Abstract
Obesity is an endemic problem in the United States and elsewhere, and data indicate that in addition to overconsumption, exposure to specific chemicals enhances obesity. CYP2B metabolizes multiple endo- and xenobiotics, and recent data suggests that repression of Cyp2b activity increases dyslipidemia and age-onset obesity, especially in males. To investigate the role played by Cyp2b in lipid homeostasis and obesity, we treated wildtype and Cyp2b-null mice with a normal (ND) or 60% high-fat diet (HFD) for 10 weeks and determined metabolic and molecular changes. Male HFD-fed Cyp2b-null mice weigh 15% more than HFD-fed wildtype mice, primarily due to an increase in white adipose tissue (WAT); however, Cyp2b-null female mice did not demonstrate greater body mass or WAT. Serum parameters indicate increased ketosis, leptin and cholesterol in HFD-fed Cyp2b-null male mice compared to HFD-fed wildtype mice. Liver triglycerides and liver:serum triglyceride ratios were higher than their similarly treated wildtype counterparts in Cyp2b-null male mice, indicating a role for Cyp2b in fatty acid metabolism regardless of diet. Furthermore, RNAseq demonstrates that hepatic gene expression in ND-fed Cyp2b-null male mice is similar to HFD-fed WT male mice, suggestive of fatty liver disease progression and a role for Cyp2b in lipid homeostasis. Females did not show as demonstrative changes in liver health, and significantly fewer changes in gene expression, as well as gene expression associated with liver disease. Overall our data indicates that the repression or inhibition of CYP2B may exacerbate metabolic disorders and cause obesity by perturbing fatty acid metabolism, especially in males.
Keywords: Non-alcoholic fatty liver disease (NAFLD), P450, Cyp2b, Triglycerides, RNAseq, obesity
Graphical abstract
Introduction
According to the National Center for Health Statistics (NCHS) more than 40% of adults and 18.5% youth are obese in United States as of 2015-2016 [1]. Factors that cause obesity include poor diet, changes in lifestyle, genetics, and the environment [2–4]. The primary problem is excess food coupled with inactivity. However, data indicates there are other anthropogenic factors, including environmental toxicants that exacerbate obesity by modulating the use and allocation of nutrient resources leading to increased white adipose tissue and obesity [4–7]. The terms “obesogen” or “metabolic disruptor” [4, 5, 7] refer to a new subclass of endocrine disruptors that perturb metabolic signaling, energy, and lipid homeostasis [8, 9]. In turn, the Organization for Economic Cooperation and Development (OECD) considers metabolic disorders one of three critical areas in toxicology in addition to testicular dysgenesis and autism spectral disorders [9].
Chemical exposures shown to cause obesity include perinatal exposure to bisphenol A that increases lipogenic gene expression and hepatic steatosis in male mice [10]. Tributyltin activation of PPARμ-RXR induces obesity primarily through increased adipocyte differentiation [11, 12], and TCDD activation of AhR induces obesity and steatosis through increased uptake of fatty acids [13–15]. Inactivation of the xenobiotic receptor, the constitutive androstane receptor (CAR), and activation of the pregnane X-receptor (PXR) are also associated with obesity [16–18]. Perturbations in hepatic Cytochrome P450 (CYP) activity were recently associated with lipid accumulation [19–23] and the inhibition of all hepatic CYP activity was associated with increased hepatic lipid accumulation [20]. In constrast, knockouts of Cy2e1 and Cyp3a members were associated with reduced lipid accumulation and weight gain (only in females in Cyp3a-null mice) [23, 24].
CYPs, primarily in families 1-3, metabolize pharmaceuticals [25], environmental pollutants [26] and endobiotics such as fatty acids [27], bile acids and steroids [28]. Several CYP1-3 members are crucial in fatty acid metabolism. For example, CYP3A4 metabolizes arachidonic acid to to 13-hydroxyeicosatrienoic acid (HETE), 10-HETE and 7-HETE [29], and anandamide to several metabolites including 5,6-epoxyeicosatrienoic acid ethanolamide, with high affinity for the cannabinoid-2 (CB-2) receptor [30]. CYP2J2 metabolizes arachidonic acid to epoxyecosatrienoic acid (EET) in cardiomyocytes [31], and some Cyp2b members metabolize arachidonic acid with high affinity [32, 33]. For example, Cyp2b19 expressed in mouse keratinocytes, epoxygenate arachidonic acid to 11,12- and 14,15-EET that play a vital role in cornification of epidermal cells [34, 35].
Several recent studies implicate Cyp2b in obesity. For example, Cyp2b members are inducible by anti-obesity transcription factors (CAR; FoxA2) [36–38]. CAR is a key regulator of Cyp2b, including Cyp2b9 and Cyp2b10 [39–41], and CAR activation by TCPOBOP ameliorates obesity, diabetic activity and fatty liver disease in ob/ob mice, but not ob/ob mice null for CAR [18]. FoxA2, which is activated by fasting and fatty acids and inhibited by insulin, is a potent inducer of Cyp2b9 [38, 42, 43]. FoxA2-null mice are age-dependent obese [44]. Thus, Cyp2b expression is induced by CAR and Foxa2 during nutritional or metabolic stress, potentially as a protective mechanism from NAFLD. Furthermore, cytochrome P450 oxidoreductase (HRN-null) conditional knockout mice that lack all liver CYP activity, activate CAR and induce Cyp2b mRNA during treatment with polyunsaturated fatty acid (PUFA)-rich sunflower oil [20]. Similarly, treatment with soybean oil (rich in 51% linoleic acid) resulted in weight gain, increased white adipose tissue and glucose intolerance in association with a significant increase in Cyp2b9 and 13 [45]. Cyp2b9 is also the most highly induced gene in a couple of diet-induced obesity studies [22, 46]. Last, observations in our laboratory following production of a Cyp2b RNAi-based knockdown mouse model on a FVB strain background [47] show increases in body weight and adiposity with age and a reduced propensity to eliminate PUFA-rich corn oil [48]. Taken together, the Cyp2b enzymes are responsive to high-fat diets and associated with obesity. Therefore, we predict that lack of Cyp2b either by gene knockout (as described here) or repression by inhibitors coupled with a high-fat diet will perturb lipid metabolism, increase body weight and lead to the development of obesity.
Murine Cyp2b members are expressed in several tissues [47]. For example, Cyp2b19 is primarily expressed in the skin and involved in keratinocyte development [32, 34]. Cyp2b23 is primarily expressed in neonate liver [49]. Cyp2b13 is primarily expressed in adult liver [49]. Cyp2b10 is primarily expressed in adult liver and highly inducible [36, 49], and Cyp2b9 is expressed in several tissues including liver, brain, and to a lesser degree kidney and lungs [49, 50]. In addition, both Cyp2b9 and Cyp2b13 are highly female predominant due to sexually dimorphic regulation by HNF4α and growth hormone [51, 52]}[39, 53].
We used our newly developed Cyp2b9/10/13-null (Cyp2b-null) mice missing the primarily hepatic Cyp2b members, Cyp2b9, Cyp2b10 and Cyp2b13 to demonstrate the significance of Cyp2b on diet-induced obesity and hepatic lipid metabolism. We treated Cyp2b-null mice with a 60% high-fat diet for 10-weeks, and examined their susceptibility to diet-induced obesity compared to wild-type (WT) mice. Physiological changes such as body weight, organ weight, glucose tolerance, serum lipids, and specific metabolic hormones were examined. In addition, RNAseq was performed to determine differential gene expression and investigate which gene ontologies and pathways were perturbed. Our data demonstrates a role for perturbations in Cyp2b activity in obesity and lipid metabolism.
Materials and Methods:
High-fat diet treatment of Cyp2b-null mice:
All mice studies followed National Institute of Health guidelines for humane use of research animals and were pre-approved by Clemson University’s Institutional Animal Care and Use Committee (IACUC). Cyp2b-null mice were developed using Crispr/Cas9 technology on C57B16/J (B6) background mice as described earlier [52]. Briefly, the liver predominant Cyp2b genes, Cyp2b10, Cyp2b13 and Cyp2b9 present as tandem repeat sequences on chromosome seven and were targeted individually using small guide RNA sequences (sgRNA). This led to the deletion of 287 kb lacking these three primarily hepatic Cyp2b genes (Supplementary Material-1; SM-1). Homozygous knock-out mice were verified using primers published previously [52].
Wildtype C57BL/6J were purchased from The Jackson Laboratory at 3 weeks of age (Bar Harbor, ME, USA), and acclimated for 6-weeks prior to the dietary treatments. Nine-ten week old male and female mice (n=9) from WT and Cyp2b-null mice were divided into normal diet (ND; 2018S-Envigo Teklad Diet, Madison, WI) and high-fat diet (HFD; Envigo, TD.06414 Adjusted calorie diet) groups. ND provided 3.1 Kcal/g with 18% of kilocalories from fat, 24% protein, and 58% carbohydrate. HFD provided 5.1 Kcal/g with 60.3% of kilocalories from fat (37% saturated, 47% monounsaturated, 16% polyunsaturated fat), 18.4% protein, and 21.3% carbohydrates.
Feed consumption was measured every other day and mice were weighed every week to determine their weight gain. Fasting plasma glucose concentrations were determined on weeks 2, 5 and 8 with an Alphatrek-2 glucometer (AlphaTRAK, Chicago IL USA) following a 4-5 hour fast. Fasting Glucose Tolerance Tests (GTT) were performed on weeks 5 and 8 and an insulin tolerance test (ITT) was performed on week 9 [54]. For the GTT the mice were fasted for 5 hr and then injected i.p. with 1g/kg of their bodyweight with D-glucose (Sigma Ultra, St. Louis MO USA). Blood glucose levels were measured using the Alphatrek-2 glucometer in 20-30 min intervals for 2 hours per protocol [54] and described by us previously [24].
At the end of the study, mice were anesthetized, blood was collected by heart puncture, and mice euthanized by carbon dioxide asphyxiation [48]. Organs such as liver, kidney, inguinal and renal white adipose tissue, and brain were excised and weighed. Livers were dissected into five fractions and snap frozen in liquid nitrogen for microsome preparation and RNA extraction. Frozen liver fractions were stored at −80°C. A portion of the liver was stored in 10% formalin (Fisher, Fair Lawn NJ USA) for histopathological analyses. A timeline of experimental procedures performed on the mice is provided (SM-2).
