Abstract
Post-translational modifications (PTM) decorate proteins to provide functional heterogeneity to an existing proteome. The large number of known PTMs highlights the many ways that cells can modify their proteins to respond to diverse stimuli. Recently, PTMs have begun to receive increased interest because new sensitive proteomics workflows and structural methodologies now allow researchers to obtain large-scale, in-depth and unbiased information concerning PTM type and site localization. However, few PTMs have been extensively assessed for functional consequences, leaving a large knowledge gap concerning the inner workings of the cell. Here, we review understanding of N-𝜀-lysine acetylation in bacteria, a PTM that was largely ignored in bacteria until a decade ago. Acetylation is a modification that can dramatically change the function of a protein through alteration of its properties, including hydrophobicity, solubility, and surface properties, all of which may influence protein conformation and interactions with substrates, cofactors and other macromolecules. Most bacteria carry genes predicted to encode the lysine acetyltransferases and lysine deacetylases that add and remove acetylations, respectively. Many bacteria also exhibit acetylation activities that do not depend on an enzyme, but instead on direct transfer of acetyl groups from the central metabolites acetyl coenzyme A or acetyl phosphate. Regardless of mechanism, most central metabolic enzymes possess lysines that are acetylated in a regulated fashion and many of these regulated sites are conserved across the spectrum of bacterial phylogeny. The interconnectedness of acetylation and central metabolism suggests that acetylation may be a response to nutrient availability or the energy status of the cell. However, this and other hypotheses related to acetylation remain untested.
Keywords: acetylation, lysine acetyltransferase, bacteria, mass spectrometry, proteomics
Introduction
The two competing theories for the origin of life (metabolism-first and gene-first) suggest that abiotic chemical reactions were the progenitors of biotic life. These abiotic chemical reactions ultimately formed the basis for metabolism and provided building blocks for the synthesis of both RNA and protein. Over millions of years, cells formed and evolved and, in doing so, generated the diversity of life we observe today. Across phylogeny, the core metabolic pathways necessary for life have been well conserved (Peregrín-Alvarez et al., 2009). In these core pathways, anabolism and catabolism generate a multitude of different metabolites. The potential chemical reactions in which these metabolites can participate greatly exceed cellular metabolic requirements. Therefore, the cell has evolved diverse mechanisms to control these “extra-metabolic” compounds to limit their toxicity or the damage that they impart upon macromolecular structures via non-enzymatic reactions within the cell (Dickinson and Chang, 2011). While cells have evolved means of controlling these “extra-metabolic” compounds, it is thought that accumulation of the resultant damage may be a prototypic driver of aging (Golubev et al., 2017).
Despite the constant possibility of damage due to their own metabolism, bacteria have survived and evolved to adapt to various niches. However, these environments are rarely constant outside of a laboratory setting, and bacteria will often face adverse conditions. In these niches, one of the major challenges is acquisition of nutrients necessary to grow and divide. While some nutrients like carbon may be readily abundant, other nutrients can be scarce, such as iron sequestered by a host (Hood and Skaar, 2012). These conditions may change suddenly, so the bacteria must respond accordingly or perish. It is well established that cells respond to environmental challenges with appropriate regulation of their transcriptional and translational programs (Gottesman, 2017). Whereas transcription and translation are time- and energy-intensive processes, post-translational control over the function of existing proteins can rapidly alter cellular physiology and permit fitness or survival in otherwise suboptimal or lethal conditions (Castaño-Cerezo et al., 2014; Lynch and Marinov, 2015). While small ligands may allosterically regulate proteins, diverse covalent chemical modifications of the existing proteome also occur; these may or may not be reversible (Martin, 2014). Researchers have identified hundreds of chemical modifications of the proteome; these are called post-translational modifications (PTMs). These PTMs can occur on a variety of protein residues and understanding how these modifications affect protein structure and function and whether they interact with one another is critical to generating a complete regulatory picture of the cell.
Many PTMs occur on lysines within proteins, including methylation (Lanouette et al., 2014), propionylation (Chen et al., 2007), butyrylation (Chen et al., 2007), crotonylation (Tan et al., 2011; Sabari et al., 2015), succinylation (Zhang et al., 2011), malonylation (Du et al., 2011; Peng et al., 2011), glutarylation (Tan et al., 2014), 2-hydroxyisobutyrylation (Dai et al., 2014), pupylation (Becker and Darwin, 2017), and N𝜀-lysine acetylation, which is becoming recognized as a prominent modification that is conserved across phylogeny (Kim et al., 2006; Choudhary et al., 2009). While lysine acetylation is best understood as an activating modification of eukaryotic histones (Hebbes et al., 1988), extensive study into the role of lysine acetylation in bacteria only began about a decade ago. Acetylation of certain lysines may have a clear consequence, such as inhibition of enzyme activity due to modification of an active site (Nakayasu et al., 2017), but the functional importance or lack thereof of many acetyllysine modifications are more difficult to discern. To uncover the role of lysine acetylation in these unclear cases, it is helpful to use a model bacterium (e.g., Escherichia coli) with a vast knowledgebase of pathways, protein structure-function relationships, and physiology.
This review will detail what we know concerning lysine acetylation in bacteria with some attention paid to eukaryotic acetylation for context. We will begin by describing the different types of acetylation, the mechanisms that result in acetylation of lysines, and some known effects of lysine acetylation on protein function. We will discuss the importance of mass spectrometry to identify and quantify acetylated proteins. Furthermore, we will explore the possibility that acetylation could be a driver of protein and organismal evolution. Finally, with our deepening understanding of acetylation, we will discuss a possible future for this new field.
Acetylation – From Small Molecules to Proteins
Acetylation is simply the transfer of an acetyl group (CH3CO) onto a molecule. The acetyl group can react with a variety of atoms or functional groups on a target molecule. The atom to which the acetyl group is attached is usually denoted in the name of either the final molecule or the enzyme that performs the acetylation. Acetylation can occur with thiol groups (sulfur), hydroxyl groups (oxygen), and often amino groups (nitrogen).
Acetylation reactions can target small molecules and metabolites, or they can occur on proteins. Multiple metabolic pathways require the transfer of acetyl groups from one metabolite to the next. For example, N-acetylglutamate synthase catalyzes acetylation of L-glutamate, the first step in arginine biosynthesis (Cunin et al., 1986). Acetylation of multiple aminoglycoside antibiotics has been reported to inactivate their bactericidal or bacteriostatic activity (Kawabe et al., 1975; Shaw and Hopwood, 1976; Garneau-Tsodikova and Labby, 2016), and acetylation of the antibiotic chloramphenicol disables its ability to inhibit translation (Bulkley et al., 2010). The chitin of insects, plants, and fungi and the peptidoglycan of the bacterial cell wall are composed of complex polysaccharides (Bernard et al., 2011). These polysaccharides often contain monomers such as N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc), which are monosaccharide derivatives acetylated on the primary amine (N), as their names suggest. However, these can be further acetylated on hydroxyl groups to increase resistance to degradative enzymes, such as lysozyme and autolysins (Bernard et al., 2011).
The most widely studied protein acetylations occur on amino groups, although acetylations on serine, threonine, and histidine residues of proteins also have been detected (Mukherjee et al., 2006, 2007; Paquette et al., 2012; Lee et al., 2015). Two distinct mechanisms for acetylation of amino groups of proteins can occur: lysine acetylation (N𝜀-acetylation) and N-terminal protein acetylation (Nα-acetylation). Lysine acetylation will be discussed in more detail below. Nα-acetylation is very common in eukaryotes, but rare in bacteria with only 47 proteins detected as N-terminally acetylated in E. coli, 117 identified in Pseudomonas aeruginosa, and 145 identified in Acinetobacter baumannii (Ouidir et al., 2015; Kentache et al., 2016; Brown et al., 2017). Acetylation is observed either on the free amino group of methionine or on the exposed amino acid after cleavage of the N-terminal methionine. The modification is usually co-translational in eukaryotes but post-translational in bacteria, mitochondria, and chloroplasts, as the methionine first must be deformylated (Polevoda and Sherman, 2003). Nα-acetylation has been shown to alter protein stability in eukaryotes and is typically irreversible (Helbig et al., 2010; Helsens et al., 2011). E. coli encodes three proteins that have demonstrated Nα-acetyltransferase activity: RimI, RimJ, and RimL. Interestingly, the only known targets of RimI, RimJ, and RimL are the ribosomal proteins S5, L12, and S18, respectively (Yoshikawa et al., 1987; Tanaka et al., 1989). The exact physiological effects of Nα-acetylation in E. coli are not known. There is evidence that RimJ plays a role in ribosome assembly, but it is not known whether this phenotype requires the Nα-acetyltransferase activity of RimJ (Yoshikawa et al., 1987; Roy-Chaudhuri et al., 2008). RimJ also plays a role in transcriptional regulation of the P pilus of uropathogenic E. coli, but the specific target and whether Nα-acetyltransferase activity is required remain unknown (White-Ziegler and Low, 1992; White-Ziegler et al., 2002). One defined role for Nα-acetylation comes from the ESAT-6 virulence factor of Mycobacterium tuberculosis. Cleavage of the N-terminal methionine of ESAT-6 reveals a threonine, which is then acetylated on the amino terminus. This acetylation alters protein–protein interactions with the binding partner CFP-10, which attenuates virulence (Okkels et al., 2004;Lange et al., 2014; Mba Medie et al., 2014).
Until recently, the only known internal residue that could be acetylated was lysine. However, recent work from a few groups found that a family of bacterial effector molecules from certain pathogenic species can acetylate serine, threonine, and histidine in addition to lysine residues. In some Yersinia species, YopJ can act as an acetyltransferase to modify serine and threonine residues of kinases in mammalian host cells to subvert host immunity (Mukherjee et al., 2006, 2007; Mittal et al., 2010; Paquette et al., 2012). Similarly, HopZ3, a YopJ family member from the plant pathogen Pseudomonas syringae, can acetylate plant proteins on lysine, serine, threonine, and histidine residues, as well as bacterial effector proteins AvrB and AvrB3 (Lee et al., 2015). In both cases, the acetylation by YopJ or HopZ3 prevents effective immune signaling, which results in successful bacterial infection. This does not appear to be an exclusively inter-kingdom modification, because O-acetylation of serine and threonine was found to occur on a proteome-wide scale in M. tuberculosis (Birhanu et al., 2017). Finally, O-acetylation of a single serine (S608) in acetyl-CoA synthetase (Acs) regulates the ability of a neighboring lysine (K610) to be acetylated in Streptomyces lividans (VanDrisse and Escalante-Semerena, 2018). The discovery of these unusual acetylated residues opens new questions concerning the prevalence of these modifications and their potential interaction with other PTMs.
N𝜀-Lysine Acetylation
N𝜀-lysine acetylation (hereafter acetylation) is the donation of an acetyl group onto the epsilon amino group of a lysine sidechain (Figure 1). Acetylation increases the size of the side chain and neutralizes the positive charge (+1 to 0) when the pH is less than the pKa of the specific lysine residue. Losing the positive charge and increasing the size of lysine disrupts salt bridges and introduces steric bulk that can alter protein–protein interactions, protein–DNA interactions, stability, and enzymatic activity (Glozak et al., 2005; Yang and Seto, 2008a; Xiong and Guan, 2012). This was first and extensively studied in the context of the histone code of eukaryotes (Phillips, 1963), where acetylation of the lysine rich C-terminal tail reduces protein–DNA binding. This remodels chromatin to activate gene transcription (Allfrey et al., 1964; Gershey et al., 1968; Vidali et al., 1968; Hebbes et al., 1988). However, recent mass spectrometric studies of acetylation have detected acetylation in all three domains of life (Zhao et al., 2003; Marsh et al., 2005; Spange et al., 2009; Zhang et al., 2009). The consortium of acetylated proteins is often called the acetylome. The first bacterial acetylome was reported in 2008 (Yu et al., 2008); since then >50 bacterial acetylomes have been reported (Christensen et al., 2019). This accumulation of bacterial acetylomes has resulted from advances in mass spectrometry-based proteomic approaches.
FIGURE 1.
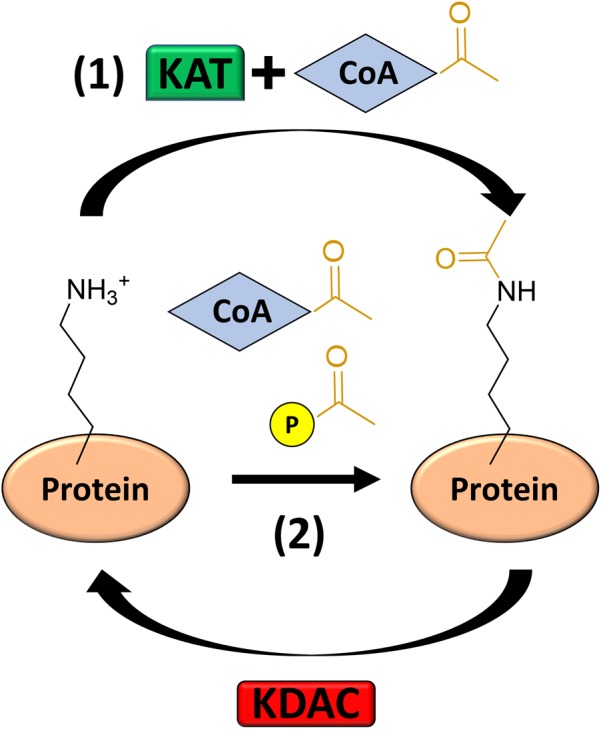
Mechanisms of Acetylation. Acetylation can be catalyzed (1) by a lysine acetyltransferase (KAT) using acetyl-CoA as the acetyl donor or (2) non-enzymatically by acetyl phosphate or acetyl-CoA. Some, but not all, acetylations can be reversed by a lysine deacetylase (KDAC).
Mass Spectrometry-Based Proteomics to Study Bacterial Acetylomes
Due to its high sensitivity and high-throughput, high-resolution mass spectrometry has emerged as a powerful technique to study lysine acetylation (Doll and Burlingame, 2015). For example, one can profile the global lysine acetylome of an organism, identifying acetyllysine (Kac) sites within a given proteome. Alternatively, one can use targeted approaches to examine specific acetyllysine sites of biological interest with high sensitivity and quantitative accuracy (Kuo and Andrews, 2013).
To compare the acetylome under different conditions, studies can be designed and mass spectrometric workflows can be optimized to not only identify Kac sites, but also to detect dynamic changes in terms of relative acetylation abundance over time, in isogenic sets of mutants, or in response to different treatments. These methods have been reviewed previously (Carabetta and Cristea, 2017). Such quantitative studies can be quite impactful and relevant. For example, two studies have provided insight into the dynamic changes that occur in response to the nutritional capacity of the bacterial environment (Kuhn et al., 2014; Schilling et al., 2019). Another study characterized the acetylome of drug resistant Salmonella strains, identifying associations between acetylation and drug resistance (Li L. et al., 2018). A recent review described how bacterial protein acetylation might mediate bacterial virulence (Ren et al., 2017). Quantitative mass spectrometric analysis of acetylation sites also has proven helpful in identifying novel lysine acetyltransferases (KATs) (Christensen et al., 2018; Carabetta et al., 2019).
In most mass spectrometry-based workflows, such as those described above, proteins are first subjected to proteolytic digestion, and peptides are immuno-enriched with anti-Kac antibodies before mass spectrometric analysis (Liu et al., 2008; Yu et al., 2008; Kuhn et al., 2014; Schilling et al., 2015, 2019; Xie et al., 2015). However, it is also possible to detect acetylation directly from bacterial whole lysates. For example, a large bacterial acetylation study was carried out by Nakayasu and co-workers to investigate the proteomes and acetylomes of 48 phylogenetically distant bacteria, identifying more than 9,000 acetylated proteins with approximately 190 acetylated proteins per organism, and more than 24,000 acetylated peptides with approximately 508 Kac per organism (Nakayasu et al., 2017). Many of the observed Kac sites were conserved across the phylogenetic spectrum, leading the authors to conclude that lysine acetylation is an evolutionarily conserved mechanism and that bacterial cells use it to regulate central metabolism (Nakayasu et al., 2017).
As mass spectrometric techniques and software have improved, the breadth of targets modified by acetylation has expanded. It is now clear that the extent of acetylation, the mechanisms by which acetylation occurs, the identities of acetylated proteins, and the locations of acetylation on those proteins can vary between organisms; yet, in most cases, there is great conservation in core cellular processes (Nakayasu et al., 2017). Such approaches have shown that acetylation occurs on proteins involved in a wide diversity of cellular process, especially central metabolism and translation (Wang et al., 2010; Weinert et al., 2013a; Baeza et al., 2014; Kuhn et al., 2014; Kosono et al., 2015; Schilling et al., 2015; Kentache et al., 2016; Birhanu et al., 2017; Jers et al., 2017; Nagano-Shoji et al., 2017; Post et al., 2017; Bontemps-Gallo et al., 2018; Gaviard et al., 2018; Sun et al., 2018; Vasileva et al., 2018; Yoshida et al., 2019). As stated above, many of these acetylations are conserved. This conservation has led some to hypothesize that the mechanisms of acetylation and the consequences of those acetylations are also conserved (Nakayasu et al., 2017).
