Abstract
Objective.
To evaluate the specificity of expression patterns of cell-free circulating microRNAs (mi-RNAs) in systemic lupus erythematosus (SLE).
Methods.
Total RNA was purified from plasma, and 45 different specific, mature miRNAs were deter-mined using quantitative reverse transcription–polymerase chain reaction assays. A total of 409 plasma samples were obtained from 364 different patients with SLE, healthy control subjects, and control subjects with other autoimmune diseases. The results in the primary cohort of 62 patients with SLE and 29 healthy control subjects were validated in 2 independent cohorts: a validation cohort comprising 68 patients with SLE and 68 healthy control subjects, and a disease control cohort comprising 20 patients with SLE (19 of whom were from the other validation cohort), 46 healthy control subjects, 38 patients with vasculitis, 18 patients with rheumatoid arthritis, and 20 immunosuppressed patients.
Results.
Seven miRNAs were statistically significantly differentially expressed in plasma from patients with SLE. The expression of miRNA-142–3p (miR-142–3p) and miR-181a was increased, and the expression of miR-106a, miR-17, miR-20a, miR-203, and miR-92a was decreased. In addition, the expression of miR-342–3p, miR-223, and miR-20a was significantly decreased in SLE patients with active nephritis. A predictive model for SLE based on 2 or 4 miRNAs differentiated patients with SLE from control subjects (76% accuracy) when validated independently (P < 2 × 10−9). Use of the 4-miRNA model provided highly significant differentiation between the SLE group and disease controls, except for those with vasculitis.
Conclusion.
Circulating miRNAs are systematically altered in SLE. A 4-miRNA signature was diagnostic of SLE, and a specific subset of miRNA profiles was associated with nephritis. All of the signature miRNAs target genes in the transforming growth factor β signaling pathways. Other targets include regulation of apoptosis, cytokine–cytokine receptors, T cell development, and cytoskeletal organization. These findings highlight possible dysregulated pathways in SLE and suggest that circulating miRNA patterns distinguish SLE from other immunoinflammatory phenotypes.
The autoimmune disease systemic lupus erythematosus (SLE) is characterized by multiple immunologic abnormalities including the presence of circulating antinuclear antibodies and a sustained type I interferon (IFN) response (1), with up-regulation of type I IFN–responsive genes (2,3) and IFNα-inducible cytokines (4). Central to this response are plasmacytoid dendritic cells (PDCs), which are activated in SLE (5) and rapidly secrete IFNα (e.g., during viral infection and in the presence of some types of immune complexes) (2,6). PDCs link the adaptive and innate immune systems, whose activation by nucleic acids via Toll-like receptors is a central event in SLE (7). Specific microRNAs (miRNAs) are one factor controlling IFNα expression by PDCs, and miRNAs also control IFNα sensitivity and cytokine receptor expression in target cells (8). Micro-RNAs are small noncoding RNAs that modulate protein translation by pairing with complementary messenger RNA bases. They regulate numerous immunologic, inflammatory, and oncogenic pathways (9). MicroRNAs are differentially expressed in peripheral blood mononuclear cells (PBMCs), immortalized B cells, and kidney biopsy specimens from patients with SLE (10–15). However, data on differentially expressed miRNAs in peripheral blood cells from patients with SLE, including associations with lupus nephritis, are discordant between different studies and different ethnic groups (10,13,15).
MicroRNAs participate in intercellular communication and circulate complexed with proteins (16,17) or sheltered in exosomes (18). Several studies have addressed the diagnostic potential of this stable population of cell-free circulating miRNAs, e.g., in cancer (19). The extent of immune dysregulation in SLE as well as the changed population of circulating subcellular particles (which may include miRNA-containing exosomes) (20–22) suggest that SLE is a condition in which specific alterations of circulating miRNA profiles are likely to be encountered. However, circulating miRNA expression signatures have not been examined and validated in independent cohorts of patients with SLE, healthy control subjects, and patients with other autoimmune conditions. Thus, we evaluated the plasma expression of a panel of miRNAs relevant to rheumatologic disease in independent cohorts of patients with SLE and control subjects, to determine potential disease-associated expression signatures and hence dysregulated genes and pathways in SLE.
PATIENTS AND METHODS
Biologic samples.
The research protocol was approved by the relevant ethics committees, and all participants provided written informed consent. Two main independent SLE cohorts were analyzed. The primary Danish exploratory cohort comprised 62 patients, and the Swedish validation cohort consisted of 68 patients. All except 1 patient, who was Asian, were white Europeans, and all fulfilled the American College of Rheumatology (ACR) criteria for SLE (23). Twenty-nine age- and sex-matched healthy control subjects (5 men and 24 women, median age 37 years [range 22–71 years]) were included in the Danish study, and 68 age- and sex-matched healthy control subjects (9 men and 59 women, median age 48 years [range 20–63 years]) were included in the Swedish study. A disease control cohort included 20 patients with SLE (19 of whom were from the Swedish cohort), 46 healthy control subjects, 38 patients with vasculitis, 18 patients with rheumatoid arthritis (RA), and 20 immunosuppressed patients (kidney transplant recipients). Finally, 40 blood samples obtained from 14 different additional patients at 2–4 different time points (6 months–1 year apart) were analyzed for correlations with disease activity. Thus, a total of 409 samples from 364 different individuals were included.
Blood samples from the Danish cohort were collected into citrate tubes, without using a tourniquet. Immediately after collection of the samples, blood cells were removed by a 2-step centrifugation protocol performed at room temperature (1,800g for 10 minutes, then 3,000g at 10 minutes) to obtain platelet-poor, cell-free plasma. Aliquots (250 μl) were snapfrozen in liquid nitrogen. Samples from the Swedish cohort were drawn into EDTA-coated tubes and left for 1 hour before 1-step centrifugation (1,600g for 10 minutes at room temperature). All samples were stored at −80°C until analyzed.
RNA isolation and miRNA profiling.
