Abstract
The recognition that only a small percentage of known human gene products are druggable using traditional modes of non-covalent ligand design, has led to a resurgence in targeted covalent inhibitors. Covalent inhibitors offer advantages over non-covalent inhibitors in engaging otherwise challenging targets. Reactive cysteine residues on proteins are a common target for covalent inhibitors, whereby the high nucleophilicity of the cysteine thiol under physiological conditions provides an ideal anchoring site for electrophilic small molecules. A chemical-proteomic platform, termed isoTOP-ABPP, allows for profiling cysteine reactivity in complex proteomes and is one of many techniques that can aid in two aspects of the covalent-inhibitor development process: (1) to identify novel functional cysteines that lead to modulation of protein function through covalent modification; and, (2) to determine cellular targets and evaluate promiscuity of electrophilic fragments, small molecules, and natural products. Herein, we discuss recent advances in isoTOPABPP and potential applications of this technology in the drug-discovery pipeline.
1. Introduction
Of the 20 proteogenic amino acids, cysteine is unique in its elevated nucleophilicity and redox sensitivity. Despite its low abundance, cysteine is highly conserved at functionally important sites [1,2]. The high nucleophilicity and redox sensitivity of the cysteine thiolate facilitates key roles in several aspects of protein function [3]: (1) active-site nucleophiles in catalysis, or resolving residues in cellular redox buffering systems [4]; (2) protein structure stabilization through disulfide bonds, and metal coordination; and, (3) regulation of protein function through post translational modifications (PTMs), such as oxidation, nitrosation, and glutathionylation [5]. Diverse protein classes, including proteases, oxidoreductases, kinases, and acyltransferases, contain reactive and functional cysteine residues [3]. Thus, the high nucleophilicity and functional importance of cysteine render this amino acid an attractive chemical handle for the development of targeted and selective covalent ligands to modulate the function of diverse proteins.
Covalent inhibitors can be categorized as reversible or irreversible depending on the target residence time. Covalent irreversible inhibitors can be further classified as either residue-specific reagents, affinity labels, or mechanism-based inhibitors, as recently described by Fast et al. [6]. Residue-specific reagents are reactive compounds with minimal noncovalent affinity to a particular binding site. General cysteine alkylating agents, such as iodoacetamide (IAA) and methylmethanthiosulfinate (MMTS), fall into this category. The potency of residue-specific reagents is generally dictated by the inherent reactivity of the electrophile, as protein modification does not rely on formation of an initial non-covalent encounter complex. As a result, these compounds generally lack selectivity and inactivate multiple targets. By contrast, affinity labels typically form an initial non-covalent complex, which increases the effective molarity of the reactive group proximal to the nucleophilic residue, and are generally more selective [7]. Potency of affinity labels is defined by the second order rate constant of inactivation, i.e., kinact/KI, which incorporates the affinity of the initial encounter complex. Optimizing the potency of an affinity label therefore involves maximizing non-covalent interactions and positioning an appropriate electrophile for optimal reaction with the nucleophilic residue on the protein. It is important to note that non-covalent interactions can contribute to the binding of some residue-specific reagents, and in a similar vein, affinity labels can display off-target effects driven solely by reactivity and not non-covalent affinity.
Covalent inhibition as a therapeutic strategy has been shown to demonstrate: (1) a long residence time and duration of action, which has been associated with efficacy [8]; (2) ability to target shallow binding pockets that are recalcitrant to non-covalent ligands [6]; and, (3) potential to circumvent resistance mechanisms, as well as the ability to selectively target disease-associated mutants as has recently been shown for KRAS G12C [9]. It is important to note that non-covalent inhibitors can also achieve the latter, as demonstrated for vermurafenib [10]. Disadvantages of covalent compounds include irreversible inhibition of off-targets leading to toxicity, as well as potential immune-related idiosyncratic adverse drug reactions [11]. Cysteine-targeted inhibitors have been approved or are in various stages of the drug-discovery pipeline including, Tecfidera (dimethyl fumarate (DMF)) [12,13], kinase inhibitors (Afatinib, Ibrutinib, Osimertinib, and Neratinib) [14], and, inhibitors of KRAS G12C [15–17].
A reactive-cysteine profiling method, known as isoTOP-ABPP, has the potential to aid in two key aspects of covalent irreversible inhibitor discovery: (1) to identify novel ligandable cysteines for covalent modulation of protein function; and, (2) to evaluate both on and off-target cysteine engagement so as to minimize risks associated with off-target drug toxicity [18]. This review will discuss recent advances in cysteine-targeted isoTOP-ABPP, followed by an overview of potential applications of isoTOP-ABPP to covalent drug discovery.
