Abstract
Cebpa encodes a transcription factor (TF) that plays an instructive role in the development of multiple myeloid lineages. The expression of Cebpa itself is finely modulated, as Cebpa is expressed at high and intermediate levels in neutrophils and macrophages respectively and downregulated in non-myeloid lineages. The cis-regulatory logic underlying the lineage-specific modulation of Cebpa’s expression level is yet to be fully characterized. Previously, we had identified 6 new cis-regulatory modules (CRMs) in a 78kb region surrounding Cebpa. We had also inferred the TFs that regulate each CRM by fitting a sequence-based thermodynamic model to a comprehensive reporter activity dataset. Here, we report the cis-regulatory logic of Cebpa CRMs at the resolution of individual binding sites. We tested the binding sites and functional roles of inferred TFs by designing and constructing mutated CRMs and comparing theoretical predictions of their activity against empirical measurements in a myeloid cell line. The enhancers were confirmed to be activated by combinations of PU.1, C/EBP family TFs, Egr1, and Gfi1 as predicted by the model. We show that silencers repress the activity of the proximal promoter in a dominant manner in G1ME cells, which are derived from the red-blood cell lineage. Dominant repression in G1ME cells can be traced to binding sites for GATA and Myb, a motif shared by all of the silencers. Finally, we demonstrate that GATA and Myb act redundantly to silence the proximal promoter. These results indicate that dominant repression is a novel mechanism for resolving hematopoietic lineages. Furthermore, Cebpa has a fail-safe cis-regulatory architecture, featuring several functionally similar CRMs, each of which contains redundant binding sites for multiple TFs. Lastly, by experimentally demonstrating the predictive ability of our sequence-based thermodynamic model, this work highlights the utility of this computational approach for understanding mammalian gene regulation.
Introduction
CCAAT/Enhancer binding protein, α (Cebpa) encodes a TF that is necessary for neutrophil development [1] as well as the specification of hepatocytes and adipocytes [2, 3]. During hematopoiesis, Cebpa is expressed in hematopoietic stem cells, granulocyte-monocyte progenitors (GMPs), neutrophils, and macrophages (http://biogps.org/gene/12606; [4, 5]). Although the most apparent hematopoietic phenotype of Cebpa−/− mice is neutropenia [1], Cebpa also has a role in specifying macrophages. Cebpa is expressed at intermediate and high levels in macrophages and neutrophils respectively and the cell-fate decision is thought to depend on the ratio of PU.1, a TF necessary for white-blood cell lineages [6], and C/EBPα expression levels [7]. Correspondingly, the cell-fate decision has been modeled as a bistable switch in which PU.1 and C/EBPα activate the mutual antagonists Egr1/2 and Gfi1 respectively [8]. Cebpa is also sufficient for specifying macrophages, since B-cells can be transdifferentiated into them by expressing Cebpa ectopically [9].
Despite its essential and pleiotropic functions, the cis regulation of Cebpa during hematopoiesis is poorly understood. C/EBPα, C/EBPβ, and C/EBPδ are known to activate Cebpa by binding to its proximal promoter [2]. Recently, ZNF143 was shown to bind and activate the promoter in a human myeloid cell line [10]. Cebpa is regulated in 32Dcl3 myeloid cells by PU.1, other Ets TFs, SCL, Gata2, Myb, and C/EBPα, which bind to an enhancer located 37kb downstream of the gene [11, 12]. It is not known whether, like other pleiotropic TFs [13], Cebpa is also regulated by multiple CRMs. More importantly, it is not understood how the regulatory contributions of these and other TFs modulate Cebpa’s gene expression during differentiation. In this study, we decode the regulatory logic of seven cis-regulatory elements (CRMs) of Cebpa at binding-site resolution during myeloid differentiation.
The deficits in our understanding of Cebpa’s regulatory logic illustrate the general challenge of decoding gene regulation of complex mammalian loci. The challenge arises from the complexity of gene regulation—genes may be regulated by multiple CRMs [13, 14] and each CRM may, in turn, be jointly regulated by several TFs exerting positive or negative influence over the target gene [15–20]. The problem of decoding regulatory logic, therefore, is one of mapping multiple inputs—TF concentrations—to a single output—the rate of transcription.
We have developed a computational approach to solve the problem of mapping multiple TF inputs to transcriptional output and decoding regulatory logic [21]. We utilize sequence-based models of transcription [16, 22–25] that simulate gene regulation by multiple TFs according to precise mechanistic rules of TF-DNA binding, competition, repression, and cooperation [17, 26–32]. The model takes estimates of TF concentrations, CRM DNA sequence, and position weight matrices as inputs and computes the resulting CRM activity as an output. Our approach does not require a priori knowledge of the identities or the regulatory roles, activation or repression, of the TFs regulating a CRM. The TFs regulating a CRM and their regulatory roles are inferred in silico by testing many alternative models, each realizing a potential regulatory scheme, against quantitative reporter data. The composition of the best fitting model then implies the regulatory roles of the TFs most congruent with the observed patterns of CRM- and cell-type-specific reporter activity. It is worth noting that this procedure not only produces a description of the TFs, their roles, and their binding sites, but also yields predictive models of CRM function.
We previously applied our approach to Cebpa in order to comprehensively decode its regulation [21] during macrophage-neutrophil differentiation. The reporter assays were carried out in PUER cells [33], which act as bipotential granulocyte-monocyte progenitors (GMPs) and can be differentiated into macrophages or neutrophils by treatment with 4-OH-tamoxifen (OHT) in the presence of IL-3 or G-CSF respectively [7, 8]. We identified 8 CRMs, of which 7 are novel, lying between -39kb and +38kb from the Cebpa transcription start site (TSS). Four CRMs, including one encompassing the +37kb enhancer identified by Guo et al. [34], acted as enhancers and upregulated reporter activity 2- to 6-fold relative to the proximal promoter of Cebpa. The remaining CRMs, appeared to behave as silencers and repressed the activity of the proximal Cebpa promoter in a dominant fashion.
Our computational analysis inferred a comprehensive map of the regulation of the Cebpa locus and suggested a novel mechanism of lineage resolution [21]. The enhancers were predicted to be activated by PU.1, C/EBP family TFs, Egr1, and Gfi1 and repressed by Myb. Surprisingly, the model predicted that the silencers exert repression through the activity of TFs strongly expressed in non-myeloid cell types, GATAs, Ebf1, and Myb. The silencing of Cebpa is necessary for the specification and maintenance of non-myeloid cell fates [9, 17, 35]. Dominant repression of the Cebpa promoter by distal silencers in non-myeloid lineages therefore might be a mechanism for resolving lineages. These inferences must however be regarded as predictions since they are yet to be verified experimentally.
Here we rigorously test the predictions of our computational models to determine the regulatory logic of Cebpa CRMs. We predicted the effect of mutations to one or more binding sites by simulating the regulation of mutated DNA in the model. The predictions were experimentally tested by synthesizing mutated CRMs and assaying their activity and by comparing against publicly available ChIP datasets. The regulatory logic of enhancers was investigated in PUER cells, representing the myeloid lineage where Cebpa is expressed robustly. The function and regulation of silencers was investigated in G1ME cells [36], which are derived from Gata1−/− mice and represent the red-blood cell lineage by virtue of being blocked at the megakaryocyte-erythrocyte progenitor (MEP) stage.
Materials and methods
Cell culture
We utilized Spi1−/− cells, expressing conditionally activable PU.1 protein, which can be differentiated into macrophages or neutrophils by PU.1 activation (PUER; [7, 8, 33]). PUER cells were routinely maintained in complete Iscove’s Modified Dulbecco’s Glutamax medium (IMDM; Gibco, 12440061) supplemented with 10% FBS, 50μM β-mercaptoethanol, 5ng/ml IL3 (Peprotech, 213-13). PUER cells were differentiated into macrophages by adding 200nM 4-hydroxy-tamoxifen (OHT; Sigma, H7904-5MG). Cells were differentiated into neutrophils by replacing IL3 with 10ng/ml Granulocyte Colony Stimulating Factor (GCSF; Peprotech, 300-23) and inducing with 100nM OHT after 48 hours. Gata1-deficient megakaryocyte-erythrocyte (G1ME) cells were routinely maintained in complete α-MEM Glutamax (Gibco, 12561056) supplemented with 20% FBS and 20ng/ml TPO (Peprotech, 315-14).
Construct design and cloning using Gibson assembly
Putative CRMs were cloned into a pGL4.10luc2 Luciferase reporter vector (Promega, E6651). The proximal promoter was introduced into the multiple cloning site (MCS) of pGL4.10luc2 between XhoI and HindIII sites. The distal CRMs were inserted between BamHI and SalI sites downstream of the SV40 late poly(A) signal. CRM sequences are provided in S1 File.
Each CRM or promoter insert was amplified from genomic DNA of C57BL/6J mice using Q5 High-Fidelity 2X Master Mix (NEB, M0492L) following the manufacturer’s instructions. The following PCR cycling conditions were used: initial denaturation of 30s at 98C, 30 cycles of 30s at 98C, 30s at 60C, and 60s at 72C, and a final extension for 10 minutes at 72C. Primers included 40bp of sequence homologous to pGL4.10luc2 (Table C in S1 Text). Gibson Assembly (GA) reactions [37] were carried out using 0.06pmol of digested vector and 0.18pmol of insert, for 60 minutes at 50C. NEB high-efficiency competent cells (NEB, E5510S) were transformed according to manufacturer’s instructions.
Transfection and Luciferase assays
PUER or G1ME cells were transfected with a reporter vector and Renilla control vector (pRL-TK, TK promoter, gift of A. Dhasarathy) in a 1:200 ratio using a 4D-Nucleofector (Lonza). PUER cells were transfected with 2.26μg total plasmid DNA in SF buffer (Lonza, V4SC-2096), using program CM134 and incubated for 24 hours prior to luminescence measurement. G1ME cells were transfected with 4.52μg total plasmid DNA in P3 buffer (Lonza, V4SP-3096), using program CM134 and incubated for 6 hours before luminescence measurement. After incubation, Firefly and Renilla luminescence were measured using the Dual-Glo Luciferase activity kit (Promega, E2920) and the DTX 880 Multimode Detector (Beckman Coulter) according to manufacturer’s instructions. Transfections were performed in at least 10 replicates. Raw luminescence data from PUER and G1ME cells are provided in S2 and S3 Datasets respectively.
