Abstract
Bacterial trans-acyltransferase polyketide synthases (trans-AT PKSs) are among the most complex known enzymes from secondary metabolism and are responsible for the biosynthesis of highly diverse bioactive polyketides. However, most of these metabolites remain uncharacterized, since trans-AT PKSs frequently Occur in poorly studied microbes and feature a remarkable array of non-canonical biosynthetic components with poorly understood functions. As a consequence, genome-guided natural product identification has been challenging. To enable de novo structural predictions for trans-AT PKS-derived polyketides, we developed the Trans-AT PKS Polyketide Predictor (TransATor). TransATor is a versatile bio- and chemoinformatics web application that suggests informative chemical structures for even highly aberrant trans-AT PKS biosynthetic gene clusters, thus permitting hypothesis-based, targeted biotechnological discovery and biosynthetic studies. We demonstrate the applicative scope in several examples, including the characterization of new variants of bioactive natural products as well as structurally novel polyketides from unusual bacterial sources.
Introduction
Trans-acyltransferase polyketide synthases (trans-AT PKSs) are extremely complex bacterial enzymes that generate a large variety of natural products.1 The structural diversity encountered in these polyketide metabolites is impressive and accompanied by a broad range of bioactivities with relevance as therapeutics2,3 and as bacterial factors of pathogenicity, symbiosis, and regulation.4–6 Trans-AT PKS biosynthetic gene clusters (BGCs) are widely distributed throughout the bacterial kingdom (Supplementary Fig. 1a-c and Supplementary Table 1).1,7,8 They are commonly present in bacterial groups distinct from those typically studied in natural product screening programs, including many uncultivated microbiota,1,9–12 but remain poorly characterized. The high frequency of architecturally novel BGCs, as well as the large number and unprecedented enzymatic components present in these PKSs,1 offer rich opportunities for metabolic discovery and biotechnology (Supplementary Fig 1b). To predict natural product structures from BGCs, powerful bioinformatic tools exist that are widely used in genomic and metagenomic analyses.13,14 However, for trans-AT PKSs, which are among the most complex and catalytically diverse of all known natural product enzymes, no tool provides reliable de novo structural predictions.
Architecturally, trans-AT PKSs consist of multiple concatenated modules harboring various functional domains.1 Each module usually elongates and often further modifies an enzyme-bound polyketide intermediate, which is then passed on to the next module. Biosynthesis is initiated by one or a few free-standing trans-acting acyltransferase enzymes (AT) that select coenzyme A-(CoA-)bound acyl building blocks, usually malonyl units, and transfer them onto an acyl carrier protein (ACP) domain present in each module. A ketosynthase (KS) domain in the same module uses this building block to elongate an incoming polyketide chain in a Claisen-like condensation reaction. Additional facultative enzymatic components, either located within the module or acting in trans, process the resulting β-keto intermediate further to a wide range of possible α- to γ-modified products that are the elongation substrates for the KS of the next module. Once the polyketide backbone is completely assembled, it is released from the PKS, typically catalyzed by a thioesterase (TE) domain. Additional tailoring enzymes can further modify the polyketide to produce the fully maturated metabolite. In many cases, trans-AT PKSs form hybrids with modular nonribosomal peptide synthetases (NRPSs).1,15–17
A related assembly line-like architecture exists for the well-studied cis-AT PKSs, which contain intramodular AT domains instead of free-standing enzymes.16 Cis-AT PKSs usually exhibit a domain and module architecture that correlates well with the polyketide core structure. This correlation, known as the PKS colinearity rule,16 permits fairly reliable structural predictions of cis-AT PKS-derived polyketides. Applied to the evolutionarily distinct and functionally much more diverse trans-AT PKSs,15 however, the colinearity concept usually generates incorrect predictions.1,15 Modules of trans-AT PKSs commonly exhibit non-canonical domain orders, novel types of domains, catalytic functions that are provided in trans or are apparently missing, and modules split between two proteins. Different module architectures can generate identical PKS modifications, or different chemistry can result from architecturally identical modules. Another idiosyncrasy is the ubiquitous presence of non-elongating modules (indicated by a KS0 in the module) that do not extend but sometimes modify the nascent polyketide backbone. As a result, some modifications can be jointly installed by two modules.1
To enable genome-based natural product mining for these challenging enzymes, we previously developed a metabolic prediction method that can be applied to even highly aberrant trans-AT PKS systems.15 It is based on a close correlation between KS sequence features and the polyketide modifications introduced by domains of the upstream PKS module, i.e., the structural features of a polyketide intermediate within the α-to-γ-region.15 At least in part, this correlation is based on a high specificity of KSs for their incoming substrates that might promote coevolution of upstream modifying domains and the accepting KS domain.18–21 By exploiting this correlation of KS sequence and polyketide moiety, a phylogenetic analysis of all KS domains present in trans-AT PKS systems reveals the collective structural information of the core polyketide. The power of this method to accurately predict canonical as well as non-canonical polyketide features has been demonstrated in various genome mining studies.11,15,22,23 To date, however, predictions require laborious KS correlations to reference trees containing a large number of KS homologs with manually assigned substrates, for which detailed biosynthetic knowledge is required.
To make such metabolic predictions accessible to a broad community, we here report the automated TransATor (Trans-AT PKS Polyketide Predictor) prediction pipeline. Using the PKS sequences as input, TransATor generates KS specificity predictions and, based on these, full structural proposals of the corresponding core polyketide. In addition to this new pipeline, we present several examples for applications that demonstrate the utility of TransATor in targeted polyketide discovery, generation of biosynthetic models, dereplication studies, and as a structure elucidation aid.
Results
Global analysis of trans-AT PKSs refines correlation rules
As a first basis for the development of an automated tool, we manually assessed the number of different modules encountered in all 54 trans-AT PKSs with characterized products (status October 2016).1 This analysis resulted in more than 160 different module types for these systems (Supplementary Fig. 2), a striking number when compared to the few module architecures known from cis-AT PKSs. In terms of module frequencies, hydroxyl- and double bond-generating modules are the most diverse and abundant module types. More interestingly, non-textbook modules that introduce different forms of β-branches and double bonds containing an α-methyl group, as well as the minimal non-elongating modules (KS0 ACP) are also amongst the most abundant module types. Since PKS modules are usually encoded by more than one gene within a BGC, we next explored whether the biosynthetic module order is reflected in the gene order. Of the 54 BGCs, only eight do not exhibit a colinear architecture (Supplementary Fig. 3). Thus, the assumption that biosynthetic genes in the BGC are ordered in a colinear fashion would in most cases permit prediction of polyketide structures.
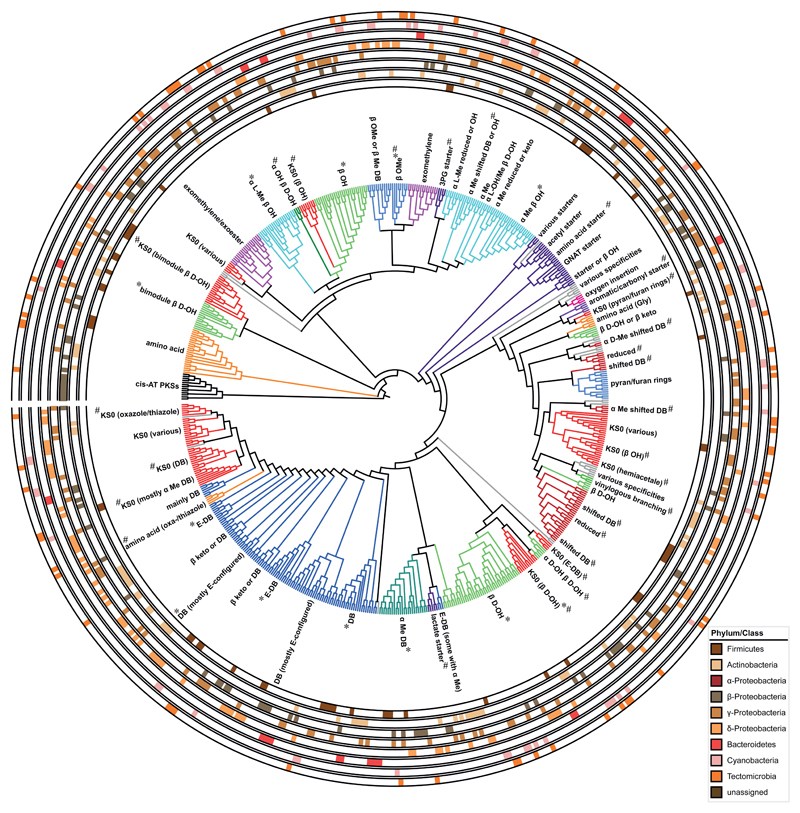
Previous phylogenetic analyses of trans-AT PKSs revealed that KSs form clades that tightly correlate with the chemical structure of their substrates,1,15 here referred to as ketide clades. To maximize the predictive resolution of this correlation, we chemically assigned and phylogenetically analyzed all 655 KS sequences from the 54 BGCs, resulting in a tree that contained >90 clades (Fig. 1, Supplementary Dataset 1 and Supplementary Tables 2-3). The analysis confirmed many previously detected ketide clades, some of which had contained only few KS sequences.15,22 Other ketide clades reported in the initial study now formed subclades (i.e., for starters, β-branches, double bonds, and α-substituted β-hydroxyl moieties) that revealed more detailed structural correlations. Some clades matched to highly differentiated stereochemical or regiochemical features (e.g., L-OH or D-OH; α,β- or β,γ-double bonds). Distinct clades also emerged that permit the distinction of β-methyl branches, β-exomethylene groups, and β-branches generated by Michael-type addition.18,24 Remarkably, separate ketide clades were also identified for various non-acetate starter units, including aromatic-, lactate-, amino acid-, aminotransferase- (AMT) and 1,3-bisphosphoglycerate-derived starters. In addition to these subclades, manual assignment of KSs resulted in the identification of ketide clades for new types of modifications, including hemiacetals, backbone-inserted oxygen, and α,β-hydroxyl groups. The latter two modifications were previously speculated to be introduced in post-PKS tailoring reactions.2,25 The presence of a KS from an α-hydroxyl clade two modules upstream of a KS from a cyclic ether clade can be used to distinguish furan from pyran-type ether moieties. Furan-type rings are putatively biosynthesized by intramolecular oxa-conjugate addition of an α-hydroxyl-group introduced two modules upstream, while pyran-type rings form by addition of a canonical hydroxyl group derived from β-ketoreduction.19 The existence of ketide clades for α,β-hydroxyl groups therefore suggests that some of these moieties are introduced by α-hydroxylation during chain elongation. As for elongating KSs, distinct ketide clades were also identified for non-elongating KSs (KS0s). The identification of clades for a wide range of common and rare biosynthetic moieties suggested remarkable predictive potential, in addition to their informative value about biosynthetic timing.
