Abstract
The cell nucleus encloses, organizes, and protects the genome. Chromatin maintains nuclear mechanical stability and shape in coordination with lamins and the cytoskeleton. Abnormal nuclear shape is a diagnostic marker for human diseases, and it can cause nuclear dysfunction. Chromatin mechanics underlies this link, as alterations to chromatin and its physical properties can disrupt or rescue nuclear shape. The cell can regulate nuclear shape through mechanotransduction pathways that sense and respond to extracellular cues, thus modulating chromatin compaction and rigidity. These findings reveal how chromatin’s physical properties can regulate cellular function and drive abnormal nuclear morphology and dysfunction in disease.
Introduction
The cell nucleus is a mechanically responsive organelle that protects and organizes the genome it encloses. Forces can directly dictate gene transcription through the physical positioning [1,2] and stretching of genes [3], as well as through force transduction that alters nuclear import and export of transcription factors [4,5]. Moreover, the nucleus must resist forces from within the cell and its microenvironment to prevent catastrophic events, such as nuclear rupture and deformations, which result in mixing nuclear and cytoplasmic contents, DNA damage, and disrupted transcription [6–8]. An essential component of this mechanical response is chromatin, i.e., the genome and associated proteins, which behaves like a spring, elastically resisting few-micron-sized deformations [9–13]. With the advent of chromosome capture and various imaging techniques, there have been numerous studies of chromatin’s spatial organization [14,15]. However, less is known about how the mechanical properties of chromatin dictate spatial organization and what role forces play in governing it, as well as the shape and stability of the nucleus as a whole. Recent research on nuclear mechanics and morphology has provided insights into the basic biophysical mechanisms underlying nuclear architecture, and consequently, the ways in which chromatin’s physical properties affect nuclear function and human diseases.
Physiological impact of defects in nuclear shape and mechanics
Abnormal nuclear shape is a pathognomonic trait that has been used as a diagnostic indicator of human disease for nearly a century, albeit without an understanding of the mechanisms of nucleus deformations and their effects on function. Cancer diagnostic assays examine cell nuclei for unusual sizes and shapes via Pap smear (cervical) [16], nuclear herniations termed “blebs” that correlate with Gleason score (prostate) [17], and aberrant shapes and orientations via quantitative histomorphometrics (breast) [18,19]. Aberrantly shaped nuclei also occur in mechanically demanding environments, such as muscle cells in heart disease (cardiomyopathy) associated with progeria and aging and muscular dystrophy [20]. The prevalence of abnormal nuclear morphology in cancers and other diseases suggests that nuclear shape is normally regulated by the cell. Furthermore, because shape stability of an object depends on its mechanical properties, these observations suggest that nuclear shape regulation and diseases in which abnormal nuclear shapes occur are linked to the mechanics of the nucleus. Indeed, nuclear mechanics play a significant role in malignant cancer cells that migrate and invade tissue; nuclear deformation is the rate-limiting step in migration [21–23], and deficient nuclear rigidity leads to ruptures and DNA damage upon migration through confined volumes [8,24,25].
Abnormal nuclear morphology disrupts nuclear function in several ways. Nuclei that are mechanically perturbed by altered lamins or chromatin are prone to rupture, which induces exchange of nuclear and cytosolic contents (including chromosomes) [6,26–32]. Loss of nuclear compartmentalization can lead to DNA damage [24,25,33], at least in part due to concurrent mislocalization of chromatin and DNA repair factors [8,34]. It has furthermore been hypothesized that nuclear rupture could activate the cGAS-STING innate immune response pathway leading to inflammation, senescence, and cancer (reviewed in [35]). Morphological disruption is also associated with changes in the overall spatial organization of chromatin and gene expression profile [28,36]. Nuclear blebs arising with ruptures correlate with decreased transcription and mRNA transport for the chromatin held within [7,37]. Abnormal shape and mechanics could also disrupt mechanotransduction, which would result in further perturbations to transcription. Together, these findings demonstrate the cell-biological importance of the physical properties of the cell nucleus – perturbations of which can lead to nuclear dysfunction – and highlight the role of altered nuclear mechanics as a factor underlying human disease.
Mechanical components dictating nuclear shape: chromatin, lamins, and the cytoskeleton
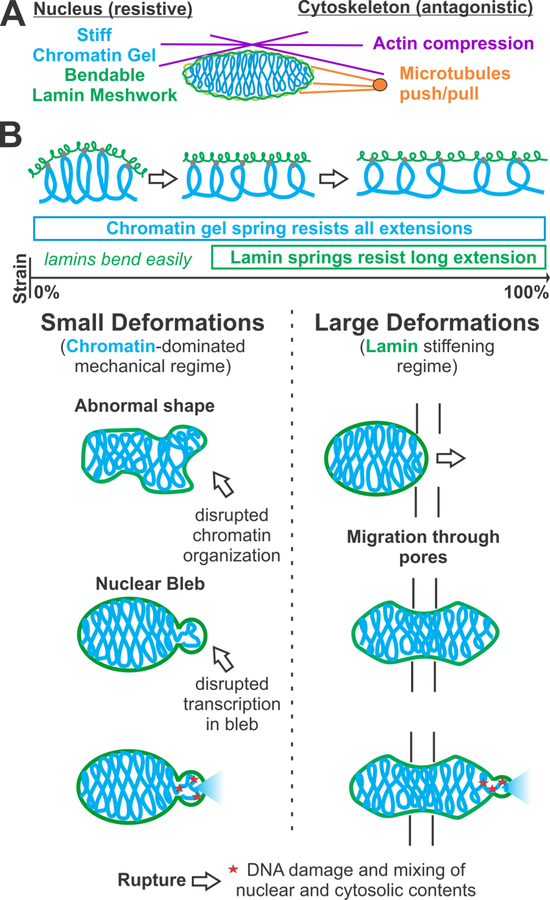
The ability of the nucleus to maintain its shape is dictated by a trio of mechanical components: chromatin, lamins, and the cytoskeleton (Figure 1A). Chromatin and lamin A are the two major resistive elements that protect the nucleus, as shown by atomic force microscopy [10,11,38], micropipette aspiration [39,40], substrate stretching [41], and micromanipulation [12,13]. However, experiments with nuclei treated with the chromatin-degrading enzyme MNase [13,42] show that lamins alone cannot maintain nuclear shape. Instead, the lamina buckles under mechanical stress when it is unsupported by chromatin. This is consistent with experiments revealing that lamins comprise a thin (10–30 nm) peripheral meshwork of highly bendable intermediate filaments with a short (few hundred nm) persistence length [43–45]. In turn, lamin networks can unfold during cell and nuclear spreading [46], but resist large stretching deformations [13,42,47]. Chromatin is a variably compacted polymer filling the nucleus [48] that interacts both with both itself (e.g., compartments and topologically associating domains, TADs seen in Hi-C [14]) and the nuclear periphery (e.g., lamin associated domains, LADs [49]). Variations in compaction may correspond to variations in chromatin stiffness [13,50] and viscoelasticity [40,51–53]. Moreover, a number of mechanical measurements demonstrate that chromatin is a stiff mechanical element [9,10,12,13,54,55]. Altogether, these data suggest a physical model of the nucleus as a semiflexible meshwork of lamins at the nuclear periphery that encloses a stiff polymeric chromatin gel [56] (Figure 1A).
