SUMMARY
Plant parasitic nematodes infect roots and trigger the formation of specialized feeding sites by substantial reprogramming of the developmental process of root cells. In this article, we describe the dynamic changes in the tomato root transcriptome during early interactions with the potato cyst nematode Globodera rostochiensis. Using amplified fragment length polymorphism‐based mRNA fingerprinting (cDNA‐AFLP), we monitored 17 600 transcript‐derived fragments (TDFs) in infected and uninfected tomato roots, 1–14 days after inoculation with nematode larvae. Six hundred and twenty‐four TDFs (3.5%) showed significant differential expression on nematode infection. We employed GenEST, a computer program which links gene expression profiles generated by cDNA‐AFLP and databases of cDNA sequences, to identify 135 tomato sequences. These sequences were grouped into eight functional categories based on the presence of genes involved in hormone regulation, plant pathogen defence response, cell cycle and cytoskeleton regulation, cell wall modification, cellular signalling, transcriptional regulation, primary metabolism and allocation. The presence of unclassified genes was also taken into consideration. This article describes the responsiveness of numerous tomato genes hitherto uncharacterized during infection with endoparasitic cyst nematodes. The analysis of transcriptome profiles allowed the sequential order of expression to be dissected for many groups of genes and the genes to be connected with the biological processes involved in compatible interactions between the plant and nematode.
INTRODUCTION
The term ‘cyst nematode’ represents a common name for members of two nematode genera: Heterodera and Globodera. The member organisms of these genera are oligophagous obligatory plant parasites. A relatively small number of cyst nematode species cause serious problems in major crops, such as potato, sugar beet and soybean. Cyst nematodes spend most of their parasitic lifetime inside the root system of a host plant, feeding on a nematode‐induced conglomerate of metabolically activated root cells, known as the syncytium. On root penetration, the preparasitic second‐stage juveniles (J2) migrate intracellularly in search of an appropriate cell to serve as a starting point for syncytium formation. For migration, J2s use a combination of mechanical force and cell wall‐degrading proteins, such as the β‐1,4‐endoglucanases (Goellner et al., 2000; Smant et al., 1998), pectase lyase (Kudla et al., 2007; Popeijus et al., 2000) and expansin (Kudla et al., 2005; Qin et al., 2004), that are produced and secreted by nematode oesophageal glands. The secreted complex protein mixture is also thought to be responsible for the initiation of the formation of the syncytium and its subsequent expansion, as well as the evasion of plant defence mechanisms (2004, 2000; Williamson and Hussey, 1996; Wubben et al., 2008). On selection of an initial syncytial cell, the infective juvenile induces cell wall dissolution between neighbouring cells. As a result, a large continuum of fused protoplasts, which may include more than 200 cells, is formed (Jones, 1981). Moreover, cyst nematode‐induced syncytia are characterized by increased cytoplasmic streaming, enlargement of the nucleus, an increased number of organelles and replacement of the central vacuole by numerous small vacuoles. The metabolically active feeding cell complex is the sole food source for this parasitic nematode.
The presence of the parasite induces drastic changes in plant gene expression (reviewed by Gheysen and Fenoll, 2002). Several molecular approaches have been exploited to pinpoint changes in plant gene expression as a result of nematode infections, including protein analysis (Rahimi et al., 1993), promoter reporter–gene fusions (Barthels et al., 1997; 2004, 1998; Goddijn et al., 1993; Goverse et al., 1998; 2003, 2004; Niebel et al., 1996; Puzio et al., 2000), in situ hybridization (Goellner et al., 2001; Niebel et al., 1993), reverse transcriptase‐polymerase chain reaction (RT‐PCR) (Karczmarek et al., 2008; Vaghchhipawala et al., 2001), differential display (2000, 1998; Vercauteren et al., 2001), microarray analysis (Alkharouf et al., 2006; Hammes et al., 2005; Ithal et al., 2007; Jammes et al., 2005; Khan et al., 2004; 2007, 2003) and serial analysis of gene expression (SAGE) (Uehara et al., 2007). The outcome of these studies has culminated in the identification of a relatively small group of genes that are involved in a wide range of nematode‐affected plant processes. Apart from genes that relate to metabolic activation, feeding site formation has been shown to be accompanied by the altered expression of genes related to other processes, including plant wounding and defence responses, modification of the cell cycle, alterations in the cytoskeleton and cell wall, and signal transduction and hormonal regulation. As a result of the inherent limitations of the techniques used, these results are diffuse and should be considered to represent the ‘tip of the iceberg’ with respect to the transcriptome of the nematode‐infected tomato root.
Oligonucleotide and cDNA microarrays with automated statistical analysis are excellent tools for the analysis of gene expression changes. Microarrays have been successfully employed in studies of many plant processes, including plant–nematode interactions. Puthoff et al. (2003) studied the mechanisms that control syncytium formation on Arabidopsis thaliana roots after infection with the sugar beet nematode (Heterodera schachtii) (BCN) and soybean cyst nematode (H. glycines) (SCN) using an oligonucleotide microarray representing about 8200 genes. The authors used two closely related cyst nematode species that exhibit the same prefeeding behaviour, but, contrary to BCN, SCN rarely reproduces on Arabidopsis roots. This approach resulted in the identification of 116 genes that were presumably altered by specific BCN parasitism, but not by penetration and migration through the root. In the interaction between SCN and soybean (Glycines max), Khan et al. (2004) showed that approximately 8% of a pool of about 1300 expressed sequence tags (ESTs) on a microarray demonstrated altered expression 2 days post‐inoculation (dpi). The authors concluded that the distribution of affected genes over the functional categories could be biased by the small size of the cDNA pool and by the origin of the soybean ESTs, some of which are derived from SCN‐resistant plants.
More recently, a global analysis of altered gene expression in soybean roots inoculated with SCN was performed using the Affymetrix GeneChip soybean genome oligonucleotide array, which can detect 35 611 soybean transcripts (Ithal et al., 2007). Surprisingly, only a small pool of genes (1.2%) showed statistically significant altered expression levels. In addition, the Affymetrix GeneChip includes probes from the SCN transcriptome (7431 transcripts) that enabled the identification of the coordinated expression of the nematode genes (approximately 24%) potentially involved in parasitism and specific stages of development.
Although the microarray approach is suitable for a broad inventory of plant processes that are altered as a result of nematode infection, it is a hybridization‐based method. As a consequence, in the case of cDNA microarrays, it is often very difficult to distinguish between expression patterns of (highly) homologous members of individual gene families. Amplified fragment length polymorphism‐based mRNA fingerprinting (cDNA‐AFLP) is an alternative technology that can be used for wide‐scale gene expression profiling. This sensitive, reproducible and highly stringent technique allows, in most cases, accurate discrimination of expression patterns of highly homologous genes (Bachem et al., 1996). Initially, this method does not depend on pre‐existing sequence information. However, when computer transcript‐derived fragment (TDF) recognition software is applied, the efficiency of in silico band identification relies on the complexity and quality of cDNA sequence databases. The GenEST software package links TDFs and ESTs (Qin et al., 2001) and enables the avoidance of the laborious processes of cloning and sequencing of individual TDFs. Using only three characteristics of individual TDFs, namely restriction endonuclease recognition sites, primer extensions and length, the corresponding ESTs can be identified. As a result, the number of bands that need to be excised, cloned and sequenced is greatly reduced. In addition, this technique is especially useful for tomato, because the available oligonucleotide microarray only covers a portion of its estimated transcriptome. cDNA‐AFLP‐based expression profiling has been successfully used to characterize plant responses to pathogen infection. The analysis of 16 000 cDNA fragments revealed 251 (1.6%) TDFs that were differentially regulated on inoculation of an Ageratum conyzoides cell culture with Agrobacterium tumefaciens (Ditt et al., 2001). A similar number of TDFs have been screened in the interaction between barley (Hordeum vulgare) and powdery mildew (Blumeria graminis f.sp. hordei), where it was determined that 3.7% of the genes were affected within 12 h after inoculation (Eckey et al., 2004). Using another forma specialis of the same fungal species, f.sp. tritici, Bruggmann et al. (2005) monitored transcriptome changes (25 000 TDFs) in epidermal and subepidermal tissue of wheat. However, most studies concentrate on a small subset of differentially expressed genes which have been subcloned and sequenced.
In this article, we describe the changes in gene expression in tomato root that occur on infection by the potato cyst nematode (PCN) Globodera rostochiensis. Infected and non‐infected root segments were collected and used as starting material for the transcriptome‐wide cDNA‐AFLP analysis. GenEST database screening was then used to identify cDNAs corresponding to relevant TDFs, and an inventory of tomato genes induced by the cyst nematode was obtained. Characterization of this set of genes provides an insight into the dynamics of the interactions between parasitic nematodes and their host plants.
RESULTS
Global view of changes in gene expression
cDNA‐AFLP analysis was used to monitor the effects of PCN infection on gene expression in tomato roots. To this end, mRNA pools from nematode‐infected root segments at four time points (1, 3, 7 and 14 dpi) were compared with those of their non‐infected equivalents (Fig. 1A). The reproducibility of the cDNA‐AFLP‐based expression profiles was verified by a second independent experiment with three primer combinations. Comparison of the autoradiograms revealed that less than 1% of bands were variable (data not shown).