Normalization of Firefly luminescence against Renilla luminescence
Well-to-well transfection efficiency variation was controlled for by normalizing Firefly luminescence against Renilla luminescence. Robust errors-in-variables (EIV) regression, implemented according to the method of Zamar [38], was used to estimate the slope, β, of the line y = βx, where y is Firefly luminescence and x is the Renilla luminescence. Briefly, β was estimated by minimizing the loss function
where, xi and yi are individual replicates of Renilla and Firefly luminescence measurements, is Tukey’s loss function with c = 4.7, and S is an estimate of the scale of the residuals. The argument of Tukey’s function is the orthogonal distance of the point (xi, yi) from the regression line. Tukey’s function is bounded for large values of t, which limits the contribution of outliers to the loss function and ensures that the slope estimate is robust to outliers. The value of S was estimated by solving the equation
where κ = 0.05 and χ(t) is Tukey’s loss function with c = 1.56. The minimization problems were solved by the sequential least-squares quadratic programming (SLSQP) algorithm of the NLOPTR package of R, with parameters xtol_rel and maxeval set to 10−7 and 1000 respectively.
95% confidence intervals were estimated by bootstrapping using the R package BOOT. 999 replicates were subsampled using the ordinary simulation and the function boot.ci was used determine confidence intervals using the basic bootstrap method.
Sequence-based thermodynamic model
We briefly describe the specific model used here to identify binding sites and make predictions. For details, see Bertolino et al. [21]. The model includes 11 TFs, C/EBPα, C/EBPδ, Egr1, Gfi1, Myb, PU.1, Jun, Myc, Ets1, Ikaros, and Fli1, that are expressed in PUER cells [8, 21] and were chosen based on differential expression between uninduced, 24 hr IL3+OHT, and 24 hr GCSF+OHT conditions. 4 TFs, which are expressed in non-myeloid cells, Ebf1, GATA(s), Elf1, E2A, were included based on the detection of their binding sites in silencer elements (see Bertolino et al. [21] for details). The regulatory roles, activation or repression, of the TFs were inferred by constructing 215 = 32, 768 alternative models realizing all possible combinations of roles. The alternative models were fit to reporter activity measurements from 46 putative CRMs from the Cebpa, Egr1, and Egr2 regions in uninduced and 24 hour IL3 and GCSF induced conditions. Hierarchical clustering was used to identify 8 models with highly consistent regulatory schemes from the 20 lowest scoring model realizations. The model utilized in this work, 81762, was representative of the low scoring models and it’s output was highly correlated with the measured reporter activity (r2 = 0.91). It is worth noting that the same model, that is the same set of TF-related parameters, was able to correctly simulate the regulation of 46 diverse CRMs in three conditions simultaneously.
Design and synthesis of mutant CRMs
Mutations to predicted TF binding sites were designed in silico with the aid of our sequence-based model of transcription [21]. A mutant binding site was created by changing each nucleotide in the wildtype site to one having the lowest frequency in the alignment matrix [39] of the cognate TF (Table A in S1 Text). The mutated CRM was then simulated in the model to predict its activity and confirm that the targeted site was lost, no new sites had been created, and the other sites were unmodified. If the mutant sequence interfered with other sites or introduced new ones, then nucleotides having the second lowest frequency in the alignment matrix were chosen at a few positions to circumvent interference. Mutant sequences were synthesized using Gibson assembly either with primers carrying the desired mutations or with synthetic dsDNA, or both (Tables B and C in S1 Text). The mutant CRMs were cloned into pGL4.10luc2 using Gibson assembly as described above. Mutant CRM sequences are provided in S1 File.
Reverse transcription real-time PCR
Total RNA was extracted using MagJet RNA kit (Thermo, K2731), and reverse transcribed using the High Capacity cDNA Transcription kit (Applied Biosystems, 4368814) following the manufacturer’s instructions. Real-Time PCR was performed using the Ssofast Evagreen Supermix (BioRad, 1725201) in a C1000 Thermal Cycler with CFX384 Real-Time System (BioRad) using the following cycling conditions: initial denaturation of 30s at 95C followed by 40 cycles of 5s at 95C and 5s at 60C. Cebpa expression relative to Hprt was computed as , where and are the threshold cycles for Cebpa and Hprt respectively. The following primers were used:
Cebpa_fwd: ACTTTCCGCGGAGCTGAG
Cebpa_rev: ATTTTTGCTCCCCCTACTCG
Hprt_fwd: ACCTCTCGAAGTGTTGGATA
Hprt_rev: CAACAACAAACTTGTCTGGA
The data are provided in S1 Dataset.
Flow cytometry
PUER cells were preincubated with Rat anti-mouse CD16/CD32 antibody (BD Biosciences Cat# 553141, RRID:AB_394656) to reduce nonspecific binding. Cells were stained with (Phycoerythrin)-CF594-conjugated anti-F4/80 (T45-2342; BD Biosciences Cat# 565613, RRID:AB_2734770) or Biotin-conjugated anti-Gr-1 (RB6-8C5; BD Biosciences Cat# 553124, RRID:AB_394640) monoclonal antibodies from BD Biosciences. Gr-1 primary incubation was followed by incubation with Allophycocyanin-conjugated Streptavidin (BD Biosciences Cat# 554067, RRID:AB_10050396). Stained cells were analyzed on a BDFACSymphony flow cytometer (BD Biosciences). Data were analyzed using FlowJo (FlowJo, LLC). Positive fraction was estimated using FlowJo’s SE Dymax method.
Immunoblotting
5 × 106 cells were lysed in RIPA buffer (Fisher Scientific) and the lysates were size-separated by 12% SDS-PAGE. Proteins were transferred to an Immobilon-FL PVDF membrane (Sigma-Aldrich). The membrane was incubated with the REVERT total protein stain (Licor) and imaged in the 700nm channel of a Licor Odyssey Fc near-infrared imaging system. Subsequently the membrane was incubated with a Rabbit anti-mouse C/EBPα (D56F10) monoclonal antibody (Cell Signaling Technology Cat# 8178, RRID:AB_11178517) and an IRDYE 800CW Donkey anti-rabbit secondary antibody (LI-COR Biosciences Cat# 925-32213, RRID:AB_2715510). The membrane was imaged in the 800nm channel to detect C/EBPα. The data were analyzed using Image Studio (LI-COR) and the total fluorescence in each band was measured and summed. Total C/EBPα fluorescence was normalized against the total fluorescence in the total protein stain.
Results
The expression of Cebpa during macrophage-neutrophil differentiation
We characterized the time course of Cebpa expression during the differentiation of PUER cells into macrophages and neutrophils. PUER cells are IL3-dependent hematopoietic progenitors derived from Spi1−/− mice and carry a transgene encoding a PU.1-Estrogen receptor fusion protein [33]. Uninduced PUER cells function like myeloid progenitors and can be induced to differentiate by treatment with 4-hydroxy-tamoxifen (OHT) into either macrophages or neutrophils in the presence of IL3 or GCSF respectively (Fig 1B, Fig A in S1 Text, and [7]). For neutrophil differentiation, IL3 medium is completely replaced with GCSF medium 48 hours prior to differentiation.
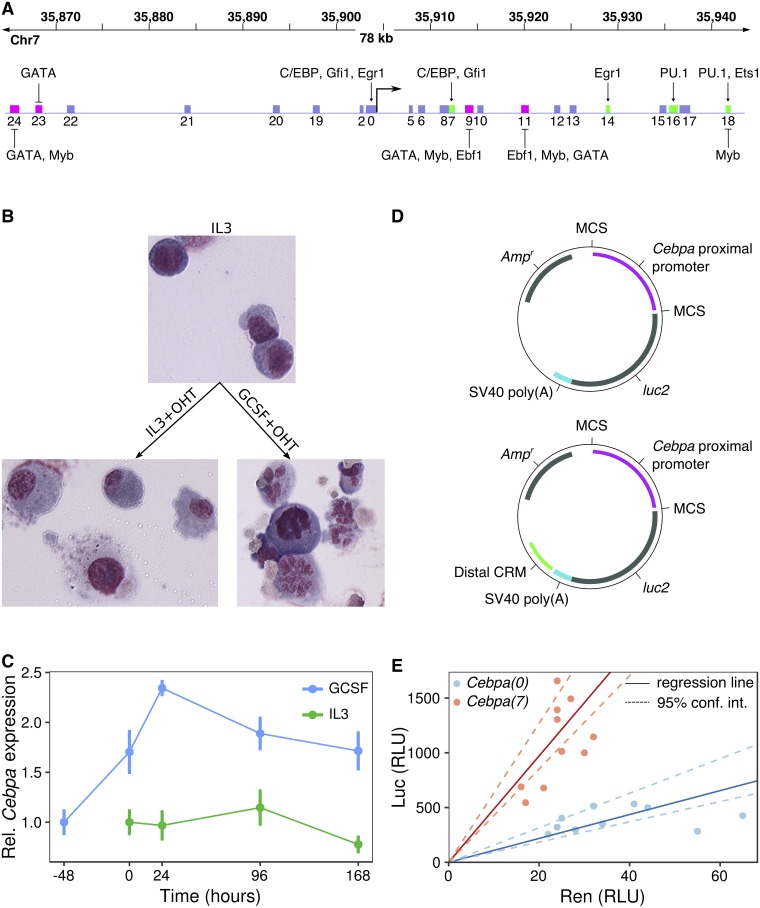
Fig 1. The regulation of Cebpa in PUER cells.
