Summary
Citrus canker is a plant disease caused by Gram‐negative bacteria from the genus Xanthomonas. The most virulent species is Xanthomonas citri ssp. citri (XAC), which attacks a wide range of citrus hosts. Differential proteomic analysis of the periplasm‐enriched fraction was performed for XAC cells grown in pathogenicity‐inducing (XAM‐M) and pathogenicity‐non‐inducing (nutrient broth) media using two‐dimensional electrophoresis combined with liquid chromatography‐tandem mass spectrometry. Amongst the 40 proteins identified, transglycosylase was detected in a highly abundant spot in XAC cells grown under inducing condition. Additional up‐regulated proteins related to cellular envelope metabolism included glucose‐1‐phosphate thymidylyltransferase, dTDP‐4‐dehydrorhamnose‐3,5‐epimerase and peptidyl‐prolyl cis–trans‐isomerase. Phosphoglucomutase and superoxide dismutase proteins, known to be involved in pathogenicity in other Xanthomonas species or organisms, were also detected. Western blot and quantitative real‐time polymerase chain reaction analyses for transglycosylase and superoxide dismutase confirmed that these proteins were up‐regulated under inducing condition, consistent with the proteomic results. Multiple spots for the 60‐kDa chaperonin and glyceraldehyde‐3‐phosphate dehydrogenase were identified, suggesting the presence of post‐translational modifications. We propose that substantial alterations in cellular envelope metabolism occur during the XAC infectious process, which are related to several aspects, from defence against reactive oxygen species to exopolysaccharide synthesis. Our results provide new candidates for virulence‐related proteins, whose abundance correlates with the induction of pathogenicity and virulence genes, such as hrpD6, hrpG, hrpB7, hpa1 and hrpX. The results present new potential targets against XAC to be investigated in further functional studies.
Keywords: cell envelope metabolism, citrus canker, differential proteomics, pathogenicity, periplasmic proteins, Xanthomonas citri ssp. citri
Introduction
Citrus canker is one of the most important diseases of citrus plants around the world and is a serious threat to the citrus industry, especially as there is no cure (Brown, 2001; Gottwald et al., 2002). The disease causes low crop productivity and leads to poor fruit quality. It is caused by the bacterium Xanthomonas citri ssp. citri (type A, XAC) (Schaad et al., 2005, 2006), which attacks all Citrus species (Brunings and Gabriel, 2003).
The XAC genome has been completely sequenced (da Silva et al., 2002), but few proteomic studies have been reported, most using total cellular extracts. The global proteome of XAC cells grown in nutrient broth (NB) medium, a rich and pathogenicity‐non‐inducing medium, has been analysed previously using two‐dimensional gel electrophoresis followed by liquid chromatography coupled with tandem mass spectrometry (2‐DE LC‐MS/MS), providing the first map of XAC expressed proteins (Soares et al., 2010). In addition, the XAC total proteome has been analysed under in vitro and in vivo infectious conditions using multidimensional protein identification technology (MudPIT), confirming the involvement of many proteins related to type III and IV secretion systems (T3SS and T4SS) and xanthan gum biosynthesis in XAC pathogenicity (Facincani et al., 2014).
Biofilm formation is an important process in bacterial pathogens and has been suggested to be required for the achievement of maximal virulence in XAC, as it plays a major role in host interactions. A comparative proteomic study using 2D difference gel electrophoresis (2D‐DIGE) was performed between XAC biofilm and planktonic cells, cultured in static XVM2, a minimal medium that mimics the plant environment (Zimaro et al., 2013). This revealed major variations in the composition of outer membrane and receptor or transporter proteins, as well as other proteins which had been shown previously to be involved in biofilm formation, such as DnaK and Ef‐Tu (Zimaro et al., 2013).
Proteomic analysis of subcellular fractions allows the identification of proteins that might not be detectable in the total proteome, especially if these proteins are not abundant or are exclusive to a cellular location. Recently, our group utilized 2D‐DIGE technology to detect changes in XAC surface proteins in response to host infection by comparison of cells grown in vivo (plant leaf, i.e. infectious condition) and in vitro (rich medium, i.e. non‐infectious control condition) (Carnielli et al., 2016). In addition to the cellular surface, the periplasm is also expected to play an important role in XAC pathogenicity, as many virulence proteins of Xanthomonas sp. are known to be transported across the periplasm towards the extracellular environment (Büttner and Bonas, 2010). A clear relationship between periplasmic proteins and bacterial pathogenesis has been reported recently for Xanthomonas oryzae pv. oryzae (Xoo) (Deng et al., 2014). In this study, a mutant strain for prc, a gene encoding a periplasmic enzyme, showed a decrease in virulence‐related periplasmic proteins and changes in the stress response of the cell envelope. However, proteomic studies with subcellular fractions of plant‐pathogenic bacteria face a major challenge to recover sufficient protein under in vivo conditions to allow 2D LC‐MS/MS analysis.
We performed a differential proteomic analysis of the XAC periplasm‐enriched fraction by comparing cells grown in vitro on pathogenicity‐inducing (XAM‐M) and pathogenicity‐non‐inducing (NB) media. The composition of XAM‐M is based on the well‐known pathogenicity‐inducing media for Xanthomonas spp.: XVM2 (Wengelnik et al., 1996) and XAM‐1 (Facincani et al., 2014). We demonstrated that XAM‐M is able to induce hrp (hypersensitive response and pathogenicity) genes in XAC. NB medium has been shown previously to be suitable for the pathogenicity‐non‐inducing condition (Moreira et al., 2015; Soares et al., 2010).
In this work, XAC cells grown in XAM‐M medium presented several proteins known to be associated with pathogenicity in either Xanthomonas species or other microorganisms, but not yet reported to be associated with XAC infection. The proteins identified here are promising targets for investigation in future functional studies.
Results
XAC periplasmic‐enriched proteome under in vitro pathogenicity induction
The growth of XAC in two different media, NB (non‐infectious condition) and XAM‐M (infectious condition), showed distinct profiles. Although the maximum culture density at an optical density at 595 nm (OD595 nm) in NB was about 1.2 after approximately 17 h, in XAM‐M, a similar density was reached only after 70 h (Fig. 1a). The ability of XAM‐M medium to induce XAC pathogenicity was confirmed by the analysis of the expression of genes known to be associated with virulence/pathogenicity. Increased transcript levels were observed for all five hrp genes tested (hrpD6, hrpG, hrpB7, hpa1, hrpX) in XAM‐M when compared with the control, NB medium, at the initial (16 h) or late (70 h) stages of growth (Fig. 1b).
Figure 1.
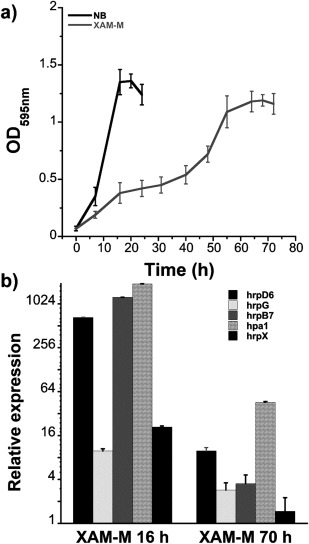
Xanthomonas citri ssp. citri (XAC) growth curves in nutrient broth (NB) and XAM‐M medium, and relative hrp mRNA expression induced by XAM‐M medium. Growth curves [optical density at 595 nm (OD595 nm) vs. time] were based on the average of four independent experiments (a); quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of hrp genes as relative mRNA expression (relative to NB, control, for 17 h) on XAM‐M for 16 h and 70 h (b). Error bars indicate standard deviation from the biological replicates.
Proteins from a periplasmic‐enriched fraction of XAC cells grown in either XAM‐M or NB up to the end of the logarithmic phase (OD595 nm ∼ 1.2) were analysed by 2‐DE (Fig. 2). Reproducibility among gel triplicates was evaluated by scatter plots, presenting a correlation greater than 0.9 (Fig. S1, see Supporting Information). Table 1 shows the XAC proteins identified in spots significantly modulated between XAM‐M and NB. Proteins from 42 differential spots showing the highest scores and having more than one matched peptide are presented in Table 1. For the majority of these spots, the experimental molecular mass and isoelectric point (pI), calculated from 2‐DE, matched the theoretical values. Twelve spots were significantly less abundant under XAM‐M, whereas 30 were more abundant. Fifteen of the latter were found exclusively under pathogenicity‐inducing conditions and corresponded to the following proteins: pyruvate kinase type II (spot 13); outer membrane hemin receptor (spot 14); inorganic pyrophosphatase (spot 16); TldD protein (spot 19); peptidyl‐prolyl cis–trans‐isomerase (PPIase, spots 20 and 21); 60‐kDa chaperonin (spots 23, 24 and 25); outer membrane protein P6 precursor (spots 26 and 29); glucose‐1‐phosphate thymidylyltransferase (spot 27); phosphoglucomutase (spot 33); elongation factor Ts (spot 34); and ATP synthase subunit β (spot 39). Matched peptides for all 79 proteins identified are included in Data S1 (see Supporting Information).
Figure 2.
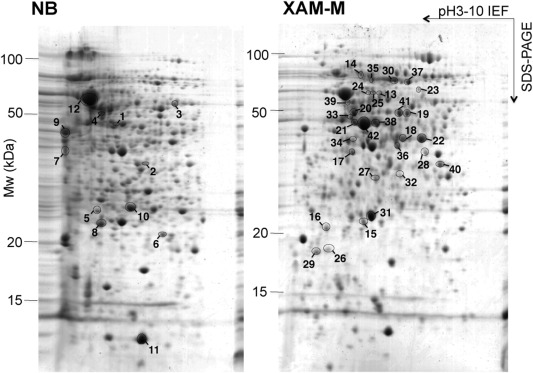
Two‐dimensional gel electrophoresis (2‐DE) of Xanthomonas citri ssp. citri (XAC) periplasmic‐enriched protein fraction. Proteins from the periplasmic‐enriched fraction of XAC grown in nutrient broth (NB) or XAM‐M medium were separated by 2‐DE. The numbered circles represent protein spots with significantly different abundance in XAM‐M relative to NB (P < 0.05). Spots with increased and decreased abundance in XAM‐M are indicated on the right (XAM‐M) and left (NB) gels, respectively. The best‐resolved central region of the 2‐DE gel is shown.
Table 1.
