Summary
The Potato virus X (PVX) triple gene block protein 3 (TGBp3), an 8‐kDa membrane binding protein, aids virus movement and induces the unfolded protein response (UPR) during PVX infection. TGBp3 was expressed from the Tobacco mosaic virus (TMV) genome (TMV‐p3), and we noted the up‐regulation of SKP1 and several endoplasmic reticulum (ER)‐resident chaperones, including the ER luminal binding protein (BiP), protein disulphide isomerase (PDI), calreticulin (CRT) and calmodulin (CAM). Local lesions were seen on leaves inoculated with TMV‐p3, but not TMV or PVX. Such lesions were the result of TGBp3‐elicited programmed cell death (PCD), as shown by an increase in reactive oxygen species, DNA fragmentation and induction of SKP1 expression. UPR‐related gene expression occurred within 8 h of TMV‐p3 inoculation and declined before the onset of PCD. TGBp3‐mediated cell death was suppressed in plants that overexpressed BiP, indicating that UPR induction by TGBp3 is a pro‐survival mechanism. Anti‐apoptotic genes Bcl‐xl, CED‐9 and Op‐IAP were expressed in transgenic plants and suppressed N gene‐mediated resistance to TMV, but failed to alleviate TGBp3‐induced PCD. However, TGBp3‐mediated cell death was reduced in SKP1‐silenced Nicotiana benthamiana plants. The combined data suggest that TGBp3 triggers the UPR and elicits PCD in plants.
Introduction
Plant RNA viruses are notorious for causing massive changes in the physical shape, protein composition and lipid composition of the endoplasmic reticulum (ER). Early and extensive reorganization of cellular membranes is necessary to create membrane bound compartments that function as the physical scaffold for viral replication complexes as well as protection against host innate immune responses (Netherton et al., 2007; Verchot, 2011). Brome mosaic virus (BMV), Tobacco etch virus (TEV), Cowpea mosaic virus (CPMV), Red clover necrotic mosaic virus (RCNMV), Grapevine fanleaf virus (GFLV) and Potato virus X (PVX) are common examples of viruses known to cause proliferation and invaginations of the ER (Carette et al., 2000, 2002; Lee and Ahlquist, 2003; Ritzenthaler et al., 2002; Schaad et al., 1997; Turner et al., 2004). In yeast, the BMV 1a protein triggers membrane lipid synthesis and alters the cell ratio of saturated to unsaturated fatty acids to increase the availability of particular unsaturated fatty acids that are bound by BMV 1a protein during virus replication (Lee and Ahlquist, 2003). In addition, in yeast, Tomato bushy stunt virus (TBSV) and Flock house virus (FHV; an insect virus) activate INO2 and INO4, which are involved in phospholipid synthesis and whose overexpression increases viral RNA accumulation (Sharma et al., 2011). The mechanisms for the stimulation of de novo membrane synthesis by CPMV, PVX and GFLV have not been elaborated.
The ER is a membrane scaffold in which proteins are synthesized, folded and assembled before they are transported via the secretory system to their final destination. However, there are cases in which proteins fail to fold properly and form aggregates that disrupt normal ER functions. Protein sensors in the ER detect the presence of malformed proteins and activate the unfolded protein response (UPR) which up‐regulates the synthesis of protein chaperones and degradation factors (Jelitto‐Van Dooren et al., 1999; Slepak et al., 2007; Urade, 2007; Xu et al., 2005). The ER‐associated degradation machinery (ERAD) is responsible for the elimination of misfolded or otherwise aberrant proteins (Meusser et al., 2005; Muller et al., 2005; Parmar and Schroder, 2012). In this scenario, the UPR is often referred to as the pro‐survival machinery that provides ER maintenance as well as protein quality control (Williams and Lipkin, 2006). However, under acute conditions, when the UPR fails to restore homeostasis, apoptosis or programmed cell death (PCD) can ensue (Martinez and Chrispeels, 2003; Urade, 2007). UPR‐related plant cell death has been widely reported to occur following chemically induced ER stress, but has not been reported as a UPR‐related outcome following plant virus infection.
Plant viruses specifically perturb the ER to promote virus infection, place a huge burden on the cell's protein synthesis and folding machinery and rarely cause cell death as a component of disease. Although the ER quality control machinery is vital to ensure the quality of cellular proteins entering the secretory system, it is also likely to ensure the quality of plant viral proteins needed for infection. Therefore, the challenge is to discover whether plant viruses usurp the UPR machinery for infection or whether the UPR machinery is a component of cellular adaptation and defence responses to ensure cell viability in the face of viral invasion. The second major challenge is to discover whether viruses can create conditions of acute ER stress which lead to PCD. An understanding of the role of UPR in plant virus disease should also facilitate the development of novel strategies to limit plant virus infection.
In plants, bZIP60 is a transcription factor that is transcriptionally induced during UPR following abiotic or biotic stress (Iwata and Koizumi, 2005a; Lu and Christopher, 2008; Martinez and Chrispeels, 2003; Tateda et al., 2008; Urade, 2007). bZIP60 contains a transmembrane domain near the C‐terminus and localizes to ER membranes. Inositol‐requiring enzyme (IRE1) is an ER‐localized sensor of ER stress that catalyses the cytoplasmic splicing of bZIP60 mRNA (called bZIP60s) (Deng et al., 2011; Iwata and Koizumi, 2012). bZIP60s lacks a transmembrane domain and localizes to the nucleus, where it elevates the expression of certain ER‐resident chaperones, such as the ER luminal binding protein (BiP), protein disulphide isomerase (PDI), calreticulin (CRT) and calmodulin (CAM) (Ellgaard and Helenius, 2003; Navazio et al., 2001; Oh et al., 2003; Seo et al., 2008; Urade, 2007; Ye et al., 2011; Ye and Verchot, 2011). Although most plant research on the UPR uses Arabidopsis, there are reports that ER stress‐inducing compounds act in tobacco to up‐regulate NtBLP‐4 (the ER luminal binding protein BiP), NtCRT and NtPDI (Denecke et al., 1991, 1995; Iwata and Koizumi, 2005b). As plant virus research is more often carried out using Nicotiana species, evidence that the mechanisms responding to ER stress are conserved between Arabidopsis and Nicotiana spp. lends further value to our studies.
BiP is highly conserved among eukaryotes and is broadly recognized as a component of the pro‐survival response to ER stress (Leborgne‐Castel et al., 1999). Specifically, BiP is a negative regulator of IRE1 and binds either competitively or separately to both IRE1 and unfolded proteins. During ER stress, BiP releases IRE1, unmasking its own substrate binding domain, which functions to bind the unfolded proteins (Iwata and Koizumi, 2012; Parmar and Schroder, 2012). BiP is unlike CRT or PDI in that its overexpression attenuates UPR by creating a larger pool of molecules that can bind IRE1 or unfolded proteins. However, it is not known whether BiP binds preferentially to malformed proteins, enhances the negative regulation of IRE1, or both, following overexpression. Other plant pro‐survival factors include SDF2, which is a target of the UPR and contributes to plant development (Schott et al., 2010). A recent study using the fungus Piriformospora indica linked ER stress induction to the vacuolar processing enzyme (VPE) and caspase‐like protease‐dependent cell death (Qiang et al., 2012).
During PVX infection, the PVX triple gene block protein 3 (TGBp3) is an 8‐kDa viral protein that is required for intercellular movement. TGBp3 has an N‐terminal transmembrane domain and a C‐terminal cytosolic domain. Mutations in either domain inhibit virus intercellular movement. TGBp3 specifically up‐regulates UPR‐related factors, including bZIP60 and ER‐resident chaperones, in Nicotiana benthamiana plants (Ye et al., 2011). Silencing NbbZIP60 suppresses BiP expression, negatively impacts virus replication in protoplasts and delays virus systemic accumulation in plants, arguing that bZIP60 and UPR contribute to plant virus infection. The exact role of bZIP60 in promoting plant virus infection is not yet known.
SKP1 is a component of the SCF (SKP1/Cullin1/F‐box protein) ubiquitin E3 ligase complex that is up‐regulated by PVX TGBp3 alongside bZIP60 and ER‐resident chaperones (BiP, CRT, CAM, PDI) (Ye et al., 2011). In Arabidopsis and N. benthamiana plants, SKP1 contributes to auxin signalling, host defence against polerovirus infection, response to jasmonates during defence and Agrobacterium tumorigenicity (Gfeller et al., 2010; Pazhouhandeh et al., 2006; Ren et al., 2005; Stuttmann et al., 2009; Xu et al., 2002; Zheng et al., 2011). Importantly, we have reported previously that SKP1 expression is at least partially controlled by bZIP60. The accumulation of SKP1 transcripts was reduced in bZIP60‐silenced plants (Ye et al., 2011). Given the broad role of SCF‐mediated protein degradation in diverse biological processes, including innate immunity and auxin signalling, it is possible that SKP1 expression might be controlled by factors other than bZIP60. Considering the preliminary data, it is also possible that TGBp3 is both an elicitor of UPR as well as of host defence (Ye et al., 2011).
In this study, we examined whether the PVX TGBp3 induces SKP1 as a pro‐survival factor or a defence‐related factor (Azevedo et al., 2006; Kadota et al., 2008). Previous investigations have employed Agrobacterium delivery of the entire PVX genome or TGBp3 alone fused to the Cauliflower mosaic virus (CaMV) 35S promoter (35S:TGBp3) to N. benthamiana leaves. Individual PVX genes were fused to the CaMV35S promoter and delivered to N. benthamiana leaves by agroinfiltration. Only the TGBp3 [and not PVX replicase, TGBp1, TGBp2 or coat protein (CP)] induced the UPR‐related NbbZIP60 and ER‐resident chaperones (BiP, CAM, CRT and PDI), as well as the defence‐related NbSKP1. TGBp3 was also fused to the Nos promoter and delivered by agroinfiltration to N. benthamiana leaves and the same set of genes was up‐regulated (Ye et al., 2011). Furthermore, the TGBp3 protein alone can cause small lesions resulting from cell death in infiltrated leaves.
The sole use of agrodelivery of 35S:TGBp3 to study its elicitor functions has certain limitations. TGBp3 expression was first noted at 2 days post‐agroinfiltration and its expression was maintained for 2–5 days. UPR and cell death are slowly induced in local areas on inoculated leaves (Ye et al., 2011). As the protein did not spread systemically, as during PVX infection, we were unable to examine the effects of TGBp3 on the whole plant physiology. Therefore, we chose TMV as a vector for the delivery of TGBp3 to N. benthamiana and N. tabacum leaves. One advantage of TMV delivery of TGBp3 is that expression occurs within hours of inoculation, making it easier to examine the early events that precede cell death. Second, we were able to examine changes in host gene expression relating to UPR and cell death in systemic tissues. In this investigation, we demonstrate that PVX TGBp3 functions both to induce UPR and elicit PCD.