2. Methods for the identification of reactive cysteines
Numerous chemical-proteomic methods currently exist for identifying reactive and functional cysteine residues. These include probes specific for cysteine PTMs, including S-sulfenylation [19–21], S-sulfination [22–24], S-nitrosation [25,26], and electrophilic lipid modifications [27], as well as probes selectively targeting cysteine residues on a defined protein class, including kinases [28], and cysteine proteases [29]. Here, we focus on a platform known as isoTOP-ABPP, which identifies a subset of reactive and functional cysteine residues independent of protein class or susceptibility to a particular PTM [1].
2.1. The isoTOP-ABPP platform for reactive-cysteine profiling
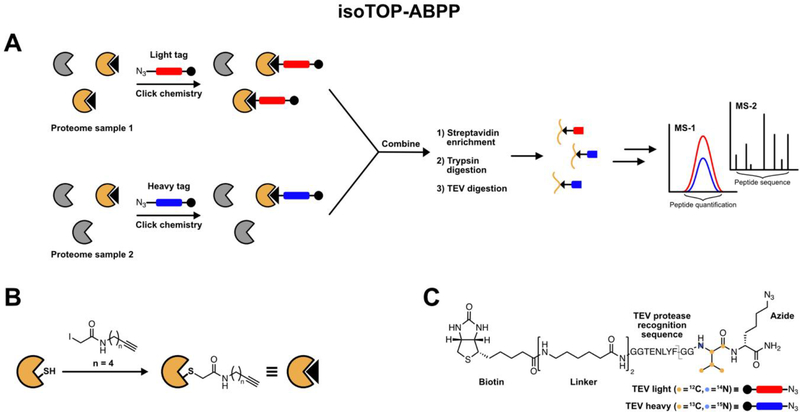
IsoTOP-ABPP is a derivative of activity-based protein profiling (ABPP), a pioneering technology for interrogating protein activity directly in complex biological systems. In general, ABPP probes contain three elements: (1) a reactive warhead for covalently labeling target proteins; (2) a reporter tag for affinity purification or fluorescence detection; and, (3) a linker to minimize steric hindrance between the reporter and reactive groups [30,31]. Early ABPP methods utilized reactive warheads targeting a specific enzyme family, such as the fluorophosphonate probe for the serine hydrolases [32]. In latter iterations, more reactive and promiscuous electrophiles were utilized [33], culminating in the use of an iodoacetamide-alkyne (IA-alkyne) probe for modification of reactive cysteines in the proteome. The isoTOP-ABPP platform couples an IA-alkyne probe with an isotopically tagged cleavable linker, enabling the selective enrichment, release, and mass-spectrometry (MS) relative quantification of IA-labeled peptides from two samples. The isoTOP-ABPP platform involves the following steps: (1) treatment of lysates with IA-alkyne to label reactive cysteines; (2) conjugation of IA-labeled cysteines in control and experimental samples to isotopically differentiated cleavable azide-biotin tags using copper-catalyzed azide-alkyne cycloaddition (CuAAC) [34]; (3) enrichment of IA-labeled proteins on streptavidin beads, followed by on-bead tryptic digestion, and linker cleavage to release IA-labeled peptides; and, (4) analysis of the resulting isotopically heavy and light peptide pairs using LC/LCMS/MS to quantify reactivity differences in two samples using light:heavy isotopic ratios [1] (Figure 1).
Figure 1:
(A) General isoTOP-ABPP workflow. Reactive cysteine residues on two proteome samples are labeled with IA-alkyne, followed by CuAAC with an isotopically heavy or light biotin-azide cleavable linker. The two lysates are combined, biotinylated proteins are enriched on streptavidin-agarose beads, and subjected to an on-bead trypsin digestion. The IA-labeled peptides are released and analyzed by LC/LC-MS/MS. Heavy and light peptide pairs are quantified by their extracted MS1 peaks. (B) IA-alkyne structure and cysteine-labeling scheme. (C) The tobacco-etch virus (TEV) protease cleavable biotin-azide tag for isoTOP-ABPP.
Limitations in the current isoTOP-ABPP platform include the low coverage of the cellular cysteinome. Since low concentrations (100 uM) of IA-alkyne are used for proteome labeling, only 1000–2000 cellular cysteines are identified in a typical analysis. The subset of cysteine residues identified are those that demonstrate high reactivity with the IA electrophile, and have been shown to be enriched in functional cysteines [1]. However, some classes of functional cysteines remain intractable to IA labeling, and are therefore not typically captured in an isoTOP-ABPP analysis. Furthermore, reduced coverage of cysteine residues from low-abundant proteins, particular those localized within subcellular organelles, could limit the potential utility of isoTOP-ABPP for certain applications. Furthermore, cell lysates for isoTOP-ABPP are not typically treated with reducing agents, thereby limiting access to proteins that are highly susceptible to oxidation or aggregation. As discussed below, the development of new cysteine-reactive electrophiles, analytical methods, and cell-based profiling approaches, can serve to overcome some of these current limitations.
2.2. Recent advances in isoTOP-ABPP methods
Since the initial development of the isoTOP-ABPP platform, various iterations to the initial workflow have been reported, including variations to: (1) the cysteine-reactive electrophile; (2) the cleavable biotin-azide tag; and, (3) the mode of heavy isotope incorporation. These recent advances are summarized herein.
