Type III secretion systems (T3SS) are widely distributed in Gram-negative microorganisms and critical for host-pathogen and host-symbiont interactions with plants and animals. Central features of the T3SS are a highly conserved set of secretion and translocation genes and contact dependence wherein host-pathogen interactions trigger effector protein delivery and serve as an inducing signal for T3SS gene expression.
Keywords: Pseudomonas aeruginosa, type III secretion, RsmA, ExsA, DeaD, Vfr
ABSTRACT
Type III secretion systems (T3SS) are widely distributed in Gram-negative microorganisms and critical for host-pathogen and host-symbiont interactions with plants and animals. Central features of the T3SS are a highly conserved set of secretion and translocation genes and contact dependence wherein host-pathogen interactions trigger effector protein delivery and serve as an inducing signal for T3SS gene expression. In addition to these conserved features, there are pathogen-specific properties that include a unique repertoire of effector genes and mechanisms to control T3SS gene expression. The Pseudomonas aeruginosa T3SS serves as a model system to understand transcriptional and posttranscriptional mechanisms involved in the control of T3SS gene expression. The central regulatory feature is a partner-switching system that controls the DNA-binding activity of ExsA, the primary regulator of T3SS gene expression. Superimposed upon the partner-switching mechanism are cyclic AMP and cyclic di-GMP signaling systems, two-component systems, global regulators, and RNA-binding proteins that have positive and negative effects on ExsA transcription and/or synthesis. In the present review, we discuss advances in our understanding of how these regulatory systems orchestrate the activation of T3SS gene expression in the context of acute infections and repression of the T3SS as P. aeruginosa adapts to and colonizes the cystic fibrosis airways.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic Gram-negative pathogen capable of causing a variety of infections in humans. Known risk factors include burn wounds, corneal scratches, catheter and ventilator usage, and cystic fibrosis (1). The virulence properties of P. aeruginosa are multifactorial and comprise adherence factors, biofilm formation, antibiotic resistance, and secreted toxins (1). One critical virulence determinant is the type III secretion system (T3SS). The T3SS is embedded in the inner membrane and used to assemble an injectisome. The injectisome is an ∼25-protein complex that spans the cell envelope and functions as a molecular syringe to translocate effector proteins into eukaryotic target cells (2). The classic effectors are ExoS, ExoT, ExoU, and ExoY (2). ExoS-secreting strains cause delayed apoptotic-like cell death, while ExoU-secreting strains cause rapid and robust host cell lysis (3, 4). Additional effectors are now appreciated and include the flagellar filament protein (FliC) (5–8), nuclear diphosphate kinase (9), and PemA/PemB (10). The translocation pore itself is sufficient to induce K+ efflux, dephosphorylate and deacetylate histone H3, and cause host cell death (11–16). The combined activities of the translocated effectors protect P. aeruginosa from phagocytic and inflammatory responses, are cytotoxic, and promote systemic dissemination. Strains defective for T3SS gene expression/function are severely attenuated for virulence in burn wound, pneumonia, neutropenic, and corneal infection models (13, 17–22).
Expression of the T3SS is tightly controlled and induced in response to a number of environmental signals, including low concentrations (micromolar) of extracellular Ca2+, serum albumin/casein, and host cell contact (23–25). Primary control of T3SS gene expression is through direct activation of transcription by ExsA, an AraC family transcription factor. Positioned upstream of ExsA is a complex regulatory network involving cyclic AMP and cyclic di-GMP signaling systems, two-component systems, global regulators, and RNA-binding proteins. In this review, we highlight the emerging theme that many of the upstream regulatory events, either directly or indirectly, control exsA transcription and/or translation.
ExsA AND CONTROL OF ExsA DNA-BINDING ACTIVITY
ExsA is the master regulator of T3SS gene expression.
The P. aeruginosa T3SS regulon consists of ∼40 genes encoding regulatory functions, the secretion and translocation machinery, effectors, and effector-specific chaperones (23). Most of the genes are organized into five operons and clustered in the genome at a common location. The effector genes and their associated chaperones are scattered throughout the chromosome. All of the known T3SS genes are activated by the master regulator ExsA, a member of the AraC/XylS family of transcription factors (17, 26–30). The ExsA consensus binding site is AaAAAnwnMygrCynnnmYTGayAk, centered ∼45 bp upstream of the transcription start site for each of the 10 ExsA-dependent promoters (28, 31). For a more thorough review of ExsA DNA-binding properties and the mechanism of transcription activation, see the paper by Diaz et al. (32).
Control of ExsA activity by a partner-switching mechanism.
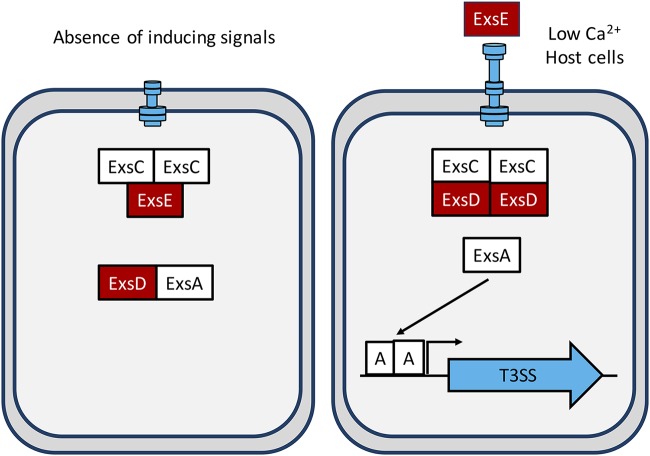
In the absence of inducing signals (low Ca2+, serum, and host cell contact), the injectisome is expressed at a low basal level and exists in a quiescent state (33, 34). Both of those features are critical because the injectisome is the sensor of inducing signals and responds by converting to a secretion-competent state through a poorly defined mechanism (33). Secretion competency indirectly activates T3SS gene expression through a partner-switching mechanism. The components of the partner-switching mechanism are the activator ExsA, the antiactivator ExsD, the antiantiactivator ExsC, and the secreted/translocated ExsE protein (Fig. 1). In the absence of inducing signals, the secreted substrate ExsE remains cytoplasmic in a 1:2 complex with ExsC (35), and ExsA is sequestered by ExsD in a 1:1 complex (36). Inducing signals lead to the secretion/translocation of ExsE through the injectisome (34, 37, 38). The decrease in cytosolic levels of ExsE triggers partner switching wherein ExsC preferentially binds ExsD in a 2:2 complex, resulting in the release of ExsA (Fig. 1) (35, 36, 39–41). Liberated ExsA then binds to target promoters and recruits RNA polymerase to activate the T3SS regulon (28, 42, 43). Partner switching is likely driven by affinity differences in the protein-protein interactions (ExsE-ExsC > ExsD-ExsE > ExsA-ExsD, in decreasing order of affinity). While certainly true for the ExsE-ExsC (18 nM) and ExsD-ExsC (1 nM) interactions (35), the affinity of the ExsA-ExsD complex is unknown but predicted to be weaker than that for ExsD-ExsC. Partner-switching mechanisms similar to the P. aeruginosa system also regulate T3SS gene expression in Photorhabdus luminescens (44, 45), Aeromonas hydrophila (46–48), Vibrio parahaemolyticus (49–52), and Vibrio alginolyticus (53).
FIG 1.
The T3SS partner-switching mechanism that controls the DNA-binding activity of ExsA. Negative regulators are in red, and positive regulators are in white.
In addition to partner-switching-mediated control of ExsA activity, PtrA may also directly bind to and inhibit ExsA activity. PtrA is a copper-sensing protein upregulated in the presence of copper (54). Although one group found that PtrA interacts with ExsA to inhibit T3SS gene expression (54), a second group found that PtrA is a periplasmic protein important for copper tolerance and that PtrA does not interact with ExsA or play a role in the control of T3SS gene expression (55). To the best of our knowledge, no studies have observed direct repression of the T3SS by copper.
TRANSCIPTIONAL CONTROL OF ExsA
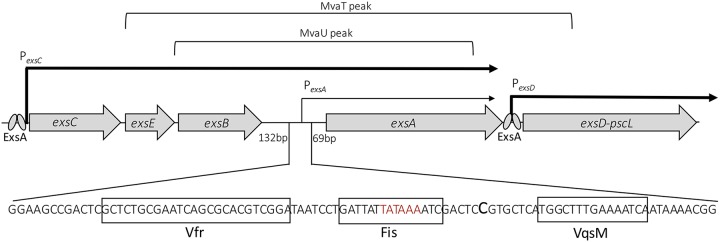
Transcription of exsA is driven from two distinct promoters. For many years, the PexsC promoter, which produces the exsCEBA polycistronic mRNA, was the only known mechanism of generating exsA transcript (Fig. 2) (27). A recent genome-wide screen for transcription start sites identified a novel transcript originating 100 nucleotides upstream of the exsA start codon (56) (Fig. 2). The same start site was identified by 5′ rapid amplification of cDNA ends (RACE) within two nucleotides (30). The newly discovered promoter, designated PexsA, generates a monocistronic exsA transcript. The PexsC promoter is ∼400 times more active than PexsA and thus makes a larger contribution to exsA transcript levels under inducing conditions. ExsA itself does not bind to or regulate PexsA promoter activity (30). Nevertheless, the PexsA promoter is subject to significant regulatory control, stimulated by Vfr, Fis, and VqsM and silenced by the histone-like proteins MvaT and MvaU (30, 57–59).
FIG 2.
Map of the T3SS regulatory locus and locations of the PexsC and PexsA promoter regions. The binding sites for Vfr (30), Fis (58), and VqsM (57) are boxed. Brackets indicate the MvaT and MvaU binding regions based on ChIP-chip data (78). The PexsA transcription start site is indicated with a large C, and the −10 region is in red.
Control of PexsA promoter activity.
Vfr is a global regulator of P. aeruginosa virulence homologous to Escherichia coli cAMP receptor protein (CRP) (60). The cAMP-Vfr signaling system activates exotoxin A production (60, 61), type IV pilus biosynthesis (62), the las quorum sensing (QS) system (63), and T3SS gene expression (64) and inhibits flagellar gene expression (65). Coordinated regulation of the T3SS with type IV pili is noteworthy because the associated adherence function is necessary for intimate adherence to host cells and translocation of the T3SS effectors. Although the primary effect of calcium chelation is through activation of secretion, the activity of the cAMP-Vfr signaling system is enhanced by EGTA (presumably through calcium chelation) (64). Because cAMP production is also stimulated by high osmolarity (64), cAMP-Vfr signaling contributes to T3SS gene control in response to hyperosmotic stress (34, 66). Vfr directly binds to and stimulates PexsA promoter activity (30). The Vfr binding site is centered ∼42 bp upstream of the PexsA promoter transcription start site (Fig. 2). Curiously, disruption of the PexsA Vfr binding site on the chromosome results in a more severe phenotype than does deletion of vfr itself (30). Whether the mutation disrupts other DNA-binding proteins that control PexsA promoter activity, RNA-binding proteins that interact with the longer exsCEBA mRNA to regulate ExsA translation (see below), or exsCEBA mRNA stability is unclear.