A. A 78kb region surrounding the Cebpa TSS is shown. The boxes represent putative CRMs previously identified using evolutionary conservation and analyzed using sequence-based thermodynamic modeling [21]. Green and magenta boxes represent enhancers and silencers respectively. The activators and repressors inferred by the model are indicated. B. Wright Giemsa stains of PUER cells in uninduced IL3 (top), 7-day OHT-induced IL3 (bottom left), and 7-day OHT-induced GCSF (bottom right) conditions. Uninduced cells have a blast morphology with high nucleocytoplasmic ratio. Cells induced in IL3 conditions have a vacuolated cytoplasm and low nucleocytoplasmic ratio, while induction in GCSF results in cells with segmented nuclei. C. Time series of the ratio of Cebpa and Hprt expression measured by RT-RTPCR during the differentiation of PUER cells. Relative expression has been normalized to average relative expression in uninduced PUER cells. -48 hours and 0 hour points are both measurements from uninduced cells. With the exception of 96 hours GCSF+OHT, for which N = 2, N ≥ 3. Error bars show standard error. D. Schematics of reporter vectors, based on the pGL4 backbone (Promega), which contain the Cebpa promoter immediately upstream of luc2 either with (below) or without (above) a distal CRM located downstream of the SV40 Poly(A) signal. E. Normalization of Firefly luminescence against Renilla luminescence to correct for sample-to-sample variation in transfection efficiency. Points are independent Firefly and Renilla luminescence measurements for Cebpa(0) (blue) and Cebpa(7) (red). Luminescence is reported in relative luminescence units (RLUs). The ratio of Firefly and Renilla luminescence was estimated as the slope of the best-fit line (solid) determined by robust errors-in-variable (EIV) regression. Dashed lines represent the 95% confidence interval for slope determined by bootstrapping (see Methods).
We measured Cebpa gene expression relative to Hprt or Gapdh using RT-RTPCR in uninduced PUER cells and at four time points during a 7-day course of differentiation in IL3 and GCSF conditions (Fig 1C). Overall, Cebpa is expressed two-fold higher in GCSF than in IL3 conditions (Wilcoxon rank sum test after pooling time points, N = 13, p = 2.99 × 10−6). Cebpa expression increases 70% during the 48 hour pretreatment with GCSF, and another 40% after the first 24 hours of GCSF+OHT treatment. Thereafter, the expression level declines gradually over time to revert to pre-OHT levels at day 7. In contrast, the expression level remains relatively constant after OHT treatment in IL3 conditions. C/EBPα protein displays the same expression pattern as the mRNA (Fig B in S1 Text). The increased expression of Cebpa during neutrophil differentiation is consistent with the essential role that C/EBPα plays in neutrophil development and previous analyses of PUER differentiation [7, 8]. These data also indicate that most of the regulatory modulation of Cebpa expression occurs during the first 24 hours of differentiation.
The activity pattern of Cebpa enhancers
We had previously identified four enhancers of Cebpa in a screen utilizing evolutionary conservation and reporter assays [21, Fig 1A]. Three of four enhancers were novel, while one enhancer, Cebpa(18), overlapped with a known enhancer located 37kb downstream of the Cebpa TSS [11, 34, 40]. Prior to dissecting the cis-regulatory logic of these newly identified enhancers, we validated their activity in PUER cells using a statistically robust procedure for measuring reporter activity that we have developed recently (see Methods).
In transient reporter assays, transfection efficiency can vary over an order of magnitude from sample to sample [41]. In our reporter data, we observed a 2-4 fold variation in luminescence from sample-to-sample (Fig 1E). The prevalent method of correcting for transfection efficiency variation is to co-transfect an independent reporter, such as the Renilla Luciferase expressed from a constitutive promoter, along with the CRM reporter being assayed. Firefly luminescence is then normalized to Renilla luminescence to control for sample-to-sample variation in transfection efficiency. Normalizing by taking the ratio of Firefly and Renilla luminescence is statistically unsound since it weights low- and high-luminescence replicates equally even though the latter produce more reliable estimates of normalized reporter activity.
In our method, we utilize linear regression to determine the normalized activity as the slope of the best fit line (Fig 1E), and hence avoid weighting all points equally. Ordinary least squares regression assumes that the values of the independent variable, Renilla luminescence in our case, are known exactly and don’t include random errors. Since Renilla luminescence is itself a random variable in transient assays, we use robust errors-in-variables (EIV) regression [38, 42] instead. The estimation of the slope and intercept is rendered insensitive to outliers by utilizing a bounded loss function [38]. Furthermore, the loss function is a sum of the squares of the scaled orthogonal distance of each data point from the line, and hence leads to the minimization of errors in both variables, instead of just the dependent variable. Finally, we performed reporter assays in 10 replicates in order to boost statistical power.
We tested the four previously identified enhancers [21] using this statistically robust methodology. In all reporter data presented in this manuscript, the reporter vectors either carry the Cebpa proximal promoter alone (Cebpa(0)) or in combination with one of the distal CRMs (Fig 1D). We denote the vector carrying a CRM along with the promoter as Cebpa(X), where X is the CRM number. The comparison of the CRM-bearing reporter with Cebpa(0) allows us to discriminate enhancing or silencing CRMs from neutral ones. Reporter activity was assayed in uninduced conditions and 24 hours after the addition of OHT in either IL3 or GCSF conditions, when the difference in Cebpa expression between the two treatments is the largest (Fig 1C).
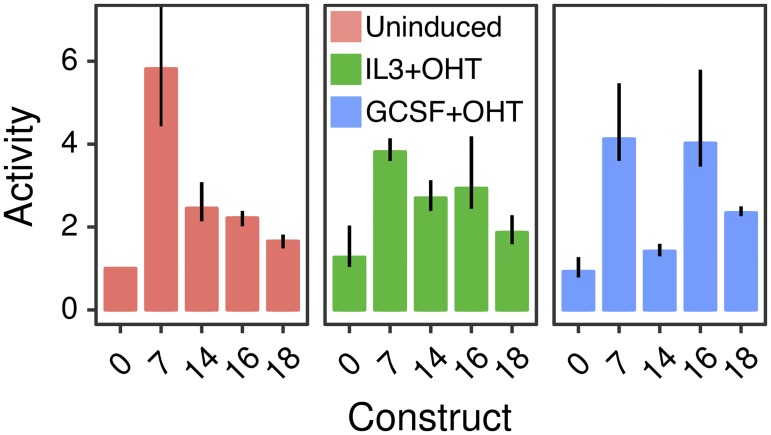
All four enhancers upregulated the activity of the promoter robustly and also exhibited cell-type specific patterns of activity (Fig 2). Cebpa(7) is the strongest enhancer in uninduced conditions, upregulating activity ∼6-fold relative to Cebpa(0). Cebpa(7)’s enhancing effect is moderated somewhat to 3-fold and 4.5-fold in induced IL3 and GCSF conditions respectively. Cebpa(14) has a qualitatively similar activity pattern as Cebpa(7), providing the greatest activation, ∼2.5-fold, in uninduced conditions. Cebpa(16) and Cebpa(18), in contrast, have the greatest activity in induced conditions. Cebpa(16) upregulates the proximal promoter 4.1-fold in induced GCSF conditions compared to ∼2-fold in uninduced conditions. Similarly, Cebpa(18) provides the greatest upregulation of ∼2.5-fold in induced GCSF conditions.
Fig 2. Relative activity of Cebpa enhancers in PUER cells.
Cebpa(0) is the construct bearing the Cebpa proximal promoter alone, while the others carry the indicated distal CRM in addition to the proximal promoter. Bar plots show the ratio of construct activity in each condition to Cebpa(0) activity in uninduced conditions. Each CRM’s activity was assayed in uninduced (red), 24 hours IL3+OHT (green), and 24 hours GCSF+OHT (blue) conditions. Reporter assays were performed in 10 replicates. Error bars are 95% confidence intervals. The error bar for Cebpa(7) extends to 15.4. Regression plots corresponding to each bar are shown in Fig C in S1 Text.
Although these activity patterns are largely consistent with our previous measurements [21], some quantitative differences were observed. For example, Cebpa(7) upregulates activity ∼6-fold instead of ∼4-fold as observed previously. These differences likely stem from two sources. First, we used the pGL4 vector backbone instead of pGL3 since the former has many fewer predicted binding sites for mammalian TFs, minimizing confounding effects from spurious TF binding. Secondly, we measured luminescence in 10 replicates and analyzed the data with robust EIV regression. Both of these modifications should result in more accurate estimates of reporter activity than before.
The cis-regulatory logic of Cebpa enhancers at binding-site resolution
Having rigorously validated the novel enhancers, we next decoded their cis-regulatory logic by mutating binding sites predicted by sequence-based models of gene regulation [21]. We provide a brief description of the model here and refer the reader to Bertolino et al. [21] for implementation details and equations. A schematic of the model is provided in Fig D in S1 Text. Given the DNA sequence of a CRM, the TFs regulating the CRM, and estimates of TF concentrations in one or more conditions, our model predicts the rate of transcription in each condition. The model utilizes position weight matrices (PWMs) to identify binding sites and to compute their binding affinity relative to the consensus site [43]. The model then determines the occupancy of each site by its TF “thermodynamically” [22, 24], that is, by enumerating all possible configurations in which the identified sites may be bound. The occupancy of a site in a given configuration takes into account potential cooperative and competitive interactions between TFs. The model implements position dependent repression, or quenching [26, 44, 45], by reducing the site occupancy of activators bound in a ∼150bp neighborhood of repressor sites. The total strength of a CRM’s interaction with the polymerase holoenzyme complex is determined by computing a weighted sum of individual activator sites’ occupancies, using activation efficiencies as weights. In the penultimate step, crucial for correctly modeling silencers, the model allows for repression over long distances by reducing the interaction strength as a function of repressor site occupancy. In the last step, transcription initiation is modeled as an enzymatic reaction, in which greater interaction strength results in higher transcription rates. In summary, the model utilizes well-known biophysical principles and phenomenological rules to predict CRM activity from DNA sequence.