Figure 1. Chromatin is a major contributor to nuclear mechanics and shape along with lamins and the cytoskeleton.
(A) The major protective mechanical components of the nucleus that aid nuclear shape stability are chromatin (blue), which is a stiff polymer gel, and lamins (green), which are an intermixed meshwork of easily bendable intermediate filaments of lamin A, B1, B2, and C. The cytoskeleton components actin (purple) and microtubules (orange) antagonize nuclear shape stability, although actin and vimentin (not shown) can also aid stability. (B) Top: Schematic showing differential force response regimes arising due to geometric considerations for lamins (a 2D meshwork) and chromatin (a 3D gel). For short, few-micron deformations (i.e., small strains), the chromatin gel acts as a spring that resists stretching, while lamins contribute little as they bend easily until they are aligned with the tension axis. Longer deformations, for which the lamins are aligned with the force, generate lamin-A-based strain stiffening. Chromatin continues to resist stretching of the nucleus at long deformations as a secondary component. Bottom: Abnormal nuclear shape and blebs are small deformations, occurring in the regime dominated by chromatin. During migration through pores, the nucleus extends many microns (>3 µm) into a deformation regime that necessitates and activates lamin A resistance to maintain shape stability. Failure to maintain shape stability in either condition can result in disruption of transcription through chromatin disorganization or blebs that inhibit it. Either shape disruption can result in nuclear ruptures that lead to DNA damage and loss of nuclear compartmentalization.
Chromatin and lamins have distinct contributions to nuclear mechanics, as detailed by micromanipulation force measurements [13]. Chromatin acts as an elastic spring that dominates the force response to small deformations (few µm), while the lamin A meshwork deforms easily for small extensions and stiffens to resist large deformations (Figure 1B) [13,42]. These two regimes reflect the geometry of the cell nucleus [42]. Differential force response is biologically important because almost all cell nuclei undergo small deformations which are resisted by chromatin, while generally only nuclei in mechanically demanding tissues and microenvironments have high lamin A levels [5]. Overall, the interdependence of chromatin and lamin organization, as well as extranuclear factors, makes understanding nuclear morphology and mechanics a rich and complex physical biology problem.
The cytoskeleton is external to the nucleus, but it is nonetheless an important contributor to nuclear shape maintenance that can both antagonize and promote nuclear stability. In its antagonistic role, actin-based confinement or compression lead to abnormal shape and rupture of perturbed nuclei or models of diseased nuclei [27,28,30,31,57]. Similarly, microtubules, along with dynein motors, exert forces that can deform or rupture the nucleus [33,55,58–60]. In contrast, the intermediate filament vimentin has a protective role essential for nuclear positional stability [61] and for perinuclear stiffness, which impedes 3D motility [62]. Additionally, actin cap cables can stabilize nuclear shape [63] in coordination with microtubules [64]. Altogether, the balance of the trio of lamins, chromatin, and the cytoskeleton tightly regulates nuclear morphology, and the perturbation of any one element can lead to global nuclear shape dysfunction.
Lamin perturbations impact nuclear shape via chromatin
Lamin perturbations are well known to induce abnormal nuclear morphology, but the physical basis for this is not well understood. Progerin, a misprocessed, permanently farnesylated lamin A mutant, causes the premature aging disease progeria and disrupts nuclear morphology [65]. Further studies show that depletion of either lamin A or lamin B results in similar abnormal morphologies and blebbing of cell nuclei [27,32,41]. While all of these alterations of lamins perturb nuclear morphology, each has a different effect on lamin-based nuclear mechanics. Lamin A mutant progerin increases nuclear stiffness, while lamin A depletion decreases stiffness, and lamin B depletion leads to either no change or increased rigidity, depending on lamin A content [5,13,41,66]. Moreover, lamins provide little contribution to the mechanical response to deformations on the small length scales of blebs and typical nuclear deformations [13,56]. Instead, lamin A is more important in cells migrating through pores where the nucleus is highly strained and compressed [22,24,25] (Figure 1B). These observations challenge the idea that blebs and other nuclear shape aberrations arise from the biophysical properties of lamins alone.