Fis is a nucleoid-associated protein that binds to and bends DNA (67, 68). T3SS gene expression and T3SS-dependent cytotoxicity are reduced in a fis transposon insertion mutant (58). Although a Fis binding site is located between the Vfr binding site and the −10 region of the PexsA promoter (Fig. 2), PexsA promoter activity is unaffected in the fis::Tn mutant. Instead, the decrease in T3SS gene expression correlates with reduced PexsC promoter activity. While the precise mechanism of control by Fis is unclear, there are a number of interesting connections between Fis and previously described regulators of the T3SS. Translation of Fis is inhibited by the small noncoding (sRNA) RgsA through direct base pairing with the fis mRNA (69). RpoS is essential for rgsA transcription, consistent with previous data showing that RpoS inhibits T3SS gene expression (70). Base pairing between RgsA and the fis mRNA is dependent upon the RNA chaperone Hfq and is consistent with recent data suggesting that Hfq inhibits T3SS gene expression (71). Finally, activation of the GacAS two-component system inhibits T3SS gene expression (72–74). While the primary effect of GacAS signaling is reduced cAMP-Vfr signaling and ExsA translation (75) (see below), rgsA transcription is indirectly stimulated by the GacAS two-component system (69).
VqsM is an AraC family transcriptional regulator which activates the las quorum sensing system and T3SS gene expression (57, 76). The reported VqsM binding site is located downstream of the transcription start site for the PexsA promoter (30, 56, 57). Given the location of the binding site, it seems unlikely that VqsM activates the PexsA promoter, raising the possibility of a second VqsM-dependent promoter that contributes to exsA transcription. The relative contribution of VqsM to T3SS gene expression appears to be strain specific, as vqsM is not found in all P. aeruginosa strains (77).
MvaT and MvaU are members of the histone-like nucleoid structuring (H-NS) family of global transcription repressors. H-NS proteins oligomerize on the DNA and silence gene expression by competing with transcription factors and/or trapping/occluding RNA polymerase (RNAP). A chromatin immunoprecipitation with microarray technology (ChIP-chip) study identified MvaT and MvaU binding sites in the PexsA promoter region (Fig. 2) (78). Whereas strains lacking mvaT or mvaU demonstrate elevated PexsA promoter activity and T3SS gene expression, overexpression of either MvaT or MvaU inhibits T3SS gene expression (59). Although an mvaTU double mutant is lethal, the depletion of mvaU by CRISPR interference in an mvaT mutant results in significant stimulation of PexsA promoter activity, leading to the conclusion that MvaT and MvaU have redundant roles in silencing T3SS gene expression. The proposed model is that MvaT and MvaU bind the PexsA promoter region and inhibit transcription until an activator overrides silencing (59). The best candidate is Vfr, as it appears to be required for PexsA transcription regardless of whether mvaT and mvaU are present or absent (59). It remains possible, however, that Fis, VqsM, or an unknown factor also competes with MvaT/MvaU to override the silencing activity.
The ChIP-chip data did not show MvaT or MvaU binding to promoter regions of any other known regulator of exsA transcription, including vfr (59, 78). MvaT and MvaU also inhibit transcription of the small RNAs RsmY and RsmZ involved in sequestration of RsmA (59, 78, 79). Although the net effect might liberate RsmA, which has a positive effect on ExsA translation (discussed below) (74, 75, 78), regulation of rsmYZ transcription by MvaT/MvaU does not appear to have a significant effect on T3SS gene expression.
Control of PexsC promoter activity.
The PexsC promoter is primarily activated by ExsA (27), resulting in a positive feedback loop with most exsA-containing transcript generated under inducing conditions originating from the PexsC promoter (Fig. 2), which is ∼400-fold more active than the PexsA promoter (30). Full activation of PexsC promoter activity is dependent upon PsrA, a TetR family member (80). PsrA is activated by long-chain fatty acids (LCFAs) such as oleate (81) and antimicrobial peptides (82). PsrA binds PexsC with relatively low affinity compared to its own promoter (80). LCFAs activate psrA transcription by directly binding to PsrA and preventing self-repression (81). When LCFAs are at low levels, there is excess PsrA to activate PexsC; however, when LCFA levels are high enough to bind all of the PsrA in the cell, PsrA-dependent transcription is inhibited (Fig. 3).
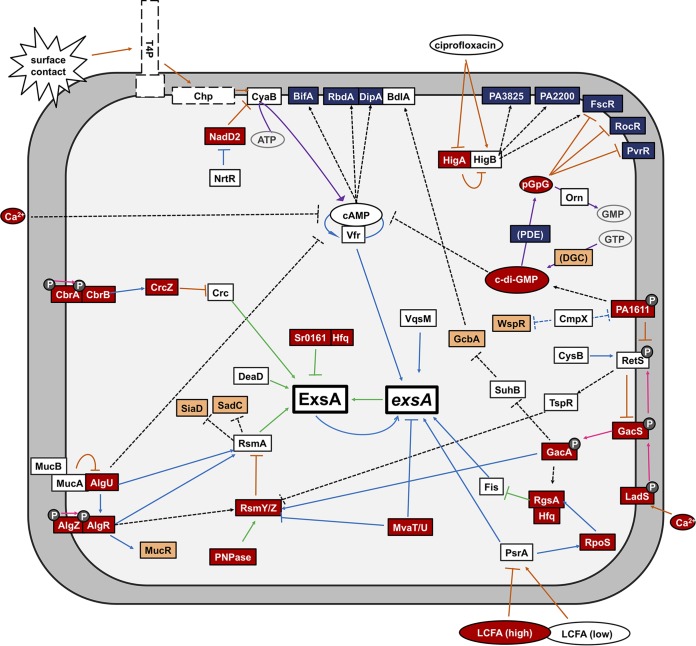
FIG 3.
Regulatory pathways that control T3SS gene expression. Gene products that stimulate T3SS gene expression are shown in white, while those that inhibit are red. Diguanylate cyclases (DGC) that synthesize c-di-GMP are orange, and phosphodiesterases (PDE) that degrade c-di-GMP are blue. P indicates a histidine kinase or response regulator. Blue lines signify transcriptional regulation, green lines signify posttranscriptional regulation, orange lines signify control of protein activity, purple lines signify enzymatic activity, pink lines signify phosphotransfer activity, and black dashed lines are links with indirect or unknown mechanisms.
TRANSLATIONAL CONTROL OF ExsA
Following transcription, the exsA mRNA is subject to additional levels of regulatory control at the posttranscriptional level. Translation of exsA is positively regulated by the RNA-binding proteins RsmA and DeaD and negatively regulated by at least one small noncoding RNA (sRNA 0161), presumably with the assistance of Hfq (71, 72, 75, 83).
Translational control by RsmA.
The Rsm system controls a critical lifestyle switch by inversely controlling phenotypes that favor acute infection or chronic colonization (84). The primary effector of the system is RsmA. Determinants positively controlled by RsmA include the cAMP-Vfr signaling system, type IV pili, and T3SS gene expression. Conversely, RsmA inhibits biofilm production, quorum sensing, the type VI secretion system (T6SS), and hydrogen cyanide (HCN) production (79, 85–88). An rsmA mutant has reduced colonization in an acute infection model, partially explained by a loss of T3SS gene expression and type IV pili, and enhanced persistence in a chronic infection model (89). This mirrors the general observation that P. aeruginosa isolates from chronic cystic fibrosis infections are defective for T3SS gene expression (90, 91). Although the significance of this observation is unclear, it suggests that T3SS gene expression is negatively selected against in the context of chronic colonization.
RsmA is an ortholog of E. coli CsrA, where it was identified as a carbon source regulator (85, 92). The CsrA family of proteins bind target mRNAs and negatively or positively modulate translation efficiency and/or stability (93). RsmA binds target mRNAs through the recognition of GGA motifs presented in the loop portion of stem-loop structures (94). For repressed targets, the RsmA binding site typically overlaps the ribosome binding site (RBS). RsmA stimulates T3SS gene expression through at least two mechanisms (Fig. 3). The first promotes cAMP-Vfr signaling and impacts T3SS gene expression through PexsA promoter activity. The transcription of both vfr and cyaB is significantly reduced in the absence of rsmA, although the mechanism of control remains unknown (72). CyaB is the primary adenylate cyclase involved in the generation of cytoplasmic cAMP (64). The second stimulatory function of RsmA enhances ExsA translation ∼2- to 3-fold (75). Although a 2- to 3-fold change in translation efficiency may appear trivial, small changes in any member of the partner-switching mechanism (ExsA, ExsD, ExsC, or ExsE) are likely sufficient to trigger significant changes in T3SS gene expression (95). It is not known whether RsmA functions directly or indirectly to stimulate Vfr and ExsA translation. Another potential explanation for a loss of T3SS gene expression in an rsmA mutant is elevated levels of c-di-GMP, which appears to antagonize cAMP-Vfr signaling (described below) (96, 97). The combined effects of RsmA on exsA transcription and translation are substantial, as rsmA and exsA mutants are equally defective for T3SS gene expression (75).
RsmA activity is controlled by several “decoy” sRNAs, including RsmV, RsmW, RsmY, and RsmZ, with RsmY and RsmZ playing the most prominent roles. Each sRNA has multiple RsmA binding sites (86, 94, 98–102). High levels of the sRNAs result in the sequestration of RsmA from target mRNAs and reduced T3SS gene expression. Cellular levels of RsmY and RsmZ are controlled by the GacAS two-component regulatory system (Fig. 3). GacS is an unusual histidine kinase with H1/D1/H2 (autophosphorylating histidine, receiver aspartate, and second histidine, respectively) domains (103). GacA is a response regulator (RR) whose sole function is activation of rsmY and rsmZ transcription following phosphorylation by GacS (74). Phosphotransfer from GacS to GacA is regulated by several additional histidine kinases. The RetS hybrid histidine kinase positively regulates the T3SS by dimerizing with the GacS H1 domain and blocking GacS autophosphorylation and by dephosphorylating GacS, preventing phosphotransfer to GacA (104–106). PA1611, another hybrid histidine kinase, counteracts RetS-mediated interference by forming heterodimers with RetS (107). The LadS histidine kinase activates GacS by transphosphorylating the GacS H2 domain (103). LadS is activated by extracellular calcium leading to the induction of rsmYZ transcription and sequestration of RsmA. Reduced RsmA availability results in an inhibition of T3SS gene expression and expression of genes associated with chronic colonization (108).
