Abstract
Objective
We tested whether genetic variants near fatty acid desaturases gene (FADS) cluster, which were recently identified to be signatures of adaptation to fish-rich and n-3 polyunsaturated fatty acids (PUFAs)-rich diet, interacted with these dietary factors on change in body mass index (BMI).
Design
Three FADS variants were examined for gene-diet interactions on long-term (~10 years) changes in BMI and body weight in four prospective cohort studies.
Setting
Population based study.
Participants
11 323 women from the Nurses’ Health Study (NHS), 6833 men from the Health Professionals Follow-up Study (HPFS) and replicated in 6254 women from the Women’s Health Initiative (WHI) and 5 264 Chinese from the Singapore Chinese Health Study (SCHS).
Main outcomes
Long-term (~10 years) changes in BMI and body weight.
Results
In the NHS and HPFS cohorts, food-sourced n-3 PUFAs intake showed interactions with the FADS rs174570 on changes of BMI (P for interaction=0.02 in NHS, 0.05 in HPFS and 0.007 in combined). Such interactions were replicated in two independent cohorts WHI and SCHS (P for interaction=0.04 in WHI, 0.02 in SCHS and 0.001 in combined). The genetic associations of the FADS rs174570 with changes in BMI increased across the tertiles of n-3 PUFAs in all the cohorts. Fish intake also accentuated the genetic associations of the FADS rs174570 with long-term changes in BMI (pooled P for interaction=0.006). Viewed differently, long chain n-3 PUFAs intake showed stronger association with long-term changes in BMI among the rs174570 T carriers (beta=0.79 kg/m2 per g, p=3×10−5) than the rs174570 non-T carriers (beta=0.16 kg/m2 per g, p=0.08). Similar results were observed for fish intake.
Conclusions
Our hypothesis-driven analyses provide replicable evidence that long chain n-3 PUFAs and fish intakes may interact with the FADS variant on long-term weight gain. Further investigation is needed to confirm our findings in other cohorts.
Keywords: genetics, epidemiology, obesity, gene-diet interaction
Strengths and limitations of this study.
This is the first study with consistent results from four well-established prospective cohorts of different racial populations such as Caucasians and Singapore Chinese. The consistent results from these independent cohorts demonstrated the robustness of our findings.
Other major strengths include the prospective design, the large sample size, use of long-term change of body mass index and replication of the results.
Dietary fatty acids, fish and adiposity measures were self-reported, measurement errors in these variables are inevitable.
Confounding by other unmeasured or unknown factors might exist, although we have carefully adjusted for multiple dietary and lifestyle factors.
We acknowledge that the different methods in measuring anthropometric traits, genetic variants and food intake across cohorts might introduce bias in the present analyses.
Introduction
Diets rich in fish and marine fatty acids, especially long chain n-3 polyunsaturated fatty acids (PUFAs) has shown beneficial effects on cardiometabolic health.1 2 However, data from population studies on the associations between such diet and body weight are inconsistent.3 4 Emerging evidence suggests genetic variations may play a role in modifying the relation between dietary factors and body weight.5–7 For example, we previously found that high intakes of fish and long-chain n-3 PUFAs are associated with an attenuation of the genetic association with long-term weight gain based on results from three prospective cohorts of Caucasians.8
A recent study of Inuit identified genetic signatures of adaptation to diets rich in fish and n-3 PUFAs.9 The strong signals locate in a cluster of fatty acid desaturases genes (FADS) that determine PUFAs levels.9 People living in the Arctic region have been found to be genetically prone to develop obesity10 11 as survival strength for energy storage.12 13 Interestingly, the identified FADS genetic signatures of diet adaptation have been also related to adiposity in the Inuit population.9 Of note, due to long-standing selection pressure, the identified FADS signatures differ in frequency of selective allele across various populations such as Europeans and Asians,14 in coincidence with varying levels of fish/marine fatty acids consumption, and adiposity patterns in these populations.15 We therefore hypothesised that the genetic signatures might interact with fish and marine PUFAs intakes on body weight.14
The present study tested the interactions between n-3 PUFAs and fish intakes and variants in FADS gene cluster, genetic signatures of adaptation to fish-rich and n-3 PUFAs-rich diet, in relation to long-term changes in body mass index (BMI) in two US prospective cohorts: the Nurses’ Health Study (NHS) and the Health Professionals Follow-up Study (HPFS). We replicated the findings in two independent, prospective cohorts the Women’s Health Initiative (WHI, US) and the Singapore Chinese Health Study (SCHS).
Methods
Discovery cohorts
Nurses’ health study
The NHS began in 1976, when 121 700 female registered nurses aged 30–55 y residing in 11 states were recruited to complete a baseline questionnaire about their lifestyle and medical history.16 The current analysis baseline was set in 1990 for the NHS. We included 11 323 women of European ancestry. Informed consent was obtained from all participants. The DNA extraction methods, quality control measures, single-nucleotide polymorphism (SNPs) genotyping and imputation when performed have been described in detail elsewhere.17–23 All participants with baseline long chain n-3 PUFAs and fish consumptions and covariates data, baseline and endpoint BMI data and genotyping data available based on previous GWASs were included.17–22 The study protocol was approved by the institutional review boards of Brigham and Women’s Hospital and Harvard School of Public Health.
Health professionals follow-up study
The HPFS was initiated in 1986, and was composed of 51 529 male dentists, pharmacists, veterinarians, optometrists, osteopathic physicians and podiatrists, aged 40–75 years at baseline. The male participants returned a baseline questionnaire about detailed medical history, lifestyle and usual diet.24 In the current analysis, we used 1990 as baseline in the HPFS, when the earliest complete dietary data were collected. Our analysis included 6833 men whose genotype data were available. Informed consent was obtained from all participants. The DNA extraction methods, quality control measures, SNPs genotyping and imputation when performed have been described in detail elsewhere.17–23 All participants with baseline long chain n-3 PUFAs and fish consumptions and covariates data, baseline and endpoint BMI data and genotyping data available based on previous GWASs were included.17–22 The study protocol was also approved by the institutional review boards of Brigham and Women’s Hospital and Harvard School of Public Health.
Replication cohorts
WHI
The Women’s Health Initiative (WHI) is a large, multiethnic, 40-centre study funded by the National Heart, Lung and Blood Institute that focuses on strategies for preventing heart disease, breast and colorectal cancer and osteoporotic fractures in postmenopausal women. A full description of the WHI study is presented elsewhere.25 26 For the analyses, all participants with baseline long chain n-3 PUFAs and fish consumption and covariate data, baseline and endpoint BMI dataand genotyping data available based on previous GWASs were included. Finally, we included 6 254 Caucasians women who participated in the WHI clinical trial studies at baseline (1994–1998) and at sixth-year follow-up and for whom DNA was measured. The genomic DNA samples were processed according to standard Affymetrix procedures for processing of the assay. The Affymetrix Human SNP Array V.6.0 (Affymetrix, Inc Santa Clara, California, USA) was used for genome wide SNP genotyping. Human subjects review committees at each participating institution reviewed and approved the study, and all women gave written informed consent.
