Abstract
Swine represent the only livestock with an established invariant natural killer T (iNKT) cell-CD1d system. Here, we exploited the fact that pig iNKT cells can be purified using a mouse CD1d tetramer reagent to establish their T cell receptor (TCR) repertoire by next generation sequencing. CD1d tetramer-positive pig cells predominantly expressed an invariant Vα-Jα rearrangement, without non-template nucleotide diversity, homologous to the Vα24-Jα18 and Vα14-Jα18 rearrangements of human and murine iNKT cells. The co-expressed β chain used a Vβ segment homologous to the semivariant Vβ11 and Vβ8.2 segments of murine and human iNKT cell receptors. Molecular modeling found that contacts within CD1d and CDR1α that underlie fine specificity differences between mouse and human iNKT cells are conserved between pigs and humans, indicating that the response of porcine and human iNKT cells to CD1d-restricted antigens may be similar. Accordingly, pigs, which are an important species for diverse fields of biomedical research, may be useful for developing human-based iNKT cell therapies for cancer, infectious diseases, and other disorders. Our study also sequenced the expressed TCR repertoire of conventional porcine αβ T cells (Tconv), which identified 48 Vα, 50 Jα, 18 Vβ, and 18 Jβ sequences, most of which correspond to human gene segments. These findings provide information on the αβ TCR usage of pigs, which is understudied and deserves further attention.
Keywords: T cell receptors, NKT cells, tetramer, repertoire, swine
Introduction
CD1d-restricted invariant natural killer T (iNKT) cells are an innate-like T cell subset expressed by some, but not all mammals. Unlike conventional αβ T cells (Tconv) that bind peptide antigens, iNKT cells recognize self and exogenous lipids with an α-anomerically-linked sugar presented by CD1d proteins expressed on the surface of various antigen-presenting cells and non-hematopoietic cells (1). iNKT cells interact with CD1d-bound antigen through a highly conserved invariant T cell receptor (TCR) that is a product of a canonical rearrangement between gene segments Vα14 and Jα18 in mice (2), Vα14 and Jα18 in rats (3), and Vα24 and Jα18 in humans (2, 4, 5). iNKT cell α chains are paired with either Vβ8.2, Vβ7, or Vβ2 in mice (6, 7) or Vβ11 in humans (5). The invariant α chain interacts with antigen via the complementarity-determining region (CDR) 1α and CDR3α, while the β chain makes stabilizing contacts with CD1d and modulates the affinity of the TCR for the antigen/CD1d complex (8, 9). A number of microorganisms contain lipid antigens that bind CD1d and activate iNKT cells (10–17). iNKT cells can also be activated with the prototypical ligand α-galactosylceramide (α-GalCer) that was originally isolated from the marine sponge Agelas mauritianus (18). Accordingly, fluorescently labeled CD1d tetramers or multimers loaded with α-GalCer analogs can be used to visualize and purify iNKT cells by flow cytometry (19).
Because CD1d is a non-polymorphic molecule, mouse CD1d (mCD1d)/α-GalCer tetramer cross-reacts with human iNKT cells and vice versa (20, 21). In addition, mouse and human CD1d/α-GalCer tetramers have been found to cross-react with porcine iNKT cells (22–24). We took advantage of this phenomenon to purify porcine iNKT cells and establish their receptor repertoire using RNA sequencing (RNA-seq). Our results show that porcine iNKT cell α and β chains are highly homologous to their human counterparts, including the critical CDR3α sequence. Molecular modeling found that several contacts which distinguish mouse and human iNKT cell TCR-antigen-CD1d interactions are conserved between pigs and humans. Accordingly, swine may be useful for testing of iNKT cell agonists for human use, especially as pigs are more similar to humans than mice with regard to iNKT cell frequency and tissue distribution (25). Also like humans, pigs possess a full complement of CD1 molecules (CD1a, CD1b, CD1c, CD1d, CD1e), some of which can present lipid antigens that may activate iNKT cells or other innate-like lymphocyte subsets (26, 27), while mice only express two copies of CD1d, one of which is non-functional in some strains (28).
The current study also examined the expressed α and β chain usage of Tconv. Our RNA-sequencing approach identified a large number of V and J segments, many of which overlapped with sequences discovered in previous studies that used traditional cloning techniques to identify TCR α- or β-chains. We also detected V and J segments that have not been previously described, which should be useful for understanding porcine TCR α and β chain usage in a variety of contexts, such as during infections, and for porcine models of cancer and xenotransplantation.
Materials and methods
iNKT cell expansion and purification
Peripheral blood (10 ml per pig) was collected from the jugular vein of eight 4- to 6-week old Hampshire, Yorkshire, Chester White, Duroc, and Landrace crossbred pigs of mixed sex that were maintained under standard husbandry conditions at the University of Florida’s swine unit. Blood was collected in heparinized vacutainers (BD Biosciences, San Jose, CA) in accordance with the University of Florida’s Institutional Animal Care and Use Committee under protocol 201509134. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-PaqueTM PREMIUM (GE Healthcare Bio-Sciences Corp., Uppsala, Sweden) as previously described (25). Cells were seeded in U-bottomed 96-well cell culture plates (BD Falcon, Multiwell Cell Culture Plate) at a density of 5×105 PBMC/well in 200 µl of RPMI 1640 (containing 10% fetal bovine serum and 1% Penicillin/Streptomycin) with DMSO or 1 µg/ml α-GalCer and cultured at 37°C with 5% CO2 for 7 days without the addition of exogenous cytokines. After culture, PBMCs were harvested and incubated at 4°C for 10 min with 10 µg rat IgG (Sigma-Aldrich, Saint Louis, MO) to block Fc receptor binding. Cells were then surface stained for 30 min at 4°C with fluorescein isothiocyanate-labelled antibody to CD3ε (clone BB23–8E6–8C8, BD Biosciences) and phycoerythrin-labelled mouse CD1d tetramer, unloaded or loaded with the α-GalCer analog PBS57 provided by the National Institutes of Health Tetramer Core Facility. Cells were washed in PBS and counted using a BD Accuri C6 flow cytometer as previously described (29). PBMC samples incubated with α-GalCer were sorted for iNKT cells (CD3ε+CD1d tetramer+) and Tconv (CD3ε+CD1d tetramer−) using a Sony SH800 cell sorter. At least 1×105 iNKT cells and 5×105 conventional T cells from each pig were collected with a purity of >90%.
Sequencing of the TCR repertoire
For each of two donor preparations, a total of 6×105 iNKT cells and at least 2.5×106 Tconv from 4 pigs were pooled, pelleted and lysed with RNA lysis buffer from Quick-RNA™ MiniPrep (ZYMO Research, Irvine, CA) to extract RNA. RNA quantity and purity were measured with an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA). mRNA was isolated using poly-T beads that hybridizes to the RNA poly-A tag (mRNA-seq kit from Bioo Scientific Corporation, Austin, TX) and sheared to ~600bp fragments using magnesium ions and high temperature. Fragments were then converted to double stranded cDNA using random primers. Two universal sequences (adaptor 1 and 2, compatible with Illumina sequencing instruments) were ligated to both ends of the cDNA molecule with unique molecular identifiers (UMIs). Nested PCR was then performed with 3’-primers binding to a segment of the constant region and the 5’-universal adapter to enrich the library for TCR transcripts. This generated Illumina compatible amplicons with greater than 90% specificity to the TCR transcripts. The library was sequenced using the Illumina NextSeq 500. Raw data is available in the Sequence Read Archive at https://trace.ncbi.nlm.nih.gov/Traces/sra/, accession number SRP156281.
Bioinformatic analysis of TCR amplicons
The UMIs in the sequenced reads were used to remove PCR duplicates. Annotations were compiled from the IMGT (http://www.imgt.org/) and EST (https://www.ncbi.nlm.nih.gov/nucest) databases, as well as data from previous swine T cell sequencing experiments performed in house. Sequences with gaps in annotation were manually curated while sequences with stop codons were dropped. All non-redundant TCR-segment sequences were grouped into sets of Vs, Js, Ds and Cs. V-J pairs, CDR3 sequences and V-CDR3-J combinations were tabulated from the data and used to catalog the restricted usage of segments. Sequences recovered by the above process were compared to those in the human V and J IMGT database and to genomic and cDNA sequences in GenBank. Jα sequences also were compared with pig Jα sequences annotated in IMGT while Vβ and Jβ sequences were correlated to published and unpublished cDNA sequences generously provided by Dr. John Butler (30). Based on these comparisons, the nomenclature used for the porcine Vα, Vβ, and Jβ genes was adopted from the human classification, with the prefix “p” for porcine added to indicate provisional nomenclature. Jα genes were named according to the IMGT database. Phylogenetic analysis of the porcine iNKT cell repertoire was performed using Blast searches of the human, mouse, and rat genome databases of the National Center for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov/Blast.cgi) with the nucleotide sequence of the of the pig pTRAV10, TRAJ18*01, and pTRBV25 gene segments as well as porcine CD1D. Phylogenetic trees were constructed according to the maximum likelihood method using Mega X (https://www.megasoftware.net).