Besides predicting the activity from sequence when the regulating TFs are known, this modeling framework can also be used to learn which TFs regulate a particular CRM and whether they act as activators or repressors. This is achieved by constructing an ensemble of models realizing all possible combinations of the regulatory roles of a set of candidate TFs and identifying which model realization best fits the empirical reporter activity data [21]. Whether a particular TF is predicted to act as an activator or repressor is implicit in the combination of regulatory roles represented in the best fitting model. The TFs predicted to regulate each CRM, as well as their binding sites, can be inferred by analyzing the utilization of TFs in the occupancy and interaction-strength calculations of the best fitting model.
Using this reverse engineering methodology, we had inferred a comprehensive map of Cebpa CRM regulation at binding-site resolution (Fig 1A). These inferences, implicit in the internal composition of the best-fit model for each CRM, constitute a set of hypotheses about the cis-regulatory logic of Cebpa. In order to place the decoded logic on a firm empirical footing, we sought to test these hypotheses by site-directed mutagenesis. The interpretation of site-directed mutagenesis experiments can be challenging because deletions change binding-site spacing while substitutions have the potential to introduce new binding sites. Having CRM models capable of predicting transcription rate from DNA sequence allowed us to circumvent these limitations. For each binding site to be tested, we designed substitutions to abolish binding by choosing the nucleotide least favored at each position according to the PWM of the cognate TF [39]. The mutated sequences were simulated in the model to predict their activity. The simulations allowed us to ensure that the mutations did not create any new binding sites for the TFs represented in the model. We tested the decoded logic by synthesizing the mutant CRMs (see Methods), assaying their activity in uninduced and induced PUER cells, and comparing with the theoretical prediction. In what follows, we describe the inferred cis-regulatory logic, the predicted effect of mutations, and the empirical results for each enhancer.
Cebpa(7)
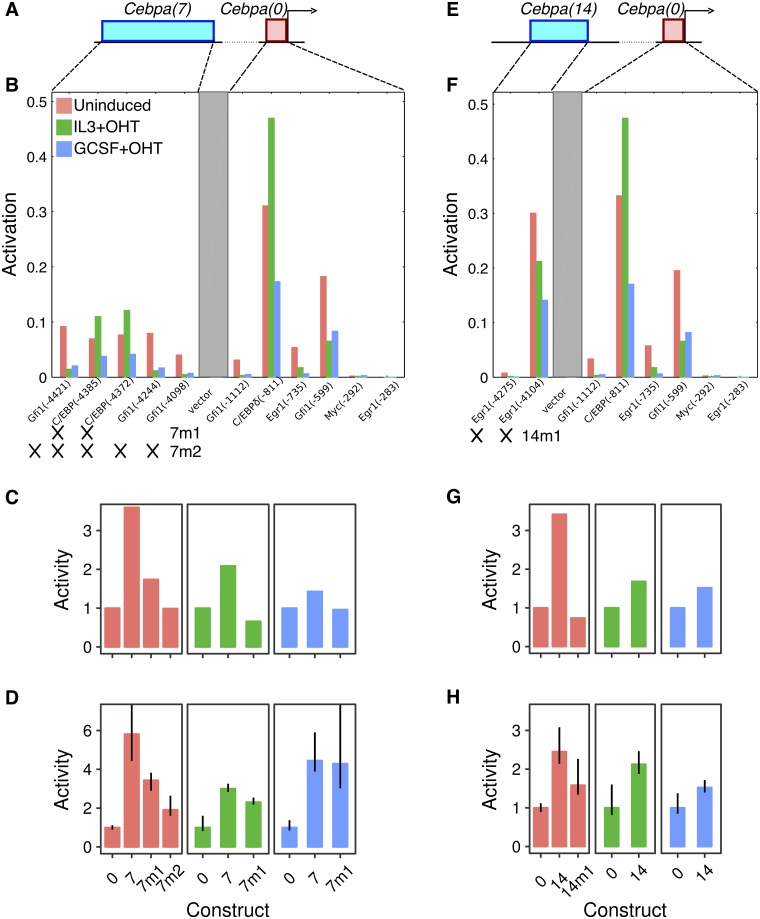
In the best-fit model, the upregulation of Cebpa(7) over Cebpa(0) results from activation provided by C/EBP family TFs and Gfi1, which bind 2 and 3 sites respectively (Fig 3A and 3B). We tested the predicted sites of the C/EBP family TFs first since C/EBP TFs are known to regulate the proximal promoter [2] and the +37kb enhancer [11]. We designed a mutant CRM, Cebpa(7m1), which lacks C/EBP sites and is predicted to have half the activity of Cebpa(7) in uninduced conditions when simulated in our model (Fig 3B and 3C). Next, we synthesized Cebpa(7m1) and assayed its activity in both uninduced and induced conditions in PUER cells. We compare fold-change relative to the proximal promoter, Cebpa(0), since the absolute scale of the reporter data used to fit the model in Bertolino et al. [21] is different owing to the use of a different vector backbone and luminometer. We observed a ∼40% reduction of activity in uninduced conditions, matching the model’s prediction and confirming the activation of Cebpa(7) by C/EBP family TFs (Fig 3C and 3D). The activity was also reduced in induced IL3 conditions, although not to the same extent as was predicted by the model. The model predicts a slight reduction of activity in induced GCSF conditions which is not observed experimentally.
Fig 3. The regulatory logic of Cebpa(7) and Cebpa(14).
A–D. Cebpa(7). E–H. Cebpa(14). A, E. Schematics of the construct design showing a distal CRM (blue) and the Cebpa proximal promoter (red). B, F. Activity of each TF activator site predicted by the sequence-based model for each construct. The activity is the amount by which an individual site reduces the activation energy barrier [21] and depends on the occupancy of the site and the efficiency of the bound activator. Sites occurring in the CRM and proximal promoter are shown. The gray box is intervening vector sequence. The x-axis shows each binding site modeled and the position of its 5’ end in the reporter construct relative to the 3’ end of the proximal promoter in parentheses. 7m1, 7m2 (panel B), and 14m1 (panel F) refer to mutant constructs tested experimentally. Crosses indicate the sites mutated in each construct. C, G. Wildtype and mutant CRM activity predicted by the model in silico. D, H. Experimentally measured activity of wildtype and synthesized mutant CRMs. Both predicted and measured activity levels have been normalized to the activity of Cebpa(0) in each condition. Reporter assays were performed in 10 replicates. Error bars are 95% confidence intervals. Regression plots corresponding to each bar are shown in Fig E in S1 Text.
The Cebpa(7m1) data also suggested that Gfi1 or other as yet unidentified sites are functional since the C/EBP sites did not account for the entirety of Cebpa(7) activity. We tested the contribution of Gfi1 sites to the residual activity of Cebpa(7m1) by designing a second mutant, Cebpa(7m2), lacking all Gfi1 and C/EBP binding sites (Fig 3B). Simulations predicted that Cebpa(7m2) completely lacked activity in the uninduced condition (Fig 3C). We observed a further ∼45% reduction in activity compared to Cebpa(7m1), so that Cebpa(7m2)’s activity was three-fold lower than that of the wildtype CRM (Fig 3D). This result confirms the cis-regulatory scheme of C/EBP and Gfi1 activation inferred by the model, although residual upregulation of Cebpa(7m2) suggests that as yet unknown TFs also contribute to the activity of Cebpa(7).
Cebpa(14)
We had inferred that Cebpa(14) is activated exclusively by Egr1, which binds the CRM at two predicted sites (Fig 3E and 3F). Consistent with regulation by a single factor, Egr1, the activity pattern of Cebpa(14) (Fig 2) matches that of Egr1 [21], having the lowest expression in induced GCSF conditions. We designed a mutant CRM, Cebpa(14m1), which lacks both Egr1 sites. Simulation of Cebpa(14m1) predicted a reversion of activity to the level of the proximal promoter (Fig 3G). Experimentally, we observed a reduction of ∼35% (Fig 3H), demonstrating the functionality of the Egr1 sites and suggesting that other TFs not represented in the model might also activate Cebpa(14).
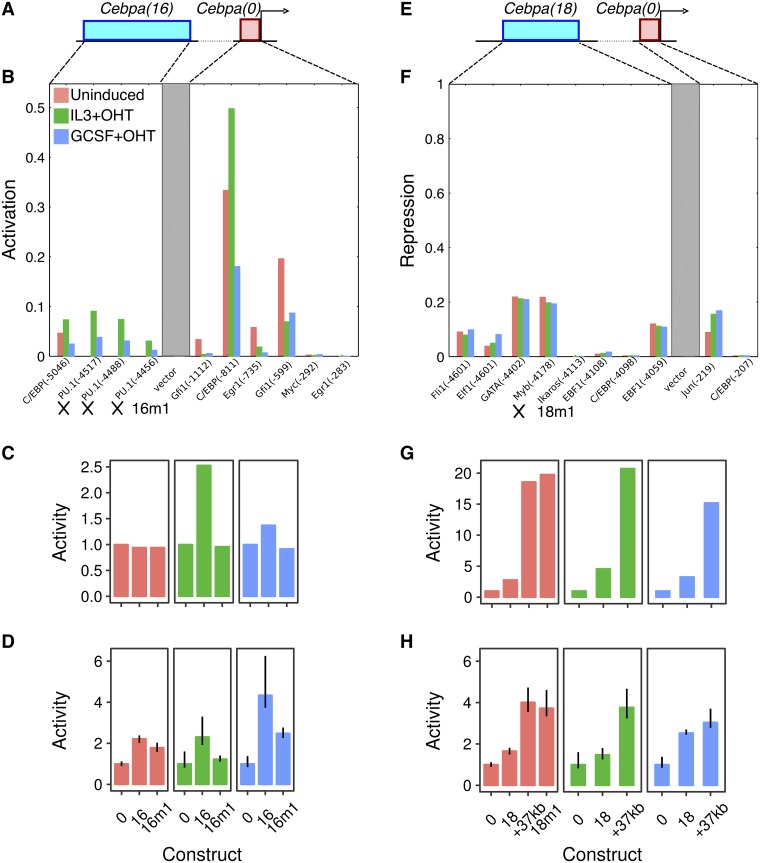
Cebpa(16)
The model for enhancer Cebpa(16) utilizes 4 activator binding sites, 3 for PU.1 and 1 for C/EBP family TFs (Fig 4A and 4B). Activation by PU.1 is consistent with the preferential upregulation of Cebpa(16) in induced conditions (Fig 2), when the PU.1-estrogen receptor fusion protein is expected to be localized to the nuclei. A mutant enhancer lacking the PU.1 sites, Cebpa(16m1), was predicted to lack enhancing activity in induced conditions, while being expressed at the same level as wildtype in uninduced conditions (Fig 4C). Experimentally, Cebpa(16m1) behaved as predicted, with an activity nearly half of Cebpa(16) and indistinguishable from that of the proximal promoter in induced IL3 conditions (Fig 4D). There was a much smaller reduction in uninduced conditions so that the activity of Cebpa(16m1) was statistically indistinguishable from that of the wildtype enhancer. The activity of Cebpa(16m1) was also ∼43% lower than that of Cebpa(16) in the induced GCSF condition, although residual upregulation relative to the proximal promoter likely implies that the C/EBP site is also functional.