SCHS cohort
The design of SCHS has been previously described in detail.27 Briefly, between 1993 and 1998, 63 257 Chinese men and women between ages of 45 and 74 years living in Singapore were enrolled into the cohort study.28 Two follow-up interviews were conducted via telephone among surviving participants between 1999 and 2004, and again between 2006 and 2010 to update information on body weight, selected lifestyle factors and medical history. All participants have given informed consent. The study was approved by the Institutional Review Boards of the National University of Singapore and the University of Pittsburgh, and the study was carried out in accordance with the approved guidelines. All participants with baseline long chain n-3 PUFAs and fish consumptions and covariates data, baseline and endpoint BMI data available were included. Among these participants, genome-wide genotyping for 2615 incident diabetes cases and 2615 matched controls was performed at the Genome Institute of the Singapore according to the manufacturer’s recommendations using an Affymetrix ASI (Asian) Axiom array. Genotype calling was performed by the Affymetrix Corporation.29 Genome-wide genotyping for 717 incident myocardial infarction cases and 644 controls was performed for SCHS samples using the Illumina HumanOmni ZhongHua-8 Bead Chip.30 Among these two case–control studies nested within the cohort, 5264 subjects with genotyping data had weight reported at both baseline and follow-up two interviews, and were included in this analysis.
Assessment of measures of BMI
Height and body weight were assessed by questionnaire at baseline, and weight information was requested on follow-up questionnaire in all four cohorts. Self-reported weights were highly correlated with directly measured values (r=0.97 in HPFS and NHS) in a validation study.31 BMI was calculated as body weight (kg)/height (m2). As defined previously,8 the long-term changes in BMI was calculated as changes in BMI from 1990 to 2000 in the NHS and HPFS cohorts,32 and from baseline (1993) to sixth-year follow-up in the WHI,25 26 and from baseline (1998) to second follow-up (2004) in the SCHS.
Assessment of diets and other covariates
Questionnaires were used to collect information on a medical history and diet/lifestyle factors in all four cohorts. Total fish, n-3 PUFAs, supplemental use of fish oil, alcohol, sugar sweetened beverages, fried food intakes and other dietary factors at baseline were assessed by validated food frequency questionnaires (FFQ) in the NHS and HPFS.33 34 A 165-item validated semiquantitative FFQ was used to collect dietary data and supplemental use of fish oil in the SCHS.28 Dietary data and supplemental use of fish oil were obtained from a self-administered baseline 122-items validated FFQ in the WHI.35 Alternate health eating index was previously calculated in the NHS, HPFS,36 WHI and SCHS. Physical activity was expressed as metabolic equivalents per week by incorporating the reported time spent on various activities, and the intensity level of each activity. The validity of the self-reported physical activity data has been described previously in the NHS and HPFS.37 In the WHI, an estimated metabolic equivalent (MET) level for each type of activity was assigned from a compendium of activities.38 Physical activity was assessed using eight continuous categories ranging from never to 31 hours or more in an average week spent doing strenuous sports; vigorous work; and moderate activities in the SCHS.27
FADS variants selection and genotyping
Three of the six FADS SNPs reported in a recent scan of Inuit genomes for signatures of adaptation9 were derived from genome-wide scans available in the NHS, HPFS. We assumed that each SNP in the panel acts independently in an additive manner. We coded the SNPs as following: rs174570 (TT=2, TC=1, CC=0); rs174602 (TT=2, TC=1, CC=0); rs7115739 (TT=2, TG=1, GG=0). The FADS rs174570 was extracted from GWAS data in the WHI and SCHS cohorts for replication (see online supplementary table 1).
bmjopen-2018-022877supp001.pdf (495.2KB, pdf)
Patient and public involvement
Neither patients nor public were involved.
Statistical analyses
We examined the associations of the FADS variants (rs174570, rs174602, rs7115739) with adiposity measures and long-term changes in BMI using multiple linear regression model. Interactions between the FADS variants (rs174570, rs174602, rs7115739) and baseline fish intake and total or food-sourced long chain n-3 PUFAs intakes on long-term changes in BMI were tested by including a multiplicative interaction term in the models in the NHS and HPFS. The significant results for rs174570 were replicated in the WHI and SCHS. Potential confounders considered in multivariable models were age, baseline physical activity, baseline television watching, baseline smoking, baseline alcohol intake, baseline alternate healthy eating index and baseline total energy intake, sugar sweetened beverages (if available), fried food intake (if available). We further tested the genetic associations with long-term changes in BMI according to long chain n-3 PUFAs and fish intakes, and associations of long chain n-3 PUFAs and fish intakes with long-term changes in BMI according to the FADS genotypes using multiple linear regression model after adjustment of potential confounders. Linear trend across categories of long chain n-3 PUFAs and fish intakes was quantified with a Wald test for linear trend by assigning the median value to each category and modelling it as a continuous variable.39 Results across cohorts were pooled with inverse variance weighted meta-analyses by fixed effects models (p≥0.05 for heterogeneity between studies).40 The individual participant data from the NHS and HPFS cohorts were pooled to generate the predicted 10-year changes in body weight according to the FADS genotypes. Hardy-Weinberg equilibrium was tested using Χ2 test. All reported p values are nominal and two sided. Statistical analyses were performed in SAS V.9.3 (SAS Institute, Cary, North Carolina, USA).
Results
Baseline characteristics of all participants in the NHS, HPFS, WHI and SCHS cohorts
Table 1 shows the baseline characteristics for all participants in the NHS, HPFS, WHI and SCHS cohorts. The present study included 11 323 women with genetic data from the NHS cohort, 6833 men with genetic data from the HPFS cohort, 6254 women from the WHI and 5 264 Chinese from the SCHS. The distribution of the FADS genetic variants in the four cohorts is shown in online supplementary table 1. χ2 test showed that the FADS rs174570 is in Hardy-Weinberg equilibrium. We did not observe any significant genetic association between the FADS rs174570 genotype and baseline BMI, and long-term changes in BMI in the three US cohorts (p>0.05). However, we found that the FADS genotype was significantly associated with baseline BMI in the SCHS (p=0.002) (see online supplementary table 2).
