SUMMARY
In many plant–bacterial interactions, loss of the type III secretion system (T3SS) severely reduces bacterial growth, symptom causation and suppression of defences in host plants. In the present study of Xanthomonas campestris pv. campestris (Xcc), Xcc strain B305 grew better than strain B186 in Arabidopsis thaliana after hydathode inoculation, and B305 strains mutated to the loss of T3SS (ΔhrcC and/or ΔhrpE; also ΔhrcCΔflgBC) grew similarly to wild‐type B305 in Arabidopsis leaves. Unlike Xcc strain B186, wild‐type B305 was relatively inefficient in secreting the exogenous T3S effector AvrBsT, but ΔhrcC and/or ΔhrpE attenuated the disease symptoms caused by Xcc B305, showing that the partially compromised T3SS of this strain still promotes necrotic leaf symptoms. In contrast with the T3SS‐dependent defence suppression that has been observed for some other plant pathogenic bacteria, the Xcc B186 and B305 wild‐type strains (which are virulent on Arabidopsis) caused greater elicitation of host PR‐1 and PR‐5 expression and callose deposition in comparison with their respective T3SS mutants. A defence‐suppressing/virulence‐enhancing activity of the Xcc T3SS effector suite was detectable when co‐inoculation with wild‐type Xcc B186 increased the growth of ΔhrcC Xcc, but this activity did not prevent the above defence elicitation. Experiments using T3SS mutants and Arabidopsis fls2 mutants suggested that FLS2 does not play a prominent role in restriction of the examined Xcc strains. However, ectopic overexpression of the Pseudomonas syringae effector AvrPto promoted in planta growth of wild‐type and ΔhrcC Xcc. In summary, the T3SS components or effector suite from virulent Xcc strains elicit some host defence responses, but suppress other defences and stimulate more severe disease symptoms, AvrPto‐disruptable elements other than FLS2 apparently contribute to the host restriction of Xcc, and in some virulent Xcc strains the T3SS is not absolutely required for wild‐type levels of bacterial growth within the plant.
INTRODUCTION
Xanthomonas campestris pv. campestris (Xcc) causes black rot disease in cruciferous plants, and is considered to be one of the most economically significant diseases of cruciferous crops worldwide (Alvarez, 2000; Williams, 1980). Arabidopsis thaliana is a natural host for pathogenic Xcc (Simpson and Johnson, 1990), making the Xcc–Arabidopsis system an attractive model for study. Unlike many other well‐studied Pseudomonas and Xanthomonas plant pathogens, Xcc is a vascular pathogen that typically enters the plant via the hydathodes or wounds (Alvarez, 2000; Hugouvieux et al., 1998; Lu et al., 2008; Schroth et al., 1991). Hence, differences can be anticipated in the modes of virulence employed by Xcc as opposed to these other bacterial pathogens.
Bacterial type III secretion systems (T3SSs) mediate elaborate interactions with hosts by translocating effector proteins into the cytosol of eukaryotic host cells (Buttner and He, 2009; da Cunha et al., 2007; Galan and Wolf‐Watz, 2006; McCann and Guttman, 2008). The primary known contribution of these effectors to the virulence of plant pathogens is host defence suppression. In Pseudomonas and Xanthomonas plant pathogens, effector proteins delivered by the T3SS suppress host defence responses by manipulating a variety of host pathways (Block et al., 2008; da Cunha et al., 2007; He et al., 2007a; Kay and Bonas, 2009; 2008, 2009b; Zhou and Chai, 2008). In Xcc, it is known that the T3SS plays an essential role in pathogenicity, and the gene cluster that encodes the core components of the Xcc T3S apparatus has been described (Arlat et al., 1991; Cornelis and Van Gijsegem, 2000; Kamoun and Kado, 1990; Kay and Bonas, 2009; Qian et al., 2005; da Silva et al., 2002; White et al., 2009). However, some plant pathogenic Xylella and Pectobacterium bacteria lack any detectable T3SS, yet are successful pathogens (Kim HS et al., 2009a; Lambais et al., 2000; Van Sluys et al., 2002).
Relevant to the present study, the hrcC gene encodes the core outer membrane component of the T3SS in diverse bacteria, and hrcC mutants in Pseudomonas and Xanthomonas have been demonstrated to be deficient in T3S (Bonas, 1994; Keshavarzi et al., 2004; Roine et al., 1997). The HrpE pilin is a major Hrp pilus component that is unique to xanthomonads (Cornelis and Van Gijsegem, 2000; Weber and Koebnik, 2005). This T3SS pilus serves as a conduit for the transfer of bacterial effector proteins into the plant cell cytosol, and is required, for example, for productive interaction of Xanthomonas campestris pv. vesicatoria (Xcv) with pepper host plants (Casper‐Lindley et al., 2002; Weber et al., 2005).
One of the first indications that a primary role of bacterial T3SS effectors is host defence suppression came from microscopy studies with T3SS mutants of Xcv, which caused much greater plant cell wall thickening and papilla formation than did wild‐type bacteria (1995, 1998). In subsequent studies, when wild‐type and ΔhrcC mutants of Xcv were mix‐inoculated into pepper host plants, the bacterial population of the ΔhrcC mutant was significantly larger than after inoculation with the ΔhrcC mutant only (Keshavarzi et al., 2004). T3S‐dependent suppression of basal defence responses triggered by bacterial lipopolysaccharides was demonstrated (Keshavarzi et al., 2004). Individual Xanthomonas T3S effector proteins have also been shown to suppress defence (2008, 2009b; Metz et al., 2005). Xanthomonas bacteria can also suppress plant defences independent of the T3SS, such as via xanthan gum or cyclic β‐(1,2) glucans (Rigano et al., 2007; Yun et al., 2006).
Xanthomonas T3S effectors can play other roles in pathogenicity (for example, AvrBs3 acts as a plant transcription factor that provokes the developmental reprogramming of host cells; Kay and Bonas, 2009; White et al., 2009). Recently, it has been demonstrated that several Xcc T3SS effectors, including XopXccN and AvrXccC, are required for full virulence (2008, 2009; Wang et al., 2007). By contrast, deletion of eight known avr genes in Xcc had no detectable effect on pathogenicity (Castaneda et al., 2005). Many additional hrp gene‐ or T3SS‐associated regulators of Xcc virulence systems have been defined recently (Chao et al., 2008; Jiang et al., 2006; Wang et al., 2008; Wei et al., 2007). Two avr proteins, AvrXccFM and AvrACXcc8004, can induce an atypical hypersensitive response (HR), i.e. HR in vascular tissues (Castaneda et al., 2005; Xu et al., 2008; see, for comparison, (Godard et al., 2000). Hence, the Xanthomonas T3SS and effectors are starting to be understood. However, these studies did not focus on the defence‐activating activity of the T3SS effector suite in virulent bacteria.
In addition to the primary T3SS, the present study tested the flagellum secretion system. The flagellum spans the bacterial membrane, and distal components are secreted through the base of the structure via a specialized T3SS (Macnab, 1999). There are examples in which the flagellum‐associated secretion system is used as a virulence protein export machine (Journet et al., 2005). FlgB and FlgC are among five proteins that make up the rod structure of the flagellum; hence, the deletion of flgB and flgC genes should block the secretion of any distal components and flagellum‐secreted virulence effectors (Aldridge and Hughes, 2002).
Microbe‐associated molecular patterns (MAMPs, also called PAMPs), such as flagellin, have been characterized as broadly conserved molecular patterns that are used by host innate immune systems to recognize microbes (Ausubel, 2005; Jones and Dangl, 2006; Nurnberger et al., 2004). Our group has demonstrated previously that some Xcc strains (including B305) express a flagellin that is detectable by the FLS2 flagellin receptor, whereas other Xcc strains (including B186) express a flagellin that is not detectable by Arabidopsis FLS2 (Sun et al., 2006). However, we found that isogenic Xcc flagellin (fliC) gene replacement strains expressing eliciting or noneliciting flagellins grew similarly in Arabidopsis leaves that carried a functional FLS2. Resistance against Xcc in Arabidopsis was enhanced if purified eliciting flagellin was used to trigger FLS2‐dependent responses 1 day prior to Xcc infection, but the FLS2 system had no detectable effect on disease outcome when naive plants were infected by Xcc that expressed a detectable flagellin. We hypothesized that Xcc bacteria might actively suppress flagellin‐elicited host defences (Sun et al., 2006); this has been shown for Pseudomonas syringae bacteria (Gohre et al., 2008; Shan et al., 2008; de Torres et al., 2006; Xiang et al., 2008).
Initially hypothesizing that the secretion systems of Xcc help to suppress flagellin‐triggered plant basal defences, the present study disrupted type III Hrp and Flg secretion systems in Xcc strains B94, B186 and/or B305. We observed that Xcc strain B305 is much less efficient than strain B186 in secreting the Xcv T3S effector AvrBsT. Intriguingly, after hydathode inoculation, Xcc B305 mutants disrupted for T3SS grew to population sizes in plants similar to those of the wild‐type strain. Loss of T3SS delayed and reduced symptom formation on plants. Moreover, for multiple Xcc strains, the induction of host basal defence was reduced rather than increased when the T3SS was knocked out. Wild‐type Xcc strains, such as B186, are virulent on Arabidopsis Col‐0 and grow with no detectable elicitation of an HR, yet produce T3SS components and/or effectors that induce as well as suppress host defence responses.
RESULTS
Loss of T3SS imposes different effects on bacterial growth in Arabidopsis leaves among three different Xcc strains
Previously studied P. syringae and X. campestris pathovars carrying a mutated hrcC are deficient in T3S, exhibit loss of pathogenicity on host plants and fail to induce an HR on resistant hosts (Alfano and Collmer, 1996, 2004; Keshavarzi et al., 2004). We used homologous recombination to construct unmarked hrcC gene deletion mutants (ΔhrcC) of Xcc strains B94, B186 and B305, precisely deleting from the start to the stop codon of the open reading frame. To corroborate the essential role of T3SS in Xcc virulence (Arlat et al., 1991; Kamoun and Kado, 1990), these ΔhrcC and wild‐type strains were spray inoculated onto Arabidopsis leaves after the induction of guttation droplets. The B186 ΔhrcC strain exhibited reduced population levels as expected, but the B305 ΔhrcC and B94 ΔhrcC strains grew to population levels similar to those of the isogenic wild‐type parent strains (Fig. 1A). When bacteria were introduced directly into the leaf interior by vacuum infiltration, the ΔhrcC mutation caused reduced bacterial populations for the B94 and B186 strains, but the B305 strain once again grew to similar levels with or without hrcC (Fig. 1B). Deletion of hrcC was confirmed by Southern blot (Fig. S1, see Supporting Information). Loss of hrcC function of all three Xcc strains was also confirmed by complementation with hrcC (data not shown) and by other means (see following sections). Hydathode invasion after spray inoculation was indicated by the classic Xcc V‐shaped lesions formed at leaf margins (Fig. S2, see Supporting Information). Based on the results of Figs 1A,B, strains B186 and B305 were chosen for more in‐depth study.
