Abstract
Different species respond differently to severe injury, such as limb loss. In species that regenerate, limb loss is met with complete restoration of the limbs’ form and function, whereas in mammals the amputated limb’s stump heals and scars. In in vitro studies, electrical stimulation (EStim) has been shown to promote cell migration, and osteo- and chondrogenesis. In in vivo studies, after limb amputation, EStim causes significant new bone, cartilage and vessel growth. Here, in a rat model, the stumps of amputated rat limbs were exposed to EStim, and we measured extracellular matrix (ECM) deposition, macrophage distribution, cell proliferation and gene expression changes at early (3 and 7 days) and later stages (28 days). We found that EStim caused differences in ECM deposition, with less condensed collagen fibrils, and modified macrophage response by changing M1 to M2 macrophage ratio. The number of proliferating cells was increased in EStim treated stumps 7 days after amputation, and transcriptome data strongly supported our histological findings, with activated gene pathways known to play key roles in embryonic development and regeneration. In conclusion, our findings support the hypothesis that EStim shifts injury response from healing/scarring towards regeneration. A better understanding of if and how EStim controls these changes, could lead to strategies that replace scarring with regeneration.
Subject terms: Morphogenesis, Developmental biology
Introduction
Limb regeneration – an organisms’ ability to regrow a healthy normally functioning limb - has intrigued scientists for ages (reviewed in1). It is well known that some vertebrates, such as salamanders, frogs and zebra fish are able to regrow partial or complete appendages and organs2, however, mammals’ endogenous ability to regenerate is limited to a few exceptions like the terminal phalanges in marsupials and rodents, and distal fingertips in young children (reviewed in3,4). Most adult human tissues do not regenerate, instead, after injury, healing occurs governed by a fibro proliferative response leading to scar tissue formation (reviewed in5). Whether and to what degree a tissue “heals and scars” versus “regenerates” depends on the tissues’ and organs’ inherent regenerative ability, the type and extent of the lesion, and local and systemic physical and chemical signals that regulate these processes (reviewed in6).
In response to injury both non-regenerative and regenerative amputation wounds undergo distinct overlapping cascades of events in which hemostasis and inflammatory response occur first, in both (reviewed in7). During the inflammatory phase of healing, damaged and dead cells are phagocytized, and cell migration and mitosis are induced by platelet-derived growth factors (reviewed in8). In case of regenerative response, the amputation site is invaded by macrophages as well as pro- and anti-inflammatory cytokines at the same time. In contrast in regeneration-incompetent wounds, the pro-inflammatory response occurs first and is then followed by the anti-inflammatory response9,10. Additionally, cellular proliferation is essential in early phases of regeneration, in which case epithelial cells cover the surface of wounds in the first 24 hours, with an epidermal layer of cells11.
In healing/scarring injury response, following hemostasis and the inflammation phases, proliferation takes over in which case collagen deposition, new vascularization and an increase in fibroblast activity occur (reviewed in12). The deposition of collagen and fibronectin form a provisional extracellular matrix (ECM), and proliferating epithelial cells cover the wounded tissues (reviewed in13). Besides providing structural support to which migrating cells adhere, ECM facilitates cell growth during homeostasis and tissue repair (reviewed in14). In regenerative injury response, progenitor cells are activated and accumulate beneath the newly created epithelial cell layer forming a blastema (reviewed in15). To promote successful appendage regeneration, provisional ECM, deposited during blastema formation, is rich in collagen type III and subpopulations of fibroblasts16. The ECM deposited in the blastema contains patterning information for the regrowing amputated part17. Once the blastema reaches a critical size, it flattens and cells begin to differentiate and patterning begins to form the new structures (reviewed in18). In contrast in non-regenerative amputation wounds a blastema is not formed, instead the tissues undergo maturation and remodeling, whereby collagen is restructured along tension lines to form scar tissue and nonessential cells experience apoptosis (reviewed in8).
Recent literature indicates that bioelectric signaling may play an important role in both healing/scarring and regeneration processes19,20. For example, mammals’ wounds display a positive polarity throughout the healing process, whereas in regenerating amphibians the polarity is initially positive and quickly changes to negative with the peak voltage occurring at the time of maximum cellular proliferation (reviewed in21). Intrigued by the possibility of recreating this bioelectric environment, seen in regenerating amphibians, researchers have tried manipulating bioelectric fields to induce regeneration in adult mammal tissues. As early as 1972 Becker and Spadaro hypothesized that changing wound polarity by applying exogenous electrical stimulation (EStim) in mammals could improve wound healing and perhaps even induce a regenerative response22. In his landmark article, published in 1972 in the journal Nature, Becker reported “blastema formation, new bone, bone marrow, cartilage, nerve, skin, muscle, and epiphyseal plate formation” in amputated rat limbs treated with low voltage direct current electrical stimulation. Based on these findings he concluded “regenerative growth can be restored in mammals by application of the appropriate levels of electrical stimulations”23.
Since then, several studies have shown that EStim can affect the healing process (reviewed in24). Jang et al., demonstrated that the use of external EStim enhanced regenerative activities at different stages of wound healing. They reported up-regulation of inflammatory macrophages and vascular endothelial growth factor (VEGF) in the inflammatory phase; increased cell proliferation markers and protein expression of type III collagen, TGF-β, Integrin α5 and Mmp2 in the proliferative phase; and increased expression of fibronectin and types I and IV collagen during the remodeling phase25. Supporting these findings, in vitro studies demonstrate that EStim influences cellular functions, such as migration, proliferation and differentiation, that play a central role in regeneration26–31.
In recent in vivo studies we reproduced Becker’s original experiment and while we did not observe blastema formation we did demonstrate that low voltage direct current EStim treatment induced significant growth of bone, cartilage and vessels in the same amputated rat limb model32. Having shown this positive effect of EStim, we then questioned if EStim modified important mechanisms, after injury, that support regeneration, which in turn suppress scar formation. In other words, we asked if EStim shifted the balance in the injury response away from healing/scarring towards regeneration. To answer this, we conducted a series of studies in the same rat limb amputation model in which we investigated the effects of EStim on ECM deposition, macrophage distribution, cell proliferation and gene expression changes in EStim treated and non-treated rat limb stump tissues, in early (3 and 7 days) and later stages (28 days) after amputation.
Results
Collagen/Extracellular matrix deposition
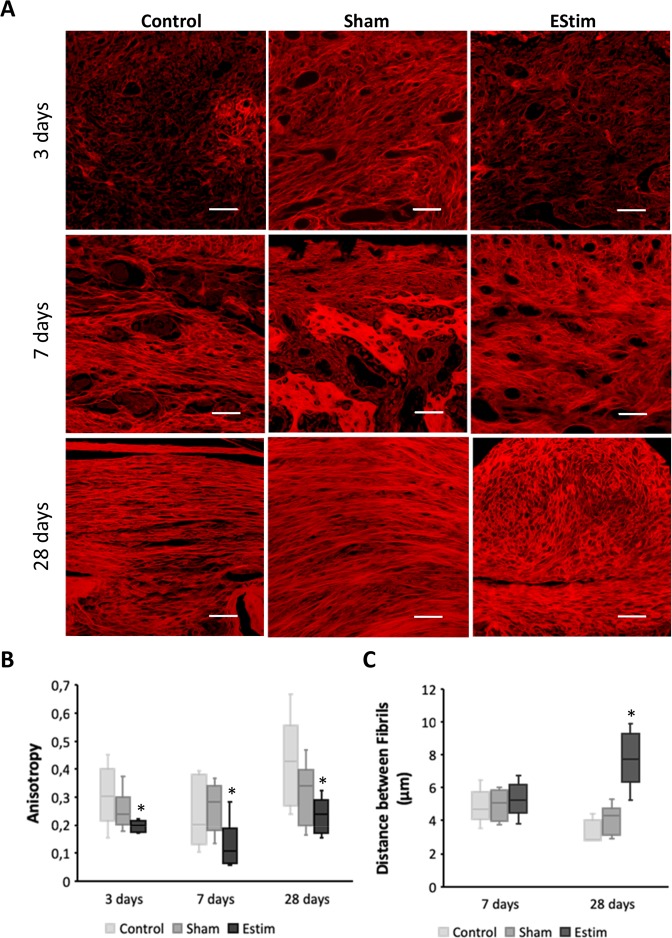
Extracellular matrix (ECM) remodeling, i.e. ECM degradation, synthesis, reassembly and modification, is a critical distinguishing characteristic between tissue healing/scarring and regeneration. We evaluated differences in collagen fibril distribution in stump tissue at 3-, 7- and 28- days post amputation by measuring fibril orientation (anisotropy) and the distance between fibrils (condensation).
