Abstract
Background
Our previous study revealed that PLAGL2 or POFUT1 can promote tumorigenesis and maintain significant positive correlations in colorectal cancer (CRC). However, the mechanism leading to the co-expression and the underlying functional and biological implications remain unclear.
Methods
Clinical tumor tissues and TCGA dataset were utilized to analyze the co-expression of PLAGL2 and POFUT1. Luciferase reporter assays, specially made bidirectional promoter vectors and ectopic expression of 3’UTR were employed to study the mechanisms of co-expression. In vitro and in vivo assays were performed to further confirm the oncogenic function of both. The sphere formation assay, immunofluorescence, Western blot and qRT-PCR were performed to investigate the effect of both genes in colorectal cancer stem cells (CSCs).
Findings
PLAGL2 and POFUT1 maintained co-expression in CRC (r = 0.91, p < .0001). An evolutionarily conserved bidirectional promoter, rather than post-transcriptional regulation by competing endogenous RNAs, caused the co-expression of PLAGL2 and POFUT1 in CRC. The bidirectional gene pair PLAGL2/POFUT1 was subverted in CRC and acted synergistically to promote colorectal tumorigenesis by maintaining stemness of colorectal cancer stem cells through the Wnt and Notch pathways. Finally, PLAGL2 and POFUT1 share transcription factor binding sites, and introducing mutations into promoter regions with shared transcription regulatory elements led to a decrease in the PLAGL2/POFUT1 promoter activity in both directions.
Interpretation
Our team identified for the first time a bidirectional promoter pair oncogene, PLAGL2-POFUT1, in CRC. The two genes synergistically promote the progression of CRC and affect the characteristics of CSCs, which can offer promising intervention targets for clinicians and researchers.
Fund
National Nature Science Foundation of China, the Hunan province projects of Postgraduate Independent Exploration and Innovation of Central South University.
Keywords: PLAGL2, POFUT1, Co-expression, Bidirectional promoter, Colorectal cancer, Cancer stem cell
Research in Context.
Evidence before this study
The long arm of chromosome 20 (20q) is frequently amplified in colorectal cancer (CRC), and multiple genes located along it contribute to oncogenic progression. In a previous study, we confirmed that the adjacent genes PLAGL2 and POFUT1 on 20q11.21 can promote tumorigenesis and maintain a co-expression pattern in CRC. However, the mechanism leading to the co-expression and the underlying functional and biological implications remain elusive.
Added value of this study
PLAGL2 and POFUT1 maintain significant positive correlations in CRC. An evolutionarily conserved bidirectional promoter, rather than post-transcriptional regulation, caused this co-expression pattern. PLAGL2 and POFUT1 can act synergistically to promote CRC by maintaining stemness of colorectal cancer stem cells. The anti-cancer effect of combined PLAGL2 and POFUT1 inhibition was much stronger than with the inhibition of either alone. Modifying or understanding the promoter shared by both may be an alternative strategy for the simultaneous intervention of transcription of PLAGL2 and POFUT1 in CRC.
Implications of all the available evidence
Our team identified for the first time a bidirectional promoter pair oncogene, consisting of PLAGL2 and POFUT1 in CRC. The two genes are simultaneously highly expressed in CRC and synergistically affect the characteristics of colorectal cancer stem cells, which can offer promising intervention targets in treating CRC for clinicians and researchers.
Alt-text: Unlabelled Box
1. Introduction
Thirty years have passed since the first report that gain/amplification of the long arm of chromosome 20 (20q) is closely associated with colorectal cancer (CRC) [1]. Since then, many studies have found that a gain of 20q is observed in >65% of CRCs and is associated with poor outcome [2]. In addition, DNA copy number amplification (CNA) of 20q frequently activates multiple oncogenes located on the arm. For example, the well-established oncogenes BCL2L1, AURKA and TPX2 map to regions of genomic amplification on 20q11.21, 20q13.2 and 20q11, respectively, and are overexpressed and play crucial roles in CRC initiation and progression [3,4]. In our previous study [5], integrated genomic analyses revealed that the most frequently upregulated genes in colorectal cancer are located on chromosome 20q. Among these genes, we further identified two genes, Pleomorphic adenoma gene like-2 (PLAGL2) and Protein O-Fucosyltransferase 1 (POFUT1), that are driven by copy number amplification and exhibit cancer-causing potential in CRC [6,7].
PLAGL2 belongs to the PLAG gene family of C2H2 zinc finger transcriptional factors, which can modulate cellular functions [8]. As with its homologous gene PLAGL1, PLAGL2 can function as an oncogene in several cancers. For example, PLAGL2 is targeted for amplification and overexpression in gliomas; it regulates Wnt signaling to impede differentiation of glioma stem cells [9]. PLAGL2 expression is significantly higher in colorectal cancer tissues and can serve as prognostic factor of patient outcome [10]. Mechanistically, research by us [11] and others [12] showed that in CRC, PLAGL2, acting as a transcription factor can bind to the promoter region of the Wnt receptor gene and promote aberrant activation of Wnt/β-catenin pathway. Compared to the PLAGL2 gene, the studies on POFUT1 are relatively narrow and small, POFUT1 encodes a member of the glycosyltransferase O-Fuc family. Its catalysis of the fucosylation of Notch receptor epidermal growth factor (EGF)-like repeats is required for optimal ligand binding and canonical Notch signaling induced by DLL1 or JAGGED1 [13]. POFTU1 plays an important role in Dowling-Degos disease, and we confirmed that POFUT1 can promote colorectal cancer development through the activation of Notch1 signaling [6,14].
Until now, all research on PLAGL2 and POFUT1 has studied them independently, and both appear to function at different cellular levels with no crosstalk. In our previous research [5], we found that PLAGL2 and POFUT1 can maintain significant positive correlations in various human cancer tissues, especially in CRC tissues, for which the Pearson coefficient (r) was >0.90, P < .0001. Based on bioinformatics analysis [5], we speculate that there are two important reasons for this co-expression feature: the first reason is that a bidirectional promoter exists, considering that PLAGL2 and POFUT1 are arranged head-to-head on opposite strands with <300 base pairs separating their transcription start sites; this configuration of gene pairs has previously been termed a “bidirectional promoter gene pair”, and as such, these genes show more correlation in expression [15,16]. Another cause of co-expression is a competitive endogenous RNA (ceRNA) mechanism, considering that the RNAs of PLAGL2 and POFUT1 have long 3’UTR lengths (3954 bp and 4006 bp, respectively) and share many of the same microRNAs [5,7]. The ceRNA hypothesis is based on the observations that RNAs containing common microRNA response elements (MREs) in 3’UTRs can regulate each other and maintain co-expression by competing for shared microRNAs, as reported in extensive research [17]. In this article, we will prove the exact mechanisms leading to the significant co-expression of PLAGL2 and POFUT1 through a series of experiments and explore the underlying biological implication of this co-expression feature in colorectal cancer.
2. Materials and methods
2.1. Cell culture and clinical material
Human CRC cell lines (SW620, SW480) were cultured with L15 medium (KeyGEN BioTECH, Nanjing, China), supplemented with 10% fetal bovine serum (FBS) (Biological Industries, Israel). HCT116 and HT-29 cells were cultured in McCoy's5A medium (KeyGEN BioTECH, Nanjing, China) with 10% FBS. human normal human colon mucosal epithelial cell line NCM460 was maintained in RPMI 1640 medium (Gibco) supplemented with 10% FBS and antibiotics. Cells were grown in a 5% CO2 cell culture incubator at 37 °C. Biological tissues were obtained from patients treated at the Third Affiliated Hospital of Central South University (Changsha China) with informed consent and approval of the Center for Medical Ethics Central South University.
2.2. RNA extraction and real-time PCR
For real-time PCR analyses (qRT-PCR), total RNA was extracted with TRIzol (Invitrogen), and cDNA was synthesized using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO) following the manufacturer's recommendations. qPCR assays were performed by using KOD SYBR® qPCR Mix (TOYOBO) on LightCycler® 480II System (Roche) according to the manufacturer's instructions. Treated samples were normalized to controls with the ∆∆Ct formula using GAPDH as an endogenous control. The primers used in this study can be found in Supplementary table 1.
2.3. CSC self-renewal and differentiation assay
Sphere formation assays were performed by plating 500–1000 cells into 6- or 24- well ultra-low cluster plates in serum-free DMEM/F12 (1:1) medium (Gibco) containing growth factors (EGF 20 ng/ml and bFGF 10 ng/ml; PeproTech), 2% B-27 supplement (Gibco), 2 μg/ml of 0.2% heparin.
(Solarbio) and 1% P/S. Sphere formation was observed at 7–15 days, and images were captured using an inverted microscope system (Olympus, IX73). For differentiation assays, the colonospheres of CSCs were plated on plastic plates in medium containing 2% fetal bovine serum (FBS) for 24–120 h.
2.4. Bioinformatics analyses
TCGA and GEO dataset data used in this article were downloaded from the corresponding official website. The web-based database Chipbase v2.0 [18] was employed to preliminarily assess the correlation between genes in CRC tissues. UCSC genome browser was adopted to analyze the PLAGL2-POFUT1 intergenic region and determine whether both genes compose a bidirectional gene pair.
2.5. Western blot, immunofluorescence and antibodies
The protein extraction and Western blot (WB) details can be found in our previous study [6,7]. Immunofluorescence (IF) was performed by using Immunol Fluorescence Staining Kit with kFluor594-Labeled Goat Anti-Rabbit IgG (H + L) and kFluor555-Labeled Goat anti-Mouse IgG (H + L) (Keygen Biotech) according to the manual. IF Images were captured using an inverted microscope system (Olympus, IX73) and confocal microscope (Zeiss LSM800). The antibodies used in this article are listed as follows: PLAGL2 (Abcam or ProteinTech), POFUT1 (ProteinTech), GAPDH (Abcam), CD44v6 (Invitrogen), HES5(Abcam), HES1(Abcam), Cleaved Notch1 (Cell Signaling Technology), β-Catenin (Cell Signaling Technology), Non-phospho (Active) β-Catenin (Cell Signaling Technology), MYC (ProteinTech), Cyclin D1 (ProteinTech), Wnt6 (Abcam), OCT4 (Abcm) and Trefoil Factor 3 (Abcam).
2.6. Plasmid constructs and transfection
All of the plasmids including TOPflash/FOPflash used in this study were constructed by and purchased from Suzhou GenePharma Co., Ltd., China. To perform the luciferase reporter assay for promoter activity, 179 bp and 89 bp PLAGL2 fragments were cloned into a promoterless pGL3 Luciferase Reporter Vector (Promega) in two orientations upstream of the luciferase reporter gene. Mutant forms of the 179 bp promoter fragment were prepared by site-directed mutagenesis (Supplementary. Table 2). The 3’UTR of PLAGL2 was subcloned into the psiCHECK-2 vector. For EGFP-promoter -mCherry vector construction, sequence fragments were obtained by PCR; gene fragments were connected with T4 DNA ligase and pUC57 vector digested with EcoRV, followed by transforming the ligation product into competent cells. Recombinant plasmid sequencing was performed for verification following plasmid extraction. For plasmid transfection, cells were seeded in 24-well dishes, and transfection was performed 24 h later with Lipofectamine 2000 (Invitrogen). Promoter luciferase reporter vectors and Renilla luciferase vector (Prl-SV40 or pRL-TK, Promega) were co-transfected into CRC cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations.