RNA sequencing (RNAseq):
Liver samples were stored in RNAlater stabilization solution (Invitrogen) at −80°C. RNA was extracted from mouse livers using an RNeasy Plus Universal kit (Qiagen) per the manufacturer’s instructions and quantified on a NanoDrop 8000 Spectrophotometer. An Agilent 2100 Bioanalyzer was used to assess the RNA integrity number (RIN). Samples with a RIN > 8 were determined to be of high quality and used for next generation sequencing. Libraries were prepared with the TruSeq Total RNA Library Prep kit. Samples were sequenced to an average sequencing depth of 20,000,000 read pairs with a 2 × 125 paired-end module using an Illumina HiSeq2500 for males and a 2 × 150 paired-end module using a NovaSeq 6000 for females. Quality metrics were checked using FastQC on all samples sequenced, and Trimmomatic was used to trim low quality bases. Trimmed reads were aligned to the Mus musculus reference genome (GCF_000001635.25_GRCm38.p5) using GSNAP, and 99.7% of the trimmed reads aligned. Subread feature counts software found reads that aligned with known genes. Raw read counts and EdgeR were used to determine differential gene expression [55]. Genes in which the lowest replicate number of samples have less than one count per million of expression were ascertained to be low coverage genes and were filtered out of the analysis. Samples were then normalized to the scale of their library sizes. Genes were considered differentially expressed if their adjusted p-value and false discovery rate were less than 0.05 and 0.1 respectively as performed previously [56]. Series GSE120761 containing the RNAseq data has been uploaded to GEO.
Heatmap hierarchical cluster analysis by Euclidiean distance using Ward’s method was performed with Heatmap.2 in R (http://www.r-project.org/). Venn diagrams were created using Venn Diagram Plotter (omics.pnl.gov). GOSeq, a GO term enrichment analysis program was used to adjust for gene length and expression bias [57]. Significantly (p < 0.05) enriched GO terms were visualized in Revigo, which reduces enriched term redundancy and displays the remaining enriched GO terms in a scatterplot [58]. Differentially expressed genes were annotated using InterPro [59], and genes with a log2FC > 1.0 were entered into KEGG Mapper (http://www.genome.jp/kegg/) to determine and visualize biochemical pathways perturbed by a HFD or a lack of Cyp2b enzymes [60].
Histopathological analysis:
Following necropsy, a clean slice of liver was placed in 10% formalin (Fisher). Hematoxylin and Eosin (H&E) staining was performed at Colorado Histoprep (Fort Collins, CO USA) and Oil Red O staining was performed with frozen samples at Baylor College of Medicine’s Comparative Pathology Laboratory (Houston, TX USA) using standard protocols [18, 61]. Histopathological scoring was performed blind by a veterinary pathologist at Colorado Histoprep (Fort Collins, CO USA).
Serum lipid panel:
Blood samples were collected by heart puncture and incubated at room temperature for 30 min followed by centrifugation at 6000 rpm for 10 minutes. Serum samples were transferred into fresh tubes and 100μ1 aliquots were shipped on dry ice to Baylor College of Medicine’s Comparative Pathology Laboratory (Houston, TX) for determination of nonfasting serum cholesterol, triglycerides, alanine aminotransferase (ALT), high density lipoprotein (HDL), low density lipoprotein (LDL), and very low density lipoprotein (VLDL) with a Beckman-Coulter AU480 analyzer and the appropriate Beckman-Coulter biochemical kits according to the manufacturer’s instructions.
Serum concentrations of adiponectin, leptin, β-hydroxybutyrate and liver triglycerides:
Serum adiponectin and leptin concentrations were determined using an EIA kit from Bertin Pharma (Montigny Le Bretonneux, FR). Serum free fatty acids (FFA) and β-hydroxybutyrate (B-OHB) concentrations were determined using fluorescent and colorimetric kits, respectively, from Cayman Chemical Co (Ann Arbor, MI). Liver triglyceride concentrations were quantified following organic extraction [24] with a colorimetric kit (Cayman Chemical).
Tests of Statistical Significance:
Data are presented as mean + SEM (n = 9) except serum lipids (n = 5). Statistical analysis was performed by one-way ANOVA followed by Fisher’s LSD as the post-hoc test when comparing more than two groups. Student’s t-tests were used when comparing two groups. Statistical analysis was performed using Graphpad Prism version 6 (La Jolla, CA USA). A p-value ≤ 0.05 was considered statistically significant.
Results:
High-fat diet increases body and white adipose tissue weight:
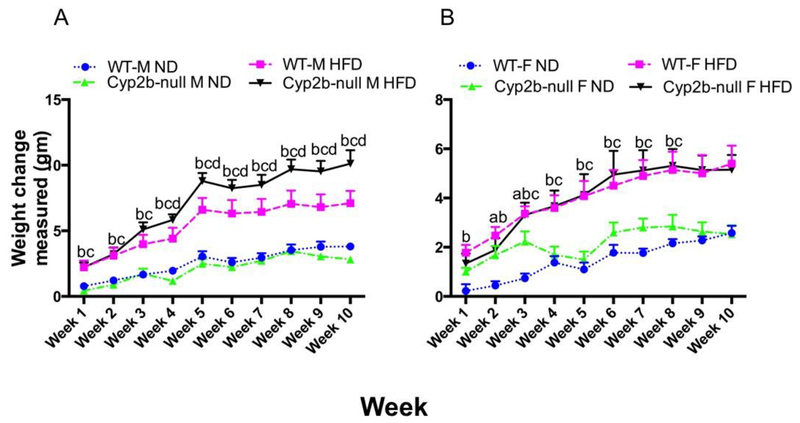
HFD-fed Cyp2b-null male mice weighed significantly more than HFD-fed WT male mice (Fig. 1A). The weight of HFD-fed Cyp2b-null male mice was significantly greater than HFD-fed WT mice after only 4-weeks and these mice stayed heavier for the rest of the study period (Fig. 1A). Much of this weight gain is from increased white adipose tissue, which increased 55% more in HFD-fed Cyp2b-null mice than HFD-fed WT mice (Table 1A). Cyp2b-null mice had similar caloric intake compared to their WT counterparts when examined by diet (HFD or ND) (SM-3). Therefore, alterations in dietary intake do not explain increased weight gain in Cyp2b-null mice.
Fig. 1: Changes in body weight during the 10-weeks of dietary treatments.
Body weight of A) male and B) female, WT, and Cyp2b-null mice were monitored during the 10-week feeding study. Male but not female Cyp2b-null mice show increased weight during HFD treatments. Data are represented as mean ± SEM. Statistical significance was determined by one-way ANOVA followed by Fisher’s LSD as the post-hoc test (n= 8-9). An ‘a’ indicates WT-ND are different than Cyp2b-null-ND, ‘b’ indicates WT-ND are different than WT-HFD, ‘c’ indicates Cyp2b-null-ND are different than Cyp2b-null-HFD, ‘d’ indicates WT-HFD are different than Cyp2b-null-HFD.
Table 1:
Organ weights determined in WT and Cyp2b9/10/13-null mice after 10-weeks of dietary treatment in male (A) and female (B) mice.
| A | |||||
|---|---|---|---|---|---|
| Mouse model | Body weight | WAT | Liver | Kidney | Brain |
| WT-NDM | 26.48±0.57 | 0.51±0.06 | 1.23±0.03 | 0.34±0.01 | 0.48±0.03 |
| WT-HFD M | 30.43±0.92b** | 1.96±0.22b*** | 1.06±0.04b* | 0.37±0.01 | 0.44±0.00 |
| Cyp2b-null ND M | 25.75±0.39 | 0.69±0.06 | 1.08±0.03a* | 0.31±0.01 | 0.46±0.01 |
| Cyp2b-null HFD M | 34.12±0.88c***d* | 3.04±0.19c***d*** | 1.08±0.04 | 0.37±0.02c** | 0.43±0.01 |
| B | |||||
| Mouse model | Body weight | WAT | Liver | Kidney | Brain |
| WT-NDF | 20.47±0.44 | 0.36±0.06 | 0.87±0.03 | 0.26±0.01 | 0.44±0.00 |
| WT-HFD F | 23.17±0.87b* | 0.95±0.15b* | 0.84±0.03 | 0.26±0.01 | 0.45±0.00 |
| Cyp2b-null ND F | 20.29±0.22 | 0.33±0.03 | 0.83±0.02 | 0.25±0.01 | 0.45±0.00 |
| Cyp2b-null HFD F | 24.15±0.93c** | 1.22±0.24c** | 0.87±0.04 | 0.28±0.01c | 0.45±0.01 |
Data represented as mean (g) +/− SEM (n = 8/9). Statistical significance determined by one-way ANOVA followed by Fisher’s LSD as the post-hoc test.
indicates WT ND different than Cyp2b9/10/13-null ND
indicates WT ND different than WT HFD
indicates Cyp2b9/10/13-null ND different than Cyp2b9/10/13-null HFD
indicates WT HFD different than Cyp2b9/10/13-null HFD,
No asterisk indicates a p-value ≤ 0.05,
indicates a p-value ≤ 0.01,
indicates a p-value ≤ 0.0001 and
indicates a p-value ≤ 0.00001.
Cyp2b-null and WT female mice fed a HFD gained significantly more weight and had higher white adipose tissue mass compared to mice fed a ND. However, the Cyp2b-null genotype had no significant effect on body weight gain or WAT mass in females (Fig. 1B; Table 1B). Overall, the loss of Cyp2b9, Cyp2b10, and Cyp2b13 led to diet-induced obesity in male, but not female mice.
High-fat diet decreases glucose tolerance:
Fasting plasma glucose levels were determined during weeks 5 and 8. ND-fed Cyp2b-null male mice show a 20% increase in fasting glucose concentrations compared to ND-fed WT mice on both weeks 5 and 8 (SM-4). HFD did not significantly increase fasting glucose with the exception of Cyp2b-null females fed a HFD compared to Cyp2b-null females fed a ND on week 8.
