Abstract
Current paradigms of cell intrinsic immunity to RNA viruses center on virus-triggered inducible antiviral responses initiated by RIG-I-like receptors (RLRs) or Toll-like receptors (TLRs) that sense pathogen-associated molecular patterns, and signal downstream through interferon regulatory factors (IRFs), transcription factors that induce synthesis of type I and type III interferons (IFNs)1. RNA viruses have evolved sophisticated strategies to disrupt these signaling pathways and evade elimination by cells, attesting to their importance2. Less attention has been paid how IRFs maintain basal levels of protection against viruses. Here, we depleted antiviral factors linked to RLR and TLR signaling in order to map critical host pathways restricting positive-strand RNA virus replication in immortalized hepatocytes and identified an unexpected role for IRF1. We show constitutively expressed IRF1 acts independently of MAVS, IRF3, and STAT1-dependent signaling to provide intrinsic antiviral protection in actinomycin D-treated cells. IRF1 localizes to the nucleus, where it maintains basal transcription of a suite of antiviral genes that protect against multiple pathogenic RNA viruses, including hepatitis A and C viruses (HAV and HCV), dengue virus (DENV) and Zika virus (ZIKV). Our findings reveal an unappreciated layer of hepatocyte intrinsic immunity to these positive-strand RNA viruses, and identify previously unrecognized antiviral effector genes.
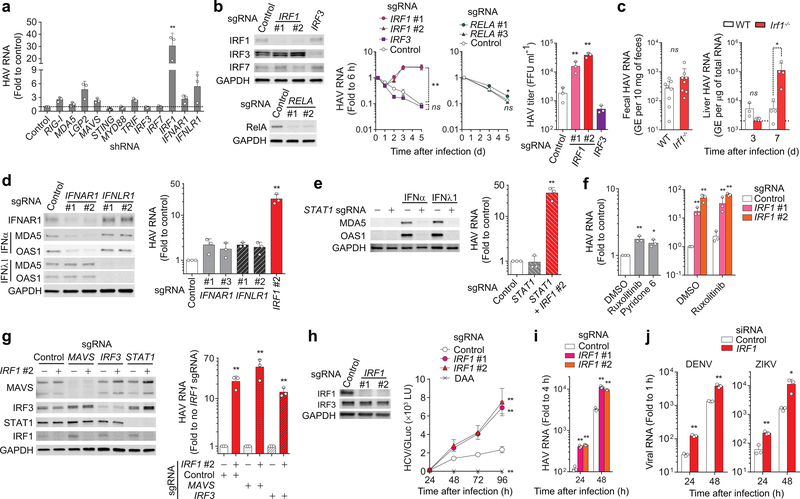
To map host antiviral pathways, we studied immortalized adult human hepatocytes. PH5CH8 cells express RLRs and TLRs similar to hepatocytes in vivo, and induce strong IFN and proinflammatory cytokine responses when infected with RNA viruses3–5. We depleted antiviral factors linked to RLRs and TLRs by transducing cells with lentiviral vectors expressing short-hairpin RNAs (shRNAs) (Supplementary Table 1), and assessed the impact on replication of HAV, an hepatotropic human picornavirus that causes acute inflammatory liver disease6. Surprisingly, depleting RLRs (RIG-I, MDA5, LGP2), signaling adaptors (MAVS, STING, MYD88, TRIF), and transcription factors (IRF3, IRF7) involved in inducible IFN responses, as well as IFN receptors (IFNAR1, IFNLR1), resulted in only small increases in HAV replication, whereas depleting IRF1 enhanced HAV RNA levels 30-fold (Fig. 1a and Supplementary Fig. 1a-f). We confirmed the marked increase in HAV replication resulting from IRF1 depletion in CRISPR/Cas9-generated PH5CH8 knockout cell pools transduced with different single-guide RNAs (sgRNAs) (IRF1#1 and IRF1#2) (Fig. 1b). In contrast, knocking out IRF3, or depleting both IRF3 and IRF7, had little effect on replication (Fig. 1b and Supplementary Fig. 2a). Thus, IRF1 is significantly more active than IRF3 in restricting HAV infection in these cells. Genetically-deficient Irf1−/− mice also shed more HAV in feces and had more viral RNA in the liver than either Irf3−/− or wild-type mice 7d after virus challenge, although HAV did not establish persistent infection as it does in Ifnar1−/− mice7 (Fig. 1c and Supplementary Fig. 2b). IRF1 may promote IFN-γ signaling, class I MHC expression and T cell activation in vivo8,9. Such effects are unlikely to cause enhancement of HAV replication in Irf1−/− mice, however, as previous studies show that neither IFN-γ receptor knockout nor the absence of functional T cells renders C57BL/6 mice permissive for infection7. Taken collectively, these results suggest IRF1 restricts viral replication in hepatocytes.
Figure 1. IRF1 restricts RNA virus infections in hepatocytes.
(a) Intracellular HAV RNA on 5 d.p.i. in PH5CH8 cells transduced with lentivirus expressing shRNAs targeting different genes. **p < 0.001 vs. control [two-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test]. (b) Kinetics of HAV RNA replication over 5 days in PH5CH8 cells expressing IRF1 vs. IRF3 vs. RELA sgRNAs. *p<0.05, **p<0.01 vs. control (two-way ANOVA with Dunnett’s multiple comparisons test). Immunoblots of IRF1, IRF3, IRF7, and RelA in the knockout cells are shown on left. Viral titers on 5 days p.i. are shown on the right. **p<0.01 vs. control (one-way ANOVA with Dunnett’s multiple comparisons test). (c) Fecal HAV shedding on days 5 and 7 postinoculation (left, data are pooled from 2 different time points), and intrahepatic HAV RNA on days 3 and 7 (right, each symbol = one animal) in wildtype (WT) vs. Irf1−/− C57BL/6 mice. *p<0.05 vs. WT (two-sided unpaired Mann-Whitney test). (d) HAV RNA on 5 d.p.i. in PH5CH8 cells expressing IFNAR1 or IFNLR1 sgRNAs vs. IRF1 sgRNA. Immunoblots of IFNAR1 and ISGs (MDA5 and OAS1) induced either by recombinant IFN-α (100 U ml−1 for 24 h) or IFN-λ (10 ng ml−1) in these knockout cells are shown on left. **p<0.01 vs. control (one-way ANOVA with Dunnett’s multiple comparisons test). (e) HAV RNA on 5 d.p.i. in PH5CH8 cells expressing STAT1 sgRNA and both STAT1 and IRF1 sgRNAs (right). **p<0.01 vs. control (one-way ANOVA with Dunnett’s multiple comparisons test). Immunoblots showing the absence of ISG expression in response to type I and type III IFNs (left). (f) HAV RNA on 5 d.p.i. in PH5CH8 cells in continued presence of JAK inhibitors, 3 μM ruxolitinib or 0.3 μM pyridone 6 (left). *p<0.05, **p<0.01 vs. control (one-way ANOVA with Dunnett’s multiple comparisons test or two-sided t-test). Knocking out IRF1 enhanced HAV replication in the presence of ruxolitinib (right). *p<0.05, **p<0.01 vs. control (two-sided unpaired t-test). (g) Effect of double-knockout of IRF1 in the absence of MAVS or IRF3 expression on HAV replication. Relative HAV RNA levels on 5 d.p.i., normalized to those without IRF1 sgRNA were set to 1 (right). Immunoblots are shown on left. **p<0.01 vs. control (two-sided unpaired t-test). (h) Immunoblots of IRF1 in control and IRF1 knockout Huh-7.5 cells (left). GLuc secreted from Huh-7.5 cells infected with JFH1-QL/GLuc virus (103 FFU ml−1) over ensuing 96 h (right). **p<0.01 vs. control (two-way ANOVA with Dunnett’s multiple comparisons test). (i) HAV RNA levels in IRF1 sgRNA-expressing vs. control Huh-7.5 cells infected at an m.o.i. of 1 over ensuing 48 h. **p < 0.01 vs. control (two-sided unpaired t-test). (j) DENV and ZIKV RNA levels in IRF1 vs. control siRNA-transfected Huh-7.5 cells infected at an m.o.i. of 1 over ensuing 48 h. **p<0.01 vs. control (two-sided unpaired t-test). Data are means ± s.d. from 3 independent experiments (a,b,d-h, j) or from 3 technical replicates representative of 2 independent experiments (c,i). The precise p-values are shown in Supplementary Table 9.
