Abstract
The family of MOBs (monopolar spindle-one-binder proteins) is highly conserved in the eukaryotic kingdom. MOBs represent globular scaffold proteins without any known enzymatic activities. They can act as signal transducers in essential intracellular pathways. MOBs have diverse cancer-associated cellular functions through regulatory interactions with members of the NDR/LATS kinase family. By forming additional complexes with serine/threonine protein kinases of the germinal centre kinase families, other enzymes and scaffolding factors, MOBs appear to be linked to an even broader disease spectrum. Here, we review our current understanding of this emerging protein family, with emphases on post-translational modifications, protein-protein interactions, and cellular processes that are possibly linked to cancer and other diseases. In particular, we summarise the roles of MOBs as core components of the Hippo tissue growth and regeneration pathway.
Keywords: Mps one binder, Hippo pathway, protein kinase, signal transduction, phosphorylation, protein-protein interactions, structure biology, STK38, NDR, LATS, MST, STRIPAK
1. Introduction
The family of MOBs (monopolar spindle-one-binder proteins) is highly conserved in eukaryotes [1,2,3,4]. To our knowledge, at least two different MOBs have been found in every eukaryote analysed so far. For example, in unicellular organisms such as yeast the MOB proteins Mob1p and Mob2p are expressed by two independent genes [5]. In multicellular organisms such as flies, at least four different MOBs, termed dMOBs (Drosophila MOB proteins), have been reported [4]. In humans, as many as seven different MOB proteins, termed hMOBs (human MOB proteins), are encoded by different gene loci. Since each hMOB has been given several names over the years, we have simplified the nomenclature of hMOBs as follows [1]: hMOB1A (UniProtKB: Q9H8S9; also termed MOBKL1B, MOBK1B, Mob1a, MOB1α, Mob4b, MATS1 and C2orf6), hMOB1B (UniProtKB: Q7L9L4; also termed MOBKL1A, Mob1b, Mob4a and MATS2), hMOB2 (UniProtKB: Q70IA6; also termed MOBKL2, Mob2 and HCCA2), hMOB3A (UniProtKB: Q96BX8; also termed MOBKL2A, Mob3A, MOB-LAK, MOB2A, Mob1C), hMOB3B (UniProtKB: Q86TA1; MOBKL2B, Mob3B, MOB2B, Mob1D and C9orf35), hMOB3C (UniProtKB: Q70IA8; also termed MOBKL2C, Mob3C, MOB2C and Mob1E) and hMOB4 (UniProtKB: Q9Y3A3; also termed MOBKL3, Class II mMOB1, MOB3, Phocein/PHOCN, PREI3, 2C4D, and CGI-95). Given the very high identity between hMOB1A and hMOB1B (Figure 1), we refer to hMOB1A/B also as hMOB1 in this review.
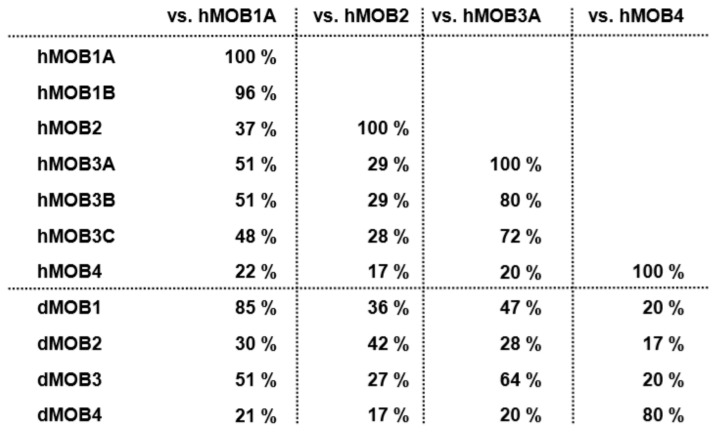
Figure 1.
Primary sequence identities of the fly and human MOBs. The identities between primary sequences are displayed. Identities were defined using EMBOSS needle for pairwise alignments (https://www.ebi.ac.uk/Tools/psa/emboss_needle/). The UniProtKB nomenclature for hMOBs can be found in the introduction section of the main text. The UniProtKB names for dMOBs are as follows [4]: dMOB1 (aka Mats and CG13852)–Q95RA8, dMOB2 (aka CG11711)–Q8IQG1, dMOB3 (aka CG4946)–Q9VL13, and dMOB4 (aka CG3403)–Q7K0E3.
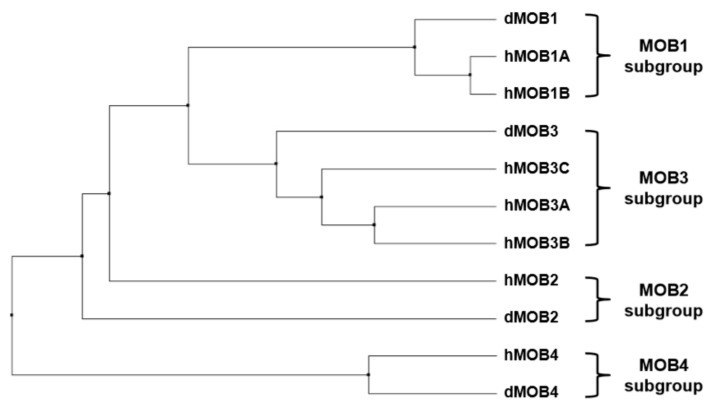
In comparing the sequence identities of MOB proteins expressed in fly and human cells (Figure 1), it becomes apparent that hMOB1 is highly conserved in flies. dMOB1 is 85% identical to hMOB1 and functionally conserved, since exogenous expression of hMOB1A rescues the phenotypes associated with dMOB1 loss-of-function [6,7]. Among other MOBs, hMOB1 is the most closely related to hMOB3A/B/C and dMOB3 (Figure 1). hMOB2 is not as well conserved as hMOB1, since hMOB2 aligns similarly with dMOB1 and dMOB2. hMOB3A is very similar to hMOB3B and hMOB3C, being also 64% identical to dMOB3. hMOB4 is 80% identical to dMOB4, and hMOB4 and dMOB4 do not show any significant identity overlaps with other MOBs (Figure 1). A phylogenetic tree analysis of the dMOBs and hMOBs showed that hMOB1A, hMOB1B and dMOB1 cluster together into one subgroup (Figure 2). Likewise, hMOB2 and dMOB2, as well as hMOB3A, hMOB3B, hMOB3C and dMOB3 form separate subgroups, respectively. It is noteworthy that hMOB4 and dMOB4 also form a subgroup of their own, representing the most distant subgroup of MOBs (Figure 2). However, in spite of these striking sequence similarities, it is currently unknown whether loss-of-function of dMOB2, dMOB3 or dMOB4 can be functionally compensated by the expression of the corresponding hMOB family member. Nevertheless, research covering the past two decades has uncovered important aspects of MOBs that seem to be universally valid for all members of the MOB family. In summary, it was found that MOBs mainly function as intracellular co-regulatory proteins. In particular MOBs can directly bind to protein kinases, and thereby influence the activities of their binding partners. As such, before discussing MOBs in flies and mammals, we will provide a brief introductory background on MOBs studied in unicellular organisms such as yeast.
Figure 2.
Phylogenetic relationships within the fly and human MOB protein family. The phylogenetic tree was defined using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) together with the Jalview 2.10.5 software using phylogenetic calculation based on the neighbour-joining method. The UniProtKB nomenclature for the analysed proteins is defined in the legend of Figure 1.
More than two decades ago Mob1p, the first MOB (monopolar spindle-one-binder) protein, was described in budding yeast [1,5,8]. It turned out that Mob1p is a central component of the mitotic exit network (MEN) and the septation initiation network (SIN) of budding and fission yeast, respectively (summarised in refs. [8,9,10,11,12]). Mechanistically, budding yeast Mob1p in complex with the Dbf2p protein kinase (a budding yeast counterpart of mammalian NDR/LATS kinases [13]) regulates the release of phosphatase Cdc14p to promote exit from mitosis through a series of dephosphorylation events. Similarly, fission yeast Mob1p bound to Sid2p (a fission yeast counterpart of mammalian NDR/LATS kinases) controls the Clp1p phosphatase (the equivalent of Cdc14p) to support the SIN. Mob2p, another member of the MOB family [1], was uncovered together with Mob1p in budding yeast [5]. Subsequent research revealed that Mob2p coordinates morphogenesis networks in budding and fission yeast [1,13], as well as fungal species [14]. In budding yeast, Mob2p in complex with Cbk1p (the second NDR/LATS kinase expressed in budding yeast [13]) controls the RAM (regulation of Ace2p activity and cellular morphogenesis) signalling network. In fission yeast, Mob2p associated with Orb6p (the second NDR/LATS kinase in fission yeast) coordinates a similar morphogenesis network.
In summary, yeast cells express two different MOB proteins, Mob1p and Mob2p. While Mob1p controls mitotic exit, Mob2p is a regulator of cell morphogenesis and polarized growth. Mob1p and Mob2p function in complex with different members of the NDR/LATS kinase family. Depending on the yeast species Mob1p binds to Dbf2p or Sid2p whereas Mob2p associates with Cbk1p or Orb6p, respectively. Consequently, in yeast Mob1p and Mob2p act as part of independent and non-interchangeable complexes containing different NDR/LATS kinases in order to regulate diverse cellular processes (please consult refs. [1,13] for a more detailed discussion of these points).
2. An Overview of MOBs in Drosophila melanogaster
Since 2005, MOBs have been studied in multicellular organisms. In contrast to yeast, fly cells encode four different MOB proteins [4]. Lai and colleagues found that dMOB1 (also termed Mats for MOB as tumour suppressor) is an essential regulator of cell proliferation and cell death in fruit flies [7]. Follow up studies established dMOB1 as a core component of the Hippo tissue growth control pathway (for more details see subchapter 4 of this review). Briefly, as a core component of the Hippo pathway, dMOB1 acts as a co-activator of the Warts protein kinase [15,16,17], one of two NDR/LATS kinases in D. melanogaster [13]. Intriguingly, dMOB1 also genetically interacts with Tricornered (Trc) [4], the second NDR/LATS kinase in flies [13]. Thus, dMOB1 does not bind specifically to a single NDR/LATS kinase, as observed for yeast Mob1p [1]. In addition, dMOB1 can form a complex with the Hippo (Hpo) protein kinase [18], with Hpo being able to phosphorylate dMOB1 [18] as well as Warts and Trc [19]. The importance of these protein-protein interactions and phosphorylation events is discussed in subchapters 4, 5 and 6. Noteworthy, dMOB1 can further play a role in mitosis (summarised in refs. [1,20,21]).
Unlike those of dMOB1, the biological functions of dMOB2, dMOB3 and dMOB4 are yet to be completely understood. Based on the work by the Adler laboratory, it seems that dMOB2 can play a role in wing hair morphogenesis, possibly by forming a complex with Trc [4]. However, the precise mechanism of action remains unknown. Moreover, dMOB2 supports the development of photoreceptor cells [22] and the growth of larval neuromuscular junctions in flies [23]. It is possible that these neurological roles are linked to the association of dMOB2 with Tricornered [4]. So far, only Trc has been established as a bona fide binding partner of dMOB2.
Interestingly, like dMOB1 and dMOB2, dMOB3 can also genetically interact with Trc [4], but any function or direct binding partners of dMOB3 have remained elusive. Similar to dMOB2, dMOB4 has been linked to a neurological function. More precisely, dMOB4 was found to play a role as a regulator of neurite branching [24]. Furthermore, dMOB4 depletion in fly cells results in defective focusing of kinetochore fibres in mitosis [25]. Possibly some of these roles of dMOB4 could be linked to dMOB4 being part of the Striatin-interacting phosphatase and kinase (STRIPAK) complex (see subchapters 6 and 7 for more details). Yet, these possible connections have yet to be experimentally explored.
In summary, based on current evidence it is tempting to conclude that, unlike in yeast, individual dMOBs can interact (at least genetically) with Warts and Trc, the two NDR/LATS kinases expressed in fly cells. Conversely, individual NDR/LATS kinases can interact with different dMOBs. Therefore, it appears that in multicellular organisms such as flies, the binding of MOBs is not restricted to a unique member of the NDR/LATS kinase family.
3. An Overview of MOBs in Human Cells
hMOB1A and hMOB1B are also known as hMOB1, since hMOB1B is 96% identical to hMOB1A (see Figure 1 and refs. [1,3,26]). hMOB1A and hMOB1B are mainly cytoplasmic proteins [27,28]. However, upon targeting to the plasma membrane of mammalian cells hMOB1 can trigger a binding-dependent activation of the human NDR/LATS kinases [27,29]. hMOB1 can form complexes with NDR1 (aka STK38), NDR2 (aka STK38L), LATS1 and LATS2, all four human NDR/LATS kinases [1,13]. These interactions are mediated through one unique and highly conserved domain in NDR/LATS kinases [29]. Like dMOB1 [18], hMOB1 can also bind to the MST1/2 kinases [30], the human counterparts of Hpo [31,32,33,34]. The significance and regulation of these interactions is discussed in much detail in subchapters 5 and 6. hMOB1 can further associate with other proteins, although the importance of these additional interactions is yet to be defined (see subchapter 6). In cases whereby a new hMOB1 binding partner has been validated by conventional interaction assays, we have included the discussion of this novel aspect in the appropriate subchapters. Nevertheless, the protein-protein interactions of hMOB1 with NDR/LATS kinases is the best understood. For a summary of the cellular roles of hMOB1 please consult subchapter 7 and ref. [1].
In contrast to hMOB1, a significant portion of hMOB2 is nuclear [27,28]. Intriguingly, hMOB2 forms a complex with NDR1/2, while hMOB2 neither associates with LATS1/2 [1,35] nor MST1/2 [36]. hMOB2 can compete with hMOB1 for NDR1/2 binding, since hMOB2 interacts with the same domain on NDR1/2 as hMOB1 [27,28]. However, the binding modes of hMOB1 and hMOB2 to NDR1/2 seem to differ [1,35]. hMOB2 can also interact with RAD50, a core component of the MRE11-RAD50-NBS1 (MRN) DNA damage sensor complex, thereby playing a role in cell cycle-related DNA damage signalling [37]. Whether the hMOB2/RAD50 complex is linked to NDR1/2 signalling is yet to be defined [38]. For a summary of the cellular roles of hMOB2, see subchapter 7 and refs. [1,38].
