Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV) is capable of inducing a storm of proinflammatory cytokines. In this study, we show that the SARS-CoV open reading frame 3a (ORF3a) accessory protein activates the NLRP3 inflammasome by promoting TNF receptor-associated factor 3 (TRAF3)–mediated ubiquitination of apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC). SARS-CoV and its ORF3a protein were found to be potent activators of pro–IL-1β gene transcription and protein maturation, the 2 signals required for activation of the NLRP3 inflammasome. ORF3a induced pro–IL-1β transcription through activation of NF-κB, which was mediated by TRAF3-dependent ubiquitination and processing of p105. ORF3a-induced elevation of IL-1β secretion was independent of its ion channel activity or absent in melanoma 2 but required NLRP3, ASC, and TRAF3. ORF3a interacted with TRAF3 and ASC, colocalized with them in discrete punctate structures in the cytoplasm, and facilitated ASC speck formation. TRAF3-dependent K63-linked ubiquitination of ASC was more pronounced in SARS-CoV–infected cells or when ORF3a was expressed. Taken together, our findings reveal a new mechanism by which SARS-CoV ORF3a protein activates NF-κB and the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of p105 and ASC.—Siu, K.-L., Yuen, K.-S., Castaño-Rodriguez, C., Ye, Z.-W., Yeung, M.-L., Fung, S.-Y., Yuan, S., Chan, C.-P., Yuen, K.-Y., Enjuanes, L., Jin, D.-Y. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC.
Keywords: SARS coronavirus
Both severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome CoV (MERS-CoV) cause a severe and highly lethal respiratory disease in humans characterized by a prominent proinflammatory response (1). A robust elevation of IL-1β was seen during early infection of SARS-CoV (2, 3). IL-1β is a key proinflammatory cytokine generated from pro–IL-1β through proteolytic cleavage by caspase 1, which is activated by multiprotein complexes termed inflammasomes. Inflammasomes assemble around discrete sensor proteins such as NLRP3 and absent in melanoma 2 (AIM2), which are activated as a result of their recognition of pathogen-associated molecular patterns (4). Upon activation, NLRP3 recruits the essential adaptor protein apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) to bind and activate caspase 1 (5). Full activation of the NLRP3 inflammasome requires signal 1, which stimulates pro–IL-1β gene transcription, and signal 2, which promotes pro–IL-1β protein cleavage (6).
Proinflammatory cytokines are double-edged swords that not only mobilize host defense but also drive pathologic inflammation (7). Inflammation can play both antiviral and proviral roles during viral infection. On one hand, it is part of the innate antiviral response that restricts viral replication and infection. On the other hand, it facilitates viral dissemination by releasing a large number of virions. In addition, infiltration of myeloid cells at the site of inflammation results in the spread of viral infection to dendritic cells and macrophages that carry the virus to other places in vivo (7). To facilitate their own infection and to evade host detection, viruses have developed different strategies to modulate inflammasome activation (7, 8). The first example of a viral activator of the NLRP3 inflammasome was found in influenza viruses, in which M2 ion channel protein augments IL-1β maturation (9). Likewise, encephalomyocarditis virus was also shown to activate the NLRP3 inflammasome through its ion channel protein 2B (10). Notably, both M2 and 2B proteins are only sufficient to activate the NLRP3 inflammasome in the presence of signal 1, which induces pro–IL-1β gene transcription. Subsequently, several other viroporins were also found to activate the NLRP3 inflammasome (11). Additional viral inducers of NLRP3 inflammasome activation such as influenza A virus PB1-F2 protein, which is not known to be a viroporin, were also reported (12). It is noteworthy that PB1-F2 induces IL-1β secretion only when signal 1 is activated by LPS.
SARS-CoV might also activate the inflammasome to facilitate viral dissemination and to cause disease. It encodes 3 ion channel proteins: E, open reading frame 3a (ORF3a), and ORF8a (13–15). E and ORF3a are collectively required for viral replication and virulence (16). Particularly, E protein is a virulence factor that activates the NLRP3 inflammasome (13, 17). In light of this, we asked whether ORF3a might also promote virus spreading and infection through inflammasome activation. ORF3a, also known as X1, is a sodium or calcium ion channel protein encoded by the region between S and E (14, 18). It is a lineage-specific accessory protein with 3 transmembrane domains, and it is localized to the Golgi complex (19, 20). ORF3a expression in infected cells is relatively abundant, and anti-ORF3a is also detectable in infected individuals (19). Analysis of a recombinant ORF3a-deficient virus revealed that ORF3a is nonessential for in vitro and in vivo replication but still contributes to viral pathogenesis (21). However, a virus lacking ORF3a and E was nonviable, indicating the complementary function of E and ORF3a (16). Expression of ORF3a induces NF-κB activation, chemokine production, Golgi fragmentation, endoplasmic reticulum stress, accumulation of intracellular vesicles, and cell death (20, 22, 23). Its ion channel activity is required for its proapoptotic property (24). ORF3a is O-glycosylated (25) and interacts with caveolin (26). It down-regulates type I IFN signaling (22) but up-regulates fibrinogen secretion, which promotes the cytokine storm (27). Its induction of chemokines such as IL-8 is mediated through NF-κB (28). Because NF-κB is also the key transcription factor that activates the pro–IL-1β promoter (29), it was of great interest to see whether ORF3a might sufficiently activate both signals required for activation of the NLRP3 inflammasome. To our surprise, although ORF3a was indeed capable of inducing both pro–IL-1β gene transcription and IL-1β protein secretion, this was independent of its ion channel activity. ORF3a was found to associate with TNF receptor-associated factor 3 (TRAF3) and facilitate TRAF3-mediated ubiquitination and activation of p105 and ASC. On one hand, ORF3a promoted p105 ubiquitination and processing, NF-κB activation, and pro–IL-1β gene transcription, providing signal 1 for inflammasome activation. On the other hand, ORF3a associated with TRAF3 and ASC to induce K63-lined polyubiquitination of ASC, leading to the activation of signal 2. Thus, ORF3a suffices to stimulate both signals for NLRP3 inflammasome activation.
MATERIALS AND METHODS
Plasmids
Expression plasmids for pro–IL-1β, NLRP3, and ASC were kindly provided by Pascal Schneider (University of Lausanne, Switzerland) (30). Expression plasmid for procaspase was a gift from Guy Salvesen (Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA, USA). Expression plasmid for cylindromatosis (CYLD) was provided by Shao-Cong Sun (MS Anderson Cancer Center, Houston, TX, USA) (31). Expression plasmids for ubiquitin and its K48 and K63 mutants were provided by James Chen (Southwestern Medical Center, Dallas, TX, USA) (32). Expression plasmid for deubiquitinating enzyme A (DUBA) was subcloned from IMAGE clone 5241560. Expression plasmids for A20 as well as other expression and reporter constructs have been previously described (33–41). ORF3a cDNA was amplified and cloned from cells infected with the GZ50 strain of SARS-CoV (33). Point mutants of ORF3a were constructed by QuikChange XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA).
The bacterial artificial chromosome (BAC) encoding recombinant ORF3a-deficient SARS-CoV was constructed from a previously generated full-length infectious cDNA clone (42). The ORF3a gene deletion was introduced by overlap extension PCR. The first PCR was performed using oligonucleotides 5′-GGCGACATTTCAGGCATTAACGC-3′ and 5′-CATCATAAAGTTATGGGTTCTAGGATTATAAGTTCGTTTATGTGTAATGTAATTTGACACCC-3′. This resulted in a deletion between nt 25270 and 25668 of the SARS-CoV genome. The deletion affects most parts of the ORF3a gene, which does not overlap with the ORF3b gene, disrupts the ATG start codon of the ORF3a gene, and includes 2 point mutations at positions 25673 and 25683 introducing 2 stop codons. A PCR product from nt 24937 to 25694 of the SARS-CoV genome was generated. The second PCR was performed using oligonucleotides 5′-CGAACTTATAATCTAGAACCCATAACTTTATGATGCC-3′ and 5′-CATCATAAATTGGATCCACTGCTGGATTAGCAACTCCTG-3′. The latter primer introduces a point mutation at position 26042, which disrupts a potential initiation codon. A PCR product from nt 25261 to 26060 of the SARS-CoV genome was generated. Both overlapping products were used as templates for PCR amplification using primers 5′-GGCGACATTTCAGGCATTAACGC-3′ and 5′-CATCATAAATTGGATCCACTGCTGGATTAGCAACTCCTG-3′. The final PCR product was cloned into the BAC infectious clone through an intermediate plasmid containing nt 18404–26049 of the SARS-CoV genome (16, 43).
Cell culture, transfection, and infection
Human embryonic kidney 293 (HEK293), A549, and Huh7 cells were grown in DMEM with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in a humidified chamber supplemented with 5% carbon dioxide. HEK293 cells were transfected with GeneJuice (MilliporeSigma, Burlington, MA, USA), and A549 cells were transfected with Lipofectamine 3000 (Thermo Fisher Scientific). Tohoku Hospital Pediatrics-1 (THP-1) cells were maintained in Roswell Park Memorial Institute 1640 medium and transfected with GeneXPLus (American Type Culture Collection, Manassas, VA, USA). Transfection of THP-1 with poly(deoxyadenylic-deoxythymidylic) [poly(dA:dT)] was achieved with Lipofectamine 2000 (Thermo Fisher Sceintific).
Viruses were produced by transfection of the infectious clones into Vero-E6 cells (42, 43). Multiplicity of infection (MOI) was determined as previously described (33, 44). Viral load was measured by real-time quantitative RT-PCR (qRT-PCR) analysis of viral RNA as previously described (33, 34). Angiotensin-converting enzyme 2 (ACE2)–expressing THP-1 cells were infected with SARS-CoV, SARS-CoV-Δ3a, or SARS-CoV-ΔE at an MOI of 0.01 for 1 h at 37°C. Unbound viruses were washed away. Conditioned medium and cell lysates were harvested 24 h post infection. ACE2 expression was reconstituted in HEK293 and TRAF3−/− HEK293 cells before infection with SARS-CoV. All infectious materials were handled in a Biosafety Level 3 facility.