2.7. Dual-luciferase reporter assay and dual fluorescent protein detection
All cells were lysed 24 or 48 h after transfection by dispensing 100 μl 1× passive lysis buffer (PLB) of the Dual-Luciferase® Reporter Assay System (cat. E1910, Promega) into each well of a 24-well plate. Then, 20 μl of PLB lysate/well was used for the luciferase reporter assay using the same kit and according to the manufacturer's instructions. The luciferase activity of reporter plasmids was normalized to the Renilla luciferase activity. For cells transfected with the EGFP-promoter –mCherry plasmid, EGFP and mCherry were arranged in a head-to-head fashion, as were PLAGL2 and POFU1, and dual fluorescent protein detection was performed by using an inverted microscope system (Olympus, IX73).
2.8. Lentiviral vector and transfection
Lentiviral vectors carrying PLAGL2, POFUT1, PLAGL2–3’UTR1 (the front 2000 bp of all 3’UTRs) and PlAGL2–3’UTR2 (the back 1948 bp) cDNA, and the lentiviral shRNA targeting human PLAGL2 and POFUT1 were purchased from Shanghai GenePharma Co., Ltd. Control groups were transduced with lentivirus carrying empty vector (EV or NC) or negative control shRNA (shNC) with a non-targeting RNA sequence. More details can be found in our previous study [6,7,11]. Briefly, all were cloned into the vector that was mostly synthetized by Genepharma, Shanghai, China. Virus packaging was performed in HEK293T cells. To create lentivirus-transduced lines, the cells were infected with virus and polybrene, and stable cell lines were selected with treatment with 4 μg/ml puromycin after 48 or 72 h transfection. The efficiency in different cells was determined by qRT-PCR and WB. the shRNA sequences are listed in Supplementary table 1.
2.9. Cell invasion assays
Cell invasion assays were performed using Transwell chambers (24-well, 8.0-μm pore membranes) (Corning) coated with matrix proteins according to the manufacturer's protocol (BD Biosciences). Cells (1–2 × 105) were counted and then seeded in the upper chamber with serum-free medium plus 0.5% bovine serum albumin (BSA). The chemoattractant used in lower chamber was medium plus 10% FBS. After 24 h~30 h incubation and removal of the noninvading cells, the cells were fixed with formaldehyde and stained with 0.5% crystal violet for 30 min.
2.10. Cell proliferation assays
The Cell-Light™ EdU Apollo®567 In Vitro Imaging Kit (Guangzhou RiboBio, China) was used to observe the proliferation rate of cancer cells according to the manufacturer's instructions. Anchorage-independent growth assays were performed in 6-well plates. The bottom layer was covered with 1.5 ml of 1.2% agar in medium supplemented with 10% FBS and allowed to solidify. The next day, cells were seeded on top in 1 ml of 0.6% agar in medium containing 10% FBS. The number and size of clones were counted after 10 and 20 days.
2.11. Proliferation and metastasis assays in vivo
Subcutaneous xenografts were performed as previously described [6,7]. For in vivo metastasis assays, CRC cells or cells dissociated from spheres were injected intravenously via the tail vein. After 6–7 weeks, all mice were sacrificed and all organs were removed for examination. Lungs and livers were harvested, sectioned and stained with H&E staining. All animal experimental procedures used in this study were approved by the Animal Ethics Committee of Central South University.
2.12. Copy number variation (CNV) detection using the AccuCopy® assay
DNA was isolated from tissues and cells using a Qiagen kit according to the manufacturer's instructions (Qiagen). AccuCopy assays were used to assess the copy number value of PLAGL1 and POFTU1 in selected tissues and cells. The AccuCopy assay was developed based on multiplex competitive amplification by Genesky Biotechnologies (Shanghai, China). The basic molecular principle and statistical analysis of AccuCopy was well described by Du et al. [19]. The copy numbers of 4 DNA fragments amplified from both ends of DNA region of PLAGL2 and POFUT1 were assessed, and the details about the DNA fragments (including primers, length, the gene region in chr. and the value) for the AccuCopy® assay can be found in Supplementary table 3.
2.13. Statistical analysis
Statistical computations were performed using GraphPad Prism 7. Experimental data are presented as the mean ± SD, violin plots and box-whisker plots. Box-whisker plots depict mean, 1st and 3rd quartiles and min/max. Correlation between two genes mRNA levels were assessed by Pearson correlation calculations choosing a two-tailed P value. The two-tailed paired Student's t-test was used to assess the comparisons between groups. The quantitative analysis of protein was performed by Image J software. p < .05 was considered statistically significant.
3. Results
3.1. PLAGL2 and POFUT1 maintain significantly correlated expression in CRC
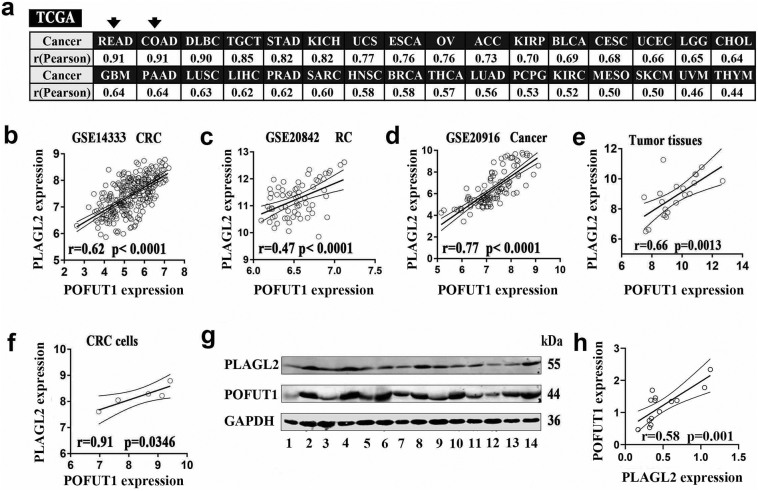
Based on our previous study [5], we further confirmed that PLAGL2 and POFUT1 maintain positive expression correlation in 32 TCGA human cancer tissues (Fig. 1a), especially in colon and rectum tissues, for which the Pearson correlation coefficient (r) =0.91 (p < .0001) (Supplementary table 4). To exclude the impact of TCGA as a single database, we expanded our analysis to published, large-scale human colorectal cancer datasets (3 GEO datasets: GSE14333, GSE20842, GSE20916). We again observed a very strong positive correlation between the expression of PLAGL2 and that of POFUT1 (p < .0001) (Fig. 1b, c and d). Then, 21 cancerous tissues from our hospital and 5 cell lines in our lab were subjected to RT-PCR assays, and an identical pattern was observed in our CRC cohort (Fig. 1e and f). Additionally, 7 pairs of colorectal carcinoma tissues and adjacent normal tissue were randomly selected for Western blot analyses, and the results confirmed that PLAGL2 and POFUT1 maintained co-expression at the protein level (Fig. 1g and h). It is noteworthy that the Candidate Cancer Gene Database (CCGD) [20] showed that transposon-based forward genetic screens in mice identified PLAGL2 and POFUT1 as potential cancer drivers in CRC, and both received the same ranking (Supplementary table 5), which, to a certain extent, suggests that the co-expression may have potential physiological significance and deserves in-depth study.
Fig. 1.
PLAGL2 and POFUT1 are positively correlated in human tumors.
(a) The TCGA dataset showed that PLAGL2 was co-expressed with POFUT1 in 32types of human cancer tissues; the Pearson correlation coefficient in colorectal tissues ranked first and second. The abbreviated names of all cancers are listed here; full names and co-expression analysis can be found in Supplemental Table 4 or TCGA website (https://cancergenome.nih.gov/) and ChIPBase v2.0 (http://rna.sysu.edu.cn/chipbase/).
(b) PLAGL2 co-expressed with POFUT1 in 292 colorectal tumor tissues (GEO: GSE14333) and (c) 65 rectal tumor tissues (GEO: GSE 20842) and (d) 111 colorectal tumor tissues (GEO: GSE20916).
(e) Correlation of PLAGL2 and POFUT1 mRNA levels analyzed by RT-PCR in 21 colorectal cancer tissues.
(f) Correlation of PLAGL2 and POFUT1 mRNA levels analyzed by RT-PCR in 5 colorectal cancer cells.
(g) Seven pairs of colorectal carcinoma tissues and adjacent normal tissues were randomly selected for Western blot analyses. (odd numbers represent paired colorectal cancer tissues and even numbers represent paired adjacent tissues of colorectal cancer)
(h) Correlation of PLAGL2 and POFUT1 protein levels analyzed by Western blot in 7 pairs of colorectal carcinoma tissues and adjacent normal tissues.
3.2. PLGAL2 and POFUT1 are jointly regulated by an evolutionarily conserved bidirectional promoter
We next aimed to identify the mechanisms underlying the co-expression of both genes. In our previous study [5], we found that PLAGL2 and POFUT1 have many characteristics unique to bidirectional promoter pair [16,21], including arrangement in a head-to-head (adjacent 5’ends) configuration on opposite strands of DNA with an 89 bp (<1000 bp) intervening sequence; an evolutionarily conserved intergenic region; similar tissue specificity of expression; overlap with a putative CpG island; lack of a typical TATA box; and shared transcription factor binding sites. Here, using Ensembl genome browser 95 and WebLogo [22], we extracted a DNA sequence (approximately 500 bp) for genomic alignments, which covers a putative bidirectional promoter flanked by parts of the first exons of both genes. The results showed that these DNA sequences were highly similar among different species, including in 12 primates, 30 or 88 mammals (Supplementary table 6 and Supplementary. Fig. 5a).Taken together, these observations strongly suggest that the PLGAL2 and POFUT1 genes are jointly regulated by an evolutionary conserved bidirectional promoter which have been reported to lead to correlated expression [21,23].
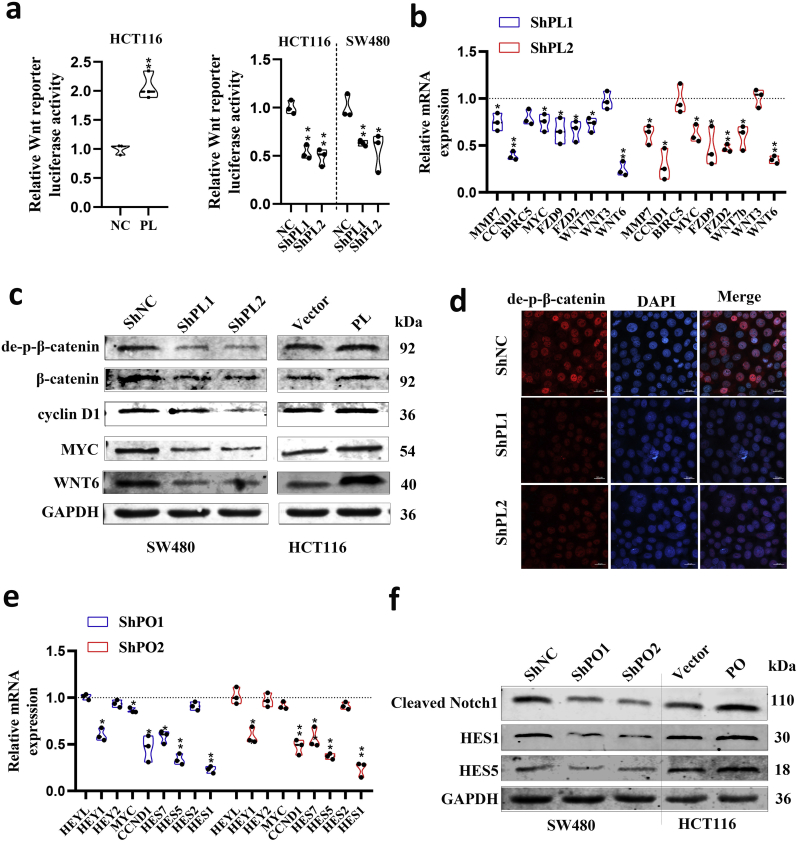
Fig. 5.