Structural modeling
The nucleotide sequence of porcine pTRAV10 was submitted to NCBI Open Reading Frame Finder to determine the amino acid sequence. The parameters were set to search for ATG and alternative initiation codons. An open reading frame was determined that codes for a protein of 92 amino acids in length. Sequencing results of pTRBV25 translated to a protein sequence of 26 amino acids in length, which is too short to build a reliable model. Therefore, the amino acid sequence was submitted to ExPASy Blast to search the UniProt database for proteins of similar sequence. The submitted sequence shared 100% identity with residues 64–95 of pig T-cell receptor Vβ25 (Uniprot identifier: I3LEA5). Sequence alignment of this sequence with the human TCR Vβ11 segment (Protein Data Bank [PDB]: 2PO6) determined these proteins share 76% sequence identity. Subsequently, the I3LEA5 sequence was determined an appropriate representation of full length porcine pTRBV25.
A model of porcine iNKT cell receptor (pTRAV10-TRAJ18*01, pTRBV25 segments) in complex with CD1d-bound α-GalCer was generated in SWISS-MODEL using the structure of human iNKT cell receptor (Vα24-Jα18, Vβ11 segments) in complex with CD1d-α-GalCer as a template. The human and mouse structures were obtained from RCSB PDB under the respective identifiers 2PO6 (https://www.rcsb.org/structure/2po6) and 3HE6 (https://www.rcsb.org/structure/3HE6). The porcine β2-microgloblulin structure was obtained from PDB under the identifier 5NQ0 (https://www.rcsb.org/structure/5NQ0) and its interactions with CD1d were modeled using the human structure. The porcine CD1d, pTRAV10, and pTRBV25 models were superimposed on the template crystal structure of the human complex and α-carbon root mean square deviation (rmsd) values calculated in COOT (31). The α-GalCer glycolipid was docked between the subunits based on the binding of α-GalCer in the human complex. The interface surface areas between each of the components were determined in PDB ePISA (http://www.ebi.ac.uk/pdbe/pisa/).
Results
Enrichment of iNKT cells from pig peripheral blood
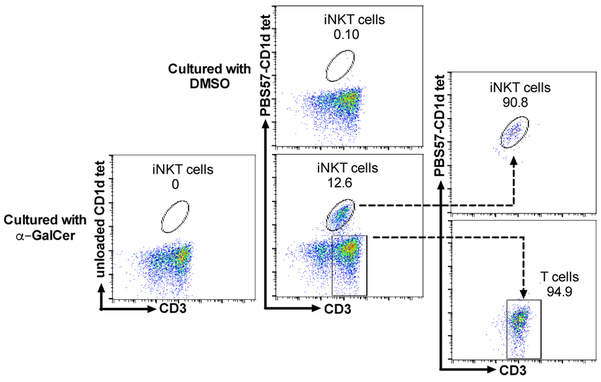
The proportion of iNKT cells in pig blood typically ranges between 0.01 and 1% of total lymphocytes (23, 25). To collect enough iNKT cells for TCR sequencing, PBMCs from 8 pigs were cultured with α-GalCer for 7 days which expanded iNKT cells by an average of 155-fold (Supplementary Table 1). Addition of exogenous cytokines was not required to expand pig iNKT cells, which is consistent with a previous report (23). iNKT cells were identified using anti-porcine CD3ε antibody and mCD1d tetramer loaded with the α-GalCer analog PBS57, which cross-reacts with the porcine invariant TCR (22, 24, 25, 32–35). iNKT cells and Tconv were respectively distinguished as CD3ε+CD1d tetramer + and CD3ε+CD1d tetramer − cells. An example of two-color flow cytometric analysis of one representative PBMC preparation cultured with DMSO or α-GalCer is shown (Figure 1). iNKT cells were not detected in α-GalCer-cultured PBMCs stained with the unloaded CD1d tetramer, which demonstrates the specificity of porcine invariant TCR for the mouse CD1d tetramer-antigen complex. For iNKT cell purification, two donor preparations were produced by pooling α-GalCer-cultured PBMCs from 4 pigs each. FACS sorting was used to isolate approximately 6×105 iNKT cells and 2.5×106 Tconv from each pool with >90% purity (Figure 1).
Figure 1.
Acquisition of porcine iNKT cells and conventional αβ T cells (Tconv) for TCR sequencing. Peripheral blood mononuclear cells (PBMCs) were labeled with PBS57-loaded or unloaded CD1d tetramer and anti-CD3 antibody seven days after culture with vehicle (DMSO) or α-GalCer. PBMCs cultured with α-GalCer were FACS sorted into iNKT cell (CD3ε+CD1d tetramer+) and Tconv (CD3ε+CD1d tetramer−) populations to a purity of >90% according to the post-sort analysis.
T cell receptor repertoire analysis
The objective was to compare the TCR α and β chains expressed by mCD1d tetramer positive and negative T cells to determine whether porcine iNKT cells are biased for specific α and β TCR genes compared to Tconv. However, annotation of the porcine TCR repertoire is incomplete, leading to the concern that porcine iNKT cells could potentially use previously undescribed TCR α or β chains. Therefore, an approach was taken to use recombination signal sequences (RSSs) to map sequence reads generated from the Tconv and enriched for TCR transcripts by PCR, to porcine V and J segments complied from Sus scrofa NCBI assembly Sscrofa11.1. Using this strategy, it was anticipated that invariant chains expressed by iNKT cells would be found on multiple Tconv clones. A total of 187,881 CDR3α and 136,529 CDR3β unique sequences were identified from the two pooled Tconv samples. These were used to identify 48 Vα sequences, 50 Jα sequences, 18 Vβ sequences, and 18 Jβ sequences. The complete collection of sequences are provided in Supplementary Table 2 and the raw data are available in the Sequence Read Archive at https://trace.ncbi.nlm.nih.gov/Traces/sra/, accession number SRP156281.
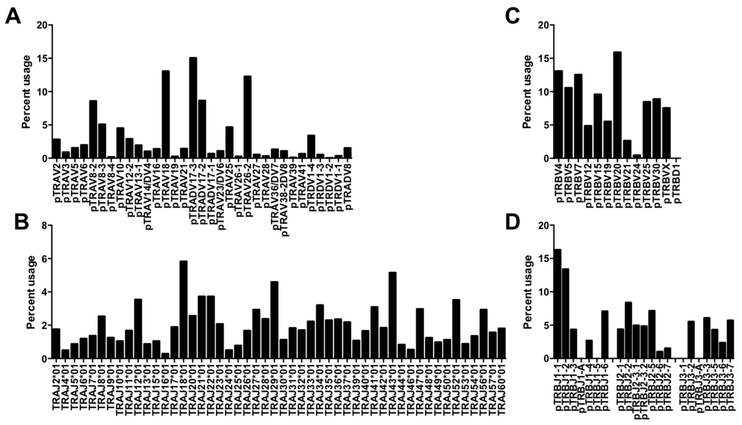
All 48 of the Vα sequences that we identified were among 33 TRAV segments deposited in Genbank by Yamamoto et al. (36) (Table 1). Because porcine Vα genes are not annotated in IMGT, segment naming was based on similarity with consensus sequences of human TRAV genes with nucleotide identities of ≥75%. The prefix “p” for porcine was added to indicate provisional nomenclature. Thirty-two of our sequences are homologous to human genes. Some human segments aligned with more than one pig V sequence that could not be distinguished from each other due to our relatively short V segment reads. We detected one Vα gene (pTRADV8) that aligned with a previously cloned porcine TCR α/δ gene with no orthologous segment in humans, rodents, or other artiodactyls. Examination of the expressed Vα repertoire found that five segments, pTRAV8–2, pTRAV18, pTRADV17–3, pTRADV17–2, and pTRAV26–2, were used preferentially and together accounted for ~58% of the TRAV segments expressed (Figure 2A). We did not detect segments homologous to human TRAV pseudogenes TRAV8–5, TRAV11, TRAV15, TRAV31, TRAV32, suggesting that these are also pseudogenes in pigs. However, a TRAV segment homologous to the human pseudogene TRAV28 was detected, albeit at low levels (0.33%). Interestingly, we did not detect a homolog to TRAV1–2 that is used by the invariant TCR α chain of human mucosal associated-invariant T (MAIT) cells (37). We were also unable to detect MAIT cells from pig blood using mouse or macaque MR1 tetramers (data not shown).
Table 1.