Figure 1.
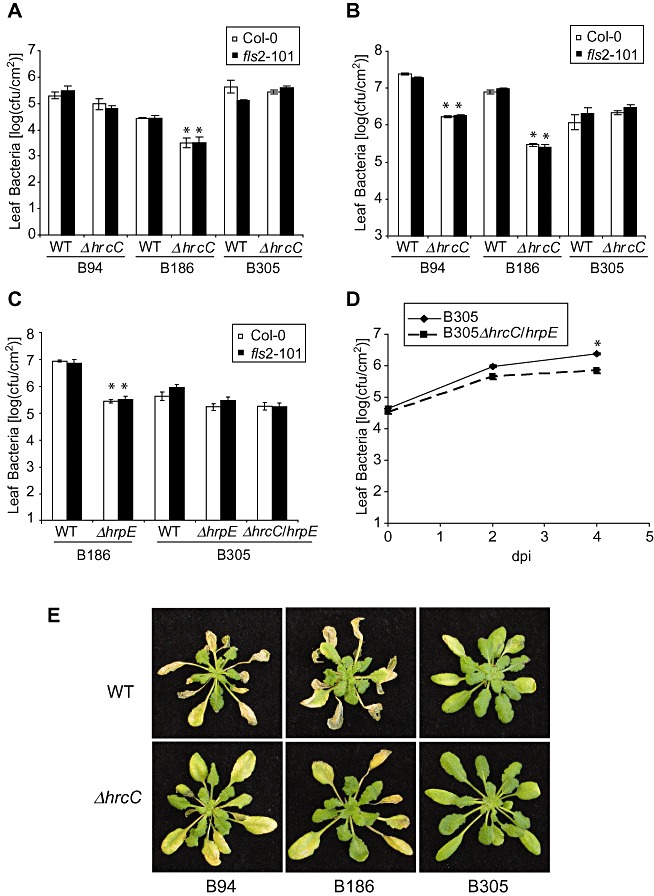
Deletion of hrcC or hrpE causes different in planta bacterial growth phenotypes of Xanthomonas campestris pv. campestris (Xcc) B94, B186 and B305. (A) Xcc population sizes within Arabidopsis leaves 4 days after hydathode inoculation (inoculation by spraying bacteria onto rosette leaves of guttating plants). Bars are means ± standard error. *Significant difference from the wild‐type version of the same strain on the same host genotype (t‐test, P < 0.05). WT, wild‐type strain; ΔhrcC, isogenic strain with unmarked deletion of hrcC gene. This and all other experiments in all figures were repeated with similar results on at least two separate dates unless specifically noted. (B, C) Xcc leaf populations 3 days after inoculation of Arabidopsis by vacuum infiltration. (D) In planta growth curve of B305 ΔhrcC/hrpE and wild‐type B305 after inoculation by vacuum infiltration into Arabidopsis Col‐0 leaves (dpi, days post‐inoculation). (A–D) Mean ± standard error of mean; bar fill in (A), (B) and (C) indicates plant genotype. cfu, colony‐forming units. (E) Representative leaf disease symptoms 7 days after vacuum infiltration. Xcc strain genotype noted below and to the left of the photographs.
To further confirm the finding of T3SS‐independent growth, a separate T3SS gene (hrpE) was deleted from Xcc strains B186 and B305, and a double mutant of B305 was also constructed. On plant inoculation by vacuum infiltration, the ΔhrpE mutation greatly reduced the virulence of B186, but caused significantly less effect on the growth of B305 (Fig. 1C). Similar growth of Xcc B305 with or without hrcC was observed in three different Arabidopsis ecotypes (Fig. 1A and Fig. S3, see Supporting Information). As noted in a previous study, wild‐type B305 grew less well than other wild‐type Xcc after vacuum infiltration into Arabidopsis (Sun et al., 2006; and compare Fig. 1A,B). The experiment reported in Fig. 1D examined this and, although a 10‐fold or greater increase in bacterial populations occurred for B305 strains even after vacuum infiltration, a difference was observed between B305 and the B305 ΔhrcCΔhrpE mutant. The results after spray inoculation of guttating plants (as in Fig. 1A) may be of more interest, where wild‐type B305 reproducibly established larger populations than B186 in Arabidopsis leaves. By spray inoculation, loss of the TTSS reproducibly caused no significant reduction in bacterial population growth of B305 within leaves.
We also made and tested B305 strains with deletion of the flagellar basal body flgB/C genes that have an impact on the secretion of flagellar and other proteins (Journet et al., 2005), as well as a B305 ΔhrcCΔflgB/C double mutant to disrupt both predicted T3SSs of Xcc. These strains also established leaf populations similar to those of wild‐type B305 in Arabidopsis Col‐0 leaves (Fig. S4, see Supporting Information). The phenotypes of the bacterial strains used in this study are summarized briefly in Table S1 (see Supporting Information).
Loss of T3SS attenuates disease symptoms caused by Xcc
Although the loss of T3SS function had less effect on the in planta growth of Xcc B305 than B186, lesion formation by B305 T3SS mutants was attenuated. Under one type of standard experimental conditions, disease symptoms became visible on Arabidopsis leaves 4 days after vacuum infiltration of wild‐type B305, whereas disease symptoms caused by B305 ΔhrcC were not evident until 7 days after inoculation (Fig. 1E, and data not shown). The behaviour of Xcc wild‐type and ΔhrcC strains was also tested for compatible interaction with Nicotiana benthamiana plants and, 5 days after syringe infiltration into leaf mesophyll, the B305 wild‐type and ΔhrcC strains again caused markedly different disease symptoms (Fig. 2A). Xcc B305 caused complete chlorosis/necrosis in the infiltrated leaf area, whereas B305 ΔhrcC produced only a few small water‐soaked spots in the inoculated leaf area. The virulence of the B305 ΔhrcC mutant was restored when a plasmid‐borne wild‐type hrcC gene was introduced into the ΔhrcC strain (Fig. 2A); B186 ΔhrcC was also complemented for growth in plants when a plasmid carrying wild‐type hrcC was added (data not shown). The severe chlorosis/necrosis caused by B305 was unlikely to be the consequence of an HR, because these symptoms only appeared 3–4 days after inoculation. Similarly, no rapid HR was observed after inoculation of Arabidopsis with Xcc B305 (1, 3). B186 ΔhrcC grew to much lower levels than wild‐type B186 in N. benthamiana, whereas a much smaller reduction in the growth of B305 ΔhrcC was observed relative to wild‐type B305 (Fig. 2B). In these N. benthamiana mesophyll inoculation experiments, B305 grew relatively better than B186 (Fig. 2B), in contrast with the Arabidopsis mesophyll inoculation results of Fig. 1B,C. The population size of B305 ΔhrcC increased more than 100‐fold in N. benthamiana plant leaves 3 days after inoculation (Fig. 2C). Overall, the above results suggest that the Xcc B305 T3SS effector suite or components promote leaf necrosis during compatible interactions and speed the development of disease symptoms, but, in contrast with most previously studied plant pathogenic proteobacteria, in planta growth of Xcc B305 is substantially less dependent on the T3SS.
Figure 2.
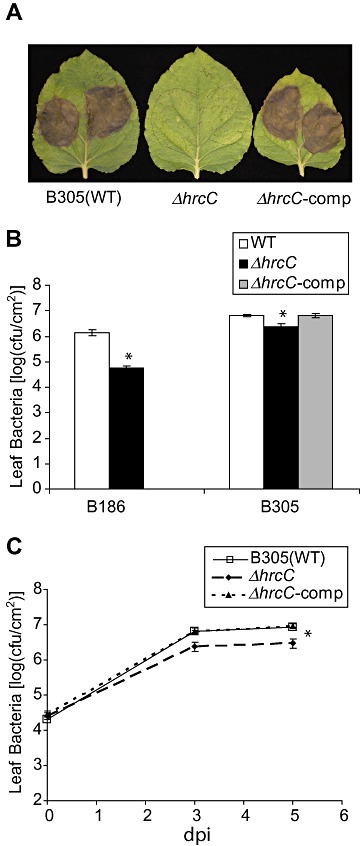
Virulence of (leaf symptoms caused by) Xanthomonas campestris pv. campestris (Xcc) B305 on Nicotiana benthamiana is greatly attenuated by the loss of the hrcC gene despite sustained growth in planta. (A) Necrosis caused by Xcc B305 strains 5 days after syringe infiltration of the same strain onto N. benthamiana leaf. No tissue collapse was evident until at least 3 days after inoculation. WT, wild‐type B305; ΔhrcC, B305ΔhrcC; ΔhrcC‐comp, B305ΔhrcC carrying a plasmid with wild‐type hrcC locus under the control of the native promoter. (B) Xcc populations in syringe‐infiltrated N. benthamiana leaves. (C) Xcc B305 populations in syringe‐infiltrated N. benthamiana leaves; dpi, days post‐inoculation. (B, C) Data are mean ± standard error; bar fill and x‐axis, or line and symbol type, denote bacterial genotypes as described in (A). *Significant difference from the wild‐type version of the same strain (t‐test, P < 0.05). cfu, colony‐forming units.
Figure 3.
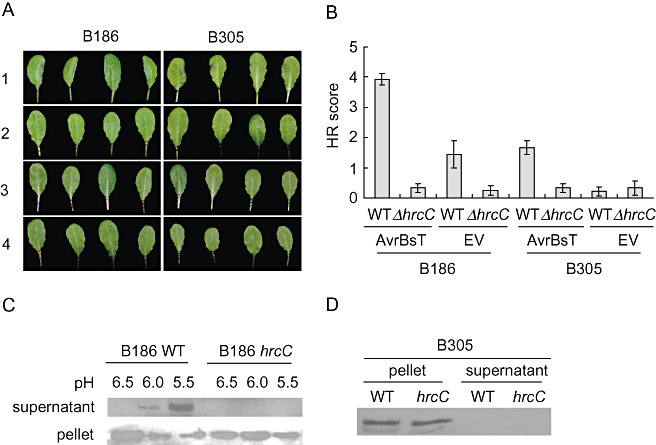
Type III secretion (T3S) effector AvrBsT is secreted efficiently by Xanthomonas campestris pv. campestris (Xcc) B186, but inefficiently by Xcc B305, and is not detectably secreted in ΔhrcC mutants. (A) Hypersensitive response (HR) tissue collapse 24 h after inoculation with Xcc strains as noted [four leaves are shown for each Xcc strain; inoculation at optical density at 600 nm (OD600) = 0.3]. 1, wild‐type strains with the AvrBsT‐HA construct; 2, ΔhrcC strains with the AvrBsT‐HA construct; 3, wild‐type strains with the pDD62 empty vector (EV); 4, ΔhrcC strains with EV. (B) HR scores for leaves inoculated as in (A). Scores were recorded for at least eight leaves per strain; means ± standard error are shown. (C, D) Western blotting for the presence of AvrBsT‐HA detected using anti‐haemagglutinin (HA) antibody after growth of Xcc B186 with the AvrBsT‐HA construct (C) in hrp‐inducing culture medium (SMMXC, pH 6.5–5.5 as noted) or B305 with the AvrBsT‐HA construct (D) in SMMXC medium, pH 5.5.
