Abstract
Background
Single omic analyses have provided some insight into the basis of lung function in children with asthma, but the underlying biologic pathways are still poorly understood.
Methods
Weighted gene coexpression network analysis (WGCNA) was used to identify modules of coregulated gene transcripts and metabolites in blood among 325 children with asthma from the Genetic Epidemiology of Asthma in Costa Rica study. The biology of modules associated with lung function as measured by FEV1, the FEV1/FVC ratio, bronchodilator response, and airway responsiveness to methacholine was explored. Significantly correlated gene-metabolite module pairs were then identified, and their constituent features were analyzed for biologic pathway enrichments.
Results
WGCNA clustered 25,060 gene probes and 8,185 metabolite features into eight gene modules and eight metabolite modules, where four and six, respectively, were associated with lung function (P ≤ .05). The gene modules were enriched for immune, mitotic, and metabolic processes and asthma-associated microRNA targets. The metabolite modules were enriched for lipid and amino acid metabolism. Integration of correlated gene-metabolite modules expanded the single omic findings, linking the FEV1/FVC ratio with ORMDL3 and dysregulated lipid metabolism. This finding was replicated in an independent population.
Conclusions
The results of this hypothesis-generating study suggest a mechanistic basis for multiple asthma genes, including ORMDL3, and a role for lipid metabolism. They demonstrate that integrating multiple omic technologies may provide a more informative picture of asthmatic lung function biology than single omic analyses.
Key Words: asthma, integrative omics, lung function, metabolome, transcriptome
Abbreviations: BDR, bronchodilator response; CAMP, Childhood Asthma Management Program; HILIC, hydrophilic interaction liquid chromatography; MS, mass spectrometry; QC, quality control; SNP, single-nucleotide polymorphism; WGCNA, weighted gene coexpression network analysis
Asthma, a disorder characterized by variable and reversible airway obstruction, hyperresponsiveness, and inflammation, represents one of the most common chronic conditions among children and adults worldwide.1, 2 Asthmatic lung function abnormalities are present early in life,3, 4 track through childhood and adulthood,5, 6 and are strong determinants of disease exacerbations and severity.7, 8
Reduced lung function in patients with asthma is thought to emerge from complex gene-environment interactions.9 Advances in high-throughput technologies allow us to explore such interactions at the level of the epigenome, genome, transcriptome, proteome, and metabolome. Combining the transcriptome, which reflects genomic activity, with the metabolome, which is sensitive to environmental influences and closely related to phenotype, may be particularly informative. Although previous studies have investigated metabolomic and transcriptomic profiles of asthma separately, to date only two studies, with limited sample sizes, have integrated the two omes together in humans.10, 11 Relative to the use of single omics technologies, this integrative approach demonstrated increased predictive ability for asthma and its subtypes, and greater biologic insights. Consequently, integrative omics represents an exciting new avenue in asthma research.12
Currently, there are no analytic standards for integrative omics. However, network medicine, a rapidly emerging field that moves away from reductionist methodologies to combine systems biology and network science, represents a particularly promising approach. It provides a holistic methodology to better understand disease through the identification and investigation of nonlinear relationships and networks of interacting components. This provides insights into these conditions beyond the level of a single gene or omic platform. Weighted gene coexpression network analysis (WGCNA) is a network method for identifying clusters or modules or highly correlated variables (eg, genes, metabolites) that are likely to be coregulated, or working together in a biologically coherent fashion. A module can then be summarized as a single unit, which can be correlated with phenotypes or other modules of interest.
The aim of this study was to conduct an integrated analysis of the blood transcriptome and metabolome among children with asthma participating in the Genetic Epidemiology of Asthma in Costa Rica13 cohort to identify biologically informative networks of genes and metabolites associated with asthmatic lung function. The Genetic Epidemiology of Asthma in Costa Rica cohort recruited children with mild-to-moderate asthma from the Central Valley of Costa Rica. This area represents a Hispanic population isolate which is genetically homogenous and has one of the highest prevalences of asthma in the world (24% in children),14 making it uniquely suited for the exploration of the integrative omic underpinnings of asthmatic lung function. In particular, the study focuses on FEV1 and FEV1/FVC ratios, which are thought to mediate the association between early life characteristics and asthma.15
Methods
Study Population
This integrative omic study was nested within the Genetic Epidemiology of Asthma in Costa Rica Study,13 which recruited children 6 to 14 years of age with mild-to-moderate asthma and their parents from the Central Valley of Costa Rica. Children were eligible if they had physician-diagnosed asthma and at least two episodes of respiratory symptoms or asthma attacks in the prior year, and a high probability of having six or more great-grandparents born in the Central Valley of Costa Rica.16, 17 A total of 1,165 children with asthma were enrolled in the original study. All children completed a protocol at enrollment, including questionnaires, spirometry, and collection of blood when children were not exacerbated. Most blood samples were processed within 4 h; RNA was extracted and stored in PAXgene tubes (QIAgen). Genome-wide single-nucleotide polymorphism (SNP) genotyping and RNA expression profiles were generated for a subset of the children with suitable samples. Genotype data were obtained with TaqMan real-time polymerase chain reaction with an ABI Prism 7900 machine (Applied Biosystems).18 Standard manufacturer-recommended polymerase chain reaction conditions were used. Children were prioritized for metabolomic profiling if they had both genome-wide genetic and genome-wide expression data, with the goal of conducting integrative omic analyses. Children with both metabolomic and transcriptomic profiling were included in the current study. Written parental and participating child consent was obtained. The study was approved by the Partners Human Research Committee at Brigham and Women’s Hospital (Boston, MA; protocol No. 2000-P-001130/55) and the Hospital Nacional de Niños (San José, Costa Rica).
Lung Function
At enrollment, baseline lung function was investigated by spirometry (FEV1 and FEV1/FVC ratio), bronchodilator response (BDR) (percentage difference in FEV1 from baseline after inhaled albuterol), and airway responsiveness to methacholine (determined as the provocative dose of methacholine resulting in a 20% drop in FEV1 from baseline) (e-Appendix 1).