Pig TRAV genes
| Pig TRAV segments† |
Number of distinct sequences |
Human homologue† |
Nucleotide identity (%) human/pig |
|---|---|---|---|
| pTRAV2 | 1 | TRAV2 | 90 |
| pTRAV3 | 1 | TRAV3 | 83 |
| pTRAV5 | 1 | TRAV5 | 79 |
| pTRAV6 | 1 | TRAV6 | 81 |
| pTRAV8-2 | 2 | TRAV8-2 | 83 |
| pTRAV8-3 | 2 | TRAV8-3 | 86 |
| pTRAV8-4 | 1 | TRAV8-4 | 85 |
| pTRAV10 | 1 | TRAV10 | 85 |
| pTRAV12-2 | 2 | TRAV12-2 | 84 |
| pTRAV13-1 | 1 | TRAV13-1 | 86 |
| pTRAV14/DV4 | 2 | TRAV14/DV4 | 82 |
| pTRAV16 | 1 | TRAV16 | 83 |
| pTRAV18 | 2 | TRAV18 | 75 |
| pTRAV19 | 1 | TRAV19 | 82 |
| pTRAV21 | 1 | TRAV21 | 83 |
| pTRADV17-3 | 3 | TRAV22 | 84 |
| pTRADV17-2 | 3 | TRAV22 | 85 |
| pTRADV17-1 | 1 | TRAV22 | 82 |
| pTRAV23/DV6 | 2 | TRAV23/DV6 | 85 |
| pTRAV25 | 1 | TRAV25 | 85 |
| pTRAV26-1 | 1 | TRAV26-1 | 79 |
| pTRAV26-2 | 1 | TRAV26-2 | 84 |
| pTRAV27 | 1 | TRAV27 | 83 |
| pTRAV28 | 1 | TRAV28 | 82 |
| pTRAV36/DV7 | 2 | TRAV36/DV7 | 75 |
| pTRAV38-2DV8 | 1 | TRAV38-2DV8 | 82 |
| pTRAV39 | 1 | TRAV39 | 81 |
| pTRAV41 | 1 | TRAV41 | 83 |
| pTRDV1-4 | 5 | TRDV1 | 78 |
| pTRDV1-3 | 1 | TRDV1 | 78 |
| pTRDV1-2 | 1 | TRDV1 | 77 |
| pTRDV1-1 | 1 | TRDV1 | 79 |
| pTRADV8 | 1 | - | - |
Sequences obtained from the NCBI database
Figure 2.
Percent usage of (A) Vα, (B) Jα, (C) Vβ, and (D) Jβ gene segments among porcine Tconv from peripheral blood.
Analysis of Jα gene segment usage found that all 50 sequences detected were among 61 Jα gene segments previously identified in the pig germline sequence and annotated in IMGT according to their human TRAJ orthologs (Table 2). Thus, designations for the Jα segments were adapted from IMGT (38). All of the Jα sequences aligned with human TRAJ genes with nucleotide identities of ≥78%. The frequency of Jα segment usage ranged between 0.28 and 5.82%, with TRAJ18*01, TRAJ29*01, and TRAJ43*01 representing the most frequently used segments (Figure 2B). Orthologs to human Jα pseudogenes, TRAJ51 and TRAJ55, were not detected, although we did identify a segment corresponding to the TRAJ60 pseudogene, which is functional in pigs (38). We also found Jα segments that corresponded with human TRAJ2, TRAJ25, and TRAJ35, which in humans contain open reading frames with non-canonical RSS and J-region segments that result in non-functional products (38). We did not detect sequences that aligned with TRAJ1*01, TRAJ19*01, TRAJ38*01, TRAJ45*01, TRAJ51*01, TRAJ55*01, TRAJ59*01, and TRAJ61*01, which are predicted to be non-functional in pigs (38), although expression of TRAJ38*01, TRAJ45*01, and TRAJ61*01 has been reported by Yamamoto et al. (39).
Table 2.
Pig TRAJ genes
| Pig TRAJ segments‡ |
Human homologue† | Nucleotide identity (%) human/pig |
|---|---|---|
| TRAJ2*01 | TRAJ2 | 78 |
| TRAJ4*01 | TRAJ4 | 84 |
| TRAJ5*01 | TRAJ5 | 83 |
| TRAJ6*01 | TRAJ6 | 86 |
| TRAJ7*01 | TRAJ7 | 89 |
| TRAJ8*01 | TRAJ8 | 82 |
| TRAJ9*01 | TRAJ9 | 88 |
| TRAJ10*01 | TRAJ10 | 81 |
| TRAJ11*01 | TRAJ11 | 90 |
| TRAJ12*01 | TRAJ12 | 90 |
| TRAJ13*01 | TRAJ13 | 83 |
| TRAJ15*01 | TRAJ15 | 83 |
| TRAJ16*01 | TRAJ16 | 88 |
| TRAJ17*01 | TRAJ17 | 86 |
| TRAJ18*01 | TRAJ18 | 86 |
| TRAJ20*01 | TRAJ20 | 85 |
| TRAJ21*01 | TRAJ21 | 79 |
| TRAJ22*01 | TRAJ22 | 81 |
| TRAJ23*01 | TRAJ23 | 90 |
| TRAJ24*01 | TRAJ24 | 86 |
| TRAJ25*01 | TRAJ25 | 93 |
| TRAJ26*01 | TRAJ26 | 80 |
| TRAJ27*01 | TRAJ27 | 90 |
| TRAJ28*01 | TRAJ28 | 88 |
| TRAJ29*01 | TRAJ29 | 88 |
| TRAJ30*01 | TRAJ30 | 83 |
| TRAJ31*01 | TRAJ31 | 90 |
| TRAJ32*01 | TRAJ32 | 87 |
| TRAJ33*01 | TRAJ33 | 95 |
| TRAJ34*01 | TRAJ34 | 86 |
| TRAJ35*01 | TRAJ35 | 83 |
| TRAJ36*01 | TRAJ36 | 85 |
| TRAJ37*01 | TRAJ37 | 84 |
| TRAJ39*01 | TRAJ39 | 89 |
| TRAJ40*01 | TRAJ40 | 86 |
| TRAJ41*01 | TRAJ41 | 83 |
| TRAJ42*01 | TRAJ42 | 88 |
| TRAJ43*01 | TRAJ43 | 89 |
| TRAJ44*01 | TRAJ44 | 78 |
| TRAJ46*01 | TRAJ46 | 90 |
| TRAJ47*01 | TRAJ47 | 86 |
| TRAJ48*01 | TRAJ48 | 84 |
| TRAJ49*01 | TRAJ49 | 86 |
| TRAJ50*01 | TRAJ50 | 81 |
| TRAJ52*01 | TRAJ52 | 91 |
| TRAJ53*01 | TRAJ53 | 92 |
| TRAJ54*01 | TRAJ54 | 92 |
| TRAJ56*01 | TRAJ56 | 84 |
| TRAJ57*01 | TRAJ57 | 88 |
| TRAJ60*01 | TRAJ60 | 81 |
Sequences obtained from the NCBI database and annotated according to IMGT
Sequences obtained from the NCBI database
Examination of the Vβ repertoire identified 18 sequences that overlapped with 12 of 19 previously reported pig TRBV gene groups (Table 3). Designations for the Vβ segments were adapted from nomenclature published by Butler et al. (30) and Eguchi-Ogawa et al. (40). Human orthologs for the pig Vβ sequences were identified, with sequence similarities of ≥75%. The exception was a sequence corresponding to the previously identified clone gT203 that belongs to the gene group pTRBVX, which is not related to any human family. The human homologs of the pig Vβ genes we identified are functional with the exception of pTRBV21, which is a pseudogene in humans due to a frameshift in the leader sequence (38). The most highly expressed porcine TRBV genes were pTRBV4, pTRBV5, pTRBV7, and pTRBV20 that belong to the Vβ supergroups V, III, VI, and I respectively (Figure 2C) (30).
Table 3.
Pig TRBV genes
| Pig TRBV segments† |
Number of distinct sequences |
Human homologue† | Nucleotide identity (%) human/pig |
|---|---|---|---|
| pTRBV4 | 1 | TRBV4-1 | 84 |
| pTRBV5 | 2 | TRBV5-4 | 76 |
| pTRBV7 | 1 | TRBV7-6 | 86 |
| pTRBV12 | 2 | TRBV12-3 | 84 |
| pTRBV15 | 1 | TRBV15 | 79 |
| pTRBV19 | 1 | TRBV19 | 82 |
| pTRBV20 | 3 | TRBV20-1 | 75 |
| pTRBV21 | 1 | TRBV21-1 | 78 |
| pTRBV24 | 2 | TRBV24-1 | 76 |
| pTRBV25 | 1 | TRBV25-1 | 79 |
| pTRBV30 | 1 | TRBV30 | 85 |
| pTRBVX | 1 | - | - |
| pTRBD1 | 1 | TRBD1 | - |
Sequences obtained from the NCBI database
Finally, we identified sequences corresponding to 17 of 21 TRBJ segments encoded in the porcine genome (40) (Table 4). As in sheep and cattle, the porcine genome carries three TRDB-J-C clusters. The clusters at either end of the TRDB-J-C-containing region respectively correspond to human TRDB-J-C1 and TRDB-J-C2. The center structure is a mixture of the end clusters and appears to have arisen from the crossover of the end clusters in the ancestors of pigs (40). Our analysis found that most sequences reported to be expressed by pigs were also expressed in the current study (30, 40, 41), with the exception of TRBJ1–5 from the first cluster and TRBJ3–1 from the last cluster. Also undetectable were TRBJ1-A and TRBJ3-A that contain alterations in the structure of their RSSs that may affect their expression (40). TRBJ1–1 and TRBJ1–2 segments that are on the edge of the first cluster, were preferentially used (Figure 2D). Human homologs were identified for 8 of the pig TRBJ segments by sequence similarity of ≥76% (Table 4). Among the segments without human orthologs, Blast searches in Genbank found that porcine TRBJ1–1, TRBJ1–2, and TRBJ2–2 most closely align with sheep TRBJ1–1, TRBJ1–2, and TRBJ3–2, respectively, while pig TRBJ2–6 and TRBJ3–2 were most similar to TRBJ2–5 and TRBJ2–2 in camels at the nucleotide level.
