Abstract
The gut microbiome is intimately related to human health, but it is not yet known which functional activities are driven by specific microbes’ ecological configurations or transcription. We report a large-scale investigation of 372 human fecal metatranscriptomes and 929 metagenomes from a subset of 308 men in the Health Professionals Follow-up Study. We identified a metatranscriptomic “core” universally transcribed over time and across participants, often by different microbes. In contrast to the housekeeping functions enriched in this core, a “variable” metatranscriptome included specialized pathways that were differentially expressed both across participants and among microbes. Finally, longitudinal metagenomic profiles allowed ecological interaction network reconstruction, which remained stable over the six-month timespan, as did strain tracking within and between participants. These results provide an initial characterization of human fecal microbial ecology into core, subject-specific, microbe-specific, and temporally-variable transcription, and they differentiate metagenomically versus metatranscriptomically informative aspects of the human fecal microbiome.
Keywords: microbiome, gut, metagenome, metatranscriptome, ecology
Introduction
Alterations in the human gut microbiome have been implicated in a wide range of complex, chronic conditions including inflammatory bowel disease (IBD), obesity, diabetes, cancer, and cardiovascular disease1,2. There is an appreciable body of work on the metagenomic potential of fecal communities3–6, yet little information is available regarding transcriptional activity of the microbiome. The metatranscriptome represents a link between the metagenome and community phenotype, and surveying its molecular activity is important to understanding the functional ecology of the human gut microbiome.
Metatranscriptomics has most commonly been applied to ecological profiles of environmental microbial populations. For instance, deep sequencing of marine bacterioplankton RNA established transcript inventories, uncovered gene expression trends among metabolic generalists and specialists, and identified patterns of substrate use and elemental cycling in the ocean ecosystem7. Early human fecal metatranscriptomics suggested subject-specific relationships between microbiome transcripts and gene copy number, differing across biological functions8. Our own pilot work with the cohort studied here introduced protocols for integrating metatranscriptomic sampling into large scale epidemiological studies, demonstrating that metatranscriptional profiles are less variable than fecal taxonomic profiles but more individualized than metagenomic function9.
Previous studies have not, however, surveyed the human fecal metatranscriptome at sufficient scale to identify areas in which it is uniquely informative relative to the underlying metagenome. It is not yet clear, for example, which human conditions are associated with specific microbes in the gut, versus their metagenomic functional profiles, versus metatranscriptomic activity10. Culture-independent fecal microbial transcription has been used in only a few cases to date to identify causal mechanisms in health outcomes, such as strain-specific variation in Eggerthella lenta expression influencing the efficacy of cardiac therapy11. Integrated metagenomics and metatranscriptomics have also been used in molecular diagnostics for cancer risk and transplant rejection12. Correspondingly, it remains to be determined what short- or long-term health-linked exposures can be assessed using fecal metagenomes or metatranscriptomes in prospective cohorts.
To address these gaps, we interrogated the human fecal metagenome and metatranscriptome in 929 and 372 samples, respectively, collected at up to four time points each from 308 healthy senior men participating in the Men’s Lifestyle Validation Study (MLVS), nested within the Health Professionals Follow-up Study (HPFS)13. This manuscript provides an overview of these communities’ molecular biology and microbial ecology; a companion paper (Mehta in review) investigates the stability of features in such data for human population epidemiology. We differentiated “core” versus variably transcribed functions, assigned them to specific microbes, and assessed differences between subjects cross-sectionally and within subjects over time. Finally, ecological co-occurrence and strain diversity were assessed metagenomically, both remaining strikingly stable over time and the latter comparing near-identically between this and an independent population from the Human Microbiome Project (HMP). Together, these findings thus provide an in-depth large-scale exploration of the human fecal metatranscriptome in a species-specific context.
Results
Meta’omic taxonomic and functional profiling
We generated taxonomic and functional profiles of 308 participants’ stool microbiome samples at up to four time points each from DNA (n=929) and RNA (n=372) reads using MetaPhlAn214 and HUMAnN215 (Fig. 1 and Methods). From the former, a total of 468 microbial species were detected, with individual samples containing 72 ± 13 (mean ± s.d.) species. HUMAnN2 identified 1,569,171 unique UniRef90 gene families in metagenomes and 602,896 in metatranscriptomes (Supplementary Table 1). Overall, 75.3% of all DNA reads and 64.1% of all RNA reads were assignable to UniRef90 gene families by HUMAnN2; of these, 54.8% and 58.1% UniRef90 gene families possessed functional characterization, respectively, and finally 10.7% and 13.2% of characterized gene families were assignable to MetaCyc pathways16. Intriguingly, an average of 69% and 85.4% UniRef90 relative abundances for metagenomes and metatranscriptomes, respectively, were attributable to gene families lacking biochemical characterization. Stool sample collection, sequence data generation, and quality control are described in Methods.
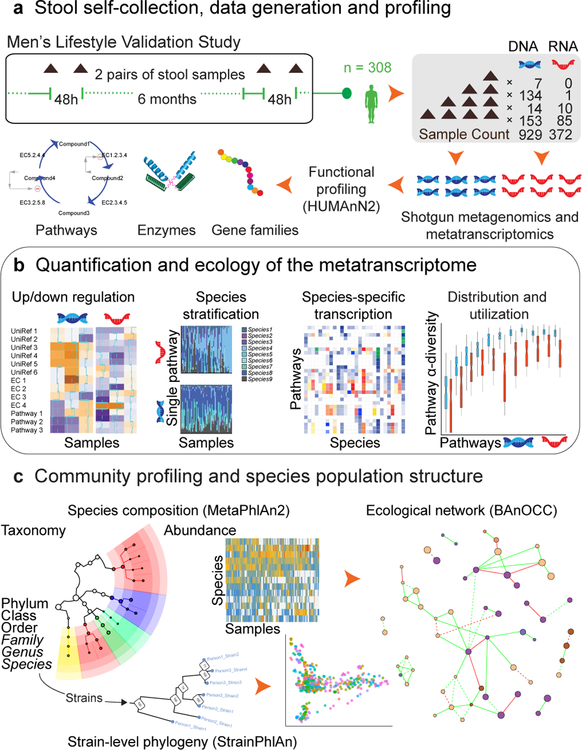
Figure 1. Metatranscriptomic and metagenomic taxonomic and functional profile of a prospective human cohort.