Table 1.
Baseline characteristics of all participants in the NHS, HPFS, WHI and SCHS cohorts
| NHS* | HPFS | WHI | SCHS | |
| n=11 323 | n=6833 | n=6254 | n=5264 | |
| Age (year) | 57±9 | 57±11 | 68±5 | 56±7 |
| Female (%) | 100 | 0 | 100 | 58.7 |
| Body weight (kg) | 70.1±14.9 | 82.8±12.5 | 73.7±15.0 | 60.3±9.8 |
| Body mass index (kg/m2) | 26.2±5.1 | 25.9±3.3 | 28.3±5.5 | 23.4±3.3 |
| Alcohol consumption (g/day) | 5.14±9.23 | 10.97±15.05 | 6.00±11.96 | 1.97±8.02 |
| Physical activity (MET-h/week) | 19.3±22.1 | 36.9±39.5 | 11.6±13.1 | 0.5±1.0† |
| Television watching (h/week) | 17.5±14.8 | 10.5±8.2 | / | 2.2±0.8 |
| Current smokers (n, (%)) | 1557 (13.8) | 493 (7.3) | 407 (15.0) | 1364 (20.0) |
| Total energy intake (kcal/day) | 1766±502 | 1949±578 | 1602±654 | 1606±573 |
| Alternative health eating index score | 53.4±10.8 | 53.8±11.4 | 53.5±10.6 | 55.8±8.2 |
| Sugar sweetened beverage intake (servings/day) | 0.13±0.39 | 0.23±0.48 | 0.39±0.82 | 0.69±2.40‡ |
| Total fried food (servings/day) | 0.12±0.20 | 0.22±0.28 | / | / |
| Fish intake (servings/day) | 0.31±0.29 | 0.33±0.30 | 0.23±0.20 | 0.16±0.07 |
| Food-sourced EPA (g/day) | 0.08±0.14 | 0.12±0.20 | 0.04±0.04 | / |
| Food-sourced DHA (g/day) | 0.17±0.14 | 0.22±0.19 | 0.07±0.07 | / |
| Food-sourced EPA+DHA (g/day) | 0.23±0.19 | 0.31±0.25 | 0.11±0.10 | 0.33±0.20 |
| Total EPA+DHA (g/day) | 0.26±0.27 | 0.35±0.37 | 0.38±0.48 | / |
Data on BMI, long chain n-3 PUFAs and fish consumptions were assessed at baseline in the NHS (1990), the HPFS (1990), the WHI (1994–1998) and the SCHS (1993–1998), respectively. Television watching assessed in 1992 for NHS and in 1990 for HPFS.
EPA: 20:5 n-3; DHA: 22:6 n-3; MET denotes metabolic equivalents. Total EPA+DHA includes food-sourced and supplemental EPA+DHA.
*Plus-minus values are means±SD.
†Hours per week of moderate activity in the SCHS.
‡Glasses per week of soda intake in the SCHS.
DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; HPFS, Health Professionals Follow-up Study; MET, metabolic equivalent; NHS, Nurses’ Health Study; PUFA, polyunsaturated fatty acids; SCHS, Singapore Chinese Health Study; WHI, Women’s Health Initiative.
Genetic associations with long-term changes in BMI according to LC n-3 PUFAs/fish intakes
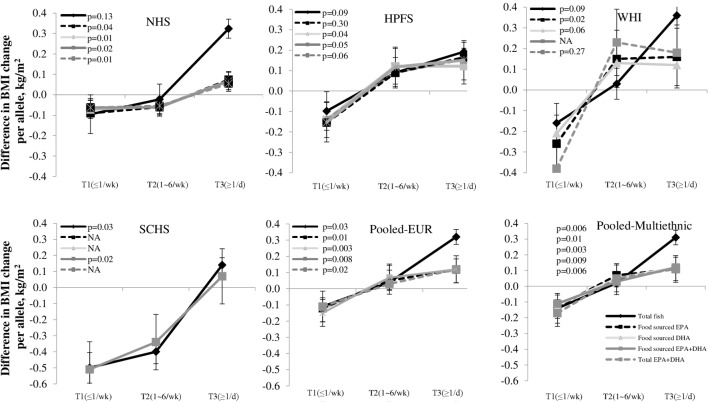
We first tested interactions between the FADS genetic variants (rs174570, rs174602, rs7115739) and intakes of variously sourced long chain n-3 PUFAs and fish in the NHS and HPFS cohorts. We found that only FADS rs174570 (C/T, with T as the common allele in Inuit, but rare allele in Europeans and Asians) showed significant interaction with long chain (LC) n-3 PUFAs/fish intakes. Food-sourced n-3 PUFAs (eicosapentaenoic acid (EPA)+docosahexaenoic acid (DHA)) intake consistently magnified the genetic association with long-term changes in BMI (P for interaction=0.02 in NHS, 0.05 in HPFS and 0.007 in combined cohorts) (figure 1). We successfully replicated our results in the WHI cohort (P for interaction=0.04) and the SCHS cohort (P for interaction=0.02).
Figure 1.
Genetic variant of FADS rs174570, long chain n-3 PUFAs and fish intakes and long-term BMI changes ES (95% CI) values are β coefficients for interaction between the FADS variant rs174570 (additive model) and diets from results of the NHS, HPFS, WHI and SCHS cohorts. Data on BMI, long chain n-3 PUFAs (food sourced EPA+DHA and total EPA+DHA (food and supplemental use)) and fish consumptions were assessed at baseline in the NHS (1990), the HPFS (1990), the WHI (1994–1998) and the SCHS (1993–1998), respectively. Data on follow-up BMI was assessed in 2000 in the NHS and HPFS, in the sixth follow-up year in the WHI, and from 2006 to 2010 in the SCHS, respectively. Long-term BMI changes were calculated based on the changes in BMI from baseline to follow-up year in the four cohorts, respectively. The multiple linear regression model was used to test the FADS variant-diets interaction by including a multiplicative interaction term in the models after adjustment for age, source of genotyping data, baseline BMI, smoking, alcohol intake, physical activity, total energy intake, alternate healthy eating index, television watching, sugar sweetened beverage, fried food consumption. The results were pooled by means of fixed effects meta-analyses (p≥0.05 for heterogeneity between studies). BMI, body mass index; DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; ES, effect size; FADS, fatty acid desaturases gene; HPFS, Health Professionals Follow-up Study; NHS, Nurses’ Health Study; SCHS, Singapore Chinese Health Study; WHI, Women’s Health Initiative.
