Abstract
As peripheral blood contains fluctuated levels of circulating cell-free mitochondrial DNA (ccf mtDNA), we aimed to evaluate ccf mtDNA as a biomarker for diagnosis and prognosis of epithelial ovarian cancer (EOC). In the present study, we recruited 165 EOC patients and 60 healthy women. Quantitative RT-PCR was applied to amplify 79-bp and 230-bp fragments of the mitochondrial 16 s RNA gene in sera of these participants. MtDNA integrity was defined as the ratio of long to short mtDNA fragments. We observed that the levels of mtDNA79 and mtDNA230 were significantly increased (P = .0001), whereas the mtDNA integrity (P = .0001) was decreased in EOC patients compared with those in healthy controls. MtDNA79 showed a sensitivity of 90.3% and a specificity of 81.7% (AUC = 0.900) to discriminate EOC from healthy controls. Moreover, the amounts of mtDNA79 (P = .0001, P = .012, P = .039) and mtDNA230 (P = .0001, P = .042) continuously raised from healthy controls over FIGO I-II to FIGO III and IV, with highest levels of mtDNA79 (P = .0001) and mtDNA230 (P = .0001) in FIGO III and IV. Increasing levels of mtDNA79 (P = .003, P = .0001) and mtDNA230 (P = .041, P = .0001) were also associated with lymph node metastasis and CA125 values. The higher levels of mtDNA79 (P = .0001; HR 3.2, 95%CI:1.6–6.3) and mtDNA230 (borderline P = .048, HR 0.9, 95%CI:0.9–1.0) also correlated with poor patients' overall survival, of which mtDNA79 could act as an independent factor for overall survival. Our data show a significant association of increasing levels of ccf mtDNA with EOC progress and poor prognosis.
Introduction
Epithelial ovarian cancer (EOC) is the second most malignant cancer in women. The high mortality of EOC patients is mainly owing to the first diagnosis at advanced stages [1]. Therefore, circulating nucleic-acid based biomarkers are emerging approaches in the exploration of noninvasive assays for an improved screening of EOC and other cancer types. Regarding studies on circulating DNA, previous publications mainly focused on the detection of circulating genomic DNA in cancer patients, but the potential of mitochondrial DNA (mtDNA) as diagnostic and prognostic molecular markers have also been reported [2].
MtDNA exists up to even thousands of copies in the energy-producing center of mitochondria. It is a uniquely circular, double-stranded DNA molecule, which contains a significant amount of unmethylated DNA. MtDNA causes inflammatory processes via activating pattern recognition receptor [3], [4]. Besides, mtDNA may crosstalk directly with activated leukocytes and be involved in anti-microbial activities [3]. Upon its release under mechanical, chemical or oxidative stress into various body fluids, alterations of the circulating cell-free (ccf) mtDNA content have been shown to correlate with a broad range of clinical conditions, including trauma severity [5], HIV infection and inflammation [6], cellular damage induced by exposure to chronic low-dose radiation [7], exposure to toxic/carcinogenic chemicals [8], aging [9], Alzheimer's [10] and Parkinson's [11] disease, multiple sclerosis [12] and cardiovascular disease [13]. A change in the amount of ccf mtDNA has also been reported in a variety of cancer types, such as head and neck cancer [14], prostate cancer [15], lung cancer [16], clear cell renal cell carcinoma [17]. In EOC patients, the levels of ccf mtDNA in plasma were reported to be elevated compared with healthy women and patients with benign ovarian diseases, indicating its diagnostic value [18]. Moreover, the predictive relevance of ccf mtDNA was also reported. Its levels significantly decreased in plasma of EOC patients after 6 cycles of chemotherapy [19].
The present study is designed to determine whether the ccf mtDNA content and its integrity could provide valuable information for the early detection and prognosis of EOC. We amplified 79 bp and 230 bp fragments of the mitochondrial 16 s RNA gene in the sera of two populations of 75 Chinese and 90 German EOC patients, and compared their levels with 60 healthy women. The short fragments (<200 bp) and the long fragments (>200 bp) are supposed to represent the mtDNA cleaved by apoptosis or degraded by macrophages and mostly mtDNA from necrosis, respectively [8]. MtDNA integrity was defined as the ratio of long to short mtDNA fragments. In the current study, our measurements showed a significant association of ccf mtDNA levels with progress and prognosis of EOC.
Materials and Methods
Patients and Healthy Donors
The present study included 165 randomly chosen serum samples from EOC patients, of whom 90 patients were treated at the Department of Gynecology, the University Medical Center Hamburg Eppendorf (Hamburg, Germany), and 75 patients were from the Department of Gynecology, the Affiliated Hospital of Medical School of Ningbo University (Ningbo, China), for histologically confirmed International Federation of Gynecology and Obstetrics (FIGO) stages I-IV. Serum samples of EOC patients from Germany and China were collected directly before surgery from February 2010 to April 2013 and from January 2011 to May 2017, respectively, and were stored in nuclease-free tubes at −80 °C. Median ages of EOC patients from Germany and China were 63 (range 30–89) and 62 (range 25–84) years, respectively. Follow-up information was available for 146 patients, of whom 19 Chinese and 26 German patients were dead. The median follow-up was 18 (range 1–77) months and 14 (rang 1–38) months for the German and Chinese patient cohorts, respectively. Table 1 shows the clinicopathological variables of German and Chinese EOC patients. In addition, the serum samples of 60 age-matched healthy women (median age of 60, range 37–78) with no history of any cancer and in good health based on the results of wellness testing in 2017 were selected and obtained from the Physical Examination Center, the Affiliated Hospital of Medical School of Ningbo University (Ningbo, China). All recruited patients and healthy women gave written informed consent to access their blood samples. Blood collection and experiments were performed in compliance with the Helsinki Declaration, and were approved by the ethics committee (Ethik-Kommission der Ärztekammer Hamburg, Hamburg) and the Clinical Research Ethics Committee of Medical School of Ningbo University.
Table 1.
