Abstract
Study Objectives
Daytime sleepiness is a consequence of inadequate sleep, sleep–wake control disorder, or other medical conditions. Population variability in prevalence of daytime sleepiness is likely due to genetic and biological factors as well as social and environmental influences. DNA methylation (DNAm) potentially influences multiple health outcomes. Here, we explored the association between DNAm and daytime sleepiness quantified by the Epworth Sleepiness Scale (ESS).
Methods
We performed multi-ethnic and ethnic-specific epigenome-wide association studies for DNAm and ESS in the Multi-Ethnic Study of Atherosclerosis (MESA; n = 619) and the Cardiovascular Health Study (n = 483), with cross-study replication and meta-analysis. Genetic variants near ESS-associated DNAm were analyzed for methylation quantitative trait loci and followed with replication of genotype-sleepiness associations in the UK Biobank.
Results
In MESA only, we detected four DNAm-ESS associations: one across all race/ethnic groups; three in African-Americans (AA) only. Two of the MESA AA associations, in genes KCTD5 and RXRA, nominally replicated in CHS (p-value < 0.05). In the AA meta-analysis, we detected 14 DNAm-ESS associations (FDR q-value < 0.05, top association p-value = 4.26 × 10−8). Three DNAm sites mapped to genes (CPLX3, GFAP, and C7orf50) with biological relevance. We also found evidence for associations with DNAm sites in RAI1, a gene associated with sleep and circadian phenotypes. UK Biobank follow-up analyses detected SNPs in RAI1, RXRA, and CPLX3 with nominal sleepiness associations.
Conclusions
We identified methylation sites in multiple genes possibly implicated in daytime sleepiness. Most significant DNAm-ESS associations were specific to AA. Future work is needed to identify mechanisms driving ancestry-specific methylation effects.
Keywords: diversity, epigenetics, race/ethnic heterogeneity, excessive daytime sleepiness, sleep–wake, methylation, genomics
Statement of Significance.
Excessive daytime sleepiness is associated with negative health outcomes such as reduction in quality of life, increased workplace accidents, and cardiovascular mortality. There are race/ethnic disparities in excessive daytime sleepiness. However, the environmental and biological mechanisms for these differences are not yet understood. We performed an association analysis of DNA methylation (DNAm), measured in monocytes, and daytime sleepiness within a racially diverse study population. We detected numerous DNAm markers associated with daytime sleepiness in African-Americans, but only one marker was associated with daytime sleepiness across all race/ethnic groups. Future work is required to elucidate the pathways between DNAm, sleepiness, and related behavioral/environmental exposures.
Introduction
Excessive daytime sleepiness (EDS), is estimated to affect between 10% and 20% of the population [1, 2]. EDS is associated with numerous adverse clinical, behavioral and public health outcomes, including work and vehicular accidents [3–10]; reduced health-related quality of life [11–13]; cognitive and performance deficits [14, 15]; and increased rates of stroke and total and cardiovascular mortality [16, 17]. There are multiple mechanisms for EDS, including insufficient sleep duration due to behavioral, social or work-related factors; sleep disruption due to a sleep disorder (sleep apnea, periodic limb movement disorder); circadian misalignment; the presence of a primary disorder of hypersomnia that affects the central sleep–wake control processes (e.g. narcolepsy); as well as poorly understood mechanisms occurring in a variety of medical (e.g. diabetes) and psychiatric (e.g. depression) disorders [18–20]. The prevalence of EDS has increased over the past decades, likely due to an increase in working hours and availability of electronic devices [3]. A rise in EDS prevalence may also reflect the increased prevalence of obesity, which is associated with sleepiness [21, 22]. This association is hypothesized to be due to obesity-related co-morbidities that reduce sleep quality, including obstructive sleep apnea, as well as to metabolic and neuroendocrine effects of adipokines on wake-promoting neurons [23].
Although EDS is prevalent in the population, there appears to be large inter-individual differences in propensity for sleepiness following sleep deprivation, sleep fragmentation, or sleep apnea [24, 25]. Similarly, there is significant population variability in prevalence of EDS [26]. Differences in sleepiness, including vulnerability or resilience to sleep-disrupting influences, have been suggested to be due to genetic and other biological differences, although social and environmental influences also likely play a role [27, 28]. Generally, the bases for differences in sleepiness between populations are not well understood.
Epigenetic modifications are increasingly recognized to mediate the impact of environmental influences on gene expression and on prevalence and severity of a wide range of health outcomes, including neuropsychiatric, metabolic and cardiovascular diseases. The most studied epigenetic marker, DNA methylation (DNAm), occurs when a methyl group is added to a cytosine that is followed by guanine on the genome (a “CpG” site). Changes in DNAm occur in response to a wide range of exposures, many of which are associated with sleep and sleepiness, such as obesity, diet, and stress [29–35]. Therefore, we postulated that changes in DNAm would associate with variation in sleepiness and these changes may be population-specific, providing insights into underlying susceptibility to sleepiness that may be partially attributed to environmental, and possibly genetic, factors. To the best of our knowledge, there has been no reported study on DNAm and sleepiness.
In this article, we performed an Epigenome-Wide Association Study (EWAS) of daytime sleepiness quantified by the Epworth Sleepiness Scale (ESS) in the Multi-Ethnic Study of Atherosclerosis (MESA) and in the Cardiovascular Health Study (CHS). We leveraged the racial/ethnic diversity of the sample to explore potential differences in EWAS associations by background groups. We further used the MESA dataset to study potential mechanisms underlying the associations. First, using gene-expression analysis in MESA, studying whether DNAm associated with sleepiness by modifying gene expression; and second, by identifying methylation quantitative trait loci (cis-meQTLs) near genes harboring associated methylation sites to suggest whether DNAm affects sleepiness by modifying genetic effects. Candidate genetic associations were carried forward for replication analysis in the UK Biobank.
Methods
MESA study sample
The study population consisted of participants from the MESA, a prospective, longitudinal cohort study established to study factors associated with the development of cardiovascular disease. MESA clinic visits were first performed between 2000 and 2002 in six field centers across the United States when participants were free of known cardiovascular disease [36]. The subset of MESA individuals in this study is composed of those who participated in a sleep examination conducted in conjunction with MESA Exam 5 (described in detail previously [26, 36]), and who also participated in the MESA DNAm study [37]. The blood draws for the methylation study were obtained during MESA Exam 5 (2010–2012) [38] on a random subset of MESA participants at four of the six field centers: John Hopkins University, University of Minnesota, Columbia University, and Wake Forest University. A total of 623 individuals were available with both sleep data and DNAm data. After excluding four individuals due to missing ESS scores, the final study sample included 619 individuals: 132 African-Americans (AAs), 202 Hispanic Americans (HAs), and 285 European Americans (EAs), where race/ethnicities were self-reported. The study was approved by the Institutional Review Boards of each participating site, and participants provided written informed consent, including the use of genetic data.
DNAm collection and processing in MESA
Methods for collection and assays for DNAm have been described previously [38]. In brief, peripheral blood was separated into mononuclear cells (CD14+) within two hours of collection. DNAm in monocytes was measured using the Illumina HumanMethylation450 BeadChip. Residual cell contamination in the monocyte data was assessed using Gene Set Enrichment Analysis, providing enrichment scores for neutrophils, B cells, T cells, and Natural Killer cells from Gene Expression data collected using the Illumina HumanHT-12 v4 Expression BeadChip and Illumina Bead Array Reader [38, 39]. The DNAm data underwent quality control tests prior to analysis using the lumi Bioconductor package [40]. These included color bias adjustments via smooth quantile normalization, median background adjustment, standard quantile adjustment, checks for potential sex or race mismatches, and outlier detection via multidimensional plots. Additional details on preprocessing of the DNAm data can be found in Liu et al. [37]. Following DNAm preprocessing, each of 484 882 methylation probes for each person has a Beta-value, representing the proportion of methylated monocytes at that site and person.
We further prepared the methylation data for analysis based on meta information and sample-specific characteristics, as follows. First, we excluded 61 219 probes that were within 10bp of a single nucleotide polymorphism (SNP) with a minor allele frequency greater than 1% in European, African, or American populations from the 1000 genomes reference data [41]. This was done to exclude loci where variation is solely due to genetic polymorphisms. We also removed any non-cg methylation probes and probes on sex chromosomes. We then removed any cross-reactive probes as defined by Chen et al. [42], leaving us with 399 526 DNAm probes to analyze. We used the software program ComBat [43] on these CpG sites to remove any signal due to technical artifacts of chip and position on chip effects while maintaining correlation with our primary set of covariates (self-reported ancestry, recruitment site, sex, age, and residual cell type enrichment). ComBat was run on the M-values (logit transformed Beta-values [44], see Supplementary Methods). After ComBat, the data was transformed back to the Beta-Value scale for analysis.