Figure 1. Phylogenetic separation of trans-AT PKS KS domains into ketide clades.
Cladogram of 655 trans-AT PKS KS domains from 54 characterized trans-AT PKS BGCs. Cis-AT PKS KS domains from the erythromycin PKS were used as outgroup. * indicates ketide clades that can be further subdivided into sub-clades. # indicates ketide clades that are first described in this study. The color code indicates similar ketide clade types. Outer circles around the central cladogram indicate the presence of a KS within the phyla with characterized trans-AT PKS BGCs. DB: double bonds; 3PG: 3-phosphoglycerate derived starters.
Development of the TransATor bio-/chemoinformatics pipeline
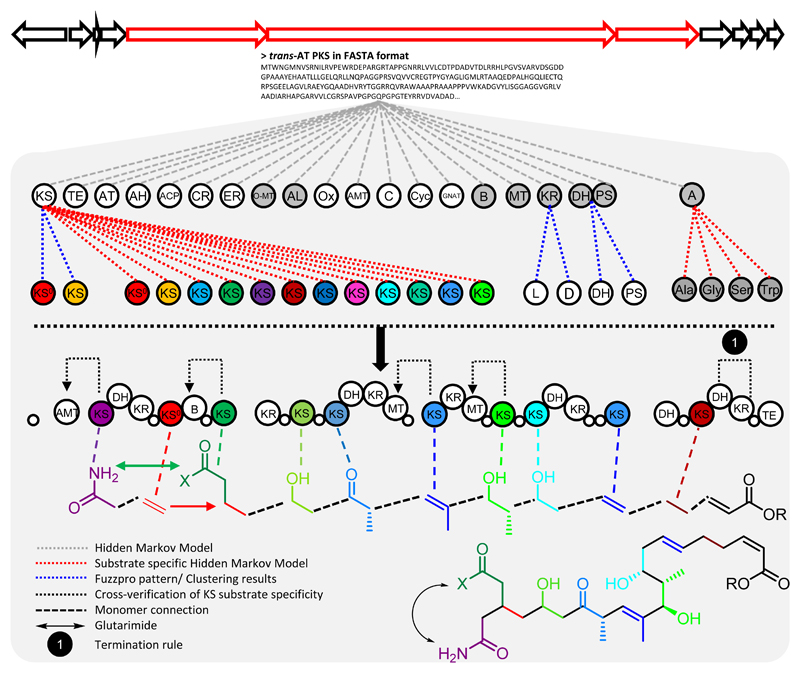
On the basis of the ketide clade assignments, we developed TransATor (Fig. 2). With a concatenated PKS protein sequence in FASTA format as input, the software annotates every defined KS clade and all other common PKS domains by comparison to custom-built Hidden Markov Models (HMMs).26 In addition to the domain set classically identified in available BGC annotation tools, TransATor detects further domains predominantly present in trans-AT PKSs, e.g., acyl-hydrolase (AH), enoyl-CoA hydratase (ECH; syn. crotonase), branching (B), C3-acyltransferase (FkbH), methyltransferase (FkbM), aminotransferase (AMT) domains, and Baeyer-Villiger monooxygenases (OX). The search for EMBOSS fuzzpro patterns in KR domains is used to predict the absolute hydroxyl stereochemistry (Supplementary Fig. 4). Adenylation domain specificity predictions (NRPSpredictor227) were implemented to account for NRPS-PKS hybrid annotations. Ultimately, a Java program generates the predicted polyketide structure based on the annotated core PKS proteins making use of the Chemistry Development Kit (CDK) (Fig. 2).28 First, the core structure is generated based on the ketide classification of all KS sequences. In a second pass, the linear polyketide is modified based on contextual rules. If applicable, amino acid side chains are added, as determined by NRPSpredictor2. Since the trans-AT PKS correlation rule is based on the correlation of a KS sequence with the modification introduced into the nascent polyketide by the module upstream of a KS, ketide moieties cannot be predicted in the absence of a downstream KS after the final module or preceding NRPS- or cis-AT PKS modules.1 In such cases, cis-AT colinearity rules are used. TransATor can be remotely accessed through a web interface (http://transator.ethz.ch) written in Java, which relies on BioJS components to display the KS clade, domain annotations, and stereochemical information. An interactive HTML5 page is used for the user-friendly visualization of the output (Fig. 3). The predicted chemical structures are shown as images and as SMILES (Fig. 3b). To estimate the level of confidence for each monomer assembled to the final predicted structure, a grey scale (black: high confidence, grey: low confidence) is used for the structural representation of the predicted polyketide. Biosynthetic domains are displayed in an annotation panel with the top five hits of the KS specificity prediction shown for each KS domain in the panel as well as in a table, including HMMs e-values and predicted substrates (Fig. 3c). The top hit is used for structural prediction. Sequence information of enzymatic domains and fuzzpro patterns can be retrieved by clicking on the respective annotation.
Figure 2. TransATor workflow.
Outline of the TransATor pipeline for protein-based analysis of trans-AT PKS BGCs. Core PKS domains are annotated and KS substrate specificities predicted based on HMMs. EMBOSS fuzzpro patterns are used to predict the absolute configuration of hydroxylated carbon atoms. CDK is used to construct the polyketide structure based on the PKS annotation. AH: acyl-hydrolase, CR: crotonase (syn: enoyl-CoA hydratase), ER: enoyl-reductase, O-MT: O-methyl-transferase, AL: acyl-ligase, GNAT: GCN5-related N-acetyltransferase, B: branching domain, MT: methyl-transferase, KR: ketoreductase, KS0: non-elongating KS, DH: dehydratase, PS: pyran synthase, A: adenylation domain, Cyc: cyclase, Ox: oxidase, small white circle: ACP. L: l-configured hydroxyl group, D: d-configured hydroxyl group. AMT: aminotransferase. C: condensation domain.
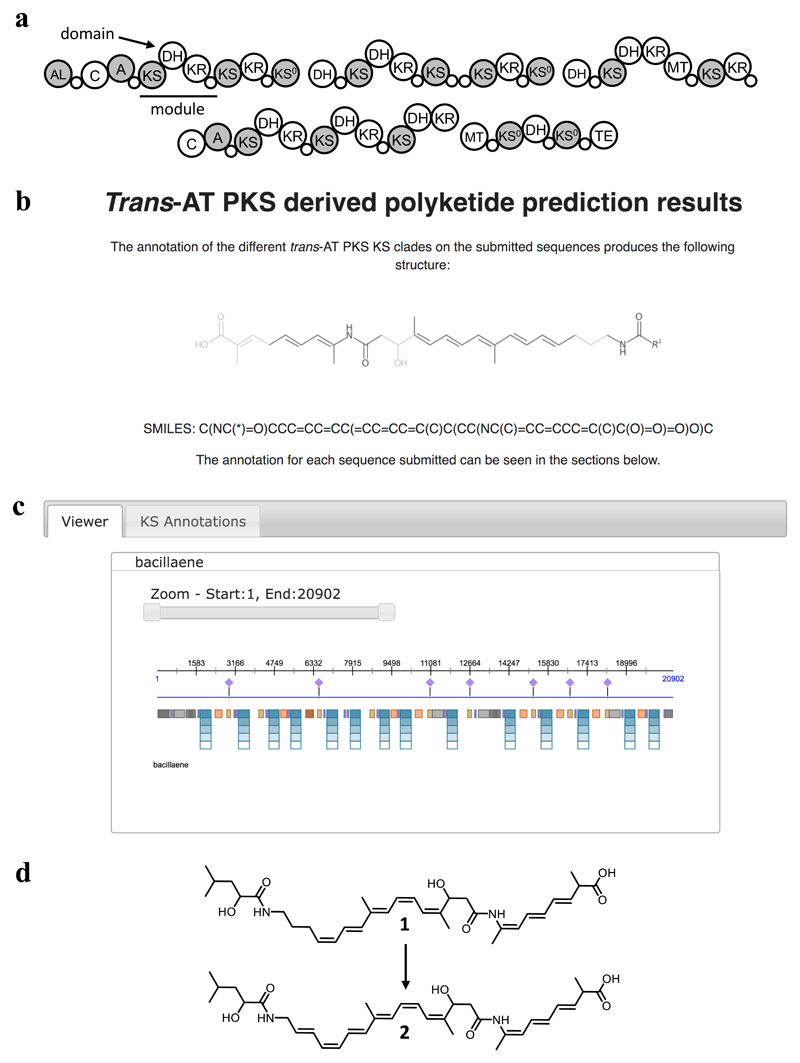
Figure 3. Example of a TransATor result.
