Abstract
Mammalian chromosome duplication progresses in a precise order and is subject to constraints that are often relaxed in developmental disorders and malignancies. Molecular information about the regulation of DNA replication at the chromatin level is lacking because protein complexes that initiate replication seem to bind chromatin indiscriminately. High-throughput sequencing and mathematical modelling have yielded detailed genome-wide replication initiation maps. Combining these maps and models with functional genetic analyses suggests that distinct DNA–protein interactions at subgroups of replication initiation sites (replication origins) modulate the ubiquitous replication machinery and supports an emerging model that delineates how indiscriminate DNA-binding patterns translate into a consistent, organized replication programme.
Before each cell division, mitotic cells create exact duplicates of their entire genome to ensure transmission of complete sets of genetic and epigenetic information to daughter cells. Genomic DNA replicates at distinct nuclear locations during specific time intervals, and the decision about where and when DNA synthesis occurs has a crucial role in ensuring genetic stability. Cells that exhibit incomplete or excessive DNA replication often suffer deleterious consequences in the form of mutations and structural variations1,2. The preponderance of cancer-predisposing mutations within DNA replication genes3–5 also suggests that the orderly progression of DNA replication is essential to cancer prevention.
Given the relatively large sizes of eukaryotic genomes, complete genome duplication requires DNA replication to start concomitantly at numerous locations (replication origins), which results in many discrete chromosomal loci replicating in parallel6,7. Indeed, some metazoan embryos initiate replication during early stages of development from abundant origin sites that are selected with limited or no specificity and can completely duplicate their genomes within minutes8,9. Differentiated somatic metazoan cells, however, exhibit slower DNA synthesis and initiate replication from fewer origins that are activated intermittently, each origin initiating replication in only a proportion of cell cycles8–11. In these cells, adjacent groups of replicating chromosomal sequences (replicons), each containing a replication origin, start DNA synthesis within a short time frame — probably within minutes of each other — thereby forming megabase-scale ‘replication-timing domains’ (REFS 11,12) (FIG. 1). Although not all origins are activated in every cell during each cell cycle, at the cell population level, replication-timing domains are activated in a consistent order and share distinct chromatin interactions6,7,11,12. Metazoan replication domains often overlap with topologically associated domains (TADs)7, which divide the genome into subnuclear compartments and are similar in size to replication domains12. The marked association between replication-timing domains and TADs suggests that the timing of replication initiation reflects a fundamental structural property of chromatin.
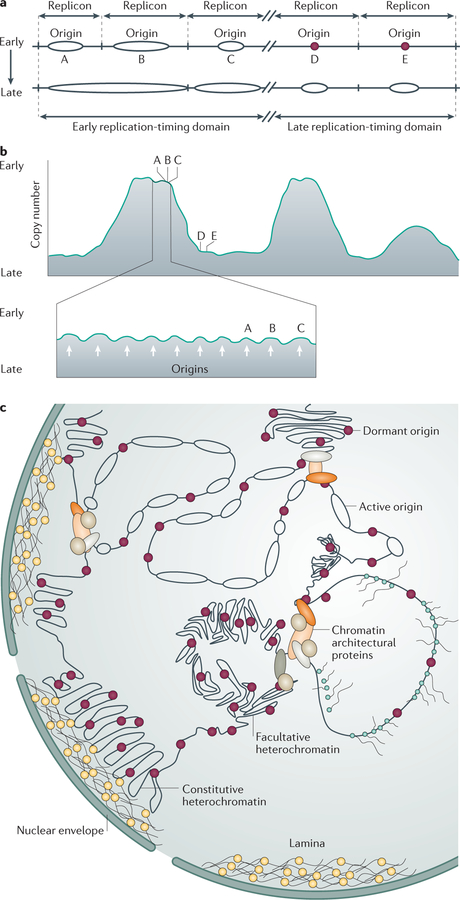
Figure 1. Organization of replication origins, replicons and replication-timing domains.
a | An illustration of a portion of a eukaryotic chromosome depicting five replicons, each defined by an origin of replication (A, B, C, D and E). Replication starts at different times during the S phase of the cell cycle. Origins A, B and C start replication first, concomitantly or in immediate succession, followed later by the initiation of DNA replication from origins D and E. The replicons containing origins A, B and C form an early replication-timing domain, and the replicons containing origins D and E form a late replication-timing domain. b | An example of possible replication-timing measurements revealing large megabase-sized replication-timing domains. Chromatin domains with similar replication timing (top panel) exhibit a similar copy number in an asynchronous cell population, with early-replicating domains (such as the domain including replicons A, B and C) containing more copies than late-replicating domains (such as the domain including replicons D and E). High-resolution mapping of replication timing (bottom panel) reveals subpeaks (indicated by white arrows) in large replication domains that correspond to individual replicons driven by distinct replication origins (for example, the subpeaks for origins A, B and C are visible within the large early-replicating domain). c | Nuclear distribution of replicons and replication domains. Replicating DNA (represented by ellipses) tends to cluster, with each replication cluster containing adjacent, concomitantly replicating regions with several active origins. Early-replicating clusters usually associate with open chromatin (euchromatin; typically in the centre of the nucleus). Late-replicating clusters (here shown containing origins that have not yet started replication, depicted as dark red circles) associate with silent, transcriptionally inactive chromatin (facultative and constitutive heterochromatin; typically near the nuclear envelope). Highly transcribed chromatin often replicates early during S phase, but replication rarely initiates within actively transcribed regions.
As rapid and accurate genome duplication can proceed in early embryos in an apparently indiscriminate order8, distinct initiation sites and organized replication-timing domains are probably not required merely to complete DNA synthesis. Rather, consistent replication origins and organization of specific replication-timing domains might facilitate higher-order chromatin interactions7,10 that help to coordinate replication with transcription and chromosome condensation. Recent studies using whole-genome analyses and mathematical models11,13–19 have aimed to elucidate how chromatin-associated molecular interactions signal the initiation of DNA replication at replication origins to form the basis for timely, complete and orderly genome duplication.
Our understanding of the metazoan signalling cascades that lead to the activation of replication origins is limited because metazoan replication origins do not share strong consensus DNA sequences that could anchor origin-specific DNA–protein interactions. Ubiquitous chromatin-bound pre-replication complexes required for DNA replication initiation (BOX 1) bind to both replication origin and non-origin sequences with equal efficiency in vitro20. In agreement, chromatin association studies of replication machinery components in both Drosophila melanogaster21 and mammalian cells22,23 have not revealed strong sequence specificity despite preferential binding to chromatin features characteristic of replication origins. In addition, whole-genome and single-fibre analyses indicate that, in many metazoan loci, replication initiates alternately within clusters of adjacent origins such that each cell in a population uses slightly different combinations of replication origins6,24. It was suggested that this flexibility might help to coordinate DNA replication with transcription and other nuclear processes in a cell type-specific manner, as well as to facilitate recovery when replication is perturbed (see REF. 25 and references therein). Both the flexibility in establishing replication initiation sites and the lack of DNA consensus sequences raise the challenge of understanding how nonspecific chromatin interactions can lead to accurate initiation at consistent origins.
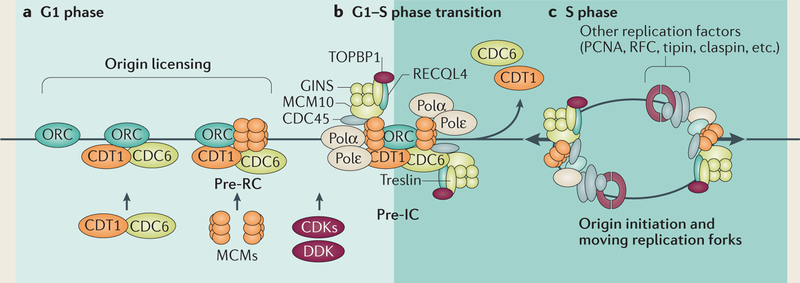
Box 1 |. Replication initiation mechanics.
The initiation of eukaryotic DNA replication comprises two distinct steps9,28,38. The first step occurs shortly after the completion of cell division and involves the assembly of pre-replication complexes (pre-RCs) that include replicative DNA helicase components (a process known as replication origin ‘licensing’; see the figure, part a). The replication origin licensing step requires binding of an origin recognition complex (ORC) to potential initiation sites and subsequent recruitment of a helicase (minichromosome maintenance 2–7 (MCM2–7)), which is mediated by the interaction of the licensing factors cell division control protein 6 (CDC6) and CDT1 with the chromatin-bound ORC. The resulting pre-RC includes an inactive form of the replicative helicase. In the second step (see the figure, part b), some pre-RCs recruit additional components to form larger pre-initiation complexes (pre-ICs) that activate the helicases and associate with DNA polymerases to start replication (a process known as origin activation). Pre-ICs are initiated by the recruitment and/or phosphorylation of additional proteins by DBF4-dependent kinase (DDK) and cyclin-dependent kinases (CDKs). Substrates of DDK and CDKs — including MCM10, RECQ-like DNA helicase type 4 (RECQL4), treslin and DNA topoisomerase 2-binding protein 1 (TOPBP1) — mediate the association of chromatin-bound MCM2–7 with additional components, such as CDC45 and GINS (go-ichi-ni-san), to form an active helicase6,38,170. Concomitantly, DNA polymerases associate with the active helicase complex and catalyse DNA synthesis (see the figure, part c). As replication progresses, pre-RCs dissociate from passively replicated origins, and high CDK activity prevents the assembly of new complexes6,38,171–174. The first step, origin licensing, is inhibited by CDKs and therefore only occurs when CDK levels are low during the G1 stage of the cell cycle. The second step, origin activation, requires CDK-mediated phosphorylation and occurs when CDK levels are high. As high CDK activity levels prevent further association of pre-RC components with DNA, no origins are licensed after replication initiates during the S phase of the cell cycle. The activation of replication origin complexes by CDKs couples genome duplication with cell-cycle signalling. Inhibition of the licensing step by the same CDKs ensures that the entire genome is duplicated only once during each cell division cycle.
PCNA, proliferating cell nuclear antigen; RFC, replication factor C; tipin, TIMELESS-interacting protein.
This Review describes the complex regulation of DNA replication in metazoans, focusing on recent insights into the genetic and epigenetic features of eukaryotic replication origins and the molecular interactions taking place at replication initiation sites. We discuss genome-wide mapping, functional analyses and mathematical simulations in a wide range of species, addressing the seemingly contradictory relationships — evident primarily in mammalian cells — between nonspecific interactions at replication origins and well-ordered, coordinated genome duplication. The mechanics of replication origin activation by the replication machinery and the distribution of replication-timing domains will not be discussed in detail, as these subjects have been recently summarized elsewhere25–28.
Characteristics of replication origins
Beyond consensus: properties of replication origin sequences.