Luciferase reporter assay and protein analysis
Dual luciferase reporter assay, coimmunoprecipitation, and Western blotting were performed as previously described (33, 34). Relative luciferase activity in arbitrary units was calculated by normalizing firefly luciferase activity to Renilla luciferase activity recovered from the same cell lysate. Nuclear fractionation was performed by using a cell fractionation kit supplied by Abcam (Cambridge, United Kingdom). NLRP3 inflammasome activation was measured as described by Wang et al. (45). An oligonucleotide inhibitor of AIM2 named A151 was synthesized by Integrated DNA Technologies (Coralville, IA, USA). Reagents for human IL-1β ELISA were purchased from R&D Systems (Minneapolis, MN, USA). The amounts of IL-1β in the conditioned media were determined as per the manufacturer’s protocol.
Mouse anti-FLAG (clone M2) was purchased from MilliporeSigma. Mouse anti-V5 was from Thermo Fisher Scientific. Mouse anti-HA (clone Y11), anti-TRAF3 and anti–β-tubulin as well as rabbit anti-TRAF6 were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Goat anti-T7, mouse anti-ASC, and rabbit anti-FLAG were from Abcam. Rabbit anti-ASC was bought from Cell Signaling Technology (Danvers, MA, USA). Rabbit antibodies against SARS-CoV N protein were from Novus Biologicals (Centennial, CO, USA). The therapeutic antibody Canakinumab, which blocks IL-1β, was from Creative Biolabs (Shirley, NY, USA).
qRT-PCR
qRT-PCR was carried out essentially as previously described (46, 47). In brief, total RNA was extracted with the RNeasy Mini Kit (Qiagen, Hilden, Germany). cDNA synthesis was achieved with PrimeScript reverse transcription reagent kit with gDNA eraser (Takara Bio, Kusatsu, Japan). Real-time PCR was performed with SYBR Premix Ex Taq Reagents (Takara) in a StepOne real-time PCR system (Thermo Fisher Scientific). Primers were 5′-TCAGCCAATCTTCATTGCTC-3′ (forward) and 5′-GCCATCAGCTTCAAAGAACA-3′ (reverse) for IL-1β mRNA, 5′-CCTAAGGGAGTCCCAGTCCT-3′ (forward) and 5′-TTTTCAAGCTGGCTTTTCGT-3′ (reverse) for ASC mRNA, 5′-TCTGCTCATCACCACGAGAC-3′ (forward) and 5′-CTTGGGCCTCATCAGAGAAG-3′ (reverse) for NLRP3 mRNA, 5′-TAGCGCCTCACGTGTGTTAG-3′ (forward) and 5′-TTGAAGCGTGTTGATCTTCG-3′ (reverse) for AIM2 mRNA, 5′-CTCGACGTCATTTGGGAGAT-3′ (forward) and 5′-ACAGTTTGGGCCACAAAGAC-3′ (reverse) for TRAF3 mRNA, and 5′-TGACACTGGCAAAACAATGCA-3′ (forward) and 5′-GGTCCTTTTCACCAGCAA GCT-3′ (reverse) for human hypoxanthine-guanine phosphoribosyl transferase (HGPRT) mRNA. The normalized value of each sample was derived from the relative quantity of target mRNA divided by the relative quantity of HGPRT mRNA. Relative mRNA expression was derived from 2−ΔΔCt by use of the comparative threshold cycle (ΔΔCt) method.
Construction of TRAF3−/− and TRAF6−/− cells
TRAF3−/− and TRAF6−/− HEK293 cells were constructed by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) editing (48, 49). Guide RNA (gRNA)–Cas9 coexpression plasmid PX459 was a gift from Feng Zhang (McGovern Institute, Cambridge, MA, USA) (48). gRNA sequences for disruption of TRAF3 and TRAF6 loci were 5′-AGCCCGAAGCAGACCGAGTG-3′ (TRAF3-1), 5′-GAAAGACCTGCGAGACCACG-3′ (TRAF3-2), 5′-GTGTGTGTGTTACTTATAA-3′ (TRAF6-1), and 5′-CTTATTGATTTTATGATGC-3′ (TRAF6-2).
HEK293 cells were transiently transfected with plasmids PX459-gRNA-TRAF3-1 plus PX459-gRNA-TRAF3-2 for TRAF3 knockout and with plasmids PX459-gRNA-TRAF6-1 plus PX459-gRNA-TRAF6-2 for TRAF6 knockout. A total of 2 gRNAs were introduced for each locus. At 2 d after transfection, cells were selected in DMEM with 3 mg/ml of puromycin for 3 d. Puromycin was then removed, and cells were allowed to recover for an additional 7 d. Clones plausibly derived from stably transfected cells appeared and were picked by a filter paper presoaked with trypsin-EDTA solution. The paper with cells attached was then transferred to a 6-well plate filled with DMEM containing 10% fetal bovine serum. Cells were allowed to recover and grow for an additional 4 d. Phenotypic verification of the survived clones was performed by Western blotting.
Potential off-target loci of the TRAF3 and TRAF6 gRNAs were predicted by the CHOPCHOP server (http://chopchop.cbu.uib.no/). The top 6–predicted off-target sites for each group of gRNAs were sequenced in HEK293-TRAF3−/− and HEK293-TRAF6−/− cells by Sanger sequencing. The predicted off-target sites were chromosome (chr) 1: 180937934, chr18: 37711090, chr1: 26540372, chr10: 127753518, chr14: 26477465, and chrl13: 9035927 for TRAF3 gRNAs, as well as chr11: 36501562, chr10: 14842691, chr10: 59137886, chr10: 80877608, chr11: 38946885, and chr11: 39287646 for TRAF6 gRNAs. No mutation was found in any of these predicted sites.
RNA interference
The RNA interference (RNAi) experiment was performed as previously described (45, 47). Short interfering RNAs (siRNAs) were transfected into THP-1 cells with Lipofectamine 2000 48 h before infection with SARS-CoV. Supernatant was collected 24 h after infection for measurement of IL-1β by ELISA. Knockdown efficiency of all siRNAs was prevalidated by qRT-PCR analysis of target mRNA. The sequences of the siRNAs were 5′-CAGGUUUGACUAUCUGUUCU-3′ for siNLRP3, 5′-UUUCAGCUUGACUUAGUGGCUUUGG-3′ for siAIM2, 5′-GAUGCGGAAGCUCUUCAGUUUCA-3′ for siASC, and 5′-GAAGGUUUCCUUGUUGCAGAAUGAA-3′ for siTRAF3.
RESULTS
ORF3a and SARS-CoV activate the NLRP3 inflammasome
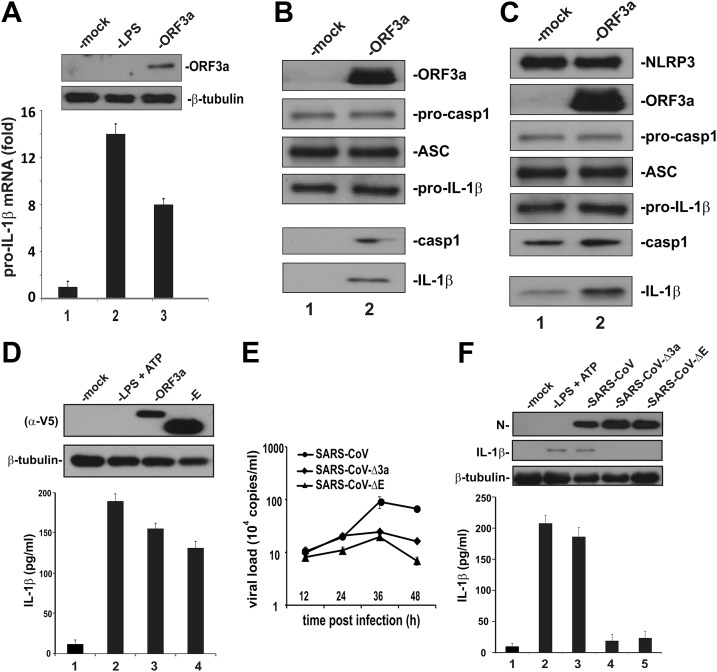
ORF3a is known to activate NF-κB (28), which is one of the key transcription factors driving pro–IL-1β gene expression (29). To interrogate whether ORF3a might induce pro–IL-1β transcription, we expressed ORF3a in human monocytic THP-1 cells. The level of pro–IL-1β transcript was elevated about 8-fold in ORF3a-expressing THP-1 cells (Fig. 1A, bar 3 vs. 1), indicating that expression of ORF3a was sufficient to induce pro–IL-1β gene expression. In light of the recent finding that E protein, which is another ion channel protein encoded by SARS-CoV, is capable of activating the NLRP3 inflammasome (17), we next investigated whether ORF3a could also activate IL-1β secretion in HEK293 cells in which the expression of key factors were reconstituted. Similar reconstitution approaches have been commonly used for mechanistic study of inflammasome activation (5). When ORF3a was expressed in HEK293 cells ectopically expressing pro-caspase 1, pro–IL-1β, ASC, the activation of caspase 1 in the cell lysate, and the production of mature IL-1β in the conditioned medium was detected (Fig. 1B, lane 2 vs. 1). In cells that also expressed NLRP3, ORF3a could also activate caspase 1 and increase IL-1β secretion over the basal level (Fig. 1C, lane 2 vs. 1). Consistently, ELISA detection of secreted IL-1β indicated that the stimulatory effect of ORF3a on IL-1β maturation was comparable to that of SARS-CoV E protein (Fig. 1D, lane 3 vs. 4), which has previously been shown to be capable of inducing inflammasome activation (13, 17). Collectively, our results support the concept that ORF3a activates pro–IL-1β gene transcription and protein maturation, the 2 signals required for optimal activation of the NLRP3 inflammasome.
Figure 1.