Figure 1.
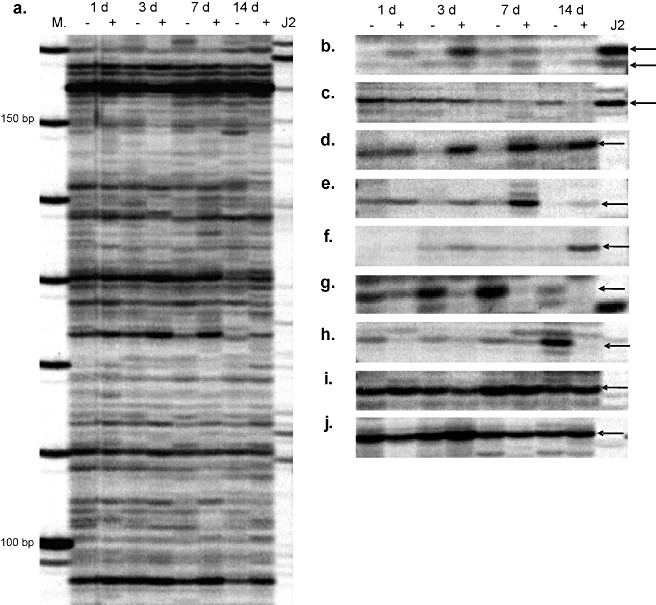
Tomato root segment amplified fragment length polymorphism‐based mRNA fingerprinting (cDNA‐AFLP) display after infection with preparasitic second‐stage juveniles (J2s) of the potato cyst nematode Globodera rostochiensis. Total mRNA extracts from nematode‐infected root segments (≈1 cm in length) were digested with AseI and TaqI and amplified using 2 × 2 selective nucleotides. (A) A section of a polyacrylamide gel displaying transcript‐derived fragments (TDFs) amplified with primers AseI‐GG and TaqI‐CC. (B, C) Root segment‐derived TDFs co‐migrating with fragments from preparasitic J2s (arrowheads). (D–F) Example of plant‐derived TDFs presumed to be up‐regulated in infected root segments (arrowheads; D, lines 3 d and 7 d; E, lines 3 d, 7 d and 14 d; F, lines 3 d and 14 d). (G, H) Example of plant‐derived TDFs presumed to be down‐regulated in potato cyst nematode‐infected root segments (arrowheads). (I, J) Examples of plant‐derived TDFs with expression levels presumed to be essentially unaffected by nematode infection and plant root development (arrowheads). 1 d, 3 d, 7 d and 14 d, days after infection; +, −, infected with potato cyst nematodes and corresponding non‐infected root segments; J2, cDNA‐AFLP profile based on RNA isolated from preparasitic J2s in the absence of plant tissue; M, fragment size marker (in bp).
All possible primer combinations with 2 × 2 selective nucleotides (AseI‐NN and TaqI‐NN) resulted in the profiling of about 17 600 TDFs. On visual inspection, 624 (3.5%) were identified as being up‐regulated. Up‐regulation is defined herein as having a clearly higher band intensity in infected roots relative to corresponding non‐infected controls at one or more time points (e.g. Fig. 1D–F). A similar quantity of bands was identified as being down‐regulated (e.g. Fig. 1G, H). In this analysis, down‐regulated mRNA fragments were not taken into consideration. Examples of TDFs that are essentially unaffected by nematode infection and/or plant root development are shown in Fig. 1I, J.
Distinguishing between transcripts of plant and nematode origin
In this research, our intention was to investigate the plant response to nematode invasion and feeding site induction, and not the developmental changes of the invading pathogen itself. As it is not feasible to remove young parasitic nematodes from plant roots without seriously affecting the mRNA pools, it was necessary to identify alternative methods to reduce the nematode background. To this end, we searched for over‐ and under‐represented sequence motifs in the nematode Caenorhabditis elegans and in tomato. In C. elegans, the relative abundance of the TA motif is very low, and only marginally low in tomato, whereas the CG motif is significantly under‐represented in tomato (Karlin and Mrázek, 1997). This observation for C. elegans is also observed for G. rostochiensis. As a consequence, AseI (AT↓TAAT) was selected as a rare cutter. To moderate the restriction frequency by the frequent cutter, we selected TaqI (T↓CGA). To confirm the plant–nematode discrimination capability of the chosen restriction enzymes, we performed GenEST analysis using 11 983 ESTs from J2s of G. rostochiensis (from GenBank). Only 758 (6%) EST sequences harboured both AseI and TaqI restriction sites, giving rise to 852 virtual TDFs with lengths in the range 80–330 bp. As a control and to provide visualization of the effect of DNA motif‐based restriction endonuclease selection, mRNA from infective J2s was analysed. Although the number of nematode‐derived TDFs was relatively low (Fig. 1, lane ‘J2’), there are some root segment‐derived TDFs that co‐migrate with an mRNA fragment from preparasitic nematode J2s (Fig. 1B, C). This necessitated the rejection of 0.7% of TDFs. Seven and 14 dpi, the nematodes can reach the J3 or J3/J4 stages, and therefore new TDFs may appear. Although these nematode‐derived bands cannot be subtracted using the control J2 lane, we assume that the quantity of such bands is negligible.
Linking nematode‐affected tomato TDFs to corresponding ESTs
The TDFs can be linked to EST databases using a computer program called GenEST (Qin et al., 2001). The TDFs are characterized by their restriction enzyme recognition sites, their size and 2 × 2 extension nucleotides at 3′ primer ends. GenEST was used for the identification of TDFs which are up‐regulated as a result of PCN infection. For 624 TDFs—each typified by three independent identifiers—257 540 entries from GenBank (national Center for Biotechnology Information, NCBI) and 41 425 from the Tomato Gene Index (The Computational Biology and Functional Genomics Laboratory at the Dana‐Farber Cancer Institute and Harvard School of Public Health) were searched. For 135 TDFs (21.5%), one or more corresponding tomato ESTs were identified (see Supporting Information).
Classification and cluster analysis
The blastx algorithm was used to link the identified EST sequences to known proteins. In total, 111 (82%) nematode‐induced ESTs were closely matched (E < 10−5) to non‐redundant database entries. These transcripts were classified into eight functional categories: ‘phytohormone related’; ‘wound, stress and defence response’; ‘cell cycle and cytoskeleton’; ‘cell wall modification’; ‘signalling’; ‘transcription regulation’; ‘metabolism and allocation’; and ‘unclassified’ (Fig. 2). The entire list of 135 up‐regulated genes, including their up‐regulation profiles and predicted functions, is provided in Table S1 (see Supporting Information).
Figure 2.
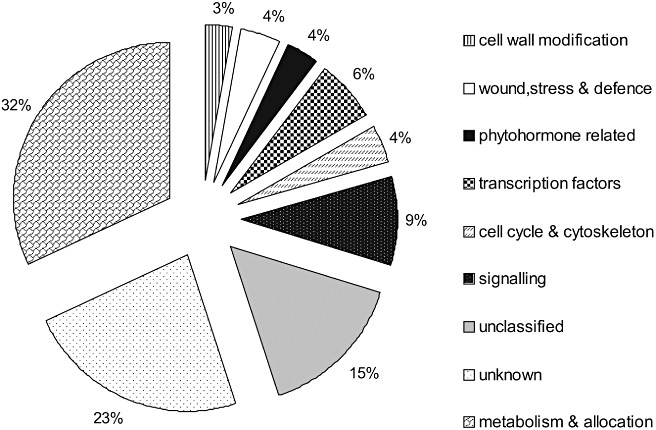
Functional categories of the identified nematode‐induced tomato genes.
A large number of differentially expressed genes appear to be involved in primary and secondary plant metabolism and allocation (45 TDFs, 32%), in accordance with the function of the syncytium as a nutrient source for the nematode. Genes belonging to this group encode proteins with possible functions, for example, in the metabolism of sucrose (sucrose phosphate synthase), aromatic amino acid or fatty acid biosynthesis (4‐hydroxyphenylpyruvate dioxygenase and acyl thioesterase, respectively) and electron transport (cytochrome P450). The second group includes unclassified genes (22 TDFs, 15%) which are involved in processes such as catalysis with ATP binding (26S proteasome AAA‐ATPase, RAD23 and CID7 protein). The next group consists of 13 TDFs (∼9%) involved in signal transduction and represented, for example, by genes encoding guanosine triphosphate (GTP)‐binding protein, putative kinase, myo‐inositol monophosphatase and calmodulin‐like protein. About 6% (nine TDFs) of identified genes differentially regulated in the infected roots are involved in regulation of transcription. About 4% of the up‐regulated transcripts belong to categories that we have termed ‘phytohormone related’, ‘wound, stress and defence response’ and ‘cell cycle and cytoskeleton’ (Table 1). These groups include about five to six TDFs per category. Our study provides additional insight into the details of phytohormone synthesis and signalling in the developing syncytium and neighbouring tissues. Although our data are consistent with previous studies that suggest an important role for auxin and ethylene in syncytium development, we have identified genes that have not been shown previously to be involved in the response to plant parasitic nematodes [e.g. endoplasmic reticulum (ER) auxin binding protein 1, ethylene‐binding protein EIL1/EIL2 and auxin and ethylene cross‐talk marker (AER)]. The smallest functional category is ‘cell wall modification,’ represented by four TDFs (∼3%) (Table 1). These are typically induced during nematode syncytium formation (endoglucanase genes and pectinesterase). Figure 2 summarizes these data and illustrates the nematode‐induced plant genes in all functional categories at 1, 3, 7 and 14 dpi.
