Summary
The growing impact of phloem‐limited pathogens on high‐value crops has led to a renewed interest in understanding how they cause disease. Although these pathogens cause substantial crop losses, many are poorly characterized. In this review, we present examples of phloem‐limited pathogens that include intracellular bacteria with and without cell walls, and viruses. Phloem‐limited pathogens have small genomes and lack many genes required for core metabolic processes, which is, in part, an adaptation to the unique phloem environment. For each pathogen class, we present multiple case studies to highlight aspects of disease caused by phloem‐limited pathogens. The pathogens presented include Candidatus Liberibacter asiaticus (citrus greening), Arsenophonus bacteria, Serratia marcescens (cucurbit yellow vine disease), Candidatus Phytoplasma asteris (Aster Yellows Witches’ Broom), Spiroplasma kunkelii, Potato leafroll virus and Citrus tristeza virus. We focus on commonalities in the virulence strategies of these pathogens, and aim to stimulate new discussions in the hope that widely applicable disease management strategies can be found.
Keywords: bacteria, insect vector, pathogen, phloem limited, phytoplasma, spiroplasma, virus
Introduction
Phloem‐limited agricultural pathogens are spreading at an alarming rate, enhanced by warming climates and increasingly interconnected agricultural systems. Current treatment methods often do not specifically target phloem‐limited pathogens, and are frequently preventative rather than curative (Table S2, see Supporting Information). Phloem‐limited pathogens include walled intracellular bacteria, intracellular bacteria without cell walls (Mollicutes) and viruses (Bové and Garnier, 2002; Fletcher and Wayadanda, 2002; Hogenhout et al., 2008).
Phloem‐limited pathogens represent a significant research challenge because they are difficult to detect within plants, and infected plants exhibit variable symptoms that develop slowly. Moreover, these pathogens have complex infection cycles involving both plant hosts and insect vectors (tritrophic interactions). Most phloem‐limited bacteria remain uncultured in vitro, meaning that Koch's postulates cannot be fulfilled, and the bacterial species are designated by the preface ‘Candidatus’. Nevertheless, many have been identified as causative disease agents, using either processes developed for viruses or sequence‐based identification (Bos, 1981; Fredricks and Relman, 1996).
The plant response to pathogens can be divided into microbe‐associated molecular pattern (MAMP)‐triggered immunity (MTI) and effector‐triggered immunity (ETI). MAMPs are slow‐evolving molecules associated with core microbial processes, for example bacterial flagellin (Monaghan and Zipfel, 2012). Damage‐associated molecular patterns (DAMPs) are endogenous signals that result from wounding insect damage, and can induce or amplify immune responses (Wu et al., 2014). Both MAMPs and DAMPs are recognized by pattern recognition receptors, which are commonly receptor‐like kinases (RLKs). Effectors are fast‐evolving molecules associated with infection processes, for example HopZ, and are recognized by nucleotide‐binding leucine‐rich repeat (NB‐LRR) proteins encoded by Resistance (R) genes (Dangl et al., 2013; Hogenhout et al., 2009; Schreiber et al., 2016). The hypersensitive response (HR) is linked to ETI, and is often characterized by localized cell death that is thought to limit pathogen spread. MTI and ETI processes are probably interconnected, and basal disease resistance has been described as a combination of MTI and weak ETI minus the susceptibility caused by pathogen effectors (Bellincampi et al., 2014; Jones and Dangl, 2006; Thomma et al., 2011). Disease resistance in plants is typically studied in non‐vascular tissues, and it is unclear whether MTI and ETI also occur in the phloem. In addition, plants use endogenous RNA‐interference (RNAi) processes to specifically target viral pathogens, and we refer the reader to several excellent reviews on this topic (Duan et al., 2012; Wang et al., 2012).
Pathogens use effector molecules to mould the host environment to suit them; they can (i) block host immune responses, (ii) promote host processes favourable to the pathogen, and/or (iii) reprogram host development in ways that benefit the pathogen. As a result of their intracellular nature, phloem‐limited pathogens probably have different effector delivery methods and use effectors for different purposes than the more widely studied extracellular pathogens.
We first introduce the role of the phloem within the plant, with a particular focus on phloem‐localized transport and defence processes. We also briefly review the insect vectors of phloem‐limited pathogens before moving on to discuss several well‐characterized phloem‐limited pathogens from different classes. These bacteria, Mollicutes and viruses were chosen for the extent of their impact on agricultural systems, the breadth of the literature available or to highlight a particular aspect of disease associated with phloem‐limited pathogens. Throughout, we emphasize how pathogens interact with their hosts to promote virulence.
The Phloem: Transporter of Nutrients and Coordinator of Defence
The phloem is a microaerophilic environment rich in sugars and nutrients, and an environmental niche for plant pathogens (Fig. 1) (Demmig‐Adams et al., 2014; van Dongen et al., 2003; Fatima and Senthil‐Kumar, 2015). Transport through the phloem is directional from sugar‐producing (photosynthetic) source leaves to growing or storage sink tissues that consume sugars (De Schepper et al., 2013; Knoblauch and Peters, 2013). Long‐distance transport through the phloem is thought to be driven by osmotically generated hydrostatic pressure (Schulz et al., 2009; Turgeon, 2010), but the physical aspects of long‐distance phloem transport remain poorly characterized (Knoblauch and Peters, 2010).
Figure 1.
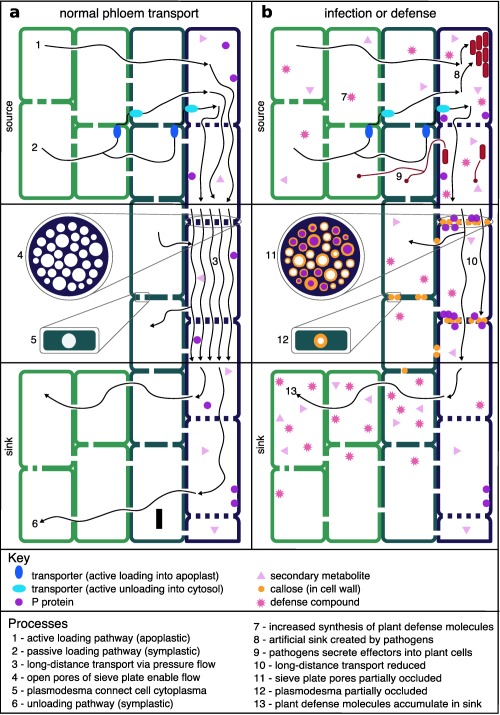
Normal phloem transport (a) and disruption of transport during infection or defence (b). Green cells are mesophyll, teal cells are companion cells, purple cells are phloem cells and all gaps between cells are plasmodesmata, except for sieve plate pores between phloem cells. Numbered generalized processes are shown in the figure and described in the processes section for representative pathogens. The temporal order of infection and defence processes in the phloem remain unclear (see text for details).
The phloem transports both signalling and defence molecules long distances, including hormones, RNA and proteins (van Bel et al., 2013; Dinant and Lemoine, 2010). Phloem transport may not be selective, and only a few molecules have been shown to function in sink tissues (Atkins et al., 2011; Haywood et al., 2005; Paultre et al., 2016). Many of the hormones carried by the phloem are involved in systemic defence processes, with jasmonates and salicylic acid being two well‐studied examples (Fu and Dong, 2013; Wasternack and Hause, 2013). There are two main systemic defence processes caused by phloem‐transported signals: (i) systemic acquired resistance (SAR); and (ii) systemic wound response (SWR) (Gao et al., 2015). Multiple signals have been associated with each of these processes, including hormones, lipid‐derived molecules and reactive oxygen species (ROS) (Gaupels and Vlot, 2012). Electrophysiological changes can occur as a result of many triggers, including insect herbivory, and are another form of defence signalling. These signals rapidly propagate throughout the plant via the phloem and are linked to calcium fluxes (van Bel et al., 2014; Hedrich et al., 2016). Calcium is also involved in other defence processes, such as sieve pore occlusion and local defence signalling cascades, which are not well understood at a mechanistic level (van Bel et al., 2014; Furch et al., 2009; Zhang et al., 2014).
Phloem‐specific defence responses remain poorly characterized because of the difficulty in studying phloem‐specific processes (Fig. 1) (Gaupels and Vlot, 2012; Knoblauch and Peters, 2010). Phloem‐localized proteins, such as forisomes and P proteins, are thought to rapidly seal sieve plates after damage (Batailler et al., 2012; Ernst et al., 2012). Forisomes are only found in legumes, expand in size in response to increased Ca2+ in injured sieve tubes and are able to partially occlude phloem tubes in a reversible manner (Hafke et al., 2009; Knoblauch et al., 2012; Peters et al., 2010). In‐depth studies of SEOR (sieve element occlusion‐related) P proteins, however, showed no evidence that sieve tubes were plugged or that phloem translocation was stopped (Knoblauch et al., 2014). The true physiological role of P proteins therefore remains unclear, and further study is needed. Mobile peptide signals in the phloem, such as systemin, act to propagate defence signals and have been implicated in multiple systemic defence processes (Gaupels and Vlot, 2012). Although the function of many of these proteins and peptides remains unclear, their induction or presence is often used in the study of phloem diseases.
Callose deposition at sieve plates and companion cell plasmodesmata (PD) is an important phloem‐localized response to wounding and pathogens (Hao et al., 2008; Millet et al., 2010; Zavaliev et al., 2011). Although the reported timing of callose deposition varies, it is considered to be a slower process than P protein accumulation at sieve plates (Knoblauch and Peters, 2010; Voigt, 2014). Callose deposits are thought to limit pathogen dispersal, and are considered to be part of MTI (Hao et al., 2008; Luna et al., 2011). Deposits do not completely seal openings, and are important components of plant viral defence (Brunkard et al., 2013; Zavaliev et al., 2011). At present, it remains unclear whether callose deposition in the phloem is a component of MTI. Many studies of phloem‐localized pathogens, however, use callose deposition as a diagnostic indication of disease (Koh et al., 2012).
Secondary plant metabolites, including glucosinolates and pyrrolizidine alkaloids, act to protect plants from herbivores and pests (De Schepper et al., 2013; Savage et al., 2016). Many of these metabolites are inducibly synthesized in response to wounding, and glucosinolates are a particularly well‐studied example of this (Bekaert et al., 2012; Textor and Gershenzon, 2009). Some phloem‐localized pathogens target synthesis or accumulation of secondary metabolites, because their insect vectors are susceptible to them.
Global alterations in resource distribution can be triggered by altered source–sink relationships caused by herbivory and infection (Gómez et al., 2010; Savage et al., 2016). These alterations also lead to the accumulation of signalling molecules and defence compounds in phloem sinks (Arnold et al., 2004; Savage et al., 2016). One example of redistribution is the growth response, in which infected plants increase photosynthesis in healthy leaves and activate dormant meristems (Järemo and Palmqvist, 2001; Lebon et al., 2014). Redistribution can also be used to compartmentalize portions of the plant or withdraw resources from affected aerial tissues (sequestering) (Appel et al., 2012; Frost and Hunter, 2008; Gómez et al., 2010). These strategies make resources unavailable to herbivores, and can allow later regrowth. There is some debate as to whether large‐scale resource reallocations represent energetically efficient defence strategies, but it is clear that pathogens can have a significant effect on plant source–sink allocations (Demmig‐Adams et al., 2014; Huot et al., 2014).
Insect Vectors: Gateway into the Phloem and Alternative Hosts
The insect vectors of phloem‐limited pathogens feed on phloem sap by inserting their stylets into sieve elements. This allows pathogens to directly enter the phloem, bypassing numerous barriers and defence mechanisms within the plant. Key aspects of pathogen transmission are linked to uptake and retention by the insect. The terms used to describe the characteristics of uptake and retention were developed when studying viral pathogens and are used inconsistently in the literature when discussing non‐viral, insect‐transmitted pathogens. For the purposes of this review, we define the three main terms applied to insect‐transmitted pathogens as follows: (i) circulative: pathogens can cross insect cell membranes and be carried internally; (ii) persistent: long feeding times are required for pathogen uptake and pathogens are maintained internally throughout the lifespan of the insect; and (iii) propagative: pathogens replicate in the insect. Phloem‐limited pathogens, with the exception of some viruses, are often circulative. All phloem‐limited pathogens require long feeding times for uptake by the insect, and are internally maintained by the insect for at least a few days (semi‐persistent) or until the end of its life (persistent). Some phloem‐limited pathogens replicate in the insect vector (propagative), whereas others are protected from degradation in insects, but only replicate in plants (non‐propagative) (Gray et al., 2014; Ng and Zhou, 2015; Perilla‐Henao and Casteel, 2016; Rosen et al., 2015). As a result of the difficulties in studying tritrophic interactions, transmission characteristics remain unclear for many phloem‐limited pathogens.
Plant defence responses to phloem‐feeding insects include local wound responses that block the flow of phloem sap (Will et al., 2009, 2013). Aphids are used as model phloem‐feeding insects, and their saliva has been shown to contain effectors that act to repress plant defence responses (Bos et al., 2010; Hogenhout and Bos, 2011; Pitino and Hogenhout, 2013). In resistant plants, aphid effectors are recognized and induce local and systemic defence responses consisting of a combination of MTI, ETI and SWR. At present, it is unclear which mechanisms are more important or how they function together in field conditions (Giordanengo et al., 2010; Will and van Bel, 2008; Züst et al., 2016). By locally removing sugars from the phloem, phloem‐feeding insects create artificial sinks where they are feeding, which disrupts carbohydrate partitioning in the plant. The presence and strength of other sinks, either formed by the plant or created by pathogens, can affect how much of the host's resources can be obtained by the phloem‐feeding insect (Heard and Buchanan, 1998; Inbar et al., 1995; Larson and Whitham, 1997; Savage et al., 2016). Disruption of normal sink–source relationships by insect vectors can contribute to symptom development.
Insects are especially important vectors for plant viruses, and multiple viruses are often transmitted by the same insect species (Gray et al., 2014; Gray and Banerjee, 1999). For phloem‐limited bacteria, the insect–vector relationship and pathogen niche is a polyphyletic trait, indicating that it has evolved independently multiple times (Orlovskis et al., 2015; Perilla‐Henao and Casteel, 2016). Many insect vectors also carry endosymbionts (Akman Gunduz and Douglas, 2009; Łukasik et al., 2013), which provide them with nutrients and protection, and, in some cases, are closely related to phloem‐limited pathogens. Once within the plant, many phloem‐limited bacteria are able to alter the infected plant, such that the insect vector is attracted to it, and will move the bacteria to a new host (Mann et al., 2012; Mas et al., 2014).
Walled Phloem‐Limited Bacteria
Walled phloem‐limited bacterial pathogens are gammaproteobacteria from several different taxa. The insect vector host range appears to be the limiting factor determining which plant species can be infected. These pathogens are primarily found in phloem sieve tubes, but, in some species, they are also present in parenchyma. Phloem‐limited bacterial pathogens have reduced genomes, and have often have lost core metabolic pathways in favour of importers to obtain products made by the plant. Interestingly, these pathogenic bacteria are often closely related to endosymbionts. Much of the experimental work has been carried out in non‐host systems, such as Nicotiana spp. and periwinkle, which have been found to be more tractable than the host plants of phloem‐limited bacteria (Bové and Garnier, 2002). We discuss three examples of these pathogens that illustrate infection mechanics, as well as different evolutionary paths leading to pathogenesis by phloem‐limited microbes.
Liberibacter bacteria: turning the citrus immune system against itself
Citrus greening or Huanglongbing (HLB) is a disease that affects all economically important citrus species, and some close citrus relatives (Tables 1, S1, see Supporting Information). Candidatus Liberibacter asiaticus (CLas) is the primary causative agent of HLB, but Candidatus Liberibacter americanus (CLam) and Candidatus Liberibacter africanus (CLaf) also cause disease in some areas (Bové, 2006; da Graca et al., 2016; Haapalainen, 2014). The characteristics of CLas insect transmission remain unclear; current evidence suggests that CLas is propagative in nymphs, but non‐propagative in adults (Canale et al., 2017; Inoue et al., 2009; Pelz‐Stelinski et al., 2010).