Results
PVX, but not TMV, induces the expression of UPR‐related chaperones and SKP1
Agrodelivery of infectious TMV or PVX genomic cDNAs [optical density (OD) = 0.5] was employed for virus inoculation of N. benthamiana plants. We also co‐inoculated plants with TMV plus PVX. Systemic infection occurred within 7–10 days post‐inoculation (dpi). The upper leaves of TMV‐infected plants showed curling, mosaic disease and dark green islands. The upper leaves of PVX‐infected plants showed less dramatic changes in leaf shape, mosaic disease and necrotic flecks (Fig. 1A). Plants that were co‐infected with TMV and PVX were stunted and showed severe mosaic and chlorosis on the adjoining leaves. Newly emerging leaves were severely curled and necrotic (Fig. 1A). Western blot analysis detected higher levels of TMV CP in co‐infected plants. PVX CP levels seemed to be unaltered by co‐infection (Fig. 1B).
Figure 1.
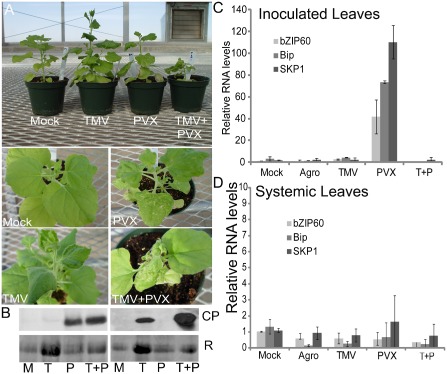
Plants inoculated with Tobacco mosaic virus (TMV) and Potato virus X (PVX). (A) Plants at 21 days post‐inoculation (dpi) following agrodelivery [optical density (OD) = 0.5] of virus infection clones. Plants are presented in a row to show contrasting changes in development. The images of each plant individually show specific symptoms on the systemically infected leaves. (B) Immunoblots at the top of the figure confirm the presence of TMV or PVX coat protein (CP) in systemically infected leaves at 14 dpi. The left panel was treated with PVX CP antisera and the panel on the right was treated with TMV CP antisera. Membranes were stained with Coomassie blue and bands (R) represent the total protein loaded on the gel. M, mock; T, TMV; P, PVX; T+P, TMV + PVX. (C, D) Semi‐quantitative real‐time polymerase chain reaction (RT‐PCR) was conducted to verify the relative levels of endogenous RNA accumulation in infected leaves at 2 and 21 dpi.
RNA was extracted from the virus‐inoculated leaves at 2 dpi and systemically infected leaves at 21 dpi (when necrotic symptoms were evident). Quantitative real‐time polymerase chain reaction (qRT‐PCR)‐based quantification of transcripts was used to monitor the accumulation of UPR‐related genes and NbSKP1 at various times. Consistent with a previous report describing PVX infection, there were significant increases in the levels of bZIP60, BiP and SKP1 transcripts in PVX‐inoculated leaves (Ye et al., 2011). This contrasted with the negligible changes in expression following treatment with buffer only (mock), A. tumefaciens expressing the empty binary vector (Agro) or TMV alone (Fig. 1C). Interestingly, leaves infected with both TMV and PVX showed no changes in host gene expression, indicating that TMV had a dominant suppressive effect (Fig. 1C).
Expression levels of the UPR genes and SKP1 showed little change in leaves that were systemically infected with PVX, indicating that these genes are early components of disease (Fig. 1D). Importantly, we failed to detect necrosis in inoculated leaves following PVX, TMV or PVX plus TMV infection. However, the upper leaves of PVX‐infected plants showed necrotic flecks and the emerging leaves of PVX plus TMV‐infected plants showed severe necrosis. These data suggest that UPR is induced prior to cell death and might contribute to a pro‐survival mechanism that limits PCD.
We introduced PVX TGBp1, TGBp2 and CP genes into the TMV genome and confirmed the expression of the PVX proteins by immunoblot analysis at 5 dpi (although TGBp2 expression was confirmed by Coomassie staining; Fig. S1, see Supporting Information). These PVX genes failed to induce the expression of ER chaperones or NbSKP1 when they were expressed from TMV vector (Fig. S1). Similar negative outcomes were reported when they were fused directly to the 35S promoter and delivered by agroinfiltration (Ye et al., 2011).
TMV delivering TGB3, but not TGB2, induces UPR‐related chaperones and SKP1
N. benthamiana plants were inoculated with 30 μg/mL purified TMV, TMV‐p2, TMV‐p3 (expressing TGBp2 or TGBp3), TMV‐p3Dm1 (mutation of TGBp3) or PVX. TMV‐p2 was employed as a control to demonstrate whether TGBp3 is a specific molecular trigger of the host UPR signalling pathway or if another PVX‐encoded, low‐M r, ER‐localized protein (the 12‐kDa TGBp2) can provide the same function. TMV‐p3Dm1 was another control harbouring a deletion in TGBp3 that eliminates the N‐terminal transmembrane domain, causing the cytosolic accumulation of TGBp3 (Ju et al., 2008).
Within the first 8 h post‐inoculation (hpi), higher gene expression levels were evident in N. benthamiana infected with TMV‐p3 than with TMV, TMV‐p2, TMV‐p3Dm1 or PVX (Fig. 2). BiP and PDI expression increased 30–35‐fold within the first 8 hpi. CRT and CAM showed about a 15‐fold induction (Fig. 2). Transcript accumulation was monitored until 20 hpi and the levels declined steeply (data not shown), suggesting that up‐regulation of ER‐resident chaperones occurs early. There was a modest increase in BiP transcript abundance among plants that were inoculated with TMV, PVX, TMV‐p3Dm1 or TMV‐p2. By comparison, the dramatic up‐regulation of ER‐resident chaperones by TGBp3 indicates that an intact TGBp3 is necessary to trigger the expression of the ER quality control machinery.
Figure 2.
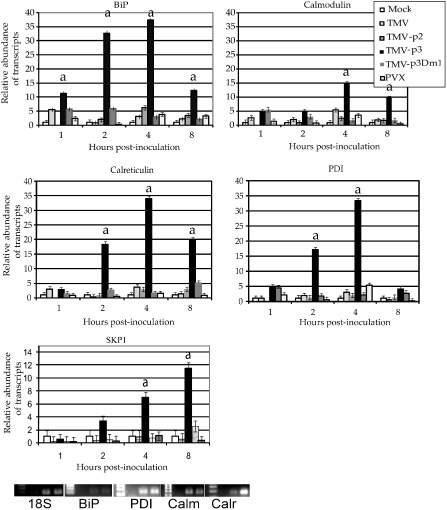
Quantitative real‐time polymerase chain reaction (qRT‐PCR) analysis of BiP, PDI, CRT, and CAM transcript abundance. The plots show the relative abundance of transcripts for each virus treatment at each time point. The title for each plot identifies the host gene and each virus is indicated in the box key. Analysis of variance (ANOVA) was used to determine the significance of each virus treatment for transcript accumulation. A significance level of 0.05 was used to judge statistical significance. The ‘a’ above the TMV‐p3 bars indicates the occasions on which the abundance of host transcripts induced by TMV‐p3 was significantly greater than that of all other viral treatments at that time point. Occasionally, TMV, TMV‐p3Dm1 and PVX showed minor, but significant, differences from the mock control, but these were often not equal to the effects of TMV‐p3, as noted in the figure. The bottom panel shows ethidium bromide‐stained gels containing PCR products prepared using each primer set. Each gel has four lanes: a size marker and RT‐PCR using buffer (no RNA) control and extracted plant RNAs from healthy and virus‐infected samples. The gels show that the PCR primers produce single bands of a discrete size necessary for qRT‐PCR analysis. PVX, Potato virus X; TMV, Tobacco mosaic virus.
To verify TGBp3 expression, a 6 × histidine (His) tag was fused to TGBp3 in the TMV and PVX genomes and immunoblot analysis was carried out. TGBp3His protein accumulation was much lower in PVX‐p3His inoculated (5 dpi) and systemically infected (8–10 dpi) leaves (Fig. S2A, lanes 2–5, see Supporting Information) than in TMV‐p3‐infected leaves (Fig. S2, lanes 6–9). Longer film exposure was required to reveal a faint TGBp3 band in the PVX‐infected samples (Fig. S2A). Agroinfiltrated leaves delivering 35S:TGBp3His were harvested at the same time points. The levels of TGBp3His in the TMV‐p3‐inoculated leaves were comparable or slightly greater than those detected in agroinfiltrated leaves (Fig. S2, lanes 12 and 13). We also showed that TMV‐p3His does not affect the level of TMV CP accumulation in systemic leaves (Fig. S2B). Taken together, TMV expressing TGBp3 induces the same set of host genes as reported in a previous study using PVX and the agrodelivery of TGBp3 alone (Fig. 2). Although TMV has a trans‐dominant suppressive effect on PVX‐induced gene expression, it has no detectable impact on the ability of TGBp3 to function as an elicitor when it is expressed from the TMV genome (Fig. 2).
TMV‐p3 triggers PCD in tobacco plants
Previously, we have reported small lesions that resemble a hypersensitive response (HR) in agroinfiltrated leaves expressing 35S:TGBp3 (Ye et al., 2011). Here, local HR‐like lesions occurred in TMV‐p3‐infected N. benthamiana and N. tabacum [N‐gene recessive (nn)] (Plants lacking the N‐gene which confers TMV resistance) leaves by 3 dpi. Over time, leaves showed total collapse. TMV‐p3 disease in the distal leaves of N. benthamiana plants was severe, with veinal and apical necrosis (Fig. 3A), whereas TMV and TMV‐p2 infection produced mosaic symptoms.
Figure 3.