Table 1.
Genes up‐regulated in potato cyst nematode (PCN)‐infected tomato roots representing the four functional categories with, presumably, the largest roles in the complex host plant reaction (phytohormone‐related genes; wound, stress and defence response; cell cycle and cytoskeleton; cell wall modification), revealed by amplified fragment length polymorphism‐based mRNA fingerprinting (cDNA‐AFLP) analysis.
| TDF identifier | Corresponding EST/cDNA | Homologous protein (Acc No) | Homologue description | E value | Expression profile | |||
|---|---|---|---|---|---|---|---|---|
| 1 d | 3 d | 7 d | 14 d | |||||
| Phytohormone‐related genes | ||||||||
| A+AA/T+CC/201 | BP890137 | Q8LLM2 | Auxin and ethylene cross‐talk marker (Nicotiana tabacum) | 8E‐60 | + | = | = | + |
| A+GT/T+GA/232 | AF204783 | Q9FR34 | Ripening‐regulated protein DDTFR8 (Solanum lycopersicum) | 0 | + | + | + | = |
| A+GA/T+CT/227 | TC170291 | AAR87866 | Ethylene‐binding protein EIL1/EIL2 (S. lycopersicum) | 0 | + | + | + | + |
| A+TG/T+AA/144 | L21194 | Q43710 | Proteinase inhibitor type II TR8 (ARPI) (S. lycopersicum) | E‐129 | = | + | + | + |
| A+TG/T+AA/160 | TC170423 | Q9ZRX6 | Endoplasmic reticulum auxin binding protein 1 (S. lycopersicum) | 7E‐118 | − | − | + | + |
| Wound, stress and defence response genes | ||||||||
| A+GT/T+GA/225 | Y08804 | P04284 | PR protein (PR1B1) (S. lycopersicum) | 0 | + | = | = | = |
| A+TG/T+AC/249 | BP911027 | Q94AJ9 | Putative heat shock factor protein hsf8 (Arabidopsis thaliana) | 3E‐25 | + | = | = | = |
| A+CA/T+CA/121 | AW932313 | Q42393 | Wound‐induced protein Sn‐1 (Capsicum annuum) | 1E‐34 | + | + | = | = |
| A+CC/T+TG/193 | ES890646 | Q84XQ4 | NtPRp27‐like protein (S. tuberosum) | 2E‐100 | = | + | = | = |
| A+GC/T+CT/103 | TC205399 | Q9LT39 | Polygalacturonase inhibitor‐like protein (A. thaliana) | E‐143 | = | + | + | = |
| A+AG/T+GA/153 | TC190722 | Q68GS0 | SKP1 (N. tabacum) | 5E‐34 | = | = | + | − |
| Cell cycle and cytoskeleton genes | ||||||||
| A+GA/T+AA/123 | BP881672 | Q10QY3 | Tubulin‐tyrosine ligase family protein (Oryza sativa) | 1E‐46 | + | + | + | = |
| A+TT/T+TA/261 | TC182218 | Q6Z1D5 | Putative microtubial‐binding protein (O. sativa) | 2E‐49 | + | + | + | + |
| A+GA/T+CT/226 | AW622695 | Q9LQU7 | ATMAP70‐4 (microtubule‐associated protein 70‐4) (A. thaliana) | 7E‐72 | + | + | + | + |
| A+CT/T+AA/89 | TC172386 | Q9XGI4 | Cyclin A2 (S. lycopersicum) | 0 | = | = | + | = |
| A+AA/T+CC/187 | BP894342 | Q2XTD8 | Histone H2B‐like (S. tuberosum) | 1E‐51 | = | = | + | + |
| A+AA/T+CC/284 | TC189297 | Q7F0M8 | Putative CRK1 protein (cdc2‐related kinase 1) (O. sativa) | 3E‐32 | = | = | + | + |
| Cell wall modification genes | ||||||||
| A+CA/T+AA/222 | TC176937 | O22256 | Putative pectinesterase (At2g47550) (A. thaliana) | 1E‐174 | + | = | = | = |
| A+CG/T+AT/163 | BE433664 | Q9LXD9 | Putative pectin methylesterase (F17I14_50) (A. thaliana) | 2.00E‐18 | = | + | = | = |
| A+CA/T+AA/216 | Y11268 | O04972 | Endo‐1,4‐β‐d‐glucanase (cel 7) (S. lycopersicum) | 0 | x | x | + | ? |
| A+GT/T+AA/322 | TC128121 | Q42875 | Endo‐1,4‐β‐glucanase (cel4) (S. lycopersicum) | 0 | = | = | + | + |
Samples were analysed at 1, 3, 7 and 14 days post‐inoculation. +, up‐regulation; −, down‐regulation; =, equivalent expression in infected and non‐infected tissue; x, no expression; ?, no result. A full list of the identified genes in all functional categories is included in the Supporting Information.
According to the induction profile, the plant genes up‐regulated on nematode infection can be divided into two groups: (i) primary up‐regulated genes (up‐regulation observed at 1 dpi and later); and (ii) secondary up‐regulated genes (induced at 3 dpi and later). In all functional categories, both induction profiles were represented. The first group is presumably related to mechanical damaging of the tissue, pathogen recognition and syncytium initiation, whereas the second group is most probably involved in developmental programme execution and feeding site maintenance. The first group is mostly represented by phytohormone‐related genes. Almost 60% of genes from this functional category respond very early (at 1 dpi and/or between 1 and 3 dpi) during nematode infection. This corresponds to the time at which PCN is actively burrowing and inducing the formation of the syncytium. The genes in the ‘wound, stress and defence response’ category and the ‘cell cycle and cytoskeleton’ category were equally involved in primary and secondary processes. In other categories, the prevailing genes were secondary induced genes during the period in which parasitism of the nematode is established at 3–14 dpi. Figure 3 shows the changes in the percentage of genes belonging to certain functional categories over the measured time points.
Figure 3.
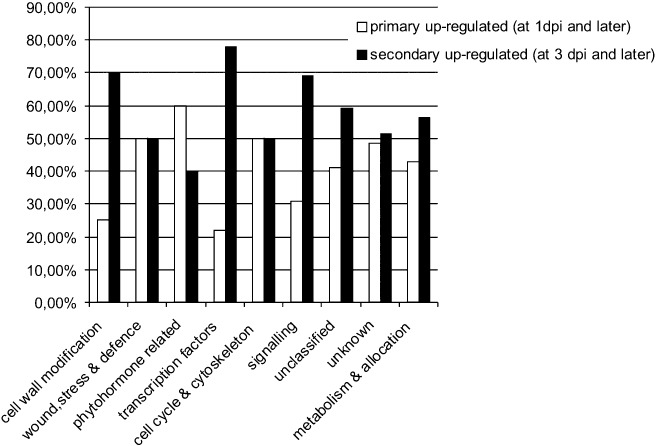
Comparison of the quantity of up‐regulated primary genes at 1 day post‐inoculation (dpi) and up‐regulated secondary genes at 3 dpi and later within functional categories.
The assignment of four time points for the monitoring of tomato root transcriptome changes allowed us to broaden the analysis beyond the primary and secondary events. Five categories show the existence of a ‘second wave’ of gene up‐regulation at 7 dpi (Fig. 4). These gene categories are ‘cell cycle and cytoskeleton’, ‘transcription regulation’, ‘cell wall modification’, ‘phytohormone‐related’ and ‘metabolism and allocation’. We propose that the ‘second wave’ includes genes specifically involved in the function of the syncytium. Unexpectedly, this process is not accompanied by a decrease in gene induction complexity.
Figure 4.
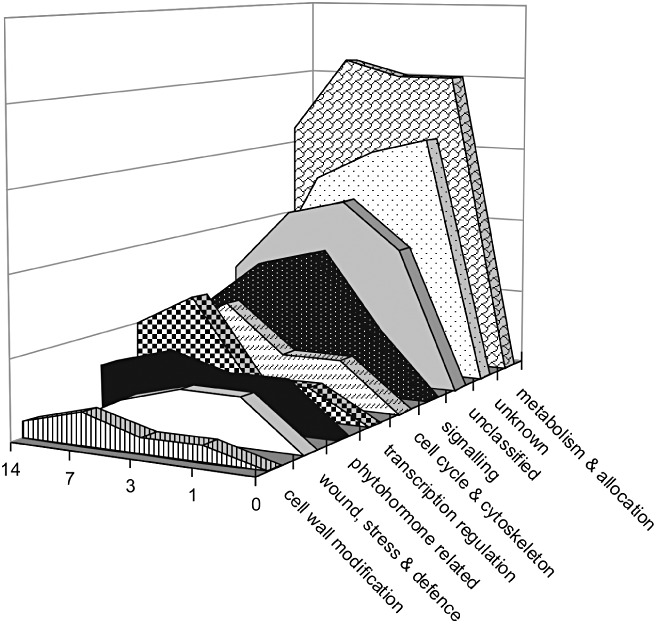
An induction profile showing the existence of a rapid expression at 1 day post‐inoculation (dpi) and a ‘second wave’ of gene up‐regulation at 7 dpi. 0, 1, 3, 7 and 14, dpi.
