We have entered an era when studies of the gut microbiota are transitioning from basic questions of composition and host effects to understanding the microbial molecules that underlie compositional shifts and mediate health and disease processes. The importance of the gut Bacteroidales to human health and disease and their potential as a source of engineered live biotherapeutics make these bacteria of particular interest for in-depth mechanistic study. However, there are still barriers to the genetic analysis of diverse Bacteroidales strains, limiting our ability to study important host and community phenotypes identified in these strains. Here, we have overcome many of these obstacles by constructing a series of vectors that allow easy genetic manipulation in diverse gut Bacteroides and Parabacteroides strains. These constructs fill a critical need and allow streamlined allelic replacement in diverse gut Bacteroidales, including the growing number of multiantibiotic-resistant strains present in the modern-day human intestine.
KEYWORDS: Bacteroides, microbiota, mutant construction, vectors
ABSTRACT
Studies of the gut microbiota have dramatically increased in recent years as the importance of this microbial ecosystem to human health and disease is better appreciated. The Bacteroidales are the most abundant order of bacteria in the healthy human gut and induce both health-promoting and disease-promoting effects. There are more than 55 species of gut Bacteroidales with extensive intraspecies genetic diversity, especially in regions involved in the synthesis of molecules that interact with other bacteria, the host, and the diet. This property necessitates the study of diverse species and strains. In recent years, the genetic toolkit to study these bacteria has greatly expanded, but we still lack a facile system for creating deletion mutants and allelic replacements in diverse strains, especially with the rapid increase in resistance to the two antibiotics used for genetic manipulation. Here, we present a new versatile and highly efficient vector suite that allows the creation of allelic deletions and replacements in multiresistant strains of Bacteroides and Parabacteroides using a gain-of-function system based on polysaccharide utilization. These vectors also allow for easy counterselection independent of creating a mutant background strain, using a toxin from a type VI secretion system of Bacteroides fragilis. Toxin production during counterselection is induced with one of two different molecules, providing flexibility based on strain phenotypes. This family of vectors greatly facilitates functional genetic analyses and extends the range of gut Bacteroidales strains that can be genetically modified to include multiresistant strains that are currently genetically intractable with existing genetic tools.
INTRODUCTION
The human gut microbiota is a diverse and dense microbial ecosystem that plays an essential role in health and development (1, 2). Large-scale metagenomic, metatranscriptomic, and metabolomic studies are revealing the molecular repertoire of this complex community (3, 4), yet functional genetic, phenotypic, and mechanistic studies lag far behind and are essential to understand the complex interactions within this microbial ecosystem and how these microbes interface with their host (3, 4).
The Bacteroidales are an order of Gram-negative bacteria that includes the abundant gut genera Bacteroides, Parabacteroides, and Prevotella, which collectively include more than 55 identified human gut species (5, 6). These bacteria display remarkable stability and substantial resilience to temporary perturbations, with many strains colonizing for decades (7–9). The majority of genetic and phenotypic studies of the gut Bacteroidales have analyzed only three strains: Bacteroides fragilis NCTC 9343, B. fragilis 638R, and B. thetaiotaomicron VPI 5482 (10, 11). However, Bacteroidales species have remarkable genome plasticity, with extensive within-species genetic diversity and metabolic abilities and large pangenomes (6, 12, 13). Many of the regions involved in the synthesis of outer surface and secreted molecules that are likely to interface with other microbes or the host are in heterogeneous regions of a species (12, 14–20). This heterogeneity highlights the need for functional genetic studies of diverse human isolates.
Bacteroides species are intrinsically resistant to many of the antibiotics commonly used in the laboratory and now display increasing resistance to many antibiotics used in clinical settings (21–28). Due to pervasive horizontal gene transfer of antibiotic resistance genes, many Bacteroides and Parabacteroides isolates from American and European subjects now display resistance to tetracycline and erythromycin, the two commonly used antibiotics for genetic selection (25, 29–31). Chloramphenicol and cefoxitin resistance genes have been used to select for replicative plasmids in B. fragilis, B. vulgatus, and B. thetaiotaomicron strains (29, 32, 33) and sometimes function for chromosomal integrations in select genetic backgrounds (29, 31, 34–36). However, cefoxitin and carbapenems are clinically relevant antibiotics and should ideally be avoided in the laboratory to prevent the spread of resistance genes (37).
Construction of markerless gene knockouts or other types of mutations in bacteria can be carried out through lambda red recombineering or CRISPR-Cas9-based systems (38–40). However, these methods often require strain-specific modifications; rely on efficient transformation, which is often impossible or very inefficient in Bacteroidales; and may lead to off-target mutations (38–42). Allelic replacement in Bacteroides is a two-step process. First, a suicide vector is transferred via conjugation and integrated into the chromosome via homology-based recombination of regions flanking the target deletion and selected via a gain-of-function phenotype, typically antibiotic resistance. This limits the tractable strains to those sensitive to erythromycin or tetracycline and therefore excludes a large number of isolates. Genomes that undergo double-crossover events result in the excision of the vector and typically are identified by one of three different methods. The first is replica plating, which has the advantages that there is no need for a genetic mutation to first be made in the background strain and that the resulting strains therefore have no additional underlying genetic mutation. The disadvantage is that this is a more labor-intensive and slow process, requiring the analysis of thousands of colonies. A second method uses counterselection where a genetic mutation of thyA, encoding thymidylate synthetase, is made in the background strain (64). thyA is placed on the suicide vector, and double-crossover resolvents are selected by plating on trimethoprim, which kills thyA-positive cells. A third method is similar to the thyA procedure but requires the creation of a background mutant strain of tdk, encoding thymidine synthase, with the selection of resolvents by plating on 5-fluoro-2′-deoxyuridine (50). Other counterselection systems readily used in proteobacteria, such as sacB and rpsL (42), do not function in Bacteroides. Recently, a new counterselection marker was implemented based on an allele of the phenylalanyl-tRNA synthetase (pheS*), lethal in the presence of p-chloro-phenylalanine (p-Cl-Phe) (43). This system functions in some strains but not others, requires growth on minimal medium, and often results in significant background growth.
Here, we created a new vector family for genetic modifications in gut Bacteroidales. These vectors allow easy allelic replacement without first making a mutant background strain for counterselection. In addition, we created vectors where the gain of function is the ability to utilize a polysaccharide rather than antibiotic resistance, greatly increasing the range of strains that can be genetically manipulated to include multiresistant strains. We provide numerous examples of these vectors functioning in diverse Bacteroidales species, regardless of the antibiotic resistance profile.
RESULTS AND DISCUSSION
An inulin selection cassette for use in antibiotic-resistant Bacteroidales strains.