Disruption of Xcc T3SS does not reveal a detectable impact of host FLS2‐mediated basal immunity
Previous work has shown that Xcc strains B305 and B94 express a flagellin that elicits defence responses in Arabidopsis Col‐0, whereas the flagellin of Xcc B186 is noneliciting (Sun et al., 2006). In that study, an Xcc B186 gene replacement strain expressing a defence‐eliciting flagellin infected Arabidopsis similarly to wild‐type B186 that produces noneliciting flagellin (Sun et al., 2006). In the present study, the hypothesis that one or more T3SS effectors of Xcc suppress FLS2‐mediated defences was examined. Each of the tested Xcc strains grew similarly in Arabidopsis Col (wild‐type FLS2 +) and fls2‐101 mutant plants (Fig. 1A–C). In particular, no contribution of FLS2 to the restriction of Xcc growth became evident after disruption of the T3SS in ΔhrcC or ΔhrpE strains.
Xcc B305 is less efficient than B186 in secreting T3S effectors
The avrBsT‐dependent HR elicited in Arabidopsis ecotype Pi‐0 by Xcc strains can be used to confirm the presence of a functioning T3SS (Cunnac et al., 2007). To further test the T3SS functionality of wild‐type and mutant strains of Xcc B186 and B305, an avrBsT construct or empty vector pDD62 was electroporated into the wild‐type and ΔhrcC mutant strains. Successful transformation was confirmed by Western blot analyses (Fig. 3C,D). Four‐ to six‐week‐old Pi‐0 plants were used for macroscopic HR analyses. The severity of HR was photographed and scored at 24 h after the leaves had been syringe inoculated with different Xcc strains at high cell density [optical density at 600 nm (OD600) = 0.3] (Fig. 3A,B). Xcc B186 expressing AvrBsT‐haemagglutinin (AvrBsT‐HA) elicited severe HRs, whereas B186 with pDD62 did not. Xcc B186 ΔhrcC expressing AvrBsT‐HA lost the ability to cause an HR. Compared to Xcc B186 with AvrBsT‐HA, Xcc B305 with AvrBsT‐HA caused an evident, but less severe, HR with subtle tissue collapse on most inoculated leaves (Fig. 3A,B), suggesting that the T3SS of Xcc B305 is functional but possibly less efficient in delivering AvrBsT‐HA into Arabidopsis cells than that of B186. In vitro secretion assays were performed for these Xcc strains to confirm the secretion of AvrBsT‐HA. In these independent experiments, the hrp‐inducing SMMXC medium (MMXC plus 100 µg/mL bovine serum albumin) at different pH values (Wang et al., 2007) was used to culture Xcc and to trigger the secretion of T3S effectors. For Xcc B186 expressing AvrBsT‐HA, Western blot analyses demonstrated that the AvrBsT effector was not detectably secreted into SMMXC medium at pH 6.5, but the amount of AvrBsT‐HA in the supernatant increased as the pH of the medium was lowered from 6.0 to 5.5 (Fig. 3C). AvrBsT‐HA was not detectable in the supernatant of the Xcc B186 ΔhrcC mutant at any pH, indicating that the hrcC deletion completely disrupted the T3SS of B186 (Fig. 3C). In contrast, for Xcc B305, AvrBsT‐HA was detected only in cell pellets and, in repeated experiments, was not detectable in the supernatant, even when the wild‐type strain with AvrBsT‐HA was cultured in SMMXC medium at pH 5.5 (Fig. 3D).
The set of host defences disrupted by Pseudomonas effector AvrPto contribute to the restriction of Xcc growth
AvrPto is an effector protein from P. syringae pv. tomato that disrupts host defences by disrupting the activity of at least one and probably more MAMP receptors (FLS2, EFR, possibly others including the co‐receptor BAK1; Shan et al., 2008; 2008, 2011). In the absence of a detectable effect of FLS2 on Xcc (see previous section), ectopic AvrPto expression was used to determine whether the broader suite of AvrPto‐sensitive host defence components had an impact on Xcc. Transgenic plants were used in which AvrPto was conditionally overexpressed 1 day prior to inoculation under the control of a dexamethasone (Dex)‐inducible promoter (Aoyama and Chua, 1997; Hauck et al., 2003). Leaf populations of the Xcc B186 wild‐type and ΔhrcC strains grew to approximately 20‐fold and 50‐fold higher levels, respectively, when AvrPto was overexpressed, in comparison with the same strains in Dex‐sprayed plants lacking the PDex‐avrPto construct or in mock‐sprayed PDex‐avrPto transgenic plants (Fig. 4A). Induced AvrPto expression also increased significantly the leaf population sizes of B305 wild‐type and ΔhrcC strains (Fig. 4B). Hence, the AvrPto effector assisted the growth of Xcc B186 and B305, even though mutational removal of only FLS2 did not.
Figure 4.
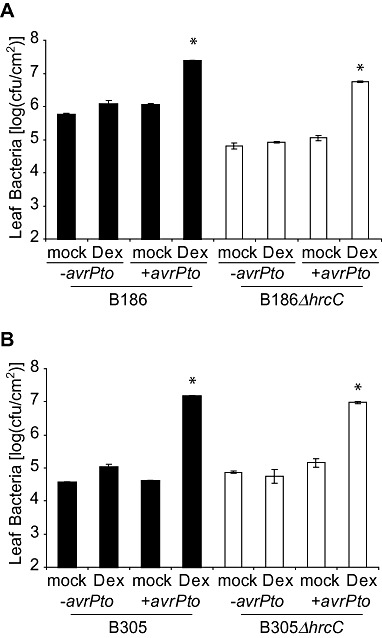
Dexamethasone‐inducible expression of avrPto in Arabidopsis promotes the in planta growth of Xanthomonas campestris pv. campestris (Xcc) wild‐type and ΔhrcC strains after vacuum infiltration. (A) Population sizes of Xcc B186 wild‐type and ΔhrcC strains in transgenic plants carrying avrPto under the control of a dexamethasone‐inducible promoter (+avrPto), or nontransgenic Col gl‐1 plants (‐avrPto), after spraying plants with dexamethasone (Dex) or with buffer (mock) 24 h prior to inoculation. (B) Population sizes of Xcc B305 wild‐type and ΔhrcC strains in an experiment similar to that in (A). All data are means ± standard error. *Significant difference from the same strain on ‐avrPto plants (t‐test, P < 0.05). cfu, colony‐forming units.
The T3SS components or effector suite in Xcc induces host basal defence responses while promoting virulence
For compatible interactions of plants with virulent P. syringae and Xcv bacteria, the T3S effector suite has been shown to suppress plant basal defences, such as callose deposition and pathogenesis‐related (PR) gene expression, that are triggered by MAMP detection (Alfano and Collmer, 2004; Block et al., 2008; Speth et al., 2007). We examined callose deposition in plant leaves after Xcc inoculation to determine whether the Xcc T3S effector suite successfully suppresses plant basal defences. Infiltration of flg22 peptide as a positive control induced callose deposition in Arabidopsis Col‐0 leaves, as expected (Fig. 5A). Wild‐type Xcc B186 and B305 strains also induced callose deposition (Fig. 5A,B). In contrast with T3SS‐deficient mutants of P. syringae pv. tomato and Xcv, which elicit more callose deposition than do isogenic wild‐type strains (e.g. Brown et al., 1995; Hauck et al., 2003), Xcc B186 ΔhrcC partially lost the ability to induce callose deposition (Fig. 5A,B). Equally interesting, the B305 wild‐type and ΔhrcC strains that grow to similar levels within leaves had a similar ability to trigger callose deposition (Fig. 5A,B). Similar callose deposition was observed on wild‐type and fls2‐101 plants for Xcc B186 and B305 strains and their derivative ΔhrcC strains (Fig. S5, see Supporting Information). This latter result suggests that Xcc elicitors other than flagellin are primarily responsible for the triggering of callose deposition in Arabidopsis.
Figure 5.
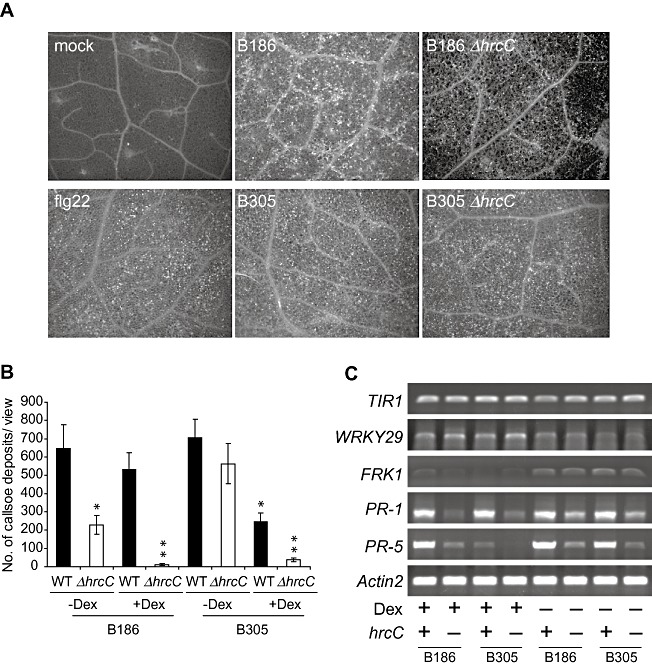
Elicitation of Arabidopsis basal defence responses by Xanthomonas campestris pv. campestris (Xcc) is impaired, not elevated, when the hrcC gene is deleted. (A) Representative images of callose deposition in leaves 24 h after infiltration of Arabidopsis Col‐0 leaves with flg22 peptide or with Xcc strains of the indicated genotype. (B) Quantification of callose deposits per field of view 24 h after infiltration of Xcc strains into Arabidopsis Col‐0 plants carrying avrPto under the control of a dexamethasone‐inducible promoter, activated in some samples by dexamethasone application 24 h prior to bacterial inoculation. Top and bottom lines of the x‐axis annotation indicate the bacterial genotype, middle line indicates plant treatment (–Dex, mock treatment; +Dex, with dexamethasone treatment). Mean ± standard error and statistical significance from other plants treated with the same bacterial strain (t‐test, P < 0.05) are shown; asterisk above ΔhrcC—Dex is for difference from wild‐type (WT)—Dex; asterisk above WT +Dex is for difference from WT—Dex; double asterisk above ΔhrcC+Dex is for difference from both ΔhrcC—Dex and WT +Dex. (C) Abundance of the indicated mRNAs 24 h after the treatments described in (B). For hrcC genotypes, + indicates wild‐type hrcC+ and—indicates ΔhrcC. Equivalent tissue quantities were extracted and assayed using semiquantitative reverse transcription‐polymerase chain reaction (RT‐PCR); Actin2 served as a control for mRNA equivalence.