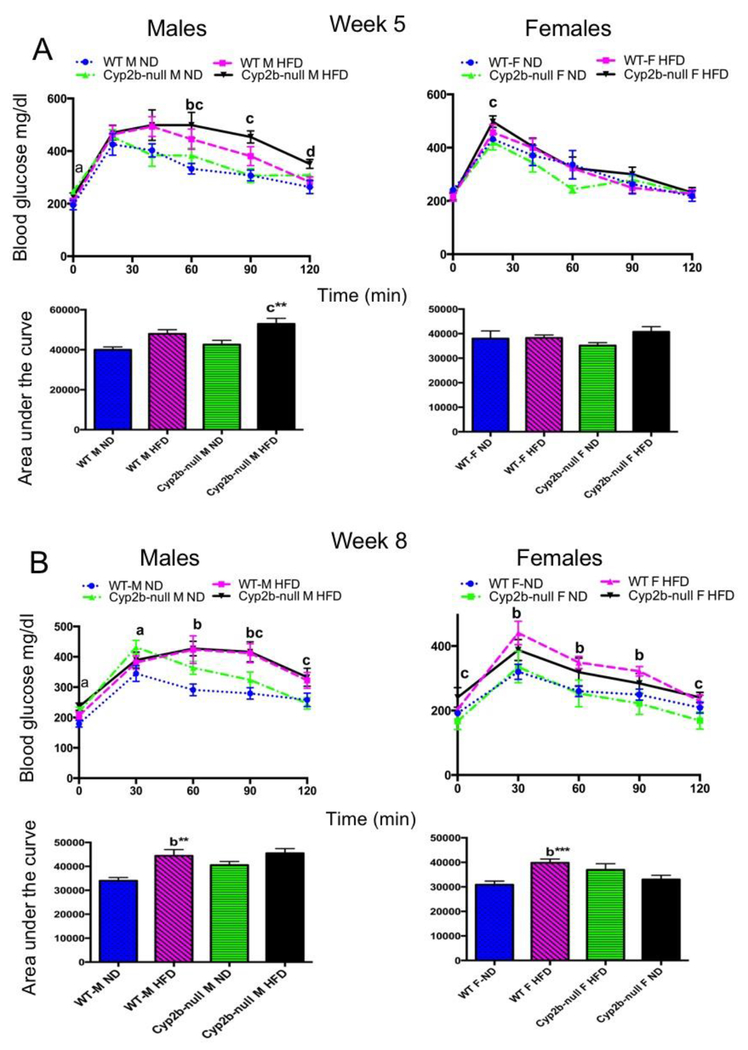
Glucose tolerance tests (GTT) were performed to determine if Cyp2b-null mice are deficient in their ability to respond to a glucose challenge as a biomarker of metabolic disease. Cyp2b-null mice showed some minor differences in serum glucose clearance time; however, most were due to the HFD and not genotype (Fig. 2). We also performed insulin tolerance tests on ND-fed WT and Cyp2b-null male mice due to the minor differences in the blood glucose levels on week 8 at 30-minute intervals, but there were no significant differences in insulin sensitivity (SM-5).
Fig. 2: Glucose tolerance tests do not show significant differences between WT and Cyp2b-null mice:
Glucose tolerance tests were performed on weeks 5 (A) and 8 (B) as described in the Materials and Methods. Results are shown for males and females over the time course of the assay and as area under the curve (AUC). Data are presented as mean ± SEM. Statistical significance was determined by one-way ANOVA followed by Fisher’s LSD as the post-hoc test (n= 8-9). ** Indicates a p-value ≤ 0.01. An ‘a’ indicates WT-ND are different than Cyp2b-null-ND, ‘b’ indicates WT-ND are different than WT-HFD, ‘c’ indicates Cyp2b-null-ND are different than Cyp2b-null-HFD, ‘d’ indicates WT-HFD are different than Cyp2b-null-HFD.
Increased hepatic triglycerides in Cyp2b-null mice:
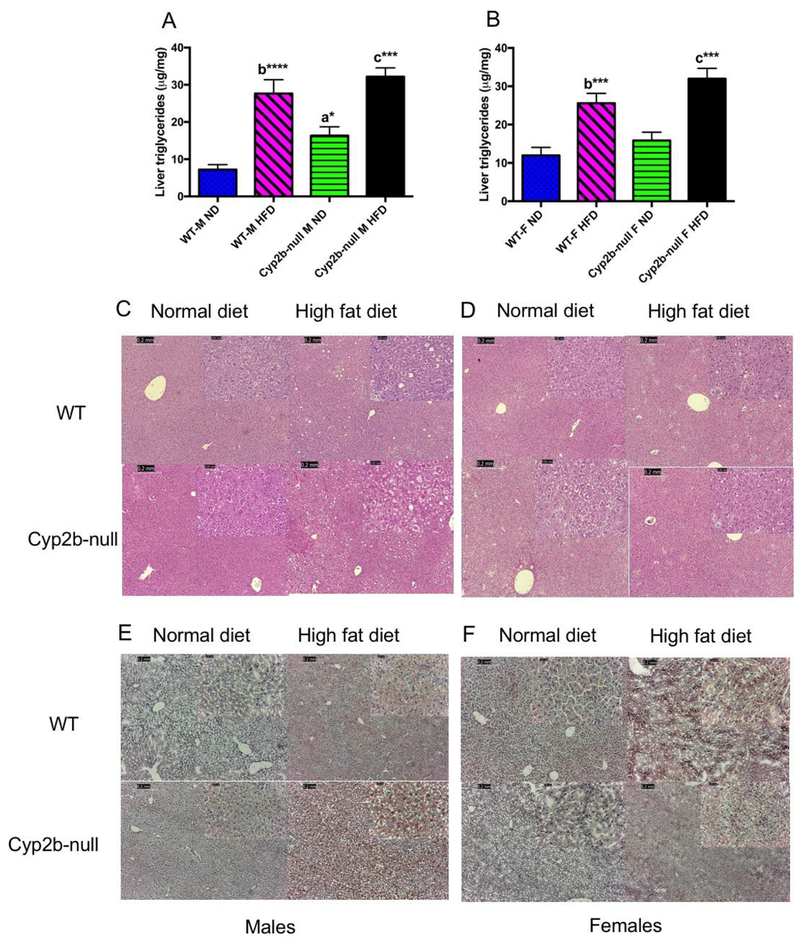
Total liver triglycerides increased 2.3X in the ND-fed Cyp2b-null male mice compared to ND-fed WT males (Fig. 3A). HFD exacerbated liver triglyceride concentrations; however, there was no significant difference between HFD-fed Cyp2b-null male mice and HFD-fed WT male mice (Fig. 3A). HFD-fed female mice showed significant increases in liver triglycerides compared to ND-fed mice; however, genotype did not significantly alter liver triglycerides in females (Fig. 3B). Despite increased liver triglycerides in both HFD-fed groups and ND-fed Cyp2b-null males, liver weight was decreased in all of these groups compared to ND-fed WT mice for unknown reasons (Table 1A). Histopathological analysis indicated mild morphological changes indicative of fat deposition in male ND-fed Cyp2b-null, HFD-fed WT, and HFD-fed Cyp2b-null mice either by H&E or Oil Red O. H&E staining showed mild vacuolization in some HFD-fed male mice of both genotypes with slightly greater vacuolization in some HFD-fed Cyp2b-null mice than HFD-fed WT mice, but no significant changes within the female mice (Fig. 3C–D). Oil Red O staining was absent or weak in male and female ND treatments with minimal staining in ND-fed Cyp2b-null males, which corroborates the total liver triglyceride data. HFD-fed Cyp2b-null male mice showed mildly greater Oil Red O staining than HFD-fed WT mice (Fig. 3E). Female mice only showed mild staining following a HFD with no difference between genotypes (Fig. 3F). Overall, the data suggests that liver triglycerides are slightly higher in Cyp2b-null male mice than correspondingly treated WT male mice.
Fig. 3: Liver triglyceride concentrations are significantly increased in Cyp2b-null mice.
Liver triglycerides were extracted and determined as described in the Materials and Methods using commercial kits. Data are presented as mean ± SEM for males (A) and females (B). Statistical significance was determined by one-way ANOVA followed by Fisher’s LSD as the post-hoc test (n= 8-9). * indicates a p-value ≤0.05, *** indicates p-value ≤0.001 and **** indicates p-value ≤ 0.0001. An ‘a’ indicates WT-ND are different than Cyp2b-null-ND, ‘b’ indicates WT-ND are different than WT-HFD, ‘c’ indicates Cyp2b-null-ND are different than Cyp2b-null-HFD, ‘d’ indicates WT-HFD different than Cyp2b-null-HFD. H&E staining was performed in male (C) and female (D) mice with increases in vacuolization in HFD-fed males. Oil Red O staining was also performed in male (E) and female (F) mice and indicate weak liver lipid staining in all ND treatments and greater triglyceride staining in HFD-fed Cyp2b-null males relative to their HFD-fed WT counterparts.
Serum lipids were perturbed in Cyp2b-null mice:
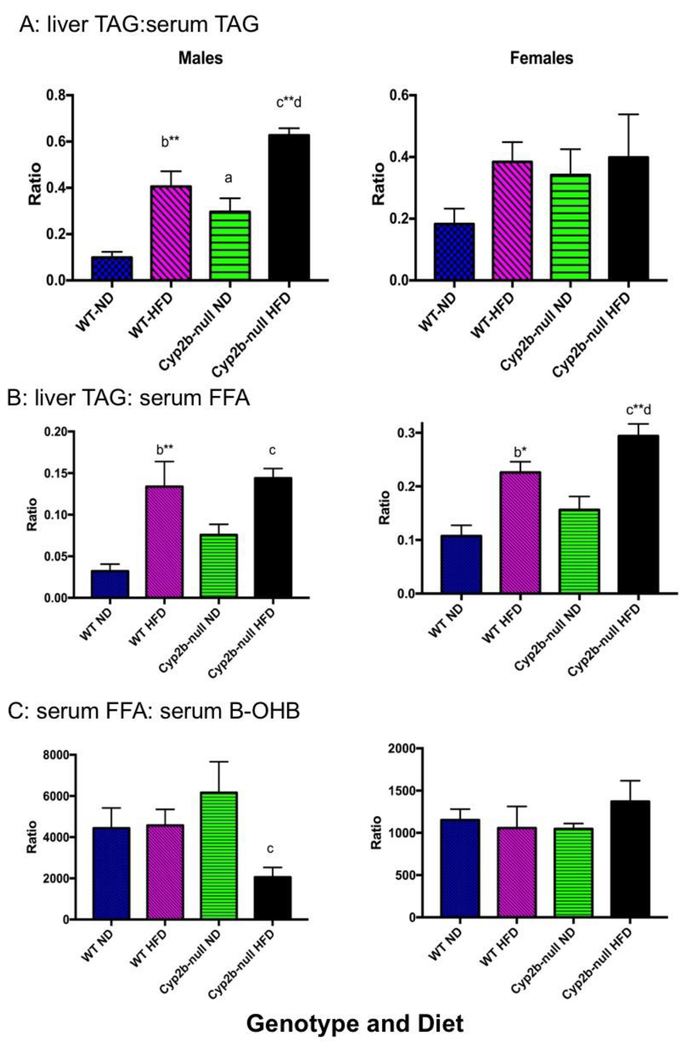
Serum triglyceride levels were significantly decreased in Cyp2b-null male mice compared to WT mice following ND (28%) or HFD (25%) treatments (Table 2), in contrast to increased liver triglyceride concentrations (Fig. 3). The ratio of liver:serum triglycerides was 3X greater in ND-fed Cyp2b-null males than ND-fed WT males, 4X greater in HFD-fed WT males than ND-fed WT males, and 1.54X greater in HFD-fed Cyp2b-null males than HFD-fed WT males (Fig. 4), demonstrating that a Cyp2b-null genotype or a HFD increased the retention of liver triglycerides, and the Cyp2b-null genotype enhanced liver triglyceride retention induced by a HFD. This indicates either decreased metabolism and distribution of hepatic lipids or increased uptake of lipids into the liver.