Xanthomonas citri ssp. citri (XAC) periplasm‐enriched fraction proteins identified by two‐dimensional gel electrophoresis (2‐DE) followed by liquid chromatography coupled with tandem mass spectrometry (LC‐MS/MS) in non‐inducing (nutrient broth, NB) and in vitro pathogenicity‐inducing (XAM‐M) conditions (P < 0.05).
| Spot | Condition (more abundant spot) | Open reading frame (ORF) | Protein ID (exclusive peptide count)* |
Theoretical
†
MW/pI |
Experimental
‡
MW/pI |
Mascot score | Matched peptide | Sequence coverage (%) | Category § | Cellular location ¶ | Spot relative abundance (XAM‐M/NB)** |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NB | XAC0002 | DNA polymerase III subunit β (0) | 40.8/5.4 | 47.4/5.7 | 49 | 2 | 4 | III | C | Unique |
| 2 | NB | XAC0818 | Ribokinase (2) | 32.3/5.5 | 33.6/6.1 | 79 | 2 | 9 | I | C | 0.2 |
| 3 | NB | XAC2546 | Ketoglutarate semialdehyde dehydrogenase (2) | 54.8/6.1 | 56.7/6.7 | 46 | 6 | 8 | I | C | 0.2 |
| 4 | NB | XAC3556 | Aminopeptidase A/I (10) | 51.5/5.2 | 52.5/5.4 | 396 | 10 | 21 | III | C/P | 0.5 |
| XAC1204 | Alanyl dipeptidyl peptidase (8) | 79.7/6.2 | 304 | 11 | 16 | III | P/IM | ||||
| XAC3651 | ATP synthase subunit α (5) | 55.4/5.3 | 104 | 9 | 19 | I | IM | ||||
| XAC2378 | Conserved hypothetical protein (0) | 48.8/5.2 | 53 | 2 | 5 | VIII | C | ||||
| 5 | NB | XAC1838 | Enolase‐phosphatase (2) | 25.8/5.1 | 25.0/5.3 | 50 | 3 | 14 | II | U | 0.4 |
| 6 | NB | XAC0554 | Putative NADH dehydrogenase/NAD(P)H nitroreductase (5) | 21.4/5.8 | 21.4/6.5 | 152 | 9 | 12 | IX | U | 0.3 |
| 7 | NB | XAC1012 | Outer membrane protein (12) | 40.0/4.6 | 34.4/4.2 | 177 | 10 | 26 | IV | OM | 0.2 |
| XAC3556 | Aminopeptidase A/I (2) | 51.5/5.2 | 104 | 2 | 5 | III | C/P | ||||
| XAC0104 | Metalloprotease (2) | 37.8/4.8 | 43 | 2 | 3 | III | S | ||||
| 8 | NB | XAC1078 | ATP‐dependent Clp protease proteolytic subunit (13) | 22.8/5.4 | 236 | 16 | 46 | III | C | 0.4 | |
| XAC2067 | Keto‐hydroxyglutarate‐aldolase/keto‐deoxy‐phosphogluconate aldolase (2) | 22.9/5.2 | 23.0/5.3 | 37 | 4 | 28 | I | C | |||
| 9 | NB | XAC1012 | Outer membrane protein (18) | 39.6/4.6 | 40.9/4.2 | 423 | 18 | 44 | IV | OM | 0.3 |
| 10* | NB | XAC0957 | Elongation factor Tu | 43.3/5.5 | 498 | 20 | 28 | III | C | 0.2 | |
| XAC1348 | Acetoacetyl‐CoA thiolase | 40.1/6.3 | 25.5/5.8 | 229 | 7 | 16 | II | P/IM | |||
| XAC0868 | Hypothetical protein XAC0868 | 28.2/9.1 | 92 | 4 | 12 | VIII | P/S | ||||
| 11* | NB | XAC0541 | 10‐kDa chaperonin (9) | 10.0/5.8 | 11.0/6.0 | 238 | 13 | 94 | III | C | 0.3 |
| 12* | NB | XAC0542 | 60‐kDa chaperonin (32) | 57.1/5.1 | 58.6/5.2 | 2584 | 58 | 68 | III | C | 0.8 |
| 13 | XAM‐M | XAC3345 | Pyruvate kinase type II | 54.1/5.6 | 58.6/5.8 | 42 | 2 | 1 | I | U | Unique |
| 14 | XAM‐M | XAC0823 | Outer membrane hemin receptor | 86.2/6.2 | 75.0/5.5 | 23 | 3 | 3 | V | OM | Unique |
| 15* | XAM‐M | XAC0025 | Xanthomonas conserved hypothetical protein | 22.2/5.7 | 22.3/5.6 | 347 | 11 | 22 | VIII | P | 2.5 |
| XAC3583 | dTDP‐4‐dehydrorhamnose‐3,5‐epimerase | 20.5/5.4 | 153 | 6 | 25 | IV | U | ||||
| 16 | XAM‐M | XAC3442 | Inorganic pyrophosphatase | 19.9/4.9 | 21.6/4.8 | 213 | 9 | 28 | I | C | Unique |
| XAC0296 | Monoxygenase | 52.3/7.9 | 24 | 2 | 1 | I | U | ||||
| 17 | XAM‐M | XAC0470 | Phosphoribosylaminoimidazole‐succinocarboxamide synthase | 34.6/5.2 | 32.7/5.3 | 46 | 4 | 10 | II | C | 16.2 |
| XAC1421 | Elongation factor Ts | 31.1/5.2 | 46 | 2 | 6 | III | C | ||||
| 18 | XAM‐M | XAC3352 | Glyceraldehyde‐3‐phosphate dehydrogenase | 36.2/6.0 | 404 | 25 | 45 | I | C/S | 3.7 | |
| XAC0124 | Fructose‐1,6‐bisphosphatase class 1 | 36.8/5.7 | 80 | 5 | 19 | I | C | ||||
| XAC4367 | Glycerophosphoryl diester phosphodiesterase | 39.8/6.0 | 37.2/6.2 | 72 | 4 | 13 | III | P | |||
| XAC2638 | Hypothetical protein | 37.2/6.2 | 20 | 2 | 3 | VIII | U | ||||
| 19 | XAM‐M | XAC0120 | TldD protein | 48.8/6.0 | 49.1/6.3 | 94 | 4 | 6 | III | C | Unique |
| XAC3352 | Glyceraldehyde‐3‐phosphate dehydrogenase | 36.2/6.0 | 43 | 2 | 4 | I | C | ||||
| 20 | XAM‐M | XAC0865 | Peptidyl‐prolyl cis–trans‐isomerase | 50.1/5.4 | 941 | 36 | 38 | III | P | Unique | |
| XAC0120 | TldD protein | 48.8/6.0 | 48.5/5.3 | 112 | 2 | 6 | III | C | |||
| XAC3851 | Conserved hypothetical protein | 50.0/5.3 | 104 | 3 | 5 | VIII | U | ||||
| 21 | XAM‐M | XAC0865 | Peptidyl‐prolyl cis–trans‐isomerase | 50.1/5.4 | 293 | 13 | 25 | III | P | Unique | |
| XACb0007/XAC3225 | Lytic murein transglycosylase/transglycosylase | 46.2/5.9 | 44.3/5.3 | 293 | 10 | 14 | IV | IM | |||
| XAC3851 | Conserved hypothetical protein | 50.0/5.3 | 129 | 5 | 11 | VIII | U | ||||
| XAC2965 | UDP‐N‐acetylglucosamine 1‐carboxyvinyltransferase | 44.6/5.3 | 69 | 4 | 7 | IV | C | ||||
| 22* | XAM‐M | XAC3352 | Glyceraldehyde‐3‐phosphate dehydrogenase (9) | 36.2/6.0 | 742 | 40 | 47 | I | C/S | 2.9 | |
| XACb0007/XAC3225 | Lytic murein transglycosylase/transglycosylase (0) | 46.2/5.9 | 37.0/6.6 | 230 | 8 | 25 | IV | IM | |||
| XAC0865 | Peptidyl‐prolyl cis–trans‐isomerase (0) | 50.1/5.4 | 43 | 2 | 6 | III | P | ||||
| 23 | XAM‐M | XAC0542 | 60‐kDa chaperonin (10) | 57.1/5.1 | 61.8/6.5 | 580 | 15 | 25 | III | C | Unique |
| 24 | XAM‐M | XAC0542 | 60‐kDa chaperonin (4) | 57.1/5.1 | 59.3/5.6 | 91 | 3 | 10 | III | C | Unique |
| 25 | XAM‐M | XAC0542 | 60‐kDa chaperonin (2) | 57.1/5.1 | 59.1/5.7 | 59 | 2 | 10 | III | C | Unique |
| 26 | XAM‐M | XAC3141 | Outer membrane protein P6 precursor (6) | 19.8/7.6 | 19.2/4.9 | 110 | 8 | 43 | IV | OM | Unique |
| XAC3510 | Peptide deformylase (0) | 19.2/4.8 | 42 | 2 | 8 | III | U | ||||
| 27 | XAM‐M | XAC3584 | Glucose‐1‐phosphate thymidylyltransferase (7) | 34.1/5.8 | 29.4/5.8 | 138 | 8 | 26 | IV | C | Unique |
| XAC1321 | Periplasmic protease (6) | 53.9/7.8 | 118 | 7 | 17 | III | P | ||||
| 28 | XAM‐M | XAC1017 | ABC transporter sulfate‐binding protein (2) | 37.8/6.7 | 33.0/6.7 | 93 | 2 | 5 | V | P | 9.0 |
| 29 | XAM‐M | XAC3141 | Outer membrane protein P6 precursor (6) | 19.8/7.6 | 18.9/4.6 | 90 | 8 | 43 | IV | OM | Unique |
| 30 | XAM‐M | XAC3372 | Transketolase 1 (6) | 72.7/5.6 | 69.8/6.1 | 114 | 5 | 7 | I | C | 9.2 |
| 31 | XAM‐M | XAC2386 | Superoxidase dismutase (12) | 22.7/5.5 | 23.0/5.7 | 303 | 16 | 71 | VII | P/S | 1.8 |
| 32 | XAM‐M | XAC1321 | Periplasmic protease (2) | 53.9/7.8 | 29.8/6.2 | 74 | 2 | 4 | III | P | 2.2 |
| 33 | XAM‐M | XAC3579 | Phosphoglucomutase (8) | 49.3/5.2 | 46.5/5.3 | 146 | 8 | 26 | VII | C | Unique |
| XACb0007/XAC3225 | Lytic murein transglycosylase/transglycosylase (3) | 46.2/5.9 | 123 | 5 | 10 | IV | IM | ||||
| 34 | XAM‐M | XAC1421 | Elongation factor Ts (4) | 31.1/5.2 | 36.7/5.3 | 89 | 4 | 10 | III | C | Unique |
| 35 | XAM‐M | XAC2999 | Peptidase (2) | 79.2/5.8 | 70.4/5.7 | 63 | 4 | 4 | III | S | 6.1 |
| 36 | XAM‐M | XAC1434 | Hypothetical protein XAC1434 (9) | 38.7/5.9 | 34.7/6.2 | 159 | 10 | 29 | VIII | P | 6.6 |
| 37 | XAM‐M | XAC1262 | Conserved hypothetical protein (3) | 63.4/5.9 | 67.7/6.4 | 67 | 5 | 9 | VIII | S | 2.3 |
| 38 | XAM‐M | XACb0007/XAC3225 | Lytic murein transglycosylase/transglycosylase (4) | 46.2/5.9 | 43.8/5.8 | 140 | 6 | 10 | IV | IM | 6.6 |
| 39 | XAM‐M | XAC3649 | ATP synthase subunit β (3) | 51.0/5.2 | 54.0/5.2 | 74 | 4 | 7 | I | IM | Unique |
| XAC0542 | 60‐kDa chaperonin (3) | 57.1/5.1 | 65 | 2 | 4 | III | C | ||||
| 40 | XAM‐M | XAC3235 | Succinyl‐CoA synthetase subunit α (11) | 29.8/6.4 | 18.6/6.9 | 336 | 12 | 37 | I | C/S | 2.7 |
| 41 | XAM‐M | XAC3309 | Aminopeptidase | 48.9/5.9 | 49.2/6.2 | 386 | 22 | 43 | III | S | 15.3 |
| XAC0120 | TldD protein | 48.8/6.0 | 41 | 2 | 4 | III | C | ||||
| 42 | XAM‐M | XACb0007/XAC3225 | Lytic murein transglycosylase/transglycosylase (11) | 46.2/5.9 | 42.9/5.6 | 522 | 17 | 24 | IV | IM | 163.3 |
| XAC0002 | DNA polymerase III subunit β (2) | 40.8/5.4 | 48 | 2 | 4 | III | C | ||||
*Proteins identified using the Mascot/XAC database and National Center for Biotechnology Information (NCBI); in parentheses are indicated the exclusive peptide counts determined for some spots using the software Scaffold™ (Proteome Software Inc., Portland, OR, USA) for 100% protein identification probability. Spots with asterisks (10, 11, 12, 15, 22) were isolated from both NB and XAM‐M conditions (as determined by ImageMaster 2D Platinum software, GE Healthcare, Uppsala, Sweden) and the same protein (in bold) was identified for both spots.
