Abstract
The Genes for Good study uses social media to engage a large, diverse participant pool in genetics research and education. Health history and daily tracking surveys are administered through a Facebook application, and participants who complete a minimum number of surveys are mailed a saliva sample kit (“spit kit”) to collect DNA for genotyping. As of March 2019, we engaged >80,000 individuals, sent spit kits to >32,000 individuals who met minimum participation requirements, and collected >27,000 spit kits. Participants come from all 50 states and include a diversity of ancestral backgrounds. Rates of important chronic health indicators are consistent with those estimated for the general U.S. population using more traditional study designs. However, our sample is younger and contains a greater percentage of females than the general population. As one means of verifying data quality, we have replicated genome-wide association studies (GWASs) for exemplar traits, such as asthma, diabetes, body mass index (BMI), and pigmentation. The flexible framework of the web application makes it relatively simple to add new questionnaires and for other researchers to collaborate. We anticipate that the study sample will continue to grow and that future analyses may further capitalize on the strengths of the longitudinal data in combination with genetic information.
Keywords: genome-wide association study, social media, direct to participant research, participant engagement, population study, complex traits, genotyping array, body mass index, diabetes, asthma
Introduction
More than 10,000 genetic loci have been successfully linked to common and complex diseases.1 In previous decades, the major challenge for human genetic studies was the cost and complexity of the genotyping itself; however, researchers now face the bigger hurdle of obtaining large enough samples that also include useful, linked medical and health data. The study designs typically used to collect such data are expensive and often exclude individuals based on location or demographics. We reasoned that using social media platforms would not only allow us to recruit a large population cohort, but also help us to reach populations that might not typically participate in genetic studies due to the time commitment or distance to a research center. Potential advantages of social media-based study designs include the ability to reach diverse populations and the ability to engage participants in research over time. Potential concerns include representativeness and the ability of this approach to reproduce findings obtained using more traditional designs.
We present a new study design to take advantage of recent developments in health survey methods using social media and widespread interest in direct-to-consumer genetic testing.2, 3 Genes for Good is an ongoing, large-scale study of health, genetic, and behavioral information. We aim to engage tens of thousands of individuals in research through a Facebook application, reducing the expense of traditional epidemiologic designs and the exclusivity and high socioeconomic status associated with current direct-to-consumer efforts.4
Our model of using social media for genetic research invites participants to complete online health assessments at their convenience, as has been successfully applied in numerous studies of health, behavior,5 and psychology,6 including studies of rare genetic diseases (J.E. Abiad et al., 2018, ACMG Ann. Clin. Genet. Meeting, abstract), childbirth preferences,7 and prediction of personality traits.6 When a consenting participant has completed a minimum number of health history and health tracking surveys, they are mailed a spit kit to collect DNA for analysis. After genotyping, we test genetic variants for association with health, disease, and environmental information collected through online assessments.
In this paper, we demonstrate that the Genes for Good study model is a viable complement to more traditional research study designs. The phenotypic and genotypic data we have collected thus far appear valid and reliable. Further, the incentive structure of Genes for Good—namely, altruism combined with the return of survey response summaries and genetic data to participants—is effective, as demonstrated by exponential recruitment from all 50 US states. Importantly, the recruitment happened organically, with participants publicizing the study through their own networks, without relying on paid advertising. We briefly explored the use of study recruitment websites (such as ResearchMatch8), but only several hundred participants were recruited this way. We also saw large influxes of participants after online articles appeared in Reddit and Buzzfeed (Web Resources). While resources still go toward answering questions about the study and resolving technical issues, efficient participant recruitment and engagement allowed us to dedicate a larger fraction of resources to sample collection, processing, and downstream analyses. The long-term goals of the study fall broadly into five main categories: (1) to identify novel genetic loci associated with a variety of phenotypes, (2) to longitudinally track an array of health and behavioral measures, (3) to enable genotype-first study designs (such as detailed phenotypic assessments of participants with naturally occurring knockout variants), (4) to educate participants and make the data available to them, and (5) to encourage data sharing among researchers. Here, we present our study design and methods, as well as initial findings about our sample demographics and important health indicators.
One particular advantage of hosting our study on social media is that we can reach participants in an environment that many already visit regularly as part of their daily routines. Social media use in the US has dramatically increased in the last decade—rising from 7% in 2005 to more than 65% in 2015 according to the Pew Research Center (see Web Resources)—and so we have the potential to reach a majority of the US population through our application. In the last few years, several research groups have recognized the major advantages social media offers: flexible timing, the possibility of incentives and reminders, and the ability to reach non-urban communities. There has already been substantial success in recruiting for studies via Facebook9 as well as in using it to prevent loss-to-follow-up.10 Further, the flexible framework of Genes for Good allows us and other research groups to continue adding new surveys and activities to address future research questions. Our study takes advantage of the opportunity for repeated contact that social media offers and represents the first large genetic study of tens of thousands of individuals conducted via Facebook.
Considering their ubiquity and ease of use, social media and mobile devices as research tools are important avenues to explore further.11 However, we recognize some of the potential disadvantages we are likely to face: (1) inaccurate data, (2) low response rate,12 (3) high attrition, and (4) a sample limited to those who have a Facebook account. In the first year of the study, we prioritized testing and combatting several of these expected limitations. With the aforementioned challenges in mind, we implemented various methods to assess the quality of our data. First, we looked at common diseases and phenotypes to validate our results—and thus our approach to data collection—by comparing them to prior findings from traditional research and meta-analysis designs. When expected phenotypic relationships hold true, such as that between BMI and type 2 diabetes, we gain confidence in the quality of the survey responses we are collecting. Additionally, we assessed the quality of the genetic data by replicating findings from genome-wide association studies (GWASs) for a variety of traits that are known to have genetic components, such as diabetes, asthma, BMI, hair color, and eye color, confirming that our data yields the expected signals. We also examined rates of chronic health conditions, such as hypertension and diabetes, to explore how our study participants compare to the overall U.S. population.
Material and Methods
We have implemented a large, IRB-approved genetic study using social media. Participants must be at least 18 years old, live in the US, and have a Facebook account. They are recruited via snowball sampling, i.e., by finding our Genes for Good Facebook application through friends, family, and social media connections. Once a person has consented, they are invited to complete online health history assessments at their convenience. The surveys consist of health history questionnaires, daily tracking surveys, and an optional health conditions module in which participants can list other conditions that they have. Once they have completed a minimum number of required questionnaires, they are mailed a spit kit to collect DNA for analysis. The cost of each participant is about $80, which includes postage, DNA extraction, and genotyping; there is essentially no cost associated with recruitment or data collection. Throughout the course of the study, we have typically employed 2–3 full-time staff (study coordinator, developers), several graduate and undergraduate students, and a part-time administrative assistant to assist with sending and receiving spit kits.
Genetic Analysis
DNA is genotyped at ∼600,000 SNPs using either the Illumina Infinium CoreExome-24 v.1.0 or v.1.1 arrays, which include both nonsynonymous exonic variants and a panel of common genome-wide markers (see Web Resources). The standard set of markers on the array is augmented with missense, loss-of-function, and potential lipid- and myocardial infarction-associated variants identified in the HUNT whole-genome sequencing and whole-exome sequencing projects;13 height-associated variants from GIANT;14 potential stop-gain variants in 96 genes at loci potentially implicated in type 2 diabetes, blood lipid levels, Alzheimer disease, nicotine/alcohol metabolism, and several others with mutations implicated in serious but treatable health conditions; complex trait-associated variants in the EBI/NHGRI GWAS catalog;1 a random subset of Neanderthal SNPs from the 1000 Genomes Project;15 ancestry informative markers identified by Paschou et al. that were highly correlated with the principal components of Human Genome Diversity Project samples;16 and pain-related variants proposed by Dr. Chad Brummett of the University of Michigan Division of Pain Research. Genotypes at an additional >30 million variants in the 1000 Genomes Phase 3 panel17 are imputed using Minimac3.18 After quality control, local genetic ancestry is estimated using RFMix,19 global ancestry with ADMIXTURE,20 and principal components analysis performed with TRACE,21 using the Human Genome Diversity Project samples as a reference panel22 for all three analyses. We provide each Genes for Good participant with a section in the app to view these estimates of genetic ancestry on the sample they provided.