IRF1 is known to induce type I IFN gene expression10, mediate type III IFN expression downstream of peroxisomal MAVS11, and exert broad antiviral effector activity12. However, knocking out receptors for type I or III IFN (IFNAR1 and IFNLR1) enhanced HAV infection less than 3-fold (Fig. 1d). Moreover, knocking out STAT1, thereby abolishing both type I and type III IFN signaling (Fig. 1e, left panel), caused no increase in HAV replication, whereas additionally knocking out IRF1 increased replication over 20-fold (Fig. 1e, right panel). Similarly, pharmacological inhibition of Janus kinases (JAK1/2), key components of IFN-induced JAK/STAT signaling, enhanced viral replication only 2-fold and failed to blunt increases in replication caused by IRF1 knockout (Fig. 1f). Collectively, these data show IRF1 restricts HAV replication independently of IFN signaling.
IRF1 expression is regulated in an NF-κB-dependent manner by RLR-mediated activation of the adaptor protein MAVS5,11,13. However, knocking out RELA did not enhance HAV replication (Fig. 1b), nor did prior MAVS (or IRF3) knockout lessen increases resulting from IRF1 knockout (Fig. 1g). Moreover, IRF1 knockout did not diminish Sendai virus (SeV)-induced IFN-β promoter activity or IFN-stimulated gene (ISG) expression, whereas these RIG-I-dependent responses were impaired in knockout IRF3-sgRNA cells (Supplementary Fig. 2c). Similarly, antiviral responses triggered by MAVS overexpression required IRF3 but not IRF1 (Supplementary Fig. 2d). Thus, IRF1 restricts HAV infection independently of RELA and MAVS signaling.
Whereas only IRF1 knockout enhanced infection with cell-free HAV (Fig. 1b), knocking out either IRF1 or IRF3 enhanced replication of electroporated synthetic HAV RNA (Supplementary Fig. 2e). IRF1 and IRF3 knockout resulted in equivalent and additive increases up to 3 days post-tranfection, but IRF1 knockout (both IRF1-sgRNA#1 and IRF1-sgRNA#2) had a greater effect at 5 days when de novo infection with newly replicated virus accounted for continuing increases in RNA abundance. Electroporated RNA, but not cell-free virus infection, also stimulated IRF3-dependent ISG expression (Supplementary Fig. 2f), likely reflecting greater immediate cytoplasmic delivery of viral RNA. Collectively, these results show IRF1 and IRF3 act non-redundantly, with IRF1 mediating protection against an early, post-entry step in HAV infection that does not elicit detectable IRF3 responses.
IRF1 depletion also promoted replication of HAV, as well as HCV, DENV, and ZIKV, all members of the Flaviviridae family in Huh-7.5 cells, a human hepatoma cell line deficient in RIG-I and TLR3 signaling3,14 (Fig. 1h-j and Supplementary Fig. 3a). The absence of IFN responses in Huh-7.5 cells is supported by a lack of enhancement of HCV, HAV, or DENV replication following ruxolitinib treatment (Supplementary Fig. 3b). IRF1 thus restricts replication of multiple pathogenic positive-strand RNA viruses in hepatocyte-derived cells. IRF1 depletion enhanced replication of transfected HCV RNA more than depleting IRF3, RLRs, MAVS, or IFN receptors in PH5CH8 cells (Supplementary Fig. 4a-e), and as with HAV, its impact on HCV was not reduced by pharmacologic blockade of IFN signaling (Supplementary Fig. 4f).
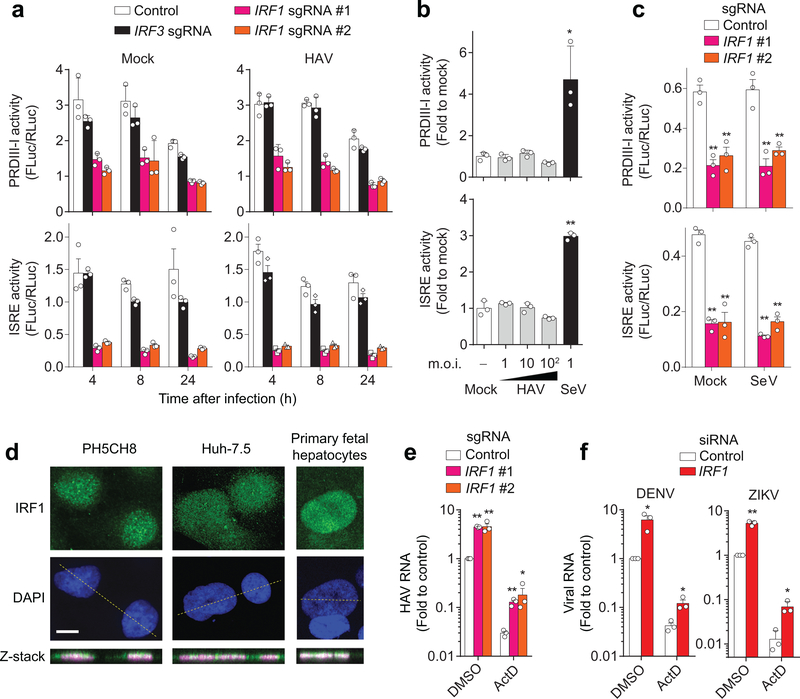
IRF1 protein abundance was not increased in HAV-infected PH5CH8 cells (Supplementary Fig. 5a), and high multiplicity infection failed to stimulate IRF1-responsive PRDIII-I and ISRE promoter elements (Fig. 2a, b)15–17. However, knocking out IRF1 markedly reduced the basal activities of these promoters in both PH5CH8 and Huh-7.5 cells (Fig. 2a,c), whereas ruxolitinib inhibition of JAK/STAT signaling did not (Supplementary Fig. 5b). Collectively, these results suggest that basal expression of IRF1 provides intrinsic antiviral protection by maintaining constitutive transcription of antiviral genes, which is consistent with the nuclear localization of IRF1 in uninfected PH5CH8 and Huh-7.5 cells, and primary human fetal hepatocytes (Fig. 2d and Supplementary Fig. 5c). Further supporting this hypothesis, IRF1 depletion boosted replication of HAV, DENV, and ZIKV in the absence of cellular transcription in actinomycin D-treated Huh-7.5 cells (Fig. 2e, f and Supplementary Fig. 5d).