PLAGL2 and POFUT1 can activate WNT and Notch pathways, respectively.
(a)TCF reporter activity was evaluated through the β-catenin responsive TOPflash reporter in empty vector control (NC) and PLAGL2-expressing HCT116 cells or in HCT116 and SW480 cells stably transduced with lentiviral vectors carrying negative control shRNA (shNC) and PLAGL2-specific small hairpin RNAs (ShPL1 and ShPL2) *p < .05, **p < .01.
(b) qRT-PCR analyses of the expression of Wnt signaling pathway related (WNT3, WNT6, WNT7B, FZD2 and FZD9) and targeted genes (MYC, CCND1, BIRC5 and MMP7) after PLAGL2 knockdown in SW620 cells. Control group set to 1 (*p < .05, ** p < .01).
(c) Western blot analyses of related Wnt protein Wnt 6, downstream effectors of the Wnt signaling pathway including non-phospho (active) β-Catenin and total β-catenin, and accepted canonical targets including cyclin D1 and c-Myc in PLAGL2-silenced or PLAGL2-overexpressing CRC cells.
(d) Immunofluorescence of de-phospho-β -catenin in PLAGL2-silenced SW480 cells.
(e) qRT-PCR analyses of the expression of Notch signaling pathway target genes after POFUT1 knockdown in SW480 cells. Control group set to 1 (*p < .05, ** p < .01).
(f) Western blot analyses of cleaved Notch1 (the Notch1 intracellular domain), and accepted canonical targets include HES1 and HES5 in POFUT1-silenced or POFUT1-overexpressing CRC cells.
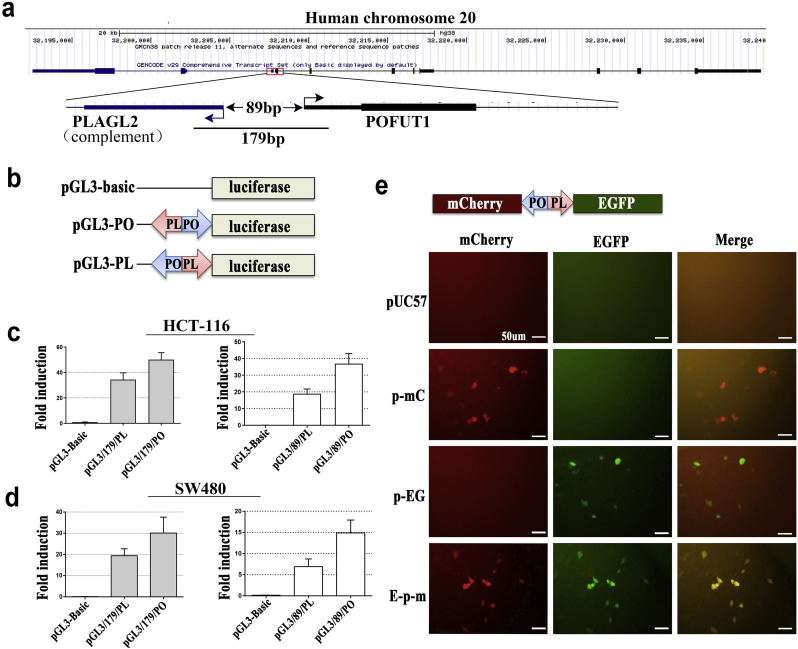
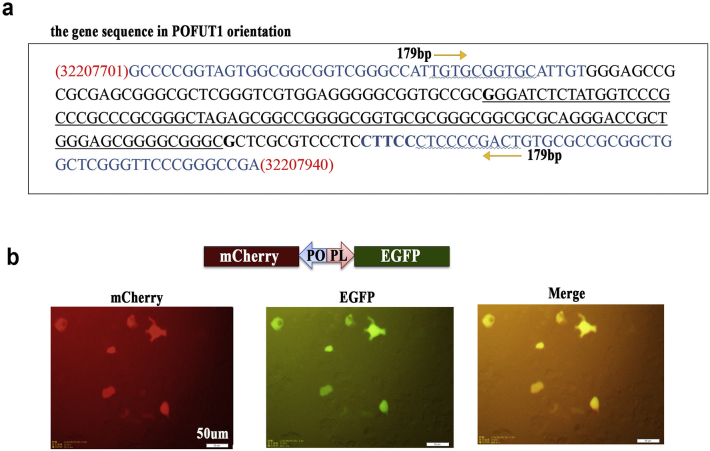
To determine whether the human PLAGL2/POFUT1 intergenic region can serve as a true functional bidirectional promoter, we cloned a 179 bp fragment (Fig. 2a and Supplementary. Fig. 1a) that contained the 89 bp putative bidirectional promoter, flanked by parts of the first exons of both genes, into the promoterless pGL3-Basic vector upstream of the luciferase coding region in two orientations (Fig. 2b). Transient transfection assays together with pRL-TK in HCT-116 and SW-480 cells revealed that the genomic fragment of 179 bp was sufficient for the expression of the firefly luciferase gene regardless of its orientation (Fig. 2c and d). Furthermore, an identical pattern was observed for the 89 bp putative core bidirectional promoter despite decreased promoter activity. All of these findings indicated that the regulation of PLAGL2 and POFUT1 expression could be coordinated through the bidirectional promoter. To further verify that the intergenic region can simultaneously drive the expression of two genes, the same 179 bp fragment was cloned into a specially made promoterless pUC57 vector, which contained green and red fluorescent protein reporter genes in opposite orientations. The resulting construct was transfected into HCT-116 and SW-480 cells. Fluorescence microscopy revealed that both genes were expressed simultaneously (Fig. 2e and Supplementary. Fig. 1b). Taken together, all these results showed that PLGAL2 and POFUT1 are jointly regulated by a true bidirectional promoter.
Fig. 2.
Human PLAGL2 and POFUT1 genes are concertedly regulated by a genuine bidirectional promoter.
(a) Structure of human PLAGL2 and POFUT1 genes (derived from UCSC Genome Browser on Human) and schematic representation of an intervening region between both genes. The bent arrows indicate the transcription direction and PLAGL2 located on complementary strand.
(b) Schematic representation of the ntervening gene fragment between both genes being cloned into the promoterless pGL3-Basic vector upstream of the luciferase coding region in two orientations.
(c) and (d) Promoter activity of the POFUT1-PLAGL2 intergenic region (179 bp is flanked by parts of the first exons of both genes, 89 bp is the putative bidirectional promoter) in HCT116 and SW-480 cells transfected with promoterless pGL3-Basic vector. PGL3-PO plasmid contains the POFUT1/PLAGL2 promoter region in the POFUT1 orientation, and PGL3-PL contains the same region in the PLAGL2 orientation.
(e) Fluorescence microscopy analysis of simultaneous expression of the fluorescent proteins mCherry (red) and EGFP (green) driven by the 179 bp POFUT1/PLAGL2 promoter region in HCT116 cells. pUC57 represents HCT116 cells transfected with an empty pUC57 vector, and p-mC and p-EG represent control vectors, which contain both reporter genes but with the unidirectional promoter. E-p-m represent the POFUT1/PLAGL2 promoter region cloned into pUC57 vector containing reporter genes of mCherry and EGFP (scale bar:50 μm). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Co-expression of PLAGL2 and POFUT1 cannot be maintained through the 3′ untranslated regions
As discussed in the introduction, we hypothesized that in addition to the bidirectional promoter, the cross-regulation of competing endogenous RNAs (ceRNAs) at the posttranscriptional level may contribute to the co-expression of the two genes [5,7]. To investigate this hypothesis, the PLAGL2 gene with only one transcript was selected for 3’UTR overexpression analysis. The PLAGL2 3’UTR was cloned as two separate fragments (PL-3’UTR1 and PL-3’UTR2) due to its large size (Fig. 3a). The results demonstrated that ectopic overexpression of PLAGL2 3’UTRs in SW620 and HCT-116 cells led to a small level of upregulation of POFTU1 mRNA. In contrast, the mRNA of PLAGL2 itself is more susceptible to 3’UTR than POFUT1(Fig. 3b). Although the mRNA level of POFUT1 can be slightly regulated by the 3’UTRs of PLAGL2, there was no consistent protein change across all colorectal cancer cells (Fig. 3c and d). In addition, we constructed and transfected a luciferase construct tagged with PL-3’UTR2 (PL-3’UTR2-luc) in SW620 cells (Fig. 3a). ShRNA-mediated POFUT1 knockdown did not significantly reduce PL-3’UTR2-luc activity, suggesting that the transcript of POFUT1 also cannot have an impact on PLAGL2 3’UTRs (Fig. 3e). Finally, enforced PLAGL2 and POFUT1 expression (close to 100-fold and 200-fold, respectively, in HCT-116 cells and close to 20-fold and 80-fold, respectively, in HT29 cells) only led to slight upregulation of POFUT1 and PLAGL2 mRNA (approximately 2-fold in HCT116 and 1.5-fold in HT29) (Fig. 3f). More importantly, Western blot analysis further confirmed that POFUT1 and PLAGL2 proteins were overexpressed in HT29 and HCT116 cells stably transduced with lentiviral vectors carrying cDNA, but there was no protein change of PLAGL2 in the POFUT1-overexpressing cells and of POFUT1 in the PLAGL2-overexpressing cells across the two colorectal cancer cell lines (Fig. 3g). Previous studies have confirmed that ectopic overexpression of ceRNA 3’UTRs of one gene in cells can led to a marked upregulation of both 3’UTR-luc activity and endogenous protein levels [24]. All these findings indicate that mRNA modifications in the 3’UTR may not be the main reason for co-expression of both genes at the mRNA level. Taken together, we believe that mediation by common promoter sequences is a key mechanism explaining the co-expression of these neighboring genes.
Fig. 3.
PLAGL2 and POFUT1 maintenance of co-expression may not be through the 3′-untranslated regions of both genes.
(a) Schematic outlining the two fragments PLAGL2 3’ UTR1 and 3’UTR2 used for overexpression and luciferase experiments.
(b) qRT-PCR of lentivirus-mediated PLAGL2 3’UTR overexpression, and the POFUT1 and PLAGL2 expression in response to overexpression of PLAGL2 3’UTR in HCT-116 and SW620 cells.
(c) WB showing POFUT1 protein in response to overexpression of PLAGL2 3’UTRs in SW620 and HCT-116 cells.
(d) Quantification of (C).
(e) Luciferase activity in SW620 cells cotransfected with luciferase-PLAGL2 3’UTR2 reporter construct and ShRNA against POFUT1.
(f) qRT-PCR in lentivirus-mediated PLAGL2 and POFUT1 overexpression and cross regulation in HT-29 and HCT-116 cells.
(g) Western blot in lentivirus-mediated PLAGL2 and POFUT1 overexpression and cross regulation in HT-29 and HCT-116 cells. EV: empty vector.