Several additional factors that control T3SS gene expression are linked to the Rsm system. Polynucleotide phosphorylase (PNPase) inhibits T3SS gene expression by directly stabilizing RsmY and RsmZ, leading to enhanced RsmA sequestration (109). TspR, a protein of unknown function, is encoded immediately downstream of retS and indirectly activated by RetS (110). T3SS gene expression and ExsA translation are reduced in a tspR mutant and restored in a tspR mutant lacking rsmY and rsmZ (110). TspR appears to stimulate T3SS gene expression by suppressing rsmY and rsmZ transcription, potentially via effects on RetS. CysB is a LysR family transcription factor that contributes to T3SS gene expression by controlling retS transcription (111). Although E. coli CysB activates sulfate assimilation, retS transcription is unaffected by sulfate or cysteine addition (111).
Translational control by the RNA helicase DeaD.
DEAD proteins are ubiquitous ATP-dependent RNA helicases with a conserved Asp-Glu-Ala-Asp (DEAD) motif. Roles of DEAD box proteins include ribosome biogenesis, translation initiation, RNA decay, and growth promotion at low temperature (112). P. aeruginosa deaD was identified by screening transposon libraries for defects in T3SS activity (113, 114). The lack of T3SS gene expression in a deaD mutant results from reduced exsA translation (2- to 3-fold) without impacting mRNA stability (114). Although the P. aeruginosa genome contains 7 DEAD box helicases, only deaD is essential for T3SS gene expression (114). The native exsA RBS is required for DeaD-mediated activation, and Mfold predictions suggest that the mRNA adopts a confirmation that partially occludes the RBS (114). Purified DeaD is sufficient to stimulate exsA translation in vitro leading to a model wherein DeaD functions by altering the mRNA structure and enhancing ribosomal access. The RsmA and DeaD requirements for exsA translation have not been fully defined, but both appear to be essential, as rsmA or deaD provided in trans is unable to complement the deaD or rsmA mutants, respectively (114).
Translational control by sRNA 0161 and Hfq.
Whereas RsmY and RsmZ are sRNAs that modulate protein activity (i.e., RsmA and RsmF), other sRNAs regulate gene expression by imperfectly base pairing with the target mRNAs (115). These sRNAs (typically 50 to 300 nucleotides [nt]) often base pair with the mRNA at or near the RBS and block translation by preventing ribosome binding. In Gram-negative bacteria, the RNA chaperone Hfq is usually required for sRNA function and/or stability. A high-throughput global sRNA target identification by ligation and sequencing (Hi-GRIL-seq) screen identified sRNAs and their cognate target mRNAs in P. aeruginosa (71). sRNA Sr0161 was found to inhibit exsA translation, and overexpression of Sr0161 reduced T3SS gene expression (71). Other sRNAs target Fis and RpoS, both of which contribute to T3SS gene expression (69, 116).
Translational control by Crc.
The Crc protein controls the availability of enzymes and transporters involved in the utilization of secondary carbon sources. A crc mutant has reduced T3SS expression, swimming, swarming, twitching, and initial biofilm formation. A microarray study identified 428 differentially controlled genes in a crc mutant, including several with described roles in T3SS gene control (117). The primary explanation for reduced T3SS gene expression is that Crc appears to stimulate ExsA translation (117). Although Crc and Hfq can function together to repress the translation of target genes (118–123), hfq and crc mutants have opposite phenotypes for T3SS gene expression (117, 122). It is unlikely, therefore, that Hfq and Crc work together to directly control the T3SS. Like RsmA, Crc activity is controlled by an sRNA, CrcZ, which acts by sequestering Crc from target mRNAs (124). The expression of crcZ is controlled by the CbrAB two-component system in response to different carbon sources (124). This has potential implications to previous studies showing that metabolic activity controls T3SS gene expression (34).
SECOND MESSENGERS
Second messengers are intracellular signaling molecules produced in response to a “first messenger,” usually an extracellular signaling molecule. The second messengers cAMP and bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) serve as regulatory switches that alter cell growth, motility, lifestyle, and virulence (125, 126). Whereas cAMP directly binds to and activates Vfr to promote T3SS gene expression (60), c-di-GMP inhibits the T3SS.
cAMP.
As discussed above, cAMP allosterically activates the DNA-binding activity of Vfr, leading to enhanced transcription of target promoters, including PexsA (30, 64, 127). cAMP is synthesized by two adenylate cyclases, CyaA and CyaB (64, 128). CyaA is located in the cytoplasm and plays a minor role in cAMP synthesis. Most cAMP is synthesized by the inner membrane-anchored CyaB (64, 129). cAMP homeostasis is maintained by CpdA, a phosphodiesterase that degrades cAMP to 5′-AMP (128). cAMP production is regulated by type IV pili and the Chp chemotaxis-like system (130, 131). Mechanosensation of surfaces by type IV pili is thought to activate the Chp system, stimulate CyaB activity, and induce expression of Vfr-dependent surface-associated virulence phenotypes (Fig. 3) (132–134).
NrtR is a transcription factor that represses itself and nadD2, a nicotinate mononucleotide adenylyltransferase gene. NrtR is required for T3SS gene expression, infection of HeLa cells, and colonization in an acute murine pneumonia model (113, 135). Loss of T3SS gene expression in the nrtR mutant results from a reduction in cAMP levels. The current model proposes that NadD2 directly interacts with and inhibits CyaB activity (135).
c-di-GMP.
c-di-GMP regulates polysaccharide synthesis, biofilm formation, adherence, and virulence factors and controls the transition between planktonic and sessile lifestyles (acute to chronic infection) (136–138). Expression of the T3SS is inhibited by elevated levels of c-di-GMP (125, 139–141). c-di-GMP is synthesized from two molecules of GTP by diguanylate cyclases (DGC) and degraded to either pGpG or two GMPs by phosphodiesterases (PDEs). pGpG is hydrolyzed into two GMP molecules by oligoribonuclease (Orn) (142, 143). Because pGpG inhibits the activity of some PDEs (e.g., RocR) (142, 143), a loss of Orn leads to an increase in cellular c-di-GMP and inhibition of the T3SS (144).
Overexpression of some DGCs inhibits cAMP-Vfr-dependent reporter activity, and overexpression of some PDEs stimulates reporter activity. DGCs with potential relevance to T3SS gene control include WspR, SiaD, SadC, MucR, and GcbA, and implicated PDEs include PvrR, RocR, FcsR, PA2200, PA3825, NbdA, BifA, DipA, and RbdA (96, 113, 126, 141–143, 145–152) (Fig. 3). The relative contribution of each has been challenging to sort out. The observation that c-di-GMP and cAMP levels are inversely related to one another likely accounts for the effect of c-di-GMP on T3SS gene expression (125, 126). The mechanistic basis for this reciprocal relationship is unclear but does not involve c-di-GMP-mediated changes to cyaAB or vfr transcription or translation or alteration of CyaA, CyaB, or CdpA activity (125). Involvement of the Chp system in cAMP production may provide a link, as both ChpA and PilK are putative c-di-GMP binding proteins (153).
SuhB is an inositol monophosphatase essential for virulence in a murine model (113, 154). SuhB inhibits the DGC GcbA, resulting in decreased c-di-GMP levels (Fig. 3) (146). SuhB also inhibits the expression of GacA (113). Because RsmY and RsmZ levels are increased in a suhB mutant, ExsA translation is dependent on the presence of suhB.
The HigB-HigA toxin/antitoxin system stimulates T3SS gene expression when cells are exposed to ciprofloxacin (Fig. 3) (148, 155). Deletion of the HigA antitoxin results in higher levels of free HigB, reduced c-di-GMP, and increased T3SS gene expression (155). Free HigB results in increased levels of three PDEs (FcsR, PA2200, and PA3825). The deletion of all three PDEs results in elevated c-di-GMP and inhibition of the T3SS phenotype (148). This is contrary to the findings from another study, however, which showed that ciprofloxacin treatment inhibits the T3SS (156).
Activation of MucA/AlgU signaling inhibits T3SS gene expression.
Chronic colonization of the cystic fibrosis airways is associated with mucoid conversion and loss of T3SS gene expression (90, 91, 157, 158). The mucoid phenotype results from alginate overproduction and is controlled by the MucA/AlgU regulatory cascade. MucA is an anti-sigma factor that sequesters the AlgU sigma factor (159). Inactivating mutations in mucA are common in chronically adapted P. aeruginosa isolated from cystic fibrosis lungs, and those mutations result in activation of the AlgU regulon (158). The AlgU regulon includes genes required for alginate biosynthesis and algR, which encodes a response regulator that controls alginate production, type IV pili, and virulence (158, 160). AlgR is part of a two-component system with AlgZ serving as the histidine kinase (161). cAMP-Vfr-dependent signaling and the Rsm system are both altered in mucA mutants (75, 162–164). AlgR and AlgU directly activate rsmA expression (165, 166), and AlgR increases RsmY and RsmZ expression through an undefined mechanism (75). The net effect of stimulating rsmA, rsmY, and rsmZ transcription is enhanced sequestration of RsmA (75). The lack of T3SS gene expression in a mucA mutant, therefore, results from reduced RsmA availability, which decreases Vfr-dependent PexsA transcription and RsmA-stimulated translation of ExsA, as described above (Fig. 3). The T3SS defect in strains lacking mucA can be largely suppressed by increasing cAMP-Vfr signaling and the levels of free RsmA (75). AlgR also directly activates mucR, a DGC, but the relative contribution of this to T3SS gene control has not been examined (152). MucB, also required for T3SS gene expression, is a periplasmic protein that binds to and protects MucA from proteolytic cleavage by AlgW (113, 167, 168). Although mucoid conversion is associated with the onset of irreversible colonization of the cystic fibrosis airways, the finding that the losses of cAMP-Vfr signaling and T3SS gene expression are coincident events suggests that the selective advantage conferred to mucA mutants in the cystic fibrosis airways may extend beyond mucoidy.
MISSING LINKS
Several genes/pathways that affect T3SS gene expression lack obvious connections to known mechanisms of T3SS gene control. The realization that most upstream regulators impact exsA transcription and/or ExsA synthesis through cAMP-Vfr, the RsmA system, and/or c-di-GMP provides a framework to rapidly evaluate regulators of unknown function.
PtrB, which is activated by degradation of the PtrR repressor following DNA damage, is a repressor of T3SS gene expression (169). Overexpression of efflux pumps MexCD-OprJ and MexEF-OprN, as well as PtrC, also inhibits T3SS gene expression (170, 171). The spermine/spermidine uptake transporter is required for T3SS gene expression (172). Spermine and spermidine are polyamines that can be utilized by P. aeruginosa as sole carbon and nitrogen sources (173, 174). While spermidine can be synthesized by P. aeruginosa, spermine is only found in eukaryotes. Mutants in the spermidine uptake transporter, but not synthesis genes, are deficient for T3SS gene expression, indicating that spermine and spermidine could be host signals that activate the T3SS (172). This is consistent with the finding that the addition of exogenous spermidine or spermine increases T3SS gene expression and cytotoxicity (172). The cAMP-Vfr and Rsm systems are unaffected in cells lacking the spermine transporter, suggesting that T3SS activation occurs through a novel mechanism.
