Abstract
Sry-Related HMG-BOX-4 (SOX4) is a developmental transcription factor that is overexpressed in as many as 23% of bladder cancer patients; however, the role of SOX4 in bladder cancer tumorigenesis is not yet well understood. Given the many roles of SOX4 in embryonic development and the context-dependent regulation of gene expression, in this study, we sought to determine the role of SOX4 in bladder cancer and to identify SOX4-regulated genes that may contribute to tumorigenesis. For this purpose, we employed a CRISPR interference (CRISPRi) method to transcriptionally repress SOX4 expression in T24 bladder cancer cell lines, 'rescued' these cell lines with the lentiviral-mediated expression of SOX4, and performed whole genome expression profiling. The cells in which SOX4 was knocked down (T24-SOX4-KD) exhibited decreased invasive capabilities, but no changes in migration or proliferation, whereas rescue experiments with SOX4 lentiviral vector restored the invasive phenotype. Gene expression profiling revealed 173 high confidence SOX4-regulated genes, including WNT5a as a potential target of repression by SOX4. Treatment of the T24-SOX4-KD cells with a WNT5a antagonist restored the invasive phenotype observed in the T24-scramble control cells and the SOX4 lentiviral-rescued cells. High WNT5a expression was associated with a decreased invasion and WNT5a expression inversely correlated with SOX4 expression, suggesting that SOX4 can negatively regulate WNT5a levels either directly or indirectly and that WNT5a likely plays a protective role against invasion in bladder cancer cells.
Keywords: SOX4, bladder cancer, CRISPR interference, invasion, WNT5A
Introduction
Urothelial carcinoma of the bladder is the 6th most common type of cancer in the United States. Bladder cancer disproportionately affects more males than females and risk factors include smoking, as well as other environmental and occupational exposures (1,2). There are also data to suggest that a diet poorly supplemented with fruits and vegetables is linked to the incidence of bladder cancer, but is not necessarily a risk factor (3).
Bladder cancer is typically grouped into two main pathological classes: Non-muscle invasive (NMIBC) and muscle invasive (MIBC). While histological and pathological grading and staging systems have existed for some time, recent advances in genomic sequencing have lent insight into molecular characterizations that stratify patients into various subtypes based on genetic markers, such as amplifications, mutations and deletions (4-7). It is hoped that these new classifications might lend insights into prognosis or new subtype-specific treatment regimens.
One of the most commonly amplified and overexpressed genes in bladder cancer is the Sry-Related HMG-BOX-4 (SOX4) transcription factor (6,8). SOX4 is responsible for regulating a number of genes implicated in cellular development and differentiation (9). SOX4 has both transcriptional activation and repressive roles, either alone or in combination with other transcription factors, that vary according to tissue type and context (10-13). Some of the most well established SOX4 target genes include DICER, TEA domain transcription factor 2 (TEAD2), tubulin beta 3 class III (TUBB3) and tenascin C (TNC) (14-19). SOX4 is located on chromosome 6p22, a genomic locus that also represents one of the most significant focal amplifications in bladder cancer and affects a number of different genes, including SOX4, inhibitor of DNA binding 4, HLH protein (ID4), CDK5 regulatory subunit associated protein 1 like 1 (CDKAL1), E2F transcription factor 3 (E2F3) and membrane bound o-acyltransferase domain containing 1 (MBOAT1) (20,21).
Nevertheless, the exact role of SOX4 expression in various tumors, including bladder cancer, has not yet been determined and a comprehensive model of SOX4 function remains elusive. Increased SOX4 expression is associated with several other cancer types (22) and in some cases, its expression levels have been shown to increase with an increasing tumor grade (23). The majority of in vitro studies have associated the aberrant expression of SOX4 with the transformation ability of cell lines, tumorigenicity and the induction of a mesenchymal phenotype (22,23). However, some contradictory data have shown higher SOX4 levels associated with the stabilization of p53, cell cycle arrest and increased apoptosis, suggesting a possible context-specific tumor suppressive arm of SOX4 (24-27). Although SOX4 overexpression has been implicated in a variety of different cancer types (22,23), its downstream targets, mechanisms of action and functional consequences, as well as clinical prognoses of patients exhibiting SOX4 overexpression vary amongst tumor subtypes (17,24,28) and conflicting results have been obtained (28,29). As a result, there is growing consensus that the role of SOX4 is context-dependent, and the role of SOX4 in bladder cancer, similar to other tumor types, is thus not well defined.
In this study, we investigated the role of SOX4 expression in the T24 bladder cancer cell line by transcriptionally repressing SOX4 expression using a CRISPR-interference (CRISPRi) approach (30) to assess the functional effects on migration, invasion and proliferation. We also re-established SOX4 expression in the T24 cell line in which SOX4 was knocked down (T24-SOX4-KD cells) and identified a set of 173 high-confidence SOX4-regulated genes. Specifically, we demonstrate that SOX4 knockdown induces WNT5a expression and that a high WNT5a expression in T24-SOX4-KD cells is associated with the decreased invasive ability of bladder cancer cells.
Materials and methods
Cell culture, cell lines and reagents
The bladder cancer cell lines, 5637 (HTB-9), HT1376 (CRL-1472), TCCSUP (HTB5), T24 (HTB-4) and SW780 (CRL-2169), were obtained from the American Type Culture Collection (ATCC). The 5637 cells were maintained in RPMI, the T24, HT1376 and SW780 cells in DMEM, and the TCCSUP cells in MEM growth media. All media were supplemented with 10% FBS (cat. no. 900-108; Gemini Bio), 1% L-glutamine (cat. no. 25030081; Thermo Fisher Scientific) and 1% penicillin-streptomycin (cat. no. 15140122; Thermo Fisher Scientific). The cells were cultured in a 37°C incubator with humidified atmosphere of 5% CO2. Parental T24 cells and subsequent cell lines used to generate stable T24 cells were genetically authenticated using STR profiling by Bio-Synthesis Inc., an Accredited Human Cell Line Genotyping Service company. The WNT5a antagonist, BOX5, was purchased from EMD Millipore (cat. no. 681673) and used as previously described (31).
Generation of stable T24 cell lines in which SOX4 was knocked down or re-expressed
Plasmid pHR-SFFV-KRAB-dCas9-P2A-mCherry was a gift from Dr Jonathan Weissman, UCSF (plasmid #60954; Addgene). SOX4-specific small guide RNAs (sgRNAs) were designed using the CRISPR design tool from Zhang Lab (http://crispr.mit.edu/) and validated using NCBI BLAST for non-specific targets. Scrambled or SOX4-TSS targeted sgRNAs were designed, annealed and ligated into the lentiviral construct pLKO.1-puro U6 sgRNA BfuAI large stuffer (a gift from Dr Scot Wolfe, University of Massachusetts Medical School; plasmid #52628; Addgene). The T24 cells were seeded at a density of 2×105/well in a 6-well plate and 24 h later were spinfected at 500 × g for 90 min at 32°C with pHR-SFFV-KRAB-dCas9-P2A-mCherry and grown for 1 week in a 37°C incubator with humidified atmosphere of 5% CO2. The cells were then sorted for pure mCherry-positive cells at Emory's Flow Cytometry Core on a BD FACSAria II (BD Biosciences) to establish the stable T24-KRAB-dCas9-P2A-mCherry cell line. These stable cells were seeded into a 6-well plate and transduced via spinfection as described above with either scrambled sgRNA pLKO.1-puro U6 sgRNA BfuAI large stuffer, or a pool of 7 sgRNAs targeting the SOX4 transcription start site (TSS). The infected cells were selected with puromycin (2 µg/ml) for 48 h following infection to create stable T24-KRAB-dCas9-P2A-mCherry-SOX4-sgRNA and stable T24- KRAB-dCas9-P2A-mCherry-Scrambled-sgRNA, hereafter referred to as T24-SOX4-KD and T24-Scr, respectively.