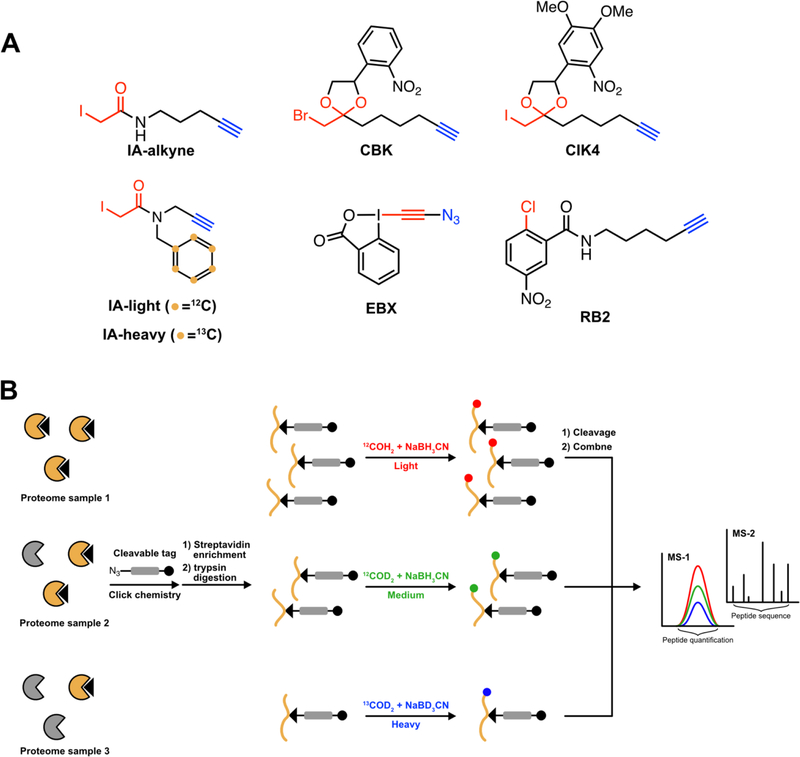
IA-alkyne is widely used as reactive warhead in ABPP [1,35,36], however other electrophiles have also been incorporated into the isoTOP-ABPP workflow. A photocaged bromomethyl ketone (CBK) [37], and iodomethyl ketone (CIK4) [38] were shown to have lower cytotoxicity compared to IA-alkyne and were used to profile reactive cysteines in live cells with high spatial and temporal control. CBK was used to monitor changes in cysteine reactivity in A431 cells in response to the epidermal growth factor (EGF) stimulated release of reactive oxygen species [37]. Alternatives to halo acetamide electrophiles have also been developed, including aryl halides such p-chloronitrobenzene [39], and hypervalent iodine regents, such as ethynyl benziodoxolone (EBX) [40].
The initial isoTOP-ABPP platform utilized isotopically labeled, protease-cleavable biotinazide tags for CuAAC-mediated conjugation to probe-labeled proteins. Due to the advent of a wide-variety of cleavable chemistries, isotopically tagged variants of chemically cleavable [41] and photocleavable [42,43] biotin-azide tags have been developed and shown to be compatible with the isoTOP-ABPP workflow.
Lastly, approaches to incorporate isotopic labels into the isoTOP-ABPP workflow have been explored, including: (1) stable incorporation of amino acids in cell culture (SILAC) to incorporate isotopes into the proteomes under evaluation; (2) isotopically tagged cysteine-reactive probes; (3) isotopically light and heavy biotin-azide linkers; and, (4) post-digest peptide labeling using reductive demethylation (ReDiMe) or isobaric tags (iTRAQ, TMT). Incorporation of isotopic labels into the cysteine-reactive probe was accomplished through synthesis of isotopically differentiated iodoacetamide-alkyne probes containing a 12C6 or 13C6 benzyl moiety, termed IA-alkyne light (IAL) or IA-alkyne heavy (IAH), respectively [44]. IAL and IAH can be obtained through a more facile synthesis, and using less expensive starting materials compared to the isotopic biotin-azide tags [44]. IAL and IAH also support the profiling of reversible cysteine modifications within the same sample in a work flow similar to the OxICAT method [45]. Combining TOP-ABPP with reductive dimethylation (ReDiMe) results in a method called rdTOPABPP [42]. rdTOP-ABPP was shown to be comparable to several commercially available linkers for site of identification, and has the added benefit of supporting triplex quantitative experiments [42]. Multiplexed thiol reactivity profiling (MTRP) cysteine profiling with isobaric tags for relative and absolute quantization (iTRAQ) labeling allows for quantitative site of labeling to be performed with up to eight samples in parallel [43]. Although MTRP reagents are more expensive than those used for rdTOP-ABPP, MTRP requires less sample input when highly multiplexed comparisons are required for an experiment.