Given that replication origins function as nucleation points for pre-replication complexes, considerable effort has been directed at mapping origins in various organisms and cell types, as well as identifying distinct sequence and chromatin features that delineate replication origins. High-throughput sequencing analyses have yielded detailed genome-wide maps of replication initiation events and identified some characteristic features of metazoan replication origins11,14–17,24,29. However, these mapping studies have failed to identify sequence-specific DNA–protein interactions characteristic of all replication origins that could provide a mechanistic explanation for orderly mitotic genome duplication. Instead, genome-wide replication initiation maps and functional genetic analyses support a model in which overall indiscriminate DNA-binding patterns and combinatorial DNA–protein interactions at groups of origins translate into a consistent, organized replication programme.
A regulatory framework for the replication initiation process was postulated by the replicon model30, which proposes that the ability to start replication at particular chromosomal locations requires specific DNA sequences known as replicators. In this model, replicators bind to initiators, which are protein complexes that recruit elements of the cellular replication machinery to chromatin. Indeed, replicators can be identified in prokaryotes and eukaryotes on the basis of their ability to facilitate replication when removed from their chromosomal context and cloned into plasmids (bacterial and yeast replicators)31–33 or inserted at ectopic chromosomal sites (metazoan replicators)34–37. Initiators have also been identified in many prokaryotes and eukaryotes on the basis of their ability to bind to replicators and facilitate DNA replication initiation6,7,10. In eukaryotes, the initiator anchor — termed the origin recognition complex (ORC) — is a highly conserved complex that binds to chromatin and recruits pre-replication complex components during the G1 phase of the cell cycle6,38 (BOX 1). An ORC is essential for the replication licensing process that renders chromosomes competent for replication, which is consistent with the role of the ORC as an initiator. In yeast, the ORC binds to replication origins in a sequence-specific manner essential for DNA replication initiation38,39. The metazoan ORC is also required for initiation of DNA replication, but metazoan ORC proteins do not exhibit sequence-specific DNA binding in vitro, and they also bind to non-origin regions on chromosomes20,22,23,40. The actual initiation of DNA replication is mediated by the replicative minichromosome maintenance (MCM) helicases, and the frequency and timing of initiation events is determined by the abundance of MCM complexes on chromatin18,41, suggesting that MCM complexes might also partially fulfil the roles of initiators. The lack of sequence specificity of ORC and MCM binding seemingly contradicts the sequence-specific ability of metazoan replicators to initiate replication when inserted at ectopic chromosomal sites34–37,42,43. This suggests that nonspecific interactions with the ubiquitous pre-replication complexes provide necessary, but not sufficient, support for initiation of DNA replication in metazoans.
Metazoan initiation sites share some genetic features10,24 but do not exhibit robust sequence similarities or a common ‘consensus’ sequence motif that specifies origin activity6,10,17,44,45. For example, replication origins are preferentially located near transcriptional start sites (TSSs), CpG islands (CGIs), regions of DNase hypersensitivity, asymmetric G-rich sequences, TAD borders or origin G-rich repeated elements (OGREs)11,14,24,46 (FIG. 2). A group of OGREs colocalize with CGIs and are associated with two flanking replication origins24. OGREs can potentially form G-quadruplex structures, which exhibit enriched association with replication origins24, but G-quadruplexes are overall more abundant than replication origins47, and many origins do not contain G-quadruplexes46,48. Replication origins are also enriched in asymmetric (A:T- and G:C-skewed) tracts, which might facilitate DNA unwinding and DNA– protein interactions, whereas they are depleted of purine-rich structures that can form DNA triplexes11. Consistent with a role for G:C-rich sequences in delineating replication initiation sites, detailed single-nucleotide polymorphism (SNP) analyses of phased genomes indicate a genetic link between strand asymmetry and replication origin activity11.
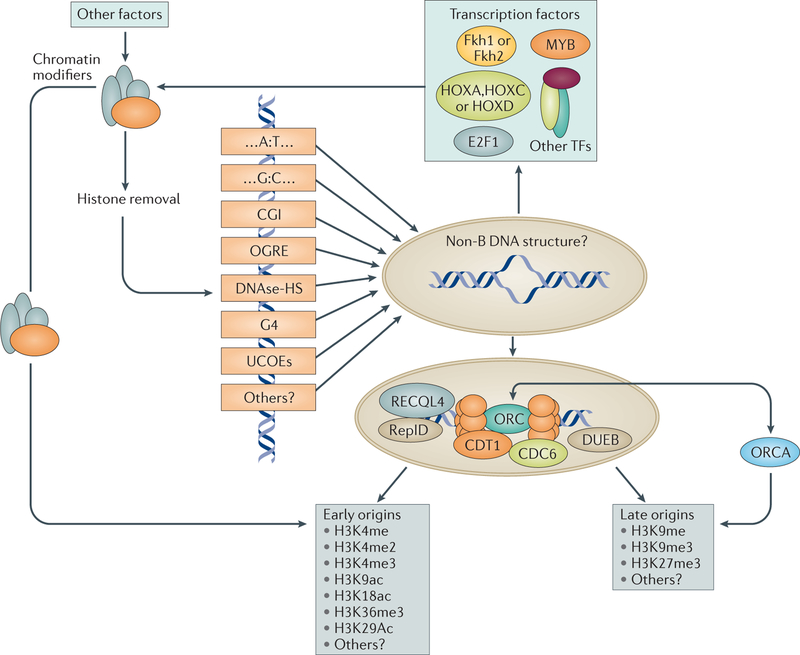
Figure 2. Genetic and epigenetic features of replication origins.
Although there are no strong consensus DNA sequences to anchor origins in metazoans, whole-genome sequence analyses have revealed several motifs that are present near or at origin sites. These include A:T- and G:C-rich motifs, CpG islands (CGIs), origin G-rich repeated elements (OGREs), DNase 1-hypersensitive sites (DNase-HS), quadruplex-like structures (G4) and ubiquitous chromatin-opening elements (UCOEs), which might increase the propensity of origins to unwind and adopt a non-B DNA structure. Groups of replication origins interact with proteins from the pre-replication complex and with proteins located at a subset of replication origins (such as replication-initiation determinant (RepID), DNA-unwinding element-binding protein B (DUEB) and RECQ-like DNA helicase type 4 (RECQL4)). Such interactions may also facilitate binding of transcription factors (TFs) such as forkhead box protein 1 (Fkh1), Fkh2, MYB, homeobox protein A (HOXA), HOXC, HOXD and E2F1) or other proteins that may regulate chromatin structure locally. Origins in early-replicating, open chromatin are preferentially enriched with euchromatin markers (such as H3K4 monomethylation (H3K4me), H3K4 dimethyation (H3K4me2), H3K4 trimethylation (H3K4me3), H3K9 acetylation (H3K9ac), H3K18ac, H3K36me3 and H3K29ac). Cell type-specific, late-replicating origins often associate with heterochromatin markers (such as H3K9me3, H3K27me3 and ORC-associated protein (ORCA), an ORC-binding protein that interacts with H3K9me3).
Although OGREs and TSSs associate with replication origins, it is unclear whether these genomic elements are required for replication initiation, or whether they share a preference for similar chromatin features (such as enhanced chromatin accessibility) with origins. For example, although G-quadruplexes and origins colocalize, a causal relationship between G-quadruplex formation and origin activity was not observed in a SNP analysis of phased human genomes11. Given that G-quadruplexes can interfere with the replication process49, the frequent presence of G-quadruplexes near replication origins might reflect a different selective factor, such as negative selection against both G-quadruplexes and origins in gene bodies47 and/or a selective evolutionary pressure that destabilizes G-quadruplexes formed at origin-distal regions50. In another example, although replication origins strongly associate with TSSs, colocalization depends on transcriptional activity. Replication origins are enriched in TSSs of moderately transcribed genes but are depleted of TSSs of highly transcribed genes14. This finding suggests that interactions at TSSs are not strictly required for initiation, which can often occur at downstream sequences. The exclusion of initiation from highly transcribed TSSs might reflect steric hindrances that prevent colocalization of highly active transcription initiation complexes and replication initiation complexes.
Chromatin structural features associated with replication origins.
As mentioned above, the lack of a single, clear consensus sequence that facilitates replication origin activity contrasts with sequence-specific replication initiation activity exhibited by several replication origins at ectopic chromosomal sites6,34–37,42. These apparently contradictory observations might be reconciled if replicators modulate chromatin accessibility for the replication machinery rather than directly mediating initiation. For instance, replicators could facilitate initiation by affecting particular histone modifications36,51–53, creating a chromosomal context permissive for both transcriptional activity and replication initiation10,54,55 (FIG. 2). One example of sequences that create permissive environments are ubiquitous chromatin-opening elements (UCOEs)56, which are used in gene therapy because they are resistant to heterochromatin-mediated silencing55. UCOEs are characterized by CGIs spanning dual, divergently transcribed promoters of housekeeping genes, and a highly permissive chromatin structure (characterized by histone H3 and H4 hyperacetylation, histone H3 lysine 4 trimethylation (H3K4me3) or lack of histone H1). Interestingly, UCOEs frequently colocalize with replication origins53. However, well-defined replication origins can be mapped in heterochromatin (see REFS 6,24,48,57 and references therein), suggesting that the ability to create an open chromatin environment is not a universal determinant of origin activity.
Recent genome-wide analyses have identified distinct DNA and chromatin modifications that associate with origin activity15,24,29,58–61. Early-replicating origins associate with euchromatic histone modifications (such as H3K4 methylation (H3K4me), H3K4me2, H3K4me3, H3K9 acetylation (H3K9ac), H3K18ac, H3K36me3 and H3K29ac), whereas late replication is associated with repressive chromatin modifications (such as H3 and H4 hypoacetylation, H3K9me, H3K9me3 and H3K27me3)17,62 (FIG. 2). Consistent with a role in genome organization, origin associations with chromatin modifications vary with cell differentiation status8,10 and might reflect coordination between replication and transcription. Replication origins that initiate replication in many cell types are preferentially associated with unmethylated CGIs and with the euchromatin markers H3K4me3 and H3K9ac, whereas origins that initiate replication only in distinct cell types or lineages preferentially colocalize with the heterochromatin marker H3K9me3 (REFS 48,57,63). Interestingly, DNase 1-hypersensitive sites are enriched in regions of enhanced origin activity and preferentially interact with ORCs22,23, but do not mark origins locally48.
Specific histone modifiers affect global origin activity. In mammalian cells, methylation of histone H4 lysine 20 by KMT5A (also known as PRSET7 or SET8) promotes loading of pre-replication complexes onto chromatin to facilitate replication licensing64, as the H4K20me2 interacts with the bromo-adjacent homology (BAH) domain of ORC1 (REF. 65). The histone acetyltransferase HBO1 facilitates initiation of DNA replication66, and DOT1L catalyses H3K79 methylation, preventing re-replication58. In D. melanogaster, the histone acetyltransferases CREB-binding protein (Cbp) and Chameau (the homologue of human HBO1) regulate both DNA replication initiation and developmental oogenesis67, and methylation of H4K20 is crucial for genomic stability, as it prevents DNA damage in late-replicating origins68. This observation suggests that chromatin modifiers may play a part in coordinating replication and transcription during developmental transitions25. In plants, H3K27 methylation is required to induce re-replication69.