Acetyltransferases in Bacteria
Multiple families of acetyltransferases have been discovered across phylogeny. Many of these families were discovered in relation to histone acetylation in eukaryotes. Two major families of acetyltransferases are the Gcn5-related N-acetyltransferases (GNATs) and MYST families (Yang et al., 1996; Vetting et al., 2005). Two smaller groups of acetyltransferases known as the p300/CBP family and the SRC family also exist, as well as the unique acetyltransferases TAFII250, TFIIIC, Rtt109, and CLOCK (Bannister and Kouzarides, 1996; Mizzen et al., 1996; Spencer et al., 1997; Marmorstein and Roth, 2001). The kinetic mechanisms of these acetyltransferases vary. While GNATs form a ternary complex allowing a direct transfer of the acetyl group, the MYST family can either use this ternary complex mechanism or utilize a ping-pong mechanism that generates an acetylated enzyme intermediate (Yan et al., 2002). In contrast to these larger families, the p300/CBP family has been shown to use a sequential mechanism called the Theorell-Chance mechanism (Liu et al., 2008). Since GNATs are the predominant family present in bacteria, we will focus on them.
The GNATs are present across bacterial phylogeny (Neuwald and Landsman, 1997) and modify a wide range of substrates, including not only proteins, but also small molecules or metabolites (Shaw et al., 1993; Davies and Wright, 1997; Vetting et al., 2005). GNATs are generally small proteins that bind acetyl-CoA (AcCoA). They have low sequence identity, but share a characteristic fold by which they are identified (Favrot et al., 2016). The number of GNATs contained in a given genome can vary wildly with Listeria monocytogenes containing ∼14 GNATs, but Streptomyces lividans containing ∼72 GNATs (Hentchel and Escalante-Semerena, 2015).
The lysine acetylation reaction commonly occurs in a ternary complex between the substrate, AcCoA, and the GNAT, or more specifically KAT. There are three key steps to this acetylation reaction: (1) deprotonation of the target lysine, (2) nucleophilic attack of the lysine on the carbonyl carbon of the acetyl group of AcCoA, and (3) protonation and dissociation of CoA (Vetting et al., 2005). Within the catalytic pocket of the KAT, there is often a catalytic glutamate that acts as a general base to deprotonate the target lysine. In the namesake Gcn5 enzyme of yeast, this glutamate is E173, which can be found conserved in many of the bacterial GNATs (Tanner et al., 1999; Trievel et al., 1999; Liszczak et al., 2011) and some MYST family members in eukaryotes (Yan et al., 2002; Berndsen et al., 2007). However, in the absence of this conserved glutamate, it is hypothesized that a series of residues form a water channel or “proton wire” whereby protons are carried away from active site to other residues or to the solvent (Dyda et al., 2000). Once the lysine is deprotonated, it can perform a nucleophilic attack on the carbonyl carbon of AcCoA, which is bound to the KAT via a positively charged patch that coordinates the CoA moiety via its pyrophosphate and pantothenate moieties (Dutnall et al., 1998; Lin et al., 1999; Tanner et al., 1999; Marmorstein and Roth, 2001; Liu et al., 2008). The newly acetylated substrate dissociates from the KAT, but for the CoA to dissociate, it must first be protonated. Often, a tyrosine in the active site donates a proton to the thiolate of CoA.
In bacteria, GNATs were first characterized as aminoglycoside N-acetyltransferases (Wright and Ladak, 1997; Wolf et al., 1998). Except those that belong to the YopJ effector protein superfamily (Lewis et al., 2011; Ma and Ma, 2016), currently every KAT discovered in bacteria belongs to the GNAT family (Latrasse et al., 2008; Thao and Escalante-Semerena, 2011; Dancy and Cole, 2015). In E. coli, five KATs have been identified with YfiQ (also called Pat, PatZ, or Pka) being the most heavily studied (Wang et al., 2010). Homologs of YfiQ exist in many bacterial species, as described below, but additional KATs with distinctly different structures have been identified, including homologs of the other KATs in E. coli: RimI, YiaC, YjaB, and PhnO (Christensen et al., 2018).
Deacetylases in Bacteria
To make a PTM versatile and energetically efficient, the ability to add and remove the PTM is essential. Lysine acetylation can be reversed via the action of a lysine deacetylase (KDAC). Two types of deacetylases are known: NAD+-dependent sirtuins (Blander and Guarente, 2004) and Zn2+-dependent deacetylases (Marmorstein, 2001; Gregoretti et al., 2004; Yang and Seto, 2008b; Lombardi et al., 2011). Putative homologs of both families are encoded by bacteria, but only a few have been shown to function as true deacetylases (Hildmann et al., 2007; Hentchel and Escalante-Semerena, 2015).
The Zn2+-dependent deacetylases are simple hydrolases that cleave the acetyl group from the lysine to release acetate. The reaction requires that a conserved histidine residue act as a general base to activate a metal-bound water that attacks the carbonyl of the acetyl group. This class of deacetylase has yet to be found in E. coli, but AcuC of Bacillus subtilis (Gardner et al., 2006) and LdaA of Rhodopseudomonas palustris (Crosby et al., 2010) are family members. Enzymes of the hydrolase family can be inhibited by the addition of butyrate (Davie, 2003).
Sirtuins bind NAD+ through a Rossmann fold domain (Sanders et al., 2010) and use the NAD+ as a co-substrate to remove acetyl groups from proteins; the products are then deacetylated lysine, nicotinamide (NAM) and 2′-O-acetyl-ADP-ribose. Since NAD+ levels are linked to nutritional status of the cell, it is thought that sirtuins allow the cell to recognize these changes and adjust acetylation accordingly (Wimpenny and Firth, 1972; Frye, 2000). Furthermore, sirtuins may regulate their own activity by producing NAM. NAM inhibits non-competitively by condensing with an ADP-ribose-like intermediate formed during the deacetylation reaction; this condensation blocks the progression of the reaction (Denu, 2003) at physiological levels (Wimpenny and Firth, 1972; Bitterman et al., 2002; Jackson et al., 2003; Sauve and Schramm, 2003; Zhao et al., 2004; Avalos et al., 2005). NADH also competitively inhibits the reaction at physiological levels (Schmidt et al., 2004; Guarente, 2005; Porcu and Chiarugi, 2005).
In many members of the Enterobacteriaceae, the sirtuin CobB exists as two isoforms. In Salmonella enterica and E. coli, a shorter isoform (CobBS) and the longer isoform (CobBL) are transcribed from two different transcription start sites (Tucker and Escalante-Semerena, 2010; Umehara et al., 2018). These isoforms differ by an N-terminal amphipathic stretch of 37 amino acids. Though the purpose of this N-terminal extension is unknown, both forms of the CobB protein were shown to be biologically active as deacetylases in S. enterica (Tucker and Escalante-Semerena, 2010).
While sirtuins are classically thought of as deacetylases, it appears that they may have a broader specificity than originally thought. Sirtuins in the mitochondria have been reported to desuccinylate (Du et al., 2011; Colak et al., 2013), demalonylate (Du et al., 2011; Peng et al., 2011; Park et al., 2013), dipropionate (Smith and Denu, 2007), deglutarylate (Tan et al., 2014), decrotonylate (Bao et al., 2014), and debutyrylate (Smith and Denu, 2007) lysines in addition to deacetylate. Certain human sirtuins, such as SIRT6, have a preference for removal of long-chain acyl groups from lysines. These long-chain acyl groups can be a long as 16 carbons, as in the case of palmitoylation (Feldman et al., 2013). Sirtuins of archaeon Archaeoglobus fulgidus have similarly been shown to desuccinylate and demyristoylate peptides (Ringel et al., 2014). Bacterial sirtuins have similar abilities to desuccinylate (Colak et al., 2013), dipropionate (Garrity et al., 2007; Sun et al., 2016b), and possibly debutyrylate (Xu et al., 2018a) lysines. Many of these PTMs occur on the same lysine, possibly implicating sirtuins as a general cleanup enzyme for certain lysines. Further complicating the possible role of sirtuins, there is evidence that sirtuins can ADP-ribosylate proteins using NAD+ as a substrate (Frye, 1999).
Effects of Enzymatic Acetylation/Deacetylation on Physiology and Pathogenesis
Several studies have explored the role of enzymatic acetylation on bacterial physiology, often through reverse genetics approaches where the deacetylase, acetyltransferase, or both are deleted or overexpressed. In this section, we will focus on CobB and YfiQ of E. coli and their homologs in S. enterica, as they have been studied the most extensively.
The earliest and thus most comprehensively understood roles for YfiQ and CobB are their opposing effects on the acetylation status of Acs, an enzyme that irreversibly converts acetate into AcCoA via two half reactions. Acs requires lysine 609 (K609) for the first half-reaction, which forms an acetyl-AMP intermediate (Starai et al., 2002). Thus, mutation or modification of this residue leads to a catalytically inactive enzyme (Starai et al., 2002; Starai and Escalante-Semerena, 2004a,b; Sun et al., 2016b). Indeed, an epistasis analysis of Δacs, ΔcobB, and ΔyfiQ mutants showed that regulation of Acs is essential for growth on acetate. In both S. enterica and E. coli, Δacs mutants cannot grow on less than 10 mM acetate as the sole carbon source. This is due to the low affinity of the alternative Pta-AckA pathway for acetate. A ΔcobB mutant phenocopies a Δacs mutant, while the double ΔyfiQΔcobB or ΔpatΔcobB mutants can grow on less than 10 mM acetate (Starai et al., 2003; Castaño-Cerezo et al., 2011). These results confirm that YfiQ/Pat-dependent acetylation of Acs shuts down acetate utilization through Acs and that deacetylation by CobB restores that activity (Starai et al., 2002; Castaño-Cerezo et al., 2011). Acs was found to be regulated in the exact same fashion in more than 15 organisms tested, ranging from bacteria to humans (Table 1) (Kim and Yang, 2011). Beyond Acs, this reversible lysine acetylation has been extended to other AMP-forming acyl-CoA synthetases, which are proteins that share a similar structure and the same conserved acetylated lysine found in Acs. R. palustris possesses 40 of these Acs-like proteins; 10 of them are reversibly acetylated on this conserved lysine (Crosby et al., 2010, 2012). As in Acs, acetylation inactivates these enzymes.
Table 1.
Proteins assessed for regulation by acetylation.
| Organism | Target | Target lysine(s) | Acetylation | Deacetylation | Acetylation | Cite |
|---|---|---|---|---|---|---|
| protein | mechanism | mechanism | effect | |||
| Actinosynnema mirum | Acs | 620 | AmiPatA and 23 homologs | Not determined | Inhibits activity | Lu et al., 2017 |
| Bacillus subtilis | Acs | 549 | AcuA | AcuC, SrtN | Abolishes activity | Gardner et al., 2006; Gardner and Escalante-Semerena, 2008, 2009 |
| Eno | 339, 390 | In vitro AcP | Not determined | Abolishes activity | Nakayasu et al., 2017 | |
| HBsu | 3, 18, 41, 80 | YfmK | Not determined | Decreases DNA binding | Carabetta et al., 2019 | |
| TufA | 42 | Not AcuA, not AcP | AcuC, SrtN | No effect | Suzuki et al., 2019 | |
| MreB | 240 | Not determined | Not determined | Reduces cell length, width, and peptidoglycan thickness | Carabetta et al., 2016 | |
| CshA | 244, 296 | Unknown | Not determined | Enhances induction of SigX and SigM-dependent genes | Ogura and Asai, 2016; Ogura and Kanesaki, 2018 | |
| Borrelia burgdorferi | GapA | Not determined | AcP | Not determined | Inhibits activity | Bontemps-Gallo et al., 2018 |
| LDH | Not determined | AcP | Not determined | Mixed results | Bontemps-Gallo et al., 2018 | |
| Corynebacterium glutamicum | PEPC | 653 | Not determined | NCgl0616 | Inhibits activity | Nagano-Shoji et al., 2017 |
| Escherichia coli | ArgRS | 126, 408 | AcP | CobB | Impairs tRNA charging | Ye et al., 2017 |
| CRP | 100 | AcP | Not determined | Reduces interaction with RNAP, enhances protein stability | Davis et al., 2018 | |
| LeuRS | 619, 624, 809 | AcP | CobB | Impairs tRNA charging | Ye et al., 2017 | |
| Mdh | 99, 140 | AcP | CobB for 140, none for 99 | Cooperatively increases activity | Fan et al., 2015; Venkat et al., 2017b | |
| RpoA | 291 | AcP | Not determined | Reduces cpxP transcription | Lima et al., 2012 | |
| TyrRS | 85, 235, 238 | AcP | CobB | Impairs tRNA charging | Venkat et al., 2017a | |
| RcsB | 154 | AcP | CobB | Enhances migration, impairs acid survival | Hu et al., 2013; Castaño-Cerezo et al., 2014 | |
| TopA | 13, 45, 346, 488 | AcP | CobB | Reduces relaxation activity by inhibiting DNA binding and cleavage activity | Zhou et al., 2017, 2018 | |
| DnaA | 178 | AcP, YfiQ | CobB | Inhibits ATP binding | Zhang et al., 2016 | |
| DnaA | 243 | AcP, YfiQ | CobB | Inhibits oriC binding at low affinity sites | Li S. et al., 2017 | |
| CheY | 92, 109 | Acs, AcCoA | Acs, Pta, CobB | Inhibits interaction with CheA, FliM, and CheZ Activates CheY, promotes clockwise rotation |
Barak and Eisenbach, 2001; Barak et al., 2006; Li R. et al., 2010; Liarzi et al., 2010 | |
| YfiQ | 146, 149, 391, 446, 635, 819 | AcCoA | CobB | Favors formation of octamers | de Diego Puente et al., 2015 | |
| Eno | 342, 393 | In vitro AcP | Not determined | Abolishes activity | Nakayasu et al., 2017 | |
| AceA | 308 | Not determined | CobB | Inhibits activity | Castaño-Cerezo et al., 2014 | |
| AlaRS | 73 | Not determined | CobB | Inhibits activity | Umehara et al., 2018 | |
| Mat | 12 lysines | Not determined | CobB (9 lysines susceptible) | Inhibits activity, may affect dimerization | Sun et al., 2016a | |
| NhoA | 214, 281 | Not determined | CobB | Inhibits activity | Zhang et al., 2013 | |
| RcsB | 180 | SePat, EcYfiQ | SeCobB, EcCobB | Inhibits DNA binding | Thao et al., 2010 | |
| Acs | 609 | YfiQ | CobB | Inhibits activity by preventing first half reaction | Zhao et al., 2004; Castaño-Cerezo et al., 2011, 2014; | |
| protein | mechanism | mechanism | effect | |||
| RNase II | 501 | YfiQ | CobB | Inhibits RNase II activity due to reduced substrate binding | Song et al., 2016 | |
| RNase R | 544 | YfiQ | NONE – not CobB | Destabilizes protein | Liang et al., 2011; Liang and Deutscher, 2012 | |
| RpoA | 298 | YfiQ | Not determined | Enhances cpxP transcription | Lima et al., 2011 | |
| Micromonospora aurantiaca | Acs | 619 | MaKat | Not determined | Inhibits activity | Xu et al., 2014 |
| Mycobacterium bovis BCG | Acs | 616 | KATbcg (BCG_1055) | Not determined | Abolishes activity | Nambi et al., 2013 |
| FadD13 | 487 | KATbcg (BCG_1055) | Not determined | Inhibits activity | Nambi et al., 2013 | |
| Mycobacterium smegmatis | FadD33 | 511 | MsPat | MSMEG_5175 | Abolishes activity | Vergnolle et al., 2013 |
| MbtA | 546 | MsPat | Rv1151c | Inhibits activity | Vergnolle et al., 2016 | |
| PrpE | 586 | MsPat | Not determined | Abolishes activity | Xu et al., 2018b | |
| MSMEG_4207 (USP) | 104 | MtPatA, MsPatA | Not determined | Unknown | Nambi et al., 2010 | |
| Ku | 29, 40 | Not determined | Not determined | Correlates with impaired NHEJ activity | Zhou et al., 2015 | |
| HupB | 86 | Not determined | Not determined | Prevents small colony variant formation | Sakatos et al., 2018 | |
| Acs | 589 | PatA | SrtN | Impairs growth on acetate | Hayden et al., 2013 | |
| Mycobacterium tuberculosis | DUSP16 (eukaryotic) | 55 | Eis | Not determined | Inhibits activation of JNK pathway | Kim et al., 2012 |
| Histone H3 (eukaryotic) | Not determined | Eis | Not determined | Enhances binding to the IL-10 promoter | Duan et al., 2016 | |
| HupB | 32 | Eis | Rv1151c | Inhibits DNA binding | Ghosh et al., 2016; Green et al., 2018 | |
| Acs | 617 | MsPat | Rv1151c, MSMEG_5175 | Abolishes activity | Xu et al., 2011; Nambi et al., 2013; Noy et al., 2014 | |
| MbtA | 542 | MsPat | Rv1151c | Inhibits activity | Vergnolle et al., 2016 | |
| DosR | 182 | MtPat | Rv1151c | Inhibits DNA binding | Bi et al., 2018; Yang et al., 2018 | |
| FadD2, FadD4, FadD5, FadD10, FadD12, FadD13, FadD22, FadD35 | 551, 525, 519, 519, 523, 487, 480, 529 | MtPat | MSMEG_5175 | Impairs palmitoyl-AMP synthesis activity for FadD2, FadD5, and FadD15 | Nambi et al., 2013 | |
| PtpB | 224 | MtPat | Rv1151c | Decreases reaction rate due to reduced Vmax | Singhal et al., 2015 | |
| HspX | 64, 78, 85 | Not determined | Not determined | Reduced immunogenicity | Liu et al., 2014 | |
| ICL1 (Rv0467) | 322 331 392 |
Not determined | Not determined | Inhibits activity (K322) Enhances activity (K331) Enhances activity (K392) |
