Abstract
Type 1 diabetes (T1D) results from the progressive destruction of pancreatic β-cells in a process mediated primarily by T lymphocytes. The T1D research community has made dramatic progress in understanding the genetic basis of the disease as well as in the development of standardized autoantibody assays that inform both disease risk and progression. Despite these advances, there remains a paucity of robust and accepted biomarkers that can effectively inform on the activity of T cells during the natural history of the disease or in response to treatment. In this article, we discuss biomarker development and validation efforts for evaluation of T-cell responses in patients with and at risk for T1D as well as emerging technologies. It is expected that with systematic planning and execution of a well-conceived biomarker development pipeline, T-cell–related biomarkers would rapidly accelerate disease progression monitoring efforts and the evaluation of intervention therapies in T1D.
Introduction
Type 1 diabetes (T1D) is a T-cell–mediated autoimmune disease, wherein both CD4+ and CD8+ T cells are believed to orchestrate the killing of insulin-producing β-cells. These cellular subsets are dynamic during the disease process following interactions with host tissues and innate immune cell subsets and are thought to fluctuate in number, function, and tissue distribution during the pathogenesis of T1D. While multiple immunoregulatory defects contribute to a collective loss of immune tolerance, there remains an outstanding need to monitor T cells during T1D pathogenesis, which thus represents the focus of this work.
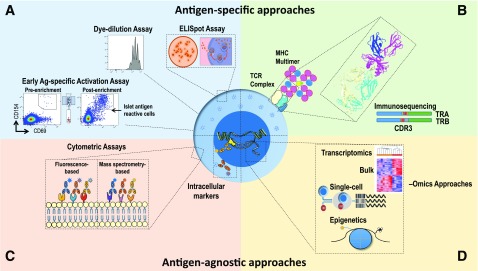
The role of T cells as essential cellular constituents of disease progression has motivated research consortium efforts to develop T-cell biomarkers in T1D, with attention to two broad classes of markers, namely, 1) antigen specific (i.e., captured by assays that measure the number and/or function of T cells specific for β-cell autoantigens) and 2) antigen agnostic (i.e., involving assays that measure T-cell attributes without accounting for the specificity conferred by the T-cell receptor [TCR]) (Fig. 1). In addition, the phenotypes of antigen-specific or antigen-agnostic T cells are only beginning to be fully evaluated with newer technologies, including single-cell approaches that may shed light on the pathophysiological mechanisms underlying the disease. For example, the value of extensive transcriptomic and cellular phenotyping is shown in the CD8+ T-cell exhaustion markers in vasculitis (1), and similar studies are ongoing related to T1D (JDRF Biomarker Working Group and Core for Assay Validation, The Environmental Determinants of Diabetes in the Young [TEDDY], INNODIA, Type 1 Diabetes TrialNet).
Figure 1.
Methods for assessing T-cell biomarkers in T1D. Experimental approaches include assays for assessing both antigen-specific (A and B) and antigen-agnostic features of T cells (C and D). A: Assays for monitoring antigen-specific T-cell activation, proliferation, and cytokine production. B: HLA class I or II multimers loaded with autoantigenic peptides facilitate the detection, phenotyping, and downstream molecular analysis of antigen-specific T cells. Shown is a rendering of the 1E6 TCR recognizing a preproinsulin peptide in the HLA-A*0201 binding groove (85). Immunosequencing of the TRA and TRB genes encoding the V (blue), D-J (red/yellow and gray), and C (green) regions of the TCR-α and TCR-β chains, respectively, facilitates characterization of the TCR reactivity antigen-binding pocket, as determined from the highly polymorphic TRB complementarity-determining region 3 (CDR3; red/yellow) or by complete α/β-chain pairing. C: Flow cytometric approaches employing antibodies conjugated to fluorescent molecules or metals (via mass cytometry) can be used to phenotype a large array of surface and intracellular markers. D: Both bulk- and single-cell technologies facilitate phenotypic, transcriptional, and epigenetic profiling of T cells. Recent advances now facilitate integration of these methodologies at the single-cell resolution, providing high-parameter T-cell biomarkers with molecular resolution.
Despite significant collective efforts to date from investigators and their funding agencies, there remains a need within the scientific community to adequately develop and widely implement validated T-cell biomarkers and fit-for-purpose assays for numerous applications monitoring T1D progression, onset, and response to therapy. The reasons for this deficiency are multifold. First, the detection of antigen-specific autoreactive T cells has been technically challenging because these cells migrate among blood, secondary lymphoid organs, and insulitic lesions, with frequencies in peripheral circulation often below 10 per million T cells (2). Second, autoreactive T cells are often characterized by low-avidity interactions between the islet peptide/HLA complex and TCR, making their isolation or enumeration challenging (3–5). Third, T cells that are reactive to the same β-cell autoantigens may be found in control subjects without diabetes and, therefore, precise definition of their phenotypes becomes essential for understanding their function in the dynamic states preceding overt clinical disease (6). Until recently, the lack of sophisticated technologies had precluded deep analyses of T-cell subsets to identify pathways, networks, and TCR repertoire characteristics that are able to represent meaningful immune alterations for clinical contexts. Finally, there appears to be significant heterogeneity among individuals within T1D that may be driven by complex genetic risk factors, age, and other variables and may affect the progression through disease stages as well as responses to therapies. The heterogeneity is manifest at the tissue level in terms of the frequency and identity of cellular infiltrates in the islets and other histopathological findings from human pancreas tissues from individuals with T1D available through the Network for Pancreatic Organ Donors with Diabetes (nPOD) program and other collections (7).
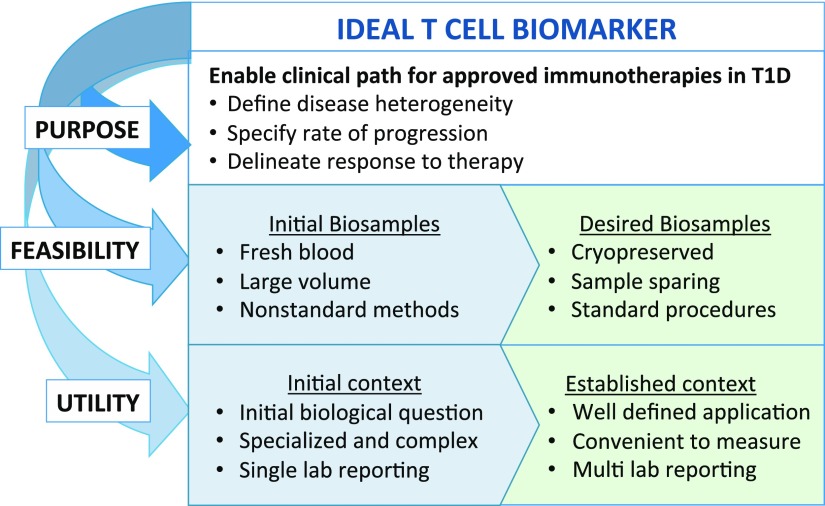
Successful development of T-cell biomarkers requires a multifaceted assessment of their purpose, feasibility, and utility (Fig. 2). T-cell biomarker research is fueled by the need to address unresolved questions in the T1D research community. This includes predicting the rate of disease progression at all stages: from high genetic risk to single-autoantibody positive (pre-stage 1) to development of two or more autoantibodies (stage 1) and then development of dysglycemia (stage 2) and ultimately to clinical onset (stage 3) (8). Biomarkers are also needed to identify subjects for evaluating therapies (stratification markers for use in clinical trials) and for early assessment of response(s) to therapy (pharmacodynamic markers). To achieve feasibility, sample requirement is a serious practical consideration for T-cell biomarkers, particularly in pediatric cohorts. Blood volumes for routine collection are inherently limited, which impacts the capacity to detect rare populations of T cells. While disease processes that directly mediate T1D presumably occur within the pancreas, frozen peripheral blood mononuclear cells (PBMC) are the primary sample type that is available for widespread analysis and clinical trial monitoring. Established sample processing protocols must be applicable to cryopreserved PBMC to accommodate batch processing and analysis in clinical trials with multiple participating sites. Ultimately, in order for T-cell biomarkers to achieve broad utility, the assays by which they are measured must be transitioned from “boutique” status requiring specialized expertise into optimized and validated assays for widespread adoption and clinical application.
