Abstract
Chronic pain is highly prevalent worldwide and represents a significant socioeconomic and public health burden. Several aspects of chronic pain, for example back pain and a severity-related phenotype ‘chronic pain grade’, have been shown previously to be complex heritable traits with a polygenic component. Additional pain-related phenotypes capturing aspects of an individual’s overall sensitivity to experiencing and reporting chronic pain have also been suggested as a focus for investigation. We made use of a measure of the number of sites of chronic pain in individuals within the UK general population. This measure, termed Multisite Chronic Pain (MCP), is a complex trait and its genetic architecture has not previously been investigated. To address this, we carried out a large-scale genome-wide association study (GWAS) of MCP in ~380,000 UK Biobank participants. Our findings were consistent with MCP having a significant polygenic component, with a Single Nucleotide Polymorphism (SNP) heritability of 10.2%. In total 76 independent lead SNPs at 39 risk loci were associated with MCP. Additional gene-level association analyses identified neurogenesis, synaptic plasticity, nervous system development, cell-cycle progression and apoptosis genes as enriched for genetic association with MCP. Genetic correlations were observed between MCP and a range of psychiatric, autoimmune and anthropometric traits, including major depressive disorder (MDD), asthma and Body Mass Index (BMI). Furthermore, in Mendelian randomisation (MR) analyses a causal effect of MCP on MDD was observed. Additionally, a polygenic risk score (PRS) for MCP was found to significantly predict chronic widespread pain (pain all over the body), indicating the existence of genetic variants contributing to both of these pain phenotypes. Overall, our findings support the proposition that chronic pain involves a strong nervous system component with implications for our understanding of the physiology of chronic pain. These discoveries may also inform the future development of novel treatment approaches.
Author summary
Chronic pain is common worldwide and imposes a significant burden from a public health and socioeconomic perspective. The reasons why some individuals develop chronic pain and others do not are not fully understood. In this study we searched for genetic variants associated with chronic pain in a large general-population cohort. We also assessed how this genetic variation was correlated with a range of other diseases and traits, such as depression and BMI, and we tested for causal relationships between depression and chronic pain. We found that chronic pain was associated with several genes involved in brain function and development and was correlated with mental health and autoimmune traits (including depression, PTSD and asthma). We also found evidence for causal relationships between chronic pain and major depressive disorder. This work provides new insights into the genetics and underlying biology of chronic pain and may help to inform new treatment strategies.
Introduction
Chronic pain, conventionally defined as pain lasting longer than 3 months, has high global prevalence (~30%; [1]), imposes a significant socioeconomic burden, and contributes to excess mortality [2,3]. It is often associated with both specific and non-specific medical conditions such as cancers, HIV/AIDS, fibromyalgia and musculoskeletal conditions [4–6], and can be classified according to different grading systems, such as the Von Korff chronic pain grade [7]. Several aspects of chronic pain, such as chronic pain grade and back pain, have been studied from the genetic point of view, and several have been shown to be complex traits with moderate heritability [3,8]. In part due to the heterogeneity of pain assessment and pain experience, there are very few large-scale genetic studies of chronic pain and no genome-wide significant genetic variants have yet been identified [9,10].
Chronic pain and chronic pain disorders are often comorbid with psychiatric and neurodevelopmental disorders, including Major Depressive Disorder (MDD) [11]. The immune and nervous systems play a central role in chronic pain development and maintenance [12,13]. Similarly, obesity and chronic pain are often comorbid, with extrinsic factors such as sleep disturbance also impacting on chronic pain [14,15]. Altered sleep quality and reduced circadian rhythmicity are also common in those with chronic pain [16]. Chronic pain is also a common component of many neurological diseases [17].
The relationship between injury and other peripheral insult, consequent acute pain and the subsequent development of chronic pain has not been fully explained. Not everyone who undergoes major surgery or is badly injured will develop chronic pain, for example [18], and the degree of joint damage in osteoarthritis is not related to chronic pain severity [19]. Conversely, Complex Regional Pain Syndrome (CRPS) can be incited by minor peripheral insult such as insertion of a needle (reviewed by Denk, McMahon and Tracey, 2014). Structural and functional changes in the brain and spinal cord are associated with the development and maintenance of chronic pain, and affective brain regions are involved in chronic pain perception (this is in contrast to acute pain and even to prolonged acute pain experience) [20–24]. It is also unlikely that there are legitimate cut-off points or thresholds for localised and widespread chronic pain, with pain instead existing on a “continuum of widespreadness” [25]. It may, therefore, be more valuable and powerful to examine measures of chronic pain as complex neuropathological traits in themselves, rather than just to study disorders and conditions with chronic pain as a main feature or pain experienced only in specific bodily locations. Our aim in this study was predicated on the idea that predisposing biological processes might influence how many sites are affected in individuals that experience any chronic pain, and we carried out a genome-wide association study of number of chronic pain sites to look for predisposing loci, assess the degree of genetic overlap with related traits and disorders and generate insights into the genetic architecture of chronic pain.
Results
Genome-wide association study
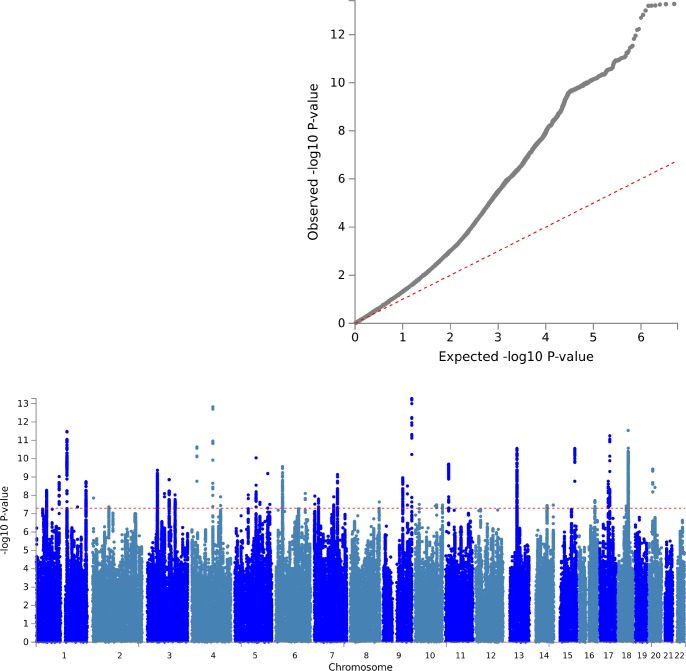
To identify genetic risk loci influencing Multisite Chronic Pain (MCP), we performed a GWAS with adjustment for age, sex and genotyping array using BOLT-LMM (see Methods). No evidence was found for inflation of the test statistics due to hidden population stratification (λGC = 1.26; after adjustment for sample size λGC1000 = 1.001). LD-score regression (LDSR) analysis was consistent with a polygenic contribution to MCP (LDSR intercept = 1.0249, SE 0.0274; Fig 1) [26] and yielded a Single Nucleotide Polymorphism (SNP) heritability estimate of 10.2%. BOLT-LMM gave a similar SNP heritability estimate (pseudo-h2 = 10.3%). In total, 1, 748 SNPs associated with MCP level at genome-wide significance (p < 5 x 10−8) were identified. Conditional analysis of the association signals at each locus revealed 76 independent genome-wide significant lead SNPs across 39 risk loci located on chromosomes 1–11, 13–18 and 20 (Table 1). Sensitivity analysis additionally adjusting for BMI did not significantly alter these association analysis results.
Fig 1. Manhattan plot & QQ plot for MCP GWAS.
A: SNP associations across chromosomes 1–22 are displayed. Genome-wide significance (a p value of 5 x 10−8, ~ 7.3 on the -log10 scale) is indicated by the dashed red line. B: Observed versus expected GWAS p values on the -log10 scale are shown.
Table 1. Genomic risk loci.