Table 2:
Serum lipid levels in Cyp2b9/10/13-null compared to WT mice treated with normal diet (ND) or high fat diet (HFD) in male (A) and female (B) mice.
| A | ||||
|---|---|---|---|---|
| Lipid panel | WT ND M | WT HFD M | Cyp2b-null ND M | Cyp2b-null HFD M |
| ALT | 17.00 ± 1.48 | 15.00 ± 0.55 | 17.80 ± 0.97 | 14.00 ± 1.00 |
| β-OHB | 0.188 ± 0.015 | 0.162 ± 0.022 | 0.120 ± 0.015 | 0.365 ± 0.087c***d** |
| Cholesterol | 103.80 ± 1.63 | 170.4 ± 7.00b** | 118.60 ± 6.27a* | 182.75 ± 3.61c**d |
| Fatty acids | 833.7 ± 97.3 | 740.2 ± 93.0 | 739.3 ± 36.6 | 751.4 ± 43.2 |
| HDL | 97.00 ± 2.21 | 150.76 ± 6.24b** | 114.02 ± 4.45a* | 168.60 ± 4.94c**d* |
| LDL | 10.98 ± 0.58 | 26.34 ± 2.46b* | 16.34 ± 1.09 | 30.20 +/− 1.57c |
| Triglycerides | 80.40 ± 7.88 | 69.60 ± 3.96b | 58.00 ± 5.93a** | 52.25 ± 3.15d* |
| VLDL | 16.04 ± 1.59 | 13.94 ± 0.80 | 11.56 ± 1.19a | 10.45 ± 0.61 |
| B | ||||
| Lipid panel | WT ND F | WT HFD F | Cyp2b-null ND F | Cyp2b-null ND F |
| ALT | 20.20 ± 1.69 | 37.20 ± 8.49 | 17.80 ± 0.80 | 17.80 ± 0.80 |
| β-OHB | 0.325 ± 0.076 | 0.360 ± 0.061 | 0.328 ± 0.098 | 0.268 ± 0.046 |
| Cholesterol | 76.40 ± 3.70 | 98.00 ± 12.63 | 77.40 ± 1.94 | 77.40 ± 1.94 |
| Fatty acids | 374.0 ± 8.19 | 380.4 ± 27.2 | 343.9 ± 20.5 | 366.5 ± 30.6 |
| HDL | 72.48 ± 4.09 | 54.18 ± 22.92 | 77.36 ± 2.57 | 77.36 ± 2.57 |
| LDL | 8.68 ± 1.82 | 8.58 ± 3.24 | 10.20 ± 1.46 | 10.20 ±1.46 |
| Triglycerides | 80.80 ± 17.74 | 78.40 ± 18.58 | 57.40 ± 12.70 | 57.40 ± 12.70 |
| VLDL | 16.16 ± 3.53 | 11.46 ± 2.52 | 15.68 ± 3.75 | 20.48 ± 3.55 |
Data represented as mean +/− SEM (n = 5). All units expressed as mg/dl except β-OHB (mM), fatty acids (μM) and ALT (U/L). Statistical significance was determined by one-way ANOVA followed by Fisher’s LSD as the post-hoc test.
indicates WT ND different than Cyp2b9/10/13-null ND.
indicates WT ND different than WT HFD
indicates Cyp2b9/10/13-null ND different than Cyp2b9/10/13-null HFD.
indicates WT HFD different than Cyp2b9/10/13-null HFD,
No asterisk indicates a p-value ≤ 0.05 and
indicates a p-value ≤ 0.01,
indicates a p-value of ≤ 0.0001.
Fig. 4: Liver triglyceride:serum triglyceride (A), liver triglyceride:serum free fatty acid (B), and serum free fatty acidrserum b-hydroxybutyrate (C) ratios in ND and HFD-fed mice.
Data are presented as mean ± SEM. Statistical significance was determined by one-way ANOVA followed by Fisher’s LSD as the post-hoc test (n= 8-9). * indicates a p-value ≤0.05, *** indicates p-value ≤0.001 and **** indicates p-value ≤ 0.0001. An ‘a’ indicates WT-ND are different than Cyp2b-null-ND, ‘b’ indicates WT-ND are different than WT-HFD, ‘c’ indicates Cyp2b-null-ND are different than Cyp2b-null-HFD, ‘d’ indicates WT-HFD different than Cyp2b-null-HFD.
Female mice showed a similar decrease (29%) to males in serum triglycerides in ND-fed Cyp2b-null mice compared to ND-fed WT mice (Table 2); however, this was not statistically significant. In contrast to males, HFD-fed Cyp2b-null female mice showed a 31% increase in serum triglycerides compared to HFD-fed WT mice. Changes in liver and serum triglycerides were proportional in females, which is different than males, and this was further demonstrated by the liver:serum triglyceride ratios that were not different between WT and Cyp2b-null female mice (Fig. 4A).
In contrast to serum triglycerides, free fatty acids (FFA) were not perturbed significantly across groups (Table 2). This relative stability ensured that serum TAG:serum FFA ratios were not altered across groups (data not shown). However, the relative stability of FFA provides another example of the differences in liver triglyceride to serum lipid ratios across treatment groups as both males and females show large increases in liver TAG:serum FFA ratio with females showing greater relative increases in HFD-fed Cyp2b-null mice (Fig. 4B).
Serum cholesterol and HDL increased 14% and 18%, respectively, in ND-fed Cyp2b-null male mice compared to ND-fed WT male mice. Serum cholesterol and HDL increased 8% and 12%, respectively, in HFD-fed Cyp2b-null male mice compared to HFD-fed WT male mice (Table 2A); thus Cyp2b-null male mice consistently present higher cholesterol concentrations. For example, HFD-fed Cyp2b-null male mice had 2X higher cholesterol than HFD-fed male WT mice, indicating a significant role Cyp2b in serum cholesterol concentrations. Despite lower weight gain and white adipose tissue mass, females showed greater changes in serum cholesterol and HDL concentrations. Most changes were associated with a combination of diet and the loss of Cyp2b (Table 2B). There were no significant changes in LDL or VLDL serum levels; only in HDL serum concentrations. In addition, ALT, a biomarker of liver damage, was increased only in Cyp2b-null females fed a HFD, indicating that these mice suffered some liver damage such as apoptosis or necrosis in addition to the abnormally high serum cholesterol. It is interesting that the HFD-fed Cyp2b-null female mice are not showing greater obesity than HFD-fed WT mice, but several other parameters are perturbed.
β-hydroxybutyate (B-OHB) is generated by the liver from fatty acids during prolonged exercise or starvation to meet energy requirements and compensate for lower carbohydrate levels. B-OHB can be utilized as an energy source by active tissues such as brain and muscles. HFD-fed Cyp2b-null males showed a 2.3-fold increase in B-OHB levels compared to HFD-fed WT males and a 3-fold increase compared to ND-fed Cyp2b-null males (Table 2A). The increase in B-OHB also fueled the decrease in the serum FFA/B-OHB ratio observed in HFD-fed Cyp2b-null male mice as serum fatty acid levels were relatively stable between groups, but ketones were increased by the combination of genotype and a HFD (Fig. 4C). Serum triglyceride/B-OHB ratio followed the same pattern (data not shown).
Serum concentrations of adiponectin and leptin:
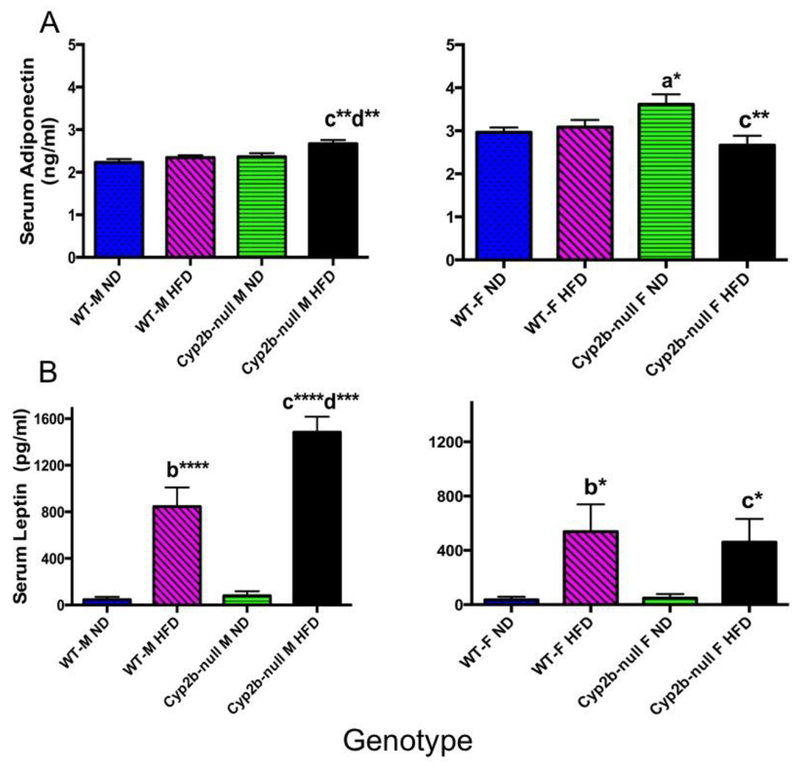
Serum adiponectin concentrations were significantly altered in several groups (Fig. 5). All significant differences in adiponectin serum concentrations were moderate and ranged from 13-25% different compared to controls. Serum leptin concentrations were greatly perturbed by a HFD, and this was exacerabated in HFD-fed Cyp2b-null male mice (Fig. 5), indicating leptin resistance in association with obesity in HFD-fed Cyp2b-null male mice. Leptin resistance indicates a disease state and the potential that the brain thinks the individual is starving [62]. The significant increase in leptin is associated with an increase in B-OHB and could be related to a significant increase in WAT in the HFD-fed Cyp2b-null males that provide fatty acids to fuel ketosis.
Fig. 5: Serum adiponectin and leptin concentrations in WT and Cyp2b-null mice.
Commercial kits were used to determine (A) adiponectin and (B) leptin concentrations in serum samples. Data are presented as mean ± SEM. Statistical significance was determined by oneway ANOVA followed by Fisher’s LSD as the post-hoc test (n= 8-9). * Indicates a p-value ≤ 0.05, ** indicates a p-value ≤0.01, *** indicates a p-value ≤0.001 and **** indicates p-value ≤ 0.0001. An ‘a’ indicates WT-ND are different than Cyp2b-null-ND, ‘b’ indicates WT-ND are different than WT-HFD, ‘c’ indicates Cyp2b-null-ND are different than Cyp2b-null-HFD, ‘d’ indicates WT-HFD are different than Cyp2b-null-HFD.
