Abstract
Orthogonal confirmation of next-generation sequencing (NGS)-detected germline variants is standard practice, although published studies have suggested that confirmation of the highest-quality calls may not always be necessary. The key question is how laboratories can establish criteria that consistently identify those NGS calls that require confirmation. Most prior studies addressing this question have had limitations: they have been generally of small scale, omitted statistical justification, and explored limited aspects of underlying data. The rigorous definition of criteria that separate high-accuracy NGS calls from those that may or may not be true remains a crucial issue. We analyzed five reference samples and over 80,000 patient specimens from two laboratories. Quality metrics were examined for approximately 200,000 NGS calls with orthogonal data, including 1662 false positives. A classification algorithm used these data to identify a battery of criteria that flag 100% of false positives as requiring confirmation (CI lower bound, 98.5% to 99.8%, depending on variant type) while minimizing the number of flagged true positives. These criteria identify false positives that the previously published criteria miss. Sampling analysis showed that smaller data sets resulted in less effective criteria. Our methodology for determining test- and laboratory-specific criteria can be generalized into a practical approach that can be used by laboratories to reduce the cost and time burdens of confirmation without affecting clinical accuracy.
The use of orthogonal assays (eg, Sanger sequencing) to confirm variants identified with next-generation sequencing (NGS) is standard practice in many laboratories to reduce the risk for delivering false-positive (FP) results. Clinical NGS tests can inform significant medical decisions,1, 2 and therefore confirmation is recommended by medical practice guidelines,3, 4 although the details are generally left up to the laboratory.3, 5 Because clinical NGS methods often emphasize sensitivity (to avoid missing clinically important variants), FP rates can be elevated compared with those in research NGS.6 Moreover, pathogenic variants are often technically challenging (eg, many are located within repetitive or complex regions), which can further increase FP rates.7, 8, 9 Confirmation assays have a monetary cost, however, and also increase the time needed to deliver results, a critical factor in many clinical situations.
Published studies examining this issue have concluded that confirmation of the highest-quality NGS calls may not always be necessary.10, 11, 12, 13, 14 Some of these studies10, 12 propose specific criteria for separating high-confidence true-positive (TP) variant calls from those that are possibly FPs. These criteria differ from those used in filtering, the separate process of removing calls confidently believed to be false or unsupportable. The remaining intermediate-confidence calls are those that benefit from confirmatory assays, additional data review, or both to determine which are TPs and which are FPs. Unfortunately, these prior studies are generally small, in some cases proposing criteria using only one or five example FPs (Table 1). The presence of few FPs may seem reassuring but leads to significant limitations in these studies. First, because their statistical power to characterize the FP population is limited, these studies do not address the question of whether future FPs are likely to resemble the few observed in the study. Quite possibly, additional FPs could be different and thus missed by the proposed criteria. Second, most of these studies use identical data sets for training and evaluating the proposed criteria, likely making the results subject to overfitting.15 Third, few of these studies provide statistical justification. Finally, all of these prior studies use data from individual laboratories and do not examine whether the methodologies can be generalized.
Table 1.
Summary of the Data Sets
| Source | Variant type | Samples | Unique variants | Variant calls | TPs | FPs | FDR, % | Total calls | Total FPs | FP sensitivity, % | CI lower bound, % |
|---|---|---|---|---|---|---|---|---|---|---|---|
| This study: Lab 1 | SNVs | GIAB | 27,202 | 136,146 | 135,945 | 201 | 0.15 | ||||
| Patients∗† | 2840 | 3699 | 3689 | 10 | 0.27 | 139,845 | 211 | 100 | 98.9 | ||
| Indels | GIAB | 3715 | 15,574 | 14,594 | 980 | 6.29 | |||||
| Patients∗† | 1749 | 2274 | 2262 | 12 | 0.53 | 17,848 | 992 | 100 | 99.8 | ||
| This study: Lab 2 | SNVs | GIAB | 5816 | 29,148 | 29,110 | 38 | 0.13 | ||||
| Patients∗‡ | 4359 | 4934 | 4804 | 130 | 2.63 | 34,082 | 168 | 100 | 98.5 | ||
| Indels | GIAB | 1185 | 3617 | 3343 | 274 | 7.58 | |||||
| Patients∗ | 267 | 389 | 372 | 17 | 4.37 | 4006 | 291 | 100 | 99.1 | ||
| Strom et al10 | SNVs | Patients§¶ | - | 108 | 107 | 1 | 0.93 | 108 | 1 | 100 | 5.1 |
| Baudhuin et al11 | SNVs | Patients‖ | 380 | 797 | 797 | 0 | 0 | ||||
| 1KG‖∗∗ | 736 | 736 | 736 | 0 | 0 | 1533 | 0 | N/A | N/A | ||
| Indels | Patients‖†† | 63 | 122 | 122 | 0 | 0 | |||||
| 1KG‖∗∗ | 26 | 26 | 26 | 0 | 0 | 148 | 0 | N/A | N/A | ||
| Mu et al12 | SNVs | Patients¶ | - | 6912 | 6818 | 94 | 1.36 | 6912 | 94 | 100 | 97.4 |
| Indels | Patients¶ | - | 933 | 928 | 5 | 0.54 | 933 | 5 | 100 | 62.1 | |
| van den Akker et al14 | SNVs | Patients¶‡‡ | 3044 | 5829 | 5524 | 305 | 5.23 | 5829 | 305 | 100 | 99.2 |
| Indels | Patients¶‡‡ | 526 | 1350 | 1142 | 208 | 15.41 | 1350 | 208 | 100 | 98.8 |
1KG, 1000 Genomes Project; FDR, false discovery rate [calculated as FPs/(FPs + TPs)]; FP, false positive; FP sensitivity, the fraction of FPs captured using the study's proposed criteria; GIAB, Genome in a Bottle Consortium; Indel, insertion or deletion; SNV, single-nucleotide variant; unique variant, a particular alteration which may be present in one or more individuals; TP, true positive.
Clinical and GIAB data were combined in the final analysis. GIAB data included on- and off-target calls.
Manual review removed certain FPs (particularly indels) and thus reduced FDRs in the Lab 1 clinical data.
Many of the clinical FPs were systematic errors in the OTOA and CFTR genes, which were tested in many patients.
The authors did not provide a count of unique variants. For the van den Akker study, it was calculated from the data provided.
CIs were calculated based on data from the publication. No such statistics were provided by the study authors.
The lack of FPs may have been a result of aggressive filtering, which can remove clinical TPs as well as FPs.
Only unique 1KG variants were analyzed. The results from the updated 1KG data mentioned in this article are described.
Most of the indels in patients were intronic and homopolymer associated. These are generally not clinically significant.
The relatively high FDRs in this data set may have been a result of under-filtering, which can also affect CIs.
We examined the role of confirmation using a set of variant calls much larger than those published previously. Our own sequences of five reference samples characterized by the Genome in a Bottle Consortium (GIAB)16, 17, 18 were combined with confirmatory data from over 80,000 clinical tests. The methodology was applied in two clinical laboratories that use similar but not identical NGS methods. Similar to prior studies, high-confidence NGS calls that do not benefit from confirmation were identified. However, a battery of criteria was found to be necessary to capture all FPs, in contrast with the one or two metrics used by most of the prior studies. Indeed, the specific criteria proposed in prior studies10, 11, 12 miss FPs in our much larger data set. Observations of a variant as a TP were found to say little about its likelihood of being an FP in a different sample or NGS run, which indicates that prior confirmations can be an ineffective quality metric. Approaches such as ours can be used by any clinical laboratory to provide efficient, effective, and statistically justified criteria for prioritizing variant calls for confirmation.
Materials and Methods
Eight component data sets from two laboratories (Table 1) were compiled following the process illustrated in Figure 1. Key aspects of the used methodology are summarized in Table 2 and are detailed here and in the Discussion. In addition to results obtained through clinical testing, five GIAB DNA specimens were sequenced: NA12878, NA24385, NA24143, NA24149, and NA24631 (Coriell Institute, Camden, NJ). Replicates of the GIAB samples were included. NGS was performed on both the GIAB and the clinical specimens by each laboratory using Illumina (San Diego, CA) 2 × 150 bp reads as described previously.8, 19, 20 Seven (Lab 1) and three (Lab 2) custom hybridization–based assays were used, each targeting 100 to 1000 genes. Clinically reportable target regions included, with some exceptions, protein-coding exons plus immediate flanking sequences (10 to 20 bp on each side). Mean coverage across targets was 300 to 1000× or more, depending on the sample, assay, and laboratory. All data used in this study passed stringent quality control at the sample and run levels.
