Summary
Xanthomonas citri ssp. citri (X. citri) is the causal agent of Asiatic citrus canker, a disease that seriously affects most commercially important Citrus species worldwide. We have identified previously a natural variant, X. citri AT, that triggers a host‐specific defence response in Citrus limon. However, the mechanisms involved in this canker disease resistance are unknown. In this work, the defence response induced by X. citri AT was assessed by transcriptomic, physiological and ultrastructural analyses, and the effects on bacterial biofilm formation were monitored in parallel. We show that X. citri AT triggers a hypersensitive response associated with the interference of biofilm development and arrest of bacterial growth in C. limon. This plant response involves an extensive transcriptional reprogramming, setting in motion cell wall reinforcement, the oxidative burst and the accumulation of salicylic acid (SA) and phenolic compounds. Ultrastructural analyses revealed subcellular changes involving the activation of autophagy‐associated vacuolar processes. Our findings show the activation of SA‐dependent defence in response to X. citri AT and suggest a coordinated regulation between the SA and flavonoid pathways, which is associated with autophagy mechanisms that control pathogen invasion in C. limon. Furthermore, this defence response protects C. limon plants from disease on subsequent challenges by pathogenic X. citri. This knowledge will allow the rational exploitation of the plant immune system as a biotechnological approach for the management of the disease.
Keywords: autophagy, biofilm formation, biological control, citrus canker resistance, hypersensitive response, salicylic acid, secondary metabolites
Introduction
Xanthomonas citri ssp. citri (X. citri) strain A is the causative agent of Asiatic citrus canker, a disease that seriously affects most commercially important Citrus spp. worldwide (Vojnov et al., 2010). In South America, other phylogenetically different canker‐causing Xanthomonas have been identified, belonging to X. fuscans ssp. aurantifolii (X. aurantifolii) strains B and C (Schaad et al., 2006, 2005). However, X. aurantifolii B strain could not be isolated from the field after Asiatic citrus canker became endemic in 2002 (Chiesa et al., 2013), and X. aurantifolii C strain has a host range restricted to Mexican lime (Citrus aurantifolia) in some citrus‐producing areas in Brazil (Graham et al., 2004). Therefore, B and C strains are not a serious threat in the field.
Xanthomonas citri ssp. citri is a hemibiotrophic pathogen that grows and persists as epiphytes, forming biofilms on the host surface prior to endophytic colonization of the intercellular spaces of the mesophyll tissue through natural openings, such as stomata, or through wounds (Rigano et al., 2007). A balance between biofilm formation and bacterial dispersion is essential to enhance the epiphytic persistence of bacteria prior to colonization and to circumvent the plant defence response (Favaro et al., 2014; Vojnov and Marano, 2015).
The host defence response is composed of complex and highly regulated molecular networks, which can be triggered by the perception of either conserved pathogen‐associated molecular patterns (PAMPs) or race‐specific pathogen effectors (Jones and Dangl, 2006; Macho and Zipfel, 2015). In Citrus spp., the first level of defence triggered by X. citri has been associated with early molecular changes in gene expression, particularly linked to the production of reactive oxygen species (ROS) (Cernadas et al., 2008; Enrique et al., 2011). However, in most cases, X. citri disrupts PAMP‐triggered immunity (PTI) and produces the disease. In the last decade, different molecular and genetic approaches, including comparative genomics and mutants, have been followed to identify X. citri virulence factors or effectors involved in the suppression of PTI, leading to canker development. Recently, we have shown that xanthan, the major exopolysaccharide secreted by Xanthomonas spp., promotes C. limon susceptibility to X. citri by suppressing hydrogen peroxide (H2O2) accumulation (Enrique et al., 2011). The pthA4 gene encoding type III‐secreted transcriptional activator‐like (TAL) effector is another well‐known pathogenicity effector of canker‐causing Xanthomonas that contributes to host susceptibility (Duan et al., 1999; Shiotani et al., 2007). Deletion of the pthA4 gene has been shown to reduce the bacterial population and abolish the ability of the pathogen to cause canker disease (Duan et al., 1999; Soprano et al., 2013).
The second level of plant immunity is triggered in many plant–pathogen interactions when specific effectors secreted by the pathogen can be recognized by plant resistance (R) proteins, activating effector‐triggered immunity (ETI) (Jones and Dangl, 2006). However, no R gene has been identified in citrus to date. Several types of citrus and closely related genera, including ‘Chinese’ citron (C. medica), calamondin (C. mitis Blanco), Yuzu (C. ichangensis × C. reticulata var. austera) and ‘Nagami’ kumquat (Fortunella margarita), have been reported to be fully resistant to X. citri, suggesting a specific recognition of avirulence effectors (Chen et al., 2012; Deng et al., 2010; Khalaf et al., 2011; Lee et al., 2009). In this regard, transcriptional responses to X. citri in ‘Nagami’ kumquat include the induction of defence‐related genes, particularly those implicated in the hypersensitive response (HR) associated with rapid programmed cell death (PCD), a process that restricts the spread of the pathogen and prevents disease development (Khalaf et al., 2011). In Arabidopsis, HR‐PCD induced by avirulent (hemi)biotrophic pathogens is associated with the activation of autophagy, an intracellular membrane trafficking pathway with substantial roles in both promotion and control of vacuole‐mediated cell death (Teh and Hofius, 2014). The formation of autophagy vesicles is mediated by autophagy‐related (ATG) proteins. In particular, the conversion from free ATG8 to ATG8‐phosphatidylethanolamine (ATG8‐PE) adducts has been reported to be a biochemical marker for the monitoring of autophagy processes (Hofius et al., 2009; Teh and Hofius, 2014). Nevertheless, in canker‐resistant genotypes, the mechanisms underlying HR‐PCD remain obscure.
Xanthomonas citri ssp. citri natural variants with restricted host range have been isolated worldwide. Two of these variants, named A* and Aw, have a host range restricted to C. aurantifolia and C. macrophylla and induce HR‐like reactions in C. paradisi and C. sinensis (Sun et al., 2004; Vernière et al., 1998). This HR‐like phenotype is correlated with the presence of the xopAG (syn. avrGf1) effector gene, identified in all Aw strains and in three A* strains (Escalon et al., 2013; Rybak et al., 2009). However, the signalling pathways involved in these HR‐like responses remain to be elucidated.
Recently, we have characterized a new variant of X. citri, named AT, which shares more than 90% genetic similarity with the type A pathogenic strain X. citri T. Despite this high similarity, the host range of this variant is restricted to C. aurantifolia and C. clementina. In C. limon, this strain triggers an atypical chlorotic phenotype associated with a host‐specific defence response (Chiesa et al., 2013).
In this work, we assess the molecular and cellular events underlying the response of C. limon to X. citri AT. We show that this variant triggers an HR‐PCD associated with the interference of biofilm development and the activation of autophagy‐related vacuolar processes. The defence response involves cell wall reinforcement, the accumulation of phenolic compounds and the induction of the salicylic acid (SA) signalling pathway. Moreover, pre‐inoculation with X. citri AT confers resistance to the pathogenic strain X. citri T.
Results
Biofilm formation is impaired and bacterial growth is arrested in the C. limon–X. citri AT interaction
Proper biofilm formation is a requirement for the achievement of maximal X. citri virulence (Malamud et al., 2013), and the ability of canker‐resistant Citrus spp. to interfere with this process has been reported in ‘Okitsu’ mandarin (Favaro et al., 2014). In this work, we examined whether the impaired ability of X. citri AT to cause disease in C. limon was associated with its inability to develop biofilms. Interestingly, no significant differences were observed between X. citri AT and the pathogenic X. citri T strain in either initial adhesion (1–3 h) or biofilm development (15–24 h) to polystyrene microplates (Fig. S1, see Supporting Information). Next, green fluorescent protein (GFP)‐tagged X. citri strains were inoculated in young C. limon leaves and the ability of X. citri AT to develop biofilms and bacterial growth was monitored. Up to 2 days post‐inoculation (dpi), epiphytic growth of both X. citri strains was similar on C. limon leaves (data not shown). At 7 dpi, biofilm formation was seen only with X. citri T and not with X. citri AT (Fig. 1a). By contrast, both strains developed biofilms on C. clementina leaves (Fig. 1a). Here, both bacterial aggregates showed a three‐dimensional structure on ZX‐axis‐projected images with the formation of compact microcolonies (Fig. 1a). These are similar structures to those reported previously for X. citri biofilms formed on the susceptible genotypes C. limon and C. clementina (Favaro et al., 2014; Rigano et al., 2007). Moreover, inoculation of both strains onto C. clementina leaves led to the development of cankerous lesions at 20 dpi (data not shown). Nevertheless, X. citri AT and T elicited different macroscopic symptoms in C. limon leaves at 20 dpi; X. citri AT induced discrete black spots, phenotypically different from the canker lesions caused by X. citri T (Fig. 1b). Trypan blue staining revealed that X. citri AT induced the cell death response at 48 hpi in C. limon, whereas no cell death was observed after inoculation of X. citri T over the monitored period (Fig. 1c).
Figure 1.
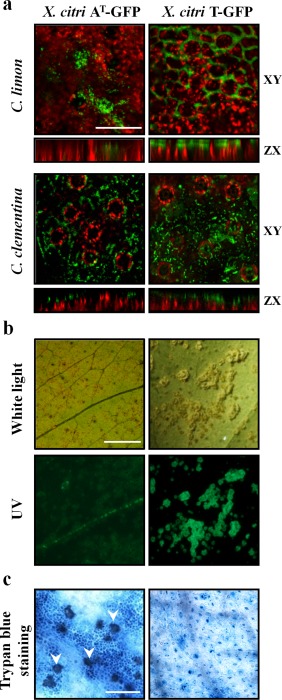
Host‐specific response triggered by Xanthomonas citri ssp. citri (X. citri) strain AT. (a) Biofilm formation on Citrus limon and C. clementina leaves at 7 days post‐inoculation (dpi). Red chlorophyll fluorescence and green signals from green fluorescent protein (GFP)‐tagged X. citri strains are shown. XY and ZX are the XY‐ and ZX‐axis‐projected images, respectively. Scale bar, 50 µm. (b) Macroscopic symptoms in C. limon leaves at 20 dpi. Leaves were photographed under white and UV light. Scale bar, 10 mm. (c) Microscopic cell death phenotype (arrows) observed at 48 h post‐inoculation. Scale bar, 150 µm.