†Theoretical MW/pI obtained from the NCBI database.
‡Experimental MW/pI calculated from the position on the two‐dimensional gel by Image Master Platinum software (GE Healthcare).
§Protein clusters according to their annotated function (da Silva et al., 2002).
¶Predicted cellular location of proteins by PredSi, SignalP 2.0 and SecretomeP 2.0. P, C, IM and OM correspond to periplasm, cytoplasm, inner membrane and outer membrane, respectively. U, unknown, could not be identified. S is used for proteins secreted to the extracellular medium as suggested by SecretomeP 2.0.
**Relative abundance (arbitrary units) for matched spots was calculated as the ratio of the volume percentage average of pathogenicity‐inducing (XAM‐M medium) and pathogenicity‐non‐inducing (NB medium) conditions. ‘Unique’ spots were detected in only one of the two conditions (NB or XAM‐M).
Additional proteins identified in spots not exclusive to, but with increased abundance in, XAM‐M were phosphoribosylaminoimidazole‐succinocarboxamide synthase, elongation factor Ts, aminopeptidase, transketolase 1, ABC transporter sulfate‐binding protein and lytic murein transglycosylase/transglycosylase. The last protein showed the highest XAM‐M/NB ratio (spot 42, Table 1).
The proteins identified were clustered according to their annotated function (da Silva et al., 2002; Soares et al., 2010) into the following classes: (I) intermediary metabolism; (II) biosynthesis of small molecules; (III) macromolecule metabolism; (IV) cell structure; (V) cellular processes; (VI) mobile genetic elements; (VII) pathogenicity, virulence and adaptation; (VIII) hypothetical; (IX) open reading frames (ORFs) with undefined category. Six, one, seven, four, two, zero, three, zero and one protein(s) clustered to each of these categories, respectively (Table 1). Two proteins detected in spots with higher relative abundance in XAM‐M were classified as XAC pathogenicity, virulence, and adaptation (class VII): phosphoglucomutase XAC3579 (spot 33) and superoxide dismutase (SOD) XAC2386 (spot 31) (Fig. 2a; Table 1).
Proteins showing higher abundance in NB samples (spots 1–12) included the outer membrane protein XAC1012 and the enolase‐phosphatase XAC1838 (Table 1, Fig. 2b), and none belonged to class VII.
Expression analysis
Among the proteins identified in spots with relatively higher abundance in XAM‐M, two proteins stand out: SOD, a class VII protein (spot 31), and transglycosylase, detected at the most prominent spot 42 in XAM‐M (Table 1, Fig. 3). The XAM‐M/NB ratios for the two spots were 1.8 and 163, respectively (Table 1). The higher expression in XAM‐M of SOD and spot 42 protein(s) was validated by western blot using the periplasmic‐enriched fraction of XAC cells and anti‐SOD2 (Abcam) or antibodies raised against gel‐extracted protein(s) from spot 42. An interesting result is related to spot 42, which was not detected in NB (Fig. 4a).
Figure 3.
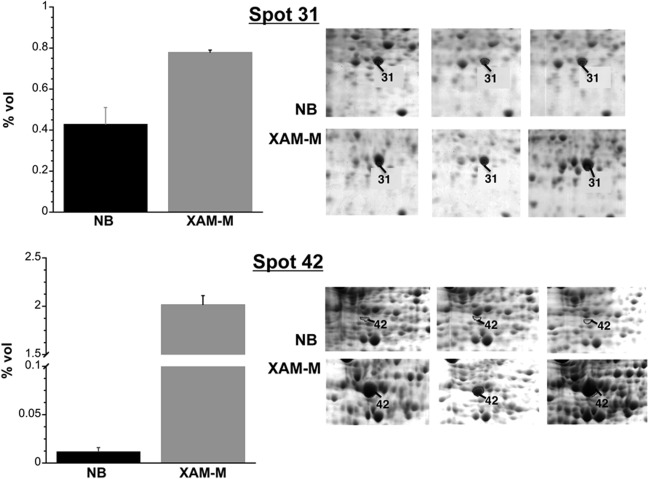
Differential abundance of superoxide dismutase (SOD) and spot 42. Data represent the mean of three independent experiments, and error bars indicate standard deviation. Details of the two‐dimensional gel electrophoresis (2‐DE) gel triplicates are shown in the right panels.
Figure 4.
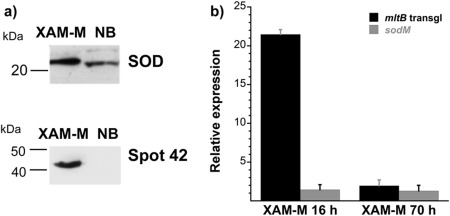
Expression analysis of superoxide dismutase (SOD) and spot 42 protein(s)/MltB. Immunoblot for SOD and spot 42 proteins using the respective antibodies (a); quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of the relative mRNA expression (relative to NB, control, at 17 h) for Xanthomonas citri ssp. citri (XAC) transglycosylase (mltB) and SOD (sodM) on XAM‐M for 16 and 70 h (b). Error bars indicate standard deviation from biological replicates. Molecular mass markers are indicated on the left of the gels in (a).
The higher gene expression on XAM‐M for SOD (sodM, XAC2386) and transglycosylase (mltB, XAC3225) was confirmed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR), which showed relative expression of about one‐ and two‐fold for sodM and two‐ and 20‐fold for mltB at 70 and 16 h of growth, respectively(Fig. 4b).
These results confirmed that SOD and transglycosylase were up‐regulated when XAC was grown on XAM‐M, consistent with the proteomic results.
Discussion
XAC pathogenicity: plant‐mimicking medium and periplasmic proteins
In this work, an in vitro approach was used to identify, for the first time, proteins from the cell periplasm related to the pathogenicity of XAC, the most aggressive agent of citrus canker. Differential proteins were identified between cells growing under steady non‐infectious conditions in NB rich medium and under infection‐mimicking conditions in XAM‐M medium.
XAM‐M is a variant of XAM‐1 medium (Facincani et al., 2014) and includes MgSO4 and casamino acids, similar to the original XVM2 (Wengelnik et al., 1996). Casamino acids have been shown previously to be necessary for the induction of some hrp genes, such as hrpF (Jiang et al., 2013; Schulte and Bonas, 1992).
Here XAM‐M medium was demonstrated to increase the expression of hrp genes when compared with NB medium (Fig. 1b), indicating that it is a plant‐mimicking medium capable of inducing XAC pathogenicity genes in vitro. Although hrp genes expression was higher at the initial phase of growth in XAM‐M (16 h, Fig. 1b), proteomic analysis was performed in cells at the late logarithmic phase because a greater number of cells can be recovered at this stage, increasing the feasibility of subproteomic analysis. This time point also corresponds to the third day of infection in plants, when the recovered bacterial cell mass is sufficient for further analysis (Facincani et al., 2014). At the end of the logarithmic phase, the hrp genes tested (hrpD6, hrpG, hrpB7, hpa1 and hrpX) were still more highly expressed than in the control (Fig. 1b), indicative of the longer term effects of induction.
The hrp genes analysed in this work are involved in the hypersensitive response of Xanthomonas spp. to non‐hosts and in the pathogenicity response to hosts, and are known to be induced in plant‐mimicking hrp‐inducing medium (Rossier et al., 1999). The hrpD6 and hpa1 genes were highly up‐regulated when XAC was grown in Citrus sinensis (Jacob et al., 2014), and hrpG and hrpX were described to be critical for the pathogenicity of X. axonopodis pv. citri (Büttner and Bonas, 2010; Guo et al., 2011). In addition, the hrpB operon has been reported to be related to HrcT expression, a key component of the T3SS in Xanthomonas spp. (Liu et al., 2014). The corresponding Hrp proteins were not detected in our differential study, possibly because they may have a diverse cell location, as in membranes for example, or are not detectable by our approach, requiring further protein purification (Weber et al., 2005).
The proteomic analysis in this study was focused on the periplasmic fraction, a cell compartment that is expected to play an important role in plant–pathogen interactions involving Gram‐negative bacteria. Several proteins identified in this work have been found previously in the periplasm of other Gram‐negative bacteria, among them SOD and PPIase (Wu et al., 2013).