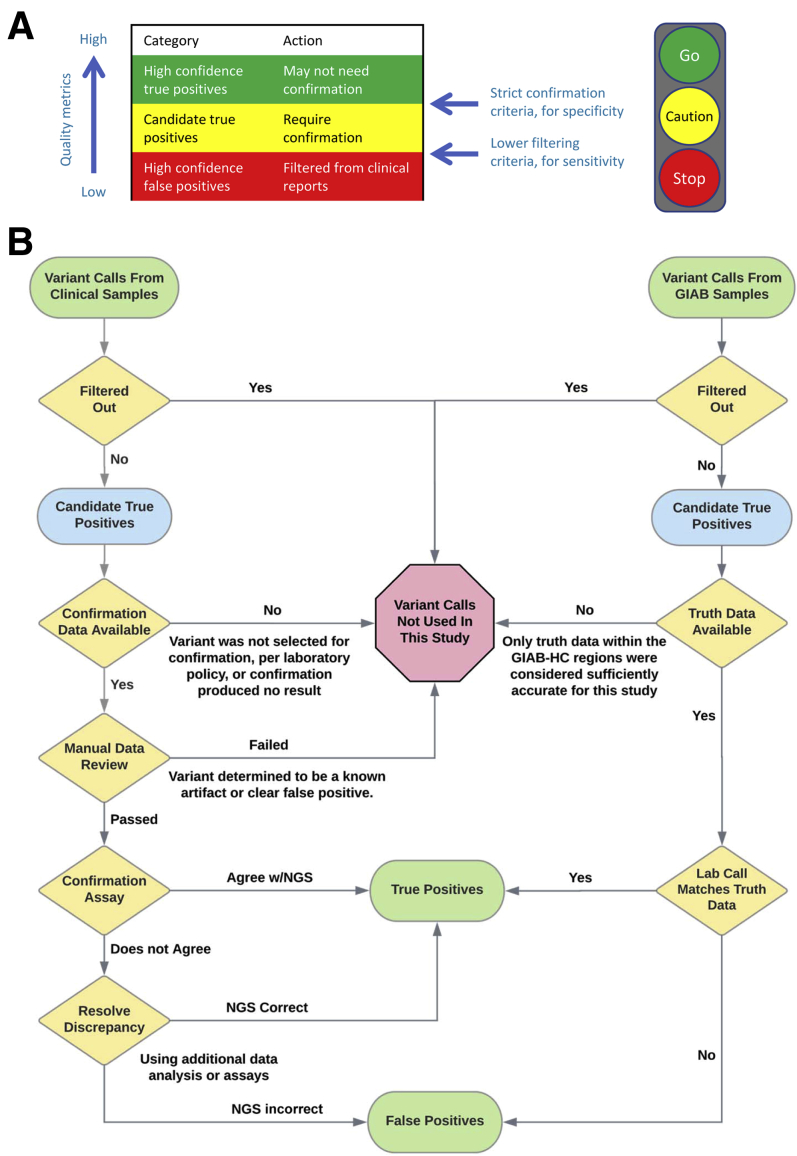
Figure 1.
Study methodology. A: Variant calls can be classified as high-confidence true positive (green) and intermediate-confidence (yellow), using strict thresholds intended to maximize the specificity of the high-confidence set. The intermediate set will contain a mixture of true- and false-positive calls. The study objective was to rigorously determine test-specific criteria that distinguish these two categories. Variant calls that are confidently false positives (red) are typically filtered out using different criteria that emphasize sensitivity. The analogy to a traffic light is illustrative. B: Process for collecting true positive and false positive variant calls for both the clinical and Genome in a Bottle Consortium (GIAB) specimens. Each laboratory's clinically validated next-generation sequencing (NGS) assays, bioinformatics pipelines, and filtering criteria were used for both specimen types. Single-nucleotide variants and indels were collected as shown. Copy number and structural variants were excluded from this study, as were any variants with an unknown confirmation status. Filtering and manual review processes were designed to remove clearly false variant calls but not those considered even potentially true. Manual review was used only with the Lab 1 clinical data. HC, high-confidence.
Table 2.
Key Aspects of Methodology Used in This Study
| Aspect of methodology used | Rationale (see text for details) |
|---|---|
| Large data sets used for both SNVs and indels | Provides confidence in the resulting criteria and helps minimize overfitting. Appropriate sizes determined by CI calculations (below). |
| Both clinical and reference (GIAB) samples used | Greatly increases the size and diversity of the data sets, particularly in FPs. |
| Same quality filtering thresholds as used in clinical practice | Confirmation criteria can depend on filtering criteria. Using lower filters would add many FPs to the study but could result in the selection of ineffective or biased confirmation criteria. |
| Separate filtering and confirmation thresholds used | Allows high sensitivity (by keeping variants of marginal quality) and high specificity (by subjecting these variants to confirmation). |
| On- and off-target variant calls analyzed in GIAB samples | Further increases the data set size and diversity. Off-target calls were subject to the same quality filters as were on-target calls. |
| Indels and SNVs analyzed separately | Indels and SNVs can have different quality determinants. An adequate population of each was required to achieve statistical significance. |
| Partial matches considered FPs | Zygosity errors and incorrect diploid genotypes do occur and can be as clinically important as “pure” FPs. |
| Algorithm selects criteria primarily by their ability to flag FPs | Other algorithms equally value classification of TPs, which may result in biased criteria, particularly as TPs greatly outnumber FPs. |
| Multiple quality metrics used to flag possible FPs | Various call-specific metrics (eg, quality scores, read depth, allele balance, strand bias) and genomic annotations (eg, repeats, segmental duplications) proved crucial. |
| Key metric: fraction of FPs flagged (FP sensitivity) | Other metrics, including test PPV and overall classifier accuracy, can be uninformative or misleading in the evaluation of confirmation criteria. |
| Requirement of 100% FP sensitivity on training data | Clinically appropriate. Resulting criteria will be effective on any subset of the training data (clinical or GIAB, on-target or off-target, etc.) |
| Statistical significance metric: CI on FP sensitivity | Rigorously indicates validity of the resulting criteria: eg, flagging 100% of 125 FPs demonstrates ≥98% FP sensitivity at P = 0.05. Smaller data sets (eg, 50 FPs) resulted in ineffective criteria. |
| Separate training and test sets used (cross validation) | In conjunction with large data sets, cross-validation is a crucial step to avoid overfitting, which can otherwise result in ineffective criteria. |
| Prior confirmation of a variant was not used as a quality metric | Successful confirmations of a particular variant can indicate little about whether future calls of that same variant are true or false. |
| All variants outside of GIAB-HC regions require confirmation | Outside of these regions, too few confirmatory data are available to prove whether the criteria are effective. |
| Laboratory- and workflow-specific criteria | Effective confirmation criteria can vary based on numerous details of a test's methodology and its target genes. Changes can necessitate revalidation of confirmation criteria. |
FP, false positive; GIAB, Genome in a Bottle; GIAB-HC, regions in which high-confidence truth data are available from the GIAB specimens (unrelated to confidence in our own calls or to on/off-target regions); indel, insertion or deletion; PPV, positive predictive value; SNV, single-nucleotide variant; TP, true positive.
Both laboratories' bioinformatics pipelines have been described previously,8, 19, 20 although after publishing those descriptions, the laboratories implemented the Genome Analysis Toolkit (GATK) software Haplotype Caller21, 22 version 3.6 (Lab 1) or 3.7 (Lab 2). Lab 1 used a battery of criteria (Supplemental Table S1) to filter out clearly erroneous variants and to generate warnings on other variants that received manual review. Variants that failed review were also removed. To deliver high sensitivity, this process was conservative: Variants considered possibly true, despite warnings (eg, relatively low depth or allele balance) were subjected to confirmation. Compared with Lab 1, Lab 2 used simpler filters (quality-depth score <4 and Fisher strand bias score >40) and limited manual review, resulting in a broader selection of variants being subjected to confirmation. Copy number and structural variants were excluded. As is typical for hybridization-based NGS, regions neighboring clinical targets received read coverage. In the GIAB specimens, variant calls within these off-target regions were used as long as they passed the same quality filters as on-target calls and were within a set distance of a target (300 bp for Lab 1 and 50 bp for Lab 2; parameters established in the clinical pipelines and not changed for this study). This requirement prevented large numbers of very low–coverage calls from being considered, although the quality filters remove most such calls in any case. In the clinical specimens, off-target calls were used because confirmatory data were unavailable.
Confirmation of clinical samples was performed by Lab 1 using Sanger (Thermo Fisher Scientific, Waltham, MA) or PacBio (Pacific Biosciences, Menlo Park, CA) amplicon sequencing. Lab 1 validated23 the PacBio circular consensus sequencing method24 specifically for use in confirmation. This method provides high accuracy25 and has been successfully applied in other clinical genetics testing.26 Lab 2 used only Sanger confirmation. When the results of a confirmation assay and NGS disagreed, both results were manually reviewed, and if the reason for the disagreement was unclear, additional rounds of confirmation using different primers or assays were performed. There were few putatively mosaic variants in this study; those present were considered TPs if confirmed and FPs if refuted by an adequately sensitive assay. Variants for which confirmation could not produce a confident answer (TP or FP) were not used in this study.
The reference calls from GIAB18 software version 3.3.2 were used as truth data to confirm variants identified by each laboratory's sequencing of the GIAB samples. Manual review of these samples was not performed. VCFeval27 software version 3.7.1 was used to compare each laboratory's calls to the GIAB truth data to determine which laboratory calls were TPs and which were FPs. VCFeval can match variant calls even when the same diploid sequence is represented in different ways (spelling differences), an important factor in comparing insertions and deletions (indels) and complex variants.27, 28, 29 VCFeval also detects partial matches—that is, zygosity errors (falsely calling a homozygous variant as heterozygous or vice versa) or heterozygous sites at which one of the called alleles is correct and one is not. These cases, reported by VCFeval as “FP_CA,” were considered FPs in this study (see Discussion). This study did not examine false negatives in detail.