Transcriptomic Profiling
Whole-blood gene expression profiles were generated with 47,009 probes from the HumanHT-12 v4 Expression BeadChip (Illumina, Inc.) that passed stringent and commonly used quality control (QC) metrics.19 Expression data were log2 transformed and quantile normalized as a single batch using the lumiT and lumiN functions, respectively, from the R package lumi (version 2.22; The R Foundation). Principal components of gene expression were generated using the getPCAFunc function from the R package iCheck (version 0.6). Prior to downstream statistical analyses, we applied a standard nonspecific variance filter to the expression data using the nsFilter function from the R package genefilter (version 1.52). Probes not annotated with a valid Entrez gene identifier or Human Genome Organization gene symbol and probes with interquartile ranges of expression variance below the 50th percentile were removed to select only the most informative probes.20 Data were then collapsed to a single probe per gene based on the largest interquartile range of expression variance.19
Metabolomic Profiling
Plasma samples were shipped from the sample repository to the Broad Institute (Cambridge, MA) on dry ice for metabolomic profiling. Samples were thawed on ice for subaliquoting for each of the metabolomic methods and then refrozen on dry ice and stored at −80°C until analysis. Four liquid chromatography-tandem mass spectrometry (MS) platforms measured complementary sets of metabolite classes: (1) hydrophilic interaction liquid chromatography (HILIC)-positive platform, amines and polar metabolites that ionize in the positive ion mode using HILIC and MS analyses; (2) HILIC-negative platform, central metabolites and polar metabolites that ionize in the negative ion mode using HILIC with an amine column and targeted MS; (3) C8 platform, polar and nonpolar lipids using reverse-phase chromatography and full-scan MS; and (4) C18 chromatography platform, free fatty acids, bile acids, and metabolites of intermediate polarity using reverse chromatography with a T3 ultra performance liquid chromatography column and MS analyses in the negative ion mode (e-Appendix 1).
QC was performed using previously described methods21: metabolite features with a signal-to-noise ratio < 10 were considered unquantifiable and excluded, as were features with undetectable/missing levels for > 10% of the sample. All remaining missing values were imputed with the median peak intensity for that feature across the whole population. Features with a coefficient of variance in the QC samples > 25% were excluded to ensure good technical reproducibility. The mean coefficient of variance of the remaining features was 13.1% (interquartile range, 8.7%-17.1%). Features were indexed by mass/charge ratio, retention time, and ion spectra for subsequent annotation. Features were analyzed as measured liquid chromatography-tandem mass spectrometry peak areas and were log transformed and Pareto scaled prior to analysis. This data processing pipeline has previously been used for multiple peer-reviewed publications.16, 21, 22, 23
Of 18,064 measured metabolite features, 8,185 passed QC and data processing procedures (e-Appendix 1). Of these, 574 could be reliably annotated to known metabolites by matching measured mass/charge ratio and retention time with authentic reference standards. Reference compounds were spiked into biologic samples to mitigate any matrix effects. MS/MS data were not acquired during the profiling to facilitate acquisition of sufficient numbers of data points across peaks for precise quantitation. To confirm identification during analytic runs, synthetic mixtures of standards and pooled QC samples were analyzed.
To reflect the fact that only a small proportion of metabolites could be annotated, both known and unknown metabolite features were included in the analysis. This allowed the capture of all relationships between the features with no missing links, which would be inevitable if only named metabolites were included, thereby reflecting the whole metabolome. Furthermore, annotation of metabolites is ongoing; metabolites identified as important in this analysis will be prioritized for annotation so they can be available for future analyses in this population.
Network Identification and Association With Lung Function Traits
WGCNA24 was used to identify transcriptomic and metabolomic networks based on correlation patterns. The correlation matrix quantifies interconnectedness between features (genes and metabolite features) and assigns them to coexpression modules. Features that did not show high enough coexpression metrics with any module were excluded from further analysis. Highly correlated modules were then merged using a cut height (ie, Euclidean distance between clusters) of 0.75 for the transcriptomic data and 0.70 for the metabolomic data. These cut-heights were chosen using an iterative process to identify an optimal number of adequately sized modules for analysis.
Modules were summarized by an eigenvector (based on the first principal component of each module) for each participant. Associations between the modules with lung function traits were computed using multivariate regression models with adjustment for potential confounders (age, sex, height, weight, and treatment regimen [daily use for persistent systems vs only intermittent use for acute symptoms]). For the modules nominally significantly (P ≤ .05) associated with at least one lung function phenotype, the hubs were identified. Hubs are the features that are most highly connected within a module, and therefore drive module formation. WGCNA computes a module membership value and associated P value for each feature within a module, which is a measure of how connected or coexpressed that feature is with others within the same module. Features with a module membership P value that retained significance after Bonferroni correction were considered to be hubs.
Pathway Enrichment Analysis
For the transcriptomic data, pathway enrichment analysis was performed using the g:GOSt tool within the g:Profiler25 web server (BIIT Group).
Metabolomic pathway analysis was performed using MetaboAnalyst v.3.026 (XiaLab at McGill University). Metabolomic pathway analysis was limited to those metabolites that could be assigned Human Metabolome Database identification; therefore, it was used here as a hypothesis-generating tool (e-Appendix 1).
Integrated Omics Analysis
Relationships between the WGCNA-generated transcriptomic and metabolomic modules were explored by computing the correlation and corresponding P value between the eigenvector of the two sets of modules. Constituent hub features of correlated pairs were submitted for integrated pathway analysis using IMPaLA: Integrated Molecular Pathway Level Analysis to identify pathways simultaneously dysregulated on both a transcriptional and metabolic level (e-Appendix 1). These pathways were then prioritized for further follow-up.
Replication
Replication of select findings was performed in a comparable childhood cohort of patients with mild-to-moderate asthma who were phenotyped in the same way as the Costa Rican cohort. The Childhood Asthma Management Program (CAMP) is a multicenter, randomized, double-masked, clinical trial designed to determine the long-term (approximately 16.5 years of follow-up) effects of three inhaled treatments (placebo, nedocromil, or budesonide) for mild-to-moderate asthma in children 5 to 12 years of age at baseline.27 A follow-up study to the primary trial extracted blood samples from 620 CAMP subjects at early adulthood (after trial completion) for gene expression profiling19 using the HumanHT-12 chip and the same protocols and methods as the Costa Rican study. Genome-wide SNP genotyping on the HumanHap550v3 BeadChip (Illumina, Inc.) was also available for these children.28 A total of 207 children additionally had metabolomic profiling conducted on the same blood samples at the Broad Institute. Metabolomic profiling was conducted on serum at the Broad Institute (e-Appendix 1). A total of 14,587 metabolite features including 324 named metabolites passed the sample QC and data processing pipeline as applied to the Costa Rican metabolomics data. A total of 222 named metabolites were common to the CAMP and Costa Rican populations. The CAMP study was approved by the institutional review board of Partners Healthcare (Partners Human Research Committee; protocol No. 1999-P-001549/29), all eight CAMP clinical centers, and the CAMP Data Coordinating Center. Each child’s parent or guardian signed a consent statement; the clinics also obtained each child’s assent.
Results
Baseline Characteristics
Of 1,165 children with asthma from the Costa Rican cohort, 328 had RNA available for transcriptomic profiling and sufficient plasma for metabolomic profiling. Initial clustering based on the transcriptomic data profiles identified three outliers which formed a distinct cluster that could be separated from the rest of the population using a cut-height of 0.9 (e-Fig 1). No metabolomic outliers were detected, and in total 325 children were included in all analyses (e-Fig 2). All children were Hispanic/Latino, 139 (42.8%) were girls, the mean age was 9.1 years (range, 4.5-13.3 years), and 90% reported asthma controller or reliever treatment (Table 1). Although distinct, there was significant correlation between the measured lung function phenotypes (e-Table 1). The 325 children with omics profiling data were representative of the entire population of 1,165 children (e-Table 2).
