Abstract
Primordial germ cells (PGCs) must complete a complex and dynamic developmental program during embryogenesis to establish the germline. This process is highly conserved and involves a diverse array of tasks required of PGCs, including migration, survival, sex differentiation, and extensive epigenetic reprogramming. A common theme across many organisms is that PGC success is heterogeneous: only a portion of all PGCs complete all these steps while many other PGCs are eliminated from further germline contribution. The differences that distinguish successful PGCs as a population are not well understood. Here, we examine variation that exists in PGCs as they navigate the many stages of this developmental journey. We explore potential sources of PGC heterogeneity and their potential implications in affecting germ cell behaviors. Lastly, we discuss the potential for PGC development to function as a multistage selection process that assesses heterogeneity in PGCs to refine germline quality.
1. Introduction
Sexual reproduction culminates with the fusion of two gametes to generate a zygote, briefly returning life to a fleeting, single-celled state. This simple cellular arithmetic, however, belies the momentous journeys undertaken by each gametic partner to enable this union. As we consider the epic reproductive development that precedes the production of this unicellular zygote, it is most appropriate that our focus begin, humbly, at the level of the single cell. Though the rise of metazoans introduced the dazzling complexity of diversified cell types and tissues, all multicellular systems are still fundamentally reducible to single cells—the smallest unit of biological structure.
While recent advances in single-cell technologies promise to reveal powerful insights about the identities of individual cells that compose a lineage or tissue, much of biology has begun by asking questions of single cells. From Anton van Leeuwenhoek at his microscope to Paul Ehrlich and his histological dyes, we have long analyzed differences that exist from cell to individual cell. The advantage of whole-mount or section immunofluorescence imaging lies in the ability to resolve multiple cells in a tissue while preserving individual differences for measurement (Levsky & Singer, 2003). Such powerful resolution permits a more comprehensive understanding of cellular interactions and a sensitivity to cell-to-cell variation. In contrast, analytical methods like Western blots, PCR, or bulk sequencing provide only population-level insights under the assumption that behavior is uniform. Even within single lineages, it is increasingly understood that significant variation can exist among seemingly identical cells (Altschuler & Wu, 2010; Raj & van Oudenaarden, 2008) and that this heterogeneity can have far-ranging functional consequences.
With this principle in mind, we can begin tracing the steps that generate gametes—themselves single-celled carriers of the genome. Each germ cell must endure a lengthy and complex development that begins shortly after fertilization when the germline is set aside from the somatic lineages with the formation of primordial germ cells (PGCs). The path from PGC to gamete is highly conserved, demonstrating the evolutionary importance of reproductive development (Nieuwkoop & Sutasurya, 1979, 1981). Early PGC development is especially dynamic and requires transition through differentiated states, interaction with diverse cellular environments, and processing a multitude of signals. This complexity can amplify differences among individual cells to yield varied cell fates. For germ cells that are tasked with creating gametes, those that survive this developmental crucible secure the ultimate biological prize: propagation of their genetic identity.
The concept of selective events acting on variation within a population has long been appreciated in biology, albeit on the level of organisms in Darwinian natural selection. These same principles are also applicable at the cellular level as well, and we argue that they can govern the fate of PGC heterogeneity during development (Buss, 1988; Laird, Chang, Weissman, & Lauzon, 2005; Weissman, 2015). The diversity of challenges posed to PGCs in this period may function as developmental selection that acts upon cellular variation in the germline in a manner analogous to natural selection with organisms. Here, we consider how heterogeneity in PGCs provides source material for selection by PGC development. This review presents a comprehensive perspective on early events in germ cell development with a particular emphasis on how heterogeneity manifests in germ cells, how it arises, and its impact on the germline and reproductive fitness.
2. Heterogeneous phenotypes of primordial germ cells
Heterogeneity is a fundamental property of biological systems consisting of multiple units such as cells. Even though cells may share a common lineage, it is increasingly recognized that substantial variation exists even within cell types. As our ability to measure cellular phenotypes and behaviors improves, we can distinguish rare cell subpopulations in tissues such the intestinal crypt (Grün et al., 2015) as well as canvass the broad diversity that exists in chaotic intratumoral settings (McGranahan & Swanton, 2017). In hematopoiesis, these subpopulations can represent increasingly smaller organizational units that possess distinctive behaviors and fates (Schroeder, 2010).
While heterogeneity has been studied in many homeostatic contexts such as the adult stem cells of the intestine or blood, its role in germline development is a unique and interesting consideration. The establishment of PGCs at the outset of embryogenesis, as the first lineage in many different organisms, mandates a lengthy process of development through which a subset of germ cells become gametes. The dynamic and multifaceted challenges encountered by PGCs during fetal development present numerous opportunities to assess heterogeneity that may exist in nascent PGCs. As PGCs go on to establish the germline and generate gametes, how heterogeneity is resolved by developmental selection can prove especially influential for transgenerational inheritance.
This section aims to establish a conceptual framework to define and identify heterogeneity as it pertains to the germline and reproductive fitness (see Box 1). We will examine evidence for heterogeneity in specific aspects of PGC development in several model organisms to uncover general principles that underlie the unique challenges and adaptations utilized by germ cells to address variation (schematized in Fig. 2).
BOX 1. Clarification of terminology.
Cell type:
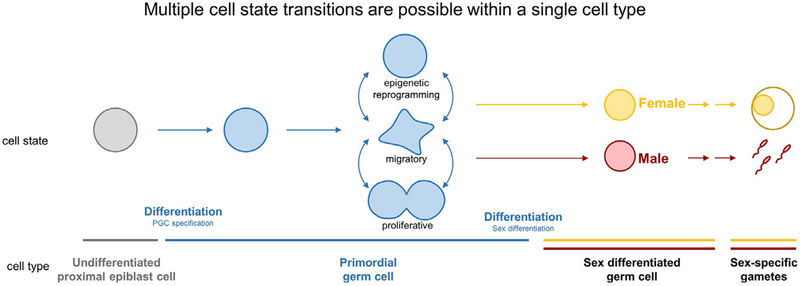
Although cell type has been defined by many criteria such as morphology, location, lineage history, and distinct transcriptional profile (Altschuler & Wu, 2010; Muller-Sieburg, Sieburg, Bernitz, & Cattarossi, 2012; Seoane, 2017), we treat the germ cell as a functional entity theoretically capable of giving rise to gametes of either sex (Fig. 1). This definition encompasses changes in morphology, location, and gene expression involved in PGC development, as well as the different modes of specification and routes of PGC migration between organisms. We define PGCs as the committed but yet sexually undifferentiated embryonic precursor of the gamete. Although we acknowledge that PGC embryonic origins may have evolved multiple times, we draw comparisons between PGCs in different organisms based on their operational similarities and parallel constraints.
Cell state:
A similarly nebulous term, we consider cell state to be a stable but transient set of activities, a potency, a distinct transcriptional profile, and/or a set of epigenetic marks that defines a subpopulation of cells within a particular cell type. Similar to the poised, active, and silenced states of embryonic stem cells associated with distinct sets of epigenetic marks and gene expression (Young, 2011), we consider that PGCs can occupy different states as well as transition back and forth between them. Cell state and cell type are interdependent; PGCs are a cell type, but PGCs in the embryo are a collection of cells in multiple states. PGC differentiation to male gonocyte or oocyte involves a change of cell type that may occur through a series of cell state transitions.
Heterogeneity (of cells):
Synonymous with cell individuality and cell diversity, cell heterogeneity is treated here as a biological difference between cells of the same type (as opposed to noise that arises from experimental or technical factors). Heterogeneity can be detected as distinct cellular behaviors, including morphology or function (such as proliferation, capacity for differentiation, or migration);these behaviors may be reflected in transcriptional state or epigenetic profile. Determinants or causes of cellular heterogeneity may include genetic changes, epigenetic changes (which is relevant for PGCs more than other cells), non-genetic changes that may or may not be stochastic, or extrinsically induced changes.
Fig. 2.
Readouts of PGC Heterogeneity. Snapshots of the reproductive life cycle evidence potential readouts of heterogeneity. These readouts can be broadly surveyed across many model organisms. Key phases of potential heterogeneity occur during specification, migration, proliferation, sex determination, survival, differentiation, and gametogenesis. Specific readouts may be applicable several cell states.
2.1. Primordial germ cell specification and heterogeneity
This section considers the potential for heterogeneity to arise between PGCs during their specification, as well as specific examples. Among metazoans, there are two modes by which PGCs are specified: preformation and induction. Also termed epistasis, preformation confers a germline fate by cytoplasmic inheritance of maternally derived organelles composed of proteins and RNAs termed germ plasm, germ granules, polar or P granules; mechanisms of germline preformation have been reviewed extensively by others (Extavour, 2003; Saitou, Payer, O’Carroll, Ohinata, & Surani, 2005; Solter, 2016). Of the model organisms commonly studied, Caenorhabditis elegans, Drosophila melanogaster, Xenopus laevis, and Danio rerio all generate PGCs through early preformation. By contrast, mammals, urodele amphibians and crickets specify germline by induction from somatic cells, which involves signaling from the surrounding niche that leads to a binary fate decision (Ewen-Campen, Donoughe, Clarke, & Extavour, 2013; Lawson et al., 1999; Nieuwkoop, 1947; Tam & Zhou, 1996); underlying molecular mechanisms are well reviewed elsewhere (Solter, 2016).
Does the mode of specification limit the extent of heterogeneity that is possible between PGCs? It is not unreasonable to assume that the germ plasm promotes homogeneity from the inception of the PGC lineage by preformation. The inclusion of proteins and transcripts in the germ plasm regulates the levels of both in the cytoplasm, particularly since PGC specification precedes zygotic transcription in model organisms with preformation, and transcription remains suppressed in nascent PGCs even after beginning in the soma (Strome & Lehmann, 2007). It might be expected that germ plasm promotes PGC homogeneity on a continual basis given the capacity for movement through specialized cytoplasmic bridges between germ cells (Extavour, 2003). During PGC specification, it has been proposed that niche functions to shield PGCs from somatic fate signals; this idea is reinforced by the observation that PGCs arise via preformation or induction paradigms in sequestered locations, such as the posterior pole in fly embryos (Eddy, 1975), near the extraembryonic tissues in mammals and birds (Ginsburg & Eyal-Giladi, 1986, 1987; Ginsburg, Snow, & McLaren, 1990), or on the periphery of zebrafish embryos (Raz, 2002). However, in the context of this discussion, we might speculate that the niche for specification instead protects PGCs from signals that would promote heterogeneity. On the other hand, the inductive mode is arguably more conducive to producing heterogeneous PGCs since the process of fate conferral by signaling can occur over a more protracted period of time in principle. The comparative effects of these two mechanisms of PGC fate determination may have relevance in an evolutionary context. Examining patterns of PGC specification across many species precipitated the notion that preformation is derived whereas induction is ancestral (Extavour, 2003). We might ask, then—if germ plasm indeed promotes uniformity, whereas induction promotes greater diversity of PGCs—what benefit does reduced heterogeneity confer upon those species that acquired preformation?
2.1.1. PGC specification by preformation
We discuss several examples of heterogeneous behaviors of PGCs associated with specification by preformation. In this mode of development, germline establishment requires asymmetrical distribution of germ plasm in the oocyte and zygote (Hird, Paulsen, & Strome, 1996), followed by asymmetric inheritance during early divisions into the germline precursors and PGCs (Eddy, 1975; Mahowald, 1962, 2001). However, subsequent development of PGCs involves a switch from asymmetric to symmetric inheritance of germ plasm to daughter cells. A key question is whether minor asymmetries in either the allocation of germ plasm between founder PGCs or the subsequent allocation to PGC daughter cells have phenotypic consequences. A recent study in Drosophila suggests that indeed some founder PGCs are short-changed in their inheritance. Quantifiable variation was observed in the levels of maternal mRNAs Nanos and Wunen associated with the germ plasm between individual PGCs. Importantly, the levels of both were not arbitrary, but highly correlated with the probability of subsequent germ cell death, suggesting that Wunen is an indicator of germ plasm quality (Slaidina & Lehmann, 2017). Similarly, in Caenorhabditis elegans, the asymmetric segregation of cytoplasmic determinants known as P granules occurs in the P4 ancestor cell of the two PGCs known as Z2 and Z3 (Sulston, Schierenberg, White, & Thomson, 1983). Surprisingly, PGCs were specified and fertility was maintained in a mutant with symmetric distribution of P granules, suggesting that the P granules themselves are not sufficient for the worm germline (Gallo, Wang, Motegi, & Seydoux, 2010).
Further insight into the critical components of germ plasm for conferring a germ cell fate and potentially generating diversity of PGCs comes from recent work in Drosophila. The germ plasm component oskar is both necessary and sufficient for specifying PGCs in flies, and plays a deterministic role in the number of precursor pole cells (Ephrussi & Lehmann, 1992; Ewen-Campen, Srouji, Schwager, & Extavour, 2012; Smith, Wilson, & Macdonald, 1992). A surprising function of one isoform of oskar is to recruit mitochondria to nascent PGCs via the actin cytoskeleton; a reduction in the number of PGCs and diminished downstream oogenesis in mutants lacking this oskar long isoform implicates mitochondrial inheritance and number in the competence and normal development of PGCs (Hurd et al., 2016). In zebrafish, the maternal gene bucky ball functions in a parallel manner to oskar via interactions with microtubule networks to assemble the germ plasm, which involves aggregating mitochondria into a structure known as the Balbiani body. Ectopic PGCs are produced by ectopic expression of bucky ball, but how they may differ from properly localized germ cells is unclear (Bontems et al., 2009; Marlow, 2015). Similarly, it is not known how mis-localized PGCs that result from ectopic expression of oskar in flies might be different than those specified according to established patterning, but an avenue of inquiry will be interrogate potential differences in mitochondria between individual PGCs as related to subsequent fate and function.