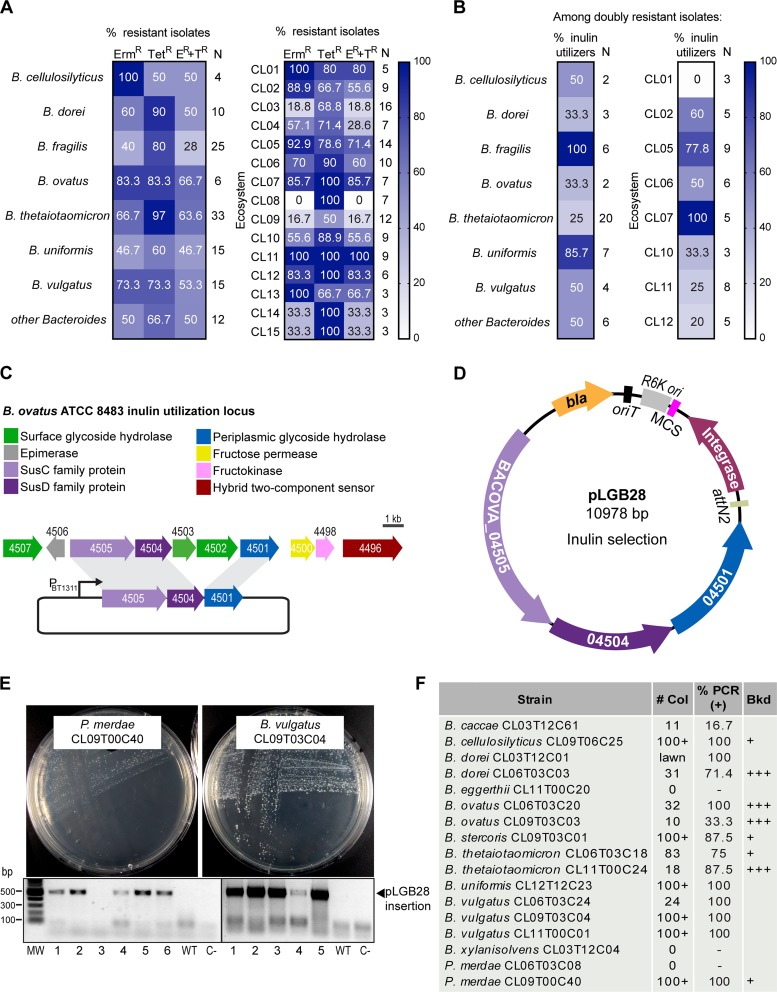
Analyses of recent human fecal isolates of Bacteroidales with interesting phenotypes revealed that many are resistant to both tetracycline and erythromycin (60, 65) and therefore are not amenable to genetic manipulation using available genetic tools for these bacteria. We evaluated tetracycline and erythromycin resistance among 120 Bacteroides strains from our collection isolated between 2009 and 2011 from the stool of 15 healthy U.S.-resident adults with no history of gastrointestinal diseases (Fig. 1A) (18). For each subject/ecosystem (named CL01 to CL15), the stated numbers of Bacteroides isolates analyzed are listed (Fig. 1A) (18). On average, 59% of the isolates from these combined ecosystems are resistant to erythromycin, and 74% are resistant to tetracycline, while 51% are doubly resistant. In particular, 63% of B. ovatus and 67% of B. thetaiotaomicron isolates from these ecosystems are doubly resistant. The incidence of resistance varies widely depending on the ecosystem (Fig. 1A), with isolates from ecosystems CL03, CL08, and CL14 displaying low levels of double resistance, while ecosystems CL01, CL05, CL07, CL11, and CL12 are almost entirely composed of doubly resistant isolates. These results are consistent with other reports of widespread resistance to tetracycline and erythromycin in recent Bacteroides and Parabacteroides isolates (22, 23, 31).
FIG 1.
A minimal inulin utilization locus can be used for selection of integrants in many Bacteroidales species. (A) Percentage of tetracycline- and erythromycin-resistant Bacteroides strains from the Comstock laboratory ecosystems strain collection. Numbers in the right column (N) indicate the number of strains tested. (B) Percentage of inulin utilizers from the doubly resistant (Tetr Ermr) Bacteroides strains. Numbers in the right column (N) indicate the number of strains tested. Ecosystems with fewer than three doubly resistant strains were excluded. (C) Inulin utilization locus of B. ovatus ATCC 8483. PBT1311 is the strong constitutive promoter from the BT_1311 sigma factor of B. thetaiotaomicron VPI 5482 used to drive the expression of the three-gene inulin utilization locus. (D) Integration vector pLGB28 integrates at chromosomal attBT2 sites and allows for inulin utilization selection. bla, β-lactamase gene conferring ampicillin and carbenicillin resistance in E. coli S17; oriT, RP4 origin of transfer; RK6, replication origin in E. coli λpir; MCS, multiple-cloning site; Integrase, IntN2 from NBU2; attN2, recognition and integration sequence. (E) Growth of the indicated strains on M9S-inulin plates to select for integration of pLGB28. Ethidium bromide-stained agarose gels show PCR amplification of the 500-bp fragment corresponding to integration of pLGB28 at attBT2. Six clones are shown per strain. WT, wild type; C−, no-DNA control; MW, molecular weight marker (Quick-Load Purple 1-kb Plus DNA ladder; NEB). (F) Results of the conjugation of pLGB28 into 17 different doubly resistant (Tetr Ermr), inulin-nonutilizing Bacteroides and Parabacteroides strains from panel B. # Col, number of colonies obtained per 13.75 ml of mating culture mix; %PCR (+), percentage of clones that were PCR positive as described above for panel E; Bkd, amount of background growth in the inulin selection plate; +, slight hazy background with distinct colonies (as observed in panel E, left); +++, strong background growth (as observed in Fig. 3B, left).
To overcome the genetic intractability of these strains, we designed a nonantibiotic selection marker that confers the ability to utilize inulin as a carbon source. To test if inulin selection would be useful in the doubly antibiotic-resistant isolates, we screened 57 of these strains for growth on defined, minimal inulin medium (M9S-inulin) (Fig. 1B) and observed that 28 (49%) are inulin utilizers. Excluding B. fragilis and B. uniformis, the majority of doubly resistant isolates of other Bacteroides species (72%) lack the ability to grow on inulin and are therefore theoretically amenable to inulin selection.
The choice of conferring inulin utilization for gain-of-function selection of cointegrates is based on the fact that inulin is a simple linear polysaccharide ([β1,2] fructose polymer) that does not require surface digestion (20, 44). Therefore, the inulin utilization locus is smaller than most polysaccharide utilization loci (PULs), which typically span 20 to 60 kb (10, 45, 46), and includes several dispensable genes (Fig. 1C) (20, 44, 47). The absence of extracellular digestion prevents background growth of nontransconjugant cells (44). We constructed a three-gene inulin utilization cassette from the inulin locus of B. ovatus ATCC 8483. It comprises genes encoding the outer membrane SusC-like TonB-dependent inulin transporter (BACOVA_04505), its associated inulin-binding surface lipoprotein (SusD-like; BACOVA_04504), and the periplasmic glycoside hydrolase (BACOVA_04501) (44, 48) (Fig. 1C). The three genes were placed under the control of a strong constitutive promoter from the B. thetaiotaomicron VPI 5482 housekeeping sigma factor PBT1311 (49). This inulin selection cassette (6.7 kb) was placed into pNBU2 (50), replacing the erythromycin cassette, to create pLGB28 (Fig. 1D). pNBU2 is a pir-dependent suicide vector which replicates in certain Escherichia coli strains but must integrate for maintenance in Bacteroides species. pNBU2 encodes an IntN2 tyrosine recombinase that catalyzes the integration of the vector at a chromosomal attBT2 site (50, 51). Most Bacteroides strains have two attBT2 sites, located at the 3′ end of the two tRNASer genes (51). Double-integration events are not expected, as the integration of the plasmid disrupts the tRNA gene (51).