(a) Bacillaene PKS. (b) Dihydrobacillaene structure predicted by TransATor. Confidence of prediction is indicated by grey scale representation with high-confidence annotations shown in black and lower-confidence annotations in grey. (c) TransATor-based PKS/NRPS annotation output. The "viewer mode" shows the annotated PKS/NRPS proteins. Colored boxes: HMM-based annotations. Top five hits for KS substrates are shown as turquoise bars. Information (e-value, specificity and if applicable stereochemical information) on all annotated domains can be obtained by hovering over the respective domain. A detailed list of the top five KS hits is displayed in the "KS annotations" view. The top hit is used for the generation of the predicted structure. Diamonds: KR-based hydroxyl group stereochemistry prediction. (d) Structures of dihydrobacillaene (1) and bacillaene (2).
Verification of the TransATor workflow
To test the accuracy of the web application, we used several trans-AT PKS sequences with known polyketide products. Figure 3 shows the output generated for the trans-AT PKS responsible for dihydrobacillaene (1) biosynthesis as an example. Dihydrobacillaene, the direct product of the bacillaene PKS, is converted to bacillaene (2) in a tailoring reaction.29,30 The actual structure of 1 contains several non-canonical features like the presence of shifted double bonds installed at the β,γ-position by different mechanisms,31,32 a fully saturated moiety introduced by module 2 that according to textbook colinearity rules (KS-DH-KR-ACP) would generate a double bond, and a β-branch for module 6 without optional domains that would classically match to a keto function. All moieties as well as the two NRPS building blocks of 1 were correctly predicted by TransATor. The only difference between the predicted and actual product of the bacillaene PKS results from the colinearity rule-based prediction of the last incorporated moiety and the ambiguous acyl starter. The comparison between a representative selection of TransATor-based structure predictions and the reported structure for a given pathway revealed >90% accuracy of TransATor predictions (Supplementary Table 4) for PKSs in the initial training data set and >82% for trans-AT PKS-derived polyketides not part of the initial training set (BGCs published between 2017 and Nov. 2018), notably outperforming antiSMASH4.0 and PRISM3-based predictions (Supplementary Table 5).
Using TransATor as a dereplication and structure elucidation aid
To assess the potential of TransATor for applications in natural product research, we next focused on uncharacterized trans-AT PKSs. An investigation of BGCs in chemically poorly explored taxa suggested the Oceanospirillales bacterium Gynuella sunhinyii YC6258, isolated from the rhizosphere of a Carex sp. plant in a marine tidal flat, as a putatively rich polyketide producer. Its genome contains the remarkable number of six trans-AT PKS BGCs. Analysis of one of the BGCs (Supplementary Table 6) resulted in a small but high-confidence substructure corresponding to the N-terminal section of the PKS (Supplementary Fig. 5), whereas several KSs of the C-terminal part could not be unambiguously assigned. Subjecting the high-confidence part (3) to a substructure search in the natural product database AntiMarin33,34 provided tartrolons as the only hits (Supplementary Fig. 5). Comparison with the characterized tartrolon BGC indeed pointed to the production of a tartrolon-like polyketide. Tartrolons show antibacterial activities and were isolated from bacteria of various phyla, including the γ-proteobacterial shipworm symbiont Teredinibacter turnerae, the source of the first described tartrolon BGC.35 To test whether the predicted polyketide fragment is indeed responsible for the production of tartrolon-type compounds, G. sunshinyii was cultivated for chemical analysis. In total, three candidate compounds that either directly matched the mass of tartrolon D or differed by two or four Da were identified and subsequently isolated. Their structures were determined by 2D NMR experiments (Supplementary Fig. 6-7, Supplementary Note Fig. 1-15 and Supplementary Note Table 1), revealing the three tartrolons 4, 5, and 6 (Supplementary Fig. 5). While 4 is identical to tartrolon D, the new congeners 5 and 6 differ by substitutions of one or both of the C9 and C9’ keto groups with a hydroxyl group and were hence named tartrolon F and G, respectively. 4, 5 and 6 showed antibacterial activity against Bacillus subtilis (MIC values: 0.38 μM for tartrolon D, 0.38 μM for tartrolon F and 1.9 μM for tartrolon G). The isolation of tartrolon-like polyketides from another unusual producer shows that TransATor can be used for early-stage in silico dereplication studies, to identify new variants of polyketides of interest, and to pinpoint new producers for difficult-to-access polyketides.
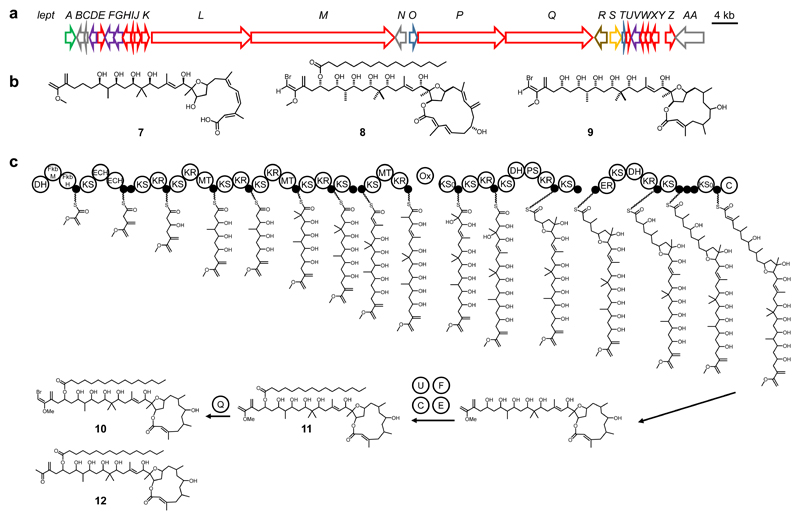
As the next test case, we mined genomes of the Institut Pasteur's cyanobacterial strain collection (PCC collection) for the presence of trans-AT PKS BGCs.36 This analysis revealed a 91 kb trans-AT PKS gene cluster (termed lept cluster) in the genome of Leptolyngbya sp. PCC 7375 (Fig. 4a and Supplementary Table 7). We subjected this cluster to TransATor analysis and used the predicted structure (7) for a similarity search in natural product libraries. AntiMarin hits suggested partial similarity to two polyketides: the potent brine shrimp toxin phormidolide37 (8) and oscillariolide (9), (Fig. 4b), an inhibitor of starfish egg development.38 However, the predicted compound differed from 8 and 9 in the size and structure of the macrocycle, suggesting a new polyketide core.
Figure 4. Model for leptolyngbyalide biosynthesis.
(a) The lept BGC. Red: PKS genes, green: transporter genes, grey: hypothetical genes, purple: genes involved in lipid metabolism, brown: oxygenase genes, yellow: halogenase genes. (b) Comparison between TransATor-based structure prediction (7) and the AntiMarin library hits phormidolide (8) and oscillariolide (9). (c) Proposed model for the biosynthesis of isolated leptolyngbyalides A-C (10-12). Letters in white balls for the post-PKS steps refer to protein names according to Supplementary Table 7.
To test the prediction, we analyzed Leptolyngbya sp. PCC 7375 cultures guided by the predicted mass, chemical composition, and the putative presence of a halogen atom in the polyketide, as indicated by a halogenase gene in the BGC. Only after three months of cultivation, three candidate compounds 10–12 were identified and subjected to chromatographic separation. MS and NMR data suggested 10 to be a monobrominated polyketide that we named leptolyngbyalide A, while leptolyngbyalides B (11) and C (12) lack bromine, and 12 additionally differs in the exchange of an enolether moiety for a methyl ketone (Fig. 4c, Supplementary Fig. 8-9, Supplementary Note Fig. 16-32, and Supplementary Note Table 2). Unexpectedly, 10–12 did not feature the predicted 12-membered macrolactone ring but the 14-membered oscillariolide macrolactone moiety. To address this discrepancy, we conducted PCR experiments to test for the presence of an additional PKS module that might have been missed during genome assembly (Supplementary Fig. 10). However, the PCR data did not support this hypothesis, suggesting that one module is used iteratively to give rise to the oscillariolide-like macrolactone ring. Since no BGC has been identified for oscillariolide, it is unknown whether its PKS also contains this aberrant feature. Due to the low amounts of 10–12 in the culture, the absolute configuration could not be determined to verify the TransATor-based stereochemical predictions. The TransATor output of the lept and the related phor PKSs resulted in virtually the same predicted configurations for all nine shared hydroxyl-bearing stereocenters of phormidolide and leptolyngbyalide (Supplementary Table 8). To our surprise, however, these predicted configurations were opposite to those reported for phormidolide.39 Reanalysis of the published data for phormidolide36,39 showed that the Mosher ester analysis was interpreted opposite to the convention.37,39 These observations suggest that the stereochemistry might need to be revised (Supplementary Fig. 11).