Taken together, these results indicate that X. citri AT is able to form microcolonies and develop biofilms on both non‐biotic and certain biotic surfaces, and suggest that it is the induction of defence responses specifically in C. limon that interferes with biofilm development and the arrest of bacterial growth.
A distinct set of C. limon genes mediates canker resistance
Transcriptome analysis was performed to gain insight into the molecular mechanisms mediating the cell death phenotype observed in C. limon plants inoculated with X. citri AT. Leaves were inoculated with bacterial suspensions of both X. citri strains and samples were harvested at 48 h post‐inoculation (hpi). Differential gene expression analysis identified 1079 up‐regulated and 1832 down‐regulated genes in the interaction with X. citri AT [fold change ≥ 2 in inoculated vs. non‐treated plants; false discovery rate (FDR) ≤ 5%]. A lower but substantial number of genes were differentially expressed in the compatible C. limon–X. citri T interaction (869 and 1036 up‐ and down‐regulated, respectively) (Table S2, see Supporting Information). Comparison of both transcriptomic responses revealed that an important number of genes were specifically expressed only by one of the two bacteria (Fig. S2, see Supporting Information). In particular, 1455 genes (461 and 994 up‐ and down‐regulated, respectively) were unique to the X. citri AT response (Table S3, see Supporting Information). Functional analysis identified 104 gene ontology (GO) categories statistically enriched in the X. citri AT interaction and 62 in the X. citri T interaction. Again, as observed at the transcript level, we found a number of biological processes that were distinct between the two interactions (Tables S4 and S5, see Supporting Information). The most significant 30 categories according to REVIGO (Supek et al., 2011) are shown in Table 1.
Table 1.
Representative subset of the gene ontology (GO) ‘biological process’ terms of Citrus limon–Xanthomonas citri ssp. citri (X. citri) strains.
| GO term | Description | Adjusted P value | |
|---|---|---|---|
| X. citri AT | |||
| Up‐regulated | GO:0006790 | Sulfur compound metabolic process | 8.69E‐05 |
| GO:0009813 | Flavonoid biosynthetic process | 3.62E‐04 | |
| GO:0009636 | Response to toxic substance | 3.80E‐04 | |
| GO:0045454 | Cell redox homeostasis | 7.01E‐04 | |
| GO:0009072 | Aromatic amino acid family metabolic process | 1.61E‐03 | |
| GO:0008299 | Isoprenoid biosynthetic process | 2.47E‐03 | |
| GO:0009751 | Response to salicylic acid | 2.47E‐03 | |
| GO:0015031 | Protein transport | 5.80E‐03 | |
| GO:0006081 | Cellular aldehyde metabolic process | 1.07E‐02 | |
| GO:0006820 | Anion transport | 1.24E‐02 | |
| GO:0009073 | Aromatic amino acid family biosynthetic process | 1.45E‐02 | |
| GO:0009814 | Defence response, incompatible interaction | 1.47E‐02 | |
| GO:0019760 | Glucosinolate metabolic process | 1.60E‐02 | |
| GO:0009407 | Toxin catabolic process | 1.83E‐02 | |
| GO:0009873 | Ethylene‐activated signalling pathway | 2.53E‐02 | |
| GO:0015833 | Peptide transport | 2.74E‐02 | |
| GO:0052543 | Callose deposition in cell wall | 2.88E‐02 | |
| GO:0015694 | Mercury ion transport | 2.88E‐02 | |
| GO:0046246 | Terpene biosynthetic process | 2.88E‐02 | |
| GO:0006801 | Superoxide metabolic process | 3.41E‐02 | |
| GO:0052386 | Cell wall thickening | 3.41E‐02 | |
| GO:0009627 | Systemic acquired resistance | 3.55E‐02 | |
| GO:0009915 | Phloem sucrose loading | 3.75E‐02 | |
| GO:0043090 | Amino acid import | 3.75E‐02 | |
| GO:0016042 | Lipid catabolic process | 4.05E‐02 | |
| GO:0016070 | RNA metabolic process | 4.35E‐02 | |
| GO:0043043 | Peptide biosynthetic process | 4.93E‐02 | |
| GO:0009805 | Coumarin biosynthetic process | 4.94E‐02 | |
| Down‐regulated | GO:0010154 | Fruit development | 1.45E‐08 |
| GO:0009755 | Hormone‐mediated signalling pathway | 1.71E‐06 | |
| GO:0009639 | Response to red or far‐red light | 1.52E‐05 | |
| GO:0008544 | Epidermis development | 1.35E‐04 | |
| GO:0015979 | Photosynthesis | 1.96E‐04 | |
| GO:0007018 | Microtubule‐based movement | 3.11E‐04 | |
| GO:0042493 | Response to drug | 3.11E‐04 | |
| GO:0050790 | Regulation of catalytic activity | 3.23E‐04 | |
| GO:0030163 | Protein catabolic process | 5.54E‐04 | |
| GO:0048589 | Developmental growth | 1.04E‐03 | |
| GO:0006163 | Purine nucleotide metabolic process | 1.09E‐03 | |
| GO:0048229 | Gametophyte development | 1.25E‐03 | |
| GO:0000902 | Cell morphogenesis | 1.90E‐03 | |
| GO:0009808 | Lignin metabolic process | 2.62E‐03 | |
| GO:0010015 | Root morphogenesis | 2.74E‐03 | |
| GO:0007010 | Cytoskeleton organization | 3.24E‐03 | |
| GO:0009555 | Pollen development | 3.80E‐03 | |
| GO:0006855 | Drug transmembrane transport | 4.39E‐03 | |
| GO:0006096 | Glycolytic process | 5.32E‐03 | |
| GO:0006508 | Proteolysis | 5.38E‐03 | |
| GO:0016568 | Chromatin modification | 6.99E‐03 | |
| GO:0006260 | DNA replication | 7.99E‐03 | |
| GO:0032268 | Regulation of cellular protein metabolic process | 7.99E‐03 | |
| GO:0006164 | Purine nucleotide biosynthetic process | 8.63E‐03 | |
| GO:0048827 | Phyllome development | 1.27E‐02 | |
| GO:0006006 | Glucose metabolic process | 1.37E‐02 | |
| GO:0010118 | Stomatal movement | 1.72E‐02 | |
| GO:0010158 | Abaxial cell fate specification | 1.86E‐02 | |
| GO:0010588 | Cotyledon vascular tissue pattern formation | 2.47E‐02 | |
| GO:0010205 | Photoinhibition | 2.47E‐02 | |
| X. citri T | |||
| Up‐regulated | GO:0009408 | Response to heat | 2.13E‐11 |
| GO:0007047 | Cell wall organization | 9.58E‐05 | |
| GO:0042744 | Hydrogen peroxide catabolic process | 1.81E‐04 | |
| GO:0009698 | Phenylpropanoid metabolic process | 1.37E‐03 | |
| GO:0006949 | Syncytium formation | 8.58E‐03 | |
| GO:0016052 | Carbohydrate catabolic process | 9.09E‐03 | |
| GO:0009827 | Plant‐type cell wall modification | 1.40E‐02 | |
| GO:0006412 | Translation | 1.50E‐02 | |
| GO:0042547 | Cell wall modification involved in multidimensional cell growth | 2.84E‐02 | |
| GO:0032508 | DNA duplex unwinding | 2.84E‐02 | |
| GO:0009620 | Response to fungus | 3.17E‐02 | |
| GO:0009807 | Lignan biosynthetic process | 4.48E‐02 | |
| Down‐regulated | GO:0006811 | Ion transport | 1.22E‐10 |
| GO:0006812 | Cation transport | 5.09E‐09 | |
| GO:0030003 | Cellular cation homeostasis | 3.09E‐08 | |
| GO:0006351 | Transcription, DNA dependent | 2.27E‐06 | |
| GO:0007166 | Cell surface receptor signalling pathway | 2.94E‐06 | |
| GO:0016070 | RNA metabolic process | 5.15E‐06 | |
| GO:0006721 | Terpenoid metabolic process | 4.77E‐04 | |
| GO:0006814 | Sodium ion transport | 6.66E‐04 | |
| GO:0009723 | Response to ethylene stimulus | 8.29E‐04 | |
| GO:0010386 | Lateral root formation | 1.33E‐03 | |
| GO:0010104 | Regulation of ethylene‐mediated signalling pathway | 1.86E‐03 | |
| GO:0051607 | Defence response to virus | 4.16E‐03 | |
| GO:0008610 | Lipid biosynthetic process | 6.65E‐03 | |
| GO:0006281 | DNA repair | 8.06E‐03 | |
| GO:0010380 | Regulation of chlorophyll biosynthetic process | 1.19E‐02 | |
| GO:0010017 | Red or far‐red light signalling pathway | 1.20E‐02 | |
| GO:0006021 | Inositol biosynthetic process | 1.51E‐02 | |
| GO:0009308 | Amine metabolic process | 1.67E‐02 | |
| GO:0009944 | Polarity specification of adaxial/abaxial axis | 2.24E‐02 | |
| GO:0005992 | Trehalose biosynthetic process | 2.69E‐02 | |
| GO:0019752 | Carboxylic acid metabolic process | 2.99E‐02 | |
| GO:0009690 | Cytokinin metabolic process | 3.14E‐02 | |
| GO:0010586 | miRNA metabolic process | 3.19E‐02 | |
| GO:0009968 | Negative regulation of signal transduction | 3.37E‐02 | |
| GO:0045017 | Glycerolipid biosynthetic process | 3.37E‐02 | |
| GO:0009735 | Response to cytokinin stimulus | 4.16E‐02 | |
| GO:0048573 | Photoperiodism, flowering | 4.45E‐02 | |
| GO:0051762 | Sesquiterpene biosynthetic process | 4.72E‐02 | |
First 30 significant GO terms summarized by REVIGO (Supek et al., 2011) are shown (adjusted P value ≤ 0.05). ‘Up‐regulated’: GO terms enriched in the up‐regulated genes. ‘Down‐regulated’: GO terms enriched in the down‐regulated genes.
These data indicate that both strains trigger an important rearrangement of the C. limon transcriptome and, although some of these responses are shared by the two interactions, there are distinct responses that are exclusively triggered by X. citri AT. Some of these processes were studied in detail using a combination of molecular, physiological and ultrastructural analyses.