Figure 2.
Process considerations for developing informative T-cell biomarkers. Biomarker development begins by defining the purpose of the biomarker and then assessing feasibility and utility. The boxes in blue (left) indicate features of candidate T-cell biomarkers and the assays used to detect them that are in development, whereas green boxes (right) highlight the key features required of validated T-cell biomarkers and associated assays. Feasibility includes considerations for sample-sparing assays utilizing cryopreserved biobanked samples.
There are some useful roadmaps we can use in this process. Notably, preceding the era of T-cell assay optimization, robust immunologic methodologies were successfully developed and established for the detection of autoantibodies (9). This process started with reproducibility testing of the assay/methodology and transitioned into multisite validation and implementation. In the sections that follow, we discuss the current landscape of candidate T-cell biomarkers and propose a systematic pipeline to serve as a guide by which such biomarkers may be further developed.
Candidate T-Cell Biomarkers in T1D
We define a candidate biomarker as a readout of an optimized assay that has been replicated in more than one laboratory. A summary of candidate T-cell biomarkers is presented in Tables 1 and 2. Promising features of T cells, measured primarily in independent laboratories, and pending further replication and validation as biomarkers, are summarized in Supplementary Table 1.
Table 1.
Antigen-agnostic and antigen-specific candidate T-cell biomarkers for T1D
| Specificity | Candidate biomarker | Observation | Assay | Cell number (PBMC) | Replicated |
Proposed next steps | Reference | |
|---|---|---|---|---|---|---|---|---|
| Same laboratory | Independent laboratory | |||||||
| Antigen nonspecific (agnostic) | CD4 Tfh cell frequency | Increased frequency of memory Tfh in peripheral blood in at-risk progressors and new-onset and long-standing T1D. More Tfh in individuals with multiple autoantibodies. Definition of Tfh varies by study with regard to PD-1 and ICOS inclusion. | Flow cytometry | 1–5 × 106 | Yes | Yes | Standardize definition of Tfh; biomarker validation | (10,65–68) |
| CD4 Treg transcript signature | Expression of 37 gene transcript panel in purified, stimulated Tregs or PBMC distinguishes new-onset T1D, long-standing T1D, and T2D from healthy control subjects; predicts C-peptide decline over time. | Nanostring expression analysis | 5–10 × 103 | Yes | Ongoing | Further biomarker validation in independent laboratory | (15) | |
| CD4 Teff resistance to suppression | Resistance of CD4 Teff (CD4+CD25−) from established T1D to Treg suppression compared with Teff from healthy control subjects. | Flow cytometry | 2 × 107 | Yes | Yes | Replication of shorter assay in independent laboratory; biomarker validation | (69–71) | |
| FOXP3+IFN-γ+ Tregs | Increased frequency of Helios− FOXP3+IFNγ+ adaptive Tregs in T1D compared with healthy control subjects. | Flow cytometry | 1–5 × 106 | Yes | Yes | Biomarker validation | (72,73) | |
| CD4 T-cell IL-2 response | Decreased IL-2 pSTAT5 signaling in CD4 Treg and CD25+ and CD25− memory T cells compared with healthy control subjects. Impacted by genotype at IL2RA and PTPN2 genes. Results in reduced FOXP3 stability and reduced frequency of memory and activated Tregs in high vs. low IL-2 T1D responders. | Flow cytometry | 5 × 106 | Yes; technically reproducible and stable over time | Yes | Biomarker validation | (12–14) | |
| IL-17+ T-cell frequency | Increased frequency of CD4 naive or memory T cells and CD8 T cells that are IL-17+ upon stimulation in recent-onset and long-standing T1D. Impacted by IL2RA genotype in at-risk subjects. | Flow cytometry, ELISA, PCR | 1 × 107 | Yes | Yes | Standardization of assay; biomarker validation | (68,74–76) | |
| Antigen specific | Islet-specific CD8 T-cell frequency | Increased frequency of class I islet Mmr+ CD8 T cells in peripheral blood or pancreas in recent-onset or established T1D compared with healthy control subjects. Increased frequency of islet Mmr+ CD8 T cells associated with recurrent autoimmunity following islet transplant. | Class I multimers | 1–5 × 106 | Yes | Yes | Clinical validation; expansion to include additional HLA types | (3,16,18,19) |
| Islet-specific CD4 T-cell frequency and phenotype | Increased frequency of islet-specific CD4 T cells to classic or posttranslationally modified islet epitopes in peripheral blood in recent-onset or established T1D compared with healthy control subjects. Higher frequency of islet CD4 T cells associated with shorter disease duration. Islet CD4 T cells with an effector memory phenotype correlated with insulin antibody titer. | Class II tetramers, antigen-stimulated proliferation | 5–10 × 106 | Yes | Yes; proliferation assays not rigorously tested in multiple laboratories | Standardization of assay; clinical validation; prioritization of available epitopes; expansion to include additional HLA types | (20–26,49,77–79) | |
| Inflammatory islet-specific T-cell signature | Increased frequency of IFN-γ+ islet antigen-stimulated T cells in new-onset and established T1D compared with healthy control subjects that had IL-10+ T-cell response. Reduction in IFN-γ+ T cells in new-onset T1D during the first year post-diagnosis. Higher frequency IFN-γ+ T cells in children than adults. IFN-γ+ islet antigen specificities differed in children vs. adults and with disease duration. | ELISpot | 2 × 107 | Yes | Yes | Optimize for use with cryopreserved cells; standardize assay; clinical validation | (5,28–33,49) | |
All biomarkers listed have been tested in stage 3 T1D. Teff, effector T cells; T2D, type 2 diabetes; pSTAT5, phosphorylated STAT5; Mmr, multimer; ND, not determined.
Table 2.
