsid protein pVIII forms the body of the phage, and its copy number is dependent on the length of the genome: an Ff phage has approximately 2,700 copies of pVIII per virion, calculated from the known quaternary structure and the length of Ff phages. pVIII is a small, α‐helical protein, with the positively charged C‐terminus in the core of the virion.
The minor capsid proteins cap either terminus of the virion. pVII and pIX cap the leading (emergent) terminus of the nascent virion, while pIII and pVI cap the terminal end. In Ff phages, the minor capsid proteins are present in an equimolar stoichiometry 29 of five copies per virion as evidenced in labelling pIII with ZnS quantum dots 30. Taken together, each cap of the filamentous phage probably conforms to the fivefold symmetry of the major capsid filament—although there are no high‐resolution data to confirm this assumption. This arrangement is not universal among filamentous phages, with the Pseudomonas Pf3 phage having only four capsid proteins.
In the minor capsid proteins, the N‐terminal half of pIII forms two receptor‐binding domains termed N1 (TolA binding) and N2 (pilus binding) 31, while the C‐terminal domain interacts with the hydrophobic protein pVI. This pIII:pVI interaction is required for both the stability of the virion and the release of the nascent virion from the host cell 32, 33, 34. The proteins pVII and pIX are both small and hydrophobic, with predicted α‐helical structures. They form a protein complex which interacts with phage DNA, packaging the signal hairpin to initiate the assembly of the virion, and thereby forming a cap on the leading terminus of the nascent virion 35. The specific roles played by these proteins within the phage life cycle are elaborated later in this review.
Filamentous phages influence the virulence of bacterial pathogens
Whereas other types of phages are pathogens of their bacterial host, killing the bacterium during egress, filamentous phages infecting E. coli are episomal replicating phages that impose only a modest burden on the host. Thus, their relationship is more one of cooperation, with the host providing for delivery of phages throughout the environment. The biology of these ride‐share phages will be covered in detail in the final sections. To understand the benefits to the host bacterium, several well‐studied phage–host scenarios are considered where the phage requires relatively little from its host, yet contributes significantly to its virulence and therefore its evolutionary fitness.
Phages that make Vibrio capable of cholera
The filamentous phages of Vibrio have garnered great attention as they are intimately linked to the evolution of toxigenic strains of Vibrio cholerae. The major means by which cholera is caused by V. cholerae is secretion of the cholera toxin: an oligomeric protein encoded by two genes that are carried by the temperate bacteriophage CTX. Cholera toxin provides an advantage to the bacterial host, as it promotes profuse diarrhoea in humans which results in the dissemination of the pathogen. Infection of a non‐pathogenic strain with CTXφ results in a toxigenic strain 36, 37. Furthermore, many other Vibrio phages and prophages interact with the CTXφ phage to promote the horizontal transfer of the cholera toxin genes (reviewed in detail by 36). The host receptor for the CTXφ phage is the toxin‐coregulated (TCP) type IV bundle‐forming pilus, which itself is an important virulence factor. Controversially, the TCP pilus itself was previously described to be located on a large (40 kb) mobile element called VPI which was described as a filamentous prophage 38, but this was later shown to be incorrect; VPI is now regarded as an independent transmissible mobile genetic element, transferred via generalized transduction 39, 40, 41, 42. Recent assessment of prophages in diverse marine populations of Vibrio spp. suggests that filamentous prophages are numerous and widespread and play comprehensive roles in evolution through shaping niche adaptation and emergence of novel pathogenic strains emerging from environmental Vibrio communities 43, 44, 45, 46. Recently, a prophage with a similar genomic arrangement to CTXφ has been described in the human pathogen Acinetobacter baylyi (and A. baumannii) genome, though no homologous CTX toxin is present 47.
Phages that convince bacteria that altruistic death is a good thing
Pseudomonas aeruginosa harbours a group of related filamentous phages called the “Pf1‐like” phages: Pf1, Pf4, Pf5 Pf6 and Pf7. The Pf1‐like phages have been isolated from different strains of P. aeruginosa and are thought to be strain‐specific variants of an ancestral prophage, with specificity now due to strain‐specific variation of the type IV pili receptors 48, 49. These Pf1‐like prophages are widespread among P. aeruginosa strains 48 and contribute to various aspects of host cell physiology. During biofilm growth, Pf4 genes are among the most upregulated with biofilm growth leading to the release of 100–1,000 times more phage virions 50. This increase in expression of phage genes was also observed when P. aeruginosa was grown in anaerobic conditions mimicking a chronic late‐stage cystic fibrosis lung 51, suggesting a cooperative role in the maintenance of infection. Pf4 plays a critical role in the overall structure, organized remodelling and seeding of mature biofilms. Filamentous phages present in the biofilm matrix self‐organize into a viscous liquid‐crystal‐like arrangement providing the bacteria with increased surface attachment, and resistance to both desiccation and antibiotics 52. Amidst biofilm microcolonies, a spatially and temporally organized Pf4‐dependent cell death occurs leading to the remodelling and seeding of the biofilm.
Removal of the Pf4 prophage drastically reduces the development and stability of biofilm, which ultimately reduces virulence in mouse pneumonia model 53, 54. Furthermore, cells seeded from the Pf4‐dependent remodelling of biofilms showed increased tendency to form “small colony variants”, which showed higher levels of phage filaments on their surface and showed an increased attachment to surfaces and have been associated with pathogenicity 55.
The production of Pf4 has been linked to the maintenance of long‐term chronic infections typical of P. aeruginosa infection. In a murine pneumonia model, phage production promoted a chronic lung biofilm infection, reducing the bacterial invasion of the host epithelial cells, and reducing the host inflammation and immune response to the infection. Combined with the increased antibiotic resistances observed in Pf4‐producing biofilms, it seems the production of Pf4 by P. aeruginosa causes the bacteria to favour persistent infection over invasive infection which may be cleared by the animal host 52, 56.
Phages that infect plant pathogens
Given the global uncertainty around food security, there is a major push for understanding how phages can shape microbial communities to disfavour bacterial pathogens on crop plants. The Xanthomonas phage Xf infects the rice leaf blight pathogen Xanthomonas oryzae and was one of the earliest filamentous phages described 57. Beyond the initial physical description, little is known about this Xanthomonas phage. Other Xanthomonas phages, Cf, Cf1t/Cf1c and Cf16, were isolated from the citrus canker pathogen Xanthomonas citri in the 1980s. Cf1c was the first discovered example of a filamentous phage integrating into the host genome 58, 59, 60. The Lf phage was later isolated from cruciferous vegetable black rot pathogen Xanthomonas campestris and was used as a cloning vehicle for the integration of DNA into X. campestris. Recently, the complete genome sequence of a “UK variant” of the Lf phage has been deposited in NCBI (GenBank: MH206184.1), along with four new Xanthomonas phages: Lf2, isolated form Xanthomonas campestris pv. campestris; Xv2, isolated from Xanthomonas campestris pv. vesicatora; Xf190, isolated from X. oryzae; and Xf409, isolated from Xanthomonas oryzae pv. Oryzicola; only XF109 has been described in the literature 61.
Screening another agricultural pathogen, Ralstonia solanacearum, revealed phages that can be loosely grouped into two families: the RSM1 and related phages 62. These phages can integrate into their host genome and reside as prophages 63. The RSM1‐like phages (including RSM3, and Rs551) have been implicated in cooperatively trading away virulence to acquire drug resistance in a plant disease model. This the phage does by reducing Ralstonia's production of extracellular polysaccharide, reducing twitching motility, increasing cell aggregation and reducing the expression of key virulence genes 64, 65, 66, 67, 68, 69, 70. Given the phage modifies the host to ameliorate virulence, they have been proposed as biocontrol agents 64. RSS1 phages increase the virulence of their Ralstonia host in a plant disease model, by increasing extracellular polysaccharide production, twitching motility, and the expression of some virulence genes 71. It has been suggested that virulence‐enhancing RSS1 phages are actually “superinfective” forms of the non‐virulence‐enhancing RSS0 prophage 63. Prophage recombination on host chromosomes is common, and RS603 appears to be a hybrid phage with elements similar to the RSM1 and RSS1‐like phages 72, whereas RS611 seems to be a hybrid of RSS1 and RSS0 73.
Filamentous phages that impact marine microbial communities
In an era of climate change, where marine environments are under inarguable physical stress, filamentous phages isolated from marine bacterial species that shape their population structure are under intense investigation. The phage f327 was isolated from an arctic sea ice Pseudoalteromonas strain, and homologous phages to f327 are widespread through arctic sea ice. Carriage of the phage slows growth somewhat, but enhances the motility and chemotaxis of the host. This has been suggested as beneficial for the survival of the host in sea ice environments by preventing population “overblooming” during the relatively nutrient‐rich summer period 74. In the case of phage SW1 isolated from the deep‐sea bacterium Shewanella piezotolerans, the phage genes and virion secretion are induced at low temperatures and presence of the phage results in differential transcription of many genes in response to low temperature and high pressure including flagella genes which results in a reduced motility 75, 76, 77. A modified form of SW1 has been engineered into a low‐temperature inducible expression vector for biotechnology applications 78. Given the recent identification of filamentous phages from the hyperthermophile Thermus thermophilus 79, equivalent engineering could also generate high‐temperature expression vectors for biotechnology applications.
Diagnostic features in filamentous phage genome sequences
With a few key exceptions, the currently described filamentous phages have been isolated from a small handful of species from the gammaproteobacterial genera Escherichia (and Salmonella), Pseudomonas, Xanthomonas, Pseudoalteromonas, Yersinia, Shewanella and Stenotrophomonas; and from the betaproteobacterial genera Ralstonia, and Neisseria (Table 1). With the benefit of a substantial body of work that defines the structural and regulatory genes that contribute to filamentous phage biology, genome sequences can now be mapped and studied comparatively (Fig 2A).
Table 1.
Filamentous phages
| Phage | Host | Additional Information # | References |
|---|---|---|---|
| Ff | Escherichia coli | Inovirus ICTV type species. AKA M13, f1, fd. Host receptor—F “sex” pilus. Non‐integrative episomal replication. 900 × 6 nm | 195 |
| If1 | Escherichia coli | Unassigned genus. Host receptor—I pilus. Non‐integrative episomal replication. 900 × 6 nm | 196 |
| IKe | Salmonella typhimurium and Escherichia coli | Lineavirus ICTV type species. Host receptor—N or I2 pilus. Non‐integrative episomal replication. 1,000 × 6 nm | 197 198 199 |
| I22 | Escherichia coli | Lineavirus. Host receptor—N, I2 or P pilus. Non‐integrative episomal replication. 1,000 × 6 nm | 199 200 |
| CUS‐1 | Escherichia coli | Prophage. RF observed but no RF sequence available. Associated with pathogenic strains. Imparts slight fitness advantage to host during mammalian infection—unknown mechanism | 168 169 |
| Ypf/CUS‐2 | Yersinia pestis | Prophage. Very similar to CUS‐1. RF observed but no RF sequence available. Associated with pathogenic strains. Imparts slight fitness advantage to host during mammalian infection—unknown mechanism. 1,200 × 8 nm | 170 201 |
| CTXφ | Vibrio cholera | Unassigned genus. Host receptor—toxin‐coregulated type IV bundle‐forming pilus. Associated with pathogenic strains. Encodes cholera toxin A/B. Typically requires a satellite prophage or prophage duplication to produce infective phage virions. | 36 202 203 |
| RS1 | Vibrio cholera | Satellite prophage depends on CTXφ, KSF1 or VJG. | 36 204 205 |
| fs2 | Vibrio cholera | Saetivirus ICTV type species. Non‐integrative episomal replication. | 191 206 |
| TLC | Vibrio cholera | Satellite prophage depends on fs2. Promotes the integration of CTXφ. | 37 |
| VFJ | Vibrio cholera | Saetivirus. Non‐integrative episomal replication. Looks like a mosaic of fs2 and VEJ or VCY. Host shows inhibition of flagellum formation and had slightly increased antibiotic resistance through an unknown mechanism. 1,400 × 7 nm | 207 |
| VCY | Vibrio cholera | Unassigned genus. Integrates into host genome. Associated with environmental Vibrio isolates. 1,800 × 7 nm | 44 |
| KSF1 | Vibrio cholera | Unassigned genus. Host receptor—MSHA type IV pili. 1,200 × 7 nm | 208 |