Xie et al., 2015; Bi et al., 2017 | |
| Histone H3 (eukaryotic) | 9, 14 | Rv3423.1 | Not determined | Inhibits DNA binding by histone H3 | Jose et al., 2016 | |
| Myxococcus xanthus | Acs | 622 | MxKat | Not determined | Inhibits activity | Liu et al., 2015 |
| Neisseria gonorrhoeae | PilT | 117 | AcP | Not determined | May contribute to membrane association, | Hockenberry et al., 2018 |
| protein | mechanism | mechanism | effect | |||
| alters microcolony formation | ||||||
| Porphyromonas gingivalis | RprY | Not determined | PgPat | CobB | Inhibits DNA binding | Li Y. et al., 2018 |
| pro-RgpB | 247, 248 | VimA, PG1842 | Not determined | Permits proper processing of pro-RgpB | Mishra et al., 2018 | |
| Rhodobacter sphaeroides | FnrL | 175, 213, 223 | AcP | RsCobB | Impairs transcriptional activation | Wei et al., 2017 |
| Rhodopseudomonas palustris | FadD | 546 | RpPat | Not determined | Inhibits activity | Crosby et al., 2012 |
| HcsA | 524 | RpPat | LdaA | Inhibits activity | Crosby et al., 2012 | |
| LcsA | 499 | RpPat | LdaA | Inhibits activity | Crosby et al., 2012 | |
| PimA | 534 | RpPat | Not determined | Inhibits activity | Crosby and Escalante-Semerena, 2014 | |
| Acs | 606 | RpPat, KatA | SrtN and LdaA | Inhibits activity | Crosby et al., 2010, 2012 | |
| AliA | 532 | RpPat, KatA | SrtN and LdaA | Inhibits activity | Crosby et al., 2010, 2012 | |
| BadA | 512 | RpPat, KatA | SrtN and LdaA | Inhibits activity | Crosby et al., 2010, 2012 | |
| FcsA | 496 | RpPat, KatA | LdaA | Inhibits activity | Crosby et al., 2012 | |
| HbaA | 503 | RpPat, KatA | SrtN and LdaA | Inhibits activity | Crosby et al., 2010, 2012 | |
| IbuA | 539 | RpPat, KatA | LdaA | Inhibits activity | Crosby et al., 2012 | |
| PrpE | 598 | RpPat, KatA | LdaA | Inhibits activity | Crosby et al., 2012 | |
| Saccharomyces cerevisiae | Pck1p | 514 | Esa1 | Sir2 | Enhances activity | Lin et al., 2009 |
| Saccharopolyspora erythraea | AcsA2 | 611 | AcP | Not determined | Inhibits activity by reducing affinity for acetate | You et al., 2014 |
| AcsA1, AcsA2, AcsA3 | 620, 628, 615 | AcuA | SrtN | Inhibits activity | You et al., 2014, 2017 | |
| GlnA1 | 179, 357 | AcuA | SrtN | Enhances GlnA1 interaction with GlnR, which enhances DNA binding affinity | You et al., 2016 | |
| GlnA4 | 319 | AcuA | SrtN | Inhibits activity | You et al., 2016 | |
| Salmonella enterica | AceA | 308 | Pat | CobB | Inhibits activity | Wang et al., 2010; Crosby et al., 2012 |
| AceK | 72, 83, 553 | Pat | CobB | Enhances activation of ICDH | Wang et al., 2010; Crosby et al., 2012 | |
| GapA | 331 | Pat | CobB | Enhances glycolytic activity, decreases gluconeogenesis activity | Wang et al., 2010; Crosby et al., 2012 | |
| HilD | 297 | Pat | Not CobB | Enhances stability, inhibits DNA binding | Sang et al., 2016, 2017 | |
| PhoP | 201 | Pat | CobB | Inhibits DNA binding | Ren et al., 2016 | |
| PrpE | 592 | Pat | CobB | Abolishes activity | Garrity et al., 2007 | |
| Lrp | 36 | Pat | CobB | Inhibits DNA binding | Qin et al., 2016 | |
| Acs | 609 | Pat | CobB | Inhibits activity by preventing first half reaction | Starai et al., 2002, 2005; Starai and Escalante-Semerena, 2004b | |
| Charged aminoacyl tRNAs | Amino terminus | TacT | N/A | Inhibits translation | Cheverton et al., 2016; VanDrisse et al., 2017 | |
| protein | mechanism | mechanism | effect | |||
| TacA | 44 | TacT | CobB | Enhances TacT activity | VanDrisse et al., 2017 | |
| Streptomyces coelicolor | Acs | 610 | Not determined | CobB1 | Inhibits activity | Mikulik et al., 2012 |
| GlnR | 142, 153, 159, 200 | Not determined | CobB2 | Alters DNA binding | Amin et al., 2016 | |
| Streptomyces griseus | StrM | 70 | SGR1683, EcYfiQ | Not determined | Inhibits activity | Ishigaki et al., 2017 |
| Streptomyces lividans | Aacs | 617 | SlPat | SeCobB | Inhibits activity | Tucker and Escalante-Semerena, 2013 |
| Acs | 610 | SlPatA, SlPatB | SlSrtA | Inhibits activity | Tucker and Escalante-Semerena, 2013; VanDrisse and Escalante-Semerena, 2018 | |
| Streptomyces venezuelae | Acs | 665 | SvePatA | Not determined | Inhibits activity | Lu et al., 2017 |
| Sulfolobus solfataricus | Alba | 16 | Pat | Sir2 | Inhibits DNA binding | Marsh et al., 2005 |
| Thermus thermophilus | IPMS | 332 | AcCoA | TT_C0104 | Inhibits activity | Yoshida et al., 2019 |
| Vibrio cholerae | Acs | 609 | YfiQ | CobB | Inhibits activity | Liimatta et al., 2018 |
| Yersinia pestis | PhoP | Not determined | YfiQ | CobB | Not determined | Liu et al., 2018 |
Other proteins have been shown to be targets of both YfiQ and CobB. For example, K501 of RNase II from E. coli is a target of YfiQ and CobB (Song et al., 2016). Acetylation of K501 by YfiQ inhibits substrate binding, which results in reduced exoribonuclease activity. Treatment of RNase II with CobB restores this activity. YfiQ and CobB also reversibly acetylate K201 of PhoP from S. enterica. A member of a two-component regulatory system that responds to low Mg2+ and acid stress, Pat-dependent acetylation of K201 decreases the affinity of PhoP for its DNA binding site and thus reduces expression of PhoP-regulated genes (Ren et al., 2015, 2016). CobB restores DNA binding by deacetylating PhoP. Finally, the highly conserved global transcriptional regulator Lrp (Brinkman et al., 2003; Martínez-Antonio and Collado-Vides, 2003) has been shown to be acetylated on K36 in S. enterica, which prevents DNA binding (Qin et al., 2016). CobB treatment permits DNA binding of Lrp.
While the mechanistic role of reversible lysine acetylation on Acs and a few other proteins is established, multiple phenotypes have been found for Δpat and ΔcobB mutants in S. enterica that are not directly attributed to acetylation of a specific enzyme, but are consistent with the opposing effects of these enzymes. For example, compared to wild-type cells, a Δpat mutant of S. enterica grows more slowly on minimal glucose but more rapidly on minimal citrate, whereas a ΔcobB mutant has the exact opposite phenotype (Wang et al., 2010). The Δpat mutant has reduced flux through glycolysis and the TCA cycle, while a ΔcobB mutant again has the opposite phenotype and promotes gluconeogenesis (Wang et al., 2010). For the latter phenotype, a ΔcobBΔpat double mutant behaves like a Δpat mutant, suggesting that CobB is dispensable in the absence of acetylation via Pat, as expected if the role of CobB was simply to deacetylate Pat-dependent acetylations. Taken together, these results support the hypothesis that Pat and CobB somehow regulate central metabolism. However, the Pat- and CobB-sensitive lysines responsible for these behaviors are not known. Acetylation by Pat was reported on isocitrate lyase (AceA), isocitrate dehydrogenase (Icd), and isocitrate dehydrogenase kinase/phosphatase (AceK), and this acetylation was shown to reduce activity for each protein (Wang et al., 2010); however, these results were called into question when another group could not confirm these findings (Crosby et al., 2012).
YfiQ/Pat, CobB, and their homologs also appear to be involved in certain stress responses. In contrast to the metabolic examples above, some of these effects might occur by modifying different lysines, as deletion of either enzyme sometimes does not result in opposite phenotypes. For example, when exposed to acid stress, both E. coli and Yersinia pestis have reduced survival when either yfiQ or cobB are deleted; in contrast, S. enterica survives better when pat is deleted and more poorly when cobB is deleted (Castaño-Cerezo et al., 2014; Ren et al., 2015; Liu et al., 2018). When exposed to heat stress, survival of E. coli diminishes when yfiQ is deleted but increases when cobB is deleted (Ma and Wood, 2011). In contrast, Y. pestis survival is diminished in both ΔcobB and ΔyfiQ mutants when exposed to heat, cold, or high salt stress (Liu et al., 2018). When exposed to reactive oxygen species, the E. coli ΔcobB mutant has enhanced survival, whereas the ΔyfiQ mutant behaves like its wild-type parent. In a more distantly related organism, M. tuberculosis, loss of its sirtuin (Rv1151c) promotes growth in acidic pH (Bi et al., 2017). Finally, the acetyltransferase PatA in Mycobacterium smegmatis acetylates the Universal Stress Protein (MSMEG_4207) (Nambi et al., 2010). Taken together, the results of these studies suggest a common role for acetylation in stress responses. The pleiotropic phenotypes of ΔyfiQ/pat and ΔcobB mutants provide evidence that the lysines they acetylate and deacetylate control a diverse set of general stress pathways.
The Regulator of Capsule Synthesis B (RcsB) can be reversibly acetylated through YfiQ and CobB in vitro. RcsB, a member of the two-component response regulator family, is a global transcriptional regulator of E. coli and S. enterica, regulating approximately 5 and 20% of their respective genomes (Prüss et al., 2006; Wang et al., 2007). RcsB promotes capsule biosynthesis and biofilm formation, while impairing transcription of genes required for motility. In E. coli and S. enterica, RcsB is acetylated on multiple residues. One of those lysines (K180) can be acetylated in vitro in a YfiQ/Pat-dependent manner and deacetylated by CobB (Thao et al., 2010). K180 is found in the RcsB DNA binding motif, and acetylation of this residue reduces DNA binding, which can then be reversed by CobB. However, there is no evidence to support YfiQ/Pat as the cause of K180 acetylation in vivo, as mutants that delete these KATs have no obvious motility defect (Hu et al., 2013). In contrast, two independent studies found CobB to be an inhibitor of migration in E. coli (Castaño-Cerezo et al., 2014; Gallego-Jara et al., 2017). A ΔcobB mutant has more numerous and longer flagella than the parent strain, which could partially be explained by the ability of CobB to deacetylate RcsB (Gallego-Jara et al., 2017).
Pat has been strongly implicated in S. enterica survival and infection. Pat enhances invasion of the bacteria into HeLa cells and enhances survival of the bacteria within macrophages (Sang et al., 2016). Pat also helps promote survival under acid stress, which may be partially responsible for the macrophage survival phenotype (Ren et al., 2015). Importantly, expression of the Salmonella pathogenicity island (SPI-1) that is required for virulence is enhanced by Pat (Sang et al., 2016). SPI-1 is controlled by transcription factor HilD, and acetylation on K297 of HilD by Pat was suggested to stabilize the protein, which could enhance SPI-1 transcription. The role of Pat in virulence was further established by infecting mice with Δpat and ΔcobB mutants. The Δpat mutants were hypovirulent and caused reduced mortality, implicating Pat and therefore acetylation as necessary for infection (Sang et al., 2016). However, ΔcobB mutants had little effect. The effect by Pat might be due, at least in part, to its ability to acetylate of K201 of PhoP, as a K201Q variant that mimics the constitutively acetylated isoform were hypovirulent (Ren et al., 2016). Like S. enterica, a ΔyfiQ mutant of Yersinia pestis, the causative agent of plague, is hypovirulent in mice (Liu et al., 2018), but unlike S. enterica, the ΔcobB mutant is equally attenuated.
The above examples demonstrate how mechanistically similar machinery has evolved in different organisms to suit their specific niches. In many cases, YfiQ/Pat and CobB act antagonistically to reversibly acetylate a specific lysine. However, in response to certain stresses, loss of either YfiQ/Pat or CobB can be detrimental to the survival of the bacterium. The pathogenicity data from S. enterica and Y. pestis suggest that, while some commonalities may exist between the requirements for acetylation in certain scenarios, there are many cases where YfiQ/Pat and CobB do not always act in opposition. These observations provide strong evidence that these enzymes can and do act independently and should not be considered a system.
Non-Enzymatic Acetylation Via AcP and AcCoA
Whereas KAT-catalyzed acetylation has been known for some time, evidence has accumulated showing that non-enzymatic acetylation is also possible. In vitro, AcCoA and the bacterial metabolite acetyl phosphate (AcP) can non-enzymatically acetylate lysine residues on many proteins. In vivo, the impact that AcP has on the acetylome has been assessed in multiple bacteria. In the cases of E. coli (Weinert et al., 2013a; Kuhn et al., 2014), B. subtilis (Kosono et al., 2015; Suzuki et al., 2019), and Neisseria gonorrhoeae (Post et al., 2017), AcP is the predominant acetyl donor.
Chemical reactions are the basis of all life. While enzymatic catalysis is a primary driver in the kinds of substrates used and products formed, the possible chemical reactions of those substrates and products greatly exceeds those that are catalyzed (Golubev et al., 2017). Many of these uncatalyzed reactions are thought to be deleterious to the cell, causing macromolecular damage, such as oxidation of DNA or proteins. Often, the damage can be caused by products of the cell’s own metabolism, such as reactive oxygen species (Madian and Regnier, 2010). Indeed, non-enzymatic modification has been implicated in multiple human diseases (Brownlee, 2005). As cells have evolved alongside this damaging potential of their own metabolism, it is not surprising that repair pathways exist to mitigate this damage. Perhaps cells have evolved ways to utilize these extra-metabolic occurrences as means to regulate their cellular processes. Specifically, in this context, we will consider acetylation via two acetyl donors, AcCoA and AcP.
The ability of AcCoA and AcP to non-enzymatically modify proteins is not a new idea. AcP has been shown to chemically acetylate amine, thiol, and hydroxyl groups (Di Sabato and Jencks, 1961; Jencks, 1969). In the 1970s, it was discovered that AcP could non-enzymatically acetylate histones, albumin, and polylysine (Ramponi et al., 1975). Due to its high energy nature (ΔG° of -43.1 kJ/mol), which is even greater than that of AcCoA (ΔG° of -31.4 kJ/mol), AcP readily hydrolyzes in water and can acetylate without the need for an enzyme (Koshland, 1952; Voet and Voet, 1990). Furthermore, the concentrations needed to achieve non-enzymatic acetylation in vitro are comparable to or below physiological levels, which in E. coli are around 300–500 μM for AcCoA and between 200 μM and 3 mM for AcP (Vallari and Rock, 1985; Klein et al., 2007; Ramos-Montañez et al., 2010). In vitro acetylation reactions are often carried out at basic pH. Basic pH promotes deacetylation of lysine, which may prime the lysine for acetylation, but also increases the rate at which AcP and AcCoA dissociate (Koshland, 1952; Di Sabato and Jencks, 1961). Susceptibility to non-enzymatic acetylation differs for each lysine and each acetyl donor used. Using glutamate dehydrogenase (GDH) as an example, it was shown a single lysine (e.g., K503) could be acetylated at different rates using two different acetyl donors, AcP and AcCoA. When comparing acetylation by a single acetyl donor, multiple lysines (e.g., K110, K415, and K503) on GDH were acetylated at different rates from one another. These in vitro acetylation reactions with AcP and AcCoA showed that acetylation rates of these specific lysines can vary by three orders of magnitude. Furthermore, some lysines were completely non-reactive under these conditions. These findings suggest that regulation of acetylation depends on the environmental context and thus reactivity of the lysine (Baeza et al., 2015).
Assessment of non-enzymatic acetylation in vivo by AcCoA is difficult. In most cases, it is an essential molecule that if depleted would result in lack of viability. However, attempts to recreate the mitochondrial environment have shown that AcCoA could acetylate proteins at physiological levels in vivo (Wagner and Payne, 2013). Furthermore, attempts to reduce the levels of AcCoA, such as by draining acetyl groups into an acetyl group polymer such as polyhydroxybutyrate, do not avoid any contribution by KATs (Hu, 2013). Work in an organism where AcCoA can be non-essential, such as the reduced genome bacterium Borrelia burgdorferi that solely synthesizes AcCoA to generate mevalonate, a precursor of cell wall lipid I, might be a useful reductionist system to determine whether this truly occurs in vivo (Bontemps-Gallo et al., 2018). Without knowing all possible KATs and eliminating them, it is hard to truly ever say whether an AcCoA-dependent acetylation is truly non-enzymatic in vivo.