A) 308 participants from the Men’s Lifestyle Validation Study (MLVS), embedded within the Health Professionals Follow-up Study (HPFS) prospective cohort13, provided a target of four stool samples each. These were self-collected in two pairs, six months apart, with each pair spanning 2–3 days. This yielded 929 total metagenomes and 372 metatranscriptomes, sequenced using previously published protocols9 and functionally profiled using HUMAnN215. B) To estimate gene family, enzyme class, and pathway relative transcription, RNA abundances were normalized to corresponding DNA abundances. We then evaluated “core” (prevalently transcribed) and variable transcriptional elements, in addition to the ecological and phylogenetic diversity of metatranscription and carriage of functional elements among species. C) Taxonomic profiles were determined using MetaPhlAn214 from both DNA and RNA data (for RNA viruses). These were also used for ecological interaction network reconstruction25 with BAnOCC (Schwager in review) (http://huttenhower.sph.harvard.edu/banocc) and for strain tracking with StrainPhlAn28.
Prior to investigating the metatranscriptome, we compared the metagenome-based taxonomic profile of this older cohort to previous population studies (Supplementary Fig. 1), since the mean age of our participants was 69 ± 6 years. As in earlier studies with comparable populations and protocols17,18, and in contrast to younger cohorts with different sample handling methodology3, Firmicutes were generally prevalent and abundant, in contrast to previous studies of comparable but smaller populations such as ELDERMET18,19 (Supplementary Fig. 2). Additionally, a small number of both DNA and RNA viruses were quantified confidently by MetaPhlAn2, which is likely an underestimate of the gut virome diversity since our extraction protocol did not enrich for virus-like particles. Although gut viral ecology is more difficult to analyze than that of the bacteriome due to inadequate viral reference sequences20, these methods allow for some incidental analysis of DNA phage and RNA plant viruses in human fecal metagenomes and metatranscriptomes. (See Supplementary Information).
As a first indication of important differences between fecal metagenomic and metatranscriptomic profiles, each sample’s metatranscriptome contained, on average, 5.4% of the total pool of gene families observed across the dataset; metagenomes averaged >10% of the pool (Supplementary Table 1). This indicates that, as in a single organism’s genome, only a subset of fecal functional potential is active under the circumstances captured by a typical sample. Technical factors played a minor role in this, since although RNA was sequenced at slightly shallower depth (Supplementary Dataset 1), rarefaction indicates that metagenomic taxa, functions, and metatranscriptomic functions were well-saturated at these depths (Supplementary Fig. 3). Biological factors appeared to dominate, since transcribed elements should typically be at most those also observed metagenomically. Ecologically, this is also in agreement with our previous observation that the metatranscriptome is more variable than the metagenome9.
Core and variable fecal metatranscriptomes differ from the metagenome
To identify important pathways expressed (and not just metagenomically encoded) by microbes in the human gut, we delineated “core” and “variable” portions of the fecal metatranscriptome (Fig. 2; Supplementary Datasets 2 and 3). The former was defined as a set of prevalently transcribed pathways that was robust to sequencing depth and specific prevalence threshold (Supplementary Fig. 4). From 289 pathways with detectable transcription in at least two samples, 81 (28%) were core by this definition, 48 of which had a mean DNA-normalized transcript abundance >1 when transcribed (see Methods for quantification of metatranscriptional activity). This was remarkably smaller than a similarly defined core metagenome in this cohort: from 407 pathways with detectable DNA in at least two samples, 182 (45%) were similarly prevalent, even though there were almost three times more metagenomes than metatranscriptomes. It is noteworthy that neither GC content nor ORF length had effects on transcription ratios (see Supplementary Information and Supplementary Fig. 5). This suggests that gene expression rather than gene abundance underlies qualitative and quantitative differences among metatranscriptomes.
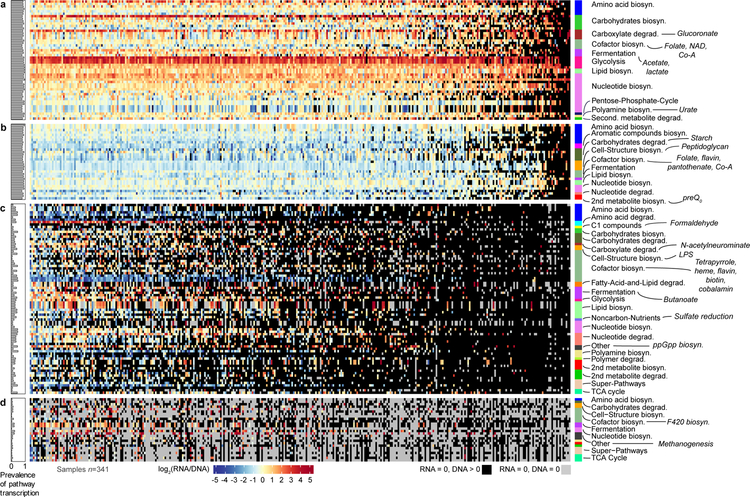
Figure 2. Core and variable metatranscriptomes of the stool microbiome.
DNA-normalized transcript abundances for 239 gut microbiome pathways with detectable RNA in >10 of the 341 metatranscriptomes, collected from 96 MLVS participants. Samples (columns) were sorted left to right based on decreasing number of transcribed pathways per sample. A) Core metatranscriptome pathways (transcribed in >80% of samples) with RNA:DNA transcription ratio >1. B) Low-expression core metatranscriptome pathways with transcript abundance detectable in >80% of samples but an RNA:DNA ratio <1. C) Variably metatranscribed pathways detected in DNA but below detection in at least half of RNA samples, and D) variably metatranscribed pathways below detection in DNA (and matching RNA) in 30%−80% of the 341 samples. Several pathways representative of functional categories are annotated, and the complete annotation of all pathway names and definitions are in Supplementary Fig. 6A–D. Thirty-eight pathways that did not fall into either of the four sections based on these criteria are in Supplementary Fig. 6E. The distribution range of pathways with the overall 30 highest and 30 lowest mean DNA-normalized transcript abundances among the 341 metatranscriptome samples are in Supplementary Fig. 6F. The grey color represents pathways that were below detection in both DNA and RNA in a given sample; the black color represents pathways that were detected in DNA but below detection in RNA.
Unlike the core metagenome, which includes a variety of host-adapted microbial community features3, the core metatranscriptome was enriched mainly for housekeeping functions. Nineteen nucleotide biosynthesis pathways, 19 glycolysis and carbohydrate metabolism pathways, and 15 amino acid biosynthesis pathways accounted for the majority of core metatranscribed pathways. Note that as annotated in MetaCyc, glycolysis represents an umbrella term that also includes anaerobic fermentation, not an indicator of aerobic respiration. Glycolysis in this sense had the highest transcript abundance (mean 8.30 ± s.d. 5.69) and, together with three nucleotide metabolism pathways, was over-transcribed (relative to DNA abundance) in virtually all metatranscriptome samples (Figs. 2A–B and Supplementary Fig. 6).