The extraction method using lysozyme and sucrose was similar to that used previously for Pseudomonas aeruginosa (Imperi et al., 2009), which was considered to be the best among the three methods analysed as it provides the highest ratio of periplasmic to cytoplasmic proteins. However, a large number of cytoplasmic proteins (39.6%) were still found (Imperi et al., 2009). Likewise, previous studies with periplasmic proteins have demonstrated that contamination with cytoplasmic proteins is often observed (Agudo et al., 2004; Carniel and Hinnebusch, 2012; Montigiani et al., 2002). It is important to realize, however, that some proteins need to pass through the periplasm to become incorporated in the outer membrane or to be transported to the extracellular milieu, which could explain their transient presence in the periplasmic space (Imperi et al., 2009). It is also possible that some of these proteins may have multiple in vivo locations, with diverse and still unknown biological functions, such as the commonly described ‘moonlighting’ proteins (Mani et al., 2014).
Many XAC proteins found in this study with increased relative abundance under infectious conditions share enzymatic roles in the cell envelope, as shown in Fig. 5 and discussed below.
Figure 5.
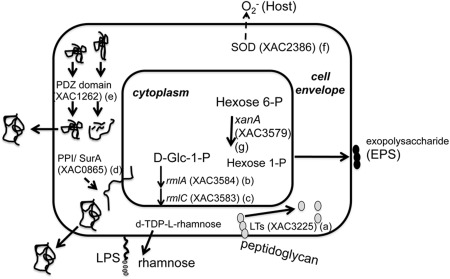
Hypothetical alterations in the cell envelope of Xanthomonas citri ssp. citri (XAC) cells grown under in vitro infectious condition deduced from the periplasmic proteome approach. The enzymatic roles of some proteins positively modulated by XAM‐M and referred to as a–g in the Discussion section are represented. The localization of XAC enzymes is not precisely known.
XAC alters carbohydrate metabolism at the cell wall under pathogenicity induction
Transglycosylase or lytic transglycosylase (LT), XAC3225/XACb0007 (Fig. 5a), catalyses exo‐ or endo‐specific cleavage at the β‐1,4‐glycosidic bond between N‐acetylmuramic acid (MurNAc) and N‐acetyl‐d‐glucosamine (GlcNAc) of peptidoglycan, a protective heteropolymeric macromolecule of the bacterial cell wall (Koraimann, 2003). A large number of enzymes are involved in the synthesis and turnover of this macromolecule (Koraimann, 2003; Typas et al., 2012). Cell wall biosynthesis must be tightly coordinated with its remodelling, as the loss of envelope integrity would lead to lysis and death (Johnson et al., 2013). Spaces in peptidoglycan created by LTs are important for the accommodation of structures, such as secretion systems, flagella and pili, whose unexpected presence causes the cell envelope to expand, requiring rearrangement of the bacterial membrane and cell wall (Koraimann, 2003). Some LTs have been suggested to be involved in secretion systems in bacterial pathogens (Büttner and Bonas, 2010; Hausner et al., 2013). Transglycosylase (XAC3225, mltB gene) has been suggested to be involved in XAC pathogenesis, because its expression is exclusively found during XAC infection in planta (Laia et al., 2008). In addition, a mutant strain in this gene showed impaired in vivo growth in relation to the wild‐type strain (Laia et al., 2008).
Interestingly, although the higher relative transglycosylase expression in XAM‐M was confirmed by western blot and qRT‐PCR assays (Fig. 4), no detectable expression was found for this enzyme in the periplasmic‐enriched fraction of X. fuscans aurantifolii types B (XauB) and C (XauC) (results not shown). These are causative agents of cancrosis, related to a milder form of citrus canker (XauB) and a sole citrus host (XauC). Despite this, XauB and XauC genomes carry genes with 96% identity to XAC3225 from XAC, annotated as T3SS hopAJ‐like proteins. It remains unclear from these results whether peptidoglycan metabolism could be related to different forms of the disease, including aspects such as XAC infectiveness and/or the wide range of hosts, and therefore an understanding of the role of XAC3225 on XAC pathogenicity still demands further studies.
Glucose‐1‐phosphate thymidylyltransferase XAC3584 (Fig. 5b), named as RmlA (EC 2.7.7.24), and dTDP‐4‐dehydrorhamnose‐3,5‐epimerase XAC3583 (Fig. 5c), named as RmlC (EC 5.1.3.13), are enzymes which, together with RmlB and RmlD, are involved in the biosynthesis of dTDP‐l‐rhamnose, the sugar‐nucleotide donor for l‐rhamnose moieties in the cell wall (Glaser and Kornfeld, 1961). All are considered as potential targets for drug design. l‐Rhamnose is a 6‐deoxyhexose rare in nature, but frequently found in the O‐antigen form of the lipopolysaccharides (LPSs) of Gram‐negative bacteria, including XAC (Casabuono et al., 2011). LPSs are important virulence factors for plant‐pathogenic bacteria and are recognized as the major pathogen‐associated molecular patterns (PAMPs) that are responsible for their antigenic properties (Newman et al., 2007). Chromosomal mutation in the rmlC gene of some P. aeruginosa serotypes results in LPS core truncation and loss of pathogenicity (Rahim et al., 2000). Indeed, biogenesis of LPS at the cell envelope has been shown to be important for the XAC infection process, as LPS‐related mutants have been reported to show reduced bacterial virulence (Petrocelli et al., 2012).
XAC up‐regulates proteins required for full virulence, stress response and extracellular polysaccharide (EPS) biosynthesis under pathogenicity condition
PPIases are known to be involved in cell envelope homeostasis and other cell envelope functions in Gram‐negative bacteria. In this study, a putative PPIase, XAC0865 (Fig. 5d), was detected in the XAM‐M sample. The analysis of its amino acid sequence by blast showed the presence of a SurA conserved domain, and 57% similarity to the SurA N‐terminal domain of the corresponding Escherichia coli protein. In this bacterium, the periplasmic chaperone PPIase SurA is required for the cis–trans isomerization of prolyl residues for folding the outer membrane and secreted proteins. This process is necessary because these proteins translocate across the cytoplasmic membrane in a mostly unfolded state (Lazar and Kolter, 1996). Interestingly, outer membrane proteins XAC0823 and XAC3141 were found uniquely in XAM‐M samples (Table 1). SurA probably plays a similar role in other Gram‐negative bacteria, and may be a valuable drug target against Gram‐negative pathogens (Behrens‐Kneip, 2010). It is also involved in pathogenicity, and is required for full virulence of E. coli, Salmonella and Shigella spp. (Behrens‐Kneip, 2010). In Xanthomonas campestris pv. campestris Xcc 8004, a PPIase encoded by XC2699 has been reported to show 49% similarity to the Mip protein (Zang et al., 2007), an important virulence factor for Legionella pneumophila (Cianciotto et al., 1989). It has been shown that this protein is required for full virulence, probably because of its role in exopolysaccharide production and its activity on extracellular proteases. Moreover, PPIase has been related to hrp genes in Xoo (Robin et al., 2014) and to XAC–citrus interaction (Swaroopa Rani and Podile, 2014).
PDZ domain‐containing proteases (HtrA family) are known to play important roles in bacterial cells by modulating disease pathogenesis and cell envelope stress responses (Clausen et al., 2002). XAC1262 (Fig. 5e), formerly annotated as a conserved hypothetical protein, was increased on XAM‐M and encodes a homologue of a protease with the C‐terminal PDZ domain of X. citri pv. citri (Aw12879 strain). Inactivation of a putative HtrA family periplasmic protease (prc gene) from Xoo led to virulence attenuation in vivo and reduced expression of 34 periplasmic proteins associated with proteolysis, macromolecule biosynthesis, carbohydrate or energy metabolism, signal transduction and protein translocation or folding (Deng et al., 2014). It has also been suggested that Prc contributes to Xoo bacterial virulence by acting as a periplasmic modulator of the cell envelope stress responses and by regulating the expression of EPSs (Deng et al., 2014).
Among the proteins detected in the XAM‐M sample, two have been classified previously as being pathogenicity related (class VII): SOD (Fig. 5f) and phosphoglucomutase (Fig. 5g). SOD XAC2386, a metalloprotein which catalyses the dismutation of to H2O2 and O2, has been identified as a virulence factor located in the periplasm of Salmonella enterica serovar Typhimurium (Fang et al., 1999) and Listeria monocytogenes (Hess et al., 1997), and is probably essential for viability in Xcc (Smith et al., 1996). In our study, SOD XAC2386 was found to be up‐regulated in XAM‐M (Fig. 5f) and is probably involved in defence against reactive oxygen species (ROS), which are commonly produced by plants against pathogens during the infection and colonization processes. It has been shown recently that SODs are expressed in the extracellular matrix wall‐bound fraction during the interaction of citrus with XAC (Swaroopa Rani and Podile, 2014). Here, SOD expression was increased in infectious XAC cells relative to non‐infectious cells, as shown by western blot and qRT‐PCR (Fig. 4), corroborating the proteomic analysis.
Phosphoglucomutase XAC3579, xanA, is a bifunctional enzyme that can reroute the metabolic flux originating from glucose‐6‐phosphate and fructose‐6‐phosphate towards the biosynthesis of cell surface glycoconjugates and xanthan, an exopolysaccharide, which is known to play a role in the Xanthomonas–host interaction (Fig. 5g). The enzyme is involved in the conversion of 6‐phosphate hexoses to their corresponding 1‐phosphates, which provides the UDP‐glucose and GDP‐mannose required for the biosynthesis of LPS and xanthan in the Xcc cellular envelope (Köplin et al., 1992). This enzyme has been referred to as a perfect target for the metabolic control of the biosynthesis of these cell surface polysaccharides (Musa et al., 2013), and its involvement in XAC pathogenicity has been confirmed recently by our group (Goto et al., 2016).
