Abstract
Glutathione transferases (GSTs) are multifunctional enzymes that play important roles in plant development and responses to biotic and abiotic stress. However, a systematic analysis of GST family members in Brassica napus has not yet been reported. In this study, we identified 179 full-length GST genes in B. napus, 44.2% of which are clustered on various chromosomes. In addition, we identified 141 duplicated GST gene pairs in B. napus. Molecular evolutionary analysis showed that speciation and whole-genome triplication played important roles in the divergence of the B. napus GST duplicated genes. Transcriptome analysis of 21 tissues at different developmental stages showed that 47.6% of duplicated GST gene pairs have divergent expression patterns, perhaps due to structural divergence. We constructed a GST gene coexpression network with genes encoding various transcription factors (NAC, MYB, WRKY and bZIP) and identified six modules, including genes expressed during late seed development (after 40 days; BnGSTU19, BnGSTU20 and BnGSTZ1) and in the seed coat (BnGSTF6 and BnGSTF12), stamen and anther (BnGSTF8), root and stem (BnGSTU21), leaves and funiculus, as well as during the late stage of pericarp development (after 40 days; BnGSTU12 and BnGSTF2) and in the radicle during seed germination (BnGSTF14, BnGSTU1, BnGSTU28, and BnGSTZ1). These findings lay the foundation for elucidating the roles of GSTs in B. napus.
Subject terms: Gene regulatory networks, Transcriptomics
Introduction
Glutathione transferases (GSTs) are multifunctional enzymes that are widely distributed in various organisms. GSTs contain an N-terminal domain and a C-terminal domain. The N-terminal domain includes a catalytic residue for glutathione (GSH) binding and catalysis, whereas the less conserved C-terminal domain, comprising five or six major helices, binds hydrophobic substrates and determines GST specificity and activity1. GSTs are divided into 14 classes: phi (GSTF), tau (GSTU), theta (GSTT), zeta (GSTZ), lambda (GSTL), metaxin, hemerythrin (GSTH), iota (GSTI), glutathione-dependent dehydroascorbate reductase (DHAR), tetrachloro hydroquinone dehalogenase (TCHQD), γ-subunit classes of the eukaryotic translation elongation factor 1B (EF1Bγ), glutathionyl hydroquinone reductase (GHR), GSTs with two thioredoxins (GST2N), and microsomal prostaglandin E synthase type 2 (mPGES2)2,3. The phi and tau classes are the most abundant classes in plants. The GSTL and DHAR classes are monomeric, with no GSH-conjugating activity towards xenobiotic substrates and function in redox homeostasis4,5. GSTI has been identified only in non-vascular plants, algae and cyanobacteria, and appears to have been lost during the evolution of most terrestrial plants3,6.
GSTs were initially found to function in herbicide detoxification7. Their xenobiotic detoxification function can be detected using the 1-chloro-2,4-dinitrobenzene (CDNB) assay, in which the chloro group of CDNB is substituted by glutathione2. GSTs catalyze the conjunction of electrophilic substrates with the tripeptide GSH. Chronopoulou et al. (2017) reviewed the roles of GSTs based on recent progress in plant proteomics, genomics and transcriptomics analyses. In addition to their catalytic activities, GSTs function as non-catalytic proteins by binding to flavonoids and anthocyanins and transporting them from the cytoplasm into the central vacuole8–10. GSTs can bind to the phytohormones auxin11 and cytokinin12, which are involved in plant development. GSTs can also bind to porphyrinogens13 and oxylipins14, thereby protecting plant cells from oxidative stress. In addition, plant GSTs play important roles in responses to biotic and abiotic stresses, such as pathogen attack15,16, heavy metals17, drought18 and salt19, as well as salicylic acid signaling16. However, the detailed mechanisms are currently unclear.
Brassica napus is an allopolyploid species that formed via hybridization of the diploid species B. oleracea and B. rapa. The model dicot Arabidopsis thaliana has been reported to contain 55 GSTs in eight classes, although the EF1Bγ, GHR, metaxin, GSTH and GST2N classes were not included in the analysis2, whereas there are 101 GSTs in soybean20, 81 in poplar21, 42 in potato22 and 99 in sorghum23, 75 in B. rapa24 and 65 in B. oleracea25. The duplicated GST genes in soybean were formed by whole-genome duplication (WGD)19, whereas the expansion of tau and phi GSTs in Capsella rubella mainly occurred through tandem gene duplication26. In B. rapa, WGD and tandem duplication played the main role in the expansion of GSTs23. However, to date, a systematic analysis of the GST family in B. napus has not been reported. The availability of whole-genome sequences for B. rapa27, B. oleracea28, and B. napus29 provides valuable resources for studying the GST family in Brassica species. In this study, we identified GSTs of all 13 classes in B. napus and analyzed their evolution and syntenic relationships. We also evaluated their expression patterns in different tissues and in response to biotic stresses. Our results shed light on this important gene family in the crop B. napus.
Materials and Methods
Identification and nomenclature of GSTs in B. napus
We identified GSTs from all 13 classes for analysis. The resulting sequences for 66 proteins from A. thaliana (http://www.arabidopsis.org/)2, 59 from rice (http://rice.plantbiology.msu.edu/)30, 81 from Populus trichocarpa (https://phytozome.jgi.doe.gov/)21, 37 from Physcomitrella patens (http://plants.ensembl.org/index.html)3, and 575 from various animals, fungi, and bacteria retrieved from Lan et al.21 were obtained (Supplementary Table 1) and used as query sequences to identify GST proteins in B. napus v5.0 (http://www.genoscope.cns.fr/brassicanapus/)29 by BLASTP analysis, with an e-value of 1e-10. In addition, GST_N and GST_C domains were identified by searching the Pfam database (http://Pfam.sanger.ac.uk/) with e-value of 1.0 and the conserved domain database (CDD) at NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd). GSTs without conserved domains were excluded from the analysis. For nomenclature, the prefix ‘Bn’ for B. napus was used, followed by GST and a unique number, such as BnGST1. Gene structures and motifs were analyzed using TBtools (https://github.com/CJ-Chen/TBtools)31. To explore the evolution of the GSTs in Brassica, the GST sequences from B. rapa v 1.527 and B. oleracea v2.1 (http://plants.ensembl.org/index.html) were obtained as described above.
Phylogeny of the GSTs
GST protein sequences from B. napus were aligned using MAFFT version 732 with default parameters, and phylogenic trees were constructed using the maximum likelihood method using PHYML 3.0 with the Jones, Taylor and Thornton model with 100 bootstrap replicates33. FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) was used to visualize the phylogenetic trees.
Distribution of GST genes, gene duplication, molecular evolutionary and Pearson correlation analysis
The distribution of GST genes on B. napus chromosomes was displayed using MapChart version 3.0. The GSTs were clustered together on the chromosomes: two or more GSTs separated by no more than three genes were referred to as a GST gene cluster. To identify the form of gene duplication, 101,040 B. napus gene sequences were aligned using BLASTp, with an e-value of 1e-10. MCScanX with default values was used to classify the duplication patterns of the GSTs into segmental, tandem, proximal, and dispersed duplications34. In addition, syntenic blocks with at least ten genes in the Brassica genome were identified. Duplicate gene pairs were identified according to the following criteria35: candidate duplicate gene pairs were located in syntenic blocks, and duplicate gene pairs were grouped together in the phylogenetic tree. KaKs_Calculator 2.036 was used to calculate the ratio of the nonsynonymous substitution rate (Ka) to the synonymous substitution rate (Ks), and the ω (Ka/Ks) value between paralogous gene pairs was determined using the MYN (Modified YN) model. Divergence time was inferred using the formula T = Ks/2R, where R is 1.5 × 10−8 synonymous substitutions per site per year37. The Pearson correlation coefficients among duplicated gene pairs were calculate using the cor command in R.