For the GWAS of Genes for Good participants’ BMI, the BMI measurements were calculated from the height and weight survey in the app, which was derived from height and weight questionnaires available from PhenX Toolkit.23 Weight measurements for the first several thousand genotyped participants were bottom-coded at 80 lb and top-coded at 251 lb; then, the top-coded value was changed to 381 lb partway through the study to capture a greater range of variation. For participants that were pregnant at the time of answering the survey, we used their pre-pregnancy weight obtained from the same survey. The BMI values were then regressed on sex, age, array chip version, and the first five principal components; the residuals were inverse-normal transformed in order to compare effect size estimates to the largest published meta-analysis of BMI24 and to reduce the impact of extreme observations. We used the SAIGE software25 to run a mixed model GWAS, accounting for sample relatedness and population structure. Polygenic risk scores were calculated using PLINK.26
Participant Engagement
We provide participants with several ways to interact with both their own data and the research study as a whole. After each health history survey is completed, we provide charts summarizing the information, in some cases comparing each participant’s answers to the Genes for Good study population (example in Figure 7). Similarly, for daily tracking surveys, we generate summaries of each participant’s health behavior over time as well as summary statistics for the entire study (example in Figure 8). In addition to providing this ongoing feedback and summary of the survey responses, we also offer participants who submit a sample a breakdown of their genetic ancestry; the current version includes seven continental human populations (Europe, Africa, East Asia, Central/South Asia, West Asia/North Africa, Americas, and Oceania), and results are served in the form of a global ancestry estimate, local ancestry inference, and principal components analysis using the methods described previously (RFMIX, ADMIXTURE, TRACE). Before seeing their estimates of genetic ancestry, they are required to watch a short video on how to interpret their results. Participants can also download their array and imputed genotypes.
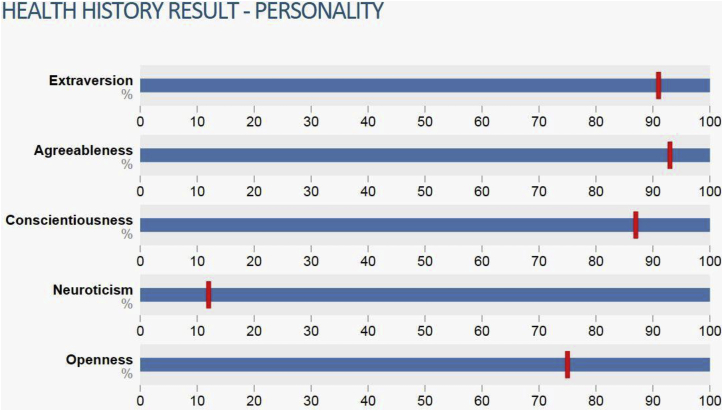
Figure 7.
Example Health History Result
An example of how participants’ results to the personality survey are displayed within the Genes for Good app. The bars show this participant’s percentile scores on the five personality attributes measured by the survey.
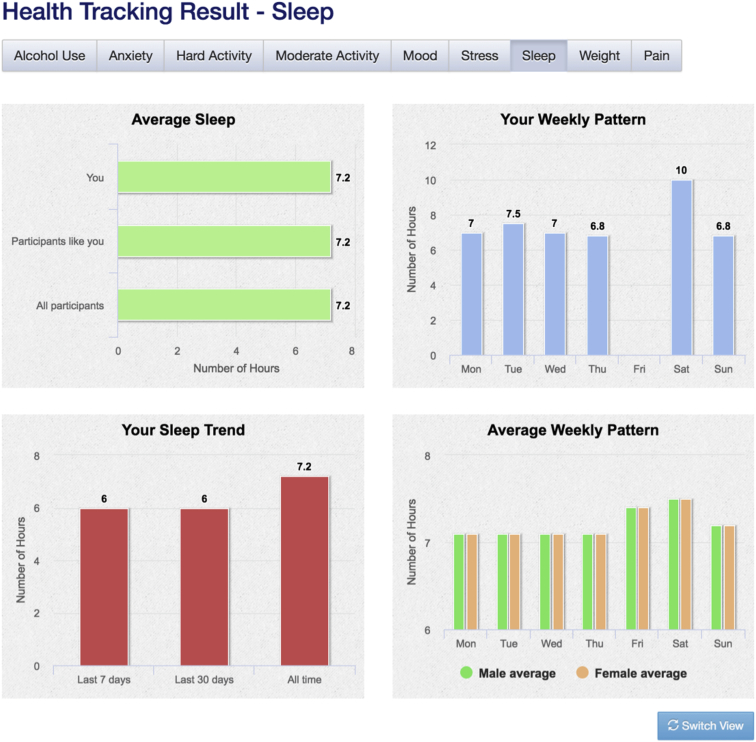
Figure 8.
Example Daily Tracking Result
An example of how participants’ answers to the daily sleep tracking survey are displayed, showing (A) average hours of sleep for this participant, compared to other participants of the same age range and sex, and to all other Genes for Good participants, (B) average hours of sleep reported for different days of the week when this participant has taken the survey, (C) average hours of sleep over the past 7 days, past 30 days, and over all responses from this participant, and (D) average hours of sleep reported for different days of the week for all Genes for Good participants stratified by sex.
Privacy and Data Security
All Genes for Good data are divided into two classes: (1) personally identifiable information, such as email addresses, Facebook user IDs, and physical mailing addresses; and (2) research information, such as survey responses and genetic data. Each class of data is stored in a distinct relational database and served from a distinct server. Extracts for outside researchers include only research-specific data. We plan to ask participants to allow use of their mailing address to link to information such as geocode pollution, built environment (for instance, the number of fast food outlets or public parks within a certain radius of one’s home), and census tract data. In these cases, the participants’ physical address would still be withheld from external collaborators, but variables generated using addresses could be shared upon request.
The privacy of Genes for Good data is monitored by the University of Michigan Institutional Review Board. All genetic and survey results are stored in a secure server on campus that is not directly connected to the public internet, and DNA samples are stored in physically secure spaces with restricted access. In addition, all archived data are de-identified to protect subject privacy including participants’ demographic summary and genetic information. Even though Genes for Good uses Facebook to authenticate login, Facebook does not access information we collect through the app and we do not use participant’s social media postings and connections in our research. We make efforts to communicate with participants about the extensive measures we take in ensuring the privacy of their data and to ease their worries about using social media as a platform for genetic research.
All communication to and from the application is encrypted. Participants are authenticated using a Facebook account and Facebook’s OAuth implementation, ensuring that participants have access only to their own data once inside the application. Communication with Facebook servers is limited to authentication only; although Facebook receives and retains information about which Facebook accounts have accessed the Genes for Good app, all other information provided by participants is provided directly to Genes for Good servers. Facebook cannot see any of the data entered by participants.
Once participants have their genetic data analyzed, they are notified that they may access results inside the app with a Results Access Code, a randomly generated alphanumeric code that must be requested by the participant and will be delivered to the email address on the participant’s Genes for Good profile. Participant genotype data is processed internally on University of Michigan servers and is distributed to participants upon request via Box, a secure third-party file-sharing platform. Participants may request their raw genotypes as often as they like from within the genetic results section of the app. Each request compresses and uploads raw genotype data and supplementary information to a private, password-protected Box account directory. For security purposes, all requested genotypes automatically expire from Box servers 3 days after being uploaded.
Results
Since the launch of Genes for Good on January 19th, 2015 (Martin Luther King Jr. Day), we have seen steadily increasing participant recruitment and consistent use of the Facebook application. Genes for Good now has enough participants to begin conducting meaningful analyses with the data. As of March 2019, 117,652 participants had tried the app, with 81,110 signing the electronic consent form. Consenting users have completed more than 2.9 million surveys, answering >22 million questions. Genes for Good has mailed 33,427 spit kits to eligible participants, of which 27,470 have been returned (as of March 2019). The genetic data freeze used for this paper contains data from 20,232 participants whose genotypes passed quality control checks as of mid-2018.
Sample Characteristics and Phenotypes
Participants were recruited successfully from all 50 states, with areas of peak participant density roughly overlapping with major US population centers (Figure 1). About 90% of users have residential addresses outside of Michigan. Compared to the US population, our sample is younger (Genes for Good median age of 33, US adult median age of 44) and enriched for females (74% of participants are women, compared to 51% for US adults, Table 1). Our sample closely resembles the US population on household income, although it is enriched for individuals from middle-income households with an annual income of $35,000–$100,000; Table 2). In contrast, the majority of the participants in the research cohort from 23andMe are from households with an annual income more than $100,000 (J.Y. Tung et al., 2011, ASHG, abstract). To confirm the quality of the data collected from our sample, we also compared disease rates to those in the general US population (Table 3). In looking at important risk factors for cardiovascular disease, we observed relatively similar rates of high cholesterol, hypertension, and smoking. However, our sample had lower rates of disease outcomes such as stroke and myocardial infarction. Our genotype data freeze contained 20,232 individuals, of which 76.3% were non-Hispanic white, 3.8% Asian, 2.7% African American, 8.8% multi-racial/other, and 8.3% Hispanic/Latino as determined by self-report through our demographics survey.