3.4. Bidirectional gene pair PLAGL2/POFUT1 are subverted in CRC and act synergistically to promote colorectal tumorigenesis
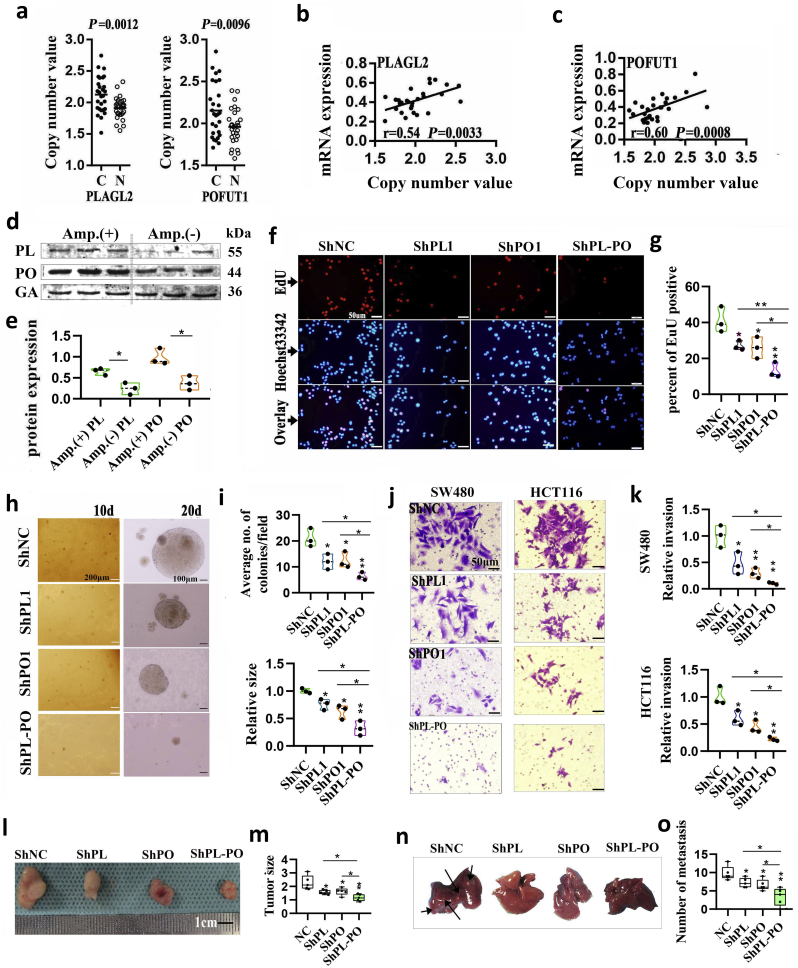
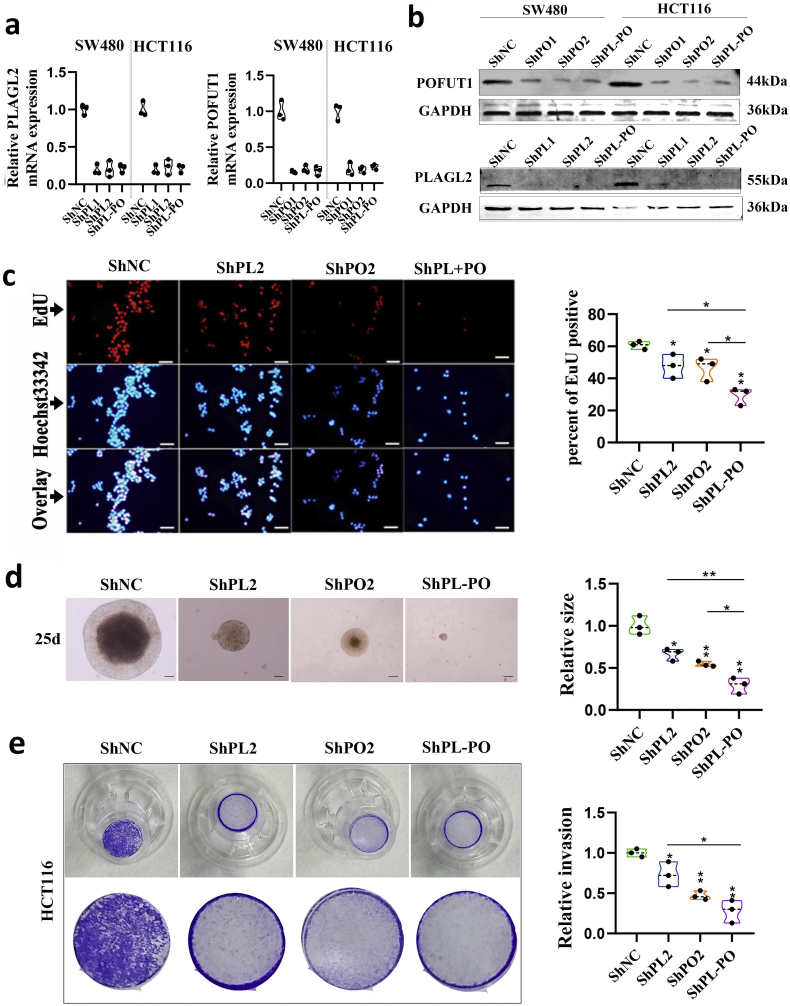
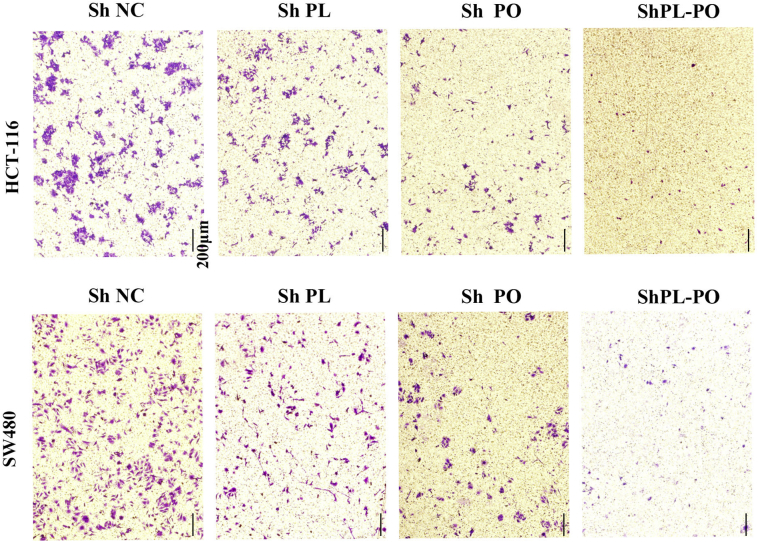
The co-expression pattern of PLAGL2 and POFUT1 in varieties of tissues may result in a fine-tuning of homeostasis. However, in previous research, we analyzed TCGA colorectal cancer data and found that the bidirectional gene pair PLAGL2/POFUT1 was subverted and owned overexpression driven by copy number amplification (both genes showed significant CNA–mRNA correlation, p < .0001) [5], Here, 14 pairs of tumor and adjacent tissues were randomly selected for AccuCopy copy number analysis; the result found that tumor tissues had significant copy number gain (Fig. 4a). Furthermore, similar to the findings with the TCGA-CRC samples, the mRNA expression of both genes was positively correlated with DNA copy number (Fig. 4b and c). Additionally, the tissues with CNA showed higher POFUT1 and PLAGL2 protein expression (p < .05; Fig. 4d and e). The simultaneous overexpression pattern of PLAGL2 and POFUT1 likely had a synergistic role in CRC, so we firmly believe that when PLAGL2 inhibition was combined with POFUT1 inhibition, the effect on tumor regression was much stronger than with either inhibition alone. To test this hypothesis, we simultaneous silenced PLAGL2 and POFUT1 in HCT116 and SW480 cells with lentiviral vectors carrying PLAGL2- and POFUT1-specific small hairpin RNAs (shPLAGL2 and shPOFUT1); silencing of both or one gene was confirmed by qRT-PCR and Western blot (Supplementary. Fig. 2 a and 2b). The proliferation activity of CRC cells was measured by the number of EdU positive cells. Simultaneous silencing of PLAGL2 and POFUT1 in SW480 cells significantly inhibited cell viability and growth relative to control or separate group values (Fig. 4f, Fig. 4g and Supplementary. Fig. 2c). Consistent with the EdU incorporation rate, shRNA-mediated suppression of PLAGL2 and POFUT1 more significantly reduced anchorage-independent growth of HCT 116 cells (Fig. 4h, Fig. 4i and Supplementary. Fig. 2d). Next, we assessed the effect of both genes on the invasive capability of cells, a hallmark characteristic of malignant CRC cells. As shown in Fig. 4j, Fig. 4k, Supplementary. Fig. 2e and Supplementary. Fig. 3, silencing of PLAGL2 and POFUT1 in SW480 and HCT 116 cells more significantly reduced their invasive capacity through Matrigel. Furthermore, when assessing the tumorigenic ability of PLAGL2 and POFUT1 in vivo, the PLAGL2 and POFUT1 knockdown group exhibited markedly reduced size and weight of HCT116 xenografts in nude mice and the number of metastatic lesions in the liver compared with control and individual groups (Fig. 4l Fig. 4m, Fig. 4n and Fig. 4o). These in vivo findings are consistent with the above in vitro assay indicating that combined PLAGL2 /POFUT1 inhibition has greater antitumor effects.
Fig. 4.
PLAGL2 and POFUT1 owned overexpression driven by CNA and combined PLAGL2 /POFUT1 inhibition have greater effects on opposing tumors.
(a) AccuCopy copy number analysis of copy number of PLAGL2 (including PLAGL2–1 and PLAGL2–2) and POFUT1 (including POFUT1–1 and POFUT1–2) in 14 pair colorectal tissues.
(b and c) The correlation between copy number value of PLAGL2–2 and PLAGL2 mRNA expression and POFUT1–2 and POFUT1 mRNA expression.
(d) Comparison of PLAGL2 and POFUT1 protein expression between CRC tissues without CNA (Amp. (−)) and CRC tissues with amplified (Amp (+)). PL: PLAGL2; PO: POFUT1; GA: GAPDH.
(e) Quantification of (d).
(f) The EdU proliferation assay was performed after shRNA-mediated suppression of POFUT1 or/and PLAGL2 in SW480 cells; shown here are the results for the ShPLAGL2–1 and ShPOFUT1–1 sequences (scale bar:50 μM).
(g) Quantification of (B).
(h) Suppressing PLAGL2 or/and POFUT1 expression reduce anchorage-independent growth in HCT116 cells, shown here are the results for the ShPLAGL2–1 and ShPOFUT1–1 sequences. (scale bar:200 μm (left), 100 μm (right)); d: day.
(i) Quantification of (D), upper showing average number of colonies after cultivation for 10 days and down showing relative size of colonies after cultivation for 20 days; the shNC group (NC) was set to 1.
(j) Representative images of invasion assay showing control group (NC) and shRNA-mediated suppression of POFUT1 or/and PLAGL2 in SW480 and HCT116 cells; shown here are the results for the ShPLAGL2–1 and ShPOFUT1–1 sequences (scale bar:50 μm).
(k) Quantification of (j)
(l and m) shRNA-mediated suppression of POFUT1 or/and PLAGL2 can decelerated growth of HCT116 xenografts in nude mice (n = 5).
(n and o) Representative macroscopic appearances of liver metastasis are shown (some tumor nodules were indicated by black arrows). (HCT116 cells was used in this assay). (scale bar:100 μm) (n = 5).