Beyond local chromatin modifications, replication origin activity is associated with distal interactions between origins and DNA elements that facilitate chromatin remodelling (FIG. 3). Such elements include a 40-kb region upstream of the HBB replication origin70, the promoter of the Chinese hamster Dhfr locus71 and an enhancer of the TH2 locus72. In addition, replicator sequences themselves can affect chromatin structure. For example, replicators from the HBB and LMNB2 loci can prevent transcriptional silencing by facilitating interaction between a locus control region and a chromatin remodelling complex52,54,70. The genome-wide correlation between TADs and replication-timing domains7,12 also implies that long-range chromatin associations have an impact on the activation of replication origins. Proteins that regulate chromatin interactions, such as cohesins73 and RIF1 (REFS 50,74,75), affect origin distribution, as reflected in the temporal distribution of replication initiation events.
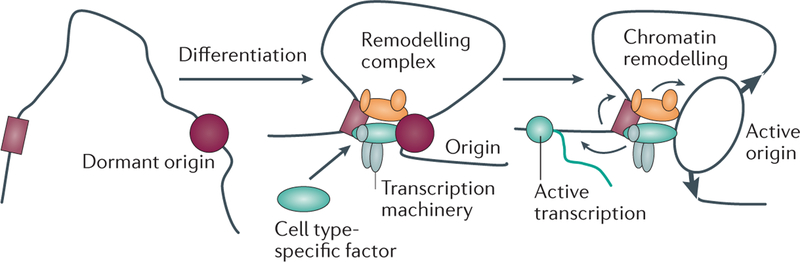
Figure 3. Long-range chromatin interactions can regulate origin activation and replication timing.
Cell type-specific transcription and/or chromatin-modifier complexes can bind both an origin of replication and a locus control region of a distant gene (dark red rectangle), modulating both the expression of a tissue-specific gene and replication initiation. Origin activation by distal interactions may enable local chromatin remodelling and the establishment of a more open chromatin configuration.
Intermittent initiation, origin ‘dormancy’ and the determinants of origin choice.
The replication initiation programme exhibits remarkable flexibility, and many origins initiate DNA replication at disparate frequencies in different cell lineages14,16,24,48. In Saccharomyces cerevisiae, potential origins can initiate replication on plasmids, but origin activity can be suppressed by the chromosomal context76,77. The timing of ORC binding in yeast origins also correlates with the efficiency of initiation. Stable binding of ORCs in the G2 phase of the cell cycle, accompanied by nucleosome remodelling, is associated with efficient origins, whereas in the G1 phase ORCs transiently bind to origins that exhibit less frequent initiation76.
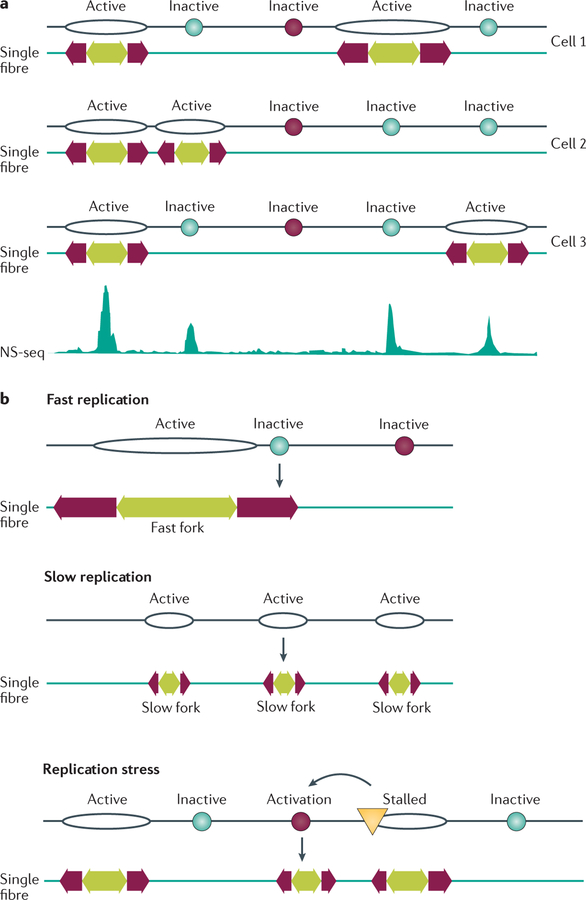
In metazoan cells, most origins do not initiate DNA replication in all cell cycles (that is, they are not constitutive origins6) but exhibit flexible initiation and are active in only a proportion of cell cycles6,10,28,57,78. Initiation frequency can vary with chromosomal context, as exemplified by reduced initiation frequency in an LMNB2 replicator transferred to an ectopic location43. Both constitutive and flexible origins are detectable in whole-genome analyses (FIG. 4), and many common origins identified in several cell lines are likely to be flexible origins. Origin choice may explain the observation that inter-origin distances measured by whole-genome sequencing are shorter than those measured by single-fibre analyses6 (FIG. 4). Common and cell type-specific origins exhibit distinct epigenetic signatures48, which is consistent with the differentiation-and developmental-stage-dependent reorganization of replication domains7,79.
Figure 4. Replication origin choice and origin dormancy.
Constitutive origins (depicted as black oval-shaped outlines) initiate replication at all times in all cells; flexible origins (turquoise circles) initiate replication intermittently with initiation frequencies that vary from cell to cell; and dormant origins (dark red circles) are licensed to initiate replication but never, or very rarely, initiate during an unperturbed cell cycle. Dormant origins can be activated, however, if they reside next to a stalled or damaged replication fork (yellow triangle). a | Top: initiation from both constitutive and flexible origins shown on three hypothetical fibres, with each cell exhibiting a unique pattern of initiation. Experimentally, single-fibre analyses indicate the selection of initiation sites from the pool of flexible origins. Bottom: a population-based assay based on the abundance of nascent strands (nascent strand sequencing (NS-seq)) captures short, newly replicated DNA from all origins. Overall, the mean distances between population-based origin peaks are shorter than those measured on single fibres, indicating origin choice. b | Initiation rates can increase when DNA synthesis slows or when replication forks stall owing to DNA damage. Fast replication is associated with few origins on average because some potential initiation sites are passively replicated by adjacent replication forks before their own replication is actively initiated. Slow replication can increase the frequency of initiation from flexible origins, resulting in shorter inter-origin distances detected on single fibres. In addition, perturbed replication can activate dormant origins that do not normally initiate replication during unperturbed DNA synthesis.
By contrast, dormant origins are not detectable in whole-genome analyses and might be activated only if replication from adjacent origins is stalled (FIGS 4,5). Activation of such novel dormant origins was demonstrated following DNA damage at the Chinese hamster Gnai3 locus80 and in association with repeat expansion in human fragile sites (reviewed in REFS 81, 82). In other cases, DNA damage and slow replication fork progression increase the frequency of initiation from flexible origins83,84.
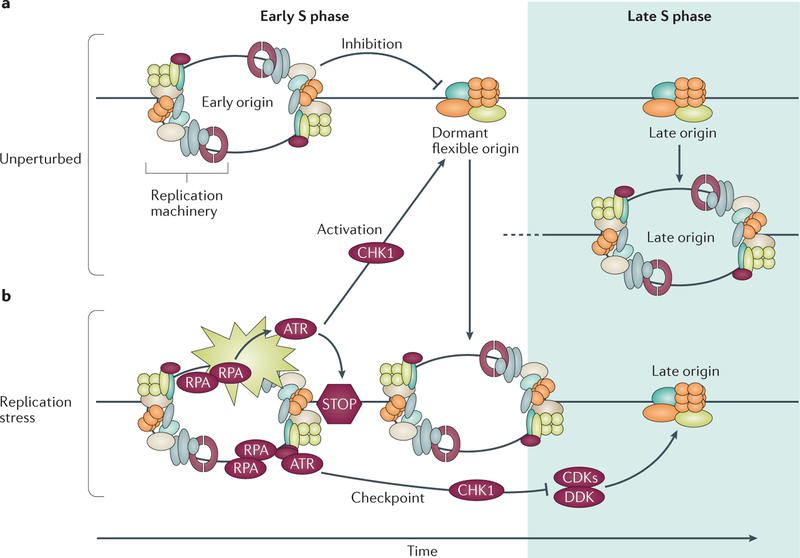
Figure 5. Origin inhibition and activation in unperturbed cells and after replication stress.
a | Only approximately 10% of origins are typically used in an unperturbed cell cycle; most origins remain dormant and are replicated passively by replication forks emanating from adjacent origins. b | Stalled replication forks contain replication protein A (RPA)-coated single-stranded DNA that is sensed by the checkpoint kinase ataxia telangiectasia and RAD3-related protein (ATR), activating a signalling cascade (indicated by the beige star shape) that prevents initiation at late replication origins through checkpoint kinase 1 (CHK1)-mediated inhibition of phosphorylation events catalysed by cyclin-dependent kinases (CDKs) and DBF4-dependent kinase (DDK). Concomitantly, CHK1 activation permits replication initiation from origins (dormant or non-activated flexible origins) located near the stalled forks to complete DNA replication locally.
Origin flexibility is also observed within each cell population, as potential origins and pre-replication complexes are more numerous than observed replication initiation sites6,7,41,85. Components of the pre-replication machinery, such as MCM proteins, are in excess, and more MCMs are loaded onto chromatin during the G1 phase than are used during replication6,7,41. In yeast, an ORC might be able to load several MCM helicases, with more MCMs on early-replicating regions18,41. The excess (dormant) MCM-loaded origins will not initiate replication during an unperturbed S phase but will do so when adjacent replication forks stall78,86,87 (FIG. 5). In metazoans, proteins involved in the cellular response to DNA damage — including Fanconi anaemia group I protein (FANCI)88 and MMS and UV-sensitive protein 81 (MUS81)83, which are both associated with the Fanconi anaemia pathway — can modulate dormant origin activation. FANCI, which was shown to bind some replication origins, affects dormant origin activation that is inhibited when FANCI is phosphorylated by ataxia telangiectasia and RAD3-related protein (ATR)88. MUS81 modulates rates of replication fork progression and the frequency of initiation from flexible replication origins during unperturbed growth83, and promotes efficient replication after exposure to DNA damage89. The interactions between modulators of initiation frequency and cellular pathways that respond to DNA damage highlight the potential role of replication origin choice in maintaining genomic stability.
Molecular interactions at origins
DNA–protein interactions that activate or repress initiation.
Genome-wide analyses have shown that subsets of replication origins are differentially regulated and have distinct genetic and epigenetic features. As interactions with the ubiquitous pre-replication machinery are not sequence specific, the decision of whether or not an origin will be activated under specific circumstances may require distinct replicator-interacting proteins at particular groups of origins6,7,10,90. Metazoan replication origins can therefore be divided into categories, each associated with a specific modifier protein (or protein combination) that determines origin usage according to cell type and developmental stage. Accordingly, tissue-specific replication programmes would then reflect the activity of several origin-binding factors, each interacting with a subgroup of origins that exhibit common features, such as association with distinct transcriptional regulatory regions or nuclear locations. Sequence-specific origin-binding proteins and associations with distinct histones might also regulate cis-acting distal genomic elements8,10,91–93 by forming chromatin loops that determine where and when replication initiates and probably coordinate replication with transcription.