The pooled analyses of the 3 US (Caucasian) samples or all four cohorts showed that high intakes of food-sourced n-3 PUFAs intake (P for interaction=0.008 and 0.009, respectively) significantly accentuated the genetic association of the FADS genotypes with long-term changes in BMI (figure 2). No significant heterogeneity in the interaction effect was observed among these cohorts. Differences in long-term changes of BMI per T allele were −0.105 (SE 0.067), 0.027 (SE 0.064) and 0.120 (SE 0.067) kg/m2 across three tertiles of food-sourced n-3 PUFAs in pooled results from all the four cohorts.
Figure 2.
Genetic association of FADS variant rs174570 with long-term BMI change according to long chain n-3 PUFAs and fish intakes Pooled-EUR: data from NHS, HPFS and WHI were pooled. Pooled Multiethnic: data from NHS, HPFS, WHI and SCHS were pooled. Data are β coefficients±SE Numbers of participants across three categories (≤1/wk/1~6/wk/≥1/d) of fish intake in the NHS, HPFS, WHI and SCHS are 1618/8465/1239, 977/5108/748, 894/4675/684 and 752/3935/576, respectively. Frequency of fish intake: ≤1 serving per week, 1~6 servings per week and 1 serving per day data on BMI, long chain n-3 PUFAs (food sourced EPA+DHA and total EPA+DHA (food and supplemental use)) and fish consumptions were assessed at baseline in the NHS (1990), the HPFS (1990), the WHI (1994–1998) and the SCHS (1993–1998), respectively. Data on follow-up BMI was assessed in 2000 in the NHS and HPFS, in the sixth follow-up year in the WHI, and from 2006 to 2010 in the SCHS, respectively. The multiple linear regression model was used to test the genetic association of the FADS variant (additive model) with long-term changes in BMI by frequency of fish intake and tertiles of LC fatty acids after adjustment for age, source of genotyping data, baseline BMI, smoking, alcohol intake, physical activity, total energy intake, alternate healthy eating index, television watching, sugar sweetened beverage, fried food consumption. The results were pooled by means of fixed effects meta-analyses (p≥0.05 for heterogeneity between studies). BMI, body mass index; DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; FADS, fatty acid desaturases gene; HPFS, Health Professionals Follow-up Study; NHS, Nurses’ Health Study; SCHS, Singapore Chinese Health Study; WHI, Women’s Health Initiative.
Individual food-sourced n-3 PUFAs such as EPA (pooled P for interaction=0.01) and DHA (pooled P for interaction=0.003) showed similar interaction patterns; and the interactions remained significant when supplemented n-3 PUFAs were considered (pooled P for interaction=0.007) (figure 2).
In addition, fish intake showed similar, though less significant, interaction patterns with the FADS genotype on long-term changes in BMI in the NHS (P for interaction=0.16), HPFS (P for interaction=0.09), WHI (P for interaction=0.09), SCHS (P for interaction=0.03) and combined results (pooled P for interaction=0.006), and the differences in BMI changes per T allele were −0.096 (SE 0.071), 0.041 (SE 0.052) and 0.251 (SE 0.151) kg/m2 across three categories (≤1 serving/week, 1~6 servings/weekand ≥1 serving/day) of fish intake in combined results from all the four cohorts.
In addition, we did not observe significant interaction between two other genetic variants in FADS cluster (rs174602 and rs7115739) and long chain n-3 PUFAs/fish intakes in relation to long-term changes in BMI in the NHS and HPFS cohorts. Similar interactions for long-term changes in body weight were observed (see online supplementary table 3 and 4).
Long chain n-3 PUFAs/fish intakes and long-term changes in BMI according to the FADS genotype
We found that individuals who consumed the highest food-sourced n-3 PUFAs (EPA+DHA; T3) had significantly greater increase of BMI (mean±SE = 0.74±0.06, kg/m2) than did those who consumed the lowest (T1) (mean±SE=0.39±0.07, kg/m2) among the T allele carriers, whereas the corresponding BMI changes were 0.68±0.03 kg/m2 and 0.49±0.03 kg/m2, respectively, among the non-T carriers in four cohorts combined (table 2 and see online supplementary table 5). Similarly, we observed different associations between fish intake and BMI changes among the T allele carriers (p=1.5×10−6) and non-carriers (p=0.01) in the pooled results from these US cohorts. No significant heterogeneity in the interaction effect was observed among the cohorts.
Table 2.
Associations of long chain n-3 PUFAs and fish intakes with long-term changes in BMI according to FADS genotypes
| FADS genotypes | Three categories of long chain n-3 PUFAs and fish intakes | P for trend | P for interaction* | |||
| Total fish, serving/day | ≤1/wk | 1~6/wk | ≥1/d | |||
| NHS | Non-T carriers | 0.82±0.06 | 0.98±0.04 | 1.15±0.13 | 0.006 | 0.03 |
| T carriers | 0.73±0.11 | 0.95±0.08 | 1.55±0.25 | 0.0007 | ||
| HPFS | Non-T carriers | 0.43±0.05 | 0.52±0.04 | 0.59±0.12 | 0.73 | 0.03 |
| T carriers | 0.21±0.11 | 0.52±0.07 | 0.79±0.22 | 0.02 | ||
| WHI | Non-T carriers | 0.11±0.08 | 0.28±0.06 | 0.28±0.34 | 0.04 | 0.09 |
| T carriers | 0.02±0.15 | 0.29±0.11 | 0.94±0.67 | 0.01 | ||
| SCHS | Non-T carriers | −3.08±0.19 | −3.00±0.17 | −3.35±0.18 | 0.32 | 0.01 |
| T carriers | −3.61±0.17 | −3.10±0.15 | −3.25±0.17 | 0.13 | ||
| Pooled* | Non-T carriers | 0.50±0.03 | 0.67±0.03 | 0.81±0.08 | 0.01 | 0.0007 |
| T carriers | 0.38±0.07 | 0.63±0.05 | 1.11±0.16 | 2×10-−4 | ||
| Food-sourced EPA+DHA, g/day | Tertile1 | Tertile2 | Tertile3 | |||
| NHS | Non-T carriers | 0.79±0.06 | 0.92±0.05 | 1.11±0.06 | 0.01 | 0.005 |
| T carriers | 0.71±0.10 | 0.84±0.11 | 1.19±0.11 | 0.0001 | ||
| HPFS | Non-T carriers | 0.46±0.05 | 0.53±0.05 | 0.49±0.05 | 0.79 | 0.02 |
| T carriers | 0.23±0.11 | 0.48±0.10 | 0.58±0.09 | 0.02 | ||
| WHI | Non-T carriers | 0.02±0.08 | 0.21±0.08 | 0.41±0.08 | 0.06 | 0.04 |
| T carriers | −0.03±0.15 | 0.29±0.14 | 0.35±0.15 | 0.004 | ||
| SCHS | Non-T carriers | −3.32±0.17 | −3.15±0.18 | −2.99±0.17 | 0.16 | 0.035 |
| T carriers | −3.55±0.16 | −3.34±0.16 | −3.05±0.16 | 0.02 | ||
| Pooled* | Non-T carriers | 0.49±0.03 | 0.64±0.03 | 0.68±0.03 | 0.01 | 0.0003 |
| T carriers | 0.39±0.07 | 0.57±0.06 | 0.74±0.06 | 1.5×10−6 | ||
| Total EPA+DHA, g/day | Tertile1 | Tertile2 | Tertile3 | |||
| NHS | Non-T carriers | 0.79±0.06 | 0.94±0.05 | 1.08±0.06 | 0.8 | 0.01 |
| T carriers | 0.72±0.11 | 0.87±0.10 | 1.16±0.11 | 0.02 | ||
| HPFS | Non-T carriers | 0.47±0.05 | 0.53±0.05 | 0.49±0.05 | 0.88 | 0.13 |
| T carriers | 0.23±0.10 | 0.50±0.09 | 0.57±0.10 | 0.16 | ||
| WHI | Non-T carriers | 0.39±0.10 | 0.04±0.08 | 0.23±0.10 | 0.42 | 0.27 |
| T carriers | 0.04±0.18 | 0.28±0.15 | 0.28±0.16 | 0.84 | ||
| Pooled* | Non-T carriers | 0.57±0.04 | 0.62±0.03 | 0.67±0.04 | 0.65 | 0.005 |
| T carriers | 0.39±0.07 | 0.60±0.06 | 0.74±0.07 | 0.01 | ||
Data are means±SE for long term changes in BMI.