The heat shock protein DnaK is required for acute virulence and T3SS gene expression (113, 154, 175). Elastase activity, exotoxin A secretion, swimming, swarming, and twitching motility are also diminished in a dnaK mutant, leading to the possibility that the phenotypes reflect a defect in cAMP-Vfr signaling (175). DnaK colocalizes in the periplasm with the nitrite reductase NirS (176). NirS inhibits c-di-GMP production and is required for T3SS gene expression (177).
THERAPEUTICS AND INHIBITORS OF T3SS GENE EXPRESSION
The essential role of the T3SS in the context of acute infection makes it an attractive target for therapeutic development. Targetable events include T3SS gene expression, injectisome assembly, secretory activity, effector translocation, and effector activity. Because T3SS gene expression is coupled with secretory activity by partner switching, drugs that inhibit assembly or secretion should also significantly reduce T3SS gene expression.
N-Hydroxybenzimidazoles were originally identified using a structure-based approach as inhibitors of AraC proteins (178). Several N-hydroxybenzimidazoles with 50% inhibitory concentrations (IC50s) in the low-micromolar range inhibit the DNA-binding activity of ExsA in vitro and T3SS-dependent toxicity toward macrophages (179). The N-hydroxybenzimidazoles interact with the carboxy-terminal DNA-binding domain of ExsA to inhibit DNA-binding activity (180). N-Hydroxybenzimidazoles also inhibit the DNA-binding activity of ExsA orthologs from Yersinia pestis, Aeromonas hydrophila, Photorhabdus luminescens, and Vibrio parahaemolyticus (180). Although active against ExsA in vitro, N-hydroxybenzimidazoles do not inhibit ExsA-dependent promoter activity in vivo when using either P. aeruginosa or E. coli reporter strains. The significant reduction in T3SS-dependent toxicity observed in coculture experiments with Chinese hamster ovary cells, therefore, is enigmatic. It is interesting to note that N-hydroxybenzimidazoles display broad-spectrum activity against distantly related AraC proteins (178, 181) and that the P. aeruginosa genome has >50 AraC family members. The protective activity of N-hydroxybenzimidazoles may result from effects on multiple AraC proteins or off-target effects.
As discussed above, spermidine transport is necessary for T3SS gene expression (172). Evidence for the efficacy of targeting spermidine was shown using a rhodamine 101-spermine conjugate that inhibits spermidine uptake, T3SS gene expression, cytotoxicity toward HeLa cells, and virulence in mice (182). This was followed by generating a spermidine IgG1 monoclonal antibody. The antibody also prevents spermidine-dependent activation of the T3SS, protects A549 cells for toxicity, lowers spermidine levels in mouse serum, and protects mice from P. aeruginosa lung infection (183).
Although the macrolide azithromycin does not kill planktonic P. aeruginosa at typically achieved clinical concentrations, sublethal doses inhibit quorum sensing and biofilm formation and increase T3SS gene expression (184–188). The biofilm and T3SS phenotypes likely result from inhibition of rsmY and rsmZ expression by azithromycin, leading to an increase in RsmA availability (189). Elevated T3SS gene expression is consistent with the findings that azithromycin treatment results in greater cytotoxicity when P. aeruginosa is cocultured with J774.A1 cells and increased mortality in mice following pretreatment with macrolides (190, 191). Azithromycin has positive therapeutic benefits for patients with chronic P. aeruginosa pulmonary disease, possibly by reducing biofilm formation (192–194). The efficacy of azithromycin in chronic infection may also reflect the tendency of chronic P. aeruginosa isolates to be defective in T3SS function (90, 91).
Several plant phenolic compounds (salicylic acid, its precursors, and their analogues) inhibit or activate T3SS gene expression (195). The compound TS027 inhibits exsA transcription likely through effects on ExsA translation via the Rsm system (195). Compound TS103 activates the T3SS, probably by inhibiting rsmYZ transcription and increasing RsmA availability. Coumarin is a phenolic compound that inhibits T3SS gene expression during planktonic growth by an unknown mechanism (196).
Other inhibitors of the T3SS include the anticancer drug cisplatin (197), salicyclidene acylhydrazide INP0341 (198), (−)-hopeaphenol (199), a selection of synthetic cyclic peptomers (200), and the small-molecule inhibitor fluorothiazinon (197, 201). Many of these compounds protect eukaryotic cells from T3SS-induced cytotoxicity (198, 199, 201).
CONCLUSIONS
The T3SS represents a mechanism to protect P. aeruginosa from predators ranging from amoebae to human neutrophils. Activation by the partner-switching cascade appears to be simplistically brilliant. Host cell contact is a highly relevant signal and triggers immediate induction of T3SS gene expression. It is puzzling, therefore, that so many additional resources are dedicated to controlling the T3SS. One potential explanation is that the signaling systems positioned above the partner-switching mechanism fine-tune T3SS gene expression. Why fine-tuning would be required is not obvious, as the requirement to express the T3SS gene expression seems straightforward: activate when a threat is present and inhibit when the threat has passed. Another explanation may relate to the bistability of T3SS gene expression. Single-cell experiments with fluorescent reporters have revealed that only a subset of P. aeruginosa cells within a population induce T3SS gene expression in response to either low calcium or cultured mammalian cells (34, 95, 202, 203). This phenomenon, referred to as bistability, results from stochastic fluctuations in the production and/or activity of a transcription factor (204). Bistable expression of the T3SS likely results from the combined effects of ExsA autoregulating its own transcription, ExsA-dependent control of the ExsD anti-activator, and the partner-switching mechanism. Bistable expression of the T3SS may promote social cheating. Social cheating occurs when certain members of a population produce a beneficial trait, such as T3SS-mediated killing of immune cells, and other members (cheaters) take advantage without incurring the costs associated with producing the benefit (205). A recent study found that ∼80% of P. aeruginosa cells express the T3SS in an acute murine infection model (206). In competition experiments, wild-type (wt) P. aeruginosa has a competitive advantage over a strain overexpressing the T3SS and a disadvantage against T3SS-negative strains (206). The fitness cost associated with T3SS gene expression may be energetic and/or related to expression of immunogenic molecules, such as the needle complex tip protein PcrV (207). When T3SS-defective strains are provided in significant excess over wt organisms, the disadvantage to wt organisms is negated (206). These findings may explain why T3SS-negative bacteria can be isolated from acute infections (208, 209). Cheater populations usually become unstable when the proportion of producers to cheaters becomes excessive. Fine-tuning T3SS gene expression in response to a wide range of environmental signals may establish an optimal bistable population and afford the greatest fitness advantage for cells that both express and repress T3SS gene expression.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grant R01 AI055042-13 to T.L.Y. E.A.W.M. was supported by National Institutes of Health training grant 5T32 AI007511-23.
REFERENCES
- 1.Gellatly SL, Hancock REW. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 2.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fleiszig SM, Wiener-Kronish JP, Miyazaki H, Vallas V, Mostov KE, Kanada D, Sawa T, Yen TS, Frank DW. 1997. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect Immun 65:579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulert GS, Feltman H, Rabin SDP, Martin CG, Battle SE, Rello J, Hauser AR. 2003. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital‐acquired pneumonia. J Infect Dis 188:1695–1706. doi: 10.1086/379372. [DOI] [PubMed] [Google Scholar]
- 5.Franchi L, Stoolman J, Kanneganti T-D, Verma A, Ramphal R, Núñez G. 2007. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur J Immunol 37:3030–3039. doi: 10.1002/eji.200737532. [DOI] [PubMed] [Google Scholar]
- 6.Galle M, Schotte P, Haegman M, Wullaert A, Yang HJ, Jin S, Beyaert R. 2008. The Pseudomonas aeruginosa type III secretion system plays a dual role in the regulation of caspase-1 mediated IL-1β maturation. J Cell Mol Med 12:1767–1776. doi: 10.1111/j.1582-4934.2007.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. 2007. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med 204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ince D, Sutterwala FS, Yahr TL. 2015. Secretion of flagellar proteins by the Pseudomonas aeruginosa type III secretion-injectisome system. J Bacteriol 197:2003–2011. doi: 10.1128/JB.00030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neeld D, Jin Y, Bichsel C, Jia J, Guo J, Bai F, Wu W, Ha U-H, Terada N, Jin S. 2014. Pseudomonas aeruginosa injects NDK into host cells through a type III secretion system. Microbiology 160:1417–1426. doi: 10.1099/mic.0.078139-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burstein D, Satanower S, Simovitch M, Belnik Y, Zehavi M, Yerushalmi G, Ben-Aroya S, Pupko T, Banin E. 2015. Novel type III effectors in Pseudomonas aeruginosa. mBio 6:e00161-15. doi: 10.1128/mBio.00161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dacheux D, Goure J, Chabert J, Usson Y, Attree I. 2001. Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol Microbiol 40:76–85. doi: 10.1046/j.1365-2958.2001.02368.x. [DOI] [PubMed] [Google Scholar]
- 12.Goure J, Pastor A, Faudry E, Chabert J, Dessen A, Attree I. 2004. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect Immun 72:4741–4750. doi: 10.1128/IAI.72.8.4741-4750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee VT, Smith RS, Tümmler B, Lory S. 2005. Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infect Immun 73:1695–1705. doi: 10.1128/IAI.73.3.1695-1705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vance RE, Rietsch A, Mekalanos JJ. 2005. Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect Immun 73:1706–1713. doi: 10.1128/IAI.73.3.1706-1713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galle M, Carpentier I, Beyaert R. 2012. Structure and function of the type III secretion system of Pseudomonas aeruginosa. Curr Protein Pept Sci 13:831–842. doi: 10.2174/138920312804871210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dortet L, Lombardi C, Cretin F, Dessen A, Filloux A. 2018. Pore-forming activity of the Pseudomonas aeruginosa type III secretion system translocon alters the host epigenome. Nat Microbiol 3:378–386. doi: 10.1038/s41564-018-0109-7. [DOI] [PubMed] [Google Scholar]