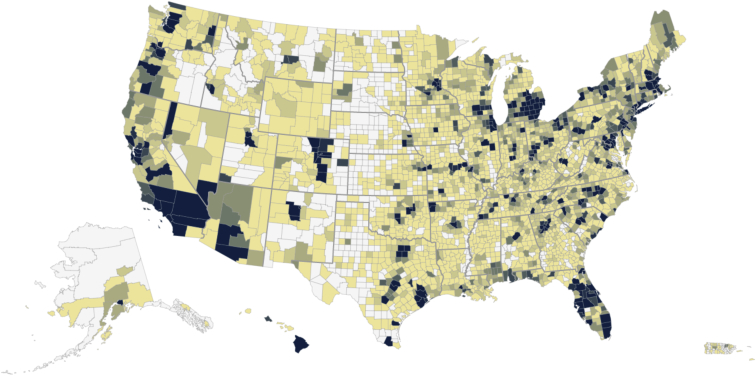
Figure 1.
Geographic Distribution
The geographic distribution of Genes for Good participants as of October 2017. The colors indicate the number of participants who have logged into the app from that county, with darker colors representing higher density.
Table 1.
Demographics
| Genes for Gooda | US Populationb | Facebook-Using Populationc | |
|---|---|---|---|
| Age | |||
| Median, years | 33 | 44d | |
| 18–24 | 17.0% | 13.2% | 19.5% |
| 25–34 | 37.1% | 17.1% | 27.0% |
| 35–44 | 21.6% | 16.4% | 19.6% |
| 45–54 | 11.9% | 18.3% | 16.5% |
| 55+ | 12.4% | 35.5% | 17.4% |
| Sex | |||
| Male | 25.9% | 49.2% | 49% |
| Female | 74.1% | 50.8% | 51% |
Data source for our study data is based on all valid responses as of August 9th, 2017
Data for US population from the 2010 U.S. Census51
Data for Facebook population from Statistica (see Web Resources)
Median age of US persons over age 18 reported in the US 2010 Census
Table 2.
Income Distribution
| Income Category | Genes for Good (%) | US Populationa(%) | 23andMeb(%) |
|---|---|---|---|
| Less than $35,000 | 28.0 | 30.2 | 10.2 |
| $35,000 to $50,000 | 18.9 | 12.9 | 7.2 |
| $50,000 to $75,000 | 19.8 | 17.0 | 13.9 |
| $75,000 to $100,000 | 14.5 | 12.3 | 14.7 |
| More than $100,000 | 18.9 | 27.7 | 54.0 |
Distribution of household income among Genes for Good participants based on answers to the demographics survey as of August 9, 2017, compared to the general US population.
Data from US Census Table H-1727
Data describing 23andMe research cohort approximated from 2011 ASHG poster (J.Y. Tung et al., 2011, ASHG, abstract)
Table 3.
Chronic Health Indicators in Study Sample Compared to Overall US Population
| Genes for Gooda | US Populationb | |
|---|---|---|
| BMI, mean, kg/m2 | 29.80 | 29.38 |
| Underweight (BMI < 18.5) | 1.9% | 1.6% |
| Normal weight (BMI 18.5–24.9) | 31.6% | 27.2% |
| Overweight (BMI 25–29.9) | 26.0% | 31.6% |
| Obese (BMI ≥ 30) | 40.4% | 39.7% |
| High cholesterol | 26.1% | 29.3% |
| Hypertension | 24.9% | 29% |
| Previous stroke | 1.3% | 2.9% |
| Previous MI | 1.5% | 4.5% |
| Diabetes (type 1 or 2) | 6.5% | 9.3% |
| Current smoker | 17.0% | 15.1% |
Data source for our study data is based on all valid responses as of August 9th, 2017
Data from nationally representative samples to determine US rates of obesity (see CDC: National Health and Nutrition Examination Survey Data in Web Resources), high cholesterol, hypertension,28 stroke,29 MI, diabetes, and smoking30
In addition to the phenotype information collected from survey responses, 12,216 participants have reported 64,401 cases of 3,067 health conditions in an optional section of the app that allows participants to search for and report disorders using the Systematized Nomenclature of Medicine (SNOMED) dictionary.31 These participant-entered data show that Genes for Good has attracted an unusually high proportion of individuals with certain rare diseases, like Ehlers-Danlos syndrome (565 cases or 0.93% of GfG participants compared to ∼0.02% prevalence worldwide) (see GeneReviews in Web Resources). The 5 most commonly reported disorders were generalized anxiety disorder (1,803 cases), asthma (1,389), hypothyroidism (941), depressive disorder (920), and migraine (918). Higher BMI was associated with increased risk for all 5 conditions in logistic regression of each of the five traits on BMI, sex, and age (odds ratios of 1.02, 1.03, 1.04, 1.01, 1.03 per unit higher BMI, p values of 7.6 × 10−9, 2.1 × 10−20, 3.9 × 10−24, 1.5 × 10−4, 6.3 × 10−14).
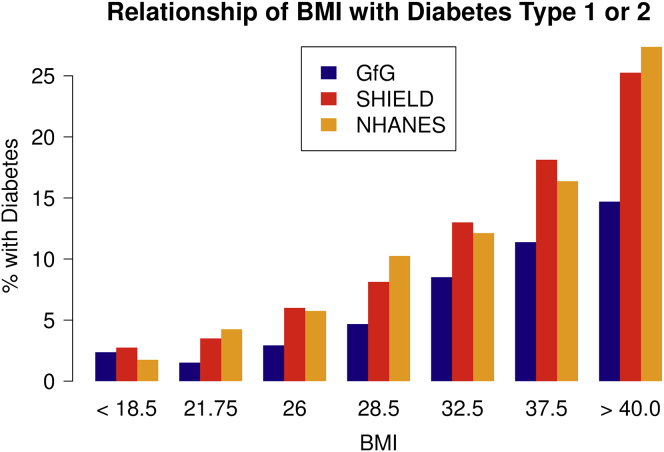
To evaluate the quality of our data, we used our survey data to verify known phenotypic relationships. Taking diabetes as an example, we analyzed the association of the disease with BMI. Given the rapidly increasing prevalence of diabetes in the US, this is a particularly important outcome to examine. Over the past three decades, the number of diagnosed Americans has more than tripled, from 5.6 million in 1980 to 21 million in 2012 (see CDC: National Diabetes Statistics Report in Web Resources). And because about one-third of diabetics are undiagnosed, national survey statistics consistently underestimate the true prevalence of diabetes (see CDC: National Diabetes Statistics Report in Web Resources). We compared rates of diabetes in our sample, within each BMI bracket, to those reported from nationally representative samples32 and found a similar trend of increasing diabetes prevalence as BMI increased (Figure 2). We further explored this relationship by calculating the estimated effect of BMI on diabetes status, adjusting for age, sex, and race, using NHANES and Genes for Good data separately. We found that the relationship between BMI and diabetes was comparable between studies (95% CI for odds ratio per 1-unit increase in BMI, NHANES: 1.07–1.10; 95% CI, GFG: 1.08–1.10). When comparing simple correlation coefficients between BMI and diabetes status across studies, we found no notable difference between Genes for Good and NHANES (rGFG = 0.18, rNHANES = 0.19, p = 0.83). Though our sample is quite different from NHANES in terms of wealth, age distribution, and ethnic diversity, we observe similar trends in both cohorts when comparing diabetes-affected case subjects and control subjects: diabetics typically have higher rates of obesity, higher age, lower income, and lower education (Table S1).
Figure 2.
Relationship between BMI and Diabetes Rates in Participants Is Consistent with that Seen in the General US Population
Type 2 diabetes is a phenotype of particular interest because of its increasing prevalence, impact on cardiovascular health, and relatively well-characterized genetics. Here, we have compared the rates of diabetes in Genes for Good participants to the rates found in the nationally representative studies SHIELD and NHANES.32
Genetic Associations
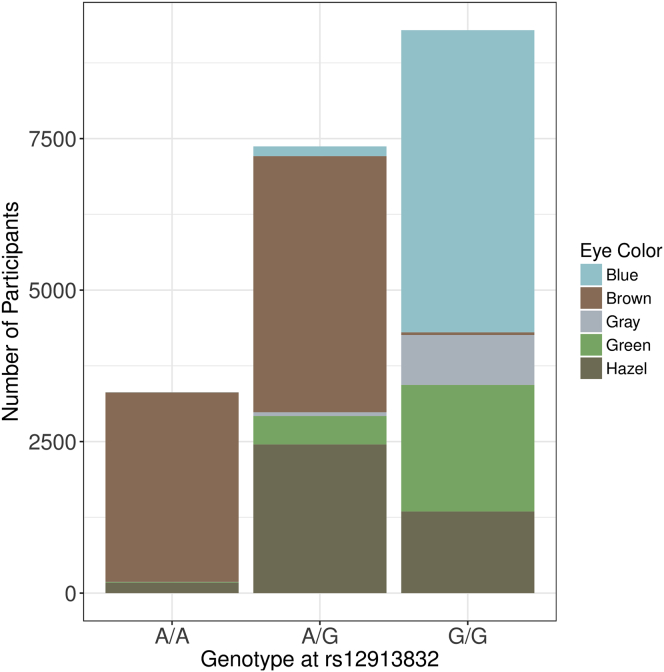
To validate the quality of our self-reported phenotypes, we analyzed a data freeze of 20,232 genotypes to see whether we could replicate known genetic associations. We first analyzed traits related to pigmentation and BMI, because these traits are known to have strong genetic factors. For example, most variation in eye color is determined by six SNPs in HERC2 and OCA2.33 Figure 3 shows the number of participants with each combination of eye color and genotype at one of the SNPs with the strongest association signal, rs12913832. We observed strong evidence of association between eye color and genotype (X2 = 15,599, df = 8, p = 10−3376, n = 19,974), and the direction of effects is consistent with what was previously reported. Other pigmentation traits like hair color, skin sun response, and hair texture are also consistent with prior studies. Table S2 shows detailed GWAS results, and Table S3 compares our results to several larger studies. We show that Genes for Good replicates the top pigmentation associations in prior studies at least nominally (p < 0.05) and frequently does so at genome-wide significance (p < 5 × 10−8).
