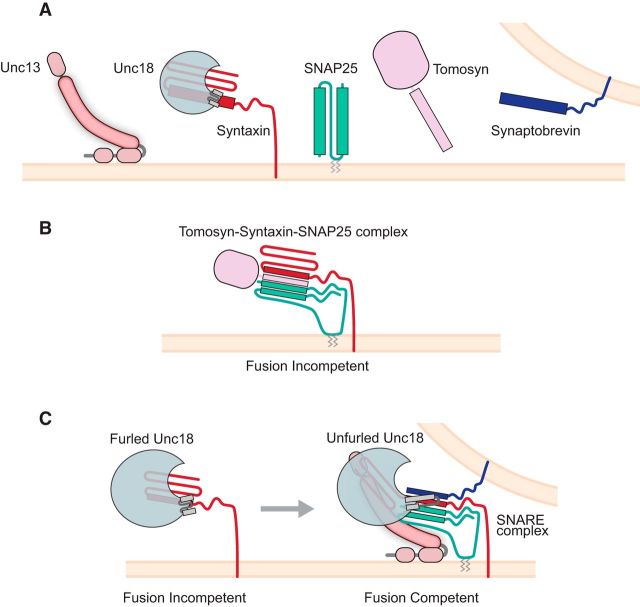
Neurotransmitter release at synapses is mediated by the fusion of synaptic vesicles with the plasma membrane. Fusion is mediated by the interaction of three SNARE proteins (soluble NSF-attachment receptor): synaptobrevin in the vesicle membrane and syntaxin and SNAP-25 in the plasma membrane (Figure 1C). Full assembly of the SNARE complex pulls the vesicle to the plasma membrane and overcomes the energy barrier for fusion (Melia et al., 2002). The formation of the SNARE complex is called “priming.” In addition to SNAREs, priming also involves the accessory proteins Unc18 (called UNC-18 in Caenorhabditis elegans and Munc18 in mice), Unc13 (called UNC-13 in C. elegans and Munc13 in mice), and tomosyn (Fig. 1A). Unc18 and Unc13 proteins align SNAREs during complex assembly (Lai et al., 2017), whereas tomosyn inhibits vesicle priming (Gracheva et al., 2006). Despite numerous studies, the precise molecular steps of synaptic vesicle priming are largely unknown.
Figure 1.
Unc18 in vesicle priming. A, Diagram representation of the proteins discussed. B, Tomosyn forms dead-end tomosyn-SNARE complex with syntaxin and SNAP25. C, Unc18 adopts two conformations. In a fusion incompetent state, furled Unc18 binds closed syntaxin. In the second conformation, unfurled Unc18 nucleates the assembly of the SNARE complex in a fusion competent state.
The Unc18 proteins act during calcium-mediated fusions and are members of the SM superfamily of cytosolic proteins, which are required for membrane fusion during different membrane trafficking steps in eukaryotic cells. Unc18 adopts two conformations. In one conformation, Unc18 forms a clamp on syntaxin, locking it in a closed, fusion incompetent, state (Misura et al., 2000). This interaction is necessary for trafficking syntaxin to the plasma membrane (Medine et al., 2007). At the synaptic terminal, syntaxin opens to permit SNARE assembly (Richmond et al., 2001). In Unc18's second conformation, a “helical hairpin” in domain 3a is extended (Hu et al., 2011) (Fig. 1C). A conserved proline in Munc18 (P335) forms the hinge point of the hairpin bend, producing an “unfurled loop conformation.” The role of this conformation is not entirely understood, but a P335A mutation designed to favor this helical extension enhances Unc18 activity in liposome fusion assays (Parisotto et al., 2014) and caused an increase in exocytosis in secretory cells (Han et al., 2014; Munch et al., 2016). Biochemical evidence suggests also that P335A is a gain-of-function mutation that favors Unc18 binding to synaptobrevin (Xu et al., 2010; Parisotto et al., 2014). Furthermore, expressing the P335A mutation in chromaffin cells increased the primed vesicle pool without affecting the kinetics or calcium dependence of fusion (Munch et al., 2016). These data strongly support a model where domain 3a plays a central role in catalyzing nucleation of the SNARE complex in vesicle priming. This hypothesis is consistent with the recent x-ray structure of the distantly related protein Vps33 with its cognate SNARE proteins (Baker et al., 2015). However, these studies did not answer two important questions. First, does the Munc18 P335A mutation alter priming in neurons? Second, does the mutation give rise to altered behavior?
The nematode C. elegans is a well-established genetic organism for examining molecular mechanisms in synaptic function. To investigate the role of the unfurled loop conformation in the context of a whole organism, Park et al. (2017) introduced the corresponding gain-of-function mutation (P334A) into worm UNC-18. To do so, the authors overexpressed UNC-18 (P334A) from an extrachromosomal array, or modified the endogenous locus using CRISPR-Cas9. Such animals exhibited increased locomotion speed and enhanced neurotransmitter release in a pharmacological assay. Electrophysiological recordings indicated an increase in tonic miniature currents at neuromuscular junctions. Expression of Munc18 P335A in mouse neurons also augmented exocytosis. Together, these data support the hypothesis that Unc18 in the extended hairpin configuration is a positive regulator of synaptic transmission, likely by increasing vesicle priming (Fig. 1C).