Figure 2. IRF1 constitutively activates basal transcription of PRDIII-I- and ISRE-dependent antiviral genes.
(a) Dual luciferase reporter analysis of 4×PRDIII-I-Luc (top panels) and ISRE-Luc (bottom panels) activities in mock- (left panels) and HAV-infected (right panels) PH5CH8 cells. Promoter activities in IRF1-sgRNA (#1, #2) vs. control or IRF3 sgRNA-expressing cells differed significantly (p < 0.01, two-way ANOVA with Dunnett’s multiple comparisons test). (b) Dose-response analysis of PRDIII-I (top) and ISRE (bottom) activities in HAV and SeV-infected, wildtype PH5CH8 cells at the indicated m.o.i. *p<0.05, **p<0.01 vs. mock (one-way ANOVA with Dunnett’s multiple comparisons test). (c) Dual luciferase reporter analysis of 4×PRDIII-I-Luc (top) and ISRE-Luc (bottom) activities in mock-infected Huh-7.5 cells. Note that SeV does not activate these promoters in Huh-7.5 cells. **p<0.01 vs. control (two-way ANOVA with Dunnett’s multiple comparisons test). (d) Nuclear localization of IRF1 in two different hepatic cell lines and primary human fetal hepatocytes. Data are representative of 2 independent experiments. Scale bar, 20 μm. (e) HAV RNA at 24 h p.i. in Huh-7.5 cells expressing IRF1 sgRNA pre-treated with actinomycin D (ActD, 5 μg ml−1) for 30 min before infection. **p<0.01, *p<0.05 vs. control (two-sided unpaired t-test). (f) DENV RNA at 18 h p.i. or ZIKV RNA at 24 h p.i. in IRF1-depleted Huh-7.5 cells pre-treated with actinomycin D (ActD, 5 μg ml−1) for 30 min before infection. *p<0.05, **p<0.01 vs. control (two-sided unpaired t-test). Data are means ± s.d. from 3 technical replicates representative of 2 independent experiments (a-d) or from 3 independent experiments (e, f). The precise p-values are shown in Supplementary Table 9.
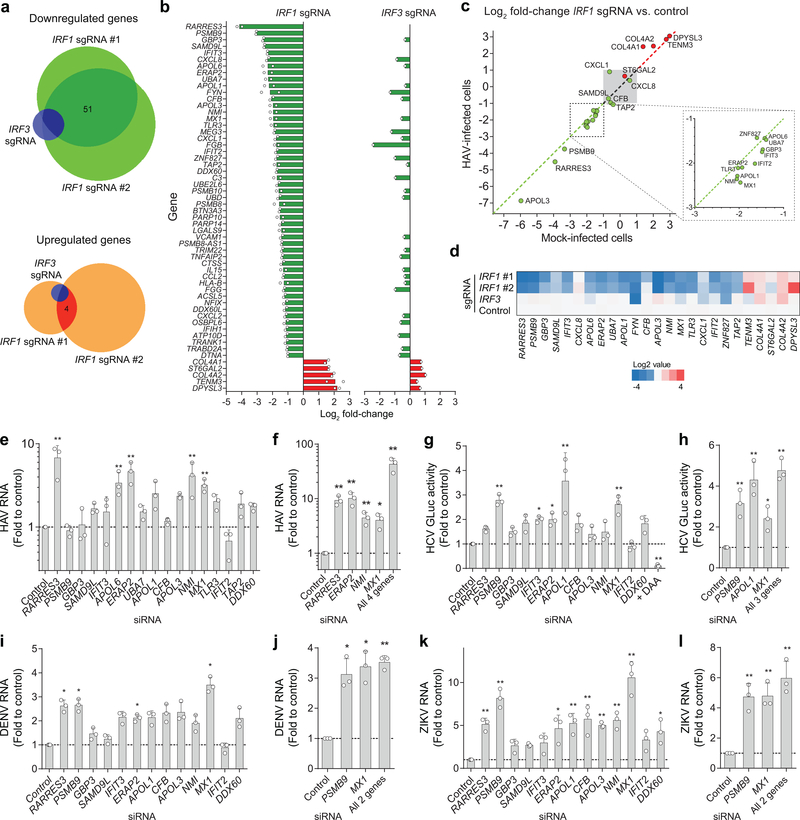
To identify specific IRF1-regulated antiviral effectors, we compared the transcriptomes of HAV-infected IRF1 and IRF3 knockout PH5CH8 cells (Fig. 3a). Changes in transcript abundance from cells expressing control sgRNA were highly congruent in two independent IRF1 knockout cell lines, with 51 genes commonly downregulated >2-fold (Spearman’s r=0.814) (Fig. 3a,b, Supplementary Fig. 6a, b, and Supplementary Table 5). Notably, these genes included known viral sensors (IFIH1, TLR3), IFN-regulated antiviral effectors (MX1, IFIT2, IFIT3), chemotactic factors (CCL2, CXCL1, CXCL2, CXCL8), and components of the immunoproteasome (PSMB8, PSMB9, PSMB10), in addition to multiple genes with no previously recognized antiviral function (Supplementary Tables 5, 6). Only 3 of these transcripts were downregulated >2-fold in IRF3-sgRNA cells (Fig. 3b, Supplementary Table 7).
Figure 3. Shared and distinct antiviral activities of IRF1 effector genes identified by high-throughput RNA sequencing.