The characteristics of patients with EOC.
| EOC patients | Chinese |
Germany |
Total |
|---|---|---|---|
| 75 (100%) | 90 (100%) | 165 (100%) | |
| Age (median) | 62 (25-84 years) | 63 (30-89 years) | 62 (25-89 years) |
| Follow-up time (median) | 14 (1-38 months) | 18 (1-77 months) | 16 (1-77 months) |
| Histology | |||
| Serious | 41 (54.7%) | 74 (82.3%) | 115 (69.7%) |
| Endometrial | 9 (12.0%) | 10 (11.1%) | 19 (11.5%) |
| Other subtypes | 11 (14.7%) | 3 (3.3%) | 14 (8.5%) |
| Unknown | 14 (18.6%) | 3 (3.3%) | 17 (10.3%) |
| FIGO stage | |||
| I | 12 (16.0%) | 18 (20.0%) | 30 (18.2%) |
| II | 15 (20.0%) | 5 (5.5%) | 20 (12.1%) |
| III | 27 (36.0%) | 51 (56.7%) | 78 (47.3%) |
| IV | 21 (28.0%) | 15 (16.7%) | 36 (21.8%) |
| Unknown | 0 (0.0%) | 1 (1.1%) | 1 (0.6%) |
| Lymph node status | |||
| N0 | 30 (40.0%) | 27 (30.0%) | 57 (34.5%) |
| N1 | 36 (48.0%) | 45 (50.0%) | 81 (49.1%) |
| Unknown | 9 (12.0%) | 18 (20.0%) | 27 (16.4%) |
| Grading | |||
| G1-2 | 18 (24.0%) | 29 (32.2%) | 47 (28.5%) |
| G3 | 37 (49.3%) | 58 (64.4%) | 95 (57.6%) |
| Unknown | 20 (26.7%) | 3 (3.4%) | 23 (13.9%) |
| Postoperative tumor residue | |||
| Tumor free | 45 (60.0%) | 59 (65.5%) | 104 (63.0%) |
| < 1 cm | 25 (33.3%) | 15 (16.7%) | 40 (24.2%) |
| > 1 cm | 5 (6.7%) | 9 (10.0%) | 14 (8.5%) |
| Unknown | 0 (0.0%) | 7 (7.8%) | 7 (4.3%) |
| Survival status | |||
| Dead | 19 (25.3%) | 26 (28.9%) | 45 (27.3%) |
| Alive | 47 (62.7%) | 54 (60.0%) | 101 (61.2%) |
| Unknown | 9 (12.0%) | 10 (11.1%) | 19 (11.5%) |
| Recurrent status | |||
| Yes | 2 (2.7%) | 37 (41.1%) | 39 (23.6%) |
| No | 3 (4.0%) | 47 (52.2%) | 50 (30.3%) |
| Unknown | 70 (93.3%) | 6 (6.7%) | 76 (46.1%) |
| CA125 (U/mL) | |||
| <65 | 17 (22.7%) | 11 (12.2%) | 28 (17.0%) |
| ≥65 | 58 (77.3%) | 65 (72.2%) | 123 (74.5%) |
| Unknown | 0 (0.0%) | 14 (15.6%) | 14 (8.5%) |
DNA Extraction
Total circulating DNA was extracted from 250 μL serum using the NucleoSpin DNA Plasma/Serum Kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions. The concentrations of extracted DNA were measured on a NanoDrop-1000 Spectrophotometer (Thermo Scientific, Waltham, MA USA) before the storage under −20 °C.
Quantitative Real-Time PCR
Two primer sets specific for the mitochondrial ribosomal 16 s-RNA were designed as described in the previous study [8]. The primer pair, mtDNA79 amplified a 79-bp DNA fragment and the primer pair, mtDNA230 amplified a 230-bp DNA fragment. The sequence of the forward primer for both, mtDNA79 and mtDNA230 was 5′-AGCCGCTATTAAAGGTTCG-3′. The sequences of the reverse primers specific for mtDNA79 and mtDNA230 were 5′-CCTGGATTACTCCGGTCTGA-3′ and 5′-GGGCTCTGCCATCTTAACAA-3′, respectively. The degree of the mitochondrial fragmentation was defined as the mtDNA integrity, which was calculated by the ratio of the relative amounts of mtDNA230 to mtDNA79. For the normalization of real-time PCR data, 3 house-keeping genes, including GAPDH DNA, 36B4 DNA and β-globin DNA were amplified in the pre-experiment to test their variations in the serum samples of 25 healthy controls and 25 EOC patients. After comparing the standard deviations of Cq values of GAPDH DNA, 36B4 DNA and β-globin between 25 healthy controls and 25 EOC patients, we chose GAPDH DNA with the forward primer 5′-CCCCACACACATGCACTTACC-3′ and the reverse primer 5′-CCTAGTCCCAGGGCTTTGATT-3′ to normalize the PCR data in our study based on the smallest standard deviation. In the serum samples of German and Chinese EOC patients as well as healthy women, we calculated a mean Cq value of 26.03 (s.d. = 1.88), 25.59 (s.d. = 2.13) and 25.82 (s.d. = 1.76), respectively, for GAPDH. Real-time PCR was performed in triplicate on a 7500 Real-Time PCR System (Applied Biosystems, USA). Each 10 μL reaction solution consisted of 1 μL DNA, 5 μL SYBR Green buffer (Invitrogen, Scotland) and 0.2 μL forward/reverse primer. The PCR conditions were 95 °C for 15 min, followed by 40 cycles at 95 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s. A melting curve analysis of 95 °C for 15 s, 60 °C for 15 s and 95 °C for 15 s at the end of each run was applied to confirm the specificity of the PCR products.
Data Normalization and Statistical Analyses
The relative amounts of mtDNA fragments were calculated and evaluated by the ΔCq method as follows: ΔCq = mean value Cq (GAPDH) – mean value Cq (mtDNA79/mtDNA230), and the relative mtDNA amounts corresponded to the value of 2(ΔCq).