As shown in Fig. 1 there were important differences in fibril orientation between the EStim treated, control and sham stump tissues.
Figure 1.
Extracellular matrix (ECM) deposition at the distal end of limb stumps for all groups at different time points. (A) Representative images of Picro Sirius Red stained collagen network at the distal end of limb stumps for control, sham and EStim groups at 3, 7, and 28 days post amputation (Scale bar = 100 µm). (B) Graph showing lower anisotropy (parallelism between fibrils) in electrically stimulated stump tissue compared to non-treated stumps, at all the time points evaluated. (C) Graph showing distance between fibrils measured at days 7 and 28 post-amputation. The greatest distance between fibrils was shown in EStim treated tissue at day 28. *p ≤ 0.05.
Whereas in sham and control stumps fibers appear tightly packed into parallel fibril layers over the stump, in EStim treated stumps fibrils appear less organized and distributed haphazardly with more space in-between fibrils (Fig. 1A). This difference in ECM deposition is evident in the stump tissues as early as 3 days post amputation. Quantification of collagen fibril parallelism (anisotropy) confirmed that EStim significantly (p < 0.05) decreased fibril anisotropy, compared to sham and control stump tissues (Fig. 1B). Moreover, the distance between fibrils was significantly (p < 0.05) greater in EStim treated stump tissues compared to both control groups at 28 days post amputation (Fig. 1C).
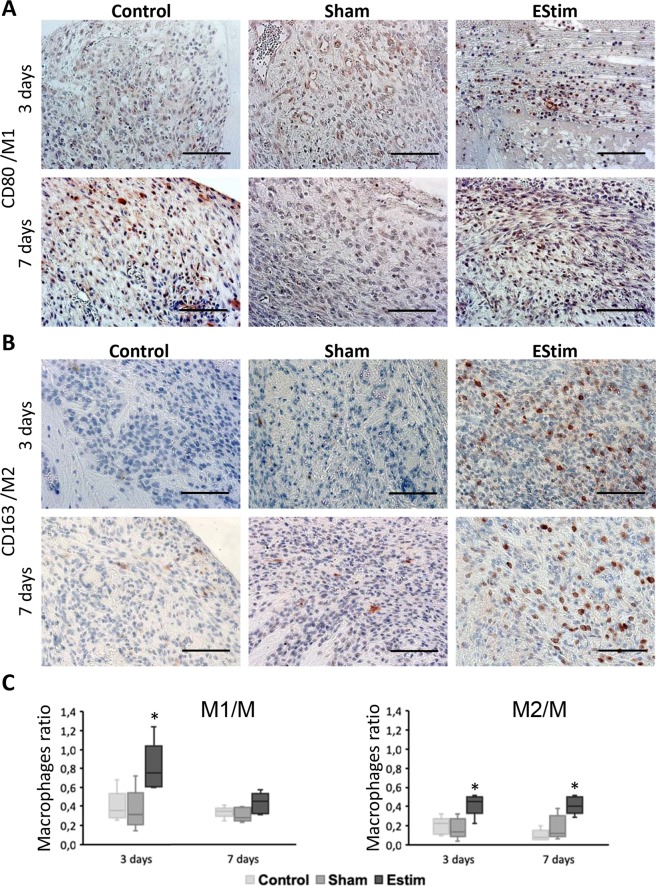
Macrophage number and distribution
Immunohistochemical analysis of M1, M2 and M macrophages at early time points (3 and 7 days) after amputation indicated that EStim impacts the distribution, density and the constitution of macrophage populations in the distal area of the limb stump (Figs 2 and S1). Macrophages were distributed heterogeneously across the entire distal area of the stump tissue. In EStim treated animals more positively stained cells were detected in tissues adjacent to the bone, on the right and left sides, and in the bone marrow cavity. When different types of macrophages were considered within the same sample, all types were distributed similarly, however, the number of CD68 positive cells (M) was, as expected, higher than CD80 (M1) and CD163 (M2) positive cells (Fig. 2A,B).
Figure 2.
Immunohistochemistry analysis of CD80 (marker for M1 macrophages) and CD163 (marker for M2 macrophages) in limb stumps at 3 and 7 days post-amputation. (A) Representative images of immunohistochemical staining showing higher incidence of M1 macrophages at day 3 in EStim treated tissues (Scale bar = 100 µm). (B) Representative images showing higher incidence of M2 macrophages at days 3 and 7 in EStim treated tissues (Scale bar = 100 µm). (C) Ratio of M1 and M2 macrophages (normalized to M macrophages) at the distal end of the stump for all groups tested at 3 and 7 days post-amputation; *p < 0.05.
While EStim exposure did not affect the number of CD68 positive cells (M) at the 3- and 7-day time point measurements (Supplementary Fig. S1), it did change the ratio between M1 and M2 macrophage populations. The number of M1 (pro-inflammatory) macrophages was significantly (p < 0.05) higher in EStim treated animals at day 3, and the number of M2 (anti-inflammatory) cells was significantly (p < 0.05) elevated at both 3 and 7 days. While the number of M1 and M2 macrophages remained the same between the 3- and 7-day measurements in control and sham treated animals, in EStim treated animals the number of M1, but not M2 cells, decreased significantly (p < 0.05) from days 3 to 7 (Fig. 2C).
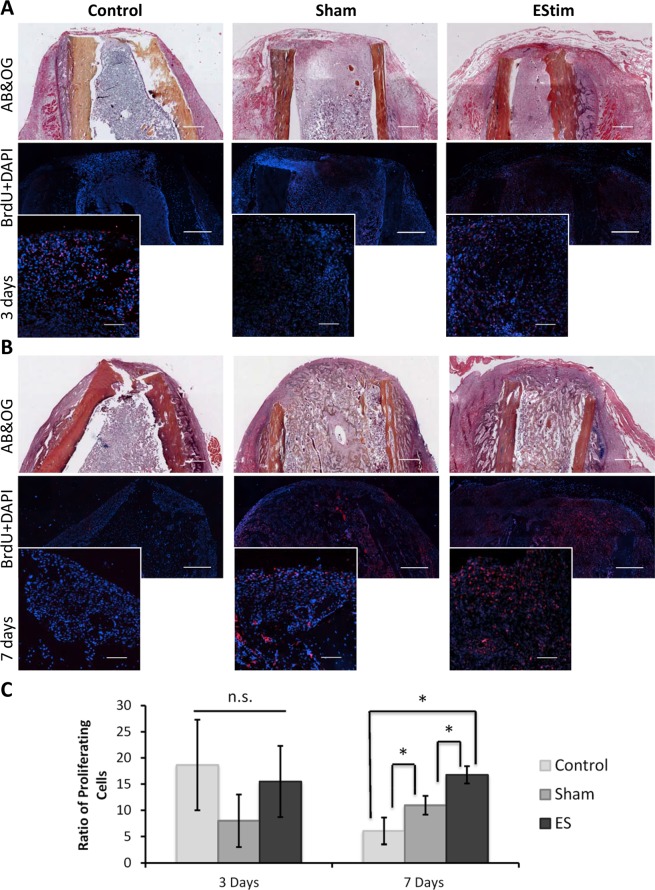
Cell proliferation
Analysis of stained tissues revealed numerous BrdU-positive cells in the stump tissues, with a higher number of proliferating cells at the distal end of the stumps. Positively stained cells were evenly distributed among groups at day 3, with notably higher concentrations of stained cells in the area of the bone marrow (Fig. 3A,C). At day 7 positively stained cells were evenly distributed in the stump tissues of sham and control animals, whereas in EStim treated animals, cells were concentrated in the area surrounding the EStim device electrodes (Fig. 3B). No difference in the number of proliferating cells was observed between EStim treated and non-treated animals 3 days after amputation (Fig. 3C). However, at day 7, the number of proliferative cells was significantly (p < 0.05) higher in EStim treated stumps, compared to controls and sham (Fig. 3C). Between days 3 and 7 there was a significant decrease in the number of proliferating cells in the control samples, and a slight increase in the sham samples, while there was no change in the EStim treated stumps.
