Summary
Xanthomonas oryzae pv. oryzicola (Xoc) causes bacterial leaf streak in rice, which is a destructive disease worldwide. Xoc virulence factors are regulated by diffusible signal factor (DSF) and the global regulator Clp. In this study, we have demonstrated that asnB (XOC_3054), encoding an asparagine synthetase, is a novel virulence‐related gene regulated by both DSF and Clp in Xoc. A sequence analysis revealed that AsnB is highly conserved in Xanthomonas. An asnB mutation in Xoc dramatically impaired pathogen virulence and growth rate in host rice, but did not affect the ability to trigger the hypersensitive response in nonhost (plant) tobacco. Compared with the wild‐type strain, the asnB deletion mutant was unable to grow in basic MMX (–) medium (a minimal medium without ammonium sulphate as the nitrogen source) with or without 10 tested nitrogen sources, except asparagine. The disruption of asnB impaired pathogen resistance to oxidative stress and reduced the transcriptional expression of oxyR, katA and katG, which encode three important proteins responsible for hydrogen peroxide (H 2 O 2) sensing and detoxification in Xanthomonas in the presence of H 2 O 2, and nine important known Xoc virulence‐related genes in plant cell‐mimicking medium. Furthermore, the asnB mutation did not affect extracellular protease activity, extracellular polysaccharide production, motility or chemotaxis. Taken together, our results demonstrate the role of asnB in Xanthomonas for the first time.
Introduction
Xanthomonas is a large genus of Gram‐negative bacteria with the typical characteristic of yellow pigment. This genus consists of a number of phytopathogenic bacteria that infect more than 400 plant hosts, including a wide variety of economically important plants, such as rice, citrus, banana, cabbage and bean (Ryan et al., 2011; Watt et al., 2005). One member of this genus, Xanthomonas oryzae pv. oryzicola (Xoc), is the causal agent of bacterial leaf streak (BLS), which is a serious bacterial disease of rice in Asia and parts of Africa (Niño‐Liu et al., 2006; Zhao et al., 2011). Xoc penetrates the leaf mainly through stomata, multiplies in the substomatal cavity and colonizes the parenchymal apoplast, causing interveinal lesions (Wang et al., 2007). Xoc can also gain access through wounds, but, in this case, it remains restricted to the mesophyll tissue apoplast and does not invade the xylem (Niño‐Liu et al., 2006). To date, the virulence mechanisms of Xoc are not entirely clear, and control measures for BLS are poorly developed.
Quorum sensing (QS) is a sophisticated cell–cell communication mechanism that relies on small signal molecules to control bacterial behaviour and coordinate gene expression in a cell density‐dependent manner (Mole et al., 2007). In Gram‐negative bacteria, the N‐acyl homoserine lactones (AHLs) are a major class of signal molecules. AHLs are composed of a homoserine lactone ring carrying C4–C18 acyl chains and are thought to function as interspecific signalling molecules (Danhorn and Fuqua, 2007; Miller and Bassler, 2001). Bacterial behaviours or phenotypes generally controlled by AHL signals include virulence, symbiosis, biofilm formation, motility, conjugation, competence, antibiotic production and sporulation (Danhorn and Fuqua, 2007; Goo et al., 2010; Miller and Bassler, 2001).
In contrast with other Gram‐negative bacteria, Xanthomonas does not produce AHL, but has evolved to produce a different type of QS signal, called the diffusible signal factor (DSF). DSF is significantly different from AHL in chemical structure (Barber et al., 1997; Chatterjee and Sonti, 2002; Tang et al., 1991). The DSF structure has been identified as cis‐11‐methyl‐2‐dodecenoic acid in X. campestris pv. campestris (causal agent of crucifer black rot) (Wang et al., 2004). The QS signals have been identified as BDSF (cis‐dodecenoic acid) and CDSF (a novel unsaturated fatty acid, which is otherwise identical to DSF, except for the double bond between C5 and C6) in X. oryzae pv. oryzae (Xoo). A recent study has suggested that BDSF is the major in vivo signal when Xoo infects rice (Professor Yawen He, Shanghai Jiao Tong University, Shanghai, personal communication). Furthermore, DSF signals have also been identified in Xylella fastidiosa (causal agent of citrus variegated chlorosis) and the Burkholderia cepacia complex (a pathogen that particularly affects cystic fibrosis patients) (Boon et al., 2008; Chatterjee et al., 2008). Similar to AHL, DSF has been linked to the regulation of virulence, motility, toxin production, aerobic respiration, biofilm dispersal, extracellular enzymes and extracellular polysaccharide (EPS) production in DSF‐producing bacteria (Deng et al., 2011; He and Zhang, 2008).
Studies have shown that the regulation of pathogenicity factor (rpf) gene cluster (core genes including rpfF, rpfC and rpfG) is responsible for DSF production and signal transduction (Chatterjee and Sonti, 2002; He et al., 2006; Siciliano et al., 2006; Tang et al., 1991). rpfF encodes a protein similar to enoyl‐CoA hydratase, which catalyses DSF synthesis. RpfC (sensor protein) and RpfG (response regulator), consisting of a unique two‐component regulatory system, are responsible for extracellular DSF sensing and intracellular signal transduction, which leads to the regulation of downstream gene expression.
A transcriptional factor, cyclic adenosine monophosphate (cAMP) receptor protein‐like protein (Clp), is known as a master regulator in the X. campestris pv. campestris DSF signalling network. Clp regulates the expression of genes belonging to the DSF regulon, such as those encoding extracellular enzymes, components of type II and type III secretion systems, and genes involved in EPS synthesis (Büttner and Bonas, 2010; He et al., 2006, 2007). However, Clp is not involved in DSF‐dependent regulation of biofilm formation (He et al., 2007).
In a previous study, we reported that DSF‐meditated QS is required for Xoc pathogenicity on host rice, and revealed 48 differentially expressed intracellular proteins between the wild‐type strain and QS‐defective mutants using the two‐dimensional method (Zhao et al., 2011). Among the 48 proteins, 18 were identified using mass spectrometry and predicted to be involved in nitrogen transfer, protein folding, resistance to oxidative stress and flagellar synthesis (Zhao et al., 2011). We in‐frame deleted each of the 18 genes to investigate the role of these 18 DSF‐regulated proteins in Xoc virulence. Here, we report the identification and functional characterization of one of the 18 genes, asnB (XOC_3054), which encodes the enzyme that converts aspartic acid to asparagine in Xoc. Our data show that asnB is involved in aspartate metabolism, resistance to oxidative stress, virulence and the regulation of the expression of nine known important virulence‐related genes, and is regulated by both DSF‐mediating QS and the global regulator Clp in Xoc. Our results provide a foundation for further molecular understanding of DSF‐mediated proteins and their functions in the Xoc–rice interaction. This is the first report of a functional analysis of asnB in Xanthomonas.
Results
asnB organization in the genome, promoter predication and mutant confirmation
Figure 1A shows asnB organization in the available genome of strain BLS256. The asnB nucleotide size was 1692 bp. Xoryp_15010 (2124 bp) and Xoryp_15015 (1008 bp), which encode the receptor and a hypothetical protein, respectively, were located upstream of asnB, whereas Xoryp_15025 and Xory_15030, which encode succinyl‐diaminopimelate desuccinylase and a hypothetical protein, respectively, were located downstream of asnB. Furthermore, the intergenic regions between Xoryp_15010 and Xoryp_15015, Xoryp_15015 and asnB, asnB and Xoryp_15025, and Xoryp_15025 and Xoryp_15030 were 476, 70, 294 and 6 bp, respectively.
Figure 1.