Fortunately, modulation or complete elimination of AcP levels is much simpler and less detrimental to many bacteria, which greatly facilitates studying AcP-dependent acetylation. In E. coli and many other bacteria, AcP is generated as an intermediate of the acetate fermentation pathway catalyzed by the reversible enzymes acetate kinase (AckA) and phosphotransacetylase (Pta) (Rose et al., 1954). Pta catalyzes the conversion of AcCoA and inorganic phosphate to AcP and free CoA. AckA then converts AcP into acetate by transferring the phosphoryl group to ADP to generate one molecule of ATP (Figure 2). In the presence of high acetate concentrations (>10 mM), the pathway can run in reverse, synthesizing AcCoA from acetate, ATP, and CoA (Brown et al., 1977; Kumari et al., 1995). By genetically manipulating this pathway, the levels of AcP and thus acetylation can be altered. For example, if cells are grown on glucose, the cells ferment acetate and generate AcP as an intermediate. If one prevents the conversion of AcCoA to AcP by deleting Pta or the entire Pta AckA pathway, AcP cannot be generated. When these mutants are analyzed by western blot, the intensity of the acetylated proteins is greatly reduced to background levels (Weinert et al., 2013a; Kuhn et al., 2014). It is worth noting that acetylation can still be detected by mass spectrometry in Δpta and Δpta ackA mutants, but the relative amount of acetylation and number of acetylated lysines is greatly reduced. On the other hand, by deleting ackA, AcP accumulates to 10–20 mM (Klein et al., 2007; Wolfe, 2010; Weinert et al., 2013a), which can be seen via western blot as a greatly intensified acetylation profile relative to wild-type cells (Weinert et al., 2013a; Kuhn et al., 2014). Mass spectrometry confirms that many more unique lysines on many more unique proteins become acetylated in this mutant, and those lysines that are detected as acetylated in wild-type become further acetylated as the proportion of the total pool of each unique peptide shifts toward acetylated (Weinert et al., 2013a; Kuhn et al., 2014).
FIGURE 2.
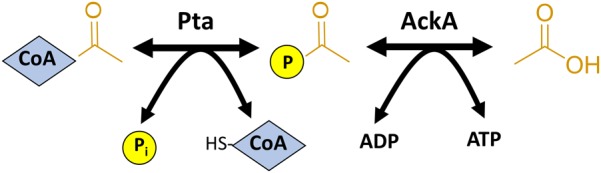
The Pta-AckA acetate fermentation pathway generates AcP as an intermediate. Phosphotransacetylase (Pta) converts AcCoA to AcP by substituting CoA for inorganic phosphate (Pi). Acetate kinase (AckA) converts AcP to acetate, generating an ATP in the process. This pathway is reversible.
Targets of Non-Enzymatic, AcP-Dependent Acetylation
To date, only a handful of groups have tried to directly determine the impact of acetylation by AcP on the acetylome of an organism (Weinert et al., 2013a, 2017; Kuhn et al., 2014; Post et al., 2017; Bontemps-Gallo et al., 2018; Reverdy et al., 2018). This has been achieved by mass spectrometry analysis of wild-type cells and comparing the acetylated proteins to acetylated proteins cells where the levels of AcP have been manipulated, usually by deleting ackA to accumulate AcP in cells or by deleting pta to prevent synthesis of AcP. Due to the prevalence of AcP-dependent acetylation, especially under conditions when cells are exposed to high carbon concentrations, much of the acetylation detected in wild type bacteria is likely a result of acetylation by AcP (Schilling et al., 2015).
These studies result in large data sets with hundreds of lysines acetylated from multiple organisms. Often, to get a picture of what processes these acetylated lysines may be involved in, the modified proteins can be grouped by keyword enrichment, gene ontology term enrichment, and Kegg pathway enrichment. Across these studies and grouping strategies, the modified proteins fall into multiple processes, but the two most common are translation and central metabolism.
From the current E. coli data sets, it appears that most of the ribosomal subunits, most of the aminoacyl-tRNA ligases, and all of the elongation factors are acetylated on multiple lysines in an AcP-dependent manner. In contrast, AcP acetylates only one lysine on one initiation factor (Kuhn et al., 2014; Schilling et al., 2015). Whether any of these acetylations impact the rate or fidelity of translation has yet to be determined. However, it has been shown that charging of tRNAs can be inhibited by AcP-dependent acetylation of certain aminoacyl-tRNA ligases (Venkat et al., 2017a; Ye et al., 2017).
Similarly, most central metabolic enzymes are acetylated in an AcP-dependent manner; this is true for all of the glycolytic pathways of E. coli and B. subtilis (i.e., Embden–Meyerhof–Parnas, Pentose Phosphate, and Entner–Doudoroff). Currently, the functional consequences of most of these acetylations remain largely undetermined. However, it is reported that AcP-dependent acetylation abolishes the activity of the glycolytic enzyme enolase from both E. coli and B. subtilis (Nakayasu et al., 2017) and inhibits the activity of glyceraldehyde 3-phosphate dehydrogenase A from Borrelia burgdorferi (Bontemps-Gallo et al., 2018). The TCA cycle is also highly acetylated by AcP. Thus far, malate dehydrogenase is the rare example of a central metabolic enzyme whose activity is enhanced by AcP-dependent acetylation (Venkat et al., 2017b).
Structural Determinants of Non-Enzymatic Acetylation
While non-enzymatic acetylation does not require a KAT, analysis of the environments that contain acetylated lysines suggest that the mechanism by which this enzyme-independent reaction occurs has many parallels with enzymatic acetylation (Figure 3). While the KAT serves as an AcCoA-binding protein to properly position the acetyl group near the target lysine, non-enzymatic acetylation requires that the protein being acetylated coordinates the CoA group for AcCoA or the phosphate moiety for AcP (Kuhn et al., 2014). Often, the coordinating amino acids are positively charged (Arg and Lys), which make ionic bonds, or polar amino acids that make hydrogen bonds via hydroxyl groups (Ser, Thr, and Tyr) or side chain amides (Gln and Asn) (Kuhn et al., 2014). In contrast to KAT-dependent acetylation, where a glutamate from the KAT typically extracts a proton from the lysine to be acetylated, during non-enzymatic acetylation, the non-enzymatic process requires an internal glutamate or a water molecule (Okanishi et al., 2013; Kuhn et al., 2014). It may be expected that lysines acetylated non-enzymatically may have a reduced pKa, and thus more propensity to lose a proton. However, the pKa has not been found to correlate with reactivity (Baeza et al., 2015). Like KATs and KDACs, there is no clear motif that is predictive of whether a lysine is susceptible to chemical acetylation. This was shown by using AcP to acetylate a peptide library with the sequence GXKZGC, where X and Z are each a standard amino acid with the exception of cysteine, on a SAMDI chip. This method showed that almost every peptide could be acetylated, but selectivity and specificity could be achieved by adding magnesium and salt (Kuhn et al., 2014). The SAMDI results indicated adjacent arginines and lysines favored acetylation; in vivo, however, there is a propensity for glutamates and aspartates to be adjacent to the lysines acetylated by AcP. Similar results were found for AcCoA, where glutamates were found near lysines in vivo (Baeza et al., 2015). For both AcP and AcCoA, there is a tendency for susceptible lysines to be surface-exposed (Kuhn et al., 2014; Baeza et al., 2015). However, due to their relatively small size, AcP and AcCoA can access buried lysines that a KAT could not (Kuhn et al., 2014). Thus, protein structures could have evolved to utilize non-enzymatic acetylation for roles that are impossible for enzymatic acetylation.
FIGURE 3.
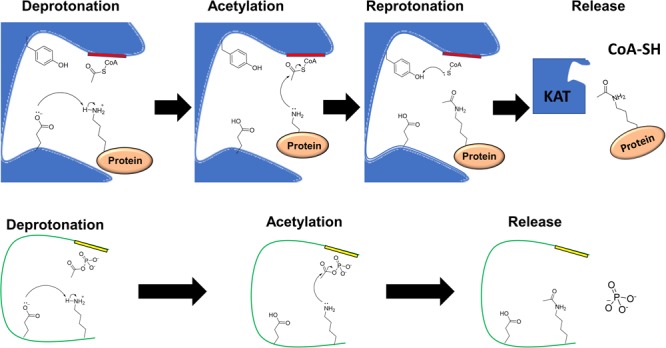
Catalytic mechanisms of enzymatic and non-enzymatic acetylation. (Top) In the enzymatic mechanism, a lysine to be acetylated binds at the acceptor site of a lysine acetyltransferase (KAT) and AcCoA binds at the donor site of the KAT called the P-loop (red) with consensus motif Gln/Arg-x-x-Gly-x-Gly/Ala (Salah Ud-Din et al., 2016). A catalytic glutamate deprotonates the epsilon amino group of the target lysine. The lysine performs a nucleophilic attack on the carbonyl carbon of AcCoA, resulting in acetylation of the lysine. The CoA group becomes protonated by a tyrosine, which regenerates the KAT and facilitates the release of the free CoA and the target protein. (Bottom) In this example of non-enzymatic acetylation, AcP is bound through its negatively charged phosphoryl group to a neighboring patch (yellow) that contains positively charged residues and/or residues that can form hydrogen bonds. In this example, the lysine is deprotonated by a glutamate on the same protein. The lysine performs a nucleophilic attack on the carbonyl carbon of AcP resulting in an acetyllysine. Inorganic phosphate is released as a byproduct of the reaction. A mechanism similar to this can be considered for non-enzymatic AcCoA-dependent acetylation.
Regulation of AcP-Dependent Acetylation – a Consequence of Fermentation
When the glucose consumption rate depletes free CoA faster than it can be recycled, E. coli will ferment acetate from AcCoA via the Pta-AckA pathway (Figure 4). This occurs even in the presence of oxygen in a process called overflow metabolism, akin to a safety valve that regenerates the limiting pools of CoA (Hollywood and Doelle, 1976; Andersen and von Meyenburg, 1980). However, the production of AcP by Pta is faster than the conversion of AcP to acetate by AckA, leaving a pool of the intermediate AcP (Klein et al., 2007; Keating et al., 2008). Thus, when E. coli cells are grown in the presence of excess carbon such as glucose, the bacteria ferment acetate and produce AcP. Thus, ameliorating the need for cells to ferment reduces acetylation. Such an example is when cells lacking the major glucose transporter EIICBglc are grown in medium supplemented with glucose. These mutants must assimilate glucose through alternative glucose transporters such as the galactose permease GalP. The use of these alternative transporters reduces the rate of glucose flux into the cell. As such, the cells also produce less acetate and, as expected, have reduced acetylation (Steinsiek and Bettenbrock, 2012; Schilling et al., 2015).
FIGURE 4.
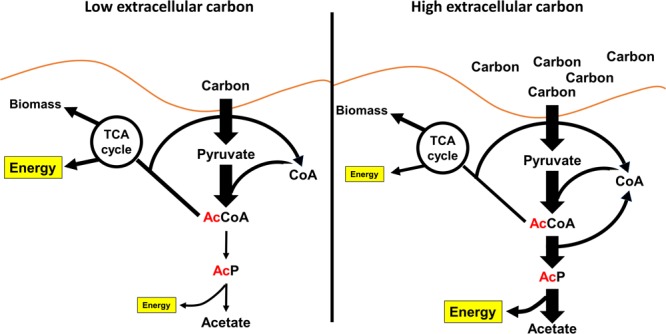
AcP is made as a consequence of overflow metabolism. (Left) When bacteria like E. coli are growing on low concentrations of carbon, much of the carbon is put into the TCA cycle, lipid metabolism, and metabolite biosynthesis. Acetate production is very low. (Right) When the concentration of carbon increases, the flux of carbon through glycolysis depletes the limiting pool of free CoA. To continue to consume carbon, E. coli can regenerate free CoA by fermenting acetate. This results in the production of AcP.
This phenomenon is not exclusive to glucose. Lactate, fructose, and xylose can also potently induce acetylation in E. coli as assessed by anti-acetyllysine western blot (Schilling et al., 2015, 2019; Christensen, 2019). Mass spectrometric analysis has determined the acetylomes of cells grown in xylose, a pentose sugar, and cells grown in glucose, a hexose sugar (Schilling et al., 2019). Glucose and xylose are catabolized through different metabolic pathways; however, each pathway results in acetate fermentation. As such, it may not be surprising that, regardless of carbon source, almost every acetylated lysine was similarly regulated in glucose or xylose. The one exception was xylose isomerase, a protein necessary for catabolism of xylose, which had two lysines differentially acetylated in glucose and xylose. These results suggest that cells regulate AcP-dependent acetylation simply by regulating overflow metabolism and production of AcP.
One other key physiological shift that causes accumulation of acetylations is cessation of growth, such as entry into stationary phase. For both E. coli and N. gonorrhoeae, acetylation increased as the culture entered stationary phase either naturally or when artificially induced (Weinert et al., 2013a; Kuhn et al., 2014; Schilling et al., 2015; Post et al., 2017). We recently determined why acetylation accumulates after E. coli cultures enter into stationary phase. E. coli cultures grown in glucose-supplemented peptide-based media consume both amino acids and glucose during exponential growth; however, most of the glucose is consumed after the culture enters into stationary phase (Christensen et al., 2017).
Compared to exponential phase, two facets of stationary phase make it the perfect growth phase to promote acetylation when excess carbon is present: reduced protein synthesis and reduced respiration. In stationary phase, rates of growth cannot be sustained due to depletion of a limiting nutrient (Reeve et al., 1984). Thus, cells reduce rates of transcription and translation, which greatly reduces the generation of new proteins (Navarro Llorens et al., 2010). Since the rate of protein biosynthesis is reduced, fewer TCA cycle intermediates are used to generate new biomass. This will cause a bottleneck at the TCA cycle, and carbon must enter other pathways. In conjunction with reduced translation, total cellular protein becomes further reduced due to increased protein turnover. When E. coli enters stationary phase, aerobic metabolism is repressed (Nyström, 2004; Navarro Llorens et al., 2010). As such, the carbon flux is driven toward fermentation. The combination of a reduced proteome in stationary phase and the continued metabolism of a fermentable carbon source increase the probability that any susceptible lysine will become acetylated.
In conclusion, data thus far suggests that non-enzymatic AcP-dependent acetylation is an unavoidable consequence of fermentation that accumulates on an aging proteome. Since AcP is made as an intermediate of fermentation, the only means of regulating acetylation would be by regulating fermentation, deacetylation of acetyllysines with a KDAC, and/or evolving protein structures to optimally favor/disfavor acetylation of specific lysines. Because acetate fermentation has evolved as a key mechanism that allows many bacteria to regenerate the limited pool of CoA, this would suggest that E. coli has evolved over millions of years to cope with or utilize this global modification.
A Possible Evolutionary History of Acetylation and Because of Acetylation
Because nearly all life has evolved to utilize activated acetate as a key molecule in metabolism, all organisms must inevitably cope with the potential for protein acetylation. To understand the physiological relevance of acetylation, it is helpful to first imagine how acetylation evolved. As proposed previously, AcP is a relatively simple molecule compared to ATP or AcCoA with higher energy, and as such is likely to be a more ancient energy currency (Ninfa et al., 2000; Wolfe, 2010). Due to the well-understood effects of phosphorylation by AcP, it was proposed that proteins like response regulators could have evolved to accept phosphoryl groups from AcP, and only later would the cell have evolved phosphatases to remove those phosphoryl groups. Finally, some phosphatases would evolve to become kinases to fine-tune these pathways (Wolfe, 2010).
Similarly, the ability for AcP to acetylate proteins also could be a driver of evolution. Acetylation of a catalytic lysine can be extremely detrimental to protein function, and inability to remove these acetylations could result in a dead enzyme. Thus, cells must have evolved deacetylases to ensure that a given lysine would be unmodified despite the presence of endogenously produced reactive metabolites. Contrary to the phosphorylation example, the acetyltransferases are not homologous to the deacetylases and must have evolved independently. Perhaps these acetyltransferases evolved after the development of their primary substrate, AcCoA.
Each acetylated lysine has the potential to be regulatory. Whether these acetylations are due to non-enzymatic mechanisms or catalyzed enzymatically, life must balance the possibility of detrimental acetylation with benign or beneficial acetylations. The best ways to ensure that only optimal lysines are acetylated under a given condition would be to either (1) evolve a lysine deacetylase to remove deleterious modifications or (2) evolve proteins such that they do not have critical lysines susceptible to acetylation. For E. coli, almost all acetylated linear peptides are susceptible to CobB (AbouElfetouh et al., 2015), but not all acetylated E. coli proteins are CobB-sensitive in vivo (Kuhn et al., 2014). The CobB-sensitive lysines tend to be surface-exposed (AbouElfetouh et al., 2015). Indeed, for a lysine to be deacetylated, it almost certainly cannot be buried within the protein structure, unless the acetylated residue becomes accessible through “protein breathing,” large scale conformational changes of the protein structure (Kossiakoff, 1986). However, the extent of breathing in vivo is probably restricted due to molecular crowding in the cytoplasm (Makowski et al., 2008). Thus, the protein must be structured in such a way to favor deacetylation of the target lysine without disrupting the activity of the protein.