The statistical analyses were performed using the SPSS software package, version 22.0 (SPSS Inc., Chicago, IL, USA). Assuming nonparametric distribution of the data, univariate analyses of the Mann Whitney U test of two independent variables (e.g. healthy controls vs. EOC) were used. For the comparison of >2 independent variables (e.g. healthy controls vs. G1–2 vs. G3, healthy controls vs. FIGO I-II vs. FIGO III vs. FIGO IV, healthy controls vs. N0 vs. N1, healthy controls vs. tumor free vs. tumor residue), relative expression data were firstly Ln transformed in order to obtain normally distributed data, and then statistical differences were evaluated using ANOVA with Tukey's HSD test for all pairwise comparisons that correct for experiment-wise error rate. In order to estimate the optimal sensitivity and specificity of the mtDNA fragments to discriminate two groups of different clinical pathological parameters, receiver operator characteristic (ROC) analysis was carried out. For the correlation analyses, bivariate analyses of the Spearman-Rho test were applied considering the nonparametric distribution of the data. Univariate and multivariate analyses were performed for prognostic factors of patients' overall survival and recurrence-free survival using the Cox regression model. Kaplan-Meier plots were drawn to estimate patients' overall survival, and the log-rank test was applied. Missing data were handled by pairwise deletion. P < .05 was considered statistically significant. All P values are two sided.
Results
Quantification of Serum mtDNA79 and mtDNA230 Fragments
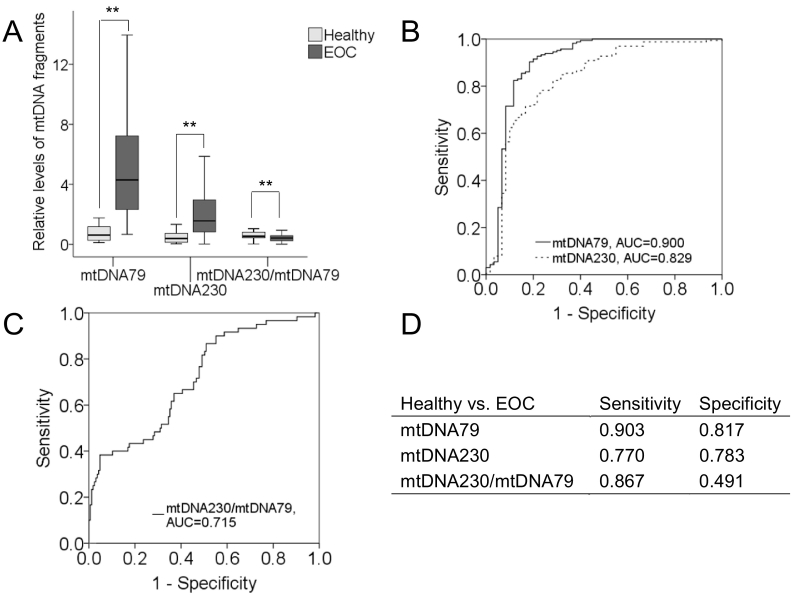
The amount of total DNA and the relative levels of the ccf mtDNA fragments (mtDNA79, mtDNA230) were quantified in the serum samples of 90 German and 75 Chinese patients with EOC, and 60 age-matched healthy women by a NanoDrop Spectrophotometer and SYBR qRT-PCR, respectively. The amplification and melting curves of GAPDH, mtDNA79 and mtDNA230 products by qPCR are shown in Supplementary Figure S1. The levels of total DNA (P = .436), mtDNA79 (P = .377), mtDNA230 (P = .842) and integrity (P = .187) showed no statistically significant difference between German and Chinese EOC patients (Supplementary Figure S2). In addition, no difference of serum DNA amounts was observed between healthy controls and EOC patients (Supplementary Figure S3). However, the serum levels of ccf mtDNA79 (P = .0001) and mtDNA230 (P = .0001) were significantly higher, whereas the levels of the mtDNA integrity (mtDNA230/mtDNA79, P = .0001) were strikingly lower in EOC patients than those in healthy women (Figure 1A). AUC (area under the curve) values of the mtDNA79 and mtDNA230 fragments to differentiate EOC patients from healthy controls were 0.900 and 0.829, respectively (Figure 1B), while that of integrity was 0.715 (Figure 1C), showing the diagnostic power of mtDNA79, mtDNA230 and the mtDNA integrity to discriminate EOC patients from healthy women. Sensitivities and specificities of the mtDNA fragments and the mtDNA integrity were determined by the highest Youden index (sensitivity+specificity-1), whereby ccf mtDNA79 could best discriminate between EOC patients and healthy controls with a sensitivity of 90.3% and a specificity of 81.7% (Figure 1D).
Figure 1.
Quantification of mtDNA79, mtDNA230 and the mtDNA integrity in the serum of patients with EOC and healthy women. (A) The relative amounts of the mtDNA79 and mtDNA230 fragments were quantified by qRT-PCR. The mtDNA integrity refers to the ratio of mtDNA230 to mtDNA79. The box plots compare the relative levels of mtDNA79, mtDNA230 and the mtDNA integrity in the serum of healthy women (n = 60) and patients with EOC (n = 165). ROC analyses show the profiles of sensitivities and specificities of (B) mtDNA79, mtDNA230 and (C) the mtDNA integrity to distinguish healthy control group from EOC cohort. (D) Summarization of sensitivities and specificities of mtDNA79, mtDNA230 and the mtDNA integrity. **P < .01.