Figure 3.
Eye Color Distribution
Distribution of eye color among participants with different genotypes at rs12913832 (the top signal when performing GWAS using blue eye color in Genes for Good participants), a marker in HERC2 known to play a role in eye color determination.
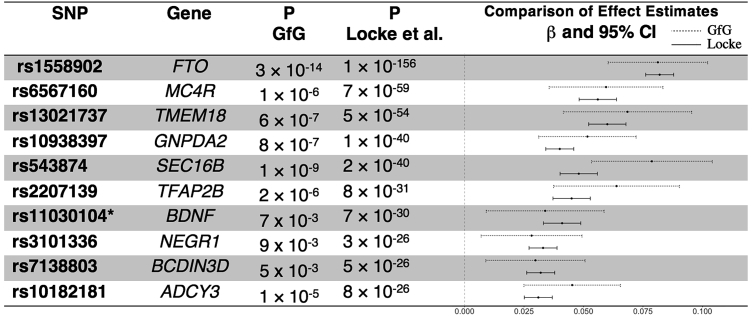
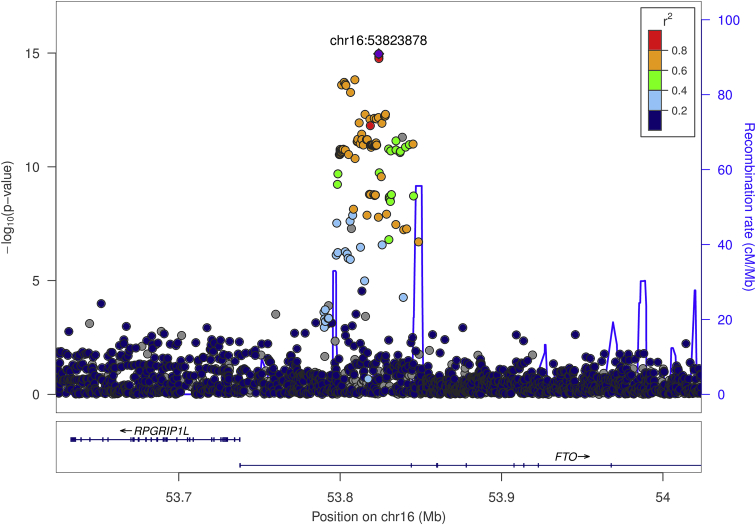
We next compared results for a mixed model GWAS of BMI, using measurements obtained from the height and weight health history survey, to results from the GIANT consortium.24 We obtained effect sizes consistent with those published for the top ten GIANT loci. We also obtained nominally significant (p < 0.05) association results at all ten loci. Figure 4 summarizes the comparison of our results with published GIANT results, showing consistency of direction of effect, magnitude, and relative significance (Figure 5 shows regional association in our top signal, at FTO). Given the relatively small sample size of our data, our effect estimates necessarily have wider confidence limits compared to the meta-analysis. However, the meta-analysis point estimates are contained within these limits for nearly every SNP, which provide evidence that self-reported phenotypes collected within our cohort are reliable.
Figure 4.
Effect Size Estimates of a GWAS for BMI in Our Study Sample Compared to Findings from a Meta-analysis
We compare effect estimates from Genes for Good to published findings from the Locke et al. meta-analysis of BMI GWAS.24 Specifically, we looked at the top ten reported signals and were able to replicate all of these effects in direction and nominal significance (p < 0.05). The forest plot on the right compares effect size estimates across studies; the dashed lines represent the confidence intervals around the Genes for Good estimates, while the solid lines represent results from Locke et al. Given the relatively small sample size available in this data freeze, our estimates have fairly wide confidence limits. However, Locke’s estimates are completely contained within our limits for eight of ten SNPs. Asterisk indicates imputed variant.
Figure 5.
LocusZoom Plot Showing Single-Variant Association Results for BMI in FTO
This result is consistent with other studies that reported their strongest evidence for association in this gene. The effect size at the nearby SNP rs1558902 (0.081) was consistent with the effect size (0.081) reported previously in Locke et al.24
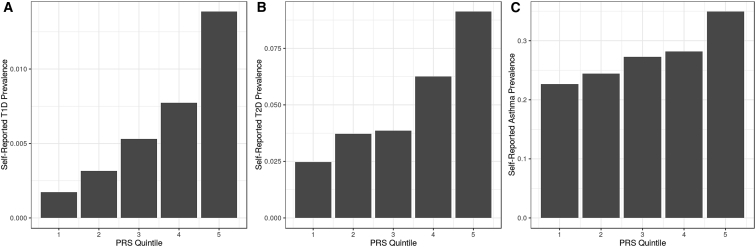
We next expanded our comparison of GWAS results obtained with Genes for Good data to include the traits of type 1 diabetes, type 2 diabetes, and asthma. For all traits except asthma, our association signals are consistent with reports from published large GWASs and show some significant hits (Tables S2, S3, and S4; Figure S1). Our asthma analysis did not give any genome-wide significant results, but when we examined the 18 SNPs associated with asthma in the study of Demenais et al.,34 we found that all had a consistent direction of effect in Genes for Good data but with smaller effect sizes (Table S4). Our asthma-affected case and control subjects were defined based on answers to “Was your asthma ever confirmed by a doctor?” with 4,378 case subjects and 11,715 control subjects reported. Given the large proportion of case subjects (27.2%), we believe that some individuals who answered “yes” did not meet the standard for an asthma diagnosis used in Demenais et al.34 A similar observation has been made in other studies of self-reported phenotypes—for example, in a study of psoriasis including data from 23andMe customers, it was estimated that only ∼36% of individuals who self-reported having psoriasis met the criteria used in clinical studies, diluting association signals and effect size estimates.35 We did an adjustment proposed by Duffy et al. to account for the apparent over-reporting of case subjects.36 We also did a power calculation at the 0.05 significance level to determine our ability to replicate the findings in Demenais et al. and estimated that we should replicate approximately 7 of 18 SNPs (summing estimated power across 18 variants gives expected number of 6.8 replicated signals). After the Duffy adjustment, more than half of our odds ratios were closer to the effect sizes reported in Demenais et al., though some odds ratios were overcorrected to have effect sizes larger than those reported in Demenais et al. As our power calculation suggested, we were able to replicate 7 of the 18 SNPs at the 0.05 significance level (Table S4).34, 35 Reassuringly, we also found that, when we calculated polygenic risk scores (PRS) for type 1 and type 2 diabetes using publicly available GWAS summary statistics,37, 38 PRS for type 2 diabetes was strongly associated with self-reported type 2 diabetes status (OR increase per PRS quintile = 1.47; p = 7.63 × 10−37) and that PRS for type 1 diabetes PRS was strongly associated with self-reported type 1 diabetes status (OR increase per PRS quintile = 1.66; p = 5.13 × 10−9) (Figure 6). We found similar support for an association between asthma PRS and self-reported asthma (OR increase per PRS quintile = 1.16; p = 3.17 × 10−26) (Figure 6).
Figure 6.
Prevalence for Self-Reported Type 1 and Type 2 Diabetes across Polygenic Risk Score Quintiles (Five Bins of Equal Sample Size)
An increase in the genetic risk score is associated with increasing prevalence of disease. We also evaluated associations between polygenic risk score quintile and type 1 diabetes, type 2 diabetes, and asthma status, adjusted for age and sex. We found that all three self-reported traits were significantly associated with calculated PRS quintile (pT1D = 5.13 × 10−9, pT2D = 7.63 × 10−37, pasthma = 3.17 × 10−26).