Fig 4. The regulatory logic of Cebpa(16) and Cebpa(18).
A–D. Cebpa(16). E–H. Cebpa(18). A, E. Schematics of the construct design showing a distal CRM (blue) and the Cebpa proximal promoter (red). B. Activity of each TF activator site predicted by the sequence-based model for Cebpa(16). 16m1 refers to the mutant construct for testing PU.1 sites (crosses). See the legend of Fig 3B and 3F for details of the calculations, axes, and legend. F. Activity of each TF repressor site predicted by the sequence-based model for Cebpa(18). The repressive activity is the fraction by which the repressor reduces the interaction strength, which results in a higher activation energy barrier. The repressive activity depends on the occupancy of the repressor site and the efficiency of long-range repression of the bound repressor [21]. 18m1 refers to the mutant construct for testing the Myb site (cross). See the legend of Fig 3B and 3F for details of the axes and legend. C, G. Wildtype and mutant CRM activity predicted by the model in silico. D, H Experimentally measured activity of wildtype and synthesized mutant CRMs. Both predicted and measured activity levels have been normalized to the activity of Cebpa(0) in each condition. Reporter assays were performed in 10 replicates. Error bars are 95% confidence intervals. Regression plots for Cebpa(16m1) are shown in Fig E in S1 Text. Regression plots for the +37kb enhancer and Cebpa(18m1) are shown in Fig G in S1 Text.
Cebpa(18)
We had inferred that Cebpa(18) is a PU.1-responsive enhancer with additional activator binding sites for Ets1, Myc, and Gfi1 (Fig F in S1 Text). Cebpa(18) (chr7: 35,156,509–35,157,149) encompasses the +37kb Cebpa enhancer previously identified by Guo et al. [34] (chr7: 35,156,536–35,156,974). Site-directed mutagenesis experiments against Ets/PU.1 sites conducted by Cooper et al. [11] independently validated these model predictions in a different cell line, 32Dcl3 myeloid cells.
Given that Cebpa(18) is 201bp longer than the +37kb enhancer, we next investigated whether the extra sequences had any function or not. As a first step, we simulated the +37kb enhancer in our model. The model predicted that the activity of the +37kb enhancer is 7- and 4.5-fold higher than that of Cebpa(18) in the uninduced and induced IL3 conditions respectively (Fig 4G), suggesting that the extra sequence has a repressive function. We tested the activity of the +37kb enhancer in PUER cells and observed a ∼2.5-fold increase relative to Cebpa(18) in both uninduced and induced IL3 conditions (Fig 4H), confirming a repressive role for the extra sequence.
We analyzed the repressors predicted by the model to pinpoint the TFs and binding sites responsible for moderating Cebpa(18)’s activity. The model had inferred five active repressor sites in Cebpa(18), Fli1, Elf1, GATA, Myb, and Ebf1 (Fig 4E and 4F). Of these five, only two, Myb and Ebf1, are unique to Cebpa(18), lying in the extra 201bp of sequence. Of the two TFs, Myb is more likely to mediate the repressive effects since Ebf1 is not expressed in myeloid cells [46]. To test the function of Myb, we simulated a mutant CRM lacking the Myb site, Cebpa(18m1), with the model. The model predicted that Cebpa(18m1) has a much higher level of activity that is indistinguishable from that of the +37kb enhancer (Fig 4G). This is indeed how the mutant enhancer behaved in experiment. Cebpa(18m1)’s activity was derepressed relative to Cebpa(18) and indistinguishable from that of the +37kb enhancer (Fig 4H), with the caveat that the model overestimated the quantitative magnitude of Myb’s repression. Since Myb is downregulated in induced PUER cells [21], this result suggests that the upregulation of Cebpa enhancers in induced conditions is a consequence not just of a gain in activation by PU.1, but also a loss of repression by Myb.
Validation against ChIP-seq datasets
Having validated the binding sites predicted by the model, we checked in publicly available genome-wide TF binding datasets whether the predicted TFs bind to Cebpa CRMs. We compiled a set of ChIP-seq datasets for C/EBP family TFs, PU.1, Gfi1, Egr1, and Myb in myeloid cell types (Fig M in S1 Text). C/EBP peaks were detected in the proximal promoter, Cebpa(7), and Cebpa(16). PU.1 peaks were detected in Cebpa(16) and Cebpa(18). Gfi1, Myb, and Egr1 bind to Cebpa(7), Cebpa(18), and the proximal promoter respectively. We were unable to verify just one prediction, the binding of Egr1 to Cebpa(14), with the available datasets. This discrepancy could be a result of cell-type specific binding of Egr1 since the ChIP data in question (GSM881139) are from Dendritic cells [47] and not GMPs. Taken together, the TF binding data strongly support the model’s predictions.
To summarize, we have decoded the cis-regulatory logic of four Cebpa enhancers at the resolution of individual binding sites. In all cases, the model’s predictions were borne out by experiment. The identified TFs and their sites are likely the most important regulators of Cebpa during macrophage-neutrophil differentiation since they account for most of the CRM activity. The investigated TFs do not however account for all of the CRM activity, suggesting that other TFs not represented in the model also perhaps regulate Cebpa. The overall picture that emerges is that C/EBP family TFs, Gfi1, and Egr1 support Cebpa’s expression in uninduced or progenitor conditions by binding to Cebpa(7) and Cebpa(14). Activation in induced conditions is provided via Cebpa(16) and Cebpa(18) by increased PU.1 activation and a loss of Myb repression.
The role of novel silencer elements in hematopoietic lineage resolution
Our previous analysis had revealed CRMs that, when placed in the reporter vector along with the Cebpa promoter, reduced the activity of the construct to levels lower than that of the promoter alone [21]. This mode of action is consistent with the definition of silencers [44, 48]. The reduction of activity to levels lower than that of the promoter alone implies that the repressors binding to these silencers act in a dominant manner, similar to long-range repression observed in Drosophila [49, 50]. Furthermore, the CRMs in question, Cebpa(9), Cebpa(11), Cebpa(23), and Cebpa(24), lie 9–40kb away from the Cebpa TSS (Fig 1A), implying that dominant repression occurs over long distances. Our analysis had inferred that the silencers were repressed by GATA family TFs, Ebf1, and Myb (Fig 1A and [21]). Gata1/Gata2 and Ebf1 play key roles in the specification of the red-blood cell and B-cell lineages respectively [28, 46], while Myb has been implicated in megakaryocyte development [51]. These inferences are supported by evidence that Gata2 binds to Cebpa(11) and Cebpa(24) in G1ME cells (Fig M in S1 Text), which are blocked at the MEP stage and can be differentiated into erythrocytes [36, 52].
These observations motivated the hypothesis that was the subject of our subsequent experiments. We hypothesized that dominant repression mediated via distal silencers is a mechanism for resolving hematopoietic lineages. We tested this hypothesis by 1) checking whether the silencers do, in fact, exert dominant repression in a non-myeloid cell type, and 2) determining whether the silencing is attributable to the predicted repressors.
The activity pattern of Cebpa silencers
Before testing the activity of the silencers in a non-myeloid cell type, we measured their activity in PUER cells using the statistically rigorous methodology we developed for analyzing reporter data. In PUER cells, all of the silencers were either neutral or had weak enhancing activity (Fig H in S1 Text). This result implies that previous observations of reduced activity in PUER cells [21] were likely artifacts of low sample size or statistically unsound normalization. Furthermore, this result implies that these CRMs do not silence Cebpa in a myeloid background.