Figure 3.
BrdU analysis of cell proliferation in the distal end of the stump in control, sham and EStim treated samples. (A) Top and middle rows represent overview of day 3 post-amputation control, sham and EStim stump sections stained with AB&OG and anti-BrdU antibody (red) counterstained with DAPI (blue) respectively (4×; Scale bar = 500 µm). Bottom row shows high magnification images of BrdU stainings (20×; Scale bar = 50 µm). (B) Representative images of stump sections from control, sham and EStim groups 7 days post-amputation. Top and middle rows show an overview of samples stained with AB&OG and anti-BrdU antibody (red) and counterstained with DAPI (blue) respectively (4×; Scale bar = 500 µm). Bottom row displays high magnification images (20×; Scale bar = 100 µm); (C) Ratio of proliferative cells measured at the distal end of the stump at days 3 and 7 post-amputation. n.s. - non significant (p ≥ 0.05); *p < 0.05.
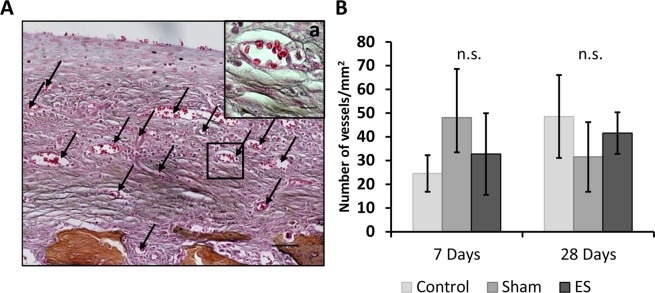
New vessel growth
Manual count of new vessels formed at days 7 and 28 in the distal area of the stumps revealed similar values in the 3 groups (Fig. 4).
Figure 4.
New vessel growth in the distal area of the EStim rat limb stumps 28 days post amputation. (A) Representative image displaying vascular ingrowth in the distal end of the EStim stump at day 28 post-amputation. (AB&OG staining; Scale bar = 100 µm). a – High magnification (40×; Scale bar = 10 µm) of new vessel. (B) Graph indicating density of new vessels at the distal end of the stumps in all 3 groups 7- and 28- days post amputation. n.s. - non significant (p ≥ 0.05).
Gene expression
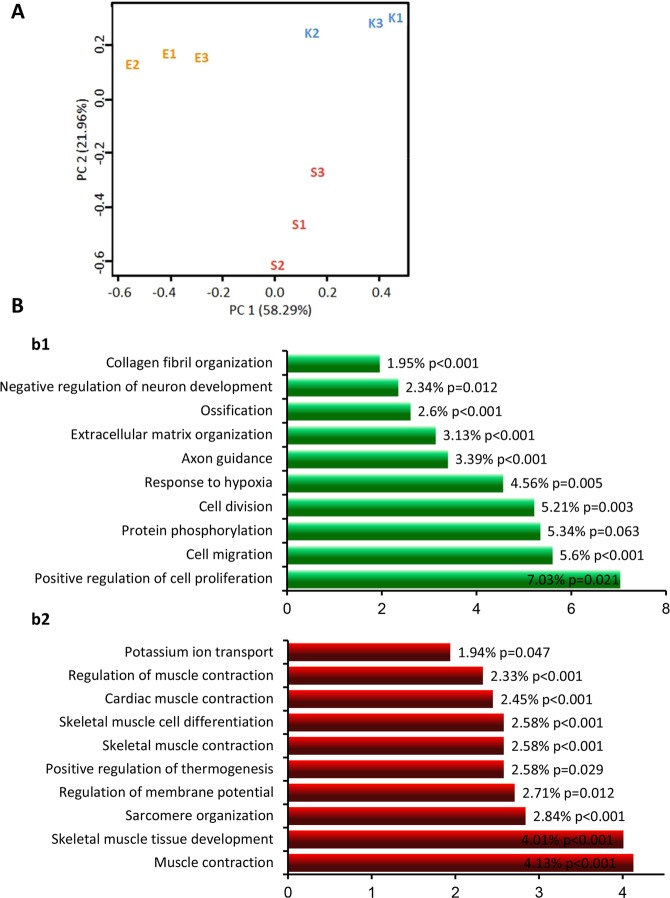
To characterize EStim induced molecular changes we performed deep RNA sequencing comparing transcriptomes in the 3 groups of limb stump tissues. Three biological replicates were analyzed for each condition and between 25 mln and 60 mln reads were generated for each sample (Supplementary Table S1). Normalized counts were used in principal-component analysis (PCA) for each biological replicate, and we found that sham- and EStim treated stump tissues differed greatly from the control samples (Fig. 5A). The number of differentially expressed genes (DEGs) was determined after multiple tests corrected the p value. The Q value was set at a false discovery rate (FDR) of 0.05, and differentially expressed genes were considered to be those transcripts passing this FDR. When comparing sham versus control tissues, there were a total of 406 differentially expressed genes. This number was dramatically increased when comparing the EStim versus the sham tissues; with a total of 1284 differentially expressed genes (all differentially expressed transcripts are provided in Supplementary Tables S1 and S2, representative examples are shown in Table 1).
Figure 5.
mRNA expression profiles in EStim treated limb stumps and non-treated controls (control and sham) are distinct. (A) Principle component analysis of normalized read counts showed strong effect of EStim treatment. Biological replicates for EStim (E1, E2, E3), sham (S1, S2, S3) and control (K1, K2, K3) samples are shown. (B) Top-10 significantly enriched biological processes in EStim and sham transcriptomes (FunReach analysis). Each the EStim and sham transcriptomes were tested against Rattus Norvegicus UniProt database, the top 10 most highly represented biological processes and the percentage of proteins predicted to belong to each category are shown for EStim (b1) and sham (b2) samples. The numbers shown at the right side of each bar indicated the % of genes (proteins) involved in each term and p-Value.
Table 1.
Examples of genes up- regulated in EStim.
| Gene | Gene name | Log2 Fold change | p-value |
|---|---|---|---|
| Mmp13 | Matrix Metalloproteinase 13 (Collagenase 3) | 3.422626 | 5.08E-34 |
| Vsig4 | V-Set and Immunoglobulin Domain-Containing Protein 4 | 3.147901 | 1.12E-11 |
| Sox10 | SRY-Box 10 | 2.856724 | 4.24E-11 |
| Pla2g5 | Phospholipase A2 Group V | 2.507993 | 1.45E-10 |
| Lif | Leukemia Inhibitory Factor | 2.406112 | 1.94E-07 |
| Mmp9 | Matrix Metalloproteinase-9 | 2.254128 | 5.13E-10 |
| Mmp3 | Matrix Metalloproteinase 3 | 2.170442 | 6.06E-10 |
| Shroom2 | Shroom Family Member 2, Apical-Like Protein | 2.007132 | 3.83E-09 |
| Foxc2 | Forkhead Box C2 (MFH-1, Mesenchyme Forkhead 1) | 1.962337 | 2.32E-05 |
| Dhh | Desert Hedgehog | 1.794296 | 3.23E-06 |
| Gsc | Homeobox Protein Goosecoid | 1.761111 | 0.000539 |
| Lhx2 | LIM Homeobox 2 | 1.736685 | 0.000146 |
| Dlx3 | Distal-Less Homeobox 3 | 1.734016 | 1.15E-06 |
| Wnt7b | Wnt Family Member 7B | 1.588366 | 8.13E-05 |
| Apln | Apelin | 1.504759 | 7.07E-06 |
| Lbh | Limb Bud and Heart Development | 1.476744 | 1.40E-07 |
| Hoxb2 | Homeobox B2 | 1.382072 | 0.000284 |
| Nrarp | Notch-regulated ankyrin repeat protein | 1.374706 | 0.00047 |
| Meox1 | Mesenchyme Homeobox 1 | 1.371066 | 1.35E-06 |
| Gli1 | Glioma-Associated Oncogene Homolog 1 | 1.307916 | 4.02E-06 |
| Wnt5a | Wnt Family Member 5A | 1.3033 | 7.28E-06 |
| Wnt5b | Wnt Family Member 5B | 1.214129 | 0.000162 |
| Mki67 | Marker of Proliferation Ki-67 | 0.985305 | 0.000111 |
For the subsequent analysis we focused on comparing the EStim and sham datasets.