Table 1.
Pathogens and diseases discussed in this review.
| Pathogen | Disease | Host | Vector | References |
|---|---|---|---|---|
| Walled phloem‐limited bacteria | ||||
| Candidatus Liberibacter asiaticus, Candidatus Liberibacter americanus, Candidatus Liberibacter africanus | Citrus greening, Huanglongbing (HLB) | Citrus L. – all economically important citrus species, as well as close citrus relatives | Psyllids: Diaphorina citri, Trioza eritreae | Haapalainen (2014); Wang and Trivedi (2013) |
| Candidatus Phlomobacter fragariae | Marginal chlorosis of strawberry | Fragaria × ananassa | Planthopper: Cixius wagneri | Danet et al., (2003); Nourrisseau et al. (1993) |
| Candidatus Arsenophonus phytopathogenicus | Low‐sugar syndrome (‘Basses richesses’) of sugar beet | Beta vulgaris ssp. vulgaris | Planthopper: Pentastiridius leporinus | Bressan et al. (2012); Sémétey et al. (2007) |
| Serratia marcescens | Cucurbit yellow vine disease | Cucurbitaceae sp. | Squash bug: Anasa tristis | Bruton et al. (2003); Rascoe et al. (2003) |
| Wall‐less phloem‐limited bacteria | ||||
| Candidatus Phytoplasma asteris/Aster Yellows Witches' Broom (AY‐WB), Aster yellows 16SrI‐A subgroup | Aster Yellows (AY) | Daucus carota ssp. sativus, Allium sepa L., Lactuca sativa L., Apium graveolens, Asteraceae | Leafhopper: Macrosteles quadrilineatus | Bai et al. (2006); Bertaccini et al. (2014) |
| Candidatus Phytoplasma asteris/Onion Yellows (OY), 16SrI‐B subgroup | Onion Yellows (OY) | Allium sepa L., Catharanthus roseus, Asteraceae | Leafhopper: Macrosteles striifrons | Bertaccini et al. (2014); Miyahara et al. (1982) |
| Candidatus Phytoplasma vitis/Flavescence dorée (FD), Elm Yellows 16SrV‐C and 16SrV‐D subgroup | Grapevine yellows: Flavescence dorée (FD) | Vitis vinifera | Leafhopper: Scaphoideus titanus | Bertaccini et al. (2014)* |
| Candidatus Phytoplasma solani/Bois Noir (BN), Stolbur 16SrXII‐A subgroup | Grapevine Yellows: Bois Noir (BN) | Vitis vinifera, wild hosts include Convolvulus arvensis L., Urtica dioica L. | Leafhopper: Hyalesthes obsoletus Signoret | Bertaccini et al. (2014) † |
| Candidatus Phytoplasma mali/Apple proliferation (AP), Apple proliferation 16SrX‐A subgroup | Apple proliferation | Malus domestica, Prunus domestica, Prunus avium, Prunus armeniaca, Corylus spp.; wild hosts include Cynodon dactylon, Convolvulus arvensis | Psyllids: Cacopsylla costalis, C. mali, C. melanoneura; Leafhopper: Fiebierella florii | Bertaccini et al. (2014) ‡ |
| Spiroplasma kunkelii | Corn stunt | Zea genus: Z. maydis, Z. perennis, Z. mays mexicana, Z. diploperennis, Z. luxurians | Leafhoppers: Dalbulus maidis, D. eliminatus, Exitianus exitiosus, Graminella nigrifons, Stirellus bicolor | Whitcomb et al. (1986) § |
| Spiroplasma citri | Citrus stubborn; Brittle root disease of horseradish | Citrus L., Amoracia rusticana Brassica spp., wild hosts include Vinca rosea, Sisymbrium irio, Raphanus raphanistrum L. | Leafhoppers: Circulifer tenellus, C. haematoceps, Scaphytopius nitridus, S. delongi | Fletcher et al. (1981); Saglio et al. (1973) |
| Phloem‐limited viruses | ||||
| Citrus tristeza virus (CTV) | Citrus tristeza, Seedling Yellows | Citrus L. – all economically important citrus species | Aphids: Toxoptera citricida, Aphis gossypii, Aphis spiraecola, Toxoptera aurantii | Bar‐Joseph et al. (1989); Moreno et al. (2008) |
| Potato leafroll virus (PLRV) | Potato leaf roll | Solanaceae including Solanum tuberosum spp. | Aphids: Myzus persicae | Taliansky et al. (2003) ¶ |
| Squash leaf curl virus (SLCV) | Squash leaf curl | Cucurbitaceae, Leguminosae, Solanaceae, Euphorbiaceae | Whitefly: Bemisia tabaci | Cohen et al. (1983)** |
This table is not intended to be exhaustive, and further host and vector species, as well as diseases, may be associated with these pathogens.
*CABI ISC datasheet 7642 (http://www.cabi.org/isc/datasheet/7642).
†CABI ISC datasheet 7642 (http://www.cabi.org/isc/datasheet/7642).
‡CABI ISC datasheet 6502 (http://www.cabi.org/isc/datasheet/6502).
§CABI ISC datasheet 50978 (http://www.cabi.org/isc/datasheet/50978).
¶Harrison, B.D. (1984) CMI/AAB Descriptions of Plant Viruses. Potato leafroll virus 291 (no. 36 revised) (http://www.dpvweb.net/dpv/showdpv.php?dpvno = 291).
**Duffus, J.E. and Stenger, D.C. (1998) CMI/AAB Descriptions of Plant Viruses, Squash leaf curl virus 358 (http://www.dpvweb.net/dpv/showdpv.php?dpvno = 358).
Candidatus Liberibacter (CL) genomes are small, and have microsyntenous orthologous regions with their plant endosymbiont relatives Sinorhizobium meliloti, Bradyrhizobium japonicum and Agrobacterium tumefaciens (Kuykendall et al., 2012). CLas is only able to metabolize a limited set of sugars, and probably uses exogenous carbon sources from phloem sap to generate energy. CLas also appears to be adapted to the microaerophilic environment of the phloem; its genome contains multiple components necessary for aerobic respiration (Duan et al., 2009; Wang and Trivedi, 2013). CLas has no restriction‐modification system, and therefore contains multiple prophage regions integrated into its genome, which are differentially expressed in different CLas hosts (Fleites et al., 2014; Zhang et al., 2011). These prophage regions contain peroxidase genes that improve growth in culture and act as secreted effectors to counter host ROS, when expressed in the non‐pathogenic and culturable CLas relative Liberibacter crescens (Jain et al., 2015). Multiple prophage regions have been observed in CLas, CLam and Candidatus Liberibacter solanacearum, and it is thought that these regions allow gene rearrangement in CL species (Duan et al., 2009; Lin et al., 2011; Wulff et al., 2014).
Many of the candidate MAMPs identified in the CL genome are similar to known MAMPs from extracellular bacterial pathogenesis systems (Mott et al., 2014; Segonzac and Zipfel, 2011). On the basis of these studies, MAMP perception is generally thought to happen at the cell surface, but there is some evidence that CL MAMPs are recognized (Hao et al., 2013; Kim et al., 2009). Transcriptional analyses in citrus identified RLKs induced in CLas‐infected plants, suggesting that citrus host cells might traffic CLas MAMPs to the cell surface or use an intermediate signalling molecule (Aritua et al., 2013; Mafra et al., 2013). CLas encodes known MAMPs, such as lipopolysaccharides (LPS) and flagellin (source of flg22). Transgenic expression of the CLas flg22 peptide resulted in callose deposition, but not cell death, making it a weaker MAMP than the flg22 of other plant pathogens (Zou et al., 2012). There is also a fimbrial low‐molecular‐weight protein (flp) pilus system, which is probably involved in tight adherence. Interestingly, the flp pilus is present in CLas, but not in L. crescens (Leonard et al., 2012).
Examination of the processes required for effector delivery showed that CLas lacks a type 3 secretion system (T3SS) (Duan et al., 2009; Galán and Wolf‐Watz, 2006). Instead, CLas has a type 1 secretion system (T1SS), which is another one‐step secretion system important for pathogenesis (Charkowski et al., 2012; Kanonenberg et al., 2013). Multiple ABC transporters have been identified in CLas, and are thought to be involved in outer membrane biogenesis, drug resistance and DNA excision (Li et al., 2012). One of these is a T1SS that may secrete serralysin, which has been shown to contribute to pathogenesis in many bacteria, including Serratia marcescens (Ishii et al., 2014; Li et al., 2012; Maeda and Morihara, 1995). CLas contains some components of T2SS and T4SS; type 4 pili are used by Xylella spp. to block xylem flow (Duan et al., 2009; De La Fuente et al., 2008). T5SS (autotransporters) have also been identified in CLas, and have been shown to localize to the cell surface (Hao et al., 2013). Bacterial effector candidates are often identified by the presence of a signal peptide domain, which directs the proteins to the secretory pathway. As it is unknown what delivery system CLas uses, the pool of proteins with putative secretion signals was computationally screened for potential effector candidates; one candidate was shown to cause cell death in Nicotiana benthamiana (Pitino et al., 2016).
HLB infection primarily affects source–sink relationships, hormone pathways and nutrient distribution within plants, which are all processes for which a functional phloem is essential (Martinelli et al., 2012; Zhao et al., 2013). There are no known resistant citrus varieties or scion–rootstock combinations, although some are more susceptible than others (Fan et al., 2013; Folimonova et al., 2009). Some tested citrus relatives are resistant to HLB, but it remains unclear whether this is a result of plant processes or non‐colonization by HLB vectors (Ramadugu et al., 2016).
Sieve tube occlusion appears to be a primary means of defence against HLB. Blocked sieve elements are thought to kill CLas cells (Trivedi et al., 2009), but this defence mechanism could also be the cause of the disrupted photoassimilate movement seen in CLas‐infected plants (Fan et al., 2013; Kim et al., 2009; Koh et al., 2012). The P protein PP2 is induced by HLB, and callose deposition reduces sieve pore size in infected plants (Kim et al., 2009; Koh et al., 2012). Phloem transport is less affected in CLas‐tolerant citrus varieties, even though susceptible and tolerant varieties have similar signs of HLB infection and defence responses (Fan et al., 2013). Transcriptome profiling shows increased expression levels of genes involved in callose deposition and cell wall breakdown in susceptible varieties, but tolerant varieties have increased expression levels of NBS‐LRR, pathogenesis‐related (PR) and RLK genes (Mafra et al., 2013; Wang et al., 2016). These findings could indicate that susceptible varieties establish callose defences too slowly to prevent pathogen spread, or that ETI is activated more rapidly in tolerant varieties. Induced defence processes in citrus do not appear to be able to restrict HLB spread throughout the plant, possibly because citrus is unable to effectively employ both MTI and ETI against CLas (Canales et al., 2016; Kim et al., 2009; Nwugo et al., 2013; Zou et al., 2012). Another hypothesis is suggested by the early presence of CLas in roots; this colonization may lead to a reservoir of pathogens that can no longer be controlled by plant defence processes (Johnson et al., 2014).
Multiple approaches have been attempted in order to grow CLas cells in the laboratory. For example, the addition of citrus juice, co‐cultivation with insect feeder cells and co‐cultivation with Actinobacteria from citrus have all been reported to improve CLas cultivation success (Davis et al., 2008; Fontaine‐Bodin et al., 2011; Parker et al., 2014). One group developed Liber A agar medium, which includes potassium phosphate, citrus vein extract (CVE) and NADP. The CLas and CLam colonies grown on Liber A were inoculated into young citrus plants, and caused HLB‐like symptoms (Sechler et al., 2009). Although promising, this method has many precise requirements that are not yet understood well enough to enable large‐scale culture of CL species.
Arsenophonus bacteria: from insect endosymbionts to plant pathogens
Marginal chlorosis of strawberry and low‐sugar syndrome of sugar beet are both caused by gammaproteobacteria in the Arsenophonus clade (Bressan, 2014; Sémétey et al., 2007) (Tables 1, S1). The disease‐causing agents are Candidatus Phlomobacter fragariae (CPhfr, marginal chlorosis) and Candidatus Arsenophonus phytopathogenicus (CArph, low‐sugar syndrome), both of which are transmitted by ciixid planthoppers (Bressan et al., 2009; Danet et al., 2003; Zreik et al., 1998). CArph has also been associated with strawberry marginal chlorosis, and can be transmitted to sugar beet by the CPhfr insect vector. Both marginal chlorosis of strawberry and low‐sugar syndrome of sugar beet can also be caused by stolbur phytoplasmas. Co‐infection by CPhfr or CArph and stolbur phytoplasmas has rarely been observed. It remains unclear what role these phytoplasmas play in natural infection systems, and whether they contribute host susceptibility to CPhfr or CArph (Danet et al., 2003; Sémétey et al., 2007).
Both CArph and CPhfr have low genetic diversity, indicating that they are recently emerged plant pathogens (Salar et al., 2010). Other bacteria in the Arsenophonus clade are facultative or secondary insect endosymbionts, which are thought to help insects resist parasites and withstand heat stress (Montllor et al., 2002; Oliver et al., 2003). CArph and CPhfr appear to have independently evolved the ability to infect plants (Bressan et al., 2012; Sémétey et al., 2007). The evolution of insect‐associated bacteria to insect‐vectored plant pathogens is thought to be one way in which phloem‐limited pathogens arise (insect‐first evolution) (Nadarasah and Stavrinides, 2011).
Serratia marcescens: from generalist to specialist
Cucurbit yellow vine disease (CYVD) affects all cucurbits (Tables 1, S1), and is caused by the gammaproteobacterium Serratia marcescens, a generalist bacterium identified in environmental samples and as a pathogen of humans and insects (Mahlen, 2011). The causative agents, CYVD‐causing strains of S. marcescens (CCS), are unable to use the same substrates as other S. marcescens strains, indicating that these strains are distinct and probably adapted to the phloem environment (Rascoe et al., 2003). CCS is vectored by the squash bug Anasa tristis, which feeds on and damages multiple plant organs, including leaves, xylem and phloem (Beard, 1940; Bonjour et al., 1991; Neal, 1993). These generalist feeding habits and large‐scale damage to plant organs distinguish A. tristis from most phloem‐localized pathogen vectors and, indeed, A. tristis was not known to vector plant pathogens before CCS was identified (Bruton et al., 2003). It is unclear where in the vector CCS is maintained, and research has shown CCS can overwinter in dormant A. tristis (Pair et al., 2004; Purcell and Finlay, 1979; Wayadande et al., 2005). Other S. marcescens strains are also plant pathogens, but are not insect vectored or phloem limited (Gillis et al., 2014; Ovcharenko et al., 2010; Wang et al., 2014). These strains, and the generalist nature of A. tristis, raise the intriguing possibility that CCS is an example in which a pathogen first infected plants before evolving to associate with an insect vector and becoming phloem limited (plant‐first evolution) (Nadarasah and Stavrinides, 2011).
Genomic analysis of CCS revealed multiple genes contributing to surface structures that may be involved in pathogenesis. These include rhamnose synthesis pathway genes, which may play a role in adhesion, phosphatase AmsI, which is probably involved in producing extracellular polysaccharide, and surA isomerase, which is important for Salmonella enterica infection processes (Petersen and Tisa, 2013; Zhang et al., 2005). In addition, CCS has type 1 fimbrial pilus genes, which appear to be part of a horizontally transferred genome island (Zhang et al., 2005). Secreted proteases, such as serralysin, have been implicated in S. marcescens virulence, but have not been characterized in CCS (Ishii et al., 2014; Petersen and Tisa, 2013). Similarly, biofilm formation using fimbrial genes and quorum sensing has been shown to be important for the pathogenesis of other S. marcescens strains in non‐plant hosts (Labbate et al., 2007; Shanks et al., 2007). It is possible that CCS uses similar processes to adhere to phloem cells and block phloem sap flow.