Data in (d)–(o) presented as violin plots from at least two independent experiments with triplicates. Each plot represents the mean from one independent experiment with triplicates. *p < .05, **p < .01. The *at the vertex of the bar graph is compared to the control group.
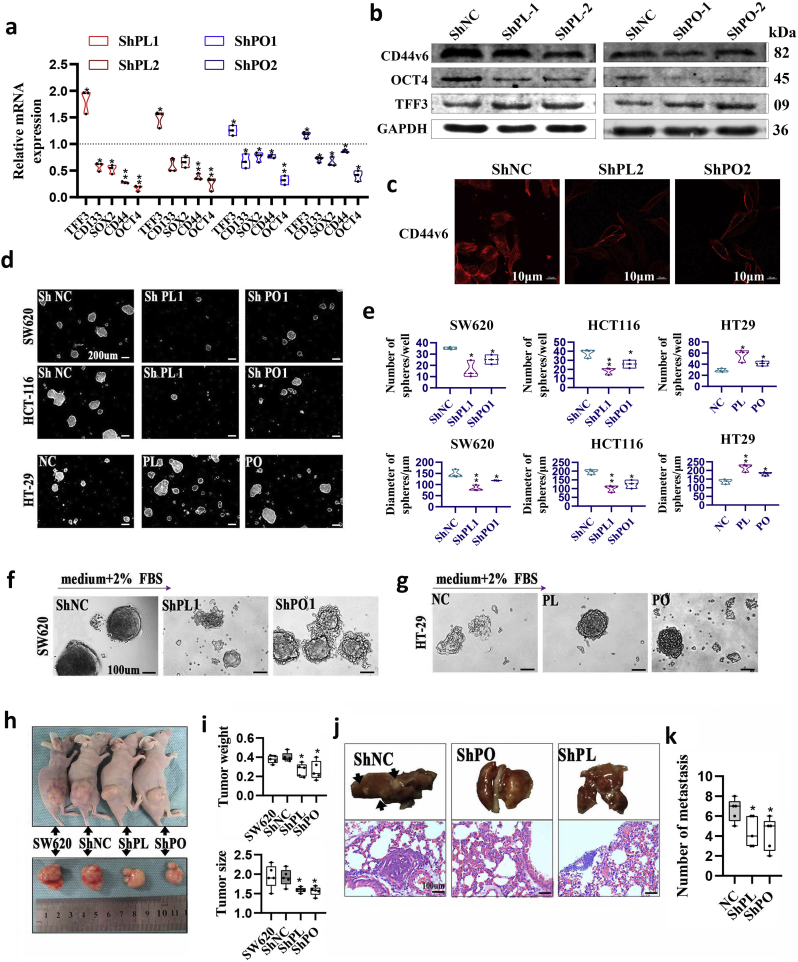
3.5. PLAGL2 and POFUT1 can act synergistically to maintain stemness of colorectal cancer stem cells through the Wnt and Notch pathways
Bidirectional promoter gene pairs with co-expression tend to fall into the same functional categories and identical biological pathways [16,21]. We asked whether this was true of PLAGL2/POFUT1. Interestingly, previous studies by ourselves and others have found that PLAGL2 and POFUT1 may individually activate the Wnt and Notch pathways in human cancer [6,11,25,26]. We further confirmed this conclusion in cell-based systems using both gain and loss strategies. A TOP/FOP luciferase reporter assay revealed that introduction of PLAGL2 into HCT116 cells enhanced their TCF-dependent TOP flash reporter activity. Conversely, PLAGL2-silenced cells exhibited reduced β-catenin/TCF transcription activity in HCT116 and SW480 cells (Fig. 5a). qRT-PCR revealed that the mRNA levels of multiple Wnt pathway components, such as CCND1 and Wnt6, were substantially downregulated in SW480 cells in which PLAGL2 was suppressed (Fig. 5b). Consistently, the protein level of the Wnt protein Wnt 6, downstream effectors of the Wnt signaling pathway, including non-phospho (active) β-catenin and total β-catenin, and accepted canonical targets include cyclin D1 and c-Myc significantly decreased in PLAGL2-silenced CRC cells. Overexpression of PLALG2 had the opposite effect on the level of all these proteins (Fig. 5c). Additionally, in SW480 colon cancer cells, accumulation of nuclear β-catenin decreased when PLAGL2 was suppressed (Fig. 5d). All these findings further confirmed that PLAGL2 can activate the canonical Wnt/β-catenin signaling pathway. Based on our previous study, we further confirmed that POFUT1 expression is necessary for Notch signaling. qRT-PCR revealed that multiple transcripts of Notch pathway target genes such as HES1, HES5 and CCND1 were repressed in CRC cells with inactivation of the POFUT1 (Fig. 5e). Consistently, Western bloting revealed that enforced POFUT1 expression also conferred markedly increased protein levels of cleaved Notch1 (the Notch1 intracellular domain), HES1 and HES5. Conversely, shRNA-mediated suppression of POFUT1 reduced the expression levels of these proteins (Fig. 5f).
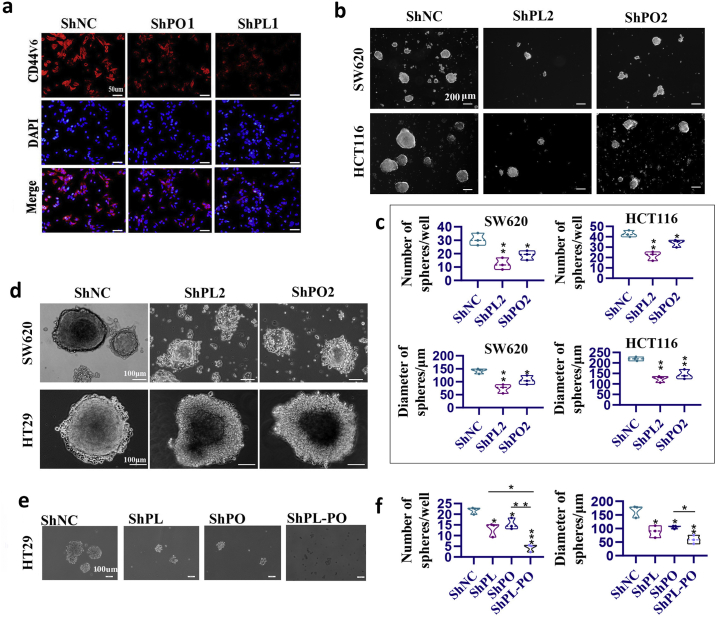
Wnt and Notch pathways are the two pivotal pathways with crosstalk in regulating intestinal epithelial homeostasis [27]. Notably, both pathways can crosstalk and are emerging as key players of colorectal CSC regulation [28]. They cooperate and are integrated in CSCs in promoting tumorigenic potential by maintaining stemness, enhancing self-renewal activity and impairing differentiation [29]. Consistently, KEGG analysis of a publicly available CRC dataset (TCGA) revealed that “Regulating pluripotency of stem cells” was significantly enriched [5]. Therefore, it can be inferred PLAGL2 and POFUT1 have synergetic effects on the regulation of stemness. qRT-PCR revealed that mRNA transcripts of stemness-related markers, including CD133, SOX2, CD44 and OCT4, were significantly downregulated when PLAGL2 or POFTU1 was silenced. The opposite trend was observed with differentiation markers such as TFF3 (Fig. 6a). An identical trend was observed in WB assays; suppressing PLAGL2 or POFUT1 expression in SW480 cells was sufficient to induce differentiation (TFF3) and suppress stem cell marker (CD44v6 and OCT4) expression (Fig. 6b). In line with WB observation, immunofluorescence assays showed that shRNA-mediated knockdown of the endogenously expressed PLAGL2 or POFUT1 in SW620 cells results in downregulation of CD44V6, which is a classic surface marker of colorectal CSCs (Fig. 6c and Supplementary. Fig. 4a). To further investigate how both influence the stem cell-like properties and impedes differentiation of CSCs, we next examined the ability of CRC cells to form spheres in ultralow attachment 6-well plates with growth factor; this ability is an important property of CSCs [30]. The number and diameter of spheres formed by CRC cells were significantly reduced in the PLAGL2- and POFUT1-deleted group, but increased expression of the two genes in cells led to a significantly higher number of tumor spheres (Figure6d, Fig. 6e and Supplementary. Fig. 4b and Supplementary. Fig. 4c). Furthermore, the colon spheres were cultured under differentiation-inducing conditions (medium containing 2% FBS). The shRNA group was rapidly differentiated by plating cells on plastic plates, but serum-induced morphological and molecular differentiation was prevented in the PLAGL2- and POFUT1-expressing group (Figure6f, Fig. 6g and Supplementary. Fig. 4d). In addition, subcutaneous and intravenous injections of cells dissociated from these spheres derived from the shRNA group significantly inhibited tumor formation (Fig. 6h and i) and invasion (Fig. 6j and k) in nude mice. We also tested whether simultaneous silencing of both genes had a more obvious effect on CSC self-renewal activity, as measured by colonsphere formation of HT29 cells. The result demonstrated that suppressing PLAGL2 and POFUT1 significantly yielded fewer and smaller multipotent spheres (Supplementary. Fig. 4e and Supplementary. Fig. 4f). Together, these data suggest that PLAGL2 and POFUT1 were involved in colorectal cancer by converging on the identical pathway of enhancing self-renewal activity and impairing differentiation of CSCs.
Fig. 6.
PLAGL2 and POFUT1 can influence stem cell-like properties and impede differentiation of CSCs.
(a) qRT-PCR analyses of stem and differentiation markers after PLAGL2 and POFUT1 knockdown in SW480 cells. Control group set to 1 (*p < .05, ** p < .01).
(b) Western blot analyses of stem and differentiation markers after PLAGL2 and POFUT1 knockdown in SW480 cells (*p < .05, ** p < .01).
(c) The immunofluorescence assay shows the expression of stem marker CD44V6 protein after PLAGL2 and POFUT1 knockdown in SW620 cells; shown here is the result for ShPLAGL2–2 and ShPOFUT1–2 sequence (scale bar:10 μm).
(d) Representative images of spheres from the SW620 and HCT116 cell lines (PLAGL2 or POFUT1 knockdown, here lists the result about ShPLAGL2–1 and ShPOFUT1–1 sequence) and HT29 cell (PLAGL2 or POFUT1 overexpressed) cultured in a serum-free medium system. (200 μm)
(e) Quantification of (d) (*p < .05, ** p < .01).
(f and g) Representative images showing that suppressing PLAGL2 and POFUT1 can promote differentiation (the sphere rapidly adopted a flattened cell morphology) and attenuate their spheres in the presence of medium plus 2% FBS. A reverse trend was observed in PLAGL2 and POFUT1-expressing cell (scale bar:100 μm).
(h and i) shRNA-mediated suppression of POFUT1 or PLAGL2 can decelerate growth of SW620 sphere xenografts in nude mice (n = 5). Violin plots in the right panel presented quantification (*p < .05). The weight unit of the tumor is grams.
(j and k) Representative macroscopic appearances of lung metastasis are shown (some tumor nodules were indicated by black arrows), representative hematoxylin-eosin stained images of lung metastasis lesions in three different groups (×100), Violin plots in right panel presented quantification. (SW620 cells dissociated from these spheres were used in this assay and *p < .05) (scale bar:100 μm).