Other notable core metatranscriptome pathways included the non-oxidative pentose phosphate cycle (to support nucleic acid synthesis), breakdown of carboxylates, synthesis of cofactors (folate, flavin, pantothenate, and Co-A), unsaturated fatty acids, and microbial recycling of uric acid (the end product of purine breakdown) to salvage nitrogen. Conversely, cell wall peptidoglycan and phospholipids, and amino acid synthesis were pathways with the lowest transcript abundance in the core metatranscriptome. Interestingly, the core metatranscriptome also included synthesis of preQ0 and queosine, pleiotropic bacterial metabolites21 that the human host salvages for regulating translational efficiency and fidelity22.
In contrast to the limited core set of metatranscriptomic pathways, the variable metatranscriptome comprised 95 functionally diverse pathways well-detected in DNA but below detection in at least half of RNA samples (Fig. 2C). 36 of these had no detectable RNA in greater than two-thirds of metatranscriptome samples. The bulk of these pathways are involved in biosynthesis: various amino acids, long fatty acids, terpenoids, polyamines, cofactors (NAD, heme and tetrapyrolle), and the (p)ppGpp alarmone23. A smaller number were pathways involved in degradation of various alcohols, sugars, formaldehyde, and sulfate reduction. Finally, 26 pathways were below detection in most DNA (and matching RNA) samples. When transcribed, however, some of these pathways had the highest transcript abundance; e.g. methanogenesis and factor 420 biosynthesis (Fig. 2D and Supplementary Fig. 6F), suggesting that metagenomically rare but overtranscribed pathways may be uniquely responsible for subject-specific gut microbial bioactivity.
Fecal microbiome pathways are transcribed by a limited subset of microbes encoding them metagenomically
Using paired metagenomic and metatranscriptomic functional profiles, we next assessed the relationship between fecal microbes that tend to carry pathways metagenomically versus those that express them metatranscriptomically (Fig. 3 and Supplementary Fig. 7). At a global level, these two aspects of functional diversity corresponded (in large part since only microbes carrying a pathway can express it), with three main groups of pathways dominating the functional diversity of these samples. First, housekeeping functions (nucleotide, carbohydrate, amino acid metabolism, etc.) were carried by virtually all stool species (high contributional alpha diversity); they were also actively transcribed by abundant and prevalent microbes, predominantly Bacteroides, Eubacterium, and Ruminococcus spp (Fig. 3A, bottom cluster). A second pathway cluster (Fig. 3A, top left) with modest contributional alpha diversity was dominated by F. prausnitzii, a particularly prevalent organism in this cohort that, when abundant, tended to contribute the majority of all pathways it encodes (synthesis of various amino acids, non-oxidative pentose phosphate pathway, putrescine synthesis, etc.) The third, low diversity, cluster (Fig. 3A, top right) was encoded by limited numbers of opportunists, e.g. E. coli, Sutterella, Enterobacter, and Enterococcus spp. It included pathways such as synthesis of the enterobactin siderophore, lipid A, and sulfate reduction. Pyruvate fermentation to acetate and lactate, synthesis of glycogen, and tetrapyrrole were examples of pathways with fairly even metagenomic and metatranscriptomic species contributions. Thus in overall summary, the same microbes were principal contributors to RNA and DNA abundances in roughly one third of pathways. For the majority of pathways, however, overtranscription was observed from members of either the Bacteroidetes (mainly for pathways contributing to active growth in the gut) or the Proteobacteria (for the subset of generalists’ pathways upregulated in feces) relative to the baseline level of metagenomic carriage (e.g. by the Firmicutes).
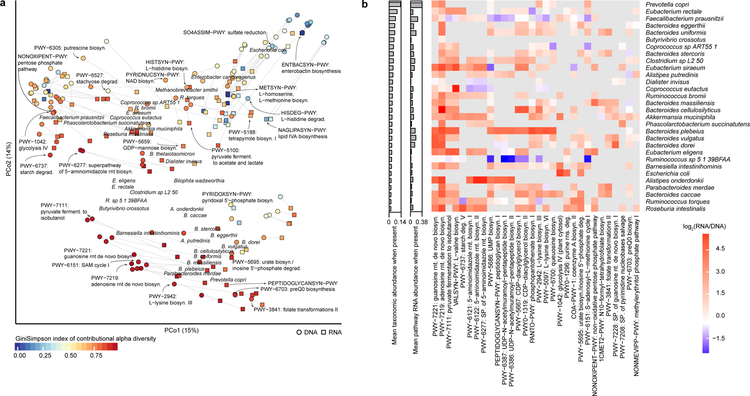
Figure 3. The gut metatranscriptome is personalized and broadly taxonomically distributed.
A) Structure of the stool metagenome and metatranscriptome as contributed by diverse species. Principal coordinates analysis of pathways with microbial species’ contributions to their DNA and RNA abundances using Bray-Curtis dissimilarity, with a biplot overlay indicating centroids of abundant species’ contributions. Each pathway is thus denoted by two points, summarizing the organisms contributing them metagenomically (averaged over 913 samples from 307 participants) and metatranscriptomically (341 samples from 96 participants), and a subset of examples are labeled. The resulting joint ordination indicates broad agreement between species carrying (metagenomically) and expressing (metatranscriptomically) groups of pathways in the fecal microbiome. B) Transcription ratios of 30 pathways that were most prevalently transcribed among the top 30 species, using the same datasets as in A. Pathways for which DNA or RNA were not detected in a given species are grey. A given pathway-species combination in the heatmap represents the transcript abundance averaged over all samples that measured a non-zero RNA/DNA ratio for that species. Only pathway-species combinations in at least 5 samples (from a total of 341) were considered. Columns in the heatmap were ordered based on average linkage clustering on a Euclidean distance matrix of log2 pathway transcription ratios.