If an acetylated lysine blocks protein function and is not accessible to CobB or another deacetylase, the cell must replace that protein by degrading it and synthesizing an entirely new protein. Not all lysines can be acetylated, but those that do become modified can be acetylated at different rates (Baeza et al., 2015; Schilling et al., 2015). As buried lysines would be refractory to CobB, one would also expect that acetylation of these lysines is rarely enzymatic due to the inability for an enzyme to access the lysine. Thus, evolution may have selected for molecular environments that disfavor acetylation of these buried and other critical lysines. However, any changes made over evolution that would disfavor acetylation had to be balanced with a protein sequence that permits proper folding and activity.
Future Thoughts
The study of acetylation is a fast growing field with technological advances in analytical methodologies, such as mass spectrometry, providing a greater depth of analysis. The report of each new acetylome opens more avenues to explore. While some acetylations are certain to be organism-specific, there are bound to be many conserved acetyllysines in critical processes, as we have already observed in central metabolism and translation. Each of these acetyllysines must result from one or more of the recognized acetylation mechanisms, either enzymatic or non-enzymatic. In turn, these mechanisms may themselves be regulated dynamically with the potential for signaling cascades akin to phosphorylation.
Researchers in this new field have two main routes that they can follow. They can investigate the effect of acetylation on individual sites on individual proteins or they can study the effect of global acetylation. The former route can take advantage of the numerous acetylome studies that have yielded data sets containing hundreds of acetylated proteins modified on thousands of unique lysines. The problem is how to prioritize the most functionally relevant acetylation sites. Typically, researchers prioritize by choosing lysines in provocative locations on proteins with easily assessed phenotypes or activities. However, it should be noted that genetic mimic/genetic ablative mutants are often used to compare extreme stoichiometries, i.e., a mimic of the 100% acetylated state versus a mimic of the 100% unacetylated state. The same is true of proteins acetylated by non-canonical amino acid substitution.
Another way to prioritize is to determine the stoichiometry of each acetylated lysine (Baeza et al., 2014; Nakayasu et al., 2014; Weinert et al., 2015, 2017; Meyer et al., 2016; Miyagi, 2017; Wei et al., 2018). A lysine with a large fold change but a low basal stoichiometry may be a less physiologically important acetylation event compared to one with a smaller fold change but a higher basal stoichiometry. Unfortunately, obtaining the stoichiometry of the acetylome is a non-trivial technical challenge for all but a few groups. To make matters worse, most of the existing analyses were performed on highly mutagenized laboratory strains grown under conditions that make physiologically relevant interpretations difficult.
Whereas site-specific or protein-specific investigations can provide lessons concerning the manner in which acetylation influences protein structure/function relationships, such focused studies cannot explain the global effect of simultaneous low stoichiometric acetylation of large numbers of lysines, as occurs by non-enzymatic acetylation. These acetylations greatly expand the acetylome, neutralizing thousands of positive charges, yet the impact of these modifications remains unclear. Furthermore, acetylation is not the only global modification. The proteome can be acylated using many other central metabolites that may compete for the same lysines (Peng et al., 2011; Zhang et al., 2011; Colak et al., 2013; Wagner and Payne, 2013; Weinert et al., 2013b; Kosono et al., 2015; Singhal et al., 2015; Yang et al., 2015; Meyer et al., 2016; Mizuno et al., 2016; Sun et al., 2016b; Basisty et al., 2018; Gaviard et al., 2018; Vasileva et al., 2018; Wei et al., 2018). The potential crosstalk amongst these acylations and between these acylations and other PTMs presents an important challenge as we attempt to truly understand the inner workings of the cell.
Since lysines can be acylated without the use of an enzyme, we would suspect that bacteria would have evolved to utilize this otherwise “wasteful” process. We propose that global acylation may serve many functions. For example, acylations may serve as a feedback mechanism to slow an excessive carbon flux. Acylations could protect proteins against damaging modifications, such as carbonylation, that result in protein misfolding and aggregation (Dalle-Donne et al., 2003; Maisonneuve et al., 2008). Acylations may be less severe modifications that could protect proteins against such harmful lesions. It may also be possible for acetylation to serve as a method for cells to store two carbon subunits. While there are designated carbon storage polymers, such as starch in plants or glycogen in animals, fungi, and bacteria, perhaps acetylation is a primordial form of carbon storage.
Finally, researchers should remember that heterologous expression of a protein in E. coli or any other bacterium carries the possibility that the purified protein preparation may be a heterogeneous mixture decorated by various acylations. Indeed, at least one group has reported that recombinant insulin produced in E. coli is acetylated (Szewczak et al., 2015). The potential for acylations to influence efficacy has great implications for the pharmaceutical industry, but also for any researcher who uses a heterologous system to express their protein of interest. Depending on the protein and the conditions under which the host bacteria are grown, the modified protein may behave in a manner that is not consistent with expectations.
Author Contributions
DC, BS, and AW outlined the review. DC wrote the first draft and designed the figures. XX, NB, JB, and SM helped to complete the review, especially sections focused on structural biology and mass spectrometry. AW did the final edit. BS and AW oversaw the entire process of this study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge Dr. Misty Kuhn and Kristen Jew of the San Francisco State University for their critical reading of this review.
Footnotes
Funding. We would like to acknowledge our funding as follows: R01 GM066130 (NIGMS to AW), R01 AI108255 (NIAID subcontract to AW), and R01 AI108255 (NIAID to BS). We also acknowledge support from the NIH shared instrumentation grant for the TripleTOF system at the Buck Institute (1S10 OD016281). The Life Science Biomedical Technology Research resource was primarily supported by the National Institutes of Health, the National Institute of General Medical Sciences (NIGMS) through a Biomedical Technology Research Resource P41 grant (P41GM111244), and by the DOE Office of Biological and Environmental Research (KP1605010). As a National Synchrotron Light Source II facility resource at the Brookhaven National Laboratory, work performed at the LSBR was supported in part by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences Program under contract number and DE-SC0012704 (KC0401040).
References
- AbouElfetouh A., Kuhn M. L., Hu L. I., Scholle M. D., Sorensen D. J., Sahu A. K., et al. (2015). The E. coli sirtuin CobB shows no preference for enzymatic and nonenzymatic lysine acetylation substrate sites. Microbiologyopen 4 66–83. 10.1002/mbo3.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey V. G., Faulkner R., Mirsky A. E. (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 51 786–794. 10.1073/pnas.51.5.786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin R., Franz-Wachtel M., Tiffert Y., Heberer M., Meky M., Ahmed Y., et al. (2016). Post-translational serine/threonine phosphorylation and lysine acetylation: a novel regulatory aspect of the global nitrogen response regulator GlnR in S. coelicolor M145. Front. Mol. Biosci. 3:38. 10.3389/fmolb.2016.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K. B., von Meyenburg K. (1980). Are growth rates of Escherichia coli in batch cultures limited by respiration? J. Bacteriol. 144 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos J. L., Bever K. M., Wolberger C. (2005). Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol. Cell 17 855–868. 10.1016/j.molcel.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Baeza J., Dowell J. A., Smallegan M. J., Fan J., Amador-Noguez D., Khan Z., et al. (2014). Stoichiometry of site-specific lysine acetylation in an entire proteome. J. Biol. Chem. 289 21326–21338. 10.1074/jbc.M114.581843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J., Smallegan M. J., Denu J. M. (2015). Site-specific reactivity of nonenzymatic lysine acetylation. ACS Chem. Biol. 10 122–128. 10.1021/cb500848p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J., Kouzarides T. (1996). The CBP co-activator is a histone acetyltransferase. Nature 384 641–643. 10.1038/384641a0 [DOI] [PubMed] [Google Scholar]
- Bao X., Wang Y., Li X., Li X. M., Liu Z., Yang T., et al. (2014). Identification of ’erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. eLife 3:e02999. 10.7554/eLife.02999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak R., Eisenbach M. (2001). Acetylation of the response regulator, CheY, is involved in bacterial chemotaxis. Mol. Microbiol. 40 731–743. 10.1046/j.1365-2958.2001.02425.x [DOI] [PubMed] [Google Scholar]
- Barak R., Yan J., Shainskaya A., Eisenbach M. (2006). The chemotaxis response regulator CheY can catalyze its own acetylation. J. Mol. Biol. 359 251–265. 10.1016/j.jmb.2006.03.033 [DOI] [PubMed] [Google Scholar]
- Basisty N., Meyer J. G., Wei L., Gibson B. W., Schilling B. (2018). Simultaneous quantification of the acetylome and succinylome by ’one-pot’ affinity enrichment. Proteomics 18:e1800123. 10.1002/pmic.201800123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S. H., Darwin K. H. (2017). Bacterial proteasomes: mechanistic and functional insights. Microbiol. Mol. Biol. Rev. 81:e00036-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard E., Rolain T., Courtin P., Guillot A., Langella P., Hols P., et al. (2011). Characterization of O-acetylation of N-acetylglucosamine: a novel structural variation of bacterial peptidoglycan. J. Biol. Chem. 286 23950–23958. 10.1074/jbc.M111.241414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen C. E., Albaugh B. N., Tan S., Denu J. M. (2007). Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry 46 623–629. 10.1021/bi602513x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J., Gou Z., Zhou F., Chen Y., Gan J., Liu J., et al. (2018). Acetylation of lysine 182 inhibits the ability of Mycobacterium tuberculosis DosR to bind DNA and regulate gene expression during hypoxia. Emerg. Microbes Infect. 7:108. 10.1038/s41426-018-0112-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J., Wang Y., Yu H., Qian X., Wang H., Liu J., et al. (2017). Modulation of central carbon metabolism by acetylation of isocitrate lyase in Mycobacterium tuberculosis. Sci. Rep. 7:44826. 10.1038/srep44826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birhanu A. G., Yimer S. A., Holm-Hansen C., Norheim G., Aseffa A., Abebe M., et al. (2017). N𝜀- and O-acetylation in Mycobacterium tuberculosis Lineage 7 and Lineage 4 strains: proteins involved in bioenergetics, virulence and antimicrobial resistance are acetylated. J. Proteome Res. 16 4045–4059. 10.1021/acs.jproteome.7b00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman K. J., Anderson R. M., Cohen H. Y., Latorre-Esteves M., Sinclair D. A. (2002). Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast SIR2 and human SIRT1. J. Biol. Chem. 277 45099–45107. 10.1074/jbc.m205670200 [DOI] [PubMed] [Google Scholar]
- Blander G., Guarente L. (2004). The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73 417–435. 10.1146/annurev.biochem.73.011303.073651 [DOI] [PubMed] [Google Scholar]
- Bontemps-Gallo S., Gaviard C., Richards C. L., Kentache T., Raffel S. J., Lawrence K. A., et al. (2018). Global profiling of lysine acetylation in Borrelia burgdorferi B31 reveals its role in central metabolism. Front. Microbiol. 9:2036. 10.3389/fmicb.2018.02036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman A. B., Ettema T. J., de Vos W. M., van der Oost J. (2003). The Lrp family of transcriptional regulators. Mol. Microbiol. 48 287–294. 10.1046/j.1365-2958.2003.03442.x [DOI] [PubMed] [Google Scholar]
- Brown C. W., Sridhara V., Boutz D. R., Person M. D., Marcotte E. M., Barrick J. E., et al. (2017). Large-scale analysis of post-translational modifications in E. coli under glucose-limiting conditions. BMC Genomics 18:301. 10.1186/s12864-017-3676-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. D., Jones-Mortimer M. C., Kornberg H. L. (1977). The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J. Gen. Microbiol. 102 327–336. 10.1099/00221287-102-2-327 [DOI] [PubMed] [Google Scholar]
- Brownlee M. (2005). The pathobiology of diabetic complications: a unifying mechanism. Diabetes Metab. Res. Rev. 54 1615–1625. 10.2337/diabetes.54.6.1615 [DOI] [PubMed] [Google Scholar]
- Bulkley D., Innis C. A., Blaha G., Steitz T. A. (2010). Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 107 17158–17163. 10.1073/pnas.1008685107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabetta V. J., Cristea I. M. (2017). Regulation, function, and detection of protein acetylation in bacteria. J. Bacteriol. 199:e00107-17. 10.1128/JB.00107-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabetta V. J., Greco T. M., Cristea I. M., Dubnau D. (2019). YfmK is an N𝜀-lysine acetyltransferase that directly acetylates the histone-like protein HBsu in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 116 3752–3757. 10.1073/pnas.1815511116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabetta V. J., Greco T. M., Tanner A. W., Cristea I. M., Dubnau D. (2016). Temporal regulation of the Bacillus subtilis acetylome and evidence for a role of MreB acetylation in cell wall growth. mSystems 1:e00005-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Cerezo S., Bernal V., Blanco-Catalá J., Iborra J. L., Cánovas M. (2011). cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol. Microbiol. 82 1110–1128. 10.1111/j.1365-2958.2011.07873.x [DOI] [PubMed] [Google Scholar]
- Castaño-Cerezo S., Bernal V., Post H., Fuhrer T., Cappadona S., Sánchez-Díaz N. C., et al. (2014). Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol. Syst. Biol. 10:762. 10.15252/msb.20145227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S. C., et al. (2007). Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6 812–819. 10.1074/mcp.m700021-mcp200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverton A. M., Gollan B., Przydacz M., Wong C. T., Mylona A., Hare S. A., et al. (2016). A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol. Cell 63 86–96. 10.1016/j.molcel.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., et al. (2009). Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325 834–840. 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- Christensen D. G. (2019). Regulation of Acetyl Phosphate-Dependent Acetylation and Identification of Novel Lysine Acetyltransferases in Escherichia coli. Doctoral Dissertation, Loyola University Chicago; Chicago, IL. [Google Scholar]
- Christensen D. G., Baumgartner J. T., Xie X., Jew K. M., Basisty N., Schilling B., et al. (2019). Mechanisms, detection, and relevance of protein acetylation in prokaryotes. mBio 10:e02708-18. 10.1128/mBio.02708-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen D. G., Meyer J. G., Baumgartner J. T., D’Souza A. K., Nelson W. C., Payne S. H., et al. (2018). Identification of novel protein lysine acetyltransferases in Escherichia coli. mBio 9:e01905-18. 10.1128/mBio.01905-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen D. G., Orr J. S., Rao C. V., Wolfe A. J. (2017). Increasing growth yield and decreasing acetylation in Escherichia coli by optimizing the carbon-to-magnesium ratio in peptide-based media. Appl. Environ. Microbiol. 83:e03034-16. 10.1128/AEM.03034-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak G., Xie Z., Zhu A. Y., Dai L., Lu Z., Zhang Y., et al. (2013). Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol. Cell. Proteomics 12 3509–3520. 10.1074/mcp.M113.031567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby H. A., Escalante-Semerena J. C. (2014). The acetylation motif in AMP-forming Acyl coenzyme A synthetases contains residues critical for acetylation and recognition by the protein acetyltransferase Pat of Rhodopseudomonas palustris. J. Bacteriol. 196 1496–1504. 10.1128/JB.00004-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby H. A., Heiniger E. K., Harwood C. S., Escalante-Semerena J. C. (2010). Reversible N epsilon-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris. Mol. Microbiol. 76 874–888. 10.1111/j.1365-2958.2010.07127.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby H. A., Pelletier D. A., Hurst G. B., Escalante-Semerena J. C. (2012). System-wide studies of N-lysine acetylation in Rhodopseudomonas palustris reveal substrate specificity of protein acetyltransferases. J. Biol. Chem. 287 15590–15601. 10.1074/jbc.M112.352104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunin R., Glansdorff N., Piérard A., Stalon V. (1986). Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50 314–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Peng C., Montellier E., Lu Z., Chen Y., Ishii H., et al. (2014). Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 10 365–370. 10.1038/nchembio.1497 [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I., Giustarini D., Colombo R., Rossi R., Milzani A. (2003). Protein carbonylation in human diseases. Trends Mol. Med. 9 169–176. 10.1016/s1471-4914(03)00031-5 [DOI] [PubMed] [Google Scholar]