We conjugally transferred pLGB28 into 17 doubly resistant Bacteroides and Parabacteroides strains unable to utilize inulin as a sole carbon source. We obtained inulin-utilizing integrants for 14 of these strains from nine different species (Fig. 1E and F). Four strains displayed some background growth, which required restreaking of single colonies for isolation. As inulin selection is not bactericidal, we recommend that integrants always be restreaked for isolation. For the majority of strains, there were numerous integrants with little or no background growth. Notably, we easily obtained transconjugants in Parabacteroides merdae CL09T00C40 (PmCL09), indicating that these protocols and vector series can be used in different families of Bacteroidales. Interestingly, both attBT2 sites in PmCL09 have two base pair differences relative to the attBT2 sequence of pNBU2 and Bacteroides chromosomes (51), indicating some promiscuity for IntN2-mediated recombination. To verify integration at the attBT2 sites, we designed PCR primers that anneal upstream of the recombination site (forward primer) and inside pNBU2 (reverse primer). Most integrates in the majority of strains had the insertion at one of the two tRNASer loci. However, in B. ovatus CL09T03C03 and B. caccae CL03T12C61, PCR revealed that most inulin-utilizing clones did not have an insertion of the plasmid in tRNASer. Importantly, these data show that selection of integrants using inulin selection is a feasible alternative to antibiotic selection for most doubly resistant Bacteroides species.
The aTC-inducible ssBfe1 is a highly effective counterselection system.
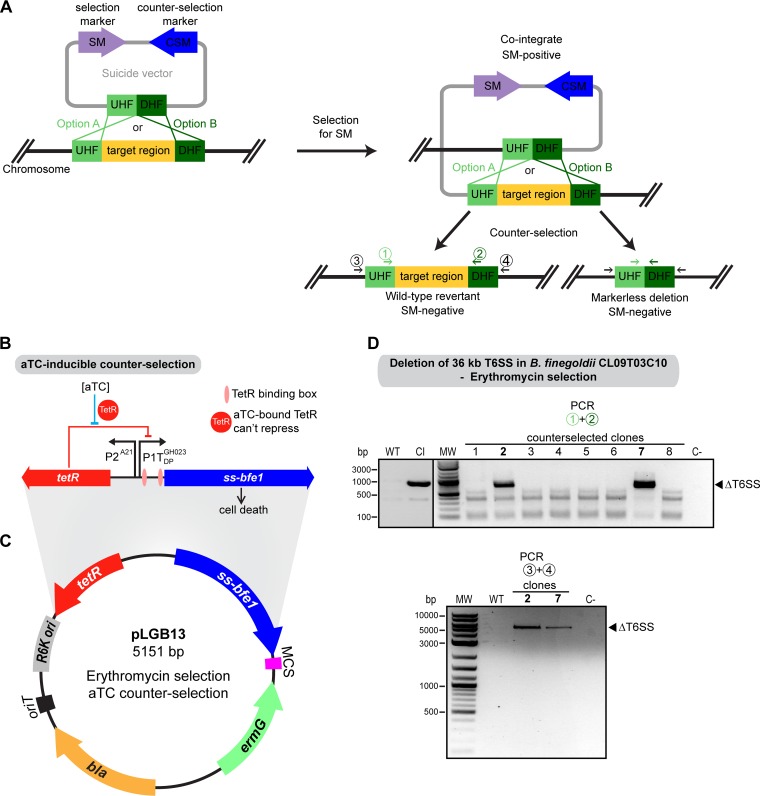
Two-step allelic exchange (Fig. 2A) involves an intermediate cointegrate step where the suicide vector containing the genes for positive selection is integrated into the chromosome at the target site, followed by growth in nonselective medium and plating with selection for cells that have undergone the second recombination (cross-out) event. Selection of double-crossover resolvents using a lethal toxin encoded on the plasmid to kill cointegrates should be an effective counterselection system. Three essential features of such a counterselection system are the low frequency of toxin-resistant escapees, broad host range of toxicity, and tight regulation of the toxin (produced only during the counterselection step). The highly toxic effector Bfe1 from the B. fragilis 638R type VI secretion system (T6SS), which mediates antibacterial antagonism against a wide range of Bacteroidales species (52), was chosen for counterselection. Lim et al. showed that Bfe1 can be endogenously expressed in B. thetaiotaomicron VPI 5482 and kill this strain when targeted to the periplasm using an N-terminal localization signal sequence (ssBfe1) (53). When placed under the tightly regulated anhydrotetracycline (aTC)-inducible promoter developed for Bacteroides by Lim et al. (Fig. 2B), periplasmic Bfe1 expression leads to rapid cell death, with no evidence of escapees (53). Furthermore, although Bacteroides species have been shown to commonly harbor islands of immunity genes to multiple T6SS effectors (54), the bfi1 (BF638R_1987) immunity gene to Bfe1 (52) is absent from these islands. tBLASTn searches of 724 available Bacteroides and Parabacteroides genomes revealed that only 35 B. fragilis strains have bfi1, all of them as part of a T6SS cluster containing bfe1 (complete genomes and whole-genome shotgun contigs in GenBank, along with our in-house-sequenced strains).
FIG 2.
Bfe1-based counterselection induced by aTC is highly effective. (A) Diagram of the two-step allelic replacement process. UHF, upstream homology flank; DHF, downstream homology flank. (B) aTC-inducible ssBfe1 counterselection cassette (53). (C) Vector pLGB13. bla, OriT, RK6, and MCS are described in the Fig. 1D legend. ermG, erythromycin resistance gene. (D) PCR verification of the counterselected clones for deletion of the 36-kb T6SS region of BfiCL09. WT, wild type; CI, cointegrate; C−, no-DNA control; MW, molecular weight marker. (Top) PCR using primers 1 and 2; (bottom) PCR using primers 3 and 4, as indicated in panel A.
As this system met all the criteria for successful counterselection, we made a counterselection suicide vector. Starting from the aTC-inducible pNBU2_erm-TetR-P1T_DP-GH023 integration vector (53), we removed the attBT2 site and its cognate integrase gene, necessitating integration by homologous recombination. The ssbfe1 cassette was added under the control of the aTC promoter. The multiple-cloning site for insertion of DNA for homology-based recombination was retained (pLGB13) (Fig. 2B and C).
To test pLGB13 for creating chromosomal deletions, we made a construct to delete the 36-kb GA1 T6SS locus (HMPREF1057_01517 to HMPREF1057_01551) of B. finegoldii CL09T03C10 (BfiCL09) (Fig. 2D). All colonies obtained after counterselection (on aTC selection plates) were erythromycin sensitive, indicating loss of the vector. PCR verification of eight clones indicated that six were wild-type revertants and that two had the desired 36-kb deletion (clones 2 and 7). To further verify this large deletion, we designed a second set of primers that anneal to the chromosome in the regions upstream and downstream of the flanking DNA cloned into pLGB13 (primers 3 and 4) (Fig. 2A). A PCR with these primers in clones 2 and 7 yielded the expected amplicon size (5.2 kb), whereas a cointegrate or a wild-type revertant would produce no amplicon (Fig. 2D). This pLGB13 counterselection system has now been successfully used in the laboratory to make deletions and allelic replacements in strains of B. uniformis, B. thetaiotaomicron, and B. vulgatus.