Defence response to X. citri AT is associated with cell wall reinforcement and the accumulation of phenolic compounds
Different genes related to cell wall modification were regulated in response to both X. citri strains, probably influencing the final outcome of each interaction. In C. limon, X. citri AT down‐regulates genes such as xyloglucan:xyloglucosyl transferases XTH6 and XTH16 (repressed by 2.8‐fold), involved in cell wall loosening, cellulose synthases CESA7 and CSLC12 (repressed by 3.8‐ and three‐fold, respectively), pectinesterases, such as PME3 and SKS6 (repressed by 4.1 and 3.8‐fold, respectively), and β‐1,3‐glucanases (repressed by 3.3‐fold), suggesting an active reinforcement of the plant cell wall through the increase in pectin methyl esterification and callose deposition (Table S3). Conversely, X. citri T up‐regulates expansin genes, such as βEXP2 and EXPA4 (induced by 64‐ and 2.7‐fold, respectively), which promote the weakening of the plant cell wall, resulting in cell enlargement (hypertrophy) and division (hyperplasia) required for canker development (Cernadas et al., 2008; Fu et al., 2012) (Table S3). βEXP2 expression was also analysed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR), confirming the microarray results (Table S6, see Supporting Information). Functional analysis using GO also revealed that categories such as ‘cell wall thickening’ (adjusted P value, 3.4 × 10−2) and ‘defence response by callose deposition in cell wall’ (adjusted P value, 2.5 × 10−2) were enriched in up‐regulated genes by X. citri AT (Table 1). However, the category ‘cell wall modification involved in multidimensional cell growth’ (adjusted P value, 2.8 × 10−2) was enriched in the C. limon–X. citri T interaction (Table 1). Taken together, these results suggest that the C. limon response to X. citri AT is associated with fortification of the cell wall, limiting the growth and spread of the bacteria.
The category ‘flavonoid biosynthetic process’ (adjusted P value, 3.6 × 10−4) was enriched in genes up‐regulated by X. citri AT, suggesting that the biosynthesis of this secondary metabolite is fostered in this interaction (Table 1). In particular, genes such as phenylalanine ammonia lyase (PAL1) (induced by 2.9‐fold), chalcone synthase (CHS1) (induced by four‐fold), flavanone 3‐hydroxylase (F3H) (induced by two‐fold), flavonol 3′‐hydroxylase (F3′H) (induced by three‐fold), downy mildew resistant 6 (DMR6) (induced by 5.5‐fold) and anthocyanidin‐3‐O‐glucosyltransferase (3GT) (induced by 2.3‐fold) were all up‐regulated only in response to X. citri AT (Table S3). The same trend was observed by qRT‐PCR for PAL1 and CHS1, as observed in Fig. 2a. Confirming these results, histological assays showed the accumulation of bright green fluorescent polyphenolic compounds, particularly on the abaxial side and around stomata, in X. citri AT‐inoculated leaves at 48 hpi (Fig. 2b). Notably, this accumulation was higher at 7 dpi, particularly surrounding dead cells (Figs 2b and S3, see Supporting Information). By contrast, leaves inoculated with X. citri T did not show the accumulation of phenolic compounds, as indicated by the homogeneous red fluorescence along the tissue, generated by the autofluorescence of chlorophyll (Fig. 2b). Moreover, spectrophotometric determinations confirmed that the content of flavonoids and anthocyanins increased significantly in response to X. citri AT, supporting the idea that phenolic compounds are implicated in this host‐specific defence response (Fig. 2c).
Figure 2.
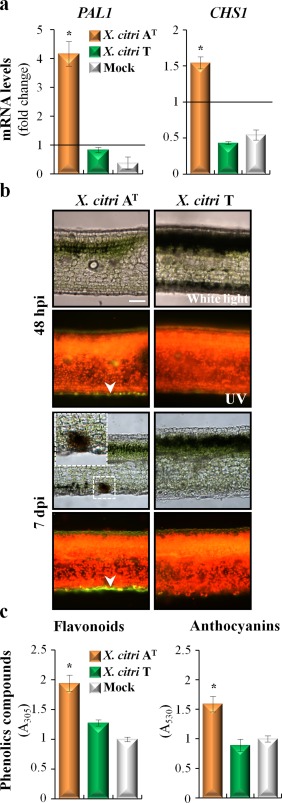
Phenolic compounds are involved in the Citrus limon response to Xanthomonas citri ssp. citri (X. citri) strain AT. (a) Quantitative reverse transcription‐polymerase chain reaction analysis of phenylalanine ammonia lyase (PAL1) and chalcone synthase (CHS1) mRNAs measured at 48 h post‐inoculation (hpi). The relative gene expression (ΔΔCt) fold change of mRNA levels was performed considering non‐treated plants as reference samples and a histone H4 transcript as an endogenous control. Values are expressed as means ± standard deviation (SD) from three independent biological replicates. The dataset marked with an asterisk is significantly different as assessed by Tukey's test (P < 0.05). (b) Light microscopic images of lemon leaves inoculated with X. citri strains. Leaves were photographed at 48 hpi and 7 days post‐inoculation (dpi) under white and UV light. Green fluorescent polyphenolic compounds (arrows) and red chlorophyll fluorescence are observed. The presence of discrete black spots in the C. limon–X. citri AT interaction is shown enlarged in the top inset. Scale bar, 10 µm. (c) Spectrophotometric determination of flavonoids and anthocyanins at 48 hpi. Values are expressed as means ± SD. Each sample consists of 10 leaf discs (0.5 cm in diameter) obtained from two shoots of three different plants, and 10 biological replicates were performed. The dataset marked with an asterisk is significantly different as assessed by Tukey's test (P < 0.05). A, absorbance.
X. citri AT down‐regulates genes related to ROS scavenging and photosynthesis
The production of ROS is one of the earliest cellular responses following successful pathogen recognition. Apoplastic generation of superoxide ( ) or its dismutation product H2O2 can cause strengthening of the plant cell walls, mediation of signalling for gene activation and promotion of HR‐PCD (Chi et al., 2013). It has been observed previously that the C. limon–X. citri AT interaction leads to an increased production of H2O2 (Chiesa et al., 2013), suggesting the deployment of a bona fide defence response leading to canker resistance. Several ROS‐related genes regulated in response to X. citri AT inoculation were found in this work, indicating that redox homeostasis is altered in the plant cell (Table S2). Microarray data indicated that a respiratory burst NADPH‐oxidase homologue to Arabidopsis RBOHD, the main enzyme responsible for the oxidative burst on pathogen infection (Kadota et al., 2014), was induced 5.2‐fold, a result that was confirmed by qRT‐PCR (Tables S2 and S6). In addition, copper/zinc superoxide dismutase (SOD2) and its chaperone (CCS) increased their expression 2.3‐fold, whereas ROS scavengers, such as catalases (CAT3) or peroxidases (PER64, PER68), were down‐regulated (Table S3). A different redox response was observed in leaves inoculated with the pathogenic X. citri T. Although the expression of the RBOHD gene was also slightly up‐regulated, this induction was two‐fold lower than in the X. citri AT interaction (Table S2). Moreover, the ‘hydrogen peroxide catabolic process’ was enriched (adjusted P value, 1.8 × 10−4) (Table 1).
In addition, the GO category related to ‘photosynthesis’ was enriched in the down‐regulated genes in response to X. citri AT (adjusted P value, 1.9 × 10−4; Table 1). This category includes genes encoding thylakoid proteins, such as the light‐harvesting complex (LHCB6 and LCHB1.4), components of the oxygen‐evolving complex of photosystem II (PSBO‐1 and PSBO‐2) and genes involved in the Calvin cycle (RBCS1A, RBCS2B, FBA1 and RCA) (Table S3). This rapid down‐regulation of photosynthesis may also be associated with the high level of ROS production shown in the C. limon–X. citri AT interaction, as demonstrated in ETI responses (Liu et al., 2007; Shapiguzov et al., 2012).
SA is involved in the local defence response induced by X. citri AT
SA is thought to act with ROS in a feed‐forward loop, promoting HR‐PCD, as demonstrated in defence responses against (hemi)biotrophic pathogen infections (Mammarella et al., 2014; Wrzaczek et al., 2013). Interestingly, functional analysis identified the GO category ‘response to salicylic acid’ enriched in the up‐regulated genes unique to the C. limon–X. citri AT interaction (adjusted P value, 2.4 × 10−3; Table 1). Belonging to this category, genes involved in SA biosynthesis, signalling and response were up‐regulated. For instance, as described previously, the expression of PAL1 was induced 2.9‐fold, and genes involved in the biosynthesis of methylsalicylate, such as S‐adenosylmethionine‐dependent methyltransferases (SAMT and BSMT1), were induced 2.4 and 4.4‐fold, respectively. The same tendency was found for the key regulator of SA signalling, nonexpressor of pathogenesis‐related genes 1 (NPR1, 2.6‐fold), the transcription factor WRKY70 (4.4‐fold) and the pathogenesis‐related (PR) genes PR1 (32.7‐fold) and PR4 (14.1‐fold) (Table S3). The induction of the first three key genes of the SA pathway was confirmed by qRT‐PCR (Fig. 3a).
Figure 3.
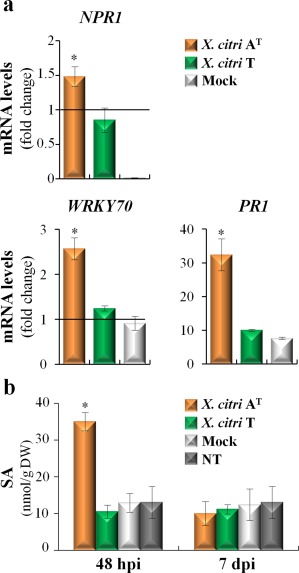
Xanthomonas citri ssp. citri (X. citri) strain AT triggers the accumulation of salicylic acid (SA) in Citrus limon. (a) Quantitative reverse transcription‐polymerase chain reaction analysis of NPR1 (nonexpressor of pathogenesis‐related genes 1), WRKY70 transcription factor and pathogenesis‐related (PR1) mRNAs was measured at 48 h post‐inoculation (hpi). The relative gene expression (ΔΔCt) fold change of mRNA levels was performed considering non‐treated plants (NT) as reference samples and the histone H4 transcript as an endogenous control. Values are expressed as means ± standard deviation (SD) from three independent biological replicates. The dataset marked with an asterisk is significantly different as assessed by Tukey's test (P < 0.05). (b) Analysis of SA through liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) performed at 48 hpi and 7 days post‐inoculation (dpi). Values are expressed as means ± SD from three independent biological replicates. The dataset marked with an asterisk is significantly different as assessed by Tukey's test (P < 0.05). DW, dry weight.
