Summary
Endocrine therapy (ET) is the standard of care for estrogen receptor-positive (ER+) breast cancers. Despite its efficacy, ∼40% of women relapse with ET-resistant (ETR) disease. A global transcription analysis in ETR cells reveals a downregulation of the neutral and basic amino acid transporter SLC6A14 governed by enhanced miR-23b-3p expression, resulting in impaired amino acid metabolism. This altered amino acid metabolism in ETR cells is supported by the activation of autophagy and the enhanced import of acidic amino acids (aspartate and glutamate) mediated by the SLC1A2 transporter. The clinical significance of these findings is validated by multiple orthogonal approaches in a large cohort of ET-treated patients, in patient-derived xenografts, and in in vivo experiments. Targeting these amino acid metabolic dependencies resensitizes ETR cells to therapy and impairs the aggressive features of ETR cells, offering predictive biomarkers and potential targetable pathways to be exploited to combat or delay ETR in ER+ breast cancers.
Keywords: endocrine therapy, resistance, metabolic reprogramming, estrogen receptor, amino acid transporters, aspartate, glutamate, miRNA, SLCs
Graphical Abstract
Highlights
-
•
ETR cells show high miR-23b-3p that reduces SLC6A14 and amino acids upload
-
•
ETR cells promote autophagy and aspartate and glutamate import via SLC1A2
-
•
Aspartate and glutamate fuel anabolic and catabolic pathways in ETR breast cancers
-
•
Targeting amino acid metabolic reprogramming is effective in ETR cells
Bacci et al. find that endocrine-resistant ER+ breast cancers are characterized by enhanced miR-23b-3p, autophagy activation, and import of aspartate and glutamate that fuel catabolic and anabolic pathways, which are essential for their aggressive features. The molecular players involved in this metabolic scenario are of clinical significance and have prognostic and predictive value.
Introduction
The majority of breast tumors are positive for estrogen receptor α (called hereafter ER) and/or progesterone receptor (PR) and negative for human epidermal growth factor receptor 2 (HER2). Since these tumors are dependent on estrogen for growth and survival, inhibiting this dependency with endocrine therapies (ETs) is the standard of care for these patients. ET agents include selective ER modulators (e.g., tamoxifen) which compete with estrogen for ER binding; selective ER downregulators (e.g., fulvestrant) that bind to and destabilize ER, inducing its degradation; and aromatase inhibitors (AIs; e.g., letrozole, anastrozole) which block the conversion of androgens to estrogens lowering estrogen levels. Over 50% of the ET-treated patients show an initial clinical benefit, but ∼40% experience de novo or acquired ET resistance (ETR). Therefore, there remains an urgent need for more effective therapeutic strategies. We have previously demonstrated that microRNA (miRNA)-dependent regulation of the expression of key metabolic genes enhances glucose-dependent metabolic plasticity in ETR cells (Bacci et al., 2016) and that impairing this reprogramming resensitizes ETR cells to therapy (Morandi and Indraccolo, 2017). Concomitantly, a number of metabolic stimuli (e.g., nutrients, hormones, cytokines) modulate the miRNA expression, thus establishing a functional association, which can be altered to perturb energy homeostasis, as in the case of many tumors (Dumortier et al., 2013).
Here, we performed genome-wide expression profiling of ETR long-term estrogen-deprived (LTED) ER+ breast cancer cells that reveals a deregulation of amino acid transporters. Notably, the mechanism that sustains amino acid metabolism in ETR is not exclusively associated with estrogen deprivation but concurs with fulvestrant and tamoxifen resistance, thus highlighting the potential metabolic vulnerabilities to be exploited for either targeting or prognostic purposes.
Results
Genome Profiling Reveals a Deregulated miR-23b-3p/SLC6A14 Axis in ETR Cells with Prognostic Value in ER+ Breast Cancer
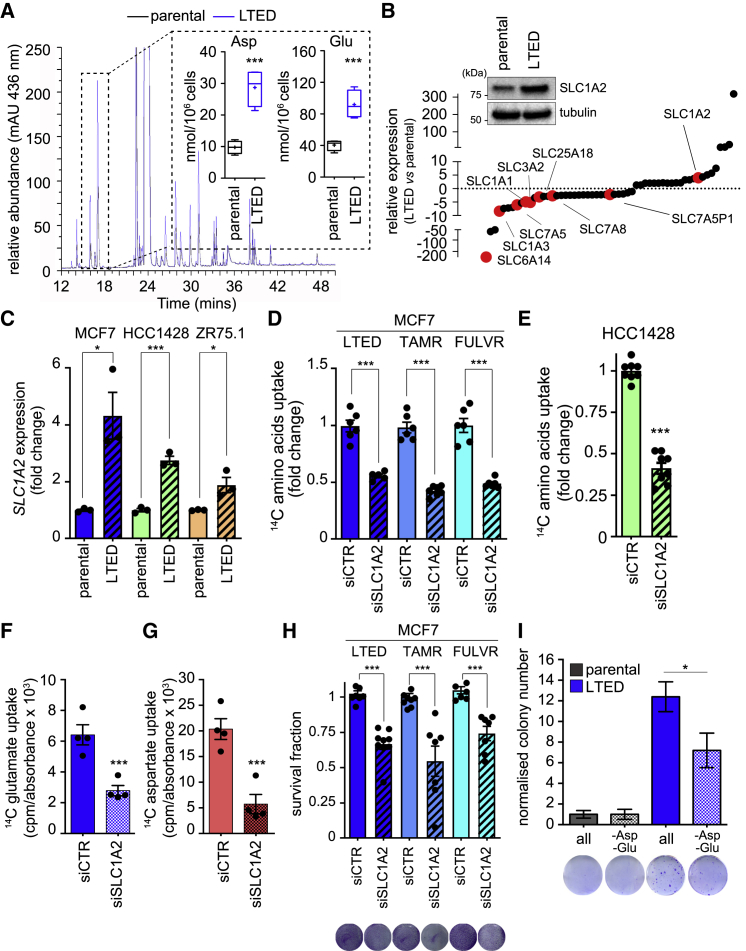
To address the interconnection between miRNAs and genes associated with estrogen deprivation, we analyzed the global gene and miRNA expression profile from 3 independent experiments. We used an isogenic model in which parental MCF7 cells in the presence of estrogen (E2) were used to model a patient at primary diagnosis, MCF7 in the absence of E2 to mimic response to an AI, and finally LTED MCF7 cells to model relapse on AIs. Statistical analysis (1-way ANOVA, Benjamini-Hochberg correction followed by Student-Newman-Keuls post hoc tests; false discovery rate [FDR] <0.05, fold-change >2 and <−2) revealed 62 miRNAs and 3,568 mRNAs significantly deregulated in MCF7-LTED versus MCF7, with or without E2. Within the parental MCF7 cells, 603 genes and 14 miRNAs were found differently regulated upon E2 treatment (Table S1). Supervised hierarchical cluster analysis shows comparable clustering of both miRNA and gene expression data, with a clear separation between MCF7-LTED samples and the parental MCF7 cells subdivided into E2-treated and short-term E2-deprived samples (Figure 1A). Although E2 treatment had, as expected, a significant impact on gene expression (Table S1), we focused on the differences between LTED and parental MCF7 cells either in the presence or absence of E2 addition to model a clinical scenario that is independent of circulating E2 levels or prior endocrine agent administration, such as that of a post-menopausal patient who relapses after AI treatment (Table S1).
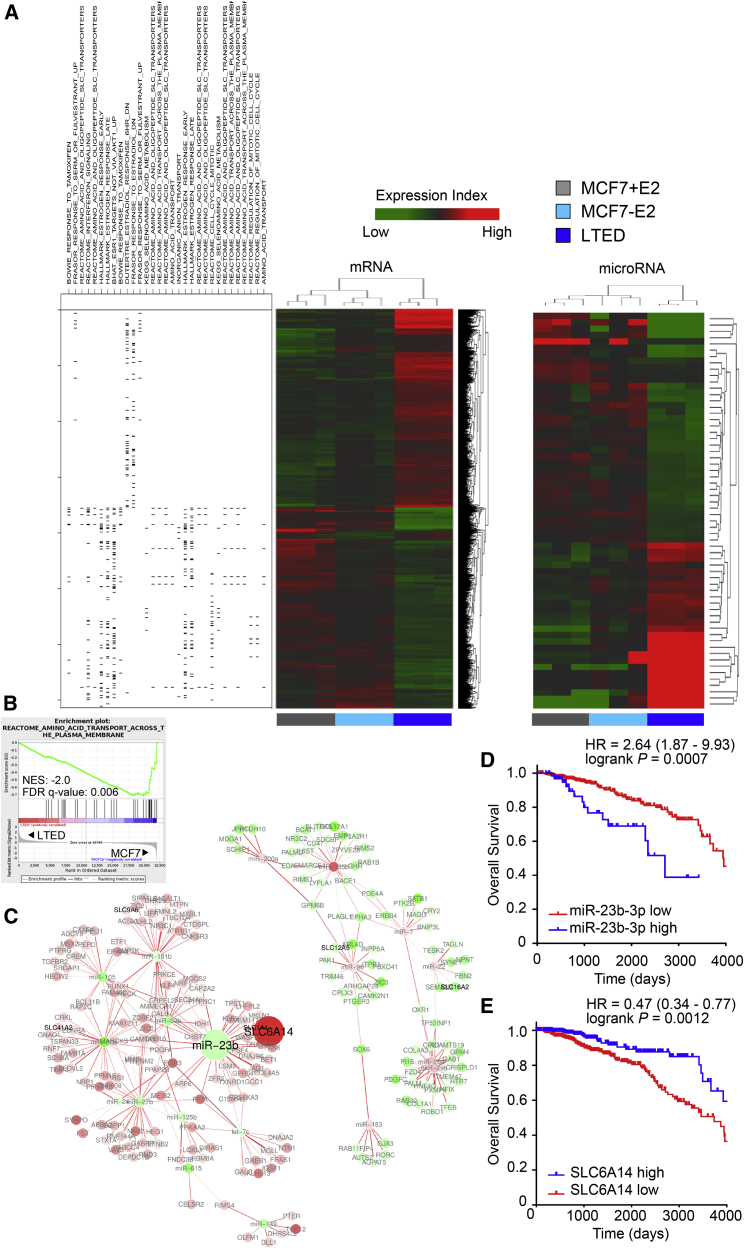
Figure 1.
Global Gene Expression and miRNA Analysis Reveals a Deregulated miR-23b-3p/SLC6A14 Node in LTED Compared to Parental MCF7 Cells
(A) Supervised hierarchical clustering of RNA transcripts (left) and miRNAs (right) of 3 biological replicates of MCF7, MCF7 deprived from estrogen (E2) for 3 days, and LTED cells that were subjected to gene or miRNA expression profiling using an Agilent Technologies assay. Differentially expressed genes (n = 3,568; ANOVA) and miRNAs (n = 62; ANOVA) were used for heatmap generation. High and low expression is normalized to the average expression across all of the samples. On the left side, association of the genes in the heatmap with leading edge genes resulting from GSEA analysis on the specified gene sets is identified.
(B) Gene set enrichment analysis enrichment plot of amino acid transport dataset showing a negative association between the MSigDb M188 dataset and the LTED gene expression profile. NES, normalized enrichment score.
(C) Anti-correlation network of differentially expressed miRNAs and mRNAs. Edge color is proportional to the strength of the anti-correlation (white to red = low to high). Node color is proportional to the LTED versus MCF7 log fold change (red, downregulated; green, upregulated). Solute carrier (SLC) genes are labeled in black; other miRNAs or mRNAs are labeled in gray. miR-23b and SLC6A14 nodes are highlighted as large circles.
(D and E) Kaplan-Meier analysis of overall survival of BRCA TCGA cohort of ER+ patients divided into high and low expressing, as described in Method Details, for miR-23b-3p expression (D: TGCA, high expressing, n = 68; low expressing, n = 475) or SLC6A14 (E: TGCA, high expressing, n = 257; low expressing, n = 467).
Using gene set enrichment analysis (GSEA) (Subramanian et al., 2005) (Table S2), many of the differentially expressed gene sets in LTED versus MCF7 cells are related to metabolic pathways (Figure 1A). In particular, the LTED profile is negatively correlated with the gene set associated with amino acid transporters and metabolism (Figure 1B). Since miRNAs exert their role by binding to the target mRNAs and impairing their translation, their expression profiles are expected to be inversely correlated. Therefore, we created a correlative network based on gene and miRNA expression profiles to identify potential deregulated miRNA-mRNA nodes (Figure 1C). Since the identification of metabolic dependencies could be of therapeutic and predictive value in ER+ breast cancer and GSEA analysis revealed deregulated amino acid transport associated with LTED, we focused our attention on a metabolic-related network involving the solute carrier (SLC) family 6 member 14 (SLC6A14), a sodium-chloride-dependent amino acid transporter for all of the amino acids, with the exception of glutamate and aspartate (Babu et al., 2015). The SLC family plays a significant role in mediating amino acid transport across the plasma membrane (Hediger et al., 2004). Among the miRNAs reported to correlate with SLC6A14 (Figure S1A) was miR-23b-3p, which, as expected, showed an inverse correlation (Bisognin et al., 2012). Furthermore, miR-23b-3p was significantly upregulated in LTED cells compared to the parental MCF7 cells, independently of E2 treatment (Table S1). The clinical relevance of the deregulated miR-23b-3p/SLC6A14 node observed was validated in retrospective clinical data. High expression of miR-23b-3p correlates with reduced survival in ER+ breast cancer patients (hazard ratio [HR] = 2.64, log-rank p = 0.0007, n = 543; Figure 1D). Conversely, high expression of SLC6A14 identifies a subset of good-prognosis ER+ breast cancer patients (HR = 0.47, log-rank p = 0.0012, n = 724; Figure 1E). Similar results were obtained in an independent cohort of ER+ breast cancers (Figures S1B and S1C). A correlation analysis between miR-23b-3p and SLC6A14 was performed on the TCGA patients’ dataset selected based on higher (upper quartile) and lower (lower quartile) expression levels of miR-23b-3p (n = 104). The correlation is significant and negative, and, crucially, SLC6A14 expression is completely lost in the high miR-23b-3p-expressing tumors (Figure S1D).
miR-23b-3p and SLC6A14 Expression Is Deregulated in ETR Models
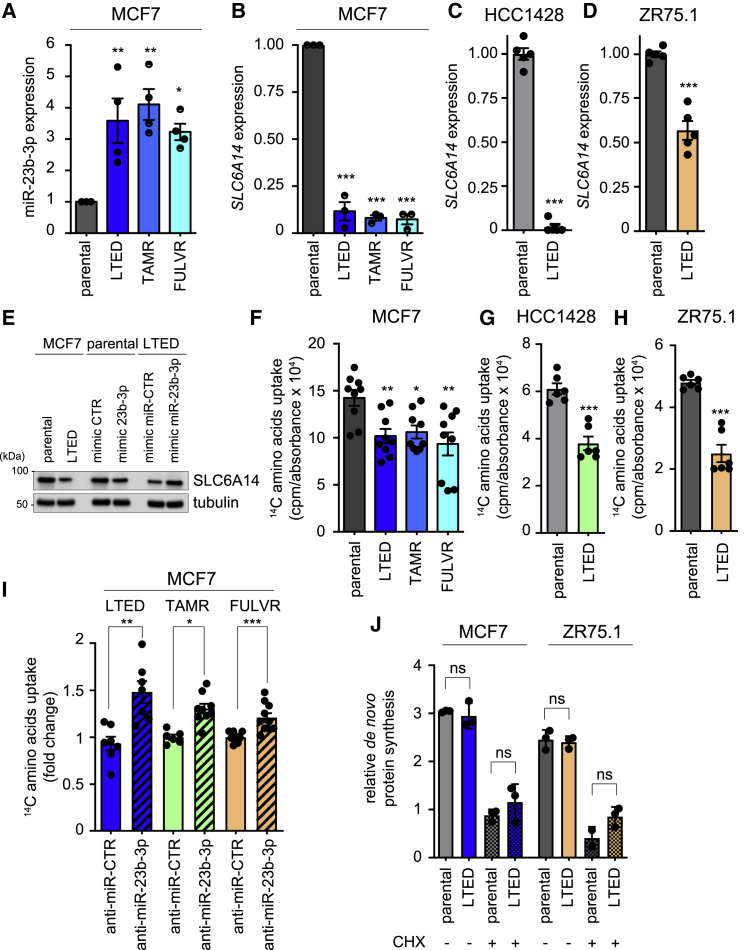
To determine whether the deregulation of the miR-23b-3p/SLC6A14 expression axis was a prerequisite of cells that are resistant to E2 deprivation or more generally a feature of ETR cells, we evaluated their expression in the LTED cells and in the MCF7-TAMR and MCF7-FULVR cell lines that are MCF7 derivatives, mimicking the acquired resistance to tamoxifen and fulvestrant, respectively. qRT-PCR analysis revealed a significant increase in the expression of miR-23b-3p (Figure 2A) and the reduced expression of SLC6A14 (Figures 2B–2D) in all of the ETR lines when compared to their parental counterpart. The miR-23b-3p mimic construct transfected into parental cells (Figure S2A) induced SLC6A14 downregulation (Figure 2E), whereas the miR-23b-3p inhibitor transfected into LTED cells (Figure S2B) led to an SLC6A14 increase, thereby demonstrating the functional relation between miR-23b-3p and SLC6A14 (Figure 2E). Consistent with reduced SLC6A14 expression, all of the ETR cells displayed a reduced uptake of exogenous 14C-labeled amino acids when compared to the parental cell counterparts (Figures 2F–2H). This impairment in amino acids uptake could be reversed by downregulating miR-23b-3p expression (Figures 2I and S2B). However, despite the reduced amino acids uptake, no significant difference was observed in de novo protein synthesis (Figure 2J), suggesting no major impact on protein availability to support ETR cell growth. Moreover, anti-miR-23b-3p treatment slightly but significantly impaired LTED and TAMR MCF7 cell survival (Figures S2C and S2D).
Figure 2.
miR-23b-3p/SLC6A14 Axis Is Deregulated in Different ETR Cells
(A–D) ETR-derived and parental MCF7 (A and B), HCC1428 (C), and ZR75.1 (D) cells were subjected to qRT-PCR analysis using the assays described in the figure. Relative expression is shown using the parental cells as comparator. Data represent means ± SEMs. One-way ANOVA; Dunnett corrected; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Each dot represents a biological replicate.
(E) Total protein lysates from MCF7 and LTED cells transfected with the oligos as described in the figure for 72 h were subjected to western blot analysis, as indicated.
(F–H) 14C-amino acids mixture upload was measured in ETR-derived and parental MCF7 (F), HCC1428 (G), and ZR75.1 (H) cells. The relative upload capacity is shown using parental cells as comparator. Data represent means ± SEMs. One-way ANOVA; Dunnett corrected; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Each dot represents a biological replicate.