Based on their similarities hMOB3A, hMOB3B and hMOB3C are sometimes also referred to as hMOB3 [39]. Like hMOB1, hMOB3s are mainly cytoplasmic proteins [28,40]. The best-understood binding partner of hMOB3s is the MST1 kinase [39]. Given the high sequence similarities between hMOB3s and hMOB1 (Figure 1), it was rather surprising that hMOB3s do not bind to any NDR/LATS kinase [35]. Nonetheless, large-scale interactome studies suggest that hMOB3s may have other binding partners in addition to MST1/2 (see ref. [1] and this review). Our current understanding of the cellular roles of hMOB3s is summarised in subchapter 7.
hMOB4 (aka Phocein) is best known as part of the multi-component STRIPAK (Striatin-interacting phosphatase and kinase) complex [41,42,43,44]. hMOB4 can bind to two distinct regions of Striatin [45]. Besides the Striatin-associated hMOB4, the core STRIPAK complex contains serine/threonine protein phosphatase 2 (PP2A) subunits such as PP2Ac and PP2A-A, as well as the Striatins as PP2A regulatory subunits. STRIPAK also encompasses CCM3 (cerebral cavernous malformation 3; also known as PDCD10) as well as the MST3 (aka STK24), MST4 (aka MASK or STK26) and STK25 (aka YSK1 or SOK1) protein kinases, the three members of the GCKIII (germinal centre kinase III) subfamily of Sterile 20 kinases [46]. In addition, the core complex contains STRIP1/2 (aka FAM40A/B). Two mutually exclusive STRIPAK complexes have been defined based on additional components such as CTTNBP2-like adaptors, SLMAP (sarcolemmal membrane-associated protein) or others [47]. However, although the STRIPAK complex has been linked to the Hippo pathway (see subchapter 4), the precise role(s) of hMOB4 in the STRIPAK complex has remained of a more speculative nature. In this regard, hMOB4 has been reported to form a complex with MST4 in a phosphorylation-dependent manner [48], an aspect that is discussed briefly in subchapter 6. The cellular functions of hMOB4 are discussed in subchapter 7.
4. An Overview of MOBs and the Hippo Pathway
The Hippo tissue growth control and regeneration pathway represents a key signalling cascade in flies and mammals [16,49,50,51]. By co-ordinating death, growth, proliferation, and differentiation on the cellular level the Hippo pathway controls organ growth, tissue homeostasis and regeneration. Hence, the deregulation of Hippo signalling has been linked to serious human diseases, such as cancers [50,52,53]. At the molecular level, a conserved Hippo core cassette is fundamental for the regulation of the co-transcriptional regulators YAP and TAZ, two main effectors of the Hippo pathway [51,54]. To date, the MST1/2 (aka STK4/3; Hpo in D. melanogaster) and LATS1/2 (aka Warts in flies) protein kinases are the best understood members of the Hippo core cassette. In this regard, MOB1 acts as central signal transducers in the Hippo core cassette. Upon activation of the Hippo pathway MST1/2 phosphorylate LATS1/2 and MOB1, thereby supporting the formation of an active MOB1/LATS complex (see subchapter 6 for molecular details), which is essential for development and tissue growth control [6]. Activated LATS1/2 in complex with MOB1 then phosphorylate YAP/TAZ on different Ser/Thr residues, thereby inhibiting nuclear activities of YAP/TAZ through cytoplasmic retention and/or degradation of phosphorylated YAP/TAZ [51,54]. Markedly, the NDR1/2 protein kinases (aka STK38 and STK38L; the closest relatives of LATS1/2 [13]), can also phosphorylate and thereby constrain YAP1 to the cytoplasm [55,56]. Similar to LATS1/2, NDR1/2 can stably associate with MOB1 [1,6] and are direct effector substrates of MST1/2 [55,57]. As already mentioned, MST1/2 also phosphorylate MOB1 [30]. Hence, LATS1/2, NDR1/2 and MOB1 represent well-established substrates of MST1/2 [1,55,57,58]. Taken together, the core of the Hippo pathway comprises distinct kinases, such as MST1/2, LATS1/2 and NDR1/2, which act together with the fundamental signal adaptor MOB1.
By directly interacting with the MST1/2, LATS1/2 and NDR1/2 kinases (aka Hpo, Warts and Trc in flies) MOB1 functions as a key signal transducer in the Hippo pathway. However, although dMOB1 and MOB1 are essential in flies and mice [6,7,59], hMOB1 appears to be dispensable in human cells, at least in transformed HEK293A cells [60]. hMOB1A/B double-knockout (DKO) cells were viable in spite of drastically impaired LATS1/2-mediated YAP/TAZ phosphorylation upon serum deprivation [60]. Very similar to LATS1/2 DKO cells, YAP/TAZ remained nuclear and co-transcriptionally active in hMOB1A/B DKO cells. Even more puzzling, hMOB1 phosphorylation by MST1/2 does not seem to be required for LATS1/2 activation in HEK293A cells [60], although biochemical evidence strongly suggests that MST1/2-mediated phosphorylation of hMOB1 is needed for LATS1/2 binding (see subchapters 5 and 6). To make things even more complicated, hMOB1 can associate with other intracellular regulatory proteins besides binding to kinases of the Hippo core cassette (see subchapter 6). Therefore, it is quite possible that hMOB1 as a central Hippo component is acting in diverse cancer-associated cellular processes.
Until recently [37], NDR1/2 were the only reported binding partners of hMOB2 [1,35]. More specifically, it was documented that hMOB2 could compete with hMOB1 for NDR1/2 binding, with hMOB2 counteracting hMOB1 as a co-activator of NDR1/2 [1,35,61]. In this context, Zhang et al. studied Hippo signalling in a hMOB2 knockout hepatocellular carcinoma (HCC) cell line [62]. They found that MST1/2-mediated phosphorylation of NDR1/2 was induced, while MST1/2-mediated phosphorylations of hMOB1 and LATS1/2 were decreased upon hMOB2 knockout [62]. LATS1/2-mediated phosphorylation of YAP was also reduced in hMOB2 knockout cells, while MOB2 overexpression resulted in opposite effects [62]. Subsequently, the authors concluded that loss of hMOB2 can favour hMOB1 binding to NDR1/2, thereby reducing the pool of hMOB1 available for LATS1/2 binding. Conversely, hMOB2 overexpression may reduce hMOB1 binding to NDR1/2, thereby freeing hMOB1 to bind to LATS, resulting in the activation of Hippo signalling upstream of YAP [62]. However, this study [62] did not examine whether this molecular reprogramming of binding partners could be a possible consequence of cell cycle effects upon hMOB2 loss-of-function. Given that loss of hMOB2 can trigger a p53-dependent G1/S cell cycle arrest [37] and that NDR/LATS are associated with cell cycle progression [20,21], it will certainly be necessary to re-examine these HCC-based hMOB2 knockout cells to ensure the changes in NDR1/2, LATS1/2, and hMOB1 phosphorylation upon hMOB2 loss [62] are not merely reflecting indirect effects triggered by an underlying cell cycle arrest/delay.
Like hMOB1 and hMOB2, the group of hMOB3 signal transducers has been linked to the Hippo pathway. It was found that hMOB3s can directly bind to MST1, while they do not bind to any member of the NDR/LATS kinase family [35,39]. Considering that hMOB3s share high sequence similarities with hMOB1 (Figure 1) this was unexpected. Nevertheless, hMOB3s have at least one aspect in common with hMOB1, namely the direct binding to the MST1/2 kinases. More specifically, hMOB3s require two conserved positively charged residues to bind to MST1 [39], with both positively charged residues also being essential for the formation of a stable complex between hMOB1 and MST1/2 [6]. However, current evidence suggests that hMOB3 binding to MST1 is inhibitory and hMOB3s can act upstream of apoptotic MST1 signalling [39], while hMOB1 seems to mainly function downstream of MST1/2 [1,30,51]. Therefore, it is possible that hMOB3 competes with hMOB1 for MST1/2 binding, like hMOB2 is competing with hMOB1 for NDR1/2 binding, but these competitive interactions are likely to involve different interaction domains (see subchapter 6).
Intriguingly, hMOB4 has likewise been connected with the Hippo pathway. In fly and human cells, the hMOB4-containing supramolecular STRIPAK complexes [43,63] have been implicated in Hippo signalling on diverse regulatory levels [48,64,65,66,67,68,69]. However, to our knowledge, the specific contributions of hMOB4 to the crosstalk between Hippo and STRIPAK signalling remain undetermined. Nevertheless, the MST4 kinase, a relative of MST1/2 [46], was shown to form a complex with hMOB4, with the overall structure of the hMOB4/MST4 complex resembling the hMOB1/MST1 complex [48]. But one of the key residues (Lys105) that promotes MOB1 binding to MST1 [6] is not conserved in hMOB4 (Figure 3). Notably, the phospho-Ser/Thr binding motif of hMOB1 [70] seems to be present in hMOB4, while key residues such as Glu51 of MOB1 that mediate NDR/LATS binding [6,71,72] are not conserved in hMOB4 (Figure 3). Therefore, the binding mode of MST4 and other GCKIIIs to hMOB4 is yet to be completely understood [73].
Figure 3.
Comparison of the primary protein sequences of human and fly MOBs. Primary protein sequences were aligned using Clustal Omega as defined in the legend of Figure 2. The sites of Thr12, Tyr26 and Thr35 phosphorylations of hMOB1A are highlighted in light blue. Important interaction sites of hMOB1A that were verified by different experimental approaches are displayed in yellow. The key residues of the phospho-Ser/Thr (pSer/Thr) binding pocket of hMOB1A are shown in green. For more details on key residues of hMOB1 that are involved in kinase binding please consult refs. [3,6,70,71,72,78,92,95]. The UniProtKB nomenclature for the analysed proteins is defined in the legends of Figure 1 and Figure 2.
Different members of the Ste20-like kinase family can phosphorylate, and thereby activate, NDR/LATS kinases (summarised in ref. [57]). Briefly, MST1/2 phosphorylate NDR/LATS in human cells. Moreover, MAP4K (mitogen-activated protein kinase kinase kinase kinase)-type kinases can regulate NDR/LATS kinases through phosphorylation. In addition, MST3, MST4 and STK25, all members of the GCKIII subfamily of Ste20-like kinases [46], can phosphorylate NDR kinases in human and fly cells [57,74]. Strikingly, hMOB1 can bind to MST1/2, while hMOB4 is part of the GCKIII-containing STRIPAK complex. In this regard, the MAP4K-type kinase MINK1 was also identified as a component of the STRIPAK complex [75]. Consequently, it will be very interesting to properly map the binding patterns of hMOBs across the Ste20-like kinase family. Potentially, an array of different Ste20-like kinases is regulated by and/or regulates diverse hMOBs upstream of NDR/LATS signalling.
5. Post-Translational Modifications (PTMs) of MOBs
As already exemplified by the description of the Hippo pathway, signal transduction cascades transmit extra- and intra- cellular inputs through a broad array of protein kinases [76]. Thus, the phosphorylation status of signal transducers and effectors is finely orchestrated by interactions between kinases and phosphatases to achieve pathway regulation. Consequently, it is not surprising that MOBs can be regulated by phosphorylation events. In this subchapter, we summarise current knowledge on specific phosphorylation events of MOBs. Where appropriate, we also mention PTMs other than phosphorylations.
About two decades ago, yeast Mob1p was already described as a phospho-protein [5] that can be phosphorylated by Cdc15p (the yeast counterpart of the fly Hippo kinase) [77]. The phosphorylation of dMOB1 by Hippo was subsequently reported in fly cells [18]. However, Avruch and colleagues were the first to define the specific phosphorylation events that linked Hippo kinases such as MST1/2 with the regulation of hMOB1 [30,58]. More precisely, hMOB1 is specifically phosphorylated on Thr12 and Thr35 by MST1/2, thereby supporting complex formation between hMOB1 and NDR/LATS kinases [30]. Considering that the then available crystal structure of hMOB1A did not cover the N-terminal 32 residues [3], the precise molecular mechanism remained poorly understood for a long time [1]. However, Kim et al. recently deciphered the high-resolution crystal structure of full-length hMOB1 in its auto-inhibited form, and further reported the structural composition of a complex between hMOB1 and the N-terminal regulatory domain (NTR) of LATS1 [71]. These structures enabled the Hakoshima laboratory to define how hMOB1 is released from its auto-inhibitory conformation [71]. They found that hMOB1 phosphorylation on Thr12 and Thr35 accelerates the dissociation of the Switch helix, a long flexible positively-charged linker, from the LATS1-binding surface within hMOB1. Consequently, they uncovered a “pull-the-string” mechanism through which hMOB1 is rendered accessible for LATS1 binding [71], a model supported by an independent recent study [78]. This discovery was significant, since it helped to understand an important aspect of the molecular regulation of the core of the Hippo pathway [51,72].
Of note: in addition to the phosphorylation of Thr12 and Thr35 by MST1/2 [30], other PTMs of hMOB1, such as the phosphorylation of Tyr26 and Ser38, have been reported [79]. While the function of Ser38 phosphorylation has remained unclear, Tyr26 phosphorylation of hMOB1 has very recently been attributed an important role in tyrosine kinase signalling [80]. Gutkind and colleagues found that focal adhesion kinase (FAK) phosphorylates hMOB1 on Tyr26, thereby preventing the formation of a functional hMOB1/LATS complex and thus causing Hippo signalling to remain inactive [80]. It remains to be tested whether this new exciting regulatory mechanism of hMOB1 by Tyr26 phosphorylation also affects hMOB1/NDR complex formation. In addition, one should note that Ser23 of hMOB1 has also been observed [79], with Ser23 of hMOB1 representing a possible ATM target site [81], suggesting a possible link of hMOB1 to the DNA damage response (DDR). Last, but not least, it has been reported that GSK3β can destabilize hMOB1 by phosphorylating serine 146 of hMOB1 in the context of neurite outgrowth downstream of the PTEN-GSK3β axis [82]. Therefore, hMOB1 can be regulated by different phosphorylation events performed by diverse kinases.
In contrast to hMOB1, the only reported PTMs for hMOB2 are modifications with ubiquitin (or NEDD8) at Lys23, Lys32 and Lys131 [83,84]. The precise nature and functions of these three PTMs are currently unknown. In addition, the hMOB2 protein contains several putative ATM phosphorylation sites, which possibly are linked to the DDR function of MOB2 [81]. However, phosphorylation of hMOB2 has yet to be documented.
Similar to hMOB1, several phosphorylations of hMOB3s have been observed. Phosphorylations of hMOB3A on Thr15, Thr26 and Ser38, hMOB3B on Thr25 and Thr77, and hMOB3C on Thr14, Thr25 and Ser37 have been documented [79]. Ser37 of hMOB3C may represent an ATM targeting site [81]. Of note, the sequence motifs surrounding Thr15 and Ser38 of hMOB3A are quite similar to Thr12 and Thr35 of hMOB1 (Figure 3), hence raising the possibility that hMOB3A is possibly phosphorylated on Thr15 and/or Ser38 by MST1/2 in a similar fashion as reported for hMOB1 [30]. However, regulatory roles for any of these putative phosphorylation events are yet to be reported.