The re-expression of SOX4 was performed in the T24-SOX4-KD background as described above. Briefly, we used our pHR-UBQ-HA-SOX4-IRES-eYFP-LlU3 lentiviral vector as previously described (16,32) to transduce the T24-SOX4-KD cells. We performed a dual sort for pure mCherry-positive and YFP-positive cells at Emory's Flow Cytometry Core on a BD FACSAria II to create stable T24-SOX4-KD+YFP-HA-SOX4 cells, hereafter referred to as T24-SOX4-Rescue.
Cell migration and invasion assay
Cell invasion was evaluated using a Boyden Chamber assay. A total of 1.25×105 cells were seeded in 2 ml of serum-free and antibiotic-free DMEM media in the top Boyden chamber containing Matrigel-coated 8-µm pore membranes (cat. no. 354481; Corning, Inc.) and 2.5 ml of complete DMEM (supplemented with 10% FBS cat. no. 900-108; Gemini Bio), 1% penicillin-streptomycin (cat. no. 15140122) and 1% L-glutamine (cat. no. 25030081) (both from Thermo Fisher Scientific) in the bottom chamber as a chemoattractant. Following incubation for 24 h at 37°C, non-invaded cells in the upper chamber were aspirated and membranes then fixed and stained in 0.5% crystal violet for 5 min, then washed 3 times for 1 min in ddH2O and washed for 3 min on a shaker at room temperature. ddH2O was aspirated and membranes allowed to dry for 2 h in a cell culture biosafety cabinet. The membranes were then visualized under an upright confocal microscope using ×40 magnification on a Nikon Eclipse Ti-S inverted microscope (Nikon Instruments). Representative images from 2-3 random fields were taken for each chamber. Cells were counted using Fiji open source analysis software (https://fiji.sc/). Each sample was assayed in triplicate in three independent experiments.
Cell migration was evaluated using a scratch-wound assay. A total of 3.75×105 cells were seeded in each well of a 12-well plate and allowed to grow to confluency for 24 h. The media were aspirated and a scratch was made using a sterile 200 µl pipette tip and fresh media was added. Images were acquired at time zero (baseline), and at 6, 12, 18 h and 24 h on a Biotek Lionheart widefield microscope at ×40 magnification.
MTT assay
To evaluate the proliferation of T24 cells expressing KRAB-dCas9-P2A-mCherry with SOX4 sgRNAs and scrambled controls, an MTT assay (cat. no. 30-1010K; ATCC) was performed by seeding the 5×103 cells/well into 96-well plates. The cells were analyzed daily as per the manufacturer's protocol for 5 consecutive days. The plates were read daily on a Biotek SYNERGY HT microplate reader. Each sample was assayed in triplicate in three independent experiments.
RNA extraction and microarray analysis
Total RNA was isolated from the cultured cells using QIAshredder (cat. no. 79654) and RNeasy (cat. no. 74104) kits as per the manufacturer's protocol (all from Qiagen). RNA concentrations were measured using a NanoDrop Spectrophotometer (mode #ND-1000). RNA samples were processed at the Emory Integrated Genomics Core Facility for quality control analysis and analyzed using the Affymetrix Clariom D Genechip platform. Total RNA from four independent control samples (T24-KRAB-dCas9-P2A-mCherry-Scrambled sgRNA), three independent SOX4 knockdown samples expressing T24-KRA B-dCas9-P2A-mCherry-SOX4-sgRNA and three independent SOX4-Rescue samples expressing T24-KRAB-dCas9-P2 A-mCherry-SOX4-sgRNA+YFP-HA-SOX4 cell lines were analyzed. The gene level signal was generated by RMA normalization. Differential gene expression was determined using the samr package (33,34) in R Bioconductor (35) with 500 permutations, a minimum fold change of 1.5-fold, and a median false discovery rate (FDR) <0.05. Sample microarray data are available on the GEO database (accession no. GSE118572). Pathway analysis was performed using Ingenuity Pathway Analysis on 1,487 significantly differentially expressed genes for T24-SOX4-KD vs. T24-scrambled. Differentially expressed genes were similarly calculated for T24-SOX4-Rescue vs. T24-SOX4-KD (identifying 561 genes) and pathway analysis was performed on the intersection of 173 differential genes from the two comparisons. Hierarchical clustering was performed using Cluster (36) and Java Treeview v1.1.4r3 (37) using complete linkage clustering of uncentered correlation.
Western blot analysis
The cells were washed twice with 1X PBS and harvested with RIPA lysis buffer (cat. no. R0278) containing protease inhibitors (cat. no. P8340) (both from Sigma-Aldrich; Merck KGaA) and phosphatase inhibitors (cat. no. 4906845001; Roche). Whole cell lysates were centrifuged at 14,000 × g for 10 min at 4°C. Supernatants were transferred to fresh tubes and protein concentration was quantified using the Pierce Bradford protein assay (cat. no. 23225; Thermo Fisher Scientific). A total of 30 µg of protein was analyzed on a 10% SDS-polyacrylamide gel by SDS-PAGE electrophoresis and transferred to a PVDF membrane (cat. no. 1620177; Bio-Rad). The membranes were blocked in 1X TBS buffer containing 5% BSA (cat. no. 700-100P; Gemini Bio) and 0.001% Tween-20 (cat. no. BP337-500; Thermo Fisher Scientific) for 1 h at room temperature, and then incubated with primary antibodies (SOX4 rabbit anti-human polyclonal, 1:1,000, cat. no. ab80261, Abcam; ZEB1 rabbit anti-human polyclonal, 1:1,000, cat. no. 3396s, Cell Signaling Technology; E-cadherin rabbit anti-human polyclonal, 1:1,000, cat. no. 3195s, Cell Signaling Technology, N-cadherin rabbit anti-human polyclonal, 1:1,000, cat. no. sc-7939, Santa Cruz Biotechnology; vimentin rabbit anti-human polyclonal, 1:1,000, cat. no. sc-5565, Santa Cruz Biotechnology; CRISPR/Cas9 mouse anti-human monoclonal, 1:500, cat. no. A-9000-100, Epigentek; β-actin rabbit anti-human polyclonal, 1:3,000, cat. no. 3700s, Cell Signaling Technology; GAPDH rabbit anti-human polyclonal, 1:1,000, cat. no. 2118s, Cell Signaling Technology) overnight at 4°C. The blots were washed with TBST three times for 5 min each and incubated with secondary antibodies (anti-mouse IgG, 1:2,000, cat. no. 7076S, Cell Signaling Technology; or anti-rabbit IgG, 1:3,000, cat. no. ab6721, Abcam) for 1 h at room temperature. Signals were visualized using SuperSignal West Pico PLUS chemiluminescence substrate (cat. no. 34580, Thermo Fisher Scientific). Densitometry of the bands was performed using Fiji open source analysis software (https://fiji.sc/).
Reverse transcription-quantitative PCR (RT-qPCR)
The cells were harvested by treatment with 0.25% Trypsin. Total RNA was isolated using the Qiagen QIAshredder (cat. no. 79654) and RNeasy (cat. no. 74104) per the manufacturer's protocol and treated with on-column DNAse digestion to remove possible contaminating genomic DNA. All RNA was converted to cDNA using the iScript cDNA Sythesis kit containing a mixture of RNase H + MMLV reverse transcriptase (cat. no. 1708891; Bio-Rad). RT-qPCR was performed on a Bio-Rad Model CFX Connect Real-Time System (Bio-Rad). The primer sequences for SOX4, transmembrane 7 superfamily member 2 (TM7SF2), 7-dehydrocholesterol reductase (DHCR7), mevalonate diphosphate decarboxylase (MVD), Wnt Family Member 5A WNT5a, TNC, indoleamine 2,3-dioxygenase 1 (IDO1) and 18s are listed below. Relative expression levels were normalized to 18s using the ΔΔCq method (38). Primers are listed as follows (5′→3′): IDO forward, TTGCTAAAGGCGCTGTTGGA and reverse, GTC TGATAGCTGGGGGTTGC; 18s forward, CGGCTACCA CATCCAAGGAA and reverse, GCTGGAATTACCGCGGCT; TM2SF2 forward, CTGCCTCATCAATGGGCTTG and reverse, GAGGTAGAAGTAGGGCAGCAG; DHCR7 forward, GAG GTGTGCGCAGGACTTTA and reverse, TGGCTTTGGGAA TGTTGGGT; MVD forward, ATCAAGTACTGGGGCAA GCG and reverse, TTCAGCCAAATCCGGTCCTC; WNT5a forward, CGCCCAGGTTGTAATTGAAG and reverse, GCA TGTGGTCCTGATACAAGT; TNC forward, AGCATCCGGAC CAAAACCAT and reverse, CCGATGCCATCCAGGAAACT; IDO1 forward, TTGCTAAAGGCGCTGTTGGA and reverse, GTCTGATAGCTGGGGGTTGC; SOX4 forward, CCGAGCT GGTGCAAGACC and reverse, CCACACCATGAAGGCGTTC.