2.1.2. PGC specification by induction
Of the organisms that specify their germline through induction and signaling from somatic cells, Mus musculus is one of the most studied. Despite this fact, much remains unknown about how heterogeneity may impact murine reproductive potential. Early fate mapping studies of single cells from E4.5 the mouse embryo produced an important negative result: no single cell of the inner cell mass exclusively gave rise to PGCs (Gardner & Rossant, 1979). Mouse PGCs are induced in the epiblast by BMPs from the surrounding extraembryonic tissues (Fujiwara, Dunn, & Hogan, 2001; Lawson et al., 1999; Ying, Liu, Marble, Lawson, & Zhao, 2000). The earliest known marker of the PGC fate is Blimp1 (also known as Prdm1), which first appears in the epiblast at E6.25. Although other genes of PGC commitment do not come on until ~E7, lineage tracing with Blimp1-Cre suggests that all cells in the region of the allantois that have expressed Blimp1 become PGCs (Ohinata et al., 2005). This approach does not distinguish between cells that descend from the first to express Blimp1 and those derived from later onset of Blimp1 in subsequent recruits from the epiblast, hence the number of PGCs specified directly from epiblast in mice remains elusive. Several groups have attempted to identify the number of founder PGCs which actually contribute to germ cell lineages. Examination of the earliest Blimp1 transcript found four to eight Blimp1-positive cells in most proximal epiblast which grow into a tight cluster of approximately 20 cells at the midstreak (MS) stage (Saitou et al., 2005). Approaches using retroviruses or generating chimeras with different-colored embryonic stem cells delivered to the blastocyst have estimated that all PGCs derive from a founder population of three or four cells (Soriano & Jaenisch, 1986; Ueno, Turnbull, & Weissman, 2009). It is unclear whether the growth of cell number from 4 to 5 to the observed BLIMP1 + cells in the epiblast to the ~40 PGCs observed at e7 (Ginsburg et al., 1990) occurs through induction of new cells into the germline from the epiblast, or proliferation of the previously induced 4–5 PGCs. The mouse germline undoubtedly derives from a narrow set of ancestral founders, and the yet unclear size of this founder population has potential implications for heterogeneity of PGCs (Zheng, Luebeck, Byers, & Moolgavkar, 2005).
Further insight into the question of whether all PGC founders are equivalent will emerge from more sophisticated lineage tracing of the germline, such as genetic barcoding. A glimpse into potential heterogeneity comes from knockout studies. Genetic ablation of Blimp1 leads to the specification of PGCs with incomplete repression of somatic genes and activation of pluripotency genes, but this phenotype varies in severity depending on the mouse genetic background (Avilion et al., 2003; Yabuta, Kurimoto, Ohinata, Seki, & Saitou, 2006). These results raise the possibility that variable levels of Blimp1 between early mouse PGCs could lead to differences in their capacity to fully realize their fate, which could potentially impact their behavior and function. More recently, the homeobox transcription factor Otx2 was identified as an earlier repressor of the PGC fate that turns off 12–24h before the appearance of Blimp1. Whereas overexpression of Otx2 decreases the number of PGCs, the absence of Otx2 augments the number of PGCs, indicating a negative correlation between PGCs and Otx2 expression (Zhang et al., 2018). Although these experiments lacked the sensitivity to detect low levels of Otx2, the question remains whether PGC precursors that incompletely extinguish Otx2 become PGCs with low levels of somatic gene expression that parlay into measurable differences in behavior (Laird, 2018).
2.2. Primordial germ cell migration and heterogeneity
In many organisms, PGCs are specified far from their permanent tissue niche, and must walk a long distance through growing and developing somatic compartments to find the gonad primordia, which will furnish a niche that promotes their survival and differentiation. To accomplish this feat, PGCs must be responsive to external migration cues as well as able to navigate their way through temporary migratory niches, recognize the gonad niche, and stop migrating when they arrive. Navigation of these challenges by each individual germ cell may amplify initially small differences, such that the migratory process reveals heterogeneity.
From the single germline progenitor P4 in C. elegans emerges the two PGCs: Z2 and Z3 (Sulston et al., 1983). While Z2 and Z3 themselves do not migrate per se, their development includes two relevant processes. First, cells of the somatic niche derived from Z1 and Z4 (called the distal tip cells) undertake a migration to reach the PGCs. Distal tip cells signal through Notch pathway to control proliferation of the germline (Rohrschneider & Nance, 2013), although how migration shapes their signaling abilities is unknown. Second, development of the surrounding embryo pulls the PGCs from the surface to the center of the embryo in order to reach their destination (Rohrschneider & Nance, 2013). Analogous to PGCs in other species, it remains unknown which mechanisms signal cessation to the moving cells (in this case the somatic gonad progenitors Z1 and Z4), although the presence of PGCs is required for the final positioning of Z1 and Z4 (Rohrschneider & Nance, 2013).
D. melanogaster PGCs, or pole cells, are specified in the posterior extreme of the embryo. Their migration begins by passive conveyance to the interior of the embryo during gastrulation. Upon reaching the gut epithelium, the PGCs exhibit amoeboid migratory movements triggered by gut tissue (Grubb, 2006); it is not known whether these gut-derived signals may vary depending on location or intrinsic signaling capacities of PGCs. However, functional studies of individual PGCs and their ability to respond to cues or enact an amoeboid migratory phenotype could begin to address this question. Active migration through the mesoderm niche takes PGCs to the forming gonads (Santos & Lehmann, 2004). Bifurcation from the midline toward the opposing gonads involves repulsive as well as attractive cues and takes individual PGCs on varying path lengths at varying speeds, but the extent or significance of this potential heterogeneity is not known.
The study of Zebrafish PGC migration reveals several interesting windows where heterogeneity may play a role in the development of the germline. After being specified in four random locations along the developmental axis, Zebrafish PGC morphology is smooth and round (Richardson & Lehmann, 2010). PGCs in Zebrafish must undergo a round of mitosis and a round of polarization before active migration initiates (Blaser et al., 2005), at which time they begin a directed, active movement under the guidance of chemokines (Raz & Reichman-Fried, 2006). This awakening occurs on an individual cell basis, emphasizing the potential for heterogeneity among the initial cell divisions and polarization processes (Raz & Reichman-Fried, 2006). Migratory initiation depends on the capability to mount an entirely new transcriptional program (Richardson & Lehmann, 2010), such that variations in transcription may correlate with variations in migration. Zebrafish PGCs exhibit a curious cycling of active “running” migration when they are polarized, and a “tumbling” pausing phase where they lose cell polarity and are able to make course corrections (Reichman-Fried, Minina, & Raz, 2004). The ability of an individual PGC to cycle properly between these stages is a necessity for survival, as those failing to complete these exercises become ectopic and undergo apoptosis outside of the range of survival signals. Interestingly, Zebrafish PGCs switch between individualized migration in initial phases to movement toward the gonads in clusters following aggregation at concentrated locations of the chemoattractant Sdf-1. This later stage of group migration may serve to mitigate differences in chemokine responsiveness or motility that arise earlier in Zebrafish PGC development.
Mouse PGCs begin their migration from their site of specification on the posterior side of the epiblast near the base of the extraembryonic structure known as the allantois (Ginsburg et al., 1990). This 3–4 day journey takes PGCs from the allantois, through the primitive gut and surrounding mesentery, and into the gonadal ridge. Migration is not synchronous, but spread out in space and time; some PGCs “lead” migration while others “lag” behind. Time-lapse imaging captures the individualistic behavior of PGCs as well as the fraction that migrate improperly, failing to reach the next mile-post or moving into ectopic locations (McDole et al., 2018; Molyneaux, Stallock, Schaible, & Wylie, 2001). Also evidenced in the live movies are differing levels of activity from E9.0 to E9.5, when germ cells are active and motile, but not yet exhibiting the directed migration which causes them to exit the gut epithelium. What distinguishes the earliest “pioneer” germ cells first exiting the gut from other germ cells that follow them is a heterogeneity profile as of yet unknown (Cantú, Altshuler-Keylin, & Laird, 2016; Gomperts, Garcia-Castro, Wylie, & Heasman, 1994). Another point at which heterogeneous PGC behaviors emerge is during the last stages of migration at E10.5, when approximately 10% of PGCs remain outside of the gonads, with little hope of completing their migration as determined by their persistence at E13.5 (Cantú et al., 2016; D.H. Nguyen & D.J. Laird, unpublished data). Motility of mouse PGCs is regulated by multiple signaling mechanisms, including the non-canonical Wnt signaling pathway via Wnt5a (Laird, Altshuler-Keylin, Kissner, Zhou, & Anderson, 2011), which attenuates the mitogenic beta-catenin dependent arm of the pathway (Cantú et al., 2016). Although migratory mouse PGC behavior is influenced by signals from different niches, which certainly promotes heterogeneous migratory outcomes, there also appears to be intrinsic heterogeneity of PGCs.
Cultured ex vivo in defined conditions, mouse PGCs mounted variable signaling responses to cues such as Wnt5a (Cantú et al., 2016). PGCs from embryos have also been observed to be heterogeneous in expression of important signaling receptors such as cKit, which together with Kit ligand regulate migration and survival (Morita-Fujimura, Tokitake, & Matsui, 2009). Finally, it is also possible that as the mesentery niche changes and develops around them, the PGCs in the lagging group lose contact with the proper migratory signals which would enable them to move forward. Even more intriguing is the idea that migration acts as a selection metric, to prevent germ cells with deleterious or detrimental heterogeneities from reaching the gonad and colonizing. This will be discussed more in the final section of this chapter.
2.3. Proliferation of primordial germ cells and heterogeneity
PGC development involves the expansion of a small handful of founder cells to a cohort of sex-undifferentiated germ cells in the gonads, which range in number from tens in flies and worms to thousands in mice. The proliferative histories of individual PGCs have not been examined. However, discrepancies in the number of self-renewing divisions could potentially amplify any inherent heterogeneity between PGCs by over-representing or under-representing particular founders. Alternatively, unequal divisions could disproportionately increase the opportunity for replication-associated damage in subsets of germ cells, thereby introducing new heterogeneity.
We present examples in several organisms of heterogeneous behavior of PGCs during proliferation. The two C. elegans founder PGCs Z2 and Z3 defer proliferation until larval stages, and rely upon regulation by somatic gonad precursors (Kimble & White, 1981). Germline development is acutely responsive to nutritional cues in worms (Hubbard, Korta, & Dalfo, 2013). Upon hatching, unless larvae eat, they enter a state of L1 developmental arrest called diapause, accompanied by an arrest in the proliferation of PGCs and their somatic support cells (Fukuyama, Rougvie, & Rothman, 2006). During L1 diapause, PGCs halt their cell cycle in G2 in a precisely controlled process under the auspices of Pten and the insulin/IGF-like signaling pathway. Release from L1 diapause and G2 arrest occurs upon feeding and involves cell cycle re-entry of only one of the two PGCs, which is believed to reflect communication between the PGCs via yet unknown mechanisms (Fukuyama et al., 2006). Although it is further unknown which of the two PGCs proliferates and the basis for this decision, this proliferative asymmetry in the C. elegans germline could amplify any heterogeneity that exists between Z2 and Z3.
Similarly, the regulation of Drosophila PGCs during development involves differential proliferation that could introduce or augment pre-existing heterogeneity. The 12 PGCs that arrive at each of the gonads double their number every 24 h during the first and second instar larval stages before curbing their expansion. Mutants possessing severely reduced numbers of embryonic PGCs—such as oskar and germcell-less which average at 2—reach the middle of 3rd larval instar with the correct number of PGCs. These mutants revealed a cross-talk between PGCs and somatic cells of the ovary via EGF signaling that senses and adjusts the number of PGCs (Gilboa & Lehmann, 2006). As in other organs, the size of the PGC pool in the larval fly ovary relies upon Hippo signaling between PGCs and somatic cells that form their niche (Sarikaya & Extavour, 2015). These compensatory mechanisms for PGC proliferation indicate that the Drosophila germline can be built from different numbers of founder PGCs and through differing numbers of divisions.
Such mechanisms for detecting and adjusting cell numbers similarly govern the proliferation of PGCs in chicken, where compensatory expansion was observed after treatment with the anti-mitogen busulfan (Lee et al., 2013). In mice, PGCs were deemed to undergo approximately eight divisions based on a population doubling time of 16 h, with compensation for early losses in the number of PGCs occurring during the period of expansion in the fetal gonads (Tam & Snow, 1981). During their migration, the proliferation of PGCs in the mouse embryo increases with each successive niche through which they transit (Cantú et al., 2016). PGC mitosis is regulated by beta-catenin-dependent Wnt signaling pathway but instructed by the niche. This extrinsic and niche-specific control of PGC proliferation, coupled with the asynchrony in PGC migration, raises the possibility that those migrating first or fastest would undergo more cycles of expansion, leading to a clonal dominance over those migrating last or slowest (Cantú et al., 2016). Although the number of cell divisions has been proposed to be deterministic in a number of processes—including the decision to enter meiosis (McLaren, 2003; McLaren & Southee, 1997; Ohkubo, Shirayoshi, & Nakatsuji, 1996), the probability of acquiring mutations (Ohno, 2018), and the quality of the oocyte (Henderson & Edwards, 1968)—the functional impact of proliferative history on the germline and gamete remains to be determined. The advent of molecular “flight recorders” that track cell divisions in vivo hold promise for revealing the extent and the consequences of heterogeneous proliferation in PGCs.
2.4. Primordial germ cell survival and heterogeneity
As PGCs progress through fetal development, many fail to complete the entire journey and are eliminated from contributing to the future germline. This elimination occurs mainly via developmentally programmed apoptosis at several stages in the fetal period. Importantly, this apoptosis is separate from PGC death in response to cytotoxic insults, such as irradiation or chemical exposure. Consistent PGC apoptosis during development suggests that apoptosis may act as a selective event. The PGCs that do survive fetal development represent the founding population that establishes the adult germline. Differences in PGC survival during this fetal period are therefore critical to determining the composition of gamete-producing cells in adulthood. Given that only a subset of the initial PGC population completes fetal development, survival represents a heterogeneous outcome that may reflect important differences between survivors and eliminated PGCs that are relevant to germ cell function.
Apoptosis is an invariant feature of C. elegans embryo development, with 131 out of 1090 somatic cells eliminated in a lineage-specific manner (Gartner, Boag, & Blackwell, 2008). In the germline, apoptosis occurs exclusively in the adult during oogenesis and eliminates a variable number of potential oocytes that does not exceed 50% (Gumienny, Lambie, Hartwieg, Horvitz, & Hengartner, 1999). Importantly, germline apoptosis differs from somatic apoptosis by virtue of the single oocyte lineage containing two variably sized but distinctly fated populations: surviving versus eliminated cells. In contrast, somatic apoptosis reproducibly eliminates precisely the same 131 cells. Oocyte apoptosis can be classified into two categories: stress-induced and physiological. Stress-induced apoptosis occurs in response to insults such as environmental stress, DNA damage, or bacterial infection. In contrast, physiological apoptosis affects a fraction of cells during oogenesis in the absence of any stress and differs from stress-induced apoptosis by not relying on pro-apoptotic BH3 proteins. Physiological apoptosis is restricted to oocytes in late pachytene that occupy a particular region of the gonad. Inhibition of germline apoptosis results in poorer quality oocytes as characterized by decreased egg viability and difficulty producing full-sized oocytes (Andux & Ellis, 2008). These reproductive defects may be due to intrinsic faults in the normally eliminated population that are not present in the oocytes that survive physiological apoptosis. Alternatively, apoptosis may not reflect intrinsic cellular differences between oocytes in the survivor versus dying populations but instead functions as a resource-management strategy. C. elegans germ cells are linked as a syncytia, allowing apoptotic cells to act as nurse cells and direct their cytoplasmic contents toward surviving cells (Wolke, Jezuit, & Priess, 2007). These two models are non-exclusive; cellular differences stemming from oocyte heterogeneity can serve as the basis for determining whether an oocyte acquires a nurse cell or survivor fate.