SNP data
For consenting individuals, DNA was extracted from whole blood and genotyped on Affymetrix 6.0 GWAS array. Standard quality control methods for SNP- and sample-level quality were applied, including the exclusion of participants and SNPs with over 5% missing call rates. This resulted in 895 289 genotyped variants in 615 individuals with DNAm data. For downstream analysis, we excluded SNPs with minor allele frequency (MAF) less than 1%. Further details on the genotype and quality control can be found in Vargas et al. [45]. We calculated the top 5 principal components (PCs) from the Linkage-Disequilibrium pruned set SNPs with MAF≥5%. PCs were calculated in the combined sample and also in each ethnic-specific group. There were four AA individuals that had missing SNP information; therefore, the PCs were imputed with the mean value of the respective PC in the AA subset that had SNP information.
Sleep assessments
As part of the MESA Sleep Exam (2010–2013), participants completed standardized questionnaires and underwent single night in-home polysomnography (Compumedics Somte Systems, Abbotsville, Australia, AU0) and 7-day wrist actigraphy (Philips-Respironics Spectrum, Murrysville, PA), as described before [26]. The primary sleep measure was daytime sleepiness as quantified by the ESS, an 8-item validated instrument that asks the individual to assess likelihood of dozing off in a variety of daily activities using a 4 point (0–3) scale [46]. ESS scores vary from 0 to 24, with higher scores denoting more sleepiness [46]. ESS was assessed within one year of blood draw for DNAm. We identified additional sleep phenotypes/exposures that might be associated with both DNAm and ESS, potentially acting as mediators or confounders of any DNAm and ESS association. These measures were: insomnia/insomnia symptoms (report of doctor-diagnosed insomnia or the Women’s Health Initiative (WHI) Insomnia Rating Scale [47], a validated scale varying from 0 to 20); Apnea-Hypopnea Index [AHI] (sum of all apneas and hypopneas associated with 3% or more oxygen desaturation divided by total sleep time by polysomnography); overnight hypoxemia (percentage of sleep time t with oxyhemoglobin saturation less than 90% [Per90]); and sleep duration (average sleep duration over the 7 monitoring nights by actigraphy as described before [26]).
Covariates
Age, sex and race/ethnicity were self-reported. Other behavioral, socioeconomic and lifestyle exposures that may confound sleepiness-methylation associations were also assessed, as follows. Alcohol use was self-reported (yes/no). Smoking status was classified by ever/former/never. Depressive symptoms were based on responses to the Center for Epidemiological Studies (CES) depression scale [48]. Socioeconomic variables were defined by having less than a college education and mother having less than a college education; these were added because they were previously shown in MESA to be associated with methylation in inflammatory genes [49]. Body mass index (BMI) was calculated from weight and height measured at MESA Exam 5. Dietary variables were derived from a food frequency questionnaire: long chain score (sum of Omega fatty acids mg/d), total fats, carbohydrates, and Total Alternative Healthy Eating Index - 2010 (Total AHEI-2010) [50]. Antidepressant and antipsychotic medications usage, potentially influencing sleepiness, were identified from medication inventories.
The CHS
The CHS is a population-based cohort study of risk factors for coronary heart disease and stroke in adults ≥65 years conducted across four field centers [51]. The original predominantly European ancestry cohort of 5201 persons was recruited in 1989–1990 from random samples of the Medicare eligibility lists; subsequently, an additional predominantly AA cohort of 687 persons was enrolled for a total sample of 5888. CHS was approved by institutional review committees at each field center and individuals in the present analysis had available DNA and provided informed consent for use of genetic information. DNAm was measured on 336 self-reported European American and 329 AA participants at study year 5. The samples were randomly selected among participants without coronary heart disease, congestive heart failure, peripheral vascular disease, valvular heart disease, stroke or transient ischemic attack at study baseline or lack of available DNA. Sleepiness was assessed at year 6 using the ESS.
Methylation measurements were performed at the Institute for Translational Genomics and Population Sciences at the Harbor-UCLA Medical Center—Los Angeles Biomedical Research Institute using the Infinium HumanMethylation450 BeadChip (Illumina Inc, San Diego, CA). Quality control was performed in the minfi R package [52–54] (version 1.12.0, http://www.bioconductor.org/packages/release/bioc/html/minfi.html). Samples were removed for any of the following: low median intensities of below 10.5 (log2) across the methylated and unmethylated channels; a proportion of probes falling detection of greater than 0.5%; QC probes falling greater than 3SD from the mean; sex-check mismatches; or failed concordance with prior genotyping or >0.5% of probes with a detection p-value > .01. In total, 11 samples were removed for sample QC resulting in a sample of 323 European-ancestry and 326 AA samples. Methylation values were normalized using the SWAN quantile normalization method [54]. Since white blood cell proportions were not directly measured in CHS they were estimated from the methylation data using the Houseman method [55].
Statistical analysis
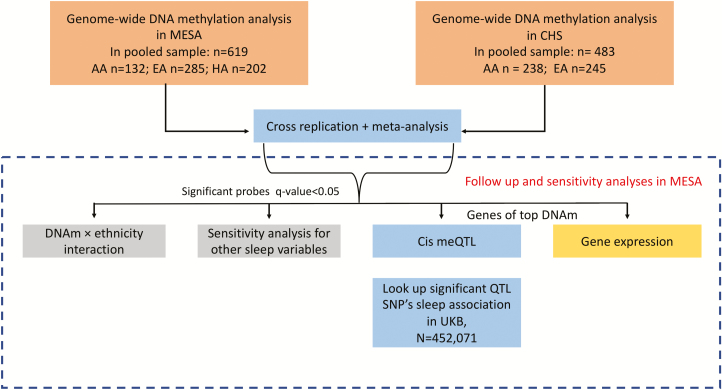
Our complete analysis approach is depicted in Figure 1. We first describe our discovery analyses, which includes a meta-analysis, followed by sensitivity, gene expression, local methylation expressive quantitative trait loci (cis-meQTL), and replication analyses.
Figure 1.
Analysis workflow for assessing associations between DNAm and ESS.
Discovery analysis for methylation-ESS association
The primary analysis outcome was ESS score, square-root transformed to achieve approximate normality. It was analyzed in a linear regression on normalized DNAm Beta-values, treated as exposures. The primary analyses were adjusted for sex, age, residual cell type enrichment (in MESA) or estimated white blood cell counts (in CHS), recruitment site, and the top 5 genetic PCs. CHS analyses were further adjusted for current smoking status (in MESA, smoking status was adjusted in sensitivity analyses, as described below). We performed discovery analysis via two different approaches. The first approach focused exclusively on each cohort (MESA, CHS), followed by cross-replication, if associations were detected. We conducted analyses in both the combined multi-ethnic sample, and in self-reported race/ethnic group-specific analyses. In MESA, combined analyses adjusted for PCs calculated in the overall group, as well as for a covariate for self-reported ethnicity (EA, AA, or HA). In group-specific analyses, we adjusted for their specific PCs. In CHS, group-specific analyses were combined in inverse-variance fixed-effects meta-analysis. We evaluated heterogeneity across race/ethnic-specific results by fitting a linear model in MESA using the overall sample with self-reported race/ethnicity interaction terms with DNAm and testing these interaction terms using an F-test via analysis of variance (ANOVA). For analyses reported in MESA only, we report replication p-values in the appropriate race/ethnic group (or combined) in CHS. Replication p-values are computed as the one-sided p-value, guided by the direction of association in MESA. This approach has been shown to be more powerful and have better control of type 1 error compared with two-sided p-values for replication [56].
The second discovery approach was a meta-analysis combining both CHS AA and EA with their respective similar MESA groups, as well as a meta-analysis across all groups from both studies. The meta-analysis was done using inverse variance weighted, fixed-effects approach.
To account for multiple testing, we controlled the false discovery rate (FDR) at the 5% level in each of the multi-ethnic and race/ethnic group-specific analyses. An association was deemed as significant if the q-value was less than 0.05 [57].