Total EPA+DHA include food-sourced and supplemental EPA+DHA.
Numbers of T carriers/Non-T carriers in the NHS, HPFS, WHI and SCHS are 1698/9625, 1025/5808, 876/5378, and 1842/3422, respectively.
Data on BMI, long chain n-3 PUFAs and fish consumptions were assessed at baseline in the NHS (1990), the HPFS (1990), the WHI (1994–1998) and the SCHS (1993–1998), respectively.
Data on follow-up BMI was assessed in 2000 in the NHS and HPFS, in the sixth follow-up year in the WHI, and from 2006 to 2010 in the SCHS, respectively.
Long-term BMI changes were calculated based on the changes in BMI from baseline to follow-up year in the four cohorts, respectively.
The multiple linear regression model was used to test the associations of long chain n-3 PUFAs and fish intakes with long-term changes in BMI by FADS genotypes after adjustment for age, source of genotyping data, baseline BMI, smoking, alcohol intake, physical activity, total energy intake, alternate healthy eating index, television watching, sugar sweetened beverage, fried food consumption.
The results were pooled by means of fixed effects meta-analyses (p≥0.05 for heterogeneity between studies).
*P for interaction was generated from dominant model of FADS rs174570 (CC vs CT+TT).
BMI, body mass index; DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; ES, effect size; FADS, fatty acid desaturases gene; HPFS, Health Professionals Follow-up Study; NHS, Nurses’ Health Study; PUFA, polyunsaturated fatty acid; SCHS, Singapore Chinese Health Study; WHI, Women’s Health Initiative.
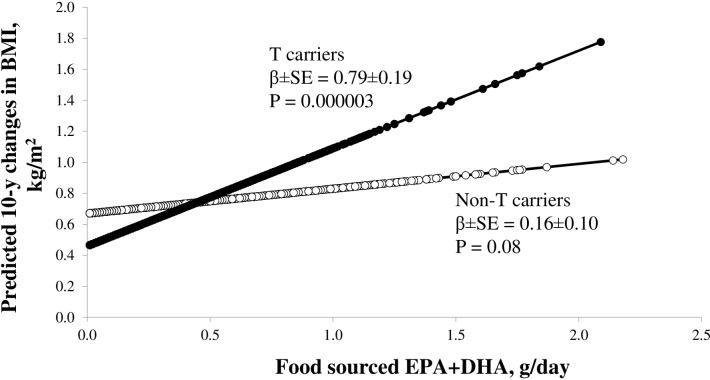
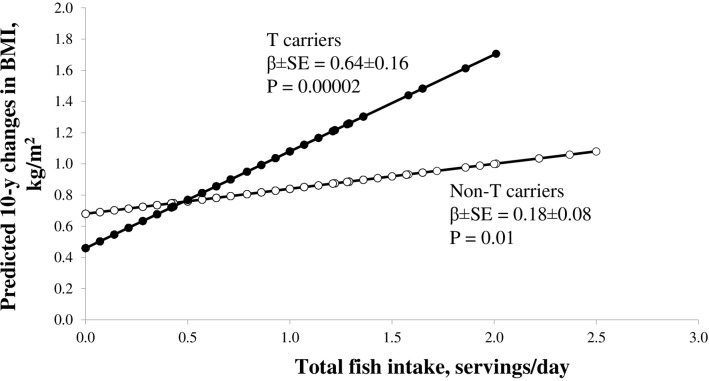
Figure 3 presents the predicted long-term changes in BMI from food-sourced n-3 PUFAs and fish intake according to the T carriers and the non-T carriers. Results from the NHS, HPFS and WHI cohorts consistently showed that the associations of food-sourced n-3 PUFAs and fish intakes with long-term changes in BMI were stronger among the T carriers than those among the non-T carriers. In the pooled results, the beta±SE for associations of food-sourced n-3 PUFAs (figure 3) and fish intake (figure 4) with long-term changes in BMI were 0.79±0.19 kg/m2 per g (p=0.000003) and 0.64±0.16 kg/m2 per serving (p=0.00002) among the T carriers, and whereas the corresponding beta±SE were 0.16±0.10 kg/m2 per g (p=0.08) and 0.18±0.08 kg/m2 per serving (p=0.01) among the non-T carriers.
Figure 3.
Predicted long-term changes in BMI from long chain n-3 PUFAs intake according to FADS genotypes numbers of T carriers/Non-T carriers in the NHS and HPFS are 1698/9625 and 1025/5808, respectively. Black circles for T allele carriers and open circle for non-T-carriers. The multiple linear regression model was used to test the associations of long chain n-3 PUFAs intake with long-term changes in BMI according to FADS genotypes after adjustment for age, source of genotyping data, baseline BMI, smoking, alcohol intake, physical activity, total energy intake, alternate healthy eating index, television watching, sugar sweetened beverage, fried food consumption. The individual participant data from the NHS and HPFS cohorts were pooled. BMI, body mass index; DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; FADS, fatty acid desaturases gene; HPFS, Health Professionals Follow-up Study; NHS, Nurses’ Health Study; PUFA, polyunsaturated fatty acid; HPFS, Health Professionals Follow-up Study; NHS, Nurses’ Health Study.