Last but not least, specific PTMs of hMOB4 are also of potential interest. In this regard, Ser147 and Tyr141 phosphorylations of hMOB4 have been observed [79]. Ser147 phosphorylation of hMOB4 is possibly performed by the DDR-linked ATM kinase [81], proposing that hMOB4 is linked to DNA damage signalling. However, no regulatory roles for any of these phosphorylation events have been described so far.
6. Protein-Protein Interactions of MOBs
Given that MOBs do not have any known enzymatic activities, one wonders how they contribute to intracellular signalling. Based on the currently available knowledge, the answer to this question is that MOBs are globular scaffold proteins that can regulate other factors such as protein kinases by direct protein-protein interactions [1]. Therefore, we will focus in this subchapter on summarising how MOBs form protein complexes and thereby help to regulate molecular machineries.
As already mentioned, hMOB1 can bind directly to MST1/2, LATS1/2 and NDR1/2. Interestingly, these interactions can occur in a phosphorylation-dependent manner. However, before discussing this important aspect in much detail, one should first note that hMOB1 can also associate with other signalling complexes. Current evidence suggests that hMOB1 can bind to at least seven apparently mutually exclusive and independent proteins in a sometimes phosphorylation-dependent fashion: (1) the NDR1/2 kinases, (2) the LATS1/2 kinases, (3) the MST1/2 kinases, (4) the PP6 phosphatase module, (5) the atypical guanine nucleotide exchange factors 6 to 8 (DOCK6-8), (6) the ubiquitin ligase Praja2, and (7) yet poorly characterized proteins, including leucine rich repeats and calponin homology domain containing (LRCH1-4) proteins and cytokine receptor like factor 3 (CRLF3) [1,6,30,65,78,85,86]. Notably, Xiong et al. found that the phosphorylation-dependent binding of hMOB1 to the PP6 and DOCK6-8 complexes appears to differ from that elucidated for protein kinase binding, with hMOB1 preferentially interacting with MST1/2 and NDR/LATS, and not the DOCK6-8 and PP6 modules [78]. Future investigations are needed to properly understand the underlying cellular functions of complexes containing hMOB1 and non-kinase binding partners such as PP6, DOCK6-8 and others.
While hMOB1 can bind to a variety of components, the complex formations with MST1/2 and NDR/LATS kinases are the only protein-protein interactions understood in detail. In Hippo core signalling, hMOB1 acts as a central signal adaptor helping to regulate NDR/LATS activation [1,6,13,71,72,87,88]. On the one hand, a stretch composed of highly conserved hydrophobic and positively charged residues N-terminal of the catalytic domains of NDR/LATS kinases, termed N-terminal regulatory domain (NTR), is essential for hMOB1 binding to NDR/LATS kinases to promote their activities. On the other hand, a conserved cluster of negatively charged residues on hMOB1 is needed for the interaction with NDR/LATS [1,3,6,13,28,71,72,87,88]. MST1/2 phosphorylation of hMOB1 on Thr12 and Thr35 regulates hMOB1 binding to the NTRs of NDR/LATS [6,30,71,72], although hMOB1 can also from a complex with NDR1/2 in a phosphorylation-independent manner [6]. In summary, hMOB1 binds to NDR/LATS kinases through highly conserved residues on hMOB1 and NDR/LATS kinases in a manner that can be regulated by MST1/2 phosphorylation of hMOB1.
hMOB1 binding to NDR/LATS has multiple functions. First, hMOB1 binding helps to stimulate auto-phosphorylation on a Ser residue located in the T-loop (also known as activation segment) of NDR/LATS kinases (summarised in [1,13,87,89]). Second, MOB1 also acts as an allosteric activator of NDR/LATS by mediating MST1/2-mediated trans-phosphorylation of a Thr residue located in the hydrophobic motif (HM) outside of the catalytic domain of NDR/LATS [6,28,78,87,88,89,90,91]. Third, it was biochemically shown that stable hMOB1 binding to MST2 can function as an important step in activating the MST1/2-LATS1/2 kinase cascade [72,92], although stable MOB1/MST1/2 binding seems dispensable for development and tissue growth control [6,90] and a stable hMOB1/MST1/2 complex is not required for efficient MST1/2-mediated phosphorylation of hMOB1 [6]. Since some of these hMOB1 functions appear to contradict each other, Manning and Harvey [93] proposed a unifying model, wherein hMOB1 acts before and after MST1/2-mediated phosphorylation of LATS1/2 and hMOB1. Notably, this model is similar, but still different, from the model proposed for NDR1/2 activation by MST1/2 and hMOB1 [6,88]. In this regard, somewhat surprisingly, T-loop and HM phosphorylations of LATS1/2 can also occur without increased MST1/2-mediated phosphorylation of hMOB1 on Thr12/Thr35 [94], suggesting that hMOB1/LATS complex formation can be dispensable for efficient LATS1/2 phosphorylation in certain settings. Nevertheless, hMOB1/LATS complex formation appears to be essential for development and tissue growth control [6].
Taken together, these data show collectively that hMOB1/NDR and hMOB1/LATS complexes are not equal, thereby allowing cells to process diverse biological inputs through diverse hMOB1 signalling. hMOB1 as an adapter protein can help to differentially regulate NDR/LATS kinases by concurrently forming complexes with NDR/LATS and their upstream activator kinases such as MST1/2 and by allosterically activating the auto- and trans-phosphorylation activities of NDR/LATS.
Interestingly, recent structural and biochemical analyses of the catalytic domain of NDR1, lacking the entire NTR domain, have suggested that the regulation of NDR1 activity by the elongated activation segment and by hMOB1 binding may represent mechanistically distinct events [89]. This notion is supported by the biochemical and structural dissection of the yeast Cbk1p-Mob2p complex [95]. However, much more work is needed to fully understand how the regulation of NDR/LATS kinases is orchestrated by hMOB1 binding to the NTR, activation segment and HM phosphorylations–three molecular events that possibly represent three distinct regulatory levels. In particular, the role of MST1/2-mediated phosphorylation of hMOB1 is yet to be completely understood with regard to the regulation of NDR1/2, in particular when considering that phospho-hMOB1 displays a much higher affinity for NDR1/2 than unphosphorylated hMOB1 [6,78], while LATS1/2 can only bind to phospho-MOB1 [6].
In contrast to NDR1/2 kinases, hMOB1 seems to associate with LATS1/2 and MST1/2 in a purely phospho-dependent manner [6,65,69,70,71,72,78,92]. In the case of MST1/2, the direct interactions involve several auto-phosphorylation sites in MST1/2 that help to recruit hMOB1 [51,65,69,72,78,92]. Noteworthy, these interactions involve a phospho-Ser/Thr binding pocket of hMOB1, encompassing the positively charged residues Lys153, Arg154 and Arg157 of hMOB1 [70]. These three key residues appear to be conserved in hMOB1 and hMOB4, while hMOB3s have only two conserved positively charged residues (Figure 3). However, phospho-peptide binding properties of hMOB2, hMOB3s and hMOB4 are yet to be reported.
Based on the structure of a short fragment of MST2 (371 to 400) bound to hMOB1 lacking the first 50 amino acids, Ni et al. [72] defined two MOB1/MST2 interfaces, namely one supportive binding site (residues 390 to 398 of MST2) and one main phospho-Thr binding site, centring around phosphorylated Thr378 of MST2 and the phospho-Ser/Thr binding pocket of hMOB1. However, other important sites involved in hMOB1 binding on MST2, such as phospho-Thr349, phospho-Thr356, and phospho-Thr364, were not examined on the structural level [72]. This notion [72] that Thr378 of MST2 (Thr380 of MST1, respectively) is not the sole main interaction site on hMOB1 is supported by other studies [78,92]. At least six different phospho-threonine sites on MST1 (Thr329, Thr340, Thr353, Thr367, Thr380 and Thr387) contribute to hMOB1 binding [78,92]. Intriguingly, Pro106 of hMOB1 contributes to phospho-Thr binding on MST1 [72,78,92], and the positively charged Lys104 and Lys105 of hMOB1 are crucial for MST1/2 binding, while being expendable for NDR/LATS binding [6]. Thus, it is quite possible that Lys104/Lys105 together with Pro106 of hMOB1 may bond with multiple negatively charged phosphorylated Thr residues on MST1/2. Notably, the Lys104/Lys105/Pro106 motif of hMOB1 is conserved in dMOB1, hMOB3s and dMOB3 (Figure 3). Alterations of Arg108 and Lys109 in hMOB3A, corresponding to Lys104 and Lys105 in hMOB1 (Figure 3), abolish MST1 binding [39], suggesting that the Lys104/Lys105/Pro106 binding motif of hMOB1 is functionally conserved.
Notably, phospho-Thr378 of MST2 can also bind to SLMAP [68], in addition to hMOB1 [69], and a T-loop mutation of MST2 reduces the MST2/SLMAP interaction [65] in the context of STRIPAK-Hippo signalling. Thus, the multisite auto-phosphorylation of the MST1/2 linkers appears to function as a molecular platform able to integrate different Hippo signalling events [68,69].
In contrast to MST1/2, the LATS1/2 kinases must not be phosphorylated to bind to phospho-MOB1 [6,71,72]. Nevertheless, the phosphorylation of LATS1/2 on serine 690 and 653, respectively (corresponding to threonine 74 and 75 in NDR1 and NDR2), are very likely to represent another regulatory layer. LATS1 S690A, LATS2 S653A, NDR1 T74A and NDR2 T75A mutants display diminished binding to MOB1 [27,28,29,87,88,96,97] and the crystal structure of phospho-MOB1 bound to a LATS1 fragment highlighted the importance of serine 690 of LATS1 for MOB1 binding [72]. In this regard, hMOB1 must be phosphorylated to bind to LATS1/2 [6,71,72], with Asp63 of hMOB1 being essential and providing the selectivity for LATS1/2 binding [6]. In this regard, the analysis of hMOB1 mutants with alterations of Lys153 and Arg154 revealed that the phospho-Ser/Thr binding interface of hMOB1 is important for MST1/2 as well as LATS1/2 binding [6]. Consequently, Lys153 and Arg154 of hMOB1 are very likely to represent central residues in a more general phospho-Ser/Thr binding domain as already proposed by Rock et al. [70]. Notably, Lys153 and Arg154 of hMOB1 can also contribute to Praja2 binding in the context of hMOB1 proteolysis [86].
In addition, the interactions of hMOB1 with NDR/LATS kinases can be modulated by Tyr26 phosphorylation of hMOB1 [80] and hMOB2 counteracting hMOB1 binding to NDR1/2 [35,62]. Comparable to hMOB2, the LIM domain protein TRIP6 can compete with MOB1 for binding to the NTR of LATS1/2, thereby negatively interfering with LATS1/2 activation by hMOB1 [98]. Similarly, phosphatidic acid-related lipid signalling can regulate the Hippo pathway by directly interacting with LATS1/2, thus disrupting hMOB1/LATS complex formation [99]. In this context, one should note that inactivation of the PTEN lipid phosphatase can interfere with the interaction of hMOB1 and LATS1/2 in gastric cancer [100]. Moreover, the Angiomotin scaffolds can support hMOB1-mediated LATS1/2 auto-phosphorylation [101].
The best understood binding partners of hMOB2, hMOB3s and hMOB4 were already mentioned in subchapters 3 and 4. Briefly, two binding partners of hMOB2 have been established: the NDR1/2 kinases [1,35] and the RAD50 scaffold [37]. However, the physiological significance of these interactions is yet to be fully defined [38], with hMOB2 possibly also having indirect effects on LATS1/2 signalling by competing with hMOB1 binding to LATS1/2 [35,62].
hMOB3s have one established binding partner: the MST1 kinase [36]. In response to apoptotic stimuli, hMOB3s bind to MST1 through conserved positively charged residues, thereby negatively regulating apoptotic MST1 signalling in glioblastoma multiforme cells by inhibiting the MST1 cleavage-based activation process [36]. In this regard, it is crucial to note that hMOB3s do not function like hMOB1 in the Hippo pathway as proposed recently [102]. While hMOB3s and hMOB1 share similar primary sequences (Figure 1 and Figure 3), they differ significantly in their binding partners, and hence their potential to regulate the core of the Hippo pathway. Nevertheless, one cannot rule out the possibility that hMOB3 and hMOB1 may compete for MST1/2 binding, similar as already reported for the competition of hMOB2 and hMOB1 for NDR1/2 binding [1,35].
As mentioned, hMOB4 is a core member of the STRIPAK complex, known to function as a regulator of Hippo signalling [43,63]. Recently, it was reported that hMOB4 forms a complex with MST4 [48], a distant relative of MST1/2 [46] and a known component of the STRIPAK complex [73]. It was proposed that MST4 and hMOB4 may disrupt assembly of the hMOB1/MST1 complex through alternative pairing [48]; however, this model has yet to be explored in more detail.
Taken together, we have started to understand the mechanistic importance of protein-protein interactions involving hMOBs. However, some of our current working models have yet to be consolidated by future studies, ranging from more structure biology to the testing of the significance of selected protein-protein interactions in adequate biological model systems. In particular, we need to comprehend how phosphorylation-triggered interactions of hMOBs help to coordinate the specificity and robustness of essential signalling cascades. In this context, it will be vital to understand the potential functional contribution of the abilities of hMOBs to engage with multiple ligands.
7. Cancer-Associated Cellular Functions of MOBs
7.1. Roles in Mitosis and Cell Cycle Progression
Since we are only briefly summarising here the mitotic functions of hMOB1, for a detailed overview of the cell cycle roles of hMOB1 see refs. [20,21]. Initially, a kinase-targeting RNAi screen found that the NDR1 kinase, a binding partner of hMOB1 and hMOB2 [1], is possibly involved in regulating spindle orientation [103]. Yao and colleagues subsequently discovered a new level of hMOB-mediated NDR1 regulation in mitosis [61]. More precisely, PLK1 phosphorylates NDR1 at Thr7, Thr183 and Thr407 upon mitotic entry, thereby eliciting PLK1-dependent suppression of NDR1 activity to ensure correct spindle orientation in mitosis [61]. Mechanistically, PLK1-mediated phosphorylation of NDR1 switches NDR1 binding from hMOB1 to hMOB2, thereby interfering with the binding of hMOB1 to NDR1 and subsequent NDR1 activation [61]. The involvement of hMOB1 in mitosis is further supported by the finding that hMOB1 knockdown displays a negative genetic interaction with the PTTG1 gene [104], with PTTG1 encoding Securin, a well-established regulator of chromosome segregation [105]. Noteworthy, hMOB1 can localise to kinetochore structures of mitotic cells [106] and contributes to cell abscission and centriole re-joining after telophase and cytokinesis [107].