| VfO3K6 | Vibrio parahaemolyticus | Unassigned genus. AKA f237 or pO3K6. Episomal replication. Associated with pathogenic strains. 2,500 × 8 nm | 74 209 |
| VfO4K68 | Vibrio parahaemolyticus | Unassigned genus. Derivative of VfO3K6 potentially carrying novel toxin gene. Episomal replication 1,300 × 6 nm | 74 |
| Vf33 | Vibrio parahaemolyticus | Unassigned genus. AKA Vf12. Integrates in to host DNA. 1,400 × 7 nm | 210 |
| fs1 | Vibrio cholera | Fibrovirus ICTV type species. Host receptor—MSHA type IV pilus. Integrates into host genome. 1,000–1,200 × 7 nm | 191 211 |
| VSK | Vibrio cholera | Considered a variant of fs1 | 191 |
| VSKK | Vibrio cholera | Considered a variant of fs1 | 191 |
| VEJ | Vibrio cholera | Considered a variant of fs1. Host receptor—MSHA type IV pilus. Recombination with CTXφ allows horizontal transfer of cholera toxin genes. | 184 191 |
| VGJ | Vibrio cholera | Fibrovirus. Host receptor—MSHA type IV pilus. Integrates into host genome. Recombination with CTXφ allows horizontal transfer of cholera toxin genes. 1,000 × 7 nm | 89 185 |
| Pf1 | Pseudomonas aeruginosa (PAK) | Unassigned genus. Episomal replication Host receptor—PAK type IV pili. Virion inhibits Candida albicans growth via sequestering iron. 2,000 × 6 nm | 91 212 213 |
| Pf3 | Pseudomonas aeruginosa | Unassigned genus. Episomal replication. Host receptor—RP4 conjugative pilus. Non‐integrative episomal replication. 700 × 6 nm | 91 214 |
| Pf4 | Pseudomonas aeruginosa (PAO1) | Prophage. Integrates into host genome. RF observed but no RF sequence available. Implicated in host virulence via biofilm remodelling and dispersal mediated by host cell death; and the formation of virulent small colony variants (SCV). Virion inhibits Aspergillus fumigatus metabolism and Candida albicans growth via sequestering iron. Predicted length 37,000 × 6 nm | 53 55 213 215 |
| Pf5 | Pseudomonas aeruginosa (PA14) | Prophage. Integrates into host genome. RF observed but no sequence available. | 216 |
| Pf6 | Pseudomonas aeruginosa (PAO1‐MPAO1) | Pf4 variant from the PAO1‐MPAO1 strain. Inserted at different locus to Pf4 and containing two additional genes encoding protein kinases. AKA RGP42. | 49 217 |
| RSM1 | Ralstonia solanacearum | Habenivirus ICTV type species. Host receptor—probably type IV pili. Integrates into host genome. Decreases host virulence. Increases host cell aggregation. 1,400 × 10 nm | 68 69 70 178 191 |
| RSM3 | Ralstonia solanacearum | Habenivirus. Host receptor—probably type IV pili. Integrates into host genome. Decreases host virulence, growth rate, extracellular polysaccharide production, motility, and expression of some virulence genes. Increases host cell aggregation and antibiotic resistance. Proposed biocontrol agent. | 64 65 68 |
| RS603 | Ralstonia solanacearum | Habenivirus. Only RF episomal form described (lacks integrase from RSM1/3). Appears to be a hybrid of RSM1/3 and RSS1/0. 1,120 × 8 nm | 72 |
| RS551 | Ralstonia solanacearum | Not classified (probably Habenivirus). Decreases host virulence, extracellular polysaccharide production, motility. Integrates into host genome. 1,200 × 7 nm | 66 67 |
| RSS1 | Ralstonia solanacearum | Unassigned genus. Host receptor—probably type IV pili. Increases host virulence, extracellular polysaccharide production, motility and expression of some virulence genes. May be an episomal “superinfective” form of RSS0. 1,100 × 10 nm | 68 69 70 71 178 |
| RSS0 | Ralstonia solanacearum | Not classified. Very similar to RSS1 with additional ORF encoding potential DNA‐binding regulator and an attP site. | 62 |
| RS611 | Ralstonia solanacearum | Not classified. Appears to be a hybrid of RSS1 and RSS0 with a deletion of two ORFs. 1,120 × 8 nm | 73 |
| p12J | Ralstonia pickettii | Not classified. Unclear if the deposited sequence is a phage or prophage sequence | 218 |
| PE226 | Ralstonia solanacearum | Not classified. Only RF episomal form described. 1,050 × 6–9 nm | 87 |
| Xf109 | Xanthomonas oryzae | Unassigned genus. Integrates into host genome. 1,210 × 8 nm | 61 |
| Xf409 | Xanthomonas oryzae | Not classified. similar to Xf109 | * |
| Lf | Xanthomonas campestris | Not classified. Complete genome of “UK variant” available. Suggested to be integrative, though not conclusively demonstrated. 1,000 × 8 nm | 219 |
| Lf2 | Xanthomonas campestris | Not classified | * |
| Xv2 | Xanthomonas campestris | Not classified | * |
| Xf | Xanthomonas oryzae | Not classified. No sequence information. 977 × 8 nm | 57 |
| Cf | Xanthomonas citri | Not classified. No sequence information. 1,000 nm long | 220 |
| Cf1t | Xanthomonas citri | Not classified. Similar to Cf. Integrates into host genome | 58 59 |
| Cf1c | Xanthomonas citri | Unassigned genus. Variant of Cf1t. Forms clear plaques. Sequence available. | 221 |
| Cf16 | Xanthomonas citri | Not classified. Integrates into host genome | 222 |
| XacF1 | Xanthomonas citri | Not classified. Integrates into host genome. Lowers host EPS production, motility, and growth. Host shows reduced virulence in plant disease model. 600 nm long | 223 224 |
| MDAφ | Neisseria meningitidis | Not classified. AKA Nf1‐A. Host receptor—probably type IV pili. Integrates into host genome. Presence of prophage correlates with hypervirulent invasive strains. Increases bacterial host attachment to epithelial cells. 1,200 nm long | 177 225 |
| Nf1 | Neisseria meningitidis | Not classified. Prophage | 171 |
| Nf2 | Neisseria meningitidis | Not classified. Prophage | 171 |
| Nf3 | Neisseria meningitidis | Not classified. Prophage | 171 |
| Nf4 | Neisseria gonorrhoeae | Not classified. Prophage | 171 |
| Ngo6 | Neisseria gonorrhoeae | Not classified. Virus derived from synthetic phagemid containing the Nf4‐G2 prophage. Reported to infect diverse proteobacterial species. | 226 |
| CRA | Acinetobacter baylyi | Not classified. Prophage. RF observed but no RF sequence available. Host receptor—probably competency pilus. Phage inhibits natural competency of cells | 47 |
| SHP1 | Stenotrophomonas maltophilia | Unassigned genus. AKA PSH1. Only episomal RF reported. 2,100 × 15 nm | 227 |
| SHP2 | Stenotrophomonas maltophilia | Not classified. Only episomal RF reported. 800 × 10 nm | 228 |
| SMA6 | Stenotrophomonas maltophilia | Unassigned genus. Integrates into host genome | 229 |
| SMA7 | Stenotrophomonas maltophilia | Unassigned genus. Integrates into host genome | 229 |
| SMA9 | Stenotrophomonas maltophilia | Unassigned genus | 88 |
| f327 | Pseudoalteromonas sp. BSi20327 | Not classified. AKA pSM327. Only RF described. Decreases host growth rate. Increases motility and chemotaxis. Widely distributed in arctic sea ice samples. 1,500 × 14 nm | 230 |
| SW1 | Shewanella piezotolerans | Not classified. Integrates into genome. Phage replication and genes expression induced at low temperatures. Seems to have a role in flagella regulation. | 76 77 231 |
| OH3 | Thermus thermophilus | Unassigned genus. Only RF episomal observed. 830 × 8 nm | 79 |
| OH16 | Thermus thermophilus | Not classified. Like OH3 but with an additional transposase. Only genome sequence—no description | * |
| PH75 | Thermus thermophilus | Not classified. Only protein sequence of major capsid protein reported. | 232 |
| B5 | Propionibacterium freudenreichii | Unassigned genus. Only RF episomal form described 620 × 12 nm | 233 |
| CAK1 | Clostridium beijerinckii | Not classified. Only RF observed. Infectivity not demonstrated. No genome sequence. 1,000 × 5–8 nm | 234 |
| NP‐2014 | Environmental | DNA sequenced during human virion project from amniotic fluid—classed as “Ralstonia phage” though no rational for this naming is given | * |
| WW‐nAnB | Environmental | DNA isolated from raw sewage. Previously detected in faecal samples but incorrectly described as “non‐A, non‐B hepatitis” | 235 |
This should not be considered a complete list of filamentous phages/prophages. * No literature publicly available, only genome sequence available. # Virion sizes are as reported in relative literature. Different methodologies may result in difference in measurements (particularly with respect to the width measurements).
Figure 2. Diversity of filamentous phage genomes.

(A) Schematic representation of filamentous phage genomes: for each gene identified in the genome, the putative function is noted either based on experimental evidence, inferred from sequence homology, or based on conserved domain predictions. Scale bare represents genome size in nucleotide base pairs. (B) Protein sequence similarity network plot of all predicted open‐reading frames from 56 filamentous phage genomes. The great proportion of orphan proteins in this plot demonstrates that at the protein sequence level, there is a very high degree of diversity in filamentous phages. Each circle node represents a sequence, and each connecting line represents a BLAST score better than 1e‐5. Identical proteins are collapsed into one circle with the size representing the number of proteins denoted. Representative species are coloured as shown, and the identity of the Ff proteins is annotated in the plot.
Across the various filamentous phage lineages, only the pI proteins stand out as being highly conserved and fully diagnostic (Fig 2B). The pI proteins have a conserved Zot domain (Pfam PF05707 Zot domain) at their N‐terminus. This domain was named for the pI homologue in the Vibrio CTXφ phage called the Zonula Occludens Toxin, which is essential for the assembly and export of CTXφ phage virions, and has been implicated in the virulence of pathogenic V. cholerae strains by increasing intestinal permeability through binding to tight junctions (zonula occludens) between small intestine epithelial cells 80, 81. While the toxin itself carries the conserved Zot domain in its N‐terminus, the Zot domain is not the toxic component: the C‐terminal domain is cleaved from the Zot protein, and it is this non‐conserved, C‐terminal fragment that intoxicates the human epithelial cells 82, 83, 84, 85, 86. Unfortunately, the “Zonula Occludens Toxin” automated annotation that has now attached itself to entries for most filamentous phages has resulted in incorrect assumptions about toxin activity in newly annotated phage genomes 87, 88.
Phylogenetic analysis of the conserved pI proteins shows that they provide a basis for phage classification (Fig 3). The phylogeny also demonstrates that based on the current ICTV threshold for the classification of filamentous phage genera, there are many other clades that could be subject to future classification. In the simplest sense, grouping filamentous phages based on their pI proteins leads to distinct clades. For example, the Escherichia phages Ff, I22, IKe and If1, the Vibrio phages fs2 and VFJ phages group together as a single clade. This clade contains the phage genera Saetivirus, Inovirus and Lineavirus, and all the members are non‐integrative episomal phages. A second clade is formed by the Vibrio VfO36K/f237, VCY and KSF1 on one branch and a group containing the Pseudomonas “Pf1‐like” phages (Pf1, Pf4, Pf5) and the Vibrio CTXφ phage on another branch. All the Neisseria prophages are in a large diverse third clade with the Xanthomonas Cf1c‐like and Xf109/Xf409 phages; the Stenotrophomonas phages SMA6, SHP2 and PSH1; the Ralstonia RSS1 family phages (RSS1, RSS0, p12J, PE226); and the Pseudomonas episomal phage Pf3. A fourth clade is formed with the Ralstonia “RSM1‐like” phages (RSM1, RSM3, RS603) of the genera Habenivirus, the Stenotrophomonas phages SMA7 and SMA9 and the Xanthomonas phages Lf and Lf2 on one branch, and the Yersinia/Escherichia CUS phages (CUS‐1, Ypf/CUS‐2) on a separate branch. A clade encompassing the Fibrovirus genus contains the Vibrio phages fs1, VSK, VSKK, VEJ, VGJ and Vf33, as well as the Shewanella phage SW1. A more distantly related branch contains the Thermus phages OH3 and OH16 and the Gram‐positive Propionibacterium phage B5.
Figure 3. Phylogenetic tree of filamentous phages.
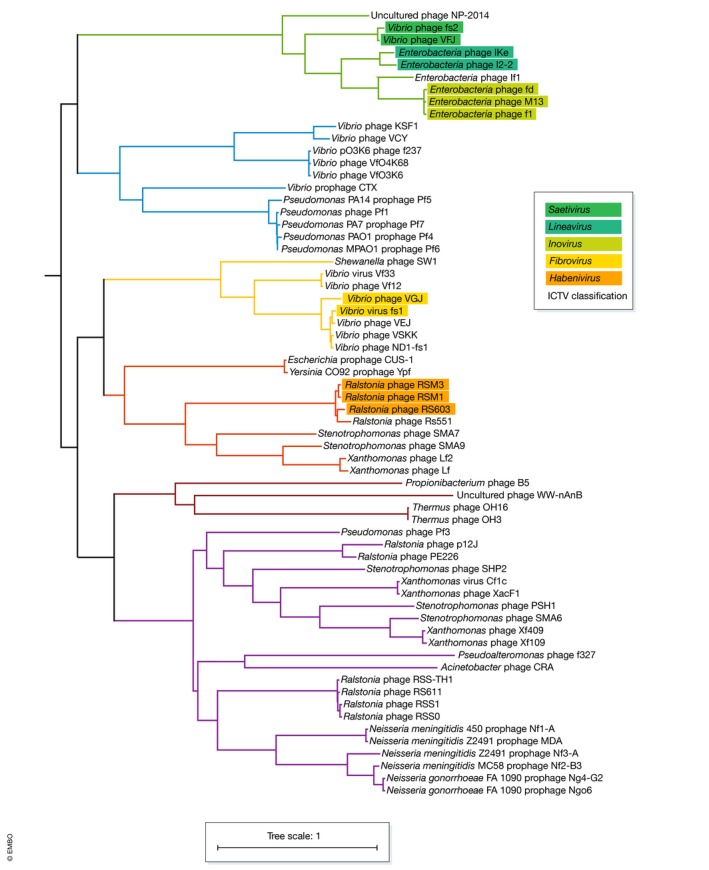
Phylogenetic tree built of the conserved pI homologues of known filamentous phages and prophages. Alignments were calculated with mafft generated (L‐INS‐i option), and sites for tree inference chosen using trimal (automated1). The tree was calculated with RAxML “PROTGAMMAAUTO” criteria (final model LG) and “autoMRE” bootstrap convergence test and midpoint rooted 236, 237, 238. Clades are coloured as described in the text, and leaves are coloured based on their ICTV genera classification.
Filamentous phage life cycle: infection
Almost all of the described filamentous phages infect Gram‐negative hosts, and thus must traverse two membrane barriers. While the cell surface receptor is unknown for the majority of described filamentous phages, where it is known the receptor has been shown to be a pilus of some description. For example, Ff and related phages use the F sex/conjugation pilus (Fig 4A); various Vibrio spp. phages use either the toxin‐coregulated type 4 pili or the mannose‐sensitive haemagglutinin type 4 pili; Pseudomonas phages Pf1 and Pf3 use the PAK type 4 pili and conjugative RP4 pilus, respectively; Xanthomonas phage Cf uses an uncharacterized type 4 pilus; and Acinetobacter phage CRAφ utilizes a competence pilus 47, 89, 90, 91, 92, 93. While commonly referred to as “pili”, the F‐pilus and the various type 4 pili are evolutionally, mechanistically and structurally unrelated; the common feature important for phage entry into its bacterial host seems to be simply the retractile function of the pilus.
Figure 4. Lifecycle of the archetypical filamentous Ff phage.