| Genomic Locus | rsID | chr | pos | Nearest Gene | A1 | A2 | MAF | r2 | beta | se | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs10888692 | 1 | 50991473 | FAF1 | C | G | 0.4301 | 1 | -0.0143 | 0.0025 | 5.30E-09 |
| 2 | rs197422 | 1 | 112000000 | KCND3 | C | A | 0.3794 | 1 | -0.015 | 0.0025 | 2.00E-09 |
| 3 | rs59898460 | 1 | 150000000 | LINC00568 | T | C | 0.4044 | 1 | 0.0169 | 0.0025 | 9.20E-12 |
| 4 | rs12071912 | 1 | 243000000 | RP11-261C10.3 | C | T | 0.3163 | 1 | -0.0153 | 0.0026 | 5.30E-09 |
| 5 | rs4852567 | 2 | 80703379 | CTNNA2 | A | G | 0.2834 | 1 | 0.0149 | 0.0027 | 4.30E-08 |
| 6 | rs7628207 | 3 | 49754970 | RNF123:AMIGO3:GMPPB | T | C | 0.1766 | 1 | 0.0195 | 0.0032 | 8.40E-10 |
| 7 | rs28428925 | 3 | 107000000 | BBX | G | A | 0.1365 | 1 | -0.0214 | 0.0035 | 1.40E-09 |
| 8 | rs6770476 | 3 | 136000000 | STAG1 | C | T | 0.289 | 1 | -0.0154 | 0.0027 | 9.40E-09 |
| 9 | rs34811474 | 4 | 25408838 | ANAPC4 | G | A | 0.2285 | 1 | 0.0192 | 0.0029 | 2.70E-11 |
| 10 | rs13135092 | 4 | 103000000 | SLC39A8 | A | G | 0.08071 | 1 | -0.0328 | 0.0044 | 1.50E-13 |
| 11 | rs13136239 | 4 | 141000000 | MAML3 | G | A | 0.3508 | 1 | 0.0141 | 0.0026 | 3.60E-08 |
| 12 | rs6869446 | 5 | 65570607 | RP11-305P14.1 | T | C | 0.3861 | 1 | -0.0144 | 0.0025 | 9.50E-09 |
| 13 | rs1976423 | 5 | 104000000 | RP11-6N13.1 | A | C | 0.4968 | 1 | -0.014 | 0.0024 | 8.20E-09 |
| 14 | rs17474406 | 5 | 123000000 | CEP120 | G | A | 0.01805 | 1 | -0.0492 | 0.0088 | 2.40E-08 |
| 15 | rs1946247 | 5 | 161000000 | GABRB2 | T | G | 0.1389 | 1 | -0.019 | 0.0035 | 4.90E-08 |
| 16 | rs11751591 | 6 | 33794215 | MLN | G | A | 0.1516 | 1 | 0.0214 | 0.0034 | 2.70E-10 |
| 17 | rs6907508 | 6 | 34592090 | C6orf106 | A | G | 0.1146 | 1 | -0.0217 | 0.0038 | 1.10E-08 |
| 18 | rs6926377 | 6 | 145000000 | UTRN | A | C | 0.294 | 1 | -0.0155 | 0.0027 | 7.90E-09 |
| 19 | rs10259354 | 7 | 3487414 | SDK1 | G | A | 0.2983 | 1 | 0.0147 | 0.0026 | 3.00E-08 |
| 20 | rs7798894 | 7 | 21552995 | SP4 | A | T | 0.2888 | 1 | 0.0153 | 0.0027 | 1.60E-08 |
| 21 | rs6966540 | 7 | 95727967 | DYNC1I1 | T | C | 0.3762 | 1 | -0.0139 | 0.0025 | 3.30E-08 |
| 22 | rs12537376 | 7 | 114000000 | FOXP2 | A | G | 0.3969 | 1 | 0.0151 | 0.0025 | 1.70E-09 |
| 23 | rs11786084 | 8 | 143000000 | AC138647.1 | G | A | 0.3328 | 1 | -0.0145 | 0.0026 | 2.30E-08 |
| 24 | rs10992729 | 9 | 96181075 | Y_RNA | C | T | 0.3344 | 1 | 0.0158 | 0.0026 | 1.10E-09 |
| 25 | rs6478241 | 9 | 119000000 | ASTN2 | A | G | 0.365 | 1 | 0.0149 | 0.0025 | 3.10E-09 |
| 26 | 9:140251458_G_A | 9 | 140000000 | EXD3 | G | A | 0.123 | 1 | -0.0277 | 0.0037 | 5.30E-14 |
| 27 | rs2183271 | 10 | 21957229 | MLLT10 | T | C | 0.3578 | 1 | -0.014 | 0.0025 | 3.10E-08 |
| 28 | rs11599236 | 10 | 106000000 | SORCS3 | T | C | 0.4058 | 1 | 0.0138 | 0.0025 | 3.30E-08 |
| 29 | rs12765185 | 10 | 135000000 | KNDC1 | T | A | 0.2669 | 1 | -0.0151 | 0.0027 | 3.90E-08 |
| 30 | rs61883178 | 11 | 16317779 | SOX6 | C | A | 0.1696 | 1 | -0.0208 | 0.0033 | 2.00E-10 |
| 31 | rs1443914 | 13 | 53917230 | AL450423.1 | T | C | 0.475 | 1 | 0.0162 | 0.0024 | 2.80E-11 |
| 32 | rs12435797 | 14 | 73797669 | NUMB | G | T | 0.1859 | 1 | -0.0173 | 0.0031 | 3.70E-08 |
| 33 | rs2006281 | 14 | 104000000 | CTD-2134A5.4 | C | T | 0.4981 | 1 | 0.0135 | 0.0024 | 3.40E-08 |
| 34 | rs2386584 | 15 | 91539572 | PRC1 | T | G | 0.3835 | 1 | -0.0166 | 0.0025 | 2.80E-11 |
| 35 | rs285026 | 16 | 77100089 | MON1B | G | T | 0.4297 | 1 | -0.0138 | 0.0025 | 1.90E-08 |
| 36 | rs11871043 | 17 | 43172849 | NMT1 | T | C | 0.4213 | 1 | 0.0149 | 0.0025 | 1.70E-09 |
| 37 | rs11079993 | 17 | 50301552 | snoZ178 | G | T | 0.3825 | 1 | -0.0173 | 0.0025 | 5.70E-12 |
| 38 | rs62098013 | 18 | 50863861 | DCC | G | A | 0.3631 | 1 | -0.0169 | 0.0026 | 4.00E-11 |
| 39 | rs2424248 | 20 | 19650324 | SLC24A3 | G | A | 0.1255 | 1 | 0.023 | 0.0037 | 3.70E-10 |
Genomic risk loci are as defined by FUMA. Genomic Locus = numeric label (1–39), rsID = SNP rsID label, chr = chromosome, pos = position in base-pairs, Nearest Gene = nearest mapped gene, A1 = effect allele, A2 = non-effect allele, MAF = minor allele frequency (MAF here refers to A1 frequency as all values are < 0.5 i.e. A1 is the minor allele as well as the effect allele), r2 = imputation r-squared value, beta = association beta value, se = standard error of beta, P = P value of association (GWAS P value).
Post-GWAS analyses including gene expression and gene-level association testing was carried out using FUMA. Gene-level association tests (MAGMA gene-based test) revealed 113 genes across 39 genomic risk loci significantly associated with MCP (S1–S3 Figs), including genes with roles in neuronal adhesion and guidance, regulation of neural development and neurotransmitter receptor function.
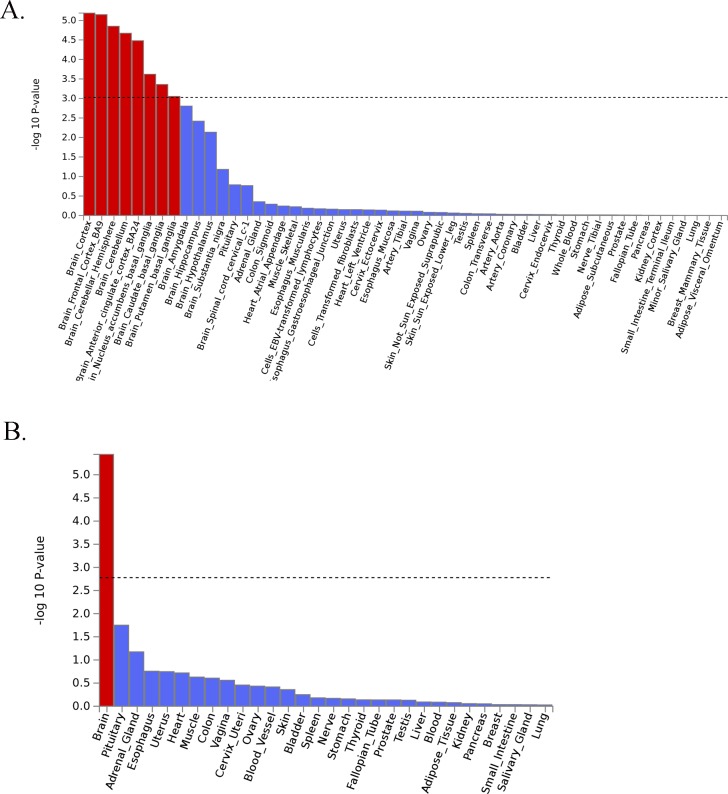
Analysis of Gene Ontology (GO) annotations revealed 3 significant categories (Table 2: Bonferroni-corrected p < 0.05). The significant categories were enriched for terms including neurogenesis and synaptic plasticity, DCC-mediated attractive signalling, neuron projection guidance and central nervous system neuron differentiation, amongst others. Genes of interest (n = 35) designated based on gene-level association tests and on annotation of genes at the identified genomic loci (see S1 Text) are listed in S2 Table. Analysis of tissue-level expression showed significant enrichment of brain-expressed genes, particularly in the cortex and cerebellum (Fig 2),
Table 2. GO annotations.
| Gene Set | N genes | Beta | SE | SE | P | Pbon |
|---|---|---|---|---|---|---|
| GO_bp:go_neuron_projection_guidance | 195 | 0.335 | 0.0341 | 0.0711 | 1.25E-06 | 0.013361 |
| Curated_gene_sets:reactome_dcc_mediated_attractive_signaling | 13 | 1.45 | 0.0381 | 0.313 | 1.94E-06 | 0.020616 |
| GO_bp:go_central_nervous_system_neuron_differentiation | 158 | 0.362 | 0.0331 | 0.0811 | 4.05E-06 | 0.043154 |
Significant GO annotations (ranked by p value) are shown. Beta = beta coefficient value from the FUMA MAGMA gene-set analyses for this Gene Ontology (GO) gene set, SE = standard error of beta, Pbon = Bonferroni-corrected p value. ‘GO_bp’ and ‘Curated_gene_sets’ refers to Gene Ontology categories biological processes and curated gene sets respectively [27].
Fig 2.
A) GteX Output–General Tissues. B) GteX Output–Detailed Tissues. Fig 2A and 2B. GTeX output–General Tissues and Detailed Tissues.
Genetic correlations
Genetic correlations between MCP and 22 traits were estimated via LD-score regression using ldsc [28]. The psychiatric phenotype most significantly genetically correlated with MCP was MDD (Table 3: rg = 0.53, pFDR = 1.69e-78) while the largest significant genetic correlation coefficient was for MCP and depressive symptoms (Table 3: rg = 0.59, pFDR = 6.19e-65). MCP was also positively genetically correlated with neuroticism (Table 3: rg = 0.40), anxiety (Table 3: rg = 0.49), schizophrenia (Table 3:rg = 0.10), cross-disorder psychiatric phenotype (Table 3: rg = 0.13) and PTSD (Table 3: rg = 0.41). Significant negative genetic correlations were observed between MCP and subjective well-being (Table 3: rg = -0.36), autism spectrum disorder (Table 3: ASD; rg = -0.10) and between MCP and anorexia nervosa (Table 3: AN; rg = -0.06). There was no significant genetic correlation between MCP and Bipolar disorder (Table 3: BD; PFDR > 0.05). In relation to the immune-related disorders, rheumatoid arthritis (Table 3: rg = 0.16) and asthma (Table 3: rg = 0.22) were significantly positively genetically correlated with MCP, as was primary biliary cholangitis (Table 3: rg = 0.10), while systemic lupus erythematosus (SLE), ulcerative colitis and Crohn disease were not (PFDR > 0.05). BMI was significantly genetically correlated with MCP (Table 3: rg = 0.31), while low relative amplitude, a circadian rhythmicity phenotype, exhibited a significant negative genetic correlation with MCP (Table 3: rg = -0.30). There was no correlation between Parkinson disease and MCP (PFDR > 0.05). Non-significant genetic correlation results are shown in S3 Table.