Verification of cDNA‐AFLP transcript profiles by semi‐quantitative RT‐PCR
To verify the cDNA‐AFLP transcript profiles, semi‐quantitative RT‐PCR was performed for two constitutively expressed genes and three differentially expressed genes. Total RNA from the same biological replicates as used for cDNA‐AFLP was employed for RT‐PCR. The expression levels of ubiquitin and cellulase 2 (Sl‐cel2), a tomato cellulase family member that has been shown to be unaffected by PCN infection (Karczmarek et al., 2008), exhibited similar band intensities at all time points, irrespective of whether RNA was analysed using cDNA‐AFLP or RT‐PCR (Fig. 5). Similarly, the expression profiles of three transcripts that have been suggested to be up‐regulated—an early and continuously (1–14 dpi) up‐regulated unknown protein, an early and transiently (1–3 dpi) up‐regulated protein kinase, and Sl‐cel7 (another cellulase family member) with a delayed up‐regulation (7 dpi)—were analysed. As shown in Fig. 5, semi‐quantitative RT‐PCR‐generated profiles were qualitatively in agreement with those produced by cDNA‐AFLP.
Figure 5.
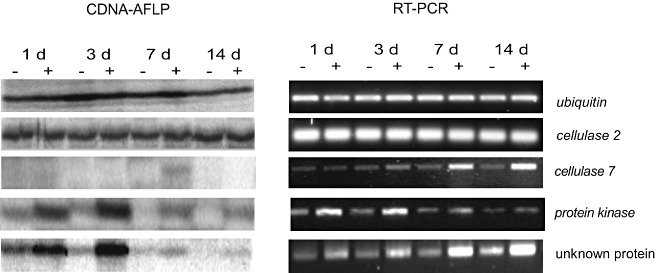
Comparison of expression patterns of selected genes between amplified fragment length polymorphism‐based mRNA fingerprinting (cDNA‐AFLP) and reverse transcriptase‐polymerase chain reaction (RT‐PCR) analysis.
DISCUSSION
The identification of genes regulated in nematode feeding sites represents a major challenge in understanding how nematodes suppress basal plant defence and alter root development to generate and maintain their feeding structure—the syncytium. This complex interaction is not solely restricted to molecular changes at the feeding site itself, but also relates to processes in neighbouring cells of the syncytium, as well as the systemic reaction of the plant. In order to provide novel insights into the complex nature of PCN–tomato interactions, a genome‐wide gene expression analysis was conducted. In this article, we present a cDNA‐AFLP analysis which represents a comprehensive and exhaustive review of dynamic changes in the tomato root transcriptome on infection with PCN (G. rostochiensis). In order to characterize the PCN infection, reacting plant gene time points for analysis were established at 1, 3, 7 and 14 dpi. The expression profiles of a total of 17 600 TDFs were analysed. This number represents the maximum resolution of cDNA‐AFLP obtained using primers with two selective nucleotides and the given restriction enzymes. As shown in the simulation in the Supporting Information, this represents about 27% of the transcriptome (mainly caused by a lack of restriction sites or inappropriate TDF size). In our opinion, this is the main shortcoming of an otherwise very powerful technique. A subset of 624 TDFs (3.5%) was assigned as representing genes which were up‐regulated as a result of cyst nematode infection. Tomato cDNA sequence databases are incomplete and restriction sites for one or both of the endonucleases used in this study are occasionally not present in sequences which are not full sized. As a result, only 21.5% (135) of the TDFs could be linked to the corresponding tomato sequence.
cDNA‐AFLP has been used to study host plant responses in several other plant–pathogen interactions. In the case of microbial infections (van der Biezen et al., 2000; Durrant et al., 2000; Zhang et al., 2003), the proportion of TDFs with altered abundance (including down‐regulated genes) was found to be lower than in the present study, where 3.5% were found to be up‐regulated, and ranged from 0.4 to 3.3%. It is probable that a large proportion of induced genes are related to the developmental modification of differentiated root cells in the syncytium, which represents a very complex structure. Moreover, the neighbouring cells also show developmental changes different from those occurring in the syncytium itself.
The changes occurring in the transcriptome during the interaction between the plant and nematode have, in turn, been studied using microarrays. One of these experiments analysed the expression profiles at three time points (2, 5 and 10 dpi), which represent different stages of SCN (Heterodera glycines) parasitism and development in soybean roots. An Affymetrix GeneChip of the soybean genome was used for this investigation (Ithal et al., 2007). Among the 35 611 soybean transcripts monitored, the authors identified 429 (1.2%) statistically significant differentially expressed genes (375 up‐regulated and 54 down‐regulated). Another recent experiment in which an Affymetrix soybean genome GeneChip was used to analyse changes in the expression profile of plant genes on SCN infection was reported by Puthoff et al. (2007). The authors employed small segments of soybean roots (1–3 mm) that were colonized by SCN at 8, 12 and 16 dpi, and identified 4616 transcripts with significant alterations in expression (Q value ≤0.05) in response to SCN infection. Within this set, 1404 transcripts increased by more than twofold, and 739 transcripts decreased by more than twofold. A direct comparison of our results with this microarray study cannot be made because of differences in the pathosystems employed, time point selection and tissue sampling methods. Although a number of genes highly similar to those identified by Puthoff et al. were found in our study (such as, for example, calmodulin‐like protein, cytochrome P450 and receptor‐like serine/threonine kinase), there are a number of different genes in their register (e.g. auxin‐transport protein—EIR1, ethylene response sensor and expansin gene family members). Possibly, their closest homologues were not identified in our cDNA‐AFLP analysis because they lack the restriction sites for AseI and TaqI or the TDFs were too small or too large for visualization by cDNA‐AFLP.
In this investigation, the greatest challenge is to correlate the differentially expressed genes to feeding site function. To do this, we observed the chronological order of gene expression changes. The penetration of a nematode and the development of a feeding site result in wounding and mechanical stress of plant tissue. Plants usually recognize the intruder and react by switching on a rapid and potent defence response. Changes in gene expression regulated by the wound and defence response have been observed in many plant–nematode interactions. The regulation of these genes differs in the level and timing of their expression. General up‐regulation of plant defence responses is activated very early, and expression levels remain high during the period of syncytium expansion. For example, the potato CAT gene, induced by Globodera pallida, is up‐regulated from 2 to 35 dpi (Niebel et al., 1995). Accordingly, in our study, wound, stress and defence‐related genes were identified. These include genes coding for PR‐1 and heat shock proteins, wound‐induced protein Sn‐1 and polygalacturonase inhibitor‐like protein, which were generally induced at 1 and/or 3 dpi (Table 1). Interestingly, in our study, wound, stress and defence‐related genes are induced until 7 dpi. At the next time point of 14 dpi, these genes are generally expressed at the same level as in uninfected tomato roots of the same age. The design of our experiments, to some extent, hampers the monitoring of recently postulated local suppression of host defence signalling during compatible interaction (Jammes et al., 2005; Wubben et al., 2008). This suppression, however, seems to be less pronounced in the case of cyst nematodes than in root knot nematodes (Ithal et al., 2007). Moreover, we expect the existence of some species‐specific defence reactions (including up‐regulation and its active suppression), such as induction of the tomato PR‐1 protein (Y08804) transcript, which is homologous to PR‐1 of A. thaliana. The expression of this gene remains at a constant level in H. schachtii‐infected Arabidopsis roots (Wubben et al., 2008).