These three major patterns in the functional structure of the fecal metatranscriptome are explained in greater detail by the pathways most commonly expressed by abundant microbes. First, all species shared enrichments for housekeeping functions (e.g. nucleotide synthesis, fermentation, etc.) (Fig. 3B, left columns). A set of anaerobic biosynthesis pathways were mainly expressed by Firmicutes from the upper left cluster of the ordination (Fig. 3B, middle columns). Transcription of cell structure, secondary metabolites, and co-factor synthesis pathways was characteristic of Bacteroides spp (Fig. 3B, right columns). Finally, the Enterobacteriaceae were not abundant in most subjects, so few of their pathways were prevalent enough to appear in those selected for visualization, but the subset of their large pangenome upregulated in feces overlapped to a degree with the anaerobic metabolism expressed by the Firmicutes (Fig. 3B, middle columns).
Many pathways are transcribed by few organisms per community, even when broadly encoded metagenomically
Finally, we noted that per-microbe pathway expression is often very different among individual hosts than is metagenomic pathway carriage, indicating that these two molecular measurements may have distinct applications in human population studies (Fig. 4). Overall, metagenomic richness (number of contributing species) generally exceeded metatranscriptomic richness, as expected. As above, a subset of pathways were both broadly encoded and expressed (high diversity), with relatively few differences between individuals, and these were again mainly housekeeping functions. However, at the other end of the spectrum, many pathways of greater biochemical interest were transcribed by only one or a few species, even when diversely carried metagenomically. An extreme example of low transcriptional diversity pathways was methanogenesis, encoded and transcribed solely by Methanobrevibacter smithii; that this class of metatranscriptomic functions may indicate keystone processes and their associated microbes in the gut.
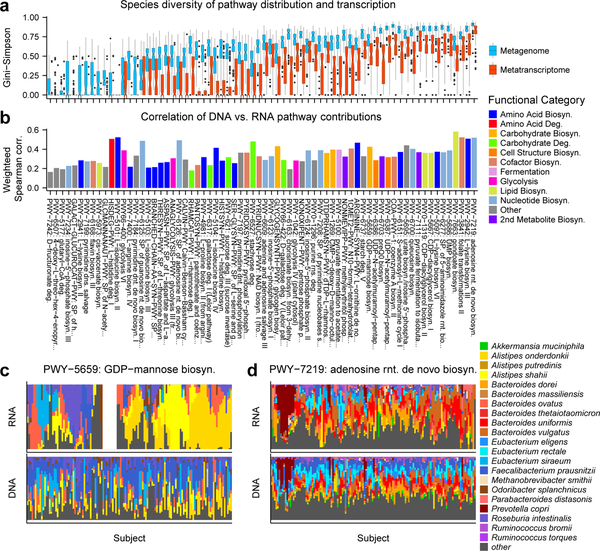
Figure 4. Transcriptional landscape of the stool microbiome.
A) Distributions of alpha diversity (Gini-Simpson index) for the species-specific metagenomic and metatranscriptomic contributions to each pathway, for 70 non-redundant pathways with the highest community level RNA abundances, averaged across 341 metatranscriptomes from 96 participants. Pathways were sorted by the sum of the median metagenomic alpha diversity and the weighted Spearman correlation from B. Boxplot whiskers represent 1.5 times the inter-quartile range from the first and third quartiles. B) Concordance of metagenomic potential with metatranscriptomic activity (metagenome-weighted mean of per-species Spearman correlations; see Methods). Metatranscriptomic diversity is, as expected, consistently lower than metagenomes, with pathways carried by only a few organisms also more differentially transcribed. Metagenomic potential (bottom) and metatranscriptomic activity (top) for example pathways with differing ecological structure, specifically C) GDP-mannose biosynthesis and D) adenosine ribonucleotide de novo biosynthesis. Abundances were normalized within each pathway for 189 subject-week pairs, from 96 participants. Subjects were ordered to emphasize blocks of subjects with similar metatranscriptomic profiles (see Methods). The top 8 (C) and top 15 (D) species in terms of their mean metatranscriptomic contribution to the pathways in C and D are shown for clarity. Examples show transcriptional ecologies that either differ strikingly from (C) or generally mirror (D) their metagenomic diversity.
To better understand this phenomenon, and to expand our previous observation of basal transcription in a substantial fraction of gut pathways9, we next identified pathways for which microbes’ transcriptional activities mirrored their metagenomic carriage (Fig. 4B). For this, we used the weighted mean of the Spearman correlation between taxonomically stratified metagenomic and metatranscriptomic profiles (see Methods). To emphasize the correlation of the principal species encoding each pathway, correlation coefficients were weighted by the mean metagenomic potential of the species. High values indicated that species-level RNA abundances closely follow DNA abundances, while low values indicated departure from basal expression. In general, carbohydrate metabolism and nucleotide biosynthesis particularly tended to be enriched for high correlations, while amino acid biosynthesis was enriched among low correlations. Low correlations indicate more context-specific, variable expression of the pathway, consistent with the generally amino acid-rich environment of the gut. Cofactor biosynthesis pathways were roughly in the middle of this range, perhaps due to the diversity of compounds produced with these functions and their variable availability in the gut. For example, pantothenate is found in most foods and easily salvaged from the gut environment, while folate transformations are critical for C1 metabolism24, making this pathway more widely transcribed when present in species.
Notably, pathways with similar metagenomic contributions were not necessarily similarly transcribed (Fig. 4C–D). Core pathways were both metagenomically and metatranscriptomically diverse, carried and expressed by many organisms per community (Fig. 4D and Supplementary Fig. 8). Typical variable pathways, however, even when broadly distributed metagenomically, were often transcribed by one or few organisms per individual. Transcribing organisms were often neither the most abundant nor the same species across individuals (Fig. 4C). Similar patterns were observed for L−isoleucine biosynthesis III (PWY-5103), L−tryptophan biosynthesis (TRPSYN-PWY), degradation of various sugars (stachyose (PWY-6527), sucrose (PWY-621), rhamnose (RHAMCAT−PWY)), non-oxidative pentose phosphate pathway, preQ0 biosynthesis (PWY−6703), and others (Supplementary Fig. 8). This finding highlights another aspect of inter-individual diversity in the microbiome: different microbes may activate shared pathways among individuals, with as-yet-unknown functional specializations and consequences.