The availability of multiple chemical probes, linkers and quantification methods, allow for tailoring the isoTOP-ABPP workflow for each desired application. Further advancements in chemical-probe and linker development, coupled with improvements in MS instrumentation and data-analysis software, will serve to increase the number of cysteine identifications from a complex proteome.
3. Applications of reactive cysteine profiling in drug discovery
To expand the protein targets amenable to covalent inhibition, it is necessary to globally interrogate the proteome for reactive cysteines that upon covalent modification, afford a functional outcome. Furthermore, given the toxicity risks associated with covalent and irreversible modification of off targets, it is imperative to extensively evaluate target engagement of covalent ligands within physiologically relevant proteomes. As described below, isoTOP-ABPP can aid in both these aspects of covalent ligand development.
3.1. Identification of reactive and functional cysteines for covalent targeting
Despite the wealth of genetic information acquired from genome sequencing efforts, only ~2% of predicted human gene products are currently targeted by small-molecule drugs, and only 10–15% percent of human gene products are thought to be druggable [46]. ABPP, and associated technologies, can be powerful tools to identify functionally important and ligandable sites present in different disease states [43]. In particular, isoTOP-ABPP has been utilized to identify reactive cysteines that modulate protein function upon covalent modification. One of the first applications of the isoTOP-ABPP platform was to identify and rank cysteines by reactivity, thereby demonstrating that cysteine reactivity is highly predictive of functionality [1]. Although active-site cysteines on proteases and oxidoreductases are well annotated, allosteric regulatory cysteines within proteomes are poorly characterized. IsoTOP-ABPP has been applied to identify non-active-site cysteines that are susceptible to PTMs, including oxidation [47], S-nitrosation [48], modification by lipid derived electrophiles [36], and zinc chelation [35]. Biochemical characterization of these non-catalytic cysteine residues has shown that modification results in modulation of protein function. The ability of these cysteines to regulate protein function render them putative target sites for designing covalent inhibitors. However, since modification with a small molecule is not a direct mimic of a specific cysteine PTM, covalent binding may not necessarily phenocopy the function of the endogenous PTM.
IsoTOP-ABPP can also identify cysteine residues whose reactivity is elevated under disease conditions. For example, Bar-Peled et al. applied isoTOP-ABPP to identify druggable cysteines in KEAP1-mutant non-small-cell lung cancers [49], and Martell et al. applied isoTOPABPP to identify changes in cysteine reactivity associated with impaired insulin signaling in C. elegans. These studies lay the groundwork for future applications of isoTOP-ABPP to compare cysteine-reactivity changes in healthy and diseased systems and aid in identifying upregulated cysteine residues that could be explored as potential targets for covalent inhibitor development.
3.2. Screening the potency and selectivity of covalent ligands
Covalent ligands of various modalities have been reported, including: (1) low-molecular weight reactive fragments; (2) drug-like small molecules with embedded electrophiles; and, (3) structurally complex electrophilic natural products. IsoTOP-ABPP provides a potential tool to evaluate the protein targets of covalent ligands.
3.2.1. Covalent fragment-based screening
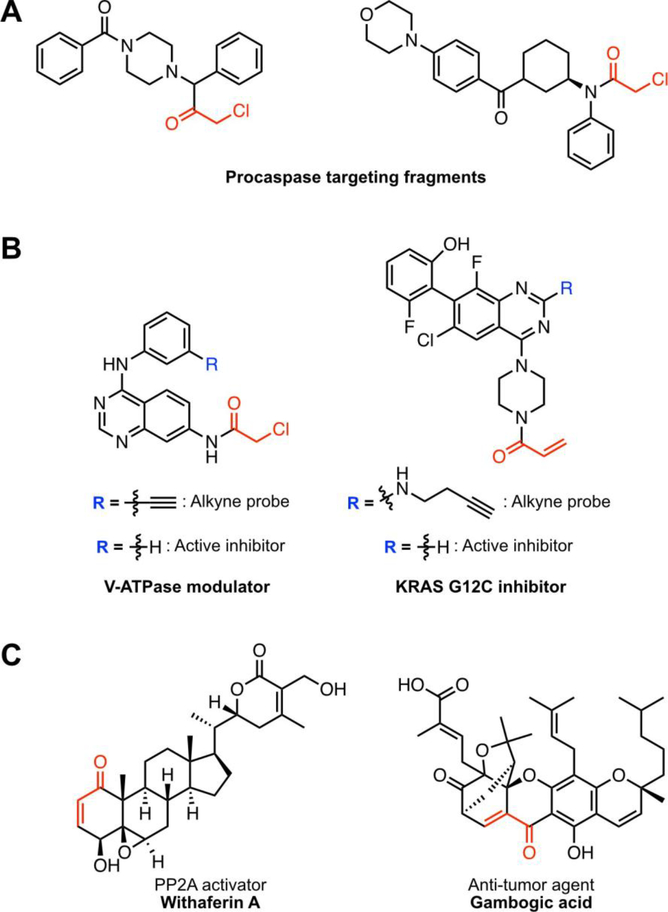