Promoter analysis
The promoter sequences in the regions 1500 bp upstream of the coding sequences were obtained, and the cis-acting elements were analyzed using the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)38 and the PLACE database39.
Expression analysis of BnGSTs during development and in response to biotic stresses in B. napus
Transcriptome sequencing was performed using 21 different B. napus tissues (root, stem, leaves, bud, funiculus, anthocaulus, anther, calyx, capillament, petal, stamen, pistil, shoot apex, silique pericarp, seed, seed coat, cotyledon, episperm, endopleura and embryo) at different stages of development; the sequencing datasets were deposited in NCBI under BioProject ID PRJCA001246. Sequencing libraries were generated using an Illumina RNA Library Prep Kit following the manufacturer’s recommendations. Sequencing reads were aligned to the B. napus reference genome and assembled using TopHat 2.0.0 and Cufflinks with default parameters40. Gene expression levels were estimated using FPKM (fragments per kilobase of exon per million mapped fragments). BnGST expression levels were obtained in response to treatment with Sclerotinia sclerotiorum41 and Leptosphaeria maculans42. A heatmap was generated using the R package pheatmap.
Weighted gene coexpression network analysis (WGCNA)
A gene coexpression network was constructed using the WGCNA package in R43. The expression levels in different tissues were log-transformed via log2 (FPKM + 1), and genes with low expression levels (maximum log2 (FPKM + 1) <4) were filtered out. Genes involved in responses to pathogens were also used to construct the network. The settings used were as follows: minModuleSize = 30, maxBlockSize = 6000, reassignThreshold = 0, mergeCutHeight = 0.25, TOMType = “unsigned”. Genes with WGCNA edge weight >0.1 were displayed using Cytoscape 3.6.144.
Results
Identification of GSTs in B. napus
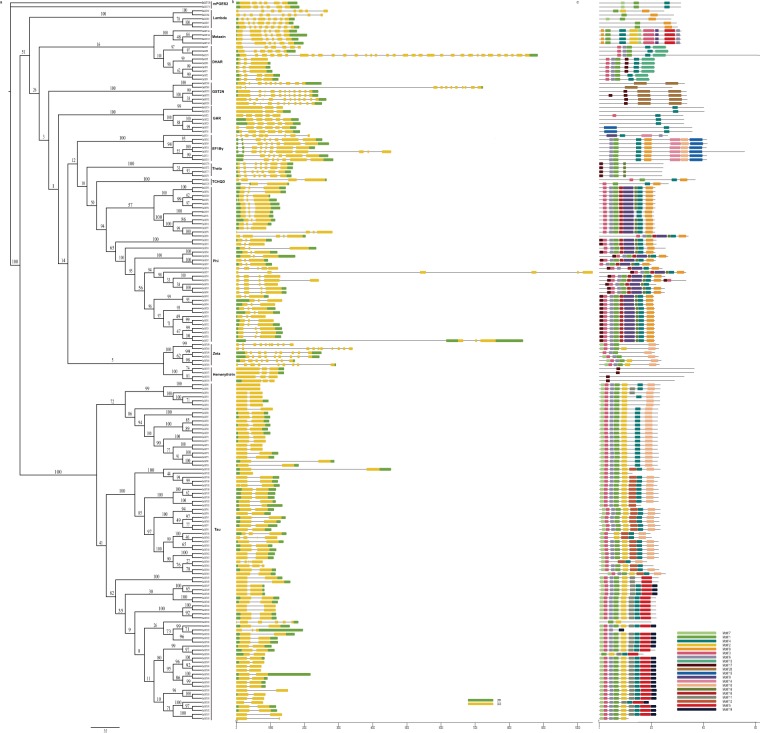
To identify GST proteins in various Brassica species, we performed BLASTP with an e-value of 1e-10. After searching for conserved GST-N and GST-C domains using the Pfam database and CDD at NCBI, we identified 179 full-length genes encoding GST proteins in B. napus (Supplementary Table 2). To classify the B. napus GSTs, we examined their phylogenetic relationships using the maximum likelihood method. Based on the maximum likelihood tree, the 179 GSTs were divided into 13 classes, including mPGES2, GST2N, hemerythrin (GSTH), zeta (GSTZ), EF1Bγ, theta (GSTT), TCHQD, DHAR, GSTF, metaxin (MTX), lambda (GSTL), GHR and tau (GSTU) (Fig. 1a). The phi and tau classes were the largest, with 39 and 84 members, respectively. mPGES2, GST2N, GSTH, GSTZ, EF1Bγ, GSTT, TCHQD, DHAR, MTX, GSTL, and GHR included 2, 6, 4, 6, 7, 4, 2, 9, 4, 5, and 7 members, respectively. We also identified 85 GST genes in B. oleracea and B. rapa (Supplementary Table 2). The phi and tau classes of GSTs in B. oleracea and B. rapa were also the largest, accounting for 72.6% and 70.6% of all GSTs, respectively. The proportion of all GST genes in B. napus, B. oleracea, and B. rapa were 0.21%, 0.18%, and 0.15%, respectively. There was no correlation between genome size and the number of GST genes.
Figure 1.
Phylogenetic analysis (a), gene structures (b) and gene motifs (c) of GSTs in B. napus.
The gene structures and positions of these GSTs were conserved (Fig. 1b). Most (76.9%) phi class GSTs had three exons, and 73 of the 82 tau (89.0%) GSTs had two exons. GST2N, zeta and lambda GSTs had approximately 10 exons, while EF1Bγ and theta GSTs had seven exons, and metaxin GSTs had six exons. The gene motifs of the GSTs were also conserved (Fig. 1c).
In addition to the full-length GSTs, 20, 11, and 7 GST fragments containing partial N- and C-domains were found in B. napus, B. oleracea, and B. rapa, respectively (Supplementary Table 3). The fragments encoded putative polypeptides ranging from 38 to 418 amino acids. We considered these GST fragments to be pseudogenes.
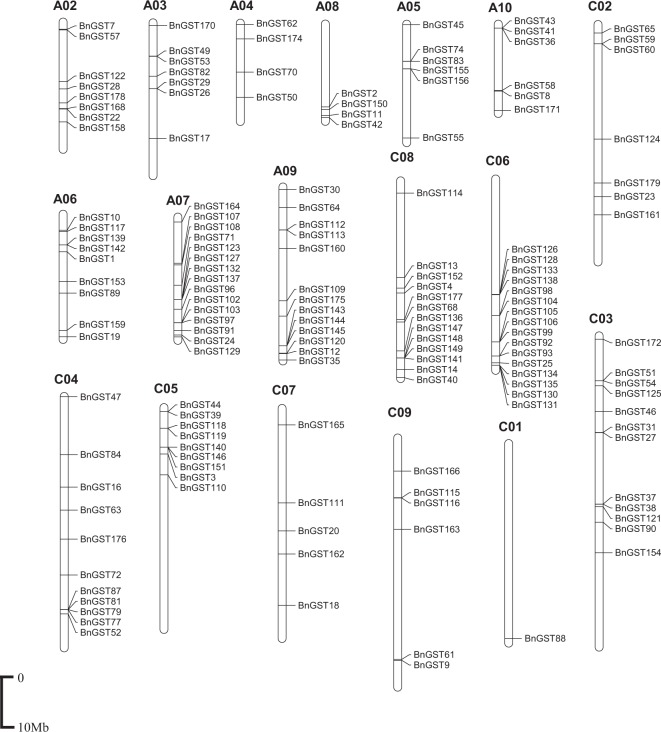
Chromosomal distribution
We investigated the chromosomal distribution of GST genes in the Brassica species. In total, 152 of the 179 full-length GSTs were distributed on all 19 B. napus chromosomes except chromosome A1 (72 on the An genome and 80 on the Cn genome), while the 27 other GSTs were assigned to random chromosomes (13 on the An genome and 14 on the Cn genome) (Fig. 2). Full-length B. oleracea and B. rapa GSTs were found on all chromosomes. The GST fragments were found on 8 of the 19 B. napus chromosomes, 6 of the 9 B. oleracea chromosomes and 4 of the 10 B. rapa chromosomes. Notably, these GSTs are unevenly distributed on the chromosomes. High-density regions harboring GSTs were discovered on chromosome A7, A9, C3, C4, C5, C6 and C8 in B. napus (Fig. 2). In B. oleracea, chromosomes C4 and C6 contained the most GST genes (15), whereas C1 contained only one GST. In B. rapa, chromosome A7 and A9 contained the most GST genes, whereas A1 contained only one GST (Supplementary Table 2).