Nematode feeding cell induction and development are connected with the alteration of existing developmental programmes in the plant root. This has consequences in changes in plant gene expression level, particularly alterations in expression of phytohormone‐related genes. It has been reported by several researchers that the plant hormones auxin and ethylene appear to mediate the induction and morphogenesis of cyst nematode syncytia (Glazer et al., 1986; Goverse et al., 2000; Hutangura et al., 1999; Karczmarek et al., 2004; Wubben et al., 2001). In these studies, the authors observed a local and transient increase in auxin perception. This indicates the local accumulation of auxin caused by nematode activity. Consistent with these observations, we detected the up‐regulation of five genes responsive to plant hormones or involved in their biosynthesis and perception (Table 1). For example, we observed the induction of auxin action markers, such as auxin‐inducible proteinase inhibitor (L21194). This gene is induced at 3 dpi and then continuously expressed during the entire infection period. This suggests the involvement of auxin in the induction and development of the nematode feeding site and surrounding cells. Another example is ER auxin binding protein 1 (TC170423), whose expression increases in the later stage of syncytium development (7–14 dpi). The expression change of this auxin‐related gene suggests, in this case, a dominance of developmental control over the rapid stress response. Ethylene is another interesting phytohormone which could play a crucial role in nematode syncytium induction and development. Like auxin, ethylene is involved in many plant developmental processes. Enhanced rates of ethylene production are observed during germination, flower development, pollination, leaf and floral abscission, and fruit ripening. It can also be activated during plant stimulation by various biotic and abiotic stresses (Williamson, 1950; Yang and Hoffman, 1984). An increase in ethylene synthesis has been demonstrated previously during root knot nematode infection in tomato (Glazer et al., 1985). Moreover, experiments with ethylene‐overproducing mutants have demonstrated that nematodes prefer ethylene‐overproducing A. thaliana mutant roots which produce larger syncytia and females (Goverse et al., 2000). In these mutants, the onset of enhanced cell wall degradation on nematode infection was also observed, and this process was found to be similar to cell wall dissolution during fruit ripening. The ripening‐regulated protein DDTFR8 (AF204783) identified in our experiment supports this proposal. Unfortunately, most of the ACC oxidase (ACO) and ACC synthase (ACS) gene family members do not have AseI and TaqI restriction sites. This prevented us from detecting the up‐regulation of any genes involved in ethylene synthesis. These genes are differentially activated by environmental stimuli (Barry et al., 1996; Oetiker et al., 1997). Puthoff et al. (2007) encountered the same difficulties in investigations in which a prevalence of down‐regulation among genes involved in ethylene synthesis or genes responding to this hormone was observed. It is proposed that, in the early stages of nematode infection, ethylene plays a role in the pathogen defence response. It later mediates the proliferation of the syncytium by inducing the modification of cell shape via changes in the cytoskeleton and the induction of endoreduplication. It has also been suggested that many of the cell wall‐modifying proteins involved in nematode syncytium induction and development may be regulated by ethylene (Goverse et al., 2000). Our results indicate new auxin‐ and ethylene‐related genes that provide clues to the role of these plant hormones in feeding site formation and subsequent development.
Many genes induced by the cyst nematode are involved in cell wall modification, as the syncytium is formed by the disassembly of cell walls and the incorporation of hundreds of neighbouring cells. Several studies on changes in plant gene expression as a result of nematode infection have presented a large list of gene families encoding cell wall‐modifying proteins that play roles in nematode feeding cell formation (Ithal et al., 2007; Jammes et al., 2005; 2007, 2003; Tucker et al., 2007; Vercauteren et al., 2002; Wieczorek et al., 2006). We identified the differential regulation of five plant cell wall‐modifying genes, including pectinesterase (TC176937), two endo‐1,4‐beta‐d‐glucanases (Egase), namely Sl‐cel4 and Sl‐cel7 (TC128121 and Y11268, respectively), and pectin methylesterase (BE433664) (Table 1). Karczmarek et al. (2008) provided evidence of the induction of two of eight described tomato endo‐β‐1,4‐glucanases, Sl‐cel7 and Sl‐cel9Cl, on infection with PCN. Our results agree with these previous investigations and demonstrate the induction of tomato Sl‐cel7 at 7 dpi. Unfortunately, the expression of Sl‐cel9Cl could not be determined in our experiment, because it generated a TDF (A+TA/T+GT/1760) which is too large for detection with cDNA‐AFLP.
In the category of ‘transcription factors’, four times more genes are induced as secondary genes (3–14 dpi) than primary genes (1 dpi). This suggests a crucial involvement of transcription regulators induced later in the reprogramming of the developmental processes in plant roots after nematode infection, as well as direct involvement in biotic and abiotic stress responses. Our analyses revealed two MYB domain‐containing transcription factors (MYB177‐DB715805 and MYB12‐TC172990), which are often involved in the stress response.
In the functional category of genes involved in signalling, there were 13 candidates identified in our screen, which are mostly induced from 3 dpi onwards. This indicates that these genes are not only required for initiation and development, but also for maintenance and correct function of the syncytium throughout the monitored period. Included in this group are protein kinases (TC127795, TC189895), which could be involved in linking a signalling component of early activated genes from other categories, such as hormone‐related and wound‐ and stress‐induced categories, and also calmodulin‐like protein (TC182046), which participates in signal transduction mechanisms associated with defence in response to mechanical wounding and insect feeding (Reymond et al., 2000).
The category of ‘metabolism and allocation’ is the largest category and contains proteins related to primary and secondary metabolism, as well as proteins which are possibly involved in the intercellular and intracellular transportation of diverse molecules. The first subcategory contains enzymes putatively belonging to the flavonoid pathway (e.g. flavonol synthase, AW222405). This pathway has diverse physiological functions and is also known to react in response to biotic stress (Winken‐Shirley, 2002), as well as playing a role in developmental processes (Taylor and Grotewold, 2005). The second subcategory is represented by genes encoding proteins engaged in vesicle transport (e.g. amino acid transporter family protein BE461455). This may reflect the observed early activation of vesicular transport in the developing syncytium (Golinowski et al., 1996) and the functional specialization of the feeding site.
The formation of the feeding site is known to be accompanied by cell cycle activation. Both giant cells and syncytium, which are formed by root knot nematode and cyst nematode, respectively, induce a rapid and potent reactivation of the cell cycle. Microscopy suggests that mitosis does not occur within the cyst nematode‐induced feeding cell, and it has been proposed that nematodes induce cycles of DNA endoreduplication by shunting the M phase (Niebel et al., 1996). In our cDNA‐AFLP analysis, cell cycle and cytoskeleton genes, including, for example, cyclin A2 (TC172386), cdc2‐related kinase 1 protein (CRK1, TC189297), histone H2B‐like (BP894342) and microtubule‐associated protein (AW622695), have been found to be up‐regulated early (at 1 dpi, primary up‐regulated group) and later (3–14 dpi, secondary up‐regulated group) during infection (Table 1). Moreover, it is known that actin and tubulin cytoskeletons are strongly disrupted in syncytia, and exogenic stabilization of the cytoskeleton blocks feeding site development (de Almeida‐Engler et al., 2004). This demonstrates the complexity of the interplay between the nematode and plant cell cycle machinery during the formation of the syncytium. Secondary activation of a large proportion of genes in this category (e.g. cyclin A2, histone H2B‐like and CRK1) is consistent with the importance of cell cycle progression throughout the developmental period (in neighbouring cells), and not only at the initiation phase (de Almeida‐Engler et al., 1999). The variability in the activity of genes in the ‘cell cycle and cytoskeleton’ category can also be explained by cell cycle phase‐specific expression, giving an unequal distribution of a given gene activity within the tissue (e.g. cdc2bAt; de Almeida‐Engler et al., 1999). This could result in inadvertent detection of gene up‐regulation at a given time point in relatively small tissue fragments. The cytoskeleton genes belong to the same functional category. Some of these cytoskeleton proteins can be engaged in cytoplasm and vesicle dynamics in syncytial cells, which are very pronounced in feeding site development (Golinowski et al., 1996).
In this article, we have presented an analysis of a large number of tomato genes expressed in response to infection by cyst nematode. This approach has provided a unique insight into the dynamics of the tomato transcriptome. Our data are consistent with previous studies conducted on cyst nematode–plant interactions, in that we observed the regulation of expression levels of genes involved in hormone responses, cell wall modification, the cell cycle and cytoskeleton, wound, stress and defence, and metabolism. Some of the genes identified have been reported previously to play a role in plant–nematode interactions. This demonstrates the reliability of the method used. Several additional genes are reported here for the first time. This demonstrates the power of cDNA‐AFLP as an alternative or complementary method with respect to microarrays. The diversity of up‐regulated genes having different temporal expression profiles clearly shows that syncytium formation and maintenance are very complex processes, and that more analyses are needed to identify the most important molecular relationships. Moreover, the collected dataset can be compared with new releases of databases in the future. When the tomato genome has been sequenced completely, our understanding of a given process will be improved by the addition of newly identified genes to the list. Another challenge in the analysis of known genes is the determination of their spatial expression patterns. This should clarify the characteristics of cells selected as targets for the initiation of syncytium growth, as well as the determination of the interactions between neighbouring cells and developing syncytium. As a consequence, we expect that these results will enrich the palette of tools which may be used in the development of new strategies for plant protection.
EXPERIMENTAL PROCEDURES
Plant material
Tomato seeds (Solanum lycopersicum cv. Moneymaker) were surface sterilized and grown in vitro in square Petri dishes (120 × 120 × 17 mm) containing 1.5% B5 medium (Gamborg basal salt mixture supplemented with 2% sucrose and 1.5% agar, pH 6.2). Each plate contained 10 seeds. Plates were tilted vertically and kept in the dark at 24 °C for 7–10 days.
Nematode culture
Surface sterilization of G. rostochiensis cysts was performed by rinsing for 15 s in 90% ethanol, followed by incubation for 7–9 min in 1.3% commercial bleach. After washing the cysts three times in sterile tap water, they were rehydrated in tap water and retained in the dark at 18 °C for 3 days. Subsequently, the original tap water was replaced by filter‐sterilized potato root diffusate and the cysts were incubated for an additional 5 days. Freshly hatched preparasitic J2s were successively incubated in antibiotic solutions as described by Goverse et al. (1999). Finally, surface‐sterile juveniles were counted and suspended in sterile tap water.