- 17.Finck-Barbançon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish JP, Fleiszig SM, Wu C, Mende-Mueller L, Frank DW. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol 25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 18.Hauser AR, Kang PJ, Engel JN. 1998. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol 27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 19.Sawa T, Ohara M, Kurahashi K, Twining SS, Frank DW, Doroques DB, Long T, Gropper MA, Wiener-Kronish JP. 1998. In vitro cellular toxicity predicts Pseudomonas aeruginosa virulence in lung infections. Infect Immun 66:3242–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holder IA, Neely AN, Frank DW. 2001. Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns 27:129–130. doi: 10.1016/S0305-4179(00)00142-X. [DOI] [PubMed] [Google Scholar]
- 21.Lee EJ, Cowell BA, Evans DJ, Fleiszig S. 2003. Contribution of ExsA-regulated factors to corneal infection by cytotoxic and invasive Pseudomonas aeruginosa in a murine scarification model. Invest Ophthalmol Vis Sci 44:3892–3898. doi: 10.1167/iovs.02-1302. [DOI] [PubMed] [Google Scholar]
- 22.Koh AY, Priebe GP, Pier GB. 2005. Virulence of Pseudomonas aeruginosa in a murine model of gastrointestinal colonization and dissemination in neutropenia. Infect Immun 73:2262–2272. doi: 10.1128/IAI.73.4.2262-2272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank DW. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol 26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 24.Vallis AJ, Yahr TL, Barbieri JT, Frank DW. 1999. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun 67:914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Ahn K, Min S, Jia J, Ha U, Wu D, Jin S. 2005. Factors triggering type III secretion in Pseudomonas aeruginosa. Microbiology 151:3575–3587. doi: 10.1099/mic.0.28277-0. [DOI] [PubMed] [Google Scholar]
- 26.Frank DW, Iglewski BH. 1991. Cloning and sequence analysis of a trans-regulatory locus required for exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol 173:6460–6468. doi: 10.1128/jb.173.20.6460-6468.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yahr TL, Frank DW. 1994. Transcriptional organization of the trans-regulatory locus which controls exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol 176:3832–3838. doi: 10.1128/jb.176.13.3832-3838.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hovey AK, Frank DW. 1995. Analysis of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol 177:4427–4436. doi: 10.1128/jb.177.15.4427-4436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yahr TL, Hovey AK, Kulich SM, Frank DW. 1995. Transcriptional analysis of the Pseudomonas aeruginosa exoenzyme S structural gene. J Bacteriol 177:1169–1178. doi: 10.1128/jb.177.5.1169-1178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsden AE, Intile PJ, Schulmeyer KH, Simmons-Patterson ER, Urbanowski ML, Wolfgang MC, Yahr L. 2016. Vfr directly activates exsA transcription to regulate expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 198:1442–1450. doi: 10.1128/JB.00049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brutinel ED, Vakulskas CA, Brady KM, Yahr TL. 2008. Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol 68:657–671. doi: 10.1111/j.1365-2958.2008.06179.x. [DOI] [PubMed] [Google Scholar]
- 32.Diaz MR, King JM, Yahr TL. 2011. Intrinsic and extrinsic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Front Microbiol 2:89. doi: 10.3389/fmicb.2011.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCaw ML, Lykken GL, Singh PK, Yahr TL. 2002. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol Microbiol 46:1123–1133. doi: 10.1046/j.1365-2958.2002.03228.x. [DOI] [PubMed] [Google Scholar]
- 34.Rietsch A, Mekalanos JJ. 2006. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol Microbiol 59:807–820. doi: 10.1111/j.1365-2958.2005.04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng Z, Chen G, Joshi S, Brutinel ED, Yahr TL, Chen L. 2007. Biochemical characterization of a regulatory cascade controlling transcription of the Pseudomonas aeruginosa type III secretion system. J Biol Chem 282:6136–6142. doi: 10.1074/jbc.M611664200. [DOI] [PubMed] [Google Scholar]
- 36.Thibault J, Faudry E, Ebel C, Attree I, Elsen S. 2009. Anti-activator ExsD forms a 1:1 complex with ExsA to inhibit transcription of type III secretion operons. J Biol Chem 284:15762–15770. doi: 10.1074/jbc.M109.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urbanowski ML, Lykken GL, Yahr TL. 2005. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc Natl Acad Sci U S A 102:9930–9935. doi: 10.1073/pnas.0504405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dasgupta N, Lykken GL, Wolfgang MC, Yahr TL. 2004. A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol 53:297–308. doi: 10.1111/j.1365-2958.2004.04128.x. [DOI] [PubMed] [Google Scholar]
- 40.Lykken GL, Chen G, Brutinel ED, Chen L, Yahr TL. 2006. Characterization of ExsC and ExsD self-association and heterocomplex formation. J Bacteriol 188:6832–6840. doi: 10.1128/JB.00884-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brutinel ED, Vakulskas CA, Yahr TL. 2010. ExsD inhibits expression of the Pseudomonas aeruginosa type III secretion system by disrupting ExsA self-association and DNA binding activity. J Bacteriol 192:1479–1486. doi: 10.1128/JB.01457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brutinel ED, Yahr TL. 2008. Control of gene expression by type III secretory activity. Curr Opin Microbiol 11:128–133. doi: 10.1016/j.mib.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marsden AE, Schubot FD, Yahr TL. 2014. Self-association is required for occupation of adjacent binding sites in Pseudomonas aeruginosa type III secretion system promoters. J Bacteriol 196:3546–3555. doi: 10.1128/JB.01969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waterfield NR, Daborn PJ, Ffrench-Constant RH. 2002. Genomic islands in Photorhabdus. Trends Microbiol 10:541–545. doi: 10.1016/S0966-842X(02)02463-0. [DOI] [PubMed] [Google Scholar]
- 45.Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles J-F, Dassa E, Derose R, Derzelle S, Freyssinet G, Gaudriault S, Médigue C, Lanois A, Powell K, Siguier P, Vincent R, Wingate V, Zouine M, Glaser P, Boemare N, Danchin A, Kunst F. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol 21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- 46.Yu HB, Rao PSS, Lee HC, Vilches S, Merino S, Tomas JM, Leung KY. 2004. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect Immun 72:1248–1256. doi: 10.1128/IAI.72.3.1248-1256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vilches S, Urgell C, Merino S, Chacón MR, Soler L, Castro-Escarpulli G, Figueras MJ, Tomás JM. 2004. Complete type III secretion system of a mesophilic Aeromonas hydrophila strain. Appl Environ Microbiol 70:6914–6919. doi: 10.1128/AEM.70.11.6914-6919.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vilches S, Jimenez N, Tomás JM, Merino S. 2009. Aeromonas hydrophila AH-3 type III secretion system expression and regulatory network. Appl Environ Microbiol 75:6382–6392. doi: 10.1128/AEM.00222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, Shah DH, Konkel ME, Call DR. 2008. Type III secretion system 1 genes in Vibrio parahaemolyticus are positively regulated by ExsA and negatively regulated by ExsD. Mol Microbiol 69:747–764. doi: 10.1111/j.1365-2958.2008.06326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X, Konkel ME, Call DR. 2010. Regulation of type III secretion system 1 gene expression in Vibrio parahaemolyticus is dependent on interactions between ExsA, ExsC, and ExsD. Virulence 1:260–272. doi: 10.4161/viru.1.4.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kodama T, Yamazaki C, Park K-S, Akeda Y, Iida T, Honda T. 2010. Transcription of Vibrio parahaemolyticus T3SS1 genes is regulated by a dual regulation system consisting of the ExsACDE regulatory cascade and H-NS. FEMS Microbiol Lett 311:10. doi: 10.1111/j.1574-6968.2010.02066.x. [DOI] [PubMed] [Google Scholar]
- 52.Erwin DP, Nydam SD, Call DR. 2012. Vibrio parahaemolyticus ExsE is requisite for initial adhesion and subsequent type III secretion system 1-dependent autophagy in HeLa cells. Microbiology 158:2303–2314. doi: 10.1099/mic.0.059931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, Lu S-Y, Orfe LH, Ren C-H, Hu C-Q, Call DR, Avillan JJ, Zhao Z. 2016. ExsE is a negative regulator for T3SS gene expression in Vibrio alginolyticus. Front Cell Infect Microbiol 6:177. doi: 10.3389/fcimb.2016.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ha U-H, Kim J, Badrane H, Jia J, Baker HV, Wu D, Jin S. 2004. An in vivo inducible gene of Pseudomonas aeruginosa encodes an anti-ExsA to suppress the type III secretion system. Mol Microbiol 54:307–320. doi: 10.1111/j.1365-2958.2004.04282.x. [DOI] [PubMed] [Google Scholar]
- 55.Elsen S, Ragno M, Attree I. 2011. PtrA is a periplasmic protein involved in Cu tolerance in Pseudomonas aeruginosa. J Bacteriol 193:3376–3378. doi: 10.1128/JB.00159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, Greenberg EP, Sorek R, Lory S. 2012. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog 8:e1002945. doi: 10.1371/journal.ppat.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang H, Deng X, Li X, Ye Y, Wu M. 2014. Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res 42:10307–10320. doi: 10.1093/nar/gku586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng X, Li M, Pan X, Zheng R, Liu C, Chen F, Liu X, Cheng Z, Jin S, Wu W. 2017. Fis regulates type III secretion system by influencing the transcription of exsA in Pseudomonas aeruginosa strain PA14. Front Microbiol 8:669. doi: 10.3389/fmicb.2017.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams McMackin EA, Marsden AE, Yahr TL. 2019. H-NS family members MvaT and MvaU regulate the Pseudomonas aeruginosa type III secretion system. J Bacteriol 201:e00054-19. doi: 10.1128/JB.00054-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.West SEH, Sample AK, Runyen-Janecky LJ. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J Bacteriol 176:7532–7542. doi: 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davinic M, Carty NL, Colmer-Hamood JA, San Francisco M, Hamood AN. 2009. Role of Vfr in regulating exotoxin A production by Pseudomonas aeruginosa. Microbiology 155:2265–2273. doi: 10.1099/mic.0.028373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beatson SA, Whitchurch CB, Sargent JL, Levesque RC, Mattick JS. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J Bacteriol 184:3605–3613. doi: 10.1128/JB.184.13.3605-3613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Albus AM, Pesci EC, Runyen-Janecky LJ, West SE, Iglewski BH. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J Bacteriol 179:3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolfgang MC, Lee VT, Gilmore ME, Lory S. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell 4:253–263. doi: 10.1016/S1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 65.Dasgupta N, Ferrell EP, Kanack KJ, West SEH, Ramphal R. 2002. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is sigma70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J Bacteriol 184:5240–5250. doi: 10.1128/JB.184.19.5240-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aspedon A, Palmer K, Whiteley M. 2006. Microarray analysis of the osmotic stress response in Pseudomonas aeruginosa. J Bacteriol 188:2721–2725. doi: 10.1128/JB.188.7.2721-2725.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koch C, Vandekerckhove J, Kahmann R. 1988. Escherichia coli host factor for site-specific DNA inversion: cloning and characterization of the fis gene. Proc Natl Acad Sci U S A 85:4237–4241. doi: 10.1073/pnas.85.12.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider R, Lurz R, Lüder G, Tolksdorf C, Travers A, Muskhelishvili G. 2001. An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res 29:5107–5114. doi: 10.1093/nar/29.24.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu P, Wang Y, Zhang Y, Hu Y, Thompson KM, Chen S. 2016. RpoS-dependent sRNA RgsA regulates Fis and AcpP in Pseudomonas aeruginosa. Mol Microbiol 102:244–259. doi: 10.1111/mmi.13458. [DOI] [PubMed] [Google Scholar]