Activation of IL-1β production by SARS-CoV and its ORF3a protein. A) Induction of pro–IL-1β transcript by ORF3a. THP-1 cells were either mock-transfected with an empty vector or transfected with an ORF3a expression vector for 24 h. Cells in the control group were treated with 10 ng/ml of LPS for 8 h. The IL-1β transcript was analyzed by qRT-PCR. The level of IL-1β mRNA was relative to that of HGPRT mRNA. Bars represent the means of 3 biologic replicates, and error bars indicate sd. B, C) Induction of IL-1β secretion by ORF3a in a HEK293 reconstitution system. HEK293 cells were transfected with expression plasmids for procaspase 1, ASC and pro–IL-1β. Cells were mock-transfected with an empty vector or cotransfected with ORF3a plasmid alone (B) or NLRP3 + ORF3a plasmids (C) for 24 h. Cell lysates and conditioned medium was collected. Secreted IL-1β in the conditioned medium was immunoprecipitated with anti-FLAG antibody. Cell lysates (upper) and precipitates (lower) were probed by Western blotting. Pro–IL-1β expression was driven by a cytomegalovirus promoter and was therefore nonresponsive to ORF3a. Procasp1, procaspase 1; casp1, caspase 1 (p20). D) Induction of IL-1β secretion by SARS-CoV ORF3a and E proteins in THP-1 cells. THP-1 cells were transfected with ORF3a and E-expression plasmids for 24 h. Cell lysates were collected and probed by Western blotting. Secreted IL-1β in the conditioned medium was analyzed by ELISA. As a positive control, cells were also treated with 10 ng/ml LPS and 5 mM ATP. Both ORF3a and E proteins were V5 tagged and therefore detected with anti-V5 (alpha-V5). E) Virus growth kinetics. THP-1 cells were inoculated with SARS-CoV, SARS-CoV-Δ3a, or SARS-CoV-ΔE at an MOI of 0.01. Samples were harvested at the indicated time points, and viral load was determined by qRT-PCR. Data points represent means of triplicates, and error bars indicate sd. F) Induction of IL-1β secretion by SARS-CoV in THP-1 cells. THP-1 cells were infected with SARS-CoV, SARS-CoV-Δ3a, or SARS-CoV-ΔE (MOI = 0.01) for 24 h. Cell lysates were collected and probed by Western blotting. Secreted IL-1β in the conditioned medium was analyzed by ELISA.
The construction of an infectious clone of SARS-CoV in a BAC provided a tool for genetic analysis of the role of ORF3a in NLRP3 inflammasome activation (42, 43). To this end, ORF3a-deficient SARS-CoV (SARS-CoV-Δ3a) was made and compared with the wild-type (WT) virus and another E-deficient mutant (SARS-CoV-ΔE). The titers of SARS-CoV-Δ3a and SARS-CoV-ΔE viruses were only slightly lower than that of WT at 36 and 48 h post infection (Fig. 1E). Because the titers of the 3 viruses were similar at 24 h post infection (Fig. 1E), we collected cell lysates for analysis of IL-1β secretion within 24 h post infection throughout our whole study. Generally consistent with a previous finding (17), infection of THP-1 cells with SARS-CoV induced the expression and maturation of pro–IL-1β (Fig. 1F, lane 3 vs. 1). Similar to SARS-CoV-ΔE, SARS-CoV-Δ3a lost the ability to induce IL-1β secretion (Fig. 1F, lanes 4, and 5 vs. 3). These results demonstrated the requirement of ORF3a for inflammasome activation by SARS-CoV.
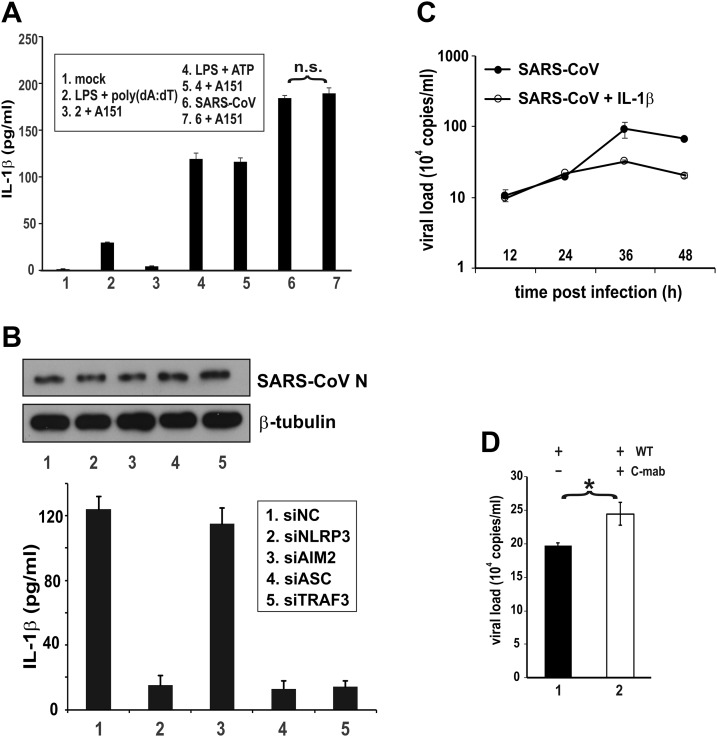
In addition to NLRP3, AIM2 is another upstream sensor that can also activate the inflammasome (4). To investigate the role of AIM2 in SARS-CoV–induced inflammasome activation, a specific oligonucleotide inhibitor of AIM2 known as A151 (50) was employed. As expected, treatment with A151 prevented the induction of IL-1β secretion by AIM2 ligand poly(dA:dT) but had no influence on the effect of ATP (Fig. 2A, bar 3 vs. 2 and bar 5 vs. 4), which is known to activate NLRP3 but not AIM2 (6, 51). In the same setting, A151 did not affect the induction of IL-1β secretion by SARS-CoV (Fig. 2A, bar 7 vs. 6), suggesting that it was not likely mediated through AIM2. Consistent with this, siRNA knockdown of AIM2 had no influence on SARS-CoV–induced IL-1β secretion (Fig. 2B, bar 3 vs. 1). In contrast, compromising NLRP3, ASC, or TRAF3 blunted the stimulatory effect of SARS-CoV in the same setting (Fig. 2B, bar 2, 4, and 5 vs. 1). Whereas ASC is a key adaptor for inflammasome activation (5), TRAF3 is a ubiquitin ligase that induces ASC ubiquitination and activation (52). Thus, our results demonstrated the specific requirements of NLRP3 and ASC for inflammasome activation by SARS-CoV.
Figure 2.
SARS-CoV–induced activation of IL-1β production requires NLRP3, ASC, and TRAF3. A) Sensitivity of SARS-CoV–induced IL-1β secretion to AIM2 inhibitor. THP-1 cells were treated, transfected, or infected with the indicated stimuli for 24 h; cells in groups 2 and 3 were transfected with 1 μg/ml poly(dA:dT) for 3 h. Cells in groups 3, 5, and 7 were also treated with 1 μM A151. Secreted IL-1β in the conditioned medium was measured by ELISA. The difference between the indicated groups was not significant (n.s.) by Student’s t test. B) Sensitivity of SARS-CoV–induced IL-1β secretion to NLRP3, AIM2, ASC, and TRAF3 knockdown. THP-1 cells were transfected with the indicated siRNA for 48 h and then infected with SARS-CoV for 24 h. Secreted IL-1β in the conditioned medium was measured by ELISA. siNC: negative control siRNA. C) Effect of IL-1β on SARS-CoV replication. Huh7 cells were infected with SARS-CoV at an MOI of 0.01. Cells were mock-treated or treated with 10 ng/ml recombinant IL-1β. Samples were harvested at the indicated time points for qRT-PCR analysis of viral load. D) Effect of anti–IL-1β on SARS-CoV replication. Huh7 cells were infected with SARS-CoV WT at an MOI of 0.01 for 24 h. Cells were mock-treated or treated with 10 ng/ml Canakinumab (C-mab). Viral load was measured by qRT-PCR. The differences between the indicated groups were statistically significant Student’s t test. *P < 0.05.
Inflammasome activation may play a critical role in the control of viral replication (8, 53). Indeed, treatment of SARS-CoV–infected Huh7 cells with IL-1β had a moderate inhibitory effect on viral replication at 36 and 48 h post infection (Fig. 2C). In keeping with this, treatment of cells with an IL-1β blocking antibody named Canakinumab mildly increased the replication of SARS-CoV WT (Fig. 2D, bar 2 vs. 1). Thus, inflammasome activation could exert a weak suppressive effect on SARS-CoV replication.
ORF3a activates NF-κB by facilitating p50 maturation
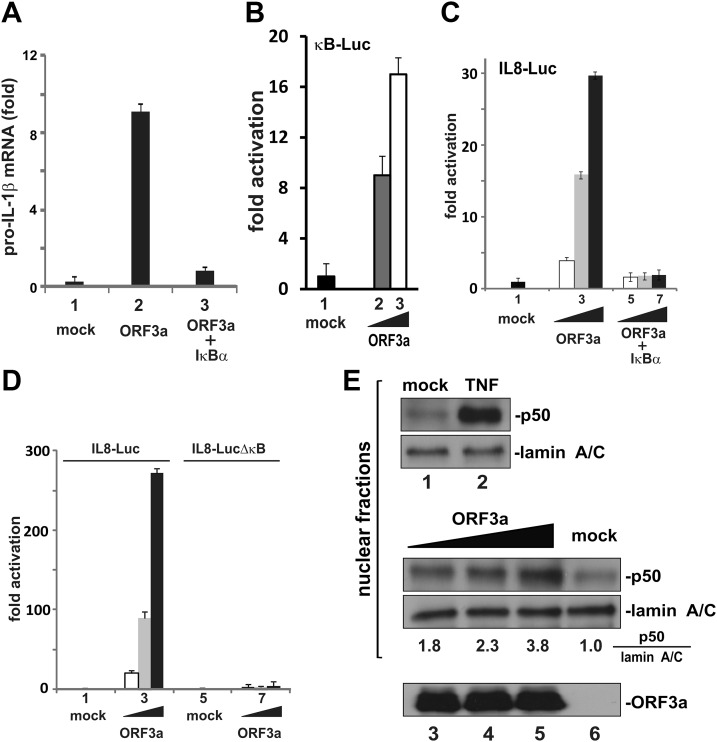
The activation of pro–IL-1β gene transcription by ORF3a was found to be mediated through NF-κB because the activity was abrogated in the presence of a dominant active S32A S36A mutant of IκBα, also known as IκBα superrepressor (Fig. 3A, bar 3 vs. 2). Indeed, ORF3a activated NF-κB–dependent expression of a luciferase reporter in a dose-dependent fashion (Fig. 3B, bars 2 and 3 vs. 1). Consistent with a previous report (28), ORF3a-induced activation of IL-8 promoter was also prevented by the IκBα superrepressor or by deletion of κB elements in the promoter region (Fig. 3C, D). Because a suitable IL-1β–Luc reporter plasmid driven by NF-κB was not available, the IL-8–Luc reporter, which was highly responsive to ORF3a in an NF-κB–dependent manner, was employed in our subsequent mechanistic studies.
Figure 3.