Inter-microbial species interactions are stable in the stool ecosystem of older men
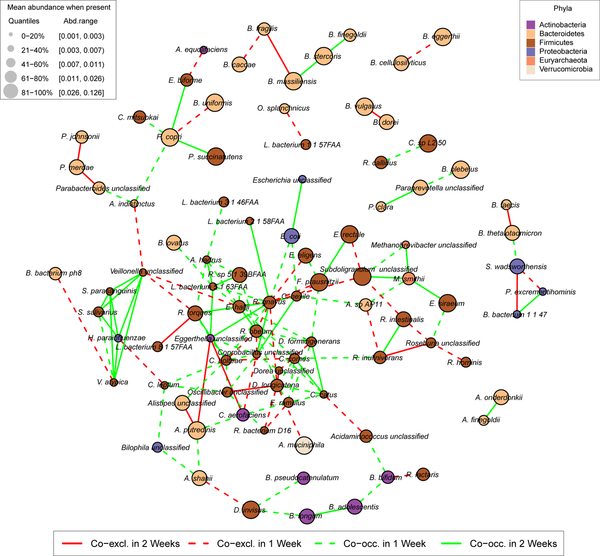
To leverage this cohort’s taxonomic profiles independently of their metatranscriptomes, we also inferred metagenomic microbial interaction networks25 using the BAnOCC Bayesian framework (see Methods, Fig. 5 and Supplementary Fig. 9). We generated one network per time point, allowing the stability of co-occurrence and -exclusion relationships to be determined. Negative associations between Bacteroides and Ruminococcaceae or Prevotellaceae observed previosly25 were not recapitulated in this population, and Firmicutes species predominantly co-occurred with other members of the phylum. This is consistent with the observation that co-occurrence/-exclusion among microbial community members is not based solely on phylogenetic relatedness, but also on how microbes complement each other functionally26.
Figure 5. Ecological interactions in the gut microbiome.
Significant co-variation and co-exclusion relationships among 104 species in 913 stool metagenomes from 307 MLVS participants. Each node represents a species and edges correspond to significant interactions inferred by BAnOCC (see Methods). Stool microbiome taxonomic profiles were averaged within each subject for the first and second collection pairs (separated by 6 months). Interactions in at least one time point are included here. No alternating associations (positive at one time and negative in another) were detected. 95% credible interval criteria was used to assess significance, and only estimated absolute correlations with effect sizes >=0.15 are reported. Networks for individual time points are in Supplementary Fig. 9.
Genetic divergence patterns of stool-associated bacterial strains is species-specific and preserved among host populations
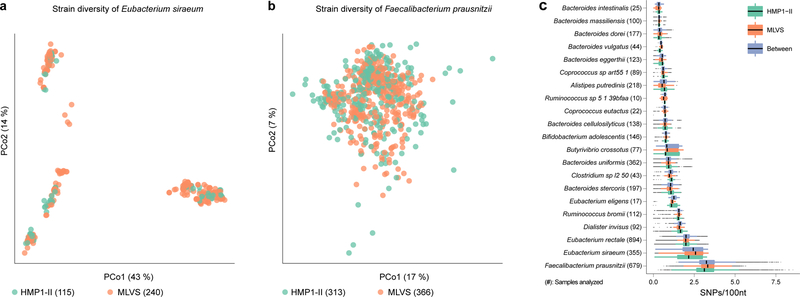
Finally, we assessed strain-level variation in stool bacterial populations using StrainPhlAn (see Methods) and provided a comparison of species population structure between the MLVS and the Human Microbiome Project3,27 cohorts (Fig. 6). Twenty-one species had sufficient genomic coverage for reliable strain identification from metagenomes in both cohorts. Eubacterium siraeum, for example, demonstrated discrete strain clustering indicative of a clonal population, with strains from the same individual remaining near-identical over the sampling period. Conversely, nucleotide variation among Faecalibacterium prausnitzii strains did not reveal a discernible pattern and was characterized by the highest median and widest range of nucleotide substitution rates, consistent with extreme genomic diversity whereby each strain can be substantially distinct from another. Several species, including Bacteroides stercoris, B. uniformis, and Butyrivibrio crossotus presented an intermediate structure with weak clonal propagation of a potential outgroup (Supplementary Fig. 10).
Figure 6. Species-specific patterns of evolutionary divergence within species preserved across cohorts.
Panels show strain-level diversity within A) Eubacterium siraeum and B) Faecalibacterium prausnitzii. Each point represents one sample’s strain, ordinated by principal coordinate analysis of sequence dissimilarity (Kimura Two-Parameter distance). C) Pairwise nucleotide substitution rates within and between cohorts for 21 out of 30 species in Fig. 3 with sufficient prevalence in both cohorts for informative comparison. Lines represent median values, points denote outliers outside 1.5 times the interquartile range. All numbers in parenthesis are sample counts in which indicated strains were above limit of detection; from a total of 913 MLVS stool metagenomes and 553 HMP stool metagenomes (from 253 male and female HMP participants) that were analyzed with StrainPhlAn.
Remarkably, nucleotide substitution rates were near-identical between the two unrelated cohorts (Fig. 6C). The average ratio of between-cohort to within-microbe divergence was 1.0±0.03 (mean±s.d.), indicating that bacterial population structure is consistent across host populations. Previous studies have shown that human gut microbes can diverge between geographically and genetically distinct host populations28. However, the existence of such closely related microbial strains between independent North American cohorts with very different age ranges (18–40 vs. 65–81) suggests that specific microbial strains (or related strain groups) may be a useful, stable feature to assay during epidemiological studies.
Discussion
The present study has provided an overview of the fecal metatranscriptome in a prospective, large-scale cohort of elderly males; identified core and variably transcribed pathways; delineated how these differ from metagenomic functional potential; and ascribed them to specific contributing organisms. Finally, paired metagenomes in this study also allowed species-specific ecological interaction networks to be reconstructed, which proved stable over time, as did strain tracking within species. This stability, in combination with the commonality of strain-level microbial population structures between cohorts, suggests that they might represent particularly effective measurement targets for the microbiome in population studies. Together, these findings extend our earlier pilot study in eight individuals9 and accompany epidemiological work (Mehta et al, in press) to integrate metatranscriptomics into population studies.
It is evident from our results that the metatranscriptome is more temporally dynamic, context-sensitive, and species-specific than the metagenome. The observed incongruence between species abundance and transcriptional activity agrees with an earlier study of taxa-function relationships in the human fecal metatranscriptome29. This is expected, as transcription is a highly variable process even among cells of the same species under steady-state conditions30. Heterogeneity among species’ transcriptional contributions in otherwise similar metagenomes, however, may be influenced by many factors, including nutrient availability, preferential utilization, or xenobiotics; temporal differences in environmental sensing that stagger response to stimuli among species; and metabolic dependence that drives cross-feeding of intermediate metabolites through community members31,32. This extends the typical model of transcriptional behavior from individual microbial or metazoan cell populations to niche ecology.