Somewhat unexpectedly, we observed that in our type 2 diabetes results the signal at CDKAL1 was stronger than at TCF7L2, which is typically the top signal reported for type 2 diabetes GWASs. Hypothesizing that this might be due to the younger age of Genes for Good participants, we split the Genes for Good data at the median age to test for changes in diabetes risk between the below-median age and above-median age groups for the TCF7L2 and CKDAL1 variants (median age = 32; casesBelow-Median = 65, controlsBelow-Median = 8,385; casesAbove-Median = 722, controlsAbove-Median = 7,728). Although we saw a trend to a larger diabetes risk for carriers of the TCF7L2 variant rs7903146 in the above-median group (ORBelow-Median = 1.21, ORAbove-Median = 1.34), we saw the same trend for carriers of the CDKAL1 variant rs7756992 (OR Below-Median = 1.04, ORAbove-Median = 1.37). Regardless, the differences between the below-median and above-median age groups for both SNPs were not significant (p > 0.05).
Discussion
We set out to recruit a large, diverse sample of engaged volunteers that might provide information about the diverse US population. For each volunteer, we used surveys to collect health and behavioral data that might inform a variety of genomic research studies. With rapid and inexpensive recruitment, we have quickly developed a participant pool with which to validate the quality of the data. We are optimistic about our ability to obtain the large sample size required for valid genetic association studies of complex diseases and behaviors. With our current analysis of 20,232 individuals, we have successfully validated several known genotype-phenotype relationships and contributed to several consortium meta-analyses.39, 40, 41, 42
We have good representation with respect to geography, age, and gender, though our sample does have some noticeable differences from a sample of random U.S. adults. One characteristic that presents both an opportunity and a challenge is the younger age of Genes for Good participants compared to the US adult population. While a younger demographic may be more interesting for some measures (behavioral data, activity levels), it will be less useful for others (age-associated cancers and development of other late-onset chronic disease). We do see slightly lower rates of the chronic conditions examined here compared to the general US population, which we attribute to the lower average age of our participants; even if participants have the relevant risk factors, they may not have had the time to develop those long-term outcomes. For instance, we see much lower rates of heart attack in our participants despite comparable hypertension rates, and we see lower rates of type 2 diabetes despite comparable BMI (Figure 2). At the same time, Genes for Good’s recruitment strategy may have led to an enrichment of individuals with certain rare diseases like Ehlers-Danlos syndrome, perhaps because of network effects within these communities.
Most participants completed the minimum number of health history surveys required to receive a spit kit (15 surveys), with many going well above that number. Completion of daily tracking surveys was modest, with most genotyped participants completing only the minimum number required to obtain a spit kit. None of our surveys are mandatory and it is certainly possible that participants will avoid surveys that are more onerous or which they are not comfortable with, introducing ascertainment biases (for example, individuals who are not skilled at reasoning puzzles might choose to skip the reasoning). The most completed surveys were generally those that appear higher in the list of available surveys within our app (Figure S2; Figure S3 provides additional details of survey completion rates).
Another challenge we face is that our sample is heavily skewed female. While targeted recruitment in the future may bring the gender distribution into balance, we also recognize the immediate potential to conduct a large-scale study of women’s health and have implemented relevant survey measures regarding polycystic ovarian syndrome and pregnancy outcomes.
Genetic Information, Privacy, and Ethics
There are a number of incentives for participation in Genes for Good besides the altruistic contribution and potential positive impact of genetics research on society. First, we provide interactive graphs and visualizations by which users can compare their survey responses to those of other participants (examples in Figures 7 and 8). Second, Genes for Good allows participants to view estimates of their genetic ancestry and download their raw genetic data, which some have argued should be the fundamental right of participants who contribute DNA to research.38 When downloading genetic data, we require participants to review a short slide show explaining that the data we generate are suitable for a research study but do not meet the standards used for clinical genetic tests. We emphasize that, compared to the data used in clinical tests, research data might be more susceptible to error. Around 70% of participants with genotypes available have requested a download link for their raw genetic data, which we provide in 23andMe format, a format known to be widely accepted at third-party interpretation sites. Many participants have told us they upload their data to third-party sites to obtain more detailed ancestry estimates, find DNA relatives, and even seek health interpretation. A recent review paper43 investigating reactions to a clinical genetic risk assessment concluded that in general, patients do not engage in risk-reducing behavior after receiving information about genetic predisposition. We expect that Genes for Good participants are unlikely to base major health or life decisions on the research-grade data we have returned. In addition, we will continue to develop Genes for Good web-based software applications to promote literacy of individuals about their genetic information.
Along with raw genetic data, we also return to participants their genetic ancestry information based on DNA analysis. The primary anticipated risk of the return of ancestry information is the discovery or suspicion of non-paternity and/or secret adoption by participants, i.e., discovering that one’s ancestry is inconsistent with what the participant knows about the ancestry of their supposedly biological parents. This has the potential to cause emotional or psychological stress on participants and their families, and we provide education about this risk during the informed consent.
Significance and Future Directions
The online platform implemented in Genes for Good is a viable study design for population-based genetic research. Now in the study’s fourth year, we have already had great success in recruitment, health history survey analysis, and genetic analysis. We are currently exploring the more than 300 phenotypes collected so far and continue to participate in ongoing collaborations. As the sample size grows, our power to detect novel associations and our ability to contribute more meaningful data to researchers will increase.
The flexibility of the study design and our ongoing relationship with participants also makes it possible to implement new methods of data collection with relative ease. Additional data collection techniques are being developed and validated in a wide array of studies, including wireless sensors for continuous collection of data related to physical activity,44, 45 heart rate,46 body temperature, sleep,47 and GPS location logging to infer habits and environmental exposures.48 These measures and more are currently available through a combination of smartphone and wrist sensors (e.g., FitBit), and many more wireless sensors exist for more specialized tasks (e.g., breathalyzers, insulin levels, QT interval). These and other novel data collection methods are developing rapidly, holding great promise in the near future for the efficient collection of large quantities of precise longitudinal data with minimal participant burden. The implementation of such devices would facilitate the collection of tracking data within Genes for Good.
Having verified the quality of our data and several known associations with particular loci, we are now poised to begin exploring new genotypic-phenotypic relationships, such as those with behavioral and health tracking information. Research in other settings with Genes for Good data show that our results are consistent with those of prior studies. Liu et al.49 show that a PRS calculated from SSGAC’s educational attainment data is effective in predicting 4% of the trait variance, which is consistent with previously reported out-of-sample predictive power for educational attainment.50 We are also working to streamline data sharing methods to facilitate collaborations with other researchers. Finally, we are actively developing new tools to provide participants with meaningful data summaries at the personal and study level. We believe that these steps will keep participants engaged and invested in the genetic research and will also help encourage longitudinal survey completions.
As we seek opportunities for long-term funding of the study, we are currently not collecting spit kits from new participants. Although enrollment has decreased since we stopped offering spit kits (we currently collect only health survey responses), interest remains high, as evidenced by the email inquiries we receive on a weekly basis. We plan to collect and genotype additional samples when future funding becomes available; when doing so, we expect to implement several changes to study protocol that will solve issues observed throughout the course of the study. For example, we noticed that survey completion correlates with the order that the survey appears on the app homepage (Figure S2); we have recently randomized the order of survey display to remedy this.
Conflicts of Interest
G.R.A. is currently an employee of Regeneron Pharmaceuticals and the beneficiary of stock options and grants in Regeneron. Previously, he served on scientific advisory boards for 23andMe, Regeneron Pharmaceuticals, and Helix.
Acknowledgments
This research has been conducted using the UK Biobank Resource under application number 24460 (specifically, calculation of PRS for type 1 diabetes and asthma was conducted using GWAS results from UK Biobank). This study was supported by University of Michigan discretionary funds. Administrative support: Irene Felicetti, Stephanie Bachoura, Samantha Bachoura, Laura Baker. IT support: Sean Caron. UM sequencing core: Robert Lyons, Susan Dagenais, Christopher Krebs, David Erdody.
Published: June 13, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.05.006.
Contributor Information
Anita Pandit, Email: anitapan@umich.edu.
Gonçalo R. Abecasis, Email: goncalo@umich.edu.