Sensitivity analysis for methylation-ESS associations
We next identified behavioral, socioeconomic and lifestyle exposures, including variation in sleep and sleep disturbances, that may influence the sleepiness-methylation associations and further adjusted for these factors in sensitivity analyses conducted in significant probe-trait primary associations. Covariates considered for the sensitivity analysis were: BMI, sleep duration (actigraphy-based), insomnia (both doctor diagnosed and the WHI insomnia rating scale), AHI, nocturnal hypoxemia [Per90], alcohol use, smoking status, CES depression scale, having less than a college education, mother having less than a college education, and dietary variables, as defined earlier. We also examined moderate sleep apnea (AHI > 15), short sleep (<5 hours), and overnight hypoxemia (Per90 > 5%). Complete details for this sensitivity analysis are provided in the Supplementary Material.
In another sensitivity analysis, we repeated analyses in MESA for the top association, after excluding individuals who reported use of antidepressant or antipsychotic medications.
Screening for cis-meQTLs in MESA
We hypothesized that genetic associations underlie some of the DNAm-ESS associations. Due to our limited sample size, we implemented a screening process on genetic variants in the CpG region, followed by a replication analysis of the SNP-sleepiness association in an independent cohort (UK Biobank).
First, we mapped all DNAm sites that passed the significance threshold (q-value < 0.05) to their respective genes to assess if genetic variants in that gene were associated with both ESS and DNAm. Specifically, for each gene that a top DNAm site mapped to, we performed association analysis between genotyped SNPs within 100kb of the gene (as exposures) and the associated DNAm site (as an outcome) adjusting for age, sex, top five PCs, study site, and residual cell type. This identified local (cis) meQTLs (screening step 1). SNPs with associations at p-value < 0.01 were next assessed for their association with sleepiness, with and without additional adjustment for the DNAm (screening step 2). These two screening steps were performed on the MESA subsample of the race/ethnic group in which the DNAm-ESS association was detected, and in all groups if the association was detected in the combined analysis. SNPs that were associated with ESS with p-value < 0.05 were carried forward to independent replication analysis (see below) of the SNP-sleepiness association in the UK Biobank [58]. We also report SNPs-ESS associations in MESA race/ethnic groups that did not identify the original DNAm-ESS association, while noting that such associations may not be detected due to lower sample size and power.
Replication analysis of cis-meQTL-sleepiness association in the UK Biobank
The UK Biobank is a large prospective study for a wide range of genetics and health outcomes in over 500 000 participants aged 40–69 years recruited from 2006 to 2010 in the United Kingdom [59]. Genotyping and imputation data are described in Bycroft et al. (2017) [60]. Sleepiness was assessed by self-reported responses to the question “How likely are you to dose off or fall asleep during the daytime when you don’t mean to? (e.g. when working, reading or driving)” with the options of “Never/rarely”, “sometimes”, “often”, “all of the time”, “do not know”, and “prefer not to answer.” For SNPs passing the cis-meQTL screening process in MESA, association analysis was performed in 452 071 individuals adjusted for age, sex, genotyping array, 10 PCs and genetic relatedness matrix.
Gene expression analysis
For genes corresponding to the DNAm sites with FDR q-value < 0.05, we used gene expression data in MESA to study the evidence for association between DNAm and gene expression (of the same gene), and gene expression association with ESS. Gene expression profiling and processing are described in detail in Liu et al. [37]. In brief, gene expression was measured using Illumina HumanHT-12 v4 Expression BeadChip array in mononuclear cells, on the same participants with DNAm measures. We performed (1) association analysis of DNAm and gene expression, using all individuals with available data (234 AA, 386 HA, and 582 EA individuals), with ESS as the outcome and DNAm as exposure; and (2) association analysis of gene expression (as exposure) and squared-root ESS (as the outcome) in the set of individuals that had both gene expression and ESS measured. Complete details are provided in the Supplementary Material.
Results
Study characteristics
The MESA sample consisted of 619 individuals (53% female) with a mean age of 68 years, with 132 AA, 202 HA, and 285 EA individuals (Table 1). The mean ESS was 6.0 and 14% were classified as having EDS (ESS > 10), with the highest prevalence of EDS among AAs (18%; p = 0.07). Sleep duration was significantly shorter in the AA group compared to the other groups (p-value < 0.001). Mean AHI was 19.6, and 46% of the sample had moderate or more severe sleep apnea (AHI > 15). Across groups, sleep apnea was more prevalent and severe in the HA group (Table 1). Significant differences were observed for BMI, having a college education, smoking status, sleep apnea, sleep duration, dietary long-chain score, and alcohol consumption by race/ethnic group (Table 1). We provide information on the median of these variables as well as the number of missing values in Supplementary Table 1.
Table 1.
MESA study participants characteristics
| Overall | EA | AA | HA | p-value | |
|---|---|---|---|---|---|
| Total | 619 | 285 | 132 | 202 | |
| Age (years) | 68.72 (9.24) | 69.58 (9.55) | 69.05 (8.90) | 67.29 (8.88) | 0.024 |
| Sex, males (%) | 290 (46.8%) | 138 (48.4%) | 53 (40.2%) | 99 (49.0%) | 0.219 |
| Recruitment site (%) | <0.001 | ||||
| Columbia University | 193 (31.2%) | 38 (13.3%) | 51 (38.6%) | 104 (51.5%) | |
| John Hopkins University | 165 (26.7%) | 86 (30.2%) | 79 (59.8%) | 0 (0.0%) | |
| University of Minnesota | 236 (38.1%) | 138 (48.4%) | 0 (0.0%) | 98 (48.5%) | |
| Wake Forest University | 25 (4.0%) | 23 (8.1%) | 2 (1.5%) | 0 (0.0%) | |
| ESS Score | 6.05 (4.04) | 5.93 (3.78) | 6.74 (4.45) | 5.76 (4.10) | 0.075 |
| Excessive Daytime Sleepiness (ESS > 10) | 85 (13.7%) | 38 (13.3%) | 24 (18.2%) | 23 (11.4%) | 0.204 |
| BMI (kg/m2) | 29.79 (5.55) | 28.96 (5.55) | 30.73 (5.51) | 30.35 (5.43) | 0.002 |
| AHI (events/hour) | 19.59 (18.79) | 19.12 (19.60) | 17.68 (17.77) | 21.45 (18.25) | 0.184 |
| Moderate to Severe Apnea (AHI > 15) | 267 (46.05%) | 109 (42.2%) | 53 (41.7%) | 105 (53.8%) | 0.027 |
| Nocturnal Hypoxemia (%time < 90% saturation) | 4.22 (10.28) | 5.25 (12.48) | 3.34 (9.44) | 3.43 (6.97) | 0.097 |
| Nocturnal Hypoxemia ≥ 5% | 108 (18.6%) | 56 (21.7%) | 18 (14.2%) | 34 (17.4%) | 0.177 |
| Sleep Duration (minutes) | 391.42 (79.83) | 410.20 (71.77) | 360.48 (87.11) | 385.13 (78.76) | <0.001 |
| Less than 5 hours of sleep a night | 73 (11.9%) | 19 (6.8%) | 32 (24.6%) | 22 (11.0%) | <0.001 |
| Less than a college education (%) | 204 (33.0%) | 54 (18.9%) | 42 (32.1%) | 108 (53.5%) | <0.001 |
| Mother having less than a college education (%) | 485 (80.8%) | 197 (69.9%) | 107 (82.9%) | 181 (95.8%) | <0.001 |
| Doctor Diagnosed Insomnia | 34 (5.5%) | 11 (3.9%) | 9 (6.8%) | 14 (6.9%) | 0.262 |
| WHI Insomnia Rating Scale | 7.65 (4.68) | 7.44 (4.63) | 7.67 (4.51) | 7.95 (4.86) | 0.494 |
| CES Depression Scale | 8.33 (7.34) | 7.82 (6.93) | 8.20 (7.21) | 9.15 (7.94) | 0.144 |
| Currently Drink Alcohol | 286 (46.3%) | 167 (58.6%) | 53 (40.2%) | 66 (32.8%) | <0.001 |
| Smoking Status | 0.005 | ||||
| Never | 242 (39.2%) | 102 (35.8%) | 50 (37.9%) | 90 (44.8%) | |
| Former | 325 (52.6%) | 163 (57.2%) | 62 (47.0%) | 100 (49.8%) | |
| Current | 51 (8.3%) | 20 (7.0%) | 20 (15.2%) | 11 (5.5%) | |
| Nutrients: Total Fat (g) | 72.97 (44.14) | 73.06 (38.90) | 79.11 (53.50) | 68.68 (44.13) | 0.113 |
| Nutrients: Total Carbohydrates (g) | 215.13 (115.39) | 207.70 (90.12) | 226.91 (144.91) | 218.06 (125.33) | 0.264 |
| Long chain fatty acid score (mg/day) | 2.56 (2.36) | 2.23 (2.03) | 3.32 (2.80) | 2.53 (2.36) | <0.001 |
| Total AHEI-2010 | 54.00 (10.48) | 54.85 (10.70) | 52.88 (11.43) | 53.51 (9.38) | 0.148 |
| Antipsychotic medications | 4 (0.6%) | 1 (0.4%) | 1 (0.8%) | 2 (1.0%) | 0.675 |
| Non-tricyclic Antidepressants (other than MAOI) | 79 (12.8%) | 57 (20.1%) | 4 (3.0%) | 18 (9.0%) | <0.001 |
| Tricyclic antidepressants | 12 (1.9%) | 7 (2.5%) | 2 (1.5%) | 3 (1.5%) | 0.689 |
Data presented as mean (SD) or n (%). CES depression scale originated from the Center for Epidemiological Studies. AHEI is the Alternative Healthy Eating Index. WHI is the Women’s Health Initiative, and the scale ranges from 0 to 20. Test of difference were either done via regression (continuous variables) or chi-squared test (categorical variable).