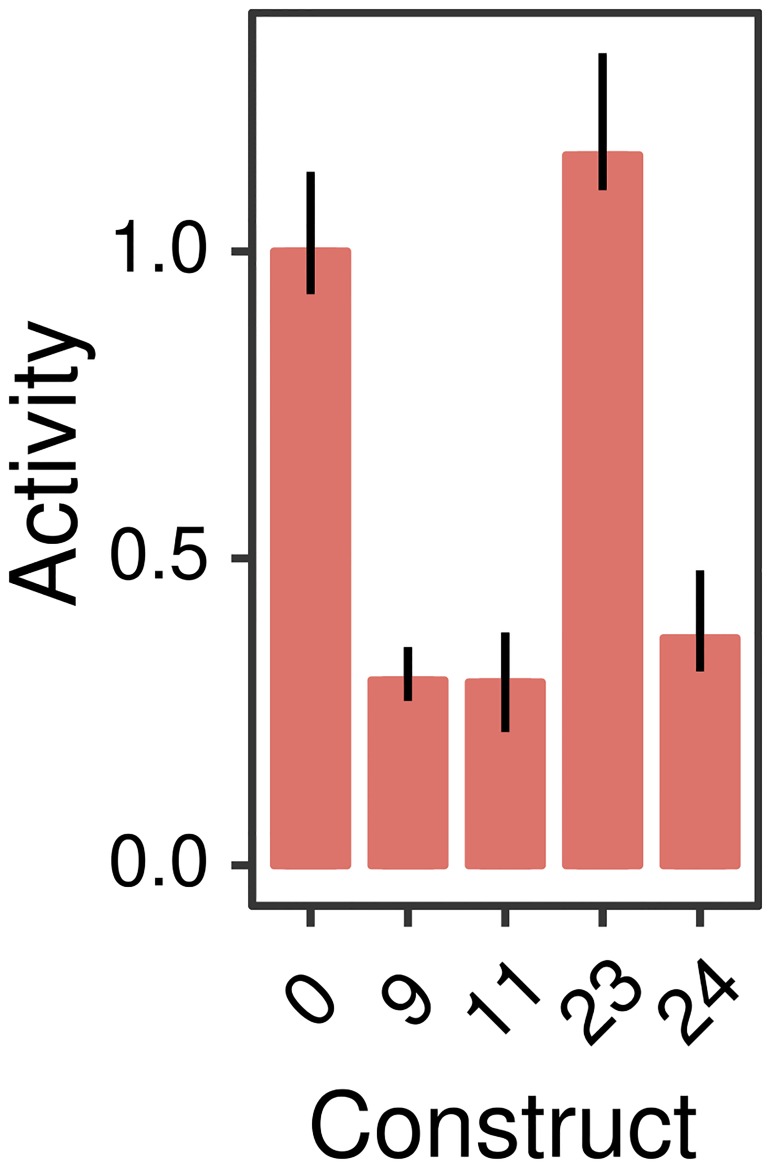
Next, we tested the function of silencers in G1ME cells, representative of the red-blood cell lineage. We chose the red-blood cell lineage since Gata2 is known to bind the silencers Cebpa(11) and Cebpa(24) in G1ME cells (Fig M in S1 Text). Even though we could not detect Cebpa expression in G1ME cells (Fig K in S1 Text), the Cebpa promoter had detectable activity (Fig 5). This suggested that additional repression is required to completely silence Cebpa. Reporter vectors carrying Cebpa(9), Cebpa(11), Cebpa(24) in addition to the promoter had ∼3-fold lower activity compared to the promoter alone (Fig 5). The silencers, therefore, while being inert in myeloid cells, repress the Cebpa proximal promoter in the red-blood cell lineage.
Fig 5. Relative activity of Cebpa silencers in G1ME cells.
Cebpa(0) is the construct bearing the Cebpa proximal promoter alone, while the others carry the indicated distal CRM in addition to the proximal promoter. Bar plots show the ratio of each construct’s activity to Cebpa(0). Reporter assays were performed in 10 replicates. Error bars are 95% confidence intervals. Regression plots corresponding to each bar are shown in Fig J in S1 Text.
GATA and Myb repress the Cebpa proximal promoter in a dominant and redundant fashion
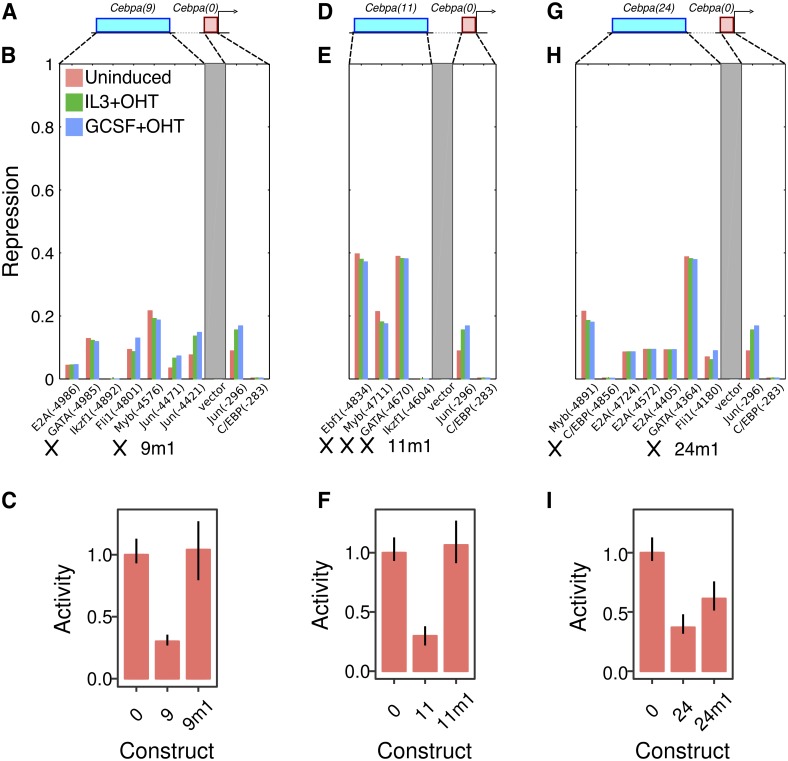
We decoded the cis-regulatory logic of the silencers using the same model-guided strategy as was employed for the enhancers. In contrast to the enhancers, each of which had a distinctive regulatory scheme, the validated silencers shared a common regulatory motif. All three silencers had GATA and Myb sites, which were predicted to be among the most active in each silencer (Fig 6B, 6E and 6H). GATA family TFs and Myb are plausible repressors of Cebpa. Knocking down Gata2 leads to the derepression of Cebpa in G1ME cells [53], while we have demonstrated that Myb represses Cebpa(18) (Fig 4G and 4H). We synthesized mutants CRMs—Cebpa(9m1), Cebpa(11m1), and Cebpa(24m1)—lacking binding sites for both TFs (Fig 6B, 6E and 6H). Cebpa(11m1) carried additional mutations in an Ebf1 site but was functionally equivalent to a GATA/Myb mutant since Ebf1 is not expressed in the red-blood cell lineage (http://biogps.org/gene/13591; [52, 54]). As predicted by the model, the mutant CRMs were derepressed relative to wildtype and, in the case of Cebpa(9m1) and Cebpa(11m1), their activity was indistinguishable from that of Cebpa(0) (Fig 6C, 6F and 6I). This implies that the silencing can be attributed specifically to GATA and Myb, which account for the entirety of the effect in two of three silencers.
Fig 6. Silencing relies on a GATA/Myb motif shared by functional Cebpa silencers.
A–C. Cebpa(9). D–F. Cebpa(11). G–I. Cebpa(24). A, D, G. Schematics of the construct design showing a distal CRM (blue) and the Cebpa proximal promoter (red). B, E, H. The activity of the TF repressor sites predicted by the model for each silencer. See the legend of Fig 4F for details of the calculations, axes, and legend. 9m1, 11m1, and 24m1 refer to mutant CRMs and crosses indicate mutated sites. C, F, I. Experimentally measured activity of wildtype and synthesized mutant silencers in G1ME cells. Activity levels have been normalized to the activity of Cebpa(0). Reporter assays were performed in 10 replicates. Error bars are 95% confidence intervals. Regression plots corresponding to each bar are shown in Fig L in S1 Text.
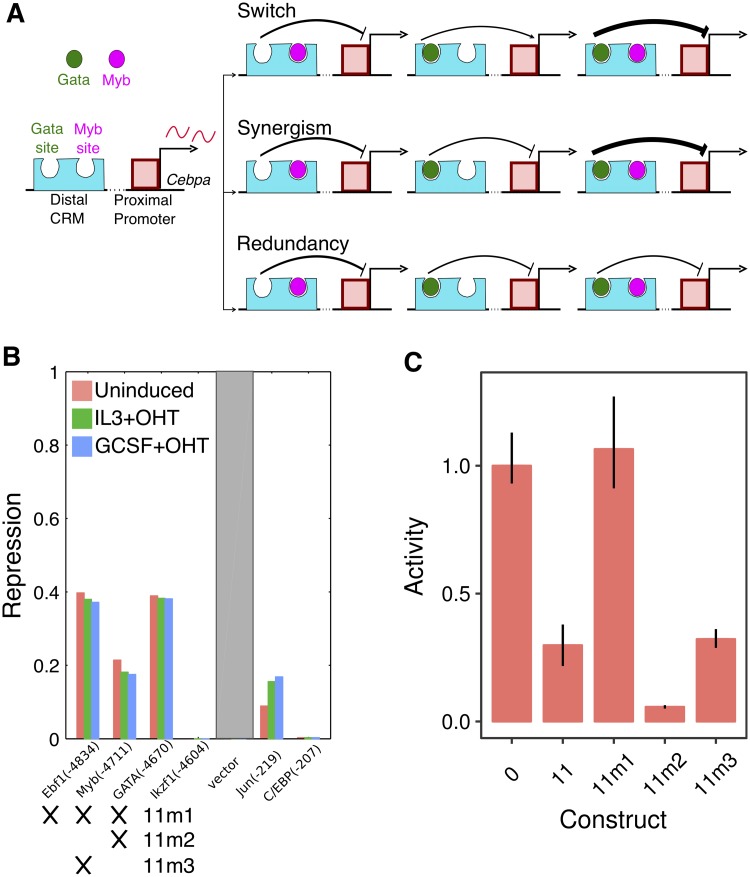
Next, we investigated how GATA and Myb jointly repress the activity of the Cebpa proximal promoter. We considered three hypotheses and tested them by mutating GATA and Myb sites individually in Cebpa(11) (Fig 7A). First, it is possible that GATA and Myb repress the proximal promoter redundantly [55], so that only one functional site is sufficient to achieve silencing. The second possibility is that of synergism [17, 56, 57], in which GATA and Myb would have much greater silencing activity together than individually. The third possibility is that of context-dependent role switching, when a TF switches its role when bound near a second TF. For example, in the Drosophila blastoderm, the repressor Hunchback activates gene expression when bound near Bicoid [58, 59]. We tested two new constructs, Cebpa(11m2) and Cebpa(11m3), which carry impaired GATA or Myb sites respectively. Both of the constructs carrying only one functional repressor site were able to silence the proximal promoter (Fig 7C). The expression of Cebpa(11m2) is lower than that of Cebpa(11), which could be interpreted to imply an activating role for GATA. However, if GATA were an activator, one would expect a loss of repression in Cebpa(11m3), which has a functional GATA site. Since this is not the case, we favor the explanation that perhaps, even though Myb is a more potent repressor, GATA competes with Myb and limits the overall repression to that achieved by GATA alone. This would help explain why the activities of Cebpa(11m3) Cebpa(11) are indistinguishable. The maintenance of repression in the single mutants supports the hypothesis that GATA and Myb are capable of repressing the promoter individually and act redundantly.