(a) The Venn diagram showing numbers of genes with expression changes of ≥ 2-fold for each knockout. (b) List of genes reduced >2-fold in IRF1 sgRNA-expressing cells, in comparison with IRF3 sgRNA-expressing cells. Values shown are means of fold changes of genes expressed in cells transduced with 2 independent IRF1 sgRNAs (left) or an IRF3 sgRNA (right). See Supplementary Tables 5–7 for more details. (c) Validation of RNA-seq results by RT-qPCR assays of RNA extracted from noninfected vs. HAV-infected PH5CH8 cells. Scatter plots show the ratio of abundance of indicated genes between IRF1 vs. control sgRNA-expressing PH5CH8 cells in HAV-infected (y-axis) and noninfected cells (x-axis). (d) Heatmap showing the relative abundance of indicated genes in noninfected PH5CH8 cells determined by RT-qPCR. (e) Relative HAV RNA abundance on 5 d.p.i. of PH5CH8 transfected with siRNA targeting different IRF1 effector genes. **p<0.01 vs. control. (f) Independent validation of the siRNA results and the combination of 4 siRNAs. *p<0.05, **p<0.01 vs. control. (g) Relative GLuc activity on 3 d.p.i. of HCV-infected Huh-7.5 cells. *p<0.05, **p<0.01 vs. control. (h) Independent validation of the siRNA results and the combination of 3 siRNAs. *p<0.05, **p<0.01 vs. control. (i) Relative DENV RNA on 24 h p.i. of infected Huh-7.5 cells. *p<0.05 vs. control. (j) Independent validation of the siRNA results and the combination of 2 siRNAs. *p<0.05, **p<0.01 vs. control. (k) ZIKV RNA abundance on 24 h p.i. of infected Huh-7.5 cells. *p<0.05, **p<0.01 vs. control. (l) Independent validation of the siRNA results and the combination of 2 siRNAs. **p<0.01 vs. control. Data are means ± s.d. from 3 independent experiments (e-g, i-l) or from 3 technical replicates representative of 2 independent experiments (h). p-values were derived using one-way ANOVA with Dunnett’s multiple comparisons test (e, g, i-l) or two-sided unpaired t-test (f, h). The precise p-values are shown in Supplementary Table 9.
We focused on the 18 genes most downregulated in IRF1 knockout cells. With the exception of CXCL8, RT-qPCR confirmed >2-fold reductions in basal expression of each in IRF1 knockout PH5CH8 cells (Fig. 3c,d). Basal PSMB9, NMI, and TLR3 protein abundances were also reduced, and TLR3 sensing of poly(I:C) lost after IRF1 knockout (Supplementary Fig. 7a, b). Importantly, the impact of IRF1 knockout on transcript levels was equivalent in HAV-infected and uninfected cells (Spearman r = 0.944–0.963, p<0.001, Fig. 3c). Thus, IRF1 restricts HAV replication by driving constitutive, basal transcription of antiviral effector genes. Importantly, each of these genes is expressed basally in primary human hepatocytes and hepatoblasts18 (Supplementary Fig. 7c).
Transfecting PH5CH8 cells with siRNA pools (Supplementary Table 4 and Supplementary Fig. 7e) targeting RARRES3, APOL6, ERAP2, NMI, or MX1 enhanced HAV replication >3-fold (Fig. 3e). Good correlation between knockdown efficiency of individual siRNAs and replication enhancement validated these results for all but APOL6 (Supplementary Fig. 7f). Importantly, simultaneously silencing RARRES3, ERAP2, NMI, and MX1 boosted replication ~40-fold (Fig. 3f), recapitulating the phenotype of IRF1 knockout cells (Fig. 1b). Similar experiments in Huh-7.5 cells demonstrated that different subsets of basally IRF1-regulated genes restrict replication of HCV (PSMB9, APOL1, and MX1), and DENV and ZIKV (PSMB9 and MX1) (Fig. 3g-l). Overexpression confirmed the antiviral activities of PSMB9 against HCV and the flaviviruses, as well as the HCV-specific antiviral activity of APOL1 (Supplementary Fig. 7g-i). Thus, IRF1 basally regulates suites of genes that, in various combinations, restrict replication of different positive-strand RNA viruses. Silencing these genes did not affect cell proliferation (Supplementary Fig. 7j).
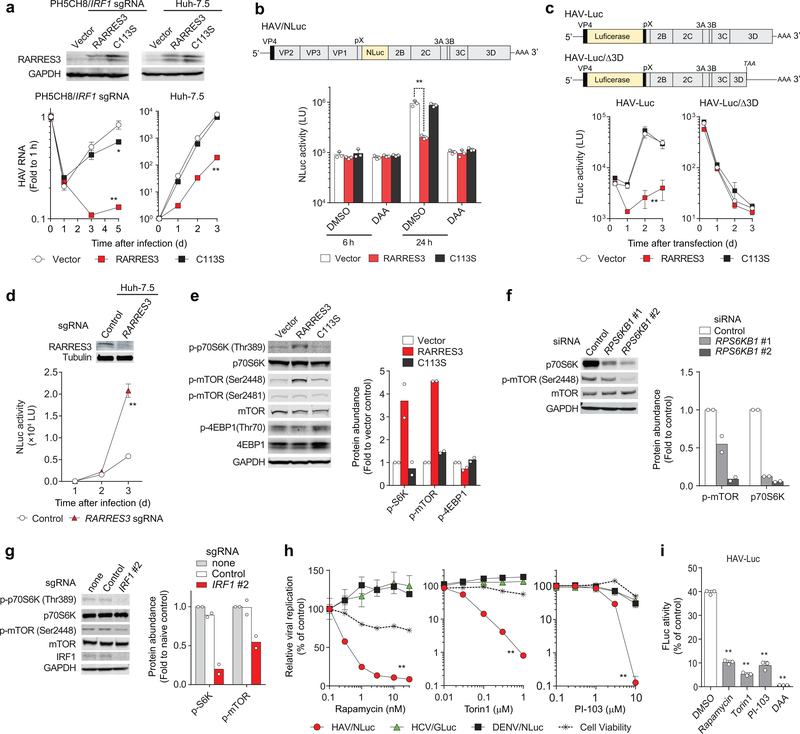
RARRES3, the gene most downregulated by IRF1 knockout and most active in restricting HAV (Fig. 3e, f), encodes a single-pass transmembrane protein with acyl transferase activity19. Although shown previously to modestly limit poliovirus replication12, RARRES3 is not recognized as an important restriction factor for any virus. IFN-γ induced the accumulation of nuclear IRF1 and restricted HAV replication in an IRF1-dependent manner in Huh-7.5 cells, and silencing RARRES3 partially attenuated this suppressive effect of IFN-γ (Supplementary Figs. 5c and 7k,l). Moreover, expressing catalytically-active RARRES3 in IRF1 knockout cells ablated HAV replication, whereas a Cys113-Ser mutant (C113S) lacking acyl transferase activity did not (Fig. 4a). Similar results were obtained in Huh-7.5 cells (Fig. 4a). Although its paralog, PLA2G16 (52% amino acid identity), is a pro-viral entry factor for several picornaviruses20, RARRES3 inhibited neither entry nor translation of a nanoluciferase-expressing HAV (HM175/18f-NLuc, ‘HAV/NLuc’, Fig. 4b) whereas it blocked replication of a subgenomic RNA replicon (Fig. 4c). Huh-7.5 cells knocked out for RARRES3 demonstrated enhanced replication of HAV/NLuc virus (Fig. 4d). While potent, the antiviral action of RARRES3 was specific to HAV, as overexpression did not restrict replication of HCV, DENV, or human rhinovirus 14 (HRV-14) (Supplementary Fig. 8a).