Components of large chromatin complexes that regulate transcription and mitotic segregation affect the selection of eukaryotic replication origins (TABLE 1). In yeast, binding of the transcription factor Ars-binding factor 1 (Abf1) to autonomously replicating sequence 1 (Ars1) seems to alter the chromatin structure and the activity of origins94. The evolutionarily conserved transcription factors forkhead box protein 1 (Fkh1) and Fkh2 interact with distinct groups of yeast replication origin (for example, Fkh1 interacts with early origins) and regulate replication initiation timing by recruiting the initiation factor cell division control protein 45 (Cdc45)95,96. Notably, Fkh1 and Fkh2 cluster replication initiation events in the yeast nucleus to facilitate long-range chromatin interactions, suggesting a link between spatial organization of the genome and origin selection95,96.
Table 1 |.
DNA–protein interactions at groups of replication origins
| Name | Definition | Species | Interaction | Refs |
|---|---|---|---|---|
| Abf1 | ARS-binding factor 1 | Saccharomyces cerevisiae | Interacts with yeast replication origins in a sequence-specific manner | 94 |
| Fkh1 and Fkh2 | Forkhead box protein 1 and forkhead box protein 2 | Saccharomyces cerevisiae | Bind a subset of yeast replication origins (Fkh1 only binds origins that initiate replication early), regulate initiation timing by clustering and participate in long-range chromatin interactions | 96 |
| RB–E2F complex | A multiprotein complex containing a heterodimeric E2F transcription factor and an RB family member | Drosophila melanogaster | Interacts with ORCs and selectively binds replication origins at the chorion gene locus | 105 |
| MYB–MUVB | Part of the DREAM–MYB–MUVB complex containing RB | Drosophila melanogaster | Interacts with ORCs, binds amplified replication origins and recruits histone acetylases | 106 |
| XRCC5 | X-ray repair cross-complementing protein 5 (also known as KU80) | Human | Interacts with primate replication origins in the G1 phase and with replication proteins | 175 |
| HOXC13 | Homeobox protein HOXC13 | Human | Binds the human LMNB2 replication origin of the gene encoding human lamin and interacts with the pre-replication complex in a cell-cycle-dependent manner | 120 |
| DUEB | DNA-unwinding element-binding protein B | Human | Binds DNA-unwinding elements at replication origins in the human MYC, LMNB2 and SCA10 loci, and interacts with pre-replication complexes at those sites | 176 |
| ORCA | Origin recognition complex-associated protein | Human | Associates with human ORCs and stabilizes them on chromatin, and binds HP1 and H3K9me3 | 132 |
| RepID | Replication-initiation determinant | Human | Binds to a group of human replication origins and is essential for their activation | 115 |
| LMO2 | LIM domain only protein 2 (also known as rhombotin 2) | Human | Interacts with replication proteins and associates with some origins at promoters in human erythroleukaemic cells | 177 |
| H3K79me2 | Dimethylated histone H3 lysine 79 | Human | Binds a group of human replication origins and is required for limiting replication such that it occurs only once per cell cycle | 58 |
| GRWD1 | Glutamate-rich WD40-repeat-containing protein 1 | Human | Binds histones, CDT1 and CDC6 in human cells, colocalizes with CDC6 on chromatin and is required for MCM loading | 117 |
| HBO1 | Histone acetyltransferase binding to ORC1 (also known as KAT7) | Human | Interacts with human CDT1 to activate replication origins | 60, 112 |
| RECQL4 | RECQ-like DNA helicase type 4 | Human | Recruited to the origins at the human LMNB2, HBB and CSF2 loci, and is required for origin binding of MCM10 and CTF4 | 110, 111 |
| BRPF3 | Bromodomain and PHD-finger-containing protein 3 | Human | Interacts with HBO1 to activate replication origins in open chromatin regions (for example, near TSSs) via histone acetylation | 60 |
ARS, autonomously replicating sequence; CDC6, cell division control protein 6; CSF2, colony stimulating factor 2; CTF4, chromosome transmission fidelity protein 4 (also known as DNA polymerase α-binding protein); DREAM, DRE-antagonist modulator (also known as calsenilin); HBB, haemoglobin subunit-β; HP1, heterochromatin protein 1; LMNB2, lamin B2; MCM, minichromosome maintenance; MUVB, Drosophila orthologue of human RB; ORC, origin recognition complex; PHD, PH domain; RB, retinoblastoma-associated protein; SCA10, spinocerebellar ataxia type 10 protein (also known as ataxin 10); TSS, transcription start site.
Sequence-specific interactions that recruit pre-replication complexes, possibly via an RNA molecule, are observed in other unicellular eukaryotes. For instance, the Tetrahymena thermophila ORC interacts with an integral noncoding RNA subunit that can bind to its corresponding DNA target in the ribosomal DNA (rDNA) replication origin97. This interaction regulates differential replication of the rDNA locus during sequential stages of the Tetrahymena thermophila life cycle.
In metazoans, site-specific interactions at replication origins are known to regulate the replication of DNA tumour viruses, which use components of the cellular replication machinery for genome propagation. Such viruses rely on viral origin-specific DNA–protein interactions (for example, papillomavirus proteins E1 and E2, Epstein–Barr virus (EBV) nuclear antigen 1 (EBNA1)) to load cellular pre-replication complexes and initiate DNA replication98,99. Proteins involved in these interactions also tether viral genomes to cellular chromatin via chromatin modifiers100,101. For example, papillomavirus E2 interacts with Bromodomain-containing protein 4 (BRD4), a chromatin-binding protein and transcription activator102,103, and with the DNA helicase CHLIR1 (also known as DDX11)104.
In D. melanogaster, follicle cells undergo a specialized amplification process in which a few specific origins repeatedly initiate DNA replication, leading to amplification of the chorion gene clusters involved in eggshell synthesis. E2F1 and the D. melanogaster orthologue of retinoblastoma-associated protein (RB) form a complex that selectively binds to the replication origins in the chorion locus105. The large MYB–MUVB complex binds to amplified D. melanogaster origins to recruit histone acetylases106, whereas tethering of the histone deacetylase Rpd3 (also known as HDAC1) at the chorion gene clusters inhibits amplification107. The transcription factor MYB binds to ORC2 and ORC6, and may modulate origin amplification by controlling ORC2 localization106.
Direct effects of transcription factors on origin localization are also evident in frog egg systems. Injection of a hybrid protein comprising the yeast transcriptional activator Gal4 and part of the herpes simplex virus protein VP16 (Gal–VP16) into Xenopus laevis eggs led to site-specific replication of a recombinant DNA containing the myc TATA box next to Gal4-binding sites8. Although binding of transcription factors located initiation events, active transcription was not required for the formation of site-specific origins, suggesting that transcription factors might facilitate site-specific initiation by recruiting other proteins, including histone-modifying enzymes.
In mammalian cells, examples of DNA–protein interactions at specific origins include the binding of DNA-unwinding element-binding protein B (DUEB; also known as D-tyrosyl-tRNA(Tyr) deacylase 1) to the MYC replicator, which results in CDC45 recruitment to form pre-initiation complexes108,109. The RECQ helicases RECQ1 and RECQ4 also associate with replication origins110, and are required for unperturbed initiation and replication110,111. Origins also interact with the histone acetylase HBO1, which is a co-activator of the licensing factor CDT1 (REFS 112–114). The DDB1–CUL4-associated factor RepID (also known as PHIP or DCAF14) is essential for DNA replication initiation within a particular origin group115. RepID participates in a long-range interaction between the replication origin and the human HBB locus control region, which regulates both transcription and replication patterns54,70,116 Glutamate-rich WD40-repeat-containing protein 1 (GRWD1), which also interacts with a subset of replication origins, colocalizes with the CDC6 component of the pre-replication complex and is required for MCM loading onto chromatin117. These observations support the notion that distinct DNA–protein interactions at specific replicator groups dictate replication initiation. Interestingly, both RepID and GRWD1 interact with DDB1–CUL4 (REFS 118,119), suggesting that this ubiquitin E3 ligase complex may influence specific origins.
Site-specific interactions at subsets of replication origins might also have other functions that are unrelated to transcriptional activity. For instance, H3K79me2 associates with a distinct proportion of replication initiation sites and can mark replicated chromatin during S phase58. H3K79me2 is not required for DNA replication initiation, but this modification prevents re-replication from occurring within a single cell cycle.
The selection of replication initiation events might also be regulated by differentiation-specific transcription factors such as the homeobox protein HOXC13, which interacts with the LMNB2 origin120. HOXC13 binding facilitates subsequent ORC4 recruitment to the origins of human LMNB2, TOP1 and MCM4. Other HOXB orthologues (such as HOXD13 and HOXA11) bind to the pre-replication complex component CDC6, along with human LMNB2 and origins near MYC, FMR1, MCM4 and TOP1 (REF. 121).
DNA–protein interactions that selectively affect replication timing.
Organization of the genome into replication-timing domains requires activation of replication origins at adjacent replicons concomitantly or in rapid succession. Chromatin modifications, distal chromatin interactions and several nuclear-scaffold structural components affect the distribution of replication initiation events and of replication factories throughout the nucleus6,7,57. Transcription factors95 and chromatin modifiers76, which associate with chromatin as DNA synthesis progresses and have a crucial role in chromatin re-assembly following replication122, differentially affect activation timing in subsets of yeast origins. In D. melanogaster, H4K20 methylation is required for complete replication and prevents the accumulation of DNA damage lesions in late-replicating regions68. In mammals, the interactions of histone-modifying enzymes and transcription factors with chromatin correlate with changes in replication timing79,123. Dynamic replication order, however, is subject to some constraints and is partially regulated by cis-acting elements that dictate the time of replication in genomic regions ranging in size from single loci to entire chromosomes52,84,91,124,125. For example, cis-acting elements that determine replication timing on a whole-chromosome level encode monoallelically expressed long noncoding RNAs that regulate both replication timing and mitotic chromosome stability in mammals91.
Sequence-specific interactions can also modulate distal interactions that affect origin activity and might coordinate replication with transcription. Overexpression of the yeast transcription factors Fkh1 and Fkh2 advances initiation of many origins, resulting in increased initiation in S phase96. In Schizosaccharomyces pombe, the heterochromatin protein 1 (HP1) homologue Swi6 associates with ORCs, Cdc18 (the homologue of human CDC6 homologue) and the DBF4-dependent kinase (DDK) homologue Hsk1, and is required for centromere replication126,127. Swi6 is also involved in pericentromeric heterochromatin formation and maintenance, and may have both positive and negative roles in centromere replication126. By recruiting DDK to pericentromeric heterochromatin, Swi6 promotes DNA replication initiation. However, as disruption of the Swi6–Cdc18 complex accelerates centromere replication126, Swi6 may negatively regulate replication timing, although the mechanistic details of this interaction are currently unclear.