Candidate T-cell biomarkers for treatment and response to therapy
| Specificity | Therapy | Candidate biomarker | Observation | Assay | Cell number (PBMC) | Replicated | Stage T1D | Proposed next steps | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Antigen nonspecific (agnostic) | Abatacept (CTLA-4-Ig) | CD4 Tcm frequency or CD4 naive T cells/Tcm ratio; altered expression of T-cell costimulatory molecules. | Increased frequency of CD4 Tcm or decrease in ratio of CD4 naive T cells/CD4 Tcm correlates with subsequent increased rate of C-peptide decline in placebo group; attenuated in treatment group. Altered expression of T-cell costimulatory ligands ICOSLG, CD40, and CD58 in responders vs. nonresponders. | Flow cytometry; whole-blood RNA-Seq | 1–5 × 106 | Initial confirmation | 3 | Confirmation in follow-on clinical study (stage 1 T1D) using same agent; variation over time and in relation to demographics and disease subtype. | (38,43) |
| Teplizumab (anti-CD3) | CD8 T-cell exhaustion | Increase in frequency of CD8 T cells with exhaustion phenotype (EOMES+,TIGIT+KLRG1+) in responders vs. nonresponders; decrease in CD4 Tem and increase in CD8 Tcm in responders. | Flow cytometry | 1–5 × 106 | Replicated RNA-Seq and flow cytometry | 3 | Confirmation ongoing in follow-on clinical study (stage 2 T1D) using same agent; variation over time and in relation to demographics and disease subtype | (39,80) | |
| Rituximab (anti-CD20) | CD3 and CD4 T-cell frequency | Increased frequency of CD3/CD4 T cells at week 26 post-treatment in subjects with progressive loss of C-peptide. | Flow cytometry | 1–5 × 106 | Replicated RNA-Seq and flow cytometry | 3 | Confirmation in follow-on clinical study using same agent with abatacept (stage 1 T1D); variation over time and in relation to demographics and disease subtype | (42) | |
| Antithymocyte globulin + G-CSF | CD4/CD8 T-cell ratio | Correlation of CD4/CD8 T-cell ratio with response to therapy (change in C-peptide AUC at 6 months and 12 months). | Flow cytometry | 1–5 × 106 | No | 3 | Confirmation in follow-on clinical study using same agent; variation over time and in relation to demographics and disease subtype | (40) | |
| Alefacept (LFA-3-Ig) | CD4 and CD8 naive T cells, Teff, and Treg frequency and phenotype | Increase in frequency of CD4 naive T cells and decrease in frequency in CD4 Tcm, CD4 Tem, and CD8 Tcm. Increase in ratio of Treg/Tcm for CD4 and CD8. Increase in PD-1+CD4+ Tem. | Flow cytometry | 1–5 × 106 | No | 3 | Identification of biomarkers of response; confirmation in follow-on clinical study using biosimilar; variation over time and in relation to demographics and disease subtype | (46) | |
| Antigen specific | BHT-3021 plasmid-encoded proinsulin | Proinsulin CD8 T-cell frequency | Reduced frequency of class I proinsulin Mmr+ CD8 T-cell frequency over the course of plasmid vaccination correlated with improved C-peptide levels in treatment group vs. placebo. | Flow cytometry | 1 × 107 | No | 3 to late stage 3 | Confirmation in follow-on clinical study utilizing same antigen | (45) |
| Nasal insulin | Frequency of proinsulin-responsive T cells | Reduced frequency of proinsulin-stimulated proliferation in treated autoantibody-positive at-risk subjects; reduced frequency of proinsulin IFN-γ+ T cells in treated recent-onset T1D subjects. | Proliferation and ELISpot | 2 × 107 | Yes | Pre–stage 1 to 1; 3 | Validation in large clinical study utilizing same antigen | (81,82) | |
| Teplizumab (anti-CD3) | Islet antigen CD8 T-cell frequency and phenotype | Increased frequency of InsB and GAD65 Mmr+ CD8 T cells 3 months post-treatment with a change in phenotype from naive to CD45RA+ Tem. | Flow cytometry | 2 × 107 | No | 3 | Confirmation ongoing in follow-on clinical study (stage 2 T1D) using same agent | (18) | |
| GAD-alum | Frequency, proliferation, and cytokine secretion of GAD65-responsive CD8 T cells | Increased proliferation of T cells to GAD65 in treated recent-onset T1D vs. placebo; increase in IFN-γ and IL-4+ T cells in response to GAD65. Cytokine secretion in response to GAD65 increased compared with placebo and favored Th2 cytokines in subjects with lower change in C-peptide. | Flow cytometry | 2 × 107 | No | 3 | Confirmation in follow-on clinical study using same agent | (83,84) | |
| Intradermal proinsulin peptide | Islet-specific CD8 T-cell frequency and phenotype | Reduced frequency of class I proinsulin, InsB, and IA-2 Mmr+ CD8 T cells with a CD57+ antigen-experienced phenotype in subjects with preserved C-peptide compared with placebo; increase in IL-10+ proinsulin CD4 T cells in high-frequency treated subjects and increase in FOXP3 levels on memory Tregs in responders. | Flow cytometry; ELISpot | 2 × 107 | No | 3 | Confirmation in follow-on clinical study utilizing same antigen | (44) |
Mmr, multimer; RNA-Seq, RNA sequencing; Tcm, central memory T cells; Teff, effector T cells; Tem, effector memory T cells.
Candidate Antigen-Agnostic T-Cell Biomarkers
Assays that measure features of T cells in an antigen-agnostic manner generally require fewer cells and have lower variability when compared with what is typically observed for antigen-specific assays. These methodologies commonly include cytometric profiling and in vitro functional assays.
Several candidate antigen-agnostic biomarkers have been replicated in multiple laboratories or have undergone additional optimization for use as clinical biomarkers in assay cores established for biomarker validation testing. Many of these candidate biomarkers have been shown to discriminate patients with T1D from healthy control subjects, including frequencies of a number of T-cell subsets as well as markers of immunoregulation and IL-2 responsiveness (Table 1). Fewer antigen-agnostic biomarkers have been demonstrated to be associated with disease progression or in defining subtypes of patients, but there have been a number of findings that are promising and noteworthy. An increased frequency of T follicular helper cells (Tfh) has been reported in the peripheral blood of patients with T1D versus healthy control subjects, and this frequency has been negatively correlated with C-peptide levels in recent-onset T1D (10). Tfh cells have a surface marker profile characterized by expression ICOS, PD-1, and CXCR5 and are involved in B-cell activation and differentiation within germinal centers. A second functional biomarker, CD4+ T-cell hyporesponsiveness to IL-2, has been identified by reduced phosphorylation of STAT5 and is consistent with the association of the CD25 (IL2RA) gene with T1D (11,12). Deficient IL-2 responses are significant for T1D because regulatory T cells (Tregs) are dependent on this cytokine for survival and metabolic fitness. Although the frequencies of Tregs overall are not different in patients and healthy control subjects, the fitness of these immune regulatory cells may be affected in the diseased state: individuals with low CD4+ T-cell IL-2 signaling have a reduction in frequency of memory and activated Tregs compared with T1D subjects who have high IL-2 signaling (13). IL-2 hyporesponsiveness is correlated with FOXP3 instability in thymic and peripheral Tregs in T1D (12,14). Although this functional abnormality cannot be distinguished by the surface phenotype of the cells, it may have utility in stratifying patients for immune therapies that target this pathway. Interestingly, Pesenacker et al. (15) recently reported that Treg gene signatures were significantly altered in T1D patients when compared with healthy control subjects and those with type 2 diabetes. Treg gene signature–based algorithms accurately predicted the rate of C-peptide decline in new-onset T1D patients enrolled in T1DAL (Inducing Remission in New Onset T1DM With Alefacept) and START (Study of Thymoglobulin to Arrest Type 1 Diabetes) trials (15).
To date, the utility of transcriptional and high-dimensional cytometric analyses of T-cell subsets as a prognostic biomarker for risk and rate of T1D progression are largely unknown. Studies to evaluate these technologies and to confirm discovery-level biomarkers with samples from large T1D consortia (e.g., including TEDDY, Type 1 Diabetes TrialNet, and the Immune Tolerance Network [ITN]) are ongoing.
Candidate Antigen-Specific T-Cell Biomarkers
Islet antigen-specific T cells represent potential T-cell biomarkers in T1D, but to be useful, features of these cells need to be differentiated from those found in healthy control subjects that are reactive with the same epitopes (6,16). Several distinct assays have been implemented to quantitate and characterize antigen-specific T cells in peripheral blood, including proliferation assays, HLA class I multimers and class II tetramers (peptide–MHC complexes coupled to fluorophores or quantum dots [Qdots]), activation-based assays, and ELISpot (detection of cytokine responses to defined HLA class I or class II binding peptides) (Table 1). However, for the reasons described, detection of antigen-specific T cells is technically challenging, often limiting their utility. The performance of antigen specific T-cell assays in T1D is discussed below.