Tissue sample collection and RNA isolation
Surface‐sterile preparasitic J2s were used to infect primary tomato roots directly behind the root tip (100–150 J2s per root). Infection sites were marked at the back of the Petri dish. The corresponding root positions were marked for non‐inoculated controls in separate plates. The plates were sealed with Nesco film (Carl Rath GmbH & Co, Karlsruhe, Germany) and incubated in the dark at 18 °C. At time points 1, 3, 7 and 14 dpi, tomato root segments (length, ≈1 cm) from both infected (as detected by microscopy) and uninfected roots were collected and placed in RNase‐free tubes (≈400 mg of fresh weight per sample). Fresh root segments were immediately frozen in liquid N2 and stored at −80 °C. For total RNA isolation, aliquots of about 100 mg of frozen tomato root segments were incubated in 0.75 mL TRIzol Reagent (Gibco/Life Technologies, Breda, The Netherlands). Subsequently, glass beads (425–600 µm) were added, and the root tissue was homogenized in a Silamat S5 shaker (six times for 30 s for each sample). The integrity of the total RNA was verified on a denaturing agarose gel, and the total RNA concentrations were measured spectrophotometrically. Dynabeads Oligo (dT)25 (Dynal A. S., Oslo, Norway) were used to isolate mRNA from each of the samples.
cDNA‐AFLP
cDNA‐AFLP was performed according to the procedure of Bachem et al. (1996). Briefly, cDNA was synthesized using the SuperScript Choice System (Gibco/Life Technologies). The double‐stranded cDNA was subsequently digested using the restriction endonucleases AseI and TaqI at 37 °C and 65 °C, respectively. Restriction fragments were ligated to AseI and TaqI adaptors (for the adaptor sequences, see Bachem et al., 1998). Pre‐amplification of the primary template was performed in 30 cycles using the following primers: 5′‐CTCGTAGACTGCGTACCTAAT‐3′ (AseI standard primer) and 5′‐GATGAGTCCTGACCGA‐3′ (TaqI standard primer). The amplification products were verified on 1% agarose gel and diluted 10‐fold.
In a touchdown PCR, the secondary template was amplified using 5′‐shortened AseI and TaqI standard primers (Bachem et al., 1998), each with an extension of two selective nucleotides (AseI‐NN and TaqI‐NN). cDNA fragments (often indicated as TDFs) were visualized by autoradiography. A 30–330‐bp AFLP ladder (Gibco/Life Technologies) was used to provide fragment size references on a 50‐cm gel.
In some cases, differentially expressed TDFs were marked, excised from the gel and re‐amplified using Advantage 2 Polymerase mix (BD Biosciences/Franklin Lakes/NJ/USA). The amplified fragments were verified on 2% agarose gel and subsequently cloned into a pCR2.1‐TOPO vector (Invitrogen/Carlsbad/CA/USA).
TDF analysis
To link cDNA sequence data with TDFs as revealed by cDNA‐AFLP, a computer program called GenEST was used (Qin et al., 2001). Each TDF is characterized by the following features: (i) the fragment is the result of restriction by AseI and TaqI; (ii) the restriction sites are flanked by 2 × 2 nucleotides that match the chosen selective nucleotides; and (iii) the size of the TDF is known. GenEST uses this information to perform the digestion of ESTs in silico with AseI and TaqI. Subsequently, GenEST searches for fragments of matching length with the appropriate selective nucleotide‐matching nucleotides.
The EST [sequences were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/dbEST/) and Gene Indices (http://compbio.dfci.harvard.edu/tgi/)] databases are available in FASTA format. Each virtual TDF sequence, predicted by GenEST, was compared against all sequences in the non‐redundant database using the blastx program (http://blast.ncbi.nlm.gov/Blast.cgi).
Semi‐quantitative RT‐PCR
To validate the cDNA‐AFLP expression profiles, semi‐quantitative RT‐PCR was performed for five genes of interest. Total RNA, mRNA and cDNA were prepared by the same procedure as used for cDNA‐AFLP analysis. Ubiquitin—a constitutively expressed gene—was used for normalization (Hoffman et al., 1991). cDNA was amplified in a multiplex PCR reaction containing primers specific for the gene of interest and ubiquitin primers. The PCR products were analysed on a 1% agarose gel. The list of primers used in RT‐PCR is provided in Table S2 (see Supporting Information).
Supporting information
Supporting text GenEST statistics on tomato cDNAs digested with AseI–TaqI restriction enzymes.
Table S1 The entire list of 135 genes induced in tomato roots upon infection with potato cyst nematode including their up‐regulation profiles and their predicted functions. All up‐regulated genes were classified into functional categories and primary induced (from 1 dpi) or secondary induced subcategories (up‐regulated form 3 dpi onwards).
Table S2 Semi‐quantitative reverse transcriptase‐polymerase chain reaction (RT‐PCR) primers used to verify gene expression profiles generated by amplified fragment length polymorphism‐based mRNA fingerprinting (cDNA‐AFLP) in tomato roots infected with Globodera rostochiensis at 1, 3, 7 and 14 days post‐inoculation.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank Professor W. Golinowski from the Department of Botany at Warsaw University of Life Sciences for valuable suggestions and ideas. This work was supported by European Union grant NONEMA (QLK5‐1999‐1501) and Warsaw University of Life Sciences rector's grant.
REFERENCES
- Alkharouf, N.W. , Klink, V.P. , Chouikha, I.B. , Beard, H.S. , MacDonald, M.H. , Meyer, S. , Knap, H.T. , Khan, R. and Matthews, B.F. (2006) Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta, 224, 838–853. [DOI] [PubMed] [Google Scholar]
- De Almeida‐Engler, J. , De Vleesschauwer, V. , Burssens, S. , Celenza, J.L. Jr , Inze, D. , Van Montagu, M. , Engler, G. and Gheysen, G. (1999) Molecular markers and cell cycle inhibitors show the importance of cell cycle progression in nematode‐induced galls and syncytia. Plant Cell, 11, 793–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Almeida‐Engler, J. , Van Poucke, K. , Karimi, M. , De Groodt, R. , Gheysen, G. , Engler, G. and Gheysen, G. (2004) Dynamic cytoskeleton rearrangements in giant cells and syncytia of nematode‐infected roots. Plant J. 38, 12–26. [DOI] [PubMed] [Google Scholar]
- Bachem, Ch.W.B. , Oomen, R.J.F.J. and Visser, R.G.F. (1998) Transcript imaging with cDNA‐AFLP: a step‐by‐step protocol. Plant Mol. Biol. Rep. 16, 157–173. [Google Scholar]
- Bachem, Ch.W.B. , Van Der Hoeven, R.S. , De Bruijn, S.M. , Vreugdenhil, D. , Zabeau, M. and Visser, R.G.F. (1996) Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 9, 745–753. [DOI] [PubMed] [Google Scholar]
- Barry, C.S. , Blume, B. , Bouzayen, M. , Cooper, W. , Hamilton, A.J. and Grierson, D. (1996) Differential expression of the 1‐aminocyclopropane‐1‐carboxylate oxidase gene family in tomato. Plant J. 9, 525–535. [DOI] [PubMed] [Google Scholar]
- Barthels, N. , Van Der Lee, F.M. , Klap, J. , Goddijn, O.J.M. , Karimi, M. , Puzio, P. , Grundler, F.M. , Ohl, S.A. , Lindsey, K. , Robertson, L. , Robertson, W.M. , Van Montagu, M. , Gheysen, G. and Sijmons, P.C. (1997) Regulatory sequences of Arabidopsis drive reporter gene expression in nematode feeding structures. Plant Cell, 9, 2119–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Biezen, A. , Juwana, H. , Parker, J.E. and Jones, J.D.G. (2000) cDNA‐AFLP display for the isolation of Peronospora parasitica genes expressed during infection in Arabidopsis thaliana . Mol. Plant–Microbe Interact. 13, 895–898. [DOI] [PubMed] [Google Scholar]
- Bruggmann, R. , Abderhalden, O. , Reymond, P. and Dudler, R. (2005) Analysis of epidermis and mesophyll‐specific transcript accumulation in powdery mildew‐inoculated wheat leaves. Plant Mol. Biol. 58, 247–267. [DOI] [PubMed] [Google Scholar]
- Davis, E.L. , Hussey, R.S. and Baum, T.J. (2004) Getting to the roots of parasitism by nematodes. Trends Parasitol. 20, 134–141. [DOI] [PubMed] [Google Scholar]