Activation of NF-κB by ORF3a. A) Activation of pro–IL-1β gene transcription by ORF3a requires NF-κB. Coexpression of IκB-α superrepressor (i.e., S32A S36A mutant) with ORF3a in THP-1 cells erased ORF3a-induced elevation of the pro–IL-1β transcript. The mean ± sd from 3 biologic replicates is presented. B) Activation of κB-Luc by ORF3a in HEK293 cells. C) Coexpression of the IκB-α superrepressor with ORF3a in HEK293 cells blunted ORF3a-induced activation of the IL-8 promoter. Dual luciferase assay was performed. Normalized results represent 3 biologic replicates, and error bars indicate sd. D) Deletion of the κB element from the IL-8 promoter prevented its activation by ORF3a in HEK293 cells. E) ORF3a facilitated p50 processing in HEK293 cells. Cells were transfected with increasing doses of ORF3a plasmid. Cells in the control group were treated with 30 ng/ml of TNF-α. Cell fractionation was performed, and nuclear fractions were collected. Proteins were detected by Western blotting. Quantitative analysis of the relative amount of p50 normalized to that of lamin A/C was obtained by densitometry, and the ratios are provided below the gel panel. ORF3a in the cytoplasm was also probed.
We assessed the steady-state levels of different NF-κB and IκB proteins in ORF3a-expressing HEK293 cells and noticed the accumulation of prominent amounts of p50 in the nucleus (Fig. 3E, lanes 3–5 vs. 6). The p65-p50 heterodimer is the most common and best-studied version of NF-κB. The p50 subunit was proteolytically processed from the p105 precursor residing in the cytoplasm (54). Plausibly, ORF3a might affect p105 processing, leading to activation of the canonical NF-κB signaling pathway.
Identification and characterization of a TRAF binding domain in ORF3a
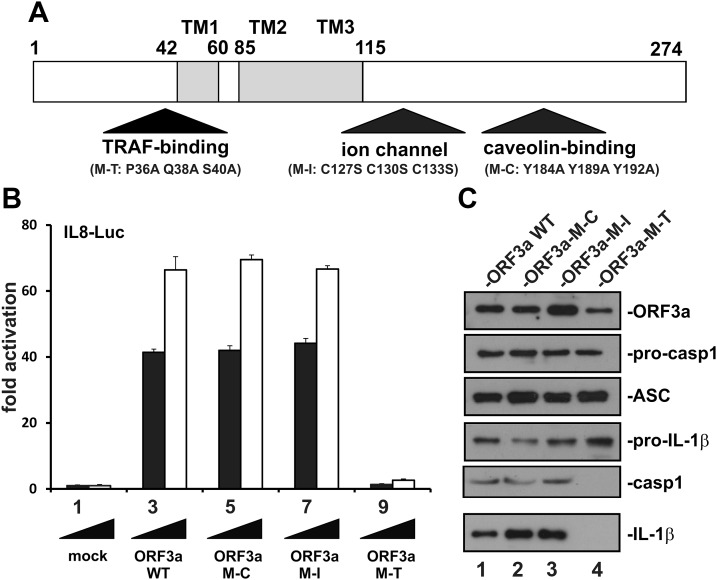
ORF3a contains an ion channel domain and a caveolin binding domain (14, 26). Consistent with the requirement of TRAF3 for SARS-CoV–induced activation of inflammasome (Fig. 2B), our bioinformatic analysis revealed another TRAF binding domain resembling that in LMP1 oncoprotein of Epstein-Barr virus and other TRAF-binding proteins (55). A TRAF-binding motif, PLQAS, consistent with the consensus sequence PxQx(T/S/D) in which x could be any residue, was located at aa 36–40 of ORF3a. To define their necessity for the NF-κB- and inflammasome-activating activity of ORF3a, 3 point mutants designated M-T, M-I, and M-C were created to disrupt the TRAF binding, ion channel, and caveolin binding domains, respectively (Fig. 4A). In M-T, 3 conserved residues (P, Q, and S) in the TRAF-binding motif were changed into A. In M-I, 3 conserved C residues required for ion channel activity (14, 24) were changed into S. Finally, 3 conserved Y residues in the caveolin-binding motif were changed into A in M-C.
Figure 4.
Definition of a TRAF binding domain in ORF3a required for activation of NF-κB and IL-1β secretion. A) Functional domains in ORF3a. Point mutants (M-T, M-I, and M-C) in which the TRAF-binding, ion channel and caveolin binding domains are individually disrupted are indicated. TM, transmembrane domain. B) Requirement of TRAF binding domain for ORF3a-induced activation of IL-8 promoter. Increasing doses of ORF3a (WT and mutants) were expressed for 24 h in HEK293 cells. Dual-luciferase reporter assay was performed. Bars represent the means of 3 biologic replicates, and error bars indicate sd. C) Requirement of the TRAF binding domain for ORF3a-induced secretion of IL-1β in HEK293 reconstitution system. Pro–IL-1β expression was driven by a cytomegalovirus promoter and therefore nonresponsive to ORF3a. Procasp1, procaspase 1; casp1, caspase 1 (p20).
Interestingly, the M-C and M-I mutants were fully competent in the activation of NF-κB and the NLRP3 inflammasome (Fig. 4B, bars 5–8 vs. 3 and 4; Fig. 4C, lanes 2 and 3 vs. 1). In sharp contrast, the M-T mutant lost its ability to activate either IL-8–Luc or IL-1β secretion (Fig. 4B, bars 9 and 10 vs. 3 and 4; Fig. 4C, lane 4 vs. 1). In other words, TRAF binding was essential for NF-κB and NLRP3 inflammasome activation by ORF3a. Surprisingly, neither caveolin binding nor the ion channel was required for this activity.
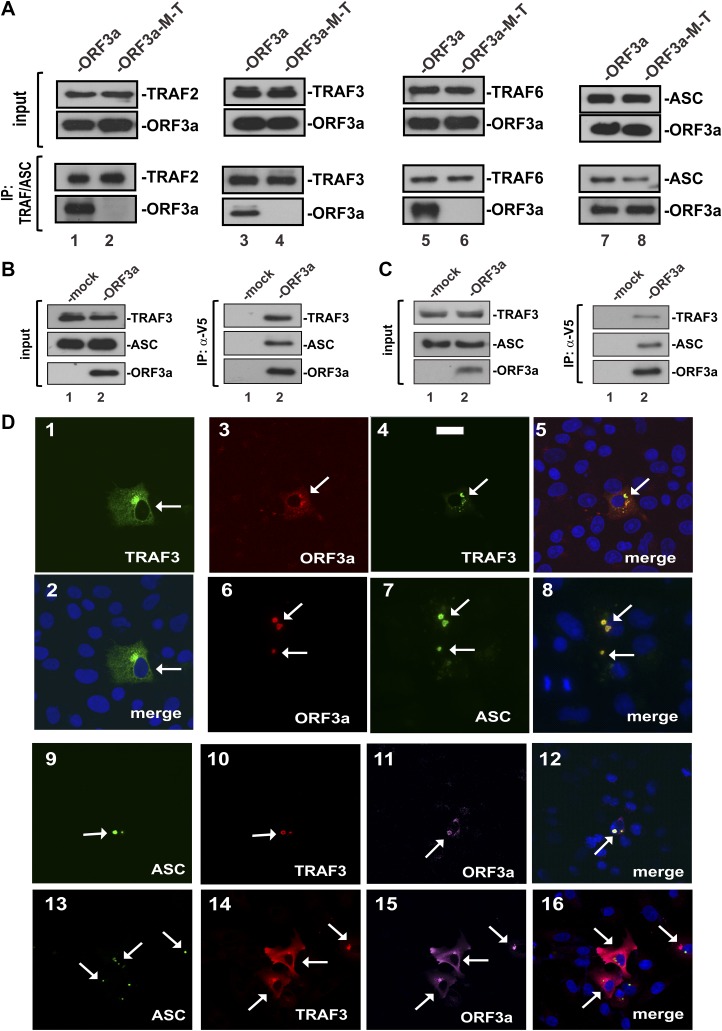
To verify the binding of ORF3a to TRAF proteins, we performed coimmunoprecipitation with lysates of HEK293 cells ectopically expressing differentially tagged ORF3a and TRAF proteins. A total of 3 representative TRAF proteins, which are known to be critically involved in NF-κB activation (54), were analyzed in our experiment. When we immunoprecipitated TRAF2, 3, and 6 with an anti-FLAG antibody, ORF3a but not its M-T mutant was detected in the precipitate (Fig. 5A, lanes 1, 3, and 5 vs. 2, 4, and 6). These results indicated that ORF3a was capable of interacting with TRAF2, 3, and 6, but this ability was abrogated in the M-T mutant. Because TRAF proteins are also known to interact with and activate ASC (52, 56), we also examined the association of ORF3a with ASC. To our surprise, both ORF3a and its M-T mutant could bind with ASC (Fig. 5A, lanes 7 and 8). This suggested that ORF3a likely interacted with ASC directly through a separate domain.
Figure 5.
Association of ORF3a with TRAF3 and ASC. A) The TRAF binding domain is required for the association of ORF3a with TRAF2, 3, and 6 and ASC. The indicated proteins were expressed in HEK293 cells for 24 h. Cell lysates were collected for immunoprecipitation (IP). TRAF proteins were precipitated with mouse anti-FLAG. ASC was pulled down with mouse anti-ASC. Input lysates and precipitates were analyzed by Western blotting. TRAF proteins were detected with rabbit anti-FLAG. ASC was probed with rabbit anti-ASC. B) Reciprocal immunoprecipitation and Western blotting. ORF3a was expressed in HEK293 cells for 24 h. V5-tagged ORF3a was immunoprecipitated with anti-V5. TRAF3 was probed with mouse anti-TRAF3. ASC was detected with rabbit anti-ASC. C) Association of ORF3a with endogenous TRAF3 and ASC. THP-1 cells were transfected with ORF3a plasmid. Immunoprecipitation of ORF3a was performed with anti-V5 (a-V5). Precipitates were probed for endogenous TRAF3 and ASC as above. D) Colocalization of ORF3a with TRAF3 and ASC. The indicated proteins were ectopically expressed in A549 cells for 24 h. Cells were fixed and stained for the overexpressed proteins using anti-tag antibodies. TRAF3 was stained with anti-FLAG. ORF3a was probed with anti-V5. ASC was detected with anti-T7. Fluorescent signals of different colors were merged with nuclear morphology in blue revealed by DAPI staining. Transfected cells are highlighted by arrows. Scale bar, 20 µm.
Reciprocally, the presence of TRAF3 and ASC in the ORF3a-containing immunoprecipitate also lent support to the interaction of ORF3a with TRAF3 and ASC (Fig. 5B, lane 2 vs. 1). Furthermore, when the same experiment was repeated in ORF3a-expressing THP-1 cells, endogenous TRAF3 and ASC were also found to be associated with ORF3a (Fig. 5C, lane 2 vs. 1).