TransATor-based discovery of a new polyketide scaffold
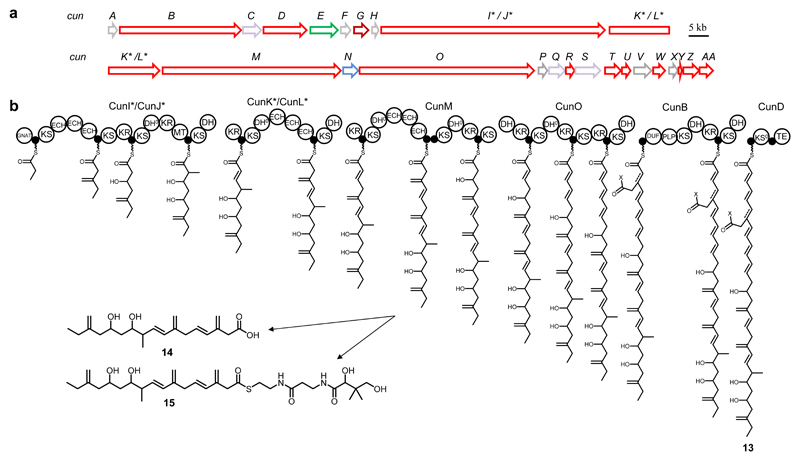
To test whether TransATor can be used to discover natural products with novel skeletons from chemically unexplored bacterial groups, we selected a trans-AT PKS from a sponge-derived Aquimarina sp. bacterium, a member of the phylum Bacteroidetes. Several KS sequences from trans-AT PKSs were previously PCR-amplified from Aquimarina strains isolated from Irciniidae sponges by homology-based gene targeting.40 We therefore selected such trans-AT PKS-positive strains for genome sequencing. One of the identified trans-AT PKS BGCs (here termed cun) stood out (Fig. 5a and Supplementary Table 9), as it was conserved among several strains and contained a module with two non-canonical domains (DUF PLP), of which homologs participate in the formation of a sulfur heterocycle pharmacophore in leinamycin-type compounds.41 Apart from this feature, however, the cun and leinamycin PKSs show little resemblance. To conduct targeted isolation of the predicted polyketide (13) (Fig. 5), we cultivated Aquimarina spp. Aq349 and Aq78. However, despite extensive variation of cultivation conditions, we were unable to identify a metabolite at the predicted mass and chemical composition range. Since the predicted structure contains three exomethylene groups that would generate characteristic NMR shifts, we aimed to identify the metabolite using an NMR-guided isolation strategy. These efforts resulted in the purification of two candidate natural products 14 and 15 (Fig. 5b). Surprisingly, structure elucidation by 2D NMR experiments (Supplementary Fig. 12, Supplementary Note Fig. 33-42, and Supplementary Note Tables 3-4) revealed that 14 and 15 are in perfect structural agreement with the TransATor-based prediction up to module six, but are lacking the remaining polyketide portion that would correspond to PKS modules 7 to 16. The compounds, wich were produced by both strains, were named cuniculene 6A (14) and 6B (15), with the numbering chosen to indicate an early release from PKS module 6. An intriguing feature of 15 is the unique (to our knowledge) panthetheinyl moiety, likely due to non-canonical release from the PKS by phosphate ester cleavage of the phosphopanthetheinyl arm. The presence of the acyltransferase-like gene cunR (PSI-blast) directly downstream of the cun region encoding PKS module 6 indicates that the release is programmed rather than spontaneous. The biological importance of this mechanism, which would inactivate the PKS module, is currently unknown. However, the isolation of a biosynthetic intermediate that is in perfect agreement with the predicted structure shows that TransATor can be employed to target novel trans-AT PKS scaffolds despite the high frequency of unprecedented biosynthetic features in such enzymes.
Figure 5. Model for cuniculene biosynthesis.
(a) Conserved Aquimarina cun BGC (here from Aquimarina sp. Ap349). Red: PKS genes, green: transporter genes, grey: hypothetical genes, light purple: aminotransferase genes/cysteine desulfurase, brown: oxygenase genes, blue: acyl-transferase-like gene. (b) Proposed model for cuniculene biosynthesis, predicted structure (13) and isolated compounds 14-15. The biosynthetic model is based exclusively on TransATor predictions, with 13 displaying the core structure predicted by TransATor. The isolated compounds 14 and 15 are in perfect agreement with TransATor predictions. * genes split between two contigs, putatively encoding one PKS protein.
Discussion
Known natural products generated by trans-AT PKSs exhibit a wide range of structures and bioactivities. These include antibiotics,2,42 anticancer drug candidates,43,44 pathogenicity factors of human5,45,46 and agricultural relevance,6,47 plant protectants,48 bacterial regulators,4 and tools used in cell biology.49 According to genomic and metagenomic data, however, much of the chemical diversity encoded by trans-AT PKS BGCs resides in chemically unexplored bacteria and therefore remains uncharacterized (Supplementary Fig. 1a-c). Recent studies have also revealed such trans-AT PKS clusters in human microbiota and pathogens, such as clostridia and Streptococcus mutans, raising important questions about their products and relevance for health.9,50 Additionally, trans-AT PKSs from uncultivated microbiota likely generate many, if not most, bioactive polyketides identified in marine invertebrate-based drug screening programs.12,25 Considering the rich opportunities provided by massively increasing DNA datasets, there is thus a need for robust automated assignments of chemical structures to trans-AT PKSs. Due to the extraordinary architectural diversity and non-colinear enzymology of these assembly lines (i.e., not adhering to cis-AT colinearity rules), such a tool was not available among the existing BGC annotation pipelines.
Here, we filled this gap by developing TransATor, a pipeline and web application for the functional assignment of trans-AT PKS sequences. This tool permits the structure prediction of polyketide products, thus aiding (meta)genome annotation, natural product discovery, and in silico dereplication. In addition, it provides insights needed for the generation of biosynthetic hypotheses, which is often challenging for these enzymes. The first publication of ketide clades that connects KS sequences with polyketide moieties describes 140 KSs and 16 clades.15 Since then, structural resolution has increased simultaneously with the number of characterized PKSs (655 KS sequences and > 90 clades identified in this work). It is reasonable to expect that third-generation sequencing in combination with TransATor-guided isolation and characterization of novel polyketides will further improve the predictive resolution in an iterative process. Although the level of accuracy in TransATor-based predictions is generally high for the examples provided in this study, ketide clades for rare substrate moieties are poorly populated (e.g. KSs for α,β-hydroxylated moieties, hemiacetals, vinylogous chain branching, or oxygen insertion). Adding further KS sequences to such clades will likely increase the reliability of TransATor-based annotations and reveal new clades. There are only a few clades that are populated by KS sequences with more than a single predicted substrate type (Fig. 1). With increasing numbers of annotated KS sequences, chemically uniform subgroups might emerge, as previously observed for other clades.15,22 Considering the diversity of trans-AT PKS modules, it is likely that the discovery of further BGCs and their polyketides will require expansion of the initial training data set by additional clades. To continuously improve TransATor, updated phylogenetic trees and new cladification patterns can be implemented into the software.
The workflow demonstrated here consists of three integral parts. First, a PKS product is predicted using TransATor. Second, the predicted structure is used for natural product database searches and third, information about the predicted polyketide scaffold is used for the targeted isolation, purification, and structure elucidation of the corresponding polyketide. As exemplified in this study, relevant information about the predicted polyketide that guides the isolation process can either be the chemical composition, predicted mass range, physico-chemical properties, or characteristic NMR shifts. TransATor assumes colinearity in the order of translated protein sequences used as input. In some cases, a non-colinear order of PKS proteins prevents TransATor from accurately predicting a structure (e.g. starter in the middle of the polyketide or a shifted double bond preceeding a canonical double bond). In cases where TransATor recognizes such conflicts, a troubleshooting guide in the tutorial will provide possible solutions.
The number of characterized modular PKS BGCs is lower than the number of complex polyketides that are present in natural product databases. Many of these biosynthetically unassigned polyketides exhibit structural features typical for a trans-AT PKS origin.25,33,34 Thus, the strategy described here, i.e., using the TransATor-based structure predictions to search in chemical databases, might outcompete homology-based searches on the gene cluster level for dereplication studies. Using the predictions generated by TransATor for database searches, formal assignments of gene clusters to known polyketides can be made by in silico dereplication and subsequently experimentally confirmed.
An important application of high biotechnological value is the targeted discovery of novel producers of pharmaceutically relevant polyketides for which the producer is either unknown, genetically not accessible, uncultivated, or yields only minute amounts of the drug candidate. In these cases, the identification of a new producer of the polyketide of interest could make challenging multi-step chemical syntheses obsolete. These and other applications should make TransATor a valuable community tool.