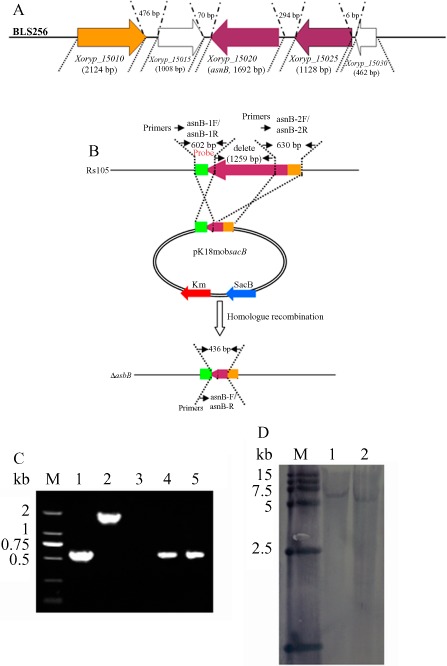
Physical map of asnB organization (A), mutant construction (B) and molecular confirmation (C, D) in Xanthomonas oryzae pv. oryzicola. (A) asnB organization in the available genome of strain BLS256. The nucleotide size of asnB is 1692 bp. Xoryp_15010 (2124 bp) and Xoryp_15015 (1008 bp), which encode an iron receptor and a hypothetical protein, respectively, were located upstream of asnB, whereas Xoryp_15025 and Xory_15030, which encode succinyl‐diaminopimelate desuccinylase and a hypothetical protein, respectively, were located downstream of asnB. (B) The gene deletion scheme. The 602‐bp (amplified by asnB‐1F/asnB‐1R) (Table 1) and 630‐bp (amplified by asnB‐2F/2R) (Table 1) DNA fragments were used as the 5′ and 3′ fragments for homologous recombination, respectively. The internal 1259‐bp DNA fragment was deleted in the asnB mutant. The asnB‐F/R primer (Table 1) was used for molecular confirmation of the asnB mutant. If the 1259‐bp internal fragment of asnB was successfully deleted, a 436‐bp DNA fragment would be amplified from the asnB mutant. (C) Polymerase chain reaction (PCR) confirmation of the asnB mutant. Owing to deletion of the asnB 1259‐bp internal fragment, only an approximately 0.5‐kb DNA fragment was amplified from the asnB deletion mutant (lanes 4 and 5). M, mark; 1, pKS‐A, positive control; 2, Rs105 wild‐type strain; 3, H2O, negative control; 4 and 5, ΔasnB. (D) Southern blotting analysis of the asnB deletion mutant. The 602‐bp fragment was used as the probe for Southern blotting. A 6.5‐kb DNA fragment was detected in the Rs105 wild‐type strain (lane 1), whereas only an approximately 5.0‐kb fragment was obtained in the asnB deletion mutant (lane 2) owing to in‐frame deletion of the 1259‐bp fragment.
A 335‐bp upstream fragment of the asnB 5′ sequence was selected for promoter prediction at the Neural Network Promoter Prediction website (http://www.fruitfly.org/seq_tools/promoter.html). The results showed that two promoter sequences from −263 to −213 (with a score of 0.51) and −211 to −161 (with a score of 0.54) were predicted.
The scheme for asnB mutant construction and confirmation is shown in Fig. 1B. As shown in Fig. 1C, a 1.7‐kb DNA fragment was amplified from the wild‐type strain, whereas only a 0.4‐kb DNA fragment was amplified in the asnB deletion mutant (lanes 4 and 5) because of the in‐frame deletion of the 1.3‐kb fragment from the asnB gene (Fig. 1B). This result shows correction of the asnB in‐frame deletion. To confirm this result, we used the 602‐bp DNA fragment amplified by asnB‐1F/asnB‐1R as a probe (Fig. 1B) for Southern blot analysis. As shown in Fig. 1D, a 6.5‐kb DNA fragment was detected in the Rs105 wild‐type strain, whereas only a 5.0‐kb fragment was obtained from the asnB deletion mutant owing to in‐frame deletion of the 1.3‐kb fragment. This result was consistent with that of polymerase chain reaction (PCR) amplification. Taken together, these results show the correct construction of the asnB deletion mutant in Xoc.
asnB is required for Xoc pathogenicity in host rice, but is not necessary for the triggering of the hypersensitive response (HR) in nonhost (plant) tobacco
To investigate the role of asnB in Xoc virulence, we generated the asnB in‐frame deletion mutant ΔasnB in a wild‐type background. As shown in Fig. 2A,B,D, ΔasnB exhibited a significant decrease in virulence in rice seedlings and adult rice leaves, whereas the complemented strain ΔasnB(asnB) (Fig. 2A,B,D) displayed the same virulence as the wild‐type. This result indicates that asnB is required for Xoc pathogenicity on host rice. In the following experiment, we aimed to understand whether asnB is also required for the triggering of HR (programmed cell death) in nonhost (plant) tobacco. The asnB mutation did not affect the ability of the wild‐type to trigger HR (Fig. 2C), indicating that asnB is not involved in the triggering of HR in nonhost (plant) tobacco. This result also suggests the diverse functions of asnB.
Figure 2.
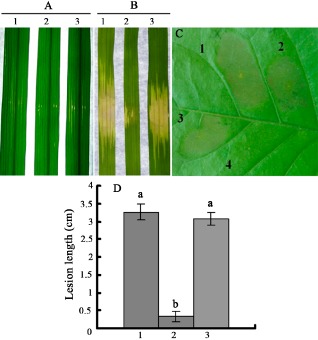
Pathogenicity test and hypersensitive response (HR) assay of the Xanthomonas oryzae pv. oryzicola (Xoc) strains in rice and tobacco, respectively. (A) Lesion lengths on the leaves of adult susceptible rice (cv. Shanyou63, 2 months old) inoculated with the Xoc strains. (B) Lesion lengths on the rice seedling leaves (cv. Shanyou63, 2 weeks old) by infiltration with the Xoc strains. (C) Phenotypes of HR in nonhost (plant) tobacco by inoculation with Xoc strains. (D) Calculated lesion lengths on the leaves of adult susceptible rice (cv. Shanyou63, 2 months old) inoculated with the Xoc strains. 1, Rs105 wild‐type strain; 2, asnB deletion mutant ΔasnB; 3, Rs105 asnB complemented strain ΔasnB(asnB); 4, negative control (double‐distilled H2O). Three replicates were used for each treatment, and the experiment was repeated three times. Vertical bars represent standard errors. Different letters above the data bars indicate a significant difference between the wild‐type strain and asnB deletion mutant or asnB complemented strain (P < 0.05; t‐test).
The AsnB mutation lowers the growth rate of the wild‐type rice strain
The growth rate of ΔasnB in nutrient‐rich broth (NB), nutrient‐limiting broth (MMX) and rice tissue was determined to understand the relationship between the growth rate of ΔasnB and its virulence deficiency. The asnB mutant exhibited the wild‐type growth rate in NB medium (data not shown). However, the ability to grow in MMX medium was abolished by the mutation (Fig. 3A). The complemented strain restored the wild‐type growth rate under the same conditions (Fig. 3A). Interestingly, the growth rate of ΔasnB in infected rice tissue decreased, but was not abolished, when compared with the complemented and wild‐type strains (Fig. 3B). This result indicates that the virulence deficiency of the asnB deletion mutant is, at least partially, caused by the growth decrease in rice tissue.
Figure 3.
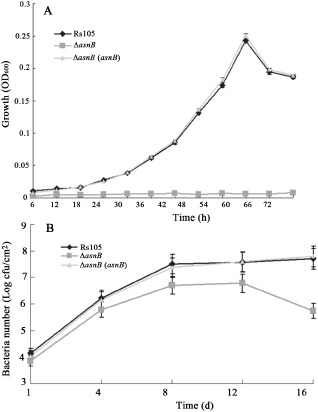
Growth rate of Xanthomonas oryzae pv. oryzicola (Xoc) strains in MMX basic medium and rice leaves. (A) Growth rate of Xoc wild‐type strain Rs105, the asnB deletion mutant ΔasnB and its complemented strain ΔasnB(asnB) in MMX basic medium. OD600 nm, optical density at 600 nm. (B) Growth rate of Xoc wild‐type strain Rs105, the asnB deletion mutant ΔasnB and its complemented strain ΔasnB(asnB) in adult‐susceptible rice leaves (cv. Shanyou63) during infection. cfu, colony‐forming unit. Three replicates for each treatment were used, and the experiment was repeated three times. Vertical bars represent standard errors.
The growth deficiency of the asnB mutant in MMX is restored by supplementation with the rice leaf extract
The results in Fig. 3 show that the asnB mutation abolished the growth ability of the wild‐type in MMX minimal medium, but only decreased the growth rate in rice leaves compared with that in the wild‐type strain. To confirm this result, we investigated whether the growth deficiency of the asnB mutant in MMX medium could be restored by supplementation with the rice leaf extract. As shown in Fig. 4, the asnB deletion mutant in MMX medium was restored to growth in the presence of rice tissue. As the concentration of rice tissues supplemented with MMX increased, growth restoration of the asnB deletion mutant also increased. In MMX medium supplemented with a 3% final concentration of rice tissue, growth of the asnB deletion mutant was significantly higher than that of the control (MMX broth). These findings provide support for the growth determination results of the asnB deletion mutant with rice leaves (Fig. 3B). Taken together, these results suggest that a component(s) from rice leaves might restore the asparagine metabolism defect in the asnB mutant.
Figure 4.