Lamins A, B, and C are interconnected with chromatin, which can cause downstream effects that impact the biophysical properties of the cell nucleus. Disruption of proteins that tether chromatin to the nuclear envelope results in abnormalities in nuclear fluctuations, overall shape, and chromatin organization [55,67–71]. Depletion of lamin A can disrupt lamin associated domains (LADs) of chromatin and change chromatin’s spatial localization [72–75]. Mutant lamin A progerin causes disruption of chromatin connections to the lamina and induces decreased heterochromatin and chromatin softening [7,76–79]. Depletion of lamin B1 also decreases chromatin connections to the periphery [75] and decreases heterochromatin [30,80]. It is significant that each of these downstream chromatin perturbations – loss of connections to the periphery, decondensation, and decreased heterochromatin content – can independently induce abnormal nuclear morphology (as discussed below). Moreover, in many of these lamin-perturbed cases, normal nuclear morphologies can be rescued by restoring heterochromatin to normal levels [30,81]. This suggests that heterochromatin mechanics may dominate the regulation of nuclear shape, and that lamin defects may induce blebs indirectly through their downstream effects on chromatin and its anchoring to the nuclear periphery.
Chromatin is a key regulator of nuclear shape
Various chromatin perturbations induce blebbing, ruptures, and other abnormal nuclear morphologies, independent of altering lamins. Histone perturbations and modifications are a major class of such chromatin alterations. Overexpression of HMGN5 disrupts histone linker H1, which results in decreased chromatin compaction, nuclear stability, and rigidity, and consequently, increased nuclear blebbing [54]. HMGN5 overexpression in transgenic mice leads to cardiac defects and premature death, demonstrating the physiological importance of chromatin-based nuclear mechanics and morphology. Alterations in the amount of compact heterochromatin and decompact euchromatin are also commonly found in cancers and other diseases [82]. Chromatin histone modifications that broadly increase euchromatin or decrease heterochromatin result in weakened chromatin-based nuclear rigidity, abnormal nuclear morphology, and nuclear ruptures [30]. Increases in heterochromatin, in contrast, can rescue nuclear shape and rigidity. Rescue by heterochromatin has been demonstrated for both chromatin and lamin perturbations, including in cells with excess histone acetylation, lamin B1 depletion, or mutant lamin A progerin overexpression, as well as in Hutchinson-Gilford progeria syndrome patient cells [30,81]. Similarly, chromatin condensation coincides with bleb healing and reabsorption into the nuclear body [29]. Additionally, loss of heterochromatin at the nuclear periphery, a common occurrence in lamin-perturbed nuclei, by depletion or mutation of heterochromatin-lamin tethering protein PRR14 causes abnormal nuclear morphology [71]. Regulators of histone modifications, such as WDR5, which promotes the euchromatin mark H3K4me3, can also regulate nuclear deformability, independent of transcription [83]. Thus, changes to histone modification state of chromatin through different molecular mechanisms are sufficient to disrupt or restore nuclear morphology without requiring lamin alterations (Table 1).
Table 1. Summary of comparison of lamin- and chromatin-based perturbations that change nuclear morphology.
Lamin perturbations that disturb nuclear shape, but can either strengthen, weaken, or not change the nucleus. The commonality is that they disrupt chromatin in some fashion. Increasing stiffness by heterochromatin formation stabilizes nuclear shape in lamin perturbations. Chromatin perturbations that soften the nucleus destabilize nuclear shape without the need to alter lamins.
| Lamin perturbations that result in abnormal nuclear morphology | ||||
|---|---|---|---|---|
| Type | Mechanics | Effect on chromatin | Shape rescue via chromatin | |
| Lamin A depletion | weaker ~50% | decreased chromatin-lamina attachments [72–75] | (not tested) | |
| Progerin, mutant lamin A | stiffer 100% | decreased chromatin-lamina attachments and decreased heterochromatin [7,76–79]; softer chromatin by 50% [76] | increased heterochromatin [30,81] | |
| Lamin B depletion | no change or stiffer (lamin A amount dependent) | decreased chromatin-lamina attachments [75]; decreased heterochromatin [30,80] | Increased heterochromatin [30,81] | |
|
Chromatin perturbations that result in abnormal nuclear morphology | ||||
| Type | Mechanics | Chromatin perturbation | Lamins | Citations |
| HMGN5 overexpression | weaker ~40% | H1 (linker histone 1) disruption, chromatin decondensation | no change | (Furusawa et al., 2015) [54] |
| broad histone acetylation | 35% weaker chromatin; lamin A stiffening unchanged | increased euchromatin (H3K9ac, H4K5ac) | no change | (Stephens et al., 2018a) [30] |
| broad histone demethylation | 35% weaker chromatin; lamin A stiffening unchanged | decreased heterochromatin (H3K9me2,3, H3K27me3) | no change | (Stephens et al., 2018a) [30] |
| WDR5/RbBP5 | correlated to weaker ~60% | increased euchromatin H3K4me3, activated in 3D, but not 2D, culture | no change | (Wang et al., 2018) [83] |
| PRR14 depletion | (not tested) | chromatin/HP1-lamin A tethering protein, loss of heterochromatin at periphery | no change | (Poleshko et al., 2013) [71] |
| BRG1 depletion | (not tested) | SWI/SNF ATPase chromatin-remodeler | lamin grooves | (Imbalzano et al., 2013) [85] |
| p53 depletion | (not tested) | chromatin protein associated with cancer | no change | (Yang et al., 2017) [84] |
| Rb depletion | (not tested) | chromatin protein associated with cancer | no change | (Yang et al., 2017) [84] |
| p63 depletion | (not tested) | transcription factor, decreased heterochromatin & HP1 | decreased lamin expr. | (Rapisarda et al., 2013) [86] |
| miR-29b blockade | (not tested) | cancer-relevant microRNA that modulates DNA methylation | - | (Kriegel et al., 2018) [87] |
| NOP53 depletion | (not tested) | p53 interacting protein | - | (Lee et al., 2018) [88] |
| BAF depletion | weaker, in vitro | depletes chromatin bridging during nuclear formation | no change reported | (Samwer et al., 2017) [89] |
Aside from cases with altered histones, nuclear rupture has also been reported for RPE-1 cells depleted of well-known cancer-related chromatin proteins Rb and p53, due to enlarged nuclei [84]. Depletion of the SWI/SNF chromatin-remodeling ATPase BRG1 causes abnormal nuclear morphology in breast epithelia MCF10A cells, due to BRG1-related changes in internal nuclear forces [85]. This mimics the abnormal nuclear shape seen in more metastatic breast cancers [18]. Depletion of other cancer-relevant molecules, particularly p63 [86], miR29-b [87], and NOP53 [88], can also induce abnormal nuclear shape, although these perturbations may have effects beyond those on chromatin. Even prior to nuclear formation, chromatin mechanics regulates shape; barrier-to-autointegration factor (BAF) stiffens chromatin by bridging chromatin sites, which inhibits the formation of micronuclei [89]. It remains to be determined whether the other diverse types of chromatin-chromatin bridging factors, such as loop-forming cohesin [90,91] and phase-separating HP1α [92,93], help stabilize chromatin-based nuclear mechanical response. These studies (summarized in Table 1) reveal that biophysical properties of chromatin – spatial organization, structure, and rigidity – are paramount in maintaining normal nuclear shape and function throughout the cell cycle.