S. pombe Taz1, an orthologue of mammalian proteins telomeric repeat-binding factor 1 (TRF1) and TRF2, is required for telomere maintenance and plays a major part in replication timing128. Taz1 binds to internal chromosomal repeats in a sequence-specific manner and forms facultative heterochromatin islands, promoting late replication of nearby origins by preventing early DDK-dependent loading of Sld3 (the yeast homologue of the human treslin) on origins, while concomitantly repressing gene expression129. The S. pombe centromeric protein Shugoshin 2 also regulates replication timing at telomeres by limiting Sld3 loading130.
Hp1, a structural component of heterochromatin, is required for both very late centromeric DNA replication and early pericentromeric DNA replication in D. melanogaster131. Another heterochromatin-binding protein, ORC-associated protein (ORCA; also known as LRWD1), interacts with the pre-replication complex ORC1 in mammalian cells. ORCA stabilizes ORC binding to chromatin, and binds to and regulates both H3K9me3 and HP1 (REF. 132); these observations are consistent with the hypothesis that ORCA binds to a group of late replication origins in heterochromatin.
The telomere-binding protein Rif1 is involved in regulating replication timing in S. pombe28,50,75,133. Rif1 is a structural component of the nucleus that also associates with chromatin loops50 and coats late-replicating domains50,74,75,133. Rif1 also mediates cell-cycle signalling by promoting dephosphorylation of the replicative MCM helicase to dynamically regulate DNA replication initiation. Specifically, Rif1 targets protein phosphatase 1 to Mcm4, a component of the MCM helicase, preventing its phosphorylation by Hsk1 to delay the activation of the helicase and ensure appropriate replication timing75.
Similarly to its yeast counterpart, mammalian RIF1 acts as a nuclear organizer, coats late replication domains and interacts with a group of late-replicating regions to delay replication74. RIF1 modulates the frequency of chromatin interactions in late-replicating domains, restricting interactions during the G1 phase of the cell cycle. In both human and mouse cells, RIF1 depletion alters replication-timing patterns and, in particular, advances the replication timing of chromosomal regions that normally replicate mid-S phase74,133. RIF1 also affects the size of chromatin loops133, supporting the notion that distal chromatin interactions have a role in establishing replication-timing patterns.
Replication origin locations are sufficient to determine replication timing without a separate ‘timing factor’.
Distinct chromatin modifications, which are often altered during development, can associate with regions that exhibit particular replication timing. Although replication proceeds randomly in the inactive X chromosome134, replication-timing domains are clearly delineated in most chromosomes, and these domains often correspond to particular TADs7. Both TADs and replication timing are cell type-specific in mammalian cells. TADs are described as stable regulatory units of replication timing; early DNA synthesis begins within transcription-rich TADs and subsequently progresses into neighbouring later-replicating and transcription-poor TADs10. Thus, replication domains that exhibit early replication tend to be gene-rich, associated with active chromatin markers and located near the nuclear interior7. Late-replicating regions are enriched in heterochromatin, in areas corresponding to lamin-associated domains, and tend to be located near the nuclear periphery7. The temporal order of chromosome replication correlates with biases in evolutionary mutation rates135,136. Replication-timing domains are conserved in syntenic regions137–139 and affect the frequency of somatic copy number alterations in cancer cells136. The frequently observed variation in replication-timing domains in cancer and developmental disorders2,27,140,141 might indicate that the order of replication helps to ensure stable genetic and epigenetic inheritance.
Consistent with the role of chromatin in determining replication timing, a large proportion of the human genome exhibits changes in the order of replication during nuclear reorganization associated with differentiation and development7,12,27. The precise timing of origin activation within replication-timing domains is determined anew after each mitotic cell division142, facilitating dynamic and flexible changes in replication order27. One possible mechanism underlying rapid changes in replication order is the dynamic distribution of one or more limiting replication factors that can preferentially associate with potential origins in accessible chromatin during the G1 phase of the cell cycle143,144. These limiting factors can be sequentially released from origins after replication, subsequently binding to and facilitating initiation from later-replicating origins. In concordance with this model, recent simulation studies based on mathematical models13,18,19 suggest that the locations of replication origins and the relative efficiency of initiation at these locations are sufficient to determine the dynamic high-level organization of replication domains.
In one simulation study13, global replication order was accurately predicted in human cells using a model that had two variables: an ‘initiation probability landscape’, corresponding to a known distribution of replication origins determined by DNase 1-hypersensitive sites, and the assumption that replication initiation is restricted by the availability of a single rate-limiting activator. The model was able to predict replication-timing domains without assuming any particular spatial genomic organization, which suggests that the distribution of DNase 1-hypersensitive sites contains sufficient information to contribute to genome conformation, possibly through histone displacement by transcription factors. This model implies that replication timing does not require a separate regulatory mechanism distinct from the location of initiation events and is ultimately determined by chromatin structure, which itself is determined by DNA sequences through the distribution of genes and other regulatory elements.
A second simulation study19, based on a model assuming spontaneous stochastic initiation within euchromatin and facultative heterochromatin, was also able to predict the general progression of DNA replication on the basis of the distribution of chromatin zone sizes and higher chromatin organization. Importantly, the second model predicted a three-dimensional spatial organization of replication events by assuming concomitant inhibition of replication initiation at distances below the size of chromatin loops and by taking into account a domino-like effect in which replication at a particular origin would induce initiation from adjacent origins. The model also predicted a transition between early and late S phase with both spontaneous and induced initiation in heterochromatin. In concordance, a third simulation study18 based on a model relying on the density of MCM replicative helicases, assuming a high level of MCMs at early origins, predicted replication timing in yeast with high accuracy.
Together, these models predict that the timing of origin activation could be determined on the basis of the locations of origins competing for a limiting factor. As these models did not assume the existence of an independent chromatin feature that determines replication time, they support the notion that a higher density of origin activation is sufficient to increase the chance of early replication in regions containing those origins. Hence, the replication-timing programme seems to be determined by local interactions that signal to licensed replication origins, which are in turn activated by a combination of sequence-specific DNA–protein interactions and chromatin modifications. Thus, the results of the simulation studies imply that tissue-and differentiation-specific replication-timing programmes are dictated by chromatin features that determine the efficiency of initiation, combined with the constraints that limit replication initiation such that it only occurs once per cell cycle.
Cells might require a consistent replication-timing programme to establish and maintain specific nuclear compartments, such as heterochromatin. For example, late replication of heterochromatin might prevent rapid, massive chromatin decondensation and re-condensation, which could affect gene expression, alter the structural integrity of the nucleus and lead to DNA damage145. Within heterochromatin, however, replication can proceed randomly with no apparent replication-timing domains, as exemplified by the inactive human X chromosome134. This observation again suggests that a consistent replication programme is required neither for completion of the replication process nor to establish heterochromatin, but rather to coordinate replication with other chromatin transactions, primarily transcription. A consistent replication-timing pattern that coordinates with chromatin transactions requires origin activation to be co-regulated with programmed changes in chromosome packaging. The simulation studies discussed above imply that replication origin distribution is sufficient to dictate replication timing. Thus, molecular interactions that determine the likelihood of origin activation can affect both the spatial and temporal properties of the replication landscape.
The central role of origin choice
The remarkable flexibility of DNA replication is facilitated by an excess of potential replication origins. Excess origins permit flexible replication times and also enable alterations in initiation frequency. For example, replication initiation patterns change during transcriptional activation in lymphocyte immunoglobulin genes123,146. Overall, only half of active origins exhibit consistent genome-wide initiation frequency across many cell lines, whereas a large proportion of potential origins exhibit cell type-specific initiation16,48.
Genomic stability in the face of replication stress seems to hinge on the excess of potential origins and pre-replication complexes. Potential replication origins that do not initiate replication during unperturbed growth can do so to complete the duplication of DNA sequences adjacent to collapsed or stalled replication forks or when nucleotide pools are depleted78,80,86,147 (FIG. 5). A role for excess origins is evident from the slow replication and loss of genomic stability that occur when the number of potential pre-replication complexes is reduced, for example in a mouse model harbouring a mutation in the gene encoding the replicative helicase MCM4 subunit147,148. Similarly, RepID-depleted cells, which do not initiate replication from a proportion of origins, exhibit diminished initiation frequency, slower replication fork elongation and frequent replication fork stalling events115 Initiation from origins adjacent to stalled replication forks is activated by checkpoint signalling cascades that concomitantly inhibit DNA replication initiation in distal origins that are programmed to replicate later82,83,89,149–151 Signalling cascades activated upon replication fork stalling (such as those involving checkpoint kinase 1 (CHK1), CDC25, cyclin-dependent kinases (CDKs) and DDK) are also involved in modulating the activation of origins during unperturbed growth6,83.
Conversely, replication rate could dictate the frequency of initiation events81,86,150. For instance, cells that exhibit low levels of DNA topoisomerase 1 (which is encoded by TOP1)152 or are deficient in the repair endo-nuclease MUS81 (REF. 83) exhibit slower replication fork progression that is compensated for by increased replication initiation frequency. Mapping of replication origins in MUS81-deficient cells identified the same pool of replication origins as in MUS81-proficient cells83, suggesting that the increased initiation frequency in cells with slow replication reflects the accrued activation of origins that exhibit a low initiation frequency when replication speeds up153,154. In other examples, however, perturbed replication induces novel replication initiation events in regions that do not initiate replication in the absence of perturbations80,153. In these cases, the pool of flexible origins was not sufficient to compensate for replication fork stalling, and so replication initiated at novel locations distinct from flexible origins, probably dormant (or cryptic) origins that are never used during non-perturbed growth. In other examples, exposure to drugs that inhibit histone deacetylases87 and depletion of RepID115 reduce initiation frequency and induce replication fork asymmetry, which often indicates chromosome breakage123,152. Locally, mutations in an adjacent replication origin can affect the expansion dynamics of triplet repeats in the rare fragile site FMR1 (REFS 82,155). Hence, replication origins that are activated only in particular cell subtypes might confer a clear evolutionary advantage in facilitating specialized transcription and differentiation programmes.
Supporting the notion that excess replication origins might be used to complete DNA synthesis when replication is perturbed, regions that contain fewer replication origins are highly susceptible to genomic instability, including DNA breakage. For example, replication origin scarcity is a hallmark of common fragile sites, which correspond to slow replication zones in yeast. In these regions, reduced replication origin abundance increases the frequency of chromosome loss156 and can trigger the DNA damage response82,157–161. Chromosome fragility can also be associated with early-replicating regions (early-replicating fragile sites162) that might reflect interference between transcription and replication. Dynamic changes in replication origin use that are associated with gene expression might allow cells to minimize such interference.