SA quantification showed that its concentration increased three‐fold in X. citri AT‐inoculated leaves at 48 hpi when compared with control samples (Fig. 3b). This increase was not observed at 7 dpi (Fig. 3b), which is also associated with the return to basal levels of WRKY70 and PR1 gene expression (data not shown). Interestingly, at this time point, biofilm development began to decline (Fig. 1a), suggesting a temporal regulation of SA signalling in the defence response against X. citri AT. No increases in SA levels were observed in X. citri T‐inoculated leaves (Fig. 3b).
Subcellular analysis suggests autophagy‐mediated vacuolar cell death events in X. citri AT‐inoculated C. limon leaves
In this work, it was shown that X. citri AT triggers a host‐specific defence response associated with HR‐PCD. To further characterize the subcellular changes induced by X. citri AT, samples from bacteria‐inoculated leaves were analysed by transmission electron microscopy (TEM). Immediately after inoculation of C. limon (0 hpi), tissues did not present any cellular change (Fig. 4a,g). At 48 hpi, X. citri T‐inoculated samples showed the presence of bacteria colonizing the leaf surface (Fig. 4b) and invading the mesophyll cells (Fig. 4c). At 7 dpi, bacteria were present within the damaged mesophyll cells (Fig. 4d), and became more abundant in the intercellular space at 20 dpi (Fig. 4e), when canker symptoms were already visible (Fig. 4f). Moreover, consistent with the ability of X. citri T and AT to develop canker on C. clementina, ultrastructural changes associated with host cell wall dissolution and cell disruption were observed (Fig. S4, see Supporting Information).
Figure 4.
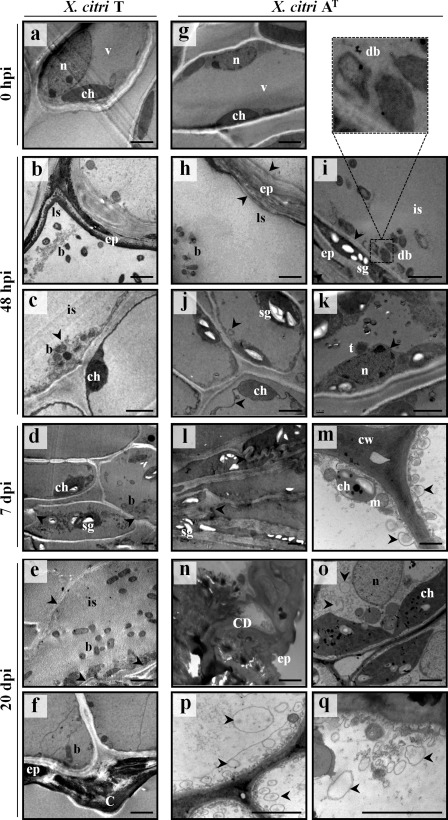
Ultrastructural features of Citrus limon leaves inoculated with Xanthomonas citri ssp. citri (X. citri) strains. (a, g) At 0 h post‐inoculation (hpi), the nucleus, vacuole and chloroplast are intact. (b) Bacteria are localized on the leaf surface and (c) within the mesophyll cells. (d) Bacteria colonizing mesophyll tissue. (e) Bacteria in the intercellular space. Arrows, electron‐dense multitextured materials. (f) Breakdown of epidermal tissue and canker formation. (h) Bacteria colonizing the leaf surface and (i) the intercellular spaces. Arrows, epidermal tissue collapse. Top panel shows the magnification of degenerated bacteria. (j) Arrows, vacuole membrane invaginations. (k) Arrow, perforations of nuclear envelope. (l) Arrows, cellular collapse. (m) Arrows, autophagosome‐like vesicles. (n) Cell death and accumulation of electron‐dense multitextured materials. (o–q) Arrows, autophagosome‐like vesicles. Scale bar, 2 µm. b, bacteria; C, canker; CD, cell death; ch, chloroplast; cw, cell wall; db, degenerated bacteria; ep, epidermis; is, intercellular space; ls, leaf surface; m, mitochondria; n, nucleus; sg, starch granules; t, tubular extensions; v, vacuole.
By contrast, although X. citri AT‐inoculated C. limon leaves showed dispersed bacteria on the leaf surface (Fig. 4h) and in the intercellular space (Fig. 4i), the epidermal pavement cells were empty and tightly cemented at 48 hpi, suggesting cellular collapse that is characteristic of HR vacuolar cell death (van Doorn et al., 2011; Hatsugai et al., 2009; Rojo et al., 2004). Associated with these processes, higher magnification images showed bacteria with an irregular cell shape undergoing degenerative processes (Fig. 4i). Furthermore, mesophyll cells showed vacuole membrane invaginations, suggesting a loss of vacuole turgor (Fig. 4j), and a perforated nuclear envelope wrapped by tubular extensions (Fig. 4k), all features of vacuolar cell death (van Doorn et al., 2011).
Next, at 7 dpi, collapsed mesophyll cells were observed, suggesting the rupture of the tonoplast and the release of the vacuolar content (Fig. 4l). However, mesophyll cells with intact chloroplasts showed the presence of autophagosome‐like vesicles (2.1 ± 0.05 autophagosomes per cell) (Fig. 4m). The formation of autophagosomes (double‐membrane vesicles) is a hallmark of the activation of the authophagy‐mediated pathway (van Doorn et al., 2011). At 20 dpi, TEM analysis exhibited similar results to those obtained previously, showing two types of mesophyll cell response. Although some were dead, with thickening of the cell wall and the accumulation of electron‐dense multitextured materials filling the intercellular space (Fig. 4n), others showed signs of chloroplast enlargement (Fig. 4o) and an increase in the number and size of autophagosomes (3.5 ± 0.12 autophagosomes per cell) (Fig. 4p,q), suggesting that an active autophagy‐regulated mechanism could be involved in the restriction of the spread of HR‐PCD. In addition, immunodetection of ATG8 proteins revealed a marked accumulation of free ATG8 isoforms and ATG8‐PE adducts in X. citri AT‐inoculated C. limon leaves, and an increase in the conversion of ATG8 to ATG8‐PE shows an active autophagic mechanism triggered by this bacterium (Fig. 5).
Figure 5.
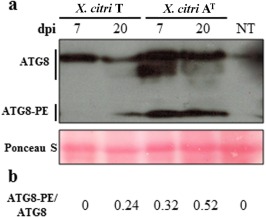
Immunoblot assay to detect free ATG8 and ATG8‐phosphatidylethanolamine (PE) adduct in Citrus limon inoculated with Xanthomonas citri ssp. citri (X. citri) strains. (a) Total protein was extracted from the inoculated tissues at 7 and 20 days post‐inoculation (dpi) and subjected to sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) in the presence of urea, followed by immunoblot analyses with ATG8 antibody. The full lines locate the groups of free ATG8 isoforms and ATG8‐PE adducts. Protein profiles in the lower panels were detected by Ponceau S staining of a polyvinylidene difluoride (PVDF) membrane. The experiment was repeated twice using three independent biological replicates, and a representative image is shown. (b) The ATG8‐PE/ATG8 ratios were determined by the inmunodetection experiment shown in (a). NT, non‐treated leaves.
Taken together, these results suggest that X. citri AT induces HR‐PCD in C. limon, mediated by vacuolar cell death associated with autophagy. At early times post‐inoculation, these autophagic processes would prevent bacterial colonization through vacuolar cell death, but, later, may restrict the spread of the cell death process itself.
X. citri AT protects C. limon from canker development
In order to investigate whether the host response triggered by X. citri AT is able to induce plant protection to the pathogenic strain X. citri T, young C. limon leaves were pre‐inoculated by cotton swab with bacterial suspensions of X. citri AT‐GFP. At 48 hpi, the leaves were challenged with X. citri T‐GFP by spraying (Fig. 6a). A significant reduction in canker development was observed in leaves pre‐inoculated with X. citri AT‐GFP when compared with mock‐inoculated leaves (Fig. 6b). Similar results were obtained when pre‐inoculation with X. citri AT‐GFP was performed by spraying (data not shown). In a new assay, both bacteria were co‐inoculated onto C. limon in equal amounts. Under these conditions, cankerous lesions were observed (Fig. 6c), indicating that bacterial competition is not the determinant of the X. citri AT‐induced protection observed previously.
Figure 6.
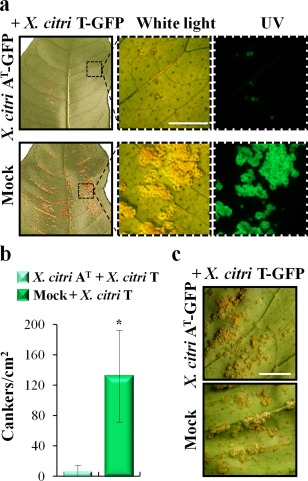
Pre‐inoculation with Xanthomonas citri ssp. citri (X. citri) strain AT protects Citrus limon from canker disease. (a) Phenotypic response of lemon leaves pre‐inoculated with X. citri AT tagged with green fluorescent protein (GFP) or mock‐inoculated by cotton swab. At 48 h post‐inoculation, the leaves were subsequently challenged, via spraying, with the pathogenic X. citri T‐GFP strain. Sections from the left panels are shown magnified in the right panels. Leaves were photographed under white and UV light. Scale bar, 10 mm. (b) Number of canker lesions per square centimetre in pre‐inoculated leaves at 20 days post‐inoculation (dpi). Values are expressed as means ± standard deviation (SD) from three independent biological replicates, each involving three different plants and five different leaves per plant. The dataset marked with an asterisk is significantly different as assessed by Student's t‐test (P < 0.05). (c) Canker symptoms, developed at 20 dpi, of lemon leaves co‐inoculated with equal amounts of both bacterial strains by cotton swab. Scale bar, 10 mm.
These data suggest that X. citri AT triggers a defence response that protects C. limon from canker disease.