Correlation of the ccf mtDNA Fragments and Integrity with Clinicopathological Parameters
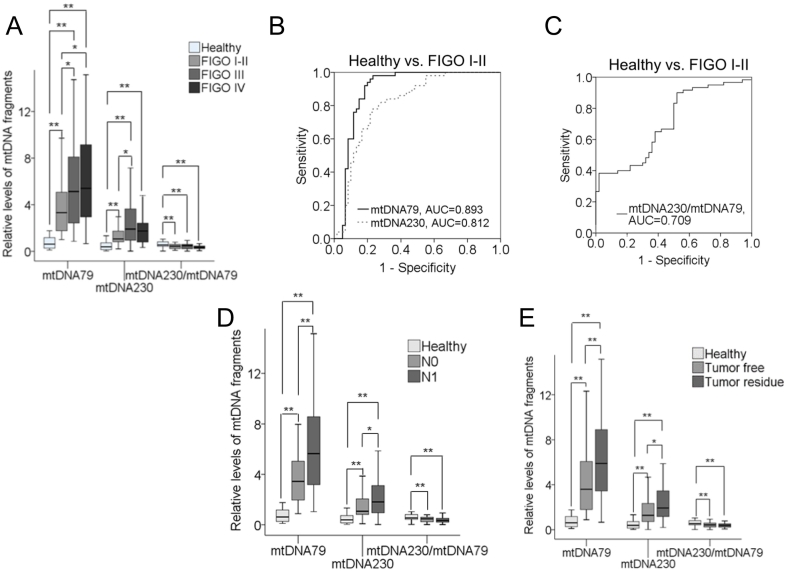
The above findings provoked us to evaluate whether the changes of the levels of mtDNA79, mtDNA230 and the mtDNA integrity correlate with the clinicopathological features, including grading, FIGO stages, lymph node status and postoperative tumor residue. Supplementary Table S1 summarizes the p-values of the mtDNA variables between different patient subgroups and the healthy control group. High levels of ccf mtDNA79 (P = .0001, P = .0001) and ccf mtDNA230 (P = .0001, P = .0001) as well as a low mtDNA integrity (P = .003, P = .001) significantly correlated with lower (G1–2) and higher (G3) grading compared with healthy controls, but no difference of the levels of mtDNA79, mtDNA230 and the mtDNA integrity could be observed between these subgroups (Supplementary Table S1). In respect to the FIGO stages, as shown in Figure 2A, there were significant continuous increases in the levels of mtDNA79 (P = .0001, P = .012, P = .039) and mtDNA230 (P = .0001, P = .042) from healthy women over the patients with FIGO stage I-II to the patients with FIGO stage III and IV, suggesting a significant association of the levels of mtDNA79 and mtDNA230 with tumor progression. Highest levels of mtDNA79 (P = .0001) and mtDNA230 (P = .0001) could be detected in FIGO III and IV stages suggesting a correlation with an increasing metastatic potential, which involves peritoneal and distant metastases. Intriguingly, the levels of mtDNA79, mtDNA230 and the integrity could discriminate healthy women from patients with FIGO I-II with the AUCs of 0.893, 0.812 and 0.709, respectively (Figure 2, B and C). These findings are also supported by the significantly higher levels of ccf mtDNA79 in lymph node-positive EOC than in lymph node-negative EOC (P = .003). The levels of ccf mtDNA230 could also differentiate between the two lymph node statuses, but only with a borderline significance (P = .041). As expected, the levels of the mtDNA integrity (P = .002, P = .0001) decreased from healthy controls to lymph node-negative and -positive EOC (Figure 2D). Additionally, a close examination was also applied with the parameter of postoperative tumor residue. Interestingly, the levels of ccf mtDNA79 (P = .001) and mtDNA230 (P = .013) were significantly higher in EOC patients with a tumor residue than in patients without a tumor residue (Figure 2E).
Figure 2.
Associations of mtDNA79, mtDNA230 and the mtDNA integrity with FIGO stages, lymph node status and postoperative tumor residue. (A) The box plots compare the levels of mtDNA79, mtDNA230 and the mtDNA integrity in healthy women (n = 60) and the patients with FIGO I-II (n = 50), III (n = 78) and IV (n = 36). ROC analyses show the profiles of sensitivities and specificities of (B) mtDNA79, mtDNA230 and (C) the mtDNA integrity to distinguish healthy control group from patients with FIGO I-II. (D) The box plots compare the levels of mtDNA79, mtDNA230 and the mtDNA integrity in the serum of healthy women (n = 60), lymph-node negative patients (N0, n = 57) and lymph-node positive patients (N1, n = 81). (E) The box plots compare the levels of mtDNA79, mtDNA230 and the mtDNA integrity in the serum of healthy women (n = 60), EOC patients without (n = 104) and with (n = 54) tumor residue after operation. *P < .05, **P < .01.
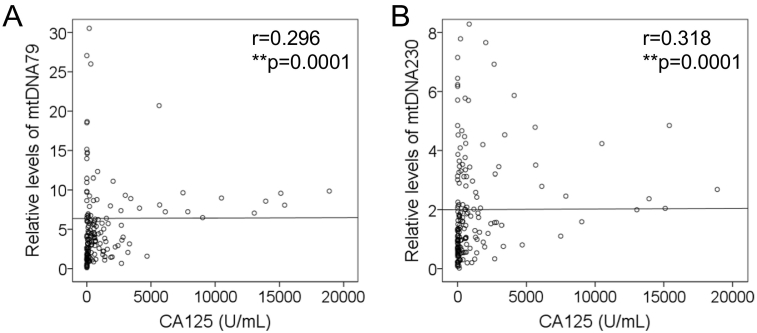
Serum assessment of the tumor marker CA125 (carbohydrate antigen 125) is currently the standard of care to perform diagnosis, to follow response to treatment, and to predict prognosis of EOC patients. In order to analyze whether the levels of mtDNA79 and mtDNA230 are associated with the values of CA125, correlation analyses of the Spearman-Rho test were performed. As CA125 values were not available in the cohort of healthy women, we could only carry out this statistical evaluation with the data derived from the serum samples of EOC patients. As shown in Figure 3, A and B, there were positive correlations of mtDNA79 (r = 0.296, P = .0001) and mtDNA230 (r = 0.318, P = .0001) with the values of CA125.
Figure 3.
Correlations of mtDNA79 and mtDNA230 with tumor marker CA125. The scatter plots show the positive correlations of the relative amounts of (A) mtDNA79 and (B) mtDNA230 with the CA125 values of patients with EOC (n = 151). **P < .01.