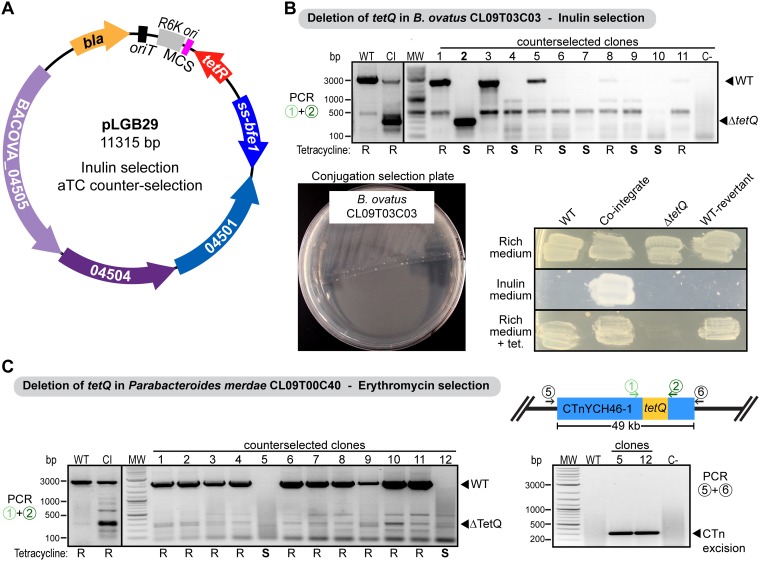
pLGB13 requires that the strain to be mutated is erythromycin sensitive. To extend this counterselection system to erythromycin-resistant strains, we replaced ermG of pLGB13 with the inulin utilization cassette, creating plasmid pLGB29 (Fig. 3A). As a proof of concept for inulin selection with homology-based recombination, we cloned into pLGB29 the 1-kb regions upstream and downstream of the tetQ tetracycline resistance gene of B. ovatus CL09T03C03 (BoCL09) (AA414_04155). Cointegrates were obtained for BoCL09 (Fig. 3B); however, there was background growth such that clones needed to be streaked onto inulin plates twice to obtain true cointegrates. We verified the cointegrates using PCR primers that anneal within the homology flanks (depicted in Fig. 2A as primer pair 1 and 2), such that amplification from the cointegrate should yield both the wild-type tetQ band (2.2 kb) as well as the shorter product representing the deletion (300 bp) (Fig. 3B).
FIG 3.
Deletion of the tetracycline resistance gene tetQ. (A) Allelic replacement vector pLGB29, using inulin selection for cointegrates. bla, oriT, RK6, and MCS are described in the Fig. 1D legend. (B) PCR verification and growth on plates of the counterselected clones for the tetQ deletion in BoCL09. An inulin selection plate for the mating of BoCL09T03C03, using homology-based integration of pLGB29 containing 1 kb of DNA on each side of the tetQ gene, is shown. S, sensitive; R, resistant. (C, left) Ethidium bromide-stained agarose gel showing PCR amplicons of the resolvent for the tetQ deletion in PmCL09 using pLGB13 (erythromycin sensitive). (Right) PCR amplicon to verify excision of the whole CTnYCH46-1 (HMPREF1078_01847 to HMPREF1078_01893). Primers anneal upstream and downstream of the 49-kb conjugative transposon. WT, wild type; CI, cointegrate. C−, no-DNA control; MW, molecular weight marker.
For comparison of inulin selection against traditional erythromycin selection, we cloned the same 1-kb regions upstream and downstream of tetQ into pLGB13 (erythromycin selection) to delete this gene in PmCL09 (erythromycin sensitive; HMPREF1078_01857). Cointegrates of BoCL09ΩpLGB29 (inulin) or PmCL09ΩpLGB13 (erythromycin) were grown in nonselective broth and plated onto medium containing 40 ng/ml aTC. None of the aTC-selected colonies grew on either inulin (BoCL09) or erythromycin (PmCL09), indicating plasmid excision. However, PCR analysis and growth on plates containing tetracycline, whose resistance is encoded by the targeted deletion, indicated that most clones were wild-type revertants (Fig. 3B and C). In 1 of 30 resolvents tested for BoCL09, we identified a mutant that produced the expected 300-bp PCR band corresponding to the tetQ deletion (clone 2) (Fig. 3B). Interestingly, five resolvents displayed no deletion band or the wild-type band (clones 4, 6, 7, 9, and 10) (Fig. 3B). Inspection of the genomic context of the tetQ gene in both strains revealed that it is part of a 49-kb conjugative transposon (CTn) with 99% sequence identity to CTnYCH46-1 that belongs to the CTn341 family (55). The excision and transfer of CTn341 (and CTnYCH46-1) are induced by tetracycline (and its derivative aTC), mediated through the two-component regulatory system RteAB (56). Therefore, selection using aTC induced the excision of the whole CTnYCH46-1 during counterselection. We observed a similar phenomenon in PmCL09, where the majority of colonies after counterselection were wild-type revertants and two were CTnYCH46-1 excisions (clones 5 and 12) (Fig. 3C). We used PCR with primers annealing upstream and downstream of this 49-kb element and confirmed the excision of CTnYCH46-1 (HMPREF1078_01847 to HMPREF1078_01893) in these clones (Fig. 3C). Therefore, although the aTC-ssBfe1 cassette is highly efficient for counterselection, it is not ideal for deletion of genes contained on tetracycline-inducible CTns. Since aTC induction also precludes the use of tetracycline as a selection marker, which would be desired in tetracycline-sensitive erythromycin-resistant strains, we devised a rhamnose-inducible alternative to be used in a tetracycline selection vector (pLGB30) (Fig. 4B) or an inulin selection vector (pLGB31) (Fig. 4F), described below. pLGB31 can also be used for allelic replacement of genes contained in the tetracycline-inducible CTn.
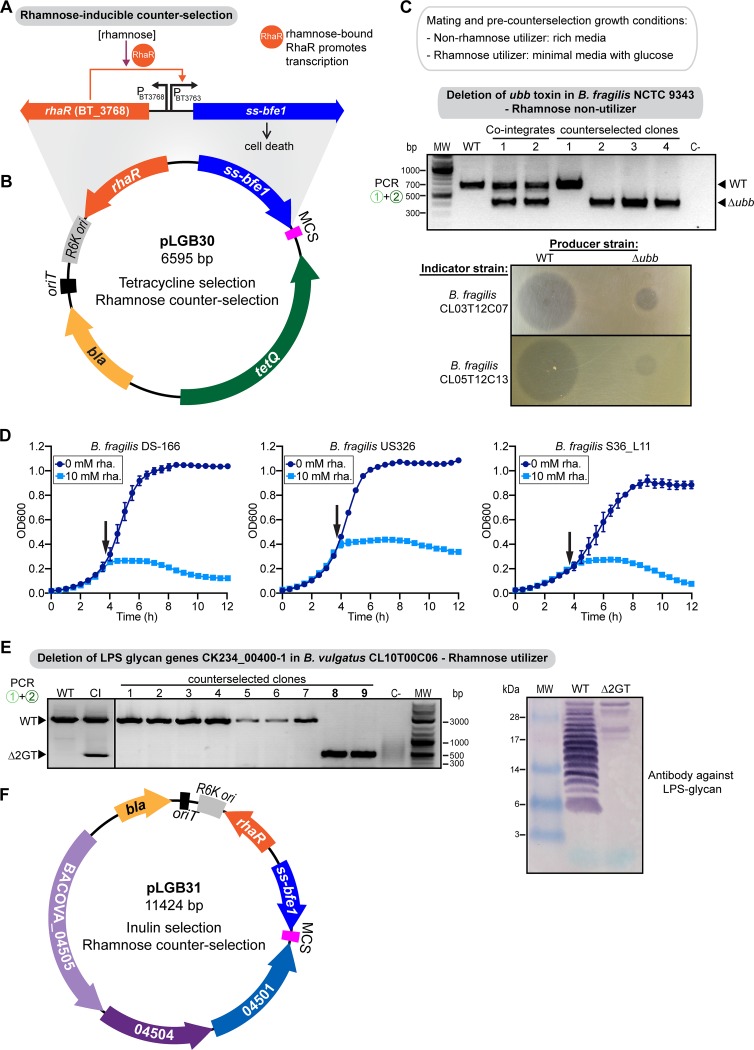
FIG 4.