Table 1.
Epidemiologic and Clinical Characteristics of 325 Children With Asthma
| Epidemiologic and Clinical Characteristics | Value |
|---|---|
| Sex, No. (%), male | 190 (58.5) |
| Age at recruitment, ya | 9.1 (1.8) |
| Height, cm | 132.1 (11.5) |
| Weight, kg | 32.5 (11.3) |
| BMI, kg/m2 | 18.1 (3.9) |
| Controller treatment, No. (%), yes | 297 (91.4) |
| Baseline FEV1, L | 1.8 (0.5) |
| Baseline FEV1/FVC ratio, % | 86.5 (7.7) |
| Postbronchodilator FEV1/FVC ratio, % | 89.1 (6.7) |
| Airway responsiveness measured by methacholine challenge, PD20, μmol | 1.7 (2.1) |
| FEV11 bronchodilator response, % | 5.0 (8.7) |
Values are mean (SD) or as otherwise indicated. Controller treatment = receiving regular treatment for the control of chronic symptoms in the form of oral or inhaled steroids, prednisone, long-acting inhaled B2 agonists, or leukotriene inhibitors/modifiers; FEV11 bronchodilator response = bronchodilator response as % of baseline FEV1; PD20 = 20% drop in FEV1.
Lung function testing and blood draw performed at recruitment.
Transcriptomic Network Analysis
WGCNA clustered the 25,060 gene probes into 39 different modules. After merging highly correlated modules using a cut-height of 0.75 (e-Fig 3), a total of eight modules, spanning 19,581 genes, were characterized (e-Table 3). A nominal significance threshold of P ≤ .05 was used to capture all potentially biologically interesting relationships. Six modules were significantly correlated with at least one measure of lung function (e-Fig 4). Four of these modules retained significance at a 95% CI after adjustment for age, weight, height, sex, and asthma treatment (Table 2).
Table 2.
Association Between Gene Modules and Measures of Lung Function After Adjustment for Confounders
| Transcriptomic Module | Trait | β (95% CI) | P Value |
|---|---|---|---|
| Dark green, adaptive immunity | Post-BD FEV1/FVC ratio | −0.0009 (−0.0018 to 4.7 × 10−5) | .064 |
| Dark gray, cell cycle | Baseline FEV1, L | −0.0233 (−0.1696 to 0.123) | .755 |
| Baseline FEV1/FVC ratio | 0.0013 (0.0005 to 0.0021) | .002a | |
| Post-BD FEV1/FVC ratio | 0.0014 (0.0005 to 0.0023) | .004a | |
| Dark olive green, asthma microRNAs | Post-BD FEV1/FVC ratio | −0.0005 (−0.0014 to 0.0004) | .29 |
| FEV1 BD response | 0.0008 (0.0001 to 0.0015) | .023a | |
| Pink, translational | Baseline FEV1/FVC ratio | 0.0009 (0.0001 to 0.0017) | .031a |
| Plum, innate immunity | PD20 | 0.0078 (0.0041 to 0.0116) | < .001 |
| Baseline FEV1/FVC ratio | −0.0008 (−0.0016 to 1.5 × 10−5) | .055 | |
| Post-BD FEV1/FVC ratio | −0.001 (−0.0019 to −0.0001) | .027a | |
| Yellow, transcriptional | FEV1 BD response | −0.0007 (−0.0014 to 3.0 × 10−5) | .062 |
Adjusted for age, height, weight, sex, and treatment regimen. BD = bronchodilator. See Table 1 legend for expansion of other abbreviation.
Significant at 95% confidence.
Lung function-associated modules were enriched for distinct groups of defined biologic processes (Table 3): the dark green and plum modules for immune processes (renamed the adaptive immunity and innate immunity modules, respectively), the dark gray modules for cell cycle and mitotic processes (cell cycle module), the dark olive green module for asthma-related microRNA targets sites (asthma microRNAs module; g:Profiler allows functional interpretation of gene lists in the context of computationally predicted microRNA target sites from the MicroCosm database), the pink modules for processes relating to translation (translational module), and the yellow module for multiple transcriptional and metabolic processes (transcriptional module) (Table 3).
Table 3.
Top Gene Ontology-Defined BPs in Six Transcriptomic Modules Correlated With Lung Function Traits
| Module | P Value | |
|---|---|---|
| Dark green, adaptive immunity | Graft-vs-host disease (keg) | < .001 |
| Natural killer cell mediated cytotoxicity (keg) | < .001 | |
| Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell (rea) | < .001 | |
| Natural killer cell mediated immunity (BP) | .013 | |
| Antigen processing and presentation (keg) | .029 | |
| Cellular defense response (BP) | .043 | |
| Dark gray, cell cycle | Mitotic cell cycle process (BP) | < .001 |
| Cell Cycle (rea) | < .001 | |
| DNA metabolic process (BP) | < .001 | |
| Cell cycle (keg) | < .001 | |
| Histone kinase activity (MF) | < .001 | |
| RHO GTPases Activate Formins (rea) | < .001 | |
| Protein localization to chromosome, centromeric region (BP) | < .001 | |
| G1/S-Specific Transcription (rea) | < .001 | |
| Chromosome condensation (BP) | .014 | |
| Proteasome-mediated ubiquitin-dependent protein catabolic process (BP) | .017 | |
| Oocyte meiosis (keg) | .018 | |
| Signal transduction by p53 class mediator (BP) | .026 | |
| SUMOylation of DNA replication proteins (rea) | .027 | |
| G0 and Early G1 (rea) | .037 | |
| Dark olive green, asthma microRNAs | MI:hsa-miR-423-5p (mi) | < .001 |
| MI:hsa-miR-615-5p (mi) | < .001 | |
| MI:hsa-miR-564 (mi) | .025 | |
| MI:hsa-miR-339-5p (mi) | .028 | |
| MI:hsa-miR-214 (mi) | .034 | |
| MI:hsa-miR-331-3p (mi) | .036 | |
| Poly(A) RNA binding (MF) | .043 | |
| MI:hsa-miR-874 (mi) | .046 | |
| Pink, translational | RNA binding (MF) | < .001 |
| RNA processing (BP) | < .001 | |
| Ribonucleoprotein complex biogenesis (BP) | < .001 | |
| Translation (BP) | < .001 | |
| Mitochondrion organization (BP) | .022 | |
| Structural constituent of ribosome (MF) | .044 | |
| Negative regulation of translational initiation in response to stress (BP) | .046 | |
| DNA metabolic process (BP) | .05 | |
| Plum, innate immunity | Innate immune system (rea) | < .001 |
| Antimicrobial humoral response (BP) | < .001 | |
| Heparin binding (MF) | < .001 | |
| Serine-type endopeptidase activity (MF) | < .001 | |
| Extracellular matrix organization (rea) | .011 | |
| Yellow, transcriptional | RNA binding (MF) | < .001 |
| mRNA metabolic process (BP) | < .001 | |
| Cotranslational protein targeting to membrane (BP) | < .001 | |
| Structural constituent of ribosome (MF) | < .001 | |
| Protein transporter activity (MF) | < .001 | |
| Macromolecular complex subunit organization (BP) | < .001 | |
| Mitochondrion organization (BP) | < .001 | |
| Posttranscriptional regulation of gene expression (BP) | < .001 | |
| Multi-organism metabolic process (BP) | .027 | |
| Ribonucleoprotein complex localization (BP) | .033 | |
| Viral transcription (BP) | .04 | |
BP = biologic process; keg = KEGG-defined biologic pathway; MF = molecular function; mi = regulatory motif in miRBase microRNA; rea = reactome-defined biologic pathway.