Knockdown of hMOB4 can also cause mitotic spindle defects in human cells [108]. Similarly, dMOB4 depletion results in mitotic defects in fly cells [25]. However, it is currently not known whether this function is related to the MOB4-containing STRIPAK complex. In contrast to hMOB1 and hMOB4, the depletion of hMOB2 results in a G1/S cell cycle arrest [37,109]. More specifically, hMOB2 knockdown triggers a p53-dependent G1/S cell cycle arrest [37] that is discussed in more detail in subchapter 7.2. hMOB3s have no known role in mitosis.
7.2. Roles in the DNA Damage Response (DDR)
Considering the possible involvement of NDR1/2 in the DDR [38], one would predict that regulators of NDR1/2, such as hMOB1 [1], may also play a role in the DDR. Interestingly, this seems to be the case, since hMOB1 knockdown appears to be sufficient to cause spontaneous DNA double-strand break formation [110] and increased γH2AX phosphorylation in human cells [111]. The single knockdown of hMOB3A or hMOB4 can also result in increased γH2AX phosphorylation in cells [111], while hMOB3C knockdown cells seem to display an increased homologous recombination repair frequency [112]. Collectively, these reports propose that hMOB1, hMOB3 and hMOB4 are possibly linked to DNA damage signalling. However, it is yet to be established whether any of these possible DDR functions are linked to binding to kinases of the Hippo core, such as NDR/LATS, or components of the STRIPAK complex.
Regarding hMOB2 we have a somewhat clearer understanding regarding the DDR [38]. Given that a genome wide screen for novel putative DDR factors identified hMOB2 (aka HCCA2) as a potential candidate [113], we sought to understand how hMOB2 might function as part of the DDR. Subsequent experiments uncovered that loss of hMOB2 causes the accumulation of DNA damage, which activates the DDR kinases ATM and CHK2, consequently activating a p53 dependent G1/S cell cycle checkpoint in the absence of any exogenously induced DNA damage [37]. Complementary experiments showed that hMOB2 promotes cell survival and G1/S cell cycle arrest upon exposure to diverse DNA damaging agents [37]. Unexpectedly, hMOB2 seems to act in the DDR independent of NDR signalling [37], therefore, the biological significance of the hMOB2/NDR complex remains poorly understood [38]. Mechanistically, hMOB2 can interact with RAD50, facilitating the recruitment of the MRE11-RAD50-NBS1 (MRN) DNA damage sensor complex and activated ATM to DNA damaged chromatin [37]. However, this mechanistic link does not appear to be relevant in all DDR settings, suggesting that additional mechanisms should be considered [38].
Of note, other independent studies support the notion that hMOB2 represents a new player in the DDR. hMOB2 knockdown cells displayed an increased sensitivity to exogenously induced DNA-damage and a defective G2/M cell cycle checkpoint in response to ionizing radiation (IR) [113]. Furthermore, patient-derived fibroblasts with MOB2 loss-of-function exhibited increased susceptibility to exogenously induced DNA-damage [114]. hMOB2 knockdown in non-small cell lung cancer (NSCLC) cell lines is also a promising strategy to increase the susceptibility to IR in the context of radio-sensitization [115]. In addition, the search for mutator alleles which increase the rate of germline mutations has revealed the hMob2 gene as one of the top hits [116]. Thus, current evidence collectively suggests that the involvement of MOB2 in the response to DNA damage is important to maintain genomic stability. hMOB2 can be part of the DDR by playing roles in DDR kinase signalling, cell survival and cell cycle checkpoints upon exposure to DNA damage. However, we have yet to understand how hMOB2 operates as a DDR protein on the molecular level [38]. Furthermore, we must comprehend which types of DNA damage repair mechanism(s) are linked to hMOB2 and whether this knowledge can be clinically exploited for precision medicine.
7.3. Apoptosis and Autophagy
hMOB1 has been linked to apoptotic signalling [1]. hMOB1 as a co-activator of human NDR kinases plays an important role in the formation of a MST1-NDR-hMOB1 complex, thereby supporting MST1-mediated phosphorylation of NDR1/2 upon the induction of apoptosis [91,117]. Interestingly, the apoptotic role of hMOB1 was further linked to cell fate decisions in response to autophagic stress conditions. Precisely, upon investigating the activation of NDR1 in macroautophagy, we observed that NDR1 is stimulated in a hMOB1-dependent manner upon autophagy induction [118]. Considering further that NDR1′s kinase activity also plays an important role in mitophagy, the removal of damaged mitochondria by selective autophagy [119], it is likely that hMOB1 as a co-activator of NDR1/2 possibly functions in different autophagic processes. Taken together, hMOB1 seems to assist NDR1/2 to coordinate autophagic and apoptotic events.
7.4. Centrosome Biology
By functioning as a co-activator of human NDR kinases, hMOB1 can also play a role in centrosome duplication [28]. However, although hMOB1 can localise to centrosomes [106,120], we do not know whether the centrosomal pool of hMOB1 is the main supporter of centrosome duplication. Based on data from a large-scale screen hMOB1 appears to bind to DIPA [121,122], which is of potential interest, considering that DIPA was found on centrosomes [123]. Moreover, the NDR2 kinase was recently connected with ciliogenesis, a cellular process that utilises the mature mother centriole at the base of cilia extrusions [124,125]. Considering further that active (phosphorylated) NDR1/2 can localise to the mother centriole in a cell cycle dependent fashion [28], it is quite tempting to speculate that hMOB1 as a main regulator of NDR1/2 is linked to the ciliogenesis. This notion is supported by additional lines of evidence:
First, upon studying different canine models for retinal degeneration, it was found that hMOB1 expression was specifically elevated in the early retinal degeneration model [126], an autosomal recessive disorder caused by a NDR2 mutation [127]. Possibly this reflects the cellular attempt to compensate for NDR2 loss-of-function. Second, murine NDR1/2 kinases are important for regulation of proliferation of terminally differentiated cells in the retina [128]. Third, increased levels of phospho-Thr35 MOB1 were observed in a fish model for photoreceptor ciliogenesis and retinal development [129]. Fourth, MOB1 accumulates at the basal bodies and MOB1 depletion delays ciliogenesis in Tetrahymena [130]. Therefore, MOB1 appears to represent a Hippo component that provides a link between the Hippo pathway and ciliogenesis in the context of diseases such as retinal degeneration and possibly also polycystic kidney disease [131,132,133,134,135,136,137,138,139].
7.5. MOBs and RAS Onco-Proteins
Oncogenic Ras signalling occurs frequently in many human cancers. However, no effective targeted therapies are currently available to treat patients suffering from Ras-driven tumours. In this regard, a systematic screen uncovered YAP1, a key effector of Hippo signalling [17,140], as a significant component of oncogenic RAS signalling [141]. Specifically, YAP1 overexpression is sufficient to rescue cell viability in a KRAS-dependent fashion [141]. Interestingly, the same screen also identified hMOB2 and hMOB3C as possible factors in oncogenic RAS signalling [141]. hMOB2 overexpression could decrease cell viability in a KRAS-dependent manner, while hMOB3C overexpression restored cell viability without any obvious effects on MEK and PI3K signalling [141]. Another genome-wide RNAi screen identified a synthetic lethal interaction between hMOB3A loss-of-function and oncogenic KRAS [142]. Perhaps the RAS-related function of hMOB2 is linked to hMOB2/NDR complex formation, since NDR1/2 were recently linked to oncogenic RAS signalling [119,143]. Future research is warranted to probe these interesting links.
8. MOBs and Cancer
8.1. Lung cancer
hMOB1A mRNA levels are low in human lung cancer samples [144]. Consequently, Suzuki and colleagues studied inducible conditional MOB1 knockout mice in the context of lung physiology and cancer, revealing that mice with postnatally MOB1 loss-of-function did not develop spontaneous lung adenocarcinomas [145]. Unexpectedly, urethane treatment-induced lung tumour formation was decreased upon postnatal MOB1 deletion [145], suggesting that MOB1 rather functions as a cancer-promoting factor in this setting. Thus, it will be very interesting to elucidate which functional protein-protein interaction(s) involving MOB1 perform this pro-cancer role. Noteworthy, hMOB3B seems to interact with NT5C2 [121]. Given that NT5C2 has been used as a prognostic marker in lung cancer [146], it is tempting to speculate that hMOB3s might be of clinical relevance in lung cancer.
8.2. Pancreatic Cancer
Chen et al. found that miR-181c can directly and selectively repress hMOB1, MST1, LATS2, and SAV1 expression in human pancreatic cancer cells [147]. Inhibition of miR-181c increased the protein levels of hMOB1 and other Hippo components. In contrast, overexpression of miR-181c suppressed them, resulting in increased pancreatic cancer cell survival and chemoresistance [147]. Moreover, deregulated expression of the long non-coding RNA (lncRNA) UCA1 has been implicated in diverse human cancers, such as pancreatic cancer. UCA1 can interact with hMOB1, LATS1, and YAP to form shielding composites, thereby allowing YAP activation in pancreatic cancer cells [148]. Thus, deregulation of hMOB1 as part of the Hippo pathway can play a significant role in the progression of pancreatic cancer. Of note, expression levels of hMOB4 were found elevated in pancreatic cancer samples [48]. Thus, diverse hMOBs are of potential interest in clinical studies of pancreatic cancer.
8.3. Liver Cancer
The Thr12 phosphorylation levels of hMOB1 were significantly decreased in human liver cancer samples [94]. Mice with liver-specific Mob1 null alleles are prone to develop liver cancers [149]. Given that this phenotype is the most severe among mutant mice lacking a Hippo signalling component in the liver [150], it is fair to conclude that MOB1 constitutes a critical hub of Hippo signalling in livers and possibly also other tissues. In this regard, it is noteworthy that increased expression of hMOB2 mRNA was associated with hepatocellular carcinoma development and progression [151]. However, due to the lack of MOB2 loss-of-function animal models it is currently unclear whether deregulated MOB2 expression can drive liver cancer development.
Given that co-infections with hepatitis delta virus (HDV) and hepatitis B virus (HBV) accelerate the development of hepatocellular carcinoma (HCC) [152], one should also note that hMOB1 can associate with CCDC85B (aka DIPA) [121,122], which is known to play a role in HDV replication [153]. hMOB1 also interacts with the hepatitis C virus (HCV) non-structural protein NS5A, although no relationship between the hMOB1A-NS5A interaction and HCV replication has been observed [154]. Nevertheless, the hMOB1-binding partners LATS1/2 represent host kinases that can phosphorylate NS5A at a highly conserved residue required for optimal HCV replication [155].
8.4. Haematological Malignancies
By studying the effects of 3-deazaneplanocin A (DZNep), a histone methyltransferase EZH2 inhibitor, on human T-cell acute lymphoid leukaemia (T-ALL) cells, Shen et al. found that DZNep treatment reduced histone methylation in hMOB1 promoters, resulting in an upregulation of hMOB1 expression associated with inhibited growth of T-ALL cells [156]. Thus, MOB1 levels may represent an indicative marker for DZNep treatments of T-ALL patients.
As mentioned, hMOB3 may interact with NT5C2 [121]. Given that NT5C2 expression levels can serve as a prognostic marker in haematological malignancies [157,158] and NT5C2 mutations were linked to ALL [159], it is therefore tempting to deduce that a link between hMOB3s and NT5C2 might be of clinical significance with regard to haematological cancers. In this regard, one should further note that altered expression levels of hMOB3A and hMOB3B were observed in mantle cell lymphoma [160,161]. However, animal studies that functionally link the deregulation of MOB3 with haematological malignancies are missing to date.
8.5. Breast and Ovarian Cancers
Kim et al. found that hMOB1 knockdown decreased the viability of breast cancer cell lines [162], suggesting that hMOB1 is supporting the survival of breast cancer cells. Changes in the methylation of the hMOB1 promoter were observed in triple negative breast cancer cells [163]. Likewise, a systematic study across cancer cell lines revealed that MOB1 knockdown results in decreased viability of ovarian cancer cells [164], suggesting that hMOB1 is supporting the survival of ovarian cancer cells. Of note, single knockdown of hMOB3A, hMOB3B or hMOB3C also decreased the viability of breast cancer cells [162].
8.6. Colon Cancer
The mRNA levels of hMOB1A are low in human colorectal cancer samples [165]. Recently, a study of mice with intestinal epithelial cell-specific depletion of MOB1 uncovered a link between MOB1 and intestinal homeostasis through the regulation of Wnt signalling [166]. Considering that the disruption of intestinal homeostasis by the deregulation of other Hippo components can result in colon cancer [167], it is quite likely that loss of MOB1 as a central component of the Hippo pathway is also associated with colon cancer development and/or progression. Of note, single knockdown of hMOB3A, hMOB3B or hMOB3C in human colon cancer cells displays a negative genetic interaction with the PTEN phosphatase [104].
8.7. Prostate Cancer
A single nucleotide polymorphisms (SNP) in the hMob2 gene has been revealed to be a prostate cancer susceptibility loci [168]. The mRNA levels of hMOB3B were significantly decreased in prostate cancer, suggesting that hMOB3B may act as a tumour suppressor in prostate cancer [169]. Moreover, the hMOB3B promoter region is also hypermethylated in a significant number of prostate cancer samples [170]. Thus, loss of hMOB3B might promote prostate cancer development and/or progression.
8.8. Glioblastoma
On the one hand, it was found that decreased hMOB1 levels help to sustain glioblastoma growth [86]. On the other hand, the protein levels of hMOB3s are significantly upregulated in samples derived from patients suffering from glioblastoma multiforme [36]. Therefore, it appears that hMOB1 and hMOB3s may be considered in clinical assessments of glioblastoma samples.
8.9. Human Cancer Cell Lines
Based on genome-scale loss-of-functions screens using a broad spectrum of diverse human cancer cell lines, one is tempted to conclude that the single knockdown or knockout of hMOB1A, hMOB1B, hMOB3A, hMOB3B, hMOB3C or hMOB4 can decrease the survival of some cancer cells [171,172,173,174]. However, these findings have yet to be validated, and even more importantly, are most likely limited in their nature, since possible compensatory redundancies in the MOB1 and MOB3 groups were not examined. Of note, the status of hMOB3B together with a few other markers can serve as predictor of acquired and intrinsic resistance to epidermal growth factor receptor tyrosine kinase inhibitors, including gefitinib, erlotinib, and afatinib [175].
9. MOBs and Other Diseases
9.1. Neurobiology
MOB2 loss-of-function impairs the correct neuronal positioning within the developing cortex, thereby associating MOB2 inactivation with a disorder such as periventricular nodular heterotopia [114]. Moreover, MOB2 plays a part in PKA-mediated signalling in astrocytes [176]. Similarly, MOB2 plays a neurological function in invertebrates, since dMOB2 expression is necessary and sufficient to regulate the growth of larval neuromuscular junctions in flies [23].