Fig 7. GATA and Myb repress the Cebpa proximal promoter redundantly.
A. Three potential hypotheses for the combined silencing of the promoter by GATA and Myb. B. The activity of the TF repressor sites predicted by the model for each silencer. See the legend of Fig 4F for details of the calculations, axes, and legend. 11m1, 11m2, and 11m3 refer to tested mutant CRMs and crosses indicate mutated sites. C. Experimentally measured activity of wildtype and synthesized mutant silencers in G1ME cells. Activity levels have been normalized to the activity of Cebpa(0). Reporter assays were performed in 10 replicates. Error bars are 95% confidence intervals. Regression plots corresponding to each bar are shown in Fig L in S1 Text.
In summary, we have shown that three of four putative silencers are capable of attenuating the activity of the Cebpa proximal promoter in a dominant manner. Dominant repression only occurs in the red-blood cell lineage and the CRMs do not silence the proximal promoter in myeloid cells. Lastly, the silencing activity is attributable to a regulatory motif shared by all three silencers—GATA and Myb sites that act redundantly.
Discussion
We have comprehensively analyzed the regulation of 7 CRMs neighboring Cebpa at the resolution of individual binding sites. In the process of doing so, we have also verified the predictive ability of a thermodynamic model of mammalian gene regulation that we developed recently [21]. It is worth noting that prior to our efforts, thermodynamic modeling was limited to Drosophila gene regulation [16, 22, 24, 60–64], with a single gene, even-skipped, as the focus of most of the work. Our model is closely related to its Drosophila counterparts, incorporating just one additional mechanism, long-distance dominant repression, lacking in the latter. The ability of models with shared mechanisms of gene regulation to predict reporter activity in these divergent species supports the view that the rules of transcriptional regulation are universal.
Long-distance dominant repression by non-myeloid TFs, GATA [65], Myb [51], and Ebf1 [46], was required to correctly model silencers [21]. This led us to hypothesize that long-distance repression by silencer-bound TFs is necessary for quenching Cebpa expression in non-myeloid lineages. The hypothesis predicts that the silencers of Cebpa would have much lower activity in the myeloid lineage, where the gene is expressed, than in non-myeloid ones, where it is not expressed. When we assayed the activity of the silencers in PUER cells, which belong to the myeloid lineage, we found they they acted as enhancers or neutral elements (Fig H in S1 Text), confirming part of the prediction. In contrast, 3 of 4 putative silencers downregulated promoter activity ∼3-fold in G1ME cells (Fig 5) belonging to the megakaryocyte-erythrocyte lineage, confirming the rest of the prediction. Furthermore, silencing by distal elements appears to be necessary for quenching Cebpa expression in G1ME cells since the promoter has detectable activity (Fig 5), even though Cebpa expression is not detectable in G1ME cells (Fig K in S1 Text). The necessity of silencing for completely quenching Cebpa expression in the red-blood cell lineage suggests that long-distance repression is a novel mechanism for resolving hematopoietic lineages.
Detailed analysis of the regulatory logic of silencers revealed two layers of redundancy in their function. First, structural similarity underlies the functional equivalence of all three silencers. All silencers contain the same regulatory motif, a pair of GATA and Myb sites, that mediates dominant repression of the Cebpa promoter (Fig 6). Second, the regulatory motif itself is structured redundantly since mutating either GATA or Myb alone is not sufficient for relieving dominant silencing (Fig 7). We did not find evidence for synergy, where the combined effect of the two sites is greater than the sum of individual effects, implying that GATA and Myb function redundantly. The presence of multiple functionally equivalent and structurally homologous silencers in the locus suggests that the regulatory architecture of distal silencing is similar to that of distal activation by multiple redundant enhancers [66, 67]. Redundant enhancers have been shown to ensure robust and precise gene expression [50, 68, 69], leading us to speculate that redundant silencing might serve a similar function.
The activation of Cebpa CRMs in myeloid cells also occurs in a redundant and overlapping regulatory arrangement reminiscent of shadow enhancers in Drosophila [66]. All enhancers are simultaneously co-active in nearly all conditions tested in PUER cells. The sole exception is CRM18, for which we could not detect statistically significant upregulation in induced IL3 (macrophage) conditions (Fig 2). The coactive enhancers of Cebpa share common regulators. Cebpa(16) (Fig 4B) and Cebpa(18) (Fig F in S1 Text) are activated by PU.1 and other ETS factors, while CRM7 and CRM16 are activated by C/EBPα. CRM14 is the exception with predicted and verified binding sites for Egr1 (Fig 3F) unique to itself.
The overall picture that emerges from our analysis of Cebpa enhancers and silencers is that of a distributed and specialized control scheme (Fig 1). CRMs distributed over an ∼80kb region specialize in either activation or repression. Specialization is a departure from cis-regulatory organization of Drosophila segmentation genes, whose enhancers are capable of both activation and repression [31, 49, 50, 70]. Although this arrangement could be evolutionary happenstance, it is also possible that it serves a functional purpose. Despite the antagonism between Gata1/Gata2 and Cebpa that we ([21] and Fig 7) and others [52] have demonstrated, Gata2 and Cebpa are known to be co-expressed in eosinophils, basophils, and mast cells [71]. The regulatory logic of Cebpa, therefore, must allow for expression even in the presence of Gata2 protein. We propose that separable activation and silencing allows Cebpa to be expressed at intermediate levels in eosinophils, basophils, and mast cells. Under this hypothesis, the enhancers are active in all GMP-derived cells, while Gata2-dependent silencers are active in MEPs and the subset of myeloid cells where Gata2 is expressed. The quenching of Cebpa expression in the red-blood cell lineage is the combined result of the induction of silencing by Gata2/Myb and a lack of activation. In Gata2-expressing myeloid cells, both enhancers and silencers are simultaneously active, resulting in an intermediate level of expression of Cebpa. The hypothesis makes the readily testable prediction that both the enhancers and silencers of Cebpa should be active in eosinophils, basophils, and mast cells.
Supporting information
Supporting figures and tables.
(PDF)
CRM sequences are provided in the FASTA format.
(TXT)
Threshold cycle values of Cebpa (CebpaCq), Gapdh (GAPDHCq), and Hprt (HprtCq) are provided. Column 1: time in hours. Column 2: replicate number. Column 3: cytokine treatment. Column 4: whether cells were treated with OHT or not (true/false).
(CSV)
Luc/Ren are luminescence measurements from each well. Construct indicates the CRM/promoter being assayed, where “0” is promoter by itself. OHT is true or false for induced or uninduced conditions respectively. The “note” column indicates whether the construct is wildtype and, if not, which sites were mutated.
(CSV)
Luc/Ren are luminescence measurements from each well. Construct indicates the CRM/promoter being assayed, where “0” is promoter by itself. OHT is true or false for induced or uninduced conditions respectively. The “note” column indicates whether the construct is wildtype and, if not, which sites were mutated.