To date, antigen-specific T-cell assays have identified CD4 and/or CD8 T cells that recognize epitopes derived from insulin/proinsulin/preproinsulin, GAD65 (glutamic acid decarboxylase), IA-2 (islet antigen 2), ZnT8 (zinc transporter 8), IGRP (islet-specific glucose-6-phosphatase catalytic subunit-related protein), chromogranin A, IAPP (islet amyloid polypeptide), and GRP78 (glucose-regulated protein 78) in T1D subjects (2). In general, these peptides were discovered by their elution off of HLA molecules associated with genetic risk for T1D (e.g., DR4) as well as their expression on class I HLA molecules on β-cells. Some (e.g., ZnT8, preproinsulin) but not all (e.g., GAD65, chromogranin) peptides are uniquely expressed in β-cells. This concept of antigen distribution among various cell types and tissues may prove to be important when considering the relationship between antigen persistence and the maintenance of immunologic memory within the adaptive immune system. Recently discovered posttranscriptionally modified, hybrid, and alternatively spliced/translated islet epitopes represent a new frontier for the application of antigen-specific assays in the context of disease monitoring and interventions (17). The emergence of T cells with reactivity to these modified epitopes provides a conceptual basis for how higher-affinity T cells may emerge to promote β-cell autoreactivity following cellular stress events. However, given the large number of antigens and epitopes that have been identified, development of standardized antigen-specific T-cell biomarkers and assays will require coordinated developmental work within the community, as well as novel computational approaches to prioritize which antigens and epitopes will be investigated and advanced as biomarkers.
Several candidate antigen-specific T-cell biomarkers have been replicated in independent laboratories. Multiple studies have detected increased frequencies of CD8 and CD4 islet-specific T cells in subjects with T1D compared with healthy control subjects using HLA class I multimers and class II tetramers, proliferation, and activation assays (3,16,18–26). Higher frequencies of antigen-specific T cells have also been observed in T1D subjects with shorter disease duration and in children versus adult T1D patients (5,20,24,27–29). Studies using ELISpot assays have identified an inflammatory signature characterized by increased frequencies of IFN-γ+ T cells in patients with T1D as compared with healthy control subjects, in whom IL-10 was the dominant cytokine secreted from PBMCs (5,28–34).
Pairing flow-based approaches to identify antigen-specific T cells with platforms such as mass cytometry (35,36) and single-cell transcript and TCR analyses (26,37), including newer technologies such as nucleotide-based barcoded sequences conjugated to antibodies or HLA multimers, have the potential to provide robust insights into the phenotypes and TCR clonotypes of antigen-specific T cells in cross-sectional or longitudinal sample sets. These types of combinatorial assessments may also generate the next generation of refined T-cell biomarkers.
Clinical Response T-Cell Candidate Biomarkers
Candidate T-cell biomarkers indicative of treatment effect or response to therapy are emerging from integrated mechanistic studies (Table 2). Even in trials that did not meet their primary clinical end points, informative assays continue to reveal information on the underlying biology of the disease and the alterations that occur upon therapeutic intervention. Both antigen-agnostic and antigen-specific candidate biomarkers have been identified in trial samples, with the latter primarily utilized in the context of antigen-specific immunotherapies. Clinical trials in T1D using abatacept (CTLA-4-Ig), teplizumab (anti-CD3), rituximab (anti-CD20), or therapy with low-dose antithymocyte globulin (ATG) alone or in combination with G-CSF have revealed changes in T-cell frequency or exhaustion that correlate with stabilization of C-peptide levels or the rate of C-peptide decline (38–42). Whole-blood transcriptome analysis of abatacept-treated new-onset T1D patients revealed altered expression of the T-cell costimulatory molecules ICOSLG, CD40, and CD58 in responders (i.e., subjects where C-peptide secretion was transiently preserved) versus nonresponders (43). Similarly, reduced frequencies of proinsulin class I multimer+ CD8 T cells were observed in subjects with preserved C-peptide levels compared with placebo in clinical trials targeting proinsulin with a DNA vaccine or intradermal proinsulin peptide (44,45).
Despite various mechanisms of action for different drugs, the concept that positive therapeutic outcomes will be associated with Treg enhancement and the depletion or disabling of effector T-cell populations is emerging as a generalized paradigm. As an example of the latter, signs of CD8+ T-cell exhaustion were shown to correlate with functional responses to teplizumab and correlate with long-term preservation of C-peptide at the end of a 2-year study period (39). Conversely, in more than one T1D immunotherapy trial, enhancing the number (absolute or relative) or function of Tregs correlated with a beneficial effect on disease progression. ATG and alefacept (LFA-3-Ig), for example, depleted the number of circulating CD4 and CD8 memory and effector T cells with relative preservation of Tregs (40,46). However, increased Treg numbers alone was not sufficient for clinical benefit, as also shown in the IL-2/rapamycin trial in which a decline in C-peptide was seen and attributed to increased NK cells and effector T cells in spite of increased Tregs (47). Given the known interplay between Treg and NK-cell homeostasis and the recent identification of NK-cell signatures during T1D progression in TEDDY cohort participants, examination of T-cell, and particularly Treg, biomarkers in the context of NK-cell frequency, activation, and function may prove beneficial following immunoregulatory interventions (E. McKinney, "TEDDY transcriptomics: patterns of progression in T1D," presented at the Immunology of Diabetes Society Congress, London, U.K., 2018, unpublished observations).
T-cell analytes that are relatively stable in an individual in a longitudinal manner yet variable among individuals represent desirable biomarkers. As a result, variability observed within an individual during the natural history of disease or in response to therapy can be attributed to the disease process or to the specific therapeutic agent(s) applied. Achieving a deeper understanding of the most relevant T-cell changes for the ascertainment of treatment and/or therapeutic effect in response to immunotherapies will require dense data sampling and analysis of relevant clinical samples in a harmonized fashion.
Collaborative Workshops and Biomarker Validation
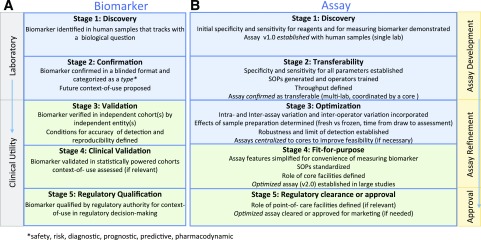
The reproducibility in detection of biomarkers is directly reliant on the optimization and transferability of the assays measuring them (i.e., similar results should be obtained in multiple laboratories using different sample sets collected in the same harmonized fashion). Toward this goal, Fig. 3 outlines steps of a proposed comprehensive pipeline for the development of T1D-relevant biomarkers in the T1D community, including T-cell biomarkers and their associated assays. In T1D, candidate T-cell biomarkers that are antigen agnostic have advanced the farthest through validation processes using clinical samples. When establishing a fit-for-purpose assay during biomarker validation, every effort should be made to comply with internationally recognized standards, when they exist, such as the guidance of the International Council for Harmonisation (https://www.ich.org/products/guidelines.html), ISO/IEC 17025 (https://www.iso.org/standard/66912.html), or the European Medicines Agency reflection paper for laboratories that perform the analysis or evaluation of clinical trial samples (https://www.ema.europa.eu/documents/regulatory-procedural-guideline/reflection-paper-laboratories-perform-analysis-evaluation-clinical-trial-samples_en.pdf) (49). Various workshops have provided an invaluable assessment of these assays and are needed on an ongoing basis to maintain the quality of immune cellular measurements through large-scale validation.