Figure 2.
Chromosomal distribution of GSTs in B. napus.
In total, 31 GST clusters containing 88 GSTs were found on 13 of the 19 B. napus chromosomes, representing 44.2% of GST genes. Chromosome C6, containing 15 GST genes, harbored the most clusters (5). Most of the clusters contained two genes, whereas cluster 18 on chromosome C5 contained six genes, and cluster 7 on A7 and cluster 12 on A10 each contained five genes. Finally, 15 clusters containing 43 GSTs (44.8%) and 12 clusters containing 33 GSTs (35.9%) were found in B. oleracea and B. rapa, respectively.
Phylogenetic and synteny analysis of GSTs in A. thaliana, B. rapa, B. oleracea, and B. napus
We constructed a phylogenetic tree based on the 66 A. thaliana, 85 B. rapa, 85 B. oleracea, and 179 B. napus GSTs (Supplementary Fig. 1). Metaxin and mPGES2 appeared to be ancient classes of GSTs. Indeed, metaxins have previously been reported to be an ancient GST superfamily45. The mPGES2 GSTs, comprising two members, were evolutionarily distant from the other groups2. The remaining GST proteins were divided into 11 classes: GSTF, GSTT, EF1Bγ, GSTZ, DHAR, TCHQD, GST2N, GHR, GSTL, GSTH, and GSTU. The tau class of GSTs was the largest, followed by the phi class. AtGSTF2 had the most orthologs in Brassica, including ten in B. napus, five in B. rapa, and five in B. oleracea.
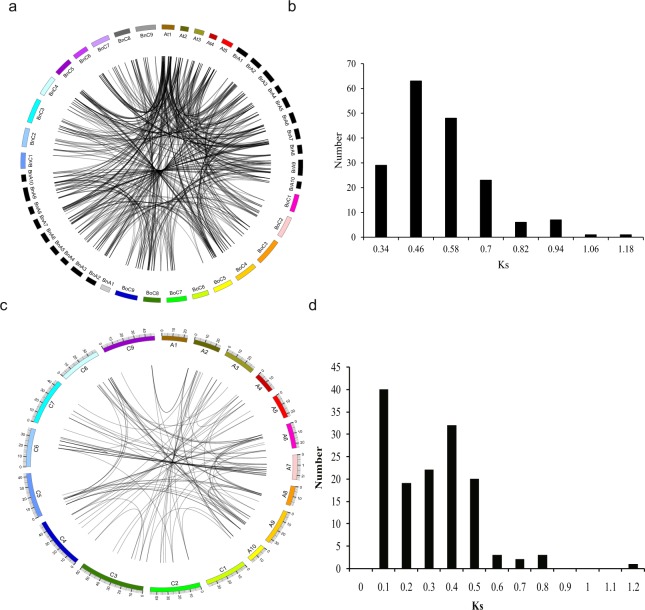
To explore the evolution of the GSTs in Brassica, we constructed synteny maps of GSTs in A. thaliana, B. oleracea, B. rapa, and B. napus (Fig. 3a) and determined the retention or loss patterns of orthologous genes based on their syntenic relationships. We detected orthologs for 55 A. thaliana GST genes in B. napus, B. rapa, and B. oleracea. Based on the syntenic relationship between A. thaliana and B. napus, a total of 163 collinear gene pairs were identified, including 6, 22, 7, 13, 2, 2, 1 and 1 GST gene in A. thaliana with 1, 2, 3, 4, 5, 6, 8 and 10 orthologs in B. napus, respectively (Supplementary Table 4). In addition, we identified 61 collinear gene pairs between A. thaliana and B. oleracea: 26, 10, 2, 1, and 1 GST genes in A. thaliana have 1, 2, 3, 4, and 5 orthologs in B. oleracea, respectively. In total, 79 collinear gene pairs were found between A. thaliana and B. rapa: 25, 15, 5, 1, and 1 A. thaliana gene has 1, 2, 3, 4, and 5 orthologs in B. rapa, respectively.
Figure 3.
Duplicated and orthologous genes in Brassica. (a) circle plot of orthologous genes in A. thaliana (At), B. oleracea (Bo), B. rapa (Br), and B. napus (Bn); (b) density of Ks values of GST gene pairs between B. napus and A. thaliana; (c) duplicate gene pairs in B. napus; (d) density of Ks values of duplicated GST genes in B. napus.
Homeologous gene pairs in B. napus were identified based on high sequence similarity of pairs of genes and location on homeologous chromosomes. These 199 GSTs in B. napus (179 full-length GSTs and 20 GST fragments) were found as 74 homeologous gene pairs on homeologous chromosomes (Supplementary Table 4). According to the synteny analysis in B. napus and its diploid progenitors B. oleracea and B. rapa, a total of 79 gene pairs maintained their relative positions between the An subgenome of B. napus and the Ar genome of B. rapa, and 62 gene pairs maintained their relative positions between the Cn subgenome of B. napus and Co genome of B. oleracea.
The expansion of GST genes in B. napus
To understand the mechanism underlying the expansion of the GST family in B. napus, we examined the types of GST gene duplication. Of the 101,040 genes in the B. napus genome, 3422 genes (3.4%) appeared to have undergone tandem duplication and 61,310 (60.7%) genes had undergone segmental duplication (Supplementary Table 5). We found that 146 of 199 GSTs (73.4%, P > 0.05%) were derived from segmental duplication, a number slightly larger than the average percentage at the whole-genome level (60.7%). Therefore, it appears that segmental duplication played an important role in the expansion of the GST family in B. napus. In addition, we examined the GST gene expansion patterns in B. oleracea and B. rapa, finding that most BoGSTs (56.2%; 54/96) and BrGSTs (63.0%;58/92) were derived from segmental duplication, followed by tandem duplication (19.8% in B. oleracea and 21.7% in B. rapa) (Supplementary Table 5).
The orthologous GST gene pairs between B. napus and A. thaliana were used to estimate the Ks value. The Ks values for all orthologous gene pairs ranged from 0.2266 to 1.0967, with an average of 0.4857 (Fig. 3b). The divergence time ranged from 7.55 MYA to 36.56 MYA, with an average value of 15.89 MYA. These results indicated that GSTs of B. napus diverged from A. thaliana ~16 MYA, which was consistent with the recent whole-genome triplication event that occurred approximately 9–15 MYA, or even 28 MYA46.
Among the GSTs in the three Brassica species examined, we identified 141 duplicate gene pairs in B. napus (Fig. 3c). We estimated the timing of the whole-genome duplication (WGD) event based on the distribution of Ks values, which ranged from 0.0064 to 1.1531 and averaged 0.2612. The corresponding duplication time ranged from 0.21 to 38.44 MYA, with an average value of 8.7 MYA (Supplementary Table 5). Two peaks of Ks values were observed in B. napus: one peak (0–0.1) represented the duplication time of these genes, which occurred during the formation of B. napus 7500–12,500 years ago (Fig. 3d), and the other peak (0.3–0.4), representing a duplication time of ~10 MYA, corresponded to the Brassica whole-genome triplication event (9–15 MYA). Therefore, the processes of B. napus speciation and Brassica whole-genome triplication likely played important roles in the divergence of the GST duplicated genes in B. napus.