The CHS sample consisted of 483 participants (63% females) with 238 AA and 245 EA (Table 2). The mean age was 74 years and the mean ESS was 6.0 with 15% being classified as having EDS, with similar trends for a higher prevalence of EDS among AAs (Table 2).
Table 2.
CHS study participants characteristics
| Overall | AA | EA | p-value | |
|---|---|---|---|---|
| Total | 483 | 238 | 245 | |
| Age, years | 74.03 (5.3) | 72.79 (5.4) | 75.23(4.9) | <0.001 |
| Sex, males (%) | 177 (37%) | 82 (34%) | 95 (38%) | 0.370 |
| Epworth Sleepiness Scale Score | 5.98 (4.1) | 6.38 (4.4) | 5.58 (3.8) | 0.037 |
| Excessive Daytime Sleepiness (ESS > 10) | 72 (15%) | 43 (18%) | 29 (12%) | 0.073 |
| BMI (kg/m2) | 27.8 (5) | 29.0 (4.9) | 26.7 (4.8) | <0.001 |
| Currently drink alcohol | 209 (43%) | 97 (41%) | 112 (46%) | 0.310 |
| Smoking status | 0.071 | |||
| Never | 216 (45%) | 108 (45%) | 108 (44%) | |
| Former | 207 (43%) | 93 (39%) | 114 (47%) | |
| Current | 60 (12%) | 37 (16%) | 23 (9%) |
Data is presented as mean (SD) or n (%).
Associations between sleepiness and DNAm
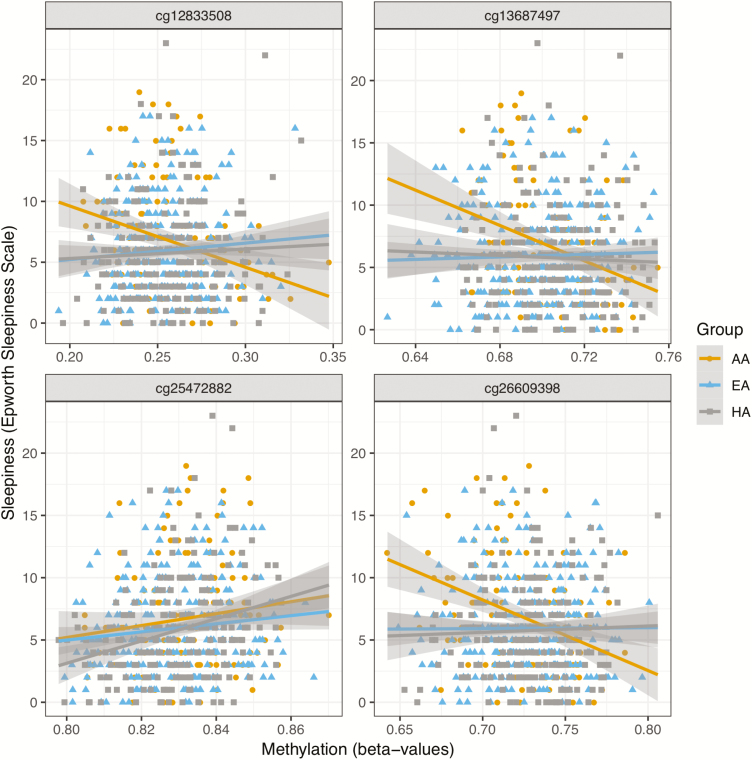
We report histograms of p-values from the various analyses as well as inflation factors in Supplementary Figure 1. The genomic inflation ranged from 0.97 (CHS combined) to 1.09 (MESA AA). Results from the discovery analysis of each cohort with cross-replication in the other cohort are reported in Table 3. This analysis identified four significant associations (FDR q-value < 0.05), all observed in MESA. A single DNAm-ESS association was detected in the combined analysis across MESA race/ethnic groups at cg25472882 located near gene C7orf50 (p-value = 6.59 × 10–8). There were three DNAm-ESS associations observed in the AA group: cg12833508 (p-value = 1.38 × 10–7), cg26609398 (p-value = 2.33 × 10–7), and cg13687497 (p-value = 3.66 × 10–7) located near genes AP1S3, KCTD5, and RXRA, respectively (Table 3). For each of these AA-specific associations, tests of interaction between race/ethnic group and DNAm on the transformed ESS scores conducted in the overall MESA sample were significant (Table 3). The race/ethnic associations between DNAm in these four sites are visualized in Figure 2. Replication (one-sided) p-values in CHS are reported in Table 3. Two DNAm sites were nominally associated with ESS in CHS: cg26609398 (replication p-value = 0.028), and cg13687497 (replication p-value = 0.045). Considering DNAm sites with FDR q-value < 0.1, there were two significant associations in DNAm sites within RAI1, a gene associated with several sleep and circadian phenotypes [61, 62]. Therefore, we further investigated these DNAm sites in follow-up analyses.
Table 3.
Methylation sites associated (FDR q-value ≤ 0.05) with the Epworth Sleepiness Scale (square-root transformed) in MESA either in AA (n = 132) or in the combined analysis (n = 619) along with their association in CHS AA (n = 238) or CHS combined (n = 483)
| Probe | CHR | Location | Analyses race/ethnic group | Gene | Effect estimate in MESA | MESA p-valuea |
MESA q-valuea |
MESA interaction p-valueb |
Effect estimate in CHSc | p-value in CHSc | p-value meta analysisd |
|---|---|---|---|---|---|---|---|---|---|---|---|
| cg25472882 | 7 | 1112040 | Combined | C7orf50 | 15.26 | 6.59e-08 | 0.0237 | 0.312 | −1.77 | 0.49 | 0.00125 |
| cg12833508 | 2 | 224701802 | AA | AP1S3 | −17.31 | 1.38e-07 | 0.0393 | 0.00021 | 1.19 | 0.54 | 1.42e-02 |
| cg26609398 | 16 | 2752235 | AA | KCTD5 | −14.88 | 2.33e-07 | 0.0412 | 0.00067 | −3.30 | 0.028 | 4.95e-06 |
| cg13687497 | 9 | 137249839 | AA | RXRA | −20.98 | 3.66e-07 | 0.0393 | 0.00024 | −3.90 | 0.045 | 2.63e-05 |
aModels adjusted for age, sex, residual cell type enrichment, site of recruitment, and top five principal components calculated within the group listed in the analysis column. If Analyses column indicates ‘Combined’, adjustment was also made for self-reported race/ethnicity.
bModels adjusted for age, sex, residual cell type enrichment, recruitment site, top five principal components in the overall sample, and self-reported race/ethnicity when testing for interaction between DNAm Beta-value and self-reported race/ethnicity in overall sample.
cAssociation results in respective CHS analysis. p-value is based on one-sided p-values informed by the direction of effect.
d p-value from meta-analysis of MESA and CHS. Genomic locations are provided in genome build 37.
Figure 2.