XAC displays multiple forms and/or atypical cellular location for classical proteins under infectious conditions
Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) XAC 3352, gapA, is known to participate in the glycolytic pathway and has been predicted to be secreted and/or located in the cytoplasm (Table 1). A number of studies in other bacteria have indicated that GAPDH can occur in cellular locations other than the cytoplasm, so that its recurrent presence in the XAM‐M sample may not be a result of cytoplasmic contamination. Our results suggest, for the first time, that the detection of this enzyme correlates with the induction of known pathogenicity factors (hrp genes). This enzyme has been considered as a virulence factor found on the outer surface or as a secretory product in a number of pathogenic organisms (Aguilera et al., 2012; Seidler, 2013). Furthermore, GAPDH is emerging as one of a group of so‐called ‘moonlighting’ proteins that can exhibit a wide range of biological roles, even in eukaryotic cells (Henderson and Martin, 2014). GAPDH has been demonstrated to be a protein associated with the cell envelopes of Aeromonas hydrophila (Villamón et al., 2003) and Streptococcus pyogenes (Jin et al., 2011), in which the enzyme was described to mediate adhesion to the human host. Inactivation of Xcc GAPDH resulted in impairment of bacterial growth and virulence in the host plant, revealing that GAPDH is required for EPS production and full pathogenicity in Xcc (Lu et al., 2009). The process by which GAPDH is exported and attached to the outer surface of cells remains unknown (Seidler, 2013). Although XAC has only one copy of GAPDH in its genome, our 2‐DE analysis showed that, in XAC cells grown in XAM‐M, GAPDH was present in at least two major forms with different pI (Table 1), probably as a result of post‐translational modifications (PTMs). Recent findings have reported that fructose‐1,6‐bisphosphatase, another typical cytoplasmic enzyme found here in the XAM‐M sample (Table 1), is required for the virulence of Leishmania major in macrophages and mice (Naderer et al., 2006). This protein was found to be located in the periplasm of the eukaryote Saccharomyces cerevisiae during prolonged glucose starvation, despite the absence of signal sequences that are required for classical secretion, indicating secretion mediated by non‐classical pathways (Giardina and Chiang, 2013). These authors mentioned that bacteria and other microorganisms secrete a large number of signal‐less proteins during infection, and that the unusual locations of intracellular proteins might be a result of additional roles unrelated to their well‐known functions. These authors also referred to GAPDH and enolase as glycolytic enzymes with participation in host–pathogen interaction.
The 60‐kDa chaperonin (GroEL, Hsp60 or Cnp60) showed multiple pI forms in XAM‐M samples (e.g. spots 23, 24 and 25; Table 1 and Fig. 2), whereas only one of them was detected in NB. Bacterial heat shock proteins (Hsps) are produced abundantly during the course of most microbial infections, and their roles in protein folding and secretion have been well characterized. In L. pneumophila, Hsp60 is uniquely located in the periplasm and on the bacterial surface, and surface‐associated Hsp60 has been shown to promote attachment and invasion in HeLa cells (Hoffman and Garduno, 1999). Avirulent strains of L. pneumophila were defective in localizing Hsp60 to their surfaces, and were 1000‐fold less capable of invading HeLa cells (Hoffman and Garduno, 1999). GroEL has been demonstrated to be predominantly cytoplasmic and membrane bound in Clostridium difficile, and GroEL‐specific antibodies partially inhibit the attachment of C. difficile cells, suggesting a role for GroEL in cell adherence (Hennequin et al., 2001). Recent findings on candidate adhesins of plant‐pathogenic Xanthomonadaceae include polysaccharide and protein structures such as chaperone/usher pili and other outer membrane adhesins (Mhedbi‐Hajri et al., 2011). In Mycobacterium tuberculosis, Hsp60 is necessary for virulence and has many non‐folding (non‐chaperone) roles, such as secreted signalling molecules and bacterial cell wall components, among others (Stokes, 2013). Additional biological roles of this highly conserved protein are apparently essential for bacterial survival in the host (Henderson et al., 2013).
Evidence for phosphorylation of the 60‐kDa chaperonin has been reported for mycobacterial GroEL (Kumar et al., 2009), but there are no reports for XAC. Phosphorylation events in Xcc have been reported (Musa et al., 2013), together with the participation of prokaryote kinases in host–pathogen interactions involving other bacterial species (Kyriakis, 2014). In addition to GAPDH and 60‐kDa chaperonin, transglycosylase was also detected in this study in several spots in XAM‐M (Table 1, Fig. 2), suggesting the involvement of PTMs.
DnaK and 60‐kDa chaperonin have been found recently at the cellular surface of infectious XAC and have been suggested to be associated with PTMs (Carnielli et al., 2016). However, more experiments are necessary to investigate PTMs on the XAC proteins reported in this study.
Other proteins found in infectious XAC cells
Our results show that proteins such as phosphoribosylaminoimidazole‐succinocarboxamide synthase (XAC0470), transketolase (XAC3372), peptidase (XAC2999) and aminopeptidase (XAC3309) were detected in spots with the highest relative abundances in XAM‐M (16.2, 9.2, 6.1 and 15.3, respectively; Table 1). Although no evident relation with XAC pathogenicity was found for these proteins in the literature, they emerge as interesting XAC targets to be investigated in further functional studies.
Conclusion
The in vitro approach utilized here showed to be a feasible method for mimicking cells under infectious condition in order to detect low‐abundant XAC proteins of the periplasmic subproteomic fraction, as reported in this work for the first time. Proteins with diverse enzymatic roles, from carbohydrate metabolism to the depletion of reactive oxygen species, have been highlighted as promising targets to be investigated in further functional studies. Our results show that the adaptation of XAC to the infectious condition has a significant impact on its envelope metabolic activity, leading to drastic alterations in the abundance and/or location of many enzymes, most not reported previously as being involved in XAC pathogenicity. Possible PTMs of proteins typically known to be cytosol located, found here in the XAC periplasm‐enriched fraction, probably play a role in XAC infectivity. Some proteins reported here are increasingly being mentioned in the literature as ‘moonlighting’ proteins in other organisms, and this interesting feature deserves further investigation in XAC.
Experimental Procedures
Bacterial strains, culture media and growth conditions
The bacterium used in this study was XAC strain 306 (da Silva et al., 2002). XauB 11122 and XauC 10535 (Moreira et al., 2010) were only used for western blot analysis (results not shown). The bacteria were maintained in the rich liquid medium NB (5 g/L peptone and 3 g/L beef extract; Difco™ NB, Sparks, MD, USA) containing 16% (v/v) glycerol, at −80 °C, and were plated on NA (NB with 15 g/L agar; Difco™ NA) at 28 °C. XAC cultures for both growth curves and proteomic analyses were incubated at 200 rpm and 28 °C in either liquid NB medium, a pathogenicity non‐inducing medium used as a control, or in XAM‐M, a pathogenicity‐inducing medium. XAM‐M medium is composed of 7.57 mm (NH4)2SO4, 33.06 mm KH2PO4, 60.28 mm K2HPO4, 1.7 mm sodium citrate (C6H5Na3O7.2H2O), 1 mm MgSO4, 0.03% (w/v) casamino acids, 10 mm fructose, 10 mm sucrose and 1 mg/mL bovine serum albumin (BSA) (Sigma, St. Louis, MO, USA), adjusted to pH 5.4.
XAC growth curves in XAM‐M and NB
XAC was initially grown in NA and then inoculated into 80 mL of NB to determine the end of the logarithmic phase of growth. At this point, two samples of 20 mL of culture were centrifuged (10 000 g, 20 min, 4 °C) and the cells were used to inoculate 400 mL of NB and 400 mL of XAM‐M. Cultures were incubated at 200 rpm and 28 °C. Cell growth was monitored by the measurement of OD595 nm. Similar conditions were used for XAC growth in NB and XAM‐M in the qRT‐PCR analysis. Normalized bacterial suspensions were used to inoculate XAC in 20 mL NB for 17 h, and on 20 mL XAM‐M for 16 and 70 h, followed by incubation at 200 rpm and 28 °C.
Subcellular fractionation
Proteomic assays were performed using XAC cultures grown in 400 mL of NB and XAM‐M, and samples were collected at the end of the logarithmic phase. Samples were collected from experimental duplicates and biological triplicates. A modified lysozyme/ethylenediaminetetraacetic acid (lysozyme/EDTA) spheroplasting method (Hu et al., 1995; Imperi et al., 2009) was used to obtain XAC proteins from the periplasmic‐enriched fractions. Briefly, after centrifugation, the cells were first washed with a buffer composed of 10 mm Tris‐HCl (pH 8), 0.9% (w/v) NaCl, 1 mm EDTA (pH 8) and 10 µL/mL Protease Inhibitor Mix (GE Healthcare, Piscataway, NJ, USA), and then suspended in a buffer containing 10 mm Tris‐HCl (pH 8), 20% (w/v) sucrose, 1 mm EDTA (pH 8), 10 µL/mL Protease Inhibitor Mix and 3 mg/mL lysozyme (Sigma). After incubation for 1 h on ice, followed by centrifugation (11 000 g, 30 min, 4 ºC) of the spheroplasts, cells deprived of their cell wall, the proteins present in the supernatant (periplasmic‐enriched fraction) were obtained by precipitation with cold 10% trichloroacetic acid (TCA) in acetone for 30 min. Following centrifugation (11 000 g, 10 min, 4 °C), the proteins were washed with 70% (v/v) cold ethanol and suspended in a solution containing 7 m urea, 2 m thiourea, 4% (w/v) 3‐[(3‐Cholamidopropyl)dimethylammonio]‐1‐propanesulfonate (CHAPS), 40 mm dithiothreitol (DTT), 1 mm EDTA (pH 8) and 10 µL/mL Protease Inhibitor Mix. Protein concentration was estimated using the Bradford reagent (BioRad, Hercules, CA, USA) (Bradford, 1976), with BSA as the standard. Samples were purified using a 2‐D Clean‐Up kit (GE Healthcare).
2‐DE
2‐DE was performed as described previously (Felício et al., 2011), with minor modifications. The proteins were separated by isoelectric focusing (IEF) using a linear pH 3–10 Immobiline™ DryStrip (13 cm in length, GE Healthcare) and the IPGphor™ system (GE Healthcare). Approximately 260 μg of protein were suspended in 250 µL of buffer containing 8 m urea, 4% (w/v) CHAPS, 50 µL DeStreak™ Rehydration Solution (GE Healthcare) and 1.25 µL IPG Buffer (pH 3–10). IEF was conducted using the following programme: 100 V for 1 h, 500 V for 1 h, 1000 V up 800 Vh, 8000 V up 11 300 Vh, 8000 V up 2900 Vh and 100 V for 10 h, with a total of 16 600 Vh. After equilibration, the protein samples were separated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) (16 × 15 cm2 gel size) using Tris‐glycine as the running buffer (Laemmli, 1970) and a BenchMark™ Protein Ladder (Invitrogen Life Technologies, Carlsbad, CA, USA) as molecular mass standard. The gels were stained with Coomassie brilliant blue R‐250 (CBB R‐250).