Ka/Ks value <1 indicates that a gene pair has experienced negative selection, whereas Ka/Ks >1 indicates positive selection and Ka/Ks = 1 indicates neutral selection. The Ka/Ks ratios for most GST collinear gene pairs were <1, except for the gene pair BnGST129 and BnaGST139 (Ka/K >1). These results indicated that most genes have experienced negative selection, whereas BnGST129 and BnGST139 experienced positive selection (Supplementary Table 6).
Extensive changes in exon–intron structure between duplicate gene pairs
We identified 141 duplicate gene pairs, 131 of which contained full-length GSTs in each duplicate. Of the 131 full-length duplicate gene pairs, 28 pairs (21.4%) showed different numbers of exons (Supplementary Table 6). In 45 other duplicates (34.3%), the number of exons was the same, but the lengths of one or more exons differed. Thus, 55.7% of the duplicated genes exhibited obvious structural divergence.
Identification of cis-acting elements in GST genes
We next characterized the cis-acting elements in promoter regions of the BnGST genes. Various abiotic- and biotic-stress related cis-elements were identified. The abiotic-stress-related cis-acting elements included light-responsive elements (G-box and I-box), abscisic acid-responsive elements (ABRE, ACGT box), GA-responsive element (GARE), auxin-responsive element (AuxRE), dehydration-responsive element (DRE), ethylene-responsive element (ERE), heat-responsive element (HSE), low temperature-responsive element (LTRE), and sugar-response element (SRE). GSTs might also be involved in responses to ammonium, copper, sulfur, and phosphate. We also identified biotic-stress-related cis-elements, such as ethylene- and pathogen-responsive element (GCC-box), and wounding- and pathogen-responsive element (W box). In addition, there were various tissue-specific cis-regulatory elements, including the seed-specific (embryo and endosperm) element AACA motif, the floral organ element CArG and the xylem-specific element AC box. Finally, many predicted transcription factor (TF) binding sites were found, such as NAC, MYB, WRKY and bZIP binding sites (Supplementary Table 7).
Expression of GSTs in different B. napus tissues
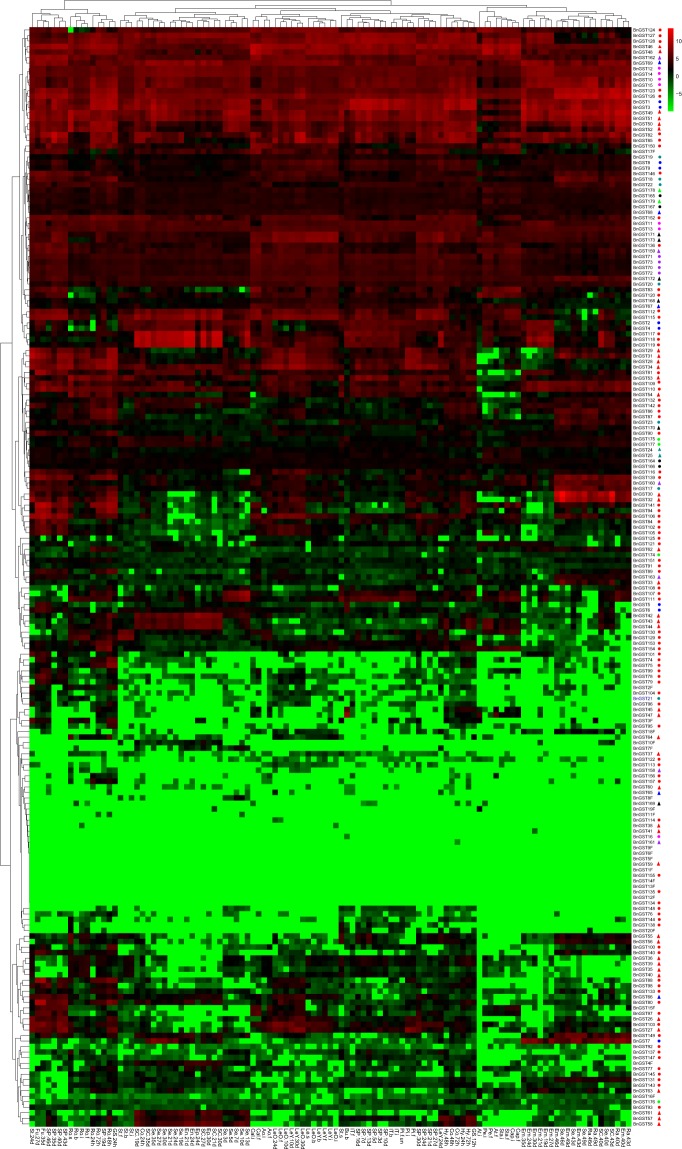
To explore the expression patterns of GSTs in B. napus, we analyzed their expression levels in 21 different tissues at different developmental stages, including root, stem, leaf, bud, funiculus, anthocaulus, anther, calyx, capillament, petal, stamen, pistil, shoot apex, silique pericarp, seed, seed coat, cotyledon, episperm, endopleura, and embryo tissue. Of the 199 GSTs, 58 were expressed in all tissues at different developmental stages, whereas 51 GSTs, including 16 GST fragments, exhibited almost no expression (Fig. 4). The 90 remaining GSTs were expressed in specific tissues. Most GST fragments were not expressed any tissues and were excluded from further expression analysis. Several genes were expressed in all tissues except petals, capillaments, stamens, and anthers, including BnGST28, BnGST29, BnGST31 and BnGST34 (GSTF3), BnGST53 and BnGST54 (GSTF10), BnGST81 (GSTU4), BnGST86 and BnGST87 (GSTU7), BnGST109 and BnGST110 (GSTU13), BnGST132 (GSTU22), and BnGST142 (GSTU25). BnGST57, BnGST58 and BnGST61 (GSTF12) were expressed during early seed development and seed coat formation (before day 40), while BnGST149 (GSTU25) was expressed during all stages of seed development and seed coat formation.
Figure 4.
Expression patterns of all 199 GSTs in 21 different tissues at different developmental stages in B. napus. The color bar represents log2 expression levels (FPKM). The group information for B. napus GSTs is indicated: red, blue, magenta, dark cyan, purple, black, and green circles represent tau, DHAR, EF1Bγ, GHR, theta, metaxin, hemerythin class GSTs; red, blue, dark cyan, purple, black, green triangles represent phi, lambda, TCHQD, zeta, GST2N and mPEGS2 class GSTs.
Of the 131 full-length GST duplicate pairs, five were almost undetectable in any tissue, and the remaining 126 showed expression in some or all tissues. The Pearson correlation coefficient (with a cutoff of 0.6) was used to investigate expression patterns of duplicated GST gene pairs. In total, 39 of the 126 (30.9%) were expressed in all tissues, and the correlation coefficients for the expression levels of 25 duplicated gene pairs were more than 0.6, which showed that the expression patterns of these gene pairs were similar (Supplementary Table 6). The 77 (61.1%) remaining duplicate GST pairs showed different expression patterns, which were classified into four groups: (I) one copy of each duplicate was expressed in all tissues and the other was not expressed in any tissue; (II) one copy was expressed in all tissues and the other was expressed in a specific tissue; (III) one copy was expressed in a specific tissue and the other was not expressed in any tissue; and (IV) both duplicates were expressed in specific tissues. Group I contained 7 GST pairs, group II contained 10, group III contained 17, and group IV contained 53 GST pairs. The expression pattern of all duplicated gene pairs in group I, 8 of 10 duplication gene pairs in group II, 16 of 17 in group III, and 15 of 53 in group IV were different. In total, 60 of 126 expressed GST duplicated gene pairs (47.6%) had different expression patterns, indicating that these duplicated GST genes have undergone rapid divergence (Supplementary Table 6). In addition, 37 of 60 (61.7%) gene pairs showed structural divergence.