Consistent with the immunoprecipitation results, ORF3a was found to colocalize substantially with TRAF3 and ASC in A549 cells to cytoplasmic punctate structures (Fig. 5D). A549 cells are lung adenocarcinoma cells derived from alveolar basal epithelium, which can be infected by MERS-CoV. They have been widely used as a model for lung cell biology (57) and were therefore employed for the analysis of ORF3a, TRAF3, and ASC localization in our study. TRAF3 was concentrated in a discrete cytoplasmic subdomain in A549 cells in the absence of ORF3a (Fig. 5D1, 2). Both TRAF3 and ASC were also found in the cytoplasmic dots that also harbored ORF3a (Fig. 5D3–16). These dots were generally consistent with the formation of ASC specks required for full activation of inflammasome (58).
TRAF3 is required for ORF3a-induced activation of NF-κB
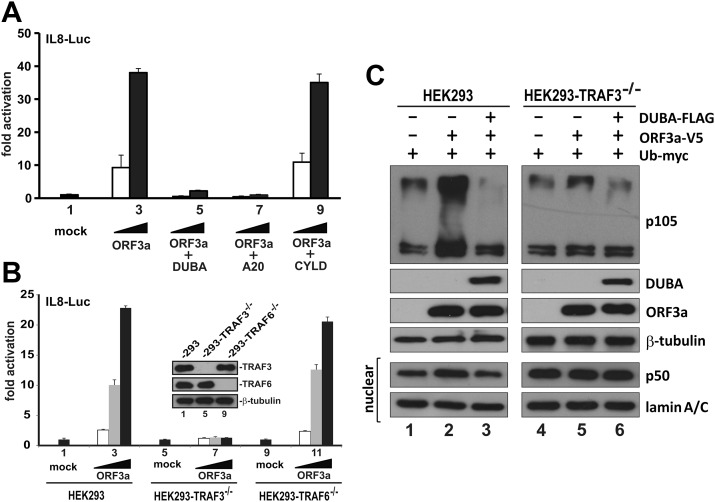
To determine which TRAF proteins might be important in ORF3a-induced activation of NF-κB, we harnessed 3 deubiquitinases that are known to deubiquitinate and inhibit TRAF proteins with some specificity. DUBA is specific for TRAF3 (59). A20 acts on both TRAF3 and TRAF6 (60, 61). TRAF2 is a substrate of CYLD, but CYLD also inhibits TRAF6 to a lesser extent (31, 62). When it was coexpressed with CYLD, ORF3a could still activate IL-8–Luc reporter expression equally well (Fig. 6A, bars 8 and 9 vs. 2 and 3). In contrast, the NF-κB–activating activity of ORF3a on IL-8–Luc was abolished when either DUBA or A20 was expressed (Fig. 6A, bars 4–7 vs. 2 and 3). Taken together, whereas TRAF2 was dispensable for ORF3a-induced activation of NF-κB, TRAF3 was probably required for this activity. Because CYLD is a weak inhibitor of TRAF6, it remained to be determined whether TRAF6 activity might also be essential for the activation of NF-κB by ORF3a.
Figure 6.
ORF3a-induced activation of NF-κB requires TRAF3. A) Effect of TRAF deubiquitinases on ORF3a-induced activation of IL-8 promoter in HEK293 cells. B) Loss of ORF3a activity on IL-8 promoter in TRAF3−/− HEK293 cells. Expression of TRAF3 and TRAF6 proteins was verified by Western blotting (inset). Similar results were also obtained from 2 other clones of TRAF3−/− and TRAF6−/− HEK293 cells. C) TRAF3-dependent enhancement of p105 ubiquitination and processing by ORF3a. HEK293 and TRAF3−/− HEK293 cells were transfected with the indicated combinations of expression plasmids for DUBA, ORF3a, and ubiquitin (Ub). Cells were harvested for nuclear fractionation (lower 2 panels) and Western blot analysis 24 h post-transfection.
CRISPR/Cas9 editing provides a new tool for loss-of-function studies in mammalian cells (48). This technology was used to construct TRAF3−/− and TRAF6−/− HEK293 cells (Fig. 6B, inset). The activation of NF-κB by ORF3a as measured by the reporter activity of IL-8–Luc was totally unaffected in TRAF6−/− HEK293 cells (Fig. 6B, bars 10–12 vs. 2–4). However, this activity of ORF3a was eliminated in TRAF3−/− HEK293 cells (Fig. 6B, bars 6–8 vs. 2–4). Hence, TRAF3 but not TRAF6 or TRAF2 was required for ORF3a-induced activation of NF-κB.
We demonstrated the accumulation of p50 upon expression of ORF3a (Fig. 3E). To shed light on p105 ubiquitination and processing as well as the requirement of TRAF3 for this modification, we overexpressed ubiquitin, ORF3a, and DUBA in HEK293 and TRAF3−/− HEK293 cells. In keeping with earlier results, expression of ORF3a resulted in the accumulation of p50 in the nuclear fraction (Fig. 6C, lane 2 vs. 1). This accumulation was abrogated by the expression of DUBA deubiquitinase or in TRAF3−/− cells (Fig. 6C, lane 3 vs. 1 and lane 5 vs. 4). Consistently, the ubiquitination ladder of p105 was also more prominent when ORF3a was expressed and the phenotype was not seen in the presence of DUBA or when TRAF3 was knocked out (Fig. 6C). Taken together, our results supported the notion that ORF3a augmented TRAF3-dependent ubiquitination and consequent processing of p105.
ORF3a activates the NLRP3 inflammasome by promoting TRAF3-mediated ASC ubiquitination
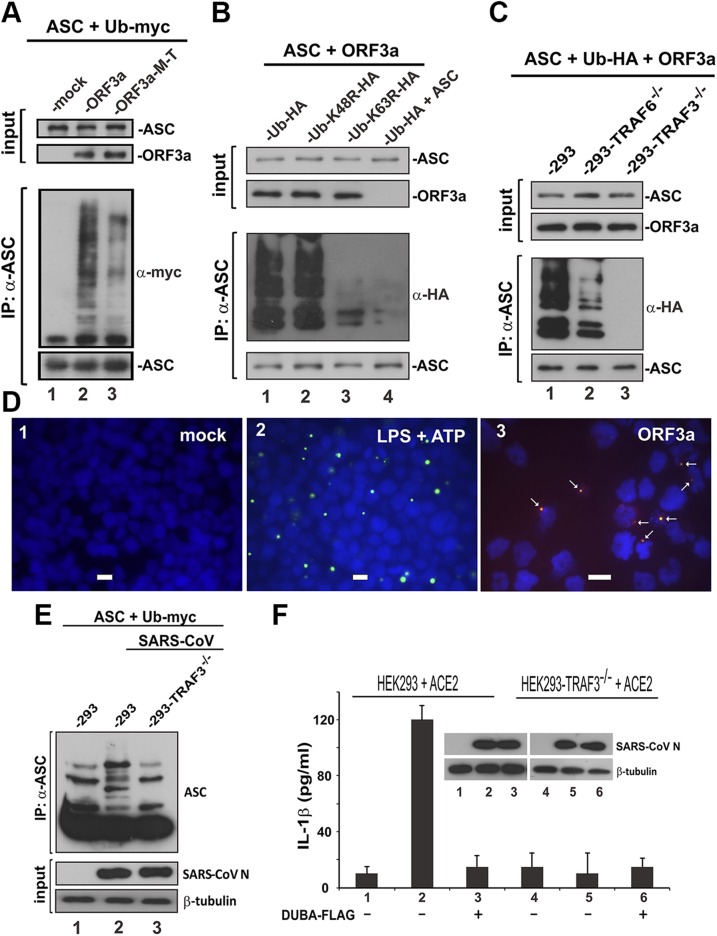
As previously described, we showed the association of ORF3a with TRAF3 and ASC. Because TRAF3 is a ubiquitin ligase catalyzing ASC ubiquitination (52), we asked whether and how ORF3a might affect polyubiquitination of ASC. An in vivo polyubiquitination assay was performed in HEK293 cells. Polyubiquitinated ASC was immunoprecipitated using anti-ASC and probed with anti-myc recognizing myc-tagged ubiquitin. A prominent polyubiquitination ladder indicated that expression of ORF3a sufficiently induced polyubiquitination of ASC (Fig. 7A, lane 2 vs. 1). We also noted that ASC polyubiquitination was less pronounced when the M-T mutant of ORF3a was expressed (Fig. 7A, lane 3 vs. 2), in support of the requirement of TRAF binding for optimal induction of ASC ubiquitination by ORF3a.
Figure 7.
Promotion of TRAF3-dependent ubiquitination of ASC by ORF3a. A) ORF3a promoted ASC polyubiquitination. The indicated proteins were expressed in HEK293 cells for 24 h. Cell lysates were collected, and ASC was immunoprecipitated (IP); a-ASC, anti-ASC; a-myc, anti-myc. B) ORF3a promoted K63-linked polyubiquitination of ASC; a-HA, anti-HA. C) ORF3a-facilitated polyubiquitination of ASC was not seen in TRAF3−/− HEK293 cells. D) ORF3a augmented ASC speck formation. THP-1 cells were transfected with ORF3a expression plasmid for 24 h. Cells in the positive control group (panel 2) were treated with 10 ng/ml LPS and 5 mM ATP. Cells were spun down onto the slide by centrifuging at 1000 rpm and then fixed with ice-cold 50% methanol/50% acetone. Cells were stained for ORF3a with anti-V5 (red) and for endogenous ASC (green). Nuclear morphology was revealed by DAPI (blue). Fluorescent signals were merged, and colocalization of ORF3a and ASC appears in yellow. Scale bars, 20 μM. E) ASC ubiquitination in infected cells. HEK293 and TRAF3−/− HEK293 cells were transfected with expression plasmids for ubiquitin (Ub), SARS-CoV receptor ACE2, and ASC for 24 h. Cells were infected with SARS-CoV for 1 h and harvested for the immunoprecipitation of ASC at 24 h postinfection. F) IL-1β secretion from infected cells. HEK293 and TRAF3−/− HEK293 cells were transfected with ACE2 expression plasmid. DUBA was also expressed in cells in groups 3 and 6. Cells were infected with SARS-CoV for 1 h at 24 h post-transfection. Then, cell lysates and conditioned media were collected at 24 h post infection for the Western blot analysis and ELISA, respectively.