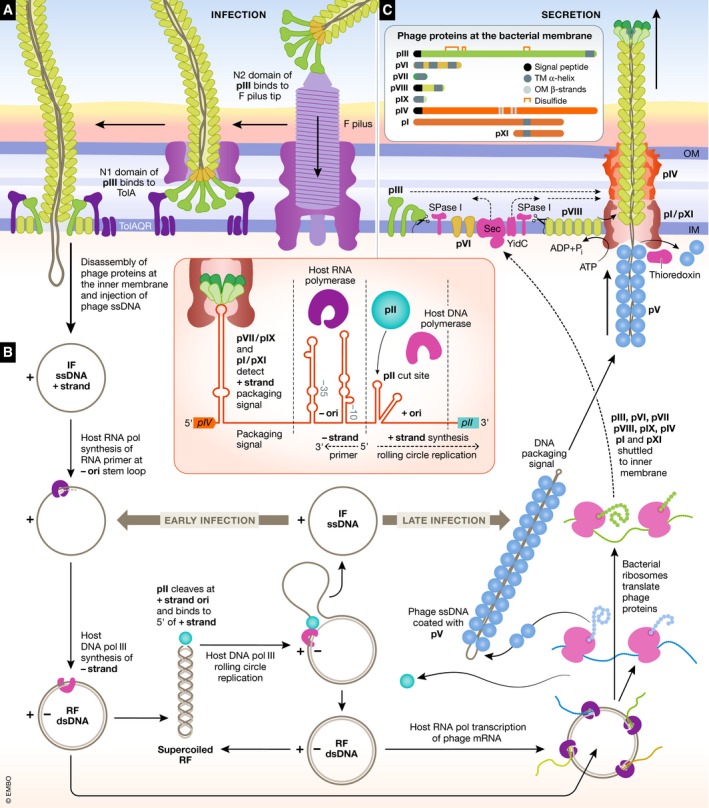
(A) In the initial stage of phage attachment, the N2 domain of pIII binds to the tip of the F‐pilus on the surface of the bacterial cell. Upon F‐pilus retraction, the pIII/pVI terminus of the phage would be brought into periplasm of the host cell. The N1 domain of pIII binds to the host protein TolA in the TolQRA complex in the inner membrane. The next stage, which has not been characterized, would need to result in phage disassembly and injection into the cytoplasm of the ssDNA genome termed the “infective form” (IF). (B) Phage replication ensues through recruitment of the host RNA polymerase to a hairpin at the negative (−) origin of replication, resulting in synthesis of a short RNA primer. The positive (+) strand is then extended by the host DNA polymerase III, generating a double‐stranded phage genome termed the “replicative form” (RF). Early in the infection, this can serve as a template for host RNA polymerase to generate phage mRNA, to be translated into phage proteins. The phage protein pII binds to the + origin of replication and nicks the + strand, and the resulting 3′ end is extended by host DNA polymerase III displacing the “old” + strand. Upon one full cycle, pII cleaves and ligates the + strands resulting in a single‐stranded IF and a double‐stranded RF. The RF can then undergo multiple rounds of rolling circle replication to replicate the phage genome and also serve as a template for transcription and translation of phage proteins. Later in the infection, single‐stranded IF is coated by phage protein pV, leaving the packaging signal‐free in preparation for secretion. (inset) A schematic representation of the phage intergenic region containing the packaging signal, the – origin of replication and the + origin of replication is shown. (C) Structural phage proteins and phage proteins required for assembly and secretion are shuttled to the inner membrane and processed by the SecYEG, YidC and signal peptidase machinery. The packaging signal hairpin of the pV‐coated ssDNA is bound by the minor capsid proteins pVII and pIX and recognized by the pI/pXI IM assembly proteins. As the emerging ssDNA traverses the inner membrane, pV is removed and replaced by the membrane‐embedded major capsid protein pVIII. As pVIII is added to the emerging phage, the tip is forced outwards through the oligomeric secretin‐gated channel pIV. The terminal phage capsid proteins pIII and pVI detect and cap the end of the nascent phage allowing its release from the host cell. Host proteins are represented as various shades of purple. Phage proteins involved in DNA replication and packaging are represented by shades of blue. Phage proteins involved in secretion are shown as shades of orange. Structural phage proteins are shown as shades of green. (inset) Phage proteins which interact with the bacterial membranes are shown. Topogenic signal peptides and transmembrane regions are annotated.
Docking onto the host cell pilus is mediated through the pIII protein (Fig 4A). Electron microscopy observations suggest that the phages bind to the tip of the pilus, in a tip‐to‐tip fashion 94. This has been experimentally confirmed in the case of CTXφ, where pIII binding to the pilus tip subunit (called the minor pilin, TcpB) of the toxin‐coregulated type 4 pilus in Vibrio cholerae has been directly demonstrated 95. It is speculated that binding of the phage to the pilus induces signalling to retract the pilus, bringing the phage towards the host cell surface 96. Upon pilus retraction, the pIII containing terminus of the virion is thereby brought into the periplasmic space, to engage a secondary receptor in the periplasm of the host. It seems most likely that this entry event would be through the pilus pore, and the spatial constraints of the pore would allow this. In the cases investigated so far, the secondary receptor is always the inner‐membrane‐anchored protein TolA, which extends out into the periplasm (Fig 4A). TolA is a component of the TolQRA complex, a nanomachine element in the “Tol‐Pal” system that controls membrane integrity and invagination during cell division 97, 98, 99, 100. While this primary function of TolA may not be relevant to phage entry, the primary function of TolA is essential; thus, the TolQRA secondary receptor is highly conserved, hence ever available for filamentous phage infection 101.
Upon pilus binding and retraction, the N2 domain of protein pIII appears to have a crucial role in assisting the infection process. An experiment where the N2 domain of pIII was recombinantly expressed in host bacteria prevented F‐pilus extension and locked the pilus in a retracted state through an unknown mechanism 102, and it has been proposed that this allows the phage to securely traverse the outer membrane and disassemble into the inner membrane without F‐pilus extension or subsequent infection with other phages interfering with the process 19. Once the N1 domain of pIII is uncovered and brought into the periplasm via the retraction of the pilus, it binds to the C‐terminal domain “III” of TolA. Beyond the pIII‐TolA binding event, very little is known of subsequent infection steps or how the virion DNA traverses the host cell inner membrane. The extreme C‐terminus of pIII is predicted to contain a transmembrane α‐helix and two short amphipathic α‐helices which are essential for phage infection, and pIII has been shown to have pore‐forming properties in artificial lipid bilayers 103, suggesting that the C‐terminus of pIII inserts into the membrane creating a pore that could allow phage DNA access to the host cytoplasm. The major capsid proteins end up embedded in the inner membrane, with their N‐termini on the periplasmic side of the membrane, potentially reused for packaging new phage particles 104, 105, 106, suggesting there is some kind of ordered disassembly of the virion capsid at the inner membrane that drives the phage DNA into the host cytoplasm.
Filamentous phage life cycle: genome replication
For Ff phages, episomal replication of the ssDNA genome (Fig 4B) is a well‐characterized process and early experiments on Ff genome replication provided seminal information for our current understanding of rolling circle DNA replication, a mechanism relevant to understanding bacterial plasmid replication, the amplification of various virial genomes, and the replication of mitochondrial DNA in at least some species of eukaryotes 107, 108.
As shown in Fig 4B, the filamentous phage genome is injected into the host cell cytoplasm as single‐stranded circular DNA referred to as the infective form (IF). Replication of the phage genome is largely controlled by the intergenic sequence (IG) located between the gIV and gII genes, which contain the + and – strand origins of replication and the packaging signal that all form double‐stranded hairpins in the ssDNA genome. Filamentous phage genome replication is entirely dependent on the core bacterial DNA replication machinery. Upon entry, the host RNA polymerase σ70 holoenzyme binds to the – strand origin hairpin, which mimics a bacterial −35 and −10 promoter sequence, with an affinity much higher than a typical bacterial promoter 109, 110. RNA polymerase begins to synthesize RNA on the ssDNA template but stalls and backtracks at a section of the genome and dissociates leaving a short RNA primer (18–20 nt long) which is extended by the host DNA polymerase III holoenzyme to generate the – strand of the genome and the double‐stranded replicative form (RF) of the phage genome 111, 112. The RF can also be synthesized from other regions of the genome but with drastically lowered efficiency 113. The RF is further processed by the host gyrase to form a supercoiled RF 114, 115.
There are three crucial aspects to the initial function of the RF. Firstly, it serves as a template for the transcription of the initial mRNA transcripts encoding the phage proteins including pII and pX, which are required for the amplification of the phage genome. Secondly, it serves as a template for the replication of the RF. Thirdly, it serves as a template for replication of the IF. Amplification of the RF and IF through rolling circle replication is mediated by the phage protein pII.
Protein pII is a strand transferase which binds to the newly synthesized supercoiled RF at the + strand origin of replication, cleaves the + strand and attaches to the 5′ end 115, 116, 117. The free 3′ end can now serve as a primer for the host DNA polymerase holoenzyme to synthesize a new + strand, displacing the original + strand as it progresses. In Ff, these pII‐mediated steps are also dependent on the host replicative Rep helicase and integration host factor, whereas some Vibrio and Pseudomonas Pf phages use the hosts alternate DNA repair helicase UvrD and the histone‐like HU proteins 118, 119, 120. Once the replication completes a full circle, pII cleaves and cyclizes the free ends resulting in a ssDNA IF and dsDNA RF 116. Early in the infection cycle, the IF is converted into new RF, as described above, until approximately 50 RF copies are present in the host 121, whereas the RF can serve as a template for further rolling circle replication generated IFs and as a template for the transcription of phage mRNA transcripts. As the number of RFs templated for transcription increases, so too does the number of phage proteins present in the host cell. Late in the infection when the level of pV protein reaches a critical number, it forms dimers and binds at the bottom of the hairpin formed by the packaging signal in the IF DNA and begins to coat the entire length (except the packaging signal) of the DNA, with one antiparallel ssDNA strand bound by each side of the dimer forming a long helical DNA–protein complex containing approximately 1,600 copies of pV 122, 123. pV also plays a role in timing the infection cycle in the host and coordinating the level of IF for packaging. Late in the infection, the increasing levels of pV directly inhibit both the synthesis of the negative strand and the translation of the pII and pX proteins, which results in the accumulation of IF DNA 124, 125. Although pII inhibition is dispensable for a successful infection cycle 126, pX protein—which is identical to the C‐terminal third of pII and translated from an internal start codon within the pII gene—appears to play an additional, though unclear, regulatory role in the levels ration of IF and RF 127, 128.
Despite this seemingly costly exercise in DNA replication by the phage, it really is a cooperative use of host resources. Most filamentous phages are produced rapidly in an initial stage lasting less than 10 host cell generations, with each bacterial cell producing around 200 phages per generation, after which time the host settles into a stable state where the phage genome is only replicated at very low levels and very few phages are produced 121. By contrast, within a few minutes of infection the classic E. coli tailed phage T4 converts the host cell into a factory, with the sole purpose of producing phage particles. The T4 genome contains more than 270 genes, with many of them encoding the machinery required for the hostile takeover of the host cell. T4 immediately inhibits host DNA replication, transcription and translation and completely remodels the host metabolism to favour phage production. The host DNA is degraded and recycled into phage DNA. Within 20–30 min, the host is actively lysed and 100–200 T4 virions are released (for a review of T4 host interactions, see 129).
Filamentous phage life cycle: phage egress
In contrast to tailed phages, the physical dimensions of filamentous phages would presumably prevent their assembly within the cell; as such, the virion is assembled at the bacterial cell envelope, with the maturing phage actively secreted through the cell envelope in a non‐lytic manner. The single‐stranded DNA genome appears to lack any significant Watson–Crick base pairing apart from a hairpin at one terminus, called the packaging signal 24. The newly replicated ssDNA IF genome is coated with the DNA‐binding protein pV in the cytoplasm, and this serves to stabilize and expose the “packaging signal” (Fig 4B and C) that will target the DNA–protein complex to the inner membrane of the host cell (Fig 4C). The trans‐envelope export complex is comprised of two phage proteins: pI and pXI (pXI is translated from an internal start site in the pI gene and is identical to the transmembrane and C‐terminal periplasmic third of pI). These two phage proteins, pI and pXI, each have a single transmembrane domain to anchor them to the host cell inner membrane, and thereby form one half of a secretion complex, equivalent but not homologous to a bacterial type 2 secretion system. Intriguingly, the translocation channel across the outer membrane utilized by filamentous phages is a protein of the secretin protein family, referred to as pIV (Fig 2B), with the secretin protein family also forming the exit channel of type 4 pili as well as type 2 secretion systems. The phage secretion machinery is remarkable in its apparent simplicity; whereas the T2SS and type 4 pili require many periplasmic or envelope proteins to assemble their pilus, the filamentous phage secretion machinery assembles a more complex DNA–protein hybrid filament with only two (or three) proteins.
The pI protein is embedded in the membrane by a signal anchor domain, leaving the N‐terminal ~250 residues in the bacterial cytoplasm and the C‐terminal ~80 residues in the periplasm, and the signal anchor domain of pXI results in the remainder of the protein exposed to the periplasm. Being (initially) signal‐anchored proteins, both pI and pXI require the bacterial Sec machinery for insertion into the membrane 130. The functional benefit of pXI to the phage is unclear, and while both pI and pXI are essential for Ff phage export 131, 132, the internal start site generating protein pXI is not a conserved feature. The cytoplasmic domain of pI is predicted to act as an ATPase, powering the assembly and transport of the phages through the envelope. This is supported by the finding that Ff phage assembly is dependent on ATP hydrolysis 133.
pI and pXI form a complex with the outer membrane protein pIV in the absence of other phage proteins or DNA, suggesting that this is a genuine secretion nanomachine representing a pre‐initiation step of phage morphogenesis 134. Like other secretins, pIV resides in the outer membrane 130, though the mechanism of secretin assembly into the outer membrane remains unknown 135. Low‐resolution cryo‐electron microscopy of purified pIV showed that the protein forms ring‐like structures with an outer ring diameter of approximately 13.5 nm and an inner (channel) diameter of approximately 6 nm. This channel diameter would be just sufficient to allow passage of the filamentous phage particle, which has a diameter of 6 nm. Although no clear symmetry could be observed, nanogold labelling suggested that there are 14 subunits per secretin complex 136. From recent near‐atomic resolution cryo‐electron microscopy models of secretins from the type 2 secretion system 137, 138, 139 and previous mutagenesis experiments on the pIV gate regions 140, we can extrapolate some information about the structure and function of pIV. Secretins form homo‐oligomeric complexes comprised of 15 copies of the secretin subunit. The outer membrane channel is formed by extended β‐strands (four from each subunit) with two β‐hairpins folded upwards into the lumen of the barrel to form a β‐barrel cone forming an internal gate. A recent structure of the T3SS with the gates in an open state showed that the two β‐hairpins forming the internal gate straighten and move approximately 40 Å upwards against the outer β‐barrel 141; these are the same regions identified as “leaky” gate mutants in a mutagenesis screen of pIV 140. Thus, it is plausible that the pIV secretin gate would open in a similar fashion during phage filament extrusion.