Prognostic Value of Increased Levels of the mtDNA Fragments and mtDNA Integrity
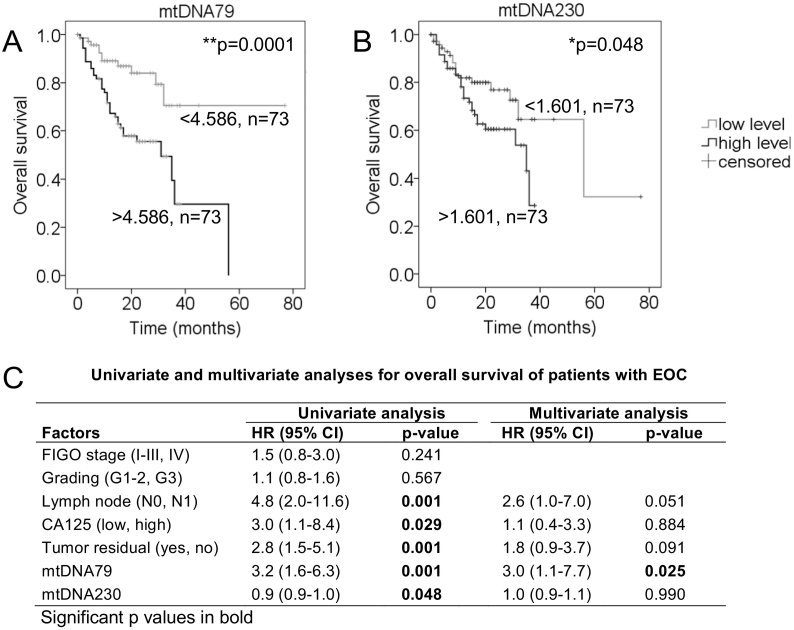
In order to evaluate the prognostic potential of mtDNA fragments and integrity, Kaplan-Meier and log-rank models were carried out in EOC patients. The median follow-up time was 16 (range 1–77) months, while the median patients' overall and recurrence-free survival times were 16 and 14 months, respectively. Median values of mtDNA79, mtDNA230 and the mtDNA integrity were used to group the EOC samples according to low and high mtDNA levels. High levels of mtDNA79 (P = .0001, n = 73, Figure 4A) and mtDNA230 (P = .048, n = 73, Figure 4B) statistically correlated with poor patients' overall survival. Further univariate analyses with the Cox proportional hazards showed that besides mtDNA79 (HR: 3.2, 95%CI: 1.6–6.3) and mtDNA230 (HR: 0.9, 95%CI: 0.9–1.0), lymph node status (HR: 4.8, 95%CI: 2.0–11.6), CA125 (HR: 3.0, 95%CI: 1.1–8.4), tumor residue (HR:2.8, 95%CI: 1.5–5.1) were prognostic factors (Figure 4C). Furthermore, closer multivariate Cox analyses with the factors of mtDNA79, mtDNA230, lymph node status, CA125 and tumor residue were carried out. Among these factors, only ccf mtDNA79 acted as an independent predictor of patients' overall survival in EOC patients (HR: 3.0, 95%CI: 1.1–7.7, P = .025, Figure 4C). Again, these findings show the more prominent role of ccf mtDNA79 than mtDNA230 in EOC diagnostics and prognostics. The mtDNA integrity showed no prognostic significance in our analyses (Supplementary Figure S4). Moreover, no correlation of mtDNA79, mtDNA230 and the mtDNA integrity could be observed with recurrence-free survival (data not shown).
Figure 4.
Association of mtDNA79 and mtDNA230 with patients' overall survival in patients with EOC. The median values of mtDNA79 and mtDNA230 were used to group the EOC samples according to low (n = 73) and high (n = 73) mtDNA levels. Univariate Kaplan–Meier curves are related to low and high levels of (A) mtDNA79 and (B) mtDNA230 for patients' overall survival. (C) Clinical pathological factors including FIGO stage, Grading, lymph node status, CA125 values, tumor residue, mtDNA79 and mtDNA230 were enrolled in the uni- and multi- variate analyses for the evaluation of patients' overall survival. The table shows the HR and the corresponding 95% CI of each factor in the uni- and multivariate analyses. *P < .05, **P < .01.
Validation of the Results in High-Grade EOC Patients
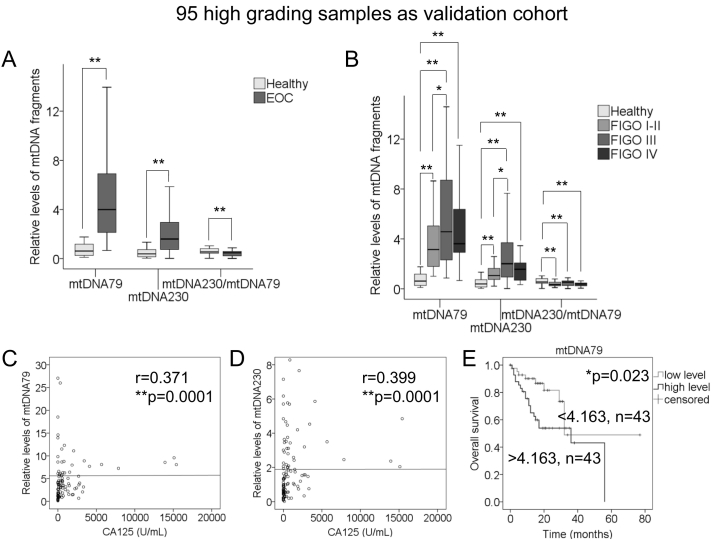
Since high-grade EOC is the most common and serious histological subtype, we further evaluated the mtDNA data in the subgroup of 95 patients with grading 3 (G3). The detailed characteristics of EOC patients with G3 are summarized in Supplementary Table S2. Compared with the above results, we found similar results to those of the whole patient cohort. Consistent with the observations in Figure 1A derived from the whole EOC patient cohort, the relative amounts of mtDNA79 (P = .0001), mtDNA230 (P = .0001) were significantly elevated, and the mtDNA integrity (P = .0001) was decreased in the serum samples of EOC patients with G3 compared with healthy controls (Figure 5A). Moreover, in the G3 group, as shown in Figure 5B, the levels of mtDNA79 and mtDNA230 were higher in the single FIGO I-II, III and IV stages than those in healthy controls, with highest levels of both mtDNA79 (P = .0001) and mtDNA230 (P = .0001) in both FIGO III and IV stages. As expected, the levels of the mtDNA integrity were significantly decreased in the subgroups of FIGO I-II, III and IV compared with healthy women (Supplementary Table S3). These findings on FIGO stages in the G3 group confirmed our results derived from the whole patient cohort. Regarding the lymph node status, the levels of mtDNA79 were increased (P = .001) in the subgroup of lymph node-positive but not -negative EOC, while the levels of mtDNA230 (P = .0001, P = .0001) were increased and the mtDNA integrity (P = .034, P = .0001) decreased in both subgroups of lymph node-negative and -positive stages (Supplementary Table S3). Regarding the tumor residue status after surgery, there were no differences of the 3 variables between the subgroups of tumor free and tumor residue (Supplementary Table S3).
Figure 5.