Because TRAF3-induced K63-linked ubiquitination of ASC is required for NLRP3 inflammasome activation (52), we verified the type of ASC ubiquitination in ORF3a-expressing cells. A prominent ASC polyubiquitination ladder was evident only when ubiquitin WT and its K48R mutant was expressed (Fig. 7B, lanes 1 and 2 vs. 4). The K63R mutant of ubiquitin could not support ORF3a-induced polyubiquitination of ASC (Fig. 7B, lane 3 vs. 1 and 2). These results were consistent with the model in which ORF3a interacts with TRAF3 and ASC to facilitate TRAF3-mediated K63-linked ubiquitination of ASC.
To determine the requirement of TRAF3 for ORF3a-induced ubiquitination of ASC, the polyubiquitination assay was repeated in TRAF6−/− and TRAF3−/− HEK293 cells. The polyubiquitination ladder of ASC was only slightly less prominent in TRAF6−/− cells but completely disappeared in TRAF3−/− cells (Fig. 7C, lanes 2 and 3 vs. 1). In other words, TRAF3 but not TRAF6 was essential for ORF3a-induced ubiquitination of ASC, a critical step in NLRP3 inflammasome activation.
ASC speck formation is a critical step in NLRP3 inflammasome activation (58, 63). Indeed, treatment of THP-1 cells with LPS and ATP resulted in the visualization of prominent ASC specks (Fig. 7D2). With this in mind, we sought to explore the functional outcome of ORF3a-induced ASC ubiquitination. In line with the results obtained from A549 cells (Fig. 5D), expression of ORF3a in THP-1 cells led to the formation of ASC specks (Fig. 7D3) to which ORF3a and ASC colocalized. These results were compatible with the model that SARS-CoV ORF3a facilitates the assembly of ASC specks required for optimal activation of the NLRP3 inflammasome.
To verify ASC ubiquitination in SARS-CoV–infected cells, we reconstituted the expression of ACE2 receptor for SARS-CoV in HEK293 cells and their TRAF3−/− mutants. Indeed, when ASC was immunoprecipitated from these ACE2-reconstituted cells infected with SARS-CoV, ubiquitinated forms of ASC were observed only in HEK293 and not TRAF3−/− HEK293 cells (Fig. 7E, lane 2 vs. 1 and 3). Furthermore, IL-1β secretion was robust in SARS-CoV–infected HEK293 cells but not the TRAF3−/− counterparts or when DUBA was also expressed (Fig. 7F, bar 2 vs. 3 and 5). Thus, SARS-CoV induced the ubiquitination of ASC and secretion of IL-1β in a TRAF3-dependent manner.
DISCUSSION
In this study, we reported activation of the NLRP3 inflammasome by SARS-CoV ORF3a protein. ORF3a sufficiently activated both pro–IL-1β gene expression and IL-1β secretion (Fig. 1A–D). This notion was further supported by our infection study, in which an impairment in pro–IL-1β gene transcription and protein maturation was seen in cells infected with SARS-CoV-Δ3a mutant virus (Fig. 1F). The inflammasome-activating property of SARS-CoV required NLRP3 and ASC, but not AIM2 (Fig. 2A, B). In addition, SARS-CoV replication was moderately affected when cells were treated with IL-1β but augmented upon addition of an IL-1β blocking antibody (Fig. 2C, D). ORF3a induced pro–IL-1β gene transcription through NF-κB (Fig. 3A). It promoted ubiquitination and processing of p105 in the cytosol (Fig. 6C), leading to the accumulation of p50 in the nucleus (Fig. 3E). A TRAF binding domain was identified in ORF3a (Fig. 4). TRAF3 was found to be required for ORF3a-induced activation of NF-κB and the NLRP3 inflammasome (Figs. 4–6). ORF3a interacted with TRAF3 and ASC to mediate K63-linked polyubiquitination of ASC, leading to the formation of ASC specks required for NLRP3 inflammasome activation (Figs. 5 and 7). Importantly, ASC ubiquitination and IL-1β secretion occurred in a TRAF3-dependent manner in infected cells (Fig. 7). These findings were summarized in our working model (Fig. 8).
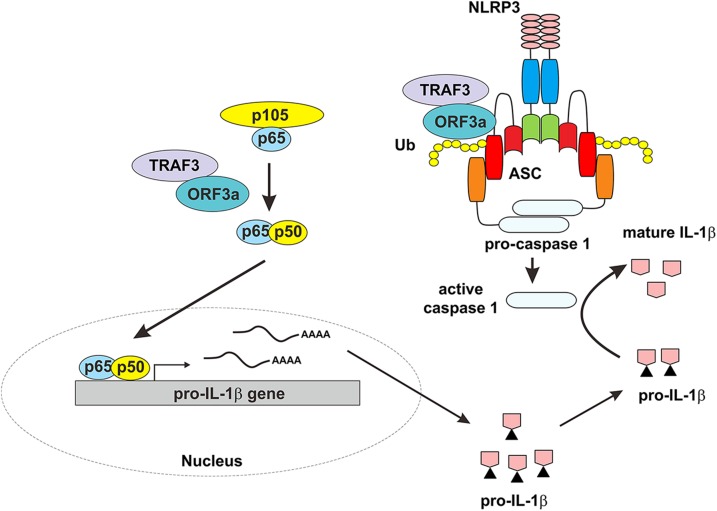
Figure 8.
A working model for ORF3a-induced activation of the NLRP3 inflammasome. On one hand, ORF3a interacts with TRAF3 to activate NF-κB, resulting in transcription of the pro–IL-1β gene. On the other hand, ORF3a interacts with TRAF3 to promote ASC ubiquitination, leading to activation of caspase 1 and IL-1β maturation. ORF3a sufficiently activates both signals required for NLRP3 inflammasome activation. Ub, ubiquitin.
Induction of a proinflammatory cytokine storm is a hallmark of SARS-CoV and MERS-CoV infection (1, 64). Understanding the mechanism by which these and other highly pathogenic human viruses potently induce the expression of proinflammatory cytokines might provide the clues for therapeutic intervention. In contrast to previously identified viral activators of the NLRP3 inflammasome, including influenza A virus M2 and PB1-F2 (9, 12), which can only function when pro–IL-1β gene transcription is activated by stimuli such as LPS, SARS-CoV ORF3a is capable of activating pro–IL-1β gene transcription and protein maturation, the 2 signals required for NLRP3 inflammasome activation. Mechanistically, ORF3a fulfills this function by activating NF-κB and TRAF3-dependent ubiquitination of ASC, which is necessary for NLRP3 inflammasome assembly (52). ORF3a interacts with ASC, and expression of ORF3a alone in the absence of NLRP3 could sufficiently activate IL-1β secretion. This raises the possibility that ORF3a could act as a scaffold for ASC speck formation and inflammasome activation to some extent. Our RNAi experiment demonstrated the requirement of NLRP3 and ASC for SARS-CoV–induced inflammasome activation (Fig. 2B). Our inhibitor test also suggested that SARS-CoV–induced inflammasome activation is not likely mediated by AIM2 (Fig. 2A). However, it will still be of interest to determine whether ORF3a might also promote ASC inflammasome activation mediated by other upstream sensors such as NLRP1 and pyrin. Results from 1 recent study conducted in reconstituted HEK293T cells are generally consistent with our finding that ORF3a activates NLRP3, but multiple mechanisms, including ASC-independent activation, were suggested in that study (65). Further investigations are required to clarify whether ORF3a might activate the inflammasome through other pathways. SARS-CoV E protein is another activator of the NLRP3 inflammasome, and IL-1β production is also compromised in SARS-CoV-ΔE (Fig. 1E and Ref. 17). Whereas the remaining activity for NLRP3 inflammasome activation in cells infected with SARS-CoV-ΔE might be ascribed to ORF3a, the lack of NLRP3 inflammasome activation by E in cells infected with SARS-CoV-Δ3a could be a result of no activation of pro–IL-1β gene transcription or the difference in experimental setting. The nonviable phenotype of the double-knockout virus indicated that ORF3a and E might jointly exert a major impact on viral replication (16). Further investigations are required to elucidate how ORF3a cooperates with E and other viral proteins to execute full induction of the cytokine storm. The activation of the inflammasome by SARS-CoV occurred in HEK293 cells even without full reconstitution of the expression of procaspase 1, ASC, and pro–IL-1β (Fig. 7F). Apparently, SARS-CoV is a more potent and promiscuous activator of inflammasome than its ORF3a protein alone. Inflammasome activation by SARS-CoV is thought to have both proviral and antiviral roles. It might contribute to innate antiviral response, the viral induction of cytokine storm, and pathogenic inflammation. Indeed, whereas IL-1β had a modest antiviral effect on SARS-CoV (Fig. 2C), anti–IL-1β exhibited a weak stimulatory effect (Fig. 2D). The positive effect of inflammasome activation on SARS-CoV dissemination in vivo and the exact biologic significance of ORF3a-induced NLRP3 inflammasome in SARS-CoV biology and pathogenesis remain to be elucidated in future study.
Most of the previously characterized viral activators of the NLRP3 inflammasome are ion channel proteins (11). Although ORF3a was originally thought to be an activator of the NLRP3 inflammasome because it is an ion channel protein, the ion channel mutant (M-I) of ORF3a was fully competent in the activation of NF-κB and the NLRP3 inflammasome. This provides an example of how viral ion channel proteins might activate inflammasomes in an ion channel–independent manner. Because the ORF3a-M-I mutant is also deficient for oligomerization (14), our results also suggested that oligomerization of ORF3a is dispensable for the activation of NF-κB or the NLRP3 inflammasome. Opposite to our finding, SARS-CoV ORF3a has recently been shown to activate the NLRP3 inflammasome in an ion channel–dependent manner (66). Surprisingly, the same ORF3a-M-I mutant was found to have lost the ability to activate inflammasome in mouse bone marrow–derived macrophages in the other study (66). However, whether that mutant was robustly expressed in their experimental setting remains to be established. Nevertheless, further investigations to be conducted in the same experimental system are required to resolve this discrepancy.