ND-fed Cyp2b-null mice have similar gene expression profiles compared to HFD-fed WT mice:
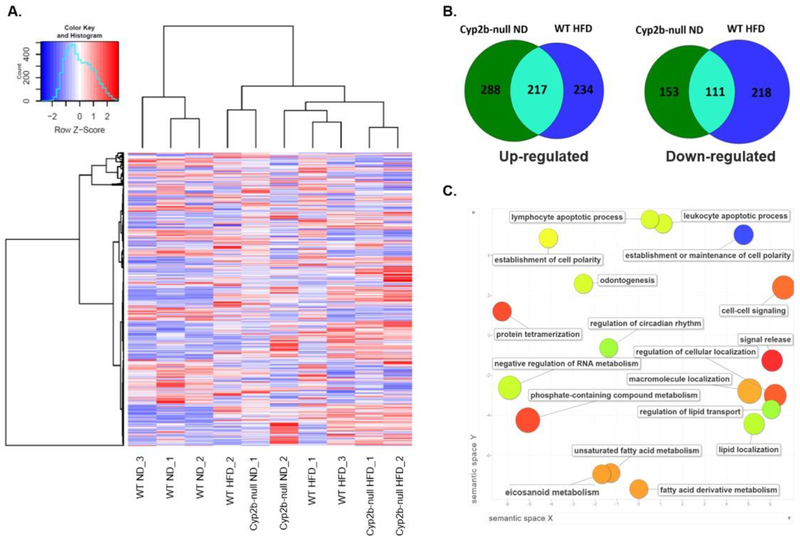
RNAseq was performed on male and female liver samples to compare and test why only male Cyp2b-null mice showed both significant weight gain and significantly increased hepatic lipids (Fig. 1,3). Analysis of global gene expression using hierarchical clustering demonstrates that male samples cluster into two different groups: one cluster consisting of WT-ND mice, and the other cluster consisting of Cyp2b-null ND and WT-HFD mice grouped among each other with Cyp2b-null HFD mice in their own subgroup. The clustering of HFD-fed WT mice and ND-fed Cyp2b-null male mice into the same clade (Fig. 6A) suggests similar changes in gene expression. Venn diagrams (Fig. 6B) confirm that within the liver the Cyp2b-null genotype with no change in diet causes comparable transcriptional effects to that of WT mice fed a HFD. Cyp2b-null ND and WT HFD mice share the same 217 significantly (p < 0.05) up-regulated genes and 111 down-regulated when compared to WT ND mice as the control. These two experimental groups share over 40% of each group’s total number of differentially expressed genes, indicating that the Cyp2b-null genotype causes effects similar to that of a HFD-treatment potentially due to increased liver triglycerides in the ND-fed Cyp2b-null male mice (Fig. 3).
Fig. 6: Cyp2b-null male mice fed a ND display a similar gene expression profile to WT mice fed a HFD.
RNAseq was performed on liver samples from WT and Cyp2b-null mice fed either a ND or HFD. (A) Heat map showing log2-transformed, Z-score scaled RNA-Seq expression of 500 genes with the highest variance between treatment groups. Red and blue color intensity indicate gene up- or down-regulation, respectively. Dendrogram clustering on the x-axis indicates sample similarity, whereas dendrogram clustering on the y-axis groups genes by expression profile across samples. (B) Venn diagrams of shared up- and down-regulated differentially expressed genes (p<0.05) between Cyp2b-null ND-fed and WT HFD-fed mice compared to WT ND mice. (C) GO term enrichment analysis summary using Revigo [58] for significantly up-regulated genes in Cyp2b-null ND mice compared to WT ND mice. The scatterplot contains enriched GO terms from the biological process class that remain after term redundancy is reduced and are displayed in a two-dimensional space where semantically similar GO terms are positioned closer together within the plot. Each circle represents an enriched GO term; the cooler the color of a term, the more significantly (p < 0.05) associated that term is with the group of genes being studied. Circle size indicates the frequency of the GO term in the underlying GO database, i.e. circles of more general terms are larger.
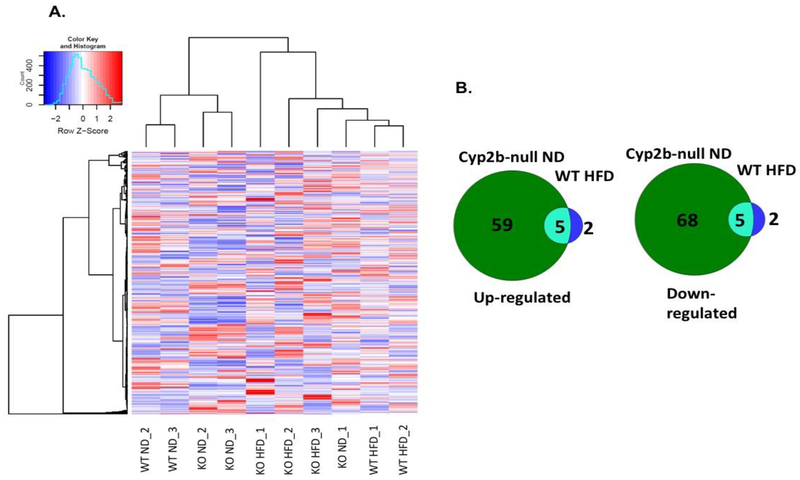
Female samples clustered in a different fashion than males. ND-fed WT and Cyp2b-null mice clustered together and separately from most of the HFD-fed mice (Fig. 7A) instead of ND-fed Cyp2b-null mice clustering with HFD-fed WT mice as seen in male groups (Fig. 6B). There is little gene expression overlap between ND-fed Cyp2b-null and HFD-fed WT female mice (Fig. 7B), in part because there are very few differentially expressed genes in HFD-fed WT female mice compared to ND-fed WT female mice (only 7; SM-6) and a significant drop in differentially expressed genes between ND-fed WT and ND-fed Cyp2b-null female mice compared to males (Fig. 7B, SM6-9). For example, in ND-fed groups, 506 genes were induced in Cyp2b-null male mice, but only 65 in Cyp2b-null female mice when compared to ND-fed WT mice with 15 genes overlapping between males and females. 264 genes were down-regulated in Cyp2b-null male mice, but only 73 were down-regulated in females with 19 overlapping (SM-8,10). Most of the differences in hierarchical clustering are due to diet in females (SM-6). This demonstrates the lack of response to the Cyp2b knockout and HFD in female mice compared to male mice (Fig. 7; SM6-8), similar to our physiological data.
Fig. 7: Cyp2b-null female mice demonstrate relatively fewer gene expression changes.
RNAseq was performed on liver samples from WT and Cyp2b-null mice fed either a ND or HFD. (A) Heat map showing log2-transformed, Z-score scaled RNA-Seq expression of 500 genes with the highest variance between treatment groups. Red and blue color intensity indicate gene up- or down-regulation, respectively. Dendrogram clustering on the x-axis indicates sample similarity, whereas dendrogram clustering on the y-axis groups genes by expression profile across samples. (B) Venn diagrams of shared up- and down-regulated differentially expressed genes (p<0.05) between Cyp2b-null ND-fed and WT HFD-fed mice compared to WT ND mice.
Interestingly, the most highly induced gene by the HFD in WT male mice was Cyp2b9 (SM-6). Previous diet-induced obesity (DIO) studies have also shown Cyp2b9 to be the most highly induced gene [46, 63]. Among the list of shared genes by Cyp2b-null ND and WT HFD male mice, up-regulated genes with the highest logFC in both groups include Cyp2a4 (logFC: Cyp2b-null ND 6.99, WT HFD 1.53), Cyp2b9 (LogFC: Cyp2b-null ND 3.60, WT HFD 6.65), and NADH-ubiquinone oxidoreductase chain 3 (ND3) (logFC: Cyp2b-null ND 3.33, WT HFD 3.67) (SM-7). Cyp2b9 is probably highly induced in Cyp2b-null mice as a compensatory mechanism as the latter half of the Cyp2b9 gene was not eliminated by Crispr/Cas9 and is therefore most likely under control of the CAR promoter found at the front of the Cyp2b gene cluster (SM-1)[52]. A Western blot confirmed that no Cyp2b’s were induced and instead deleted in the Cyp2b-null mice as expected (SM-11). Genes down-regulated in males and females when comparing the various treatments are also available in Supplementary Material (SM 6-9) under a separate tab.
Significantly (p < 0.05) enriched up-regulated gene ontology (GO) terms for male ND-fed Cyp2b-null mice in comparison to ND-fed WT mice were visualized in Revigo (Fig. 6C). Within the biological process class, ND-fed Cyp2b-null mice have enriched terms such as regulation of lipid transport, lipid localization, unsaturated fatty acid metabolism, eicosanoid metabolism, cell-cell signaling, circadian rhythms, and establishment or maintenance of cell polarity. Many of these terms are associated with NAFLD and perturbations in lipid metabolism [46, 64, 65]. Down-regulated GO terms for this group were mainly associated with protein stability and folding (SM-12), possibly indicating endoplasmic reticulum stress via lipotoxicity from intermediate lipid metabolites or perturbations in the endoplasmic reticulum due to the loss of CYPs [66]. There were no significantly enriched GO terms when comparing ND-fed Cyp2b-null and ND-fed WT mice in females (SM-12).
There are not as many differentially expressed genes when comparing HFD-fed Cyp2b-null to HFD-fed WT mice in males or females (SM-9). In males, there are several genes involved in triglyceride accumulation and lipid metabolism, including circadian genes associated with liver lipid metabolism, which is expected given the greater obesity and liver triglycerides in the HFD-fed Cyp2b-null mice than the HFD-fed WT mice. In addition, genes involved in tissue growth and repair and cytoskeletal remodeling are perturbed that when combined with excess serum leptin suggests that HFD-fed Cyp2b-null mice are more susceptible to tissue damage or in the early stages or fibrosis [67–69]. In females, estrogen and thyroid metabolism, steroid hormone biosynthesis, endoplasmic reticulum protein processing, and pancreatic secretion were primarily perturbed, and only a few of these are directly linked to obesity, energy metabolism, and none of the multigene pathways perturbed are linked to fibrosis.
There were 646 KEGG pathway perturbations caused by a lack of Cyp2b (comparing WT-ND to Cyp2b-null-ND) in male mice, but only 190 in female mice (Table 3). KEGG pathway analysis for both up- and down-regulated differentially expressed genes in ND-fed Cyp2b-null ND compared to ND-fed WT mice (log2FC > 1.0) revealed 42 metabolic genes with altered expression in males, but only 14 in females. Pathways perturbed include PI3K-Akt signaling, protein processing in the endoplasmic reticulum, retinol metabolism, arachidonic acid metabolism, insulin resistance, glycerolipid metabolism, and fatty acid elongation within the endoplasmic reticulum (SM-13; Table 3). Many of these pathways are also crucial in fatty acid metabolism and the development of NAFLD [64, 70]. The greater transcriptional (numbers of genes and pathways) effects perturbed in ND-fed Cyp2b-null and HFD-fed WT male mice compared to female mice provides a clue as to why males are more susceptible, and females more resistant, to obesity (Table 4,5).