Online Methods
Statistical analysis
Metadata for the relative distribution of classes of polyketides were retrieved from all polyketide BGCs identified in the antiSMASH database. The relative abundance of trans-AT PKS gene clusters from sequenced genomes is based on the metadata published by O’Brien et al. (2014) and the antiSMASH database.7,51 All published trans-AT PKS BGCs (excluding NRPS-PKS hybrid BGCs which only harbor one trans-AT PKS module e.g. didemnin PKS) were retrieved from Helfrich and Piel (2016) and Piel (2010).1 Analysis on module architectures and co-linearity of biosynthetic genes in trans-AT PKS BGCs are based on the biosynthetic models reviewed and, if applicable, revised by Helfrich and Piel (2016) and Piel (2010).1
Phylogenetic analysis of KS sequences
655 trans-AT PKS KS amino acid sequences of all characterized 54 trans-AT PKS BGCs1 were, if deposited, retrieved from public databases. Two cis-AT PKS KS sequences from the erythromycin PKS were used as outgroup. Sequence alignments were either conducted using the MUSCLE or MAFFT algorithm, manually refined and re-aligned. In order to find the optimal phylogenetic representation, the “Best protein model ML and NJ” was computed in MEGA 6.4. For comparison, a RAxML tree was constructed from both MUSCLE and MAFFT alignments. The phylogenetic representations were compared with sequence similarity networks (https://efi.igb.illinois.edu/efi-est/). Most phylogenetic trees resulted in a comparable level of clade resolution; the trees shown in this paper were generated from MUSCLE alignments followed by phylogenetic analysis using ML trees and 1000 bootstrap replicates. The LG substitution model with Frequencies (+F) was used with rates and patterns set to gamma distribution with invariant sites (G+I) and gap treatment to complete deletion. Ketide clades were assigned manually and the resulting tree was exported in Newick format and uploaded into iTOL. Graphical representation was conducted in iTOL.52
HMM models of PKS domains commonly present in trans-AT PKS gene clusters
Alignments of individual domains were generated from sequences retrieved from our in house database of annotated trans-AT PKS BGCs, the MIBiG database or GenBank.53 Sequences were aligned using the MUSCLE algorithm, and the resulting alignment was manually refined. HMM models were generated using the HMMER3 suite. Cutoff values for the HMM models were determined manually by comparing characterized PKS with the TransATor output at different cutoffs for each domain.
EMBOSS fuzzpro patterns
The KS phylogeny-based stereochemical predictions are verified by an EMBOSS fuzzpro pattern (EMBOSS:6.6.0.0) in the KR domain upstream of the module that harbors the KS domain relevant for the prediction of the monomer in question. The relevant fuzzpro pattern is detected approximately 80 amino acids downstream of the general KR motif: GG[ALVMTGS]G [GRYDAVTHSK][LIV]G. The detection of the motif GXXHXAXXXXD indicates the formation of a D-OH group (fuzzpro [GDPSERVIHKNAT]x(3)[GVTA][IVAL][VHILF][HYFVQY][SIATGMLCFNV][AVTPS][GLIMRP]x(3)D). In the absence of the characteristic aspartate residue, the introduced hydroxyl group is L-configured.
TransATor software development
The program is composed of two applications: a core, mainly focused on sequence annotation, and a second application handling molecule rendering and user interaction. The core, written in Python 2.8, takes a query in FASTA format. It runs HMMER, NRPSpredictor2, EMBOSS Fuzzpro against the query, and collects the results. The results are consolidated into a format wide enough to include all types of annotations, and written to disk as a ".features" file. The HMMER3 suite was used to pre-train the profile HMM models based on manually retrieved sequences which were aligned using the MUSCLE algorithm. Models for the following domains were created: KS (90 clades, 655 sequences), KR (248 sequences), DH (149 sequences), ER (12 sequences), MT (82 sequences), O-MT (11 sequences), A (142 sequences), TE (28 sequences), Enoyl-CoA hydratase (ECH; 20 sequences), epimerase (E; 15 sequences), condensation (C; 189 sequences), ACP (460 sequences), GNAT (8 sequences), flavin-dependent monooxygenase (FMO; 6 sequences), AT/AH (47 sequences), acyl ligase (AL; 3 sequences), cyclase (Cyc; 10 sequences), pyran synthase (PS; 12 sequences) and branching domains (5 sequences).
The second application is written in Java 1.8 and has two main functions: molecule rendering and user interaction. The user interaction is built in two parts: A Servlet handling user input of the sequence query, and a REST API providing the results. Two input formats are supported: a textbox, and a file upload. After a successful submission of the input query a "results" page is served. This page polls a REST API for a PNG picture of the predicted polyketide molecular structure, SMILES data thereof and the data for the visualization of the annotation clustering. The BioJS library (https://biojs.net/) is used for said visualization.
The molecule rendering starts by running the core with the query, and parsing the resulting ".features" file. The annotations describing PKS domains (of HMMER origin) are filtered so that only annotations with a score value above a fixed threshold remain. The threshold for HMMER-based annotations was optimized for every domain annotation.
The annotations are clustered by their location in the query protein sequence with DBSCAN (version 1.1-3):54 non-overlapping regions are tolerated up to a distance of 40 amino acids. Within each cluster, the annotations are sorted by predicted score, and the annotation with the highest score is selected for further processing. This method of selecting relevant annotations from a regional cluster has also been referred to as "Best Match Cascade" in the literature.55
The molecule rendering code processes the filtered and selected annotations in the order they appear in the query sequence. Depending on the type of annotation, different actions are taken. The most important type of domain annotations for the prediction of the molecular structure are elongating ketosynthase domains (KS): they determine the next monomer to be incorporated into the growing polyketide structure. For them a function implementing said elongation is called. All other domains leading up to a KS domain provide additional information relevant to the elongation step and therefore are stored for later processing. This information is later used to modify a predicted monomer before its incorporation into the growing polyketide. We call this step “preprocessing”. E.g. preprocessing of an amino acid monomer is conducted as follows: A base structure which does not include the amino acid specific side chain, is modified by a "NRPSMonomerProcessor" that adds the corresponding chain as predicted by NRPSpredictor2 before it is incorporated. After each elongating step, this list of non KS domain annotations is cleared.
The algorithm for the elongation of the molecular prediction works by merging CDK (version 2.0) IAtomContainer typed objects at special points denoted by so called "pseudo atoms". These atoms do not represent real atoms, but can be incorporated into structures as metadata. We use two pseudo atoms: "R1" and "R2". “R1” denotes where the monomer should be attached to the growing polyketide structure prediction, and "R2" denotes where the following monomer will be attached. The algorithm looks for the “R2” pseudo atom in the growing polyketide and for the R1 pseudo atom in the monomer to be added. It then removes these pseudo atoms with their corresponding bonds and connects the C-atoms to which the pseudo atoms were connected to each other instead. Finally, the implicit hydrogen atom count is adjusted to the number of hydrogens present at the carbon atoms involved in the novel bond.
KS domains that correlated with furan and pyran rings as their predicted substrates: Forming the ring requires the modification of a monomer which was incorporated two monomers upstream. When such a KS domain is processed, the algorithm registers a "PostProcessor" which is run after it has processed all annotations in a linear fashion. In this case, a "CyclizationPostProcessor" finds the two atoms involved in the ring forming reaction across multiple monomers and connects them accordingly.
However, relying on substrate specificity of the incoming intermediates for the final structure prediction also bears a problem. The KS domains select for the structure created, or modified by the upstream domains, and hence cannot be used for the prediction of modifications downstream of the last KS domain. To predict the effects of these domains, we use a co-linearity rule-based approach. We determine the set of the non-KS domain types that follow the last elongating KS domain. For each set, a fixed mapping to a monomer is defined that is then incorporated at the termination boundaries.
A similar problem occurs in hybrid PKS-NRPS BGCs: The effects of the domains between a KS domain and a NRPS module, cannot be predicted with the KS substrate specificity, and is handled in the same way as described above.
Finally, the individual monomers in the predicted structure are colored in a grey scale according to the confidence of the algorithm for the predicted monomer substrate of the corresponding KS. A combination of e-value of the top hit and the difference of the e-value of the top hit in comparison to the second top hit is used as a basis for the color coding.
Organisms and culture conditions
Gynuella sunshinyii YC6258 was cultured in marine broth 2216 in 1 L ultra-high yield flasks for 3 days at 30 °C at 120 rpm. Leptolyngbya sp. PCC 7375 was cultured in 40 mL ASNIII medium at room temperature for 2 up to 8.5 months exposed to a 13 h-11 h light-dark cycle at 20 μmol photon.m-2.s-1. Aquimarina sp. 78 and Aquimarina sp. 349 were cultured in marine broth 2216 (BD Difco™) in 1 L ultra-high yield™ flasks (Thomson) for 2 or 3 days at 25 °C at 160 rpm. Cultures volumes were scaled up to determine the structure of the compounds.
Genome sequencing and annotation
Aquimarina sp. Aq78 and Aquimarina sp. 349 were retrieved from the marine sponge Sarcotragus spinosulus as described previously.56 Prior to whole genome sequencing, genomic DNA was extracted from a pure culture grown in marine broth for five days at 19 °C using the Wizard Genomic DNA Purification kit (Promega Corporation, Madison, WI, USA). Paired-end sequence reads (125 bp) were generated using an Illumina HiSeq2500 platform at BaseClear (Leiden, The Netherlands). FASTQ sequence files were generated using the Illumina Casava pipeline version 1.8.3. Sequencing output was 287 Mb consisting of 2 x 128 bp quality-filtered paired-end reads, resulting in a predicted genome coverage of 48X. Adapter sequences were trimmed and the reads assembled using the CLC Genomics Workbench version 7.0.4. Initial annotation was performed with the RAST (Rapid Annotation using Subsystem Technology) server, version 2.0.57 Aquimarina sp. Aq78 shares 98.98% 16S rRNA gene identity with type strain Aquimarina macrocephali JAMB N27, isolated from marine sediment.58
The genome sequences of Leptolyngbya sp. PCC 7375 (ALVN00000000) and G. sunshinyii YC6258 (NZ_CP007142.1) were obtained previously.36,59
PCR-based verification of the lept BGC
Polymerase chain reactions (PCRs) were performed with Phusion polymerase (Thermo Fischer Scientific) and GC buffer. The reagents for a 50 μL reaction were mixed (Phusion HC buffer (5X) 10 μl, dNTPs (10 mM each) 2 μL, primers (10 μM each) 2 μL each, DMSO 1.5 μL, phusion polymerase 0.5 μL, DNA 20-100 ng and the thermocycler program set as follows: initial denaturation 2 min at 98 °C, denaturation 15 sec at 98 °C, annealing 20 sec at 68 °C, extension 3 min at 72 °C, 35 cycles.