An excess of replication origins might be beneficial for two reasons. First, excess origins provide a mechanistic basis for flexible initiation that synchronizes with cell type-specific gene expression. Second, excess origins can be used as ‘backup’ origins in cases for which there is a need to rescue stalled replication forks during perturbed growth. Identifying the determinants of replication origin activation is crucial because unregulated changes in replication initiation frequency can lead to genomic instability and DNA damage, such as that caused by oncogene-facilitated DNA over-replication163. The pace of replication fork progression directly determines the cellular response to stalled replication forks. Consistent with a role in maintaining genomic stability, mutations in components of the pre-replication complex can cause developmental syndromes164. In addition, deregulated expression of pre-replication complex components, driven by transcription factors such as E2F1 and MYC, is associated with a high risk of re-replication and cancer upon co-occurrence with inactivation of cell-cycle check-points that maintain genomic stability165–167. Oncogene activation also induces fragile sites, and activation patterns are oncogene specific (for example, RAS induces a set of fragile sites that is different from that induced by cyclin E)168, suggesting that over-replication overwhelms the machinery that maintains replication equilibrium in sites with fewer origins.
Perspectives and future directions
The original replicon model30, which proposes that interactions between replicators and initiators activate DNA replication, still provides the fundamental framework for understanding the regulation of chromosome replication. However, the diverse interactions that dictate replication initiation sites and timing for distinct sets of origins provide an additional regulatory layer to ensure orderly and accurate eukaryotic genome duplication. The recent observations summarized in this Review imply that diverse DNA–protein interactions contribute, possibly in a combinatorial manner, to the activation of pre-replication complexes and the initiation of DNA replication at distinct cis-acting sequences. The assumption that replication initiation patterns emerge from combinations of diverse sequence-specific DNA–protein interactions could resolve the apparent paradoxical finding of consistent initiation patterns at specific DNA elements that lack a single consensus protein-binding sequence. As combinatorial DNA–protein interactions can be modified by cellular circumstances (such as gene expression, developmental stage, and external and internal stress), the capacity to initiate replication at additional origins when necessary provides cells with the ability to adapt to a changing environment by modifying the replication programme.
Nevertheless, the following questions remain. First, it is still unclear whether replicators establish the location of initiation events by interacting directly with the replication machinery or by creating a chromatin environment that is conducive to initiation. The recently observed colocalization of TADs with replication-timing domains supports the latter12, but direct DNA–protein interactions that facilitate initiation could still determine the locations of replication initiation events locally. Second, the mechanics of establishing the locations and timing of initiation remain to be clarified. In particular, emerging observations suggest that some components of the replication machinery associate with late-replicating origins only after early origins complete replication28,169, implying that cells contain several subsets of potential licensed replication origins at different stages of assembly. To understand replication-timing patterns, it would be crucial to study how origins are assigned to each origin group: a group of early-replicating origins, associated with pre-replication complexes that are ready to start DNA replication at the onset of S phase, and groups of later-replicating origins, associated with some components of pre-replication complexes but awaiting one or more critical factors. Future studies should also ask if the assignment of potential origins to the earliest or subsequent replicating groups could be accomplished solely on the basis of the site and the initiation frequency from each origin, without an independent ‘replication-timing factor’, as deduced from simulation studies13,19. If replication timing is ingrained in the location of initiation events, DNA–protein interactions at the origin level can provide a critical determinant underlying the abrupt changes in the replication-timing programme that are observed upon differentiation, activation of a gene expression programme or environmental stress.
Notably, the need for consistent origins and a clear replication-timing programme has not yet been clarified, as data from embryonic systems8 and the inactive X chromosome134 show that specific localization of replication initiation events and a structured replication-timing programme are not required for genome duplication. The past decade has revealed striking correlations between replication timing and chromatin structure at the megabase-scale domain level7. The strict temporal regulation of DNA replication and the consistency of initiation events also suggest that temporal and spatial regulation of the initiation of DNA replication help to coordinate DNA replication with gene expression. A consistent replication-timing programme might also be required to establish and maintain specific nuclear compartments, such as heterochromatin. The next challenge will be to better understand the detailed mechanisms linking replication initiation and the whole-genome replication-timing programme. For example, it would be pertinent to characterize the actual effect of RIF1 (and other proteins with similar roles) on modulating replication timing for a proportion of replication domains. Another challenge will involve investigating how the properties of replication domain structures reconcile with reports on genetic determinants of replication timing that are embedded in DNA sequences that delay or advance replication timing directly. Also, although mathematical models suggest that replication timing is stochastically determined, future studies should ask whether the replication initiation landscape that affects timing patterns is associated with specific chromatin marks that have yet to be identified. Finally, the recent realization that proteins involved in the progression of unperturbed replication also play a part in the cellular response to DNA damage6 lends new urgency to studies aimed at understanding the molecular basis of genome duplication. Such dual roles in modulating both perturbed and unperturbed replication are perhaps not surprising, but a complete understanding of the role of cell-cycle checkpoint signalling in regulating replication patterns might help us to exploit the regulatory interactions that modulate replication for therapeutic purposes.
Replication origins
Locations on chromatin from which replication forks emanate.
Replicons
Units of chromatin that are replicated from a single origin.
Replication-timing domain
A region on chromatin that exhibits a uniform replication time from multiple adjacent replicons.
Topologically associated domains
Discrete units of chromatin that tend to exhibit internal, rather than external, chromatin interactions that probably reflect distinct folding patterns.
Single-fibre analyses
Studies of chromosomal dynamics that are based on visualization of labelled, stretched single DNA molecules (for example, on microscope slides). Single-fibre replication analyses visualize the incorporation of nucleotide analogues into chromatin fibres.
Replicon model
A model proposing that replication initiates through an interaction between cis-acting elements (replicators) and trans-acting factors (initiators).
Replicators
Genetic elements that are required for the initiation of DNA replication from a particular chromosomal site.
Initiators
Proteins, or protein complexes, that interact with a replicator and are required for initiation of DNA replication.
CpG islands
Stretches of nucleotides (longer than 200 bp) that exhibit a high abundance of CpG dinucleotides.
G-quadruplex structures
Stacked, planar structures formed by intramolecular interactions among DNA sequences that contain multiple stretches of four guanines.
DNA triplexes
Non-B helix-type structures that contain a third strand.
Phased genomes
Assembled genome sequences that identify alleles of maternal and paternal chromosomes.
Negative selection
An evolutionary process by which genetic elements with potentially deleterious effects are removed from a population.
Constitutive origins
DNA replication origins that initiate replication in all cells.
Flexible origins
DNA replication origins that initiate replication in some cells but not in all cells.
Dormant origins
DNA replication origins that undergo replication licensing but do not initiate replication during a typical, unperturbed cell cycle.
Acknowledgements
The authors thank H. Fu, K Utani, S. Jang, J. Murai, F. E. Indig and A. B. Marks for critical reading of the manuscript, and would like to apologize to colleagues whose primary work could not be cited directly owing to space limitations. Work in the authors’ laboratory is funded by the intramural programme of the Centre for Cancer Research, National Cancer Institute, US National Institutes of Health.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Inaki K & Liu ET Structural mutations in cancer: mechanistic and functional insights. Trends Genet 28, 550–559 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Abbas T, Keaton MA & Dutta A Genomic instability in cancer. Cold Spring Harb. Perspect. Biol 5, a012914 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosh RM Jr. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 13, 542–558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sousa FG et al. Alterations of DNA repair genes in the NCI-60 cell lines and their predictive value for anticancer drug activity. DNA Repair (Amst.) 28, 107–115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lange SS, Takata K & Wood RD DNA polymerases and cancer. Nat. Rev. Cancer 11, 96–110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fragkos M, Ganier O, Coulombe P & Mechali M DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol 16, 360–374 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Rivera-Mulia JC & Gilbert DM Replicating large genomes: divide and conquer. Mol. Cell 62, 756–765 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mechali M Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell Biol 11, 728–738 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N & Oda M Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem 79, 89–130 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Aladjem MI Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. Genet 8, 588–600 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Bartholdy B, Mukhopadhyay R, Lajugie J, Aladjem MI & Bouhassira EE Allele-specific analysis of DNA replication origins in mammalian cells. Nat. Commun 6, 7051 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pope BD et al. Topologically associating domains are stable units of replication-timing regulation. Nature 515, 402–405 (2014).This study showed that megabase-scale chromatin domains, which exhibit almost synchronous replication during S phase, align with chromatin domains that favour internal chromatin interactions, suggesting that the order of replication delineates specific chromosomal structures.
- 13.Gindin Y, Valenzuela MS, Aladjem MI, Meltzer PS & Bilke S A chromatin structure-based model accurately predicts DNA replication timing in human cells. Mol. Syst. Biol 10, 722 (2014).This simulation study based on a simple model of replication kinetics was able to predict experimental replication-timing profiles using DNAse 1-hypersensitive sites as an approximation of initiation sites, necessitating no explicit timing information.
- 14.Martin MM et al. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res 21, 1822–1832 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marks AB, Smith OK & Aladjem MI Replication origins: determinants or consequences of nuclear organization? Curr. Opin. Genet. Dev 37, 67–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besnard E et al. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol 19, 837–844 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Mechali M, Yoshida K, Coulombe P & Pasero P Genetic and epigenetic determinants of DNA replication origins, position and activation. Curr. Opin. Genet. Dev 23, 124–131 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Das SP et al. Replication timing is regulated by the number of MCMs loaded at origins. Genome Res 25, 1886–1892 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lob D et al. 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression. Nat. Commun 7, 11207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vashee S et al. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev 17, 1894–1908 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacAlpine HK, Gordan R, Powell SK, Hartemink AJ & MacAlpine DM Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20, 201–211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miotto B, Ji Z & Struhl K Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc. Natl Acad. Sci. USA 113, E4810–E4819 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dellino GI et al. Genome-wide mapping of human DNA-replication origins: levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res 23, 1–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cayrou C et al. The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res 25, 1873–1885 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivera-Mulia JC & Gilbert DM Replication timing and transcriptional control: beyond cause and effect — part III. Curr. Opin. Cell Biol 40, 168–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prioleau MN & MacAlpine DM DNA replication origins — where do we begin? Genes Dev 30, 1683–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhind N & Gilbert DM DNA replication timing. Cold Spring Harb. Perspect. Med 3, 1–26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renard-Guillet C, Kanoh Y, Shirahige K & Masai H Temporal and spatial regulation of eukaryotic DNA replication: from regulated initiation to genome-scale timing program. Semin. Cell Dev. Biol 30, 110–120 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Eaton ML et al. Chromatin signatures of the Drosophila replication program. Genome Res 21, 164–174 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacob F, Brenner J & Cuzin F On the regulation of DNA replication in bacteria. Cold Spring Harb. Symp. Quant. Biol 28, 329–348 (1963).The replicon model described in this paper continues to form the basis of our current thinking on the regulation of genome duplication, although some modifications have been made.