The analysis of the five GIAB specimens was restricted to the sample-specific high-confidence regions (GIAB-HC), which are annotated by the GIAB consortium to indicate where their reference data have high accuracy.18 The GIAB-HC regions span 88% to 90% of the genome of each GIAB sample and cover most exons, introns, and intergenic regions, an improvement compared with older versions of the GIAB reference data16 for which there was a greater bias toward “easy” regions.30 The GIAB-HC designation is unrelated to any quality assessment of data produced by our laboratories. Indeed, the NGS assays produced both high- and low-quality variant calls within and outside the GIAB-HC regions. The GIAB-HC designation is also unrelated to whether calls were on- or off-target: Most (not all) of the clinical targets were within the GIAB-HC regions, as were most off-target calls. Because the majority of the confirmatory data lay within the GIAB-HC regions, however, our confirmation criteria were selected by focusing within these regions (see Discussion). Doing so required extrapolating the GIAB-HC regions to patient specimens, for which the union of the five GIAB-HC files was used—that is, if a region was considered GIAB-HC in any of the five GIAB specimens, it was considered GIAB-HC in all patients. This approach prevented specific low-confidence calls in the reference data of a particular GIAB specimen(s) from inappropriately annotating that site as low confidence in general.
Approximately two dozen quality metrics (Supplemental Table S2) were examined individually. The most useful metrics included different ways of measuring read depth (Supplemental Table S2), allele balance for heterozygous and homozygous calls, multiple quality scores, various indicators of strand bias, aspects of the variant call itself, and aspects of the genomic context. We manually chose candidate thresholds for each quantitative metric. Both the metrics and thresholds could be specific to a laboratory and variant type, but usually were not. Metrics for each variant call were then turned into discrete flags.
To delineate technically challenging genomic regions, stratification BED (browser extensible data) files produced by the Global Alliance for Genomics and Health Benchmarking Workgroup were used.28 These regions were padded by 10 bp on each side to ensure that all affected positions were appropriately annotated. These BED files were grouped as follows: i) repeats combined homopolymers and short tandem repeats, ii) segdups (segmental duplications) included larger regions with homologous copies in the GRCh37 reference genome, and iii) unmappable regions were those in which the NGS reads could not map uniquely. Specific definitions are provided in Supplemental Table S2. The unmappable and segdup regions largely overlapped—but both were generally distinct from the (short) repeats.
These data were passed into a heuristic algorithm that selected a combination of flags for the final battery of criteria. The Python version 2.7 code for this algorithm is available (https://github.com/slincoln/flagem). Details of the algorithm are provided in Supplemental Appendix S1.
CIs were computed at 95% using the Jeffreys method. Both the Wilson score method and the tolerance interval method (as mentioned in the guidelines from the Association for Molecular Pathology and the College of American Pathologists29, 31) are included in Figure 2 for comparison.
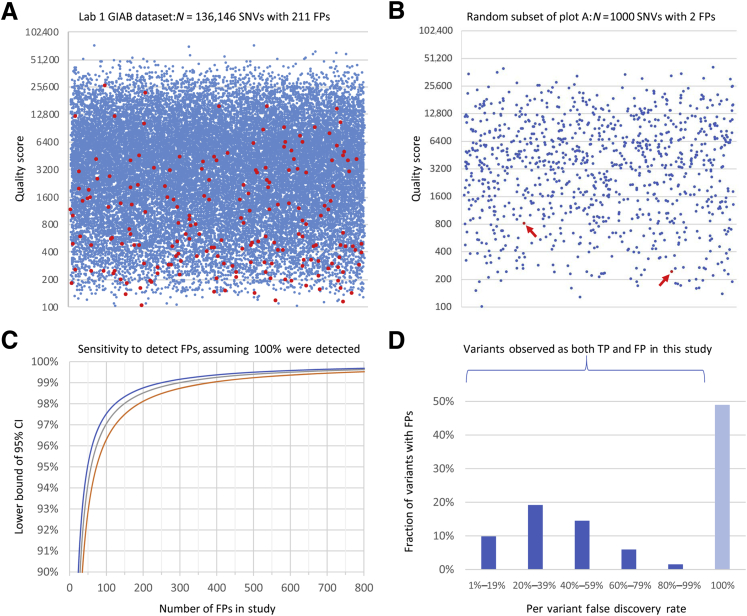
Figure 2.
Individual quality metric results and statistics. A: Variant call quality scores (QUAL, y axis) for true-positive (TP; blue) and false-positive (FP; red) single-nucleotide variant (SNV) calls in the Lab 1 Genome in a Bottle Consortium (GIAB) data set. The x axis position of each point is randomly assigned. To make density changes visible, a random selection of one-fifth of the TP calls was plotted along with all FPs. In the large data set, some FPs had quite high QUAL scores, demonstrating that this metric is inadequate alone. Corresponding histograms using the full data set without down-sampling are shown in Supplemental Figure S1. B: Random sample of 1000 data points from the same data set plotted in A. All points are displayed. Arrows indicate the two FPs present. One thousand such random samples were generated, and compared with the full data set in A, many would lead to quite different conclusions about the effectiveness of QUAL thresholds. C: Lower bound of the 95% CI on the fraction of FPs flagged (y axis) as a function of the number of FPs used to determine criteria (x axis) assuming that 100% success is observed. The y axis range is 90% to 100%. This calculation used the Jeffreys method (blue), the Wilson score method (red), and the tolerance interval method (gray). All methods produce generally similar results and indicate the validity of any study such as this. For example, using the Jeffreys method, flagging 49 of 49 FPs shows 100% effectiveness, with a CI of 95% to 100%. Many prior studies did not achieve this level of statistical significance (Table 1). Consistent with these CI calculations, small data sets indeed resulted in ineffective criteria (see Results). D: Histogram of per-variant false-discovery rates (FDRs; x axis) for all variants that were observed more than once in the Lab 1 data set, and for which one or more of those calls was an FP. SNVs and insertions and deletions (indels) are combined. An FDR of 100% indicates a fully systematic FP (insofar as we can measure); an FDR of 0% indicates a consistent TP (not shown in this graph). Each unique variant (ie, a genetic alteration that may be present in multiple individuals) is counted once. The y axis range is 0% to 50% of variants. Approximately half of all variants that were FPs were also correctly called as TPs in a different specimen(s) or run(s). Examples of this were observed in both the clinical and the GIAB specimens and included both SNVs and indels. Lab 2 results were similar. Many of these variants have low per-variant FDRs, which usually but not always are correctly called. Repeated TP observations of such a variant provide little information about the accuracy of any following observation of that same variant. This study was underpowered to measure FDRs near 0% or 100%, and many more of these variants may exist than are shown here.
Results
The data sets included almost 14,000 NGS variant calls subjected to confirmation during clinical testing, and more than 184,000 calls with high-quality truth data from the GIAB samples (Table 1). A total of 1662 FPs were observed. To initially characterize the data, the false-discovery rate (FDR) of calls in each set was calculated [FPs/(FPs + TPs)]. Note that FDR is 1 minus the analytic positive predictive value (FDR is also called positive percent agreement), a metric recommended in the guidelines from the Association for Molecular Pathology and the College of American Pathologists, which consider it preferable to specificity (ie, the specific calculation) for describing multigene sequencing tests.29 As expected, the FDR of indels was considerably higher than that of single-nucleotide variants (SNVs), and manual review had reduced FDRs in the Lab 1 clinical data compared with those in the GIAB data. FDRs in the GIAB samples were comparable between laboratories.
Analysis of Individual Quality Metrics from Lab 1
A variety of metrics (Supplemental Tables S2 and S3, Supplemental Figures S1–S4) were examined to determine which were informative for identifying (or flagging) FPs. SNVs and indels were analyzed separately, as were the GIAB and clinical samples. For each metric and threshold, the fraction of FPs and TPs flagged—these indicate the value and cost, respectively, of each potential flag—were computed. FDR, which indicates how likely flagged variants are to be FPs, was also computed. A number of informative metrics were identified, although no single metric proved adequate alone. For example, consider the quality score (QUAL) for SNVs in the GIAB data (Figure 2, A and B). This metric was used by Strom et al10 to identify SNVs that require confirmation, although Strom had only a single FP to use in defining thresholds (Supplemental Figure S5). In the much larger data set, no threshold value for QUAL could be chosen that flagged all, or even most, of the FPs without also flagging many TPs. Indeed, some FPs had quite high QUAL scores. A QUAL cutoff of 800, for example, flagged only 56% of FPs but also flagged 17% of TPs. These flagged variants had an FDR of 0.28%, indicating that this subset was not highly enriched for errors compared with the overall FDR of 0.15%. Indels fared similarly: 55% of FPs and 18% of TPs had a QUAL of <800, with an FDR of 16% compared with the baseline rate of 6.3%.
This observation was also true of the clinical data set, in which 39% of FP SNVs and 2.9% of TPs had a QUAL of <800. These variants had a 3.6% FDR compared with 0.16% overall—20-fold enrichment—although most low-QUAL calls were still TPs. There is little precision in these measurements, however, because the clinical data set contained only 10 FP SNVs, and only 4 had QUAL<800. Flagging all 10 FP SNVs required a QUAL threshold of 6400 (using logarithmic binning), which flagged almost 30% of TPs. Overfitting would certainly be an issue when analyzing the clinical data set alone: If the single FP SNV with the highest QUAL score (5516) had been absent from these data, then thresholds would be driven by the next, much lower, score (2567). There is little statistical confidence in any such threshold: If 10 of 10 FPs are flagged, the point estimate is 100% FP sensitivity, but the lower bound of the CI is only 78% (Figure 2C), demonstrating that thresholds determined using such a small data set could miss many FPs.
Other metrics were similarly analyzed; allele balance was found to be the most informative, followed by strand bias, the presence of nearby variant calls, and certain characteristics of the variant itself (particularly, whether it was het-alt, meaning a heterozygous call in which neither allele is present in the GRCh37 reference genome). All of these criteria had limitations, however. Some provided strong indications of a call being an FP but flagged few such variants—strand bias was one example. More commonly, these criteria captured many TPs in addition to FPs yet still missed FPs at useful thresholds, similar to QUAL.