Bfe1-based counterselection induced by rhamnose. (A) Rhamnose-inducible ssBfe1 counterselection cassette. (B) Allelic replacement vector pLGB30 using tetracycline selection of cointegrates. bla, oriT, RK6, and MCS are described in the Fig. 1D legend. tetQ, tetracycline resistance gene. (C, top) PCR verification of the cointegrates and counterselected clones for the ubb deletion in Bf9343, using primers 1 and 2, as indicated in the Fig. 2A legend. WT, wild type; CI, cointegrate; C−, no-DNA control; MW, molecular weight marker. (Bottom) Overlay assays to test BfUbb inhibitory activity by wild-type or Δubb Bf9343, using the specified strains as indicators and producers. (D) Growth curves of the specified B. fragilis strains containing a chromosomally integrated rhamnose-inducible ssbfe1. Black arrows indicate the timing of rhamnose addition to the culture. The means and SEM from 3 biological replicates per treatment are plotted. (E, left) PCR verification of the counterselected clones for the deletion of CK234_00400-1 in BvCL10, using primers 1 and 2, as indicated in the Fig. 2A legend. (Right) Western immunoblotting of whole-cell lysates probed with antiserum raised to WT BvCL10 adsorbed with the ΔCK234_00400-1 mutant. (F) Allelic replacement vector pLGB31 using inulin selection. bla, oriT, RK6, and MCS are described in the Fig. 1D legend.
The rhamnose-inducible ssBfe1 system.
Mimee et al. showed that the RhaR (BT_3768) transcriptional activator from B. thetaiotaomicron VPI 5482 can be used to drive robust activation of the BT_3763 (rhaK) promoter (Fig. 4A) (57, 58). We placed ssbfe1 under the control of this promoter cassette, creating suicide vectors pLGB30 (tetracycline selection) (Fig. 4B) and pLGB31 (inulin selection) (Fig. 4F). As some strains of Bacteroidales are able to grow with rhamnose as the sole carbon source and others are not, we analyzed whether this system would function in strains with each of these phenotypes. We first tested this system in B. fragilis NCTC 9343 (Bf9343) (Tets), which does not have the ability to grow using rhamnose as the sole carbon source and whose genome does not harbor a described rhamnose utilization locus. Using pLGB30, we made a construct to delete the gene encoding the antimicrobial toxin BfUbb (BF9343_3779), which inhibits the growth of other B. fragilis strains (59). Despite the inability of Bf9343 to utilize rhamnose, the system drove the expression of ssbfe1 to high-enough levels that only double-crossover resolvents grew on the rhamnose counterselection plates, and all these colonies lost pLGB30, as they were not tetracycline resistant. Therefore, Bf9343 cells have some ability to take up rhamnose, although they do not have a described RhaT permease (58). PCR analysis of four resolvents identified three with the correct ubb deletion (clones 2, 3, and 4) (Fig. 4C). We verified this deletion phenotypically by testing for killing activity using the spot agar overlay assay (Fig. 4C). Wild-type Bf9343 produced a large zone of growth inhibition for the two tested BfUbb-sensitive strains, whereas the Bf9343 Δubb strain lost this ability. A small inhibition zone was observed, corresponding to the size of the original bacterial spot, demonstrating that Bf9343 produces an additional antimicrobial toxin, as is typical of B. fragilis strains (59, 60). These analyses show that pLGB30 allows for easy creation of double-crossover mutants in erythromycin-resistant strains where tetracycline selection of cointegrates is necessary.
To determine if this vector can be widely used in non-rhamnose-utilizer strains, we carried out BLAST searches of all 724 available Bacteroides and Parabacteroides genomes. We used as queries the rhamnose symporter (BT_3765), rhamnulose-1-phosphate aldolase (BT_3766), and rhamnulose kinase (BT_3763) genes from B. thetaiotaomicron VPI 5482 (58). All Bacteroides and Parabacteroides genomes from all species had >90% identity hits, indicating that rhamnose utilization is a conserved trait. The exceptions were all 167 strains of B. fragilis. To establish if rhamnose induction of ssBfe1 would function for counterselection in other strains of B. fragilis, which homology-based searches suggest are also unable to utilize rhamnose, we made a pNBU2-derivative chromosomal integration vector carrying the RhaR-ssbfe1 cassette. We integrated this construct into three B. fragilis strains and monitored their growth with or without added rhamnose (Fig. 4D). For all three strains, rhamnose addition led to a very rapid growth arrest, with no evidence of resumed growth up to 8 h after toxin induction. Therefore, even though B. fragilis strains are unable to utilize inulin as a sole carbon source, this rhamnose-inducible toxin system functions broadly in this species.
Next, we tested this counterselection system in a rhamnose-utilizing strain, B. vulgatus CL10T00C06 (BvCL10). Unlike B. fragilis, BoCL09 has a rhamnose permease as well as a rhamnose two-component signaling system, likely leading to induction of the rhaK promoter at much lower concentrations of rhamnose than for a rhamnose nonutilizer. Therefore, all steps prior to counterselection were performed using defined M9S-glucose medium, as small amounts of rhamnose in BHIS plates or supplemented basal medium (SBM) broth could induce premature expression of the toxin. We made a construct in pLGB30 to delete the genes CK234_00400 and CK234_00401 (CK234_00400-1). These genes encode glycosyltransferases involved in the synthesis of the lipopolysaccharide (LPS) glycan (15). Counterselection on M9S-rhamnose plates yielded single colonies, all tetracycline sensitive. PCR analysis demonstrated that of 10 resolvents analyzed, 2 were deletion mutants (clones 8 and 9) (Fig. 4E). We further verified the phenotype of this deletion mutant by Western immunoblotting of whole-cell lysates using an antiserum specific to the LPS glycan of BvCL10 (15). The antibody reactivity to this glycan is lost in the ΔCK234_00400-1 mutant (Fig. 4E), providing phenotypic confirmation of the deletion.
To further increase the breadth of strains that can be mutated and the genetic pool to include allelic replacement of genes in tetracycline-inducible CTns, we created pLGB31 (Fig. 4F). This vector is similar to pLGB29 in that cointegrates are selected by the acquisition of inulin utilization but with rhamnose as the inducer of ssbfe1 for selection of double-crossover resolvents.
In summary, we have created a versatile family of vectors and protocols to greatly facilitate genetic manipulation of diverse Bacteroides and Parabacteroides strains. These vectors allow for allelic deletions and replacements in the increasingly abundant antibiotic-resistant strains that are currently not amenable to genetic manipulation with existing genetic tools. Routine mutant construction times are reduced from several weeks to less than 2 weeks, and the steps are much less labor-intensive than protocols without counterselection. In addition, there is no need to create a mutant background strain for counterselection. Bfe1 is a broad-range and potent toxin, and we have not encountered instances of spontaneous resistance mutations in any species. This is an easily modifiable system where the ssBfe1 toxin gene could be exchanged for a different GA3 T6SS toxin gene for genetic manipulation in B. fragilis strains, such as 638R, that encode the cognate immunity protein Bfi1. The versatility of two different inducers of Bfe1 increases the breadth of strains and genes that can be genetically manipulated.