Drosophila germ cell development shares many conserved elements with higher-order organisms such as migration. Drosophila PGCs, also termed pole cells, that fail to migrate accurately undergo programmed cell death in the early embryo (Coffman et al., 2002). As many as half of the initial cohort of PGCs mismigrate and are ultimately eliminated (Underwood, Caulton, Allis, & Mahowald, 1980). Ectopic PGC removal does not require apoptosis genes grim, reaper, or hid and is caspase-independent (Sano, Renault, & Lehmann, 2005). Several studies have demonstrated that elimination of ectopic PGCs instead requires a pathway involving both p53 and out (Coffman et al., 2002; Yamada, Davis, & Coffman, 2008). Overexpression of p53 induces programmed cell death only in ectopic PGCs, suggesting that the threshold for elimination varies across subpopulations of PGCs that differ by migratory success. Differentiation is another criterion that can distinguish surviving PGCs from eliminated ones. In studying Nanos (nos), an evolutionarily conserved protein critical for many aspects of germline development, Hayashi et al. discovered that nos-mutant PGCs inappropriately acquire somatic identities and are subsequently removed through apoptosis (Hayashi, Hayashi, & Kobayashi, 2004). Interestingly, despite the central role of nanos in securing germline identity, nos-mutant PGCs only display a partially penetrant phenotype of somatic expression (Schaner, Deshpande, Schedl, & Kelly, 2003), suggesting that heterogeneity exists among PGCs in regard to retaining somatic competence. Given the role of nanos in regulating histone modifications, heterogeneous chromatin states among individual cells may explain this variation in differentiation propensity. To summarize, programmed cell death in Drosophila operates through both an apoptosis-independent and dependent mechanism, with apoptosis being associated with PGCs that inappropriately differentiate to the somatic lineage.
In Zebrafish, PGC apoptosis also is linked to migration, with disrupted migration leading to apoptosis. Weidinger et al. injected morpholinos targeting dead end mRNA, a component of germ plasm in zebrafish, and observed an increase in ectopic PGCs that would ultimately be eliminated by apoptosis (Weidinger et al., 2003). Apoptosis was preceded by down-regulation of key germ cell maintenance genes such as nos-1, indicating that mismigration disrupts germ cell identity in ectopic PGCs, leading to downstream apoptosis. Apoptosis is the eventual fate of ectopic PGCs in other migratory mutants (Köprunner, Thisse, Thisse, & Raz, 2001; Ramasamy, Wang, Quach, & Sampath, 2006) and may also be due to loss of nos-1 in ectopic PGCs. Importantly, in these migratory mutants some PGCs do correctly localize to the gonads, maintain nos-1 and its protein product Nos1, and survive, suggesting that correct migration supports the maintenance of germ cell identity and provides a survival advantage.
PGC survival in mice follows a similar paradigm to that in Drosophila and Zebrafish. Correct PGC migration is tightly linked to PGC survival. This coupling is primarily accomplished through genes that have dual roles in chemotaxis as well as survival. In mice, mutants for Dead-end (Dnd) and KitL (Steel) result in increased numbers of ectopic PGCs, which subsequently undergo apoptosis (Runyan et al., 2006; Youngren et al., 2005). KitL is downregulated in the midline after PGCs migrate through the tissue by E10.5, enforcing a narrow pro-survival niche (Runyan et al., 2006). The stringent connection between survival and migratory signals resolves migratory heterogeneity in favor of PGCs that can accurately navigate to the gonad. Because ectopic PGCs can give rise to germ cell tumors, this linking of migratory success to survival can prevent deleterious differentiation from mismigrated PGCs (Runyan, Gu, Shoemaker, Looijenga, & Wylie, 2008).
After reaching the gonads, mouse PGC apoptosis occurs primarily through a Bax-mediated mechanism peaking by E13.5 in both males and females (Coucouvanis, Sherwood, Carswell-Crumpton, Spack, & Jones, 1993; Wang, Nakane, & Koji, 1998). PGCs begin to upregulate transcripts of Bax, increasing their apoptotic potential between E10.5 through E13.5 (Stallock, Molyneaux, Schaible, Knudson, & Wylie, 2003). The Bcl-2 family members such as Bcl-x (Rucker et al., 2000) further regulate PGC survival. Variation in the expression and balancing of these apoptotic and survival pathways can modulate cell death propensity and could give rise to the apoptotic fraction observed even in wild-type backgrounds. Interestingly, successful differentiation is associated with increased survival. Knockout of Nanos3, a germline transcription factor important for germline differentiation, results in PGC depletion by Bax-dependent and -independent mechanisms (Suzuki & Saga, 2008). However, a subset of Nanos3 null PGCs are capable of surviving and differentiating in male and female adult gonads, suggesting that a subset of PGCs is more robust and capable of survival despite the loss of pro-differentiation components.
While studies of mutant PGCs have illuminated much of the differentiation and survival networks operating in the fetal period, the fraction of wild-type PGCs that undergo apoptosis indicate that not all PGCs are equivalent in their likelihood to contribute to gametogenesis. The identities of apoptotic germ cells on a wild-type background are beginning to be understood through techniques such as single-cell RNA sequencing and clonal labeling, both of which can elucidate subpopulation-level differences among germ cells that bulk analyses cannot. In particular, clonal labeling allows for direct comparisons among potentially heterogeneous clonal subpopulations. Multicolor clonal labeling revealed that apoptosis in the fetal male germline is clonal, suggesting that mitotically heritable differences among subpopulations—particularly in the expression of epigenetically regulated genes that control sex differentiation—can determine whether germ cells survive or are eliminated (Nguyen & Laird, 2019).
2.5. Differentiation of PGCs and heterogeneity
After specification, the next major differentiation event in PGC development is sex differentiation. Of the common model organisms, C. elegans is unique in featuring hermaphroditic gametogenesis, although genera-principles regarding somatic regulation of germline differentiation can apply. For example, C. elegans somatic cells at the distal tips of the gonads maintain mitotic potential, and meiosis only occurs as germ cells move away (Ellis, 2008). In Drosophila and mice, the somatic gonadal sexual identity significantly influences the sex differentiation of resident germ cells (Casper & Van Doren, 2006; Ewen & Koopman, 2010). This section will primarily discuss differentiation of male and female germ cells in mice, with a focus on meiotic initiation and mitotic arrest.
Upon arriving in the gonad, PGCs undergo sexually dimorphic differentiation into the highly specialized male and female lineages. The signal for meiotic initiation in mice is retinoic acid (RA), which is secreted by somatic cells in the mesonephros (Bowles et al., 2006; Koubova et al., 2006). As RA must diffuse to germ cells in the ovary, the diffusion gradient could produce variation in both strength and timing of RA signaling. Meiosis in the mouse ovary is observed to occur in an anterior to posterior wave that may reflect asynchronous initiation (Menke, Koubova, & Page, 2003). Interestingly, this anteroposterior wave does not align with the axis of diffusion between the mesonephros and the ovary, suggesting that other signals may modulate RA signaling within the ovary. RA has also been discovered to be produced by ovarian cells (Bowles et al., 2016; Childs, Cowan, Kinnell, Anderson, & Saunders, 2011) and the Aldh1a1 enzyme. Meiotic progression involves Stra8, which is activated by Dmrt binding to the Stra8 promoter (Matson et al., 2010). Stra8 mutants display greatly reduced follicle formation, but a small population of Dmrt1-null oocytes can continue to form functional follicles and express the appropriate markers of later meiosis, including Stra8, Sycp3, and Msy2. This meiotically competent subpopulation represents more robust oocytes that do not require Dmrt1 to complete meiosis, possibly due to a compensatory meiotic program that is only activated in a subset of oocytes. Double knockout mice that lack both Stra8 and Msx1/2, another activator of Stra8, also contain a subpopulation of oocytes that can still complete meiosis (Le Bouffant et al., 2011). The variation in meiotic robustness suggests that heterogeneity exists during female sex differentiation to endow certain oocytes with greater meiotic potential.
Male sex differentiation in mice is initiated at E12 by supportive somatic cells in the nascent testis called Sertoli cells (Ohta et al., 2012). Sertoli cell primarily regulates production of female-inhibitory and male-inducing factors such as Fgf9 and Cyp26b1 to suppress the female meiotic fate (Bowles et al., 2010, 2006). Male germ cells continue to proliferate in the testis but undergo mitotic arrest as early as E12.5. This timing is asynchronous, as some germ cells are still proliferating by E14.5 (Western, Miles, van den Bergen, Burton, & Sinclair, 2008). Mitotic arrest is accompanied by expression of Nanos2, a key male sex differentiation marker (Suzuki & Saga, 2008). Delayed mitotic arrest is associated with aberrant maintenance of pluripotency, decreased Nanos2, and an increased susceptibility for teratoma formation (Dawson et al., 2018; Heaney et al., 2012). Heterogeneity in the timing of mitotic arrest can expose later-arresting male germ cells to these deleterious fates. Mitotic arrest is part of many processes regulated by complex networks of secreted factors such as Tgfβ and Nodal (Spiller, Burnet, & Bowles, 2017; Spiller, Koopman, & Bowles, 2017) active in germ cells during this period. Heterogeneous expression of these factors as well as of their receptors on germ cells can significantly modulate the differentiation states of male germ cells, leading to vastly asynchronous or divergent cell fates.
An overarching theme during sex differentiation is the variation in both position and timing of these events. Many secreted factors regulate this process in males and females, and the precise signaling environments remain to be defined. Heterogeneous progression through sex differentiation can also be due to PGCs themselves differing in their capacity to respond to differentiation cues. Sex differentiation has been shown to be facilitated by epigenetically regulated genes that are activated by demethylation (Hill et al., 2018). Individual variation in the extent of demethylation particularly at these loci could lead to highly variable differentiation potentials. Thus, initial cellular differences in epigenetic state could be amplified as epigenetically heterogeneous subpopulations of PGCs proceed through sex differentiation asynchronously, or fail to do so at all. The later impact of differentiation heterogeneity can be significant, affecting reproductive and tumorigenic potential (Hunt & Hassold, 2008).
3. Determinants of heterogeneity
In this section, we consider the potential causes for diverse behaviors among PGCs described above. We separate these causes into those which are genetically encoded, those which arise from traditional epigenetic marks such as DNA methylation, chromatin modifications, or small RNAs, and a third and separate category of non-genetic determinants (summarized in Fig. 3). Among these sources, we distinguish between those which are permanent (and therefore heritable to all cellular progeny), those which are transient and do not cause similar behaviors in cellular progeny, and those determinants which are semi-permanent, or lasting in daughter cells but not for infinite cellular generations. Finally, we discuss mechanisms that mitigate variation between germ cells, or buffers of heterogeneity. For an outstanding taxonomy of the sources of heterogeneity, refer to Huang (2009).
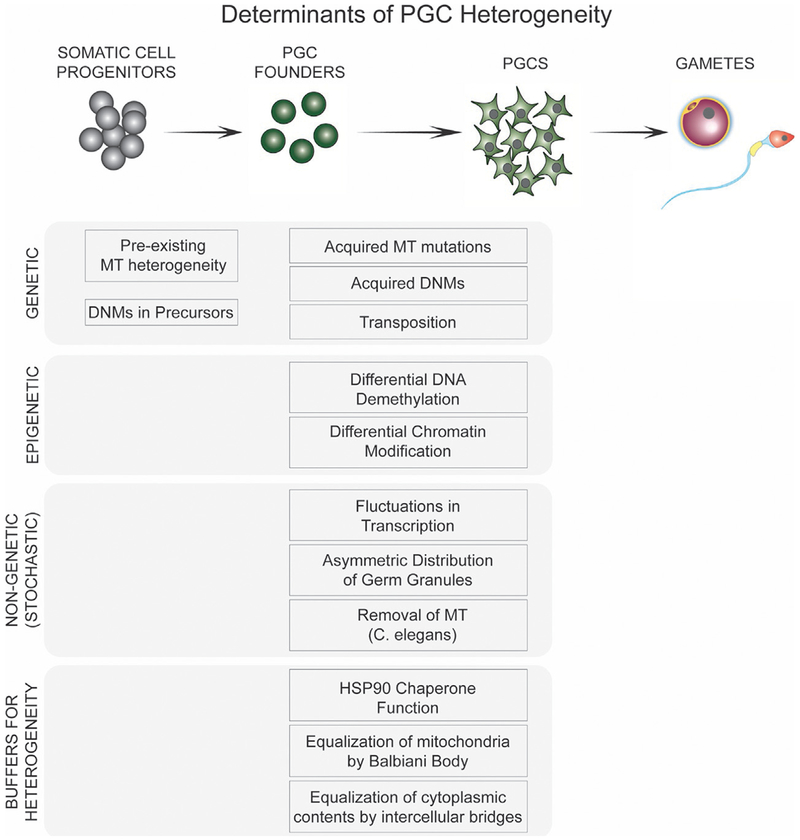
Fig. 3.
Determinants of PGC Heterogeneity. For the various cell types, different underlying cell states can be causes for heterogeneity, impacting cell type transition as well as impacting the fitness of progeny.
3.1. Genetic determinants
3.1.1. Acquired genetic changes
De novo mutations (DNMs) in the DNA sequence typically originate during cell division. Although DNMs occur in all cells, only those in the germline are inherited to offspring. The Darwinian model of evolution by random mutation, causing a phenotypic change which undergoes natural selection in fact originates in germ cells, or potentially in the early embryo of animals such as mice which induce PGCs from somatic cells. Although the diversity of multicellular organisms is living testament to DNMs in the germline, the rate of mutation is low: approximately 10−9 to 10−10 mutations per nucleotide pair per cell division in eukaryotic cells (Arana & Kunkel, 2010; Drake, Charlesworth, Charlesworth, & Crow, 1998). Studies across mice and humans suggest that DNMs arise in the germline at an even lower frequency than in somatic cells (Kohler et al., 1991; Milholland et al., 2017; Walter, Intano, McCarrey, McMahan, & Walter, 1998).