More than 80% of the Lab 1 GIAB variant calls that were analyzed were off target. These calls all passed the standard quality filters (Supplemental Table S1) and most had high read depth (>50×; Supplemental Table S3). Despite passing QC, some calls had relatively low coverage (10× to 50×) or were in complex regions (repeats, high GC%, etc). Such issues are present but less common within clinical targets, and including these additional examples allowed thresholds to be established in a data-based manner. Nonetheless, the off-target data appeared representative: Indel FDRs were similar both on and off target (5.3% vs. 6.4%, respectively). SNV FDRs differed somewhat (0.04% vs. 0.17%) but remained low both on and off target. Repeats accounted for half of the GIAB indels (both on and off target) and accounted for approximately 15% of the clinical indels. Repeats also accounted for many of the indel FPs across the study, although the vast majority of repeat-associated calls were correct (TPs), reflecting the high genetic variability of these sequences.
Of the variants observed more than once, approximately half of those with at least one FP call were also correctly called as TPs in one or more different run(s) (Figure 2D). This was true for both SNVs and indels. It was even sometimes true across replicates of the same sample, a situation made possible because partial match errors were considered FPs in this analysis (see Materials and Methods). Many examples were found in the GIAB specimens, a result attributed to the increased power to observe such variants in these data (Supplemental Appendix S1). Clinical examples were observed as well. Case-by-case review suggests that the root causes of this behavior are varied and sometimes complex—partial match errors accounted for approximately half of these cases. Given the limited power to detect such variants, many more may exist. Historical confirmation performance may be an ineffective quality metric in these data sets.
Combining Metrics
Because no single metric proved adequate, it was investigated whether using multiple metrics might be more effective. One precedent of this approach is a study that suggested that requiring a depth of >100 and an allele balance between 40% and 60% would identify variants that do not require confirmation.12 In our data set, which was much larger than the one in the study by Mu et al,12 this was not the case: 29 FP indels and 7 FP SNVs failed to be flagged as requiring confirmation using Mu's criteria. Nonetheless, a larger battery of metrics might prove effective.
A heuristic algorithm was developed to explore this hypothesis. Briefly, this algorithm incrementally adds flags to a proposed set with the primary aim of capturing 100% of FPs using the combination of flags. This algorithm secondarily prioritizes minimizing the number of TPs also captured. Criteria were separately chosen for SNVs and indels. Variants in the GIAB and clinical samples were combined in this analysis to increase both the number and diversity of FPs available. This analysis was restricted to the GIAB-HC regions because of the limited amount of confirmatory data outside of these regions (see Discussion). To minimize overfitting, Monte Carlo cross-validation—running the algorithm hundreds of times—in each iteration, choosing flags (training) using two-thirds of the data (chosen at random) and testing these flags using the remaining one-third, was performed. The flags selected by each iteration and the specific variants that led to differences among iterations (ie, depending on whether the variant was randomly assigned to the training or the test set) were examined. This review highlights the importance of particular implementation details of our algorithm (Supplemental Appendix S1).
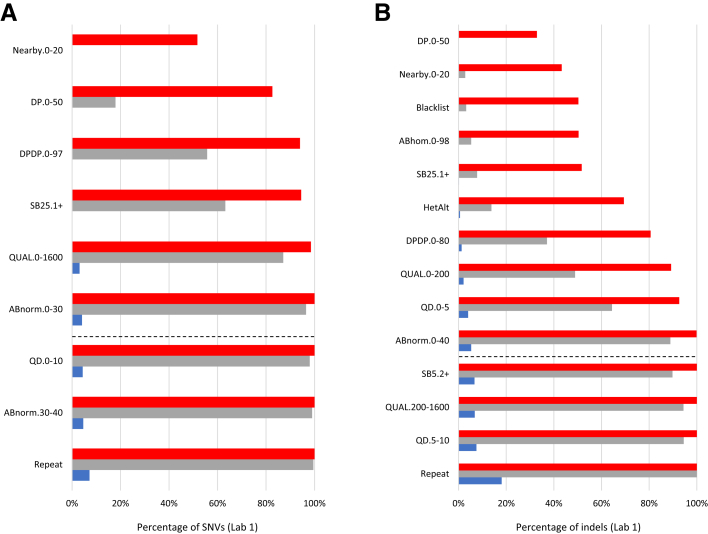
The final selected criteria are shown in Figure 3. Combined, these criteria flagged 100% of all 201 FP SNVs (CI, 98.9%–100%) and 100% of all 987 FP indels (CI, 99.8%–100%). Only 4.1% of clinical (ie, non-GIAB) TP SNVs and 6.7% of TP indels were flagged by the same criteria. Many FPs received multiple flags, providing redundancy. Adding redundancy to the algorithm as an explicit objective increased the fraction of TPs flagged to 6.8% (SNVs) and 18% (indels). This increase was largely due to the addition of the repeat flag in this step (Supplemental Appendix S1).
Figure 3.
Combining flags. These plots show the cumulative effect (top to bottom) of sequentially combining flags chosen by our algorithm. A and B: Single-nucleotide variants (SNVs) (A) and insertions and deletions (indels; B) in the Lab 1 data set. Red indicates the fraction of all false positives (FPs) captured; blue, the fraction of clinical true positives (TPs); gray, the fraction of FPs captured by two or more flags. The dashed lines illustrate the flags needed to capture 100% of the FPs using at least one flag each. To be conservative, the full set of flags shown here was used (maximizing double coverage) and, in particular, required confirmation of all repeat-associated calls. Note that in the indel analysis, higher QUAL and QD thresholds were required to maximize double coverage than were required to achieve 100% capture of variants by a single flag each. The flags include: DP, read depth, specifically GT_DP; DP/DP, ratio of GT_DP to INFO_DP; SB5 and SB25, strand bias metrics; QUAL, quality score; QD, quality-depth score; ABnorm, allele balance for heterozygotes, normalized to be within 0.0 to 0.5; ABhom, allele balance for homozygotes; HetAlt, heterozygous call for which neither allele is in the GRCh37 reference genome; Repeat, variant call within a homopolymer or short tandem repeat.
The importance of data set size was similarly assessed. For example, when the cross-validation was changed to use only 50 FPs in training, then 100% of indel iterations and 71% of SNV iterations produced criteria that failed to flag at least some FPs in the test data. Between 2.0% and 6.0% of FP SNVs and between 1.5% and 8.4% of FP indels were missed by the poorest performing 25% of data sets. These rates were consistent with the CI calculations (Figure 2C), which predicted that criteria established using such small data sets will capture between 95% and 100% of FPs and will accomplish that 19 of 20 times. Further reducing the number of training FPs to 20 produced criteria that uniformly failed; however, 125 FP SNVs or 250 FP indels performed far better.
Application of These Methods to Lab 2 Data
It was examined whether a similar approach would work for the Lab 2 data sets, which were produced using somewhat different NGS methods. As of this analysis, Lab 2 had sequenced only one GIAB sample (NA12878) in addition to compiling clinical confirmation data. One consequence of this limitation was that clinical FPs played a larger role in determining both criteria and CIs. This is not problematic, although clinical confirmation data can have significant biases, such as overrepresentation of recurrent variants. Lab 2's combined (clinical and GIAB) data set was nonetheless diverse.
Individual metrics were first analyzed (Supplemental Table S2), with observations of general similarities with those of Lab 1. Allele balance was the most informative criterion for separating TPs from FPs, with quality score, read depth, and strand bias also showing utility. Het-alt calls, variants with others nearby, and repeat-associated variants were often but not always FPs. As for Lab 1, none of these criteria was adequate alone. Flags were combined (Supplemental Figure S6) and were able to capture 100% of FPs with CIs of 98.5% to 100% (SNVs) and 99.1% to 100% (indels). These criteria flagged 13.2% of TP SNVs and 15.4% of TP indels. Requiring double coverage increased these rates to 19.6% and 29.8%, respectively.
In comparison with Lab 1 criteria, the criteria chosen for Lab 2 appeared equally effective although less efficient—a greater fraction of TPs were flagged as requiring confirmation. One reason was that fewer “bad” variant calls had been removed before confirmation, a result of Lab 2's different filtering and (for clinical specimens) manual review processes. This difference does not indicate an accuracy problem for Lab 2, but it resulted in broader confirmation criteria. This observation reinforced the belief that confirmation criteria can vary based on filtering thresholds, and supported the approach of first establishing (and validating) filters before establishing confirmation criteria. These results also suggest next steps for Lab 2: Tightening up filtering thresholds (where possible without impacting sensitivity) could further reduce confirmation workload by removing unambiguous FPs, similar to Lab 1's process. Reducing the number of FPs in this study, however, would make the CIs wider (ie, less confident), reflecting an important design aspect of studies such as ours, which depends on the set of FPs provided (see Discussion). The four additional GIAB samples would likely address this issue by providing additional useful FPs.
Discussion
This study investigated whether a large and diverse data set could be used to develop statistically robust criteria to guide the application of confirmation assays in clinical genetic testing. The data sets combined clinical confirmation results with data from the sequencing of GIAB specimens, allowing for the two data types to complement each other—a key aspect of our methodology (Table 2). Similar to prior studies,10, 11, 12, 13, 14 this method identifies intermediate-confidence calls that require confirmation (ie, to determine which are TPs and which are FPs) from calls that are high-confidence TPs from NGS alone and do not benefit from confirmation. It cannot be guaranteed that the criteria chosen by our method (or any method) will capture all FPs. However, the probability of missing an FP is below a measurable level in data sets containing, collectively, almost 200,000 diverse variant calls with confirmatory data.