Figure 4. IRF1-regulated RARRES3 acyl transferase restricts HAV replication by downregulating mTOR.
(a) Lentivirus transduction of catalytically-active RARRES3 restricts HAV infection in PH5CH8 cells expressing IRF1 sgRNA #2 (left panels) or Huh-7.5 cells (right panels). RARRES3, but not its catalytically-inactive RARRES3/C113S, inhibited HAV infection in both cell lines. *p<0.05, **p<0.01 vs. vector control (two-way ANOVA with Dunnett’s multiple comparisons test). (b) Huh-7.5 cells stably expressing indicated lentiviral vectors were challenged with HAV carrying NanoLuc (NLuc) with 30 μM 2’CMA (DAA) or vehicle (DMSO). NLuc activities at indicated time points post-infection are shown. **p<0.01 (two-way ANOVA with Dunnett’s multiple comparisons test). (c) Transfection of subgenomic HAV-Luc RNA or its replication-incompetent mutant (Δ3D) in Huh-7.5 cells expressing wild-type RARRES3 versus RARRES3/C113S. **p<0.01 vs. vector control (two-way ANOVA with Dunnett’s multiple comparisons test). (d) Infection of HAV/NLuc in Huh-7.5 cells expressing RARRES3 sgRNA. Immunoblots are shown on top. **p<0.01 vs. vector control (two-way ANOVA with Dunnett’s multiple comparisons test). (e) Steady-state levels of mTOR-related factors in Huh-7.5 cells stably expressing RARRES3 and RARRES3/C113S. (f) Immunoblots of P70S6K siRNA-transfected Huh-7.5/RARRES3 cells. (g) Phosphorylation of p70S6K and mTOR in Huh-7.5 cells expressing IRF1 sgRNA. (h) Impacts of mTOR inhibitors on HAV/NLuc vs. HCV/GLuc vs. DENV/NLuc replication and the cell viability. Inhibitory effects of all these inhibitors on HAV/NLuc vs. other reporter viruses differed significantly. **p<0.01 (two-way ANOVA with Dunnett’s multiple comparisons test). (i) Inhibition of transfected subgenomic HAV-Luc RNA replication in Huh-7.5 cells by the mTOR inhibitors. DAA, 30 μM 2’CMA. **p<0.01 vs. DMSO control (one-way ANOVA with Dunnett’s multiple comparisons test). Data are means ± s.d. from 3 independent experiments (b, d, i) or from 3 technical replicates representative of 2 (a, e-h) or 3 independent experiments (c). The precise p-values are shown in Supplementary Table 9.
The acyl transferase activity of RARRES3 may exert pleiotropic effects on cellular signaling pathways, including the PI3K/Akt/mTOR axis21,22. RARRES3 overexpression induced phosphorylation of p70S6K at Thr-389 in an acyl transferase-dependent manner, downregulating mTOR by catalyzing its phosphorylation at Ser-244823,24 and reducing mTOR-dependent phosphorylation of 4EBP1 at Thr-70 (Fig. 4e,f). Consistent with this, both p70S6K and mTOR phosphorylation were reduced in IRF1 knockout cells (Fig. 4g). Pharmacologic inhibition of mTOR also inhibited HAV, but not HCV or DENV replication (Fig. 4h, i). Thus, while we cannot exclude additional antiviral actions, RARRES3 likely exerts an antiviral effect by downregulating mTOR. Despite its phospholipase activity19, overexpressing RARRES3 resulted only in minimal increases in phosphoinositide PI(3,4,5)P3, and no other changes among 211 lipid species (Supplementary Fig. 8b).
These data establish RARRES3 as an important IRF1-regulated HAV restriction factor. Of the other three genes with major restriction activity against HAV (Fig. 3e), only MX1 is well known for its antiviral activities. NMI was suggested previously to promote degradation of IRF7 and to function as a proviral, negative regulator of IFN responses25. ERAP2, an endosomal aminopeptidase, contributes to T cell responses by generating class I HLA-binding peptides, but it has no known cell-intrinsic antiviral activity. Interestingly, we found that PSMB9, a component of the immunoproteasome that is also involved in antigen processing26, provides basal antiviral protection against HCV and the flaviviruses, DENV and ZIKV (Fig. 3g-l and Supplementary Fig. 7h-i). Additional studies are needed, but these results point to potential unrecognized intrinsic antiviral functions of antigen processing machinery and may help to explain active suppression of the immunoproteasome by many viruses26,27. Thus, although many of the basally IRF1-regulated genes we identified have been linked to IFN responses previously, only a minority (for example, MX1 and IFIT3) have well-established direct antiviral function28.
Our data show differences in the key IRF1-regulated genes that basally restrict replication of various positive-strand RNA viruses (Fig. 3e-l). Differences may also exist between mammalian species, perhaps reflecting evolutionary history with viruses. Mice (Mus musculus) are not naturally permissive for HAV infection, due to overwhelming virus control by MAVS and IRF3/IRF7-mediated transcriptional responses7. Nonetheless, HAV replication is enhanced in Irf1−/− mice early after infection (Fig. 1c), even though orthologs of two of the four IRF1-regulated genes that most restrict HAV replication in human hepatocytes, RARRES3 and ERAP2, do not exist in mice (Fig. 3e,f).
IRF1 has been shown previously to contribute to the basal expression of dozens of IFN-γ inducible pro-inflammatory and antimicrobial genes in macrophages29, but its crucial role in basally regulating genes that restrict virus replication has not been appreciated. Our data show that the constitutive expression of IRF1 maintains basal transcription of suites of genes, some with no previously known antiviral function, that provide immediate defense against virus invasion of hepatocytes. Since IRF1 also mediates early protection against alphaviruses in muscle cells independently of IFNs30, it may act similarly in nonhepatic tissues. Little attention has been paid previously to this aspect of IRF1-regulated cell-intrinsic immunity. Further elucidating the mechanisms by which IRF1-regulated restriction factors act to intrinsically limit virus replication may provide fresh directions for host-directed antiviral therapies.
METHODS
Cells
Huh-7.5 human hepatoma cells and PH5CH8 immortalized human hepatocytes were mycoplasma-free and cultured in Dulbecco’s modified Eagle’s medium (DMEM), High Glucose supplemented with 10% fetal bovine serum (FBS), 1×Penicillin-Streptomycin, 1×GlutaMAX™-I and 1×MEM Non-Essential Amino Acids Solution (Thermo Fisher Scientific), as previously described31,32.