The observed rate of mutation is the output of an equation that includes incurred errors, DNA repair, and elimination of cells. For studies that infer germline mutations by examining progeny, gamete fitness is also included in the rate calculation. The recent generation of exome and whole genome data in human families has provided precise measurements of germline DNMs, with single-nucleotide variations (SNVs) arising at 1.2 × 10−8 per nucleotide per generation (Besenbacher et al., 2015; Campbell et al., 2012; Conrad et al., 2011; Jónsson et al., 2017; Kong et al., 2012; Lynch, 2010; Michaelson et al., 2012; Rahbari et al., 2016; Roach et al., 2010; Ségurel, Wyman, & Przeworski, 2014; Wong et al., 2016). The rate of germline mutation in humans as well as non-human primates (Pfeifer, 2017) is nearly an order of magnitude greater than that measured in mice (5.4 × 10−9 pernt/generation; Uchimura et al., 2015), birds (4.6 × 10−9/nt/generation; Smeds, Qvarnstrom, & Ellegren, 2016), or Drosophila (2.8 × 10−9/nt/generation; Keightley, Ness, Halligan, & Haddrill, 2014). This difference most likely stems from the greater number of cell divisions that occur in the human germline and specifically in the spermatogonial stem cells, which divide throughout life, in contrast to oocytes which cease replication in the fetus (Crow, 2000). Indeed, sequencing of parents and offspring revealed that the majority of human DNMs arise in the paternal germline and increase with paternal age by 1–2 per year (Campbell et al., 2012; Jónsson et al., 2017; Kong et al., 2012; Michaelson et al., 2012; Rahbari et al., 2016). However, with an estimated 10 cellular divisions from the zygote to PGC in mammals (Rahbari et al., 2016), the theoretical number of incurred mutations across the genome of each PGC ranges from ~2 to 15 for mice and ~4 to 38 for humans; hence DNMs do not account for the extent of heterogeneity in cellular behavior that is observed.
A second source of acquired mutations in PGCs is the mitochondrial genome. Separate from the nuclear genome, the circular mitochondrial DNA molecule produces 13 mRNAs, which all encode oxidative phosphorylation components, as well as 2 rRNAs and 22 tRNAs needed for mitochondrial translation of these mRNAs (Falkenberg, Larsson, & Gustafsson, 2007). Critically, the rate of mutation of the mitochondrial genome is 2- to 20-fold greater than the nuclear genome, due to replication errors (Allio, Donega, Galtier, & Nabholz, 2017). The presence of more than one mitochondrial haplotype within the same cell, or heteroplasmy, arises in cells because fidelity of the mitochondrial polymerase is poor and varies depending on biological condition; for example, the presence of reactive oxygen species (ROS) decreases the fidelity of the mitochondrial polymerase, while the presence of a proofreading subunit increases fidelity (Longley, Nguyen, Kunkel, & Copeland, 2001; Pinz, Shibutani, & Bogenhagen, 1995). Although mitochondrial replication is paused in the early mouse embryo until the time of gastrulation (Cao et al., 2007; Wai, Teoli, & Shoubridge, 2008), in the course of PGC development, estimates suggest the number of mitochondria per cell increases from 100 at E7.5 to over 6000 at E14.5 (Wai et al., 2008). Given the size of the mitochondrial genome, mutation rate, and rate of mitochondrial expansion, approximately 35 mitochondrial DNMs would accumulate by E14.5 in mice, although this does not account for unequal replication. However, as with nuclear mutations, this rate of acquired mutation in the mitochondrial genome is likely a very small contribution, at best, to observed heterogeneity of PGCs.
A third potential source of acquired mutation in PGCs is through retrotransposition. Transposable elements comprise a significant fraction of the genome and many of the evolutionarily young Alu and autonomous L1 (or Long Interspersed, LINE-1) elements remain active and capable of re-inserting into new sites in the genome (Kazazian & Moran, 2017). During development, the period of epigenetic reprogramming in PGCs de-silences transposable elements, leading to their expression and potential for transposition in the window of development before transposons are silenced by piRNAs (Brennecke et al., 2007; Teixeira et al., 2017). In humans, new L1 and Alu integrations were detected in ~1/100 and 1/20 births, respectively (Hancks & Kazazian, 2012). This rate is even higher in mice, where new L1 insertions occur in 1/8 births (Richardson et al., 2017). However, on a cellular level, given its rate of occurrence, transposition again makes an exceedingly minor contribution to PGC heterogeneity.
3.1.2. Pre-existing genetic changes
A special case for animals that specify their germline by induction is the potential for prior genetic mutations as a source for heterogeneity. In mice, DNMs arising in the epiblast would lead to mosaicism among the founder PGCs as well as in somatic lineages. Cells in the mouse epiblast undergo rapid divisions—measuring as little as 3h (Snow, 1977)—hence providing opportunity for errors in replication. Furnishing evidence of the elevated potential for damage and repair during this period of rapid division in the early mouse embryo, PGCs and cells of the epiblast have been shown to be hypersensitive to genotoxic stress as measured by DNA breaks, Atm and p53-dependent apoptosis (Heyer, MacAuley, Behrendtsen, & Werb, 2000). From genome sequencing of human families, it has been imputed that approximately 10 divisions occur between the zygote and the specification of PGCs (Ohno, 2018; Rahbari et al., 2016); and this number is probably similar in mice. Detection of DNMs in offspring that co-exist at low levels in the blood of one parent provides evidence that replication errors in cells of the early embryo before PGC specification are present in a subset of somatic and germline derivatives. Data in humans estimate that 0.2—0.6 DNMs accumulate per cell division in the pre-PGC embryo (Rahbari et al., 2016), which would suggest a theoretical maximum of six pre-existing mutations among PGC founders could contribute to heterogeneity.
A second type of pre-existing genetic mutation that could contribute to heterogeneity among PGCs is mitochondrial heteroplasmy (Mishra & Chan, 2014). Mitochondrial heteroplasmy and mutations in mitochondrial (mt) DNA arise in oocyte maturation due to the inherently higher error rate in mitochondrial polymerase (Allio et al., 2017; Khrapko et al., 1997; Madsen, Ghivizzani, & Hauswirth, 1993). During early embryogenesis, the replication of mtDNA and fission of mitochondria ceases (~E1.0—E3.0 in mice) (Cao et al., 2007; Wai et al., 2008), so that each cell division segregates existing mitochondria and produces an increasingly purer pool of mtDNA. Known as a mitochondrial bottleneck, this segregation limits the diversity of mitochondrial genomes inherited from the oocyte (Mishra & Chan, 2014; Stewart & Chinnery, 2015). As PGC specification in mice occurs during this bottleneck, the founders inherit different mtDNA cohorts. Replication of mitochondria resumes after PGC specification, amplifying the heterogeneity between cells that was created during the bottleneck. Together these phenomena may reveal mtDNA mutations by altered PGC phenotypes.
The potential for revealing a mutation in a mitochondrial genome increases as heteroplasmy from the oocyte trends to homoplasmy (similarity in mtDNA) in PGCs. Preexisting or de novo mtDNA mutations would be inherited in a minority of PGCs during specification or could arise during PGC development; either way, those cells with a high concentration of mitochondria bearing deleterious mutations would exhibit altered phenotypes. Concomitant with the onset of PGC migration, metabolism within the embryo shifts from glycolysis to oxidative phosphorylation, which provides a potential source of selective pressure for cells unable to make the switch (Folmes &Terzic, 2014). Deleterious mitochondrial DNA mutations within PGCs may be revealed as compromised ability to initiate oxidative phosphorylation or the bioenergetic response required for migration. Little is known about the cellular or molecular mechanisms for mitochondrial organelle quality control and selection, but the migratory time-point may be particularly important for early embryonic metabolism. A new area of inquiry concerns the selective destruction of mitochondria in PGCs by mechanisms such as mitophagy. Recent work describes an unprecedented means for removing mitochondria from PGCs in C. elegans. This unique cellular surgical procedure is carried out by somatic endoderm cells via extrusion of cytoplasmic lobes from the PGCs (Abdu, Maniscalco, Heddleston, Chew, & Nance, 2016). Although speculative, the effect could serve to concentrate other contents such as germ granules or to remove faulty mitochondria in order to increase cellular fitness, reduce oxidative damage, or alter metabolic capacity of PGCs.
3.1.3. Epigenetic determinants
Having focused on the genetic determinants of heterogeneity, we turn our attention to the determinants not directly linked to the genome itself (Brock, Chang, & Huang, 2009). Our interest in the epigenetic determinants of heterogeneity encompasses both transcription regulation through modification of histone and chromatin structure, as well as the molecular states defined by covalent modifications of DNA. We note that epigenetic determinants have the capacity for altering PGC states semi-permanently, as many of the mechanisms below are preserved through cell divisions.
Reprogramming of the epigenome occurs in two main stages in the fetal germline: first, during PGC migration during which methylation is maintained at specific regions; and second, after gonadal colonization when epigenetic marks are erased to create a hypomethylated epigenetic ground state (Hackett & Surani, 2012; Seisenberger et al., 2012). At day 8 of mouse development, PGCs begin a significant loss of genome-wide DNA methylation that is considered the first of three waves of demethylation before E13 (Bohacek & Mansuy, 2015; Hackett & Surani, 2012; Hajkova et al., 2002; Seisenberger et al., 2012). Changes in histone modifications of PGCs occur starting at E9.0 include loss of histone H3-lysine (K) 9 dimethylation and increase in H3K27 trimethylation (Seki et al., 2005, 2007). These epigenetic changes occur during migration (Nikolic, Volarevic, Armstrong, Lako, & Stojkovic, 2016) and before sex differentiation, and are thought to carry out the following: ensure that the germline genome is ready to receive parent-of-origin specific epigenetic marks; erase potential epimutations incurred in early development; and confer a state of pluripotency for the embryo (Hajkova et al., 2002; Seki et al., 2007).
As epigenomic studies to date have analyzed PGCs in bulk, the variation in epigenetic states of single cells is not known. In principle, differences in the timing of epigenetic reprogramming or failure to lose cytosine methylation or histone marks—epimutations—could lead to PGC heterogeneities. Such differences could be manifested as altered expression of genes critical to differentiation. In support of this idea, PGC maturation genes such as Mvh (Vasa) and the meiotic component Sycp3 are regulated by methylation at their promoters (Maatouk et al., 2006) and would be sensitive to epi-mutations. Recent studies have shown that efficient sex differentiation is dependent upon demethylation at the loci of 45 germline reprogramming-responsive genes (GRRs; Hill et al., 2018). Any individual cell differences in the extent of demethylation at these GRRs could stratify PGCs by differentiation capacity and produce greater population variation as individual PGCs heterogeneously differentiate.
Demethylation during epigenetic reprogramming can also relieve inhibition of transposon sequences that were previously repressed by methylation. Retrotransposition activity from these unsuppressed elements can lead to genomic disruption and inappropriate gene expression, further exacerbating initial variation among germ cell subpopulations. Accordingly, germ cells that fail to repress transposons (Kuramochi-Miyagawa et al., 2004; Malki, van der Heijden, O’Donnell, Martin, & Bortvin, 2014; Soper et al., 2008) arrest prior to gametogenesis and are eliminated, indicating that dysregulated transposons are stringently selected against. Single-cell assessments of transposon expression have identified LINE-1 elements as differentially expressed within both mouse and human PGCs at individual timepoints, which could reflect cell-to-cell variability in repressive methylation (Nguyen & Laird, 2019; Reznik et al., 2019).
A second mechanism of non-genomic transcriptional regulation to be addressed as a potential source of PGC heterogeneity is small non-coding RNAs, including micro RNAs (miRNAs), endogenous small interfering RNAs (endo-siRNAs), and piwi-interacting RNAs (piRNAs). In D. melanogaster and D. rerio, miRNAs function to silence germline enriched mRNAs in cells which are destined to be a part of a somatic fate (Banisch, Goudarzi, & Raz, 2012). In the mouse, the suppression of the miRNA let-7 by lin-28 is necessary for expression of critical PGC specification genes like Blimp1/Prdm1, and without lin-28 the germline fails to form (West et al., 2009); this kind of binary signal requirement during specification does not invite much heterogeneity. However, it is not known whether incremental differences of line-28 can lead to phenotypic consequences. Recent work has begun to characterize miRNA expression in the mouse at later developmental times such as sex differentiation, finding different sexually and developmentally regulated populations of miRNAs (Fernández-Pérez, Brieño-Enríquez, Isoler-Alcaraz, Larriba, & Del Mazo, 2018). Examining miRNA heterogeneity at a single cell level is becoming possible with new technology (Hagemann-Jensen, Abdullayev, Sandberg, & Faridani, 2018), and variability in the expression of miRNAs has recently been speculated to lead to non-genetic cell-to-cell heterogeneity outside of the germline (Wang et al., 2019). Since dysregulation of miRNA expression has been linked to testicular cancer (McIver, Roman, Nixon, & McLaughlin, 2012), the possibility of heterogeneous expression of miRNAs in the early germ cell populations merits exploration.
piRNAs are small RNAs that are nearly exclusive to the germ line which silence transposable elements in the genome (Banisch et al., 2012). Although transposition events are rare and therefore not a significant source of germ cell heterogeneity, heterogeneous expression of transposable elements such as intracisternal-A particle (IAP) family of endogenous retroviruses could lead to phenotypic readouts. Recently identified additional functions for IAPs in regulating the transcription of neighboring genes (Vasiliauskaitė et al., 2018) raise the possibility that the de-repression of IAPs could lead to inappropriate gene expression in PGCs as well as their derivative gametes.
The wave of demethylation which germ cells undergo during their development potentially enables transposable elements relocation to deleterious effect. During this period, endo-siRNAs are considered a secondary defense to silence transposable elements by degrading transcripts. Endo-siRNAs have been studied in C. elegans, D. melanogaster, and mouse, and also function to prevent viral infection (Banisch et al., 2012). By nature, endo-siRNAs are heterogeneous in their response to an infection or transposition event, as these occur on a cell-to-cell basis. However, the ability of an individual germ cell to mount an endo-siRNAs has not yet been investigated, as the contribution of endo-siRNA to the suppression of transposons is just beginning to be appreciated (Berrens et al., 2017).