Table 3. Genetic correlations between MCP and multiple traits.
| Trait | rg | se | z | h2 | Ph2 (fdr) | source | PMID | Category | p | P (fdr-corrected) |
|---|---|---|---|---|---|---|---|---|---|---|
| MDD | 0.53 | 0.03 | 18.92 | 0.077 | 1.25E-47 | PGC | 29700475 | psychiatric | 7.68E-80 | 1.69E-78 |
| Depressive symptoms | 0.59 | 0.03 | 17.16 | 0.047 | 6.87E-29 | ld_hub | 27089181 | psychiatric | 5.63E-66 | 6.19E-65 |
| BMI | 0.31 | 0.02 | 15.69 | 0.138 | 5.42E-59 | GIANT consortium | 25673413 | anthropometric | 1.90E-55 | 1.39E-54 |
| Neuroticism | 0.4 | 0.03 | 11.9 | 0.089 | 3.66E-26 | ld_hub | 27089181 | personality | 1.24E-32 | 6.82E-32 |
| Subjective well being | -0.36 | 0.04 | -8.94 | 0.025 | 2.77E-32 | ld_hub | 27089181 | psychiatric | 3.78E-19 | 1.66E-18 |
| Low Relative Amplitude | -0.3 | 0.05 | -6.37 | 0.053 | 3.03E-13 | In-house analysis | 30120083 | circadian | 1.91E-10 | 7.00E-10 |
| Rheumatoid Arthritis | 0.16 | 0.03 | 4.7 | 0.160 | 7.41E-08 | ld_hub | 24390342 | autoimmune | 2.64E-06 | 8.30E-06 |
| Anxiety (Case-Control) | 0.49 | 0.11 | 4.53 | 0.081 | 0.00405 | PGC | 26754954 | psychiatric | 5.91E-06 | 1.63E-05 |
| Schizophrenia | 0.1 | 0.03 | 4.08 | 0.443 | 6.56E-79 | PGC | 25056061 | psychiatric | 4.50E-05 | 1.10E-04 |
| Asthma | 0.22 | 0.06 | 3.63 | 0.123 | 3.53E-06 | ld_hub | 17611496 | autoimmune | 3.00E-04 | 6.60E-04 |
| PGC cross-disorder analysis | 0.13 | 0.04 | 3.54 | 0.172 | 7.89E-36 | ld_hub | 23453885 | psychiatric | 4.00E-04 | 8.00E-04 |
| PTSD (European Ancestry) | 0.41 | 0.12 | 3.28 | 0.097 | 0.030855 | PGC | 28439101 | psychiatric | 0.001047 | 1.92E-03 |
| Autism spectrum disorder | -0.1 | 0.04 | -2.22 | 0.451 | 9.38E-17 | ld_hub | NA | psychiatric | 0.026 | 0.0443 |
| Primary biliary cirrhosis | 0.1 | 0.04 | 2.17 | 0.376 | 1.11E-08 | ld_hub | 26394269 | autoimmune | 0.03 | 0.047 |
| Anorexia Nervosa | -0.06 | 0.03 | -2.14 | 0.556 | 2.18E-63 | ld_hub | 24514567 | psychiatric | 0.032 | 0.0471 |
rg = genetic correlation coefficient value, se = standard error of correlation value, z = z value, h2 = SNP-heritability value, ph2(fdr) = p value (FDR-corrected) for SNP-heritability, source = source of GWAS summary statistics, PMID = PubMed ID of associated paper (if applicable), p = p value for genetic correlation coefficient, p(fdr) = FDR-corrected p value for genetic correlation coefficient.
Mendelian randomisation of MCP and major depressive disorder
Mendelian Randomisation with Robust Adjusted Profile Score (MR-RAPS) analysis was performed to investigate causal relationships between MDD and MCP, first with MDD as the exposure and MCP as the outcome. QQ plots, leave-one out versus t-value plots (S4 Fig) and Anderson-Darling/ Shapiro-Wilk test p values indicated that models without dispersion were best-fitting (S4 Table rows 1–3, pAD > 0.05, pSW > 0.05). Effects of outliers (idiosyncratic pleiotropy) are not ameliorated in models with dispersion despite robust regression (S4D, S4E and S4F Fig right-hand panels). The model allowing the greatest amelioration of pleiotropy is one without over-dispersion and with a Tukey loss function (S4 Table: row 3, S4C Fig). This indicates idiosyncratic pleiotropy (pleiotropy in some but not all instruments), i.e. that a subset of instruments may affect MCP through pathways other than via MDD (the exposure). The causal effect of MDD on MCP is positive and significant at beta = 0.019 and p = 0.0006, but the diagnostic plots show a ‘swapping’ of sign for the causal estimate (S4 Fig), suggesting that there is not a truly significant causal effect of MDD on MCP.
MR-RAPS analyses were then carried out with MCP as the exposure and MDD as the outcome. Models with dispersion are a better fit than those without (S5A, S5B, S5C vs S5D, S5E and S5F Fig, S5 Table: rows 4–6, pAD > 0.05, pSW > 0.05, pτ << 0.05). This indicates that effectively all instruments are pleiotropic (affecting MDD through pathways other than via MCP). The causal effect of MCP on MDD is positive and significant at beta = 0.16 and p = 0.047.
Overall, this analysis suggests a causal effect of MCP on MDD.
Relationship between multisite chronic pain and chronic widespread pain
Polygenic Risk Score (PRS) analyses were carried out to examine the relationship between MCP and chronic widespread pain in UK Biobank. Increasing MCP PRS value was significantly associated with having chronic pain all over the body (S6 Table: p = 1.45 x 10−109), with each per-standard-deviation increase in PRS associated with a 63% increase in the odds of having chronic widespread pain.
A secondary GWAS of chronic widespread pain (CWP) was carried out, the results from which were used in LD score regression analysis to determine the genetic correlation between CWP and MCP. This was found to be large (rg = 0.83) and significant (p = 2.45 x 10−54). A lookup analysis was also carried out using the CWP GWAS summary statistics, and >90% of SNPs showed consistent direction of effect between MCP and CWP (S7 Table). In addition, a paired t-test of MCP versus CWP effect values showed that they are not significantly different overall (t = -1.82, p = 0.07).
LocusZoom plots
LocusZoom plots for independent, genome-wide significant loci, calculated according to the supplementary methods detailed in S1 Text, are shown in S6 Fig.
Discussion
We identified 76 independent genome-wide significant SNPs associated with MCP across 39 loci. The genes of interest had diverse functions, but many were implicated in nervous-system development, neural connectivity and neurogenesis.
Genes of interest identified in GWAS of MCP
Potentially interesting genes included DCC (Deleted in Colorectal Cancer a.k.a. DCC netrin 1 receptor) which encodes DCC, the receptor for the guidance cue netrin 1, which is important for nervous-system development [29]. SDK1 (Sidekick Cell Adhesion molecule 1) is implicated in HIV-related nephropathy in humans [30] and synaptic connectivity in vertebrates [31], and ASTN2 (Astrotactin 2) is involved in glial-guided neuronal migration during development of cortical mammalian brain regions [32].
MAML3 (Mastermind-Like Transcriptional coactivator 3) is a key component of the Notch signalling pathway [33,34], which regulates development and maintenance of a range of cell and tissue types in metazoans. During neurogenesis in development the inhibition of Notch signalling by Numb promotes neural differentiation [35]. Numb is encoded by NUMB (Endocytic Adaptor Protein), which was also associated with MCP. In the adult brain Notch signalling has been implicated in CNS plasticity across the lifespan [35].
CTNNA2 (Catenin Alpha 2) encodes a protein involved in cell-cell adhesion [36], found to play a role in synapse morphogenesis and plasticity [37,38]. CEP120 (Centrosomal Protein 120) encodes Cep120, vital for Interkinetic Nuclear Migration (INM) in neural progenitor cells of the cortex [39]. KNDC1 (Kinase Non-Catalytic C-Lobe Domain Containing 1) encodes v-KIND in mice, linked to neural morphogenesis in the cortex [40], and KNDC1 in humans, linked to neuronal dendrite development and cell senescence [41]. SOX6 (SRY-Box 6) is part of the Sox gene family, first characterised in mouse and human testis-determining gene Sry [42] and encoding transcription factors involved in a range of developmental processes [43,44]. SOX6 may be involved in development of skeletal muscle [43], maintenance of brain neural stem cells [45] and cortical interneuron development [46], and variants in this gene have been associated with bone mineral density in both white and Chinese populations [47]. CA10 (Carbonic Anhydrase 10) is predominantly expressed in the CNS, encoding a protein involved in development and maintenance of synapses [48]. DYNC1I1 (Dynein Cytoplasmic 1 Intermediate Chain 1) encodes a subunit of cytoplasmic dynein, a motor protein which plays a role in cargo transport along microtubules, including in the function of neuronal cells [49]. UTRN (Utrophin) is a homologue of Duchenne Muscular Dystrophy gene (DMD), encoding utrophin protein which is localised to the neuromuscular junction (NMJ) [50]. Utrophin has also been implicated in neutrophil activation [51], dystrophin-associated-protein (DPC)-like complex formation in the brain [52], and is expressed during early foetal brain development in neurons and astrocytes [53].
FOXP2 encodes a member of the FOX family of transcription factors, which are thought to regulate expression of hundreds of genes in both adult and foetal tissue, including the brain [54]. These transcription factors may play an important role in brain development, neurogenesis, signal transmission and synaptic plasticity [55]. FOXP2 is essential for normal speech and language development [56]. GABRB2 encodes a GABA (gamma-aminobutyric acid) type A receptor beta subunit. These pentameric chloride channels mediate fast inhibitory synaptic transmission and are extremely important for network function in many brain regions, with the b2 subunit forming part of the most widely expressed receptor across the mammalian brain [57,58].
Another group of genes associated with MCP were linked to cell-cycle progression, DNA replication and apoptosis such as EXD3 (Exonuclease 3’-5’ Domain Containing 3), which encodes a protein involved in maintaining DNA fidelity during replication (‘proof-reading’) [59]. BBX (HMG-Box Containing protein 2) encodes an HMG (high mobility group) box-containing protein necessary for cell-cycle progression from G1 to S phase [60]. STAG1 (Cohesin Subunit SA-1) encodes a cohesin-complex component–cohesin ensures sister chromatids are organised together until prometaphase [61–63]. ANAPC4 (Anaphase Promoting Complex Subunit 4) encodes a protein making up the anaphase promoting complex (APC), an essential ubiquitin ligase for eukaryotic cell-cycle progression [64]. PRC1 (Protein Regulator of Cytokinesis 1) is involved in the regulation of cytokinesis [65], the final stage of the cell cycle. Y RNA (Small Non-Coding RNA, Ro-Associated Y3) encodes a small non-coding Y RNA. These RNAs have been implicated in a wide range of processes, including cell stress response, DNA replication initiation and RNA stability [66]. FAM120A (Oxidative Stress-Associated Src Activator) encodes an RNA-binding protein which regulated Src-kinase activity during oxidative stress-induced apoptosis [67]. The protein encoded by MON1B (MON1 Homolog B, Secretory Trafficking Associated) is necessary for clearance of cell ‘corpses’ following apoptosis, with defects associated with autoimmune pathology [68]. FAF1 (Fas Associated Factor 1) encodes a protein which binds the Fas antigen to initiate or facilitate apoptosis, amongst a wide range of other biological processes (including neuronal cell survival) [69].
Several MCP associated genes have been previously implicated in diseases such as Brugada Syndrome 9 and Spinal ataxia 19 & 22 (KCND3) [70–72], Systemic lupus erythematosus (SLE) (Y RNAs) [66], Joubert syndrome 31 and short-rib thoracic dysplasia 13 (CEP120) [73], Amyotrophic lateral sclerosis (ALS) (FAF1) [74], Urbach-Wiethe disease (ECM1) [75,76], mental retardation and other cohesinopathies such as Cornelia de Lange Syndrome (STAG1) [77,78], split hand/ split foot malformation (DYNC1I1) [79,80], and a wide range of cancers (PRC1) [81]. Other disorders found to involve MCP-related genes include schizophrenia (FOXP2 and GABRB2) [82–88], intellectual disability and epilepsy (GABRB2) [89], and neuroleptic-induced tardive dyskinesia (GABRB2) [90].