Oligonucleotides used in this study
| Primer name | Primer sequence |
|---|---|
| KS13_forw. | TGGAGAATTCTCCATTTTACGTAAATACTACGTGCCG |
| KS14_rev | ACCAATATTGCCAGATTCGTCGGAATAATAGTCCCTC |
| KS14_forw. | GCCTATCATTAACGGTAGGCCGAGGCGGCGCAT |
| KS15_rev. | AGCTCCCCACTGCCCATAGATCTTGTTTGGATG |
| KS15_for. | AGGTGCGCCGAGCCGCTGTCAGTTCCTTTG |
| C_rev. | TGTGAATGTCAAATAACAGGCTCTGGCGTTCCTTCTGA |
Isolation and structure elucidation of tartrolons D, F and G (4-6)
A 2 L culture of G. sunshinyii was centrifuged, and the pellet was extracted with acetone. The extract was dissolved in MeCN, and separated by reversed-phase HPLC (Phenomonex Luna 5 μm C18, 10 x 250 mm, UV detection at λ = 220 nm, r.t.) eluting with 5% MeCN for 5 min, then a gradient from 5% MeCN to 100% MeCN for 30 min, and 100% MeCN for 25 min. The fraction eluting between 46-49 min was further purified by reversed-phase HPLC (Phenomenex Luna 5 μm Phenyl-Hexyl, 10 x 250 mm, UV detection at λ = 220 nm, r.t.) eluting with 65% MeCN to yield 0.9 mg of compound 4 and 1.3 mg of compound 5. In addition, the fraction eluting between 52-55 min in the first chromatographic separation step was further purified by reversed-phase HPLC (Phenomenex Luna 5 μm Phenyl-Hexyl, 10 x 250 mm, UV detection at λ = 220 nm, r.t.) eluting with 65% MeCN to yield 1.0 mg of compound 6.
Compound 4 had a molecular formula of C44H68O14, which was suggested by HR-LC-ESI-MS (m/z 843.4475, [M+Na]+, Δ-2.6 mmu). Comparison of the 1H NMR spectrum of compound 4 with the literature values of tartrolon D revealed its identity.
Compound 5 had a molecular formula of C44H70O14, which was suggested by HR-LC-ESI-MS (m/z 845.4635, [M+Na]+, Δ-2.3 mmu). Analysis of the 1H NMR spectrum and MS2 data suggested structural asymmetry consisting of a tartrolon D monomer and a highly similar second monomer that form a heterodimer. Units a-c were deduced by COSY data. Unit a was connected to unit b by HMBC correlations from H-7, H-8, H-10, and H-11 to C-9, and from H-10 to C-8. The hemiketal moiety in unit b was formed by HMBC correlations from H-4, H-5, H-7, and H-22 to C-3. Unit b was connected to unit c by HMBC correlations from H-2 to C-1, C-3, and C-4, from H-4 to C-2, and from H-20' to C-1. The hemiketal moiety in unit c and the connection between unit a and unit c were determined in the same manner.
14E (14'E) geometry was determined by NOESY correlations between H-13 (H-13') and H-15 (H-15'), and between H-14 (H-14') and H-16 (H-16'). 16Z (16'Z) geometry was determined by the small coupling constant (3JH16,H17 (3JH16',H17') = 10.8 Hz) and NOESY correlation between H-16 (H-16') and H-17 (H-17').
Compound 6 had a molecular formula of C44H72O14, which was suggested by HR-LC-ESI-MS (m/z 847.4791, [M+Na]+, Δ -2.3 mmu). Analysis of the 1H NMR spectrum and MS2 data suggested that the structure was the homodimer of the novel half of compound 5. The structure was confirmed by 2D NMR.
Isolation and structure elucidation of leptolyngbyalides A-C (10-12)
A three month old, 1.6 L culture of the cyanobacterium Leptolyngbya sp. PCC 7375 was filtered, and the pellet was extracted with acetone and MeOH overnight. The extract was concentrated in vacuo, and the residue was resuspended in 90% MeOH and extracted with n-hexane. The 90% MeOH layer was concentrated and separated by reversed-phase HPLC (Phenomenex Luna 5C8, 10 x 250 mm, UV detection at λ = 220 nm, r.t.) and eluted with 5% MeCN for 5 min, then a gradient from 5% MeCN to 100% MeCN for 30 min, and 100% MeCN for 25 min. The fractions eluting after 50 min were combined and further separated by reversed-phase HPLC (Phenomenex Luna 5C18, 10 x 250 mm, UV detection at λ = 220 nm, at room temperature) eluted with 100% MeCN to afford fractions 1 to 12. 10-12 were purified from fractions 2, 8 and 9 by reversed-phase HPLC (Phenomenex Luna 5C18, 10 x 250 mm, UV detection at λ = 220 nm, r.t.) eluted with 95% MeOH.
Compound 12 had a molecular formula of C56H98O12, which was suggested by HR-LC-ESI-MS (m/z 985.6940, [M+Na]+, Δ-1.1 mmu). Analysis of the 1H NMR spectrum in conjunction with the HSQC data revealed the presence of six singlet methyls, three doublet methyls, one ethyl group, nine oxygenated methines, and one exomethylene. Units a-g were deduced by COSY data. The acetyl moiety was attached to unit a via an exomethylene which was determined by HMBC correlations from H-28 to C-29, C-30, and C-40, from H-31 to C-29 and C-30, and from H-40 to C-28, C-29, and C-30. Unit a and unit c were connected via unit b on the basis of HMBC correlations from H-23 to C-25 and C-39, from H-25 to C-23 and C-39, from H-26 to C-24, from OH-23 to C-23 and C-24, from OH-25 to C-24 and C-25, and from H-39 to C-23 and C-25. Unit c was connected to unit d via the dimethyl quaternary carbon, which was elucidated by HMBC correlations from H-19 to C-21 and C-38, from H-21 to C-19 and C-38, from OH-19 to C-19 and C-20, from OH-21 to C-20 and C-21, from H-37 to C-19, C-20, C-21, and C-38, and from H-38 to C-19, C-20, C-21, and C-37. Unit d and unit e were connected by HMBC correlations from H-15 to C-17, from H-16 to C-18 and C-36, from H-18 to C-16, C-17, and C-36, from H-19 to C-17, and from H-36 to C-16, C-17, and C-18. Unit e and unit f were connected via a methyl hydroxyl tetrahydrofuran by HMBC correlations from H-11 to C-14, from H-12 to C-14, from H-13 to C-14 and C-35, from H-15 to C-13 and C-14, and from H-35 to C-13, C-14, and C-15. Unit f was connected to unit g by HMBC correlations from H-8 to C-10, from H-10 to C-9 and C-34, and from H-34 to C-10. The macrolide was formed between the other terminus of unit f and unit g, which was determined by HMBC correlations from H-2 to C-1, C-3, C-4, and C-32, from H-4 to C-2, C-3, and C-32, from H-32 to C-2, C-3, and C-4, and from H-18 to C-1. Finally, the fatty acid was attached to C-27 as revealed by HMBC correlation from H-27 to C-41. The length of the fatty acid was deduced from the molecular formula.
The 2E geometry was determined by NOESY correlation between H-2 and H-4. The 16E geometry was determined by NOESY correlations between H-15 and H-36, and between H-16 and H-18. The relative stereochemistry of the ether ring was elucidated by NOESY correlations between H-35 and H-18, between H-18 and H-12b, and between H-12b and H-11.
Compound 11 had a molecular formula of C57H100O12, which was suggested by HR-LC-ESI-MS (m/z 999.7105, [M+Na]+, Δ -0.2 mmu). Analysis of the 1H NMR spectrum in conjunction with HSQC data suggested the absence of the acetyl group in compound 12 but the additional oxygenated methyl and an exomethylene. HMBC correlations from H-28 to C-29, C-30, and C-40, from H-31 to C-29 and C-30, from H-40 to C-28, C-29, and C-30, and from H-57 to C-30 determined the substructure, which was different from compound 12.
Compound 10 had a molecular formula of C57H99O12Br, which was suggested by HR-LC-ESI-MS (m/z 1077.6213, [M+Na]+, Δ +0.1 mmu). Analysis of the 1H NMR spectrum in conjunction with the molecular formula and HSQC data suggested that compared to compound 11, compound 10 lacked one hydrogen but gained one bromide at the terminal exomethylene moiety. 2D NMR data confirmed the structure. The length of the fatty acid was deduced from the molecular formula and MS2 fragmentation patterns.