(I) 14C-amino acids mixture upload was measured in ETR-derived MCF7 cells transfected for 72 h with either anti-miR-CTR or anti-miR-23b-3p oligos. The relative upload capacity is shown using anti-miR-CTR transfected cells as comparator. Data represent means ± SEMs. Student’s t test; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Each dot represents a biological replicate.
(J) LTED derivatives and parental counterparts of MCF7 and ZR75.1 cells were subjected to protein synthesis assay, as described in Method Details. No differences were observed between parental and LTED cells in the absence or presence of the protein synthesis inhibitor cycloheximide (CHX, 50 μg/mL for 30 min). Data represent means ± SEMs. One-way ANOVA; Dunnett corrected; ns, not significant.
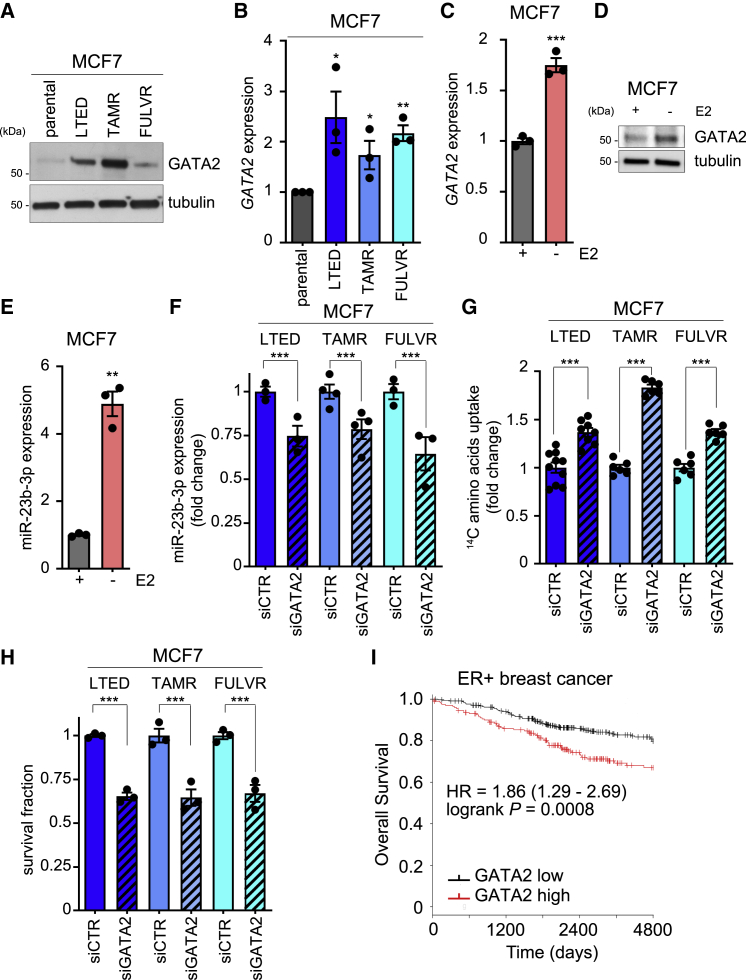
GATA2 Is Enhanced during ETR and Controls miR-23b-3p-Dependent Amino Acids Upload
GATA2 is a transcription factor involved in embryonic development, self-renewal, and stemness found overexpressed and involved in cancer (Rodriguez-Bravo et al., 2017). GATA2 expression is negatively controlled by E2 (Yang et al., 2017), and miR-23b-3p is a putative target of GATA2 (JASPAR Predicted Transcription Factor Targets, http://amp.pharm.mssm.edu/Harmonizome). GATA2 expression was significantly enhanced in ETR cells both at the protein and mRNA levels (Figures 3A and 3B). E2 deprivation induced enhanced GATA2 expression (Figures 3C and 3D) and the subsequent increase in miR-23b-3p expression (Figure 3E) in the parental cells, thus reinforcing the functional link between E2 deprivation, GATA2, and miR-23b-3p that characterized E2-deprived and endocrine agent-treated cells. Silencing GATA2 in all of the high-expressing ETR cells (Figure S3A) resulted in reduced miR-23b-3p expression (Figure 3F), enhanced amino acids uptake (Figure 3G), and impaired cell survival (Figure 3H). The clinical significance of GATA2 in ER+ breast cancer was validated in a large retrospective breast cancer patient cohort in which GATA2 higher expression identifies a subset of ER+ breast cancers characterized by poorer prognosis (HR = 1.86, log-rank p = 0.0008, n = 548; Figure 3I).
Figure 3.
GATA2 Is Enhanced during ETR and Controls miR-23b-3p-Dependent Amino Acids Upload
(A) Total protein lysates from ETR-derived and parental MCF7 cells were analyzed by western blotting with the antibody indicated.
(B) ETR-derived and parental MCF7 cells were subjected to qRT-PCR for GATA2 analysis. Relative expression is shown using the parental cells as comparator. Data represent means ± SEMs. One-way ANOVA; Dunnett corrected; ∗p < 0.05; ∗∗p < 0.01. Each dot represents a biological replicate.
(C–E) Parental MCF7 cells were 3-day E2 deprived and subjected to either qRT-PCR (C and E) or western blot (D) analyses, as indicated in the figure. Data represent means ± SEMs. Student’s t test; ∗∗p < 0.01; ∗∗∗p < 0.001.
(F–H) ETR-MCF7 cell derivatives transfected with non-targeting small interfering RNA (siRNA) (siCTR) or siRNA targeting GATA2 (siGATA2) for 72 h were subjected to qRT-PCR analysis for miR-23b-3p expression (F), 14C-amino acids upload capacity (G), or cell viability assay using crystal violet (H). siCTR transfected cells were used as comparator. Data represent means ± SEMs. Student’s t test; ∗∗∗p < 0.001.
(I) Kaplan-Meier analysis of overall survival of ER+ breast cancer patients divided into high and low GATA2 expressing, as described in Method Details. Hazard ratio (HR) and log-rank Mantel-Cox p values are shown.
Enhanced Autophagic Flux Is Essential for ETR Cell Survival
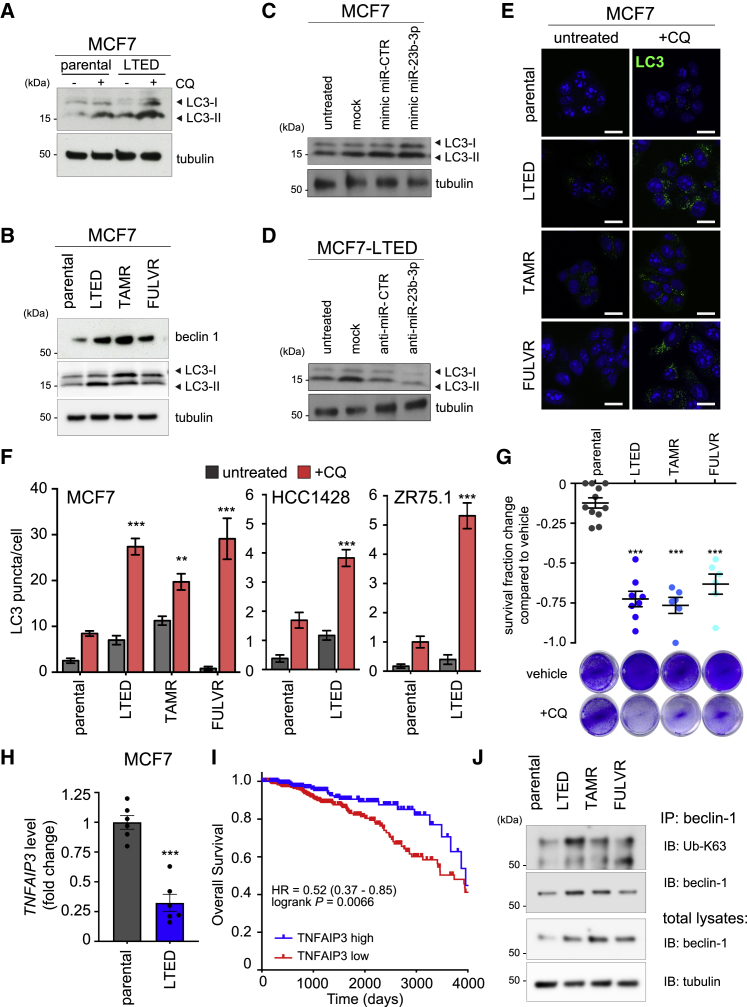
Despite the reduced amino acids uptake in ETR cells, we did not observe a significant reduction in cell growth, protein synthesis, or protein content between LTED and parental cells (Figures 2J, S4A, and S4B). Therefore, we hypothesized that LTED cells may compensate for decreased amino acids uptake by activating macro-autophagy, hereafter referred as autophagy. Autophagy allows a cell to recycle dispensable and/or dysfunctional components to fuel catabolic and anabolic processes, including protein synthesis (Galluzzi et al., 2015). Moreover, SLC6A14 downregulation has been reported to induce autophagy activation (Coothankandaswamy et al., 2016). Western blot analysis revealed that LTED cells have increased levels of the microtubule-associated protein 1A-1B-light chain 3 (LC3), an established marker of autophagy activation, which was further enhanced by chloroquine (CQ) administration (Figure 4A). CQ is a lysosomotropic agent that prevents endosomal acidification, hence inhibiting lysosomal hydrolases and preventing autophagosomal fusion and degradation. Since autophagy is a dynamic process, acute CQ administration results in the accumulation of autophagosomes, and the observed LC3-II increase is thus directly correlated with the autophagic flux (Yoshii and Mizushima, 2017). Additional regulators of autophagy induction include mammalian target of rapamycin (mTOR) inhibition and beclin-1 expression, which functions as the central scaffold protein for autophagosome formation. The 3 MCF7 ETR derivatives showed increased levels of beclin-1 and LC3-II expression (Figure 4B). The autophagy activation described is functionally linked to miR-23b-3p expression, since ectopic miR-23b-3p overexpression in parental MCF7 cells enhanced LC3-II (Figure 4C), whereas the miR-23b-3p inhibitor reduced that of LTED cells (Figure 4D). In addition, ETR cells showed an increased number of LC3 puncta in the presence of CQ, when compared to parental cells (Figures 4E and 4F). Comparable results were obtained in ETR models of ZR75.1 and HCC1428 cells when compared to their parental counterparts (Figures 4F, S4C, and S4D). Based upon this observation, longer CQ treatment (120 h) or silencing ATG7, a key component in the autophagosome formation, selectively impaired cell survival in ETR cells (Figures 4G and S4E–S4H), but not in the parental cells, indicating that targeting autophagy may resensitize ETR cells to endocrine agents.
Figure 4.
ETR Cells Rely on miR-23b-3p-Dependent Autophagy Activation for Cell Survival
(A) LTED and parental MCF7 cells were subjected to 25 μmol/L chloroquine (CQ) for 16 h. Total protein lysates were analyzed by western blotting with the antibody indicated.
(B) Total protein lysates from ETR and parental MCF7 cells were subjected to western blotting analysis with the antibodies indicated.
(C and D) Total protein lysates from MCF7 (C) and LTED (D) cells transfected with the oligos as described in the figure for 72 h were subjected to western blot analysis, as indicated.
(E and F) MCF7 ETR and parental (E) cells were treated with 25 μmol/L of CQ for 16 h and subjected to confocal analysis (E, green: LC3; blue: TO-PRO-3, nuclei). Comparable analysis was performed for HCC1428-LTED (Figure S4C), ZR75.1-LTED (Figure S4D), and parental counterparts, and LC3 puncta were quantified (F) as described in Method Details. Representative images are shown; scale bar, 10 μm. Data represent means ± SEMs. Two-way ANOVA; ∗∗p < 0.01; ∗∗∗p < 0.001.
(G) ETR-derived and MCF7 parental cells were grown either in the absence (vehicle) or presence of 50 μmol/L CQ treatment for 120 h before subjecting them to cell viability assay. Data are presented as fold change survival fraction of CQ-treated versus vehicle-treated cells. Representative images are shown. Data represent means ± SEMs and were compared to parental cells using 1-way ANOVA; Dunnett corrected; ∗∗∗p < 0.001.
(H) LTED and MCF7 parental cells were subjected to qRT-PCR using the assay indicated. Data represent means ± SEMs. Student’s t test; ∗∗∗p < 0.001.
(I) Kaplan-Meier analysis of overall survival of BRCA The Cancer Genome Atlas (TCGA) cohort of ER+ patients divided into high and low expressing, as described in Method Details for TNFAIP3 expression (high expressing, n = 231; low expressing, n = 493). The HR and log-rank Mantel-Cox p value are shown.
(J) Total protein lysates from ETR and parental MCF7 cells were subjected to immunoprecipitation (IP) using an anti-beclin-1 antibody and then subjected to immunoblotting (IB), as reported in the figure (top). Total lysates were also subjected to western blot analysis, as indicated (bottom).
An additional indication that autophagy may play a key role in ETR comes from the report that miR-23b-3p acts as a regulator of beclin-1 (Gozuacik et al., 2017). Ubiquitination of beclin-1 on Lys63 (K63) promotes autophagy, and this K63 ubiquitination is controlled by the balanced action of the ubiquitinating enzyme tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6) and the deubiquitinating enzyme TNFAIP3 (also known as A20) (Shi and Kehrl, 2010). Our analysis revealed that TNFAIP3 is a predicted target gene of miR-23b-3p (using miRGator3.0 [Cho et al., 2013], a positive score was found in TargetScan, miRNAorg, PITA, PicTar, and miRDB). This interaction has also been validated by cross-linking immunoprecipitation (Kishore et al., 2011). Gene expression data (0.48-fold compared to MCF7−E2 and 0.29-fold compared to MCF7+E2) and qRT-PCR analysis demonstrated a reduction in the expression of TNFAIP3 in the LTED cells (Figure 4H). This reduced expression is of clinical significance as low TNFAIP3 expression identifies a poor prognosis subset of patients in independent studies (HR = 0.52, log-rank p = 0.0066; Figures 4I and S1E). In support of these data, immunoprecipitation and western blot demonstrated an increase in not only the beclin-1 levels in the 3 ETR lines (see also Figure 4B) but also the level of K63-ubiquinated beclin-1, thus sustaining autophagy in the ETR cells (Figure 4J).
Increased Levels of Intracellular Aspartate and Glutamate Sustain the Aggressive Phenotype of ETR Cells
Despite the demonstration that ER+ cells are dependent on the SLC6A14 transporter (Babu et al., 2015) and that all ETR cell lines show a substantial reduction in SLC6A14 expression (Figures 2B–2D), it was notable that ETR cells show only a modest reduction in amino acids uptake (Figures 2F–2H) and no significant difference in de novo protein synthesis (Figure 2J), suggesting no major impact on protein availability to support ETR cell growth (Figures S4A and S4B). To determine whether the maintenance of de novo protein synthesis was accompanied by an imbalance in the intracellular amino acid levels, we analyzed their relative intracellular concentration in the LTED and parental MCF7 cells. High-performance liquid chromatography (HPLC) analysis revealed a significant increase in the intracellular levels of aspartate and glutamate in MCF7-LTED cells compared to parental cells (Figure 5A), and, moreover, the levels of aspartate and glutamate were unaffected by acute CQ treatment (Figure S4I). These analyses indicate that the increased levels of aspartate and glutamate in LTED cells were not dependent on the enhanced autophagic flux. By contrast, a general decrease in essential amino acid intracellular levels was induced by CQ (Figure S4I). We therefore hypothesized that an acidic amino acid transporter may be expressed at higher levels in the LTED model to provide the increased levels of aspartate and glutamate. Gene expression analysis of LTED and parental cells revealed 57 significantly deregulated SLC genes (Figure 5B), 9 of which encode amino acid transporters. Of these, 8 were downregulated in the LTED compared to parental cells, including SLC6A14. The exception was SLC1A2, a sodium-dependent high-affinity glutamate/aspartate transporter (3.4-fold increased in LTED cells compared to MCF7−E2 and 4.1-fold compared to MCF7+E2). Western blot and qRT-PCR analyses confirmed SLC1A2 increased expression in the LTED cells compared to their corresponding parental line (Figures 5B and 5C), an observation that is independent of E2 stimulation (Table S1). SLC1A2 has a predicted GATA2 binding site at the promoter region (JASPAR Predicted Transcription Factor Targets, http://amp.pharm.mssm.edu/Harmonizome), and GATA2 silencing reduced SLC1A2 expression levels (Figure S3B). SLC1A2 silencing further reduced the amino acids uptake of ETR cells (Figures 5D and 5E), which was already diminished compared to parental cells (Figures 2F–2H). Furthermore, SLC1A2 silencing impaired the incorporation by MCF7-LTED cells of 14C-glutamate (Figure 5F) and 14C-aspartate (Figure 5G), thus reducing ETR cell survival (Figure 5H) and MCF7-LTED invasion (Figure S5A), an aggressive feature of LTED cells. In addition, LTED colony formation ability was reduced by selective aspartate and glutamate deprivation (Figure 5I), thus reinforcing the notion that the selective upload of aspartate and glutamate drives the more “aggressive phenotype” of LTED cells.
Figure 5.
Aspartate and Glutamate Intracellular Levels Sustain the Aggressive Phenotype of ETR Cells
(A) Intracellular amino acids were extracted from LTED and parental MCF7 cells. Amino acid derivatization was performed using 4-N,N-dimethylaminoazobenzene-4′-sulfonyl chloride (DABS). DABS-amino acids were detected at visible light wavelengths using HPLC. A representative chromatogram is reported, and the quantification of aspartate (Asp) and glutamate (Glu) from 4 biological replicates is represented in a box and whiskers graph (mean, 25th percentile, and 75th percentile are shown; + indicates the median of the values). Student’s t test; ∗∗∗p < 0.001.
(B) Gene expression analysis of the significantly deregulated SLC transporters in LTED cells when compared to parental cells. Of the 57 significantly deregulated SLC genes, 9 amino acid transporters are shown in red. Each dot represents the mean value of the fold change derived by the analysis of 3 biological replicates (see Figure 1). Western blot analysis of total lysates from parental and LTED MCF7 cells for SLC1A2 is reported in the inset.
(C) LTED and parental MCF7, HCC1428, and ZR75.1 cells were subjected to qRT-PCR using the assay indicated. Data represent means ± SEMs, n = 3. Student’s t test; ∗p < 0.05; ∗∗∗p < 0.001.