2‐DE image acquisition and data analysis
Three 2‐DE gels were examined for each condition, using three independent bacterial cultures. Images of the stained gels were acquired using an ImageScanner™II (GE Healthcare) at 300 dpi resolution and were analysed by ImageMaster™ 2D Platinum 7.0 software (GE Healthcare), wherein the intensity values, experimental molecular weights and pI were estimated for each protein spot. For quantification, the percentage of spot volume criterion was chosen, which is automatically calculated by ImageMaster software considering 100% as the sum of the volume of all spots detected in each gel. Reproducibility among the three gels for each condition (NB or XAM‐M) was assessed by scatter plots which were provided by ImageMaster software analysis, matching the spot pools of one gel, chosen as a reference, against each of the other two gels. Correlation values above 0.9 were considered to be significant. Match analysis between NB and XAM‐M reference gels was performed in an automatic mode, followed by one‐way analysis of variance (ANOVA) statistical analysis of the difference in percentage volume for the corresponding spots.
Only spots with significant differential abundance between XAC in XAM‐M and XAC in NB (P < 0.05), present in three gels of the same condition (replicate), were analysed by MS.
In‐gel digestion and LC‐MS/MS
The spots were excised and digested in gel with Trypsin Gold (Promega, Madison, WI, USA), according to a previously described procedure (Hanna et al., 2000). Trypsin‐digested samples were then analysed using previously described procedures (Aragão et al., 2012; Paes Leme et al., 2012). Briefly, the digested peptides were separated on a C18 column 1.7 μm BEH130 (100 µm × 100 mm, Waters), using a gradient of 2%–90% acetonitrile in 0.1% formic acid, over 10 min. An RP‐nanoUPLC instrument (nanoAcquity, Waters, Milford, MA, USA) coupled to a Q‐TOF Ultima mass spectrometer (Waters) was employed, which is equipped with a nanoelectrospray source operated at a flow rate of 0.6 µL/min. The nanoelectrospray voltage was 3.5 kV, the cone voltage was 30 V and the source temperature was 100 °C. The instrument was operated in top three mode.
Data analysis and processing
MS data were analysed as described previously (Aragão et al., 2012). Spectra were acquired using MassLynx v.4.1 software, and raw data files were converted to peak list format (mgf) using Mascot Distiller v.2.3.2.0 (Matrix Science Ltd.). A search was performed using the Mascot v.2.3.01 engine (Matrix Science Ltd., Boston, MA, USA) against the XAC (strain 306) genome database in the National Center for Biotechnology Information (NCBI) (Accession number NC_003919, 5.4 Mb; 43 427 sequences from one chromosome and two plasmids). Search parameters included carbamidomethylation as a fixed modification, oxidation of methionine as a variable modification, one trypsin missed cleavage and tolerance of 0.1 Da for precursor and fragment ions.
To increase the confidence of protein identification, Mascot output was loaded into Scaffold™ Q+ (Proteome Software Inc., Portland, OR, USA) (Searle, 2010). Peptide identifications were accepted if they could be established at greater than 95% probability. Protein identifications were accepted if they showed greater than 99% probability and contained at least two identified peptides.
Production of antibodies against the major XAC spot in XAM‐M
The 43‐kDa spot, named as spot 42 and strongly expressed under pathogenicity‐inducing condition, was excised from 12 2‐DE gels in order to generate polyclonal antibodies and validate its differential expression using western blot analysis. The antibodies were produced using the following procedure (approved by the Animal Ethics Committee at UFSCar, Brazil, protocol n° 018/2009). Gel fragments were pooled and crushed in 400 µL of ultra‐pure water and the resulting solution was divided into two parts. The first dose involved immunization of two mice by intradermal injection. After 49 days, another dose was injected in a similar manner. Ten days later, the mice were sacrificed and their blood was collected, coagulated and then centrifuged at 8,000 g for 20 min at room temperature. The resulting antiserum was used in western blot analysis.
Western blot analysis
Validation of differential expression was performed by western blot for SOD and for spot 42 protein(s) in XAC cells grown in XAM‐M or NB medium. Equal amounts (60 µg) of proteins from the periplasm‐enriched fraction were separated (in duplicate) by SDS‐PAGE 14% (Laemmli, 1970). One gel was stained with CBB R‐250 and the other was electroblotted onto a nitrocellulose membrane (Hybond‐C Extra, GE Healthcare). The blot was then stained with 0.5% Ponceau S (Sigma) in 0.1% acetic acid to verify equal loading in each lane (Pedras and Minic, 2012). After destaining in water, the membrane was incubated for 5 min (three times) in TBST containing 20 mm Tris, pH 7.4, 0.5 m NaCl and 0.05% v/v Tween‐20 (Sigma), and overnight in 9% non‐fat milk in TBST, washed three times in this buffer and incubated for 2 h with antiserum raised in rabbit against human SOD2 (ab 13533, Abcam, Cambridge, MA, USA) diluted 1 : 5000. The mouse antiserum containing polyclonal antibodies against spot 42 protein(s) was diluted to 1 : 1000. The primary antibodies were detected with anti‐rabbit horseradish peroxidase (HRP) conjugate (ECL Western Blotting, GE Healthcare) diluted to 1 : 5000 and anti‐mouse HRP conjugate, respectively, using a luminol‐based method (ECL Western Blotting/Hyperfilm ECL kit, GE Healthcare).
qRT‐PCR analysis
XAC cells were grown in XAM‐M or NB in a manner similar to that used for the proteomic assay (Fig. 1a), and total RNA was extracted using the QIAGEN RNeasy Protect Bacteria Mini Kit (Qiagen, Valencia, CA, USA). Bacterial suspensions were adjusted to OD595 = 0.3 [108 colony‐forming units (CFU)/mL] and genomic DNA was removed with Qiagen's RNase‐Free DNase. The integrity, purity and concentration of RNA samples were evaluated using a Qubit RNA BR Assay Kit (Invitrogen, Frederick, Maryland, USA) and an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). The first strand of complementary DNA (cDNA) was synthesized from 1 µg of total RNA using a Revertaid™ H Minus First Strand cDNA Synthesis kit (Fermentas, Vilnius, Baltic, Lithuania). Primer Express 3.0 software (Applied Biosystems, Foster City, CA, USA) was used to design the primers for the genes mltB (XAC3225) and sodM (XAC2386). The remaining primers used in this study have been reported previously in Jacob et al. (2011, 2014). Three endogenous genes whose expression stability was accessed using the Expression Suite Software v1.0.3 (Applied Biosystems) were used (atpD, rpoB and gyrB) as controls. The general features of the primers used in this study are described in Table S1 (see Supporting information).
The qRT‐PCRs were performed using SYBR Green Power Master Mix (Applied Biosystems) and were run on an Applied Biosystems 7500 Real‐Time PCR System. Three biological and technical replicates were used for all reactions. The final reaction consisted of 1 × SYBR Green Power Master Mix, 80 ng cDNA and 300 nm of all forward and reverse primers (300/300), except the transglycosylase gene primers (mltB), for which 600 nm of forward and reverse primers (600/600) were used (Table S1). Amplification efficiency was calculated through a standard curve generated using cDNA (1 : 2) serial dilutions. Relative expression was calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Scatter plots indicating the relationship of replicate gels produced from three independent biological experiments for Xanthomonas citri ssp. citri (XAC) on nutrient broth (NB) (a) or XAM‐M (b).
Data S1 Xanthomonas citri ssp. citri (XAC) proteins identified by mass spectrometry (P < 0.05) based on the XAC306 database (Accession number NC_003919). Matched peptides are indicated in bold/underlined. The number of different peptides with the same sequence is shown in parentheses.
Table S1 Primers used in this study for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR).
Acknowledgements
Financial support was mainly provided by Fundação de Amparo à Pesquisa do Estado de São Paulo ‐ FAPESP, Brazil (Young Investigator Grant, Process 2007/50910‐2) and partially by Fundecitrus (Fundo de Defesa da Citricultura ‐ Araraquara, SP, Brazil). We gratefully acknowledge Dr Ana Paula Felicio and Dr Adilson José da Silva for their assistance and discussions, Dr Flávio Henrique Silva and Dr Gilberto Moraes for accessibility to their laboratories, and Dr S. Ferreira for a general revision of the manuscript. We also thank the Mass Spectrometry Laboratory at the Brazilian National Biosciences Laboratory (LNBio), CNPEM, Campinas, Brazil, especially Adriana Franco Paes Leme for mass spectrometry analyses, and T. Dombroski and R. R. Domingues for technical support. J.A., F.d.S.Z. and C.M.C. received fellowships from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), Fundecitrus and FAPESP. The authors declare no financial/commercial conflicts of interest.