3.6. Mutations of specific transcription factor binding sites shared by PLAGL2 and POFUT1 can simultaneously influence the transcription of two genes
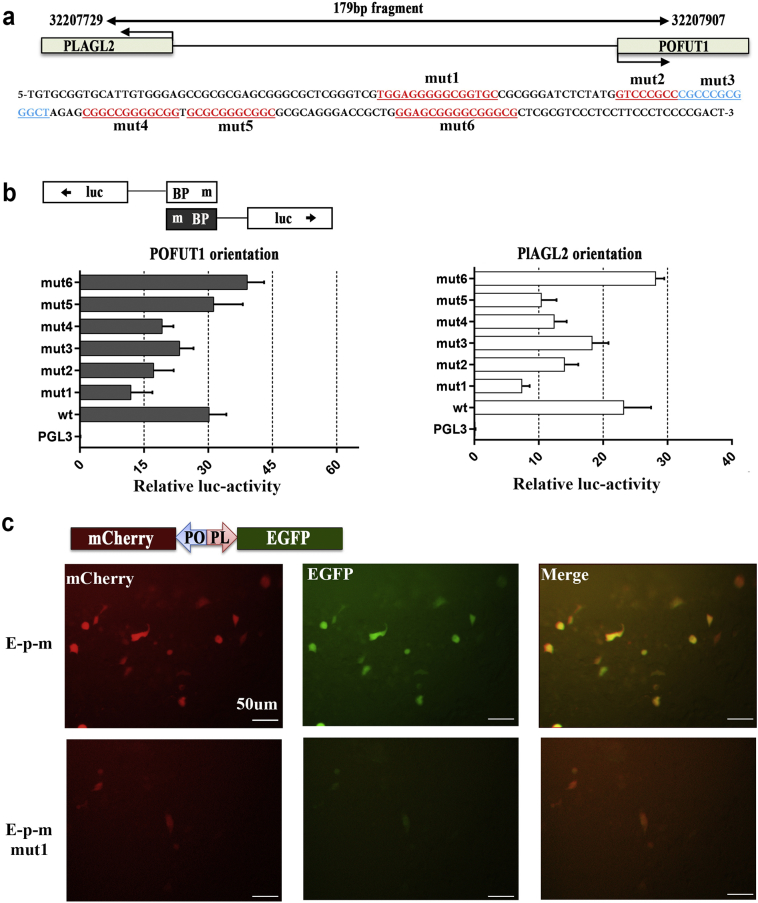
Such promising results drew our attention to develop novel and highly effective methods of dual PLAGL2 and POFUT1 targets. Co-expression of bidirectional promoter gene pairs is often due to shared transcription factors binding sites, and mutations in shared transcription factor binding sites can simultaneously change the expression of both [23,31]. Therefore, studying the transcription factor binding sites shared by PLAGL2 and POFUT1 may provide a good therapeutic target for the simultaneous intervention of overexpression of PLAGL2 and POFUT1 in the future. The MatInspector program [32] was used to identify potential binding sites for transcription factors that could regulate the expression of both genes. As shown in Supplementary. Table 7, 101 cis-regulatory elements distributed in the positive and negative chain were identified, 77 of which were identified with the highest confidence (core similarity = 1). Remarkably, many frequently occurring transcriptional factors that bind to the PLAGL2/POFUT1 bidirectional loci, including EGR1, sp1, sp3, E2F4, E2F1 and the AP-2 family, have been reported to be over-represented in genuine bidirectional promoters [16,21,33]. To further elucidate whether the two closely adjacent genes in divergent orientations could be co-regulated by shared transcription factors binding to a specific bidirectional promoter region, the 101 factors were organized according to their binding site on the evolutionarily conserved 179 bp sequence (Supplementary. Fig. 5a and b). We found approximately 6 concentrated regions that are highly evolutionarily conserved where many transcription factors are targeted. We then introduced 6 mutations (mut) into these regions (Fig. 7a and Supplementary table 2). The wild type and mutant 179 bp fragments were cloned into PGL3-basic in both orientations 5′ to the luciferase gene and transfected into HCT-116 cells. As shown in Fig. 7b, mut 1–4 decreased promoter activity in both directions; mut 5 did so only in one direction; and mut 6 increased promoter activities in either direction. The different result obtained with mut 5 may be because some transcription factors targeting this region can only regulate the expression of one gene in one orientation; similar results were found in another study [34], in which E2F1 and E2F4 only mediated the expression of one gene, whereas Sp1 drove gene expression in either orientation. Additionally, considering the mutations in mut1 led to a significant decrease in the PLGAL2/POFUT1 promoter activity in both directions, the 179 bp fragment with or without mut1 was cloned between genes of the mCherry and EGFP fluorescent proteins. The resulting construct was transfected into HCT116 cells, and the cells were visualized by fluorescence microscopy (Fig. 7c). We found that both genes were expressed simultaneously, but the expression in the mut1 group was much weaker than that of the wild group. All these results indicated that modifying or editing transcriptional binding sites in common promoter sequence could offer promising therapeutic approaches to simultaneously suppress PLAGL2 and POFUT1 expression and thus treat CRC by eliminating cancer stem cells.
Fig. 7.
Mutations in shared transcription factor binding sites in the PLAGL2/POFUT1 bidirectional promoter region lead to decreased promoter activity in both directions.
(a) Schematic outlining the 6 mutations in the 179 bp promoter sequence (more details can be found in Supplementary Table 2).
(a) Schematic outlining the 6 mutations in the 179 bp promoter sequence (more details can be found in Supplementary Table 2).
(b) Activity of mutated and wild-type forms of the PLAGL2/POFUT1 promoter region in HCT116 cells.
(c) Fluorescence microscopy analysis of simultaneous expression of the fluorescent proteins mCherry (red) and EGFP (green) driven by the 179 bp POFUT1/PLAGL2 promoter region with or without mut1 mutation in HCT116 cells. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
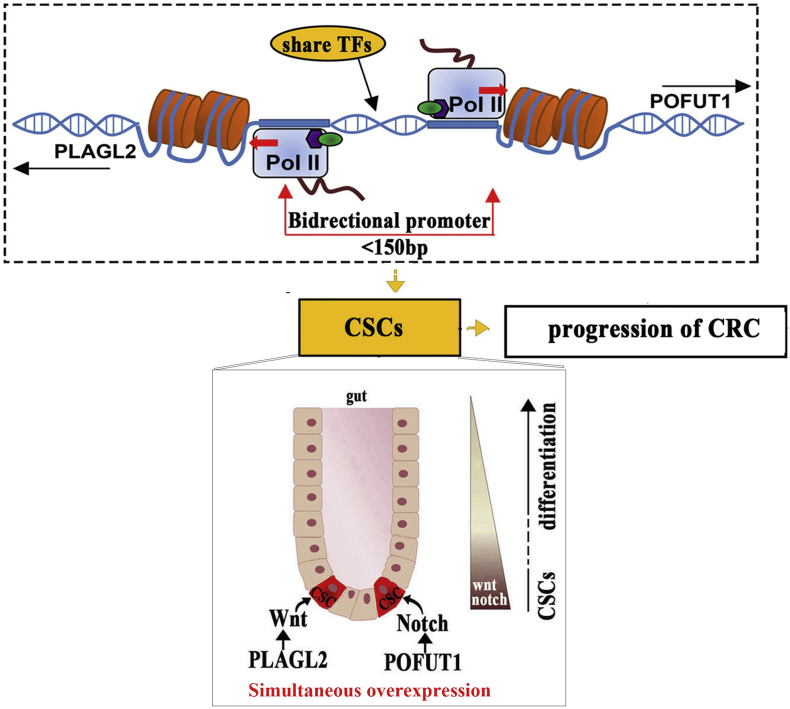
Accumulating evidence indicates that gene order in eukaryotic genomes is not completely random and that genes within the same genomic neighborhoods often maintain co-expression and synergistically participate in important physiological functions [35]. Especially some two adjacent genes whose transcription start sites are neighboring and directed away from each other can form bidirectional gene pairs, which have the potential to participate in the development of cancer [16,23]. In existing literature, direct exploration of the connection between bidirectional gene pairs and CRC is still relatively scarce. Here, we reported such a bidirectional gene pair, PLAGL2-POFUT1, which is driven by copy number amplification, maintains co-expression and is collaboratively involved in colorectal cancer by maintaining stemness in CSCs (Fig. 8).
Fig. 8.
Model of bidirectional gene pair PLAGL2-POFUT1 maintaining coexpression and collaboratively involved in colorectal cancer by enhancing stemness in CSCs.
Previous studies found that neighboring genes in the eukaryotic genome have a tendency to be concurrently expressed; however, the underlying mechanism is yet unclear. One potential key mechanism is the sharing of regulatory elements, such as transcription factors, promoters, and enhancers, especially for genes that form head-to-head pairs, which may be subject to bidirectional expression regulated by common promoter sequences or a bidirectional promoter [[35], [36], [37]]. The bioinformatic analyses in our study revealed that the intergenic PLAGL2/POFUT1 region exhibits factors characteristic of a bidirectional promoter. We then presented experimental evidence that the intergenic region can act as a bidirectional promoter. Identical classic experimental methods for confirming bidirectional promoters have been reported in other studies [31,38]. Computational estimates published by other researchers suggest that nearly 10% of the coding gene promoters are arranged in such a manner, and such architecture is a common and conserved feature across many species, indicating this maintenance is beneficial and functionally important. However, no >20 of these gene pairs have been identified and characterized in detail [39]. In addition, the expression correlation between PLAGL2 and POFUT1 is probably explained by the bidirectional promoter, however the expression of POFUT1 and PLAGL2 also correlated with the expression of other genes, especially the genes located on 20q with mRNA overexpression driven by copy number amplification, such as POFUT1 co-expressed with TM9SF4 (r = 0.82), DDX27 (r = 0.83) and POFUT1 with YTHDF1(r = 0.81) [5]. The mechanism behind that correlation is currently unclear. TM9SF4, DDX26 and YTHDF1 have also been shown to be closely related to colorectal cancer in recent studies [[40], [41], [42], [43]]. Therefore, this co-expression pattern may provide a basis for the synergistic participation of these genes in colorectal cancer.
In addition to having coordinated expression patterns, these gene pairs tend to fall into the same functional categories, such as regulating DNA repair gene, chromatin stability and participating in the same pathway [35,36]. The common function does not mean that the two genes are located in the same organelle, co-localized, or participate in a completely consistent biological pathway and regulate each other, but both can ultimately perform identical physiological and pathological processes. For example, the human PSENEN and U2AF1L4 genes are concertedly regulated by a genuine bidirectional promoter, and the proteins encoded by the PSENEN and U2AF1L4 can be necessary for regulation of T-cell activity [31]. SIRT3 and PSMD13 also belong to a bidirectional promoter gene, and both have association with aging [44]. Using the database of spliced-expressed sequence tags, Elnitski LL [23] found that highly significant enrichment of bidirectional promoters in genes was implicated in somatic cancer, which suggests that bidirectional promoter pairs have a propensity of being involved in cancer. Our study and others have found that PLAGL2 can activate the Wnt signaling pathway [9,12,23] and the glycosylation of POFUT1 is essential for the Notch pathway [13,45]. Wnt and Notch signaling are the two most important pathways in colorectal cancer [[46], [47], [48]] and crosstalk between them initiates tumorigenesis mainly through disrupting the balance between cancer stem cell proliferation and differentiation [27,46,47]. CSC self-renewal and differentiation assays and other methods were used in our study and preliminarily revealed that PLAGL2 and POFUT1 can promote the renewal and impede the differentiation of colorectal cancer stem cells. In agreement with our results, many studies have suggested that PLAGL2 or POFTU1 plays a pivotal role in fine-tuning progenitor cell proliferation and differentiation. POFUT1 loss induces the binary choice of proliferating progenitors between differentiation toward secretory or absorptive cell lineages and the altered proliferation of crypt progenitor cells [49]; the rescue of POFUT1 expression in knockdown cells restores Notch signaling activation and a normal course of C2C12 differentiation [50]. PLAGL2 mediates Wnt signaling to impair differentiation in neural stem cells, enhances stem cell fate and activates expression of ASCL2 in intestinal epithelial cells [9,51]. Additionally, PLAGL2 can result in the activation of Rac1 [52], which controls nuclear localization of β-catenin during canonical Wnt signaling, as well as in colon cancer progression [53,54].