MATERIALS AND METHODS
Media and growth conditions.
E. coli strains were grown aerobically at 37°C in LB medium, with 100 μg/ml carbenicillin added when indicated. Bacteroides and Parabacteroides strains were routinely grown at 37°C under anaerobic conditions on supplemented basal medium (SBM) (liquid cultures) (61) and brain heart infusion plates supplemented with 5 mg/liter hemin and 2.5 μg/liter vitamin K1 (BHIS). When necessary for selection, M9S plates were used, which are M9 minimal medium (62) supplemented with 50 mg/liter l-cysteine, 5 mg/liter hemin, 2.5 μg/liter vitamin K1, 2 mg/liter FeSO4 · 7H2O, 5 μg/liter vitamin B12, and 0.7% agarose. The carbon source for these plates was either 0.25% (wt/vol) glucose (M9S-glucose) or 0.4% (wt/vol) inulin (M9S-inulin) (inulin from chicory; Sigma-Aldrich). Antibiotics were used at the following concentrations, where appropriate: 5 μg/ml erythromycin, 200 μg/ml gentamicin, and 6 μg/ml tetracycline. Anhydrotetracycline (aTC) was added at 40 or 100 ng/ml, while l-(+)-rhamnose was used at 10 mM. For growth curves, bacteria from a culture grown overnight in broth were diluted 1:200 in fresh medium, allowed to enter exponential growth, and diluted again to an optical density at 600 nm (OD600) of 0.02. Growth curves were carried out in flat-bottom 96-well plates, with 200 μl per well, under anaerobic conditions at 37°C. OD600 values were measured every 30 min with an Eon high-performance microplate spectrophotometer (BioTek Instruments). A total of 10 mM rhamnose was added to the corresponding wells after 3 h 35 min of incubation. The means and standard errors of the means (SEM) from 3 biological replicates per treatment were plotted in Prism 8 for macOS (GraphPad Software).
Plasmid construction.
Table S1 in the supplemental material summarizes the plasmids and construction methods used in this study. Phusion high-fidelity DNA polymerase or Q5 high-fidelity DNA polymerase was used for PCR cloning steps (New England Biolabs [NEB]), and all restriction endonucleases were high-fidelity restriction endonucleases from NEB. All plasmid assembly reactions were carried out using NEBuilder HiFi DNA assembly master mix (NEB). Oligonucleotides used for plasmid construction and PCR strain verification are listed in Table S2. Whole-plasmid sequencing was performed at the Massachusetts General Hospital CCIB DNA Core. The inulin utilization cassette causes a fitness defect in E. coli, leading to colonies that are smaller and more mucoid than the wild type and take 24 h to appear. These should be preferentially picked over the faster-appearing, nonmucoid colonies, which are likely to carry loss-of-function mutations. In order to avoid accumulation of mutations in the inulin plasmid, we recommend minimizing propagation steps and working on a Tn10-negative cloning strain.
Plasmids and construction details. Download Table S1, PDF file, 0.04 MB (46.4KB, pdf) .
Copyright © 2019 García-Bayona and Comstock.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S2, PDF file, 0.03 MB (32.5KB, pdf) .
Copyright © 2019 García-Bayona and Comstock.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conjugation and selection conditions.
Parental strains and strains constructed in this study are listed in Table S3. Plasmids were transformed into the donor E. coli strain S17-λpir and conjugated into Bacteroides or Parabacteroides as described previously (31, 63), with some modifications. Briefly, donor and recipient strains were grown on liquid medium, 25 ml and 2.5 ml, respectively. When the recipient strain reached an OD600 of between 0.1 and 0.2 (0.05 for B. fragilis) and the donor strain reached an OD600 of 0.2 to 0.6, both strains were mixed and pelleted by centrifugation at 9,000 × g for 10 min. The pellet was resuspended in 100 μl SBM, spotted directly in the center of a prewarmed BHIS plate, and incubated at 37°C aerobically for 15 to 18 h. The bacteria from the mating spot were streaked onto the appropriate selection plates (BHIS plus erythromycin, BHIS plus tetracycline, or M9S-inulin) containing gentamicin (one plate had half the mating spot, another plate had a quarter, and the last quarter of the mating spot was streaked for isolation in the third plate). For rhamnose counterselection using pLGB30 for rhamnose-utilizing strains, cointegrates were selected using M9S-glucose plates with gentamicin and tetracycline. Plates were incubated anaerobically. Colonies were picked after 2 to 3 days (BHIS and M9S-glucose) or 3 to 4 days (M9S-inulin) and restreaked for isolation. PCRs for strain verification were performed using Phusion DNA polymerase with the oligonucleotides listed in Table S2.
Bacteroidales strains used or created in this study. Download Table S3, PDF file, 0.04 MB (46.1KB, pdf) .
Copyright © 2019 García-Bayona and Comstock.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Counterselection conditions.
After confirmation of cointegrates via PCR (Table S2), each strain was grown overnight in 6 ml SBM, diluted 1:100 in fresh SBM, and incubated for 6 to 8 h. Aliquots of 50, 5, 1, and 0.2 μl were plated onto BHIS plates containing 40 or 100 ng/ml aTC or 10 mM rhamnose, depending on the selection method. We have found that cointegrates can be grown from plates in nonselective medium for as little as an hour before plating for counterselection, eliminating the need for culture overnight. For rhamnose counterselection in rhamnose-utilizing strains, bacteria were grown in M9S-glucose broth and plated onto M9S-rhamnose (10 mM). After 2 to 4 days, single colonies were restreaked and analyzed by PCR to confirm the loss of the selection marker (Table S2).
Agar overlay assays.
Agar spot overlay assays were carried out as previously described (30). Briefly, 5 μl of the toxin-producing strain or mutant was spotted onto plates and incubated overnight anaerobically. Cells were removed, and the plates were exposed to chloroform vapor to kill remaining bacteria. The overlay strain was grown to exponential phase, added to 4 ml warm BHIS with 0.8% agar, and poured on top of the chloroform-treated plate. Plates were incubated overnight anaerobically before imaging.
Western blot analyses.
Western immunoblot analyses of exponential-phase cell lysates were performed as described previously, using antiserum prepared in rabbits to whole-cell B. vulgatus CL10T00C06, and subjected to antibody adsorption to B. vulgatus CL10T00C06 ΔCK234_00400-1 (15).
Data availability.
The vectors created in this study can be acquired from the Addgene repository (ID numbers 126617 to 126621; https://www.addgene.org/browse/article/28203359/).
ACKNOWLEDGMENTS
We thank Michael Coyne and Juan Vasquez for discussions and critical comments on the manuscript. We are grateful to members of the Comstock lab for technical assistance, in particular Hongxia Bao for identifying P. merdae tractable strains. We thank Andrew Goodman for providing vector pNBU2_erm-TetR-P1T_DP-GH023 and Mark Mimee for multiple vectors and helpful discussions.
This work was supported by Public Health Service grant R01AI120633 from the National Institutes of Health/National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflicts of interest.