(D–G) MCF7 ETR derivatives (D), HCC1428-LTED (E), and MCF7-LTED (F and G) were silenced using the non-targeting control (siCTR) or siRNAs targeting SLC1A2 (siSLC1A2) for 72 h. 14C-amino acid (D and E), 14C-glutamate (F), and 14C-aspartate (G) uptake was measured, and the radioactive signal normalized on protein content is shown, using siCTR-treated cells as comparators. Data represent means ± SEMs. One-way ANOVA; Dunnett corrected; ∗∗∗p < 0.001.
(H) Complementarily, cell viability was measured using crystal violet staining and reported as fold change using parental cells as comparator. Data represent means ± SEMs. Student’s t test; ∗∗∗p < 0.001.
(I) ETR and parental cells were subjected to colony formation assay either in the presence of all of the amino acids (all) or in the absence of both aspartate and glutamate (−Asp, −Glu), as described in Method Details. Data represent means ± SEMs of 3 biological replicates. Two-way ANOVA; Bonferroni corrected; ∗p < 0.05.
Aspartate and Glutamate Confer Metabolic Plasticity to ETR Cells
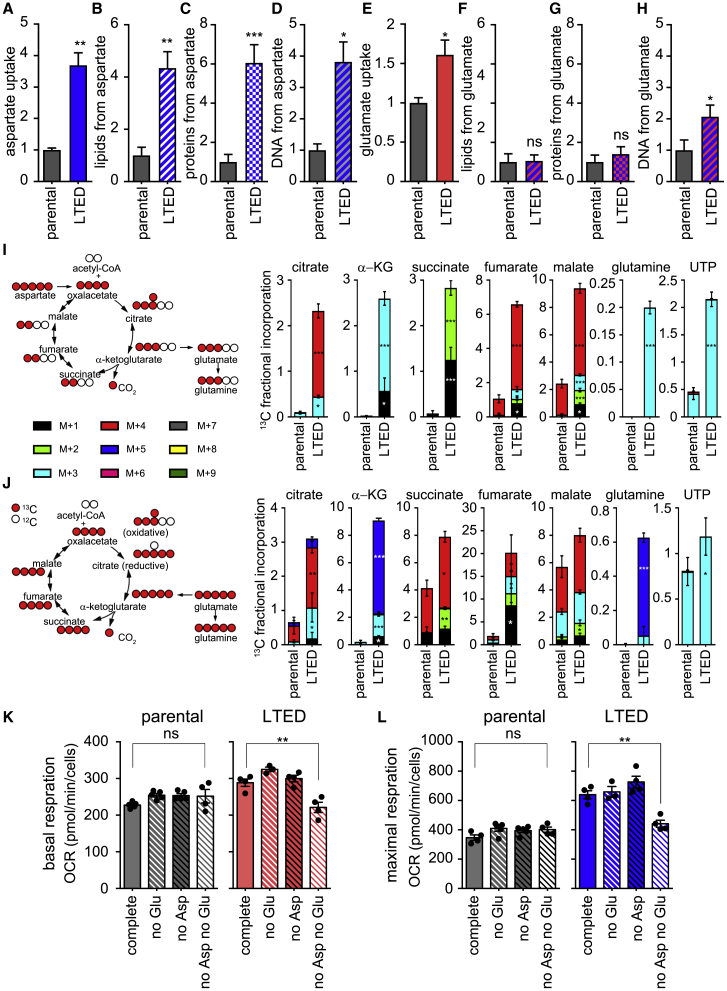
To identify the fate of the uploaded aspartate and glutamate, we performed a radioactive tracing analysis. The enhanced incorporation of radioactive aspartate observed in ETR cells (Figures 6A and S6A) was paralleled by the increased radioactive labeling of lipids (Figure 6B), proteins (Figure 6C), and DNA (Figure 6D) in ETR cells (Figures S6B–S6D). Conversely, the enhanced incorporation of glutamate in ETR cells (Figures 6E and S6E) was paralleled only by a significant increase in radioactive DNA labeling (Figure 6H), although a tendency toward enhanced radioactive lipids and proteins was observed in ETR cells (Figures 6F, 6G, S6F, and S6G). To quantify the contribution of aspartate and glutamate to the LTED metabolism, cells were cultured in medium containing uniformly (U) labeled 13C-aspartate (Figure 6I) or 13C-glutamate (Figure 6J), and the 13C-labeling of the tricarboxylic acid (TCA) cycle intermediates, together with metabolites that could act as precursors of anaplerotic reactions (e.g., uridine-5′-triphosphate [UTP] for DNA synthesis and glutamine for protein synthesis), were analyzed by liquid chromatography-mass spectrometry (LC-MS). When U-13C-aspartate was used, significant increases in 13C-labeled α-ketoglutarate (α-KG), succinate, malate, oxaloacetate, and citrate were observed in LTED cells when compared to the parental cell counterpart (Figure 6I). This was paralleled by a significant labeling of UTP and glutamine, indicating that aspartate is a major source of carbons for the TCA cycle and this could fuel anabolic pathways. Isotopologues analysis revealed that the TCA cycle in LTED cells occurs predominantly via canonical cycle activity, since a considerable fraction of citrate was 4 atoms of 13C, whereas the fraction of 5 atoms is limited, suggesting that reductive carboxylation of α-KG, although occurring in LTED cells, is not their main feature (Figure 6I). Comparable results were obtained when U-13C-aspartate was used in combination with (unlabeled) 12C-glutamate (Figure S6I), suggesting that the ability of aspartate to fuel the TCA cycle and subsequent anabolic and catabolic pathways is independent of the presence of glutamate. When U-13C-glutamate was used, significant increases in 13C-labeled α-KG, succinate, malate, oxaloacetate, and citrate were observed in LTED cells when compared to the parental cell counterpart (Figure 6J). This was paralleled by a significant labeling of UTP and glutamine, indicating that glutamate is a source of carbons for the TCA cycle and that can fuel anabolic pathways (Figure 6J). Similar to aspartate, comparable results were obtained when U-13C-glutamate was used in combination with 12C-aspartate (Figure S6J).
Figure 6.
Metabolic Plasticity of ETR Cells Is Sustained by Aspartate and Glutamate
(A–D) LTED and MCF7 cells were subjected to 14C radioactive aspartate uptake (A) or cultured for 24 h in a medium containing radioactive aspartate. Lipids, proteins, or DNA were extracted in parallel and radioactive signal measured to monitor the amount of aspartate that is incorporated into lipids (B), proteins (C), and DNA (D), as described in Method Details. Each value was normalized on protein content.
(E–H) LTED and MCF7 cells were subjected to 14C radioactive glutamate uptake (E) or cultured for 24 h in a medium containing radioactive glutamate. Lipids, proteins, or DNA were extracted in parallel and the radioactive signal was measured to monitor the amount of glutamate that is incorporated into lipids (F), proteins (G), and DNA (H), as described in Method Details. Each value was normalized on protein content. Data represent means ± SEMs, n = 3. Student’s t test; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ns, not significant.
(I and J) Schematic overview of the metabolism of downstream 13C-labeled aspartate and fluxes and relative incorporation of 13C carbons derived from aspartate in the metabolites (I). Schematic overview of metabolism of downstream 13C-labeled glutamate and fluxes and relative incorporation of 13C carbons derived from glutamate in the metabolites (J). Two-way ANOVA, Sidak corrected; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ns, not significant.
(K and L) Parental and LTED MCF7 cells were subjected to Seahorse XFe96 Mito Stress Test analysis and oxygen consumption rate (OCR) was measured in real time in the presence or absence of the indicated amino acids. Basal (K) and maximal (L) respiration was calculated as described in Method Details, based on the OCR after the administration of the ATP synthase inhibitor oligomycin, the proton uncoupler carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP), and the respiratory complex I inhibitor rotenone, together with the respiratory complex III inhibitor antimycin A. Data represent means ± SEMs. One-way ANOVA; Dunnett’s corrected; ∗∗p < 0.01.
As ETR cells have enhanced metabolic plasticity (Morandi and Indraccolo, 2017), we next addressed whether their increased aspartate and glutamate uptake was used, not only for anabolism but also as a source of energy. As assessed using the Seahorse XFe Mito Stress Test (Figures 6K, 6L, S6K, and S6L), there were no significant differences in basal and maximal respiration by monitoring the oxygen consumption rate (OCR) of parental MCF7 cells either in the presence or absence of single or both amino acids. By contrast, concomitant deprivation of aspartate and glutamate in ETR cells (Figures 6K, 6L, S6K, and S6L) significantly impaired their basal and maximal respiration. However, the withdrawal of each single amino acid was not sufficient to affect the OCR levels of ETR cells, suggesting that ETR cells can replenish the TCA cycle and subsequent electron transport chain with either amino acid, when the other is absent. This is in line with the results of the metabolomic analysis using U-13C-glutamate and U-13C-asparate (Figure 6I and 6J).
Increased Aspartate and Glutamate Levels in Patient-Derived Xenografts (PDXs) Correlate with ETR, and Impairing Their Transport Reduces the Metastatic Potential of ETR Cells In Vivo
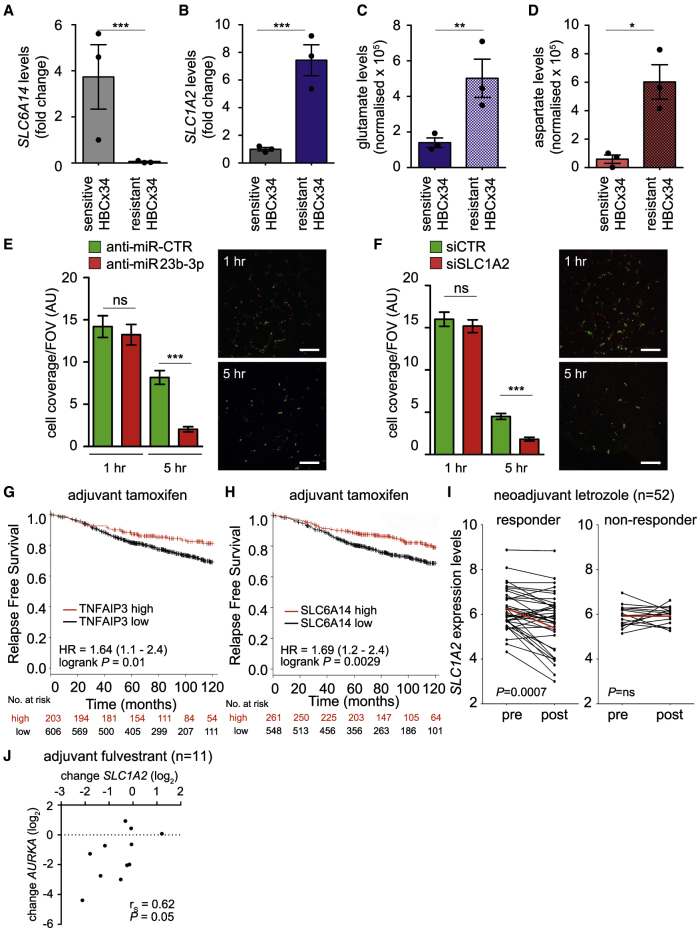
To further corroborate the role of the amino acid transporters in our model, we monitored SLC6A14 and SLC1A2 in an established ER+ breast cancer PDX model (HBCx34 TAMR), which has acquired resistance to tamoxifen treatment in vivo (Cottu et al., 2014). qRT-PCR analysis revealed a significant reduction in SLC6A14 expression and a concomitant increase in SLC1A2 in the TAMR PDXs when compared to the parental PDXs (Figures 7A and 7B). This differential SLC expression was accompanied by enhanced intratumoral levels of glutamate and aspartate, assessed using gas chromatography-MS (GC-MS) analysis (Figures 7C and 7D). Since we and others have previously reported that ETR cells display enhanced invasive abilities in vitro and in vivo (Bacci et al., 2016, Nguyen et al., 2015, Liu et al., 2014), we assessed whether impairing amino acid metabolism and availability, by targeting SLC1A2 or miR-23b-3p, had an impact on the invasive phenotype of the ETR cell lines, as observed in vitro (Figures S5A and S5B). To directly assess the role of the −3p isoform of the miR-23b, we could not stably express the pre-miR-23b construct, as this results in the downregulation of both the −3p or −5p isoforms. Therefore, we transfected cells with either a control non-targeting miRNA (anti-miR-CTR) or an anti-miR-23b-3p in a short-term in vivo assay, which has been previously published to reflect the metastatic potential of breast cancer cells (Murugaesu et al., 2014). Since metastatic burden resulted from the ability of the cells to survive in the circulation and to colonize secondary sites, transfected cells were labeled with cell tracker dyes, mixed in a 1:1 ratio, and injected via the tail vein into SCID mice. Imaging the lungs 1 h post-injection confirmed that an equal number of cells had been inoculated. An examination of the lungs 5 h post-injection revealed that a reduced number of anti-miR-23b-3p-treated cells were retained in the lungs compared to the anti-miR-CTR transfected cells, indicating that miR-23b-3p expression is required for efficient survival in the circulation and retention in the lungs (Figures 7E and S7A). Equivalent results were obtained in a dye swap experiment (Figure S7B). Crucially, comparable data were obtained by targeting SLC1A2 (Figure 7F and S7C), indicating that both miR-23b-3p and SLC1A2 are responsible for the increased invasive ability of the ETR cells and that their targeting could reduce the aggressiveness of ETR breast cancer.
Figure 7.
Increased Aspartate and Glutamate Levels in PDX Correlate with ETR, and Impairing Their Transport Reduces the Metastatic Potential of ETR Cells
(A–D) ET (i.e., tamoxifen)-sensitive and tamoxifen-resistant PDX (HBCx34) were obtained from ER+ breast cancer specimens previously characterized as described in Method Details. Tumor tissues were excised, and total RNA was extracted and subjected to qRT-PCR using the assay to detect SLC6A14 (A) and SLC1A2 (B). Each dot represents an independent mouse. Alternatively, tumor tissues were extracted and subjected to GC-MS (as detailed in Method Details) to evaluate the intratumoral levels of glutamate (C) and aspartate (D). Data represent means ± SEMs. Student’s t test; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(E and F) MCF7-LTED cells were transfected with either anti-miR-23b-3p or a non-targeting control (anti-miR-CTR, E). Cells were labeled with CellTracker dyes (green for anti-miR-CTR and red for anti-miR-23b-3p) before injecting a mixture of anti-miR-23b-3p and anti-miR-CTR transfected cells in a 1:1 ratio into the tail vein of SCID mice (n = 4 per group), as described in Method Details. Similarly, SLC1A2 silenced (red) and siCTR transfected cells (green) were injected (F). The lungs were recovered at the time indicated and imaged to analyze the number of fluorescent cells that colonize the lungs. Data shown are means for tumor cell coverage per field of view (FOV); n = 4 mice per group per time point ± SEMs. Representative images are shown; scale bar, 200 μm. ∗∗∗p < 0.001.
(G and H) Kaplan-Meier analysis of relapse-free survival of a curated cohort of ER+ patients treated with tamoxifen in the adjuvant setting and divided into high and low expressing as described in Method Details for TNFAIP3 (G) and SLC6A14 expression (H). HR and log-rank Mantel-Cox p values are shown.
(I) Correlation of SLC1A2 expression with response to AI in 52 paired ER+ breast cancer samples pre- and post-2-week letrozole treatment (Miller et al., 2007). A significant decrease in SLC1A2 expression was observed in the responder group, but no difference was found in the non-responder group (Wilcoxon paired t test). ns, not significant. Each dot represents the value derived by a patient’s specimen. Red lines connect the mean values pre- and post-letrozole treatment.
(J) Correlation of the change in the SLC1A2 and AURKA expression levels following adjuvant fulvestrant treatment (Geo: GSE33658, n = 11; Spearman correlation and p value are indicated in the figure).
Finally, we evaluated whether the molecular players differentially expressed in ETR cells versus the parental counterpart and involved in the described reprogramming could have prognostic and predictive value for ET-treated ER+ breast cancers. We used a curated dataset that combined ∼800 ER+ breast cancer patients (from independent retrospective studies) who had been treated with adjuvant tamoxifen. Relapse-free survival was used as a surrogate for therapy response. Kaplan-Meier analysis revealed that patients characterized by lower levels of TNFAIP3 (HR = 1.64, p = 0.01, n = 809; Figure 7G) or SLC6A14 (HR = 1.69, p = 0.0029, n = 809; Figures 7G and 7H) showed poorer relapse-free survival when compared with higher-expressing tumors. Multivariate Cox regression analysis revealed that the prognostic value is independent of proliferation (Table S3). The predictive value of SLC1A2 in response and resistance to ET was validated by in silico analysis, which was performed by retrieving publicly available gene expression data from the biopsies of 52 ER+ breast cancer patients taken before and after 2 weeks of neoadjuvant letrozole treatment (Miller et al., 2007). The patients were divided into responders and non-responders, defined by a >50% and a <50% reduction, respectively, in tumor volume following a further 3 months of letrozole treatment. A pairwise comparison shows a significant decrease in SLC1A2 expression after 2 weeks of letrozole treatment in the responder cohort but not in the non-responder cohort (p = 0.0007; Figure 7I). The relevance of the players involved in response to selective downregulators of ER was finally validated in a small cohort of ER+ breast cancer patients who received adjuvant fulvestrant. Since the clinical outcome was not available, we used the changes in the proliferation-related gene AURKA as a surrogate for therapy response. The evaluation of proliferation using Ki67 staining has been shown to predict poor long-term disease outcome (Forbes et al., 2008). Changes in the expression levels of SLC1A2 induced by fulvestrant treatment were positively correlated with those of AURKA (rs = 0.62, p = 0.05, n = 11; Figure 7J), suggesting that SLC1A2 expression levels are linked to fulvestrant-induced changes in proliferation (i.e., response to therapy).
These data support a role for amino acid metabolic reprogramming in ETR breast cancers, and the predictive value of the molecular and metabolic players identified should be validated in clinical prospective (neoadjuvant) trials.
Discussion
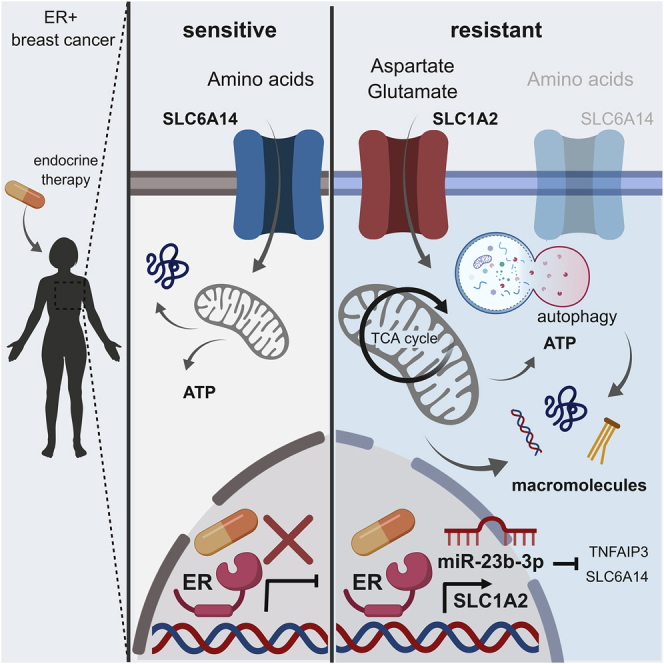
The data presented in this study highlight the amino acid metabolic pathway as an important determinant of resistance to ET in ER+ breast cancers. In particular, molecular reprogramming occurs and sustains the catabolic and anabolic processes of ETR breast cancer cells by (1) activating autophagy and (2) modulating the expression of amino acid transporters to selectively import certain amino acids (in this case, aspartate and glutamate).