Metabolomic Network Analysis
WGCNA clustered 8,185 metabolite features (including 574 annotated metabolites) into 44 different modules. After merging (cut-height 0.7) (e-Fig 5), there were eight modules including 7,473 metabolite features (excluding the gray module) (e-Table 4). Six modules were correlated at a nominal significance level of 95% with at least one measure of lung function (e-Fig 6), and five modules were robust to confounder adjustment (P ≤ .05) (Table 4).
Table 4.
Association Between Metabolite Modules and Lung Function Traits After Adjustment for Confounders
| Metabolite Module | Trait | β (95% CI) | P Value |
|---|---|---|---|
| Red | Baseline FEV1/FVC ratio | −0.001 (−0.0018 to −0.0002) | .019a |
| Post-BD FEV1/FVC ratio | −0.0014 (−0.0023 to −0.0004) | .004a | |
| Purple | Baseline FEV1/FVC ratio | 0.0009 (0.0001 to 0.0017) | .028a |
| Medium purple, lipid | Baseline FEV1 | −0.1386 (−0.2808 to 0.0036) | .057 |
| Dark red | Baseline FEV1/FVC ratio | 0.0009 (0.0001 to 0.0017) | .029a |
| FEV1 BD response | −0.0007 (−0.0014 to 1.75 × 10−5) | .057 | |
| Light cyan | Baseline FEV1/FVC ratio | −0.0008 (−0.0016 to 3.45 × 10−5) | .062 |
| FEV1 BD response | 0.0007 (1.65 × 10−5 to 0.0014) | .046a | |
| Tan, purine | PD20 | −0.0065 (−0.0103 to −0.0027) | .001a |
| Baseline FEV1/FVC ratio | 0.0009 (0.0001 to 0.0017) | .028a | |
| Post-BD FEV1/FVC ratio | 0.0017 (0.0007 to 0.0026) | .001a |
All pathways with a nominally significant P < .10 are reported for hypothesis-generating purposes. The medium purple module (renamed the lipid module) was enriched for glycosylphosphatidylinositol anchor biosynthesis (P = .034), sphingolipid metabolism (P = .061), glycerolipid metabolism (P = .077), and glycerophospholipid metabolism (P = .093). The tan module was enriched for caffeine (P = .017) and purine metabolism (P = .075) (Table 5).
Table 5.
Enriched Biologic Pathways and Processes in Six Metabolomic Modules Correlated With Lung Function Traits
| Module | Trait Association | No. of Metabolite Features | No. of Hub Features | Enriched Pathways Among Known Metabolites |
|---|---|---|---|---|
| Red, ubiquinone | Baseline FEV1/FVC ratioa; post-BD FEV1/FVC ratioa | 449 | 412 | No enriched pathways: known metabolites— Ubiquinone-2; C18:0 Mono Acyl Glycerol; (S)-2-Propylpiperidine; Polyoxyethylene (600) monoricinoleate |
| Medium purple, lipid | Baseline FEV1 | 537 | 518 | GPI-anchor biosynthesis (P = .034); sphingolipid metabolism (P = .061); glycerolipid metabolism (P = .077); glycerophopholipid metabolism (P = .093) |
| Tan, purine | PD20; baseline FEV1/FVC ratio; post-BD FEV1/FVC ratio | 147 | 141 | Caffeine metabolism (P = .017); purine metabolism (P = .075) |
| Dark red | Baseline FEV1/FVC ratioa; baseline FEV1 | 93 | 85 | No enriched pathways; no known metabolites |
| Light cyan | Baseline FEV1/FVC ratio; baseline FEV1a | 111 | 98 | No enriched pathways: known metabolites—methyl dihydrojasmonate |
| Purple | Baseline FEV1/FVC ratioa | 411 | 330 | No enriched pathways: known metabolites—tetradecyl sulfate |
Integrated Omics Analysis
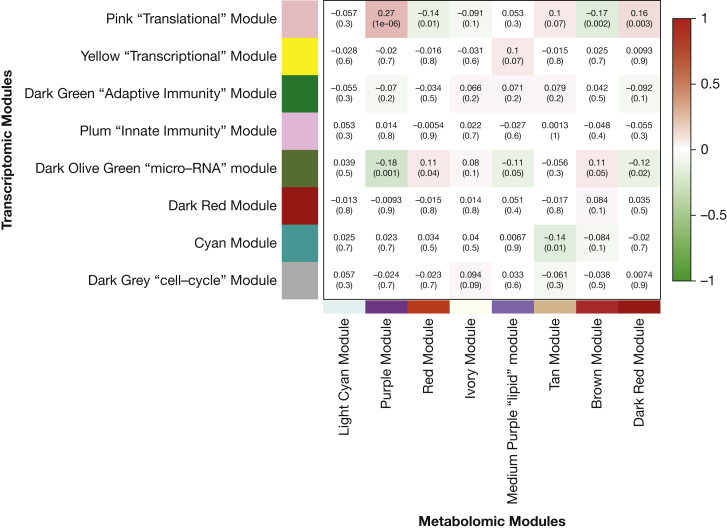
There were 10 nominally significant (P ≤ .05) associations between metabolite modules and gene modules based on their eigenvector (Fig 1). Of these, seven were between modules that both correlated with lung function phenotypes.
Figure 1.
Correlation between the eigenvectors of the transcriptomic and metabolomics modules. Correlation coefficients are shown for each module-trait pair with the associated P value in parenthesis; colors indicate direction of association and darker colors indicate more significant associations.
Integrated analyses of the asthma microRNAs transcriptomic and lipid metabolomic modules, which correlated with FEV1 and the FEV1/FVC ratio, identified 186 pathways including 51 pathways robust to false discovery rate correction (top pathways in Table 6, full list in e-Table 5). Most were for the asthma microRNAs module, most of which relate to immune regulation. This integrative analysis extends on the findings of the metabolomic analysis, where sphingolipid, glycerolipid, and glycerophospholipid metabolism were all enriched by identifying the upstream hub genes, including ORMDL3, SLC1A2, and AKR1B1, driving the dysregulation of these pathways.