Mobile genetic elements probably contributed to the acquisition of the aforementioned genes, a hypothesis supported by the over‐representation of a transposase in CCS (Zhang et al., 2005). In addition, an onion‐infecting S. marcescens strain contains a potentially pathogenesis‐promoting mobile genetic element (Ovcharenko et al., 2010). These findings suggest that the versatility of S. marcescens strains is a result of mobile genetic elements, and that CCS could have specialized in this fashion.
Wall‐Less Phloem‐Limited Bacteria: Candidatus Phytoplasma and Spiroplasma Species
Mollicutes are a class of obligate parasitic bacteria distinguished from other bacteria by their lack of cell walls and small size; they have small genomes and limited metabolic capacities (Bai et al., 2004b; Razin et al., 1998; Woese, 1987). Within the Mollicutes, there are two major clades, referred to as the AAA clade and the SEM clade. Mollicutes live in and on a variety of animal and plant hosts, and many are pathogenic; for example, the human pathogen Mycoplasma pneumonia is a Mollicute. Many Mollicutes have a disproportionate number of repetitive elements for their genome size. These are used to vary cell surface antigens and promote pathogenesis in animal hosts (Bai et al., 2006; Rocha and Blanchard, 2002). There are two groups of insect‐transmitted plant‐pathogenic Mollicutes: phytoplasmas and spiroplasmas (Ammar et al., 2004; Gasparich, 2010; Orlovskis et al., 2015). Phytoplasmas are a monophyletic genus (Candidatus Phytoplasma, CPh) in the AAA clade, whereas spiroplasmas are a genus in the SEM clade (Bai et al., 2004b).
CPh species cause disease in hundreds of economically important plants, have many different shapes and are difficult to culture in laboratory settings (Bai et al., 2006; Lee et al., 2000). They can be transmitted by multiple insect species within the leafhopper, planthopper and psyllid hemipteran insect groups (Garnier et al., 2001; Orlovskis et al., 2015). CPh species are grouped by their 16S rDNA sequence, which remains the sole identifier for many known phytoplasmal pathogens (Bertaccini et al., 2014). CPh pathogens probably use a phytoplasma‐specific pathway to generate energy that could play a role in pathogenesis (Bai et al., 2006; Kube et al., 2012; Saigo et al., 2014). They are thought to adhere to cell surfaces, like other mycoplasmas, and may move through the phloem passively, like viruses (Christensen et al., 2005; Lefol et al., 1993; Razin, 1999). Adhesion may involve actin, and appears to result in the rearrangement of ultrastructures within the sieve elements (Buxa et al., 2015; Musetti et al., 2016). The CPh outer membrane is largely composed of immunodominant membrane proteins (IDPs) of largely unknown function (Kakizawa et al., 2006), although the IDP antigenic membrane protein (Amp) appears to be involved in uptake and internalization by the insect vector (Rashidi et al., 2015). Uniquely, CPh plant pathogens are able to manipulate plant development to cause distinctive phenotypes, such as shoot proliferation and flower virescence. They do this via secreted effector proteins that are able to leave the phloem and target conserved plant transcription factor proteins (Bai et al., 2009; Hoshi et al., 2009; MacLean et al., 2011).
Spiroplasma species are one of the most widespread insect endosymbionts (Shokal et al., 2016). There are over 50 spiroplasma species, all of which have the characteristic spiral shape and are motile (Bové, 1997; Zhao et al., 2004). Endosymbiotic spiroplasmas alter insect immune responses, affect pathogen and endosymbiont titres and selectively kill male insects (Hayashi et al., 2016; Herren and Lemaitre, 2011; Shokal et al., 2016). There are only three known phytopathogenic spiroplasmas, all of which are vectored by leafhoppers (Davis et al., 1979; Orlovskis et al., 2015). Spiroplasmal genomes are less reduced than phytoplasmal genomes: they have more biosynthesis, transcriptional regulation, cell envelope and DNA‐binding genes (Bai and Hogenhout, 2002). Although their genomes are less reduced, spiroplasmas are auxotrophs for sterols, fatty acids and phospholipids, and use a phosphotransferase system to import sugars (Bai et al., 2006; Razin et al., 1998).
We discuss two Mollicutes: a CPh species with well‐characterized effectors, and one spiroplasma that remains an agricultural problem.
Candidatus Phytoplasma asteris strains: pathogens that mould their hosts
The phytoplasma strain Aster Yellows Witches’ Broom (AY‐WB) is a member of the 16SrI‐A subgroup of Candidatus Phytoplasma asteris (CPhas) (Tables 1, S1) (Bai et al., 2006; Lee et al., 2000; Zhang et al., 2004). Within CPhas, genome sizes vary widely, indicating a high degree of genome plasticity. AY‐WB has a small genome without many metabolic processes, but with high repetitive DNA content (Bai et al., 2006). These repetitive regions contain membrane‐targeted sequences involved in membrane‐linked processes, and are probably used to vary the AY‐WB cell surface in different hosts and environments (Bai et al., 2006). AY‐WB has been shown to lengthen the lifespan and improve the fertility of its leafhopper vector (Beanland et al., 2000; Murral et al., 1996).
AY‐WB secretes effectors, such as SAP11 and SAP54, into plant tissues beyond the phloem. SAP11 has a nuclear localization signal, and is found in the nuclei of non‐phloem cells (Bai et al., 2009; Lu et al., 2014). In AY‐WB‐infected Arabidopsis thaliana, SAP11 binds to and destabilizes multiple class II TCP transcription factors, which affects the jasmonic acid (JA) synthesis pathway, weakening plant defences against the insect vector (Sugio et al., 2011, 2014). Destabilizing this set of transcription factors also affects leaf morphogenesis, causing some of the phytoplasma‐induced developmental phenotypes (Lu et al., 2014; Sugio et al., 2011). SAP11 also appears to induce phosphate starvation pathways (Lu et al., 2014).
SAP54 is the AY‐WB effector responsible for the altered flower morphology (virescence and phyllody) seen in infected Arabidopsis (MacLean et al., 2011). It degrades MADS‐domain transcription factor family proteins, such as APETALA1, which are essential floral development regulators (Sugio et al., 2014). Plants with these leaf‐flowers are more attractive to the AY‐WB leafhopper vector, and improve phytoplasmal transmission (MacLean et al., 2014; Orlovskis and Hogenhout, 2016).
In plants infected by the CPhas strain Onion Yellows (OY), TENGU protein was found in apical buds, indicating that it is transported out of the phloem, like the SAPs. It acts to down‐regulate auxin‐responsive genes, including AUXIN RESPONSE FACTOR 6 (ARF6) and ARF8, which regulate floral development and are linked to JA (Hoshi et al., 2009). This indicates that TENGU could regulate both auxin and JA, as well as play a role in the disease‐related altered growth and floral development phenotypes (Minato et al., 2014). Phytoplasma‐infected plants have been termed ‘zombie plants’, because extensive developmental and morphological changes render them sterile (MacLean et al., 2014).
A recent study used multiple complex media to grow phytoplasma strains, including a CPhas strain, from infected grapevine tissue. The strains were found to have highly specific growth requirements, including microaerophilic conditions, high salt concentration and a sterol‐binding antifungal (Contaldo et al., 2016). As with the methods used to culture CL species, the growth requirements for CPh species are not yet well understood. Further study on phloem‐limited bacteria and Mollicutes might benefit from a simulated phloem environment, an approach which has been proven to be successful with uncultivatable environmental bacteria (Kaeberlein et al., 2002; Zengler et al., 2002).The ability to culture these microbes would facilitate advances in genomics, species classification and molecular manipulation of these pathogens.
Spiroplasma kunkelii: a Mollicute with high virulence and low genetic diversity
Spiroplasma kunkelii causes corn stunt disease, and is transmitted by leafhopper insects (Tables 1, S1) (Carloni et al., 2011; Davis et al., 1972; Whitcomb et al., 1986). Extended maize growth periods and the ability of these vectors to overwinter mean that the pathogen can remain present throughout the year (Hruska et al., 1996; Summers et al., 2004). Maize varieties resistant to corn stunt have been bred, but this resistance is short lived. It is unclear why maize resistance is quickly overcome, as genomic analysis has shown that S. kunkelii isolates have low genetic diversity (Carpane et al., 2013). Corn stunt is linked to magnesium metabolism: the symptoms are similar to magnesium deficiency, magnesium is involved in S. kunkelii localization and infected plants seem to be unable to process high magnesium concentrations (Nome et al., 2009).
Two S. kunkelii genes probably involved in insect transmission or plant pathogenicity were identified in a comparative study using AY‐WB: (i) PNPase, a virulence factor regulator in S. enterica; and (ii) CBF, a plasmid replication enhancer (Bai et al., 2004b). The PNPase could be involved in the alteration of gene expression depending on the host environment, whereas CBF could regulate plasmids that may contain virulence factors (Oshima et al., 2002; Razin et al., 1998). One such plasmid (pSKU146) in S. kunkelii carries an adhesin (SARP1), parts of a T4SS and may be involved in genetic exchange (Davis et al., 2005). In Spiroplasma citri, SARP1 acts to attach the pathogen to the insect gut, and contains a conserved Mollicute adhesion motif important in CPhas strain adhesion (Berg et al., 2001; Neriya et al., 2014). Spiroplasma citri probably uses membrane proteins to adhere to insect cells, and has undergone large‐scale genome rearrangement that allows it to be transmissible (Fletcher et al., 1998). Four traE genes are present in S. kunkelii, and their protein sequences are highly similar to the VirB4 domain involved in T4SS pathways (Bai et al., 2004a; Censini et al., 1996; Zatyka and Thomas, 1998). Spiroplasma kunkelii also has fimbriae and pili, and may have morphologically distinct tips that could also be involved in orientation and attachment to host surfaces (Ammar et al., 2004; Özbek et al., 2003). Furthermore, analysis of the S. kunkelii genome sequence found multiple ABC systems, which contribute to virulence in bacterial and fungal pathosystems (Zhao et al., 2004).
Phloem‐Limited Viruses
Viruses use the vasculature to systematically infect the plant, and PD to move between cells. Viral movement is promoted by movement proteins (MPs) and coat proteins (CPs) (Hipper et al., 2013). MPs have several functions within the host plant, including the modification of PD to permit viral genomes and proteins to move between cells. Viruses can move as encapsidated particles or ribonucleoprotein complexes, and many viruses move in multiple forms (Oparka and Cruz, 2000; Solovyev et al., 2012; Verchot‐Lubicz et al., 2010). Although viral replication is tightly linked with movement of the virus (Heinlein, 2015), we focus on viral movement because it appears to play a more important role in restricting viruses to the phloem.
Viral cell‐to‐cell movement through the PD allows the virus to move from the initial site of infection to adjacent cells, and then eventually to the vasculature (long distance) (Heinlein, 2015; Hipper et al., 2013; Oparka and Cruz, 2000). Long‐distance movement through the phloem follows the normal source‐to‐sink movement of sugars, and allows viruses to be trafficked in all directions from the point of entry. Virus loading appears to be possible in all vein classes, whereas virus unloading seems to be limited to major veins in sink tissues (Hipper et al., 2013). Host factors can facilitate or block viral movement, but the mechanism of these processes is not well understood (Ueki and Citovsky, 2007; Wang, 2015). The difficulty of studying phloem‐specific processes has meant that long‐distance viral movement remains poorly characterized. Indeed, many aspects of viral movement are not fully elucidated, and much of what is known only applies to specific systems.
Viral defence processes in plants include RNAi and HR, both of which limit viral movement. Many viral proteins first thought to be involved in systemic spread, including the potyviral HC‐Pro, are, in fact, RNAi suppressors (Taliansky et al., 2008; Ueki and Citovsky, 2007). These suppressors have also been shown to disrupt plant signalling systems, perhaps preventing the activation of systemic viral defences (Alvarado and Scholthof, 2012; Melnyk et al., 2011).
Phloem‐restricted viruses are able to move long distances, but appear to be unable to leave the vasculature and to move cell to cell. The mechanism of this limitation remains unclear, although it is probably a result of a combination of host and viral factors. Plants can be fully resistant to some phloem‐limited viruses, indicating that these pathogens are more readily perceived or controlled than other phloem‐limited pathogens.
We discuss Potato leafroll virus, which has been developed as a model system to study the movement processes of phloem‐restricted viruses. We also discuss Citrus tristeza virus, a phloem‐limited pathogen that citrus is able to resist.
Potato leafroll virus: limiting its own movement
Potato leafroll virus (PLRV) is a positive‐sense RNA Polerovirus (family Luteoviridae) that forms icosahedral virids (Tables 1, S1). Luteovirids are all transmitted by aphids and retained in the phloem. They can move locally and long distance in the phloem, but not between non‐vascular cells or from non‐vascular cells into vascular cells (Taliansky et al., 2003).
PLRV encodes a 17‐kDa MP (MP17/P4) that can localize to PD in some cell types, suggesting that it assists in the movement of PLRV through PD (Link et al., 2011; Vogel et al., 2007). Without MP17, PLRV is either unable to systemically infect or has severely reduced systemic infection ability, confirming that MP17 contributes to viral movement (Lee et al., 2002). The constitutive expression of MP17 at low levels in Arabidopsis increases sucrose efflux from source leaves, as well as overall biomass production, but the expression of MP17 at high levels impairs sucrose efflux, leading to high accumulation in source leaves and reduced vegetative growth (Hofius et al., 2001; Kronberg et al., 2007). These results indicate that MP17 interferes with PD transport, although its effects could be caused by defence processes in response to the presence of MP17 at PD rather than a direct effect on the source–sink system (Rinne et al., 2005).
Two hypotheses for the phloem limitation of viruses have been proposed: (i) host silencing machinery outside the vasculature prevents the virus from leaving; or (ii) the virus does not encode MPs that allow the virus to leave. The first hypothesis was tested using plants expressing the strong RNAi suppressor HC‐Pro. In these plants, the number of PLRV‐infected cells increases, but PLRV is still physically restricted to the phloem (Savenkov and Valkonen, 2001). This suggests that phloem limitation of PLRV is not caused by host silencing. The second hypothesis is supported by co‐infection experiments with PLRV and the potyvirus Potato virus A (PVA), which is not restricted to the vasculature. Co‐infection allows PLRV to exit the phloem and infect all leaf types, indicating that MPs in PVA can complement the movement deficiencies of PLRV. Moreover, these findings indicate that certain co‐infections can remove phloem limitation, a process not seen with other phloem‐limited pathogens (Savenkov and Valkonen, 2001).
Recent work has conclusively established that the phloem‐limiting factor in PLRV is a result of the features of another PLRV MP. PLRV is able to move through the phloem without MP17, instead using its CP and a translational readthrough product (RTP or P3/P5) (Kaplan et al., 2007; Peter et al., 2008). The RTP produces a protein fusion of CP with ORF5, one portion of which is necessary for aphid transmission, and another portion of which is necessary for phloem retention (DeBlasio et al., 2015; Peter et al., 2009). Deletion or mutation of key sections of the RTP portion required for phloem retention allows PLRV to exit the phloem and establish infection in mesophyll tissues (Chavez et al., 2012; Kelley et al., 2009; Peter et al., 2009). Mutated RTPs in the related luteoviruses Beet western yellows virus (BWYV) and Barley yellow dwarf virus (BYDV‐PAV) have reduced viral movement, reduced systemic infection efficiency and accumulate to lower titres (Brault et al., 1995; Chay et al., 1996; Mutterer et al., 1999). RTP has two forms: (i) a non‐incorporated form, which seems to restrict PLRV to the phloem; and (ii) an incorporated form, which replaces CP subunits, protrudes from virions and appears to be necessary for movement into mature tissues (DeBlasio et al., 2015; Peter et al., 2009).