The structural features of bidirectional promoters imply that the two genes are simultaneously regulated by specialized transcription factors. In this article, the introduction of mutations at the specific transcription factor binding site shared by PLAGL2 and POFUT1 affects the simultaneous transcription of two genes, which remind us that modifying or editing transcriptional binding sites or promoters by genome-editing technology such as CRISPR/Cas9 may be a strategy for the simultaneous intervention of overexpression of PLAGL2 and POFUT1 in the future. The reasons are chiefly as follows. Firstly, mut 1–4 decreased promoter activity in both directions, while mut 6 increased the activities in either direction, suggesting that the high expression of PLAGL2 and POFUT1 in colorectal cancer may be partly due to the increased expression of carcinogenic transcription factors and the decreased expression of anticancer transcription factors shared by both, such as transcription factors E2F4 contributing to CRC [55], which not only bind to the PLAGL2/POFUT1 bidirectional loci but also correlate significantly with POFUT1 and PLAGL2 in colon and rectal cancer (p < .001) [5]. However, another transcription factor AP-2alpha, that is shared by both, has a tumor suppressor role and down-regulated expression in CRC; moreover, AP-2alpha maintains a strong inverse relationship to PLAGL2 and POFUT1 in CRC (Pearson correlation coefficient r = −0.614 and r = −0.647, respectively) (cBioPortal for Cancer Genomics, colorectal adenocarcinoma: TCGA, Provisional [56]). In addition, some transcription factors are overactive in human cancer, which makes them targets for the development of anticancer drugs [57]. The functions of transcription factors are implemented by regulating the expression of other genes, so modifying carcinogenic transcriptional binding sites by CRISPR/Cas9 can attenuate their carcinogenicity without affecting the normal physiological function of the two genes. Interestingly, TFAP2C, identified as a transcriptional factor regulating PLAGL2 and POFUT1 in our previous study [5], was recently reported to participate in a gene regulatory network controlling cell growth and differentiation and to promote stemness and chemotherapeutic resistance in colorectal cancer [58,59], which is consistent with PLAGL2 and POFUT1. So, blocking TFAPC from binding to the PLAGL2/POFUT1 bidirectional loci may weaken its role in CRC. Certainly, we believe that deletion or modification of the base sequence of the entire promoter rather than the whole gene by gene editing technology can also completely inhibit the expression of both genes.
In summary, we first identified that two human oncogenes, PLAGL2 and POFUT1, maintain a significantly positive correlation in colorectal cancer. Mechanistically, they are concertedly regulated by a genuine bidirectional promoter rather than a ceRNA mechanism. This bidirectional promoter gene pair promotes progression of cancer by enhancing self-renewal and impeding the differentiation of colorectal cancer stem cells. Simultaneous suppressing the PLAGL2 and POFUT1 expression can significantly reduce CRC cell proliferation, invasion and self-renewal activity in vitro and inhibit murine xenograft tumor growth and liver metastasis in vivo. The introduction of mutations in bidirectional regions led to a decrease in the PLAGL2/POFUT1 promoter activity in both directions, which is worth to investigating, as understanding regulatory cues controlling bidirectional promoter pair PLAGL2-POFUT1 in CRC could offer promising intervention targets for clinicians and researchers.
The following are the supplementary data related to this article.
Supplemental Fig. 1.
Human PLAGL2 and POFUT1 genes are concertedly regulated by a genuine bidirectional promoter. (a) The 179 bp intergenic sequence between POFUT1 and PLAGL2 used in this article. The sequence on the horizontal line represents the 89 bp putative bidirectional promoter (POFUT1 orientation). (b) Simultaneous expression of the fluorescent proteins mCherry (red) and EGFP (green) driven by the 179 bp POFUT1/PLAGL2 promoter region in SW480 cells.
Supplemental Fig. 2.
Combined PLAGL2 /POFUT1 inhibition have greater effects on opposing tumor. (a and b) HCT116 and SW480 were stably transduced with lentiviral vectors carrying PLAGL2 or /and POFUT1-specific small hairpin RNAs. The control group contained lentiviral vectors carrying negative control shRNA (shNC). Silencing of both or single genes was confirmed by RT-PCR (a) and Western blot analysis (b). (c) The EdU proliferation assay was performed after shRNA-mediated suppression of POFUT1 or/and PLAGL2 in SW480 cells. Violin plots in the right panel present quantification. Shown here are the results for the ShPLAGL2–2 and ShPOFUT1–2 sequences (scale bar:50 μM). (d) Suppressing PLAGL2 or/and POFUT1 expression reduces anchorage-independent growth in HCT116 cells. Violin plots in the right panel present quantification. Shown here are the results for the ShPLAGL2–2 and ShPOFUT1–2 sequences (scale bar:100 μm); d: day. (e) Representative images of invasion assays showing HCT116 and SW480 cells stably transduced with lentiviral vectors carrying PLAGL2 or /and POFUT1-specific small hairpin RNAs after 36–72 h induction with 10% FBS followed by crystal violet (0.1%) staining. Violin plots in the right panel presented quantification. Shown here are the results for the ShPLAGL2–2 and ShPOFUT1–2 sequences.
Supplemental Fig. 3.
Representative images of invasion assays showing control group (NC) and shRNA-mediated suppression of POFUT1 or/and PLAGL2 in SW480 and HCT116 cells. (scale bar:200 μm).
Supplemental Fig. 4.
PLAGL2 and POFUT1 can influence stem cell-like properties and impede differentiation of CSCs. (a) The immunofluorescence assay shows the expression of stem marker CD44V6 protein after PLAGL2 and POFUT1 knockdown in SW620 cells; shown here are the results for the ShPLAGL2–1 and ShPOFUT1–1 sequences (scale bar:50 μm). (b) Representative images of spheres from the SW620 and HCT116 cell lines (PLAGL2 or POFUT1 knockdown; shown here are the results for the ShPLAGL2–2 and ShPOFUT1–2 sequences) cultured in a serum-free medium system (200 μm). (c) Quantification of (b) (*p < .05, **p < .01). (d) Representative images showing that suppressing PLAGL2 and POFUT1 can promote differentiation (the sphere rapidly adopted a flattened cell morphology) and attenuate their spheres in the presence of medium plus 2% FBS (scale bar:100 μm). (e) Representative images of spheres from the HT29 cell line (PLAGL2 or/and POFUT1 knockdown) cultured in a serum-free medium system; shown here are the results for the ShPLAGL2–1 and ShPOFUT1–1 sequences (200 μm). (f) Quantification of (e) (*p < .05, **p < .01, ***p < .001).
Supplemental Fig. 5.
PLAGL2 and POFUT1 share transcription factor binding sites in the highly evolutionarily conserved bidirectional promoter region. (a) The 179 bp promoter region is highly evolutionarily conserved using the UCSC Genome Browser and Ensembl genome browser 96(more DNA sequence details can be found in Supplemental Table 6). The consensus sequence was generated using WebLogo (http://weblogo.berkeley.edu/). (b) From top to bottom: (top) schematic figure of the 179 bp sequence that contained the 89 bp putative bidirectional promoter flanked by parts of the first exons of PLAGL2 and POFUT1. The sequence marked by red colour and horizontal lines representes 6 mutation regions (more details on mutations can be found in Supplemental Table 2). (middle) The 101 transcription factor biding sites predicted by The MatInspector program are depicted according to the binding site on the 179 bp. The horizontal axis represented the 179 bp, and the Y-axis representes the 101 transcription factors biding to the179bp. The red line indicates the binding site from start position to end position of each transcription factor. The rectangular boxes of different colors in the figure indicate the six regions in which the transcription factors mainly bind, such as gray, which represents the binding sites of transcription factors such as EGR1, ZNF263 and Sp4 (bottom). Based on the six regions in which the transcription factors are mainly bound, we constructed six mutants, mut1–6. Some representative transcription factors were predicted by MatInspector and JASPAR 2018 (http://jaspar.genereg.net/analysis).
All the primers used in this article were listed below.
The wild-type sequence of 179bp promoter and 6 mutated sequence (here only list the sequence of PLAGL2 orientation).
The details about the DNA fragment for copy nubmer analysis.
Correlation analysis results of PLAGL2 and POFUT1 in TCGA cancer tissues.
The (Candidate Cancer Gene Database) CCGD identified PLAGL2 and POFUT1 as a candidate driver in colorectal cancer and both have identical ranking.
The promoter sequence between PLAGL2 and POFUT1 (here list 215bp sequence of upstream of POFUT1 in 12 primates), and DNA sequence of 20:32207380-32207941 was carried for genomic alignments by using Ensembl genome browser 95.
List of transcription factor binding sites predicted in the human PLAGL2/POFUT1 intergenic regions of 179bp. (Analyzed by MatInspector programme).
Acknowledgments
Acknowledgements
We thank Suzhou GenePharma Co., Ltd. for technical support of this study.
Funding sources
This work was supported by the National Nature Science Foundation of China, No. 81773130, the Fundamental research funds for the Central Universities of Central South University (the Hunan province projects of Postgraduate Independent Exploration and Innovation of Central South University, No. 2018zzts050), and the New Xiangya Talent Projects of the Third Xiangya Hospital of Central South University, No. JY201508. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of interests
The authors declare no conflicts of interest that pertain to this work.
Authors' contributions
Daojiang Li, Nanpeng Li, Yuheng Du, Chunxing Yang, YangBai, Zhicai Feng, Chen Su, Runliu Wu, Shenglei Song, Peicheng Yan and Miao Chen mainly finished experiments, performed statistical analysis and draft the manuscript. Arad Jain, Lihua Huang and Yi Zhang participated literature search, experiments and data collection; Changwei Lin and Miao Chen mainly collected tumor tissues; Daojiang Li, Changwei Lin, Xiaorong Li contributed in funding, designed the study and coordination.