Figure 3.
Proposed stages of development for T1D biomarkers and assays (consensus view of the authors). Biomarkers and their associated assays have parallel and independent lines of development, ideally converging at the stage of biomarker validation using fit-for-purpose assays for reliable use by the scientific community. A: A biomarker must successfully pass through stage 3 to be considered validated for research purposes. If a biomarker is a candidate for regulatory decision-making, subsequent stages of development (stages 4–5) must be completed. B: All assays should ideally achieve fit-for-purpose status (stage 4) for widespread use to measure a validated biomarker. In specific instances, where an assay has achieved approval for marketing purposes, it must be cleared by regulatory bodies (stage 5). SOPs, standard operating procedures.
In previous and ongoing efforts, sharing of blinded replicate samples between laboratories has been used to establish the reproducibility of biomarker detection and the sensitivity and specificity of assays measuring them. The ITN and Type 1 Diabetes TrialNet have conducted workshops to evaluate the transferability of various antigen-specific T-cell–based assays and the reproducibility of biomarker detection in a blinded fashion. These workshops showed the highest sensitivity and specificity for a cellular proliferation (immunoblot) assay and for ELISpot measurements, although optimization for wide utility involving small sample volumes or frozen biosamples is yet to be achieved. These challenges have stalled the progress of these assays through the optimization stage of the pipeline (Fig. 3 and Table 1) (50,51). The performance of class II MHC tetramers has been modest in workshop testing. In formal validation efforts by the Immunology of Diabetes Society, detection of antigen-specific CD8+ and CD4+ T cells with HLA class I multimers and class II tetramers showed good reproducibility in individual laboratories but were somewhat variable for replicate samples measured in different laboratories (52–54), limiting their feasibility as easily assayable pharmacodynamic markers for antigen-specific therapies and challenging their development to “fit-for-purpose” status as well. Thus, there continues to be a dearth of widely usable, optimized assays for antigen-specific biomarkers in T1D. In related efforts, the effects of sample preparation on T-cell assay outcomes (e.g., fresh versus frozen and time to sample assessment) has been carefully studied by the Immunology of Diabetes Society and will continue to be evaluated with new assays for the detection of novel biomarkers (55,56). Workshop efforts are being increasingly embraced by the T1D biomarker community via centralized facilities (57), which should significantly help with go/no-go decisions along a development pipeline, like the one proposed herein (Fig. 3). A summary of key challenges and considerations for progress in this field is presented in Table 3.
Table 3.
Challenges impeding progress toward the development of effective T-cell biomarkers in T1D
| Challenges | Potential solutions and technological advances needed |
|---|---|
| Biological | |
| High repertoire diversity and low precursor frequency of autoreactive T cells in peripheral blood | Develop or improve assays capable of measuring the complex mixture of autoreactive T cells |
| Implement new technologies and approaches for identifying pathogenic signatures, including high-dimensional flow cytometry, mass cytometry, and barcoded antibodies or pMHC multimers for use in scRNA-Seq approaches | |
| Develop sensitive molecular biomarkers capable of detecting signatures of autoreactive T cells, including TCR immunosequencing | |
| Large numbers of genetic risk variants impacting cellular function | Create isogenic cellular systems to identify causative SNPs and elucidate their impact on T-cell function |
| Employ well-characterized biobanks with genotype-selectable donor samples | |
| High degree of heterogeneity in T-cell phenotypes among subjects with T1D | Conduct functional testing on subjects with defined phenotypic profiles |
| Define and control for covariates leading to heterogeneity in T-cell responses | |
| Design and conduct interventional trials using targeted populations with mechanistic outcomes | |
| Transient or variable autoreactivity over the natural history of the disease | Build robust longitudinal and interventional cohorts with sufficient clinical samples |
| Process | |
| Low sample volumes in peripheral blood of pediatric samples | Work toward miniaturizing functional assays |
| Develop surrogate markers of autoreactivity that do not require large sample volumes | |
| Need for measures that correlate T-cell autoreactivity with endogenous β-cell mass and/or function | Develop assays capable of detecting signals from autoreactive T cells in circulation reflective of ongoing pathology within T1D islets |
| Characterize the degree of overlap between tissues and peripheral blood signatures | |
| Need to understand the pathogenic potential of T-cell subsets or reactivities | Create biomimetic devices to model the islet:immune microenvironment |
| Employ new technologies to test the function of antigen-specific T cells in viable pancreatic tissue sections | |
| Need for assay reproducibility and interoperability | Employ independent validation cores and sample resources capable of repeating assays to test reproducibility and robustness |
| Incentivize replication testing | |
| Paradigms | |
| Focus on limited epitopes from known autoantigens | Consider nonnative peptides, hybrid peptides, posttranslationally modified peptides |
| Implement novel high-throughput unbiased peptide screens | |
| Implement novel computational approaches to model peptides capable of activating T cells through the TCR:MHC complex | |
| Consider alternate concepts to explain origins of autoreactivity | Improve understanding of endogenous stress response and host response to commensal bacteria and viral agents, for example |
| Focus on classical T1D pathogenesis | Broaden studies to include longitudinal studies of T cells in cancer subjects receiving immune checkpoint inhibitors |
| Understand autoreactivity emanating from rare genetic variants with high penetrance of T1D | |
| T-cell–centric approaches | Broaden studies to better understand T cell:B cell and T cell:APC interactions |
| Understand exogenous signals that can break T-cell tolerance | |
| Heavy focus on the pathogenic features of T-cell autoreactivity in subjects with known genetic risk | Better understand the principles related to the mechanisms by which the MHC class II haplotype of DR15-DQ6 influences the T-cell repertoire and leads to dominant protection from disease |
APC, antigen-presenting cell; pMHC, peptide MHC; scRNA-Seq, single-cell RNA sequencing; SNPs, single nucleotide polymorphisms.
Future Prospects With Genomics
TCR Immunosequencing as a Biomarker
An area that straddles the antigen-specific and antigen-agnostic biomarker space is TCR immunosequencing. Each T-cell clone expresses a unique TCR to recognize an antigen and thus TCR sequences can be used as a surrogate for individual T-cell and clonotypic measurements. Advantages of TCR biomarkers include the absence of a requirement to have live T cells for assays, minimal intra- and interassay variations, and the capacity to detect extremely infrequent T cells. Two strategies have been pursued in the field thus far to utilize TCR sequences as part of disease-specific biomarkers. First, TCR repertoire diversity in the blood has shown differences between T1D patients and control subjects (Supplementary Table 1) (57,58). Second, disease-specific TCR clonotypes have been identified within the target organ (25,59,60) or in islet antigen-specific T cells (34,37,61–63) that are commonly and/or exclusively detected in T1D patients. Of importance, prevalent TCR clonotypes in the pancreas and pancreatic lymph nodes are detected in peripheral blood of the same donor and of other T1D patients (16,59,64), and longitudinal studies have demonstrated consistent presence of antigen-specific TCR clonotypes over time in subjects (37,61).