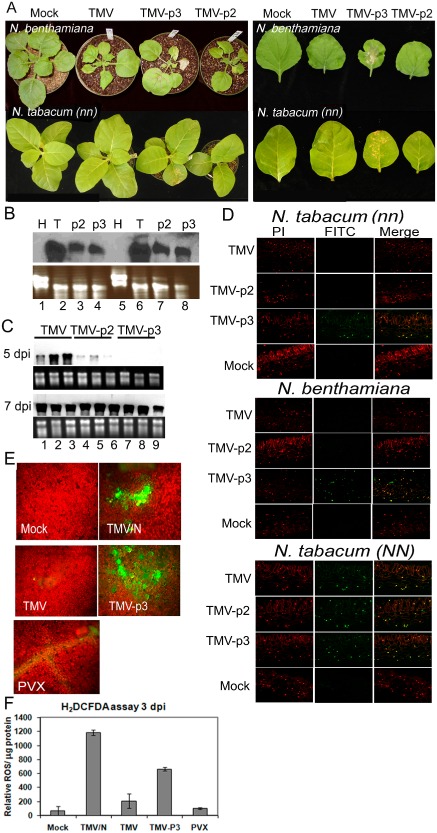
Programmed cell death (PCD) is induced by TMV‐p3, but not TMV or TMV‐p2, in Nicotiana benthamiana and N. tabacum. (A) Inoculated N. benthamiana and N. tabacum whole infected plants and inoculated leaves at 7 days post‐inoculation (dpi). Real‐time polymerase chain reaction (RT‐PCR) was used to amplify the TMV coat protein (CP) gene, using RNA extracts from upper noninoculated leaves; it confirms the systemic spread of TMV and TMV‐p2, but not TMV‐p3 (data not shown). (B) Northern analysis shows TMV (T), TMV‐p2 (p2) and TMV‐p3 (p3) accumulation in BY2 protoplasts harvested 24 h post‐inoculation. Blots were probed with TMV CP cDNA. RNA from healthy protoplasts (H) was extracted and analysed alongside virus‐infected samples. The ethidium bromide‐stained gel shows ribosomal RNA below the Northern blot. (C) Northern analysis of TMV‐, TMV‐p2‐ and TMV‐p3‐infected N. benthamiana. Upper leaves of inoculated plants were harvested 5 and 7 dpi. The ethidium bromide‐stained gel shows ribosomal RNA below the Northern blots. (D) TUNEL (terminal deoxynucleotidyl transferase‐mediated UTP nick end labelling) assays of virus‐inoculated leaves. Propidium iodide (PI) stains nuclei red. Fluorescein isothiocyanate (FITC) labels nuclei green in cells showing PCD. Merged images show yellow where the green and red signals overlap. (E, F) Virus‐ and mock‐inoculated N. benthamiana leaves treated with 2′,7′‐dichlorofluorescein diacetate (H2DCFDA) dye at 3 dpi, prior to the onset of necrosis. The green fluorescence seen in the leaf segments expressing triple gene block protein 3 (TGBp3) is an indication of reactive oxygen species (ROS) production. Epifluorescence microscopy shows a red background and green fluorescence caused by H2DCFDA histochemical staining. H2DCFDA fluorescence was quantified spectrophotometrically. The Bradford reagent was used to quantify the total protein in each sample. Data from independent measurements were averaged and expressed as relative fluorescence units (RFU) per microgram of protein. Data represent the averages of n = 5 leaves. PVX, Potato virus X; TMV, Tobacco mosaic virus.
In mammals and in yeast, prolonged ER stress is linked to apoptosis. In plants, abiotic or biotic factors can induce prolonged ER stress, resulting in PCD. Although evidence links the Arabidopsis BAX inhibitor 1 (AtBI) and VPE/caspase 1‐like activity to PCD, the molecular basis of ER stress‐induced PCD is not understood (Lam, 2005; Qiang et al., 2012; Tabas and Ron, 2011; Watanabe and Lam, 2008b). With regard to TMV‐p3 infection, the HR‐like lesions might be the outcome of ER stress‐related PCD, activated innate immune responses or enhanced virus titre causing more severe disease pathology. To test the latter possibility, Northern analysis was used to compare virus in synchronously infected tobacco BY‐2 (nn) protoplasts at 24 hpi. The levels of TMV RNA were significantly higher than those of TMV‐p2 and TMV‐p3 in protoplasts, pointing to a negative impact of TGBp2 and TGBp3 (Fig. 3B). Furthermore, Northern analysis showed that the amount of TMV‐p2 and TMV‐p3 RNA in the upper leaves was considerably less than that of TMV RNA at 5 dpi, although the levels were comparable by 7 dpi (Fig. 3C). Thus, TGBp2 and TGBp3 failed to exacerbate TMV, but had the opposite affect by suppressing RNA accumulation and delaying the systemic spread of TMV. Thus, the areas of cell death seen during TMV‐p3 infection are probably a result of TGBp3 acting as a molecular activator of host defences.
N. tabacum and N. benthamiana leaves (nn) produced positive TUNEL (terminal deoxynucleotidyl transferase‐mediated UTP nick end labelling) reactions at 4 dpi as a result of TMV‐p3, but not TMV or TMV‐p2, infection (Fig. 3D), indicating that TGBp3 is a PCD elicitor in plants lacking the N gene. Positive TUNEL staining of control N. tabacum N‐gene diploid (NN) shows PCD as a result of TMV, TMV‐p2 and TMV‐p3 viruses, which is consistent with the N gene‐mediated response to TMV infection (Fig. 3D).
Reactive oxygen species (ROS) production is another well‐established hallmark of PCD. 2′,7′‐Dichlorofluorescein diacetate (H2DCFDA) staining produced green fluorescence as a result of ROS production as early as 1 dpi in TMV‐p3‐, but not in TMV‐ or PVX‐infected, N. benthamiana leaves (Fig. 3E). As a control, N gene‐expressing transgenic N. benthamiana was inoculated with TMV, and we noted local sites containing green fluorescence, as expected of an N gene‐mediated HR response (Erickson et al., 1999). The fluorometric values, representing ROS production at 3 dpi, were normalized against the total amount of protein in each leaf extract (Fig. 3F). The bar graph in Fig. 3F shows significant levels of ROS in TMV‐p3‐ and TMV‐infected nontransgenic and N gene‐expressing transgenic N. benthamiana. ROS levels in mock‐, TMV‐ and PVX‐inoculated N. benthamiana leaves were comparably low.
UPR reduces PCD in TMV‐p3‐infected plants
BiP is a component of the ER quality control machinery and protects plants from ER stress, as seen in BiP‐overexpressing transgenic tobacco plants (Leborgne‐Castel et al., 1999; Parmar and Schroder, 2012). Furthermore, co‐delivery of the N. benthamiana BLP‐4 homologue and 35S:TGBp3 via agroinfiltration prevented PCD on inoculated leaves (Ye et al., 2011). BiP expression was about 50% higher in leaves infiltrated with A. tumefaciens alone, but was three‐fold higher in NbBLP‐4‐infiltrated leaves than in buffer‐treated leaves, and this was sufficient to alleviate TGBp3‐induced cell death in a previously reported investigation (Ye et al., 2011; Ye, unpublished data). In the current study, N. benthamiana leaves were agroinfiltrated with NbBLP‐4 and then inoculated with TMV, PVX or TMV‐p3 (Ye et al., 2011) (Fig. 4A). TMV‐p3‐infected leaves treated with buffer or A. tumefaciens alone showed typical HR‐like lesions (Fig. 4A). However, such lesions were absent from TMV‐p3‐infected leaves that overexpressed NbBLP‐4. All leaves infected with either TMV or PVX showed mosaic disease. Co‐delivery of TMV or PVX with A. tumefaciens containing an empty vector or NbBLP‐4 caused yellowing (Fig. 4A).
Figure 4.
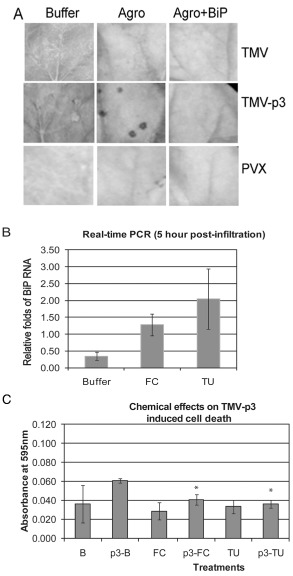
Overexpression of endoplasmic reticulum luminal binding protein (BiP) following agrodelivery and chemical treatment. (A) Nicotiana benthamiana leaves were infiltrated with buffer, Agrobacterium tumefaciens carrying an empty pBI121 (Agro) or pBI‐NbBLP‐4 (Agro+BiP) and inoculate with TMV, TMV‐p3, PVX, or TMV plus PVX. Lesions appeared at 3–5 days post‐inoculation (dpi). Three‐fold overexpression was confirmed by immunoblot analysis (unpublished data and Ye et al., 2011). (B) N. benthamiana leaves were infiltrated with Murashige and Skoog (MS) medium as a control, 20 μm fusicoccin (FC) or 10 μg/mL tunicamycin (TU) prepared in MS medium. Quantitative real‐time polymerase chain reaction (qRT‐PCR) confirmed elevated NbBLP‐4 transcripts. (C) Quantification of Evans blue‐stained TMV‐p3‐infected and chemically treated leaves. Buffer (B, which is MS medium) or TMV‐p3 (p3)‐infected samples were treated with FC or TU. The ‘*’ indicates the results of a t‐test comparison of samples treated with TMV‐p3 and buffer versus those treated with TMV‐p3 plus chemicals to determine the effects of the chemicals on virus infection (P < 0.05). The ‘*’ indicates that FC and TU treatments influence TMV‐p3‐related cell death. A t‐test comparison of chemical and buffer treatments for healthy (no virus) samples found no effect (P < 0.05).
We employed pharmacological agents to induce BiP expression prior to TMV‐p3 inoculation. If BiP is antagonistic to TGBp3‐induced cell death, chemically induced BiP expression should reduce PCD. Fusicoccin (FC) and tunicamycin (TU) are commonly used chemical inducers of ER stress (Kamauchi et al., 2005; Liu et al., 2007). FC specifically binds to proteins of the 14‐3‐3 gene family and FC‐treated cells show increased pathogenesis‐related (PR) gene expression (Pike et al., 2005; Roberts and Bowles, 1999). TU inhibits N‐linked protein glycosylation in the ER and enhances the expression of UPR‐related chaperones (including BiP) and ER‐associated degradation proteins (Martinez and Chrispeels, 2003; Roberts and Bowles, 1999). Several studies have linked BiP induction to PR protein expression (although the relationship is not well defined), suggesting that the effects of TU and FC on plant gene expression overlap (Jelitto‐Van Dooren et al., 1999). Leaves were treated with TU or FC and, by 5 hpi, the NbBLP‐4 transcript levels were elevated (Fig. 4B). Leaf segments were harvested from the infiltrated areas at 3 dpi and stained with Evans blue to detect PCD. The average absorbance values following buffer or chemical treatment alone were not significantly different (Fig. 4C). Thus, the FC and TU concentrations employed were sufficient to up‐regulate BiP without causing cell death (Fig. 4B,C).
Absorbance values for TMV‐p3‐infected leaves were two‐fold greater than those of leaf segments treated with buffer alone, and the increase was statistically significant (Fig. 4C). TMV‐p3‐infected leaf segments treated with FC or TU showed significant reduction in leaf PCD when compared with leaves treated with TMV‐p3 alone. As both FC and TU treatment of plant tissues increased gene expression of ER‐resident chaperones needed to restore normal ER function, these data show that TU or FC treatment is sufficient to counter TGBp3‐induced defence responses. These data show a correlation between an early increase in BiP gene expression, as well as other UPR‐related genes, and reduced PCD.
In summary, these data reveal that increased levels of BiP did not cause the PCD elicited by TGBp3 expression. It is more likely that the early induction of UPR‐related BiP served to limit PCD and this is probably important to aid virus infection. We have reported previously that TGBp3 elicits the expression of bZIP60 transcription factor and ER‐resident chaperones, and that the silencing of bZIP60 is deleterious to PVX infection (Ye et al., 2011; Ye and Verchot, 2011). Although the expression of UPR‐related ER chaperones increased prior to the onset of cell death, both agrodelivery of BiP and chemically induced endogenous BiP expression served to limit the amount of cell death caused by TGBp3. Thus, the work presented in this study, combined with previous research, argues that TGBp3 is both an inducer of UPR‐related gene expression and a cell death elicitor.