Of the 31 GST clusters, 29 contained full-length GSTs: Clusters 6 and 9 contained fragments and were excluded from expression analysis. Marked divergence in gene expression was also observed among GST family members (Supplementary Fig. 2). GSTs in Cluster 4, 11, 13, and 16 were not expressed in any tissue. BnGST49 in Cluster 1 and BnGST51 in Cluster 14 were expressed in all tissues, while BnGST53 in Cluster 1 and BnGST54 in Cluster 14 were expressed in all tissues except petal, capillament, stamen, and anther tissue. Some GSTs in Cluster3, 10, 20, 21 and 30 were expressed in all tissues, while other GSTs were barely expressed in any tissue. Among Cluster 15 genes, BnGST31 (GSTF3) was not expressed in petal, capillament, stamen, or anther tissue, while BnGST27 (GSTF2) showed little or no expression during seed development in tissues such as seed, seed coat, episperm, endopleura, cotyledon, and embryo in addition to petal, capillament, stamen, and anther tissue.
Construction of an expression network
Constructing coexpression networks is an effective way to identify clusters of genes with similar functions. In the current study, no module was identified when we analyzed GSTs expressed in tissues at different developmental stages, revealing the divergent functions of GSTs. TFs including NAC, MYB, WRKY and bZIP TFs are thought to regulate the expression of GSTs. To compare the expression patterns of the genes for these TFs and GSTs, six modules were identified using WGCNA with a power of 10. Genes in the turquoise module were expressed during the late stage of seed development (after 40 days), including embryo and radical development. For example, BnGST7 (DHAR3), BnGST123 and BnGST126 (GSTU19), BnGST128 (GSTU20), BnGST131 (GSTU21), and BnGST160 (GSTZ1) shared similar expression patterns with the TF genes NAC32, NAC60, NAC89, WRKY32, and ABI5. BnGST10 and BnGST15 (EF1Bγ1) shared similar expression patterns with WRKY2 and MYB3 (Supplementary Table 8, Fig. 5, Table 1). Genes in the brown module, which are involved in seed coat development, including BnGST42, BnGST43, and BnGST44 (GSTF6), BnGST58 and BnGST61 (GSTF12) and BnGST117 (GSTU17), were coexpressed with MYB5, MYB56, MYB61, MYB118, TTG2, and TT2. BnGST47 (GSTF8), the gene in the green module, which is involved in stamen and anther development, was coexpressed with MYB3, MYB21, MYB101, MYB108, MYB57, MYB78, and MYB117. BnGST129 (GSTU21), the gene in the yellow module, was involved in root and stem development and was coexpressed with MYB103, MYB46, MYB69, MYB85, NAC73, NAC12, and NAC66. Genes in the blue module were primarily expressed in leaves, the funiculus and during the late stage of pericarp development (after 40 days). Most of these GSTs were coexpressed with WRKY TFs. For example, BnGST102, BnGST103, and BnGST105 (GSTU12) and BnGST27 (GSTF2) were coexpressed with WRKY25, WRKY26, and WRKY33. In addition, the expression patterns of BnGST168 and BnGST171 (GST_2_N) and BnGST173 (GSTH1) were similar to that of NAC35. Finally, genes in the red module, which are expressed in the radical development during seed germination, including BnGST64 (GSTF14), BnGST74 and BnGST75 (GSTU1), BnGST156 (GSTU28), and BnGST157 (GSTZ1) shared similar expression patterns with WRKY34, WRKY36, WRKY40, WRKY65, and WRKY72.
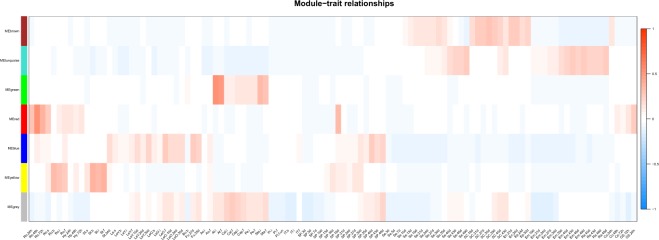
Figure 5.
Module-trait relationships detected by GST and transcription factor (NAC, MYB, WRKY and bZIP) coexpression networks in B. napus. Each row represents a module and each column represents a tissue. The color scale shows the correlation between each module and trait from −1 (blue) to 1 (red). GS, germinate seed; Ro, root; Hy, hypocotyl; St, stem; Ao, anthocaulus; Le, leaf; Bu, bud; Fu, fulicus; Ao, anthocaulus; At, anther; IT, shoot apex; Cal, calyx; Cap, capillament; Pe, petal; Sta, stamen; Pi, pistil; SP, silique; Se, seed; SC, seed coat; Em, embryo; En, endopleura; Ep, episperm; Ra; Co, cotyledon. For stages, s, seedling stage; b, bud stage; i, initial flowering stage; and f, full-bloom stage. The time after seed germination is indicated as 24, 48, and 72 h. The number of days after pollination (DAP) is indicated as 3, 5, 7, 10, 13, 19, 21, 30, 40, and 46 d.
Table 1.
Coexpressed GST and transcription factor (NAC, MYB, WRKY and bZIP) genes in various in B. napus tissues.
| Module | GST Genes | Tissue | TF Genes |
|---|---|---|---|
| Turquoise | BnGST7 (DHAR3), BnGST123 and BnGST126 (GSTU19), BnGST128 (GSTU20), BnGST131 (GSTU21) and BnGST160 (GSTZ1) | Seed | NAC32, NAC60, NAC89, WRKY32, ABI5 |
| BnGST10, BnGST15 (EF1Bγ1) | Seed | WRKY2 and MYB3 | |
| Brown | BnGST42, BnGST43 and BnGST44 (GSTF6), BnGST58 and BnGST61 (GSTF12), BnGST117 (GSTU17) | Seed coat | MYB5, MYB56, MYB61, MYB118, TTG2 and TT2 |
| Green | BnGST47 (GSTF8) | Stamen and anther | MYB3, MYB21, MYB101, MYB108, MYB57, MYB78, MYB117 |
| Yellow | BnGST129 (GSTU21) | Root and stem | MYB103, MYB46, MYB69, MYB85, NAC73, NAC12, NAC66 |
| Blue | BnGST102, BnGST103 and BnGST105 (GSTU12), BnGST27 (GSTF2) | Leaf, funiculus and late-stage pericarp Leaf, funiculus and late-stage pericarp | WRKY25, WRKY26, WRKY33 |
| BnGST168 and BnGST171 (GST_2_N), BnGST173 (GSTH1) | NAC35 | ||
| Red | BnGST64 (GSTF14), BnGST74 and BnGST75 (GSTU1), BnGST156 (GSTU28), BnGST157 (GSTZ1) | Radicle during seed germination | WRKY34, WRKY36, WRKY40, WRKY65, WRKY72 |
We also analyzed the expression patterns of 99 GST duplicated pairs (log2 (FPKM + 1) >4) using WGCNA. In total, 49 of 97 (50.5%) GST duplicated gene pairs were in different modules, which showed that these duplicated gene pairs likely had different functions (Supplementary Table 8). From these results and the correlation analysis of expression levels of GST duplicated gene pairs above, we conclude that GST duplicated gene pairs have divergent roles in the growth and development of B. napus.
Expression of GSTs in response to biotic stress
Blackleg disease (caused by L. maculans) and white stem rot (caused by S. sclerotiorum) are the most serious diseases of B. napus. We therefore examined the expression patterns of the GSTs in response to these pathogens (Supplementary Figs 3 and 4). The most pathogen-responsive genes included BnGST101 and BnGST102 (GSTU12), BnGST26 (GSTF2), BnGST29 (GSTF3), BnGST66 (GSTL1), BnGST81 (GSTU4), BnGST88 (GSTU8), BnGST99 (GSTU11), BnGST110 (GSTU13), BnGST139 (GSTU24), and BnGST141 (GSTU25). In addition, BnGST44 (GSTF6), BnGST58, and BnGST61 (GSTF12) appeared to be important for resistance to L. maculans.