Native pathways for regulating nuclear morphology through chromatin
Recent experiments have begun to reveal how chromatin is natively regulated to maintain nuclear morphology. While there are several known mechanisms for regulation of the cytoskeleton and the lamina that modulate nuclear shape [46,69,94–97], these alterations are often upstream of chromatin modifications. Recent studies have demonstrated that cells can also directly regulate chromatin compaction through a native pathway. External mechanical stimuli trigger mechanotransduction through mechanosensitive ion channels in the plasma membrane [98–100], leading to chromatin condensation and heterochromatin formation [81,94,101,102]. Mechanotransduction via mechanosensitive ion channels can increase heterochromatin levels and chromatin-based nuclear rigidity, while concurrently rescuing nuclear shape in lamin-perturbed, chromatin-pertubed, and disease model cells (e.g., progeria and breast cancer) [81]. This provides a native chromatin regulation pathway for the cell to sense and respond to the extracellular environment in order to protect nuclear shape and organization.
Similarly, chromatin condensation and compaction can be regulated through extracellular osmotic changes [103,104], compression [105], substrate micropatterning [97,106], cell substrate stretching [94,101,102,107], changes in charge composition [81], and cell migration [36,108,109]. Notably, heterochromatin levels increase during cell migration, and migration through pores can be blocked by the concurrent increase in nuclear rigidity [110]. Similar to nuclear rigidity changes due to lamin A increases [22], this increase in chromatin-based rigidity could protect nuclei from ruptures that occur when cells migrate through narrow pores [24,25] or block migration through pores entirely, thus stopping cancerous invasion [21,83,110]. Altogether, cells possess a variety of native mechanisms for regulating nuclear shape through rigidity linked to the underlying chromatin compaction state.
Biophysical modeling of nuclear shape
Despite knowing many of the biological factors involved in nuclear morphology, the biophysical mechanisms by which chromatin, lamins, and the cytoskeleton regulate nuclear morphology remain unclear. Physical modeling may fill the gaps in our mechanistic understanding of how these three components cooperate and compete to maintain or disrupt nuclear morphology. Several existing models for nuclear shape and rupture focus exclusively on the role of lamins [111,112], consider only osmotic pressure associated with the nucleoplasm [113], or treat chromatin as a viscoelastic material that is secondary to the lamina [114]. Other studies incorporate more robust models for chromatin but primarily apply to specific experimental conditions [42,115] or do not extensively explore the physical role of chromatin and its linkages to the lamina [115]. Most of these models omit the cytoskeleton. Each of these models illustrates how the mechanical properties of different cellular components may regulate nuclear morphology, yet altogether, they provide an incomplete mechanistic picture. Figure 1 depicts key concepts that could be elucidated by further modeling efforts. Such efforts are likely to both inform and be informed by continuing experiments in this developing field.
Conclusion
The tight interplay between chromatin’s genetic regulation, compaction, spatial organization, and mechanics controls nuclear function. Chromatin is a stiff polymer gel that fills the nucleus, providing the nucleus with a robust mechanical response complementing the strain stiffening of the nuclear lamina. These two nuclear components, along with the cytoskeleton, shape the nucleus and the genome within. An imbalance between these three components can induce abnormal nuclear shape, which can disrupt chromatin organization and transcription, cause nuclear rupture, and increase DNA damage. We have emphasized that chromatin-based mechanics is an underlying mechanism of abnormal nuclear morphology. Furthermore, emerging data reveal that extracellular stimuli sensed by the cell can regulate chromatin mechanics, and thus, shape through modulating histone modification state. Chromatin itself, through its structure and mechanics, is emerging as key factor that determines normal nuclear function, as well as dysfunction in a variety of disease contexts.
Many intriguing questions remain regarding the connections between chromatin organization, nuclear shape, and nuclear function. Are there specific histone modifications and chromatin remodeling factors that are particularly important in governing nuclear shape? Can we develop a detailed understanding of how shape impacts chromatin organization, e.g., via Hi-C experiments? There are also many interesting questions about function, such as how do chromatin modifications that control nuclear shape affect transcription? And how do we separate the effects chromatin modifications have on mechanics, organization, biochemistry, and transcription from each other? As we have begun to see for cell migration and DNA damage response, addressing these questions will provide insight into broader questions of how chromatin organization and nuclear shape impact cellular functions, e.g., the cell cycle, development, homeostasis, and tissue self-organization. Study of nuclear shape and mechanoregulation may reveal new therapeutic targets across a range of diseases spanning cancers, dystrophies, progerias, aging, and more.
Acknowledgements
A.D.S. is supported by NIH grant K99GM123195. This work was also supported by NIH grants GM105847, CA193419 (PS-ON), and by subcontract, DK107980 (4DN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brickner JH, Walter P: Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol 2004, 2:e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddy KL, Zullo JM, Bertolino E, Singh H: Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 2008, 452:243–247. [DOI] [PubMed] [Google Scholar]
- 3.Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, Wang N: Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater 2016, 15:1287–1296. *Stress applied to the cell surface with a magnetic bead propagated force into the nucleus and stretched chromatin, causing an increase in transcription of stretched genes. Thus, external stresses can directly regulate gene expression.