Tissues for processing fetal liver cells were provided by the accredited nonprofit corporation Advanced Biosciences Resources, Inc. (ABR) and obtained from fetuses between 19–21 weeks gestation during elective terminations of pregnancy. Tissues were collected with written informed consent from all donors and in accordance with the US Food and Drug Administration CFR Part 1271 Good Tissue Practices regulations. Tissue processing, and hepatoblasts isolation and culture were previously described32. The use of commercially procured fetal liver cells was determined by the University of North Carolina at Chapel Hill Institutional Review Board to be exempt from review.
HAV infectious challenge in genetically-modified mice
Mice were bred and housed at the University of North Carolina at Chapel Hill in accordance with the policies and guidelines of the Institutional Animal Care and Use Committee. C57BL/6, Ifnar1−/−, Irf3−/− and Irf1−/− mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were intravenously inoculated with the indicated virus inocula at 6–10 weeks of age, as described previously7. Mice were housed in individual cages for collection of fecal pellets with periodic collection of serum samples. Tissues were harvested at necropsy and stored in RNAlater (Thermo Fisher Scientific, Maltham, MA), or snap frozen on dry ice and kept at −80 °C until processed for RNA extraction. All experiments involving mice were approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill.
Reagents and antibodies
miR-122 mimics were synthesized by Dharmacon and transfected by electroporation as miRNA/miRNA* duplexes as described33. Puromycin, blasticidin, and ruxolitinib were purchased from Invivogen. Pyridone 6 was from Millipore. Recombinant human IFN-λ1 and IFN-α, actinomycin D were from Sigma-Aldrich. Recombinant human IFN-γ was from Peprotech. PSI-7977 (Sofosbuvir) was from Chemscene and 2’CMA (2’-C-methyladenosine) was from Santa Cruz Biotechnology. Cell viability was determined using Cell Counting Kit-8 (DOJINDO, Japan) on 96-well plates according to the manufacturer’s protocol.
Primary antibodies to IRF1 (1:500 dilution, #8478), IRF7 (1:500 dilution, #13014), IFIT1 (1:500 dilution, #14769), STAT1 (1:500 dilution, #14994), STING (1:500 dilution, #13647), MyD88 (1:500 dilution, #4283), TLR3 (1:500 dilution, #6961), NF-κB p65 (RelA; 1:500 dilution, #8242), mTOR (1:500 dilution, #2983), phospho-mTOR (Ser-2448) (1:500 dilution, #5536), phospho-mTOR (Ser-2481) (1:500 dilution, #2974), p70S6K (1:1,000 dilution, #2708), phospho-p70S6K (Thr-389) (1:1,000 dilution, #9234), 4E-BP1 (1:1,000 dilution, #9644), and phospho-4E-BP1 (Thr-70) (1:1,000 dilution, #9455) were from Cell Signaling Technology; IRF3 (1:200 dilution, sc-9082) and OAS1 (F-3; 1:100 dilution, sc-374656) were from Santa Cruz Biotechnology; LGP2 (1:500, ab67270) was from Abcam; GAPDH was from Thermo Fisher Scientific (Clone 6C5; 1:10,000 dilution, AM4300) or Wako (Clone 5A12; 1:4,000 dilution, 016–25523); RIG-I (Clone Alme-1; 1:1,000 dilution, ALX-804–849), Cardif (MAVS; 1:2,000 dilution, ALX-210–929) and MDA5 (1:1,000 dilution, ALX-210–935-C100) were from Enzo life sciences; Actin (Clone AC-74; 1:40,000 dilution, A2228), α-tubulin (Clone DM1A; 1:15,000 dilution, T6199) and IL28RA (IFNLR1; 1:500 dilution, AV48070) were from Sigma; IFNAR1 (1:2,000 dilution, A304–290A) and NMI (1:4,000 dilution, A300–551A) were from Bethyl Laboratories; and LMP2 (PSMB9; 1:400 dilution; 14544–1-AP), APOL1 (1:500 dilution; 11486-AP) and RARRES3 (1:800 dilution; 12065–1-AP) were from Proteintech. IRDye 680 or 800 secondary antibodies including #926–32211, #926–32212, #926–32214, #926–68020 and #926–68073 (1:12,000) were from LI-COR.
Viruses
High-titer HAV (HM175/18f strain) stock was mycoplasma-free and prepared as described before34. HAV infection was performed at an m.o.i of 10. Sendai virus (Cantell strain) was obtained from Charles River Laboratories and was inoculated at 50 units ml−1, unless otherwise indicated. Infection with HCV carrying Gaussia luciferase (GLuc) reporter was described previously32. Dengue virus (DENV) serotype 2 (o1Sa-054 strain) and Zika virus (ZIKV, MR-766 and AB-59 strains) were propagated in Vero, C6/36, or Huh-7.5 cells as described35 and inoculated at an m.o.i. of 1.
HM175/18f-NLuc reporter virus
The pHM175/18f-NLuc plasmid was created by PCR amplifying the NLuc ORF using pNL1.1 plasmid (Promega) as template and primers containing the tri-glycine sequence flanked by XbaI and BamHI restriction sites. This PCR product was digested with the above enzymes and ligated into similarly digested pSK-2A-Zeo-2B plasmid36 to obtain pSK-2A-NLuc-2B plasmid. This plasmid was further digested with SacI/PflMI to release the the entire 2A-NLuc-2B fragment and ligated into a similarly digested HM175/18f parental plasmid37 to obtain pHM175/18f-NLuc.
DENV/NLuc reporter virus
Plasmids encoding capsid and subgenomic RNA containing NS1–5 region fused with a NanoLuc reporter flanked by 5’ and 3’untranslated RNAs derived from DENV1 (D1/Hu/Saitama/NIID100/2014 strain) and prME derived from DENV2 (o1Sa-054 strain) were transfected into 293T cells and infectious virons secreted into the supernatant fluids harvested in accordance with previously described methods38.
Other plasmids
pJFH1-QL containing the cell culture–adaptive mutation Q221L in the NS3 helicase, pJFH1/GND, pH77S.3, pH77D, pT7–18f, pHAV-Luc, pHAV-LucΔ3D were described previously32,34,39. The lentiviral transfer plasmids encoding IRF1 effector genes (RARRES3, -PSMB9, and –APOL1) were created by PCR amplifying the host genes using cDNA derived from PH5CH8 cell total RNA as template and primers flanked by XbaI and PstI or NheI restriction sites. The PCR products were digested with the above enzymes and ligated into similarly digested pCSII-EF-MCSII plasmid to obtain pCSII-EF-RARRES3, -PSMB9, and -APOL1. A point mutation in pCSII-EF-RARRES3/C113S was introduced by primer-directed mutagenesis of the sequence spanning the XbaI and PstI sites. The firefly luciferase reporter vectors including pIFNβ-Luc, p4×PRDIII-I-Luc, as well as the Renilla luciferase control reporter vector pRL-TK were described previously3,31.