Table 4.
Pig TRBJ genes
| Pig TRBJ segments† |
Number of distinct sequences |
Human homologue† |
Nucleotide identity (%) human/pig |
|---|---|---|---|
| pTRBJ1-1 | 1 | - | - |
| pTRBJ1-2 | 1 | - | - |
| pTRBJ1-3 | 1 | TRBJ1-3 | 80 |
| pTRBJ1-4 | 1 | TRBJ1-4 | 81 |
| pTRBJ1-6 | 1 | TRBJ1-6 | 79 |
| pTRBJ2-1 | 1 | TRBJ2-1 | 80 |
| pTRBJ2-2 | 2 | - | - |
| pTRBJ2-3.1 | 1 | TRBJ2-3 | 80 |
| pTRBJ2-3.2 | 1 | TRBJ2-3 | 84 |
| pTRBJ2-5 | 1 | TRBJ2-5 | 87 |
| pTRBJ2-6 | 1 | - | - |
| pTRBJ2-7 | 1 | TRBJ2-7 | 76 |
| pTRBJ3-2 | 1 | - | - |
| pTRBJ3-3 | 1 | TRBJ2-3 | 87 |
| pTRBJ3-5 | 1 | TRBJ2-5 | 89 |
| pTRBJ3-6 | 1 | TRBJ2-6 | 80 |
| pTRBJ3-7 | 1 | TRBJ2-7 | 83 |
Sequences obtained from the NCBI database
Vβ/Jβ rearrangements
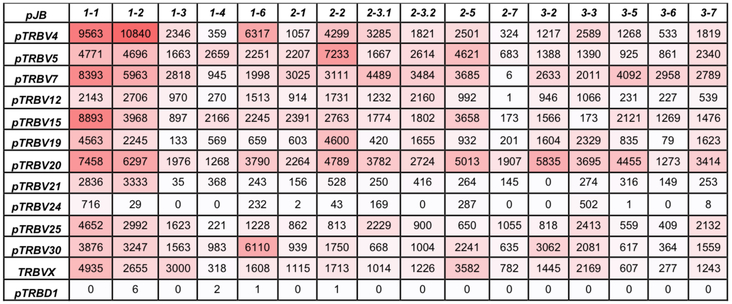
The expression of Vβ/Jβ combinations was examined to establish whether particular rearrangements were favored. Of note, TRBJ1–1 and TRBJ1–2 were used in many rearrangements with TRBV4, TRBV7, TRBV15, and TRBV20, which accounted for 16.3% of the repertoire (Figure 3). Also interesting is the combination of TRBJ3–6 and TRBV7 that accounted for 35.2% of the TRBJ3–6 usage. The TRBJ2–7 segment was seldom combined with 5’ TRBV genes although these segments are used at high frequencies with other Jβ segments. The pTRBVX group, which has no human homolog, was used most frequently with TRBJ1–1, TRBJ2–5, and TRBJ3–3 among the first, second and third TRDB-J-C clusters, respectively.
Figure 3.
The relationship between porcine TRBV and TRBJ usage in porcine TCR rearrangements in peripheral T cells. Boxes contain the number of reads for each V-J combination. Intensity of red shading corresponds to the number of reads in each box.
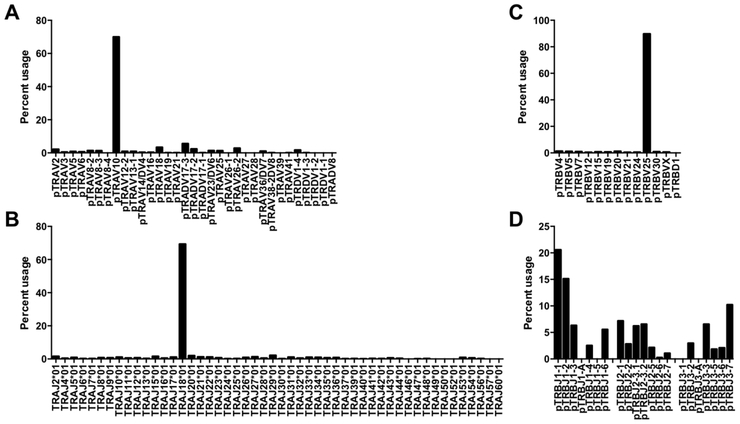
TCR usage by porcine iNKT cells
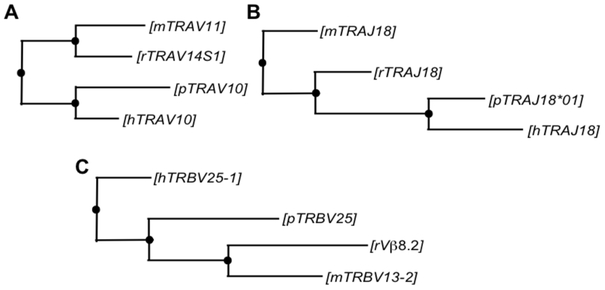
In contrast to Tconv, porcine iNKT cells mostly used a single combination of Vα, Jα, and Vβ segments: pTRAV10, TRAJ18*01, and pTRBV25, respectively (Figure 4A-C). Blast searches were performed to compare the homology of these genes to TCR segments in humans, mice and rats that are species with known iNKT cell receptor usage. pTRAV10 was highly homologous to segments encoding TRAV10 (Vα24) in humans, Trav11 (Vα14) in mice, and Trav14S1 (Vα14) in rats, which are the canonical Vα segments used by iNKT cells in these species (Table 5). The best alignments for TRAJ18*01 were TRAJ18, Traj18, and Traj18, the Jα18 segments respectively used by the human, rat and mouse invariant α chain. pTRBV25 was most similar to human TRBV25–1 (Vβ11), mouse Trbv13–2 (Vβ8.2) and rat Vβ8.2, the canonical Vβ segments used by iNKT cells in these species. Sequence alignments were more similar between pig and human gene segments than between humans and mice and rats (Table 5). Furthermore, phylogenetic trees constructed using the maximum likelihood method indicate that porcine invariant TCR chains are more evolutionarily related to human than rodent TCR chains (Figure 5). Porcine CD1d is also more homologous to human than rodent CD1d (Table 5).
Figure 4.
Percent usage of (A) Vα, (B) Jα, (C) Vβ, and (D) Jβ gene segments among porcine iNKT cells from peripheral blood.
Table 5.
Comparison of identity between porcine iNKT cell TCR [nucleotide (nt)] and CD1D proteins [amino acid (aa)] and the corresponding TCR genes or proteins in human, mouse, and rat
| Human | Mouse | Rat | |
|---|---|---|---|
| pTRAV10 | TRAV10 | Trav11 | Trav14S1 |
| Accession # | NC_000014.9 | NC_000080.6 | AB041999.1 |
| nt identity | 282/333 (85%) | 228/309 (74%) | 214/290 (74%) |
| TRAJ18*01 | TRAJ18 | Traj18 | Traj18 |
| Accession # | NC_000014.9 | NC_000080.6 | DQ340291.1 |
| nt identity | 74/86 (86%) | 69/87 (79%) | 49/59 (83%) |
| pTRBV25 | TRBV25-1 | Trbv13-2 | Vβ8.2 |
| Accession # | NC_000007.14 | NC_000072.6 | X14973.1 |
| nt identity | 98/132 (74%) | 70/98 (71%) | 66/89 (74%) |
| CD1D | CD1D | Cd1d | CD1d1 |
| Accession # | BC027926.1 | NM_007640.2 | NP_058775.1 |
| aa identity | 213/313 (68%) | 200/345 (58%) | 199/345 (58%) |
Accession numbers and nucleotide identities, and amino acid identities were obtained from the NCBI database. Nucleotide and amino acid identities displayed as the number and percentage of overlapping nucleotides and amino acids, respectively.
Figure 5.
Phylogenetic trees comparing (A) TRAV, (B) TRAJ, and (C) TRBV genes expressed by pig (p), human (h), mouse (m), and rat (r) iNKT cells. Phylogenetic trees were constructed by the neighbor-joining method using nucleotide sequences from the current study compared to the following sources in the Genbank database; Homsap: TRAV10 (NC_000014.9), TRAJ18 (NC_000014.9), TRBV25-1 (NC_000007.14); Musmus (C57BL/6J): Trav11 (NC_000080.6), Traj18 (NC_000080.6), Trbv13-2 (NC_000072.6); Rattus-Norway rat: Trav14S1 (AB041999.1), Traj18 (DQ340291.1), Vβ8.2 (X14973.1).