Validation of the results in the G3 subgroup. (A) The box plots compare the relative levels of mtDNA79, mtDNA230 and the mtDNA integrity in the serum of healthy women (n = 60) and patients with G3 (n = 95). (B) The box plots compare the levels of mtDNA79, mtDNA230 and the mtDNA integrity in healthy women (n = 60) and in the patients with FIGO I-II (n = 25), III (n = 51) and IV (n = 19) confined in the G3 cohort. The scatter plots show the positive correlations of the relative amounts of (C) mtDNA79 and (D) mtDNA230 with the CA125 values of patients with G3 (n = 88). (E) The median values of mtDNA79 were used to group the EOC samples according to low (n = 43) and high (n = 43) mtDNA levels. Univariate Kaplan–Meier curves are related to low and high levels of mtDNA79 for patients' overall survival. *P < .05, **P < .01.
In addition, in line with our findings above on the correlations of mtDNA79 and mtDNA230 with CA125 values in Figure 3, and mtDNA79 with patients' overall survival in Figure 4A, we observed similar significant correlations between mtDNA79 and CA125 values (r = 0.371, P = .0001, Figure 5C), and between mtDNA230 and CA125 values (r = 0.399, P = .0001, Figure 5D), as well as between higher levels of mtDNA79 and poor patients' overall survival (P = .023, Figure 5E). No correlations of mtDNA230 and the mtDNA integrity could be observed with patients' overall survival (P = .052, P = .504, respectively) and recurrence-free survival in the G3 group (data not shown).
Discussion
In the present study, we show that the serum mtDNA content (mtDNA79 and mtDNA230 fragments) was significantly increased (P = .0001, P = .0001) in EOC patients compared with healthy women, of which mtDNA79 distinguished EOC patients from healthy women with a sensitivity of 90.3% and a specificity of 81.7%. Highest levels of both, mtDNA79 and mtDNA230 were associated with advanced pathological features and metastatic potential (peritoneal and distant metastasis), including FIGO III and IV, lymph node invasion and postoperative tumor residue, as well as poor patients' overall survival. The mtDNA integrity was significantly decreased (P = .0001) in EOC cohort compared with healthy controls, indicating particularly an association between an increase in small mtDNA fragments with tumor progression.
Our EOC patient cohort consisted of 2 populations of 90 German and 75 Chinese individuals, in whom the contents of total DNA, mtDNA79, mtDNA230 and the mtDNA integrity showed no differences, indicating a consistent presence of mtDNA in human blood between both races. Based on these data, serum mtDNA was analyzed and compared with the patients' pathological parameters in both populations together. In our investigations, increased mtDNA79 and mtDNA230 levels, as well as the decreased mtDNA integrity in the serum of EOC patients had diagnostic value in discriminating EOC from healthy controls. To date, there are only a few studies dealing with the quantification of ccf mtDNA in EOC patients. In line to our findings, by the quantification of mitochondrial DNA encoded ATPase (MTATP) 8 gene, Zachariah et al. demonstrated that the quantities of plasma mtDNA in EOC were elevated compared with a healthy control group, and had the capacity to discriminate between EOC patients and healthy individuals with a sensitivity of 63% and a specificity of 62% [18]. We show that mtDNA79 was even able to distinguish between EOC patients and healthy controls with a sensitivity of 90% and a specificity of 82%. These authors did not also find an association between their measurements of mtDNA with the patients' pathological parameters. These differences may be explained by that they analyzed a low number of EOC patients, and thus, much smaller patient subgroups. Similar with our findings on the correlation of high mtDNA levels with advanced FIGO stages, through the quantification of mitochondrial cytochrome b gene), Kalavska et al. reported that patients with stage I had significantly lower plasma mtDNA compared to patients with stage II-IV [19]. However, in contrast to our study, which considered early FIGO I-II, FIGO III with peritoneal metastasis and FIGO IV with distant metastasis separately, the study by Kalavska et al. merged FIGO II with FIGO III-IV. Importantly, in uni- and multivariate analyses, we observed correlations of higher levels of mtDNA79 (HR 3.2, 95%CI 1.6–6.3) and mtDNA230 (HR 0.9, 95%CI 0.9–1.0) with poor patients' overall survival, of which mtDNA79 could serve as an independent factor in estimating patients' overall survival, indicating its prognostic value in EOC.
Besides pelvic examination and transvaginal ultrasound, current diagnostic methods for detection and monitoring of EOC mainly include measurement of serum biomarker CA125. However, these methods are not sufficiently specific to diagnose EOC at an early stage, since e.g., the sensitivity of CA125 increases during cancer progression. CA125 values are only elevated in approximately 50% of FIGO I, but 70% to 90% of advanced FIGO stages [20]. In particular, the significant association of increasing levels of mtDNA79 and mtDNA230 with those of CA125 values detected in our study shows the relevance of these ccf mtDNA fragments in advanced FIGO stages. Furthermore, our findings suggest that quantification of the mtDNA79 and mtDNA230 yields in addition to the measurements of CA125 values may contribute to the improvement of standard of care in performing diagnosis, following response to treatment, and predicting prognosis of EOC patients. Amazingly, we also found that the levels of ccf mtDNA79 (P = .001) and mtDNA230 (P = .013) were significantly higher in EOC patients with a postoperative tumor residue than in patients without a tumor residue. Although mtDNA was quantified in preoperative serum samples, this correlation of mtDNA levels with tumor residue after surgery could point to a higher tumor load and heterogeneity.
We also evaluated our observations from the whole patient cohort in the high grading subgroup, the most serious EOC, and found similar data. In the high grading subgroup, highest levels of mtDNA79 and mtDNA230 could also be detected in advanced FIGO stage and were associated with increasing CA125 values. Moreover, higher levels of mtDNA79 were related to poor patients' overall survival, indicating the prognostic relevance of the mtDNA79 fragment. These reproducible findings confirm the increase in mtDNA levels as a risk factor in EOC. Advanced EOC harbored significantly higher mtDNA levels, suggesting an excessive release of mtDNA into the blood circulation in aggressive EOC. This phenomenon might be explained by the fact that in advanced malignancies the abundant cell death of tumor and surrounding tissues may induce more mtDNA shed into the bloodstream.