Web Resources
Box compliance with HIPAA guidelines, https://www.box.com/industries/healthcare
BuzzFeed, https://www.buzzfeed.com/virginiahughes/a-new-facebook-app-wants-to-test-your-dna
CDC: National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States, https://www.cdc.gov/diabetes/pdfs/data/statistics/national-diabetes-statistics-report.pdf
CDC: National Health and Nutrition Examination Survey, https://wwwn.cdc.gov/Nchs/Nhanes/Search/DataPage.aspx?Component=Examination&CycleBeginYear=2015
GeneReviews, Levy, H.P. (2018). Hypermobile Ehlers-Danlos Syndrome, https://www.ncbi.nlm.nih.gov/books/NBK1279/
Genes for Good Facebook application, https://app.genesforgood.org
Genes for Good full text survey, https://genesforgood.org/for_researchers
Genes for Good informational website, https://www.genesforgood.org
Illumina Infinium CoreExome BeadChip, https://www.illumina.com/content/dam/illumina-marketing/documents/products/datasheets/datasheet_human_core_exome_beadchip.pdf
Pew Research Center, Social Networking Usage (2005-2015), https://www.pewinternet.org/2015/2010/2008/social-networking-usage-2005-2015
Reddit, https://www.reddit.com/r/freebies/comments/67v9c5/free_dna_test_from_the_university_of_michigan/
Statista: Distribution of Facebook Users, https://www.statista.com/statistics/187041/us-user-age-distribution-on-facebook
Statista: Number of Facebook Users, https://www.statista.com/statistics/398136/us-facebook-user-age-groups
Supplemental Data
References
- 1.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]; Welter, D., MacArthur, J., Morales, J., Burdett, T., Hall, P., Junkins, H., Klemm, A., Flicek, P., Manolio, T., Hindorff, L., and Parkinson, H. (2014). The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001-D1006. [DOI] [PMC free article] [PubMed]
- 2.Stoeklé H.C., Mamzer-Bruneel M.F., Vogt G., Hervé C. 23andMe: a new two-sided data-banking market model. BMC Med. Ethics. 2016;17:19. doi: 10.1186/s12910-016-0101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stoekle, H.C., Mamzer-Bruneel, M.F., Vogt, G., and Herve, C. (2016). 23andMe: a new two-sided data-banking market model. BMC Med. Ethics 17, 19. [DOI] [PMC free article] [PubMed]
- 3.Royal C.D., Novembre J., Fullerton S.M., Goldstein D.B., Long J.C., Bamshad M.J., Clark A.G. Inferring genetic ancestry: opportunities, challenges, and implications. Am. J. Hum. Genet. 2010;86:661–673. doi: 10.1016/j.ajhg.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Royal, C.D., Novembre, J., Fullerton, S.M., Goldstein, D.B., Long, J.C., Bamshad, M.J., and Clark, A.G. (2010). Inferring genetic ancestry: opportunities, challenges, and implications. Am. J. Hum. Genet. 86, 661-673. [DOI] [PMC free article] [PubMed]
- 4.Agurs-Collins T., Ferrer R., Ottenbacher A., Waters E.A., O’Connell M.E., Hamilton J.G. Public Awareness of Direct-to-Consumer Genetic Tests: Findings from the 2013 U.S. Health Information National Trends Survey. J. Cancer Educ. 2015;30:799–807. doi: 10.1007/s13187-014-0784-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Agurs-Collins, T., Ferrer, R., Ottenbacher, A., Waters, E.A., O’Connell, M.E., and Hamilton, J.G. (2015). Public Awareness of Direct-to-Consumer Genetic Tests: Findings from the 2013 U.S. Health Information National Trends Survey. J. Cancer Educ. 30, 799-807. [DOI] [PMC free article] [PubMed]
- 5.Pedersen E.R., Kurz J. Using Facebook for Health-related Research Study Recruitment and Program Delivery. Curr Opin Psychol. 2016;9:38–43. doi: 10.1016/j.copsyc.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pedersen, E.R., and Kurz, J. (2016). Using Facebook for Health-related Research Study Recruitment and Program Delivery. Curr Opin Psychol 9, 38-43. [DOI] [PMC free article] [PubMed]
- 6.Kosinski M., Matz S.C., Gosling S.D., Popov V., Stillwell D. Facebook as a research tool for the social sciences: Opportunities, challenges, ethical considerations, and practical guidelines. Am. Psychol. 2015;70:543–556. doi: 10.1037/a0039210. [DOI] [PubMed] [Google Scholar]; Kosinski, M., Matz, S.C., Gosling, S.D., Popov, V., and Stillwell, D. (2015). Facebook as a research tool for the social sciences: Opportunities, challenges, ethical considerations, and practical guidelines. Am. Psychol. 70, 543-556. [DOI] [PubMed]
- 7.Arcia A. Facebook Advertisements for Inexpensive Participant Recruitment Among Women in Early Pregnancy. Health Educ. Behav. 2014;41:237–241. doi: 10.1177/1090198113504414. [DOI] [PMC free article] [PubMed] [Google Scholar]; Arcia, A. (2014). Facebook Advertisements for Inexpensive Participant Recruitment Among Women in Early Pregnancy. Health Educ. Behav. 41, 237-241. [DOI] [PMC free article] [PubMed]
- 8.Harris P.A., Scott K.W., Lebo L., Hassan N., Lightner C., Pulley J. ResearchMatch: a national registry to recruit volunteers for clinical research. Acad. Med. 2012;87:66–73. doi: 10.1097/ACM.0b013e31823ab7d2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harris, P.A., Scott, K.W., Lebo, L., Hassan, N., Lightner, C., and Pulley, J. (2012). ResearchMatch: a national registry to recruit volunteers for clinical research. Acad. Med. 87, 66-73. [DOI] [PMC free article] [PubMed]
- 9.Fenner Y., Garland S.M., Moore E.E., Jayasinghe Y., Fletcher A., Tabrizi S.N., Gunasekaran B., Wark J.D. Web-based recruiting for health research using a social networking site: an exploratory study. J. Med. Internet Res. 2012;14:e20. doi: 10.2196/jmir.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fenner, Y., Garland, S.M., Moore, E.E., Jayasinghe, Y., Fletcher, A., Tabrizi, S.N., Gunasekaran, B., and Wark, J.D. (2012). Web-based recruiting for health research using a social networking site: an exploratory study. J. Med. Internet Res. 14, e20. [DOI] [PMC free article] [PubMed]
- 10.Mychasiuk R., Benzies K. Facebook: an effective tool for participant retention in longitudinal research. Child Care Health Dev. 2012;38:753–756. doi: 10.1111/j.1365-2214.2011.01326.x. [DOI] [PubMed] [Google Scholar]; Mychasiuk, R., and Benzies, K. (2012). Facebook: an effective tool for participant retention in longitudinal research. Child Care Health Dev. 38, 753-756. [DOI] [PubMed]
- 11.Steinhubl S.R., Muse E.D., Topol E.J. The emerging field of mobile health. Sci. Transl. Med. 2015;7:283rv3. doi: 10.1126/scitranslmed.aaa3487. [DOI] [PMC free article] [PubMed] [Google Scholar]; Steinhubl, S.R., Muse, E.D., and Topol, E.J. (2015). The emerging field of mobile health. Sci. Transl. Med. 7, 283rv3. [DOI] [PMC free article] [PubMed]
- 12.Kapp J.M., Peters C., Oliver D.P. Research recruitment using Facebook advertising: big potential, big challenges. J. Cancer Educ. 2013;28:134–137. doi: 10.1007/s13187-012-0443-z. [DOI] [PubMed] [Google Scholar]; Kapp, J.M., Peters, C., and Oliver, D.P. (2013). Research recruitment using Facebook advertising: big potential, big challenges. J. Cancer Educ. 28, 134-137. [DOI] [PubMed]
- 13.Krokstad S., Langhammer A., Hveem K., Holmen T.L., Midthjell K., Stene T.R., Bratberg G., Heggland J., Holmen J. Cohort Profile: the HUNT Study, Norway. Int. J. Epidemiol. 2013;42:968–977. doi: 10.1093/ije/dys095. [DOI] [PubMed] [Google Scholar]; Krokstad, S., Langhammer, A., Hveem, K., Holmen, T.L., Midthjell, K., Stene, T.R., Bratberg, G., Heggland, J., and Holmen, J. (2013). Cohort Profile: the HUNT Study, Norway. Int. J. Epidemiol. 42, 968-977. [DOI] [PubMed]
- 14.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z., Electronic Medical Records and Genomics (eMEMERGEGE) Consortium. MIGen Consortium. PAGEGE Consortium. LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wood, A.R., Esko, T., Yang, J., Vedantam, S., Pers, T.H., Gustafsson, S., Chu, A.Y., Estrada, K., Luan, J., Kutalik, Z., et al.; Electronic Medical Records and Genomics (eMEMERGEGE) Consortium; MIGen Consortium; PAGEGE Consortium; LifeLines Cohort Study (2014). Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173-1186. [DOI] [PMC free article] [PubMed]
- 15.Sankararaman S., Mallick S., Dannemann M., Prüfer K., Kelso J., Pääbo S., Patterson N., Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sankararaman, S., Mallick, S., Dannemann, M., Prufer, K., Kelso, J., Paabo, S., Patterson, N., and Reich, D. (2014). The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354-357. [DOI] [PMC free article] [PubMed]