A feature of the metatranscriptome, which is critical for human microbiome population studies, was the propensity of different organisms to appear as primary transcribers of pathways between individuals. In relating metagenomic features to the metatranscriptome, we observed only 44% of the “core” metagenomic potential (81 transcribed pathways out of 182 prevalent metagenomically) to be transcribed in the cohort. In combination with previous studies of “core” metagenomics17,33,34, this suggests a functional ecological model in which a prevalent metagenome encoding substantial redundancy is distributed among many microbes per individual, with the microbes containing this core varying among hosts. Transcription at any one time or in any given environment is then typically dominated by one or a few members. This model of microbial ecology would be analogous to silencing versus upregulation of distinct portions of the human genome among cell types, which also consists of a “core” underlying DNA genome with long-term (epigenetics, instead of phylogeny) and short-term (transcriptional) regulatory mechanisms35.
In addition to these insights into metatranscriptional activity, stool taxonomic profiles in the MLVS cohort remained diverse and stable, in agreement with previous studies of the microbiome in the elderly19. Few profiles of this unique ecosystem have yet been generated, however, and those that do exist tend to employ widely varied technical characteristics and study design, prohibiting direct comparisons. ELDERMET36, for example, posited a trend toward Bacteroidetes dominance in aging, but this replicated neither across technologies within this cohort nor in our MLVS data. As MLVS data are drawn from within the broader HPFS, for which several decades of dietary and environmental data are available, we anticipate that future studies focusing on these detailed metadata will further detail microbial links to lifestyle and nutrition.
A challenge going forward will thus be to identify epidemiological contexts in which metatranscriptomic features are specifically informative, and the appropriate ways in which to measure them. This might include, for example, tests for health outcomes that are predicted uniquely by metagenomic activities. These may, based on this study’s results, be functionally consistent but contributed by different microbes across individuals, even when not differentially represented in underlying metagenomes. It also remains to associate metatranscriptomic responses with detailed information on immediate lifestyle exposures, such as recent diet, to determine temporal responses of the metatranscriptome to key environmental perturbations. Ultimately, if there exist health outcomes for which causal molecular mechanisms are uniquely detectable in the fecal metatranscriptome, its functional profile will need to be better characterized and integrated as a measurement in human epidemiological population studies.
Methods
MLVS Cohort, stool sample collection, shotgun sequencing and quality control.
The Health Professionals Follow-Up Study (HPFS) is a prospective cohort study aimed at investigating the determinants of men’s health, into which 51,529 U.S. men aged 40–75 were recruited in 1986 and subsequently followed biennially13. For this analysis, we used data from a sub-study of the HPFS, the Men’s Lifestyle Validation Study (MLVS), in which 308 participants provided up to two pairs each of self-collected stool samples from consecutive bowel movements, during 2012. The second pair of samples was collected approximately six months after the first. The median time between consecutive bowel movements for a pair of samples was 48 hours; collection dates are in Table S2. At the time of collection, age of participants ranged between 65 and 81 years. Cohort details, sample collection and immediate ex-situ conservation of metagenomic and metatranscriptomic components, laboratory handling, and paired-end (100 × 100 nt) shotgun sequencing of RNA and DNA are detailed in the companion manuscript (Mehta et al., in press) and in our pilot study9, respectively. Study protocol 22067–102 titled “Men’s Lifestyle Validation Study and Microbiome Correlation” was approved by the Harvard Chan School of Public Health Institutional Review Board, and informed consent was obtained from all participants.
The gut microbiome, as captured by stool, was sampled from 308 male participants (ages 65–81) within the MLVS sub-cohort of the HPFS (Fig. 1). Each participant provided up to two pairs of self-collected stool samples from consecutive bowel movements; with the second pair of samples collected approximately six months after the first. DNA was extracted from all 929 resulting samples, in addition to RNA from a subset of 372 samples spanning 96 participants. Illumina HiSeq sequencing yielded a total of 4.5 Tnt of paired-end reads (100×100 nt). This included an average of 3.8 Gnt ± 1.5 Gnt (mean ± s.d. Giga nucleotides) before quality filtering (see below) and 1.9 Gnt ± 0.7 Gnt afterward per metagenome, and 3.0 Gnt ± 2.4 Gnt and 1.3 Gnt ± 1.0 Gnt before and after quality control for metatranscriptomes. Forty-one samples (16 DNA and 25 RNA) had <1M reads after quality filtering and were excluded from further analysis. Thus, the final datasets analyzed comprised 913 metagenomes and 347 metatranscriptomes.
Taxonomic and functional profiling of metagenomic and metatranscriptomic samples.
Sequence reads were passed through the KneadData v0.3 quality control pipeline (http://huttenhower.sph.harvard.edu/kneaddata), which incorporates the Trimmomatic37 and BMTagger38 filtering and decontamination algorithms to remove low quality read bases (thresholding Phred quality score at <20) and remove reads of human origin, respectively. Trimmed non-human reads shorter than 70 nt were discarded. Taxonomic profiling was performed using the MetaPhlAn2 classifier14, which relies on approximately 1 M clade-specific marker genes derived from 17,000 microbial genomes (corresponding to >7,500 bacterial, viral, archaeal, and eukaryotic species) to unambiguously classify metagenomic reads to taxonomies and yield relative abundances of taxa identified in the sample. We quality controlled taxonomic profiles by requiring at least 10% of clade-specific markers to recruit at least 1 read per kilobase (RPK) for inclusion in subsequent analyses. In addition to DNA, RNA (cDNA) reads were also analyzed with MetaPhlAn2 to quantify RNA viruses.
Metagenomes and metatranscriptomes were functionally profiled using HUMAnN215 to quantify genes and pathways (http://huttenhower.sph.harvard.edu/humann2). Briefly, for each sample, taxonomic profiling is used to identify detectable organisms. Reads are recruited to sample-specific pangenomes including all gene families in any detected microbes using Bowtie239. Unmapped reads are aligned against UniRef9040 using DIAMOND translated search41. Hits are counted per gene family and normalized for length and alignment quality. For calculating abundances from reads that map to more than one reference sequence, search hits are weighted by significance (alignment quality, gene length, and gene coverage). UniRef90 abundances from both the nucleotide and protein levels were then i) mapped to level 4 Enzyme Commission (EC) nomenclature and ii) combined into structured pathways from MetaCyc16. We used the MinPath42 and gap filling options in HUMAnN2 version 0.8.0. For the purposes of functional profiling, each read can be i) mapped to a specific organism’s characterized gene family in one or more known pathways, ii) mapped to a characterized protein family (without assignment to a specific organism), iii) mapped to an uncharacterized gene family (not in any pathway), or iv) not mapped to any gene family. HUMAnN2 refers to these as i) species-specific, ii) unclassified, iii) unintegrated, and iv) unmapped reads, respectively. Per sample breakdown of HUMAnN2 mapping categories (i.e. mapped, unclassified, unintegrated, and unmapped RPKs) are in provided in Supplementary Dataset 5. Reads mapped only at the amino acid level are not used when calculating specific taxa’s functional contributions. Instead, only reads mapped (unambiguously) at the nucleotide level are included in these totals.