Regarding the mtDNA integrity, which is determined by the ratio of mtDNA230 to mtDNA79, compared with the healthy controls, EOC patients showed lower levels of the mtDNA integrity. Our findings show that there was more abundant mtDNA79 than mtDNA230 circulating in the serum of EOC patients, implying an active apoptotic phenomenon in EOC. However, the mtDNA integrity could not differentiate between less and more aggressive EOC, and had no relationships with CA125 values and patients' overall survival, indicating that the mtDNA integrity rather reflects the state and source of mtDNA than pathological conditions. So far, the pathophysiological mechanisms leading to the increase in ccf mtDNA in EOC remain largely enigmatic, but it is hypothesized that the excessive release by apoptotic or necrotic cells and the reduced clearance by inflammatory cells during tumor development may contribute to the altered mtDNA content. Additionally, ccf mtDNA not only circulates in a cell-free form, but also in a particle-associated form [21], which protects mtDNA from digestion by DNase.
Although there are increasing publications reporting the possible usefulness of circulating mtDNA content as diagnostic or prognostic factor in other tumor types, such as non-Hodgkin lymphoma [22], lung cancer [16], pancreatic cancer [23], breast cancer [24], prostate cancer [25], hepatocellular carcinoma [26], renal cell carcinoma [27] or glioma [28], prior to the application of ccf mtDNA as a noninvasive biomarker in clinic, large-scaled multicenter studies with long follow-up data are necessary to confirm the clinical relevance of ccf mtDNA.
In summary, in our study, we show that the abnormal high levels of serum mtDNA79 and mtDNA230 are accompanied by advanced EOC, and related to poor patients' overall survival. These results provide an alternative to utilize ccf mtDNA as a noninvasive tool for the diagnosis and prognosis of EOC.
The following are the supplementary data related to this article.
The amplification and melting curves of GAPDH, mtDNA79 and mtDNA230. The figures show the amplification curves of GAPDH (A), mtDNA79 (C) and mtDNA230 (E) fragments. The figures show the melting curves of GAPDH (B), mtDNA79 (D) and mtDNA230 (F) products.
Comparison of total serum DNA concentrations, and the relative levels of mtDNA79, mtDNA230 and the mtDNA integrity in the German and Chinese patients with EOC. The box plots compare (A) the amounts of total serum DNA, and mtDNA79, mtDNA230 and (B) the mtDNA integrity in the serum of German (n=90) and Chinese (n=75) patients with EOC. NS, no significance.
Comparison of total serum DNA concentrations in the serum of patients with EOC and healthy women. The figure shows the amounts of total serum DNA in the serum of patients with EOC (n=165) and healthy women (n=60). NS, no significance.
Relationships of the mtDNA integrity with patients’ overall survival in patients with EOC. The median value of the mtDNA integrity was used to group the EOC samples according to low (n=73) and high (n=73) levels. Univariate Kaplan-Meier curves of the high and low mtDNA integrity levels show no significant difference. NS, no significance.
Comparison of the levels of mtDNA fragments in 165 patients with EOC
The characteristics of 95 high grade EOC patients
Comparison of the levels of mtDNA fragments in 95 high grade EOC patients
Acknowledgements
This study was supported by the research grants from the Natural Science Foundation of Zhejiang (LQ18H200001), the Non-profit Technology Research Program of Zhejiang (LGF18H160006), the Non-profit Technology Research Program of Ningbo (2019C50040), the Natural Science Foundation of Ningbo (2018A610204), the Scientific Innovation Team Project of Ningbo (2017C110019) and the K.C. Wong Magna Fund in Ningbo University.
Contributor Information
Xiaodan Meng, Email: mengxiaodan@nbu.edu.cn.
Zhaohui Gong, Email: zhaohui@ncri.org.cn.
References
- 1.Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014;384:1376–1388. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 2.Tuboly E, McLlroy D, Briggs G, Lott N, Balogh ZJ. Clinical implications and pathological associations of circulating mitochondrial DNA. Front Biosci (Landmark Ed) 2017;22:1011–1022. doi: 10.2741/4530. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, Vandenabeele P. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011;32:157–164. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Lam NY, Rainer TH, Chiu RW, Joynt GM, Lo YM. Plasma mitochondrial DNA concentrations after trauma. Clin Chem. 2004;50:213–216. doi: 10.1373/clinchem.2003.025783. [DOI] [PubMed] [Google Scholar]
- 6.Arshad O, Gadawska I, Sattha B, Cote HCF, Hsieh AYY. Elevated cell-free mitochondrial DNA in filtered plasma Is Associated With HIV Infection and Inflammation. J Acquir Immune Defic Syndr. 2018;78:111–118. doi: 10.1097/QAI.0000000000001650. [DOI] [PubMed] [Google Scholar]
- 7.Borghini A, Mercuri A, Turchi S, Chiesa MR, Piccaluga E, Andreassi MG. Increased circulating cell-free DNA levels and mtDNA fragments in interventional cardiologists occupationally exposed to low levels of ionizing radiation. Environ Mol Mutagen. 2015;56:293–300. doi: 10.1002/em.21917. [DOI] [PubMed] [Google Scholar]
- 8.Budnik LT, Kloth S, Baur X, Preisser AM, Schwarzenbach H. Circulating mitochondrial DNA as biomarker linking environmental chemical exposure to early preclinical lesions elevation of mtDNA in human serum after exposure to carcinogenic halo-alkane-based pesticides. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinti M, Cevenini E, Nasi M, De Biasi S, Salvioli S, Monti D, Benatti S, Gibellini L, Cotichini R, Stazi MA. Circulating mitochondrial DNA increases with age and is a familiar trait: Implications for "inflamm-aging". Eur J Immunol. 2014;44:1552–1562. doi: 10.1002/eji.201343921. [DOI] [PubMed] [Google Scholar]
- 10.Podlesniy P, Figueiro-Silva J, Llado A, Antonell A, Sanchez-Valle R, Alcolea D, Lleo A, Molinuevo JL, Serra N, Trullas R. Low cerebrospinal fluid concentration of mitochondrial DNA in preclinical Alzheimer disease. Ann Neurol. 2013;74:655–668. doi: 10.1002/ana.23955. [DOI] [PubMed] [Google Scholar]