- 70.Hogardt M, Roeder M, Schreff AM, Eberl L, Heesemann J. 2004. Expression of Pseudomonas aeruginosa exoS is controlled by quorum sensing and RpoS. Microbiology 150:843–851. doi: 10.1099/mic.0.26703-0. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y-F, Han K, Chandler CE, Tjaden B, Ernst RK, Lory S. 2017. Probing the sRNA regulatory landscape of P. aeruginosa: post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility. Mol Microbiol 106:919–937. doi: 10.1111/mmi.13857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mulcahy H, O’Callaghan J, O’Grady EP, Adams C, O’Gara F. 2006. The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect Immun 74:3012–3015. doi: 10.1128/IAI.74.5.3012-3015.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.González N, Heeb S, Valverde C, Kay E, Reimmann C, Junier T, Haas D. 2008. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics 9:167. doi: 10.1186/1471-2164-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Intile PJ, Diaz MR, Urbanowski ML, Wolfgang MC, Yahr TL. 2014. The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 196:357–366. doi: 10.1128/JB.01199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dong Y-H, Zhang X-F, Xu J-L, Tan A-T, Zhang L-H. 2005. VqsM, a novel AraC-type global regulator of quorum-sensing signalling and virulence in Pseudomonas aeruginosa. Mol Microbiol 58:552–564. doi: 10.1111/j.1365-2958.2005.04851.x. [DOI] [PubMed] [Google Scholar]
- 77.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman F. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Castang S, McManus HR, Turner KH, Dove SL. 2008. H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci 105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol 72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shen DK, Filopon D, Kuhn L, Polack B, Toussaint B. 2006. PsrA is a positive transcriptional regulator of the type III secretion system in Pseudomonas aeruginosa. Infect Immun 74:1121–1129. doi: 10.1128/IAI.74.2.1121-1129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kang Y, Lunin VV, Skarina T, Savchenko A, Schurr MJ, Hoang TT. 2009. The long-chain fatty acid sensor, PsrA, modulates the expression of rpoS and the type III secretion exsCEBA operon in Pseudomonas aeruginosa. Mol Microbiol 73:120–136. doi: 10.1111/j.1365-2958.2009.06757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock R. 2008. Induction by cationic antimicrobial peptides and involvement in intrinsic polymyxin and antimicrobial peptide resistance, biofilm formation, and swarming motility of PsrA in Pseudomonas aeruginosa. J Bacteriol 190:5624–5634. doi: 10.1128/JB.00594-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 84.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A 103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pessi G, Williams F, Hindle Z, Heurlier K, Holden MT, Cámara M, Haas D, Williams P. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J Bacteriol 183:6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heurlier K, Williams F, Heeb S, Dormond C, Pessi G, Singer D, Cámara M, Williams P, Haas D. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J Bacteriol 186:2936–2945. doi: 10.1128/JB.186.10.2936-2945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.O’Grady EP, Mulcahy H, O’Callaghan J, Adams C, O’Gara F. 2006. Pseudomonas aeruginosa infection of airway epithelial cells modulates expression of Kruppel-like factors 2 and 6 via RsmA-mediated regulation of type III exoenzymes S and Y. Infect. Immun 74:5893–5902. doi: 10.1128/IAI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mulcahy H, O’Callaghan J, O’Grady EP, Maciá MD, Borrell N, Gómez C, Casey PG, Hill C, Adams C, Gahan CGM, Oliver A, O’Gara F. 2008. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect Immun 76:632–638. doi: 10.1128/IAI.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jain M, Ramirez D, Seshadri R, Cullina JF, Powers CA, Schulert GS, Bar-Meir M, Sullivan CL, McColley SA, Hauser AR. 2004. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J Clin Microbiol 42:5229–5237. doi: 10.1128/JCM.42.11.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jain M, Bar-Meir M, McColley S, Cullina J, Potter E, Powers C, Prickett M, Seshadri R, Jovanovic B, Petrocheilou A, King JD, Hauser AR. 2008. Evolution of Pseudomonas aeruginosa type III secretion in cystic fibrosis: a paradigm of chronic infection. Transl Res 152:257–264. doi: 10.1016/j.trsl.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol 175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Babitzke P, Romeo T. 2007. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol 10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 94.Schulmeyer KH, Diaz MR, Bair TB, Sanders W, Gode CJ, Laederach A, Wolfgang MC, Yahr TL. 2016. Primary and secondary sequence structure requirements for recognition and discrimination of target RNAs by Pseudomonas aeruginosa RsmA and RsmF. J Bacteriol 198:2458–2469. doi: 10.1128/JB.00343-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Urbanowski ML, Brutinel ED, Yahr TL. 2007. Translocation of ExsE into Chinese hamster ovary cells is required for transcriptional induction of the Pseudomonas aeruginosa type III secretion system. Infect Immun 75:4432–4439. doi: 10.1128/IAI.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moscoso JA, Jaeger T, Valentini M, Hui K, Jenal U, Filloux A. 2014. The diguanylate cyclase SadC is a central player in Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. J Bacteriol 196:4081–4088. doi: 10.1128/JB.01850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Colley B, Dederer V, Carnell M, Kjelleberg S, Rice SA, Klebensberger J. 2016. SiaA/D interconnects c-di-GMP and RsmA signaling to coordinate cellular aggregation of Pseudomonas aeruginosa in response to environmental conditions. Front Microbiol 7:179. doi: 10.3389/fmicb.2016.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dubey AK, Baker CS, Romeo T, Babitzke P. 2005. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain F-T. 2007. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol 14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 100.Janssen KH, Diaz MR, Golden M, Graham JW, Sanders W, Wolfgang MC, Yahr TL. 2018. Functional analyses of the RsmY and RsmZ small noncoding regulatory RNAs in Pseudomonas aeruginosa. J Bacteriol 200:e00736-17. doi: 10.1128/JB.00736-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Valverde C, Heeb S, Keel C, Haas D. 2003. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol Microbiol 50:1361–1379. doi: 10.1046/j.1365-2958.2003.03774.x. [DOI] [PubMed] [Google Scholar]
- 102.Kay E, Humair B, Dénervaud V, Riedel K, Spahr S, Eberl L, Valverde C, Haas D. 2006. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J Bacteriol 188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chambonnier G, Roux L, Redelberger D, Fadel F, Filloux A, Sivaneson M, de Bentzmann S, Bordi C. 2016. The hybrid histidine kinase LadS forms a multicomponent signal transduction system with the GacS/GacA two-component system in Pseudomonas aeruginosa. PLoS Genet 12:e1006032. doi: 10.1371/journal.pgen.1006032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Laskowski MA, Osborn E, Kazmierczak BI. 2004. A novel sensor kinase-response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa. Mol Microbiol 54:1090–1103. doi: 10.1111/j.1365-2958.2004.04331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev 23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Francis VI, Waters EM, Finton-James SE, Gori A, Kadioglu A, Brown AR, Porter SL. 2018. Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa. Nat Commun 9:2219. doi: 10.1038/s41467-018-04640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kong W, Chen L, Zhao J, Shen T, Surette MG, Shen L, Duan K. 2013. Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol Microbiol 88:784–797. doi: 10.1111/mmi.12223. [DOI] [PubMed] [Google Scholar]
- 108.Broder UN, Jaeger T, Jenal U. 2017. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat Microbiol 2:16184. doi: 10.1038/nmicrobiol.2016.184. [DOI] [PubMed] [Google Scholar]
- 109.Chen R, Weng Y, Zhu F, Jin Y, Liu C, Pan X, Xia B, Cheng Z, Jin S, Wu W. 2016. Polynucleotide phosphorylase regulates multiple virulence factors and the stabilities of small RNAs RsmY/Z in Pseudomonas aeruginosa. Front Microbiol 7:247. doi: 10.3389/fmicb.2016.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhu M, Zhao J, Kang H, Kong W, Liang H, Wu M, Liang H. 2016. Modulation of type III secretion system in Pseudomonas aeruginosa: involvement of the PA4857 gene product. Front Microbiol 7:7. doi: 10.3389/fmicb.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Song Y, Yang C, Chen G, Zhang Y, Seng Z, Cai Z, Zhang C, Yang L, Gan J, Haihua L. 2019. Molecular insights into the master regulator CysB-mediated bacterial virulence in Pseudomonas aeruginosa. Mol Microbiol 111:1195–1210. doi: 10.1111/mmi.14200. [DOI] [PubMed] [Google Scholar]
- 112.Khemici V, Linder P. 2016. RNA helicases in bacteria. Curr Opin Microbiol 30:58–66. doi: 10.1016/j.mib.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 113.Li K, Xu C, Jin Y, Sun Z, Liu C, Shi J, Chen G, Chen R, Jin S, Wu W. 2013. SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. mBio 4:e00419-13. doi: 10.1128/mBio.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Intile PJ, Balzer GJ, Wolfgang MC, Yahr TL. 2015. The RNA helicase DeaD stimulates ExsA translation to promote expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 197:2664–2674. doi: 10.1128/JB.00231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu P, Wang Y, Hu Y, Chen S. 2018. RgsA, an RpoS-dependent sRNA, negatively regulates rpoS expression in Pseudomonas aeruginosa. Microbiology 164:716–724. doi: 10.1099/mic.0.000632. [DOI] [PubMed] [Google Scholar]