To reveal the types of processes regulated by EStim, we grouped the enriched pathways considering “large-scale functions.” Most of the transcripts up-regulated by EStim, were related to anatomical structure morphogenesis and development, animal organ and tissue development, extracellular matrix organization, cell differentiation, mitosis and migration (Figs 5B, 6 and Table S2). Most of the transcripts down-regulated by EStim were related to cell energy and metabolisms as well as muscle system process, muscle contraction and muscle structure development (Figs 5B, 6 and Supplementary Table S2).
Figure 6.
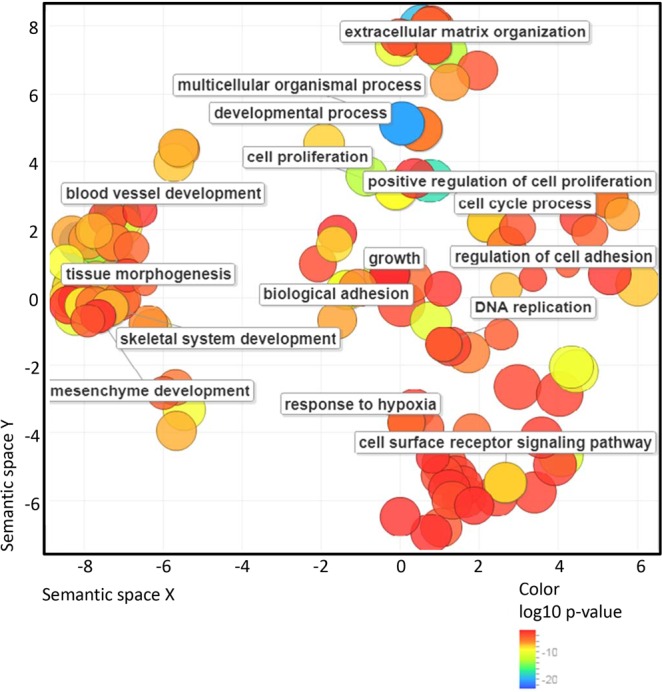
Gene Ontology (GO) analysis of up-regulated in EStim genes using REVIGO. The scatter plot shows the cluster representatives (terms remaining after reducing redundancy) in a two-dimensional space derived by applying multi-dimensional scaling to a matrix of GO terms semantic similarities. Bubble color indicates the p-value (legend in lower right-hand corner).
Biological processes enrichment analysis against the UniProt database revealed that genes, significantly (p < 0.05) upregulated in EStim treated stump tissue, are involved in positive regulation of cell proliferation, negative regulation of apoptosis, cell migration, cell division, ECM organization, ossification, axon guidance and others (Fig. 5B). Conversely, among downregulated genes in EStim treated stumps, enrichment was shown for those involved in muscle contraction, skeletal muscle development, regulation of membrane potential, skeletal muscle differentiation, glycogen metabolic process, glycogen biosynthesis and others (Figs 5B, 6 and Supplementary Tables S1, S2). Thus, our gene expression analysis revealed profound transcriptional remodeling of EStim treated stump tissues and suggests numerous pathway targets for subsequent study and therapeutic interventions.
Discussion
Limb amputation in mammals triggers a series of events that ultimately result in healing and fibrotic scar formation (reviewed in12); with some exceptions such as mouse digit tip amputation and, human fingertip which under special conditions have been shown to regenerate.
In a previous study, in an amputated rat forelimb model, we demonstrated that instead of eliciting scar formation, direct current EStim stimulated a regenerative response resulting in new bone, cartilage and vessel formation32. Regenerative response to amputation is strongly dependent on a tightly controlled inflammatory response, direct recruitment and proliferation of resident stem cells and regulation of biochemical, biophysical and biomechanical signals between cells and the ECM (reviewed in33). To evaluate if EStim is able to shift the injury response from healing/scarring toward regeneration, we designed this in vivo study to assess how EStim affects extracellular matrix deposition, macrophage response, cell proliferation and gene expression, all cellular functions known to play an important role in both of these processes.
Following injury in species capable of regenerating limbs, the ECM in local remaining tissues is broken down and replaced with a new regeneration-specific ECM34. Differences in matrix metalloproteinases (Mmps), responsible for ECM remodeling have been shown in regeneration-competent Axolotl and regeneration-deficient Xenopus froglets35. Of note, mammal and amphibian myoblasts responded similarly when stimulated with ECM substrates, suggesting that the key to inciting regenerative repair may be preserved in these species36. For most injuries, fibroblasts and myofibroblasts interact to produce extracellular matrix, mainly in the form of collagen, which ultimately forms the bulk of a mature scar37. In a skin wound model, Whitby DJ, and Ferguson MW described scar tissue in wound beds of adult mice as being covered by collagen fibrils, laid in parallel dense bundles. In contrast in fetus’, they found that collagen fibrils were deposited in a reticular fashion, almost identical to the surrounding dermis, and healing was scar-free38. In our electrically stimulated stump tissues, the ECM structure appeared to have less condensed fibrils, forming a net-like structure, similar to the reticular structure of fetus ECM. Conversely, in sham and control stump tissues, not exposed to EStim, the ECM was densely packed and appeared more like that seen in scar tissue. During healing, histolysis, in which ECM is degraded to release dermal fibroblasts, is mainly mediated by Mmps39. In regenerative injury response, proteinases are activated to degrade ECM and release cells for regeneration; Mmp-8, -9, and -10 are found to be up-regulated throughout blastema formation and Mmp-13 is increased in stage 2 of epidermal wound closure (histolysis/dedifferentiation)35. These observations coincided with what we saw in our EStim treated stump tissues, that had elevated expression of various matrix metalloproteinase genes (Mmp3, Mmp9 and Mmp13) and genes associated with ECM remodeling. While little is known about how EStim effects collagen deposition and alignment in vivo, in vitro studies have shown that electrical fields can effect extracellular protein structures40.
There are many reports in the literature describing the important role that macrophages play in tissue regeneration and wound healing. In experiments in which macrophages were depleted in axolotl and mice models both healing and regeneration were compromised41–44. In response to injury, M1 (pro-inflammatory) and M2 (anti-inflammatory) macrophages are regulated differently in healing versus regeneration; while M1 and M2 peak at the same time during regeneration, in non-regenerative tissues M1 response comes first followed by the rise of M29,10,43. In the present study we saw a simultaneous increase in the ratio of M1 and M2 in electrically stimulated stump tissue at day 3 post amputation, then a sustained higher ratio in M2 macrophages at day 7. Our transcriptome data supported these observations in that the expression of Vsig4 gene, responsible for M1 inhibition45 and Pla2g5, involved in M2 development46, were significantly increased in EStim treated stumps. In in vitro experiments, others have demonstrated that externally applied EStim affects macrophage migration, phenotype and plasticity28,47,48. However, little is known about the effect EStim has on macrophages in vivo. McLeand and Verge showed that exposing nerves to electrical stimulation shifted macrophage phenotype from M1 (pro-inflammatory) toward predominantly M2 (anti-inflammatory) in demyelinated nerves29.