Discussion
X. citri AT triggers a recognition event interfering with biofilm development in C. limon
In this work, we demonstrate that X. citri AT is able to develop biofilms on C. clementina and to cause disease. The presence of X. citri AT inside the damaged C. clementina mesophyll cells implies the ability of these bacteria to dissolve the host cell wall and disrupt the cell, inducing similar morphological changes as pathogenic X. citri T. These ultrastructural modifications during citrus canker development have been well reported in X. citri‐inoculated Mexican lime samples (Lee et al., 2009). However, although X. citri AT is able to colonize the leaf surface and the intercellular spaces of the mesophyll tissue, it fails to develop a mature biofilm structure in C. limon. The presence of degenerated X. citri AT bacteria near the cell wall might be linked to the release of vacuolar hydrolytic enzymes during the HR‐PCD response, affecting biofilm development. Altogether, these results indicate that bacterial biofilm formation constitutes not only a virulence factor of canker‐causing Xanthomonas, but its disruption could also be used as a marker of the canker resistance response.
X. citri AT triggers an HR‐PCD response which is associated with elevated levels of flavonoids and SA
The phenotypes triggered by X. citri strains in C. limon are associated with extensive transcriptional reprogramming. An important degree of commonality between the two interactions with different outcome is found, which may be related to the similar genetic backgrounds of the two strains (Chiesa et al., 2013). Therefore, this common subset of genes could account for the PTI basal response, as reported previously between C. sinensis–X. aurantifolii C and C. sinensis–X. citri interactions (Cernadas et al., 2008). The most striking differences are observed in the number of unique genes regulated during X. citri AT infection when compared with the response to pathogenic X. citri T. From the total of the unique genes considered to be differentially expressed, nearly 76% correspond to the X. citri AT‐triggered response. In a similar way, X. aurantifolii C induces a greater number of defence‐related genes than X. citri infection in C. sinensis, suggesting that the amplitude of this response is sufficient to halt X. aurantifolii C growth and to establish an effective HR (Cernadas et al., 2008). In contrast, a relatively small number of defence‐related genes were up‐regulated in the partially resistant ‘Meiwa’ kumquat cultivar to X. citri, when compared with susceptible C. sinensis (Fu et al., 2012). Overall, our results are consistent with the contention that the C. limon defence response to X. citri AT is governed by the recognition of bacterial effectors. Another remarkable feature of the resistance response is that the number of repressed genes is doubled when compared with the susceptible response (pathogenic interaction). The down‐regulation of genes coding for development and photosynthesis proteins, coupled with the up‐regulation of genes coding for defence proteins, points to a possible cross‐talk between these biological processes in the infected tissue. This regulation would allow a better management of the energy resources, as reported previously in other interactions that are known to be mediated by ETI (Bilgin et al., 2010; Karpinski et al., 2013).
The maintenance of host cell wall integrity in response to X. citri AT, through the repression of the xyloglucan‐cellulose network and the production of highly methyl‐esterified pectins, may protect it from bacterial enzymatic degradation. The fact that the increase in PME activity leads to enhanced Pseudomonas syringae susceptibility in Arabidopsis reinforces this idea (Bethke et al., 2014). Interestingly, X. aurantifolii C also down‐regulates the XTH genes in C. sinensis (Cernadas et al., 2008), suggesting that this repression plays a protective role in the defence response, limiting pathogen invasion. Furthermore, thickening of the cell wall by increased callose deposition is the first barrier, not only to X. citri infection in citrus plants (Enrique et al., 2011), but also in other plant–bacterial interactions (Hauck et al., 2003; Voigt, 2014; Yun et al., 2006). In agreement with these results, the repression of β‐1,3‐glucanase and the reinforcement of the cell wall were observed in response to X. citri AT, which are correlated with the beginning of cell death and the restriction of bacterial colonization in C. limon.
The repression of cellulose biosynthesis genes and the lignin biosynthetic pathways may lead to the accumulation of secondary metabolites. In Arabidopsis, cellulose synthase (CESA7)‐deficient mutants showed increased resistance to a broad range of pathogens through the up‐regulation of defence‐related genes, including those involved in the accumulation of antimicrobial secondary metabolites (Hernandez‐Blanco et al., 2007). Moreover, Vanholme et al. (2012) proposed that a reduced flow of the lignin biosynthesis pathway may lead to a higher availability of substrates for the biosynthesis of phenolic compounds. In our work, GO analysis reveals that, although the category of ‘lignin biosynthetic process’ is not significantly represented, several categories related to phenylpropanoid pathways are enriched in the genes up‐regulated by X. citri AT. Accordingly, at early times post‐inoculation, an increase in antimicrobial phenylpropanoids, including flavonoids and anthocyanins, is observed. Phenolic deposits have also been reported around the HR lesions triggered by X. citri in the ‘Nagami’ kumquat and calamondin resistant plants (Chen et al., 2012).
Over recent years, significant progress has been made to understand the role of SA in the regulation of the plant defence response to pathogen attack (Fu and Dong, 2013; Kazan and Lyons, 2014). However, there are no data on the activation of SA‐dependent defence in response to X. citri in citrus. Here, we show an accumulation of SA at early times of X. citri AT inoculation, which is correlated with the beginning of HR‐PCD in C. limon. Interestingly, at later stages of the defence response, although SA decreases to basal levels, the accumulation of phenolic compounds continues to increase. In Arabidopsis and maize, it has been proposed that flavones act as signalling molecules modulating the SA levels under abiotic and biotic stress conditions (Falcone et al., 2015; Pourcel et al., 2013). Our results show a temporal regulation of SA in the X. citri AT resistance response, and suggest a coordinated regulation between SA and flavonoid pathways.
HR triggered by X. citri AT involves autophagy‐associated vacuolar processes protecting the plant from canker development
Autophagy has emerged as a central process in the regulation of pathogen‐triggered HR. In recent years, several studies in model plants have shown that defence‐related autophagy is involved in both cell survival (pro‐survival; avoiding the spread of HR) and cell death (pro‐death) (Hofius et al., 2011; Lenz et al., 2011; Seay and Dinesh‐Kumar, 2005; Teh and Hofius, 2014; Zhou et al., 2014). However, the mechanism governing this molecular switch is not well understood. In this work, we provide different lines of evidence indicating that X. citri AT triggers an ETI‐like response in C. limon which correlates with autophagic processes, which are temporally regulated. In recent years, it has been shown that SA signals play an important role in the induction of autophagy, which, in turn, operates as a negative regulator of SA‐dependent signalling, restricting the spread of HR‐PCD. In Arabidopsis, ATG mutants show an increase in SA levels leading to the ETI‐associated spread of PCD during P. syringae effector AvrRPM1 challenge. These results suggest that autophagy is a critical mechanism for the control of HR‐mediated PCD (Liu et al., 2005; Xia et al., 2013; Yoshimoto et al., 2009). According to our results, the formation of autophagosome‐like vesicles in survival cells and the reduction of the SA level at 7 dpi suggest that autophagy‐associated vacuolar processes may also regulate the spread of cell death.
In Arabidopsis, the vacuole‐mediated PCD triggered by the P. syringae effector AvrRPM1 is associated with two different pathways: proteasome‐regulated membrane fusion and the activation of vacuolar processing enzyme (VPE)‐dependent defences (Hatsugai et al., 2009; Rojo et al., 2004). In this regard, we observed that vacuole‐mediated cell death in X. citri AT‐induced HR‐PCD in C. limon coincides with the up‐regulation of γ‐VPE and ATG8f genes (Tables S3 and S6). The accumulation of transcripts of the ATG8 gene family has been reported in pathogen‐infected Arabidopsis plants, and they are widely used to monitor the temporal regulation and subcellular dynamics of autophagy processes (Hofius et al., 2011; Kabbage et al., 2013; Yoshimoto et al., 2004). In addition, an increase in the conversion of ATG8 to ATG8‐PE suggests an active autophagic mechanism triggered by X. citri AT.
In conclusion, our results suggest that X. citri AT‐triggered HR is mediated by a vacuolar membrane collapse that releases the antimicrobial content into the cytoplasm, causing cell death. Although, in X. citri AT, we were unable to detect the presence of the xopAG effector gene (data not shown), other bacterial effectors should be involved in triggering HR‐PCD in C. limon.
To our knowledge, this is the first report of the molecular mechanisms involved in HR induction by X. citri variants in commercially important citrus species, setting our results as a novel study to exploit the plant immune system as a biotechnological approach for the management of the disease. Moreover, the fact that pre‐inoculation with X. citri AT confers resistance to pathogenic X. citri establishes the basis for the eventual biological control of citrus canker.
Experimental Procedures
Plant material, bacterial strains and pathogenicity assays
One‐year‐old ‘Eureka’ lemon [C. limon (L.) Burm. f.] plants grafted onto Troyer citrange and ‘Clemenules’ mandarin (C. clementina Hort. ex Tan.) plants grafted onto Poncirus trifoliata were kept under controlled conditions in a growth chamber. New shoots of approximately 1 cm in length, with at least five leaves, were selected for pathogenicity assays after pruning the plants. All the leaves on a new shoot were considered to be of the same ontological age (Favaro et al., 2014).
Xanthomonas citri ssp. citri strains were transformed by electroporation with plasmid pMP2444 expressing GFP (Rigano et al., 2007). Bacterial suspensions [109 colony‐forming units (cfu)/mL] were prepared in 10 mm MgCl2 and inoculated by spraying or cotton swab onto 15‐day‐old leaves of the new shoots. A 10 mm MgCl2 solution was used as mock inoculation. Inoculated plants were maintained for 30 days in a growth chamber as reported previously (Enrique et al., 2011). Disease progression was phenotypically monitored using an MVX10 stereomicroscope and photographed under white and UV light (520 nm). The canker lesions were quantified per square centimetre using Image J software (v1.41; National Institutes of Health, Bethesda, MD, USA).
The images in Figs 1, 2, 4 and 5 are representative results from three independent biological replicates, each involving three different plants and three different leaves per plant.
Biofilm analysis
Bacterial adhesion and biofilm formation in vitro were performed as described previously (Rigano et al., 2007).
Biofilm formation in vivo was examined using GFP‐tagged X. citri strains and an inverted confocal laser scanning microscope as described previously (Favaro et al., 2014). Simulated three‐dimensional images and sections were generated by the software Nikon EZ‐C1 3.9 Free Viewer (Nikon Instruments Inc., Melville, NY, USA).