Table 3:
Comparison of the number of up- and down-regulated genes within specific KEGG pathways of male and female Cyp2b-null ND-fed mice.
| KEGG Pathway | Male Cyp2b-null ND vs WT ND | Female Cyp2b-null ND vs WT ND |
|---|---|---|
| PI3K-Akt signaling | 11 | 5 |
| Protein processing in endoplasmic reticulum | 10 | 4 |
| Retinol metabolism | 7 | 2 |
| MAPK signaling | 7 | 3 |
| Glycerophospholipid metabolism | 7 | 2 |
| Apoptosis | 6 | 4 |
| Drug metabolism | 6 | 1 |
| Thyroid hormone signaling | 6 | 1 |
| Insulin resistance/signaling | 5 | 4 |
| PUFA metabolism | 5 | 1 |
| Circadian rhythm | 4 | 3 |
| NAFLD | 4 | 1 |
| Calcium signaling | 3 | 1 |
| FoxO signaling | 2 | 3 |
| Cholesterol metabolism | 1 | 3 |
| Sphingolipid metabolism | 1 | 2 |
| Total Perturbations | 646 | 190 |
Table 4:
Comparison of the number of up- and down-regulated genes within specific KEGG pathways.
| KEGG Pathway | Cyp2b-null ND vs WT ND | WT HFD vs WT ND | Cyp2b-null HFD vs WT HFD |
|---|---|---|---|
| PI3K-Akt signaling | 11 | 12 | 2 |
| Protein processing in endoplasmic reticulum | 10 | 1 | 0 |
| Retinol metabolism | 7 | 13 | 1 |
| MAPK signaling | 7 | 12 | 1 |
| Glycerophospholipid metabolism | 7 | 1 | 1 |
| Hepatocellular carcinoma | 6 | 11 | 1 |
| Drug metabolism | 6 | 9 | 0 |
| Thyroid hormone signaling | 6 | 5 | 1 |
| Apoptosis | 6 | 1 | 0 |
| PUFA metabolism | 5 | 10 | 3 |
| Insulin signaling/resistance | 5 | 3 | 2 |
| Circadian rhythm | 4 | 2 | 3 |
| NAFLD | 4 | 2 | 0 |
| Phospholipase D signaling | 4 | 0 | 0 |
| Glutathione metabolism | 2 | 12 | 0 |
| Cholesterol metabolism | 1 | 4 | 2 |
| Total Perturbations | 646 | 620 | 75 |
Table 5:
Comparison of the number of up- and down-regulated genes in female mice within specific KEGG pathways.
| KEGG Pathway | Cyp2b-null ND vs WT ND | WT HFD vs WT ND | Cyp2b-null HFD vs WT HFD |
|---|---|---|---|
| PI3K-Akt signaling | 5 | 0 | 0 |
| Protein processing in endoplasmic reticulum | 4 | 0 | 6 |
| Insulin resistance/signaling | 4 | 0 | 0 |
| Apoptosis | 4 | 0 | 0 |
| FoxO signaling | 3 | 0 | 0 |
| Circadian rhythm | 3 | 0 | 0 |
| Cholesterol metabolism | 3 | 1 | 0 |
| MAPK signaling | 3 | 0 | 1 |
| Sphingolipid metabolism | 2 | 0 | 0 |
| Glycerophospholipid metabolism | 2 | 0 | 0 |
| Retinol metabolism | 2 | 0 | 1 |
| Calcium signaling | 1 | 0 | 2 |
| Drug metabolism | 1 | 1 | 0 |
| NAFLD | 1 | 0 | 0 |
| Total Perturbations | 190 | 13 | 49 |
Discussion
Male, but not female Cyp2b-null mice are diet-induced obese. Male Cyp2b-null mice fed a HFD gained 15% more body weight than male WT mice fed a HFD primarily due to a 55% increase in WAT mass. Diet-induced obesity and genotype-specific diet-induced obesity are more difficult to measure in B6 female mice than male mice because the female mice are much less susceptible to obesity and diabetes than male mice (or human females) [71, 72]. Previous observations indicated that Cyp2b-KD mice developed age-onset obesity in males with a significant lesser effect in females. However, this work was an observation from aging mice (age-onset obesity); not a tightly planned experiment examining diet-induced obesity [48]. Furthermore, the loss of Cyp2b, through either RNAi or Crispr/Cas9 [47, 52], causes obesity independent of strain as it occurs in FVB (Cyp2b-KD) and B6 (Cyp2b-null) mice. Overall, the loss of the primarily hepatic Cyp2b members increases WAT that in turn leads to obesity in males and suggests that inhibition of these enzymes by environmental chemicals or pharmaceuticals may have similar consequences.
Liver triglyceride concentrations were significantly increased in ND-fed Cyp2b-null mice compared to their WT male counterparts. Increases in liver triglycerides were accompanied by decreases in serum triglycerides in ND- and HFD-fed Cyp2b-null mice. In turn, both liver:serum triglyceride ratios and liver TAG:serum FFA ratios were greatly perturbed, and this includes comparisons between HFD-fed WT and HFD-fed Cyp2b-null mice. which indicates a crucial role for Cyp2b in liver triglyceride levels or distribution. Overall, triglyceride distribution was greatly perturbed in ND-fed Cyp2b-null mice, HFD-fed WT mice, and even more so in HFD-fed Cyp2b-null mice.
It is noteworthy that ND-fed Cyp2b-null male mice retain triglycerides in their liver and display a similar gene expression profile to the HFD-fed WT male mice, indicating similar early stage liver disease and compensatory changes due to increased lipids. A survey of the key biomarker genes altered in ND-fed Cyp2b-null mice such as Cyps and transporters suggests activation of transcription factors such as Foxa2, LXR, FXRb, and potentially Rev-Erb, RAR, ROR and CAR/PXR in males [36, 38, 43, 73–75]. All of these transcription factors response to changes in lipid distribution or energy allocation. Hierarchical clustering, direct comparisons by Venn diagrams, shared GO terms, and similarly perturbed KEGG pathways all present evidence that ND-fed Cyp2b-null male mice have similar gene expression changes to HFD-fed WT mice. For example, both HFD-fed WT and ND-fed Cyp2b-null male mice show enriched terms for lipid transport, lipid localization, unsaturated fatty acid metabolism, and eicosanoid metabolism, clearly indicating perturbations in lipid utilization and metabolism. However, it should be noted that there are also some differences based on KEGG pathway analysis. For example, ND-fed Cyp2b-null male mice show perturbations in glycerolipid metabolism, apoptosis, and endoplasmic reticulum protein processing that are not manifested in WT-HFD mice; whereas WT-HFD mice show perturbed hepatocellular carcinoma and glutathione metabolism that is not manifested in Cyp2b-null-ND mice. Cyp2b induction by CAR is partially responsible for subsequent CAR-initiated Nrf2 activation [76], and therefore Cyp2b-null mice may not activate protective mechanisms from oxidative stress such as glutathione metabolism as well as WT mice. It is also interesting that the apoptotic and hepatocellular carcinoma responses are reversed, suggesting Cyp2b-null mice may be less susceptible to cancer even with the increase in liver triglycerides. Overall, the similarities in differential gene expression outweigh the differences, and provide data associating the loss of hepatic Cyp2b members to liver triglycerides and obesity in males.
This was not the case in females. Significantly fewer genes were altered in females, especially following a HFD and in turn ND-fed Cyp2b-null females did not cluster with HFD-fed WT females. Female mice are generally considered partially resistant to obesity except post-menopausally [17, 71]. This resistance was also observed during this study and demonstrates that Cyp2b enzymes such as Cyp2b9 and Cyp2b13 ,expressed at much greater levels in females [51, 52]}[39], are not the reason for the sexual dimorphism in obesity. It is still possible that some of the transcription factors that regulate Cyp2b and are predominantly expressed in females such as CAR, PXR, and FoxA2 [17, 38, 39, 77] are in part responsible for the murine female resistance to obesity.
HFD-fed Cyp2b-null females were the only mice with increased serum triglycerides and these mice also present with higher ALT levels, suggesting that the combination of a HFD and Cyp2b-knockout caused some liver damage in females. In contrast to males, Cyp2b-null females did not show significant perturbations in liver:serum ratios probably because neither serum nor liver triglycerides were perturbed as much in females, but also because both serum and liver triglycerides went up in HFD-fed Cyp2b-null female mice. Ratios are often used to increase the sensitivity of data from two physiological systems (liver:serum; TAG:HDL) or sexually dimorphic comparison (steroid metabolites)[52, 78]. Therefore, we also examined liver triglyceride:serum FFA ratios, and found they varied significantly in HFD-fed Cyp2b-null female mice.
Serum cholesterol and HDL were also significantly increased in Cyp2b-null male mice compared to WT male mice. Increased serum cholesterol coupled with decreased serum triglycerides was observed previously in diet induced obesity studies performed in B6 mice [79], and recent studies indicate that high cholesterol is associated with progressive NAFLD and a potential marker for NASH [80, 81]. ALT is also associated with HDL, NAFLD and metabolic syndrome in men, but not women [82]. Overall, multiple serum and liver lipid parameters that are associated with NAFLD and NASH were perturbed [83, 84], indicating that Cyp2b-null male mice are progressing towards hepatic steatosis and NASH. However, the lack of changes in LDL and VLDL suggest that accompanying cardiovascular disease may not be a chronic issue [78, 85]. Histopathology did not show any pathological changes indicative of significant liver injury, although liver lipids were increased by a HFD and exacerbated by Cyp2b-knockout as measured by Oil Red O. However, our research was performed for only 10-weeks and recent work indicates B6 mice are relatively resistant to fatty liver disease and take upwards of 22-weeks to develop NAFLD [86]. Therefore this study was not performed for long enough to fully assess progressive NAFLD or NASH, but primarily obesity and early stage steatosis. Long-term (22-weeks) or better yet, methionine-choline deficient dietary studies are needed to determine if the Cy2b-null mice may be susceptible to NASH.
HFD consumption increases leptin levels and leads to the development of leptin resistance ultimately perturbing satiety and energy expenditure regulation [87]. Leptin was greatly increased by a HFD and more so in Cyp2b-null male mice fed a HFD, indicating leptin resistance and perturbed energy utilization in these mice, especially the HFD-fed Cyp2b-null mice (Fig. 5). Serum adiponectin and B-OHB, a biomarker of ketogenesis, were also significantly increased in HFD-fed Cyp2b-null male mice, but not females (Fig. 5). B-OHB is increased from the breakdown of fatty acids in response to starvation and consistent with higher WAT and leptin levels [62]. Taken together, WAT, liver triglycerides, serum cholesterol, leptin, and B-OHB levels were all increased in Cyp2b-null mice, primarily in males, indicating an unhealthy lipid state with increased ketosis and β-oxidation sometimes regardless of dietary treatment. This combined with perturbations in transcription associated with lipid metabolism, distribution and homeostasis demonstrates a role for Cyp2b members in lipid metabolism and obesity.