Chromatin immunoprecipitation followed by deep sequencing identified the hMOB3A gene as a target for the NRF1 (Nuclear respiratory factor 1), a transcription factor linked to neurodegenerative diseases [177]. Recombinant hMOB3A and hMOB3B can bind to β-amyloid, suggesting that hMOB3s are possibly linked to Alzheimer disease and other atrophies [178]. Interestingly, the same study found that hMOB1 can also associate with β-amyloid [178].
hMOB4 is mainly expressed in the central and peripheral nervous system [179]. Striatin-1, a core component of the STRIPAK complex, appears to require hMOB4 binding to fulfil its neurological function [180]. This notion is supported by the observation that dMOB4 also plays a role in the regulation of axonal transport, neurite elongation, and synapse formation in flies [24].
Taken together, each member of the MOB family seems to have at least one neurological function, with the neurological roles of MOB2 and MOB4 being the best understood.
9.2. Immunity and Virology
Quite likely hMOB1, based on its associations with NDR/LATS and MST1/2 kinases [1] is linked to immunological functions as summarised in ref. [57]. In addition, it was reported that the conserved Legionella pneumophila effector kinase LegK7 can phosphorylate MOB1 and thereby mimic MST1/2 activation to shut down the Hippo effectors YAP/TAZ, resulting in the promotion of bacterial growth and infection [181]. In this regard, the other hMOBs are possibly also associated with immune signalling in health and disease. More precisely, a SNP in the hMOB2 gene is associated with altered cytokine levels, in particular interleukin 9 (IL-9) levels [182]. SNPs in the hMOB3B gene are associated with rheumatoid arthritis [183], osteoarthritis [184] and asthma [185]. Moreover, a genome-wide RNAi screen identified hMOB4 as a possible regulator of cytokine secretion, since IL-8 levels were decreased upon hMOB4 knockdown [186].
9.3. Diverse Disease Spectrum
In addition to links to neurological and immune diseases, the deregulation of hMOBs has also been associated with other sicknesses as briefly summarised below. MOB1A/B-deficient mouse epidermis cannot be engrafted successfully onto donor mice, thereby influencing skin engraftment efficiency [187], suggesting a crucial role of MOB1 in the skin. Mice with chondrocyte-specific MOB1A/B-deficiency display impaired chondrocyte homeostasis, leading to chondrodysplasia, thereby linking MOB1 loss-of-function with a hereditary skeletal disorder [188]. Furthermore, an autosomal SNP of hMob2 is associated with type 2 diabetes [189,190]. Last but not least, it is notable that the dysregulation of the hMOB4-containing STRIPAK complex correlates with a broad range of human diseases including, diabetes, autism, cardiac disease and cerebral cavernous malformation (CCM) in addition to cancer [43,44]. However, it remains to be determined the degree to which hMOB4 is essential to support fundamental cellular processes such as cell proliferation, growth, differentiation, migration and apoptosis.
10. Conclusions and Future Outlook
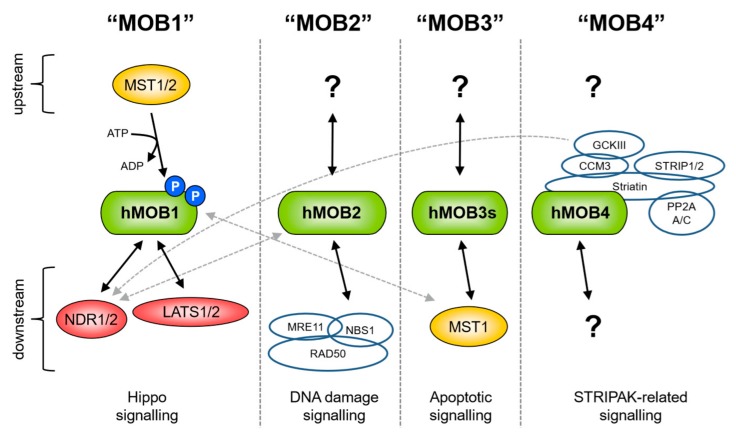
In summary, yeast cells express two different MOBs that act in independent and non-interchangeable complexes formed with members of the conserved NDR/LATS kinase family. In contrast, in multicellular organisms such as flies and humans, MOBs do not exclusively bind to a unique NDR/LATS kinase. Fly cells express at least four different dMOBs, while the human genome encodes at least seven different hMOB genes. hMOBs have been implicated in different crucial intracellular functions through forming complexes with diverse binding partners (Figure 4; see also subchapter 6). By associating with members of the NDR/LATS kinase family, hMOB1 has central roles in Hippo signalling (Figure 4, left panel). Importantly, hMOB1 also binds directly to MST1/2 as part of the Hippo core cassette (Figure 4, left panel). By forming a complex with the MRN DNA damage sensor complex, hMOB2 has been linked to DNA damage signalling (Figure 4, middle left panel). Similar to hMOB1, hMOB3s can associate with MST1 in the context of apoptotic signalling, but unlike hMOB1, hMOB3s do not interact with NDR/LATS kinases (Figure 4, middle right panel). Last but not least, hMOB4 has been documented as a core member of the STRIPAK complex (Figure 4, right panel).
Figure 4.
The four branches of MOB control. The “MOB1 branch”: In human cells, hMOB1 phosphorylated by MST1/2 promotes the activation of NDR1/2 and LATS1/2 in the context of Hippo signalling and the regulation of other important cellular functions [1,55,57]. The “MOB2 branch”: hMOB2 can interact with the MRN (Mre11-Rad50-Nbs1) DNA damage sensor complex, thereby supporting DNA damage and consequently cell cycle signalling [37,38]. The “MOB3 branch”: hMOB3s form a complex with MST1, in so doing regulating apoptotic signalling [39]. The “MOB4 branch”: hMOB4 as part of the STRIPAK (Striatin-interacting phosphatase and kinase) complex is likely to support the diverse cellular roles of the STRIPAK complex [43,44]. Notably, it is possible that these MOB branches do not signal in isolation, but rather may be interlinked as highlighted by dashed grey lines. For example, hMOB2 may connect with the “MOB1 branch” by competing with hMOB1 for NDR1/2 binding [35,62], the hMOB3s may link with the “MOB1 branch” through competing with hMOB1 for MST1/2 binding [6,39], or hMOB4 as part of the GCKIII-containing STRIPAK complex may connect with the “MOB1 branch” through GCKIII-mediated phosphorylation of NDR1/2 (see subchapter 4). Noteworthy, Hippo and STRIPAK signalling have already been found to be interconnected (see subchapter 4), although, to our knowledge, the specific roles of hMOB4 have not been defined. hMOBs are in green. MST1/2 and NDR/LATS kinases are in yellow and red, respectively. Established interactions are highlighted by black lines, while putative interconnections between “MOB branches” are shown by dashed grey lines. Phosphorylations are indicated by “P” in blue.
Although the four branches of MOB signalling can be depicted as four separate signal transduction cascades, one should note that it is quite likely that they are interconnected (Figure 4, see dashed grey lines). For example, it is known that hMOB2 can oppose certain functions of the hMOB1/NDR complex by competing with hMOB1 for NDR1/2 binding. Hence, it could well be that hMOB2 is interlinked with the Hippo pathway through interfering with hMOB1-regulated processes. Another example is the possible connection of hMOB3s with the Hippo pathway. Perhaps hMOB3s can compete with hMOB1 for MST1/2 binding and thereby meddle with hMOB1-regulated Hippo signalling. Furthermore, hMOB4 as part of the STRIPAK complexes may help to connect the GCKIII upstream kinases with NDR1/2 as members of the Hippo pathway. As is apparent from these examples, it is conceivable that hMOB2, hMOB3s and hMOB4 are interconnected with hMOB1-regulated Hippo signalling to some degree (Figure 4, see dashed grey lines). Certainly, future research is warranted to address these predicted crosstalks between different hMOB-containing complexes.
Some characteristics of MOB proteins are conserved from yeast to man, but MOB signalling is surely more complex in multicellular organisms. Therefore, one future challenge is to fully comprehend and appreciate this complexity. Despite great progress in understanding how MOBs can help to regulate their binding partners, much more remains to be accomplished in this regard. For instance, more research is needed to fully understand how hMOBs are fine-tuning the activities of NDR/LATS kinases, with particular emphasis on delineating the in vivo implications of MST1/2-mediated phosphorylation of hMOB1. In this context, we also must understand which protein-protein interactions of hMOBs are essential on the cellular and organismal levels. For example, multiple binding partners of hMOB1 have been reported, but we have only begun to appreciate the physiological importance of a fraction of these hMOB1-focused complexes. The molecular and biological understanding of the significance of hMOB2/NDR, hMOB2/MRN, hMOB3/MST1 and hMOB4/STRIPAK complexes has also remained a pressing question that is yet to be dissected in much detail. Possibly, forthcoming studies on MOBs from organisms such as fungi, parasites, bacteria and plants [1,181,191,192] will help to provide breakthroughs in this regard; studies of yeast MOBs have initially led the way [1,5,8].
Taken together, future research is needed to improve the structural and biochemical understanding of hMOB-regulated signalling in order to fully elucidate the molecular mechanisms underlying human diseases, such as cancer, that are linked to the deregulation of hMOBs. In this regard, research into the cellular functions of MOBs is also timely and relevant. The biological roles of MOBs must likewise be addressed in appropriate animal models, such as transgenic mice. To date, only MOB1 knock-out animals have been reported. To our knowledge, transgenic mice allowing for studies of the physiological functions of MOB2, MOB3s and MOB4 are currently not available. Hopefully studies of disease-relevant cell biological roles of MOBs will soon be complemented by appropriate loss-of-function animal models. Many research avenues must be pursued to adequately describe the complexity of MOB signalling in multicellular organisms. Undoubtedly, exciting new discoveries related to MOBs are to be expected over the coming years.
Acknowledgments
We thank Y. Kulaberoglu and J. Lisztwan for their critical input on this manuscript. This research was funded by the Wellcome Trust grant number 090090/Z/09/Z, BBSRC grant number BB/I021248/1, Worldwide Cancer Research (formerly AICR) grant number 11-0634, UCL Cancer Research UK Centre funding, and the National Institute for Health Research University College London Hospitals Biomedical Research Centre. R.G. is supported by the Research Council of the Republic of Turkey (TUBITAK grant 119S007).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hergovich A. MOB control: Reviewing a conserved family of kinase regulators. Cell Signal. 2011;23:1433–1440. doi: 10.1016/j.cellsig.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow A., Hao Y., Yang X. Molecular characterization of human homologs of yeast MOB1. Int. J. Cancer. 2010;126:2079–2089. doi: 10.1002/ijc.24878. [DOI] [PubMed] [Google Scholar]
- 3.Stavridi E.S., Harris K.G., Huyen Y., Bothos J., Verwoerd P.M., Stayrook S.E., Pavletich N.P., Jeffrey P.D., Luca F.C. Crystal structure of a human Mob1 protein: Toward understanding Mob-regulated cell cycle pathways. Structure. 2003;11:1163–1170. doi: 10.1016/S0969-2126(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 4.He Y., Emoto K., Fang X., Ren N., Tian X., Jan Y.N., Adler P.N. Drosophila Mob family proteins interact with the related tricornered (Trc) and warts (Wts) kinases. Mol. Biol. Cell. 2005;16:4139–4152. doi: 10.1091/mbc.e05-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luca F.C., Winey M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulaberoglu Y., Lin K., Holder M., Gai Z., Gomez M., Assefa Shifa B., Mavis M., Hoa L., Sharif A.A.D., Lujan C., et al. Stable MOB1 interaction with Hippo/MST is not essential for development and tissue growth control. Nat. Commun. 2017;8:695. doi: 10.1038/s41467-017-00795-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai Z.C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L.L., Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 8.Bardin A.J., Amon A. Men and sin: What’s the difference? Nat. Rev. Mol. Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- 9.Meitinger F., Palani S., Pereira G. The power of MEN in cytokinesis. Cell Cycle. 2012;11:219–228. doi: 10.4161/cc.11.2.18857. [DOI] [PubMed] [Google Scholar]
- 10.Baro B., Queralt E., Monje-Casas F. Regulation of Mitotic Exit in Saccharomyces cerevisiae. Methods Mol. Biol. 2017;1505:3–17. doi: 10.1007/978-1-4939-6502-1_1. [DOI] [PubMed] [Google Scholar]
- 11.Simanis V. Pombe’s thirteen–control of fission yeast cell division by the septation initiation network. J. Cell Sci. 2015;128:1465–1474. doi: 10.1242/jcs.094821. [DOI] [PubMed] [Google Scholar]
- 12.Hotz M., Barral Y. The Mitotic Exit Network: New turns on old pathways. Trends Cell Biol. 2014;24:145–152. doi: 10.1016/j.tcb.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Hergovich A., Stegert M.R., Schmitz D., Hemmings B.A. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- 14.Saputo S., Chabrier-Rosello Y., Luca F.C., Kumar A., Krysan D.J. The RAM network in pathogenic fungi. Eukaryot Cell. 2012;11:708–717. doi: 10.1128/EC.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staley B.K., Irvine K.D. Hippo signaling in Drosophila: Recent advances and insights. Dev. Dyn. 2012;241:3–15. doi: 10.1002/dvdy.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun S., Irvine K.D. Cellular Organization and Cytoskeletal Regulation of the Hippo Signaling Network. Trends Cell Biol. 2016;26:694–704. doi: 10.1016/j.tcb.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misra J.R., Irvine K.D. The Hippo Signaling Network and Its Biological Functions. Annu. Rev. Genet. 2018;52:65–87. doi: 10.1146/annurev-genet-120417-031621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei X., Shimizu T., Lai Z.C. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emoto K., Parrish J.Z., Jan L.Y., Jan Y.N. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- 20.Hergovich A. Hippo Signaling in Mitosis: An Updated View in Light of the MEN Pathway. Methods Mol. Biol. 2017;1505:265–277. doi: 10.1007/978-1-4939-6502-1_19. [DOI] [PubMed] [Google Scholar]
- 21.Hergovich A., Hemmings B.A. Hippo signalling in the G2/M cell cycle phase: Lessons learned from the yeast MEN and SIN pathways. Semin. Cell Dev. Biol. 2012;23:794–802. doi: 10.1016/j.semcdb.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L.Y., Lin C.H., Fan S.S. Function of Drosophila mob2 in photoreceptor morphogenesis. Cell Tissue Res. 2009;338:377–389. doi: 10.1007/s00441-009-0878-7. [DOI] [PubMed] [Google Scholar]