Figure 4.
Predicted long-term changes in BMI from fish intake according to FADS genotypes numbers of T carriers/Non-T carriers in the NHS and HPFS are 1698/9625 and 1025/5808, respectively. Black circles for T allele carriers and open circle for non-T-carriers. The multiple linear regression model was used to test the associations of and fish intake with long-term changes in BMI according to FADS genotypes after adjustment for age, source of genotyping data, baseline BMI, smoking, alcohol intake, physical activity, total energy intake, alternate healthy eating index, television watching, sugar sweetened beverage, fried food consumption. The individual participant data from the NHS and HPFS cohorts were pooled. BMI, body mass index; FADS, fatty acid desaturases gene; HPFS, Health Professionals Follow-up Study; NHS, Nurses’ Health Study.
Discussion
In four large prospective cohorts of the US and Chinese populations, our hypothesis-driven analyses showed reproducible evidence that long chain n-3 PUFAs and fish intakes accentuated the genetic association of the FADS genotypes with long-term changes in BMI. In addition, our results showed that the FADS rs174570 T allele carriers gained more weight than the non-carriers when they had higher long chain n-3 PUFAs and fish intakes.
Large prospective cohort studies examining the associations of fish or n-3 PUFAs with body weight and obesity risk have generated conflicting results.3 4 In addition, several randomised controlled trials supported the protective effects of fish, fish oils, or/and n-3 PUFAs intake on weight-loss,41–43 but the benefit was not evident in other trials.44–46 The results from the current study lend support to our hypothesis that the heterogeneous associations between fish or n-3 PUFAs and body weight might be at least partly due to gene-diet interactions.
We found that the genetic associations between the FADS rs174570 and long-term BMI change were stronger along with increasing intakes of long chain n-3 PUFAs and fish. Viewed from a different angle, the magnitude of associations of fish and long chain n-3 PUFAs intakes with BMI changes varied among individuals with different genotypes. The FADS rs174570 was recently identified from a study of the Inuit, who had high fish/n-3 PUFAs intakes.9 The high frequency of the T allele in Inuit reflects genetic adaptation to the special fish-rich and n-3 PUFA-rich diet. Interestingly, the identified FADS genetic signatures of diet adaptation have been also related to adiposity in this population. Our data indicated that the signature allele (T) was related diffrently to weight changes (decrease or increase), depending on the levels of fish/n-3 PUFAs intakes. In people with high fish/n-3 PUFAs intakes, carrying the signature allele predisposed to greater weight gain and an increased risk of obesity; while carriers of this allele tended to have less body weight when they are exposed to a diet low in fish and n-3 PUFAs.
We found that individual food-sourced n-3 PUFAs such as EPA and DHA showed similar interaction patterns in relation to long-term changes in BMI; and the interactions also remained significant when supplemented n-3 PUFAs were considered. In addition, our results indicated that the interactions of fish/n-3 PUFAs intakes and the FADS genotype were persistent across different racial populations such as Europeans and Asians. Our data suggest that the interactions between n-3 PUFAs and the FADS genotype is robust for fatty acids from various sources.
The mechanisms underlying the observed gene-diet interactions remain unclear. However, such interactions are biologically plausible. It has long been known that the FADS genes such as FADS1 and FADS2 encode delta-5 and delta-6 desaturases, respectively, which are the important rate-limiting steps in the endogenous formation of long-chain PUFA such as EPA and DHA from linoleic acid (n-6) and α-linolenic acid (n-3).47 The selected allele of FADS rs174570 is significantly associated with an increase in the concentration of n-3 fatty acids upstream in the n-3 synthesis pathway.47 Further, it has been reported that dietary n-3 PUFAs might regulate adipocyte FADS expression and function.48 In addition, storage of energy and body fat is very important for the Arctic population, who are regularly exposed to extremely low temperatures and fishes rich in n-3 PUFAs.12 13 Under natural selection, these people are genetically prone to high fish intake to keep body fat.10 11 Therefore, it’s not surprising that high fish or n-3 PUFAs intake accentuated genetic susceptibility to obesity among people carrying selective FADS signature.49 50 Our findings support the view that extra n-3 PUFAs may not have much benefit for Europeans with selective FADS signature.9 14
Strengths
Several strengths of this study merit mention. To our knowledge, this is the first study with consistent results from four well-established prospective cohorts of different racial populations such as Caucasians and Singapore Chinese. The consistent results from these independent cohorts demonstrated the robustness of our findings. Other major strengths include the prospective design, the large sample size, use of long-term change of BMI and replication of the results. Although we prospectively analysed the data, we cannot exclude the possibility of reverse causality as this is a study on dietary intake and BMI or weight change from the baseline, which by default builds in the starting point (ie, the cross sectional association).
Limitations
However, several limitations need to be acknowledged. First, dietary fatty acids, fish and adiposity measures were self-reported, measurement errors in these variables are inevitable. Despite this, the food frequency questionnaires and adiposity measures data have been well validated.28 31 33–35 Second, confounding by other unmeasured or unknown factors might exist, although we have carefully adjusted for multiple dietary and lifestyle factors. Third, a causal relation among long chain n-3 PUFAs and fish consumption, and adiposity cannot be inferred from an observational study. Fourth, all subjects with genetic data were selected in each cohort. The source of genotyping data was diverse (eg, subcohort, case–control studies), therefore, subject selection might be a major source of bias. Fifth, we acknowledge that the different methods in measuring anthropometric traits, genetic variants and food intake across cohorts might introduce bias in the present analyses. Finally, the participants included in our study were middle aged and older adults of Caucasians in the USA and Chinese in Singapore, and it is unknown whether our findings could be generalised to other demographic or ethnic groups.
Conclusions
In summary, our data provide reproducible evidence from four multiethnic cohorts that high long chain n-3 PUFAs and fish intakes accentuate the genetic association of the FADS with adiposity. These findings emphasise the importance of considering precision nutritional interventions on prevention and treatment of obesity. We acknowledge that these results are hypothesis-generating and need to be confirmed in additional cohorts.