Other Genomics Approaches
A number of exciting technologies are now available to the T1D community to rapidly advance T-cell biomarker discovery, including the expanding –omics platforms and the creation of high-dimensional data sets. Notably, these opportunities emanate from significant advances in single-cell RNA sequencing, epigenetic assessments (e.g., DNA methylation; Assay for Transposase-Accessible Chromatin, ATACseq), molecular profiling of antigen-specific T cells (26,37), and high-parameter mass cytometry of islet MHC multimer–stained cells (35,36), among others. These technologies have not been widely tested in T1D for purposes of prediction or in clinical trial settings to date, but increasing numbers of clinical and tissue-based studies have begun to include these novel technologies as part of measured parameters. As progress continues and data assimilates, cross-expertise collaborative efforts will become vital in the biomarker field, with necessary involvement of systems and computational biology for the analysis of large multidimensional data sets.
Conclusions
T cells play a central role in T1D pathogenesis, and therefore, validated T-cell biomarkers will undoubtedly expedite the clinical path toward approved immunotherapies for T1D. The T1D Biomarker Working Group and the associated Core for Assay Validation (www.t1dbiomarkers.org) have been committed in recent years to moving promising candidate biomarkers out of the discovery realm into confirmation and validation testing via a collaborative and coordinated process. Despite years of dedicated attempts to establish T-cell biomarkers for the prediction and monitoring of T1D, the identification and acquisition of sample biobanks from large-scale repositories and/or longitudinal studies (e.g., TEDDY, Type 1 Diabetes TrialNet, T1D Exchange) (38–42) coupled with modern technologies and biostatistical and machine learning approaches now make success more likely than ever. As a community, we must now validate the most promising T-cell biomarkers and assays through large, harmonized studies and publish standard operating procedures for widespread use. Once these stages are accomplished, T-cell biomarkers should also be evaluated in relation to other immune cell populations, such as NK cells, B cells, macrophages, and dendritic cells, which may inform the mechanisms underlying the breach of T-cell tolerance in T1D. The establishment of clinically validated T-cell biomarkers for T1D is an achievable reality that will require ongoing commitment and integrated efforts by key stakeholders including, scientists, clinicians, industry partners, regulators, and funding agencies.
Supplementary Material
Article Information
Acknowledgments. The authors would like to thank all members of the JDRF Biomarker Working Group for championing collaborative biomarker studies in T1D and JDRF science staff who have contributed to discussions. The authors are grateful for the editorial assistance of Catherine McCaffrey (JDRF) and Amanda Posgai (University of Florida). We sincerely regret if references to prior work were omitted to meet editorial guidelines.
Funding. Research efforts related to the contents of this Perspective are supported by JDRF and the National Institutes of Health (P01 AI42288, R01 DK106191, and HIRN UC4 DK104194 to T.M.B.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-0119/-/DC1.
S.A. and K.C. contributed equally to this work.
References
- 1.McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 2015;523:612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roep BO, Peakman M. Antigen targets of type 1 diabetes autoimmunity. Cold Spring Harb Perspect Med 2012;2:a007781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Unger WW, Velthuis J, Abreu JR, et al. Discovery of low-affinity preproinsulin epitopes and detection of autoreactive CD8 T-cells using combinatorial MHC multimers. J Autoimmun 2011;37:151–159 [DOI] [PubMed] [Google Scholar]
- 4.Yang J, Chow IT, Sosinowski T, et al. Autoreactive T cells specific for insulin B:11-23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc Natl Acad Sci U S A 2014;111:14840–14845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scotto M, Afonso G, Østerbye T, et al. HLA-B7-restricted islet epitopes are differentially recognized in type 1 diabetic children and adults and form weak peptide-HLA complexes. Diabetes 2012;61:2546–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Duque S, Azoury M, Colli M, et al. Conventional and neo-antigenic peptides presented by β cells are targeted by circulating naïve CD8+ T cells in type 1 diabetic and healthy donors. Cell Metab 2018;28:946–960.e6 [DOI] [PubMed]
- 7.Leete P, Willcox A, Krogvold L, et al. Differential insulitic profiles determine the extent of β cell destruction and the age at onset of type 1 diabetes. Diabetes 2016;65:1362–1369 [DOI] [PubMed] [Google Scholar]
- 8.Insel RA, Dunne JL, Atkinson MA, et al. Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care 2015;38:1964–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bingley PJ, Williams AJ, Colman PG, et al.; T1DGC . Measurement of islet cell antibodies in the Type 1 Diabetes Genetics Consortium: efforts to harmonize procedures among the laboratories. Clin Trials 2010;7(Suppl.):S56–S64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X, Shi Y, Cai Y, et al. Inhibition of increased circulating Tfh cell by anti-CD20 monoclonal antibody in patients with type 1 diabetes. PLoS One 2013;8:e79858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garg G, Tyler JR, Yang JH, et al. Type 1 diabetes-associated IL2RA variation lowers IL-2 signaling and contributes to diminished CD4+CD25+ regulatory T cell function. J Immunol 2012;188:4644–4653 [DOI] [PMC free article] [PubMed]
- 12.Long SA, Cerosaletti K, Bollyky PL, et al. Defects in IL-2R signaling contribute to diminished maintenance of FOXP3 expression in CD4+CD25+ regulatory T-cells of type 1 diabetic subjects. Diabetes 2010;59:407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang JH, Cutler AJ, Ferreira RC, et al. Natural variation in interleukin-2 sensitivity influences regulatory T-cell frequency and function in individuals with long-standing type 1 diabetes. Diabetes 2015;64:3891–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerosaletti K, Schneider A, Schwedhelm K, et al. Multiple autoimmune-associated variants confer decreased IL-2R signaling in CD4+ CD25(hi) T cells of type 1 diabetic and multiple sclerosis patients. PLoS One 2013;8:e83811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pesenacker AM, Chen V, Gillies J, et al. Treg gene signatures predict and measure type 1 diabetes trajectory. JCI Insight 2019;4:123879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Culina S, Lalanne AI, Afonso G, et al.; ImMaDiab Study Group . Islet-reactive CD8+ T cell frequencies in the pancreas, but not in blood, distinguish type 1 diabetic patients from healthy donors. Sci Immunol 2018;3:eaao4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.James EA, Pietropaolo M, Mamula MJ. Immune recognition of β-cells: neoepitopes as key players in the loss of tolerance. Diabetes 2018;67:1035–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cernea S, Herold KC. Monitoring of antigen-specific CD8 T cells in patients with type 1 diabetes treated with antiCD3 monoclonal antibodies. Clin Immunol 2010;134:121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinkse GG, Tysma OH, Bergen CA, et al. Autoreactive CD8 T cells associated with beta cell destruction in type 1 diabetes. Proc Natl Acad Sci U S A 2005;102:18425–18430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spanier JA, Sahli NL, Wilson JC, et al. Increased effector memory insulin-specific CD4+ T cells correlate with insulin autoantibodies in patients with recent-onset type 1 diabetes. Diabetes 2017;66:3051–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGinty JW, Chow IT, Greenbaum C, Odegard J, Kwok WW, James EA. Recognition of posttranslationally modified GAD65 epitopes in subjects with type 1 diabetes. Diabetes 2014;63:3033–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chow IT, Yang J, Gates TJ, et al. Assessment of CD4+ T cell responses to glutamic acid decarboxylase 65 using DQ8 tetramers reveals a pathogenic role of GAD65 121-140 and GAD65 250-266 in T1D development. PLoS One 2014;9:e112882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oling V, Marttila J, Ilonen J, et al. GAD65- and proinsulin-specific CD4+ T-cells detected by MHC class II tetramers in peripheral blood of type 1 diabetes patients and at-risk subjects. J Autoimmun 2005;25:235–243 [DOI] [PubMed] [Google Scholar]