- 4.Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J: Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature 2013, 497:507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. : Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 2013, 341:1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EM, Verstraeten VL, van Steensel MA, Marcelis CL, et al. : Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum Mol Genet 2011, 20:4175–4186. [DOI] [PubMed] [Google Scholar]
- 7.Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, et al. : The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev 2008, 22:3409–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia Y, Ivanovska IL, Zhu K, Smith L, Irianto J, Pfeifer CR, Alvey CM, Ji J, Liu D, Cho S, et al. : Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J Cell Biol 2018. **Experiments with blunt and sharp AFM tips show that nuclear rupture occurs at sites of high mechanical stress, which results in DNA damage. This DNA damage is a result of loss of nuclear DNA repair factors to the cytoplasm. Such high stresses can occur during migration through pores and nuclear blebbing, which can occur during cancerous invasion or other disease processes.
- 9.Chalut KJ, Hopfler M, Lautenschlager F, Boyde L, Chan CJ, Ekpenyong A, Martinez-Arias A, Guck J: Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells. Biophys J 2012, 103:2060–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haase K, Macadangdang JK, Edrington CH, Cuerrier CM, Hadjiantoniou S, Harden JL, Skerjanc IS, Pelling AE: Extracellular Forces Cause the Nucleus to Deform in a Highly Controlled Anisotropic Manner. Sci Rep 2016, 6:21300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krause M, Te Riet J, Wolf K: Probing the compressibility of tumor cell nuclei by combined atomic force-confocal microscopy. Phys Biol 2013, 10:065002. [DOI] [PubMed] [Google Scholar]
- 12.Shimamoto Y, Tamura S, Masumoto H, Maeshima K: Nucleosome-nucleosome interactions via histone tails and linker DNA regulate nuclear rigidity. Mol Biol Cell 2017, 28:1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF: Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell 2017, 28:1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker J, Mirny L: The 3D Genome as Moderator of Chromosomal Communication. Cell 2016, 164:1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser J, Williamson I, Bickmore WA, Dostie J: An Overview of Genome Organization and How We Got There: from FISH to Hi-C. Microbiol Mol Biol Rev 2015, 79:347–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papanicolaou GN, Traut HF: The diagnostic value of vaginal smears in carcinoma of the uterus. 1941. Arch Pathol Lab Med 1997, 121:211–224. [PubMed] [Google Scholar]
- 17.Helfand BT, Wang Y, Pfleghaar K, Shimi T, Taimen P, Shumaker DK: Chromosomal regions associated with prostate cancer risk localize to lamin B-deficient microdomains and exhibit reduced gene transcription. J Pathol 2012, 226:735–745. [DOI] [PubMed] [Google Scholar]
- 18.Lu C, Romo-Bucheli D, Wang X, Janowczyk A, Ganesan S, Gilmore H, Rimm D, Madabhushi A: Nuclear shape and orientation features from H&E images predict survival in early-stage estrogen receptor-positive breast cancers. Lab Invest 2018. [DOI] [PMC free article] [PubMed]
- 19.Radhakrishnan A, Damodaran K, Soylemezoglu AC, Uhler C, Shivashankar GV: Machine Learning for Nuclear Mechano-Morphometric Biomarkers in Cancer Diagnosis. Sci Rep 2017, 7:17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butin-Israeli V, Adam SA, Goldman AE, Goldman RD: Nuclear lamin functions and disease. Trends Genet 2012, 28:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson PM, Denais C, Bakshi MC, Lammerding J: Nuclear deformability constitutes a rate-limiting step during cell migration in 3-D environments. Cell Mol Bioeng 2014, 7:293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PC, Ivanovska IL, Discher DE: Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol 2014, 204:669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P: Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol 2013, 201:1069–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J: Nuclear envelope rupture and repair during cancer cell migration. Science 2016, 352:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Dumenil AM, Manel N, et al. : ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 2016, 352:359–362. [DOI] [PubMed] [Google Scholar]
- 26.Chen NY, Kim P, Weston TA, Edillo L, Tu Y, Fong LG, Young SG: Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. Proc Natl Acad Sci U S A 2018, 115:10100–10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatch EM, Hetzer MW: Nuclear envelope rupture is induced by actin-based nucleus confinement. J Cell Biol 2016, 215:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Berre M, Aubertin J, Piel M: Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr Biol (Camb) 2012, 4:1406–1414. [DOI] [PubMed] [Google Scholar]
- 29.Robijns J, Molenberghs F, Sieprath T, Corne TD, Verschuuren M, De Vos WH: In silico synchronization reveals regulators of nuclear ruptures in lamin A/C deficient model cells. Sci Rep 2016, 6:30325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens AD, Liu PZ, Banigan EJ, Almassalha LM, Backman V, Adam SA, Goldman RD, Marko JF: Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol Biol Cell 2018, 29:220–233. **Altering chromatin-based nuclear stiffness via histone modification state is sufficient to induce abnormal nuclear morphology in wild type cells and rescue normal shape in lamin mutants that mimic aging (mutant lamin A progerin) and senescence (lamin B1 depletion). This shows that chromatin is a dominant component determining nuclear shape.
- 31.Tamiello C, Kamps MA, van den Wijngaard A, Verstraeten VL, Baaijens FP, Broers JL, Bouten CC: Soft substrates normalize nuclear morphology and prevent nuclear rupture in fibroblasts from a laminopathy patient with compound heterozygous LMNA mutations. Nucleus 2013, 4:61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vargas JD, Hatch EM, Anderson DJ, Hetzer MW: Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus 2012, 3:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Earle AJ, Kirby TJ, Fedorchak GR, Isermann P, Patel J, Iruvanti S, Bonne G, Wallrath LL, Lammerding J: Mutant lamins cause mechanically-induced nuclear envelope rupture, DNA damage, and DNA-PK activation in muscle. bioRxiv 2018. [DOI] [PMC free article] [PubMed]
- 34.Pfeifer CR, Xia Y, Zhu K, Liu D, Irianto J, Garcia VMM, Millan LMS, Niese B, Harding S, Deviri D, et al. : Constricted migration increases DNA damage and independently represses cell cycle. Mol Biol Cell 2018, 29:1948–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, Chen ZJ: The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med 2018, 215:1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobson EC, Perry JK, Long DS, Olins AL, Olins DE, Wright BE, Vickers MH, O’Sullivan JM: Migration through a small pore disrupts inactive chromatin organization in neutrophil-like cells. BMC Biology 2018, 16:142. **Neutrophils undergoing confined migration lose heterochromatic Hi-C contacts, while active euchromatic contacts are comparatively conserved. This suggests that heterochromatin absorbs the mechanical stresses during migration. This study also details how the nucleus preserves or alters the gene expression profile when the genome undergoes physical stress.