- 16.Paschou P., Lewis J., Javed A., Drineas P. Ancestry informative markers for fine-scale individual assignment to worldwide populations. J. Med. Genet. 2010;47:835–847. doi: 10.1136/jmg.2010.078212. [DOI] [PubMed] [Google Scholar]; Paschou, P., Lewis, J., Javed, A., and Drineas, P. (2010). Ancestry informative markers for fine-scale individual assignment to worldwide populations. J. Med. Genet. 47, 835-847. [DOI] [PubMed]
- 17.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]; Auton, A., Brooks, L.D., Durbin, R.M., Garrison, E.P., Kang, H.M., Korbel, J.O., Marchini, J.L., McCarthy, S., McVean, G.A., and Abecasis, G.R.; 1000 Genomes Project Consortium (2015). A global reference for human genetic variation. Nature 526, 68-74. [DOI] [PMC free article] [PubMed]
- 18.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]; Das, S., Forer, L., Schonherr, S., Sidore, C., Locke, A.E., Kwong, A., Vrieze, S.I., Chew, E.Y., Levy, S., McGue, M., et al. (2016). Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284-1287. [DOI] [PMC free article] [PubMed]
- 19.Maples B.K., Gravel S., Kenny E.E., Bustamante C.D. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Maples, B.K., Gravel, S., Kenny, E.E., and Bustamante, C.D. (2013). RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 93, 278-288. [DOI] [PMC free article] [PubMed]
- 20.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Alexander, D.H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655-1664. [DOI] [PMC free article] [PubMed]
- 21.Wang C., Zhan X., Liang L., Abecasis G.R., Lin X. Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am. J. Hum. Genet. 2015;96:926–937. doi: 10.1016/j.ajhg.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, C., Zhan, X., Liang, L., Abecasis, G.R., and Lin, X. (2015). Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am. J. Hum. Genet. 96, 926-937. [DOI] [PMC free article] [PubMed]
- 22.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Myers R.M. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]; Li, J.Z., Absher, D.M., Tang, H., Southwick, A.M., Casto, A.M., Ramachandran, S., Cann, H.M., Barsh, G.S., Feldman, M., Cavalli-Sforza, L.L., and Myers, R.M. (2008). Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100-1104. [DOI] [PubMed]
- 23.Hamilton C.M., Strader L.C., Pratt J.G., Maiese D., Hendershot T., Kwok R.K., Hammond J.A., Huggins W., Jackman D., Pan H. The PhenX Toolkit: get the most from your measures. Am. J. Epidemiol. 2011;174:253–260. doi: 10.1093/aje/kwr193. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hamilton, C.M., Strader, L.C., Pratt, J.G., Maiese, D., Hendershot, T., Kwok, R.K., Hammond, J.A., Huggins, W., Jackman, D., Pan, H., et al. (2011). The PhenX Toolkit: get the most from your measures. Am. J. Epidemiol. 174, 253-260. [DOI] [PMC free article] [PubMed]
- 24.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]; Locke, A.E., Kahali, B., Berndt, S.I., Justice, A.E., Pers, T.H., Day, F.R., Powell, C., Vedantam, S., Buchkovich, M.L., Yang, J., et al.; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium (2015). Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197-206. [DOI] [PMC free article] [PubMed]
- 25.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhou, W., Nielsen, J.B., Fritsche, L.G., Dey, R., Gabrielsen, M.E., Wolford, B.N., LeFaive, J., VandeHaar, P., Gagliano, S.A., Gifford, A., et al. (2018). Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50, 1335-1341. [DOI] [PMC free article] [PubMed]
- 26.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chang, C.C., Chow, C.C., Tellier, L.C., Vattikuti, S., Purcell, S.M., and Lee, J.J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7. [DOI] [PMC free article] [PubMed]
- 27.Semega J.L., Fontenot K.R., Kollar M.A. U.S. Government Printing Office; Washington, DC: 2017. Households by Total Money Income, Race, and Hispanic Origin of Householder: 1967 to 2016. In US Census Bureau, Current Population Reports, P60-259, Income and Poverty in the United States: 2016; pp. 23–29. [Google Scholar]; Semega, J.L., Fontenot, K.R., and Kollar, M.A. (2017). Households by Total Money Income, Race, and Hispanic Origin of Householder: 1967 to 2016. In US Census Bureau, Current Population Reports, P60-259, Income and Poverty in the United States: 2016. (Washington, DC, U.S. Government Printing Office), pp 23-29.
- 28.Nwankwo T., Yoon S.S., Burt V., Gu Q. Hypertension among adults in the United States: National Health and Nutrition Examination Survey, 2011-2012. NCHS Data Brief. 2013:1–8. [PubMed] [Google Scholar]; Nwankwo, T., Yoon, S.S., Burt, V., and Gu, Q. (2013). Hypertension among adults in the United States: National Health and Nutrition Examination Survey, 2011-2012. NCHS Data Brief 1-8. [PubMed]
- 29.Mozaffarian D., Benjamin E.J., Go A.S., Arnett D.K., Blaha M.J., Cushman M., de Ferranti S., Després J.P., Fullerton H.J., Howard V.J., American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–e322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]; Mozaffarian, D., Benjamin, E.J., Go, A.S., Arnett, D.K., Blaha, M.J., Cushman, M., de Ferranti, S., Despres, J.P., Fullerton, H.J., Howard, V.J., et al.; American Heart Association Statistics Committee and Stroke Statistics Subcommittee (2015). Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation 131, e29-e322. [DOI] [PubMed]
- 30.Ward, B.W., Clarke, T.C., Nugent, C.N., and Schiller, J.S. (2016). Early Release of Selected Estimates Based on Data From the 2015 National Health Interview Survey. National Center for Health Statistics, May 2016 https://www.cdc.gov/nchs/data/nhis/earlyrelease/earlyrelease201605.pdf.
- 31.Lee D., Cornet R., Lau F., de Keizer N. A survey of SNOMED CT implementations. J. Biomed. Inform. 2013;46:87–96. doi: 10.1016/j.jbi.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, D., Cornet, R., Lau, F., and de Keizer, N. (2013). A survey of SNOMED CT implementations. J. Biomed. Inform. 46, 87-96. [DOI] [PMC free article] [PubMed]
- 32.Bays H.E., Chapman R.H., Grandy S., SHIELD Investigators’ Group The relationship of body mass index to diabetes mellitus, hypertension and dyslipidaemia: comparison of data from two national surveys. Int. J. Clin. Pract. 2007;61:737–747. doi: 10.1111/j.1742-1241.2007.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bays, H.E., Chapman, R.H., and Grandy, S.; SHIELD Investigators’ Group (2007). The relationship of body mass index to diabetes mellitus, hypertension and dyslipidaemia: comparison of data from two national surveys. Int. J. Clin. Pract. 61, 737-747. [DOI] [PMC free article] [PubMed]
- 33.Liu F., van Duijn K., Vingerling J.R., Hofman A., Uitterlinden A.G., Janssens A.C., Kayser M. Eye color and the prediction of complex phenotypes from genotypes. Curr. Biol. 2009;19:R192–R193. doi: 10.1016/j.cub.2009.01.027. [DOI] [PubMed] [Google Scholar]; Liu, F., van Duijn, K., Vingerling, J.R., Hofman, A., Uitterlinden, A.G., Janssens, A.C., and Kayser, M. (2009). Eye color and the prediction of complex phenotypes from genotypes. Curr. Biol. 19, R192-R193. [DOI] [PubMed]
- 34.Demenais F., Margaritte-Jeannin P., Barnes K.C., Cookson W.O.C., Altmüller J., Ang W., Barr R.G., Beaty T.H., Becker A.B., Beilby J., Australian Asthma Genetics Consortium (AAGC) collaborators Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat. Genet. 2018;50:42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demenais, F., Margaritte-Jeannin, P., Barnes, K.C., Cookson, W.O.C., Altmuller, J., Ang, W., Barr, R.G., Beaty, T.H., Becker, A.B., Beilby, J., et al.; Australian Asthma Genetics Consortium (AAGC) collaborators (2018). Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat. Genet. 50, 42-53. [DOI] [PMC free article] [PubMed]
- 35.Tsoi L.C., Stuart P.E., Tian C., Gudjonsson J.E., Das S., Zawistowski M., Ellinghaus E., Barker J.N., Chandran V., Dand N. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat. Commun. 2017;8:15382. doi: 10.1038/ncomms15382. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsoi, L.C., Stuart, P.E., Tian, C., Gudjonsson, J.E., Das, S., Zawistowski, M., Ellinghaus, E., Barker, J.N., Chandran, V., Dand, N., et al. (2017). Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat. Commun. 8, 15382. [DOI] [PMC free article] [PubMed]