Statistical analysis
All statistical analysis of the experiments were performed from three independent experiments. Graphs are represented by the mean ± standard error bars. All data were evaluated using a one-way ANOVA with α = 0.05 followed by Tukey's post hoc test for multiple comparisons. A P-value <0.05 was considered to indicate a statistically significant difference. The PRISM software suite (version 6.00 for Mac OS X, GraphPad Software; www.graphpad.com) was utilized for statistical analysis.
Cancer genomics data mining
All cancer genomics data mining was performed using the cBioPortal website (http://www.cbioportal.org) and the TCGA Cell 2017 dataset for BLCA (6). SOX4 mRNA Z-score plots were generated on cBioPortal against clinical parameters and between SOX4 and WNT5A. Linear regression fits were obtained using default methods and Pearson's correlation coefficients were computed.
Results
Expression of SOX4 in bladder cancer patients and bladder cancer cell lines
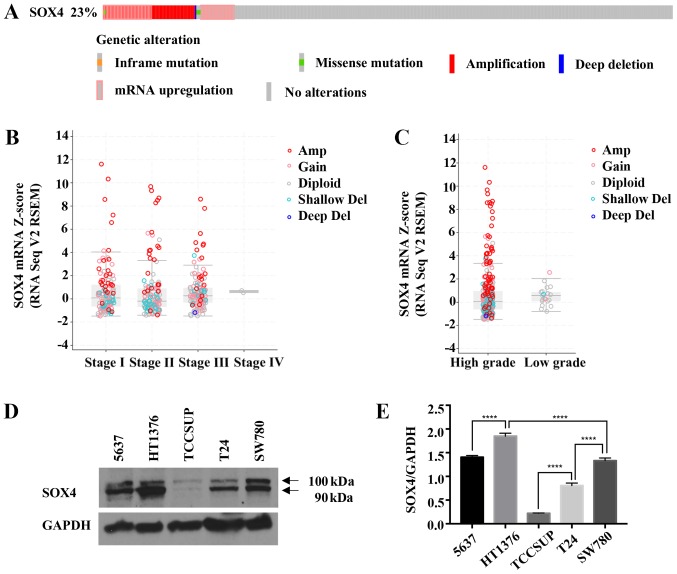
We queried The Cancer Genome Atlas (TCGA) bladder cancer dataset via cBioPortal (http://www.cbioportal.org) (39,40) for SOX4 using the TCGA Cell 2017 dataset (6) and observed that the SOX4 gene has either copy number amplifications or an increased mRNA expression in 23% (93/404) of bladder cancer patients (Fig. 1A). We selected the TCGA Cell, 2017 dataset as it is the most current and largest dataset available in cBioPortal that contains mRNA gene expression data and copy number data that includes SOX4. Cancer Outlier Profiling Analysis (COPA) (41-43) on Oncomine (http://www.oncomine.org) indicated SOX4 amplification in the top 1% of genes for BLCA in the TCGA dataset. The high outlier expression of SOX4 was observed with the increasing tumor stage (Fig. 1B) and tumor grade (Fig. 1C). To determine how representative SOX4 levels are in bladder cancer cell lines, we performed western blot analysis of the 5637, HT1376, TCCSUP, T24 and SW780 cells (Fig. 1D and E). The data indicated that these bladder cancer cell lines recapitulated the range of genetic alterations and SOX4 expression levels observed in bladder cancer patients.
Figure 1.
Increased SOX4 expression in patients and in bladder cancer cell lines. (A) cBioPortal data from the Cell, 2017 dataset (6) showing copy number alterations and mRNA expression levels (+2.0) in 93 out of 404 (23%) patients. (B) SOX4 mRNA levels for different stages of BLCA in the TCGA dataset. Amplified samples are shown in red, and deletions are in blue, and diploid are in gray. (C) SOX4 mRNA levels for TCGA BLCA samples for low grade and high grade samples. Sample colors are the same as in (B and D). Western blot analysis of bladder cancer cell lines showing varying degrees of SOX4 protein expression. (E) Quantification of SOX4 protein levels. ****P<0.0001.
Generation of stable T24 bladder cancer cells in which SOX4 was knocked down using CRISPRi
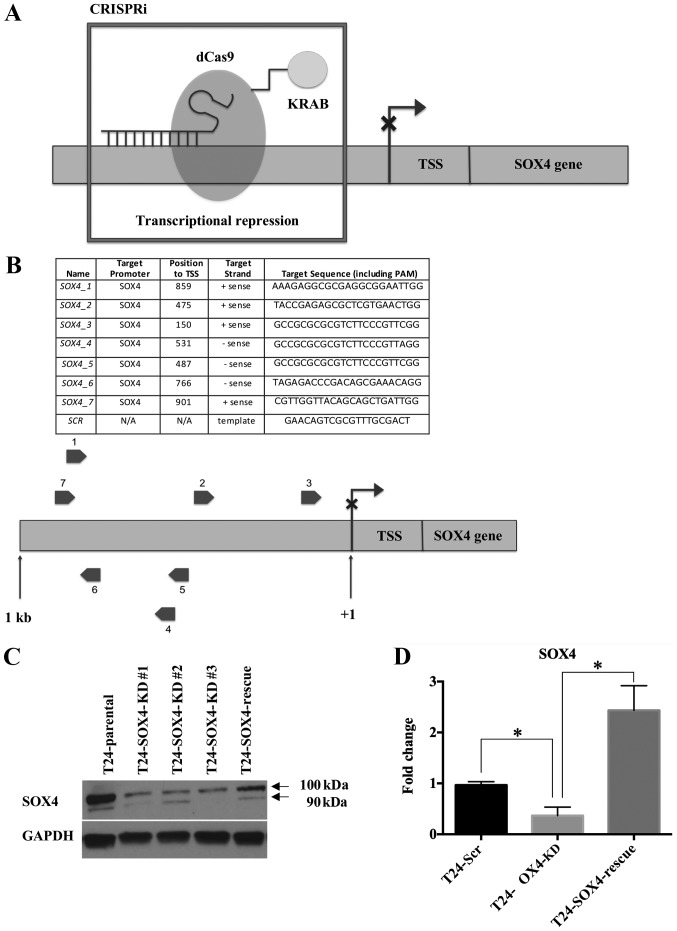
To better understand the function of SOX4 in bladder cancer cells with a high expression and/or amplification of SOX4, we performed CRISPRi as previously described (44-46) to induce the stable repression of SOX4 mRNA expression by targeting sgRNA's upstream of the SOX4 transcription start site (TSS) (Fig. 2A). This approach uses a catalytically inactivated Cas9 enzyme (dCas9) fused to the KRAB repressor domain (KRAB-dCas9). In this manner, the sgRNAs and KRAB- dCas9 act as an RNA-guided DNA binding domain that can both block RNA polymerase and also induce heterochromatin at the SOX4 TSS. We chose to use the T24 cells four our model system as they represent SOX4 overexpression without the genomic amplification of SOX4. The other bladder cancer cell lines (5637, HT1376 and SW780) not only overexpressed SOX4, but also contained genomic amplifications of the SOX4 locus, which presents technical challenges for the CRISPRi system. Briefly, we stably transduced the T24 cells with lentiviral KRAB-dCas9 and 7 sgRNAs, and sorted for positive mCherry subpopulations by flow cytometry to produce T24-SOX4-KD cells (see Materials and methods) (Fig. S1). Cas9 expression in the T24 cell lines was confirmed by western blot analysis using Cas9 antibody (Fig. S2). We designed 7 sgRNAs targeting both the sense and antisense strands corresponding to sites 150 bp-901 bp upstream of the SOX4 TSS (Fig. 2B). We prepared lentivirus containing a scrambled sgRNA to create stable T24-Scr negative control cells (Fig. 2B). In addition, we transduced the T24-SOX4-KD cells with YFP-HA-SOX4 lentiviral constructs to produce stable T24-SOX4-Rescue cells that express SOX4 in the presence of the KRAB-dCas9 and sgRNAs (Fig. S1). To confirm SOX4 knockdown, we performed western blot analysis on T24 cell lysates (Fig. 2C), and collected total RNA and performed RT-qPCR analysis of RNA from the T24-Scr control, T24-SOX4-KD and T24-SOX4-Rescue cells (Fig. 2D).