A Total RNA Purification Kit (Norgen Biotek) was used to purify RNA (including miRNA) from 100 μl of plasma (24), according to the manufacturer’s instructions, with the following minor modifications: 10 mM dithiothreitol and Caenorhabditis elegans synthetic miRNA-39 (miR-39), miR-54, and miR-238 (TAG Copenhagen A/S), each at 1.7 pM, were added to a volume of lysis buffer (from the RNA purification kit) sufficient for all of the samples. This volume was then aliquotted into 2-ml portions and kept at −20°C until used. One microliter of RNase inhibitor (20 units/μl; Applied Biosystems) was added to every elution tube before elution of RNA. Purified RNA was kept at −20°C for up to 2 weeks before being used for reverse transcription (RT). Reverse transcription was performed using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). The RT primer mix consisted of equal volumes of each of 48 different 5× RT miRNA-specific stem-loop primers (see Supplementary Tables 2 and 3, available on the Arthritis & Rheumatism web site at http://onlinelibrary.wiley.com/doi/10.1002/art.37890/abstract). The RT reaction volume was 10 μl; 1 μl of MultiScribe Reverse Transcriptase (Invitrogen), 3 μl of RT primer mix, 1 μl of 10× buffer, 0.2 μl of 100 mM dNTPs, 0.15 μl of RNAse inhibitor, and 4.65 μl of the purified RNA were used.
Reverse transcription was performed on an Applied Biosystems 2720 Thermal Cycler, using standard protocols. Specific target amplification was accomplished using TaqMan PreAmp Master Mix and TaqMan assay mix (Applied Biosystems) consisting of equal volumes of the 48 different 20× assays diluted with 1× TE buffer to a final concentration of 0.2×. The preamplification mixture (10 μl) contained 2.5 μl of diluted complementary DNA (diluted 1:3 with H2O) mixed with 5 μl of 2× TaqMan PreAmp Master Mix and 2.5 μl of the 0.2× TaqMan MicroRNA Assay mix. Preamplification was performed at 95°C for 10 minutes, followed by 16 cycles at 95°C for 15 seconds and at 60°C for 4 minutes, and a hold at 4°C. Preamplified samples (diluted 1:5 with H2O) and TaqMan MicroRNA 20× assays were applied to primed 96.96 Dynamic Array chips using loading and assay reagents according to the manufacturer’s instructions (Fluidigm). All miRNA assays were performed in duplicate.
Real-time polymerase chain reaction (PCR) was performed with a BioMark Real-Time PCR System (Fluidigm) using single probe (FAM-labeled MGB, ROX reference dye) settings and 40 cycles. The default data acquisition protocol (GE 96 × 96) of the instrument was used. Data were processed using Fluidigm Real-Time PCR Analysis software (version 3.0.1) with the autodetector setting. Conventional quantitative real-time RT-PCR was performed using an Applied Biosystems ViiA 7 system with 8 TaqMan MicroRNA assays (hsa-miR-17, hsa-miR-20a, hsa-miR-92a, hsa-miR-142–3p, hsa-miR-146a, hsa-miR-181a, hsa-miR-181b, and cel-miR-54) in 384-well plates.
Data handling and statistical analysis.
Average raw quantification cycle (Cq) values of >30 were removed from all data sets. Average Cq values of the duplicate analysis of each miRNA were then subtracted from the average Cq value of the 3 cel-miRNAs for that particular sample, yielding the ΔCq values used in further analyses. Thus, samples with high cycle values (low expression) have lower ΔCq values than samples with low cycle values (higher expression). All ΔCq values were then row-normalized to correct for variations in total input RNA. The average value of 26 miRNAs that were detected in all samples (miR-106a, miR-125a-3p, miR-132, miR-142–3p, miR-146a, miR-146b, miR-150, miR-155, miR-15a, miR-16, miR-17, miR-181b, miR-184, miR-196a, miR-203, miR-20a, miR-21, miR-221, miR-223, miR-24, miR-342–3p, miR-34a, miR-383, miR-409–3p, miR-638, miR-92a) was subtracted from all miRNA ΔCq values in each sample. These rownormalized expression values were used for the statistical analyses.
Expression graphs, receiver operating characteristic (ROC) curves, and ΔCq values were analyzed using GraphPad Prism version 5.04 software. Statistical significance between groups was determined by unpaired t-test, Mann-Whitney U test, or one-way analysis of variance, with Dunnett’s multiple comparison test as appropriate. P values less than 0.05 were considered significant. Correction for multiple testing was performed using false discovery rates (FDRs) (25). Correlations were evaluated by Spearman’s correlation analysis. For principal components analysis (PCA), the PASW Statistics 18 program (IBM SPSS) was used. Diagnostic accuracy was estimated by leave-one-out cross-validation. Diagnostic performance was evaluated using 2 × 2 contingency tables and Fisher’s exact test.
For the 4 top-performing miRNAs, we derived a risk probability score, p, by logistic regression using Stata version 11.2, where p = 1/(1/ex + 1) and x is the sum of input miRNA ΔCq values, each multiplied with specific coefficients. Unsupervised hierarchical clustering was performed with Partek Genomics Suite 6.6 with the default settings that use Euclidian distance with average linkage. Class comparisons of all mi-RNAs in the 2 main cohorts were performed with BRB-ArrayTools version 4.1.0 (http://linus.nci.nih.gov/BRB-ArrayTools.html). For analysis of KEGG pathway enrichment in specific miRNA target genes, microT-3.0 prediction software (http://diana.cslab.ece.ntua.gr/pathways/) was used.
RESULTS
Expression of circulating miRNA in SLE.