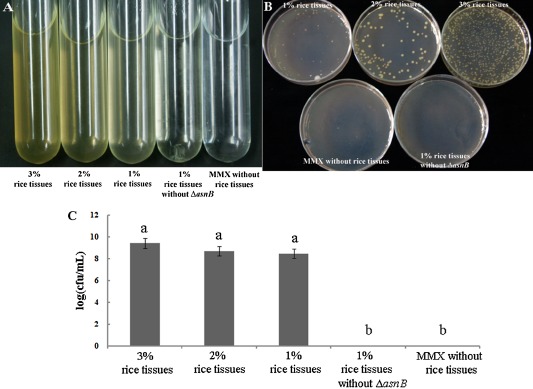
Growth of the asnB mutant in MMX minimal medium was restored in the presence of rice tissue. (A) Representative photographs of growth of the asnB mutant in liquid MMX with or without different final concentrations of rice tissue (1%, 2% and 3%). (B) Representative photographs of growth of the asnB mutant on MMX plates with or without different final concentrations of rice tissue (1%, 2% and 3%). (C) Calculated data for growth of the asnB mutant on MMX plates with or without different final concentrations of rice tissues (1%, 2% and 3%). The 1% rice tissue without the asnB mutant, and ΔasnB and MMX without rice tissue, but containing ΔasnB, were used as negative controls in all experiments. Three replicates per treatment were used, and the experiment was repeated three times. Vertical bars represent standard errors. Different letters above the bars indicate a significant difference between the rice tissue treatments and the control (P < 0.05; t‐test). cfu, colony‐forming unit.
AsnB is involved in aspartate metabolism in Xoc
asnB encodes the enzyme that converts aspartic acid into asparagine in bacteria. To confirm that the AsnB protein in Xoc is involved in aspartate metabolism, we tested the growth rate of ΔasnB in MMX with or without aspartic acid or asparagine. We clearly observed that ΔasnB was unable to grow in minimal medium (Fig. 3A). However, ΔasnB displayed the wild‐type growth rate when asparagine, but not aspartic acid, was added to the MMX medium, indicating that asnB is involved in aspartate metabolism. To further investigate whether asnB is involved in the metabolism of other nitrogen sources, the growth rate of ΔasnB in MMX (–) medium (a minimal medium without ammonium sulphate as the nitrogen source) was assessed when the medium was amended with other nitrogen sources. As shown in Fig. 5, the ability of ΔasnB to grow in MMX (–) medium containing peptone and yeast extract was partially restored. Other nitrogen sources (glutamine, glutamate, urea, potassium nitrate and alanine) did not restore the growth rate of ΔasnB under the same conditions. Taken together, these results indicate that asnB is involved in aspartate metabolism.
Figure 5.
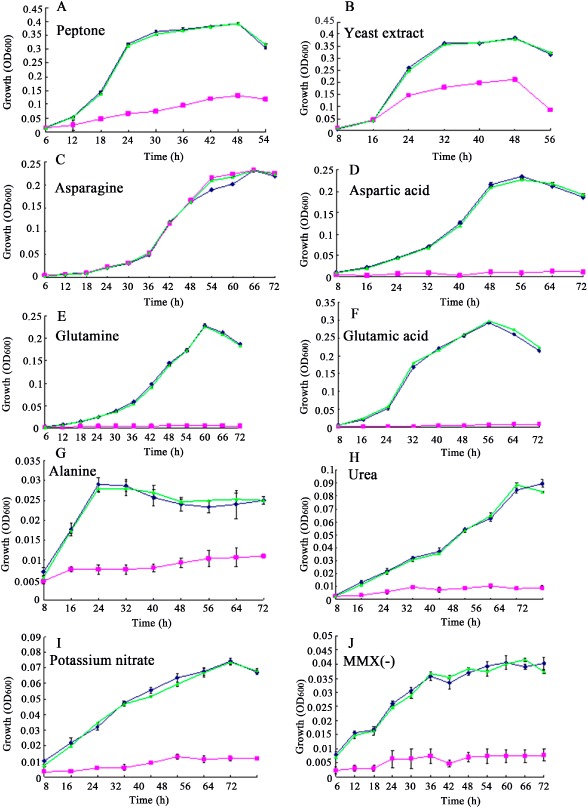
Growth rate of Xanthomonas oryzae pv. oryzicola (Xoc) strains in MMX (–) basic medium supplemented with different nitrogen sources. The Rs105 wild‐type strain (blue), asnB deletion mutant (pink) and corresponding complemented strain (green) were tested with 10 nitrogen sources to evaluate the role of asnB in Xoc nitrogen utilization. MMX (–), MMX medium without ammonium sulphate. Three replicates for each treatment were used, and the experiment was repeated three times. Vertical bars represent standard errors.
The asnB mutation impairs bacterial resistance to hydrogen peroxide (H 2 O 2) and reduces the expression of three genes responsible for H 2 O 2 sensing and detoxification in Xoc
Sensing, detoxification and adaptation to oxidative stress play critical roles during successful pathogen infection and pathogenesis in Xanthomonas (Charoenlap et al., 2011). We conducted the following experiments to understand the role of asnB in resistance to H2O2. As shown in Fig. 6, the asnB mutant clearly displayed a greater sensitivity than the Rs105 wild‐type strain and ΔasnB(asnB) complemented strain to H2O2. Furthermore, the expression of three important genes (kagA, katG and oxyR), which are responsible for H2O2 sensing and detoxification, clearly decreased in the asnB mutant compared with that in the Rs105 wild‐type strain in the presence of 0.1 m H2O2 (Fig. 7A). However, we observed that the expression of only one gene (katG) decreased remarkably in the asnB mutant compared with that in the wild‐type strain in NB without H2O2 (Fig. 7B)
Figure 6.
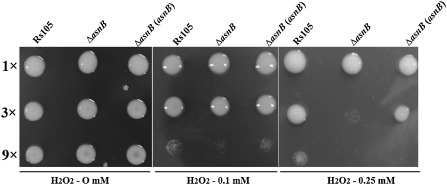
asnB mutations impair resistance to H2O2 in Xanthomonas oryzae pv. oryzicola (Xoc). Xoc strains, including the wild‐type strain Rs105, the asnB deletion mutant ΔasnB and its complemented strain ΔasnB(asnB), were grown on nutrient broth (NB) agar plates with 0 mm H2O2 (A), 0.1 mm H2O2 (B) or 0.25 mm H2O2 (C). Three replicates for each treatment were used, and the experiment was repeated three times.
Figure 7.
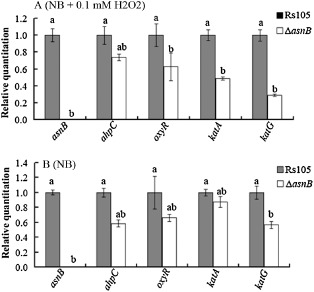
asnB mutation reduces KatA, katG and oxyR transcriptional expression in Xanthomonas oryzae pv. oryzicola in the presence of hydrogen peroxide (H2O2). Total RNA was extracted from strains Rs105 and ΔasnB and cultured on nutrient broth (NB) plus 0.1 mm H2O2 (A) and NB (B). Three replicates for each treatment were used, and the experiment was repeated three times. Vertical bars represent standard errors. Different letters above the bars indicate a significant difference between the wild‐type strain and asnB deletion mutant (P < 0.05; t‐test).
asnB is regulated by QS and the Clp global regulator
In a previous proteomic study, it was evident that AsnB is regulated by DSF‐mediated QS in Xoc (Zhao et al., 2011). To understand whether asnB is regulated by DSF at the mRNA level, the asnB transcriptional levels in the Rs105 wild‐type strain and ΔrpfF DSF‐deficient strain were measured. As shown in Fig. 8, the transcriptional level of the asnB rpfF mutant decreased relative to that in the Rs105 wild‐type strain. Furthermore, the asnB transcriptional level decreased in the clp mutant (Fig. 8). These results indicate that asnB is positively regulated by both DSF and the Clp global regulator.
Figure 8.