Examination of the ten most frequently expressed CDR3α sequences showed that porcine iNKT cells predominantly use a single V-J rearrangement (pTRAV10-TRAJ18*01) and that the amino acid sequence of this junction was highly restricted. Most of the CDR3α sequences identified contained the single amino acid combination cVVGDRGSRLGRLYf (Table 6), which is highly homologous to CDR3α sequences used by mouse (cVVGDRGSALGRLHf) and human (cVVSDRGSTLGRLYf) iNKT cell receptors. All of the porcine CDR3α segments analyzed were 15 amino acids in length and the V-J junction consisted of a single amino acid that was usually glycine. In contrast, Tconv used a wide variety of CDR3α lengths (Table 7). Unlike the invariant TCRα chain, the TCRβ chain of iNKT cells used all 17 Jβ segments expressed by Tconv and with a similar relative frequency (Figure 4D versus Figure 2D). They also displayed a large variation in N nucleotides indicating that, like rodents and humans, the CDR3β does not contribute to the structural specificity of the invariant TCR.
Table 6.
Ten most frequently used CDR3α sequences from porcine iNKT cells
| pTRAV | pTRAJ | CDR3 sequence | Frequency |
|---|---|---|---|
| pTRAV10 | TRAJ18*01 | cVVGDRGSRLGRLYf | 53.62% |
| pTRAV10 | TRAJ18*01 | cVGGDRGSRLGRLYf | 1.14% |
| pTRAV10 | TRAJ18*01 | cVVADRGSRLGRLYf | 1.09% |
| pTRAV10 | TRAJ18*01 | cVVVDRGSRLGRLYf | 1.01% |
| pTRAV10 | TRAJ18*01 | cVVCDRGSRLGRLYf | 0.97% |
| pTRAV10 | TRAJ18*01 | cVVGDRGSSLGRLYf | 0.96% |
| pTRAV10 | TRAJ18*01 | cVVDDRGSRLGRLYf | 0.80% |
| pTRAV10 | TRAJ18*01 | cVVGDRGSRLGRLFf | 0.69% |
| pTRAV10 | TRAJ18*01 | cVVGNRGSRLGRLYf | 0.67% |
| pTRAV10 | TRAJ18*01 | cVVGDRGSRLGRLYl | 0.54% |
Table 7.
Ten most frequently used CDR3α sequences from porcine Tconv
| pTRAV | pTRAJ | CDR3 sequence | Frequency |
|---|---|---|---|
| pTRAV26-2 | TRAJ21*01 | cIGVPYNTNRLYf | 1.49% |
| pTRADV17-3 | TRAJ29*01 | cAGASRQLVf | 0.83% |
| pTRADV17-3 | TRAJ29*01 | cAGASRQLVf | 0.57% |
| pTRADV8 | TRAJ43*01 | cALRNNDLRf | 0.42% |
| pTRAV8-3 | TRAJ11*01 | cAVSDPGNSGYSKLTf | 0.37% |
| pTRAV26-2 | TRAJ43*01 | cILRNNDLRf | 0.63% |
| pTRADV17-3 | TRAJ35*01 | cAGELPNSGGVLHf | 0.35% |
| pTRAV26-2 | TRAJ2*01 | cILIGATTGKLIf | 0.29% |
| pTRAV26-2 | TRAJ12*01 | cILPNDGGYKWIf | 0.27% |
| pTRAV18 | TRAJ34*01 | cAVSVSYDKLIf | 0.24% |
Among purified mCD1d tetramer positive cells, non-canonical sequences accounted for 30% of TRAV segments, 31% of TRAJ segments, and 10% of TRBV segments (Figure 4A-C). These sequences may have originated from Tconv contamination during the sorting process (Figure 1) and/or from cells that recognize α-GalCer by non-canonical TCRs. To address the second possibility, we searched for but failed to discover enrichment of sequences homologs to non-canonical mouse TCR α and β segments reported to expand when murine iNKT cells are cultured with α-GalCer (42, 43). Nevertheless, pigs may express alternative non-canonical TCRs capable of binding PBS57-loaded CD1d tetramers.
Homology modeling of antigen recognition
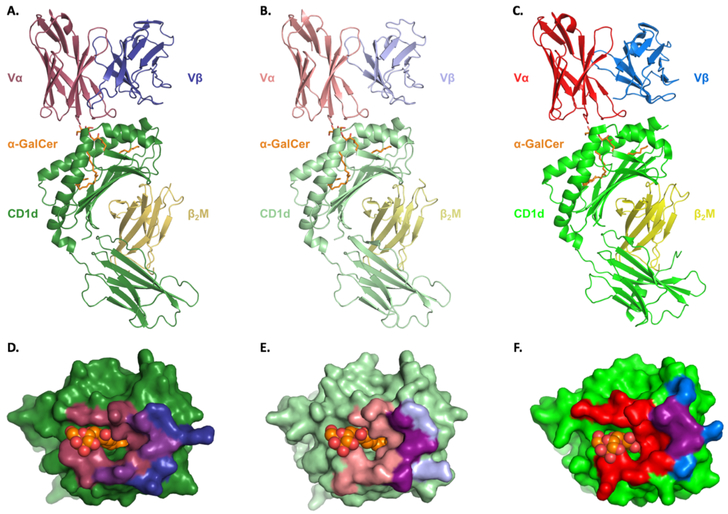
Molecular modeling of the porcine iNKT-CD1d/α-GalCer complex suggests that the overall structure and interface surface areas are similar to its mouse and human counterparts (Figure 6). Superposition of the modeled porcine components CD1d, pTRAV10, and pTRBV25 with the human template complex respectively results in α-carbon rmsd values of 0.08, 0.08, and 0.32Å (Supplementary Table 3), indicating that the three-dimensional structures are essentially the same. Like other species, porcine CD1d is composed of α1, α2, and α3 domains. The α1 and α2 domains combine to form a hydrophobic antigen-binding groove that further separates into two connected pockets, A’ and F’, which bind the sphingoloid base and fatty acid chain of α-GalCer, respectively. CD1d contact sites at residues Arg79, Asp80, Glu83, and Asp151 (Arg106, Asp107, Glu110, and Asp178 in porcine CD1d) that are known to mediate α-GalCer-induced iNKT cell activation are conserved in pigs (Supplementary Table 4) (44–46). Three CD1d-mediated interactions, at positions 84, 89, and 150, that are non-conserved between mouse and human are also non-conserved between mouse and pig (47). Among these, only Phe84 (Phe111 in porcine CD1d) is conserved between pig and human. This residue contributes to the different specificities that mouse and human iNKT cells possess for various CD1d-presented antigens, including α-GalCer (48).
Figure 6.
Molecular modeling of the iNKT TCR interacting with the CD1d-α-GalCer complex in (A) human, (B) pig, and (C) mouse. CD1d, green; β2m, yellow; α-GalCer, orange; iNKT TCR α and β chain, red and blue, respectively. Footprint of the iNKT TCR on the surface of (D) human, (E) pig and, (F) mouse CD1d-α-GalCer. Color coding as in A-C.
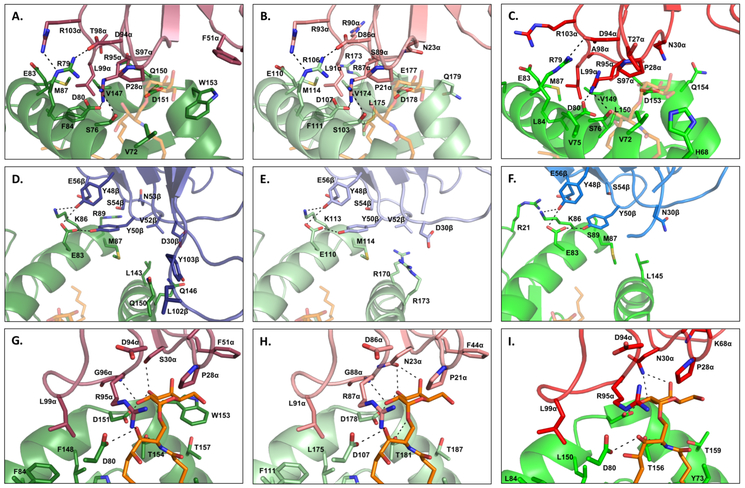
Contacts at the pig TCR-αGalCer-CD1d interface include interactions that are conserved and different between the mouse and human complexes. CDR2β contacts with CD1d at positions 48, 50, and 56 that are conserved between mouse Vβ8.2 and human Vβ11, are also conserved in pig TRBV25 (Figure 7D-F). Contact sites in CD1d that interface with CDR2β are conserved in mice, humans and pigs, with the exception that interactions between Arg89 in human CD1d and Asn53 at the tip of the Vβ11 CDR2β loop are lost in pigs and mice due to respective alterations to Gly116 and Ser89. CDR3α residues that contact CD1d are also mostly conserved (Figure 7A-C). However, two H-bonds and one van der Waal bond between Gln150 of CD1d and Thr98 of CDR3α in humans are lost in pigs and mice due to alterations to Arg98 and Ala98, respectively. Contact sites at Pro28 and Asn30 of mouse CDR1α, which mediate interactions with carbon atoms and hydroxyl groups of the α-GalCer glycosyl head, are conserved in pigs (Figure 7G-I). Pro28 is conserved in humans, but Asn30 is altered to Ser30. Asn30 forms hydrogen bonds with both the 3’-OH and 4’-OH moieties of α-GalCer, while Ser30 bonds only with the 3’-OH moiety. Position 29 is altered to phenylalanine in humans, which enables the CDR1α loop to make additional contacts with 4’-OH and C6 of α-GalCer. This position is conserved between pigs and humans.