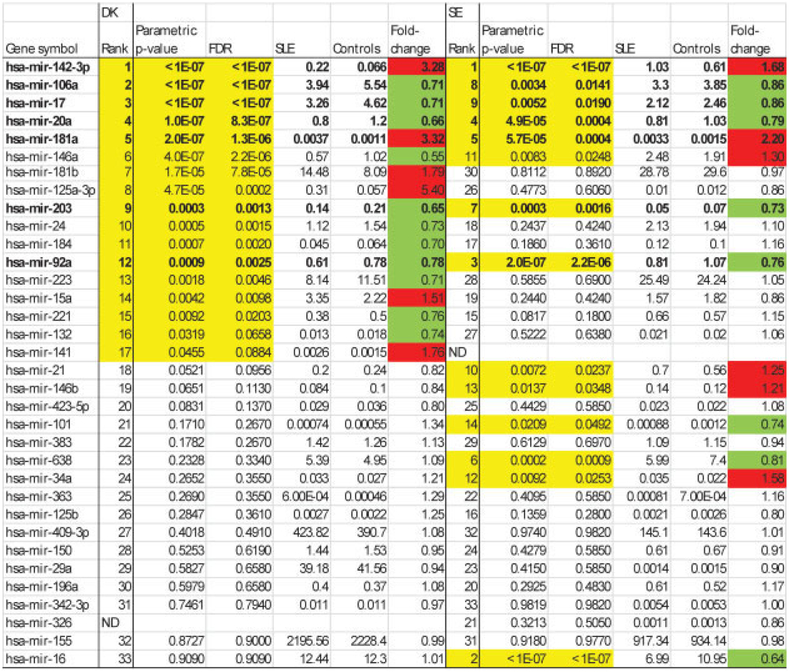
The characteristics of the patients with SLE in the exploratory (Danish) cohort and the validation (Swedish) cohort are shown in Table 1. The 2 cohorts were ethnically and racially homogeneous and comparable. We analyzed the expression of 45 mature miRNAs relevant to rheumatic disease (see Supplementary Table 1, available on the Arthritis & Rheumatism web site at http://onlinelibrary.wiley.com/doi/10.1002/art.37890/abstract) in total RNA isolated from plasma and compared the results in patients with SLE with those in age- and sex-matched healthy control subjects. Graphs showing the expression of all miRNAs appear in Supplementary Figure 1, available on the Arthritis & Rheumatism web site at http://onlinelibrary.wiley.com/doi/10.1002/art.37890/abstract. Significant differences were observed (Figures 1 and 2). In the Danish cohort, 33 of the 45 assayed miRNAs (see Supplementary Table 1) were detectable (Cq values ≤30), and 15 of these 33 miRNAs were shown to be significantly (FDR <0.05) differentially expressed (Figure 1).
Table 1.
Characteristics of the study populations from the Danish and Swedish SLE cohorts*
| Characteristic | Exploratory cohort | Validation cohort |
|---|---|---|
| Recruitment area | Copenhagen, Denmark | Lund, Sweden |
| Control subjects | ||
| Age, median (range) years | 37 (22–71) | 48 (20–63) |
| No. men/no. women | 5/24 | 9/59 |
| Patients with SLE | ||
| Age, median (range) years | 40 (21–76) | 51 (20–84) |
| No. men/no. women | 6/56 | 9/59 |
| Disease duration, median (range) years | 10 (0–37) | 13 (0–49) |
| Disease manifestations† | ||
| Renal disease | 9(15) | 12(17) |
| Vasculitis | 4(6) | 2(3) |
| Arthritis | 6 (10) | 8 (12) |
| Rash | 2(3) | 5 (7) |
| Alopecia | 4(6) | 4(6) |
| Mucosal ulcers | 5(8) | 3 (4) |
| Serositis | 2(3) | 0 (0) |
| Leukopenia | 8(13) | 3 (4) |
| Thrombocytopenia | 2(3) | 1 (1) |
| Visual disturbance | 0 | 1 (1) |
| Fever | 0 | 1 (1) |
| SLEDAI, mean ± SD (range) | 5 ± 4 (0–18) | 3 ± 4 (0–14) |
| Autoantibodies and complement† | ||
| Anti-double-stranded DNA | 30 (48) | 15 (22) |
| Low C3 or C4 level | 42 (68) | 19 (28) |
| Medication† | ||
| Prednisolone ≤7.5 mg/day | 14 (23) | 36 (53) |
| Prednisolone >7.5 mg/day | 11 (18) | 14 (20) |
| Antimalarials | 13 (21) | 46 (67) |
| Azathioprine, MTX, or MMF | 27 (44) | 31 (45) |
| SLICC/ACR Damage Index, mean ± SD (range) | 1 ± 2 (0–8) | 2 ± 2 (0–10) |
| Antiphospholipid syndrome | 15 (24) | 19 (29) |
| History of thrombosis (arterial/venous) | 15/10 (24/16) | 13/20 (19/29) |
The exploratory cohort comprised 62 patients with systemic lupus erythematosus (SLE) and 29 control subjects. The validation cohort comprised 68 patients with SLE and 68 control subjects. Clinical manifestations were defined and scored according to the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI). Except where indicated otherwise, values are the number (%). MTX = methotrexate; MMF = mycophenolate mofetil; SLICC/ACR = Systemic Lupus International Collaborating Clinics/American College of Rheumatology.
At study entry.
Figure 1.
MicroRNA (miRNA) expression in the exploratory (Danish; DK) and validation (Swedish; SE) cohorts, ranked according to P values for univariate testing in the Danish cohort. Values in the columns headed SLE and Controls are the geometric means of ratios. The fold change is the ratio of expression in patients with systemic lupus erythematosus (SLE) versus controls. All P values less than 0.05 are shown in yellow. The red highlighting indicates significant up-regulation, and the green highlighting indicates significant down-regulation. The 7 miRNAs that were consistently differentially regulated in the 2 cohorts are shown in boldface. FDR = false-discovery rate; ND = not detected.
Figure 2.