Unc13 proteins are large proteins associated with the plasma membrane that are also thought to be involved in SNARE complex formation. The central MUN domain of Unc13 interacts with the amino terminus of syntaxin, and expression of a constitutively open form of syntaxin bypasses secretion defects of unc-13 mutants in C. elegans (Richmond et al., 2001), suggesting that Unc13 opens syntaxin. Interestingly, the UNC-18 (P334A) also partially suppresses locomotor defects in unc-13 mutants and bypasses the requirement for Unc13 in in vitro fusion assays. One possible explanation for the ability of this mutation to bypass the Unc13 requirement is that Unc13 normally promotes this conformational change in Unc18. Unc18 interacts physically and genetically with Unc13 (Sassa et al., 1999), which suggests that Unc13 might directly facilitate the transition from furled to an unfurled loop conformation. Alternatively, Unc13's participation in the Unc18 conformational switch might be an indirect consequence of Unc13's role in the closed-to-open syntaxin transition.
Another important implication of the “unfurled” Unc18 mutant bypassing the requirement for Unc13 involves SNARE complex assembly. To fuse membranes, SNARE proteins must nucleate assembly by aligning their helices in a parallel configuration (Sutton et al., 1998). SNAREs can also form dead-end antiparallel configurations, suggesting that a parallel orientation is not ensured (Weninger et al., 2003). Recently, Unc13 has been shown to orient SNARE proteins during pairing, so that synaptobrevin binds in a parallel orientation (Lai et al., 2017). Because unfurled Munc18 favors binding to synaptobrevin (Xu et al., 2010; Parisotto et al., 2014), it is possible that unfurled Unc18 restores function by facilitating correct synaptobrevin orientation during assembly. Consistent with this idea, Park et al. (2017) found that the Munc18 P335A mutation increased SNARE assembly and Munc18 binding to the SNARE complex.
In contrast, unfurled Unc18 does not seem to play a role in relieving SNARE inhibition by the tomosyn. Tomosyn has an N-terminal β-propeller region, followed by a C-terminal region containing a synaptobrevin-like SNARE motif. Tomosyn blocks incorporation of synaptobrevin into the SNARE complex by occupying the same binding site. Tomosyn forms dead-end tomosyn-SNARE complex with syntaxin and SNAP25 (Hatsuzawa et al., 2003) (Fig. 1B). When overexpressed, tomosyn causes a significant reduction in neurotransmitter release; and loss of tomosyn enhances exocytosis by increasing the number of primed vesicles (Gracheva et al., 2007). Park et al. (2017) found that Unc18 and tomosyn function in parallel: a double mutant lacking tomosyn and also expressing UNC-18 (P334A) further increases neurotransmitter release, demonstrating that UNC-18 (P334A) does not solely act by removing inhibition by tomosyn. Additionally, strong synergy in rescuing secretion defects of unc-13 mutants was observed in triple mutants lacking Unc13 and tomosyn and expressing the unfurled Unc18. An alternative interpretation for these findings is that the unfurled Unc18 can displace tomosyn from dead-end complexes allowing synaptobrevin binding. This would facilitate Unc18's role catalyzing the nucleation of the SNARE complex in vesicle priming. This hypothesis is consistent with biochemical interactions between Unc18 and synaptobrevin (Xu et al., 2010; Parisotto et al., 2014; Sitarska et al., 2017) and structural studies (Baker et al., 2015).
For years, a mechanistic understanding of Unc18's role in neurotransmitter release has been lacking. With the identification of the unfurled loop conformation as central for the postdocking role of Unc18 in cells, the reconstitution of Unc18 functions in vitro (Parisotto et al., 2014; Munch et al., 2016), the structural data that show Unc18 as a template for SNARE complex assembly (Hu et al., 2011; Baker et al., 2015), and the important contribution by Park et al. (2017), this is finally starting to change. Park et al. (2017) show compelling evidence that an Unc18 conformational change has physiological implications in SNARE priming (Fig. 1C). Mutations in the human Unc18 have been associated with intellectual disability and early infantile epileptic encephalopathies (Barcia et al., 2014; Park et al., 2017), opening new venues to understand the implications of priming in neurological disorders.
Footnotes
Editor's Note: These short reviews of recent JNeurosci articles, written exclusively by students or postdoctoral fellows, summarize the important findings of the paper and provide additional insight and commentary. If the authors of the highlighted article have written a response to the Journal Club, the response can be found by viewing the Journal Club at www.jneurosci.org. For more information on the format, review process, and purpose of Journal Club articles, please see http://jneurosci.org/content/preparing-manuscript#journalclub.
I thank Mark Palfreyman, Wayne Davis, and Robert Hobson for comments on the manuscript.
The authors declare no competing financial interests.