Figure 7.
iNKT cell receptor contacts with CD1d in human (A, D, G), pig (B, E, H), and mouse (C, F, I). Images A-C describe interactions between Vα and CD1d. Images D-F describe interactions between Vβ and CD1d. Images G-I describe interactions between Vα and CD1d with α-GalCer. Hydrogen bonds are shown as black dashed lines.
Discussion
Although a previous report predicted the porcine invariant Vα segment based on homology to the TRAV10 sequence used by human iNKT cells (49), the current study is the first, to our knowledge, to establish the porcine iNKT cell receptor and compare its sequence and conformation to mouse and human iNKT cell receptors. As previously described, pigs express a population of CD3ε+CD1d tetramer+ cells that share similar characteristics to iNKT cells in rodents and primates, including that they rapidly produce IFNγ in response to activation with α-GalCer analogs and that once stimulated, they transactivate a wide range of innate and adaptive immune responses (50). Another similarity is that CD1d is necessary for porcine iNKT cell development as CD1d knockout swine do not possess CD1d tetramer positive cells (32). Pig iNKT cells express high levels of the transcription factor promyelocytic leukemia zinc finger, which is required for iNKT cell development and innate-like functions in other species (24). Furthermore, pig iNKT cell frequency in blood and tissues closely resembles the distribution of iNKT cells in human tissues (25). The current work found that pig iNKT cells predominantly express a single Vα, Jα, and Vβ segment. The sequences are highly homologous to invariant chain receptors expressed by mice, rats, cotton rats, and humans (3, 5, 51–54), which are other species for which iNKT cell receptor sequences have been established. Phylogenetic trees constructed using the maximum likelihood algorithm showed that the Vα, Vβ, and Jα segments of pig and human iNKT cell receptors were more related to each other than the corresponding mouse and rat segments, which reflects the relative similarity in genomic sequences between these species (55).
Previously published crystal structures of mouse and human iNKT cell receptor- α-GalCer-CD1d complexes have demonstrated the evolutionarily conserved nature of this interaction. Our in-silico model of the porcine iNKT TCR/CD1d/α-GalCer complex identified most of the same highly conserved interactions, including critical CD1d contact sites at positions 79, 80, 83, and 151 (positions 106, 107, 110, and 178 in porcine CD1d) that are involved in TCR recognition of α-GalCer (44–46). The only contact site conserved between pig and mouse and not conserved between pig and human is Asn30 in CDR1α that forms hydrogen bonds with the 3’-OH and 4’-OH moieties of α-GalCer (56). The Ser30 modification in humans lacks the hydrogen bond with 4’-OH, which may weaken the iNKT TCR/CD1d/α-GalCer interaction compared to mice and pigs. The human complex also contains two contact sites that are missing in mice and pigs (Thr98 in CDR3α and Arg89 in CD1d), which increases the stability of TCR interactions. Contacts conserved between humans and pigs, but not mice, include Phe29 that provides interactions with 4’-OH and C6 of α-GalCer. Another difference is position 84 that is altered from Leu84 in mouse CD1d to Phe84 in humans and Phe111 in pigs. In mice, Leu84 is able to form a roof over the F’ pocket before TCR engagement. This structure is created when the hydrocarbon chains of various lipid antigens, including α-GalCer, orientate CD1d sidechains at Leu84, Val149 and Leu150 to an optimal alignment for docking with TCR CDR3α residue Leu99. Consequently, the TCR does not expend energy to keep the roof closed upon engagement, which results in a more stable complex with a reduced TCR dissociation rate (57). Not all iNKT cell ligands pre-form the F’ roof in mice, and those ligands that do not preform the F’ roof have much faster TCR off rates than ligands that do (58). The F’ roof is unable to pre-form when Leu84 is altered to Phe84, which probably underlies why the TCR dissociation rates are more similar between iNKT cell agonists when they bind human CD1d compared to mouse CD1d (48). Conservation of Phe84 (Phe111 in porcine CD1d) between pigs and humans suggest that swine also have a narrower range of TCR dissociation rates for different CD1d-presented antigens.
To our knowledge, this study represents the first use of RNA-seq to characterize the TCR repertoire of swine, and thus provides a useful opportunity to compare this approach to previous studies that used traditional cloning and DNA sequencing. A characteristic of our method is that it sometimes generated several sequences that aligned with one human TRAV or TRBV segment (Tables 1 & 3). We cannot determine whether this is because of polymorphisms or due to homologous gene segments that were indistinguishable from each other. The latter scenario is more likely because our method involves sequencing 135 nucleotides in one direction starting from the C region. This yields full-length J segments that can be unambiguously mapped to the genome, but only partial V segments that are sometimes less than 30 nucleotides in length. We mapped the V segments to the Sus scrofa genome when reliable RSS could be identified at the 3’ end of the V segment. However, because of the relatively short V segment reads and incomplete annotation of the pig genome, we could not disambiguate exons that were similar to each other. Nor could we be sure if we are missing any segments.
Previous characterization of porcine TRAV and TRAJ gene expression comes from ~100 independent cDNA clones generated by Yamamoto et al., from the thymus of a 1 month-old Large White piglet and the peripheral blood of a 5 month-old Clawn minipig (36, 39). From the resulting clones, 33 Vα segments and 44 Jα segments were identified as well as V-J CDR3 sequences. The authors found corresponding human segments for all but one of these Vα segments (designated Vα01), which is a sequence that more closely aligns with mouse and horse TRDV clones (36). With the exception of Vα16, the current study identified the same collection of TRAV gene segments as previously reported, including the Vα01 sequence (designated pTRADV8). In addition, we identified sequences corresponding to 11 human TRAV genes which had not been previously detected, including TRAV10 that is used by human iNKT cells. We also detected 41 of the 44 TRAJ genes previously identified (39), as well as nine extra TRAJ sequences that included the TRAJ18 segment expressed by iNKT cells.
Previous information about the expressed porcine TRB repertoire also comes from a small number of studies. Barron et al. used pig renal graft infiltrates to identify 12 TRBJ segments, two TRBD segments and 19 TRBV segments, including a TRBV subfamily (designated TRBVX) that corresponds to a Vβ segment in the mouse but not the human genome (59). In another study, Butler et al. examined more than 300 clones isolated from pig thymocytes and peripheral blood lymphocytes and identified 19 TRBV groups and two (subsequently discovered to be three) groups of J segments (30). Watanabe et al. identified many of the same clones from pig thymocytes and peripheral blood lymphocytes (41). The TRBV segments detected in the current study corresponded to 12 TRBV groups from the above described reports, including sequences that matched the TRBVX group. All 17 TRBJ segments that we identified have been previously described. Only four TRBJ segments encoded in the pig germline were not detected; two non-functional genes (TRBJ1-A and TRBJ3-A) and two genes (TRBJ1–5 and TRBJ3–1) that only Butler et al. previously detected and at very low frequencies (30).
Several similarities exist between our dataset and the cloning-based studies described above. They include that: i) almost all V and J segments identified in previous studies were detected within our collection of sequences (with the exception of the Vβ cluster where only 12 of 19 previously reported groups were detected). ii) We also did not detect predicted TCR pseudogenes. iii) The relative usage of Vα, Vβ, and Jβ segments in the current work was correlated to their frequency in previous publications; we found a high (r2 = 0.75, p value <0.0001), medium (r2 = 0.47, p value <0.0001) and low (r2 = 0.27, p value = 0.023) correlation in Jβ, Vα and Vβ segment usage between our results and the frequency of Jβ, Vα and Vβ segments respectively reported by Eguchi-Ogawa et al. (40), Yamamoto et al. (36), and Butler et al. (30). The high correlation in Jβ gene usage was likely because these segments are particularly sensitive to non-random rearrangement according to their chromosomal location (30, 36, 40, 60). Indeed, we found a closely matched pattern of Jβ segment usage in our results compared with a previous compilation of publicly available porcine TRBJ cDNA and EST sequences (Figures 2D & 4D vs. (40)). No correlation was found for Jα segments (Jα compared to (39): r2 = 0.03, p value = 0.1988), which is not surprising as usage of these genes is thought to be random (39, 61). (iv) Lastly, we confirmed the findings of Butler et al. that some Vβ/Jβ combinations are non-randomly expressed, such as TRBJ1–1 and TRBJ1–2 with TRBV4 and TRBV7, respectively (30). This is consistent with reports that TCRβ chain repertoires contain public clonotypes that are shared across individuals. It has been shown that the observed repertoire overlap between any two individuals is several thousand-fold larger than the potential diversity of CDR3 sequences and that there is a bias towards sequences using specific Vβ and Jβ segment combinations (62–64).
In summary, our study characterized the expressed αβ TCR repertoire of pig peripheral blood T cells using RNA-seq, which provides an efficient, unbiased method that does not require prior knowledge of the V or J segments. The high-throughput nature of this approach allows for the simultaneous sequencing of both α and β chains. It is also is sensitive enough to detect TCRs expressed by rare T cell populations, such as the iNKT cell receptor, which was identified in this study. Analysis of the porcine invariant TCR found that it closely resembles the mouse, but particularly the human receptor, including for its predicted interaction with antigen-loaded CD1d molecules. Thus, pigs may be useful for determining the efficacy of various glycolipid ligands for human use. There is also potential to use iNKT cell agonists for veterinary applications including as vaccine adjuvants and anti-microbial agents, which has recently been explored in swine with encouraging results (25, 33–35).