Table 6.
Results of the Integrated Pathway Enrichment Analysis Based on the Genes and Metabolites of Correlated Lung Function-Associated Transcriptomic and Metabolomics Modules
| Gene Module | Metabolite Module | Pathway Name | No. of Overlapping Genes | No. of Overlapping Metabolites | Joint P Value |
|---|---|---|---|---|---|
| Dark olive green, asthma microRNAs | Medium purple, lipid | Immune System | 409 | 4 | 9.38 × 10−11,a |
| Membrane Trafficking | 152 | 1 | 1.46 × 10−8,a | ||
| B cell receptor signaling pathway - Homo sapiens (human) | 31 | 1 | 5.12 × 10−8,a | ||
| Adipocytokine signaling pathway - Homo sapiens (human) | 23 | 2 | 6.64 × 10−7,a | ||
| Signalling by NGF | 106 | 2 | 1.03 × 10−6,a | ||
| Adaptive Immune System | 168 | 3 | 1.42 × 10−6,a | ||
| TNF alpha Signaling Pathway | 33 | 1 | 1.47 × 10−6,a | ||
| Neurotrophin signaling pathway - Homo sapiens (human) | 34 | 2 | 1.84 × 10−6,a | ||
| Insulin resistance - Homo sapiens (human) | 27 | 3 | 2.23 × 10−6,a | ||
| Innate Immune System | 276 | 2 | 3.88 × 10−6,a | ||
| Dark olive green, asthma microRNAs | Red, ubiquinone | TCA cycle | 12 | 1 | 4.53 × 10−4 |
| Citrate cycle | 12 | 1 | 7.98 × 10−4 | ||
| The citric acid (TCA) cycle and respiratory electron transport | 40 | 1 | 3.74 × 10−3 | ||
| Respiratory electron transport_ ATP synthesis by chemiosmotic coupling_ and heat production by uncoupling proteins | 27 | 1 | 7.12 × 10−3 | ||
| Sulfide oxidation to sulfate | 2 | 1 | 9.24 × 10−3 | ||
| Oxidative phosphorylation - Homo sapiens (human) | 27 | 1 | .010 | ||
| Respiratory electron transport | 20 | 1 | .012 | ||
| Electron Transport Chain | 19 | 1 | .013 | ||
| Degradation of cysteine and homocysteine | 2 | 1 | .035 | ||
| Pyrimidine metabolism | 6 | 1 | .043 | ||
| Pink, translational | Red, ubiquinone | Respiratory electron transport | 17 | 1 | 5.06 × 10−4,a |
| The citric acid (TCA) cycle and respiratory electron transport | 26 | 1 | 9.86 × 10−4 | ||
| Respiratory electron transport_ ATP synthesis by chemiosmotic coupling_ and heat production by uncoupling proteins | 18 | 1 | 2.06 × 10−3 | ||
| Citrate cycle | 7 | 1 | 2.91 × 10−3 | ||
| Oxidative phosphorylation - Homo sapiens (human) | 18 | 1 | 2.96 × 10−3 | ||
| Electron Transport Chain | 14 | 1 | 2.97 × 10−3 | ||
| TCA cycle | 6 | 1 | 5.96 × 10−3 | ||
| Arginine Proline metabolism | 8 | 1 | .020 | ||
| Ubiquinone and other terpenoid-quinone biosynthesis - Homo sapiens (human) | 2 | 1 | .027 | ||
| Sulfur amino acid metabolism | 4 | 1 | .029 |
Top ten pathways/processes enriched for both genes and metabolites are shown. Dark red metabolite module contained no known metabolites and so could not be included in the analyses.
Retains significance when applying the Benjamini-Hochberg false discovery rate.
The asthma microRNA module was also correlated with the red ubiquinone-containing metabolite module, and both were associated with the FEV1/FVC ratio (Table 6). Integration of these modules identified a large number of pathways involved in cellular respiration; however, none were significant after correction for multiple testing.
Cellular respiration was also captured by the integration of the translational gene module and the red metabolite module, which both associated with the FEV1/FVC ratio.
ORMDL3
Given the role of ORMDL3 as a hub gene in the asthma microRNAs transcriptomic module, we used genotype data available for the Costa Rican subjects to identify SNPs that may influence the expression of this gene and which also associate with the lipid metabolite module. Three relevant SNPs were available in the Costa Rican population (rs2872507, rs7216389, and rs8079416). Although all three associated with at least one of the lipids in the lipid metabolite module, the strongest results were observed for rs8079416, which was associated with 165 of the 537 metabolites (31%) comprising the module, and with the module eigenvector (P = .024), with an increasing number of C alleles associated with lower levels of these lipids (e-Fig 7). Importantly, the C allele was also strongly associated with increased expression of ORMDL3 in this population (additive model P = .0006), which provides a further link between ORMDL3 and dysregulation of lipid metabolism.
Replication of the Integrated Analysis
To explore the validity of the nominally significant integrative findings, a replication analysis was conducted, focusing on the relationship between the lung function-associated transcriptomic asthma microRNAs and metabolomic lipid modules. Whole genome transcriptomic profiling and metabolomic data were available on blood samples from 207 participants from the CAMP cohort (e-Appendix 1, e-Table 6).29
A summary score based on the first principal component was generated using the expression levels in CAMP of 3,926 hub genes from the Costa Rican asthma microRNAs module. A significant association was observed between this score and FEV1 BDR in a regression model adjusting for age, sex, height, weight, and treatment group (P = .027). Metabolomic profiling information was available for 564 named metabolites, including 30 of the named metabolites identified in the lipid metabolomic module. A score based on the first principal component of these 30 metabolites had a borderline significant association with baseline FEV1 (P = .073). Furthermore, there was evidence of a correlation between the transcriptomic and metabolomic modules (r = 0.13, P = .065). Finally, it was again shown that the C allele of the ORMDL3 SNP rs8079416 was strongly associated with increased expression of ORMDL3 (P = 5.2 × 10−10), and with four of the investigated lipids, thereby providing a measure of replication of the Costa Rican metabolomics, transcriptomic, and integrative omic findings.
Discussion
This study represents one of the first integrative omics studies of children with asthma. A network approach was taken to identify and integrate modules of highly coregulated genes and metabolites that correlate with lung function metrics. Specifically, this included metrics related to FEV1 and the FEV1/FVC ratio, which associate with childhood asthma severity and have been shown to be predictive of future lung function.15 Interrogation of the biologic pathways and processes underlying these modules allowed the exploration of asthmatic lung function mechanisms. These findings strengthen the evidence for the role of sphingolipids, lipids, and fatty acids and provide novel evidence suggesting these effects may be partly driven by an SNP that influences ORMDL3 expression, rs8079416. In sum, these results demonstrate the potential of network-based methods for integrating large-scale omic datasets in a biologically meaningful way. Network-based approaches may be particularly useful as a screening step to identify the most important components of these highly dimensional omic datasets and how they work together across hierarchical levels. Such knowledge could underlie the development of improved biomarkers and precision medicine initiatives.