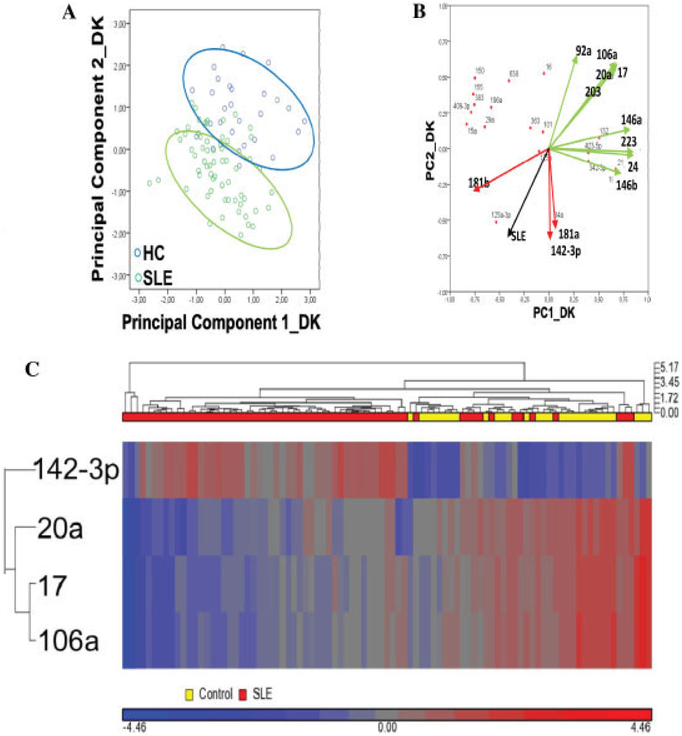
Expression of plasma miRNAs in the exploratory (Danish) cohort. A, Diagram of principal components analysis, showing the distribution of patients with SLE and healthy controls (HCs) according to the first 2 principal components (PC1 and PC2). B, Loadings plot showing the contribution of individual miRNAs to PC1 and PC2. Significantly down-regulated miRNAs are shown in green, and significantly up-regulated miRNAs are shown in red (see Figure 1). The black vector shows the PC variation with regard to having/not having SLE. C, Unsupervised hierarchical clustering based on expression of the 4 top-performing miRNAs. Data are scaled to mean = 0 and SD = 1 to enable comparison across miRNAs. See Figure 1 for other definitions.
The main contributors to differences between patients with SLE and healthy control subjects were demonstrated by the PCA (Figure 2A), in which the first 2 principal components together accounted for ~75% of the data variation. The loadings plot (Figure 2B) showed the relationship between miRNA expression and the subspace dimension and indicated that the downregulated miR-17/miR-20a/miR-106a cluster contributed similarly to the variation. Likewise, miR-142–3p and miR-181a, both of which were increased >3-fold, contributed in parallel. Unsupervised hierarchical clustering using the 4 top-performing miRNAs (Figure 2C) illustrated the differentiation between patients and controls and the concordant expression of miR-17, miR-20a, and miR-106a.
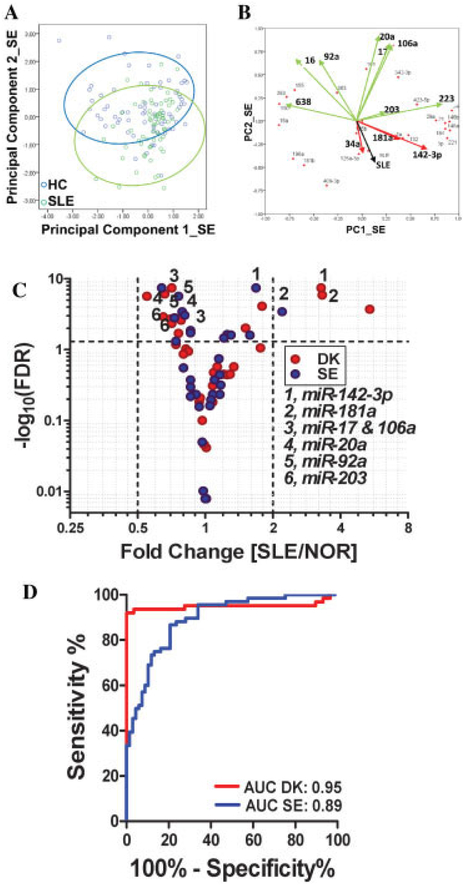
We next examined miRNA profiles in the validation (Swedish) cohort. Except for the use of EDTA plasma and the performance of 1 instead of 2 centrifugation steps for plasma preparation, these samples were processed and analyzed exactly as in the Danish cohort. The data are summarized in Figure 1. The results validated findings in the exploratory cohort regarding 7 significantly changed miRNAs (Figure 1), i.e., an increase in the expression of miR-142–3p and miR-181a and down-regulation of miR-17, miR-20a, miR-92a, miR-106a, and miR-203, with no difference between patients with SLE and healthy control subjects for 10 other miRNAs. Additionally, 15 miRNAs behaved differently in the 2 cohorts (see Supplementary Table 3, available on the Arthritis & Rheumatism web site at http://onlinelibrary.wiley.com/doi/10.1002/art.37890/abstract). Figures 3A and B show the results of PCA and loadings plot analysis of these data, respectively. In the validation cohort, the main miRNA contributors to sample variability were shown to be the same as those in the exploratory cohort. Figure 3C illustrates validation of significant miRNAs by summarizing the FDRs and fold changes in the data from the exploratory and validation cohorts.
Figure 3.
Expression of plasma microRNAs (miRNAs; miR) in the validation (Swedish; SE) cohort and comparison with the exploratory cohort. A, Diagram of principal components analysis, showing partial differentiation between patients with systemic lupus erythematosus (SLE) and healthy controls. B, Loadings plot showing the contribution of individual miRNAs to PC1 and PC2. Significantly down-regulated miRNAs are shown in green, and significantly up-regulated miRNAs are shown in red. The black vector shows the PC variation with regard to having/not having SLE. C, Volcano plot showing the expression of 33 miRNAs in plasma in the Danish (DK) and SE cohorts. The 7 numbered miRNAs are those that were consistently and significantly changed in the DK and the SE cohorts. The broken horizontal line corresponds to a false discovery rate (FDR) of 0.05. D, Receiver operating characteristic curves (plots of true-positive versus false-positive rate for different cutoffs of the test) for the DK and SE cohorts, using the difference between the change in quantification cycle values for miR-106a and miR-142–3p as a classifier. NOR = normal; AUC = area under the curve (see Figure 2 for other definitions).