While some phages encode their own secretin (pIV), most do not (Fig 2B). Nonetheless, it seems likely that secretin‐mediated egress is a common feature for the filamentous phages, given two well‐studied cases where phages were shown to share the host cell secretion for their own travel out of the bacterial cell. When CTXφ is secreted from V. cholerae, it uses the endogenous T2SS secretin EspD for phage secretion, and this process is independent of the T2SS inner membrane machinery 142, 143. When MDAφ infects N. meningitidis, it uses the endogenous type 4 pilus secretin PilQ for phage secretion 144. It is assumed that the majority of other filamentous phages lacking a dedicated secretin use a similar mechanism. This ride sharing by filamentous phages contrasts with the process of host cell protein secretion, wherein secretins normally require highly organized interactions with their cognate inner membrane machinery 145, 146, 147. This suggests that evolution has driven phage pI proteins to present periplasmic domains that mimic and/or displace secretin‐binding domains of the cognate bacterial secretion systems.
The Ff structural proteins are initially integrated in to the bacterial IM prior to assembly. In a past era of pioneering work on how bacteria target and assemble membrane proteins, filamentous phage coat proteins were used as models and helped drive our understanding of membrane protein biogenesis in E. coli 148, 149, 150, 151, 152. The “procoat protein” pVIII has a signal sequence to engage with the Sec/YidC machinery of the host cell, and was therefore used as a model protein to dissect the role of the targeting pathways and membrane translocation events in E. coli 149, 153, 154. The phage proteins also have sequences predicted as transmembrane domains; initially perplexing, since the phage contains no lipids, it is now clear that the coat proteins use a mode of integration into the inner membrane in order to coalesce together and to co‐translocationally displace the pV (DNA‐binding protein) from the ssDNA, and thereby coat the DNA to create the filamentous phage capsid 23. How the coat proteins are extracted out of the inner membrane during virion assembly is unknown, but an active area of investigation (see also Box 1).
Box 1: In need of answers.
The structure of both termini of the Ff virion—Although various methods have resolved the super structure of the virion shaft and several crystal structures of domains of the minor capsid pIII exist, the tertiary and quaternary structures of the pVII:pIX and pIII:pVI caps and how they interface with the helical shaft remain unknown. Modern advances in electron microscopy may lead to advanced in our understanding of the virion structure.
Assembly of the virion—How do the capsid proteins transition from their inner membrane embedded to their structural virion forms? How are they extracted from the membrane, and what initiates the assembly? Is the assembly reaction actively driven by the pI ATPase, akin to the pilus of the type 4 pili?
Egress through the bacterial OM—do all filamentous phages use secretins for secretion across the OM? Do those lacking an endogenous OM secretin pores all hijack the bacterial secretion systems to exit the cells as seen in CTXφ and the T2SS in V. cholerae?
The “origin” of filamentous phages and bacterial secretins—The origin of filamentous phages is directly linked to the evolution of secretins and thus the T2SS, T3SS and the T4P. So, are filamentous phages the progenitors of bacterial secretin systems or simply hijacking established systems?
How widespread are filamentous phages in nature? In the current review, we have provided evidence suggesting filamentous prophages are distributed widely throughout the bacterial and archaeal kingdoms, though only very few have been experimentally validated. A more systematic approach to identifying filamentous phages and prophages both experimentally and in bacterial and metagenomic sequence data will elucidate our understanding of these fascinating systems.
Filamentous phage assembly is initiated by the minor capsid proteins pVII and pIX which are small hydrophobic proteins, integrated into the bacterial inner membrane. These proteins bind to the exposed packaging signal of the phage DNA, thereby forming the cap of the virion to initiate assembly of the emergent phage 35, 155, 156, 157. During the early stages of phage infection, the major capsid protein pVIII becomes one of the most abundant proteins in the cytoplasm with upwards of 4 million copies per cell 121. The protein may (e.g. E. coli) or may not (e.g. P. aeruginosa) contain an N‐terminal signal peptide, but its integration into the inner membrane is mediated by the YidC translocon 158, 159, 160, 161. The signal peptide of pVIII is cleaved by the host cell signal peptidase, leaving the membrane‐embedded capsid with its N‐terminus in the periplasm and the C‐terminus in the cytosol 149, 150, 162. Accumulation of the local concentration of pVIII capsid protein in the inner membrane leads to protein oligomerization 163, 164 driving phage filament assembly (Fig 4C). Precisely how the assembly of the coat protein subunits is mediated is unknown, but models for the transition of membrane‐embedded pVIII into virion filaments have been proposed and reviewed recently 23, 165.
Once the entirety of the phage DNA is coated in the major capsid protein, it must be released from the inner membrane and capped with the minor capsid proteins pIII and pVI, which form a stable complex at the base of the virion 166. If pIII of pVI is deleted, the virion cannot be released from the host cell and the filament continues to grow integrating multiple copies of the phage DNA into the growing filament 32. As well as being responsible for host cell receptor binding and injections of the phage DNA during the infection process, pIII plays a key role in the release of mature phages. pIII is targeted to the inner membrane via its N‐terminal signal peptide, which is then cleaved leaving pIII embedded in the membrane via a C‐terminal transmembrane helix 33, 167. The C‐terminal domain of pIII is implicated in binding to and thereby releasing the newly assembled phages. It is thought significant rearrangements must occur in pIII to achieve this, but how the pIII/pVI capping complex detects the end of the phage and induces this final release of the virion is not known 32.
Prophages and discovery science
Many filamentous phages can integrate into the host chromosome and be replicated along with the bacterial genome during cell division. In addition to providing clues as to the details of phage–host interactions, this feature means that filamentous phages can be discovered through genome and metagenome surveys of diverse environments. Filamentous prophage integration (Fig 5 and Table 2) can be mediated by one of two methods: using host recombinases XerC and XerD or using a phage‐encoded recombinase.
Figure 5. Methods of filamentous phage host chromosome integration and excision.
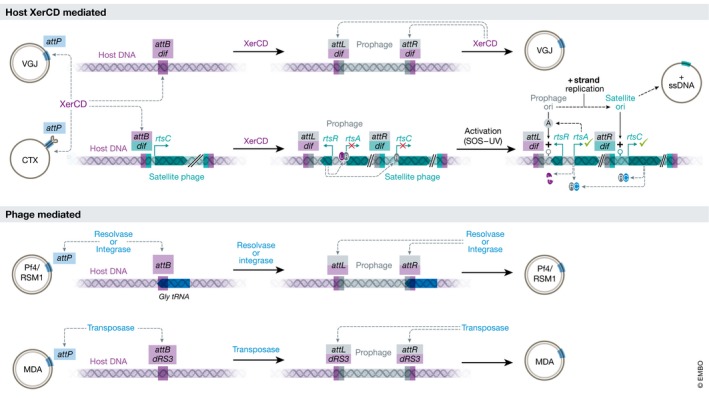
Methods for filamentous prophage integration into the host chromosome are shown. Top: Host‐mediated (XerCD) integration via two methods. Vibrio phage VJG uses a reversible integration—a dsDNA RF phage genome with an attP site is recognized by the host XerCD recombinase which mediates homologous recombination at the dif site on the host chromosome. The prophage can be excised by XerCD‐mediated recombination at the resulting attL and attR sites. Vibrio phage CTXφ uses an irreversible integration—XerCD recognizes an attP site formed by a hairpin in the ssDNA phage genome and mediates homologous recombination at the dif site on the host chromosome (and typically a satellite phage). Due to nature of the attP hairpin, the resulting AttL site on the prophage is defective and thus cannot be excised by XerCD. Replication of the resulting prophage is inactivated by a regulatory loop involving the phage‐encoded repressor RtsR (R), the host repressor LexA (L) and the satellite activator RtsC (C). Upon activation by the host SOS response, LexA is degraded and the positive regulator RtsC is produced and binds to the RtsR repressor allowing expression of the phage replication protein RtsA (functionally equivalent of pII). RtsA binds to the + ori on the prophage genome and acts in an analogous way to that of pII on RF circular DNA. The resulting phage ssDNA is amplified and packaged as described in Fig 4B. In examples of phage‐mediated integration (bottom), Pseudomonas phage Pf4 uses a phage‐encoded integrase to reversibly integrate itself into the Gly tRNA site of the host chromosome and Ralstonia phage RSM1 uses a phage‐encoded resolvase to reversibly integrate into Ser tRNA site on the host chromosome, while Neisseria phage MDAφ uses a phage‐mediated transposase to integrate at a 20‐bp repeat region (dRS3) on the host chromosome.
Table 2.
Filamentous prophage integration methods
| Integration method | Host integration site attB | Examplesa |
|---|---|---|
| Host XerC/XerD (reversible) | dif | VGJ, TLC (satellite), VEJ, VSK, Vf33, fs2, Lf, Cf1c, Cf16, RSS0 |
| Host XerC/XerD (irreversible) | dif | CTX, CUS‐1, Ypf |
| Integrase—tyrosine recombinase | Gly tRNA (Met‐tRNA) | Pf4, Pf5, Pf7 (Pf6) |
| Resolvase—serine recombinase | Ser tRNA | RSM1, RSM3 |
| Transposase | Variable—dRS3 repeat | MDAφ |
This is not a complete list of phages using the listed method. Not all experimentally validated (may be based on attP sequence homology or presence of recombinase on phage/prophage genome).
Integrated prophages pay their way through contributions to virulence and other advantageous phenotypes. An E. coli prophage called CUS‐1 is correlated with invasive extraintestinal pathogenic E. coli strains, and the prophage encodes puvA, which was identified as contributing to bacterial virulence in a rat disease model 168, 169. An almost identical phage, Ypf/CUS‐2, has been described in Yersinia pestis, where again the prophage is associated with virulent plague strains and disruption of the prophage resulted in reduced virulence in mice 170.
Many prophages thought to encode filamentous phages have been described in Neisseria gonorrhoeae and Neisseria meningitidis strains 144, 171, 172, 173. Whole‐genome sequence analysis of four Neisseria species found 12 complete prophages and 11 incomplete prophages. These various prophage elements have been implicated in plasticity of Neisseria genomes, and assisting the massive chromosomal rearrangements observed between strains 171, 173, 174, 175. For N. meningitidis, the presence of one of these prophages, Nf1‐A, has been assigned Meningococcal Disease Associated (MDA) because it was one of the only loci correlated with hypervirulence 144, 176. It was later shown that carriage of the prophage does not increase the virulence of the septicaemic phase of the disease in a human tissue disease model, but that phage secretion increases biofilm formation and colonization of epithelial cells 177. In this scenario, the MDAφ virion behaves analogously to type IV pili, with many virions remaining associated with the bacterial surface and promoting bacteria–bacteria interactions.
Our current understanding of prophage integration into bacterial chromosomes
The host‐mediated XerC and XerD site‐specific recombination is the most well‐characterized integration process, because of studies focused on understanding the method of integration in Vibrio filamentous phages. The endogenous role of the recombinases XerC and XerD is to ensure the segregation of two bacterial circular chromosomes during genome replication, by catalysing recombination between two dif (deletion‐induced filamentation) resolving dimeric chromosomes formed during DNA replication 178. Filamentous phages have hijacked this system by containing a dif‐like site (termed attP) on their genome (the bacterial chromosome site for integration, which is typically a dif site is termed the attB site) (Fig 5).
The Vibrio phage VGJ genome contains an attP site, which XerCD can bind to in the dsDNA RF of the phage genome and catalyse recombination between the attP and attB sites. This integration is reversible because the prophage is flanked by functional attL and attR sites 179, 180. However, the Vibrio phage CTXφ genome contains two adjacent “defective” attB sites in an inverted orientation. In the ssDNA IF, these sites can form a forked hairpin that results in a functional attB site. XerCD can catalyse recombination with the bacterial attB site 181, 182. Integration of the CTXφ ssDNA template leaves a Holliday junction intermediate that must be resolved by the host DNA replication machinery through an unclear process. The prophage cannot be excised as the att hairpin cannot form on the chromosome. Thus, once CTXφ has been integrated as a prophage, it must undergo rolling circle replication on the chromosome to form new phage genomes for amplification and virion assembly. This poses a problem when terminating rolling circle replication, as to reach the positive strand origin for termination the pII/DNA polymerase would have to replicate the entire bacterial chromosome. Consequently, the CTXφ prophage is always found integrated adjacent to a second Vibrio prophage, a satellite phage (a prophage lacking phage morphogenesis genes which uses a second phage to produce virions), or in a duplicated form, where it utilizes the neighbouring prophage positive strand origin to terminate and release ssDNA phage genomes. A consequence of this is that the resulting phages contain fragments of the second prophage 183 (Fig 5).
The CTXφ prophage can be extracted under specific conditions—if CTXφ infects a strain already harbouring a reversibly integrated prophage such as VGJ. CTXφ can integrate adjacent to the VGJ prophage (using the dif/att site of the prophage) resulting in a VGJ‐CTX module flanked by two intact functional att sites which can subsequently be used by the host Xer recombinase machinery to extract a hybrid VGJ‐CTX genome that can undergo rolling circle replication and be packaged into functional virions containing the VGJ‐CTX hybrid genome with the VGJ phage proteins 179, 180, 184, 185. This interplay of Vibrio prophage integration, chimera formation, excision and potential horizontal transfer results in the diverse concatenated prophage arrays observed at dif integration sites in Vibrio genomes 186, 187.
There are three distinct examples of integration methods using filamentous phage‐encoded recombinase, though there are limited mechanistic insights. Pseudomonas Pf4 and Pf5 phages are integrated into the chromosome at a site encoding a tRNAGly, whereas Pf6 is integrated at the tRNAMet site. These phages encode a tyrosine recombinase of the phage integrase family, protein which presumably facilitates this integration. The Ralstonia RSM phages integrate into the tRNASer site and encode their own serine recombinase of the resolvase/invertase family, which presumably facilitates this integration. Similar prophages are detected in the genomes of other species of Burkholderiaceae, such as the human pathogen Burkholderia pseudomallei 188. The Neisseria prophages may have a more promiscuous integration mechanism, where they are integrated into a short repeat region called dRS3; in N. meningitidis, there are more than 250 of these sites present on the chromosome. The Nf typically encode a Piv/MooV family transposase, and it is thought that this mediates the insertion and excision (transposition) of the prophage genome via a method similar to IS110 transposons 171, 173.
Prophages as a means to discover new Ff sequences (and new Ff applications)
As the pI N‐terminal Zot domain (Pfam PF05707) seems to be characteristic of and unique to filamentous phages and prophages, this sequence signature can be used to gauge the prevalence of undocumented filamentous phages in genome and metagenome sequence data. There are approximately 2,300 proteins with PF05707 domains present in the UniProtKB (2018_09) database with almost all of them (> 99%) associated with prokaryotic genomes and likely from filamentous phage origins. There are representatives across a wide range of prokaryotes including various phyla from Gram‐negative and Gram‐positive bacteria, as well as Archaea. Thus, there are potentially thousands of uncharacterized filamentous prophages present within the prokaryotic genomic repertoire (Fig 6). Furthermore, searching the Joint Genome Institutes Integrated Microbial Genomes and Microbiomes database returns more than 300,000 genes from metagenome sequences predicted to contain the PF05707 domains.