The transcriptomic network analysis revealed six gene modules that associated with lung function measures: airway obstruction, airway responsiveness to methacholine, and BDR. Pathway analysis determined these modules were enriched for distinct processes with biologically plausible relationships with lung function, including adaptive and innate immunity.
Mitosis and processes relating to cell division characterized the cell cycle transcriptomic module. Interestingly, a previous WGCNA study of gene expression data from asthmatic airway epithelial cells also identified a mitosis module characterized by PTTG1, BIRC5, NCAPG, CDCA2, and FANCI.30 All these genes were hubs within our cell cycle module. Dysregulation of cell division can alter the composition of the airway epithelium, initiate airway remodeling, and lead to chronic airway obstruction and more severe asthma.31 Intriguingly, the cell cycle module was significantly associated with the FEV1/FVC ratio pre- and postbronchodilator, but not with FEV1. It has been shown that children with asthma can have an abnormal FEV1/FVC ratio despite a normal FEV1 and FVC; this condition, known as dysanapsis, occurs when growth in lung volume and airway length outpaces the increase in airway caliber.32 This may explain the association with a module enriched for dysregulated cell cycle processes in children.
The asthma microRNAs gene module, based on microRNA target sites, was enriched for seven microRNA regulatory motifs, which have previously been implicated in lung function and asthma severity: hsa-miRNA-339-5p has been shown to be differentially expressed in the blood of asthmatics,33 and serum miR-331-3p has been associated with the FEV1/FVC ratio in childhood asthma.34 Additionally, expression of miR-874 has been shown to be upregulated in patients with allergic rhinitis,35 whereas miR-874 and miR-423 have both been demonstrated to be upregulated in response to antigens in mouse models.36 Crucially, in an independent population, a summary score based on the expression levels of the genes of this module was again associated with FEV1 BDR.
Analysis of the metabolomic profiles from the same children at the same time point also identified six modules associated with the metrics of FEV1 and the FEV1/FVC ratio. The lipid metabolite module is of particular interest because lipid mediators influence asthmatic airway inflammation, in which the conversion of arachidonic acid in membrane phospholipids to eicosanoids, leukotrienes, and prostaglandins results in bronchoconstriction and inflammation.37 It has also been shown that essential omega n-3 fatty acids in foods and oily fish, such as eicosapentanoic acid and docosahexaenoic acid, are capable of displacing arachidonic acid from the cell membrane, promoting resolution of inflammation and dampening of airway hyperresponsiveness.38, 39 Both glycosylphosphatidylinositol and glycerophospholipids are key constituents of cell membranes, and the identification of these pathways in a metabolite module associated with FEV1 further highlights the importance of fatty acids and lipid mediators in the obstructive, reduced lung function characterizing asthma pathology.
Sphingolipids have also been implicated in asthma pathogenesis.40 ORMDL3, one of the most replicated genes for childhood asthma,41 is thought to exert its effect through the sphingolipid metabolism pathway via negative regulation of serine palmitoyltransferase, which catalyzes the initial step of sphingolipid biosynthesis.41 In mouse models, knockout of serine palmitoyltransferase has been shown to result in alterations in de novo lung epithelial tissue sphingolipid biosynthesis and an increase in inflammation and airway hyperresponsiveness.42 However, direct evidence of an association in human studies is lacking.
In the integrated omics analysis, the lipid metabolite module correlated with the asthma microRNAs transcriptomic module that included ORMDL3, providing evidence for a mechanistic connection between ORMDL3, microRNA regulatory motifs, and sphingolipid metabolism in asthma. Furthermore, we were able to demonstrate an SNP within the 17q21 locus, rs8079416, which has previously been associated with asthma,43 was significantly associated with both the expression of ORMDL3 and with the lipid metabolomic module and its constituent metabolites. This highlights the potential of multilevel integrative omics to elaborate how SNPs can impact metabolite levels though transcriptomic regulation. The asthma microRNAs module also encompassed several other asthma genes which, like ORMDL3, map to the 17q21 locus, including CCL21 and GSDMB. This suggests variants in this region systematically contribute to the pathogenesis of asthma through a dysregulation of sphingolipid metabolism, potentially because of the binding of these genes to the regulatory microRNAs. Intriguingly, the asthma microRNAs module also included CRISPLD2, previously identified as an asthma candidate gene because of its role as a regulator of the antiinflammatory effects of glucocorticoids in the airway smooth muscle.44 Sphingolipids have been shown to mediate the effects of glucocorticoids,45 further supporting the observed link between the asthma microRNAs and the lipid modules and suggesting a possible connection between CRISPLD2 and sphingolipid metabolism. Full replication and validation of these findings was not possible because there was incomplete crossover between the known metabolites measured in Costa Rica and CAMP, and the crossover between the unknown metabolites cannot be computed. However, in the independent CAMP population, there was evidence that a lung function-associated lipid module correlated with this module in blood samples taken at the same time point. Furthermore, there was evidence that rs8079416 was again associated with increased expression of ORMDL3 and a number of pertinent lipids. This adds weight to the theory that the genes enriched for microRNA targets may be acting on lung function through the dysregulation of lipid metabolism.
To date, relatively few studies have attempted to combine metabolomic and transcriptomic data in any human disease, and no analytic gold standards exist. However, statistical integration using pathway and network-based approaches has shown promising results.46 This study demonstrated relationships between transcriptomic and metabolomic modules, which were generated independently and which were independently associated with lung function. The integrative analysis enhanced single omics analysis and improved pathway recovery, capturing associations that may be missed based on a significance threshold when a single omics data type is analyzed. The downstream metabolites anchored the transcriptome,46 providing a functional readout of the changes taking place at a transcriptional level. Furthermore, we were able to identify an upstream SNP which may be driving these results. This makes these transcriptomic changes more biologically interpretable and provide mechanistic evidence to support the role of common asthma genes such as ORMDL3.
A major strength of this study included the innovative use of validated30 network methods to perform, for the first time, integrative omics in a large, well-characterized cohort of children with asthma, allowing the simultaneous exploration of multiple clinically relevant phenotypic characteristics of lung function. Blood is well suited for future clinical translation because mounting research suggests that plasma is an excellent resource for metabolic profiling in asthma, and it is known that gene expression patterns in peripheral blood show systematic changes when asthma exacerbations occur.47 However, it is difficult to determine how the mixture of different cell types in whole blood may affect gene expression.
There are some limitations to this study. Both known and unknown metabolites are required to build a complete metabolomic network. In this study, only 7% of metabolite features could be annotated to known metabolites. Subsequent pathway enrichment analysis was therefore limited; therefore, additional dysregulated metabolomic pathways within the modules could not be identified. Although there is likely some redundancy in the metabolite features, we consider the inclusion of all features necessary to generate the truest metabolic network possible. If only named metabolites were included in an unsupervised clustering approach, such as WGCNA, multiple connections would be missed, leading to metabolites not being assigned to their optimal module. Although nominal significance thresholds were used to consider module relationships, reflecting the exploratory nature of the analysis, the biologic plausibility of the reported results renders them worthy of further exploration.