Callose deposition assays were also performed after Xcc infiltration into Arabidopsis leaves that overexpressed the P. syringae effector AvrPto. Consistent with published findings for P. syringae bacteria, AvrPto reduced callose deposition in plants infiltrated with Xcc B305, B30 5ΔhrcC and B186 ΔhrcC strains. The callose deposition caused by Xcc B186 remained prominent in plants that expressed AvrPto (Fig. 5B).
The expression of PR and MAMP‐induced genes (van Loon et al., 2006; Navarro et al., 2004) was monitored using semiquantitative reverse transcription‐polymerase chain reaction (RT‐PCR). Once again, unlike P. syringae or Xcv T3SS mutants, Xcc ΔhrcC mutants exhibited distinctly less, rather than more, elicitation of PR‐1 and PR‐5 gene expression in comparison with that elicited by wild‐type Xcc (Fig. 5C). The presence/absence of hrcC did not alter significantly the expression of the MAMP‐inducible WRKY29 and FRK1 genes. Consistent with callose deposition assays, AvrPto expression suppressed PR‐5 gene expression induced by Xcc B305, but did not detectably suppress Xcc B186‐induced PR gene expression. Returning to the primary observation, during compatible interactions with virulent Xcc strain B186, Xcc T3SS components or the T3S effector suite caused more, rather than less, induction of benchmark basal defence responses, such as callose deposition and PR‐1 or PR‐5 gene expression.
Co‐inoculation with wild‐type Xcc B186 increases the growth of ΔhrcC Xcc
As noted previously, wild‐type strains of Xcv have not only been shown to inhibit basal defences in pepper, but also to partially restore the multiplication of isogenic nonpathogenic ΔhrcC bacteria on co‐inoculation into plant leaves (Keshavarzi et al., 2004). We traced the in planta growth of kanamycin‐resistant Xcc B186 ΔhrcC and B305 ΔhrcC strains in Arabidopsis on co‐inoculation with wild‐type and kanamycin‐sensitive Xcc B186. Bacterial growth assays indicated that wild‐type B186 assisted the multiplication of B186 ΔhrcC and B305 ΔhrcC (Fig. 6A,B). Co‐inoculation with B186 ΔhrcC did not increase the growth of B305 ΔhrcC (Fig. 6B). A greater growth increase was observed when the inoculum contained a 10:1 rather than a 1:10 ratio of wild‐type to ΔhrcC bacteria (Fig. 6A vs. Fig. 6C). Hence, the Xcc T3SS or its effector suite causes a greater induction of defence responses (such as callose and PR genes; Fig. 5) and more host leaf necrosis (1, 2). The Xcc T3SS effector suite also, in at least some instances, contributes demonstrably to bacterial growth (1, 6).
Figure 6.
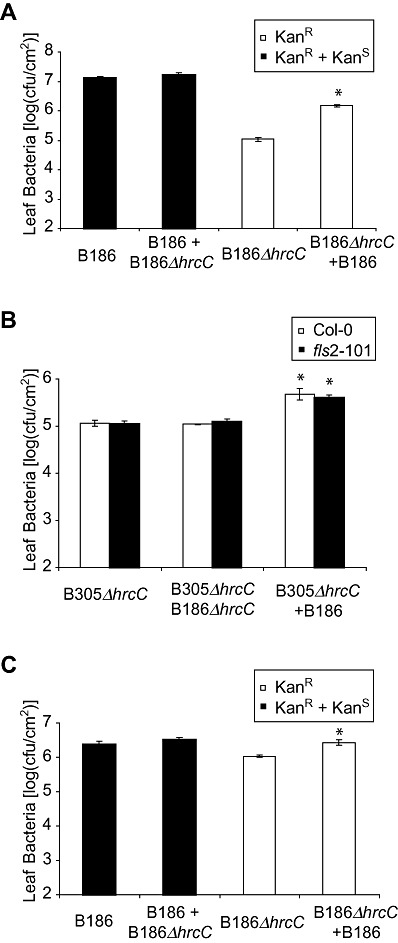
Co‐inoculation of Arabidopsis with wild‐type (WT) Xanthomonas campestris pv. campestris (Xcc) B186 promotes the in planta growth of Xcc ΔhrcC strains. x‐axis labels denote bacteria that were applied to the plant; data are means ± standard error. *Significant difference between population sizes of the first stain listed in the presence/absence of the second strain listed (t‐test, P < 0.05). In (A) and (C), open bars show the main result (leaf population of Xcc ΔhrcC strain carrying a KanR plasmid in the absence or presence of co‐inoculated WT Xcc B186), and filled bars show control data (leaf population of all KanS and/or KanR Xcc bacteria). (A) Leaf population of B186ΔhrcC 3 days after mix‐inoculation [at an optical density at 600 nm (OD600) = 0.0005, low cell density] with WT B186 (at OD600= 0.005, high cell density). (B) Leaf population of B305ΔhrcC 3 days after mix‐inoculation (at OD600= 0.0005, low cell density) with WT B186 or B186ΔhrcC (inoculated at OD600= 0.005, high cell density). In (B), open bars show data for growth in WT Arabidopsis Col‐0 and filled bars show data for a Col‐0 fls2 – mutant. (C) Leaf population of B186ΔhrcC 3 days after mix‐inoculation (at OD600= 0.005, high cell density) with WT B186 (at OD600= 0.0005, low cell density). cfu, colony‐forming units.
DISCUSSION
Pseudomonas, Ralstonia and Xanthomonas mutants deficient in hrcC or other core components of the T3SS consistently exhibit a near‐complete loss of pathogenicity on host plants (Alfano and Collmer, 1996, 2004; Kamoun and Kado, 1990; Lindgren et al., 1986; Rahme et al., 1991; see also Coburn et al., 2007). In contrast, although Xcc strain B186 was strongly dependent on the T3SS for in planta growth, we found that the loss of T3SS core structural genes (hrcC or hrpE) had a distinctly minor impact on the in planta growth of Xcc B305. However, Xcc B305 ΔhrcC or ΔhrpE strains exhibited a delay and overall reduction in disease symptom development in Arabidopsis and N. benthamiana. The ΔhrcC disease phenotype was restored to wild‐type levels if a full‐length hrcC gene was introduced into the mutant strain. B305 ΔhrcCΔflgBC double mutants also grew like the wild‐type strain when vacuum infiltrated into the leaf, suggesting that the in planta growth of Xcc B305 ΔhrcC could not be attributed to the alternative secretion of T3S effectors via the flagellar biosynthetic apparatus, as has been observed in some systems (Journet et al., 2005). Some plant pathogenic bacteria have multiple T3S systems. It would be interesting to determine whether Xcc B305 carries other secretion systems that substitute for the canonical T3SS in contributing to in planta growth.
The differences in symptom development between wild‐type B305 and B305 ΔhrcC indicate that the T3SS of wild‐type B305 is sufficiently functional to secrete effectors that promote disease causation in mesophyll cells. However, direct assays for AvrBsT protein secretion and tests for AvrBsT‐dependent HR (Fig. 3) indicated that, in contrast with Xcc B186, wild‐type B305 carries a defective or differently regulated T3SS, which is consistent with the relative independence of B305 from T3SS noted in the preceding paragraph. We also observed that wild‐type B305 and B305 ΔhrcC triggered a similar amount of callose deposition (Fig. 5), and that the populations of B305 and B305 ΔhrcC were similar in AvrPto‐expressing Arabidopsis plants (Fig. 4B), whereas this was not the case for Xcc B186 (4, 5). Previous studies have shown that some T3SS‐independent factors, such as the major exopolysaccharide xanthan gum or cyclic β‐(1,2) glucans in Xcc, suppress host defences and induce plant susceptibility (Rigano et al., 2007; Yun et al., 2006). These findings are consistent with our observations in suggesting that, at least in some strains, factors other than T3S effectors can contribute significantly to Xcc virulence.
The in planta growth of B305 was comparable with that of B186 ΔhrcC after vacuum infiltration, and B305 was less efficient in secreting the Xcv AvrBsT effector. These observations might suggest that B305 is a universally less virulent strain. However, given the fact that the population size of B305 was equal to or larger than that of B186 after spray inoculation onto Arabidopsis leaves (Fig. 1A; see also Sun et al., 2006), other explanations cannot be ruled out. It is known that some vascular pathogens, such as Xylella fastidiosa and certain strains of Pectobacterium carotovorum and Pectobacterium wasabiae, have no T3SS (Kim et al., 2009a; Lambais et al., 2000; Van Sluys et al., 2002). For vascular pathogens, a T3SS may be less essential in the vessel‐colonizing phase of a vascular lifestyle, where a smaller proportion of the bacterial population is in contact with ‘living’ plant cells. This hypothesis is consistent with our observations. In some cases, the in planta bacterial population of B305 was significantly higher than that of Xcc B305 T3SS mutants after pressure infiltration into leaf mesophyll, despite no differences after hydathode inoculation into Arabidopsis. In addition, Xcc B305 was more virulent than Xcc B186 after hydathode inoculation, but this was reversed when bacteria were introduced into the entire mesophyll by vacuum infiltration. Pathogens such as Xcc are likely to exhibit only partial overlap between the mechanisms required for vascular and mesophyll colonization, with strain‐to‐strain differences apparent. Significant strain‐to‐strain differences in virulence mechanisms may be an increasingly common observation in pathogenesis research given that, for example, 15%–20% of the genes in Xcc pathovars are absent or highly divergent among strains (He et al., 2007b; Lu et al., 2008). Hence, the Xcc B305/B186 contrast may be useful for future studies examining the specific structures, secreted molecules, regulatory pathways and other features that allow bacteria to successfully invade through hydathodes and colonize vascular tissues.
In the present study, co‐inoculation with wild‐type Xcc B186 increased the in planta growth of Xcc ΔhrcC mutants, revealing some defence suppression activity by the wild‐type strain. This observation is consistent with many previous studies (Alfano and Collmer, 2004; Block et al., 2008; Brown et al., 1995; Jakobek et al., 1993; Speth et al., 2007). However, in contrast with many of these previous studies with other bacteria, wild‐type Xcc B305 and B186 elicited more host callose deposition and PR gene expression than did their T3SS‐defective mutants, on a compatible host. Similarly, in a compatible interaction between turnip and Xcc strain 8004, wild‐type bacteria induced a host β‐1,3‐glucanase gene more strongly than did an hrp mutant (Newman et al., 1994). This phenomenon was also observed in two recent studies published after the present report was submitted (Oh et al., 2010; Rong et al., 2010).