(CSV)
Acknowledgments
PUER cells were a gift of R. Dahl. G1ME cells were a gift of M. Weiss. We thank R. Dahl, D. Darland, T. Rhen, S. Nechaev, and A. Dhasarathy for discussion and technical assistance and T. Rhen for comments on the manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the National Science Foundation (https://www.nsf.gov) under Grant Nos. 1615916 and IIA- 1355466, project UND0019821 (North Dakota EPSCoR) to M. The funding agency had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A. 1997;94(2):569–74. 10.1073/pnas.94.2.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Legraverend C, Antonson P, Flodby P, Xanthopoulos KG. High level activity of the mouse CCAAT/enhancer binding protein (C/EBP alpha) gene promoter involves autoregulation and several ubiquitous transcription factors. Nucleic Acids Res. 1993;21(8):1735–42. 10.1093/nar/21.8.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 2002;16(1):22–6. 10.1101/gad.948702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016;44(D1):D313–6. 10.1093/nar/gkv1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, et al. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5 10.1186/1745-7580-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–7. 10.1126/science.8079170 [DOI] [PubMed] [Google Scholar]
- 7. Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4(10):1029–36 [DOI] [PubMed] [Google Scholar]
- 8. Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126(4):755–66. 10.1016/j.cell.2006.06.052 [DOI] [PubMed] [Google Scholar]
- 9. Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–76. 10.1016/S0092-8674(04)00419-2 [DOI] [PubMed] [Google Scholar]
- 10. Gonzalez D, Luyten A, Bartholdy B, Zhou Q, Kardosova M, Ebralidze A, et al. ZNF143 protein is an important regulator of the myeloid transcription factor C/EBP±. J Biol Chem. 2017;292(46):18924–18936. 10.1074/jbc.M117.811109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cooper S, Guo H, Friedman AD. The +37 kb Cebpa Enhancer Is Critical for Cebpa Myeloid Gene Expression and Contains Functional Sites that Bind SCL, GATA2, C/EBP±, PU.1, and Additional Ets Factors. PLoS One. 2015;10(5):e0126385 10.1371/journal.pone.0126385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo H, Cooper S, Friedman AD. In Vivo Deletion of the Cebpa +37 kb Enhancer Markedly Reduces Cebpa mRNA in Myeloid Progenitors but Not in Non-Hematopoietic Tissues to Impair Granulopoiesis. PLoS One. 2016;11(3):e0150809 10.1371/journal.pone.0150809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilson NK, Calero-Nieto FJ, Ferreira R, Göttgens B. Transcriptional regulation of haematopoietic transcription factors. Stem Cell Res Ther. 2011;2(1):6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613–26 [DOI] [PubMed] [Google Scholar]
- 15. Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. The EMBO Journal. 1992;11:4047–4057. 10.1002/j.1460-2075.1992.tb05498.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Janssens H, Hou S, Jaeger J, Kim A, Myasnikova E, Sharp D, et al. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even-skipped gene. Nature Genetics. 2006;38:1159–1165 [DOI] [PubMed] [Google Scholar]
- 17. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. ENCODE Project Consortium, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489(7414):91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330(6012):1775–87. 10.1126/science.1196914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bertolino E, Reinitz J, Manu. The analysis of novel distal Cebpa enhancers and silencers using a transcriptional model reveals the complex regulatory logic of hematopoietic lineage specification. Dev Biol. 2016;413(1):128–44. 10.1016/j.ydbio.2016.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540 [DOI] [PubMed] [Google Scholar]
- 23. White MA, Parker DS, Barolo S, Cohen BA. A model of spatially restricted transcription in opposing gradients of activators and repressors. Mol Syst Biol. 2012;8:614 10.1038/msb.2012.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim AR, Martinez C, Ionides J, Ramos AF, Ludwig MZ, Ogawa N, et al. Rearrangements of 2.5 kilobases of noncoding DNA from the Drosophila even-skipped locus define predictive rules of genomic cis-regulatory logic. PLoS Genet. 2013;9(2):e1003243 10.1371/journal.pgen.1003243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sayal R, Dresch JM, Pushel I, Taylor BR, Arnosti DN. Quantitative perturbation-based analysis of gene expression predicts enhancer activity in early Drosophila embryo. Elife. 2016;5 10.7554/eLife.08445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arnosti D, Gray S, Barolo S, Zhou J, Levine M. The gap protein Knirps mediates both quenching and direct repression in the Drosophila embryo. The EMBO Journal. 1996;15:3659–3666. 10.1002/j.1460-2075.1996.tb00735.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cantor AB, Orkin SH. Hematopoietic development: a balancing act. Curr Opin Genet Dev. 2001;11(5):513–9. 10.1016/S0959-437X(00)00226-4 [DOI] [PubMed] [Google Scholar]
- 28. Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21(21):3368–76 [DOI] [PubMed] [Google Scholar]
- 29. He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26(1):37–42. 10.1101/gad.173930.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kulkarni MM, Arnosti DN. cis-Regulatory logic of short-range transcriptional repression in Drosophila melanogaster. Molecular and Cellular Biology. 2005;25:3411–3420. 10.1128/MCB.25.9.3411-3420.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Small S, Arnosti DN, Levine M. Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development. 1993;119:767–772. [PubMed] [Google Scholar]
- 32. Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Developmental Biology. 1996;175:314–324. 10.1006/dbio.1996.0117 [DOI] [PubMed] [Google Scholar]
- 33. Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF, et al. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17(5):665–76. 10.1016/S1074-7613(02)00452-1 [DOI] [PubMed] [Google Scholar]
- 34. Guo H, Ma O, Speck NA, Friedman AD. Runx1 deletion or dominant inhibition reduces Cebpa transcription via conserved promoter and distal enhancer sites to favor monopoiesis over granulopoiesis. Blood. 2012;119(19):4408–18. 10.1182/blood-2011-12-397091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chou ST, Khandros E, Bailey LC, Nichols KE, Vakoc CR, Yao Y, et al. Graded repression of PU.1/Sfpi1 gene transcription by GATA factors regulates hematopoietic cell fate. Blood. 2009;114(5):983–94. 10.1182/blood-2009-03-207944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stachura DL, Chou ST, Weiss MJ. Early block to erythromegakaryocytic development conferred by loss of transcription factor GATA-1. Blood. 2006;107(1):87–97. 10.1182/blood-2005-07-2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–5 [DOI] [PubMed] [Google Scholar]
- 38. Zamar RH. Robust Estimation in the Errors-in-Variables Model. Biometrika. 1989;76(1):149–160. 10.1093/biomet/76.1.149 [DOI] [Google Scholar]
- 39. Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. 10.1093/bioinformatics/15.7.563 [DOI] [PubMed] [Google Scholar]
- 40. Guo H, Ma O, Friedman AD. The Cebpa +37-kb enhancer directs transgene expression to myeloid progenitors and to long-term hematopoietic stem cells. J Leukoc Biol. 2014;96(3):419–26. 10.1189/jlb.2AB0314-145R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jacobs JL, Dinman JD. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32(20):e160 10.1093/nar/gnh157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Casella G, Berger RL. Statistical Inference. 2nd ed Duxbury Press; 2001. [Google Scholar]
- 43. Berg OG, von Hippel PH. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J Mol Biol. 1987;193(4):723–50. 10.1016/0022-2836(87)90354-8 [DOI] [PubMed] [Google Scholar]
- 44. Ogbourne S, Antalis TM. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J. 1998;331 (Pt 1):1–14. 10.1042/bj3310001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24(21):3712–23. 10.1038/sj.emboj.7600834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pongubala JMR, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9(2):203–15. 10.1038/ni1555 [DOI] [PubMed] [Google Scholar]
- 47. Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47(5):810–22. 10.1016/j.molcel.2012.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Davidson EH. The Regulatory Genome: Gene Regulatory Networks In Development And Evolution. Academic Press; 2006. [Google Scholar]
- 49. Dunipace L, Ozdemir A, Stathopoulos A. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development. 2011. 10.1242/dev.069146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci U S A. 2011;108(33):13570–5. 10.1073/pnas.1109873108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14(6):843–53. 10.1016/j.devcel.2008.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Doré LC, Chlon TM, Brown CD, White KP, Crispino JD. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012;119(16):3724–33. 10.1182/blood-2011-09-380634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang Z, Dore LC, Li Z, Orkin SH, Feng G, Lin S, et al. GATA-2 reinforces megakaryocyte development in the absence of GATA-1. Mol Cell Biol. 2009;29(18):5168–80. 10.1128/MCB.00482-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hagman J, Belanger C, Travis A, Turck CW, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7(5):760–73. 10.1101/gad.7.5.760 [DOI] [PubMed] [Google Scholar]
- 55. Delneri D, Gardner DC, Oliver SG. Analysis of the seven-member AAD gene set demonstrates that genetic redundancy in yeast may be more apparent than real. Genetics. 1999;153(4):1591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. [DOI] [PubMed] [Google Scholar]
- 57. Akasaka T, van Lohuizen M, van der Lugt N, Mizutani-Koseki Y, Kanno M, Taniguchi M, et al. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development. 2001;128(9):1587–97. [DOI] [PubMed] [Google Scholar]
- 58. Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes and Development. 1991;5:827–839. 10.1101/gad.5.5.827 [DOI] [PubMed] [Google Scholar]
- 59. Simpson-Brose M, Treisman J, Desplan C. Synergy between the Hunchback and Bicoid morphogens is required for anterior patterning in Drosophila. Cell. 1994;78:855–865. 10.1016/S0092-8674(94)90622-X [DOI] [PubMed] [Google Scholar]
- 60. Reinitz J, Hou S, Sharp DH. Transcriptional control in Drosophila. ComPlexUs. 2003;1:54–64. 10.1159/000070462 [DOI] [Google Scholar]
- 61. Zinzen RP, Senger K, Levine M, Papatsenko D. Computational models for neurogenic gene expression in the Drosophila embryo. Current Biology. 2006;16:1358–1365. 10.1016/j.cub.2006.05.044 [DOI] [PubMed] [Google Scholar]
- 62. Kazemian M, Blatti C, Richards A, McCutchan M, Wakabayashi-Ito N, Hammonds AS, et al. Quantitative analysis of the Drosophila segmentation regulatory network using pattern generating potentials. PLoS Biol. 2010;8(8). 10.1371/journal.pbio.1000456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Samee MAH, Sinha S. Quantitative modeling of a gene’s expression from its intergenic sequence. PLoS Comput Biol. 2014;10(3):e1003467 10.1371/journal.pcbi.1003467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barr KA, Reinitz J. A sequence level model of an intact locus predicts the location and function of nonadditive enhancers. PLoS One. 2017;12(7):e0180861 10.1371/journal.pone.0180861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Doré LC, Crispino JD. Transcription factor networks in erythroid cell and megakaryocyte development. Blood. 2011;118(2):231–9. 10.1182/blood-2011-04-285981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321(5894):1314 10.1126/science.1160631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466(7305):490–3. 10.1038/nature09158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20(17):1562–7. 10.1016/j.cub.2010.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Perry MW, Bothma JP, Luu RD, Levine M. Precision of hunchback expression in the Drosophila embryo. Curr Biol. 2012;22(23):2247–52. 10.1016/j.cub.2012.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gray S, Szymanski P, Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes and Development. 1994;8:1829–1838. 10.1101/gad.8.15.1829 [DOI] [PubMed] [Google Scholar]
- 71. Iwasaki H, Mizuno Si, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, et al. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20(21):3010–21. 10.1101/gad.1493506 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting figures and tables.
(PDF)
CRM sequences are provided in the FASTA format.
(TXT)
Threshold cycle values of Cebpa (CebpaCq), Gapdh (GAPDHCq), and Hprt (HprtCq) are provided. Column 1: time in hours. Column 2: replicate number. Column 3: cytokine treatment. Column 4: whether cells were treated with OHT or not (true/false).
(CSV)
Luc/Ren are luminescence measurements from each well. Construct indicates the CRM/promoter being assayed, where “0” is promoter by itself. OHT is true or false for induced or uninduced conditions respectively. The “note” column indicates whether the construct is wildtype and, if not, which sites were mutated.
(CSV)
Luc/Ren are luminescence measurements from each well. Construct indicates the CRM/promoter being assayed, where “0” is promoter by itself. OHT is true or false for induced or uninduced conditions respectively. The “note” column indicates whether the construct is wildtype and, if not, which sites were mutated.
(CSV)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.