These results differ from those of prior studies in important ways. Neither quality score (as suggested by Strom et al10) nor the combination of allele balance and read depth (as suggested by Mu et al12) captured all of the FPs in our data sets even when thresholds were reoptimized. Although the criteria must be established independently for each NGS workflow—and indeed, workflows varied among these studies—this discrepancy may have resulted from the small data sets used in the prior studies as well as the studies' lack of separate training and test data sets. These limitations may have left these studies underpowered and subject to overfitting, which can impact the effectiveness of the chosen criteria. A study by Baudhuin et al11 reported no FPs, a seemingly excellent result. However, this study and others6, 12, 14 showed that both FPs and TPs are abundant within the intermediate-confidence calls produced by current NGS methods. Aggressive filtering thus improves specificity at the expense of sensitivity. Indeed, a separate study showed that the methods used by Baudhuin had sensitivity limitations.7 The two-threshold model (Figure 1) helps to address this issue.
In these data sets, a battery of criteria was required to flag FPs, consistent with metrics recommended in the guidelines from the Association for Molecular Pathology and the College of American Pathologists.29 This result is intuitive given that a variety of underlying factors can result in NGS FPs and different FPs indeed have different properties. In theory, a single quality score that captures all of these factors could be simpler to use than a battery of criteria. Unfortunately, no quality score produced by current variant callers that can identify FPs without also capturing many TPs is known. Other studies agree.14, 21, 32, 33
A recent study by van den Akker et al14 was somewhat similar to this one: The authors used a supervised learning framework, used multiple quality metrics, and analyzed a larger data set than was used in prior studies (albeit smaller than this). There were important differences, however: Van den Akker considered only one relatively small gene panel and omitted certain challenging regions of those genes. By contrast, over 2000 genes were examined here and off-target regions included. Van den Akker's logistic regression approach effectively creates (another) arbitrary quality score by mathematically combining metrics. Here, individual thresholds on metrics recommended by guidelines were preferred,29 which makes these results understandable and easily implementable and avoids the statistical issues associated with mathematical combinations of highly correlated inputs.
Also, the objective of the heuristic algorithm, which focuses on the detection of FPs, was preferred, as opposed to machine learning methods that optimize overall prediction accuracy, an approach that equally values the classification of TPs. Detecting FPs is clearly of paramount clinical importance. Moreover, in highly imbalanced data sets (ie, every data set in Table 1, in which TPs vastly outnumber FPs), such approaches can produce classifiers that work far better on the majority class (TPs) than on the minority (FPs) unless specific corrections are implemented.34 Overall performance metrics (eg, prediction accuracy) also become uninformative with imbalanced data. For example, in the combined data sets (N = 194,119), an algorithm could show 99.6% prediction accuracy yet still miss half of the 1662 FPs. The FP-centric approach used minimizes these issues.
Another important aspect of this methodology, not used in prior studies, was the inclusion of partial match errors, per recommendations.28 These errors include both zygosity differences and incorrect diploid genotypes. Both of these error types can have significant clinical implications, and both are often resolved by confirmation. It is important to distinguish partial match errors from “spelling differences,” a different issue in which sequences are correct but are described in a noncanonical way.29 This study ignored spelling differences but considered partial match errors to be as important as pure FPs (ie, cases in which a variant was called but none is actually present). Partial match errors represented approximately half of the FPs in our GIAB data sets. Many partial match errors occurred in repeats, and thus they were less common in the clinical data sets (approximately 10% of FPs).
There were numerous benefits to using the large data sets generated. These not only provided a diverse set of FPs for training (ie, selecting criteria) but also allowed for the establishment of separate test sets to properly validate these criteria, identify outliers, and minimize overfitting. Using all five GIAB specimens also reduced the risk for biases resulting from the fact that variant callers are often trained, and may exhibit superior performance, on one of these specimens (NA12878).30 Furthermore, the large data sets allowed computing statistical measures of confidence, a crucial part of any laboratory validation study, albeit one that is not always used.29 The statistical metric used (CI lower bound on fraction of FPs flagged) was inspired by recent guidelines.29, 31 It uses the size of the FP data set as a proxy for whether those data are likely to be adequately representative and diverse for use in setting robust criteria. The CI bound does not directly measure diversity, however, and can be artificially inflated by under-filtering, as described later in this section. More sophisticated statistical approaches are ideal topics for future work. However, metrics or statistics based on net accuracy of the classifier, which can be uninformative, as described earlier in this section, or on positive predictive value, which can be misleading because variants of intermediate quality become diluted by the large number of high-confidence TPs, should be used with caution.
A key question is how large a data set is adequate. For example, if certain criteria are shown to flag 49 of 49 FPs, these criteria are ostensibly 100% effective. However, they have been statistically demonstrated to flag only between 95% and 100% of FPs at P = 0.05 (ie, 19 of 20 times) (Figure 2C). This issue was found not to be hypothetical: Small training data sets (eg, 50 FPs) indeed resulted in poorly performing criteria, and the corresponding CI bound (≥95.1%) was considered inadequate. Our laboratories' tests require high specificity, and 100% FP detection with CI lower bounds between 98.5% and 99.8% (Table 1) was achieved by using hundreds of example FPs. Such bounds are likely appropriate for panel and exome tests, which have an increased risk for producing FPs compared with single-gene tests. Indels deserve careful attention, as FPs are more likely to appear pathogenic compared with FP SNVs.
Obtaining a large number of FPs for study can be challenging, and care was taken not to do so in artificial and problematic ways. For example, many FPs could be added by simply lowering filtering thresholds (underfiltering). However, the resulting criteria might be effective only in flagging clearly erroneous calls as opposed to accurately defining the intermediate-confidence set (Figure 1) for which confirmation matters most. This limitation would be particularly problematic when using machine-learning algorithms that best classify the largest input data subsets.34 For example, van den Akker's FDRs are quite high14 compared with those in our data sets and those of prior studies (Table 1), suggesting that many low-quality FPs may have been included. Underfiltering also will artificially tighten CIs by counting obvious FPs, which could be misleading. A similarly problematic approach would be to run many samples containing the same FP variants. To avoid such problems, this study used the same filtering thresholds that our laboratories had previously validated for use in clinical practice, and both the number of unique variants and the number of variant calls were considered when designing this study (Table 1). The GIAB specimens provided a great deal of data, eliminating potential incentives to increase the data set in problematic ways.
One might argue that this study artificially increased the number of FPs by including off-target regions. To the contrary, it was considered valuable to deliberately challenge this approach by adding off-target calls, some of which present technical challenges that are present but less frequent in coding exons. Nonetheless, the off-target data appeared reasonably representative (see Results). Because 100% of the combined GIAB and clinical FPs were required to have been flagged, these criteria worked for both on- and off-target calls, as well as both GIAB and clinical specimens. Quality metrics for clinical, on-target GIAB, and off-target GIAB variant calls were examined separately (Supplemental Table S3), with observations of similarities and expected differences resulting from: i) data set sizes, ii) selection bias (in general, only clinically reportable variants were subject to confirmation in the clinical specimens), and iii) manual review (applied to the Lab 1 clinical specimens but not the GIAB samples).
This study had important limitations. The truth data provided by the GIAB consortium were used for confirmation in the GIAB specimens. These calls are highly accurate but imperfect.18 In addition, when the Coriell GIAB specimens are sequenced (as done here), there can be genetic differences compared with the original DNA samples used to develop the truth data. In general, these issues will cause the methodology to establish broader criteria than might otherwise be necessary, increasing the number of TPs flagged. Outliers (FPs with unusually high confidence in our laboratory data) were examined in the data from the GIAB consortium to ascertain whether those results appeared correct. No sites in the GIAB specimens were confirmed using an orthogonal assay, although it may be valuable to do so in future studies.
A further limitation resulted from the focus on GIAB-HC regions, in which there were the largest numbers of variant calls with confirmatory data. The GIAB-HC designation does not indicate confidence in our own laboratories' data (Materials and Methods), although there was an important bias to recognize. The small fraction of the human genome (10% to 12%) that lies outside of the GIAB-HC regions comprises sites for which the GIAB consortium, using multiple platforms and extensive analysis, could not confidently determine the true sequences.16 These are the hardest regions of the genome to sequence, and they present diverse challenges that increase error rates. Criteria that identify FPs within the GIAB-HC regions might not be effective outside of those regions, and there were few data with which to examine this. It was concluded that clinically relevant variants outside of the GIAB-HC regions need to be confirmed, regardless of other quality metrics. As a consequence, variants in segmentally duplicated regions (which are usually not GIAB-HC) require confirmation. Within the Lab 1 data set, 7.0% of clinically reported SNVs and 15% of clinical indels were thus flagged, in addition to those variants described in the Results. Additional statistically valid studies would be required to determine which variant calls outside of the GIAB-HC regions could forego confirmation in the future.
The American College of Medical Genetics guideline on NGS recommends that laboratories have “extensive experience with NGS technology and be sufficiently aware of the pitfalls … before deciding that result confirmation with orthogonal technology can be eliminated.”3,p.6 This methodology provides a practical and rigorous way to follow this recommendation and to ensure that the experience (ie, data) is based on a laboratory's own specific methodologies and test targets, not NGS in general. Laboratories using the same sequencing instrument and variant caller (eg, GATK) may still need different confirmation criteria owing to the many subtle differences among tests (particularly bioinformatics). Determining whether universal, interlaboratory criteria are feasible would require additional and extensive study. Consistent with guidelines,3, 5, 29 these data suggest that each laboratory should validate its own confirmation criteria and that revalidation of these criteria should accompany any significant process change (including filtering changes).