Given the reduced macroscopic damage caused by T3SS‐defective mutants, even in cases in which these mutants achieved similar leaf population levels, the simplest explanation for the greater elicitation of host defences by the T3SS‐defective mutants is that the T3SS components or effectors of Xcc contribute directly to the elicitation of host responses as well as to the suppression of host defences. However, other alternative explanations for the data obtained cannot be ruled out. In many cases, defences are more evident in plants in which bacteria are multiplying at a higher rate, causing more damage and/or producing more T3SS‐independent elicitors. This explanation may be more relevant to the concept of the host recognition of ‘patterns of pathogenesis’—that the immune system responds to PAMPs in the context of additional signals (Vance et al., 2009). A recent study on Xcc 8004 has suggested that effector‐triggered immunity contributes to the induction of defence responses in Arabidopsis, such as callose deposition and PR gene expression—a third explanation of this phenomenon (Rong et al., 2010). Other studies have also demonstrated that several T3S effectors have quantitative (weak) avirulence activities on their susceptible hosts (Vinatzer et al., 2006). Our findings and those of others apparently demonstrate that, as bacterial pathogens and their hosts co‐evolve, strains can be present whose T3SS effector suite plays a greater or lesser role in facilitating growth within hosts, and in eliciting rather than suppressing defences. Individual effectors are well known to serve as virulence factors or as strong avirulence factors depending on the host genotype, but there are also effectors that are only weak avirulence factors or trigger an atypical HR. It is interesting to consider that a mild defence‐eliciting activity of effectors may provide adaptive utility to the pathogen.
The loss of disease symptomatology despite relatively unaltered bacterial titres, observed for Xcc B305, provides a unique form of evidence for the contribution of a portion of the Xcc T3SS effector suite to disease symptom development. More typically, loss of symptom‐inducing effectors also causes a major decrease in bacterial growth within plants (White et al., 2009). The T3S effector suite of Xcc has been partially predicted and studied (Castaneda et al., 2005; 2008, 2009; Qian et al., 2005; Wang et al., 2007; Xu et al., 2008). It would be interesting to discover the Xcc T3S effectors that act as lesion‐promoting factors, and to determine whether these are the same as or different from the Xcc effectors that elicit defence responses.
The plant FLS2 receptor activates biologically significant defences in response to the flagellins of many bacteria, yet some Xcc strains cause similar disease whether or not they have a flagellin that is detectable by Arabidopsis FLS2 (Chinchilla et al., 2006; Forsyth et al., 2010; Gomez‐Gomez and Boller, 2000, 2002; Pfund et al., 2004; Sun et al., 2006). In the present study, we observed that FLS2‐mediated bacterial growth restriction was not unmasked when the T3SS was knocked out, suggesting that T3S effectors are not the primary reason why FLS2 plays a less detectable role in defence against Xcc than it does, for example, with P. syringae pv. tomato (Zipfel et al., 2004). Previous work has shown that Arabidopsis FLS2‐mediated defences can restrict Xcc growth if they are activated with flagellin 1 day prior to Xcc infection, but, on naive Arabidopsis, the presence or absence of FLS2 does not have a detectable impact on the outcome of Xcc infections (Sun et al., 2006). With no evidence for strong FLS2 suppression via these Xcc secretion systems, other hypotheses for the lack of FLS2 impact on Xcc merit testing. Xcc strains may not make or expose sufficient flagellin within host leaves to elicit an adequate defence response; non‐T3S compounds [such as cyclic β‐(1,2) glucans] may suppress FLS2‐mediated defences; or the plant vasculature (the most typical route for Xcc infection) may be less sensitive to flagellin than is the leaf mesophyll.
AvrPto is a widely conserved T3S effector of Pseudomonas species, and can suppress multiple host defences during compatible interactions (Hauck et al., 2003; He et al., 2006). AvrPto binds directly FLS2, EFR and possibly other MAMP receptors or co‐receptor proteins for defence suppression (Shan et al., 2008; 2008, 2011). Induced expression of avrPto in transgenic Arabidopsis plants has been reported to enhance the growth of a P. syringae pv. tomato strain DC3000 ΔhrcC mutant, but to have no significant growth‐promoting effect on wild‐type P. syringae pv. tomato strain DC3000 that expresses an endogenous avrPto (Hauck et al., 2003). In the present study, we found that the expression of avrPto greatly benefited the growth of both wild‐type and ΔhrcC mutant strains of Xcc. This suggests that FLS2‐independent defence pathways targeted by AvrPto play a significant role in the Xcc–host interaction and restrict Xcc growth in the host mesophyll, as also indicated recently for Xcv (Kim et al., 2009b).
AvrPto allowed greater growth of wild‐type B186, but did not suppress significantly the callose or PR gene mRNA levels elicited by wild‐type B186. This indicates that callose deposition and expression of PR‐1 and PR‐5 are not the major defence responses that restrict the in planta growth of Xcc B186. Hence, caution may be advisable in the use of callose deposition as a marker for PAMP‐triggered immunity in the study of Xcc–Arabidopsis interactions. Because the induction of these responses was reduced by ΔhrcC mutation of B186, B186 apparently carries T3S effectors or structural components that elicit defences in Arabidopsis via mechanisms that are not inhibited by AvrPto. AvrPto has been predicted to inhibit MAMP (PAMP) perception rather than T3S effector‐triggered immunity (Jones and Dangl, 2006). It is probable that MAMPs from the T3SS mutants elicit basal defences that can be inhibited by AvrPto (callose accumulation, expression of FRK1 and weak expression of PR genes), whereas T3S effectors from Xcc B186 apparently elicit defences (more callose deposition and stronger PR gene expression) via additional pathways that are not inhibited by AvrPto.
To summarize, the contributions of the T3SS were found to differ in certain respects for Xcc relative to other plant pathogenic bacteria, and to differ between wild‐type Xcc strains. The T3SS is required for the in planta bacterial growth of some, but not all, Xcc strains, whereas its loss significantly attenuates disease symptom causation in all examined cases. The impacts of FLS2 on defence against wild‐type Xcc have not been detected previously and, in the present study, disruption of the T3SS did not reveal any latent effects of FLS2‐mediated defences that are suppressed by the Xcc T3SS. The contribution of ectopic AvrPto expression to Xcc growth within plants suggests that one or more host factors targeted by AvrPto, other than FLS2, contribute significantly to the restriction of Xcc growth. Lastly, the T3SS components or effector suite in virulent Xcc induces host basal defence responses while promoting virulence.
EXPERIMENTAL PROCEDURES
Xcc strains
Xanthomonas campestris pv. campestris wild‐type strains, originally from the collection of Dr Norman Schaad, were the kind gift of Dr Wesley Chun and Dr Al Poplawsky at the University of Idaho, Moscow, ID, USA. Xcc B94 was collected from field mustard, Xcc B186 from cauliflower and the source of Xcc B305 was not recorded.
Molecular biology methods
Standard molecular biology methods were used (Ausubel et al., 1997) unless noted. For DNA blot analyses, genomic DNA (500 ng) from Xcc strains was digested with SacII or XhoI and probed with a 32P‐labelled PCR product generated with the primer set hrcC‐probe‐F/hrcC‐probe‐R, with final washing of the blot in 0.1 × standard saline citrate (SSC), 0.1% sodium dodecylsulphate (SDS) at 65 °C.
Construction of Xcc mutant strains with hrcC, hrpE and flgB/C single, double and triple gene deletions using nonmarker homologous recombination
DNA was isolated from different Xcc strains using a genomic DNA isolation kit (Promega, Madison, WI, USA). Two fragments, approximately 1 kb in length, upstream and ending at the start codon of the hrcC gene, or downstream and beginning at the stop codon of the hrcC gene, were independently PCR amplified from Xcc genomic DNA using Pfu polymerase with the respective primer sets hrcC‐5′‐F/hrcC‐del‐R and hrcC‐del‐F/hrcC‐3′‐R with underlined BamHI and XhoI restriction sites, respectively (Table S2, see Supporting Information). These two PCR products were gel purified and added together into a splice overlap PCR (the pairs of ‘del’ primers carry complementary sequences and will anneal). The resultant PCR fragment carrying flanking regions of the hrcC gene, but lacking the hrcC open reading frame, was cloned into the pUFR80 suicide vector (Castaneda et al., 2005), a sacB suicide vector that allows the generation of precise unmarked chromosomal gene replacements/deletions in Gram‐negative bacteria (Ried and Collmer, 1987). Construction and screening of Xcc ΔhrcC mutants were performed following the procedures described by Sun et al. (2006). Briefly, pUFR80‐ΔhrcC plasmids were electroporated into Xcc and subjected to kanamycin selection. Xcc transformant single colonies with kanamycin resistance were picked and cultured in nutrient yeast glycerol (NYG) medium (Daniels et al., 1984) overnight with no kanamycin, and then spread onto nutrient yeast glycerol agar (NYGA) plates with 5% sucrose to select sucrose‐insensitive clones. The gene deletion genotype of kanamycin‐sensitive/sucrose‐insensitive Xcc colonies was confirmed by colony PCR and by sequencing PCR products, as well as by Southern blot analyses. The same strategy was applied to construct precise open reading frame deletion strains for hrpE and flgB/C, except that BamHI and HindIII restriction sites were created by PCR for deletion fragments (Table S2). For double mutants, the ΔhrcC single mutant strain was used to construct the second deletion. For complementation experiments, the full‐length hrcC gene, including the 5′ and 3′ regulatory sequences (687 and 225 bp, respectively, for hrcC), was amplified by PCR using the respective primer sets hrcC‐comp‐F/hrcC‐comp‐R (Table S2). Hence, the hrcC complementation construct carried short segments of adjacent open reading frames from separate operons, but no other full‐length genes. The hrcC complementation PCR fragments were cloned into the wide‐host‐range vector pVSP61 (Loper and Lindow, 1987) and electroporated into the specified Xcc strains.