Figure 2.
CRISPRi methods and confirmed SOX4 knockdown. (A) CRISPRi model with KRAB effector domain to transcriptionally silence SOX4 at sites upstream of the transcription start site (TSS). (B) Seven sgRNAs targeting the SOX4 TSS at both sense and antisense strands and a scrambled sgRNA control are shown. (C) Western blot analysis confirmed SOX4 knockdown and SOX4 overexpression at the protein level. (D) RT-qPCR data confirmed decreased SOX4 mRNA expression in T24-SOX4-KD cells and increased SOX4 mRNA in T24-SOX4-Rescue cells compared to the T24-Scr controls. *P<0.05. CRISPRi, CRISPR interference.
Knockdown of SOX4 in T24 cells suppreses cellular invasion
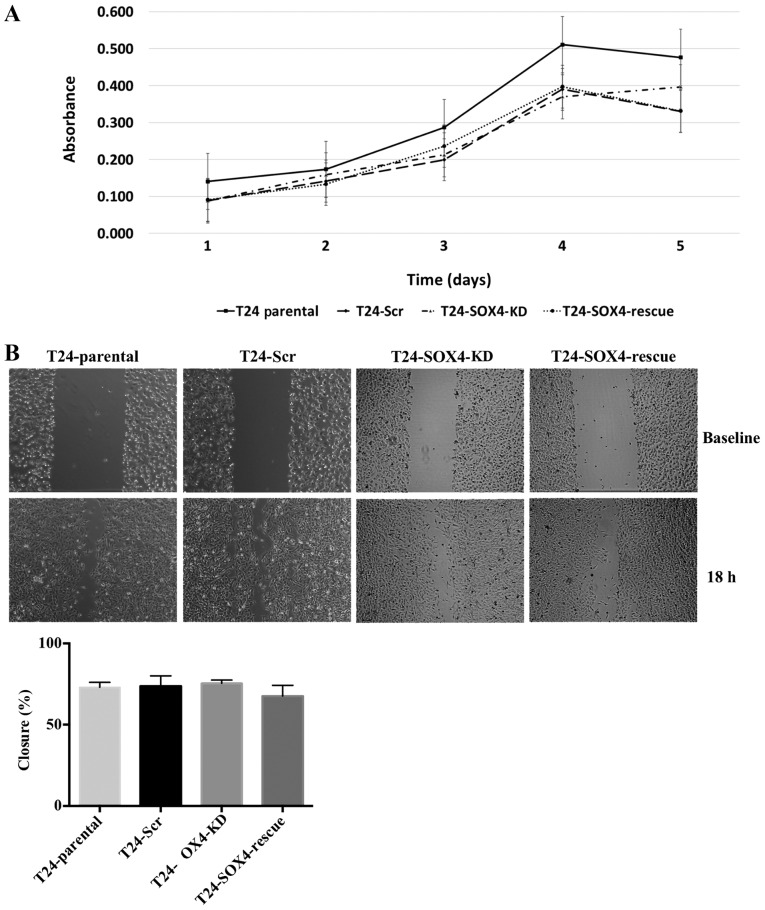
Previous studies have shown that the loss of SOX4 decreases the proliferation of a variety of cell lines (47,48). In this study, To investigate the role of SOX4 in the proliferation of T24 bladder cancer cell lines, we examined the hypothesis that T24-SOX4-KD would proliferate slower than T24-Scr control. We performed MTT assays to assess the changes in the proliferation of the T24-Parental, T24-Scr, T24-SOX4-KD and T24-SOX4-Rescue cell lines. We observed that our T24-SOX4-KD cells did not exhibit any significant changes in proliferation compared to the T24-Scr control (Fig. 3A). Moreover, the re-expression of SOX4 by transducing T24-SOX4-KD with lentiviral SOX4 did not alter the proliferation rates compared to the T24-Scr or T24-SOX4-KD cells. These data indicate that SOX4 expression levels do not have a substantial effect on the proliferation of T24 cells, which is consistent with previous findings on 5637 bladder cell lines (20).
Figure 3.
SOX4 knockdown does not alter T24 bladder cancer cell proliferation or migration but does decrease invasiveness. (A) Five-day MTT assay showed no significant changes in proliferation as a result of SOX4 knockdown or overexpression compared to scrambled control. (B) Scratch wound assay indicated no significant changes in migratory patterns across all cell lines. These images are representative of one experiment and statistical analysis was performed using data from three independent experiments.
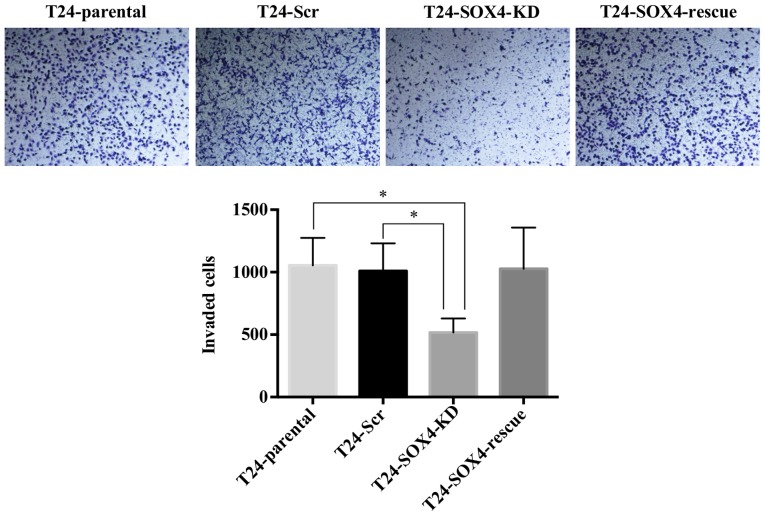
SOX4 has been shown to induce various cellular changes related to invasion, migration, and epithelial to mesenchymal transition (EMT) in other cell types (49,50). However, western blot analysis indicated no changes in the canonical EMT markers, ZEB1, E-Cadherin, N-Cadherin, or vimentin (Fig. S2) as a result of SOX4 knockdown in T24 cells. While we did not detect vimentin in the western blots, it was highly expressed in all samples at the mRNA level based on our microarray data (see below), and exhibited no significant changes. We nevertheless examined the effects of SOX4 knockdown on cellular migration and invasion. Although we observed no changes in migration by scratch-wound assay in the T24-SOX4-KD cells (Fig. 3B), we did observe that the T24-SOX4-KD cells exhibited a decreased invasion compared to the T24-Scr control cells that approached significance by ANOVA (P=0.0646) (Fig. 4). Moreover, the re-expression of SOX4 in the T24-SOX4-Rescue cells restored the invasive capabilities to levels similar to those of the T24-Scr controls, but had no effect on migration (Figs. 3B and 4).