The pathogenic processes of PLRV are less well characterized. Comparison of wild‐type and mutant PLRV strains in N. benthamiana plants did not show altered host protein stability or expression level, indicating that PLRV proteins do not significantly modulate host protein processes during infection (DeBlasio et al., 2015). Multiple wild potato relatives have PLRV resistance loci, for example Solanum tuberosum (Kelley et al., 2009; Marczewski et al., 2004; Novy et al., 2007). In Solanum tuberosum ssp. andigena, resistance was mapped to the upper arm of chromosome V, which contains a known cluster of disease resistance genes (Velásquez et al., 2007). This resistance locus was subsequently identified in other potato varieties (Mihovilovich et al., 2014).
Citrus tristeza virus: recognized by the citrus immune system
Citrus tristeza virus (CTV) is a filamentous, single‐stranded, positive‐sense RNA virus (Moreno et al., 2008) (Tables 1, S1). CTV encodes 12 open reading frames (ORFs) with poorly defined functions. Genetic approaches deleting one or several of these ORFs have demonstrated that full CTV virulence requires proteins for replication, movement and suppression of the host RNAi machinery (Albiach‐Marti, 2013; Pérez‐Clemente et al., 2015). The gene p33 may be an MP, and is required for systemic infection in some citrus species, together with p18 and p13 (Bak and Folimonova, 2015). These three genes appear to be unique to CTV, and may have played a role in increasing the CTV host range (Bak and Folimonova, 2015; Tatineni et al., 2008). The p33 protein is also required for superinfection exclusion (SIE), in which an established viral infection interferes with later infection by closely related viruses (Folimonova, 2012). This process leads to complex spatial and temporal viral infection patterns, which could be important in field infection systems. The p33 protein is involved in systemic SIE, but not cellular SIE (Bergua et al., 2014), and this distinction between cellular and systemic SIE has been seen with human immunodeficiency virus (HIV) in animals (Nethe et al., 2005). These results provide evidence that SIE is a virus‐controlled process, and could be used in the development of viral management strategies. Other genes potentially involved in CTV movement include p6, p20 and the protein components of CTV particle coats (Dolja et al., 2006; Tatineni et al., 2008).
CTV pathogenesis varies depending on the genotype of the host (Dawson et al., 2013). Some citrus species are fully resistant to CTV, unlike CLas, and may achieve resistance by specifically inhibiting viral movement (Albiach‐Marti et al., 2004). One region conferring resistance has been characterized, and found to contain R genes and many retrotransposons, indicating that CTV resistance could function using canonical ETI processes (Bernet et al., 2004; Rai, 2006). Citrus resistance to CTV has been shown to involve both salicylic acid signalling and RNAi, both of which are suppressed by the CTV genes p20 and p23 (Gómez‐Muñoz et al., 2016).
Conclusions/Emerging Themes
The unique environment they inhabit shapes phloem‐limited pathogens. Within the phloem, pathogens have access to the entirety of the plant, including its metabolic output and its means of limiting damage. Insect vectors are key to the access of this central hub, because they directly transmit phloem‐limited pathogens into it. Pathogen, plant and vector form a complex tritrophic system that is difficult to study in its entirety.
Although each phloem‐limited pathogen is unique, the symptoms caused by phloem‐limited pathogens are similar (Table S1). As the diseases caused by phloem‐limited pathogens progress, characteristic symptoms, such as chlorotic leaves and small, bitter fruits, become evident. In most cases, the causal links between the virulence strategies of phloem‐limited pathogens and disease symptoms are not well understood.
Plant hosts of phloem‐limited pathogens can become aware of their presence from the moment the insect vector begins to feed. Insect saliva and wounding can result in phloem blocks and SWR, which the insect will attempt to counter with its own effectors. Phloem‐limited pathogens can also elicit plant defence responses, including MTI, ETI and SAR. Although the phloem is the conduit for many defence‐related compounds, it is unclear how pathogen recognition within the phloem would occur; some studies indicate that Ca2+ signalling could play an important role. Similarly, intracellular pathogen recognition remains poorly characterized in plants. It is possible that phloem‐localized pathogens are able to evade most plant defences merely by being delivered intracellularly. Some of these aspects could be the cause of the slow progression of disease for most phloem‐limited pathogens.
The interaction between phytoplasmas and host defences is of particular interest, because multiple plant species are able to spontaneously recover from phytoplasmal infection. The phytoplasma bois noir (BN) in grapevine (Tables 1, S1) establishes a carbohydrate sink at its infection site, potentially by co‐regulating sucrose transport and cleavage (Santi et al., 2013a). Spontaneously recovered grapevines appear to have restored carbohydrate allocation and increased capacity for both sucrose transport and defence signalling (Santi et al., 2013b). A system using Flavescence dorée phytoplasma (FD) in beans (Vicia faba) showed that phytoplasmas trigger Ca2+ signalling, which, in turn, leads to sieve tube occlusion via forisomes (Musetti et al., 2013). When apple trees spontaneously recover from apple proliferation (Tables 1, S1), symptoms are no longer seen in the crown, but the pathogen is still present in the roots. These recovered trees have higher levels of Ca2+ and H2O2 in the phloem, as well as increased callose and phloem protein accumulation, which could inhibit re‐colonization of the aerial tissues (Musetti et al., 2004, 2010). These studies indicate that spontaneous recovery involves both the re‐establishment of the sink–source system and the use of plant defence mechanisms effectively.
In field settings, many phloem‐limited pathogens can co‐infect plants, which can produce complex infection dynamics and result in genetic exchange. Genomic analysis of phloem‐limited pathogens consistently shows hallmarks of gene transfer and rearrangement, which are facilitated by repetitive regions and plasmids (Bai et al., 2006; Saillard et al., 2008). Phytoplasmas and spiroplasmas can co‐infect both insect and plant hosts, and it has been suggested that they horizontally transfer virulence genes (Bai et al., 2004b; Davis et al., 2005). In viruses, genetic exchange can lead to the production of infectious viral reassortants, such as in the case of the geminiviruses Cucurbit leaf curl virus and Squash leaf curl virus (Brown et al., 2002). Phloem‐limited pathogens can also interact with non‐pathogenic species in their insect vectors; for example, CL species have been shown to acquire genes from Profftella endosymbionts in the psyllid Diaphorina citri (Nakabachi et al., 2013). Genetic exchange has the potential to alter infectivity, increase host and vector range, and could be an important driver of pathogenesis for phloem‐limited pathogens.
Continually improving techniques and analyses have given us more information on these pathogens than ever before. By focusing on commonalities between these pathogens, and on the unique environment that these pathogens share, we will be able to make more progress in the management of these diseases. For example, the development of methods to maintain high rates of phloem transport in infected plants, by disrupting host defences that have evolved to restrict phloem transport around infections, could help to combat many of the phloem‐limited pathogens described in this review. Another useful approach would be to develop tractable systems for each of these pathogen categories. Although some progress on this has been made, well‐understood models would enable more rapid progress to be made with newly emergent pathogens. With these diseases becoming more prevalent worldwide, disease containment focusing on population management is no longer sufficient. New research approaches across traditional boundaries will be needed to develop disease treatments for the modern era.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Symptoms of the diseases discussed in this review.
Table S2 Management strategies for the diseases discussed in this review.
ACKNOWLEDGEMENTS
We would like to thank Saskia Hogenhout, William O. Dawson and an anonymous reviewer for their insightful comments and advice. Research on plant immunity in the Lewis laboratory was supported by the United States Department of Agriculture (USDA) Agricultural Research Service (ARS) 5335‐21000‐040‐00D (J.D.L.) and by the USDA ARS Research Associate Program Class of 2015 (J.D.L.). The authors have no conflicts of interest to declare.
REFERENCES
- Akman Gunduz, E. and Douglas, A. (2009) Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc. R. Soc. B: Biol. Sci. 276, 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiach‐Marti, M.R. (2013) The complex genetics of Citrus tristeza virus In: Current Issues in Molecular Virology – Viral Genetics and Biotechnological Applications. (Romanowski, V., ed.), pp. 1–25. Rijeka: InTech. [Google Scholar]
- Albiach‐Marti, M.R. , Grosser, J.W. , Gowda, S. , Mawassi, M. , Satyanarayana, T. , Garnsey, S.M. and Dawson, W.O. (2004) Citrus tristeza virus replicates and forms infectious virions in protoplasts of resistant citrus relatives. Mol. Breed. 14, 117–128. [Google Scholar]
- Alvarado, V.Y. and Scholthof, H.B. (2012) AGO2: a new Argonaute compromising plant virus accumulation. Front. Plant Sci. 2, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammar, E.D. , Fulton, D. , Bai, X. , Meulia, T. and Hogenhout, S.A. (2004) An attachment tip and pili‐like structures in insect‐ and plant‐pathogenic spiroplasmas of the class Mollicutes. Arch. Microbiol. 181, 97–105. [DOI] [PubMed] [Google Scholar]
- Appel, H.M. , Arnold, T.M. and Schultz, J.C. (2012) Effects of jasmonic acid, branching and girdling on carbon and nitrogen transport in poplar. New Phytol. 195, 419–426. [DOI] [PubMed] [Google Scholar]
- Aritua, V. , Achor, D. , Gmitter, F.G. , Albrigo, G. and Wang, N. (2013) Transcriptional and microscopic analyses of citrus stem and root responses to Candidatus Liberibacter asiaticus infection. PLoS One, 8, e73742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, T. , Appel, H. , Patel, V. , Stocum, E. , Kavalier, A. and Schultz, J. (2004) Carbohydrate translocation determines the phenolic content of Populus foliage: a test of the sink–source model of plant defense. New Phytol. 164, 157–164. [DOI] [PubMed] [Google Scholar]
- Atkins, C.A. , Smith, P.M.C. and Rodriguez‐Medina, C. (2011) Macromolecules in phloem exudates—a review. Protoplasma, 248, 165–172. [DOI] [PubMed] [Google Scholar]
- Bai, X. and Hogenhout, S.A. (2002) A genome sequence survey of the mollicute corn stunt spiroplasma Spiroplasma kunkelii . FEMS Microbiol. Lett. 210, 7–17. [DOI] [PubMed] [Google Scholar]
- Bai, X. , Fazzolari, T. and Hogenhout, S.A. (2004a) Identification and characterization of traE genes of Spiroplasma kunkelii . Gene, 336, 81–91. [DOI] [PubMed] [Google Scholar]
- Bai, X. , Zhang, J. , Holford, I.R. and Hogenhout, S.A. (2004b) Comparative genomics identifies genes shared by distantly related insect‐transmitted plant pathogenic mollicutes. FEMS Microbiol. Lett. 235, 249–258. [DOI] [PubMed] [Google Scholar]
- Bai, X. , Zhang, J. , Ewing, A. , Miller, S.A. , Jancso Radek, A. , Shevchenko, D.V. , Tsukerman, K. , Walunas, T. , Lapidus, A. , Campbell, J.W. and Hogenhout, S.A. (2006) Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188, 3682–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, X. , Correa, V.R. , Toruño, T.Y. , Ammar, E.D. , Kamoun, S. and Hogenhout, S.A. (2009) AY‐WB phytoplasma secretes a protein that targets plant cell nuclei. Mol. Plant–Microbe Interact. 22, 18–30. [DOI] [PubMed] [Google Scholar]
- Bak, A. and Folimonova, S.Y. (2015) The conundrum of a unique protein encoded by Citrus tristeza virus that is dispensable for infection of most hosts yet shows characteristics of a viral movement protein. Virology, 485, 86–95. [DOI] [PubMed] [Google Scholar]
- Bar‐Joseph, M. , Marcus, R. and Lee, R.F. (1989) The continuous challenge of Citrus tristeza virus control. Annu. Rev. Phytopathol. 27, 291–316. [Google Scholar]
- Batailler, B. , Lemaître, T. , Vilaine, F. , Sanchez, C. , Renard, D. , Cayla, T. , Beneteau, J. and Dinant, S. (2012) Soluble and filamentous proteins in Arabidopsis sieve elements. Plant. Cell Environ. 35, 1258–1273. [DOI] [PubMed] [Google Scholar]
- Beanland, L. , Hoy, C.W. , Miller, S.A. and Nault, L.R. (2000) Influence of Aster Yellows Phytoplasma on the fitness of Aster Leafhopper (Homoptera: Cicadellidae). Ann. Entomol. Soc. Am. 93, 271–276. [Google Scholar]
- Beard, R.L. (1940) The biology of Anasa tristis DeGeer with particular reference to the tachinid parasite Trichopoda pennipes Fabr. Connect. Agric. Exp. Sta. Bull. 440, 595–680. [Google Scholar]
- Bekaert, M. , Edger, P.P. , Hudson, C.M. , Pires, J.C. and Conant, G.C. (2012) Metabolic and evolutionary costs of herbivory defense: systems biology of glucosinolate synthesis. New Phytol. 196, 596–605. [DOI] [PubMed] [Google Scholar]
- van Bel, A.J.E. , Helariutta, Y. , Thompson, G.A. , Ton, J. , Dinant, S. , Ding, B. and Patrick, J.W. (2013) Phloem: the integrative avenue for resource distribution, signaling, and defense. Front. Plant Sci. 4, 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel, A.J.E. , Furch, A.C.U. , Will, T. , Buxa, S.V. , Musetti, R. and Hafke, J.B. (2014) Spread the news: systemic dissemination and local impact of Ca2+ signals along the phloem pathway. J. Exp. Bot. 65, 1761–1787. [DOI] [PubMed] [Google Scholar]