3.1.4. Non-genetic determinants
Distinct from alterations to the DNA or epigenetic mechanisms discussed above, non-genetic determinants can lead to measurable differences between cells of a single type. Bacteriologists studying genetically identical, clonal populations cultured in uniform conditions first described “non-genetic cell individuality” based on cell-to-cell variations in viral yield (known as burst size; Delbruck, 1945) and chemotactic behavior (Spudich & Koshland, 1976). These heterogeneities were ascribed to the interaction of stochastic fluctuations in molecules within the cell (Spudich & Koshland, 1976). In the field of cancer biology, non-genetic causes of cellular heterogeneity have been parsed into “population noise” and “temporal noise.” Temporal noise derives from the fluctuations of protein levels within a cell over short durations. Population noise, on the other hand, reflects different states of cells at a given moment. The broad assumption is that for genetically identical cells, population noise merely reflects temporal noise, and that all cells experience a similar profile of fluctuations over time (Brock et al., 2009).
One such generator of fluctuation may be the very nature of transcription, which was revealed to be pulsatile, occurring in bursts that emerge at locus-specific rates (Chubb, Trcek, Shenoy, & Singer, 2006; Raj, Peskin, Tranchina, Vargas, & Tyagi, 2006; Suter et al., 2011). In germ cells, stochastic variation due to transcription may be particularly relevant, given that an elevated rate of global transcription was recently reported in mouse fetal germ cells (Percharde, Wong, & Ramalho-Santos, 2017). This result raises the possibility that non-deterministic effects due to timing and transcription may be magnified in the developing germline, although there are currently no studies that have rigorously compared the absolute levels of transcripts in germ cells across time to capture such cycles. It is not yet clear how variation in levels and rate of transcription relates to the level of proteins, although imminent developments in technologies for single cell proteomics will be revealing.
A key assumption of the stochastic processes that generate fluctuations in molecules is that they are not permanent; although a cell may remain in a particular state, its daughter cells will not remember that state. Remarkably, there are documented instances of cells in culture maintaining levels of particular proteins through mitosis. In one example, variations in the levels of apoptotic regulators were maintained through 1–2 cellular generations of human mammary epithelial cells, so that the response to extrinsic death signals was similar between mother and daughter cells (Spencer, Gaudet, Albeck, Burke, & Sorger, 2009). In the early mouse embryo, the levels of cMYC are variable between cells, and inherited through divisions. As cMyc levels determine the capacity of pluripotent cells in the epiblast to outcompete their neighbors by inducing programmed cell death, this observation indicates that cell fitness states are remembered transiently through mechanisms that remain to be elucidated (Diaz-Diaz et al., 2017). We can conceive of scenarios by which non-epigenetic, stochastic heterogeneity could lead to permanent genetic changes; for example, variability in the levels of DNA repair machinery could render some cells more susceptible or resistant to acquired mutations.
A final non-genetic source of heterogeneity we consider is the germ granule. A highly conserved feature of germ cells, these cytoplasmic granules consist of aggregates of mRNA, small RNA, and proteins (Seydoux & Braun, 2006). In C. elegans, Drosophila, and Xenopus, these granules (known as polar granules, P granules or germ plasm) segregate to PGCs during specification and are required for specification and function of germ cells (Gallo et al., 2010; Hay, Ackerman, Barbel, Jan, &Jan, 1988; Strome & Wood, 1983). In mammals, PGCs synthesize components of germ granules rather than inherit them prior to specification, although these germ granules are then passed on with each cell division. Mammalian germ granules resemble processing bodies in somatic cells, which contain mRNA decay machinery. By delivering granule-associated RNAs that can regulate gene expression, germ granules can produce heritable changes in recipient cells independent of changes to DNA sequence, DNA methylation, or chromatin state. Heterogeneity could be imparted by the composition of the germ granules that are transmitted. The contents of germ granules can be modulated according to different environmental conditions as a stress response mechanism (Snee & Macdonald, 2004, 2009). Alternatively, the number of germ granules can be asymmetrically divided between two daughter cells. In Drosophila, the amount of germ plasm as measured by wun2 mRNA levels can vary within a field of cells, with central cells receiving more germ plasm than peripheral cells (Slaidina & Lehmann, 2017). This asymmetry provides central cells with relatively higher amounts of maternal mRNA and is demonstrated to provide a survival advantage over peripheral cells.
3.2. Extrinsic sources of heterogeneity: Environment, niche, and timing
The necessity of the tissue niche for proper development of cells has been widely recognized across biology, and the germline niche is no exception. Unlike the well-characterized cooperative signaling and maintenance of cell fate of the adult Drosophila stem cell niche, the notion that PGCs receive instruction from their niche during specification, migration, and colonization is less established. Across many organisms, the developing embryo changes drastically from the time of PGC specification to the time that functional gametes are produced. The idea that the landscape with which the developing germ cells must interact changes constantly is not new. It is known that fly PGCs receive different levels of signaling from different niches (Stepanik et al., 2016). How the changing niche may contribute to heterogeneity is only just beginning to be appreciated.
PGC migration in mice exhibits spread over space and time. Egress of PGCs from the hindgut into the surrounding mesoderm occurs asynchronously, with “pioneers” emerging first; chemotaxis toward the gonads occurs at varying speeds; and colonization of the gonads takes place over >24h, with late arrivals remaining in the mesentery (Molyneaux et al., 2001). A recent study found that the rate of proliferation of PGCs increases during migration and is regulated by the successive niches through which they transit (Cantú et al., 2016). The progressive increase of mitotic signals along the migratory route implies that those PGCs that move through the series of niches first would gain a proliferative advantage. Upon reaching the gonads, the earlier or faster PGCs would have generated a larger number of clonal progeny as compared to the later or slower migrants. It is here that the heterogeneity of PGCs may be influenced by the environment, as cells which are capable of efficient migration are rewarded by enhanced clonal expansion.
3.3. Buffers of heterogeneity
In contrast to the determinants discussed above, which potentially generate and reinforce differences between individual cells, we now consider the opposite: those mechanisms that could mitigate differences between PGCs. Although we propose that a certain level of PGC heterogeneity could increase the overall fitness of the germline through selection (as discussed below), there are several ways that germ cell diversity could be unfavorable. First, genetic mutations or epi-mutations in PGCs that potentially promote the initiation, growth, or metastasis of tumors by rendering PGCs more resistant to apoptosis, more proliferative, or able to migrate would be dangerous in gametes. Such PGC variants could out-compete other PGCs and even homogenize the gamete pool. Second, the phenotypes that increase the fitness of a PGC are not necessarily the same phenotypes that benefit a gamete or embryo. Finally, too much tolerance for heterogeneity may have consequences for the overall fertility of the organism over a lifetime, such as early loss of fertility or lack of consistency of gametes. In this light, we point to evidence for mechanisms that buffer heterogeneity in PGCs.
Hsp90 is a highly conserved chaperone that assists with the folding of proteins. It has been dubbed an “evolutionary capacitor” because its absence in a number of organisms leads to enhanced phenotypic variability that is believed to arise from cryptic protein variants that are normally degraded or chaperoned by Hsp90 (Rutherford & Lindquist, 1998; Specchia et al., 2010). Most cell types express Hsp90b and elevate expression in stressful conditions, but an A isoform is exclusive to germ cells (Nishikawa & Kinjo, 2018). Hsp90a was further linked to germ cell development through its interaction with the germ plasm component and piRNA biogenesis factor Piwi (Gangaraju et al., 2010). We posit that Hsp90a is a likely candidate to buffer heterogeneity in PGCs during their development.
Intercellular bridges are a deeply conserved cellular structure found in germ cells at various points in their development. Formed by incomplete cytokinesis, the bridges stably connect sister germ cells into “cysts” and permit the passage of cytoplasm and organelles such as mitochondria (Greenbaum, Iwamori, Buchold, & Matzuk, 2011). Intercellular bridges coordinate differentiation and allow sharing of resources between cells, particularly through self-sacrifice, when one member of a cyst commits apoptosis (Lei & Spradling, 2016). Although the formation of intercellular bridges between sister cells limits the potential difference between cells of a cyst, we note their potential to buffer phenotypic consequences of heterogeneity that arises in PGCs such as acquired DNMs or epi-mutations. The increased geographic spread of cysts compared to single PGCs also raises the possibility that they buffer differences introduced by the niche.
4. Selection and heterogeneity
Natural selection is an elegant force of evolution, working slowly through time to shape and change organisms to better suit their needs in changing environments and pressures. This concept has implications for species that reproduce and share heritable characteristics with their offspring. As organisms reproduce, their offspring inherit different traits with stochastic variation; if a random trait is beneficial to the offspring, it will have a competitive advantage, and potentially pass this trait along through its germline. Through many of the causes of heterogeneity that we have already discussed in this review, changes can be wrought within an organism. Changes within somatic cells, while impactful for the organism itself, pale in comparison to the multigenerational effect that heterogeneous changes can have if the germline is altered.
Whether through a genetic or non-genetic transmission, discrete traits are brought to prominence over time if placed under enough positive selective pressure. Natural selection within a population results from interaction between organisms and their environment, as the environment creates selection pressure among heterogeneous individuals, resulting in increased fitness of the population. Less fit animals are phased out of prominence as natural selection leads to increase in the frequency of certain alleles over time.
We propose that like natural selection acts at the organismal and population level, there is a similar kind of selection operating at the cellular level (Buss, 1988; Laird et al., 2005). At both the population and cellular level, alleles subjected to positive selection increase in frequency. Over time, variants are selected due to a competitive advantage they convey to potentially produce an organism or cell with greater fitness. The diversity of PGC development is a special, comprehensive series of selection events during which multiple types of fitnesses can be examined. We will consider how the totality of these developmental events act selectively on PGCs and discuss their implications on reproductive fitness.
4.1. Defining selection in the developing germline
In developmental selection, each stage of germ cell development provides an opportunity for the emergence of underlying heterogeneity to produce different outcomes for subpopulations of germ cells. As previously discussed, both non-genetic and genetic variation among individual cells can present distinct cell identities or states for selection to act upon. We consider a selective process to be one which alters the proportions of PGC subpopulations during development—by mechanisms such as apoptosis, proliferation, migration, or differentiation. Here, we will briefly explicate what we consider a “selected cell” and the processes by which this selection occurs.
Selection can be either negative or positive with respect to gametogenic potential. In negative selection, a germ cell subpopulation is functionally eliminated from further contributing to gametogenesis. In positive selection, a germ cell subpopulation is increased relative to other subpopulations. For the developing germline, we will consider how key events during the fetal period can potentially select for certain subpopulations and discuss the impact on the germline. For example, migration is one of the first challenges a PGC faces after specification at E7.0. During migration, PGCs must respond to precise chemotactic cues to remain on course as they navigate to the gonads. Owing to the distinctive signaling environments presented by each niche along the migratory route, the precise timing of migration may also represent another form of selection. Damaged or unfit PGCs might migrate inefficiently, resulting in reduced colonization of the gonad by their cellular progeny, which corresponds to negative selection. If PGCs that arrive first in the gonad receive different signals than later arriving PGCs, this could alter differentiation on the basis of migratory speed. Additionally, the duration of these differentiation steps could vary as early migrants would be exposed to signals for a longer period of time compared to late arrivals.
In contrast to spatially based selection during PGC migration, selection can also occur during differentiation. After reaching the gonad, PGCs continue maturation by epigenetic reprogramming as well as sex differentiation. The concomitant increase in the expression of apoptotic genes such as Bax sensitizes PGCs toward elimination. Differentiation downregulates these genes and protects successfully differentiated PGCs from apoptosis. PGCs are additionally eliminated based on clonal lineage, implying that apoptotic selection can work on heritable qualities such as epigenetic permissiveness for differentiation (Nguyen & Laird, 2019). The balancing of these two programs selects for differentiated PGCs and selects against PGCs that are incapable or inefficient in differentiating.
Selection can also occur through proliferation, rather than elimination. Germ cell subpopulations that are more proliferative will increase their proportional representation within the germline with each successive division. This may also induce competition for niches or access to signaling factors that continue the tightly regulated differentiation program essential for further germ cell development into optimal gamete-producing cells. Earlier studies utilizing clonal labeling have noted that a small subset of clonal populations in the fetal period is responsible for producing the small number of large monoclonal areas in testes that actually contribute to spermatogenesis (Ueno et al., 2009). Later spermatogenesis in the adult has also been observed to originate from a limited number of spermatogonial stem cell clones (Kanatsu-Shinohara, Naoki, & Shinohara, 2016). In examining fetal populations of germ cells, Lei and Spradling found clone sizes to be variable (Lei & Spradling, 2013), although a longitudinal study would clarify if larger clones resulted from or continued to enjoy a proliferative advantage relative to other germ cell clones.
4.2. How does PGC fitness relate to fitness of gametes or individuals?
While heterogeneities and selective pressure are seen during all phases of germline development, we have thus limited our discussion to competition among fellow cells of the same type. Considering how each competitive cell contributes to the gametes as a whole, providing variation and heterogeneity within a single organism, leads us to speculate on how PGC selection may have an impact on the rate of evolution. Heterogeneity within the germline may exert effects not only within one or two generations, but could also shape the population of a species on a longer evolutionary scale. The fine processes of generating and selecting upon PGC heterogeneity in the embryo may influence the processes of natural selection or genetic drift.
The relationship between processes of germline development and processes of evolution has been considered by others with PGC specification. Broad surveys of modes of germline formation across phylogeny have driven the conclusion that PGC preformation is not ancestral but rather has evolved convergently in many species (reviewed in Extavour, 2003; Johnson, Richardson, Bachvarova, & Crother, 2011). While one study found that species that form their germline through preformation enjoy a faster rate of sequence evolution (Evans, Wade, Chapman, Johnson, & Loose, 2014), follow-up work found instead that species with PGC specification by preformation and PGC induction were equally represented among species with rapidly evolving genomes (Whittle & Extavour, 2016). Since the rate of evolution may be influenced by the heterogeneity of the germline, perhaps organisms with less constraint on their PGCs and a more stringent selection would promote variation within the germline and faster rate of evolution through generations.
If selection during the fetal period can have multi-generational effect, then the fitness of cells which will contribute to these population effects must be multifaceted in their ability to respond to changing environments. Tests of germ cell selection may occur at many developmental timepoints to ensure greatest cell competency; germ cells that excel in one characteristic might be less fit for others. Take, for example, cells which excel dramatically at migration. How might supermigrators such as these compete in later developmental tests? Previous work has shown that as oocytes mature, their order of entry into meiosis corresponds to the order of follicle maturation (Hirshfield, 1992; Polani & Crolla, 1991), and later differentiation follows a “first in, first out” order of follicle maturation and ovulation (Reizel et al., 2012). While supermigrators have not been directly linked to order of entry into meiosis, one might speculate that the earliest migrants will also be the first to enter meiosis, and thus the first to initiate folliculogenesis (Arora et al., 2016). However, many of the earliest follicles release before puberty occurs, causing them to die due to a lack of supportive hormones environment (McGee, 2000). In this instance, a cell overly specialized for migration may fail later developmental checkpoints. The shifting selective pressures of germ cell development ensure that the fittest cells are the germ cells competent at facing many developmental challenges.