Figure 4.
SOX4 knockdown decreases the invasion of T24 cells. Boyden chamber invasion assay showed a decreased invasive ability of T24-SOX4-KD cells and increased invasion in T24-SOX4-KD-Rescue. These images are representative of one experiment and statistical analysis was performed using data from three independent experiments. *P<0.05.
CRISPRi knockdown of SOX4 and gene expression analysis
To further understand global transcriptome changes as a result of SOX4 knockdown, we analyzed total RNA from the T24-SOX4-KD cells, T24-Scr and T24-SOX4-Rescue cells using Affymetrix Clariom D microarrays. Whole genome expression profiling analysis identified 1,487 genes significantly affected by SOX4 knockdown (FDR 0.05) compared to the T24-Scr cells (Table SI), and 561 genes significantly impacted by SOX4 re-expression (Table SII, GEO accession no. GSE118572). Ingenuity Pathway Analysis (IPA) between T24-Scr and T24-SOX4 knockdown revealed significantly upregulated osteoarthritis and Wnt/β-catenin signaling (Fig. S3A). Notably, the most significantly downregulated pathways in this analysis were associated with cholesterol metabolism (Fig. S3A). Our RT-qPCR validation of a selection of these cholesterol related genes confirmed the microarray data (Fig. S3B). To the best of our knowledge, SOX4 has not been previously associated with cholesterol biosynthesis pathways, and thus this remains an area for future investigation.
We also observed TNC as the most significantly upregulated gene in SOX4-KD cell lines and validated this finding by RT-qPCR (Fig. S3C). This potential regulation is supported by our prior data showing TNC as a SOX4 target gene in LNCaP prostate cancer cells (16), although in those cells we observed that SOX4 positively regulates TNC expression rather than represses TNC. This context-dependent difference in SOX4 activity suggests an opposite form of regulation in bladder cancer cell lines that could be due to the availability of other binding partners at the TNC promoter. Additionally, we observed a decrease in the IDO1 mRNA levels upon SOX4 knockdown (Fig. S3C). IDO1 is critical for immune system evasion in many cancers (51) and recent clinical trials of IDO1 blockers have shown promise in bladder cancer (52).
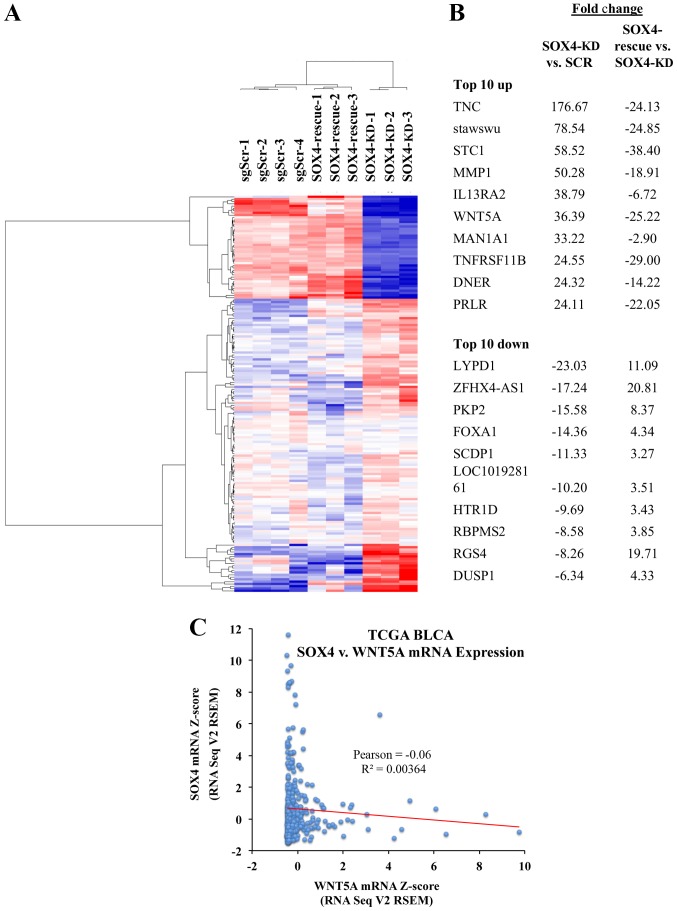
In order to identify genes regulated by SOX4 with high confidence, we compared the gene expression patterns of the T24-SOX4-KD, T24-Scr and T24-SOX4-Rescue cells. We identified 173 high-confidence genes regulated in opposite directions by SOX4 knockdown and re-expression (Fig. 5A) (for a complete list see Table SIII). The top 10 upregulated and downregulated genes are shown in Fig. 5B. We also noted that in the TCGA BLCA samples with a high SOX4 expression, the WNT5A levels are low and vice versa (Fig. 5C). While the negative Pearson's correlation (r=-0.06) was not significant (P=0.226), this may be due to the fact that the SOX4 regulation of WNT5A is likely indirect, and there are intermediary factors necessary for the regulation of WNT5A by SOX4. Some of the most significantly upregulated pathways via IPA analysis were also in the Wnt/β-catenin signaling and osteoarthritis pathways (Table I).
Figure 5.
Analysis of microarray data from SOX4 knockdown and re-expression. (A) Heatmap of 173 genes regulated as a result of SOX4 knockdown and re-expression in T24 cell lines. (B) List of top 10 upregulated and downregulated genes from the 173 gene data set. (C) Plot of WNT5A expression vs. SOX4 mRNA expression in the TCGA dataset indicating low WNT5A expression when SOX4 is high and low SOX4 expression when WNT5A is high.
Table I.
IPA analysis of statistically significant upregulated and downregulated pathways in T24 SOX4-KD cell lines compared to T24-SOX4-Rescue cell lines.
| ↓ or ↑ | P-value | Ingenuity canonical pathways |
|---|---|---|
| – | 2.29E-06 | Axonal guidance signaling |
| – | 1.78E-04 | Role of osteoblasts, osteoclasts and chondrocytes in rheumatoid arthritis |
| – | 4.07E-04 | Human embryonic stem cell pluripotency |
| ↓ | 1.41E-03 | Basal cell carcinoma signaling |
| ↓ | 1.45E-03 | Colorectal cancer metastasis signaling |
| – | 1.78E-03 | Role of Wnt/GSK-3 beta signaling in the pathogenesis of influenza |
| – | 2.24E-03 | Inhibition of matrix metalloproteases |
| – | 2.88E-03 | Ovarian cancer signaling |
| ↑ | 3.09E-03 | Osteoarthritis pathway |
| ↓ | 6.17E-03 | Wnt/β-catenin signaling |
| – | 7.94E-03 | PCP pathway |
| – | 8.71E-03 | Wnt/Ca2+ pathway |
| – | 8.91E-03 | Regulation of the epithelial-mesenchymal transition pathway |
| – | 9.12E-03 | Role of NANOG in mammalian embryonic stem cell pluripotency |
↓ or ↑ indicate down- or upregulation, respectively.