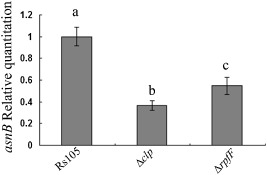
asnB transcription is positively regulated by diffusible signal factor (DSF) and the Clp global regulator. Differential asnB expression was confirmed by relative quantitative real‐time polymerase chain reaction (PCR). Total RNA was extracted from Xanthomonas oryzae pv. oryzicola (Xoc) strains Rs105, ΔrpfF (DSF‐deficient strain) (Zhao et al., 2011) and Δclp (Zhao et al., 2012) at the mid‐stage of growth [optical density at 600 nm (OD600 nm) = 1.6]. Three replicates for each treatment were used, and the experiment was repeated three times. Vertical bars represent standard errors. Different letters above the bars indicate a significant difference between the wild‐type strain and ΔrpfF or Δclp (P < 0.05; t‐test)
Transcriptional expression of nine known virulence‐associated genes is reduced in the asnB deletion mutant in plant cell‐mimicking XOM3 medium, but not in NB medium
To date, a series of important virulence‐associated genes, including purF (encodes amidophosphoribosyltransferase), wxolB (encodes putative glycosyltransferase), pgk (encodes phosphoglycerate kinase), thrC (encodes threonine synthase), gapA (encodes glyceraldehyde‐3‐phosphate dehydrogenase, type I), tal‐C10c‐like (encodes a transcriptional activator‐like TAL effector), trpA (encodes tryptophan synthase α subunit), dsbC (encodes disulphide isomerase), rfbA (encodes glucose‐1‐phosphate thymidylyltransferase) and atmR (encodes aminotransferase), has been identified and analysed in Xoc (Guo et al., 2012; Wang et al., 2007). A corresponding quantitative real‐time reverse transcription‐PCR (qRT‐PCR) experiment was performed between the wild‐type and asnB deletion mutant when both were cultivated in NB and plant cell‐mimicking medium (XOM3) to investigate whether asnB has a regulatory role for transcriptional expression of these known virulence‐associated genes in Xoc. We observed that deletion of asnB reduced significantly the transcriptional expression of the nine known virulence‐associated genes (purF, wxocB, pgk, gapA, thrC, tal‐C10c‐like, trpA, rfbA and atmR) in plant cell‐mimicking XOM3 medium (Fig. 9B), whereas all selected genes exhibited wild‐type expression levels in NB medium (Fig. 8, 9A).
Figure 9.
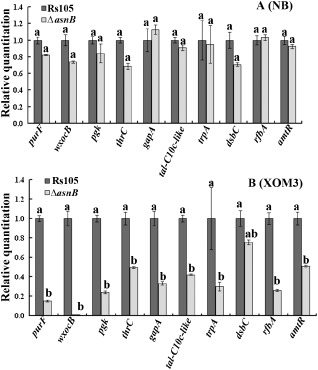
Transcriptional expression of 10 known Xanthomonas oryzae pv. oryzicola (Xoc)virulence‐associated genes in the asnB deletion mutant and wild‐type strain. (A) Comparison of transcriptional expression of 10 known Xoc‐associated genes cultivated in nutrient‐rich broth (NB) between the asnB deletion mutant and the wild‐type strain. Total RNA was extracted from the Rs105 wild‐type strain of Xoc and ΔasnB (asnB deletion mutant) at the mid‐stage of growth [optical density at 600 nm (OD600 nm) = 1.6]. (B) Comparison of the transcriptional expression of 10 known Xoc‐associated genes cultivated in plant cell‐mimicking broth (XOM3) between the epv deletion mutant and the Rs105 wild‐type strain. The Xoc strains were preincubated in NB medium overnight, resuspended at OD600 nm = 2.0 in XOM3 medium and washed twice. Then, 2 mL of the bacterial suspension was inoculated into 50 mL of XOM3 plant cell‐mimicking broth (pH 6.5) at 28 °C for 16 h (Li et al., 2011). Total RNA was extracted from the Rs105 wild‐type strain of Xoc and ΔasnB (asnB deletion mutant). Three replicates for each treatment were used, and the experiment was repeated three times. Vertical bars represent standard errors. Different letters above the bars indicate a significant difference between the wild‐type strain and asnB deletion mutant (P < 0.05; t‐test).
AsnB is not involved in motility, chemotaxis, EPS or protease production in Xoc
Motility, chemotaxis, EPS and proteases are associated with virulence in Xanthomonas (Chen et al., 2009; He et al., 2006; Tang et al., 1991; Yin et al., 2011). Because the asnB mutation in Xoc led to lost virulence ability in the wild‐type strain in host rice, it was reasonable to investigate whether or not the asnB mutation affected these virulence‐associated functions that result in virulence deficiency of the asnB mutant in rice (detailed methods were provided in part B, C and D of Supplementary File 1). Interestingly, the asnB mutation did not affect EPS production (Fig. S1‐2A, see Supporting Information), protease activity (Fig. S1‐2B), chemotaxis (Fig. S1‐3) or motility (Fig. S2‐7, see Supporting Information) in Xoc.
Discussion
In this study, we demonstrated, for the first time, that asnB, encoding an asparagine synthetase, is involved in aspartate metabolism, resistance to H2O2 and virulence in Xoc. Two families of asparagine synthetases have been identified in bacteria (Yoshida et al., 1999). One is the AsnA family, such as the AsnA of Escherichia coli, whose members are found in prokaryotes, such as E. coli and Klebsiella aerogenes (1980; 1982). Members of the AsnA family are only able to use ammonia as the amino group donor (Yoshida et al., 1999). The other is the AsnB family, such as the AsnB protein in E. coli (Hughes et al., 1997; Ren and Liu, 2006). Compared with the AsnA family, AsnB family members are found in both prokaryotes and eukaryotes, and are able to use both glutamine and ammonia as nitrogen donors, but glutamine is preferred (Hughes et al., 1997; Ren and Liu, 2006). Escherichia coli and K. aerogenes have two asparagine synthetase genes, asnA and asnB, and the presence of either ensures sufficient asparagine biosynthesis, whereas disruption of both causes asparagine auxotrophy (Humbert and Simoni, 1980; Reitzer and Magasanik, 1982; Yoshida et al., 1999). A bioinformatic analysis revealed that no AsnA homologue was detected in the Xoc genome. Further analysis indicated that the AsnA protein is absent in all published Xanthomonas species genomes, unlike for E. coli. However, a blastp search revealed that a number of Xanthomonas species, including Xoo, X. axonopodis pv. citri, X. campestris pv. vesicatoria, X. fuscans, X. vesicatoria, X. gardneri and X. campestris pv. campestris, contain a protein with >95% amino acid identity and similar size to AsnB found in Xoc. Xanthomonas‐related species, such as Xylella fastidiosa and Stenotrophomonas maltophilia, also contain a protein with 80%–90% amino acid identity and a similar size to AsnB of Xoc. Taken together, these results indicate that AsnB is highly conserved in Xanthomonas and the related genera Xylella and Stenotrophomonas. However, the role(s) of the AsnB homologue in these bacterial species remains to be investigated.
Another defined asparagine synthetase (Xoc_2999) is found in the Xoc strain BLS256 genome, named AsnB2. A domain analysis of AsnB2 (564 amino acids) using the Pfam database (http://pfam.sanger.ac.uk/) showed that AsnB2 contains two domains (Fig. S2‐1): one is the glutamine amidotransferase_7 domain, at positions 50–170, and the other is the asparagine synthase domain, at positions 214–458. These two domains were also detected in the AsnB protein. An alignment analysis showed only 33% similarity between AsnB and AsnB2 at the amino acid level.
Furthermore, similar to AsnB, AsnB2 was also highly conserved, with 95%–99% similarity at the amino acid level to its counterparts in several available Xanthomonas genomes, including Xoo strain KACC 10331 (AE013598.1), X. campestris pv. vesicatoria strain 85‐10 (NC_007508.1), X. campestris pv. campestris strain B100 (NC_010688.1) and X. perforans strain 91–118 (NZ_AEQW01000228.1).
The asnB mutation in Mycobacterium smegmatis leads to sensitivity of the wild‐type strains to multiple antibiotics, including rifampicin, erythromycin, novobiocin and fusidic acid (Ren and Liu, 2006). However, our results showed that the asnB and asnB2 mutations (Fig. S2‐2) in Xoc did not impair bacterial resistance to rifampicin, gentamicin and kanamycin (Figs S1‐1 and S2‐3; detailed method was provided in part A of Supplementary File 1). These results indicate that the role of asnB and asnB2 in bacterial resistance to antibiotics might be diverse in different bacterial species. Exogenous supplementation of asparagine into minimal medium as the sole nitrogen source facilitated the growth rate of the Xoc asnB and asnB2 mutants (Figs 3 and S2‐4), which was significantly different from the observation in M. smegmatis (Ren and Liu, 2006). These results also indicate that both asnB and asnB2 are involved in aspartate metabolism in Xoc. Furthermore, the addition of other nitrogen sources did not restore the growth rate of the asnB mutant to the wild‐type level in the corresponding medium (Fig. 5), indicating that AsnB might not be related to the metabolism of other nitrogen sources in Xoc under in vitro conditions.