These selective processes that comprise germ cell development provide an opportunity for selfish cells to take advantage. Selection favoring a subpopulation of germ cells can eliminate aberrant cells to improve PGC fitness, but it also is vulnerable to cheating. For example, mutations in the FGF receptors can increase FGF ligand binding affinity to confer a proliferative advantage in spermatogenesis. This would lead to clonal expansion of FGFR mutants during development and increased representation in the gamete pool (Goriely, McVean, Röjmyr, Ingemarsson, & Wilkie, 2003). While advantageous for germ cell success, these mutations are detrimental to the organism and can lead to disorders in somatic tissues such as in Apert syndrome (Glaser et al., 2003). The potential for germ cell cheating perhaps illustrates the utility of a multipartite developmental examination of various germ cell behaviors and phenotypes. A single mutation that provides an advantage in one stage may be selected against in another. This cumulative developmental selection would secure diversity in the gamete pool, which is essential for variation at the organismal level to respond to environmental and evolutionary challenges.
5. Conclusion
The arduous journey to establish the germline is defined by variation—both in the challenges faced by PGCs as well as the heterogeneity of the PGCs themselves. The diversity of events during PGC development creates a gauntlet that is ideally positioned to assess heterogeneity in PGCs as they begin to expand and differentiate to establish the eventual germline. Applying the principles of natural selection to the level of germ cells, we see that multiple selective opportunities exist to alter the PGC population that continues onto gametogenesis. The ultimate effect that PGC selection can have on reproductive fitness and even the fitness of organisms will be a fascinating area of study.
The advent of many single-cell analytical techniques now enables us to more comprehensively describe the variation within PGCs. Early attempts at morphological phenotyping are bolstered by single-cell transcriptomics, epigenomics, and single-molecule imaging. In addition to evaluating differences among PGCs at each stage of development, complex barcoding can also extend our understanding how these differences change over developmental time. This is especially relevant to the germline given the dynamic challenges faced by PGCs and the varied capabilities assessed throughout their entire journey. Combining these technologies with a longitudinal perspective will clarify how each event during PGC development contributes to improving gamete quality.
Heterogeneity is a ubiquitous property of biology, but this is especially amplified when a population is faced with making a highly regulated and complex transition such as PGCs in the early embryo. Recent advances in reproductive technology have enabled the generation of mouse germ cells in vitro that produce gametes capable offertilization at exceedingly low efficiency (Hikabe et al., 2016; Ishikura et al., 2016). These germ cells do not undergo the same complex in vivo development and it remains to be seen how bypassing this lengthy process affects the fidelity of their development. Understanding the events and criteria of selection during early embryonic PGC development can improve how we evaluate in vitro-derived gametes to ensure the highest quality ones will be chosen.
Fig. 1.
PGCs transit through multiple cell states during development but remain a single cell type defined by an uncommitted sexual identity. Cell states are not necessarily discrete nor mutually exclusive. We consider germ cells to encompass all cell types with gametogenic potential, including PGCs and sex-differentiated oogonia and prospermatogonia.
References
- Abdu Y, Maniscalco C, Heddleston JM, Chew T-L, & Nance J (2016). Developmentally programmed germ cell remodelling by endodermal cell cannibalism. Nature Cell Biology, 18, 1302 EP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allio R, Donega S, Galtier N, & Nabholz B (2017). Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: Implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Molecular Biology and Evolution, 34, 2762–2772. 10.1093/molbev/msx197. [DOI] [PubMed] [Google Scholar]
- Altschuler SJ, & Wu LF (2010). Cellular heterogeneity: Do differences make a difference? Cell, 141, 559–563. 10.1016/jxell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andux S, & Ellis RE (2008). Apoptosis maintains oocyte quality in aging Caenorhabditis elegans females. PLoS Genetics, 4, e1000295 10.1371/journal.pgen.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana ME, &Kunkel TA (2010). Mutatorphenotypes due to DNAreplicationin delity. Seminars in Cancer Biology, 20, 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Abby E, Ross ADJ, Cantu AV, Kissner MD, Castro V, et al. (2016). Meiotic onset is reliant on spatial distribution but independent of germ cell number in the mouse ovary. Journal of Cell Science, 129, 2493–2499. 10.1242/jcs.189910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, & Lovell-Badge R (2003). Multipotent cell lineages in early mouse development depend on SOX2 function. Genes & Development, 17, 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banisch TU, Goudarzi M, & Raz E (2012). Small RNAs in germ cell development. Current Topics in Developmental Biology, 99, 79–113. 10.1016/B978-0-12-387038-4.00004-5. [DOI] [PubMed] [Google Scholar]
- Berrens RV, Andrews S, Spensberger D, Santos F, Dean W, Gould P, et al. (2017). An endosiRNA-based repression mechanism counteracts transposon activation during global DNA demethylation in embryonic stem cells. Cell Stem Cell, 21, 694–703.e7. 10.1016/j.stem.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besenbacher S, Liu S, Izarzugaza JMG, Grove J, Belling K, Bork-Jensen J, et al. (2015). Novel variation and de novo mutation rates in population-wide de novo assembled Danish trios. Nature Communications, 6, 5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser H, Eisenbeiss S, Neumann M, Reichman-Fried M, Thisse B, Thisse C, et al. (2005). Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. Journal of Cell Science, 118, 4027–4038. 10.1242/jcs.02522. [DOI] [PubMed] [Google Scholar]
- Bohacek J, & Mansuy IM (2015). Molecular insights into transgenerational non-genetic inheritance of acquired behaviours. Nature Reviews. Genetics, 16, 641–652. 10.1038/nrg3964. [DOI] [PubMed] [Google Scholar]
- Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, Mullins MC, et al. (2009). Bucky ball organizes germ plasm assembly in zebrafish. Current Biology, 19, 414–422. 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- Bowles J, Feng C-W,Miles K, Ineson J, Spiller C, &Koopman P (2016).ALDH1A1 provides a source of meiosis-inducing retinoic acid in mouse fetal ovaries. Nature Communications, 7, 10845 10.1038/ncomms10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Feng C-W, Spiller C, Davidson T-L, Jackson A, & Koopman P (2010). FGF9 suppresses meiosis and promotes male germ cell fate in mice. Developmental Cell, 19, 440–449. 10.1016/j.devcel.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, et al. (2006). Retinoid signaling determines germ cell fate in mice. Science, 312, 596–600. 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell, 128, 1089–1103. 10.1016/jxell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brock A, Chang H, & Huang S (2009). Non-genetic heterogeneity—A mutation-independent driving force for the somatic evolution of tumours. Nature Reviews. Genetics, 10, 336–342. 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- Buss LW (1988). The evolution of individuality. Princeton: Princeton University Press; 10.1515/9781400858712. [DOI] [Google Scholar]
- Campbell CD, Chong JX, Malig M, Ko A, Dumont BL, Han L, et al. (2012). Estimating the human mutation rate using autozygosity in a founder population. Nature Genetics, 44, 1277–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu AV, Altshuler-Keylin S, & Laird DJ (2016). Discrete somatic niches coordinate proliferation and migration of primordial germ cells via Wnt signaling. The Journal of Cell Biology, 214, 215–229. 10.1083/jcb.201511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Shitara H, Horii T, Nagao Y, Imai H,Abe K, et al. (2007). The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nature Genetics, 39, 386–390. 10.1038/ng1970. [DOI] [PubMed] [Google Scholar]
- Casper A, & Van Doren M (2006). The control of sexual identity in the Drosophila germline. Development, 133, 2783–2791. [DOI] [PubMed] [Google Scholar]
- Childs AJ, Cowan G, Kinnell HL, Anderson RA, & Saunders PTK (2011). Retinoic acid signalling and the control of meiotic entry in the human fetal gonad. PLoS One, 6, e20249 10.1371/journal.pone.0020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, & Singer RH (2006). Transcriptional pulsing of a developmental gene. Current Biology, 16, 1018–1025. 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman CR, Strohm RC, Oakley FD, Yamada Y, Przychodzin D, & Boswell RE (2002). Identification of X-linked genes required for migration and programmed cell death of Drosophila melanogaster germ cells. Genetics, 162, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, et al. (2011). Variation in genome-wide mutation rates within and between human families. Nature Genetics, 43, 712–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucouvanis EC, Sherwood SW, Carswell-Crumpton C, Spack EG, & Jones PP (1993). Evidence that the mechanism of prenatal germ cell death in the mouse is apoptosis. Experimental Cell Research, 209, 238–247. 10.1006/excr.1993.1307. [DOI] [PubMed] [Google Scholar]
- Crow JF (2000). The origins, patterns and implications of human spontaneous mutation. Nature Reviews. Genetics, 1, 40–47. [DOI] [PubMed] [Google Scholar]
- Dawson EP, Lanza DG, Webster NJ, Benton SM, Suetake I, & Heaney JD (2018). Delayed male germ cell sex-specification permits transition into embryonal carcinoma cells with features of primed pluripotency. Development, 145 dev156612. 10.1242/dev.156612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbruck M (1945). The burst size distribution in the growth of bacterial viruses (Bacteriophages). Journal of Bacteriology, 50, 131–135. [DOI] [PubMed] [Google Scholar]
- Diaz-Diaz C, Fernandez de Manuel L, Jimenez-Carretero D, Montoya MC, Clavería C,& Torres M (2017). Pluripotency surveillance by Myc-driven competitive elimination of differentiating cells. Developmental Cell 42 10.1016/j.devcel.2017.08.011.585-599.e4. [DOI] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, & Crow JF (1998). Rates of spontaneous mutation. Genetics, 148, 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy EM (1975). Germ plasm and the differentiation of germ cell line. International Review of Cytology, 43, 229–280. [DOI] [PubMed] [Google Scholar]
- Ellis RE (2008). Sex determination in the Caenorhabditis elegans germ line. Current Topics in Developmental Biology, 83, 41–64. 10.1016/S0070-2153(08)00402-X. [DOI] [PubMed] [Google Scholar]
- Ephrussi A,& Lehmann R (1992). Induction of germ cell formation by oskar. Nature, 358, 387–392. 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- Evans T, Wade CM, Chapman FA, Johnson AD, & Loose M (2014). Acquisition of germ plasm accelerates vertebrate evolution. Science, 344, 200–203. 10.1126/science.1249325. [DOI] [PubMed] [Google Scholar]
- Ewen KA, & Koopman P (2010). Mouse germ cell development: From specification to sex determination. Molecular and Cellular Endocrinology, 323, 76–93. 10.1016/j.mce.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B, Donoughe S, Clarke DN, &Extavour CG (2013). Germ cell specification requires zygotic mechanisms rather than germ plasm in a basally branching insect. Current Biology, 23(10), 835–842. [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B, Srouji JR, Schwager EE, &Extavour CG (2012). Oskar predates the evolution of germ plasm in insects. Current Biology, 22, 2278–2283. [DOI] [PubMed] [Google Scholar]
- Extavour CG (2003). Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development, 130, 5869–5884. 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Larsson N-G, & Gustafsson CM (2007). DNA replication and transcription in mammalian mitochondria. Annual Review of Biochemistry, 76, 679–699. 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- Fernández-Pérez D, Brieño-Enríquez MA, Isoler-Alcaraz J, Larriba E, & Del Mazo J (2018). MicroRNA dynamics at the onset of primordial germ and somatic cell sex differentiation during mouse embryonic gonad development. RNA, 24, 287–303. 10.1261/rna.062869.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CDL, & Terzic A (2014). Metabolic determinants of embryonic development and stem cell fate. Reproduction, Fertility, and Development, 27, 82–88. 10.1071/RD14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Dunn NR, & Hogan BL (2001). Bone morphogenetic protein 4 in the extraembryonic mesoderm is required for allantois development and the localization and survival of primordial germ cells in the mouse. PNAS, 98, 13739–13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama M, Rougvie AE, & Rothman JH (2006). C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Current Biology, 16, 773–779. 10.1016/jxub.2006.02.073. [DOI] [PubMed] [Google Scholar]
- Gallo CM, Wang JT, Motegi F, & Seydoux G (2010). Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science, 330, 1685–1689. 10.1126/science.1193697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Yin H, Weiner MM, Wang J, Huang XA, & Lin H (2010). Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nature Genetics, 43, 153–158. 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RL, & Rossant J (1979). Investigation of the fate of 4–5 day post-coitum mouse inner cell mass cells by blastocyst injection. Journal of Embryology and Experimental Morphology, 52, 141–152. [PubMed] [Google Scholar]
- Gartner A, Boag PR, & Blackwell TK (2008). Germline survival and apoptosis In WormBook (Ed.), The C. elegans Research Community. WormBook; https://doi.org/10.1895/wormbook.1.145.1 . https://doi.org/10.1895/wormbook.1.145.1http://www.wormbook.org. http://www.wormbook.org . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa L, & Lehmann R (2006). Soma-germline interactions coordinate homeostasis and growth in the Drosophila gonad. Nature, 443, 97–100. 10.1038/nature05068. [DOI] [PubMed] [Google Scholar]
- Ginsburg M, & Eyal-Giladi H (1986). Temporal and spatial aspects of the gradual migration of primordial germ cells from the epiblast into the germinal crescent in the avian embryo. Journal of Embryology and Experimental Morphology, 95, 53–71. [PubMed] [Google Scholar]
- Ginsburg M, &Eyal-Giladi H (1987). Primordialgerm cells of the young chickblastoderm originate from the central zone of the area pellucida irrespective of the embryo-forming process. Development, 101, 209–219. [DOI] [PubMed] [Google Scholar]
- Ginsburg M, Snow MH, & McLaren A (1990). Primordial germ cells in the mouse embryo during gastrulation. Development, 110, 521–528. [DOI] [PubMed] [Google Scholar]
- Glaser RL, Broman KW, Schulman RL, Eskenazi B, Wyrobek AJ, &Jabs EW (2003). The paternal-age effect in Apert syndrome is due, in part, to the increased frequency of mutations in sperm. American Journal of Human Genetics, 73, 939–947. 10.1086/378419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomperts M, Garcia-Castro M, Wylie C, & Heasman J (1994). Interactions between primordial germ cells play a role in their migration in mouse embryos. Development, 120, 135–141. [DOI] [PubMed] [Google Scholar]