Several GWASs of chronic pain at specific body sites, of specific pain types such as neuropathic pain, and of diseases and disorders where chronic pain is a defining symptom, have been carried out previously (reviewed by [10], [91]). DCC and SOX5 (which jointly functions with SOX6 in chondrogenesis) have been associated with chronic back pain [92], GABRB3 (encoding one of three beta subunits of the GABA A receptor along with GABRB2) has been associated with migraine and fibromyalgia [10], and ASTN2 and SLC24A3 have been associated with migraine [10,93]
Overall, this indicated that MCP, a chronic pain phenotype, involves structural and functional changes to the brain, including impact upon neurogenesis and synaptic plasticity both during development and in adulthood. Also implicated was regulation of cell-cycle progression and apoptosis. This is also supported by GO categories DCC-mediated attractive signalling, neuron projection guidance and CNS neuron differentiation being significantly associated with MCP. There was also evidence of pleiotropy, with genes associated with a range of neurodegenerative, psychiatric, developmental and autoimmune disease traits, as well as being associated with MCP.
Genetic correlations
Chronic pain and chronic pain disorders are often comorbid with psychiatric and neurodevelopmental disorders [11]. This has been observed for Major Depressive Disorder (MDD) [8,94], post-traumatic stress-disorder (PTSD) [95–99], schizophrenia [100–102] and bipolar disorder (BD) [94,103]. There are also reported differences in the perception of pain and interoception (sensing and integration of bodily signals) for people with schizophrenia [104,105], anorexia nervosa (AN) [106–108] and autism spectrum disorders (ASD) [109,110], with some evidence of an increase in pain thresholds for AN and ASD.
There is significant cross-talk between the immune system and nervous system in nociception and sensitisation leading to chronic pain [12,13], and many autoimmune disorders cause or have been associated with chronic pain including neuroinflammation implicated in development of neuropathic pain [111].
Similarly, obesity and chronic pain are often comorbid, with extrinstic factors such as MDD and sleep disturbance also impacting on chronic pain [14,15]. Obesity and related chronic inflammation may affect chronic pain [112], and adipose tissue is metabolically active in ways that can affect pain perception and inflammation [113–115].
Sleep changes and loss of circadian rhythm is common in those with chronic pain [16], and myriad chronic diseases, including chronic pain, have shown diurnal patterns in symptom severity, intensity and mortality [116,117]. Chronic pain is also a common component of many neurological diseases, particularly Parkinson’s disease [17], and disorders such as Multiple Sclerosis and migraines are considered neurological in nature.
MCP showed moderate positive genetic correlation with a range of psychiatric disorders including MDD, SCZ, and PTSD, along with traits anxiety and neuroticism. The magnitude of genetic correlation between MCP and MDD was similar to that shown for von Korff chronic pain grade (a chronic pain phenotype) and MDD by McIntosh et al via a mixed-modelling approach (ρ = 0.53) [8]. This is in line with previous observations of association and indicates that shared genetic risk factors exist between MCP and a range of psychiatric disorders, most notably MDD, and that the genetic correlation between MCP and MDD matches with that between MDD and von Korff CPG, a validated chronic-pain questionnaire-derived phenotype [7].
Autoimmune disorders rheumatoid arthritis, asthma and primary biliary cholangitis showed positive genetic correlation with MCP. However, gastrointestinal autoimmune disorders UC, IBD and Crohn’s Disease did not. This suggests separate genetic variation and mechanisms underlying chronic pain associated with these autoimmune disorders compared to those outwith the digestive system. Pain related to inflammatory bowel diseases may represent something less ‘chronic’ and more ‘on-going acute’, as stricture, abscesses and partial or complete obstruction of the small bowel result in pain [118]. Structural and functional brain changes associated with the transition to chronic pain may also play a less central role in gastrointestinal autoimmune disorder-associated pain, due to potential for the enteric nervous system (ENS) to act independently from the CNS, and the role of the gut-brain axis (GBA) [119,120].
There was significant negative genetic correlation between low relative amplitude, a circadian rhythmicity phenotype indicating poor rhythmicity [121]. Opposing direction of effect of genetic variants on MCP versus low RA may mean that insomnia and other sleep difficulties (for which low RA represents a proxy phenotype) associated with MCP are due to environmental and lifestyle factors related to chronic pain, rather than shared genetic factors predisposing to increased risk for both traits. There was also significant negative genetic correlation between MCP and both AN and ASD, which may be linked to changes in interoception and atypical pain experience seen in individuals with these conditions [106–110], and may suggest a genetic basis for increased pain thresholds.
SNP heritability of MCP
LDSR analyses gave a heritability estimate of 10.2% for MCP, lower than the pseudo-h2 estimate of 10.3% given by BOLT-LMM. this suggests SNP-heritability (h2) of MCP to be roughly-10%, slightly lower than an estimate of ‘any chronic pain’ of 16%, and markedly lower than a heritability estimate of 30% for ‘severe chronic pain’ derived from a pedigree-based analyses [3].
Causal associations between MDD and MCP
Mendelian randomisation analyses indicated a causal effect of MCP on MDD, with widespread pleiotropy and a less significant causal estimate value for MCP as the exposure–this suggests most instruments for MCP are pleiotropic, affecting MDD through pathways other than directly through MCP. In contrast, only a small subset of instruments for MDD as the exposure were found to be pleiotropic.
Relationship between MCP and CWP
It has been argued that CWP and other clinical syndromes involving chronic pain all over the body represent the upper end of a spectrum of centralisation of pain, or the extreme of a chronic pain state [122]. It has also been argued that there are not “natural cut-off points” when it comes to chronic widespread pain versus localised chronic pain [25]. In support of this view, the MCP PRS was significantly associated with increased odds of having chronic pain all over the body/ CWP, suggesting that chronic widespread pain may in fact represent the upper end of a spectrum of ‘widespreadness’ of chronic pain, as previously suggested [25,122], and that there are likely to be genetic variants that predispose both to MCP and to CWP.
Conclusions & limitations
Multisite chronic pain (MCP), a chronic pain phenotype defined as the number of sites at which chronic pain is experienced, is a complex trait with moderate heritability. To date, this study represents the largest GWAS of any chronic pain phenotype and elucidates potential underlying mechanisms of chronic pain development. Substantial genetic correlations with a range of psychiatric, personality, autoimmune, anthropometric and circadian traits were identified.
The genes potentially associated with MCP implicated neurogenesis, neuronal development and neural connectivity, along with cell-cycle and apoptotic processes, and expression was primarily within brain tissues. This is in line with theories of functional and structural changes to the brain contributing to development of chronic pain [21,24,123–125], and may also explain the genetic correlations observed. A causal effect of MCP on MDD was identified.
Although the phenotype was based on self-report, this study was very large in size and so likely had sufficient power to detect genetic variation associated with MCP. Replication of SNP associations was not possible due to the nature of chronic pain phenotyping and available cohort sizes, but several genes significantly associated with MCP have been previously associated with chronic pain conditions including chronic back pain, migraine and fibromyalgia, and genetic risk for MCP was found to be significantly associated with chronic widespread pain.
Methods
We carried out a GWAS of Multisite Chronic Pain (MCP), a derived chronic pain phenotype, in 387,649 UK Biobank participants (Table 4). UK Biobank is a general-population cohort of roughly 0.5 million participants aged 40–79 recruited across the UK between 2006 and 2010. Details on phenotyping, follow-up and genotyping have been described in detail elsewhere [126].
Table 4. Demographics of those included in BOLT-LMM GWAS of MCP.
| chronic pain sites | male (N) | female (N) | male (%) | female (%) | age (mean) | total (N) | total (%) |
|---|---|---|---|---|---|---|---|
| 0 | 105474 | 113148 | 48.2 | 51.8 | 56.71 | 218622 | 56.40 |
| 1 | 42734 | 49984 | 46.1 | 53.9 | 57.03 | 92718 | 23.92 |
| 2 | 18612 | 26000 | 41.7 | 58.3 | 57.29 | 44612 | 11.51 |
| 3 | 7771 | 12376 | 38.6 | 61.4 | 57.65 | 20147 | 5.20 |
| 4 | 2970 | 5319 | 35.8 | 64.2 | 57.48 | 8289 | 2.14 |
| 5 | 780 | 1723 | 31.2 | 68.8 | 56.53 | 2503 | 0.65 |
| 6 | 181 | 471 | 27.8 | 72.2 | 56.20 | 652 | 0.17 |
| 7 | 34 | 72 | 32.1 | 67.9 | 56.17 | 106 | 0.03 |
| total | 178556 | 209093 | NA | NA | 56.91 | 387649 | NA |
Phenotype definition and GWAS
During the baseline investigations, UK Biobank participants were asked via a touchscreen questionnaire about “pain types experienced in the last month” (field ID 6159), with possible answers: ‘None of the above’; ‘Prefer not to answer’; pain at seven different body sites (head, face, neck/shoulder, back, stomach/abdomen, hip, knee); or ‘all over the body’. The seven individual body-site pain options were not mutually exclusive and participants could choose as many as they felt appropriate. Where patients reported recent pain at one or more body sites, or all over the body, they were additionally asked (category ID 100048) whether this pain had lasted for 3 months or longer. Those who chose ‘all over the body’ could not also select from the seven individual body sites.
Multisite Chronic Pain (MCP) was defined as the sum of body sites at which chronic pain (at least 3 months duration) was recorded: 0 to 7 sites. Those who answered that they had chronic pain ‘all over the body’ were excluded from the GWAS as there is some evidence that this phenotype relating to widespread pain can be substantially different from more localised chronic pain [94] and should not, therefore, be considered a logical extension of the multisite scale. 10,000 randomly-selected individuals reporting no chronic pain were excluded from the GWAS to use as controls in subsequent polygenic risk score (PRS) analyses.
SNPs with an imputation quality score of less than 0.3, Minor Allele Frequency (MAF) < 0.01 and Hardy-Weinberg equilibrium (HWE) test p < 10−6 were removed from the analyses. Participants whose self-reported sex did not match their genetically-determined sex, those who had putative sex-chromosome aneuploidy, those considered outliers due to missing heterozygosity, those with more than 10% missing genetic data and those who were not of self-reported white British ancestry were excluded from analyses.
An autosomal GWAS was run using BOLT-LMM [127], with the outcome variable, MCP, modelled as a linear quantitative trait under an infinitesimal model, and the model adjusted for age, sex and chip (genotyping array). Related individuals are included and accounted for, as are any population stratification effects, via use of a genetic relatedness matrix as part of the BOLT-LMM analysis [127]. The SNP-level summary statistics from the GWAS output were analysed using FUMA [128], which implements a number of the functions from MAGMA (gene-based association testing, gene-set analyses) [129]. Tissue expression (GTEx) analysis [130] and Gene Ontology [27] and ANNOVAR [131] annotation analysis with default settings was used to characterise lead SNPs further. LocusZoom [132] was used to plot association results at higher resolution (N = 47) (S1 Text). Genomic risk loci were identified using the definition deployed by FUMA [128].