- 117.Dong Y-H, Zhang X-F, Zhang L-H. 2013. The global regulator Crc plays a multifaceted role in modulation of type III secretion system in Pseudomonas aeruginosa. Microbiologyopen 2:161–172. doi: 10.1002/mbo3.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Milojevic T, Grishkovskaya I, Sonnleitner E, Djinovic-Carugo K, Bläsi U. 2013. The Pseudomonas aeruginosa catabolite repression control protein Crc is devoid of RNA binding activity. PLoS One 8:e64609. doi: 10.1371/journal.pone.0064609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sonnleitner E, Bläsi U. 2014. Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet 10:e1004440. doi: 10.1371/journal.pgen.1004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang N, Ding S, Chen F, Zhang X, Xia Y, Di H, Cao Q, Deng X, Wu M, Wong CCL, Tian X-X, Yang C-G, Zhao J, Lan L. 2015. The Crc protein participates in down-regulation of the Lon gene to promote rhamnolipid production and rhl quorum sensing in Pseudomonas aeruginosa. Mol Microbiol 96:526–547. doi: 10.1111/mmi.12954. [DOI] [PubMed] [Google Scholar]
- 121.Sonnleitner E, Prindl K, Bläsi U. 2017. The Pseudomonas aeruginosa CrcZ RNA interferes with Hfq-mediated riboregulation. PLoS One 12:e0180887. doi: 10.1371/journal.pone.0180887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sonnleitner E, Wulf A, Campagne S, Pei X-Y, Wolfinger MT, Forlani G, Prindl K, Abdou L, Resch A, Allain F-T, Luisi BF, Urlaub H, Bläsi U. 2018. Interplay between the catabolite repression control protein Crc, Hfq and RNA in Hfq-dependent translational regulation in Pseudomonas aeruginosa. Nucleic Acids Res 46:1470–1485. doi: 10.1093/nar/gkx1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pei XY, Dendooven T, Sonnleitner E, Chen S, Bläsi U, Luisi BF. 2019. Architectural principles for Hfq/Crc-mediated regulation of gene expression. Elife 8:e43158. doi: 10.7554/eLife.43158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sonnleitner E, Abdou L, Haas D. 2009. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 106:21866–21871. doi: 10.1073/pnas.pnas.0910308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Almblad H, Harrison JJ, Rybtke M, Groizeleau J, Givskov M, Parsek MR, Tolker-Nielsen T. 2015. The cyclic AMP-Vfr signaling pathway in Pseudomonas aeruginosa is inhibited by cyclic di-GMP. J Bacteriol 197:2190–2200. doi: 10.1128/JB.00193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Almblad H, Rybtke M, Hendiani S, Andersen JB, Givskov M, Tolker-Nielsen T. 2019. High levels of cAMP inhibit Pseudomonas aeruginosa biofilm formation through reduction of the c-di-GMP content. Microbiology 165:324–333. doi: 10.1099/mic.0.000772. [DOI] [PubMed] [Google Scholar]
- 127.Fuchs EL, Brutinel ED, Jones AK, Fulcher NB, Urbanowski ML, Yahr TL, Wolfgang MC. 2010. The Pseudomonas aeruginosa Vfr regulator controls global virulence factor expression through cyclic AMP-dependent and -independent mechanisms. J Bacteriol 192:3553–3564. doi: 10.1128/JB.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fuchs EL, Brutinel ED, Klem ER, Fehr AR, Yahr TL, Wolfgang MC. 2010. In vitro and in vivo characterization of the Pseudomonas aeruginosa cyclic AMP (cAMP) phosphodiesterase CpdA, required for cAMP homeostasis and virulence factor regulation. J Bacteriol 192:2779–2790. doi: 10.1128/JB.00168-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Siegel LS, Hylemon PB, Phibbs PV. 1977. Cyclic adenosine 3′,5′-monophosphate levels and activities of adenylate cyclase and cyclic adenosine 3′,5′-monophosphate phosphodiesterase in Pseudomonas and Bacteroides. J Bacteriol 129:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, Bertrand JJ, Semmler ABT, Mellick AS, Martin PR, Alm RA, Hobbs M, Beatson SA, Huang B, Nguyen L, Commolli JC, Engel JN, Darzins A, Mattick JS. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol 52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 131.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. 2015. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. 2010. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol 76:889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Inclan YF, Huseby MJ, Engel JN. 2011. FimL regulates cAMP Synthesis in Pseudomonas aeruginosa. PLoS One 6:e15867. doi: 10.1371/journal.pone.0015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GCL, O’Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6:e02456-14. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jin Y, Zhang M, Zhu F, Peng Q, Weng Y, Zhao Q, Liu C, Bai F, Cheng Z, Jin S, Wu W. 2019. NrtR regulates the type III secretion system through cAMP-Vfr pathway in Pseudomonas aeruginosa. Front Microbiol 10:85. doi: 10.3389/fmicb.2019.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 137.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kuchma SL, Connolly JP, O'Toole GA. 2005. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J Bacteriol 187:1441–1454. doi: 10.1128/JB.187.4.1441-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol 13:3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 142.Orr MW, Donaldson GP, Severin GB, Wang J, Sintim HO, Waters CM, Lee VT. 2015. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci U S A 112:E5048–E5057. doi: 10.1073/pnas.1507245112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cohen D, Mechold U, Nevenzal H, Yarmiyhu Y, Randall TE, Bay DC, Rich JD, Parsek MR, Kaever V, Harrison JJ, Banin E. 2015. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:11359–11364. doi: 10.1073/pnas.1421450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen G, Zhao Q, Zhu F, Chen R, Jin Y, Liu C, Pan X, Jin S, Wu W, Cheng Z. 2016. Oligoribonuclease is required for the type III secretion system and pathogenesis of Pseudomonas aeruginosa. Microbiol Res 188:90–96. doi: 10.1016/j.micres.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 145.Petrova OE, Cherny KE, Sauer K. 2015. The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. J Bacteriol 197:174–187. doi: 10.1128/JB.02244-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li K, Yang G, Debru AB, Li P, Zong L, Li P, Xu T, Wu W, Jin S, Bao Q. 2017. SuhB regulates the motile-sessile switch in Pseudomonas aeruginosa through the Gac/Rsm pathway and c-di-GMP signaling. Front Microbiol 8:1045. doi: 10.3389/fmicb.2017.01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rossello J, Lima A, Gil M, Rodríguez Duarte J, Correa A, Carvalho PC, Kierbel A, Durán R. 2017. The EAL-domain protein FcsR regulates flagella, chemotaxis and type III secretion system in Pseudomonas aeruginosa by a phosphodiesterase independent mechanism. Sci Rep 7:10281. doi: 10.1038/s41598-017-09926-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang Y, Xia B, Li M, Shi J, Long Y, Jin Y, Bai F, Cheng Z, Jin S, Wu W. 2018. HigB reciprocally controls biofilm formation and the expression of type III secretion system genes through influencing the intracellular c-di-GMP level in Pseudomonas aeruginosa. Toxins (Basel) 10:424. doi: 10.3390/toxins10110424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol 191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A 109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roy AB, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol 194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kong W, Zhao J, Kang H, Zhu M, Zhou T, Deng X, Liang H. 2015. ChIP-seq reveals the global regulator AlgR mediating cyclic di-GMP synthesis in Pseudomonas aeruginosa. Nucleic Acids Res 43:8268–8282. doi: 10.1093/nar/gkv747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Düvel J, Bertinetti D, Möller S, Schwede F, Morr M, Wissing J, Radamm L, Zimmermann B, Genieser H-G, Jänsch L, Herberg FW, Häussler S. 2012. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods 88:229–236. doi: 10.1016/j.mimet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 154.Yasuda M, Nagata S, Yamane S, Kunikata C, Kida Y, Kuwano K, Suezawa C, Okuda J. 2017. Pseudomonas aeruginosa serA gene is required for bacterial translocation through Caco-2 cell monolayers. PLoS One 12:e0169367. doi: 10.1371/journal.pone.0169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li M, Long Y, Liu Y, Liu Y, Chen R, Shi J, Zhang L, Jin Y, Yang L, Bai F, Jin S, Cheng Z, Wu W. 2016. HigB of Pseudomonas aeruginosa enhances killing of phagocytes by up-regulating the type III secretion system in ciprofloxacin induced persister cells. Front Cell Infect Microbiol 6:125. doi: 10.3389/fcimb.2016.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cirz RT, O’Neill BM, Hammond JA, Head SR, Romesberg FE. 2006. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol 188:7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Govan JRW, Nelson JW. 1992. Microbiology of lung infection in cystic fibrosis. Br Med Bull 48:912–930. doi: 10.1093/oxfordjournals.bmb.a072585. [DOI] [PubMed] [Google Scholar]
- 158.Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A 90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xie ZD, Hershberger CD, Shankar S, Ye RW, Chakrabarty AM. 1996. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol 178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lizewski SE, Schurr JR, Jackson DW, Frisk A, Carterson AJ, Schurr MJ. 2004. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J Bacteriol 186:5672–5684. doi: 10.1128/JB.186.17.5672-5684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Whitchurch CB, Alm RA, Mattick JS. 1996. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 93:9839–9843. doi: 10.1073/pnas.93.18.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rau MH, Hansen SK, Johansen HK, Thomsen LE, Workman CT, Nielsen KF, Jelsbak L, Høiby N, Yang L, Molin S. 2010. Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ Microbiol 12:1643–1658. doi: 10.1111/j.1462-2920.2010.02211.x. [DOI] [PubMed] [Google Scholar]
- 163.Jones AK, Fulcher NB, Balzer GJ, Urbanowski ML, Pritchett CL, Schurr MJ, Yahr TL, Wolfgang MC. 2010. Activation of the Pseudomonas aeruginosa AlgU regulon through mucA mutation inhibits cyclic AMP/Vfr signaling. J Bacteriol 192:5709–5717. doi: 10.1128/JB.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pritchett CL, Little AS, Okkotsu Y, Frisk A, Cody WL, Covey CR, Schurr MJ. 2015. Expression analysis of the Pseudomonas aeruginosa AlgZR two-component regulatory system. J Bacteriol 197:736–748. doi: 10.1128/JB.02290-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stacey SD, Pritchett CL. 2016. Pseudomonas aeruginosa AlgU contributes to posttranscriptional activity by increasing rsmA expression in a mucA22 strain. J Bacteriol 198:1812–1826. doi: 10.1128/JB.00133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Stacey SD, Williams DA, Pritchett CL. 2017. The Pseudomonas aeruginosa two-component regulator AlgR directly activates rsmA expression in a phosphorylation independent manner. J Bacteriol 199:e00048-17. doi: 10.1128/JB.00048-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mathee K, McPherson CJ, Ohman DE. 1997. Posttranslational control of the algT (algU)-encoded σ22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J Bacteriol 179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cezairliyan BO, Sauer RT. 2009. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol 72:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wu W, Jin S. 2005. PtrB of Pseudomonas aeruginosa suppresses the type III secretion system under the stress of DNA damage. J Bacteriol 187:6058–6068. doi: 10.1128/JB.187.17.6058-6068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Linares JF, López JA, Camafeita E, Albar JP, Rojo F, Martínez JL. 2005. Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J Bacteriol 187:1384–1391. doi: 10.1128/JB.187.4.1384-1391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jin Y, Yang H, Qiao M, Jin S. 2011. MexT regulates the type III secretion system through MexS and PtrC in Pseudomonas aeruginosa. J Bacteriol 193:399–410. doi: 10.1128/JB.01079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhou L, Wang J, Zhang L-H. 2007. Modulation of bacterial type III secretion system by a spermidine transporter dependent signaling pathway. PLoS One 2:e1291. doi: 10.1371/journal.pone.0001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lu C-D, Itoh Y, Nakada Y, Jiang Y. 2002. Functional analysis and regulation of the divergent spuABCDEFGH-spuI operons for polyamine uptake and utilization in Pseudomonas aeruginosa PAO1. J Bacteriol 184:3765–3773. doi: 10.1128/JB.184.14.3765-3773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nishijyo T, Haas D, Itoh Y. 2001. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol Microbiol 40:917–931. doi: 10.1046/j.1365-2958.2001.02435.x. [DOI] [PubMed] [Google Scholar]