Plant inoculation and pathogen growth assays
All strains were grown at 28 °C on NYGA medium. For hydathode inoculation, 3‐week‐old Arabidopsis Col‐0 and fls2‐101 mutant plants were placed in a dew chamber (wall, 10 °C; water, 28 °C; air, 22 °C) for 4–5 h until guttation drops formed at the edges of the leaves. Bacteria at approximately 1 × 109 colony‐forming units (cfu)/mL in 10 mm MgCl2/0.04% Silwet L‐77 were then gently misted onto the leaves, and the plants were returned to their normal growth chamber and covered with transparent domes to maintain increased humidity for 2 days. To assess bacterial populations in hydathode‐infected leaves, four entire leaves of similar size were removed 4 days after inoculation, surface sterilized in 70% ethanol for 45 s and rinsed three times with sterile distilled water. Leaf discs were then punched from surface‐sterilized leaves before grinding in 10 mm MgCl2, and extracts were serially diluted on NYGA plates as described above. Triplicate samples were tested (12 leaves per treatment). For vacuum infiltration, a fresh culture of bacteria was scraped off the plate, resuspended in 10 mm MgCl2 with 0.005% Silwet L‐77 (Lehle Seeds, Round Rock, TX, USA) to 106 cfu/mL and vacuum infiltrated into leaves. The same concentration of Silwet L‐77 was included in all mock treatments. Three days after infection, leaf discs from four different leaves were combined and ground in 10 mm MgCl2 with a microcentrifuge tube plastic pestle, with triplicate replication (12 leaves in total). The samples were vortex mixed, diluted 1:10 serially, plated on NYGA solid medium and colonies were counted after 2 days of growth at 28 °C. Colonies with a non‐Xcc morphology were only rarely observed. For syringe inoculation in callose deposition assays, a fresh culture of bacteria was scraped off the plate, resuspended in 10 mm MgCl2 to 108 cfu/mL and infiltrated into leaves by syringe infiltration using a 1‐cm3 plastic syringe with no needle. Unless otherwise noted, bacteria were applied to plant leaves by vacuum infiltration and populations were sampled 3 days after inoculation. For the experiments in Fig. 6, the unmarked Xcc ΔhrcC strains were electroporated with pVSP61 plasmid (no insert) with a kanamycin resistance gene to allow the differentiation of wild‐type (KanS) and ΔhrcC mutant (KanR) strains. One‐way analysis of variance was performed using Minitab (release 14).
Macroscopic HR assays
Arabidopsis thaliana Pi‐0 plants were grown for 4–6 weeks at 22 °C with a 12‐h light/12‐h dark cycle, and fully expanded leaves were used for macroscopic HR analyses. Wild‐type and mutant strains of Xcc B186 and B305 with either pDD62(empty vector) or pDD62‐avrBsT‐HA (Cunnac et al., 2007) were cultured in NYG medium overnight and resuspended in 1 mm MgCl2 at OD600= 0.3 for inoculation. Leaf mesophyll tissue was infiltrated using a syringe with no needle. The severity of macroscopic HR tissue collapse was scored at 24 h after inoculation according to the following scale: 0, no collapse; 1, hints of collapse, leaf looks normal but inoculated region is slightly less turgid when touched on the edge; 2, obvious but subtle tissue collapse, inoculated areas retaining original green colour and somewhat turgid; 3, intermediate or patchy, some areas grey–green and collapsed; 4, strong response, entire inoculated area grey–green and collapsed; 5, severe, entire inoculated area thin and drying out.
Secretion assays and Western blot analysis
Secretion assays were performed as described previously with minor modifications (Rossier et al., 1999; Wang et al., 2007). The wild‐type and hrcC mutant Xcc with AvrBsT‐HA were cultured in NYG medium for 24 h. After washing twice with 1 mm MgCl2, the cells were diluted in 10 mL of the hrp‐inducing secretion medium SMMXC to OD600= 0.4, and cultured at 28 °C for another 5 h. The total culture (0.2 mL) was precipitated for 1 h on ice with 10% trichloroacetic acid (TCA). After centrifugation at 15 000 g for 40 min at 4 °C, protein precipitates were washed with ethanol and resuspended in 40 µL of Laemmli buffer. The supernatant was collected after centrifugation at 4000 g for 10 min at 4 °C, and then filtered through 0.45‐µm filters. The supernatant was precipitated with TCA and resuspended in Laemmli buffer (1/200 v/v). Proteins were separated by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and detected by immunoblot using anti‐HA antibody.
RNA isolation and RT‐PCR
Three leaves of Col gl‐1 and avrPto transgenic plants (Hauck et al., 2003) were collected 1 day after pressure inoculation of Xcc B186, B305 wild‐type and mutant strains for RNA isolation. Total RNA was isolated and subjected to on‐column DNase digestion using an RNeasy mini kit according to the protocol provided (Qiagen, Valencia, CA, USA). cDNA was synthesized by reverse transcriptase Superscript III (Invitrogen, Carlsbad, CA, USA) using 2 µg of total RNA as template. One microlitre of cDNA product from RT reactions was used as template for PCR performed with GoTaq polymerase (Promega, Madison, WI, USA). The primer sets used to track the expression of different genes are listed in Table S2. PCR products were separated by gel electrophoresis and stained with ethidium bromide before viewing.
Callose deposition assay
For callose studies, bacteria were applied by syringe inoculation at 108 cfu/mL in 10 mm MgCl2, and flg22 peptide (Gomez‐Gomez et al., 1999) was applied as a 10 µm solution in water. Callose staining was performed 24 h after bacterial inoculation, as described previously (Gomez‐Gomez et al., 1999). Briefly, the bacteria‐infiltrated leaves were detached from the plants and fixed overnight in 1% glutaraldehyde, cleared and dehydrated with 100% ethanol. The leaves were sequentially rehydrated with 50% ethanol and 67 mm K2HPO4, and stained with 0.01% aniline blue for callose observation. Callose deposits were examined with an Olympus BX60 photomicroscope (Tokyo, Japan) with a 4′,6‐diamidino‐2‐phenylindole (DAPI) fluorescence filter. The number of callose deposits was quantified (typically six replicates per treatment) with IMAGE PRO PLUS software (Media Cybernetics, Bethesda, MD, USA).
Supporting information
Fig. S1 Southern blot analysis indicates that Xanthomonas campestris pv. campestris (Xcc) B94, B186 and B305 contain one copy of hrcC. M, marker; WT, wild‐type.
Fig. S2 Symptom differences on Arabidopsis leaves after spray inoculation with Pseudomonas syringae pv. tomato (Pst) DC3000 (A) and Xanthomonas campestris pv. campestris (Xcc) strains (B). Note that DC3000 causes disease symptoms in the midst of leaves primarily through stomates, whereas Xcc causes disease symptoms at the margins of leaves primarily through hydathodes.
Fig. S3 Similar growth of Xanthomonas campestris pv. campestris (Xcc) strain B305 with or without a functional hrcC in the leaves of Arabidopsis ecotype Ws and Ler plants. All data are means ± standard error.
Fig. S4 In vacuum‐infiltrated Arabidopsis leaves, growth of Xanthomonas campestris pv. campestris (Xcc) B305 was slightly impaired by Δ hrcC, but not detectably impaired by Δ hrpE, Δ flgB/C, Δ tatC single, double or triple gene deletions. All data are means ± standard error. Asterisks above the bars indicate significantly different from wild‐type (WT) B305 on same plant genotype (analysis of variance, P < 0.05). (A) Population sizes of B305 and mutants, as noted, in vacuum‐infiltrated Arabidopsis Col‐0 and Col‐0 fls2‐101 leaves. (B) Growth of B305 and mutants in Arabidopsis leaves (typically at least 10‐fold within 2 days after inoculation) after vacuum infiltration.
Fig. S5 Elicitation of Arabidopsis basal defence responses by Xanthomonas campestris pv. campestris (Xcc) was impaired when the hrcC gene was deleted. Xcc infiltration elicits callose deposits in fls2‐101 leaves, whereas flg22 treatment does not.
Table S1 Mutant strains used in this study, with phenotypes noted.
Table S2 Primers for gene knockout, complementation and reverse transcription‐polymerase chain reaction (RT‐PCR) experiments.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank Muthusubramanian Venkateshwaran for help in constructing the Xcc ΔhrcC strains, Zhe Sun (China Agricultural University, Beijing, China) for assistance in bacterial growth assays, and Caitilyn Allen, Jian Yao and anonymous reviewers for helpful suggestions. We also thank Mary Beth Mudgett (Stanford University, Stanford, CA, USA) for providing the pDD62‐avrBsT‐HA construct and Arabidopsis Pi‐0 seeds, Sheng Yang He (Michigan State University, East Lansing, MI, USA), David Mackey (Ohio State University, Columbus, OH, USA) and Jen Sheen (Harvard University, Cambridge, MA, USA) for providing transgenic plants, Al Poplawsky and Wesley Chun (University of Idaho, Moscow, ID, USA) for the Xcc strains, Dean Gabriel (University of Florida, Gainesville, FL, USA) for providing the pUFR80 vector before publication, and Tom German (University of Wisconsin–Madison, Madison, WI, USA) for Nicotiana benthamiana plants. This project was supported by Department of Energy Grant DE‐FG02‐02ER15342 to A.F.B., and by the 973 program 2011CB100700 of the Ministry of Science and Technology of China and National Natural Science Foundation of China (NSFC) grant 30971893 to W.S.