Histochemical and TEM assays
Cell death was visualized in C. limon leaves after staining with lactophenol–trypan blue, as described previously (Koch and Slusarenko, 1990). Autofluorescence of phenolic compounds was observed by fluorescence microscopy (excitation at 450–490 nm, emission at 520 nm) (Chen et al., 2012) using free‐hand leaf sections (Lux et al., 2005). Observations were performed with an Olympus BX50F4 microscope (Olympus Optical Ltd. Company, Shinjuku, Tokyo, Japan).
For TEM experiments, leaf pieces (2 × 3 mm2) were fixed in 4% (v/v) glutaraldehyde in phosphate buffer (1.8 g/L NaH2PO4, 23.25 g/L Na2HPO4.7H2O and 5 g/L NaCl, pH 7.4) for 24 h at 4 °C, and processed according to standard protocols. Sections were examined with a transmission electron microscope (JEOL‐100CXII, Tokyo, Japan) at an accelerating voltage of 80 kV, and digital images were recorded with a ES1000W CCD digital camera (Gatan Inc., Pleasanton, CA, USA). The number of autophagosomes per cell in C. limon leaves was quantified at 7 and 20 days post‐inoculation with X. citri AT. At least 30 TEM images for each time point were used, which were representative of three independent experiments.
RNA preparation
Total RNA from C. limon leaves (4 g) was ground in liquid nitrogen and homogenized in 15 mL extraction buffer [200 mm Tris‐HCl, pH 8.5; 200 mm sucrose; 30 mm magnesium acetate; 60 mm KCl; 0.5% (w/v) polyvinylpyrrolidone; 0.5% (w/v) sodium deoxycolate; 1% (w/v) sodium dodecylsulfate (SDS); 1% (w/v) sodium‐n‐lauroylsarcosine; 10 mm ethylenediaminetetraacetic acid (EDTA); 2% (v/v) β‐mercaptoethanol]. The extraction procedure was performed as described previously (Marano and Carrillo, 1992). RNA samples were purified over Qiagen RNeasy mini‐columns (Hilden, Germany).
Microarray experiments
Five inoculated leaves were randomly harvested at 48 hpi from three different plants and considered as independent biological replicates. Three biological replicates were performed.
RNA samples were amplified using the Amino Allyl MessageAmp™ II aRNA amplification kit (Applied Biosystems, Carlsbad, CA, USA). Reverse transcription, cDNA purification, dye coupling and fluorescent cDNA purification were performed according to the manufacturer's instructions. A citrus microarray developed by the Interdisciplinary Center for Biotechnology Research (ICBR) of the University of Florida and Agilent Technologies Inc. (Palo Alto, CA, USA) was used. This microarray contains 44 000 probes based on citrus expressed sequence tags (ESTs) from Rutaceae (Febres et al., 2012). Microarray hybridization was performed according to the manufacturer's instructions (Agilent Gene Expression Hybridization Kit, Agilent Technologies Inc.). The slides were scanned with GenePix Pro 4000B and analysed with GenePix6.0 software (Axon Instruments, Sunnyvale, CA, USA). Those features with a background‐subtracted intensity lower than two‐fold of the local background intensity in the two channels were discarded. Raw data were normalized as described in Martinez‐Godoy et al. (2008). Only features with valid data in the three replicates were considered for further analysis.
Microarray data analysis
The identification of differentially expressed genes was performed using a significance analysis of microarrays (SAM) test (Tusher et al., 2001). A 5% FDR and two‐fold expression cut‐off were considered to determine the up‐ and down‐regulated genes. Functional analysis was carried out using FatiGO (Babelomics 4.0; Medina et al., 2010), considering as statistically significant those GO terms having an adjusted P value of ≤0.05. Microarray data were deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE78013.
qRT‐PCR
The qRT‐PCRs were performed according to Enrique et al. (2011). Reactions were carried out with real‐time PCR master mix (Biodynamics SRL, Buenos Aires, Argentina) and monitored in a Mastercycler® ep realplex system (Eppendorf, Hamburg, Germany). The primers used in the experiments are listed in Table S1 (Mafra et al., 2012) (see Supporting Information). Transcript levels were normalized against histone H4 (Shiotani et al., 2007) using the ΔΔCt method (Livak and Schmittgen, 2001). Non‐treated (NT) lemon leaves served as the reference sample.
Quantification of UV‐absorbing compounds and SA
Spectrophotometric determination of phenolic compounds was performed according to Mazza et al. (2000). Ten leaf discs (approximately 100 mg of fresh tissue) per sample were detached from C. limon plants at 48 hpi with each X. citri strain. The samples were homogenized in extraction solvent [99% (v/v) methanol in HCl] and incubated for 48 h at −20 °C. For flavonoid compounds, the absorbance of the extracts was read at 305 nm (A305). For anthocyanin determination, the samples were re‐extracted with 0.6 mL of chloroform and 0.3 mL of distilled water, vortexed and centrifuged (2 min at 3000 g). The upper aqueous phases were collected and the anthocyanins were quantified by measurement of the absorbance at 530 nm (A530), as described in Falcone et al. (2010). Mock inoculation with 10 mm MgCl2 of C. limon leaves served as the reference sample.
For SA quantification, leaf samples were obtained as described for microarray experiments. Free SA was extracted from freeze‐dried leaves (200 mg) homogenized in 5 mL of ultrapure water. Five microlitres of 50 ng [2H4]‐SA were added before extraction as an internal standard. Samples were vortexed and centrifuged (15 min at 5000 g). The supernatant was recovered. The pH was adjusted to 2.8 with acetic acid, and partitioned twice with an equal amount of diethyl ether (Durgbanshi et al., 2005). The samples were evaporated to dryness at 35 °C, dissolved in 1.5 mL of methanol, filtered and evaporated at 35 °C. The dried extracts were dissolved in 50 µL of methanol and samples (10 µL) were injected directly into a liquid chromatograph (LC) coupled with electrospray tandem mass spectrometry (MS/MS, Quattro Ultima, Micromass, Manchester, UK). The liquid chromatographic and mass spectrometric analyses were performed according to Castillo et al. (2013).
Immunoblot analysis
Citrus limon leaves (100 mg) were ground in liquid nitrogen and homogenized in 500 µL of sample loading buffer [60 mm Tris, 10% (v/v) glycerol, 180 mm β‐mercaptoethanol, 0.003% (w/v) bromophenol blue and 2% (w/v) SDS, pH 6.8]. The samples were boiled for 10 min at 100 °C prior to loading to the gels. Total protein samples (40 µg) were separated by urea‐sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) [12% (w/v) acrylamide, 6 m urea] and electrophoretically transferred onto pre‐wetted polyvinylidene difluoride membranes (PVDF‐Immun‐Blot®, BioRad, Hercules, CA, USA). Immunoblotting was performed using a polyclonal antibody with reactivity against Arabidopsis ATG8 (1 : 2000 dilution) (Agrisera, Vännäs, Sweden; Álvarez et al., 2012), and then visualized using a peroxidase‐conjugated goat anti‐rabbit immunoglobulin G (IgG) (1 : 10 000 dilution) and Pierce™ ECL Western Blotting Substrate (Thermo Scientific, Petaluma, CA, USA) according to the manufacturers. Quantification of free ATG8 and ATG8‐PE bands was performed by ImageJ analysis (v1.41; National Institutes of Health) of the immunoblot normalized against the Rubisco protein transferred to the membrane, as detected by Ponceau staining.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Bacterial adhesion and biofilm formation on an inert plastic surface.
Fig. S2 Venn diagrams representing the distribution of regulated transcripts in Citrus limon after Xanthomonas citri ssp. citri (X. citri) inoculation.
Fig. S3 Phenolic compound accumulation surrounding the hypersensitive response associated with rapid programmed cell death (HR‐PCD) induced by Xanthomonas citri ssp. citri AT.
Fig. S4 Transmission electron microscopy of Citrus clementina leaves inoculated with Xanthomonas citri ssp. citri (X. citri) strains.
Table S1 List of all oligonucleotide primers used for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis.
Table S2 Microarray expression data for up‐ and down‐regulated genes in response to Xanthomonas citri ssp. citri (X. citri) strains in Citrus limon leaves.
Table S3 Comparison of microarray expression data for up‐ and down‐regulated genes in Citrus limon leaves in response to Xanthomonas citri ssp. citri (X. citri) strains.
Table S4 Gene ontology (GO) ‘biological process’ terms enriched in the differentially expressed genes unique to the Citrus limon–Xanthomonas citri ssp. citri strain AT interaction.
Table S5 Gene ontology (GO) ‘biological process’ terms enriched in the differentially expressed genes unique to the Citrus limon–Xanthomonas citri ssp. citri strain T interaction.
Table S6 Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of genes involved in the defence and pathogenesis response to Xanthomonas citri ssp. citri (X. citri) strains.
Acknowledgements
This work was principally supported by the Agencia Nacional de Promoción Científica y Tecnológica (PICT‐2011‐1833) to M.R.M. and by a grant from the Florida Citrus Research and Development Foundation to F.G.G. and M.R.M. M.A.C., A.A.V., A.P.C., M.P.F. and M.R.M. are Career Investigators of the Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET). The authors thank J. M. Dow and G. Gudesblat for critical review of the manuscript.
Contributor Information
José Gadea, Email: jgadeav@ibmcp.upv.es.