Fragment-based ligand discovery (FBLD) utilizes low-molecular weight fragments that generally conform to the rule of three [50]; molecular mass > 300 Da, up to three hydrogen bond donors and acceptors, and calculated logP ≤ 3. Early covalent FBLD screens used covalent tethering approaches in which a target protein containing a reactive cysteine reacts with a library of disulfide-containing fragments [51]. This disulfide-tethering approach was recently used to identify covalent ligands for the oncogenic KRAS G12C mutant, which allosterically attenuates GTP affinity [15]. In a more global and untargeted approach, Backus et al. used isoTOP-ABPP to assess the proteome reactivity of a 52-member fragment library containing chloroacetamide and acrylamide electrophiles [52]. The analysis was performed in a competitive format, whereby a proteome is treated with a covalent fragment prior to treatment with IA-alkyne, and a decrease in IA-alkyne labeling is indicative in ligand binding. Of the 700 ligandable cysteines identified, 535 were found on proteins which had no known ligands in DrugBank, representing classes of proteins classically considered to be undruggable, including transcription factors, and adaptor proteins [52]. Among the ligands screened were two fragments that covalently modified pro-caspases [52] (Figure 3). Although the identified fragments are typically promiscuous and show low affinity, further chemical elaboration has the potential to yield potent and selective small molecules for these traditionally undruggable targets.
Figure 3:
Covalent ligand discoveries aided by isoTOP-ABPP (A) covalent fragments targeting procaspases (B) drug-like small-molecules targeting V-ATPase and KRAS G12C, and (C) electrophilic natural products. Electrophiles are highlighted in red.
3.2.2. Drug-like small-molecule screening
Competitive isoTOP-ABPP has also been applied to drug-like electrophilic compounds. Dimethyl fumarate (DMF) is an electrophilic, immunomodulatory drug believed to function by covalently modifying cysteine residues. Blewett et al. found that DMF covalently modified conserved cysteines in the non-catalytic domain of protein kinase Cθ (PKCθ) and disrupted PKCθ-CD28 association during T-cell activation [12]. T-cells expressing a cysteine mutant of PKCθ showed impaired activation, however, DMF treatment of these mutant-expressing cells showed a further reduction in activation, suggesting that DMF exhibits polypharmacology, and likely acts by concurrently targeting multiple cellular cysteines. Similarly, isoTOP-ABPP was used to demonstrate the high selectivity of a chloroacetamide-bearing quinazolinone for the vacuolar H+ ATPase (V-ATPase) [53]. In a variation of competitive isoTOP-ABPP, a desthiobiotin-linked IA probe was used to determine target engagement of a quinazoline-based KRAS G12C inhibitor [16] (Figure 3). Lastly, Whitby et al. used isoTOP-ABPP to investigate proteome labeling by reactive metabolites generated in vivo upon treatment with the hepatotoxic drugs, acetaminophen, troglitazone, clozapine, and tienilic acid [54]. These studies demonstrate the utility of isoTOPABPP to investigate both target occupancy and promiscuity of drug-like small molecules.
3.2.3. Electrophilic natural-product screening
Natural products (NPs) exhibit structurally complex scaffolds that often demonstrate exquisite target selectivity [55], and often contain cysteine-targeting electrophilic motifs, including Michael acceptors and epoxides [43]. Typically, an alkyne variant of a covalent ligand can be used to assess target occupancy, however, the complexity of NP total synthesis and limited information of structure activity relationships, complicate the use of alkyne-tagged natural product analogs. Competitive isoTOP-ABPP, whereby proteome treatment with the unmodified NP is subsequently followed by addition of IA-alkyne, has been applied to identify putative protein targets of several natural products including licochalcone A, celastrol, and curcumin [40,56,57] Grossman et al. assessed the proteome reactivity of withaferin A, an electrophilic natural produce known to exhibit cancer anti-proliferative activity [58]. Withaferin A was shown to activate the tumor suppressor phosphatase PP2A, and a covalent fragment screen was used to identify a more synthetically tractable small molecule that recapitulates the anti-proliferative activity of withaferin A (Figure 3) [58]. Similarly, MTRP was used to map sites of labeling of several electrophilic natural products, including gambogic acid, diverse α,β-unsaturated γ-lactones, and acetylbritannilactone [43].