Figure 6. Ff discovery through gene signatures in prokaryote (host) chromosomes.
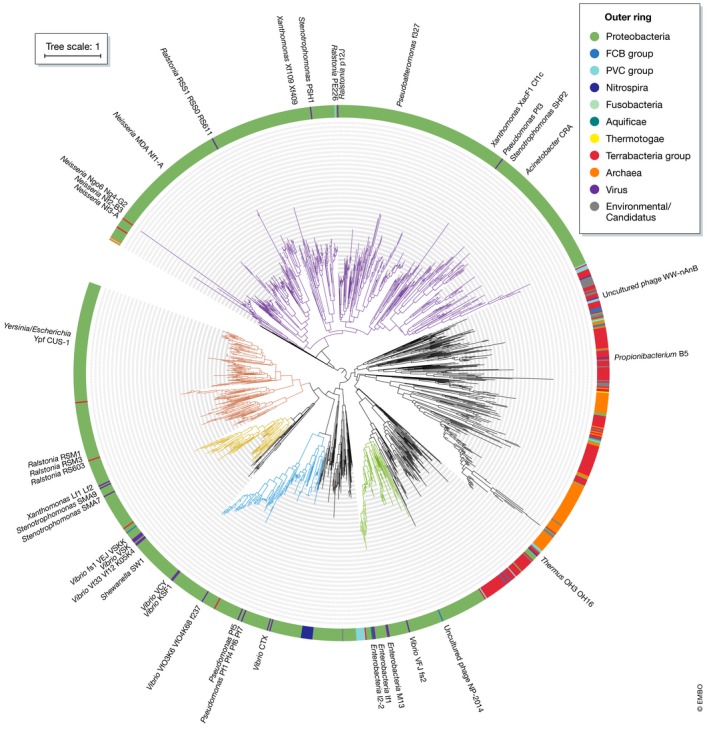
Zot domains are widely distributed throughout prokaryotic organisms including Gram‐negative, Gram‐positive and Archaeal organisms. Proteins from the UniRef90 database (representing 2,205 UniProtKB entries) with predicted “Zot” domains (PF05707) are represented in a phylogenetic tree. The taxonomic kingdom (or bacterial superphylum) is indicated in the outer ring. Branches/clades containing the known filamentous phages are coloured according to clades described in Fig 3. PF05707 domains were aligned and a tree was built with RAxML “PROTGAMMAAUTO” criteria (final model used = BLOSUM62) and “autoMRE_ING” bootstrap convergence test and midpoint rooted.
Our appreciation of this diversity of filamentous phages heralds an exciting phase of further discovery, about their biology and their evolution (see also Box 1). A massive restructuring of the taxonomy of phages, undertaken by the International Committee on Taxonomy of Viruses (ICTV), put emphasis on genome sequence information 189, 190, 191. This led to a complete restructuring of the Inoviridae family into seven genera, but left the bulk of the described filamentous phages as “unclassified” or “unassigned”, again suggesting that we are only seeing a small cross section of the diversity of these useful and enigmatic viruses. The previous Plectrovirus genus containing the mollicute‐infecting rod‐shaped viruses has been divided into the genera Plectrovirus (containing the Acholeplasma virus L51) and the Vespertiliovirus (containing the Spiroplasma viruses). The previously defined genus Inovirus containing the “classic” filamentous phages has been divided into five new genera: Inovirus, Habenivirus, Fibrovirus, Lineavirus and Saetivirus; and many of the previously classified species have been shifted into an “unclassified” genus category. More recently, there has been a further push away from morphological taxonomy, to integrate more metagenomic data into viral taxonomy and develop a universal method to classify viruses 192, 193, 194. Appling these methods to the Inoviridae family results in three unrelated groups representing the Plectrovirus genus, the Vespertiliovirus genus and the third diverse group comprising all the related classic filamentous phages described in this review, with Saetivirus, Lineavirus and Inovirus in one clade, acutely separated from a clade formed by Fibrovirus, Habenivirus and other currently unassigned groups 192.
Given how beneficial filamentous phages have been to our understanding of fundamental aspects of bacterial cell biology (e.g. membrane biogenesis, protein secretion, DNA recombination and replication) and developments in biotechnology, as well as their crucial roles in promoting bacterial virulence and shaping bacterial communities, this new age of filamentous phage discovery and characterization is one of promise and possibility.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank Rebecca Bamert, Rhys Dunstan, Christopher Stubenrauch and Mike McDonald for discussion and critical comments on the manuscript. The authors’ work in this area is supported by the Department of Education and Training|Australian Research Council (ARC) (FL130100038). I.D.H. is an ARC Laureate Postdoctoral Fellow, and T.L. is an ARC Australian Laureate Fellow.
EMBO Reports (2019) 20: e47427
See the Glossary for abbreviations used in this article.
Contributor Information
Iain D Hay, Email: iain.hay@auckland.ac.nz.
Trevor Lithgow, Email: trevor.lithgow@monash.edu.
References
- 1. Edwards RA, Rohwer F (2005) Viral metagenomics. Nat Rev Microbiol 3: 504–510 [DOI] [PubMed] [Google Scholar]
- 2. Hatfull GF, Hendrix RW (2011) Bacteriophages and their genomes. Curr Opin Virol 1: 298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thurber RV (2009) Current insights into phage biodiversity and biogeography. Curr Opin Microbiol 12: 582–587 [DOI] [PubMed] [Google Scholar]
- 4. Weinbauer MG (2004) Ecology of prokaryotic viruses. FEMS Microbiol Rev 28: 127–181 [DOI] [PubMed] [Google Scholar]
- 5. Casjens SR, Hendrix RW (2015) Bacteriophage lambda: early pioneer and still relevant. Virology 479–480: 310–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu B (2014) Bacteriophage T7 DNA polymerase ‐ sequenase. Front Microbiol 5: 181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang W, Li Y, Wang Y, Shi C, Li C, Li Q, Linhardt RJ (2018) Bacteriophage T7 transcription system: an enabling tool in synthetic biology. Biotechnol Adv 36: 2129–2137 [DOI] [PubMed] [Google Scholar]
- 8. Sanger F, Coulson AR, Friedmann T, Air GM, Barrell BG, Brown NL, Fiddes JC, Hutchison CA III, Slocombe PM, Smith M (1978) The nucleotide sequence of bacteriophage phiX174. J Mol Biol 125: 225–246 [DOI] [PubMed] [Google Scholar]
- 9. Eisenberg S, Griffith J, Kornberg A (1977) phiX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc Natl Acad Sci USA 74: 3198–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith HO, Hutchison CA III, Pfannkoch C, Venter JC (2003) Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci USA 100: 15440–15445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pichon X, Lagha M, Mueller F, Bertrand E (2018) A growing toolbox to image gene expression in single cells: sensitive approaches for demanding challenges. Mol Cell 71: 468–480 [DOI] [PubMed] [Google Scholar]
- 12. Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K (2008) Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol 190: 300–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA (2014) Exploiting CRISPR‐Cas nucleases to produce sequence‐specific antimicrobials. Nat Biotechnol 32: 1146–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hofschneider PH (1963) Untersuchungen uber kleine E. coli K 12 bakteriophagen 1 Und 2 mitteilung. Z Naturforsch Pt B 18: 203–210 [Google Scholar]
- 15. Marvin DA, Hoffmann‐Berling H (1963) Physical and chemical properties of two new small bacteriophages. Nature 197: 517 [Google Scholar]
- 16. Zinder ND, Valentine RC, Roger M, Stoeckenius W (1963) F1, a rod‐shaped male‐specific bacteriophage that contains DNA. Virology 20: 638–640 [DOI] [PubMed] [Google Scholar]
- 17. Loeb T (1960) Isolation of a bacteriophage specific for the F plus and Hfr mating types of Escherichia coli K‐12. Science 131: 932–933 [DOI] [PubMed] [Google Scholar]
- 18. Messing J (1993) M13 cloning vehicles. Their contribution to DNA sequencing. Methods Mol Biol 23: 9–22 [DOI] [PubMed] [Google Scholar]
- 19. Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, Russel M (2011) Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr Issues Mol Biol 13: 51–76 [PubMed] [Google Scholar]
- 20. Smith GP (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228: 1315–1317 [DOI] [PubMed] [Google Scholar]
- 21. McCafferty J, Griffiths AD, Winter G, Chiswell DJ (1990) Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348: 552–554 [DOI] [PubMed] [Google Scholar]
- 22. Parmley SF, Smith GP (1988) Antibody‐selectable filamentous fd phage vectors: affinity purification of target genes. Gene 73: 305–318 [DOI] [PubMed] [Google Scholar]
- 23. Marvin DA, Symmons MF, Straus SK (2014) Structure and assembly of filamentous bacteriophages. Prog Biophys Mol Biol 114: 80–122 [DOI] [PubMed] [Google Scholar]
- 24. Day LA, Marzec CJ, Reisberg SA, Casadevall A (1988) DNA packing in filamentous bacteriophages. Annu Rev Biophys Biophys Chem 17: 509–539 [DOI] [PubMed] [Google Scholar]
- 25. Greenwood J, Hunter GJ, Perham RN (1991) Regulation of filamentous bacteriophage length by modification of electrostatic interactions between coat protein and DNA. J Mol Biol 217: 223–227 [DOI] [PubMed] [Google Scholar]
- 26. Symmons MF, Welsh LC, Nave C, Marvin DA, Perham RN (1995) Matching electrostatic charge between DNA and coat protein in filamentous bacteriophage. Fibre diffraction of charge‐deletion mutants. J Mol Biol 245: 86–91 [DOI] [PubMed] [Google Scholar]
- 27. Liu DJ, Day LA (1994) Pf1 virus structure: helical coat protein and DNA with paraxial phosphates. Science 265: 671–674 [DOI] [PubMed] [Google Scholar]
- 28. Tsuboi M, Tsunoda M, Overman SA, Benevides JM, Thomas GJ Jr (2010) A structural model for the single‐stranded DNA genome of filamentous bacteriophage Pf1. Biochemistry 49: 1737–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grant RA, Lin TC, Konigsberg W, Webster RE (1981) Structure of the filamentous bacteriophage fl. Location of the A, C, and D minor coat proteins. J Biol Chem 256: 539–546 [PubMed] [Google Scholar]
- 30. Lee SW, Mao C, Flynn CE, Belcher AM (2002) Ordering of quantum dots using genetically engineered viruses. Science 296: 892–895 [DOI] [PubMed] [Google Scholar]
- 31. Holliger P, Riechmann L, Williams RL (1999) Crystal structure of the two N‐terminal domains of g3p from filamentous phage fd at 1.9 A: evidence for conformational lability. J Mol Biol 288: 649–657 [DOI] [PubMed] [Google Scholar]
- 32. Rakonjac J, Feng J, Model P (1999) Filamentous phage are released from the bacterial membrane by a two‐step mechanism involving a short C‐terminal fragment of pIII. J Mol Biol 289: 1253–1265 [DOI] [PubMed] [Google Scholar]
- 33. Endemann H, Model P (1995) Location of filamentous phage minor coat proteins in phage and in infected cells. J Mol Biol 250: 496–506 [DOI] [PubMed] [Google Scholar]
- 34. Bennett NJ, Gagic D, Sutherland‐Smith AJ, Rakonjac J (2011) Characterization of a dual‐function domain that mediates membrane insertion and excision of Ff filamentous bacteriophage. J Mol Biol 411: 972–985 [DOI] [PubMed] [Google Scholar]
- 35. Russel M, Model P (1989) Genetic analysis of the filamentous bacteriophage packaging signal and of the proteins that interact with it. J Virol 63: 3284–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Faruque SM, Mekalanos JJ (2012) Phage‐bacterial interactions in the evolution of toxigenic Vibrio cholerae . Virulence 3: 556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hassan F, Kamruzzaman M, Mekalanos JJ, Faruque SM (2010) Satellite phage TLCphi enables toxigenic conversion by CTX phage through dif site alteration. Nature 467: 982–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karaolis DK, Somara S, Maneval DR Jr, Johnson JA, Kaper JB (1999) A bacteriophage encoding a pathogenicity island, a type‐IV pilus and a phage receptor in cholera bacteria. Nature 399: 375–379 [DOI] [PubMed] [Google Scholar]
- 39. Faruque SM, Zhu J, Asadulghani Kamruzzaman M, Mekalanos JJ (2003) Examination of diverse toxin‐coregulated pilus‐positive Vibrio cholerae strains fails to demonstrate evidence for Vibrio pathogenicity island phage. Infect Immun 71: 2993–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Murphy RA, Boyd EF (2008) Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J Bacteriol 190: 636–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O'Shea YA, Boyd EF (2002) Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP‐T1 generalized transduction. FEMS Microbiol Lett 214: 153–157 [DOI] [PubMed] [Google Scholar]
- 42. Rajanna C, Wang J, Zhang D, Xu Z, Ali A, Hou YM, Karaolis DK (2003) The vibrio pathogenicity island of epidemic Vibrio cholerae forms precise extrachromosomal circular excision products. J Bacteriol 185: 6893–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Castillo D, Kauffman K, Hussain F, Kalatzis P, Rorbo N, Polz MF, Middelboe M (2018) Widespread distribution of prophage‐encoded virulence factors in marine Vibrio communities. Sci Rep 8: 9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xue H, Xu Y, Boucher Y, Polz MF (2012) High frequency of a novel filamentous phage, VCY phi, within an environmental Vibrio cholerae population. Appl Environ Microbiol 78: 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weynberg KD, Voolstra CR, Neave MJ, Buerger P, van Oppen MJ (2015) From cholera to corals: viruses as drivers of virulence in a major coral bacterial pathogen. Sci Rep 5: 17889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sakib SN, Reddi G, Almagro‐Moreno S (2018) Environmental role of pathogenic traits in Vibrio cholerae . J Bacteriol 200: e00795‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Renda BA, Chan C, Parent KN, Barrick JE (2016) Emergence of a competence‐reducing filamentous phage from the genome of acinetobacter baylyi ADP1. J Bacteriol 198: 3209–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Knezevic P, Voet M, Lavigne R (2015) Prevalence of Pf1‐like (pro)phage genetic elements among Pseudomonas aeruginosa isolates. Virology 483: 64–71 [DOI] [PubMed] [Google Scholar]
- 49. Mai‐Prochnow A, Hui JG, Kjelleberg S, Rakonjac J, McDougald D, Rice SA (2015) Big things in small packages: the genetics of filamentous phage and effects on fitness of their host. FEMS Microbiol Rev 39: 465–487 [DOI] [PubMed] [Google Scholar]
- 50. Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP (2001) Gene expression in Pseudomonas aeruginosa biofilms. Nature 413: 860–864 [DOI] [PubMed] [Google Scholar]