WNT5a antagonist restores the invasive ability of the T24-SOX4-KD cell line
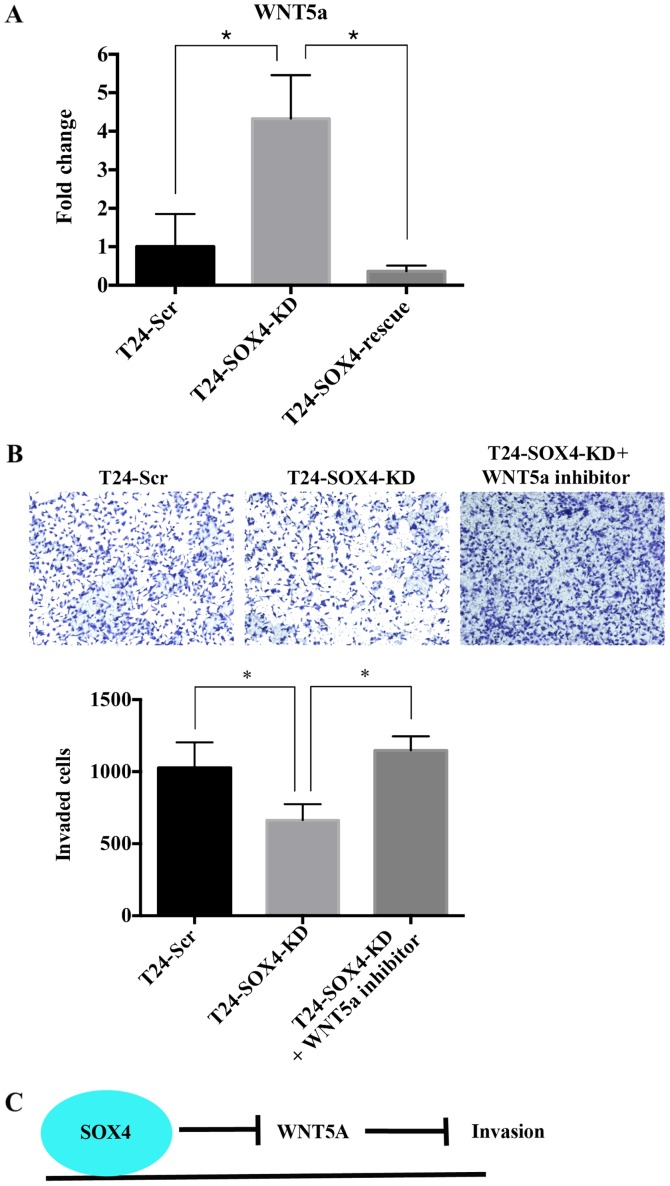
The top 10 putative SOX4-regulated genes included WNT5a, a non-canonical Wnt pathway ligand and a component of both Wnt/β-catenin signaling and osteoarthritis pathways. Moreover, WNT5a is one of the most statistically significant upregulated genes as a result of SOX4 knockdown and significantly downregulated upon the re-expression of SOX4. WNT5a expression has been shown to decrease the migratory or invasive characteristics in FTC-133 thyroid cell lines and EJ bladder cancer cell lines (53,54), a derivative of T24 cells. In this study, we confirmed the high expression of WNT5a in T24-SOX4-KD cell lines compared to the T24-Scr and T24-SOX4-Rescue cell lines by RT-qPCR (Fig. 6A), and hypothesized that WNT5a may mediate the effects of SOX4 by inhibiting the cellular invasion of T24 cells. We examined this hypothesis by treating the T24-SOX4-KD cells with a WNT5a peptide antagonist (200 µM), BOX5, for 24 h as previously described (31). Treatment of the T24-SOX4-KD cells with the WNT5a antagonist significantly increased invasiveness relative to the water vehicle-treated T24-SOX4-KD cells to levels comparable to those of the T24-Scr cells (P<0.05 by ANOVA) (Fig. 6B). These data suggest that SOX4 may inhibit WNT5a expression in T24 cells directly or indirectly, and that high WNT5a levels inhibit invasion in T24 bladder cancer cell lines (Fig. 6C).
Figure 6.
WNT5a antagonist restores invasive ability in T24-SOX4-KD cell line. (A) RT-qPCR validation of microarray for WNT5a mRNA expression across our T24 bladder cancer cell line samples. (B) Boyden chamber invasion assay showed the increased invasive ability of T24-SOX-KD cells treated with 200 µM of WNT5a antagonist. (C) Summary hypothesis of the effects of SOX4 on WNT5a and invasion. *P<0.05. These images are representative of 1 experiment and statistical analysis was performed using data from 3 independent experiment.
Discussion
While the precise function of SOX4 in bladder cancer is not yet well understood, the observation that it is overexpressed in as many as 23% of bladder cancer patients strongly supports the case for continued research into the role of SOX4 in bladder cancer.
While it has been well established that SOX4 expression is increased in a number of bladder cancer patients, some studies are in disagreement regarding associations between SOX4 expression levels and tumorigenicity, and tumor stage or grade (28,29), and there is not yet a consensus as to whether SOX4 expression is tumor-protective or tumor-promoting in bladder cancer patients. Tissue microarray analysis of 309 transitional cell carcinoma supported an oncogenic role for SOX4, since a high SOX4 expression was associated with a worse patient survival and was enriched in muscle-invasive patients (29). These data were contradicted by the tissue microarray from Aaboe et al, which showed that although SOX4 may be an early event in tumorigenesis, there was no association between SOX4 expression levels and tumor stage (28). Moreover, patients with a strong SOX4 expression, either cytoplasmic or nuclear, exhibited increased survival, suggesting a tumor-suppressive role (28). Discrepancies between these two immunohistochemical studies may be due to the fact that they used different SOX4 antibodies, and the possibility of cross-reactivity with other SOX family proteins.
In this study, we aimed to elucidate the role of SOX4 and to identify high confidence SOX4-regulated genes in T24 bladder cancer cells. We used a CRISPRi + re-expression system to identify 173 high confidence SOX4 regulated genes by whole genome expression profiling. Some of the most significantly altered pathways included the basal cell carcinoma, colorectal cancer metastasis, WNT/β-catenin and osteoarthritis pathways. Although neither SOX4 knockdown nor re-expression affected the proliferation rates of T24 cell lines compared to the scramble controls, we did observe a marked decrease in Matrigel invasion as a result of SOX4-KD and a restored invasion upon SOX4 re-expression. Whole genome expression profiling suggested that the non-canonical WNT5a pathway may play a critical role as a mediator of the effects of SOX4 on invasion.
Surprisingly, IPA analysis of genes significantly altered between the scramble control and the SOX4 KD group revealed that the most significantly downregulated pathway was the regulation of cholesterol biosynthesis. The most significantly downregulated genes in this group included TM7SF2, DHCR7 and MVD. However, the expression of this pathway was not rescued in the SOX4-overexpressed groups. There are a number of potential reasons for the lack of rescue for these genes. For example, the HA-tag fused to the amino-terminus of SOX4 could interfere with putative SOX4 binding partners, and the availability of co-activators may be limiting such that the reintroduction of SOX4 alone may not be sufficient to restore expression. This represents a limitation to our study not only for cholesterol biosynthesis pathway genes, but also for other genes putatively regulated by SOX4. Notably, previous studies support cholesterol biosynthesis in maintaining the mesenchymal state (55,56) and many of the enzymes affected by SOX4 also affect geranylgeranylation, which is critical for activity of Rac and Rho family GTPases (57), and thus this observation represents an area of great interest for further research.
A previous study with SOX4 in prostate cancer cells identified TNC as a direct target of SOX4 (16). Notably, the microarray data in this study revealed TNC as the most significantly upregulated gene in response to SOX4 KD, and the TNC levels decreased significantly when rescued with SOX4. These findings stand in contrast to our previous observations in LNCaP prostate cancer cells that suggest that SOX4 positively drives TNC expression (16), but are consistent with the finding that TNC is a target of SOX4 and that SOX4 represses TNC in human mammary epithelial HMLE cells (58). This observation may be due to context/cell line dependent differences in transcriptional networks and the availability of co-factors that in one context function as activators and then as repressors in another context.
Of note, our data corroborate previous results (28) using the transient expression of SOX4 in SOX4-null HU609 bladder cancer cell lines. The most prominent genes upregulated by SOX4 in that study included ZNF195, EFNA4 (EPHA4) and CGI-62. Our data confirmed the SOX4 positive regulation of both ZNF195 and EFNA4 (EPHA4) and the repression of NRP2 (Table SI) (28). However, we did not observe increased cell death as a result of SOX4 expression.