- Bellincampi, D. , Cervone, F. and Lionetti, V. (2014) Plant cell wall dynamics and wall‐related susceptibility in plant–pathogen interactions. Front. Plant Sci. 5, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, M. , Melcher, U. and Fletcher, J. (2001) Characterization of Spiroplasma citri adhesion related protein SARP1, which contains a domain of a novel family designated sarpin. Gene, 275, 57–64. [DOI] [PubMed] [Google Scholar]
- Bergua, M. , Zwart, M.P. , El‐Mohtar, C. , Shilts, T. , Elena, S.F. and Folimonova, S.Y. (2014) A viral protein mediates superinfection exclusion at the whole‐organism level but is not required for exclusion at the cellular level. J. Virol. 88, 11 327–11 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernet, G.P. , Bretó, M.P. and Asins, M.J. (2004) Expressed sequence enrichment for candidate gene analysis of Citrus tristeza virus resistance. Theor. Appl. Genet. 108, 592–602. [DOI] [PubMed] [Google Scholar]
- Bertaccini, A. , Duduk, B. , Paltrinieri, S. and Contaldo, N. (2014) Phytoplasmas and phytoplasma diseases: a severe threat to agriculture. Am. J. Plant Sci. 5, 1763–1788. [Google Scholar]
- Bonjour, E.L. , Fargo, W.S. , Webster, J.A. , Richardson, P.E. and Brusewitz, G.H. (1991) Probing behavior comparisons of Squash Bugs (Heteroptera: Coreidae) on Cucurbit hosts. Environ. Entomol. 20, 143–149. [Google Scholar]
- Bos, J.I.B. , Prince, D. , Pitino, M. , Maffei, M.E. , Win, J. and Hogenhout, S.A. (2010) A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (Green Peach Aphid). PLoS Genet. 6, e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, L. (1981) Hundred years of Koch's Postulates and the history of etiology in plant virus research. Neth. J. Pl. Path. 87, 91–110. [Google Scholar]
- Bové, J.M. (1997) Spiroplasmas: infectious agents of plants, arthropods and vertebrates. Wien. Klin. Wochenschr. 109, 604–612. [PubMed] [Google Scholar]
- Bové, J.M. (2006) Huanglongbing: a destructive, newly‐emerging, century‐old disease of citrus. J. Plant Pathol. 88, 7–37. [Google Scholar]
- Bové, J.M. and Garnier, M. (2002) Phloem‐ and xylem‐restricted plant pathogenic bacteria. Plant Sci. 163, 1083–1098. [Google Scholar]
- Brault, V. , van den Heuvel, J.F. , Verbeek, M. , Ziegler‐Graff, V. , Reutenauer, A. , Herrbach, E. , Garaud, J.C. , Guilley, H. , Richards, K. and Jonard, G. (1995) Aphid transmission of Beet western yellows luteovirus requires the minor capsid read‐through protein P74. EMBO J. 14, 650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan, A. (2014) Emergence and evolution of Arsenophonus bacteria as insect‐vectored plant pathogens. Infect. Genet. Evol. 22, 81–90. [DOI] [PubMed] [Google Scholar]
- Bressan, A. , Sémétey, O. , Arneodo, J. , Lherminier, J. and Boudon‐Padieu, E. (2009) Vector transmission of a plant‐pathogenic bacterium in the Arsenophonus clade sharing ecological traits with facultative insect endosymbionts. Phytopathology, 99, 1289–1296. [DOI] [PubMed] [Google Scholar]
- Bressan, A. , Terlizzi, F. and Credi, R. (2012) Independent origins of vectored plant pathogenic bacteria from arthropod‐associated Arsenophonus endosymbionts. Microb. Ecol. 63, 628–638. [DOI] [PubMed] [Google Scholar]
- Brown, J.K. , Idris, A.M. , Alteri, C. and Stenger, D.C. (2002) Emergence of a new Cucurbit‐infecting Begomovirus species capable of forming viable reassortants with related viruses in the Squash leaf curl virus cluster. Phytopathology, 92, 734–742. [DOI] [PubMed] [Google Scholar]
- Brunkard, J.O. , Runkel, A.M. and Zambryski, P.C. (2013) Plasmodesmata dynamics are coordinated by intracellular signaling pathways. Curr. Opin. Plant Biol. 16, 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruton, B.D. , Mitchell, F. , Fletcher, J. , Pair, S.D. , Wayadande, A. , Melcher, U. , Brady, J. , Bextine, B. and Popham, T.W. (2003) Serratia marcescens, a phloem‐colonizing, Squash Bug‐transmitted bacterium: causal agent of Cucurbit yellow vine disease. Plant Dis. 87, 937–944. [DOI] [PubMed] [Google Scholar]
- Buxa, S.V. , Degola, F. , Polizzotto, R. , De Marco, F. , Loschi, A. , Kogel, K.H. , di Toppi, L.S. , Bel, A.J.E. and van Musetti, R. (2015) Phytoplasma infection in tomato is associated with re‐organization of plasma membrane, ER stacks, and actin filaments in sieve elements. Front. Plant Sci. 6, 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canale, M.C. , Tomaseto, A.F. , Haddad, M. D L. , Coletta‐Filho, H.D. and Lopes, J.S. (2017) Latency and persistence of Candidatus Liberibacter asiaticus in its psyllid vector, Diaphorina citri Kuwayama (Hemiptera: Liviidae). Phytopathology. 107, 264–272. [DOI] [PubMed] [Google Scholar]
- Canales, E. , Coll, Y. , Hernández, I. , Portieles, R. , Rodríguez García, M. , López, Y. , Aranguren, M. , Alonso, E. , Delgado, R. , Luis, M. , Batista, L. , Paredes, C. , Rodríguez, M. , Pujol, M. , Ochagavia, M.E. , Falcón, V. , Terauchi, R. , Matsumura, H. , Ayra‐Pardo, C. , Llauger, R. , Pérez Mdel, C. , Núñez, M. , Borrusch, M.S. , Walton, J.D. , Silva, Y. , Pimentel, E. , Borroto, C. and Borrás‐Hidalgo, O. (2016) “ Candidatus Liberibacter asiaticus”, causal agent of citrus Huanglongbing, is reduced by treatment with brassinosteroids. PLoS One, 11, e0146223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carloni, E. , Virla, E. , Paradell, S. , Carpane, P. , Nome, C. , Laguna, I. and Giménez Pecci, M.P. (2011) Exitianus obscurinervis (Hemiptera: Cicadellidae), a new experimental vector of Spiroplasma kunkelii . J. Econ. Entomol. 104, 1793–1799. [DOI] [PubMed] [Google Scholar]
- Carpane, P. , Melcher, U. , Wayadande, A. , de la Paz Gimenez Pecci, M. , Laguna, G. , Dolezal, W. and Fletcher, J. (2013) An analysis of the genomic variability of the phytopathogenic mollicute Spiroplasma kunkelii . Phytopathology, 103, 129–134. [DOI] [PubMed] [Google Scholar]
- Censini, S. , Lange, C. , Xiang, Z. , Crabtree, J.E. , Ghiara, P. , Borodovsky, M. , Rappuoli, R. and Covacci, A. (1996) cag, a pathogenicity island of Helicobacter pylori, encodes type I‐specific and disease‐associated virulence factors. Proc. Natl. Acad. Sci. USA, 93, 14 648–14 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charkowski, A. , Blanco, C. , Condemine, G. , Expert, D. , Franza, T. , Hayes, C. , Hugouvieux‐Cotte‐Pattat, N. , López Solanilla, E. , Low, D. , Moleleki, L. , Pirhonen, M. , Pitman, A. , Perna, N. , Reverchon, S. , Rodríguez Palenzuela, P. , San Francisco, M. , Toth, I. , Tsuyumu, S. , van der Waals, J. , van der Wolf, J. , Van Gijsegem, F. , Yang, C.H. and Yedidia, I. (2012) The role of secretion systems and small molecules in soft‐rot Enterobacteriaceae pathogenicity. Annu. Rev. Phytopathol. 50, 425–449. [DOI] [PubMed] [Google Scholar]
- Chavez, J.D. , Cilia, M. , Weisbrod, C.R. , Ju, H.J. , Eng, J.K. , Gray, S.M. and Bruce, J.E. (2012) Cross‐linking measurements of the Potato leafroll virus reveal protein interaction topologies required for virion stability, aphid transmission, and virus–plant interactions. J. Proteome Res. 11, 2968–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chay, C.A. , Gunasinge, U.B. , Dinesh‐Kumar, S.P. , Miller, W.A. and Gray, S.M. (1996) Aphid transmission and systemic plant infection determinants of Barley yellow dwarf luteovirus‐PAV are contained in the coat protein readthrough domain and 17‐kDa protein, respectively. Virology, 219, 57–65. [DOI] [PubMed] [Google Scholar]
- Christensen, N.M. , Axelsen, K.B. , Nicolaisen, M. and Schulz, A. (2005) Phytoplasmas and their interactions with hosts. Trends Plant Sci. 10, 526–535. [DOI] [PubMed] [Google Scholar]
- Cohen, S. , Duffus, J.E. , Larsen, R.C. , Liu, H.Y. and Flock, R.A. (1983) Purification, serology, and vector relationships of Squash leaf curl virus, a Whitefly‐transmitted Geminivirus . Phytopathology, 73, 1669–1673. [Google Scholar]
- Contaldo, N. , Satta, E. , Zambon, Y. , Paltrinieri, S. and Bertaccini, A. (2016) Development and evaluation of different complex media for phytoplasma isolation and growth. J. Microbiol. Methods, 127, 105–110. [DOI] [PubMed] [Google Scholar]
- Danet, J.L. , Foissac, X. , Zreik, L. , Salar, P. , Verdin, E. , Nourrisseau, J.G. and Garnier, M. (2003) “ Candidatus Phlomobacter fragariae” is the prevalent agent of Marginal chlorosis of strawberry in French production fields and is transmitted by the Planthopper Cixius wagneri (China). Phytopathology, 93, 644–649. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. , Horvath, D.M. and Staskawicz, B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, M.J. , Mondal, S.N. , Chen, H. , Rogers, M.E. and Brlansky, R.H. (2008) Co‐cultivation of “Candidatus Liberibacter asiaticus” with Actinobacteria from citrus with Huanglongbing. Plant Dis. 92, 1547–1550. [DOI] [PubMed] [Google Scholar]
- Davis, R.E. , Worley, J.F. , Whitcomb, R.F. , Ishijima, T. and Steere, R.L. (1972) Helical filaments produced by a Mycoplasma‐like organism associated with corn stunt disease. Science, 176, 521–523. [DOI] [PubMed] [Google Scholar]
- Davis, R.E. , Lee, I.M. and Basciano, L.K. (1979) Spiroplasmas: serological grouping of strains associated with plants and insects. Can. J. Microbiol. 25, 861–866. [DOI] [PubMed] [Google Scholar]
- Davis, R.E. , Dally, E.L. , Jomantiene, R. , Zhao, Y. , Roe, B. , Lin, S. and Shao, J. (2005) Cryptic plasmid pSKU146 from the wall‐less plant pathogen Spiroplasma kunkelii encodes an adhesin and components of a type IV translocation‐related conjugation system. Plasmid, 53, 179–190. [DOI] [PubMed] [Google Scholar]
- Dawson, W.O. , Garnsey, S.M. , Tatineni, S. , Folimonova, S.Y. , Harper, S.J. and Gowda, S. (2013) Citrus tristeza virus–host interactions. Front. Microbiol. 4, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBlasio, S.L. , Johnson, R. , Sweeney, M.M. , Karasev, A. , Gray, S.M. , MacCoss, M.J. and Cilia, M. (2015) Potato leafroll virus structural proteins manipulate overlapping, yet distinct protein interaction networks during infection. Proteomics, 15, 2098–2112. [DOI] [PubMed] [Google Scholar]
- De La Fuente, L. , Burr, T.J. and Hoch, H.C. (2008) Autoaggregation of Xylella fastidiosa cells is influenced by type I and type IV pili. Appl. Environ. Microbiol. 74, 5579–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig‐Adams, B. , Stewart, J.J. and Adams, W.W. (2014) Multiple feedbacks between chloroplast and whole plant in the context of plant adaptation and acclimation to the environment. Philos. Trans. R. Soc. London B: Biol. Sci. 369, 20130244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schepper, V. , De Swaef, T. , Bauweraerts, I. and Steppe, K. (2013) Phloem transport: a review of mechanisms and controls. J. Exp. Bot. 64, 4839–4850. [DOI] [PubMed] [Google Scholar]
- Dinant, S. and Lemoine, R. (2010) The phloem pathway: new issues and old debates. C. R. Biol. 333, 307–319. [DOI] [PubMed] [Google Scholar]
- Dolja, V.V. , Kreuze, J.F. and Valkonen, J.P.T. (2006) Comparative and functional genomics of closteroviruses. Virus Res. 117, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen, J.T. , Schurr, U. , Pfister, M. and Geigenberger, P. (2003) Phloem metabolism and function have to cope with low internal oxygen. Plant Physiol. 131, 1529–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, C.G. , Wang, C.H. and Guo, H.S. (2012) Application of RNA silencing to plant disease resistance. Silence, 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Y. , Zhou, L. , Hall, D.G. , Li, W. , Doddapaneni, H. , Lin, H. , Liu, L. , Vahling, C.M. , Gabriel, D.W. , Williams, K.P. , Dickerman, A. , Sun, Y. and Gottwald, T. (2009) Complete genome sequence of citrus huanglongbing bacterium, “Candidatus Liberibacter asiaticus” obtained through metagenomics. Mol. Plant–Microbe Interact. 22, 1011–1020. [DOI] [PubMed] [Google Scholar]
- Ernst, A.M. , Jekat, S.B. , Zielonka, S. , Müller, B. , Neumann, U. , Rüping, B. , Twyman, R.M. , Krzyzanek, V. , Prüfer, D. and Noll, G.A. (2012) Sieve element occlusion (SEO) genes encode structural phloem proteins involved in wound sealing of the phloem. Proc. Natl. Acad. Sci. 109, E1980–E1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , Chen, C. , Achor, D.S. , Brlansky, R.H. , Li, Z.G. and Gmitter, F.G. (2013) Differential anatomical responses of tolerant and susceptible citrus species to the infection of “Candidatus Liberibacter asiaticus”. Physiol. Mol. Plant Pathol. 83, 69–74. [Google Scholar]
- Fatima, U. and Senthil‐Kumar, M. (2015) Plant and pathogen nutrient acquisition strategies. Front. Plant Sci. 6, 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleites, L.A. , Jain, M. , Zhang, S. and Gabriel, D.W. (2014) “ Candidatus Liberibacter asiaticus” prophage late genes may limit host range and culturability. Appl. Environ. Microbiol. 80, 6023–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, J. and Wayadanda, A. (2002) Fastidious vascular‐colonizing bacteria. Plant Health Instr. DOI: 10.1094/PHI‐I‐2002‐1218‐02 [Google Scholar]
- Fletcher, J. , Wayadande, A. , Melcher, U. and Ye, F. (1998) The phytopathogenic mollicute–insect vector interface: a closer look. Phytopathology, 88, 1351–1358. [DOI] [PubMed] [Google Scholar]
- Fletcher, J. , Schultz, G.A. , Davis, R.E. , Eastman, C.E. and Goodman, R.M. (1981) Brittle root disease of horseradish: evidence for an etiological role of Spiroplasma citri . Phytopathology, 71, 1073. [Google Scholar]
- Folimonova, S.Y. (2012) Superinfection exclusion is an active virus‐controlled function that requires a specific viral protein. J. Virol. 86, 5554–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folimonova, S.Y. , Robertson, C.J. , Garnsey, S.M. , Gowda, S. and Dawson, W.O. (2009) Examination of the responses of different genotypes of citrus to huanglongbing (citrus greening) under different conditions. Phytopathology, 99, 1346–1354. [DOI] [PubMed] [Google Scholar]
- Fontaine‐Bodin, L. , Fabre, S. , Gatineau, F. and Dollet, M. (2011) In vitro culture of the fastidious bacteria Candidatus Liberibacter asiaticus in association with insect feeder cells. In: 2nd International Research Conference on Huanglongbing, Orlando, USA, January 10–14, 2011.