This study originated from the analysis of extensively characterized ER+ cells adapted to study ETR that we along with others have demonstrated to be clinically relevant (Martin et al., 2003, Martin et al., 2012, Nguyen et al., 2015, Sanchez et al., 2011, Morandi et al., 2013, Fox et al., 2013, Plaza-Menacho et al., 2010, Liu et al., 2014). In addition, in vitro findings were validated using PDX, lung retention assay, and retrospective clinical data derived from ET-treated patients.
miR-23b is a member of the miR-23b/-27b/-24-1 cluster, and its deregulation has been reported in several types of cancer, although no conclusive results to date have provided a link between its role as an oncomiR or as a tumor suppressor. miR-23b-enhanced expression levels were found in renal cell carcinomas (Zaman et al., 2012), lymphomas (Li et al., 2013), non-small-cell lung cancers (Begum et al., 2015), and pancreatic cancers (Chen et al., 2017). Conversely, reduced levels were found to be associated with colon cancer (Zhang et al., 2011) and multiple myeloma (Fulciniti et al., 2016). Reduced levels of miR-23b were found to promote the chemoresistance of gastric cancers (An et al., 2015) and radioresistance in pancreatic cancers (Wang et al., 2013). An additional layer of complexity may come from the −5p and −3p isoforms that could have independent and/or opposing effects in different tissue contexts. Since some of the above-mentioned studies did not dissect which of the isoforms were predominant in the cohort of tumors analyzed, some of the discrepancies observed may derive from such bias. In this study, enhanced miR-23b-3p expression characterizes ETR breast cancer cells and a subset of breast cancer patients who show a worse prognosis. Since stable knockdown of miR-23b-3p in vivo could be achieved only by concomitant downmodulation of the −5p, we have opted for a short-term assay (i.e., quantification of residing cells in the lung after tail vein injection). Lung retention assay has some limitations and does not recapitulate the complex mechanism of the full metastatic cascade. However, we have carefully selected the time point of the analysis such that the ability of a given cell to be retained is likely due to the ability to survive and to colonize the lung. The priming event that leads to miR-23b-3p-enhanced expression is the upregulation of the transcription factor GATA2 mediated by ER signaling disengagement. GATA2 has been reported to favor breast cancer progression (Wang et al., 2012) and to regulate chemotherapy resistance and tumorigenicity in prostate cancer (Vidal et al., 2015). Crucially, GATA2 silencing impairs the miR-23b-3p-dependent amino acid transport regulation and ETR cell survival and has a prognostic value in ER+ breast cancer.
miR-23b-3p-enhanced levels in ETR cells are paralleled by a significant decrease in its target SLC6A14, an amino acid transporter that has been demonstrated to be an ER-dependent gene, and its expression was associated exclusively with ER+ breast cancers. Moreover, SLC6A14 targeting is sufficient to induce autophagy-dependent cell death in ER+ breast cancer models (Karunakaran et al., 2011). Since the reduced expression levels of SLC6A14 are compatible with ER+ ETR cell survival, it is plausible that SLC6A14 downregulation may have been caused by a miR-23b-3p-induced adaptation, thus establishing a metabolic phenotype of the ETR cells that is characterized by enhanced pro-survival autophagy and the reliance on another amino acid transporter to compensate for the reduction in amino acid import caused by SLC6A14 impairment. ETR cells and parental cells show comparable cell growth rates and protein synthesis, although the import of amino acids is reduced in ETR cells. We hypothesized that autophagy could compensate for the reduced amino acids import. However, amino acids uptake, although significantly reduced, was not abolished, suggesting that other amino acid transporters may still function in ETR cells. It has been reported that glutamine can contribute to ETR cell survival in an MYC-dependent mechanism (Shajahan-Haq et al., 2014) and that autophagy activation could support tamoxifen resistance (Cook et al., 2011). Moreover, tamoxifen resistance could be supported by importing leucine in an LLGL2-dependent fashion (Saito et al., 2019). Our data revealed that SLC1A2 expression and the subsequent increases in the intracellular levels of aspartate and glutamate and uptake in ETR cells and PDX models confer a catabolic and anabolic advantage to ETR breast cancers.
Our analyses show that both autophagy and aspartate and glutamate uploaded together establish a metabolic condition that sustains resistance to ET. Although this phenomenon should warrant further investigation, a potential explanation of the need for both autophagy and selective amino acid import may come from the reduction in essential amino acid levels that we have observed following autophagy inhibition. In this context, the aspartate and glutamate that are not affected by autophagy inhibition could be used to derive all of the other non-essential amino acids. In addition, glutamate and aspartate import could mitigate the effects caused by a severe decrease in the intracellular levels of amino acids, an established activator of autophagy (Galluzzi et al., 2014), to modulate the flux. This promotes a pro-survival autophagy, without the activation of a pro-apoptotic autophagic flux, which has been reported after the acute targeting of SLC6A14. In addition, in silico analysis of the METABRIC database (Pereira et al., 2016) shows that SLC1A2 and SLC6A14 are not concomitantly overexpressed or amplified in breast cancers and may be mutually exclusive (Figure S7D). It is interesting to note that SLC1A2 expression has been reported to play a role in different cancers. SLC1A2 undergoes a genomic breakpoint in its 5′ region and forms a fusion gene with CD44 in gastric and colorectal cancers (Tao et al., 2011, Shinmura et al., 2015). The CD44-SLC1A2 aberration induces higher glutamate intracellular levels, and SLC1A2 targeting resensitizes tumors to chemotherapy (Tao et al., 2011). Similarly, the enhanced levels of SLC1A2 in the ETR breast cancer models resulted in a significant increase in intracellular and intratumoral aspartate and glutamate levels. Once targeted, ETR cellular aggressive features are reduced and cells are resensitized to ET. Although we did not investigate the potential fusion gene in the ETR models analyzed, gene expression profiling revealed that CD44 expression is upregulated in LTED cells (7.3-fold when compared to parental MCF7), and therefore it would be interesting to investigate whether a CD44-SLC1A2 fusion may also occur in ETR breast cancer models. The role of aspartate in cancer has been recently explained by the seminal work of the Vander Heiden group (Sullivan et al., 2015). Initially, Sullivan et al. (2015) demonstrated that in oxidative phosphorylation (OXPHOS)-proficient cells, respiration serves to fuel electron acceptors for aspartate synthesis, and aspartate supplementation is sufficient to sustain proliferation in OXPHOS-deficient cells (Tao et al., 2011). More recently, 2 independent studies revealed that aspartate availability is a metabolic limitation that tumors experience and that overcoming this limitation, either via aspartate upload via SLC1A3 (Garcia-Bermudez et al., 2018) or via the conversion of asparagine into aspartate (Sullivan et al., 2018), is advantageous for tumor growth. Previous studies have reported that metformin anti-tumor activity correlates with a diminution of NAD+ and aspartate in tumors (Gui et al., 2016), and these findings indicate that aspartate limitation is required for the anti-tumor efficacy of metformin and, broadly, other biguanides. We have previously reported that metformin was ineffective in AI-resistant models (Bacci et al., 2016), and this can now be explained by the enhanced SLC1A2-dependent aspartate-boosting mechanism that maintains aspartate homeostasis. Therefore, lowering aspartate intratumoral levels may be of benefit in ETR. However, as confirmed in our study, SLC1A2 can also serve for glutamate import, and only the concomitant deprivation of both amino acids was able to reduce the mitochondrial potential and OCR of the resistant cells. However, even if the glutamate contribution seems lower than that of aspartate by 14C tracing experiments, flux analysis using U-13C-glutamate revealed a role for glutamate in anabolism, which is able to sustain not only DNA biosynthetic pathways but also lipid and protein biosynthesis. This anabolic supporting role of aspartate and glutamate is in line with previous reports showing that the majority of carbon mass in cells is derived from amino acids rather than glucose or glutamine (Hosios et al., 2016).
In conclusion, the experimental and clinical data herein presented support a role for amino acid metabolism and availability in promoting ETR. The findings provide mechanistic insights into the metabolic deregulation of ETR and a series of molecular players that could be useful in identifying a signature for breast cancer patients with an increased risk of ETR. This is particularly pertinent given that the majority of ER+ breast cancer patients will undergo ET and that the identification of resistant patients is an important and still unmet clinical issue.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-tubulin | Sigma | Cat# T5168; RRID: AB_477579 |
| Rabbit polyclonal anti-GATA2 | Cell signaling | Cat# 4595; RRID: AB_2108579 |
| Mouse monoclonal anti-SLC1A2 | Santa Cruz Biotechnology | Cat# 365634; RRID: AB_10844832 |
| Rabbit polyclonal anti-SLC6A14 | Abcam | Cat# ab99102; RRID: AB_10696963 |
| Rabbit polyclonal anti-beclin1 | Thermo Fisher Scientific | Cat# PA1-16857; RRID:AB_568459 |
| Rabbit polyclonal anti-LC3 | Thermo Fisher Scientific | Cat# PA1-16930; RRID: AB_2281384 |
| Mouse monoclonal anti-ubiquitin-K63 | Thermo Fischer Scientific | Cat# 14-6077-80; RRID:AB_1257214 |
| Mouse monoclonal anti-APG7 | Santa Cruz Biotechnology | Cat# sc-376212 RRID:AB_10988418 |
| Mouse monoclonal anti-beclin1 | Santa Cruz Biotechnology | Cat# sc-48341 RRID:AB_626745 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 488 conjugate | Thermo Fisher Scientific | Cat# A-11034; RRID:AB_2576217 |
| Goat Anti-Rabbit IgG (H+L) Highly Cross-adsorbed Antibody, Alexa Fluor 633 conjugate | Thermo Fisher Scientific | Cat# A-21071; RRID:AB_141419 |
| Biological Samples | ||
| Patient-derived xenografts HBCx34 | Prof. Elisabetta Marangoni | Cottu et al., 2014 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Methanol | Sigma | Cat# 900688-1 |
| Acetonitrile | Sigma | Cat# 34851 |
| 4-N,N-dimethylaminoazobenzene-4’-sulfonyl chloride | Supelco | Cat# 502219 |
| KH2PO4 | Sigma | Cat# P3786 |
| Amino acids standards | Sigma | Cat# A9781 |
| Chloroform | Sigma | Cat# 34854-1 |
| Water | Merck | Cat# 102699-1000 |
| Norvaline | Sigma | Cat# 53721 |
| Methoxyamine hydrochloride | Sigma | Cat# 226904 |
| Pyridine | Thermo Fisher Scientific | Cat# 25104 |
| N-Methyl-N-(trimethylsilyl)trifluoroacetamide with 1% trimethylchlorosilane | Sigma | Cat# 69478 |
| Chloroquine diphosphate | Sigma | Cat# C6628 |
| Glutamine | Sigma | Cat# G7513 |
| RPMI 1640 medium | GIBCO | Cat# 11835-063 |
| DMEM 1X | GIBCO | Cat# A14430-01 |
| 14C-Glutamic acid | Perkin Elmer | Cat# NEC290E050UC |
| 14C-Aspartic acid | Perkin Elmer | Cat# NEC268E050UC |
| 14C-Amino Acid Mixture | Perkin Elmer | Cat# NEC850E050UC |
| 13C-Glutamic acid | Sigma | Cat# 604860 |
| 13C-Aspartic acid | Cambridge Isotope Laboratories | Cat# CNLM-544-H-0.25 |
| D-Glutamic acid | Sigma | Cat# G1001 |
| L-Aspartic acid | Sigma | Cat# A9256 |
| Z-4-hydroxytamoxifen | Sigma | Cat# H7904 |
| 17-β-estradiol | Sigma | Cat# E2758 |
| Fulvestrant | Sigma | Cat# I4409 |
| Lipofectamine RNAiMAX | Thermo Fisher Scientific | Cat# 13778-150 |
| Opti-MEM | GIBCO | Cat# 31985062 |
| Fetal Bovine Serum | Euroclone | Cat# ECS0180L |
| Charcoal/Dextran Treated FBS (DCC) | Hyclone | Cat# SH30068.03 |
| Protease inhibitor cocktail | Sigma | Cat# P8340 |
| Matrigel Matrix | BD Biosciences | Cat# 356234 |
| Protein A/G-PLUS Agarose beads | Santa Cruz Biotechnology | Cat# sc-2003 |
| DiffQuick solution | BD Biosciences | Cat# 726443 |
| Laemmli Sample Buffer | Bio-Rad | Cat# 161-0737 |
| Clarity Western ECL Substrate | Bio-Rad | Cat# 170-5061 |
| XFe Seahorse RPMI medium | Agilent Technologies | Cat# 103336-100 |
| Critical Commercial Assays | ||
| TaqMan Universal Mastermix no UNG | Applied Biosystems | Cat# 4440040 |
| QuantiTect Reverse Transcription Kit | QIAGEN | Cat# 205311 |
| TaqMan MicroRNA Reverse Transcription Kit | Applied Biosystems | Cat# 43665967 |
| miRNeasy Mini Kit | QIAGEN | Cat# 217004 |
| SosAdvanced Universal Sybr Green Supermix | Bio-Rad | Cat# 1725270 |
| Cayman Chemicals Protein Synthesis Assay | Cayman Chemical | Cat# 601100 |
| Seahorse XF Cell Mito Stress Kit | Agilent Technologies | Cat# 103015-100 |
| Human genome microarray | Agilent Technologies | Cat# G4851A |
| Human miRNA microarray v16 | Agilent Technologies | Cat# G4870A |
| Deposited Data | ||
| mRNA and miRNA microarray data | This paper | GEO: GSE120931 |
| Gene expression data | Rème et al., 2013 | ArrayExpress: E-MTAB-365 |
| The Cancer Genome Atlas (TGCA) | https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga | RRID:SCR_003193 |
| Gene expression data | Loi et al., 2007 | GEO: GSE6532 |
| Gene expression data | Dedeurwaerder et al., 2011 | GEO: GSE20711 |
| Gene expression data | Desmedt et al., 2007 | GEO: GSE7390 |
| Gene expression data | Sabatier et al., 2011 | GEO: GSE21653 |
| Gene expression data | Wang et al., 2005 | GEO: GSE2034 |
| Gene expression data | Sotiriou et al., 2006 | GEO: GSE2990 |
| Gene expression data | Symmans et al., 2010 | GEO: GSE17705 |
| Gene expression data | Zhang et al., 2009 | GEO: GSE12093 |
| Gene expression data | Loi et al., 2008 | GEO: GSE9195 |
| Gene expression data | Miller et al., 2005 | GEO: GSE3494 |
| Gene expression data | Karn et al., 2010 | GEO: GSE4611 |
| Gene expression data | Nagalla et al., 2013 | GEO: GSE45255 |
| Gene expression data | Minn et al., 2005 | GEO: GSE2603 |
| Gene expression data | Desmedt et al., 2009 | GEO: GSE16391 |
| Gene expression data | Clarke et al., 2013 | GEO: GSE42568 |
| Gene expression data | Filipits et al., 2011 | GEO: GSE26971 |
| Gene expression data | Li et al., 2010 | GEO: GSE19615 |
| Gene expression data | Miller et al., 2007 | GEO: GSE5462 |
| Gene expression data | Massarweh et al., 2011 | GEO: GSE33658 |
| Gene expression data | Pereira et al., 2016 | METABRIC |
| Experimental Models: Cell Lines | ||
| MCF7 | ATCC | Cat# HTB-22 |
| ZR75.1 | ATCC | Cat# CRL-1500 |
| HCC1428 | ATCC | Cat# CRL-2327 |
| LTED Cells | Bacci et al., 2016 | N/A |
| MCF7-TAMR | This paper | N/A |
| MCF7-FULVR | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| BALB/c mice | Charles River Laboratories | Cat# 236CB17SCID |
| NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) | Simões et al., 2015 | N/A |
| Oligonucleotides | ||
| anti-miR-23b-3p | Ambion | Cat# MH10711 |
| siSLC1A2 | Sigma | Cat# Hs01_00109453 |
| siCTR | Sigma | Cat# SIC001 |
| anti-miR-negative control (anti-miR-CTR) | Ambion | Cat# AM17011 |
| siGATA2 | Sigma | Cat# Hs01_00106117 |
| siATG7_71 | Sigma | Cat# Hs02_00341471 |
| siATG7_49 | Sigma | Cat# Hs01_00077649 |
| siATG7_48 | Sigma | Cat# Hs01_00077648 |
| SLC6A14 (Hs00924564_m1) | Applied Biosystems | Cat# 4448892 |
| SLC1A2 (Hs01102423_m1) | Applied Biosystems | Cat# 4448892 |
| GATA2 (Hs00231119_m1) | Applied Biosystems | Cat# 4453320 |
| GAPDH (Hs02786624_g1) | Applied Biosystems | Cat# 4331182 |
| ACTB (Id:Hs99999903_m1) | Applied Biosystems | Cat# 4331182 |
| miR-23b-3-p (000400) | Applied Biosystems | Cat# 4427975 |
| RNU48 (001006) | Applied Biosystems | Cat# 4427975 |
| U6 (001973) | Applied Biosystems | Cat# 4427975 |
| TNFAIP3 precast primer (HsaCID0012648) | Bio-Rad | Cat# 10025636 |
| Beclin-1 precast primer (HsaCID0016032) | Bio-Rad | Cat# 10025636 |
| GAPDH precast primer (HsaCED0038674) | Bio-Rad | Cat# 10025636 |
| Software and Algorithms | ||
| Masshunter Quantitative Analysis Software | Agilent Technologies | RRID: SCR_015040 |
| Masshunter Qualitative Analysis | Agilent Technologies | RRID:SCR_016657 |
| Fihen Metabolomics RTL library | Agilent Technologies | Cat# G1676AA |
| 7500 Real-Time PCR Software | Applied Biosystems | RRID:SCR_014596 |
| Thermo XCalibur Quan Browser software | Thermo Fisher Scientific | RRID: SCR_014593 |
| FiJi | https://fiji.sc | N/A |
| GraphPad Prism v8.0c | https://www.graphpad.com/ | N/A |
| KM plotter | Györffy et al., 2010, Lánczky et al., 2016 | http://kmplot.com/analysis/ |
| cBioPortal | https://www.cbioportal.org/ | RRID:SCR_014555 |
| Firebrowse | http://firebrowse.org/ | N/A |
| Seahorse Wave software | Agilent Technologies | RRID:SCR_014526 |
| GeneSpring GX v.14.8 software | Agilent Technologies | RRID:SCR_010972 |
| R Statistical Software | https://www.r-project.org/ | N/A |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be made available upon reasonable request by the Lead Contact, Andrea Morandi (andrea.morandi@unifi.it).