In amphibian regeneration, interactions between immune- and stem cells have been shown to play an important role43. Stem cell migration and proliferation are essential in blastema formation and limb regeneration15,18. Our results showed that in addition to increasing the M2 population, EStim treatment also increased the rate of cell proliferation in stump tissue at day 7. These findings were supported by gene expression analysis that showed an upregulation of several proliferation marker genes (mki67, Prr11, Fam111a, Mybl2, Mcm10, E2f8, Asf1b, Recql4, Cdh1 etc.) in EStim samples. In fact, we found that the most enriched set of genes were those associated with cell proliferation. While, in this study we did not focus on identifying the origin of the proliferating cells in the distal stump tissues, we can speculate that they could have come from bone marrow derived MSC. This speculation is supported by reports in previous literature and our own data, that show the concentration of BrdU-labeled cells located in the bone marrow niche of EStim treated stumps and activation of ossification gene expression associated with EStim exposure24–26,30,31,49–52. The mechanisms by which EStim affect MSC proliferation are not known, however its involvement in cytoskeletal components, cellular microenvironment, ions and small molecular transport53 are the focus of several investigations. Our findings here and those of others, showing important changes in gene expression related to membrane potential, potassium ion transport and activity of sodium ion trans-membrane transporters, support the hypothesis that EStim acts by regulating ion and molecule transport through the cell membrane21,27,54–56. The observed increase of BrdU-positive stained cells in sham-treated stumps could have been caused by mechanical stimulation due to the presence of the metallic electrode (inactive device) in the bone marrow cavity, as MSCs and other cells have been shown to be highly mechanosensitive57.
In regeneration there is an initial increase in local stem cell populations and ECM remodeling, followed by new vessel growth15. The new vessels are crucial for the nourishment and waste removal required in the important growth of newly formed tissues characteristic in regeneration8. In a previous study we demonstrated that EStim increased new vessel formation in the distal end of limb stumps32. In the present experiments we found that vessel density in EStim treated stump tissue was higher than the controls, however not significantly. The limited number of samples in these studies could be the reason for this variation. Despite this, our RNA profile analysis showed up-regulation of the vascularization-controlling genes (Foxc2, Apln, Nrarp, Shroom2) in EStim treated samples.
From the time of injury until the successful healing/scarring or regeneration, surviving cells display a diversity of behaviors, including; death, growth, dedifferentiation, adhesion, communication, migration, mitosis, extracellular matrix remodeling, and differentiation1, all of which correlate with changes in transcription58. We observed important differences in gene expression profiles between EStim treated and non-treated stump tissues. In our EStim treated stump tissues Wnt signaling pathway members (wnt5a, wnt5b and wnt7b), Homeobox (Hox) genes (Gsc, Lhx2, Hoxb2, Meox1) and Hedgehog pathway genes (Gli1 and Dhh), were up-regulated. It is known that in axolotl limb regeneration as well as in mammal limb development, Wnt, Hox and other signaling pathways play an important role by forming expression gradients involved in determining axial identity59. Wnt signaling has been shown to have important functions during mammalian limb bud initiation, limb outgrowth, early limb patterning, and later in limb morphogenesis events60. Hox genes have been reported to encode homeodomain-containing transcription factors that determine cell and tissue identities in the embryo during development and are very conservative in evolution that justify the importance of their role in development61. Hedgehog pathway genes (Gli1 and Dhh), also activated in our EStim treated stump tissues, are known to encode signaling molecules that play a role in morphogenesis regulation62,63. Moreover, a number of genes involved in cell cycle regulation, stem cell differentiation and proliferation were activated in EStim treated tissues. The majority of genes, down-regulated in EStim treated tissues, were those involved in cell energy metabolism as well as skeletal muscle homeostasis, that correlate with findings reported in axolotl regeneration. It was shown that Axolotl blastema cells actively repress a muscle program by suppressing genes similar to those we observed to be suppressed64 in our tissue samples. This suggests that EStim activates/suppresses defined pathways and thus shifts tissue injury response from one characteristic of healing/scarring toward that seen in development and/or regeneration.
Regeneration is known to be time dependent, regulated by mechanisms coordinating transcription within and across cell types at different stages of appendage regeneration65. While in this study we did not focus on how EStim affects temporal and positional expression gradients, this is an important goal in our ongoing and future studies in order to answer the question how EStim acts to create a pro-regenerative environment.
Despite the promising results obtained in the present experiments, it is important to consider some limitations. First, in order to reproduce the model Becker used in his 1972 study, animals were operated at the same young age. It is well known that wound healing ability declines with age66,67, therefore in future studies it will be important to compare the observed effects EStim has on healing in aged animals. Second, due to several technical limitations related to the limb amputation model and ethical concerns (such as bleeding, self-mutilation, infection, dislodging of the EStim device, and additional animal suffering) in the present study skin was sutured over the stump after surgery. Aware that this could negatively impact the regenerative response68 skin was excluded from this analysis. Third, the effect EStim has on macrophage polarization was analyzed by means of M1- and M2- marker staining. Markers used in this study are widely recognized for their distinct affinities to specific proteins present in M1 and M2 cells, still the possible presence of intermediately polarized M1/M2 cells69 could have interfered with our results.
In summary, our data supports the hypothesis that EStim shifts injury response, in our amputated rat limb model, from healing/scarring toward regeneration. This effect appears to be in the early inflammatory response where it stimulates cell proliferation and ECM remodeling. EStim induces profound changes in gene expression, including activation of genes involved in morphogenesis and development. Additional studies are needed to spatially and temporally characterize this tissue reaction to electricity and to elucidate which cellular players orchestrate it. A greater understanding of these underlying mechanisms will bring us closer to the possibility of being able to control cellular functions, which could later be translated into new treatments that promote regenerative responses.
Material and Methods
All animal experiments were performed in accordance with guidelines established by our animal care and oversight committee at the Johann Wolfgang Goethe-University in Frankfurt am Main, Germany and were approved by the Veterinary Department of the Regional Council in Darmstadt, Germany (Regierungspräsidium Darmstadt, Veterinärdezernat, Wilhelminenstraße 1–3) (Project No. FU/1114).
To assess the effects of EStim on limb tissue regeneration, the right forelimbs of 60 male Sprague Dawley rats (Charles River Labs Int., Germany) (age = 5 weeks; weight = 100–150 g) were amputated and the limb stumps were treated with: 1) Electrical stimulation (implanted functioning EStim device) (n = 20), 2) No electrical stimulation (implanted deactivated device – sham control) (n = 20), and 3) No electrical stimulation (no device – control) (n = 20). Table 2 shows the treatment, the number and the distribution of animals in their respective groups.
Table 2.
Treatment, number and distribution of animals into groups.
| Group | Treatment | Time point (days) | Number of animals | Analysis |
|---|---|---|---|---|
| Control | No | 3 | 5 | Histology, IHC |
| 7 | 10 | Histology, IHC, deep RNA-seq | ||
| 28 | 5 | Histology, IHC | ||
| Sham | Disabled device | 3 | 5 | Histology, IHC |
| 7 | 10 | Histology, IHC, deep RNA-seq | ||
| 28 | 5 | Histology, IHC | ||
| EStim | Active device | 3 | 5 | Histology, IHC |
| 7 | 10 | Histology, IHC, deep RNA-seq | ||
| 28 | 5 | Histology, IHC |
Limb amputation and EStim device implantation surgeries
Electrical stimulation was applied using a purpose-built bimetallic device and surgeries were performed as described previously32. Animals were euthanized (CO2 inhalation) at 3-, 7- and 28- days post amputation, weighed, and the limb stumps were collected and carefully examined macro- and microscopically. In the macroscopic examination, the skin covering the limb stumps of the EStim and sham-treated animals was carefully incised and the EStim device and corresponding electrodes were controlled for integrity and carefully removed. In addition, all limbs were inspected for signs of infection and/or tumors. The specimens were fixed in Zinc-Formal-Fixx, (Thermo Electron, USA) for 24hrs and stored for subsequent histomorphometric and immunohistological analysis or stored in liquid nitrogen for subsequent RNA isolation.
Tissue preparation
Fixed samples were decalcified in 10% EDTA/TRIS-HCl (pH 7.4) for 14 days and were embedded in paraffin. Longitudinal sections (5 µm thick) were taken parallel to the long axis of the humerus for all samples from all groups and timepoints. Sections were selected based on the following criteria; presence of stump tissue from side to side, absence of damaged tissues, presence of humerus bone’s medullary cavity and compact bone on both sides of the cavity (Supplementary Fig. S2). Selected sections were stained according to the specific parameters analyzed, as described below. Analysis and quantitative evaluations were performed using light and fluorescent microscopy (Ti-E, Nikon GmbH, Germany) and image analysis software (NIS-Elements 4.4, Nikon GmbH, Germany). For all staining and analysis, a minimum of 3 sections per limb, and 5 limbs per groups for each time point, were used.