Discussion
Duplication and evolution of GST genes in Brassica
In our study, we identified 179, 85 and 85 full-length GST genes in B. napus, B. oleracea and B. rapa, respectively. Khan et al. (2018) identified 75 GSTs in eight classes for B. rapa, but did not include the mPEGS2, GHR, metaxin, GSTH and GST2N classes24. Vijayakumar et al. (2016) found 65 B. oleracea GSTs that were divided into 11 classes, but did not include the metaxin and GSTH classes25. We have included all 13 GST classes in our analyses, so the numbers of GSTs in B. oleracea and B. rapa were higher than those in previous studies. Whole-genome triplication (WGT) has occurred in Brassica species46. Given that there were 66 GST genes in A. thaliana, after WGT more than 198 GST genes should be found in B. rapa and B. oleracea. In our study, 85 BrGSTs and 85 BoGSTs were found, which indicates that duplicated genes might have been lost after WGT. About 35% genes have been lost via deletion after the divergence of A. thaliana and Brassica47. In total 79 collinear gene pairs were found between A. thaliana and B. rapa, whereas 61 collinear gene pairs were found between A. thaliana and B. oleracea, perhaps due to assembly errors in the currently available B. oleracea genome information. With the release of new B. rapa and B. oleracea reference genomes based on Nanopore technology48, we should be able to detect the retention or loss of gene families more accurately to avoid the false observation due to the incompleteness of the reference genome or annotation errors.
B. napus, an allopolyploid, was formed by hybridization between the diploid progenitors B. rapa and B. oleracea (U 1935). The number of GSTs (179) in B. napus was roughly equal to the sum of B. rapa (85) and B. oleracea (85). The synteny analysis between B. napus and its diploid progenitors B. oleracea and B. rapa indicated that most GST genes in Brassica were located in the syntenic regions, with 79 gene pairs shared between the An subgenome of B. napus and the Ar genome of B. rapa, and 62 gene pairs shared between the Cn subgenome of B. napus and the Co genome of B. oleracea. However, we still find that some GST genes in diploid progenitors were lost. The progenitor species had different numbers of chromosomes, and the A and C subgenomes have undergone rearrangements; thus, genes could be lost in the process of polyploidization.
GSTF6 and GSTF12 are essential for seed coat development
In the current study, coexpression network analysis suggested that BnGSTF6 and BnGSTF12 are involved in seed coat development. Indeed, GSTF12 has been shown to play important roles in seed and fruit coloration in plants. In Arabidopsis, AtGSTF12 (TT19) functions as a carrier that transports anthocyanin from the cytosol to tonoplasts49. Anthocyanin is responsible for the red/purple color of flowers, leaves, fruits and seeds. In strawberry, RAP (Reduced Anthocyanin in Petioles), a homolog of AtGSTF12, alters foliage and fruit color50. Here, we found that the TF genes MYB5, MYB61, MYB118, MYB107, and TT2 (MYB123) were coexpressed with BnGSTF6 and BnGSTF12. These TFs are responsible for seed coat development. Medicago truncatula MYB5 mutants have darker seed coats than wild-type plants51. In Arabidopsis, TT2 is responsible for proanthocyanid in accumulation in developing seeds, as TT2 mutant seed coats are golden yellow52. MYB118 represses endosperm maturation in Arabidopsis seeds53. MYB107 positively regulates suberin synthesis in the seed coat, as myb107 seeds are darker than wild type54. The detailed roles of BnGSTF6, BnGSTF12, and the TFs coexpressed with these GSTs in B. napus should be elucidated in the future.
GSTs plays pleiotropic roles in plants
In addition to its role in seed coat development, BnGSTF12 appears to play an important role in the response of B. napus to pathogen attack. GSTF12 transcripts accumulate in A. thaliana in response to Verticillium dahliae infection55. GSTs play pleiotropic roles in improving plant tolerance to adverse environment conditions. Hodgkin56 defined seven types of pleiotropy and their underlying mechanisms, including artefactual pleiotropy, secondary pleiotropy, adoptive pleiotropy, parsimonious pleiotropy, opportunistic pleiotropy, combination pleiotropy and unifying pleiotropy. In poplar, alternative splicing at the REVOLUTA locus is responsible for the pleiotropic effects of this TF, which is associated with fungal resistance, leaf drop, and cellulose content57. In combination pleiotropy, a gene interacts with a variety of partners, which could alter its biochemical activity56. Gene duplication and the mutation of its regulatory partners play a leading role in pleiotropy58. In Arabidopsis, CBF1 regulates UGT79B2 and UGT79B3 in response to low temperatures, and other TFs might regulate UGT79B2 in response to other environmental conditions59. In rice, the TF IDEAL PLANT ARCHITECTURE 1 (IPA1) enhances yield by increasing grains per panicle through activating the DEP1 promoter and enhances immunity through binding to WRKY4560,61. Indeed, the promoter regions of the BnGSTFs include many cis-elements associated with abiotic and biotic stress responses. The pleiotropic roles of BnGSTF12 in seed development and biotic stress tolerance might be regulated by different TFs. Due to the lack of an adequate number of samples, we were unable to perform coexpression analysis of genes involved in disease resistance.
Expressional divergence of duplicate GST genes
Polyploidy and WGD are widespread in nature and are considered to be the main forces driving speciation and plant evolution. Most duplicated genes have arisen from WGD; studying the fate of duplicated genes is important for understanding plant evolution62. In the current study, 141 duplicate gene pairs were identified, including 10 containing pseudogenes. Duplicate genes produced by WGD are often lost or nonfunctional63. The Ka/Ks values of all duplicate genes except BnaGST129 and BnaGST139 were <1, indicating that the duplicate BnaGST genes have undergone extensive purifying selection. This idea is consistent with the finding that retained duplicate genes have experienced strong purifying selection64,65.
Functional and expressional divergence are important properties of retained duplicate genes. Indeed, 60.3% of full-length duplicated BnGST genes and most gene clusters showed divergent expression patterns, indicating that the duplicate gene pairs have undergone subfunctionalization or neofunctionalization. Expression bias or divergence has been observed in cotton (Gossypium raimondii), as approximately 93% of gene pairs are differentially expressed on a tissue basis64. Divergent expression between duplicates can arise due to the presence of different cis-regulatory elements63 or divergent microRNA binding sites66.
Epigenetic modification also has an important effect on expressional variation67. Structural divergence (differences in exon-intron structure) are prevalent in duplicate genes and can generate proteins with distinct biochemical functions68. A significant relationship between expressional and structural divergence has been observed in the bovine (Bos taurus) genome69. In the current study, 64.6% of duplicate gene pairs exhibited structural divergence, which might play an important role in the evolution of duplicate genes.
GSTs play multiple roles in plant development and stress responses. These genes could have important biotechnological applications through gene pyramiding and co-engineering70. A GST gene in maize was found to be associated with resistance to three plant diseases71. Ten GST genes were significantly upregulated after infection by Botryosphaeria dothidea in poplar72. Therefore, GSTs could be widely used for the improvement of disease resistance in plants. The overaccumulation of flavonoids can enhance tolerance to multiple stresses73: the Arabidopsis ugt79b2/b3 double mutant exhibits reduced anthocyanin accumulation and increased sensitivity to stress59. Therefore, novel plant lines could be produced that overexpress GST genes. Perhaps novel crop varieties with enhanced biotic and abiotic stress tolerance could be generated through the overexpression of GSTs, likely leading to increased crop production.