While the effects of SOX4 on canonical WNT/β-catenin signaling have been extensively studied, the effects of SOX4 on non-canonical WNT signaling are less clear. In this study, we identified WNT5a as one of the most significantly regulated genes affected in T24 SOX4 knockdown (+36.39 fold) and T24-SOX4-Rescue (-25.21 fold) cell lines, which we confirmed via RT-qPCR. Importantly, treatment of the T24-SOX4-KD cells with WNT5a antagonist restored the invasive phenotype to levels comparable to those of the T24-Scr cells. The WNT5a signaling pathway has two well established arms: The planar cell polarity and Ca2+ signaling pathways (59). Further research is required to evaluate which arm of the WNT5a pathway is active in T24-SOX4-KD cells.
Our observation that WNT5a may have a tumor-suppressive effect by means of decreasing invasion is consistent with previous studies on thyroid carcinoma cells in which WNT5a inhibited migration, invasion and proliferation (53). Similarly, WNT5a has been shown to impair the migration of breast epithelial cells (60). Moreover, patients with an increased WNT5a expression in Dukes B colon carcinomas exhibit improved 5- and 10-year survival rates compared to patients with the loss of WNT5a (61). Similarly, prostate cancer patients with low-grade localized disease and high WNT5a expression post-surgery have been shown to have much better outcomes than patients with a low WNT5a expression (62). Consistent with our data on SOX4-KD and SOX4-Rescue cells, the siRNA-mediated knockdown of WNT5a has been shown to increase the invasive ability of LNCAP and 22RV1 prostate cancer cells (62).
In contrast to our results, a previous study demonstrated that the SOXC family of transcription factors (SOX4, SOX11 and SOX12) positively regulate WNT5a expression in mouse growth plate chondrocytes, although this was mostly driven by SOX11 and no direct regulation was demonstrated (63). Notably, our microarray data revealed no significant gene expression changes in SOX11 or SOX12, suggesting that SOX4 can regulate WNT5a without changes in other SOXC family members. Promoter sequence analysis did not identify any obvious SOX4 binding sites in WNT5a regulatory regions, and thus it is likely that SOX4 may indirectly regulate WNT5a expression. Further studies are required to adequately evaluate the mechanism by which SOX4 regulates WNT5a either directly or indirectly.
In conclusion, our findings suggest a mechanism through which SOX4 contributes to overall tumor aggressiveness in bladder cancer by modulating cellular invasion. Taken together, these data provide further evidence of a tumor-promoting role for SOX4 and a tumor suppressive mechanism of WNT5a, and suggest a novel mechanism of SOX4 regulation of non-canonical Wnt signaling. While the effects of SOX4 on activated canonical WNT signaling through β-catenin have been well established (64), to the best of our knowledge, this is the first study to demonstrate that SOX4 may repress non-canonical WNT5a signaling in bladder cancer cells. Additional research is needed to further clarify the role of SOX4 in bladder cancer.
Supplementary Data
Acknowledgments
The research reported in this publication was supported in part by the Emory Integrated Genomics Core (EIGC) Shared Resource of Winship Cancer Institute of Emory University and NIH/NCI under award number UL1TR002378. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding
This study was funded by NIH grant no. U01 CA217875-01. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data, in the writing of the manuscript, and in the decision to publish the results.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the GEO repository, accession no. GSE118572 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118572).
Authors' contributions
JDM and CSM were involved in the conceptualization of the study. JDM was involved in the methodology. JDM and CSM provided the software. JDM, ZL and CSM were involved in data validation. JDM and ZL were involved in formal analysis. JDM, HK and ZL performed the experiments and data generation. JDM, HHK and ZL were involved in data curation. JDM was involved in the writing of the original draft preparation. JDM and CSM were involved in the writing, reviewing and editing of the manuscript. JDM and HK were involved in visualization. CSM supervised the study and JDM was involved in project administration. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, Kassouf W, Kiemeney LA, La Vecchia C, Shariat S, et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63:234–241. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 2.Hashim D, Boffetta P. Occupational and environmental exposures and cancers in developing countries. Ann Glob Health. 2014;80:393–411. doi: 10.1016/j.aogh.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Smith ND, Prasad SM, Patel AR, Weiner AB, Pariser JJ, Razmaria A, Maene C, Schuble T, Pierce B, Steinberg GD. Bladder Cancer Mortality in the United States: A Geographic and Temporal Analysis of Socioeconomic and Environmental Factors. J Urol. 2016;195:290–296. doi: 10.1016/j.juro.2015.07.091. [DOI] [PubMed] [Google Scholar]
- 4.Hurst CD, Knowles MA. Bladder cancer: Multi-omic profiling refines the molecular view. Nat Rev Clin Oncol. 2018;15:203–204. doi: 10.1038/nrclinonc.2017.195. [DOI] [PubMed] [Google Scholar]
- 5.Aine M, Eriksson P, Liedberg F, Höglund M, Sjödahl G. On molecular classification of bladder cancer: Out of one, many. Eur Urol. 2015;68:921–923. doi: 10.1016/j.eururo.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 6.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA, Akbani R, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2018;174:1033. doi: 10.1016/j.cell.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sjödahl G, Eriksson P, Liedberg F, Höglund M. Molecular classification of urothelial carcinoma: Global mRNA classification versus tumour-cell phenotype classification. J Pathol. 2017;242:113–125. doi: 10.1002/path.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Network TCGAR. Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harley V, Lefebvre V. Twenty Sox, twenty years. The International Int J Biochem Cell Biol. 2010;42:376–377. doi: 10.1016/j.biocel.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Aue G, Du Y, Cleveland SM, Smith SB, Davé UP, Liu D, Weniger MA, Metais JY, Jenkins NA, Copeland NG, et al. Sox4 cooperates with PU.1 haploinsufficiency in murine myeloid leukemia. Blood. 2011;118:4674–4681. doi: 10.1182/blood-2011-04-351528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikushima H, Todo T, Ino Y, Takahashi M, Saito N, Miyazawa K, Miyazono K. Glioma-initiating cells retain their tumorigenicity through integration of the Sox axis and Oct4 protein. J Biol Chem. 2011;286:41434–41441. doi: 10.1074/jbc.M111.300863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol. 2004;24:8834–8846. doi: 10.1128/MCB.24.20.8834-8846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dy P, Penzo-Méndez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins–Sox4, Sox11 and Sox12 - exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jauch R, Ng CK, Narasimhan K, Kolatkar PR. The crystal structure of the Sox4 HMG domain-DNA complex suggests a mechanism for positional interdependence in DNA recognition. Biochem J. 2012;443:39–47. doi: 10.1042/BJ20111768. [DOI] [PubMed] [Google Scholar]
- 16.Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25:5626–5639. doi: 10.1038/sj.onc.1209566. [DOI] [PubMed] [Google Scholar]