Genetic correlation analysis
Genetic correlations between MCP and 22 complex traits selected on the basis of prior phenotypic association evidence were calculated using linkage disequilibrium score regression (LDSR) analyses [28], implemented either using the ‘ldsc’ package [28] and downloaded publicly-available summary statistics and summary statistics from in-house analyses or using LD Hub [133]. LD Hub datasets from the categories Psychiatric, Personality, Autoimmune and Neurological were selected and datasets with the attached warning note ‘Caution: using this data may yield less robust results due to minor departure from LD structure’ were excluded from the analyses. Where multiple GWAS datasets were available for the same trait, the one with the largest sample size and/or European ancestry was retained with priority given to European ancestry.
Mendelian randomisation analysis of MCP and major depressive disorder
Mendelian randomisation analysis was carried out with MR-RAPS (MR-Robust Adjusted Profile Score; [134] using the R package ‘mr-raps’. This method is appropriate when doing MR analysis of phenotypes that are moderately genetically correlated and likely to share some pleiotropic risk loci. MDD was chosen for MR analysis as this disorder represents an important and common comorbidity with chronic pain [2,8,135]. Summary statistics from the most recent MDD GWAS meta-analysis [136], with UK Biobank and 23andMe results removed, were harmonised with MCP GWAS summary statistics following guidelines [137] as closely as possible with the available data. Bi-allelic SNPs shared between the two datasets were identified and harmonised (by ‘flipping’) with respect to the strand used to designate alleles. Reciprocal MR analysis was carried out using subsets of SNPs associated with each of the exposure traits (MCP and MDD) at p < 10−5. This threshold is an order of magnitude lower than suggested as part of the MR-RAPS method [134] and was chosen in order to attempt to account for ‘winner’s curse’, as independently selecting and then testing association for instruments in separate GWAS datasets was not possible in this study. The harmonisation process also involved ensuring that the effect allele was trait-increasing in the exposure trait, and that the effect allele matched between the exposure and the outcome. These selected subsets of variants were then LD-pruned at a threshold of r2 < 0.01 using command-line PLINK using ‘indep-pairwise’ with a 50-SNP window and sliding window of 5 SNPs [138]. This resulted in a set of 200 instruments for MCP as the exposure, and a set of 99 instruments for MDD as the exposure.
PRS prediction of chronic widespread pain
Those who reported chronic pain all over the body were excluded from the MCP GWAS analyses above. This is because chronic pain all over the body, taken as a proxy for chronic widespread pain (CWP), may be a different clinical syndrome from more localised chronic pain, and does not necessarily directly reflect chronic pain at 7 bodily sites. To investigate the relationship between CWP and MCP, a polygenic risk score (PRS) approach was taken.
A PRS was constructed for MCP in individuals who reported chronic pain all over the body (n = 6,815; these individuals had all been excluded from the MCP GWAS), and in controls (n = 10,000 individuals reporting no chronic pain at any site, also excluded from the MCP GWAS). The PRS was calculated using SNPs associated with MCP at p < 0.01, weighting by MCP GWAS effect size (GWAS β) for each SNP. A standardised PRS (based on Z-scores) was used in all analyses, constructed by dividing the calculated PRS by its standard deviation across all samples. The ability of the standardised PRS to predict chronic widespread pain status was investigated in logistic regression models adjusted for age, sex, genotyping array and the first 8 genetic principal components.
Individual-level data are available via application to UK Biobank. Multisite chronic pain GWAS summary statistics are available via contacting the authors and will be submitted to UK Biobank for publication at their website.
Supporting information
Supplementary methods and background information on defining genes of interest, MR-RAPS and LocusZoom.
(DOCX)
Further information on genomic risk loci as identified by FUMA is shown, including locus size in terms of base-pairs (Size(kb)), number of SNP associations within the locus range (#SNPs), number of genes mapped to the locus (#mapped genes) and the number of genes physically located within the locus (#genes physically located in loci).
(TIF)
Results of the MAGMA gene-based test results implemented via FUMA are shown, with the SNPs with the top 10 most-significant gene associations (by Bonferroni-corrected gene-based test p value) labelled. Significance (a Bonferroni-corrected p-value of less than ~6 on the -log10 scale) is indicated by the dashed red line.
(TIF)
Observed versus expected gene-based test p values on the -log10 scale are shown.
(TIF)
Quantile-Quantile plots (left-hand panels), and leave-one-out beta estimate versus t-value plots (right-hand panels) for each of the six models fitted during MR-RAPS analysis with MDD as the exposure are shown (A-F).
(PDF)
Quantile-Quantile plots (left-hand panels), and leave-one-out beta estimate versus t-value plots (right-hand panels) for each of the six models fitted during MR-RAPS analysis with MCP as the exposure are shown (A-F).
(PDF)
Plots of the 46 SNP regions +/- 1 mega-base pairs flanking the region are shown. Mb = mega-base pairs, cM = centimorgans, -log10(p-value) refers to GWAS p value on -log10 scale. Lower panel shows genes in the plotted region. Lead SNP is marked with a purple diamond point and labelled with rsID.
(PDF)
Six different regression models fitted during MR-RAPS analysis and their corresponding S1 or S2 Figs label (A-F) are shown. L2 = L2 loss function, huber = Huber loss function, tukey = Tukey loss function.
(PDF)
Genes of interest as determined via Supplementary Methods. Note that this is distinct from MAGMA gene-based test results (N significant genes there = 113).
(DOCX)
(DOCX)
MR results for MDD-exposure. Β refers to the causal effect, SE (β) and P (β) to the standard error and p value of β, P (AD) to the Anderson-Darling test of normality p value, P (SW) to the Shapiro-Wilk test of normality p value, tau to the over-dispersion statistic size and P (τ) to the p value. C.F = corresponding QQ plot panel for the model. P (τ) was calculated from the tau estimate and its standard error [139]. The row of the table corresponding to the regression model found to be best-fitting is in bold.
(DOCX)
MR results for chronic pain-exposure. Β refers to the causal effect, SE (β) and P (β) to the standard error and p value of β, P (AD) to the Anderson-Darling test of normality p value, P (SW) to the Shapiro-Wilk test of normality p value, τ to the over-dispersion statistic size and P (τ) to the p value. P (τ) was calculated from the τ estimate and its standard error [139]The row of the table corresponding to the regression model found to be of best fit is in bold.
(DOCX)
Regression beta coefficient values (Estimate), odds ratios (OR), and P values. The reference level for ‘sex’ is set to female, PRS = z-polygenic risk score.
(DOCX)
GL = Genomic Locus, Chr = chromosome, pos = position, base pairs, mcp_A2 = other allele (MCP GWAS), mcp_A1 = effect allele (MCP GWAS), mcp_beta = effect (beta) (MCP GWAS), mcp_se = standard error of beta (MCP GWAS), cwp_A1 = effect allele (CWP GWAS), cwp_A2 = other allele (CWP GWAS), cwp_beta = effect (beta) (CWP GWAS), cwp_se = standard error of the beta (CWP GWAS), cwp_gwas_p = gwas P value (CWP GWAS).
(DOCX)
Acknowledgments
We thank all participants in the UK Biobank study. UK Biobank was established by the Wellcome Trust, Medical Research Council, Department of Health, Scottish Government and Northwest Regional Development Agency.
Data Availability
Individual-level UK Biobank data are available upon application to UK Biobank (https://www.ukbiobank.ac.uk/register-apply/). GWAS summary statistics for the Multisite Chronic Pain GWAS are available for download at http://dx.doi.org/10.5525/gla.researchdata.822.