Main Conclusion
Coexpression network analysis of GSTs and genes encoding various transcription factors (NAC, MYB, WRKY and bZIP) points to different roles of GST during development in B. napus.
Supplementary information
Supplementary Figure 1 Phylogenetic tree of A. thaliana (At), B. oleracea (Bo), B. rapa (Br), and B. napus (Bn) GST sequences
Supplementary Figure 2 Expression patterns of BnGST clusters in 21 different tissues at different developmental stages in B. napus
Supplementary Figure 3 Expression patterns of BnGSTs in response to Leptosphaeria maculans in B. napus
Supplementary Figure 4 Expression patterns of BnGSTs in response to Sclerotinia sclerotiorum in B. napus
Supplementary Table 1 GST protein sequences used to identify GST candidates in B. napus
Supplementary Table 2 GST protein identified in Brassica species
Supplementary Table 3 GST fragments identified in Brassica species
Supplementary Table 4 GST orthologous genes in A. thaliana, B. napus, B. oleracea and B.rapa
Supplementary Table 5 Gene duplication pattern in B. napus, B. oleracea and B. rapa
Supplementary Table 6 The Ka/Ks values, duplicaiton time, gene structures and expresion patterns for duplicated GST gene pairs in B. napus
Supplementary Table 7 Cis-elements of GSTs in B. napus
Supplementary Table 8 Coexpressed genes in various B. napus tissues
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31701460), National Postdoctoral Program for Innovative Talents (BX201700201), China Postdoctoral Science Foundation Funded Project (2017M622945) and the “111” Project (B12006).
Author Contributions
Conceived and designed the research: J.L. Performed the experiments: Y.Z., R.L. and M.Z. Analyzed the data: L.W., Y.Z., R.L. and A.L. Drew the graph: A.Z., W.X. Modified the manuscript: K.L., J.L. Wrote the paper: L.W.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lijuan Wei, Yan Zhu and Ruiying Liu contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45744-5.
References
- 1.Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. The Biochemical journal. 2001;360:1–16. doi: 10.1042/bj3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dixon DP, Edwards R. Glutathione transferases. The Arabidopsis book. 2010;8:e0131. doi: 10.1199/tab.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu YJ, Han XM, Ren LL, Yang HL, Zeng QY. Functional divergence of the glutathione S-transferase supergene family in Physcomitrella patens reveals complex patterns of large gene family evolution in land plants. Plant physiology. 2013;161:773–786. doi: 10.1104/pp.112.205815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan C, Lam HM. A putative lambda class glutathione S-transferase enhances plant survival under salinity stress. Plant & cell physiology. 2014;55:570–579. doi: 10.1093/pcp/pct201. [DOI] [PubMed] [Google Scholar]
- 5.Lallement PA, et al. Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants. Biochemical Journal. 2014;462:39–52. doi: 10.1042/Bj20140390. [DOI] [PubMed] [Google Scholar]
- 6.Lallement PA, Brouwer B, Keech O, Hecker A, Rouhier N. The still mysterious roles of cysteine-containing glutathione transferases in plants. Frontiers in pharmacology. 2014;5:192. doi: 10.3389/fphar.2014.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards, R. & Dixon, D. P. The role of glutathione transferases in herbicide metabolism. In: Cobb, A. H. & Kirkwood, R. C., editors. Herbicides and their mechanisms of action. Sheffield, UK: Sheffield Academic Press 1, 38–71 (2000).
- 8.Kitamura S, Shikazono N, Tanaka A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant Journal. 2004;37:104–114. doi: 10.1046/j.1365-313X.2003.01943.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J. Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci. 2015;20:576–585. doi: 10.1016/j.tplants.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Marrs KA, Alfenito MR, Lloyd AM, Walbot V. A glutathione-S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature. 1995;375:397–400. doi: 10.1038/375397a0. [DOI] [PubMed] [Google Scholar]
- 11.Smith AP, et al. Arabidopsis AtGSTF2 is regulated by ethylene and auxin, and encodes a glutathione S-transferase that interacts with flavonoids. Plant Journal. 2003;36:433–442. doi: 10.1046/j.1365-313X.2003.01890.x. [DOI] [PubMed] [Google Scholar]
- 12.Gonneau J, Mornet R, Laloue M. A Nicotiana plumbaginifolia protein labeled with an azido cytokinin agonist is a glutathione S-transferase. Physiol Plantarum. 1998;103:114–124. doi: 10.1034/j.1399-3054.1998.1030114.x. [DOI] [Google Scholar]
- 13.Dixon DP, et al. Binding and glutathione conjugation of porphyrinogens by plant glutathione transferases. The Journal of biological chemistry. 2008;283:20268–20276. doi: 10.1074/jbc.M802026200. [DOI] [PubMed] [Google Scholar]
- 14.Dixon DP, Edwards R. Selective binding of glutathione conjugates of fatty acid derivatives by plant glutathione transferases. The Journal of biological chemistry. 2009;284:21249–21256. doi: 10.1074/jbc.M109.020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen Mi, Zhao Da-Ke, Qiao Qin, Liu Lei, Wang Jun-Ling, Cao Guan-Hua, Li Tao, Zhao Zhi-Wei. Identification of Glutathione S-Transferase (GST) Genes from a Dark Septate Endophytic Fungus (Exophiala pisciphila) and Their Expression Patterns under Varied Metals Stress. PLOS ONE. 2015;10(4):e0123418. doi: 10.1371/journal.pone.0123418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sappl PG, et al. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. The Plant journal: for cell and molecular biology. 2009;58:53–68. doi: 10.1111/j.1365-313X.2008.03761.x. [DOI] [PubMed] [Google Scholar]
- 17.Gunning V, et al. Arabidopsis glutathione transferases U24 and U25 exhibit a range of detoxification activities with the environmental pollutant and explosive, 2,4,6-trinitrotoluene. Plant physiology. 2014;165:854–865. doi: 10.1104/pp.114.237180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, et al. Transgenic Arabidopsis plants expressing tomato glutathione S-transferase showed enhanced resistance to salt and drought Stress. PloS one. 2015;10:e0136960. doi: 10.1371/journal.pone.0136960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma R, Sahoo A, Devendran R, Jain M. Over-expression of a rice tau class glutathione s-transferase gene improves tolerance to salinity and oxidative stresses in Arabidopsis. PloS one. 2014;9:e92900. doi: 10.1371/journal.pone.0092900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu HJ, et al. Divergence in enzymatic activities in the soybean GST supergene family provides new insight into the evolutionary dynamics of whole-genome duplicates. Molecular biology and evolution. 2015;32:2844–2859. doi: 10.1093/molbev/msv156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan T, et al. Extensive functional diversification of the populus glutathione S-transferase supergene family. The Plant cell. 2009;21:3749–3766. doi: 10.1105/tpc.109.070219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding, N. et al. Identification and analysis of glutathione S-transferase gene family in sweet potato reveal divergent GST-mediated networks in aboveground and underground tissues in response to abiotic stresses. BMC plant biology17, 10.1186/S12870-017-1179-Z (2017). [DOI] [PMC free article] [PubMed]
- 23.Chi YH, et al. Expansion Mechanisms and Functional Divergence of the Glutathione S-Transferase Family in Sorghum and Other Higher Plants. DNA Res. 2011;18:1–16. doi: 10.1093/dnares/dsq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan N, Hu CM, Amjad Khan W, Hou X. Genome-wide identification, classification, and expression divergence of glutathione-transferase family in Brassica rapa under multiple hormone treatments. BioMed research international. 2018;2018:6023457. doi: 10.1155/2018/6023457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vijayakumar Harshavardhanan, Thamilarasan Senthil, Shanmugam Ashokraj, Natarajan Sathishkumar, Jung Hee-Jeong, Park Jong-In, Kim HyeRan, Chung Mi-Young, Nou Ill-Sup. Glutathione Transferases Superfamily: Cold-Inducible Expression of Distinct GST Genes in Brassica oleracea. International Journal of Molecular Sciences. 2016;17(8):1211. doi: 10.3390/ijms17081211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He G, et al. Genome-wide analysis of the glutathione S-transferase gene family in Capsella rubella: identification, expression, and biochemical functions. Frontiers in plant science. 2016;7:1325. doi: 10.3389/fpls.2016.01325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang XW, et al. The genome of the mesopolyploid crop species Brassica rapa. Nature genetics. 2011;43:1035–U1157. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 28.Parkin IA, et al. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome biology. 2014;15:R77. doi: 10.1186/gb-2014-15-6-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chalhoub B, et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 30.Jain M, Ghanashyam C, Bhattacharjee A. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC genomics. 2010;11:73. doi: 10.1186/1471-2164-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, Xia R, Chen H, He Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv. 2018 doi: 10.1101/289660. [DOI] [Google Scholar]