- Fredricks, D.N. and Relman, D.A. (1996) Sequence‐based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin. Microbiol. Rev. 9, 18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost, C.J. and Hunter, M.D. (2008) Herbivore‐induced shifts in carbon and nitrogen allocation in red oak seedlings. New Phytol. 178, 835–845. [DOI] [PubMed] [Google Scholar]
- Fu, Z.Q. and Dong, X. (2013) Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. [DOI] [PubMed] [Google Scholar]
- Furch, A.C.U. , van Bel, A.J.E. , Fricker, M.D. , Felle, H.H. , Fuchs, M. and Hafke, J.B. (2009) Sieve element Ca2+ channels as relay stations between remote stimuli and sieve tube occlusion in Vicia faba . Plant Cell, 21, 2118–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán, J.E. and Wolf‐Watz, H. (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature, 444, 567–573. [DOI] [PubMed] [Google Scholar]
- Gao, Q.M. , Zhu, S. , Kachroo, P. and Kachroo, A. (2015) Signal regulators of systemic acquired resistance. Front. Plant Sci. 6, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier, M. , Foissac, X. , Gaurivaud, P. , Laigret, F. , Renaudin, J. , Saillard, C. and Bové, J.M. (2001) Mycoplasmas, plants, insect vectors: a matrimonial triangle. C. R. Seances Acad. Sci. III 324, 923–928. [DOI] [PubMed] [Google Scholar]
- Gasparich, G.E. (2010) Spiroplasmas and phytoplasmas: microbes associated with plant hosts. Biologicals, 38, 193–203. [DOI] [PubMed] [Google Scholar]
- Gaupels, F. and Vlot, A.C. (2012) Plant defense and long‐distance signaling in the phloem In: Phloem: Molecular Cell Biology, Systemic Communication, Biotic Interactions, (Thompson, G. A. and van Bel, A. J. E., eds.) pp. 227–247. Oxford: Wiley. [Google Scholar]
- Gillis, A. , Rodríguez, M. and Santana, M.A. (2014) Serratia marcescens associated with bell pepper (Capsicum annuum L.) soft‐rot disease under greenhouse conditions. Eur. J. Plant Pathol. 138, 1–8. [Google Scholar]
- Giordanengo, P. , Brunissen, L. , Rusterucci, C. , Vincent, C. , van Bel, A. , Dinant, S. , Girousse, C. , Faucher, M. and Bonnemain, J.L. (2010) Compatible plant–aphid interactions: how aphids manipulate plant responses. C. R. Biol. 333, 516–523. [DOI] [PubMed] [Google Scholar]
- Gómez, S. , Ferrieri, R.A. , Schueller, M. and Orians, C.M. (2010) Methyl jasmonate elicits rapid changes in carbon and nitrogen dynamics in tomato. New Phytol. 188, 835–844. [DOI] [PubMed] [Google Scholar]
- Gómez‐Muñoz, N. , Velázquez, K. , Vives, M.C. , Ruiz‐Ruiz, S. , Pina, J.A. , Flores, R. , Moreno, P. and Guerri, J. (2016) The resistance of sour orange to Citrus tristeza virus is mediated by both the salicylic acid and the RNA silencing defense pathways. Mol. Plant Pathol. DOI: 10.111/mpp.12488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Graca, J.V. , Douhan, G.W. , Halbert, S.E. , Keremane, M.L. , Lee, R.F. , Vidalakis, G. and Zhao, H. (2016) Huanglongbing: an overview of a complex pathosystem ravaging the world's citrus. J. Integr. Plant Biol. 58, 373–387. [DOI] [PubMed] [Google Scholar]
- Gray, S. , Cilia, M. and Ghanim, M. (2014) Chapter Four – Circulative, “nonpropagative” virus transmission: an orchestra of virus‐, insect‐, and plant‐derived instruments. Adv. Virus Res. 89, 141–199. [DOI] [PubMed] [Google Scholar]
- Gray, S.M. and Banerjee, N. (1999) Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 63, 128–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapalainen, M. (2014) Biology and epidemics of Candidatus Liberibacter species, psyllid‐transmitted plant‐pathogenic bacteria. Ann. Appl. Biol. 165, 172–198. [Google Scholar]
- Hafke, J.B. , Furch, A.C.U. , Fricker, M.D. and van Bel, A.J.E. (2009) Forisome dispersion in Vicia faba is triggered by Ca2+ hotspots created by concerted action of diverse Ca2+ channels in sieve elements. Plant Signal Behav. 4, 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, G. , Boyle, M. , Zhou, L. and Duan, Y. (2013) The intracellular citrus huanglongbing bacterium, “Candidatus Liberibacter asiaticus” encodes two novel autotransporters. PLoS One, 8, e68921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, P. , Liu, C. , Wang, Y. , Chen, R. , Tang, M. , Du, B. , Zhu, L. and He, G. (2008) Herbivore‐induced callose deposition on the sieve plates of rice: an important mechanism for host resistance. Plant Physiol. 146, 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, M. , Watanabe, M. , Yukuhiro, F. , Nomura, M. and Kageyama, D. (2016) A nightmare for males? A maternally transmitted male‐killing bacterium and strong female bias in a green lacewing population. PLoS One, 11, e0155794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood, V. , Yu, T.S. , Huang, N.C. and Lucas, W.J. (2005) Phloem long‐distance trafficking of GIBBERELLIC ACID‐INSENSITIVE RNA regulates leaf development. Plant J. 42, 49–68. [DOI] [PubMed] [Google Scholar]
- Heard, S.B. and Buchanan, C.K. (1998) Larval performance and association within and between two species of hackberry nipple gall insects, Pachypsylla spp. (Homoptera: Psyllidae). Am. Midl. Nat. 140, 351–357. [Google Scholar]
- Hedrich, R. , Salvador‐Recatalà, V. and Dreyer, I. (2016) Electrical wiring and long‐distance plant communication. Trends Plant Sci. 21, 376–387. [DOI] [PubMed] [Google Scholar]
- Heinlein, M. (2015) Plant virus replication and movement. Virology, 479–480, 657–671. [DOI] [PubMed] [Google Scholar]
- Herren, J.K. and Lemaitre, B. (2011) Spiroplasma and host immunity: activation of humoral immune responses increases endosymbiont load and susceptibility to certain Gram‐negative bacterial pathogens in Drosophila melanogaster . Cell. Microbiol. 13, 1385–1396. [DOI] [PubMed] [Google Scholar]
- Hipper, C. , Brault, V. , Ziegler‐Graff, V. and Revers, F. (2013) Viral and cellular factors involved in phloem transport of plant viruses. Front. Plant Sci. 4, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius, D. , Herbers, K. , Melzer, M. , Omid, A. , Tacke, E. , Wolf, S. and Sonnewald, U. (2001) Evidence for expression level‐dependent modulation of carbohydrate status and viral resistance by the Potato leafroll virus movement protein in transgenic tobacco plants. Plant J. 28, 529–543. [DOI] [PubMed] [Google Scholar]
- Hogenhout, S.A. and Bos, J.I. (2011) Effector proteins that modulate plant–insect interactions. Curr. Opin. Plant Biol. 14, 422–428. [DOI] [PubMed] [Google Scholar]
- Hogenhout, S.A. , Ammar, E.D. , Whitfield, A.E. and Redinbaugh, M.G. (2008) Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46, 327–359. [DOI] [PubMed] [Google Scholar]
- Hogenhout, S.A. , Van der Hoorn, R.A.L. , Terauchi, R. and Kamoun, S. (2009) Emerging concepts in effector biology of plant‐associated organisms. Mol. Plant–Microbe Interact. 22, 115–122. [DOI] [PubMed] [Google Scholar]
- Hoshi, A. , Oshima, K. , Kakizawa, S. , Ishii, Y. , Ozeki, J. , Hashimoto, M. , Komatsu, K. , Kagiwada, S. , Yamaji, Y. and Namba, S. (2009) A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc. Natl. Acad. Sci. USA, 106, 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruska, A.J. , Gladstone, S.M. and Obando, R. (1996) Epidemic roller coaster: maize stunt disease in Nicaragua. Am. Entomol. 42, 248–252. [Google Scholar]
- Huot, B. , Yao, J. , Montgomery, B.L. and He, S.Y. (2014) Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant. 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar, M. , Eshel, A. and Wool, D. (1995) Interspecific competition among phloem‐feeding insects mediated by induced host‐plant sinks. Ecology, 76, 1506–1515. [Google Scholar]
- Inoue, H. , Ohnishi, J. , Ito, T. , Tomimura, K. , Miyata, S. , Iwanami, T. and Ashihara, W. (2009) Enhanced proliferation and efficient transmission of Candidatus Liberibacter asiaticus by adult Diaphorina citri after acquisition feeding in the nymphal stage. Ann. Appl. Biol. 155, 29–36. [Google Scholar]
- Ishii, K. , Adachi, T. , Hamamoto, H. and Sekimizu, K. (2014) Serratia marcescens suppresses host cellular immunity via the production of an adhesion‐inhibitory factor against immunosurveillance cells. J. Biol. Chem. 289, 5876–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, M. , Fleites, L. and Gabriel, D.W. (2015) Prophage encoded peroxidase in “Candidatus Liberibacter asiaticus” is a secreted effector that suppresses plant defenses. Mol. Plant–Microbe Interact. 28, 1330–1337. [DOI] [PubMed] [Google Scholar]
- Järemo, J. and Palmqvist, E. (2001) Plant compensatory growth: a conquering strategy in plant–herbivore interactions? Evol. Ecol. 15, 91–102. [Google Scholar]
- Johnson, E.G. , Wu, J. , Bright, D.B. and Graham, J.H. (2014) Association of “Candidatus Liberibacter asiaticus” root infection, but not phloem plugging with root loss on huanglongbing‐affected trees prior to appearance of foliar symptoms. Plant Pathol. 63, 290–298. [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nat. Rev. 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kaeberlein, T. , Lewis, K. and Epstein, S.S. (2002) Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science, 296, 1127–1129. [DOI] [PubMed] [Google Scholar]
- Kakizawa, S. , Oshima, K. and Namba, S. (2006) Diversity and functional importance of phytoplasma membrane proteins. Trends Microbiol. 14, 254–256. [DOI] [PubMed] [Google Scholar]
- Kanonenberg, K. , Schwarz, C.K.W. and Schmitt, L. (2013) Type I secretion systems – a story of appendices. Res. Microbiol. 164, 596–604. [DOI] [PubMed] [Google Scholar]
- Kaplan, I.B. , Lee, L. , Ripoll, D.R. , Palukaitis, P. , Gildow, F. and Gray, S.M. (2007) Point mutations in the Potato leafroll virus major capsid protein alter virion stability and aphid transmission. J. Gen. Virol. 88, 1821–1830. [DOI] [PubMed] [Google Scholar]
- Kelley, K.B. , Whitworth, J.L. and Novy, R.G. (2009) Mapping of the Potato leafroll virus resistance gene, Rlr etb, from Solanum etuberosum identifies interchromosomal translocations among its E‐genome chromosomes 4 and 9 relative to the A‐genome of Solanum L. sect. Petota . Mol. Breed. 23, 489–500. [Google Scholar]
- Kim, J.S. , Sagaram, U.S. , Burns, J.K. , Li, J.L. and Wang, N. (2009) Response of sweet orange (Citrus sinensis) to “Candidatus Liberibacter asiaticus” infection: microscopy and microarray analyses. Phytopathology, 99, 50–57. [DOI] [PubMed] [Google Scholar]
- Knoblauch, M. and Peters, W.S. (2010) Münch, morphology, microfluidics – our structural problem with the phloem. Plant. Cell Environ. 33, 1439–1452. [DOI] [PubMed] [Google Scholar]
- Knoblauch, M. and Peters, W.S. (2013) Long‐distance translocation of photosynthates: a primer. Photosynth. Res. 117, 189–196. [DOI] [PubMed] [Google Scholar]
- Knoblauch, M. , Stubenrauch, M. , van Bel, A.J.E. and Peters, W.S. (2012) Forisome performance in artificial sieve tubes. Plant. Cell Environ. 35, 1419–1427. [DOI] [PubMed] [Google Scholar]
- Knoblauch, M. , Froelich, D.R. , Pickard, W.F. and Peters, W.S. (2014) SEORious business: structural proteins in sieve tubes and their involvement in sieve element occlusion. J. Exp. Bot. 65, 1879–1893. [DOI] [PubMed] [Google Scholar]
- Koh, E.J. , Zhou, L. , Williams, D.S. , Park, J. , Ding, N. , Duan, Y.P. and Kang, B.H. (2012) Callose deposition in the phloem plasmodesmata and inhibition of phloem transport in citrus leaves infected with “Candidatus Liberibacter asiaticus”. Protoplasma, 249, 687–697. [DOI] [PubMed] [Google Scholar]
- Kronberg, K. , Vogel, F. , Rutten, T. , Hajirezaei, M.R. , Sonnewald, U. and Hofius, D. (2007) The silver lining of a viral agent: increasing seed yield and harvest index in Arabidopsis by ectopic expression of the Potato leafroll virus movement protein. Plant Physiol. 145, 905–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kube, M. , Mitrovic, J. , Duduk, B. , Rabus, R. and Seemüller, E. (2012) Current view on phytoplasma genomes and encoded metabolism. Sci. World J. 2012, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuykendall, L.D. , Shao, J.Y. and Hartung, J.S. (2012) Conservation of gene order and content in the circular chromosomes of “Candidatus Liberibacter asiaticus” and other Rhizobiales . PLoS One, 7, e34673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbate, M. , Zhu, H. , Thung, L. , Bandara, R. , Larsen, M.R. , Willcox, M.D.P. , Givskov, M. , Rice, S.A. and Kjelleberg, S. (2007) Quorum‐sensing regulation of adhesion in Serratia marcescens MG1 is surface dependent. J. Bacteriol. 189, 2702–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson, K.C. and Whitham, T.G. (1997) Competition between gall aphids and natural plant sinks: plant architecture affects resistance to galling. Oecologia, 109, 575–582. [DOI] [PubMed] [Google Scholar]
- Lebon, A. , Mailleret, L. , Dumont, Y. and Grognard, F. (2014) Direct and apparent compensation in plant–herbivore interactions. Ecol. Modell. 290, 192–203. [Google Scholar]
- Lee, I.M. , Davis, R.E. and Gundersen‐Rindal, D.E. (2000) Phytoplasma: phytopathogenic Mollicutes. Annu. Rev. Microbiol. 54, 221–255. [DOI] [PubMed] [Google Scholar]
- Lee, L. , Palukaitis, P. and Gray, S.M. (2002) Host‐dependent requirement for the Potato leafroll virus 17‐kDa protein in virus movement. Mol. Plant–Microbe Interact. 15, 1086–1094. [DOI] [PubMed] [Google Scholar]
- Lefol, C. , Caudwell, A. , Lherminier, J. and Larrue, J. (1993) Attachment of the Flavescence dorée pathogen (MLO) to leafhopper vectors and other insects. Ann. Appl. Biol. 123, 611–622. [Google Scholar]
- Leonard, M.T. , Fagen, J.R. , Davis‐Richardson, A.G. , Davis, M.J. and Triplett, E.W. (2012) Complete genome sequence of Liberibacter crescens BT‐1. Stand. Genomic Sci. 7, 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Cong, Q. , Pei, J. , Kinch, L.N. and Grishin, N.V. (2012) The ABC transporters in Candidatus Liberibacter asiaticus. Proteins, 80, 2614–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. , Lou, B. , Glynn, J.M. , Doddapaneni, H. , Civerolo, E.L. , Chen, C. , Duan, Y. , Zhou, L. and Vahling, C.M. (2011) The complete genome sequence of “Candidatus Liberibacter solanacearum”, the bacterium associated with potato zebra chip disease. PLoS One, 6, e19135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, K. , Vogel, F. and Sonnewald, U. (2011) PD trafficking of Potato leafroll virus movement protein in Arabidopsis depends on site‐specific protein phosphorylation. Front. Plant Sci. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y.T. , Li, M.Y. , Cheng, K.T. , Tan, C.M. , Su, L.W. , Lin, W.Y. , Shih, H.T. , Chiou, T.J. and Yang, J.Y. (2014) Transgenic plants that express the phytoplasma effector SAP11 show altered phosphate starvation and defense responses. Plant. Physiol. 164, 1456–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łukasik, P. , van Asch, M. , Guo, H. , Ferrari, J. , Charles, J. and Godfray, H. (2013) Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol. Lett. 16, 214–218. [DOI] [PubMed] [Google Scholar]