The difference between the ΔCq values of the 2 most significant miRNAs (miR-142–3p and miR-106a) was used to classify samples (Figure 3D). In leave-one-out cross-validation, the classification accuracy was 89% (positive predictive value 100%, negative predictive value 74%) and the P value (by Fisher’s exact test) was 1 × 10−15 in the exploratory cohort. In the validation cohort, the area under the curve (AUC) of the ROC curve was 0.89 (versus 0.95 in the exploratory cohort), and the classification performance was 76% (P = 2 × 10−9), without optimizing cutoff scores.
MiRNA expression, disease activity, and complications.
No specific miRNAs common to both cohorts correlated significantly with Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) scores (26). Notably, miR-181a correlated with both the SLEdAI (r = 0.48, P < 0.0001) and the Lupus International Collaborating Clinics (SLICC)/ACR Damage Index (27) (r = −0.39, P = 0.0014) in the Swedish cohort but not in the Danish cohort. Both cohorts consisted primarily of patients with mild-to-moderate disease activity (Table 1). In consecutive samples using a panel of 7 miRNAs, including 3 of the 4 top-performing miRNAs from the primary analyses, we also observed no correlation between disease activity and miRNA expression in individual patients. However, when analyzing the consecutive data as a whole, we did observe a correlation (r = −0.35/0.38, P = 0.02) between the SLEDAI score and decreased expression of miR-142–3p and increased expression of miR-181b. In accordance with previous studies (28), we did not observe correlations between any miRNA and patient age.
We examined the possibility that immunosuppressive treatment by itself would affect miRNA levels. Expression of the 4 best-performing miRNAs (miR-142–3p, miR-106a, miR-17, and miR-20a) was compared between patients who were not receiving immunosuppressive treatment (n = 23 in the Danish cohort, n = 15 in the Swedish cohort) and those receiving treatment with any of the following: azathioprine, methotrexate, mycophenylate mofetil, or hydroxychloroquine. No statistically significant differences were observed between these 2 groups in either cohort (data not shown). Thus, the dysregulation of these miRNAs in SLE was independent of immunosuppressive treatment. Active nephritis at the time of sampling was significantly associated (FDR <0.05) with decreased levels of miR-342–3p, miR-223, and miR-20a, in both the Danish cohort (9 of 62 patients) and the Swedish cohort (12 of 68 patients). For cardiovascular disease, we observed associations only in the Swedish cohort (31 of 68 patients with a history of myocardial infarction, arterial thrombosis, or venous thrombosis); in this cohort, vascular events were highly significantly associated (P < 0.0001, FDR <0.0035) with increased levels of miR-150, miR-155, and miR-383. All 3 of these miRNAs also correlated with platelet activation (C1q binding; r = 0.35–0.36), the SLICC (r = 0.38–0.49), expression of type I IFN signature proteins in platelets (29), i.e., IFN-induced transmembrane protein 1 (r = 0.34–0.54), and protein kinase, IFN-inducible double-stranded RNA-dependent activator (r = 0.51–0.63).
MicroRNA expression in patients with SLE and disease controls.
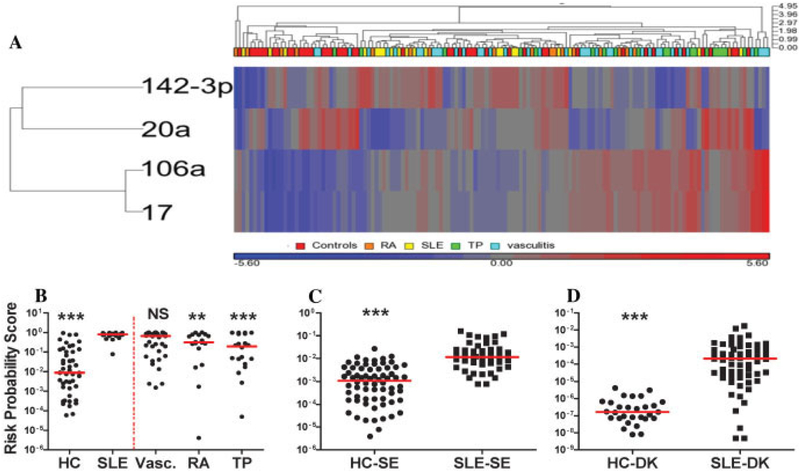
Samples obtained from patients with SLE (n = 20; 19 of the samples were from patients in the above-described Swedish cohort) were analyzed together with those from disease controls (Figure 4). The results confirmed a significant decrease in the expression of miR-20a and miR-92a (and a trend toward decreased expression of miR-203) and an increase in miR-142–3p expression in patients with SLE relative to healthy control subjects. MicroRNA-181a was not consistently detected, and the expression of miR-17 and miR-106a was not significantly changed (see Supplementary Table 4, available on the Arthritis & Rheumatism web site at http://onlinelibrary.wiley.com/doi/10.1002/art.37890/abstract). However, 3 miRNAs that were decreased in only 1 of the Danish/Swedish cohorts (miR-146a in the Danish cohort; miR-16 and miR-101 in the Swedish cohort) were all significantly down-regulated the SLE samples in this data set. Subsequently, a risk probability score based on the 4 top-performing miRNAs validated in the Danish and Swedish cohorts was developed by logistic regression in the 20 patients with SLE and 46 healthy control subjects in this training sample set (Figure 4B) (see also Supplementary Table 5, available on the Arthritis & Rheumatism web site at http://onlinelibrary.wiley.com/doi/10.1002/art.37890/abstract). This score showed highly significant (P < 0.006) differentiation between patients with SLE and healthy control subjects and between patients with SLE and the collective group of patients with other diseases (RA, vasculitis, and kidney transplant). The AUC for the risk probability score was 0.67. The risk score also significantly discriminated the SLE group from the RA and kidney transplant groups (Figure 4B) but not from the vasculitis group. The risk score was tested in the Swedish cohort and validated in the Danish cohort. In each instance, scores in patients with SLE were significantly (P < 0.0001) higher than those in healthy control subjects (Figures 4C and D).