- Dancy B. M., Cole P. A. (2015). Protein lysine acetylation by p300/CBP. Chem. Rev. 115 2419–2452. 10.1021/cr500452k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie J. R. (2003). Inhibition of histone deacetylase activity by butyrate. J. Nutr. 133 2485S–2493S. 10.1093/jn/133.7.2485S [DOI] [PubMed] [Google Scholar]
- Davies J., Wright G. D. (1997). Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 5 234–240. 10.1016/s0966-842x(97)01033-0 [DOI] [PubMed] [Google Scholar]
- Davis R., Écija-Conesa A., Gallego-Jara J., de Diego T., Filippova E. V., Kuffel G., et al. (2018). An acetylatable lysine controls CRP function in E. coli. Mol. Microbiol. 107 116–131. 10.1111/mmi.13874 [DOI] [PubMed] [Google Scholar]
- de Diego Puente T., Gallego-Jara J., Castaño-Cerezo S., Bernal Sánchez V., Fernández Espín V., García de la Torre J., et al. (2015). The protein acetyltransferase PatZ from Escherichia coli is regulated by autoacetylation-induced oligomerization. J. Biol. Chem. 290 23077–23093. 10.1074/jbc.M115.649806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denu J. M. (2003). Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem. Sci. 28 41–48. 10.1016/s0968-0004(02)00005-1 [DOI] [PubMed] [Google Scholar]
- Di Sabato G., Jencks W. P. (1961). Mechanism and catalysis of reactions of acyl phosphates. I. Nucleophilic reactions. J. Am. Chem. Soc. 83 4393–4400. 10.1021/bi201921a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson B. C., Chang C. J. (2011). Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol. 7 504–511. 10.1038/nchembio.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll S., Burlingame A. L. (2015). Mass spectrometry-based detection and assignment of protein posttranslational modifications. ACS Chem. Biol. 10 63–71. 10.1021/cb500904b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Zhou Y., Su X., Yu J. J., Khan S., Jiang H., et al. (2011). Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334 806–809. 10.1126/science.1207861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L., Yi M., Chen J., Li S., Chen W. (2016). Mycobacterium tuberculosis EIS gene inhibits macrophage autophagy through up-regulation of IL-10 by increasing the acetylation of histone H3. Biochem. Biophys. Res. Commun. 473 1229–1234. 10.1016/j.bbrc.2016.04.045 [DOI] [PubMed] [Google Scholar]
- Dutnall R. N., Tafrov S. T., Sternglanz R., Ramakrishnan V. (1998). Structure of the yeast histone acetyltransferase Hat1: insights into substrate specificity and implications for the Gcn5-related N-acetyltransferase superfamily. Cold Spring Harb. Symp. Quant. Biol. 63 501–507. [DOI] [PubMed] [Google Scholar]
- Dyda F., Klein D. C., Hickman A. B. (2000). GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 29 81–103. 10.1146/annurev.biophys.29.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Xiong H., Reynolds N. M., Söll D. (2015). Rationally evolving tRNAPyl for efficient incorporation of noncanonical amino acids. Nucleic Acids Res. 43:e156. 10.1093/nar/gkv800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favrot L., Blanchard J. S., Vergnolle O. (2016). Bacterial GCN5-Related N-Acetyltransferases: from resistance to regulation. Biochemistry 55 989–1002. 10.1021/acs.biochem.5b01269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman J. L., Baeza J., Denu J. M. (2013). Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 288 31350–31356. 10.1074/jbc.C113.511261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye R. A. (1999). Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260 273–279. 10.1006/bbrc.1999.0897 [DOI] [PubMed] [Google Scholar]
- Frye R. A. (2000). Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273 793–798. 10.1006/bbrc.2000.3000 [DOI] [PubMed] [Google Scholar]
- Gallego-Jara J., Écija Conesa A., de Diego Puente T., Lozano Terol G., Cánovas Díaz M. (2017). Characterization of CobB kinetics and inhibition by nicotinamide. PLoS One 12:e0189689. 10.1371/journal.pone.0189689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J. G., Escalante-Semerena J. C. (2008). Biochemical and mutational analyses of AcuA, the acetyltransferase enzyme that controls the activity of the acetyl coenzyme a synthetase (AcsA) in Bacillus subtilis. J. Bacteriol. 190 5132–5136. 10.1128/JB.00340-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J. G., Escalante-Semerena J. C. (2009). In Bacillus subtilis, the sirtuin protein deacetylase, encoded by the srtN gene (formerly yhdZ), and functions encoded by the acuABC genes control the activity of acetyl coenzyme A synthetase. J. Bacteriol. 191 1749–1755. 10.1128/JB.01674-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J. G., Grundy F. J., Henkin T. M., Escalante-Semerena J. C. (2006). Control of acetyl-coenzyme A synthetase (AcsA) activity by acetylation/deacetylation without NAD(+) involvement in Bacillus subtilis. J. Bacteriol. 188 5460–5468. 10.1128/jb.00215-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau-Tsodikova S., Labby K. J. (2016). Mechanisms of resistance to aminoglycoside Antibiotics: overview and perspectives. Medchemcomm 7 11–27. 10.1039/c5md00344j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity J., Gardner J. G., Hawse W., Wolberger C., Escalante-Semerena J. C. (2007). N-lysine propionylation controls the activity of propionyl-CoA synthetase. J. Biol. Chem. 282 30239–30245. 10.1074/jbc.m704409200 [DOI] [PubMed] [Google Scholar]
- Gaviard C., Broutin I., Cosette P., Dé E., Jouenne T., Hardouin J. (2018). Lysine succinylation and acetylation in Pseudomonas aeruginosa. J. Proteome Res. 17 2449–2459. 10.1021/acs.jproteome.8b00210 [DOI] [PubMed] [Google Scholar]
- Gershey E. L., Vidali G., Allfrey V. G. (1968). Chemical studies of histone acetylation. The occurrence of epsilon-N-acetyllysine in the f2a1 histone. J. Biol. Chem. 243 5018–5022. [PubMed] [Google Scholar]
- Ghosh S., Padmanabhan B., Anand C., Nagaraja V. (2016). Lysine acetylation of the Mycobacterium tuberculosis HU protein modulates its DNA binding and genome organization. Mol. Microbiol. 100 577–588. 10.1111/mmi.13339 [DOI] [PubMed] [Google Scholar]
- Glozak M. A., Sengupta N., Zhang X., Seto E. (2005). Acetylation and deacetylation of non-histone proteins. Gene 363 15–23. 10.1016/j.gene.2005.09.010 [DOI] [PubMed] [Google Scholar]
- Golubev A., Hanson A. D., Gladyshev V. N. (2017). Non-enzymatic molecular damage as a prototypic driver of aging. J. Biol. Chem. 292 6029–6038. 10.1074/jbc.R116.751164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. (2017). Stress reduction, bacterial style. J. Bacteriol. 199:e00433-17. 10.1128/JB.00433-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K. D., Biswas T., Pang A. H., Willby M. J., Reed M. S., Stuchlik O., et al. (2018). Acetylation by Eis and deacetylation by Rv1151c of Mycobacterium tuberculosis HupB: biochemical and structural insight. Biochemistry 57 781–790. 10.1021/acs.biochem.7b01089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti I. V., Lee Y. M., Goodson H. V. (2004). Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338 17–31. 10.1016/j.jmb.2004.02.006 [DOI] [PubMed] [Google Scholar]
- Guarente L. (2005). Calorie restriction and SIR2 genes–towards a mechanism. Mech. Ageing Dev. 126 923–928. 10.1016/j.mad.2005.03.013 [DOI] [PubMed] [Google Scholar]
- Hayden J. D., Brown L. R., Gunawardena H. P., Perkowski E. F., Chen X., Braunstein M. (2013). Reversible acetylation regulates acetate and propionate metabolism in Mycobacterium smegmatis. Microbiology 159 1986–1999. 10.1099/mic.0.068585-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T. R., Thorne A. W., Crane-Robinson C. (1988). A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7 1395–1402. 10.1002/j.1460-2075.1988.tb02956.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig A. O., Gauci S., Raijmakers R., van Breukelen B., Slijper M., Mohammed S., et al. (2010). Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol. Cell. Proteomics 9 928–939. 10.1074/mcp.M900463-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsens K., Van Damme P., Degroeve S., Martens L., Arnesen T., Vandekerckhove J., et al. (2011). Bioinformatics analysis of a Saccharomyces cerevisiae N-terminal proteome provides evidence of alternative translation initiation and post-translational N-terminal acetylation. J. Proteome Res. 10 3578–3589. 10.1021/pr2002325 [DOI] [PubMed] [Google Scholar]
- Hentchel K. L., Escalante-Semerena J. C. (2015). Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol. Mol. Biol. Rev. 79 321–346. 10.1128/MMBR.00020-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildmann C., Riester D., Schwienhorst A. (2007). Histone deacetylases–an important class of cellular regulators with a variety of functions. Appl. Microbiol. Biotechnol. 75 487–497. 10.1007/s00253-007-0911-2 [DOI] [PubMed] [Google Scholar]
- Hockenberry A. M., Post D. M. B., Rhodes K. A., Apicella M., So M. (2018). Perturbing the acetylation status of the Type IV pilus retraction motor, PilT, reduces Neisseria gonorrhoeae viability. Mol. Microbiol. 110 677–688. 10.1111/mmi.13979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollywood N., Doelle H. W. (1976). Effect of specific growth rate and glucose concentration on growth and glucose metabolism of Escherichia coli K-12. Microbios 17 23–33. [PubMed] [Google Scholar]
- Hood M. I., Skaar E. P. (2012). Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10 525–537. 10.1038/nrmicro2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L. (2013). Understanding the Posttranslational Regulation of the Response Regulator RcsB and Acetyl Phosphate as an Acetyl Group Donor in E. coli. Doctoral Dissertation, Loyola University Chicago; Chicago, IL. [Google Scholar]
- Hu L. I., Chi B. K., Kuhn M. L., Filippova E. V., Walker-Peddakotla A. J., Bäsell K., et al. (2013). Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J. Bacteriol. 195 4174–4186. 10.1128/JB.00383-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y., Akanuma G., Yoshida M., Horinouchi S., Kosono S., Ohnishi Y. (2017). Protein acetylation involved in streptomycin biosynthesis in Streptomyces griseus. J. Proteomics 155 63–72. 10.1016/j.jprot.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Jackson M. D., Schmidt M. T., Oppenheimer N. J., Denu J. M. (2003). Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J. Biol. Chem. 278 50985–50998. 10.1074/jbc.m306552200 [DOI] [PubMed] [Google Scholar]
- Jencks W. P. (1969). Catalysis in Chemistry and Enzymology. New York, NY: McGraw-Hill Book Company. [Google Scholar]
- Jers C., Ravikumar V., Lezyk M., Sultan A., Sjöling Å., Wai S. N.et al. (2017). The global acetylome of the human pathogen Vibrio cholerae V52 reveals lysine acetylation of major transcriptional regulators. Front. Cell. Infect. Microbiol. 7:537. 10.3389/fcimb.2017.00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose L., Ramachandran R., Bhagavat R., Gomez R. L., Chandran A., Raghunandanan S., et al. (2016). Hypothetical protein Rv3423.1 of Mycobacterium tuberculosis is a histone acetyltransferase. FEBS J. 283 265–281. 10.1111/febs.13566 [DOI] [PubMed] [Google Scholar]
- Kawabe H., Kondo S., Umezawa H., Mitsuhashi S. (1975). R factor-mediated aminoglycoside antibiotic resistance in Pseudomonas aeruginosa: a new aminoglycoside 6′-N-acetyltransferase. Antimicrob. Agents Chemother. 7 494–499. 10.1128/aac.7.5.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating D. H., Shulla A., Klein A. H., Wolfe A. J. (2008). Optimized two-dimensional thin layer chromatography to monitor the intracellular concentration of acetyl phosphate and other small phosphorylated molecules. Biol. Proced. Online 10 36–46. 10.1251/bpo141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentache T., Jouenne T., Dé E., Hardouin J. (2016). Proteomic characterization of Nα- and N𝜀-acetylation in Acinetobacter baumannii. J. Proteomics 144 148–158. 10.1016/j.jprot.2016.05.021 [DOI] [PubMed] [Google Scholar]
- Kim G. W., Yang X. J. (2011). Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem. Sci. 36 211–220. 10.1016/j.tibs.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Kim K. H., An D. R., Song J., Yoon J. Y., Kim H. S., Yoon H. J., et al. (2012). Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. Proc. Natl. Acad. Sci. U.S.A. 109 7729–7734. 10.1073/pnas.1120251109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., et al. (2006). Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23 607–618. 10.1016/j.molcel.2006.06.026 [DOI] [PubMed] [Google Scholar]
- Klein A. H., Shulla A., Reimann S. A., Keating D. H., Wolfe A. J. (2007). The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J. Bacteriol. 189 5574–5581. 10.1128/jb.00564-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Jr. (1952). Effect of catalysts on the hydrolysis of acetyl phosphate. Nucleophilic displacement mechanisms in enzymatic reactions. J. Am. Chem. Soc. 74 2286–2292. 10.1021/ja01129a035 [DOI] [Google Scholar]
- Kosono S., Tamura M., Suzuki S., Kawamura Y., Yoshida A., Nishiyama M., et al. (2015). Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLoS One 10:e0131169. 10.1371/journal.pone.0131169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossiakoff A. A. (1986). Protein dynamics investigated by neutron diffraction. Methods Enzymol. 131 433–447. 10.1016/0076-6879(86)31051-6 [DOI] [PubMed] [Google Scholar]
- Kuhn M. L., Zemaitaitis B., Hu L. I., Sahu A., Sorensen D., Minasov G., et al. (2014). Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One 9:e94816. 10.1371/journal.pone.0094816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S., Tishel R., Eisenbach M., Wolfe A. J. (1995). Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 177 2878–2886. 10.1128/jb.177.10.2878-2886.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo Y. M., Andrews A. J. (2013). Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3. PLoS One 8:e54896. 10.1371/journal.pone.0054896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S., Rosenkrands I., Stein R., Andersen P., Kaufmann S. H., Jungblut P. R. (2014). Analysis of protein species differentiation among mycobacterial low-Mr-secreted proteins by narrow pH range Immobiline gel 2-DE-MALDI-MS. J. Proteomics 97 235–244. 10.1016/j.jprot.2013.06.036 [DOI] [PubMed] [Google Scholar]
- Lanouette S., Mongeon V., Figeys D., Couture J. F. (2014). The functional diversity of protein lysine methylation. Mol. Syst. Biol. 10:724. 10.1002/msb.134974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D., Benhamed M., Henry Y., Domenichini S., Kim W., Zhou D. X., et al. (2008). The MYST histone acetyltransferases are essential for gametophyte development in Arabidopsis. BMC Plant Biol. 8:121. 10.1186/1471-2229-8-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Manning A. J., Wolfgeher D., Jelenska J., Cavanaugh K. A., Xu H., et al. (2015). Acetylation of an NB-LRR plant immune-effector complex suppresses immunity. Cell Rep. 13 1670–1682. 10.1016/j.celrep.2015.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Lee A., Ma W., Zhou H., Guttman D. S., Desveaux D. (2011). The YopJ superfamily in plant-associated bacteria. Mol. Plant Pathol. 12 928–937. 10.1111/j.1364-3703.2011.00719.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang W., Zhang R., Xu J., Wang R., Wang L., et al. (2018). First acetyl-proteome pro?ling of Salmonella Typhimurium revealed involvement of lysine acetylation in drug resistance. Vet. Microbiol. 226 1–8. 10.1016/j.vetmic.2018.09.024 [DOI] [PubMed] [Google Scholar]
- Li R., Gu J., Chen Y. Y., Xiao C. L., Wang L. W., Zhang Z. P., et al. (2010). CobB regulates Escherichia coli chemotaxis by deacetylating the response regulator CheY. Mol. Microbiol. 76 1162–1174. 10.1111/j.1365-2958.2010.07125.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang Q., Xu Z., Yao Y. F. (2017). Acetylation of lysine 243 inhibits the oriC binding ability of DnaA in Escherichia coli. Front. Microbiol. 8:699. 10.3389/fmicb.2017.00699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Krishnan K., Duncan M. J. (2018). Post-translational regulation of a Porphyromonas gingivalis regulator. J. Oral Microbiol. 10:1487743. 10.1080/20002297.2018.1487743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Deutscher M. P. (2012). Post-translational modification of RNase R is regulated by stress-dependent reduction in the acetylating enzyme Pka (YfiQ). RNA 18 37–41. 10.1261/rna.030213.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Malhotra A., Deutscher M. P. (2011). Acetylation regulates the stability of a bacterial protein: growth stage-dependent modification of RNase R. Mol. Cell 44 160–166. 10.1016/j.molcel.2011.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liarzi O., Barak R., Bronner V., Dines M., Sagi Y., Shainskaya A., et al. (2010). Acetylation represses the binding of CheY to its target proteins. Mol. Microbiol. 76 932–943. 10.1111/j.1365-2958.2010.07148.x [DOI] [PubMed] [Google Scholar]
- Liimatta K., Flaherty E., Ro G., Nguyen D. K., Prado C., Purdy A. E. (2018). A putative acetylation system in Vibrio cholerae modulates virulence in arthropod hosts. Appl. Environ. Microbiol. 84:e01113-18. 10.1128/AEM.01113-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima B. P., Antelmann H., Gronau K., Chi B. K., Becher D., Brinsmade S. R., et al. (2011). Involvement of protein acetylation in glucose-induced transcription of a stress-responsive promoter. Mol. Microbiol. 81 1190–1204. 10.1111/j.1365-2958.2011.07742.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima B. P., Thanh Huyen T. T., Bäsell K., Becher D., Antelmann H., Wolfe A. J. (2012). Inhibition of acetyl phosphate-dependent transcription by an acetylatable lysine on RNA polymerase. J. Biol. Chem. 287 32147–32160. 10.1074/jbc.M112.365502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Fletcher C. M., Zhou J., Allis C. D., Wagner G. (1999). Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A. Nature 400 86–89. 10.1038/21922 [DOI] [PubMed] [Google Scholar]