Viral RNA transcription and transfection
In vitro transcription of HAV or HCV RNA was carried out using T7 RiboMAX™ Express Large Scale RNA Production System (Promega) as per manufacturer’s protocol. Transfection of viral RNA was performed in a Gene Pulser Xcell Total System (Bio-Rad) as previously described33 or using TransIT®-mRNA Transfection Kit (Mirus) for HAV-Luc RNA as described32.
Lentivirus production and transduction
For shRNA lentivirus production, shRNA plasmids obtained from Sigma-Aldrich listed in Supplementary Table 1 were co-transfected with MISSION Lentiviral Packaging Mix (SHP001) into 293FT cells and the supernatant fluids harvested at 48 h and 72 h were filtered through a 0.22 μm syringe filter. Production of sgRNA CRISPR/Cas9 lentivirus was carried out by co-transfecting sgRNA expressing vectors listed in Supplementary Table 2 and 3rd Generation Packaging System Mix (LV053, abm Inc., Canada). Lentivirus transduction was performed by supplementation of 8 μg ml−1 polybrene, followed by antibiotic selection with 6 μg ml−1 puromycin for single knockout derivatives, or 6 μg ml−1 puromycin plus 5 μg ml−1 blasticidin for double-knockout derivatives. We used antibiotic-resistant bulk cell populations for experiments to avoid clonal biases.
RNA extraction and quantitative RT-PCR
Total RNA extraction was performed with the RNeasy mini Kit (Qiagen). Detection of HAV genome RNA was carried out by a two-step quantitative RT-PCR analysis with the SuperScript®III First-Strand Synthesis System (Thermo Fisher Scientific) and iTaq SYBR Green Supermix (Bio-Rad) or alternatively Thunderbird SYBR qPCR Mix (TOYOBO) using specific primers 5’-GGTAGGCTACGGGTGAAAC-3’ and 5’-AACAACTCACCAATATCCGC-3’. HCV RNA level was determined as previously described32. Quantification of IRF1 target genes was performed with the primer pairs listed in Supplementary Table 3. DENV and ZIKV RNA levels were quantified using specific primer pairs targeting DENV genome RNA, 5’-ACACCACAGAGTTCCATTACAGA-3’ and 5’-CATCTCATTAAAGTCGAGGCC-3’, or ZIKV genome RNA, 5’-AARTACACATACCARAACAAAGTGGT-3’ and 5’- TCCRCTCCCYCTYTGGTCTTG-3’ respectively, using RNA-directTM SYBR Green Realtime PCR Master Mix (TOYOBO).
Phospholipid preparation
Methods for comprehensive phospholipids (PLs) analysis were described previously40,41. Briefly, total PLs were extracted from the culture cells with the Bligh-Dyer method. An aliquot of the lower/organic phase was evaporated to dryness under N2, and the residue was dissolved in methanol for LC/MS/MS measurements of PC and PE. To analyze PA, PS, PI and PIPs, another aliquot of the same lipid extract was added with an equal volume of methanol before being loaded onto a DEAE cellulose column (Santa Cruz Biotechnology) pre-equilibrated with chloroform. After successive washes with chloroform/methanol (1:1, v/v), the acidic PLs were eluted with chloroform/methanol/HCl/water (12:12:1:1, v/v), followed by evaporation to dryness to give a residue, which was resolved in methanol. The resultant fraction was subjected to a methylation reaction with TMS-diazomethane before LC/MS/MS analysis42.
Mass spectrometric analyses
LC-electrospray ionization-MS/MS analysis was performed with an UltiMate 3000 LC system (Thermo-Fisher Scientific) equipped with HTC PAL autosampler (CTC Analytics). A 10 μL aliquot of the lipid samples was injected and the lipids were separated on Waters X-Bridge C18 column (3.5 μm, 150 mm × 1.0 mm i.d.) at room temperature (25°C) using a gradient solvent system as follows: mobile phase A (isopropanol/methanol/water (5/1/4 v/v/v) supplemented with 5 mM ammonium formate and 0.05% ammonium hydroxide)/mobile phase B (isopropanol supplemented with 5 mM ammonium formate and 0.05% ammonium hydroxide) ratios of 70%/30% (0 min), 50%/50% (2 min), 20%/80% (13 min), 5%/95% (15–30 min), 95%/5% (31–35 min) and 70%/30% (35–45 min). Flow rate was 20 μL/min. PLs species was measured by the selected reaction monitoring (SRM) in positive ion mode with a triple-stage quadrupole mass spectrometer (TSQ Vantage AM, Thermo-Fisher Scientific). The characteristic fragments of individual PLs were detected by the product ion scan (MS/MS mode). Chromatographic peak areas were used for comparative quantitation of each molecular species (e.g. 38:6, 40:6) in a given class of the phospholipids (e.g. PA, PC).
Immunoblots
Western blotting was performed with standard methods. Odyssey Infrared Imaging System (Li-COR Biosciences, Lincoln) was used for visualization.
RNA interference
siRNA pools listed in Supplementary Table 4 were obtained from Dharmacon or Thermo Fisher Scientific and transfected into cells using siLentfect Lipid Reagent (Bio-Rad) or Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Luciferase assay
Gaussia luciferase (GLuc) analysis of HCV replication and dual luciferase assay to analyze transcriptional induction were performed as described previously31,32. NanoLuc activity was measured using Nano-Glo Luciferase Assay System (Promega) as per the manufacture’s protocol. For virus replication assays, medium was replaced with fresh medium containing chemical inhibitors at 1 h after inoculation.
RNA-sequencing
RNA purity was assessed with a Nanodrop 2000 (Thermo Fisher Scientific) and integrity was determined with an Agilent 2100 Bioanalyzer (Agilent). RNA integrity and sequencing quality were comparable for all samples. Sequencing was performed on an Illumina HiSeq 2000 platform. RNA sequences were aligned to hg38 using STAR v2.4.2a43, genes were quantified using SalmonBeta-0.4.244, and differential expression was determined using DESeq245. Gene ontology enrichment analysis was performed using DAVID 6.8.
Laser-scanning confocal microscopy
Cells grown on an 8-well chamber slide (Falcon) was fixed with 4% paraformaldehyde and permeabilized with 0.25% Triton X-100. The cell monolayer was then incubated with rabbit anti-IRF-1 antibody (1:50 dilution, #8478, Cell Signaling Technology) at 4°C overnight, followed by a secondary antibody, goat anti-rabbit Alexa Fluor 488 (1:200 dilution, Thermo Fisher Scientific). Nuclei were counterstained with DAPI. Images were collected using a Leica DMIRB Inverted Microscope in the Michael Hooker Microscopy Facility of the University of North Carolina.