Figure 4.
Expression of miRNAs and risk probability scores in patients with SLE and disease controls. A, Unsupervised hierarchical clustering using the 4 top-performing miRNAs from the exploratory and validation cohorts, healthy controls (HCs), patients with SLE, patients with vasculitis (Vasc.), patients with rheumatoid arthritis (RA), and kidney transplant recipients (TP). B-D, Risk probability scores, derived after applying a logistic regression model based on the 4 top-performing miRNAs in the SLE/HC samples in the disease control sample set (left of the broken red line in B). B, Disease controls training set. C, Swedish cohort (19 healthy controls were removed because they were used in the data set shown in B). D, Danish cohort. Symbols represent individual data points; horizontal lines show the mean. ** = P < 0.01; *** = P < 0.001 versus SLE. NS = not significant (see Figure 3 for other definitions).
Targeted pathway analysis.
The most prominently targeted pathways of miR-181a are the transforming growth factor β (TGFβ) signaling and the cytokine-cytokine receptor pathways. The gene for TGFβ receptor type I (TGFBR1) is also targeted (along with genes involved in cytoskeletal organization) by the other highly significantly up-regulated miRNA (miR-142–3p). The down-regulated miR-17–92 cluster and its paralogs also regulate genes, prominently BMPR2, in the TGFβ signaling pathway. Thus, TGFβ signaling emerged as a common target for all consistently differentially regulated miRNAs in our study. The targeted genes were TGFBR1/2, ACVR2A/2B, SMAD6/7, SMURF1, BMPR2, and MAPK1 (ERK1/2). Another major pathway targeted by miR-17 is regulation of the actin cytoskeleton (7 genes targeted). The only significantly down-regulated miRNA outside of the miR-17–92 family was miR-203. This miRNA targets genes in the MAPK signaling and cytokine-cytokine receptor pathways and many genes involved in focal adhesion and tight junctions.
DISCUSSION
To date, this is the largest study of circulating cell-free miRNA in SLE using 2 independent cohorts. Seven miRNAs were differentially expressed, and the expression of 10 miRNAs was unchanged; these observations were completely consistent between the 2 cohorts. Expression of the remaining 16 miRNAs was different in the 2 cohorts, which may be partially explained by differences in sample processing (e.g., more platelets in the validation cohort). Overall, however, the findings suggest that predictable changes in the profiles of miRNA released to the circulation may characterize systemic autoimmunity and may include patterns specific for SLE.
The sources and states of miRNA in the circulation are heterogeneous. Most miRNA originates from blood cells, including platelets, and endothelial cells (28,30). Aberrations of miRNA in circulating cells have been shown in SLE, but data are inconsistent (13,15). This may be attributable to small numbers of investigated samples, different ethnic groups, and different cell types studied. Also, miRNA in the circulation is found in vesicles (exosomes, microparticles) and/or in miRNA-protein complexes. The types of vehicles for the miRNAs in the present study are not known. This would be interesting to explore in future work, using differential centrifugation or immunomagnetic isolation of vesicles or specific miRNA-binding proteins.
With regard to cell-free circulating miRNA, our group previously demonstrated the stability of plasma miRNA after several freeze-thaw cycles and the reproducibility of identical, independently processed samples (24). In the present study, we observed higher expression of miR-142–3p and miR-181a and lower expression of miR-17, miR-20a, miR-106a, miR-92a, and miR-203 in SLE. Microarray profiling of circulating miRNAs in a Chinese SLE population showed up-regulation of 19 different miRNAs and down-regulation of 32 different miRNAs (31). The observed up-regulation of miR-142–3p and down-regulation of miR-92a are consistent with our results, while the other miRNAs that were significantly increased or decreased in that study were not included in our panel. Our data also are consistent with up-regulation of miR-142–3p (15,29) and miR-181a (15) and down-regulation of miR-17 (13) and miR-20a (15) in B cells and PBMCs from patients with SLE. Additionally, a recent study showed that circulating miR-142–3p expression was also specifically increased in patients with systemic sclerosis, to levels above those observed in patients with SLE (32).
Conversely, CD4+ T cells in patients with SLE contain decreased miR-142–3p/5p (33). Increased miR-142–3p expression in cell-free SLE plasma may thus be caused by increased cellular release of this miRNA through increased exocytosis (leading to decreased intracellular content) and/or normal exocytosis of cells containing increased miRNA. MicroRNA-181a, which is present at low levels in plasma, was up-regulated >2-fold in both cohorts. This miRNA is important for hematopoietic cell differentiation (34,35), increases the fraction of B-lineage cells (36), and is also expressed by endothelial cells (37). Our results do not confirm the results of a study in which miR-181a expression was decreased in the peripheral blood of pediatric patients with SLE (38).
Except for miR-203, all of the consistently downregulated miRNAs observed in the present study belong to the polycistronic miR-17–92 family and its paralogs. These are known oncogenic, antiapoptotic, and immunoinflammation-modulating miRNAs (39). Thus, miR-17, miR-20a, and miR-92a regulate apoptosis by repressing Bim and PTEN. Together with miR-106a, miR-17 and miR-20a also control monocytopoiesis (40) and regulate regulatory T cells (targeting CREB1 and TGFBR2, among others) (41). Down-regulation of these miRNAs that display homologous seed sequences supports the functional significance of our findings and would be expected to accelerate apoptosis and inhibit monocytopoiesis (39). Increased apoptosis and impaired clearance are central to the pathogenesis of SLE (42–44).
Cellular miR-146a and miR-125a have been associated with lupus nephritis and activation of cytokine pathways in SLE (10,15), but dysregulation of these miRNAs in PBMCs from patients with SLE has not been confirmed (14,15,31). Our data showed that the level of circulating miR-146a was significantly decreased in the exploratory cohort and was significantly increased in the validation cohort, while expression of miR-125a-3p was increased in one cohort and unchanged in the other. The increased miR-155 expression observed in activated lymphocytes from patients with SLE (45) is not supported by the circulating levels observed in a previous study (31), in accordance with our data.