Supplementary Material
Acknowledgments:
The National Institutes of Health Tetramer Core Facility provided the CD1d tetramers under the contract HHSN272201300006C. The authors are grateful for the assistance of Dr. John Butler for kindly providing his library of Vβ sequences.
Funding: This work was supported by the United States Department of Agriculture Grant 2016–09448 (JPD) and National Institutes of Health Grant HD092286 (JPD). CL is supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under University of Florida Clinical and Translational Science Awards TL1TR001428 and UL1TR001427.
Abbreviations used in this paper:
- α-GalCer
α-galactosylceramide
- CDR
complementarity determining region
- iNKT cell
invariant natural killer T cell
- MAIT
mucosal associated-invariant T
- NCBI
National Center for Biotechnology Information
- PBMC
peripheral blood mononuclear cell
- PDB
Protein Data Bank
- RNA-seq
RNA sequencing
- RSS
recombination signal sequence
- Tconv
conventional porcine αβ T cells
- TCR
T cell receptor
- UMI
unique molecular identifier
Footnotes
Disclosures: Both RS and AJ are co-founders of Girihlet Inc., which has licensed the TCR sequencing technology from Icahn School of Medicine at Mount Sinai, with the goal of developing it as a commercial product.
References
- 1.Porcelli SA, and Modlin RL. 1999. The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu. Rev. Immunol 17: 297–329. [DOI] [PubMed] [Google Scholar]
- 2.Lantz O, and Bendelac A. 1994. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4–8- T cells in mice and humans. J. Exp. Med 180: 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuura A, Kinebuchi M, Chen HZ, Katabami S, Shimizu T, Hashimoto Y, Kikuchi K, and Sato N. 2000. NKT cells in the rat: organ-specific distribution of NK T cells expressing distinct V alpha 14 chains. J. Immunol 164: 3140–3148. [DOI] [PubMed] [Google Scholar]
- 4.Exley M, Garcia J, Balk SP, and Porcelli S. 1997. Requirements for CD1d recognition by human invariant Valpha24+ CD4-CD8- T cells. J. Exp. Med 186: 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dellabona P, Padovan E, Casorati G, Brockhaus M, and Lanzavecchia A. 1994. An invariant V alpha 24-J alpha Q/V beta 11 T cell receptor is expressed in all individuals by clonally expanded CD4–8- T cells. J. Exp. Med 180: 1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohteki T, and MacDonald HR. 1996. Stringent V beta requirement for the development of NK1.1+ T cell receptor-alpha/beta+ cells in mouse liver. J. Exp. Med 183: 1277–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arase H, Arase N, Ogasawara K, Good RA, and Onoe K. 1992. An NK1.1+ CD4+8- single-positive thymocyte subpopulation that expresses a highly skewed T-cell antigen receptor V beta family. Proc. Natl. Acad. Sci. USA 89: 6506–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macho-Fernandez E, and Brigl M. 2015. The Extended Family of CD1d-Restricted NKT Cells: Sifting through a Mixed Bag of TCRs, Antigens, and Functions. Front. Immunol 6: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan PJ, Brigl M, and Brenner MB. 2013. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol 13: 101–117. [DOI] [PubMed] [Google Scholar]
- 10.Sriram V, Du W, Gervay-Hague J, and Brutkiewicz RR. 2005. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur. J. Immunol 35: 1692–1701. [DOI] [PubMed] [Google Scholar]
- 11.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, Khurana A, Hoebe K, Behar SM, Beutler B, Wilson IA, Tsuji M, Sellati TJ, Wong CH, and Kronenberg M. 2006. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat. Immunol 7: 978–986. [DOI] [PubMed] [Google Scholar]
- 12.Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X, Li Y, Imamura M, Kaneko Y, Okawara A, Miyazaki Y, Gomez-Velasco A, Rogers P, Dahesh S, Uchiyama S, Khurana A, Kawahara K, Yesilkaya H, Andrew PW, Wong CH, Kawakami K, Nizet V, Besra GS, Tsuji M, Zajonc DM, and Kronenberg M. 2011. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat. Immunol 12: 966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wieland Brown LC, Penaranda C, Kashyap PC, Williams BB, Clardy J, Kronenberg M, Sonnenburg JL, Comstock LE, Bluestone JA, and Fischbach MA. 2013. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS. Biol 11: e1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Bohmer G, Prandi J, Mori L, Puzo G, and De Libero G. 2004. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J. Exp. Med 199: 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamora-Chimal J, Fernandez-Figueroa EA, Ruiz-Remigio A, Wilkins-Rodriguez AA, Delgado-Dominguez J, Salaiza-Suazo N, Gutierrez-Kobeh L, and Becker I. 2017. NKT cell activation by Leishmania mexicana LPG: Description of a novel pathway. Immunobiology 222: 454–462. [DOI] [PubMed] [Google Scholar]
- 16.Lotter H, Gonzalez-Roldan N, Lindner B, Winau F, Isibasi A, Moreno-Lafont M, Ulmer AJ, Holst O, Tannich E, and Jacobs T. 2009. Natural killer T cells activated by a lipopeptidophosphoglycan from Entamoeba histolytica are critically important to control amebic liver abscess. PLoS. Pathog 5: e1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albacker LA, Chaudhary V, Chang YJ, Kim HY, Chuang YT, Pichavant M, DeKruyff RH, Savage PB, and Umetsu DT. 2013. Invariant natural killer T cells recognize a fungal glycosphingolipid that can induce airway hyperreactivity. Nat. Med 19: 1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, and Taniguchi M. 1997. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science 278: 1626–1629. [DOI] [PubMed] [Google Scholar]
- 19.Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, and Kronenberg M. 2000. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J. Exp. Med 192: 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karadimitris A, Gadola S, Altamirano M, Brown D, Woolfson A, Klenerman P, Chen JL, Koezuka Y, Roberts IA, Price DA, Dusheiko G, Milstein C, Fersht A, Luzzatto L, and Cerundolo V. 2001. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc. Natl. Acad. Sci. USA 98: 3294–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brossay L, Chioda M, Burdin N, Koezuka Y, Casorati G, Dellabona P, and Kronenberg M. 1998. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J. Exp. Med 188: 1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang G, Artiaga BL, Lewis ST, and Driver JP. 2017. Characterizing porcine invariant natural killer T cells: A comparative study with NK cells and T cells. Dev. Comp. Immunol 76: 343–351. [DOI] [PubMed] [Google Scholar]
- 23.Renukaradhya GJ, Manickam C, Khatri M, Rauf A, Li X, Tsuji M, Rajashekara G, and Dwivedi V. 2011. Functional invariant NKT cells in pig lungs regulate the airway hyperreactivity: a potential animal model. J. Clin. Immunol 31: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thierry A, Robin A, Giraud S, Minouflet S, Barra A, Bridoux F, Hauet T, Touchard G, Herbelin A, and Gombert JM. 2012. Identification of invariant natural killer T cells in porcine peripheral blood. Vet. Immunol. Immunop 149: 272–279. [DOI] [PubMed] [Google Scholar]
- 25.Artiaga BL, Whitener RL, Staples CR, and Driver JP. 2014. Adjuvant effects of therapeutic glycolipids administered to a cohort of NKT cell-diverse pigs. Vet. Immunol. Immunop 162: 1–13. [DOI] [PubMed] [Google Scholar]
- 26.Eguchi-Ogawa T, Morozumi T, Tanaka M, Shinkai H, Okumura N, Suzuki K, Awata T, and Uenishi H. 2007. Analysis of the genomic structure of the porcine CD1 gene cluster. Genomics 89: 248–261. [DOI] [PubMed] [Google Scholar]
- 27.Fox LM, Miksanek J, May NA, Scharf L, Lockridge JL, Veerapen N, Besra GS, Adams EJ, Hudson AW, and Gumperz JE. 2013. Expression of CD1c enhances human invariant NKT cell activation by α-GalCer. Cancer. Immun 13: 9. [PMC free article] [PubMed] [Google Scholar]
- 28.Park SH, Roark JH, and Bendelac A. 1998. Tissue-specific recognition of mouse CD1 molecules. J. Immunol 160: 3128–3134. [PubMed] [Google Scholar]
- 29.Hirano M, Guo P, McCurley N, Schorpp M, Das S, Boehm T, and Cooper MD. 2013. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature 501: 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler JE, Wertz N, Sun J, and Sacco RE. 2005. Comparison of the expressed porcine Vbeta and Jbeta repertoire of thymocytes and peripheral T cells. Immunology 114: 184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, and Cowtan K (2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 32.Yang G, Artiaga BL, Hackmann TJ, Samuel MS, Walters EM, Salek-Ardakani S, and Driver JP. 2015. Targeted disruption of CD1d prevents NKT cell development in pigs. Mamm. Genome 26: 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dwivedi V, Manickam C, Dhakal S, Binjawadagi B, Ouyang K, Hiremath J, Khatri M, Hague JG, Lee CW, and Renukaradhya GJ. 2016. Adjuvant effects of invariant NKT cell ligand potentiates the innate and adaptive immunity to an inactivated H1N1 swine influenza virus vaccine in pigs. Vet. Microbiol 186: 157–163. [DOI] [PubMed] [Google Scholar]