Statistical Analysis
Unless noted otherwise, all between-group comparisons were carried out by ANOVA or t test using Prism 6.0 software (GraphPad Software, Inc.). The p values were calculated from 3 biological replicates unless otherwise indicated. In some experiments designed to validate earlier conclusions using orthogonal approaches, we carried out 2 independent experiments, each with 3 technical replicates. These few exceptions are noted in the legends.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. RNA-sequencing data has been deposited with GEO (GSE114916).
Supplementary Material
Acknowledgements
The authors thank C.M. Rice, S. Inoue and N. Kato for reagents, M. Soloway for bioinformatics support, M. Chua for technical assistance, and H. Dansako, B.B. Queliconi and W.J. Zuercher for helpful discussions. This work was supported in part by JSPS KAKENHI [Grant Numbers, JP16H07462, JP17H05070 (DY), JP18K05987 (AH-Y), JP17K08870, JP15K19109 (TH)], AMED (JP18jk0210014 to AH-Y, JP18fk0108035 to TH, and JP16fk0210108 to MK), and NIH grants R01-AI103083 (SML), U19-AI109965 (SML) and R01-AI131685 (SML and JKW).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Yoneyama M, Onomoto K, Jogi M, Akaboshi T & Fujita T Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol 32, 48–53 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Chan YK & Gack MU Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol 14, 360–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li K, Chen Z, Kato N, Gale M Jr. & Lemon SM Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J Biol Chem 280, 16739–16747 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Woodson SE & Holbrook MR Infection of hepatocytes with 17-D vaccine-strain yellow fever virus induces a strong pro-inflammatory host response. J Gen Virol 92, 2262–2271 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Feng H et al. NLRX1 promotes immediate IRF1-directed antiviral responses by limiting dsRNA-activated translational inhibition mediated by PKR. Nat Immunol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemon SM, Ott JJ, Van Damme P & Shouval D Type A viral hepatitis: A summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J Hepatol (2017). [DOI] [PubMed] [Google Scholar]
- 7.Hirai-Yuki A et al. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science 353, 1541–1545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taki S et al. Multistage regulation of Th1-type immune responses by the transcription factor IRF-1. Immunity 6, 673–679 (1997). [DOI] [PubMed] [Google Scholar]
- 9.White LC et al. Regulation of LMP2 and TAP1 genes by IRF-1 explains the paucity of CD8+ T cells in IRF-1−/− mice. Immunity 5, 365–376 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Fujita T, Kimura Y, Miyamoto M, Barsoumian EL & Taniguchi T Induction of endogenous IFN-alpha and IFN-beta genes by a regulatory transcription factor, IRF-1. Nature 337, 270–272 (1989). [DOI] [PubMed] [Google Scholar]
- 11.Odendall C et al. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol 15, 717–726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoggins JW et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixit E et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumpter R Jr. et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79, 2689–2699 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leblanc JF, Cohen L, Rodrigues M & Hiscott J Synergism between distinct enhanson domains in viral induction of the human beta interferon gene. Mol Cell Biol 10, 3987–3993 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyamoto M et al. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell 54, 903–913 (1988). [DOI] [PubMed] [Google Scholar]
- 17.Tanaka N, Kawakami T & Taniguchi T Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol Cell Biol 13, 4531–4538 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oikawa T et al. Model of fibrolamellar hepatocellular carcinomas reveals striking enrichment in cancer stem cells. Nat Commun 6, 8070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uyama T, Jin XH, Tsuboi K, Tonai T & Ueda N Characterization of the human tumor suppressors TIG3 and HRASLS2 as phospholipid-metabolizing enzymes. Biochim Biophys Acta 1791, 1114–1124 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Staring J et al. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature (2017). [DOI] [PubMed] [Google Scholar]
- 21.Hsu TH et al. Involvement of RARRES3 in the regulation of Wnt proteins acylation and signaling activities in human breast cancer cells. Cell Death Diff 22, 801–814 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ou CC et al. Downregulation of HER2 by RIG1 involves the PI3K/Akt pathway in ovarian cancer cells. Carcinogenesis 29, 299–306 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Chiang GG & Abraham RT Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem 280, 25485–25490 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Figueiredo VC, Markworth JF & Cameron-Smith D Considerations on mTOR regulation at serine 2448: implications for muscle metabolism studies. Cell Mol Life Sci 74, 2537–2545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J et al. Negative regulation of Nmi on virus-triggered type I IFN production by targeting IRF7. J Immunol (Baltimore, Md. : 1950) 191, 3393–3399 (2013). [DOI] [PubMed] [Google Scholar]
- 26.McCarthy MK & Weinberg JB The immunoproteasome and viral infection: a complex regulator of inflammation. Frontiers in microbiology 6, 21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verweij MC et al. Viral inhibition of the transporter associated with antigen processing (TAP): a striking example of functional convergent evolution. PLoS Pathog 11, e1004743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoggins JW et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langlais D, Barreiro LB & Gros P The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J Exp Med 213, 585–603 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair S, Poddar S, Shimak RM & Diamond MS Interferon regulatory factor-1 (IRF-1) protects against chikungunya virus induced immunopathology by restricting infection in muscle cells. J Virol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dansako H et al. Class A scavenger receptor 1 (MSR1) restricts hepatitis C virus replication by mediating toll-like receptor 3 recognition of viral RNAs produced in neighboring cells. PLoS Pathog 9, e1003345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamane D et al. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med 20, 927–935 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamane D et al. Differential hepatitis C virus RNA target site selection and host factor activities of naturally occurring miR-122 3’ variants. Nucleic Acids Res (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Z et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496, 367–371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hishiki T et al. Interferon-mediated ISG15 conjugation restricts dengue virus 2 replication. Biochem Biophys Res Commun 448, 95–100 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Beard MR, Cohen L, Lemon SM & Martin A Characterization of recombinant hepatitis A virus genomes containing exogenous sequences at the 2A/2B junction. J Virol 75, 1414–1426 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Binn LN et al. Primary isolation and serial passage of hepatitis A virus strains in primate cell cultures. J Clin Microbiol 20, 28–33 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuda M et al. High-throughput neutralization assay for multiple flaviviruses based on single-round infectious particles using dengue virus type 1 reporter replicon. Sci Rep 8, 16624 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi M & Lemon SM Replication of subgenomic hepatitis A virus RNAs expressing firefly luciferase is enhanced by mutations associated with adaptation of virus to growth in cultured cells. J Virol 76, 1171–1180 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baba T et al. Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. J Biol Chem 289, 11497–11511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imae R et al. LYCAT, a homologue of C. elegans acl-8, acl-9, and acl-10, determines the fatty acid composition of phosphatidylinositol in mice. J Lipid Res 53, 335–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kielkowska A et al. A new approach to measuring phosphoinositides in cells by mass spectrometry. Adv Biol Regul 54, 131–141 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information. RNA-sequencing data has been deposited with GEO (GSE114916).