Several unsaturated fatty acids are metabolized by Cyp2b members including arachidonic acid [34, 88], anandamide [89], and potentially linoleic acid [20, 29]. Some data indicates high affinity for fatty acids [34]; however, most Cyp2b-mediated metabolism likely occurs under high fat diet conditions or directly after a meal high in PUFA [20, 89]. It is interesting to consider that a Cyp2b-mediated product of PUFA metabolism such as an epoxyecosatrienoic acid produced from AA or anandamide, may perform signaling functions that mediate lipid uptake, distribution, or metabolism in the liver, WAT, or other tissues following a diet high in PUFAs.
In summary, the data presented indicates that the hepatic Cyp2b genes are important in fatty acid metabolism. Cyp2b-null mice are diet-induced obese and show greater susceptibility to obesity than WT mice. HFD-fed Cyp2b-null male mice present increased WAT mass, liver triglycerides, serum cholesterol, leptin, and B-OHB. In addition, the increased accumulation of liver triglycerides and decreased serum triglycerides in ND- and HFD-fed Cyp2b-null male mice suggests difficulty in allocating and utilizing fatty acids. The ND-fed Cyp2b-null male mice also showed a similar gene expression profile to the HFD-fed WT male mice as demonstrated by RNAseq, indicating increased lipids and perturbed lipid signaling regardless of diet in the Cyp2b-null mice. Taken together, this data indicates a role of Cyp2b in fatty acid metabolism and obesity, and therefore, it is certainly possible that chemical inhibition of Cyp2b members by dietary fats, pesticides, plasticizers, and pharmaceuticals could elicit similar effects on lipid metabolism and elicit obesity.
Supplementary Material
Acknowledgements:
This work was supported by NIEHS grant R15ES017321. Transcriptomoic support was provided by Dr. Rooksana E. Noorai through the Clemson University Genomics and Bioinformatics Facility, NIH COBRE grant P20GM109094.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare that they have no conflicts of interest with the contents of this article
References:
- [1].Hales CM, Carroll MD, Fryar CD, Ogden CL. Prevalence of obesity among adults and youth: United States, 2015-2016. NCHS data brief 2017;288. [PubMed] [Google Scholar]
- [2].Romieu I, Dossus L, Barquera S, Blottière H,M, Franks PW, Gunter M, et al. Energy balance and obesity: what are the main drivers? Cancer Causes Control. 2017;28:247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Choquet H, Meyre D. Genetics of Obesity: What have we Learned? Curr Genomics. 2011;12:169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Grun F, Blumberg B. Minireview: the case for obesogens. Mol Endocrinol. 2009;23:1127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hatch EE, Nelson JW, Stahlhut RW, Webster TF. Association of endocrine disruptors and obesity: Perspectives from epidemiologic studies. Int J Androl. 2010;33:324–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sharp D Environmental toxins, a potential risk factor for diabetes among Canadian Aboriginals. Int J Circumpolar Health. 2009;68:316–26. [DOI] [PubMed] [Google Scholar]
- [7].Wahlang B, Falkner KC, Gregory B, Ansert D, Young D, Conklin DJ, et al. Polychlorinated biphenyl 153 is a diet-dependent obesogen that worsens nonalcoholic fatty liver disease in male C57BL6/J mice. J Nutr Biochem. 2013;24:1587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Riu A, Grimaldi M, le Maire A, Bey G, Phillips K, Boulahtouf A, et al. Peroxysome proliferator-activated receptor γ is a target for halogenated analogues of bisphenol-A. Environ Health Perspect. 2011;119:1227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].LeBlanc GA, Norris DO, Kloas W, Kullman SW, Baldwin WS, Greally JM. Detailed Review Paper on the State of the Science on Novel In Vitro and In Vivo Screening and Testing Methods and Endpoints for Evaluating Endocdrine Disruptors Series on Testing & Assessment: No 178. Paris: Organisation for Economic Co-operation and Development; 2012. p. 213. [Google Scholar]
- [10].Marmugi A, Ducheix S, Lasserre F, Polizzi A, Paris A, Priymenko N, et al. Low doses of bisphenol A induce gene expression related to lipid synthesis and trigger triglyceride accumulation in adult mouse liver. Hepatology. 2012;55:395–407. [DOI] [PubMed] [Google Scholar]
- [11].Chamorro-García R, Sahu M, Abbey RJ, Laude J, Pham N, B B. Transgenerational inheritance of increased fat depot size, stem cell reprogramming, and hepatic steatosis elicited by prenatal exposure to the obesogen tributyltin in mice. Environ Health Perspect. 2013;121:359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].le Maire A, Grimaldi M, Roecklin D, Dagnino S, Vivat-Hannah V, Balaguer P, et al. Activation of RXR-PPAR heterodimers by organotin environmental endocrine disruptors. EMBO Reports. 2009;10:367–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Angrish MM, Mets BD, Jones AD, Zacharewski TR. Dietary fat is a lipid source in 2,3,7,8-tetrachlorodibenzo-ρ-dioxin (TCDD)-elicited hepatic steatosis in C57BL/6 mice. Toxicol Sci. 2012;128:377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee JH, Wada T, Febbraio M, He J, Matsubara T, Lee MJ, et al. A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology 2010;139:653–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chang JW, Chen HL, Su HJ, Lee CC. Abdominal Obesity and Insulin Resistance in People Exposed to Moderate-to-High Levels of Dioxin. PLoS One. 2016;11:e0145818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Spruiell K, Richardson RM, Cullen JM, Awumey EM, Gonzalez FJ, Gyamfi MA. Role of pregnane X receptor in obesity and glucose homeostasis in male mice. J Biol Chem. 2014;289:3244–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Spruiell K, Jones DZ, Cullen JM, Awumey EM, Gonzalez FJ, Gyamfi MA. Role of human pregnane X receptor in high fat diet-induced obesity in pre-menopausal female mice. Biochem Pharmacol. 2014;89:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dong B, Saha PK, Huang W, Chen W, Abu-Elheiga LA, Wakil SJ, et al. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci U S A. 2009;106:18831–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang XJ, Chamberlain M, Vassieva O, Henderson CJ, Wolf CR. Relationship between hepatic phenotype and changes in gene expression in cytochrome P450 reductase (POR) null mice. Biochem J. 2005;388:857–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Finn RD, Henderson CJ, Scott CL, Wolf CR. Unsaturated fatty acid regulation of cytochrome P450 expression via a CAR-dependent pathway. Biochem J. 2009;417:43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hoek-van den Hil EF, van Schothorst EM, van der Stelt I, Swarts HJM, Venema D, Sailer M, et al. Quercetin decreases high-fat diet induced body weight gain and accumulation of hepatic and circulating lipids in mice. Genes Nutr 2014;9:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hoek-van den Hil EF, van Schothorst EM, van der Stelt I, Swarts HJ, van Vliet M, Amolo T, et al. Direct comparison of metabolic health effects of the flavonoids quercetin, hesperetin, epicatechin, apigenin and anthocyanins in high-fat-diet-fed mice. Genes Nutr. 2015;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zong H, Armoni M, Harel C, Karnieli E, Pessin JE. Cytochrome P-450 CYP2E1 knockout mice are protected against high-fat diet-induced obesity and insulin resistance. Am J Physiol Endocrinol Metab 2012;302:E532–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kumar R, Litoff EJ, Boswell WT, Baldwin WS. High fat diet induced obesity is mitigated in Cyp3a-null female mice. Chem-Biol Interact. 2018;289:129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–41. [DOI] [PubMed] [Google Scholar]
- [26].Foxenberg RJ, McGarrigle BP, Knaak JB, Kostyniak PJ, Olson JR. Human hepatic cytochrome P450-specific metabolism of parathion and chlorpyrifos. Drug Metab Dispos. 2007;35:189–93. [DOI] [PubMed] [Google Scholar]
- [27].Arnold C, Markovic M, Blossey K, Wallukat G, Fischer R, Dechend R, et al. Arachidonic acid-metabolizing cytochrome P450 enzymes are targets of {omega}−3 fatty acids. J Biol Chem. 2010;285:32720–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Waxman DJ. Interactions of hepatic cytochromes P-450 with steroid hormones: Regioselectivity and stereoselectivity of steroid metabolism and hormonal regulation of rat P-450 enzyme expression. Biochem Pharmcol. 1988;37:71–84. [DOI] [PubMed] [Google Scholar]
- [29].Bylund J, Kunz T, Valmsen K, Oliw EH. Cytochromes P450 with bisallylic hydroxylation activity on arachidonic and linoleic acids studied with human recombinant enzymes and with human and rat liver microsomes. J Pharmacol Exp Ther. 1998;284:51–60. [PubMed] [Google Scholar]
- [30].Pratt-Hyatt M, Zhang H, Snider NT, Hollenberg PF. Effects of a commonly occurring genetic polymorphism of human CYP3A4 (I118V) on the metabolism of anandamide. Drug Metab Dispos. 2010;38:2075–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wu S, Moomaw CR, Tomer KB, Falck JR, Zeldin DC. Molecular Cloning and Expression of CYP2J2, a Human Cytochrome P450 Arachidonic Acid Epoxygenase Highly Expressed in Heart. J Biol Chem. 1996;271:3460–8. [DOI] [PubMed] [Google Scholar]
- [32].Keeney DS, Skinner C, Travers JB, Capdevila JH, Nanney LB, King LE Jr., et al. Differentiating keratinocytes express a novel cytochrome P450 enzyme, Cyp2b19, having arachidonate monoxygenase activity. . J Biol Chem. 1998;273:32071–9. [DOI] [PubMed] [Google Scholar]
- [33].Capdevila JH, Karara A, Waxman DJ, Martin MV, Falck JR, Guengerich FP. Cytochrome P-450 enzyme-specific control of the regio- and enantiofacial selectivity of the microsomal arachidonic acid epoxygenase. J Biol Chem. 1990;265:10865–71. [PubMed] [Google Scholar]
- [34].Du L, Yermalitsky V, Ladd PA, Capdevila JH, Mernaugh R, Keeney DS. Evidence that cytochrome P450 CYP2B19 is the major source of epoxyeicosatrienoic acids in mouse skin. Arch Biochem Biophys. 2005;435:125–33. [DOI] [PubMed] [Google Scholar]
- [35].Ladd PA, Du L, Capdevila JH, Mernaugh R, Keeney DS. Epoxyeicosatrienoic acids activate transglutaminases in situ and induce cornification of epidermal keratinocytes. J Biol Chem. 2003;278:35184–92. [DOI] [PubMed] [Google Scholar]
- [36].Hernandez JP, Mota LC, Baldwin WS. Activation of CAR and PXR by dietary, environmental and occupational chemicals alters drug metabolism, intermediary metabolism, and cell proliferation. Curr Pharmacog Personal Med. 2009;7:81–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–3. [DOI] [PubMed] [Google Scholar]
- [38].Hashita T, Sakuma T, Akada M, Nakajima A, Yamahara H, Ito S, et al. Forkhead box A2-mediated regulation of female-predominant expression of the mouse Cyp2b9 gene. Drug Metab Dispos. 2008;36:1080–7. [DOI] [PubMed] [Google Scholar]