- 51. Platt MD, Schurr MJ, Sauer K, Vazquez G, Kukavica‐Ibrulj I, Potvin E, Levesque RC, Fedynak A, Brinkman FS, Schurr J et al (2008) Proteomic, microarray, and signature‐tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late‐stage cystic fibrosis airway conditions. J Bacteriol 190: 2739–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Secor PR, Sweere JM, Michaels LA, Malkovskiy AV, Lazzareschi D, Katznelson E, Rajadas J, Birnbaum ME, Arrigoni A, Braun KR et al (2015) Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe 18: 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rice SA, Tan CH, Mikkelsen PJ, Kung V, Woo J, Tay M, Hauser A, McDougald D, Webb JS, Kjelleberg S (2009) The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J 3: 271–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Webb JS, Thompson LS, James S, Charlton T, Tolker‐Nielsen T, Koch B, Givskov M, Kjelleberg S (2003) Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol 185: 4585–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Webb JS, Lau M, Kjelleberg S (2004) Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol 186: 8066–8073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Secor PR, Sass G, Nazik H, Stevens DA (2017) Effect of acute predation with bacteriophage on intermicrobial aggression by Pseudomonas aeruginosa . PLoS ONE 12: e0179659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kuo TT, Huang TC, Chow TY (1969) A filamentous bacteriophage from Xanthomonas oryzae . Virology 39: 548–555 [DOI] [PubMed] [Google Scholar]
- 58. Kuo TT, Lin YH, Huang CM, Chang SF, Dai H, Feng TY (1987) The lysogenic cycle of the filamentous phage Cflt from Xanthomonas campestris pv. citri. Virology 156: 305–312 [DOI] [PubMed] [Google Scholar]
- 59. Kuo TT, Chao YS, Lin YH, Lin BY, Liu LF, Feng TY (1987) Integration of the DNA of filamentous bacteriophage Cflt into the chromosomal DNA of its host. J Virol 61: 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kuo TT, Chiang CC, Chen SY, Lin JH, Kuo JL (1994) A long lytic cycle in filamentous phage Cf1tv infecting Xanthomonas campestris pv. citri. Arch Virol 135: 253–264 [DOI] [PubMed] [Google Scholar]
- 61. Yeh TY (2017) Complete nucleotide sequence of a new filamentous phage, Xf109, which integrates its genome into the chromosomal DNA of Xanthomonas oryzae . Arch Virol 162: 567–572 [DOI] [PubMed] [Google Scholar]
- 62. Askora A, Yamada T (2015) Two different evolutionary lines of filamentous phages in Ralstonia solanacearum: their effects on bacterial virulence. Front Genet 6: 217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yamada T (2013) Filamentous phages of Ralstonia solanacearum: double‐edged swords for pathogenic bacteria. Front Microbiol 4: 325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Addy HS, Askora A, Kawasaki T, Fujie M, Yamada T (2012) Utilization of filamentous phage phi RSM3 to control bacterial wilt caused by Ralstonia solanacearum . Plant Dis 96: 1204–1209 [DOI] [PubMed] [Google Scholar]
- 65. Addy HS, Askora A, Kawasaki T, Fujie M, Yamada T (2012) Loss of virulence of the phytopathogen Ralstonia solanacearum through infection by phiRSM filamentous phages. Phytopathology 102: 469–477 [DOI] [PubMed] [Google Scholar]
- 66. Ahmad AA, Stulberg MJ, Huang Q (2017) Prophage Rs551 and its repressor gene orf14 reduce virulence and increase competitive fitness of its Ralstonia solanacearum carrier strain UW551. Front Microbiol 8: 2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ahmad AA, Stulberg MJ, Mershon JP, Mollov DS, Huang Q (2017) Molecular and biological characterization of varphiRs551, a filamentous bacteriophage isolated from a race 3 biovar 2 strain of Ralstonia solanacearum . PLoS ONE 12: e0185034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Askora A, Kawasaki T, Usami S, Fujie M, Yamada T (2009) Host recognition and integration of filamentous phage phiRSM in the phytopathogen, Ralstonia solanacearum . Virology 384: 69–76 [DOI] [PubMed] [Google Scholar]
- 69. Kawasaki T, Nagata S, Fujiwara A, Satsuma H, Fujie M, Usami S, Yamada T (2007) Genomic characterization of the filamentous integrative bacteriophages {phi}RSS1 and {phi}RSM1, which infect Ralstonia solanacearum . J Bacteriol 189: 5792–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Narulita E, Addy HS, Kawasaki T, Fujie M, Yamada T (2016) The involvement of the PilQ secretin of type IV pili in phage infection in Ralstonia solanacearum . Biochem Biophys Res Commun 469: 868–872 [DOI] [PubMed] [Google Scholar]
- 71. Addy HS, Askora A, Kawasaki T, Fujie M, Yamada T (2012) The filamentous phage varphiRSS1 enhances virulence of phytopathogenic Ralstonia solanacearum on tomato. Phytopathology 102: 244–251 [DOI] [PubMed] [Google Scholar]
- 72. Van TT, Yoshida S, Miki K, Kondo A, Kamei K (2014) Genomic characterization of varphiRS603, a filamentous bacteriophage that is infectious to the phytopathogen Ralstonia solanacearum . Microbiol Immunol 58: 697–700 [DOI] [PubMed] [Google Scholar]
- 73. Van TT, Yoshida S, Miki K, Kondo A, Kamei K (2015) Complete genome sequence of a filamentous bacteriophage, RS611, that infects the phytopathogen Ralstonia solanacearum . Arch Virol 160: 865–867 [DOI] [PubMed] [Google Scholar]
- 74. Chan B, Miyamoto H, Taniguchi H, Yoshida S (2002) Isolation and genetic characterization of a novel filamentous bacteriophage, a deleted form of phage f237, from a pandemic Vibrio parahaemolyticus O4:K68 strain. Microbiol Immunol 46: 565–569 [DOI] [PubMed] [Google Scholar]
- 75. Jian H, Xiong L, Xu G, Xiao X (2016) Filamentous phage SW1 is active and influences the transcriptome of the host at high‐pressure and low‐temperature. Environ Microbiol Rep 8: 358–362 [DOI] [PubMed] [Google Scholar]
- 76. Jian H, Xu J, Xiao X, Wang F (2012) Dynamic modulation of DNA replication and gene transcription in deep‐sea filamentous phage SW1 in response to changes of host growth and temperature. PLoS ONE 7: e41578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang F, Wang F, Li Q, Xiao X (2007) A novel filamentous phage from the deep‐sea bacterium Shewanella piezotolerans WP3 is induced at low temperature. J Bacteriol 189: 7151–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang XW, Jian HH, Wang FP (2015) pSW2, a novel low‐temperature‐inducible gene expression vector based on a filamentous phage of the deep‐sea bacterium Shewanella piezotolerans WP3. Appl Environ Microbiol 81: 5519–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nagayoshi Y, Kumagae K, Mori K, Tashiro K, Nakamura A, Fujino Y, Hiromasa Y, Iwamoto T, Kuhara S, Ohshima T et al (2016) Physiological properties and genome structure of the hyperthermophilic filamentous phage phiOH3 which infects Thermus thermophilus HB8. Front Microbiol 7: 50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fasano A, Baudry B, Pumplin DW, Wasserman SS, Tall BD, Ketley JM, Kaper JB (1991) Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junctions. Proc Natl Acad Sci USA 88: 5242–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Perez‐Reytor D, Jana V, Pavez L, Navarrete P, Garcia K (2018) Accessory toxins of Vibrio pathogens and their role in epithelial disruption during infection. Front Microbiol 9: 2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Baudry B, Fasano A, Ketley J, Kaper JB (1992) Cloning of a gene (zot) encoding a new toxin produced by Vibrio cholerae . Infect Immun 60: 428–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Di Pierro M, Lu R, Uzzau S, Wang W, Margaretten K, Pazzani C, Maimone F, Fasano A (2001) Zonula occludens toxin structure‐function analysis. Identification of the fragment biologically active on tight junctions and of the zonulin receptor binding domain. J Biol Chem 276: 19160–19165 [DOI] [PubMed] [Google Scholar]
- 84. Goldblum SE, Rai U, Tripathi A, Thakar M, De Leo L, Di Toro N, Not T, Ramachandran R, Puche AC, Hollenberg MD et al (2011) The active Zot domain (aa 288‐293) increases ZO‐1 and myosin 1C serine/threonine phosphorylation, alters interaction between ZO‐1 and its binding partners, and induces tight junction disassembly through proteinase activated receptor 2 activation. FASEB J 25: 144–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Uzzau S, Cappuccinelli P, Fasano A (1999) Expression of Vibrio cholerae zonula occludens toxin and analysis of its subcellular localization. Microb Pathog 27: 377–385 [DOI] [PubMed] [Google Scholar]
- 86. Schmidt E, Kelly SM, van der Walle CF (2007) Tight junction modulation and biochemical characterisation of the zonula occludens toxin C‐and N‐termini. FEBS Lett 581: 2974–2980 [DOI] [PubMed] [Google Scholar]
- 87. Murugaiyan S, Bae JY, Wu J, Lee SD, Um HY, Choi HK, Chung E, Lee JH, Lee SW (2011) Characterization of filamentous bacteriophage PE226 infecting Ralstonia solanacearum strains. J Appl Microbiol 110: 296–303 [DOI] [PubMed] [Google Scholar]
- 88. Hagemann M, Hasse D, Berg G (2006) Detection of a phage genome carrying a zonula occludens like toxin gene (zot) in clinical isolates of Stenotrophomonas maltophilia . Arch Microbiol 185: 449–458 [DOI] [PubMed] [Google Scholar]
- 89. Campos J, Martinez E, Suzarte E, Rodriguez BL, Marrero K, Silva Y, Ledon T, del Sol R, Fando R (2003) VGJ phi, a novel filamentous phage of Vibrio cholerae, integrates into the same chromosomal site as CTX phi. J Bacteriol 185: 5685–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Deng LW, Malik P, Perham RN (1999) Interaction of the globular domains of pIII protein of filamentous bacteriophage fd with the F‐pilus of Escherichia coli . Virology 253: 271–277 [DOI] [PubMed] [Google Scholar]
- 91. Holland SJ, Sanz C, Perham RN (2006) Identification and specificity of pilus adsorption proteins of filamentous bacteriophages infecting Pseudomonas aeruginosa . Virology 345: 540–548 [DOI] [PubMed] [Google Scholar]
- 92. Jouravleva EA, McDonald GA, Marsh JW, Taylor RK, Boesman‐Finkelstein M, Finkelstein RA (1998) The Vibrio cholerae mannose‐sensitive hemagglutinin is the receptor for a filamentous bacteriophage from V. cholerae O139. Infect Immun 66: 2535–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yang YC, Chou CP, Kuo TT, Lin SH, Yang MK (2004) PilR enhances the sensitivity of Xanthomonas axonopodis pv. citri to the infection of filamentous bacteriophage Cf. Curr Microbiol 48: 251–261 [DOI] [PubMed] [Google Scholar]
- 94. Jacobson A (1972) Role of F pili in the penetration of bacteriophage fl. J Virol 10: 835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gutierrez MA (2017) The minor pilin TcpB is located at the tip of the toxin co‐regulated pilus of Vibrio cholerae and is the receptor for the filamentous phage CTXφ. In Department of Molecular Biology and Biochemistry, Faculty of Science Simon Fraser University
- 96. Clarke M, Maddera L, Harris RL, Silverman PM (2008) F‐pili dynamics by live‐cell imaging. Proc Natl Acad Sci USA 105: 17978–17981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Godlewska R, Wisniewska K, Pietras Z, Jagusztyn‐Krynicka EK (2009) Peptidoglycan‐associated lipoprotein (Pal) of Gram‐negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol Lett 298: 1–11 [DOI] [PubMed] [Google Scholar]
- 98. Click EM, Webster RE (1997) Filamentous phage infection: required interactions with the TolA protein. J Bacteriol 179: 6464–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Heilpern AJ, Waldor MK (2000) CTXphi infection of Vibrio cholerae requires the tolQRA gene products. J Bacteriol 182: 1739–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Riechmann L, Holliger P (1997) The C‐terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli . Cell 90: 351–360 [DOI] [PubMed] [Google Scholar]
- 101. Holliger P, Riechmann L (1997) A conserved infection pathway for filamentous bacteriophages is suggested by the structure of the membrane penetration domain of the minor coat protein g3p from phage fd. Structure 5: 265–275 [DOI] [PubMed] [Google Scholar]
- 102. Boeke JD, Model P, Zinder ND (1982) Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol Gen Genet 186: 185–192 [DOI] [PubMed] [Google Scholar]
- 103. Glaser‐Wuttke G, Keppner J, Rasched I (1989) Pore‐forming properties of the adsorption protein of filamentous phage fd. Biochim Biophys Acta 985: 239–247 [DOI] [PubMed] [Google Scholar]
- 104. Click EM, Webster RE (1998) The TolQRA proteins are required for membrane insertion of the major capsid protein of the filamentous phage f1 during infection. J Bacteriol 180: 1723–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Smilowitz H (1974) Bacteriophage f1 infection: fate of the parental major coat protein. J Virol 13: 94–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Armstrong J, Hewitt JA, Perham RN (1983) Chemical modification of the coat protein in bacteriophage fd and orientation of the virion during assembly and disassembly. EMBO J 2: 1641–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Baas PD (1985) DNA replication of single‐stranded Escherichia coli DNA phages. Biochim Biophys Acta 825: 111–139 [DOI] [PubMed] [Google Scholar]
- 108. Ruiz‐Maso JA, Macho NC, Bordanaba‐Ruiseco L, Espinosa M, Coll M, Del Solar G (2015) Plasmid rolling‐circle replication. Microbiol Spectr 3: PLAS‐0035‐2014 [DOI] [PubMed] [Google Scholar]
- 109. Higashitani A, Higashitani N, Horiuchi K (1997) Minus‐strand origin of filamentous phage versus transcriptional promoters in recognition of RNA polymerase. Proc Natl Acad Sci USA 94: 2909–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Higashitani N, Higashitani A, Guan ZW, Horiuchi K (1996) Recognition mechanisms of the minus‐strand origin of phage f1 by Escherichia coli RNA polymerase. Genes Cells 1: 829–841 [DOI] [PubMed] [Google Scholar]
- 111. Zenkin N, Naryshkina T, Kuznedelov K, Severinov K (2006) The mechanism of DNA replication primer synthesis by RNA polymerase. Nature 439: 617–620 [DOI] [PubMed] [Google Scholar]