- Lin Y. Y., Lu J. Y., Zhang J., Walter W., Dang W., Wan J., et al. (2009). Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136 1073–1084. 10.1016/j.cell.2009.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszczak G., Arnesen T., Marmorstein R. (2011). Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. J. Biol. Chem. 286 37002–37010. 10.1074/jbc.M111.282863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Yang M., Wang X., Yang S., Gu J., Zhou J., et al. (2014). Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis. Mol. Cell. Proteomics 13 3352–3366. 10.1074/mcp.M114.041962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Tan Y., Cao S., Zhao H., Fang H., Yang X., et al. (2018). Protein acetylation mediated by YfiQ and CobB is involved in the virulence and stress response of Yersinia pestis. Infect. Immun. 86:e00224-18. 10.1128/IAI.00224-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang L., Zhao K., Thompson P. R., Hwang Y., Marmorstein R., et al. (2008). The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451 846–850. 10.1038/nature06546 [DOI] [PubMed] [Google Scholar]
- Liu X. X., Liu W. B., Ye B. C. (2015). Regulation of a protein acetyltransferase in Myxococcus xanthus by the coenzyme NADP. J. Bacteriol. 198 623–632. 10.1128/JB.00661-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi P. M., Cole K. E., Dowling D. P., Christianson D. W. (2011). Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr. Opin. Struct. Biol. 21 735–743. 10.1016/j.sbi.2011.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. X., Liu X. X., Liu W. B., Ye B. C. (2017). Identification and characterization of two types of amino acid-regulated acetyltransferases in actinobacteria. Biosci. Rep. 37:BSR20170157. 10.1042/BSR20170157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Marinov G. K. (2015). The bioenergetic costs of a gene. Proc. Natl. Acad. Sci. U.S.A. 112 15690–15695. 10.1073/pnas.1514974112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K. W., Ma W. (2016). YopJ family effectors promote bacterial infection through a unique acetyltransferase activity. Microbiol. Mol. Biol. Rev. 80 1011–1027. 10.1128/mmbr.00032-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Wood T. K. (2011). Protein acetylation in prokaryotes increases stress resistance. Biochem. Biophys. Res. Commun. 410 846–851. 10.1016/j.bbrc.2011.06.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madian A. G., Regnier F. E. (2010). Proteomic identification of carbonylated proteins and their oxidation sites. J. Proteome Res. 9 3766–3780. 10.1021/pr1002609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E., Fraysse L., Lignon S., Capron L., Dukan S. (2008). Carbonylated proteins are detectable only in a degradation-resistant aggregate state in Escherichia coli. J. Bacteriol. 190 6609–6614. 10.1128/JB.00588-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L., Rodi D. J., Mandava S., Minh D. D., Gore D. B., Fischetti R. F. (2008). Molecular crowding inhibits intramolecular breathing motions in proteins. J. Mol. Biol. 375 529–546. 10.1016/j.jmb.2007.07.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R. (2001). Structure of histone deacetylases: insights into substrate recognition and catalysis. Structure 9 1127–1133. 10.1016/s0969-2126(01)00690-6 [DOI] [PubMed] [Google Scholar]
- Marmorstein R., Roth S. Y. (2001). Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev. 11 155–161. 10.1016/s0959-437x(00)00173-8 [DOI] [PubMed] [Google Scholar]
- Marsh V. L., Peak-Chew S. Y., Bell S. D. (2005). Sir2 and the acetyltransferase, Pat, regulate the archaeal chromatin protein, Alba. J. Biol. Chem. 280 21122–21128. 10.1074/jbc.m501280200 [DOI] [PubMed] [Google Scholar]
- Martin B. L. (ed.) (2014). “Regulation by covalent modification,” in eLS (Midsomer Norton: JWS Ltd.). [Google Scholar]
- Martínez-Antonio A., Collado-Vides J. (2003). Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6 482–489. 10.1016/j.mib.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Mba Medie F., Champion M. M., Williams E. A., Champion P. A. (2014). Homeostasis of N-α-terminal acetylation of EsxA correlates with virulence in Mycobacterium marinum. Infect. Immun. 82 4572–4586. 10.1128/IAI.02153-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. G., D’Souza A. K., Sorensen D. J., Rardin M. J., Wolfe A. J., Gibson B. W., et al. (2016). Quantification of lysine acetylation and succinylation stoichiometry in proteins using mass spectrometric data-independent acquisitions (SWATH). J. Am. Soc. Mass Spectrom. 27 1758–1771. 10.1007/s13361-016-1476-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulik K., Felsberg J., Kudrnáèová E., Bezoušková S., Setinová D., Stodùlková E., et al. (2012). CobB1 deacetylase activity in Streptomyces coelicolor. Biochem. Cell Biol. 90 179–187. 10.1139/o11-086 [DOI] [PubMed] [Google Scholar]
- Mishra A., Roy F., Dou Y., Zhang K., Tang H., Fletcher H. M. (2018). Role of acetyltransferase PG1842 in gingipain biogenesis in Porphyromonas gingivalis. J. Bacteriol. 200:e00385-18. 10.1128/JB.00385-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R., Peak-Chew S. Y., Sade R. S., Vallis Y., McMahon H. T. (2010). The acetyltransferase activity of the bacterial toxin YopJ of Yersinia is activated by eukaryotic host cell inositol hexakisphosphate. J. Biol. Chem. 285 19927–19934. 10.1074/jbc.M110.126581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi M. (2017). Site-specific quantification of lysine acetylation using isotopic labeling. Methods Enzymol. 586 85–95. 10.1016/bs.mie.2016.09.029 [DOI] [PubMed] [Google Scholar]
- Mizuno Y., Nagano-Shoji M., Kubo S., Kawamura Y., Yoshida A., Kawasaki H., et al. (2016). Altered acetylation and succinylation profiles in Corynebacterium glutamicum in response to conditions inducing glutamate overproduction. Microbiologyopen 5 152–173. 10.1002/mbo3.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen C. A., Yang X. J., Kokubo T., Brownell J. E., Bannister A. J., Owen-Hughes T., et al. (1996). The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87 1261–1270. 10.1016/s0092-8674(00)81821-8 [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Hao Y. H., Orth K. (2007). A newly discovered post-translational modification–the acetylation of serine and threonine residues. Trends Biochem. Sci. 32 210–216. 10.1016/j.tibs.2007.03.007 [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Keitany G., Li Y., Wang Y., Ball H. L., Goldsmith E. J., et al. (2006). Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312 1211–1214. 10.1126/science.1126867 [DOI] [PubMed] [Google Scholar]
- Nagano-Shoji M., Hamamoto Y., Mizuno Y., Yamada A., Kikuchi M., Shirouzu M., et al. (2017). Characterization of lysine acetylation of a phosphoenolpyruvate carboxylase involved in glutamate overproduction in Corynebacterium glutamicum. Mol. Microbiol. 104 677–689. 10.1111/mmi.13658 [DOI] [PubMed] [Google Scholar]
- Nakayasu E. S., Burnet M. C., Walukiewicz H. E., Wilkins C. S., Shukla A. K., Brooks S., et al. (2017). Ancient regulatory role of lysine acetylation in central metabolism. mBio 8:e01894-17. 10.1128/mBio.01894-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu E. S., Wu S., Sydor M. A., Shukla A. K., Weitz K. K., Moore R. J., et al. (2014). A method to determine lysine acetylation stoichiometries. Int. J. Proteomics 2014:730725. 10.1155/2014/730725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambi S., Basu N., Visweswariah S. S. (2010). cAMP-regulated protein lysine acetylases in Mycobacteria. J. Biol. Chem. 285 24313–24323. 10.1074/jbc.M110.118398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambi S., Gupta K., Bhattacharyya M., Ramakrishnan P., Ravikumar V., Siddiqui N., et al. (2013). Cyclic AMP-dependent protein lysine acylation in mycobacteria regulates fatty acid and propionate metabolism. J. Biol. Chem. 288 14114–14124. 10.1074/jbc.M113.463992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro Llorens J. M., Tormo A., Martínez-García E. (2010). Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 34 476–495. 10.1111/j.1574-6976.2010.00213.x [DOI] [PubMed] [Google Scholar]
- Neuwald A. F., Landsman D. (1997). GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22 154–155. 10.1016/s0968-0004(97)01034-7 [DOI] [PubMed] [Google Scholar]
- Ninfa A. J., Jiang P., Atkinson M. R., Peliska J. A. (2000). Integration of antagonistic signals in the regulation of nitrogen assimilation in Escherichia coli. Curr. Top. Cell. Regul. 36 31–75. [DOI] [PubMed] [Google Scholar]
- Noy T., Xu H., Blanchard J. S. (2014). Acetylation of acetyl-CoA synthetase from Mycobacterium tuberculosis leads to specific inactivation of the adenylation reaction. Arch. Biochem. Biophys. 550–551 42–49. 10.1016/j.abb.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström T. (2004). Stationary-phase physiology. Annu. Rev. Microbiol. 58 161–181. 10.1146/annurev.micro.58.030603.123818 [DOI] [PubMed] [Google Scholar]
- Ogura M., Asai K. (2016). Glucose induces ECF sigma factor genes, sigX and sigM, independent of cognate anti-sigma factors through acetylation of CshA in Bacillus subtilis. Front. Microbiol. 7:1918 10.3389/fmicb.2016.01918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M., Kanesaki Y. (2018). Newly identified nucleoid-associated-like protein YlxR regulates metabolic gene expression in Bacillus subtilis. mSphere 3:e00501-18. 10.1128/mSphere.00501-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okanishi H., Kim K., Masui R., Kuramitsu S. (2013). Acetylome with structural mapping reveals the significance of lysine acetylation in Thermus thermophilus. J. Proteome Res. 12 3952–3968. 10.1021/pr400245k [DOI] [PubMed] [Google Scholar]
- Okkels L. M., Müller E. C., Schmid M., Rosenkrands I., Kaufmann S. H., Andersen P., et al. (2004). CFP10 discriminates between nonacetylated and acetylated ESAT-6 of Mycobacterium tuberculosis by differential interaction. Proteomics 4 2954–2960. 10.1002/pmic.200400906 [DOI] [PubMed] [Google Scholar]
- Ouidir T., Jarnier F., Cosette P., Jouenne T., Hardouin J. (2015). Characterization of N-terminal protein modifications in Pseudomonas aeruginosa PA14. J. Proteomics 114 214–225. 10.1016/j.jprot.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Paquette N., Conlon J., Sweet C., Rus F., Wilson L., Pereira A., et al. (2012). Serine/threonine acetylation of TGFβ-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc. Natl. Acad. Sci. U.S.A. 109 12710–12715. 10.1073/pnas.1008203109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Chen Y., Tishkoff D. X., Peng C., Tan M., Dai L., et al. (2013). SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50 919–930. 10.1016/j.molcel.2013.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C., Lu Z., Xie Z., Cheng Z., Chen Y., Tan M., et al. (2011). The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics 10:M111.012658. 10.1074/mcp.M111.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peregrín-Alvarez J. M., Sanford C., Parkinson J. (2009). The conservation and evolutionary modularity of metabolism. Genome Biol. 10:R63. 10.1186/gb-2009-10-6-r63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D. M. (1963). The presence of acetyl groups of histones. Biochem. J. 87 258–263. 10.1042/bj0870258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B., Sherman F. (2003). N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 325 595–622. 10.1016/s0022-2836(02)01269-x [DOI] [PubMed] [Google Scholar]
- Porcu M., Chiarugi A. (2005). The emerging therapeutic potential of sirtuin-interacting drugs: from cell death to lifespan extension. Trends Pharmacol. Sci. 26 94–103. 10.1016/j.tips.2004.12.009 [DOI] [PubMed] [Google Scholar]
- Post D. M. B., Schilling B., Reinders L. M., D’Souza A. K., Ketterer M. R., Kiel S. J., et al. (2017). Identification and characterization of AckA-dependent protein acetylation in Neisseria gonorrhoeae. PLoS One 12:e0179621. 10.1371/journal.pone.0179621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüss B. M., Besemann C., Denton A., Wolfe A. J. (2006). A complex transcription network controls the early stages of biofilm development by Escherichia coli. J. Bacteriol. 188 3731–3739. 10.1128/jb.01780-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin R., Sang Y., Ren J., Zhang Q., Li S., Cui Z., et al. (2016). The bacterial two-hybrid system uncovers the involvement of acetylation in regulating of Lrp Activity in Salmonella Typhimurium. Front. Microbiol. 7:1864. 10.3389/fmicb.2016.01864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Montañez S., Kazmierczak K. M., Hentchel K. L., Winkler M. E. (2010). Instability of ackA (acetate kinase) mutations and their effects on acetyl phosphate and ATP amounts in Streptococcus pneumoniae D39. J. Bacteriol. 192 6390–6400. 10.1128/JB.00995-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramponi G., Manao G., Camici G. (1975). Nonenzymatic acetylation of histones with acetyl phosphate and acetyl adenylate. Biochemistry 14 2681–2685. 10.1021/bi00683a018 [DOI] [PubMed] [Google Scholar]
- Reeve C. A., Bockman A. T., Matin A. (1984). Role of protein degradation in the survival of carbon-starved Escherichia coli and Salmonella Typhimurium. J. Bacteriol. 157 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Sang Y., Lu J., Yao Y. F. (2017). Protein acetylation and its role in bacterial virulence. Trends Microbiol. 25 768–779. 10.1016/j.tim.2017.04.001 [DOI] [PubMed] [Google Scholar]
- Ren J., Sang Y., Ni J., Tao J., Lu J., Zhao M., et al. (2015). Acetylation regulates survival of Salmonella enterica serovar Typhimurium under acid stress. Appl. Environ. Microbiol. 81 5675–5682. 10.1128/AEM.01009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Sang Y., Tan Y., Tao J., Ni J., Liu S., et al. (2016). Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PLoS Pathog. 12:e1005458. 10.1371/journal.ppat.1005458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdy A., Chen Y., Hunter E., Gozzi K., Chai Y. (2018). Protein lysine acetylation plays a regulatory role in Bacillus subtilis multicellularity. PLoS One 13:e0204687. 10.1371/journal.pone.0204687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel A. E., Roman C., Wolberger C. (2014). Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2. Protein Sci. 23 1686–1697. 10.1002/pro.2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose I. A., Grunberg-Manago M., Korey S. R., Ochoa S. (1954). Enzymatic phosphorylation of acetate. J. Biol. Chem. 211 737–756. [PubMed] [Google Scholar]
- Roy-Chaudhuri B., Kirthi N., Kelley T., Culver G. M. (2008). Suppression of a cold-sensitive mutation in ribosomal protein S5 reveals a role for RimJ in ribosome biogenesis. Mol. Microbiol. 68 1547–1559. 10.1111/j.1365-2958.2008.06252.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari B. R., Tang Z., Huang H., Yong-Gonzalez V., Molina H., Kong H. E., et al. (2015). Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58 203–215. 10.1016/j.molcel.2015.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakatos A., Babunovic G. H., Chase M. R., Dills A., Leszyk J., Rosebrock T., et al. (2018). Posttranslational modification of a histone-like protein regulates phenotypic resistance to isoniazid in mycobacteria. Sci. Adv. 4:eaao1478. 10.1126/sciadv.aao1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah Ud-Din A. I., Tikhomirova A., Roujeinikova A. (2016). Structure and functional diversity of GCN5-related N-acetyltransferases (GNAT). Int. J. Mol. Sci. 17:E1018. 10.3390/ijms17071018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders B. D., Jackson B., Marmorstein R. (2010). Structural basis for sirtuin function: what we know and what we don’t. Biochim. Biophys. Acta 1804 1604–1616. 10.1016/j.bbapap.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y., Ren J., Ni J., Tao J., Lu J., Yao Y. F. (2016). Protein acetylation is involved in Salmonella enterica serovar Typhimurium virulence. J. Infect. Dis. 213 1836–1845. 10.1093/infdis/jiw028 [DOI] [PubMed] [Google Scholar]
- Sang Y., Ren J., Qin R., Liu S., Cui Z., Cheng S., et al. (2017). Acetylation regulating protein stability and DNA-binding ability of HilD, thus modulating Salmonella Typhimurium virulence. J. Infect. Dis. 216 1018–1026. 10.1093/infdis/jix102 [DOI] [PubMed] [Google Scholar]
- Sauve A. A., Schramm V. L. (2003). Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry 42 9249–9256. 10.1021/bi034959l [DOI] [PubMed] [Google Scholar]
- Schilling B., Basisty N., Christensen D. G., Sorensen D., Orr J. S., Wolfe A. J., et al. (2019). Global lysine acetylation in Escherichia coli results from growth conditions that favor acetate fermentation. J. Bacteriol. 201:e00768-18. 10.1128/JB.00768-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B., Christensen D., Davis R., Sahu A. K., Hu L. I., Walker-Peddakotla A., et al. (2015). Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol. Microbiol. 98 847–863. 10.1111/mmi.13161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. T., Smith B. C., Jackson M. D., Denu J. M. (2004). Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J. Biol. Chem. 279 40122–40129. 10.1074/jbc.m407484200 [DOI] [PubMed] [Google Scholar]
- Shaw K. J., Rather P. N., Hare R. S., Miller G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57 138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V., Hopwood D. A. (1976). Chloramphenicol acetylation in Streptomyces. J. Gen. Microbiol. 94 159–166. 10.1099/00221287-94-1-159 [DOI] [PubMed] [Google Scholar]
- Singhal A., Arora G., Virmani R., Kundu P., Khanna T., Sajid A., et al. (2015). Systematic analysis of mycobacterial acylation reveals first example of acylation-mediated regulation of enzyme activity of a bacterial phosphatase. J. Biol. Chem. 290 26218–26234. 10.1074/jbc.M115.687269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B. C., Denu J. M. (2007). Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. J. Biol. Chem. 282 37256–37265. 10.1074/jbc.m707878200 [DOI] [PubMed] [Google Scholar]