Animal anti‐apoptotic genes alleviate TMV‐ and TMV‐p2‐, but not TMV‐p3‐related, necrosis
In animal cells, IRE1 is linked to ER stress‐induced apoptosis via two mechanisms. First, IRE1 binds Bak and Bax, which contribute to the mitochondrial apoptotic machinery. Two other ER stress pathways overlap in their ability to activate CHOP‐induced apoptosis. CHOP suppresses the pro‐survival protein Bcl‐2, which prevents apoptosis by down‐regulating oxidative stress. Transgenic N. tabacum [N‐gene diploid (NN)] plants expressing the oxidative stress/apoptotic inhibitors Caenorhabditis elegans CED‐9, baculovirus Op‐IAP and chicken Bcl‐xl have been reported previously to alleviate PCD induced by Tomato spotted wilt virus (TSWV), Cucumber mosaic virus (CMV) and the necrotrophic fungus Sclerotinia sclerotiorum (Dickman et al., 2001; Kim et al., 2008; Xu et al., 2004). Thus, we and others have shown that these genes can regulate cell death processes in plants in a manner that is functionally conserved across kingdoms. Therefore, we employed these same transgenic plants to determine whether these cytoprotective genes play a role in modulating the PCD pathways instigated by TMV‐p3 (Fig. 5).
Figure 5.
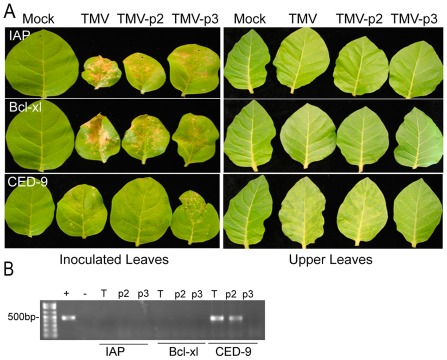
Programmed cell death (PCD) on transgenic N. tabacum (N‐gene diploid) plants inoculated with TMV, TMV‐p2 and TMV‐p3. (A) CED‐9 alleviates TMV‐ and TMV‐p2‐, but not TMV‐p3‐induced, hypersensitive response (HR)‐like PCD at 7 days post‐inoculation (dpi). By 14 dpi, systemic symptoms occur in CED‐9 transgenic plants infected with TMV or TMV‐p2, but not TMV‐p3. (B) Real‐time polymerase chain reaction (RT‐PCR) analysis detecting TMV and coat protein (CP) RNA in total RNA extracted from the upper leaves of virus‐inoculated Op‐IAP and CED‐9 transgenic plants at 14 dpi. TMV, Tobacco mosaic virus.
TMV‐p3 produced HR‐like lesions on N. tabacum (nn), as well as on CED‐9, IAP and Bcl‐xl transgenic plants (Figs 3 and 5A). Interestingly, CED‐9 suppressed N gene‐mediated PCD induced by TMV or TMV‐p2, but had no effect on TMV‐p3. TMV and TMV‐p2 viruses spread systemically causing mosaic symptoms in the upper leaves of CED‐9 transgenic plants, which was confirmed by RT‐PCR (Fig. 5A,B). TMV and TMV‐p2 produced typical necrotic lesions on Op‐IAP and Bcl‐xl transgenic leaves, which are indicative of N gene‐mediated PCD triggered by TMV infection (Fig. 5A). Thus, the PCD induced by the PVX TGBp3 is independent of N gene‐mediated resistance pathways affected by these anti‐apoptotic genes. Based on these outcomes, TGBp3 either elicits an immune response independent of its role in ER stress induction, or TGBp3‐induced PCD involves a mechanism that depends on prolonged ER stress induction.
TGBp3‐induced PCD is compromised on SKP1‐silenced plants
A virus‐induced gene silencing (VIGS) assay was employed using A. tumefaciens harbouring recombinant Tobacco rattle virus (TRV) containing fragments of NbSKP1 to knockdown endogenous host transcripts. This experimental approach has been employed previously, and we reported that PVX‐GFP produced fewer infection foci on SKP1‐silenced N. benthamiana plants than on buffer or TRV‐pretreated leaves, and systemic infection was delayed. We also reported higher GFP fluorescence in the upper leaves, although the PVX CP was unaltered (Ye et al., 2011). In this study, semiquantitative RT‐PCR was conducted at 10–14 dpi using RNA extracted from the upper leaves, and the endogenous NbSKP1 transcripts were reduced by 60%. As expected, SKP1 expression was similar among plants pretreated with buffer or TRV alone (Fig. 6B) (Ye et al., 2011).
Figure 6.
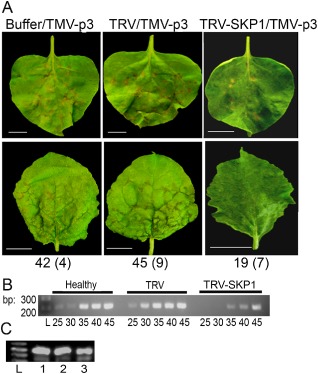
Virus‐induced gene silencing (VIGS) assay using recombinant Tobacco rattle virus (TRV) and TRV‐SKP1 to knock down host gene expression. (A) Silenced leaves were inoculated with TMV‐p3. The top panels show hypersensitive response (HR) lesions on inoculated leaves and the bottom panels show systemically infected leaves. The numbers at the bottom of the panels represent the average number of necrotic lesions on TMV‐p3‐inoculated leaves at 5 days post‐inoculation (dpi). The numbers in parentheses are the standard deviations. (B) Semi‐quantitative real‐time polymerase chain reaction (RT‐PCR) detecting SKP1 in buffer‐treated (Healthy), TRV‐ or TRV‐SKP1‐treated plants conducted using RNA extracted from the upper leaves at 14 dpi. The data show partial suppression of SKP1 transcripts by the TRV‐SKP1 silencing vector. (C) Ethidium bromide‐stained 1% agarose gel containing RT‐PCR products detecting TMV coat protein (CP) RNA in systemically infected leaves at 14 dpi. Lanes 1, 2 and 3 represent mock‐treated, TRV‐treated and TRV‐SKP1‐treated leaves prior to TMV‐p3 inoculation. TMV, Tobacco mosaic virus.
The TMV‐p3 PCD was significantly compromised in SKP1‐silenced N. benthamiana leaves. NbSKP1‐silenced leaves showed fewer TMV‐p3‐related lesions (average of 18 lesions per leaf; n = 10 leaves), and veinal necrosis in the systemically infected tissues was absent. In contrast, buffer‐ or TRV‐pretreated leaves showed numerous lesions (average of 42 lesions per leaf; n = 10 leaves) and severe veinal necrosis following TMV‐p3 inoculation (Fig. 6A). RT‐PCR detected TMV RNAs in the upper leaves, verifying the systemic accumulation of TMV‐p3 (Fig. 6C).
SKP1 proteins in yeast, humans and plants are components of the SCF‐type E3 ubiquitin ligase complex associated with protein degradation. Azevedo et al. (2006) and Liu et al. (2002) have shown that N or Rx gene‐mediated virus resistance is compromised by knockout of NbSKP1, indicating a role for the ubiquitin–proteasome pathway in plant defence. Considering the loss of PCD on TMV‐p3‐inoculated NbSKP1‐silenced leaves, these data suggest that TGBp3 either elicits an independent SKP1‐linked immune response, or that TGBp3 induces an ER stress response in which SKP1 is an essential component involved in PCD regulation.
Discussion
The TMV vector was employed to study the ability of PVX TGBp3 to activate the host UPR‐related gene expression and to induce PCD. TMV has been proven to be a highly efficient and reliable protein expression system that is compatible with agroinfection for the production of foreign epitopes and recombinant proteins in plants, the expression of reporter genes in single cells and whole plants, and for high‐throughput experimentation (Lindbo, 2007a,b; Liu and Kearney, 2010), without causing severe tissue toxicity or cell death. Using the TMV expression vector, we reported TGBp3‐induced host gene expression within hours of delivery. We also determined that the level of TGBp3 expression from the TMV vector was comparable with the levels reported using the agroinfiltration of 35S:TGBp3 plasmids. However, agrodelivery of 35S:TGBp3 plasmids can take several days to obtain adequate expression of the foreign gene to study activities, making TMV a more robust expression system (Ye et al., 2011, 2012). We have reported previously that PVX TGBp3 specifically induces the expression of bZIP60, ER‐resident chaperones and SKP1 during PVX infection (Ye et al., 2011). Interestingly, TMV has a trans‐dominant suppressive effect on PVX host gene induction. These data suggest that interactions occur between the two replicating viruses that limit signal transduction events triggered during PVX infection. Regardless of the virus interactions, TMV was still an adequate vector for the study of TGBp3‐induced host gene expression. TGBp3 clearly is an adequate elicitor of host gene expression when expressed directly from the CaMV 35S promoter following agroinfiltration or from a viral vector, such as TMV.
The UPR is an early response to virus infection and preceded the onset of cell death in this study. It is possible that the UPR effectively delays the host immune responses, including PCD, and enables PVX to establish infection. The ER‐resident proteins BiP, PDI, CRT and CAM were specifically induced within the first 8 h following TMV‐p3 inoculation, and cell death was observed when these genes were down‐regulated in PVX‐infected leaves. These data also confirm the specific requirement of an intact TGBp3 to elicit the expression of key ER‐resident chaperones which represent a molecular signature of the UPR. Mutations eliminating the membrane binding domain of TGBp3 failed to stimulate gene expression. Although plant virus‐induced ER stress is largely unexplored in plants, research using chemical and osmotic stressors have linked bZIP60‐controlled expression of ER‐resident chaperones, such as BiP, to the protection of cells from the toxic accumulation of malformed proteins (Tardif et al., 2004; Zhang and Kaufman, 2006). The main function of BiP is to regain protein folding capacity and restore ER homeostasis. It has also been reported that the overexpression of BiP in transgenic plants or by agroinfiltration suppresses PCD. In this study, agroinfiltration of NbBLP‐4 eliminated TMV‐p3‐induced PCD, which is consistent with a cytoprotective function. BiP was also up‐regulated by chemical inducers of ER stress, such as TU or FC, indicating that TGBp3 has a similar ability to induce the UPR in plants (Costa et al., 2008). Moreover, PCD caused by TMV‐p3 infection was reduced (as evidenced by lower absorbance values following Evans blue staining) in chemically primed leaves that showed higher levels of BiP expression. These combined data lend support to the interpretation that BiP plays a cytoprotective role in virus‐infected leaves. These data support the reasonable speculation that the ER‐resident PVX TGBp3 protein triggers host pro‐survival genes to create an environment that favours virus infection and cell‐to‐cell spread by delaying the onset of PCD.