Isolation and structure elucidation of compounds cuniculene 6A and 6B (14-15)
A 3 L culture of Aquimarina sp. Aq78 (3 days at 25 °C) was centrifuged, and the supernatant was extracted with EtOAc. The extract was dissolved in MeCN, and separated by reversed-phase HPLC (Phenomonex Luna 5 μm C18, 20 x 250 mm, UV detection at λ = 220 nm, r.t.) eluted with 5% MeCN for 5 min, then a gradient from 5% MeCN to 100% MeCN for 30 min, and 100% MeCN for 25 min. The fraction eluting between 22-26 min was further purified by reversed-phase HPLC (Phenomenex Luna 5 μm Phenyl-Hexyl, 10 x 250 mm, UV detection at λ = 220 nm, r.t.) eluted with 45% MeCN to yield compound 14. A 3 L culture of Aquimarina sp. Aq78 (2 days at 25 °C) was centrifuged, and the supernatant was extracted with EtOAc. The extract was dissolved in MeCN, and separated by reversed-phase HPLC (Phenomonex Kinetex 5 μm C18, 10 x 250 mm, UV detection at λ = 220 nm, r.t.) eluted with 40% MeCN. The fraction eluting between 44-46 min was collected on ice and quickly concentrated afterwards. The fraction was further purified by reversed-phase HPLC (Phenomenex Luna 5 μm Phenyl-Hexyl, 10 x 250 mm, UV detection at λ = 220 nm, r.t.) eluted with 40% MeCN. The fraction between 46-50 min was collected on ice and dried as soon as possible to yield compound 15. In each step, all fractions were measured by 1H NMR to detect the fraction containing exomethylenes.
Compound 14 had a molecular formula of C21H32O4, which was suggested by HR-LC-ESI-MS (m/z 371.2196, [M+Na]+, Δ-0.3 mmu). The 1H NMR spectrum and HSQC data of compound 8 revealed the presence of one triplet methyl, one doublet methyl, two oxygenated methines, four protons attached to sp2 carbons, and three exomethylenes. The three units a-c were deduced from the COSY spectrum. Unit a and unit b were connected by HMBC correlations from H-14 to C-15, C-16, and C-21, from H-16 to C-14, C-15, and C-21, from H-17 to C-15, and from H-21 to C-14, C-15, and C-16. Unit b and unit c were connected by HMBC correlations from H-5 to C-7, from H-6 to C-7, C-8, and C-19, from H-8 to C-6, C-7, and C-19, from H-9 to C-7, and from H-19 to C-6 and C-8. The structure of the other side of unit c was determined by HMBC correlations from H-2 to C-1, C-3, C-4, and C-18, from H-4 to C-2, C-3, and C-18, from H-5 to C-3, and from H-18 to C-2 and C-4. 4E and 8E geometry was determined by the large coupling constants (3JH4,H5 = 15.8 Hz, 3JH8,H9 = 15.8 Hz).
Compound 15 had a molecular formula of C32H52O7N2S1, which was suggested by HR-LC-ESI-MS (m/z 609.3577, [M+H]+, Δ+0.3 mmu). The 1H NMR spectrum and HSQC data of compound 15 revealed the presence of one triplet methyl, one doublet methyl, two singlet methyls, three oxygenated methines, one oxygenated methylene, four protons attached to sp2 carbons, and three exomethylenes. Six units a-f and the presence of two hydroxymethines were deduced by the interpretation of the COSY spectrum. Unit a and unit b were connected by HMBC correlations from H-14 to C-15, from H-16 to C-15 and C-21, and from H-17 to C-15. Unit b and unit c were connected by HMBC correlation from H-20 to C-11. Unit c and unit d were connected by HMBC correlations from H-6 to C-7, C-8, and C-19. Unit e and unit f were connected by HMBC correlations from H-7' and H-10' to C-8'. The terminus of unit f was determined by HMBC correlations from H-3' to C-2' and C-4', from OH-3' to C-2' and C-4', from H-12' to C-1', C-2', C-3', and C-13', and from H-13' to C-1', C-2', C-3', and C-12'. Finally, unit d and unit e were connected by HMBC correlations from H-2 to C-1, C-3, C-4, and C-18, from H-18 to C-2 and C-4, and from H-11' to C-1. The presence of sulfur atom between C-1 and C-11' were determined by the chemical formula, and the chemical values of C-1 and C-11'. 4E and 8E geometry was determined by the large coupling constants (3JH4,H5 = 15.9 Hz, 3JH8,H9 = 15.9 Hz).
Antibacterial assays
Overnight culture of Arthrobacter crystallopoietes, Bacillus subtilis, Escherechia coli, and Pseudomonas putida grown in LB were diluted to an OD600 = 0.01 with LB. 200 μl aliquots were then pipetted into each well of flat-bottom 96 well plates. Compounds were added to the wells in a serial dilution along with a negative and positive control (methanol and ampicillin). The plates were incubated at 30 °C for 24 hours after which OD600 was measured. The respective minimum inhibitory concentration was determined as the lowest concentration at which there was no observed growth (OD600 ≤ 0.02).
Instrumentation
NMR spectra were recorded on a Bruker Avance III spectrometer equipped with a cold probe at 500 MHz and 600 MHz for 1H NMR and 125 MHz and 150 MHz for 13C NMR. Chemical shifts were referenced to the solvent peaks at δH 7.27 and δC 77.23 for chloroform-d, δH 3.31 and δC 49.15 methanol-d4, and δH 2.50 and δC 39.51 for DMSO-d6. UPLC-HESI (HR-LC-ESI-MS) mass spectrometry was performed on a Thermo Scientific Q Exactive mass spectrometer coupled to a Dionex Ultimate 3000 UPLC system.
Supplementary Material
Acknowledgements
JP acknowledges funding by the ERC (ERC Advanced Project SynPlex), the SNF (NRP 72 "Antimicrobial resistance", 407240_167051), and the DFG Research Unit 854. RC acknowledges funding by the Portuguese Foundation for Science and Technology (FCT) through the research grants PTDC/BIA-MIC/3865/2012 and PTDC/MAR-BIO/1547/2014. MG thanks the Institut Pasteur funding for the Action Ciblée-Collection. CS and PM acknowledge generous core funding by the European Molecular Biology Laboratory – European Bioinformatics Institute (EMBL-EBI).
Footnotes
Data availability
GenBank: The Aquimarina sp. Aq349 and Aq78 genome sequence harboring the cuniculene BGC has been submitted to the European Nucleotide Archive (accession numbers OMKB01000001-OMKB01000022 and OMKF01000001-OMKF01000170). The leptolyngbyalide BGC (BK010645) from Leptolyngbyia sp. PCC 7375, and the tartrolon BGC (BK010667) from G. sunshinyii were deposited in GenBank. MIBiG: The lepolyngbyalide (BGC0001835), tartrolon (BGC0001836) and cuniculene (BGC0001855) biosynthetic gene clusters were deposited in the MIBiG database. The TransATor web server is freely accessible at http://transator.ethz.ch. Code for TransATor pipeline/web application is available at https://github.com/pcm32/transator-container.
Author contributions
EJNH, CS, PM and JP designed research, EJNH, MR, AB, RAM, and JP performed bioinformatic analyses, EJNH performed statistical analyses, EJNH compiled training dataset for TransATor, EJNH defined biosynthetic rules, EJNH generated biosynthetic models, PM designed the bioinformatics/chemoinformatics pipeline, PM and AD programmed TransATor, EJNH and AD validated TransATor and predicted compound structures, RU isolated compounds and performed structure elucidation, MG, RC, and GC provided genome sequences and bacterial strains, EJNH and JP wrote the manuscript with the help of all authors.