- 18.Jafarnejad SM, Ardekani GS, Ghaffari M, Martinka M, Li G. Sox4-mediated Dicer expression is critical for suppression of melanoma cell invasion. Oncogene. 2013;32:2131–2139. doi: 10.1038/onc.2012.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen H, Morrison CD, Zhang J, Underwood W, III, Yang N, Frangou C, Eng K, Head K, Bollag RJ, Kavuri SK, et al. 6p22.3 amplification as a biomarker and potential therapeutic target of advanced stage bladder cancer. Oncotarget. 2013;4:2124–2134. doi: 10.18632/oncotarget.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Q, Hoffmann MJ, Hartmann FH, Schulz WA. Amplification and overexpression of the ID4 gene at 6p22.3 in bladder cancer. Mol Cancer. 2005;4:16. doi: 10.1186/1476-4598-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: Friend or foe? Oncogene. 2013;32:3397–3409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 23.Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye DL, et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 24.Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T, Zhang HY, Gong WL, Yu M, Man JH, et al. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci USA. 2009;106:3788–3793. doi: 10.1073/pnas.0810147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hur EH, Hur W, Choi JY, Kim IK, Kim HY, Yoon SK, Rhim H. Functional identification of the pro-apoptotic effector domain in human Sox4. Biochem Biophys Res Commun. 2004;325:59–67. doi: 10.1016/j.bbrc.2004.09.215. [DOI] [PubMed] [Google Scholar]
- 26.Hur W, Rhim H, Jung CK, Kim JD, Bae SH, Jang JW, Yang JM, Oh ST, Kim DG, Wang HJ, et al. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: Clinical implication and functional analysis in vitro. Carcinogenesis. 2010;31:1298–1307. doi: 10.1093/carcin/bgq072. [DOI] [PubMed] [Google Scholar]
- 27.Ahn SG, Cho GH, Jeong SY, Rhim H, Choi JY, Kim IK. Identification of cDNAs for Sox-4, an HMG-Box protein, and a novel human homolog of yeast splicing factor SSF-1 differentially regulated during apoptosis induced by prostaglandin A2/delta12-PGJ2 in Hep3B cells. Biochem Biophys Res Commun. 1999;260:216–221. doi: 10.1006/bbrc.1999.0856. [DOI] [PubMed] [Google Scholar]
- 28.Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sørensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjøt L, Ørntoft T. SOX4 expression in bladder carcinoma: Clinical aspects and in vitro functional characterization. Cancer Res. 2006;66:3434–3442. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- 29.Shen H, Blijlevens M, Yang N, Frangou C, Wilson KE, Xu B, Zhang Y, Zhang L, Morrison CD, Shepherd L, et al. Sox4 expression confers bladder cancer stem cell properties and predicts for poor patient outcome. Int J Biol Sci. 2015;11:1363–1375. doi: 10.7150/ijbs.13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenei V, Sherwood V, Howlin J, Linnskog R, Säfholm A, Axelsson L, Andersson T. A t-butyloxycarbonyl-modified Wnt5a-derived hexapeptide functions as a potent antagonist of Wnt5a-dependent melanoma cell invasion. Proc Natl Acad Sci USA. 2009;106:19473–19478. doi: 10.1073/pnas.0909409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai YH, Cheng J, Cheng D, Feasel ME, Beste KD, Peng J, Nusrat A, Moreno CS. SOX4 interacts with plakoglobin in a Wnt3a-dependent manner in prostate cancer cells. BMC Cell Biol. 2011;12:50. doi: 10.1186/1471-2121-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- 34.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBio-Portal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubin MA, Chinnaiyan AM. Bioinformatics approach leads to the discovery of the TMPRSS2:ETS gene fusion in prostate cancer. Lab Invest. 2006;86:1099–1102. doi: 10.1038/labinvest.3700477. [DOI] [PubMed] [Google Scholar]
- 42.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 43.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 44.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kearns NA, Genga RMJ, Enuameh MS, Garber M, Wolfe SA, Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai W, Xu X, Li S, Ma J, Shi Q, Guo S, Liu L, Guo W, Xu P, He Y, et al. SOX4 promotes proliferative signals by regulating glycolysis through AKT activation in melanoma cells. J Invest Dermatol. 2017;137:2407–2416. doi: 10.1016/j.jid.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J, Xu J, Zhang R. MicroRNA-539 inhibits colorectal cancer progression by directly targeting SOX4. Oncol Lett. 2018;16:2693–2700. doi: 10.3892/ol.2018.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, Feng J, Zhang Y, Gao H, Liu DX, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72:4597–4608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Chen P, Zu L, Liu B, Wang M, Zhou Q. MicroRNA-338-3p suppresses metastasis of lung cancer cells by targeting the EMT regulator Sox4. Am J Cancer Res. 2016;6:127–140. [PMC free article] [PubMed] [Google Scholar]
- 51.Zhai L, Ladomersky E, Lenzen A, Nguyen B, Patel R, Lauing KL, Wu M, Wainwright DA. IDO1 in cancer: A Gemini of immune checkpoints. Cell Mol Immunol. 2018;15:447–457. doi: 10.1038/cmi.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Article CD. Blocking IDO1 helps shrink bladder, cervical tumors. Cancer Discov. 2018;8:OF3. doi: 10.1158/2159-8290.CD-NB2017-167. [DOI] [PubMed] [Google Scholar]
- 53.Kremenevskaja N, von Wasielewski R, Rao AS, Schöfl C, Andersson T, Brabant G. Wnt-5a has tumor suppressor activity in thyroid carcinoma. Oncogene. 2005;24:2144–2154. doi: 10.1038/sj.onc.1208370. [DOI] [PubMed] [Google Scholar]
- 54.Cao Y, Wang X, Xu C, Gao Z, Zhou H, Wang Y, Cao R, Liu T, Liu T. 4-HPR impairs bladder cancer cell migration and invasion by interfering with the Wnt5a/JNK and Wnt5a/MMP-2 signaling pathways. Oncol Lett. 2016;12:1833–1839. doi: 10.3892/ol.2016.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen Z, Zhu D, Liu J, Chen J, Liu Y, Hu C, Li Z, Li Y. 27-Hydroxycholesterol induces invasion and migration of breast cancer cells by increasing MMP9 and generating EMT through activation of STAT-3. Environ Toxicol Pharmacol. 2017;51:1–8. doi: 10.1016/j.etap.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 56.Marwarha G, Raza S, Hammer K, Ghribi O. 27-hydroxycho-lesterol: A novel player in molecular carcinogenesis of breast and prostate cancer. Chem Phys Lipids. 2017;207:108–126. doi: 10.1016/j.chemphyslip.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 57.Roy M, Kung HJ, Ghosh PM. Statins and prostate cancer: role of cholesterol inhibition vs. prevention of small GTP-binding proteins. Am J Cancer Res. 2011;1:542–561. [PMC free article] [PubMed] [Google Scholar]
- 58.Vervoort SJ, Lourenço AR, Tufegdzic Vidakovic A, Mocholi E, Sandoval JL, Rueda OM, Frederiks C, Pals C, Peeters JG, Caldas C, et al. SOX4 can redirect TGF-beta-mediated SMAD3-transcriptional output in a context-dependent manner to promote tumorigenesis. Nucleic Acids Res. 2018;46:9578–9590. doi: 10.1093/nar/gky755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishita M, Enomoto M, Yamagata K, Minami Y. Cell/tissue-tropic functions of Wnt5a signaling in normal and cancer cells. Trends Cell Biol. 2010;20:346–354. doi: 10.1016/j.tcb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 60.Säfholm A, Leandersson K, Dejmek J, Nielsen CK, Villoutreix BO, Andersson T. A formylated hexapeptide ligand mimics the ability of Wnt-5a to impair migration of human breast epithelial cells. J Biol Chem. 2006;281:2740–2749. doi: 10.1074/jbc.M508386200. [DOI] [PubMed] [Google Scholar]
- 61.Dejmek J, Dejmek A, Säfholm A, Sjölander A, Andersson T. Wnt-5a protein expression in primary dukes B colon cancers identifies a subgroup of patients with good prognosis. Cancer Res. 2005;65:9142–9146. doi: 10.1158/0008-5472.CAN-05-1710. [DOI] [PubMed] [Google Scholar]
- 62.Syed Khaja AS, Helczynski L, Edsjö A, Ehrnström R, Lindgren A, Ulmert D, Andersson T, Bjartell A. Elevated level of Wnt5a protein in localized prostate cancer tissue is associated with better outcome. PLoS One. 2011;6:e26539. doi: 10.1371/journal.pone.0026539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kato K, Bhattaram P, Penzo-Méndez A, Gadi A, Lefebvre V. SOXC transcription factors induce cartilage growth plate formation in mouse embryos by promoting noncanonical WNT signaling. J Bone Miner Res. 2015;30:1560–1571. doi: 10.1002/jbmr.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee AK, Ahn SG, Yoon JH, Kim SA. Sox4 stimulates β-catenin activity through induction of CK2. Oncol Rep. 2011;25:559–565. doi: 10.3892/or.2010.1091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the GEO repository, accession no. GSE118572 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118572).