ECM collagen deposition measurements
An important distinguishing characteristic between healing/scarring and regeneration tissue is the content and distribution of collagen fibers. In order to measure these morphological structures in the limb stumps, tissue sections were deparaffinized and hydrated, colored with Picro Sirius Red (0.1% of Sirius Red in saturated aqueous picric acid) (Sigma-Aldrich, Germany) for 1 hour, washed two times in 0.5% acetic acid solution, dehydrated in three changes of 100% ethanol, cleared in xylene and mounted. Images were visualized using fluorescence microscopy (Ti-E, Nikon GmbH, Germany)70. To evaluate the distribution of collagen fibrils, the orientation (anisotropy), and the distance between fibrils (condensation), image analysis software (ImageJ; NIS-Elements 4.4, Nikon GmbH, Germany) was used. A total of three sections per animal, and five samples per group/timepoint were analyzed. To quantify the parallel distribution of collagen fibers (anisotropy), “FibrilTool” plug-in for ImageJ software was used as described by71. Anisotropy was measured in a region of interest (ROI) measuring 250 µm × 250 µm, located at the distal end of the stump in the middle of medular cavity (Supplementary Fig. S2), and the mean value for 5 animals was calculated and used to represent each group/time point. The distance between fibrils (condensation) was measured in 20 randomly chosen locations within the same ROI, and the mean values for each animal was calculated. These values were used to calculate the mean value for 5 animals, to represent each group/time.
Macrophage number and distribution measurements
To quantify different macrophage phenotypes present in the rat limb stumps, tissue sections were stained with primary specific antibodies against (1) general and non-activated macrophages (M - CD68 marker; MCA341GA; BIO-RAD Laboratories; GmbH Germany), (2) pro-inflammatory macrophages (M1 - CD80 marker; AA270-321; Antibodies online GmbH, Germany), and (3) anti-inflammatory macrophages (M2 - CD163 marker; MCA342R; BIO-RAD Laboratories GmbH, Germany). Briefly, tissue sections were deparaffinized, rehydrated and trypsin (CD68 and CD163 antibodies) or heat (CD 80 antibody, Citrate buffer pH6) antigen retrieval was performed before staining with antibodies. For signal detection, an EnVision + System-HRP (AEC) kit (Dako, Germany) was applied for sections stained with CD163 and CD68 antibodies or an EXPOSE Rabbit HRP-AEC detection IHC Kit (Abcam, Germany) was used for sections stained with CD80 antibody. Finally, a counterstain with hematoxylin was performed. An Isotype identical (IgG1) non-specific mouse antibody served as a negative control (eBioscience, Germany). Three slides per animal were analyzed using light microscopy (at 10×) (Ti-E, Nikon GmbH, Germany) and image analysis software (NIS-Elements 4.4, Nikon GmbH, Germany). Five standardized regions of interest (ROIs – 420 µm × 315 µm) (at 20x magnification) were selected in the distal area of the stump. Stained macrophages were manually counted by two independent trained examiners, blinded to the group makeup, and similar results were obtained. After obtaining the number of CD80+ cells, the ratio of cells expressing M1 phenotype was obtained by dividing it by the number of CD68+ cells for each ROI. The same calculations were done to obtain the ratio of M2 macrophages (CD163+ cells/CD63+ cells). Mean values were obtained for each ROI and each animal, and were used to statistically analyze the differences among groups at each timepoint. The presence of the CD68 surface marker designates a macrophage phenotype, which is a common marker for both M1 and M2 states and other cells from monocyte lineage, therefore the sum of M1 and M2 is not necessarily equal to the number of CD68+ cells72.
Cell proliferation measurements
Cell proliferation was assessed by immunohistochemical detection of incorporated Bromdesoxyuridin (BrdU) at the distal end of the limb stumps. Animals received a peritoneal injection of BrdU labeling reagent (Invitrogen, eBiosciences, Germany) 1 day prior to being euthanized. After sample processing, as described above, paraffin sections were deparaffinized and treated with 2 M HCl solution at 37 °C for 20 min and 0.05% trypsin for 30 min at 37 °C. Endogenous peroxidase activity was blocked by 3% H2O2 solution, after which 7% goat serum, 1% BSA in TBS buffer blocking solution was applied for 1 hour. Staining was performed at +4 °C overnight with primary anti-BrdU antibody (MoBU-1, BioLegend, Germany, 1:100). Secondary antibodies (ab150128, Abcam, Germany, 1:500) were applied for 1 hour at room temperature and tissue sections were covered with DAPI-containing Fluoroshield Mounting Medium (ab104139, Abcam, Germany). For each group, tissue sections from three animals were analyzed under a fluorescence microscope using NIS software. Quantification: Three regions of interest (ROI) located at the left, center, and right of the distal stumps (at 20x magnification) were selected. Positive BrdU- and DAPI-stained cells were thresholded, and for each ROI the area with BrdU-positive cells was normalized to the total (DAPI-stained) area of cells to obtain the fraction of proliferative cells in each ROI. The mean value of the 3 ROIs was calculated for each animal and the values of 3 animals per group were used for subsequent statistical analysis.
New vessel growth measurements
Tissue sections used in the ECM measurements were also used for new vessel growth measurements, using bright field microscopy. Quantification of new vessels present in the distal stump tissue was performed on five samples from each group at days 7 and 28 post-amputation. Two trained examiners (blinded to the group composition) counted new vessels manually in defined ROIs, located in the distal area of the stump tissue, of five samples. The number of vessels was normalized to the dimensions of the evaluated area (ROI). The density of newly-formed vessels in each sample was obtained, mean values were calculated for each group and these were statistically analyzed.
Gene expression measurements
Preparation and sequencing of RNA-seq libraries
Total RNA was extracted with Trizol reagent (Invitrogen) according to the manufacturers’ protocol from 3 samples from each group collected 7 days post-amputation. mRNA was purified with NEXTflex Poly(A) beads (Bioo Scientific). RNA libraries were prepared with NEXTflex® Rapid Directional qRNA-Seq™ Kit (Bioo Scientific) following the manufacturer’s protocol and sequenced on an Illumina HiSeq2500 machine.
Differential gene expression analysis
Illumina sequencing reads were mapped to the rat genome assembly Rnor_6.0 using STAR v.2.5.3 (https://www.ncbi.nlm.nih.gov/pubmed/23104886) and gene annotations from Ensembl release 90. Gene counts were generated by the “quantMode GeneCounts” option in STAR. Count normalization was performed with RUVSeq software73 with k = 4, and differential gene expression analysis was carried out with edgeR package74. Significantly differential expression genes were selected based on a corrected P-value of 0.05 as the threshold. GO term enrichment analysis of differentially expressed genes was performed with GOrilla75 and functional enrichment analysis against the UniProt database was performed with the FunReach analysis tool76,77. Revigo web server was used for visualization of enriched GO terms by means of a semantic similarity-based scatterplot78.
Statistical analysis
For comparisons among and between groups, Kruskal-Wallis-Test with multiple Conover-Iman-Comparison (Bonferroni-Holm-correction) was applied on BiAS for WindowsTM version 11.0 software (http://www.bias-online.de). Data are presented as mean ± SD and a significance level was set at p < 0.05.
Supplementary information
Acknowledgements
This study was supported in part by the Friedrichsheim Foundation (Stiftung Friedrichsheim) based in Frankfurt/Main, Germany.
Author Contributions
Conceptualization, J.H.B., D.H. and L.L.; Methodology, K.M.C.O. and L.L; Investigation, K.M.C.O., E.B., L.P., S.K., M.E.L., Z.H., M.B.B. and L.L.; Formal Analysis, K.M.C.O., E.B. and L.L.; Writing – Original Draft, K.M.C.O. and L.L.; Writing – Review&Editing, K.M.C.O., J.H.B., D.H. and L.L.; Supervision, J.H.B. and L.L; Project Administration, L.L.
Data Availability
The Illumina sequencing data were deposited to the NCBI Sequence Read Archieve under BioProject Accession Number PRJNA532925 (SRA entries SRR8902417-SRR8902425).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47389-w.
References
- 1.Carlson, B. M. Principles of regenerative biology (Elsevier/Academic, Amsterdam, London, 2007).