These results had specific implications for the current New York State guidelines, under which confirmation may be waived after “at least 10 positive samples per target gene” have been confirmed.4 In these data, many FP variants were also called as TPs (Figure 2D), an issue not examined by prior studies. Examples were found in both the GIAB and clinical data from both laboratories. These variants run a high risk for being confirmed TPs in a series of tests and then, after confirmation is no longer considered necessary, being called falsely. Moreover, in these data sets, different variants within a gene often exhibited remarkably different properties that correlated with remarkably different FDRs. Observing some variants within a gene as TPs provides little information about whether other variants within that gene are FPs. In summary, these results argue against using the New York criteria for confirmation with multigene sequencing tests.
This methodology does not address other roles that confirmation assays serve, such as verifying the identity of a specimen or determining the exact structure of certain variants. Furthermore, this framework does not directly address conflicts between the results of NGS and confirmation assays. As Beck and Mullikin13 elegantly showed, naively assuming that a confirmation assay is always correct can introduce more errors than confirmation corrects. This study also does not address the important issue of setting filtering criteria to ensure sensitivity. Laboratories need to address these issues separately. Finally, note that this approach is not necessarily optimal at minimizing the number of TPs that would receive confirmation. Instead, it is deliberately conservative and designed to prevent FPs from escaping confirmation.
These data show that criteria can be established to limit confirmation assays to a small fraction of variants without any measurable effect on analytic specificity. This study shows that a large and diverse data set is required to accomplish this with confidence, and that the specifics of how criteria are chosen can have a substantial impact on their effectiveness (Table 2). Limiting confirmation assays in this careful manner may help to reduce costs and improve the turnaround time of clinical genetics testing without compromising quality.
Acknowledgments
We thank Invitae and the Partners HealthCare Laboratory for laboratory staff in molecular medicine, who produced the data used in this study; the collaborators in the Genome in a Bottle Consortium for their excellent work on the reference specimens used; Nancy Jacoby for editorial help on the manuscript; and Tina Hambuch and Katya Kosheleva for insightful comments.
Footnotes
Supported by NIH grants U01HG006500, U01HG008676, and U19HD077671 (M.S.L. and H.L.R.), the Partners HealthCare Laboratory for Molecular Medicine (M.S.L. and H.L.R.), and funding from individual employers.
Disclosures: C.-F.L., M.S.L., S.E.L., R.L.N., J.P., H.L.R., V.H.R., and R.T. work for laboratories that provide clinical genetic testing services. S.E.L., R.L.N., J.P., V.H.R., and R.T. own stock and/or stock options in Invitae. S.E.L. owns stock in Illumina and Thermo Fisher Scientific.
Certain commercial equipment, instruments, or materials are identified in this article only to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
The use of deidentified clinical data in this study was subject to the requirements of the IRB of each laboratory. The GIAB specimens were used under the terms of their material transfer agreements.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2018.10.009.
Supplemental Data
Various quality metrics for single-nucleotide variants (SNVs) in the Genome in a Bottle Consortium (GIAB) samples sequenced by Lab 1. Plots within this figure are numbered y.z, where y (a letter) indicates the panel corresponding to a particular metric, and z, when used, indicates a subplot within that panel. The panel letters correspond to the letters of each metric defined in Supplemental Table S2. Certain metrics were not applicable to a particular laboratory or variant type. When that was the case, the associated letter was skipped in this figure, to keep the letter to metric assignments consistent between all figures and tables. The rightmost column in Supplemental Table S2 describes these differences between data sets. Lab 1 used metrics G and H to measure strand bias (Supplemental Figures S1 and S2), whereas strand bias metrics I and J were used by Lab 2 (Supplemental Table S3). Metric M applies only to insertions and deletions (Supplemental Figures S2 and S4). Metric L was measured only in the GIAB samples (Supplemental Figures S1 and S2). This analysis is restricted to the high-confidence regions (GIAB-HC) that had accurate confirmatory data. On- and off-target regions are included. Note that both the GIAB-HC and on/off-target designations are unrelated to quality assessment of the sequencing data (Materials and Methods). Most clinical targets are within the GIAB-HC regions. A–M: Various quantitative quality metrics, each with four subplots: i) the percentages of true positives (TPs) and false positives (FPs) captured within the indicated range, indicating the value (in FPs) and cost (in TPs) of using this metric as a criterion for identifying potential FPs (top left); ii) the false discovery rate (FDR) of calls within each range [calculated as FPs/(TPs + FPs)], indicating the extent of erroneous calls within this range (top right); iii) the cumulative percentage of FPs and TPs flagged at each threshold, indicating the value (in FPs) and cost (in TPs) of using the metric at that threshold (bottom left); and iv) the cumulative FDR of all flagged variant calls at each threshold, indicating how erroneous calls are at this threshold (bottom right). The cumulative plots are summed left to right for metrics for which a higher number indicates a more confident call (eg, read depth), and are summed right to left in cases in which lower values indicate higher confidence (eg, strand bias). Note that both the x and y axes are adapted for each metric to make useful differences clear. The x axis in some cases is highly nonlinear. The y axis is always a percentage but is scaled differently in each plot to fit the data. N and O: Various binary metrics (those metrics that are either on or off for any variant call but which do not have a quantitative score), each with two subplots: i) the percentage of TPs and FPs captured by each flag, indicating the value (in FPs) and cost (in TPs) of the flag (left); and ii) the FDR of calls with each flag, the ratio of FPs divided by TPs plus FPs, indicating the extent of erroneous calls with this flag (right). P: Per-variant FDRs, which address whether variants observed (obs) as FPs (at least once) in this data set are also TPs in different samples or runs. These plots are similar to those in Figure 2D. Q: The application of the specific criteria from Mu et al12 and Strom et al10 to these data. The specific definition is in Supplemental Table S2. Similar plots from Lab 2 are not shown, although the results for each flag and threshold in both laboratories are summarized in Supplemental Table S3. The flags include: DP, read depth, specifically GT_DP; DP/DP, ratio of GT_DP to INFO_DP; SB5 and SB25, strand bias metrics; QUAL, quality score; QD, quality-depth score; ABhom, allele balance for heterozygotes, normalized to be within 0.0 to 0.5; Abhom, allele balance for homozygotes; HetAlt, heterozygous call for which neither allele is in the GRCh37 reference genome; Repeat, variant call within a homopolymer or short tandem repeat.
Various quality metrics for insertions and deletions (indels) in the Genome in a Bottle Consortium (GIAB) samples sequenced by Lab 1. This figure follows the same format as Supplemental Figure S1. The flags include: DP, read depth, specifically GT_DP; DP/DP, ratio of GT_DP to INFO_DP; SB5 and SB25, strand bias metrics; QUAL, quality score; QD, quality-depth score; ABnorm, allele balance for heterozygotes, normalized to be within 0.0 to 0.5; ABhom, allele balance for homozygotes; HetAlt, heterozygous call for which neither allele is in the GRCh37 reference genome; Repeat, variant call within a homopolymer or short tandem repeat.
Various quality metrics for single-nucleotide variants (SNVs) in the clinical samples sequenced by Lab 1. This figure follows the same format as does Supplemental Figure S1. This analysis is restricted to the on-target regions in which confirmatory data on selected clinical variants were obtained (Figure 1). Both Genome in a Bottle Consortium high-confidence regions (GIAB-HC) and non-HC regions are included. Note that both the GIAB-HC and on/off-target designations are unrelated to quality assessment of the sequencing data (Materials and Methods). Most clinical targets are within the GIAB-HC regions. QUAL, variant call quality scores; obs, observed. The flags include: DP, read depth, specifically GT_DP; DP/DP, ratio of GT_DP to INFO_DP; SB5 and SB25, strand bias metrics; QUAL, quality score; QD, quality-depth score; ABnorm, allele balance for heterozygotes, normalized to be within 0.0 to 0.5; ABhom, allele balance for homozygotes; HetAlt, heterozygous call for which neither allele is in the GRCh37 reference genome; Repeat, variant call within a homopolymer or short tandem repeat.
Various quality metrics for insertions and deletions (indels) in the clinical samples sequenced by Lab 1. This figure follows the same format as Supplemental Figure S1. This analysis is restricted to the on-target regions in which confirmatory data for selected clinical variants were obtained (Figure 1). Both Genome in a Bottle Consortium high-confidence regions (GIAB-HC) and non-HC regions are included. Note that both the GIAB-HC and on/off-target designations are unrelated to quality assessment of our sequencing data (Materials and Methods). Most clinical targets are within the GIAB-HC regions. QUAL, variant call quality scores. The flags include: DP, read depth, specifically GT_DP; DP/DP, ratio of GT_DP to INFO_DP; SB5 and SB25, strand bias metrics; QUAL, quality score; QD, quality-depth score; ABnorm, allele balance for heterozygotes, normalized to be within 0.0 to 0.5; ABhom, allele balance for homozygotes; HetAlt, heterozygous call for which neither allele is in the GRCh37 reference genome; Repeat, variant call within a homopolymer or short tandem repeat.