- Davis, E.L. , Hussey, R.S. , Baum, T.J. , Schots, A. , Rosso, M.N. and Abad, P. (2000) Nematode parasitism genes. Annu. Rev. Phytopathol. 38, 365–396. [DOI] [PubMed] [Google Scholar]
- Ditt, R.F. , Nester, E.W. and Comai, L. (2001) Plant gene expression response to Agrobacterium tumefaciens . Proc. Natl. Acad. Sci. USA, 98, 10954–10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant, W.E. , Rowland, O. , Piedras, P. , Hammond‐Kosack, K.E. and Jones, J.D.G. (2000) cDNA‐AFLP reveals a striking overlap in race‐specific resistance and wound response gene expression profiles. Plant Cell, 12, 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckey, C. , Korrel, M. , Leib, K. , Biedenkopf, D. , Jansen, C. , Langen, G. and Kogel, K.H. (2004) Identification of powdery mildew‐induced barley genes by cDNA‐AFLP: functional assessment of an early expressed MAP kinase. Plant Mol. Biol. 55, 1–15. [DOI] [PubMed] [Google Scholar]
- Favery, B. , Chelysheva, L.A. , Lebris, M. , Jammes, F. , Marmagne, A. , De Almeida‐Engler, J. , Lecomte, P. , Vaury, C. , Arkowitz, R.A. and Abad, P. (2004) Arabidopsis forming AtFH6 is a plasma membrane‐associated protein up‐regulated in giant cells induced by parasitic nematodes. Plant Cell, 16, 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery, B. , Lecompte, P. , Gil, N. , Bechtold, N. , Bouchez, D. , Dalmasso, A. and Abad, P. (1998) RPE, a plant gene involved in early developmental steps of nematode feeding cells. EMBO J. 17, 6799–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen, G. and Fenoll, C. (2002) Gene expression in nematode feeding sites. Annu. Rev. Phytopathol. 40, 191–219. [DOI] [PubMed] [Google Scholar]
- Glazer, I. , Epstein, E. , Orion, D. and Apelbaum, A. (1986) Interactions between auxin and ethylene in root‐knot nematode (Meloidogyne javanica) infected tomato roots. Physiol. Mol. Plant Pathol. 28, 171–179. [Google Scholar]
- Glazer, I. , Orion, D. and Apelbaum, A. (1985) Effect of inhibitors and stimulators of ethylene production on gall development in Meloidogyne javanica infected tomato roots. J. Nematol. 17, 145–149. [PMC free article] [PubMed] [Google Scholar]
- Goddijn, O.J.M. , Lindsey, K. , Van Der Lee, F.M. , Klap, J.C. and Sijmons, P.C. (1993) Differential gene expression in nematode‐induced feeding structures of transgenic plants harbouring promoter‐gusA fusion constructs. Plant J. 4, 863–873. [DOI] [PubMed] [Google Scholar]
- Goellner, M. , Smant, G. , De Boer, J.M. , Baum, T. and Davis, E.L. (2000) Isolation of β‐1,4‐endoglucanase genes of Globodera tabacum and their expression during parasitism. J. Nematol. 32, 154–165. [PMC free article] [PubMed] [Google Scholar]
- Goellner, M. , Wang, X. and Davis, E.L. (2001) Endo‐β‐1,4‐glucanase expression in compatible plant–nematode interactions. Plant Cell, 13, 2241–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golinowski, W. , Grundler, F.M.W. and Sobczak, M. (1996) Changes in the structure of Arabidopsis thaliana during female development of the plant‐parasitic nematode Heterodera schachtii . Protoplasma, 194, 103–116. [Google Scholar]
- Goverse, A. , Biesheuvel, J. , Wijers, G.‐J. , Gommers, F.J. , Bakker, J. , Schots, A. and Helder, J. (1998) In planta monitoring of the activity of two ‘constructive’ promoters, CaMV 35S and TR2′, in developing feeding cells induced by Globodera rostochiensis using GFP in combination with confocal laser scanning microscopy. Physiol. Mol. Plant Pathol. 52, 275–284. [Google Scholar]
- Goverse, A. , Overmars, H. , Engelbertink, J. , Schots, A. , Bakker, J. and Helder, J. (2000) Both induction and morphogenesis of cyst nematode feeding cells are mediated by auxin. Mol. Plant–Microbe Interact. 13, 1121–1129. [DOI] [PubMed] [Google Scholar]
- Goverse, A. , Rouppe van der Voort, J. , Roppe van der Voort, C. , Kavelaars, A. , Smant, G. , Schots, A. , Bakker, J. and Helder, J. (1999) Naturally induced secretions of the potato cyst nematode co‐stimulate the proliferation of both tobacco leaf protoplasts and human peripheral blood mononuclear cells. Mol. Plant–Microbe Interact. 12, 872–881. [DOI] [PubMed] [Google Scholar]
- Hammes, U.Z. , Schachtman, D.P. , Berg, R.H. , Nielsen, E. , Koch, W. , McIntyre, L.M. and Taylor, Ch.G. (2005) Nematode‐induced changes of transporter gene expression in Arabidopsis roots. Mol. Plant–Microbe Interact. 18, 1247–1257. [DOI] [PubMed] [Google Scholar]
- Hermsmeier, D. , Hart, J.K. , Byzova, M. , Rodermel, S.R. and Baum, T.J. (2000) Changes in mRNA abundance with Heterodera schachtii‐infected roots of Arabidopsis thaliana . Mol. Plant–Microbe Interact. 13, 309–315. [DOI] [PubMed] [Google Scholar]
- Hermsmeier, D. , Mazarei, M. and Baum, T.J. (1998) Differential display analysis of the early compatible interaction between soybean and the soybean cyst nematode. Mol. Plant–Microbe Interact. 11, 1258–1263. [Google Scholar]
- Hoffman, N.E. , Ko, K. , Milkowski, D. and Pichersky, E. (1991) Isolation and characterization of tomato cDNA and genomic clones encoding the ubiquitin gene ubi3. Plant Mol. Biol. 6, 1189–1201. [DOI] [PubMed] [Google Scholar]
- Hutangura, P. , Mathesius, U. , Jones, M.G.K. and Rolfe, B.G. (1999) Auxin induction is a trigger for root gall formation caused by root‐knot nematodes in white clover and is associated with the activation of the flavonoid pathway. Aust. J. Plant Physiol. 26, 221–231. [Google Scholar]
- Ithal, N. , Recknor, J. , Nettleton, D. , Hearne, L. , Maier, T. , Baum, T.J. and Mitchum, M.G. (2007) Parallel genome‐wide expression profiling of host and pathogen during soybean cyst nematode infection of soybean. Mol. Plant–Microbe Interact. 20, 293–305. [DOI] [PubMed] [Google Scholar]
- Jammes, F. , Lecomte, P. , De Almeida‐Engler, J. , Bitton, F. , Martin‐Magniette, M.‐L. , Renou, J.P. , Abad, P. and Favery, B. (2005) Genome‐wide expression profiling of the host response to root‐knot nematode infection in Arabidopsis. Plant J. 44, 447–458. [DOI] [PubMed] [Google Scholar]
- Jones, M.G.K. (1981) Host cell responses to endoparasitic nematode attack: structure and function of giant cells and syncytia. Ann. Appl. Biol. 97, 353–372. [Google Scholar]
- Karczmarek, A. , Fudali, S. , Lichocka, M. , Sobczak, M. , Kurek, W. , Janakowski, S. , Roosien, J. , Golinowski, W. , Bakker, J. , Goverse, A. and Helder, J. (2008) Expression of two functionally distinct plant endo‐β‐1,4‐glucanases is essential for the compatible interaction between potato cyst nematode and its host. Mol. Plant–Microbe Interact. 21, 791–798. [DOI] [PubMed] [Google Scholar]
- Karczmarek, A. , Overmars, H. , Helder, J. and Goverse, A. (2004) Feeding cell development by cyst and root‐knot nematodes involves a similar early, local and transient activation of a specific auxin‐inducible promoter element. Mol. Plant Pathol. 5, 343–346. [DOI] [PubMed] [Google Scholar]
- Karlin, S. and Mrázek, J. (1997) Compositional differences within and between eukaryotic genomes. Proc. Natl. Acad. Sci. USA, 94, 10227–10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, R. , Alkharouf, N. , Beard, H. , MacDonald, M. , Chouikha, I. , Meyer, S. , Grefenstette, J. , Knap, H. and Matthews, B. (2004) Microarray analysis of gene expression in soybean roots susceptible to the soybean cyst nematode two days post invasion. J. Nematol. 36, 241–248. [PMC free article] [PubMed] [Google Scholar]
- Kudla, U. , Milac, A.L. , Qin, L. , Overmars, H. , Roze, E. , Holterman, M. , Petrescu, A.J. , Goverse, A. , Bakker, J. , Helder, J. and Smant, G. (2007) Structural and functional characterization of a novel, host penetration‐related pectate lyase from the potato cyst nematode Globodera rostochiensis . Mol. Plant Pathol. 8, 293–305. [DOI] [PubMed] [Google Scholar]
- Kudla, U. , Qin, L. , Milac, A. , Kielak, A. , Maissen, C. , Overmars, H. , Popeijus, H. , Roze, E. , Petrescu, A. , Smant, G. , Bakker, J. and Helder, J. (2005) Origin, distribution and 3D‐modeling of Gr‐EXPB1, an expansin from the potato cyst nematode Globodera rostochiensis . FEBS Lett. 579, 2451–2457. [DOI] [PubMed] [Google Scholar]