- 37.Bercht Pfleghaar K, Taimen P, Butin-Israeli V, Shimi T, Langer-Freitag S, Markaki Y, Goldman AE, Wehnert M, Goldman RD: Gene-rich chromosomal regions are preferentially localized in the lamin B deficient nuclear blebs of atypical progeria cells. Nucleus 2015, 6:66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schape J, Prausse S, Radmacher M, Stick R: Influence of lamin A on the mechanical properties of amphibian oocyte nuclei measured by atomic force microscopy. Biophys J 2009, 96:4319–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dahl KN, Kahn SM, Wilson KL, Discher DE: The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci 2004, 117:4779–4786. [DOI] [PubMed] [Google Scholar]
- 40.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE: Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci U S A 2007, 104:15619–15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT: Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem 2006, 281:25768–25780. [DOI] [PubMed] [Google Scholar]
- 42.Banigan EJ, Stephens AD, Marko JF: Mechanics and Buckling of Biopolymeric Shells and Cell Nuclei. Biophys J 2017, 113:1654–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Forster F, Hyman AA, Plitzko JM, Baumeister W: Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 2016, 351:969–972. [DOI] [PubMed] [Google Scholar]
- 44.Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD: Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell 2015, 26:4075–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD, Medalia O: The molecular architecture of lamins in somatic cells. Nature 2017, 543:261–264. **Cryo-EM studies of the lamina show that lamins A, B1, B2, and C comprise distinct but intertwined meshworks of lamin intermediate filaments. These filaments are thin (several nm) and flexible with a short persistence length of order of 100’s of nm.
- 46.Li Y, Lovett D, Zhang Q, Neelam S, Kuchibhotla RA, Zhu R, Gundersen GG, Lele TP, Dickinson RB: Moving Cell Boundaries Drive Nuclear Shaping during Cell Spreading. Biophys J 2015, 109:670–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panorchan P, Schafer BW, Wirtz D, Tseng Y: Nuclear envelope breakdown requires overcoming the mechanical integrity of the nuclear lamina. J Biol Chem 2004, 279:43462–43467. [DOI] [PubMed] [Google Scholar]
- 48.Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC: ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 2017, 357. [DOI] [PMC free article] [PubMed]
- 49.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. : Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453:948–951. [DOI] [PubMed] [Google Scholar]
- 50.Ghosh S, Seelbinder B, Henderson J, Watts R, Veress A, NEU CP: Deformation microscopy for dynamic intracellular and intranuclear mapping of mechanics with high spatiotemporal resolution. bioRxiv 2018:403261. [DOI] [PMC free article] [PubMed]
- 51.Dahl KN, Engler AJ, Pajerowski JD, Discher DE: Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys J 2005, 89:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Vries AH, Krenn BE, van Driel R, Subramaniam V, Kanger JS: Direct observation of nanomechanical properties of chromatin in living cells. Nano Lett 2007, 7:1424–1427. [DOI] [PubMed] [Google Scholar]
- 53.Guilak F, Tedrow JR, Burgkart R: Viscoelastic properties of the cell nucleus. Biochem Biophys Res Commun 2000, 269:781–786. [DOI] [PubMed] [Google Scholar]
- 54.Furusawa T, Rochman M, Taher L, Dimitriadis EK, Nagashima K, Anderson S, Bustin M: Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat Commun 2015, 6:6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schreiner SM, Koo PK, Zhao Y, Mochrie SG, King MC: The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun 2015, 6:7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephens AD, Banigan EJ, Marko JF: Separate roles for chromatin and lamins in nuclear mechanics. Nucleus 2017:1–6. [DOI] [PMC free article] [PubMed]
- 57.Tocco VJ, Li Y, Christopher KG, Matthews JH, Aggarwal V, Paschall L, Luesch H, Licht JD, Dickinson RB, Lele TP: The nucleus is irreversibly shaped by motion of cell boundaries in cancer and non-cancer cells. J Cell Physiol 2017. **The nucleus requires the actin cytoskeleton to induce abnormal nuclear morphology in models of breast cancer cells, but it does not elastically rebound shape when actin stress fibers are severed. This suggests that mechanical stresses can permanently reorganize nuclear components in these cells.
- 58.Chu FY, Haley SC, Zidovska A: On the origin of shape fluctuations of the cell nucleus. Proc Natl Acad Sci U S A 2017, 114:10338–10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP: Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science 2014, 344:527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Penfield L, Wysolmerski B, Mauro M, Farhadifar R, Martinez MA, Biggs R, Wu HY, Broberg C, Needleman D, Bahmanyar S: Dynein-pulling forces counteract lamin-mediated nuclear stability during nuclear envelope repair. Mol Biol Cell 2018. [DOI] [PMC free article] [PubMed]
- 61.Neelam S, Chancellor TJ, Li Y, Nickerson JA, Roux KJ, Dickinson RB, Lele TP: Direct force probe reveals the mechanics of nuclear homeostasis in the mammalian cell. Proc Natl Acad Sci U S A 2015, 112:5720–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patteson AE, Pogoda K, Byfield FJ, Charrier EE, Galie PA, Deptuła P, Bucki R, Janmey PA: Loss of vimentin intermediate filaments decreases peri-nuclear stiffness and enhances cell motility through confined spaces. bioRxiv 2018.