Four ESS-DNAm associations detected in MESA and reported in Table 3. The bottom left panel displays an association that was detected in the (combined) MESA sample, while the other panels display associations that were detected only in the AA group.
In a second discovery step, we performed meta-analysis of all DNAm for the two cohorts, by race/ethnic groups and across all groups (Table 4). Fourteen associations (FDR q-value ≤ 0.05) were detected, all in the meta-analysis of MESA AA and CHS AA. Of these, eight DNAm sites had p-value < 0.05 in a test for interaction of DNAm and self-reported race/ethnicity (Supplementary Table 2-Sheet 2). The top associated DNAm site was cg18371750 (meta p-value = 4.26 × 10–8) located in the HRH2 gene. In 13 of the 14 DNAm sites, decreased methylation was correlated with an increase in ESS (Table 4).
Table 4.
Methylation sites associated (FDR q-value ≤ 0.05) with the Epworth Sleepiness Scale (square-root transformed) in the meta-analysis of MESA AA (n = 132) and CHS AA (n = 238)
| Probe | CHR. | Location | Gene | Effect estimate in MESA AAa | MESA p-valuea |
Effect estimate in CHS AA | p-value in CHS AA | p-value in meta-analysis | q-value in meta-analysis |
|---|---|---|---|---|---|---|---|---|---|
| cg18371750 | 5 | 175112799 | HRH2 | −22.94 | 1.02E-02 | −22.47 | 2.65E-06 | 4.26E-08 | 0.0138 |
| cg26130090 | 12 | 54615724 | −7.85 | 2.82E-05 | −6.16 | 7.89E-04 | 1.26E-07 | 0.0164 | |
| cg23574298 | 2 | 55382186 | −17.11 | 1.21E-03 | −16.16 | 6.05E-05 | 1.53E-07 | 0.0164 | |
| cg01904985 | 15 | 75117658 | CPLX3/LMAN1L | −18.19 | 1.24E-04 | −14.90 | 2.92E-04 | 2.55E-07 | 0.0195 |
| cg02374944 | 15 | 91463093 | MAN2A2 | −12.36 | 1.27E-05 | −9.06 | 1.83E-03 | 3.02E-07 | 0.0195 |
| cg13588265 | 17 | 1382042 | MYO1C | −14.66 | 4.37E-03 | −13.70 | 5.96E-05 | 6.21E-07 | 0.0335 |
| cg01769243 | 17 | 42992917 | GFAP | −13.19 | 3.06E-04 | −10.96 | 5.69E-04 | 8.62E-07 | 0.0398 |
| cg11335203 | 1 | 6398594 | ACOT7 | −14.23 | 2.65E-02 | −15.19 | 3.89E-05 | 2.14E-06 | 0.0494 |
| cg03737424 | 16 | 67550511 | −11.84 | 1.53E-04 | −9.42 | 3.71E-03 | 2.12E-06 | 0.0494 | |
| cg00181327 | 13 | 31439278 | −17.86 | 7.61E-04 | −15.58 | 4.74E-04 | 1.16E-06 | 0.0425 | |
| cg00583046 | 6 | 31024990 | HCG22 | −11.07 | 5.24E-05 | −8.36 | 1.92E-03 | 1.18E-06 | 0.0425 |
| cg00331852 | 16 | 88723987 | MVD | −12.34 | 4.69E-05 | −9.03 | 4.11E-03 | 1.98E-06 | 0.0494 |
| cg13776718 | 9 | 139617752 | FAM69B | −9.97 | 1.97E-03 | −8.74 | 2.06E-04 | 1.83E-06 | 0.0494 |
| cg21883754 | 19 | 54926437 | TTYH1 | 21.19 | 1.09E-01 | 21.88 | 1.12E-05 | 1.89E-06 | 0.0494 |
aModels adjusted for age, sex, residual cell type enrichment, site of recruitment, and top five principal components calculated in AA.
Genomic location is provided in genome build 37.
Supplementary Table 2-Sheet 1 reports association results from all MESA and CHS race/ethnic groups for all associations reported in Tables 3 and 4. Supplementary Tables 3 and 4 report results from sensitivity analysis. Overall, the analysis did not highlight any phenotype as driving any of the observed DNAm-ESS associations, with the exception that the ESS association with cg03737424 significantly changed in the model with the sensitivity variables (permutation p-value = 0.03). Removing medication users from the analyses resulted in little difference in results.
Cis-meQTL analysis and UK Biobank replication
We performed cis-meQTL analysis for the DNAm reported in Tables 3 and 4, which mapped to 14 genes. Across the genes, we tested 877 SNPs. Of these, 20 SNPs passed screening step 1 (p-value < 0.01; Supplementary Table 5), three of which passed screening step 2. Rs9896285 of RAI1 was associated with ESS both in the model with and without adjustment to DNAm at the site of interest. In contrast, rs10785870 of RXRA and rs7495739 of LMAN1L/CPLX3 were associated with ESS only in the model that was unadjusted for DNAm at the relevant site, suggesting that DNAm mediated an association between these SNPs and sleepiness.
These SNPs were carried forward for testing the SNP-sleepiness association in the UK Biobank data set of European Ancestry individuals (Supplementary Table 6). An associated was deemed replicated if it had a replication p-value threshold < 0.05/3 = 0.017, were replication p-value is a one-sided p-value guided by the direction of association in the discovery sample. Only rs9896285 in RAI1 replicated with p-value = 6.5 × 10−5. Notably, the two other associations were nominally significant, with replication p-values 0.024 and 0.038.
Gene expression analysis
Comprehensive results are provided in Supplementary Tables 7 and 8. Briefly, we considered six genes corresponding to some of DNAm sites reported in Tables 3 and 4, and that also had expressed transcripts in the gene expression data set. Of the two DNAm sites associated with RAI1, cg14720773 had p-value = 0.05 in the combined cohort in association with expression of one of the RAI1 transcripts, but the expression of this transcript was not associated with ESS. In contrast, cg27208169 had p-value = 0.04 in the MESA AA analysis with a second RAI1 transcript, and increased expression of this transcript was associated with lower ESS (p-value = 0.002 in the combined cohort). None of the other associations were nominally significant.
Summary statistics and code availability
Code and results have been deposited on GitHub (https://github.com/tamartsi/sleepiness_methylation).
Discussion
This study reports for the first time a DNAm association analysis in a multi-ethnic population designed to detect DNAm markers associated with daytime sleepiness. We performed association analyses in two cohorts, MESA and CHS, using both a cross-replication approach and a meta-analysis, examining analyzing data for each race/ethnicity group (AA, EA, and HA) individually as well as by pooling groups. In the cross-replication analysis, two associated DNA sites discovered in MESA AAs (cg26609398, KCTD5 gene, and cg13687497, RXRA gene) replicated in CHS AAs. In the meta-analysis, we discovered additional 14 DNAm associations in 11 genes in CHS and MESA AAs. At a higher statistical threshold, we also identified DNAm sites in the RAI1 gene (FDR adjusted p-value < 0.1). Some of the genes that we identified have strong biological relevance and were previously reported as associated with several sleep traits [61–66].
Our study focused on the multi-ethnic epigenetic bases of daytime sleepiness. Several studies have shown that even after accounting for sleep duration and sleep disorders, sleepiness is lower in EA compared to other ethnic groups [20, 26, 67–69]. Compared to EA groups, AAs also have a higher prevalence of both short and long sleep durations and sleep apnea syndrome (defined by an elevated AHI plus daytime sleepiness [26]) and in the larger MESA cohort, AA had higher ESS than EA [26]. Race/ethnicity differences in sleep patterns and susceptibility to EDS appear early in life. AA children have been shown to have shorter sleep duration than white children as early as 1 year of age [70]. In children with sleep apnea treated with adenotonsillectomy, AA children have higher sleepiness scores than EA children both at baseline and following treatment [71]. Here, in our study of a middle-aged to older community multi-racial sample, we found significant epigenetic findings mostly in the AA samples of two independent cohorts. The specificity of the findings in the AA sample could reflect differences in underlying genetic architecture that influence susceptibility to methylation or differences in environmental exposures, such as air pollution, or stress that differentially affect different groups [72–75]. We address this hypothesis using sensitivity analyses that explored potential explanatory effects due to sleep variation and other lifestyle exposures. These analyses mostly did not find any phenotype explaining the association, when comparing the results to those obtained via permutation, with the exception of one DNAm-ESS association. However, these measures do not characterize lifetime exposures nor included precise measurements of the social or physical environments, emphasizing the need for additional research that links key aspects of an individual’s environmental and sociological exposures with epigenetic markers and health conditions such as sleepiness as important routes for understanding health disparities.