María R. Marano, Email: marano@ibr-conicet.gov.ar
References
- Álvarez, C. , García, I. , Moreno, I. , Pérez‐Pérez, M.E. , Crespo, J.L. , Romero, L.C. and Gotor, C. (2012) Cysteine‐generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell, 24, 4621–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke, G. , Grundman, R.E. , Sreekanta, S. , Truman, W. , Katagiri, F. and Glazebrook, J. (2014) Arabidopsis pectin methylesterases contribute to immunity against Pseudomonas syringae . Plant Physiol. 164, 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin, D.D. , Zavala, J.A. , Zhu, J. , Clough, S.J. , Ort, D.R. and De Lucia, E.H. (2010) Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 33, 1597–1613. [DOI] [PubMed] [Google Scholar]
- Castillo, P. , Escalante, M. , Gallardo, M. , Alemano, S. and Abdala, G. (2013) Effects of bacterial single inoculation and co‐inoculation on growth and phytohormone production of sunflower seedlings under water stress. Acta Physiol. Plant. 35, 2299–2309. [Google Scholar]
- Cernadas, R.A. , Camillo, L.R. and Benedetti, C.E. (2008) Transcriptional analysis of the sweet orange interaction with the citrus canker pathogens Xanthomonas axonopodis pv. citri and Xanthomonas axonopodis pv. aurantifolii . Mol. Plant Pathol. 9, 609–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P.S. , Wang, L.Y. , Chen, Y.J. , Tzeng, K.C. , Chang, S.C. , Chung, K.R. and Lee, M.H. (2012) Understanding cellular defence in kumquat and calamondin to citrus canker caused by Xanthomonas citri subsp. citri . Physiol. Mol. Plant Pathol. 79, 1–12. [Google Scholar]
- Chi, Y.H. , Paeng, S.K. , Kim, M.J. , Hwang, G.Y. , Melencion, S.M.B. , Oh, H.T. and Lee, S.Y. (2013) Redox‐dependent functional switching of plant proteins accompanying with their structural changes. Front. Plant Sci. 4, 277. doi: 10.3389/fpls.2013.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa, M.A. , Siciliano, M.F. , Ornella, L. , Roeschlin, R.A. , Favaro, M.A. , Delgado, N.P. , Sendín, L.N. , Orce, I.G. , Ploper, L.D. , Vojnov, A.A. , Vacas, J.G. , Filippone, M.P. , Castagnaro, A.P. and Marano, M.R. (2013) Characterization of a variant of Xanthomonas citri subsp. citri that triggers a host‐specific defense response. Phytopathology, 103, 555–564. [DOI] [PubMed] [Google Scholar]
- Deng, Z.N. , Xu, L. , Li, D.Z. , Long, G.Y. , Liu, L.P. , Fang, F. and Shu, G.P. (2010) Screening citrus genotypes for resistance to canker disease (Xanthomonas axonopodis pv. citri). Plant Breeding, 129, 341–345. [Google Scholar]
- van Doorn, W.G. , Beers, E.P. , Dangl, J.L. , Franklin‐Tong, V.E. , Gallois, P. , Hara‐Nishimura, I. , Jones, A.M. , Kawai‐Yamada, M. , Lam, E. , Mundy, J. , Mur, L.A. , Petersen, M. , Smertenko, A. , Taliansky, M. , Van Breusegem, F. , Wolpert, T. , Woltering, E. , Zhivotovsky, B. and Bozhkov, P.V. (2011) Morphological classification of plant cell deaths. Cell Death Differ. 18, 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Y.P. , Castaneda, A. , Zhao, G. , Erdos, G. and Gabriel, D.W. (1999) Expression of a single, host‐specific, bacterial pathogenicity gene in plant cells elicits division, enlargement, and cell death. Mol. Plant–Microbe Interact. 12, 556–560. [Google Scholar]
- Durgbanshi, A. , Arbona, V. , Pozo, O. , Miersch, O. , Sancho, J.V. and Gomez‐Cadenas, A. (2005) Simultaneous determination of multiple phytohormones in plant extracts by liquid chromatography‐electrospray tandem mass spectrometry. J. Agric. Food Chem. 53, 8437–8442. [DOI] [PubMed] [Google Scholar]
- Enrique, R. , Siciliano, F. , Favaro, M.A. , Gerhardt, N. , Roeschlin, R. , Rigano, L. , Sendín, L. , Castagnaro, A. , Vojnov, A. and Marano, M.R. (2011) Novel demonstration of RNAi in citrus reveals importance of citrus callose synthase in defence against Xanthomonas citri subsp. citri . Plant Biotechnol. J. 9, 394–407. [DOI] [PubMed] [Google Scholar]
- Escalon, A. , Javegny, S. , Vernière, C. , Noel, L.D. , Vital, K. , Poussier, S. , Hajri, A. , Boureau, T. , Pruvost, O. , Arlat, M. and Gagnevin, L. (2013) Variations in type III effector repertoires, pathological phenotypes and host range of Xanthomonas citri pv. citri pathotypes. Mol. Plant Pathol. 14, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone, M.L.F. , Rius, S. , Emiliani, J. , Pourcel, L. , Feller, A.J. , Morohashi, K. , Casati, P. and Grotewold, E. (2010) Cloning and characterization of an UV‐B‐inducible maize flavonol synthase. Plant J. 62, 77–91. [DOI] [PubMed] [Google Scholar]
- Falcone, M.L.F. , Emiliani, J. , Rodriguez, E.J. , Campos‐Bermudez, V.A. , Grotewold, E. and Casati, P. (2015) The identification of maize and Arabidopsis type I flavone synthases links flavones with hormones and biotic interactions. Plant Physiol. 169, 1090–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro, M.A. , Micheloud, N.G. , Roeschlin, R.A. , Chiesa, M.A. , Castagnaro, A.P. , Vojnov, A.A. , Gmitter, F.G. Jr. , Gadea, J. , Rista, L.M. , Gariglio, N.F. and Marano, M.R. (2014) Surface barriers of mandarin ‘Okitsu’ leaves make a major contribution to canker disease resistance. Phytopathology, 104, 970–976. [DOI] [PubMed] [Google Scholar]
- Febres, V.J. , Khalaf, A. , Gmitter, F.G. Jr. and Moore, G.A. (2012) Evaluating gene expression responses of citrus to two types of defense inducers using a newly developed citrus Agilent microarray. Acta Hortic. 929, 59–64. [Google Scholar]
- Fu, X.Z. , Gong, X.Q. , Zhang, Y.X. , Wang, Y. and Liu, J.H. (2012) Different transcriptional response to Xanthomonas citri subsp. citri between kumquat and sweet orange with contrasting canker tolerance. PLoS One, 7, e41790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Z.Q. and Dong, X. (2013) Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. [DOI] [PubMed] [Google Scholar]
- Graham, J.H. , Gottwald, T.R. , Cubero, J. and Achor, D.S. (2004) Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 1, 1–15. [DOI] [PubMed] [Google Scholar]
- Hatsugai, N. , Iwasaki, S. , Tamura, K. , Kondo, M. , Fuji, K. , Ogasawara, K. , Nishimura, M. and Hara‐Nishimura, I. (2009) A novel membrane fusion‐mediated plant immunity against bacterial pathogens. Genes Dev. 23, 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck, P. , Thilmony, R. and He, S.Y. (2003) A Pseudomonas syringae type III effector suppresses cell wall‐based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA, 100, 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez‐Blanco, C. , Feng, D.X. , Hu, J. , Sanchez‐Vallet, A. , Deslandes, L. , Llorente, F. , Berrocal‐Lobo, M. , Keller, H. , Barlet, X. , Sanchez‐Rodriguez, C. , Anderson, L.K. , Somerville, S. , Marco, Y. and Molina, A. (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell, 19, 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius, D. , Schultz‐Larsen, T. , Joensen, J. , Tsitsigiannis, D.I. , Petersen, N.H.T. , Mattsson, O. , Jørgensen, L.B. , Jones, J.D.G. , Mundy, J. and Petersen, M. (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell, 37, 773–783. [DOI] [PubMed] [Google Scholar]
- Hofius, D. , Munch, D. , Bressendorff, S. , Mundy, J. and Petersen, M. (2011) Role of autophagy in disease resistance and hypersensitive response‐associated cell death. Cell Death Differ. 18, 1257–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kabbage, M. , Williams, B. and Dickman, M.B. (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum . PLoS Pathog. 9, e1003287. doi: 10.1371/journal.ppat.1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota, Y. , Sklenar, J. , Derbyshire, P. , Stransfeld, L. , Asai, S. , Ntoukakis, V. , Jones, J.D. , Shirasu, K. , Menke, F. , Jones, A. and Zipfel, C. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR‐associated kinase BIK1 during plant immunity. Mol. Cell, 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Karpinski, S. , Szechynska‐Hebda, M. , Wituszynska, W. and Burdiak, P. (2013) Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 36, 736–744. [DOI] [PubMed] [Google Scholar]
- Kazan, K. and Lyons, R. (2014) Intervention of phytohormone pathways by pathogen effectors. Plant Cell, 26, 2285–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaf, A.A. , Gmitter, F.G. Jr. , Conesa, A. , Dopazo, J. and Moore, G.A. (2011) Fortunella margarita transcriptional reprogramming triggered by Xanthomonas citri subsp. citri . BMC Plant Biol. 159. doi: 10.1186/1471-2229-11-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, E. and Slusarenko, A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell, 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I.J. , Kim, K.W. , Hyun, J.W. , Lee, Y.H. and Park, E.W. (2009) Comparative ultrastructure of nonwounded Mexican lime and Yuzu leaves infected with the citrus canker bacterium Xanthomonas citri pv. citri . Microsci. Res. Technol. 72, 507–516. [DOI] [PubMed] [Google Scholar]
- Lenz, H.D. , Haller, E. , Melzer, E. , Kober, K. , Wurster, K. , Stahl, M. , Bassham, D.C. , Vierstra, R.D. , Parker, J.E. , Bautor, J. , Molina, A. , Escudero, V. , Shindo, T. , van der Hoorn, R.A.L. , Gust, A.A. and Nürnberger, T. (2011) Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66, 818–830. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Czymmek, K. , Talloczy, Z. , Levine, B. and Dinesh‐Kumar, S.P. (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell, 121, 567–577. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Ren, D. , Pike, S. , Pallardy, S. , Gassmann, W. and Zhang, S. (2007) Chloroplast‐generated reactive oxygen species are involved in hypersensitive response‐like cell death mediated by a mitogen‐activated protein kinase cascade. Plant J. 51, 941–954. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(–Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lux, A. , Morita, S. , Abe, J. and Ito, K. (2005) An improved method for clearing and staining free‐hand sections and whole‐mount samples. Ann. Bot. 96, 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho, A.P. and Zipfel, C. (2015) Targeting of plant pattern recognition receptor‐triggered immunity by bacterial type‐III secretion system effectors. Curr. Opin. Microbiol. 23, 14–22. [DOI] [PubMed] [Google Scholar]