Funding Statement
RJS is supported by a UKRI Innovation- HDR-UK Fellowship (MR/S003061/1). JW is supported by the JMAS Sim Fellowship for depression research from the Royal College of Physicians of Edinburgh (173558). AF is supported by an MRC Doctoral Training Programme Studentship at the University of Glasgow (MR/K501335/1). KJAJ is supported by an MRC Doctoral Training Programme Studentship at the Universities of Glasgow and Edinburgh. DJS acknowledges the support of a Lister Prize Fellowship (173096) and the MRC Mental Health Data Pathfinder Award (MC_PC_17217). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Elzahaf RA, Tashani OA, Unsworth BA, Johnson MI. The prevalence of chronic pain with an analysis of countries with a Human Development Index less than 0.9: a systematic review without meta-analysis. Curr Med Res Opin. 2012;28(7):1221–9. 10.1185/03007995.2012.703132 [DOI] [PubMed] [Google Scholar]
- 2.Vos T, Barber RM, Bell B, Bertozzi-Villa A, Biryukov S, Bolliger I, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hocking LJ, Morris AD, Dominiczak AF, Porteous DJ, Smith BH. Heritability of chronic pain in 2195 extended families. Eur J Pain (United Kingdom). 2012;16(7):1053–63. [DOI] [PubMed] [Google Scholar]
- 4.Greene SA. Chronic Pain: Pathophysiology and Treatment Implications. Top Companion Anim Med [Internet]. 2010;25(1):5–9. Available from: 10.1053/j.tcam.2009.10.009 [DOI] [PubMed] [Google Scholar]
- 5.Merskey H, Bogduk N. Classification of Chronic Pain. IASP Pain Terminology. 1994. 240 p. [Google Scholar]
- 6.Vellucci R. Heterogeneity of chronic pain. Clinical Drug Investigation. 2012. [DOI] [PubMed] [Google Scholar]
- 7.Von Korff M, Ormel J, Keefe FJ, Dworkin SF. Grading the severity of chronic pain. Pain [Internet]. 1992;50(1092):133–49. Available from: http://www.ncbi.nlm.nih.gov/pubmed/1408309 [DOI] [PubMed] [Google Scholar]
- 8.McIntosh AM, Hall LS, Zeng Y, Adams MJ, Gibson J, Wigmore E, et al. Genetic and Environmental Risk for Chronic Pain and the Contribution of Risk Variants for Major Depressive Disorder: A Family-Based Mixed-Model Analysis. PLoS Med. 2016;13(8):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mogil JS. Pain genetics: Past, present and future. Trends Genet [Internet]. 2012;28(6):258–66. Available from: 10.1016/j.tig.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 10.Zorina-Lichtenwalter K, Meloto CB, Khoury S, Diatchenko L. Genetic predictors of human chronic pain conditions. Neuroscience [Internet]. 2016;338:36–62. Available from: 10.1016/j.neuroscience.2016.04.041 [DOI] [PubMed] [Google Scholar]
- 11.Gureje O, Von Korff M, Kola L, Demyttenaere K, He Y, Posada-Villa J, et al. The relation between multiple pains and mental disorders: Results from the World Mental Health Surveys. Pain. 2008;135(1–2):82–91. 10.1016/j.pain.2007.05.005 [DOI] [PubMed] [Google Scholar]
- 12.Kwiatkowski K, Mika J. The importance of chemokines in neuropathic pain development and opioid analgesic potency. Pharmacol Reports. 2018;70(4):821–30. [DOI] [PubMed] [Google Scholar]
- 13.Pinho-Ribeiro FA, Verri WA, Chiu IM. Nociceptor Sensory Neuron–Immune Interactions in Pain and Inflammation. Trends Immunol [Internet]. 2017;38(1):5–19. Available from: 10.1016/j.it.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okifuji A, Hare BD. The association between chronic pain and obesity. J Pain Res. 2015;8:399–408. 10.2147/JPR.S55598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paley CA, Johnson MI. Physical activity to reduce systemic inflammation associated with chronic pain and obesity a narrative review. Clin J Pain. 2016;32(4):365–70. 10.1097/AJP.0000000000000258 [DOI] [PubMed] [Google Scholar]
- 16.Alföldi P, Wiklund T, Gerdle B. Comorbid insomnia in patients with chronic pain: A study based on the Swedish quality registry for pain rehabilitation (SQRP). Disabil Rehabil. 2014;36(20):1661–9. 10.3109/09638288.2013.864712 [DOI] [PubMed] [Google Scholar]
- 17.Borsook D. Neurological diseases and pain. Brain. 2012;135(2):320–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denk F, McMahon SB, Tracey I. Pain vulnerability: A neurobiological perspective. Nat Neurosci [Internet]. 2014;17(2):192–200. Available from: 10.1038/nn.3628 [DOI] [PubMed] [Google Scholar]
- 19.Trouvin A, Perrot S. Pain in osteoarthritis. Implications for optimal management. Jt Bone Spine. 2018;85(4):429–34. [DOI] [PubMed] [Google Scholar]
- 20.Hashmi JA, Baliki MN, Huang L, Baria AT, Torbey S, Hermann KM, et al. Shape shifting pain: Chronification of back pain shifts brain representation from nociceptive to emotional circuits. Brain. 2013;136(9):2751–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baliki MN, Apkarian AV. Nociception, Pain, Negative Moods, and Behavior Selection. Neuron [Internet]. 2015;87(3):474–91. Available from: 10.1016/j.neuron.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansour AR, Baliki MN, Huang L, Torbey S, Herrmann KM, Schnitzer TJ, et al. Brain white matter structural properties predict transition to chronic pain. Pain. 2013;154(10):2160–8. 10.1016/j.pain.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bliss TVP, Collingridge GL, Kaang BK, Zhuo M. Synaptic plasticity in the anterior cingulate cortex in acute and chronic pain. Nat Rev Neurosci. 2016;17(8):485–96. 10.1038/nrn.2016.68 [DOI] [PubMed] [Google Scholar]
- 24.Baliki MN, Mansour AR, Baria AT, Apkarian AV. Functional reorganization of the default mode network across chronic pain conditions. PLoS One. 2014;9(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamaleri Y, Natvig B, Ihlebaek CM, Benth JS, Bruusgaard D. Number of pain sites is associated with demographic, lifestyle, and health-related factors in the general population. Eur J Pain. 2008;12(6):742–8. 10.1016/j.ejpain.2007.11.005 [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Weedon MN, Purcell S, Lettre G, Estrada K, Willer CJ, et al. Genomic inflation factors under polygenic inheritance. 2011;(July 2010):807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashburner Michael, Ball CA Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nature. 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulik-Sullivan B, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–5. 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manitt C, Mimee A, Eng C, Pokinko M, Stroh T, Cooper HM, et al. The Netrin Receptor DCC Is Required in the Pubertal Organization of Mesocortical Dopamine Circuitry. J Neurosci. 2011;31(23):8381–94. 10.1523/JNEUROSCI.0606-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaufman L, Yang G, Hayashi K, Ashby JR, Huang L, Ross MJ, et al. The homophilic adhesion molecule sidekick-1 contributes to augmented podocyte aggregation in HIV-associated nephropathy. FASEB J Off Publ Fed Am Soc Exp Biol. 2007;21(7):1367–75. [DOI] [PubMed] [Google Scholar]
- 31.Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451(7177):465–9. 10.1038/nature06469 [DOI] [PubMed] [Google Scholar]
- 32.Wilson PM, Fryer RH, Fang Y, Hatten ME. Astn2, A Novel Member of the Astrotactin Gene Family, Regulates the Trafficking of ASTN1 during Glial-Guided Neuronal Migration. J Neurosci. 2010;30(25):8529–40. 10.1523/JNEUROSCI.0032-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138(17):3593–612. 10.1242/dev.063610 [DOI] [PubMed] [Google Scholar]
- 34.Kitagawa M. Notch signalling in the nucleus: Roles of Mastermind-like (MAML) transcriptional coactivators. J Biochem. 2015;159(3):287–94. 10.1093/jb/mvv123 [DOI] [PubMed] [Google Scholar]
- 35.Ables JL, Breunig JJ, Eisch AJ, Rakic P. Not(ch) just development: Notch signalling in the adult brain. Nat Rev Neurosci. 2011;12(5):269–83. 10.1038/nrn3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janssens B, Goossens S, Staes K, Gilbert B, van Hengel J, Colpaert C, et al. alphaT-catenin: a novel tissue-specific beta-catenin-binding protein mediating strong cell-cell adhesion. J Cell Sci. 2001;114(Pt 17):3177–88. [DOI] [PubMed] [Google Scholar]
- 37.Arikkath J, Reichardt LF. Cadherins and catenins at synapses: roles in synaptogenesis and synaptic plasticity. Trends Neurosci. 2008;31(9):487–94. 10.1016/j.tins.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arikkath J, Peng I-F, Gie Ng Y, Israely I, Liu X, Ullian EM, et al. -Catenin Regulates Spine and Synapse Morphogenesis and Function in Hippocampal Neurons during Development. J Neurosci. 2009;29(17):5435–42. 10.1523/JNEUROSCI.0835-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerrier S, Polleux F. The Ups and Downs of Neural Progenitors: Cep120 and TACCs Control Interkinetic Nuclear Migration. Neuron. 2007;56(1):1–3. 10.1016/j.neuron.2007.09.019 [DOI] [PubMed] [Google Scholar]
- 40.Hayashi K, Furuya A, Sakamaki Y, Akagi T, Shinoda Y, Sadakata T, et al. The brain-specific RasGEF very-KIND is required for normal dendritic growth in cerebellar granule cells and proper motor coordination. PLoS One. 2017;12(3):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji J, Hao Z, Liu H, Liu Y, Liu J, Lin B, et al. Effect of KNDC1 overexpression on the senescence of human umbilical vein endothelial cells. Mol Med Rep. 2018;17(5):7037–44. 10.3892/mmr.2018.8775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346(1):240. [DOI] [PubMed] [Google Scholar]
- 43.Cohen-Baraka O, Hagiwaraa N, Arltb MF, Hortona JP, Brilliant MH. Cloning, characterization and chromosome mapping of the human SOX6 gene. Gene. 2001;265:157–64. [DOI] [PubMed] [Google Scholar]
- 44.Denny P, Swift S, Brand N, Dabhade N, Barton P, Ashworth A. A conserved family of genes related to the testis determining gene, SRY. Nucleic Acids Res. 1992;20(11):2887 10.1093/nar/20.11.2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurtsdotter I, Topcic D, Karlén A, Singla B, Hagey DW, Bergsland M, et al. SOX5/6/21 prevent oncogene-driven transformation of brain stem cells. Cancer Res. 2017;77(18):4985–97. 10.1158/0008-5472.CAN-17-0704 [DOI] [PubMed] [Google Scholar]
- 46.Batista-Brito R, Rossignol E, Hjerling-Leffler J, Denaxa M, Wegner M, Lefebvre V, et al. The Cell-Intrinsic Requirement of Sox6 for Cortical Interneuron Development. Neuron. 2009;63(4):466–81. 10.1016/j.neuron.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang TL, Guo Y, Liu YJ, Shen H, Liu YZ, Lei SF, et al. Genetic variants in the SOX6 gene are associated with bone mineral density in both Caucasian and Chinese populations. Osteoporos Int. 2012;23(2):781–7. 10.1007/s00198-011-1626-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sterky FH, Trotter JH, Lee S-J, Recktenwald C V., Xiao Du BZ, Zhou P, et al. Carbonic anhydrase-related protein CA10 is an evolutionarily conserved pan-neurexin ligand. Proc Natl Acad Sci. 2017;114(14):E2984–E2984. 10.1073/pnas.1703198114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldstein LSB, Yang Z. Microtubule-Based Transport Systems In Neurons: The Roles of Kinesins and Dyneins. Annu Rev Neurosci. 2000;23:39–71. 10.1146/annurev.neuro.23.1.39 [DOI] [PubMed] [Google Scholar]
- 50.Blake DJ, Tinsley JM, Davies KE. Utrophin: A Structural and Functional Comparison to Dystrophin. Brain Pathol. 1996;6:37–47. [DOI] [PubMed] [Google Scholar]
- 51.Cerecedo D, Cisneros B, Gómez P, Galván IJ. Distribution of dystrophin- and utrophin-associated protein complexes during activation of human neutrophils. Exp Hematol. 2010;38(8):618–28. 10.1016/j.exphem.2010.04.010 [DOI] [PubMed] [Google Scholar]