A few of our sleepiness-associated DNAm sites are within genes that have strong evidence for associations with sleep-related processes. In the MESA discovery analysis, the most significant DNAm association was in the combined MESA cohort (cg25472882, p-value = 6.59 × 2−8, CHS replication p-value = 0.75), in gene C7orf50. Among sub-Saharan Africans [76] different DNAm sites in this gene were associated with type 2 Diabetes, a condition commonly associated with sleep problems. Epigenetic analysis of cord blood samples showed that epigenetic changes in this gene also were associated with serum immunoglobulin E in children [77].
Three DNAm associations were discovered in MESA AA only, and two of these has replication p-value < 0.05 in CHS. One of these associations was in the KCTD5 gene (cg26609398 p-value 2.33 × 10–7, CHS AA replication p-value = 0.028). KCTD5 is a mammalian ortholog of the Drosophila gene Inc, a gene expressed in multiple sleep associated regions in the central nervous system and shown to play a fundamental role in sleep in Drosophila by interacting with Cullin-3 ubiquitin ligase (Cul3). When mutated, both genes (KCTD5 and Cul3) produce similar phenotypes characterized by markedly shortened and fragmented sleep together with altered synaptic structure and impaired synaptic transmission in relevant neuronal circuits [63]. This suggests that alterations in protein dynamics (e.g. ubiquitination) might disrupt synaptic function and impact sleep. Future research is needed to examine how epigenetic changes in KCTD5 and similar genes influence gene expression and core elements of the sleep homeostat, with initial work indicating potential protein interactions with the circadian genes FBXW11 and BTRC [78].
In MESA AA and replicated in CHS AA, DNA methylation association was also observed for cg13687497, within the RXRA gene, a retinoic acid receptor involved in lipid metabolism and cellular senescence [79–81]. Genes involved in retinoic acid pathways may be particularly relevant to sleepiness phenotypes given the central role of retinoic acid in central nervous system signaling pathways. Interestingly, two of our associated methylation sites (at a less significant statistical threshold) were within RAI1, which is induced by retinoic acid. RAI1 encodes a protein that is highly expressed in neuronal tissues and is involved in early neural differentiation and transcriptional regulation of circadian clock components [62, 64–66]. Variants in this gene have been associated with circadian rhythm disruption, several abnormal sleep traits, neurobehavioral problems and obesity [64, 66]. In addition, SNPs in RAI1 have been found to be associated with sleep apnea [61]. In addition to our DNAm-sleepiness association, we identified additional evidence supporting a role of RAI1 in sleepiness: (1) expression of RAI1 in mononuclear cells was associated with ESS in MESA; (2) our cis-meQTL analysis detected a SNP in RAI1 influencing sleepiness in an EA population from the UK Biobank. Moreover, our analysis in MESA suggests that the SNP rs9896285 has both direct and indirect (through DNAm) effects on sleepiness in MESA AAs. The nonsignificant associations in MESA HAs and EAs could relate to differences in genetic architecture, environmental exposures or to lower power, which is a result of both low sample size and lower allele frequency in EAs and HAs compared to AAs (0.04 and 0.11 in MESA EAs and HAs, respectively, compared to 0.28 in MESA AAs). More work is needed to further dissect the association of retinoic acid pathway genes with sleepiness.
In the meta-analysis of MESA and CHS AAs, we discovered 14 DNAm associations with ESS, revealing relevant biological mechanism associated with sleepiness. The most significant association was in the Histamine Receptor H2 (HRH2) gene (cg18371750, AA meta p-value = 4.3 × 10−8). The histaminergic system plays a central role in modulating brain activity, including alertness. H2 receptors are found in a wide variety of tissues, including neurons, where cross-talk with H1 receptors modulate complex signaling pathways [82]. Genetic variability in this gene is associated with some neurological diseases (e.g. Parkinson, autism [82–84]) and in pain reception in knockout mice [85]. Another association was in the CPLX3/LMAN1L genes region (cg01904985, p-value = 2.5 × 10−7). CPLX3 is involved in light sensitization and in thalamo-cortical integration, which are key for sleep regulation [86, 87]. Both genes are highly expressed in brain tissue (see Supplementary Figure 2). Our cis-meQTL analysis identified rs7495739 associated with these genes which predicted methylation, was associated with ESS in MESA AAs (p-value = 0.034), and was also nominally associated with sleepiness in UK Biobank whites (replication p-value = 0.038). Another gene highlighted by the meta-analysis is the Glial Fibrillary Acidic Protein (GFAP, cg01769243, p-value = 8.6 × 10−7), which is also highly expressed in brain tissues (Supplementary Figure 3), and is associated with nervous system disease [88].
Cedernaes et al. [89] recently reported numerous associations between experimental sleep loss in 15 individuals and DNAm in adipose tissue. Of the 148 DMRs reported (Supplementary Table 2 in Cedernaes et al.), seven DNAm sites (cg25265360, cg05481452, cg01771651, cg25265360, cg23781467, cg20815339, and cg01168201) were within 5000 bp of a DNAm site associated with ESS in our analysis of MESA AA at the FDR < 0.1 level. Interestingly, Cedernaes et al. reported that most of the differentially methylated regions following sleep loss were hypermethylated, while in our analysis, sleepiness was mostly associated with hypomethylation in the top associated DNAm sites. We studied whether this could be due to a global pattern of hypomethylation among participants with EDS in MESA. We computed mean global methylation for all participants and compared the distribution of these means between individuals with EDS, and others. However, the mean and median global methylation averages were nearly identical between the groups (Supplementary Figure 4). There are multiple differences in the study by Cedernaes et al. [89] and our, including differences in sleep exposures, the age of study participants and tissue assayed (adipose tissue vs blood).
A unique feature of this study is its multi-ethnic sample. In MESA alone, we identified three DNAm-sleepiness associations that are only present in AAs, and one association in the combined population. Two of the AA-specific associations replicated in CHS. We further identified 14 AA-specific associations in the meta-analysis of MESA and CHS. DNAm differences in some of these sites may explain some of the differences in sleepiness between AAs and EAs, highlighting potential mechanisms to explore in studies of health disparities. A limitation is that we had small sample sizes in the group-specific analyses. Despite this, we could directly replicate two of the associations discovered in MESA AAs. Lack of further replication may relate to many factors, including the older sample of CHS participants, biases and misclassification in self-reported sleepiness assessment [90], and potential difference in their exposure history compared to MESA participants. Another limitation is the time difference in measurement of DNAm, and sleep study. Participants filled the ESS questionnaire within a year following the blood draw for DNAm, and both DNAm and ESS may vary over this time frame. In addition, we examined DNAm within monocytes, while sleep is organized in the brain. Despite this, we identified associated DNAm sites near genes with potential neurological impact. A limitation which is shared by many epigenome-wide association studies, is the lack of identified exposure explaining the change in methylation and the group-specificity of the associations. We did study potential mechanism by considering cis-meQTLs analysis and a sensitivity analysis adjusting for various, plausible, exposure variable (sleep, lifestyle, and nutrition variables) which may be independently associated with either DNAm, ESS, or both, but these analyses did not lead to strong hypotheses about causal pathways.
In conclusion, we identified multiple DNAm sites associated with sleepiness. For the majority of these sites, sleepiness was associated with decreased methylation. Some of our top DNAm sites are near genes known to be associated with critical neuronal processes that influence sleep. Taken together, our findings suggest that these sleepiness-associated DNAm sites are related to genes that have a biologically meaningful function with regards to sleep. The significance of our findings, which were mostly in AAs, identifies biological areas that may contribute to the observed differences in sleepiness between AAs and EAs, providing an avenue for further investigation, both genetic and environmental, of health disparities. As mentioned in the introduction, poor sleep outcomes are associated with negative life outcomes and a recent review presented extensive evidence that obstructive sleep apnea and aging share various biological pathways [91]. Future studies with larger sample sizes may elucidate these genomic pathways and how they relate to potential biological or sociological patterns that are being reflected in excessive sleepiness.