Experimental Model and Subject Details
Cell Culture
MCF7, ZR75.1 and HCC1428 human female breast cancer cells were obtained from ATCC and cultured in phenol red-free RPMI 1640 medium (GIBCO #11835-063) supplemented with 10% fetal bovine serum (FBS, Euroclone #ECS0180L) 2 mmol/L glutamine (Sigma #G7513) and 1 nmol/L 17β-estradiol (E2, Sigma #E2758). The corresponding LTED (long term estrogen deprived) derivatives were maintained in sterol-deprived medium in phenol red–free RPMI1640 medium containing 10% dextran charcoal-stripped FBS (Hyclone #SH30068.03) and 2 mmol/L glutamine (DCC medium). Tamoxifen- (TAMR) and fulvestrant- (FULVR) resistant cells were cultured in DCC medium supplemented with 1 μmol/L 4-OH tamoxifen (Sigma #H7904) or 100 nmol/L fulvestrant (Sigma #I4409), respectively. Cells were short tandem repeat tested, amplified, stocked, routinely subjected to mycoplasma testing and once thawed were kept in culture for a maximum of 20 passages.
Mouse models and care
5- to 6-week-old female BALB/c mice were purchased from Charles River Laboratories (236CB17SCID) and used for tail vein injection experiments. Animal work was carried out under the Project licenses 1002/2017-PR and was approved by the Ministero della Salute. All animals were monitored by staff from the Centro Stabulazione Animali da Laboratorio (CESAL) for signs of ill health. Mice were housed under sterile conditions (five or less animals per cage) with ad libitium access to food and water.
For patient-derived xenografts (PDX), mouse studies were conducted in accordance with the UK Home Office Animals (Scientific Procedures) Act 1986, using 8- to 10-week-old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) female mice at the University of Manchester. HBCx34 and HBCx34 TAMR PDXs have been established as previously described (Cottu et al., 2014). Serial passaging of the PDX was carried out by implanting small fragments of the tumor subcutaneously into dorsal flanks of NSG mice. HBCx34 breast cancer estrogen-dependent PDXs were administered with 8 μg/ml of E2 in drinking water at all times. HBCx34 TAMR PDXs were treated for the entire duration of the experiments in the presence of tamoxifen (10 mg/kg/day, oral gavage). Experiments were performed using PDX tumors between passages 5 and 8. Animal weight and tumor size were measured twice a week. Tumor chunks were snap frozen and further processed for either RNA extraction and qRT-PCR analysis or GC-MS.
Method Details
Cell viability assay
Sensitive and resistant cells were seeded into 12 well plates in either standard conditions (see cell culture) or experimental conditions such as in the presence of 25 μmol/l of the autophagy inhibitor CQ (Sigma #C6628). 5 days post seeding cells, plates were stained with crystal violet (triphenylmethane dye 4-[(4-dimethylaminophenyl)-phenyl-methyl]-N,N-dymethyl-alanine; Sigma #548-62-9), dried overnight and the crystal violet within the adherent cells solubilized with 500 μl/well of 2% SDS. The absorbance at 595 nm was evaluated using a microplate reader.
MTT assay
Sensitive and resistant cells transfected using siRNA against ATG7 were seeded into 24 well plates in standard medium (see cell culture). 48 hours after transfection cells were subjected to the colorimetric non-radioactive quantification of cell proliferation and viability using MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) assay (0,2mg/ml, Sigma #M5655). MTT was added into the medium of each well and after 2 hours at 37°C, the cells were extensively washed with PBS, allow to dry and then 200 μL of DMSO was added to resuspended the amount of MTT within the cells and 100 μL was subjected to absorbance wavelength evaluation at 565 nm using a microplate reader.
Colony formation assay
To evaluate clonogenicity of breast cancer cells under stress conditions, 0.5-1 × 103 cells were seeded per well in a 6 well plate in either the recommended medium conditions or experimental conditions such as amino acids deprivation. 15 days post seeding, the colonies were fixed with 4% formaldehyde and stained with crystal violet staining solution. Air-dried plates were scanned at 600 dpi and images analyzed using FiJi.
Transwell invasion assay
Invasion assay was performed using 8-μm-pore transwells (Corning #3428) coated with 10 μg/cm2 of reconstituted standard formulation Matrigel (BD Biosciences #356334). Transfected cells (using anti-miR-23b-3p, siSLC1A2 and relative controls oligos) were serum starved for 6 hours and then 1 × 105 cells seeded onto the upper chamber of the transwell in serum-free medium and allow to invade overnight toward complete medium. Air-dried membranes stained with DiffQuick solution (BD Biosciences #726443) were imaged and invasion was evaluated by counting the cells migrated to the lower surface of the filters (six randomly chosen fields).
Protein de novo synthesis assay
To analyze protein de novo synthesis rate in resistant cells compared to parental cells, 3 × 104 cells were seeded per well in a 96 well clear bottom black plate (Corning #3603). The following day, the culture media were removed and cells were incubated for 30 minutes in fresh culture medium containing O-Propargyl-Puromycin (OPP) with or without 50 μg/ml cycloheximide. OPP incorporates into the C terminus of translating polypeptide chains, thereby stopping translation. The truncated C-terminal alkyne-labeled proteins can subsequently be detected via copper-catalyzed click chemistry (Liu et al., 2012). Cells were then processed for detection of protein synthesis according to the protocol of Cayman Chemicals Protein Synthesis assay (Cayman Chemicals #601100).
RNAi transfection
Cells were transfected with 15 nmol/L anti-miR-23b-3p (MH10711, Ambion), with 45 nmol/L siSLC1A2 (Hs01_00109453, Sigma), siGATA2 (Hs01_00106117, Sigma), or siATG7 (Hs02_00341471, Hs01_00077649, or Hs01_00077648, Sigma) and respective negative controls (AM17011, Ambion; SIC001, Sigma) using Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific #13778-150) and Opti-MEM (GIBCO #31985062) accordingly to manufacturer’s instructions and analysis was performed 3 days after transfection.
Radioactive assays
Breast cancer cells were treated as described in the figures for 3 days (see RNAi transfection). Aspartic acid, glutamic acid or amino acids uptake was evaluated incubating the cells with uptake buffer solution (140 mmol/L NaCl, 20 mmol/L HEPES/Na, 2.5 mmol/L MgSO4, 1 mmol/L CaCl2, and 5 mmol/L KCl, pH 7.4) containing [U-14C] radioactive metabolites for 15 minutes. Cells were subsequently washed with ice cold PBS and lysed with 0.1 mol/L NaOH.
To analyze the incorporation of radioactive amino acids into proteins, lipids or DNA, culture media were changed to a deprived-DMEM medium (GIBCO #A14430-01) with or without non-radioactive amino acids in the presence of 10% DCC, 11 mM glucose, 2 mmol/L glutamine and then supplemented with 1 μCi 14C-aspartic acid (Perkin Elmer # NEC268E050UC) or 1.5 μCi 14C-glutamic acid (Perkin Elmer # NEC850E050UC) 24 hours prior to the end of the experiment. For radioactive incorporation into proteins, cells were washed in ice cold PBS and then resuspended in 20% trichloroacetic acid, placed on ice for 30 minutes and centrifuged (10000 rpm for 10 minutes at 4°C). Pellet was resuspended in dH2O, transferred to a scintillation vial and counted on the scintillation counter. For fatty acids synthesis analysis, cells were washed three times in ice cold PBS and then lysed in methanol. Samples were first resuspended in 4 volumes of a CHCl3:MeOH (1:1) solution and then an additional volume of dH2O was added. The solution was then centrifuged at 1000 rpm for 5 minutes at room temperature. The lower phase was collected, transferred to a scintillation vial and counted on the scintillation counter. For DNA incorporation analysis, cells were washed and then resuspended in Tris-Hcl 50mM/EDTA 100mM/SDS 0.5%/proteinase K lysis buffer. Samples were added to one volume of phenol:chloroform:isoamyl alcohol (25:24:1), mixed and centrifuged (10000 rpm for 10 minutes at room temperature). The upper aqueous phase was collected, resuspended in 1/10 volume of 3 M NH4OAc and 2 volumes of ethanol and incubated for 3 hours at −80°C. The precipitated DNA was pelleted (10000 rpm for 10 minutes at room temperature), washed with 70% ethanol and allowed to dry. The pellet was resuspended in dH2O and transferred to a scintillation vial and counted on the scintillation counter. All the radioactive signals were normalized on protein content.
Immunoblotting and immunoprecipitation
Cells were lysed using cold RIPA buffer (50 mM Tris Hcl pH7.5, 150 mM NaCL, 1% Nonidet P-40,2 mM EGTA, 1 mM sodium ortovanadate, 100 mM NaF) supplemented with protease inhibitors (Sigma # P8340) and protein concentrations of lysate were measured by Bradford method (Bio-Rad Protein Assay #500-006), 20-40 μg of total proteins were loaded in precast SDS-PAGE gels (Biorad #456-8096) and then transferred onto nitrocellulose membrane by Trans-Blot Turbo Transfer Pack (Biorad #1704157). The immunoblots were incubated in non-fat dry milk 2%, tween 20 0.05% in PBS at room temperature and probed with primary and appropriate secondary antibodies. The antibodies used in immunoblotting were GATA2 (Cell Signaling, #4595S) and SLC1A2 (Santa Cruz Biotechnology, #sc-365634), SLC6A14 (Abcam, #ab99102), beclin-1 (Invitrogen, #PA1-16857), LC3 (Invitrogen, #PA1-16930) and ubiquitin-K63 (Thermo Fisher, #14-6077-80), APG7 (Santa Cruz Biotechnology, #sc-376212), tubulin (Sigma, #T5168).
Immunoprecipitation was performed using the Protein A/G-PLUS Agarose beads (Santa Cruz Biotechnology #sc-2003) according to the manufacturer’s instructions. Briefly, 300 μg of cell lysate were incubated overnight with beclin-1 antibody (Santa Cruz Biotechnology, sc-48341) prior to incubating the protein:antibody complex with the beads. Immunoprecipitated proteins:antibody:beads complexes were dissociated using Laemmli buffer (Bio-Rad #161-0737) and subjected to SDS-PAGE.
Immunofluorescence
Glass coverslip (Nunc LabTek® Chambered CoverGlass, Thermo Scientific)-plated cells were fixed with 4% formaldehyde for 1 hours, then permeabilized with 0.1% Triton in PBS before incubation with primary antibodies. Primary antibodies were diluted in blocking solution 1/400 (1% BSA and 2% serum in PBS) and incubated overnight at 4°C. Secondary antibodies conjugated with Alexa Fluor 488 (Life Technologies, goat anti-rabbit #A-11034) or 633 (Life Technologies, goat anti-rabbit #A-21071) were diluted in blocking solution (1/1000) and incubated for 1 hour at room temperature. All fluorescence samples were examined at room temperature using a microscope (TCS SP5; Leica) with lasers exciting at 488, 543, and 633 nm (Leica). Lasers and spectral detection bands were chosen for the optimal imaging of Alexa Fluor 488 and 633 signals. PMT levels were set using control samples. Multicolor images were collected simultaneously in two or three channels. Images were taken using a 20x/NA/air 40x/1.25 NA oil HCX Plan Apo and 63x, 1.4–0.6 NA, oil, HCX Plan APO lens. Images were captured using the Leica LAS-AF image acquisition software. Overlays were generated using LAS-AF software. Photo montages were generated using LAS-AF, but were not further processed. The quantification was performed using FiJi: an outline was drawn around each cell and spots were counted in at least 50 cells derived from three representative 63x images from 3 independent experiments (one per experiment).
Real Time RT-PCR (qRT-PCR) and miRNA analysis
Total RNA, including small RNAs, was extracted using miRNeasy (QIAGEN #217004), quantified and 500 ng were reverse transcribed using the QuantiTect high capacity cDNA reverse transcription kit (QIAGEN #205311), for mRNA analysis, or 10 ng using Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems #43665967), for miRNA analysis. qRT-PCR was done using a 7500 Fast Real Time PCR system (Applied Biosystems) using Taqman gene/microRNA assays (Applied Biosystems), respectively. The probes used in the work are: SLC6A14 (Hs00924564_m1), SLC1A2 (Hs01102423_m1), GATA2 (Hs00231119_m1) and endogenous controls GAPDH (Hs02786624_g1) and ACTB (Hs99999903_m1)) for mRNA analysis. For microRNA assay the probes used are miR-23b-3p (ID:000400), and endogenous control RNU 48 (ID:001006) and U6 (ID:001973). TNFAIP3 (HsaCID0012648, Bio-Rad) and beclin-1 (HsaCID0016032, Bio-Rad) mRNA expression analysis was performed using SYBR green dye precast primers (Bio-Rad) and normalized on the results obtained using GAPDH (HsaCED0038674, Bio-Rad). The relative quantity was determined using ΔΔCt, according to the manufacturer’s instructions (Applied Biosystems).
Seahorse XF MitoStress Test
Cells were seeded in XFe96 cell culture plates with 1.5-2 × 104 cells per well in 80 μL of culture medium and incubated at 37°C (5-8 technical replicates). 24 hours post seeding medium was replaced with 180 μL XF base medium supplemented with 25 mM glucose (Sigma #G8644), 2 mM glutamine, 1 mM sodium pyruvate (Sigma #S8636). Cells were incubated for 1 hour at 37°C in atmospheric CO2 to allow the cells to pre-equilibrate with the XF base medium. An accurate titration with the uncoupler FCCP was performed for each cell type. Together with OCR measurement, values of ECAR were also recorded. Addition of Aspartate (0.15 mM, Sigma #A9256), Glutamate (0.13 mM Sigma #G1001), the ATP synthase inhibitor oligomycin (0.8 μM), the proton uncoupler FCCP (0.5-1 μM), the respiratory complex I inhibitor rotenone (1 μM), and the respiratory complex III inhibitor antimycin A (1 μM) was carried out at the times indicated. Protein quantification was used to normalize the results. Basal respiration is calculated as last rate measurement before injection – non-mitochondrial respiration rate. Maximal respiration is calculated as the maximum rate measurement after FCCP injection – non-mitochondrial respiration.
High-performance liquid chromatography (HPLC)
Amino acids extraction was performed as reported by Nemkov et al. (2015). Briefly, cell pellets were extracted immediately before analysis in ice-cold lysis/extraction buffer (methanol/acetonitrile/water, 5/3/2 v/v, Sigma) at a ratio of 2 × 106 cells/ml. Tubes were first agitated at 4°C for 30 minutes and then centrifuged at 4°C, 10000 g for 10 minutes. The recovered supernatants were subjected to precolumn derivatization of amino acids using 4-N,N-dimethylaminoazobenzene-4’-sulfonyl chloride (Supelco #502219) following the manufacture instructions (DABS, Sigma). DABS-amino acids were detected at visible light wavelengths (436nm) using a DIONEX P680-UVD170U HPLC system (column: Kinetex 5μm C18 100 Å LC column 250x4.6mm; mobile phase: A = KH2PO4 25mM pH 6.8 (Sigma), B = acetonitrile (Sigma #34851); gradient program (time:%B): 0-5 min: 20%; 5-9 min: 25%; 9-15 min: 25%; 15-18 min 30%; 18-22 30%; 22-27 35%; 27-35 60%; 35-43 70%; flow rate: 0.8ml/min; injection volume: 10μl). A reference amino acids spectrum was obtained by titrating a mixture of amino acids of known concentration (Sigma # A9781).
Gas chromatography–MS (GC-MS)
Snap frozen tissue samples were minced and subjected to extraction using a mixture of CHCl3:MeOH:H2O (Sigma #34854-1 and #900688-1, Merck #102699-1000) (1:1:1). Minced tissues were quenched with 0.4 mL ice cold methanol and an equal volume of water containing 1 μg norvaline (Sigma #53721), used as internal standard. One volume of chloroform was added, and the samples were vortexed at 4°C for 30 minutes. Samples were centrifuged at 3000 g for 10 minutes, and the aqueous phase was collected in a new tube and evaporated at room temperature. Dried polar metabolites were dissolved in 60 μL of 2% methoxyamine hydrochloride (Sigma #226904) in pyridine (25104 Thermo), and held at 30°C for 2 hours. After dissolution and reaction, 90 μL MSTFA + 1% TMCS (69478-10x Sigma) were added and samples were incubated at 37°C for 60 minutes. Gas chromatographic runs were performed with helium as carrier gas at 0.6 mL/min. The split inlet temperature was set to 250°C and the injection volume of 1 μL. A split ratio of 1:10 was used. The GC oven temperature ramp was from 60°C to 325°C at 10°C/min. The data acquisition rate was 10 Hz. For the Quadrupole, an EI source (70 eV) was used, and full-scan spectra (mass range from 50 to 600) were recorded in the positive ion mode. The ion source and transfer line temperatures were set, respectively, to 250°C and 290°C. The MassHunter data processing tool (Agilent) was used to obtain a global metabolic profiling. Fihen Metabolomics RTL library (Agilent G1676AA)
13C-tracing experiments using liquid-chromatography-MS (LC-MS)
Parental and LTED MCF7 cells were incubated for 48 hours with 0.15 mM [U-13C]-aspartic acid (Cambridge Isotope Laboratories) or 0.13 mM [U-13C]-L-glutamic acid (Sigma Aldrich). The cells were washed once with ice cold 0.9% NaCl solution. The metabolite extraction was performed using 80% methanol. After 5 minutes of incubation cells were scraped and collected in a new tube. Following a centrifugation at 20000 g for 10 minutes at 4°C, the supernatant was transferred to a new vial for MS analysis. Pellet was used for protein quantification. 5 μl of each sample was loaded into a Dionex UltiMate 3000 LC System (Thermo Scientific Bremen) equipped with a C-18 column (Acquity UPLC -HSS T3 1. 8 μm; 2.1 × 150 mm, Waters) coupled to a Q Exactive Orbitrap mass spectrometer (Thermo Scientific) operating in negative ion mode. A step gradient was carried out using solvent A (10 mM TBA and 15 mM acetic acid) and solvent B (100% methanol). The gradient started with 0% of solvent B and 100% solvent A and remained at 0% B until 2 minutes post injection. A linear gradient to 37% B was carried out until 7 minutes and increased to 41% until 14 minutes. Between 14 and 26 minutes the gradient increased to 100% of B and remained at 100% B for 4 minutes. At 30 minutes the gradient returned to 0% B. The chromatography was stopped at 40 minutes. The flow was kept constant at 250 μL/min and the column was placed at 25°C throughout the analysis. The MS operated in full scan - SIM (negative mode) using a spray voltage of 3.2 kV, capillary temperature of 320°C, sheath gas at 10.0, auxiliary gas at 5.0. For full scan - SIM mode, AGC target was set at 1e6 using a resolution of 70.000, with a maximum IT of 256 ms. For the data analyses we integrated the peak areas using the Thermo XCalibur Quan Browser software (Thermo Scientific).