- 112. Zenkin N, Severinov K (2004) The role of RNA polymerase sigma subunit in promoter‐independent initiation of transcription. Proc Natl Acad Sci USA 101: 4396–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kim MH, Hines JC, Ray DS (1981) Viable deletions of the M13 complementary strand origin. Proc Natl Acad Sci USA 78: 6784–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Meyer TF, Geider K (1979) Bacteriophage fd gene II‐protein. II. Specific cleavage and relaxation of supercoiled RF from filamentous phages. J Biol Chem 254: 12642–12646 [PubMed] [Google Scholar]
- 115. Horiuchi K (1997) Initiation mechanisms in replication of filamentous phage DNA. Genes Cells 2: 425–432 [DOI] [PubMed] [Google Scholar]
- 116. Asano S, Higashitani A, Horiuchi K (1999) Filamentous phage replication initiator protein gpII forms a covalent complex with the 5′ end of the nick it introduced. Nucleic Acids Res 27: 1882–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Meyer TF, Geider K (1982) Enzymatic synthesis of bacteriophage fd viral DNA. Nature 296: 828–832 [DOI] [PubMed] [Google Scholar]
- 118. Martinez E, Campos‐Gomez J (2016) Pf filamentous phage requires UvrD for replication in Pseudomonas aeruginosa . mSphere 1: e00104–e00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Martinez E, Campos‐Gomez J, Barre FX (2016) CTXvarphi: exploring new alternatives in host factor‐mediated filamentous phage replications. Bacteriophage 6: e1128512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Martinez E, Paly E, Barre FX (2015) CTXphi replication depends on the histone‐like HU protein and the UvrD helicase. PLoS Genet 11: e1005256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lerner TJ, Model P (1981) The “steady state” of coliphage f1: DNA synthesis late in infection. Virology 115: 282–294 [DOI] [PubMed] [Google Scholar]
- 122. Guan Y, Zhang H, Wang AH (1995) Electrostatic potential distribution of the gene V protein from Ff phage facilitates cooperative DNA binding: a model of the GVP‐ssDNA complex. Protein Sci 4: 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Alberts B, Frey L, Delius H (1972) Isolation and characterization of gene 5 protein of filamentous bacterial viruses. J Mol Biol 68: 139–152 [DOI] [PubMed] [Google Scholar]
- 124. Fulford W, Model P (1988) Bacteriophage f1 DNA replication genes. II. The roles of gene V protein and gene II protein in complementary strand synthesis. J Mol Biol 203: 39–48 [DOI] [PubMed] [Google Scholar]
- 125. Michel B, Zinder ND (1989) Translational repression in bacteriophage f1: characterization of the gene V protein target on the gene II mRNA. Proc Natl Acad Sci USA 86: 4002–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zaman GJ, Kaan AM, Schoenmakers JG, Konings RN (1992) Gene V protein‐mediated translational regulation of the synthesis of gene II protein of the filamentous bacteriophage M13: a dispensable function of the filamentous‐phage genome. J Bacteriol 174: 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Fulford W, Model P (1988) Regulation of bacteriophage f1 DNA replication. I. New functions for genes II and X. J Mol Biol 203: 49–62 [DOI] [PubMed] [Google Scholar]
- 128. Kokoska RJ, Steege DA (1998) Appropriate expression of filamentous phage f1 DNA replication genes II and X requires RNase E‐dependent processing and separate mRNAs. J Bacteriol 180: 3245–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kutter E, Bryan D, Ray G, Brewster E, Blasdel B, Guttman B (2018) From host to phage metabolism: hot tales of phage T4's takeover of E. coli . Viruses 10: E387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Rapoza MP, Webster RE (1993) The filamentous bacteriophage assembly proteins require the bacterial SecA protein for correct localization to the membrane. J Bacteriol 175: 1856–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Haigh NG, Webster RE (1999) The pI and pXI assembly proteins serve separate and essential roles in filamentous phage assembly. J Mol Biol 293: 1017–1027 [DOI] [PubMed] [Google Scholar]
- 132. Rapoza MP, Webster RE (1995) The products of gene I and the overlapping in‐frame gene XI are required for filamentous phage assembly. J Mol Biol 248: 627–638 [DOI] [PubMed] [Google Scholar]
- 133. Feng JN, Russel M, Model P (1997) A permeabilized cell system that assembles filamentous bacteriophage. Proc Natl Acad Sci USA 94: 4068–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Feng JN, Model P, Russel M (1999) A trans‐envelope protein complex needed for filamentous phage assembly and export. Mol Microbiol 34: 745–755 [DOI] [PubMed] [Google Scholar]
- 135. Dunstan RA, Hay ID, Wilksch JJ, Schittenhelm RB, Purcell AW, Clark J, Costin A, Ramm G, Strugnell RA, Lithgow T (2015) Assembly of the secretion pores GspD, Wza and CsgG into bacterial outer membranes does not require the Omp85 proteins BamA or TamA. Mol Microbiol 97: 616–629 [DOI] [PubMed] [Google Scholar]
- 136. Opalka N, Beckmann R, Boisset N, Simon MN, Russel M, Darst SA (2003) Structure of the filamentous phage pIV multimer by cryo‐electron microscopy. J Mol Biol 325: 461–470 [DOI] [PubMed] [Google Scholar]
- 137. Hay ID, Belousoff MJ, Dunstan RA, Bamert RS, Lithgow T (2018) Structure and membrane topography of the vibrio‐type secretin complex from the type 2 secretion system of enteropathogenic Escherichia coli . J Bacteriol 200: e00521–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hay ID, Belousoff MJ, Lithgow T (2017) Structural basis of type 2 secretion system engagement between the inner and outer bacterial membranes. MBio 8: e01344–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Yan Z, Yin M, Xu D, Zhu Y, Li X (2017) Structural insights into the secretin translocation channel in the type II secretion system. Nat Struct Mol Biol 24: 177–183 [DOI] [PubMed] [Google Scholar]
- 140. Spagnuolo J, Opalka N, Wen WX, Gagic D, Chabaud E, Bellini P, Bennett MD, Norris GE, Darst SA, Russel M et al (2010) Identification of the gate regions in the primary structure of the secretin pIV. Mol Microbiol 76: 133–150 [DOI] [PubMed] [Google Scholar]
- 141. Hu J, Worrall LJ, Hong C, Vuckovic M, Atkinson CE, Caveney N, Yu Z, Strynadka NCJ (2018) Cryo‐EM analysis of the T3S injectisome reveals the structure of the needle and open secretin. Nat Commun 9: 3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Davis BM, Lawson EH, Sandkvist M, Ali A, Sozhamannan S, Waldor MK (2000) Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXphi. Science 288: 333–335 [DOI] [PubMed] [Google Scholar]
- 143. Davis BM, Waldor MK (2003) Filamentous phages linked to virulence of Vibrio cholerae . Curr Opin Microbiol 6: 35–42 [DOI] [PubMed] [Google Scholar]
- 144. Bille E, Zahar JR, Perrin A, Morelle S, Kriz P, Jolley KA, Maiden MC, Dervin C, Nassif X, Tinsley CR (2005) A chromosomally integrated bacteriophage in invasive meningococci. J Exp Med 201: 1905–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Daefler S, Russel M, Model P (1997) Module swaps between related translocator proteins pIV(f1), pIV(IKe) and PulD: identification of a specificity domain. J Mol Biol 266: 978–992 [DOI] [PubMed] [Google Scholar]
- 146. de Groot A, Koster M, Gerard‐Vincent M, Gerritse G, Lazdunski A, Tommassen J, Filloux A (2001) Exchange of Xcp (Gsp) secretion machineries between Pseudomonas aeruginosa and Pseudomonas alcaligenes: species specificity unrelated to substrate recognition. J Bacteriol 183: 959–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Russel M (1993) Protein‐protein interactions during filamentous phage assembly. J Mol Biol 231: 689–697 [DOI] [PubMed] [Google Scholar]
- 148. Rohrer J, Kuhn A (1990) The function of a leader peptide in translocating charged amino acyl residues across a membrane. Science 250: 1418–1421 [DOI] [PubMed] [Google Scholar]
- 149. Cao G, Dalbey RE (1994) Translocation of N‐terminal tails across the plasma membrane. EMBO J 13: 4662–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kiefer D, Hu X, Dalbey R, Kuhn A (1997) Negatively charged amino acid residues play an active role in orienting the Sec‐independent Pf3 coat protein in the Escherichia coli inner membrane. EMBO J 16: 2197–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Kuhn A (1995) Major coat proteins of bacteriophage Pf3 and M13 as model systems for Sec‐independent protein transport. FEMS Microbiol Rev 17: 185–190 [DOI] [PubMed] [Google Scholar]
- 152. Wickner W (1988) Mechanisms of membrane assembly: general lessons from the study of M13 coat protein and Escherichia coli leader peptidase. Biochemistry 27: 1081–1086 [DOI] [PubMed] [Google Scholar]
- 153. Xie K, Dalbey RE (2008) Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol 6: 234–244 [DOI] [PubMed] [Google Scholar]
- 154. de Gier JW, Scotti PA, Saaf A, Valent QA, Kuhn A, Luirink J, von Heijne G (1998) Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli . Proc Natl Acad Sci USA 95: 14646–14651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Grant RA, Lin TC, Webster RE, Konigsberg W (1981) Structure of filamentous bacteriophage: isolation, characterization, and localization of the minor coat proteins and orientation of the DNA. Prog Clin Biol Res 64: 413–428 [PubMed] [Google Scholar]
- 156. Lopez J, Webster RE (1985) Assembly site of bacteriophage f1 corresponds to adhesion zones between the inner and outer membranes of the host cell. J Bacteriol 163: 1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Simons GF, Konings RN, Schoenmakers JG (1981) Genes VI, VII, and IX of phage M13 code for minor capsid proteins of the virion. Proc Natl Acad Sci USA 78: 4194–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Samuelson JC, Chen M, Jiang F, Moller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE (2000) YidC mediates membrane protein insertion in bacteria. Nature 406: 637–641 [DOI] [PubMed] [Google Scholar]
- 159. Samuelson JC, Jiang F, Yi L, Chen M, de Gier JW, Kuhn A, Dalbey RE (2001) Function of YidC for the insertion of M13 procoat protein in Escherichia coli: translocation of mutants that show differences in their membrane potential dependence and Sec requirement. J Biol Chem 276: 34847–34852 [DOI] [PubMed] [Google Scholar]
- 160. Serek J, Bauer‐Manz G, Struhalla G, van den Berg L, Kiefer D, Dalbey R, Kuhn A (2004) Escherichia coli YidC is a membrane insertase for Sec‐independent proteins. EMBO J 23: 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Klenner C, Yuan J, Dalbey RE, Kuhn A (2008) The Pf3 coat protein contacts TM1 and TM3 of YidC during membrane biogenesis. FEBS Lett 582: 3967–3972 [DOI] [PubMed] [Google Scholar]
- 162. Cao G, Kuhn A, Dalbey RE (1995) The translocation of negatively charged residues across the membrane is driven by the electrochemical potential: evidence for an electrophoresis‐like membrane transfer mechanism. EMBO J 14: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Haigh NG, Webster RE (1998) The major coat protein of filamentous bacteriophage f1 specifically pairs in the bacterial cytoplasmic membrane. J Mol Biol 279: 19–29 [DOI] [PubMed] [Google Scholar]
- 164. Nagler C, Nagler G, Kuhn A (2007) Cysteine residues in the transmembrane regions of M13 procoat protein suggest that oligomeric coat proteins assemble onto phage progeny. J Bacteriol 189: 2897–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Straus SK, Bo HE (2018) Filamentous bacteriophage proteins and assembly. Subcell Biochem 88: 261–279 [DOI] [PubMed] [Google Scholar]
- 166. Gailus V, Rasched I (1994) The adsorption protein of bacteriophage fd and its neighbour minor coat protein build a structural entity. Eur J Biochem 222: 927–931 [DOI] [PubMed] [Google Scholar]
- 167. Boeke JD, Model P (1982) A prokaryotic membrane anchor sequence: carboxyl terminus of bacteriophage f1 gene III protein retains it in the membrane. Proc Natl Acad Sci USA 79: 5200–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Gonzalez MD, Lichtensteiger CA, Caughlan R, Vimr ER (2002) Conserved filamentous prophage in Escherichia coli O18:K1:H7 and Yersinia pestis biovar orientalis. J Bacteriol 184: 6050–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Gonzalez MD, Lichtensteiger CA, Vimr ER (2001) Adaptation of signature‐tagged mutagenesis to Escherichia coli K1 and the infant‐rat model of invasive disease. FEMS Microbiol Lett 198: 125–128 [DOI] [PubMed] [Google Scholar]
- 170. Derbise A, Chenal‐Francisque V, Pouillot F, Fayolle C, Prevost MC, Medigue C, Hinnebusch BJ, Carniel E (2007) A horizontally acquired filamentous phage contributes to the pathogenicity of the plague bacillus. Mol Microbiol 63: 1145–1157 [DOI] [PubMed] [Google Scholar]
- 171. Kawai M, Uchiyama I, Kobayashi I (2005) Genome comparison in silico in Neisseria suggests integration of filamentous bacteriophages by their own transposase. DNA Res 12: 389–401 [DOI] [PubMed] [Google Scholar]
- 172. Piekarowicz A, Majchrzak M, Klyz A, Adamczyk‐Poplawska M (2006) Analysis of the filamentous bacteriophage genomes integrated into Neisseria gonorrhoeae FA1090 chromosome. Pol J Microbiol 55: 251–260 [PubMed] [Google Scholar]
- 173. Skaar EP, Lecuyer B, Lenich AG, Lazio MP, Perkins‐Balding D, Seifert HS, Karls AC (2005) Analysis of the Piv recombinase‐related gene family of Neisseria gonorrhoeae . J Bacteriol 187: 1276–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Gibbs CP, Meyer TF (1996) Genome plasticity in Neisseria gonorrhoeae . FEMS Microbiol Lett 145: 173–179 [DOI] [PubMed] [Google Scholar]