4. Conclusions
The unique properties of cysteine, including nucleophilicy, redox susceptibility and polarizability, facilitates a central role for this amino acid in protein structure, function and regulation. Targeting disease-relevant cysteines can be a fruitful strategy to overcome some of the limitations of non-covalent drugs, including in the targeting of classically undruggable sites. IsoTOP-ABPP is one of many chemical-proteomic platforms that can aid in identifying novel ligandable and functional cysteines, revealing reactive cysteines upregulated in disease, and elucidating the selectivity of electrophilic fragments, drug-like molecules, and natural products. Limitations in current isoTOP-ABPP approaches include the following: (1) only a subset of cellular cysteines are identified, thereby limiting the ability to assess compound selectivity across the entire cellular cysteinome; and, (2) typically cell lysates are treated under non-reducing conditions, which could result in oxidation or aggregation of a subset of proteins with highly reactive cysteines. Therefore isoTOP-ABPP techniques need to continue to mature in order to provide important exploratory tools for the drug-development process.
Figure 2:
Recent modifications to the isoTOP-ABPP platform (A) New cysteine reactive chemical probes. Electrophiles are highlighted in red, reporting handles in blue, and isotopic labels in orange. (B) Triplex rdTOP-ABPP workflow.
Acknowledgments
This work was financially supported by NIH grants 1R01GM117004 and 1R01GM118431-01A1 to E. W. We thank Professor Paul Thompson (UMass Medical School) for comments and critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing declared.
References
Papers of particular interest, published within the period of review have been highlighted as: special interest (*) or outstanding interest (**)
- 1.**.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF: Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468:790–795.First study to use isoTOP-ABPP to quantify cysteine reactivity in a complex proteome. This study demonstrated that cysteine reactivity is highly correlated with functionality.
- 2.Marino SM, Gladyshev VN: Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J Mol Biol 2010, 404:902–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giles NM, Giles GI, Jacob C: Multiple roles of cysteine in biocatalysis. Biochem Biophys Res Commun 2003, 300:1–4. [DOI] [PubMed] [Google Scholar]
- 4.Truong TH, Carroll KS: Redox regulation of epidermal growth factor receptor signaling through cysteine oxidation. Biochemistry 2012, 51:9954–9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Carroll KS, Liebler DC: The Expanding Landscape of the Thiol Redox Proteome. Mol Cell Proteomics 2016, 15:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. *.Tuley A, Fast W: The Taxonomy of Covalent Inhibitors. Biochemistry 2018, 57:3326–3337.Outlines the taxonomy of covalent inhibitors based off their mode of action. Examples of bioactive compounds in each category are also provided.
- 7.Schwartz PA, Kuzmic P, Solowiej J, Bergqvist S, Bolanos B, Almaden C, Nagata A, Ryan K, Feng J, Dalvie D, et al. : Covalent EGFR inhibitor analysis reveals importance of reversible interactions to potency and mechanisms of drug resistance. Proc Natl Acad Sci U S A 2014, 111:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copeland RA: The drug-target residence time model: a 10-year retrospective. Nat Rev Drug Discov 2016, 15:87–95. [DOI] [PubMed] [Google Scholar]
- 9. *.Visscher M, Arkin MR, Dansen TB: Covalent targeting of acquired cysteines in cancer. Curr Opin Chem Biol 2016, 30:61–67.Surveys the most common mutations which result in acquired cysteines in cancer. In addition to giving relevant examples of cysteine targeted therapeutics, the authors describe how oncogenic acquired cysteine mutations can be used to personalize cancer treatment.
- 10.Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, Hirth P: Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov 2012, 11:873–886. [DOI] [PubMed] [Google Scholar]
- 11.Johnson DS, Weerapana E, Cravatt BF: Strategies for discovering and derisking covalent, irreversible enzyme inhibitors. Future Med Chem 2010, 2:949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.* Blewett MM, Xie J, Zaro BW, Backus KM, Altman A, Teijaro JR, Cravatt BF: Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci Signal 2016, 9:rs10.Authors used competitive isoTPP-ABPP to profile the DMF sensitivity of 2400 cysteine residues in human T cells and shed light on the previously unknown mechanism of action of DMF.
- 13.Kornberg MD, Bhargava P, Kim PM, Putluri V, Snowman AM, Putluri N, Calabresi PA, Snyder SH: Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 2018, 360:449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. *.Chaikuad A, Koch P, Laufer SA, Knapp S: The Cysteinome of Protein Kinases as a Target in Drug Development. Angew Chem Int Ed Engl 2018, 57:4372–4385.Review which outlines the challenges and opportunities in targeting cystine with covalent kinase inhibitors. The authors discuss strategies and considerations when optimizing both non-covalent and covalent interactions.
- 15. *.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM: K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013, 503:548–551.Authors use a covalent fragment screen to identify compounds which bind to the oncogenic K-Ras G12C mutation. This study is the first example of a compound designed to covalently target an acquired cysteine mutation.