- 36.Duffy S.W., Warwick J., Williams A.R.W., Keshavarz H., Kaffashian F., Rohan T.E., Nili F., Sadeghi-Hassanabadi A. A simple model for potential use with a misclassified binary outcome in epidemiology. J. Epidemiol. Community Health. 2004;58:712–717. doi: 10.1136/jech.2003.010546. [DOI] [PMC free article] [PubMed] [Google Scholar]; Duffy, S.W., Warwick, J., Williams, A.R.W., Keshavarz, H., Kaffashian, F., Rohan, T.E., Nili, F., and Sadeghi-Hassanabadi, A. (2004). A simple model for potential use with a misclassified binary outcome in epidemiology. J. Epidemiol. Community Health 58, 712-717. [DOI] [PMC free article] [PubMed]
- 37.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bycroft, C., Freeman, C., Petkova, D., Band, G., Elliott, L.T., Sharp, K., Motyer, A., Vukcevic, D., Delaneau, O., O’Connell, J., et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203-209. [DOI] [PMC free article] [PubMed]
- 38.Lunshof J.E., Church G.M., Prainsack B. Information access. Raw personal data: providing access. Science. 2014;343:373–374. doi: 10.1126/science.1249382. [DOI] [PubMed] [Google Scholar]; Lunshof, J.E., Church, G.M., and Prainsack, B. (2014). Information access. Raw personal data: providing access. Science 343, 373-374. [DOI] [PubMed]
- 39.Jiang Y., Chen S., McGuire D., Chen F., Liu M., Iacono W.G., Hewitt J.K., Hokanson J.E., Krauter K., Laakso M. Proper conditional analysis in the presence of missing data: Application to large scale meta-analysis of tobacco use phenotypes. PLoS Genet. 2018;14:e1007452. doi: 10.1371/journal.pgen.1007452. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jiang, Y., Chen, S., McGuire, D., Chen, F., Liu, M., Iacono, W.G., Hewitt, J.K., Hokanson, J.E., Krauter, K., Laakso, M., et al. (2018). Proper conditional analysis in the presence of missing data: Application to large scale meta-analysis of tobacco use phenotypes. PLoS Genet. 14, e1007452. [DOI] [PMC free article] [PubMed]
- 40.Zhan X., Chen S., Jiang Y., Liu M., Iacono W.G., Hewitt J.K., Hokanson J.E., Krauter K., Laakso M., Li K.W. Association Analysis and Meta-Analysis of Multi-allelic Variants for Large Scale Sequence Data. bioRxiv. 2017 doi: 10.3390/genes11050586. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhan, X., Chen, S., Jiang, Y., Liu, M., Iacono, W.G., Hewitt, J.K., Hokanson, J.E., Krauter, K., Laakso, M., Li, K.W., et al. (2017). Association Analysis and Meta-Analysis of Multi-allelic Variants for Large Scale Sequence Data. bioRxiv 10.1101/197913. [DOI] [PMC free article] [PubMed]
- 41.Liu M., Jiang Y., Wedow R., Li Y., Brazel D.M., Chen F., Datta G., Davila-Velderrain J., McGuire D., Tian C., 23andMe Research Team. HUNT All-In Psychiatry Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu, M., Jiang, Y., Wedow, R., Li, Y., Brazel, D.M., Chen, F., Datta, G., Davila-Velderrain, J., McGuire, D., Tian, C., et al.; 23andMe Research Team; HUNT All-In Psychiatry (2019). Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237-244. [DOI] [PMC free article] [PubMed]
- 42.Sanchez-Roige S., Fontanillas P., Elson S.L., Pandit A., Schmidt E.M., Foerster J.R., Abecasis G.R., Gray J.C., de Wit H., 23andMe Research Team Genome-wide association study of delay discounting in 23,217 adult research participants of European ancestry. Nat. Neurosci. 2018;21:16–18. doi: 10.1038/s41593-017-0032-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sanchez-Roige, S., Fontanillas, P., Elson, S.L., Pandit, A., Schmidt, E.M., Foerster, J.R., Abecasis, G.R., Gray, J.C., de Wit, H., et al.; 23andMe Research Team (2018). Genome-wide association study of delay discounting in 23,217 adult research participants of European ancestry. Nat. Neurosci. 21, 16-18. [DOI] [PMC free article] [PubMed]
- 43.Hollands G.J., French D.P., Griffin S.J., Prevost A.T., Sutton S., King S., Marteau T.M. The impact of communicating genetic risks of disease on risk-reducing health behaviour: systematic review with meta-analysis. BMJ. 2016;352:i1102. doi: 10.1136/bmj.i1102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hollands, G.J., French, D.P., Griffin, S.J., Prevost, A.T., Sutton, S., King, S., and Marteau, T.M. (2016). The impact of communicating genetic risks of disease on risk-reducing health behaviour: systematic review with meta-analysis. BMJ 352, i1102. [DOI] [PMC free article] [PubMed]
- 44.Dobkin B.H., Dorsch A. The promise of mHealth: daily activity monitoring and outcome assessments by wearable sensors. Neurorehabil. Neural Repair. 2011;25:788–798. doi: 10.1177/1545968311425908. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dobkin, B.H., and Dorsch, A. (2011). The promise of mHealth: daily activity monitoring and outcome assessments by wearable sensors. Neurorehabil. Neural Repair 25, 788-798. [DOI] [PMC free article] [PubMed]
- 45.Appelboom G., Camacho E., Abraham M.E., Bruce S.S., Dumont E.L., Zacharia B.E., D’Amico R., Slomian J., Reginster J.Y., Bruyère O., Connolly E.S., Jr. Smart wearable body sensors for patient self-assessment and monitoring. Arch. Public Health. 2014;72:28. doi: 10.1186/2049-3258-72-28. [DOI] [PMC free article] [PubMed] [Google Scholar]; Appelboom, G., Camacho, E., Abraham, M.E., Bruce, S.S., Dumont, E.L., Zacharia, B.E., D’Amico, R., Slomian, J., Reginster, J.Y., Bruyere, O., and Connolly, E.S., Jr. (2014). Smart wearable body sensors for patient self-assessment and monitoring. Arch. Public Health 72, 28. [DOI] [PMC free article] [PubMed]
- 46.El-Amrawy F., Nounou M.I. Are Currently Available Wearable Devices for Activity Tracking and Heart Rate Monitoring Accurate, Precise, and Medically Beneficial? Healthc. Inform. Res. 2015;21:315–320. doi: 10.4258/hir.2015.21.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]; El-Amrawy, F., and Nounou, M.I. (2015). Are Currently Available Wearable Devices for Activity Tracking and Heart Rate Monitoring Accurate, Precise, and Medically Beneficial? Healthc. Inform. Res. 21, 315-320. [DOI] [PMC free article] [PubMed]
- 47.Montgomery-Downs H.E., Insana S.P., Bond J.A. Movement toward a novel activity monitoring device. Sleep Breath. 2012;16:913–917. doi: 10.1007/s11325-011-0585-y. [DOI] [PubMed] [Google Scholar]; Montgomery-Downs, H.E., Insana, S.P., and Bond, J.A. (2012). Movement toward a novel activity monitoring device. Sleep Breath. 16, 913-917. [DOI] [PubMed]
- 48.Glasgow M.L., Rudra C.B., Yoo E.H., Demirbas M., Merriman J., Nayak P., Crabtree-Ide C., Szpiro A.A., Rudra A., Wactawski-Wende J., Mu L. Using smartphones to collect time-activity data for long-term personal-level air pollution exposure assessment. J. Expo. Sci. Environ. Epidemiol. 2016;26:356–364. doi: 10.1038/jes.2014.78. [DOI] [PubMed] [Google Scholar]; Glasgow, M.L., Rudra, C.B., Yoo, E.H., Demirbas, M., Merriman, J., Nayak, P., Crabtree-Ide, C., Szpiro, A.A., Rudra, A., Wactawski-Wende, J., and Mu, L. (2016). Using smartphones to collect time-activity data for long-term personal-level air pollution exposure assessment. J. Expo. Sci. Environ. Epidemiol. 26, 356-364. [DOI] [PubMed]
- 49.Liu M., Rea-Sandin G., Foerster J., Fritsche L., Brieger K., Clark C., Li K., Pandit A., Zajac G., Abecasis G.R., Vrieze S. Validating Online Measures of Cognitive Ability in Genes for Good, a Genetic Study of Health and Behavior. Assessment. 2017 doi: 10.1177/1073191117744048. 1073191117744048. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu, M., Rea-Sandin, G., Foerster, J., Fritsche, L., Brieger, K., Clark, C., Li, K., Pandit, A., Zajac, G., Abecasis, G.R., and Vrieze, S. (2017). Validating Online Measures of Cognitive Ability in Genes for Good, a Genetic Study of Health and Behavior. Assessment. 1073191117744048. [DOI] [PMC free article] [PubMed]
- 50.Branigan A.R., McCallum K.J., Freese J. Variation in the Heritability of Educational Attainment: An International Meta-Analysis. Soc. Forces. 2013;92:109–140. [Google Scholar]; Branigan, A.R., McCallum, K.J., and Freese, J. (2013). Variation in the Heritability of Educational Attainment: An International Meta-Analysis. Soc. Forces 92, 109-140.
- 51.Howden L.M., Meyer J.A. Census Briefs, C2010BR-03. U.S. Census Bureau; Washington, D.C.: 2011. Age and Sex Composition: 2010; pp. 1–16.https://www.census.gov/prod/cen2010/briefs/c2010br-2003.pdf [Google Scholar]; Howden, L.M., and Meyer, J.A. (2011). Age and Sex Composition: 2010. In Census Briefs, C2010BR-03 (Washington, D.C.: U.S. Census Bureau), pp. 1-16, https://www.census.gov/prod/cen2010/briefs/c2010br-2003.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.