REFERENCES
- Aldridge, P. and Hughes, K.T. (2002) Regulation of flagellar assembly. Curr. Opin. Microbiol. 5, 160–165. [DOI] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer, A. (1996) Bacterial pathogens in plants: life up against the wall. Plant Cell, 8, 1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. [DOI] [PubMed] [Google Scholar]
- Alvarez, A.M. (2000) Black rot of crucifers In: Mechanisms of Resistance to Plant Diseases (Slusarenko A., Fraser R.S.S. and van Loon L.C., eds), pp. 21–52. Dordrecht: Kluwer. [Google Scholar]
- Aoyama, T. and Chua, N.‐H. (1997) A glucocorticoid‐mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Arlat, M. , Gough, C.L. , Barber, C.E. , Boucher, C. and Daniels, M.J. (1991) Xanthomonas campestris contains a cluster of hrp genes related to the larger hrp cluster of Pseudomonas solanacearum . Mol. Plant–Microbe Interact. 4, 593–601. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M. (2005) Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6, 973–979. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M. , Brent, R. , Kingston, R.E. , Moore, D.D. , Seidman, J.G. , Smith, J.A. and Struhl, K. (1997) Current Protocols in Molecular Biology. New York: John Wiley & Sons. [Google Scholar]
- Block, A. , Li, G. , Fu, Z.Q. and Alfano, J.R. (2008) Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 11, 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonas, U. (1994) hrp genes of phytopathogenic bacteria. Curr. Top. Microbiol. Immunol. 192, 79–98. [DOI] [PubMed] [Google Scholar]
- Brown, I. , Mansfield, J. and Bonas, U. (1995) Hrp genes in Xanthomonas campestris pv. vesicatoria determine ability to suppress papilla deposition in pepper mesophyll cells. Mol. Plant–Microbe Interact. 8, 825–836. [Google Scholar]
- Brown, I. , Trethowan, J. , Kerry, M. , Mansfield, J. and Bolwell, G.P. (1998) Localization of components of the oxidative cross‐linking of glycoproteins and of callose synthesis in papillae formed during the interaction between non‐pathogenic strains of Xanthomonas campestris and French bean mesophyll cells. Plant J. 15, 333–344. [Google Scholar]
- Buttner, D. and He, S.Y. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper‐Lindley, C. , Dahlbeck, D. , Clark, E.T. and Staskawicz, B.J. (2002) Direct biochemical evidence for type III secretion‐dependent translocation of the AvrBs2 effector protein into plant cells. Proc. Natl. Acad. Sci. USA, 99, 8336–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda, A. , Reddy, J.D. , El‐Yacoubi, B. and Gabriel, D.W. (2005) Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol. Plant–Microbe Interact. 18, 1306–1317. [DOI] [PubMed] [Google Scholar]
- Chao, N.X. , Wei, K. , Chen, Q. , Meng, Q.L. , Tang, D.J. , He, Y.Q. , Lu, G.T. , Jiang, B.L. , Liang, X.X. , Feng, J.X. , Chen, B. and Tang, J.L. (2008) The rsmA‐like gene rsmA(Xcc) of Xanthomonas campestris pv. campestris is involved in the control of various cellular processes, including pathogenesis. Mol. Plant–Microbe Interact. 21, 411–423. [DOI] [PubMed] [Google Scholar]
- Chinchilla, D. , Bauer, Z. , Regenass, M. , Boller, T. and Felix, G. (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell, 18, 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn, B. , Sekirov, I. and Finlay, B.B. (2007) Type III secretion systems and disease. Clin. Microbiol. Rev. 20, 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis, G.R. and Van Gijsegem, F. (2000) Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54, 735–774. [DOI] [PubMed] [Google Scholar]
- da Cunha, L. , Sreerekha, M.V. and Mackey, D. (2007) Defense suppression by virulence effectors of bacterial phytopathogens. Curr. Opin. Plant Biol. 10, 349–357. [DOI] [PubMed] [Google Scholar]
- Cunnac, S. , Wilson, A. , Nuwer, J. , Kirik, A. , Baranage, G. and Mudgett, M.B. (2007) A conserved carboxylesterase is a suppressor of avrbst‐elicited resistance in Arabidopsis. Plant Cell, 19, 688–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, M.J. , Barber, C.E. , Turner, D.C. , Cleary, S.G. and Sawzyc, M.K. (1984) Isolation of mutants of Xanthomonas campestris pv. campestris showing altered pathogenicity. J. Gen. Microbiol. 130, 2447–2455. [Google Scholar]
- Forsyth, A. , Mansfield, J.W. , Grabov, N. , de Torres, M. , Sinapidou, E. and Grant, M.R. (2010) Genetic dissection of basal resistance to Pseudomonas syringae pv. phaseolicola in accessions of Arabidopsis. Mol. Plant–Microbe Interact. 23, 1545–1552. [DOI] [PubMed] [Google Scholar]
- Galan, J.E. and Wolf‐Watz, H. (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature, 444, 567–573. [DOI] [PubMed] [Google Scholar]
- Godard, F. , Lummerzheim, M. , Saindrenan, P. , Balague, C. and Roby, D. (2000) hxc2, an Arabidopsis mutant with an altered hypersensitive response to Xanthomonas campestris pv. campestris . Plant J. 24, 749–761. [DOI] [PubMed] [Google Scholar]
- Gohre, V. , Spallek, T. , Haweker, H. , Mersmann, S. , Mentzel, T. , Boller, T. , de Torres, M. , Mansfield, J.W. and Robatzek, S. (2008) Plant pattern‐recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18, 1824–1832. [DOI] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. and Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. and Boller, T. (2002) Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 7, 251–256. [DOI] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. , Felix, G. and Boller, T. (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana . Plant J. 18, 277–284. [DOI] [PubMed] [Google Scholar]
- Hauck, P. , Thilmony, R. and He, S.Y. (2003) A Pseudomonas syringae type III effector suppresses cell wall‐based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA, 100, 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, P. , Shan, L. , Lin, N.‐C. , Martin, G.B. , Kemmerling, B. , Nurnberger, T. and Sheen, J. (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell, 125, 563–575. [DOI] [PubMed] [Google Scholar]
- He, P. , Shan, L. and Sheen, J. (2007a) Elicitation and suppression of microbe‐associated molecular pattern‐triggered immunity in plant–microbe interactions. Cell. Microbiol. 9, 1385–1396. [DOI] [PubMed] [Google Scholar]
- He, Y.Q. , Zhang, L. , Jiang, B.L. , Zhang, Z.C. , Xu, R.Q. , Tang, D.J. , Qin, J. , Jiang, W. , Zhang, X. , Liao, J. , Cao, J.R. , Zhang, S.S. , Wei, M.L. , Liang, X.X. , Lu, G.T. , Feng, J.X. , Chen, B. , Cheng, J. and Tang, J.L. (2007b) Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris . Genome Biol. 8, R218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux, V. , Barber, C.E. and Daniels, M.J. (1998) Entry of Xanthomonas campestris pv. campestris into hydathodes of Arabidopsis thaliana leaves: a system for studying early infection events in bacterial pathogenesis. Mol. Plant–Microbe Interact. 11, 537–543. [DOI] [PubMed] [Google Scholar]
- Jakobek, J.L. , Smith, J.A. and Lindgren, P.B. (1993) Suppression of bean defense responses by Pseudomonas syringae . Plant Cell, 5, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, B. , Xu, R. , Li, X. , Wei, H. , Bai, F. , Hu, X. , He, Y. and Tang, J. (2006) Construction and characterization of a hrpG mutant rendering constitutive expression of hrp genes in Xanthomonas campestris pv. campestris . Prog. Nat. Sci. 16, 480–485. [Google Scholar]
- Jiang, B.‐L. , He, Y.‐Q. , Cen, W.‐J. , Wei, H.‐Y. , Jiang, G.‐F. , Jiang, W. , Hang, X.‐H. , Feng, J.‐X. , Lu, G.‐T. , Tang, D.‐J. and Tang, J.‐L. (2008) The type III secretion effector XopXccN of Xanthomonas campestris pv. campestris is required for full virulence. Res. Microbiol. 159, 216–220. [DOI] [PubMed] [Google Scholar]
- Jiang, W. , Jiang, B.L. , Xu, R.Q. , Huang, J.D. , Wei, H.Y. , Jiang, G.F. , Cen, W.J. , Liu, J. , Ge, Y.Y. , Li, G.H. , Su, L.L. , Hang, X.H. , Tang, D.J. , Lu, G.T. , Feng, J.X. , He, Y.Q. and Tang, J.L. (2009) Identification of six type III effector genes with the PIP box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity. Mol. Plant–Microbe Interact. 22, 1401–1411. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Journet, L. , Hughes, K.T. and Cornelis, G.R. (2005) Type III secretion: a secretory pathway serving both motility and virulence (review). Mol. Membr. Biol. 22, 41–50. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. and Kado, C.I. (1990) A plant‐inducible gene of Xanthomonas campestris pv. campestris encodes an exocellular component required for growth in the host and hypersensitivity on nonhosts. J. Bacteriol. 172, 5165–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, S. and Bonas, U. (2009) How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 12, 37–43. [DOI] [PubMed] [Google Scholar]
- Keshavarzi, M. , Soylu, S. , Brown, I. , Bonas, U. , Nicole, M. , Rossiter, J. and Mansfield, J. (2004) Basal defenses induced in pepper by lipopolysaccharides are suppressed by Xanthomonas campestris pv. vesicatoria . Mol. Plant–Microbe Interact. 17, 805–815. [DOI] [PubMed] [Google Scholar]
- Kim, J.G. , Taylor, K.W. , Hotson, A. , Keegan, M. , Schmelz, E.A. and Mudgett, M.B. (2008) XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas‐infected tomato leaves. Plant Cell, 20, 1915–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.S. , Ma, B. , Perna, N.T. and Charkowski, A.O. (2009a) Phylogeny and virulence of naturally occurring type III secretion system‐deficient Pectobacterium strains. Appl. Environ. Microbiol. 75, 4539–4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.G. , Li, X. , Roden, J.A. , Taylor, K.W. , Aakre, C.D. , Su, B. , Lalonde, S. , Kirik, A. , Chen, Y. , Baranage, G. , McLane, H. , Martin, G.B. and Mudgett, M.B. (2009b) Xanthomonas T3S effector XopN suppresses PAMP‐triggered immunity and interacts with a tomato atypical receptor‐like kinase and TFT1. Plant Cell, 21, 1305–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambais, M.R. , Goldman, M.H. , Camargo, L.E. and Goldman, G.H. (2000) A genomic approach to the understanding of Xylella fastidiosa pathogenicity. Curr. Opin. Microbiol. 3, 459–462. [DOI] [PubMed] [Google Scholar]
- Lindgren, P.B. , Peet, R.C. and Panopoulos, N.J. (1986) Gene cluster of Pseudomonas syringae pv. ‘phaseolicola’ controls pathogenicity of bean plants and hypersensitivity on nonhost plants. J. Bacteriol. 168, 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Loper, J.E. and Lindow, S.E. (1987) Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv syringae on bean leaf surfaces. Phytopathology, 77, 1449–1454. [Google Scholar]
- Lu, H. , Patil, P. , Van Sluys, M.A. , White, F.F. , Ryan, R.P. , Dow, J.M. , Rabinowicz, P. , Salzberg, S.L. , Leach, J.E. , Sonti, R. , Brendel, V. and Bogdanove, A.J. (2008) Acquisition and evolution of plant pathogenesis‐associated gene clusters and candidate determinants of tissue‐specificity in Xanthomonas . PloS One, 3, e3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab, R.M. (1999) The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 181, 7149–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann, H.C. and Guttman, D.S. (2008) Evolution of the type III secretion system and its effectors in plant–microbe interactions. New Phytol. 177, 33–47. [DOI] [PubMed] [Google Scholar]