- 23.Campbell M., Ganetzky B. Identification of Mob2, a novel regulator of larval neuromuscular junction morphology, in natural populations of Drosophila melanogaster. Genetics. 2013;195:915–926. doi: 10.1534/genetics.113.156562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulte J., Sepp K.J., Jorquera R.A., Wu C., Song Y., Hong P., Littleton J.T. DMob4/Phocein regulates synapse formation, axonal transport, and microtubule organization. J. Neurosci. 2010;30:5189–5203. doi: 10.1523/JNEUROSCI.5823-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trammell M.A., Mahoney N.M., Agard D.A., Vale R.D. Mob4 plays a role in spindle focusing in Drosophila S2 cells. J. Cell Sci. 2008;121:1284–1292. doi: 10.1242/jcs.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devroe E., Erdjument-Bromage H., Tempst P., Silver P.A. Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. J. Biol. Chem. 2004;279:24444–24451. doi: 10.1074/jbc.M401999200. [DOI] [PubMed] [Google Scholar]
- 27.Hergovich A., Bichsel S.J., Hemmings B.A. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol. Cell Biol. 2005;25:8259–8272. doi: 10.1128/MCB.25.18.8259-8272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hergovich A., Kohler R.S., Schmitz D., Vichalkovski A., Cornils H., Hemmings B.A. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr. Biol. 2009;19:1692–1702. doi: 10.1016/j.cub.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 29.Hergovich A., Schmitz D., Hemmings B.A. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem. Biophys. Res. Commun. 2006;345:50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- 30.Praskova M., Xia F., Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harvey K., Tapon N. The Salvador-Warts-Hippo pathway–an emerging tumour-suppressor network. Nat. Rev. Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 32.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 33.Saucedo L.J., Edgar B.A. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- 34.Thompson B.J., Sahai E. MST kinases in development and disease. J. Cell Biol. 2015;210:871–882. doi: 10.1083/jcb.201507005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohler R.S., Schmitz D., Cornils H., Hemmings B.A., Hergovich A. Differential NDR/LATS interactions with the human MOB family reveal a negative role for human MOB2 in the regulation of human NDR kinases. Mol. Cell Biol. 2010;30:4507–4520. doi: 10.1128/MCB.00150-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang F., Gill J., Ficht X., Barthlott T., Cornils H., Schmitz-Rohmer D., Hynx D., Zhou D., Zhang L., Xue G., et al. The kinases NDR1/2 act downstream of the Hippo homolog MST1 to mediate both egress of thymocytes from the thymus and lymphocyte motility. Sci. Signal. 2015;8:ra100. doi: 10.1126/scisignal.aab2425. [DOI] [PubMed] [Google Scholar]
- 37.Gomez V., Gundogdu R., Gomez M., Hoa L., Panchal N., O’Driscoll M., Hergovich A. Regulation of DNA damage responses and cell cycle progression by hMOB2. Cell Signal. 2015;27:326–339. doi: 10.1016/j.cellsig.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gundogdu R., Hergovich A. The Possible Crosstalk of MOB2 With NDR1/2 Kinases in Cell Cycle and DNA Damage Signaling. J. Cell Signal. 2016;1:125. doi: 10.4172/2576-1471.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang F., Zhang L., Xue G., Hynx D., Wang Y., Cron P.D., Hundsrucker C., Hergovich A., Frank S., Hemmings B.A., et al. hMOB3 modulates MST1 apoptotic signaling and supports tumor growth in glioblastoma multiforme. Cancer Res. 2014;74:3779–3789. doi: 10.1158/0008-5472.CAN-13-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson J.C., Wellenreuther R., Poustka A., Pepperkok R., Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno C.S., Lane W.S., Pallas D.C. A mammalian homolog of yeast MOB1 is both a member and a putative substrate of striatin family-protein phosphatase 2A complexes. J. Biol. Chem. 2001;276:24253–24260. doi: 10.1074/jbc.M102398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baillat G., Moqrich A., Castets F., Baude A., Bailly Y., Benmerah A., Monneron A. Molecular cloning and characterization of phocein, a protein found from the Golgi complex to dendritic spines. Mol. Biol. Cell. 2001;12:663–673. doi: 10.1091/mbc.12.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi Z., Jiao S., Zhou Z. STRIPAK complexes in cell signaling and cancer. Oncogene. 2016;35:4549–4557. doi: 10.1038/onc.2016.9. [DOI] [PubMed] [Google Scholar]
- 44.Hwang J., Pallas D.C. STRIPAK complexes: Structure, biological function, and involvement in human diseases. Int. J. Biochem. Cell Biol. 2014;47:118–148. doi: 10.1016/j.biocel.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gordon J., Hwang J., Carrier K.J., Jones C.A., Kern Q.L., Moreno C.S., Karas R.H., Pallas D.C. Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20-Like kinase Mst3. BMC Biochem. 2011;12:54. doi: 10.1186/1471-2091-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dan I., Watanabe N.M., Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/S0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 47.Goudreault M., D’Ambrosio L.M., Kean M.J., Mullin M.J., Larsen B.G., Sanchez A., Chaudhry S., Chen G.I., Sicheri F., Nesvizhskii A.I., et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell Proteom. 2009;8:157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen M., Zhang H., Shi Z., Li Y., Zhang X., Gao Z., Zhou L., Ma J., Xu Q., Guan J., et al. The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer. J. Biol. Chem. 2018;293:14455–14469. doi: 10.1074/jbc.RA118.003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson R., Halder G. The two faces of Hippo: Targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu F.X., Zhao B., Guan K.L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng Z., Moroishi T., Guan K.L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan D. The hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harvey K.F., Zhang X., Thomas D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 54.Zanconato F., Cordenonsi M., Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hergovich A. The Roles of NDR Protein Kinases in Hippo Signalling. Genes. 2016;7 doi: 10.3390/genes7050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., Tang F., Terracciano L., Hynx D., Kohler R., Bichet S., Hess D., Cron P., Hemmings B.A., Hergovich A., et al. NDR functions as a physiological YAP1 kinase in the intestinal epithelium. Curr. Biol. 2015;25:296–305. doi: 10.1016/j.cub.2014.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharif A.A.D., Hergovich A. The NDR/LATS protein kinases in immunology and cancer biology. Semin. Cancer Biol. 2018;48:104–114. doi: 10.1016/j.semcancer.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 58.Gomez M., Kulaberoglu Y., Hergovich A. MST1/2 Kinase Assays Using Recombinant Proteins. Methods Mol. Biol. 2019;1893:319–331. doi: 10.1007/978-1-4939-8910-2_24. [DOI] [PubMed] [Google Scholar]
- 59.Nishio M., Hamada K., Kawahara K., Sasaki M., Noguchi F., Chiba S., Mizuno K., Suzuki S.O., Dong Y., Tokuda M., et al. Cancer susceptibility and embryonic lethality in Mob1a/1b double-mutant mice. J. Clin. Invest. 2012;122:4505–4518. doi: 10.1172/JCI63735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plouffe S.W., Meng Z., Lin K.C., Lin B., Hong A.W., Chun J.V., Guan K.L. Characterization of Hippo Pathway Components by Gene Inactivation. Mol. Cell. 2016;64:993–1008. doi: 10.1016/j.molcel.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan M., Chu L., Qin B., Wang Z., Liu X., Jin C., Zhang G., Gomez M., Hergovich A., Chen Z., et al. Regulation of NDR1 activity by PLK1 ensures proper spindle orientation in mitosis. Sci. Rep. 2015;5:10449. doi: 10.1038/srep10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang W., Shen J., Gu F., Zhang Y., Wu W., Weng J., Liao Y., Deng Z., Yuan Q., Zheng L., et al. Monopolar spindle-one-binder protein 2 regulates the activity of large tumor suppressor/yes-associated protein to inhibit the motility of SMMC-7721 hepatocellular carcinoma cells. Oncol. Lett. 2018;15:5375–5383. doi: 10.3892/ol.2018.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang Y., Chen M., Zhou L., Ma J., Li Y., Zhang H., Shi Z., Xu Q., Zhang X., Gao Z., et al. Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling. Cell Discov. 2019;5:3. doi: 10.1038/s41421-018-0077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ribeiro P.S., Josue F., Wepf A., Wehr M.C., Rinner O., Kelly G., Tapon N., Gstaiger M. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 65.Couzens A.L., Knight J.D., Kean M.J., Teo G., Weiss A., Dunham W.H., Lin Z.Y., Bagshaw R.D., Sicheri F., Pawson T., et al. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal. 2013;6:rs15. doi: 10.1126/scisignal.2004712. [DOI] [PubMed] [Google Scholar]
- 66.Liu B., Zheng Y., Yin F., Yu J., Silverman N., Pan D. Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila. Cell. 2016;164:406–419. doi: 10.1016/j.cell.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sakuma C., Saito Y., Umehara T., Kamimura K., Maeda N., Mosca T.J., Miura M., Chihara T. The Strip-Hippo Pathway Regulates Synaptic Terminal Formation by Modulating Actin Organization at the Drosophila Neuromuscular Synapses. Cell Rep. 2016;16:2289–2297. doi: 10.1016/j.celrep.2016.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bae S.J., Ni L., Osinski A., Tomchick D.R., Brautigam C.A., Luo X. SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK. Elife. 2017;6 doi: 10.7554/eLife.30278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng Y., Liu B., Wang L., Lei H., Pulgar Prieto K.D., Pan D. Homeostatic Control of Hpo/MST Kinase Activity through Autophosphorylation-Dependent Recruitment of the STRIPAK PP2A Phosphatase Complex. Cell Rep. 2017;21:3612–3623. doi: 10.1016/j.celrep.2017.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rock J.M., Lim D., Stach L., Ogrodowicz R.W., Keck J.M., Jones M.H., Wong C.C., Yates J.R., Winey M., Smerdon S.J., et al. Activation of the yeast Hippo pathway by phosphorylation-dependent assembly of signaling complexes. Science. 2013;340:871–875. doi: 10.1126/science.1235822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim S.Y., Tachioka Y., Mori T., Hakoshima T. Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway. Sci. Rep. 2016;6:28488. doi: 10.1038/srep28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ni L., Zheng Y., Hara M., Pan D., Luo X. Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling. Genes Dev. 2015;29:1416–1431. doi: 10.1101/gad.264929.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sugden P.H., McGuffin L.J., Clerk A. SOcK, MiSTs, MASK and STicKs: The GCKIII (germinal centre kinase III) kinases and their heterologous protein-protein interactions. Biochem. J. 2013;454:13–30. doi: 10.1042/BJ20130219. [DOI] [PubMed] [Google Scholar]
- 74.Poon C.L.C., Liu W., Song Y., Gomez M., Kulaberoglu Y., Zhang X., Xu W., Veraksa A., Hergovich A., Ghabrial A., et al. A Hippo-like Signaling Pathway Controls Tracheal Morphogenesis in Drosophila melanogaster. Dev. Cell. 2018;47:564–575. doi: 10.1016/j.devcel.2018.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hyodo T., Ito S., Hasegawa H., Asano E., Maeda M., Urano T., Takahashi M., Hamaguchi M., Senga T. Misshapen-like kinase 1 (MINK1) is a novel component of striatin-interacting phosphatase and kinase (STRIPAK) and is required for the completion of cytokinesis. J. Biol. Chem. 2012;287:25019–25029. doi: 10.1074/jbc.M112.372342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 77.Mah A.S., Jang J., Deshaies R.J. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA. 2001;98:7325–7330. doi: 10.1073/pnas.141098998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xiong S., Couzens A.L., Kean M.J., Mao D.Y., Guettler S., Kurinov I., Gingras A.C., Sicheri F. Regulation of Protein Interactions by Mps One Binder (MOB1) Phosphorylation. Mol. Cell Proteom. 2017;16:1111–1125. doi: 10.1074/mcp.M117.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feng X., Arang N., Rigiracciolo D.C., Lee J.S., Yeerna H., Wang Z., Lubrano S., Kishore A., Pachter J.A., Konig G.M., et al. A Platform of Synthetic Lethal Gene Interaction Networks Reveals that the GNAQ Uveal Melanoma Oncogene Controls the Hippo Pathway through FAK. Cancer Cell. 2019;35:457–472. doi: 10.1016/j.ccell.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wong Y.H., Lee T.Y., Liang H.K., Huang C.M., Wang T.Y., Yang Y.H., Chu C.H., Huang H.D., Ko M.T., Hwang J.K. KinasePhos 2.0: A web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 2007;35:W588–W594. doi: 10.1093/nar/gkm322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song Z., Han X., Zou H., Zhang B., Ding Y., Xu X., Zeng J., Liu J., Gong A. PTEN-GSK3beta-MOB1 axis controls neurite outgrowth in vitro and in vivo. Cell Mol. Life Sci. 2018;75:4445–4464. doi: 10.1007/s00018-018-2890-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wagner S.A., Beli P., Weinert B.T., Scholz C., Kelstrup C.D., Young C., Nielsen M.L., Olsen J.V., Brakebusch C., Choudhary C. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell Proteom. 2012;11:1578–1585. doi: 10.1074/mcp.M112.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Akimov V., Barrio-Hernandez I., Hansen S.V.F., Hallenborg P., Pedersen A.K., Bekker-Jensen D.B., Puglia M., Christensen S.D.K., Vanselow J.T., Nielsen M.M., et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018;25:631–640. doi: 10.1038/s41594-018-0084-y. [DOI] [PubMed] [Google Scholar]