An alternative explanation is that severe ER stress caused by TMV‐p3 induces UPR‐related cell death. There have been reports of UPR‐stimulating chemicals, such as TU, causing prolonged ER stress, which leads to cell death in plants (Costa et al., 2008; Crosti et al., 2001; Watanabe and Lam, 2008b). However, a molecular mechanism of ER stress‐related death has not been elaborated in plants. In mammalian systems, severe stress can lead to IRE1‐dependent cell death, and there are two relevant pathways that can lead to such an outcome. IRE1 has endoribonuclease activity and can degrade membrane‐associated mRNAs through a process known as IRE1‐dependent decay (RIDD) (Tabas and Ron, 2011). In our investigation, we noted a role for bZIP60 which lies downstream of IRE1 (Iwata and Koizumi, 2012; Iwata et al., 2009), but future research should focus on the potential role of IRE1 in regulating plant virus infection. RIDD has not yet been explored in plants as a mechanism for RNA degradation, but evidence in Fig. 2 that TMV‐p3 mRNAs accumulate to a lower level than TMV in inoculated and systemic leaves could be explained by such a mechanism. Given that TGBp3 lies in the ER and appears to stimulate the expression of the IRE1 major downstream effector bZIP60 (Ye et al., 2011), it is also reasonable to consider that IRE1 RNAse activity might contribute to the reduced TMV‐p3 accumulation seen in inoculated and systemic leaves (Fig. 2B,C). It is possible that RIDD may help to defend cells against TGBp3‐induced ER stress by degrading the ER‐associated viral mRNAs. In mammalian cells, RIDD‐related mRNA decay is directly linked to apoptosis (Tabas and Ron, 2011). Given that plant cells lack an apoptotic mechanism for cell death, we can only speculate about the possibility of a RIDD‐linked PCD in plants.
The second link between IRE1 and cell death in mammalian cells involves the interaction of IRE1 with Bak and Bax, which are proteins involved in the mitochondrial pathway of apoptosis (Tabas and Ron, 2011). Bak and Bax mediate mitochondrial permeabilization and apoptosis, and are controlled by factors that are sequestered by Bcl‐2 to enhance cell survival. Although Bcl‐2‐ and Bax‐related machinery is not known to exist in plants, previous work from our laboratory has shown that the overexpression of Bcl‐2 family members in plants can enhance cell survival in response to certain pathogens, suggesting that component mechanisms of ER stress‐mediated cell death are conserved among plant and animal systems (Chae et al., 2003; Chen et al., 2003, 2004; Dickman et al., 2001; Khanna et al., 2007). Evidence that the overexpression of the Bcl‐2 family members Bcl‐xl and CED‐9 in plants inhibits oxidative stress‐induced PCD is consistent with transkingdom function, and suggests an overlap (conceptual and biochemical) in PCD pathways in plants and animals, even though the specific gene products modulating such responses differ (Chen et al., 2003; Dickman et al., 2001; Xu et al., 2004). The observation that CED‐9, Bcl‐xl and Op‐IAP overexpression had no impact on TGBp3‐induced cell death points to an alternative mechanism for ROS production and PCD, which is not regulated by these genes. This is also similar to the situation in mammalian PCD, where cell death regulators have distinct targets.
Another highly conserved protein that suppresses ER stress‐mediated cell death is BI‐1, which is a protein that mainly resides in the ER. In mammalian cells, BI‐1 down‐regulates Bax‐related cell death and, in Arabidopsis, BI‐1 has a role in down‐regulating ER stress‐related PCD (Chae et al., 2003; Watanabe and Lam, 2006, 2008b). It has been proposed recently that AtBI‐1 is a survival factor that is activated during ER stress, in parallel with the UPR‐related genes BiP and CRT, to enhance survival. However, under severe stress, where misfolded proteins accumulate, calcium released from the ER can activate ROS and cell death in a manner that is not preventable by AtBI‐1 (Watanabe and Lam, 2008a). Although we did not explore a role for BI‐1 in TMV‐p3 infection, further research could examine whether BI‐1 is a critical factor in TGBp3‐induced PCD.
Notably, PVX TGBp3 elicits HR‐like local lesions on TMV‐p3‐inoculated leaves. These local lesions exhibit the hallmarks of PCD, including DNA fragmentation, the oxidative burst and positive staining of dead cells with Evans blue dye. Interestingly, TMV‐p3, but not TMV‐p2, induces HR‐like PCD, even though both PVX TGBp2 and TGBp3 proteins reside in the ER and cause delayed TMV accumulation in protoplasts and plants. Based on the results of TMV and A. tumefaciens delivery, we conclude that TGBp3 itself is a specific elicitor of UPR‐related gene expression and host defences. It is worth noting that, during normal PVX infection, TGBp3 is expressed at low levels from a bicistronic subgenomic RNA and by translational read‐through of the upstream TGBp2 open reading frame (ORF) (Verchot et al., 1998). Either the low levels of TGBp3 produced during PVX infection prevent or limit the induction of HR‐like defences, or TGBp2 may act as a guard molecule that prevents TGBp3 from binding a cellular receptor that triggers host defence responses. Further experiments are needed to explore the functions of TGBp2 and TGBp3 interactions.
We reported previously that TGBp3 induces SKP1 expression during PVX infection and following the agrodelivery of 35S:TGBp3‐containing plasmids to N. benthamiana plants (Ye et al., 2011). Interestingly, NbSKP1 suppression resulted in fewer TMV‐p3‐related local lesions on inoculated leaves and the loss of systemic HR. This is reminiscent of the studies of Liu et al. (2002, 2004) and Azevedo et al. (2006), which demonstrated that suppression of SKP1 compromised N and Rx gene‐mediated resistance to TMV and PVX, and enabled the virus to spread to the upper leaves.
SKP1 is a core subunit of SCF (SKP1/Cullin1/F‐box protein) E3 ubiquitin ligase which directs the ubiquitination of targets as a signal for 26S proteasome degradation. SKP1 is an adapter that links CUL1 and the F‐box protein (Cheng et al., 2011; Smalle and Vierstra, 2004). With regard to ERAD, the cellular mechanism for the elimination of misfolded proteins from the ER involves destruction by the cytoplasmic ubiquitin–proteasome system. Hrd1p/Der3p provides the essential ubiquitin ligase activity for proteolytic breakdown of misfolded proteins in mammalian and yeast cells, and has been identified recently in plant cells (Meusser et al., 2005; Su et al., 2011; Zhang and Kaufman, 2006). Hrd1/Der3p is located in the ER and acts to ubiquitinate substrates to enable their dislocation and subsequent degradation. SKP1 is not reported to be a component of the Hrd1p/Der3p complex and therefore its link to TGBp3‐induced ER stress is intriguing. We have shown previously that silencing of bZIP60 can suppress SKP1 expression, which suggests a link between bZIP60‐related UPR and the SCF complex (Ye et al., 2011). Importantly, silencing of bZIP60 failed to eliminate SKP1 expression, which could indicate that SKP1 expression is controlled by other factors that might be induced by TGBp3. It is also possible that TGBp3 functions to dislodge other host proteins for degradation in a manner that is independent of the Hrd1/Der3p complex, but involves cytoplasmic ubiquitination. Such a mechanism has been reported involving the HIV Vpu protein (Meusser et al., 2005; Nomaguchi et al., 2008; Schubert et al., 1998). Vpu is similar to TGBp3 in that it is a low‐molecular‐weight protein with a single transmembrane domain and resides in the ER. Vpu triggers the degradation of the host CD4 protein, which is otherwise a stable protein that has a single membrane spanning domain. Vpu binds CD4 and has a cytoplasmic tail which, on phosphorylation, mimics substrates of the SCFβTrCP E3 complex. Vpu attracts the SCFβTrCP E3 complex to ubiquitinate the CD4 receptor protein, which is then degraded by the 26S proteasome (Meusser et al., 2005; Schubert et al., 1998). CD4 is a cell surface receptor that is degraded in the ER by HIV‐1 Vpu, and whose degradation is vital for virion release. It is possible to consider that TGBp3, which is an essential factor in virus egress, has analogous functions to Vpu and targets host substrate proteins to the SCF complex for ubiquitination and degradation. However, further research is needed to identify host factors that might interact with TGBp3, and whether TGBp3 participates in such a mechanism to ensure virus spread by the suppression of host proteins (Malim and Emerman, 2008; Nomaguchi et al., 2008).
Several plant viruses have mechanisms to subvert the ubiquitination and proteasome degradation machinery via interactions with SKP1. The polerovirus Beet western yellows virus (BWYV) P0 protein has an F‐box protein motif and binds SKP1 (Pazhouhandeh et al., 2006). In yeast, the BWYV P0 protein has been shown to bind ASK1 and ASK2, which are the yeast homologues of SKP1. Mutations that interrupt the ability of P0 to interact with SKP1 result in systemic necrosis, suggesting that the P0–SKP1 complex modulates PCD during virus infection. P0 is also a silencing suppressor protein and has been suggested to present target components of the silencing machinery for proteasomal degradation through its interactions with SKP1 (Pazhouhandeh et al., 2006). Faba bean necrotic yellows virus (FBNYV) is a single‐strand DNA virus and encodes a protein named ‘Clink’, which is an F‐box‐like protein that binds both SKP1 and pRB (Aronson et al., 2000). pRB is essential for viral DNA replication and is involved in cell cycle progression. Clink might target pRB for ubiquitin‐mediated degradation, but it is also possible that Clink presents another substrate or is itself a target (Aronson et al., 2000). The benyvirus Beet necrotic yellow vein virus (BNYVV) P25 protein interacts with an F‐box protein in Arabidopsis (Thiel et al., 2012). There is evidence in yeast that the BNYVV P25 protein is a competitor of ASK–F‐box protein binding. However, there are insufficient data to date to determine whether it functions in plants to present a host protein substrate for degradation or to disrupt certain SCF complexes. Thus, viral proteins commonly interact with components of the SCF complex, but it is not known whether such interactions are designed to target certain cellular defences for degradation or to compromise immune responses that depend on the SCF complex. Thus, it is of interest that PVX TGBp3 enhances SKP1 and that silencing of SKP1 compromises PCD. Although there are no data to date to indicate a PVX protein that binds directly to components of the SCF complex, these data, combined with the BWYV, FBNYV and BNYVV examples, suggest that there might be a common viral strategy to manipulate or alter the ubiquitin‐mediated degradation machinery to promote plant virus infection. Moreover, each of these viral protein interactions with components of the SCF complex is linked to an HR‐like cell death response (Aronson et al., 2000; Pazhouhandeh et al., 2006; Thiel et al., 2012). In this investigation, silencing of NbSKP1 abolishes PCD during TMV‐p3 infection. These data suggest that TGBp3‐induced changes in NbSKP1 gene expression are also linked to the regulation of a resistance response. In conclusion, these analyses provide a strong basis for further studies to examine the role of the SCF complex in PVX disease and resistance.