- 63.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D: A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci U S A 2009, 106:19017–19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramdas NM, Shivashankar GV: Cytoskeletal control of nuclear morphology and chromatin organization. J Mol Biol 2015, 427:695–706. [DOI] [PubMed] [Google Scholar]
- 65.Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, et al. : Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A 2004, 101:8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shin JW, Spinler KR, Swift J, Chasis JA, Mohandas N, Discher DE: Lamins regulate cell trafficking and lineage maturation of adult human hematopoietic cells. Proc Natl Acad Sci U S A 2013, 110:18892–18897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Falk M, Feodorova Y, Naumova N, Imakaev M, Lajoie BR, Leonhardt H, Joffe B, Dekker J, Fudenberg G, Solovei I, et al. : Heterochromatin drives organization of conventional and inverted nuclei. bioRxiv 2018.
- 68.Kandert S, Luke Y, Kleinhenz T, Neumann S, Lu W, Jaeger VM, Munck M, Wehnert M, Muller CR, Zhou Z, et al. : Nesprin-2 giant safeguards nuclear envelope architecture in LMNA S143F progeria cells. Hum Mol Genet 2007, 16:2944–2959. [DOI] [PubMed] [Google Scholar]
- 69.Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT: Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J Cell Biol 2005, 170:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luke Y, Zaim H, Karakesisoglou I, Jaeger VM, Sellin L, Lu W, Schneider M, Neumann S, Beijer A, Munck M, et al. : Nesprin-2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J Cell Sci 2008, 121:1887–1898. [DOI] [PubMed] [Google Scholar]
- 71.Poleshko A, Mansfield KM, Burlingame CC, Andrake MD, Shah NR, Katz RA: The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep 2013, 5:292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL: Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol 2015, 208:33–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luperchio TR, Sauria ME, Wong X, Gaillard M-C, Tsang P, Pekrun K, Ach RA, Yamada NA, Taylor J, Reddy K: Chromosome Conformation Paints Reveal The Role Of Lamina Association In Genome Organization And Regulation. bioRxiv 2017.
- 74.Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. : LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013, 152:584–598. [DOI] [PubMed] [Google Scholar]
- 75.Zheng X, Hu J, Yue S, Kristiani L, Kim M, Sauria M, Taylor J, Kim Y, Zheng Y: Lamins Organize the Global Three-Dimensional Genome from the Nuclear Periphery. Mol Cell 2018, 71:802–815 e807. *Loss of lamins has drastic impact on chromatin organization through its tethering to the lamina. Without lamins, lamin associated domains (LADs) of chromatin detach and decondense, causing disruption of 3D genome organization and transcription.
- 76.Booth EA, Spagnol ST, Alcoser TA, Dahl KN: Nuclear stiffening and chromatin softening with progerin expression leads to an attenuated nuclear response to force. Soft Matter 2015, 11:6412–6418. [DOI] [PubMed] [Google Scholar]
- 77.McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K: Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res 2013, 23:260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, et al. : Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A 2006, 103:8703–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taimen P, Pfleghaar K, Shimi T, Moller D, Ben-Harush K, Erdos MR, Adam SA, Herrmann H, Medalia O, Collins FS, et al. : A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc Natl Acad Sci U S A 2009, 106:20788–20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Camps J, Wangsa D, Falke M, Brown M, Case CM, Erdos MR, Ried T: Loss of lamin B1 results in prolongation of S phase and decondensation of chromosome territories. FASEB J 2014, 28:3423–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stephens AD, Liu PZ, Kandula V, Chen H, Almassalha LM, Backman V, O’Halloran T, Adam SA, Goldman RD, Banigan EJ, et al. : Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. bioRxiv 2018.
- 82.Morgan MA, Shilatifard A: Chromatin signatures of cancer. Genes Dev 2015, 29:238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang P, Dreger M, Madrazo E, Williams CJ, Samaniego R, Hodson NW, Monroy F, Baena E, Sanchez-Mateos P, Hurlstone A, et al. : WDR5 modulates cell motility and morphology and controls nuclear changes induced by a 3D environment. Proc Natl Acad Sci U S A 2018, 115:8581–8586. *WDR5, a main regulator of the H3K4 methyltransferase, is responsible for modulating nuclear stiffness through this euchromatin histone modification in response to the cell sensing a 3D, but not 2D, environment.
- 84.Yang Z, Maciejowski J, de Lange T: Nuclear Envelope Rupture Is Enhanced by Loss of p53 or Rb. Mol Cancer Res 2017, 15:1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Imbalzano KM, Cohet N, Wu Q, Underwood JM, Imbalzano AN, Nickerson JA: Nuclear shape changes are induced by knockdown of the SWI/SNF ATPase BRG1 and are independent of cytoskeletal connections. PLoS One 2013, 8:e55628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rapisarda V, Malashchuk I, Asamaowei IE, Poterlowicz K, Fessing MY, Sharov AA, Karakesisoglou I, Botchkarev VA, Mardaryev A: p63 Transcription Factor Regulates Nuclear Shape and Expression of Nuclear Envelope-Associated Genes in Epidermal Keratinocytes. J Invest Dermatol 2017, 137:2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kriegel AJ, Terhune SS, Greene AS, Noon KR, Pereckas MS, Liang M: Isomer-specific effect of microRNA miR-29b on nuclear morphology. J Biol Chem 2018, 293:14080–14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee S, Ahn YM, Kim JY, Cho YE, Park JH: Downregulation of NOP53 Ribosome Biogenesis Factor Leads to Abnormal Nuclear Division and Chromosomal Instability in Human Cervical Cancer Cells. Pathol Oncol Res 2018. [DOI] [PubMed]
- 89.Samwer M, Schneider MWG, Hoefler R, Schmalhorst PS, Jude JG, Zuber J, Gerlich DW: DNA Cross-Bridging Shapes a Single Nucleus from a Set of Mitotic Chromosomes. Cell 2017, 170:956–972 e923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. : Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171:305–320 e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, C HH, Mirny L, et al. : Two independent modes of chromatin organization revealed by cohesin removal. Nature 2017, 551:51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ: Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 2017, 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH: Phase separation drives heterochromatin domain formation. Nature 2017, 547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Le HQ, Ghatak S, Yeung CY, Tellkamp F, Gunschmann C, Dieterich C, Yeroslaviz A, Habermann B, Pombo A, Niessen CM, et al. : Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol 2016, 18:864–875. **Stretching forces induce compaction of chromatin and gene silencing by a mechanotransduction pathway that involves actin, myosin, and emerin. These forces ultimately trigger EZH2 which upregulates H3K27me3 heterochromatic marks. This pathway illustrates that the regulation of cell nuclear mechanics is important in morphogenesis for the timing of lineage commitment.