The requirement of K63-linked ubiquitination of ASC for optimal inflammasome activation has previously been demonstrated (52). TRAF3-mediated K63-linked ubiquitination of ASC is thought to facilitate ASC aggregation and speck formation in the cytoplasm. The strategy to facilitate TRAF3-induced ubiquitination of ASC through interaction with TRAF3 and ASC could also be used by other viruses and viral proteins. For example, we have previously shown the interaction of SARS-CoV and MERS-CoV M proteins with TRAF3 (33–35). It will therefore be of interest to see whether these 2 viral proteins might also modulate TRAF3-mediated ubiquitination of ASC. We demonstrated the first example of viral activation of ASC in this work. Plausibly, this could also be a common viral strategy to usurp host inflammasomes for viral benefits. In support of this notion, ASC-null mice were highly susceptible to West Nile virus infection (67). Mechanistically, how ORF3a activates TRAF3-mediated ASC ubiquitination remains to be determined. ORF3a likely used a separate domain to interact with ASC. Further analysis is warranted for the possibility that ORF3a adapts ASC to TRAF3.
Consistent with the idea that ORF3a activates pro–IL-1β gene transcription through NF-κB, the p50 subunit of NF-κB was found to accumulate in the nucleus of ORF3a-expressing cells. p50 is proteolytically processed from p105 when the latter is ubiquitinated by E3 ubiquitin ligases such as KPC1 (68, 69). Although TRAF3 is not known to mediate p105 ubiquitination, this possibility merits reexamination in light of our finding that TRAF3 is also required for ORF3a-induced activation of NF-κB. Reciprocally, it will also be of interest to see whether ORF3a might interact with KPC1 and modulate its activation of p105 processing.
The interaction between ORF3a and TRAF3 is required for ORF3a-induced activation of NF-κB and the NLRP3 inflammasome. The key residues for this interaction were mapped, and their essentiality was shown by mutational analysis. Peptide mimetics based on these residues are potentially useful modulators of NF-κB and NLRP3 inflammasome activation. In addition, small-molecule compounds that perturb this interaction could be further analyzed for antiviral or anti-inflammatory effect.
ACKNOWLEDGMENTS
The authors thank Lilong Jia (Department of Microbiology, The University of Hong Kong) for assistance with the infection study. This work was supported by the Hong Kong Health and Medical Research Fund (13121032, 14130822, and HKM-15-M01), Hong Kong Research Grants Council (T11-707/15-R, C7011-15R, and 17124415), Government of Spain (BIO2013-42869-R), U.S. National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases (2P01AI060699-06A1), and European Commission (Innovative Medicines Initiative Grant Agreement 115760). C.C.-R. received a fellowship from Fundacion La Caixa. K.-L.S., K.-S.Y., and C.C.-R. are co-first authors. The authors declare no conflicts of interest.
Glossary
- ACE2
angiotensin-converting enzyme 2
- AIM2
absent in melanoma 2
- ASC
apoptosis-associated speck-like protein containing a caspase recruitment domain
- BAC
bacterial artificial chromosome
- Cas9
CRISPR-associated protein 9
- chr
chromosome
- CRISPR
clustered regularly interspaced short palindromic repeats
- CYLD
cylindromatosis
- DUBA
deubiquitinating enzyme A
- gRNA
guide RNA
- HEK293
human embryonic kidney 293
- HGPRT
hypoxanthine-guanine phosphoribosyltransferase
- MERS-CoV
Middle East respiratory syndrome coronavirus
- MOI
multiplicity of infection
- ORF
open reading frame
- poly(dA:dT)
poly(deoxyadenylic-deoxythymidylic)
- qRT-PCR
quantitative RT-PCR
- RNAi
RNA interference
- SARS-CoV
severe acute respiratory syndrome coronavirus
- siRNA
short interfering RNA
- THP-1
Tohoku Hospital Pediatrics-1
- TRAF
TNF receptor-associated factor
- WT
wild type
AUTHOR CONTRIBUTIONS
K.-L. Siu, K.-S. Yuen, C. Castaño-Rodriguez, Z.-W. Ye, M.-L. Yeung, C.-P. Chan, K.-Y. Yuen, L. Enjuanes, and D.-Y. Jin conceptualized and designed the study; K.-L. Siu and K.-S. Yuen performed most experiments with help from Z.-W. Ye and S.-Y. Fung; C. Castaño-Rodriguez and L. Enjuanes created and characterized ORF3a-deficient and E-deficient viruses; Z.-W. Ye, M.-L. Yeung, and S. Yuan performed the infection study; all authors contributed to data analysis; and D.-Y. Jin wrote the manuscript with input from all authors.
REFERENCES
- 1.Chan J. F., Lau S. K., To K. K., Cheng V. C., Woo P. C., Yuen K. Y. (2015) Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 28, 465–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang B. S., Chan K. H., Cheng V. C., Woo P. C., Lau S. K., Lam C. C., Chan T. L., Wu A. K., Hung I. F., Leung S. Y., Yuen K. Y. (2005) Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J. Virol. 79, 6180–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Lang A., Baas T., Teal T., Leijten L. M., Rain B., Osterhaus A. D., Haagmans B. L., Katze M. G. (2007) Functional genomics highlights differential induction of antiviral pathways in the lungs of SARS-CoV-infected macaques. PLoS Pathog. 3, e112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broz P., Dixit V. M. (2016) Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 [DOI] [PubMed] [Google Scholar]
- 5.Agostini L., Martinon F., Burns K., McDermott M. F., Hawkins P. N., Tschopp J. (2004) NALP3 forms an IL-1β-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 20, 319–325 [DOI] [PubMed] [Google Scholar]
- 6.Haneklaus M., O’Neill L. A. (2015) NLRP3 at the interface of metabolism and inflammation. Immunol. Rev. 265, 53–62 [DOI] [PubMed] [Google Scholar]
- 7.Gram A. M., Frenkel J., Ressing M. E. (2012) Inflammasomes and viruses: cellular defence versus viral offence. J. Gen. Virol. 93, 2063–2075 [DOI] [PubMed] [Google Scholar]
- 8.Stewart M. K., Cookson B. T. (2016) Evasion and interference: intracellular pathogens modulate caspase-dependent inflammatory responses. Nat. Rev. Microbiol. 14, 346–359 [DOI] [PubMed] [Google Scholar]
- 9.Ichinohe T., Pang I. K., Iwasaki A. (2010) Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 11, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito M., Yanagi Y., Ichinohe T. (2012) Encephalomyocarditis virus viroporin 2B activates NLRP3 inflammasome. PLoS Pathog. 8, e1002857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H. C., Jin Y., Zhi X. Y., Yan D., Sun S. Q. (2015) NLRP3 inflammasome activation by viroporins of animal viruses. Viruses 7, 3380–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAuley J. L., Tate M. D., MacKenzie-Kludas C. J., Pinar A., Zeng W., Stutz A., Latz E., Brown L. E., Mansell A. (2013) Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathog. 9, e1003392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nieto-Torres J. L., DeDiego M. L., Verdiá-Báguena C., Jimenez-Guardeño J. M., Regla-Nava J. A., Fernandez-Delgado R., Castaño-Rodriguez C., Alcaraz A., Torres J., Aguilella V. M., Enjuanes L. (2014) Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 10, e1004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu W., Zheng B. J., Xu K., Schwarz W., Du L., Wong C. K., Chen J., Duan S., Deubel V., Sun B. (2006) Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. USA 103, 12540–12545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C. C., Krüger J., Sramala I., Hsu H. J., Henklein P., Chen Y. M., Fischer W. B. (2011) ORF8a of SARS-CoV forms an ion channel: experiments and molecular dynamics simulations. Biochim. Biophys. Acta 1808, 572–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castaño-Rodriguez C., Honrubia J. M., Gutiérrez-Álvarez J., DeDiego M. L., Nieto-Torres J. L., Jimenez-Guardeño J. M., Regla-Nava J. A., Fernandez-Delgado R., Verdia-Báguena C., Queralt-Martín M., Kochan G., Perlman S., Aguilella V. M., Sola I., Enjuanes L. (2018) Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. MBio 9, e02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nieto-Torres J. L., Verdiá-Báguena C., Jimenez-Guardeño J. M., Regla-Nava J. A., Castaño-Rodriguez C., Fernandez-Delgado R., Torres J., Aguilella V. M., Enjuanes L. (2015) Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology 485, 330–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minakshi R., Padhan K., Rehman S., Hassan M. I., Ahmad F. (2014) The SARS Coronavirus 3a protein binds calcium in its cytoplasmic domain. Virus Res. 191, 180–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan Y. J., Lim S. G., Hong W. (2006) Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antiviral Res. 72, 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narayanan K., Huang C., Makino S. (2008) SARS coronavirus accessory proteins. Virus Res. 133, 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yount B., Roberts R. S., Sims A. C., Deming D., Frieman M. B., Sparks J., Denison M. R., Davis N., Baric R. S. (2005) Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79, 14909–14922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. (2009) The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One 4, e8342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freundt E. C., Yu L., Goldsmith C. S., Welsh S., Cheng A., Yount B., Liu W., Frieman M. B., Buchholz U. J., Screaton G. R., Lippincott-Schwartz J., Zaki S. R., Xu X. N., Baric R. S., Subbarao K., Lenardo M. J. (2010) The open reading frame 3a protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death. J. Virol. 84, 1097–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan C. M., Tsoi H., Chan W. M., Zhai S., Wong C. O., Yao X., Chan W. Y., Tsui S. K., Chan H. Y. (2009) The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 41, 2232–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oostra M., de Haan C. A., de Groot R. J., Rottier P. J. (2006) Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 80, 2326–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padhan K., Tanwar C., Hussain A., Hui P. Y., Lee M. Y., Cheung C. Y., Peiris J. S., Jameel S. (2007) Severe acute respiratory syndrome coronavirus Orf3a protein interacts with caveolin. J. Gen. Virol. 88, 3067–3077 [DOI] [PubMed] [Google Scholar]