The asnB mutant of the Gram‐positive bacterium Bacillus subtilis shows poorer growth than that of the wild‐type strain (Yoshida et al., 1999), which was also observed in Xoc when the asnB mutant was grown in MMX basic medium. These results indicate that the contribution of asnB to bacterial growth might be conserved in Bacillus and Xanthomonas. Interestingly, the growth rate of the asnB mutant in rice leaves decreased, but was not deficient (Fig. 3B), which is clearly different from the asnB mutant in MMX basic medium. This result indicates that another mechanism(s) might be involved in aspartate metabolism when Xoc is grown in rice leaves. Interestingly, supplementation with the rice leaf extracts restored the growth deficiency of the asnB mutant in MMX basic medium (Fig. 4), indicating that a component(s) from rice leaves might restore the asnB mutant defect in aspartate metabolism, resulting in restored growth ability in the host rice. Whether this component(s) was identical to asparagine should be investigated further. More interestingly, supplementation with complex nitrogen sources (peptone and yeast extract) partially restored the growth ability of the asnB mutant. This indicates that Xoc might possess other asnB‐independent mechanism(s) involved, at least partially, in aspartate metabolism (Fig. 5), which requires further investigation.
Oxidative stress plays a crucial role in plant defence against invasion and colonization by pathogens (Baker and Orlandi, 1995). To survive and proliferate, pathogenic bacteria must overcome the plant defence response (Charoenlap et al., 2011). Four proteins in Xanthomonas, including KatA (encoding a monofunctional catalase), KatG (encoding a catalase‐peroxidase), AhpC (alkyl hydroperoxide reductase) and OxyR (encoding an H2O2 sensor and a LysR‐type transcriptional regulator) are involved in the bacterial H2O2 sensing and protective system (Charoenlap et al., 2011; Jittawuttipoka et al., 2009). Furthermore, the X. campestris pv. campestris katA, oxyR, and ahpC mutations impair pathogen virulence in Chinese radish (Raphanus sativus) (Charoenlap et al., 2011). Similarly, the disruption of oxyR impairs bacterial virulence in Xoc (2012). In the present study, the Xoc asnB mutation impaired bacterial resistance to H2O2 and virulence (Figs 2 and 6), whereas the asnB2 mutation did not impair wild‐type antioxidative ability or virulence (Fig. S2‐5 and S2‐6), indicating that asnB, not asnB2, plays a critical role in antioxidative ability and virulence in Xoc. Furthermore, katA, katG and oxyR expression in the presence of H2O2, which mimics the Xoc initial infection condition in rice cells, was positively regulated by asnB (Fig. 7). We assumed that the reduction in katA, katG and oxyR expression in the asnB mutant might contribute to the sensitivity of the asnB mutant to an increase in H2O2 (Fig. 6) and virulence deficiency (Fig. 2) in Xoc. However, further investigation is needed to clarify how asnB affects the expression of these genes.
Current evidence indicates that the global transcriptional regulator Clp is located downstream of the RpfC/RpfG two‐component system in the DSF signalling pathway (He et al., 2007; Mole et al., 2007), which is involved in the regulation of virulence, resistance to oxidative stress, extracellular enzymes and EPS biosynthesis (He et al., 2007; Zhao et al., 2011). In the present study, the asnB mutant displayed similar virulence and resistance phenotypes to those of oxidative stress in the Xoc clp mutant reported by Zhao et al. (2012). These overlapping functions of clp and asnB indicate that a hierarchical relationship may exist between clp and asnB in the DSF signalling pathway in Xoc. qRT‐PCR showed that asnB transcriptional expression was regulated by clp in Xoc. Taken together, these results indicate that asnB might be located downstream of clp. Furthermore, Crp regulates the expression of downstream genes in E. coli by binding directly to the motif (TGTGANNNNNNTCACA) of the target gene promoter (Zheng et al., 2004). Interestingly, an imperfect Clp (Crp‐like protein) binding motif (TGCGGGTTTTTTTATG), spaced 68 bp away from the ‘ATG’ asnB start codon, was found in the asnB promoter region, indicating that Clp might regulate asnB expression by binding directly to its promoter in Xoc.
In addition, two transcriptional regulators in X. campestris pv. campestris, named Zur and FhrR (a TetR‐type transcription factor), are located downstream of Clp and are involved in the regulation of a wide range of Clp regulons (He et al., 2007). Whether Clp regulates asnB expression via Zur or in a FhrR‐dependent manner should be investigated further in Xoc.
Because the asnB mutant simply showed a decrease in growth in rice leaves, but was not deficient, the contribution of asnB to virulence was only partially dependent on growth. This also indicates that other unknown mechanism(s) might contribute to virulence in the asnB mutant. We observed that the expression of nine important known virulence‐related genes was reduced in the asnB mutant in XOM3 plant cell‐mimicking medium (Fig. 9), indicating that asnB has a wide impact on virulence‐related gene expression. More importantly, seven of these nine genes, including pgk, thrC, gapA, tal‐C10c‐like, trpA, rfbA and atmR, have been described as being responsible for Xoc virulence, but not as being involved in the induction of HR in nonhost (plant) tobacco, which was consistent with the result of asnB during HR induction.
HR, EPS production, motility, protease activity and chemotaxis are important virulence‐associated functions in Xoc (Chen et al., 2009; 1999; Zou et al., 2006). However, we found that the asnB mutation did not impair these virulence‐associated functions (Figs 2, S1‐2, S1‐3 and S2‐7), indicating that the contribution of asnB to virulence was independent of HR, EPS, motility, protease activity and chemotaxis in Xoc.
In summary, we hypothesize that asnB is regulated by a DSF‐dependent QS located downstream of the Clp global regulator. The ability of this protein to contribute to pathogen virulence was dependent, at least partially, on the regulation of bacterial antioxidative ability and the expression of several critical virulence‐related genes in Xoc.
Experimental Procedures
Strains, plasmids and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were cultivated at 37 °C in Luria–Bertani (LB) medium or on LB agar plates. Unless specified otherwise, Xanthomonas strains were grown at 28 °C in NB medium (beef extract, 3 g/L; yeast extract, 1 g/L; polypeptone, 5 g/L; sucrose, 10 g/L) or on nutrient agar (NA). MMX [glucose, 5 g/L; (NH4)2SO4, 2 g/L; MgSO4·7H2O, 0.2 g/L; K2HPO4, 4 g/L; KH2PO4, 6 g/L; trisodium citrate, 1 g/L; pH 7.0] was used as minimal medium. Antibiotics were added to the medium at the following final concentrations when required: 100 μg/mL ampicillin and 50 μg/mL kanamycin (Km) for E. coli and 100 μg/mL rifampicin (Rif) and 50 μg/mL Km for Xanthomonas campestris 8523 (pKLN55) and Xoc.
Table 1.