- 95.Li Y, Chu JS, Kurpinski K, Li X, Bautista DM, Yang L, Sung KL, Li S: Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys J 2011, 100:1902–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Makhija E, Jokhun DS, Shivashankar GV: Nuclear deformability and telomere dynamics are regulated by cell geometric constraints. Proc Natl Acad Sci U S A 2016, 113:E32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Versaevel M, Grevesse T, Gabriele S: Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat Commun 2012, 3:671. [DOI] [PubMed] [Google Scholar]
- 98.Guilak F, Zell RA, Erickson GR, Grande DA, Rubin CT, McLeod KJ, Donahue HJ: Mechanically induced calcium waves in articular chondrocytes are inhibited by gadolinium and amiloride. J Orthop Res 1999, 17:421–429. [DOI] [PubMed] [Google Scholar]
- 99.Hayakawa K, Tatsumi H, Sokabe M: Actin stress fibers transmit and focus force to activate mechanosensitive channels. J Cell Sci 2008, 121:496–503. [DOI] [PubMed] [Google Scholar]
- 100.Kim TJ, Joo C, Seong J, Vafabakhsh R, Botvinick EL, Berns MW, Palmer AE, Wang N, Ha T, Jakobsson E, et al. : Distinct mechanisms regulating mechanical force-induced Ca(2)(+) signals at the plasma membrane and the ER in human MSCs. Elife 2015, 4:e04876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heo SJ, Driscoll TP, Thorpe SD, Nerurkar NL, Baker BM, Yang MT, Chen CS, Lee DA, Mauck RL: Differentiation alters stem cell nuclear architecture, mechanics, and mechano-sensitivity. Elife 2016, 5. **In differentiating mesenchymal stem cells, physical deformations of cells via substrate stretching increases heterochromatin formation faster than is possible with normal biochemical signaling. The mechanism is mechanosensitive ion channels in the plasma membrane that trigger a calcium signaling cascade that ultimately results in increased heterochromatin, which stiffens the nucleus.
- 102.Heo SJ, Thorpe SD, Driscoll TP, Duncan RL, Lee DA, Mauck RL: Biophysical Regulation of Chromatin Architecture Instills a Mechanical Memory in Mesenchymal Stem Cells. Sci Rep 2015, 5:16895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Kupper K, Joffe B, Thormeyer T, von Hase J, et al. : Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res 2006, 14:707–733. [DOI] [PubMed] [Google Scholar]
- 104.Irianto J, Swift J, Martins RP, McPhail GD, Knight MM, Discher DE, Lee DA: Osmotic challenge drives rapid and reversible chromatin condensation in chondrocytes. Biophys J 2013, 104:759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Damodaran K, Venkatachalapathy S, Alisafaei F, Radhakrishnan AV, Sharma Jokhun D, Shenoy VB, Shivashankar GV: Compressive force induces reversible chromatin condensation and cell geometry dependent transcriptional response. Mol Biol Cell 2018:mbcE18040256. [DOI] [PMC free article] [PubMed]
- 106.Jain N, Iyer KV, Kumar A, Shivashankar GV: Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci U S A 2013, 110:11349–11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gilbert HT, Mallikarjun V, Dobre O, Jackson MR, Pedley R, Gilmore AP, Richardson SM, Swift J: Nuclear decoupling is part of a rapid protein-level cellular response to high-intensity mechanical loading. bioRxiv 2018:317404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gerlitz G, Bustin M: Efficient cell migration requires global chromatin condensation. J Cell Sci 2010, 123:2207–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Segal T, Salmon-Divon M, Gerlitz G: The Heterochromatin Landscape in Migrating Cells and the Importance of H3K27me3 for Associated Transcriptome Alterations. Cells 2018, 7:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Heureaux-Torres J, Luker KE, Haley H, Pirone M, Lee LM, Herrera Y, Luker GD, Liu AP: The effect of mechanosensitive channel MscL expression in cancer cells on 3D confined migration. APL Bioengineering 2018, 2:032001. **Expression of a constitutively active mechanosensitive ion channel (MscL) disrupts metastasis in vivo and migration through small pores in vitro.
- 111.Funkhouser CM, Sknepnek R, Shimi T, Goldman AE, Goldman RD, Olvera de la Cruz M : Mechanical model of blebbing in nuclear lamin meshworks. Proc Natl Acad Sci U S A 2013, 110:3248–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wren NS, Zhong Z, Schwartz RS, Dahl KN: Modeling nuclear blebs in a nucleoskeleton of independent filament networks. Cell Mol Bioeng 2012, 5:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim D-H, Li B, Si F, Phillip JM, Wirtz D, Sun SX: Volume regulation and shape bifurcation in the cell nucleus. Journal of Cell Science 2015, 128:3375–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deviri D, Discher DE, Safran SA: Rupture Dynamics and Chromatin Herniation in Deformed Nuclei. Biophys J 2017, 113:1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cao X, Moeendarbary E, Isermann P, Davidson PM, Wang X, Chen MB, Burkart AK, Lammerding J, Kamm RD, Shenoy VB: A Chemomechanical Model for Nuclear Morphology and Stresses during Cell Transendothelial Migration. Biophys J 2016, 111:1541–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]