Footnotes
Citation García-Bayona L, Comstock LE. 2019. Streamlined genetic manipulation of diverse Bacteroides and Parabacteroides isolates from the human gut microbiota. mBio 10:e01762-19. https://doi.org/10.1128/mBio.01762-19.
Contributor Information
Katherine P. Lemon, The Forsyth Institute.
Jean-Marc Ghigo, Institut Pasteur.
Eugene Chang, University of Chicago.
REFERENCES
- 1.Kundu P, Blacher E, Elinav E, Pettersson S. 2017. Our gut microbiome: the evolving inner self. Cell 171:1481–1493. doi: 10.1016/j.cell.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 2.McKenney PT, Pamer EG. 2015. From hype to hope: the gut microbiota in enteric infectious disease. Cell 163:1326–1332. doi: 10.1016/j.cell.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heintz-Buschart A, Wilmes P. 2018. Human gut microbiome: function matters. Trends Microbiol 26:563–574. doi: 10.1016/j.tim.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Taroncher-Oldenburg G, Jones S, Blaser M, Bonneau R, Christey P, Clemente JC, Elinav E, Ghedin E, Huttenhower C, Kelly D, Kyle D, Littman D, Maiti A, Maue A, Olle B, Segal L, van Hylckama Vlieg JET, Wang J. 2018. Translating microbiome futures. Nat Biotechnol 36:1037–1042. doi: 10.1038/nbt.4287. [DOI] [PubMed] [Google Scholar]
- 5.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, Armanini F, Beghini F, Manghi P, Tett A, Ghensi P, Collado MC, Rice BL, DuLong C, Morgan XC, Golden CD, Quince C, Huttenhower C, Segata N. 2019. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176:649.e20–662.e20. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta RS, Abu-Ali GS, Drew DA, Lloyd-Price J, Subramanian A, Lochhead P, Joshi AD, Ivey KL, Khalili H, Brown GT, DuLong C, Song M, Nguyen LH, Mallick H, Rimm EB, Izard J, Huttenhower C, Chan AT. 2018. Stability of the human faecal microbiome in a cohort of adult men. Nat Microbiol 3:347–355. doi: 10.1038/s41564-017-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG, McDonald D, Franzosa EA, Knight R, White O, Huttenhower C. 2017. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550:61–66. doi: 10.1038/nature24485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wexler AG, Goodman AL. 2017. An insider’s perspective: Bacteroides as a window into the microbiome. Nat Microbiol 2:17026. doi: 10.1038/nmicrobiol.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salyers AA, Bonheyo G, Shoemaker NB. 2000. Starting a new genetic system: lessons from Bacteroides. Methods 20:35–46. doi: 10.1006/meth.1999.0903. [DOI] [PubMed] [Google Scholar]
- 12.Lange A, Beier S, Steimle A, Autenrieth IB, Huson DH, Frick J-S. 2016. Extensive mobilome-driven genome diversification in mouse gut-associated Bacteroides vulgatus mpk. Genome Biol Evol 8:1197–1207. doi: 10.1093/gbe/evw070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Husain F, Tang K, Veeranagouda Y, Boente R, Patrick S, Blakely G, Wexler HM. 14 November 2017. Novel large-scale chromosomal transfer in Bacteroides fragilis contributes to its pan-genome and rapid environmental adaptation. Microb Genomics doi: 10.1099/mgen.0.000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coyne MJ, Comstock LE. 2019. Type VI secretion systems and the gut microbiota. Microbiol Spectr 7:PSIB-0009-2018. doi: 10.1128/microbiolspec.PSIB-0009-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McEneany VL, Coyne MJ, Chatzidaki-Livanis M, Comstock LE. 2018. Acquisition of MACPF domain-encoding genes is the main contributor to LPS glycan diversity in gut Bacteroides species. ISME J 12:2919–2928. doi: 10.1038/s41396-018-0244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coyne MJ, Roelofs KG, Comstock LE. 2016. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17:58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coyne MJ, Zitomersky NL, McGuire AM, Earl AM, Comstock LE. 2014. Evidence of extensive DNA transfer between Bacteroidales species within the human gut. mBio 5:e01305-14. doi: 10.1128/mBio.01305-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zitomersky NL, Coyne MJ, Comstock LE. 2011. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect Immun 79:2012–2020. doi: 10.1128/IAI.01348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casterline BW, Hecht AL, Choi VM, Bubeck Wardenburg J. 2017. The Bacteroides fragilis pathogenicity island links virulence and strain competition. Gut Microbes 8:374–383. doi: 10.1080/19490976.2017.1290758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joglekar P, Sonnenburg ED, Higginbottom SK, Earle KA, Morland C, Shapiro-Ward S, Bolam DN, Sonnenburg JL. 2018. Genetic variation of the SusC/SusD homologs from a polysaccharide utilization locus underlies divergent fructan specificities and functional adaptation in Bacteroides thetaiotaomicron strains. mSphere 3:e00185-18. doi: 10.1128/mSphereDirect.00185-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boente RF, Ferreira LQ, Falcão LS, Miranda KR, Guimarães PLS, Santos-Filho J, Vieira JMBD, Barroso DE, Emond J-P, Ferreira EO, Paula GR, Domingues RMCP. 2010. Detection of resistance genes and susceptibility patterns in Bacteroides and Parabacteroides strains. Anaerobe 16:190–194. doi: 10.1016/j.anaerobe.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Boyanova L, Kolarov R, Mitov I. 2015. Recent evolution of antibiotic resistance in the anaerobes as compared to previous decades. Anaerobe 31:4–10. doi: 10.1016/j.anaerobe.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Snydman DR, Jacobus NV, McDermott LA, Goldstein EJC, Harrell L, Jenkins SG, Newton D, Patel R, Hecht DW. 2017. Trends in antimicrobial resistance among Bacteroides species and Parabacteroides species in the United States from 2010–2012 with comparison to 2008–2009. Anaerobe 43:21–26. doi: 10.1016/j.anaerobe.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Hastey CJ, Boyd H, Schuetz AN, Anderson K, Citron DM, Dzink-Fox J, Hackel M, Hecht DW, Jacobus NV, Jenkins SG, Karlsson M, Knapp CC, Koeth LM, Wexler H, Roe-Carpenter DE. 2016. Changes in the antibiotic susceptibility of anaerobic bacteria from 2007–2009 to 2010–2012 based on the CLSI methodology. Anaerobe 42:27–30. doi: 10.1016/j.anaerobe.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whittle G, Shoemaker NB, Salyers AA. 2002. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell Mol Life Sci 59:2044–2054. doi: 10.1007/s000180200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy E, Urbán E, Nord CE. 2011. Antimicrobial susceptibility of Bacteroides fragilis group isolates in Europe: 20 years of experience. Clin Microbiol Infect 17:371–379. doi: 10.1111/j.1469-0691.2010.03256.x. [DOI] [PubMed] [Google Scholar]
- 27.Wexler HM. 2007. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecht DW. 2006. Anaerobes: antibiotic resistance, clinical significance, and the role of susceptibility testing. Anaerobe 12:115–121. doi: 10.1016/j.anaerobe.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Bacic MK, Smith CJ. 2008. Laboratory maintenance and cultivation of Bacteroides species. Curr Protoc Microbiol Chapter 13:Unit 13C.1. doi: 10.1002/9780471729259.mc13c01s9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roelofs KG, Coyne MJ, Gentyala RR, Chatzidaki-Livanis M, Comstock LE. 2016. Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. mBio 7:e01055-16. doi: 10.1128/mBio.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salyers AA, Shoemaker N, Cooper A, D’Elia J, Shipman JA. 1999. 8 genetic methods for Bacteroides species. Methods Microbiol 29:229–249. doi: 10.1016/S0580-9517(08)70119-3. [DOI] [Google Scholar]