- Metz, M. , Dahlbeck, D. , Morales, C.Q. , Al Sady, B. , Clark, E.T. and Staskawicz, B.J. (2005) The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana . Plant J. 41, 801–814. [DOI] [PubMed] [Google Scholar]
- Navarro, L. , Zipfel, C. , Rowland, O. , Keller, I. , Robatzek, S. , Boller, T. and Jones, J.D. (2004) The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene‐dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, M.A. , Conrads‐Strauch, J. , Scofield, G. , Daniels, M.J. and Dow, J.M. (1994) Defense‐related gene induction in Brassica campestris in response to defined mutants of Xanthomonas campestris with altered pathogenicity. Mol. Plant–Microbe Interact. 7, 553–563. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T. , Brunner, F. , Kemmerling, B. and Plater, L. (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Oh, H.S. , Park, D.H. and Collmer, A. (2010) Components of the Pseudomonas syringae Type III secretion system can suppress and may elicit plant innate immunity. Mol. Plant–Microbe Interact. 23, 727–739. [DOI] [PubMed] [Google Scholar]
- Pfund, C. , Tans‐Kersten, J. , Dunning, F.M. , Alonso, J.M. , Ecker, J.R. , Allen, C. and Bent, A.F. (2004) Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana . Mol. Plant–Microbe Interact. 17, 696–706. [DOI] [PubMed] [Google Scholar]
- Qian, W. , Jia, Y. , Ren, S.‐X. , He, Y.‐Q. , Feng, J.‐X. , Lu, L.‐F. , Sun, Q. , Ying, G. , Tang, D.‐J. , Tang, H. , Wu, W. , Hao, P. , Wang, L. , Jiang, B.‐L. , Zeng, S. , Gu, W.‐Y. , Lu, G. , Rong, L. , Tian, Y. , Yao, Z. , Fu, G. , Chen, B. , Fang, R. , Qiang, B. , Chen, Z. , Zhao, G.‐P. , Tang, J.‐L. and He, C. (2005) Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris . Genome Res. 15, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme, L.G. , Mindrinos, M.N. and Panopoulos, N.J. (1991) Genetic and transcriptional organization of the hrp cluster of Pseudomonas syringae pv. phaseolicola . J. Bacteriol. 173, 575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried, J.L. and Collmer, A. (1987) An nptI‐sacB‐sacR cartridge for constructing directed, unmarked mutations in Gram‐negative bacteria by marker exchange‐eviction mutagenesis. Gene, 57, 239–246. [DOI] [PubMed] [Google Scholar]
- Rigano, L.A. , Payette, C. , Brouillard, G. , Marano, M.R. , Abramowicz, L. , Torres, P.S. , Yun, M. , Castagnaro, A.P. , Oirdi, M.E. , Dufour, V. , Malamud, F. , Dow, J.M. , Bouarab, K. and Vojnov, A.A. (2007) Bacterial cyclic beta‐(1,2)‐glucan acts in systemic suppression of plant immune responses. Plant Cell, 19, 2077–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roine, E. , Wei, W. , Yuan, J. , Nurmiaho‐Lassila, E.L. , Kalkkinen, N. , Romantschuk, M. and He, S.Y. (1997) Hrp pilus: an hrp‐dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA, 94, 3459–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, W. , Feng, F. , Zhou, J.M. and He, C.Z. (2010) Effector‐triggered innate immunity contributes Arabidopsis resistance to Xanthomonas campestris . Mol. Plant Pathol. 11, 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossier, O. , Wengelnik, K. , Hahn, K. and Bonas, U. (1999) The Xanthomonas Hrp type III system secretes proteins from plant and mammalian bacterial pathogens. Proc. Natl. Acad. Sci. USA, 96, 9368–9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroth, M.N. , Hildebrand, D.C. and Panopoulos, N. (1991) Phytopathogenic Pseudomonads and related plant‐associated Pseudomonads In: The Prokaryotes, 2nd edn. (Balows A., Truper H.G., Dworkin M., Harder W. and Schleifer K.‐H., eds), pp. 3104–3131. New York: Springer Verlag. [Google Scholar]
- Shan, L. , He, P. , Li, J. , Heese, A. , Peck, S.C. , Nurnberger, T. , Martin, G.B. and Sheen, J. (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor‐signaling complexes and impede plant immunity. Cell Host Microbe, 4, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva, A.C. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J. , Ferreira, R.C. , Ferro, M.I. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G. , Lemos, M.V. , Locali, E.C. , Machado, M.A. , Madeira, A.M. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A. , Silva, C. , de Souza, R.F. , Spinola, L.A. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Simpson, R.B. and Johnson, L.J. (1990) Arabidopsis thaliana as a host for Xanthomonas campestris pv. campestris . Mol. Plant–Microbe Interact. 3, 233–237. [Google Scholar]
- Speth, E.B. , Lee, Y.N. and He, S.Y. (2007) Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr. Opin. Plant Biol. 10, 580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W. , Dunning, F.M. , Pfund, C. , Weingarten, R. and Bent, A.F. (2006) Within‐species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2‐dependent defenses. Plant Cell, 18, 764–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres, M. , Mansfield, J.W. , Grabov, N. , Brown, I.R. , Ammouneh, H. , Tsiamis, G. , Forsyth, A. , Robatzek, S. , Grant, M. and Boch, J. (2006) Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J. 47, 368–382. [DOI] [PubMed] [Google Scholar]
- Van Sluys, M.A. , Monteiro‐Vitorello, C.B. , Camargo, L.E. , Menck, C.F. , Da Silva, A.C. , Ferro, J.A. , Oliveira, M.C. , Setubal, J.C. , Kitajima, J.P. and Simpson, A.J. (2002) Comparative genomic analysis of plant‐associated bacteria. Annu. Rev. Phytopathol. 40, 169–189. [DOI] [PubMed] [Google Scholar]
- Vance, R.E. , Isberg, R.R. and Portnoy, D.A. (2009) Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 6, 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer, B.A. , Teitzel, G.M. , Lee, M.W. , Jelenska, J. , Hotton, S. , Fairfax, K. , Jenrette, J. and Greenberg, J.T. (2006) The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non‐host plants. Mol. Microbiol. 62, 26–44. [DOI] [PubMed] [Google Scholar]
- Wang, L.F. , Tang, X.Y. and He, C.Z. (2007) The bifunctional effector AvrXccC of Xanthomonas campestris pv. campestris requires plasma membrane‐anchoring for host recognition. Mol. Plant Pathol. 8, 491–501. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Rong, W. and He, C. (2008) Two Xanthomonas extracellular polygalacturonases, PghAxc and PghBxc, are regulated by type III secretion regulators HrpX and HrpG and are required for virulence. Mol. Plant–Microbe Interact. 21, 555–563. [DOI] [PubMed] [Google Scholar]
- Weber, E. and Koebnik, R. (2005) Domain structure of HrpE, the Hrp pilus subunit of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 187, 6175–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, E. , Ojanen‐Reuhs, T. , Huguet, E. , Hause, G. , Romantschuk, M. , Korhonen, T.K. , Bonas, U. and Koebnik, R. (2005) The type III‐dependent Hrp pilus is required for productive interaction of Xanthomonas campestris pv. vesicatoria with pepper host plants. J. Bacteriol. 187, 2458–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, K. , Tang, D.J. , He, Y.Q. , Feng, J.X. , Jiang, B.L. , Lu, G.T. , Chen, B. and Tang, J.L. (2007) hpaR, a putative marR family transcriptional regulator, is positively controlled by HrpG and HrpX and involved in the pathogenesis, hypersensitive response, and extracellular protease production of Xanthomonas campestris pathovar campestris . J. Bacteriol. 189, 2055–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, F.F. , Potnis, N. , Jones, J.B. and Koebnik, R. (2009) The type III effectors of Xanthomonas . Mol. Plant Pathol. 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, P.H. (1980) Black Rot: a continuing threat to world crucifers. Plant Dis. 64, 736–742. [Google Scholar]
- Xiang, T. , Zong, N. , Zou, Y. , Wu, Y. , Zhang, J. , Xing, W. , Li, Y. , Tang, X. , Zhu, L. , Chai, J. and Zhou, J.‐M. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18, 74–80. [DOI] [PubMed] [Google Scholar]
- Xiang, T. , Zong, N. , Zhang, J. , Chen, J. , Chen, M. and Zhou, J.M. (2011) BAK1 is not a target of the Pseudomonas syringae effector AvrPto. Mol. Plant–Microbe Interact. 24, 100–107. [DOI] [PubMed] [Google Scholar]
- Xu, R.Q. , Blanvillain, S. , Feng, J.X. , Jiang, B.L. , Li, X.Z. , Wei, H.Y. , Kroj, T. , Lauber, E. , Roby, D. , Chen, B. , He, Y.Q. , Lu, G.T. , Tang, D.J. , Vasse, J. , Arlat, M. and Tang, J.L. (2008) AvrAC(Xcc8004), a type III effector with a leucine‐rich repeat domain from Xanthomonas campestris pathovar campestris confers avirulence in vascular tissues of Arabidopsis thaliana ecotype Col‐0. J. Bacteriol. 190, 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, M.H. , Torres, P.S. , El Oirdi, M. , Rigano, L.A. , Gonzalez‐Lamothe, R. , Marano, M.R. , Castagnaro, A.P. , Dankert, M.A. , Bouarab, K. and Vojnov, A. (2006) Xanthan induces plant susceptibility by suppressing callose deposition. Plant Physiol. 141, 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M. and Chai, J. (2008) Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 11, 179–185. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Southern blot analysis indicates that Xanthomonas campestris pv. campestris (Xcc) B94, B186 and B305 contain one copy of hrcC. M, marker; WT, wild‐type.
Fig. S2 Symptom differences on Arabidopsis leaves after spray inoculation with Pseudomonas syringae pv. tomato (Pst) DC3000 (A) and Xanthomonas campestris pv. campestris (Xcc) strains (B). Note that DC3000 causes disease symptoms in the midst of leaves primarily through stomates, whereas Xcc causes disease symptoms at the margins of leaves primarily through hydathodes.
Fig. S3 Similar growth of Xanthomonas campestris pv. campestris (Xcc) strain B305 with or without a functional hrcC in the leaves of Arabidopsis ecotype Ws and Ler plants. All data are means ± standard error.
Fig. S4 In vacuum‐infiltrated Arabidopsis leaves, growth of Xanthomonas campestris pv. campestris (Xcc) B305 was slightly impaired by Δ hrcC, but not detectably impaired by Δ hrpE, Δ flgB/C, Δ tatC single, double or triple gene deletions. All data are means ± standard error. Asterisks above the bars indicate significantly different from wild‐type (WT) B305 on same plant genotype (analysis of variance, P < 0.05). (A) Population sizes of B305 and mutants, as noted, in vacuum‐infiltrated Arabidopsis Col‐0 and Col‐0 fls2‐101 leaves. (B) Growth of B305 and mutants in Arabidopsis leaves (typically at least 10‐fold within 2 days after inoculation) after vacuum infiltration.
Fig. S5 Elicitation of Arabidopsis basal defence responses by Xanthomonas campestris pv. campestris (Xcc) was impaired when the hrcC gene was deleted. Xcc infiltration elicits callose deposits in fls2‐101 leaves, whereas flg22 treatment does not.
Table S1 Mutant strains used in this study, with phenotypes noted.
Table S2 Primers for gene knockout, complementation and reverse transcription‐polymerase chain reaction (RT‐PCR) experiments.
Supporting info item
Supporting info item