- 175. Spencer‐Smith R, Varkey EM, Fielder MD, Snyder LA (2012) Sequence features contributing to chromosomal rearrangements in Neisseria gonorrhoeae . PLoS ONE 7: e46023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Bille E, Ure R, Gray SJ, Kaczmarski EB, McCarthy ND, Nassif X, Maiden MC, Tinsley CR (2008) Association of a bacteriophage with meningococcal disease in young adults. PLoS ONE 3: e3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Bille E, Meyer J, Jamet A, Euphrasie D, Barnier JP, Brissac T, Larsen A, Pelissier P, Nassif X (2017) A virulence‐associated filamentous bacteriophage of Neisseria meningitidis increases host‐cell colonisation. PLoS Pathog 13: e1006495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Castillo F, Benmohamed A, Szatmari G (2017) Xer site specific recombination: double and single recombinase systems. Front Microbiol 8: 453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Das B (2014) Mechanistic insights into filamentous phage integration in Vibrio cholerae . Front Microbiol 5: 650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Das B, Bischerour J, Barre FX (2011) VGJphi integration and excision mechanisms contribute to the genetic diversity of Vibrio cholerae epidemic strains. Proc Natl Acad Sci USA 108: 2516–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Das B, Bischerour J, Val ME, Barre FX (2010) Molecular keys of the tropism of integration of the cholera toxin phage. Proc Natl Acad Sci USA 107: 4377–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, Barre FX (2005) The single‐stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae . Mol Cell 19: 559–566 [DOI] [PubMed] [Google Scholar]
- 183. McLeod SM, Kimsey HH, Davis BM, Waldor MK (2005) CTXphi and Vibrio cholerae: exploring a newly recognized type of phage‐host cell relationship. Mol Microbiol 57: 347–356 15978069 [Google Scholar]
- 184. Campos J, Martinez E, Izquierdo Y, Fando R (2010) VEJ{phi}, a novel filamentous phage of Vibrio cholerae able to transduce the cholera toxin genes. Microbiology 156: 108–115 [DOI] [PubMed] [Google Scholar]
- 185. Campos J, Martinez E, Marrero K, Silva Y, Rodriguez BL, Suzarte E, Ledon T, Fando R (2003) Novel type of specialized transduction for CTX phi or its satellite phage RS1 mediated by filamentous phage VGJ phi in Vibrio cholerae . J Bacteriol 185: 7231–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon YS, Kim DW, Lee JH et al (2009) Comparative genomics reveals mechanism for short‐term and long‐term clonal transitions in pandemic Vibrio cholerae . Proc Natl Acad Sci USA 106: 15442–15447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kim EJ, Lee CH, Nair GB, Kim DW (2015) Whole‐genome sequence comparisons reveal the evolution of Vibrio cholerae O1. Trends Microbiol 23: 479–489 [DOI] [PubMed] [Google Scholar]
- 188. Askora A, Kawasaki T, Fujie M, Yamada T (2011) Resolvase‐like serine recombinase mediates integration/excision in the bacteriophage phiRSM. J Biosci Bioeng 111: 109–116 [DOI] [PubMed] [Google Scholar]
- 189. Adams MJ, Lefkowitz EJ, King AM, Harrach B, Harrison RL, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Mushegian AR et al (2017) 50 years of the international committee on taxonomy of viruses: progress and prospects. Arch Virol 162: 1441–1446 [DOI] [PubMed] [Google Scholar]
- 190. Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB (2018) Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Res 46: D708–D717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Adriaenssens EM, Krupovic M, Knezevic P, Ackermann HW, Barylski J, Brister JR, Clokie MR, Duffy S, Dutilh BE, Edwards RA et al (2017) Taxonomy of prokaryotic viruses: 2016 update from the ICTV bacterial and archaeal viruses subcommittee. Arch Virol 162: 1153–1157 [DOI] [PubMed] [Google Scholar]
- 192. Aiewsakun P, Adriaenssens EM, Lavigne R, Kropinski AM, Simmonds P (2018) Evaluation of the genomic diversity of viruses infecting bacteria, archaea and eukaryotes using a common bioinformatic platform: steps towards a unified taxonomy. J Gen Virol 99: 1331–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Simmonds P, Adams MJ, Benko M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B et al (2017) Consensus statement: virus taxonomy in the age of metagenomics. Nat Rev Microbiol 15: 161–168 [DOI] [PubMed] [Google Scholar]
- 194. Simmonds P, Aiewsakun P (2018) Virus classification ‐ where do you draw the line? Arch Virol 163: 2037–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Rakonjac J, Russel M, Khanum S, Brooke SJ, Rajic M (2017) Filamentous phage: structure and biology. Adv Exp Med Biol 1053: 1–20 [DOI] [PubMed] [Google Scholar]
- 196. Lorenz SH, Jakob RP, Weininger U, Balbach J, Dobbek H, Schmid FX (2011) The filamentous phages fd and IF1 use different mechanisms to infect Escherichia coli . J Mol Biol 405: 989–1003 [DOI] [PubMed] [Google Scholar]
- 197. Peeters BP, Peters RM, Schoenmakers JG, Konings RN (1985) Nucleotide sequence and genetic organization of the genome of the N‐specific filamentous bacteriophage IKe. Comparison with the genome of the F‐specific filamentous phages M13, fd and f1. J Mol Biol 181: 27–39 [DOI] [PubMed] [Google Scholar]
- 198. Khatoon H (1976) Genetic properties of RM 98, an R plasmid of Salmonella which determines sensitivity to the phage IKe. Zentralbl Bakteriol Orig A 234: 294–304 [PubMed] [Google Scholar]
- 199. Bradley DE, Coetzee JN, Hedges RW (1983) IncI2 plasmids specify sensitivity to filamentous bacteriophage IKe. J Bacteriol 154: 505–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Stassen AP, Schoenmakers EF, Yu M, Schoenmakers JG, Konings RN (1992) Nucleotide sequence of the genome of the filamentous bacteriophage I2‐2: module evolution of the filamentous phage genome. J Mol Evol 34: 141–152 [DOI] [PubMed] [Google Scholar]
- 201. Chouikha I, Charrier L, Filali S, Derbise A, Carniel E (2010) Insights into the infective properties of YpfPhi, the Yersinia pestis filamentous phage. Virology 407: 43–52 [DOI] [PubMed] [Google Scholar]
- 202. Davis BM, Waldor MK (2000) CTXphi contains a hybrid genome derived from tandemly integrated elements. Proc Natl Acad Sci USA 97: 8572–8577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Waldor MK, Mekalanos JJ (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272: 1910–1914 [DOI] [PubMed] [Google Scholar]
- 204. Davis BM, Kimsey HH, Kane AV, Waldor MK (2002) A satellite phage‐encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J 21: 4240–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Faruque SM, Asadulghani Kamruzzaman M, Nandi RK, Ghosh AN, Nair GB, Mekalanos JJ, Sack DA (2002) RS1 element of Vibrio cholerae can propagate horizontally as a filamentous phage exploiting the morphogenesis genes of CTXphi. Infect Immun 70: 163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Ikema M, Honma Y (1998) A novel filamentous phage, fs‐2, of Vibrio cholerae O139. Microbiology 144(Pt 7): 1901–1906 [DOI] [PubMed] [Google Scholar]
- 207. Wang Q, Kan B, Wang R (2013) Isolation and characterization of the new mosaic filamentous phage VFJ Phi of Vibrio cholerae . PLoS ONE 8: e70934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Faruque SM, Bin Naser I, Fujihara K, Diraphat P, Chowdhury N, Kamruzzaman M, Qadri F, Yamasaki S, Ghosh AN, Mekalanos JJ (2005) Genomic sequence and receptor for the Vibrio cholerae phage KSF‐1phi: evolutionary divergence among filamentous vibriophages mediating lateral gene transfer. J Bacteriol 187: 4095–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Nasu H, Iida T, Sugahara T, Yamaichi Y, Park KS, Yokoyama K, Makino K, Shinagawa H, Honda T (2000) A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J Clin Microbiol 38: 2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Chang B, Taniguchi H, Miyamoto H, Yoshida S (1998) Filamentous bacteriophages of Vibrio parahaemolyticus as a possible clue to genetic transmission. J Bacteriol 180: 5094–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Nakasone N, Honma Y, Toma C, Yamashiro T, Iwanaga M (1998) Filamentous phage fs1 of Vibrio cholerae O139. Microbiol Immunol 42: 237–239 [DOI] [PubMed] [Google Scholar]
- 212. Hill DF, Short NJ, Perham RN, Petersen GB (1991) DNA sequence of the filamentous bacteriophage Pf1. J Mol Biol 218: 349–364 [DOI] [PubMed] [Google Scholar]
- 213. Nazik H, Joubert LM, Secor PR, Sweere JM, Bollyky PL, Sass G, Cegelski L, Stevens DA (2017) Pseudomonas phage inhibition of Candida albicans . Microbiology 163: 1568–1577 [DOI] [PubMed] [Google Scholar]
- 214. Luiten RG, Putterman DG, Schoenmakers JG, Konings RN, Day LA (1985) Nucleotide sequence of the genome of Pf3, an IncP‐1 plasmid‐specific filamentous bacteriophage of Pseudomonas aeruginosa . J Virol 56: 268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Penner JC, Ferreira JA, Secor PR, Sweere JM, Birukova MK, Joubert LM, Haagensen JA, Garcia O, Malkovskiy AV, Kaber G et al (2016) Pf4 bacteriophage produced by Pseudomonas aeruginosa inhibits Aspergillus fumigatus metabolism via iron sequestration. Microbiology 162: 1583–1594 [DOI] [PubMed] [Google Scholar]
- 216. Mooij MJ, Drenkard E, Llamas MA, Vandenbroucke‐Grauls CM, Savelkoul PH, Ausubel FM, Bitter W (2007) Characterization of the integrated filamentous phage Pf5 and its involvement in small‐colony formation. Microbiology 153: 1790–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Klockgether J, Munder A, Neugebauer J, Davenport CF, Stanke F, Larbig KD, Heeb S, Schock U, Pohl TM, Wiehlmann L et al (2010) Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol 192: 1113–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Yang F, Pecina DA, Kelly SD, Kim SH, Kemner KM, Long DT, Marsh TL (2010) Biosequestration via cooperative binding of copper by Ralstonia pickettii . Environ Technol 31: 1045–1060 [DOI] [PubMed] [Google Scholar]
- 219. Tseng YH, Lo MC, Lin KC, Pan CC, Chang RY (1990) Characterization of filamentous bacteriophage phi Lf from Xanthomonas campestris pv. campestris. J Gen Virol Pt 71(8): 1881–1884 [DOI] [PubMed] [Google Scholar]
- 220. Dai H, Chiang KS, Kuo TT (1980) Characterization of a new filamentous phage Cf from Xanthomonas‐Citri . J Gen Virol 46: 277–289 [Google Scholar]
- 221. Kuo TT, Tan MS, Su MT, Yang MK (1991) Complete nucleotide sequence of filamentous phage Cf1c from Xanthomonas campestris pv. citri. Nucleic Acids Res 19: 2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Dai H, Tsay SH, Kuo TT, Lin YH, Wu WC (1987) Neolysogenization of Xanthomonas campestris pv. citri infected with filamentous phage Cf16. Virology 156: 313–320 [DOI] [PubMed] [Google Scholar]
- 223. Ahmad AA, Askora A, Kawasaki T, Fujie M, Yamada T (2014) The filamentous phage XacF1 causes loss of virulence in Xanthomonas axonopodis pv. citri, the causative agent of citrus canker disease. Front Microbiol 5: 321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Ahmad AA, Kawabe M, Askora A, Kawasaki T, Fujie M, Yamada T (2017) Dynamic integration and excision of filamentous phage XacF1 in Xanthomonas citri pv. citri, the causative agent of citrus canker disease. FEBS Open Bio 7: 1715–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Meyer J, Brissac T, Frapy E, Omer H, Euphrasie D, Bonavita A, Nassif X, Bille E (2016) Characterization of MDAPhi, a temperate filamentous bacteriophage of Neisseria meningitidis . Microbiology 162: 268–282 [DOI] [PubMed] [Google Scholar]
- 226. Piekarowicz A, Klyz A, Majchrzak M, Szczesna E, Piechucki M, Kwiatek A, Maugel TK, Stein DC (2014) Neisseria gonorrhoeae filamentous phage NgoPhi6 is capable of infecting a variety of Gram‐negative bacteria. J Virol 88: 1002–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Liu J, Liu Q, Shen P, Huang YP (2012) Isolation and characterization of a novel filamentous phage from Stenotrophomonas maltophilia . Arch Virol 157: 1643–1650 [DOI] [PubMed] [Google Scholar]
- 228. Liu J, Chen P, Zheng C, Huang YP (2013) Characterization of maltocin P28, a novel phage tail‐like bacteriocin from Stenotrophomonas maltophilia . Appl Environ Microbiol 79: 5593–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Petrova M, Shcherbatova N, Kurakov A, Mindlin S (2014) Genomic characterization and integrative properties of phiSMA6 and phiSMA7, two novel filamentous bacteriophages of Stenotrophomonas maltophilia . Arch Virol 159: 1293–1303 [DOI] [PubMed] [Google Scholar]
- 230. Yu ZC, Chen XL, Shen QT, Zhao DL, Tang BL, Su HN, Wu ZY, Qin QL, Xie BB, Zhang XY et al (2015) Filamentous phages prevalent in Pseudoalteromonas spp. confer properties advantageous to host survival in Arctic sea ice. ISME J 9: 871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Jian H, Xiao X, Wang F (2013) Role of filamentous phage SW1 in regulating the lateral flagella of Shewanella piezotolerans strain WP3 at low temperatures. Appl Environ Microbiol 79: 7101–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Pederson DM, Welsh LC, Marvin DA, Sampson M, Perham RN, Yu M, Slater MR (2001) The protein capsid of filamentous bacteriophage PH75 from Thermus thermophilus . J Mol Biol 309: 401–421 [DOI] [PubMed] [Google Scholar]
- 233. Chopin MC, Rouault A, Ehrlich SD, Gautier M (2002) Filamentous phage active on the gram‐positive bacterium Propionibacterium freudenreichii . J Bacteriol 184: 2030–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Kim AY, Blaschek HP (1991) Isolation and characterization of a filamentous viruslike particle from Clostridium acetobutylicum NCIB 6444. J Bacteriol 173: 530–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Cantalupo PG, Calgua B, Zhao G, Hundesa A, Wier AD, Katz JP, Grabe M, Hendrix RW, Girones R, Wang D et al (2011) Raw sewage harbors diverse viral populations. MBio 2: e00180–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Capella‐Gutierrez S, Silla‐Martinez JM, Gabaldon T (2009) trimAl: a tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics 25: 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30: 1312–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]