References
- Agudo, D. , Mendoza, M.T. , Castañares, C. , Nombela, C. and Rotger, R. (2004) A proteomic approach to study Salmonella typhi periplasmic proteins altered by a lack of the DsbA thiol: disulfide isomerase. Proteomics, 4, 355–363. [DOI] [PubMed] [Google Scholar]
- Aguilera, L. , Ferreira, E. , Giménez, R. , Fernández, F.J. , Taulés, M. , Aguilar, J. , Vega, M.C. , Badia, J. and Baldomà, L. (2012) Secretion of the housekeeping protein glyceraldehyde‐3‐phosphate dehydrogenase by the LEE‐encoded type III secretion system in enteropathogenic Escherichia coli . Int. J. Biochem. Cell Biol. 44, 955–962. [DOI] [PubMed] [Google Scholar]
- Aragão, A.Z.B. , Belloni, M. , Simabuco, F.M. , Zanetti, M.R. , Yokoo, S. , Domingues, R.R. , Kawahara, R. , Pauletti, B.A. , Gonçalves, A. , Agostini, M. , Graner, E. , Coletta, R.D. , Fox, J.W. and Paes Leme AF. (2012) Novel processed form of syndecan‐1 shed from SCC‐9 cells plays a role in cell migration. PLoS One, 7, e43521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens‐Kneip, S. (2010) The role of SurA factor in outer membrane protein transport and virulence. Int. J. Med. Microbiol. 300, 421–428. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brown, K. (2001) Florida fights to stop citrus canker. Science, 292, 2275–2276. [DOI] [PubMed] [Google Scholar]
- Brunings, A.M. and Gabriel, D.W. (2003) Xanthomonas citri: breaking the surface. Mol. Plant Pathol. 4, 141–157. [DOI] [PubMed] [Google Scholar]
- Büttner, D. and Bonas, U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. [DOI] [PubMed] [Google Scholar]
- Carniel, E. and Hinnebusch, B.J. (2012) Yersinia: Systems Biology and Control. UK: Horizon Scientific Press. [Google Scholar]
- Carnielli, C.M. , Artier, J. , de Oliveira, J.C.F. and Novo‐Mansur, M.T.M. (2016) Xanthomonas citri subsp. citri surface proteome by 2D‐DIGE: ferric enterobactin receptor and other outer membrane proteins potentially involved in citric host interaction. J Proteomics 151, 251–263. [DOI] [PubMed] [Google Scholar]
- Casabuono, A. , Petrocelli, S. , Ottado, J. , Orellano, E.G. and Couto, A.S. (2011) Structural analysis and involvement in plant innate immunity of Xanthomonas axonopodis pv. citri lipopolysaccharide. J. Biol. Chem. 286, 25 628–25 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciotto, N. , Eisenstein, B. , Mody, C. , Toews, G. and Engleberg, N. (1989) A Legionella pneumophila gene encoding a species‐specific surface protein potentiates initiation of intracellular infection. Infect. Immun. 57, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, T. , Southan, C. and Ehrmann, M. (2002) The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell, 10, 443–455. [DOI] [PubMed] [Google Scholar]
- da Silva, A.C. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J. , Ferreira, R.C. , Ferro, M.I. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G. , Lemos, M.V. , Locali, E.C. , Machado, M.A. , Madeira, A.M. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A. , Silva, C. , de Souza, R.F. , Spinola, L.A. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Deng, C.Y. , Deng, A.H. , Sun, S.T. , Wang, L. , Wu, J. , Wu, Y. , Chen, X.Y. , Fang, R.X. , Wen, T.Y. and Qian, W. (2014) The periplasmic PDZ domain‐containing protein Prc modulates full virulence, envelops stress responses, and directly interacts with dipeptidyl peptidase of Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 27, 101–112. [DOI] [PubMed] [Google Scholar]
- Facincani, A.P. , Moreira, L.M. , Soares, M.R. , Ferreira, C.B. , Ferreira, R.M. , Ferro, M.I. , Ferro, J.A. , Gozzo, F.C. and de Oliveira, J.C. (2014) Comparative proteomic analysis reveals that T3SS, Tfp, and xanthan gum are key factors in initial stages of Citrus sinensis infection by Xanthomonas citri subsp. citri . Funct. Integr. Genomics, 14, 205–217. [DOI] [PubMed] [Google Scholar]
- Fang, F.C. , DeGroote, M.A. , Foster, J.W. , Bäumler, A.J. , Ochsner, U. , Testerman, T. , Bearson, S. , Giárd, J.C. , Xu, Y. , Campbell, G. and Laessig T. (1999) Virulent Salmonella typhimurium has two periplasmic Cu, Zn‐superoxide dismutases. Proc. Natl. Acad. Sci. USA, 96, 7502–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felício, A.P. , de Oliveira, E. , Odena, M.A. , Garcia, O. Jr , Bertolini, M.C. , Ferraz, L.F.C. , Ottoboni, L.M.M. and Novo, M.T.M. (2011) Differential proteomic analysis of Acidithiobacillus ferrooxidans cells maintained in contact with bornite or chalcopyrite: proteins involved with the early bacterial response. Process Biochem. 46, 770–776. [Google Scholar]
- Giardina, B.J. and Chiang, H.L. (2013) The key gluconeogenic enzyme fructose‐1,6‐bisphosphatase is secreted during prolonged glucose starvation and is internalized following glucose re‐feeding via the non‐classical secretory and internalizing pathways in Saccharomyces cerevisiae . Plant Signal Behav. 8, e24936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser, L. and Kornfeld, S. (1961) The enzymatic synthesis of thymidine‐linked sugars: II. thymidine diphosphate l‐rhamnose. J. Biol. Chem. 236, 1795–1799. [PubMed] [Google Scholar]
- Goto, L.S. , Alexandrino, A.V. , Pereira, C.M. , Martins, C.S. , Pereira, H.D'M. , Brandão‐Neto, J. and Novo‐Mansur, M.T.M. (2016) Structural and functional characterization of the phosphoglucomutase from Xanthomonas citri subsp. citri . Biochim. Biophys. Acta, 1864, 1658–1666. [DOI] [PubMed] [Google Scholar]
- Gottwald, T.R. , Graham, J.H. and Schubert, T.S. (2002) Citrus canker: the pathogen and its impact. Online. Plant Health Progress. doi: 10.1094/PHP-2002-0812-01-RV. [DOI] [Google Scholar]
- Guo, Y. , Figueiredo, F. , Jones, J. and Wang, N. (2011) HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv. citri . Mol. Plant–Microbe Interact. 24, 649–661. [DOI] [PubMed] [Google Scholar]
- Hanna, S.L. , Sherman, N.E. , Kinter, M.T. and Goldberg, J.B. (2000) Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: an analysis by 2‐D gel electrophoresis and capillary column liquid chromatography‐tandem mass spectrometry. Microbiology, 146, 2495–2508. [DOI] [PubMed] [Google Scholar]
- Hausner, J. , Hartmann, N. , Lorenz, C. and Büttner, D. (2013) The periplasmic HrpB1 protein from Xanthomonas spp. binds to peptidoglycan and to components of the type III secretion system. Appl. Environ. Microbiol. 79, 6312–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, B. and Martin, A.C. (2014) Protein moonlighting: a new factor in biology and medicine. Biochem. Soc. Trans. 42, 1671–1678. [DOI] [PubMed] [Google Scholar]
- Henderson, B. , Fares, M.A. and Lund, P.A. (2013) Chaperonin 60: a paradoxical, evolutionarily conserved protein family with multiple moonlighting functions. Biol. Rev. 88, 955–987. [DOI] [PubMed] [Google Scholar]
- Hennequin, C. , Porcheray, F. , Waligora‐Dupriet, A.J. , Collignon, A. , Barc, M.C. , Bourlioux, P. and Karjalainen, T. (2001) GroEL (Hsp60) of Clostridium difficile is involved in cell adherence. Microbiology, 147, 87–96. [DOI] [PubMed] [Google Scholar]
- Hess, J. , Dietrich, G. , Gentschev, I. , Miko, D. , Goebel, W. and Kaufmann, S. (1997) Protection against murine listeriosis by an attenuated recombinant Salmonella typhimurium vaccine strain that secretes the naturally somatic antigen superoxide dismutase. Infect. Immun. 65, 1286–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, P.S. and Garduno, R.A. (1999) Surface‐associated heat shock proteins of Legionella pneumophila and Helicobacter pylori: roles in pathogenesis and immunity. Infect. Dis. Obstet. Gynecol. 7, 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, N.T. , Hung, M.N. , Liao, C.T. and Lin, M.H. (1995) Subcellular location of XpsD, a protein required for extracellular protein secretion by Xanthomonas campestris pv. campestris . Microbiology, 141, 1395–1406. [DOI] [PubMed] [Google Scholar]
- Imperi, F. , Ciccosanti, F. , Perdomo, A.B. , Tiburzi, F. , Mancone, C. , Alonzi, T. , Ascenzi, P. , Piacentini, M. , Visca, P. and Fimia, G.M. (2009) Analysis of the periplasmic proteome of Pseudomonas aeruginosa, a metabolically versatile opportunistic pathogen. Proteomics, 9, 1901–1915. [DOI] [PubMed] [Google Scholar]
- Jacob, T.R. , Laia, M.L. , Ferro, J.A. and Ferro, M.I. (2011) Selection and validation of reference genes for gene expression studies by reverse transcription quantitative PCR in Xanthomonas citri subsp. citri during infection of Citrus sinensis . Biotechnol. Lett. 33, 1177–1184. [DOI] [PubMed] [Google Scholar]
- Jacob, T.R. , Laia, M.L. , Moreira, L.M. , Gonçalves, J.F. , Carvalho, F.M.S. , Ferro, M.I. and Ferro, J.A. (2014) Type IV secretion system is not involved in infection process in citrus. Int. J. Microbiol. 2014, 763575, 9, 2014. doi:10.1155/2014/763575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, G.F. , Jiang, B.L. , Yang, M. , Liu, S. , Liu, J. , Liang, X.X. , Bai, X.F. , Tang, D.J. , Lu, G.T. , He, Y.Q. , Yu, D.Q. and Tang, J.L. (2013) Establishment of an inducing medium for type III effector secretion in Xanthomonas campestris pv. campestris . Braz. J. Microbiol. 44, 945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H. , Agarwal, S. , Agarwal, S. and Pancholi, V. (2011) Surface export of GAPDH/SDH, a glycolytic enzyme, is essential for Streptococcus pyogenes virulence. Mbio, 2, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J.W. , Fisher, J.F. and Mobashery, S. (2013) Bacterial cell‐wall recycling. Ann. N. Y. Acad. Sci. 1277, 54–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köplin, R. , Arnold, W. , Hötte, B. , Simon, R. , Wang, G. and Pühler, A. (1992) Genetics of xanthan production in Xanthomonas campestris: the xanA and xanB genes are involved in UDP‐glucose and GDP‐mannose biosynthesis. J. Bacteriol. 174, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koraimann, G. (2003) Lytic transglycosylases in macromolecular transport systems of gram‐negative bacteria. Cell. Mol. Life Sci. 60, 2371–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, C.M.S. , Khare, G. , Srikanth, C.V. , Tyagi, A.K. , Sardesai, A.A. and Mande, S.C. (2009) Facilitated oligomerization of mycobacterial GroEL: evidence for phosphorylation‐mediated oligomerization. J. Bacteriol. 191, 6525–6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis, J.M. (2014) In the beginning, there was protein phosphorylation. J. Biol. Chem. 289, 9460–9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Laia, M.L. , Moreira, L.M. , Dezajacomo, J. , Brigati, J.B. , Ferreira, C.B. , Ferro, M.I. , Silva, A.C. , Ferro, J.A. and Oliveira, J.C. (2008) New genes of Xanthomonas citri subsp. citri involved in pathogenesis and adaptation revealed by a transposon‐based mutant library. BMC Microbiol. 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, S.W. and Kolter, R. (1996) SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 178, 1770–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.Y. , Zou, L.F. , Xue, X.B. , Cai, L.L. , Ma, W.X. , Xiong, L. , Ji, Z.Y. and Chen, G.Y. (2014) HrcT is a key component of the type III secretion system in Xanthomonas spp. and also regulates the expression of the key hrp transcriptional activator HrpX. Appl. Environ. Microbiol. 80, 3908–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu, G.T. , Xie, J.R. , Chen, L. , Hu, J.R. , An, S.Q. , Su, H.Z. , Feng, J.X. , He, Y.Q. , Jiang, B.L. , Tang, D.J. and Tang, J.L. (2009) Glyceraldehyde‐3‐phosphate dehydrogenase of Xanthomonas campestris pv. campestris is required for extracellular polysaccharide production and full virulence. Microbiology, 155, 1602–1612. [DOI] [PubMed] [Google Scholar]