- 32.Nakamura T, Yamada KD, Tomii K, Katoh K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 2018;34:2490–2492. doi: 10.1093/bioinformatics/bty121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic acids research. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu HJ, et al. Divergence in enzymatic activities in the soybean GST supergene family provides new insight into the evolutionary dynamics of whole-genome duplicates. Molecular biology and evolution. 2015;32:2844–2859. doi: 10.1093/molbev/msv156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics, proteomics & bioinformatics. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Molecular biology and evolution. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 38.Lescot M, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic acids research. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic acids research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei LJ, et al. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol J. 2016;14:1368–1380. doi: 10.1111/pbi.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becker MG, et al. Transcriptome analysis of the Brassica napus-Leptosphaeria maculans pathosystem identifies receptor, signaling and structural genes underlying plant resistance. The Plant journal: for cell and molecular biology. 2017;90:573–586. doi: 10.1111/tpj.13514. [DOI] [PubMed] [Google Scholar]
- 43.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Armstrong LC, Komiya T, Bergman BE, Mihara K, Bornstein P. Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. The Journal of biological chemistry. 1997;272:6510–6518. doi: 10.1074/jbc.272.10.6510. [DOI] [PubMed] [Google Scholar]
- 46.Cheng, F., Wu, J. & Wang, X. W. Genome triplication drove the diversification of Brassica plants. Hortic Res-England1, 10.1038/Hortres.2014.24 (2014). [DOI] [PMC free article] [PubMed]
- 47.Town CD, et al. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. The Plant cell. 2006;18:1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belser C, et al. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat Plants. 2018;4:879–+. doi: 10.1038/s41477-018-0289-4. [DOI] [PubMed] [Google Scholar]
- 49.Sun Y, Li H, Huang JR. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Molecular plant. 2012;5:387–400. doi: 10.1093/mp/ssr110. [DOI] [PubMed] [Google Scholar]
- 50.Luo H, et al. Reduced Anthocyanins in Petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. Journal of experimental botany. 2018;69:2595–2608. doi: 10.1093/jxb/ery096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu C, Jun JH, Dixon RA. MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula. Plant physiology. 2014;165:1424–1439. doi: 10.1104/pp.114.241877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. The Plant cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barthole G, et al. MYB118 represses endosperm maturation in seeds of Arabidopsis. The Plant cell. 2014;26:3519–3537. doi: 10.1105/tpc.114.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gou M, et al. The MYB107 transcription factor positively regulates suberin biosynthesis. Plant physiology. 2017;173:1045–1058. doi: 10.1104/pp.16.01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pantelides IS, Tjamos SE, Paplomatas EJ. Ethylene perception via ETR1 is required in Arabidopsis infection by Verticillium dahliae. Molecular plant pathology. 2010;11:191–202. doi: 10.1111/j.1364-3703.2009.00592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodgkin J. Seven types of pleiotropy. The International journal of developmental biology. 1998;42:501–505. [PubMed] [Google Scholar]
- 57.Porth I, et al. Extensive functional pleiotropy of REVOLUTA substantiated through forward genetics. Plant physiology. 2014;164:548–554. doi: 10.1104/pp.113.228783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guillaume F, Otto SP. Gene functional trade-offs and the evolution of pleiotropy. Genetics. 2012;192:1389–1409. doi: 10.1534/genetics.112.143214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li P, et al. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant Journal. 2017;89:85–103. doi: 10.1111/tpj.13324. [DOI] [PubMed] [Google Scholar]
- 60.Wang J, et al. A single transcription factor promotes both yield and immunity in rice. Science. 2018;136:1026–1028. doi: 10.1126/science.aat7675. [DOI] [PubMed] [Google Scholar]
- 61.Lu Z, et al. Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. The Plant cell. 2013;25:3743–3759. doi: 10.1105/tpc.113.113639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panchy N, Lehti-Shiu M, Shiu SH. Evolution of Gene Duplication in Plants. Plant physiology. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zou Cheng, Lehti-Shiu Melissa D., Thomashow Michael, Shiu Shin-Han. Evolution of Stress-Regulated Gene Expression in Duplicate Genes of Arabidopsis thaliana. PLoS Genetics. 2009;5(7):e1000581. doi: 10.1371/journal.pgen.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Renny-Byfield S, et al. Ancient gene duplicates in Gossypium (cotton) exhibit near-complete expression divergence. Genome biology and evolution. 2014;6:559–571. doi: 10.1093/gbe/evu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 66.Wang S, Adams KL. Duplicate gene divergence by changes in microRNA binding sites in Arabidopsis and Brassica. Genome biology and evolution. 2015;7:646–655. doi: 10.1093/gbe/evv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annual review of plant biology. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu G, Guo C, Shan H, Kong H. Divergence of duplicate genes in exon-intron structure. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1187–1192. doi: 10.1073/pnas.1109047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liao X, et al. Sequence, structural and expression divergence of duplicate genes in the bovine genome. PloS one. 2014;9:e102868. doi: 10.1371/journal.pone.0102868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nianiou-Obeidat I, et al. Plant glutathione transferase-mediated stress tolerance: functions and biotechnological applications. Plant cell reports. 2017;36:791–805. doi: 10.1007/s00299-017-2139-7. [DOI] [PubMed] [Google Scholar]
- 71.Wisser RJ, et al. Multivariate analysis of maize disease resistances suggests a pleiotropic genetic basis and implicates a GST gene. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7339–7344. doi: 10.1073/pnas.1011739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liao WH, et al. Identification of glutathione S-transferase genes responding to pathogen infestation in Populus tomentosa. Funct Integr Genomic. 2014;14:517–529. doi: 10.1007/s10142-014-0379-y. [DOI] [PubMed] [Google Scholar]
- 73.Nakabayashi R, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant Journal. 2014;77:367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Phylogenetic tree of A. thaliana (At), B. oleracea (Bo), B. rapa (Br), and B. napus (Bn) GST sequences
Supplementary Figure 2 Expression patterns of BnGST clusters in 21 different tissues at different developmental stages in B. napus
Supplementary Figure 3 Expression patterns of BnGSTs in response to Leptosphaeria maculans in B. napus
Supplementary Figure 4 Expression patterns of BnGSTs in response to Sclerotinia sclerotiorum in B. napus
Supplementary Table 1 GST protein sequences used to identify GST candidates in B. napus
Supplementary Table 2 GST protein identified in Brassica species
Supplementary Table 3 GST fragments identified in Brassica species
Supplementary Table 4 GST orthologous genes in A. thaliana, B. napus, B. oleracea and B.rapa
Supplementary Table 5 Gene duplication pattern in B. napus, B. oleracea and B. rapa
Supplementary Table 6 The Ka/Ks values, duplicaiton time, gene structures and expresion patterns for duplicated GST gene pairs in B. napus
Supplementary Table 7 Cis-elements of GSTs in B. napus
Supplementary Table 8 Coexpressed genes in various B. napus tissues