Lung retention assay
MCF7-LTED anti-miR-CTR, anti-miR-23b-3p, siCTR and siSLC1A2 cells were labeled with CellTracker Red CMTPX or Green CMFDA dyes (Molecular Probes), trypsinized, mixed at a 1:1 ratio, and a total of 1 × 106 cells injected into the tail veins of 5- to 6-week-old BALB/c mice. Mice were sacrificed at 1 and 5 hours post injection, and lungs were examined on a Leica TCS SP5 microscope using x10 lens. Six images were taken for each lung. Tumor cell colonization of the lung was quantified in the Fiji open platform for image analysis, by converting the red and green images into separate binary images and measuring total tumor cell coverage per field of view.
Gene and miRNA expression analysis
MCF7 cells were cultured in either the recommended medium conditions or E2 deprived for 3 days and compared to MCF7-LTED cells cultured in standard medium. Triplicate samples from 3 independent experiments were assessed for global miRNA and gene expression by Microarrays. Specifically, 9 samples were hybridized on Agilent whole human genome microarray (Agilent Technologies #G4851A), which represents 60k unique human transcripts. The same samples were hybridized also on Agilent human miRNA microarray v16 (Agilent Technologies #G4870A). One-color gene expression was performed according to the manufacturer’s procedure as previously described (Ferracin et al., 2013).
Bioinformatic analyses
Microarray data were normalized and analyzed using GeneSpring GX v.14.8 software (Agilent Technologies). Data transformation was applied to set all the negative raw values at 1.0, then the quantile normalization was applied. The probes detected in at least one sample were used for statistical analyses. Differentially expressed genes were selected to have a ≥ 2-fold expression difference between groups and an adjusted P-value ≤ 0.05 at ANOVA test, with Benjamini and Hoechberg correction. Supervised hierarchical clustering was performed for OS samples with GeneSpring clustering tool using the list of differentially expressed genes and Euclidean correlation as a measure of similarity.
The mRNA and miRNA anti-correlation network analyses described in Figure 1C were performed using the R software. Briefly, mRNAs and miRNAs found as differentially expressed in each pairwise comparison (FDR < 0.05, FC < −2 or > 2) were initially used to filter candidate interactors emerging from the PITA miRNA/target predictor tool (Kertesz et al., 2007). To ensure the identification of relevant interactors and limit the size of the network, the filtered collection of mRNA/miRNA pairs was further purged from experimentally validated pairs, according to the miRTarBase repository (Chou et al., 2018). The expression values of the remaining candidate interactors (taken from the whole set of samples rather than from those relevant for the differentially expressed analysis) were tested for mRNA/miRNA expression anti-correlation (correlation test, Benjamini-Hochberg correction, adjusted p < 0.05 and correlation < 0). The resulting networks were graphically annotated and represented using functions from the igraph R package.
Analysis of human datasets
The BRCA TCGA dataset analyzed in Figures 1A, 1B, and 4I is available in The Cancer Genome Atlas (TCGA) data portal (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). Gene and miRNA expression data and the corresponding clinical information for ER+ BRCA from TCGA provisional dataset were obtained through Firebrowse (http://firebrowse.org/) data portal. The normalized reads count for mRNA (RSEM) and mature miRNAs (RPM) were downloaded and log2 transformed. From the BRCA dataset, we selected samples with miRNA-mRNA and overall survival (OS) data. Survival analysis was performed using Kaplan-Meier curve log-rank testing, using Cutoff finder (Budczies et al., 2012) for best miR-23b-3p/SLCA6A14 high- and low-expression selection. For mRNA analysis in Figure S1C, overall survival (OS) of untreated breast cancer patients belonging to the METABRIC dataset were retrieved using Km-plotter (http://kmplot.com) (Lánczky et al., 2016). For SLC6A14 (Figure S1B), TNFAIP3 (Figure S1E) survival analysis, the curated dataset of ER+ breast cancers was created using Km-plotter and included the relapse free survival data of patients belonging to the following datasets: GSE6532 (Loi et al., 2007), GSE20711 (Dedeurwaerder et al., 2011), GSE7390 (Desmedt et al., 2007), GSE21653 (Sabatier et al., 2011), E-MTAB-365 (Rème et al., 2013), GSE2034 (Wang et al., 2005), GSE2990 (Sotiriou et al., 2006), GSE17705 (Symmans et al., 2010), GSE12093 (Zhang et al., 2009), GSE9195 (Loi et al., 2008), GSE3494 (Miller et al., 2005), GSE4611 (Karn et al., 2010), GSE45255 (Nagalla et al., 2013), GSE2603 (Minn et al., 2005), GSE16391 (Desmedt et al., 2009), GSE42568 (Clarke et al., 2013), GSE26971 (Filipits et al., 2011) and GSE19615 (Li et al., 2010).
For GATA2 (209710_at; Figure 3I) overall survival analysis the curated dataset comprises the following datasets: GSE20711, GSE7390, GSE45255, GSE42568, GSE3494. For SLC6A14 (219795_at; Figure 7G), TNFAIP3 (202643_s_at; Figure 7H) survival analysis, the curated dataset of ER+ breast cancers that have been treated with adjuvant tamoxifen was created using Km-plotter and included patients that belong to the following datasets GSE6532, GSE2990, GSE17705, GSE12093, GSE9195, GSE45255, GSE16391, GSE3494, GSE26971 and GSE19615. SLC1A2 data in 52 paired ER+ breast cancer samples pre- and post-2-week letrozole treatment (Figure 7I) were retrieved using GEO (GSE5462) (Miller et al., 2007) and normalized log2 expression values are shown either pre- or post letrozole treatment for each patient. Correlation data on patients that have been treated with adjuvant fulvestrant (Figure 7J) were from the GSE33658 dataset (Massarweh et al., 2011). SLC1A2 and AURKA log2 normalized expression levels were reported. Changes were obtained by subtracting from the expression value of tumor that have been treated with fulvestrant that of baseline expression. Correlation analysis was performed using Spearman correlation test.
Quantitation and Statistical Analysis
Statistics were performed using GraphPad Prism 8. Unless stated otherwise, all numerical data are expressed as the mean ± error of the mean (SEM) and noted in figure legends. Unless stated otherwise, comparisons between 2 groups were made using the two-tailed, unpaired Student’s t test. Comparisons between multiple groups were made using one-way analysis of variance (ANOVA), and two-way ANOVA for comparisons between multiple groups with independent variables. Bonferroni post-testing (unless otherwise stated) with a confidence interval of 95% was used for individual comparisons. Multivariate Cox Analyses on the cohort of patients analyzed in Figure 7G and 7H were generated using KM-plotter and are reported in Table S3. Statistical significance was defined as: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ns, not significant.
Data and Code Availability
mRNA and miRNA data derived from microarray analysis are deposited in NCBI Gene Expression Omnibus (GEO) database: accession number GSE120931. Feature Extraction 10.7.3.1 software (Agilent Technologies) was used to obtain the microarray raw-data and data.
The R script (mM_Net.R) for the interactor network analysis displayed in Figure 1C is available and can be downloaded (https://github.com/matteoramazzotti/TransTools).
Acknowledgments
The work of A.M. was funded by Fondazione Umberto Veronesi, Associazione Italiana Ricerca sul Cancro (AIRC), and Fondazione Cassa di Risparmio di Firenze (grant 19515 to P.C. and A.M.), and an AIRC grant (8797) to P.C. M.B. is an AIRC fellow. L.-A.M. is supported by Breast Cancer Now, working in partnership with Walk the Walk. M.M. is supported by FWO (G0D1717N) and holds an ERC consolidator grant (773208). We thank Prof. Massimo Negrini (University of Ferrara) and the facility of the Laboratorio per le Tecnologie delle Terapie Avanzate (LTTA) for performing the miRNA and gene expression analyses, Angela Subbiani for helping with cell culturing, and Prof. Clare M. Isacke (The Institute of Cancer Research) for critical reading of the manuscript.
Author Contributions
The authors contributed to this work in different capacities, described as follows. M.B. performed cell-based and biochemistry experiments, study design, data acquisition, analysis, and interpretation; N.L. performed cell-based and biochemistry experiments, Seahorse and metabolic analysis, and participated in the interpretation of the data; L.I. performed Seahorse and metabolic analysis and participated in the interpretation of the data; M.R., Q.G., and M.F. performed gene expression, clinical data, and bioinformatic analyses; S.L. performed HPLC analysis and amino acid quantification; S.R. performed the cell-based experiments; M.P. performed the GC-MS analysis; F.B. performed the xenograft mouse model; B.M.S. and E.M. generated and performed the experiments on the patient-derived xenografts; F.C., F.V., and M.M. designed and performed the LC-MS analysis and data interpretation; L.-A.M. generated the LTED cell lines; G.C. and E.G. developed the methodology, provided administrative and technical support, interpreted the data; P.C. performed the data analysis and interpretation and revised the manuscript; A.M. conceived, designed, and supervised the study; performed the cell-based and metabolic experiments; analyzed and interpreted the data; and wrote the manuscript. All of the authors reviewed the prepared manuscript.
Declaration of Interests
L.-A.M. receives academic funding from Radius Pharma, PUMA Biotechnology, and Pfizer. The other authors declare no competing interests.
Published: July 2, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.06.010.
Supplemental Information
References
- An Y., Zhang Z., Shang Y., Jiang X., Dong J., Yu P., Nie Y., Zhao Q. miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis. 2015;6:e1766. doi: 10.1038/cddis.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]; An, Y., Zhang, Z., Shang, Y., Jiang, X., Dong, J., Yu, P., Nie, Y., and Zhao, Q. (2015). miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis. 6, e1766. [DOI] [PMC free article] [PubMed]
- Babu E., Bhutia Y.D., Ramachandran S., Gnanaprakasam J.P., Prasad P.D., Thangaraju M., Ganapathy V. Deletion of the amino acid transporter Slc6a14 suppresses tumour growth in spontaneous mouse models of breast cancer. Biochem. J. 2015;469:17–23. doi: 10.1042/BJ20150437. [DOI] [PubMed] [Google Scholar]; Babu, E., Bhutia, Y.D., Ramachandran, S., Gnanaprakasam, J.P., Prasad, P.D., Thangaraju, M., and Ganapathy, V. (2015). Deletion of the amino acid transporter Slc6a14 suppresses tumour growth in spontaneous mouse models of breast cancer. Biochem. J. 469, 17-23. [DOI] [PubMed]
- Bacci M., Giannoni E., Fearns A., Ribas R., Gao Q., Taddei M.L., Pintus G., Dowsett M., Isacke C.M., Martin L.A. miR-155 drives metabolic reprogramming of ER+ breast cancer cells following long-term estrogen deprivation and predicts clinical response to aromatase inhibitors. Cancer Res. 2016;76:1615–1626. doi: 10.1158/0008-5472.CAN-15-2038. [DOI] [PubMed] [Google Scholar]; Bacci, M., Giannoni, E., Fearns, A., Ribas, R., Gao, Q., Taddei, M.L., Pintus, G., Dowsett, M., Isacke, C.M., Martin, L.A., et al. (2016). miR-155 drives metabolic reprogramming of ER+ breast cancer cells following long-term estrogen deprivation and predicts clinical response to aromatase inhibitors. Cancer Res. 76, 1615-1626. [DOI] [PubMed]
- Begum S., Hayashi M., Ogawa T., Jabboure F.J., Brait M., Izumchenko E., Tabak S., Ahrendt S.A., Westra W.H., Koch W. An integrated genome-wide approach to discover deregulated microRNAs in non-small cell lung cancer: clinical significance of miR-23b-3p deregulation. Sci. Rep. 2015;5:13236. doi: 10.1038/srep13236. [DOI] [PMC free article] [PubMed] [Google Scholar]; Begum, S., Hayashi, M., Ogawa, T., Jabboure, F.J., Brait, M., Izumchenko, E., Tabak, S., Ahrendt, S.A., Westra, W.H., Koch, W., et al. (2015). An integrated genome-wide approach to discover deregulated microRNAs in non-small cell lung cancer: clinical significance of miR-23b-3p deregulation. Sci. Rep. 5, 13236. [DOI] [PMC free article] [PubMed]
- Bisognin A., Sales G., Coppe A., Bortoluzzi S., Romualdi C. MAGIA2: from miRNA and genes expression data integrative analysis to microRNA-transcription factor mixed regulatory circuits (2012 update) Nucleic Acids Res. 2012;40(Web Server issue):W13–W21. doi: 10.1093/nar/gks460. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bisognin, A., Sales, G., Coppe, A., Bortoluzzi, S., and Romualdi, C. (2012). MAGIA2: from miRNA and genes expression data integrative analysis to microRNA-transcription factor mixed regulatory circuits (2012 update). Nucleic Acids Res. 40(Web Server issue), W13-W21. [DOI] [PMC free article] [PubMed]
- Budczies J., Klauschen F., Sinn B.V., Győrffy B., Schmitt W.D., Darb-Esfahani S., Denkert C. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]; Budczies, J., Klauschen, F., Sinn, B.V., Győrffy, B., Schmitt, W.D., Darb-Esfahani, S., and Denkert, C. (2012). Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One 7, e51862. [DOI] [PMC free article] [PubMed]
- Chen D., Wu X., Xia M., Wu F., Ding J., Jiao Y., Zhan Q., An F. Upregulated exosomic miR-23b-3p plays regulatory roles in the progression of pancreatic cancer. Oncol. Rep. 2017;38:2182–2188. doi: 10.3892/or.2017.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen, D., Wu, X., Xia, M., Wu, F., Ding, J., Jiao, Y., Zhan, Q., and An, F. (2017). Upregulated exosomic miR-23b-3p plays regulatory roles in the progression of pancreatic cancer. Oncol. Rep. 38, 2182-2188. [DOI] [PMC free article] [PubMed]
- Cho S., Jang I., Jun Y., Yoon S., Ko M., Kwon Y., Choi I., Chang H., Ryu D., Lee B. MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013;41:D252–D257. doi: 10.1093/nar/gks1168. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cho, S., Jang, I., Jun, Y., Yoon, S., Ko, M., Kwon, Y., Choi, I., Chang, H., Ryu, D., Lee, B., et al. (2013). MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 41, D252-D257. [DOI] [PMC free article] [PubMed]
- Chou C.H., Shrestha S., Yang C.D., Chang N.W., Lin Y.L., Liao K.W., Huang W.C., Sun T.H., Tu S.J., Lee W.H. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chou, C.H., Shrestha, S., Yang, C.D., Chang, N.W., Lin, Y.L., Liao, K.W., Huang, W.C., Sun, T.H., Tu, S.J., Lee, W.H., et al. (2018). miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 46 (D1), D296-D302. [DOI] [PMC free article] [PubMed]
- Clarke C., Madden S.F., Doolan P., Aherne S.T., Joyce H., O’Driscoll L., Gallagher W.M., Hennessy B.T., Moriarty M., Crown J. Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis. 2013;34:2300–2308. doi: 10.1093/carcin/bgt208. [DOI] [PubMed] [Google Scholar]; Clarke, C., Madden, S.F., Doolan, P., Aherne, S.T., Joyce, H., O’Driscoll, L., Gallagher, W.M., Hennessy, B.T., Moriarty, M., Crown, J., et al. (2013). Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis 34, 2300-2308. [DOI] [PubMed]
- Cook K.L., Shajahan A.N., Clarke R. Autophagy and endocrine resistance in breast cancer. Expert Rev. Anticancer Ther. 2011;11:1283–1294. doi: 10.1586/era.11.111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cook, K.L., Shajahan, A.N., and Clarke, R. (2011). Autophagy and endocrine resistance in breast cancer. Expert Rev. Anticancer Ther. 11, 1283-1294. [DOI] [PMC free article] [PubMed]
- Coothankandaswamy V., Cao S., Xu Y., Prasad P.D., Singh P.K., Reynolds C.P., Yang S., Ogura J., Ganapathy V., Bhutia Y.D. Amino acid transporter SLC6A14 is a novel and effective drug target for pancreatic cancer. Br. J. Pharmacol. 2016;173:3292–3306. doi: 10.1111/bph.13616. [DOI] [PMC free article] [PubMed] [Google Scholar]; Coothankandaswamy, V., Cao, S., Xu, Y., Prasad, P.D., Singh, P.K., Reynolds, C.P., Yang, S., Ogura, J., Ganapathy, V., and Bhutia, Y.D. (2016). Amino acid transporter SLC6A14 is a novel and effective drug target for pancreatic cancer. Br. J. Pharmacol. 173, 3292-3306. [DOI] [PMC free article] [PubMed]
- Cottu P., Bièche I., Assayag F., El Botty R., Chateau-Joubert S., Thuleau A., Bagarre T., Albaud B., Rapinat A., Gentien D. Acquired resistance to endocrine treatments is associated with tumor-specific molecular changes in patient-derived luminal breast cancer xenografts. Clin. Cancer Res. 2014;20:4314–4325. doi: 10.1158/1078-0432.CCR-13-3230. [DOI] [PubMed] [Google Scholar]; Cottu, P., Bieche, I., Assayag, F., El Botty, R., Chateau-Joubert, S., Thuleau, A., Bagarre, T., Albaud, B., Rapinat, A., Gentien, D., et al. (2014). Acquired resistance to endocrine treatments is associated with tumor-specific molecular changes in patient-derived luminal breast cancer xenografts. Clin. Cancer Res. 20, 4314-4325. [DOI] [PubMed]