Funding
R.T.B. was funded by National Cancer Institute (NCI) grant T32CA094880 and National Heart, Lung, and Blood Institute (NHLBI) grant R01HL113338. S.R. and T.S. were partially funded by R35 HL135818. H.W. was supported by NHLBI R01HL113338 (to S.R.) and Sleep Research Society Foundation Career Development Award 018-JP-18 (to H.W.). R.S. was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01DK107859, R01DK102696, and R01DK105072. MESA: This research was supported by the Multi-Ethnic Study of Atherosclerosis (MESA). MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR-001881, and DK063491. MESA SLEEP was funded by R01HL098433. Funding support for the Sleep Polysomnography dataset was provided by grant HL56984. The MESA Epigenomics Studies were funded by NHLBI R01HL101250, NIDDK R01 DK103531-01, R01DK103531, National Institute on Aging (NIA) R01 AG054474, and NHBLI R01 HL135009-01 to Wake Forest University Health Sciences. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, TSCI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research (DRC) grant DK063491. Funding for MESA SNP Health Association Resource (SHARe) genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, CA) and the Broad Institute of Harvard and MIT (Boston, MA) using the Affymetrix Genome-Wide Human SNP Array 6.0. CHS: Infrastructure for the CHARGE Consortium is supported in part by the National Heart, Lung, and Blood Institute grant R01HL105756. The CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants U01HL080295, U01HL130114, K08HL116640, R01HL087652, R01HL092111, R01HL103612, R01HL105756, R01HL103612, R01HL111089, R01HL116747 and R01HL120393 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 from the National Institute on Aging (NIA), Merck Foundation/Society of Epidemiologic Research as well as Laughlin Family, Alpha Phi Foundation, and John Locke Foundation. A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes’ Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest statement. S.R. receives grants from the National Institute of Health (NIH), American Sleep Medicine Foundation and Jazz Pharmaceuticals. She received consulting fees from Jazz Pharmaceuticals.
Supplementary Material
Acknowledgments
We thank the participants of the UK Biobank; replication of the SNP-sleepiness association was performed under application 6818. The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
References
- 1. Hublin C, et al. . Daytime sleepiness in an adult, Finnish population. J Intern Med. 1996;239(5):417–423. [DOI] [PubMed] [Google Scholar]
- 2. Ohayon MM. From wakefulness to excessive sleepiness: what we know and still need to know. Sleep Med Rev. 2008;12(2):129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsai SC. Excessive sleepiness. Clin Chest Med. 2010;31(2):341–351. [DOI] [PubMed] [Google Scholar]
- 4. Mullins HM, et al. . Sleepiness at work: a review and framework of how the physiology of sleepiness impacts the workplace. J Appl Psychol. 2014;99(6):1096–1112. [DOI] [PubMed] [Google Scholar]
- 5. Cohen DA, et al. . Uncovering residual effects of chronic sleep loss on human performance. Sci Transl Med. 2010;2(14):14ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Connor J, et al. . The role of driver sleepiness in car crashes: a systematic review of epidemiological studies. Accid Anal Prev. 2001;33(1):31–41. [DOI] [PubMed] [Google Scholar]
- 7. Ellen RL, et al. . Systematic review of motor vehicle crash risk in persons with sleep apnea. J Clin Sleep Med. 2006;2(2):193–200. [PubMed] [Google Scholar]
- 8. Herman J, et al. . Driver sleepiness and risk of motor vehicle crash injuries: a population-based case control study in Fiji (TRIP 12). Injury. 2014;45(3):586–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Philip P, et al. . Sleep disorders and accidental risk in a large group of regular registered highway drivers. Sleep Med. 2010;11(10):973–979. [DOI] [PubMed] [Google Scholar]
- 10. Terán-Santos J, et al. . The association between sleep apnea and the risk of traffic accidents. Cooperative Group Burgos-Santander. N Engl J Med. 1999;340(11):847–851. [DOI] [PubMed] [Google Scholar]
- 11. Briones B, et al. . Relationship between general health status, sleepiness, and sleep apnea. Am J Respir Crit Care. 1995;151:A104. [Google Scholar]
- 12. Gooneratne NS, et al. . Functional outcomes of excessive daytime sleepiness in older adults. J Am Geriatr Soc. 2003;51(5):642–649. [DOI] [PubMed] [Google Scholar]
- 13. Vinnikov D, et al. . Fatigue and sleepiness determine respiratory quality of life among veterans evaluated for sleep apnea. Health Qual Life Outcomes. 2017;15(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ohayon MM, et al. . Daytime sleepiness and cognitive impairment in the elderly population. Arch Intern Med. 2002;162(2):201–208. [DOI] [PubMed] [Google Scholar]
- 15. Merlino G, et al. . Daytime sleepiness is associated with dementia and cognitive decline in older Italian adults: a population-based study. Sleep Med. 2010;11(4):372–377. [DOI] [PubMed] [Google Scholar]
- 16. Newman AB, et al. . Daytime sleepiness predicts mortality and cardiovascular disease in older adults. The Cardiovascular Health Study Research Group. J Am Geriatr Soc. 2000;48(2):115–123. [DOI] [PubMed] [Google Scholar]
- 17. Qureshi AI, et al. . Habitual sleep patterns and risk for stroke and coronary heart disease: a 10-year follow-up from NHANES I. Neurology. 1997;48(4):904–911. [DOI] [PubMed] [Google Scholar]
- 18. Basta M, et al. . Lack of regular exercise, depression, and degree of apnea are predictors of excessive daytime sleepiness in patients with sleep apnea: sex differences. J Clin Sleep Med. 2008;4(1):19–25. [PMC free article] [PubMed] [Google Scholar]
- 19. Bixler EO, et al. . Excessive daytime sleepiness in a general population sample: the role of sleep apnea, age, obesity, diabetes, and depression. J Clin Endocrinol Metab. 2005;90(8):4510–4515. [DOI] [PubMed] [Google Scholar]
- 20. Whitney CW, et al. . Correlates of daytime sleepiness in 4578 elderly persons: the Cardiovascular Health Study. Sleep. 1998;21(1):27–36. [DOI] [PubMed] [Google Scholar]
- 21. Ng WL, et al. . The relationship between weight change and daytime sleepiness: the Sleep Heart Health Study. Sleep Med. 2017;36:109–118. [DOI] [PubMed] [Google Scholar]
- 22. Vgontzas AN, et al. . Obesity-related sleepiness and fatigue: the role of the stress system and cytokines. Ann N Y Acad Sci. 2006;1083:329–344. [DOI] [PubMed] [Google Scholar]
- 23. Panossian LA, et al. . Daytime sleepiness in obesity: mechanisms beyond obstructive sleep apnea–a review. Sleep. 2012;35(5):605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Dongen HP, et al. . Systematic interindividual differences in neurobehavioral impairment from sleep loss: evidence of trait-like differential vulnerability. Sleep. 2004;27(3):423–433. [PubMed] [Google Scholar]
- 25. Kapur VK, et al. . Sleepiness in patients with moderate to severe sleep-disordered breathing. Sleep. 2005;28(4):472–477. [DOI] [PubMed] [Google Scholar]
- 26. Chen X, et al. . Racial/ethnic differences in sleep disturbances: the multi-ethnic study of atherosclerosis (MESA). Sleep. 2015;38(6):877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cacioppo JT, et al. . Do lonely days invade the nights? Potential social modulation of sleep efficiency. Psychol Sci. 2002;13(4):384–387. [DOI] [PubMed] [Google Scholar]
- 28. Dinges DF. The state of sleep deprivation: from functional biology to functional consequences. Sleep Med Rev. 2006;10(5):303–305. [DOI] [PubMed] [Google Scholar]
- 29. Dick KJ, et al. . DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383(9933):1990–1998. [DOI] [PubMed] [Google Scholar]
- 30. Klengel T, et al. . The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology. 2014;80:115–132. [DOI] [PubMed] [Google Scholar]
- 31. Mendelson MM, et al. . Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a Mendelian randomization approach. PLoS Med. 2017;14(1):e1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saunderson EA, et al. . Stress-induced gene expression and behavior are controlled by DNA methylation and methyl donor availability in the dentate gyrus. Proc Natl Acad Sci U S A. 2016;113(17):4830–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quach A, et al. . Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY). 2017;9(2):419–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pauwels S, et al. . Dietary and supplemental maternal methyl-group donor intake and cord blood DNA methylation. Epigenetics. 2017;12(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharp GC, et al. ; Cohorts for Heart and Aging Research in Genomic Epidemiology plus (CHARGE +) methylation alcohol working group. Maternal alcohol consumption and offspring DNA methylation: findings from six general population-based birth cohorts. Epigenomics. 2018;10(1):27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bild DE, et al. . Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871–881. [DOI] [PubMed] [Google Scholar]