- 32.Smith CJ, Rogers MB, McKee ML. 1992. Heterologous gene expression in Bacteroides fragilis. Plasmid 27:141–154. doi: 10.1016/0147-619X(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 33.Park J, Salyers AA. 2011. Characterization of the Bacteroides CTnDOT regulatory protein RteC. J Bacteriol 193:91–97. doi: 10.1128/JB.01015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teixeira FL, Pauer H, Costa SB, Smith CJ, Domingues RMCP, Rocha ER, Lobo LA. 2018. Deletion of BmoR affects the expression of genes related to thiol/disulfide balance in Bacteroides fragilis. Sci Rep 8:14405. doi: 10.1038/s41598-018-32880-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betteken MI, Rocha ER, Smith CJ. 2015. Dps and DpsL mediate survival in vitro and in vivo during the prolonged oxidative stress response in Bacteroides fragilis. J Bacteriol 197:3329–3338. doi: 10.1128/JB.00342-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutanto Y, DiChiara JM, Shoemaker NB, Gardner JF, Salyers AA. 2004. Factors required in vitro for excision of the Bacteroides conjugative transposon, CTnDOT. Plasmid 52:119–130. doi: 10.1016/j.plasmid.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Ruppé É, Woerther P-L, Barbier F. 2015. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann Intensive Care 5:21. doi: 10.1186/s13613-015-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters JM, Silvis MR, Zhao D, Hawkins JS, Gross CA, Qi LS. 2015. Bacterial CRISPR: accomplishments and prospects. Curr Opin Microbiol 27:121–126. doi: 10.1016/j.mib.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selle K, Barrangou R. 2015. Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol 23:225–232. doi: 10.1016/j.tim.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 40.Arazoe T, Kondo A, Nishida K. 2018. Targeted nucleotide editing technologies for microbial metabolic engineering. Biotechnol J 13:e1700596. doi: 10.1002/biot.201700596. [DOI] [PubMed] [Google Scholar]
- 41.Ichimura M, Nakayama-Imaohji H, Wakimoto S, Morita H, Hayashi T, Kuwahara T. 2010. Efficient electrotransformation of Bacteroides fragilis. Appl Environ Microbiol 76:3325–3332. doi: 10.1128/AEM.02420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lazarus JE, Warr AR, Kuehl CJ, Giorgio RT, Davis BM, Waldor MK. 14 June 2019. A new suite of allelic exchange vectors for the scarless modification of proteobacterial genomes. Appl Environ Microbiol doi: 10.1128/AEM.00990-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kino Y, Nakayama-Imaohji H, Fujita M, Tada A, Yoneda S, Murakami K, Hashimoto M, Hayashi T, Okazaki K, Kuwahara T. 2016. Counterselection employing mutated pheS for markerless genetic deletion in Bacteroides species. Anaerobe 42:81–88. doi: 10.1016/j.anaerobe.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Rakoff-Nahoum S, Foster KR, Comstock LE. 2016. The evolution of cooperation within the gut microbiota. Nature 533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rakoff-Nahoum S, Coyne MJ, Comstock LE. 2014. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol 24:40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vingadassalom D, Kolb A, Mayer C, Rybkine T, Collatz E, Podglajen I. 2005. An unusual primary sigma factor in the Bacteroidetes phylum: primary sigma factor of Bacteroides fragilis. Mol Microbiol 56:888–902. doi: 10.1111/j.1365-2958.2005.04590.x. [DOI] [PubMed] [Google Scholar]
- 50.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. 2008. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Shoemaker NB, Wang G-R, Salyers AA. 2000. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J Bacteriol 182:3559–3571. doi: 10.1128/jb.182.12.3559-3571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. 2016. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci U S A 113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim B, Zimmermann M, Barry NA, Goodman AL. 2017. Engineered regulatory systems modulate gene expression of human commensals in the gut. Cell 169:547.e15–558.e15. doi: 10.1016/j.cell.2017.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross BD, Verster AJ, Radey MC, Schmidtke DT, Pope CE, Hoffman LR, Hajjar A, Peterson SB, Borenstein E, Mougous J. 2018. Acquired interbacterial defense systems protect against interspecies antagonism in the human gut microbiome. bioRxiv doi: 10.1101/471110. [DOI]
- 55.Bacic M, Parker AC, Stagg J, Whitley HP, Wells WG, Jacob LA, Smith CJ. 2005. Genetic and structural analysis of the Bacteroides conjugative transposon CTn341. J Bacteriol 187:2858–2869. doi: 10.1128/JB.187.8.2858-2869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peed L, Parker AC, Smith CJ. 2010. Genetic and functional analyses of the mob operon on conjugative transposon CTn341 from Bacteroides spp. J Bacteriol 192:4643–4650. doi: 10.1128/JB.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mimee M, Tucker AC, Voigt CA, Lu TK. 2015. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst 1:62–71. doi: 10.1016/j.cels.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patel EH, Paul LV, Patrick S, Abratt VR. 2008. Rhamnose catabolism in Bacteroides thetaiotaomicron is controlled by the positive transcriptional regulator RhaR. Res Microbiol 159:678–684. doi: 10.1016/j.resmic.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 59.Chatzidaki-Livanis M, Coyne MJ, Roelofs KG, Gentyala RR, Caldwell JM, Comstock LE. 2017. Gut symbiont Bacteroides fragilis secretes a eukaryotic-like ubiquitin protein that mediates intraspecies antagonism. mBio 8:e01902-17. doi: 10.1128/mBio.01902-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shumaker AM, Laclare McEneany V, Coyne MJ, Silver PA, Comstock LE. 2019. Identification of a fifth antibacterial toxin produced by a single Bacteroides fragilis strain. J Bacteriol 201:e00577-18. doi: 10.1128/JB.00577-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pantosti A, Tzianabos AO, Onderdonk AB, Kasper DL. 1991. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect Immun 59:2075–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 63.Shoemaker NB, Getty C, Guthrie EP, Salyers AA. 1986. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J Bacteriol 166:959–965. doi: 10.1128/jb.166.3.959-965.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baughn AD, Malamy MH. 2002. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc Natl Acad Sci U S A 99:4662–4667. doi: 10.1073/pnas.052710199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coyne MJ, Béchon N, Matano LM, Laclare McEneany V, Chatzidaki-Livanis M, Comstock LE. 2019. A family of anti-Bacteroidales peptide toxins wide-spread in the human gut microbiota. Nat Commun 10:3460. doi: 10.1038/s41467-019-11494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plasmids and construction details. Download Table S1, PDF file, 0.04 MB (46.4KB, pdf) .
Copyright © 2019 García-Bayona and Comstock.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S2, PDF file, 0.03 MB (32.5KB, pdf) .
Copyright © 2019 García-Bayona and Comstock.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacteroidales strains used or created in this study. Download Table S3, PDF file, 0.04 MB (46.1KB, pdf) .
Copyright © 2019 García-Bayona and Comstock.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The vectors created in this study can be acquired from the Addgene repository (ID numbers 126617 to 126621; https://www.addgene.org/browse/article/28203359/).