The association of miR-223 with lupus nephritis observed in this study is consistent with findings in renal biopsy specimens (14). We also observed that miR-342–3p expression was specifically decreased in patients with lupus nephritis; expression of this miRNA is decreased in PBMCs and Epstein-Barr virus-transformed B cells from African American patients with SLE (15).
Type I IFNs are associated with vascular disease in SLE, modulating endothelial function and regeneration (29,46,47), and miR-155/155* regulate type I IFN production by PDCs (2,6,48). In addition, miR-150 is enriched in lymphoid cells (30), and miR-155 is present in platelets (49). In the current study, strong associations of miR-150 and miR-155 with vascular disease were observed in the Swedish cohort, where these miRNAs also correlated with expression of type I IFN-regulated platelet proteins and with platelet activation. Due to less extensive centrifugation, more cellular (especially platelets) and subcellular elements may have contributed to specific miRNA profiles in the Swedish cohort samples. Therefore, the range of ΔCq values for miR-150, miR-155, and miR-383 was much larger in samples from the Swedish cohort compared with those from the Danish cohort (see Supplementary Figure 1, available on the Arthritis & Rheumatism web site at http://onlinelibrary.wiley.com/doi/10.1002/art.37890/abstract). Increased cell or platelet counts and cellular and platelet activation may thus contribute to the increased expression of miR-150 and miR-155 in patients with vascular disease. In addition, preliminary data show that plasma miRNAs distribute differently (i.e., not all are located in exosomes) upon extended ultracentrifugation (Carlsen AL, et al: unpublished observations). Additional studies are needed to establish relationships between miRNAs, cellular stimulation, and development of vascular disease in patients with SLE. Further validation of the methods used for determination of circulating miRNAs, including putative differences between plasma from blood anticoagulated with citrate or EDTA, is also required.
In conclusion, our study confirms specific expression patterns of cell-free circulating miRNA in SLE and validates the use of a “top 4” miRNA-based score for SLE risk. The extent to which these miRNA profiles may be used diagnostically, prognostically, and for monitoring purposes is yet to be determined, but our results do suggest cellular pathways involved in SLE pathogenesis and thus advance the basis for understanding the function and targeting of miRNAs in SLE.
Supplementary Material
Acknowledgments
Supported by the Danish Rheumatism Association (grant R99-A1937). Drs. Schetter and Harris’ work was supported by the Intramural Research Program of the National Cancer Institute, NIH. Drs. Lood and Bengtsson’s work was supported by the Medical Faculty at Lund University, the Alfred Österlund Foundation, the Crafoord Foundation, the Swedish Rheumatism Association, the Greta and Johan Kock Foundation, King Gustaf V’s 80-Year Foundation, the Swedish Society of Medicine, the Foundation of the Swedish National Board of Health and Welfare, and Skåne University Hospital, Lund, Sweden. Dr. Voss’ work was supported by the Danish Rheumatism Association (grant R33-A1836), the A. P. Møller Foundation, and the Region of Southern Denmark. Drs. Hellmark and Segelmark’s work was supported by the Swedish Renal Foundation and the Swedish Research Council. Dr. Jacobsen’s work was supported by the Novo Nordisk Research Foundation. Dr. Heegaard’s work was supported by the Foundation for the Advancement of Medical Science.
REFERENCES
- 1.Ronnblom L, Alm GV, Eloranta ML. Type I interferon and lupus. Curr Opin Rheumatol 2009;21:471–7. [DOI] [PubMed] [Google Scholar]
- 2.Santer DM, Wiedeman AE, Teal TH, Ghosh P, Elkon KB. Plasmacytoid dendritic cells and C1q differentially regulate inflammatory gene induction by lupus immune complexes. J Immunol 2012;188:902–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baechler EC, Gregersen PK, Behrens TW. The emerging role of interferon in human systemic lupus erythematosus. Curr Opin Immunol 2004;16:801–7. [DOI] [PubMed] [Google Scholar]
- 4.Bauer JW, Baechler EC, Petri M, Batliwalla FM, Crawford D, Ortmann WA, et al. Elevated serum levels of interferon-regulated chemokines are biomarkers for active human systemic lupus erythematosus. PLoS Med 2006;3:e491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eloranta ML, Lovgren T, Finke D, Mathsson L, Ronnelid J, Kastner B, et al. Regulation of the interferon-α production induced by RNA-containing immune complexes in plasmacytoid dendritic cells. Arthritis Rheum 2009;60:2418–27. [DOI] [PubMed] [Google Scholar]
- 6.Lood C, Gullstrand B, Truedsson L, Olin AI, Alm GV, Ronnblom L, et al. C1q inhibits immune complex-induced interferon-α production in plasmacytoid dendritic cells: a novel link between C1q deficiency and systemic lupus erythematosus pathogenesis. Arthritis Rheum 2009;60:3081–90. [DOI] [PubMed] [Google Scholar]
- 7.Lovgren T, Eloranta ML, Bave U, Alm GV, Ronnblom L. Induction of interferon-α production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum 2004;50:1861–72. [DOI] [PubMed] [Google Scholar]
- 8.Papadopoulou AS, Dooley J, Linterman MA, Pierson W, Ucar O, Kyewski B, et al. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-α receptor. Nat Immunol 2011;13:181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis 2010;31:37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum 2009;60:1065–75. [DOI] [PubMed] [Google Scholar]
- 11.Luo X, Tsai LM, Shen N, Yu D. Evidence for microRNA-mediated regulation in rheumatic diseases. Ann Rheum Dis 2010;69 Suppl 1:i30–i36. [DOI] [PubMed] [Google Scholar]
- 12.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 2006;103:12481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus 2007;16: 939–46. [DOI] [PubMed] [Google Scholar]
- 14.Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int 2009;29:749–54. [DOI] [PubMed] [Google Scholar]