Strains, plasmids and primers used in this study
| Strains, plasmids and primers | Characteristics | Source |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | Φ80 lacZΔM15, Δ(lacZYA‐argF)U169. recA1 | Laboratory collection |
| Rs105 | RifR, wild‐type strain of Xanthomonas oryzae pv. oryzicola | Laboratory collection |
| ΔrpfF | RifR, deletion of rpfF in wild‐type strain Rs105 | 2011) |
| Δclp | RifR, deletion of clp in wild‐type strain Rs105 | Zhao et al. (2011) |
| ΔasnB | RifR, deletion of asnB in wild‐type strain Rs105 | This study |
| ΔasnB2 | RifR, deletion of asnB2 in wild‐type strain Rs105 | This study |
| ΔasnB(asnB) | RifR, KmR, complemented strain of ΔasnB | This study |
| Plasmids | ||
| pK18mobsacB | KmR, allelic exchange suicide vector, sacB oriT(RP4) | Zou et al. (2011) |
| pK18SA | KmR, a 1238‐bp fusion fragment of asnB‐1, asnB‐2 gene ligated in pK18mobsacB | This study |
| pHMI | SpR, broad‐host‐range cosmid vector, pSa ori | Laboratory collection |
| pHMIAS | SpR , asnB‐H gene ligated in pHMI | This study |
| Primers | ||
| asnB‐1F | 5′‐CGGGATCCCCAAGGCATCTATCTCTCGG‐3′ | This study |
| asnB‐1R | 5′GGGGTACCGCAGACCAAGGAAGCGTATT‐3′ | This study |
| asnB‐2F | 5′‐GGGGTACCATCTCGCCATTGACCGCCAG‐3′ | This study |
| asnB‐2R | 5′‐GCTCTAGACAACGCCAGCATTCATCAGG‐3′ | This study |
| asnBH‐F | 5′‐CCAAGCTTCGCTGTATCGCACGCTGATC‐3′ | This study |
| asnBH‐R | 5′‐CGGAATTCCACGAGCAATAAGACACGCG‐3′ | This study |
| asnB2‐1F | 5′‐CGGGATCCTGTTCAGATACGCCAGCACG‐3′ | This study |
| asnB2‐1R | 5′‐GGGGTACCTGTGGACGGTGCTGATGTTC‐3′ | This study |
| asnB2‐2F | 5′‐GGGGTACCGCGAAGCGAGCAACAGTCCT‐3′ | This study |
| asnB2‐2R | 5′‐GCTCTAGACCAAGAATCCCATCGGCAAG‐3′ | This study |
Generation of the asnB deletion mutant in Xoc
The Xoc Rs105 wild‐type strain was used as the parental strain to generate the in‐frame deletion mutant via allelic homologous recombination. In‐frame deletion of asnB was performed as described previously (Zhao et al., 2011). Briefly, two asnB flanking regions were generated by PCR using the primer pairs asnB‐1F/asnB‐1R and asnB‐2F/asnB‐2R (Table 2). The asnB‐1 fragment (digested with BamHI and KpnI) and the asnB‐2 fragment (digested with KpnI and XbaI) were ligated into pK18mobsacB and digested with BamHI and XbaI. This recombinant vector, designated pK18SA, was transformed into the Rs105 wild‐type strain by electroporation. Transconjugants were selected on NA plates without sucrose, but with Rif (100 μg/mL) and Km (50 μg/mL). Positive colonies were plated onto NA plates containing 10% (w/v) sucrose and Rif (100 μg/mL) to select for resolution of the construct by a second cross‐over event. The resulting mutant, containing the asnB in‐frame deletion, was confirmed by PCR and Southern blotting (data not shown). One of the confirmed mutants, named ΔasnB, was selected for further study.
Table 2.
Primers for quantitative real‐time polymerase chain reaction (PCR)
| Gene fragment | Primer sequence for 5′ sequence | Primer sequence for 3′ sequence | Primer source | Gene source |
|---|---|---|---|---|
| 16S rRNA | 5′‐AATGGGCGCAAGCCTGATC‐3′ | 5′‐AACCACCACCTACGCACGC‐3′ | Zhao et al. (2011) | Zhao et al. (2011) |
| katA (Xoryp_4325) | 5′‐ATATCCGGAGGCAGCAAAGT‐3′ | 5′‐AATTCCTTGAACGGCGTATG‐3′ | This study | Charoenlap et al. (2011) |
| katG (Xoryp_3410) | 5′‐AGTGGTCCGTGCTGATCTTC‐3′ | 5′‐ATAGTTGTCGGCAGCGTCTT‐3′ | This study | Charoenlap et al. (2011) |
| oxyR (Xoryp_0957) | 5′‐CAGATGGCAGACATCCAGTG‐3′ | 5′‐TTCGAAGAACCTTTCGTGCT‐3′ | This study | Charoenlap et al. (2011) |
| ahpC (Xoryp_0959) | 5′‐CAGACCGATGTGGACTCCTT‐3′ | 5′‐CGACCAGTACGGTCAATTCC‐3′ | This study | Jittawuttipoka et al. (2009) |
| asnB (Xoryp_3054) | 5′‐CTTCCGTCTTATCCCACAGC‐3′ | 5′‐CTACAACCATCGCGAACTCA‐3′ | This study | This study |
| tal‐C10c‐like (Xoryp_4384) | 5′‐ACCGTGATGTGGGAACAAGATG‐3′ | 5′‐GAGGCAATAGCTCCCTCAACC‐3′ | Laboratory collection | Guo et al. (2012) |
| dsbC (Xoryp_3811) | 5′‐GCATTCAAGGATTTCGGTGCG‐3′ | 5′‐ATGGTGAGCGGGCGGTCTTC‐3′ | Laboratory collection | Guo et al. (2012) |
| wxocB (Xoryp_3864) | 5′‐CCGCGTACTGGACGATAAGT‐3′ | 5′‐TGCGATAAGAGCGTTGACAC‐3′ | Laboratory collection | Wang et al. (2007) |
| pgk (Xoryp_3589) | 5′‐CGCAGTGGACAAGTACGACA‐3′ | 5′‐CTTGCCTTCCAGAAACTCCA‐3′ | Laboratory collection | Wang et al. (2007) |
| gapA (Xoryp_3592) | 5′‐GCTGGGGTATACCGAAGACA‐3′ | 5′‐TCGTTGTCGTACCAGGACAC‐3′ | Laboratory collection | Wang et al. (2007) |
| rfbA (Xoryp_3845) | 5′‐GGCGTTGTTTCAATCCTTGT‐3′ | 5′‐AGTCGCGACCGATCAAATAC‐3′ | Laboratory collection | Guo et al. (2012) |
| atmR (Xoryp_2825) | 5′‐GTCGGTGGGCAGGTGCTGTA‐3′ | 5′‐GACGATGCGGTCGGCTTT‐3′ | Laboratory collection | Guo et al. (2012) |
| thrC (Xoryp_2208) | 5′‐ATGCGTTGGCGTTGATTCTG‐3′ | 5′‐CGATGGCCTTCTCCACCCT‐3′ | Laboratory collection | Guo et al. (2012) |
| purF (Xoryp_1105) | 5′‐CAGGATCTGGAAGACCTGGA‐3′ | 5′‐TGATGTACTCGCCGTTGAAG‐3′ | Laboratory collection | Guo et al. (2012) |
| trpA (Xoryp_2950) | 5′‐CTTGCGTACGTGTTGGAAGC‐3′ | 5′‐TCTCGATCGGGTTGAGGTAG‐3′ | Laboratory collection | Guo et al. (2012) |
Complementation of the asnB mutant
The asnB mutation was complemented as described previously (Chen et al., 2009; Zhao et al., 2011, 2012). Briefly, a 2110‐bp DNA fragment containing asnB and its predicted promoter region was amplified from the Rs105 wild‐type strain with asnBH‐F/asnBH‐R primers. The amplified fragment was digested with the HindIII/EcoRI enzyme and cloned into HindIII/EcoRI‐digested pHMI, resulting in the pHMIAS plasmid (Table 1) used for complementation. Plasmid pHMIAS was transformed into ΔasnB competent cells by electroporation. Finally, one positive complemented strain, named ΔasnB(asnB), was selected on NA plates with Rif (100 μg/mL) and Km (50 μg/mL) for further study.
Pathogenicity testing and determination of bacterial load in plants
Pathogenicity assays were conducted in a glasshouse at 22–30 °C as described previously (Zhao et al., 2011, 2012; Zou et al., 2006). Briefly, Xoc strains were cultivated in NB medium with appropriate antibiotics at 28 °C. The strains were adjusted to an optical density at 600 nm (OD600 nm) ∼ 0.1 and inoculated into leaves of 6‐week‐old rice plants (Shanyou‐63, susceptible to the pathogen) using leaf needling. Lesion lengths were measured 14 days after inoculation. Twenty‐five leaves were inoculated for each Xoc strain in each treatment. The same experiment was repeated three times.
The growth of each Xoc strain in rice leaf tissue was detected by homogenizing five inoculated leaves in 9 mL of sterile water. The leaves were cut into 6‐mm sections around the inoculation spots on days 0, 4, 8, 12 and 16 after inoculation (Lee et al., 2008). Diluted homogenates were plated onto NA plates supplemented with Rif (for the wild‐type and mutants) or Rif plus Km (for the complemented strains). The number of bacterial colonies on these plates was counted after a 2‐day incubation at 28 °C (Feng et al., 2009). Each diluted homogenate was plated onto three plates. Three replicates for each treatment were used, and the experiment was repeated three times.
H 2 O 2 resistance assay
H2O2 resistance assays were performed as described previously (Lan et al., 2010), with some modifications. NB agar plates were prepared containing H2O2 concentrations of 0, 0.1 and 0.25 mm. Xoc strains were cultured to the mid‐logarithmic phase (OD600 = 1.0) in NB medium, and three‐fold and nine‐fold dilutions were made. A 5‐μL aliquot of the initial culture and diluted cultures for each strain were spotted onto NB agar plates (in triplicate) and cultured for 36 h at 28 °C (Lan et al., 2010). The same experiment was repeated at least three times.