- Dedeurwaerder S., Desmedt C., Calonne E., Singhal S.K., Haibe-Kains B., Defrance M., Michiels S., Volkmar M., Deplus R., Luciani J. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med. 2011;3:726–741. doi: 10.1002/emmm.201100801. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dedeurwaerder, S., Desmedt, C., Calonne, E., Singhal, S.K., Haibe-Kains, B., Defrance, M., Michiels, S., Volkmar, M., Deplus, R., Luciani, J., et al. (2011). DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med. 3, 726-741. [DOI] [PMC free article] [PubMed]
- Desmedt C., Piette F., Loi S., Wang Y., Lallemand F., Haibe-Kains B., Viale G., Delorenzi M., Zhang Y., d’Assignies M.S., TRANSBIG Consortium Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]; Desmedt, C., Piette, F., Loi, S., Wang, Y., Lallemand, F., Haibe-Kains, B., Viale, G., Delorenzi, M., Zhang, Y., d’Assignies, M.S., et al.; TRANSBIG Consortium (2007). Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 13, 3207-3214. [DOI] [PubMed]
- Desmedt C., Giobbie-Hurder A., Neven P., Paridaens R., Christiaens M.R., Smeets A., Lallemand F., Haibe-Kains B., Viale G., Gelber R.D. The Gene expression Grade Index: a potential predictor of relapse for endocrine-treated breast cancer patients in the BIG 1-98 trial. BMC Med. Genomics. 2009;2:40. doi: 10.1186/1755-8794-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]; Desmedt, C., Giobbie-Hurder, A., Neven, P., Paridaens, R., Christiaens, M.R., Smeets, A., Lallemand, F., Haibe-Kains, B., Viale, G., Gelber, R.D., et al. (2009). The Gene expression Grade Index: a potential predictor of relapse for endocrine-treated breast cancer patients in the BIG 1-98 trial. BMC Med. Genomics 2, 40. [DOI] [PMC free article] [PubMed]
- Dumortier O., Hinault C., Van Obberghen E. MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 2013;18:312–324. doi: 10.1016/j.cmet.2013.06.004. [DOI] [PubMed] [Google Scholar]; Dumortier, O., Hinault, C., and Van Obberghen, E. (2013). MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 18, 312-324. [DOI] [PubMed]
- Ferracin M., Bassi C., Pedriali M., Pagotto S., D’Abundo L., Zagatti B., Corrà F., Musa G., Callegari E., Lupini L. miR-125b targets erythropoietin and its receptor and their expression correlates with metastatic potential and ERBB2/HER2 expression. Mol. Cancer. 2013;12:130. doi: 10.1186/1476-4598-12-130. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ferracin, M., Bassi, C., Pedriali, M., Pagotto, S., D’Abundo, L., Zagatti, B., Corra, F., Musa, G., Callegari, E., Lupini, L., et al. (2013). miR-125b targets erythropoietin and its receptor and their expression correlates with metastatic potential and ERBB2/HER2 expression. Mol. Cancer 12, 130. [DOI] [PMC free article] [PubMed]
- Filipits M., Rudas M., Jakesz R., Dubsky P., Fitzal F., Singer C.F., Dietze O., Greil R., Jelen A., Sevelda P., EP Investigators A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 2011;17:6012–6020. doi: 10.1158/1078-0432.CCR-11-0926. [DOI] [PubMed] [Google Scholar]; Filipits, M., Rudas, M., Jakesz, R., Dubsky, P., Fitzal, F., Singer, C.F., Dietze, O., Greil, R., Jelen, A., Sevelda, P., et al.; EP Investigators (2011). A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 17, 6012-6020. [DOI] [PubMed]
- Forbes J.F., Cuzick J., Buzdar A., Howell A., Tobias J.S., Baum M., Arimidex, Tamoxifen, Alone or in Combination (ATAC) Trialists’ Group Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9:45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]; Forbes, J.F., Cuzick, J., Buzdar, A., Howell, A., Tobias, J.S., Baum, M.; Arimidex, Tamoxifen, Alone or in Combination (ATAC) Trialists’ Group (2008). Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 9, 45-53. [DOI] [PubMed]
- Fox E.M., Kuba M.G., Miller T.W., Davies B.R., Arteaga C.L. Autocrine IGF-I/insulin receptor axis compensates for inhibition of AKT in ER-positive breast cancer cells with resistance to estrogen deprivation. Breast Cancer Res. 2013;15:R55. doi: 10.1186/bcr3449. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fox, E.M., Kuba, M.G., Miller, T.W., Davies, B.R., and Arteaga, C.L. (2013). Autocrine IGF-I/insulin receptor axis compensates for inhibition of AKT in ER-positive breast cancer cells with resistance to estrogen deprivation. Breast Cancer Res. 15, R55. [DOI] [PMC free article] [PubMed]
- Fulciniti M., Amodio N., Bandi R.L., Cagnetta A., Samur M.K., Acharya C., Prabhala R., D’Aquila P., Bellizzi D., Passarino G. miR-23b/SP1/c-myc forms a feed-forward loop supporting multiple myeloma cell growth. Blood Cancer J. 2016;6:e380. doi: 10.1038/bcj.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fulciniti, M., Amodio, N., Bandi, R.L., Cagnetta, A., Samur, M.K., Acharya, C., Prabhala, R., D’Aquila, P., Bellizzi, D., Passarino, G., et al. (2016). miR-23b/SP1/c-myc forms a feed-forward loop supporting multiple myeloma cell growth. Blood Cancer J. 6, e380. [DOI] [PMC free article] [PubMed]
- Galluzzi L., Pietrocola F., Levine B., Kroemer G. Metabolic control of autophagy. Cell. 2014;159:1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Galluzzi, L., Pietrocola, F., Levine, B., and Kroemer, G. (2014). Metabolic control of autophagy. Cell 159, 1263-1276. [DOI] [PMC free article] [PubMed]
- Galluzzi L., Pietrocola F., Bravo-San Pedro J.M., Amaravadi R.K., Baehrecke E.H., Cecconi F., Codogno P., Debnath J., Gewirtz D.A., Karantza V. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]; Galluzzi, L., Pietrocola, F., Bravo-San Pedro, J.M., Amaravadi, R.K., Baehrecke, E.H., Cecconi, F., Codogno, P., Debnath, J., Gewirtz, D.A., Karantza, V., et al. (2015). Autophagy in malignant transformation and cancer progression. EMBO J. 34, 856-880. [DOI] [PMC free article] [PubMed]
- Garcia-Bermudez J., Baudrier L., La K., Zhu X.G., Fidelin J., Sviderskiy V.O., Papagiannakopoulos T., Molina H., Snuderl M., Lewis C.A. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 2018;20:775–781. doi: 10.1038/s41556-018-0118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Garcia-Bermudez, J., Baudrier, L., La, K., Zhu, X.G., Fidelin, J., Sviderskiy, V.O., Papagiannakopoulos, T., Molina, H., Snuderl, M., Lewis, C.A., et al. (2018). Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 20, 775-781. [DOI] [PMC free article] [PubMed]
- Gozuacik D., Akkoc Y., Ozturk D.G., Kocak M. Autophagy-Regulating microRNAs and Cancer. Front. Oncol. 2017;7:65. doi: 10.3389/fonc.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gozuacik, D., Akkoc, Y., Ozturk, D.G., and Kocak, M. (2017). Autophagy-Regulating microRNAs and Cancer. Front. Oncol. 7, 65. [DOI] [PMC free article] [PubMed]
- Gui D.Y., Sullivan L.B., Luengo A., Hosios A.M., Bush L.N., Gitego N., Davidson S.M., Freinkman E., Thomas C.J., Vander Heiden M.G. Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab. 2016;24:716–727. doi: 10.1016/j.cmet.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gui, D.Y., Sullivan, L.B., Luengo, A., Hosios, A.M., Bush, L.N., Gitego, N., Davidson, S.M., Freinkman, E., Thomas, C.J., and Vander Heiden, M.G. (2016). Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab. 24, 716-727. [DOI] [PMC free article] [PubMed]
- Györffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]; Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. (2010). An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients.Breast Cancer Res Treat. 123, 725-31. [DOI] [PubMed]
- Hediger M.A., Romero M.F., Peng J.B., Rolfs A., Takanaga H., Bruford E.A. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]; Hediger, M.A., Romero, M.F., Peng, J.B., Rolfs, A., Takanaga, H., and Bruford, E.A. (2004). The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Arch. 447, 465-468. [DOI] [PubMed]
- Hosios A.M., Hecht V.C., Danai L.V., Johnson M.O., Rathmell J.C., Steinhauser M.L., Manalis S.R., Vander Heiden M.G. Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev. Cell. 2016;36:540–549. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hosios, A.M., Hecht, V.C., Danai, L.V., Johnson, M.O., Rathmell, J.C., Steinhauser, M.L., Manalis, S.R., and Vander Heiden, M.G. (2016). Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev. Cell 36, 540-549. [DOI] [PMC free article] [PubMed]
- Karn T., Metzler D., Ruckhäberle E., Hanker L., Gätje R., Solbach C., Ahr A., Schmidt M., Holtrich U., Kaufmann M., Rody A. Data-driven derivation of cutoffs from a pool of 3,030 Affymetrix arrays to stratify distinct clinical types of breast cancer. Breast Cancer Res. Treat. 2010;120:567–579. doi: 10.1007/s10549-009-0416-z. [DOI] [PubMed] [Google Scholar]; Karn, T., Metzler, D., Ruckhaberle, E., Hanker, L., Gatje, R., Solbach, C., Ahr, A., Schmidt, M., Holtrich, U., Kaufmann, M., and Rody, A. (2010). Data-driven derivation of cutoffs from a pool of 3,030 Affymetrix arrays to stratify distinct clinical types of breast cancer. Breast Cancer Res. Treat. 120, 567-579. [DOI] [PubMed]
- Karunakaran S., Ramachandran S., Coothankandaswamy V., Elangovan S., Babu E., Periyasamy-Thandavan S., Gurav A., Gnanaprakasam J.P., Singh N., Schoenlein P.V. SLC6A14 (ATB0,+) protein, a highly concentrative and broad specific amino acid transporter, is a novel and effective drug target for treatment of estrogen receptor-positive breast cancer. J. Biol. Chem. 2011;286:31830–31838. doi: 10.1074/jbc.M111.229518. [DOI] [PMC free article] [PubMed] [Google Scholar]; Karunakaran, S., Ramachandran, S., Coothankandaswamy, V., Elangovan, S., Babu, E., Periyasamy-Thandavan, S., Gurav, A., Gnanaprakasam, J.P., Singh, N., Schoenlein, P.V., et al. (2011). SLC6A14 (ATB0,+) protein, a highly concentrative and broad specific amino acid transporter, is a novel and effective drug target for treatment of estrogen receptor-positive breast cancer. J. Biol. Chem. 286, 31830-31838. [DOI] [PMC free article] [PubMed]
- Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]; Kertesz, M., Iovino, N., Unnerstall, U., Gaul, U., and Segal, E. (2007). The role of site accessibility in microRNA target recognition. Nat. Genet. 39, 1278-1284. [DOI] [PubMed]
- Kishore S., Jaskiewicz L., Burger L., Hausser J., Khorshid M., Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]; Kishore, S., Jaskiewicz, L., Burger, L., Hausser, J., Khorshid, M., and Zavolan, M. (2011). A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods 8, 559-564.. [DOI] [PubMed]
- Lánczky A., Nagy Á., Bottai G., Munkácsy G., Szabó A., Santarpia L., Győrffy B. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]; Lanczky, A., Nagy, A., Bottai, G., Munkacsy, G., Szabo, A., Santarpia, L., and Győrffy, B. (2016). miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 160, 439-446. [DOI] [PubMed]
- Li Y., Zou L., Li Q., Haibe-Kains B., Tian R., Li Y., Desmedt C., Sotiriou C., Szallasi Z., Iglehart J.D. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat. Med. 2010;16:214–218. doi: 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, Y., Zou, L., Li, Q., Haibe-Kains, B., Tian, R., Li, Y., Desmedt, C., Sotiriou, C., Szallasi, Z., Iglehart, J.D., et al. (2010). Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat. Med. 16, 214-218. [DOI] [PMC free article] [PubMed]
- Li B., Sun M., Gao F., Liu W., Yang Y., Liu H., Cheng Y., Liu C., Cai J. Up-regulated expression of miR-23a/b targeted the pro-apoptotic Fas in radiation-induced thymic lymphoma. Cell. Physiol. Biochem. 2013;32:1729–1740. doi: 10.1159/000356607. [DOI] [PubMed] [Google Scholar]; Li, B., Sun, M., Gao, F., Liu, W., Yang, Y., Liu, H., Cheng, Y., Liu, C., and Cai, J. (2013). Up-regulated expression of miR-23a/b targeted the pro-apoptotic Fas in radiation-induced thymic lymphoma. Cell. Physiol. Biochem. 32, 1729-1740. [DOI] [PubMed]
- Liu J., Xu Y., Stoleru D., Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl. Acad. Sci. USA. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu, J., Xu, Y., Stoleru, D., and Salic, A. (2012). Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl. Acad. Sci. USA 109, 413-418. [DOI] [PMC free article] [PubMed]
- Liu S., Meng X., Chen H., Liu W., Miller T., Murph M., Lu Y., Zhang F., Gagea M., Arteaga C.L. Targeting tyrosine-kinases and estrogen receptor abrogates resistance to endocrine therapy in breast cancer. Oncotarget. 2014;5:9049–9064. doi: 10.18632/oncotarget.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu, S., Meng, X., Chen, H., Liu, W., Miller, T., Murph, M., Lu, Y., Zhang, F., Gagea, M., Arteaga, C.L., et al. (2014). Targeting tyrosine-kinases and estrogen receptor abrogates resistance to endocrine therapy in breast cancer. Oncotarget 5, 9049-9064. [DOI] [PMC free article] [PubMed]
- Loi S., Haibe-Kains B., Desmedt C., Lallemand F., Tutt A.M., Gillet C., Ellis P., Harris A., Bergh J., Foekens J.A. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J. Clin. Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]; Loi, S., Haibe-Kains, B., Desmedt, C., Lallemand, F., Tutt, A.M., Gillet, C., Ellis, P., Harris, A., Bergh, J., Foekens, J.A., et al. (2007). Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J. Clin. Oncol. 25, 1239-1246. [DOI] [PubMed]
- Loi S., Haibe-Kains B., Desmedt C., Wirapati P., Lallemand F., Tutt A.M., Gillet C., Ellis P., Ryder K., Reid J.F. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]; Loi, S., Haibe-Kains, B., Desmedt, C., Wirapati, P., Lallemand, F., Tutt, A.M., Gillet, C., Ellis, P., Ryder, K., Reid, J.F., et al. (2008). Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics 9, 239. [DOI] [PMC free article] [PubMed]
- Martin L.A., Farmer I., Johnston S.R., Ali S., Marshall C., Dowsett M. Enhanced estrogen receptor (ER) alpha, ERBB2, and MAPK signal transduction pathways operate during the adaptation of MCF-7 cells to long term estrogen deprivation. J. Biol. Chem. 2003;278:30458–30468. doi: 10.1074/jbc.M305226200. [DOI] [PubMed] [Google Scholar]; Martin, L.A., Farmer, I., Johnston, S.R., Ali, S., Marshall, C., and Dowsett, M. (2003). Enhanced estrogen receptor (ER) alpha, ERBB2, and MAPK signal transduction pathways operate during the adaptation of MCF-7 cells to long term estrogen deprivation. J. Biol. Chem. 278, 30458-30468. [DOI] [PubMed]
- Martin L.A., Pancholi S., Farmer I., Guest S., Ribas R., Weigel M.T., Thornhill A.M., Ghazoui Z., A’Hern R., Evans D.B. Effectiveness and molecular interactions of the clinically active mTORC1 inhibitor everolimus in combination with tamoxifen or letrozole in vitro and in vivo. Breast Cancer Res. 2012;14:R132. doi: 10.1186/bcr3330. [DOI] [PMC free article] [PubMed] [Google Scholar]; Martin, L.A., Pancholi, S., Farmer, I., Guest, S., Ribas, R., Weigel, M.T., Thornhill, A.M., Ghazoui, Z., A’Hern, R., Evans, D.B., et al. (2012). Effectiveness and molecular interactions of the clinically active mTORC1 inhibitor everolimus in combination with tamoxifen or letrozole in vitro and in vivo. Breast Cancer Res. 14, R132. [DOI] [PMC free article] [PubMed]
- Massarweh S., Tham Y.L., Huang J., Sexton K., Weiss H., Tsimelzon A., Beyer A., Rimawi M., Cai W.Y., Hilsenbeck S. A phase II neoadjuvant trial of anastrozole, fulvestrant, and gefitinib in patients with newly diagnosed estrogen receptor positive breast cancer. Breast Cancer Res. Treat. 2011;129:819–827. doi: 10.1007/s10549-011-1679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Massarweh, S., Tham, Y.L., Huang, J., Sexton, K., Weiss, H., Tsimelzon, A., Beyer, A., Rimawi, M., Cai, W.Y., Hilsenbeck, S., et al. (2011). A phase II neoadjuvant trial of anastrozole, fulvestrant, and gefitinib in patients with newly diagnosed estrogen receptor positive breast cancer. Breast Cancer Res. Treat. 129, 819-827. [DOI] [PMC free article] [PubMed]
- Miller L.D., Smeds J., George J., Vega V.B., Vergara L., Ploner A., Pawitan Y., Hall P., Klaar S., Liu E.T., Bergh J. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Miller, L.D., Smeds, J., George, J., Vega, V.B., Vergara, L., Ploner, A., Pawitan, Y., Hall, P., Klaar, S., Liu, E.T., and Bergh, J. (2005). An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. USA 102, 13550-13555. [DOI] [PMC free article] [PubMed]
- Miller W.R., Larionov A.A., Renshaw L., Anderson T.J., White S., Murray J., Murray E., Hampton G., Walker J.R., Ho S. Changes in breast cancer transcriptional profiles after treatment with the aromatase inhibitor, letrozole. Pharmacogenet. Genomics. 2007;17:813–826. doi: 10.1097/FPC.0b013e32820b853a. [DOI] [PubMed] [Google Scholar]; Miller, W.R., Larionov, A.A., Renshaw, L., Anderson, T.J., White, S., Murray, J., Murray, E., Hampton, G., Walker, J.R., Ho, S., et al. (2007). Changes in breast cancer transcriptional profiles after treatment with the aromatase inhibitor, letrozole. Pharmacogenet. Genomics 17, 813-826. [DOI] [PubMed]