- Goriely A, McVean GAT, Rojmyr M, Ingemarsson B, & Wilkie AOM (2003). Evidence for selective advantage of pathogenic FGFR2 mutations in the male germ line. Science, 301, 643–646. 10.1126/science.1085710. [DOI] [PubMed] [Google Scholar]
- Greenbaum MP, Iwamori T, Buchold GM, & Matzuk MM (2011). Germ cell intercellular bridges. Cold Spring Harbor Perspectives in Biology, 3, a005850 a005850 10.1101/cshperspect.a005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb BJ (2006). Developmental biology, Eighth Edition. Integrative and Comparative Biology, 46(5), 652–653. 10.1093/icb/icl011. [DOI] [Google Scholar]
- Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, et al. (2015). Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature, 525, 251. [DOI] [PubMed] [Google Scholar]
- Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, & Hengartner MO (1999). Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development, 126, 1011–1022. 10.5167/uzh-1041. [DOI] [PubMed] [Google Scholar]
- Hackett JA, & Surani MA (2012). DNA methylation dynamics during the mammalian life cycle. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 368 10.1098/rstb.2011.0328. 20110328–20110328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann-Jensen M,Abdullayev I, Sandberg R, & Faridani OR (2018). Small-seqfor single-cell small-RNA sequencing. Nature Protocols, 13, 2407–2424. 10.1038/s41596-018-0049-y. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, et al. (2002). Epigenetic reprogramming in mouse primordial germ cells. Mechanisms of Development, 117, 15–23. [DOI] [PubMed] [Google Scholar]
- Hancks DC, & Kazazian HH (2012). Active human retrotransposons: Variation and disease. Current Opinion in Genetics & Development, 22(3), 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B, Ackerman L, Barbel S, Jan LY, &Jan YN (1988). Identification of a component of Drosophila polar granules. Development, 103, 625–640. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Hayashi M, & Kobayashi S (2004). Nanos suppresses somatic cell fate in Drosophila germ line. PNAS, 101, 10338 10.1073/pnas.0401647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney JD,Anderson EL,Michelson MV, Zechel JL, Conrad PA, Page DC,et al. (2012). Germ cell pluripotency, premature differentiation and susceptibility to testicular teratomas in mice. Development, 139, 1577–1586. 10.1242/dev.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson SA, & Edwards RG (1968). Chiasma frequency and maternal age in mammals. Nature, 218, 22–28. [DOI] [PubMed] [Google Scholar]
- Heyer BS, MacAuley A, Behrendtsen O, &Werb Z (2000). Hypersensitivity to DNA damage leads to increased apoptosis during early mouse development. Genes & Development, 14, 2072–2084. [PMC free article] [PubMed] [Google Scholar]
- Hikabe O, Hamazaki N, Nagamatsu G, Obata Y, Hirao Y, Hamada N, et al. (2016). Reconstitution in vitro of the entire cycle of the mouse female germ line. Nature, 539, 299–303. 10.1038/nature20104. [DOI] [PubMed] [Google Scholar]
- Hill PWS, Leitch HG, Requena CE, Sun Z, Amouroux R, Roman-Trufero M, et al. (2018). Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature, 555, 392–396. 10.1038/nature25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird SN, Paulsen JE, & Strome S (1996). Segregation of germ granules in living Caenorhabditis elegans embryos: Cell-type-specific mechanisms for cytoplasmic localisation. Development, 122, 1303–1312. [DOI] [PubMed] [Google Scholar]
- Hirshfield AN (1992). Heterogeneity of cell populations that contribute to the formation of primordial follicles in rats. Biology of Reproduction, 47, 466–472. [DOI] [PubMed] [Google Scholar]
- Huang S (2009). Non-genetic heterogeneity of cells in development: More than just noise. Development, 136, 3853–3862. 10.1242/dev.035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard EJA, Korta DZ,& Dalfo D (2013). Physiological control of germline development. Advances in Experimental Medicine and Biology, 757, 101–131. 10.1007/978-1-4614-4015-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PA, & Hassold TJ (2008). Human female meiosis: What makes a good egg go bad? Trends in Genetics, 24, 86–93. [DOI] [PubMed] [Google Scholar]
- Hurd TR, Herrmann B, Sauerwald J, Sanny J, Grosch M, & Lehmann R (2016). Long Oskar controls mitochondrial inheritance in Drosophila melanogaster. Developmental Cell, 39(5), 560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura Y , Yabuta Y , Ohta H , Hayashi K , Nakamura T , Okamoto I , et al. (2016). In-vitro derivation and propagation of spermatogonial stem cell activity from mouse pluripotent stem cells. Cell Reports, 17, 2789–2804. 10.1016/jxelrep.2016.11.026. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Richardson E, Bachvarova RF, & Crother BI (2011). Evolution of the germ line-soma relationship in vertebrate embryos. Reproduction, 141, 291–300. 10.1530/REP-10-0474. [DOI] [PubMed] [Google Scholar]
- Jonsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, Hjartarson E, et al. (2017). Parental in uence on human germline de novo mutations in 1,548 trios from Iceland. Nature, 549, 519–522. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Naoki H, & Shinohara T (2016). Nonrandom germline transmission of mouse spermatogonial stem cells. Developmental Cell, 38, 248–261. 10.1016/j.devcel.2016.07.011. [DOI] [PubMed] [Google Scholar]
- Kazazian HH, & Moran JV (2017). Mobile DNA in health and disease. The New England Journal of Medicine, 377(4), 361–370. http://doi:10.1056/NEJMra1510092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Ness RW, Halligan DL, & Haddrill PR (2014). Estimation of the spontaneous mutation rate per nucleotide site in a Drosophila melanogaster full-sib family. Genetics, 196, 313–320. 10.1534/genetics.113.158758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrapko K, Coller HA, Andre PC, Li XC, Hanekamp JS, &Thilly WG (1997). Mitochondrial mutational spectra in human cells and tissues. PAMS, 94, 13798–13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble JE, & White JG (1981). On the control of germ cell development in Caenorhabditis elegans. Developmental Biology, 81, 208–219. [DOI] [PubMed] [Google Scholar]
- Kohler SW, Provost GS, Fieck A, Kretz PL, Bullock WO, Sorge JA, et al. (1991). Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America, 88, 7958–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, et al. (2012). Rate of de novo mutations and the importance of father’s age to disease risk. Nature, 488, 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köprunner M, Thisse C, Thisse B, & Raz E (2001). A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes & Development, 15, 2877–2885. 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, & Page DC (2006). Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proceedings of the National Academy of Sciences of the United States of America, 103, 2474–2479. 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, et al. (2004). Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development, 131, 839–849. 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Laird DJ (2018). How to lose your inheritance. Nature, 562, 497–498. 10.1038/d41586-018-06849-5. [DOI] [PubMed] [Google Scholar]
- Laird DJ, Altshuler-Keylin S, Kissner MD, Zhou X, &Anderson KV (2011). Ror2 enhances polarity and directional migration of primordial germ cells. PLoS Genetics, 7, e1002428 10.1371/journal.pgen.1002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird DJ, Chang W-T, Weissman IL,& Lauzon RJ (2005). Identification of a novel gene involved in asexual organogenesis in the budding ascidian Botryllus schlosseri. Developmental Dynamics, 234, 997–1005. 10.1002/dvdy.20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, et al. (1999). Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes & Development, 13, 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bouffant R, Souquet B, Duval N, Duquenne C, Herve R, Frydman N, et al. (2011). Msx1 and Msx2 promote meiosis initiation. Development, 138, 5393–5402. 10.1242/dev.068452. [DOI] [PubMed] [Google Scholar]
- Lee H, Kim S, Park T, Rengaraj D, Park K, Lee H, et al. (2013). Compensatory proliferation of endogenous chicken primordial germ cells after elimination by busulfan treatment. Stem Cell Research & Therapy, 4, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, & Spradling AC (2013). Mouse primordial germ cells produce cysts that partially fragment prior to meiosis. Development, 140, 2075–2081. 10.1242/dev.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, & Spradling AC (2016). Mouse oocytes differentiate through organelle enrichment from sister cyst germ cells. Science, 352, 95–99. 10.1126/science.aad2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsky JM, & Singer RH (2003). Fluorescence in situ hybridization: Past, present and future. Journal of Cell Science, 116, 2833–2838. 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Nguyen D, Kunkel TA, & Copeland WC (2001). The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. The Journal of Biological Chemistry, 276, 38555–38562. 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- Lynch M (2010). Evolution of the mutation rate. Trends in Genetics, 26, 345–352. 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk DM, Kellam LD, Mann MRW, Lei H, Li E, Bartolomei MS, et al. (2006). DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development, 133, 3411–3418. 10.1242/dev.02500. [DOI] [PubMed] [Google Scholar]
- Madsen CS, Ghivizzani SC, & Hauswirth WW (1993). In vivo and in vitro evidence for slipped mispairing in mammalian mitochondria. PNAS, 90, 7671–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald AP (1962). Fine structure of pole cells and polar granules in Drosophila melanogaster. The Journal of Experimental Zoology, 151, 201–215. [Google Scholar]
- Mahowald AP (2001). Assembly of the Drosophila germplasm. International Review of Cytology, 203, 187–213. [DOI] [PubMed] [Google Scholar]
- Malki S, van der Heijden GW, O’Donnell KA, Martin SL, & Bortvin A (2014). A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Developmental Cell, 29, 521–533. 10.1016/j.devcel.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow F (2015). Primordial germ cell specification and migration. F1000Res, 16, 4 10.12688/f1000research.6995.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, & Zarkower D (2010). The mammalian Doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Developmental Cell, 19, 612–624. 10.1016/j.devcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDole K, Guignard L, Amat F, Berger A, Malandain G, Royer LA, et al. (2018). In toto imaging and reconstruction of post-implantation mouse development at the single-cell level. Cell 175 10.1016/j.cell.2018.09.031.859-876.e33. [DOI] [PubMed] [Google Scholar]
- McGee EA (2000). The regulation of apoptosis in preantral ovarian follicles. Biological Signals and Receptors, 9, 81–86. 10.1159/000014626. [DOI] [PubMed] [Google Scholar]
- McGranahan N, & Swanton C (2017). Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell, 168, 613–628. 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- McIver SC, Roman SD, Nixon B, & McLaughlin EA (2012). miRNA and mammalian male germ cells. Human Reproduction Update, 18, 44–59. 10.1093/humupd/dmr041. [DOI] [PubMed] [Google Scholar]
- McLaren A (2003). Primordial germ cells in the mouse. Developmental Biology, 262, 1–15. [DOI] [PubMed] [Google Scholar]
- McLaren A, & Southee D (1997). Entry of mouse embryonic germ cells into meiosis. Developmental Biology, 187, 107–113. 10.1006/dbio.1997.8584. [DOI] [PubMed] [Google Scholar]
- Menke DB, Koubova J, & Page DC (2003). Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Developmental Biology, 262, 303–312. [DOI] [PubMed] [Google Scholar]
- Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D,Jin X, et al. (2012). Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell, 151, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milholland B, Dong X, Zhang L, Hao X, Suh Y, & Vijg J (2017). Differences between germline and somatic mutation rates in humans and mice. Nature Communications, 8, 15183 10.1038/ncomms15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, & Chan DC (2014). Mitochondrial dynamics and inheritance during cell division, development and disease. Nature, 15, 634–646. 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux KA, Stallock J, Schaible K, &Wylie C (2001). Time-lapse analysis of living mouse germ cell migration. Developmental Biology, 240, 488–498. 10.1006/dbio.2001.0436. [DOI] [PubMed] [Google Scholar]
- Morita-Fujimura Y, Tokitake Y, &Matsui Y (2009). Heterogeneity of mouse primordial germ cells reflecting the distinct status of their differentiation, proliferation and apoptosis can be classified by the expression of cell surface proteins integrin α6 and c-Kit. Development, Growth & Differentiation, 51, 567–583. 10.1111/j.1440-169X.2009.01119.x. [DOI] [PubMed] [Google Scholar]
- Muller-Sieburg CE, Sieburg HB, Bernitz JM, & Cattarossi G (2012). Stem cell heterogeneity: Implications for aging and regenerative medicine. Blood, 119, 3900–3907. 10.1182/blood-2011-12-376749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, & Laird DJ (2019). Apoptosis in the fetal testis eliminates developmentally defective germ cell clones. bioRxiv. 10.1101/601013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD (1947). Experimental observations on the origin and determination of the germ cells, and on the development of the lateral plates and germ ridges in the urodeles. Archives Néerlandaises de Zoologie, 8(1), 1–205. [Google Scholar]
- Nieuwkoop PD, & Sutasurya LA (1979). Primordial germ cells in the chordates: Embryogenesis and phylogenesis In Volume 7 of developmental and cell biology series. Cambridge University Press; ISBN: 0521223032. CUP Archive. [Google Scholar]
- Nieuwkoop PD, & Sutasurya LA (1981). Primordial germ cells in the invertebrates: From epigenesis to preformation In Volume 10 of developmental and cell biology series. CUP Archive; ISBN: 0521221897. [Google Scholar]
- Nikolic A, Volarevic V, Armstrong L, Lako M, & Stojkovic M (2016). Primordial germ cells: Current knowledge and perspectives. Stem Cells International, 2016, 1741072–1741078. 10.1155/2016/1741072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K, & Kinjo AR (2018). Mechanism of evolution by genetic assimilation: Equivalence and independence of genetic mutation and epigenetic modulation in phenotypic expression. Biophysical Reviews, 10, 667–676. 10.1007/s12551-018-0403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, et al. (2005). Blimp1 is a critical determinant of the germ cell lineage in mice. Nature, 436, 207–213. 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Ohno M (2018). Spontaneous de novo germline mutations in humans and mice: Rates, spectra, causes and consequences. Genes & Genetic Systems, 94, 13–22. 10.1266/ggs.18-00015. [DOI] [PubMed] [Google Scholar]
- Ohkubo Y, Shirayoshi Y, & Nakatsuji N (1996). Autonomous regulation of proliferation and growth arrest in mouse primordial germ cells studied by mixed and clonal cultures. Experimental Cell Research, 222, 291–297. 10.1006/excr.1996.0037. [DOI] [PubMed] [Google Scholar]
- Ohta K, Yamamoto M, Lin Y, Hogg N, Akiyama H, Behringer RR, et al. (2012). Male differentiation of germ cells induced by embryonic age-specific Sertoli cells in mice. Biology of Reproduction, 86, 112 10.1095/biolreprod.111.095943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Wong P, & Ramalho-Santos M (2017). Global hypertranscription in the mouse embryonic germline. Cell Reports, 19, 1987–1996. 10.1016/j.celrep.2017.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer SP (2017). Direct estimate of the spontaneous germ line mutation rate in African green monkeys. Evolution, 71, 2858–2870. 10.1111/evo.13383. [DOI] [PubMed] [Google Scholar]