- 85.Mou F., Praskova M., Xia F., Van Buren D., Hock H., Avruch J., Zhou D. The Mst1 and Mst2 kinases control activation of rho family GTPases and thymic egress of mature thymocytes. J. Exp. Med. 2012;209:741–759. doi: 10.1084/jem.20111692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lignitto L., Arcella A., Sepe M., Rinaldi L., Delle Donne R., Gallo A., Stefan E., Bachmann V.A., Oliva M.A., Tiziana Storlazzi C., et al. Proteolysis of MOB1 by the ubiquitin ligase praja2 attenuates Hippo signalling and supports glioblastoma growth. Nat. Commun. 2013;4:1822. doi: 10.1038/ncomms2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hoa L., Kulaberoglu Y., Gundogdu R., Cook D., Mavis M., Gomez M., Gomez V., Hergovich A. The characterisation of LATS2 kinase regulation in Hippo-YAP signalling. Cell Signal. 2016;28:488–497. doi: 10.1016/j.cellsig.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 88.Cook D., Hoa L.Y., Gomez V., Gomez M., Hergovich A. Constitutively active NDR1-PIF kinase functions independent of MST1 and hMOB1 signalling. Cell Signal. 2014;26:1657–1667. doi: 10.1016/j.cellsig.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiong S., Lorenzen K., Couzens A.L., Templeton C.M., Rajendran D., Mao D.Y.L., Juang Y.C., Chiovitti D., Kurinov I., Guettler S., et al. Structural Basis for Auto-Inhibition of the NDR1 Kinase Domain by an Atypically Long Activation Segment. Structure. 2018;26:1101–1115. doi: 10.1016/j.str.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vrabioiu A.M., Struhl G. Fat/Dachsous Signaling Promotes Drosophila Wing Growth by Regulating the Conformational State of the NDR Kinase Warts. Dev. Cell. 2015;35:737–749. doi: 10.1016/j.devcel.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vichalkovski A., Gresko E., Cornils H., Hergovich A., Schmitz D., Hemmings B.A. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr. Biol. 2008;18:1889–1895. doi: 10.1016/j.cub.2008.10.060. [DOI] [PubMed] [Google Scholar]
- 92.Couzens A.L., Xiong S., Knight J.D.R., Mao D.Y., Guettler S., Picaud S., Kurinov I., Filippakopoulos P., Sicheri F., Gingras A.C. MOB1 Mediated Phospho-recognition in the Core Mammalian Hippo Pathway. Mol. Cell Proteom. 2017;16:1098–1110. doi: 10.1074/mcp.M116.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Manning S.A., Harvey K.F. Warts Opens Up for Activation. Dev. Cell. 2015;35:666–668. doi: 10.1016/j.devcel.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 94.Zhou D., Conrad C., Xia F., Park J.S., Payer B., Yin Y., Lauwers G.Y., Thasler W., Lee J.T., Avruch J., et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gogl G., Schneider K.D., Yeh B.J., Alam N., Nguyen Ba A.N., Moses A.M., Hetenyi C., Remenyi A., Weiss E.L. The Structure of an NDR/LATS Kinase-Mob Complex Reveals a Novel Kinase-Coactivator System and Substrate Docking Mechanism. PLoS Biol. 2015;13:e1002146. doi: 10.1371/journal.pbio.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bichsel S.J., Tamaskovic R., Stegert M.R., Hemmings B.A. Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J. Biol. Chem. 2004;279:35228–35235. doi: 10.1074/jbc.M404542200. [DOI] [PubMed] [Google Scholar]
- 97.Stegert M.R., Tamaskovic R., Bichsel S.J., Hergovich A., Hemmings B.A. Regulation of NDR2 protein kinase by multi-site phosphorylation and the S100B calcium-binding protein. J. Biol. Chem. 2004;279:23806–23812. doi: 10.1074/jbc.M402472200. [DOI] [PubMed] [Google Scholar]
- 98.Dutta S., Mana-Capelli S., Paramasivam M., Dasgupta I., Cirka H., Billiar K., McCollum D. TRIP6 inhibits Hippo signaling in response to tension at adherens junctions. EMBO Rep. 2018;19:337–350. doi: 10.15252/embr.201744777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han H., Qi R., Zhou J.J., Ta A.P., Yang B., Nakaoka H.J., Seo G., Guan K.L., Luo R., Wang W. Regulation of the Hippo Pathway by Phosphatidic Acid-Mediated Lipid-Protein Interaction. Mol. Cell. 2018;72:328–340. doi: 10.1016/j.molcel.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu W., Yang Z., Xie C., Zhu Y., Shu X., Zhang Z., Li N., Chai N., Zhang S., Wu K., et al. PTEN lipid phosphatase inactivation links the hippo and PI3K/Akt pathways to induce gastric tumorigenesis. J. Exp. Clin. Cancer Res. 2018;37:198. doi: 10.1186/s13046-018-0795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mana-Capelli S., McCollum D. Angiomotins stimulate LATS kinase autophosphorylation and act as scaffolds that promote Hippo signaling. J. Biol. Chem. 2018;293:18230–18241. doi: 10.1074/jbc.RA118.004187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Joung J., Engreitz J.M., Konermann S., Abudayyeh O.O., Verdine V.K., Aguet F., Gootenberg J.S., Sanjana N.E., Wright J.B., Fulco C.P., et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548:343–346. doi: 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matsumura S., Hamasaki M., Yamamoto T., Ebisuya M., Sato M., Nishida E., Toyoshima F. ABL1 regulates spindle orientation in adherent cells and mammalian skin. Nat. Commun. 2012;3:626. doi: 10.1038/ncomms1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vizeacoumar F.J., Arnold R., Vizeacoumar F.S., Chandrashekhar M., Buzina A., Young J.T., Kwan J.H., Sayad A., Mero P., Lawo S., et al. A negative genetic interaction map in isogenic cancer cell lines reveals cancer cell vulnerabilities. Mol. Syst. Biol. 2013;9:696. doi: 10.1038/msb.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peters J.M., Tedeschi A., Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 106.Wilmeth L.J., Shrestha S., Montano G., Rashe J., Shuster C.B. Mutual dependence of Mob1 and the chromosomal passenger complex for localization during mitosis. Mol. Biol. Cell. 2010;21:380–392. doi: 10.1091/mbc.e09-06-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Florindo C., Perdigao J., Fesquet D., Schiebel E., Pines J., Tavares A.A. Human Mob1 proteins are required for cytokinesis by controlling microtubule stability. J. Cell Sci. 2012;125:3085–3090. doi: 10.1242/jcs.097147. [DOI] [PubMed] [Google Scholar]
- 108.Rines D.R., Gomez-Ferreria M.A., Zhou Y., DeJesus P., Grob S., Batalov S., Labow M., Huesken D., Mickanin C., Hall J., et al. Whole genome functional analysis identifies novel components required for mitotic spindle integrity in human cells. Genome Biol. 2008;9:R44. doi: 10.1186/gb-2008-9-2-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leng J.J., Tan H.M., Chen K., Shen W.G., Tan J.W. Growth-inhibitory effects of MOB2 on human hepatic carcinoma cell line SMMC-7721. World J. Gastroenterol. 2012;18:7285–7289. doi: 10.3748/wjg.v18.i48.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.O’Donnell L., Panier S., Wildenhain J., Tkach J.M., Al-Hakim A., Landry M.C., Escribano-Diaz C., Szilard R.K., Young J.T., Munro M., et al. The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol. Cell. 2010;40:619–631. doi: 10.1016/j.molcel.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E., et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Adamson B., Smogorzewska A., Sigoillot F.D., King R.W., Elledge S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cotta-Ramusino C., McDonald E.R., 3rd, Hurov K., Sowa M.E., Harper J.W., Elledge S.J. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–1317. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.liO’Neill A.C., Kyrousi C., Einsiedler M., Burtscher I., Drukker M., Markie D.M., Kirk E.P., Gotz M., Robertson S.P., Cappello S. Mob2 Insufficiency Disrupts Neuronal Migration in the Developing Cortex. Front. Cell Neurosci. 2018;12:57. doi: 10.3389/fncel.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cron K.R., Zhu K., Kushwaha D.S., Hsieh G., Merzon D., Rameseder J., Chen C.C., D’Andrea A.D., Kozono D. Proteasome inhibitors block DNA repair and radiosensitize non-small cell lung cancer. PLoS ONE. 2013;8:e73710. doi: 10.1371/journal.pone.0073710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Seoighe C., Scally A. Inference of Candidate Germline Mutator Loci in Humans from Genome-Wide Haplotype Data. PLoS Genet. 2017;13:e1006549. doi: 10.1371/journal.pgen.1006549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou Y., Adolfs Y., Pijnappel W.W., Fuller S.J., Van der Schors R.C., Li K.W., Sugden P.H., Smit A.B., Hergovich A., Pasterkamp R.J. MICAL-1 is a negative regulator of MST-NDR kinase signaling and apoptosis. Mol. Cell Biol. 2011;31:3603–3615. doi: 10.1128/MCB.01389-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Joffre C., Dupont N., Hoa L., Gomez V., Pardo R., Goncalves-Pimentel C., Achard P., Bettoun A., Meunier B., Bauvy C., et al. The Pro-apoptotic STK38 Kinase Is a New Beclin1 Partner Positively Regulating Autophagy. Curr. Biol. 2015;25:2479–2492. doi: 10.1016/j.cub.2015.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bettoun A., Joffre C., Zago G., Surdez D., Vallerand D., Gundogdu R., Sharif A.A., Gomez M., Cascone I., Meunier B., et al. Mitochondrial clearance by the STK38 kinase supports oncogenic Ras-induced cell transformation. Oncotarget. 2016;7:44142–44160. doi: 10.18632/oncotarget.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bothos J., Tuttle R.L., Ottey M., Luca F.C., Halazonetis T.D. Human LATS1 is a mitotic exit network kinase. Cancer Res. 2005;65:6568–6575. doi: 10.1158/0008-5472.CAN-05-0862. [DOI] [PubMed] [Google Scholar]
- 121.Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 122.Vinayagam A., Stelzl U., Foulle R., Plassmann S., Zenkner M., Timm J., Assmus H.E., Andrade-Navarro M.A., Wanker E.E. A directed protein interaction network for investigating intracellular signal transduction. Sci. Signal. 2011;4:rs8. doi: 10.1126/scisignal.2001699. [DOI] [PubMed] [Google Scholar]
- 123.Du X., Wang Q., Hirohashi Y., Greene M.I. DIPA, which can localize to the centrosome, associates with p78/MCRS1/MSP58 and acts as a repressor of gene transcription. Exp. Mol. Pathol. 2006;81:184–190. doi: 10.1016/j.yexmp.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 124.Kim J., Lee J.E., Heynen-Genel S., Suyama E., Ono K., Lee K., Ideker T., Aza-Blanc P., Gleeson J.G. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chiba S., Amagai Y., Homma Y., Fukuda M., Mizuno K. NDR2-mediated Rabin8 phosphorylation is crucial for ciliogenesis by switching binding specificity from phosphatidylserine to Sec15. EMBO J. 2013;32:874–885. doi: 10.1038/emboj.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gardiner K.L., Downs L., Berta-Antalics A.I., Santana E., Aguirre G.D., Genini S. Photoreceptor proliferation and dysregulation of cell cycle genes in early onset inherited retinal degenerations. BMC Genom. 2016;17:221. doi: 10.1186/s12864-016-2477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Goldstein O., Kukekova A.V., Aguirre G.D., Acland G.M. Exonic SINE insertion in STK38L causes canine early retinal degeneration (erd) Genomics. 2010;96:362–368. doi: 10.1016/j.ygeno.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leger H., Santana E., Leu N.A., Smith E.T., Beltran W.A., Aguirre G.D., Luca F.C. Ndr kinases regulate retinal interneuron proliferation and homeostasis. Sci. Rep. 2018;8:12544. doi: 10.1038/s41598-018-30492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lobo G.P., Fulmer D., Guo L., Zuo X., Dang Y., Kim S.H., Su Y., George K., Obert E., Fogelgren B., et al. The exocyst is required for photoreceptor ciliogenesis and retinal development. J. Biol. Chem. 2017;292:14814–14826. doi: 10.1074/jbc.M117.795674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tavares A., Goncalves J., Florindo C., Tavares A.A., Soares H. Mob1: Defining cell polarity for proper cell division. J. Cell Sci. 2012;125:516–527. doi: 10.1242/jcs.096610. [DOI] [PubMed] [Google Scholar]
- 131.Bergmann C., Guay-Woodford L.M., Harris P.C., Horie S., Peters D.J.M., Torres V.E. Polycystic kidney disease. Nat. Rev. Dis Primers. 2018;4:50. doi: 10.1038/s41572-018-0047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Seixas C., Choi S.Y., Polgar N., Umberger N.L., East M.P., Zuo X., Moreiras H., Ghossoub R., Benmerah A., Kahn R.A., et al. Arl13b and the exocyst interact synergistically in ciliogenesis. Mol. Biol. Cell. 2016;27:308–320. doi: 10.1091/mbc.e15-02-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Happe H., van der Wal A.M., Leonhard W.N., Kunnen S.J., Breuning M.H., de Heer E., Peters D.J. Altered Hippo signalling in polycystic kidney disease. J. Pathol. 2011;224:133–142. doi: 10.1002/path.2856. [DOI] [PubMed] [Google Scholar]
- 134.Grampa V., Delous M., Zaidan M., Odye G., Thomas S., Elkhartoufi N., Filhol E., Niel O., Silbermann F., Lebreton C., et al. Novel NEK8 Mutations Cause Severe Syndromic Renal Cystic Dysplasia through YAP Dysregulation. PLoS Genet. 2016;12:e1005894. doi: 10.1371/journal.pgen.1005894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim M., Kim M., Lee M.S., Kim C.H., Lim D.S. The MST1/2-SAV1 complex of the Hippo pathway promotes ciliogenesis. Nat. Commun. 2014;5:5370. doi: 10.1038/ncomms6370. [DOI] [PubMed] [Google Scholar]
- 136.Frank V., Habbig S., Bartram M.P., Eisenberger T., Veenstra-Knol H.E., Decker C., Boorsma R.A., Gobel H., Nurnberg G., Griessmann A., et al. Mutations in NEK8 link multiple organ dysplasia with altered Hippo signalling and increased c-MYC expression. Hum. Mol. Genet. 2013;22:2177–2185. doi: 10.1093/hmg/ddt070. [DOI] [PubMed] [Google Scholar]
- 137.Habbig S., Bartram M.P., Muller R.U., Schwarz R., Andriopoulos N., Chen S., Sagmuller J.G., Hoehne M., Burst V., Liebau M.C., et al. NPHP4, a cilia-associated protein, negatively regulates the Hippo pathway. J. Cell Biol. 2011;193:633–642. doi: 10.1083/jcb.201009069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ma S., Guan K.L. Polycystic kidney disease: A Hippo connection. Genes Dev. 2018;32:737–739. doi: 10.1101/gad.316570.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cai J., Song X., Wang W., Watnick T., Pei Y., Qian F., Pan D. A RhoA-YAP-c-Myc signaling axis promotes the development of polycystic kidney disease. Genes Dev. 2018;32:781–793. doi: 10.1101/gad.315127.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Moya I.M., Halder G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019;20:211–226. doi: 10.1038/s41580-018-0086-y. [DOI] [PubMed] [Google Scholar]
- 141.Shao D.D., Xue W., Krall E.B., Bhutkar A., Piccioni F., Wang X., Schinzel A.C., Sood S., Rosenbluh J., Kim J.W., et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Luo J., Emanuele M.J., Li D., Creighton C.J., Schlabach M.R., Westbrook T.F., Wong K.K., Elledge S.J. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Grant T.J., Mehta A.K., Gupta A., Sharif A.A.D., Arora K.S., Deshpande V., Ting D.T., Bardeesy N., Ganem N.J., Hergovich A., et al. STK38L kinase ablation promotes loss of cell viability in a subset of KRAS-dependent pancreatic cancer cell lines. Oncotarget. 2017;8:78556–78572. doi: 10.18632/oncotarget.20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sasaki H., Kawano O., Endo K., Suzuki E., Yukiue H., Kobayashi Y., Yano M., Fujii Y. Human MOB1 expression in non-small-cell lung cancer. Clin. Lung Cancer. 2007;8:273–276. doi: 10.3816/CLC.2007.n.006. [DOI] [PubMed] [Google Scholar]