Quantification of metatranscriptomic functional activity
Metatranscriptomic functional activity was assessed in the 341 samples with both RNA and DNA data in a manner not unlike two-channel microarrays using RNA:DNA ratios (see RNA/DNA normalization below). Due to the compositionality of RNA and DNA measurements, the resulting ratio is relative to the mean transcript abundance of the entire microbial community. That is, a ratio of 1 implies that the pathway is transcribed at the mean transcription abundance of all pathways in the microbial community. These quotients of RNA:DNA feature abundances allowed unbiased comparison of transcript abundances of metagenomic features between samples and also provided a comparative index of over/under-transcription (relative to DNA copy number) within individual microbiome samples. Pathways that had <1 RPK (reads per kilobase) of either RNA or DNA were treated as not detected in the analyses.
RNA/DNA normalization.
Metatranscriptomic features (i.e. RNA abundances of genes, enzymes, pathways) were normalized to corresponding metagenomic features to obtain an estimate of the mean transcription abundance λ as follows:
Rf,i and Df,i are the counts, in RPK, of feature f in sample i, for the metatranscriptome and metagenome respectively. H(x) is a unit step function with threshold x, and t is the detection threshold, here set to 1 RPK. This threshold ensures that RNA and DNA abundances that are confidently quantified (>1 RPK) are included, reducing the effect of increased noise due to genes and transcripts with low sequencing coverage. Summary statistics and analyses were only performed where both the numerator and denominator are measurable, although such gene families were tracked in RNA or DNA alone, respectively. Note that a value λf,i = 1 does not imply that the feature has an equal number of copies in RNA as it does in DNA. Rather, it implies that the mean transcription abundance is equal to the global mean transcription abundance of all organisms in the community in sample i. Thus, λf,i > 1 implies a transcript abundance above the community mean, which may be different in different samples.
EC dispersion.
Co-expression of functionally-related ECs was quantified by the mean variance of the standardized EC expression log-ratios for the set of ECs contributing to a given pathway. Specifically:
where xec,i is the expression ratio for EC ec in sample i, P is a given pathway, ⟨●⟩ represents the mean, and Var[●] the variance. When the expression of functionally-related ECs is not related (i.e. uncorrelated), then this value is expected to be 1. Lower values indicate the presence of co-expression of ECs, with 0 indicating a perfect relationship.
Species-specific meta’omic concordance.
Concordance between species-level metagenomic and metatranscriptomic pathway abundances was assessed using the mean of the Spearman correlation, weighted by the mean metagenomic contribution of each species to the overall pathway abundance. After averaging across multiple samples per subject, we calculated:
where dp,s,i and rp,s,i are the relative abundances of pathway p, contributed by species s in sample i in DNA and RNA, respectively. Spearman is the Spearman correlation between two vectors, where ties are given the mean rank of the tied values, and defined to be 0 when either vector has no variance. This weighting downweights the concordance for species which do not contribute much of the pathway’s abundance, mitigating the uncertainty inherent in estimating transcript abundances of low-abundance species and genes.
Microbial ecology networks.
Ecological covariation was assessed using BAnOCC (Bayesian Analysis of Compositional Covariance), a Bayesian model for detecting significant pairwise associations in compositional data25 (http://huttenhower.sph.harvard.edu/banocc). Briefly, BAnOCC models the sequence generation process using a lognormal distribution on unobserved absolute counts and constrains the associated correlation matrix through a sparsity-inducing prior. For posterior inference, we use the 95% credible interval, i.e. a correlation estimate is considered significant if the corresponding 95% credible interval excludes zero. We estimated ecological networks for the two sampling time points independently, from within-subject means of species abundance profiles, and then overlaid the networks to assess similarities and differences between networks over the length of the sampling period.
Inference of strain-level population structure.
Strain profiling was carried out using StrainPhlAn v1.028 (http://segatalab.cibio.unitn.it/tools/strainphlan). Briefly, after mapping reads to MetaPhlAn2 species-specific markers for sufficiently abundant species in each sample, a per-sample consensus sequence is built for each marker. For each species, these are concatenated, aligned, and variants identified relative to reference. Here, pairwise evolutionary distances were calculated from these variant alignments, with the Kimura Two-Parameter distance43 for ordination analysis using R packages vegan and ggplot2.
Structure of the stool metagenome and metatranscriptome as contributed by diverse species.
The input for the joint ordination of pathways and species (Fig. 3A) was the pathway genomic and transcriptional abundance of known taxonomic provenance only, averaged over 913 metagenomes and 341 metatranscriptomes. Species contributing <1.0E-7 metagenomic relative abundance to a pathway in <5% of pathways were removed, and the same criteria were used to remove metagenomic pathways found in species. Out of 339 species that were found to contribute to 219 pathways, 223 (65.8%) species and 109 (34.2%) pathways satisfied the filtering criteria. The metatranscriptome pathway by species matrix was subset to the same 109 pathways as for metagenomes, which were contributed by 194 species, and merged with the DNA pathway by species matrices into a single input matrix.
Supplementary Material
Acknowledgments
We thank the participants in the Men’s Lifestyle Validation Study and the Human Microbiome Project who graciously contributed to this research. This work was supported by funding from STARR Cancer Consortium Award #I7-A714 to CH, NCI R01CA202704 (ATC, CH, JI), NIDDK DK098311 (ATC), and NIDDK U54DE023798 (CH). JI is further supported by Nebraska Tobacco Settlement Biomedical Research Development Funds (JI). KLI is supported by the National Health and Medical Research Council. Components of the Men’s Lifestyle Validation Study were supported by NCI U01CA152904 and UM1 CA167552. RM is supported by a Howard Hughes Medical Institute Fellowship Award. The authors are also grateful for initial pilot funding provided by B Wu and Eric Larsen.