- 34.Artiaga BL, Yang G, Hackmann TJ, Liu Q, Richt JA, Salek-Ardakani S, Castleman WL, Lednicky JA, and Driver JP. 2016. alpha-Galactosylceramide protects swine against influenza infection when administered as a vaccine adjuvant. Sci. Rep 6: 23593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Artiaga BL, Yang G, Hutchinson TE, Loeb JC, Richt JA, Lednicky JA, Salek-Ardakani S, and Driver JP. 2016. Rapid control of pandemic H1N1 influenza by targeting NKT-cells. Sci. Rep 6: 37999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto R, Uenishi H, Hatsuse H, Sato E, Awata T, Yasue H, and Takagaki Y. 2005. TRAV gene usage in pig T-cell receptor alpha cDNA. Immunogenetics 57: 219–225. [DOI] [PubMed] [Google Scholar]
- 37.Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, Bendelac A, Bonneville M, and Lantz O. 1999. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J. Exp. Med 189: 1907–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lefranc MP 2003. IMGT databases, web resources and tools for immunoglobulin and T cell receptor sequence analysis, http://imgt.cines.fr. Leukemia 17: 260–266. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto R, Uenishi H, Hatsuse H, Sato E, Awata T, Yasue H, and Takagaki Y. 2005. Jalpha-gene segment usage and the CDR3 diversity of porcine TCRalpha-chain cDNA clones from the PBL of a five-month-old pig and the thymus of a one-month-old pig. Mol. Immunol 42: 1375–1383. [DOI] [PubMed] [Google Scholar]
- 40.Eguchi-Ogawa T, Toki D, and Uenishi H. 2009. Genomic structure of the whole D-J-C clusters and the upstream region coding V segments of the TRB locus in pig. Dev. Comp. Immunol 33: 1111–1119. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe M, Iwasaki Y, Mita Y, Ota S, Yamada S, Shimizu M, and Takagaki Y. 2007. Porcine T-cell receptor beta-chain: a genomic sequence covering Dbeta1.1 to Cbeta2 gene segments and the diversity of cDNA expressed in piglets including novel alternative splicing products. Mol. Immunol 44: 2332–2343. [DOI] [PubMed] [Google Scholar]
- 42.Uldrich AP, Patel O, Cameron G, Pellicci DG, Day EB, Sullivan LC, Kyparissoudis K, Kjer-Nielsen L, Vivian JP, Cao B, Brooks AG, Williams SJ, Illarionov P, Besra GS, Turner SJ, Porcelli SA, McCluskey J, Smyth MJ, Rossjohn J, and Godfrey DI. 2011. A semi-invariant Vα10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties. Nat. Immunol 12: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiba A, Cohen N, Brigl M, Brennan PJ, Besra GS, and Brenner MB. 2009. Rapid and reliable generation of invariant natural killer T-cell lines in vitro. Immunology 128: 324–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burdin N, Brossay L, Degano M, Iijima H, Gui M, Wilson IA, and Kronenberg M. 2000. Structural requirements for antigen presentation by mouse CD1. Proc. Natl. Acad. Sci. USA 97: 10156–10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamada N, Iijima H, Kimura K, Harada M, Shimizu E, Motohashi S, Kawano T, Shinkai H, Nakayama T, Sakai T, Brossay L, Kronenberg M, and Taniguchi M. 2001. Crucial amino acid residues of mouse CD1d for glycolipid ligand presentation to V(alpha)14 NKT cells. Int. Immunol 13: 853–861. [DOI] [PubMed] [Google Scholar]
- 46.Sidobre S, Naidenko OV, Sim BC, Gascoigne NR, Garcia KC, and Kronenberg M. 2002. The V alpha 14 NKT cell TCR exhibits high-affinity binding to a glycolipid/CD1d complex. J. Immunol 169: 1340–1348. [DOI] [PubMed] [Google Scholar]
- 47.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, and Rossjohn J. 2007. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448: 44–49. [DOI] [PubMed] [Google Scholar]
- 48.Koch M, Stronge VS, Shepherd D, Gadola SD, Mathew B, Ritter G, Fersht AR, Besra GS, Schmidt RR, Jones EY, and Cerundolo V. 2005. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat. Immunol 6: 819–826. [DOI] [PubMed] [Google Scholar]
- 49.Looringh van Beeck FA, Reinink P, Hermsen R, Zajonc DM, Laven MJ, Fun A, Troskie M, Schoemaker NJ, Morar D, Lenstra JA, Vervelde L, Rutten VP, van Eden W, and Van Rhijn I. 2009. Functional CD1d and/or NKT cell invariant chain transcript in horse, pig, African elephant and guinea pig, but not in ruminants. Mol. Immunol 46: 1424–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang G, Richt JA, and Driver JP. 2017. Harnessing Invariant NKT Cells to Improve Influenza Vaccines: A Pig Perspective. Int. J. Mol. Sci 19: 10.3390/ijms19010068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makino Y, Yamagata N, Sasho T, Adachi Y, Kanno R, Koseki H, Kanno M, and Taniguchi M. 1993. Extrathymic development of V alpha 14-positive T cells. J. Exp. Med 177: 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makino Y, Koseki H, Adachi Y, Akasaka T, Tsuchida K, and Taniguchi M. 1994. Extrathymic differentiation of a T cell bearing invariant V alpha 14J alpha 281 TCR. Int. Rev. Immunol 11: 31–46. [DOI] [PubMed] [Google Scholar]
- 53.Kinebuchi M, and Matsuura A. 2004. Rat T-cell receptor TRAV11 (Valpha14) genes: further evidence of extensive multiplicity with homogeneous CDR1 and diversified CDR2 by genomic contig and cDNA analysis. Immunogenetics 55: 756–762. [DOI] [PubMed] [Google Scholar]
- 54.Fichtner AS, Paletta D, Starick L, Schumann RF, Niewiesk S, and Herrmann T. 2015. Function and expression of CD1d and invariant natural killer T-cell receptor in the cotton rat (Sigmodon hispidus). Immunology 146: 618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wernersson R, Schierup MH, Jorgensen FG, Gorodkin J, Panitz F, Staerfeldt HH, Christensen OF, Mailund T, Hornshoj H, Klein A, Wang J, Liu B, Hu S, Dong W, Li W, Wong GK, Yu J, Wang J, Bendixen C, Fredholm M, Brunak S, Yang H, and Bolund L. 2005. Pigs in sequence space: a 0.66X coverage pig genome survey based on shotgun sequencing. BMC. Genomics 6: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Sullivan LC, Kyparissoudis K, Brooks AG, Reid HH, Gras S, Lucet IS, Koh R, Smyth MJ, Mallevaey T, Matsuda JL, Gapin L, McCluskey J, Godfrey DI, and Rossjohn J. 2009. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity 31: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joyce S, Girardi E, and Zajonc DM. 2011. NKT cell ligand recognition logic: molecular basis for a synaptic duet and transmission of inflammatory effectors. J. Immunol 187: 1081–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Birkholz A, Nemcovic M, Yu ED, Girardi E, Wang J, Khurana A, Pauwels N, Farber E, Chitale S, Franck RW, Tsuji M, Howell A, Van Calenbergh S, Kronenberg M, and Zajonc DM. 2015. Lipid and Carbohydrate Modifications of alpha-Galactosylceramide Differently Influence Mouse and Human Type I Natural Killer T Cell Activation. J. Biol. Chem 290: 17206–17217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baron C, Sachs DH, and LeGuern C. 2001. A particular TCR beta variable region used by T cells infiltrating kidney transplants. J. Immunol 166: 2589–2596. [DOI] [PubMed] [Google Scholar]
- 60.Glusman G, Rowen L, Lee I, Boysen C, Roach JC, Smit AF, Wang K, Koop BF, and Hood L. 2001. Comparative genomics of the human and mouse T cell receptor loci. Immunity 15: 337–349. [DOI] [PubMed] [Google Scholar]
- 61.Davodeau F, Difilippantonio M, Roldan E, Malissen M, Casanova JL, Couedel C, Morcet JF, Merkenschlager M, Nussenzweig A, Bonneville M, and Malissen B. 2001. The tight interallelic positional coincidence that distinguishes T-cell receptor Jalpha usage does not result from homologous chromosomal pairing during ValphaJalpha rearrangement. EMBO. J 20: 4717–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venturi V, Price DA, Douek DC, and Davenport MP. 2008. The molecular basis for public T-cell responses? Nat. Rev. Immunol 8: 231–238. [DOI] [PubMed] [Google Scholar]
- 63.Robins HS, Srivastava SK, Campregher PV, Turtle CJ, Andriesen J, Riddell SR, Carlson CS, and Warren EH. 2010. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci. Transl. Med 2: 47ra64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emerson RO, DeWitt WS, Vignali M, Gravley J, Hu JK, Osborne EJ, Desmarais C, Klinger M, Carlson CS, Hansen JA, Rieder M, and Robins HS. 2017. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat. Genet 49: 659–665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.