- Minn A.J., Gupta G.P., Siegel P.M., Bos P.D., Shu W., Giri D.D., Viale A., Olshen A.B., Gerald W.L., Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]; Minn, A.J., Gupta, G.P., Siegel, P.M., Bos, P.D., Shu, W., Giri, D.D., Viale, A., Olshen, A.B., Gerald, W.L., and Massague, J. (2005). Genes that mediate breast cancer metastasis to lung. Nature 436, 518-524. [DOI] [PMC free article] [PubMed]
- Morandi A., Indraccolo S. Linking metabolic reprogramming to therapy resistance in cancer. Biochim. Biophys. Acta Rev. Cancer. 2017;1868:1–6. doi: 10.1016/j.bbcan.2016.12.004. [DOI] [PubMed] [Google Scholar]; Morandi, A., and Indraccolo, S.. (2017). Linking metabolic reprogramming to therapy resistance in cancer. Biochim. Biophys. Acta Rev. Cancer 1868, 1-6. [DOI] [PubMed]
- Morandi A., Martin L.A., Gao Q., Pancholi S., Mackay A., Robertson D., Zvelebil M., Dowsett M., Plaza-Menacho I., Isacke C.M. GDNF-RET signaling in ER-positive breast cancers is a key determinant of response and resistance to aromatase inhibitors. Cancer Res. 2013;73:3783–3795. doi: 10.1158/0008-5472.CAN-12-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]; Morandi, A., Martin, L.A., Gao, Q., Pancholi, S., Mackay, A., Robertson, D., Zvelebil, M., Dowsett, M., Plaza-Menacho, I., and Isacke, C.M. (2013). GDNF-RET signaling in ER-positive breast cancers is a key determinant of response and resistance to aromatase inhibitors. Cancer Res. 73, 3783-3795. [DOI] [PMC free article] [PubMed]
- Murugaesu N., Iravani M., van Weverwijk A., Ivetic A., Johnson D.A., Antonopoulos A., Fearns A., Jamal-Hanjani M., Sims D., Fenwick K. An in vivo functional screen identifies ST6GalNAc2 sialyltransferase as a breast cancer metastasis suppressor. Cancer Discov. 2014;4:304–317. doi: 10.1158/2159-8290.CD-13-0287. [DOI] [PubMed] [Google Scholar]; Murugaesu, N., Iravani, M., van Weverwijk, A., Ivetic, A., Johnson, D.A., Antonopoulos, A., Fearns, A., Jamal-Hanjani, M., Sims, D., Fenwick, K., et al. (2014). An in vivo functional screen identifies ST6GalNAc2 sialyltransferase as a breast cancer metastasis suppressor. Cancer Discov. 4, 304-317. [DOI] [PubMed]
- Nagalla S., Chou J.W., Willingham M.C., Ruiz J., Vaughn J.P., Dubey P., Lash T.L., Hamilton-Dutoit S.J., Bergh J., Sotiriou C. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 2013;14:R34. doi: 10.1186/gb-2013-14-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nagalla, S., Chou, J.W., Willingham, M.C., Ruiz, J., Vaughn, J.P., Dubey, P., Lash, T.L., Hamilton-Dutoit, S.J., Bergh, J., Sotiriou, C., et al. (2013). Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 14, R34. [DOI] [PMC free article] [PubMed]
- Nemkov T., D’Alessandro A., Hansen K.C. Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole orbitrap mass spectrometry. Amino Acids. 2015;47:2345–2357. doi: 10.1007/s00726-015-2019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nemkov, T., D’Alessandro, A., and Hansen, K.C. (2015). Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole orbitrap mass spectrometry. Amino Acids 47, 2345-2357. [DOI] [PMC free article] [PubMed]
- Nguyen V.T., Barozzi I., Faronato M., Lombardo Y., Steel J.H., Patel N., Darbre P., Castellano L., Győrffy B., Woodley L. Differential epigenetic reprogramming in response to specific endocrine therapies promotes cholesterol biosynthesis and cellular invasion. Nat. Commun. 2015;6:10044. doi: 10.1038/ncomms10044. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nguyen, V.T., Barozzi, I., Faronato, M., Lombardo, Y., Steel, J.H., Patel, N., Darbre, P., Castellano, L., Győrffy, B., Woodley, L., et al. (2015). Differential epigenetic reprogramming in response to specific endocrine therapies promotes cholesterol biosynthesis and cellular invasion. Nat. Commun. 6, 10044. [DOI] [PMC free article] [PubMed]
- Pereira B., Chin S.F., Rueda O.M., Vollan H.K., Provenzano E., Bardwell H.A., Pugh M., Jones L., Russell R., Sammut S.J. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pereira, B., Chin, S.F., Rueda, O.M., Vollan, H.K., Provenzano, E., Bardwell, H.A., Pugh, M., Jones, L., Russell, R., Sammut, S.J., et al. (2016). The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479. [DOI] [PMC free article] [PubMed]
- Plaza-Menacho I., Morandi A., Robertson D., Pancholi S., Drury S., Dowsett M., Martin L.A., Isacke C.M. Targeting the receptor tyrosine kinase RET sensitizes breast cancer cells to tamoxifen treatment and reveals a role for RET in endocrine resistance. Oncogene. 2010;29:4648–4657. doi: 10.1038/onc.2010.209. [DOI] [PubMed] [Google Scholar]; Plaza-Menacho, I., Morandi, A., Robertson, D., Pancholi, S., Drury, S., Dowsett, M., Martin, L.A., and Isacke, C.M. (2010). Targeting the receptor tyrosine kinase RET sensitizes breast cancer cells to tamoxifen treatment and reveals a role for RET in endocrine resistance. Oncogene 29, 4648-4657. [DOI] [PubMed]
- Rème T., Hose D., Theillet C., Klein B. Modeling risk stratification in human cancer. Bioinformatics. 2013;29:1149–1157. doi: 10.1093/bioinformatics/btt124. [DOI] [PubMed] [Google Scholar]; Reme, T., Hose, D., Theillet, C., and Klein, B. (2013). Modeling risk stratification in human cancer. Bioinformatics 29, 1149-1157. [DOI] [PubMed]
- Rodriguez-Bravo V., Carceles-Cordon M., Hoshida Y., Cordon-Cardo C., Galsky M.D., Domingo-Domenech J. The role of GATA2 in lethal prostate cancer aggressiveness. Nat. Rev. Urol. 2017;14:38–48. doi: 10.1038/nrurol.2016.225. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rodriguez-Bravo, V., Carceles-Cordon, M., Hoshida, Y., Cordon-Cardo, C., Galsky, M.D., and Domingo-Domenech, J. (2017). The role of GATA2 in lethal prostate cancer aggressiveness. Nat. Rev. Urol. 14, 38-48. [DOI] [PMC free article] [PubMed]
- Sabatier R., Finetti P., Cervera N., Lambaudie E., Esterni B., Mamessier E., Tallet A., Chabannon C., Extra J.M., Jacquemier J. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res. Treat. 2011;126:407–420. doi: 10.1007/s10549-010-0897-9. [DOI] [PubMed] [Google Scholar]; Sabatier, R., Finetti, P., Cervera, N., Lambaudie, E., Esterni, B., Mamessier, E., Tallet, A., Chabannon, C., Extra, J.M., Jacquemier, J., et al. (2011). A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res. Treat. 126, 407-420. [DOI] [PubMed]
- Saito Y., Li L., Coyaud E., Luna A., Sander C., Raught B., Asara J.M., Brown M., Muthuswamy S.K. LLGL2 rescues nutrient stress by promoting leucine uptake in ER+ breast cancer. Nature. 2019;569:275–279. doi: 10.1038/s41586-019-1126-2. [DOI] [PubMed] [Google Scholar]; Saito, Y., Li, L., Coyaud, E., Luna, A., Sander, C., Raught, B., Asara, J.M., Brown, M., and Muthuswamy, S.K. (2019). LLGL2 rescues nutrient stress by promoting leucine uptake in ER+ breast cancer. Nature 569, 275-279. [DOI] [PubMed]
- Sanchez C.G., Ma C.X., Crowder R.J., Guintoli T., Phommaly C., Gao F., Lin L., Ellis M.J. Preclinical modeling of combined phosphatidylinositol-3-kinase inhibition with endocrine therapy for estrogen receptor-positive breast cancer. Breast Cancer Res. 2011;13:R21. doi: 10.1186/bcr2833. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sanchez, C.G., Ma, C.X., Crowder, R.J., Guintoli, T., Phommaly, C., Gao, F., Lin, L., and Ellis, M.J. (2011). Preclinical modeling of combined phosphatidylinositol-3-kinase inhibition with endocrine therapy for estrogen receptor-positive breast cancer. Breast Cancer Res. 13, R21. [DOI] [PMC free article] [PubMed]
- Shajahan-Haq A.N., Cook K.L., Schwartz-Roberts J.L., Eltayeb A.E., Demas D.M., Warri A.M., Facey C.O., Hilakivi-Clarke L.A., Clarke R. MYC regulates the unfolded protein response and glucose and glutamine uptake in endocrine resistant breast cancer. Mol. Cancer. 2014;13:239. doi: 10.1186/1476-4598-13-239. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shajahan-Haq, A.N., Cook, K.L., Schwartz-Roberts, J.L., Eltayeb, A.E., Demas, D.M., Warri, A.M., Facey, C.O., Hilakivi-Clarke, L.A., and Clarke, R. (2014). MYC regulates the unfolded protein response and glucose and glutamine uptake in endocrine resistant breast cancer. Mol. Cancer 13, 239. [DOI] [PMC free article] [PubMed]
- Shi C.S., Kehrl J.H. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shi, C.S., and Kehrl, J.H. (2010). TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 3, ra42. [DOI] [PMC free article] [PubMed]
- Shinmura K., Kato H., Igarashi H., Inoue Y., Nakamura S., Du C., Kurachi K., Nakamura T., Ogawa H., Tanahashi M. CD44-SLC1A2 fusion transcripts in primary colorectal cancer. Pathol. Oncol. Res. 2015;21:759–764. doi: 10.1007/s12253-014-9887-2. [DOI] [PubMed] [Google Scholar]; Shinmura, K., Kato, H., Igarashi, H., Inoue, Y., Nakamura, S., Du, C., Kurachi, K., Nakamura, T., Ogawa, H., Tanahashi, M., et al. (2015). CD44-SLC1A2 fusion transcripts in primary colorectal cancer. Pathol. Oncol. Res. 21, 759-764. [DOI] [PubMed]
- Simões B.M., O’Brien C.S., Eyre R., Silva A., Yu L., Sarmiento-Castro A., Alférez D.G., Spence K., Santiago-Gόmez A., Chemi F. Anti-estrogen Resistance in Human Breast Tumors Is Driven by JAG1-NOTCH4-Dependent Cancer Stem Cell Activity. Cell Rep. 2015;12:1968–1977. doi: 10.1016/j.celrep.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]; Simoes, B.M., O’Brien, C.S., Eyre, R., Silva, A., Yu, L., Sarmiento-Castro, A., Alferez, D.G., Spence, K., Santiago-Gόmez, A., Chemi, F., et al. (2015). Anti-estrogen Resistance in Human Breast Tumors Is Driven by JAG1-NOTCH4-Dependent Cancer Stem Cell Activity. Cell Rep. 12, 1968-1977. [DOI] [PMC free article] [PubMed]
- Sotiriou C., Wirapati P., Loi S., Harris A., Fox S., Smeds J., Nordgren H., Farmer P., Praz V., Haibe-Kains B. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]; Sotiriou, C., Wirapati, P., Loi, S., Harris, A., Fox, S., Smeds, J., Nordgren, H., Farmer, P., Praz, V., Haibe-Kains, B., et al. (2006). Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 98, 262-272. [DOI] [PubMed]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Subramanian, A., Tamayo, P., Mootha, V.K., Mukherjee, S., Ebert, B.L., Gillette, M.A., Paulovich, A., Pomeroy, S.L., Golub, T.R., Lander, E.S., and Mesirov, J.P. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550. [DOI] [PMC free article] [PubMed]
- Sullivan L.B., Gui D.Y., Hosios A.M., Bush L.N., Freinkman E., Vander Heiden M.G. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sullivan, L.B., Gui, D.Y., Hosios, A.M., Bush, L.N., Freinkman, E., and Vander Heiden, M.G. (2015). Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 162, 552-563. [DOI] [PMC free article] [PubMed]
- Sullivan L.B., Luengo A., Danai L.V., Bush L.N., Diehl F.F., Hosios A.M., Lau A.N., Elmiligy S., Malstrom S., Lewis C.A., Vander Heiden M.G. Aspartate is an endogenous metabolic limitation for tumour growth. Nat. Cell Biol. 2018;20:782–788. doi: 10.1038/s41556-018-0125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sullivan, L.B., Luengo, A., Danai, L.V., Bush, L.N., Diehl, F.F., Hosios, A.M., Lau, A.N., Elmiligy, S., Malstrom, S., Lewis, C.A., and Vander Heiden, M.G. (2018). Aspartate is an endogenous metabolic limitation for tumour growth. Nat. Cell Biol. 20, 782-788. [DOI] [PMC free article] [PubMed]
- Symmans W.F., Hatzis C., Sotiriou C., Andre F., Peintinger F., Regitnig P., Daxenbichler G., Desmedt C., Domont J., Marth C. Genomic index of sensitivity to endocrine therapy for breast cancer. J. Clin. Oncol. 2010;28:4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]; Symmans, W.F., Hatzis, C., Sotiriou, C., Andre, F., Peintinger, F., Regitnig, P., Daxenbichler, G., Desmedt, C., Domont, J., Marth, C., et al. (2010). Genomic index of sensitivity to endocrine therapy for breast cancer. J. Clin. Oncol. 28, 4111-4119. [DOI] [PMC free article] [PubMed]
- Tao J., Deng N.T., Ramnarayanan K., Huang B., Oh H.K., Leong S.H., Lim S.S., Tan I.B., Ooi C.H., Wu J. CD44-SLC1A2 gene fusions in gastric cancer. Sci. Transl. Med. 2011;3:77ra30. doi: 10.1126/scitranslmed.3001423. [DOI] [PubMed] [Google Scholar]; Tao, J., Deng, N.T., Ramnarayanan, K., Huang, B., Oh, H.K., Leong, S.H., Lim, S.S., Tan, I.B., Ooi, C.H., Wu, J., et al. (2011). CD44-SLC1A2 gene fusions in gastric cancer. Sci. Transl. Med. 3, 77ra30. [DOI] [PubMed]
- Vidal S.J., Rodriguez-Bravo V., Quinn S.A., Rodriguez-Barrueco R., Lujambio A., Williams E., Sun X., de la Iglesia-Vicente J., Lee A., Readhead B. A targetable GATA2-IGF2 axis confers aggressiveness in lethal prostate cancer. Cancer Cell. 2015;27:223–239. doi: 10.1016/j.ccell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vidal, S.J., Rodriguez-Bravo, V., Quinn, S.A., Rodriguez-Barrueco, R., Lujambio, A., Williams, E., Sun, X., de la Iglesia-Vicente, J., Lee, A., Readhead, B., et al. (2015). A targetable GATA2-IGF2 axis confers aggressiveness in lethal prostate cancer. Cancer Cell 27, 223-239. [DOI] [PMC free article] [PubMed]
- Wang Y., Klijn J.G., Zhang Y., Sieuwerts A.M., Look M.P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M.E., Yu J. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]; Wang, Y., Klijn, J.G., Zhang, Y., Sieuwerts, A.M., Look, M.P., Yang, F., Talantov, D., Timmermans, M., Meijer-van Gelder, M.E., Yu, J., et al. (2005). Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671-679. [DOI] [PubMed]
- Wang Y., He X., Ngeow J., Eng C. GATA2 negatively regulates PTEN by preventing nuclear translocation of androgen receptor and by androgen-independent suppression of PTEN transcription in breast cancer. Hum. Mol. Genet. 2012;21:569–576. doi: 10.1093/hmg/ddr491. [DOI] [PubMed] [Google Scholar]; Wang, Y., He, X., Ngeow, J., and Eng, C. (2012). GATA2 negatively regulates PTEN by preventing nuclear translocation of androgen receptor and by androgen-independent suppression of PTEN transcription in breast cancer. Hum. Mol. Genet. 21, 569-576. [DOI] [PubMed]
- Wang P., Zhang J., Zhang L., Zhu Z., Fan J., Chen L., Zhuang L., Luo J., Chen H., Liu L. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133–1143.e12. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]; Wang, P., Zhang, J., Zhang, L., Zhu, Z., Fan, J., Chen, L., Zhuang, L., Luo, J., Chen, H., Liu, L., et al. (2013). MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology 145, 1133-1143.e12. [DOI] [PubMed]
- Yang J.A., Stires H., Belden W.J., Roepke T.A. The Arcuate Estrogen-Regulated Transcriptome: Estrogen Response Element-Dependent and -Independent Signaling of ERα in Female Mice. Endocrinology. 2017;158:612–626. doi: 10.1210/en.2016-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang, J.A., Stires, H., Belden, W.J., and Roepke, T.A. (2017). The Arcuate Estrogen-Regulated Transcriptome: Estrogen Response Element-Dependent and -Independent Signaling of ERα in Female Mice. Endocrinology 158, 612-626. [DOI] [PMC free article] [PubMed]
- Yoshii S.R., Mizushima N. Monitoring and Measuring Autophagy. Int. J. Mol. Sci. 2017;18:E1865. doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yoshii, S.R., and Mizushima, N. (2017). Monitoring and Measuring Autophagy. Int. J. Mol. Sci. 18, E1865. [DOI] [PMC free article] [PubMed]
- Zaman M.S., Thamminana S., Shahryari V., Chiyomaru T., Deng G., Saini S., Majid S., Fukuhara S., Chang I., Arora S. Inhibition of PTEN gene expression by oncogenic miR-23b-3p in renal cancer. PLoS One. 2012;7:e50203. doi: 10.1371/journal.pone.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zaman, M.S., Thamminana, S., Shahryari, V., Chiyomaru, T., Deng, G., Saini, S., Majid, S., Fukuhara, S., Chang, I., Arora, S., et al. (2012). Inhibition of PTEN gene expression by oncogenic miR-23b-3p in renal cancer. PLoS One 7, e50203. [DOI] [PMC free article] [PubMed]
- Zhang Y., Sieuwerts A.M., McGreevy M., Casey G., Cufer T., Paradiso A., Harbeck N., Span P.N., Hicks D.G., Crowe J. The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res. Treat. 2009;116:303–309. doi: 10.1007/s10549-008-0183-2. [DOI] [PubMed] [Google Scholar]; Zhang, Y., Sieuwerts, A.M., McGreevy, M., Casey, G., Cufer, T., Paradiso, A., Harbeck, N., Span, P.N., Hicks, D.G., Crowe, J., et al. (2009). The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res. Treat. 116, 303-309. [DOI] [PubMed]
- Zhang H., Hao Y., Yang J., Zhou Y., Li J., Yin S., Sun C., Ma M., Huang Y., Xi J.J. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat. Commun. 2011;2:554. doi: 10.1038/ncomms1555. [DOI] [PubMed] [Google Scholar]; Zhang, H., Hao, Y., Yang, J., Zhou, Y., Li, J., Yin, S., Sun, C., Ma, M., Huang, Y., and Xi, J.J. (2011). Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat. Commun. 2, 554. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
mRNA and miRNA data derived from microarray analysis are deposited in NCBI Gene Expression Omnibus (GEO) database: accession number GSE120931. Feature Extraction 10.7.3.1 software (Agilent Technologies) was used to obtain the microarray raw-data and data.
The R script (mM_Net.R) for the interactor network analysis displayed in Figure 1C is available and can be downloaded (https://github.com/matteoramazzotti/TransTools).