Experimental Procedures
Plasmids and bacterial strains
PVX TGBp2, TGBp3 and TGBp3Dm1 sequences were PCR amplified using primers containing added PacI and XhoI restriction sites, and then inserted into the TMV 30B infectious clone (Shivprasad et al., 1999). Agrobacterium tumefaciens LBA4404 containing pBI121‐BLP‐4 or pBI121‐TGBp3His was prepared. The NbBLP‐4 (Accession No. X60057) or TGBp3His coding sequence was inserted between the XbaI and SacI restriction sites of pBI121 plasmids (Jefferson et al., 1987; Leborgne‐Castel et al., 1999; Ye et al., 2011). Agrobacterium GV2260 containing TRV1 plus TRV2‐NbSKP1 or TRV2‐GFP was kindly provided by Dr S. Dinesh‐Kumar (Yale University, New Haven, CT, USA) (Bhattarai et al., 2007; Liu et al., 2004).
Plant materials, virus inoculation and transient gene expression assays
N. benthamiana, N. tabacum cv. Samsun NN and N. tabacum cv. Samsun nn were used. Transgenic N. benthamiana line BN3 was obtained from Dr B. Baker (University of California, Berkeley, CA, USA) and contains the N gene for TMV resistance. N. tabacum transgenic lines expressing IAP, Bcl‐xl and CED‐9 were prepared using N. tabacum cv. Glurk NN.
BY‐2 protoplasts were inoculated with 10 μg of infectious transcripts synthesized using the RiboMAX Large Scale RNA Production System (Promega Corp., Madison, WI, USA) (Ju et al., 2005). The preparation and transfection of BY‐2 protoplasts were performed as reported previously (Ju et al., 2008). Plants were inoculated with 30 μg/mL purified virus suspended in 0.01 m phosphate‐buffered saline (PBS) (pH 7.0) (Shadwick and Doran, 2007). Agroinfiltration to deliver viruses or for transient gene expression in N. benthamiana leaves was performed according to published protocols (Liu et al., 2002).
H 2 DCFDA or TUNEL staining of leaf segments
Total protein was extracted from 200‐mg leaf samples taken at 1, 3 and 5 dpi using 10 mm Tris‐HCl (pH 7.2) and quantified using the Bradford reagent (Sigma‐Aldrich, St. Louis, MO, USA). ROS activity was visualized using 50 μm H2DCFDA (Mahalingam et al., 2006). Leaf extracts were incubated with 10 μm H2DCFDA in the dark for 10 min at room temperature and fluorescence was measured using a Versa Fluor fluorometer (excitation at 460–500 nm, and emission at 510–560 nm) (Bio‐Rad Laboratories, Hercules, CA, USA). TUNEL staining of leaf segments was conducted using an In Situ Cell Death Detection Kit, Fluorescein (Boehringer Mannheim GmbH, Indianapolis, IN, USA). Leaf segments were mounted on polylysine slides, permeabilized with proteinase K for 15 min, rinsed with PBS, treated with TUNEL reaction mixture and rinsed again with PBS. Samples were treated with propidium iodide for 30 min and rinsed with PBS.
Leaf treatments with TU or FC
TMV‐p3‐inoculated N. benthamiana leaves were infiltrated with 20 μm FC or 10 μg/mL TU (Koizumi et al., 1999; Liu et al., 2002; Malerba et al., 2004, 2008) prepared in Murashige and Skoog (MS) medium. Three 1.2‐cm‐diameter leaf discs were taken at 5 dpi (Il‐Pyung, 2007), incubated in 0.25% (w/v in double‐distilled H2O) Evans blue stain for 30 min and rinsed with double‐distilled H2O. Stain was extracted using 3 mL of a solution of 50% (v/v) methanol and 1% (w/v) sodium dodecylsulphate (SDS) for 60 min at 50 °C. The intensity of staining was measured with a Wallac 1420 Multilabel Counter spectrophotometer (Perkin‐Elmer, Waltham, MA, USA) at 595 nm.
Northern and immunoblot analyses
Total RNA was extracted from systemic leaves at 3 and 5 dpi or from BY‐2 protoplasts at 24 and 48 hpi using TRIzol Reagent (Invitrogen, Grand Island, NY, USA) and loaded onto 1.2% denaturing formaldehyde gel. Northern analysis was conducted using a North‐2‐South Chemiluminescent Hybridization and Detection Kit (Pierce Biotechnology, Rockford, IL, USA). cDNA probes were prepared using the North‐2‐South Biotin Random Prime Labeling Kit and PCR amplification of the TMV CP gene.
Immunoblot analysis was carried out using an ECL Advanced Western Blotting Kit (GE Healthcare, Piscataway, NJ, USA). Total protein was extracted from agroinfiltrated leaves by grinding samples with extraction buffer [100 mm Tris‐HCl, pH 7.5, 10 mm KCl, 0.4 m sucrose, 10% glycerol, 10 μm phenylmethylsulphonylfluoride (PMSF)] and quantified using the Bradford reagent (Sigma‐Aldrich). Thirty micrograms of protein for each sample were loaded onto a 10% SDS polyacrylamide gel, electroblotted to Hybond P membrane and probed with GRP78 antibody (Affinity BioReagents, Golden, CO, USA). Densitometric analysis was performed by Alpha Ease FC software (Alpha Innotech, San Leandro, CA, USA).
qRT‐PCR analysis of virus‐infected leaves and silenced plants
Nicotiana benthamiana leaves were inoculated with TMV, TMV‐p3, TMV‐p3Dm1 and PVX. RNA was extracted from samples collected at 0, 2, 4 and 8 hpi with an SV Total RNA Isolation Kit (Promega Corp.). The first‐strand cDNA was synthesized by Superscript reverse transcriptase III (Invitrogen) using hexamer random primers. qPCR was carried out using 25‐μL reactions and 100–900 nm primers designed using the coding sequences for known genes (Ye et al., 2011). qRT‐PCRs of NbSGT1 and NbSKP1 primers were designed to anneal outside the target sequence of VIGS (Liu et al., 2002). Twenty‐five nanograms of cDNA were used to perform qPCR employing the Power SYBR Green II Master Mix and ABI 7500 PCR machine (Applied Biosystems, Foster City, CA, USA). Reactions involved 40 cycles of 95 °C for 15 s and 60 °C for 60 s. The comparative C T method was employed for relative quantification of gene expression. Validation reactions were conducted to determine the efficiencies of the target and control amplifications using 0.01, 0.1, 1, 10 and 100 ng of template cDNA. The comparative C T method employs the formula where the values of the endogenous control (18S RNA) and calibrator (constant quantity of healthy sample template) are subtracted from the target sample value to provide the ΔΔC T value. The value of represents the fold change in RNA accumulation.
Statistical analysis
All statistical analyses were conducted with PC SAS Version 9 (SAS Institute, Cary, NC, USA). The response variable was transformed using a natural logarithm function prior to the analysis as a remedy for heterogeneous variances. An analysis of variance procedure (PROC MIXED) assuming a two‐factor (sample and time) factorial arrangement in a randomized complete block design was conducted. The simple effects of sample given time and time given sample were analysed with a SLICE option in an LSMEANS statement. If significance of the simple effects was detected, the means were compared using pair‐wise t‐tests. A significance level of 0.05 was used to judge statistical significance. A single‐tailed Student's t‐test with two‐sample variances was used to compare virus‐ and buffer‐inoculated samples following each chemical treatment.
Supporting information
Fig. S1 (A) qRT‐PCR analyzing bZIP60, BiP and SKP1 transcript abundance. The plots show the relative abundance of transcripts due to each TMV, PVX, and TMV plus PVX infection. Clearly transcript abundance is higher during PVX infection. Furthermore, TMV has a dominant suppressive effect on the ability of PVX to induce host gene expression. Furthermore, we fused the PVX replicase, TGBp1, and CP to the CaMV35S promoter and delivered these plasmids to plants using agro‐infiltration. qRT‐PCR shows host gene expression is not induced by these factors. Immunoblots demonstrating the relative abundance of TGBp1 (B), PVX CP (D), and commassie stained gel detecting TGBp2 (C) when expressed from either PVX or TMV vector in infected N. benthamiana leaves. Since we lack antisera to TGBp2, commassie staining was used to identify a band that likely corresponds to TGBp2 in infected leaf extracts. Commassie stained immunoblot membranes in panels B and D show similar levels of protein in each lane.
Fig. S2 Immunoblot analysis detecting PVX TGBp3‐His protein using antisera detecting the His tag. (A) Leaf extracts from mock inoculated (M; lane 1), PVX‐TGBp3His (lanes 2–5), TMV‐p3His (lanes 6–9), and agrobacterium (lane 11) expressing TGBp3His from CaMV 35S promoter (lanes 12–13). Leaf extracts were taken from virus‐inoculated (Inoc) and systemically (Syst) infected tissues. TMV‐p3His showed higher levels of protein accumulation than PVX‐TGBp3His and required 20 s exposure to film to see abundance of protein in the top panel. To visualize TGBp3His expressed from PVX, we exposed the film for 1 min and this is shown in the middle panel. TGBp3His expression from Agrobacterium was comparable to TMV‐p3His as seen after 20 s exposure to film. Below is a commassie stained PVDF membrane photographed after electrophoretic transfer to show protein levels in each lane were comparable. (B) Immunoblot detects TMV coat protein. TMV‐p3 and TMV‐p3His were detected in systemic tissues and produced comparable levels of TMV coat protein indicating that the His tag is not deleterious to virus infection.
Acknowledgements
We thank Drs K. Mysore and S. R. Uppalapati (Samuel Roberts Noble Foundation, Ardmore, OK, USA) for technical assistance. This research was funded by the US Department of Agriculture National Research Initiative (USDA NRI) Project Number: OKL02649. There are no conflicts of interest or commercial affiliations for any authors.