- 37. Liu Y, et al. . Methylomics of gene expression in human monocytes. Hum Mol Genet. 2013;22(24):5065–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reynolds LM, et al. . Age-related variations in the methylome associated with gene expression in human monocytes and T cells. Nat Commun. 2014;5:5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Subramanian A, et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Du P, et al. . lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. [DOI] [PubMed] [Google Scholar]
- 41. Illumina450ProbeVariants.db: Annotation Package combining variant data from 1000 Genomes Project for Illumina HumanMethylation450 Bead Chip probes [computer program]. 2013. [Google Scholar]
- 42. Chen YA, et al. . Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnson WE, et al. . Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. [DOI] [PubMed] [Google Scholar]
- 44. Du P, et al. . Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinf. 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vargas JD, et al. . Common genetic variants and subclinical atherosclerosis: the Multi-Ethnic Study of Atherosclerosis (MESA). Atherosclerosis. 2016;245:230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Johns MW. A new method for measuring daytime sleepiness: the Epworth sleepiness scale. Sleep. 1991;14(6):540–545. [DOI] [PubMed] [Google Scholar]
- 47. Levine DW, et al. . Reliability and validity of the Women’s Health Initiative Insomnia Rating Scale. Psychol Assess. 2003;15(2):137–148. [DOI] [PubMed] [Google Scholar]
- 48. Radloff LS. The CES-d scale: a self-report depression scale for research in the general population. Applied Psychological Measurement 1977;1(3):385–401. [Google Scholar]
- 49. Needham BL, et al. . Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: the multi-ethnic study of atherosclerosis. Epigenetics. 2015;10(10):958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chiuve SE, et al. . Alternative dietary indices both strongly predict risk of chronic disease. J Nutr. 2012;142(6):1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fried LP, et al. . The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1(3):263–276. [DOI] [PubMed] [Google Scholar]
- 52. Aryee MJ, et al. . Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maksimovic J, et al. . SWAN: subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fortin JP, et al. . Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15(12):503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Houseman EA, et al. . DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sofer T, et al. . A powerful statistical framework for generalization testing in GWAS, with application to the HCHS/SOL. Genet Epidemiol. 2017;41(3):251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Storey JD. A direct approach to false discovery rates. J Royal Stat Soc: Series B (Statistical Methodology). 2002;64(3):479–498. [Google Scholar]
- 58. Wang H, et al. . Genome-wide association analysis of excessive daytime sleepiness identifies 42 loci that suggest phenotypic subgroups. bioRxiv. 2018:454561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sudlow C, et al. . UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bycroft C, et al. . Genome-wide genetic data on ~500,000 UK Biobank participants. bioRxiv. 2017. [Google Scholar]
- 61. Chen H, et al. . Multiethnic meta-analysis identifies RAI1 as a possible obstructive sleep apnea-related quantitative trait locus in Men. Am J Respir Cell Mol Biol. 2018;58(3):391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Smith AC, et al. . Sleep disturbance in Smith-Magenis syndrome (del 17 p11.2). Am J Med Genet. 1998;81(2):186–191. [PubMed] [Google Scholar]
- 63. Li Q, et al. . Conserved properties of Drosophila Insomniac link sleep regulation and synaptic function. PLoS Genet. 2017;13(5):e1006815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boone PM, et al. . Abnormal circadian rhythm of melatonin in Smith-Magenis syndrome patients with RAI1 point mutations. Am J Med Genet A. 2011;155A(8):2024–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cheng HY, et al. . The molecular gatekeeper Dexras1 sculpts the photic responsiveness of the mammalian circadian clock. J Neurosci. 2006;26(50):12984–12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Williams SR, et al. . Smith-Magenis syndrome results in disruption of CLOCK gene transcription and reveals an integral role for RAI1 in the maintenance of circadian rhythmicity. Am J Hum Genet. 2012;90(6):941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Durrence HH, et al. . The sleep of African Americans: a comparative review. Behav Sleep Med. 2006;4(1):29–44. [DOI] [PubMed] [Google Scholar]
- 68. Jean-Louis G, et al. . Evaluation of sleep apnea in a sample of black patients. J Clin Sleep Med. 2008;4(5):421–425. [PMC free article] [PubMed] [Google Scholar]
- 69. Redline S, et al. . Racial differences in sleep-disordered breathing in African-Americans and Caucasians. Am J Respir Crit Care Med. 1997;155(1):186–192. [DOI] [PubMed] [Google Scholar]
- 70. Cespedes EM, et al. . Television viewing, bedroom television, and sleep duration from infancy to mid-childhood. Pediatrics. 2014;133(5):e1163–e1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Paruthi S, et al. . Effect of adenotonsillectomy on parent-reported sleepiness in children with obstructive sleep apnea. Sleep. 2016;39(11):2005–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cunliffe VT. The epigenetic impacts of social stress: how does social adversity become biologically embedded? Epigenomics. 2016;8(12):1653–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Alasaari JS, et al. . Environmental stress affects DNA methylation of a CpG rich promoter region of serotonin transporter gene in a nurse cohort. PLoS One. 2012;7(9):e45813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Baccarelli A, et al. . Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179(7):572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nwanaji-Enwerem JC, et al. . Associations between long-term exposure to PM2.5 component species and blood DNA methylation age in the elderly: the VA normative aging study. Environ Int. 2017;102:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Meeks KAC, et al. . Epigenome-wide association study in whole blood on type 2 diabetes among sub-Saharan African individuals: findings from the RODAM study. Int J Epidemiol. 2018; 48(1):58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Peng C, et al. . Epigenome-wide association study of total serum immunoglobulin E in children: a life course approach. Clin Epigenetics. 2018;10:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kim TY, et al. . Substrate trapping proteomics reveals targets of the βTrCP2/FBXW11 ubiquitin ligase. Mol Cell Biol. 2015;35(1):167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kölsch H, et al. . RXRA gene variations influence Alzheimer’s disease risk and cholesterol metabolism. J Cell Mol Med. 2009;13(3):589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Peloso GM, et al. . Common genetic variation in multiple metabolic pathways influences susceptibility to low HDL-cholesterol and coronary heart disease. J Lipid Res. 2010;51(12):3524–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ma X, et al. . The nuclear receptor RXRA controls cellular senescence by regulating calcium signaling. Aging Cell. 2018;17(6):e12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Monczor F, et al. . Current knowledge and perspectives on histamine H1 and H2 receptor pharmacology: functional selectivity, receptor crosstalk, and repositioning of classic histaminergic ligands. Mol Pharmacol. 2016;90(5):640–648. [DOI] [PubMed] [Google Scholar]
- 83. García-Martín E, et al. . Genetic variability of histamine receptors in patients with Parkinson’s disease. BMC Med Genet. 2008;9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wright C, et al. . Altered expression of histamine signaling genes in autism spectrum disorder. Transl Psychiatry. 2017;7(5):e1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mobarakeh JI, et al. . Role of histamine H(1) receptor in pain perception: a study of the receptor gene knockout mice. Eur J Pharmacol. 2000;391(1-2):81–89. [DOI] [PubMed] [Google Scholar]
- 86. Babai N, et al. . Functional roles of complexin 3 and complexin 4 at mouse photoreceptor ribbon synapses. J Neurosci. 2016;36(25):6651–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Viswanathan S, et al. . Molecularly defined subplate neurons project both to thalamocortical recipient layers and thalamus. Cereb Cortex. 2017;27(10):4759–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hol EM, et al. . Neuronal expression of GFAP in patients with Alzheimer pathology and identification of novel GFAP splice forms. Mol Psychiatry. 2003;8(9):786–796. [DOI] [PubMed] [Google Scholar]
- 89. Cedernaes J, et al. . Acute sleep loss results in tissue-specific alterations in genome-wide DNA methylation state and metabolic fuel utilization in humans. Sci Adv. 2018;4(8):eaar8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Beaudreau SA, et al. ; Study of Osteoporotic Fractures. Validation of the pittsburgh sleep quality index and the epworth sleepiness scale in older black and white women. Sleep Med. 2012;13(1):36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gaspar LS, et al. . Obstructive sleep apnea and hallmarks of aging. Trends Mol Med. 2017;23(8):675–692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.