The samples were not originally collected for metabolomic and transcriptomic profiling; therefore, there is the potential that some of the methods used may have impacted on the omic profiles and subsequently the results. However, most samples were processed within 4 h of collection; additionally, both the metabolomic and transcriptomic data underwent rigorous QC and processing, which aimed to reduce noise, identify outliers, and eliminate systematic bias. There was a lack of extreme phenotypes in this population; nevertheless, a variety of studies have demonstrated that omics profiling can capture phenotypic differences within patients with nonsevere asthma.10, 11, 19, 30, 47 Children from the Central Valley of Costa Rica represent a genetically homogenous population, which may limit generalizability; however, there is abundant evidence that the results of previous genetic analyses in this population can be replicated in outbred populations from different geographic locations.29, 48, 49 Crucially, some of the most intriguing findings of this study could be replicated in the independent CAMP cohort. The ability to replicate between studies remains an ongoing issue in metabolomics because of heterogeneity in approach, technology, and the fact that no one method is capable of capturing the complete metabolome. Accordingly, the current replication is limited by the fact that not all the relevant metabolites were available in CAMP and therefore should be interpreted with caution. Furthermore, the differing biologic media used for metabolomic profiling in the two studies may have dampened the ability to replicate.50 Further replication and functional validation is still necessary and should consider targeted profiling of the most interesting findings to obtain absolute metabolite quantification. Such work is necessary before potential clinical translation of these findings into biomarkers for asthma prognosis or endotyping can be considered.
In conclusion, this hypothesis-generating study demonstrates how integrating multiple omics technologies provides a more informative picture of asthmatic lung function biology than a single omics approach and suggests that network-based methods represent viable integrative strategies.
Acknowledgments
Author contributions: R. S. K. takes full responsibility for this manuscript and its contents. R. S. K. had full access to all the data in the study and final responsibility for the decision to submit for publication. R. S. K. assisted with the conception of the project, performed the statistical analysis, and wrote the initial draft of the manuscript. B. L. C. and J. A. L.-S. assisted with the conception of the project and the statistical analysis and edited the manuscript. K. Blighe, Y. V. V., D. C. C.-C., and M. J. M. assisted and advised on the statistical analysis and edited the manuscript. J. C. C. and S. T. W. conceptualized the initial Costa Rican study, assisted with the conception of this project, and edited the manuscript. C. B. C. and K. Bullock performed the analytical experiment, advised on the biochemistry, and reviewed the manuscript.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: J. A. L.-S. is a consultant to Metabolon, Inc. None declared (R. S. K., B. L. C., K. B., Y. V. V., D. C. C.-C., M. J. M., C. B. C., K. B., J. C. C., S. T. W.).
Role of sponsors: Funding sources played no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Other contributions: We thank the participants of the Genetic Epidemiology of Asthma in Costa Rica Study and all those involved in conducting the study.
Additional information: The e-Appendix, e-Figures, and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
FUNDING/SUPPORT: The Genetics of Asthma in Costa Rica Study was supported by the National Institutes of Health [Grants HL066289, HL04370]. The metabolomics work was supported by National Heart Lung and Blood Institute (NHLBI) [Grant 1R01HL123915-01]. J. C. C.’s contribution was supported by the National Institutes of Health [Grants HL117191, HL119952]. D. C. C-C.’s contribution was supported by the National Institutes of Health [Grant K01 HL127265].
Supplementary Data
References
- 1.Eder W., Ege M.J., von Mutius E. The asthma epidemic. N Engl J Med. 2006;355(21):2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 2.Boulet L.P., FitzGerald J.M., Reddel H.K. The revised 2014 GINA strategy report: opportunities for change. Curr Opin Pulm Med. 2015;21(1):1–7. doi: 10.1097/MCP.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 3.Bisgaard H., Jensen S.M., Bønnelykke K. Interaction between asthma and lung function growth in early life. Am J Respir Crit Care Med. 2012;185(11):1183–1189. doi: 10.1164/rccm.201110-1922OC. [DOI] [PubMed] [Google Scholar]
- 4.Martinez F.D., Morgan W.J., Wright A.L., Holberg C.J., Taussig L.M. Diminished lung function as a predisposing factor for wheezing respiratory illness in infants. N Engl J Med. 1988;319(17):1112–1117. doi: 10.1056/NEJM198810273191702. [DOI] [PubMed] [Google Scholar]
- 5.McGeachie M.J., Yates K.P., Zhou X. Patterns of growth and decline in lung function in persistent childhood asthma. N Engl J Med. 2016;374(19):1842–1852. doi: 10.1056/NEJMoa1513737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Håland G., Carlsen K.C., Sandvik L. Reduced lung function at birth and the risk of asthma at 10 years of age. N Engl J Med. 2006;355(16):1682–1689. doi: 10.1056/NEJMoa052885. [DOI] [PubMed] [Google Scholar]
- 7.Chung K.F., Wenzel S.E., Brozek J.L. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43(2):343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- 8.Belgrave D.C., Buchan I., Bishop C., Lowe L., Simpson A., Custovic A. Trajectories of lung function during childhood. Am J Respir Crit Care Med. 2014;189(9):1101–1109. doi: 10.1164/rccm.201309-1700OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renz H., Conrad M., Brand S., Teich R., Garn H., Pfefferle P.I. Allergic diseases, gene–environment interactions. Allergy. 2011;66:10–12. doi: 10.1111/j.1398-9995.2011.02622.x. [DOI] [PubMed] [Google Scholar]
- 10.McGeachie M.J., Dahlin A., Qiu W. The metabolomics of asthma control: a promising link between genetics and disease. Immun Inflamm Dis. 2015;3(3):224–238. doi: 10.1002/iid3.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh A., Yamamoto M., Kam S.H. Gene-metabolite expression in blood can discriminate allergen-induced isolated early from dual asthmatic responses. PLoS One. 2013;8(7):e67907. doi: 10.1371/journal.pone.0067907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly R.S., Dahlin A., McGeachie M.J. Asthma metabolomics and the potential for integrative omics in research and the clinic. Chest. 2017;151:262–277. doi: 10.1016/j.chest.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Celedon J.C., Soto-Quiros M.E., Avila L. Significant linkage to airway responsiveness on chromosome 12q24 in families of children with asthma in Costa Rica. Hum Genet. 2007;120(5):691–699. doi: 10.1007/s00439-006-0255-5. [DOI] [PubMed] [Google Scholar]
- 14.Asher M.I., Montefort S., Bjorksten B. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC phases one and three repeat multicountry cross-sectional surveys. Lancet. 2006;368(9537):733–743. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- 15.den Dekker H.T., Sonnenschein-van der Voort A.M., de Jongste J.C. Early growth characteristics and the risk of reduced lung function and asthma: a meta-analysis of 25,000 children. J Allergy Clin Immunol. 2016;137(4):1026–1035. doi: 10.1016/j.jaci.2015.08.050. [DOI] [PubMed] [Google Scholar]