- Mafra, V. , Kubo, K.S. , Alves‐Ferreira, M. , Ribeiro‐Alves, M. , Stuart, R.M. , Boava, L.P. , Rodrigues, C.M. and Machado, M.A. (2012) Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One, 7, e31263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamud, F. , Homem, R.A. , Conforte, V.P. , Yaryura, P.M. , Castagnaro, A.P. , Marano, M.R. , do Amaral, A.M. and Vojnov, A.A. (2013) Identification and characterization of biofilm formation‐defective mutants of Xanthomonas citri subsp. citri . Microbiology, 157, 819–829. [DOI] [PubMed] [Google Scholar]
- Mammarella, N.D. , Cheng, Z. , Fu, Z.Q. , Daudi, A. , Bolwell, G.P. , Dong, X. and Ausubel, F.M. (2014) Apoplastic peroxidases are required for salicylic acid‐mediated defense against Pseudomonas syringae . Phytochemistry, 112, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marano, M.R. and Carrillo, N. (1992) Constitutive transcription and stable RNA accumulation in plastids during the conversion of chloroplasts to chromoplasts in ripening tomato fruits. Plant Physiol. 100, 1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Godoy, M.A. , Mauri, N. , Juarez, J. , Marques, M.C. , Santiago, J. , Forment, J. and Gadea, J. (2008) A genome‐wide 20 K citrus microarray for gene expression analysis. BMC Genomics, 9, 318. doi: 10.1186/1471-2164-9-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza, C.A. , Boccalandro, H.E. , Giordano, C.V. , Battista, D. , Scopel, A.L. and Ballare, C.L. (2000) Functional significance and induction by solar radiation of ultraviolet‐absorbing sunscreens in field‐grown soybean crops. Plant Physiol. 122, 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina, I. , Carbonell, J. , Pulido, L. , Madeira, S.C. , Goetz, S. , Conesa, A. , Tarraga, J. , Pascual‐Montano, A. , Nogales‐Cadenas, R. , Santoyo, J. , García, F. , Marbà, M. , Montaner, D. and Dopazo, J. (2010) Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 38, W210–213. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel, L. , Irani, N.G. , Koo, A.J.K. , Bohorquez‐Restrepo, A. , Howe, G.A. and Grotewold, E. (2013) A chemical complementation approach reveals genes and interactions of flavonoids with other pathways. Plant J. 74, 383–397. [DOI] [PubMed] [Google Scholar]
- Rigano, L.A. , Siciliano, F. , Enrique, R. , Sendín, L. , Filippone, P. , Torres, P.S. , Qüesta, J. , Dow, J.M. , Castagnaro, A.P. , Vojnov, A.A. and Marano, M.R. (2007) Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri . Mol. Plant–Microbe Interact. 20, 1222–1230. [DOI] [PubMed] [Google Scholar]
- Rojo, E. , Martin, R. , Carter, C. , Zouhar, J. , Pan, S. , Plotnikova, J. , Jin, H. , Paneque, M. , Sanchez‐Serrano, J.J. , Baker, B. , Ausubel, F.M. and Raikhel, N.V. (2004) VPEgamma exhibits a caspase‐like activity that contributes to defense against pathogens. Curr. Biol. 14, 1897–1906. [DOI] [PubMed] [Google Scholar]
- Rybak, M. , Minsavage, G.V. , Stall, R.E. and Jones, J.B. (2009) Identification of Xanthomonas citri ssp. citri host specificity genes in a heterologous expression host. Mol. Plant Pathol. 10, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, N.C. , Postnikova, E. , Lacy, G.H. , Sechler, A. , Agarkova, I. , Stromberg, P.E. , Stromberg, V.K. and Vidaver, A.K. (2006) Emended classification of xanthomonad pathogens on citrus. Syst. Appl. Microbiol. 29, 690–695. [DOI] [PubMed] [Google Scholar]
- Schaad, N.W. , Postnikova, E. , Lacy, G.H. , Sechler, A. , Agarkova, I. , Stromberg, P.E. , Stromberg, V.K. and Vidaver, A.K. (2005) Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii ssp. citri (ex Hasse) sp. nov. nom. rev. comb. nov., X. fuscans ssp. aurantifolii (ex Gabriel 1989) sp. nov. nom. rev. comb. nov., and X. alfalfae ssp. citrumelo (ex Riker and Jones) Gabriel 1989 sp. nov. nom. rev. comb. nov.; X. campestris pv. malvacearum (ex Smith 1901) Dye 1978 as X. smithii ssp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) dye 1978 as X. alfalfae ssp. alfalfae (ex Riker et al., 1935) sp. nov. nom. rev.; and “var. fuscans” of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans ssp. fuscans sp. nov. Syst. Appl. Microbiol. 28, 494–518. [DOI] [PubMed] [Google Scholar]
- Seay, M.D. and Dinesh‐Kumar, S.P. (2005) Life after death: are autophagy genes involved in cell death and survival during plant innate immune responses? Autophagy, 1, 185–186. [DOI] [PubMed] [Google Scholar]
- Shapiguzov, A. , Vainonen, J.P. , Wrzaczek, M. and Kangasjarvi, J. (2012) ROS‐talk‐how the apoplast, the chloroplast, and the nucleus get the message through. Front. Plant Sci. 3, 292. doi: 10.3389/fpls.2012.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani, H. , Fujikawa, T. , Ishihara, H. , Tsuyumu, S. and Ozaki, K. (2007) A pthA homolog from Xanthomonas axonopodis pv. citri responsible for host‐specific suppression of virulence. J. Bacteriol. 189, 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano, A.S. , Abe, V.Y. , Smetana, J.H. and Benedetti, C.E. (2013) Citrus MAF1, a repressor of RNA polymerase III, binds the Xanthomonas citri canker elicitor PthA4 and suppresses citrus canker development. Plant Physiol. 163, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Stall, R.E. , Jones, J.B. , Cubero, J. , Gottwald, T.R. , Graham, J.H. , Dixon, W.N. , Schubert, T.S. , Chaloux, P.H. and Stromberg, V.K. (2004) Detection and characterization of a new strain of citrus canker bacteria from Key/Mexican lime and alemow in South Florida. Plant Dis. 88, 1179–1188. [DOI] [PubMed] [Google Scholar]
- Supek, F. , Bošnjakn, M. , Škunca, N. and Šmuc, T. (2011) REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS One, 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh, O.K. and Hofius, D. (2014) Membrane trafficking and autophagy in pathogen‐triggered cell death and immunity. J. Exp. Bot. 65, 1297–1312. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G. , Tibshirani, R. and Chu, G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA, 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme, R. , Storme, V. , Vanholme, B. , Sundin, L. , Christensen, J.H. , Goeminne, G. , Halpin, C. , Rohde, A. , Morreel, K. and Boerjan, W. (2012) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell, 24, 3506–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernière, C. , Hartung, J.S. , Pruvost, O.P. , Civerolo, E.L. , Alvarez, A.M. , Maestri, P. and Luisetti, J. (1998) Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from Southwest Asia. Eur. J. Plant Pathol. 104, 477–487. [Google Scholar]
- Voigt, C.A. (2014) Callose‐mediated resistance to pathogenic intruders in plant defense‐related papillae. Front. Plant Sci. 5, 168. doi: 10.3389/fpls.2014.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojnov, A. and Marano, M.R. (2015) Biofilm formation and virulence in bacterial plant pathogens In: Virulence Mechanisms of Plant‐Pathogenic Bacteria (Wang N., Jones J.B., Sundin G.W., White F.F., Hogenhout S.A., Roper C., De la Fuente L. and Ham J.H., eds), pp. 21–34. St. Paul, Minnesota: The American Phytopathological Society APS Press. [Google Scholar]
- Vojnov, A.A. , do Amaral, A.M. , Dow, J.M. , Castagnaro, A.P. and Marano, M.R. (2010) Bacteria causing important diseases of citrus utilise distinct modes of pathogenesis to attack a common host. Appl. Microbiol. Biotechnol. 87, 467–477. [DOI] [PubMed] [Google Scholar]
- Wrzaczek, M. , Brosché, M. and Kangasjärvi, J. (2013) ROS signaling loops—production, perception, regulation. Curr. Opin. Plant Biol. 16, 575–582. [DOI] [PubMed] [Google Scholar]
- Xia, P. , Wang, S. , Du, Y. , Xhao, Z. , Shi, L. , Sun, L. , Huang, G. , Ye, B. , Li, C. , Dai, Z. , Hou, N. , Cheng, X. , Sun, Q. , Li, L. , Yang, X. and Fan, Z. (2013) WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 32, 2685–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, K. , Hanaoka, H. , Sato, S. , Kato, T. , Tabata, S. , Noda, T. and Ohsumia, Y. (2004) Processing of ATG8s, ubiquitin‐like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell, 16, 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, K. , Jikumaru, Y. , Kamiya, Y. , Kusano, M. , Consonni, C. , Panstruga, R. , Ohsumi, Y. and Shirasu, K. (2009) Autophagy negatively regulates cell death by controlling NPR1‐dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell, 21, 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, M.H. , Torres, P.S. , El Oirdi, M. , Rigano, L.A. , Gonzalez‐Lamothe, R. , Marano, M.R. , Castagnaro, A.P. , Dankert, M.A. , Bouarab, K. and Vojnov, A.A. (2006) Xanthan induces plant susceptibility by suppressing callose deposition. Plant Physiol. 141, 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Yu, J.Q. and Chen, Z. (2014) The perplexing role of autophagy in plant innate immune responses. Mol. Plant Pathol. 15, 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Bacterial adhesion and biofilm formation on an inert plastic surface.
Fig. S2 Venn diagrams representing the distribution of regulated transcripts in Citrus limon after Xanthomonas citri ssp. citri (X. citri) inoculation.
Fig. S3 Phenolic compound accumulation surrounding the hypersensitive response associated with rapid programmed cell death (HR‐PCD) induced by Xanthomonas citri ssp. citri AT.
Fig. S4 Transmission electron microscopy of Citrus clementina leaves inoculated with Xanthomonas citri ssp. citri (X. citri) strains.
Table S1 List of all oligonucleotide primers used for quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis.
Table S2 Microarray expression data for up‐ and down‐regulated genes in response to Xanthomonas citri ssp. citri (X. citri) strains in Citrus limon leaves.
Table S3 Comparison of microarray expression data for up‐ and down‐regulated genes in Citrus limon leaves in response to Xanthomonas citri ssp. citri (X. citri) strains.
Table S4 Gene ontology (GO) ‘biological process’ terms enriched in the differentially expressed genes unique to the Citrus limon–Xanthomonas citri ssp. citri strain AT interaction.
Table S5 Gene ontology (GO) ‘biological process’ terms enriched in the differentially expressed genes unique to the Citrus limon–Xanthomonas citri ssp. citri strain T interaction.
Table S6 Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of genes involved in the defence and pathogenesis response to Xanthomonas citri ssp. citri (X. citri) strains.