- Song L., Wang G., Malhotra A., Deutscher M. P., Liang W. (2016). Reversible acetylation on Lys501 regulates the activity of RNase II. Nucleic Acids Res. 44 1979–1988. 10.1093/nar/gkw053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spange S., Wagner T., Heinzel T., Krämer O. H. (2009). Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 41 185–198. 10.1016/j.biocel.2008.08.027 [DOI] [PubMed] [Google Scholar]
- Spencer T. E., Jenster G., Burcin M. M., Allis C. D., Zhou J., Mizzen C. A., et al. (1997). Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389 194–198. 10.1038/38304 [DOI] [PubMed] [Google Scholar]
- Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. (2002). Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298 2390–2392. 10.1126/science.1077650 [DOI] [PubMed] [Google Scholar]
- Starai V. J., Escalante-Semerena J. C. (2004a). Acetyl-coenzyme A synthetase (AMP forming). Cell. Mol. Life Sci. 61 2020–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai V. J., Escalante-Semerena J. C. (2004b). Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340 1005–1012. 10.1016/j.jmb.2004.05.010 [DOI] [PubMed] [Google Scholar]
- Starai V. J., Garrity J., Escalante-Semerena J. C. (2005). Acetate excretion during growth of Salmonella enterica on ethanolamine requires phosphotransacetylase (EutD) activity, and acetate recapture requires acetyl-CoA synthetase (Acs) and phosphotransacetylase (Pta) activities. Microbiology 151 3793–3801. 10.1099/mic.0.28156-0 [DOI] [PubMed] [Google Scholar]
- Starai V. J., Takahashi H., Boeke J. D., Escalante-Semerena J. C. (2003). Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinsiek S., Bettenbrock K. (2012). Glucose transport in Escherichia coli mutant strains with defects in sugar transport systems. J. Bacteriol. 194 5897–5908. 10.1128/JB.01502-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Guo H., Lu G., Gu J., Wang X., Zhang X. E., et al. (2016a). Lysine acetylation regulates the activity of Escherichia coli S-adenosylmethionine synthase. Acta Biochim. Biophys. Sin. 48 723–731. 10.1093/abbs/gmw066 [DOI] [PubMed] [Google Scholar]
- Sun M., Xu J., Wu Z., Zhai L., Liu C., Cheng Z., et al. (2016b). Characterization of protein lysine propionylation in Escherichia coli: global profiling, dynamic change, and enzymatic regulation. J. Proteome Res. 15 4696–4708. 10.1021/acs.jproteome.6b00798 [DOI] [PubMed] [Google Scholar]
- Sun X. L., Yang Y. H., Zhu L., Liu F. Y., Xu J. P., Huang X. W., et al. (2018). The lysine acetylome of the nematocidal bacterium Bacillus nematocida and impact of nematode on the acetylome. J. Proteomics 177 31–39. 10.1016/j.jprot.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Suzuki S., Kondo N., Yoshida M., Nishiyama M., Kosono S. (2019). Dynamic changes in lysine acetylation and succinylation of the elongation factor Tu in Bacillus subtilis. Microbiology 165 65–77. 10.1099/mic.0.000737 [DOI] [PubMed] [Google Scholar]
- Szewczak J., Bierczyñska-Krzysik A., Piejko M., Mak P., Stadnik D. (2015). Isolation and characterization of acetylated derivative of recombinant insulin Lispro produced in Escherichia coli. Pharm. Res. 32 2450–2457. 10.1007/s11095-015-1637-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Luo H., Lee S., Jin F., Yang J. S., Montellier E., et al. (2011). Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146 1016–1028. 10.1016/j.cell.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Peng C., Anderson K. A., Chhoy P., Xie Z., Dai L., et al. (2014). Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19 605–617. 10.1016/j.cmet.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Matsushita Y., Yoshikawa A., Isono K. (1989). Cloning and molecular characterization of the gene rimL which encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol. Gen. Genet. 217 289–293. 10.1007/bf02464895 [DOI] [PubMed] [Google Scholar]
- Tanner K. G., Trievel R. C., Kuo M. H., Howard R. M., Berger S. L., Allis C. D., et al. (1999). Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 274 18157–18160. 10.1074/jbc.274.26.18157 [DOI] [PubMed] [Google Scholar]
- Thao S., Chen C. S., Zhu H., Escalante-Semerena J. C. (2010). N𝜀-lysine acetylation of a bacterial transcription factor inhibits Its DNA-binding activity. PLoS One 5:e15123. 10.1371/journal.pone.0015123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao S., Escalante-Semerena J. C. (2011). Control of protein function by reversible N?-lysine acetylation in bacteria. Curr. Opin. Microbiol. 14 200–204. 10.1016/j.mib.2010.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel R. C., Rojas J. R., Sterner D. E., Venkataramani R. N., Wang L., Zhou J., et al. (1999). Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl. Acad. Sci. U.S.A. 96 8931–8936. 10.1073/pnas.96.16.8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker A. C., Escalante-Semerena J. C. (2010). Biologically active isoforms of CobB sirtuin deacetylase in Salmonella enterica and Erwinia amylovora. J. Bacteriol. 192 6200–6208. 10.1128/JB.00874-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker A. C., Escalante-Semerena J. C. (2013). Acetoacetyl-CoA synthetase activity is controlled by a protein acetyltransferase with unique domain organization in Streptomyces lividans. Mol. Microbiol. 87 152–167. 10.1111/mmi.12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara T., Kosono S., Söll D., Tamura K. (2018). Lysine acetylation regulates alanyl-tRNA synthetase activity in Escherichia coli. Genes 9:E473. 10.3390/genes9100473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallari D. S., Rock C. O. (1985). Pantothenate transport in Escherichia coli. J. Bacteriol. 162 1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDrisse C. M., Escalante-Semerena J. C. (2018). In Streptomyces lividans, acetyl-CoA synthetase activity is controlled by O-serine and N. Mol. Microbiol. 107 577–594. 10.1111/mmi.13901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDrisse C. M., Parks A. R., Escalante-Semerena J. C. (2017). A toxin involved in Salmonella persistence regulates its activity by acetylating its cognate antitoxin, a modification reversed by CobB sirtuin deacetylase. mBio 8:e00708-17. 10.1128/mBio.00708-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva D., Suzuki-Minakuchi C., Kosono S., Yoshida M., Okada K., Nojiri H. (2018). Proteome and acylome analyses of the functional interaction network between the carbazole-degradative plasmid pCAR1 and host Pseudomonas putida KT2440. Environ. Microbiol. Rep. 10 299–309. 10.1111/1758-2229.12639 [DOI] [PubMed] [Google Scholar]
- Venkat S., Gregory C., Gan Q., Fan C. (2017a). Biochemical characterization of the lysine acetylation of tyrosyl-tRNA synthetase in Escherichia coli. Chembiochem 18 1928–1934. 10.1002/cbic.201700343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat S., Gregory C., Sturges J., Gan Q., Fan C. (2017b). Studying the lysine acetylation of malate dehydrogenase. J. Mol. Biol. 429 1396–1405. 10.1016/j.jmb.2017.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle O., Xu H., Blanchard J. S. (2013). Mechanism and regulation of mycobactin fatty acyl-AMP ligase FadD33. J. Biol. Chem. 288 28116–28125. 10.1074/jbc.M113.495549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle O., Xu H., Tufariello J. M., Favrot L., Malek A. A., Jacobs W. R., et al. (2016). Post-translational acetylation of MbtA modulates mycobacterial siderophore biosynthesis. J. Biol. Chem. 291 22315–22326. 10.1074/jbc.m116.744532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetting M. W., de Carvalho L. P., Yu M., Hegde S. S., Magnet S., Roderick S. L., et al. (2005). Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433 212–226. 10.1016/j.abb.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Vidali G., Gershey E. L., Allfrey V. G. (1968). Chemical studies of histone acetylation. The distribution of epsilon-N-acetyllysine in calf thymus histones. J. Biol. Chem. 243 6361–6366. [PubMed] [Google Scholar]
- Voet D., Voet J. G. (1990). Biochemistry. New York, NY: John Wiley and Sons, Inc. [Google Scholar]
- Wagner G. R., Payne R. M. (2013). Widespread and enzyme-independent N𝜀-acetylation and N𝜀-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 288 29036–29045. 10.1074/jbc.M113.486753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., et al. (2010). Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327 1004–1007. 10.1126/science.1179687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhao Y., McClelland M., Harshey R. M. (2007). The RcsCDB signaling system and swarming motility in Salmonella enterica serovar Typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J. Bacteriol. 189 8447–8457. 10.1128/jb.01198-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Meyer J. G., Schilling B. (2018). Quantification of site-specific protein lysine acetylation and succinylation stoichiometry using data-independent acquisition mass spectrometry. J. Vis. Exp. 134:57209. 10.3791/57209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Liu T., Li X., Wang R., Zhao W., Zhao G., et al. (2017). Lysine acetylation regulates the function of the global anaerobic transcription factor FnrL in Rhodobacter sphaeroides. Mol. Microbiol. 104 278–293. 10.1111/mmi.13627 [DOI] [PubMed] [Google Scholar]
- Weinert B. T., Iesmantavicius V., Moustafa T., Schölz C., Wagner S. A., Magnes C., et al. (2015). Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 11:833. 10.15252/msb.156513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert B. T., Iesmantavicius V., Wagner S. A., Schölz C., Gummesson B., Beli P., et al. (2013a). Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell 51 265–272. 10.1016/j.molcel.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Weinert B. T., Schölz C., Wagner S. A., Iesmantavicius V., Su D., Daniel J. A., et al. (2013b). Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4 842–851. 10.1016/j.celrep.2013.07.024 [DOI] [PubMed] [Google Scholar]
- Weinert B. T., Satpathy S., Hansen B. K., Lyon D., Jensen L. J., Choudhary C. (2017). Accurate quantification of site-specific acetylation stoichiometry reveals the impact of sirtuin deacetylase CobB on the E. coli acetylome. Mol. Cell. Proteomics 16 759–769. 10.1074/mcp.M117.067587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Ziegler C. A., Black A. M., Eliades S. H., Young S., Porter K. (2002). The N-acetyltransferase RimJ responds to environmental stimuli to repress pap fimbrial transcription in Escherichia coli. J. Bacteriol. 184 4334–4342. 10.1128/jb.184.16.4334-4342.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Ziegler C. A., Low D. A. (1992). Thermoregulation of the pap operon: evidence for the involvement of RimJ, the N-terminal acetylase of ribosomal protein S5. J. Bacteriol. 174 7003–7012. 10.1128/jb.174.21.7003-7012.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimpenny J. W., Firth A. (1972). Levels of nicotinamide adenine dinucleotide and reduced nicotinamide adenine dinucleotide in facultative bacteria and the effect of oxygen. J. Bacteriol. 111 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf E., Vassilev A., Makino Y., Sali A., Nakatani Y., Burley S. K. (1998). Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell 94 439–449. [DOI] [PubMed] [Google Scholar]
- Wolfe A. J. (2010). Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr. Opin. Microbiol. 13 204–209. 10.1016/j.mib.2010.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G. D., Ladak P. (1997). Overexpression and characterization of the chromosomal aminoglycoside 6′-N-acetyltransferase from Enterococcus faecium. Antimicrob. Agents Chemother. 41 956–960. 10.1128/aac.41.5.956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Wang X., Zeng J., Zhou M., Duan X., Li Q., et al. (2015). Proteome-wide lysine acetylation profiling of the human pathogen Mycobacterium tuberculosis. Int. J. Biochem. Cell Biol. 59 193–202. 10.1016/j.biocel.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Guan K. L. (2012). Mechanistic insights into the regulation of metabolic enzymes by acetylation. J. Cell Biol. 198 155–164. 10.1083/jcb.201202056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Hegde S. S., Blanchard J. S. (2011). Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry 50 5883–5892. 10.1021/bi200156t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Y., Xu Z., Liu X., Tan M., Ye B. C. (2018a). Protein acetylation and butyrylation regulate the phenotype and metabolic shifts of the endospore-forming Clostridium acetobutylicum. Mol. Cell. Proteomics 17 1156–1169. 10.1074/mcp.RA117.000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Y., Zhao L., Liu X., Hu H., Liu P., Tan M., et al. (2018b). Characterization of the lysine acylomes and the substrates regulated by Protein Acyltransferase in Mycobacterium smegmatis. ACS Chem. Biol. 13 1588–1597. 10.1021/acschembio.8b00213 [DOI] [PubMed] [Google Scholar]
- Xu J. Y., You D., Leng P. Q., Ye B. C. (2014). Allosteric regulation of a protein acetyltransferase in Micromonospora aurantiaca by the amino acids cysteine and arginine. J. Biol. Chem. 289 27034–27045. 10.1074/jbc.M114.579078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Harper S., Speicher D. W., Marmorstein R. (2002). The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat. Struct. Biol. 9 862–869. [DOI] [PubMed] [Google Scholar]
- Yang H., Sha W., Liu Z., Tang T., Liu H., Qin L., et al. (2018). Lysine acetylation of DosR regulates the hypoxia response of Mycobacterium tuberculosis. Emerg. Microbes Infect. 7:34. 10.1038/s41426-018-0032-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Wang Y., Chen Y., Cheng Z., Gu J., Deng J., et al. (2015). Succinylome analysis reveals the involvement of lysine succinylation in metabolism in pathogenic Mycobacterium tuberculosis. Mol. Cell. Proteomics 14 796–811. 10.1074/mcp.M114.045922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. J., Ogryzko V. V., Nishikawa J., Howard B. H., Nakatani Y. (1996). A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382 319–324. 10.1038/382319a0 [DOI] [PubMed] [Google Scholar]
- Yang X. J., Seto E. (2008a). Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 31 449–461. 10.1016/j.molcel.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. J., Seto E. (2008b). The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9 206–218. 10.1038/nrm2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Ji Q. Q., Yan W., Yang F., Wang E. D. (2017). Acetylation of lysine ?-amino groups regulates aminoacyl-tRNA synthetase activity in Escherichia coli. J. Biol. Chem. 292 10709–10722. 10.1074/jbc.M116.770826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Yoshida M., Kuzuyama T., Nishiyama M., Kosono S. (2019). Protein acetylation on 2-isopropylmalate synthase from Thermus thermophilus HB27. Extremophiles 23 377–378. 10.1007/s00792-019-01090-y [DOI] [PubMed] [Google Scholar]
- Yoshikawa A., Isono S., Sheback A., Isono K. (1987). Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol. Gen. Genet. 209 481–488. 10.1007/bf00331153 [DOI] [PubMed] [Google Scholar]
- You D., Wang M. M., Ye B. C. (2017). Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and post-translational levels. Mol. Microbiol. 103 845–859. 10.1111/mmi.13595 [DOI] [PubMed] [Google Scholar]
- You D., Yao L. L., Huang D., Escalante-Semerena J. C., Ye B. C. (2014). Acetyl coenzyme A synthetase is acetylated on multiple lysine residues by a protein acetyltransferase with a single Gcn5-type N-acetyltransferase (GNAT) domain in Saccharopolyspora erythraea. J. Bacteriol. 196 3169–3178. 10.1128/JB.01961-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D., Yin B. C., Li Z. H., Zhou Y., Yu W. B., Zuo P., et al. (2016). Sirtuin-dependent reversible lysine acetylation of glutamine synthetases reveals an autofeedback loop in nitrogen metabolism. Proc. Natl. Acad. Sci. U.S.A. 113 6653–6658. 10.1073/pnas.1525654113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B. J., Kim J. A., Moon J. H., Ryu S. E., Pan J. G. (2008). The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 18 1529–1536. [PubMed] [Google Scholar]
- Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., et al. (2009). Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics 8 215–225. 10.1074/mcp.M800187-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhou A., Li S., Ni J., Tao J., Lu J., et al. (2016). Reversible lysine acetylation is involved in DNA replication initiation by regulating activities of initiator DnaA in Escherichia coli. Sci. Rep. 6:30837. 10.1038/srep30837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. F., Zhang Q., Gu J., Gong P., Wang X. D., Wang X., et al. (2013). Reversibly acetylated lysine residues play important roles in the enzymatic activity of Escherichia coli N-hydroxyarylamine O-acetyltransferase. FEBS J. 280 1966–1979. 10.1111/febs.12216 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. (2011). Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7 58–63. 10.1038/nchembio.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Chai X., Marmorstein R. (2003). Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA binding. J. Biol. Chem. 278 26071–26077. 10.1074/jbc.m303666200 [DOI] [PubMed] [Google Scholar]
- Zhao K., Harshaw R., Chai X., Marmorstein R. (2004). Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases. Proc. Natl. Acad. Sci. U.S.A. 101 8563–8568. 10.1073/pnas.0401057101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Gomez Hernandez M. E., Fernandez-Lima F., Tse-Dinh Y. C. (2018). Biochemical basis of E. coli topoisomerase I relaxation activity reduction by nonenzymatic lysine acetylation. Int. J. Mol. Sci. 19:E1439. 10.3390/ijms19051439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Zhou Y. N., Jin D. J., Tse-Dinh Y. C. (2017). Deacetylation of topoisomerase I is an important physiological function of E. coli CobB. Nucleic Acids Res. 45 5349–5358. 10.1093/nar/gkx250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Chen T., Zhou L., Fleming J., Deng J., Wang X., et al. (2015). Discovery and characterization of Ku acetylation in Mycobacterium smegmatis. FEMS Microbiol. Lett. 362:fnu051. 10.1093/femsle/fnu051 [DOI] [PubMed] [Google Scholar]