References
- Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, Hughson FM (2015) A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349:1111–1114. 10.1126/science.aac7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcia G, Chemaly N, Gobin S, Milh M, Van Bogaert P, Barnerias C, Kaminska A, Dulac O, Desguerre I, Cormier V, Boddaert N, Nabbout R (2014) Early epileptic encephalopathies associated with STXBP1 mutations: could we better delineate the phenotype? Eur J Med Genet 57:15–20. 10.1016/j.ejmg.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Gracheva EO, Burdina AO, Holgado AM, Berthelot-Grosjean M, Ackley BD, Hadwiger G, Nonet ML, Weimer RM, Richmond JE (2006) Tomosyn inhibits synaptic vesicle priming in Caenorhabditis elegans. PLoS Biol 4:e261. 10.1371/journal.pbio.0040261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracheva EO, Burdina AO, Touroutine D, Berthelot-Grosjean M, Parekh H, Richmond JE (2007) Tomosyn negatively regulates both synaptic transmitter and neuropeptide release at the C. elegans neuromuscular junction. J Physiol 585:705–709. 10.1113/jphysiol.2007.138321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GA, Park S, Bin NR, Jung CH, Kim B, Chandrasegaram P, Matsuda M, Riadi I, Han L, Sugita S (2014) A pivotal role for pro-335 in balancing the dual functions of Munc18-1 domain-3a in regulated exocytosis. J Biol Chem 289:33617–33628. 10.1074/jbc.M114.584805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsuzawa K, Lang T, Fasshauer D, Bruns D, Jahn R (2003) The R-SNARE motif of tomosyn forms SNARE core complexes with syntaxin 1 and SNAP-25 and down-regulates exocytosis. J Biol Chem 278:31159–31166. 10.1074/jbc.M305500200 [DOI] [PubMed] [Google Scholar]
- Hu SH, Christie MP, Saez NJ, Latham CF, Jarrott R, Lua LH, Collins BM, Martin JL (2011) Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation. Proc Natl Acad Sci U S A 108:1040–1045. 10.1073/pnas.0914906108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Choi UB, Leitz J, Rhee HJ, Lee C, Altas B, Zhao M, Pfuetzner RA, Wang AL, Brose N, Rhee J, Brunger AT (2017) Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron 95:591–607.e10. 10.1016/j.neuron.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medine CN, Rickman C, Chamberlain LH, Duncan RR (2007) Munc18-1 prevents the formation of ectopic SNARE complexes in living cells. J Cell Sci 120:4407–4415. 10.1242/jcs.020230 [DOI] [PubMed] [Google Scholar]
- Melia TJ, Weber T, McNew JA, Fisher LE, Johnston RJ, Parlati F, Mahal LK, Sollner TH, Rothman JE (2002) Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J Cell Biol 158:929–940. 10.1083/jcb.200112081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misura KM, Scheller RH, Weis WI (2000) Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature 404:355–362. 10.1038/35006120 [DOI] [PubMed] [Google Scholar]
- Munch AS, Kedar GH, van Weering JR, Vazquez-Sanchez S, He E, André T, Braun T, Söllner TH, Verhage M, Sørensen JB (2016) Extension of Helix 12 in Munc18-1 induces vesicle priming. J Neurosci 36:6881–6891. 10.1523/JNEUROSCI.0007-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisotto D, Pfau M, Scheutzow A, Wild K, Mayer MP, Malsam J, Sinning I, Söllner TH (2014) An extended helical conformation in domain 3a of Munc18-1 provides a template for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex assembly. J Biol Chem 289:9639–9650. 10.1074/jbc.M113.514273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Bin NR, Yu B, Wong R, Sitarska E, Sugita K, Ma K, Xu J, Tien CW, Algouneh A, Turlova E, Wang S, Siriya P, Shahid W, Kalia L, Feng ZP, Monnier PP, Sun HS, Zhen M, Gao S, et al. (2017) UNC-18 and tomosyn antagonistically control synaptic vesicle priming downstream of UNC-13 in Caenorhabditis elegans. J Neurosci 37:8797–8815. 10.1523/JNEUROSCI.0338-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Weimer RM, Jorgensen EM (2001) An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature 412:338–341. 10.1038/35085583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa T, Harada S, Ogawa H, Rand JB, Maruyama IN, Hosono R (1999) Regulation of the UNC-18-Caenorhabditis elegans syntaxin complex by UNC-13. J Neurosci 19:4772–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitarska E, Xu J, Park S, Liu X, Quade B, Stepien K, Sugita K, Brautigam CA, Sugita S, Rizo J (2017) Autoinhibition of Munc18-1 modulates synaptobrevin binding and helps to enable Munc13-dependent regulation of membrane fusion. Elife 6:e24278. 10.7554/eLife.24278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395:347–353. 10.1038/26412 [DOI] [PubMed] [Google Scholar]
- Weninger K, Bowen ME, Chu S, Brunger AT (2003) Single-molecule studies of SNARE complex assembly reveal parallel and antiparallel configurations. Proc Natl Acad Sci U S A 100:14800–14805. 10.1073/pnas.2036428100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Su L, Rizo J (2010) Binding of Munc18-1 to synaptobrevin and to the SNARE four-helix bundle. Biochemistry 49:1568–1576. 10.1021/bi9021878 [DOI] [PMC free article] [PubMed] [Google Scholar]