Variant call quality scores (QUAL; y axis) for all true-positive (TP, blue) and the single false-positive (FP; arrow) single-nucleotide variant (SNV) call in the data set from Strom et al.10 The x axis position of each point is randomly assigned. The dashed line shows Strom's proposed threshold; variant calls with quality scores above this line would not require confirmation. This graph follows the format of that in Figure 2, A and B, although the quality scores in this plot are not calibrated to the main figures (because Strom used an older version of the Genome Analysis Toolkit software). Nonetheless, no Strom-like threshold would prove effective in separating TPs and FPs in Figure 2A. Much smaller data subsets (eg, Figure 2B) would incorrectly suggest otherwise.
Cumulative effect (top to bottom) of sequentially combining flags chosen by our algorithm. A: Single-nucleotide variants (SNVs) in the Lab 2 data set. B: Insertions and deletions (indels) in the Lab 2 data set. Red bars indicate the fraction of all false positives (FPs) captured; blue bars indicate the fraction of clinical true positives. Gray bars show the fraction of FPs captured by two or more flags. The dashed lines illustrate the flags needed to capture 100% of the FPs using at least one flag each. To be conservative, the full set of flags shown here were used (double coverage). Metrics are defined in Supplemental Table S2. The flags include: DP, read depth, specifically GT_DP; DP/DP, ratio of GT_DP to INFO_DP; SB5 and SB25, strand bias metrics; QUAL, quality score; QD, quality-depth score; ABnorm, allele balance for heterozygotes, normalized to be within 0.0 to 0.5; ABhom, allele balance for homozygotes; HetAlt, heterozygous call for which neither allele is in the GRCh37 reference genome; Repeat, variant call within a homopolymer or short tandem repeat.
References
- 1.Korf B.R., Rehm H.L. New approaches to molecular diagnosis. JAMA. 2013;309:1511–1521. doi: 10.1001/jama.2013.3239. [DOI] [PubMed] [Google Scholar]
- 2.Rehm H.L. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013;14:295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rehm H.L., Bale S.J., Bayrak-Toydemir P., Berg J.S., Brown K.K., Deignan J.L., Friez M.J., Funke B.H., Hegde M.R., Lyon E., Working Group of the American College of Medical Genetics and Genomics Laboratory Quality Assurance Committee ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–747. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.New York State Department of Health . New York State Department of Health; Albany, NY: 2015. Guidelines for Validation Submissions of Next Generation Sequencing (NGS) assays under the NYS Testing Category of Genetic Testing.https://www.wadsworth.org/sites/default/files/WebDoc/2080900015/Germline_NextGen_Validation_Guidelines.pdf Available at. [Google Scholar]
- 5.Aziz N., Zhao Q., Bry L., Driscoll D.K., Funke B., Gibson J.S., Grody W.W., Hegde M.R., Hoeltge G.A., Leonard D.G.B., Merker J.D., Nagarajan R., Palicki L.A., Robetorye R.S., Schrijver I., Weck K.E., Voelkerding K.V. College of American Pathologists' laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med. 2015;139:481–493. doi: 10.5858/arpa.2014-0250-CP. [DOI] [PubMed] [Google Scholar]
- 6.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., McKenna A., Fennell T.J., Kernytsky A.M., Sivachenko A.Y., Cibulskis K., Gabriel S.B., Altshuler D., Daly M.J. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lincoln S.E., Zook J.M., Chowdhury S., Mahamdallie S., Fellowes A., Klee E.W., Truty R., Huang C., Tomson F.L., Cleveland M.H., Vallone P.M., Ding Y., Seal S., DeSilva W., Garlick R.K., Salit M., Rahman N., Kingsmore S.F., Aradhya S., Nussbaum R.L., Ferber M.J., Shirts B.H. An interlaboratory study of complex variant detection. bioRxiv. 2017:218529. [Google Scholar]
- 8.Lincoln S.E., Kobayashi Y., Anderson M.J., Yang S., Desmond A.J., Mills M.A., Nilsen G.B., Jacobs K.B., Monzon F.A., Kurian A.W., Ford J.M., Ellisen L.W. A systematic comparison of traditional and multigene panel testing for hereditary breast and ovarian cancer genes in more than 1000 patients. J Mol Diagn. 2015;17:533–544. doi: 10.1016/j.jmoldx.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Mandelker D., Schmidt R.J., Ankala A., McDonald Gibson K., Bowser M., Sharma H., Duffy E., Hegde M., Santani A., Lebo M., Funke B. Navigating highly homologous genes in a molecular diagnostic setting: a resource for clinical next-generation sequencing. Genet Med. 2016;18:1282–1289. doi: 10.1038/gim.2016.58. [DOI] [PubMed] [Google Scholar]
- 10.Strom S.P., Lee H., Das K., Vilain E., Nelson S.F., Grody W.W., Deignan J.L. Assessing the necessity of confirmatory testing for exome-sequencing results in a clinical molecular diagnostic laboratory. Genet Med. 2014;16:510–515. doi: 10.1038/gim.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baudhuin L.M., Lagerstedt S.A., Klee E.W., Fadra N., Oglesbee D., Ferber M.J. Confirming variants in next-generation sequencing panel testing by Sanger sequencing. J Mol Diagn. 2015;17:456–461. doi: 10.1016/j.jmoldx.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Mu W., Lu H.-M., Chen J., Li S., Elliott A.M. Sanger confirmation is required to achieve optimal sensitivity and specificity in next-generation sequencing panel testing. J Mol Diagn. 2016;18:923–932. doi: 10.1016/j.jmoldx.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Beck T.F., Mullikin J.C., NISC Comparative Sequencing Program. Biesecker L.G. Systematic evaluation of Sanger validation of next-generation sequencing variants. Clin Chem. 2016;62:647–654. doi: 10.1373/clinchem.2015.249623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van den Akker J., Mishne G., Zimmer A.D., Zhou A.Y. A machine learning model to determine the accuracy of variant calls in capture-based next generation sequencing. BMC Genomics. 2018;19:263. doi: 10.1186/s12864-018-4659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alpaydin E. MIT Press; Boston: 2014. Introduction to Machine Learning. [Google Scholar]
- 16.Zook J.M., Chapman B., Wang J., Mittelman D., Hofmann O., Hide W., Salit M. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32:246–251. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
- 17.Zook J.M., Catoe D., McDaniel J., Vang L., Spies N., Sidow A. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data. 2016;3:160025. doi: 10.1038/sdata.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zook J., McDaniel J., Parikh H., Heaton H., Irvine S.A., Trigg L., Truty R., McLean C.Y., De La Vega F.M., Salit M., Genome in a Bottle Consortium Reproducible integration of multiple sequencing datasets to form high-confidence SNP, indel, and reference calls for five human genome reference materials. bioRxiv. 2018:281006. [Google Scholar]
- 19.Pugh T.J., Kelly M.A., Gowrisankar S., Hynes E., Seidman M.A., Baxter S.M., Bowser M., Harrison B., Aaron D., Mahanta L.M., Lakdawala N.K., McDermott G., White E.T., Rehm H.L., Lebo M., Funke B.H. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16:601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 20.Alfares A.A., Kelly M.A., McDermott G., Funke B.H., Lebo M.S., Baxter S.B., Shen J., McLaughlin H.M., Clark E.H., Babb L.J., Cox S.W., DePalma S.R., Ho C.Y., Seidman J.G., Seidman C.E., Rehm H.L. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17:880–888. doi: 10.1038/gim.2014.205. [DOI] [PubMed] [Google Scholar]
- 21.Poplin R., Ruano-Rubio V., DePristo M.A., Fennell T.J., Carneiro M.O., Van der Auwera G.A., Kling D.E., Gauthier L.D., Levy-Moonshine A., Roazen D., Shakir K., Thibault J., Chandran S., Whelan C., Lek M., Gabriel S., Daly M.J., Neale B., MacArthur D.G., Banks E. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2017:201178. [Google Scholar]
- 22.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCalmon S, Konvicka K, Reddy N, Olivares E, Whittaker J, Kautzer C, Rosendorff A: SMRTer Confirmation: scalable clinical read-through variant confirmation using the Pacific Biosciences SMRT Sequencing Platform. American Society of Human Genetics 2016 Annual Meeting [abstract 996F], Oct. 18-22, 2016, Vancouver, BC, Canada. Bethesda, MD: American Society of Human Genetics, 2016
- 24.Travers K.J., Chin C.-S., Rank D.R., Eid J.S., Turner S.W. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 2010;38:e159. doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao W., Yang Y., Sebra R., Mendiratta G., Gaedigk A., Desnick R.J., Scott S.A. Long-read single molecule real-time full gene sequencing of cytochrome P450-2D6. Hum Mutat. 2016;37:315–323. doi: 10.1002/humu.22936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ardui S., Race V., de Ravel T., Van Esch H., Devriendt K., Matthijs G., Vermeesch J.R. Detecting AGG interruptions in females with a FMR1 premutation by long-read single-molecule sequencing: a 1 year clinical experience. Front Genet. 2018;9:150. doi: 10.3389/fgene.2018.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cleary J.G., Braithwaite R., Gaastra K., Hilbush B.S., Inglis S., Irvine S.A., Jackson A., Littin R., Rathod M., Ware D., Zook J.M., Trigg L., De La Vega F.M. Comparing variant call files for performance benchmarking of next-generation sequencing variant calling pipelines. bioRxiv. 2015:023754. [Google Scholar]
- 28.Krusche P., Trigg L., Boutros P.C., Mason C.E., De La Vega F.M., Moore B.L., Gonzalez-Porta M., Eberle M.A., Tezak Z., Labadibi S., Truty R., Asimenos G., Funke B., Fleharty M., Salit M., Zook J.M., Global Alliance for Genomics and Health Benchmarking Team Best practices for benchmarking germline small variant calls in human genomes. bioRxiv. 2018:270157. [Google Scholar]