- 2.Simon A, Tanaka EM. Limb regeneration. Wiley interdisciplinary reviews. Developmental biology. 2013;2:291–300. doi: 10.1002/wdev.73. [DOI] [PubMed] [Google Scholar]
- 3.Carlson BM. Stimulation of regeneration in mammals: pipe dream or realistic goal? Wound repair and regeneration: official publication of the Wound Healing. Society [and] the European Tissue Repair Society. 1998;6:425–433. doi: 10.1046/j.1524-475x.1998.60504.x. [DOI] [PubMed] [Google Scholar]
- 4.Yu L, et al. BMP9 stimulates joint regeneration at digit amputation wounds in mice. Nature communications. 2019;10:424. doi: 10.1038/s41467-018-08278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harty M, Neff AW, King MW, Mescher AL. Regeneration or scarring: An immunologic perspective. Dev. Dyn. 2003;226:268–279. doi: 10.1002/dvdy.10239. [DOI] [PubMed] [Google Scholar]
- 6.Sattler Susanne, Rosenthal Nadia. The neonate versus adult mammalian immune system in cardiac repair and regeneration. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2016;1863(7):1813–1821. doi: 10.1016/j.bbamcr.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Brøchner AC, Toft P. Pathophysiology of the systemic inflammatory response after major accidental trauma. Scandinavian journal of trauma, resuscitation and emergency medicine. 2009;17:43. doi: 10.1186/1757-7241-17-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinke JM, Sorg H. Wound repair and regeneration. European surgical research. Europaische chirurgische Forschung. Recherches chirurgicales europeennes. 2012;49:35–43. doi: 10.1159/000339613. [DOI] [PubMed] [Google Scholar]
- 9.Mahdavian Delavary B, van der Veer WM, van Egmond M, Niessen FB, Beelen RHJ. Macrophages in skin injury and repair. Immunobiology. 2011;216:753–762. doi: 10.1016/j.imbio.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9415–9420. doi: 10.1073/pnas.1300290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endo T, Bryant SV, Gardiner DM. A stepwise model system for limb regeneration. Developmental biology. 2004;270:135–145. doi: 10.1016/j.ydbio.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 12.Choi Y, et al. Regeneration and Regrowth Potentials of Digit Tips in Amphibians and Mammals. International journal of cell biology. 2017;2017:5312951. doi: 10.1155/2017/5312951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue M, Jackson CJ. Extracellular Matrix Reorganization During Wound Healing and Its Impact on Abnormal Scarring. Advances in Wound Care. 2015;4:119–136. doi: 10.1089/wound.2013.0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krafts KP. Tissue repair. The hidden drama. organogenesis. 2010;6:225–233. doi: 10.4161/org.6.4.12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolan CP, Dawson LA, Muneoka K. Digit Tip Regeneration. Merging Regeneration Biology with Regenerative Medicine. Stem cells translational medicine. 2018;7:262–270. doi: 10.1002/sctm.17-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrero L, Simkin J, Sammarco M, Muneoka K. Fibroblast reticular cells engineer a blastema extracellular network during digit tip regeneration in mice. Regeneration. 2017;4:69–84. doi: 10.1002/reg2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phan AQ, et al. Positional information in axolotl and mouse limb extracellular matrix is mediated via heparan sulfate and fibroblast growth factor during limb regeneration in the axolotl (Ambystoma mexicanum) Regeneration. 2015;2:182–201. doi: 10.1002/reg2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payzin-Dogru D, Whited JL. An integrative framework for salamander and mouse limb regeneration. The International journal of developmental biology. 2018;62:393–402. doi: 10.1387/ijdb.180002jw. [DOI] [PubMed] [Google Scholar]
- 19.Nuccitelli R. Endogenous electric fields in embryos during development, regeneration and wound healing. Radiation protection dosimetry. 2003;106:375–383. doi: 10.1093/oxfordjournals.rpd.a006375. [DOI] [PubMed] [Google Scholar]
- 20.McCaig CD, Song B, Rajnicek AM. Electrical dimensions in cell science. Journal of cell science. 2009;122:4267–4276. doi: 10.1242/jcs.023564. [DOI] [PubMed] [Google Scholar]
- 21.Levin M. Bioelectric mechanisms in regeneration. Unique aspects and future perspectives. Seminars in cell & developmental biology. 2009;20:543–556. doi: 10.1016/j.semcdb.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker RO, Spadaro JA. Electrical stimulation of partial limb regeneration in mammals. Bulletin of the New York Academy of Medicine. 1972;48:627–641. [PMC free article] [PubMed] [Google Scholar]
- 23.Becker RO. Stimulation of Partial Limb Regeneration in Rats. Nature. 1972;235:109–111. doi: 10.1038/235109a0. [DOI] [PubMed] [Google Scholar]
- 24.Kloth LC. Electrical stimulation for wound healing: a review of evidence from in vitro studies, animal experiments, and clinical trials. The international journal of lower extremity wounds. 2005;4:23–44. doi: 10.1177/1534734605275733. [DOI] [PubMed] [Google Scholar]
- 25.Jang Hyeon-Ki, Oh Jin, Jeong Gun-Jae, Lee Tae-Jin, Im Gwang-Bum, Lee Ju-Ro, Yoon Jeong-Kee, Kim Dong-Ik, Kim Byung-Soo, Bhang Suk, Lee Tae. A Disposable Photovoltaic Patch Controlling Cellular Microenvironment for Wound Healing. International Journal of Molecular Sciences. 2018;19(10):3025. doi: 10.3390/ijms19103025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ongaro A, et al. Pulsed electromagnetic fields stimulate osteogenic differentiation in human bone marrow and adipose tissue derived mesenchymal stem cells. Bioelectromagnetics. 2014;35:426–436. doi: 10.1002/bem.21862.. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Li M, Kang E-T, Neoh KG. Electrical stimulation of adipose-derived mesenchymal stem cells in conductive scaffolds and the roles of voltage-gated ion channels. Acta biomaterialia. 2016;32:46–56. doi: 10.1016/j.actbio.2015.12.024. [DOI] [PubMed] [Google Scholar]
- 28.Hoare Joseph I., Rajnicek Ann M., McCaig Colin D., Barker Robert N., Wilson Heather M. Electric fields are novel determinants of human macrophage functions. Journal of Leukocyte Biology. 2015;99(6):1141–1151. doi: 10.1189/jlb.3A0815-390R. [DOI] [PubMed] [Google Scholar]
- 29.McLean NA, Verge VMK. Dynamic impact of brief electrical nerve stimulation on the neural immune axis-polarization of macrophages toward a pro-repair phenotype in demyelinated peripheral nerve. Glia. 2016;64:1546–1561. doi: 10.1002/glia.23021. [DOI] [PubMed] [Google Scholar]
- 30.Mobini S, Leppik L, Thottakkattumana Parameswaran V, Barker JH. In vitro effect of direct current electrical stimulation on rat mesenchymal stem cells. PeerJ. 2017;5:e2821. doi: 10.7717/peerj.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eischen-Loges M, Oliveira KMC, Bhavsar MB, Barker JH, Leppik L. Pretreating mesenchymal stem cells with electrical stimulation causes sustained long-lasting pro-osteogenic effects. PeerJ. 2018;6:e4959. doi: 10.7717/peerj.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leppik LP, et al. Effects of electrical stimulation on rat limb regeneration, a new look at an old model. Scientific reports. 2015;5:18353. doi: 10.1038/srep18353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iismaa SE, et al. Comparative regenerative mechanisms across different mammalian tissues. NPJ Regenerative medicine. 2018;3:6. doi: 10.1038/s41536-018-0044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tassava RA, Nace JD, Wei Y. Extracellular matrix protein turnover during salamander limb regeneration. Wound repair and regeneration: official publication of the Wound Healing. Society [and] the European Tissue Repair Society. 1996;4:75–81. doi: 10.1046/j.1524-475X.1996.40113.x. [DOI] [PubMed] [Google Scholar]
- 35.Santosh N, et al. Matrix metalloproteinase expression during blastema formation in regeneration-competent versus regeneration-deficient amphibian limbs. Developmental dynamics: an official publication of the American Association of Anatomists. 2011;240:1127–1141. doi: 10.1002/dvdy.22503. [DOI] [PubMed] [Google Scholar]
- 36.Calve S, Odelberg SJ, Simon H-G. A transitional extracellular matrix instructs cell behavior during muscle regeneration. Developmental biology. 2010;344:259–271. doi: 10.1016/j.ydbio.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 38.Whitby DJ, Ferguson MW. The extracellular matrix of lip wounds in fetal, neonatal and adult mice. Development (Cambridge, England) 1991;112:651–668. doi: 10.1242/dev.112.2.651. [DOI] [PubMed] [Google Scholar]