- 27.Tan Y. J., Tham P. Y., Chan D. Z., Chou C. F., Shen S., Fielding B. C., Tan T. H., Lim S. G., Hong W. (2005) The severe acute respiratory syndrome coronavirus 3a protein up-regulates expression of fibrinogen in lung epithelial cells. J. Virol. 79, 10083–10087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanzawa N., Nishigaki K., Hayashi T., Ishii Y., Furukawa S., Niiro A., Yasui F., Kohara M., Morita K., Matsushima K., Le M. Q., Masuda T., Kannagi M. (2006) Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-kappaB activation. FEBS Lett. 580, 6807–6812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiscott J., Marois J., Garoufalis J., D’Addario M., Roulston A., Kwan I., Pepin N., Lacoste J., Nguyen H., Bensi G., Fenton M. (1993) Characterization of a functional NF-κ B site in the human interleukin 1 β promoter: evidence for a positive autoregulatory loop. Mol. Cell. Biol. 13, 6231–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou R., Yazdi A. S., Menu P., Tschopp J. (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225; erratum: 475, 122 [DOI] [PubMed] [Google Scholar]
- 31.Reiley W., Zhang M., Wu X., Granger E., Sun S. C. (2005) Regulation of the deubiquitinating enzyme CYLD by IkappaB kinase γ-dependent phosphorylation. Mol. Cell. Biol. 25, 3886–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng W., Xu M., Liu S., Sun L., Chen Z. J. (2009) Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol. Cell 36, 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siu K. L., Kok K. H., Ng M. H., Poon V. K., Yuen K. Y., Zheng B. J., Jin D. Y. (2009) Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 284, 16202–16209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siu K. L., Chan C. P., Kok K. H., Chiu-Yat Woo P., Jin D. Y. (2014) Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell. Mol. Immunol. 11, 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lui P. Y., Wong L. Y., Fung C. L., Siu K. L., Yeung M. L., Yuen K. S., Chan C. P., Woo P. C., Yuen K. Y., Jin D. Y. (2016) Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg. Microbes Infect. 5, e39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Valck D., Jin D. Y., Heyninck K., Van de Craen M., Contreras R., Fiers W., Jeang K. T., Beyaert R. (1999) The zinc finger protein A20 interacts with a novel anti-apoptotic protein which is cleaved by specific caspases. Oncogene 18, 4182–4190 [DOI] [PubMed] [Google Scholar]
- 37.Siu Y. T., Ching Y. P., Jin D. Y. (2008) Activation of TORC1 transcriptional coactivator through MEKK1-induced phosphorylation. Mol. Biol. Cell 19, 4750–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng M. H., Ho T. H., Kok K. H., Siu K. L., Li J., Jin D. Y. (2011) MIP-T3 is a negative regulator of innate type I IFN response. J. Immunol. 187, 6473–6482 [DOI] [PubMed] [Google Scholar]
- 39.Chaudhary V., Zhang S., Yuen K. S., Li C., Lui P. Y., Fung S. Y., Wang P. H., Chan C. P., Li D., Kok K. H., Liang M., Jin D. Y. (2015) Suppression of type I and type III IFN signalling by NSs protein of severe fever with thrombocytopenia syndrome virus through inhibition of STAT1 phosphorylation and activation. J. Gen. Virol. 96, 3204–3211 [DOI] [PubMed] [Google Scholar]
- 40.Lui P. Y., Wong L. R., Ho T. H., Au S. W. N., Chan C. P., Kok K. H., Jin D. Y. (2017) PACT facilitates RNA-induced activation of MDA5 by promoting MDA5 oligomerization. J. Immunol. 199, 1846–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Y., Gao W. W., Tang H. M., Deng J. J., Wong C. M., Chan C. P., Jin D. Y. (2016) β-TrCP-mediated ubiquitination and degradation of liver-enriched transcription factor CREB-H. Sci. Rep. 6, 23938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fett C., DeDiego M. L., Regla-Nava J. A., Enjuanes L., Perlman S. (2013) Complete protection against severe acute respiratory syndrome coronavirus-mediated lethal respiratory disease in aged mice by immunization with a mouse-adapted virus lacking E protein. J. Virol. 87, 6551–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almazán F., Dediego M. L., Galán C., Escors D., Alvarez E., Ortego J., Sola I., Zuñiga S., Alonso S., Moreno J. L., Nogales A., Capiscol C., Enjuanes L. (2006) Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 80, 10900–10906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeung M. L., Yao Y., Jia L., Chan J. F., Chan K. H., Cheung K. F., Chen H., Poon V. K., Tsang A. K., To K. K., Yiu M. K., Teng J. L., Chu H., Zhou J., Zhang Q., Deng W., Lau S. K., Lau J. Y., Woo P. C., Chan T. M., Yung S., Zheng B. J., Jin D. Y., Mathieson P. W., Qin C., Yuen K. Y. (2016) MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat. Microbiol. 1, 16004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang P. H., Ye Z. W., Deng J. J., Siu K. L., Gao W. W., Chaudhary V., Cheng Y., Fung S. Y., Yuen K. S., Ho T. H., Chan C. P., Zhang Y., Kok K. H., Yang W., Chan C. P., Jin D. Y. (2018) Inhibition of AIM2 inflammasome activation by a novel transcript isoform of IFI16. EMBO Rep. 19, e45737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang P. H., Fung S. Y., Gao W. W., Deng J. J., Cheng Y., Chaudhary V., Yuen K. S., Ho T. H., Chan C. P., Zhang Y., Kok K. H., Yang W., Chan C. P., Jin D. Y. (2018) A novel transcript isoform of STING that sequesters cGAMP and dominantly inhibits innate nucleic acid sensing. Nucleic Acids Res. 46, 4054–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan C. P., Yuen C. K., Cheung P. H., Fung S. Y., Lui P. Y., Chen H., Kok K. H., Jin D. Y. (2018) Antiviral activity of double-stranded RNA-binding protein PACT against influenza A virus mediated via suppression of viral RNA polymerase. FASEB J. 32, 4380–4393 [DOI] [PubMed] [Google Scholar]
- 48.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuen K. S., Chan C. P., Wong N. H., Ho C. H., Ho T. H., Lei T., Deng W., Tsao S. W., Chen H., Kok K. H., Jin D. Y. (2015) CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J. Gen. Virol. 96, 626–636 [DOI] [PubMed] [Google Scholar]
- 50.Kaminski J. J., Schattgen S. A., Tzeng T. C., Bode C., Klinman D. M., Fitzgerald K. A. (2013) Synthetic oligodeoxynucleotides containing suppressive TTAGGG motifs inhibit AIM2 inflammasome activation. J. Immunol. 191, 3876–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Place D. E., Kanneganti T. D. (2018) Recent advances in inflammasome biology. Curr. Opin. Immunol. 50, 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guan K., Wei C., Zheng Z., Song T., Wu F., Zhang Y., Cao Y., Ma S., Chen W., Xu Q., Xia W., Gu J., He X., Zhong H. (2015) MAVS promotes inflammasome activation by targeting ASC for K63-linked ubiquitination via the E3 ligase TRAF3. J. Immunol. 194, 4880–4890 [DOI] [PubMed] [Google Scholar]
- 53.Lupfer C., Malik A., Kanneganti T. D. (2015) Inflammasome control of viral infection. Curr. Opin. Virol. 12, 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oeckinghaus A., Hayden M. S., Ghosh S. (2011) Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 [DOI] [PubMed] [Google Scholar]
- 55.Ye H., Park Y. C., Kreishman M., Kieff E., Wu H. (1999) The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol. Cell 4, 321–330 [DOI] [PubMed] [Google Scholar]
- 56.Chiu H. W., Chen C. H., Chang J. N., Chen C. H., Hsu Y. H. (2016) Far-infrared promotes burn wound healing by suppressing NLRP3 inflammasome caused by enhanced autophagy. J. Mol. Med. (Berl.) 94, 809–819; erratum: 821 [DOI] [PubMed] [Google Scholar]
- 57.Gazdar A. F., Girard L., Lockwood W. W., Lam W. L., Minna J. D. (2010) Lung cancer cell lines as tools for biomedical discovery and research. J. Natl. Cancer Inst. 102, 1310–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Opdenbosch N., Gurung P., Vande Walle L., Fossoul A., Kanneganti T. D., Lamkanfi M. (2014) Activation of the NLRP1b inflammasome independently of ASC-mediated caspase-1 autoproteolysis and speck formation. Nat. Commun. 5, 3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O’Rourke K. M., Eby M., Pietras E., Cheng G., Bazan J. F., Zhang Z., Arnott D., Dixit V. M. (2007) DUBA: a deubiquitinase that regulates type I interferon production. Science 318, 1628–1632 [DOI] [PubMed] [Google Scholar]
- 60.Heyninck K., Beyaert R. (1999) The cytokine-inducible zinc finger protein A20 inhibits IL-1-induced NF-kappaB activation at the level of TRAF6. FEBS Lett. 442, 147–150 [DOI] [PubMed] [Google Scholar]
- 61.Parvatiyar K., Barber G. N., Harhaj E. W. (2010) TAX1BP1 and A20 inhibit antiviral signaling by targeting TBK1-IKKi kinases. J. Biol. Chem. 285, 14999–15009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trompouki E., Hatzivassiliou E., Tsichritzis T., Farmer H., Ashworth A., Mosialos G. (2003) CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424, 793–796 [DOI] [PubMed] [Google Scholar]
- 63.Stutz A., Horvath G. L., Monks B. G., Latz E. (2013) ASC speck formation as a readout for inflammasome activation. Methods Mol. Biol. 1040, 91–101 [DOI] [PubMed] [Google Scholar]
- 64.Menachery V. D., Mitchell H. D., Cockrell A. S., Gralinski L. E., Yount B. L., Jr., Graham R. L., McAnarney E. T., Douglas M. G., Scobey T., Beall A., Dinnon K., III, Kocher J. F., Hale A. E., Stratton K. G., Waters K. M., Baric R. S. (2017) MERS-CoV accessory ORFs play key role for infection and pathogenesis. MBio 8, e00665-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yue Y., Nabar N. R., Shi C. S., Kamenyeva O., Xiao X., Hwang I. Y., Wang M., Kehrl J. H. (2018) SARS-Coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis. 9, 904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen I. Y., Moriyama M., Chang M. F., Ichinohe T. (2019) Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 10, 50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar M., Roe K., Orillo B., Muruve D. A., Nerurkar V. R., Gale M., Jr., Verma S. (2013) Inflammasome adaptor protein Apoptosis-associated speck-like protein containing CARD (ASC) is critical for the immune response and survival in west Nile virus encephalitis. J. Virol. 87, 3655–3667; erratum: 92, e02176-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sears C., Olesen J., Rubin D., Finley D., Maniatis T. (1998) NF-κ B p105 processing via the ubiquitin-proteasome pathway. J. Biol. Chem. 273, 1409–1419 [DOI] [PubMed] [Google Scholar]
- 69.Kravtsova-Ivantsiv Y., Shomer I., Cohen-Kaplan V., Snijder B., Superti-Furga G., Gonen H., Sommer T., Ziv T., Admon A., Naroditsky I., Jbara M., Brik A., Pikarsky E., Kwon Y. T., Doweck I., Ciechanover A. (2015) KPC1-mediated ubiquitination and proteasomal processing of NF-κB1 p105 to p50 restricts tumor growth. Cell 161, 333–347 [DOI] [PubMed] [Google Scholar]