- Pinz KG, Shibutani S, &Bogenhagen DF (1995). Action of mitochondrial DNA polymerase gamma at sites of base loss or oxidative damage. The Journal of Biological Chemistry, 270, 9202–9206. [DOI] [PubMed] [Google Scholar]
- Polani PE, & Crolla JA (1991). A test of the production line hypothesis of mammalian oogenesis. Human Genetics, 88, 64–70. [DOI] [PubMed] [Google Scholar]
- Rahbari R, Wuster A, Lindsay SJ, Hardwick RJ, Alexandrov LB, Al Turki S, et al. (2016). Timing, rates and spectra of human germline mutation. Nature Genetics, 48, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, & Tyagi S (2006). Stochastic mRNA synthesis in mammalian cells. PLoS Biology, 4, e309 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, &van Oudenaarden A (2008). Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell, 135, 216–226. 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S, Wang H, Quach HNB, & Sampath K (2006). Zebrafish Staufenl and Staufen2 are required for the survival and migration of primordial germ cells. Developmental Biology, 292, 393–406. 10.1016/j.ydbio.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Raz E (2002). Primordial germ cell development in zebrafish. Seminars in Cell & Developmental Biology, 13, 489–495. [DOI] [PubMed] [Google Scholar]
- Raz E, & Reichman-Fried M (2006). Attraction rules: Germ cell migration in zebrafish. Current Opinion in Genetics & Development, 16, 355–359. 10.1016/j.gde.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Reichman-Fried M, Minina S, & Raz E (2004). Autonomous modes of behavior in primordial germ cell migration. Developmental Cell, 6, 589–596. [DOI] [PubMed] [Google Scholar]
- Reizel Y, Itzkovitz S, Adar R, Elbaz J, Jinich A, Chapal-Ilani N, et al. (2012). Cell lineage analysis of the mammalian female germline. PLoS Genetics, 8, e1002477 10.1371/journal.pgen.1002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik B, Cincotta SA, Jaszczak RG, Mateo LJ, Shen J, Cao M, et al. (2019). Heterogeneity of transposon expression and activation of the repressive network in human fetal germ cells. Development, 146 dev171157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Gerdes P, Gerhardt DJ, Sanchez-Luque FJ, Bodea G-O, Mufioz-Lopez M, et al. (2017). Heritable L1 retrotransposition in the mouse primordial germline and early embryo. Genome Research, 27(8), 1395–1405. http://doi:10.1101/gr.219022.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BE, & Lehmann R (2010). Mechanisms guiding primordial germ cell migration: Strategies from different organisms. Nature, 11, 37–49. 10.1038/nrm2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Smit AFA, Huff CD, Hubley R, Shannon PT, et al. (2010). Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science, 328, 636–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrschneider MR, & Nance J (2013). The union of somatic gonad precursors and primordial germ cells during Caenorhabditis elegans embryogenesis. Developmental Biology, 379, 139–151. https://doi.org/10.1016Zj.ydbio.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucker EB, Dierisseau P, Wagner K-U, Garrett L, Wynshaw-Boris A, Flaws JA, et al. (2000). Bcl-x and Bax regulate mouse primordial germ cell survival and apoptosis during embryogenesis. Molecular Endocrinology, 14,1038–1052. 10.1210/mend.14.7.0465. [DOI] [PubMed] [Google Scholar]
- Runyan C, Gu Y, Shoemaker A, Looijenga L, &Wylie C (2008). The distribution and behavior of extragonadal primordial germ cells in Bax mutant mice suggest a novel origin for sacrococcygeal germ cell tumors. The International Journal of Developmental Biology, 52(4), 333–344. 10.1387/ijdb.072486cr. [DOI] [PubMed] [Google Scholar]
- Runyan C, Schaible K, Molyneaux K, Wang Z, Levin L, & Wylie C (2006). Steel factor controls midline cell death of primordial germ cells and is essential for their normal proliferation and migration. Development, 133, 4861–4869. 10.1242/dev.02688. [DOI] [PubMed] [Google Scholar]
- Rutherford SL, & Lindquist S (1998). Hsp90 as a capacitor for morphological evolution. Nature, 396, 336–342. 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Saitou M, Payer B, O’Carroll D, Ohinata Y, & Surani MA (2005). Blimp1 and the emergence of the germ line during development in the mouse. Cell Cycle, 4, 1736–1740. 10.4161/ccA12.2209. [DOI] [PubMed] [Google Scholar]
- Sano H, Renault AD, & Lehmann R (2005). Control of lateral migration and germ cell elimination by the Drosophila melanogaster lipid phosphate phosphatases Wunen and Wunen 2. The Journal of Cell Biology, 171, 675 10.1083/jcb.200506038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AC, &Lehmann R (2004). Germ cell specification and migration in Drosophilaand beyond. Current Biology, 14, R578–R589. 10.1016/jxub.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Sarikaya DP, &Extavour CG (2015). The Hippo pathway regulates homeostatic growth of stem cell niche precursors in the Drosophila ovary. PLoS Genetics, 11, e1004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner CE, Deshpande G, Schedl PD, &Kelly WG (2003). A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Developmental Cell, 5, 747–757. 10.1016/S1534-5807(03)00327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T (2010). Hematopoietic stem cell heterogeneity: Subtypes, not unpredictable behavior. Cell Stem Cell, 6, 203–207. 10.1016/j.stem.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Segurel L, Wyman MJ, & Przeworski M (2014). Determinants of mutation rate variation in the human germline. Annual Review of Genomics and Human Genetics, 15, 47–70. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, et al. (2012). The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Molecular Cell, 48, 849–862. 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, &Matsui Y (2005). Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Developmental Biology, 278, 440–458. 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, et al. (2007). Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development, 134, 2627–2638. 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- Seoane J (2017). Cancer: Division hierarchy leads to cell heterogeneity. Nature, 549, 164–166. 10.1038/nature23546. [DOI] [PubMed] [Google Scholar]
- Seydoux G, & Braun RE (2006). Pathway to totipotency: Lessons from germ cells. Cell, 127, 891–904. 10.1016/jxell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Slaidina M, & Lehmann R (2017). Quantitative differences in a single maternal factor determine survival probabilities among drosophila germ cells. Current Biology, 27, 291–297. 10.1016/j.cub.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeds L, Qvarnstrom A, & Ellegren H (2016). Direct estimate of the rate of germline mutation in a bird. Genome Research, 26, 1211–1218. 10.1101/gr.204669.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Wilson JE, & Macdonald PM (1992). Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell, 70, 849–859. [DOI] [PubMed] [Google Scholar]
- Snee MJ, & Macdonald PM (2004). Live imaging of nuage and polar granules: Evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. Journal of Cell Science, 117, 2109–2120. 10.1242/jcs.01059. [DOI] [PubMed] [Google Scholar]
- Snee MJ,& Macdonald PM (2009). Dynamic organization and plasticity of sponge bodies. Developmental Dynamics, 238, 918–930. 10.1002/dvdy.21914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow MHL (1977). Gastrulation in the mouse: Growth and regionalization of the epiblast. Journal of Embryology and Experimental Morphology, 42, 293–303. [PubMed] [Google Scholar]
- Solter D (2016). Preformation versus epigenesis in early mammalian development. Current Topics in Developmental Biology, 117, 377–391. 10.1016/bs.ctdb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Soper SFC, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, et al. (2008). Mouse Maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Developmental Cell, 15, 285–297. 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P, & Jaenisch R (1986). Retroviruses as probes for mammalian development: Allocation of cells to the somatic and germ cell lineages. Cell, 46(1), 19–29. [DOI] [PubMed] [Google Scholar]
- Specchia V, Piacentini L, Tritto P, Fanti L, D’Alessandro R, Palumbo G, et al. (2010). Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature, 463, 662–665. 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, Burke JM, & Sorger PK (2009). Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature, 459, 428–432. 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller C, Burnet G, & Bowles J (2017). Regulation of fetal male germ cell development by members of the TGFβ superfamily. Stem Cell Research, 24, 174–180. 10.1016/j.scr.2017.07.016. [DOI] [PubMed] [Google Scholar]
- Spiller C, Koopman P, & Bowles J (2017). Sex determination in the mammalian germline. Annual Review of Genetics, 51, 265–285. 10.1146/annurev-genet-120215-035449. [DOI] [PubMed] [Google Scholar]
- Spudich JL, & Koshland DEJ (1976). Non-genetic individuality: Chance in the single cell. Nature, 262, 467–471. [DOI] [PubMed] [Google Scholar]
- Stallock J, Molyneaux K, Schaible K, Knudson CM, & Wylie C (2003). The pro-apoptotic gene Bax is required for the death of ectopic primordial germ cells during their migration in the mouse embryo. Development, 130, 6589–6597. 10.1242/dev.00898. [DOI] [PubMed] [Google Scholar]
- Stepanik V, Dunipace L, Bae Y-K, Macabenta F, Sun J, Trisnadi N, et al. (2016). The migrations of Drosophila muscle founders and primordial germ cells are interdependent. Development, 143, 3206–3215. 10.1242/dev.134346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB,& Chinnery PF (2015). The dynamics of mitochondrial DNA heteroplasmy: Implications for human health and disease. Nature Reviews. Genetics, 16, 530–542. 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- Strome S, & Lehmann R (2007). Germ versus soma decisions: Lessons from flies and worms. Science, 316, 392–393. 10.1126/science.1140846. [DOI] [PubMed] [Google Scholar]
- Strome S, & Wood WB (1983). Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell, 35, 15–25. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, & Thomson JN (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology, 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, & Naef F (2011). Mammalian genes are transcribed with widely different bursting kinetics. Science, 332, 472–474. 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- Suzuki A, & Saga Y (2008). Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes & Development, 22, 430–435. 10.1101/gad.1612708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP, & Snow MH (1981). Proliferation and migration of primordial germ cells during compensatory growth in mouse embryos. Journal of Embryology and Experimental Morphology, 64, 133–147. [PubMed] [Google Scholar]
- Tam PP, & Zhou SX (1996). The allocation of epiblast cells to ectodermal and germline lineages is influenced by the position of the cells in the gastrulating mouse embryo. Developmental Biology, 178(1), 124–132. [DOI] [PubMed] [Google Scholar]
- Teixeira FK, Okuniewska M, Malone CD, Coux R-X, Rio DC, & Lehmann R (2017). piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature, 552, 268–272. 10.1038/nature25018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura A, Higuchi M, Minakuchi Y, Ohno M, Toyoda A, Fujiyama A, et al. (2015). Germline mutation rates and the long-term phenotypic effects of mutation accumulation in wild-type laboratory mice and mutator mice. Genome Research, 25, 1125–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H, Turnbull BB, & Weissman IL (2009). Two-step oligoclonal development of male germ cells. PNAS, 106, 175–180. 10.1073/pnas.0810325105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood EM, Caulton JH, Allis CD, &Mahowald AP (1980). Developmental fate of pole cells in Drosophila melanogaster. Developmental Biology, 77, 303–314. 10.1016/0012-1606(80)90476-5. [DOI] [PubMed] [Google Scholar]
- Vasiliauskaitė L, Berrens RV, Ivanova I, Carrieri C, Reik W, Enright AJ, et al. (2018). Defective germline reprogramming rewires the spermatogonial transcriptome. Nature Structural & Molecular Biology, 25, 394–404. 10.1038/s41594-018-0058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai T, Teoli D,& Shoubridge EA (2008). The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nature Genetics, 40, 1484–1488. 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- Walter CA, Intano GW, McCarrey JR, McMahan CA, & Walter RB (1998). Mutation frequency declines during spermatogenesis in young mice but increases in old mice. PNAS, 95, 10015–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RA, Nakane PK, & Koji T (1998). Autonomous cell death of mouse male germ cells during fetal and postnatal period. Biology of Reproduction, 58, 1250–1256. [DOI] [PubMed] [Google Scholar]
- Wang N, Zheng J, Chen Z, Liu Y, Dura B, Kwak M, et al. (2019). Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nature Communications, 10, 1–12. 10.1038/s41467-018-07981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, et al. (2003). Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Current Biology, 13, 1429–1434. 10.1016/S0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- Weissman IL (2015). Stem cells are units of natural selection for tissue formation, for germline development, and in cancer development. PNAS, 112, 8922–8928. 10.1073/pnas.1505464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park I-H, et al. (2009). A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature, 460, 909–913. 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western PS, Miles DC, van den Bergen JA, Burton M, & Sinclair AH (2008). Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells, 26, 339–347. 10.1634/stemcells.2007-0622. [DOI] [PubMed] [Google Scholar]
- Whittle CA, & Extavour CG (2016). Refuting the hypothesis that the acquisition of germ plasm accelerates animal evolution. Nature Communications, 7, 12637 10.1038/ncomms12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolke U, Jezuit EA, & Priess JR (2007). Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development, 134, 2227–2236. 10.1242/dev.004952. [DOI] [PubMed] [Google Scholar]
- Wong WS, Solomon BD, Bodian DL, Kothiyal P, Eley G, Huddleston KC, et al. (2016). New observations on maternal age effect on germline de novo mutations. Nature Communications, 7, 10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, & Saitou M (2006). Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biology of Reproduction, 75, 705–716. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Davis KD, & Coffman CR (2008). Programmed cell death of primordial germ cells in Drosophila is regulated by p53 and the Outsiders monocarboxylate transporter. Development, 135, 207–216. 10.1242/dev.010389. [DOI] [PubMed] [Google Scholar]
- Ying Y, Liu XM, Marble A, Lawson KA, & Zhao GQ (2000). Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Molecular Endocrinology, 14, 1053–1063. [DOI] [PubMed] [Google Scholar]
- Young RA (2011). Control of the embryonic stem cell state. Cell, 144, 940–954. 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, et al. (2005). The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature, 435, 360–364. 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang M, Acampora D, Vojtek M, Yuan D, Simeone A, et al. (2018). OTX2 restricts entry to the mouse germline. Nature, 562, 595–599. 10.1038/s41586-018-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C-J, Luebeck EG, Byers B, & Moolgavkar SH (2005). On the number of founding germ cells in humans. Theoretical Biology & Medical Modelling, 2, 32 10.1186/1742-4682-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]