- 29.Roy S., Coldren C., Karunamurthy A., Kip N.S., Klee E.W., Lincoln S.E., Leon A., Pullambhatla M., Temple-Smolkin R.L., Voelkerding K.V., Wang C., Carter A.B. Standards and guidelines for validating next-generation sequencing bioinformatics pipelines: a joint recommendation of the Association for Molecular Pathology and the College of American Pathologists. J Mol Diagn. 2018;20:4–27. doi: 10.1016/j.jmoldx.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Li H., Bloom J.M., Farjoun Y., Fleharty M., Gauthier L.D., Neale B., MacArthur D. New synthetic-diploid benchmark for accurate variant calling evaluation. bioRxiv. 2017:223297. doi: 10.1038/s41592-018-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jennings L.J., Arcila M.E., Corless C., Kamel-Reid S., Lubin I.M., Pfeifer J., Temple-Smolkin R.L., Voelkerding K.V., Nikiforova M.N. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;19:341–365. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirooznia M., Kramer M., Parla J., Goes F.S., Potash J.B., McCombie W.R., Zandi P.P. Validation and assessment of variant calling pipelines for next-generation sequencing. Hum Genomics. 2014;8:14. doi: 10.1186/1479-7364-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldfeder R.L., Priest J.R., Zook J.M., Grove M.E., Waggott D., Wheeler M.T., Salit M., Ashley E.A. Medical implications of technical accuracy in genome sequencing. Genome Med. 2016;8:24. doi: 10.1186/s13073-016-0269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batista G.E.A.P.A., Prati R.C., Monard M.C. A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD Explorations Newsletter. 2004;6:20–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Various quality metrics for single-nucleotide variants (SNVs) in the Genome in a Bottle Consortium (GIAB) samples sequenced by Lab 1. Plots within this figure are numbered y.z, where y (a letter) indicates the panel corresponding to a particular metric, and z, when used, indicates a subplot within that panel. The panel letters correspond to the letters of each metric defined in Supplemental Table S2. Certain metrics were not applicable to a particular laboratory or variant type. When that was the case, the associated letter was skipped in this figure, to keep the letter to metric assignments consistent between all figures and tables. The rightmost column in Supplemental Table S2 describes these differences between data sets. Lab 1 used metrics G and H to measure strand bias (Supplemental Figures S1 and S2), whereas strand bias metrics I and J were used by Lab 2 (Supplemental Table S3). Metric M applies only to insertions and deletions (Supplemental Figures S2 and S4). Metric L was measured only in the GIAB samples (Supplemental Figures S1 and S2). This analysis is restricted to the high-confidence regions (GIAB-HC) that had accurate confirmatory data. On- and off-target regions are included. Note that both the GIAB-HC and on/off-target designations are unrelated to quality assessment of the sequencing data (Materials and Methods). Most clinical targets are within the GIAB-HC regions. A–M: Various quantitative quality metrics, each with four subplots: i) the percentages of true positives (TPs) and false positives (FPs) captured within the indicated range, indicating the value (in FPs) and cost (in TPs) of using this metric as a criterion for identifying potential FPs (top left); ii) the false discovery rate (FDR) of calls within each range [calculated as FPs/(TPs + FPs)], indicating the extent of erroneous calls within this range (top right); iii) the cumulative percentage of FPs and TPs flagged at each threshold, indicating the value (in FPs) and cost (in TPs) of using the metric at that threshold (bottom left); and iv) the cumulative FDR of all flagged variant calls at each threshold, indicating how erroneous calls are at this threshold (bottom right). The cumulative plots are summed left to right for metrics for which a higher number indicates a more confident call (eg, read depth), and are summed right to left in cases in which lower values indicate higher confidence (eg, strand bias). Note that both the x and y axes are adapted for each metric to make useful differences clear. The x axis in some cases is highly nonlinear. The y axis is always a percentage but is scaled differently in each plot to fit the data. N and O: Various binary metrics (those metrics that are either on or off for any variant call but which do not have a quantitative score), each with two subplots: i) the percentage of TPs and FPs captured by each flag, indicating the value (in FPs) and cost (in TPs) of the flag (left); and ii) the FDR of calls with each flag, the ratio of FPs divided by TPs plus FPs, indicating the extent of erroneous calls with this flag (right). P: Per-variant FDRs, which address whether variants observed (obs) as FPs (at least once) in this data set are also TPs in different samples or runs. These plots are similar to those in Figure 2D. Q: The application of the specific criteria from Mu et al12 and Strom et al10 to these data. The specific definition is in Supplemental Table S2. Similar plots from Lab 2 are not shown, although the results for each flag and threshold in both laboratories are summarized in Supplemental Table S3. The flags include: DP, read depth, specifically GT_DP; DP/DP, ratio of GT_DP to INFO_DP; SB5 and SB25, strand bias metrics; QUAL, quality score; QD, quality-depth score; ABhom, allele balance for heterozygotes, normalized to be within 0.0 to 0.5; Abhom, allele balance for homozygotes; HetAlt, heterozygous call for which neither allele is in the GRCh37 reference genome; Repeat, variant call within a homopolymer or short tandem repeat.
Various quality metrics for insertions and deletions (indels) in the Genome in a Bottle Consortium (GIAB) samples sequenced by Lab 1. This figure follows the same format as Supplemental Figure S1. The flags include: DP, read depth, specifically GT_DP; DP/DP, ratio of GT_DP to INFO_DP; SB5 and SB25, strand bias metrics; QUAL, quality score; QD, quality-depth score; ABnorm, allele balance for heterozygotes, normalized to be within 0.0 to 0.5; ABhom, allele balance for homozygotes; HetAlt, heterozygous call for which neither allele is in the GRCh37 reference genome; Repeat, variant call within a homopolymer or short tandem repeat.
Various quality metrics for single-nucleotide variants (SNVs) in the clinical samples sequenced by Lab 1. This figure follows the same format as does Supplemental Figure S1. This analysis is restricted to the on-target regions in which confirmatory data on selected clinical variants were obtained (Figure 1). Both Genome in a Bottle Consortium high-confidence regions (GIAB-HC) and non-HC regions are included. Note that both the GIAB-HC and on/off-target designations are unrelated to quality assessment of the sequencing data (Materials and Methods). Most clinical targets are within the GIAB-HC regions. QUAL, variant call quality scores; obs, observed. The flags include: DP, read depth, specifically GT_DP; DP/DP, ratio of GT_DP to INFO_DP; SB5 and SB25, strand bias metrics; QUAL, quality score; QD, quality-depth score; ABnorm, allele balance for heterozygotes, normalized to be within 0.0 to 0.5; ABhom, allele balance for homozygotes; HetAlt, heterozygous call for which neither allele is in the GRCh37 reference genome; Repeat, variant call within a homopolymer or short tandem repeat.
Various quality metrics for insertions and deletions (indels) in the clinical samples sequenced by Lab 1. This figure follows the same format as Supplemental Figure S1. This analysis is restricted to the on-target regions in which confirmatory data for selected clinical variants were obtained (Figure 1). Both Genome in a Bottle Consortium high-confidence regions (GIAB-HC) and non-HC regions are included. Note that both the GIAB-HC and on/off-target designations are unrelated to quality assessment of our sequencing data (Materials and Methods). Most clinical targets are within the GIAB-HC regions. QUAL, variant call quality scores. The flags include: DP, read depth, specifically GT_DP; DP/DP, ratio of GT_DP to INFO_DP; SB5 and SB25, strand bias metrics; QUAL, quality score; QD, quality-depth score; ABnorm, allele balance for heterozygotes, normalized to be within 0.0 to 0.5; ABhom, allele balance for homozygotes; HetAlt, heterozygous call for which neither allele is in the GRCh37 reference genome; Repeat, variant call within a homopolymer or short tandem repeat.
Variant call quality scores (QUAL; y axis) for all true-positive (TP, blue) and the single false-positive (FP; arrow) single-nucleotide variant (SNV) call in the data set from Strom et al.10 The x axis position of each point is randomly assigned. The dashed line shows Strom's proposed threshold; variant calls with quality scores above this line would not require confirmation. This graph follows the format of that in Figure 2, A and B, although the quality scores in this plot are not calibrated to the main figures (because Strom used an older version of the Genome Analysis Toolkit software). Nonetheless, no Strom-like threshold would prove effective in separating TPs and FPs in Figure 2A. Much smaller data subsets (eg, Figure 2B) would incorrectly suggest otherwise.
Cumulative effect (top to bottom) of sequentially combining flags chosen by our algorithm. A: Single-nucleotide variants (SNVs) in the Lab 2 data set. B: Insertions and deletions (indels) in the Lab 2 data set. Red bars indicate the fraction of all false positives (FPs) captured; blue bars indicate the fraction of clinical true positives. Gray bars show the fraction of FPs captured by two or more flags. The dashed lines illustrate the flags needed to capture 100% of the FPs using at least one flag each. To be conservative, the full set of flags shown here were used (double coverage). Metrics are defined in Supplemental Table S2. The flags include: DP, read depth, specifically GT_DP; DP/DP, ratio of GT_DP to INFO_DP; SB5 and SB25, strand bias metrics; QUAL, quality score; QD, quality-depth score; ABnorm, allele balance for heterozygotes, normalized to be within 0.0 to 0.5; ABhom, allele balance for homozygotes; HetAlt, heterozygous call for which neither allele is in the GRCh37 reference genome; Repeat, variant call within a homopolymer or short tandem repeat.