- 52.Blake DJ, Hawkes R, Benson MA, Beesley PW. Different dystrophin-like complexes are expressed in neurons and glia. J Cell Biol. 1999;147(3):645–57. 10.1083/jcb.147.3.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sogos V, Curto M, Reali C, Gremo F. Developmentally regulated expression and localization of dystrophin and utrophin in the human fetal brain. Mech Ageing Dev. 2002;123(5):455–62. [DOI] [PubMed] [Google Scholar]
- 54.Carlsson P, Mahlapuu M. Forkhead Transcription Factors: Key Players in Development and Metabolism. 2002;23:1–23. [DOI] [PubMed] [Google Scholar]
- 55.Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, et al. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 2011. July;7(7):e1002145 10.1371/journal.pgen.1002145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacDermot KD, Bonora E, Sykes N, Coupe A-M, Lai CSL, Vernes SC, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005. June;76(6):1074–80. 10.1086/430841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacob TC, Moss SJ, Jurd R. GABA(A) receptor trafficking and its role in the dynamic modulation of neuronal inhibition. Nat Rev Neurosci. 2008. May;9(5):331–43. 10.1038/nrn2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sigel E, Steinmann ME. Structure, Function, and Modulation of GABA A. 2012;287(48):40224–31. 10.1074/jbc.R112.386664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bębenek A, Ziuzia-Graczyk I. Fidelity of DNA replication—a matter of proofreading. Curr Genet. 2018;64(5):985–96. 10.1007/s00294-018-0820-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malarkey CS, Churchill MEA. The high mobility group box: The ultimate utility player of a cell. Trends Biochem Sci. 2012;37(12):553–62. 10.1016/j.tibs.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murayama Y, Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2014;505(7483):367–71. 10.1038/nature12867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol. 2000;150(3):405–16. 10.1083/jcb.150.3.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peters JM, Nishiyama T. Sister chromatid cohesion. Cold Spring Harb Perspect Biol. 2012;4(11):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peters JM. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7(9):644–56. 10.1038/nrm1988 [DOI] [PubMed] [Google Scholar]
- 65.Shrestha S, Wilmeth LJ, Eyer J, Shuster CB. PRC1 controls spindle polarization and recruitment of cytokinetic factors during monopolar cytokinesis. Mol Biol Cell. 2012;23(7):1196–207. 10.1091/mbc.E11-12-1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kowalski MP, Krude T. Functional roles of non-coding Y RNAs. Int J Biochem Cell Biol. 2015;66:20–9. 10.1016/j.biocel.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanaka M, Sasaki K, Kamata R, Hoshino Y, Yanagihara K, Sakai R. A Novel RNA-Binding Protein, Ossa/C9orf10, Regulates Activity of Src Kinases To Protect Cells from Oxidative Stress-Induced Apoptosis. Mol Cell Biol. 2009;29(2):402–13. 10.1128/MCB.01035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kinchen JM, Ravichandran KS. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 2010;464(7289):778–82. 10.1038/nature08853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menges CW, Altomare DA, Testa JR. FAS-associated factor 1 (FAF1): Diverse functions and implications for oncogenesis. Cell Cycle. 2009;8(16):2528–34. 10.4161/cc.8.16.9280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giudicessi JR, Ye D, Tester DJ, Crotti L, Mugione A, Nesterenko V V., et al. Transient outward current (Ito) gain-of-function mutations in the KCND3-encoded Kv4.3 potassium channel and Brugada syndrome. Hear Rhythm. 2011;8(7):1024–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee YC, Durr A, Majczenko K, Huang YH, Liu YC, Lien CC, et al. Mutations in KCND3 cause spinocerebellar ataxia type 22. Ann Neurol. 2012;72(6):859–69. 10.1002/ana.23701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duarri A, Jezierska J, Fokkens M, Meijer M, Schelhaas HJ, Den Dunnen WFA, et al. Mutations in potassium channel KCND3 cause spinocerebellar ataxia type 19. Ann Neurol. 2012;72(6):870–80. 10.1002/ana.23700 [DOI] [PubMed] [Google Scholar]
- 73.Roosing S, Romani M, Isrie M, Rosti RO, Micalizzi A, Musaev D, et al. Mutations in cep120 cause joubert syndrome as well as complex ciliopathy phenotypes. J Med Genet. 2016;53(9):608–15. 10.1136/jmedgenet-2016-103832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baron Y, Pedrioli PG, Tyagi K, Johnson C, Wood NT, Fountaine D, et al. VAPB/ALS8 interacts with FFAT-like proteins including the p97 cofactor FAF1 and the ASNA1 ATPase. BMC Biol. 2014;12(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oyama N, Chan I, Neill SM, Hamada T, South AP, Wessagowit V, et al. Autoantibodies to extracellular matrix protein 1 in lichen sclerosus. Lancet. 2003;362(9378):118–23. 10.1016/S0140-6736(03)13863-9 [DOI] [PubMed] [Google Scholar]
- 76.Hamada T, Wessagowit V, South AP, Ashton GHS, Chan I, Oyama N, et al. Extracellular matrix protein 1 gene (ECM1) mutations in lipoid proteinosis and genotype-phenotype correlation. J Invest Dermatol. 2003;120(3):345–50. 10.1046/j.1523-1747.2003.12073.x [DOI] [PubMed] [Google Scholar]
- 77.Liu J, Krantz ID. Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet. 2009;76(4):303–14. 10.1111/j.1399-0004.2009.01271.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lehalle D, Mosca-Boidron AL, Begtrup A, Boute-Benejean O, Charles P, Cho MT, et al. STAG1 mutations cause a novel cohesinopathy characterised by unspecific syndromic intellectual disability. J Med Genet. 2017;54(7):479–88. 10.1136/jmedgenet-2016-104468 [DOI] [PubMed] [Google Scholar]
- 79.Tayebi N, Jamsheer A, Flöttmann R, Sowinska-Seidler A, Doelken SC, Oehl-Jaschkowitz B, et al. Deletions of exons with regulatory activity at the DYNC1I1 locus are associated with split-hand/split-foot malformation: Array CGH screening of 134 unrelated families. Orphanet J Rare Dis. 2014;9(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roberts SH, Hughes HE, Davies SJ, Meredith AL. Bilateral split hand and split foot malformation in a boy with a de novo interstitial deletion of 7q21.3. J Med Genet. 1991;28:479–81. 10.1136/jmg.28.7.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J, Dallmayer M, Kirchner T, Musa J, Grünewald TGP. PRC1: Linking Cytokinesis, Chromosomal Instability, and Cancer Evolution. Trends in Cancer. 2018;4(1):59–73. 10.1016/j.trecan.2017.11.002 [DOI] [PubMed] [Google Scholar]
- 82.Li T, Zeng Z, Zhao Q, Wang T, Huang K, Li J, et al. FoxP2 is significantly associated with schizophrenia and major depression in the Chinese Han Population. World J Biol Psychiatry. 2013. March;14(2):146–50. 10.3109/15622975.2011.615860 [DOI] [PubMed] [Google Scholar]
- 83.Yin J, Jia N, Liu Y, Jin C, Zhang F, Yu S, et al. No association between FOXP2 rs10447760 and schizophrenia in a replication study of the Chinese Han population. Psychiatr Genet. 2018. January;1. [DOI] [PubMed] [Google Scholar]
- 84.Tolosa A, Sanjuán J, Dagnall AM, Moltó MD, Herrero N, de Frutos R. FOXP2 gene and language impairment in schizophrenia: association and epigenetic studies. BMC Med Genet. 2010. December;11(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanjuá J, Tolosa A, Gonzá Lez A JC, Aguilar EJ, Pé Rez-Tur J, Ná Jera B, María C, et al. Association between FOXP2 polymorphisms and schizophrenia with auditory hallucinations. Vol. 16, Psychiatric Genetics. 2006. [DOI] [PubMed] [Google Scholar]
- 86.Laroche F, Ramoz N, Leroy S, Fortin C, Rousselot-Paillet B, Philippe A, et al. Polymorphisms of coding trinucleotide repeats of homeogenes in neurodevelopmental psychiatric disorders. Psychiatr Genet. 2008. December;18(6):295–301. 10.1097/YPG.0b013e3283060fa5 [DOI] [PubMed] [Google Scholar]
- 87.Petryshen TL, Middleton FA, Tahl AR, Rockwell GN, Purcell S, Aldinger KA, et al. Genetic investigation of chromosome 5q GABAA receptor subunit genes in schizophrenia. Mol Psychiatry. 2005. December;10(12):1074–88. 10.1038/sj.mp.4001739 [DOI] [PubMed] [Google Scholar]
- 88.Lo W-S, Harano M, Gawlik M, Yu Z, Chen J, Pun FW, et al. GABRB2 Association with Schizophrenia: Commonalities and Differences Between Ethnic Groups and Clinical Subtypes. Biol Psychiatry. 2007. March;61(5):653–60. 10.1016/j.biopsych.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 89.Srivastava S, Cohen J, Pevsner J, Aradhya S, McKnight D, Butler E, et al. A novel variant in GABRB2 associated with intellectual disability and epilepsy. Am J Med Genet. 2014. November;164A(11):2914–21. 10.1002/ajmg.a.36714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inada T, Koga M, Ishiguro H, … YH-P, 2008 U. Pathway-based association analysis of genome-wide screening data suggest that genes associated with the γ-aminobutyric acid receptor signaling pathway are. Pharmacogenet Genomics. 2008;18(4). [DOI] [PubMed] [Google Scholar]
- 91.Zorina-Lichtenwalter K, Parisien M, Diatchenko L. Genetic studies of human neuropathic pain conditions. Pain. 2017;159(3):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suri P, Palmer MR, Tsepilov YA, Freidin MB, Boer CG, Yau MS, et al. Genome-wide meta-analysis of 158,000 individuals of European ancestry identifies three loci associated with chronic back pain. 2018;48(7):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gormley P, Anttila V, Winsvold BS, Palta P, Esko T, Hinds DA, et al. Susceptibility Loci for Migraine. Nat Genet [Internet]. 2016;48(8):16–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27322543%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5331903%5Cnhttp://www.nature.com/doifinder/10.1038/ng.3598 [Google Scholar]
- 94.Nicholl BI, Mackay D, Cullen B, Martin DJ, Ul-Haq Z, Mair FS, et al. Chronic multisite pain in major depression and bipolar disorder: cross-sectional study of 149,611 participants in UK Biobank. BMC Psychiatry. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shipherd JC, Keyes M, Jovanovic T, Ready DJ, Baltzell D, Worley V, et al. Veterans seeking treatment for posttraumatic stress disorder: What about comorbid chronic pain? J Rehabil Res Dev. 2007;44(2):153 [DOI] [PubMed] [Google Scholar]
- 96.Dunn AS, Julian T, Formolo LR, Green BN, Chicoine DR. Preliminary analysis of posttraumatic stress disorder screening within specialty clinic setting for OIF/OEF veterans seeking care for neck or back pain. J Rehabil Res Dev. 2011;48(5):493 [DOI] [PubMed] [Google Scholar]
- 97.Outcalt SD, Kroenke K, Krebs EE, Chumbler NR, Wu J, Yu Z, et al. Chronic pain and comorbid mental health conditions: independent associations of posttraumatic stress disorder and depression with pain, disability, and quality of life. J Behav Med. 2015;38(3):535–43. 10.1007/s10865-015-9628-3 [DOI] [PubMed] [Google Scholar]
- 98.Phifer J, Skelton K, Weiss T, Schwartz AC, Wingo A, Gillespie CF, et al. Pain symptomatology and pain medication use in civilian PTSD. Pain. 2011;152(10):2233–40. 10.1016/j.pain.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Akhtar E, Ballew AT, Orr WN, Mayorga A, Khan TW. The Prevalence of Post-Traumatic Stress Disorder Symptoms in Chronic Pain Patients in a Tertiary Care Setting: A Cross-Sectional Study Eeman. Psychosomatics. 2018; [DOI] [PubMed] [Google Scholar]