Footnotes
Data availability
Sequence data have been deposited in the Sequence Read Archive under BioProject accession PRJNA354235. Data from the Health Professionals Follow-up Study including metadata not included in the current manuscript but collected as a part of the MLVS can be obtained through written application. As per standard controlled access procedure, applications to use HPFS resources will be reviewed by our External Collaborators Committee for scientific aims, evaluation of the fit of the data for the proposed methodology, and verification that the proposed use meets the guidelines of the Ethics & Governance Framework and the consent that was provided by the participants. Investigators wishing to use HFPS / MLVS cohort data are asked to submit a brief (2 pages) description of the proposed project (“letter of intent”) to Dr. Eric Rimm, HPFS Director (erimm@hsph.harvard.edu).
Competing interests
The authors declare no competing financial interests.
References
- 1.O’Doherty KC, Virani A & Wilcox ES The Human Microbiome and Public Health: Social and Ethical Considerations. Am J Public Health 106, 414–420, doi: 10.2105/AJPH.2015.302989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shreiner AB, Kao JY & Young VB The gut microbiome in health and in disease. Curr Opin Gastroenterol 31, 69–75, doi: 10.1097/MOG.0000000000000139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium, H. M.P. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214, doi: 10.1038/nature11234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vatanen T et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 165, 1551, doi: 10.1016/j.cell.2016.05.056 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Le Chatelier E et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546, doi: 10.1038/nature12506 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Korpela K et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nature communications 7, 10410, doi: 10.1038/ncomms10410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satinsky BM et al. Microspatial gene expression patterns in the Amazon River Plume. Proceedings of the National Academy of Sciences of the United States of America 111, 11085–11090, doi: 10.1073/pnas.1402782111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A 107, 7503–7508, doi: 10.1073/pnas.1002355107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franzosa EA et al. Relating the metatranscriptome and metagenome of the human gut. Proceedings of the National Academy of Sciences of the United States of America 111, E2329–2338, doi: 10.1073/pnas.1319284111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segata N et al. Computational meta’omics for microbial community studies. Molecular systems biology 9, 666, doi: 10.1038/msb.2013.22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haiser HJ et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298, doi: 10.1126/science.1235872 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD & Craig DW Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nature reviews. Genetics 17, 257–271, doi: 10.1038/nrg.2016.10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan AT et al. Aspirin dose and duration of use and risk of colorectal cancer in men. Gastroenterology 134, 21–28, doi: 10.1053/j.gastro.2007.09.035 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truong DT et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nature methods 12, 902–903, doi: 10.1038/nmeth.3589 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Abubucker S et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS computational biology 8, e1002358, doi: 10.1371/journal.pcbi.1002358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caspi R et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic acids research 44, D471–480, doi: 10.1093/nar/gkv1164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin J et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65, doi: 10.1038/nature08821 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claesson MJ et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184, doi: 10.1038/nature11319 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Claesson MJ et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proceedings of the National Academy of Sciences of the United States of America 108 Suppl 1, 4586–4591, doi: 10.1073/pnas.1000097107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virgin HW The virome in mammalian physiology and disease. Cell 157, 142–150, doi: 10.1016/j.cell.2014.02.032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarty RM & Bandarian V Biosynthesis of pyrrolopyrimidines. Bioorg Chem 43, 15–25, doi: 10.1016/j.bioorg.2012.01.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinayak M & Pathak C Queuosine modification of tRNA: its divergent role in cellular machinery. Biosci Rep 30, 135–148, doi: 10.1042/BSR20090057 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T & Gerdes K Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nature reviews. Microbiology 13, 298–309, doi: 10.1038/nrmicro3448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chistoserdova L, Kalyuzhnaya MG & Lidstrom ME The expanding world of methylotrophic metabolism. Annu Rev Microbiol 63, 477–499, doi: 10.1146/annurev.micro.091208.073600 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faust K et al. Microbial co-occurrence relationships in the human microbiome. PLoS computational biology 8, e1002606, doi: 10.1371/journal.pcbi.1002606 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy R & Borenstein E Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proceedings of the National Academy of Sciences of the United States of America 110, 12804–12809, doi: 10.1073/pnas.1300926110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lloyd-Price J et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66, doi: 10.1038/nature23889 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truong DT, Tett A, Pasolli E, Huttenhower C & Segata N Microbial strain-level population structure and genetic diversity from metagenomes. Genome research, doi: 10.1101/gr.216242.116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gosalbes MJ et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One 6, e17447, doi: 10.1371/journal.pone.0017447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez A & Golding I Genetic determinants and cellular constraints in noisy gene expression. Science 342, 1188–1193, doi: 10.1126/science.1242975 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pande S et al. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. The ISME journal 8, 953–962, doi: 10.1038/ismej.2013.211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Souza G & Kost C Experimental Evolution of Metabolic Dependency in Bacteria. PLoS Genet 12, e1006364, doi: 10.1371/journal.pgen.1006364 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan XC et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome biology 13, R79, doi: 10.1186/gb-2012-13-9-r79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J et al. An integrated catalog of reference genes in the human gut microbiome. Nature biotechnology 32, 834–841, doi: 10.1038/nbt.2942 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Pimentel D Population Regulation and Genetic Feedback. Science 159, 1432–1437 (1968). [DOI] [PubMed] [Google Scholar]
- 36.O’Toole PW & Jeffery IB Gut microbiota and aging. Science 350, 1214–1215, doi: 10.1126/science.aac8469 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, doi: 10.1093/bioinformatics/btu170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Consortium HMP A framework for human microbiome research. Nature 486, 215–221, doi: 10.1038/nature11209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzek BE, Huang H, McGarvey P, Mazumder R & Wu CH UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282–1288, doi: 10.1093/bioinformatics/btm098 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Buchfink B, Xie C & Huson DH Fast and sensitive protein alignment using DIAMOND. Nature methods 12, 59–60, doi: 10.1038/nmeth.3176 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Ye Y & Doak TG A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS computational biology 5, e1000465, doi: 10.1371/journal.pcbi.1000465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura M A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16, 111–120 (1980). [DOI] [PubMed] [Google Scholar]
- 44.Wesolowska-Andersen A et al. Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2, 19, doi: 10.1186/2049-2618-2-19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shafquat A, Joice R, Simmons SL & Huttenhower C Functional and phylogenetic assembly of microbial communities in the human microbiome. Trends in microbiology 22, 261–266, doi: 10.1016/j.tim.2014.01.011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang YM & Rock CO Membrane lipid homeostasis in bacteria. Nature reviews. Microbiology 6, 222–233, doi: 10.1038/nrmicro1839 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.