- 31.Yasuda S & Hirota Y Cloning and mapping of the replication origin of Escherichia coli. Proc. Natl Acad. Sci. USA 74, 5458–5462 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eichenlaub R, Figurski D & Helinski DR Bidirection replication from a unique origin in a mini-F plasmid. Proc. Natl Acad. Sci. USA 74, 1138–1141 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Struhl K, Stinchcomb DT, Scherer S & Davis RW High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc. Natl Acad. Sci. USA 76, 1035–1039 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aladjem MI, Rodewald LW, Kolman JL & Wahl GM Genetic dissection of a mammalian replicator in the human β-globin locus. Science 281, 1005–1009 (1998).This study showed that replicators can induce sequence-specific initiation of DNA replication when placed at ectopic sites, demonstrating the regulation of DNA replication by cis-acting elements in mammalian cells.
- 35.Altman AL & Fanning E Defined sequence modules and an architectural element cooperate to promote initiation at an ectopic mammalian chromosomal replication origin. Mol. Cell. Biol 24, 4138–4150 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu G, Malott M & Leffak M Multiple functional elements comprise a mammalian chromosomal replicator. Mol. Cell. Biol 23, 1832–1842 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paixao S et al. Modular structure of the human lamin B2 replicator. Mol. Cell. Biol 24, 2958–2967 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remus D & Diffley JF Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol 21, 771–777 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Bolon YT & Bielinsky AK The spatial arrangement of ORC binding modules determines the functionality of replication origins in budding yeast. Nucleic Acids Res 34, 5069–5080 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remus D, Beall EL & Botchan MR DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC–DNA binding. EMBO J 23, 897–907 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das SP & Rhind N How and why multiple MCMs are loaded at origins of DNA replication. Bioessays 38, 613–617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashizume T & Shimizu N Dissection of mammalian replicators by a novel plasmid stability assay. J. Cell. Biochem 101, 552–565 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Guan Z et al. Decreased replication origin activity in temporal transition regions. J. Cell Biol 187, 623–635 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urban JM, Foulk MS, Casella C & Gerbi SA The hunt for origins of DNA replication in multicellular eukaryotes. F1000Prime Rep 7, 30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hyrien O Peaks cloaked in the mist: the landscape of mammalian replication origins. J. Cell Biol 208, 147–160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petryk N et al. Replication landscape of the human genome. Nat. Commun 7, 10208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huppert JL & Balasubramanian S Prevalence of quadruplexes in the human genome. Nucleic Acids Res 33, 2908–2916 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith OK et al. Distinct epigenetic features of differentiation-regulated replication origins. Epigenetics Chromatin 9, 18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherstyuk VV, Shevchenko AI & Zakian SM Epigenetic landscape for initiation of DNA replication. Chromosoma 123, 183–199 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Kanoh Y et al. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat. Struct. Mol. Biol 22, 889–897 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Chen X, Liu G & Leffak M Activation of a human chromosomal replication origin by protein tethering. Nucleic Acids Res 41, 6460–6474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu H et al. Preventing gene silencing with human replicators. Nat. Biotechnol 24, 572–576 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Conner AL & Aladjem MI The chromatin backdrop of DNA replication: lessons from genetics and genome-scale analyses. Biochim. Biophys. Acta 1819, 794–801 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang L et al. Prevention of transcriptional silencing by a replicator-binding complex consisting of SWI/SNF, MeCP1 and hnRNP C1/C2. Mol. Cell. Biol 31, 3472–3484 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majocchi S, Aritonovska E & Mermod N Epigenetic regulatory elements associate with specific histone modifications to prevent silencing of telomeric genes. Nucleic Acids Res 42, 193–204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muller-Kuller U et al. A minimal ubiquitous chromatin opening element (UCOE) effectively prevents silencing of juxtaposed heterologous promoters by epigenetic remodeling in multipotent and pluripotent stem cells. Nucleic Acids Res 43, 1577–1592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith OK & Aladjem MI Chromatin structure and replication origins: determinants of chromosome replication and nuclear organization. J. Mol. Biol 426, 3330–3341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu H et al. Methylation of histone H3 on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle. PLoS Genet 9, e1003542 (2013).This was the first report of a histone modification that specifically marks replication origins and functions to prevent re-replication in mammalian cells.
- 59.Karnani N, Taylor CM, Malhotra A & Dutta A Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol. Biol. Cell 21, 393–404 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng Y et al. BRPF3–HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J 35, 176–192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldar A et al. Deciphering DNA replication dynamics in eukaryotic cell populations in relation with their averaged chromatin conformations. Sci. Rep 6, 22469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Picard F et al. The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genet 10, e1004282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lubelsky Y et al. DNA replication and transcription programs respond to the same chromatin cues. Genome Res 24, 1102–1114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tardat M et al. The histone H4 lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol 12, 1086–1093 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Kuo AJ et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier–Gorlin syndrome. Nature 484, 115–119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iizuka M, Matsui T, Takisawa H & Smith MM Regulation of replication licensing by acetyltransferase Hbo1. Mol. Cell. Biol 26, 1098–1108 (2006).References 64–66 demonstrate that the initiation of DNA replication requires a chromatin modification, methylation of histone H4 on lysine 20, which is recognized by the ORC1 component of the pre-replication complex.
- 67.McConnell KH, Dixon M & Calvi BR The histone acetyltransferases CBP and Chameau integrate developmental and DNA replication programs in Drosophila ovarian follicle cells. Development 139, 3880–3890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y, Armstrong RL, Duronio RJ & MacAlpine DM Methylation of histone H4 lysine 20 by PR-Set7 ensures the integrity of late replicating sequence domains in Drosophila. Nucleic Acids Res 44, 7204–7218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stroud H et al. DNA methyltransferases are required to induce heterochromatic re-replication in Arabidopsis. PLoS Genet 8, e1002808 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aladjem MI et al. Participation of the human β-globin locus control region in initiation of DNA replication. Science 270, 815–819 (1995). [DOI] [PubMed] [Google Scholar]
- 71.Kalejta RF et al. Distal sequences, but not ori-β/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell 2, 797–806 (1998). [DOI] [PubMed] [Google Scholar]
- 72.Hayashida T et al. Replication initiation from a novel origin identified in the Th2 cytokine cluster locus requires a distant conserved noncoding sequence. J. Immunol 176, 5446–5454 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Guillou E et al. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev 24, 2812–2822 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Foti R et al. Nuclear architecture organized by Rif1 underpins the replication-timing program. Mol. Cell 61, 260–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hiraga S et al. Rif1 controls DNA replication by directing protein phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev 28, 372–383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoggard T, Shor E, Muller CA, Nieduszynski CA & Fox CA A link between ORC-origin binding mechanisms and origin activation time revealed in budding yeast. PLoS Genet 9, e1003798 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nieduszynski CA, Blow JJ & Donaldson AD The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription. Nucleic Acids Res 33, 2410–2420 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DePamphilis ML et al. Regulating the licensing of DNA replication origins in metazoa. Curr. Opin. Cell Biol 18, 231–239 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Hiratani I et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20, 155–169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anglana M, Apiou F, Bensimon A & Debatisse M Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114, 385–394 (2003).This study showed that the locations of replication origins are flexible and can be altered by metabolic conditions.
- 81.Debatisse M, Le Tallec B, Letessier A, Dutrillaux B & Brison O Common fragile sites: mechanisms of instability revisited. Trends Genet 28, 22–32 (2012). [DOI] [PubMed] [Google Scholar]
- 82.Gerhardt J et al. The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells. Mol. Cell 53, 19–31 (2014).The study showed that changes in replication origin usage are associated with replication fork stalling and repeat instability, which lead to repeat expansion at the FMR1 locus during early embryogenesis.
- 83.Fu H et al. The DNA repair endonuclease Mus81 facilitates fast DNA replication in the absence of exogenous damage. Nat. Commun 6, 6746 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ozeri-Galai E, Tur-Sinai M, Bester AC & Kerem B Interplay between genetic and epigenetic factors governs common fragile site instability in cancer. Cell. Mol. Life Sci 71, 4495–4506 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bielinsky AK Replication origins: why do we need so many? Cell Cycle 2, 307–309 (2003). [PubMed] [Google Scholar]
- 86.Blow JJ, Ge XQ & Jackson DA How dormant origins promote complete genome replication. Trends Biochem. Sci 36, 405–414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Conti C et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res 70, 4470–4480 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen YH et al. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol. Cell 58, 323–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Regairaz M et al. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I–DNA complexes. J. Cell Biol 195, 739–749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.DePamphilis ML Replication origins in metazoan chromosomes: fact or fiction? Bioessays 21, 5–16 (1999). [DOI] [PubMed] [Google Scholar]
- 91.Donley N, Smith L & Thayer MJ ASAR15, a cis-acting locus that controls chromosome-wide replication timing and stability of human chromosome 15. PLoS Genet 11, e1004923 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Flickinger RA Possible role of H1 histone in replication timing. Dev. Growth Differ 57, 1–9 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Schepers A & Papior P Why are we where we are? Understanding replication origins and initiation sites in eukaryotes using ChIP-approaches. Chromosome Res 18, 63–77 (2010). [DOI] [PubMed] [Google Scholar]
- 94.Marahrens Y & Stillman B A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255, 817–823 (1992). [DOI] [PubMed] [Google Scholar]
- 95.Knott SR et al. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148, 99–111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peace JM, Villwock SK, Zeytounian JL, Gan Y & Aparicio OM Quantitative BrdU immunoprecipitation method demonstrates that Fkh1 and Fkh2 are rate-limiting activators of replication origins that reprogram replication timing in G1 phase. Genome Res 26, 365–375 (2016).References 95 and 96 report a specific interaction between yeast transcription factors and replication origins that modulates the timing of replication by facilitating clustering of early-replicating origins with pre-replication complex proteins.