- Luna, E. , Pastor, V. , Robert, J. , Flors, V. , Mauch‐Mani, B. and Ton, J. (2011) Callose deposition: a multifaceted plant defense response. Mol. Plant–Microbe Interact. 24, 183–193. [DOI] [PubMed] [Google Scholar]
- MacLean, A.M. , Sugio, A. , Makarova, O.V. , Findlay, K.C. , Grieve, V.M. , Toth, R. , Nicolaisen, M. and Hogenhout, S.A. (2011) Phytoplasma effector SAP54 induces indeterminate leaf‐like flower development in Arabidopsis plants. Plant Physiol. 157, 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean, A.M. , Orlovskis, Z. , Kowitwanich, K. , Zdziarska, A.M. , Angenent, G.C. , Immink, R.G.H. and Hogenhout, S.A. (2014) Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS‐box proteins and promotes insect colonization in a RAD23‐dependent manner. PLoS Biol. 12, e1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, H. and Morihara, K. (1995) Serralysin and related bacterial proteinases. Methods Enzymol. 248, 395–413. [DOI] [PubMed] [Google Scholar]
- Mafra, V. , Martins, P.K. , Francisco, C.S. , Ribeiro‐Alves, M. , Freitas‐Astúa, J. and Machado, M.A. (2013) Candidatus Liberibacter americanus induces significant reprogramming of the transcriptome of the susceptible citrus genotype. BMC Genomics, 14, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlen, S.D. (2011) Serratia infections: from military experiments to current practice. Clin. Microbiol. Rev. 24, 755–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, R.S. , Ali, J.G. , Hermann, S.L. , Tiwari, S. , Pelz‐Stelinski, K.S. , Alborn, H.T. and Stelinski, L.L. (2012) Induced release of a plant‐defense volatile “deceptively” attracts insect vectors to plants infected with a bacterial pathogen. PLoS Pathog. 8, e1002610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczewski, W. , Flis, B. , Syller, J. , Strzelczyk‐Żyta, D. , Hennig, J. and Gebhardt, C. (2004) Two allelic or tightly linked genetic factors at the PLRV.4 locus on potato chromosome XI control resistance to Potato leafroll virus accumulation. Theor. Appl. Genet. 109, 1604–1609. [DOI] [PubMed] [Google Scholar]
- Martinelli, F. , Uratsu, S.L. , Albrecht, U. , Reagan, R.L. , Phu, M.L. , Britton, M. , Buffalo, V. , Fass, J. , Leicht, E. , Zhao, W. , Lin, D. , D'Souza, R. , Davis, C.E. , Bowman, K.D. and Dandekar, A.M. (2012) Transcriptome profiling of citrus fruit response to huanglongbing disease. PLoS One, 7, e38039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas, F. , Vereijssen, J. and Suckling, D.M. (2014) Influence of the pathogen Candidatus Liberibacter solanacearum on tomato host plant volatiles and psyllid vector settlement. J. Chem. Ecol. 40, 1197–1202. [DOI] [PubMed] [Google Scholar]
- Melnyk, C.W. , Molnar, A. and Baulcombe, D.C. (2011) Intercellular and systemic movement of RNA silencing signals. EMBO J. 30, 3553–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihovilovich, E. , Aponte, M. , Lindqvist‐Kreuze, H. and Bonierbale, M. (2014) An RGA‐derived SCAR marker linked to PLRV resistance from Solanum tuberosum ssp. andigena . Plant Mol. Biol. Rep. 32, 117–128. [Google Scholar]
- Millet, Y.A. , Danna, C.H. , Clay, N.K. , Songnuan, W. , Simon, M.D. , Werck‐Reichhart, D. and Ausubel, F.M. (2010) Innate immune responses activated in Arabidopsis roots by microbe‐associated molecular patterns. Plant Cell, 22, 973–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minato, N. , Himeno, M. , Hoshi, A. , Maejima, K. , Komatsu, K. , Takebayashi, Y. , Kasahara, H. , Yusa, A. , Yamaji, Y. , Oshima, K. , Kamiya, Y. and Namba, S. (2014) The phytoplasmal virulence factor TENGU causes plant sterility by downregulating of the jasmonic acid and auxin pathways. Sci. Rep. 4, 7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahara, K. , Matsuzaki, M. , Tanaka, K. and Sako, N. (1982) A new disease of onion caused by mycoplasma‐like organism in Japan. Jpn. J. Phytopathol. 48, 551–554. [Google Scholar]
- Monaghan, J. and Zipfel, C. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. [DOI] [PubMed] [Google Scholar]
- Montllor, C.B. , Maxmen, A. and Purcell, A.H. (2002) Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27, 189–195. [Google Scholar]
- Moreno, P. , Ambrós, S. , Albiach‐Marti, M.R. , Guerri, J. and Peña, L. (2008) Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol. Plant Pathol. 9, 251–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott, G.A. , Middleton, M.A. , Desveaux, D. and Guttman, D.S. (2014) Peptides and small molecules of the plant‐pathogen apoplastic arena. Front. Plant Sci. 5, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murral, D.J. , Nault, L.R. , Hoy, C.W. , Madden, L.V. and Miller, S.A. (1996) Effects of temperature and vector age on transmission of two Ohio strains of Aster Yellows phytoplasma by the Aster Leafhopper (Homoptera: Cicadellidae). Horticultural Entomology, 89, 1223–1232. [Google Scholar]
- Musetti, R. , di Toppi, L.S. , Ermacora, P. and Favali, M.A. (2004) Recovery in apple trees infected with the apple proliferation phytoplasma: an ultrastructural and biochemical study. Phytopathology, 94, 203–208. [DOI] [PubMed] [Google Scholar]
- Musetti, R. , Paolacci, A. , Ciaffi, M. , Tanzarella, O.A. , Polizzotto, R. , Tubaro, F. , Mizzau, M. , Ermacora, P. , Badiani, M. and Osler, R. (2010) Phloem cytochemical modification and gene expression following the recovery of apple plants from apple proliferation disease. Phytopathology, 100, 390–399. [DOI] [PubMed] [Google Scholar]
- Musetti, R. , Buxa, S.V. , De Marco, F. , Loschi, A. , Polizzotto, R. , Kogel, K.H. , Van Bel, A.J.E. (2013) Phytoplasma‐triggered Ca2+ influx is involved in sieve‐tube blockage. Mol. Plant–Microbe Interact. 26, 379–386. [DOI] [PubMed] [Google Scholar]
- Musetti, R. , Pagliari, L. , Buxa, S.V. , Degola, F. , De Marco, F. , Loschi, A. , Kogel, K.H. and van Bel, A.J.E. (2016) OHMS**: Phytoplasmas dictate changes in sieve‐element ultrastructure to accommodate their requirements for nutrition, multiplication and translocation. Plant Signal. Behav. 11, e1138191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutterer, J.D. , Stussi‐Garaud, C. , Michler, P. , Richards, K.E. , Jonard, G. and Ziegler‐Graff, V. (1999) Role of the Beet western yellows virus readthrough protein in virus movement in Nicotiana clevelandii . J. Gen. Virol. 80, 2771–2778. [DOI] [PubMed] [Google Scholar]
- Nadarasah, G. and Stavrinides, J. (2011) Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol. Rev. 35, 555–575. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A. , Nikoh, N. , Oshima, K. , Inoue, H. , Ohkuma, M. , Hongoh, Y. , Miyagishima, S.Y. , Hattori, M. and Fukatsu, T. (2013) Horizontal gene acquisition of Liberibacter plant pathogens from a bacteriome‐confined endosymbiont of their psyllid vector. PLoS One, 8, e82612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal, J.J. (1993) Xylem transport interruption by Anasa tristis feeding causes Cucurbita pepo to wilt. Entomol. Exp. Appl. 69, 195–200. [Google Scholar]
- Neriya, Y. , Maejima, K. , Nijo, T. , Tomomitsu, T. , Yusa, A. , Himeno, M. , Netsu, O. , Hamamoto, H. , Oshima, K. and Namba, S. (2014) Onion yellow phytoplasma P38 protein plays a role in adhesion to the hosts. FEMS Microbiol. Lett. 361, 115–122. [DOI] [PubMed] [Google Scholar]
- Nethe, M. , Berkhout, B. and van der Kuyl, A.C. (2005) Retroviral superinfection resistance. Retrovirology, 2, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, J.C. and Zhou, J.S. (2015) Insect vector–plant virus interactions associated with non‐circulative, semi‐persistent transmission: current perspectives and future challenges. Curr. Opin. Virol. 15, 48–55. [DOI] [PubMed] [Google Scholar]
- Nome, C. , Magalhães, P.C. , Oliveira, E. , Nome, S. and Lagune Irma, G. (2009) Differences in intracellular localization of corn stunt spiroplasmas in magnesium treated maize. Biocell, 33, 133–136. [PubMed] [Google Scholar]
- Nourrisseau, J.G. , Lansac, M. and Garnier, M. (1993) Marginal chlorosis, a new disease of strawberries associated with a bacteriumlike organism. Plant Dis. 77, 1055–1059. [Google Scholar]
- Novy, R.G. , Gillen, A.M. and Whitworth, J.L. (2007) Characterization of the expression and inheritance of Potato leafroll virus (PLRV) and Potato virus Y (PVY) resistance in three generations of germplasm derived from Solanum etuberosum . Theor. Appl. Genet. 114, 1161–1172. [DOI] [PubMed] [Google Scholar]
- Nwugo, C.C. , Duan, Y. and Lin, H. (2013) Study on citrus response to huanglongbing highlights a down‐regulation of defense‐related proteins in lemon plants upon “Ca. Liberibacter asiaticus” infection. PLoS One, 8, e67442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, K.M. , Russell, J.A. , Moran, N.A. and Hunter, M.S. (2003) Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. 100, 1803–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka, K.J. and Cruz, S.S. (2000) The great escape: phloem transport and unloading of macromolecules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 323–347. [DOI] [PubMed] [Google Scholar]
- Orlovskis, Z. and Hogenhout, S.A. (2016) A bacterial parasite effector mediates insect vector attraction in host plants independently of developmental changes. Front. Plant Sci. 7, 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlovskis, Z. , Canale, M.C. , Thole, V. , Pecher, P. , Lopes, J.R. and Hogenhout, S.A. (2015) Insect‐borne plant pathogenic bacteria: getting a ride goes beyond physical contact. Curr. Opin. Insect Sci. 9, 16–23. [DOI] [PubMed] [Google Scholar]
- Oshima, K. , Miyata, S. , Sawayanagi, T. , Kakizawa, S. , Nishigawa, H. , Jung, H.Y. , Furuki, K. , Yanazaki, M. , Suzuki, S. , Wei, W. , Kuboyama, T. , Ugaki, M. and Namba S. (2002) Minimal set of metabolic pathways suggested from the genome of Onion Yellows phytoplasma. J. Gen. Plant Pathol. 68, 225–236. [Google Scholar]
- Ovcharenko, L.P. , Voznyuk, T.M. , Zaetz, I.E. , Potopalsky, A.I. , Reva, O. and Kozyrovska, N.O. (2010) A mobile genetic element in Serratia marcescens, a causative agent of onion disease. Biopolym. Cell, 26, 279–285. [Google Scholar]
- Özbek, E.O. , Miller, S.A. , Meulia, T. and Hogenhout, S.A. (2003) Infection and replication sites of Spiroplasma kunkelii (Class: Mollicutes) in midgut and Malpighian tubules of the leafhopper Dalbulus maidis . J. Invertebr. Pathol. 82, 167–175. [DOI] [PubMed] [Google Scholar]
- Pair, S.D. , Bruton, B.D. , Mitchell, F. , Fletcher, J. , Wayadande, A. and Melcher, U. (2004) Overwintering squash bugs harbor and transmit the causal agent of cucurbit yellow vine disease. J. Econ. Entomol. 97, 74–78. [DOI] [PubMed] [Google Scholar]
- Parker, J.K. , Wisotsky, S.R. , Johnson, E.G. , Hijaz, F.M. , Killiny, N. , Hilf, M.E. and De La Fuente, L. (2014) Viability of “Candidatus Liberibacter asiaticus” prolonged by addition of citrus juice to culture medium. Phytopathology, 104, 15–26. [DOI] [PubMed] [Google Scholar]
- Paultre, D.S.G. , Gustin, M.P. , Molnar, A. and Oparka, K.J. (2016) Lost in transit: long‐distance trafficking and phloem unloading of protein signals in Arabidopsis homografts. Plant Cell, 28, 2016–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelz‐Stelinski, K.S. , Brlansky, R.H. , Ebert, T.A. and Rogers, M.E. (2010) Transmission parameters for Candidatus Liberibacter asiaticus by Asian citrus psyllid (Hemiptera: Psyllidae). J. Econ. Entomol. 103, 1531–1541. [DOI] [PubMed] [Google Scholar]
- Pérez‐Clemente, R.M. , Montoliu, A. , Vives, V. , López‐Climent, M.F. and Gómez‐Cadenas, A. (2015) Photosynthetic and antioxidant responses of Mexican lime (Citrus aurantifolia) plants to Citrus tristeza virus infection. Plant Pathol. 64, 16–24. [Google Scholar]
- Perilla‐Henao, L.M. and Casteel, C.L. (2016) Vector‐borne bacterial plant pathogens: interactions with Hemipteran insects and plants. Front. Plant Sci. 7, 1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, K.A. , Liang, D. , Palukaitis, P. and Gray, S.M. (2008) Small deletions in the Potato leafroll virus readthrough protein affect particle morphology, aphid transmission, virus movement and accumulation. J. Gen. Virol. 89, 2037–2045. [DOI] [PubMed] [Google Scholar]
- Peter, K.A. , Gildow, F. , Palukaitis, P. and Gray, S.M. (2009) The C terminus of the Polerovirus p5 readthrough domain limits virus infection to the phloem. J. Virol. 83, 5419–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, W.S. , Haffer, D. , Hanakam, C.B. , van Bel, A.J.E. and Knoblauch, M. (2010) Legume phylogeny and the evolution of a unique contractile apparatus that regulates phloem transport. Am. J. Bot. 97, 797–808. [DOI] [PubMed] [Google Scholar]
- Petersen, L.M. and Tisa, L.S. (2013) Friend or foe? A review of the mechanisms that drive Serratia towards diverse lifestyles. Can. J. Microbiol. 59, 627–640. [DOI] [PubMed] [Google Scholar]
- Pitino, M. and Hogenhout, S.A. (2013) Aphid protein effectors promote aphid colonization in a plant species‐specific manner. Mol. Plant–Microbe Interact. 26, 130–139. [DOI] [PubMed] [Google Scholar]
- Pitino, M. , Armstrong, C.M. , Cano, L.M. and Duan, Y. (2016) Transient expression of Candidatus Liberibacter asiaticus effector induces cell death in Nicotiana benthamiana . Front. Plant Sci. 7, 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, A.H. and Finlay, A. (1979) Evidence for noncirculative transmission of Pierce's disease bacterium by sharpshooter leafhoppers. Phytopathology, 69, 393–395. [Google Scholar]
- Rai, M. (2006) Refinement of the Citrus tristeza virus resistance gene (CTV) positional map in Poncirus trifoliata and generation of transgenic grapefruit (Citrus paradisi) plant lines with candidate resistance genes in this region. Plant Mol. Biol. 61, 399–414. [DOI] [PubMed] [Google Scholar]
- Ramadugu, C. , Keremane, M.L. , Halbert, S.E. , Duan, Y.P. , Roose, M.L. , Stover, E. and Lee, R.F. (2016) Long‐term field evaluation reveals Huanglongbing resistance in Citrus relatives. Plant Dis. 100, 1858–1869. [DOI] [PubMed] [Google Scholar]
- Rascoe, J. , Berg, M. , Melcher, U. , Mitchell, F.L. , Bruton, B.D. , Pair, S.D. and Fletcher, J. (2003) Identification, phylogenetic analysis, and biological characterization of Serratia marcescens strains causing Cucurbit yellow vine disease. Phytopathology, 93, 1233–1239. [DOI] [PubMed] [Google Scholar]
- Rashidi, M. , Galetto, L. , Bosco, D. , Bulgarelli, A. , Vallino, M. , Veratti, F. and Marzachì, C. (2015) Role of the major antigenic membrane protein in phytoplasma transmission by two insect vector species. BMC Microbiol. 15, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin, S. (1999) Adherence of pathogenic mycoplasmas to host cells. Biosci. Rep. 19, 367–372. [DOI] [PubMed] [Google Scholar]