References
- 1.Muleris M., Salmon R.J., Dutrillaux A.M. Characteristic chromosomal imbalances in 18 near-diploid colorectal tumors. Cancer Genet Cytogenet. 1987;29:289–301. doi: 10.1016/0165-4608(87)90239-1. [DOI] [PubMed] [Google Scholar]
- 2.Carvalho B., Postma C., Mongera S. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- 3.Sillars-Hardebol A.H., Carvalho B., Tijssen M. TPX2 and AURKA promote 20q amplicon-driven colorectal adenoma to carcinoma progression. Gut. 2012;61:1568–1575. doi: 10.1136/gutjnl-2011-301153. [DOI] [PubMed] [Google Scholar]
- 4.Ali Hassan N.Z., Mokhtar N.M., Kok Sin T. Integrated analysis of copy number variation and genome-wide expression profiling in colorectal cancer tissues. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D., Lin C., Chen M. Comprehensive bioinformatics analysis of the characterization and determination underlying mechanisms of over-expression and co-expression of genes residing on 20q in colorectal cancer. Oncotarget. 2017;8:78642–78659. doi: 10.18632/oncotarget.20204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du Y., Li D., Li N. POFUT1 promotes colorectal cancer development through the activation of Notch1 signaling. Cell Death Dis. 2018;9:995. doi: 10.1038/s41419-018-1055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su C., Li D., Li N. Studying the mechanism of PLAGL2 overexpression and its carcinogenic characteristics based on 3′-untranslated region in colorectal cancer. Int J Oncol. 2018;52:1479–1490. doi: 10.3892/ijo.2018.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Declercq J., Hensen K., Van De Ven W.J. PLAG proteins: how they influence apoptosis and cell proliferation. Annals of the New York Academy of Sciences. 2003;1010:264–265. doi: 10.1196/annals.1299.045. [DOI] [PubMed] [Google Scholar]
- 9.Zheng H., Ying H., Wiedemeyer R. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17:497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B., Lu C., Song Y.X. The role of pleomorphic adenoma gene-like 2 in gastrointestinal cancer development, progression, and prognosis. Int J Clin Exp Pathol. 2014;7:3089–3100. [PMC free article] [PubMed] [Google Scholar]
- 11.Li N., Li D., Du Y. Overexpressed PLAGL2 transcriptionally activates Wnt6 and promotes cancer development in colorectal cancer. Oncol Rep. 2019;41:875–884. doi: 10.3892/or.2018.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y.P., Guo P.T., Zhu Z. Pleomorphic adenoma gene like-2 induces epithelial-mesenchymal transition via Wnt/beta-catenin signaling pathway in human colorectal adenocarcinoma. Oncol Rep. 2017;37:1961–1970. doi: 10.3892/or.2017.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider M., Kumar V., Nordstrom L.U. Inhibition of Delta-induced notch signaling using fucose analogs. Nat Chem Biol. 2018;14:65–71. doi: 10.1038/nchembio.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Villanueva I., Gutierrez M., Hispan P. Novel POFUT1 mutation associated with hidradenitis suppurativa-Dowling-Degos disease firm up a role for notch signalling in the pathogenesis of this disorder. Br J Dermatol. 2018;178:984–986. doi: 10.1111/bjd.16264. [DOI] [PubMed] [Google Scholar]
- 15.Trinklein N.D., Aldred S.F., Hartman S.J. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakano C., Byun J.S., Di L.J. The dual lives of bidirectional promoters. Biochim Biophys Acta. 2012;1819:688–693. doi: 10.1016/j.bbagrm.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 18.Li J.H., Liu S., Zhou H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du R., Lu C., Jiang Z. Efficient typing of copy number variations in a segmental duplication-mediated rearrangement hotspot using multiplex competitive amplification. J Hum Genet. 2012;57:545–551. doi: 10.1038/jhg.2012.66. [DOI] [PubMed] [Google Scholar]
- 20.Abbott K.L., Nyre E.T., Abrahante J. The candidate Cancer gene database: a database of cancer driver genes from forward genetic screens in mice. Nucleic Acids Res. 2015;43:D844–D848. doi: 10.1093/nar/gku770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orekhova A.S., Rubtsov P.M. Bidirectional promoters in the transcription of mammalian genomes. Biochem Biokhimiia. 2013;78:335–341. doi: 10.1134/S0006297913040020. [DOI] [PubMed] [Google Scholar]
- 22.Crooks G.E., Hon G., Chandonia J.M. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M.Q., Koehly L.M., Elnitski L.L. Comprehensive annotation of bidirectional promoters identifies co-regulation among breast and ovarian cancer genes. PLoS Comput Biol. 2007;3:e72. doi: 10.1371/journal.pcbi.0030072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tay Y., Kats L., Salmena L. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X., Chen X., Zeng K. DNA-methylation-mediated silencing of miR-486-5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/beta-catenin signal pathways. Cell Death Dis. 2018;9:1037. doi: 10.1038/s41419-018-1105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chabanais J., Labrousse F., Chaunavel A. POFUT1 as a promising novel biomarker of colorectal Cancer. Cancers. 2018;10 doi: 10.3390/cancers10110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura T., Tsuchiya K., Watanabe M. Crosstalk between Wnt and notch signaling in intestinal epithelial cell fate decision. J Gastroenterol. 2007;42:705–710. doi: 10.1007/s00535-007-2087-z. [DOI] [PubMed] [Google Scholar]
- 28.Collu G.M., Hidalgo-Sastre A., Brennan K. Wnt-notch signalling crosstalk in development and disease. Cell Mol Life Sci. 2014;71:3553–3567. doi: 10.1007/s00018-014-1644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeuner A., Todaro M., Stassi G. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell. 2014;15:692–705. doi: 10.1016/j.stem.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 30.Dotse E., Bian Y. Isolation of colorectal cancer stem-like cells. Cytotechnology. 2016;68:609–619. doi: 10.1007/s10616-014-9806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Didych D.A., Shamsutdinov M.F., Smirnov N.A. Human PSENEN and U2AF1L4 genes are concertedly regulated by a genuine bidirectional promoter. Gene. 2013;515:34–41. doi: 10.1016/j.gene.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 32.Cartharius K., Frech K., Grote K. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinform (Oxford, England) 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 33.Meersseman C., Lejard V., Rebours E. Bovine TWINKLE and mitochondrial ribosomal protein L43 genes are regulated by an evolutionary conserved bidirectional promoter. Gene. 2014;537:154–163. doi: 10.1016/j.gene.2013.11.088. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa H., Tategu M., Yamauchi R. Transcriptional regulation of an evolutionary conserved intergenic region of CDT2-INTS7. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics. 2008;91:243–248. doi: 10.1016/j.ygeno.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 36.Tsai H.K., Huang P.Y., Kao C.Y. Co-expression of neighboring genes in the zebrafish (Danio rerio) genome. Int J Mol Sci. 2009;10:3658–3670. doi: 10.3390/ijms10083658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel J.H., von Heydebreck A., Purmann A. Chromosomal clustering of a human transcriptome reveals regulatory background. BMC Bioinform. 2005;6:230. doi: 10.1186/1471-2105-6-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lejard V., Rebours E., Meersseman C. Construction and validation of a novel dual reporter vector for studying mammalian bidirectional promoters. Plasmid. 2014;74:1–8. doi: 10.1016/j.plasmid.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Zhang Y., Zhang C. The gene pair PRR11 and SKA2 shares a NF-Y-regulated bidirectional promoter and contributes to lung cancer development. Biochim Biophys Acta. 2015;1849:1133–1144. doi: 10.1016/j.bbagrm.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Lozupone F., Borghi M., Marzoli F. TM9SF4 is a novel V-ATPase-interacting protein that modulates tumor pH alterations associated with drug resistance and invasiveness of colon cancer cells. Oncogene. 2015;34:5163–5174. doi: 10.1038/onc.2014.437. [DOI] [PubMed] [Google Scholar]
- 41.Tang J., Chen H., Wong C.C. DEAD-box helicase 27 promotes colorectal cancer growth and metastasis and predicts poor survival in CRC patients. Oncogene. 2018;37:3006–3021. doi: 10.1038/s41388-018-0196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han D., Liu J., Chen C. Anti-tumour immunity controlled through mRNA m(6)a methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai Y., Yang C.X., Wu R.L. YTHDF1 regulates Tumorigenicity and Cancer stem cell-like activity in human colorectal carcinoma. Front Oncol. 2019;9:12. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellizzi D., Dato S., Cavalcante P. Characterization of a bidirectional promoter shared between two human genes related to aging: SIRT3 and PSMD13. Genomics. 2007;89:143–150. doi: 10.1016/j.ygeno.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 45.Okajima T., Matsuda T. Roles of O-fucosyltransferase 1 and O-linked fucose in notch receptor function. Methods Enzymol. 2006;417:111–126. doi: 10.1016/S0076-6879(06)17009-3. [DOI] [PubMed] [Google Scholar]
- 46.Anastas J.N., Moon R.T. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 47.Ranganathan P., Weaver K.L., Capobianco A.J. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 48.Bertrand F.E., Angus C.W., Partis W.J. Developmental pathways in colon cancer: crosstalk between WNT, BMP, hedgehog and notch. Cell cycle (Georgetown, Tex) 2012;11:4344–4351. doi: 10.4161/cc.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guilmeau S., Flandez M., Bancroft L. Intestinal deletion of Pofut1 in the mouse inactivates notch signaling and causes enterocolitis. Gastroenterology. 2008;135 doi: 10.1053/j.gastro.2008.05.050. [849-860, 860.e841-846] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Der Vartanian A., Audfray A., Al Jaam B. Protein O-fucosyltransferase 1 expression impacts myogenic C2C12 cell commitment via the notch signaling pathway. Mol Cell Biol. 2015;35:391–405. doi: 10.1128/MCB.00890-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strubberg A.M., Veronese Paniagua D.A., Zhao T. The zinc finger transcription factor PLAGL2 enhances stem cell fate and activates expression of ASCL2 in intestinal epithelial cells. Stem cell reports. 2018;11:410–424. doi: 10.1016/j.stemcr.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sekiya R., Maeda M., Yuan H. PLAGL2 regulates actin cytoskeletal architecture and cell migration. Carcinogenesis. 2014;35:1993–2001. doi: 10.1093/carcin/bgu081. [DOI] [PubMed] [Google Scholar]
- 53.Wu X., Tu X., Joeng K.S. Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell. 2008;133:340–353. doi: 10.1016/j.cell.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu G., Wang Y., Huang B. A Rac1/PAK1 cascade controls beta-catenin activation in colon cancer cells. Oncogene. 2012;31:1001–1012. doi: 10.1038/onc.2011.294. [DOI] [PubMed] [Google Scholar]
- 55.Xanthoulis A., Tiniakos D.G. E2F transcription factors and digestive system malignancies: how much do we know? World J Gastroenterol. 2013;19:3189–3198. doi: 10.3748/wjg.v19.i21.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerami E., Gao J., Dogrusoz U. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darnell J.E., Jr. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 58.Wang X., Sun D., Tai J. TFAP2C promotes stemness and chemotherapeutic resistance in colorectal cancer via inactivating hippo signaling pathway. J Exp Clin Cancer Res. 2018;37:27. doi: 10.1186/s13046-018-0683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cyr A.R., Kulak M.V., Park J.M. TFAP2C governs the luminal epithelial phenotype in mammary development and carcinogenesis. Oncogene. 2015;34:436–444. doi: 10.1038/onc.2013.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All the primers used in this article were listed below.
The wild-type sequence of 179bp promoter and 6 mutated sequence (here only list the sequence of PLAGL2 orientation).
The details about the DNA fragment for copy nubmer analysis.
Correlation analysis results of PLAGL2 and POFUT1 in TCGA cancer tissues.
The (Candidate Cancer Gene Database) CCGD identified PLAGL2 and POFUT1 as a candidate driver in colorectal cancer and both have identical ranking.
The promoter sequence between PLAGL2 and POFUT1 (here list 215bp sequence of upstream of POFUT1 in 12 primates), and DNA sequence of 20:32207380-32207941 was carried for genomic alignments by using Ensembl genome browser 95.
List of transcription factor binding sites predicted in the human PLAGL2/POFUT1 intergenic regions of 179bp. (Analyzed by MatInspector programme).