- Mani, M. , Chen, C. , Amblee, V. , Liu, H. , Mathur, T. , Zwicke, G. , Zabad, S. , Patel, B. , Thakkar, J. and Jeffery, C.J. (2014) MoonProt: a database for proteins that are known to moonlight. Nucleic Acids Res. 43, D277–D282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhedbi‐Hajri, N. , Jacques, M.A. and Koebnik, R. (2011) Adhesion mechanisms of plant‐pathogenic Xanthomonadaceae In: Bacterial Adhesion (Linke D. and Goldman A., eds), pp. 71–89. Netherlands: Springer. [DOI] [PubMed] [Google Scholar]
- Montigiani, S. , Falugi, F. , Scarselli, M. , Finco, O. , Petracca, R. , Galli, G. , Mariani, M. , Manetti, R. , Agnusdei, M. , Cevenini, R. , Donati, M. , Nogarotto, R. , Norais, N. , Garaguso, I. , Nuti, S. , Saletti, G. , Rosa, D. , Ratti, G. and Grandi, G. (2002) Genomic approach for analysis of surface proteins in Chlamydia pneumoniae . Infect. Immun. 70, 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, L.M. , Almeida, N.F. , Potnis, N. , Digiampietri, L.A. , Adi, S.S. , Bortolossi, J.C. , da Silva, A.C. , da Silva, A.M. , de Moraes, F.E. , de Oliveira, J.C. , de Souza, R.F. , Facincani, A.P. , Ferraz, A.L. , Ferro, M.I. , Furlan, L.R. , Gimenez, D.F. , Jones, J.B. , Kitajima, E.W. , Laia, M.L. , Leite, R.P. Jr ., Nishiyama, M.Y. , Neto, R.J. , Nociti, L.A. , Norman, D.J. , Ostroski, E.H. , Pereira, H.A. Jr , Staskawicz, B.J. , Tezza, R.I. , Ferro, J.A. , Vinatzer, B.A. and Setubal, J.C. (2010) Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii . BMC Genomics, 11, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, L.M. , Facincani, A.P. , Ferreira, C.B. , Ferreira, R.M. , Ferro, M.I.T. , Gozzo, F.C. , de Oliveira, J.C. , Ferro, J.A. and Soares, M.R. (2015) Chemotactic signal transduction and phosphate metabolism as adaptive strategies during citrus canker induction by Xanthomonas citri . Funct. Integr. Genomics, 15, 197–210. [DOI] [PubMed] [Google Scholar]
- Musa, Y.R. , Bäsell, K. , Schatschneider, S. , Vorhölter, F.J. , Becher, D. and Niehaus, K. (2013) Dynamic protein phosphorylation during the growth of Xanthomonas campestris pv. campestris B100 revealed by a gel‐based proteomics approach. J. Biotechnol. 167, 111–122. [DOI] [PubMed] [Google Scholar]
- Naderer, T. , Ellis, M.A. , Sernee, M.F. , De Souza, D.P. , Curtis, J. , Handman, E. and McConville, M.J. (2006) Virulence of Leishmania major in macrophages and mice requires the gluconeogenic enzyme fructose‐1, 6‐bisphosphatase. Proc. Natl. Acad. Sci. USA, 103, 5502–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, M.A. , Dow, J.M. , Molinaro, A. and Parrilli, M. (2007) Invited review: priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J. Endotoxin Res. 13, 69–84. [DOI] [PubMed] [Google Scholar]
- Paes Leme, A.F. , Sherman, N.E. , Smalley, D.M. , Sizukusa, L.O. , Oliveira, A.K. , Menezes, M.C. , Fox, J.W. and Serrano, S.M. (2012) Hemorrhagic activity of HF3, a snake venom metalloproteinase: insights from the proteomic analysis of mouse skin and blood plasma. J. Proteome. Res. 11, 279–291. [DOI] [PubMed] [Google Scholar]
- Pedras, M.S.C. and Minic, Z. (2012) Differential protein expression in response to the phytoalexin brassinin allows the identification of molecular targets in the phytopathogenic fungus Alternaria brassicicola . Mol. Plant Pathol. 13, 483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocelli, S. , Tondo, M.L. , Daurelio, L.D. and Orellano, E.G. (2012) Modifications of Xanthomonas axonopodis pv. citri lipopolysaccharide affect the basal response and the virulence process during citrus canker. PLoS One, 7, e40051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim, R. , Burrows, L.L. , Monteiro, M.A. , Perry, M.B. and Lam, J.S. (2000) Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa . Microbiology, 146, 2803–2814. [DOI] [PubMed] [Google Scholar]
- Robin, G.P. , Ortiz, E. , Szurek, B. , Brizard, J.P. and Koebnik, R. (2014) Comparative proteomics reveal new HrpX‐regulated proteins of Xanthomonas oryzae pv. oryzae . J. Proteomics, 97, 256–264. [DOI] [PubMed] [Google Scholar]
- Rossier, O. , Wengelnik, K. , Hahn, K. and Bonas, U. (1999) The Xanthomonas Hrp type III system secretes proteins from plant and mammalian bacterial pathogens. Proc. Natl. Acad. Sci. USA, 96, 9368–9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, N.W. , Postnikova, E. , Lacy, G.H. , Sechler, A. , Agarkova, I. , Stromberg, P.E. , Stromberg, V.K. and Vidaver, A.K. (2005) Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp. citri (ex Hasse) sp. nov. nom. rev. comb. nov., X. fuscans subsp. aurantifolii (ex Gabriel 1989) sp. nov. nom. rev. comb. nov., and X. alfalfae subsp. citrumelo (ex Riker and Jones) Gabriel et al., 1989 sp. nov. nom. rev. comb. nov.; X. campestris pv malvacearum (ex Smith 1901) Dye 1978 as X. smithii subsp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) dye 1978 as X. alfalfae subsp. alfalfae (ex Riker et al., 1935) sp. nov. nom. rev.; and “var. fuscans” of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp. fuscans sp. nov. Syst. Appl. Microbiol. 28, 494–518. [DOI] [PubMed] [Google Scholar]
- Schaad, N.W. , Postnikova, E. , Lacy, G. , Sechler, A. , Agarkova, I. , Stromberg, P.E. , Stromberg, V.K. and Vidaver, A.K. (2006) Emended classification of xanthomonad pathogens on citrus. Syst. Appl. Microbiol. 29, 690–695. [DOI] [PubMed] [Google Scholar]
- Schulte, R. and Bonas, U. (1992) A Xanthomonas pathogenicity locus is induced by sucrose and sulfur‐containing amino acids. Plant Cell, 4, 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle, B.C. (2010) Scaffold: a bioinformatic tool for validating MS/MS‐based proteomic studies. Proteomics, 10, 1265–1269. [DOI] [PubMed] [Google Scholar]
- Seidler, N.W. (2013) GAPDH, as a virulence factor In: GAPDH: Biological Properties and Diversity, pp. 149–178. Dordrecht: Springer. [Google Scholar]
- Smith, S.G. , Wilson, T.G. , Dow, J.M. and Daniels, M.J. (1996) A gene for superoxide dismutase from Xanthomonas campestris pv. campestris and its expression during bacterial–plant interactions. Mol. Plant–Microbe Interact. 9, 584–593. [DOI] [PubMed] [Google Scholar]
- Soares, M.R. , Facincani, A.P. , Ferreira, R.M. , Moreira, L.M. , Oliveira, J.C. , Ferro, J.A. , Ferro, M.I. , Meneghini, R. and Gozzo, F.C. (2010) Proteome of the phytopathogen Xanthomonas citri subsp. citri: a global expression profile. Proteome Sci. 8, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, R.W. (2013) Mycobacterium tuberculosis chaperonin 60 paralogues contribute to virulence in tuberculosis In: Moonlighting Cell Stress Proteins in Microbial Infections. (Henderson B., ed.), pp. 123–141. Dordrecht: Springer. [Google Scholar]
- Swaroopa Rani, T. and Podile, A.R. (2014) Extracellular matrix‐associated proteome changes during non‐host resistance in citrus–Xanthomonas interactions. Physiol. Plant, 150, 565–579. [DOI] [PubMed] [Google Scholar]
- Typas, A. , Banzhaf, M. , Gross, C.A. and Vollmer, W. (2012) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villamón, E. , Villalba, V. , Nogueras, M.M. , Tomás, J.M. , Gozalbo, D. and Gil, M.L. (2003) Glyceraldehyde‐3‐phosphate dehydrogenase, a glycolytic enzyme present in the periplasm of Aeromonas hydrophila . Antonie Van Leeuwenhoek, 84, 31–38. [DOI] [PubMed] [Google Scholar]
- Weber, E. , Ojanen‐Reuhs, T. , Huguet, E. , Hause, G. , Romantschuk, M. , Korhonen, T.K. , Bonas, U. and Koebnik, R. (2005) The type III‐dependent Hrp pilus is required for productive interaction of Xanthomonas campestris pv. vesicatoria with pepper host plants. J. Bacteriol. 187, 2458–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik, K. , Marie, C. , Russel, M. and Bonas, U. (1996) Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. J. Bacteriol. 178, 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S. , Brown, R.N. , Payne, S.H. , Meng, D. , Zhao, R. , Tolić, N. , Cao, L. , Shukla, A. , Monroe, M.E. , Moore, R.J. , Lipton, M.S. and Paša‐Tolić, L. (2013) Top–down characterization of the post‐translationally modified intact periplasmic proteome from the bacterium Novosphingobium aromaticivorans . Int. J. Proteomics, 2013, 279590, 10. https://www.hindawi.com/journals/ijpro/2013/279590/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang, N. , Tang, D.J. , Wei, M.L. , He, Y.Q. , Chen, B. , Feng, J.X. , Xu, J. , Gan, Y.Q. , Jiang, B.L. and Tang, J.L. (2007) Requirement of a mip‐like gene for virulence in the phytopathogenic bacterium Xanthomonas campestris pv. campestris . Mol. Plant–Microbe Interact. 20, 21–30. [DOI] [PubMed] [Google Scholar]
- Zimaro, T. , Thomas, L. , Marondedze, C. , Garavaglia, B.S. , Gehring, C. , Ottado, J. and Gottig, N. (2013) Insights into Xanthomonas axonopodis pv. citri biofilm through proteomics. BMC Microbiol. 13, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Scatter plots indicating the relationship of replicate gels produced from three independent biological experiments for Xanthomonas citri ssp. citri (XAC) on nutrient broth (NB) (a) or XAM‐M (b).
Data S1 Xanthomonas citri ssp. citri (XAC) proteins identified by mass spectrometry (P < 0.05) based on the XAC306 database (Accession number NC_003919). Matched peptides are indicated in bold/underlined. The number of different peptides with the same sequence is shown in parentheses.
Table S1 Primers used in this study for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR).