- Razin, S. , Yogev, D. and Naot, Y. (1998) Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62, 1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne, P.L.H. , Boogaard, R. , van den Mensink, M.G.J. , Kopperud, C. , Kormelink, R. , Goldbach, R. and Schoot, C. V D. (2005) Tobacco plants respond to the constitutive expression of the Tospovirus movement protein NSM with a heat‐reversible sealing of plasmodesmata that impairs development. Plant J. 43, 688–707. [DOI] [PubMed] [Google Scholar]
- Rocha, E.P.C. and Blanchard, A. (2002) Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 30, 2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen, R. , Kanakala, S. , Kliot, A. , Cathrin Pakkianathan, B. , Farich, B.A. , Santana‐Magal, N. , Elimelech, M. , Kontsedalov, S. , Lebedev, G. , Cilia, M. and Ghanim, M. (2015) Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr. Opin. Virol. 15, 1–8. [DOI] [PubMed] [Google Scholar]
- Saglio, P. , Lhospital, M. , LaFléche, D. , Dupont, G. , Bové, J.M. , Tully, J.G. and Freundt, E.A. (1973) Spiroplasma citri gen. and sp.n.: a mycoplasma‐like organism associated with “Stubborn” disease of citrus. Int. J. Syst. Bacteriol. 23, 191–204. [Google Scholar]
- Saigo, M. , Golic, A. , Alvarez, C.E. , Andreo, C.S. , Hogenhout, S.A. , Mussi, M.A. and Drincovich, M.F. (2014) Metabolic regulation of phytoplasma malic enzyme and phosphotransacetylase supports the use of malate as an energy source in these plant pathogens. Microbiology, 160, 2794–2806. [DOI] [PubMed] [Google Scholar]
- Saillard, C. , Carle, P. , Duret‐Nurbel, S. , Henri, R. , Killiny, N. , Carrère, S. , Gouzy, J. , Bové, J.M. , Renaudin, J. and Foissac, X. (2008) The abundant extrachromosomal DNA content of the Spiroplasma citri GII3‐3X genome. BMC Genomics, 9, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salar, P. , Sémétey, O. , Danet, J.L. , Boudon‐Padieu, E. and Foissac, X. (2010) “ Candidatus Phlomobacter fragariae” and the proteobacterium associated with the low sugar content syndrome of sugar beet are related to bacteria of the Arsenophonus clade detected in hemipteran insects. Eur. J. Plant Pathol. 126, 123–127. [Google Scholar]
- Santi, S. , Grisan, S. , Pierasco, A. , De Marco, F. and Musetti, R. (2013a) Laser microdissection of grapevine leaf phloem infected by stolbur reveals site‐specific gene responses associated to sucrose transport and metabolism. Plant. Cell Environ. 36, 343–355. [DOI] [PubMed] [Google Scholar]
- Santi, S. , De Marco, F. , Polizzotto, R. , Grisan, S. and Musetti, R. (2013b) Recovery from stolbur disease in grapevine involves changes in sugar transport and metabolism. Front. Plant Sci. 4, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, J.A. , Clearwater, M.J. , Haines, D.F. , Klein, T. , Mencuccini, M. , Sevanto, S. , Turgeon, R. and Zhang, C. (2016) Allocation, stress tolerance and carbon transport in plants: how does phloem physiology affect plant ecology? Plant. Cell Environ. 39, 709–725. [DOI] [PubMed] [Google Scholar]
- Savenkov, E.I. and Valkonen, J.P. (2001) Potyviral helper‐component proteinase expressed in transgenic plants enhances titers of Potato leafroll virus but does not alleviate its phloem limitation. Virology, 283, 285–293. [DOI] [PubMed] [Google Scholar]
- Schreiber, K.J. , Baudin, M. , Hassan, J.A. and Lewis, J.D. (2016) Die another day: molecular mechanisms of effector‐triggered immunity elicited by type III secreted effector proteins. Semin. Cell Dev. Biol. 56, 124–133. [DOI] [PubMed] [Google Scholar]
- Schulz, A. , Thompson, G.A. , Schulz, A. and Thompson, G.A. (2009) Phloem structure and function In: Encyclopedia of Life Sciences. Chichester: Wiley. DOI: 10.1002/9780470015902.a0001290.pub2 [Google Scholar]
- Sechler, A. , Schuenzel, E.L. , Cooke, P. , Donnua, S. , Thaveechai, N. , Postnikova, E. , Stone, A.L. , Schneider, W.L. , Damsteegt, V.D. and Schaad, N.W. (2009) Cultivation of “Candidatus Liberibacter asiaticus”, “Ca. L. africanus”, and “Ca. L. americanus” associated with huanglongbing. Phytopathology, 99, 480–486. [DOI] [PubMed] [Google Scholar]
- Segonzac, C. and Zipfel, C. (2011) Activation of plant pattern‐recognition receptors by bacteria. Curr. Opin. Microbiol. 14, 54–61. [DOI] [PubMed] [Google Scholar]
- Sémétey, O. , Gatineau, F. , Bressan, A. and Boudon‐Padieu, E. (2007) Characterization of a γ‐3 proteobacteria responsible for the syndrome Basses richesses of sugar beet transmitted by Pentastiridius sp. (Hemiptera: Cixiidae). Phytopathology, 97, 72–78. [DOI] [PubMed] [Google Scholar]
- Shanks, R.M.Q. , Stella, N.A. , Kalivoda, E.J. , Doe, M.R. , O'Dee, D.M. , Lathrop, K.L. , Guo, F.L. and Nau, G.J. (2007) A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J. Bacteriol. 189, 7262–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokal, U. , Yadav, S. , Atri, J. , Accetta, J. , Kenney, E. , Banks, K. , Katakam, A. , Jaenike, J. and Eleftherianos, I. (2016) Effects of co‐occurring Wolbachia and Spiroplasma endosymbionts on the Drosophila immune response against insect pathogenic and non‐pathogenic bacteria. BMC Microbiol. 16, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev, A.G. , Kalinina, N.O. and Morozov, S.Y. (2012) Recent advances in research of plant virus movement mediated by triple gene block. Front. Plant Sci. 3, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, A. , Kingdom, H.N. , MacLean, A.M. , Grieve, V.M. and Hogenhout, S.A. (2011) Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. 108, E1254–E1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, A. , MacLean, A.M. and Hogenhout, S.A. (2014) The small phytoplasma virulence effector SAP11 contains distinct domains required for nuclear targeting and CIN‐TCP binding and destabilization. New Phytol. 202, 838–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers, C.G. , Newton, A.S. and Opgenorth, D.C. (2004) Overwintering of corn leafhopper, Dalbulus maidis (Homoptera: Cicadellidae), and Spiroplasma kunkelii (Mycoplasmatales: Spiroplasmataceae) in California's San Joaquin Valley. Environ. Entomol. 33, 1644–1651. [Google Scholar]
- Taliansky, M. , Mayo, M.A. and Barker, H. (2003) Potato leafroll virus: a classic pathogen shows some new tricks. Mol. Plant Pathol. 4, 81–89. [DOI] [PubMed] [Google Scholar]
- Taliansky, M. , Torrance, L. and Kalinina, N.O. (2008) Role of plant virus movement proteins. Methods Mol. Biol. 451, 33–54. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. , Robertson, C.J. , Garnsey, S.M. , Bar‐Joseph, M. , Gowda, S. and Dawson, W.O. (2008) Three genes of Citrus tristeza virus are dispensable for infection and movement throughout some varieties of citrus trees. Virology, 376, 297–307. [DOI] [PubMed] [Google Scholar]
- Textor, S. and Gershenzon, J. (2009) Herbivore induction of the glucosinolate–myrosinase defense system: major trends, biochemical bases and ecological significance. Phytochem. Rev. 8, 149–170. [Google Scholar]
- Thomma, B.P.H.J. , Nürnberger, T. and Joosten, M.H.A.J. (2011) Of PAMPs and effectors: the blurred PTI–ETI dichotomy. Plant Cell, 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, P. , Sagaram, U.S. , Kim, J.S. , Brlansky, R.H. , Rogers, M.E. , Stelinski, L.L. , Oswalt, C. and Wang, N. (2009) Quantification of viable Candidatus Liberibacter asiaticus in hosts using quantitative PCR with the aid of ethidium monoazide (EMA). Eur. J. Plant Pathol. 124, 553–563. [Google Scholar]
- Turgeon, R. (2010) The puzzle of phloem pressure. Plant Physiol. 154, 578–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki, S. and Citovsky, V. (2007) Spread throughout the plant: systemic transport of viruses In: Viral Transport in Plants, (Waigmann, E. and Heinlein, H., eds.) pp. 85–118. Berlin: Springer. [Google Scholar]
- Velásquez, A.C. , Mihovilovich, E. and Bonierbale, M. (2007) Genetic characterization and mapping of major gene resistance to Potato leafroll virus in Solanum tuberosum ssp. andigena . Theor. Appl. Genet. 114, 1051–1058. [DOI] [PubMed] [Google Scholar]
- Verchot‐Lubicz, J. , Torrance, L. , Solovyev, A.G. , Morozov, S.Y. , Jackson, A.O. and Gilmer, D. (2010) Varied movement strategies employed by triple gene block‐encoding viruses. Mol. Plant–Microbe Interact. 23, 1231–1247. [DOI] [PubMed] [Google Scholar]
- Vogel, F. , Hofius, D. and Sonnewald, U. (2007) Intracellular trafficking of Potato leafroll virus movement protein in transgenic Arabidopsis. Traffic, 8, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Voigt, C.A. (2014) Callose‐mediated resistance to pathogenic intruders in plant defense‐related papillae. Front. Plant Sci. 5, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A. (2015) Dissecting the molecular network of virus–plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 53, 45–66. [DOI] [PubMed] [Google Scholar]
- Wang, M.B. , Masuta, C. , Smith, N.A. and Shimura, H. (2012) RNA silencing and plant viral diseases. Mol. Plant–Microbe Interact. 25, 1275–1285. [DOI] [PubMed] [Google Scholar]
- Wang, N. and Trivedi, P. (2013) Citrus huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology, 103, 652–665. [DOI] [PubMed] [Google Scholar]
- Wang, X.Q. , Bi, T. , Li, X.D. , Zhang, L.Q. , Lu, S.E. , Li, X.D. and Lu, S.E. (2014) First report of Corn whorl rot caused by Serratia marcescens in China. J. Phytopathol. 163, 1059–1063. [Google Scholar]
- Wang, Y. , Zhou, L. , Yu, X. , Stover, E. , Luo, F. and Duan, Y. (2016) Transcriptome profiling of Huanglongbing (HLB) tolerant and susceptible citrus plants reveals the role of basal resistance in HLB tolerance. Front. Plant Sci. 7, 933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. and Hause, B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayadande, A. , Bruton, B. , Fletcher, J. , Pair, S. and Mitchell, F. (2005) Retention of Cucurbit yellow vine disease bacterium Serratia marcescens through transstadial molt of vector Anasa tristis (Hemiptera: Coreidae). Ann. Entomol. Soc. Am. 98, 770–774. [Google Scholar]
- Whitcomb, R.F. , Chen, T.A. , Williamson, D.L. , Liao, C. , Tully, J.G. , Bové, J.M. , Mouchès, C. , Rose, D.L. , Coan, M.E. and Clark, T.B. (1986) Characterization of the etiological agent of corn stunt disease. Int. J. Syst. Bacteriol. Int. Union Microbiol. Soc. 86, 170–178. [Google Scholar]
- Will, T. and van Bel, A.J.E. (2008) Induction as well as suppression: how aphid saliva may exert opposite effects on plant defense. Plant Signal. Behav. 3, 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, T. , Kornemann, S.R. , Furch, A.C.U. , Tjallingii, W.F. and van Bel, A.J.E. (2009) Aphid watery saliva counteracts sieve‐tube occlusion: a universal phenomenon? J. Exp. Biol. 212, 3305–3312. [DOI] [PubMed] [Google Scholar]
- Will, T. , Furch, A.C.U. and Zimmermann, M.R. (2013) How phloem‐feeding insects face the challenge of phloem‐located defenses. Front. Plant Sci. 4, 336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C.R. (1987) Bacterial evolution. Microbiol. Rev. 51, 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S. , Shan, L. and He, P. (2014) Microbial signature‐triggered plant defense responses and early signaling mechanisms. Plant Sci. 228, 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff, N. A. , Zhang, S. , Setubal, J.C. , Almeida, N.F. , Martins, E.C. , Harakava, R. , Kumar, D. , Rangel, L.T. , Foissac, X. , Bové, J.M. and Gabriel, D.W. (2014) The complete genome sequence of “Candidatus Liberibacter americanus”, associated with Citrus huanglongbing. Mol. Plant–Microbe Interact. 27, 163–176. [DOI] [PubMed] [Google Scholar]
- Zatyka, M. and Thomas, C.M. (1998) Control of genes for conjugative transfer of plasmids and other mobile elements. FEMS Microbiol. Rev. 21, 291–319. [DOI] [PubMed] [Google Scholar]
- Zavaliev, R. , Ueki, S. , Epel, B.L. and Citovsky, V. (2011) Biology of callose (β‐1,3‐glucan) turnover at plasmodesmata. Protoplasma, 248, 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengler, K. , Toledo, G. , Rappe, M. , Elkins, J. , Mathur, E.J. , Short, J.M. and Keller, M. (2002) Cultivating the uncultured. Proc. Natl. Acad. Sci. USA, 99, 15 681–15 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Hogenhout, S.A. , Nault, L.R. , Hoy, C.W. and Miller, S.A. (2004) Molecular and symptom analyses of phytoplasma strains from lettuce reveal a diverse population. Phytopathology, 94, 842–849. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Du, L. and Poovaiah, B.W. (2014) Calcium signaling and biotic defense responses in plants. Plant Signal. Behav. 9, e973818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Melcher, U. , Zhou, L. , Najar, F.Z. , Roe, B.A. and Fletcher, J. (2005) Genomic comparison of plant pathogenic and nonpathogenic Serratia marcescens strains by suppressive subtractive hybridization. Appl. Environ. Microbiol. 71, 7716–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Flores‐Cruz, Z. , Zhou, L. , Kang, B.H. , Fleites, L.A. , Gooch, M.D. , Wulff, N.A. , Davis, M.J. , Duan, Y.P. and Gabriel, D.W. (2011) “ Ca. Liberibacter asiaticus” carries an excision plasmid prophage and a chromosomally integrated prophage that becomes lytic in plant infections. Mol. Plant–Microbe. Interact. 24, 458–468. [DOI] [PubMed] [Google Scholar]
- Zhao, H. , Sun, R. , Albrecht, U. , Padmanabhan, C. , Wang, A. , Coffey, M.D. , Girke, T. , Wang, Z. , Close, T.J. , Roose, M. , Yokomi, R.K. , Folimonova, S. , Vidalakis, G. , Rouse, R. , Bowman, K.D. and Jin, H. (2013) Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus huanglongbing disease. Mol. Plant. 6, 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Wang, H. , Lin, S. , Roe, B.A. , Davis, R.E. , Hammond, R.W. , Liu, Q. and Jomantiene, R. (2004) Predicted ATP‐binding cassette systems in the phytopathogenic mollicute Spiroplasma kunkelii . Mol. Genet. Genomics, 271, 325–338. [DOI] [PubMed] [Google Scholar]
- Zou, H. , Gowda, S. , Zhou, L. , Hajeri, S. , Chen, G. and Duan, Y. (2012) The destructive citrus pathogen, “Candidatus Liberibacter asiaticus” encodes a functional flagellin characteristic of a pathogen‐associated molecular pattern. PLoS One, 7, e46447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zreik, L. , Bove, J.M. and Garnier, M. (1998) Phylogenetic characterization of the bacterium‐like organism associated with marginal chlorosis of strawberry and proposition of a Candidatus taxon for the organism, “Candidatus Phlomobacter fragariae”. Int. J. Syst. Bacteriol. 48, 257–261. [DOI] [PubMed] [Google Scholar]
- Züst, T. and Agrawal, A.A. (2016) Mechanisms and evolution of plant resistance to aphids. Nat. Plants, 2, 15 206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Symptoms of the diseases discussed in this review.
Table S2 Management strategies for the diseases discussed in this review.