Supplementary Material
Footnotes
Contributors: TH and LQ designed the study and wrote the first draft. TH analysed the data. FBH provided statistical expertise. TW, YH, DS, CF, JW, LRP, ATC, GC, IDV, H-KC, JF, XS, CCK, YF, RMvD, HCK, J-mY, KWP, and LQ were involved in data collection. TH and LQ are guarantors. All authors contributed to the interpretation of the results and critical revision of the manuscript for important intellectual content and approved the final version of the manuscript.
Funding: This study was supported by grants HL126024, HL034594, DK100383, DK091718, HL071981, HL073168, CA87969, CA49449, CA055075, HL34594, HL088521, U01HG004399, DK080140, P30DK46200, U01CA137088, U54CA155626, DK58845, DK098311, U01HG004728, EY015473, CA134958, DK70756 and DK46200 from the National Institutes of Health, with additional support for genotyping from Merck Research Laboratories, North Wales, PA. LQ is a recipient of the American Heart Association Scientist Development Award (0730094N). LRP is supported by the Arthur Ashley Williams Foundation and a Harvard Ophthalmology Scholar Award (Harvard Medical School) from the Harvard Glaucoma Center of Excellence. ATC is a Damon Runyon Cancer Foundation Clinical Investigator. The SCHS study is supported by the National Institutes of Health, USA (NCI R01 CA144034, UM1 CA182876, R01 DK080720), the Singapore National Medical Research Council (NMRC 1270/2010), the HUJ-CREATE Programme of the National Research Foundation, Singapore (Project Number 370062002) and Biomedical Research Council, Singapore.
Disclaimer: The funding sources had no role in the design or conduct of the study; collection, management, analysis, and interpretation of the data; or preparation, review, or approval of the manuscript.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data available.
Patient consent for publication: Not required.
References
- 1. Del Gobbo LC, Imamura F, Aslibekyan S, et al. ω-3 Polyunsaturated fatty acid biomarkers and coronary heart disease: pooling project of 19 cohort studies. JAMA Intern Med 2016;176:1155–66. 10.1001/jamainternmed.2016.2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hu FB, Bronner L, Willett WC, et al. Fish and omega-3 fatty acid intake and risk of coronary heart disease in women. JAMA 2002;287:1815–21. 10.1001/jama.287.14.1815 [DOI] [PubMed] [Google Scholar]
- 3. He K, Rimm EB, Merchant A, et al. Fish consumption and risk of stroke in men. JAMA 2002;288:3130–6. 10.1001/jama.288.24.3130 [DOI] [PubMed] [Google Scholar]
- 4. Iso H, Rexrode KM, Stampfer MJ, et al. Intake of fish and omega-3 fatty acids and risk of stroke in women. JAMA 2001;285:304–12. 10.1001/jama.285.3.304 [DOI] [PubMed] [Google Scholar]
- 5. Qi L, Cho YA. Gene-environment interaction and obesity. Nutr Rev 2008;66:684–94. 10.1111/j.1753-4887.2008.00128.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qi Q, Chu AY, Kang JH, et al. Fried food consumption, genetic risk, and body mass index: gene-diet interaction analysis in three US cohort studies. BMJ 2014;348:g1610 10.1136/bmj.g1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qi Q, Chu AY, Kang JH, et al. Sugar-sweetened beverages and genetic risk of obesity. N Engl J Med 2012;367:1387–96. 10.1056/NEJMoa1203039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang T, Wang T, Heianza Y, et al. Habitual consumption of long-chain n-3 PUFAs and fish attenuates genetically associated long-term weight gain. Am J Clin Nutr 2019;109:665–73. 10.1093/ajcn/nqy238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fumagalli M, Moltke I, Grarup N, et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 2015;349:1343–7. 10.1126/science.aab2319 [DOI] [PubMed] [Google Scholar]
- 10. Hegele RA, Anderson C, Young TK, et al. G-protein beta3 subunit gene splice variant and body fat distribution in Nunavut Inuit. Genome Res 1999;9:972–7. 10.1101/gr.9.10.972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hegele RA, Huff MW, Young TK. Common genomic variation in LMNA modulates indexes of obesity in Inuit. J Clin Endocrinol Metab 2001;86:2747–51. 10.1210/jc.86.6.2747 [DOI] [PubMed] [Google Scholar]
- 12. Jørgensen ME, Bjeregaard P, Borch-Johnsen K. Diabetes and impaired glucose tolerance among the inuit population of Greenland. Diabetes Care 2002;25:1766–71. 10.2337/diacare.25.10.1766 [DOI] [PubMed] [Google Scholar]
- 13. Jørgensen ME, Glümer C, Bjerregaard P, et al. Obesity and central fat pattern among Greenland Inuit and a general population of Denmark (Inter99): relationship to metabolic risk factors. Int J Obes Relat Metab Disord 2003;27:1507–15. 10.1038/sj.ijo.0802434 [DOI] [PubMed] [Google Scholar]
- 14. Tishkoff S, Genetics TS. Strength in small numbers. Science 2015;349:1282–3. 10.1126/science.aad0584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dewailly E, Blanchet C, Lemieux S, et al. n-3 Fatty acids and cardiovascular disease risk factors among the Inuit of Nunavik. Am J Clin Nutr 2001;74:464–73. 10.1093/ajcn/74.4.464 [DOI] [PubMed] [Google Scholar]
- 16. Colditz GA, Manson JE, Hankinson SE. The Nurses' Health Study: 20-year contribution to the understanding of health among women. J Womens Health 1997;6:49–62. 10.1089/jwh.1997.6.49 [DOI] [PubMed] [Google Scholar]
- 17. Cornelis MC, Monda KL, Yu K, et al. Genome-wide meta-analysis identifies regions on 7p21 (AHR) and 15q24 (CYP1A2) as determinants of habitual caffeine consumption. PLoS Genet 2011;7:e1002033 10.1371/journal.pgen.1002033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jensen MK, Pers TH, Dworzynski P, et al. Protein interaction-based genome-wide analysis of incident coronary heart disease. Circ Cardiovasc Genet 2011;4:549–56. 10.1161/CIRCGENETICS.111.960393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wiggs JL, Kang JH, Yaspan BL, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet 2011;20:4707–13. 10.1093/hmg/ddr382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 2007;39:870–4. 10.1038/ng2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qi L, Cornelis MC, Kraft P, et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum Mol Genet 2010;19:2706–15. 10.1093/hmg/ddq156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Köttgen A, Albrecht E, Teumer A, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 2013;45:145–54. 10.1038/ng.2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peters U, Jiao S, Schumacher FR, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology 2013;144:799–807. 10.1053/j.gastro.2012.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rimm EB, Giovannucci EL, Willett WC, et al. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet 1991;338:464–8. 10.1016/0140-6736(91)90542-W [DOI] [PubMed] [Google Scholar]