HR assay
Xoc strains were grown in NB medium at 28 °C with shaking at 200 rpm. Cells were pelleted at the early logarithmic phase by centrifugation at 6000 rpm. Cell pellets were suspended in water for HR or pathogenicity tests. The HR test was conducted as described previously (Zou et al., 2006). Briefly, suspensions (OD600 nm = 0.1) of Xoc strains were infiltrated into leaves of glasshouse‐grown tobacco (Nicotiana tabacum L. cv. Samsun), and the results were observed after 48 h of infiltration. If the strain had the ability to trigger HR, the phenomenon of programmed cell death would be observed around the inoculation sites on tobacco leaves.
Growth determination of ΔasnB in MMX minimal medium supplemented with different nitrogen sources
asnB in bacteria encodes the enzyme that converts aspartic acid to asparagine. To confirm that the AsnB protein in Xoc is involved in aspartate metabolism, we tested the growth rate of ΔasnB in basic medium with or without aspartic acid, asparagine or other nitrogen sources. In brief, Xoc strains were grown in NB broth at 28 °C with shaking at 200 rpm. Cells were pelleted at the early logarithmic phase (OD600 nm = 0.5) by centrifugation at 6000 rpm. Cell pellets were suspended in an equal volume of water. Then, 0.5 mL of cell suspension was inoculated into 100 mL of basic MMX medium broth with or without different nitrogen sources, including peptone, yeast extract, asparagine, aspartic acid, glutamine, glutamic acid, ammonium sulphate, urea, potassium nitrate and alanine. All inoculation broths were grown at 28 °C with shaking at 200 rpm, and the OD600 nm value was determined every 6 or 8 h until bacterial growth reached the stationary stage.
Determination of the growth status of the asnB mutant in MMX minimal medium broth supplemented with different concentrations of rice leaf extract
Rice leaves were extracted as described previously (Angela and Yoko, 2001), with some modifications. Briefly, susceptible IR24 rice leaves (500 g) at the tillering stage were selected, followed by sterilization with 75% (v/v) ethanol. After carefully milling and the addition of sterilized double‐distilled H2O, the constant volume of rice leaves was 1 L. The supernatant was collected after centrifugation (7000 rpm, 4 °C, 30 min) and was concentrated to 10 mg/mL using a vacuum freeze drier. In the following assay, the concentrated rice leaf extracts were added to MMX minimal medium at final concentrations of 1%, 2% and 3%. The asnB deletion mutant was inoculated into the prepared MMX broth with the rice leaf extracts to determine growth. The colony formation unit‐based method was used to determine bacterial growth (Zhao et al., 2012).
qRT‐PCR
qRT‐PCR was carried out using a SYBR Premix EX Tag™ II kit (TaKaRa Bio, Shiga, Japan) in an ABI PRISM 7500 Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA). As the endogenous control, 16S rRNA was used. The primer sequences are listed in Table 2. RNA was extracted from Xoc wild‐type strain Rs105, ΔrpfF, Δclp and ΔasnB at different growth stages using the RNAiso Plus Reagent (TaKaRa Bio) following the manufacturer's instructions. To remove genomic DNA, the eluted RNA samples were treated with RNase inhibitors and DNaseI (TaKaRa Bio). RNA integrity was confirmed by electrophoresis on 1.2% agarose gels. Then, 2 μg of each RNA sample was used to synthesize cDNA with a cDNA Synthesis kit (TaKaRa Bio). The same experiment was repeated three times. The asnB transcriptional levels in the wild‐type strain, ΔrpfF and Δclp were assessed and compared. The transcriptional levels of katA (Xoryp_4325), katG (Xoryp_3410), oxyR (Xoryp_0957), aphC (Xoryp_0959), wxolB (Xoryp_3864), pgk (Xoryp_3589), gapA (Xoryp_3592), tal‐C10c‐like (Xoryp_4384), dsbC (Xoryp_3811), rfbA (Xoryp_3845), atmR (Xoryp_2825), thrC (Xoryp_2208), purF (Xoryp_1105), trpA (Xoryp_2950) and gumB (Xoryp_1867) in the wild‐type strain and ΔasnB were also assessed and compared.
Data analysis
All analyses were conducted using SPSS 14.0 (SPSS Inc., Chicago, IL, USA). The hypothesis test of percentages (t‐test, P = 0.05) was used to determine significant differences in the pathogenicity test, gene expression and bacterial growth.
Supporting information
Supplementary File 1 Outlines, Results, Figures S1‐1 to S1‐3.
Supplementary File 2 Figures S2‐1 to S2‐7.
Acknowledgements
We are very grateful to Professor Gongyou Chen (Shanghai Jiaotong University, Shanghai, China) for kindly providing the pK18mobsacB suicide vector. We also thank Professor Liangcheng Du (University of Nebraska‐Lincoln, Lincoln, NE, USA) and Dr Yu Chen (Anhui Academy of Agricultural Sciences, Hefei, China) for critical editing of the manuscript. This study was supported by the National Natural Science Foundation of China (30971894, 31171810 and 31071657), the Natural Science Foundation of Jiangsu Province (No. BK2010445), Special Fund for Agro‐Scientific Research in the Public Interest (No. 200903052; No. 201003004), the Doctoral Fund of the Ministry of Education of China (No. 20100097120013) and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
References
- Angela, M. and Yoko, B.R. (2001) Differentially expressed proteins in the interaction of Xanthomonas axonopodis pv. citri with leaf extract of the host plant. Proteomics, 1, 1111–1118. [DOI] [PubMed] [Google Scholar]
- Baker, C.J. and Orlandi, E.W. (1995) Active oxygen in plant pathogenesis. Annu. Rev. Phytopathol. 33, 99–321. [DOI] [PubMed] [Google Scholar]
- Barber, C.E. , Tang, J.L. , Feng, J.X. , Pan, M.Q. , Wilson, T.J. , Slater, H. , Dow, J.M. , Williams, P. and Daniels, M.J. (1997) A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24, 555–566. [DOI] [PubMed] [Google Scholar]
- Boon, C. , Deng, Y. , Wang, L.H. , He, Y.W. , Xu, J.L. , Fan, Y. , Pan, S.Q. and Zhang, L.H. (2008) A novel DSF‐like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. J. Int. Soc. Microb. Ecol. 2, 27–36. [DOI] [PubMed] [Google Scholar]
- Büttner, D. and Bonas, U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. [DOI] [PubMed] [Google Scholar]
- Charoenlap, N. , Buranajitpakorn, S. , Duang‐Nkern, J. , Namchaiw, P. , Vattanaviboon, P. and Mongkolsuk, S. (2011) Evaluation of the virulence of Xanthomonas campestris pv. campestris mutant strains lacking functional genes in the OxyR regulon. Curr. Microbiol. 63, 232–237. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. and Sonti, R.V. (2002) rpfF mutants of Xanthomonas oryzae pv. oryzae are deficient for virulence and growth under low iron conditions. Mol. Plant–Microbe Interact. 15, 463–471. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. , Newman, K.L. and Lindow, S.E. (2008) Cell‐to‐cell signaling in Xylella fastidiosa suppresses movement and xylem vessel colonization in grape. Mol. Plant–Microbe Interact. 21, 1309–1315. [DOI] [PubMed] [Google Scholar]
- Chen, L. , Hu, B.S. , Qian, G.L. , Wang, C. , Yang, W.F. , Han, Z.C. and Liu, F.Q. (2009) Identification and molecular characterization of twin‐arginine translocation system (Tat) in Xanthomonas oryzae pv. oryzae strain PXO99. Arch. Microbiol. 191, 163–170. [DOI] [PubMed] [Google Scholar]
- Danhorn, T. and Fuqua, C. (2007) Biofilm formation by plant‐associated bacteria. Annu. Rev. Microbiol. 61, 401–422. [DOI] [PubMed] [Google Scholar]
- Deng, Y. , Wu, J.E. , Tao, F. and Zhang, L.H. (2011) Listening to a new language: DSF‐based quorum sensing in Gram‐negative bacteria. Chem. Rev. 111, 160–173. [DOI] [PubMed] [Google Scholar]
- Dharmapuri, S. and Sonti, R.V. (1999) A transposon insertion in the gumG homologue of Xanthomonas oryzae pv. oryzae causes loss of extracellular polysaccharide production and virulence. FEMS Microbiol. Lett. 179, 53–59. [DOI] [PubMed] [Google Scholar]