- 16.Wijeratne A, Xiao J, Reutter C, Furness KW, Leon R, Zia-Ebrahimi M, Cavitt RN, Strelow JM, Van Horn RD, Peng SB, et al. : Chemical Proteomic Characterization of a Covalent KRASG12C Inhibitor. ACS Med Chem Lett 2018, 9:557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, et al. : Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018, 172:578–589 e517. [DOI] [PubMed] [Google Scholar]
- 18.Lanning BR, Whitby LR, Dix MM, Douhan J, Gilbert AM, Hett EC, Johnson TO, Joslyn C, Kath JC, Niessen S, et al. : A road map to evaluate the proteome-wide selectivity of covalent kinase inhibitors. Nat Chem Biol 2014, 10:760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS: Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol 2011, 8:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta V, Carroll KS: Rational design of reversible and irreversible cysteine sulfenic acid-targeted linear C-nucleophiles. Chem Commun (Camb) 2016, 52:3414–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta V, Yang J, Liebler DC, Carroll KS: Diverse Redoxome Reactivity Profiles of Carbon Nucleophiles. J Am Chem Soc 2017, 139:5588–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo YH, Konopko AM, Borotto NB, Majmudar JD, Haynes SE, Martin BR: Profiling Protein S-Sulfination with Maleimide-Linked Probes. Chembiochem 2017, 18:2028–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akter S, Fu L, Jung Y, Conte ML, Lawson JR, Lowther WT, Sun R, Liu K, Yang J, Carroll KS: Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat Chem Biol 2018, 14:995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alcock LJ, Farrell KD, Akol MT, Jones GH, Tierney MM, Kramer HB, Pukala TL, Bernardes GJL, Perkins MV, Chalker JM: Norbornene probes for the study of cysteine oxidation. Tetrahedron 2018, 74:1220–1228. [Google Scholar]
- 25.Bechtold E, Reisz JA, Klomsiri C, Tsang AW, Wright MW, Poole LB, Furdui CM, King SB: Water-Soluble Triarylphosphines as Biomarkers for Protein S-Nitrosation. ACS Chemical Biology 2010, 5:405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Xian M: Fast reductive ligation of S-nitrosothiols. Angew Chem Int Ed Engl 2008, 47:6598–6601. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Cong Y, Quan B, Lan T, Chu X, Ye Z, Hou X, Wang C: Chemoproteomic profiling of targets of lipid-derived electrophiles by bioorthogonal aminooxy probe. Redox Biol 2017, 12:712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Browne CM, Jiang B, Ficarro SB, Doctor ZM, Johnson JL, Card JD, Sivakumaren SC, Alexander WM, Yaron T, Murphy CJ, et al. : A Chemoproteomic Strategy for Direct and Proteome-wide Covalent Inhibitor Target-site Identification. J Am Chem Soc 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hewings DS, Flygare JA, Wertz IE, Bogyo M: Activity-based probes for the multicatalytic proteasome. FEBS J 2017, 284:1540–1554. [DOI] [PubMed] [Google Scholar]
- 30.Weerapana E, Speers AE, Cravatt BF: Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat Protoc 2007, 2:1414–1425. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Wong YK, Wang J, Zhang J, Lee YM, Shen HM, Lin Q, Hua ZC: Target identification with quantitative activity based protein profiling (ABPP). Proteomics 2017, 17. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Patricelli MP, Cravatt BF: Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A 1999, 96:14694–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weerapana E, Simon GM, Cravatt BF: Disparate proteome reactivity profiles of carbon electrophiles. Nat Chem Biol 2008, 4:405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolb HC, Finn MG, Sharpless KB: Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl 2001, 40:2004–2021. [DOI] [PubMed] [Google Scholar]
- 35.Pace NJ, Weerapana E: A competitive chemical-proteomic platform to identify zinc-binding cysteines. ACS Chem Biol 2014, 9:258–265. [DOI] [PubMed] [Google Scholar]
- 36.Wang C, Weerapana E, Blewett MM, Cravatt BF: A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat Methods 2014, 11:79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. *.Abo M, Weerapana E: A Caged Electrophilic Probe for Global Analysis of Cysteine Reactivity in Living Cells. J Am Chem Soc 2015, 137:7087–7090.Use of a photocaged electrophilic probe to monitor transient cysteine oxidation events in live cells while minimizing cytotoxicity.