References
- Aronson, M.N. , Meyer, A.D. , Gyorgyey, J. , Katul, L. , Vetten, H.J. , Gronenborn, B. and Timchenko, T. (2000) Clink, a nanovirus‐encoded protein, binds both pRB and SKP1. J. Virol. 74, 2967–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo, C. , Betsuyaku, S. , Peart, J. , Takahashi, A. , Noel, L. , Sadanandom, A. , Casais, C. , Parker, J. and Shirasu, K. (2006) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 25, 2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai, K.K. , Li, Q. , Liu, Y. , Dinesh‐Kumar, S.P. and Kaloshian, I. (2007) The MI‐1‐mediated pest resistance requires Hsp90 and Sgt1. Plant Physiol. 144, 312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette, J.E. , Stuiver, M. , Van Lent, J. , Wellink, J. and Van Kammen, A. (2000) Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J. Virol. 74, 6556–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette, J.E. , van Lent, J. , MacFarlane, S.A. , Wellink, J. and van Kammen, A. (2002) Cowpea mosaic virus 32‐ and 60‐kilodalton replication proteins target and change the morphology of endoplasmic reticulum membranes. J. Virol. 76, 6293–6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, H.J. , Ke, N. , Kim, H.R. , Chen, S. , Godzik, A. , Dickman, M. and Reed, J.C. (2003) Evolutionarily conserved cytoprotection provided by Bax Inhibitor‐1 homologs from animals, plants, and yeast. Gene, 323, 101–113. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Vaghchhipawala, Z. , Li, W. , Asard, H. and Dickman, M.B. (2004) Tomato phospholipid hydroperoxide glutathione peroxidase inhibits cell death induced by Bax and oxidative stresses in yeast and plants. Plant Physiol. 135, 1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S.R. , Dunigan, D.D. and Dickman, M.B. (2003) Bcl‐2 family members inhibit oxidative stress‐induced programmed cell death in Saccharomyces cerevisiae . Free Radic. Biol. Med. 34, 1315–1325. [DOI] [PubMed] [Google Scholar]
- Cheng, Y.T. , Li, Y. , Huang, S. , Huang, Y. , Dong, X. , Zhang, Y. and Li, X. (2011) Stability of plant immune‐receptor resistance proteins is controlled by SKP1‐CULLIN1‐F‐box (SCF)‐mediated protein degradation. Proc. Natl. Acad. Sci. USA, 108, 14 694–14 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M.D. , Reis, P.A. , Valente, M.A. , Irsigler, A.S. , Carvalho, C.M. , Loureiro, M.E. , Aragão, F.J. , Boston, R.S. , Fietto, L.G. and Fontes, E.P. (2008) A new branch of endoplasmic reticulum stress signaling and the osmotic signal converge on plant‐specific asparagine‐rich proteins to promote cell death. J. Biol. Chem. 283, 20 209–20 219. [DOI] [PubMed] [Google Scholar]
- Crosti, P. , Malerba, M. and Bianchetti, R. (2001) Tunicamycin and brefeldin A induce in plant cells a programmed cell death showing apoptotic features. Protoplasma, 216, 31–38. [DOI] [PubMed] [Google Scholar]
- Denecke, J. , Goldman, M.H. , Demolder, J. , Seurinck, J. and Botterman, J. (1991) The tobacco luminal binding protein is encoded by a multigene family. Plant Cell, 3, 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke, J. , Carlsson, L.E. , Vidal, S. , Hoglund, A.S. , van Ek, B., Zeijl, M.J. , Sinjorgo, K.M. and Palva, E.T. (1995) The tobacco homolog of mammalian calreticulin is present in protein complexes in vivo. Plant Cell, 7, 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Humbert, S. , Liu, J.X. , Srivastava, R. , Rothstein, S.J. and Howell, S.H. (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA, 108, 7247–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman, M.B. , Park, Y.K. , Oltersdorf, T. , Li, W. , Clemente, T. and French, R. (2001) Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc. Natl. Acad. Sci. USA, 98, 6957–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard, L. and Helenius, A. (2003) Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–191. [DOI] [PubMed] [Google Scholar]
- Erickson, F.L. , Dinesh‐Kumar, S.P. , Holzberg, S. , Ustach, C.V. , Dutton, M. , Handley, V. , Corr, C. and Baker, B.J. (1999) Interactions between Tobacco mosaic virus and the tobacco N gene. Philos. Trans. R. Soc. London: B. Biol. Sci. 354, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller, A. , Liechti, R. and Farmer, E.E. (2010) Arabidopsis jasmonate signaling pathway. Sci. Signal 3, cm4. [DOI] [PubMed] [Google Scholar]
- Il‐Pyung, A. (2007) Disturbance of the Ca2+/calmodulin‐dependent signalling pathway is responsible for the resistance of Arabidopsis dnd1 against Pectobacterium carotovorum infection. Mol. Plant Pathol. 8, 747–759. [DOI] [PubMed] [Google Scholar]
- Iwata, Y. and Koizumi, N. (2005a) An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA, 102, 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, Y. and Koizumi, N. (2005b) Unfolded protein response followed by induction of cell death in cultured tobacco cells treated with tunicamycin. Planta, 220, 804–807. [DOI] [PubMed] [Google Scholar]
- Iwata, Y. and Koizumi, N. (2012) Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci. (Available at 10.1016/j.tplants.2012.06.014). [DOI] [PubMed] [Google Scholar]
- Iwata, Y. , Yoneda, M. , Yanagawa, Y. and Koizumi, N. (2009) Characteristics of the nuclear form of the Arabidopsis transcription factor AtbZIP60 during the endoplasmic reticulum stress response. Biosci. Biotechnol. Biochem. 73, 865–869. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. , Kavanagh, T.A. and Bevan, M.W. (1987) GUS fusions: beta‐glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelitto‐Van Dooren, E.P. , Vidal, S. and Denecke, J. (1999) Anticipating endoplasmic reticulum stress. A novel early response before pathogenesis‐related gene induction. Plant Cell, 11, 1935–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, H.J. , Samuels, T.D. , Wang, Y.S. , Blancaflor, E. , Payton, M. , Mitra, R. , Krishnamurthy, K. , Nelson, R.S. and Verchot‐Lubicz, J. (2005) The potato virus X TGBp2 movement protein associates with endoplasmic reticulum‐derived vesicles during virus infection. Plant Physiol. 138, 1877–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, H.J. , Ye, C.M. and Verchot‐Lubicz, J. (2008) Mutational analysis of Potato virus X TGBp3 links subcellular accumulation to protein turnover during virus infection. Virology, 375, 103–117. [DOI] [PubMed] [Google Scholar]
- Kadota, Y. , Amigues, B. , Ducassou, L. , Madaoui, H. , Ochsenbein, F. , Guerois, R. and Shirasu, K. (2008) Structural and functional analysis of SGT1‐HSP90 core complex required for innate immunity in plants. EMBO Rep. 9, 1209–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamauchi, S. , Nakatani, H. , Nakano, C. and Urade, R. (2005) Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana . FEBS J. 272, 3461–3476. [DOI] [PubMed] [Google Scholar]
- Khanna, H.K. , Paul, J.Y. , Harding, R.M. , Dickman, M.B. and Dale, J.L. (2007) Inhibition of Agrobacterium‐induced cell death by antiapoptotic gene expression leads to very high transformation efficiency of banana. Mol. Plant–Microbe Interact. 20, 1048–1054. [DOI] [PubMed] [Google Scholar]
- Kim, K.S. , Min, J.Y. and Dickman, M.B. (2008) Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant–Microbe Interact. 21, 605–612. [DOI] [PubMed] [Google Scholar]
- Koizumi, N. , Ujino, T. , Sano, H. and Chrispeels, M.J. (1999) Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine‐linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin‐induced unfolded protein response. Plant Physiol. 121, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E. (2005) Vacuolar proteases livening up programmed cell death. Trends Cell Biol. 15, 124–127. [DOI] [PubMed] [Google Scholar]
- Leborgne‐Castel, N. , Jelitto‐Van Dooren, E.P. , Crofts, A.J. and Denecke, J. (1999) Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell, 11, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.M. and Ahlquist, P. (2003) Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive‐strand RNA virus RNA replication protein. J. Virol. 77, 12 819–12 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo, J.A. (2007a) High‐efficiency protein expression in plants from agroinfection‐compatible Tobacco mosaic virus expression vectors. BMC Biotechnol. 7, 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo, J.A. (2007b) TRBO: a high‐efficiency tobacco mosaic virus RNA‐based overexpression vector. Plant Physiol. 145, 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.X. , Srivastava, R. , Che, P. and Howell, S.H. (2007) An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane‐associated transcription factor, bZIP28. Plant Cell, 19, 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Serino, G. , Deng, X.W. and Dinesh‐Kumar, S.P. (2002) Role of SCF ubiquitin‐ligase and the COP9 signalosome in the N gene‐mediated resistance response to Tobacco mosaic virus . Plant Cell, 14, 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Burch‐Smith, T. , Schiff, M. , Feng, S. and Dinesh‐Kumar, S.P. (2004) Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 279, 2101–2108. [DOI] [PubMed] [Google Scholar]
- Liu, Z. and Kearney, C.M. (2010) A tobamovirus expression vector for agroinfection of legumes and Nicotiana . J. Biotechnol. 147, 151–159. [DOI] [PubMed] [Google Scholar]
- Lu, D.P. and Christopher, D.A. (2008) Endoplasmic reticulum stress activates the expression of a sub‐group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana . Mol. Genet. Genomics, 280, 199–210. [DOI] [PubMed] [Google Scholar]
- Mahalingam, R. , Jambunathan, N. , Gunjan, S.K. , Faustin, E. , Weng, H. and Ayoubi, P. (2006) Analysis of oxidative signalling induced by ozone in Arabidopsis thaliana . Plant Cell Environ. 29, 1357–1371. [DOI] [PubMed] [Google Scholar]
- Malerba, M. , Cerana, R. and Crosti, P. (2004) Comparison between the effects of fusicoccin, tunicamycin, and brefeldin A on programmed cell death of cultured sycamore (Acer pseudoplatanus L.) cells. Protoplasma, 224, 61–70. [DOI] [PubMed] [Google Scholar]
- Malerba, M. , Contran, N. , Tonelli, M. , Crosti, P. and Cerana, R. (2008) Role of nitric oxide in actin depolymerization and programmed cell death induced by fusicoccin in sycamore (Acer pseudoplatanus) cultured cells. Physiol. Plant. 133, 449–457. [DOI] [PubMed] [Google Scholar]
- Malim, M.H. and Emerman, M. (2008) HIV‐1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe, 3, 388–398. [DOI] [PubMed] [Google Scholar]
- Martinez, I.M. and Chrispeels, M.J. (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell, 15, 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser, B. , Hirsch, C. , Jarosch, E. and Sommer, T. (2005) ERAD: the long road to destruction. Nat. Cell Biol. 7, 766–772. [DOI] [PubMed] [Google Scholar]
- Muller, J. , Piffanelli, P. , Devoto, A. , Miklis, M. , Elliott, C. , Ortmann, B. , Schulze‐Lefert, P. and Panstruga, R. (2005) Conserved ERAD‐like quality control of a plant polytopic membrane protein. Plant Cell, 17, 149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navazio, L. , Mariani, P. and Sanders, D. (2001) Mobilization of Ca2+ by cyclic ADP‐ribose from the endoplasmic reticulum of cauliflower florets. Plant Physiol. 125, 2129–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netherton, C. , Moffat, K. , Brooks, E. and Wileman, T. (2007) A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 70, 101–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomaguchi, M. , Fujita, M. and Adachi, A. (2008) Role of HIV‐1 Vpu protein for virus spread and pathogenesis. Microbes Infect. 10, 960–967. [DOI] [PubMed] [Google Scholar]