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.Helfrich EJ, Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2016;33:231–316. doi: 10.1039/c5np00125k. [DOI] [PubMed] [Google Scholar]
- 2.El-Sayed AK, et al. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem Biol. 2003;10:419–430. doi: 10.1016/s1074-5521(03)00091-7. [DOI] [PubMed] [Google Scholar]
- 3.Pulsawat N, Kitani S, Nihira T. Characterization of biosynthetic gene cluster for the production of virginiamycin M, a streptogramin type A antibiotic, in Streptomyces virginiae. Gene. 2007;393:31–42. doi: 10.1016/j.gene.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Ueoka R, Bortfeld-Miller M, Morinaka BI, Vorholt JA, Piel J. Toblerols, cyclopropanol-containing modulators of methylobacterial antibiosis generated by an unusual polyketide synthase. Angew Chem Int Ed Engl. 2017 doi: 10.1002/anie.201709056. [DOI] [PubMed] [Google Scholar]
- 5.Möbius N, et al. Biosynthesis of the respiratory toxin bongkrekic acid in the pathogenic bacterium Burkholderia gladioli. Chem Biol. 2012;19:1164–1174. doi: 10.1016/j.chembiol.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 6.Partida-Martinez LP, Hertweck C. A gene cluster encoding rhizoxin biosynthesis in Burkholderia rhizoxina, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem. 2007;8:41–45. doi: 10.1002/cbic.200600393. [DOI] [PubMed] [Google Scholar]
- 7.O'Brien RV, Davis RW, Khosla C, Hillenmeyer ME. Computational identification and analysis of orphan assembly-line polyketide synthases. J Antibiot. 2014;67:89–97. doi: 10.1038/ja.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blin K, M M, Kottmann R, Lee SY, Weber T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2016:555–559. doi: 10.1093/nar/gkw960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L, Hao T, Xie Z, Horsman GP, Chen Y. Genome mining unveils widespread natural product biosynthetic capacity in human oral microbe Streptococcus mutans. Sci Rep. 2016;6:37479. doi: 10.1038/srep37479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ueoka R, et al. Metabolic and evolutionary origin of actin-inhibiting polyketides from diverse organisms. Nat Chem Biol. 2015;11:705–712. doi: 10.1038/nchembio.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakabachi A, et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol. 2013;23:1478–1484. doi: 10.1016/j.cub.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Wilson MC, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 13.Blin K, et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017:36–41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skinnider MA, et al. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM) Nucleic Acids Res. 2015;43:9645–9662. doi: 10.1093/nar/gkv1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen T, et al. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- 16.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 17.Helfrich EJN, Reiter S, Piel J. Recent advances in genome-based polyketide discovery. Curr Opin Biotechnol. 2014;29:107–115. doi: 10.1016/j.copbio.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Bretschneider T, et al. Vinylogous chain branching catalysed by a dedicated polyketide synthase module. Nature. 2013;502:124–128. doi: 10.1038/nature12588. [DOI] [PubMed] [Google Scholar]
- 19.Pöplau P, Frank S, Morinaka BI, Piel J. An enzymatic domain for the formation of cyclic ethers in complex polyketides. Angew Chem Int Ed Engl. 2013;52:13215–13218. doi: 10.1002/anie.201307406. [DOI] [PubMed] [Google Scholar]
- 20.Jenner M, et al. Substrate specificity in ketosynthase domains from trans-AT polyketide synthases. Angew Chem Int Ed Engl. 2013;52:1143–1147. doi: 10.1002/anie.201207690. [DOI] [PubMed] [Google Scholar]
- 21.Jenner M, et al. Acyl-chain elongation drives ketosynthase substrate selectivity in trans-acyltransferase polyketide synthases. Angew Chem Int Ed Engl. 2015;54:1817–1821. doi: 10.1002/anie.201410219. [DOI] [PubMed] [Google Scholar]
- 22.Teta R, et al. Genome mining reveals trans-AT polyketide synthase directed antibiotic biosynthesis in the bacterial phylum bacteroidetes. ChemBioChem. 2010;11:2506–2512. doi: 10.1002/cbic.201000542. [DOI] [PubMed] [Google Scholar]
- 23.Kampa A, et al. Metagenomic natural product discovery in lichen provides evidence for specialized biosynthetic pathways in diverse symbioses. Proc Natl Acad Sci U S A. 2013;110:3129–3127. doi: 10.1073/pnas.1305867110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sundaram S, Heine D, Hertweck C. Polyketide synthase chimeras reveal key role of ketosynthase domain in chain branching. Nat Chem Biol. 2015;11:949–951. doi: 10.1038/nchembio.1932. [DOI] [PubMed] [Google Scholar]
- 25.Fisch KM, et al. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat Chem Biol. 2009;5:494–501. doi: 10.1038/nchembio.176. [DOI] [PubMed] [Google Scholar]
- 26.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Röttig M, et al. NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011;39:362–367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinbeck C, et al. Recent developments of the chemistry development kit (CDK) - an open-source java library for chemo- and bioinformatics. Curr Pharm Des. 2006;12:2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- 29.Reddick JJ, Antolak SA, Raner GM. PksS from Bacillus subtilis is a cytochrome P450 involved in bacillaene metabolism. Biochem Biophys Res Commun. 2007;358:363–367. doi: 10.1016/j.bbrc.2007.04.151. [DOI] [PubMed] [Google Scholar]
- 30.Moldenhauer J, Chen XH, Borriss R, Piel J. Biosynthesis of the antibiotic bacillaene, the product of a giant polyketide synthase complex of the trans-AT family. Angew Chem Int Ed Engl. 2007;46:8195–8197. doi: 10.1002/anie.200703386. [DOI] [PubMed] [Google Scholar]
- 31.Moldenhauer J, et al. The final steps of bacillaene biosynthesis in Bacillus amyloliquefaciens FZB42: direct evidence for β,γ dehydration by a trans-acyltransferase polyketide synthase. Angew Chem Int Ed Engl. 2010;49:1465–1467. doi: 10.1002/anie.200905468. [DOI] [PubMed] [Google Scholar]
- 32.Kusebauch B, Busch B, Scherlach K, Roth M, Hertweck C. Functionally distinct modules operate two consecutive α,β→β,γ double-bond shifts in the rhizoxin polyketide assembly line. Angew Chem Int Ed Engl. 2010;49:1460–1464. doi: 10.1002/anie.200905467. [DOI] [PubMed] [Google Scholar]
- 33.Dabb S, Blunt J, Munro M. MarinLit: Database and essential tools for the marine natural products community. Abstr Pap Am Chem S. 2014;248 [Google Scholar]
- 34.Blunt JW, Munro MHG, Laatsch H. Antimarin database. University of Canterbury; 2006. [Google Scholar]
- 35.Elshahawi SI, et al. Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills. Proc Natl Acad Sci U S A. 2013;110:295–304. doi: 10.1073/pnas.1213892110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih PM, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci U S A. 2013;110:1053–1058. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson RT, Boulanger A, Vulpanovici A, Roberts MA, Gerwick WH. Structure and absolute stereochemistry of phormidolide, a new toxic metabolite from the marine cyanobacterium Phormidium sp. J Org Chem. 2002;67:7927–7936. doi: 10.1021/jo020240s. [DOI] [PubMed] [Google Scholar]
- 38.Murakami M, Matsuda H, Makabe K, Yamaguchi K. Oscillariolide, a novel macrolide from a blue-green-alga Oscillatoria sp. Tetrahedron Lett. 1991;32:2391–2394. [Google Scholar]
- 39.Bertin MJ, et al. The phormidolide biosynthetic gene cluster: A trans-AT PKS pathway encoding a toxic macrocyclic polyketide. ChemBioChem. 2016;17:164–173. doi: 10.1002/cbic.201500467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esteves AIS, Hardoim CCP, Xavier JR, Goncalves JMS, Costa R. Molecular richness and biotechnological potential of bacteria cultured from Irciniidae sponges in the north-east Atlantic. FEMS Microbiol Ecol. 2013;85:519–536. doi: 10.1111/1574-6941.12140. [DOI] [PubMed] [Google Scholar]
- 41.Ma M, Lohman JR, Liu T, Shen B. C-S bond cleavage by a polyketide synthase domain. Proc Natl Acad Sci U S A. 2015;112:10359–10364. doi: 10.1073/pnas.1508437112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mast Y, Wohlleben W. Streptogramins - Two are better than one! Int J Med Microbiol. 2014;304:44–50. doi: 10.1016/j.ijmm.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 43.Sudek S, et al. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J Nat Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 44.Eustaquio AS, Janso JE, Ratnayake AS, O'Donnell CJ, Koehn FE. Spliceostatin hemiketal biosynthesis in Burkholderia spp. is catalyzed by an iron/α-ketoglutarate-dependent dioxygenase. Proc Natl Acad Sci U S A. 2014;111:3376–3385. doi: 10.1073/pnas.1408300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biggins JB, Ternei MA, Brady SF. Malleilactone, a polyketide synthase-derived virulence factor encoded by the cryptic secondary metabolome of Burkholderia pseudomallei group pathogens. J Am Chem Soc. 2012;134:13192–13195. doi: 10.1021/ja3052156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piel J, Hofer I, Hui D. Evidence for a symbiosis island involved in horizontal acquisition of pederin biosynthetic capabilities by the bacterial symbiont of Paederus fuscipes beetles. J Bacteriol. 2004;186:1280–1286. doi: 10.1128/JB.186.5.1280-1286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cociancich S, et al. The gyrase inhibitor albicidin consists of p-aminobenzoic acids and cyanoalanine. Nat Chem Biol. 2015;11:195–197. doi: 10.1038/nchembio.1734. [DOI] [PubMed] [Google Scholar]
- 48.Loper JE, et al. Rhizoxin analogs, orfamide A and chitinase production contribute to the toxicity of Pseudomonas protegens strain Pf-5 to Drosophila melanogaster. Environ Microbiol. 2016;11:3509–3521. doi: 10.1111/1462-2920.13369. [DOI] [PubMed] [Google Scholar]
- 49.Schneider-Poetsch T, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donia MS, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blin K, Medema MH, Kottmann R, Lee SY, Weber T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017;45:555–559. doi: 10.1093/nar/gkw960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li YF, et al. Comprehensive curation and analysis of fungal biosynthetic gene clusters of published natural products. Fungal Genet Biol. 2016;89:18–28. doi: 10.1016/j.fgb.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duan L, Xu L, Guo F, Lee J, Yan BP. A local-density based spatial clustering algorithm with noise. Inform Syst. 2007;32:978–986. [Google Scholar]
- 55.Yeats C, Redfern OC, Orengo C. A fast and automated solution for accurately resolving protein domain architectures. Bioinformatics. 2010;26:745–751. doi: 10.1093/bioinformatics/btq034. [DOI] [PubMed] [Google Scholar]
- 56.Esteves AI, Hardoim CC, Xavier JR, Goncalves JM, Costa R. Molecular richness and biotechnological potential of bacteria cultured from Irciniidae sponges in the north-east Atlantic. FEMS Microbiol Ecol. 2013;85:519–536. doi: 10.1111/1574-6941.12140. [DOI] [PubMed] [Google Scholar]
- 57.Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyazaki M, Nagano Y, Fujiwara Y, Hatada Y, Nogi Y. Aquimarina macrocephali sp. nov., isolated from sediment adjacent to sperm whale carcasses. Int J Syst Evol Microbiol. 2010;60:2298–2302. doi: 10.1099/ijs.0.018747-0. [DOI] [PubMed] [Google Scholar]
- 59.Chung EJ, Park JA, Jeon CO, Chung YR. Gynuella sunshinyii gen. nov., sp. nov., an antifungal rhizobacterium isolated from a halophyte, Carex scabrifolia Steud. Int J Syst Evol Microbiol. 2015;65:1038–1043. doi: 10.1099/ijs.0.000060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.