- 175.Okuda J, Yamane S, Nagata S, Kunikata C, Suezawa C, Yasuda M. 2017. The Pseudomonas aeruginosa dnaK gene is involved in bacterial translocation across the intestinal epithelial cell barrier. Microbiology 163:1208–1216. doi: 10.1099/mic.0.000508. [DOI] [PubMed] [Google Scholar]
- 176.Acuña JMB, Molinari G, Rohde M, Dammeyer T, Wissing J, Jänsch L, Arias S, Jahn M, Schobert M, Timmis KN, Jahn D. 2015. A periplasmic complex of the nitrite reductase NirS, the chaperone DnaK, and the flagellum protein FliC is essential for flagellum assembly and motility in Pseudomonas aeruginosa. J Bacteriol 197:3066–3075. doi: 10.1128/JB.00415-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Van Alst NE, Wellington M, Clark VL, Haidaris CG, Iglewski BH. 2009. Nitrite reductase NirS is required for type III secretion system expression and virulence in the human monocyte cell line THP-1 by Pseudomonas aeruginosa. Infect Immun 77:4446–4454. doi: 10.1128/IAI.00822-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bowser TE, Bartlett VJ, Grier MC, Verma AK, Warchol T, Levy SB, Alekshun MN. 2007. Novel anti-infection agents: small-molecule inhibitors of bacterial transcription factors. Bioorg Med Chem Lett 17:5652–5655. doi: 10.1016/j.bmcl.2007.07.072. [DOI] [PubMed] [Google Scholar]
- 179.Grier MC, Garrity-Ryan LK, Bartlett VJ, Klausner KA, Donovan PJ, Dudley C, Alekshun MN, Ken Tanaka S, Draper MP, Levy SB, Kim OK. 2010. N-Hydroxybenzimidazole inhibitors of ExsA MAR transcription factor in Pseudomonas aeruginosa: in vitro anti-virulence activity and metabolic stability. Bioorg Med Chem Lett 20:3380–3383. doi: 10.1016/j.bmcl.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 180.Marsden AE, King JM, Spies MA, Kim OK, Yahr TL. 2016. Inhibition of Pseudomonas aeruginosa ExsA DNA-binding activity by N-hydroxybenzimidazoles. Antimicrob Agents Chemother 60:766–776. doi: 10.1128/AAC.02242-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kim OK, Garrity-Ryan LK, Bartlett VJ, Grier MC, Verma AK, Medjanis G, Donatelli JE, Macone AB, Tanaka SK, Levy SB, Alekshun MN. 2009. N-Hydroxybenzimidazole inhibitors of the transcription factor LcrF in Yersinia: novel antivirulence agents. J Med Chem 52:5626–5634. doi: 10.1021/jm9006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang C, Liu X, Wang J, Zhou J, Cui Z, Zhang L-H. 2016. Design and characterization of a polyamine derivative inhibiting the expression of type III secretion system in Pseudomonas aeruginosa. Sci Rep 6:30949. doi: 10.1038/srep30949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang JJ, Wang JJ, Zhang L-H. 14 May 2018. Immunological blocking of spermidine-mediated host-pathogen communication provides effective control against Pseudomonas aeruginosa infection. Microb Biotechnol doi: 10.1111/1751-7915.13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nalca Y, Jänsch L, Bredenbruch F, Geffers R, Buer J, Häussler S. 2006. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob Agents Chemother 50:1680–1688. doi: 10.1128/AAC.50.5.1680-1688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tateda K, Comte R, Pechere JC, Köhler T, Yamaguchi K, Van Delden C. 2001. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother 45:1930–1933. doi: 10.1128/AAC.45.6.1930-1933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gillis RJ, Iglewski BH. 2004. Azithromycin retards Pseudomonas aeruginosa biofilm formation. J Clin Microbiol 42:5842–5845. doi: 10.1128/JCM.42.12.5842-5845.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hoffmann N, Lee B, Hentzer M, Rasmussen TB, Song Z, Johansen HK, Givskov M, Høiby N. 2007. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr(−/−) mice. Antimicrob Agents Chemother 51:3677–3687. doi: 10.1128/AAC.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kai T, Tateda K, Kimura S, Ishii Y, Ito H, Yoshida H, Kimura T, Yamaguchi K. 2009. A low concentration of azithromycin inhibits the mRNA expression of N-acyl homoserine lactone synthesis enzymes, upstream of lasI or rhlI, in Pseudomonas aeruginosa. Pulm Pharmacol Ther 22:483–486. doi: 10.1016/j.pupt.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 189.Pérez-Martínez I, Haas D. 2011. Azithromycin inhibits expression of the GacA-dependent small RNAs RsmY and RsmZ in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:3399–3405. doi: 10.1128/AAC.01801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nicolau DP, Banevicius MA, Nightingale CH, Quintiliani R. 1999. Beneficial effect of adjunctive azithromycin in treatment of mucoid Pseudomonas aeruginosa pneumonia in the murine model. Antimicrob Agents Chemother 43:3033–3035. doi: 10.1128/AAC.43.12.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kobayashi T, Tateda K, Matsumoto T, Miyazaki S, Watanabe A, Nukiwa T, Yamaguchi K. 2002. Macrolide-treated Pseudomonas aeruginosa induces paradoxical host responses in the lungs of mice and a high mortality rate. J Antimicrob Chemother 50:59–66. doi: 10.1093/jac/dkf048. [DOI] [PubMed] [Google Scholar]
- 192.Florescu DF, Murphy PJ, Kalil AC. 2009. Effects of prolonged use of azithromycin in patients with cystic fibrosis: a meta-analysis. Pulm Pharmacol Ther 22:467–472. doi: 10.1016/j.pupt.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 193.Yousef AA, Jaffe A. 2010. The role of azithromycin in patients with cystic fibrosis. Paediatr Respir Rev 11:108–114. doi: 10.1016/j.prrv.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 194.El Boustany P, Gachelin E, Colomban C, Cernoia J, Sudour P, Carsin A, Dubus J-C. 2019. A review of non-cystic fibrosis bronchiectasis in children with a focus on the role of long-term treatment with macrolides. Pediatr Pulmonol 54:487–496. doi: 10.1002/ppul.24252. [DOI] [PubMed] [Google Scholar]
- 195.Yamazaki A, Li J, Zeng Q, Khokhani D, Hutchins WC, Yost AC, Biddle E, Toone EJ, Chen X, Yang C-H. 2012. Derivatives of plant phenolic compound affect the type III secretion system of Pseudomonas aeruginosa via a GacS-GacA two-component signal transduction system. Antimicrob Agents Chemother 56:36–43. doi: 10.1128/AAC.00732-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang Y, Sass A, Van Acker H, Wille J, Verhasselt B, Van Nieuwerburgh F, Kaever V, Crabbé A, Coenye T. 2018. Coumarin reduces virulence and biofilm formation in Pseudomonas aeruginosa by affecting quorum sensing, type III secretion and c-di-GMP levels. Front Microbiol 9:1952. doi: 10.3389/fmicb.2018.01952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yuan M, Chua SL, Liu Y, Drautz-Moses DI, Yam JKH, Aung TT, Beuerman RW, Salido MMS, Schuster SC, Tan C-H, Givskov M, Yang L, Nielsen TE. 2018. Repurposing the anticancer drug cisplatin with the aim of developing novel Pseudomonas aeruginosa infection control agents. Beilstein J Org Chem 14:3059–3069. doi: 10.3762/bjoc.14.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Uusitalo P, Hägglund U, Rhöös E, Scherman Norberg H, Elofsson M, Sundin C. 2017. The salicylidene acylhydrazide INP0341 attenuates Pseudomonas aeruginosa virulence in vitro and in vivo. J Antibiot (Tokyo) 70:937–943. doi: 10.1038/ja.2017.64. [DOI] [PubMed] [Google Scholar]
- 199.Zetterström CE, Hasselgren J, Salin O, Davis RA, Quinn RJ, Sundin C, Elofsson M. 2013. The resveratrol tetramer (-)-hopeaphenol inhibits type III secretion in the Gram-negative pathogens Yersinia pseudotuberculosis and Pseudomonas aeruginosa. PLoS One 8:e81969. doi: 10.1371/journal.pone.0081969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lam H, Schwochert J, Lao Y, Lau T, Lloyd C, Luu J, Kooner O, Morgan J, Lokey S, Auerbuch V. 2017. Synthetic cyclic peptomers as type III secretion system inhibitors. Antimicrob Agents Chemother 61:e00060-17. doi: 10.1128/AAC.00060-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sheremet AB, Zigangirova NA, Zayakin ES, Luyksaar SI, Kapotina LN, Nesterenko LN, Kobets NV, Gintsburg AL. 2018. Small molecule inhibitor of type three secretion system belonging to a class 2,4-disubstituted-4H-[1,3,4]-thiadiazine-5-ones improves survival and decreases bacterial loads in an airway Pseudomonas aeruginosa infection in mice. Biomed Res Int 2018:5810767. doi: 10.1155/2018/5810767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hornef MW, Roggenkamp A, Geiger AM, Hogardt M, Jacobi CA, Heesemann J. 2000. Triggering the ExoS regulon of Pseudomonas aeruginosa: a GFP-reporter analysis of exoenzyme (Exo) S, ExoT and ExoU synthesis. Microb Pathog 29:329–343. doi: 10.1006/mpat.2000.0398. [DOI] [PubMed] [Google Scholar]
- 203.Zeng Q, Laiosa MD, Steeber DA, Biddle EM, Peng Q, Yang C-H. 2012. Cell individuality: the bistable gene expression of the type III secretion system in Dickeya dadantii 3937. Mol Plant Microbe Interact 25:37–47. doi: 10.1094/MPMI-05-11-0105. [DOI] [PubMed] [Google Scholar]
- 204.Dubnau D, Losick R. 2006. Bistability in bacteria. Mol Microbiol 61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 205.Strassman JE, Queller DC, Avise JC, Ayala FJ. 2011. In the light of evolution: volume v: cooperation and conflict. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 206.Czechowska K, McKeithen-Mead S, Al Moussawi K, Kazmierczak BI. 2014. Cheating by type 3 secretion system-negative Pseudomonas aeruginosa during pulmonary infection. Proc Natl Acad Sci U S A 111:7801–7806. doi: 10.1073/pnas.1400782111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sawa T, Yahr TL, Ohara M, Kurahashi K, Gropper MA, Wiener-Kronish JP, Frank DW. 1999. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat Med 5:392–398. doi: 10.1038/7391. [DOI] [PubMed] [Google Scholar]
- 208.Hauser AR, Cobb E, Bodi M, Mariscal D, Vallés J, Engel JN, Rello J. 2002. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit Care Med 30:521–528. doi: 10.1097/00003246-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 209.Ledizet M, Murray TS, Puttagunta S, Slade MD, Quagliarello VJ, Kazmierczak BI. 2012. The ability of virulence factor expression by Pseudomonas aeruginosa to predict clinical disease in hospitalized patients. PLoS One 7:e49578. doi: 10.1371/journal.pone.0049578. [DOI] [PMC free article] [PubMed] [Google Scholar]