- 38.Abo M, Bak DW, Weerapana E: Optimization of Caged Electrophiles for Improved Monitoring of Cysteine Reactivity in Living Cells. Chembiochem 2017, 18:81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon DA, Banerjee R, Webster ER, Bak DW, Wang C, Weerapana E: Investigating the proteome reactivity and selectivity of aryl halides. J Am Chem Soc 2014, 136:3330–3333. [DOI] [PubMed] [Google Scholar]
- 40.Abegg D, Frei R, Cerato L, Prasad Hari D, Wang C, Waser J, Adibekian A: Proteome-Wide Profiling of Targets of Cysteine reactive Small Molecules by Using Ethynyl Benziodoxolone Reagents. Angew Chem Int Ed Engl 2015, 54:10852–10857. [DOI] [PubMed] [Google Scholar]
- 41.Qian Y, Weerapana E: A Quantitative Mass-Spectrometry Platform to Monitor Changes in Cysteine Reactivity. Methods Mol Biol 2017, 1491:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang F, Gao J, Che J, Jia G, Wang C: A Dimethyl-Labeling-Based Strategy for Site-Specifically Quantitative Chemical Proteomics. Anal Chem 2018, 90:9576–9582. [DOI] [PubMed] [Google Scholar]
- 43.Tian C, Sun R, Liu K, Fu L, Liu X, Zhou W, Yang Y, Yang J : Multiplexed Thiol Reactivity Profiling for Target Discovery of Electrophilic Natural Products. Cell Chem Biol 2017, 24:1416–1427 e1415. [DOI] [PubMed] [Google Scholar]
- 44.Abo M, Li C, Weerapana E: Isotopically-Labeled Iodoacetamide-Alkyne Probes for Quantitative Cysteine-Reactivity Profiling. Mol Pharm 2018, 15:743–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U: Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A 2008, 105:8197–8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. *.Roberts AM, Ward CC, Nomura DK: Activity-based protein profiling for mapping and pharmacologically interrogating proteome-wide ligandable hotspots. Curr Opin Biotechnol 2017, 43:25–33.A comprehensive review discussing proteomic approaches to identify ligandable cysteine residues. Also gives examples of small molecules whose development was aided by ABPP.
- 47.Deng X, Weerapana E, Ulanovskaya O, Sun F, Liang H, Ji Q, Ye Y, Fu Y, Zhou L, Li J, et al. : Proteome-wide quantification and characterization of oxidation-sensitive cysteines in pathogenic bacteria. Cell Host Microbe 2013, 13:358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y, Wynia-Smith SL, Couvertier SM, Kalous KS, Marletta MA, Smith BC, Weerapana E: Chemoproteomic Strategy to Quantitatively Monitor Transnitrosation Uncovers Functionally Relevant S-Nitrosation Sites on Cathepsin D and HADH2. Cell Chem Biol 2016, 23:727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. *.Bar-Peled L, Kemper EK, Suciu RM, Vinogradova EV, Backus KM, Horning BD, Paul TA, Ichu TA, Svensson RU, Olucha, et al. : Chemical Proteomics Identifies Druggable Vulnerabilities in a Genetically Defined Cancer. Cell 2017, 171:696–709 e623.Authors combined chemical genetics with proteomic analysis to identify druggable cysteines in proteins regulated by NRF2 which contribute to KEAP1 mutant driven cancer cell growth.
- 50.Congreve M, Carr R, Murray C, Jhoti H: A ‘Rule of Three’ for fragment-based lead discovery? Drug Discovery Today 2003, 8:876–877. [DOI] [PubMed] [Google Scholar]
- 51.Erlanson DA, Braisted AC, Raphael DR, Randal M, Stroud RM, Gordon EM, Wells JA: Site-directed ligand discovery. Proc Natl Acad Sci U S A 2000, 97:9367–9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. **.Backus KM, Correia BE, Lum KM, Forli S, Horning BD, Gonzalez-Paez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, et al. : Proteome-wide covalent ligand discovery in native biological systems. Nature 2016, 534:570–574.Authors used a competitive ABPP approach to identify cysteine residues ligandable by covalent fragments against a complex proteome background and demonstrated that many highly reactive cysteines are present on traditionally undruggable targets.
- 53.Chen YC, Backus KM, Merkulova M, Yang C, Brown D, Cravatt BF, Zhang C: Covalent Modulators of the Vacuolar ATPase. J Am Chem Soc 2017, 139:639–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitby LR, Obach RS, Simon GM, Hayward MM, Cravatt BF: Quantitative Chemical Proteomic Profiling of the in Vivo Targets of Reactive Drug Metabolites. ACS Chem Biol 2017, 12:2040–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodrigues T, Reker D, Schneider P, Schneider G: Counting on natural products for drug design. Nat Chem 2016, 8:531–541. [DOI] [PubMed] [Google Scholar]
- 56.Roberts AM, Miyamoto DK, Huffman TR, Bateman LA, Ives AN, Akopian D, Heslin MJ, Contreras CM, Rape M, Skibola CF, et al. : Chemoproteomic Screening of Covalent Ligands Reveals UBA5 As a Novel Pancreatic Cancer Target. ACS Chem Biol 2017, 12:899–904. [DOI] [PubMed] [Google Scholar]
- 57.Zhou Y, Li W, Wang M, Zhang X, Zhang H, Tong X, Xiao Y: Competitive profiling of celastrol targets in human cervical cancer HeLa cells via quantitative chemical proteomics. Mol Biosyst 2016, 13:83–91. [DOI] [PubMed] [Google Scholar]
- 58.Grossman EA, Ward CC, Spradlin JN, Bateman LA, Huffman TR, Miyamoto DK, Kleinman JI, Nomura DK: Covalent Ligand Discovery against Druggable Hotspots Targeted by Anti-cancer Natural Products. Cell Chem Biol 2017, 24:1368–1376 e1364. [DOI] [PMC free article] [PubMed] [Google Scholar]