- 16.Kelly R.S., Virkud Y., Giorgio R., Celedon J.C., Weiss S.T., Lasky-Su J. Metabolomic profiling of lung function in Costa-Rican children with asthma. Biochim Biophys Acta. 2017;1863(6):1590–1595. doi: 10.1016/j.bbadis.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunninghake G.M., Soto-Quiros M.E., Avila L. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol. 2007;119(3):654–661. doi: 10.1016/j.jaci.2006.12.609. [DOI] [PubMed] [Google Scholar]
- 18.Himes B.E., Hunninghake G.M., Baurley J.W. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84(5):581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croteau-Chonka D.C., Qiu W., Martinez F.D. Gene expression profiling in blood provides reproducible molecular insights into asthma control. Am J Respir Crit Care Med. 2017;195(2):179–188. doi: 10.1164/rccm.201601-0107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourgon R., Gentleman R., Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci U S A. 2010;107(21):9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly R.S., Croteau-Chonka D.C., Dahlin A. Integration of metabolomic and transcriptomic networks in pregnant women reveals biological pathways and predictive signatures associated with preeclampsia. Metabolomics. 2016;13(1):7. doi: 10.1007/s11306-016-1149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayers J.R., Wu C., Clish C.B. Elevated circulating branched chain amino acids are an early event in pancreatic adenocarcinoma development. Nat Med. 2014;20(10):1193–1198. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Townsend M.K., Bao Y., Poole E.M. Impact of pre-analytic blood sample collection factors on metabolomics. Cancer Epidemiol Biomarkers Prev. 2016;25(5):823–829. doi: 10.1158/1055-9965.EPI-15-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-559. 559-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reimand J., Arak T., Adler P. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44(W1):W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia J., Wishart D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6(6):743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 27.The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials. 1999;20(1):91–120. [PubMed] [Google Scholar]
- 28.Forno E., Lasky-Su J., Himes B. Genome-wide association study of the age of onset of childhood asthma. J Allergy Clin Immunol. 2012;130(1):83–90.e84. doi: 10.1016/j.jaci.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hersh C.P., Raby B.A., Soto-Quiros M.E. Comprehensive testing of positionally cloned asthma genes in two populations. Am J Respir Crit Care Med. 2007;176(9):849–857. doi: 10.1164/rccm.200704-592OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modena D.B., Bleecker D.E., Busse D.W. Gene expression correlated to severe asthma characteristics reveals heterogeneous mechanisms of severe disease. Am J Respir Crit Care Med. 2017;195(11):1449–1463. doi: 10.1164/rccm.201607-1407OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Muhsen S., Johnson J.R., Hamid Q. Remodeling in asthma. J Allergy Clin Immunol. 2011;128(3):451–462. doi: 10.1016/j.jaci.2011.04.047. [DOI] [PubMed] [Google Scholar]
- 32.Forno E., Weiner D.J., Mullen J. Obesity and airway dysanapsis in children with and without asthma. Am J Respir Crit Care Med. 2016;195(3):314–323. doi: 10.1164/rccm.201605-1039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pietrusinska M., Pajak A., Gorski P. Preliminary studies: differences in microRNA expression in asthma and chronic obstructive pulmonary disease. Postepy Dermatol Alergol. 2016;33(4):276–280. doi: 10.5114/ada.2016.61603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kho A.T., Sharma S., Davis J.S. Circulating MicroRNAs: association with lung function in asthma. PLoS One. 2016;11(6):e0157998. doi: 10.1371/journal.pone.0157998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu G., Yang G., Zhang R. Altered microRNA expression profiles of extracellular vesicles in nasal mucus from patients with allergic rhinitis. Allergy Asthma Immunol Res. 2015;7(5):449–457. doi: 10.4168/aair.2015.7.5.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ha T.Y. The role of MicroRNAs in regulatory T cells and in the immune response. Immune Network. 2011;11(1):11–41. doi: 10.4110/in.2011.11.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dennis E.A., Norris P.C. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15(8):511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calder P.C. Omega-3 polyunsaturated fatty acids and inflammatory processes: nutrition or pharmacology? Br J Clin Pharmacol. 2013;75(3):645–662. doi: 10.1111/j.1365-2125.2012.04374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serhan C.N. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510(7503):92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levy B.D. Sphingolipids and susceptibility to asthma. N Engl J Med. 2013;369(10):976–978. doi: 10.1056/NEJMcibr1306864. [DOI] [PubMed] [Google Scholar]
- 41.Paulenda T., Draber P. The role of ORMDL proteins, guardians of cellular sphingolipids, in asthma. Allergy. 2016;71(7):918–930. doi: 10.1111/all.12877. [DOI] [PubMed] [Google Scholar]
- 42.Worgall T.S., Veerappan A., Sung B. Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity. Sci Transl Med. 2013;5(186):186ra167. doi: 10.1126/scitranslmed.3005765. [DOI] [PubMed] [Google Scholar]
- 43.Toncheva A.A., Potaczek D.P., Schedel M. Childhood asthma is associated with mutations and gene expression differences of ORMDL genes that can interact. Allergy. 2015;70(10):1288–1299. doi: 10.1111/all.12652. [DOI] [PubMed] [Google Scholar]
- 44.Himes B.E., Jiang X., Wagner P. RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PLoS One. 2014;9(6):e99625. doi: 10.1371/journal.pone.0099625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucki N.C., Sewer M.B. The interplay between bioactive sphingolipids and steroid hormones. Steroids. 2010;75(6):390–399. doi: 10.1016/j.steroids.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cavill R., Jennen D., Kleinjans J., Briedé J.J. Transcriptomic and metabolomic data integration. Brief Bioinform. 2016;17(5):891–901. doi: 10.1093/bib/bbv090. [DOI] [PubMed] [Google Scholar]
- 47.Bjornsdottir U.S., Holgate S.T., Reddy P.S. Pathways activated during human asthma exacerbation as revealed by gene expression patterns in blood. PloS One. 2011;6(7):e21902. doi: 10.1371/journal.pone.0021902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torgerson D.G., Ampleford E.J., Chiu G.Y. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du R., Litonjua A.A., Tantisira K.G. Genome-wide association study reveals class I MHC-restricted T cell-associated molecule gene (CRTAM) variants interact with vitamin D levels to affect asthma exacerbations. J Allergy Clin Immunol. 2012;129(2):368–373. doi: 10.1016/j.jaci.2011.09.034. 373.e361-e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Z., Kastenmüller G., He Y. Differences between human plasma and serum metabolite profiles. PLoS One. 2011;6(7):e21230. doi: 10.1371/journal.pone.0021230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.