- 39.Rohani MG, Parks WC. Matrix remodeling by MMPs during wound repair. Matrix biology: journal of the International Society for. Matrix Biology. 2015;44-46:113–121. doi: 10.1016/j.matbio.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen HT, et al. Electric field stimulation through a substrate influences Schwann cell and extracellular matrix structure. Journal of neural engineering. 2013;10:46011. doi: 10.1088/1741-2560/10/4/046011. [DOI] [PubMed] [Google Scholar]
- 41.Mirza R, Dipietro LA, Koh TJ. Selective and specific macrophage ablation is detrimental to wound healing in mice. The American journal of pathology. 2009;175:2454–2462. doi: 10.2353/ajpath.2009.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wynn TA, Vannella KM. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity. 2016;44:450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Godwin JW, Rosenthal N. Scar-free wound healing and regeneration in amphibians. Immunological influences on regenerative success. Differentiation; research in biological diversity. 2014;87:66–75. doi: 10.1016/j.diff.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 44.Simkin J, et al. Macrophages are required to coordinate mouse digit tip regeneration. Development (Cambridge, England) 2017;144:3907–3916. doi: 10.1242/dev.150086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, et al. VSIG4 inhibits proinflammatory macrophage activation by reprogramming mitochondrial pyruvate metabolism. Nature communications. 2017;8:1322. doi: 10.1038/s41467-017-01327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaguchi M, Firdous J, Boyce JA, Balestrieri B. ILC2 Are Activated By Macrophages through Pla2g5-Dependent Generation of Linoleic Acid and Oleic Acid. Journal of Allergy and Clinical Immunology. 2017;139:AB72. doi: 10.1016/j.jaci.2016.12.282. [DOI] [Google Scholar]
- 47.Cho MR, Thatte HS, Lee RC, Golan DE. Integrin-dependent human macrophage migration induced by oscillatory electrical stimulation. Annals of biomedical engineering. 2000;28:234–243. doi: 10.1114/1.263. [DOI] [PubMed] [Google Scholar]
- 48.Li C, Levin M, Kaplan DL. Bioelectric modulation of macrophage polarization. Scientific reports. 2016;6:21044. doi: 10.1038/srep21044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Rouabhia M, Zhang Z. Pulsed electrical stimulation benefits wound healing by activating skin fibroblasts through the TGFβ1/ERK/NF-κB axis. Biochimica et biophysica acta. 2016;1860:1551–1559. doi: 10.1016/j.bbagen.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, et al. Influence of the intensity and loading time of direct current electric field on the directional migration of rat bone marrow mesenchymal stem cells. Frontiers of medicine. 2016;10:286–296. doi: 10.1007/s11684-016-0456-9. [DOI] [PubMed] [Google Scholar]
- 51.Ceccarelli G, et al. A comparative analysis of the in vitro effects of pulsed electromagnetic field treatment on osteogenic differentiation of two different mesenchymal cell lineages. BioResearch open access. 2013;2:283–294. doi: 10.1089/biores.2013.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwon HJ, Lee GS, Chun H. Electrical stimulation drives chondrogenesis of mesenchymal stem cells in the absence of exogenous growth factors. Sci. Rep. 2016;6:39302. doi: 10.1038/srep39302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thrivikraman G, Boda SK, Basu B. Unraveling the mechanistic effects of electric field stimulation towards directing stem cell fate and function: A tissue engineering perspective. Biomaterials. 2018;150:60–86. doi: 10.1016/j.biomaterials.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 54.Khatib L, Golan DE, Cho M. Physiologic electrical stimulation provokes intracellular calcium increase mediated by phospholipase C activation in human osteoblasts. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2004;18:1903–1905. doi: 10.1096/fj.04-1814fje. [DOI] [PubMed] [Google Scholar]
- 55.Wolf-Goldberg T, Barbul A, Ben-Dov N, Korenstein R. Low electric fields induce ligand-independent activation of EGF receptor and ERK via electrochemical elevation of H(+) and ROS concentrations. Biochimica et biophysica acta. 2013;1833:1396–1408. doi: 10.1016/J.BBAMCR.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 56.Bhavsar MB, et al. Membrane potential (Vmem) measurements during mesenchymal stem cell (MSC) proliferation and osteogenic differentiation. PeerJ. 2019;7:e6341. doi: 10.7717/peerj.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delaine-Smith RM, Reilly GC. Mesenchymal stem cell responses to mechanical stimuli. Muscles, Ligaments and Tendons. Journal. 2012;2:169–180. [PMC free article] [PubMed] [Google Scholar]
- 58.Géraudie J, Ferretti P. Gene expression during amphibian limb regeneration. International review of cytology. 1998;180:1–50. doi: 10.1016/S0074-7696(08)61769-6. [DOI] [PubMed] [Google Scholar]
- 59.Alibardi L. Review: Limb regeneration in humans: Dream or reality? Annals of anatomy = Anatomischer Anzeiger: official organ of the Anatomische Gesellschaft. 2018;217:1–6. doi: 10.1016/j.aanat.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 60.Geetha-Loganathan P, Nimmagadda S, Scaal M. Wnt signaling in limb organogenesis. organogenesis. 2008;4:109–115. doi: 10.4161/org.4.2.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taniguchi Yasushi. Hox Transcription Factors: Modulators of Cell-Cell and Cell-Extracellular Matrix Adhesion. BioMed Research International. 2014;2014:1–12. doi: 10.1155/2014/591374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tickle C, Towers M. Sonic Hedgehog Signaling in Limb Development. Frontiers in cell and developmental biology. 2017;5:14. doi: 10.3389/fcell.2017.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh BN, et al. Hedgehog Signaling during Appendage Development and Regeneration. Genes. 2015;6:417–435. doi: 10.3390/genes6020417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bryant DM, et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell reports. 2017;18:762–776. doi: 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voss SR, et al. Gene expression during the first 28 days of axolotl limb regeneration I: Experimental design and global analysis of gene expression. Regeneration. 2015;2:120–136. doi: 10.1002/reg2.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bak B, Andreassen TT. The effect of aging on fracture healing in the rat. Calcified tissue international. 1989;45:292–297. doi: 10.1007/BF02556022. [DOI] [PubMed] [Google Scholar]
- 67.Soybir OC, et al. Delayed cutaneous wound healing in aged rats compared to younger ones. International wound journal. 2012;9:478–487. doi: 10.1111/j.1742-481X.2011.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Illingworth CM. Trapped fingers and amputated finger tips in children. Journal of pediatric surgery. 1974;9:853–858. doi: 10.1016/S0022-3468(74)80220-4. [DOI] [PubMed] [Google Scholar]
- 69.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nature reviews. Immunology. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vogel B, Siebert H, Hofmann U, Frantz S. Determination of collagen content within picrosirius red stained paraffin-embedded tissue sections using fluorescence microscopy. MethodsX. 2015;2:124–134. doi: 10.1016/j.mex.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boudaoud A, et al. FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nature protocols. 2014;9:457–463. doi: 10.1038/nprot.2014.024. [DOI] [PubMed] [Google Scholar]
- 72.Badylak SF, Valentin JE, Ravindra AK, McCabe GP, Stewart-Akers AM. Macrophage Phenotype as a Determinant of Biologic Scaffold Remodeling. Tissue Engineering Part A. 2008;14:1835–1842. doi: 10.1089/ten.tea.2007.0264. [DOI] [PubMed] [Google Scholar]
- 73.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nature biotechnology. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic acids research. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pathan M, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 77.Pathan M, et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. Journal of extracellular vesicles. 2017;6:1321455. doi: 10.1080/20013078.2017.1321455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina sequencing data were deposited to the NCBI Sequence Read Archieve under BioProject Accession Number PRJNA532925 (SRA entries SRR8902417-SRR8902425).