- [39].Hernandez JP, Mota LC, Huang W, Moore DD, Baldwin WS. Sexually dimorphic regulation and induction of P450s by the constitutive androstane receptor (CAR). Toxicology. 2009;256:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mota LC, Hernandez JP, Baldwin WS. CAR-null mice are sensitive to the toxic effects of parathion: Association with reduced Cytochrome P450-mediated parathion metabolism. Drug Metab Dispos. 2010;38:1582–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Oshida K, Vasani N, Jones C, Moore T, Hester S, Nesnow S, et al. Identification of chemical modulators of the constitutive activated receptor (CAR) in a gene expression compendium. Nucl Recept Signal. 2015;13:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wolfrum C, Asilmaz E, Luca E, Friedman JM, Stoffel M. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature. 2004;432:1027–32. [DOI] [PubMed] [Google Scholar]
- [43].Bochkis IM, Rubins NE, White P, Furth EE, Friedman JR, Kaestner KH. Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in ER stress. Nat Med. 2008;14:828–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bochkis IM, Shin S, Kaestner KH. Bile acid-induced inflammatory signaling in mice lacking Foxa2 in the liver leads to activation of mTOR and age-onset obesity. Mol Metab. 2013;2:447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Deol P, Evans JR, Dhahbi J, Chellappa K, Han DS, Spindler S, et al. Soybean oil is more obesogenic and diabetogenic than coconut oil and fructose in mouse: potential role for the liver. PloS One. 2015;10:e0132672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Leung A, Trac C, Du J, Natarajan R, Schones DE. Persistent chromatin modifications induced by a high fat diet. J Biol Chem. 2016;291:10446–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Damiri B, Holle E, Yu X, Baldwin WS. Lentiviral-mediated RNAi knockdown yields a novel mouse model for studying Cyp2b function. Toxicol Sci. 2012;125:368–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Damiri B, Baldwin WS. Cyp2b-knockdown Mice Poorly Metabolize Corn Oil and are Age-Dependent Obese. Lipids. 2018;53:871–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Peng L, Yoo B, Gunewardena SS, Lu H, Klaassen CD, Zhong X-b. RNA sequencing Reveals Dynamic Changes of mRNA Abundance of Cytochromes P450 and Their Alternative Transcripts during Mouse Liver Development. Drug Metab Dispos. 2012;40:1198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hersman EM, Bumpus NN. A Targeted Proteomics Approach for Profiling Murine Cytochrome P450 Expression. J Pharmacol Exp Ther. 2014;349:221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wiwi CA, Gupte M, Waxman DJ. Sexually dimorphic P450 gene expression in liver-specific hepatocyte nuclear factor 4a-deficient mice. Mol Endocrinol. 2004;18:1975–87. [DOI] [PubMed] [Google Scholar]
- [52].Kumar R, Mota LC, Litoff EJ, Rooney JP, Boswell WT, Courier E, et al. Compensatory changes in CYP expression in three different toxicology mouse models: CAR-null, Cyp3a-null, and Cyp2b9/10/13-null mice. PLOS ONE. 2017;12:e0174355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Waxman DJ, Pampori NA, Ram PA, Agrawal AK, Shapiro BH. Interpulse interval in circulating growth hormone patterns regulates sexually dimorphic expression of hepatic cytochrome P450. Proc Natl Acad Sci USA. 1991;88:6868–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ayala JE, Samuel VT, Morton GJ, Obici S, Croniger CM, Shulman GI, et al. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis Model Mech. 2010;3:525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Park BS, Mori M. Balancing false discovery and false negative rates in selection of differentially expressed genes in microarrays. Open Access Bioinformatics. 2010;2:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Supek F, Bosnjak M, Skunca N, Smuc T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One 201 l;6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, et al. InterPro in 2017—beyond protein family and domain annotations. Nucl Acids Res. 2017;45(Database issue):D190–D9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Acevedo R, Villanueva H, Parnell PG, Chapman LM, Gimenez T, Gray SL, et al. The contribution of hepatic steroid metabolism to serum estradiol and estriol concentrations in nonylphenol treated MMTVneu mice and its potential effects on breast cancer incidence and latency. J Appl Toxicol. 2005;25:339–53. [DOI] [PubMed] [Google Scholar]
- [62].Jastreboff AM, Lacadie C, Seo D, Kubat J, Van Name MA, Giannini C, et al. Leptin Is Associated With Exaggerated Brain Reward and Emotion Responses to Food Images in Adolescent Obesity. Diabetes Care. 2014;37:3061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].McGregor RA, Kwon E-Y, Shin S-K, Jung UJ, Kim E, Park JHY, et al. Time-course microarrays reveal modulation of developmental, lipid metabolism and immune gene networks in intrascapular brown adipose tissue during the development of diet-induced obesity. Int J Obes. 2013;37:1524–31. [DOI] [PubMed] [Google Scholar]
- [64].Berlanga A, Guiu-Jurado E, Porras JA, Auguet T. Molecular pathways in non-alcoholic fatty liver disease. Clin Exp Gastroenterol. 2014;7:221–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sookoian S, Pirola CJ. Systems Biology Elucidates Common Pathogenic Mechanisms between Nonalcoholic and Alcoholic-Fatty Liver Disease. PLoS ONE. 2013;8:e58895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mantzaris MD, Tsianos EV, Galaris D. Interruption of triacylglycerol synthesis in the endoplasmic reticulum is the initiating event for saturated fatty acid-induced lipotoxicity in liver cells. FEBS J. 2011;278:519–30. [DOI] [PubMed] [Google Scholar]
- [67].Sanchez-Antolín G, Almohalla-Alvarez C, Bueno P, Almansa R, Iglesias I, Rico L, et al. Evidence of Active Pro-Fibrotic Response in Blood of Patients with Cirrhosis. PLOS One. 2015;10:e0137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Pace JM, Corrado M, Missero C, Byers PH. Identification, characterization and expression analysis of a new fibrillar collagen gene, COL27A1. Matrix Biol. 2003;22:3–14. [DOI] [PubMed] [Google Scholar]
- [69].Battaller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Naa A-Y, Joa JJ, Kwona OK, Shresthaa R, Choa PJ, Kimb KM, et al. Investigation of nonalcoholic fatty liver disease-induced drug metabolism by comparative global toxicoproteomics. Toxicol Appl Pharmacol. 2018;352:28–37. [DOI] [PubMed] [Google Scholar]
- [71].Hong J, Stubbins RE, Smith RR, Harvey AE, Nunez NP. Differential susceptibility to obesity between male, female and ovariectomized female mice. Nutr J. 2009;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wade GN, Gray JM, Bartness TJ. Gonadal influences on adiposity. Int J Obes. 1985;9(Suppl l):83–92. [PubMed] [Google Scholar]
- [73].Sakai K, Fukushima H, Yamamoto Y, Ikeuchi T. A fourth subtype of retinoic acid receptor-related orphan receptors is activated by oxidized all-trans retinoic acid in medaka (Oryzias latipes). Zool Lett. 2017;3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Urquhart BL, Tirona RG, Kim RB. Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: implications for interindividual variability in response to drugs. J Clin Pharmacol. 2007;47:566–78. [DOI] [PubMed] [Google Scholar]
- [75].Thatcher JE, Isoherranen N. The role of CYP26 enzymes in retinoic acid clearance. Expert Opin Drug Metab Toxicol. 2009;5:875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Rooney JP, Oshida I, Kumar R, Baldwin WS, Corton JC. Chemical Activation of the Constitutive Androstane Receptor Leads to Activation of Oxidant-Induced Nrf2. Toxicol Sci. 2019;167:172–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ledda-Columbano GM, Pibiri M, Concas D, Molotzu F, Simbula G, Cossu C, et al. Sex difference in the proliferative response of mouse hepatocytes to treatment with the CAR ligand, TCPOBOP. Carcinogenesis. 2003;24:1059–65. [DOI] [PubMed] [Google Scholar]
- [78].Di Bonito P, Valerio G, Grugni G, Licenziati MR, Maffeis C, Manco M, et al. CARdiometabolic risk factors in overweight and obese children in ITALY (CARITALY) Study Group. Comparison of non-HDL-cholesterol versus triglycerides-to-HDL-cholesterol ratio in relation to cardiometabolic risk factors and preclinical organ damage in overweight/obese children: the CARITALY study. Nutr Metab Cardiovas Dis. 2015;25:489–94. [DOI] [PubMed] [Google Scholar]
- [79].Eisinger K, Krautbauer S, Hebei T, Schmitz G, Aslanidis C, Liebisch G, et al. Lipidomic analysis of the liver from high-fat diet induced obese mice identifies changes in multiple lipid classes. Exp Mol Pathol. 2014;97:37–43. [DOI] [PubMed] [Google Scholar]
- [80].Walenbergh SMA, Shiri-Sverdlov R. Cholesterol is a significant risk factor for non-alcoholic steatohepatitis. Exp Rev Gastronentrol Hepatol 2015;9:1343–6. [DOI] [PubMed] [Google Scholar]
- [81].Kerr TA, Davidson NO. Cholesterol and NAFLD: Renewed focus on an old villain. Hepatology. 2012;56:1995–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chen Z-w, Chen L-y, Dai H-l, Chen J-h, Fang L-z. Relationship between alanine aminotransferase levels and metabolic syndrome in nonalcoholic afatty liver disease. J Zhejiang Univ Sci B. 2008;9:616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Arguello G, Balboa E, Arrese M, Zanlungo S. Recent insights on the role of cholesterol in non-alcoholic fatty liver disease. Biochim Biophys Acta. 2015;1852:1765–78. [DOI] [PubMed] [Google Scholar]
- [84].Peng K, Mo Z, Tian G. Serum lipid abnormalities and nonalcoholic fatty liver disease in adult males. Am J Med Sci. 2017;353:236–41. [DOI] [PubMed] [Google Scholar]
- [85].Kawano Y, Cohen DE. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol. 2013;48:434–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Asgharpour A, Cazanave SC, Pacana T, Seneshaw M, Vincent R, Banini BA, et al. A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J Hepatol. 2016;66:579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kahn BB, Flier JS. Obesity and insulin resistance. J Clin Invest. 2000;106:473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bishop-Bailey D, Thomson S, Askari A, Faulkner A, Wheeler-Jones C. Lipid-metabolizing CYPs in the regulation and dysregulation of metabolism. Annu Rev Nutr. 2014;34:261–79. [DOI] [PubMed] [Google Scholar]
- [89].Sridar C, Snider NT, Hollenberg PF. Anandamide oxidation by wild-type and polymorphically expressed CYP2B6 and CYP2D6. Drug Metab Dispos. 2011;39:782–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Van der Hoeven TA, Coon MJ. Preparation and properties of partially purified cytochrome P450 and NADPH-cytochrome P450 reductase from rabbit liver microsomes. J Biol Chem. 1974;249:6302–10. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.