- Feng, J.X. , Song, Z.Z. , Duan, C.J. , Zhao, S. , Wu, Y.Q. , Wang, C. , Dow, J.M. and Tang, J.L. (2009) The xrvA gene of Xanthomonas oryzae pv. oryzae, encoding an H‐NS‐like protein, regulates virulence in rice. Microbiology, 155, 3033–3044. [DOI] [PubMed] [Google Scholar]
- Goo, E. , Kang, Y.S. , Kim, H.S. and Hwang, I. (2010) Proteomics analysis of quorum‐sensing‐dependent proteins in Burkholderia glumae . J. Proteome Res. 9, 3184–3199. [DOI] [PubMed] [Google Scholar]
- Guo, W. , Cui, Y.P. , Li, Y.R. , Che, Y.Z. , Yuan, L. , Zou, L.F. , Zou, H.S. and Chen, G.Y. (2012) Identification of seven genes of Xanthomonas oryzae pv. oryzicola potentially involved in pathogenesis in rice. Microbiology, 158, 505–518. [DOI] [PubMed] [Google Scholar]
- He, Y.W. and Zhang, L.H. (2008) Quorum sensing and virulence regulation in Xanthomonas campestris . FEMS Microbiol. Rev. 32, 842–857. [DOI] [PubMed] [Google Scholar]
- He, Y.W. , Xu, M. , Lin, K. , Ng, Y.J. , Wen, C.M. , Wang, L.H. , Liu, Z.D. , Zhang, H.B. , Dong, Y.H. , Dow, J.M. and Zhang, L.H. (2006) Genome scale analysis of diffusible signal factor regulon in Xanthomonas campestris pv. campestris: identification of novel cell–cell communication‐dependent genes and functions. Mol. Microbiol. 59, 610–622. [DOI] [PubMed] [Google Scholar]
- He, Y.W. , Ng, A.Y. , Xu, M. , Lin, K. , Wang, L.H. , Dong, Y.H. and Zhang, L.H. (2007) Xanthomonas campestris cell–cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol. Microbiol. 64, 281–292. [DOI] [PubMed] [Google Scholar]
- Hughes, C.A. , Beard, H.S. and Matthews, B.F. (1997) Molecular cloning and expression of two cDNAs encoding asparagines synthetase in soybean. Plant Mol. Biol. 33, 301–311. [DOI] [PubMed] [Google Scholar]
- Humbert, R. and Simoni, R.D. (1980) Genetic and biochemical studies demonstrating a second gene coding for asparagines synthetase in Escherichia coli . J. Bacteriol. 142, 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jittawuttipoka, T. , Buranajitpakorn, S. , Vattanaviboon, P. and Mongkolsuk, S. (2009) The catalase‐peroxidase KatG is required for virulence of Xanthomonas campestris pv. campestris in a host plant by providing protection against low levels of H2O2 . J. Bacteriol. 191, 7372–7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, L.F. , Murray, T.S. , Kazmierczak, B.I. and He, C. (2010) Pseudomonas aeruginosa OspR is an oxidative sensing regulator that affects pigment production, antibiotic resistance and dissemination during infection. Mol. Microbiol. 75, 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.W. , Jeong, K.S. , Han, S.W. , Lee, S.E. , Phee, B.K. , Hahn, T.R. and Ronald, P. (2008) The Xanthomonas oryzae pv. oryzae PhoPQ two‐component system is required for AvrXA21 activity, hrpG expression, and virulence. J. Bacteriol. 190, 2183–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.R. , Zou, H.S. , Che, Y.Z. , Cui, Y.P. , Guo, W. , Zou, L.F. , Chatterjee, S. , Biddle, E.M. , Yang, C.H. and Chen, G.Y. (2011) A novel regulatory role of HrpD6 in regulating hrp‐hrc‐hpa genes in Xanthomonas oryzae pv. oryzicola . Mol. Plant–Microbe Interact. 24, 1086–1101. [DOI] [PubMed] [Google Scholar]
- Miller, M.B. and Bassler, B.L. (2001) Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. [DOI] [PubMed] [Google Scholar]
- Mole, B.M. , Baltrus, D.A. , Dangl, J.L. and Grant, S.R. (2007) Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol. 15, 363–371. [DOI] [PubMed] [Google Scholar]
- Niño‐Liu, D.O. , Ronald, P.C. and Bogdanove, A.J. (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. [DOI] [PubMed] [Google Scholar]
- Reitzer, L.J. and Magasanik, B. (1982) Asparagine synthetase of Klebsiella aerogenes: properties and regulation of synthesis. J. Bacteriol. 151, 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, H.P. and Liu, J. (2006) AsnB is involved in natural resistance of Mycobacterium smegmatis to multiple drugs. Antimicrob. Agents Chemother. 50, 250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, R.P. , Vorhölter, F.J. , Potnis, N. , Jones, J.B. , Van Sluys, M.A. , Bogdanove, A.J. and Dow, J.M. (2011) Pathogenomics of Xanthomonas understanding bacterium–plant interactions. Nat. Rev. 9, 344–355. [DOI] [PubMed] [Google Scholar]
- Siciliano, F. , Torres, P. , Sendín, L. , Bermejo, C. , Filippone, P. , Vellice, G. , Ramallo, J. , Castagnaro, A. , Vojnov, A. and Marano, M.R. (2006) Analysis of the molecular basis of Xanthomonas axonopodis pv. citri pathogenesis in Citrus limon . Electron. J. Biotechnol. 9, 200–204. [Google Scholar]
- Tang, J.L. , Liu, Y.N. , Barber, C.E. , Dow, J.M. , Wootton, J.C. and Daniels, M.J. (1991) Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris . Mol. Gen. Genet. 226, 409–417. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Makino, S. , Subedee, A. and Bogdanove, A.J. (2007) Novel candidate virulence factors in rice pathogen Xanthomonas oryzae pv. oryzicola as revealed by mutational analysis. Appl. Environ. Microbiol. 73, 8023–8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.H. , He, Y.W. , Gao, Y.F. , Wu, J.E. , Dong, Y.H. , He, C.Z. , Wang, S.X. , Weng, L.X. , Xu, J.L. , Tay, L. , Fang, R.X. and Zhang, L.H. (2004) A bacterial cell–cell communication signal with cross‐kingdom structural analogues. Mol. Microbiol. 51, 903–912. [DOI] [PubMed] [Google Scholar]
- Watt, S.A. , Wilke, A. , Patschkowski, T. and Niehaus, K. (2005) Comprehensive analysis of the extracellular proteins from Xanthomonas campestris pv. campestris B100. Proteomics, 5, 153–167. [DOI] [PubMed] [Google Scholar]
- Yin, F.Q. , Zhao, Y.C. , Liu, C.H. , Qian, G.L. , Fan, J.Q. , Hu, B.S. and Liu, F.Q. (2011) Analysis of the flgD and flgE genes regulated by diffusible signal factor in Xanthomonas oryzae pv. oryzicola . Acta Microbiol. Sin. 51, 891–897. [PubMed] [Google Scholar]
- Yoshida, K.I. , Fujita, Y. and Ehrlich, S.D. (1999) Three asparagine synthetase genes of Bacillus subtilis . J. Bacteriol. 181, 6081–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.C. , Qian, G.L. , Yin, F.Q. , Fan, J.Q. , Zhai, Z.W. , Liu, C.H. , Hu, B.S. and Liu, F.Q. (2011) Proteomic analysis of the regulatory function of DSF‐dependent quorum sensing in Xanthomonas oryzae pv. oryzicola . Microb. Pathog. 50, 48–55. [DOI] [PubMed] [Google Scholar]
- Zhao, Y.C. , Qian, G.L. , Fan, J.Q. , Liu, C.H. , Yin, F.Q. , Zhou, Y.J. , Shen, Q. , Hu, B.S. and Liu, F.Q. (2012) Identification and characterization of a novel gene hshB in Xanthomonas oryzae pv. oryzicola co‐regulated by quorum sensing and clp . Phytopathology, 102, 252–259. [DOI] [PubMed] [Google Scholar]
- Zheng, D. , Constantinidou, C. , Hobman, J.L. and Minchin, S.D. (2004) Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32, 5874–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L.F. , Wang, X.P. , Xiang, Y. , Zhang, B. , Li, Y.R. , Xiao, Y.L. , Wang, J.S. , Walmsley, A.R. and Chen, G.Y. (2006) Elucidation of the hrp clusters of Xanthomonas oryzae pv. oryzicola that control the hypersensitive response in nonhost tobacco and pathogenicity in susceptible host rice. Appl. Environ. Microbiol. 72, 6216–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L.F. , Li, Y.R. and Chen, G.Y. (2011) A non‐marker mutagenesis strategy to generate poly‐hrp gene mutants in the rice pathogen Xanthomonas oryzae pv. oryzicola . Agric. Sci. China, 10, 1139–1150. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1 Outlines, Results, Figures S1‐1 to S1‐3.
Supplementary File 2 Figures S2‐1 to S2‐7.