- Mazarei, M. , Lennon, K.A. , Puthoff, D.P. , Rodermel, S.R. and Baum, T.J. (2003) Expression of an Arabidopsis phosphoglycerate mutase homologue is localized to apical meristems, regulated by hormones, and induced by sedentary plant‐parasitic nematodes. Plant Mol. Biol. 53, 513–530. [DOI] [PubMed] [Google Scholar]
- Mazarei, M. , Lennon, K.A. , Puthoff, D.P. , Rodermel, S.R. and Baum, T.J. (2004) Homologous soybean and Arabidopsis genes share responsiveness to cyst nematode infection. Mol. Plant Pathol. 5, 409–423. [DOI] [PubMed] [Google Scholar]
- Niebel, A. , De Almeida‐Engler, J. , Hemerly, A. , Ferreira, P. , Inzé, D. , Van Montagu, M. and Gheysen, G. (1996) Induction of cdc2a and cyc1At expression in Arabidopsis thaliana during early phases of nematode‐induced feeding cell formation. Plant J. 10, 1037–1043. [DOI] [PubMed] [Google Scholar]
- Niebel, A. , De Almeida‐Engler, J. , Tiré, C. , Engler, G. , Van Montagu, M. and Gheysen, G. (1993) Induction patterns of an extensin gene in tobacco upon nematode infection. Plant Cell, 5, 1697–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebel, A. , Heungens, K. , Brthels, N. , Inzé, D. , Van Montagu, M. and Gheysen, G. (1995) Characterization of a pathogen‐induced potato catalase and its systemic expression upon nematode and bacterial infection. Mol. Plant–Microbe Interact. 8, 371–378. [DOI] [PubMed] [Google Scholar]
- Oetiker, J.H. , Olson, D.C. , Shiu, O.Y. and Yang, S.F. (1997) Differential induction of seven 1‐aminocyclopropane‐1‐carboxylate synthase genes by elicitor in suspension cultures of tomato (Lycopersicon esculentum). Plant Mol. Biol. 34, 275–286. [DOI] [PubMed] [Google Scholar]
- Popeijus, H. , Overmars, H. , Jones, J. , Blok, V. , Goverse, A. , Helder, J. , Schots, A. , Bakker, J. and Smant, G. (2000) Enzymology—degradation of plant cell walls by a nematode. Nature, 406, 36–37. [DOI] [PubMed] [Google Scholar]
- Puthoff, D.P. , Ehrenfried, M.L. , Vinyard, B.T. and Tucker, M.L. (2007) GeneChip profiling of transcriptional responses to soybean cyst nematode, Heterodera glycines, colonization of soybean roots. J. Exp. Bot. 58, 3407–3418. [DOI] [PubMed] [Google Scholar]
- Puthoff, D.P. , Nettleton, D. , Rodermel, S.R. and Baum, T.J. (2003) Arabidopsis gene expression changes during cyst nematode parasitism revealed by statistical analyses of microarray expression profiles. Plant J. 33, 911–921. [DOI] [PubMed] [Google Scholar]
- Puzio, P.S. , Lausen, J. , Heinen, P. and Grundler, F.M.W. (2000) Promoter analysis of pyk20, a gene from Arabidopsis thaliana . Plant Sci. 157, 245–255. [DOI] [PubMed] [Google Scholar]
- Qin, L. , Kudla, U. , Roze, E.H. , Goverse, A. , Popeijus, H. , Nieuwland, J. , Overmars, H. , Jones, J.T. , Schots, A. , Smant, G. , Bakker, J. and Helder, J. (2004) Plant degradation: a nematode expansin acting on plants. Nature, 427, 30. [DOI] [PubMed] [Google Scholar]
- Qin, L. , Prins, P. , Jones, J.T. , Popeijus, H. , Smant, G. , Bakker, J. and Helder, J. (2001) GenEST, a powerful bi‐directional link between cDNA sequence data and gene expression profiles generated by cDNA‐AFLP. Nucleic Acids Res. 29, 1616–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi, S. , Perry, R.N. and Wright, D.J. (1993) Induction and detection of pathogenesis‐related proteins in leaves and roots of potato plants infected with pathotypes of Globodera pallida . Fundam. Appl. Nematol. 16, 549–556. [Google Scholar]
- Reymond, P. , Weber, H. , Damond, M. and Farmer, E.E. (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis . Plant Cell, 12, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smant, G. , Stokkermans, J.P. , Yan, Y. , De Boer, J.M. , Baum, T.J. , Wang, X. , Hussey, R.S. , Gommers, F.J. , Henrissat, B. , Davis, E.L. , Helder, J. , Schots, A. and Bakker, J. (1998) Endogenous cellulases in animals: isolation of beta‐1,4‐endoglucanase genes from two species of plant‐parasitic cyst nematodes. Proc. Natl. Acad. Sci. USA, 95, 4906–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, L.P. and Grotewold, E. (2005) Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 8, 317–323. [DOI] [PubMed] [Google Scholar]
- Tucker, M.L. , Burke, A. , Murphy, Ch. A. , Thai, V.K. and Ehrenfried, M.L. (2007) Gene expression profiles for cell wall‐modifying proteins associated with soybean cyst nematode infection, petiole abscission, root tips, flowers, apical buds, and leaves. J. Exp. Bot. 58, 3395–3406. [DOI] [PubMed] [Google Scholar]
- Uehara, T. , Sugiyama, S. and Masuta, C. (2007) Comparative serial analysis of gene expression of transcript profiles of tomato roots infected with cyst nematode. Plant Mol. Biol. 63, 185–194. [DOI] [PubMed] [Google Scholar]
- Vaghchhipawala, Z. , Bassüner, R. , Clayton, K. , Lewers, K. , Shoemaker, R. and Mackenzie, S. (2001) Modulations in gene expression and mapping of genes associated with cyst nematode infection of soybean. Mol. Plant–Microbe Interact. 14, 42–54. [DOI] [PubMed] [Google Scholar]
- Vercauteren, I. , Engler, J.D. , De Groodt, R. and Gheysen, G. (2002) An Arabidopsis thaliana pectin acetylesterase gene is up‐regulated in nematode feeding sites induced by root‐knot and cyst nematodes. Mol. Plant–Microbe Interact. 15, 404–407. [DOI] [PubMed] [Google Scholar]
- Vercauteren, I. , Van Der Schueren, E. , Van Montagu, M. and Gheysen, G. (2001) Arabidopsis thaliana genes expressed in the early compatible interaction with root‐knot nematodes. Mol. Plant–Microbe Interact. 14, 288–299. [DOI] [PubMed] [Google Scholar]
- Wieczorek, K. , Golecki, B. , Gerdes, L. , Heinen, P. , Szakasits, D. , Durachko, D.M. , Cosgrove, D.J. , Kreil, D.P. , Puzio, P.S. , Bohlmann, H. and Grundler, F.M.W. (2006) Expansins are involved in the formation of nematode‐induced syncytia in roots of Arabidopsis thaliana . Plant J. 48, 98–112. [DOI] [PubMed] [Google Scholar]
- Williamson, C.E. (1950) Ethylene, a metabolic product of diseased or injured plants. Phytopathology, 40, 205–208. [Google Scholar]
- Williamson, V.M. and Hussey, R.S. (1996) Nematode pathogenesis and resistance in plants. Plant Cell, 8, 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winken‐Shirley, B. (2002) Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223. [DOI] [PubMed] [Google Scholar]
- Wubben, M.J., II , Su, H. , Rodermel, S.R. and Baum, T.J. (2001) Susceptibility to the sugar beet cyst nematode is modulated by ethylene signal transduction in Arabidopsis thaliana . Mol. Plant–Microbe Interact. 14, 1206–1212. [DOI] [PubMed] [Google Scholar]
- Wubben, M.J.E. , Jin, J. and Baum, T.J. (2008) Cyst nematode parasitism of Arabidopsis thaliana is inhibited by salicylic acid (SA) and elicits uncoupled SA‐independent pathogenesis‐related gene expression in roots. Mol. Plant–Microbe Interact. 21, 424–432. [DOI] [PubMed] [Google Scholar]
- Yang, S.F. and Hoffman, N.E. (1984) Ethylene biosynthesis in higher plants. Annu. Rev. Plant Physiol. 35, 155–189. [Google Scholar]
- Zhang, L. , Meakin, H. and Dickinson, M. (2003) Isolation of genes expressed during compatible interaction between leaf rust (Puccinia triticina) and wheat using cDNA‐AFLP. Mol. Plant Pathol. 4, 469–477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting text GenEST statistics on tomato cDNAs digested with AseI–TaqI restriction enzymes.
Table S1 The entire list of 135 genes induced in tomato roots upon infection with potato cyst nematode including their up‐regulation profiles and their predicted functions. All up‐regulated genes were classified into functional categories and primary induced (from 1 dpi) or secondary induced subcategories (up‐regulated form 3 dpi onwards).
Table S2 Semi‐quantitative reverse transcriptase‐polymerase chain reaction (RT‐PCR) primers used to verify gene expression profiles generated by amplified fragment length polymorphism‐based mRNA fingerprinting (cDNA‐AFLP) in tomato roots infected with Globodera rostochiensis at 1, 3, 7 and 14 days post‐inoculation.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item