- 25. Anderson GL, Manson J, Wallace R, et al. Implementation of the Women’s Health Initiative study design. Ann Epidemiol 2003;13(9 Suppl):S5–17. 10.1016/S1047-2797(03)00043-7 [DOI] [PubMed] [Google Scholar]
- 26. Anon. Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Control Clin Trials 1998;19:61–109. [DOI] [PubMed] [Google Scholar]
- 27. Yuan JM, Stram DO, Arakawa K, et al. Dietary cryptoxanthin and reduced risk of lung cancer: the Singapore Chinese Health Study. Cancer Epidemiol Biomarkers Prev 2003;12:890–8. [PubMed] [Google Scholar]
- 28. Hankin JH, Stram DO, Arakawa K, et al. Singapore Chinese Health Study: development, validation, and calibration of the quantitative food frequency questionnaire. Nutr Cancer 2001;39:187–95. 10.1207/S15327914nc392_5 [DOI] [PubMed] [Google Scholar]
- 29. Chen Z, Pereira MA, Seielstad M, et al. Joint effects of known type 2 diabetes susceptibility loci in genome-wide association study of Singapore Chinese: the Singapore Chinese health study. PLoS One 2014;9:e87762 10.1371/journal.pone.0087762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dorajoo R, Sun Y, Han Y, et al. A genome-wide association study of n-3 and n-6 plasma fatty acids in a Singaporean Chinese population. Genes Nutr 2015;10:53 10.1007/s12263-015-0502-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rimm EB, Stampfer MJ, Colditz GA, et al. Validity of self-reported waist and hip circumferences in men and women. Epidemiology 1990;1:466–73. 10.1097/00001648-199011000-00009 [DOI] [PubMed] [Google Scholar]
- 32. Huang T, Zheng Y, Qi Q, et al. DNA methylation variants at HIF3A locus, B-vitamin intake, and long-term weight change: gene-diet interactions in two U.S. cohorts. Diabetes 2015;64:3146–54. 10.2337/db15-0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Willett WC, Sampson L, Stampfer MJ, et al. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am J Epidemiol 1985;122:51–65. 10.1093/oxfordjournals.aje.a114086 [DOI] [PubMed] [Google Scholar]
- 34. Rimm EB, Giovannucci EL, Stampfer MJ, et al. Reproducibility and validity of an expanded self-administered semiquantitative food frequency questionnaire among male health professionals. Am J Epidemiol 1992;135:1114–26. 10.1093/oxfordjournals.aje.a116211 [DOI] [PubMed] [Google Scholar]
- 35. Patterson RE, Kristal AR, Tinker LF, et al. Measurement characteristics of the Women’s Health Initiative food frequency questionnaire. Ann Epidemiol 1999;9:178–87. 10.1016/S1047-2797(98)00055-6 [DOI] [PubMed] [Google Scholar]
- 36. Chiuve SE, Fung TT, Rimm EB, et al. Alternative dietary indices both strongly predict risk of chronic disease. J Nutr 2012;142:1009–18. 10.3945/jn.111.157222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wolf AM, Hunter DJ, Colditz GA, et al. Reproducibility and validity of a self-administered physical activity questionnaire. Int J Epidemiol 1994;23:991–9. 10.1093/ije/23.5.991 [DOI] [PubMed] [Google Scholar]
- 38. Langer RD, White E, Lewis CE, et al. The Women’s Health Initiative Observational Study: baseline characteristics of participants and reliability of baseline measures. Ann Epidemiol 2003;13(9 Suppl):S107–21. 10.1016/S1047-2797(03)00047-4 [DOI] [PubMed] [Google Scholar]
- 39. Li Y, Hruby A, Bernstein AM, et al. Saturated Fats Compared With Unsaturated Fats and Sources of Carbohydrates in Relation to Risk of Coronary Heart Disease: A Prospective Cohort Study. J Am Coll Cardiol 2015;66:1538–48. 10.1016/j.jacc.2015.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 41. Kabir M, Skurnik G, Naour N, et al. Treatment for 2 mo with n 3 polyunsaturated fatty acids reduces adiposity and some atherogenic factors but does not improve insulin sensitivity in women with type 2 diabetes: a randomized controlled study. Am J Clin Nutr 2007;86:1670–9. 10.1093/ajcn/86.6.1670 [DOI] [PubMed] [Google Scholar]
- 42. Munro IA, Garg ML. Prior supplementation with long chain omega-3 polyunsaturated fatty acids promotes weight loss in obese adults: a double-blinded randomised controlled trial. Food Funct 2013;4:650–8. 10.1039/c3fo60038f [DOI] [PubMed] [Google Scholar]
- 43. Thorsdottir I, Tomasson H, Gunnarsdottir I, et al. Randomized trial of weight-loss-diets for young adults varying in fish and fish oil content. Int J Obes 2007;31:1560–6. 10.1038/sj.ijo.0803643 [DOI] [PubMed] [Google Scholar]
- 44. Munro IA, Garg ML. Dietary supplementation with n-3 PUFA does not promote weight loss when combined with a very-low-energy diet. Br J Nutr 2012;108:1466–74. 10.1017/S0007114511006817 [DOI] [PubMed] [Google Scholar]
- 45. Crochemore IC, Souza AF, de Souza AC, et al. ω-3 polyunsaturated fatty acid supplementation does not influence body composition, insulin resistance, and lipemia in women with type 2 diabetes and obesity. Nutr Clin Pract 2012;27:553–60. 10.1177/0884533612444535 [DOI] [PubMed] [Google Scholar]
- 46. Bender N, Portmann M, Heg Z, et al. Fish or n3-PUFA intake and body composition: a systematic review and meta-analysis. Obes Rev 2014;15:657–65. 10.1111/obr.12189 [DOI] [PubMed] [Google Scholar]
- 47. Ameur A, Enroth S, Johansson A, et al. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am J Hum Genet 2012;90:809–20. 10.1016/j.ajhg.2012.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ralston JC, Matravadia S, Gaudio N, et al. Polyunsaturated fatty acid regulation of adipocyte FADS1 and FADS2 expression and function. Obesity 2015;23:725–8. 10.1002/oby.21035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith GI, Julliand S, Reeds DN, et al. Fish oil-derived n-3 PUFA therapy increases muscle mass and function in healthy older adults. Am J Clin Nutr 2015;102:115–22. 10.3945/ajcn.114.105833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jourdan C, Kloiber S, Nieters A, et al. Gene-PUFA interactions and obesity risk. Br J Nutr 2011;106:1263–72. 10.1017/S0007114511001541 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2018-022877supp001.pdf (495.2KB, pdf)