- 24.So M, Elso CM, Tresoldi E, et al. Proinsulin C-peptide is an autoantigen in people with type 1 diabetes. Proc Natl Acad Sci U S A 2018;115:10732–10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pathiraja V, Kuehlich JP, Campbell PD, et al. Proinsulin-specific, HLA-DQ8, and HLA-DQ8-transdimer-restricted CD4+ T cells infiltrate islets in type 1 diabetes. Diabetes 2015;64:172–182 [DOI] [PubMed] [Google Scholar]
- 26.Heninger A, Eugster A, Kuehn D, et al. A divergent population of autoantigen-responsive CD4+ T cells in infants prior to β cell autoimmunity. Sci Transl Med 2017;9:eaaf8848 [DOI] [PubMed]
- 27.Martinuzzi E, Novelli G, Scotto M, et al. The frequency and immunodominance of islet-specific CD8+ T-cell responses change after type 1 diabetes diagnosis and treatment. Diabetes 2008;57:1312–1320 [DOI] [PubMed] [Google Scholar]
- 28.Arif S, Gibson VB, Nguyen V, et al. β-Cell specific T-lymphocyte response has a distinct inflammatory phenotype in children with type 1 diabetes compared with adults. Diabet Med 2017;34:419–425 [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Gu Y, Bian L, et al. Characterization of immune response to novel HLA-A2-restricted epitopes from zinc transporter 8 in type 1 diabetes. Vaccine 2016;34:854–862 [DOI] [PubMed] [Google Scholar]
- 30.Énée É, Kratzer R, Arnoux JB, et al. ZnT8 is a major CD8+ T cell-recognized autoantigen in pediatric type 1 diabetes. Diabetes 2012;61:1779–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scotto M, Afonso G, Larger E, et al. Zinc transporter (ZnT)8(186-194) is an immunodominant CD8+ T cell epitope in HLA-A2+ type 1 diabetic patients. Diabetologia 2012;55:2026–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dang M, Rockell J, Wagner R, et al. Human type 1 diabetes is associated with T cell autoimmunity to zinc transporter 8. J Immunology 2011;186:6056–6063 [DOI] [PMC free article] [PubMed]
- 33.Mallone R, Martinuzzi E, Blancou P, et al. CD8+ T-cell responses identify β-cell autoimmunity in human type 1 diabetes. Diabetes 2007;56:613–621 [DOI] [PubMed] [Google Scholar]
- 34.Nakayama M, McDaniel K, Fitzgerald-Miller L, et al. Regulatory vs. inflammatory cytokine T-cell responses to mutated insulin peptides in healthy and type 1 diabetic subjects. Proc Natl Acad Sci U S A 2015;112:4429–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laban S, Suwandi JS, van Unen V, et al. Heterogeneity of circulating CD8 T-cells specific to islet, neo-antigen and virus in patients with type 1 diabetes mellitus. PLoS One 2018;13:e0200818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogura H, Preston-Hurlburt P, Perdigoto A, et al. Identification and analysis of islet antigen-specific CD8+ T cells with T cell libraries. J Immunology 2018;201:1662–1670 [DOI] [PMC free article] [PubMed]
- 37.Cerosaletti K, Barahmand-Pour-Whitman F, Yang J, et al. Single-cell RNA sequencing reveals expanded clones of islet antigen-reactive CD4(+) T cells in peripheral blood of subjects with type 1 diabetes. J Immunology 2017;199:323–335 [DOI] [PMC free article] [PubMed]
- 38.Orban T, Beam CA, Xu P, et al.; Type 1 Diabetes TrialNet Abatacept Study Group . Reduction in CD4 central memory T-cell subset in costimulation modulator abatacept-treated patients with recent-onset type 1 diabetes is associated with slower C-peptide decline. Diabetes 2014;63:3449–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long SA, Thorpe J, DeBerg HA, et al. Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes. Sci Immunol 2016;1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haller MJ, Gitelman SE, Gottlieb PA, et al. Antithymocyte globulin plus G-CSF combination therapy leads to sustained immunomodulatory and metabolic effects in a subset of responders with established type 1 diabetes. Diabetes 2016;65:3765–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haller MJ, Gitelman SE, Gottlieb PA, et al. Anti-thymocyte globulin/G-CSF treatment preserves β cell function in patients with established type 1 diabetes. J Clin Invest 2015;125:448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linsley PS, Greenbaum CJ, Rosasco M, Presnell S, Herold KC, Dufort MJ. Elevated T cell levels in peripheral blood predict poor clinical response following rituximab treatment in new-onset type 1 diabetes. Genes Immun 2019;20:293–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linsley PS, Greenbaum CJ, Speake C, Long SA, Dufort MJ. B lymphocyte alterations accompany abatacept resistance in new-onset type 1 diabetes. JCI Insight 2019;4:126136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alhadj Ali M, Liu YF, Arif S, et al. Metabolic and immune effects of immunotherapy with proinsulin peptide in human new-onset type 1 diabetes. Sci Transl Med 2017;9:9. [DOI] [PubMed] [Google Scholar]
- 45.Roep BO, Solvason N, Gottlieb PA, et al.; BHT-3021 Investigators . Plasmid-encoded proinsulin preserves C-peptide while specifically reducing proinsulin-specific CD8⁺ T cells in type 1 diabetes. Sci Transl Med 2013;5:191ra82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rigby MR, Harris KM, Pinckney A, et al. Alefacept provides sustained clinical and immunological effects in new-onset type 1 diabetes patients. J Clin Invest 2015;125:3285–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Long SA, Rieck M, Sanda S, et al.; Diabetes TrialNet and the Immune Tolerance Network . Rapamycin/IL-2 combination therapy in patients with type 1 diabetes augments Tregs yet transiently impairs β-cell function. Diabetes 2012;61:2340–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis BH, Dasgupta A, Kussick S, Han JY, Estrellado A, Group IIW; ICSH/ICCS Working Group . Validation of cell-based fluorescence assays: practice guidelines from the ICSH and ICCS - part II - preanalytical issues. Cytometry B Clin Cytom 2013;84:286–290 [DOI] [PubMed] [Google Scholar]
- 49.Herold KC, Brooks-Worrell B, Palmer J, et al.; Type 1 Diabetes TrialNet Research Group . Validity and reproducibility of measurement of islet autoreactivity by T-cell assays in subjects with early type 1 diabetes. Diabetes 2009;58:2588–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seyfert-Margolis V, Gisler TD, Asare AL, et al. Analysis of T-cell assays to measure autoimmune responses in subjects with type 1 diabetes: results of a blinded controlled study. Diabetes 2006;55:2588–2594 [DOI] [PubMed] [Google Scholar]
- 51.James EA, Abreu JRF, McGinty JW, et al.; Immunology of Diabetes Society T Cell Workshop Committee . Combinatorial detection of autoreactive CD8+ T cells with HLA-A2 multimers: a multi-centre study by the Immunology of Diabetes Society T Cell Workshop. Diabetologia 2018;61:658–670 [DOI] [PubMed] [Google Scholar]