- 11.Pyle A, Brennan R, Kurzawa-Akanbi M, Yarnall A, Thouin A, Mollenhauer B, Burn D, Chinnery PF, Hudson G. Reduced cerebrospinal fluid mitochondrial DNA is a biomarker for early-stage Parkinson's disease. Ann Neurol. 2015;78:1000–1004. doi: 10.1002/ana.24515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowes H, Pyle A, Duddy M, Hudson G. Cell-free mitochondrial DNA in progressive multiple sclerosis. Mitochondrion. 2018;46:307–312. doi: 10.1016/j.mito.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashar FN, Zhang Y, Longchamps RJ, Lane J, Moes A, Grove ML, Mychaleckyj JC, Taylor KD, Coresh J, Rotter JI. Association of mitochondrial DNA copy number with cardiovascular disease. JAMA Cardiol. 2017;2:1247–1255. doi: 10.1001/jamacardio.2017.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang WW, Masayesva B, Zahurak M, Carvalho AL, Rosenbaum E, Mambo E, Zhou S, Minhas K, Benoit N, Westra WH. Increased mitochondrial DNA content in saliva associated with head and neck cancer. Clin Cancer Res. 2005;11:2486–2491. doi: 10.1158/1078-0432.CCR-04-2147. [DOI] [PubMed] [Google Scholar]
- 15.Mehra N, Penning M, Maas J, van Daal N, Giles RH, Voest EE. Circulating mitochondrial nucleic acids have prognostic value for survival in patients with advanced prostate cancer. Clin Cancer Res. 2007;13:421–426. doi: 10.1158/1078-0432.CCR-06-1087. [DOI] [PubMed] [Google Scholar]
- 16.Hosgood HD, 3rd, Liu CS, Rothman N, Weinstein SJ, Bonner MR, Shen M, Lim U, Virtamo J, Cheng WL, Albanes D. Mitochondrial DNA copy number and lung cancer risk in a prospective cohort study. Carcinogenesis. 2010;31:847–849. doi: 10.1093/carcin/bgq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu H, Busch J, Jung M, Rabenhorst S, Ralla B, Kilic E, Mergemeier S, Budach N, Fendler A, Jung K. Diagnostic and prognostic potential of circulating cell-free genomic and mitochondrial DNA fragments in clear cell renal cell carcinoma patients. Clin Chim Acta. 2016;452:109–119. doi: 10.1016/j.cca.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Zachariah RR, Schmid S, Buerki N, Radpour R, Holzgreve W, Zhong X. Levels of circulating cell-free nuclear and mitochondrial DNA in benign and malignant ovarian tumors. Obstet Gynecol. 2008;112:843–850. doi: 10.1097/AOG.0b013e3181867bc0. [DOI] [PubMed] [Google Scholar]
- 19.Kalavska K, Minarik T, Vlkova B, Manasova D, Kubickova M, Jurik A, Mardiak J, Sufliarsky J, Celec P, Mego M. Prognostic value of various subtypes of extracellular DNA in ovarian cancer patients. J Ovarian Res. 2018;11:85. doi: 10.1186/s13048-018-0459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meinhold-Heerlein I, Hauptmann S. The heterogeneity of ovarian cancer. Arch Gynecol Obstet. 2014;289:237–239. doi: 10.1007/s00404-013-3114-3. [DOI] [PubMed] [Google Scholar]
- 21.Chiu RW, Chan LY, Lam NY, Tsui NB, Ng EK, Rainer TH, Lo YM. Quantitative analysis of circulating mitochondrial DNA in plasma. Clin Chem. 2003;49:719–726. doi: 10.1373/49.5.719. [DOI] [PubMed] [Google Scholar]
- 22.Lan Q, Lim U, Liu CS, Weinstein SJ, Chanock S, Bonner MR, Virtamo J, Albanes D, Rothman N. A prospective study of mitochondrial DNA copy number and risk of non-Hodgkin lymphoma. Blood. 2008;112:4247–4249. doi: 10.1182/blood-2008-05-157974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch SM, Weinstein SJ, Virtamo J, Lan Q, Liu CS, Cheng WL, Rothman N, Albanes D, Stolzenberg-Solomon RZ. Mitochondrial DNA copy number and pancreatic cancer in the alpha-tocopherol beta-carotene cancer prevention study. Cancer Prev Res (Phila) 2011;4:1912–1919. doi: 10.1158/1940-6207.CAPR-11-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen J, Platek M, Mahasneh A, Ambrosone CB, Zhao H. Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion. 2010;10:62–68. doi: 10.1016/j.mito.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He WS, Bishop KS. The potential use of cell-free-circulating-tumor DNA as a biomarker for prostate cancer. Expert Rev Mol Diagn. 2016;16:839–852. doi: 10.1080/14737159.2016.1197121. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Hann HW, Wan S, Hann RS, Wang C, Lai Y, Ye X, Evans A, Myers RE, Ye Z. Cell-free circulating mitochondrial DNA content and risk of hepatocellular carcinoma in patients with chronic HBV infection. Sci Rep. 2016;6 doi: 10.1038/srep23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008;100:1104–1112. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Qu Y, Gao K, Yang Q, Shi B, Hou P, Ji M. High copy number of mitochondrial DNA (mtDNA) predicts good prognosis in glioma patients. Am J Cancer Res. 2015;5:1207–1216. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The amplification and melting curves of GAPDH, mtDNA79 and mtDNA230. The figures show the amplification curves of GAPDH (A), mtDNA79 (C) and mtDNA230 (E) fragments. The figures show the melting curves of GAPDH (B), mtDNA79 (D) and mtDNA230 (F) products.
Comparison of total serum DNA concentrations, and the relative levels of mtDNA79, mtDNA230 and the mtDNA integrity in the German and Chinese patients with EOC. The box plots compare (A) the amounts of total serum DNA, and mtDNA79, mtDNA230 and (B) the mtDNA integrity in the serum of German (n=90) and Chinese (n=75) patients with EOC. NS, no significance.
Comparison of total serum DNA concentrations in the serum of patients with EOC and healthy women. The figure shows the amounts of total serum DNA in the serum of patients with EOC (n=165) and healthy women (n=60). NS, no significance.
Relationships of the mtDNA integrity with patients’ overall survival in patients with EOC. The median value of the mtDNA integrity was used to group the EOC samples according to low (n=73) and high (n=73) levels. Univariate Kaplan-Meier curves of the high and low mtDNA integrity levels show no significant difference. NS, no significance.
Comparison of the levels of mtDNA fragments in 165 patients with EOC
The characteristics of 95 high grade EOC patients
Comparison of the levels of mtDNA fragments in 95 high grade EOC patients