- 15.Te JL, Dozmorov IM, Guthridge JM, Nguyen KL, Cavett JW, Kelly JA, et al. Identification of unique microRNA signature associated with lupus nephritis. PLoS One 2010;5:e10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13: 423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–9. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen CT, Ostergaard O, Johnsen C, Jacobsen S, Heegaard NH. Distinct features of circulating microparticles and their relationship to clinical manifestations in systemic lupus erythematosus. Arthritis Rheum 2011;63:3067–77. [DOI] [PubMed] [Google Scholar]
- 21.Distler JH, Huber LC, Hueber AJ, Reich CF III, Gay S, Distler O, et al. The release of microparticles by apoptotic cells and their effects on macrophages. Apoptosis 2005;10:731–41. [DOI] [PubMed] [Google Scholar]
- 22.Ullal AJ, Reich CF III, Clowse M, Criscione-Schreiber LG, Tochacek M, Monestier M, et al. Microparticles as antigenic targets of antibodies to DNA and nucleosomes in systemic lupus erythematosus. J Autoimmun 2011;36:173–80. [DOI] [PubMed] [Google Scholar]
- 23.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus [letter]. Arthritis Rheum 1997;40:1725. [DOI] [PubMed] [Google Scholar]
- 24.Heegaard NH, Schetter AJ, Welsh JA, Yoneda M, Bowman ED, Harris CC. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer 2012;130:1378–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995;57:289–300. [Google Scholar]
- 26.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang DH, and the Committee on Prognosis Studies in SLE. Derivation of the SLEDAI: a disease activity index for lupus patients. Arthritis Rheum 1992;35:630–40. [DOI] [PubMed] [Google Scholar]
- 27.Gladman DD, Urowitz MB, Goldsmith CH, Fortin P, Ginzler E, Gordon C, et al. The reliability of the Systemic Lupus International Collaborating Clinics/American College of Rheumatology Damage Index in patients with systemic lupus erythematosus. Arthritis Rheum 1997;40:809–13. [DOI] [PubMed] [Google Scholar]
- 28.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 2008;3:e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lood C, Amisten S, Gullstrand B, Jonsen A, Allhorn M, Truedsson L, et al. Platelet transcriptional profile and protein expression in patients with systemic lupus erythematosus: upregulation of the type I interferon system is strongly associated with vascular disease. Blood 2010;116:1951–7. [DOI] [PubMed] [Google Scholar]
- 30.Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Peng W, Ouyang X, Li W, Dai Y. Circulating micro-RNAs as candidate biomarkers in patients with systemic lupus erythematosus. Transl Res 2012;160:198–206. [DOI] [PubMed] [Google Scholar]
- 32.Makino K, Jinnin M, Kajihara I, Honda N, Sakai K, Masuguchi S, et al. Circulating miR-142–3p levels in patients with systemic sclerosis. Clin Exp Dermatol 2012;37:34–9. [DOI] [PubMed] [Google Scholar]
- 33.Ding S, Liang Y, Zhao M, Liang G, Long H, Zhao S, et al. Decreased miR-142–3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum 2012;64:2953–63. [DOI] [PubMed] [Google Scholar]
- 34.Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, et al. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol 2011;187: 6171–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 2007;129:147–61. [DOI] [PubMed] [Google Scholar]
- 36.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004;303:83–6. [DOI] [PubMed] [Google Scholar]
- 37.Kazenwadel J, Michael MZ, Harvey NL. Prox1 expression is negatively regulated by miR-181 in endothelial cells. Blood 2010; 116:2395–401. [DOI] [PubMed] [Google Scholar]
- 38.Lashine YA, Seoudi AM, Salah S, Abdelaziz AI. Expression signature of microRNA-181-a reveals its crucial role in the pathogenesis of paediatric systemic lupus erythematosus. Clin Exp Rheumatol 2011;29:351–7. [PubMed] [Google Scholar]
- 39.Olive V, Jiang I, He L. mir-17–92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol 2010;42:1348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, et al. MicroRNAs 17–5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol 2007;9:775–87. [DOI] [PubMed] [Google Scholar]
- 41.Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, et al. Molecular dissection of the miR-17–92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood 2011;118:5487–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren Y, Tang J, Mok MY, Chan AW, Wu A, Lau CS. Increased apoptotic neutrophils and macrophages and impaired macrophage phagocytic clearance of apoptotic neutrophils in systemic lupus erythematosus. Arthritis Rheum 2003;48:2888–97. [DOI] [PubMed] [Google Scholar]
- 43.Bengtsson AA, Sturfelt G, Gullstrand B, Truedsson L. Induction of apoptosis in monocytes and lymphocytes by serum from patients with systemic lupus erythematosus: an additional mechanism to increased autoantigen load? Clin Exp Immunol 2004;135:535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grossmayer GE, Munoz LE, Gaipl US, Franz S, Sheriff A, Voll RE, et al. Removal of dying cells and systemic lupus erythematosus. Mod Rheumatol 2005;15:383–90. [DOI] [PubMed] [Google Scholar]
- 45.Divekar AA, Dubey S, Gangalum PR, Singh RR. Dicer insufficiency and microRNA-155 overexpression in lupus regulatory T cells: an apparent paradox in the setting of an inflammatory milieu. J Immunol 2011;186:924–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thacker SG, Zhao W, Smith CK, Luo W, Wang H, Vivekanandan-Giri A, et al. Type I interferons modulate vascular function, repair, thrombosis and plaque progression in murine models of lupus and atherosclerosis. Arthritis Rheum 2012;64:2975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Somers EC, Zhao W, Lewis EE, Wang L, Wing JJ, Sundaram B, et al. Type I interferons are associated with subclinical markers of cardiovascular disease in a cohort of systemic lupus erythematosus patients. PLoS One 2012;7:e37000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou H, Huang X, Cui H, Luo X, Tang Y, Chen S, et al. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood 2010;116:5885–94. [DOI] [PubMed] [Google Scholar]
- 49.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol 2009;16:961–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.