- 52.James EA, Mallone R, Schloot NC, et al.; T-Cell Workshop Committee, Immunology of Diabetes Society . Immunology of Diabetes Society T-Cell Workshop: HLA class II tetramer-directed epitope validation initiative. Diabetes Metab Res Rev 2011;27:727–736 [DOI] [PubMed] [Google Scholar]
- 53.Mallone R, Scotto M, Janicki CN, et al.; T-Cell Workshop Committee, Immunology of Diabetes Society . Immunology of Diabetes Society T-Cell Workshop: HLA class I tetramer-directed epitope validation initiative T-Cell Workshop Report-HLA Class I Tetramer Validation Initiative. Diabetes Metab Res Rev 2011;27:720–726 [DOI] [PubMed] [Google Scholar]
- 54.Brooks-Worrell B, Tree T, Mannering SI, et al.; T-Cell Workshop Committee, Immunology of Diabetes Society . Comparison of cryopreservation methods on T-cell responses to islet and control antigens from type 1 diabetic patients and controls. Diabetes Metab Res Rev 2011;27:737–745 [DOI] [PubMed] [Google Scholar]
- 55.Mallone R, Mannering SI, Brooks-Worrell BM, et al.; T-Cell Workshop Committee, Immunology of Diabetes Society . Isolation and preservation of peripheral blood mononuclear cells for analysis of islet antigen-reactive T cell responses: position statement of the T-Cell Workshop Committee of the Immunology of Diabetes Society. Clin Exp Immunol 2011;163:33–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Speake C, Odegard JM. Evaluation of candidate biomarkers of type 1 diabetes via the Core for Assay Validation. Biomark Insights 2015;10(Suppl. 4):19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tong Y, Li Z, Zhang H, et al. T cell repertoire diversity is decreased in type 1 diabetes patients. Genomics Proteomics Bioinformatics 2016;14:338–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gomez-Tourino I, Kamra Y, Baptista R, Lorenc A, Peakman M. T cell receptor β-chains display abnormal shortening and repertoire sharing in type 1 diabetes. Nat Commun 2017;8:1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Codina-Busqueta E, Scholz E, Munoz-Torres PM, et al. TCR bias of in vivo expanded T cells in pancreatic islets and spleen at the onset in human type 1 diabetes. J Immunology 2011;186:3787–3797 [DOI] [PubMed]
- 60.Michels AW, Landry LG, McDaniel KA, et al. Islet-derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes 2017;66:722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eugster A, Lindner A, Catani M, et al. High diversity in the TCR repertoire of GAD65 autoantigen-specific human CD4+ T cells. J Immunology 2015;194:2531–2538 [DOI] [PubMed]
- 62.Fuchs YF, Eugster A, Dietz S, et al. CD8+ T cells specific for the islet autoantigen IGRP are restricted in their T cell receptor chain usage. Sci Rep 2017;7:44661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Estorninho M, Gibson VB, Kronenberg-Versteeg D, et al. A novel approach to tracking antigen-experienced CD4 T cells into functional compartments via tandem deep and shallow TCR clonotyping. J Immunol 2013;191:5430–5440 [DOI] [PubMed] [Google Scholar]
- 64.Seay HR, Yusko E, Rothweiler SJ, et al. Tissue distribution and clonal diversity of the T and B cell repertoire in type 1 diabetes. JCI Insight 2016;1:e88242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Viisanen T, Ihantola EL, Näntö-Salonen K, et al. Circulating CXCR5+PD-1+ICOS+ follicular t helper cells are increased close to the diagnosis of type 1 diabetes in children with multiple autoantibodies. Diabetes 2017;66:437–447 [DOI] [PubMed] [Google Scholar]
- 66.Kenefeck R, Wang CJ, Kapadi T, et al. Follicular helper T cell signature in type 1 diabetes. J Clin Invest 2015;125:292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Serr I, Fürst RW, Ott VB, et al. miRNA92a targets KLF2 and the phosphatase PTEN signaling to promote human T follicular helper precursors in T1D islet autoimmunity. Proc Natl Acad Sci U S A 2016;113:E6659–E6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferreira RC, Simons HZ, Thompson WS, et al. IL-21 production by CD4+ effector T cells and frequency of circulating follicular helper T cells are increased in type 1 diabetes patients. Diabetologia 2015;58:781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider A, Rieck M, Sanda S, Pihoker C, Greenbaum C, Buckner JH. The effector T cells of diabetic subjects are resistant to regulation via CD4+ FOXP3+ regulatory T cells. J Immunology 2008;181:7350–7355 [DOI] [PMC free article] [PubMed]
- 70.Lawson JM, Tremble J, Dayan C, et al. Increased resistance to CD4+CD25hi regulatory T cell-mediated suppression in patients with type 1 diabetes. Clin Exp Immunol 2008;154:353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Long AE, Tatum M, Mikacenic C, Buckner JH. A novel and rapid method to quantify Treg mediated suppression of CD4 T cells. J Immunol Methods 2017;449:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McClymont SA, Putnam AL, Lee MR, et al. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunology 2011;186:3918–3926 [DOI] [PMC free article] [PubMed]
- 73.Ferreira RC, Simons HZ, Thompson WS, et al. Cells with Treg-specific FOXP3 demethylation but low CD25 are prevalent in autoimmunity. J Autoimmun 2017;84:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marwaha AK, Crome SQ, Panagiotopoulos C, et al. Cutting edge: increased IL-17-secreting T cells in children with new-onset type 1 diabetes. J Immunology 2010;185:3814–3818 [DOI] [PubMed]
- 75.Marwaha AK, Panagiotopoulos C, Biggs CM, et al. Pre-diagnostic genotyping identifies T1D subjects with impaired Treg IL-2 signaling and an elevated proportion of FOXP3+IL-17+ cells. Genes Immun 2017;18:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Honkanen J, Nieminen JK, Gao R, et al. IL-17 immunity in human type 1 diabetes. J Immunology 2010;185:1959–1967 [DOI] [PubMed]
- 77.Blahnik G, Uchtenhagen H, Chow IT, et al. Analysis of pancreatic beta cell specific CD4+ T cells reveals a predominance of proinsulin specific cells. Cell Immunol 2019;335:68–75 [DOI] [PubMed] [Google Scholar]
- 78.Brooks-Worrell B, Gersuk VH, Greenbaum C, Palmer JP. Intermolecular antigen spreading occurs during the preclinical period of human type 1 diabetes. J Immunology 2001;166:5265–5270 [DOI] [PubMed]
- 79.Brooks-Worrell BM, Starkebaum GA, Greenbaum C, Palmer JP. Peripheral blood mononuclear cells of insulin-dependent diabetic patients respond to multiple islet cell proteins. J Immunology 1996;157:5668–5674 [PubMed]
- 80.Tooley JE, Vudattu N, Choi J, et al. Changes in T-cell subsets identify responders to FcR-nonbinding anti-CD3 mAb (teplizumab) in patients with type 1 diabetes. Eur J Immunol 2016;46:230–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fourlanos S, Perry C, Gellert SA, et al. Evidence that nasal insulin induces immune tolerance to insulin in adults with autoimmune diabetes. Diabetes 2011;60:1237–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harrison LC, Honeyman MC, Steele CE, et al. Pancreatic β-cell function and immune responses to insulin after administration of intranasal insulin to humans at risk for type 1 diabetes. Diabetes Care 2004;27:2348–2355 [DOI] [PubMed] [Google Scholar]
- 83.Axelsson S, Chéramy M, Hjorth M, et al. Long-lasting immune responses 4 years after GAD-alum treatment in children with type 1 diabetes. PLoS One 2011;6:e29008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hjorth M, Axelsson S, Rydén A, Faresjö M, Ludvigsson J, Casas R. GAD-alum treatment induces GAD65-specific CD4+CD25highFOXP3+ cells in type 1 diabetic patients. Clin Immunol 2011;138:117–126 [DOI] [PubMed] [Google Scholar]
- 85.Cole DK, Bulek AM, Dolton G, et al. Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. J Clin Invest 2016;126:2191–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.