- 100.de Almeida JG, Braga PE, Neto FL, Pimenta CA de M. Chronic pain and quality of life in schizophrenic patients. Rev Bras Psiquiatr. 2013;35(1):13–20. [DOI] [PubMed] [Google Scholar]
- 101.Engels G, Francke AL, Van Meijel B, Douma JG, De Kam H, Wesselink W, et al. Clinical pain in schizophrenia: A systematic review. J Pain. 2014;15(5):457–67. 10.1016/j.jpain.2013.11.005 [DOI] [PubMed] [Google Scholar]
- 102.Watson GD, Chandarana PC, Merskey H. Relationships between pain and schizophrenia. Br J Psychiatry. 1981;138(1):33–6. [DOI] [PubMed] [Google Scholar]
- 103.Stubbs B, Eggermont L, Mitchell AJ, De Hert M, Correll CU, Soundy A, et al. The prevalence of pain in bipolar disorder: A systematic review and large-scale meta-analysis. Acta Psychiatr Scand. 2015;131(2):75–88. 10.1111/acps.12325 [DOI] [PubMed] [Google Scholar]
- 104.Lévesque M, Potvin S, Marchand S, Stip E, Grignon S, Pierre L, et al. Pain Perception in Schizophrenia: Evidence of a Specific Pain Response Profile. Pain Med (United States). 2012;13(12):1571–9. [DOI] [PubMed] [Google Scholar]
- 105.Urban-Kowalczyk M, Pigońska J, Śmigielski J. Pain perception in schizophrenia: Influence of neuropeptides, cognitive disorders, and negative symptoms. Neuropsychiatr Dis Treat. 2015;11:2023–30. 10.2147/NDT.S87666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Strigo IA, Matthews SC, Simmons AN, Oberndorfer T, Klabunde M, Reinhardt LE, et al. Altered insula activation during pain anticipation in individuals recovered from anorexia nervosa: Evidence of interoceptive dysregulation. Int J Eat Disord. 2013;46(1):23–33. 10.1002/eat.22045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bär KJ, de la Cruz F, Berger S, Schultz CC, Wagner G. Structural and functional differences in the cingulate cortex relate to disease severity in anorexia nervosa. J Psychiatry Neurosci. 2015;40(4):269–79. 10.1503/jpn.140193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bischoff-Grethe A, Wierenga CE, Berner LA, Simmons AN, Bailer U, Paulus MP, et al. Neural hypersensitivity to pleasant touch in women remitted from anorexia nervosa. Transl Psychiatry. 2018;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Clarke C. Autism Spectrum Disorder and Amplified Pain. Case Rep Psychiatry. 2015;2015:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gu X, Zhou TJ, Anagnostou E, Soorya L, Kolevzon A, Hof PR, et al. Heightened brain response to pain anticipation in high-functioning adults with autism spectrum disorder. Eur J Neurosci. 2018;47(6):592–601. 10.1111/ejn.13598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ren K, Dubner R. Interactions between the immune and nervous systems in pain. Nat Med. 2010;16(11):1267–76. 10.1038/nm.2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ramesh G, Maclean AG, Philipp MT. Cytokines and chemokines at the crossroads of neuroinflammation, neurodegeneration, and neuropathic pain. Mediators Inflamm. 2013;2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Olefsky JM, Glass CK. Macrophages, Inflammation, and Insulin Resistance. Vol. 72, Annual Review of Physiology. 2010. 219–246 p. 10.1146/annurev-physiol-021909-135846 [DOI] [PubMed] [Google Scholar]
- 114.Chawla A, Nguyen KD, Goh YPS. Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol. 2011;11(11):738–49. 10.1038/nri3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hotamisligil GS, Shargill NS SB. Adipose expressionof tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science (80-). 1993;259(5091):87–91. [DOI] [PubMed] [Google Scholar]
- 116.Smolensky MH, Portaluppi F, Manfredini R, Hermida RC, Tiseo R, Sackett-Lundeen LL, et al. Diurnal and twenty-four hour patterning of human diseases: Acute and chronic common and uncommon medical conditions. Sleep Med Rev. 2015;21:12–22. 10.1016/j.smrv.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 117.Segal JP, Tresidder KA, Bhatt C, Gilron I, Ghasemlou N. Circadian control of pain and neuroinflammation. J Neurosci Res. 2018;96(6):1002–20. 10.1002/jnr.24150 [DOI] [PubMed] [Google Scholar]
- 118.Docherty MJ, Jones RCW, Wallace MS. Managing Pain in Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y). 2011;7(9):592–601. [PMC free article] [PubMed] [Google Scholar]
- 119.Carabotti M, Scirocco A, Antonietta M, Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol. 2015;28(2):203–9. [PMC free article] [PubMed] [Google Scholar]
- 120.Cryan JF, Dinan TG. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–12. 10.1038/nrn3346 [DOI] [PubMed] [Google Scholar]
- 121.Ferguson A, Lyall LM, Ward J, Strawbridge RJ, Cullen B, Graham N, et al. Genome-Wide Association Study of Circadian Rhythmicity in 71,500 UK Biobank Participants and Polygenic Association with Mood Instability. EBioMedicine. 2018;35:279–87. 10.1016/j.ebiom.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Phillips K, Clauw DJ. Central pain mechanisms in chronic pain states—Maybe it is all in their head. Best Pract Res Clin Rheumatol. 2011;25(2):141–54. 10.1016/j.berh.2011.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Apkarian AV, Hashmi JA, Baliki MN. Pain and the brain: Specificity and plasticity of the brain in clinical chronic pain. Pain. 2011;152(SUPPL.3):S49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Baliki MN, Petre B, Torbey S, Herrmann KM, Huang L, Schnitzer TJ, et al. Corticostriatal functional connectivity predicts transition to chronic back pain. Nat Neurosci [Internet]. 2012;15(8):1117–9. Available from: 10.1038/nn.3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fasick V, Spengler RN, Samankan S, Nader ND, Ignatowski TA. The hippocampus and TNF: Common links between chronic pain and depression. Neuroscience and Biobehavioral Reviews. 2015. [DOI] [PubMed] [Google Scholar]
- 126.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Loh PR, Tucker G, Bulik-Sullivan BK, Vilhjálmsson BJ, Finucane HK, Salem RM, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47(3):284–90. 10.1038/ng.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Watanabe K, Taskesen E, Van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1–10. 10.1038/s41467-016-0009-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput Biol. 2015;11(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–13. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. 2010;38(16):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. 2010;26(18):2336–7. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. LD Hub: A centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272–9. 10.1093/bioinformatics/btw613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. 2018; [Google Scholar]
- 135.Flint J, Kendler KS. The Genetics of Major Depression. Neuron. 2014;81:484–503. 10.1016/j.neuron.2014.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet [Internet]. 2018;50(May):167577 Available from: https://www.biorxiv.org/content/early/2017/07/24/167577%0Ahttp://www.ncbi.nlm.nih.gov/pubmed/29700475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hartwig FP, Davies NM, Hemani G, Smith GD. Counterfactual causation: Avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 2016;45(6):1717–26. 10.1093/ije/dyx028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Purcell S, Neale B, Todd-brown K, Thomas L, Ferreira MAR, Bender D, et al. REPORT PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. 2007;81(September):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Altman DG, Bland JM. How to obtain the confidence interval from a P value. BMJ. 2011;2090(August):1–2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods and background information on defining genes of interest, MR-RAPS and LocusZoom.
(DOCX)
Further information on genomic risk loci as identified by FUMA is shown, including locus size in terms of base-pairs (Size(kb)), number of SNP associations within the locus range (#SNPs), number of genes mapped to the locus (#mapped genes) and the number of genes physically located within the locus (#genes physically located in loci).
(TIF)
Results of the MAGMA gene-based test results implemented via FUMA are shown, with the SNPs with the top 10 most-significant gene associations (by Bonferroni-corrected gene-based test p value) labelled. Significance (a Bonferroni-corrected p-value of less than ~6 on the -log10 scale) is indicated by the dashed red line.
(TIF)
Observed versus expected gene-based test p values on the -log10 scale are shown.
(TIF)
Quantile-Quantile plots (left-hand panels), and leave-one-out beta estimate versus t-value plots (right-hand panels) for each of the six models fitted during MR-RAPS analysis with MDD as the exposure are shown (A-F).
(PDF)
Quantile-Quantile plots (left-hand panels), and leave-one-out beta estimate versus t-value plots (right-hand panels) for each of the six models fitted during MR-RAPS analysis with MCP as the exposure are shown (A-F).
(PDF)
Plots of the 46 SNP regions +/- 1 mega-base pairs flanking the region are shown. Mb = mega-base pairs, cM = centimorgans, -log10(p-value) refers to GWAS p value on -log10 scale. Lower panel shows genes in the plotted region. Lead SNP is marked with a purple diamond point and labelled with rsID.
(PDF)
Six different regression models fitted during MR-RAPS analysis and their corresponding S1 or S2 Figs label (A-F) are shown. L2 = L2 loss function, huber = Huber loss function, tukey = Tukey loss function.
(PDF)
Genes of interest as determined via Supplementary Methods. Note that this is distinct from MAGMA gene-based test results (N significant genes there = 113).
(DOCX)
(DOCX)
MR results for MDD-exposure. Β refers to the causal effect, SE (β) and P (β) to the standard error and p value of β, P (AD) to the Anderson-Darling test of normality p value, P (SW) to the Shapiro-Wilk test of normality p value, tau to the over-dispersion statistic size and P (τ) to the p value. C.F = corresponding QQ plot panel for the model. P (τ) was calculated from the tau estimate and its standard error [139]. The row of the table corresponding to the regression model found to be best-fitting is in bold.
(DOCX)
MR results for chronic pain-exposure. Β refers to the causal effect, SE (β) and P (β) to the standard error and p value of β, P (AD) to the Anderson-Darling test of normality p value, P (SW) to the Shapiro-Wilk test of normality p value, τ to the over-dispersion statistic size and P (τ) to the p value. P (τ) was calculated from the τ estimate and its standard error [139]The row of the table corresponding to the regression model found to be of best fit is in bold.
(DOCX)
Regression beta coefficient values (Estimate), odds ratios (OR), and P values. The reference level for ‘sex’ is set to female, PRS = z-polygenic risk score.
(DOCX)
GL = Genomic Locus, Chr = chromosome, pos = position, base pairs, mcp_A2 = other allele (MCP GWAS), mcp_A1 = effect allele (MCP GWAS), mcp_beta = effect (beta) (MCP GWAS), mcp_se = standard error of beta (MCP GWAS), cwp_A1 = effect allele (CWP GWAS), cwp_A2 = other allele (CWP GWAS), cwp_beta = effect (beta) (CWP GWAS), cwp_se = standard error of the beta (CWP GWAS), cwp_gwas_p = gwas P value (CWP GWAS).
(DOCX)
Data Availability Statement
Individual-level UK Biobank data are available upon application to UK Biobank (https://www.ukbiobank.ac.uk/register-apply/). GWAS summary statistics for the Multisite Chronic Pain GWAS are available for download at http://dx.doi.org/10.5525/gla.researchdata.822.