- 145.Otsubo K., Goto H., Nishio M., Kawamura K., Yanagi S., Nishie W., Sasaki T., Maehama T., Nishina H., Mimori K., et al. MOB1-YAP1/TAZ-NKX2.1 axis controls bronchioalveolar cell differentiation, adhesion and tumour formation. Oncogene. 2017;36:4201–4211. doi: 10.1038/onc.2017.58. [DOI] [PubMed] [Google Scholar]
- 146.Seve P., Mackey J.R., Isaac S., Tredan O., Souquet P.J., Perol M., Cass C., Dumontet C. cN-II expression predicts survival in patients receiving gemcitabine for advanced non-small cell lung cancer. Lung Cancer. 2005;49:363–370. doi: 10.1016/j.lungcan.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 147.Chen M., Wang M., Xu S., Guo X., Jiang J. Upregulation of miR-181c contributes to chemoresistance in pancreatic cancer by inactivating the Hippo signaling pathway. Oncotarget. 2015;6:44466–44479. doi: 10.18632/oncotarget.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang M., Zhao Y., Zhang Y., Wang D., Gu S., Feng W., Peng W., Gong A., Xu M. LncRNA UCA1 promotes migration and invasion in pancreatic cancer cells via the Hippo pathway. Biochim. Biophys. Acta Mol. Basis. Dis. 2018;1864:1770–1782. doi: 10.1016/j.bbadis.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 149.Nishio M., Sugimachi K., Goto H., Wang J., Morikawa T., Miyachi Y., Takano Y., Hikasa H., Itoh T., Suzuki S.O., et al. Dysregulated YAP1/TAZ and TGF-beta signaling mediate hepatocarcinogenesis in Mob1a/1b-deficient mice. Proc. Natl. Acad. Sci. USA. 2016;113:E71–80. doi: 10.1073/pnas.1517188113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Patel S.H., Camargo F.D., Yimlamai D. Hippo Signaling in the Liver Regulates Organ Size, Cell Fate, and Carcinogenesis. Gastroenterology. 2017;152:533–545. doi: 10.1053/j.gastro.2016.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang Z.X., Wang H.Y., Wu M.C. Identification and characterization of a novel human hepatocellular carcinoma-associated gene. Br. J. Cancer. 2001;85:1162–1167. doi: 10.1054/bjoc.2001.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shirvani-Dastgerdi E., Schwartz R.E., Ploss A. Hepatocarcinogenesis associated with hepatitis B, delta and C viruses. Curr. Opin. Virol. 2016;20:1–10. doi: 10.1016/j.coviro.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goodrum G., Pelchat M. Insight into the Contribution and Disruption of Host Processes during HDV Replication. Viruses. 2018;11 doi: 10.3390/v11010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chung H.Y., Gu M., Buehler E., MacDonald M.R., Rice C.M. Seed sequence-matched controls reveal limitations of small interfering RNA knockdown in functional and structural studies of hepatitis C virus NS5A-MOBKL1B interaction. J. Virol. 2014;88:11022–11033. doi: 10.1128/JVI.01582-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Meistermann H., Gao J., Golling S., Lamerz J., Le Pogam S., Tzouros M., Sankabathula S., Gruenbaum L., Najera I., Langen H., et al. A novel immuno-competitive capture mass spectrometry strategy for protein-protein interaction profiling reveals that LATS kinases regulate HCV replication through NS5A phosphorylation. Mol. Cell Proteom. 2014;13:3040–3048. doi: 10.1074/mcp.M113.028977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shen J., Su J., Wu D., Zhang F., Fu H., Zhou H., Xu M. Growth Inhibition Accompanied by MOB1 Upregulation in Human Acute Lymphoid Leukemia Cells by 3-Deazaneplanocin, A. Biochem. Genet. 2015;53:268–279. doi: 10.1007/s10528-015-9688-7. [DOI] [PubMed] [Google Scholar]
- 157.Galmarini C.M., Cros E., Thomas X., Jordheim L., Dumontet C. The prognostic value of cN-II and cN-III enzymes in adult acute myeloid leukemia. Haematologica. 2005;90:1699–1701. [PubMed] [Google Scholar]
- 158.Suzuki K., Sugawara T., Oyake T., Uchiyama T., Aoki Y., Tsukushi Y., Onodera S., Ito S., Murai K., Ishida Y. Clinical significance of high-Km 5’-nucleotidase (cN-II) mRNA expression in high-risk myelodysplastic syndrome. Leuk. Res. 2007;31:1343–1349. doi: 10.1016/j.leukres.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 159.Meyer J.A., Wang J., Hogan L.E., Yang J.J., Dandekar S., Patel J.P., Tang Z., Zumbo P., Li S., Zavadil J., et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat. Genet. 2013;45:290–294. doi: 10.1038/ng.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bea S., Salaverria I., Armengol L., Pinyol M., Fernandez V., Hartmann E.M., Jares P., Amador V., Hernandez L., Navarro A., et al. Uniparental disomies, homozygous deletions, amplifications, and target genes in mantle cell lymphoma revealed by integrative high-resolution whole-genome profiling. Blood. 2009;113:3059–3069. doi: 10.1182/blood-2008-07-170183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hartmann E.M., Campo E., Wright G., Lenz G., Salaverria I., Jares P., Xiao W., Braziel R.M., Rimsza L.M., Chan W.C., et al. Pathway discovery in mantle cell lymphoma by integrated analysis of high-resolution gene expression and copy number profiling. Blood. 2010;116:953–961. doi: 10.1182/blood-2010-01-263806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kim S.Y., Dunn I.F., Firestein R., Gupta P., Wardwell L., Repich K., Schinzel A.C., Wittner B., Silver S.J., Root D.E., et al. CK1epsilon is required for breast cancers dependent on beta-catenin activity. PLoS ONE. 2010;5:e8979. doi: 10.1371/journal.pone.0008979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Medina-Aguilar R., Perez-Plasencia C., Marchat L.A., Gariglio P., Garcia Mena J., Rodriguez Cuevas S., Ruiz-Garcia E., Astudillo-de la Vega H., Hernandez Juarez J., Flores-Perez A., et al. Methylation Landscape of Human Breast Cancer Cells in Response to Dietary Compound Resveratrol. PLoS ONE. 2016;11:e0157866. doi: 10.1371/journal.pone.0157866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cheung H.W., Cowley G.S., Weir B.A., Boehm J.S., Rusin S., Scott J.A., East A., Ali L.D., Lizotte P.H., Wong T.C., et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc. Natl. Acad. Sci. USA. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kosaka Y., Mimori K., Tanaka F., Inoue H., Watanabe M., Mori M. Clinical significance of the loss of MATS1 mRNA expression in colorectal cancer. Int. J. Oncol. 2007;31:333–338. doi: 10.3892/ijo.31.2.333. [DOI] [PubMed] [Google Scholar]
- 166.Bae J.S., Jeon Y., Kim S.M., Jang J.Y., Park M.K., Kim I.H., Hwang D.S., Lim D.S., Lee H. Depletion of MOB1A/B causes intestinal epithelial degeneration by suppressing Wnt activity and activating BMP/TGF-beta signaling. Cell Death Dis. 2018;9:1083. doi: 10.1038/s41419-018-1138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hong A.W., Meng Z., Guan K.L. The Hippo pathway in intestinal regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 2016;13:324–337. doi: 10.1038/nrgastro.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Schumacher F.R., Al Olama A.A., Berndt S.I., Benlloch S., Ahmed M., Saunders E.J., Dadaev T., Leongamornlert D., Anokian E., Cieza-Borrella C., et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kim E.A., Kim Y.H., Kang H.W., Yoon H.Y., Kim W.T., Kim Y.J., Yun S.J., Moon S.K., Choi Y.H., Kim I.Y., et al. Lower Levels of Human MOB3B Are Associated with Prostate Cancer Susceptibility and Aggressive Clinicopathological Characteristics. J. Korean Med. Sci. 2015;30:937–942. doi: 10.3346/jkms.2015.30.7.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Haldrup C., Mundbjerg K., Vestergaard E.M., Lamy P., Wild P., Schulz W.A., Arsov C., Visakorpi T., Borre M., Hoyer S., et al. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. J. Clin. Oncol. 2013;31:3250–3258. doi: 10.1200/JCO.2012.47.1847. [DOI] [PubMed] [Google Scholar]
- 171.Gerlinger M., Santos C.R., Spencer-Dene B., Martinez P., Endesfelder D., Burrell R.A., Vetter M., Jiang M., Saunders R.E., Kelly G., et al. Genome-wide RNA interference analysis of renal carcinoma survival regulators identifies MCT4 as a Warburg effect metabolic target. J. Pathol. 2012;227:146–156. doi: 10.1002/path.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F., Pantel S.E., et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Behan F.M., Iorio F., Picco G., Goncalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511–516. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 174.Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M., et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–576. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kim Y.R., Kim S.Y. Machine learning identifies a core gene set predictive of acquired resistance to EGFR tyrosine kinase inhibitor. J. Cancer Res. Clin. Oncol. 2018;144:1435–1444. doi: 10.1007/s00432-018-2676-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fang K.M., Liu Y.Y., Lin C.H., Fan S.S., Tsai C.H., Tzeng S.F. Mps one binder 2 gene upregulation in the stellation of astrocytes induced by cAMP-dependent pathway. J. Cell Biochem. 2012;113:3019–3028. doi: 10.1002/jcb.24180. [DOI] [PubMed] [Google Scholar]
- 177.Satoh J., Kawana N., Yamamoto Y. Pathway Analysis of ChIP-Seq-Based NRF1 Target Genes Suggests a Logical Hypothesis of their Involvement in the Pathogenesis of Neurodegenerative Diseases. Gene. Regul. Syst. Bio. 2013;7:139–152. doi: 10.4137/GRSB.S13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Olah J., Vincze O., Virok D., Simon D., Bozso Z., Tokesi N., Horvath I., Hlavanda E., Kovacs J., Magyar A., et al. Interactions of pathological hallmark proteins: Tubulin polymerization promoting protein/p25, beta-amyloid, and alpha-synuclein. J. Biol. Chem. 2011;286:34088–34100. doi: 10.1074/jbc.M111.243907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Blondeau C., Gaillard S., Ternaux J.P., Monneron A., Baude A. Expression and distribution of phocein and members of the striatin family in neurones of rat peripheral ganglia. Histochem. Cell Biol. 2003;119:131–138. doi: 10.1007/s00418-003-0503-x. [DOI] [PubMed] [Google Scholar]
- 180.Li D., Musante V., Zhou W., Picciotto M.R., Nairn A.C. Striatin-1 is a B subunit of protein phosphatase PP2A that regulates dendritic arborization and spine development in striatal neurons. J. Biol. Chem. 2018;293:11179–11194. doi: 10.1074/jbc.RA117.001519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lee P.C., Machner M.P. The Legionella Effector Kinase LegK7 Hijacks the Host Hippo Pathway to Promote Infection. Cell Host. Microbe. 2018;24:429–438. doi: 10.1016/j.chom.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ahola-Olli A.V., Wurtz P., Havulinna A.S., Aalto K., Pitkanen N., Lehtimaki T., Kahonen M., Lyytikainen L.P., Raitoharju E., Seppala I., et al. Genome-wide Association Study Identifies 27 Loci Influencing Concentrations of Circulating Cytokines and Growth Factors. Am. J. Hum. Genet. 2017;100:40–50. doi: 10.1016/j.ajhg.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Liu C., Batliwalla F., Li W., Lee A., Roubenoff R., Beckman E., Khalili H., Damle A., Kern M., Furie R., et al. Genome-wide association scan identifies candidate polymorphisms associated with differential response to anti-TNF treatment in rheumatoid arthritis. Mol. Med. 2008;14:575–581. doi: 10.2119/2008-00056.Liu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zengini E., Hatzikotoulas K., Tachmazidou I., Steinberg J., Hartwig F.P., Southam L., Hackinger S., Boer C.G., Styrkarsdottir U., Gilly A., et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat. Genet. 2018;50:549–558. doi: 10.1038/s41588-018-0079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Almoguera B., Vazquez L., Mentch F., Connolly J., Pacheco J.A., Sundaresan A.S., Peissig P.L., Linneman J.G., McCarty C.A., Crosslin D., et al. Identification of Four Novel Loci in Asthma in European American and African American Populations. Am. J. Respir Crit Care Med. 2017;195:456–463. doi: 10.1164/rccm.201604-0861OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Warner N., Burberry A., Pliakas M., McDonald C., Nunez G. A genome-wide small interfering RNA (siRNA) screen reveals nuclear factor-kappaB (NF-kappaB)-independent regulators of NOD2-induced interleukin-8 (IL-8) secretion. J. Biol. Chem. 2014;289:28213–28224. doi: 10.1074/jbc.M114.574756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nishio M., Miyachi Y., Otani J., Tane S., Omori H., Ueda F., Togashi H., Sasaki T., Mak T.W., Nakao K., et al. Hippo pathway controls cell adhesion and context-dependent cell competition to influence skin engraftment efficiency. FASEB J. 2019 doi: 10.1096/fj.201802005R. [DOI] [PubMed] [Google Scholar]
- 188.Goto H., Nishio M., To Y., Oishi T., Miyachi Y., Maehama T., Nishina H., Akiyama H., Mak T.W., Makii Y., et al. Loss of Mob1a/b in mice results in chondrodysplasia due to YAP1/TAZ-TEAD-dependent repression of SOX9. Development. 2018;145 doi: 10.1242/dev.159244. [DOI] [PubMed] [Google Scholar]
- 189.de Miguel-Yanes J.M., Shrader P., Pencina M.J., Fox C.S., Manning A.K., Grant R.W., Dupuis J., Florez J.C., D’Agostino R.B., Cupples L.A., et al. Genetic risk reclassification for type 2 diabetes by age below or above 50 years using 40 type 2 diabetes risk single nucleotide polymorphisms. Diabetes Care. 2011;34:121–125. doi: 10.2337/dc10-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hanson R.L., Guo T., Muller Y.L., Fleming J., Knowler W.C., Kobes S., Bogardus C., Baier L.J. Strong parent-of-origin effects in the association of KCNQ1 variants with type 2 diabetes in American Indians. Diabetes. 2013;62:2984–2991. doi: 10.2337/db12-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Cui X., Guo Z., Song L., Wang Y., Cheng Y. NCP1/AtMOB1A Plays Key Roles in Auxin-Mediated Arabidopsis Development. PLoS Genet. 2016;12:e1005923. doi: 10.1371/journal.pgen.1005923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Xiong J., Cui X., Yuan X., Yu X., Sun J., Gong Q. The Hippo/STE20 homolog SIK1 interacts with MOB1 to regulate cell proliferation and cell expansion in Arabidopsis. J. Exp. Bot. 2016;67:1461–1475. doi: 10.1093/jxb/erv538. [DOI] [PubMed] [Google Scholar]