- Oh, D.H. , Kwon, C.S. , Sano, H. , Chung, W.I. and Koizumi, N. (2003) Conservation between animals and plants of the cis‐acting element involved in the unfolded protein response. Biochem. Biophys. Res. Commun. 301, 225–230. [DOI] [PubMed] [Google Scholar]
- Parmar, V.M. and Schroder, M. (2012) Sensing endoplasmic reticulum stress. Adv. Exp. Med. Biol. 738, 153–168. [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh, M. , Dieterle, M. , Marrocco, K. , Lechner, E. , Berry, B. , Brault, V. , Hemmer, O. , Kretsch, T. , Richards, K.E. , Genschik, P. and Ziegler‐Graff, V. (2006) F‐box‐like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA, 103, 1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike, S.M. , Zhang, X.C. and Gassmann, W. (2005) Electrophysiological characterization of the Arabidopsis avrRpt2‐specific hypersensitive response in the absence of other bacterial signals. Plant Physiol. 138, 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang, X. , Zechmann, B. , Reitz, M.U. , Kogel, K.H. and Schafer, P. (2012) The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an endoplasmic reticulum stress‐triggered caspase‐dependent cell death. Plant Cell, 24, 794–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, C. , Pan, J. , Peng, W. , Genschik, P. , Hobbie, L. , Hellmann, H. , Estelle, M. , Gao, B. , Peng, J. , Sun, C. and Xie, D. (2005) Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 42, 514–524. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler, C. , Laporte, C. , Gaire, F. , Dunoyer, P. , Schmitt, C. , Duval, S. , Piéquet, A. , Loudes, A.M. , Rohfritsch, O. , Stussi‐Garaud, C. and Pfeiffer, P. (2002) Grapevine fanleaf virus replication occurs on endoplasmic reticulum‐derived membranes. J. Virol. 76, 8808–8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, M.R. and Bowles, D.J. (1999) Fusicoccin, 14‐3‐3 proteins, and defense responses in tomato plants. Plant Physiol. 119, 1243–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, M.C. , Jensen, P.E. and Carrington, J.C. (1997) Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum‐targeted viral protein. EMBO J. 16, 4049–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott, A. , Ravaud, S. , Keller, S. , Radzimanowski, J. , Viotti, C. , Hillmer, S. , Sinning, I. and Strahl, S. (2010) Arabidopsis stromal‐derived Factor2 (SDF2) is a crucial target of the unfolded protein response in the endoplasmic reticulum. J. Biol. Chem. 285, 18 113–18 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, U. , Anton, L.C. , Bacik, I. , Cox, J.H. , Bour, S. , Bennink, J.R. , Orlowski, M. , Strebel, K. and Yewdell, J.W. (1998) CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin‐conjugating pathway. J. Virol. 72, 2280–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, P.J. , Kim, S.G. and Park, C.M. (2008) Membrane‐bound transcription factors in plants. Trends Plant Sci. 13, 550–556. [DOI] [PubMed] [Google Scholar]
- Shadwick, F.S. and Doran, P.M. (2007) Propagation of plant viruses in hairy root cultures: a potential method for in vitro production of epitope vaccines and foreign proteins. Biotechnol. Bioeng. 96, 570–583. [DOI] [PubMed] [Google Scholar]
- Sharma, M. , Sasvari, Z. and Nagy, P.D. (2011) Inhibition of phospholipid biosynthesis decreases the activity of the tombusvirus replicase and alters the subcellular localization of replication proteins. Virology, 415, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivprasad, S. , Pogue, G.P. , Lewandowski, D.J. , Hidalgo, J. , Donson, J. , Grill, L.K. and Dawson, W.O. (1999) Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus‐based vectors. Virology, 255, 312–323. [DOI] [PubMed] [Google Scholar]
- Slepak, T.I. , Tang, M. , Slepak, V.Z. and Lai, K. (2007) Involvement of endoplasmic reticulum stress in a novel Classic Galactosemia model. Mol. Genet. Metab. 92, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J. and Vierstra, R.D. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590. [DOI] [PubMed] [Google Scholar]
- Stuttmann, J. , Lechner, E. , Guerois, R. , Parker, J.E. , Nussaume, L. , Genschik, P. and Noël, L.D. (2009) COP9 signalosome‐ and 26S proteasome‐dependent regulation of SCFTIR1 accumulation in Arabidopsis. J. Biol. Chem. 284, 7920–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, W. , Liu, Y. , Xia, Y. , Hong, Z. and Li, J. (2011) Conserved endoplasmic reticulum‐associated degradation system to eliminate mutated receptor‐like kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA, 108, 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas, I. and Ron, D. (2011) Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif, K.D. , Mori, K. , Kaufman, R.J. and Siddiqui, A. (2004) Hepatitis C virus suppresses the IRE1‐XBP1 pathway of the unfolded protein response. J. Biol. Chem. 279, 17 158–17 164. [DOI] [PubMed] [Google Scholar]
- Tateda, C. , Ozaki, R. , Onodera, Y. , Takahashi, Y. , Yamaguchi, K. , Berberich, T. , Koizumi, N. and Kusano, T. (2008) NtbZIP60, an endoplasmic reticulum‐localized transcription factor, plays a role in the defense response against bacterial pathogens in Nicotiana tabacum . J. Plant Res. 121, 603–611. [DOI] [PubMed] [Google Scholar]
- Thiel, H. , Hleibieh, K. , Gilmer, D. and Varrelmann, M. (2012) The P25 pathogenicity factor of Beet necrotic yellow vein virus targets the sugar beet 26S proteasome involved in the induction of a hypersensitive resistance response via interaction with an F‐box protein. Mol. Plant–Microbe Interact. 25, 1058–1072. [DOI] [PubMed] [Google Scholar]
- Turner, K.A. , Sit, T.L. , Callaway, A.S. , Allen, N.S. and Lommel, S.A. (2004) Red clover necrotic mosaic virus replication proteins accumulate at the endoplasmic reticulum. Virology, 320, 276–290. [DOI] [PubMed] [Google Scholar]
- Urade, R. (2007) Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 274, 1152–1171. [DOI] [PubMed] [Google Scholar]
- Verchot, J. (2011) Wrapping membranes around plant virus infection. Curr. Opin. Virol. 1, 388–395. [DOI] [PubMed] [Google Scholar]
- Verchot, J. , Angell, S.M. and Baulcombe, D.C. (1998) In vivo translation of the triple gene block of potato virus X requires two subgenomic mRNAs. J. Virol. 72, 8316–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, N. and Lam, E. (2006) Arabidopsis Bax inhibitor‐1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 45, 884–894. [DOI] [PubMed] [Google Scholar]
- Watanabe, N. and Lam, E. (2008a) Arabidopsis Bax inhibitor‐1: a rheostat for ER stress‐induced programmed cell death. Plant Signal Behav. 3, 564–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, N. and Lam, E. (2008b) BAX inhibitor‐1 modulates endoplasmic reticulum stress‐mediated programmed cell death in Arabidopsis. J. Biol. Chem. 283, 3200–3210. [DOI] [PubMed] [Google Scholar]
- Williams, B.L. and Lipkin, W.I. (2006) Endoplasmic reticulum stress and neurodegeneration in rats neonatally infected with borna disease virus. J. Virol. 80, 8613–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. , Bailly‐Maitre, B. and Reed, J.C. (2005) Endoplasmic reticulum stress: cell life and death decisions. J. Clin. Invest. 115, 2656–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Liu, F. , Lechner, E. , Genschik, P. , Crosby, W.L. , Ma, H. , Peng, W. , Huang, D. and Xie, D. (2002) The SCF(COI1) ubiquitin‐ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell, 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P. , Rogers, S.J. and Roossinck, M.J. (2004) Expression of antiapoptotic genes bcl‐xL and ced‐9 in tomato enhances tolerance to viral‐induced necrosis and abiotic stress. Proc. Natl. Acad. Sci. USA, 101, 15 805–15 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, C. and Verchot, J. (2011) Role of unfolded protein response in plant virus infection. Plant Signal Behav. 6, 1212–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, C. , Dickman, M.B. , Whitham, S.A. , Payton, M. and Verchot, J. (2011) The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 156, 741–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, C.M. , Kelly, V. , Payton, M. , Dickman, M.B. and Verchot, J. (2012) SGT1 is induced by the Potato virus X TGBp3 and enhances virus accumulation in Nicotiana benthamiana . Mol. Plant, 5, 1151–1153. [DOI] [PubMed] [Google Scholar]
- Zhang, K. and Kaufman, R.J. (2006) The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology, 66, S102–S109. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Miller, N.D. , Lewis, D.R. , Christians, M.J. , Lee, K.H. , Muday, G.K. , Spalding, E.P. and Vierstra, R.D. (2011) AUXIN UP‐REGULATED F‐BOX PROTEIN1 regulates the cross talk between auxin transport and cytokinin signaling during plant root growth. Plant Physiol. 156, 1878–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 (A) qRT‐PCR analyzing bZIP60, BiP and SKP1 transcript abundance. The plots show the relative abundance of transcripts due to each TMV, PVX, and TMV plus PVX infection. Clearly transcript abundance is higher during PVX infection. Furthermore, TMV has a dominant suppressive effect on the ability of PVX to induce host gene expression. Furthermore, we fused the PVX replicase, TGBp1, and CP to the CaMV35S promoter and delivered these plasmids to plants using agro‐infiltration. qRT‐PCR shows host gene expression is not induced by these factors. Immunoblots demonstrating the relative abundance of TGBp1 (B), PVX CP (D), and commassie stained gel detecting TGBp2 (C) when expressed from either PVX or TMV vector in infected N. benthamiana leaves. Since we lack antisera to TGBp2, commassie staining was used to identify a band that likely corresponds to TGBp2 in infected leaf extracts. Commassie stained immunoblot membranes in panels B and D show similar levels of protein in each lane.
Fig. S2 Immunoblot analysis detecting PVX TGBp3‐His protein using antisera detecting the His tag. (A) Leaf extracts from mock inoculated (M; lane 1), PVX‐TGBp3His (lanes 2–5), TMV‐p3His (lanes 6–9), and agrobacterium (lane 11) expressing TGBp3His from CaMV 35S promoter (lanes 12–13). Leaf extracts were taken from virus‐inoculated (Inoc) and systemically (Syst) infected tissues. TMV‐p3His showed higher levels of protein accumulation than PVX‐TGBp3His and required 20 s exposure to film to see abundance of protein in the top panel. To visualize TGBp3His expressed from PVX, we exposed the film for 1 min and this is shown in the middle panel. TGBp3His expression from Agrobacterium was comparable to TMV‐p3His as seen after 20 s exposure to film. Below is a commassie stained PVDF membrane photographed after electrophoretic transfer to show protein levels in each lane were comparable. (B) Immunoblot detects TMV coat protein. TMV‐p3 and TMV‐p3His were detected in systemic tissues and produced comparable levels of TMV coat protein indicating that the His tag is not deleterious to virus infection.