- 97.Donti TR, Datta S, Sandoval PY & Kapler GM Differential targeting of Tetrahymena ORC to ribosomal DNA and non-rDNA replication origins. EMBO J 28, 223–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chiang CM et al. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl Acad. Sci. USA 89, 5799–5803 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schepers A et al. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J 20, 4588–4602 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sears J, Kolman J, Wahl GM & Aiyar A Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by Epstein– Barr nuclear antigen 1. J. Virol 77, 11767–11780 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Piirsoo M, Ustav E, Mandel T, Stenlund A & Ustav M Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J 15, 1–11 (1996). [PMC free article] [PubMed] [Google Scholar]
- 102.McPhillips MG, Oliveira JG, Spindler JE, Mitra R & McBride AA Brd4 is required for E2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol 80, 9530–9543 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McPhillips MG, Ozato K & McBride AA Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol 79, 8920–8932 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Parish JL, Bean AM, Park RB & Androphy EJ ChlR1 is required for loading papillomavirus E2 onto mitotic chromosomes and viral genome maintenance. Mol. Cell 24, 867–876 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Bosco G, Du W & Orr-Weaver TL DNA replication control through interaction of E2F–RB and the origin recognition complex. Nat. Cell Biol 3, 289–295 (2001). [DOI] [PubMed] [Google Scholar]
- 106.Beall EL et al. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420, 833–837 (2002). [DOI] [PubMed] [Google Scholar]
- 107.Aggarwal BD & Calvi BR Chromatin regulates origin activity in Drosophila follicle cells. Nature 430, 372–376 (2004). [DOI] [PubMed] [Google Scholar]
- 108.Ghosh M et al. Differential binding of replication proteins across the human c-myc replicator. Mol. Cell. Biol 26, 5270–5283 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chowdhury A et al. The DNA unwinding element binding protein DUE-B interacts with Cdc45 in preinitiation complex formation. Mol. Cell. Biol 30, 1495–1507 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Thangavel S et al. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell. Biol 30, 1382–1396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Im JS et al. RecQL4 is required for the association of Mcm10 and Ctf4 with replication origins in human cells. Cell Cycle 14, 1001–1009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Miotto B & Struhl K HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev 22, 2633–2638 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miotto B & Struhl K HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by geminin. Mol. Cell 37, 57–66 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Miotto B & Struhl K JNK1 phosphorylation of Cdt1 inhibits recruitment of HBO1 histone acetylase and blocks replication licensing in response to stress. Mol. Cell 44, 62–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang Y et al. A replicator-specific binding protein essential for site-specific initiation of DNA replication in mammalian cells. Nat. Commun 7, 11748 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mahajan MC, Karmakar S & Weissman SM Control of beta globin genes. J. Cell. Biochem 102, 801–810 (2007). [DOI] [PubMed] [Google Scholar]
- 117.Sugimoto N et al. Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture. Nucleic Acids Res 43, 5898–5911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jin J, Arias EE, Chen J, Harper JW & Walter JC A family of diverse Cul4–Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721 (2006). [DOI] [PubMed] [Google Scholar]
- 119.Higa LA et al. CUL4–DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol 8, 1277–1283 (2006). [DOI] [PubMed] [Google Scholar]
- 120.Comelli L et al. The homeotic protein HOXC13 is a member of human DNA replication complexes. Cell Cycle 8, 454–459 (2009). [DOI] [PubMed] [Google Scholar]
- 121.Salsi V et al. HOXD13 binds DNA replication origins to promote origin licensing and is inhibited by geminin. Mol. Cell. Biol 29, 5775–5788 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Budhavarapu VN, Chavez M & Tyler JK How is epigenetic information maintained through DNA replication? Epigenetics Chromatin 6, 32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Demczuk A et al. Regulation of DNA replication within the immunoglobulin heavy-chain locus during B cell commitment. PLoS Biol 10, e1001360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koren A et al. Genetic variation in human DNA replication timing. Cell 159, 1015–1026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mukhopadhyay R et al. Allele-specific genome-wide profiling in human primary erythroblasts reveal replication program organization. PLoS Genet 10, e1004319 (2014).References 124 and 125 show that variations in replication timing can be associated with specific alleles, suggesting a role for cis-acting elements in the determination of replication timing.
- 126.Li PC, Chretien L,, Cote J,, Kelly TJ & Forsburg SL S. pombe replication protein Cdc18 (Cdc6) interacts with Swi6 (HP1) heterochromatin protein: region specific effects and replication timing in the centromere. Cell Cycle 10, 323–336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J & Masukata H The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat. Cell Biol 11, 357–362 (2009). [DOI] [PubMed] [Google Scholar]
- 128.Tazumi A et al. Telomere-binding protein Taz1 controls global replication timing through its localization near late replication origins in fission yeast. Genes Dev 26, 2050–2062 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zofall M, Smith DR, Mizuguchi T, Dhakshnamoorthy J & Grewal SI Taz1–shelterin promotes facultative heterochromatin assembly at chromosome-internal sites containing late replication origins. Mol. Cell 62, 862–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tashiro S et al. Shugoshin forms a specialized chromatin domain at subtelomeres that regulates transcription and replication timing. Nat. Commun 7, 10393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schwaiger M, Kohler H, Oakeley EJ, Stadler MB & Schubeler D Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res 20, 771–780 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Giri S et al. The preRC protein ORCA organizes heterochromatin by assembling histone H3 lysine 9 methyltransferases on chromatin. eLife 4, e06496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yamazaki S et al. Rif1 regulates the replication timing domains on the human genome. EMBO J 31, 3667–3677 (2012).References 74, 75 and 133 identify the RIF1 protein as a determinant of replication timing in mammalian cells and propose that it modulates the activity of the MCM replicative helicase by interfering with an essential phosphorylation step.
- 134.Koren A & McCarroll SA Random replication of the inactive X chromosome. Genome Res 24, 64–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kenigsberg E et al. The mutation spectrum in genomic late replication domains shapes mammalian GC content. Nucleic Acids Res 44, 4222–4232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.De S & Michor F DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat. Biotechnol 29, 1103–1108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ryba T et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20, 761–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hiratani I, Takebayashi S, Lu J & Gilbert DM Replication timing and transcriptional control: beyond cause and effect — part II. Curr. Opin. Genet. Dev 19, 142–149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yaffe E et al. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet 6, e1001011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thayer MJ Mammalian chromosomes contain cis-acting elements that control replication timing, mitotic condensation, and stability of entire chromosomes. Bioessays 34, 760–770 (2012).This essay describes studies showing that cis-acting elements can regulate the replication timing of entire chromosomes in cancer cells and thus affect genomic stability.
- 141.Mendez J Temporal regulation of DNA replication in mammalian cells. Crit. Rev. Biochem. Mol. Biol 44, 343–351 (2009). [DOI] [PubMed] [Google Scholar]
- 142.Wu JR & Gilbert DM A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science 271, 1270–1272 (1996). [DOI] [PubMed] [Google Scholar]
- 143.Mantiero D, Mackenzie A, Donaldson A & Zegerman P Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 30, 4805–4814 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tanaka S, Nakato R, Katou Y, Shirahige K & Araki H Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr. Biol 21, 2055–2063 (2011). [DOI] [PubMed] [Google Scholar]
- 145.Bustin M & Misteli T Nongenetic functions of the genome. Science 352, aad6933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Norio P et al. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell 20, 575–587 (2005).This study directly visualized replication initiation events on stretched DNA fibres, demonstrating that the gradual activation of replication origins correlates with increased replication time during differentiation.
- 147.Kawabata T et al. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell 41, 543–553 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Luebben SW, Kawabata T, Johnson CS, O’Sullivan MG & Shima N A concomitant loss of dormant origins and FANCC exacerbates genome instability by impairing DNA replication fork progression. Nucleic Acids Res 42, 5605–5615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shirahige K et al. Regulation of DNA-replication origins during cell-cycle progression. Nature 395, 618–621 (1998).This was the first report of a cell-cycle checkpoint that regulates the initiation of DNA replication in response to DNA damage.
- 150.Seiler JA, Conti C, Syed A, Aladjem MI & Pommier Y The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol. Cell. Biol 27, 5806–5818 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Naim V, Wilhelm T, Debatisse M & Rosselli F ERCC1 and MUS81–EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol 15, 1008–1015 (2013). [DOI] [PubMed] [Google Scholar]
- 152.Tuduri S et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol 11, 1315–1324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Courbet S et al. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455, 557–560 (2008). [DOI] [PubMed] [Google Scholar]
- 154.Ge XQ, Jackson DA & Blow JJ Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev 21, 3331–3341 (2007).This study showed that activation of excess dormant origins requires additional MCM helicase complexes and provides the first protection against deleterious replication fork stalling.
- 155.Gerhardt J et al. Cis-acting DNA sequence at a replication origin promotes repeat expansion to fragile X full mutation. J. Cell Biol 206, 599–607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dershowitz A et al. Linear derivatives of Saccharomyces cerevisiae chromosome III can be maintained in the absence of autonomously replicating sequence elements. Mol. Cell. Biol 27, 4652–4663 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shimada K, Pasero P & Gasser SM ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev 16, 3236–3252 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Letessier A et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470, 120–123 (2011). [DOI] [PubMed] [Google Scholar]
- 159.Casper AM, Nghiem P, Arlt MF & Glover TW ATR regulates fragile site stability. Cell 111, 779–789 (2002). [DOI] [PubMed] [Google Scholar]
- 160.Franchitto A & Pichierri P Replication fork recovery and regulation of common fragile sites stability. Cell. Mol. Life Sci 71, 4507–4517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Koundrioukoff S et al. Stepwise activation of the ATR signaling pathway upon increasing replication stress impacts fragile site integrity. PLoS Genet 9, e1003643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Barlow JH et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 152, 620–632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Halazonetis TD, Gorgoulis VG & Bartek J An oncogene-induced DNA damage model for cancer development. Science 319, 1352–1355 (2008). [DOI] [PubMed] [Google Scholar]
- 164.Bicknell LS et al. Mutations in the pre-replication complex cause Meier–Gorlin syndrome. Nat. Genet 43, 356–359 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dominguez-Sola D et al. Non-transcriptional control of DNA replication by c-Myc. Nature 448, 445–451 (2007). [DOI] [PubMed] [Google Scholar]
- 166.Jones RM et al. Increased replication initiation and conflicts with transcription underlie cyclin E-induced replication stress. Oncogene 32, 3744–3753 (2013). [DOI] [PubMed] [Google Scholar]
- 167.Vaziri C et al. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11, 997–1008 (2003). [DOI] [PubMed] [Google Scholar]
- 168.Miron K, Golan-Lev T, Dvir R, Ben-David E & Kerem B Oncogenes create a unique landscape of fragile sites. Nat. Commun 6, 7094 (2015). [DOI] [PubMed] [Google Scholar]
- 169.Sheu YJ, Kinney JB & Stillman B Concerted activities of Mcm4, Sld3, and Dbf4 in control of origin activation and DNA replication fork progression. Genome Res 26, 315–330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Aparicio T, Ibarra A & Mendez J Cdc45–MCM– GINS, a new power player for DNA replication. Cell Div 1, 18 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Depamphilis ML, de Renty CM, Ullah Z & Lee CY “The Octet”: eight protein kinases that control mammalian DNA replication. Front. Physiol 3, 368 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tanaka S & Araki H Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol 5, a010371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sansam CG, Goins D, Siefert JC, Clowdus EA & Sansam CL Cyclin-dependent kinase regulates the length of S phase through TICRR/TRESLIN phosphorylation. Genes Dev 29, 555–566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Boos D, Yekezare M & Diffley JF Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science 340, 981–984 (2013). [DOI] [PubMed] [Google Scholar]
- 175.Novac O, Matheos D, Araujo FD, Price GB & Zannis-Hadjopoulos M In vivo association of Ku with mammalian origins of DNA replication. Mol. Biol. Cell 12, 3386–3401 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gao Y et al. Protein phosphatase 2A and Cdc7 kinase regulate the DNA unwinding element-binding protein in replication initiation. J. Biol. Chem 289, 35987–36000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sincennes MC et al. The LMO2 oncogene regulates DNA replication in hematopoietic cells. Proc. Natl Acad. Sci. USA 113, 1393–1398 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]